Summary
DNA helicases are a class of molecular motors that catalyze processive unwinding of double stranded DNA. In spite of much study, we know relatively little about the mechanisms by which these enzymes carry out the function for which they are named. Most current views are based on inferences from crystal structures. A prominent view is that the canonical ATPase motor exerts a force on the ssDNA resulting in “pulling” the duplex across a “pin” or “wedge” in the enzyme leading to a mechanical separation of the two DNA strands. In such models, DNA base pair separation is tightly coupled to ssDNA translocation of the motors. However, recent studies of the E. coli RecBCD helicase suggest an alternative model in which DNA base pair melting and ssDNA translocation occur separately. In this view, the enzyme-DNA binding free energy is used to melt multiple DNA base pairs in an ATP-independent manner, followed by ATP-dependent translocation of the canonical motors along the newly formed ssDNA tracks. Repetition of these two steps results in processive DNA unwinding. We summarize recent evidence suggesting this mechanism for RecBCD helicase action.
Keywords: DNA helicase, translocase, kinetics, mechanism, allostery, motor protein, nuclease
1. Introduction
The term “helicase” was coined by Hoffmann-Berling nearly 40 years ago to describe a class of enzymes that catalyze the ATP-dependent unwinding of duplex DNA1. The first enzyme referred to as a helicase, Helicase I, was isolated from E. coli2, but later shown to be encoded by the TraI gene of the F episome3. In fact, the RecBCD enzyme, which is the focus of this review, was the first enzyme shown to have ATP-dependent DNA unwinding activity4, although this preceded the use of the term helicase. Helicases are ubiquitous in all organisms and have been studied intensively since their discovery; however, we know relatively little about the mechanisms by which these enzymes catalyze duplex DNA unwinding.
The focus of this review is the E. coli RecBCD helicase/nuclease that is involved in degradation of foreign DNA as well as repair of chromosomal double stranded DNA breaks that are lethal if not repaired5,6. RecBCD binds to the ends of a double stranded DNA break and initiates unwinding of the duplex using its ATP-dependent helicase activity. RecBCD also contains a single stranded (ss)DNA exonuclease activity that initially degrades the unwound ssDNA from both the 3’ and 5’ ends. If the DNA is of foreign origin (e.g., a bacteriophage), degradation will continue. However, if RecBCD is acting on E. coli chromosomal DNA, it will eventually come upon an over-represented DNA sequence, called “chi” (Chromosomal Hotspot Initiator)7,8 which signals to RecBCD that the DNA is from E. coli and should be set up for repair rather than continued to be degraded. As such, after “chi” recognition by RecBCD, only the 5’-ended ssDNA continues to be degraded resulting in a stable 3’ ended ssDNA onto which RecBCD loads a RecA filament that then initiates a homologous recombination event resulting in eventual repair of the double stranded DNA break5,6.
In spite of much research, the mechanisms by which helicases unwind duplex DNA are not understood. It is clear from studies of the SF1 helicases, Rep, UvrD and PcrA, that ATP-dependent single stranded DNA translocase activity is not sufficient to yield helicase activity9–14. Here we discuss recent evidence suggesting that processive DNA unwinding by the E. coli RecBCD helicase is a two-step process involving ATP-independent DNA melting due to the favorable free energy of the protein-DNA interaction, followed by ATP-dependent ssDNA translocation.
2. Structures of RecBCD-DNA complexes
The RecBCD hetero-trimeric enzyme contains two canonical superfamily 1 (SF1) ATPase motors. The canonical SF1A RecB motor has 3’ to 5’ ssDNA translocase activity15,16, whereas the canonical SF1B RecD motor has 5’ to 3’ ssDNA translocase activity16–19. The RecC subunit interacts with both RecB and RecD, enhances DNA unwinding processivity and is involved in chi recognition20. A crystal structure of RecBCD bound to DNA duplex ends has been solved21 and is shown in Figure 1. This structure was determined with a fully blunt-ended DNA in the absence of nucleotide cofactors (no ATP), yet four bp at the end of the duplex are melted in the RecBCD complex. This is a demonstration that RecBCD is able to melt out four bp using only its binding free energy, consistent with results from biochemical studies22. Subsequent structures with a longer DNA duplex show that six bp can be melted out by RecBCD23, consistent with DNA binding studies24,25. The 3’-ssDNA is observed to be bound to the RecB motor domain, whereas the 5’-ssDNA is directed toward the RecD motor domain, although it is not long enough to make contact with RecD in that structure21. Consistent with these findings, RecBC and RecBCD bind with highest affinity to DNA ends with pre-melted ssDNA ends. RecBC binds with highest affinity to a ss/dsDNA end possessing twin ssDNA tails that are six nucleotides long (a 3’-(dT)6 tail and a 5’-(dT)6 tail)24. However, RecBCD binds with highest affinity to a ss/dsDNA end possessing a 3’-(dT)6 tail and a 5’-(dT)10 tail24. The longer 5’-(dT)10 tail is needed to optimize interactions with the RecD subunit18,19,24,26.
Figure 1.
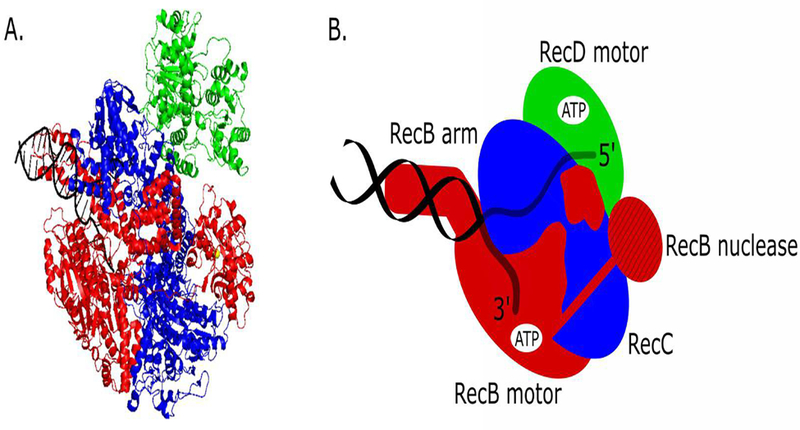
RecBCD-DNA structure. A. Crystal structure (PDB 1W36) of RecBCD bound to a DNA duplex. RecB subunit is red, RecC subunit is blue, RecD subunit is green, and DNA is black. B. Stylized RecBCD structure with the subunit colors as in A. The positions of the RecB SF1A translocase motor, the SF1B RecD translocase motor, the RecB arm domain, the RecB nuclease domain and the RecC subunit are noted. The approximate positions of the ATP binding sites within the RecB and RecD motors are shown in white. The RecB arm domain interacts with the duplex DNA ahead of the fork. The nuclease domain is attached to the RecB motor domain via a long (~70 amino acid) linker.
Other features to note in the RecBCD-DNA crystal structure are the arm domain of RecB that interacts with the duplex DNA ahead of the fork and the nuclease domain of RecB that is attached by a long (~70 amino acids) tether to the RecB motor domain and is located far from the duplex DNA at a position that would be ready to accept the unwound 3’-terminal ssDNA for degradation21.
3. A mechanical model for RecBCD-catalyzed helicase activity
Our focus is on the mechanism of DNA unwinding catalyzed by RecBCD helicase. Based on crystal structures of RecBCD bound to a DNA end, a mechanical model for how RecBCD might unwind DNA has been proposed21. Although the ssDNA directionalities of the RecB (3’ to 5’) and RecD (5’ to 3’) motors are opposite, they interact with the two unwound antiparallel strands of the DNA and thus move in the same net direction towards the duplex. Based on this, it was proposed that duplex disruption during helicase activity is the consequence of each of the two motors moving directionally along the two complementary ssDNA tails (RecB moving 3’ to 5’ and RecD moving 5’ to 3’) while simultaneously pulling the duplex DNA across a “pin” or a “wedge” within the RecC subunit21. This model implies that duplex unwinding is tightly coupled to ssDNA translocation by the two canonical SF1 motors and that helicase activity requires movement of the ssDNA translocase motors. This type of mechanical “wedge” model has been invoked as a general mechanism for helicase activity27.
However, a series of biochemical and biophysical studies15,19,26,28–30 support an alternative mechanism in which DNA melting of multiple base pairs occurs in an ATP-independent reaction and that ATP hydrolysis is used primarily for directed ssDNA translocation of the enzyme. We first summarize the evidence that leads to this model.
4. Passive vs. Active Helicases
DNA helicase mechanisms can be classified as either “passive” or “active”31–33. In a passive mechanism, the enzyme uses its uni-directional ssDNA translocation motor activity to capture and stabilize ssDNA that is formed transiently due to thermal fraying of the duplex base pairs. In an active mechanism, the helicase plays a direct role in destabilizing the base pairs. In addition to the two extremes there exists a continuous gradient of energetic contributions of the helicase to duplex unwinding34. One criteria for a passive helicase is that the rate of ssDNA translocation is expected to be much faster (≥ 7-fold) than the rate of DNA unwinding34,35. The rate of ssDNA translocation by RecBC is only ~2-fold faster than its rate of DNA unwinding28. By this and several other criteria, both RecBC and RecBCD are active helicases19,28.
5. RecBCD has three translocase activities
The RecB subunit contains the canonical superfamily 1A (SF1A) ATPase motor that displays 3’ to 5’ directional ssDNA translocation15–17,19,36 as well as a nuclease domain37. The RecD subunit contains the canonical SF1B motor that displays 5’ to 3’ ssDNA translocase activity16,17,19. The RecC subunit stimulates the helicase activity and processivity of RecB15, and is involved in “chi” recognition21. Even without the RecD motor, RecBC is a rapid and processive helicase, although slower than RecBCD15,19,36. A secondary ssDNA translocase activity, that operates within both RecBC15,28 and RecBCD19 and is controlled by the ATPase activity of the RecB motor15,28,38,39, is associated with the RecB arm region30 that is observed to contact duplex DNA in crystal structures21,40. However, this secondary translocase activity shows no polarity preference on ssDNA and thus may actually reflect a double stranded DNA translocase activity15,28,30. Hence, RecBCD possesses, not two, but three translocase activities, the RecB motor regulates its 3’ to 5’ ssDNA translocase and the secondary translocase activities, whereas the RecD motor regulates translocation only in the 5’ to 3’ direction along the 5’-ended ssDNA15,19,28.
6. RecBC and RecBCD melt out 4–6 base pairs upon binding to blunt-ended DNA in the absence of ATP
To initiate DNA unwinding, RecBCD binds to blunt or nearly blunt-ended DNA and melts (separates) 4–6 base pairs in a reaction that uses only its favorable binding free energy. i.e., it does not require ATP binding or hydrolysis21,22,24. RecBC displays the same ability24,25. The fact that DNA melting to form an initiation complex does not require ATP raises the possibility that DNA melting during processive DNA unwinding may also not require ATP28.
7. DNA Unwinding by RecBC and RecBCD occurs with a kinetic step size of ~4 base pairs
Analysis of transient DNA unwinding kinetics experiments indicates that RecBCD and RecBC catalyzed unwinding of DNA can be described by a mechanism in which DNA unwinding occurs with a “kinetic step size” of 4±1 bp. That is, 4±1 bp are unwound between successive rate-limiting steps that are repeated during the unwinding process19,26,41–43. The molecular interpretation of a kinetic step size is subject to numerous caveats44; however, one interpretation is that DNA unwinding occurs by successive steps involving rapid melting of ~ 4±1 bp, followed by a rate-limiting step41,43,45. The observation that the kinetic step size is independent of temperature and ATP concentration supports this interpretation42. This observation is also consistent with the proposal that processive DNA unwinding can be viewed as occurring in two steps, ATP-independent melting of 4–6 bp, followed by ATP-dependent ssDNA translocation along the newly formed ssDNA.
8. The same amount of ATP is hydrolyzed by RecBC during duplex DNA unwinding and ssDNA translocation
As for all processive helicases, DNA unwinding by RecBCD is coupled to ATP binding and hydrolysis. Estimates of the ATP coupling found ~2 ATP per bp unwound for RecBCD46–48 and one ATP per bp unwound for RecBC28,48 and RecBCDK177Q, where RecDK177Q is an ATP-deficient mutant of the RecD motor48. Hence DNA unwinding is coupled to hydrolysis of one ATP per motor per bp. In addition, the same amount of ATP is hydrolyzed by RecBC during processive DNA unwinding (1 ATP/bp) (ssDNA translocation plus DNA melting) as during ssDNA translocation alone (1 ATP/nucleotide translocated) 28. This suggests that most, possibly all ATP hydrolysis is used for ssDNA translocation rather than DNA melting and that DNA unwinding may occur separately form ssDNA translocation28, although it is also consistent with models in which DNA unwinding and ssDNA translocation occur simultaneously21,49.
9. Processive duplex DNA unwinding by RecBCD can be uncoupled from ssDNA translocation by the canonical RecB and RecD motors
The mechanical model for DNA helicase activity by RecBCD21 implies that processive DNA unwinding is tightly coupled to ssDNA translocation. That is, processive DNA unwinding should not occur without ssDNA translocation. Simon et al.30 recently tested whether RecBCD could unwind duplex DNA in the absence of ssDNA translocation by its canonical RecB and RecD motors. DNA duplexes were designed with a unique initiation site for RecBCD, but contained so-called “reversed polarity” (RP) linkages within each DNA strand just after the RecBCD binding site. The RP linkages consist of ssDNA that contains a single 3’−3’ phosphodiester linkage or a single 5’−5’ phosphodiester linkage33. These are positioned within each strand at the same position in the duplex DNA. Since SF1 ssDNA translocases are strictly directional, a 3’−3’ linkage will stop the translocation of a canonical 3’ to 5’ translocase motor, and a 5’−5’ linkage will stop the translocation of a canonical 5’ to 3’ translocase motor15,30. Hence, the RecB and RecD canonical motors are unable to move past the point in the duplex DNA that contains the RP linkages. Fluorophore pairs that undergo fluorescence resonance energy transfer (FRET) when in close proximity were incorporated into the DNA beyond the RP linkages in order to detect DNA unwinding. Surprisingly, RecBCD was able to processively unwind duplex DNA for at least 80 bp beyond the RP linkages with rates only half (~400 bp/s) those during unwinding of natural DNA (~800 bp/s). Although the canonical RecB and RecD motors are unable to bypass the RP linkages, another region of the RecBCD helicase must be capable of moving along and unwinding the duplex DNA in an ATP-dependent process30. Although RecBCD transiently unwinds the duplex DNA, once all of the ATP is hydrolyzed, the DNA substrate reanneals to its original state as long as the RecBCD enzyme remains bound to the DNA30. These results indicate that RecBCD, as well as RecBC, is able to unwind DNA processively without the need for translocation of the canonical RecB and RecD motors. A RecBCD-DNA intermediate that is suggested by these results is depicted in Figure 3.
Figure 3.
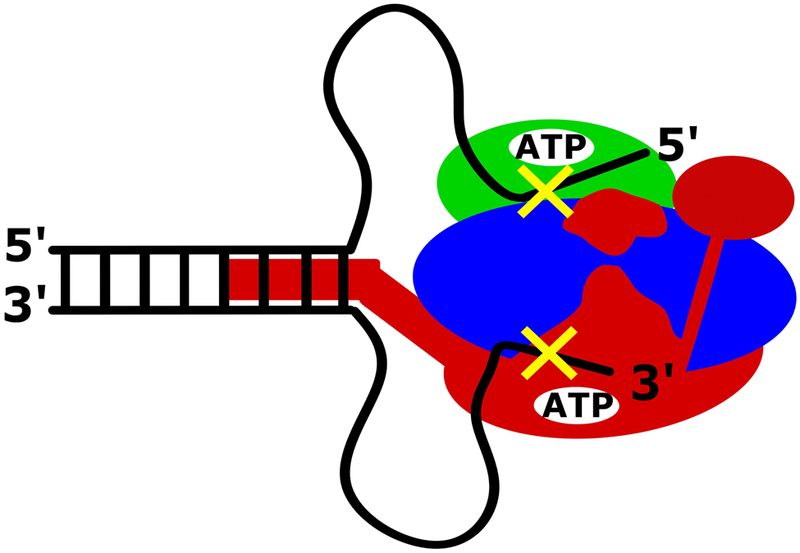
RecBCD can unwind DNA processively in the absence of ssDNA translocation by the canonical RecB and RecD motors. RecBCD can transiently unwind duplex DNA beyond the reverse polarity switches in the DNA backbone (indicated here by yellow x’s), which prevent the canonical RecB and RecD motors from translocating along the ssDNA. This activity is dependent upon the secondary translocase activity of RecBC that is fueled by the RecB ATPase activity and requires both the RecB arm domain and the RecB nuclease domain, but not nuclease activity. It is possible that the RecB arm act as a dsDNA translocase, pulling the dsDNA towards RecBCD, resulting in the formation of ssDNA loops.
10. The RecB arm may function as a double stranded DNA translocase
The ability of RecBCD to unwind DNA past the RP linkages is dependent upon an active RecB ATPase motor and the secondary translocase activity30. This is consistent with the fact that the secondary translocase activity is controlled by the RecB canonical motor15,19,28. Furthermore, a variant of RecBCD in which the RecB arm domain has been deleted has very poor helicase activity and is unable to unwind DNA beyond the RP linkages30. This suggests that the RecB arm is important for DNA unwinding and may be directly involved in the secondary translocase activity. Two other observations support the suggestion that the RecB arm may operate as a double stranded DNA translocase controlled by the RecB ATPase. The first is that the secondary translocase activity shows no polarity preference on ssDNA15. Hence movement could occur along either strand within the duplex DNA.
The second observation suggesting that the RecB arm may function as a double stranded DNA translocase is that in a crystal structure21 (see Figure 1), the RecB arm makes direct contact with the duplex DNA ahead of the fork. Additional support comes from structural studies of the B. subtilis AddAB helicase/nuclease40, that is structurally similar to E. coli RecBC. Krajewski et al.40 proposed a model for DNA unwinding that involves the AddA “arm” domain, that is similar to the RecB “arm” domain. In a crystal structure, the AddA arm and the C-terminal nuclease domain of AddB contact the duplex DNA ahead of the fork as does the RecB arm in the RecBCD-DNA structure21. Based on this it was proposed that in an ATP-coupled reaction, the AddA arm pulls on the duplex DNA in the opposite direction, but in concert with the AddA canonical motor that pulls on the 3’-ended ssDNA. This is proposed to create stress in the duplex DNA that results in base pair opening. Krajewski et al.40 further suggest that this proposed role for the arm domain may provide a basis for the RecBC secondary translocase activity that also functions within RecBCD19. This supports the suggestion that the arm domain may actually operate as a double stranded DNA translocase driven by the ATPase activity of the RecB (or AddA, or motor).
Another finding that was a major surprise was that the ability of RecBCD to unwind DNA past the RP linkages is also dependent on the RecB nuclease domain30. It is not dependent on the nuclease activity since a RecB(D1040A) mutation that knocks out nuclease activity still enables RecBD1040ACD to unwind DNA past the RP linkages. However, a complete deletion of the RecB nuclease domain to form RecBΔNucCD does not support this activity30. The molecular basis for this intriguing result remains obscure. However, since the RecB nuclease domain is attached to the RecB motor by a long (~70 amino acid) tether, and there is good evidence that the nuclease domain must detach from the position that it occupies in a crystal structure21,50,51, there are many interesting possibilities that need to be tested.
11. RecBCD is able to transiently invade duplex DNA at a junction to unwind an additional ~4 bp in an ATP-independent reaction
Recent single molecule optical trapping experiments from the Perkins lab29 have shown very interesting dynamics within a RecBCD-DNA complex even in the absence of ATP. These experiments show that RecBCD is able to invade duplex DNA to transiently unwind an additional ~4 bp in an ATP-independent reaction, even when RecBCD is bound to a fully pre-melted ss/ds DNA junction. This activity is only observed when RecBCD is bound to a pre-melted ss/ds junction with 3’-(dT)6 and 5’-(dT)10 tails. If the 5’-tail is shortened to 5’-(dT)6, the transient melting is no longer observed. Since a 5’-ssDNA tail of at least ten nucleotides (5’-(dT)10) is needed to contact the RecD motor in an initiation complex24,26, this implicates the RecD motor in this activity29. This activity is abolished upon interstrand DNA crosslinking of the first base pair within the duplex which supports the conclusion that the transient movement of RecBCD is coupled to base pair melting that initiates at the ss/ds DNA junction29. These results indicate that RecBCD is able to melt out multiple base pairs of the duplex in an ATP-independent reaction in further support of the proposal that DNA melting and directed ssDNA translocation are separate processes in processive helicase activity.
12. A model for RecBCD-catalyzed processive DNA unwinding
Based on the observations summarized above, we have proposed a mechanism for processive DNA unwinding by RecBCD28,30 that is depicted schematically in Figure 2. In this mechanism, the free energy of RecBCD binding to DNA is used to melt multiple (4–6) DNA bp as in its initiation complex. The newly formed ssDNA then provides the tracks along which the RecB and RecD motors translocate. Translocation proceeds, hydrolyzing 1 ATP per nucleotide moved per motor, until the new ss/ds DNA junction is reached. ATP binding/hydrolysis then resets the enzyme so that it can melt out another stretch of duplex DNA and continue the unwinding process. The DNA melting process might involve the type of reaction observed by Carter et al.29, such that RecBCD at a junction can transiently invade and melt ~4 bp in an ATP-independent process. Subsequent ATP-dependent ssDNA translocation of RecBCD would rectify the enzyme towards the junction so that the melting of the 4 bp becomes irreversible. Our results suggest that ATP hydrolysis by the RecB motor not only drives ssDNA translocation of that motor, but also transmits conformational changes allosterically to the RecB arm to generate a force that enables the arm region to move along the DNA ahead of the fork. This secondary (double stranded DNA?) translocase activity associated with the RecB arm within RecBC has also been implicated in the DNA melting process30. The dsDNA translocase activity might create strain during the “reeling in” of the dsDNA resulting in DNA melting to form ssDNA loops.
Figure 2.
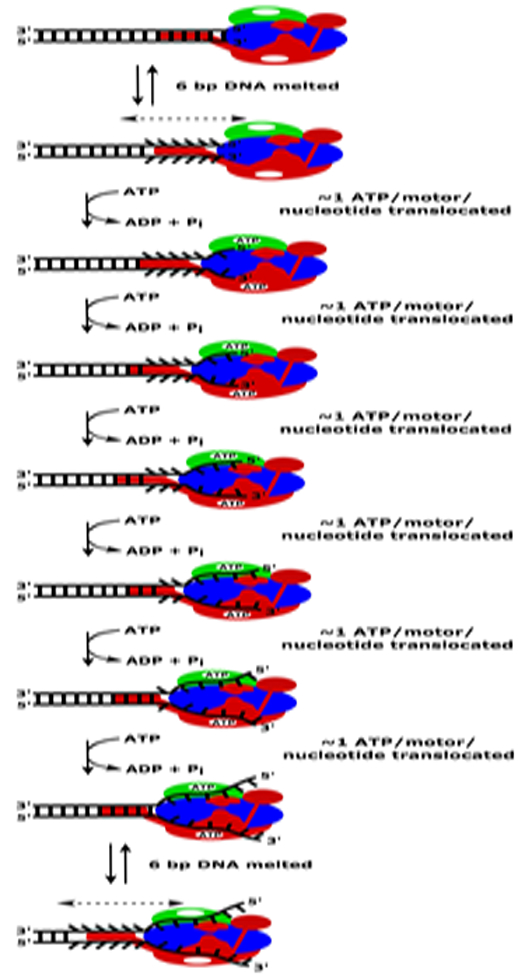
Proposed mechanism for processive DNA unwinding by RecBCD helicase. RecBCD initially binds to the blunt end of a duplex DNA and couples its favorable free energy of binding to melt 4–6 bp of the duplex DNA in an ATP-independent manner. RecBCD then translocates along the resulting ssDNA tracks hydrolyzing 1 ATP/motor/nucleotide until it reaches the new ss/ds DNA junction. ATP binding/hydrolysis resets the enzyme so that RecBCD can melt another 4–6 bp and then translocate along the newly formed ssDNA tracks. These DNA melting and ssDNA translocation steps are repeated during processive DNA unwinding.
13. Conclusions
Additional studies are needed to further test the details of this proposed mechanism. Structural studies are important in this regard, but studies of the mechanisms and dynamics of these enzyme machines will be required to address most of the details. Important information remains to be determined. Direct measurements of the distribution of step sizes during DNA unwinding are needed using single molecule approaches. How does the nuclease domain regulate DNA unwinding? How does interaction with a chi sequence allosterically regulate the RecB and RecD motors?
Might the mechanistic conclusions summarized above for the RecBCD helicase be general? Are aspects of this proposed mechanism shared by other similar helicases52, such as AdnAB53,54 and AddAB40,55. Are other SF1 helicases able to melt duplex DNA using only their binding free energy and can processive DNA unwinding be uncoupled from ssDNA translocation by their canonical SF1 motors? In this regard, there are other SF1 helicases for which it has been shown that single stranded DNA translocase activity can be separated from DNA helicase activity. The SF1 enzymes, Rep, UvrD and PcrA are rapid, directional and processive ssDNA translocases as monomers, but have no detectable helicase activity9,12,56,57 unless the DNA is under tension58. The helicase activity of these enzymes needs to be activated either by dimerization14,56,57, interaction with an accessory protein, removal of the auto-inhibitory 2B sub-domain11, or crosslinking of the 2B sub-domain into its closed conformation59. In fact, it was demonstrated 25 years ago33 that a dimeric Rep helicase is able to unwind a short duplex DNA beyond a 5’−5’ RP linkage in the 3’ ssDNA tail loading site, thus showing a similar ability to that of RecBCD30. It will be interesting to test these ideas with other helicases.
Acknowledgements
We thank the many members of the lab who contributed to our studies of DNA helicases which have been supported by grants from the National Institutes of Health (GM045948) to T.M.L. N.T.F. was supported in part by a fellowship from Sigma Chemical Co. The authors have no conflict of interest to declare.
Abbreviations:
- RP
reversed polarity.
References
- 1.Kuhn B, Abdel-Monem M, Krell H & Hoffmann-Berling H Evidence for two mechanisms for DNA unwinding catalyzed by DNA helicases. J Biol Chem 254, 11343–50 (1979). [PubMed] [Google Scholar]
- 2.Abdel-Monem M & Hoffmann-Berling H Enzymatic unwinding of DNA. 1. Purification and characterization of a DNA-dependent ATPase from Escherichia coli. Eur J Biochem 65, 431–40 (1976). [DOI] [PubMed] [Google Scholar]
- 3.Abdel-Monem M, Taucher-Scholz G & Klinkert M-Q Identification of Escherichia coli DNA helicase I as the traI gene product of the F sex factor. Proc.Natl.Acad.Sci.USA 80, 4659–4663 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mackay V & Linn S Selective inhibition of the dnase activity of the recBC enzyme by the DNA binding protein from Escherichia coli. J Biol Chem 251, 3716–9 (1976). [PubMed] [Google Scholar]
- 5.Dillingham MS & Kowalczykowski SC RecBCD Enzyme and the Repair of Double-Stranded DNA Breaks. Microbiol Mol Biol Rev 72, 642–71 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith GR How RecBCD enzyme and Chi promote DNA break repair and recombination: a molecular biologist’s view. Microbiol Mol Biol Rev 76, 217–28 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor AF, Amundsen SK & Smith GR Unexpected DNA context-dependence identifies a new determinant of Chi recombination hotspots. Nucleic Acids Res 44, 8216–28 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith GR, Kunes SM, Schultz DW, Taylor A & Triman KL Structure of chi hotspots of generalized recombination. Cell 24, 429–36 (1981). [DOI] [PubMed] [Google Scholar]
- 9.Lohman TM, Tomko EJ & Wu CG Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat Rev Mol Cell Biol 9, 391–401 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Fischer CJ, Maluf NK & Lohman TM Mechanism of ATP-dependent translocation of E.coli UvrD monomers along single-stranded DNA. J Mol Biol 344, 1287–309 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Brendza KM et al. Autoinhibition of Escherichia coli Rep monomer helicase activity by its 2B subdomain. Proc Natl Acad Sci U S A 102, 10076–81 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niedziela-Majka A, Chesnik MA, Tomko EJ & Lohman TM Bacillus stearothermophilus PcrA Monomer Is a Single-stranded DNA Translocase but Not a Processive Helicase in Vitro. J. Biol. Chem 282, 27076–27085 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Chisty LT et al. Monomeric PcrA helicase processively unwinds plasmid lengths of DNA in the presence of the initiator protein RepD. Nucleic Acids Res 41, 5010–23 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen B, Ordabayev Y, Sokoloski JE, Weiland E & Lohman TM Large domain movements upon UvrD dimerization and helicase activation. Proc Natl Acad Sci U S A 114, 12178–12183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu CG, Bradford C & Lohman TM Escherichia coli RecBC helicase has two translocase activities controlled by a single ATPase motor. Nat Struct Mol Biol 17, 1210–7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor AF & Smith GR RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature 423, 889–93 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Dillingham MS, Spies M & Kowalczykowski SC RecBCD enzyme is a bipolar DNA helicase. Nature 423, 893–7 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Dillingham MS, Webb MR & Kowalczykowski SC Bipolar DNA translocation contributes to highly processive DNA unwinding by RecBCD enzyme. J Biol Chem 280, 37069–77 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Xie F, Wu CG, Weiland E & Lohman TM Asymmetric regulation of bipolar single-stranded DNA translocation by the two motors within Escherichia coli RecBCD helicase. J Biol Chem 288, 1055–64 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilkinson M & Wigley DB Structural features of Chi recognition in AddAB with implications for RecBCD. Cell Cycle 13, 2812–20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC & Wigley DB Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature 432, 187–93 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Farah JA & Smith GR The RecBCD enzyme initiation complex for DNA unwinding: enzyme positioning and DNA opening. J Mol Biol 272, 699–715 (1997). [DOI] [PubMed] [Google Scholar]
- 23.Saikrishnan K, Griffiths SP, Cook N, Court R & Wigley DB DNA binding to RecD: role of the 1B domain in SF1B helicase activity. Embo J 27, 2222–9 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong CJ, Lucius AL & Lohman TM Energetics of DNA end binding by E.coli RecBC and RecBCD helicases indicate loop formation in the 3’-single-stranded DNA tail. J. Mol. Biol 352, 765–82 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Wong CJ & Lohman TM Kinetic control of Mg2+-dependent melting of duplex DNA ends by Escherichia coli RecBC. J Mol Biol 378, 759–75 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu CG & Lohman TM Influence of DNA end structure on the mechanism of initiation of DNA unwinding by the Escherichia coli RecBCD and RecBC helicases. J Mol Biol 382, 312–26 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhattacharyya B & Keck JL Grip it and rip it: structural mechanisms of DNA helicase substrate binding and unwinding. Protein Sci 23, 1498–507 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu CG, Xie F & Lohman TM The primary and secondary translocase activities within E. coli RecBC helicase are tightly coupled to ATP hydrolysis by the RecB motor. J Mol Biol 423, 303–14 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carter AR et al. Sequence-dependent nanometer-scale conformational dynamics of individual RecBCD-DNA complexes. Nucleic Acids Res 44, 5849–60 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon MJ, Sokoloski JE, Hao L, Weiland E & Lohman TM Processive DNA Unwinding by RecBCD Helicase in the Absence of Canonical Motor Translocation. J Mol Biol 428, 2997–3012 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lohman TM Escherichia coli DNA helicases: mechanisms of DNA unwinding. Mol Microbiol 6, 5–14 (1992). [DOI] [PubMed] [Google Scholar]
- 32.Lohman TM & Bjornson KP Mechanisms of helicase-catalyzed DNA unwinding. Annu Rev Biochem 65, 169–214 (1996). [DOI] [PubMed] [Google Scholar]
- 33.Amaratunga M & Lohman TM Escherichia coli Rep helicase unwinds DNA by an active mechanism. Biochemistry 32, 6815–6820 (1993). [DOI] [PubMed] [Google Scholar]
- 34.Betterton MD & Julicher F Opening of nucleic-acid double strands by helicases: active versus passive opening. Phys Rev E Stat Nonlin Soft Matter Phys 71, 011904 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Manosas M, Xi XG, Bensimon D & Croquette V Active and passive mechanisms of helicases. Nucleic Acids Res 38, 5518–26 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bianco PR & Kowalczykowski SC Translocation step size and mechanism of the RecBC DNA helicase. Nature 405, 368–72 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Yu M, Souaya J & Julin DA The 30-kDa C-terminal domain of the RecB protein is critical for the nuclease activity, but not the helicase activity, of the RecBCD enzyme from Escherichia coli. Proc Natl Acad Sci U S A 95, 981–6 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cho CC, Chung C & Li HW How Chi Sequence Modifies RecBCD Single-Stranded DNA Translocase Activity. Chemphyschem 19, 243–247 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Chung C & Li HW Direct observation of RecBCD helicase as single-stranded DNA translocases. J Am Chem Soc 135, 8920–5 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Krajewski WW et al. Structural basis for translocation by AddAB helicase-nuclease and its arrest at chi sites. Nature 508, 416–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lucius AL et al. DNA unwinding step-size of E. coli RecBCD helicase determined from single turnover chemical quenched-flow kinetic studies. J Mol Biol 324, 409–28 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Lucius AL & Lohman TM Effects of temperature and ATP on the kinetic mechanism and kinetic step-size for E.coli RecBCD helicase-catalyzed DNA unwinding. J. Mol. Biol 339, 751–71 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Lucius AL, Jason Wong C & Lohman TM Fluorescence stopped-flow studies of single turnover kinetics of E.coli RecBCD helicase-catalyzed DNA unwinding. J. Mol. Biol 339, 731–50 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Lucius AL, Maluf NK, Fischer CJ & Lohman TM General methods for analysis of sequential “n-step” kinetic mechanisms: application to single turnover kinetics of helicase-catalyzed DNA unwinding. Biophys J 85, 2224–39 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ali JA & Lohman TM Kinetic measurement of the step size of DNA unwinding by Escherichia coli UvrD helicase. Science 275, 377–80 (1997). [DOI] [PubMed] [Google Scholar]
- 46.Roman LJ & Kowalczykowski SC Characterization of the adenosinetriphosphatase activity of the Escherichia coli RecBCD enzyme: relationship of ATP hydrolysis to the unwinding of duplex DNA. Biochemistry 28, 2873–81 (1989). [DOI] [PubMed] [Google Scholar]
- 47.Roman LJ & Kowalczykowski SC Characterization of the helicase activity of the Escherichia coli RecBCD enzyme using a novel helicase assay. Biochemistry 28, 2863–73 (1989). [DOI] [PubMed] [Google Scholar]
- 48.Korangy F & Julin DA Efficiency of ATP hydrolysis and DNA unwinding by the RecBC enzyme from Escherichia coli. Biochemistry 33, 9552–60 (1994). [DOI] [PubMed] [Google Scholar]
- 49.Singleton MR, Dillingham MS & Wigley DB Structure and Mechanism of Helicases and Nucleic Acid Translocases. Annu Rev Biochem 76, 23–50 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Spies M & Kowalczykowski SC The RecA binding locus of RecBCD is a general domain for recruitment of DNA strand exchange proteins. Mol Cell 21, 573–80 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Taylor AF et al. Control of RecBCD enzyme activity by DNA binding- and Chi hotspot-dependent conformational changes. J Mol Biol 426, 3479–99 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wigley DB Bacterial DNA repair: recent insights into the mechanism of RecBCD, AddAB and AdnAB. Nat Rev Microbiol 11, 9–13 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Unciuleac MC & Shuman S Characterization of the mycobacterial AdnAB DNA motor provides insights into the evolution of bacterial motor-nuclease machines. J Biol Chem 285, 2632–41 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Unciuleac MC & Shuman S Double strand break unwinding and resection by the mycobacterial helicase-nuclease AdnAB in the presence of single strand DNA-binding protein (SSB). J Biol Chem 285, 34319–29 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yeeles JT, Gwynn EJ, Webb MR & Dillingham MS The AddAB helicase-nuclease catalyses rapid and processive DNA unwinding using a single Superfamily 1A motor domain. Nucleic Acids Res 39, 2271–85 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheng W, Hsieh J, Brendza KM & Lohman TME coli Rep oligomers are required to initiate DNA unwinding in vitro. J. Mol. Biol 310, 327–50 (2001). [DOI] [PubMed] [Google Scholar]
- 57.Maluf NK, Fischer CJ & Lohman TM A Dimer of Escherichia coli UvrD is the active form of the helicase in vitro. J. Mol. Biol 325, 913–35 (2003). [DOI] [PubMed] [Google Scholar]
- 58.Comstock MJ et al. Protein structure. Direct observation of structure-function relationship in a nucleic acid-processing enzyme. Science 348, 352–4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arslan S, Khafizov R, Thomas CD, Chemla YR & Ha T Protein structure. Engineering of a superhelicase through conformational control. Science 348, 344–7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]


