This work shows that the BadL protein of Rhodopseudomonas palustris has N-acetyltransferase activity and that this activity is required for the catabolism of benzoate under photosynthetic conditions in this bacterium. R. palustris occupies lignin-rich habitats, making its benzoate-degrading capability critical for the recycling of this important, energy-rich biopolymer. This work identifies the product of the BadL enzyme as acetamidobenzoates, which were needed to derepress genes encoding benzoate-degrading enzymes and proteins of the photosynthetic apparatus responsible for the generation of the proton motive force under anoxia in the presence of light. In short, acetamidobenzoates potentially coordinate the use of benzoate as a source of reducing power and carbon with the generation of a light-driven proton motive force that fuels ATP synthesis, motility, transport, and many other processes in the metabolically versatile bacterium R. palustris.
KEYWORDS: N-acetyltransferases, benzoate degradation, regulation of gene expression, small molecule acetylation
ABSTRACT
The degradation of lignin-derived aromatic compounds such as benzoate has been extensively studied in Rhodopseudomonas palustris, and the chemistry underpinning the conversion of benzoate to acetyl coenzyme A (acetyl-CoA) is well understood. Here we characterize the last unknown gene, badL, of the bad (benzoic acid degradation) cluster. BadL function is required for growth under photoheterotrophic conditions with benzoate as the organic carbon source (i.e., light plus anoxia). On the basis of bioinformatics and in vivo and in vitro data, we show that BadL, a Gcn5-related N- acetyltransferase (GNAT) (PF00583), acetylates aminobenzoates to yield acetamidobenzoates. The latter relieved repression of the badDEFGAB operon by binding to BadM, triggering the synthesis of enzymes that activate and dearomatize the benzene ring. We also show that acetamidobenzoates are required for the expression of genes encoding the photosynthetic reaction center light-harvesting complexes through a BadM-independent mechanism. The effect of acetamidobenzoates on pigment synthesis is new and different than their effect on the catabolism of benzoate.
INTRODUCTION
Lignin is the second most abundant polymer in nature, second only to cellulose. Unlike cellulose, lignin does not contain carbohydrate monomers; instead, it is comprised of phenyl derivatives (e.g., coumaryl alcohol, syryngyl alcohol, and coniferyl alcohol). Lignin is found in the cell walls of plants, and it is estimated to represent approximately 25% of the terrestrial biomass (1). Aromatic compounds released from lignin are rich in energy and carbon; hence, it is not surprising that microbes that occupy environments rich in plant materials have evolved metabolic strategies for the degradation of such compounds. The purple nonsulfur photosynthetic alphaproteobacterium Rhodopseudomonas palustris is an aquatic bacterium that can degrade aromatic compounds into central metabolites. Shown in Fig. 1A is the pathway used by R. palustris for the degradation of aromatic compoundss under anoxic conditions (2). Other aromatic compounds such as chorismate, p-coumarate, toluene, vanillate, cresol, and phenol (to name a few; see reference 3 for a complete list of compounds) feed into the benzoate catabolism pathway via 4-hydroxybenzoate, benzoate, or benzoyl-coenyme A (CoA) (2, 3).
FIG 1.
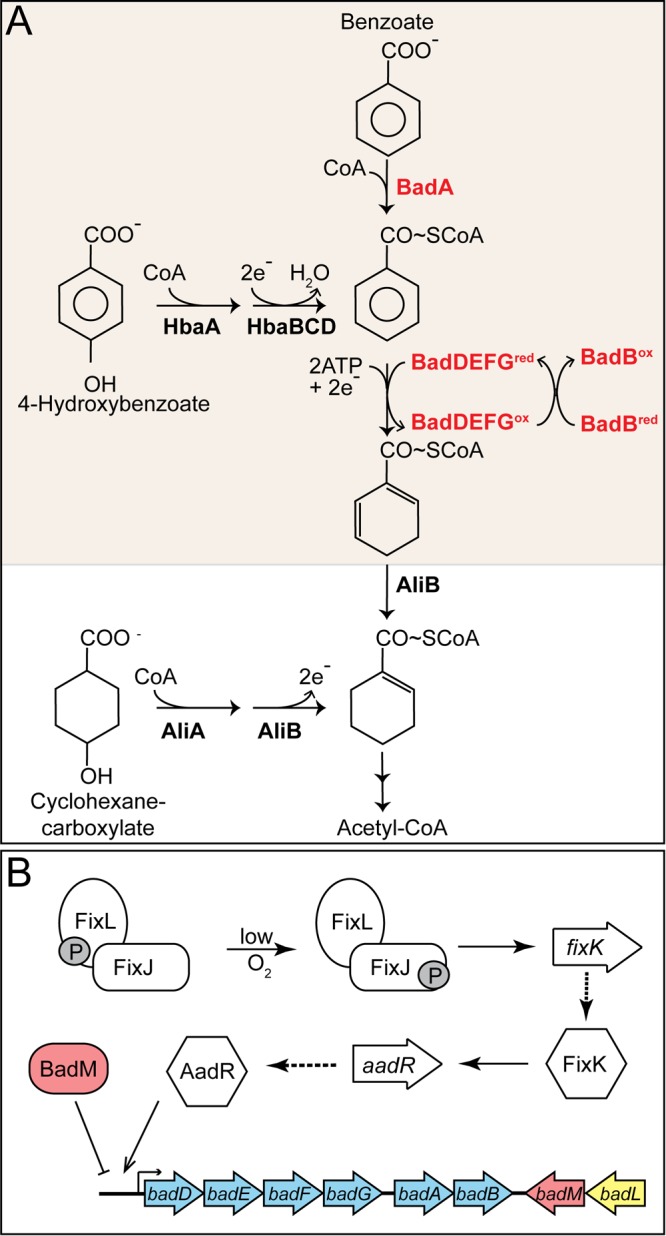
Aromatic catabolism and gene regulation in R. palustris. (A) Benzoate is activated to its CoA thioester by BadA. Hydroxybenzoate can also be converted to benzoyl-CoA in multiple steps by HbaA and HbaBC. The benzene ring of benzoyl-CoA is reduced by BadDEFG, yielding cyclohex-1,5-ene-carboxyl-CoA, which is further reduced by BadJ to cyclohex-1-ene-carboxyl-CoA. The latter is the entry point for cyclohexanecarboxylate into the pathway. Cyclohex-1-ene-carboxyl-CoA undergoes a ring reduction, ring cleavage, and several β-oxidations to eventually form acetyl-CoA. Arrows between cyclohex-1-ene-carboxyl-CoA and acetyl-CoA represent a simplified version of enzymatic reactions carried out by BadK, BadH, and BadI. BadA, benzoyl-CoA synthetase; HbaA, 4-hydroxybenzoyl-CoA synthetase; HcrABC, 4-hydroxybenzoyl-CoA reductase; BadDEFG, benzoyl-CoA reductase; BadB, ferredoxin; AliA, cyclohexancarboxyl-CoA synthetase; AliB, cyclohexancarboxyl-CoA dehydrogenase. Adapted from Harwood and Gibson (14). (B) Gene regulation of the badDEFGAB operon. Under low-oxygen conditions, phosphorylated FixL (FixL-P) relays its phosphate to FixJ, and FixJ-P activates expression of the global regulator, fixK. FixK activates aadR transcription, and AadR activates expression of the badDEFGAB operon (16). BadM is an Rrf2-like regulator that represses the badDEFGAB operon (15). The promoter region of BadM has been characterized and lies between badD and the upstream gene, badC (not shown). FixL, sensor histidine kinase; FixJ, response regulator; FixK, transcriptional regulator; AadR, Crp-like transcriptional activator; BadM, transcription factor.
Benzoate catabolism has been studied in detail in R. palustris (3–11). The first step of the pathway is catalyzed by the benzoyl-CoA synthetase (BadA) (EC 6.2.1.25) enzyme, which activates benzoate to its CoA thioester, benzoyl-CoA (Fig. 1A) (7). The subsequent ring reduction of benzoyl-CoA to cyclohex-1,5-diene-1-carboxyl-CoA is catalyzed by the two [4Fe-4S]+, two [2Fe-2S]+ ATP-dependent reductase BadDEFG enzyme (EC 1.3.7.8) (Fig. 1A) (12, 13). The oxygen labile iron-sulfur centers of the reductase are rereduced by the ferredoxin protein BadB (1). Cyclohex-1,5-diene-1-carboxyl-CoA undergoes a series of reductions, ring cleavage, and β-oxidations, releasing acetyl-CoA and CO2 (2, 3, 14).
Due to the oxygen sensitivity and energetically demanding initial steps of benzoate catabolism (i.e., BadA, BadDEFG, and BadB), the genes encoding these proteins are tightly regulated by activators and repressors (4, 15). A majority of the genes required for benzoate catabolism are clustered within the genome of R. palustris (3). Specifically, the genes coding for the benzoyl-CoA synthetase (badA), benzoyl-CoA reductase (badDEFG), and ferredoxin (badB) comprise the badDEFGAB operon (Fig. 1B). The badDEFGAB operon is activated by AadR (a Crp-type family regulator) and repressed by BadM (a Rrf2-type regulator) (4, 15). The aadR gene is additionally regulated via activation by the oxygen-sensing two-component Fix (FixL, FixJ, and FixK) system (Fig. 1B) (16). The Fix-AadR hierarchy mediates the transition from microaerobic to anaerobic growth and further ensures anoxic conditions for the iron-sulfur center proteins of BadDEFG and BadB. To date, it is not known what signals lead to BadM derepression of badDEFGAB.
Within the aromatic degradation gene cluster, a single gene (badL) remains uncharacterized and is annotated as coding for a putative N-acetyltransferase. Acetylation occurs in all domains of life and involves the transfer of an acetyl group to the α or ε amino groups of proteins (Nα or Nε) and amine groups (Nα) of small molecules (17). Commonly, protein acetylation occurs on active site lysines of proteins (Nε), which in turn modulates their activity (18–25). Small-molecule acetylation has been shown to be involved in detoxification (26), translation inhibition (27, 28), and antibiotic neutralization (29, 30).
This study identifies BadL as a small-molecule N-acetyltransferase that modifies aminobenzoates (ABAs). We show that acetylated aminobenzoates (ABAAc, also known as acetamidobenzoate) bind to BadM and that BadM/ABAAc complexes no longer repress badDEFGAB operon expression. Results of growth analyses of wild-type and mutant strains, quantitative reverse transcription-PCR (qRT-PCR), and electrophoretic mobility shift assays suggest that acetamidobenzoates play a role in the regulation of benzoate degradation in R. palustris. High-performance liquid chromatography (HPLC) coupled to tandem mass spectrometry (LC/MS/MS) data show that BadL acetylates ABAs, and electrophoretic mobility shift assays (EMSAs) show that BadM can bind all three forms of acetamidobenzoates. On the basis of our data, we conclude that BadL activity is required for R. palustris growth on benzoate, and we suggest that BadL may work as a sensing mechanism for the presence of ABAs in the environment. Surprisingly, acetamidobenzoates also affect the synthesis of light-harvesting complexes I and II of R. palustris. These results suggest that BadL may be a link between carbon utilization and the generation of a light-driven proton motive force. To our knowledge, this may be the first example of a Gcn5-type acetyltransferase connecting carbon and energy conservation in a photosynthetic bacterium.
RESULTS
BadL acetyltransferase activity is required for photoheterotrophic growth on benzoate.
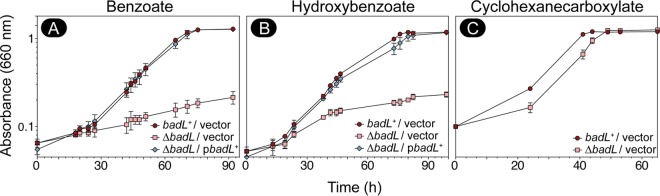
Prior to this work, the function of the putative BadL protein was unknown. Given the genomic context of the badL gene, we began investigating the function of this protein by deleting badL and screening for phenotypes related to benzoate utilization. The growth behavior of the R. palustris badL strain was assessed under photoheterotrophic conditions. An R. palustris ΔbadL strain failed to grow photosynthetically on benzoate (Fig. 2A) or 4-hydroxybenzoate (Fig. 2B) compared to badL+ controls. Notably, the ΔbadL strain grew as well as the badL+ strain on cyclohexanecarboxylate (Fig. 2C), suggesting that BadL function was necessary only for substrate activation and ring reduction (Fig. 1A, shaded area). The enzymes catalyzing the early steps of the pathway include the benzoyl-CoA synthetase (BadA) (EC 6.2.1.25), a benzoyl-CoA reductase (BadDEFG [dearomatizing]) (EC 1.3.7.8), and ferredoxin (BadB). The above-mentioned genes comprise the badDEFGAB operon of R. palustris and are tightly regulated by activation and repression (Fig. 1B). On the basis of the results shown in Fig. 2, we hypothesized that BadL played an as-yet-unidentified role in the regulation of badDEFGAB expression and that its putative function was to acylate either a protein or a small molecule.
FIG 2.
The BadL acetyltransferase is required for photoheterotrophic growth on benzoate and hydroxybenzoate. Cells were grown in biological triplicate photoheterotrophically on the carbon source (3 mM) listed above each panel, and growth was monitored at OD660. Cells lacking BadL had lower growth rates compared to badL+ controls when grown with benzoate (A) (pink squares versus red circles) or hydroxybenzoate (B) (pink squares versus red circles), but not when grown with cyclohexancarboxylate (C). Growth rates were restored when badL was reintroduced on a plasmid (blue diamonds). Each experiment was repeated in triplicate, and a representative growth curve is shown.
Deletion of badM restores photoheterotrophic growth of a ΔbadL strain on benzoate.
To address the possible involvement of BadL in badDEFGAB expression, we deleted badM in a ΔbadL strain. In the absence of BadL, growth of R. palustris was restored when badM was deleted (Fig. 3A, asterisks). A ΔbadL ΔbadM strain carrying a plasmid encoding the wild-type allele of badM failed to grow photoheterotrophically on benzoate (Fig. 3A, triangles), suggesting that somehow BadL function affected BadM DNA-binding activity.
FIG 3.
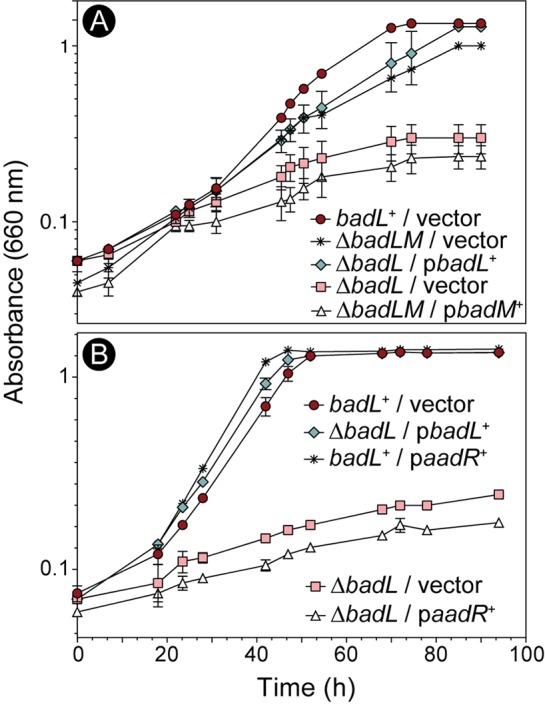
BadL-dependent phenotypes are restored upon deletion of badM, but not overexpression of aadR. Cells were grown in biological triplicate photoheterotrophically on benzoate (3 mM), and growth was monitored at OD660. (A) A ΔbadM ΔbadL strain grew similar to the badL+ strain (red circles versus asterisks), and ectopic expression of badM+ in the ΔbadM ΔbadL strain restored repression, hence a lack of growth (triangles). (B) Overexpression of aadR in a ΔbadL strain did not recover growth (white triangles). Each experiment was performed in triplicate, and a representative growth curve is shown. 4-ABA was present in the growth medium at 11 µM (2 mg/liter).
Importantly, overexpression of aadR, the activator of badDEFGAB, did not restore growth of the ΔbadL strain on benzoate (Fig. 3B, triangles), leading us to hypothesize that either BadL directly acetylated BadM, or alternatively, BadL acetylated a small-molecule effector of BadM. We note that prior to this work, small-molecule effectors of BadM DNA-binding activity were not known (15).
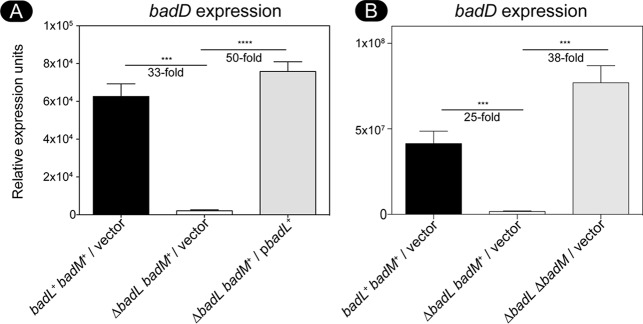
We investigated the potential role of BadL in badDEFGAB expression. For this purpose, we used qRT-PCR to monitor badDEFGAB operon expression using primers and conditions described elsewhere (15). When R. palustris was grown photoheterotrophically on succinate supplemented with benzoate, cells lacking badL had 30-fold-less badDEFGAB transcript than badL+ strains (Fig. 4A). Additionally, deletion of badM in a ΔbadL strain restored transcription of badDEFGAB under photoheterotrophic conditions with succinate plus benzoate (Fig. 4B). These results supported the idea that BadL was involved in the regulation of badDEFGAB expression, but it was unclear how.
FIG 4.
In a badL strain, badDEFGAB operon expression is reduced. Total RNA was obtained from cells grown photosynthetically with succinate and benzoate. (A) Expression of the badDEFGAB operon was assessed in badL+ and ΔbadL strains using qRT-PCR. The absence of badL led to a 30-fold decrease in badD expression. When badL was reintroduced on a plasmid, transcription increased 50-fold. (B) Expression of badDEFGAB was compared in badM+ or ΔbadM strains. Cells lacking badL had 25-fold-lower transcription of badDEFGAB. If badM was also deleted in cells lacking badL, badDEFGAB transcription was restored. Transcripts of badD were normalized to transcripts of the housekeeping gene fixJ. Cells were grown in biological triplicate, and qRT-PCR was performed in technical triplicates of each biological replicate. Error bars represent the standard deviations of the means. Values that are significantly different are indicated by a bar and asterisks as follows: ***, P value of >0.0005; ****, P value of <0.0001.
BadL homologues acetylate aminobenzoates, and the resulting acetamidobenzoates bind to BadM derepressing badDEFGAB expression.
Attempts to isolate reliably active R. palustris BadL (RpBadL) protein were unsuccessful. While RpBadL acetylated the small-molecule substrates mentioned below in subsequent experiments, the activity was inconsistent. For this reason, we used SEED viewer version 2.0 (31) to identify BadL homologues that clustered with BadM repressors. We focused our attention on BadL homologues present in Magnetospirillum magneticum (MmBadL; locus tag amb3392, 41% identity to RpBadL) and Geobacter metallireducens (GmBadL; locus tag gmet_2096, 47% identity to RpBadL) (Fig. 5A) because they shared the same genomic context and shared the highest level of identity. Both of these bacteria have been shown and predicted to degrade benzoate (32–36). Additionally, the genes coding for MmBadL or GmBadL partially complemented an R. palustris ΔbadL strain, indicating they may have similar functions in M. magneticum and G. metallireducens, respectively (Fig. 5B). MmBadL and GmBadL were successfully isolated and used in subsequent in vitro analyses. Attempts to acetylate BadM with MmBadL or GmBadL did not yield BadMAc under the conditions tested (data not shown).
FIG 5.
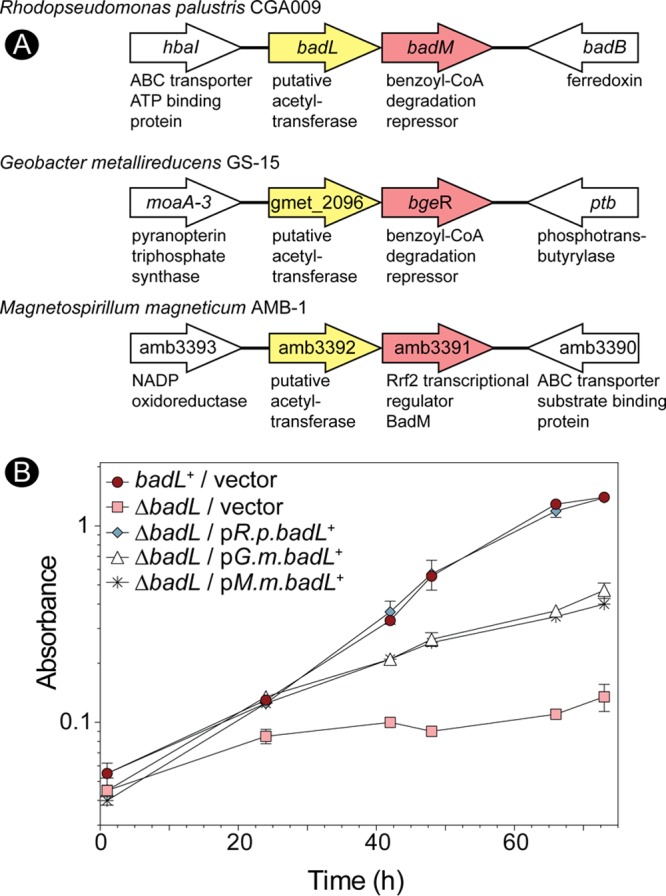
Bioinformatic analysis reveals putative badL badM gene clustering in other proteobacteria. (A) By analyzing the R. palustris badL and badM features of The SEED Viewer (pubseed.theseed.org/), other organisms were found to have similar clustering of Rrf2-type transcriptional regulators in putative operons with putative acetyltransferases. The putative BadL acetyltransferases from Geobacter metallireducens and Magnetospirillum magneticum were used for subsequent in vitro experiments. Genes surrounding the upstream and downstream badL badM regions for each organism are shown, and genes are not drawn to scale. (B) Cells were grown in biological triplicates photoheterotrophically with benzoate (3 mM). Cells lacking BadL complemented with G. metallireducens badL (G.m.badL) or M. magneticum badL (M.m.badL) had lower cell densities than badL+ controls did and recovered growth only partially.
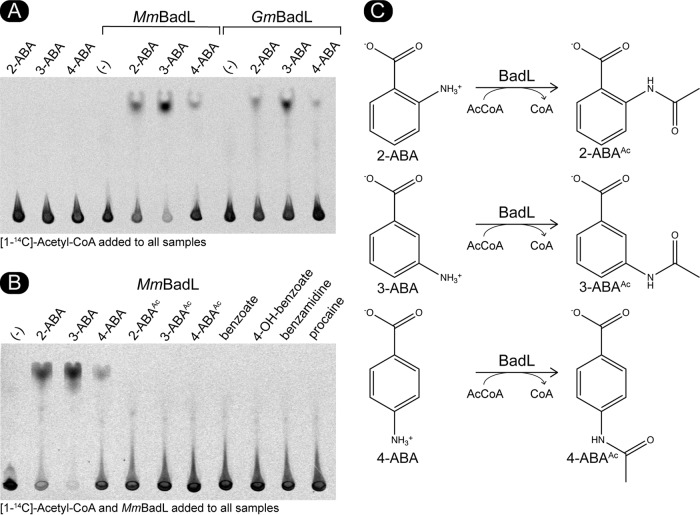
We considered the possibility that the substrate of BadL was not a protein but a small molecule. For this purpose, we incubated putative small-molecule substrates structurally related to benzoate. The transfer of radiolabeled acetyl moieties was assessed by thin-layer chromatography (TLC) using a mobile phase that resolved substrates and acetyl-CoA from products. Radiolabel distribution on the TLC was visualized by phosphor imaging. As mentioned above, we tested benzoate derivatives that could be found in the environment occupied by this bacterium. One such molecule, 4-aminobenzoate, is routinely added to the growth medium (11 µM) as a precursor of folate synthesis, because the genome of R. palustris does not code for enzymes to synthesize its own 4-aminobenzoate. Using this approach, we found that MmBadL and GmBadL acetylated 4-aminobenzoate producing 4-acetamidobenzoate (Fig. 6A). We further tested whether BadL could also acetylate 2- and 3-aminobenzoate. In fact, MmBadL and GmBadL did acetylate 2-, 3-, and 4-aminobenzoate, yielding 2-, 3-, and 4-acetamidobenzoate, albeit to different extents. These results suggested that BadL acetylated the amino group of aminobenzoates. In contrast, MmBadL and GmBadL did not acetylate benzoate, benzoates with hydroxyl group substituents at the same positions, or acetamidobenzoates (Fig. 6B). The proposed reactions and their products are shown in Fig. 6C. When analyzing unreacted [1-14C]-acetyl-CoA ([1-14C]Ac-CoA) at the point of sample application on the TLC plate, it appeared as if MmBadL had a preference for 3-ABA in vitro (Fig. 6A). We attempted to establish substrate preference by kinetic means, but unfortunately, the levels of activity of MmBadL and GmBadL were lower than the background limits of our assay; thus, we could not evaluate kinetic MmBadL and GmBadL parameters to determine a preferred substrate.
FIG 6.
BadL homologues acetylate aminobenzoates in vitro. (A) MmBadL and GmBadL (3 μg) were incubated with [1-14C]acetyl-CoA and 2-aminobenzoate (2-ABA), 3-aminobenzoate (3-ABA), or 4-aminobenzoate (4-ABA). As a negative control, 2-ABA, 3-ABA, and 4-ABA were incubated with [1-14C]Ac-CoA (three leftmost lanes). The lanes designated by (-) indicate reaction mixtures that contained BadL and [1-14C]Ac-CoA but lacked substrate. The reaction mixtures were spotted onto the gels, and acetylated products migrated away from the origin where [1-14C]-Ac-CoA remained. (B) MmBadL was incubated with various aromatic substrates as listed above each lane. Lanes labeled as 2-ABAAc, 3-ABAAc, and 4-ABAAc indicate reaction mixtures containing authentic acetylated 2-ABA, 3-ABA, and 4-ABA as described in Materials and Methods. Lane designated by (-) identify a reaction mixture where BadL was incubated with [14C-1]Ac-CoA but no substrate. The reaction mixtures were spotted onto gels, and acetylated products migrated away from the origin, where [1-14C]Ac-CoA remained. Images were acquired by exposure to a phosphor screen and subsequent imaging. (C) Proposed reaction schematic of BadL-mediated aminobenzoate acetylation.
Mass spectrometry analysis of the product of the MmBadL reaction.
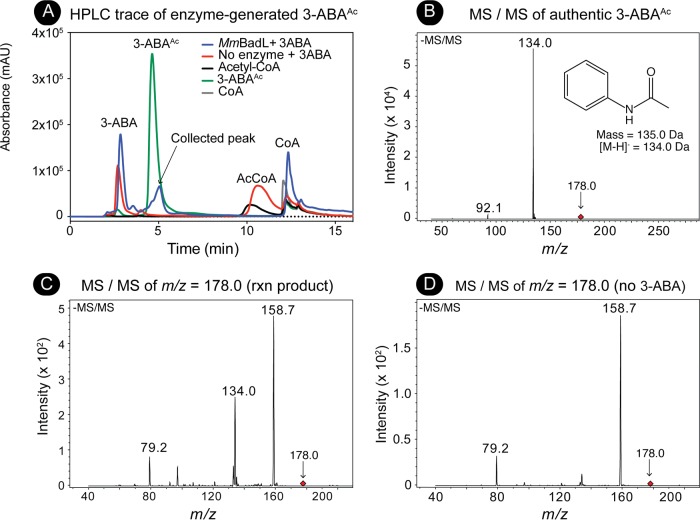
Since it appeared that MmBadL acetylated 3-ABA more efficiently and 3-ABAAc (retention time = 5.0 min) was readily resolved by reverse-phase HPLC away from 3-ABA (retention time = 2.8 min) (Fig. 7A), the products of reaction mixtures containing MmBadL and 3-ABA were analyzed by mass spectrometry (MS and MS/MS). A signal corresponding to the mass of enzyme-generated 3-ABAAc (m/z = 178.0 Da) (see Fig. S1 in the supplemental material) was fragmented (Fig. 7C) and compared to the authentic standard (Fig. 7B). MmBadL-generated and commercially available 3-ABAAc retained the same fragmentation pattern (m/z = 134.0), a fragment that corresponded to the loss of the carboxylate moiety (Fig. 7B, inset). These results confirmed the identity of the MmBadL product as 3-ABAAc. A control experiment (solvent without HPLC product) was performed, and the signals with m/z values of 79.2 and 158.7 in Fig. 7B were found to be unrelated to the product of the reaction (Fig. 7D).
FIG 7.
Isolation and identification of the MmBadL reaction product by HPLC and mass spectrometry. (A) Reaction mixtures containing MmBadL, 3-ABA, and Ac-CoA were set up as described in Materials and Methods. Protein was removed from mixtures, and material elution from a C18 kinetex column was monitored at 254 nm. Reaction mixtures containing MmBadL were compared to known standards (3-acetamidobenzoate [green peak]), and the black arrow indicated fractions that were collected for MS/MS. Absorbance is shown in milli arbitrary units (mAU) on the y axis. (B) Mass spectrometry was performed with commercially available 3-acetamidobenzoate (1 mM), which was identified by the signal with an m/z of 178.0 Da (red diamond) in the negative ion spectrum shown. The 178.0-Da compound was fragmented (negative ion shown). The inset shows the structure of the compound with a mass of 134.0 Da, which was consistent with 3-acetamidobenzoate without the carboxylic acid. (C) Mass spectrometry was performed on the fraction collected in panel A. The compound with an m/z of 178.0 Da (red diamond) corresponding to 3-acetomidobenzoate was fragmented (negative ion spectrum shown). The peak at 134.0 Da corresponds to the structure in the inset in panel B. Signals observed at m/z values of 158.7 Da and 79.2 Da were determined to be background from machine, as indicated in panel D. rxn product, reaction product.
Mass spectrometry of 3-ABAAc, 3-ABA, and BadL reaction product. Mass spectrometry (electrospray ionization [ESI]) was performed with commercially available 3-acetamidobenzoic acid or 3-aminobenzoic acid (1 mM), which was identified by the signal with an m/z of 178.0 Da or 137.1 Da, respectively. The inset shows the structure of each compound. MS/MS of BadL acetylated 3-ABA (as shown in Fig. 7) was performed on HPLC- purified product (right panel). Download FIG S1, TIF file, 9.5 MB (9.7MB, tif) .
Copyright © 2018 VanDrisse and Escalante-Semerena..
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
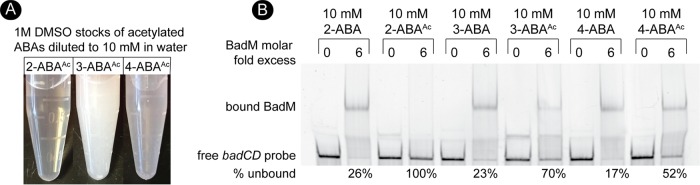
Acetamidobenzoates bind to BadM lowering its affinity for the badDEFGAB promoter.
The binding of BadM to the badDEFGAB promoter has been analyzed by others (15). We synthesized the badDEFGAB promoter region (212 nucleotides, −191 to +21 of badD ATG) with a 6-carboxyfluorescein (6-FAM) fluorescent label covalently attached to the 3′ end of the probe. Under the conditions tested, BadM bound all of the fluorescently labeled probes at sixfold molar excess, and this condition was used to test the effects of 2-, 3-, and 4-acetamidobenzoate. When BadM was incubated with 2-ABA, its binding to the badDEFGAB promoter probe did not differ from that of BadM incubated with the probe in the absence of ABAs (Fig. 8A, compare lane 2 to lane 4). However, addition of 2-acetamidobenzoate blocked binding of BadM to its DNA target (Fig. 8A, compare lane 2 to lane 6). This change was clear when the concentration of 2-acetamidobenzoate in the reaction mixture was 10 mM; when the concentration of 2-acetamidobenzoate was 5 mM, BadM/badDEFGAB promoter probe interactions were unaffected (Fig. 8B). This information helped us assess the effects of 3- and 4-acetamidobenzoate on BadM/badDEFGAB promoter probe interactions. Notably, at the concentrations necessary to affect BadM/badDEFGAB promoter probe interactions, the solubility of 3- and 4-acetoamidobenzoate was substantially reduced relative to that of 2-acetoamidobenzoate (Fig. 9A). When incubated under the same conditions, BadM/badDEFGAB promoter probe interactions changed upon incubation with 3- and 4-acetoamidobenzoate, albeit to a lesser degree than with 2-acetoamidobenzoate (Fig. 9B, see free probe band percentages). The alluded solubility issues with 3-acetoamidobenzoate and 4-acetoamidobenzoate prevented the quantification of ligand effects of BadM’s DNA-binding activity. It is possible that all three acetamidobenzoates may serve as ligands for BadM. In combination with qRT-PCR data shown in Fig. 4, these results indicate that BadL acetylates aminobenzoates, which in turn bind to BadM, leading to derepression of the badDEFGAB operon.
FIG 8.
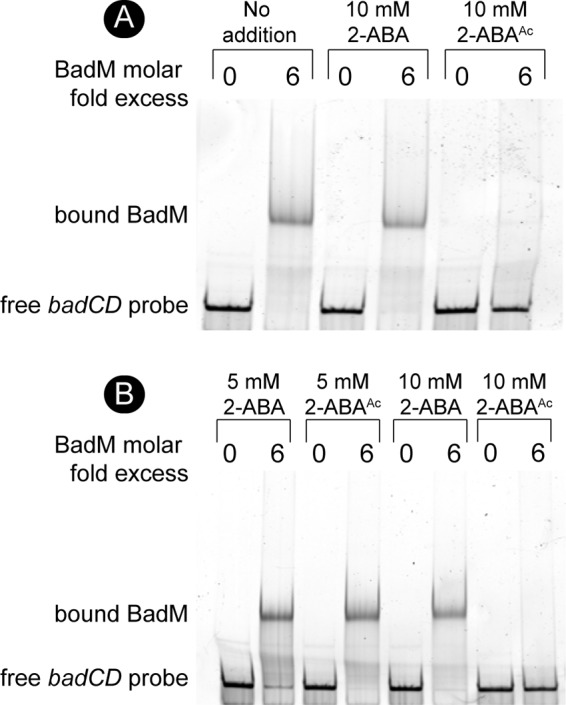
BadM DNA binding to the badDEFG promoter is reduced in the presence of acetylated 2-aminobenzoate. (A) Binding of BadM (72 nM) to the badDEFGAB promoter (12 nM) was analyzed by electrophoretic mobility shift assays using 6-FAM 5′-labeled probes. (A) Probe containing the intergenic region of badCD (212 bp, 0.303 pmol) was incubated with BadM (1.8 pmol) or without BadM. The addition of 2-aminobenzoate or 2-acetamidobenzoate is indicated above the respective lanes. (B) Binding of BadM to the badCD intergenic region as in panel A, but with different concentrations of 2-aminobenzoate or 2-acetamidobenzoate.
FIG 9.
Binding of BadM to DNA is affected by all acetamidobenzoates. (A) To meet reaction volumes, 2-, 3-, and 4-acetamidobenzoate were resuspended in 100% DMSO to a final concentration of 1 M. The images show the solubility of each chemical when diluted to 10 mM in water. (B) 6-FAM 5′-labeled probe containing the intergenic region of badCD (0.303 pmol, 12 nM) was incubated with BadM (1.8 pmol, 72 nM) or without BadM. Various acetamidobenzoates were added to reaction mixtures to the final concentrations indicated above each lane. The percentage of unbound badCD probe was analyzed by comparing the intensities of upper and lower bands of samples containing BadM. Intensities were calculated using ImageQuant software, and the percentages correspond to the lanes directly above the values.
Addition of acetamidobenzoates to benzoate medium restores growth of cells devoid of BadL.
To assess the substrate specificity of BadM in vivo, authentic 2-, 3-, or 4-acetamidobenzoate was added to cultures of a ΔbadL strain, and photoheterotropic growth with benzoate as the carbon source was compared to the response of the same strain to nonacetylated aminobenzoate. When 4-acetamidobenzoate was added to the medium, the final density of the ΔbadL cultures was similar to that of the badL+ strain, albeit growth rates were different (Fig. 10A). We note that restoration of growth of the ΔbadL strain upon addition of 4-acetamidobenzoate was not due to alternative carbon source utilization, as badL+ and ΔbadL cells failed to grow when only 4-acetamidobenzoate was added to the medium (Fig. 10B). Notably, the effect of 2- or 3-acetamidobenzoate on the growth of the ΔbadL strain was erratic and was not studied further (data not shown). Of note, the addition of 4-acetamidobenzoate decreased the lag phase of badL+ cells by 20% to 40% but did not alter the doubling time of these cells (from Fig. 10).
FIG 10.
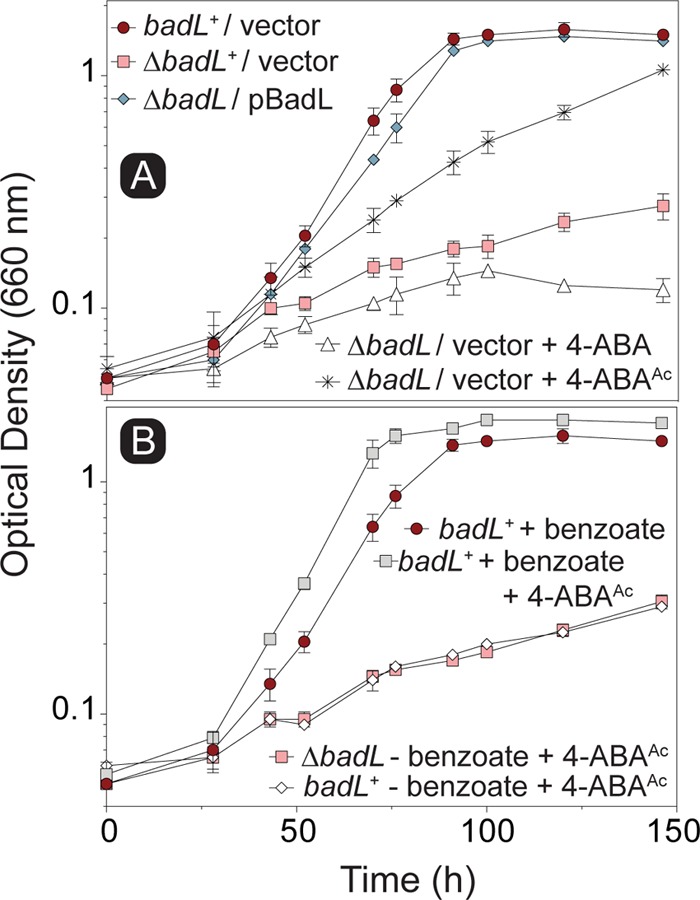
Addition of 4-acetamidobenzoate restores photoheterotrophic growth of ΔbadL cells on benzoate. Cells were grown in biological duplicates photoheterotrophically on benzoate (3 mM) with or without acetamidobenzoates (5 mM), and growth was monitored at 660 nm. (A) Growth analysis of ΔbadL cells was assessed upon the addition of 4-acetamidobenzoate. (B) To determine whether growth restoration of ΔbadL cells was due to alternative carbon source utilization, badL+ and ΔbadL cells were grown on 4-acetamidobenzoate in the presence or absence of benzoate, as indicated in the symbol key within each panel. Each experiment was performed in triplicate, and a representative growth curve is shown. Error bars represent standard deviations (SD) of a biological triplicate. 4-ABA was present in the growth medium at 11 µM (2 mg/liter).
BadL plays a role in the expression of genes encoding reaction center proteins.
We noticed a substantial difference in pigmentation between badL+ and ΔbadL strains during photoheterotrophic growth with benzoate. To determine whether this observation was due to differences in cell density or pigment synthesis, R. palustris badL+ and ΔbadL strains were grown on benzoate, and peak intensities at wavelengths corresponding to the light-harvesting 1 (LH1) reaction center complex (880 nm) and the light-harvesting 2 (LH2) reaction center complex (808 nm and 863 nm) were analyzed. Absorbance scans (A600 to A1000 [A600–1000]) of cultures of badL+ and ΔbadL strains were obtained during lag, log, and stationary growth phases. The R. palustris ΔbadL strain grew at a much lower rate with benzoate compared to the badL+ strain (9-h versus 36-h doubling time [Fig. 2]), and it was therefore ensured that comparisons between badL+ and ΔbadL were presented at the same cell densities. When the pigments of LH1 and LH2 complexes were analyzed, we found that ΔbadL cells had less pigment in both complexes than badL+ cells (Fig. 11). The lower level of pigment in the ΔbadL strain was independent of growth phase and was seen in lag, log, and stationary growth phase (Fig. 11). This lighter pigmentation was most evident in stationary-phase cells, as shown in the inset of Fig. 11.
FIG 11.
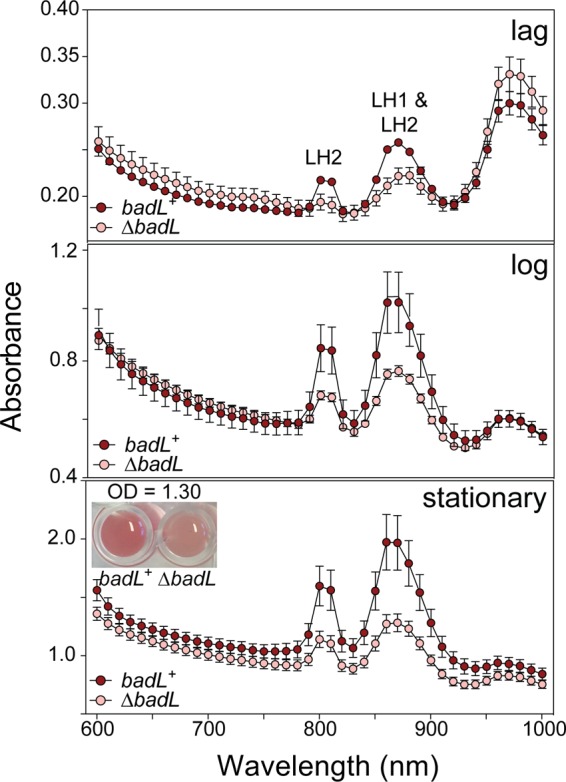
Cells lacking BadL have reduced light-harvesting reaction center complex pigments. The amount of light-harvesting 1 (LH1) and light-harvesting 2 (LH2) reaction center complexes in badL+ and ΔbadL strains was monitored spectrophotometrically during lag, log, and stationary growth phases. LH1 complexes absorb at 880 nm, and LH2 complexes absorb at 808 and 863 nm. Cells were analyzed at similar cell densities (measured at 660 nm) to ensure that differences in complexes were not due to differences in cell number. Data were obtained in triplicate on a SpectraMax instrument with absorbance readings every 10 nm. For visualization purposes, the inset in the bottom panel shows badL+ and ΔbadL cells at identical stationary-phase OD660. The experiment was repeated in biological triplicates, and error bars represent SD of a technical triplicate. 4-ABA was present in the growth medium at 11 µM (2 mg/liter).
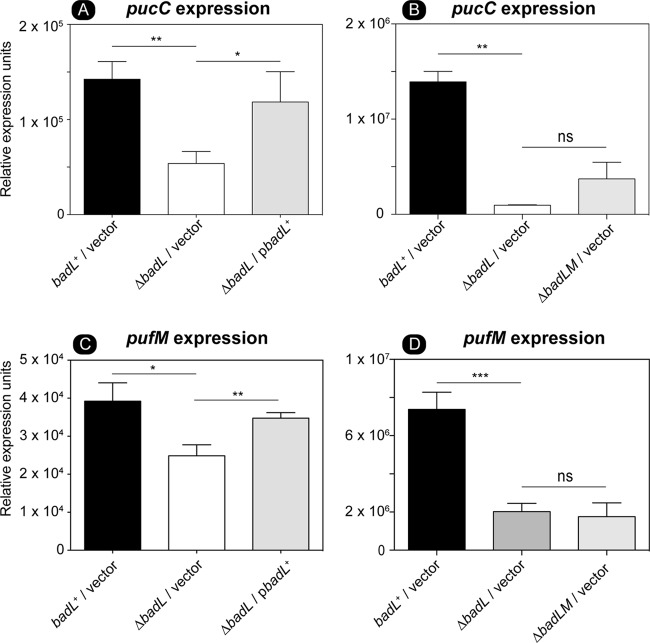
We looked further into an unexpected, possible role of BadL in the generation of light-driven proton motive force (pmf) in R. palustris. For this purpose, we used qRT-PCR to measure transcript levels of pucC and pufM. PucC is a putative chlorophyll major facilitator superfamily exporter, while PufM is a photosynthetic reaction center subunit. Subsequent experiments were performed in the presence of succinate to allow cells to grow. When cells were grown photoheterotrophically with succinate and benzoate, a strain lacking BadL had significantly lower levels of pucC and pufM transcripts compared to the levels measured in the badL+ strain (Fig. 12A and C). To determine whether or not this result was related to BadM function, pucC and pufM transcripts were analyzed in a ΔbadL ΔbadM strain. In cells lacking badL, the presence or absence of badM did not change the levels of pucC or pufM transcripts (Fig. 12B and D). In addition to qRT-PCR, spectral analysis of LH1 and LH2 complexes were obtained in badL+, ΔbadL, ΔbadM, and ΔbadL ΔbadM strains. In agreement with the qRT-PCR results, the strain carrying a deletion of badM and badL did not display increased pigment synthesis relative to the badL strain (Fig. 13A), suggesting that the reduction in pigmentation in the ΔbadL strain was unrelated to BadM function. To investigate whether acetamidobenzoates were involved in pigment synthesis or to determine whether BadL had different acetylation targets, pigment scans were obtained with badL cultures grown photoheterotrophically on benzoate supplemented with 2-, 3-, or 4-acetamidobenzoate. Absorbance readings related to LH1 and LH2 were partially restored to badL+ levels only when 4-acetamidobenzoate was added to the medium (Fig. 13B). This would suggest a new role for BadL in energy conservation or a secondary target for acetamidobenzoates that is directly or indirectly affecting pigment biosynthesis. These possibilities are under investigation.
FIG 12.
Deletion of badL lowers the level of pucC RNA transcripts, an effect that is independent of the presence or absence of BadM. Total RNA of cells grown photosynthetically with succinate plus benzoate was harvested as described in Materials and Methods. Expression of pucC was assessed in badL+, ΔbadL, or ΔbadL ΔbadM strains using qRT-PCR. (A) The absence of badL led to a decrease in pucC transcription. The gene for pucC codes for a probable major facilitator superfamily transporter for photosynthetic complex assembly. Transcription of pucC is restored when badL was provided on a plasmid. (B) pucC RNA level was not significantly different in ΔbadL and ΔbadL ΔbadM strains. (C) Deletion of badL led to a decrease in pufM transcription. The gene for pufM codes for the photosynthetic reaction center complex M. Transcription of pufM is restored with badL on a plasmid. (D) pufM RNA level was not significantly different in a ΔbadL versus ΔbadL ΔbadM strain. Values that are significantly different are indicated by a bar and asterisks as follows: **, P value of >0.005; *, P value of <0.01. Values that are not significantly different are indicated by a bar labeled ns.
FIG 13.
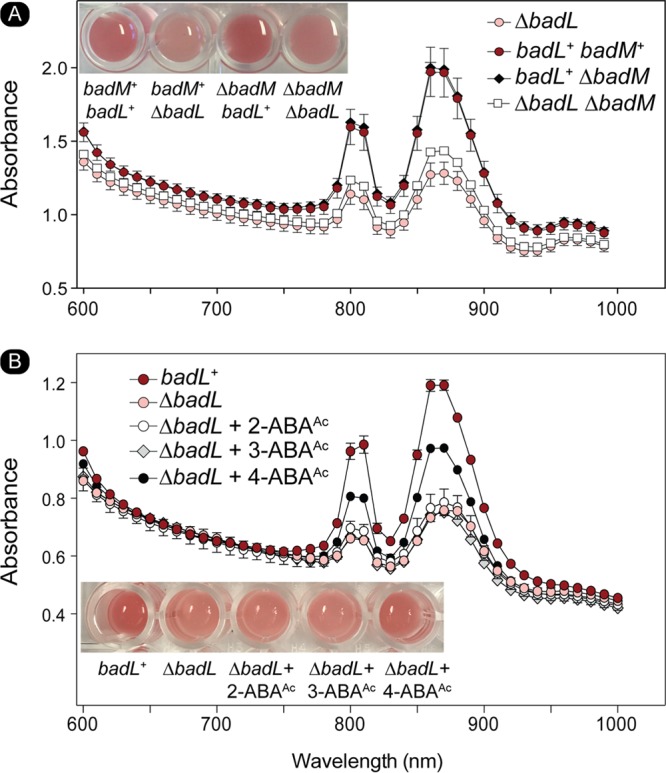
The absence of BadM does not restore pigment synthesis in a ΔbadL strain. The content of light-harvesting 1 (LH1) and light-harvesting 2 (LH2) reaction center complexes was assessed spectrophotometrically. LH1 complexes absorb at 880 nm, and LH2 complexes absorb at 808 and 863 nm. Cells were analyzed at similar cell densities (measured at OD660) to ensure that differences in complexes were not due to differences in cell numbers. Data were obtained in triplicate on a SpectraMax spectrophotometer with absorbance readings every 10 nm. (A) LH1 and LH2 complex analysis for badL+ and ΔbadL, ΔbadM, and ΔbadL ΔbadM cells. Picture inset shows the coloration of cells in a microtiter dish. (B) LH1 and LH2 complex analysis for badL+ and ΔbadL cells with either 2-, 3-, or 4-acetamidobenzoate (5 mM). 4-ABA was present in the growth medium at 11 µM (2 mg/liter).
DISCUSSION
The degradation of lignin-derived aromatics (e.g., benzoate, hydroxybenzoate) under anoxic conditions in the presence of light has been extensively studied in R. palustris. The pathway is well defined, and so are the genes encoding the enzymes required to convert such compounds to acetyl-CoA (3). In addition, elegant system-wide studies of the regulation of expression of the benzoate acid degradation (bad) genes of this bacterium have been reported (4, 15). In spite of this wealth of information, the function of one gene, badL, remained enigmatic. In genome databases, badL is annotated as encoding a homologue of the yeast Gcn5-type histone N- acetyltransferase (GNAT) (PF00583).
Here, we present evidence in support of the physiological role of the BadL protein in the degradation of benzoate and reveal an additional, unexpected role for this protein in the generation of the light-driven proton motive force of this bacterium. Importantly, we show that the role of BadL in photosynthesis is independent of its role in benzoate catabolism.
BadL is required for the expression of the genes encoding enzymes that dearomatize the benzene ring.
As shown by in vivo data (Fig. 2A and B), R. palustris requires BadL function to grow on benzoate as the source of carbon under photoheterotrophic conditions. Notably, BadL function is not required beyond the point of the pathway where intermediates have been dearomatized (Fig. 2C). On the basis of bioinformatic information (Fig. 5), we propose that the strategy of using an acetyltransferase to affect the function of the repressor is shared by Geobacter metallireducens, Magnetospirillum magneticum, and probably many other benzoate degraders. Prior to this work, it was known that AadR, a Crp-like regulator, activated transcription of the badDEFGAB operon and that AadR was itself regulated through an oxygen-sensing two-component system (16). We have shown that the repression of badDEFGAB is relieved through small-molecule acetylation and the binding of the acetylated product to BadM.
BadL function generates acetamidobenzoates, which bind to BadM, leading to badDEFGAB operon derepression.
The link between BadL function and the expression of genes encoding enzymes responsible for the activation of benzoate (EC 6.2.1.25) and its reduction to cyclohexa-1,5-diene-1-carbonyl-CoA (EC 1.3.7.8) suggested two possible functions for BadL. Either BadL modified and altered the function of BadA, BadB, BadDEFG, or BadM, or it acetylated a small molecule that would trigger the expression of the badDEFGAB genes. In vivo and in vitro evidence reported herein shows that the latter scenario is correct. BadL acetylates aminobenzoates (2-, 3-, or 4-ABA), yielding acetamidobenzoates (or 2-, 3-, or 4-ABAAc) (Fig. 6 and 7), which bind to the BadM repressor, decreasing its affinity for its binding site upstream of the badDEFGAB operon (Fig. 8); this conclusion is supported by qRT-PCR data (Fig. 4A). While the concentration (10 mM) of ABAAc required in gel shift analyses was high, other studies characterizing Rrf2 regulators (37–39) utilized similar concentrations. An Rrf2 regulator, NsrR, senses nitric oxide (NO) and controls the expression of genes for NO metabolism. In vitro gel shift analyses utilized 10 to 20 mM concentrations of NO in order to completely abolish DNA binding (38). Additionally, studies that characterized regulators that bind aromatics as ligands used concentrations ranging from 0.1 to 10 mM (40, 41). One such study analyzed MobR, a repressor for the 3-hydroxybenzoate 4-hydroxylase gene in the soil bacterium, Comamonas testosterone (40). 3-Hydroxybenzoate acted as a ligand for MobR and was required at 10 mM concentrations in gel shift analyses (40). Furthermore, consistent with the idea that acetamidobenzoates act through BadM is the fact that in the absence of BadM, BadL function is irrelevant to benzoate degradation (Fig. 4B). A model of this hypothesis is shown in Fig. 14.
FIG 14.
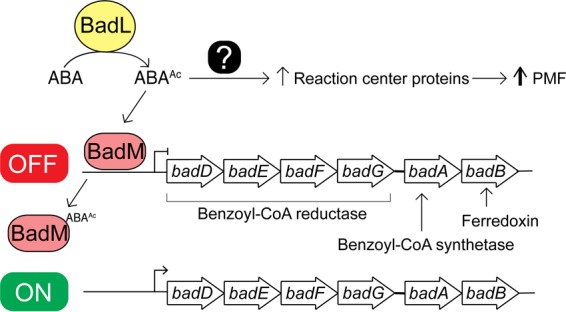
A model of the impact of acetamidobenzoates on benzoate degradation and photosynthesis. BadL acetylates aminobenzoates (ABAs) under photoheterotrophic growth. The resulting acetamidobenzoates (ABAAc) either directly or indirectly increase the presence of reaction center proteins. Additionally, ABAAc binds to the BadM repressor, triggering expression of the badDEFGAB operon. The combined activities of benzoyl-CoA synthetase (BadA), benzoyl-CoA reductase (BadDEFG), and ferredoxin (BadB) activate and dearomatize the benzene ring. PMF, proton motive force.
Phenotypic analyses validate the assigned functionality of BadL as an aminobenzoate acetyltransferase.
Results of growth behavior analyses of mutant and wild-type strains offer strong support to the conclusion that, in R. palustris, acetamidobenzoates generated by BadL are required to relieve the BadM-dependent repression of the badDEFGAB operon. In other words, in the absence of BadL, the badDEFGAB operon is not expressed unless BadM is absent (Fig. 3A and 4). The fact that certain acetamidobenzoates present in the growth medium can bypass the need for BadL (Fig. 10) validates the role of BadL in benzoate degradation. Concerning the physiological relevance of each acetamidobenzoate derivative, we cannot say for certain whether a single acetamidobenzoate is the true ligand for BadM. All three acetamidobenzoates tested altered the affinity of BadM for the badDEFGAB promoter (Fig. 8 and 9), but addition of each acetamidobenzoate to the growth medium of ΔbadL cultures led to different growth patterns. Notably, 4-acetamidobenzoate was not utilized as a carbon source (Fig. 10B), allowed the ΔbadL strain to grow photoheterotrophically on benzoate (Fig. 10A), altered DNA binding of BadM (Fig. 9), and restored pigmentation of the ΔbadL strain to wild-type levels (Fig. 13B). We would argue that in addition, 4-acetamidobenzoate may be used to allow for optimal growth of R. palustris under photosynthetic conditions.
BadL activity is required for the expression of light-harvesting proteins.
The initial observation regarding the difference in pigmentation between ΔbadL and badL+ strains was intriguing because there was no reason to suspect a link between BadL function and energy conservation at the gene expression level. Since benzoate is catabolized by R. palustris only under photosynthetic conditions, it is reasonable that benzoate catabolism and photosynthesis would be linked in this organism. Clearly, the extraction of reducing equivalents from carbon sources requires that reduced electron carriers be oxidized so they can continually participate in carbon catabolism. In microorganisms that have electron transport systems, the oxidation of reduced electron carriers results in proton extrusion with the concomitant generation of a proton motive force. Data presented in this paper (Fig. 10 to 13) uncover an unexpected role for the BadL acetyltransferase in the regulation of expression of genes encoding proteins that are later assembled into light-harvesting complexes. To our knowledge, a connection between BadL function and puc and puf gene expression has not been reported. Previously, it has been shown that puc (RPA1547) expression is modulated through the Fix oxygen-sensing histidine kinase system (16). We find it intriguing that BadL would also be required for the transcription of pucC. We have shown that this is a BadM-independent process, and it has proved to be more complex for the scope of this paper. Additional experiments to find 4-ABAAc targets with regard to puc and puf expression must be performed. These experiments could include restoration of pigmentation in ΔbadL strains by mutation or in vitro studies of interactions between 4-ABAAc and known pigment synthesis gene regulators.
Even though at this point we do not understand how BadL function is connected to energy generation, two points are worth discussing. First, we note that the observed decrease in light-harvesting 1 (LH1) and light-harvesting 2 (LH2) complexes in a ΔbadL strain is not affected by the absence of the BadM repressor (Fig. 12 and 13A), and second, 4-acetamidobenzoate partially corrects the pigmentation phenotype (Fig. 13B), consistent with the need for the newly assigned BadL function as a source of acetamidobenzoates. This unexpected new role for acetamidobenzoates in the expression of genes encoding functions involved in reaction center biosynthesis is an exciting contribution to the field that warrants further investigation.
Importance of BadL in Rhodopseudomonas palustris physiology.
From a physiological standpoint, the most intriguing question raised by this work is why does R. palustris rely on an N-acetyltransferase to control the transcription of genes required for photosynthetic growth on benzoate? One possible explanation is that aminobenzoates are very abundant in the environments occupied by R. palustris and that aminobenzoates can be readily deaminated to yield benzoate. Hence, high levels of aminobenzoates could have exerted a strong selective pressure for the evolution of a sensing mechanism that would activate the expression of genes encoding benzoate catabolic enzymes. At present, it appears as if the apparent lack of specificity of BadL for a given aminobenzoate may be an advantage, since maybe all aminobenzoates are used as a source of ammonia, yielding benzoate that can be used as carbon and energy. The connection of acetamidobenzoates to the expression of genes whose protein products are needed for the generation of a light-driven proton motive force is equally exciting, and further work in this area of R. palustris physiology will likely uncover new knowledge regarding the physiology of other bacteria that also appear to use N-acetyltransferases to regulate gene expression in response to diverse environmental stimuli.
MATERIALS AND METHODS
Chemicals, bacterial strains, culture media, and growth conditions.
All chemicals were purchased from Sigma-Aldrich with the following exceptions: [1-14C]acetyl-CoA (Moravek) (15 mCi mmol−1), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (GoldBio), isopropyl β-D-1-thiogalactopyranoside (IPTG) (GoldBio), ampicillin (GoldBio), 2-acetamidobenzoic acid (Alfa Aesar), 3- and 4-acetamidobenzoic acid (VWR). All strains and plasmids used in this study are listed in Tables S1 and S2 in the supplemental material. Escherichia coli strains DH5α (New England Biolabs) or C41(λDE3) (42) were grown on lysogeny broth (LB) (Difco) at 37°C. When used, antibiotics were added at the following concentrations: kanamycin (75 µg ml−1) and ampicillin (100 µg ml−1). All Rhodopseudomonas strains used in this study were derivatives of Rhodopseudomonas palustris CGA009 (43). For details on growth and pigment analysis of R. palustris, refer to Text S1 in the supplemental material.
Bacterial strains and plasmids used in this study. Unless otherwise noted, all plasmids and strains were constructed in this study. Download Table S1, DOCX file, 0.01 MB (14.8KB, docx) .
Copyright © 2018 VanDrisse and Escalante-Semerena..
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmids used in this study. Unless otherwise noted, all plasmids and strains were constructed in this study. Download Table S2, DOCX file, 0.02 MB (16.7KB, docx) .
Copyright © 2018 VanDrisse and Escalante-Semerena..
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental Materials and Methods. Download Text S1, PDF file, 0.1 MB (94.3KB, pdf) .
Copyright © 2018 VanDrisse and Escalante-Semerena..
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Molecular techniques.
Primers were synthesized from Integrated DNA Technologies and are listed in Table S3. Genomic DNA was synthesized using ethanol precipitation (44). DNA manipulations were performed using standard techniques (45). DNA was amplified using Phusion High-Fidelity DNA polymerase (New England Biolabs) following the manufacturer’s protocol for amplification of high-GC DNA (GC buffer and 3% dimethyl sulfoxide [DMSO] [vol/vol]). PCR products were analyzed on agarose (1% [wt/vol]) gels developed at 100 V. PCR products were purified using the Wizard SV Gel and PCR Clean-Up System (Promega), and plasmids were purified using the Wizard Plus SV Miniprep kit (Promega). DNA sequencing was performed at the Georgia Genomics Facility (Athens, GA, USA). For details on plasmid construction for overexpression in E. coli or complementation in R. palustris, refer to Text S1.
Primers used in this study. Download Table S3, DOCX file, 0.02 MB (16.3KB, docx) .
Copyright © 2018 VanDrisse and Escalante-Semerena..
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Purification of BadM and BadL proteins.
Plasmids pRpBadM8, pRpBadL1, pGmBadL1, and pMmBadL2 were transformed into E. coli C41(λDE3) pka12::kan+ (JE9314) cells. The resulting strains were grown overnight in 50 ml LB plus ampicillin. The cultures grown overnight were subcultured (1:100) into 2 liters of LB plus ampicillin and grown at 25°C to an optical density at 650 nm (OD650) of 0.5, after which transcription of genes was induced by the addition of IPTG (0.5 mM). Cells were harvested the next day by centrifugation at 6,000 × g for 15 min at 4°C in an Avanti J-2 XPI centrifuge equipped with rotor JLA-8.1000 (Beckman Coulter). Cell pellets were stored at −80°C until used. For further information on protein purifications and subsequent enzyme assays, refer to Text S1.
DNA-binding assays.
Electrophoretic mobility shift assays (EMSAs) were performed using DNA probes containing a 6-carboxyfluorescein (6-FAM) label covalently attached to the 3′ ends of synthesized PCR products. Probes were generated using primers listed in Table S3 and included the intergenic region (212 bp) of badC and badD as described previously (15). Further details can be found in Text S1.
RNA isolation.
Strains JE11529 (badL+/pBBR1-MCS2), JE13235 (ΔbadL/pBBR1-MCS2), and JE13236 (ΔbadL/pRpBadL3) were each grown in quadruplicate in 5 ml of YP medium plus kanamycin for 4 days. Cells were diluted (1:20 [vol/vol]) into 5 ml of fresh PM plus succinate and grown photosynthetically until cells reached mid-exponential phase (OD660 of ∼0.5, 24 h later). Exponentially growing cells were back diluted into 15 ml of fresh PM plus succinate to an OD660 of 0.03 and grown photosynthetically at 30 °C until the cultures reached an OD of 0.1, at which point benzoate (3 mM) and NaHCO3 (10 mM) were added to the cultures with a sterile syringe and needle (15). Cells were grown for 24 h after the addition of benzoate, and all 15 ml was harvested by centrifugation at 4,000 × g for 10 min. Supernatants were decanted, and cells were immediately centrifuged at 4,000 × g for 1 min. Excess medium was aspirated off, and cells were flash frozen and stored at −80°C until used. RNA was isolated using the RNAsnap method (46). For RNAsnap protocol and cDNA synthesis procedure, refer to Text S1.
ACKNOWLEDGMENTS
We thank Heidi Crosby for fruitful discussions and to the Proteomics and Mass Spectrometry Core Facility of the University of Georgia for the performance and analysis of LC/MS/MS.
Footnotes
Citation VanDrisse CM, Escalante-Semerena JC. 2018. Small-molecule acetylation controls the degradation of benzoate and photosynthesis in Rhodopseudomonas palustris. mBio 9:e01895-18. https://doi.org/10.1128/mBio.01895-18.
Contributor Information
Stephen Carlyle Winans, Cornell University.
Thomas Hanson, University of Delaware.
Tobias Erb, Max-Planck Institute, Marburg.
REFERENCES
- 1.Hunter CN, Daldal F, Thurnauer MC, Beatty JT (ed). 2009. Advances in photosynthesis and respiration, vol 28. The purple phototrophic bacteria. Springer, Dordrecht, The Netherlands. [Google Scholar]
- 2.Heider J, Fuchs G. 1997. Anaerobic metabolism of aromatic compounds. Eur J Biochem 243:577–596. doi: 10.1111/j.1432-1033.1997.00577.x. [DOI] [PubMed] [Google Scholar]
- 3.Harwood CS, Burchhardt G, Herrmann H, Fuchs G. 1998. Anaerobic metabolism of aromatic compounds via the benzoyl-CoA pathway. FEMS Microbiol Rev 22:439–458. doi: 10.1111/j.1574-6976.1998.tb00380.x. [DOI] [Google Scholar]
- 4.Dispensa M, Thomas CT, Kim MK, Perrotta JA, Gibson J, Harwood CS. 1992. Anaerobic growth of Rhodopseudomonas palustris on 4-hydroxybenzoate is dependent on AadR, a member of the cyclic AMP receptor protein family of transcriptional regulators. J Bacteriol 174:5803–5813. doi: 10.1128/jb.174.18.5803-5813.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dutton PL, Evans WC. 1969. The metabolism of aromatic compounds by Rhodopseudomonas palustris. A new, reductive, method of aromatic ring metabolism. Biochem J 113:525–536. doi: 10.1042/bj1130525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harwood CS, Gibson J. 1986. Uptake of benzoate by Rhodopseudomonas palustris grown anaerobically in light. J Bacteriol 165:504–509. doi: 10.1128/jb.165.2.504-509.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geissler JF, Harwood CS, Gibson J. 1988. Purification and properties of benzoate-coenzyme A ligase, a Rhodopseudomonas palustris enzyme involved in the anaerobic degradation of benzoate. J Bacteriol 170:1709–1714. doi: 10.1128/jb.170.4.1709-1714.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egland PG, Pelletier DA, Dispensa M, Gibson J, Harwood CS. 1997. A cluster of bacterial genes for anaerobic benzene ring biodegradation. Proc Natl Acad Sci U S A 94:6484–6489. doi: 10.1073/pnas.94.12.6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrison FH, Harwood CS. 2005. The pimFABCDE operon from Rhodopseudomonas palustris mediates dicarboxylic acid degradation and participates in anaerobic benzoate degradation. Microbiology 151:727–736. doi: 10.1099/mic.0.27731-0. [DOI] [PubMed] [Google Scholar]
- 10.Pelletier DA, Harwood CS. 1998. 2-Ketocyclohexanecarboxyl coenzyme A hydrolase, the ring cleavage enzyme required for anaerobic benzoate degradation by Rhodopseudomonas palustris. J Bacteriol 180:2330–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pelletier DA, Harwood CS. 2000. 2-Hydroxycyclohexanecarboxyl coenzyme A dehydrogenase, an enzyme characteristic of the anaerobic benzoate degradation pathway used by Rhodopseudomonas palustris. J Bacteriol 182:2753–2760. doi: 10.1128/JB.182.10.2753-2760.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boll M, Albracht SS, Fuchs G. 1997. Benzoyl-CoA reductase (dearomatizing), a key enzyme of anaerobic aromatic metabolism. A study of adenosinetriphosphatase activity, ATP stoichiometry of the reaction and EPR properties of the enzyme. Eur J Biochem 244:840–851. doi: 10.1111/j.1432-1033.1997.00840.x. [DOI] [PubMed] [Google Scholar]
- 13.Gibson KJ, Gibson J. 1992. Potential early intermediates in anaerobic benzoate degradation by Rhodopseudomonas palustris. Appl Environ Microbiol 58:696–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harwood CS, Gibson J. 1997. Shedding light on anaerobic benzene ring degradation: a process unique to prokaryotes? J Bacteriol 179:301–309. doi: 10.1128/jb.179.2.301-309.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirakawa H, Hirakawa Y, Greenberg EP, Harwood CS. 2015. BadR and BadM proteins transcriptionally regulate two operons needed for anaerobic benzoate degradation by Rhodopseudomonas palustris. Appl Environ Microbiol 81:4253–4262. doi: 10.1128/AEM.00377-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rey FE, Harwood CS. 2010. FixK, a global regulator of microaerobic growth, controls photosynthesis in Rhodopseudomonas palustris. Mol Microbiol 75:1007–1020. doi: 10.1111/j.1365-2958.2009.07037.x. [DOI] [PubMed] [Google Scholar]
- 17.Hentchel KL, Escalante-Semerena JC. 2015. Acylation of biomolecules in prokaryotes: a widespread strategy for the control of biological function and metabolic stress. Microbiol Mol Biol Rev 79:321–346. doi: 10.1128/MMBR.00020-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Starai VJ, Escalante-Semerena JC. 2004. Identification of the protein acetyltransferase (Pat) enzyme that acetylates acetyl-CoA synthetase in Salmonella enterica. J Mol Biol 340:1005–1012. doi: 10.1016/j.jmb.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 19.Thao S, Chen CS, Zhu H, Escalante-Semerena JC. 2010. N(epsilon)-lysine acetylation of a bacterial transcription factor inhibits its DNA-binding activity. PLoS One 5:e15123. doi: 10.1371/journal.pone.0015123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crosby HA, Heiniger EK, Harwood CS, Escalante-Semerena JC. 2010. Reversible N(epsilon)-lysine acetylation regulates the activity of acyl-CoA synthetases involved in anaerobic benzoate catabolism in Rhodopseudomonas palustris. Mol Microbiol 76:874–888. doi: 10.1111/j.1365-2958.2010.07127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tucker AC, Escalante-Semerena JC. 2013. Acetoacetyl-CoA synthetase activity is controlled by a protein acetyltransferase with unique domain organization in Streptomyces lividans. Mol Microbiol 87:152–167. doi: 10.1111/mmi.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.VanDrisse CM, Escalante-Semerena JC. 2018. In Streptomyces lividans, acetyl-CoA synthetase activity is controlled by O-serine and N(epsilon)-lysine acetylation. Mol Microbiol 107:577–594. doi: 10.1111/mmi.13901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.You D, Wang MM, Ye BC. 2017. Acetyl-CoA synthetases of Saccharopolyspora erythraea are regulated by the nitrogen response regulator GlnR at both transcriptional and post-translational levels. Mol Microbiol 103:845–859. doi: 10.1111/mmi.13595. [DOI] [PubMed] [Google Scholar]
- 24.Xu JY, You D, Leng PQ, Ye BC. 2014. Allosteric regulation of a protein acetyltransferase in Micromonospora aurantiaca by the amino acids cysteine and arginine. J Biol Chem 289:27034–27045. doi: 10.1074/jbc.M114.579078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song L, Wang G, Malhotra A, Deutscher MP, Liang W. 2016. Reversible acetylation on Lys501 regulates the activity of RNase II. Nucleic Acids Res 44:1979–1988. doi: 10.1093/nar/gkw053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hentchel KL, Escalante-Semerena JC. 2015. In Salmonella enterica, the Gcn5-related acetyltransferase MddA (formerly YncA) acetylates methionine sulfoximine and methionine sulfone, blocking their toxic effects. J Bacteriol 197:314–325. doi: 10.1128/JB.02311-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.VanDrisse CM, Parks AR, Escalante-Semerena JC. 2017. A toxin involved in Salmonella persistence regulates its activity by acetylating its cognate antitoxin, a modification reversed by CobB sirtuin deacetylase. mBio 8:e00708-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheverton AM, Gollan B, Przydacz M, Wong CT, Mylona A, Hare SA, Helaine S. 2016. A Salmonella toxin promotes persister formation through acetylation of tRNA. Mol Cell 63:86–96. doi: 10.1016/j.molcel.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burckhardt RM, Escalante-Semerena JC. 2017. In Bacillus subtilis, the SatA (formerly YyaR) acetyltransferase detoxifies streptothricin via lysine acetylation. Appl Environ Microbiol 83:e01590-17. doi: 10.1128/AEM.01590-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vetting MW, Hegde SS, Javid-Majd F, Blanchard JS, Roderick SL. 2002. Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis in complex with coenzyme A and aminoglycoside substrates. Nat Struct Biol 9:653–658. doi: 10.1038/nsb830. [DOI] [PubMed] [Google Scholar]
- 31.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crecy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, Gerdes S, Glass EM, Goesmann A, Hanson A, Iwata-Reuyl D, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N, Linke B, McHardy AC, Meyer F, Neuweger H, Olsen G, Olson R, Osterman A, Portnoy V, Pusch GD, Rodionov DA, Ruckert C, Steiner J, Stevens R, Thiele I, Vassieva O, Ye Y, Zagnitko O, Vonstein V. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res 33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carmona M, Diaz E. 2005. Iron-reducing bacteria unravel novel strategies for the anaerobic catabolism of aromatic compounds. Mol Microbiol 58:1210–1215. doi: 10.1111/j.1365-2958.2005.04937.x. [DOI] [PubMed] [Google Scholar]
- 33.Butler JE, He Q, Nevin KP, He Z, Zhou J, Lovley DR. 2007. Genomic and microarray analysis of aromatics degradation in Geobacter metallireducens and comparison to a Geobacter isolate from a contaminated field site. BMC Genomics 8:180. doi: 10.1186/1471-2164-8-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawaguchi K, Shinoda Y, Yurimoto H, Sakai Y, Kato N. 2006. Purification and characterization of benzoate-CoA ligase from Magnetospirillum sp. strain TS-6 capable of aerobic and anaerobic degradation of aromatic compounds. FEMS Microbiol Lett 257:208–213. doi: 10.1111/j.1574-6968.2006.00165.x. [DOI] [PubMed] [Google Scholar]
- 35.Shinoda Y, Akagi J, Uchihashi Y, Hiraishi A, Yukawa H, Yurimoto H, Sakai Y, Kato N. 2005. Anaerobic degradation of aromatic compounds by magnetospirillum strains: isolation and degradation genes. Biosci Biotechnol Biochem 69:1483–1491. doi: 10.1271/bbb.69.1483. [DOI] [PubMed] [Google Scholar]
- 36.Carmona M, Zamarro MT, Blázquez B, Durante-Rodríguez G, Juárez JF, Valderrama JA, Barragán MJL, García JL, Díaz E. 2009. Anaerobic catabolism of aromatic compounds: a genetic and genomic view. Microbiol Mol Biol Rev 73:71–133. doi: 10.1128/MMBR.00021-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munnoch JT, Martinez MT, Svistunenko DA, Crack JC, Le Brun NE, Hutchings MI. 2016. Characterization of a putative NsrR homologue in Streptomyces venezuelae reveals a new member of the Rrf2 superfamily. Sci Rep 6:31597. doi: 10.1038/srep31597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Isabella VM, Lapek JD Jr, Kennedy EM, Clark VL. 2009. Functional analysis of NsrR, a nitric oxide-sensing Rrf2 repressor in Neisseria gonorrhoeae. Mol Microbiol 71:227–239. doi: 10.1111/j.1365-2958.2008.06522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tucker NP, Hicks MG, Clarke TA, Crack JC, Chandra G, Le Brun NE, Dixon R, Hutchings MI. 2008. The transcriptional repressor protein NsrR senses nitric oxide directly via a [2Fe-2S] cluster. PLoS One 3:e3623. doi: 10.1371/journal.pone.0003623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshida M, Hiromoto T, Hosokawa K, Yamaguchi H, Fujiwara S. 2007. Ligand specificity of MobR, a transcriptional regulator for the 3-hydroxybenzoate hydroxylase gene of Comamonas testosteroni KH122-3s. Biochem Biophys Res Commun 362:275–280. doi: 10.1016/j.bbrc.2007.07.190. [DOI] [PubMed] [Google Scholar]
- 41.Bundy BM, Collier LS, Hoover TR, Neidle EL. 2002. Synergistic transcriptional activation by one regulatory protein in response to two metabolites. Proc Natl Acad Sci U S A 99:7693–7698. doi: 10.1073/pnas.102605799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miroux B, Walker JE. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol 260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 43.Larimer FW, Chain P, Hauser L, Lamerdin J, Malfatti S, Do L, Land ML, Pelletier DA, Beatty JT, Lang AS, Tabita FR, Gibson JL, Hanson TE, Bobst C, Torres JL, Peres C, Harrison FH, Gibson J, Harwood CS. 2004. Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris. Nat Biotechnol 22:55–61. doi: 10.1038/nbt923. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 45.Elion EA, Marina P, Yu L. 2007. Constructing recombinant DNA molecules by PCR. Curr Protoc Mol Biol Chapter 3:Unit 3.17.1–3.17.12. [DOI] [PubMed] [Google Scholar]
- 46.Stead MB, Agrawal A, Bowden KE, Nasir R, Mohanty BK, Meagher RB, Kushner SR. 2012. RNAsnap: a rapid, quantitative and inexpensive, method for isolating total RNA from bacteria. Nucleic Acids Res 40:e156. doi: 10.1093/nar/gks680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mass spectrometry of 3-ABAAc, 3-ABA, and BadL reaction product. Mass spectrometry (electrospray ionization [ESI]) was performed with commercially available 3-acetamidobenzoic acid or 3-aminobenzoic acid (1 mM), which was identified by the signal with an m/z of 178.0 Da or 137.1 Da, respectively. The inset shows the structure of each compound. MS/MS of BadL acetylated 3-ABA (as shown in Fig. 7) was performed on HPLC- purified product (right panel). Download FIG S1, TIF file, 9.5 MB (9.7MB, tif) .
Copyright © 2018 VanDrisse and Escalante-Semerena..
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacterial strains and plasmids used in this study. Unless otherwise noted, all plasmids and strains were constructed in this study. Download Table S1, DOCX file, 0.01 MB (14.8KB, docx) .
Copyright © 2018 VanDrisse and Escalante-Semerena..
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmids used in this study. Unless otherwise noted, all plasmids and strains were constructed in this study. Download Table S2, DOCX file, 0.02 MB (16.7KB, docx) .
Copyright © 2018 VanDrisse and Escalante-Semerena..
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental Materials and Methods. Download Text S1, PDF file, 0.1 MB (94.3KB, pdf) .
Copyright © 2018 VanDrisse and Escalante-Semerena..
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download Table S3, DOCX file, 0.02 MB (16.3KB, docx) .
Copyright © 2018 VanDrisse and Escalante-Semerena..
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.