Transfer of mobile genetic elements from one bacterium to another is the principal cause of the spread of antibiotic resistance. However, the dissemination of these elements in environmental contexts is poorly understood. In clinical and environmental settings, bacteria are often found living in multicellular communities encased in a matrix, a structure known as a biofilm. In this study, we examined how forming a biofilm influences the transmission of an integrative and conjugative element (ICE). Using the model Gram-positive bacterium B. subtilis, we observed that biofilm formation highly favors ICE transfer. This increase in conjugative transfer is due to the production of extracellular matrix, which creates an ideal biophysical context. Our study provides important insights into the role of the biofilm structure in driving conjugative transfer, which is of major importance since biofilm is a widely preponderant bacterial lifestyle for clinically relevant bacterial strains.
KEYWORDS: Bacillus subtilis, ICEBs1, biofilms, extracellular matrix, horizontal gene transfer
ABSTRACT
Horizontal gene transfer by integrative and conjugative elements (ICEs) is a very important mechanism for spreading antibiotic resistance in various bacterial species. In environmental and clinical settings, most bacteria form biofilms as a way to protect themselves against extracellular stress. However, much remains to be known about ICE transfer in biofilms. Using ICEBs1 from Bacillus subtilis, we show that the natural conjugation efficiency of this ICE is greatly affected by the ability of the donor and recipient to form a biofilm. ICEBs1 transfer considerably increases in biofilm, even at low donor/recipient ratios. Also, while there is a clear temporal correlation between biofilm formation and ICEBs1 transfer, biofilms do not alter the level of ICEBs1 excision in donor cells. Conjugative transfer appears to be favored by the biophysical context of biofilms. Indeed, extracellular matrix production, particularly from the recipient cells, is essential for biofilms to promote ICEBs1 transfer. Our study provides basic new knowledge on the high rate of conjugative transfer of ICEs in biofilms, a widely preponderant bacterial lifestyle in the environment, which could have a major impact on our understanding of horizontal gene transfer in natural and clinical environments.
IMPORTANCE Transfer of mobile genetic elements from one bacterium to another is the principal cause of the spread of antibiotic resistance. However, the dissemination of these elements in environmental contexts is poorly understood. In clinical and environmental settings, bacteria are often found living in multicellular communities encased in a matrix, a structure known as a biofilm. In this study, we examined how forming a biofilm influences the transmission of an integrative and conjugative element (ICE). Using the model Gram-positive bacterium B. subtilis, we observed that biofilm formation highly favors ICE transfer. This increase in conjugative transfer is due to the production of extracellular matrix, which creates an ideal biophysical context. Our study provides important insights into the role of the biofilm structure in driving conjugative transfer, which is of major importance since biofilm is a widely preponderant bacterial lifestyle for clinically relevant bacterial strains.
INTRODUCTION
Acquisition of genetic material via horizontal gene transfer (HGT) is a fundamental phenomenon for bacterial adaptation and evolution (1). Conjugation, which is regarded as the broadest and most efficient mechanism of HGT, allows bacteria to transfer genetic material such as conjugative plasmids and integrative and conjugative elements (ICEs) through direct cellular contact (2). These mobile genetic elements are autonomous since they encode their own mating apparatus. They often contain genes responsible for a wide range of functions, including virulence, antibiotic resistance, and symbiosis (3–5). Conjugative plasmids and ICEs can often transfer between different bacterial species and genera and mobilize genomic islands or plasmids that are otherwise not self-transmissible, granting these elements an extensive role in bacterial evolution (6–9).
ICEBs1 is a 20.5-kb ICE that is present in many strains of Bacillus subtilis (10, 11), a low-G+C Gram-positive bacterium that is well studied for its plant growth-promoting effect (12–14). While ICEBs1 can mobilize genetic elements lacking mobilization functions (8), whether it provides any advantage for its host cell remains unclear (15). ICEBs1 transmission is initiated in the donor cell by its excision from the 3′ end of the chromosomal trnS-leu2 gene (10). The resulting double-stranded circular intermediate undergoes rolling circle replication initiated at the origin of transfer (oriT) by the relaxase NicK, which cleaves the DNA strand to be transferred (16, 17). The nicked strand of ICEBs1 is then translocated into the recipient cells by an ICEBs1-encoded type IV secretion system (17, 18). In the recipient cell, the transferred strand is recircularized, and its complementary strand is synthesized. The circular copy of ICEBs1 then integrates at the 3′ end of trnS-leu2, the attB site of the chromosome of the recipient (19).
Interestingly, two distinct cellular pathways regulate ICEBs1 excision. One is the global DNA damage response, which is mediated via the DNA repair protein RecA, which acts as an activator of conjugation. The other pathway is the ICEBs1-encoded quorum-sensing system RapI-PhrI, consisting of RapI, an inducer of ICEBs1 excision that can be inhibited by the coexpressed oligopeptide PhrI (10), which is secreted in the extracellular environment and imported back into the cell through a permease. In this pathway, ICEBs1 excision is repressed in a community where ICEBs1-harboring cells are widely present since the extracellular PhrI level is sufficient to inhibit RapI (10).
Biofilms are microbial communities surrounded by an extracellular matrix that protects bacterial cells from external stressors such as antibiotics and heavy metals (20, 21). In the environment and during chronic infections, most bacteria live within biofilms (22). B. subtilis biofilm matrix is mostly composed of exopolysaccharides and amyloid-like fibers, synthesis of which is encoded by the epsA to -O (epsA–O) operon and the tapA-sipW-tasA operon, respectively (23). Matrix production, and thus, biofilm formation, is triggered by a variety of environmental and physiological signals, including the lipopeptide surfactin, plant polysaccharides, and a combination of glycerol and manganese (24–26). In a planktonic population, the expression of the matrix production operons epsA–O and tapA-sipW-tasA is inhibited by the transcriptional repressor SinR (27).
Studies have suggested that biofilms are hot spots for the transfer of conjugative plasmids due to the high proximity of cells within this multicellular structure, but the importance of the extracellular matrix in this process is unexplored (28, 29). Also, many of the pathogens that have acquired antibiotic resistance through conjugative elements can form biofilms (30–32). However, despite their fundamental importance in antibiotic resistance gene dissemination, ICE propagation in biofilms has not yet been examined. Here, we take advantage of the extensive knowledge of ICEBs1 and B. subtilis biofilms to evaluate the dynamics of ICEBs1 dissemination within biofilms. Using medium that does or does not induce biofilm formation as support for conjugation, we report here that natural ICEBs1 transmission is 100- to 10,000-fold more efficient when cells are forming a biofilm, even when recipient cells outnumber donor cells. However, while biofilm formation increases conjugation, it does not influence ICEBs1 excision, suggesting that its effect occurs at the contact level. Accordingly, we observed that the biofilm extracellular matrix is crucial for enhanced ICEBs1 transfer in biofilms.
RESULTS
Biofilm enhances the conjugative transfer of ICEBs1.
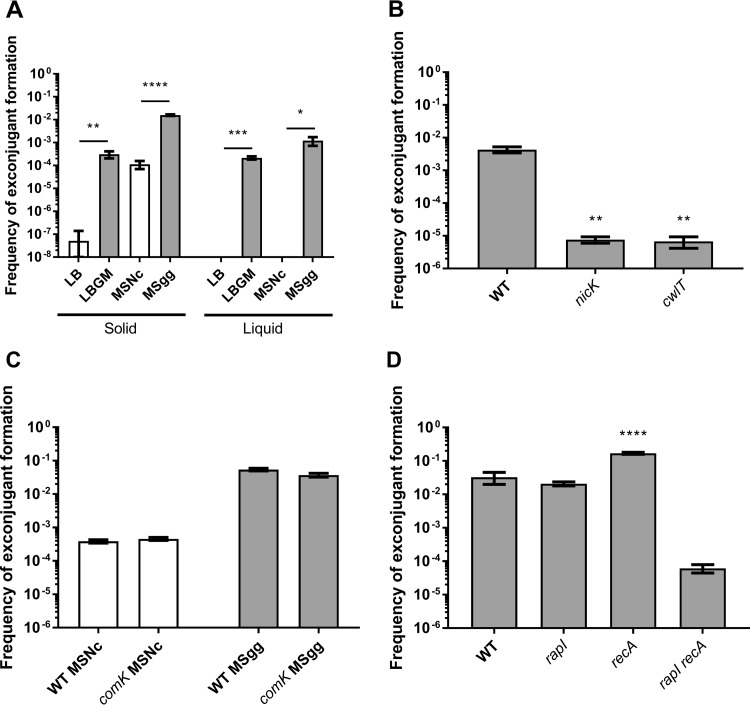
Throughout the years, many aspects of ICEBs1 regulation and transmission have been characterized by using a domesticated strain of B. subtilis, incapable of forming a biofilm (10, 33). Here, we wanted to assess the real impact of biofilm on ICEBs1 transfer, and thus, without artificial induction. Mating assays were performed using NCIB3610, an undomesticated B. subtilis strain that forms strong and well-characterized biofilms and contains a kanamycin selection marker in ICEBs1, as donor cells. Recipient cells were constructed by curing NCIB3610 of ICEBs1 (ICEBs10) as described previously (10). To discriminate between the effects of medium composition versus biofilm induction, pairs of complex rich media and defined minimal media were used as conjugative support. For each pair, one medium induces biofilm formation (LBGM and MSgg [see Materials and Methods]), while the other does not (LB and MSNc [see Materials and Methods]) (24, 25, 34) (see Fig. S1A in the supplemental material). Donor and recipient cells were mated in a 1:1 ratio, spotted, and incubated for 20 h on the various solid media. Conjugation efficiency was determined by plating serial dilutions of mating mixtures on selective media. Strikingly, we observed that the efficiency of exconjugant formation on biofilm-inducing media, compared to the conjugation on noninducing media, increased between 100-fold (minimal media; compare MSgg with MSNc) and 10,000-fold (rich media; compare LBGM with LB) (Fig. 1A). Importantly, these very high levels of transfer were obtained without the need to artificially activate ICEBs1 excision or add a DNA-damaging reagent such as mitomycin C. Similarly, formation of floating biofilms (pellicles) in liquid biofilm-inducing media induced high levels of ICEBs1 transfer, while media not inducing biofilms, in which cells are in planktonic form, showed no ICEBs1 transfer (Fig. 1A).
FIG 1.
Biofilm formation enhances ICEBs1 transfer. (A) Donor cells with a kanamycin resistance cassette inserted in ICEBs1 were mated with recipient cells bearing an intergenic chloramphenicol resistance cassette in a 1:1 ratio on non-biofilm-inducing solid and liquid media (LB and MSNc [white bars]) and biofilm-inducing solid and liquid (pellicle-inducing) media (LBGM and MSgg [gray bars]). Statistical analysis indicates a significant increase in ICEBs1 transfer when biofilm is formed (Student’s t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). (B) Conjugation-deficient donor cells (nicK and cwlT) were mated with WT cells on MSgg to assess mating efficiency. Statistical analysis shows a significant decrease in mating efficiency with the nicK and cwlT donor cells (one-way ANOVA; **, P < 0.01). (C) Transformation-deficient cells (comK) were mated on MSNc and MSgg, and mating efficiency was compared to that of WT cells. Statistical analysis shows no significant difference in mating efficiencies between comK and WT cells (Student’s t test). (D) Single and double mutant ICEBs1 activation pathway donor cells were mated with WT recipient cells on MSgg. Statistical analysis shows a significant increase of ICEBs1 mating efficiency between recA and WT donor cells, but not with the rapI mutant (one-way ANOVA; ****, P < 0.0001). While the double mutant showed a decrease in mating efficiency, that difference was not significant. For all panels, mating efficiency was measured after 20 h for solid media and 28 h for liquid media at 30°C. The results shown are representative of at least three independent experiments, and error bars represent the standard error of the mean (SEM).
LBGM and MSgg induce biofilm formation. WT (A) and sinR (B) donor and recipient cells were mated on LB (i), LBGM (ii), MSNc (iii), and MSgg (iv). Pictures were taken after incubation for 20 h at 30°C. Download FIG S1, PDF file, 0.3 MB (274.5KB, pdf) .
Copyright © 2018 Lécuyer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To confirm that increased conjugative ICEBs1 transfer explains the high level of exconjugant formation in biofilms, we performed mating assays on MSgg using nicK and cwlT deletion mutants: nicK and cwlT encode the relaxase and a cell wall hydrolase associated with ICEBs1 type IV secretion system, respectively (17, 35). As expected, transfer efficiencies dropped significantly for donors with either deletion (Fig. 1B), indicating that conjugation is the main HGT mechanism used for ICEBs1 acquisition in biofilm. Of note, on MSgg, nicK and cwlT mutants exhibited a residual frequency of exconjugant formation of 10−5: i.e., less than 1% of wild type (WT). Since extracellular DNA is a common feature of biofilm matrix, these exconjugants could result from acquisition of genetic material via transformation (27, 36). We tested this hypothesis by doing a conjugation assay with donor and recipient cells mutated for comK, the competence transcription factor of B. subtilis required for transformation (37), and nicK donor cells. As shown in Fig. S2 in the supplemental material, conjugation- and transformation-deficient cells did not produce any kan+ cat+ (exconjugants) cells on MSgg, confirming that natural transformations is responsible for approximately 1 out of 100 kan+ cat+ cells formed in biofilm. However, since the contribution of natural competency is negligible, mating assays performed with comK or WT cells showed similar efficiencies of exconjugant formation in biofilms (Fig. 1C).
nicK residual exconjugant formation is due to natural transformation. Conjugation assays were performed with WT plus WT, nicK plus WT, and nicK comK plus comK cells on MSgg. Mating efficiency was measured after 20 h at 30°C. While the nicK plus WT assay showed a low level of exconjugant formation, the combination nicK comK plus comK led to complete abolition of transfer. Results shown are representative of at least three independent experiments, and error bars represent the SEM. Download FIG S2, PDF file, 0.1 MB (59.7KB, pdf) .
Copyright © 2018 Lécuyer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In B. subtilis, RapI and RecA are both capable of lifting inhibition on ICEBs1 excision and transfer (10). To evaluate which pathway regulates ICEBs1 transfer during biofilm formation, we performed mating assays on MSgg using rapI or recA null mutants as donors. We observed that neither led to a significant decrease of mating efficiency and that recA donor cells exhibited 5- to 10-fold higher mating efficiency. These results suggest either that a third unknown ICEBs1 activation pathway is active in biofilms or that both RapI and RecA pathways are redundant in biofilms. To discriminate between these two hypotheses, we tested the mating efficiency of a rapI recA donor strain and observed a 1,000-fold decrease in mating efficiency compared to the WT (Fig. 1D). These results suggest that both ICEBs1 activation pathways are active during biofilm formation and that they are redundant in biofilm. Interestingly, the frequencies of exconjugant formation with nicK, cwlT, or the rapI recA donors were comparable (Fig. 1B and D), further strengthening the notion that rapI and recA are the only ICEBs1 activators in biofilm.
Biofilm allows for highly efficient conjugation in a low donor/recipient ratio.
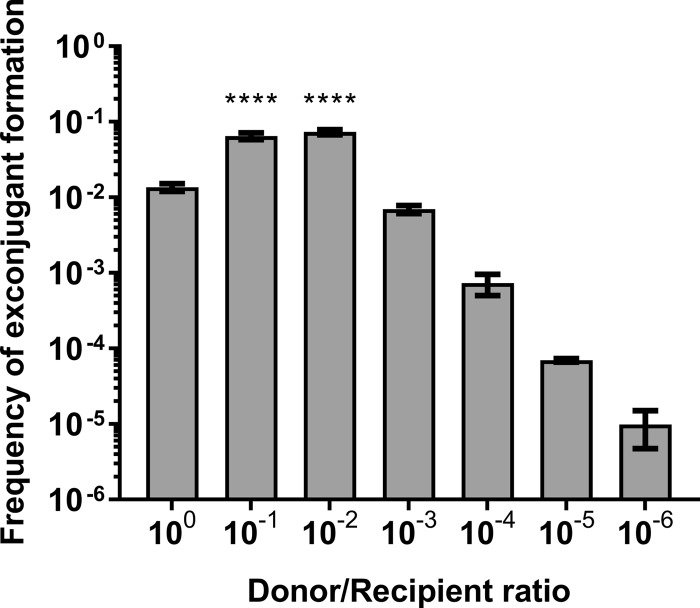
The 1:1 ratio of donor to recipient cells often used to assess conjugation in vitro is probably not frequently encountered in the environment. Therefore, we performed mating assays on MSgg using donor/recipient ratios ranging from 1:1 to 1:106. We observed that exconjugant formation frequency was at its highest when recipient cells outnumbered donor cells by 10 to 100 times, whereas similar efficiencies were obtained between the 1:1 and the 1:103 ratios and lower ratios showed decreased efficiency (Fig. 2). These observations can be partially explained by the ICEBs1-encoded quorum-sensing system RapI-PhrI, since extracellular PhrI would not be sufficient to inhibit the RapI activator upon low donor cell density in the population. Alternatively, diffusion of PhrI could be hampered in the presence of the biofilm matrix. Of note, it is possible that newly formed exconjugants transfer ICEBs1 immediately after receiving it, which would compensate for the low initial level of donor cells. These results indicate that a small population of donor cells can efficiently transfer ICEBs1 in a biofilm community.
FIG 2.
Lower donor/recipient ratio allows increased ICEBs1 transfer in biofilm. WT donor cells were diluted and mated with a fixed number of WT recipient cells on MSgg. Transfer efficiency was measured after 20 h at 30°C. Donor/recipient ratios of 1:10 and 1:100 show significantly more ICEBs1 transfer efficiency than the 1:1 ratio (one-way ANOVA; ****, P < 0.0001). The results shown are representative of at least three independent experiments, and error bars represent the SEM.
Conjugation activation and biofilm formation are simultaneous.
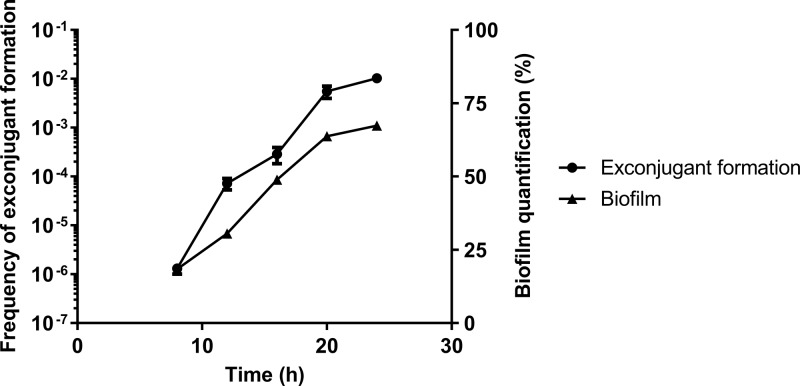
Since biofilm formation positively influences ICEBs1 transfer, we examined the timing of conjugation in relation to biofilm formation. Mating efficiency was assessed on MSgg at different time points using donor and recipient cells carrying the PtapA-yfp reporter. In this reporter gene construction, the yellow fluorescent protein (YFP) is under the control of a matrix gene promoter, thus, allowing its expression concomitantly to matrix production, which can therefore be evaluated by quantification of fluorescent cells in a population by flow cytometry (24, 38). On solid MSgg medium, we observed a steady increase in both mating efficiency and biofilm matrix induction from 8 to 20 h after inoculation (Fig. 3), indicating a temporal correlation between biofilm formation and conjugation.
FIG 3.
Biofilm formation and ICEBs1 conjugation activation are simultaneous. Recipient and donor cells harboring a PtapA-yfp fluorescent marker in the amyE locus were mated on MSgg. Biofilms were then harvested after 8, 12, 16, 20, and 24 h. For each time point, cells were used to quantify biofilm expression by fluorescence-activated cell sorter (FACS) and to assess mating efficiency. The results shown are representative of at least three independent experiments, and error bars represent the SEM.
Biofilm formation does not alter excision of ICEBs1.
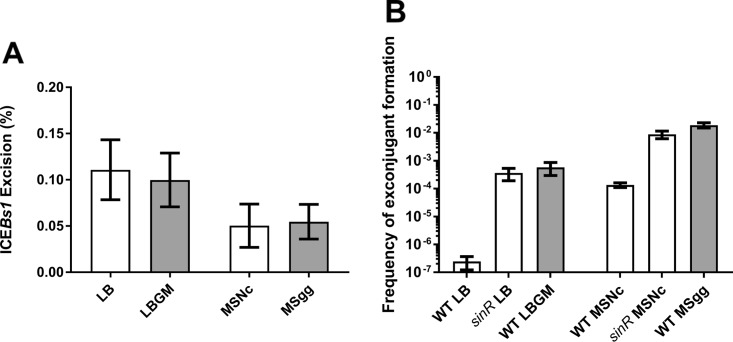
The increase of conjugation throughout biofilm formation could be due to a gradual increase in ICEBs1 excision, which is under the control of several signaling pathways. To examine this hypothesis, we determined the ICEBs1 excision level under biofilm-forming conditions by monitoring the formation of the attB site in donor cells using quantitative PCR (qPCR). Since quorum sensing can influence ICEBs1 excision, we used a 1:1 donor/recipient ratio. Accordingly, we constructed recipient cells in which an erythromycin resistance cassette was inserted at the hybridization site of one of the qPCR primers. In this context, the recipient unoccupied attB site cannot be amplified, although the site remains functional (see Fig. S3 in the supplemental material). Surprisingly, there was no significant increase of ICEBs1 excision in donor cells on biofilm-inducing media compared to noninducing media (Fig. 4A). We also followed ICEBs1 excision in donor cells over time using the same time points as the conjugation assay previously described and observed low levels of excision between 4 h and 24 h (see Fig. S4 in the supplemental material). Of note, this method does not allow us to evaluate excision rates in exconjugants, which may lead to an underestimation of the subset of cells bearing excised ICEBs1. However, these results show that biofilm formation does not alter ICEBs1 excision in donor cells.
FIG 4.
Matrix production is important for conjugation. (A) WT donor cells and ICEBs10 attB-down recipient cells were mated on LB, LBGM, MSNc, and MSgg for 20 h at 30°C, and the donor attB site was amplified by qPCR. There was no significant difference in the ICEBs1 excision rate when there was biofilm formation (Student’s t test). (B) sinR donor and recipient cells were mated on non-inducing media, and mating efficiency was compared to that of WT cells mated on non-biofilm-inducing (LB and MSNc [white bars]) and biofilm-inducing solid media (LBGM and MSgg [gray bars]). Mating efficiency was measured after 20 h at 30°C. sinR mutants led to higher ICEBs1 transfer efficiency compared to WT cells on noninducing media. For all panels, the results shown are representative of at least three independent experiments, and error bars represent the SEM.
ICEBs10 attB-down recipient cells can acquire ICEBs1, while the attB site cannot be amplified. (A) ICEBs10 attB-down (JSB18) cells were mated on MSgg with WT donor cells to assess their capacity to acquire ICEBs1, despite an erythromycin cassette inserted near the attB site. Mating efficiency was measured after 20 h at 30°C. Statistical analysis showed no significant difference in mating efficiencies between the WT recipient cells and JSB18 cells (Student’s t test). Results shown are representative of at least three independent experiments, and error bars represent the SEM. (B) The attB site of a WT recipient cell (lane 1), JSB18 (lane 2), and a 1:1 mixture using JSB18 (lane 3) as a recipient was amplified by PCR using primers P333 and P358 (see Materials and Methods) and run on a 1% agarose gel. We were able to amplify the attB site from the WT recipient cells and the 1:1 mixture but not from JSB18 alone. Download FIG S3, PDF file, 0.1 MB (71.2KB, pdf) .
Copyright © 2018 Lécuyer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Biofilm does not affect ICEBs1 excision over time. WT donor cells and ICEBs10 attB-down recipient cells were mated on MSgg, and the donor attB site was amplified by qPCR after 4, 8, 12, 16, and 20 h at 30°C. We did not detect an increase in ICEBs1 excision over time. Download FIG S4, PDF file, 0.1 MB (55.1KB, pdf) .
Copyright © 2018 Lécuyer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A second hypothesis to explain the positive effect of biofilm on conjugation is that the biophysical context provided by the extracellular matrix highly favors conjugative transfer. To test this idea, we used donor and recipient cells deleted for sinR, the transcriptional repressor of the tapA-sipW-tasA and epsA–O operons responsible for matrix production. A sinR mutant constitutively produces the biofilm matrix, even on non-biofilm-inducing media, with little effect on the upstream signaling pathways (27). This single mutation is sufficient to induce the formation robust biofilm, regardless of the media (Fig. S1B). As shown in Fig. 4B, transfer efficiency using sinR cells on both non-biofilm-inducing media was similar to that in WT cells on biofilm-inducing media. These results demonstrate that matrix production is sufficient to promote efficient transfer of ICEBs1 under non-biofilm conditions. To evaluate the importance of cell contact mediated by biofilm matrix for conjugative transfer versus the possible effects of sinR mutants or biofilm-inducing media on other cell processes, we examined ICEBs1 transfer in WT and sinR cells in planktonic (shaking) LB, LBGM, MSNc, and MSgg. Importantly, the WT in shaking biofilm-inducing media did not transfer ICEBs1 at all (LBGM) or transferred it at a lower rate than under biofilm conditions (floating pellicles) at similar donor and recipient cell densities (see Fig. S5 in the supplemental material). Also, sinR cells in all media and WT cells in MSgg rapidly clump despite agitation and show ICEBs1 transfer, suggesting that these cell aggregates mediated by matrix secretion are microenvironments favoring conjugative transfer (Fig. S5). Together, these results suggest that maximum transfer rates are obtained when biofilm matrix is produced and hold cells together.
Conjugative transfer of ICEBs1 is inefficient under shaking conditions. WT, eps tasA, and sinR donor and recipient cells were mixed and inoculated in liquid LB, LBGM, MSNc, and MSgg. Tubes were then incubated for 20 h with shaking at 30°C, and exconjugant frequency was evaluated. The results shown are representative of at least three independent experiments, and error bars represent the SEM. Download FIG S5, PDF file, 0.1 MB (55KB, pdf) .
Copyright © 2018 Lécuyer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
TasA was shown to bind cell together in the biofilm, and matrix exopolysaccharides were suggested to favor adhesion on neighboring cell chains in complex community development (39, 40). To strengthen the hypothesis that biofilm formation can provide a favorable context for conjugation by bringing cells closer or by stabilizing cell-cell contacts, we mated donor and recipient cells incapable of producing matrix (epsA–O and tasA mutants) on MSNc. We decided to emulate the binding effect provided by the extracellular matrix by adding 1% agarose, which is expected to move the matrix-deficient cells closer and stabilize their contact. We observed that addition of agarose to eps tasA mutants increased ICEBs1 transfer efficiency (see Fig. S6 in the supplemental material), suggesting that the polymer helps to stabilize the contact between cells the same way the extracellular matrix can, albeit to a lesser degree.
Polymers can act as a surrogate biofilm in absence of extracellular matrix. WT cells and epsA–O tasA (eps) cells were resuspended in 1% agarose and mated on MSNc. Mating efficiency was measured after 20 h at 30°C. Statistical analysis showed a significant increase in ICEBs1 transfer efficiency when agarose was added to eps tasA cells, but not for the WT cells (Student’s t test; ****, P < 0.0001). Results shown are representative of at least three independent experiments, and error bars represent the SEM. Download FIG S6, PDF file, 0.1 MB (62.4KB, pdf) .
Copyright © 2018 Lécuyer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Matrix production by recipient cells is important for optimum conjugation in biofilm.
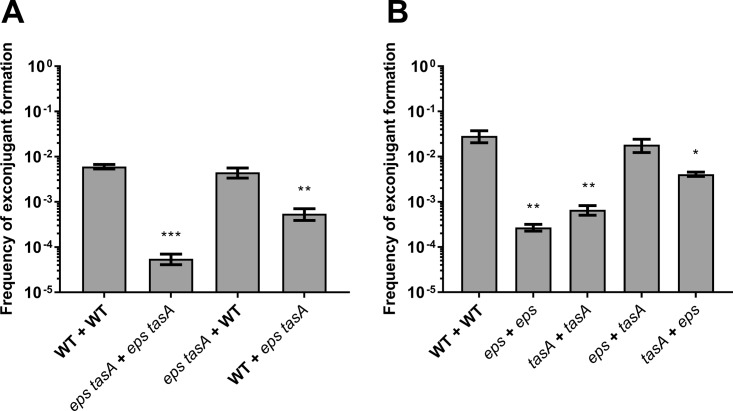
The results obtained with sinR mutants suggest that the biofilm matrix acts as a structure promoting cell-cell contacts and optimal ICEBs1 conjugative transfer. This hypothesis was further verified by carrying out mating assays with cells incapable of secreting matrix (i.e., eps tasA mutants). In B. subtilis, the epsA–O (eps) and tasA operons produce two major matrix components: exopolysaccharides and the TasA amyloid-like fibers, respectively (23). As shown in Fig. 5A, production of the extracellular matrix polymers was required for efficient ICEBs1 transfer on biofilm-inducing medium. Indeed, mating assays using mutant donor and recipient cells yielded transfer efficiencies similar to those observed on a minimal non-biofilm-inducing medium. Interestingly, while donor cells deficient for matrix production could still efficiently conjugate with WT recipient cells, mating WT donor with non-matrix-producing recipient cells significantly reduced transfer (Fig. 5A). This result suggests that the biofilm matrix production of the recipient cells is particularly important for efficient ICEBs1 transfer.
FIG 5.
Both matrix components are important for ICEBs1 conjugation. (A) Donor and recipient cells deleted for the epsA–O (eps) and tasA operons were mated together or with WT cells on MSgg. The first genotype shown represents the donor genotype, while the second represent the recipient. Statistical analysis showed that absence of matrix and nonproduction from the recipient cells reduced significantly ICEBs1 transfer efficiency (one-way ANOVA; **, P < 0.01; ***, P < 0.001). (B) Donor and recipient cells mutated for either eps or tasA operon were mated on MSgg. Statistical analysis showed that absence of either exopolysaccharides or amyloid-like fibers in both donor and recipient decreases ICEBs1 transfer significantly. However, eps donors and tasA recipients can complement each other and restore the WT level of conjugation, while tasA donors and eps recipients are significantly different from WT pairs (one-way ANOVA; *, P < 0.05; **, P < 0.01). For both panels, mating efficiency was measured after 20 h at 30°C. The results shown are representative of at least three independent experiments, and error bars represent the SEM.
To examine the importance of both components for conjugation, mating assays with donor and recipient lacking either eps or tasA were performed. As shown in Fig. 5B, both components of the matrix were instrumental for efficient ICEBs1 transfer, stressing the importance of the matrix integrity for maximal conjugation. Various reports have shown that eps and tasA mutants can complement each other extracellularly to establish biofilm both in vitro and on plant roots (23, 24). Interestingly, combination of these mutants can also restore conjugation efficiency, but only when TasA is produced by donor cells (eps mutant) and the exopolysaccharides are produced by recipient cells (tasA mutant). The reverse combination resulted in a 5-fold reduction of transfer efficiency compared to WT cells (Fig. 5B). This result confirms that the extracellular matrix is essential for ICEBs1 transfer in biofilms and that matrix production by recipient cells is essential for optimal transfer.
DISCUSSION
ICEBs1 regulation and its transfer mechanism have been thoroughly characterized in the last decade. However, as is also the case for most conjugative elements, its transfer between cells within a biofilm has not been previously studied. Here, we show that biofilm formation greatly increases conjugation of ICEBs1, allowing for high-efficiency transfer in the absence of added DNA-damaging reagents.
Using donor cells carrying nicK and cwlT deletion mutations, both unable to transfer ICEBs1, we validated that the high transfer observed in biofilm is due to conjugation events (Fig. 1B). However, we also observed a 10−5 background level of kan+ cat+ “exconjugant” cell formation using donor cells with nicK, cwlT, or rapI recA deleted, the latter being unable to excise ICEBs1 (Fig. 1B and D). Further experiments allowed us to determine that these kan+ cat+ cells arose by natural transformation, via transfer of the Kanr gene present in ICEBs1 to recipient cells or the transfer of the cat gene present in recipient cells to donor cells (Fig. S2) (data not shown). These observations suggest that natural transformation contributes to approximately 1 out of 100 HGTs observed on MSgg and thus is also fairly efficient to promote gene transfer in biofilms formed by nondomesticated strains.
The minimal biofilm-inducing medium (MSgg) provided the highest frequency of ICEBs1 exconjugant formation (i.e., 10−2). Interestingly, rapI overexpression in a domesticated, non-biofilm-inducing B. subtilis strain also gives similar transfer efficiency, suggesting that it might be the upper limit for ICEBs1 transfer in a 1:1 ratio (10, 41). This transfer efficiency, obtained without artificial activation of ICEBs1 excision, demonstrates the high mobility of this element. ICEBs1 is therefore one of the ICEs transferring at the highest rate in the Firmicutes. Indeed, Staphylococcus aureus ICE6013, an ICE closely related to ICEBs1, has a mating efficiency of around 10−5 (42). Tn916, an ICE found in a variety of Gram-positive bacteria, was reported to transfer at frequencies ranging from 10−9 to 10−4 (43, 44), while Streptococcus agalactiae TnGBS1 and TnGBS2 transfer at around 10−5 (45). Finally, Streptococcus thermophilus ICESt3 transfers at a rate of 3.4 × 10−6, and only one conjugation event was ever reported for ICESt1 (46). However, in the studies mentioned above, filter mating assays, which generally do not promote biofilm formation, were used to assess mating efficiencies. Our study demonstrates that to evaluate naturally relevant conjugation transfer of ICEs, the transfer rate within a biofilm must be examined.
Variable donor/recipient ratios ranging from 1:1 to 1:103 in the mating population did not decrease the frequency of recipient cells acquiring ICEBs1. This observation is very important, since it reflects how a genetic element, present in a small subset of an initial population, can propagate rapidly and efficiently. In fact, a ratio of 1:10 to 1:102 led to a higher mating efficiency than a ratio of 1:1 (Fig. 2). This observation was previously reported for ICEBs1 (10), but was never explored in a biofilm-related setting. This increase in efficiency could be explained by the RapI-PhrI quorum-sensing system, since a smaller amount of donor cells in the biofilm leads to a low level of PhrI in the extracellular environment, thus favoring the action of the RapI activator (10). A similar system was described in Enterococcus faecalis, where a lower donor cell density led to higher transfer efficiency of conjugative plasmids pAD1 and pAM373, which both encode a secreted conjugation inhibitor (iAD1 and iAM373, respectively) (47). However, quorum-sensing regulation of excision has been observed only in a limited subset of genetic elements. For many others, DNA damage and/or environmental conditions such as reaching stationary phase or the presence of subinhibitory concentrations of antibiotics trigger ICE excision (48–50). ICEBs1 excision is also induced by DNA damage via the recA pathway, but recA donors showed better conjugation efficiency than WT donors (Fig. 1D). A similar increase of mating efficiency with a recA donor was observed previously (51). This result can be explained by the poor growth of recA mutants, known to have a slower doubling time (52). While cells were initially mated at a 1:1 donor/recipient ratio, we observed that after 20 h, the ratio had become approximately 1:50 (see Table S1 in the supplemental material), leading to a higher mating efficiency (Fig. 2).
recA and WT donor cell counts after 20 h. Download Table S1, PDF file, 0.1 MB (95KB, pdf) .
Copyright © 2018 Lécuyer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Somewhat surprisingly, we observed that biofilm formation does not induce excision of ICEBs1 in donor cells. Despite the low excision rate (∼0.2%), transfer levels in biofilms were similar to those obtained with donor cells overexpressing rapI, for which excision rates reach approximately 90% (10, 51). Of note, ∼0.2% is more than 10 times as high as the excision rate observed in an uninduced domesticated strain (51). ICEBs1 is known to replicate in a rolling circle and can be present in multiple copies in the donor cell (16). Thus, we hypothesize that under biofilm-forming conditions, ICEBs1 rapidly reintegrates into its host chromosome following replication. The extrachromosomal copies would then be transferred to recipient cells, explaining the efficient transfer despite low excision levels. Another hypothesis underlying the high conjugative transfer of ICEBs1 in biofilm could be the presence of abundant cell chains. Indeed, ICEBs1 transfers exceptionally well through bacterial chains (53), and these structures are frequently found in biofilms, which could help propagate ICEBs1 much more efficiently (23, 34). Considering the very low excision rate and the high conjugative transfer, it is also extremely likely that a single donor cell can propagate ICEBs1 to multiple recipients in biofilms. It is also likely that once a recipient receives ICEBs1, it can immediately become a donor, further spreading it in the population. Importantly, the excision rate is not necessarily correlated with conjugation efficiency, as shown for Tn916 (43).
Many conjugative elements encode surface factors that stabilize the contact between donor and recipient cells, such as conjugative pili in Gram-negative bacteria and adhesins in Gram-positive bacteria (54, 55). While it is unknown whether ICEBs1 encodes surface factors, its low transfer efficiency in liquid compared to solid media suggests that no such factors are expressed under these conditions (56). Here, we have shown that both components of the extracellular matrix are required for the positive effect of biofilm on conjugation, suggesting that these polymers could help stabilize the donor-recipient pair and compensate for the lack of adhesion factors of ICEBs1. Other conjugative elements that are not known to encode adhesion factors, such as pCW3 and Tn916, are found in the biofilm-forming bacteria Clostridium perfringens and E. faecalis, respectively (57–59). Lack of adhesion factors in those elements could be compensated for by the ability of their host cells to form biofilms.
Experiments with single and double biofilm mutants allowed us to determine the individual importance of both matrix components in conjugation. Interestingly, matrix production from the recipient cells, but not from the donor cells, is likely essential for efficient transfer (Fig. 5A). This observation could be explained by the fact that recipients that do not produce matrix will not form cell chains, and thus, lead to less-efficient ICEBs1 transfer. It also suggests that cells within a biofilm might be able to receive ICEs from either biofilm- or non-biofilm-forming cells, making the biofilm a very receptive environment for genetic element transfer. These results allow us to better understand conjugative element dynamics in natural and clinical environments, where biofilms are ubiquitous. Biofilm matrix could thus have a considerable impact on the dissemination of mobile genetic elements, such as for the clinically important bacteria S. aureus and C. difficile, which can acquire multiple-antibiotic resistance through ICEs (60, 61).
MATERIALS AND METHODS
Strains and media.
The strains used in this study are derivatives of the ancestor strain NCIB3610 (see Table S2 in the supplemental material). The different media used for mating assays are LB (Luria Bertani; 1% tryptone, 0.5% yeast extract, 0.5% NaCl), LBGM (LB plus 1% glycerol and 0.1 mM MnCl2 [25]), MSNc (5 mM potassium phosphate buffer, pH 7, 0.1 M morpholinepropanesulfonic acid [MOPS], pH 7, 2 mM MgCl2, 0.05 mM MnCl2, 1 µM ZnCl2, 2 µM thiamine, 700 µM CaCl2, 0.2% NH4Cl, 0.5% cellobiose) (24), and MSgg (5 mM potassium phosphate buffer, pH 7, 0.1 M MOPS, pH 7, 0.025 mM FeCl3, 2 mM MgCl2, 0.05 mM MnCl2, 1 µM ZnCl2, 2 µM thiamine, 700 µM CaCl2, 0.5% glycerol, 0.5% glutamate) solidified with 1.5% agar (34). Media did not affect significantly bacterial growth, with biofilm-inducing media leading to slightly more yield compared to noninducing media (see Table S3 in the supplemental material). When needed, the following antibiotics were added to media: MLS (1 μg ml−1 erythromycin, 25 μg ml−1 lincomycin), spectinomycin (100 μg ml−1), tetracycline (10 μg ml−1), chloramphenicol (5 μg ml−1), and kanamycin (10 μg ml−1).
Bacillus subtilis strains used. Download Table S2, PDF file, 0.2 MB (162.5KB, pdf) .
Copyright © 2018 Lécuyer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Donor and recipient cell counts in different media after 20 h. Download Table S3, PDF file, 0.1 MB (104.8KB, pdf) .
Copyright © 2018 Lécuyer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strain construction.
Most strains were made by transferring genetic constructs present in domesticated strains in NCIB3610, using SPP1-mediated generalized transduction (62). JMA348 (ICEBs1::kan), CAL51 [(rapI phrI)342::kan], JMA208 (immR::cat), and CAL419 (ICEBs10 comK::cat) were kind gifts from Alan D. Grossman (Massachusetts Institute of Technology, MA), and 3610 ICEBs10 strains were cured from ICEBs1 and verified as described in reference 10. Briefly, MG9 (3610 immR::cat) was inoculated in LB, grown for 4 h, diluted at an optical density at 600 nm (OD600) of 0.01 in fresh LB, and grown overnight at 37°C. The culture was then diluted back to an OD600 of 0.01 in fresh LB and grown until the culture reached an OD600 of 1. The cells were then plated on LB agar and grown overnight at 37°C, and colonies were streaked on LB with or without chloramphenicol. Colonies that lost the resistance were then PCR verified for ICEBs1 excision with the following primers (5′→3′): P197 (GAC GAA TAT GGC AAG CCT ATG TTA C) and P198 (GGG TAT ACA ATC ATG GGT GAT CGA G).
Long-flanking homology PCR was used to insert a spectinomycin cassette between ycbU and lmrB and to create the recipient used for qPCR (JSB18). The following primers (5′→3′) were used for the ycbU-lmrB::spec insertion: P246 (CCA TTG ATG TGA AGG AAT GGG GCG TA), P247 (CGT TAC GTT ATT AGC GAG CCA GTC ATG TTT ACT TGT GGA TCG TTT TCG CCG), P248 (CAA TAA ACC CTT GCC CTC GCT ACG CCT GAA CAC TAG TCA GGG GCT TTT CA), and P249 (GGC TTA GTC CTC ACT GCA TTT GCA TC). The following primers (5′→3′) were used for the attB-down::erm deletion: P328 (CCG TTG GTC AAG CGG TTA AG), P329 (GAG GGT TGC CAG AGT TAA AGG ATC TAT TAT TGA GAT GCG GCC GAG), P330 (CGA TTA TGT CTT TTG CGC AGT CGG CGT GTG GAA AAT ACG GCT ATG GG), and P331 (AGT AAG CTT ATT CCA CCC ACT G). PCR products were then introduced in B. subtilis 168 by natural competency (63), verified by PCR, and transferred in derivatives of B. subtilis NCIB3610 by SPP1-mediated generalized transduction (62).
Mating assays.
Donor and recipient cells were grown in 3 ml LB broth at 37°C overnight, diluted at an OD600 of 1.5 in LB, and mixed at a 1:1 ratio (or the specified ratio [Fig. 2]). The mixture was then centrifuged for 3 min at 5,000 rpm. The cell pellet was resuspended in 50 µl LB, and 10 µl was dropped onto the appropriate medium and incubated for 20 h (or the time specified [Fig. 3]) at 30°C, which is the temperature at which B. subtilis biofilm grows efficiently. For mating assays in liquid, 10 µl of bacterial mixture was used to inoculate 3 ml of medium for shaking conditions, while 3.33 µl was used to inoculate 1 ml of medium found in a 24-well plate for pellicle mating; both were also done at 30°C. Pellicles were incubated for 28 rather than 20 h, since the growth dynamic is slower under nonshaking conditions. For mating assays using agarose, cells were resuspended in 50 µl of warm (55°C) 1% molecular-grade agarose before being dropped on agar medium. Cells were then collected with 1 ml LB broth and sonicated at 30% amplitude for 20 s two times for cells grown for 20 h on biofilm-inducing media. Microscopy observation allowed us to determine that sonication was sufficient to obtain single cells. Cells were then serially diluted and plated on LB with the appropriate antibiotics. Donor, recipient, and exconjugant CFU were then counted. We expressed the frequency of exconjugant formation as a function of the number of recipient CFU (number of exconjugant CFU divided by the number of recipient CFU), because the starting donor amount varied in some experiments (Fig. 2).
Flow cytometry.
Mating assays were performed on MSgg as described above, using the FL60 and FL63 strains. For each time point, three biofilms were harvested with 500 µl phosphate-buffered saline (PBS: 137 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, 2 mM KH2PO4, pH 7.4) and disrupted with up and down pipetting. Subsequent steps were performed as described previously (38). Flow cytometry analysis was performed on a BD FACSJazz (BD Biosciences).
ICEBs1 excision quantification.
ICEBs1 excision was evaluated using qPCR. Donor and recipient cells were mated on MSgg as described above, and cells were harvested and flash-frozen at the appropriate time point. Genomic DNA was extracted using the BioBasic genomic DNA extraction kit, and qPCR was performed on the attB site (created when ICEBs1 is excised from the chromosome) using the following primers (5′→3′): P358 (GCC TAC TAA ACC AGC ACA AC) and P333 (AAA GGT GGT TAA ACC CTT GG). Since the recipient strain (JSB18) contains an erythromycin resistance cassette in the hybridization site of the P333 primer, amplification is only possible for donor cells in which ICEBs1 is excised. qPCR on a chromosomal chloramphenicol resistance cassette present only in the donor genome was performed for normalization (threshold cycle [ΔCT]) of donor cells, using the following primers (5′→3′): P363 (AGA ACT GGT TAC AAT AGC GAC GGA GAG) and P366 (CCC CGA ACC ATT ATA TTT CTC TAC ATC AGA AAG G). The percentage of excision is calculated as the ΔΔCT using the culture of the ICEBs10 ylnF/yboA::Tn917::amyE::cat control strain grown under the same conditions, which is considered as being 100% excised.
Stereomicroscopy.
Donor and recipient WT and sinR cells were mated on LB, LBGM, MSNc, and MSgg as described above. Photographs of colonies were taken after 20 h at 30°C with a Leika M165 FC (Leika).
Statistical analyses.
Statistical analyses were performed using GraphPad Prism 7. Comparisons were done using Student's t test or one-way analysis of variance (ANOVA) followed by Tukey’s multiple-comparison test, both with 95% confidence intervals.
ACKNOWLEDGMENTS
We thank A. D. Grossman and R. Kolter for their kind gift of strains, A. Lavigueur and A. Maréchal for critical reading of the manuscript, and members of the Beauregard, Burrus, and Rodrigue lab for helpful discussions. This work was supported by a Discovery Grant (RGPIN-2014-04628) from the Natural Sciences and Engineering Council of Canada (NSERC) and a Team Project Grant (2019-PR-253077) from the Fond de Recherche du Québec - Nature et Technologie (FRQ-NT) to P.B.B. and by a Master Degree Fellowship (207639) from the Fond de Recherche du Québec - Nature et Technologie (FRQ-NT) to F.L.
REFERENCES
- 1.Ochman H, Lawrence JG, Groisman EA. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 2.Frost LS, Leplae R, Summers AO, Toussaint A. 2005. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol 3:722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- 3.Moreno Switt AI, den Bakker HC, Cummings CA, Rodriguez-Rivera LD, Govoni G, Raneiri ML, Degoricija L, Brown S, Hoelzer K, Peters JE, Bolchacova E, Furtado MR, Wiedmann M. 2012. Identification and characterization of novel Salmonella mobile elements involved in the dissemination of genes linked to virulence and transmission. PLoS One 7:e41247. doi: 10.1371/journal.pone.0041247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garriss G, Waldor MK, Burrus V. 2009. Mobile antibiotic resistance encoding elements promote their own diversity. PLoS Genet 5:e1000775. doi: 10.1371/journal.pgen.1000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sullivan JT, Ronson CW. 1998. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc Natl Acad Sci U S A 95:5145–5149. doi: 10.1073/pnas.95.9.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aviv G, Rahav G, Gal-Mor O. 2016. Horizontal transfer of the Salmonella enterica serovar Infantis resistance and virulence plasmid pESI to the gut microbiota of warm-blooded hosts. mBio 7:e01395-16. doi: 10.1128/mBio.01395-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamamoto M, Matsumura Y, Gomi R, Matsuda T, Tanaka M, Nagao M, Takakura S, Uemoto S, Ichiyama S. 2016. Interspecies dissemination of a mobilizable plasmid harboring blaIMP-19 and the possibility of horizontal gene transfer in a single patient. Antimicrob Agents Chemother 60:5412–5419. doi: 10.1128/AAC.00933-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee CA, Thomas J, Grossman AD. 2012. The Bacillus subtilis conjugative transposon ICEBs1 mobilizes plasmids lacking dedicated mobilization functions. J Bacteriol 194:3165–3172. doi: 10.1128/JB.00301-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douard G, Praud K, Cloeckaert A, Doublet B. 2010. The Salmonella genomic island 1 is specifically mobilized in trans by the IncA/C multidrug resistance plasmid family. PLoS One 5:e15302. doi: 10.1371/journal.pone.0015302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Auchtung JM, Lee CA, Monson RE, Lehman AP, Grossman AD. 2005. Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. Proc Natl Acad Sci U S A 102:12554–12559. doi: 10.1073/pnas.0505835102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Earl AM, Losick R, Kolter R. 2007. Bacillus subtilis genome diversity. J Bacteriol 189:1163–1170. doi: 10.1128/JB.01343-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bais HP, Fall R, Vivanco JM. 2004. Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol 134:307–319. doi: 10.1104/pp.103.028712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arkhipova TN, Veselov SU, Melentiev AI, Martynenko EV, Kudoyarova GR. 2005. Ability of bacterium Bacillus subtilis to produce cytokinins and to influence the growth and endogenous hormone content of lettuce plants. Plant Soil 272:201–209. doi: 10.1007/s11104-004-5047-x. [DOI] [Google Scholar]
- 14.Allard-Massicotte R, Tessier L, Lécuyer F, Lakshmanan V, Lucier JF, Garneau D, Caudwell L, Vlamakis H, Bais HP, Beauregard PB. 2016. Bacillus subtilis early colonization of Arabidopsis thaliana roots involves multiple chemotaxis receptors. mBio 7:e01664-16. doi: 10.1128/mBio.01664-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Auchtung JM, Aleksanyan N, Bulku A, Berkmen MB. 2016. Biology of ICEBs1, an integrative and conjugative element in Bacillus subtilis. Plasmid 86:14–25. doi: 10.1016/j.plasmid.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 16.Lee CA, Babic A, Grossman AD. 2010. Autonomous plasmid-like replication of a conjugative transposon. Mol Microbiol 75:268–279. doi: 10.1111/j.1365-2958.2009.06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee CA, Grossman AD. 2007. Identification of the origin of transfer (oriT) and DNA relaxase required for conjugation of the integrative and conjugative element ICEBs1 of Bacillus subtilis. J Bacteriol 189:7254–7261. doi: 10.1128/JB.00932-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leonetti CT, Hamada MA, Laurer SJ, Broulidakis MP, Swerdlow KJ, Lee CA, Grossman AD, Berkmen MB. 2015. Critical components of the conjugation machinery of the integrative and conjugative element ICEBs1 of Bacillus subtilis. J Bacteriol 197:2558–2567. doi: 10.1128/JB.00142-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright LD, Johnson CM, Grossman AD. 2015. Identification of a single strand origin of replication in the integrative and conjugative element ICEBs1 of Bacillus subtilis. PLoS Genet 11:e1005556. doi: 10.1371/journal.pgen.1005556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. 2010. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents 35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 21.Nocelli N, Bogino P, Banchio E, Giordano W. 2016. Roles of extracellular polysaccharides and biofilm formation in heavy metal resistance of rhizobia. Materials 9:E418. doi: 10.3390/ma9060418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 23.Branda SS, Chu F, Kearns DB, Losick R, Kolter R. 2006. A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol 59:1229–1238. doi: 10.1111/j.1365-2958.2005.05020.x. [DOI] [PubMed] [Google Scholar]
- 24.Beauregard PB, Chai Y, Vlamakis H, Losick R, Kolter R. 2013. Bacillus subtilis biofilm induction by plant polysaccharides. Proc Natl Acad Sci U S A 110:E1621–E1630. doi: 10.1073/pnas.1218984110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shemesh M, Chai Y. 2013. A combination of glycerol and manganese promotes biofilm formation in Bacillus subtilis via histidine kinase KinD signaling. J Bacteriol 195:2747–2754. doi: 10.1128/JB.00028-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopez D, Fischbach MA, Chu F, Losick R, Kolter R. 2009. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc Natl Acad Sci U S A 106:280–285. doi: 10.1073/pnas.0810940106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kearns DB, Chu F, Branda SS, Kolter R, Losick R. 2004. A master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol 55:739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- 28.Molin S, Tolker-Nielsen T. 2003. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr Opin Biotechnol 14:255–261. doi: 10.1016/S0958-1669(03)00036-3. [DOI] [PubMed] [Google Scholar]
- 29.Madsen JS, Burmølle M, Hansen LH, Sørensen SJ. 2012. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol Med Microbiol 65:183–195. doi: 10.1111/j.1574-695X.2012.00960.x. [DOI] [PubMed] [Google Scholar]
- 30.Monecke S, Coombs G, Shore AC, Coleman DC, Akpaka P, Borg M, Chow H, Ip M, Jatzwauk L, Jonas D, Kadlec K, Kearns A, Laurent F, O'Brien FG, Pearson J, Ruppelt A, Schwarz S, Scicluna E, Slickers P, Tan H-L, Weber S, Ehricht R. 2011. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS One 6:e17936. doi: 10.1371/journal.pone.0017936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Archer NK, Mazaitis MJ, Costerton JW, Leid JG, Powers ME, Shirtliff ME. 2011. Properties, regulation and roles in human disease Staphylococcus aureus biofilms. Virulence 2:445–459. doi: 10.4161/viru.2.5.17724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teschler JK, Zamorano-Sánchez D, Utada AS, Warner CJA, Wong GCL, Linington RG, Yildiz FH. 2015. Living in the matrix: assembly and control of Vibrio cholerae biofilms. Nat Rev Microbiol 13:255–268. doi: 10.1038/nrmicro3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McLoon AL, Guttenplan SB, Kearns DB, Kolter R, Losick R. 2011. Tracing the domestication of a biofilm-forming bacterium. J Bacteriol 193:2027–2034. doi: 10.1128/JB.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. 2001. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci U S A 98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeWitt T, Grossman AD. 2014. The bifunctional cell wall hydrolase CwlT is needed for conjugation of the integrative and conjugative element ICEBs1 in Bacillus subtilis and B. anthracis. J Bacteriol 196:1588–1596. doi: 10.1128/JB.00012-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 37.Sinderen D, Luttinger A, Kong L, Dubnau D, Venema G, Hamoen L. 1995. comK encodes the competence transcription factor, the key regulatory protein for competence development in Bacillus subtilis. Mol Microbiol 15:455–462. doi: 10.1111/j.1365-2958.1995.tb02259.x. [DOI] [PubMed] [Google Scholar]
- 38.Vlamakis H, Aguilar C, Losick R, Kolter R. 2008. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev 22:945–953. doi: 10.1101/gad.1645008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Gestel J, Vlamakis H, Kolter R. 2015. From cell differentiation to cell collectives: Bacillus subtilis uses division of labor to migrate. PLoS Biol 13:e1002141. doi: 10.1371/journal.pbio.1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Romero D, Aguilar C, Losick R, Kolter R. 2010. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci U S A 107:2230–2234. doi: 10.1073/pnas.0910560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Auchtung JM, Lee CA, Garrison KL, Grossman AD. 2007. Identification and characterization of the immunity repressor (ImmR) that controls the mobile genetic element ICEBs1 of Bacillus subtilis. Mol Microbiol 64:1515–1528. doi: 10.1111/j.1365-2958.2007.05748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sansevere EA, Luo X, Park JY, Yoon S, Seo KS, Robinson DA. 2017. Transposase-mediated excision, conjugative transfer, and diversity of ICE6013 elements in Staphylococcus aureus. J Bacteriol 199:e00629-16. doi: 10.1128/JB.00629-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marra D, Pethel B, Churchward GG, Scott JR. 1999. The frequency of conjugative transposition of Tn916 is not determined by the frequency of excision. J Bacteriol 181:5414–5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seier-Petersen MA, Jasni A, Aarestrup FM, Vigre H, Mullany P, Roberts AP, Agersø Y. 2014. Effect of subinhibitory concentrations of four commonly used biocides on the conjugative transfer of Tn916 in Bacillus subtilis. J Antimicrob Chemother 69:343–348. doi: 10.1093/jac/dkt370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guerillot R, Da Cunha V, Sauvage E, Bouchier C, Glaser P. 2013. Modular evolution of TnGBSs, a new family of integrative and conjugative elements associating insertion sequence transposition, plasmid replication, and conjugation for their spreading. J Bacteriol 195:1979–1990. doi: 10.1128/JB.01745-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bellanger X, Roberts AP, Morel C, Choulet F, Pavlovic G, Mullany P, Decaris B, Guedon G. 2009. Conjugative transfer of the integrative conjugative elements ICESt1 and ICESt3 from Streptococcus thermophilus. J Bacteriol 191:2764–2775. doi: 10.1128/JB.01412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bandyopadhyay A, O'Brien S, Frank KL, Dunny GM, Hu W-S. 2016. Antagonistic donor density effect conserved in multiple enterococcal conjugative plasmids. Appl Environ Microbiol 82:4537–4545. doi: 10.1128/AEM.00363-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Delavat F, Miyazaki R, Carraro N, Pradervand N, van der Meer JR. 2017. The hidden life of integrative and conjugative elements. FEMS Microbiol Rev 41:512–537. doi: 10.1093/femsre/fux008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson CM, Grossman AD. 2015. Integrative and conjugative elements (ICEs): what they do and how they work. Annu Rev Genet 49:577–601. doi: 10.1146/annurev-genet-112414-055018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poulin-Laprade D, Carraro N, Burrus V. 2015. The extended regulatory networks of SXT/R391 integrative and conjugative elements and IncA/C conjugative plasmids. Front Microbiol 6:837. doi: 10.3389/fmicb.2015.00837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee CA, Auchtung JM, Monson RE, Grossman AD. 2007. Identification and characterization of int (integrase), xis (excisionase) and chromosomal attachment sites of the integrative and conjugative element ICEBs1 of Bacillus subtilis. Mol Microbiol 66:1356–1369. doi: 10.1111/j.1365-2958.2007.06000.x. [DOI] [PubMed] [Google Scholar]
- 52.Ramírez-Guadiana FH, Barajas-Ornelas RDC, Corona-Bautista SU, Setlow P, Pedraza-Reyes M. 2016. The RecA-dependent SOS response is active and required for processing of DNA damage during Bacillus subtilis sporulation. PLoS One 11:e0150348. doi: 10.1371/journal.pone.0150348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Babic A, Berkmen MB, Lee CA, Grossman AD. 2011. Efficient gene transfer in bacterial cell chains. mBio 2:e00027-11. doi: 10.1128/mBio.00027-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bhatty M, Laverde Gomez JA, Christie PJ. 2013. The expanding bacterial type IV secretion lexicon. Res Microbiol 164:620–639. doi: 10.1016/j.resmic.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goessweiner-Mohr N, Arends K, Keller W, Grohmann E. 2013. Conjugative type IV secretion systems in Gram-positive bacteria. Plasmid 70:289–302. doi: 10.1016/j.plasmid.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson CM, Grossman AD. 2014. Identification of host genes that affect acquisition of an integrative and conjugative element in Bacillus subtilis. Mol Microbiol 93:1284–1301. doi: 10.1111/mmi.12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alvarez-Martinez CE, Christie PJ. 2009. Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev 73:775–808. doi: 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vidal JE, Shak JR, Canizalez-Roman A. 2015. The CpAL quorum sensing system regulates production of hemolysins CPA and PFO to build Clostridium perfringens biofilms. Infect Immun 83:2430–2442. doi: 10.1128/IAI.00240-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dale JL, Nilson JL, Barnes AMT, Dunny GM. 2017. Restructuring of Enterococcus faecalis biofilm architecture in response to antibiotic-induced stress. NPJ Biofilms Microbiomes 3:15. doi: 10.1038/s41522-017-0023-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haaber J, Penadés JR, Ingmer H. 2017. Transfer of antibiotic resistance in Staphylococcus aureus. Trends Microbiol 25:893–905. doi: 10.1016/j.tim.2017.05.011. [DOI] [PubMed] [Google Scholar]
- 61.Mullany P, Allan E, Roberts AP. 2015. Mobile genetic elements in Clostridium difficile and their role in genome function. Res Microbiol 166:361–367. doi: 10.1016/j.resmic.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yasbin RE, Young FE. 1974. Transduction in Bacillus subtilis by bacteriophage SPP1. J Virol 14:1343–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harwood CR, Cutting SM. 1990. Molecular biological methods for Bacillus. Wiley, Chichester, United Kingdom. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
LBGM and MSgg induce biofilm formation. WT (A) and sinR (B) donor and recipient cells were mated on LB (i), LBGM (ii), MSNc (iii), and MSgg (iv). Pictures were taken after incubation for 20 h at 30°C. Download FIG S1, PDF file, 0.3 MB (274.5KB, pdf) .
Copyright © 2018 Lécuyer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
nicK residual exconjugant formation is due to natural transformation. Conjugation assays were performed with WT plus WT, nicK plus WT, and nicK comK plus comK cells on MSgg. Mating efficiency was measured after 20 h at 30°C. While the nicK plus WT assay showed a low level of exconjugant formation, the combination nicK comK plus comK led to complete abolition of transfer. Results shown are representative of at least three independent experiments, and error bars represent the SEM. Download FIG S2, PDF file, 0.1 MB (59.7KB, pdf) .
Copyright © 2018 Lécuyer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ICEBs10 attB-down recipient cells can acquire ICEBs1, while the attB site cannot be amplified. (A) ICEBs10 attB-down (JSB18) cells were mated on MSgg with WT donor cells to assess their capacity to acquire ICEBs1, despite an erythromycin cassette inserted near the attB site. Mating efficiency was measured after 20 h at 30°C. Statistical analysis showed no significant difference in mating efficiencies between the WT recipient cells and JSB18 cells (Student’s t test). Results shown are representative of at least three independent experiments, and error bars represent the SEM. (B) The attB site of a WT recipient cell (lane 1), JSB18 (lane 2), and a 1:1 mixture using JSB18 (lane 3) as a recipient was amplified by PCR using primers P333 and P358 (see Materials and Methods) and run on a 1% agarose gel. We were able to amplify the attB site from the WT recipient cells and the 1:1 mixture but not from JSB18 alone. Download FIG S3, PDF file, 0.1 MB (71.2KB, pdf) .
Copyright © 2018 Lécuyer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Biofilm does not affect ICEBs1 excision over time. WT donor cells and ICEBs10 attB-down recipient cells were mated on MSgg, and the donor attB site was amplified by qPCR after 4, 8, 12, 16, and 20 h at 30°C. We did not detect an increase in ICEBs1 excision over time. Download FIG S4, PDF file, 0.1 MB (55.1KB, pdf) .
Copyright © 2018 Lécuyer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Conjugative transfer of ICEBs1 is inefficient under shaking conditions. WT, eps tasA, and sinR donor and recipient cells were mixed and inoculated in liquid LB, LBGM, MSNc, and MSgg. Tubes were then incubated for 20 h with shaking at 30°C, and exconjugant frequency was evaluated. The results shown are representative of at least three independent experiments, and error bars represent the SEM. Download FIG S5, PDF file, 0.1 MB (55KB, pdf) .
Copyright © 2018 Lécuyer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Polymers can act as a surrogate biofilm in absence of extracellular matrix. WT cells and epsA–O tasA (eps) cells were resuspended in 1% agarose and mated on MSNc. Mating efficiency was measured after 20 h at 30°C. Statistical analysis showed a significant increase in ICEBs1 transfer efficiency when agarose was added to eps tasA cells, but not for the WT cells (Student’s t test; ****, P < 0.0001). Results shown are representative of at least three independent experiments, and error bars represent the SEM. Download FIG S6, PDF file, 0.1 MB (62.4KB, pdf) .
Copyright © 2018 Lécuyer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
recA and WT donor cell counts after 20 h. Download Table S1, PDF file, 0.1 MB (95KB, pdf) .
Copyright © 2018 Lécuyer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacillus subtilis strains used. Download Table S2, PDF file, 0.2 MB (162.5KB, pdf) .
Copyright © 2018 Lécuyer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Donor and recipient cell counts in different media after 20 h. Download Table S3, PDF file, 0.1 MB (104.8KB, pdf) .
Copyright © 2018 Lécuyer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.