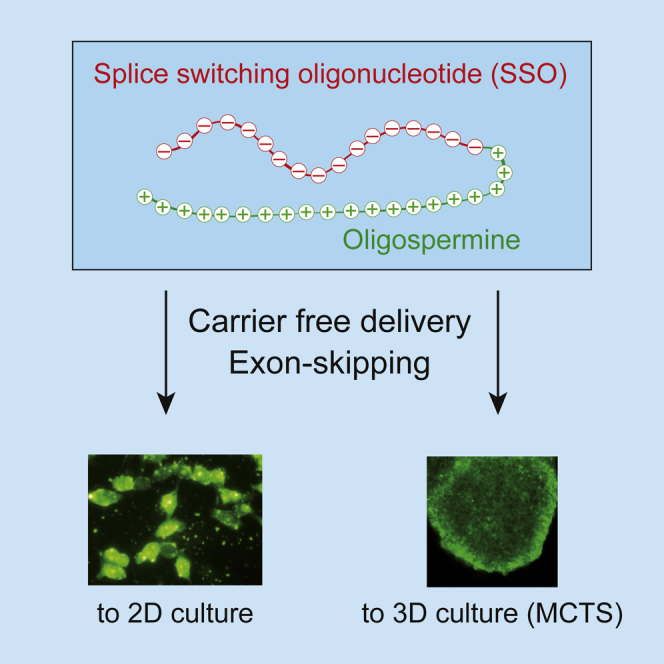
Abstract
We report the evaluation of 18-mer 2′-O-methyl-modified ribose oligonucleotides with a full-length phosphorothioate backbone chemically conjugated at the 5′ end to the oligospermine units (Sn-: n = 5, 15, 20, 25, and 30 [number of spermine units]) as splice switching oligonucleotides (SSOs). These conjugates contain, in their structure, covalently linked oligocation moieties, making them capable of penetrating cells without transfection vector. In cell culture, we observed efficient cytoplasmic and nuclear delivery of fluorescein-labeled S20-SSO by fluorescent microscopy. The SSO conjugates containing more than 15 spermine units induced significant carrier-free exon skipping at nanomolar concentration in the absence and in the presence of serum. With an increasing number of spermine units, the conjugates became slightly toxic but more active. Advantages of these molecules were particularly demonstrated in three-dimensional (3D) cell culture (multicellular tumor spheroids [MCTSs]) that mimics living tissues. Whereas vector-complexed SSOs displayed a drastically reduced splice switching in MCTS compared with the assay in monolayer culture, an efficient exon skipping without significant toxicity was observed with oligospermine-grafted SSOs (S15- and S20-SSOs) transfected without vector. It was shown, by flow cytometry and confocal microscopy, that the fluorescein-labeled S20-SSO was freely diffusing and penetrating the innermost cells of MCTS, whereas the vector-complexed SSO penetrated only the cells of the spheroid’s outer layer.
Keywords: cationic oligonucleotide-oligospermine conjugate, carrier-free oligonucleotide delivery, multicellular tumor spheroids, MCTS, splice switching oligonucleotides, SSO, exon skipping
Graphical Abstract
Introduction
Splice switching oligonucleotide (SSO) is one of the most advanced families of therapeutic oligonucleotides, being extremely valuable for the treatment of genetic diseases including Duchenne muscular dystrophy and spinal muscular atrophy.1, 2, 3, 4, 5, 6 SSOs are 15- to 30-mer oligonucleotide analogs that bind specifically and tightly to pre-mRNA targets, thus inducing an alternative splicing. A wide variety of chemically modified oligonucleotides is commercially available, possessing several desired properties for SSO drugs, for example, enhanced hybridization against RNA and improved stability against chemical and enzymatic degradations. In particular, SSOs based on the phosphorodiamidate morpholino oligonucleotide (PMO) and the 2′-methoxyethyl phosphorothioate oligonucleotide (2′-MOE/PS) led to the first approvals of SSO drugs by the US Food and Drug Administration (FDA) in 2016, even though further improvements are still required to increase their efficacy.7, 8, 9, 10 Contrary to classical drugs of low molecular weight (MW < 500), SSOs are macromolecular compounds (MW > 6 kDa) with high polarity. Their efficacy is mainly limited by their poor delivery to the target cells, and the development of an efficient, reliable, and less toxic delivery method remains the main challenge in this area. Our approach is to link covalently SSOs to a well-defined cationic vector by solid-phase chemistry.
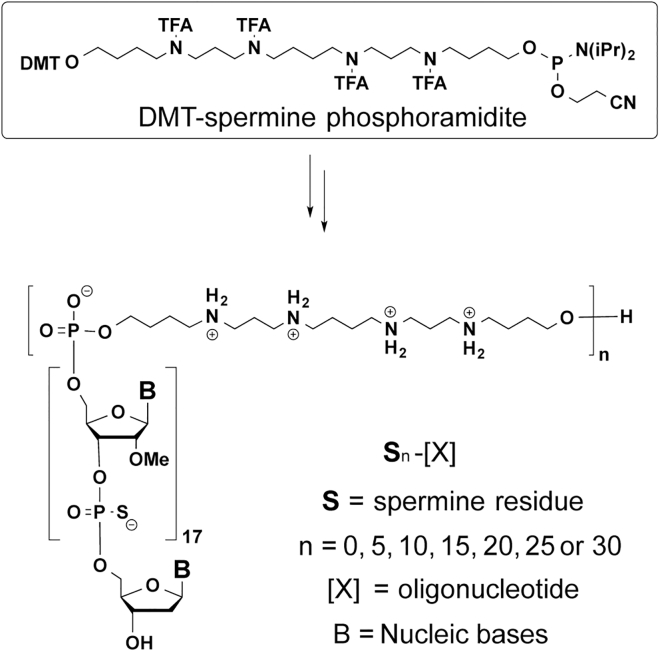
We already reported the use of DMT-spermine phosphoramidite (Figure 1) as a versatile reagent compatible with solid-phase oligonucleotide synthesis for the attachment of the desired number of spermine moieties to oligonucleotides,11, 12 which allowed us to synthesize a variety of oligospermine-oligonucleotide conjugates (zip nucleic acids [ZNAs]). When ZNAs are hybridized to their complementary strands, the cationic oligospermine tail acts as a zipper to neutralize the polyanionic internucleotidic phosphates, thus enhancing binding affinity and binding kinetics.13 These biophysical properties can be finely tuned according to the number of attached spermine units, making ZNA a versatile PCR probe.14, 15 ZNA is commercially available (number of spermine units <10) and used in numerous nucleic-acid-based diagnostic applications.16 Cationic oligospermines covalently attached to oligonucleotides can also act similarly to the polyamine-type delivery vectors. We described small interfering RNA (siRNA)-oligospermine conjugates containing 30 spermine units that induced an efficient carrier-free luciferase gene silencing.17, 18, 19, 20 Locked nucleic acid (LNA)-oligospermine conjugates with nine spermine units were also reported as active cell-permeable oligonucleotides for antisense and antigene inhibition of gene expression.21 More recently, the oligospermine with 15 spermine units was attached to cyclic RGD (cRGD)-siRNA conjugate, thus enhancing the tumor cell-specific delivery.22
Figure 1.
Solid-Phase Synthesis of Oligospermine-Oligonucleotide Conjugates (S)n-[X]
In this article, we used the same chemistry to prepare a series of oligospermine-SSO conjugates (Sn-SSOs, n = 5–30) and evaluated their splice switching efficacy without delivery vector. First, we studied time-course and concentration dependence of carrier-free cellular uptake of S20-SSO labeled with fluorescein at the 3′ end. Then, we studied the exon-skipping activity (duration and efficacy) of Sn-SSOs without vectors on a model cell line HeLa pLuc/705.23, 24 Our studies showed that the SSO conjugated to 20 spermine units was efficiently transfected into monolayer cells without any vector, and that the Sn-SSOs with more than 15 spermines units (n > 15) induced a sequence-specific exon skipping that restored luciferase expression at a significant level even 5 days after a single transfection. We finally investigated Sn-SSOs properties using multicellular tumor spheroids (MCTSs); they correspond to three-dimensional (3D) cell culture that has been suggested as a model for assessing oligonucleotide in vivo delivery.25 On the one hand, unconjugated SSOs transfected with a vector displayed a drastically reduced activity in MCTS as compared with the assay in monolayer culture. On the other hand, oligospermine conjugates, S15- and S20-SSOs, repeatedly induced high levels of carrier-free splice switching that surpassed the level of the cationic vector formulation. Cellular uptake of these conjugates in MCTS was also studied by flow cytometry and confocal microscopy.
Results
In this study, all assays were performed in HeLa pLuc/705 cells. This cell line was developed by Kole and colleagues23, 24 and used for the first time to demonstrate that appropriate oligonucleotide analogs (2′-O-methyloligoribonucleotide phosphorothioate) induce alternative splicing when delivered in cells. HeLa pLuc/705 cells were stably transfected with a recombinant plasmid (pLuc/705) carrying the luciferase gene (reporter gene) interrupted by a mutated human β-globin intron 2 (IVS2-705). The mutation in the intron causes aberrant splicing of luciferase pre-mRNA, preventing translation of luciferase. Efficient delivery of appropriate SSOs overlapping the 705 splice site induces correct splicing, and thus restores luciferase activity.
Oligonucleotide-Oligospermine Conjugates Synthesis
Structures of 18-mer oligonucleotide-oligospermine conjugates used in the present study are presented in Figure 1 and their sequences are listed in Table 1. They were synthesized as 2′-O-methyloligoribonucleotide phosphorothioate using essentially the same procedure as previously reported.20 The spermine units at the 5′ end of the oligoribonucleotide were introduced by efficient coupling (coupling yields >95%) of DMT-spermine phosphoramidite. Final conjugates were characterized by MALDI-TOF (Figures S3–S10) and/or SDS-PAGE (Figure S1).
Table 1.
Oligospermine-Oligonucleotide Conjugates
| Name | Sequence (5′ to 3′) | Yield (nmol) | [MH]+calc (Da) | [MH]+obs (Da) |
|---|---|---|---|---|
| [ON705] | CCU CUU ACC UCA GUU ACdA | 72 | 6,068 | 6,069 |
| S5-[ON705] | S5-CCU CUU ACC UCA GUU ACdA | 90 | 8,111 | 8,123 |
| S15-[ON705] | S15-CCU CUU ACC UCA GUU ACdA | 61 | 12,197 | 12,225 |
| S20-[ON705] | S20-CCU CUU ACC UCA GUU ACdA | 50 | 14,239 | 14,523 |
| S30-[ON705] | S30-CCU CUU ACC UCA GUU ACdA | 66 | 18,324 | ND |
| S15-[ON705mis]a | S15-CCU CUU ACA UCA GUU ACdA | 55 | 12,220 | 12,311 |
| S15-[ON705scr] | S15-ACU ACC CGA UAU CUC CUdC | 120 | 12,195 | ND |
| S15-[ON119] | S15-UGA GAC UUC CAC ACU GAdT | 97 | 12,313 | ND |
| [ON705]-F | CCU CUU ACC UCA GUU ACA-F | 300 | 6,685 | 6,690 |
| S20-[ON705]-F | S20-CCU CUU ACC UCA GUU ACA-F | 40 | 14,855 | 14,880 |
All capital letters in the sequences refer to 2′-O-methylribonucleotides. dN, 2′-deoxyribonucleotides; -F, fluorescein label at the 3′ end; ND, not determined.
Mismatch is underlined.
Carrier-free Splice Switching with Spermine-Grafted SSOs in Monolayer Culture
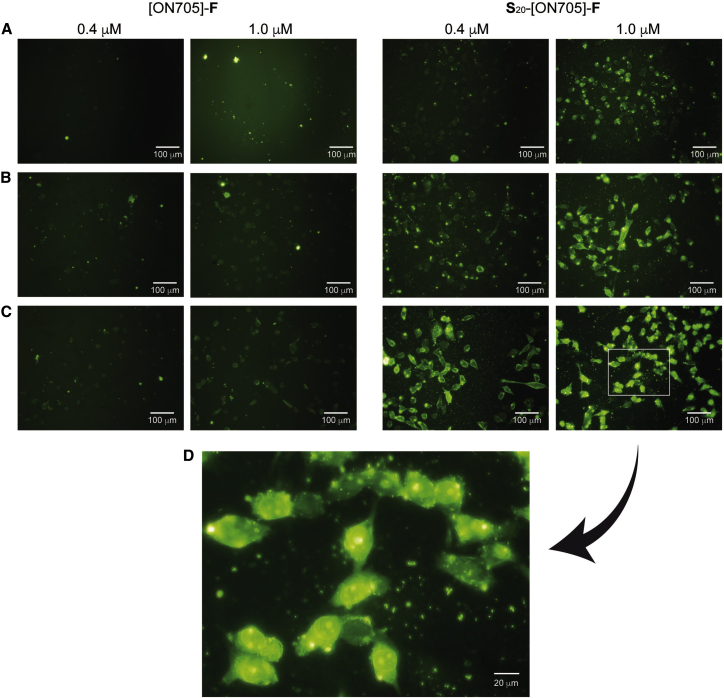
Carrier-free cellular uptake of cationic DNA oligonucleotides and transfection of spermine-grafted siRNAs have already been reported.17, 18, 19, 20 Here, we used two single-stranded fluorescein-labeled SSOs (2′-O-methyloligoribonucleotide phosphorothioate) with and without spermine units to re-evaluate their cell penetration properties (Figure 2). No significant cellular uptake in HeLa cells was observed when unconjugated [ON705]-F was incubated at 0.4 or 1.0 μM for up to 4 hr. In contrast, cellular uptake of S20-[ON705]-F was already observed at 0.4 μM in 2 hr. Transfection efficacy was progressively increased with dose and over time. All cells were transfected at 1.0 μM after 2 hr and at 0.4 μM after 4 hr. Transfected cells appeared with fluorescent nuclear edge and nucleoli concentration in diffuse green throughout their cytoplasm, highlighting the efficient cytoplasmic and nuclear delivery of S20-[ON705]-F.
Figure 2.
Carrier-free Cellular Uptake of (S)20-[ON705]-(F) in Monolayer HeLa pLuc/705 Cells
Fluorescently labeled (S)20-[ON705]-(F) were incubated at the concentrations indicated on the top of images during (A, right panels) 45 min, (B, right panels) 2 hr, and (C, right panels) 4 hr in serum-free medium. Naked [ON705]-(F) was used as a control in the same conditions (A–C, left panels). Images were taken by fluorescent microscopy using 488-nm laser. (D) Zoom of the right panel in (C).
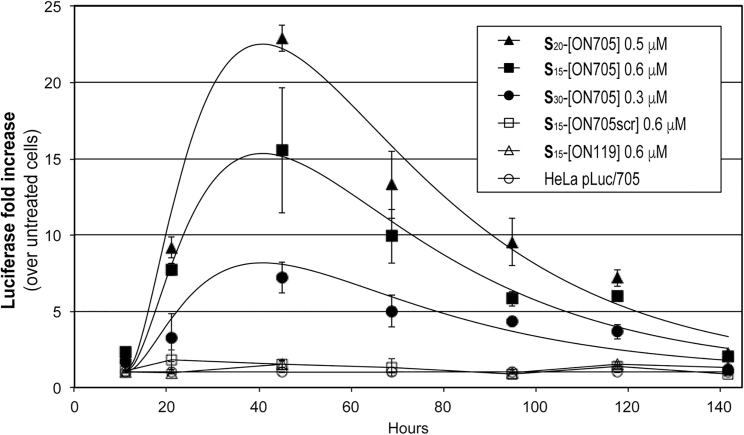
Restoration of luciferase expression was studied by harvesting cells at various times, between 10 and 144 hr after carrier-free transfection of three Sn-[ON705] (n = 15, 20, and 30) (Figure 3). For all three Sn-[ON705] tested, luciferase expression increased progressively over 45 hr before decreasing slowly. With S15-[ON705] (0.5 μM), a 15-fold increase in luciferase expression was observed after 45 hr of incubation compared with the control cells. Best results were obtained with S20-[ON705] (0.6 μM), resulting in a 22-fold increase in luciferase expression. However, increasing the number of spermine residues further proved to be detrimental because it resulted in higher levels of toxicity (see below) and lower increase in luciferase expression (7-fold for S30-[ON705] at 0.3 μM). Interestingly, with all three Sn-[ON705] (n = 15, 20, and 30), only one transfection was sufficient to observe significant luciferase expression (3- to 7-fold increase), even after 5 days. No significant luciferase expression was observed with S15-[ON705scr] or with S15-[ON119].
Figure 3.
Time-Course Profile of Carrier-free Splice Switching in Monolayer HeLa pLuc/705 Cells Using (S)n-[ON705]
(S)n-[ON705] (n = 15, 20, and 30) were added to the cells initially in serum-free DMEM, and FBS was added to 10% after 4 hr. Luciferase reporter gene expression levels were periodically measured as RLU/mg of cell proteins and normalized against the levels of untreated cells. All data are presented as mean ± SEM of n = 3 separate experiments.
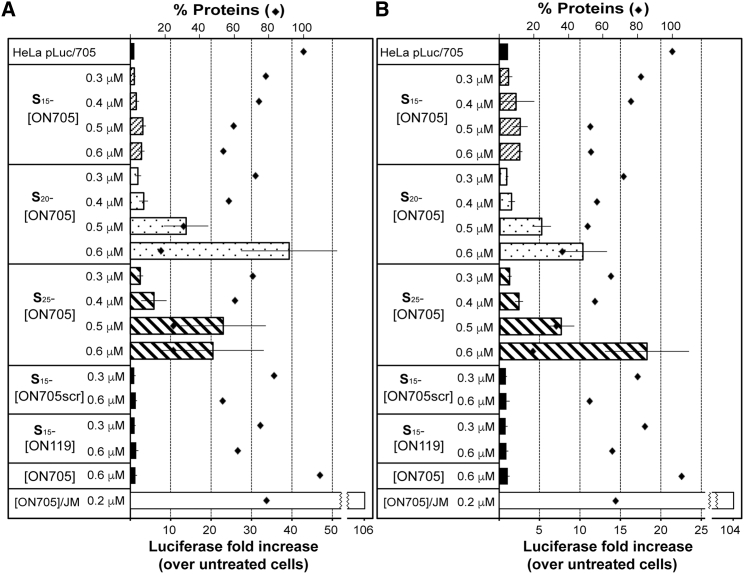
Dose-dependent splice switching activities of Sn-[ON705] (n = 15, 20, and 25) were next examined. These conjugates were initially incubated in the absence of serum for 4 hr (Figure 4A). Luciferase gene expression was measured after 48 hr (total incubation time), as well as the total cellular protein measurement. We found that a progressive increase in Sn-[ON705] (n = 15, 20, and 25) concentrations from 0.3 μM to 0.4 and 0.5 μM resulted in an enhanced restoration of luciferase expression. For example, luciferase expression increased by 2-, 3-, and 14-fold at 0.3, 0.4, and 0.5 μM, respectively, compared with untreated cells, when S20-[ON705] was employed. However, increasing the concentrations of S15-[ON705] and S25-[ON705] from 0.5 to 0.6 μM led to no significant change in luciferase expression level. Interestingly, S20-[ON705] gave better results at 0.6 μM, with a luciferase expression increased by 40-fold. As mentioned previously, luciferase expression restoration was also found to be dependent on the number of conjugated spermine units: levels of luciferase increased by 3-, 14-, and 23-fold with S15-, S20-, and S25-[ON705], respectively, at 0.5 μM. However, higher toxicity was observed with S20-[ON705] and S25-[ON705] under these conditions via total cellular protein measurement.
Figure 4.
Carrier-free Splice Switching in Monolayer HeLa pLuc/705 Cells by (S)n-[ON705] (n = 15, 20, and 25)
Luciferase activity was determined after 48 hr of incubation. (A) (S)n-[ON705] were incubated for 4 hr in serum-free conditions; then FBS (10%) was added. (B) (S)n-[ON705] were added in serum-containing DMEM (10%). The rhombi indicate the total protein measurement. All data are presented as mean ± SEM of n = 3 separate experiments. JM, JetMessenger.
The same SSOs were re-evaluated by adding the conjugate molecules directly in the presence of serum (Figure 4B). Under these conditions, luciferase restoration was slightly less efficient with Sn-[ON705] (n = 15, 20, and 25), even though they were found to be less toxic.
In both experiments in Figure 4, sequence specificity of exon skipping was clearly demonstrated, as S15-[ON705scr] and S15-[ON119] were completely inactive (S15-[ON705mis] was also shown to be inactive in a preliminary study; data not shown). Vector-assisted transfection of unconjugated [ON705] induced efficient luciferase expression restoration (106- or 104-fold increase in the absence or the presence of serum, respectively) with low toxicity at 0.2 μM.
In summary, data shown in Figures 2, 3, 4, and 5 demonstrate that the oligospermine-grafted 18-mer 2′-O-methyloligoribonucleotide phosphorothioates (Sn-[ON705]; n = 15, 20, and 25) are efficiently delivered in monolayer cells without vector, and that they induce long-lived splice switching in the presence and in the absence of serum. Even though unconjugated SSOs transfected with a vector were more efficient and less toxic under these experimental conditions, such nanoparticles should be less diffusible in vivo than molecular Sn-SSO drugs to attain the target tissue through interstitial fluid. In order to demonstrate this assumption, 3D cultured cells assays were performed, whose results are described in the following section.
Figure 5.
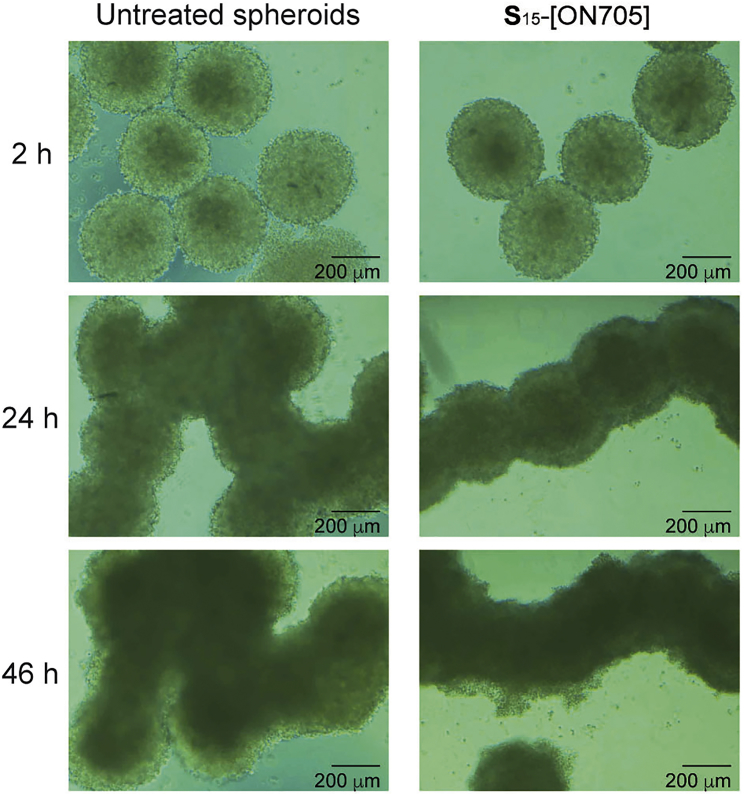
Morphology of HeLa pLuc/705 MCTS
Two-day-old spheroids were incubated with (S)15-[ON705] 0.7 μM in serum-containing DMEM (10%) for 46 hr. Images were periodically taken by microscopy. (Left) Untreated spheroids. (Right) Spheroids treated with (S)15-[ON705].
Carrier-free Splice Switching with Spermine-Grafted SSOs in MCTS
MCTS is an in vitro 3D culture that can be generated by growing cells to form complex spherical structures with 200- to 500-μm diameters. These multicellular spheroids are used as models in drug screening research because of their complexity level lying between standard in vitro two-dimensional (2D) monolayer cultures and in vivo tumors.26, 27, 28, 29 Here we studied delivery of the oligospermine-conjugated SSOs (S15- and S20-[ON705]) in 3D cell culture and found that the cationic SSOs were efficiently penetrating the cells in the inner region of spheroids to restore luciferase gene expression. Vector-assisted transfection of unconjugated [ON705] in MCTS was drastically less efficient than in 2D culture, with their cellular uptake being observed only in the outer layer of spheroids.
We first prepared HeLa pLuc/705 MCTSs using the hanging drop method.25, 30 About 2,500 cells were simply incubated in a suspended droplet of medium (25 μL) during 48 hr. The resulting MCTSs were very homogeneous in size (about 400 μm in diameter) and in shape (Figure 5). Twelve MCTSs were gathered per wells for assays. Under these conditions, untreated MCTSs (left photos in Figure 5) continued to grow over the next 46 hr and formed a chaplet-like structure by interacting with neighboring spheroids. The darkening of the central part of each spheroid can be interpreted partly by the increase of the cell density and partly by the known development of the central hypoxic and necrotic areas.26 Growth of the spheroids treated with 0.7 μM S15-[ON705] is shown in the right photos of Figure 5. These photos are indistinguishable from those of the untreated spheroids, demonstrating that spermine-grafted SSOs present no toxicity under these conditions.
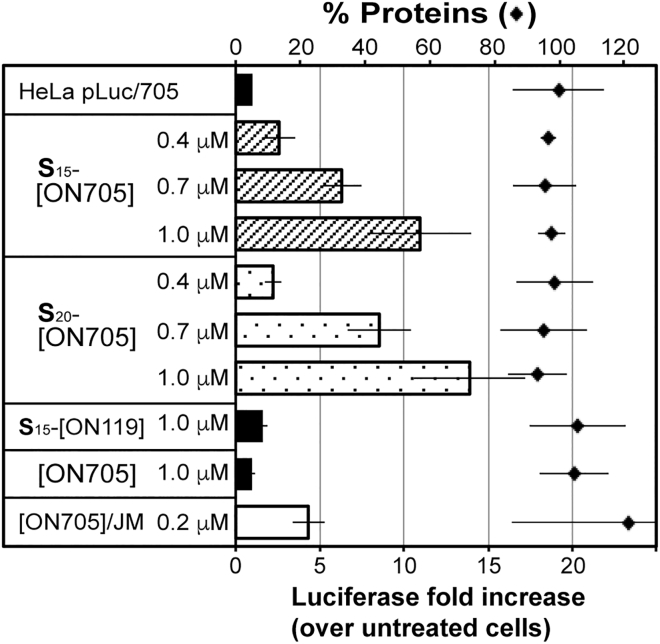
Carrier-free deliveries of S15-[ON705] and S20-[ON705] in HeLa pLuc/705 MCTSs were then evaluated by luciferase gene expression restoration. Incubation of S15-[ON705] or S20-[ON705] in increasing concentrations progressively enhanced the luciferase gene expression (Figure 6). For example, increases in luciferase expression were 3-, 6-, and 11-fold at 0.4, 0.7, and 1.0 μM, respectively, with S15-[ON705], and 2-, 8-, and 14-fold at 0.4, 0.7, and 1.0 μM with S20-[ON705]. Contrary to the aforementioned 2D culture assays, the 20-spermine units grafted SSO (S20-[ON705]) was only slightly better than the 15-spermine units grafted SSO (S15-[ON705]). As previously mentioned, no significant toxicity was observed in 3D culture, neither with S15-[ON705] nor with S20-[ON705], even at 1.0 μM, according to the total protein measurement (rhombi in Figure 6). In 3D MCTS, vector-assisted formulation of naked [ON705] induced a reduced level of luciferase expression, as compared with the results obtained in 2D culture. The increase of luciferase expression was only 4-fold, whereas it was 104-fold under the same conditions in monolayer culture.
Figure 6.
Carrier-Free Splice Switching in 3D Culture of HeLa pLuc/705 Cells by (S)15- and (S)20-[ON705]
(S)15- and (S)20-[ON705] were added to 2-day-old HeLa pLuc/705 spheroids in serum-containing DMEM (10%). Luciferase activity was determined after 48 hr of incubation. The rhombi indicate the total protein measurement. All data are presented as mean ± SEM of n = 3 separate experiments. JM, JetMessenger.
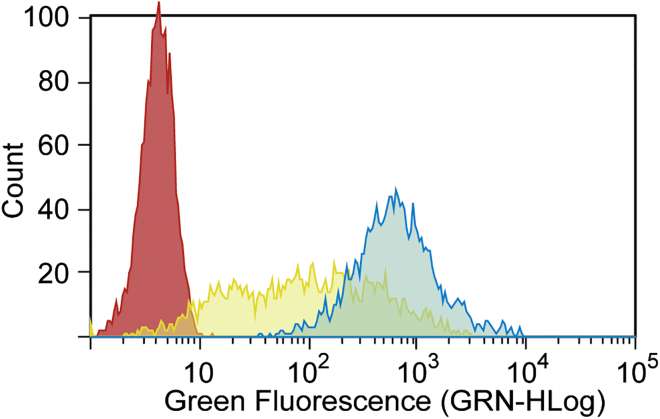
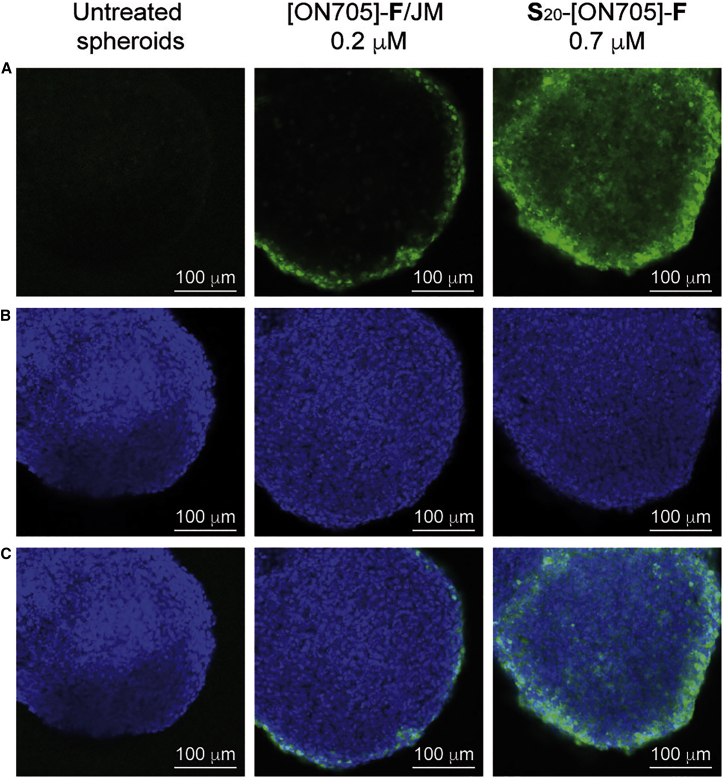
We further examined the delivery of oligospermine-oligonucleotide conjugates in spheroids by flow cytometry (Figure 7) and confocal microscopy (Figure 8). MCTSs were treated with vector-complexed [ON705]-F and S20-[ON705]-F for 48 hr. Fluorescence intensity of spheroid cells treated with [ON705]-F/JM (Figure 7, yellow) is widely spread over three orders of magnitude. The corresponding confocal microscopy images showed a green ring on the outer spheroid cells indicating that only the exterior spheroid cells were transfected. As seen in Figure S2, this vector-assisted transfection efficiency was not enhanced with higher concentration. Upon carrier-free transfection of spheroids with S20-[ON705]-F (Figure 7, blue), a distinct narrow peak corresponding to the high fluorescein active population was observed by flow cytometry. Efficient delivery of S20-[ON705]-F was confirmed by confocal microscopy because green fluorescein-active cells were observed everywhere, including in the innermost part of the spheroids.
Figure 7.
Delivery of Fluorescently Labeled SSOs in 3D Culture of HeLa pLuc/705 Cells Analyzed by Flow Cytometry
Untreated spheroids (red, control). [ON705]-(F) (0.2 μM, yellow) complexed with JM (JetMessenger) and (S)20-[ON705]-(F) (0.7 μM, blue) were added to 2-day-old HeLa pLuc/705 spheroids in serum-containing DMEM (10%). After 48 hr of incubation, cellular uptake was analyzed using a 488-nm laser.
Figure 8.
Delivery of Fluorescently Labeled SSOs in 3D Culture of HeLa pLuc/705 Cells Analyzed by Confocal Microscopy
To 2-day-old HeLa pLuc/705 spheroids in serum-containing DMEM (10%) was added [ON705]-(F) complexed with JM (JetMessenger) or (S)20-[ON705]-(F). Images were taken after 48 hr. (A) Fluorescein (488 nm). (B) Nuclei (405 nm). (C) Merge.
Discussion
In this study, we reported a new SSO-delivery method using oligospermine conjugates. An oligospermine-SSO conjugate containing 15–20 spermine units (S15- or S20-SSO) penetrates in HeLa pLuc/705 cells without vector to induce splice switching that restores luciferase expression, in the absence and in the presence of serum. Our approach takes advantage of both the cationic vector formulations and the molecule-scale delivery. Cationic vector formulations are the most popular and the most studied oligonucleotide delivery strategies.8, 9, 31, 32, 33 Oligonucleotides are complexed with cationic vectors to form nanoparticles of typically 100–200 nm and gain enhanced cell penetration properties driven by electrostatic interactions with anionic cellular heparan sulfate proteoglycans.34 However, it also raises several problems, that is, poor colloidal stability, poor biodistribution, and toxicity related to the large amount of cationic vector introduced in cells. The other emerging strategy is to use precise molecular conjugates of smaller size (<50 nm) to circumvent these difficulties. Cationic SSOs with in vivo activity were prepared by linking oligocationic structures (e.g., peptides containing eight arginines35, 36 or dendrimer with eight guanidine functions37) to the PMO that is a neutral oligonucleotide analog. In our approach, oligospermine tails containing an exact number of spermine units were attached to polyanionic SSOs so that the resulting conjugates may transfect as a single molecule by forming an intramolecular complex. Oligospermine moiety will act similarly to the polyethylenimine (PEI)-type cationic polymer vector, except that the amount of the vector used is minimized.
Our current study demonstrated efficient carrier-free transfection of SSO attached to an oligospermine tail containing only 15–20 spermine units. In our previous articles,17, 18, 19, 20 we showed that an attachment of oligospermine containing 30 spermine units was necessary for carrier-free siRNA delivery to induce gene silencing. Required spermine number in these conjugates can be primarily defined by N/P ratio (number of amino groups/number of phosphate groups). We found that a slightly higher N/P ratio was required for SSO delivery (S20-SSO: N/p = 2.1) as compared with siRNA delivery (S30-siRNA: N/p = 1.7). This observation may indicate that the S30-siRNA is structurally better organized than the S20-SSO. In fact, the SSO used in this study (i.e., [ON705]) is a single-stranded 18-mer oligonucleotide that is an about 15-nm-long chain and folds into various 3D structures; the attached oligospermine chain (S20) will interact with the negative phosphate groups and afford overall cationic entities of reduced size like a ball of wool. In contrast, siRNA is an assembly of two 21-mer RNA oligonucleotides that form an A-type double helix. The oligospermine (S30) attached to siRNA will wind onto the minor and/or the major grove, enhance duplex stability, and provide a supramolecular structure like a thread reel.
MCTS was recently proposed by Juliano and colleagues25 as a versatile tool to assess oligonucleotide delivery in 3D cells. These authors described an efficient delivery of 13-nm-size nanoparticle formulation based on human serum albumin linked to PMO-RGD peptide conjugates in MCTS and compared with their lipoplex and polyplex formulations that are inefficient. Relevance of oligospermine-conjugated SSO was evidenced by the assays performed in 3D cell culture. MCTS is a heterogeneously associated spherical cell assembly that is considered as a rational and cost-effective model to assess in vivo drug delivery efficacy. In monolayer culture cells, carrier-free SSO-oligospermine delivery was less efficient, as compared with the cationic vector-assisted transfection of unconjugated SSO. However, S20-[ON705] induced a higher level of splice switching than the vector-complexed [ON705] in MCTS. Reduced activity of SSO formulation in MCTS is related to the nanoparticle size (around 100 nm). Whereas molecular S20-[ON705] may circulate freely to attain the internal cells of spheroids, [ON705]/JetMessenger complexes will only penetrate to the outer layer cells. Indeed, homogeneous distribution of the fluorescently labeled S20-[ON705]-F in MCTS was proven by flow cytometry and by confocal microscopy, whereas the vector-complexed [ON705]-F was observed only as a bright ring by confocal microscopy, indicating local cellular uptake in accordance with the flow cytometry histogram. It should also be noted that S20-[ON705] present almost no toxicity in MCTS, whereas a dose-dependent toxicity was observed for the same conjugate in monolayer culture. Further studies are still required to determine the origin of these observations.
Our results open a new scope for oligospermine-oligonucleotide conjugates as these molecules were proven to induce splice switching spontaneously without formulation in cells. Compared with siRNA-oligospermine conjugates, SSO conjugates are about twice as small, and fewer steps are required for their manufacturing. Further improvement of splice switching potency may be achieved by structural tuning of conjugates.
Materials and Methods
Synthesis of 2′-O-Methyl Oligonucleotide Phosphorothioate-Oligospermine Conjugates
Oligonucleotide synthesis reagents were purchased from Glen Research/Eurogentec (Paris, France; reference G.R.). All reagents including the DMT-spermine phosphoramidite are commercially available. Oligonucleotides and their oligospermine conjugates were prepared on an automated Expedite 8900 nucleic acid synthesis system (GMI, USA) at 1-μmol scale using modified procedures based on the standard instrumental protocol as described previously.20 Phosphorothioate linkages were introduced using 3-((N,N-dimethylaminomethylidene)amino)-[3H]-1,2,4-dithiazole-5-thione (DDTT) sulfurizing reagent (G.R. 40-4137-51). 3′-(6-FAM) CPG (G.R. 20-2961-41) was used to introduce fluorescein label. All oligonucleotides were synthesized with final DMT-ON conditions. Coupling yields were evaluated by measuring the absorbance at 504 nm of several selected DMT fractions (e.g., the second, the last, before and after spermine couplings), which were diluted to 100 mL with 3% trichloroacetic acid (TCA) in dichloromethane as follows: DMT fractions were first diluted to 5 mL (volumetric flask) with dichloromethane before a 250-μL aliquot of the resulting solution (pipetted with a Microman, Gilson pipette equipped with a capillary piston) was transferred into another volumetric flask and completed to 5 mL with 3% TCA in dichloromethane solution. Average coupling yields were >97% for nucleotides and about 95% for spermine couplings. For oligonucleotide conjugates, cleavage, successive deprotection steps, and purification were performed according to Glen Research-reported protocols using DNA Glen-Pak columns (G.R. 60-5000-96) except that a vacuum manifold apparatus was not used. Solutions were manually added using 1-mL plastic syringes. Overall yields (Table 1) were driven from the final stock solution concentrations as measured from their absorption at 260 nm and then calculated using extinction coefficients estimated by the described empirical formula.38 MALDI-TOF mass spectra were obtained in positive mode on a Bruker Ultraflex apparatus with hydroxypicolinic acid or trihydroxyacetophenone combined, with or without diammonium citrate as matrix (Figures S3–S10).
The single-stranded [ON705] was targeted to the aberrant splice site.23, 39 Oligonucleotides with scrambled sequence ([ON705scr])40 or with one mismatch ([ON705mis]) were used as negative controls. An oligonucleotide [ON119] targeted to position 119 of β-globin intron 2 was used as an additional negative control.24
SDS-PAGE of SSOs
Four hundred pmol of single-strand oligospermine conjugates were loaded on 4%–12% Bis-Tris precast gels (Criterion XT; Bio-Rad). Electrophoresis was carried out in XT-MES denaturing running buffer during 2 hr at 75 V. Precast gels were then incubated in ethidium bromide (0.5 μg/mL) and analyzed.
Cell Culture
HeLa cells transfected with pLuc/705 were kindly provided by Dr. R. Kole.23, 24 Cells were grown in low-glucose DMEM (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% fetal bovine serum (FBS) (Perbio, Brebieres, France), 100 U/mL penicillin, 100 μg/mL streptomycin (Eurobio), and 200 μg/mL hygromycin B (Invitrogen, Carlsbad, CA, USA).
Cellular Uptake of S20-[ON705] in HeLa pLuc/705 Cells
HeLa pLuc/705 cells (2.5 × 104 per well) were seeded in four-chambered Lab-Tek plates (Ref 155383; Nunc, Rochester, NY, USA) in 500 μL of complete medium the day before their use. Cells were then incubated with 0.4 and 1.0 μM SSOs [ON705]-F and S20-[ON705]-F, respectively, both fluorescently labeled with fluorescein. Cells were observed after 45 min, 2 hr, and 4 hr in red-phenol-free DMEM by fluorescent microscopy (Axiovert 25; Zeiss, Germany).
Transfection with Cationic Oligospermine Conjugates
Twenty-four hours prior to transfection, 6 × 103 cells per well were seeded in 96-well tissue culture plates (Corning, NY, USA) in 100 μL of DMEM containing 10% FBS. The spermine-grafted SSOs were diluted up to 100 μL in serum-free or serum-containing DMEM and then added onto HeLa pLuc/705 cells after washing. After 4 hr, 10 μL of FBS was added on serum-free transfected cells. As positive controls, cells were transfected with 0.2 μM [ON705] complexed with JetMessenger (JM; Polyplus-Transfection, Illkirch, France) according to the supplier’s instructions. All cells were incubated for 48 hr at 37°C in 5% CO2 humidified atmosphere.
Splice-Switching Longevity Study
Twenty-four hours prior to transfection, 5 × 104 HeLa pLuc/705 cells per well were seeded in 24-well tissue culture plates (Corning, NY) in 0.5 mL of DMEM complete medium. Spermine-conjugated oligonucleotides S15-[ON705] (0.6 μM), S20-[ON705] (0.5 μM), S30-[ON705] (0.3 μM), S15-[ON119] (0.6 μM), and S15-[ON705scr] (0.6 μM) were diluted in 300 μL of serum-free DMEM and added to cells after washing. After 4 hr, FBS was added up to a final 10% concentration. Luciferase reporter gene expression was measured 11, 21, 45, and 69 hr after delivery with spermine-grafted SSOs. After 69 hr, transfected and non-transfected HeLa pLuc/705 cells were split in new 24-well tissue culture plates at 5 × 104 cells per well to maintain exponential growth. The restoration of luciferase gene expression was then determined 95, 118, and 142 hr after transfection.
Luciferase Activity
HeLa pLuc/705 cells were rinsed with PBS and were lysed (40 μL/well for 96-well plates and 100 μL for 24-well plates) with cell culture lysis buffer (Promega, Madison, WI, USA) at room temperature for 30 min. After collecting and centrifuging the cell lysates, the luciferase enzyme activity was quantified from supernatant lysate after addition of 50 μL of luciferin solution (Promega) with a Centro LB960 luminometer (Bertold, Thoiry, France). The protein concentration in the extract was measured by the BCA protein assay kit (Uptima; Interchim, Montluçon, France). Final luciferase activity was first measured as relative light units integrated for 10 s per milligram of cell protein (RLU/mg) and then converted to the increase of luciferase expression relative to the luciferase level of untreated HeLa pLuc/705 cells.
Generation and Transfection of Multicellular Spheroids
HeLa pLuc/705 MCTSs were generated using the hanging drop method.30 Starting from a monolayer cell culture, cells were resuspended in growth medium containing 20% methyl cellulose (Sigma) at a concentration of 1 × 105 cells/mL. Twenty-five-microliter drops containing 2.5 × 103 cells were prepared on 90-mm Petri dishes lids (Greiner, Frickenhausen, Germany) and then inverted to obtain hanging drops that were incubated at 37°C. After 48 hr, 12 MCTSs per well were transferred in non-treated 96-well plates. Single-strand oligonucleotides S15-[ON705], S20-[ON705], S15-[ON119], and [ON705] were prepared in 100 μL of complete DMEM and added to the spheroids. Control assays for MCTSs with unconjugated [ON705] transfected with JetMessenger were performed in the same way as for monolayer assays. All experiments were done in quadruplicate. After 48 hr, spheroids were washed with PBS and lysed for quantification of luciferase activity.
Flow Cytometry
Six 2-day-old spheroids per well were transferred in non-treated 96-well plates. Forty-eight hours after transfection with fluorescently labeled SSOs ([ON705]-F complexed with JetMessenger and S20-[ON705]-F), cells were trypsinized and resuspended in PBS. Single-cell suspensions were analyzed by flow cytometry on a Guava easyCyte HT Cytometer (Merck Millipore) using a 488-nm laser.
Confocal Microscopy
Two-day-old spheroids were transferred in non-treated 96-well plates for transfection with fluorescently labeled SSOs: [ON705]-F complexed with JetMessenger and S20-[ON705]-F. Cell nuclei were stained with Hoechst 33258 (Sigma-Aldrich). After fixation in 4% paraformaldehyde, transfected spheroids were mounted with Kaiser’s glycerol gelatine (Millipore, Darmstadt, Germany). Pictures were taken with a Leica TSC SPE confocal microscope (Leica Microsystems, Mannheim, Germany).
Author Contributions
Conceptualization, J.-P.B., J.-S.R., and M.K.; Methodology, M.N. and M.K.; Investigation, M.N., P.P.-L., and M.K.; Writing, M.N. and M.K.; Supervision and Project Administration, J.-P.B. and J.-S.R.
Conflicts of Interest
The authors have no conflicts of interest.
Acknowledgments
HeLa pLuc/705 cells were a generous gift of Dr. R. Kole (University of North Carolina, Chapel Hill, NC, USA). The authors thank Dr. G. Chaubet and Dr. A. Wagner for review of the manuscript and Dr. Jean-Marc Strub for mass spectrometry data of the conjugates. This work was supported by the CNRS, Université de Strasbourg, the Association Française contre les Myopathies (AFM), and the International Center for Frontier Research in Chemistry (icFRC; Université de Strasbourg, France).
Footnotes
Supplemental Information includes ten figures and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.09.027.
Contributor Information
Jean-Serge Remy, Email: remy@unistra.fr.
Mitsuharu Kotera, Email: m.kotera@unistra.fr.
Supplemental Information
References
- 1.Lim K.R.Q., Maruyama R., Yokota T. Eteplirsen in the treatment of Duchenne muscular dystrophy. Drug Des. Devel. Ther. 2017;11:533–545. doi: 10.2147/DDDT.S97635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Havens M.A., Hastings M.L. Splice-switching antisense oligonucleotides as therapeutic drugs. Nucleic Acids Res. 2016;44:6549–6563. doi: 10.1093/nar/gkw533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kole R., Krieg A.M. Exon skipping therapy for Duchenne muscular dystrophy. Adv. Drug Deliv. Rev. 2015;87:104–107. doi: 10.1016/j.addr.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Evers M.M., Toonen L.J.A., van Roon-Mom W.M.C. Antisense oligonucleotides in therapy for neurodegenerative disorders. Adv. Drug Deliv. Rev. 2015;87:90–103. doi: 10.1016/j.addr.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Kole R., Krainer A.R., Altman S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug Discov. 2012;11:125–140. doi: 10.1038/nrd3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Järver P., O’Donovan L., Gait M.J. A chemical view of oligonucleotides for exon skipping and related drug applications. Nucleic Acid Ther. 2014;24:37–47. doi: 10.1089/nat.2013.0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Godfrey C., Desviat L.R., Smedsrød B., Piétri-Rouxel F., Denti M.A., Disterer P., Lorain S., Nogales-Gadea G., Sardone V., Anwar R. Delivery is key: lessons learnt from developing splice-switching antisense therapies. EMBO Mol. Med. 2017;9:545–557. doi: 10.15252/emmm.201607199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Juliano R.L. The delivery of therapeutic oligonucleotides. Nucleic Acids Res. 2016;44:6518–6548. doi: 10.1093/nar/gkw236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krhac Levacic A., Morys S., Wagner E. Solid-phase supported design of carriers for therapeutic nucleic acid delivery. Biosci. Rep. 2017;37 doi: 10.1042/BSR20160617. BSR20160617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen X., Corey D.R. Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs. Nucleic Acids Res. 2018;46:1584–1600. doi: 10.1093/nar/gkx1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pons B., Kotera M., Zuber G., Behr J.P. Online synthesis of diblock cationic oligonucleotides for enhanced hybridization to their complementary sequence. ChemBioChem. 2006;7:1173–1176. doi: 10.1002/cbic.200600178. [DOI] [PubMed] [Google Scholar]
- 12.Voirin E., Behr J.P., Kotera M. Versatile synthesis of oligodeoxyribonucleotide-oligospermine conjugates. Nat. Protoc. 2007;2:1360–1367. doi: 10.1038/nprot.2007.177. [DOI] [PubMed] [Google Scholar]
- 13.Noir R., Kotera M., Pons B., Remy J.S., Behr J.P. Oligonucleotide-oligospermine conjugates (zip nucleic acids): a convenient means of finely tuning hybridization temperatures. J. Am. Chem. Soc. 2008;130:13500–13505. doi: 10.1021/ja804727a. [DOI] [PubMed] [Google Scholar]
- 14.Moreau V., Voirin E., Paris C., Kotera M., Nothisen M., Rémy J.S., Behr J.P., Erbacher P., Lenne-Samuel N. Zip nucleic acids: new high affinity oligonucleotides as potent primers for PCR and reverse transcription. Nucleic Acids Res. 2009;37:e130. doi: 10.1093/nar/gkp661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paris C., Moreau V., Deglane G., Voirin E., Erbacher P., Lenne-Samuel N. Zip nucleic acids are potent hydrolysis probes for quantitative PCR. Nucleic Acids Res. 2010;38:e95. doi: 10.1093/nar/gkp1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Navarro E., Serrano-Heras G., Castaño M.J., Solera J. Real-time PCR detection chemistry. Clin. Chim. Acta. 2015;439:231–250. doi: 10.1016/j.cca.2014.10.017. [DOI] [PubMed] [Google Scholar]
- 17.Nothisen M., Kotera M., Voirin E., Remy J.S., Behr J.P. Cationic siRNAs provide carrier-free gene silencing in animal cells. J. Am. Chem. Soc. 2009;131:17730–17731. doi: 10.1021/ja908017e. [DOI] [PubMed] [Google Scholar]
- 18.Paris C., Moreau V., Deglane G., Karim L., Couturier B., Bonnet M.E., Kedinger V., Messmer M., Bolcato-Bellemin A.L., Behr J.P. Conjugating phosphospermines to siRNAs for improved stability in serum, intracellular delivery and RNAi-mediated gene silencing. Mol. Pharm. 2012;9:3464–3475. doi: 10.1021/mp300278b. [DOI] [PubMed] [Google Scholar]
- 19.Perche P., Nothisen M., Bagilet J., Behr J.P., Kotera M., Remy J.S. Cell-penetrating cationic siRNA and lipophilic derivatives efficient at nanomolar concentrations in the presence of serum and albumin. J. Control. Release. 2013;170:92–98. doi: 10.1016/j.jconrel.2013.04.013. [DOI] [PubMed] [Google Scholar]
- 20.Nothisen M., Bagilet J., Behr J.P., Remy J.S., Kotera M. Structure tuning of cationic oligospermine-siRNA conjugates for carrier-free gene silencing. Mol. Pharm. 2016;13:2718–2728. doi: 10.1021/acs.molpharmaceut.6b00309. [DOI] [PubMed] [Google Scholar]
- 21.Gagnon K.T., Watts J.K., Pendergraff H.M., Montaillier C., Thai D., Potier P., Corey D.R. Antisense and antigene inhibition of gene expression by cell-permeable oligonucleotide-oligospermine conjugates. J. Am. Chem. Soc. 2011;133:8404–8407. doi: 10.1021/ja200312y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakamoto K., Akao Y., Furuichi Y., Ueno Y. Enhanced intercellular delivery of cRGD−siRNA conjugates by an additional oligospermine modification. ACS Omega. 2018;3:8226–8232. doi: 10.1021/acsomega.8b00850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dominski Z., Kole R. Restoration of correct splicing in thalassemic pre-mRNA by antisense oligonucleotides. Proc. Natl. Acad. Sci. USA. 1993;90:8673–8677. doi: 10.1073/pnas.90.18.8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang S.H., Cho M.J., Kole R. Up-regulation of luciferase gene expression with antisense oligonucleotides: implications and applications in functional assay development. Biochemistry. 1998;37:6235–6239. doi: 10.1021/bi980300h. [DOI] [PubMed] [Google Scholar]
- 25.Carver K., Ming X., Juliano R.L. Multicellular tumor spheroids as a model for assessing delivery of oligonucleotides in three dimensions. Mol. Ther. Nucleic Acids. 2014;3:e153. doi: 10.1038/mtna.2014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sutherland R.M. Cell and environment interactions in tumor microregions: the multicell spheroid model. Science. 1988;240:177–184. doi: 10.1126/science.2451290. [DOI] [PubMed] [Google Scholar]
- 27.Hirschhaeuser F., Menne H., Dittfeld C., West J., Mueller-Klieser W., Kunz-Schughart L.A. Multicellular tumor spheroids: an underestimated tool is catching up again. J. Biotechnol. 2010;148:3–15. doi: 10.1016/j.jbiotec.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 28.Thoma C.R., Zimmermann M., Agarkova I., Kelm J.M., Krek W. 3D cell culture systems modeling tumor growth determinants in cancer target discovery. Adv. Drug Deliv. Rev. 2014;69–70:29–41. doi: 10.1016/j.addr.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 29.Weiswald L.B., Bellet D., Dangles-Marie V. Spherical cancer models in tumor biology. Neoplasia. 2015;17:1–15. doi: 10.1016/j.neo.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Timmins N.E., Nielsen L.K. Generation of multicellular tumor spheroids by the hanging-drop method. In: Hauser H., Fussenegger M., editors. Tissue Engineering. Methods in Molecular Medicine. Volume 140. Humana Press; 2007. pp. 141–151. [DOI] [PubMed] [Google Scholar]
- 31.Allen T.M., Cullis P.R. Liposomal drug delivery systems: from concept to clinical applications. Adv. Drug Deliv. Rev. 2013;65:36–48. doi: 10.1016/j.addr.2012.09.037. [DOI] [PubMed] [Google Scholar]
- 32.Kanasty R., Dorkin J.R., Vegas A., Anderson D. Delivery materials for siRNA therapeutics. Nat. Mater. 2013;12:967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen J., Szoka F.C. Nucleic acid delivery: the missing pieces of the puzzle? Acc. Chem. Res. 2012;45:1153–1162. doi: 10.1021/ar3000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kopatz I., Remy J.S., Behr J.P. A model for non-viral gene delivery: through syndecan adhesion molecules and powered by actin. J. Gene Med. 2004;6:769–776. doi: 10.1002/jgm.558. [DOI] [PubMed] [Google Scholar]
- 35.Morcos P.A., Li Y., Jiang S. Vivo-Morpholinos: a non-peptide transporter delivers Morpholinos into a wide array of mouse tissues. Biotechniques. 2008;45:613–614. doi: 10.2144/000113005. 616, 618 passim. [DOI] [PubMed] [Google Scholar]
- 36.Li Y.F., Morcos P.A. Design and synthesis of dendritic molecular transporter that achieves efficient in vivo delivery of morpholino antisense oligo. Bioconjug. Chem. 2008;19:1464–1470. doi: 10.1021/bc8001437. [DOI] [PubMed] [Google Scholar]
- 37.Moulton H.M., Moulton J.D. Morpholinos and their peptide conjugates: therapeutic promise and challenge for Duchenne muscular dystrophy. Biochim. Biophys. Acta. 2010;1798:2296–2303. doi: 10.1016/j.bbamem.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 38.Brown T., Brown D.J.S. Modern machine-aided methods of oligodeoxyribonucleotide synthesis. In: Eckstein F., editor. Oligonucleotides and Analogues: A Practical Approach. IRL Press at Oxford University Press; 1991. pp. 1–24. [Google Scholar]
- 39.Sierakowska H., Sambade M.J., Agrawal S., Kole R. Repair of thalassemic human β-globin mRNA in mammalian cells by antisense oligonucleotides. Proc. Natl. Acad. Sci. USA. 1996;93:12840–12844. doi: 10.1073/pnas.93.23.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Resina S., Kole R., Travo A., Lebleu B., Thierry A.R. Switching on transgene expression by correcting aberrant splicing using multi-targeting steric-blocking oligonucleotides. J. Gene Med. 2007;9:498–510. doi: 10.1002/jgm.1044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.