Abstract
Lysine crotonylation (Kcr) is a newly discovered posttranslational modification (PTM) existing in mammals. A global crotonylome analysis was undertaken in rice (Oryza sativa L. japonica) using high accuracy nano-LC-MS/MS in combination with crotonylated peptide enrichment. A total of 1,265 lysine crotonylation sites were identified on 690 proteins in rice seedlings. Subcellular localization analysis revealed that 51% of the crotonylated proteins identified were localized in chloroplasts. The photosynthesis-associated proteins were also mostly enriched in total crotonylated proteins. In addition, a genomic localization analysis of histone Kcr by ChIP-seq was performed to assess the relevance between histone Kcr and the genome. Of the 10,923 identified peak regions, the majority (86.7%) of the enriched peaks were located in gene body, especially exons. Furthermore, the degree of histone Kcr modification was positively correlated with gene expression in genic regions. Compared with other published histone modification data, the Kcr was co-located with the active histone modifications. Interestingly, histone Kcr-facilitated expression of genes with existing active histone modifications. In addition, 77% of histone Kcr modifications overlapped with DNase hypersensitive sites (DHSs) in intergenic regions of the rice genome and might mark other cis-regulatory DNA elements that are different from IPA1, a transcription activator in rice seedlings. Overall, our results provide a comprehensive understanding of the biological functions of the crotonylome and new active histone modification in transcriptional regulation in plants.
Keywords: Epigenetics, Histones, Transcriptional Regulation, Protein Modification, Photosynthesis, crotonyl lysine, histone lysine crotonylation, Oryza sativa, post-translational modification
Precursor proteins are typically inactive and could be converted into mature functional proteins through a series of posttranslational modifications (PTMs), which modulate diverse protein properties and functions (1). PTMs have been associated with almost all known metabolic processes and cellular pathways in various ways (2, 3). The major form of PTMs is covalent addition of functional chemical groups to one or more amino acids. PTMs can greatly increase the complexity of the proteome based on the presence of multiple modification sites within a protein, each with different types. Due to specific chemical reactivity, lysine (K) is one of the most common residues that is subject to PTMs (4), such as acetylation (Kac) (5, 6), methylation (Kme) (7, 8), malonylation (Kma) (9, 10), propionylation (Kpro) (11, 12), butyrylation (Kbu) (12, 13), and succinylation (Ksucc) (10, 14). With the development of high-specific antibodies and high-resolution MS techniques, increasing numbers of lysine modifications of both histone and non-histone proteins have been identified in the proteome. These modifications include Kac (15–18), Ksucc (14, 19), Kme (20), and Kma (9).
Among the lysine modifications of the proteome, histone lysine modifications are essential for the control of gene expression by complex interactions of transcription factors binding to regulatory DNA elements, including promoters, enhancers, insulators, and silencers (21, 22). Specific histone modifications of the chromatin activate or repress regulatory DNA elements, thus playing an important role in transcriptional regulation (23). For example, H3K9ac, H3K4me3, and H3K4me2 mark active promoters, whereas repressed genes are marked by H3K27me3 or H3K9me2, and enhancers are commonly marked by H3K27ac and H3K4me1/2 (24, 25).
Lysine crotonylation (Kcr)1 is a newly discovered PTM that exists in mammals (26, 27). This histone modification has been identified in evolutionary distant eukaryotic organisms, such as yeast (Saccharomyces. cerevisiae) and invertebrate species including Caenorhabditis. elegans and Drosophila, as well as in mice and humans, suggesting that this modification is widely conserved (26). Crotonylation is a histone modification, involving a four-carbon length in the planar orientation (28). This modification neutralizes the positive charge of the ε-amino group of lysine, leading to the possibility of charge-based cis-effects on the chromatin fiber, where the increased bulk and rigidity of the crotonyl group may result in an enhanced effect (23). Therefore, histone Kcr is generally enriched in the regions of active promoters and potential enhancers in mammalian cells (26). In addition, Kcr has been shown to clearly mark autosomal testis-specific genes, which are activated in postmeiotic round spermatids (29). Recently, the global profiling of crotonylation on non-histone proteins has been reported in mammals and tobacco (28, 30, 31). Crotonylation of non-histone proteins is involved in different signaling pathways and cellular functions. To our knowledge, crotonylation of non-histone and histone proteins has rarely been reported in monocots.
Rice (Oryza sativa) is one of the most important cereal crops in the world and represents a valuable model plant for the investigation of monocots in functional genome research (32). Since crotonylated proteins have not yet been identified in rice, we initiated a systematic study to identify and investigate functional roles of the crotonylated proteins in Nipponbare, which is the first rice variety with a complete genomic sequence published (33). In this study, we obtained the crotonylome of Nipponbare using high accuracy LC-MS/MS in combination with the enrichment of crotonylated peptides from digested cell lysates and subsequent peptide identification. In total, 1,265 crotonylated sites were identified in 690 proteins in Nipponbare. In addition, we conducted a genome-wide study of histone Kcr by ChIP-seq analysis with the pan anti-Kcr and H3K14cr antibodies. This information will broaden our understanding of the biological functions influenced by histone Kcr. In short, our findings provide significant insights into the range of functions regulated by lysine crotonylation in rice.
EXPERIMENTAL PROCEDURES
Materials
O. sativa variety “Nipponbare” seeds were germinated at ambient temperature for 72 h. The germinated seeds were then sown and grown in water under greenhouse conditions (12 h light at 28 °C/12 h dark at 25 °C) with 70% humidity. Leaves of the two-week-old rice seedling were sampled for protein and ChIP-DNA isolation.
Green seedlings and albino seedlings derived from another culture of Nipponbare were grown in Murashige and Skoog plates containing 0.5% naphthylacetic acid, 3% sucrose, and 0.5% agar.
Protein Extraction, Trypsin Digestion, and HPLC Fractionation
Protein extraction, trypsin digestion and HPLC fractionation were conducted using a procedure described by Xue et al. (34). Leaf samples were ground under liquid nitrogen and sonicated three times on ice using a high intensity ultrasonic processor (Scientz) in lysis buffer (8 m urea, 1% Triton-100, 65 mm DTT, and 0.1% Protease Inhibitor Mixture). The remaining debris was removed by centrifugation (20,000 g, 4 °C, 10 min). Finally, the protein was precipitated with cold 15% TCA for 2 h at −20 °C. After centrifugation (12,000 g, 4 °C, 10 min), the supernatant was discarded. The remaining precipitate was washed three times with cold acetone. The protein was redissolved in buffer (8 m urea, 100 mm NH4CO3, pH 8.0). For digestion, the protein solution was reduced with 10 mm DTT for 1 h at 37 °C and alkylated with 20 mm iodoacetamide for 45 min at room temperature in darkness. For trypsin digestion, the protein samples were diluted by adding 100 mm NH4CO3 to reduce the urea concentration to below 2 m. The protein concentration was determined with BCA kit (P0011-1, Beyotime Biotechnology, Shanghai, China) according to the manufacturer's instructions. Finally, trypsin was added at 1:50 trypsin-to-protein mass ratio for an overnight digestion, followed by an addition of 1:100 trypsin-to-protein mass ratio for another 4-h digestion. The samples were then separated into 80 fractions by high pH reverse-phase HPLC using Agilent 300Extend C18 column (5 μm particles, 4.6 mm inner diameter, 250 mm length). Briefly, peptides were first separated with a gradient of 2% to 60% acetonitrile in 10 mm ammonium bicarbonate with a pH of 10 over 80 min. The peptides were then combined into eight fractions and dried by vacuum centrifugation.
Affinity Enrichment of Kcr Peptides and Enrichment of Lysine Peptides and LC-MS/MS Analysis
To enrich Kcr peptides, tryptic peptides dissolved in NETN buffer (100 mm NaCl, 1 mm EDTA, 50 mm Tris-HCl, 0.5% NP-40, pH 8.0) were incubated with prewashed antibody beads (PTM BioLabs, HangZhou, China PTM-501) at 4 °C overnight with gentle shaking. The beads were washed four times with NETN buffer and twice with ddH2O. The bound peptides were eluted from the beads with 0.1% TFA. The eluted fractions were combined and vacuum-dried. The resulting peptides were cleaned with C18 ZipTips (Millipore) according to the manufacturer's instructions prior to LC-MS/MS analysis. Enrichment of lysine peptides were analyzed using LC-MS/MS as described by Xue et al. (34). Briefly, peptides were dissolved in 0.1% formic acid and directly loaded onto a reversed-phase precolumn (Acclaim PepMap 100, Thermo Scientific). Peptide separation was performed using a reversed-phase analytical column (Acclaim PepMap RSLC, Thermo Scientific). The gradient was as follows: 6% to 22% solvent B (0.1% formic acid in 98% acetonitrile) for 24 min, 22% to 35% for 8 min then increased to 80% over 5 min and held at 80% for 3 min. The gradient was generated at a constant flow rate of 300 nl/min on an EASY-nLC 1000 UPLC system. The resulting peptides were analyzed by Q ExactiveTM hybrid quadrupole-Orbitrap mass spectrometer (ThermoFisher Scientific). The peptides were subjected to a nano electrospray ionization source followed by tandem mass spectrometry (MS/MS) using Q ExactiveTM (Thermo) coupled online to the UPLC. Intact peptides were detected in the Orbitrap at a resolution of 70,000. Peptides were selected for MS/MS using a normalized collision energy setting of 28; ion fragments were detected in the Orbitrap at a resolution of 17,500. A data-dependent procedure that alternated between one MS scan followed by 20 MS/MS scans was applied for the top 20 precursor ions above a threshold ion count of 2e4 in the MS survey scan with 10.0-s dynamic exclusion. The electrospray voltage applied was 2.0 kV. Automatic gain control was used to prevent overfilling of the ion trap; 5e4 ions were accumulated for generation of MS/MS spectra. The MS scans were performed over the range of 350 to 1,800 m/z.
Database Search
The resulting tandem MS data were processed using the MaxQuant with integrated Andromeda search engine (v.1.4.1.2) described by Zhou et al. (35) according to a method described by He et al. (36). Tandem mass spectra were searched against the UniProt_Oryza sativa database (UniProt Oryza sativa subsp. japonica) concatenated with the reverse decoy database. The search database consisted of the UniProt Oryza sativa subsp. japonica proteome set (including 63,195 protein sequences) downloaded from UniProtKB (http://www.uniprot.org) in July 2014. Trypsin/P was specified as the cleavage enzyme with allowances set for up to four missing cleavages, five modifications per peptide and five charges. Mass error was set to 10 ppm for precursor ions and 0.02 Da for fragment ions. Carbamidomethylation of Cys was specified as a fixed modification. Oxidation of Met, crotonylation of lysine, and acetylation on protein N-terminal were specified as variable modifications. False discovery rate thresholds for proteins, peptides, and modification sites were set at 1%. Minimum peptide length was set at 7. All the other MaxQuant parameters were set to default values and the site localization probability was set at >0.75.
The phylogenetic tree was constructed using the neighbor-joining method with MEGA Version 6.0 (37) (gaps/missing data treatment: pairwise deletion, bootstrap: 1,000). Human and mouse p300 genes were used as an outgroup. All p300 protein sequences were aligned using T-coffee (38) with default options.
Bioinformatics Analysis
Analysis of Sequences Around Crotonylated Site
The analysis of sequences around the Kcr site was performed based on Shen et al. (39). For all proteins, Motif-X was used to analyze the model of sequences constituted by amino acids in specific positions of modifier 21-mers (10 amino acids upstream and downstream of the site). And all the database protein sequences were used as background database parameters, while other parameters were set to the default. All the crotonylation substrate categories obtained after enrichment were collated along with their p values and then filtered for those categories that were enriched in at least one of the clusters with p value <0.05. This filtered p value matrix was transformed by the x = −log10 (p value). The results were visualized in a heat map generated using “heatmap.2.”
Functional Annotation of Proteins
Gene ontology (GO) annotation of the proteome was achieved with reference to the UniProt-gene ontology annotation (GOA) database (www.http://www.ebi.ac.uk/GOA/) using a procedure described by Xue et al. (34). Proteins were classified by GO annotation based on three categories: biological process, cellular component, and molecular function.
Functional Enrichment Analysis
The Encyclopedia of Genes and Genomes (KEGG) database was used to identify enriched pathways using the Functional Annotation Tool of database for annotation, visualization and integrated discovery (DAVID) against the background of Nipponbare, following a detailed procedure described by Xue et al. (34). GO annotation and enriched pathways with a corrected p value <0.05 were considered to be statistically significant.
Protein-protein Interaction Analysis
Protein-protein interaction (PPI) analysis of identified crotonylated proteins was performed using Cytoscape software (Version 3.3.0). PPI networks were obtained using the search tool for the retrieval of interacting genes/proteins (STRING) database, which uses a metric known as the “confidence score” to define interaction confidence. We fetched all interactions with a confidence score ≥0.9 (high confidence) (40). The posttranslational protein was analyzed for densely connected regions with a theoretical cluster graph generated using the molecular complex detection algorithm, which is part of the plug-in tool kit of the network analysis and visualization software Cytoscape. The highest-ranking modules containing modification sites were selected for further analysis and rendering.
Whole Protein and Histone Extraction
Whole Protein Extraction
Leaves collected from rice seedling (0.2 g) were first ground under liquid nitrogen. The tissue was then transferred to a 1.5-ml centrifuge tube in lysis buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 0.05% 2-hydroxy-1-ethanethiol, 2% SDS, 65 mm DTT, and 1 mm Protease Inhibitor Mixture). The tissue suspension was mixed for 3 h on ice. The remaining debris was removed by centrifugation (13,000 g, 4 °C, 10 min).
Histone Extraction
Two-week-old rice seedling leaves (4 g) were first ground under liquid nitrogen. The tissue was then transferred to a 50-ml centrifuge tube containing extraction buffer A (0.4 m sucrose, 10 mm Tris-HCl, pH 8.0, 10 mm MgCl2, 1% Triton-100, 5 mm 2-hydroxy-1-ethanethiol, 100 mm Protease Inhibitor Mixture). The remaining debris was collected by centrifugation (13,000 rpm 16,000 g, 4 °C, 10 min). The tissue suspension was sonicated (10 s on/10 s off, 30% power) in nuclei lysis buffer (50 mm Tris-HCl, pH 8.0, 10 mm EGTA, 1% SDS, 10 mm Protease Inhibitor Mixture) for 10 min on ice and then mixed with extraction buffer B (0.2 m HCl) for 30 min on ice. The protein was precipitated with cold 100% TCA for 10 min at −20 °C. After centrifugation at 13,000 g at 4 °C for 30 min, the supernatant was discarded. The remaining precipitate was washed three times with cold acetone. The protein was redissolved in milli-Q water.
Western Blotting and Immunofluorescence Analyses
Western blotting (WB) was performed based on a previously described method (41). Briefly, total proteins and histone proteins were extracted from two-week-old rice seedling leaves and separated by gradient (5–12%) sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) prior to detection with rabbit pan anti-Kcr antibody (1:3,000; PTM BioLabs, PTM-501), rabbit anti-H3 antibody (1:3,000; PTM BioLabs, PTM-1001), and rabbit anti-H3K14cr antibody (1:3,000; PTM BioLabs, PTM-535). Antigen-antibody complexes were detected by Fluor Chem E and AlphaView SA. Immunofluorescence analysis of mitotic chromosomes was performed using a procedure described by Gong et al. (42). Histone modifications were identified with rabbit pan anti-Kcr antibodies (PTM BioLabs, PTM-501) and rabbit anti-H3K14cr antibody (1:3,000; PTM BioLabs, PTM-535), and the goat secondary antibody (Alexa 488-conjugated anti-rabbit; Invitrogen, A11008). Chromosomes were counterstained with DAPI using Vectashield (Vector Laboratories, H-1200).
Dot Blotting
Samples (2 μl) were spotted slowly onto a dry nitrocellulose membrane at the center of a predrawn grid to minimize the area that the solution penetrates. Nonspecific sites were blocked by soaking in 5% BSA in TBS-T (20 mm Tris-HCl, 150 mm NaCl, 0.05% Tween20) for 30–60 min at room temperature. The membrane was incubated with rabbit pan anti-Kcr antibody (1:3,000; PTM BioLabs, PTM-501) dissolved in BSA/TBS-T for 30 min at room temperature and washed three times with TBS-T (3 × 5 min). The membrane was then incubated with secondary antibody conjugated with HRP (diluted according to the manufacturer's recommendation) for 30 min at room temperature. The membrane was washed three times with TBS-T (15 min × 1 and 5 min × 2) and once with TBS (20 mm Tris-HCl, 150 mm NaCl, pH 7.5) for 5 min. For visualization of the dots, the membrane was then incubated with ECL reagent for 1 min, covered with Saran-wrap, and exposed to X-ray film in the dark.
ChIP, ChIP-seq, and qPCR
ChIP experiments were performed using a pan anti-Kcr antibody (PTM BioLabs, PTM-501) and H3K14cr antibody (PTM BioLabs, PTM-535) following a published protocol (43), including methods for nuclei extraction and cleaning, chromatin digestion, precleaning of digested chromatin, and evaluation of digested chromatin. Mock treatment using normal rabbit serum served as a negative control. The DNAs identified in ChIP experiments were then used for library construction according to the protocol provided by Illumina and were sequenced using the HiSeq 2500 platform. The reads from the HiSeq analysis were mapped to the rice reference genome Tigr 7 (44) using the Bowtie program (45). Only the reads mapped to unique positions in the genome were retained to identify histone Kcr-enriched regions using model-based analysis of ChIP-Seq (MACS) with p value <1e-5 (46). The DNAs identified in ChIP experiments were also used to perform real-time quantitative PCR (qPCR) analysis according to the procedure described by Mukhopadhyay et al. (47). Input-DNA was set as the control and the following thermocycling conditions were used: initial denaturation at 95 °C for 600s, three-step amplification comprising 40 cycles of 95 °C for 10 s to 55 °C for 10 s to 72 °C for 20s. All data for published histone modifications (H3K9ac, H4K12ac, H3K4me2, H3K36me3, H3K27me3) were downloaded from the Gene Expression Omnibus at the National Center for Biotechnology Information (NCBI GEO) (48, 49). The gene expression data from RNA-seq were also published (49).
Experimental Design and Statistical Rationale
Crotonylation of O. sativa was investigated by WB and immunofluorescence analyses using two-week-old rice leaves. Crotonylated peptides were then enriched using immunoaffinity enrichment strategies and analyzed by high accuracy nanoflow LC-MS/MS. False discovery rate thresholds for proteins, peptides, and modification sites were set at 1%. Minimum peptide length was set at 7. All the other parameters in MaxQuant (v.1.4.1.2) were set to default values. The site localization probability was set at >0.75. Proteins were classified by GO annotation into three categories. GO enrichment was performed by DAVID (41) using hypergeometric tests with corresponding p values <0.05 (hypergeometric test) considered to be statistically significant. Soft Motif-X was used to analyze the model of sequences constituted by amino acids in specific positions of modifier-21-mers (10 amino acids upstream and downstream of the site) in all protein sequences. In the histone Kcr biological function study, ChIP-DNA, which was enriched by anti-Kcr and H3K14cr antibodies, was used for library construction and sequenced using the HiSeq 2500 platform. The reads that mapped to unique positions in the genome were retained to identify histone Kcr-enriched regions using MACS with p value <1e-5.
RESULTS
The Lysine Crotonylome Map of Rice is Represented by 1,265 Kcr Sites in 690 Proteins
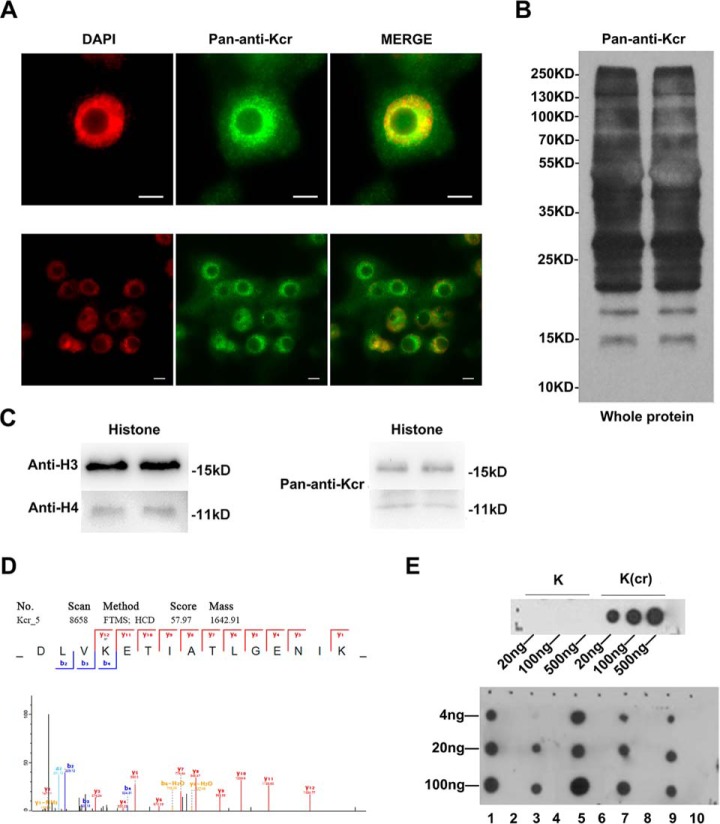
To characterize the global lysine crotonylation (Kcr) distribution in rice, an overview of Kcr modifications was obtained by immunofluorescence and WB analyses using a pan anti-Kcr antibody. Results using immunofluorescence revealed obvious distribution of lysine crotonylation in the nuclei and cytoplasm (Fig. 1A). The results from WB also showed that all proteins in the leaves of rice seedlings were widely crotonylated (Fig. 1B). Furthermore, the signals of Kcr in histone H3 (∼15 kDa) and H4 (∼11 kDa) were detected by histone WB analysis (Fig. 1C).
Fig. 1.
An overview of Kcr modifications in rice. (A) Crotonylation was detected in the nucleus and cytoplasm of two-week-old rice root by immunofluorescence using an anti-Kcr antibody (green), and nuclei were stained with DAPI (red). Scale bars: 5 μm. (B) Western blot analysis of the total protein content of rice seedling leaves showing duplicates. (C) Western blot analysis of histones of rice seedling leaves. (D) Randomly selected crotonylpeptide DLV-(crotonyl)K-ETIATL with the crotonylation site at K1099 in the elongation factor. (E) Dot-spot assay of validated crotonylated proteins. The numbers 1, 3, 5, 7, and 9 represent crotonyl peptides TIMP-(crotonyl)K-DIQLA, IQGIT-(crotonyl)K-PAIR, LEV-(crotonyl)K-EIAEIM, LAEEG-(crotonyl)K-VAIR, and DLV-(crotonyl)K-ETIATL, respectively. The numbers 2, 4, 6, 8, and 10 represent unmodified peptides TIMP-K-DIQLA, IQGIT-K-PAIR, LEV-K-EIAEIM, LAEEG-K-VAIR, and DLV-K-ETIATL, respectively.
The preliminary analysis indicated an extensive existence of Kcr in rice. A combination of Kcr antibodies and HPLC-MS/MS was used to characterize the Kcr distribution in the crotonylome of rice. To validate the MS data, we first checked the mass error of the identified peptides. The distribution of mass error of all the identified peptides was extremely close to zero, and most were less than 0.02 Da, indicating the accuracy of the MS data (supplemental Fig. 1A). The length of most peptides was distributed between 7 and 18 residues (supplemental Fig. 1B), implying the high quality of sample preparation. Tandem mass spectra were searched against the UniProt Oryza sativa database concatenated with a reverse decoy database. By dataset search, 3,250 crotonylated PSMs and 751 noncrotonylated PSMs were identified. The raw data for all crotonylated peptides have been uploaded to the ProteomeXchange Consortium (dataset identifier PXD008716). Using a false discovery rate threshold <1% for peptides, we identified 1,265 crotonylation sites with high confidence in 690 rice proteins (supplemental Table 1). Also, we searched the data with K butyrylation as a variable modification, and no butylated peptide was identified. Moreover, out of the 690 Kcr protein substrates, ∼62.8%, 19.1%, 7.8%,, and 4.6% contained one, two, three, or four Kcr sites, respectively. There were five proteins with a high crotonylation intensity containing 11 or more Kcr sites (supplemental Fig. 1C).
To validate the crotonylated proteins identified by the MS data, five crotonylated proteins, including three non-histones and two histones, were randomly selected (Fig. 1D and supplemental Fig. 2). According to sequences of the five crotonylated proteins, we compounded the crotonylated peptides and performed dot-spot assay analysis. The results showed that the polyclonal rabbit anti-crotonyl lysine antibody reacted only with the crotonylated peptides but not with the corresponding unmodified peptides (Fig. 1E). This confirmed the specificity of the polyclonal rabbit anti-crotonyl lysine antibody and verified the credibility of the crotonylated proteins identified by the MS data.
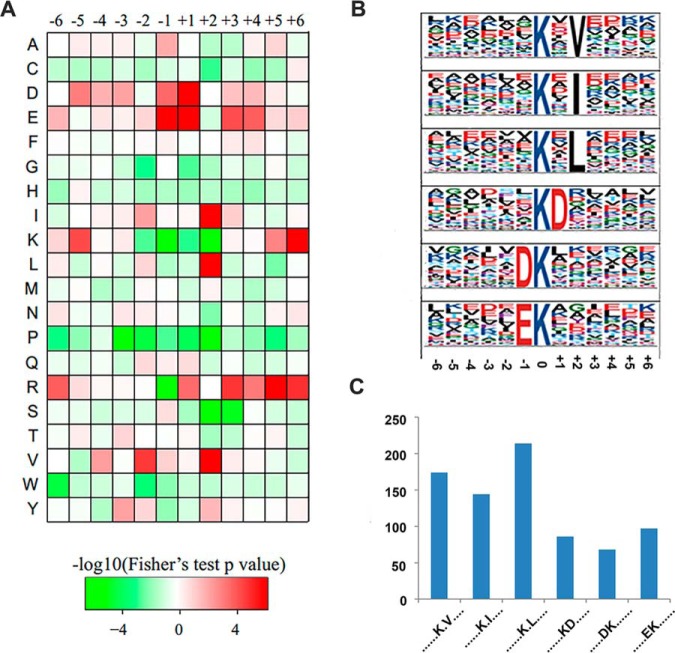
Identification of Kcr Sites Reveals Specific Motifs in Rice
To understand the properties of Kcr and to identify specific amino acids adjacent to Kcr sites, we examined the amino acid sequences flanking Kcr sites by generating a heat map (Fig. 2A). Substantial bias in amino acid distribution was observed from the −6 to +6 positions around Kcr sites in rice. Seven amino acid residues, aspartic acid (D), glutamine (E), isoleucine (I), lysine (K), leucine (L), arginine (R), and valine (V), were overrepresented in regions surrounding Kcr sites. To identify a possible consensus sequence motif around Kcr sites, we identified the sequence motifs in all of the identified Kcr sites using the Motif-X program. A total of six obviously conserved motifs (Fig. 2B), Kcr×V, Kcr×I, Kcr×L, Kcr×D, D×Kcr, and E×Kcr, were identified with different abundances (Fig. 2C), where Kcr and × indicate the crotonylated lysine and a random amino acid residue, respectively. These motifs were divided into two types according to the position of these residues around the Kcr, with one type containing a residue with aliphatic groups (V, I, or L) at the +2 position, while the second type contained a residue with acidic groups (D or E) at the −1 or +1 position.
Fig. 2.
Motif analysis of lysine crotonylation peptides. (A) Heat map of the amino acid compositions of the crotonylation sites showing the frequency of the different of amino acids around the Kcr; +1, and −1 represent the position around the Kcr. (B) Crotonylated peptide motifs and conservation of Kcr sites. The height of each letter corresponds to the frequency of that amino acid residue at that position. The 0 position K refers to the crotonylated sites. (C) Number of identified peptides containing Kcr in each motif.
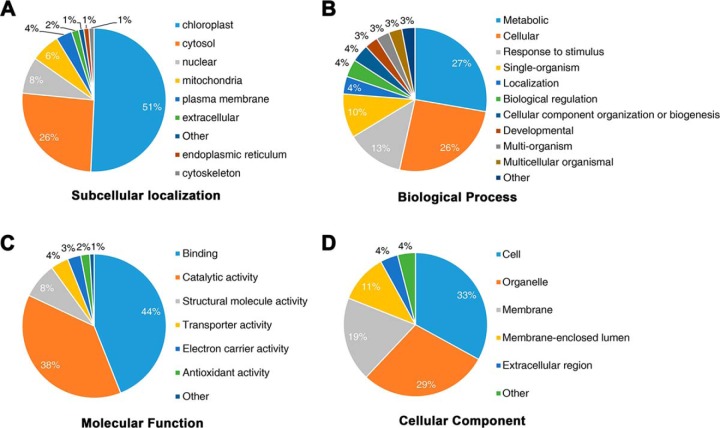
Gene Ontology Functional Annotation and Enrichment Analysis of the Crotonylated Proteins
In terms of subcellular location, most of the crotonylated proteins identified were predicted to be located in the chloroplast (51%), while few proteins were predicted to be associated with the cytoskeleton (1%), the endoplasmic reticulum (1%), or extracellularly located (2%) (Fig. 3A). The overall trend in subcellular location of the crotonylated proteins indicated the functional association with photosynthesis in rice seedlings. To further understand the potential roles of the lysine crotonylation in rice, we performed GO functional classification of all identified crotonylated proteins based on their biological processes, molecular functions and cellular components (Fig. 3). Among the 690 crotonylated proteins, we identified, 563 crotonyl-proteins were annotated for their biological processes (Fig. 3B), 564 for their molecular functions (Fig. 3C), and 568 for their cellular components (Fig. 3D), indicating that crotonylated proteins are involved in various processes with different biological functions. The GO annotations of Kcr sites in the biological processes, molecular function, and cellular components categories showed that Kcr modifications were significantly enriched in photosynthesis and its associated process (supplemental Fig. 3 and supplemental Table 2). According to the KEGG pathway analysis, the proteins involved in photosynthesis were also predominantly crotonylated (supplemental Fig. 3D). Therefore, crotonylation may be important for photosynthesis. WB analysis to verify this hypothesis revealed that the albino seedlings differed from the green seedlings derived from another culture of O. sativa variety Nipponbare (supplemental Fig. 4A). The results from WB showed a significantly lower lever of lysine crotonylation in albino seedlings (supplemental Fig. 4B). Overall, these findings indicated that photosynthesis in rice seedlings is strictly regulated by crotonylation.
Fig. 3.
Gene ontology functional classification of crotonylated proteins. Gene ontology functional classification of the identified crotonylated proteins based on (A) subcellular localization, (B) biological processes, (C) molecular function, and (D) cellular component.
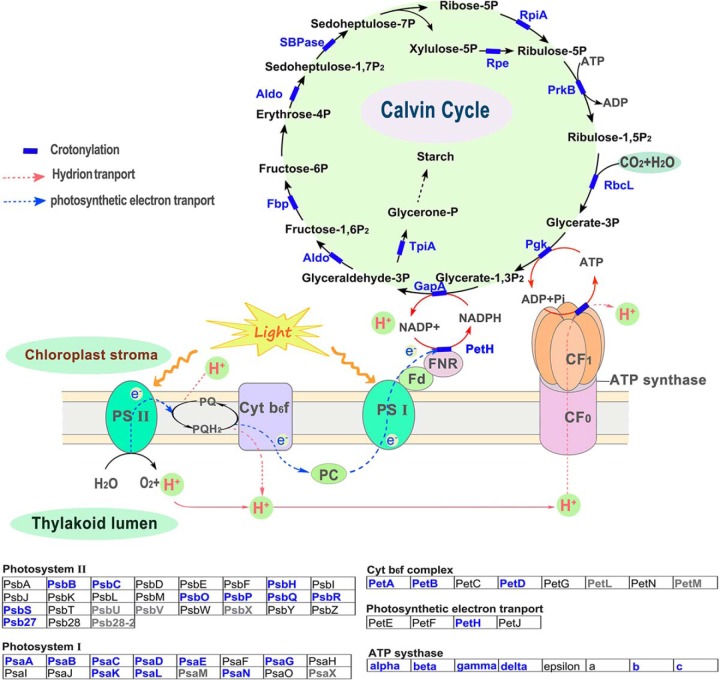
Crotonylation of Enzymes Involved in the Calvin Cycle and Photosynthesis
Photosynthesis is a very important biological process in plants. The numerous proteins and enzymes involved in the photosynthetic process are regulated by a variety of modifications, such as succinylation (50, 51) and acetylation (34, 52, 53). We mapped crotonylated proteins to components of the photosynthesis and carbon fixation pathways and identified the crotonylation of metabolic enzymes in C3 plants. Of all the enzymes involved in Calvin cycle and photosynthesis, a large proportion of metabolic enzymes were found to be crotonylated in rice (Fig. 4). Light-harvesting complex acts as a more efficient light-collecting unit than would be captured by the photosynthetic reaction center alone in higher plants. In rice, five subunits of light-harvesting complex (Lhca1/3/4 and Lhcb4/6) were identified to be crotonylated proteins by MS analysis (supplemental Table 1). Light-harvesting complex b-binding proteins form the major antenna protein complex of photosystem II in green plants, which transfers light energy to a chlorophyll A molecule at the reaction center of photosystems. The cytochrome b6f complex, which is located in the thylakoid membrane in chloroplasts, catalyzes the transfer of electrons between the two reaction complexes from photosystem II to photosystem I. LC-MS/MS analysis indicated that three subunits of the cytochrome b6f complex (Pet A/B/D), eight subunits of photosystem II (Psb B/C/O/P/Q/R/S/27), and nine subunits of photosystem I (Psa A/B/C/D/E/G/K/L/N) were crotonylated (Fig. 4 and supplemental Table 1). Based on these data, we deduced that lysine crotonylation may play a regulatory role in carbon metabolic pathways and photosynthetic organisms.
Fig. 4.
Crotonylated proteins in enriched in carbon assimilation and photosynthesis pathways in C3 plants. The identified crotonylated proteins are highlighted in blue.
Interactive Network Among Crotonylated Proteins in Rice
To further understand the regulatory role of crotonylation in photosynthesis, we analyzed PPI among the 690 crotonylated proteins identified using Cytoscape software (54). In the rice PPI network, 414 crotonylated proteins were identified as nodes, connected by 3,126 interactions identified using the STRING database (STRING database Version 10.0) (supplemental Table 3). The complete crotonylated PPI network for rice, which is shown in supplemental Fig. 5, is presented as the interactive network among crotonylated proteins in eukaryotic cells.
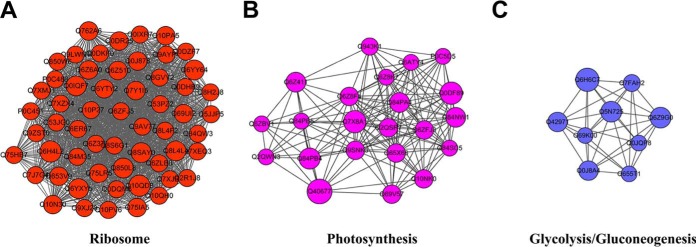
We retrieved 19 clusters of Kcr proteins from the complicated interaction network. The most enriched interaction cluster (Cluster I) was identified as ribosome-associated proteins, consisting of 57 ribosome-associated proteins with 88 Kcr sites (Fig. 5A). Cluster II consisted of 22 photosynthesis associated proteins with 67 Kcr sites (Fig. 5B), of which, 15 contained more than one Kcr site. Cluster III comprised proteins involved in glycolysis/gluconeogenesis (Fig. 5C). The PPI information suggested that lysine crotonylation in rice is involved in multiple biological processes, especially in ribosomes and photosynthesis.
Fig. 5.
Top three clusters of highly interconnected lysine-crotonylated protein networks. The network of lysine-crotonylated protein interactions (listed in protein ID names) was analyzed using the Cytoscape software (version 3.3.0). (A) Ribosome; (B) photosynthesis; (C) glycolysis/gluconeogenesis. The size of the balls represents the numbers of Kcr modifications in each figure.
Histone Kcr Sites in Rice Cells
Kcr is a conserved histone marker that has been reported in yeast and animals (26, 55). To identify histone Kcr sites in rice, we focused on crotonylated peptides derived from histones. In total, we detected seven histone Kcr sites consisting of one Kcr site in histone H2B (K32), four Kcr sites in histone H3 (K14/K56/K79/K122), and two Kcr sites in histone H4 (K31/K79) (supplemental Table 4), thus indicating that histone Kcr also exists in rice cells. Nevertheless, we found four histone Kcr sites (including H3K14/K79, H4K31/K79) in rice cells that have not yet been reported in humans, implying functional divergence of histone Kcr between plants and animals (Fig. 6). In addition, H3K14cr, one site of histone crotonylation identified by the MS data, was selected and identified in rice seedlings by immunofluorescence and WB analyses using its specific antibody (supplemental Fig. 6). The results further confirmed existence of histone Kcr in the rice genome.
Fig. 6.
The Kcr sites identified in human histones and rice histones (H3, H4 in this study). In humans, seven H3 Kcr PTM sites (H3K4/K9/K18/K23/K27/K56/K122) and three H4 Kcr PTM sites (H4K5/K8/K122) were identified. In rice, four H3 Kcr PTM sites (H3K14/K56/K79/K122) and two H4 Kcr PTM sites (H4K31/K79) were identified. The data for human Kcr sites were used from Suzuki et al. (55) and Tan et al. (26).
Genome-wide Mapping of Histone Kcr in Rice Cells
We next explored the in vivo function of histone Kcr in plants. We performed ChIP-seq analysis with the pan anti-Kcr antibody to determine the genomic distribution of histone Kcr in the rice seedling tissue consisting mainly of leaves with a small proportion of stem tissues. Two biological replicates of the ChIP-seq libraries were constructed and then sequenced using the HiSeq 2500 platform with the paired-end method. In total, ∼25 million read-pairs were obtained from the two replicate libraries (supplemental Table 5), ∼90% of which were mapped to the rice reference genome (Tigr 7). We found that 88% of enriched regions were shared between the replicates in the rice genome, indicating a high reproducibility of the ChIP-seq data experiments. The shared regions (10,923) were then subjected to further analysis as histone Kcr-enriched regions (supplemental Table 6).
Real-time quantitative PCR (qPCR) was conducted to verify the identified Kcr-enriched regions. We randomly selected 14 peak sites and 14 nonpeak sites to examine the difference in threshold cycles (ΔCt) between ChIP-DNA and Input-DNA. Of the peak sites, 13 out of 14 peak sites showed significant Kcr enrichment by showing more than one cycle for the ChIP-DNA samples compared with the Input-DNA, which was consistent with the ChIP-seq results (supplemental Table 7). Only two of the 14 nonpeak sites had a ΔCt of more than one cycle. Thus, the qPCR results confirmed the validity of the ChIP-seq data for use in further analysis.
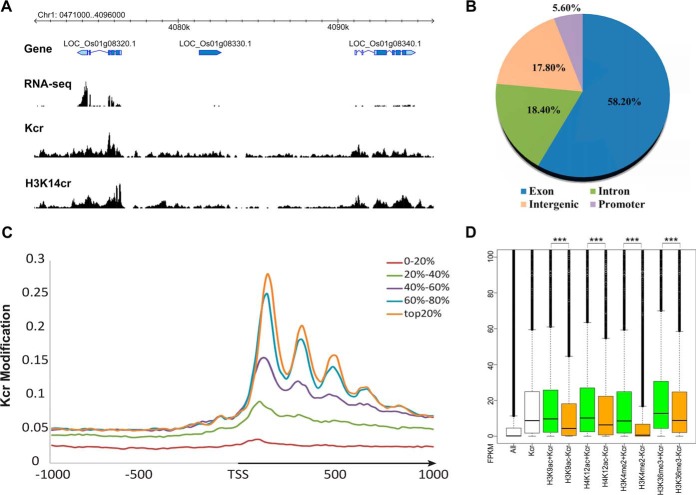
To further validate the whole genome histone Kcr data, we selected another antibody specific for H3K14cr to perform ChIP-seq analysis with two biological replicates (supplemental Table 5). With high reproducibility between the two replicates (87%), we retained 18,813 H3K14cr-enriched regions shared between the two replicates (supplemental Table 6). In total, 64.9% of Kcr-enriched regions (6,983 of 10,923, using the pan-antibody) overlapped with the H3K14cr regions (using the H3K14cr antibody). In addition, there was a clear pattern of histone Kcr co-localization with H3K14cr (Figs. 7A, 8A–8C), indicating the reliability of the ChIP data obtained using the histone Kcr pan-antibody.
Fig. 7.
The genomic distribution of histone Kcr-enriched regions. (A) Visualization of the Kcr seq locus on Chr1:407100–409600. (B) Genome-wide distribution of histone Kcr in the rice genome. The promoter was defined as the 1 kb sequence upstream of the gene transcription start site. (C) Distribution of Kcr density around differentially expressed genes. The Kcr modification was calculated by the number of reads per kilobase of the mapped genomic region. The arrow indicates the direction of transcription from the transcription start site. The rice genes were divided into five categories based the expression level from the top 20% to the bottom 20% based on published RNA-seq data from same rice seedlings at the same stage of development (49). (D) Expression comparisons of genes associated with different combinations of histone modifications. The nontranscription element gene expression values (FPKM) of each combination are indicated by box plots. All: all rice genes. Kcr: genes with Kcr modification. H3K9ac+Kcr: genes with both H3K9ac and Kcr. H3K9ac-Kcr: Genes with H3K9ac but not Kcr; the rest may be deduced by analogy. *** indicates significant differences between the two combinations (p < 2.2e-16, Kolmogorov-Smirnov test).
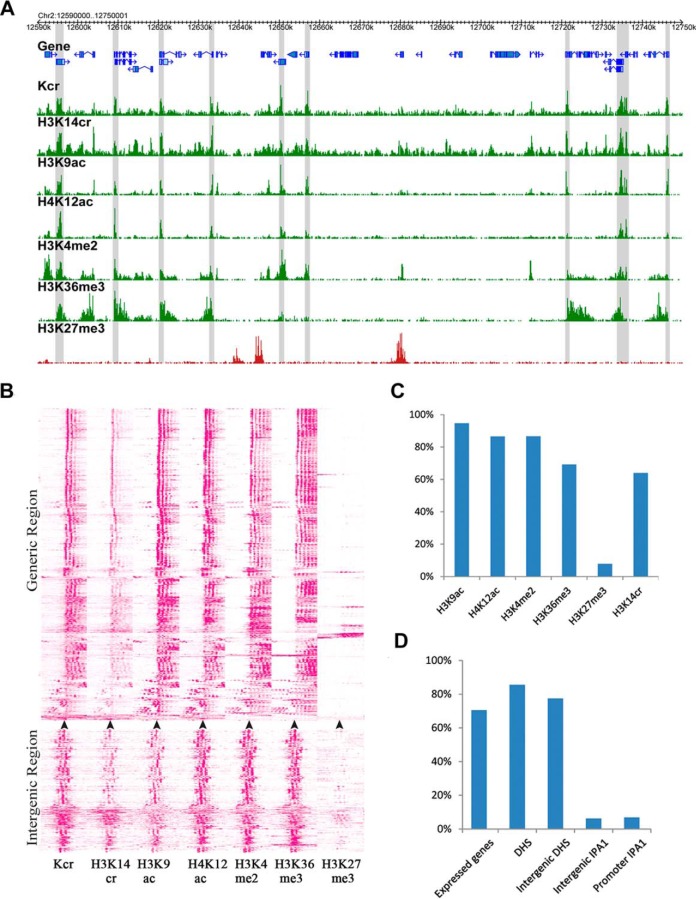
Fig. 8.
Association of histone Kcr and gene transcription in rice genome. (A) A representative region showing histone modifications (Kcr, H3K14cr, H3K9ac, H4K12ac, H3K4me2, H3K36me3, and H3K27me3) in the rice genome. The gray blocks indicate the Kcr-enriched genomic regions. Y indicates H3K14cr-enriched regions, while X indicates H3K14cr nonenriched regions in the gray block. (B) Heat map of histone Kcr and five histone modifications in generic regions and intergenic regions. The regions 1 kb upstream and downstream of the transcription start site (indicated by an arrow, for the generic regions) and the middle of Kcr regions (for the intergenic regions) were clustered based on density of corresponding histone modifications. Density was calculated by the number of reads per kilobase region per million mapped reads. (C) The percentage of histone Kcr-enriched regions overlapping with regions enriched with other histone modifications (H3K4me3, H3K4me2, H3K9ac, H3K36me3, H3K27me3, and H3K14cr). (D) The percentage of histone Kcr-enriched regions overlapping with regions enriched with functional regions (DNase I hypersensitive sites (DHSs), and IPA1-binding regions). The promoter was defined as the 1 kb sequence upstream of the transcriptional start site and the intergenic region indicates the nongeneric and nonpromoter regions.
Histone Kcr is Positively Correlated with Gene Expression in Rice
To elucidate the biological function of histone Kcr in rice, the genomic distribution of histone Kcr-enriched regions was determined. Histone Kcr sites were found mainly in expressed genes (Figs. 7A and 8D). The rice genome was characterized mainly as promoters (1-kb regions upstream of the gene transcription start site), intergenic regions, and genic regions, comprising exons and introns. We found that the majority (86.7%) of the histone Kcr regions overlapped with the genic regions, with 76.6% of peak summits of Kcr regions contained within the exon and intron regions. Strikingly, 58.2% of peak summits of the Kcr regions were located in the exon regions, while only 18.4% were found in the intron regions (Fig. 7B). The majority of Kcr regions in exons were located in the coding sequence (45.2% of 10,923 Kcr regions) and the 5′ untranslated region (UTR) (20.7% of 10,923 Kcr regions), while only 1.5% were located in the 3′UTR. Unlike humans, in which the histone Kcr peaks tend to be associated with intergenic regions and promoters (26), only 17.8% and 5.6% of the histone Kcr peaks in the rice genome were located in intergenic regions and promoters, respectively. Similarly, the majority (62.7%) of H3K14cr-antibody-specific regions were contained within the exonic (45.6%) and intronic (17.1%) regions, while 28.3 and 9% of the H3K14cr peaks were located in intergenic regions and promoters, respectively (supplemental Fig. 7A). Overall, the histone Kcr density peaked at +1 nucleosome behind the transcription start sites (TSS) and declined toward the end of the gene (Fig. 7C). In addition, we found a high correlation between gene expression and histone Kcr in genic regions, with generally higher expression levels associated with higher histone Kcr density (Fig. 7C), as well as H3K14cr (supplemental Fig. 7B). Interestingly, histone Kcr facilitated expression of genes with existing active histone modifications such as H3K9ac, H4K12ac, H3K4me2, and H3K36me3 (Fig. 7D and supplemental Fig. 8), suggesting an important biological role of histone Kcr in transcriptional regulation.
Co-occupancy Among Histone Kcr, Other Histone Modifications, and Regulatory Regions
In the rice genome, histone Kac and Kme1/2/3 play important roles in regulating gene transcription (48, 49, 56). To explore the relationship between histone Kcr and other histone modifications, we investigated co-occupancy between histone Kcr and the data for five other published histone modifications (H3K9ac, H4K12ac, H3K4me2, H3K36me3, and H3K27me3) (48, 49). With the single nucleosome resolution of the histone modification data, we found that histone Kcr tended to co-locate with histone modifications associated with active genes, such as H3K9ac, H3K12ac, H3K4me2, and H3K36me3 (Figs. 8A and 8B).
Accordingly, 95%, 87%, 87%, and 69% of histone Kcr regions overlapped with the active histone modifications H3K9ac, H3K12ac, H3K4me2, and H3K36me3, respectively (Fig. 8C). As expected, histone Kcr was not co-located with H3K27me3, which is a repressive marker of gene expression (Figs. 8A and 8B). Only 8% of histone Kcr regions overlapped with H3K27me3 regions (Fig. 8C). These results indicated a tendency for co-occupancy among Kcr and histone modifications related to active genes.
In human genomes, 68% of histone Kcr markers are active promoters and enhancers (26). Although we found a clear tendency of histone Kcr to be located in genic regions in rice, we focused on intergenic regions (17.8%) associated with histone Kcr to determine the potential of these regions as markers of enhancer functions. We explored the histone Kcr frequency in the binding regions of the transcription activator ideal plant architecture1 (IPA1), as a representative of a category of active enhancers in rice seedlings (57). Previous studies have indicated that 88% of IPA1-binding regions show DNase I hypersensitivity sites (DHSs), which mark cis-regulatory DNA elements (58). Interestingly, we found 77% of histone Kcr regions overlapped with DHSs in intergenic regions of the rice genome, while only 6% overlapped with IPA1-binding regions (Fig. 8D). These results showed that histone Kcr in intergenic regions of rice genome mark cis-regulatory DNA elements, which are not contained within the IPA1-binding regions.
Enriched Biological Functions of Genes Associated with Histone Kcr
As mentioned previously, we found that histone Kcr co-located with the four histone modifications associated with active genes. In particular, there was a 95% overlap in Kcr and H3K9ac regions. However, there were only about 10,000 histone Kcr regions (∼14.67 Mb), which is far fewer than the 26,693 H3K9ac regions (∼26.2Mb) in the rice genome. Based on these observations, we hypothesized that histone Kcr marks genes with specific functions in the rice genome. To test this hypothesis, we conducted GO analysis to identify the biological functions of genes with significantly Kcr-enriched regions. Our analysis indicated that the genes with histone Kcr were associated with macromolecule metabolism or biosynthesis processes, particularly translation (supplemental Table 8). Accordingly, we found that these genes trended to be expressed in the nucleolus, Golgi apparatus, and plant lumen. These data indicate that histone Kcr mediates epigenetic regulation of the expression of genes involved in protein synthesis.
DISCUSSION
Lysine Crotonylation Exists Widely in Dicots and Monocots
To date, non-histone Kcr has been identified in humans and tobacco (28, 30, 31), whereas this modification in the proteome is rarely reported in monocots. In this study, using Kcr peptide enrichment coupled with high accuracy LC-MS/MS, we obtained extensive data on the lysine crotonylome in rice seedlings. The present study identified a total of 1,265 Kcr sites from 690 proteins that belong to diverse functional groups and are localized in multiple cellular compartments. These data suggest that lysine crotonylation plays an important role in regulating numerous cellular processes in monocots. In dicots, 2,044 Kcr sites on 637 crotonylated proteins were identified in tobacco (31), with the majority of proteins containing two or more Kcr sites; however, 565 (81.9%) proteins identified in our study contained only one or two crotonylation sites.
In general, the specific amino acids adjacent to lysine acylation sites are highly conserved. Comparative analysis between tobacco and rice revealed that three conserved motifs identified (KD, DK, and EK) were consistent, which also exist in the crotonylated proteins of mammals (31). In addition, the major categories of biological processes and molecular functions in the GO functional classification of all rice Kcr proteins were similar to those of tobacco. In terms of subcellular localization, 51% of Kcr proteins in rice were localized to the chloroplast, which is higher than the proportion in tobacco. The KEGG pathway enrichment analysis revealed that the Kcr proteins in rice were significantly enriched in processes associated with photosynthesis, which is similar to tobacco. The rice PPI networks, including ribosome, oxidative phosphorylation, and proteasome, were also found in tobacco. However, the PPI networks in rice which are involved in photosynthesis and glycolysis were not previously found in tobacco (28). These results suggested that the major function of non-histone lysine crotonylation is evolutionarily conserved in plants, while the functions of some Kcr proteins are specific to rice.
Lysine Crotonylation Plays an Important Role in the Regulation of Cellular Physiology, Which is Similar to Lysine Acetylation
Protein lysine acetylation is another major PTM that is involved in the regulation of many metabolic pathways (59). Recently, numerous acetylation modifications of non-histone proteins have been identified in different plants, such as soybean (60), grape (61), Arabidopsis (62, 63), rice (34, 64), and wheat (53). Interestingly, in GO analysis of the rice acetylome, metabolic process-associated proteins, proteins with binding activity, and cellular process-associated proteins have been found to represent the largest group of biological process, molecular function, and cellular components, respectively (34, 64). The other important categories of biological processes are cellular process proteins, responses to stimuli, and single-organism processes. Proteins with catalytic activity are the second largest group of molecules (34, 64). The organelles, the membrane, and the macromolecular cellular components also accounted for a large proportion of the proteins. These results in the present study are strikingly similar to those previous findings. GO analysis of the rice crotonylome demonstrated the location of crotonylated proteins in diverse cellular compartments, with different molecular functions, and the involvement of various processes. These phenomena suggest that crotonylation and acetylation have some common connections in rice.
Subcellular localization analysis revealed that most acetylated proteins in rice are located in the chloroplast (34, 64), while in wheat, most are located in the cytosol (41%) and the chloroplast (36%) (53). Our data revealed that 51% of crotonylated proteins were localized in the chloroplast in rice, suggesting that these proteins are involved in photosynthesis. GO enrichment analysis of the crotonylation data showed that the biological process and molecular functions of the crotonylated proteins were significantly related to chloroplasts and photosynthesis, respectively. Many cellular components associated with photosynthesis also showed an increased tendency to be crotonylated. In accordance with these observations, the photosynthesis-associated proteins identified in the KEGG pathway analysis were also enriched. All these findings indicate that lysine crotonylation plays a key role in the regulation of photosynthesis. The PPI information suggests that rice lysine crotonylation is involved in multiple biological processes, especially in association with the ribosome and photosynthesis. Interestingly, the network of lysine acetylation also contains a subnetwork involved in photosynthesis (52). In this study, 28 crotonylated proteins were identified among the 63 proteins involved in photosynthesis (photosystem I subunits, photosystem II subunits, photosynthetic electron transport, cyt b6f complex, and ATP synthase). Compared with previous studies (64), in which 21 acetylated proteins were identified, 16 overlapping proteins were both crotonylated and acetylated. Therefore, we speculate that Kcr and Kac may play important roles in photosynthesis.
Differentiation of Histone Kcr in Mammalian and Rice Cells
In humans, seven H3 Kcr PTM sites (H3K4/K9/K18/K23/K27/K56/K122) and three H4 Kcr PTM sites (H4K5/K8/H3K12) have been identified (26, 55). Four histone Kcr sites (H3K14, H3K79, H4K31, and H4K79) identified in rice cells in this study have not been reported in humans, and only two (H3K56/K122) are similar to the human sites, indicating the existence of functional divergence in histone Kcr between plants and animals. Furthermore, genome-wide mapping of histone Kcr modification in mammalian genomes by ChIP-seq analyses revealed histone Kcr enrichment of active gene promoters and potential enhancer regions. In promoters, the strongest enrichment of histone Kcr was identified in regions flanking the transcription start sites (26, 27). Nevertheless, a genome-wide examination of histone Kcr in rice seedlings performed by ChIP-seq in this study showed that the histone Kcr modification was located predominantly within genic regions of nontranscription element genes and enriched around gene bodies, especially in exons; however, this was not the case in humans. Furthermore, Kcr is a specific marker of active sex-chromosome-linked genes in postmeiotic male germ cells in mammals (26, 29). In this study, histone Kcr was distributed evenly on the 12 chromosomes of rice without any obvious chromosome preferences (supplemental Fig. 9).
DHSs are markers of regulatory DNA and span all classes of cis-regulatory elements, including promoters, enhancers, and insulators (49, 58, 65). In the present study, we found 77% of histone Kcr regions overlapped with DHSs in intergenic regions of the rice genome, implying that histone Kcr is associated with cis-regulatory DNA elements in the rice genome. Nevertheless, we found that only 6% of histone Kcr regions overlapped with enhancer regions bound by transcription activator IPA1, which represents another difference in the characteristics of histone Kcr between mammals and rice (26). Although we did not investigate other types of enhancer, these results indicate that there is a possibility that histone Kcr does not mark enhancer regions in the rice genome.
Histone Kcr May Play an Active Role in Regulating Gene Transcription in the Rice Genome
The genome-wide examination of histone Kcr in rice (O. sativa L. japonica) seedlings in the present study revealed that the histone Kcr appeared mainly in genic regions, especially in exons. Integration with public H3K9ac, H4K12ac, H3K4me2, K3K36me3, and H3K27me3 data (48, 49) revealed that the distribution patterns of histone Kcr are similar to the histone modifications associated with active genes, implying the histone Kcr shares similar genome-wide distributions with H4K12ac, H3K4me2, K3K36me3, H3K27me3, and especially with H3K9ac. This global characterization of histone Kcr will improve our understanding of epigenetic regulation in plants and enrich our knowledge of the types of plant histone modifications.
The identification and characterization of the enzyme system that catalyzes covalent modification in specific target lysine residues is key to understand how histone modifications are regulated. The p300 protein is a histone acetyltransferase (66) catalyzing histone crotonylation in humans as well. For specific target residues, p300-catalyzed histone Kcr directly stimulates transcription to a greater degree than histone Kac in humans (23). In this study, we show that histone Kcr tends to co-localize with several histone modifications, including Kac and Kme, which raises many interesting questions. Based on the high degree of p300 conservation, we identified three homologous genes by phylogenetic trees analysis of the rice genome (supplemental Fig. 10). However, whether p300 catalyzes histone Kcr in rice and, if so, how acetylation and crotonylation is regulated in rice remain to be elucidated. Additionally, the role of histone Kcr in the regulation of histone structure and function in rice requires further investigation.
DATA AVAILABILITY
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD010376.
Supplementary Material
Acknowledgments
We thank Jiayin Pang from the University of Western Australia for the manuscript copyedited and check the grammar errors carefully. This work was supported by the National Natural Science Foundation of China [grant numbers 31571266, 31871232], the Jiangsu Provincial Natural Science Foundation [grant number BK20151309], Postgraduate Education Reform Project of Jiangsu Province [grant number KYZZ16_0493], and the Fund of Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Footnotes
 This article contains supplemental material Tables 1–8 and Figs. 1–10. The authors declare that they have no conflicts of interest.
This article contains supplemental material Tables 1–8 and Figs. 1–10. The authors declare that they have no conflicts of interest.
1 The abbreviations used are:
- Kcr
- lysine crotonylation
- PTM
- posttranslational modification
- nano-LC-MS/MS
- nanoscale liquid chromatography coupled to tandem mass spectrometry
- ChIP
- chromatin immunoprecipitation
- DHSs
- DNase hypersensitive sites
- IPA1
- ideal plant architecture1
- Kac
- lysine acetylation
- Kme
- lysine methylation
- Kma
- lysine malonylation
- Kpro
- lysine propionylation
- Kbu
- lysine butyrylation
- Ksucc
- lysine succinylation
- WB
- Western blotting
- GO
- gene ontology
- MACS
- model-based analysis of ChIP-Seq
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- PPIs
- protein-protein interaction
- qPCR
- quantitative polymerase chain reaction.
REFERENCES
- 1. Deribe Y. L., Pawson T., and Dikic I. (2010) Post-translational modifications in signal integration. Nat. Struct. Mol. Biol. 17, 666–672 [DOI] [PubMed] [Google Scholar]
- 2. Zhao Y., and Jensen O. N. (2009) Modification-specific proteomics: Strategies for characterization of post-translational modifications using enrichment techniques. Proteomics 9, 4632–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Olsen J. V., and Mann M. (2013) Status of large-scale analysis of post-translational modifications by mass spectrometry. Mol. Cell. Proteomics 12, 3444–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Azevedo C., and Saiardi A. (2016) Why always lysine? The ongoing tale of one of the most modified amino acids. Advances Biolog. Reg. 60, 144–150 [DOI] [PubMed] [Google Scholar]
- 5. Zhao S., Xu W., Jiang W., Yu W., Lin Y., Zhang T., Yao J., Zhou L., Zeng Y., Li H., Li Y., Shi J., An W., Hancock S. M., He F., Qin L., Chin J., Yang P., Chen X., Lei Q., Xiong Y., and Guan K. L. (2010) Regulation of cellular metabolism by protein lysine acetylation. Science 327, 1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tropberger P., Pott S., Keller C., Kamieniarz-Gdula K., Caron M., Richter F., Li G. H., Mittler G., Liu E. T., Bühler M., Margueron R., and Schneider R. (2013) Regulation of transcription through acetylation of H3K122 on the lateral surface of the histone octamer. Cell 152, 859–872 [DOI] [PubMed] [Google Scholar]
- 7. Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R. S., and Zhang Y. (2002) Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science 298, 1039–1043 [DOI] [PubMed] [Google Scholar]
- 8. Peach S. E., Rudomin E. L., Udeshi N. D., Carr S. A., and Jaffe J. D. (2012) Quantitative assessment of chromatin immunoprecipitation grade antibodies directed against histone modifications reveals patterns of co-occurring marks on histone protein molecules. Mol. Cell. Proteomics 11, 128–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Peng C., Lu Z. K., Xie Z. Y., Cheng Z. Y., Chen Y., Tan M. J., Luo H., Zhang Y., He W., Yang K., Zwaans B. M. M., Tishkoff D., Ho L., Lombard D., He T. C., Dai J. B., Verdin E., Ye Y., and Zhao Y. M. (2011) The first identification of lysine malonylation substrates and its regulatory enzyme. Mol. Cell. Proteomics 10, M111.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xie Z., Dai J., Dai L., Tan M., Cheng Z., Wu Y., Boeke J. D., and Zhao Y. (2012) Lysine succinylation and lysine malonylation in histones. Mol. Cell Proteomics 11, 100–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheng Z., Tang Y., Chen Y., Kim S., Liu H., Li S., Gu W., and Zhao Y. (2009) Molecular characterization of propionyllysines in non-histone proteins. Mol. Cell. Proteomics 8, 45–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang K., Chen Y., Zhang Z., and Zhao Y. (2009) Identification and verification of lysine propionylation and butyrylation in yeast core histones using PTMap software. J. Proteome Res. 8, 900–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen Y., Sprung R., Tang Y., Ball H., Sangras B., Kim S. C., Falck J. R., Peng J., Gu W., and Zhao Y. (2007) Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell. Proteomics 6, 812–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang Z., Tan M., Xie Z., Dai L., Chen Y., and Zhao Y. (2011) Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol. 7, 58–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim S. C., Sprung R., Chen Y., Xu Y., Ball H., Pei J., Cheng T., Kho Y., Xiao H., Xiao L., Grishin N. V., White M., Yang X. J., and Zhao Y. (2006) Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 23, 607–618 [DOI] [PubMed] [Google Scholar]
- 16. Jeffers V., and Sullivan W. J. Jr. (2012) Lysine acetylation is widespread on proteins of diverse function and localization in the protozoan parasite Toxoplasma gondii. Eukaryot. Cell 11, 735–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nallamilli B. R., Edelmann M. J., Zhong X., Tan F., Mujahid H., Zhang J., Nanduri B., and Peng Z. (2014) Global analysis of lysine acetylation suggests the involvement of protein acetylation in diverse biological processes in rice (Oryza sativa). PLOS One 9, e89283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fang X., Chen W., Zhao Y., Ruan S., Zhang H., Yan C., Jin L., Cao L., Zhu J., Ma H., and Cheng Z. (2015) Global analysis of lysine acetylation in strawberry leaves. Front. Plant Sci. 6, 739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li X., Hu X., Wan Y., Xie G., Li X., Chen D., Cheng Z., Yi X., Liang S., and Tan F. (2014) Systematic identification of the lysine succinylation in the protozoan parasite Toxoplasma gondii. J. Proteome Res. 13, 6087–6095 [DOI] [PubMed] [Google Scholar]
- 20. Cao X. J., and Garcia B. A. (2016) Global proteomics analysis of protein lysine methylation. Curr. Protoc. Protein Sci. 86, 24.8.1–24.8.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Perillo B., Ombra M. N., Bertoni A., Cuozzo C., Sacchetti S., Sasso A., Chiariotti L., Malorni A., Abbondanza C., and Avvedimento E. V. (2008) DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science 319, 202–206 [DOI] [PubMed] [Google Scholar]
- 22. Bannister A. J., and Kouzarides T. (2011) Regulation of chromatin by histone modifications. Cell Res. 21, 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sabari B. R., Tang Z., Huang H., Yong-Gonzalez V., Molina H., Kong H. E., Dai L., Shimada M., Cross J. R., Zhao Y., Roeder R. G., and Allis C. D. (2015) Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol. Cell 58, 203–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Heintzman N. D., Stuart R. K., Hon G., Fu Y., Ching C. W., Hawkins R. D., Barrera L. O., Van Calcar S., Qu C., Ching K. A., Wang W., Weng Z., Green R. D., Crawford G. E., and Ren B. (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311–318 [DOI] [PubMed] [Google Scholar]
- 25. Zhou V. W., Goren A., and Bernstein B. E. (2011) Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genet. 12, 7–18 [DOI] [PubMed] [Google Scholar]
- 26. Tan M., Luo H., Lee S., Jin F., Yang J. S., Montellier E., Buchou T., Cheng Z., Rousseaux S., Rajagopal N., Lu Z., Ye Z., Khochbin S., Ren B., and Zhao Y. (2011) Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146, 1016–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bao X. C., Wang Y., Li X., Li X. M., Liu Z., Yang T. P., Wong C. F., Zhang J. W., Hao Q., and Li X. D. (2014) Identification of 'erasers' for lysine crotonylated histone marks using a chemical proteomics approach. Elife 3, e02999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sun H. J., Liu X. W., Li F. F., Li W., Zhang J., Xiao Z. X., Shen L. L., Li Y., Wang F. L., and Yang J. G. (2017) First comprehensive proteome analysis of lysine crotonylation in seedling leaves of Nicotiana tabacum. Sci. Rep. 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Montellier E., Rousseaux S., Zhao Y., and Khochbin S. (2012) Histone crotonylation specifically marks the haploid male germ cell gene expression program. Bioessays 34, 187–193 [DOI] [PubMed] [Google Scholar]
- 30. Wei W., Mao A., Tang B., Zeng Q., Gao S., Liu X., Lu L., Li W., Du J. X., Li J., Wong J., and Liao L. (2017) Large-scale identification of protein crotonylation reveals its role in multiple cellular functions. J. Proteome Res. 16, 1743–1752 [DOI] [PubMed] [Google Scholar]
- 31. Xu W., Wan J., Zhan J., Li X., He H., Shi Z., and Zhang H. (2017) Global profiling of crotonylation on non-histone proteins. Cell Res. 27, 946–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shi J., Dong A., and Shen W. H. (2014) Epigenetic regulation of rice flowering and reproduction. Front. Plant Sci. 5, 803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Goff S. A., Ricke D., Lan T. H., Presting G., Wang R., Dunn M., Glazebrook J., Sessions A., Oeller P., Varma H., Hadley D., Hutchinson D., Martin C., Katagiri F., Lange B. M., Moughamer T., Xia Y., Budworth P., Zhong J., Miguel T., Paszkowski U., Zhang S., Colbert M., Sun W. L., Chen L., Cooper B., Park S., Wood T. C., Mao L., Quail P., Wing R., Dean R., Yu Y., Zharkikh A., Shen R., Sahasrabudhe S., Thomas A., Cannings R., Gutin A., Pruss D., Reid J., Tavtigian S., Mitchell J., Eldredge G., Scholl T., Miller R. M., Bhatnagar S., Adey N., Rubano T., Tusneem N., Robinson R., Feldhaus J., Macalma T., Oliphant A., and Briggs S. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp japonica). Science 296, 92–100 [DOI] [PubMed] [Google Scholar]
- 34. Xue C., Liu S., Chen C., Zhu J., Yang X. B., Zhou Y., Guo R., Liu X. Y., and Gong Z. Y. (2018) Global proteome analysis links lysine acetylation to diverse functions in Oryza sativa. Proteomics 18, 1700036. [DOI] [PubMed] [Google Scholar]
- 35. Zhou S., Yang Q., Yin C., Liu L., and Liang W. (2016) Systematic analysis of the lysine acetylome in Fusarium graminearum. BMC Genomics 17, 1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. He D., Wang Q., Li M., Damaris R. N., Yi X., Cheng Z., and Yang P. (2016) Global proteome analyses of lysine acetylation and succinylation reveal the widespread involvement of both modification in metabolism in the embryo of germinating rice seed. J. Proteome Res. 15, 879–890 [DOI] [PubMed] [Google Scholar]
- 37. Tamura K., Stecher G., Peterson D., Filipski A., and Kumar S. (2013) MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 30, 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Keller O., Kollmar M., Stanke M., and Waack S. (2011) A novel hybrid gene prediction method employing protein multiple sequence alignments. Bioinformatics 27, 757–763 [DOI] [PubMed] [Google Scholar]
- 39. Shen Z., Wang B., Luo J., Jiang K., Zhang H., Mustonen H., Puolakkainen P., Zhu J., Ye Y., and Wang S. (2016) Global-scale profiling of differential expressed lysine acetylated proteins in colorectal cancer tumors and paired liver metastases. J. Proteomics 142, 24–32 [DOI] [PubMed] [Google Scholar]
- 40. von Mering C., Huynen M., Jaeggi D., Schmidt S., Bork P., and Snel B. (2003) STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 31, 258–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nie Z., Zhou F., Li D., Lv Z., Chen J., Liu Y., Shu J., Sheng Q., Yu W., Zhang W., Jiang C., Yao Y., Yao J., Jin Y., and Zhang Y. (2013) RIP-seq of BmAgo2-associated small RNAs reveal various types of small non-coding RNAs in the silkworm, Bombyx mori. BMC Genomics 14, 661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gong Z., Yu H., Huang J., Yi C., and Gu M. (2009) Unstable transmission of rice chromosomes without functional centromeric repeats in asexual propagation. Chromosome Res. 17, 863–872 [DOI] [PubMed] [Google Scholar]
- 43. Nagaki K., Talbert P. B., Zhong C. X., Dawe R. K., Henikoff S., and Jiang J. M. (2003) Chromatin immunoprecipitation reveals that the 180-bp satellite repeat is the key functional DNA element of Arabidopsis thaliana centromeres. Genetics 163, 1221–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kawahara Y., de la Bastide M., Hamilton J. P., Kanamori H., McCombie W. R., Ouyang S., Schwartz D. C., Tanaka T., Wu J., Zhou S., Childs K. L., Davidson R. M., Lin H., Quesada-Ocampo L., Vaillancourt B., Sakai H., Lee S. S., Kim J., Numa H., Itoh T., Buell C. R., and Matsumoto T. (2013) Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Langmead B., Trapnell C., Pop M., and Salzberg S. L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., and Liu X. S. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mukhopadhyay A., Deplancke B., Walhout A. J., and Tissenbaum H. A. (2008) Chromatin immunoprecipitation (ChIP) coupled to detection by quantitative real-time PCR to study transcription factor binding to DNA in Caenorhabditis elegans. Nat. Protoc. 3, 698–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. He G., Zhu X., Elling A. A., Chen L., Wang X., Guo L., Liang M., He H., Zhang H., Chen F., Qi Y., Chen R., and Deng X. W. (2010) Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell 22, 17–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang W., Wu Y., Schnable J. C., Zeng Z., Freeling M., Crawford G. E., and Jiang J. (2012) High-resolution mapping of open chromatin in the rice genome. Genome Res. 22, 151–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jin W., and Wu F. (2016) Proteome-wide identification of lysine succinylation in the proteins of tomato (Solanum lycopersicum). PLOS One 11, e0147586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhen S. M., Deng X., Wang J., Zhu G. R., Cao H., Yuan L. L., and Yan Y. M. (2016) First comprehensive proteome analyses of lysine acetylation and succinylation in seedling leaves of Brachypodium distachyon L. Sci. Rep. 6, 31576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mo R., Yang M., Chen Z., Cheng Z., Yi X., Li C., He C., Xiong Q., Chen H., Wang Q., and Ge F. (2015) Acetylome analysis reveals the involvement of lysine acetylation in photosynthesis and carbon metabolism in the model Cyanobacterium synechocystis sp PCC 6803. J. Proteome Res. 14, 1275–1286 [DOI] [PubMed] [Google Scholar]
- 53. Zhang Y., Song L., Liang W., Mu P., Wang S., and Lin Q. (2016) Comprehensive profiling of lysine acetylproteome analysis reveals diverse functions of lysine acetylation in common wheat. Sci. Rep. 6, 21069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T. (2003) Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Suzuki Y., Horikoshi N., Kato D., and Kurumizaka H. (2016) Crystal structure of the nucleosome containing histone H3 with crotonylated lysine 122. Biochem. Bioph. Res. Co. 469, 483–489 [DOI] [PubMed] [Google Scholar]
- 56. Du Z., Li H., Wei Q., Zhao X., Wang C., Zhu Q., Yi X., Xu W., Liu X. S., Jin W., and Su Z. (2013) Genome-wide analysis of histone modifications: H3K4me2, H3K4me3, H3K9ac, and H3K27ac in Oryza sativa L. japonica. Mol. Plant 6, 1463–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lu Z., Yu H., Xiong G., Wang J., Jiao Y., Liu G., Jing Y., Meng X., Hu X., Qian Q., Fu X., Wang Y., and Li J. (2013) Genome-wide binding analysis of the transcription activator Ideal Plant Architecture1 reveals a complex network regulating rice plant architecture. Plant Cell 25, 3743–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wu Y., Zhang W., and Jiang J. (2014) Genome-wide nucleosome positioning is orchestrated by genomic regions associated with DNase I hypersensitivity in rice. PLOS Genet. 10, e1004378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liu Z., Wang Y., Gao T., Pan Z., Cheng H., Yang Q., Cheng Z., Guo A., Ren J., and Xue Y. (2014) CPLM: A database of protein lysine modifications. Nucleic Acids Res. 42, D531–D536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Smith-Hammond C. L., Swatek K. N., Johnston M. L., Thelen J. J., and Miernyk J. A. (2014) Initial description of the developing soybean seed protein Lys-N-epsilon -acetylome. J. Proteomics 96, 56–66 [DOI] [PubMed] [Google Scholar]
- 61. Melo-Braga M. N., Verano-Braga T., León I. R., Antonacci D., Nogueira F. C., Thelen J. J., Larsen M. R., and Palmisano G. (2012) Modulation of protein phosphorylation, N-glycosylation and lys-acetylation in grape (Vitis vinifera) mesocarp and exocarp owing to Lobesia botrana infection. Mol. Cell Proteomics 11, 945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Finkemeier I., Laxa M., Miguet L., Howden A. J., and Sweetlove L. J. (2011) Proteins of diverse function and subcellular location are lysine acetylated in Arabidopsis. Plant Physiol. 155, 1779–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wu X., Oh M. H., Schwarz E. M., Larue C. T., Sivaguru M., Imai B. S., Yau P. M., Ort D. R., and Huber S. C. (2011) Lysine acetylation is a widespread protein modification for diverse proteins in Arabidopsis. Plant Physiol. 155, 1769–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Xiong Y., Peng X., Cheng Z., Liu W., and Wang G. L. (2016) A comprehensive catalog of the lysine-acetylation targets in rice (Oryza sativa) based on proteomic analyses. J. Proteomics 138, 20–29 [DOI] [PubMed] [Google Scholar]
- 65. Thurman R. E., Rynes E., Humbert R., Vierstra J., Maurano M. T., Haugen E., Sheffield N. C., Stergachis A. B., Wang H., Vernot B., Garg K., John S., Sandstrom R., Bates D., Boatman L., Canfield T. K., Diegel M., Dunn D., Ebersol A. K., Frum T., Giste E., Johnson A. K., Johnson E. M., Kutyavin T., Lajoie B., Lee B. K., Lee K., London D., Lotakis D., Neph S., Neri F., Nguyen E. D., Qu H., Reynolds A. P., Roach V., Safi A., Sanchez M. E., Sanyal A., Shafer A., Simon J. M., Song L., Vong S., Weaver M., Yan Y., Zhang Z., Zhang Z., Lenhard B., Tewari M., Dorschner M. O., Hansen R. S., Navas P. A., Stamatoyannopoulos G., Iyer V. R., Lieb J. D., Sunyaev S. R., Akey J. M., Sabo P. J., Kaul R., Furey T. S., Dekker J., Crawford G. E., and Stamatoyannopoulos J. A. (2012) The accessible chromatin landscape of the human genome. Nature. 7414, 75–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kundu T. K., Palhan V. B., Wang Z., An W., Cole P. A., and Roeder R. G. (2000) Activator-dependent transcription from chromatin in vitro involving targeted histone acetylation by p300. Mol. Cell 6, 551–561 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD010376.