Significance
De novo protein synthesis in neuronal axons plays important roles in neural circuit formation, maintenance, and disease. Key to the selectivity of axonal protein synthesis is whether an mRNA is present at the right place to be translated, but the mechanisms behind axonal mRNA localization remain poorly understood. In this work, we quantitatively analyze the link between axonal β-actin mRNA trafficking and its localization patterns. By developing a single-molecule approach to live-image β-actin mRNAs in axons, we explore the biophysical drivers behind β-actin mRNA motion and uncover a mechanism for generating increased density at the axon tip by differences in motor protein-driven transport speeds. These results provide mechanistic insight into the control of local translation through mRNA trafficking.
Keywords: RNA localization, axon, β-actin, molecular beacon, single molecule
Abstract
During embryonic nervous system assembly, mRNA localization is precisely regulated in growing axons, affording subcellular autonomy by allowing controlled protein expression in space and time. Different sets of mRNAs exhibit different localization patterns across the axon. However, little is known about how mRNAs move in axons or how these patterns are generated. Here, we couple molecular beacon technology with highly inclined and laminated optical sheet microscopy to image single molecules of identified endogenous mRNA in growing axons. By combining quantitative single-molecule imaging with biophysical motion models, we show that β-actin mRNA travels mainly as single copies and exhibits different motion-type frequencies in different axonal subcompartments. We find that β-actin mRNA density is fourfold enriched in the growth cone central domain compared with the axon shaft and that a modicum of directed transport is vital for delivery of mRNA to the axon tip. Through mathematical modeling we further demonstrate that directional differences in motor-driven mRNA transport speeds are sufficient to generate β-actin mRNA enrichment at the growth cone. Our results provide insight into how mRNAs are trafficked in axons and a mechanism for generating different mRNA densities across axonal subcompartments.
Localizing mRNA to different subcellular locations is an evolutionarily conserved mechanism to control protein expression. For example, it facilitates axis patterning of the Drosophila embryo and mating-type switching in budding yeast and promotes the migration of mammalian fibroblasts (reviewed in ref. 1). In neurons across different animal phyla, mRNA trafficking into neurites plays an important role in brain development. During the establishment of neural connectivity, growing axons must navigate toward their synaptic partners by responding rapidly to molecular signals in the environment. With axons extending long distances from the soma, local translation of trafficked mRNAs delivers functional autonomy to these subcellular compartments by letting them control their own proteome. Distal neurites can thus respond quickly to particular extracellular signals with high spatiotemporal precision as well as locally mediate requirements for their growth and survival (reviewed in ref. 2). The importance of transporting mRNAs into axons is highlighted by the sheer range of roles their local translation plays in nervous system assembly, spanning axon navigation, elongation, and synapse formation (3–8). Not surprisingly, therefore, deregulated axonal mRNA trafficking and translation are linked to the pathology of various neurological disorders, including fragile X syndrome and amyotrophic lateral sclerosis (9).
mRNA localization has been demonstrated to be a primary factor determining the local proteome in neurites (10) and consequently is under meticulous control to prevent aberrant protein expression. Tight regulation of mRNA-localization patterns ensures axons are enriched with different sets of mRNAs to their cell body (11–14). mRNA-localization patterns also vary across the axon itself, where differential enrichment occurs between the sensing growth cone tip and the axon shaft according to mRNA species (12, 15). Proper targeting requires the mRNA to undergo a series of processing events. mRNA transcribed in the nucleus is packaged into large protein complexes called “ribonucleoproteins” (RNPs) through association with RNA-binding proteins (RBPs) (reviewed in ref. 16). After nuclear export, RNPs may be further remodeled, and the mRNA is trafficked to sites of translation (16). It is thought that specificity in recognition by particular RBPs through cis-acting elements in the mRNA determines where in the neuron an mRNA becomes subcellularly localized (2). In the neuron’s dendritic compartment, mRNA localization is achieved through bidirectional directed transport of RNPs interspersed with long stationary periods (see below) (17–19). Subsequent anchoring at dendritic synapses and local translation are important for plasticity and long-term memory formation (20). In contrast, the mechanism by which mRNAs become localized in the axonal compartment remains less clear. We do not know how mRNAs that localize to axons are packaged in RNPs (for example, do multiple copies of the same mRNA travel together?), nor do we know precisely how they move and how this movement links to intraaxonal mRNA localization patterns.
There are several ways by which mRNAs can be delivered to their target destinations in eukaryotic cells. The simplest mechanism is based on diffusion and entrapment and has been observed in Xenopus and Drosophila oocytes (21, 22); for example, Drosophila nanos mRNA diffuses both freely and via microtubule-dependent cytoplasmic flow and then becomes anchored posteriorly through interaction with actin filaments (21). The second mechanism is biased directed transport by motor proteins. RNPs can associate directly with motor proteins that drive processive motion along cytoskeletal tracks. In some instances, binding of opposing motors occurs, which results in bidirectional behavior (23). Here, a bias in movement frequency in one direction can drive polarized localization (24, 25). Third, the “sushi belt” model of directed transport coupled with local entrapment has been proposed to take place in neuronal dendrites (20). Rather than specifically trafficking mRNA to predetermined sites, the model proposes that mRNAs continually circulate throughout dendritic trees by motor-driven bidirectional transport before being captured at activated synapses. Such a mechanism has been directly visualized for β-actin mRNA docking at dendritic spines upon glutamate stimulation (19). Finally, localized protection of mRNAs from degradation may play a role in enforcing localization patterns (26). Although mRNA degradation has already been shown to temporally regulate local translation during axon navigation (6), how it might influence intraaxonal localization patterns is currently unknown. One way to understand the contribution of these different mechanisms for mRNA localization in the axon is by using live-cell imaging to quantify exactly how a differentially enriched mRNA species moves. As one of the best-characterized mRNAs in neurons, β-actin represents an attractive subject.
β-Actin mRNA is highly abundant in dendrites of adult neurons and is enriched in the growth cone of axons during development (27, 28). β-Actin mRNA in retinal axons is abundantly translated in vivo (29) and in vitro in response to extrinsic cues (4) and moreover is one of the most abundant basally synthesized nascent proteins (30). The cis-acting sequences in β-actin mRNA and interacting RBPs have been studied extensively (31), as have the physiological roles for localized β-actin synthesis in neurons. Here, subcellular localization of β-actin mRNA allows site-specific synthesis of new actin monomers that is thought to promote localized actin filament assembly, causing changes in cytoskeleton architecture and hence morphology. In mature dendrites, β-actin mRNA translation is associated with structural changes in dendritic spines that underlie synaptic plasticity (32). In contrast, during development axonal translation of β-actin mRNA is necessary for attractive turning in vitro and for branching both in vivo and in vitro (4, 5, 29, 33). Given high translation levels and observations of differential enrichment across distinct regions of the axon, β-actin mRNA provides an interesting example for investigating the connection between mRNA trafficking and intraaxonal mRNA-localization patterns during development.
Live imaging provides an approach to decipher the mechanisms behind mRNA trafficking in cells. Here, one can extract the mechanisms driving mRNA localization through quantitative information held in their trajectories. Single molecules of endogenous mRNA have previously been visualized in neurons through genetically encoded reporters (reviewed in ref. 34). Here, we were interested in visualizing endogenous mRNA dynamics in neuronal compartments at a single-molecule level without genetic modification. To do this, we adapted molecular beacon technology to label single molecules of endogenous, unmodified β-actin mRNA in growing Xenopus retinal ganglion cell (RGC) axons, visualized with highly inclined and laminated optical sheet (HILO) microscopy. Using this system, combined with automated tracking, biophysical motion models, and single-molecule FISH (smFISH), we characterize the stoichiometry and trafficking behavior of β-actin mRNA in axons. We show that β-actin mRNA exhibits different density distributions and motion-type frequencies within different subcompartments of the growing axon. We further demonstrate mathematically that diffusion alone cannot explain axonal mRNA trafficking to peripheral regions but that the differences between anterograde- and retrograde-directed transport speeds are sufficient to confer the growth cone-enriched localization patterns quantitatively observed.
Results
Chimeric Molecular Beacons Label Endogenous β-Actin mRNA Molecules in Axons.
To investigate how mRNAs move within growing axons, we employed an approach using molecular beacons (MBs). MBs are composed of a short antisense oligonucleotide loop complementary to the mRNA of interest followed by a GC-rich stem, placing a fluorophore and quencher in close proximity. Hybridization of the antisense loop region to the mRNA of interest generates a conformational change, releasing fluorophore from quencher and subsequently labeling the mRNA of interest. Although proposed 20 y ago (35), MBs are little used, because robust methodologies have been developed to fluorescently tag exogenous mRNA (36), as have genetically encoded alternatives such as the MS2 system (36, 37), an approach in which the mRNA of interest is engineered to contain multiple MS2-binding sites visualized through MS2 coat protein (MCP)-GFP coexpression that binds dimerically to each MS2 stem–loop. MBs provide an alternative method for investigating the movement of endogenous mRNA and have the advantage of allowing us to follow single molecules of genetically unmodified mRNA.
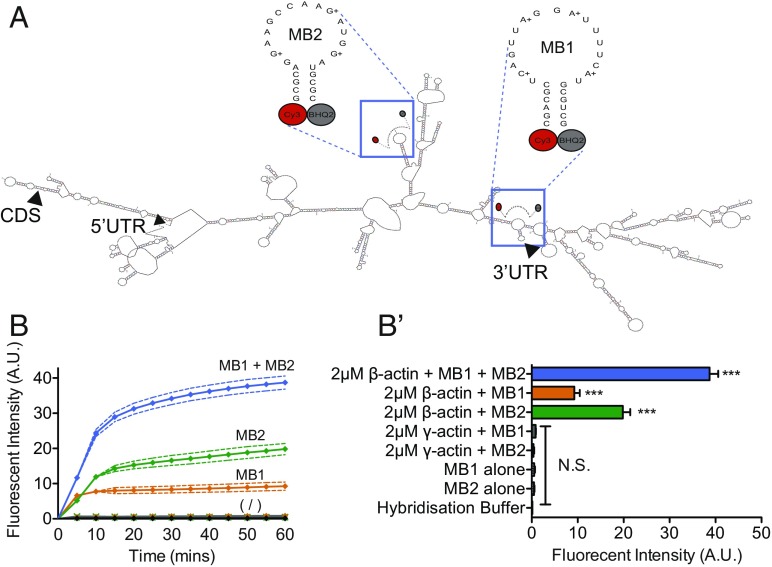
Several studies have elegantly examined the physicochemical properties of MBs (38–40) and have demonstrated their ability to mimic expected mRNA-localization patterns in living cells (41, 42). However, their suitability for live-cell imaging has yet to be inclusively validated, as MBs have not directly been shown to label an endogenous mRNA of interest within a living cellular context. We addressed this question using two MBs (MB1 and MB2) comprised of nuclease-resistant 2′O-methyl RNA/locked nucleic acid (LNA) chimeras designed to target the coding sequence of β-actin mRNA (Fig. 1A). Such chimeric design has been demonstrated to enhance both hybridization efficiency and probe specificity (43, 44). In vitro addition of full-length β-actin mRNA dramatically increased the fluorescent intensity of solutions containing MB1 and MB2, by ninefold and 20-fold, respectively (Fig. 1B). MB1 and MB2 generated different increases in fluorescence in response to the addition of β-actin mRNA, suggesting that different MB sequences have different hybridization kinetics for labeling β-actin mRNA. In contrast, γ-actin mRNA, an actin isoform with 87% coding sequence similarity to β-actin mRNA (www.xenbase.org/entry/) and differing from the MB1 target sequence by only one nucleotide, did not trigger an increase in fluorescent intensity (Fig. 1B). Moreover, while the fluorescent signal was proportional to the concentration of β-actin mRNA present, two MBs in combination increased fluorescence by more than 30-fold 60 min after the addition of β-actin mRNA compared with each MB alone without β-actin mRNA, suggesting that signal-to-noise ratio (SNR) per mRNA could be amplified by combining two MBs that target different regions of the β-actin mRNA coding sequence (Fig. 1B and SI Appendix, Fig. S1A).
Fig. 1.
Molecular beacons selectively label β-actin mRNA to single-nucleotide precision in vitro. (A) Schematic showing the predicted secondary structure of the sequences of each molecular beacon used to target β-actin mRNA (mFOLD) and the molecular beacon sequences used to target predicted single-stranded regions in the mRNA. Arrowheads indicate the start site of the 5′ UTR, coding sequence (CDS), and 3′ UTR. (B) The addition of 2-μM β-actin mRNA to 1-μM solutions of MB1 or MB2 significantly increased the fluorescent intensity of the solutions compared with MBs in hybridization solution alone and cumulatively amplified fluorescence further when 2-μM β-actin mRNA was added to 1-μM solutions of MB1 and MB2 together (MB1 + MB2). Below the symbol (/) are the changes in fluorescence over time for 1 μM MB1 alone, 1 μM MB2 alone, hybridization buffer alone, 2 μM γ-actin + MB1, and 2 μM γ-actin + MB2. In contrast, the addition of 2-μM γ-actin mRNA did not elicit a change in fluorescent intensity in solutions containing 1 μM of either MB1 or MB2. (B′) For a clearer comparison, B′ shows the fluorescent intensity of each condition at 60 min. ***P < 0.0001 unpaired Students t test; N.S., not significant; n = 3 replicates per condition.
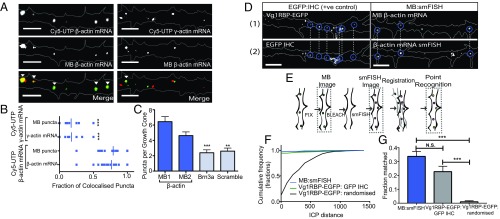
We next examined MB1 and MB2 in direct comparison with fluorescently tagged Cy5-β-actin mRNA in live RGC axons. Electroporation into eye primordia resulted in dynamic Cy3-MB puncta within axons (SI Appendix, Fig. S1B and Movie S1). Codelivery of MBs along with fluorescently tagged Cy5-UTP β-actin mRNA into RGCs by eye electroporation followed by quantitative axonal analysis revealed that on average 77 ± 20% of all Cy5-UTP β-actin mRNA puncta colocalized with MB puncta, and 80 ± 20% of the MB puncta population colocalized with Cy5-UTP β-actin mRNA (Fig. 2 A and B). The colocalized signals shared the same dynamics, moving together for the duration of the movie. In contrast, significantly less colocalization was observed between MB puncta and Cy5-UTP γ-actin mRNA (17.6 ± 6.9% and 17.1 ± 6.7% for exogenous mRNA and MB puncta, respectively, P < 0.0001 unpaired t test) (Fig. 2 A and B). We then investigated the specificity of MBs targeting β-actin mRNA further by comparing axonal expression with that of control MBs targeting (i) Brn3a mRNA that is not expressed in RGC axons (30) and (ii) a scrambled sequence that was not predicted to target other mRNAs in the genome. We found significantly decreased expression in our control MBs compared with each β-actin mRNA-targeting MB individually, indicating that while our mRNA of interest is indeed labeled, a level of background labeling may also occur (P < 0.001 and P = 0.001, unpaired t test for Brn3a- and scramble-targeting MBs, respectively) (Fig. 2C).
Fig. 2.
Molecular beacons label β-actin mRNA in growing axons. (A) MBs colocalize significantly more with exogenous fluorescently tagged β-actin mRNA than with exogenous fluorescently tagged γ-actin mRNA in growing RGC axons. (Scale bars: 5 μm.) (B) Quantification of colocalization between β-actin mRNA-targeting MBs and exogenous Cy5-UTP β-actin mRNA and Cy5-UTP γ-actin mRNA. P < 0.0001, n = 21 axons and 11 axons, respectively. (C) MBs targeting β-actin mRNA showed significantly more puncta per growth cone than control MBs targeting Brn3a mRNA or a scrambled sequence. **P = 0.0043, ***P = 0.001, unpaired t test. n = 64, 73, 82, and 86 axons for β-actin MB1, β-actin MB2, Brn3a, and the scrambled sequence, respectively. (D) MBs and smFISH puncta could be observed to mark the same place in nonsimultaneous images, similar to EGFP puncta and IHC against EGFP. (Scale bars: 5 μm.) (E) Schematic showing the workflow for the registration of nonsimultaneous images used to quantify colocalization. (F) Cumulative frequency distribution of ICP distance between matched puncta in MB and smFISH images (n = 14 axons) and in Vg1RBP-EGFP and IHC positive and randomized negative controls (n = 11 axons). (G) Quantification of colocalization based on an ICP distance threshold of 1. ***P < 0.0001, unpaired t test; N.S., not significant.
To investigate whether MBs label endogenous β-actin mRNA in RGC axons, we mapped MB puncta immediately postfixation in relation to β-actin mRNA puncta visualized after performing smFISH on the same axon. The MB signal does not survive the smFISH protocol. Nonetheless, between these steps, samples were bleached using the 561-nm laser line to avoid any possibility of residual MB signal in smFISH images (as we found smFISH worked best using the Cal590 fluorophore). By eye, MBs were commonly observed at the same sites as endogenous β-actin mRNA labeled by smFISH in the same axon between nonsimultaneous images (Fig. 2D). This co-occurrence was similar to that observed by comparing images from live Vg1RBP-EGFP puncta immediately postfixation to immunostaining against EGFP (Fig. 2D).
Because the smFISH protocol causes distortion in axon morphology, the MB and smFISH images, which were acquired at two separate time points in the protocol, were not entirely in register. Therefore, to quantify the degree of colocalization between nonsimultaneous images, we developed a MATLAB-based script for semiautomated registration as well as colocalization analysis. Our registration script aligned two nonsimultaneous images to one another based on brightfield images of the same axon pre- and post-smFISH. Initially, user identification of brightfield landmarks in the axon (such as branches) guides a point-based manual registration; then an automated iterative closest point (ICP) algorithm (45) is used for the final registration of images. The ICP-transformed distance was then employed to quantify the probability of colocalization between MB and smFISH puncta (Fig. 2E and SI Appendix, Fig. S2 A and B). As a positive control, we analyzed colocalization between the fluorescent signal of the RNA-binding protein Vg1RBP-EGFP immediately postfixation and the immunohistochemistry (IHC) signal for GFP. These data were then compared with a randomized negative control in which Vg1RBP-EGFP puncta were registered to IHC that had been randomized in the same axon. Comparison of the normalized cumulative frequency showed a similar distribution of ICP distances between matched MB and smFISH puncta to the positive control, whereas substantially larger ICP distances were observed in the randomized control (Fig. 2F). Likewise, after setting an ICP distance threshold for matched puncta as 1, and including the population of puncta that were initially identified as unmatched by the script, similar fractions of puncta were predicted to be colocalized between the MB:smFISH puncta and the positive control (33.8 ± 3.6% and 22.8 ± 3.9%, respectively) (Fig. 2F). In contrast, a significantly smaller fraction of the randomized negative control was predicted to colocalize (1.0 ± 0.8%, P < 0.001, unpaired t test). These findings are more valuable as indications of relative rather than absolute percentages of colocalization, as we cannot completely match the same axon in two nonsimultaneous images and thus likely underestimate the number of colocalized puncta in the positive control and MB:smFISH conditions. In addition, although the relatively large degree of unmatched puncta in the first MB images could suggest that some nonspecific labeling occurs, the finding that levels of colocalization are not significantly different from the positive EGFP control, and that we also observe fewer puncta after IHC against EGFP compared with immediately after fixation (the average decrease is 26 ± 8%) also indicates that protein and RNA degradation occurs after the first images are captured. Even with strict RNase-free conditions, some degradation is inevitable during such protocols. Indeed, previous studies demonstrating a high correlation between smFISH-determined transcript number and estimated mRNA levels, compare smFISH to mRNA copy number estimations calculated using techniques such as qPCR and RNA-sequencing (RNA-seq) (46–48) that predict relative rather than absolute mRNA levels and thus cannot account for mRNA degradation that might take place. Importantly, the fact that we label β-actin mRNA with single-nucleotide specificity using MBs in vitro and observe high levels of colocalization between exogenous β-actin mRNA and MBs in living axons and levels of colocalization to endogenous β-actin mRNA comparable to those in our positive control strongly suggests that we label β-actin mRNA in growing axons using MBs. In addition, we found that endogenous β-actin protein levels remain the same in MB-labeled axons and observed comparable fluorescence recovery after photobleaching using an EGFP protein synthesis reporter with MBs (SI Appendix, Fig. S3 A–E). These data suggest that MB labeling does not affect β-actin mRNA translation. During translation, however, it is possible that the MBs become detached as ribosomes progress along the mRNA. Together, these findings demonstrate that MBs provide a good method to label endogenous mRNA in axons.
Imaging β-Actin mRNA with Single-Molecule Sensitivity Reveals the Majority of β-Actin mRNAs Exist Singly in Growing Axons.
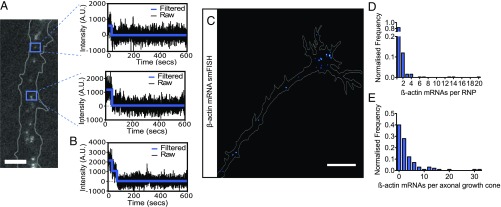
MBs have not previously been considered a technique with single-molecule sensitivity unless the mRNA of interest is labeled with multiple fluorophores (49–52). As unmodified endogenous mRNA is highly structured with few predicted single-stranded regions accessible as MB-binding sites, labeling one mRNA with multiple MBs is, in practice, possible only using engineered exogenous mRNA or through genetic modification. How then might we visualize single, unmodified mRNA molecules using a few MBs? One approach would be to increase the SNR through single-molecule microscopy methods such as HILO microscopy (53). Here, analysis of single-protein dynamics in cells can be performed using a single genetically encoded fluorophore such as GFP, which is considerably less bright and photostable than the organic fluorophores used with MBs (54, 55). Theoretically, therefore, if appropriately spaced, single mRNA molecules should also be trackable for short time periods using only one organic fluorophore with MBs. To test whether this was the case, we counted the number of fluorophores associated with each observed mRNA granule through stepwise photobleaching (56). Using a single MB (MB1), one fluorophore would equate to one mRNA molecule; thus single stepwise photobleaching events would suggest we were visualizing single molecules. Reassuringly, we were able to detect single stepwise photobleaching events, suggesting the detection of single mRNA molecules using only one fluorophore (Fig. 3A). Interestingly, some examples of multiple stepwise photobleaching events were also observed (Fig. 3B), indicating that multiple β-actin mRNAs could also exist within RNPs. It has been widely reported that fluctuations in background intensity generate noise in single-molecule photobleaching data (57). The use of the HILO microscopy approach here meant that the sizes of stepwise events depend on the plane and field of illumination. Additionally, because one cannot accurately count larger numbers of photobleaching steps using this technique (57), we decided to explore β-actin mRNA RNP stoichiometry in axons further using the smFISH approach, as described below. Further experiments investigating single-molecule sensitivity, such as a comparison of synthetic RNA-MB hybrids (49), could not be conducted due to technical limitations of microinjection into RGCs within eye explants. However, taken together, the facts that we (i) detected single stepwise photobleaching events, (ii) observed colocalization with smFISH puncta, and (iii) used a single-molecule imaging approach capable of imaging the dynamics of single fluorophores that are dimmer than the organic Cy3 fluorophores used with MBs strongly suggest that we are able to image endogenous β-actin mRNA molecules with single-molecule sensitivity using the MB approach in RGC axons.
Fig. 3.
Endogenous β-actin mRNA stoichiometry in growing axons. (A) Single stepwise photobleaching events were observed using MB1 and HILO microscopy. Black lines represent background-subtracted intensity trace of MB puncta; colored lines show the median intensity of each step after filtering. (Scale bar: 5 μm.) (B) An example of two stepwise photobleaching events in one trace. (C) Representative image of β-actin mRNA distribution using smFISH. (Scale bar: 10 μm.) (D) Frequency distribution of the number of β-actin mRNAs per RNP calculated from smFISH spot intensity distributions. (E) Variability in the number of β-actin mRNAs within axonal growth cones was observed. n = 35 RNase-A–treated axons and n = 100 untreated axons.
To further probe the stoichiometry of β-actin mRNA within axonal RNPs independently, we used smFISH (Fig. 3C). Here we counted mRNA copy number through Gaussian intensity distributions of smFISH puncta using automated image-analysis software to eliminate user bias (58). To ensure the lowest puncta intensity was not a consequence of nonspecific labeling, we used RNase A-treated axons as a negative control for background by comparing the intensity distributions in the two conditions (SI Appendix, Fig. S4A). Here, the intensity of one mRNA molecule was calculated by subtracting the background observed from the RNase A-treated population (SI Appendix, Fig. S4B). We found that the great majority of β-actin mRNAs existed singly (84%), while a smaller frequency was observed for packaging β-actin mRNA in twos (12%). However, up to 20 β-actin mRNAs per RNP could be observed occasionally (Fig. 3D). Because the brighter, higher-copy-number puncta often appeared larger, it is possible that these might constitute stress granules or p-bodies.
Our stoichiometry analysis also revealed that a large degree of cell-to-cell variability existed for the total number of β-actin mRNAs in axons, ranging from 0 to 32 molecules per axonal growth cone, with most holding fewer than four (Fig. 3E and SI Appendix, Fig. S4 D and E). However, the expected mRNA degradation that we previously observed during our smFISH protocol means that total copy number and stoichiometry may be underestimated. To test how the stoichiometry of smFISH datasets might be affected by RNA degradation during our in situ hybridization protocol, we performed binomial fitting using a relatively high arbitrary degradation constant, P = 0.5 (50% probability of degradation). Here, binomial fitting allows us to predict the frequency of number of β-actin mRNAs existing within an RNP by solving each possibility individually through an equation and then summing the frequency of expected values. For example, for two β-actin mRNAs per RNP, there would be a certain probability of observing smFISH puncta intensity representative of two, one, or zero β-actin mRNAs per RNP, depending on how many have degraded. To limit the number of equations, we performed binomial fitting for up to four β-actin mRNAs per RNP, which comprises >98% of our original observed smFISH distribution. After degradation is taken into account, the resulting distribution of β-actin mRNA smFISH stoichiometry was slightly more right-skewed, with a relatively small difference in copy number frequency from that observed (less than 20% for estimated frequency distributions of single- and double-copy-number–containing RNPs) (SI Appendix, Fig. S4C). These data together show that, even with a theoretically large degree of degradation during our smFISH protocol, most β-actin mRNAs travel singly in growing axons.
Differences in Directed Transport Are Sufficient to Drive Differential β-Actin mRNA-Localization Patterns Across Axonal Subcompartments.
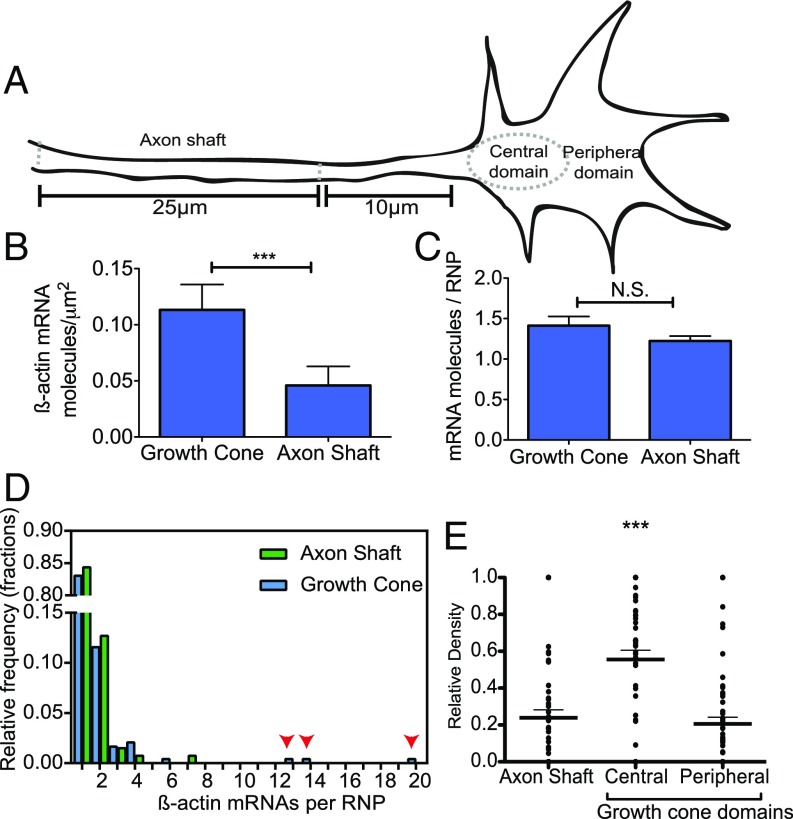
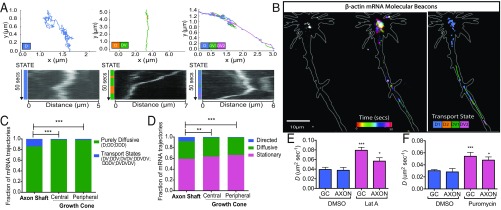
Using smFISH, we further quantitatively examined how endogenous β-actin mRNA-localization patterns might change across different axonal subcompartments (Fig. 4A). Consistently we found a greater number of mRNA molecules per square micrometer in the growth cone than in the axon shaft (Fig. 4B). No difference in mean β-actin mRNA RNP stoichiometry was observed (Fig. 4C), although more highly multiplexed RNPs tended to reside in the growth cone (arrows in Fig. 4D). Probing these localization patterns further, we found the greatest density of β-actin mRNA molecules was located in the central domain, where fourfold enrichment was observed, compared with the peripheral domain of the growth cone and axon shaft (n = 63 axons, P < 0.0001, paired t test) (Fig. 4E).
Fig. 4.
smFISH reveals that the localization patterns of β-actin mRNA vary according to axonal subcompartment. (A) Cartoon representation of the different axonal regions analyzed. (B) A comparison of the density of β-actin mRNA molecules in the growth cone and axon shaft via smFISH shows significantly increased density of β-actin mRNA in the growth cone. (C) No difference between β-actin mRNA RNP stoichiometry was observed in the different subcompartments. N.S. not significant. (D) Histogram of β-actin mRNA stoichiometry. Arrowheads indicate more highly multiplexed copy numbers in the growth cone. (E) Relative density of β-actin mRNA across subcompartments of the same axon shows significant enrichment in the central domain of the growth cone. ***P < 0.0001; paired Student’s t test; n = 63 axons.
We wondered whether mRNA trafficking might play a role in conferring central domain-enriched localization of β-actin mRNA. To address this question, we analyzed β-actin mRNA dynamics in growing axons using MBs imaged under HILO microscopy. To increase the SNR and enable longer-term imaging, we used two MBs together in these live-imaging experiments. By adapting a particle-tracking script (59), we were able to automatically detect puncta, obtain trajectories, and classify the directionality of mRNA moving anterograde or retrograde in the axon. MB-labeled β-actin mRNA puncta frequently displayed complex motions in axons. For example, puncta underwent fast directional transport interspersed with periods of pausing or switching directionality midtrajectory (Fig. 5 A and B and Movies S2–S4). The biophysical drivers behind each trafficking mode were consequently extracted using HMM–Bayes, an approach that combines hidden Markov modeling with Bayesian model selection to predict switches in diffusive and directed transport states (60). Here, puncta displacements within each β-actin mRNA trajectory are treated as a finite series of hidden motion states, with the type of motion (i.e., different diffusive or directed transport states) depending on the mean and SD of displacements within the trajectory (60). For example, diffusion, by definition, has no net directionality and thus would have a mean centered on zero, while different types of diffusion would have different SDs in displacement that vary by the level of puncta confinement. The script then uses Bayesian statistics to predict the simplest stochastic set of motion states that best fit the observed β-actin mRNA displacements within the trajectory (60).
Fig. 5.
Differential subcompartmental trafficking of β-actin mRNA. (A) Examples of complex motion trajectories displayed by β-actin mRNA in axons. Different transport modes were characterized through HMM–Bayes. D, diffusive states; DV, directed transport states. (B) β-Actin mRNA dynamics within a representative axon. (Left) Initial β-actin mRNA distribution labeled by MBs. (Center) Temporally color-coded β-actin mRNA dynamics. (Right) Annotated mRNA trajectories. (C) Significantly more trajectories undergo motion with directed transport in the axon shaft than in the central and peripheral domains of the growth cone. (D) Trajectories annotated according to the predominant motion state again show a significantly greater fraction of directed transport of β-actin mRNA in the axon shaft. ***P < 0.001, **P < 0.0025; χ2 test; n = 6, 66 axons. (E) The addition of latrunculin A significantly increased the diffusive mobility of MB β-actin mRNA puncta compared with DMSO treatment only [***P < 0.0001 and *P = 0.0454 for growth cone (GC) and axon shaft; n = 12 axons for DMSO treatment, and n =11 axons for latrunculin A]. (F) Dissociation from ribosomes by puromycin addition increased the diffusion coefficient of MB puncta in both axons and growth cones compared with wild-type untreated axons [n = 11 puromycin-treated axons and n = 66 wild-type axons, ***P < 0.0001 (growth cone) and *P = 0.0253 (axon)].
Using the HMM–Bayes approach, we found that while most β-actin mRNA puncta assumed diffusive behavior at all axonal regions analyzed, a modicum of trajectories containing directed transport states occurred in the axon shaft (14.0 ± 3.3% SEM). In contrast, directed transport was rarely observed in any region of the growth cone (central domain 1.6 ± 0.8% SEM; peripheral domain 2.0 ± 1.1% SEM). These differences resulted in significantly different motion-type frequencies in the axon shaft but not between the peripheral and central domains of the growth cone (P < 0.0001, n = 66 axons, χ2 test) (Fig. 5C). Further characterization based on the predominant motion type of each β-actin mRNA trajectory yielded similar results (Fig. 5D). Here, we additionally characterized diffusive particles as “anchored” based on the range of diffusion coefficients exhibited by stationary MB puncta in fixed axons.
While microtubule-dependent transport has been shown to be important for directed motion in axons (28, 61, 62), we wondered whether actin filaments might play a role in anchoring β-actin mRNA, as has been observed in fibroblasts (18). In support of this notion, we found that disrupting the actin cytoskeleton by latrunculin A did not prevent directed transport but instead increased the diffusive mobility of β-actin mRNA (P = 0.0387 and P < 0.0001 for axon and growth cone, respectively, unpaired t test) (Fig. 5E). Interestingly, the effect of actin destabilization on diffusion was greater in the growth cone than in the axon shaft, corresponding to increased F-actin levels at this region (63). Triggering ribosome dissociation from mRNA by the addition of puromycin (64) was also able to increase the diffusion coefficient, indicating that some MB-labeled RNPs were bound to ribosomes (P < 0.0001, unpaired t test) (Fig. 5F).
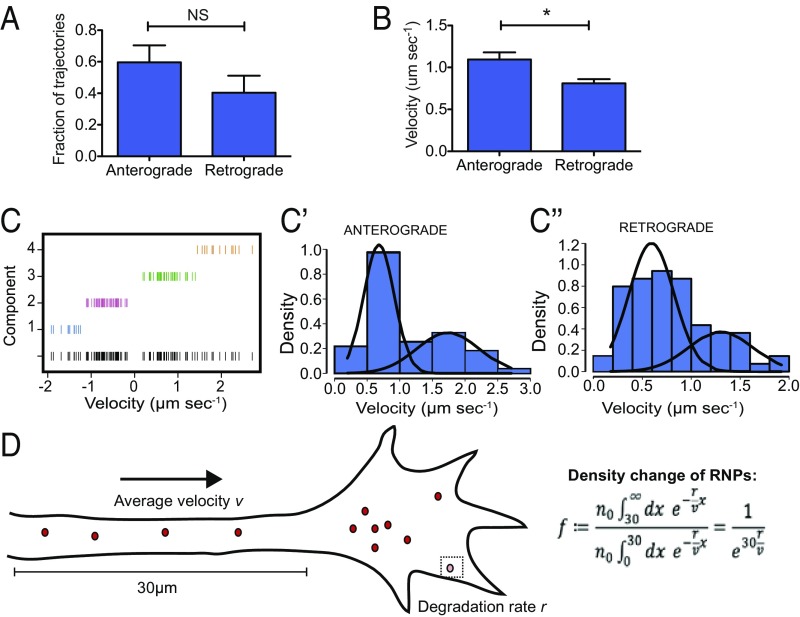
There was no significant bias in the proportion of β-actin mRNA puncta within the axon shaft traveling toward or away from the growth cone. This suggests that the enrichment of β-actin mRNA in the growth cone is not be due to an increase in the frequency of anterograde-directed transport (Fig. 6A). However, this enrichment could be fueled by the differences in the speed of directed transport we observed for mRNAs moving anterogradely versus retrogradely (1.1 ± 0.08 μm⋅s−1 and 0.81 ± 0.05 μm⋅s−1, respectively, P = 0.0167, Mann–Whitney U test) (Fig. 6B). Interestingly, velocities for the pure directed-transport component of trajectories were non-unimodal for both anterograde and retrograde transport (Hartigan’s dip test for unimodality, P = 0.96 and P = 0.70, respectively). Instead, these velocities together fitted a quadmodal distribution (bimodal anterograde and bimodal retrograde), indicating that two different transport modes drive directed β-actin mRNA transport in each direction of the axon shaft (Fig. 6C). As discussed below, these modes could represent different motors or regulatory proteins driving anterograde and retrograde axonal β-actin mRNA transport.
Fig. 6.
Directional differences in β-actin mRNA-directed transport along the axon shaft are sufficient to confer localization patterns. (A) No significant difference (NS) was found in the frequency of anterograde- versus retrograde-directed transport of β-actin mRNA in axons. (B) A comparison of the velocity of anterograde- and retrograde-directed transport states shows that β-actin mRNA moves significantly faster in the anterograde direction. (C) Multimixture modeling of Gaussian modal distributions of directed transport speeds best fits a quadmodal distribution in which velocities exhibit a bimodal distribution in the anterograde (C′) and retrograde (C′′) directions. (D) A mathematical model shows that fold change in density resulting from an anterograde bias in directed transport speeds is sufficient to predict enriched β-actin mRNA density in the central domain of the growth cone. *P = 0.0167; Mann–Whitney U test; n = 124 puncta, and n =66 axons.
Although most β-actin mRNA puncta displayed diffusive behavior, the sheer length of neuronal axons (up to 1 mm for RGC axons in the Xenopus embryos we study) means that diffusion alone may not explain how β-actin mRNA reaches the periphery. Using a rearrangement of the mean squared displacement equation for a diffusing particle, we calculated it would take an average of ∼48 d for one mRNA molecule to reach the tip of a 500-μm-long axon in the optic tract by diffusion alone (SI Appendix, S1 Mathematical Modeling). Moreover, diffusion alone could not account for the enrichment of β-actin mRNA in the growth cone. It is reasonable, therefore, to assume that directed transport must play an important role in contributing to the flux of β-actin mRNA trafficking throughout the axon shaft. To investigate how directed transport might contribute to increased β-actin mRNA density in the growth cone, we generated a mathematical model. Here, we calculated the expected fold increase in mRNA from axon shaft to growth cone through an advection-diffusion model (Fig. 6D, and SI Appendix, Fig. S5). Parameters were obtained from our quantitative imaging data showing that 3.3% of β-actin mRNA RNPs move with directed transport at a given time and that the total average moving velocity (anterograde and retrograde) of mRNAs is 0.004 ± 0.002 μm⋅s−1. In addition, we used a consensus for the β-actin mRNA half-life of 8 h from previous publications (65, 66). We found the observed velocity differences between anterograde and retrograde transport yield a 4.8 ± 3.5-fold increase in β-actin mRNA density at the growth cone (SI Appendix, S1 Mathematical Modeling). This is within the range of increase we observe through smFISH (fourfold). Together our results show that, despite relatively few mRNA molecules being actively transported at a given moment in time, directed motion provides the main flux for β-actin mRNA trafficking throughout the axon shaft and that directional disparity in trafficking speeds provides a mechanistic basis for localizing β-actin mRNA to the axon tip.
Discussion
Molecular Beacons as a Single-Molecule Method for Visualizing Endogenous mRNA in Axons.
How individual molecules move in the cytoplasm holds a trove of information about the biological processes regulating their kinetics. Previous methods for visualizing single molecules of mRNA have yielded exciting insights into the world of RNA trafficking, uncovering, for example, how particular mRNAs become localized in the Drosophila oocyte (25), depolarization-linked messenger RNP disassembly in mammalian dendrites (18), and even whether an mRNA is being translated by changes in diffusive behavior (64). So far, single-molecule techniques have focused on the idea that one mRNA molecule can be imaged in vivo only if tagged by large numbers of fluorophores. These approaches allow weak fluorescence emission from a single molecule to be multiplied, generating SNR above background associated with cellular autofluorescence and other nonspecific signals (for instance, from freely diffusing MCP-GFP complexes). For visualizing endogenous mRNA transcribed in vivo, these techniques are possible only through genetic modification and run the risk of affecting mRNA dynamics through heavy labeling. In contrast, single protein dynamics are routinely analyzed by tagging the protein of interest with only one fluorophore that is visualized using illumination techniques that increase SNR without high laser intensity or exposure time (53, 67, 68). Here we have applied this principle to mRNA. We show that endogenous β-actin mRNA is labeled in growing axons using two chimeric MBs and that a single MB is capable of capturing single mRNA dynamics via HILO microscopy.
Imaging mRNA dynamics using MBs is not without caveats, however. As previously reported (41), nonspecific signal in the nucleus was also observed in our system. Because such signal overrides MB fluorescence in the vicinity of the nucleus, it could be difficult to image single-mRNA dynamics at proximal regions of the axon without methods to prevent nuclear sequestration, for example by coupling to streptavidin (42) or facilitating siRNA-like nuclear export (69). Fortunately for the purpose of this study, the remoteness from the soma of the peripheral axonal regions analyzed meant that the nuclear signal did not obscure detection or localization patterns of mRNA puncta. Moreover, imaging the axon periphery lends itself to single-molecule imaging because of the relative sparsity of mRNA molecules in neurites and because axon length limits potential diffusive background from unbound MBs. Some studies have reported false-positive signal generated from nuclease degradation and nonspecific interactions (70, 71). We also found that a degree of background labeling occurred in our control MBs. Although we cannot rule out the possibility that a degree of false positives might remain, the use of nuclease-resistant MBs, the short time period between electroporation and imaging, and the extensive validation of the signal led us to be confident we were labeling β-actin mRNA with reasonable specificity.
Packaging mRNAs Singly in the Axon Shaft.
Using MBs and smFISH, we counted the number of β-actin mRNA molecules within axonal RNPs and found that the great majority existed singly, while less than 30% traveled in twos. Such findings are in line with studies investigating mRNA stoichiometry in dendrites (18, 72), indicating that common strategies for mRNA packaging might exist across neuronal compartments. Directed transport by motor proteins is an ATP-driven process; therefore, in terms of energy efficiency, it might seem somewhat illogical for mRNAs to travel long distances singly. The median cellular translation rate has been estimated as 140 proteins per mRNA per hour (73) and has been hypothesized to be even higher in neurites (74). Therefore careful regulation of each mRNA individually might be necessary because their cumulative translation would presumably have a potent effect at the subcellular locality. Moreover, RNA copackaging has been linked to translational repression (75, 76). As β-actin mRNA is translated in the axon shaft in addition to the growth cone (77), the transport of single mRNA molecules may additionally provide a mechanism for continual protein synthesis at a basal level by the circulation of translatable substrates.
Endogenous β-Actin mRNA Dynamics Explain Localization Patterns in Growing Axons.
In this study, we investigated how individual β-actin mRNA molecules move and are distributed across growing axons. Precise quantitation of mRNA levels allowed us to demonstrate that β-actin mRNA-localization patterns vary according to the axonal subcompartment. We observed significantly increased mRNA density in the central domain of the growth cone compared with peripheral domains and the axon shaft. Because of the growth cone’s role as the sensing “nose” of a navigating axon, it is perhaps not surprising that this region is enriched in β-actin mRNA, providing a local store for basal and cue-induced morphological changes through the local synthesis of cytoskeletal proteins. Indeed, nonquantitative observations from in situ hybridization images have previously indicated that this might be the case (28). Other studies have demonstrated that different mRNA species exhibit varying localization patterns across the axon. For example, using laser capture microdissection followed by genome-wide microarray analysis, we have previously shown that the growth cone transcriptome is enriched with different sets of mRNAs compared with the axon shaft (12). mRNA localization may be regulated by mechanisms involving degradation, e.g., by nonsense-mediated mRNA decay (6). However, the contribution of mRNA trafficking to the spatial patterns of mRNA has so far remained elusive.
mRNA trafficking in axons has previously been visualized indirectly through the dynamics of RBPs bound to fluorescent proteins (4), by the translation of fluorescent proteins conjugated to UTRs (62), and by nonspecific labeling of all RNA (29, 78). These studies have provided valuable insight into the factors regulating mRNA localization, e.g., changes in mRNA trafficking triggered by extracellular cues (4, 62). However, because one cannot follow a particular mRNA species in real time, the information about axonal mRNA trafficking that we can glean using such indirect/nonspecific methods is limited. To our knowledge, only two other studies have directly visualized trafficking of a specific mRNA in axons. In the first, mutations in the RBP TDP-43 were shown to modulate axonal trafficking of Nefl mRNA (79). However, only the direction of mRNA trafficking was analyzed, without quantitative analysis of motion type or single-molecule sensitivity. In the second study, cue-induced changes in trafficking of in vitro transcribed β-actin mRNA was investigated in axons (80). Interestingly, a bias in anterograde transport speed of mRNA was also reported. However, this study used exogenous mRNA and did not quantitatively analyze biophysical motion states. In this study, we characterized the dynamics of single endogenous β-actin mRNA molecules in growing axons. We demonstrated that while the great majority of β-actin mRNAs display diffusive behavior, a modicum of directed transport exists in the axon shaft that is a critical determinant of β-actin mRNA enrichment in the growth cone.
Our results show a motion-type distribution in growing axons highly similar to β-actin mRNA transport in other neuronal compartments and cell types (18, 81). That the great majority of mRNAs exhibit diffusive/stationary behavior over our imaging period does not, however, mean that these mRNAs display the same behavior throughout their lifetime. Our calculations demonstrated that it is highly improbable that β-actin mRNA molecules traverse the length of the axon by diffusion alone, suggesting that at one point they must undergo directed transport. In support of this view, it has recently been shown by imaging over longer acquisition times that β-actin mRNA in dendrites consistently cycles between stationary and trafficking states (19). Directed transport is thus a crucial component of axonal mRNA dynamics. We demonstrate that this component of β-actin mRNA trafficking becomes lost as mRNAs travel from the axon shaft to the growth cone. Through mathematical modeling, we further show that the differences between anterograde- and retrograde-directed transport velocities are sufficient to cause β-actin mRNA enrichment in the growth cone. In the future, it would be fascinating to determine how these differences compare with mRNA species that are enriched in the axon shaft [e.g., mRNAs encoding proteins with roles in membrane trafficking and protein folding (12)] and what relative roles mRNA transport and degradation might play in conferring differential localization patterns.
The approach we employed, HMM–Bayes, to identify switching between motion states (60) provided a unique opportunity to analyze directed transport in axons. Here, it was possible to extract the kinetic information behind pure directed transport states from trajectories containing mixtures of motion behavior. Various studies have suggested that long-range mRNA transport along the axon shaft is microtubule-dependent (28, 61, 62). Because of the highly polarized microtubule orientation in axons, anterograde and retrograde microtubule-based transport are driven by different microtubule-associated motor proteins: Plus-end–directed kinesins drive anterograde transport, and cytoplasmic dynein drives retrograde transport (82). Different motor proteins have different kinetic characteristics (83); therefore it is likely that the difference in speeds between anterograde- and retrograde-moving β-actin mRNAs is due to the properties of different motor proteins. However, it was surprising that bimodal distributions of velocity were also observed in each direction. These data suggest that different directed transport modes underlie anterograde and retrograde transport. Indeed, the combined use of kinesin-1 and kinesin-2 motor proteins has previously been observed for plus-end–directed Vg1 mRNA transport in Xenopus oocytes (84), an mRNA that shares the same RBP (Vg1RBP) for transport as β-actin mRNA. Moreover, regulating the balancing between cargo velocities driven by two different motor proteins has been shown to provide a cargo-sorting mechanism at the proximal axon shaft (85). Interestingly, these data reveal that the different velocities of kinesin-1 and kinesin-3 motors are able to regulate vesicle distribution across the axon (85), suggesting a similar mechanism may underlie the faster anterograde transport that drives increased β-actin mRNA localization in the growth cone. However, cytoplasmic dynein is the only motor protein likely to drive retrograde β-actin mRNA transport; thus multiple motor types are unlikely to explain the bimodal retrograde velocity distribution. Instead, different numbers of motor proteins attached to cargoes could cause differences in speeds (86–88). Dynein velocity can also be increased by association with the regulatory protein Lissencephaly-1 (89, 90). How motor proteins and their regulators might coordinately control directed mRNA transport in axons is an intriguing avenue for future research that would help us understand further the control of axonal mRNA-localization patterns through mRNA trafficking.
Methods
MB Design and Validation.
MB loop sequences were manually designed to bind predicted single-stranded regions on Xenopus laevis β-actin mRNA with favorable MB:mRNA stability using the intersection of two secondary structure-prediction algorithms: m-Fold (91) and OLIGOWALK (92). Sequences were confirmed to be unique to the Xenopus genome by blasting with NCBI BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and confirming unannotated gene sequences by blasting again against β-actin mRNA. The only off-target with 100% minus/plus base-pairing to MB2 (Fam83h-like) was confirmed not to be expressed in Xenopus RGC axons by performing RT-PCR using RNA extracted from isolated axons (SI Appendix, Supplementary Methods).
MBs were designed as LNA (+N)/2′-O-methyl RNA (oN) chimeras (44) with a GC-rich stem and Cy3:BHQ-2 fluorophore:quencher and were synthesized by TIB Molbiol. In vitro tests for MB specificity were performed at 37 °C in hybridization buffer (44) using a LightCycler 480 system (Roche). Sequences (5′–3′) were as follows:
MB1: Cy3oCoGoAoCoGoCoU+CoAoGoUoU+AoGoG+AoUoUoUoUoC+AoUoGoCoGoUoCoG BHQ2
MB2: Cy3oGoCoGoCoAoG+GoAoA+GoCoCoAoA+GoAoUoG+GoAoUoGoCoGoC BHQ2
Scramble: Cy3oCoCoGoCoGoGoCoGoGoA+AoAoCoUoU+AoUoA+CoA+CoUoUoAoAoCoGoCoCoGoCoGoG BHQ-2
Brn3a: Cy3oCoCoGoCoGoGoCoGoUoUoUoU+AoU+AoUoA+AoCoUoUoUoUoU+AoCoCoGoCoCoGoCoGoG BHQ-2
Electroporation and Culture.
Electroporation was performed on stage 28 eye primordia as described in ref. 93. MBs were electroporated at a concentration of 50 μM. The expression of each β-actin MB did not significantly differ when half the concentration was used (25 μM), suggesting that we were electroporating at saturating concentrations (SI Appendix, Fig. S6B). The Vg1RBP-EGFP plasmid (4) and fluorescently tagged mRNA were electroporated at a concentration of 1.85 and 1 μg/μL, respectively. Eye explant cultures were performed as described previously (94) but with the use of phenol red-free L15 medium (Life Technologies), with fixation or imaging within 24 h. Importantly, in X. laevis eye explant cultures, only the axons of RGCs grow out from the retina; other protrusions, such as dendrites, remain in the retina, as eye morphology is preserved in culture (94). As such, we were able to visualize RGC axons exclusively in fixed cells and during live imaging. This research was performed under the Animals (Scientific Procedures) Act 1986 Amendment Regulations 2012 following ethical review by the University of Cambridge Animal Welfare and Ethical Review Body.
smFISH.
Explant cultures were fixed in 4% paraformaldehyde, 4% sucrose in 1× PBS for 5 min, washed in 4% sucrose, 1× PBS for 5 min, and then were fixed further in ice-cold 100% MeOH at −20 °C for 10 min. After three 10-min washings in 1× PBS + 0.001% Triton X-100, cultures were permeabilized in 70% EtOH for 24 h at 4 °C. Cultures were subsequently rehydrated at room temperature in 1× PBS and then were preincubated for 30 min in 50% formamide, 2× SSC at 40 °C, washed three times for 5 min each washin in wash buffer (10% formamide, 2× SSC) preheated to 40 °C, and were hybridized overnight at 40 °C in 2.5 μM Stellaris RNA FISH probes with CAL Fluor Red 590 Dye (Biosearch Technologies) designed to target β-actin mRNA using the online Stellaris probe designer in hybridization buffer described in ref. 58. Cultures were then washed twice for 30 min each in washing buffer at 40 °C and twice for 5 min each washing in 2× SSC before mounting in FluorSave (Merck). Images were acquired on an Olympus IX81 inverted microscope equipped with a PerkinElmer Spinning Disk UltraVIEW VoX system and a 100× oil immersion objective with an ORCA-Flash4.0 V2 CMOS camera (Hamamatsu) using Volocity 6.3.0 software (PerkinElmer) and a 561-nm laser line at 26.5% laser intensity and 400-ms exposure time. As a negative control, the smFISH protocol was performed as above, but the culture was subjected to RNase A treatment at 37 °C for 1 h before hybridization. β-Actin mRNA stoichiometry, based on intensity distributions of smFISH puncta, was analyzed using the Spatzcells MATLAB script (58). Inverse binomial fitting to generate corrected β-actin mRNA smFISH stoichiometry after degradation was performed using Mathematica.
Live Imaging and Analysis.
Stage 28 Xenopus eye primordia were electroporated with 50 μM MB1 and MB2 and then were cultured for 18–22 h. Time-lapse movies of MB dynamics were acquired via HILO microscopy using a 561-nm laser line at 20% laser intensity with an Nikon TiiE inverted microscope using a CFI Plan Apo total internal reflection fluorescence 100× 1.49 N.A. objective (Nikon) and ILAS2-targeted laser illumination (Cairn Research). Images were acquired using a Photometrics Evolve Delta EM-CCD camera and MetaMorph software (Molecular Devices) at a frame rate of 0.25 s with 100-ms exposure time for 50 s (200 frames). For axon shaft dynamics, only tracks within 20-μm sections of the axon shaft 10 μm from the growth cone were analyzed. The central domain of the growth cone was distinguished from the peripheral domains as the dense central region under brightfield images of the axon. Some puncta observed outside the axon were easily distinguished in brightfield images as remnants of dead cells and thus were excluded from the analysis (SI Appendix, Fig. S6C). Particle tracks were obtained using plusTipTracker software (59), and a custom-made particle-tracking add-on script was developed to assess the directionality of mRNA trajectories in the axon. After noise and duplicate tracks were discarded by checking movies by eye, tracks were assessed for switching between motion states using the HMM–Bayes script (60). Experiments examining the effects of latrunculin A on MB dynamics were performed as above but with the addition of either 10 μM latrunculin A (Sigma) in 2% DMSO or 2% DMSO only into the culture medium and imaging 5–10 min after the addition. Puromycin experiments were performed using 100 μg⋅μL−1 puromycin diluted in water (Sigma) in the culture medium, and the dynamics of MBs were captured at 30–45 min.
Statistics.
Statistical tests were performed using PRISM software (GraphPad). The modality of velocity distributions for directed transport states was calculated in R using the diptest and mclust packages.
Detailed descriptions of all other experimental procedures are given in SI Appendix, Supplementary Methods.
Supplementary Material
Acknowledgments
We thank J. Gallop for use of the total internal reflection fluorescence microscope and S. Bullock for valuable discussion and critical reading of the manuscript. This work was funded by Wellcome Trust Studentship 102457/Z/13/A (to B.T-B.), Wellcome Trust Program Grant 085314/Z/08/Z (to C.E.H.), and European Research Council Advanced Grant 322817 (to C.E.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806189115/-/DCSupplemental.
References
- 1.Holt CE, Bullock SL. Subcellular mRNA localization in animal cells and why it matters. Science. 2009;326:1212–1216. doi: 10.1126/science.1176488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jung H, Yoon BC, Holt CE. Axonal mRNA localization and local protein synthesis in nervous system assembly, maintenance and repair. Nat Rev Neurosci. 2012;13:308–324. doi: 10.1038/nrn3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu KY, et al. Local translation of RhoA regulates growth cone collapse. Nature. 2005;436:1020–1024. doi: 10.1038/nature03885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leung KM, et al. Asymmetrical beta-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat Neurosci. 2006;9:1247–1256. doi: 10.1038/nn1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yao J, Sasaki Y, Wen Z, Bassell GJ, Zheng JQ. An essential role for beta-actin mRNA localization and translation in Ca2+-dependent growth cone guidance. Nat Neurosci. 2006;9:1265–1273. doi: 10.1038/nn1773. [DOI] [PubMed] [Google Scholar]
- 6.Colak D, Ji S-J, Porse BT, Jaffrey SR. Regulation of axon guidance by compartmentalized nonsense-mediated mRNA decay. Cell. 2013;153:1252–1265. doi: 10.1016/j.cell.2013.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merianda TT, Vuppalanchi D, Yoo S, Blesch A, Twiss JL. Axonal transport of neural membrane protein 35 mRNA increases axon growth. J Cell Sci. 2013;126:90–102. doi: 10.1242/jcs.107268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Batista AFR, Martínez JC, Hengst U. Intra-axonal synthesis of SNAP25 is required for the formation of presynaptic terminals. Cell Rep. 2017;20:3085–3098. doi: 10.1016/j.celrep.2017.08.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costa CJ, Willis DE. To the end of the line: Axonal mRNA transport and local translation in health and neurodegenerative disease. Dev Neurobiol. 2018;78:209–220. doi: 10.1002/dneu.22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zappulo A, et al. RNA localization is a key determinant of neurite-enriched proteome. Nat Commun. 2017;8:583. doi: 10.1038/s41467-017-00690-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor AM, et al. Axonal mRNA in uninjured and regenerating cortical mammalian axons. J Neurosci. 2009;29:4697–4707. doi: 10.1523/JNEUROSCI.6130-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zivraj KH, et al. Subcellular profiling reveals distinct and developmentally regulated repertoire of growth cone mRNAs. J Neurosci. 2010;30:15464–15478. doi: 10.1523/JNEUROSCI.1800-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gumy LF, et al. Transcriptome analysis of embryonic and adult sensory axons reveals changes in mRNA repertoire localization. RNA. 2011;17:85–98. doi: 10.1261/rna.2386111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minis A, et al. Subcellular transcriptomics-dissection of the mRNA composition in the axonal compartment of sensory neurons. Dev Neurobiol. 2014;74:365–381. doi: 10.1002/dneu.22140. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y-Y, et al. In vitro growth conditions and development affect differential distributions of RNA in axonal growth cones and shafts of cultured rat hippocampal neurons. Mol Cell Neurosci. 2014;61:141–151. doi: 10.1016/j.mcn.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 16.Müller-McNicoll M, Neugebauer KM. How cells get the message: Dynamic assembly and function of mRNA-protein complexes. Nat Rev Genet. 2013;14:275–287. doi: 10.1038/nrg3434. [DOI] [PubMed] [Google Scholar]
- 17.Dynes JL, Steward O. Dynamics of bidirectional transport of Arc mRNA in neuronal dendrites. J Comp Neurol. 2007;500:433–447. doi: 10.1002/cne.21189. [DOI] [PubMed] [Google Scholar]
- 18.Park HY, et al. Visualization of dynamics of single endogenous mRNA labeled in live mouse. Science. 2014;343:422–424. doi: 10.1126/science.1239200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoon YJ, et al. Glutamate-induced RNA localization and translation in neurons. Proc Natl Acad Sci USA. 2016;113:E6877–E6886. doi: 10.1073/pnas.1614267113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doyle M, Kiebler MA. Mechanisms of dendritic mRNA transport and its role in synaptic tagging. EMBO J. 2011;30:3540–3552. doi: 10.1038/emboj.2011.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forrest KM, Gavis ER. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr Biol. 2003;13:1159–1168. doi: 10.1016/s0960-9822(03)00451-2. [DOI] [PubMed] [Google Scholar]
- 22.Chang P, et al. Localization of RNAs to the mitochondrial cloud in Xenopus oocytes through entrapment and association with endoplasmic reticulum. Mol Biol Cell. 2004;15:4669–4681. doi: 10.1091/mbc.E04-03-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bullock SL. Messengers, motors and mysteries: Sorting of eukaryotic mRNAs by cytoskeletal transport. Biochem Soc Trans. 2011;39:1161–1165. doi: 10.1042/BST0391161. [DOI] [PubMed] [Google Scholar]
- 24.Bullock SL, Nicol A, Gross SP, Zicha D. Guidance of bidirectional motor complexes by mRNA cargoes through control of dynein number and activity. Curr Biol. 2006;16:1447–1452. doi: 10.1016/j.cub.2006.05.055. [DOI] [PubMed] [Google Scholar]
- 25.Zimyanin VL, et al. In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell. 2008;134:843–853. doi: 10.1016/j.cell.2008.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bashirullah A, Cooperstock RL, Lipshitz HD. Spatial and temporal control of RNA stability. Proc Natl Acad Sci USA. 2001;98:7025–7028. doi: 10.1073/pnas.111145698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cajigas IJ, et al. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron. 2012;74:453–466. doi: 10.1016/j.neuron.2012.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bassell GJ, et al. Sorting of beta-actin mRNA and protein to neurites and growth cones in culture. J Neurosci. 1998;18:251–265. doi: 10.1523/JNEUROSCI.18-01-00251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong HH-W, et al. RNA docking and local translation regulate site-specific axon remodeling in vivo. Neuron. 2017;95:852–868.e8. doi: 10.1016/j.neuron.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cagnetta R, Frese CK, Shigeoka T, Krijgsveld J, Holt CE. Rapid cue-specific remodeling of the nascent axonal proteome. Neuron. 2018;99:29–46.e4. doi: 10.1016/j.neuron.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Condeelis J, Singer RH. How and why does beta-actin mRNA target? Biol Cell. 2005;97:97–110. doi: 10.1042/BC20040063. [DOI] [PubMed] [Google Scholar]
- 32.Eom T, Antar LN, Singer RH, Bassell GJ. Localization of a beta-actin messenger ribonucleoprotein complex with zipcode-binding protein modulates the density of dendritic filopodia and filopodial synapses. J Neurosci. 2003;23:10433–10444. doi: 10.1523/JNEUROSCI.23-32-10433.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donnelly CJ, et al. Axonally synthesized β-actin and GAP-43 proteins support distinct modes of axonal growth. J Neurosci. 2013;33:3311–3322, and erratum (2013) 33:5878. doi: 10.1523/JNEUROSCI.1722-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buxbaum AR, Yoon YJ, Singer RH, Park HY. Single-molecule insights into mRNA dynamics in neurons. Trends Cell Biol. 2015;25:468–475. doi: 10.1016/j.tcb.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tyagi S, Kramer FR. Molecular beacons: Probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 36.Wang J, Cao LG, Wang YL, Pederson T. Localization of pre-messenger RNA at discrete nuclear sites. Proc Natl Acad Sci USA. 1991;88:7391–7395. doi: 10.1073/pnas.88.16.7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bertrand E, et al. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 38.Tyagi S, Bratu DP, Kramer FR. Multicolor molecular beacons for allele discrimination. Nat Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 39.Tsourkas A, Behlke MA, Bao G. Hybridization of 2′-O-methyl and 2′-deoxy molecular beacons to RNA and DNA targets. Nucleic Acids Res. 2002;30:5168–5174. [PMC free article] [PubMed] [Google Scholar]
- 40.Tsourkas A, Behlke MA, Rose SD, Bao G. Hybridization kinetics and thermodynamics of molecular beacons. Nucleic Acids Res. 2003;31:1319–1330. doi: 10.1093/nar/gkg212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bratu DP, Cha B-J, Mhlanga MM, Kramer FR, Tyagi S. Visualizing the distribution and transport of mRNAs in living cells. Proc Natl Acad Sci USA. 2003;100:13308–13313. doi: 10.1073/pnas.2233244100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tyagi S, Alsmadi O. Imaging native beta-actin mRNA in motile fibroblasts. Biophys J. 2004;87:4153–4162. doi: 10.1529/biophysj.104.045153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kierzek E, et al. The influence of locked nucleic acid residues on the thermodynamic properties of 2′-O-methyl RNA/RNA heteroduplexes. Nucleic Acids Res. 2005;33:5082–5093. doi: 10.1093/nar/gki789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Catrina IE, Marras SAE, Bratu DP. Tiny molecular beacons: LNA/2′-O-methyl RNA chimeric probes for imaging dynamic mRNA processes in living cells. ACS Chem Biol. 2012;7:1586–1595. doi: 10.1021/cb300178a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chui H, Rangarajan A. A new point matching algorithm for non-rigid registration. Comput Vision Image Understanding. 2003;89:114–141. [Google Scholar]
- 46.So L-H, et al. General properties of transcriptional time series in Escherichia coli. Nat Genet. 2011;43:554–560. doi: 10.1038/ng.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hebenstreit D, et al. RNA sequencing reveals two major classes of gene expression levels in metazoan cells. Mol Syst Biol. 2011;7:497. doi: 10.1038/msb.2011.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moffitt JR, et al. High-performance multiplexed fluorescence in situ hybridization in culture and tissue with matrix imprinting and clearing. Proc Natl Acad Sci USA. 2016;113:14456–14461. doi: 10.1073/pnas.1617699113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vargas DY, Raj A, Marras SAE, Kramer FR, Tyagi S. Mechanism of mRNA transport in the nucleus. Proc Natl Acad Sci USA. 2005;102:17008–17013. doi: 10.1073/pnas.0505580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang X, et al. Quantitative assessment of ratiometric bimolecular beacons as a tool for imaging single engineered RNA transcripts and measuring gene expression in living cells. Nucleic Acids Res. 2013;41:e152. doi: 10.1093/nar/gkt561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao D, et al. Single-molecule detection and tracking of RNA transcripts in living cells using phosphorothioate-optimized 2′-O-methyl RNA molecular beacons. Biomaterials. 2016;100:172–183. doi: 10.1016/j.biomaterials.2016.05.022. [DOI] [PubMed] [Google Scholar]
- 52.Chen M, et al. A molecular beacon-based approach for live-cell imaging of RNA transcripts with minimal target engineering at the single-molecule level. Sci Rep. 2017;7:1550. doi: 10.1038/s41598-017-01740-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tokunaga M, Imamoto N, Sakata-Sogawa K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat Methods. 2008;5:159–161. doi: 10.1038/nmeth1171. [DOI] [PubMed] [Google Scholar]
- 54.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 55.Dempsey GT, Vaughan JC, Chen KH, Bates M, Zhuang X. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat Methods. 2011;8:1027–1036. doi: 10.1038/nmeth.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Coffman VC, Wu J-Q. Counting protein molecules using quantitative fluorescence microscopy. Trends Biochem Sci. 2012;37:499–506. doi: 10.1016/j.tibs.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leake MC. Analytical tools for single-molecule fluorescence imaging in cellulo. Phys Chem Chem Phys. 2014;16:12635–12647. doi: 10.1039/c4cp00219a. [DOI] [PubMed] [Google Scholar]
- 58.Skinner SO, Sepúlveda LA, Xu H, Golding I. Measuring mRNA copy number in individual Escherichia coli cells using single-molecule fluorescent in situ hybridization. Nat Protoc. 2013;8:1100–1113. doi: 10.1038/nprot.2013.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Applegate KT, et al. plusTipTracker: Quantitative image analysis software for the measurement of microtubule dynamics. J Struct Biol. 2011;176:168–184. doi: 10.1016/j.jsb.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Monnier N, et al. Inferring transient particle transport dynamics in live cells. Nat Methods. 2015;12:838–840. doi: 10.1038/nmeth.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Muslimov IA, Titmus M, Koenig E, Tiedge H. Transport of neuronal BC1 RNA in Mauthner axons. J Neurosci. 2002;22:4293–4301. doi: 10.1523/JNEUROSCI.22-11-04293.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Willis DE, et al. Extracellular stimuli specifically regulate localized levels of individual neuronal mRNAs. J Cell Biol. 2007;178:965–980. doi: 10.1083/jcb.200703209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dent EW, Gertler FB. Cytoskeletal dynamics and transport in growth cone motility and axon guidance. Neuron. 2003;40:209–227. doi: 10.1016/s0896-6273(03)00633-0. [DOI] [PubMed] [Google Scholar]
- 64.Katz ZB, et al. Mapping translation ‘hot-spots’ in live cells by tracking single molecules of mRNA and ribosomes. eLife. 2016;5:e10415. doi: 10.7554/eLife.10415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dormoy-Raclet V, et al. The RNA-binding protein HuR promotes cell migration and cell invasion by stabilizing the beta-actin mRNA in a U-rich-element-dependent manner. Mol Cell Biol. 2007;27:5365–5380. doi: 10.1128/MCB.00113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sharova LV, et al. Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res. 2009;16:45–58. doi: 10.1093/dnares/dsn030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Funatsu T, Harada Y, Tokunaga M, Saito K, Yanagida T. Imaging of single fluorescent molecules and individual ATP turnovers by single myosin molecules in aqueous solution. Nature. 1995;374:555–559. doi: 10.1038/374555a0. [DOI] [PubMed] [Google Scholar]
- 68.Ritter JG, Veith R, Veenendaal A, Siebrasse JP, Kubitscheck U. Light sheet microscopy for single molecule tracking in living tissue. PLoS One. 2010;5:e11639. doi: 10.1371/journal.pone.0011639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen AK, Davydenko O, Behlke MA, Tsourkas A. Ratiometric bimolecular beacons for the sensitive detection of RNA in single living cells. Nucleic Acids Res. 2010;38:e148. doi: 10.1093/nar/gkq436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen AK, Behlke MA, Tsourkas A. Avoiding false-positive signals with nuclease-vulnerable molecular beacons in single living cells. Nucleic Acids Res. 2007;35:e105. doi: 10.1093/nar/gkm593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Su X, Zhang C, Zhao M. Discrimination of the false-positive signals of molecular beacons by combination of heat inactivation and using single walled carbon nanotubes. Biosens Bioelectron. 2011;26:3596–3601. doi: 10.1016/j.bios.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 72.Batish M, van den Bogaard P, Kramer FR, Tyagi S. Neuronal mRNAs travel singly into dendrites. Proc Natl Acad Sci USA. 2012;109:4645–4650. doi: 10.1073/pnas.1111226109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 74.Glock C, Heumüller M, Schuman EM. mRNA transport & local translation in neurons. Curr Opin Neurobiol. 2017;45:169–177. doi: 10.1016/j.conb.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 75.Krichevsky AM, Kosik KS. Neuronal RNA granules: A link between RNA localization and stimulation-dependent translation. Neuron. 2001;32:683–696. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- 76.Chekulaeva M, Hentze MW, Ephrussi A. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell. 2006;124:521–533. doi: 10.1016/j.cell.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 77.Spillane M, Ketschek A, Merianda TT, Twiss JL, Gallo G. Mitochondria coordinate sites of axon branching through localized intra-axonal protein synthesis. Cell Rep. 2013;5:1564–1575. doi: 10.1016/j.celrep.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pathak GK, Love JM, Chetta J, Shah SB. A comparative quantitative assessment of axonal and dendritic mRNA transport in maturing hippocampal neurons. PLoS One. 2013;8:e65917. doi: 10.1371/journal.pone.0065917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alami NH, et al. Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron. 2014;81:536–543. doi: 10.1016/j.neuron.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Leung KM, et al. Cue-polarized transport of β-actin mRNA depends on 3′UTR and microtubules in live growth cones. Front Cell Neurosci. 2018 doi: 10.3389/fncel.2018.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fusco D, et al. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr Biol. 2003;13:161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kapitein LC, Hoogenraad CC. Which way to go? Cytoskeletal organization and polarized transport in neurons. Mol Cell Neurosci. 2011;46:9–20. doi: 10.1016/j.mcn.2010.08.015. [DOI] [PubMed] [Google Scholar]
- 83.Hancock WO. Bidirectional cargo transport: Moving beyond tug of war. Nat Rev Mol Cell Biol. 2014;15:615–628. doi: 10.1038/nrm3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Messitt TJ, et al. Multiple kinesin motors coordinate cytoplasmic RNA transport on a subpopulation of microtubules in Xenopus oocytes. Dev Cell. 2008;15:426–436. doi: 10.1016/j.devcel.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gumy LF, et al. MAP2 defines a pre-axonal filtering zone to regulate KIF1- versus KIF5-dependent cargo transport in sensory neurons. Neuron. 2017;94:347–362.e7. doi: 10.1016/j.neuron.2017.03.046. [DOI] [PubMed] [Google Scholar]
- 86.Shtridelman Y, Cahyuti T, Townsend B, DeWitt D, Macosko JC. Force-velocity curves of motor proteins cooperating in vivo. Cell Biochem Biophys. 2008;52:19–29. doi: 10.1007/s12013-008-9021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Reis GF, et al. Molecular motor function in axonal transport in vivo probed by genetic and computational analysis in Drosophila. Mol Biol Cell. 2012;23:1700–1714. doi: 10.1091/mbc.E11-11-0938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Urnavicius L, et al. Cryo-EM shows how dynactin recruits two dyneins for faster movement. Nature. 2018;554:202–206. doi: 10.1038/nature25462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Baumbach J, et al. Lissencephaly-1 is a context-dependent regulator of the human dynein complex. eLife. 2017;6:e21768. doi: 10.7554/eLife.21768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gutierrez PA, Ackermann BE, Vershinin M, McKenney RJ. Differential effects of the dynein-regulatory factor Lissencephaly-1 on processive dynein-dynactin motility. J Biol Chem. 2017;292:12245–12255. doi: 10.1074/jbc.M117.790048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lu ZJ, Mathews DH. OligoWalk: An online siRNA design tool utilizing hybridization thermodynamics. Nucleic Acids Res. 2008;36:W104–W108. doi: 10.1093/nar/gkn250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Falk J, et al. Electroporation of cDNA/Morpholinos to targeted areas of embryonic CNS in Xenopus. BMC Dev Biol. 2007;7:107. doi: 10.1186/1471-213X-7-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Harris WA, Holt CE, Smith TA, Gallenson N. Growth cones of developing retinal cells in vivo, on culture surfaces, and in collagen matrices. J Neurosci Res. 1985;13:101–122. doi: 10.1002/jnr.490130108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.