Abstract
Maternal-effect genes are especially critical for early embryonic development after fertilization and until massive activation of the embryonic genome occurs. By applying a tandem mass tag (TMT)-labeled quantitative proteomics combined with RNA sequencing approach, the proteome of the buffalo was quantitatively analyzed during parthenogenesis of mature oocytes and the two-cell stage embryo. Of 1908 quantified proteins, 123 differed significantly. The transcriptome was analyzed eight stages (GV, MII, 2-cell, 4-cell, 8-cell, 16-cell, morula, blastocyst) of Buffalo using the RNA sequencing approach, and a total of 3567 unique genes were identified to be differently expressed between all consecutive stages of pre-implantation development. Validation of proteomics results (TUBB3, CTNNA1, CDH3, MAP2K1), which are involved in tight junction and gap junction, revealing that the maternal expression of the proteins possibly plays a role in the formation of cellular junctions firstly after parthenogenetic activation. Correlation and hierarchical analyses of transcriptional profiles and the expression of NPM2 and NLRP5 mRNA of buffalo in vitro developed oocytes and parthenogenetic embryos indicated that the “maternal-to-zygotic transition” (MZT) process might exist in the model of parthenogenesis, which is similar to a normally fertilized embryo, and may occur between the 8-cell to 16-cell stage. These data provide a rich resource for further studies on maternal proteins and genes and are conducive to improving nuclear transfer technology.
Keywords: Protein-Protein Interactions, Quantification, Cell differentiation, Fluorescence, Gene Expression, HPLC, Immunohistochemistry, Mass Spectrometry, Pathway Analysis, RNA SEQ, buffalo, maternal protein, oocyte, parthenogenesis model, single-cell RNA sequencing
An embryo exclusively relies on the maternal genome for its early development until activation of zygotic genome. Oocytes of all animals contain mRNAs and proteins that are deposited during oogenesis. The oocyte provides the specialized environment within which the zygotic nucleus initiates and executes its developmental program and as a result, also plays an obligatory role in its regulation. Maternal effects are pronounced in mammals especially. The uterine environment has a major effect on offspring development in the early stage, regulated by the mechanisms including hormone transmission (1) across the placenta and nutrient transfer (2). On the other hand, the postnatal environment is structured by the joint influence of milk quality and maternal behaviors, including time spent nursing (3, 4), nest building, maintenance (5), and offspring grooming, and the quality of milk varies in its composition, such as fats, carbohydrates, and protein (6), and in biologically active proteins that can have effects on growth and immunological (7). To fully describe maternal effects in mammals, therefore, the identification of qualitative differences in mRNA transcript and protein content of maternal genome will enhance our understanding of the molecular genetic factors essential for maternally directed control of early embryonic development.
And maternal-effect genes were first defined in Drosophila 30 years ago, and in mammals in 2000 (8, 9). Maternal genes are those genes whose products, RNA or protein, are produced or deposited in the oocyte or are present in the fertilized egg or embryo before expression of zygotic genes is initiated (10). Microarray expression studies have revealed numerous genes that are maternally expressed in vertebrates (11–20). Maternal-gene products are particularly important in early embryos after fertilization occurs and up to the point when vast activation of the embryonic genome appears. The maternal factors that are stored in the oocyte greatly affect the maternal-to-embryonic transition, including the subcellular organelles and macromolecules, and they are acquired during folliculogenesis and oogenesis. Various technological approaches have been used to identify maternal-effect genes in mammals since 2000. Several maternal-effect genes have been identified in mice using typical knockout strategies (8, 9, 21, 22). The conventional knockout method can lead to embryonic lethality, but conducted with conditional knockdown via oocyte injection and RNAi (the RNA interference pathway) can make precise exploration of maternal genes. In recent years, an increasing list of maternal factors involved in the regulation of MET has been identified. These factors include NLRP5 (official name for MATER) (9), HSF1 (8), NPM2 (21), ZARL (22), ZFP36l2 (23), BRG1 (24), PADI6 (25), CTCF (26), ATG5 (27), and AGO2 (official name for EIF2C2) (28).
Also, many maternal genes have been described whose products are involved in protecting imprinted methylation sites during preimplantation development, including DPPA3(29), ZFP57 (30), DNMT1 (31), and TRIM28 (32). Although the importance of maternal effects (MEs) is becoming widely appreciated, we know little about dynamic function and interaction between maternally expressed proteins or genes at the level of protein and transcriptome.
DNA microarray research has provided valuable information for the dynamic changes of important factors at the transcription level. However, there is a large amount of material that is transcribed and stored in oocytes that has no apparent effect on oogenesis, but that requires the regulation of embryonic development. Thus, a precise picture of the functional status of oocytes is needed and can be achieved through transcriptomic and proteomic analyses. Also, the regulation of maternally expressed proteins from maternal genome remains poorly understood. How to obtain the maternal information only? To this end, we used parthenogenetic activation of oocyte as our research model because it does not require any paternal factors, which makes it an ideal model for the study of maternal genome. Also, parthenogenetic activation of mammalian oocytes is a very important research ingredient in reproductive biology. And it is not only provides alternative embryonic resources of embryos, but also is the most important technical condition for activation of nuclear transfer recombinant embryos in somatic cell nuclear technology; Parthenogenetic activation of mammalian oocytes also are used to establish embryonic stem cell lines.
Proteomic methods have been used in the exploration of mammalian oocytes and embryos, including mouse (33–37), bovine (38–40), and pig (41–44), and the amount of proteomics data has increased. Also, transcriptomic methods are also widely used in this area of research (45–48). But these related studies were mainly focused on identifying protein expression profiles of oocytes and embryos at different developmental stages, however, to date, there is no report on the dynamic changes of maternally expressed proteins and genes from maternal genome (without paternal genome) that occur during the early stages of the embryo using the parthenogenesis as an experimental model at the level of transcription and translation for mammal.
Buffalo (Bubalus bubalis) is an important economic livestock species in Southern Asia. Buffalo was used as a tool for farming in the past, but with the rapid development of agricultural machinery, its advantages in farming have disappeared, so this species faces the risk of being eliminated. However, we are pleased to discover that buffalo have many good characteristics, such as resistance to roughage, the high value of dairy products, and good quality meat. However, its economic value is limited by its low reproductive efficiency. There are many factors which result in the low reproductive efficiency of Buffalo, for example, the beginning of suitable breeding age for buffalo is latter than cattle and cow; and the level of progesterone of female buffalo during its period of pregnancy is much lower than the cattle; and the characteristics of estrus are inconspicuous in female buffalo, and its interval of calving is longer than cattle and cow, also the female buffalo is not sensitive to the treatment of reproductive hormone. Notably, under the laboratory conditions, we found that the quantity of vesicular follicle and the oocyte suited to the IVM (in vitro maturation) collected from the buffalo ovary is less than the cattle and cow, and we found that the rate of cleavage and blastocyst in buffalo is lower than the cattle during its period of pre-implantation, which indicated that the different regulatory mechanism may exist in the development of pre-implantation, and before the activation of zygotic genome, the early development of embryo is regulated by maternal genome mainly.
The application of somatic cell nuclear transfer technology in buffalo reproduction has been reported (49–52). However, there are still many problems in nuclear transfer technology, which limits the promotion of nuclear transfer technology, such as low cloning efficiency, low embryo implantation rate, abnormal fetal development, and many abnormal gene expressions during genome development of cloned embryos, including abnormalities of DNA methylation, acetylation, abnormal expression of imprinted genes, and so on. It is well known that the improvement of reproductive technology for the domestic animal must be based on the understanding of its reproductive mechanism and characteristics. However, the current theoretical knowledge about the reproductive mechanism of this species is still not enough, and the improvement of our breeding technology is limited by this. This study made the research of maternal genome as the entry point to study the reproductive mechanism of buffalo, and this knowledge will enhance the efficacy of in vitro maturation protocols utilized in either an agricultural or clinical setting and attempts to clone domestic animals by nuclear transfer. In addition, the strategy of this research may provide a valuable reference to explore the function of the maternal genome for other species.
In this study, we used the tandem mass tag (TMT)1-labeled quantitative proteomic combined with transcriptome of oocytes/embryos to research the dynamic function of the maternal genome during the early embryonic development. And mature oocytes (metaphase II [MII]) and two-cell stage embryos (parthenogenetic cleavage [PC]) were used as a quantitative proteomic object and the oocytes and embryos from 8 stages of buffalo were used as RNA sequencing-based transcriptomic object. The aim of this study was to show the dynamic function of maternally expressed proteins or genes using the research model of parthenogenetic activation of oocyte and to identify the important role of maternally expressed proteins in early embryo development. We also applied bioinformatic methods to an analysis of the protein expression profiles by Gene Ontology (GO) classification and a mapping of the interaction control network for partial proteins. Finally, we verified the expression of differential proteins at transcription and translation levels using real-time polymerase chain reaction (RT-PCR) and Western blotting, and we analyzed protein migration and expression using changes in immunofluorescence.
EXPERIMENTAL PROCEDURES
Collection of Buffalo In Vitro Developed Oocytes and Parthenogenetic Embryos
The collection and maturation of oocytes were performed as previously reported (53). In brief, we collected buffalo ovaries from the local slaughter house and transported them to the laboratory at 25 °C in physiological saline. We washed the ovaries three times in saline solution, extracted cumulus-oocyte complexes (COCs) from 2 to 6 mm follicles in the ovaries, and selected COCs with compact cumulus-cell layers for maturation in vitro (IVM). Before mature culture, we collected the immature oocyte of germinal vesicle (GV stage).Then we washed the immature oocytes in TCM199 (Gibco, Grand Island, NY) complemented with 0.3% bovine serum albumin, and transported them to the maturation medium (TCM199 complemented with 5% estrous bovine serum and 10 μg/ml follicle-stimulating hormone (FSH). The oocytes were cultured in an incubator at 38.5 °C under 5% CO2 for 22 h–24 h. Matured oocytes (MII stage) were collected under a microscope based on a morphological evaluation; completely matured oocytes included with homogenous cytoplasm that had at least three cumulus layers and were accompanied with the first polar body. Parthenogenetic embryos were collected by chemical activation, as previously described (54). Briefly, maturated oocytes were induced by exposure to 5 mm ionomycin in embryo culture medium for 5 min and subsequent incubation in 2 mm 6-dimethylaminopurine for 3 h at 39 °C and 5% CO2. The oocytes were activated to new embryo culture medium, and two-cell stage parthenogenetic embryos were collected after 22 h–24 h, and after 2–4 days to collect embryos at the 2- to 16-cell stages, morula and blastocysts were collected at days 5 and 7. All oocytes and embryos were examined and staged under light microscopy.
RNA Sequencing and Identification of DEGs
The oocytes and embryos from 8 stages of buffalo were conducted the experimental process of transcriptional sequencing includes RNA extraction, sample detection, library construction, quality control and sequencing. We isolated total RNA from individual oocytes/embryos using ArrayPure™ Nano-scale.
RNA Purification Kit (Epicenter). We conducted Smart-Seq(55), and firstly we lysed each cell in hypotonic solution and converted poly(A)+ RNA to full-length cDNA using oligo (dT) priming and SMART template switching technology, followed by 12–18 cycles of PCR preamplification of cDNA, and the amplified cDNA was used to construct standard Illumina sequencing libraries. After that, Using Qubit2.0 for preliminary quantification and Agilent 2100 to test the insert size of the library, then insert size is expected and proceed with the next experiment. The qPCR method is used to accurately quantify the effective concentration of the library (library effective concentration >2 nm) and complete the checking. After the library is checked, and the Illumina Hiseq X ten platform is used for sequencing. The adaptor sequences and low-quality sequence reads were removed from the data sets. Raw sequences were transformed into clean reads after data processing. These clean reads were then mapped to the reference genome sequence. (ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCA/000/471/725/GCA_000471725.1_UMD_CASPUR_WB_2.0/). Only reads with a perfect match or one mismatch were further analyzed and annotated based on the reference genome. Tophat2 tools (56) soft were used to map with the reference genome. Cuffquant and Cuffnorm, which are the components of the Cufflinks software (57), were used to perform gene expression analysis. During the construction of the database, it was performed by mixing RNA from three oocytes or embryos of the same stage, to reduce the influence of cellular heterogeneity and differences between individual cell. Therefore, the sample at each stage resulted in a data set, so EBseq (58) was used to make a differently expressed analysis of genes.
Protein Extraction
Pooled groups of 1,000 buffalo maturated oocytes (MII oocytes) and 1,000 two-cell stage parthenogenetic embryos (PC embryos) were dissolved using 30 μl of lysate (a mixture of 8 m urea, 2 mm ethylenediaminetetraacetic acid, 10m M dithiothreitol, and 1% protease inhibitor) for 30 min on ice. The samples were centrifuged (12,000 × g at 4 °C for 10 min), and the supernatant was transferred to a new 1.5 ml tube for further experiment.
Digestion and Peptide Labeling Using a Tandem Mass Tag (TMT) Reagent
The extracted protein was dissolved in 45 μl of 100 mm triethyl ammonium bicarbonate, 5 μl of 2% sodium dodecyl sulfate (SDS), and 55 μl of ultrapure water. Next, we used 5 μl of 200 mm trichloroethyl phosphate for 1 h at 55 °C to reduce the sample, followed by 5 μl of 375 mm iodoacetamide for 30 min at room temperature in an environment protected by light. Each sample was then precipitated in cold acetone (1:6) at −20 °C overnight, centrifuged (12,000 × g; 4 °C; 10 min), and the supernatant was transferred to a new 1.5 ml tube and dissolved in 100 μl of 100 mm triethyl ammonium bicarbonate. Finally, 2.5 μl of Trypsin (1 μg/μl) was added to each sample for digestion at 37 °C overnight. Peptides were labeled with TMT reagents according to the manufacturer's instructions. Briefly, 41 μl of anhydrous acetonitrile (ACN) was added to the TMT reagent in each tube (0.8 mg) and dissolved for 5 min at room temperature. Next, 20 μl of each tag were added to each sample for labeling and the samples were incubated for 1 h at room temperature. We then added 8 μl of 5% hydroxylamine into each sample and incubated for 15 min at room temperature to terminate the reaction. Samples were pooled and evaporated in a vacuum; samples of MII oocytes and PC embryos were labeled with TMT- 128 and TMT-129, respectively.
Peptide Prefractionation Using High-pH Reversed-phase High-performance Liquid Chromatography
Each sample was evaporated in a vacuum and re-suspended in 50 μl of buffer A (98% ddH2O, 2% ACN; pH 10.0) and separated by high-pH reversed-phase liquid chromatography (RP-HPLC) column (2.1 × 100 mm, 3 μm, 150 Å, C18). For separation, we used buffer A and buffer B (98% ACN, 2% ddH20; pH 10.0) for a 60-min linear gradient (4–20% buffer B for 30 min, 20–95% buffer B for 25 min, and 95% buffer B for 5 min. The fractions that were eluted from the column every 1.5 min were collected, and 20 fractions were collected according to the peak intensity. The collected fractions were desalted using a ZipTips C18 column (Millipore, Billerica, MA). The processed samples were stored at −80 °C until being used for mass spectrometry analyses.
Peptide Identification by Liquid Chromatography-Tandem Mass Spectrometry Analysis
After being desalted, the fractions were evaporated with a vacuum, and 10 μl of solvent A (2% ACN and 0.1% formic acid) were used for resuspend the samples. A volume of 2 μl from each sample was loaded onto a trap column (PepMap RSLC C18 column, 50 um × 15 cm, 2 μm nanoViper, Thermo Fisher Scientific, Bremen, Germany), a maximum pressure (600 bar) was applied, and an analytical column (Acclaim® PepMap100C18 column, 0.075 × 150 mm, 3 μm, 100Å, Thermo Fisher Scientific, Bremen, Germany) was used for elution at a rate of 300 nL/min. Next, we used buffer A (2% ACN and 0.1% formic acid) and buffer B (98% ACN and 0.1% formic acid), followed by a 60-min gradient (5–40% buffer B for 45 min, 40–100% buffer B for 10 min, 100% buffer B for 5 min) to separate the peptides. An LTQ-Orbitrap Elite hybrid mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) connected to an Easy-nLC 1000 nano- liquid chromatography system (Thermo Fisher Scientific, Odense, Denmark) was used for analyses of all peptide fractions online. A data-dependent mode in the scan range of 350–1800 m/z was carried out for the mass spectrometry (MS) analyses, and the survey scans were captured at a mass resolution of 60,000 at 400 m/z by the Orbitrap analyzer. In the linear ion trap, ten of the most intense precursor ions were selected for secondary mass spectrometry analysis (MS2) under the mode of high-energy collision dissociation. The dynamic exclusion parameter included an exclusion count of 2 and an exclusion time of 40 s. Siloxane ions were used for internal calibration (m/z = 445.1200).
Raw Data Processing, Protein Quantification, and Bioinformatic Analyses
The raw data files were processed and quantified by Proteome Discoverer software v1.3.0.339 (Thermo Fisher Scientific, Massachusetts), and searched against the UniProt Bos Taurus protein database (UP000009136, 24078 sequences, release 2017_03) using the SEQUEST algorithm. The search parameters used were as follows: fixed modifications, including carbamidomethylation of cysteine (+57.02146 Da); TMT reagent adducts (+229.162932 Da) on the lysine and peptide N termini; and variable modification, including oxidation of methionine (+15.99492 Da). Mass tolerance of precursor ions was 20 ppm, and fragment ions were set to ± 0.5 Da, Enzyme specificity, trypsin. The false discovery rate (FDR) was calculated using the peptide validator and based on a decoy database search. A p value < 0.05 and a fold change >1.2 were significant.
The identified proteins were analyzed by gene ontology annotation, including by biological processes, cellular components, and molecular functions, which were based on the UniProt-GOA database (http://www.ebi.ac.uk/GOA/). The pathway analyses of differently expressed proteins were based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. The online KEGG Automatic Annotation Server (KAAS) service tools were used to annotate each protein's KEGG database description, and then to map the annotation result using other KEGG online service tools and the KEGG mapper. The interaction between differential expression protein (DEPs) groups was derived from the Search Tools for the Retrieval of Interacting Genes/Proteins (STRING) database. A high confidence (0.7) for the required interaction score and the active interaction sources including text mining, experiments, and databases were chosen to draw the protein-protein interaction map using Cytoscape 3.2.1.
Real-time Quantitative Reverse Transcription Polymerase Chain Reaction
Total RNA was extracted from oocytes or embryos at different stages (MII and PC) using an RNeasy® Micro Kit (Qiagen Co.). A Takara RNA polymerase chain reaction (PCR) kit was used to reverse-transcribe the cDNAs. Quantitative reverse transcription PCR (qRT-PCR) analyses were performed on a LightCycler 96 system (Roche, Switzerland). The reaction solution (20 μl) consisted of 1.5 μl of cDNA, 10 μl of SYBR® Premix Ex TaqTM II (Takara), 2 μl of primer mix (2 nm), and 7 μl of ddH2O. The 2−ΔΔCT method was used to calculate the relative expression levels of targeted genes. Each gene was analyzed using different sets of oocytes, and experiments were performed in three replicates. The primer sequences used for the qRT-PCR analyses are presented in supplemental Table S8.
Western Blotting Validation
TUBB3, CTNNA1, CDH3, and MAP2K1 protein expression levels were determined by a Western blotting assay. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was used to separate equal amounts of proteins from 400 oocytes or embryos, and the samples were then transferred onto a polyvinylidene difluoride (PVDF) membrane by a semidry Western blotting system (Trans-Blot® TurboTM System, Bio-Rad, Singapore). The membrane was blocked in 5% nonfat milk for 1 h at 37 °C and incubated at 4 °C with primary antibodies (1:500–2000 dilution) overnight. Membranes were washed three times using TBST buffer (a mixture of Tris-buffered saline (TBS) and Tween 20), and then membranes were incubated with secondary antibodies in TBST buffer for 1 h at 37 °C. Bands were visualized with an alkaline phosphatase detection kit (C3206, Beyotime Biotechnology Inc, Shanghai, China). The following commercially available antibodies were used as primary antibodies: anti-beta-III tubulin mouse monoclonal antibody (ab7751; Abcam, Cambridge, MA), anti-alpha 1 catenin rabbit polyclonal antibody (ab6299; Abcam), anti-MEK1 rabbit polyclonal antibody (ab32091; Abcam), anti-actin (loading control) rabbit polyclonal antibody (bs-17654R; Bioss Biotechnology Inc., Beijing, China), and anti-P-cadherin rabbit polyclonal antibody (bs-1159R; Bioss Biotechnology Inc., Beijing, China).
Immunocytochemical Analysis
The oocytes were removed from the granulosa cells and placed in 4% polyoxymethylene for 30 min at room temperature, the oocytes were transferred to a 1% Triton X-100 phosphate-buffered saline (PBS) solution at room temperature for 20min, and the oocytes were blocked with a PBS solution containing 1% BSA (albumin from bovine serum). The oocytes were mixed with a primary antibody at an effective concentration and incubated overnight at 4 °C. The oocytes were washed three times for 2 min each time with a PBS solution containing 0.1% Tween-20 and 0.01% Triton X-100. The oocytes were transferred into a diluted solution containing a secondary antibody labeled with fluorescein isothiocyanate (FITC) for 1 h at room temperature. The oocytes were then washed three times for 2 min each time with a PBS solution containing 0.1% Tween-20 and 0.01% Triton, covered with a coverslip coated in a ProLong Gold antifade reagent with DAPI (Life Technologies), and kept in the dark until an inverted fluorescence microscope was used to assess fluorescence (Olympus, IX73, Japan). As a negative control, the primary antibody was replaced with normal rabbit IgG (Immunoglobulin G) or mouse lgG.
Experimental Design and Statistical Rationale
For mRNA expression profiling, the transcriptomic analysis was conducted for eight stages for the oocytes and early parthenogenetic activated embryos of buffalo in vitro(GV, MII, 2cells, 4cells, 8cells, 16cells, morula, blastocyst) using paired-end Sequencing in Illumina Hiseq X ten platform. Quantification of gene expression levels was estimated by fragments per kilobase of transcript per million fragments mapped (59). The formula is shown as follow:
Before differential gene expression analysis, for each sequenced library, the read counts were adjusted by edgeR program package through one scaling normalized factor. Differential expression analysis of two samples was performed using the EBSeq R package. The resulting FDR (false discovery rate) were adjusted using the PPDE(posterior probability of being DE) .The FDR < 0.05 and log2(fold change) ≥1 was set as the threshold for significantly differential expression. A matrix of Pearson correlation coefficient was created using R, which was in turn used to create the heatmap. Principle component analysis (PCA) was analyzed by using R.A matrix of Pearson correlation coefficient was created using R, which was in turn used to create the heatmap. Principle component analysis (PCA) was also analyzed by using R. Genes were deemed differentially expressed between hierarchical subsequent developmental stages if they showed a p value of less than 0.05 (Negative Binomial Distribution). For protein expression profiling, a TMT-based quantitative proteomic approach was employed to screen DEPs in a total of 2000 MII stage oocytes and 2-cell stage embryos. Two biological replicates are included in the proteomics experiments. The raw data files were processed and quantified by Proteome Discoverer software v1.3.0.339 (Thermo Fisher Scientific, MA), and searched against the UniProt Bos taurus protein database (UP000009136, 24078 sequences, release 2017_03) using the SEQUEST algorithm. The false discovery rate (FDR) was calculated using the peptide validator and based on a decoy database search. A p value < 0.05 and a fold change >1.2 were significant. All quantitative data are presented as the mean ± S.E. from three independent experiments. A p value <0.05 was statistically significant. Statistical analyses were done using IBM SPSS Statistics version 17.0 and GraphPad Prism version 5.0. One-way Analysis of Variance (ANOVA) combined with Tukey's multiple comparison Test was used to calculate the significance of differences between different groups.
RESULTS
Quantitative Proteomic Analyses of Early Parthenogenetic Embryo Stages Using Tandem Mass Tags
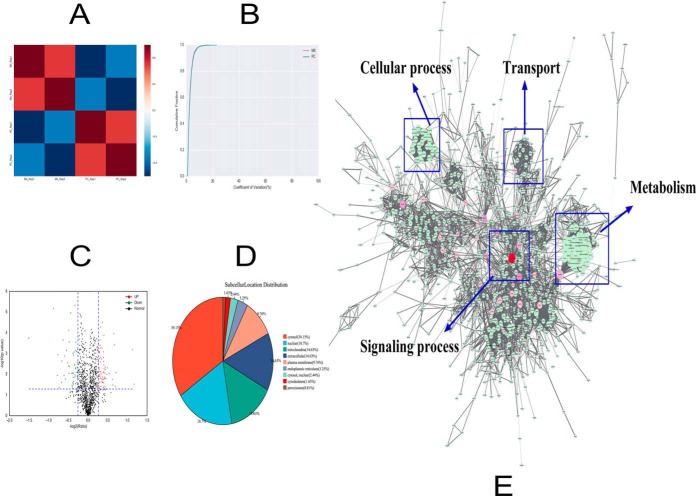
A total of 2000 oocytes or embryos from MII oocytes (matured oocyte) and the two-cell stage of the early embryo after parthenogenesis were treated with 5 mm ionomycin and 2 mm 6-dimethylaminopurine. Next, TMT-6-plex reagents were combined with liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses. Proteins isolated from the MII stage and the two-cell stage were labeled with mass tags TMT-128 and TMT-129, respectively. Finally, a total of 1908 maternally expressed proteins were identified (FDR < 1%) in two biological replicates (supplemental Table S1 and S2). We used the Pearson correlation coefficient and the coefficient of variation to analyze the correlation of protein expression levels for each sample, which demonstrated that the experiment had high reproducibility (Fig. 1A and 1B). A total of 1224 maternally expressed proteins were found in both experiments. A complete list of all identified proteins with annotation from every repeat are shown in supplemental Table S3. Compared with the data from the MII stage, 123 proteins were expressed differently (fold change > 1.2; p < 0.05); of the123 proteins, 67 proteins were up regulated and 56 proteins were downregulated (Fig. 1C). We also analyzed the subcellular structure distribution for these proteins and found that they were mainly located in the following areas of the cell: cytosol (34.15%), nucleus (18.70%), mitochondria (14.63%), extracellular matrix (14.63%), and plasma membrane (9.76%) (Fig. 1D). These findings demonstrate that the differentially expressed proteins are widely distributed in the cell. The top 10 differently maternally expressed proteins are listed in Table I, and they are sorted by fold change. A complete list of all differently expressed proteins with annotation are shown in supplemental Table S4.
Fig. 1.
A, The correlations between the protein expression levels of each sample were calculated by using the Pearson correlation coefficient. The red represents the MII stage and the blue represents the two-cell stage after parthenogenetic activation. B, The coefficient of variation (CV) is used to reflect the degree of discretization of each sample data, which is the ratio of the standard deviation of the data to the mean, expressed as a percentage (CV = [standard deviation/average value] × 100%). The horizontal axis is the CV value, and the vertical axis represents the proportion of total protein. The red line represents the MII stage and the green line represents the two-cell (that is, the parthenogenetic cleavage [PC]) stage. C, Each point in the volcanic map represents a protein; the abscissa represents the logarithmic value of the expression of a certain protein in the two samples. The green point represents the downregulation of the expressed protein, the red point represents the upregulated differential expression protein, and the black dot represents the non-differentially expressed protein. D, An analysis of the distribution of proteins in the subcellular structure found that the were mainly located in the cytosol (34.15%), nucleus (18.70%), mitochondria (14.63%), extracellular matrix (14.63%), and plasma membrane (9.76%). E, The interaction between the identified proteins visualized using Cytoscape 3.2.1 software. The color of the node is represented by the betweenness, which means the influence in the network; if the color is redder, the influence is greater. The size of the node is represented by the degree, which means the stability in the network; if the shape is bigger, the stability is stronger. Using the BinGO package, we found that the identified proteins were mainly involved in the following processes: regulation of biological processes, metabolism, transport, cellular process, and signaling process.
Table I. The top 10 differentially maternally-expressed proteins in the 2-cell stage of early embryos.
| Uniprot accession no. | Protein name | Gene symbol | MW [kDa] | Calc. pI | Fold changea | Regulationa | Subcellular location |
|---|---|---|---|---|---|---|---|
| A7E320 | E3 ubiquitin-protein ligase UHRF1 | UHRF1 | 88.28 | 8.00 | 1.40/1.38 | Up | Nucleus |
| F1MRC2 | Myosin-2 OS | MYH2 | 223.19 | 5.80 | 1.28/1.50 | Up | Cytoplasm |
| Q3SYU9 | Major vault protein | MVP | 98.86 | 5.60 | 1.28/1.22 | Up | Cytosol |
| P63097 | Guanine nucleotide-binding protein G(i) subunit alpha-1 | GNAI1 | 40.34 | 5.97 | 1.32/1.18 | Up | Plasma membrane |
| A7MBC2 | Pyridoxal-dependent decarboxylase domain-containing protein 1 | PDXDC1 | 86.56 | 5.39 | 1.23/1.25 | Up | Cytosol |
| Q3SYT9 | Poly(RC) binding protein 2 | PCBP2 | 36.78 | 7.06 | 0.71/0.82 | Down | Cytosol |
| Q3SZ86 | 28S ribosomal protein S26, mitochondrial | MRPS26 | 23.76 | 9.83 | 0.75/0.78 | Down | Mitochondrion |
| E1BHF2 | Katanin p60 ATPase-containing subunit A-like 1 | KATNAL1 | 55.35 | 6.74 | 0.76/0.84 | Down | Cytoplasm, cytoskeleton |
| G3X705 | 28S ribosomal protein S6, mitochondrial | MRPS6 | 13.77 | 9.26 | 0.80/0.81 | Down | Mitochondrion |
| A5D7N3 | Transmembrane protein 11, mitochondrial | TMEM11 | 21.46 | 7.33 | 0.80/0.83 | Down | Mitochondrion inner membrane |
aNote: The quantitative proteomic experiments were conducted twice. The fold change and regulation data were all compared with the data from MII stage. p value <0.05 and fold change >1.2 were considered significant.
We also used the Cytoscape 3.2.1 mapping software to visualize the interaction between the 1,908 identified proteins (Fig. 1E and supplemental Table S5). It can be seen from the figure that the relationship between these proteins is complicated. Using the BinGO package for gene ontology (GO) analysis, we found that the proteins were mainly involved in the regulation of biological processes, metabolism, transport, cellular processes, and signaling processes.
Gene Ontology (GO) Analysis of Differentially Expressed Proteins
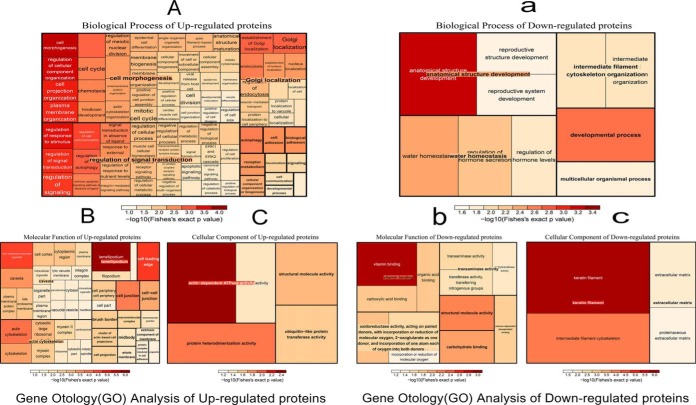
The differentially expressed proteins (upregulated and downregulated) were classified by gene ontology annotation based on three categories: biological process, cellular component, and molecular function. The GO annotation proteome was derived from the UniProt-GOA database (http://www.ebi.ac.uk/GOA/) (supplemental Table S6). After GO analysis of up-regulated proteins and an analysis of the biological processes, we found that the biological processes include plasma membrane organization, actin cytoskeleton organization, processes associated with the cell cycle, positive regulation of cell junction assembly, processes associated with the cell junctions, oocyte differentiation, and regulation of cell communication, signal transduction, and cell proliferation (Fig. 2A). The molecular function of these proteins is important in actin-dependent ATPase activity (Fig. 2B). The results of the cellular component analysis showed that these proteins were mainly as the component of caveola, lamellipodium and actin cytoskeleton (Fig. 2C).
Fig. 2.
Gene ontology (GO) analysis of differently expressed proteins. A, The biological process analysis of upregulated proteins. B, The molecular function analysis of upregulated proteins. C, The cellular component analysis of upregulated proteins. a, The biological process analysis of downregulated proteins. b, The molecular function analysis of downregulated proteins. c, The cellular component analysis of downregulated proteins.
A GO analysis was conducted on the downregulated proteins, and we found that the results of this group (41 GO terms) was much less than the upregulated proteins (273 GO terms), which may indicate that upregulated proteins are more important than the downregulated proteins during parthenogenetic development in early embryos. In an analysis of biological processes, we identified some processes such as intermediate filament cytoskeleton organization and regulation of hormone secretion (Fig. 2a). The molecular function of these proteins plays a major role in the binding of vitamins and carbohydrates, and in the activity of oxidoreductase, transaminase, and methyltransferase (Fig. 2b). Furthermore, the downregulated proteins are mainly a component of keratin filament, intermediate filament cytoskeleton, and the extracellular matrix (Fig. 2c).
KEGG Pathway Analysis of Differentially Expressed Proteins
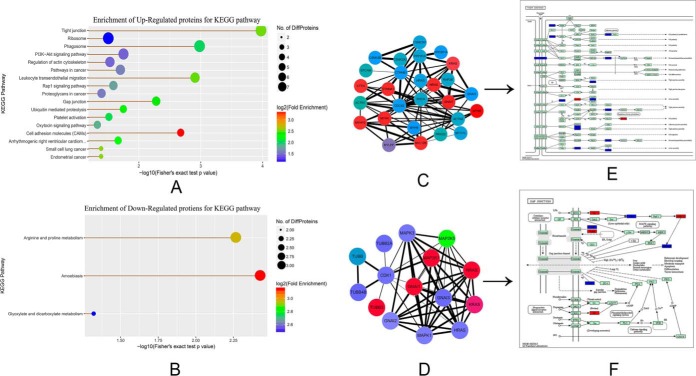
We analyzed differentially expressed proteins based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (which is shown in Fig. 3A–3B and supplemental Table S7). We found that the upregulated proteins were mainly enriched in tight junctions, phosphatidylinositol 3-kinase (P13k-Akt) signaling pathways, the regulation of actin cytoskeleton, Rap1 signaling pathways, gap junctions, and cellular adhesion molecules (CAMs); these signaling pathways may be associated with early embryonic development. Of note, we found that the number of proteins enriched in the tight junctions is the largest; the gap junctions are also significantly enriched, which is like the tight junction pathway. These revealed that these proteins might play an important role in the formation of intercellular junctions during early embryonic development. The connective complex of cells plays an important role in the polarization of cells and the formation of the blastocele, and these connections include the tight junction at the top of the cell, the gap junction on the side, and the desmosomes. We then picked out the identified proteins that were involved in the two signaling pathways and used the information to create a visualization of the interactions (Fig. 3C and 3D) and to conduct KEGG mapping (Fig. 3E and 3F). We found nine proteins (MYH8, GNAI1, MYH4, NRAS, CTNNA1, MYL12B, CTTN, MYH10, KRAS) that were upregulated in tight junctions (Fig. 3C and 3E) and five proteins (GNAI1, NRAS, TUBB3, MAP2K1, KRAS) that were upregulated in gap junctions, and we also found that MAP2K5 was downregulated in gap junctions (Fig. 3D and 3F). The time of compaction of the blastocyst was at the 8- to 16-cell stage during embryonic development in bovine (60–63). However, in this study we found that many proteins were involved in the formation of tight junctions and gap junctions at the two-cell stage of early parthenogenetic embryo development in the buffalo. We also found that the development rate of parthenogenetic oocyte cleavage was advanced by at least 6 h compared with the embryo that underwent in vitro fertilization in the actual experiment, which indicates that the time of compaction during early development of parthenogenetic embryo was earlier than the fertilized eggs. Maternal proteins may play an important role in the compaction of preimplantation embryos, and these proteins may participate in the formation of cellular junctions shortly after parthenogenetic activation. To verify our ideas, we selected four upregulated proteins in the tight junction and gap junction for subsequent validation analysis (Table II).
Fig. 3.
Statistics of regulated enrichment from KEGG pathway analysis. A–B, The KEGG pathway enrichment of differently expressed protein. The size of the node is represented by the number of the enriched proteins; if the size is larger, the number is larger. The color represents the fold-change enrichment; the darker the red color, the greater the fold change. A, The KEGG pathway enrichment of upregulated proteins. B, The KEGG pathway enrichment of downregulated proteins. The interaction visualization and KEGG mapping for tight junctions and gap junctions (C–F). The red nodes represent the upregulated proteins, and green nodes represent the downregulated proteins. C, The interaction visualization of tight junctions; D, the interaction visualization of gap junctions; E, the KEGG mapping of tight junctions; and F, the KEGG mapping of gap junctions.
Table II. Up-regulated proteins involved in both tight junction and gap junction selected for further study.
Transcriptomic Analysis
Using linearly amplified RNA from oocytes or embryos, we obtained more than 570 million sequencing reads from 8 stages of buffalo immature and matured oocytes and pre-implantation embryos (supplemental Table S9). The total numbers of detectable genes ranged from immature oocytes to blastocysts of buffalo in our research with the use of RNA-seq are shown in supplemental Table S10.This experiment identified 9112 differential expressed genes(p < 0.05) (supplemental Table S11). A total of 3567 unique genes were identified to be differently expressed between all consecutive stages of development (FPKM>0) (supplemental Table S12).
Correlation and Hierarchical Analyses of Transcriptional Profiles and the Expression of NPM2 and NLRP5 mRNA of Buffalo In Vitro Developed Oocytes and Parthenogenetic Embryos
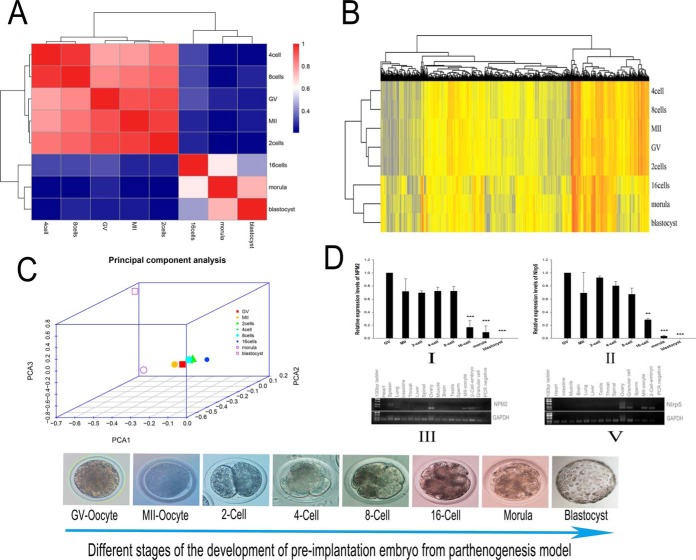
In oocytes/embryos RNA sequencing research, Pearson correlation coefficients (Fig. 4A and supplemental Table S10) and principal component analyses (PCA) (Fig. 4C) of all detected genes revealed the two distinct segmentations of buffalo pre-implantation development: the first from GV (immatured oocyte) to 8-cell,and second from 16-cell to blastocyst stage (Fig. 4A and 4C), which is consisted with the result of Hierarchical clustering of differentially expressed genes in developed oocytes and parthenogenetic embryos of buffalo (Fig. 4B). This segmentation demonstrated that parthenogenetic activation of embryonic development exists an embryonic genome activation process which is the same as the normal fertilization of embryos. Also, the EGA in buffalo occurs between 8-cells–16-cells, consistent with the time of the development of the normal fertilized embryonic development (60–63).
Fig. 4.
Correlation and hierarchical analyses of transcriptional profiles and the expression of NPM2 and NLRP5 mRNA of buffalo in vitro developed oocytes and parthenogenetic embryos. A, Heatmap of all stages from buffalo oocytes and embryonic development of parthenogenesis. The color spectrum, ranging from red through white to blue, represents Pearson correlation coefficients ranging from 1 to 0.2, indicating high to low correlations, and the correlation between the eight stages of expression can be divided into two stages: the first stage from GV to 8cells which is high correlation between this five stages, and the second stage of higher correlation is from 16cells to blastocyst. B, Hierarchical clustering of differentially expressed genes in developed oocytes and parthenogenetic embryos of buffalo. Two major clusters are shown, one consisted of the GV (immatured oocytes), MII (matured oocytes) and embryos at the 2-cell, 4-cell and 8cell stages. The second cluster is consisted of embryos at the 16cell, morula and blastocyst stages. The separation of embryos into two groups demonstrated that parthenogenetic activation of embryonic development exist an embryonic genome activation process which is the same as the normal fertilization of embryos, also the EGA in buffalo occurs between 8 cells-16 cells. C, Principal component analysis (PCA) of the transcriptomes for eight stages of buffalo in vitro developed oocytes and parthenogenetic embryos. Embryos from the same stage are shown by symbols of the same shape. PCA1, PCA2 and PCA3 represent the top three dimensions of the differentially expressed genes among the parthenogenetic embryos. D-I and II, Real-time quantitative polymerase chain reaction analysis of buffalo NPM2 and NLRP5.The mRNA expression during in vitro oocyte maturation and early embryogenesis after parthenogenetic activation (mean ± standard deviation; **, p < 0.01; ***, p < 0.001). (d-III and V). The tissue distribution of NPM2 and NLRP5 in buffalo.GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as a positive control for RNA quality.
In the quantitative proteomic study, we found two maternal proteins—NPM2 and NLRP5 (official name for MATER)—that have been previously reported (Table III). NPM2 and NLRP5 are maternal genes that are both involved in the regulation of embryonic gene activation (9, 21). Notably, expression of NPM2 and NLRP5 is upregulated at the two-cell stage after parthenogenetic activation. We used the RT-qPCR method to detect the expression of the two genes during the early stages of buffalo embryonic development, and GV (germinal vesicle) oocyte was used as control. We found that there was not a significant difference between the two genes before the 16-cell stage. Expression of the GV stage was highest compared with all other stages, and the expression during the two-cell stage was higher compared with during the MII stage; these findings were consistent with proteome data. After the eight-cell stage, the expression of the two genes was significantly decreased, and they were little expressed during the blastocyst stage (Fig. 4D-I and II). The results of the analysis of NPM2 and NLRP5 were consistent with a previous report (64, 65). In addition, we analyzed the tissue expression specificity of NPM2 and Nlrp5, and we found that they only expressed in the ovary, MII oocyte, the two-cell stage embryo, and granular cells (Fig. 4D-III and V). NLRP5 and NPM2 showed similar expression changes, indicating that the two genes may be an important maternal protein during early embryonic development in the buffalo. The analysis of quantitative proteomic study combined with research of RNA sequencing demonstrates that the “maternal-to-zygotic transition”(MZT) possibly exists in parthenogenesis similar to the fertilized embryo(maternal genome and paternal genome) and occurs between the 8-cell stage to 16-cell stage.
Table III. Information of identified maternal proteins NPM2 and NLRP5.
| Protein accession | Protein name | GN | MW[kD] | PI | Fold change | Function | References |
|---|---|---|---|---|---|---|---|
| D2DSG3 | Nucleoplasmin 2 | NPM2 | 22.6 | 5.07 | 1.32 | Chromatin remodeling | Burns et al. 2003; Inoue et al. 2011 |
| Q647I9 | NACHT, LRR and PYD domains- containing protein 5 | NLRP5 (MATER) | 120.9 | 6.93 | 1.32 | Component of the subcortical maternal complex | Tong et al. 2000; Pennetier et al. 2006 |
Proteomic Profile Validation
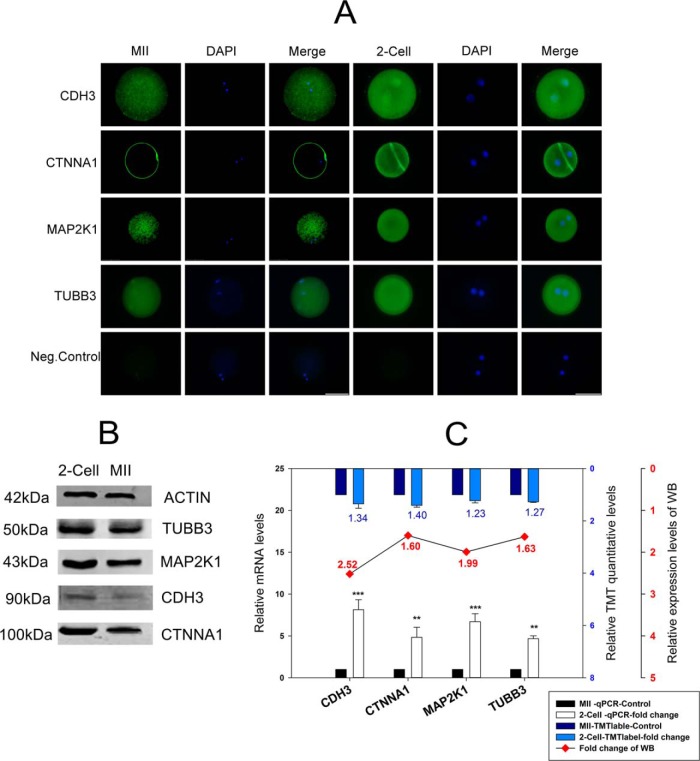
The four upregulated proteins involved in the formation of gap junctions and tight junctions were chosen for Western blotting, RT-qPCR, and cellular immunofluorescence localization analysis (Fig. 5A–5C). The results of Western blot analysis of the four proteins (CDH3, CTNNA1, MAP2K1, TUBB3) are presented in Fig. 5B. We analyzed the gray value of each lane of Western blotting using Image J software, and the results were consistent with the analyses of TMT quantification and transcription levels (Fig. 5C). All results used MII stage as a control. To further explore the potential functions of the differentially expressed proteins, we used cellular immunofluorescence localization analysis to investigate these proteins because there is little-known information on their cellular localization in the MII oocyte and the two-cell embryo. As shown in Fig. 5A, CTNNA1 was mainly expressed in the cell membrane, CDH3 was mainly expressed in cytoplasm and nucleus, and the other two proteins (MAP2K1 and TUBB3) were both mainly located in the cytoplasm. The fluorescence intensity of the four proteins in the two-cell embryo was higher compared with the MII oocyte, which indicates that the gap junctions and tight junctions in parthenogenetic embryos may regulate embryo densification processes earlier than fertilized egg embryos.
Fig. 5.
The validation of four proteins (CDH3, CTNNA1, MAP2K1, TUBB3) involved in gap and tight junctions. A, Cellular immunofluorescence localization analysis. The last line in the figure is a negative control, Bar, 50 μm; B, Western blot analysis. β-actin was used as a loading control. C, The combination chart showing the fold change for the tandem mass tag (TMT) quantitative data. The figure shows mRNA expression levels and gray values of the Western blotting. The blue columns represent the TMT quantitative data, the white columns represent mRNA expression levels, and the line chart represents the gray value of the Western blotting. All the above analyses used the MII stage as a control. The data on mRNA expression levels are represented as means ± standard deviation (S.D.); **, p < 0.01; ***, p < 0.001. The TMT quantitative data are represented as means ± S.D., and its difference standard is the fold change > 1.2, and the p value < 0.05. The fold change of the Western blotting is dependent on the gray value of each line from the MII stage and the two-cell stage of each protein.
DISCUSSION
Proteomic and transcriptomic methods have been applied to the research of oocytes and embryos in mammals, and proteomic data have been obtained that include protein expression profiles of oocytes and embryos at different developmental stages. A growing list of maternal proteins encoded by maternal-effect genes have been characterized using proteomic approaches, such as CDH1(66–68), Dnmt1(31, 69), DPPA3 (29, 70), H3.3 (71–73), NLRP5 (official name for Mater) (9, 65), NPM2 (21, 74), and NOBOX (75, 76). Various aspects of early embryonic development are regulated by these factors, such as epigenetic modifications and chromatin remodeling, onset of embryonic genome activation, maternal mRNA degradation, cell compaction and signal transduction protein translation. However, there are no reports to date that focus on expression profiles of maternally expressed proteins using parthenogenetic activation as an experimental model during early embryo development in the buffalo. Parthenogenetic embryos only contain maternal genetic information that can be used to facilitate the study of the role of the maternal genome in early embryonic development and to discover the maternally expressed genes and proteins that are involved in cleavage and cell differentiation. In addition, transplantation studies allow for an investigation of the role that the oocyte plays in nuclear donor replication; the start of this development is completely dependent on the activation of oocytes. Therefore, the study of the parthenogenesis process is also conducive to improving nuclear transfer technology.
General Aspects
In this study, we undertook a panoramic analysis of the proteomes of two-cell embryos, which is the first developmental stage after parthenogenetic activation, and made a comparison to MII oocytes using TMT-labeled quantitative proteomics. We generated two biological replicates for each stage, and we were able to identify 1908 proteins in 1908 protein clusters with high confidence (FDR < 1%). We used a Pearson correlation coefficient and a coefficient of variation for our analysis and found that the two experiments had high reproducibility (Fig. 1A and 1B). We then used Cytoscape 3.2.1 software to visualize the interactions between all of the identified proteins and, using the BinGO package, we found that the biological processes of those proteins were mainly involved in the regulation of metabolism; transport; cellular processes; signaling processes (Fig. 1E), which revealed that embryo DNA replication is rapid in the cleavage stage; and the mechanism of cleavage, which involves changes to mitochondria and surface cytoplasm, including the karyokinesis and cytokinesis. Many proteins, RNAs, and other substances that are required in the early stages of embryonic development are mainly derived from oocytes; maternal RNAs and proteins that control early embryonic development begin to synthesize and accumulate during the oocyte development, growth, and maturation in mammals. We analyzed the distribution of subcellular structures for all identified proteins and found that they were mainly located in the cytosol (34.15%), nucleus (18.70%), mitochondria (14.63%), extracellular matrix (14.63%), and plasma membrane (9.76%) (Fig. 1D). The distribution of the structures indicates that metabolism, signal transduction, and transcriptional translation are active after the activation of oocytes, which is consistent with the results of the BinGO package analysis. We successfully identified 123 differentially expressed proteins (fold change > 1.2; p < 0.05) at the two-cell stage compared with the data from the MII stage—67 of the proteins were upregulated and 56 of the proteins were downregulated (Fig. 1C). The 123 proteins were classified by gene ontology annotation, and we found that the results of the downregulated proteins (42 GO terms) were much less than the upregulated proteins (273 GO terms). This finding may indicate that upregulated proteins are more active than the downregulated proteins during the first developmental stage following parthenogenetic activation, and that two maternal proteins that have been previously reported (that is, NPM2 and NLRP5) have both identified in the present study as being upregulated (Table III). After an analysis using the KEGG database to investigate the differentially expressed proteins, we found that the upregulated proteins were mainly enriched in the tight junctions, P13k-Akt signaling pathway, regulation of actin cytoskeleton, Rap1 signaling pathway, gap junctions, and CAMs (Fig. 3A), and the downregulated proteins are related to the metabolism of arginine, proline, glyoxylate, and dicarboxylate (Fig. 3B). Notably, we found that the largest number of enriched proteins was in the tight junctions, followed by the gap junctions, which led us to infer that these proteins may play an important role in the formation of intercellular junctions during early embryonic development. We chose four upregulated proteins (CDH3, CTNNA1, MAP2K1, TUBB3) involved in tight junctions and gap junctions for validation analyses using RT-qPCR (Fig. 5C), Western blotting (Fig. 5B), and cell immunofluorescence localization (Fig. 5A), and we found that these results were consistent.
Also, we conducted the RNA sequencing-based transcriptomic analysis of the oocytes and embryos from eight stages (GV, MII, 2-cell, 4-cell, 8-cell, 16-cell, morula, blastocyst) of Buffalo, and we obtained more than 570 million sequencing reads from these stages; for the first time, it is revealed that the buffalo oocytes and early embryos expressed more than 60% of the total estimated 29,424 genes in the buffalo genome, which is a dynamic expression profile of maternal genome during pre-implantation development in Buffalo. We conducted correlation and hierarchical analysis and PCA (Principal components analysis) for the maternal transcriptional profiles of oocytes and parthenogenetic embryos in buffalo, and both results revealed that all identified genes demonstrated the two distinct segmentations during buffalo pre-implantation development: the first from the GV (immature oocyte) to the 8-cell stage and the second from the 16-cell to the blastocyst stage. Interestingly, this result consisted with the significant decrease of the two maternal genes (NPM2 and Nlrp5) between 8 and 16 cells, which found in proteomic data. In addition, to confirm the results from the transcriptomic data, we conducted quantitative real-time PCR (qRT-PCR) on 10 genes (SNAI1, LFNG, POLRMT, RBP4, RPRD1B, FOSL1, F2, CDK5R1, WWTR1, and SETD2) that upregulated in 16-cell stage using parthenogenetic embryos at the 8-cell, 16-cell, and blastocyst (n = 2) stages. We found that all these genes are upregulated in 16-cell embryos, whose change trends are consistent with those from the RNA-seq (supplemental Fig. S2).
Maternally Expressed Proteins Are Possibly Involved in the Formation of Cellular Junctions First After Parthenogenetic Activation
In the tight junctions, there were nine proteins (MYH8, GNAI1, MYH4, NRAS, CTNNA1, MYL12B, CTTN, MYH10, KRAS) that were upregulated (Fig. 3C and 3E). In the gap junctions, there were five proteins (GNAI1, NRAS, TUBB3, MAP2K1, KRAS) that were upregulated and one protein (MAP2K5) that was downregulated (Fig. 3D and 3F). Four proteins (CDH3, CTNNA1, MAP2K1, TUBB3) were validated by a combined analysis (Fig. 5C); CTNNA1 was mainly expressed in the cell membrane, CDH3 was mainly expressed in the cytoplasm and nucleus, and other two proteins (MAP2K1, TUBB3) were both located mainly in the cytoplasm (Fig. 5A). Those findings indicate that the four proteins may be involved in the biological processes of metabolism, transport, cellular processes, and signaling processes (Fig. 1E). The cellular junctions exist in the same tissue or different organizations and are formed by the adjacent cell surface and cell gap, and then organizational structure can maintain a relatively stable structure between cells.
Tight junction biogenesis regulates pattern formation during embryogenesis (77). The tight junctions begin to form after compaction occurs at the late eight-cell stage in the mouse embryo. In the present study, CDH3 and CTNNA1 are involved in the formation of tight junctions and they are upregulated during the two-cell stage (Fig. 5B–5C). CDH3 encodes a classic cadherin from the cadherin superfamily. Alternative splicing results in multiple transcript variants, at least one of which encodes a preproprotein that is proteolytically processed to generate the mature glycoprotein. CDH3 is a calcium-dependent cell-cell adhesion protein that is comprised of five extracellular cadherin repeats that make up a transmembrane region and a highly conserved cytoplasmic tail. GO annotations related to this protein include calcium ion binding, which is involved in cell junction organization and adhesion. In the present research, the result of cell immunofluorescence localization analysis revealed that CDH3 is in the cytoplasm and nucleus (Fig. 5A), which is consistent with its function. The process of tight junction membrane assembly is dependent on prior activation of the E-cadherin adhesion system. When cultured with neutralizing anti-E-cadherin antibody, embryo compaction is inhibited and membrane assembly of tight junctions is seriously disrupted and cannot target the apicolateral site of cell contact (78). Similarly, the normal assembly of tight junctions cannot develop in E-cadherin null mutant embryos (79). In addition, there is evidence that E-cadherin adhesion at compaction acts to balance tight junction proteins from turnover in case they are assembled at the membrane by their anchorage to the cytoskeleton (80). CTNNA1 encodes a member of the catenin protein family that is critical to cell adhesion, and it is located on the plasma membrane with conjoining cadherins; this finding is consistent with immunofluorescence localization (Fig. 5A). The association between catenins and cadherins forms a solid complex that is linked to the actin filament network. It seems to be critical for cadherin cell-adhesion properties, and it is associated with both E- and N-cadherins. The solid complex of E-cadherin-catenin adhesion mediates the connection between cadherins and the actin cytoskeleton at adherence junctions. We also found many upregulated proteins that are primarily involved in Golgi localization (Fig. 2A), which is like the occludin protein (an important protein for the formation of tight junctions). The ZO-1α+ isoform is transcribed and translated for the first time in the embryo; it associates with occludin at the Golgi complex, which occurs concurrently with maturation steps in occludin processing (81).
Gap junctions are essentially composed of only one protein, connexin, which is an integral membrane protein made up of four membrane-spanning domains (82). In the mouse embryo, the dominant proteins contributing to gap-junction formation are Cx32 and Cx43. Cx32 is maternally expressed in the mouse, whereas Cx43 expression appears in the embryonic genome and increases during later stages of cleavage (83–85). The onset of gap-junction coupling at the eight-cell stage is regulated developmentally, and this evidence suggests that DNA replication during the two-cell stage may be required for coordination of this process (86). In the present study, we found many maternally expressed proteins that are upregulated at the two-cell stage. At the time of assembly, Cx43 becomes phosphorylated, and stimulation of protein kinase A enhances both phosphorylation and assembly events (87). In the present study, we also found that MAP2K1 and TUBB3 were involved in gap junctions and were upregulated at the two-cell stage. MAP2K1 belongs to the dual specificity protein kinase family, which acts as a mitogen-activated protein (MAP) kinase. MAP kinases, also known as extracellular signal-regulated kinases (ERKs), act as an integration point for multiple biochemical signals; its related pathways include Trk receptor signaling, which is mediated by the MAPK pathway, and development of VEGF signaling via the VEGFR2-generic cascades. GO annotations for this gene include transferase activity, transferring phosphorus-containing groups and protein tyrosine kinase activity, and MAP2K1 may have the function of making proteins that are associated with gap junction phosphorylation, such as Cx43 and Cx32. TUBB3 encodes a class III member of the beta tubulin protein family. Beta tubulins are one of two core protein families (alpha and beta tubulins) that heterodimerize and assemble to form microtubules. Tubulin is the major constituent of microtubules, and it binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain. TUBB3 plays a critical role in proper axon guidance and maintenance. In the present study, we found that TUBB3 was in the cytoplasm, which was consistent with its role in cytoskeleton formation. Gap-junction formation between mouse blastomeres occurs for the first time during the eight-cell stage (88, 89) following synthesis of constituents during the preceding cell cycle (86, 90).
The “Maternal-to-Zygotic” Process Possibly Exists in Parthenogenesis and Occurs Between the 8-cell Stage to 16-cell Stage Like Normal Embryo
In general, the first cleavages are primarily supported by maternal factors, whereas blastocyst formation requires a combination of maternal and embryonic factors (65). NPM2 protein, also known as nucleoplasmin, is one of the crucially maternal proteins in the Xenopus oocyte nucleus (91–93). NPM2 is a nuclear chaperone protein that is expressed in oocytes. It is an acidic and highly phosphorylated protein that joins histones to form a stable pentamer complex, and it participates in nucleosome assembly (94, 95). Also, nucleoplasmin is found natively bound to histones H2A and H2B, facilitating the storage of these proteins in the oocytes (96). NPM2 may also be involved in the formation of nucleolus-like bodies during oocyte growth (97), and it is necessary for the composition of oocyte nuclear and nucleolar domains and the compaction of oocyte chromatin during the final stages of oocyte development (21). NPM2 knockout mice (NPM2-null) are infertile or subfertile, and embryos that lack NPM2 exhibit a reduced initial cleavage rate, obstruct the oocyte development to the two-cell stage, and lead to cell death 50 h after fertilization (21).
In the present experiment, TMT quantitative methods were used to determine that the expression of NPM2 is upregulated at the two-cell stage following parthenogenetic activation (Table III). We used an RT-qPCR method to detect the expression of the two genes(NPM2 and NLRP5) during the early stages of buffalo embryonic development. When compared with the GV stage, we found that there was no significant difference in the presence of NPM2 before the eight-cell stage. We also found that the expression of the GV stage was highest compared with all other stages, and the expression of the two-cell stage was higher than the MII stage, which was consistent with proteome data. Also, despite the existence of species specific differences in the precise timing of the onset of zygotic transcription in mammals, commonalties exist including evidence for a minor degree of transcriptional activity preceding the major activation of the genome in bovine, and patterns of gene expression may also be conserved, at least between the cow and the mouse, including a transient expression pattern which may represent a hallmark of chromatin remodeling during ZGA(98). After the development of the eight-cell stage, the expression of NPM2 sharply decreased, and it was not expressed in the blastocyst stage (Fig. 4D–I), which may indicate that the NPM2 protein is maternally inherited in buffalo eggs. The expression pattern of buffalo NPM2 mRNA during early embryogenesis was very similar to some known maternal effect genes that are essential for early embryonic development, such as MATER (65), ZAR1 (99, 100), and KPNA7 (101). We analyzed the tissue expression specificity of NPM2 and found that it only expressed in the ovary, MII oocyte, the two-cell stage embryo, and granular cells (Fig. 4D-III), which revealed that NPM2 may be reproduction-related maternal proteins in Buffalo.
MATER (a maternal antigen that embryos require; also known as NLRP5) is an oocyte-specific maternal effect gene that is necessary for early embryonic development beyond the two-cell stage in mice(65). In the present experiment, NLRP5 mRNA decreased strongly after the eight-cell stage, it progressively decreased during the embryo cleavage stages, and it was hardly detected in blastocysts (Fig. 4D-II), which is similar to the behavior of NPM2 and consistent with previous research(65). The mRNA expression of NPM2 and NLRP5 sharply decreased after the eight-cell stage in buffalo. We also observed a consistent decrease in NPM2 mRNA levels during early development and, more specifically, around the time of MZT, which is a period during which developmental control is transferred from maternally provided gene products to those synthesized by the zygotic genome(102). We also analyzed the tissue expression specificity of MATER and found that it only expressed in the ovary, MII oocyte, the two-cell stage embryo, and granular cells (Fig. 4D-V), which revealed that MATER might be reproduction-related maternal proteins in buffalo, like NPM2.
Also, combing with the analysis of RNA sequencing, Pearson correlation coefficients (Fig. 4A and supplemental Table S13) and principal component analyses (PCA) (Fig. 4C) of all detected genes revealed the two distinct segmentations of buffalo pre-implantation development: the first from GV (immatured oocyte) to 8-cell,and second from 16-cell to blastocyst stage (Fig. 4A and 4C), which is consisted with the result of Hierarchical clustering of differentially expressed genes in developed oocytes and parthenogenetic embryos of buffalo (Fig. 4B).
All these results demonstrate that parthenogenetic embryonic development and normal fertilization embryo development may have a similar genome activation transcription regulation process and the MZT process also possibly exists during parthenogenesis, and it may occur between the 8-cell stage to 16-cell stage in buffalo.
In conclusion, this study provided the first insights into the numerous maternally expressed proteins and genes. Our research demonstrates that the maternally expressed proteins after parthenogenetic activation are possibly involved in the formation of cell junctions firstly. Also, the two maternal proteins (NPM2, NLRP5) that have been reported in previous studies were confirmed again in the present study. Also, correlation and hierarchical analyses of transcriptome from oocytes/embryos and the expression of NPM2 and NLRP5 mRNA of buffalo in vitro developed oocytes and parthenogenetic embryos indicate that parthenogenetic embryos may exist an “MZT” like normally fertilized embryos, and that they may occur between the 8-cell stage to 16-cell stage. These data provide a rich resource for further studies on maternal proteins and genes, and they improve our understanding of the mechanisms of maternal genomics regulation and are conducive to improving nuclear transfer technology, also the strategy of this research may provide a valuable reference to explore the function of maternal genome for other species.
DATA AVAILABILITY
The MS raw data for TMT-LC-MS/MS have been deposited to the Proteome Xchange Consortium via the PRIDE partner repository (103) with the dataset identifier PXD008549.The RNA-Seq data have been deposited in NCBI's Gene Expression Omnibus (104) and are accessible through GEO Series accession number GSE108578 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE108578).
Supplementary Material
Acknowledgments
We thank Dr. Bernard A Goodman for providing language assistance and editorial help. We also appreciate Yizhi Zhong (YanLeShang Biotech, Guangxi, China) for providing the technical support of Western blotting, RT-qPCR, and cellular immunofluorescence localization analysis.
Footnotes
* This work was supported by grants from the National Natural Science Foundation of China (Grant No.31460603), the open program of key Laboratory of Buffalo Genetics, Breeding and Reproduction Technology of Guangxi Province (No.SNKF-2017-01), the Natural Science Foundation of Guangxi Province (No.2014GXNSFAA118134) and Innovation Project of Guangxi Graduate Education (No. YCBZ2017005).
 This article contains supplemental Figures and Tables.
This article contains supplemental Figures and Tables.
1 The abbreviations used are:
- TMT
- tandem mass tag
- MII
- metaphase II
- PC
- parthenogenetic cleavage
- COC
- cumulus-oocyte complex
- IVM
- maturation in vitro
- MZT
- maternal-to-zygotic transition
- GO
- Gene Ontology
- ACN
- acetonitrile
- FDR
- false discovery rate
- DEP
- differential expression protein
- PCA
- principal component analysis
- GV
- germinal vesicle.
REFERENCES
- 1. Dloniak S. M., French J. A., and Holekamp K. E. (2006) Rank-related maternal effects of androgens on behaviour in wild spotted hyaenas. Nature 440, 1190–1193 [DOI] [PubMed] [Google Scholar]
- 2. Jones H. N., Powell T. L., and Jansson T. (2007) Regulation of placental nutrient transport–a review. Placenta 28, 763–774 [DOI] [PubMed] [Google Scholar]
- 3. Ward R. (1980) Some effects of strain differences in the maternal behavior of inbred mice. Dev. Psychobiol. 13, 181–190 [DOI] [PubMed] [Google Scholar]
- 4. Brown R. E., Mathieson W. B., Stapleton J., and Neumann P. E. (1999) Maternal behavior in female C57BL/6J and DBA/2J inbred mice. Physiol. Behav. 67, 599–605 [DOI] [PubMed] [Google Scholar]
- 5. Bult A., and Lynch C. B. (1997) Nesting and fitness: lifetime reproductive success in house mice bidirectionally selected for thermoregulatory nest-building behavior. Behav. Genet. 27, 231–240 [DOI] [PubMed] [Google Scholar]
- 6. Jenness R. (1979) The composition of human milk. Semin. Perinatol. 3, 225–239 [PubMed] [Google Scholar]
- 7. Lonnerdal B. (2003) Nutritional and physiologic significance of human milk proteins. Am. J. Clin. Nutr. 77, 1537S–1543S [DOI] [PubMed] [Google Scholar]
- 8. Christians E., Davis A. A., Thomas S. D., and Benjamin I. J. (2000) Maternal effect of Hsf1 on reproductive success. Nature 407, 693–694 [DOI] [PubMed] [Google Scholar]
- 9. Tong Z. B., Gold L., Pfeifer K. E., Dorward H., Lee E., Bondy C. A., Dean J., and Nelson L. M. (2000) Mater, a maternal effect gene required for early embryonic development in mice. Nat. Genetics 26, 267–268 [DOI] [PubMed] [Google Scholar]
- 10. Matzuk M. M., and Burns K. H. (2012) Genetics of mammalian reproduction: modeling the end of the germline. Ann. Rev. Physiol. 74, 503–528 [DOI] [PubMed] [Google Scholar]
- 11. Dworkin M. B., and Dworkin-Rastl E. (1990) Functions of maternal mRNA in early development. Mol. Reproduction Develop. 26, 261–297 [DOI] [PubMed] [Google Scholar]
- 12. Schultz G. A., and Heyner S. (1992) Gene expression in pre-implantation mammalian embryos. Mutat. Res. 296, 17–31 [DOI] [PubMed] [Google Scholar]
- 13. Wassarman P. M., and Kinloch R. A. (1992) Gene expression during oogenesis in mice. Mutat. Res. 296, 3–15 [DOI] [PubMed] [Google Scholar]
- 14. Temeles G. L., Ram P. T., Rothstein J. L., and Schultz R. M. (1994) Expression patterns of novel genes during mouse preimplantation embryogenesis. Mol. Reproduction Develop. 37, 121–129 [DOI] [PubMed] [Google Scholar]
- 15. Zimmermann J. W., and Schultz R. M. (1994) Analysis of gene expression in the preimplantation mouse embryo: use of mRNA differential display. Proc. Natl. Acad. Sci. U.S.A. 91, 5456–5460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nothias J. Y., Majumder S., Kaneko K. J., and DePamphilis M. L. (1995) Regulation of gene expression at the beginning of mammalian development. J. Biol. Chem. 270, 22077–22080 [DOI] [PubMed] [Google Scholar]
- 17. Wiekowski M., Miranda M., Nothias J. Y., and DePamphilis M. L. (1997) Changes in histone synthesis and modification at the beginning of mouse development correlate with the establishment of chromatin mediated repression of transcription. J. Cell Sci. 110, 1147–1158 [DOI] [PubMed] [Google Scholar]
- 18. Pelegri F. (2003) Maternal factors in zebrafish development. Dev. Dyn. 228, 535–554 [DOI] [PubMed] [Google Scholar]
- 19. Mager J., Schultz R. M., Brunk B. P., and Bartolomei M. S. (2006) Identification of candidate maternal-effect genes through comparison of multiple microarray data sets. Mamm. Genome 17, 941–949 [DOI] [PubMed] [Google Scholar]
- 20. White J. A., and Heasman J. (2008) Maternal control of pattern formation in Xenopus laevis. J. Exp. Zool. 310, 73–84 [DOI] [PubMed] [Google Scholar]
- 21. Burns K. H., Viveiros M. M., Ren Y., Wang P., DeMayo F. J., Frail D. E., Eppig J. J., and Matzuk M. M. (2003) Roles of NPM2 in chromatin and nucleolar organization in oocytes and embryos. Science 300, 633–636 [DOI] [PubMed] [Google Scholar]
- 22. Wu X., Viveiros M. M., Eppig J. J., Bai Y., Fitzpatrick S. L., and Matzuk M. M. (2003) Zygote arrest 1 (Zar1) is a novel maternal-effect gene critical for the oocyte-to-embryo transition. Nat. Genetics 33, 187–191 [DOI] [PubMed] [Google Scholar]
- 23. Ramos S. B., Stumpo D. J., Kennington E. A., Phillips R. S., Bock C. B., Ribeiro-Neto F., and Blackshear P. J. (2004) The CCCH tandem zinc-finger protein Zfp36l2 is crucial for female fertility and early embryonic development. Development 131, 4883–4893 [DOI] [PubMed] [Google Scholar]
- 24. Bultman S. J., Gebuhr T. C., Pan H., Svoboda P., Schultz R. M., and Magnuson T. (2006) Maternal BRG1 regulates zygotic genome activation in the mouse. Genes Development 20, 1744–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yurttas P., Vitale A. M., Fitzhenry R. J., Cohen-Gould L., Wu W., Gossen J. A., and Coonrod S. A. (2008) Role for PADI6 and the cytoplasmic lattices in ribosomal storage in oocytes and translational control in the early mouse embryo. Development 135, 2627–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wan L. B., Pan H., Hannenhalli S., Cheng Y., Ma J., Fedoriw A., Lobanenkov V., Latham K. E., Schultz R. M., and Bartolomei M. S. (2008) Maternal depletion of CTCF reveals multiple functions during oocyte and preimplantation embryo development. Development 135, 2729–2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tsukamoto S., Kuma A., and Mizushima N. (2008) The role of autophagy during the oocyte-to-embryo transition. Autophagy 4, 1076–1078 [DOI] [PubMed] [Google Scholar]
- 28. Lykke-Andersen K., Gilchrist M. J., Grabarek J. B., Das P., Miska E., and Zernicka-Goetz M. (2008) Maternal Argonaute 2 is essential for early mouse development at the maternal-zygotic transition. Mol. Biol. Cell 19, 4383–4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nakamura T., Arai Y., Umehara H., Masuhara M., Kimura T., Taniguchi H., Sekimoto T., Ikawa M., Yoneda Y., Okabe M., Tanaka S., Shiota K., and Nakano T. (2007) PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat. Cell Biol. 9, 64–71 [DOI] [PubMed] [Google Scholar]
- 30. Li X., Ito M., Zhou F., Youngson N., Zuo X., Leder P., and Ferguson-Smith A. C. (2008) A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Develop. Cell 15, 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hirasawa R., Chiba H., Kaneda M., Tajima S., Li E., Jaenisch R., and Sasaki H. (2008) Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Develop. 22, 1607–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Messerschmidt D. M., de Vries W., Ito M., Solter D., Ferguson-Smith A., and Knowles B. B. (2012) Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science 335, 1499–1502 [DOI] [PubMed] [Google Scholar]
- 33. Ma M., Guo X., Wang F., Zhao C., Liu Z., Shi Z., Wang Y., Zhang P., Zhang K., Wang N., Lin M., Zhou Z., Liu J., Li Q., Wang L., Huo R., Sha J., and Zhou Q. (2008) Protein expression profile of the mouse metaphase-II oocyte. J. Proteome Res. 7, 4821–4830 [DOI] [PubMed] [Google Scholar]
- 34. Vitale A. M., Calvert M. E., Mallavarapu M., Yurttas P., Perlin J., Herr J., and Coonrod S. (2007) Proteomic profiling of murine oocyte maturation. Mol. Reproduction Develop. 74, 608–616 [DOI] [PubMed] [Google Scholar]
- 35. Zhang P., Ni X., Guo Y., Guo X., Wang Y., Zhou Z., Huo R., and Sha J. (2009) Proteomic-based identification of maternal proteins in mature mouse oocytes. BMC Genomics 10, 348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang S., Kou Z., Jing Z., Zhang Y., Guo X., Dong M., Wilmut I., and Gao S. (2010) Proteome of mouse oocytes at different developmental stages. Proc. Natl. Acad. Sci. U.S.A. 107, 17639–17644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pfeiffer M. J., Siatkowski M., Paudel Y., Balbach S. T., Baeumer N., Crosetto N., Drexler H. C., Fuellen G., and Boiani M. (2011) Proteomic analysis of mouse oocytes reveals 28 candidate factors of the “reprogrammome”. J. Proteome Res. 10, 2140–2153 [DOI] [PubMed] [Google Scholar]
- 38. Memili E., Peddinti D., Shack L. A., Nanduri B., McCarthy F., Sagirkaya H., and Burgess S. C. (2007) Bovine germinal vesicle oocyte and cumulus cell proteomics. Reproduction 133, 1107–1120 [DOI] [PubMed] [Google Scholar]
- 39. Peddinti D., Memili E., and Burgess S. C. (2010) Proteomics-based systems biology modeling of bovine germinal vesicle stage oocyte and cumulus cell interaction. PloS ONE 5, e11240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Deutsch D. R., Frohlich T., Otte K. A., Beck A., Habermann F. A., Wolf E., and Arnold G. J. (2014) Stage-specific proteome signatures in early bovine embryo development. J. Proteome Res. 13, 4363–4376 [DOI] [PubMed] [Google Scholar]
- 41. Kim J., Kim J. S., Jeon Y. J., Kim D. W., Yang T. H., Soh Y., Lee H. K., Choi N. J., Park S. B., Seo K. S., Chung H. M., Lee D. S., and Chae J. I. (2011) Identification of maturation and protein synthesis related proteins from porcine oocytes during in vitro maturation. Proteome Sci. 9, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ellederova Z., Halada P., Man P., Kubelka M., Motlik J., and Kovarova H. (2004) Protein patterns of pig oocytes during in vitro maturation. Biol. Reproduction 71, 1533–1539 [DOI] [PubMed] [Google Scholar]
- 43. Susor A., Ellederova Z., Jelinkova L., Halada P., Kavan D., Kubelka M., and Kovarova H. (2007) Proteomic analysis of porcine oocytes during in vitro maturation reveals essential role for the ubiquitin C-terminal hydrolase-L1. Reproduction 134, 559–568 [DOI] [PubMed] [Google Scholar]
- 44. Powell M. D., Manandhar G., Spate L., Sutovsky M., Zimmerman S., Sachdev S. C., Hannink M., Prather R. S., and Sutovsky P. (2010) Discovery of putative oocyte quality markers by comparative ExacTag proteomics. Proteomics. Clin. Appl. 4, 337–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yan L., Yang M., Guo H., Yang L., Wu J., Li R., Liu P., Lian Y., Zheng X., Yan J., Huang J., Li M., Wu X., Wen L., Lao K., Li R., Qiao J., and Tang F. (2013) Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Structural Mol. Biol. 20, 1131–1139 [DOI] [PubMed] [Google Scholar]
- 46. Fan X., Zhang X., Wu X., Guo H., Hu Y., Tang F., and Huang Y. (2015) Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol. 16, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Blakeley P., Fogarty N. M., del Valle I., Wamaitha S. E., Hu T. X., Elder K., Snell P., Christie L., Robson P., and Niakan K. K. (2015) Defining the three cell lineages of the human blastocyst by single-cell RNA-seq. Development 142, 3151–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shi J., Chen Q., Li X., Zheng X., Zhang Y., Qiao J., Tang F., Tao Y., Zhou Q., and Duan E. (2015) Dynamic transcriptional symmetry-breaking in pre-implantation mammalian embryo development revealed by single-cell RNA-seq. Development 142, 3468–3477 [DOI] [PubMed] [Google Scholar]
- 49. Shi D., Lu F., Wei Y., Cui K., Yang S., Wei J., and Liu Q. (2007) Buffalos (Bubalus bubalis) cloned by nuclear transfer of somatic cells. Biol. Reproduction 77, 285–291 [DOI] [PubMed] [Google Scholar]
- 50. Huang B., Cui K., Li T., Wang X., Lu F., Liu Q., da Silva F. M., and Shi D. (2010) Generation of buffalo (Bubalus bubalis) transgenic chimeric and nuclear transfer embryos using embryonic germ-like cells expressing enhanced green fluorescent protein. Reprod. Domest. Anim. 45, 103–108 [DOI] [PubMed] [Google Scholar]
- 51. Lu F., Jiang J., Li N., Zhang S., Sun H., Luo C., Wei Y., and Shi D. (2011) Effects of recipient oocyte age and interval from fusion to activation on development of buffalo (Bubalus bubalis) nuclear transfer embryos derived from fetal fibroblasts. Theriogenology 76, 967–97421752448 [Google Scholar]
- 52. Luo C., Lu F., Wang X., Wang Z., Li X., Gong F., Jiang J., Liu Q., and Shi D. (2013) Treatment of donor cells with trichostatin A improves in vitro development and reprogramming of buffalo (Bubalus bubalis) nucleus transfer embryos. Theriogenology 80, 878–886 [DOI] [PubMed] [Google Scholar]
- 53. Zhang M., Lu K. H., and Seidel G. E. (2003) Development of bovine embryos after in vitro fertilization of oocytes with flow cytometrically sorted, stained and unsorted sperm from different bulls. Theriogenology 60, 1657–1663 [DOI] [PubMed] [Google Scholar]
- 54. Lu F., Shi D., Wei J., Yang S., and Wei Y. (2005) Development of embryos reconstructed by interspecies nuclear transfer of adult fibroblasts between buffalo (Bubalus bubalis) and cattle (Bos indicus). Theriogenology 64, 1309–1319 [DOI] [PubMed] [Google Scholar]
- 55. Ramskold D., Luo S., Wang Y. C., Li R., Deng Q., Faridani O. R., Daniels G. A., Khrebtukova I., Loring J. F., Laurent L. C., Schroth G. P., and Sandberg R. (2012) Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 30, 777–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., and Salzberg S. L. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., van Baren M. J., Salzberg S. L., Wold B. J., and Pachter L. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Leng N., Dawson J. A., Thomson J. A., Ruotti V., Rissman A. I., Smits B. M., Haag J. D., Gould M. N., Stewart R. M., and Kendziorski C. (2013) EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 29, 1035–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Florea L., Song L., and Salzberg S. L. (2013) Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Res 2, 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Telford N. A., Watson A. J., and Schultz G. A. (1990) Transition from maternal to embryonic control in early mammalian development: a comparison of several species. Mol. Reproduction Development 26, 90–100 [DOI] [PubMed] [Google Scholar]
- 61. Memili E., and First N. L. (1998) Developmental changes in RNA polymerase II in bovine oocytes, early embryos, and effect of alpha-amanitin on embryo development. Mol. Reproduction Development 51, 381–389 [DOI] [PubMed] [Google Scholar]
- 62. Misirlioglu M., Page G. P., Sagirkaya H., Kaya A., Parrish J. J., First N. L., and Memili E. (2006) Dynamics of global transcriptome in bovine matured oocytes and preimplantation embryos. Proc. Natl. Acad. Sci U.S.A. 103, 18905–18910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Graf A., Krebs S., Zakhartchenko V., Schwalb B., Blum H., and Wolf E. (2014) Fine mapping of genome activation in bovine embryos by RNA sequencing. Proc. Natl. Acad. Sci. U.S.A. 111, 4139–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lingenfelter B. M., Tripurani S. K., Tejomurtula J., Smith G. W., and Yao J. (2011) Molecular cloning and expression of bovine nucleoplasmin 2 (NPM2): a maternal effect gene regulated by miR-181a. Reproductive Biol. Endocrinol. 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pennetier S., Perreau C., Uzbekova S., Thelie A., Delaleu B., Mermillod P., and Dalbies-Tran R. (2006) MATER protein expression and intracellular localization throughout folliculogenesis and preimplantation embryo development in the bovine. BMC Developmental Biol. 6, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Larue L., Ohsugi M., Hirchenhain J., and Kemler R. (1994) E-cadherin null mutant embryos fail to form a trophectoderm epithelium. Proc. Natl. Acad. Sci. U.S.A. 91, 8263–8267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Riethmacher D., Brinkmann V., and Birchmeier C. (1995) A targeted mutation in the mouse E-cadherin gene results in defective preimplantation development. Proc. Natl. Acad. Sci. U.S.A. 92, 855–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. De Vries W. N., Evsikov A. V., Haac B. E., Fancher K. S., Holbrook A. E., Kemler R., Solter D., and Knowles B. B. (2004) Maternal beta-catenin and E-cadherin in mouse development. Development 131, 4435–4445 [DOI] [PubMed] [Google Scholar]
- 69. Golding M. C., Williamson G. L., Stroud T. K., Westhusin M. E., and Long C. R. (2011) Examination of DNA methyltransferase expression in cloned embryos reveals an essential role for Dnmt1 in bovine development. Mol. Reproduction Develop. 78, 306–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bakhtari A., and Ross P. J. (2014) DPPA3 prevents cytosine hydroxymethylation of the maternal pronucleus and is required for normal development in bovine embryos. Epigenetics 9, 1271–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lin C. J., Conti M., and Ramalho-Santos M. (2013) Histone variant H3.3 maintains a decondensed chromatin state essential for mouse preimplantation development. Development 140, 3624–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Inoue A., and Zhang Y. (2014) Nucleosome assembly is required for nuclear pore complex assembly in mouse zygotes. Nat. Structural Mol. Biol. 21, 609–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wen D., Banaszynski L. A., Liu Y., Geng F., Noh K. M., Xiang J., Elemento O., Rosenwaks Z., Allis C. D., and Rafii S. (2014) Histone variant H3.3 is an essential maternal factor for oocyte reprogramming. Proc. Natl. Acad. Sci. U.S.A. 111, 7325–7330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Inoue A., Ogushi S., Saitou M., Suzuki M. G., and Aoki F. (2011) Involvement of mouse nucleoplasmin 2 in the decondensation of sperm chromatin after fertilization. Biol. Reproduction 85, 70–77 [DOI] [PubMed] [Google Scholar]
- 75. Rajkovic A., Pangas S. A., Ballow D., Suzumori N., and Matzuk M. M. (2004) NOBOX deficiency disrupts early folliculogenesis and oocyte-specific gene expression. Science 305, 1157–1159 [DOI] [PubMed] [Google Scholar]
- 76. Tripurani S. K., Lee K. B., Wang L., Wee G., Smith G. W., Lee Y. S., Latham K. E., and Yao J. (2011) A novel functional role for the oocyte-specific transcription factor newborn ovary homeobox (NOBOX) during early embryonic development in cattle. Endocrinology 152, 1013–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Fleming T. P., Ghassemifar M. R., and Sheth B. (2000) Junctional complexes in the early mammalian embryo. Sem. Reproductive Med. 18, 185–193 [DOI] [PubMed] [Google Scholar]
- 78. Fleming T. P., McConnell J., Johnson M. H., and Stevenson B. R. (1989) Development of tight junctions de novo in the mouse early embryo: control of assembly of the tight junction-specific protein, ZO-1. J. Cell Biol. 108, 1407–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ohsugi M., Larue L., Schwarz H., and Kemler R. (1997) Cell-junctional and cytoskeletal organization in mouse blastocysts lacking E-cadherin. Developmental Biol. 185, 261–271 [DOI] [PubMed] [Google Scholar]
- 80. Javed Q., Fleming T. P., Hay M., and Citi S. (1993) Tight junction protein cingulin is expressed by maternal and embryonic genomes during early mouse development. Development 117, 1145–1151 [DOI] [PubMed] [Google Scholar]
- 81. Sheth B., Fesenko I., Collins J. E., Moran B., Wild A. E., Anderson J. M., and Fleming T. P. (1997) Tight junction assembly during mouse blastocyst formation is regulated by late expression of ZO-1 alpha+ isoform. Development 124, 2027–2037 [DOI] [PubMed] [Google Scholar]
- 82. Simon A. M., and Goodenough D. A. (1998) Diverse functions of vertebrate gap junctions. Trends Cell Biol. 8, 477–483 [DOI] [PubMed] [Google Scholar]
- 83. Barron D. J., Valdimarsson G., Paul D. L., and Kidder G. M. (1989) Connexin32, a gap junction protein, is a persistent oogenetic product through preimplantation development of the mouse. Developmental Gen. 10, 318–323 [DOI] [PubMed] [Google Scholar]
- 84. Valdimarsson G., De Sousa P. A., Beyer E. C., Paul D. L., and Kidder G. M. (1991) Zygotic expression of the connexin43 gene supplies subunits for gap junction assembly during mouse preimplantation development. Mol. Reproduction Develop. 30, 18–26 [DOI] [PubMed] [Google Scholar]
- 85. De Sousa P. A., Valdimarsson G., Nicholson B. J., and Kidder G. M. (1993) Connexin trafficking and the control of gap junction assembly in mouse preimplantation embryos. Development 117, 1355–1367 [DOI] [PubMed] [Google Scholar]
- 86. Valdimarsson G., and Kidder G. M. (1995) Temporal control of gap junction assembly in preimplantation mouse embryos. J. Cell Sci. 108 (Pt 4), 1715–1722 [DOI] [PubMed] [Google Scholar]
- 87. Ogawa H., Oyamada M., Mori T., Mori M., and Shimizu H. (2000) Relationship of gap junction formation to phosphorylation of connexin43 in mouse preimplantation embryos. Mol. Reproduction Develop. 55, 393–398 [DOI] [PubMed] [Google Scholar]
- 88. Lo C. W., and Gilula N. B. (1979) Gap junctional communication in the preimplantation mouse embryo. Cell 18, 399–409 [DOI] [PubMed] [Google Scholar]
- 89. Goodall H., and Johnson M. H. (1984) The nature of intercellular coupling within the preimplantation mouse embryo. J. Embryol. Exp. Morphol. 79, 53–76 [PubMed] [Google Scholar]
- 90. McLachlin J. R., and Kidder G. M. (1986) Intercellular junctional coupling in preimplantation mouse embryos: effect of blocking transcription or translation. Developmental Biol. 117, 146–155 [DOI] [PubMed] [Google Scholar]
- 91. Laskey R. A., Honda B. M., Mills A. D., and Finch J. T. (1978) Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature 275, 416–420 [DOI] [PubMed] [Google Scholar]
- 92. Mills A. D., Laskey R. A., Black P., and De Robertis E. M. (1980) An acidic protein which assembles nucleosomes in vitro is the most abundant protein in Xenopus oocyte nuclei. J. Mol. Biol. 139, 561–568 [DOI] [PubMed] [Google Scholar]
- 93. Krohne G., and Franke W. W. (1980) Immunological identification and localization of the predominant nuclear protein of the amphibian oocyte nucleus. Proc. Natl. Acad. Sci. U.S.A. 77, 1034–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Earnshaw W. C., Honda B. M., Laskey R. A., and Thomas J. O. (1980) Assembly of nucleosomes: the reaction involving X. laevis nucleoplasmin. Cell 21, 373–383 [DOI] [PubMed] [Google Scholar]
- 95. Dilworth S. M., Black S. J., and Laskey R. A. (1987) Two complexes that contain histones are required for nucleosome assembly in vitro: role of nucleoplasmin and N1 in Xenopus egg extracts. Cell 51, 1009–1018 [DOI] [PubMed] [Google Scholar]
- 96. Philpott A., Krude T., and Laskey R. A. (2000) Nuclear chaperones. Sem. Cell Developmental Biol. 11, 7–14 [DOI] [PubMed] [Google Scholar]
- 97. Inoue A., and Aoki F. (2010) Role of the nucleoplasmin 2 C-terminal domain in the formation of nucleolus-like bodies in mouse oocytes. FASEB J. 24, 485–494 [DOI] [PubMed] [Google Scholar]
- 98. De Sousa P. A., Watson A. J., Schultz G. A., and Bilodeau-Goeseels S. (1998) Oogenetic and zygotic gene expression directing early bovine embryogenesis: a review. Mol. Reproduction Development 51, 112–121 [DOI] [PubMed] [Google Scholar]
- 99. Pennetier S., Uzbekova S., Perreau C., Papillier P., Mermillod P., and Dalbies-Tran R. (2004) Spatio-temporal expression of the germ cell marker genes MATER, ZAR1, GDF9, BMP15,andVASA in adult bovine tissues, oocytes, and preimplantation embryos. Biol. Reproduction 71, 1359–1366 [DOI] [PubMed] [Google Scholar]
- 100. Brevini T. A., Cillo F., Colleoni S., Lazzari G., Galli C., and Gandolfi F. (2004) Expression pattern of the maternal factor zygote arrest 1 (Zar1) in bovine tissues, oocytes, and embryos. Mol, Reproduction Development 69, 375–380 [DOI] [PubMed] [Google Scholar]
- 101. Tejomurtula J., Lee K. B., Tripurani S. K., Smith G. W., and Yao J. (2009) Role of importin alpha8, a new member of the importin alpha family of nuclear transport proteins, in early embryonic development in cattle. Biol. Reproduction 81, 333–342 [DOI] [PubMed] [Google Scholar]
- 102. Tadros W., and Lipshitz H. D. (2009) The maternal-to-zygotic transition: a play in two acts. Development 136, 3033–3042 [DOI] [PubMed] [Google Scholar]
- 103. Vizcaino J. A., Csordas A., del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Edgar R., Domrachev M., and Lash A. E. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS raw data for TMT-LC-MS/MS have been deposited to the Proteome Xchange Consortium via the PRIDE partner repository (103) with the dataset identifier PXD008549.The RNA-Seq data have been deposited in NCBI's Gene Expression Omnibus (104) and are accessible through GEO Series accession number GSE108578 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE108578).