Abstract
We examined how polysorbate 20 (PS20; Tween 20) and polysorbate 80 (PS80; Tween 80) affect the higher order structure of a monoclonal antibody (mAb) and its Fab and Fc fragments, using near-UV circular dichroism and 2D NMR. Both polysorbates bind to the mAb with sub-millimolar affinity. Binding causes significant changes in the tertiary structure of mAb with no changes in its secondary structure. 2D 13C-1H methyl NMR indicates that with increasing concentration of polysorbates, the Fab region showed a decrease in crosspeak volumes. In addition to volume changes, PS20 caused significant changes in the chemical shifts compared to no changes in the case of PS80. No such changes in crosspeak volumes or chemical shifts were observed in the case of Fc region, indicating that polysorbates predominantly affect the Fab region compared to the Fc region. This differential effect of polysorbates on the Fab and Fc regions was because of the lesser thermodynamic stability of the Fab compared to the Fc. These results further indicate that PS80 is the preferred polysorbate for this mAb formulation, because it offers higher protection against aggregation, causes lesser structural perturbation, and has weaker binding affinity with fewer binding sites compared to PS20.
Keywords: Proteins, Excipients, Surfactants, Stability, Protein Aggregation, Protein Structure, Protein Formulation, Stabilization, Protein Binding, Physical Characterization
INTRODUCTION
Conformational integrity of therapeutic proteins encounters numerous challenges from manufacturing through eventual injection in intended patient which include exposure to various interfaces (air-liquid, liquid-liquid, and liquid-solid). Many processes such as filtration1, storage2, freeze-thaw3, vial filling4, agitation5, lyophilization6, and final injection7 expose therapeutic proteins to denaturing interfaces resulting in significant protein aggregation. One strategy to minimize interface-induced protein aggregation is to include polysorbates, such as polysorbate 20 (PS20; Fig. 1a) or polysorbate 80 (PS80; Fig. 1b) in protein formulations. Polysorbates are a class of nonionic detergents that protect biotherapeutics against destabilizing environments at various interfaces8–13. Owing to large fatty acid aliphatic chains, critical micelle concentration (CMC) for polysorbates, which is the maximum concentration of monomeric polysorbate available in solution, is very low. The approximate CMC values for PS20 and PS80 are 55 μM and 13 μM respectively14,15. Increased hydrophobicity of longer fatty acid (mono-oleate vs. mono-laureate) underlies the lower CMC of PS80 (13 μM or 0.001% w/v) when compared with PS20 (CMC = 55 μM or 0.006% w/v). Therapeutic protein formulations contain polysorbates in the range of 0.003%-0.8% w/v16–18, for example, Humira (adalimumab) contains 0.1% w/v PS8017, Raptiva (efalizumab) contains 0.2% PS2017, and Tecentriq (atezolizumab) contains 0.8% w/v PS20 (https://www.accessdata.fda.gov/drugsatfda_docs/label/2016/761034s0001b1.pdf). Although the protection of protein biotherapeutics by polysorbates against interface-induced aggregation is well documented in literature8–11, the mechanism by which polysorbates interact with proteins is less understood.
Figure 1.
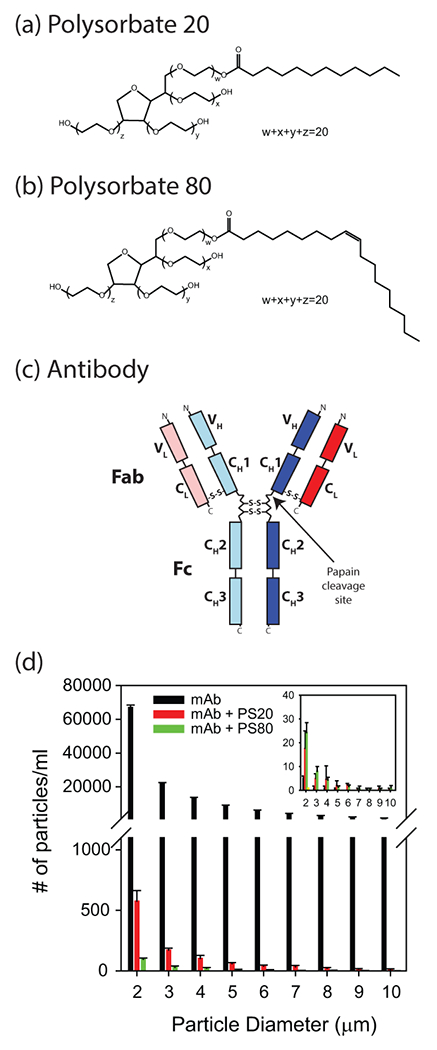
Molecular structures of (a) polysorbate 20, (b) polysorbate 80, and (c) antibody. Antibody is made up of two heavy (subscript H; blue colored) and two light (subscript L; red colored) chains, with each chain having variable (V) and constant (C) domains. Heavy chain consists of three constant domains (CH1, CH2, and CH3) and one variable domain (VH), whereas the light chain is made up of one constant domain (CL) and one variable domain (VL). (d) Both PS20 and PS80 protect the antibody against agitation-induced aggregation, as measured by the particle counts using FlowCAM technique. Inset shows the particle counts in the case of control quiescent samples.
Monoclonal antibodies (mAbs) constitute a major class of therapeutic drugs, and their market share is growing at an ever increasing pace19. Polysorbates are integral to mAb formulations to protect against interface-induced aggregation20. Therefore, a fundamental understanding of how polysorbates interact with mAbs is becoming increasingly important, in particular, do polysorbates bind to mAbs, how strong is the binding, the effect of binding on the structure and stability of mAb, and whether the mechanism by which polysorbates protect proteins against aggregation is through increasing the conformational stability of proteins. Because of their hydrophobic nature, polysorbates can bind to the hydrophobic regions in proteins. Such binding can result in either stabilization against aggregation or lead to unfolding and induce instability. This work answered some of these questions by probing how two most commonly used polysorbates PS20 and PS80 interact with a mAb of IgG1 class (obtained from Pfizer Inc) and its isolated Fab and Fc fragments using 2D NMR spectroscopy in combination with other biophysical tools. Full mAb (IgG1) is composed of two light chains and two heavy chains linked by disulfide bonds (Fig. 1c). Each light chain is composed of two domains (VL and CL), and each heavy chain is composed of four domains (VH, CH1, CH2, and CH3). Fab region, which binds to antigen, is composed of a light chain (VL and CL) and the N-terminal half of a heavy chain (VH, CH1). Fc is composed of two C-terminal halves of the heavy chains, each having CH2, CH3 domains21,22. Experimental results presented here indicate that PS20 and PS80 affect specifically the structure and stability of the Fab region of this mAb compared to its Fc region. Both PS20 and PS80 protect mAb, and its Fab and Fc fragments against agitation-induced aggregation. Compared to PS20, binding of PS80 causes lesser conformational changes in mAb and offers higher protection against agitation-induced aggregation, suggesting that PS80 to be the choice of surfactant in formulating the mAb studied here.
MATERIALS AND METHODS
Materials
Monoclonal antibody (mAb-IgG1) was provided by Pfizer Inc. (New York City, NY) and received at a concentration of 20 mg/ml in 20 mM histidine, pH 5.5 buffer with no polysorbate (histidine buffer). Fab and Fc regions were produced by cleaving the mAb with papain protease, and purified using ion exchange chromatography by Pfizer. High purity PS20 (TWEEN 20 HP-LQ-(MH), Product Code SD40271) and PS80 (TWEEN 80 HP-LQ-(MH); Product Code SD43361) were obtained from Croda (Edison, NJ). To avoid oxidation with time, freshly purchased polysorbates were aliquoted into 0.5 ml each in amber glass vials topped with nitrogen gas. Each vial was used only once during the experiments, and the remaining amount in the glass vial was discarded. All other chemicals were of reagent grade or higher, and purchased from Fisher Scientific (Pittsburgh, PA).
Agitation induced particle counting
Protein samples (1 μM without and with 2 mM polysorbates) in histidine buffer (20 mM histidine, pH 5.5) or phosphate buffer (20 mM NaH2PO4/Na2HPO4, pH 5.8 or 8.0) were used for this study. Samples (in triplicate) were agitated at 37°C for 16 hrs using orbital shaker (200 rpm). Fluid Imaging Technologies Benchtop FlowCAM® (Scarborough, ME USA) was used for counting particles between 1 μm and 10 μm (equivalent spherical diameter). The FlowCAM was fitted with a QCFC300 flow cell, 10× objective lens, and 10× collimator. A volume of 300 μl was analyzed for each sample at a flow rate of 80 μl/min. Quiescent protein samples without and with polysorbates were used as controls.
Isothermal titration calorimetry (ITC)
ITC experiments were conducted using MicroCal ITC200 (Malvern, UK) at 5°C. mAb at a concentration of 20 mg/ml in histidine buffer was titrated with 10 mM polysorbates. Except for the first injection (0.2 μl), injection volume for the remaining twelve injections was 3 μl each. Each injection duration was 6 seconds, and spacing between individual injections was 300 sec. Blank ITC thermograms (titration of polysorbate into histidine buffer without mAb) were recorded under similar conditions, and subtracted from mAb-polysorbate binding thermograms. Data was analyzed using ORIGIN® software provided with the instrument. Respective blank subtracted curves were fitted using single set multiple site model to calculate the dissociation constant (Kd).
Raman spectroscopy
mAb at 18 mg/ml was used without and with polysorbates for recording Raman scattering using a Zetasizer-Nano equipped with Raman RXN System (Malvern, UK). Each spectrum was recorded with 20 sec/transient and 50 transients were averaged to enhance the signal to noise ratio. Raman spectra of respective blanks (buffer without mAb) were recorded under identical settings and data was analyzed using Helix® software provided with the instrument.
Near-UV Circular dichroism (CD) spectroscopy
mAb at 18 mg/ml was used without and with polysorbates for recording near-UV CD spectra on a Chirascan Plus spectrometer (Applied Photophysics, UK) at 25°C. Recorded spectral wavelength range was from 250 nm to 350 nm with 2 nm band width, 1 sec signal averaging, and a step size of 1 nm.
Analytical ultracentrifugation (AUC)
Protein samples (1 mg/ml mAb in histidine buffer without and with polysorbates) were subjected to sedimentation velocity (SV-AUC) experiment on a Beckman XL-A analytical centrifuge at 25°C. 200 scans were recorded at 42,000 rpm. Protein absorbance at 280 nm was used to monitor the radially outward shift in the meniscus. Density and viscosity of the solutions were estimated using SEDNTERP. SV-AUC data was fitted using continuous sedimentation distribution model with SEDFIT software.
Size exclusion chromatography (SEC)
Protein samples (20 mg/ml without and with polysorbates) were incubated at 50°C for 20 hrs and subjected to SEC using Agilent-1100 HPLC equipped with auto-sampler. Absorbance at 280 nm was monitored to record the elution profile of protein through TskGel G3000sw column. 100 mM sodium phosphate and 100 mM sodium sulfate at pH 6.8 was used as the mobile phase with a flow rate of 1 ml/min.
2D NMR spectroscopy
Protein samples were exchanged into deuterated histidine buffer (20 mM d5-histidine (Isotec Inc; Cat# DLM-7855), pH 5.5) by multiple washings using Centricons (Millipore). Samples (20 mg/ml Fab or Fc without and with polysorbates) in 0.5 mm Shigemi tubes were subjected to 2D 13C-1H methyl NMR at 50°C using a 900 MHz NMR (Varian-Agilent). Both Fab and Fc were titrated with 0.2 mM (0.025% w/v PS20 or 0.026% w/v PS80) and 2 mM (0.25% w/v PS20 or 0.26% w/v PS80) of polysorbates. SOFAST HMQC pulse sequence from VNMRJ 4.0 Biopack was used for recording the NMR spectra after optimizing the required parameters. Spectra were collected with a spectral width of 14044.9 Hz in 1H dimension and 7239.06 Hz in 13C dimension. Number of transients in the second dimension was 128. Raw data (FID) was processed using NMRPipe software with zero filling to twice the real points, solvent filtered, apodized using SP (Sine-bell), GMB (Gaussian) and Poly (baseline correction) function. Changes in chemical shifts and peak volumes were analyzed using NMRPipe.
Chemical Denaturation Melts
For denaturation melts of the mAb, Fab, and Fc, 1 μM protein in histidine buffer (pH 5.5) was used. Guanidinium chloride (GdmCl) was used as the denaturant. Protein samples were prepared at varying GdmCl concentrations and equilibrated for 1 hr. Changes in the intrinsic protein fluorescence of aromatic amino acids (excitation at 280 nm, emission at 355 nm) were monitored as a function of increasing GdmCl concentration.
Differential scanning calorimetry (DSC)
Excess heat thermograms for thermal denaturation of mAb, Fab, and Fc were recorded using MicroCal VP-Capillary DSC (Malvern, UK) equipped with an autosampler in either histidine buffer or phosphate buffer. Protein concentration was 0.5 mg/ml. Thermograms were recorded from 25°C to 100°C with a thermal ramp of 90°C/hour. Thermograms of respective blanks (buffer without protein) were recorded under identical settings. ORIGIN® analysis software provided with the instrument was used to analyze protein denaturation thermograms.
RESULTS
PS20 and PS80 protect mAb against agitation-induced aggregation
mAb (without and with PS20 or PS80) was subjected to overnight agitation at 37°C, and counted the number of micro-particles using FlowCAM. Number of particles (> 1μm) was significantly reduced by ~100 times in the presence of PS20 when compared with no polysorbate in the solution (Fig. 1d). With PS80, the number of microparticles were much less compared to PS20, indicating that PS80 provided better protection against agitation-induced aggregation. Number of particles were less than 30 for quiescent samples (inset of Fig. 1d). This particle data clearly shows that both polysorbates PS20 and PS80 protect mAb against agitation-induced aggregation.
PS20 and PS80 bind to mAb with a sub-millimolar Kd
ITC was used for measuring binding. PS20 or PS80 was titrated with mAb in small successive installments, thereby saturating the binding sites on the protein23. The heat transfer data associated with ligand binding was fitted to obtain the dissociation constant (Kd) and the reaction stoichiometry (N) (Fig. 2). PS20 was bound to mAb with a Kd of 67.6 ± 10.0 μM (0.008 ± 0.001% w/v) and N = 5.6 ± 0.1 molecules of PS20 binding to one molecule of mAb (Fig. 2a). Correspondingly, PS80 was bound to the mAb with a Kd of 167.5 ± 36.2 μM (0.022 ± 0.005% w/v) and N = 3.1 ± 0.3 molecules of PS80 binding to one molecule of mAb (Fig. 2b). These Kd values are within the polysorbate concentration range used in protein formulations (0.003% - 0.8% w/v). The lesser number of binding sites for PS80 compared to PS20 might be due to its larger hydrophobic group (oleate) and a kink in the aliphatic sidechain because of cis-double bond causing more stearic hindrance when compared with PS20 (monolaurate).
Figure 2.
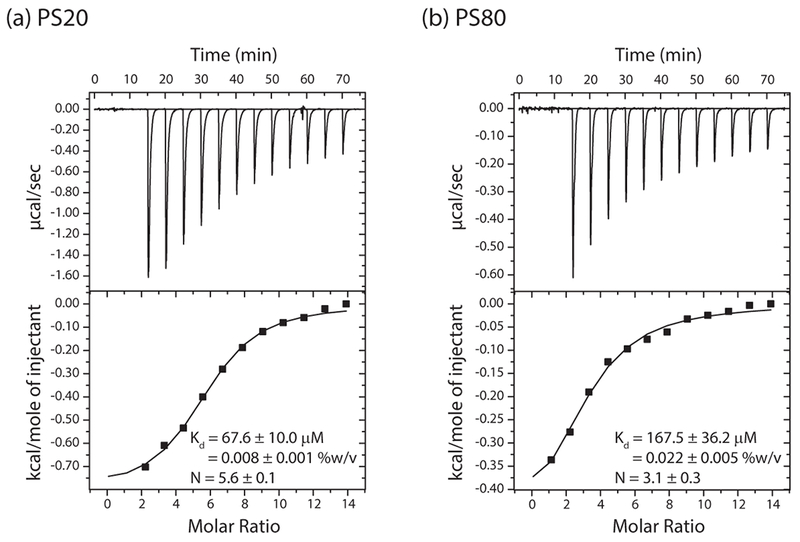
Binding of (a) PS20 and (b) PS80 to the mAb probed using isothermal titration calorimetry (ITC). The figure also shows the number of binding sites (N) and the dissociation constants (Kd) obtained from data fitting. The model assumes that all binding sites have similar Kd.
The above Kd values were obtained using the total polysorbate concentration used for isothermal titration. However, polysorbates exist as monomers as well as micelles in solution. Previously published data suggests that proteins do not bind to the polysorbate micelles because of the hydrophilic nature of the micelle surface 24. Therefore, if Kd values were to be calculated based on the available monomer concentration, the true Kd values would be much lower than the above estimated Kd values from ITC measurements. The specific binding of proteins to polysorbate monomers could also partly explain the previously observed increase in the apparent CMC of polysorbates in the presence of proteins 25, because the presence of proteins deplete the concentration of free monomers available for micelle formation by binding to the monomeric form of polysorbates.
PS20 and PS80 perturb the tertiary structure of mAb, but not its secondary structure
Raman scattering and near-UV CD were used for measuring the effect of polysorbates on the secondary and tertiary structures of mAb, respectively. Far-UV CD could not be used for these samples because histidine present in the buffer significantly absorbs in this wavelength range. Amide I region in the Raman spectrum is sensitive to changes in the secondary structure of a protein26. Antibodies exhibit a Raman amide I band at 1674 cm−1 characteristic of their β-structure (Fig. 3a)26. Addition of polysorbates did not cause significant changes in the Raman spectrum, indicating that PS20 or PS80 did not perturb the secondary structure of mAb.
Figure 3.
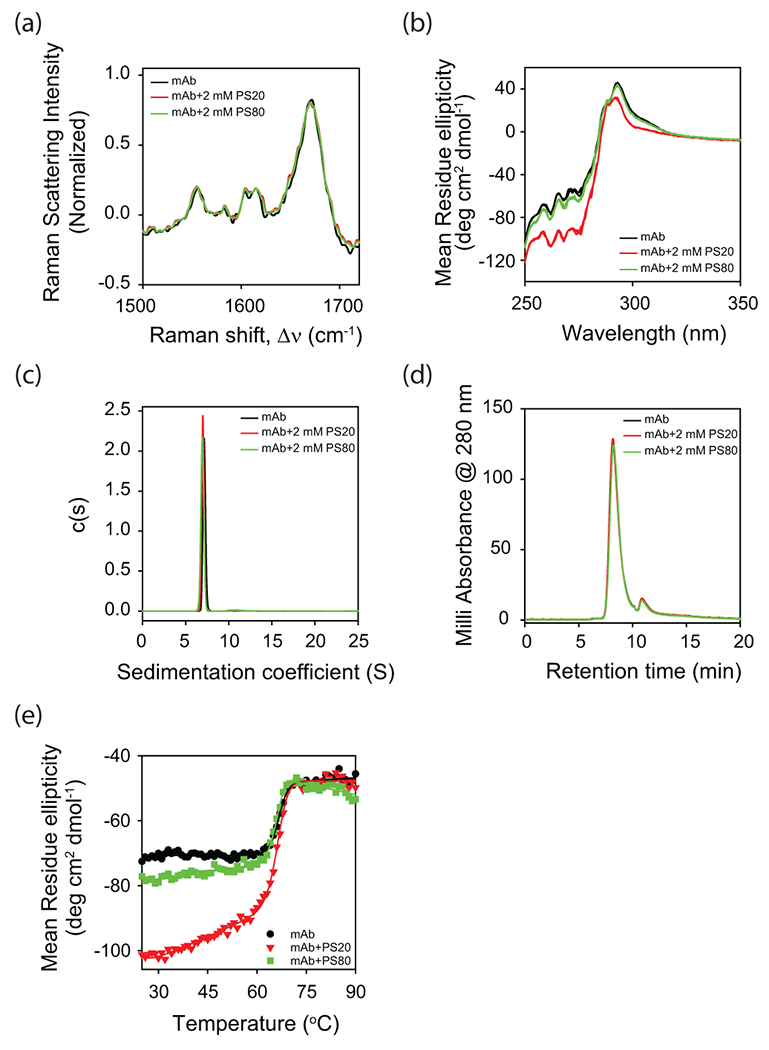
Effect of polysorbate binding on (a) the secondary structure of mAb as probed by Raman scattering, and (b) the tertiary structure of mAb as probed by near-UV CD. Both (c) analytical ultracentrifugation (AUC) and (d) size exclusion chromatography (SEC) indicate that mAb is a monomer in the absence and presence of polysorbates. The peak in the SEC chromatogram (panel d) at the elution time of 11 min correspond to the histidine present in the buffer. (e) Change in the near-UV CD signal as a function of increasing solution temperature. Both PS20 and PS80 affect the partial protein unfolding with relatively no change in the global unfolding of mAb. In all the panels, black, red, and green curves represent the data in the absence of polysorbates, with PS20, and with PS80, respectively. In panel b, triplicate data has been shown for each experimental condition to indicate the statistical significance of the changes in spectra with the addition of polysorbates.
Near-UV CD is a highly sensitive method to evaluate the impact of various buffers and processes on the tertiary structure of biotherapeutic proteins 24,27. Binding of polysorbates caused significant changes in the near-UV CD spectrum (Fig. 3b), indicating that the PS20 and PS80 affected the tertiary structure of mAb. Fig. 3b shows the triplicate data for each experimental condition to indicate the statistical significance of the changes in spectra with the addition of polysorbates. Quantitative spectral similarity was compared using the weighted spectral difference (WSD) parameter described earlier 28. Spectral variation between samples under different solution conditions (WSD = 61% for PS20 and WSD = 12% for PS80, with respect to the control sample with no polysorbates) is much higher than the spectral variation between different samples under identical solution conditions (WSD < 5%). Near-UV CD measures chirality of the structural environment around the sidechains of aromatic amino acids (Phe, Tyr, and Trp). Changes in the intensity and spectral pattern around 295 nm indicate that the environment around tryptophan residues is altered upon the addition of polysorbates. Although the pattern of spectral region around 260 nm is similar, the intensity is significantly changed upon the addition of polysorbates, which is indicative of the tertiary structural changes around tyrosines and phenylalanine residues as well. Further, mAb tertiary structure perturbation is more pronounced in the case of PS20 compared to PS80, which is consistent with the higher binding affinity and stoichiometry of PS20 when measured by ITC (Fig. 2).
It was further confirmed that the above changes in the tertiary structure of mAb upon the addition of polysorbates is not due to oligomerization. Both analytical ultracentrifugation (AUC) (Fig. 3c) and size exclusion chromatography (SEC) (Fig. 3d) showed that the mAb was a monomer in the absence and in the presence of polysorbates. These AUC and SEC data also indicated the purity of mAb samples received from Pfizer. The small peak observed around the retention time of 11 min in the SEC chromatogram is from the histidine present in the buffer.
The effect of polysorbates on the tertiary structure of mAb has also been observed in its partial unfolding with no effect on its global unfolding (Fig. 3e). The new tertiary structure that is formed in the presence of polysorbates unfolds at lower temperatures compared to global unfolding at higher temperatures, as indicated by the appearance of sloped native baseline in the presence of polysorbates (Fig. 3e). Sloped baselines in thermal melts can originate either from the presence of partially unfolded states or because the signal of the native state changes as a function of solution temperature 29–32. Since sloped native baseline was not observed in the absence of polysorbates, the sloped baselines in the presence of polysorbates can be attributed solely to the presence of partially unfolded states 32. Compared to the effect of polysorbates on partial protein unfolding, polysorbates did not significantly affect the global unfolding of mAb, as evident from minimal changes in the midpoint temperature of the main transition (close to 66°C) in the near-UV CD thermal melts (Fig. 3e).
PS20 and PS80 specifically perturb the tertiary structure of Fab region when compared to the Fc region
The main goal of this project was to determine which specific regions in the mAb interact with the polysorbates. Because of the unique papain protease site in the mAb structure (Fig. 1c), Fab and Fc regions can be individually examined. Pfizer provided us the purified Fab and Fc fragments after papain proteolysis. Changes in the tertiary structure of the Fab and Fc fragments upon the addition of polysorbates were monitored using near-UV CD in the presence of 2 mM PS20 or PS80. The 2 mM (0.25% w/v PS20 or 0.26% w/v PS80) concentration used in this study is within the concentration range (<0.8% w/v) used in current protein antibody formulations 16,17(https://www.accessdata.fda.gov/drugsatfda_docs/label/2016/761034s000lbl.pdf). Near-UV CD spectra of the Fab region was significantly altered in the presence of polysorbates (Fig. 4a). These spectral differences observed upon the addition of polysorbates were statistically significant as evident from the triplicate data sets shown for each experimental condition. The differences in the near-UV CD spectra for the three different conditions (without polysorbates, with PS20, and with PS80) are significantly higher (WSD = 76% for PS20 and WSD = 37% for PS80, with respect to the control sample with no polysorbates) compared to the spectral differences in the triplicate data for each experimental condition (WSD < 5%). In comparison, no significant changes were observed in the near-UV CD spectra of the Fc region upon the addition of polysorbates (WSD < 5%) (Fig. 4b). These spectral changes clearly indicate that both PS20 and PS80 interact preferentially with the Fab region compared to the Fc region. Further, the degree of alteration in the near-UV CD spectra was more pronounced in the case of PS20 when compared with PS80, which is consistent with the higher affinity and stoichiometry of PS20 binding to the mAb compared to PS80 (Fig. 2). Under these experimental conditions, both Fab and Fc are monomers as indicated by SEC (Figs. 4c and 4d).
Figure 4.
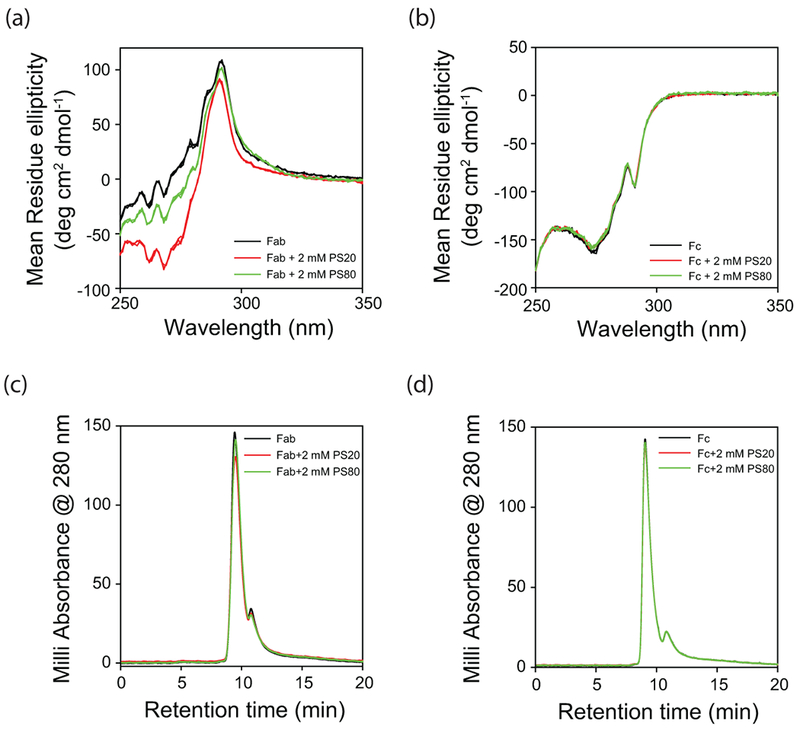
Effect of polysorbates on the tertiary structures of isolated (a) Fab and (b) Fc fragments of mAb as probed by near-UV CD. Black, red, and green curves represent the data in the absence of polysorbates, with PS20, and with PS80, respectively. Triplicate data has been shown for each experimental condition to indicate the statistical significance of the changes in spectra with the addition of polysorbates. Both Fab and Fc are monomers in the absence and presence of polysorbates as evident from the SEC chromatograms shown in panels (c) and (d). The peak in the SEC chromatogram at the elution time of 11 min correspond to the histidine present in the buffer.
2D 13C-1H methyl NMR spectroscopy suggests similar structure for isolated Fab and Fc regions as that in the full-length mAb
2D NMR spectroscopy provides high-resolution site-specific structural information that can be used to probe the effect of polysorbates on local protein regions of mAb. In general, proteins need to be labeled with NMR-sensitive nuclei such as 15N or 13C for increased resolution and sensitivity. Alternatively, 2D NMR spectra can be recorded at the natural abundance of NMR isotopes. Since 13C natural abundance is three times to that of 15N (1.1% vs 0.36%), 13C NMR spectrum can be recorded in a much shorter time. However, since an average amino acid contains 5.4 carbons, the normal 13C spectrum of an antibody (1332 residues; 7193 resonances) will be very crowded with poor spectral resolution. For improved spectral resolution, 13C-1H methyl 2D NMR spectrum is the preferred choice 33, which will have crosspeaks emanating from the sidechain methyl groups. Only six (Ala, Val, Met, Thr, Leu, and Ile) out of the twenty naturally occurring amino acids have methyl groups. 2D 13C-1H methyl NMR spectroscopy was used to probe the effect of polysorbates on the mAb structure. To further enhance the resolution, these NMR experiments were done at 50°C, which increases the tumbling rate of the molecule 33. However, owing to the large size of mAb (~150 kDa), its 2D methyl spectrum is crowded with many overlapping crosspeaks (Fig. S1). Since our goal was to determine which specific antibody regions interact with polysorbates, 2D methyl NMR spectra of isolated Fab and Fc fragments were recorded. Owing to the reduced molecular size for each fragment (~50 kDa, one third of the intact mAb), methyl 2D NMR spectra were obtained with reasonable peak dispersion for both Fab and Fc fragments (Fig. S1). More importantly, combined NMR spectra of isolated Fab and Fc regions was similar to that of the intact mAb, implying that the isolated Fab and Fc fragments retain similar structures as that in the full-length mAb.
PS20 and PS80 predominantly affect the Fab region when compared to the Fc
Fab and Fc fragments were titrated with PS20 or PS80 (0, 0.2 mM and 2 mM), and the changes in 2D 13C-1H methyl NMR spectra were shown in Figs. 5 & 6. These NMR experiments were done at 50°C. Increasing the solution temperature from 5°C to 50°C decreases the CMC of polysorbates (from 90 μM to 48 μM (1.9 times) for PS20, and from 36 μM to 12 μM (3 times) for PS80) 34. Since CMC defines the maximum monomer concentration possible at equilibrium under micelle forming conditions and since CMC decreases with temperature, the Kd values at 50°C will be higher than those measured by ITC at 5°C 35, which implies that the polysorbate-bound protein population will be lower at 50°C under NMR conditions compared to that at 5°C. Even with such decreased population of polysorbate-bound conformation of mAb, significant effects of PS20 and PS80 on mAb NMR were observed. When compared to the Fc, the NMR crosspeak volumes of Fab showed a significant decrease upon the addition of either PS20 (Fig. 5b vs 5d) or PS80 (Fig. 6b vs 6d). The decrease in NMR crosspeak volumes of Fab upon the addition of polysorbates was not an artifact of the NMR experiment or data processing, as similar decrease was not seen in the case of Fc with identical protocols. Most changes in the NMR crosspeak volumes of the Fc were random and were within the errors associated with NMR data processing and peak picking (<10%). In that sense, Fc acts as a control for Fab in NMR experiments. These results imply that PS20 and PS80 predominantly affect the Fab region compared to the Fc region.
Figure 5.
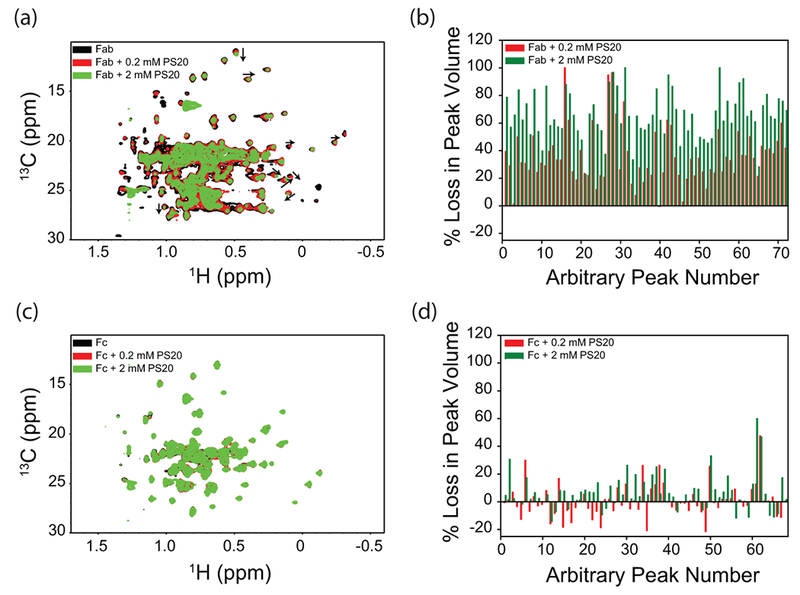
Effect of PS20 on the NMR spectra of isolated Fab and Fc fragments of mAb. Panels (a) and (c) show the NMR spectra, and panels (b) and (d) show the loss of volumes of individual crosspeaks. Peaks were arbitrarily numbered because of the unavailability of assignments. Black, red, and green represent the data in the absence of PS20, with 0.2 mM PS20, and with 2 mM PS20, respectively.
Figure 6.
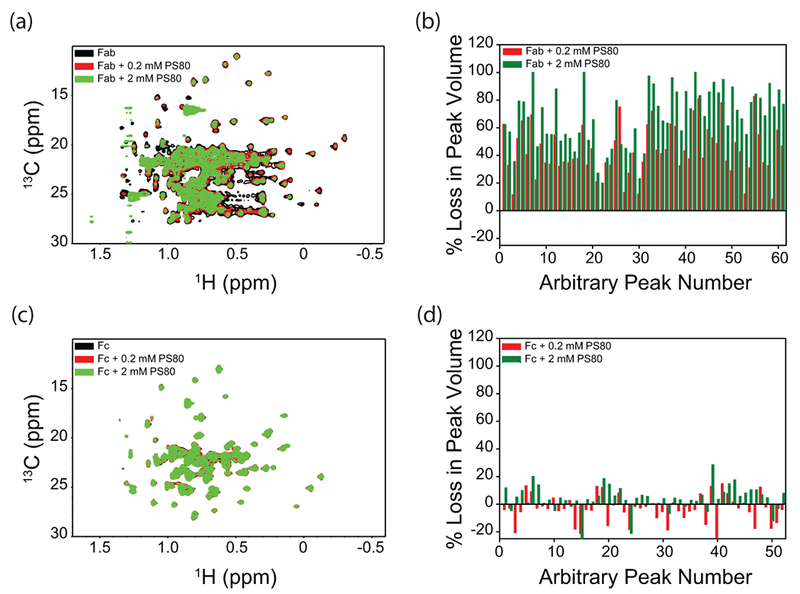
Effect of PS80 on the NMR spectra of isolated Fab and Fc fragments of mAb. Panels (a) and (c) show the NMR spectra, and panels (b) and (d) show the loss of volumes of individual crosspeaks. Peaks were arbitrarily numbered because of the unavailability of assignments. Black, red, and green represent the data in the absence of PS80, with 0.2 mM PS80, and with 2 mM PS80, respectively.
Decrease in NMR crosspeak volumes of Fab can be because of five possible reasons: (1) increase in solution viscosity upon the addition of polysorbates; (2) increase in the molecular size of Fab upon polysorbate binding; (3) irreversible protein aggregation upon the addition of polysorbates decreasing the monomer Fab concentration in solution; (4) formation of transient interactions between Fab monomers; and (5) increased protein dynamics in the presence of polysorbates. Addition of 2 mM polysorbates, which correspond to 0.25% w/v PS20 or 0.26% w/v PS80, does not increase solution viscosity 36. Further, Fc did not show decrease in crosspeak volumes with the addition of polysorbates, indicating that the observed phenomenon is specific to Fab and is not due to the changes in solvent properties. However, specific binding of polysorbates to Fab can cause an increase in microviscosity around Fab, whose effect cannot be completely ruled out. Increase in Fab size upon binding to polysorbates can be ruled out, because only a fewer number of polysorbate molecules (6 in the case of PS20 and 3 in the case of PS80) bind to mAb. No visible aggregates were observed in the NMR sample tube after the completion of the NMR experiments. In addition, the SEC chromatogram showed no oligomeric species (Fig. S2), indicating the absence of irreversible protein aggregation. However, the presence of transient oligomer formation during the NMR experiments resulting in a decrease in NMR peak volumes cannot be completely ruled out. Increased protein dynamics30,37–40 in the presence of polysorbates populating an ensemble of conformations with the average conformation close to that in the absence of polysorbates can also lead to decreased crosspeak volumes. In addition, the pattern of the volume decrease is not uniform across all the crosspeaks, indicating varying protein dynamics in various regions of the protein. Nonetheless, irrespective of the origin of the decrease in crosspeak volumes, the fact that changes were observed only in the case of Fab, but not in Fc, indicates that polysorbates affect predominantly the Fab region compared to the Fc region.
In addition to the decrease in NMR crosspeak volumes, additional effect was observed in the case of PS20. Some of the Fab crosspeaks showed a clear change in their chemical shift positions (as indicated by the black arrows in Fig. 5a and in Fig. S3a & b), whereas no such changes were observed in the case of Fc (Fig. 5c) or in the presence of PS80 (Figs. 6a, 6c, and in Fig. S3c & d). These peak shifts indicate a conformational change in the case of Fab with PS20, compared to Fab with PS80, or the Fc region with PS20 or PS80. This is quite consistent with the near-UV CD results which show significant structural changes in the Fab with the addition of PS20 (Fig. 4a), but not in Fc (Fig. 4b).
Note that the near-UV CD (Fig. 4) and 2D 13C-1H methyl NMR (Figs. 5 & 6) measure the effect of polysorbate binding on different levels of tertiary structure of proteins. Near-UV CD monitors the tertiary structure around the sidechains of aromatic amino acids (Phe, Tyr, and Trp), whereas the methyl NMR monitors the tertiary structure around the methyl sidechains of six amino acids (Ala, Val, Met, Thr, Leu and Ile) which constitute the hydrophobic core of the protein.
Polysorbates predominantly affect the Fab region because of its lesser thermodynamic stability compared to the Fc
Both Fab and Fc regions contain hydrophobic residues; however, predominant effect of polysorbates has been observed only on the structure of Fab (Figs. 4, 5 & 6). Possible explanation underlying this differential effect of polysorbates is in the intrinsic thermodynamic stabilities of Fab and Fc regions. Thermodynamically lesser stable domain is more vulnerable to structural perturbations, because of its increased vulnerability to partial unfolding, compared to a more stable domain 37–39,41,42. When the stabilities of the Fab and Fc regions were measured using chemical denaturant melts with guanidinium chloride (GdmCl) as the denaturant (Fig. 7a), Fab melted at a lower denaturant concentration compared to the Fc region, indicating that Fab is less stable than Fc. Being less stable, Fab is more prone to increased unfolding populating partially unfolded states with exposed hydrophobic residues to which polysorbates can bind, compared to the Fc region.
Figure 7.
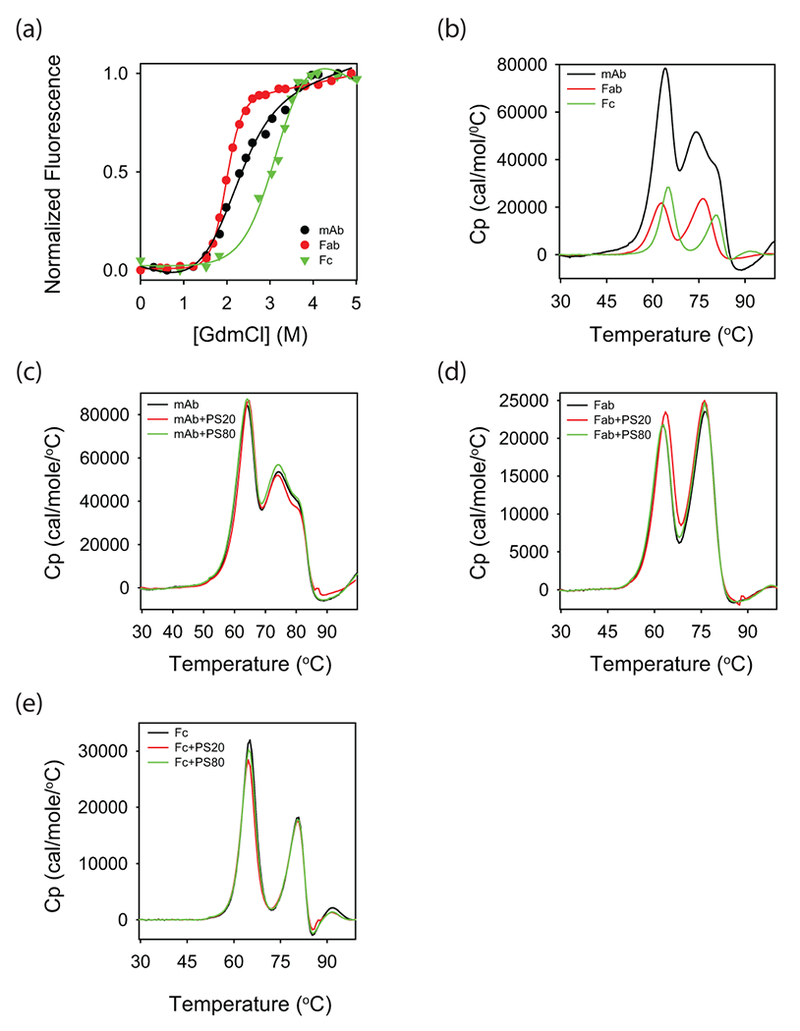
Thermodynamic stability of the mAb and its Fab and Fc fragments in the absence and presence of polysorbates. (a) Chemical denaturant melts with guanidinium chloride (GdmCl) as the denaturant. (b) DSC thermograms. In both panels a and b, black, red, and green curves correspond to that of mAb, Fab, and Fc, respectively. (c-e) Effect of polysorbates on the DSC thermograms of mAb, Fab, and Fc, respectively. In these three panels, black, red, and green curves represent the data in the absence of polysorbates, with PS20, and with PS80, respectively.
The lesser stability of Fab with respect to the Fc region might be because of its variable domain. Fab contains one variable and one constant domain, whereas Fc contains two constant domains. Recent studies have shown that the constant domains in multiple IgG1-mAbs have similar stability, and the difference in stabilities of mAbs can be accounted by the varying stabilities of variable domains43. Compared to the denaturant melts, differential scanning calorimetry (DSC) can detect the unfolding of individual domains, because of the intrinsic differences in their heat capacities43. DSC thermograms of the mAb, Fab, and Fc used in this study (Fig. 7b) indicate that the lesser stability of Fab compared to Fc is due to the decreased stability of its variable domain, which unfolds at lower temperatures around 62°C compared to the constant domain of Fab or the two constant domains of Fc. This speculation that Fab is less stable because of its variable domain needs to be further confirmed by studying the properties of individual domains or obtaining high-resolution structural and stability data on intact mAb and its Fab and Fc fragments. It is interesting to note that mAbs can sometimes show changes in the structure and dynamics of constant domains with variations in the complementarity-determining regions (CDRs) of variable domains 44, and the conformational changes can be propagated over long distances in the mAb structure 45. However, our observation that the Tm values of individual peaks in the DSC thermogram of intact mAb closely correspond to the Tm values of individual peaks in the thermograms of isolated Fab and Fc (Fig. 7b) indicates that the Fab and Fc of this mAb might have similar stability as that in the intact mAb.
Polysorbates do not significantly affect the global stability of mAb and its Fab and Fc regions
DSC was used to measure the effect of polysorbates on the global stabilities of mAb and its individual domains. DSC thermograms of mAb and its Fab and Fc fragments were similar in the absence and presence of polysorbates (Figs. 7c-e), implying that the presence of polysorbates did not significantly affect the overall stability of mAb, Fab, and Fc. This is also consistent with the near-UV CD results (Fig. 3e), which showed minimal effect of polysorbates on the global unfolding transition. Similar conclusions were drawn earlier where DSC was unable to detect the effect of polysorbates on the structural stabilities of local protein regions in mAbs35,46.
Despite lower thermodynamic stability, Fab aggregates lesser than that of Fc
Chemical denaturation melts and DSC thermograms showed that the Fab region has lower thermodynamic stability than that of Fc (Fig. 7a & b). How this difference in conformational stability translates into the aggregation propensity of Fab and Fc regions was further probed (Fig. 8). Upon overnight agitation at 37°C, Fc aggregated to a higher extent than that of Fab (Fig. 8a). Similar to mAb (Fig. 1d), both PS20 and PS80 protected Fab and Fc from agitation-induced aggregation (Figs. 8b & c). However, the extent of protection by PS20 or PS80 against protein aggregation is lesser for Fab compared to Fc. This can be understood as follows. Both PS20 and PS80 protect proteins against interface-induced aggregation by competing with proteins for interfaces. However, since PS20 and PS80 interact specifically with Fab causing its unfolding (Figs. 4–6), Fab is more prone to aggregation compared to Fc. Therefore, the observed effect of PS20 and PS80 on Fab aggregation (Fig. 8b) is the sum of decreased aggregation at interfaces and increased aggregation because of decreased conformational stability, whereas in the case of Fc, the effect of PS20 and PS80 is just the decreased aggregation at interfaces. This might be the reason why less protection was observed in the case of Fab compared to Fc.
Figure 8.
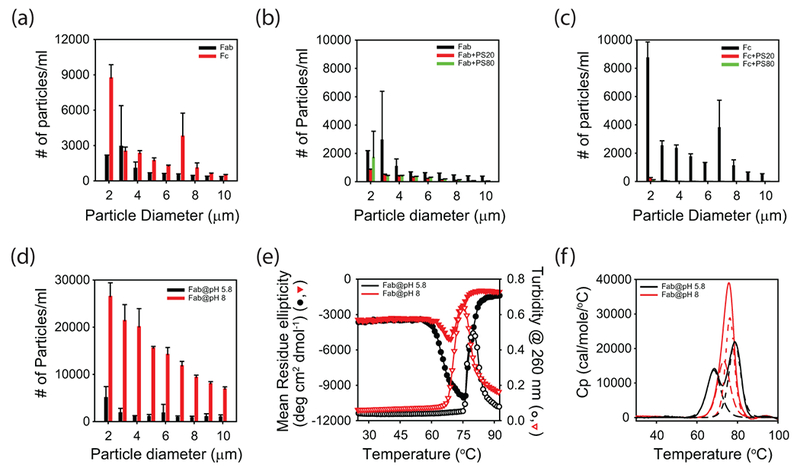
Role of conformational stability vs colloidal stability in mAb aggregation. (a) Aggregation of Fab (black) and Fc (red) regions at pH 5.5 (histidine buffer), as measured by the particle counts using FlowCAM. (b) Aggregation of Fab in the absence of polysorbates (black), with PS20 (red), and with PS80 (green), respectively, as measured by FlowCAM. (c) Aggregation of Fc in the absence of polysorbates (black), with PS20 (red), and with PS80 (green), respectively, as measured by FlowCAM. (d) Aggregation of Fab at pH 5.8 (black) and pH 8.0 (red), respectively, as measured by FlowCAM. (e) Conformational stability and aggregation of Fab at pH 5.8 (black) and pH 8.0 (red). Far-UV CD measured the change in protein conformation, whereas solution turbidity measured the aggregation. The scale on the left side of y-axis corresponds to far-UV CD, whereas the scale on the right side of y-axis corresponds to solution turbidity. (f) DSC thermograms of Fab at pH 5.8 (black) and pH 8.0 (red). Solid lines show the raw data, and the dashed lines show the deconvolution of the solid lines into unfolding of individual domains.
In addition, Fab aggregating lesser than that of Fc in the absence of PS20 or PS80 despite its lower conformational stability implies that additional physical parameters contribute to the aggregation of Fab and Fc. In addition to conformational stability, colloidal stability, although relatively less appreciated, plays a big role in protein aggregation47,48. To a large extent, electrostatic interactions between individual protein molecules resulting from the net surface charge distribution determine the colloidal stability. This is one reason why many proteins do not aggregate at pH values far away from their pI. Theoretical pI values for the Fab and Fc regions used in this study are 8.8 and 6.6, respectively. Since formulation pH was 5.5 (histidine buffer), Fab is 3.3 units below its pI, whereas Fc is 1.1 units below its pI. These values imply that Fab is relatively highly positively charged compared to Fc, resulting in higher intermolecular repulsions leading to less aggregation. To test the role of colloidal stability in Fab aggregation, aggregation of Fab at two different solution pHs (5.8 and 8.0; phosphate buffer) was examined. Both these pHs were below the theoretical pI for the Fab region. Fab aggregation was higher at pH 8.0, closer to its pI of 8.8, compared to pH 5.8 (Fig. 8d). The conformational stability of Fab at these two pH values was measured using far-UV CD thermal denaturation melts (Fig. 8e). With increase in solution temperature, far-UV CD signal initially decreased followed by an increase. Comparing these changes in the far-UV CD signal with the DSC results, the first transition during which the far-UV CD signal decreased can be attributable to structure unfolding. Turbidity was also measured simultaneously to probe the Fab aggregation at the two pH values. The second transition observed in the far-UV CD measurements during which the signal increased matched with that of the turbidity measurements, implying that the second transition in far-UV CD corresponds to the Fab aggregation. Comparing the first transition that corresponds to structural unfolding (Fig. 8e), Fab had lower Tonset of denaturation at pH 5.8 compared to the Tonset at pH 8.0. Comparing the second transition that corresponds to Fab aggregation (Fig. 8e), Fab aggregated at lower temperatures at pH 8.0 compared to pH 5.8. These results imply that despite exhibiting lower Tonset for conformational changes at pH 5.8, Fab aggregates less at pH 5.8 compared to pH 8.0, indicating that the colloidal stability is playing a major role in Fab aggregation. These results are quite consistent with recent studies which indicated that the colloidal stability can account for the lack of a clear relationship between the conformational stability and aggregation of mAbs 49,50.
To probe which domain of the Fab is affected by the pH change, it was subjected to DSC analysis at pH 5.8 and 8.0. Thermogram for Fab at pH 5.8 exhibited two peaks (solid black line in Fig. 8f). As discussed above, first peak at lower temperatures corresponds to the variable domain, whereas the second peak corresponds to the constant domain 43. However, at pH 8.0, DSC thermogram exhibited a single peak (solid red line in Fig. 8f). Since this single peak constitutes the unfolding of both the variable and constant domain, it could not be satisfactorily fitted into a two-state single Tm transition. This single DSC peak at pH 8 was satisfactorily deconvoluted into two peaks (broken red line in Fig. 8f). After deconvolution, a comparison of Tm values clearly shows that a change of pH from 5.8 to 8.0 increases the stability of the variable domain.
DISCUSSION
Polysorbates such as PS20 and PS80 are integral to antibody and other protein-based formulations to prevent protein aggregation at interfaces16. Proteins are polymers composed of various hydrophilic and hydrophobic amino acids with diverse chemical side chain moieties, and therefore are highly surface active compounds51. This causes a quick adsorption of protein molecules at the interfaces, which leads to their denaturation and aggregation51–53. Polysorbates being amphiphilic, compete with proteins for the interfaces and protect proteins from interface-induced aggregation54–61. Alternatively, polysorbates can also bind to exposed hydrophobic patches on proteins, thereby reducing the propensity of hydrophobic interactions among protein molecules leading to less aggregation9,11,24,62–65. However, the specific nature of these polysorbate-protein interactions has not yet been clearly demonstrated, in particular, whether polysorbates bind to mAbs, how strong is the binding, the effect of binding on the structure and stability of mAb, and whether the binding affects local or global protein structure. In this manuscript, an attempt has been made to address these questions on a mAb that is under drug development by Pfizer. First, it was confirmed that both PS20 and PS80 protect mAb against agitation induced aggregation (Fig. 1d). ITC data showed that both polysorbates bind to the mAb with Kd in the range of hundreds of micromolar (Fig. 2), which are in the range of polysorbate concentrations used in protein formulations. Similar attempts have been made previously on two mAbs to determine their binding affinity to polysorbates24,35. The Kd values were in the order of 1 – 10 mM, implying that the binding affinity of polysorbates to mAbs may depend on the nature of mAb. Binding of polysorbates to mAbs is not very surprising; other non-ionic surfactants used in mAb formulations have been shown to bind to mAbs with a micromolar affinity66. In our case, Raman scattering showed that binding of polysorbates has no impact on the secondary structure of mAb (Fig. 3a). On the contrary, polysorbates significantly affect the tertiary structure of mAb as indicated by the near UV-CD spectroscopy (Fig. 3b). Such changes in near-UV CD spectra upon polysorbate binding has been observed earlier in darbepoetin alfa24. Ours is the first study on using near-UV CD to determine the effect of polysorbates on the structure of a mAb. Although it can be argued how these spectral changes reflect the changes in the mAb structure, near-UV CD spectra have been shown to be very useful in determining the effect of polysorbates on the tertiary structure of proteins (both in the case of darbepoetin alfa24 and the mAb studied here). In addition to tertiary structural changes, binding of polysorbates affected the partial unfolding of local protein regions rather than the global unfolding of the entire mAb (Fig. 3e). To determine which region of mAb binds to polysorbates, isolated Fab and Fc regions were subjected to similar experiments. Near-UV CD of Fab and Fc indicated that polysorbates preferentially interact with the Fab region leading to structural changes, with relatively no effect on the Fc (Figs. 4a & 4b). To further confirm the near-UV CD results that polysorbates affect the local protein structures rather than the global protein structure, isolated Fab and Fc regions were subjected to 2D NMR spectroscopy of amino acid sidechain methyl groups. Application of NMR techniques to study antibodies has been feasible only recently33,67. Overall chemical shift pattern of the Fab remained the same with the addition of polysorbates except a few crosspeaks clearly showing chemical shift changes (Figs. 5a, 6a, and S3), indicating that polysorbates affect the structure of local protein regions rather than the global structure of Fab. In contrary to Fab, no changes were observed in the crosspeak pattern or the chemical shifts of Fc with the addition of polysorbates. Using chemical denaturation melts and DSC, an attempt was made to address why the effect of polysorbates was restricted to Fab leaving Fc unaffected. Fab of the mAb studied here is less stable than its Fc (Fig. 7a). Compared to Fc, lesser stable Fab will have increased population of partially unfolded states with hydrophobic residues exposed to solvent, which can interact with polysorbates. Similar observations were made in our earlier studies where the least stable region of a protein has been shown to be affected most by the excipients used in protein formulations37–39,41. The lesser stability of Fab is due to its variable domain (Fig. 7b). Since mAb used in this study is an IgG1, its DSC thermogram was compared with the published thermograms of three other IgG1-mAbs43 to identify the domain that unfolds at lower temperatures. Differences in the DSC thermograms of various Fabs of mAbs arise from the differences in their variable domains, and the mAb used in this study has less stable variable domain compared to the other mAbs. This observation explains why polysorbates affect predominantly the Fab region compared to the Fc region in the mAb studied here.
Impact of polysorbates on the global and local stabilities of mAb, Fab and Fc regions was also probed. Both near-UV CD (Fig. 3e) and DSC (Fig. 7c-e) showed no significant changes in the global unfolding, whereas near-UV CD (Fig. 3e) indicated the unfolding of local structural regions occurring at lower temperatures before the entire mAb unfolds at higher temperatures, as evident from the appearance of sloped native baselines with the addition of polysorbates (Fig. 3e).
It was further examined how the conformational stability of the Fab and Fc determines their aggregation behavior. Despite having lower stability, Fab aggregates lesser compared to Fc (Fig. 8a). By probing the effect of pH on the conformational stability and aggregation (Figs. 8d-f), colloidal stability seems to play a major role in mAb aggregation. Fab is less stable at pH 5.8, but aggregates less compared to that at pH 8. These results further indicate that the variations in conformational stability (controlled by intramolecular interactions) and colloidal stability (controlled by intermolecular interactions) of a protein may not be identical under different solution conditions. Similar results have been observed in two other published studies. In the first study on the aggregation behavior of ten different IgG1 antibodies49, there was no absolute trend between the conformational stability and aggregation of the IgG1 molecules, implying that colloidal stability plays a major role in IgG1 aggregation. In the second study50, changing the solution pH from 6.5 to 4.5 reduced the thermal stability of IgG1; however, less protein aggregation was observed at pH 4.5, because of the increased colloidal stability of the protein. Decreasing pH to acidic values increases the net charge on the protein surface leading to increased repulsive interactions between individual protein molecules, thus leading to an increase in the colloidal stability of the protein.
Although polysorbates protect proteins against interface-induced aggregation, they do not always lead to conformational stabilization of proteins. In the case of mAb studied here, loss in NMR crosspeak volumes was observed for Fab upon the addition of polysorbates. Normally, this type of peak volume loss is attributed to protein aggregation. However, no protein aggregation was observed in our case. Other plausible explanation underlying peak volume loss can be the presence of transient attractive intermolecular protein interactions in NMR samples. Although no significant changes were observed in the global conformational stability of mAb and its Fab and Fc fragments upon the addition of polysorbates, conformational destabilization of proteins by polysorbates have been observed in other proteins68–71. For example, PS20 has been shown to reduce the thermodynamic stability of human interferon-γ and darbepoetin24,69. Contrary evidence also exists where polysorbates can stabilize proteins. In the case of albutropin, PS20 and PS80 shifted the chemical denaturation melts to higher denaturant concentration, which resulted in increased thermodynamic stability of albutropin62,69. In addition, polysorbates have been proposed to have dual effects on the stability and aggregation of proteins depending on the nature of protein and the aggregation conditions70. Therefore, polysorbates protecting proteins against interface-induced aggregation and the effect of polysorbates on the conformational stability of proteins can be two independent phenomena. In protein formulations, polysorbates are used at a concentration where benefits of adding polysorbates to compete with proteins for interfaces outweigh the possible disadvantages involving protein conformational changes.
In summary, our study for the first time probed the detailed nature of molecular interactions between the polysorbates and mAbs using high resolution techniques such as 2D NMR. Published accounts of the application of 2D NMR techniques to study antibodies are only two years old33,67,72, and this study for the first time shows the power of simple 2D NMR techniques in probing how excipients in therapeutic formulations interact with mAbs. Future availability of NMR peak assignments for mAbs, similar to the recently solved backbone assignments for Fc 73, will further help in pinpointing the observed structural and dynamics changes to specific local regions in mAbs. In addition to showing the applicability of 2D NMR methods to study polysorbate-mAb interactions, our experimental data presented here indicate that PS80 will be the preferred choice over PS20 in formulating this mAb, because PS80 offers higher protection against aggregation, causes lesser structural perturbations, and has weaker binding affinity with fewer binding sites compared to PS20.
Supplementary Material
Figure S1. 2D 13C-1H HMQC NMR spectra of mAb (black) and its Fab (red) and Fc (green) fragments. Isolated Fab and Fc have similar structure as that in the intact mAb.
Figure S2. SEC indicates that both Fab and Fc fragments exist as monomers after NMR experiments in the presence of polysorbates.
Figure S3. Addition of PS20 caused chemical shift changes in the Fab region compared to PS80.
ACKNOWLEDGMENTS
We sincerely thank Ying Zhang, Pfizer Inc. and her team for providing us the mAb, and isolated Fab and Fc fragments in polysorbate-free buffer solutions. We thank Marguerite Arechederra, Pfizer Inc. (currently at Waters Corporation) for initiating this project. We highly appreciate the help we received from the 900 MHz Rocky Mountain NMR Facility and the Biophysics Core, University of Colorado Anschutz Medical Campus in carrying out the experiments. We acknowledge Marco Tonelli, National Magnetic Resonance Facility at Madison, University of Wisconsin for performing some of the initial NMR experiments. We also thank John Carpenter for many helpful discussions and for his critical reading of the manuscript, and John Gabrielson regarding the use of weighted spectral difference (WSD) parameter. This work was funded by a basic sciences research grant from Pfizer Inc.
ABBREVIATIONS
- CD
circular dichroism
- DSC
Differential Scanning Calorimetry
- Fab
antigen-binding fragment
- Fc
crystallizable fragment
- ITC
isothermal titration calorimetry
- Kd
dissociation constant
- mAb
monoclonal antibody
- NMR
nuclear magnetic resonance
- PS
polysorbate
- PS20
polysorbate 20
- PS80
polysorbate 80
- Tonset
onset temperature
REFERENCES
- 1.Maa YF, Hsu CC. 1998. Investigation on fouling mechanisms for recombinant human growth hormone sterile filtration. J Pharm Sci 87: 808–812. [DOI] [PubMed] [Google Scholar]
- 2.McLeod AG, Walker IR, Zheng S, Hayward CPM. 2000. Loss of factor VIII activity during storage in PVC containers due to adsorption. Haemophilia 6: 89–92. [DOI] [PubMed] [Google Scholar]
- 3.Eckhardt BM, Oeswein JQ, Bewley TA. 1991. Effect of freezing on aggregation of human growth hormone. Pharm Res 8: 1360–1364. [DOI] [PubMed] [Google Scholar]
- 4.Cromwell ME, Hilario E, Jacobson F. 2006. Protein aggregation and bioprocessing. AAPS J 8: E572–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maa YF, Hsu CC. 1997. Protein denaturation by combined effect of shear and air-liquid interface. Biotech Bioeng 54: 503–512. [DOI] [PubMed] [Google Scholar]
- 6.Dong A, Prestrelski SJ, Allison SD, Carpenter JF. 1995. Infrared spectroscopic studies of lyophilization- and temperature-induced protein aggregation. J Pharm Sci 84: 415–424. [DOI] [PubMed] [Google Scholar]
- 7.Tzannis ST, Hrushesky WJM, Wood PA, Przybycien TM. 1996. Irreversible inactivation of interleukin 2 in a pump-based delivery environment. Proc Natl Acad Sci USA 93: 5460–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nema S, Washkuhn RJ, Brendel RJ. 1997. Excipients and their use in injectable products. PDA J Pharm Sci Tech 51: 166–171. [PubMed] [Google Scholar]
- 9.Katakam M, Bell LN, Banga AK. 1995. Effect of surfactants on the physical stability of recombinant human growth hormone. J Pharm Sci 84: 713–716. [DOI] [PubMed] [Google Scholar]
- 10.Chang BS, Kendrick BS, Carpenter JF. 1996. Surface-induced denaturation of proteins during freezing and its inhibition by surfactants. J Pharm Sci 85: 1325–1330. [DOI] [PubMed] [Google Scholar]
- 11.Bam NB, Cleland JL, Yang J, Manning MC, Carpenter JF, Kelley RF, Randolph TW. 1998. Tween protects recombinant human growth hormone against agitation-induced damage via hydrophobic interactions. J Pharm Sci 87: 1554–1559. [DOI] [PubMed] [Google Scholar]
- 12.Kim HL, McAuley A, McGuire J. 2014. Protein effects on surfactant adsorption suggest the dominant mode of surfactant-mediated stabilization of protein. J Pharm Sci 103: 1337–1345. [DOI] [PubMed] [Google Scholar]
- 13.Mehta SB, Lewus R, Bee JS, Randolph TW, Carpenter JF. 2015. Gelation of a monoclonal antibody at the silicone oil-water interface and subsequent rupture of the interfacial gel results in aggregation and particle formation. J Pharm Sci 104: 1282–1290. [DOI] [PubMed] [Google Scholar]
- 14.Mittal KL. 1972. Determination of CMC of polysorbate 20 in aqueous solution by surface tension method. J Pharm Sci 61: 1334–1335. [DOI] [PubMed] [Google Scholar]
- 15.Patist A, Bhagwat SS, Penfield KW, Aikens P, Shah DO. 2000. On the measurement of critical micelle concentrations of pure and technical-grade nonionic surfactants. Journal of Surfactants and Detergents 3: 53–58. [Google Scholar]
- 16.Kerwin BA. 2008. Polysorbates 20 and 80 used in the formulation of protein biotherapeutics: structure and degradation pathways. J Pharm Sci 97: 2924–2935. [DOI] [PubMed] [Google Scholar]
- 17.Warne NW. 2011. Development of high concentration protein biopharmaceuticals: the use of platform approaches in formulation development. Eur J Pharm Biopharm 78: 208–212. [DOI] [PubMed] [Google Scholar]
- 18.Martos A, Koch W, Jiskoot W, Wuchner K, Winter G, Friess W, Hawe A. 2017. Trends on analytical characterization of polysorbates and their degradation products in biopharmaceutical formulations. J Pharm Sci 106: 1722–1735. [DOI] [PubMed] [Google Scholar]
- 19.Ecker DM, Jones SD, Levine HL. 2015. The therapeutic monoclonal antibody market. mAbs 7: 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang W, Singh S, Zeng DL, King K, Nema S. 2007. Antibody structure, instability, and formulation. J Pharm Sci 96: 1–26. [DOI] [PubMed] [Google Scholar]
- 21.Padlan EA. 1994. Anatomy of the antibody molecule. Mol Immunol 31: 169–217. [DOI] [PubMed] [Google Scholar]
- 22.Nelson DL, Cox MM. 2017. Lehninger Principles of Biochemistry. Seventh edit, W.H. Freeman and Company, New York, NY. [Google Scholar]
- 23.Freyer MW, Lewis EA. 2008. Isothermal titration calorimetry: experimental design, data analysis, and probing macromolecule/ligand binding and kinetic interactions. Methods Cell Biol 84: 79–113. [DOI] [PubMed] [Google Scholar]
- 24.Deechongkit S, Wen J, Narhi LO, Jiang Y, Park SS, Kim J, Kerwin BA. 2009. Physical and biophysical effects of polysorbate 20 and 80 on darbepoetin alfa. J Pharm Sci 98: 3200–3217. [DOI] [PubMed] [Google Scholar]
- 25.Mahler HC, Senner F, Maeder K, Mueller R. 2009. Surface activity of a monoclonal antibody. J Pharm Sci 98: 4525–4533. [DOI] [PubMed] [Google Scholar]
- 26.Wen ZQ. 2007. Raman spectroscopy of protein pharmaceuticals. J Pharm Sci 96: 2861–2878. [DOI] [PubMed] [Google Scholar]
- 27.Li CH, Nguyen X, Narhi L, Chemmalil L, Towers E, Muzammil S, Gabrielson J, Jiang Y. 2011. Applications of circular dichroism (CD) for structural analysis of proteins: qualification of near- and far-UV CD for protein higher order structural analysis. J Pharm Sci 100: 4642–4654. [DOI] [PubMed] [Google Scholar]
- 28.Dinh NN, Winn BC, Arthur KK, Gabrielson JP. 2014. Quantitative spectral comparison by weighted spectral difference for protein higher order structure confirmation. Anal Biochem 464: 60–62. [DOI] [PubMed] [Google Scholar]
- 29.Harder ME, Deinzer ML, Leid ME, Schimerlik MI. 2004. Global analysis of three-state protein unfolding data. Protein Sci 13: 2207–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Latypov RF, Cheng H, Roder NA, Zhang J, Roder H. 2006. Structural characterization of an equilibrium unfolding intermediate in cytochrome c. J Mol Biol 357: 1009–1025. [DOI] [PubMed] [Google Scholar]
- 31.Mayne L, Englander SW. 2000. Two-state vs. multistate protein unfolding studies by optical melting and hydrogen exchange. Protein Sci 9: 1873–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuwajima K (1995). Circular Dichroism In Protein stability and folding: theory and practice (Shirley B, ed.), pp. 115–135. Humana Press, Totowa, NJ. [Google Scholar]
- 33.Arbogast LW, Brinson RG, Marino JP. 2015. Mapping monoclonal antibody structure by 2D 13C NMR at natural abundance. Anal Chem 87: 3556–3561. [DOI] [PubMed] [Google Scholar]
- 34.Mohajeri E, Noudeh GD. 2012. Effect of Temperature on the Critical Micelle Concentration and Micellization Thermodynamic of Nonionic Surfactants: Polyoxyethylene Sorbitan Fatty Acid Esters. E-Journal of Chemistry 9: 2268–2274. [Google Scholar]
- 35.Hoffmann C, Blume A, Miller I, Garidel P. 2009. Insights into protein-polysorbate interactions analysed by means of isothermal titration and differential scanning calorimetry. Eur Biophys J 38: 557–568. [DOI] [PubMed] [Google Scholar]
- 36.Braun RJ, Parrott EL. 1972. Influence of viscosity and solubilization on dissolution rate. J Pharm Sci 61: 175–178. [DOI] [PubMed] [Google Scholar]
- 37.Singh SM, Cabello-Villegas J, Hutchings RL, Mallela KMG. 2010. Role of partial protein unfolding in alcohol-induced protein aggregation. Proteins 78: 2625–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh SM, Hutchings RL, Mallela KMG. 2011. Mechanisms of m-cresol-induced protein aggregation studied using a model protein cytochrome c. J Pharm Sci 100: 1679–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hutchings RL, Singh SM, Cabello-Villegas J, Mallela KMG. 2013. Effect of antimicrobial preservatives on partial protein unfolding and aggregation. J Pharm Sci 102: 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kiefhaber T, Labhardt AM, Baldwin RL. 1995. Direct NMR evidence for an intermediate preceding the rate-limiting step in the unfolding of ribonuclease a. Nature 375: 513–515. [DOI] [PubMed] [Google Scholar]
- 41.Bis RL, Singh SM, Cabello-Villegas J, Mallela KMG. 2015. Role of benzyl alcohol in the unfolding and aggregation of interferon α-2a. J Pharm Sci 104: 407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mehta SB, Bee JS, Randolph TW, Carpenter JF. 2014. Partial unfolding of a monoclonal antibody: role of a single domain in driving protein aggregation. Biochemistry 53: 3367–3377. [DOI] [PubMed] [Google Scholar]
- 43.Ionescu RM, Vlasak J, Price C, Kirchmeier M. 2008. Contribution of variable domains to the stability of humanized IgG1 monoclonal antibodies. J Pharm Sci 97: 1414–1426. [DOI] [PubMed] [Google Scholar]
- 44.Yan Y, Wei H, Fu Y, Jusuf S, Zeng M, Ludwig R, Krystek SR Jr., Chen G, Tao L, Dass TK. 2016. Isomerization and Oxidation in the Complementarity-Determining Regions of a Monoclonal Antibody: A Study of the Modification-Structure-Function Correlations by Hydrogen-Deuterium Exchange Mass Spectrometry. Anal Chem 88: 2041–2050. [DOI] [PubMed] [Google Scholar]
- 45.Majumdar R, Esfandiary R, Bishop SM, Samra HS, Middaugh CR, Volkin DB, Weis DD. 2015. Correlations between changes in conformational dynamics and physical stability in a mutant IgG1 mAb engineered for extended serum half-life. MAbs 7: 84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garidel P, Hoffmann C, Blume A. 2009. A thermodynamic analysis of the binding interaction between polysorbate 20 and 80 with human serum albumins and immunoglobulins: a contribution to understand colloidal protein stabilisation. Biophys Chem 143: 70–78. [DOI] [PubMed] [Google Scholar]
- 47.Chi EY, Krishnan S, Randolph TW, Carpenter JF. 2003. Physical stability of proteins in aqueous solution: mechanism and driving forces in nonnative protein aggregation. Pharm Res 20: 1325–1336. [DOI] [PubMed] [Google Scholar]
- 48.Goldberg DS, Bishop SM, Shah AU, Sathish HA. 2011. Formulation development of therapeutic monoclonal antibodies using high-throughput fluorescence and static light scattering techniques: role of conformational and colloidal stability. J Pharm Sci 100: 1306–1315. [DOI] [PubMed] [Google Scholar]
- 49.Thiagarajan G, Semple A, James JK, Cheung JK, Shameem M. 2016. A comparison of biophysical characterization techniques in predicting monoclonal antibody stability. mAbs 8: 1088–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kalonia C, Toprani V, Toth R, Wahome N, Gabel I, Middaugh CR, Volkin DB. 2016. Effects of Protein Conformation, Apparent Solubility, and Protein-Protein Interactions on the Rates and Mechanisms of Aggregation for an IgG1Monoclonal Antibody. J Phys Chem B 120: 7062–7075. [DOI] [PubMed] [Google Scholar]
- 51.Neurath H, Bull HB. 1938. The Surface Activity of Proteins. Chem Rev 23: 391–435. [Google Scholar]
- 52.Heitz F, Van Mau N. 2002. Protein structural changes induced by their uptake at interfaces. Biochim Biophys Acta 1597: 1–11. [DOI] [PubMed] [Google Scholar]
- 53.Sharp JS, Forrest JA, Jones RA. 2002. Surface denaturation and amyloid fibril formation of insulin at model lipid-water interfaces. Biochemistry 41: 15810–15819. [DOI] [PubMed] [Google Scholar]
- 54.Kreilgaard L, Jones LS, Randolph TW, Frokjaer S, Flink JM, Manning MC, Carpenter JF. 1998. Effect of Tween 20 on freeze-thawing- and agitation-induced aggregation of recombinant human factor XIII. J Pharm Sci 87: 1597–1603. [DOI] [PubMed] [Google Scholar]
- 55.Thirumangalathu R, Krishnan S, Ricci MS, Brems DN, Randolph TW, Carpenter JF. 2009. Silicone oil- and agitation-induced aggregation of a monoclonal antibody in aqueous solution. J Pharm Sci 98: 3167–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu L, Qi W, Schwartz DK, Randolph TW, Carpenter JF. 2013. The effects of excipients on protein aggregation during agitation: an interfacial shear rheology study. J Pharm Sci 102: 2460–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kerwin BA, Heller MC, Levin SH, Randolph TW. 1998. Effects of tween 80 and sucrose on acute short-term stability and long- term storage at −20 °C of a recombinant hemoglobin. J Pharm Sci 87: 1062–1068. [DOI] [PubMed] [Google Scholar]
- 58.Joshi O, Chu L, McGuire J, Wang DQ. 2009. Adsorption and function of recombinant Factor VIII at the air-water interface in the presence of Tween 80. J Pharm Sci 98: 3099–3107. [DOI] [PubMed] [Google Scholar]
- 59.Joshi O, McGuire J, Wang DQ. 2008. Adsorption and function of recombinant factor VIII at solid-water interfaces in the presence of Tween-80. J Pharm Sci 97: 4741–4755. [DOI] [PubMed] [Google Scholar]
- 60.Ludwig DB, Carpenter JF, Hamel JB, Randolph TW. 2010. Protein adsorption and excipient effects on kinetic stability of silicone oil emulsions. J Pharm Sci 99: 1721–1733. [DOI] [PubMed] [Google Scholar]
- 61.Petkov JT, Gurkov TD, Campbell BE, Borwankar RP. 2000. Dilatational and shear elasticity of gel-like protein layers on air/water interface. Langmuir 16: 3703–3711. [Google Scholar]
- 62.Chou DK, Krishnamurthy R, Randolph TW, Carpenter JF, Manning MC. 2005. Effects of Tween 20® and Tween 80® on the stability of Albutropin during agitation. J Pharm Sci 94: 1368–1381. [DOI] [PubMed] [Google Scholar]
- 63.Bam NB, Randolph TW, Cleland JL. 1995. Stability of protein formulations: Investigation of surfactant effects by a novel EPR spectroscopic technique. Pharm Res 12: 2–11. [DOI] [PubMed] [Google Scholar]
- 64.Bam NB, Cleland JL, Randolph TW. 1996. Molten globule intermediate of recombinant human growth hormone: stabilization with surfactants. Biotechnol Prog 12: 801–809. [DOI] [PubMed] [Google Scholar]
- 65.Jones LS, Cipolla D, Liu J, Shire SJ, Randolph TW. 1999. Investigation of protein-surfactant interactions by analytical ultracentrifugation and electron paramagnetic resonance: the use of recombinant human tissue factor as an example. Pharm Res 16: 808–812. [DOI] [PubMed] [Google Scholar]
- 66.Budyak IL, Doyle BL, Weiss IV WF. 2015. Technical decision-making with higher order structure data: Specific binding of a nonionic detergent perturbs higher order structure of a therapeutic monoclonal antibody. J Pharm Sci 104: 1543–1547. [DOI] [PubMed] [Google Scholar]
- 67.Arbogast LW, Brinson RG, Formolo T, Hoopes JT, Marino JP. 2016. 2D (1)H(N), (15)N correlated NMR methods at natural abundance for obtaining structural maps and statistical comparability of monoclonal antibodies. Pharm Res 33: 462–475. [DOI] [PubMed] [Google Scholar]
- 68.Treuheit MJ, Kosky AA, Brems DN. 2002. Inverse relationship of protein concentration and aggregation. Pharm Res 19: 511–516. [DOI] [PubMed] [Google Scholar]
- 69.Webb SD, Cleland JL, Carpenter JF, Randolph TW. 2002. A new mechanism for decreasing aggregation of recombinant human interferon-gamma by a surfactant: slowed dissolution of lyophilized formulations in a solution containing 0.03% polysorbate 20. J Pharm Sci 91: 543–558. [DOI] [PubMed] [Google Scholar]
- 70.Wang W, Wang YJ, Wang DQ. 2008. Dual effects of Tween 80 on protein stability. Int J Pharm 347: 31–38. [DOI] [PubMed] [Google Scholar]
- 71.Agarkhed M, O’Dell C, Hsieh MC, Zhang J, Goldstein J, Srivastava A. 2013. Effect of polysorbate 80 concentration on thermal and photostability of a monoclonal antibody. AAPS PharmSciTech 14: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Japelj B, Ilc G, Marusic J, Sencar J, Kuzman D, Plavec J. 2016. Biosimilar structural comparability assessment by NMR: from small proteins to monoclonal antibodies. Sci Rep 6: 32201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yagi H, Zhang Y, Yagi-Utsumi M, Yamaguchi T, Iida S, Yamaguchi Y, Kato K. 2015. Backbone (1)H, (13)C, and (15)N resonance assignments of the Fc fragment of human immunoglobulin G glycoprotein. Biomol NMR Assign 9: 257–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. 2D 13C-1H HMQC NMR spectra of mAb (black) and its Fab (red) and Fc (green) fragments. Isolated Fab and Fc have similar structure as that in the intact mAb.
Figure S2. SEC indicates that both Fab and Fc fragments exist as monomers after NMR experiments in the presence of polysorbates.
Figure S3. Addition of PS20 caused chemical shift changes in the Fab region compared to PS80.


