Abstract
Epigenetic processes are known to have powerful roles in organ development across biology. It has recently been found that some of the chromatin modulatory machinery essential for proper development plays a previously unappreciated role in the pathogenesis of cardiac disease in adults. Investigations using genetic and pharmacologic gain- and loss-of-function approaches have interrogated the function of distinct epigenetic regulators, while the increased deployment of the suite of next-generation sequencing technologies have fundamentally altered our understanding of the genomic targets of these chromatin modifiers. Here, we review recent developments in basic and translational research that have provided tantalizing clues that may be used to unlock the therapeutic potential of the epigenome in heart failure. Additionally, we provide a hypothesis to explain how signal-induced crosstalk between histone tail modifications and long non-coding RNAs triggers chromatin architectural remodeling and culminates in cardiac hypertrophy and fibrosis.
Keywords: epigenetics, cardiac hypertrophy, fibrosis, heart failure
Introduction
Advances in the pharmacologic management of serum lipid levels and surgical, device-aided, and pharmacologic abatement of myocardial ischemia/reperfusion injury have conspired with increases in obesity to exacerbate the epidemic of heart failure across the developed world. The syndrome of heart failure encompasses a spectrum of symptoms with variable manifestation in different people: impaired ability to meet the oxygen demands of the body are accompanied by impaired contractile function of the heart, myocyte hypertrophy, fibrotic deposition and fibroblast to myofibroblast transformation, and/or metabolic derangements. The scale of the heart failure clinical problem—7 million affected in the United States alone 1—combined with the multifarious nature of the pathologic mechanisms make novel molecular mechanisms of cardiac dysfunction a major target for ongoing basic and translational research.
Heart failure is a progressive condition, wherein the cumulative effects of stress to the heart are integrated to alter the function of the organ. While some aspects of cardiac dysfunction can be mitigated by targeting risk factors (e.g. blood pressure control), cellular and molecular insults change the function of the heart in a more lasting and, presently, from a drug development standpoint, intractable manner. To develop the next class of therapeutic targets for heart failure, we need to dissect the influence of pathologic stress on (a) the major classes of cells in the heart and (b) the molecular substrate for persistent cell and organ dysfunction. Towards the latter goal, emerging evidence indicates that chromatin regulatory mechanisms are mobilized following injury to establish and entrench diseased transcriptomes and phenotypes. The families of histone-modifying enzymes that write, read, and erase marks on these proteins in a signal-responsive and locus-specific manner establish the differential accessibility that allows the same genome to encode different cell types. Linking transcription with chromatin regulation, long noncoding RNAs (lncRNAs) have been revealed to exert powerful effects on cardiac cell phenotype through the regulation of gene expression. Together, these and other epigenetic mechanisms impart structure and regulation to chromatin between distinct cell types in the healthy and diseased setting. In this essay, we identify epigenomic regulation of specialized cardiac cell types like myocytes and fibroblasts as promising targets for therapeutic development.
Targeting histone acetylation for heart failure: HAT, HDAC, and BET inhibitors
Lysine is a versatile amino acid target for post-translational modifications, including methylation, acyl modifications (e.g. acetylation, crotonylation, and succinylation), and small protein isopeptide bonds (e.g. ubiquitination, NEDDylation, and SUMOylation) 2. For the purposes of this review, we focus on the acetylation of ε-amino groups of lysine residues within nucleosomal histone tails. This post-translational modification is mediated by histone acetyltransferase (HAT) enzymes and has historically been linked to gene activation, with the charge neutralization provided by acetylation promoting a more permissive environment for transcription by weakening histone:DNA associations and altering histone:histone interactions.
Findings with genetically engineered mice have suggested important roles for the p300 HAT in the control of pathological cardiac remodeling. Heterozygous deletion of p300 was shown to suppress cardiac hypertrophy in response to pressure overload, and overexpression of p300 triggered pathological hypertrophy and heart failure 3, 4. Unfortunately, efforts to advance HAT inhibitors as a therapeutic strategy for heart failure have been hampered by the lack of potent and selective pharmacological inhibitors of p300 5. However, a recent virtual screening and medicinal chemistry optimization campaign yielded A-485, an orally bioavailable small molecule inhibitor that is highly selective for p300 and the related HAT, CREB-binding protein (CBP) 6, 7. The drug-like properties of A-485 provide an excellent opportunity to assess the efficacy of HAT inhibition in pre-clinical models of pathological cardiac remodeling and thereby determine the translational potential of p300 catalytic activity inhibition for the treatment of heart failure in humans.
A bromodomain, which is an acetyl-lysine binding motif, in p300 is required for chromatin targeting of the HAT 8, 9. CBP112 and CBP30 have been developed as small molecules that target the p300 bromodomain and function as acetyl-lysine competitive inhibitors 10, 11. An impactful recent study employed proteomics and transcriptomics to quantify acetylation and mRNA and protein abundance in mouse embryonic fibroblasts after cellular p300 inhibition with A-485 versus CBP112 12. Interestingly, gene expression changes triggered by CBP112 were modest compared to those observed upon catalytic inhibition of p300 with A-485, illustrating that the bromodomain of the HAT is required for the regulation of only a subset of target genes. It will be interesting to compare the effects of A-485 and CBP112 in cardiac myocytes to determine the relative contributions of p300 catalytic activity and bromodomain function in the control of pathologic gene expression.
In contrast to HAT inhibitors, a multitude of potent and selective HDAC inhibitors are available 13. Most HDAC inhibitors possess a tripartite structure consisting of a zinc-binding group that binds the active site, a linker that mimics the lysine side chain, and a surface recognition cap that confers specificity of the compounds for HDAC enzymes. Distinct genes encode the 18 mammalian HDAC isoforms, which are either zinc dependent (HDACs 1-11) or NAD + dependent (SirT1–7) 14. The original demonstration that HDAC inhibitors could be beneficial in the heart was based on the use of trichostatin A (TSA), a Streptomyces metabolite that acts as a pan-inhibitor of zinc-dependent HDACs but does not affect the activity of NAD +-dependent sirtuins 15– 18. Subsequently, additional natural products as well as synthetic HDAC inhibitors were shown to be efficacious in animal models of heart failure, blocking pathological cardiac hypertrophy, fibrosis, and inflammation, and improving systolic and diastolic function 19.
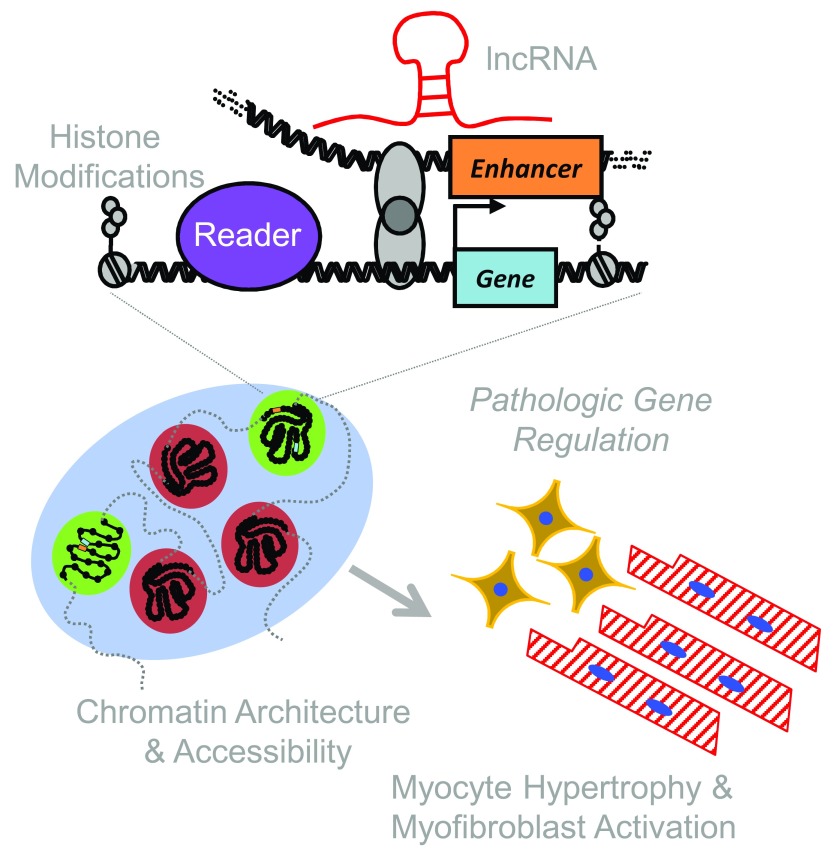
Remarkably, our understanding of the functions of HDACs in the control of epigenetic regulation of gene expression in heart failure is still extremely limited. A genome-wide evaluation of the impact of HDAC inhibition on one epigenetic mark in normal and stressed hearts was described 20. Mice were subjected to left ventricular pressure overload and were administered TSA or vehicle control for four weeks. ChIP-seq of whole heart homogenates with an anti-acetyl-H3K9/K14 antibody revealed that pressure overload broadly altered histone acetylation throughout the genome, and these changes were reversed by TSA. A paradoxical finding from this study was the profound ability of TSA to also reduce H3 acetylation at many loci. These findings suggest the possibility that HDAC activity controls HAT genomic targeting, expression, and/or function in the heart. This mode of crosstalk could explain the seemingly counterintuitive finding that inhibiting enzymes that either add (HAT) or remove (HDAC) acetyl groups can suppress pathogenic processes that contribute to the development of heart failure. Taking this a step further, it is our strong belief that HDACs mediate extensive interplay between diverse epigenetic regulators, including lncRNAs, and coordinate complex remodeling of chromatin architecture in response to pathological stress in cardiac myocytes and fibroblasts, thereby promoting hypertrophy, fibrosis, and ventricular dysfunction ( Figure 1).
Figure 1. A model for integrating histone marks, long noncoding RNAs (lncRNAs), and chromatin architecture in heart failure.
The epigenomic regulation of cardiac phenotype occurs at multiple interacting scales. Histone isoforms, post-translational modification, and nucleosome distribution influence local transcription. lncRNAs have emerged as powerful regulators of gene expression, interacting with chromatin-modifying enzymes and influencing their histone targets. Together with other chromatin regulatory proteins, histone modifications and lncRNAs establish local chromatin accessibility and global chromatin architecture, facilitating short- and long-range regulatory interactions that enable cell type-specific transcriptomes in healthy and diseased conditions.
Another way to pharmacologically target histone acetylation is through the use of BET protein inhibitors. The most well-characterized family of proteins that ‘read’ acetyl-lysine marks, without also containing catalytic domains for epigenetic modifying activity (e.g. HATs), are the bromodomain and extraterminal domain-containing (BET) proteins (BRD2, BRD3, BRD4, and BRDT). BRD4 and BRDT (testis-specific) harbor a unique carboxy-terminal domain that is able to activate RNA polymerase II (Pol II) by recruiting CDK9, a kinase component of the P-TEFb complex; CDK9 phosphorylates serine-2 of the tail of Pol II, leading to transcription elongation 21– 23.
A developing function for BET proteins, in particular BRD4, is the creation of dynamic, cell state-specific enhancers called super-enhancers (SEs). The association of BRD4 with acetyl-H3K27-containing SEs, the signaling of which to proximal promoters is believed to stabilize BRD4-containing coactivator complexes close to transcription start sites, enables P-TEFb-mediated Pol II phosphorylation and transcription elongation. JQ1, which is a small molecule inhibitor that is selective for BET bromodomains, was shown to effectively prevent and reverse cardiac hypertrophy, fibrosis, and ventricular dysfunction, in part, by suppressing the association of BRD4 with SEs associated with pro-hypertrophic and pro-fibrotic genes in the heart 24– 28. The extent to which BRD4 genomic targeting in the heart is controlled by distinct HDAC and HAT isoforms has not been determined, nor has the role of BRD4 in coupling to pathogenic lncRNAs and coordinating chromatin architecture remodeling in response to cardiac stress.
The notion of using small molecule inhibitors of epigenetic regulators to treat a chronic condition such as heart failure is often met with doubt, since the regulators to be targeted are widely expressed and mediate fundamental transcriptional mechanisms in many cell types. Nevertheless, there are four FDA-approved HDAC inhibitors, two approved DNMT inhibitors, and several other epigenetic modifying therapies in clinical development for oncologic and non-oncologic indications 29. Thus, the feasibility of using ‘epigenetic therapies’ to treat human diseases has been validated, and we believe that this approach has tremendous potential for patients suffering from the complex syndrome of heart failure.
This is also an exciting time to employ chemical biology to elucidate novel epigenetic pathways that control heart failure. No longer are we solely reliant on natural product inhibitors of epigenetic regulators, which often lack selectivity. Exhaustive and sophisticated medicinal chemistry programs in industry and academia have led to the development of highly selective and potent inhibitors of a wide array of epigenetic targets, and many of the compounds are available to the scientific community through programs such as the Structural Genomics Consortium 30. Coupling the use of these compounds with well-validated phenotypic assays, such as cell-based assays of cardiomyocyte hypertrophy or fibrosis 31, has the potential to rapidly uncover novel roles for epigenetic regulators in the control of heart failure and thus provide crucial mechanistic insights.
Long non-coding RNAs
lncRNAs are a class of RNA transcripts recently identified to be widely expressed in all tissues. Unlike several functionally and structurally well-defined species of non-coding RNAs, such as rRNA, miRNA, snoRNA, and piRNA, the very definition of lncRNAs remains arbitrary and is often applied to any RNA transcripts that have no coding capacity and are more than 200 nucleotides in length 32. Recent progress in transcriptome profiling using next-generation RNA sequencing methods has begun to uncover the enormous scale of the lncRNA products within the cardiac transcriptome and the scope of their contribution to the overall transcriptome reprogramming under physiological and pathological conditions 33– 37. In addition to lack of protein coding capacity, common features of cardiac lncRNAs also include low abundance in expression (with notable exceptions) and a lower degree of sequence conservation 33. They can be produced from intergenic regions of the genome (intergenic lncRNAs) or from different parts of known genes, such as enhancers, promoters, and exon or intron regions in either sense or anti-sense directions 38. It is clear that the transcriptomic complexity of lncRNAs matches, if not surpasses, that of coding mRNAs in the heart.
Because of the scale and complexity of cardiac lncRNAs, physiological or pathological functions for the vast majority of them still remain to be determined. However, recent evidence shows that many cardiac lncRNAs do play important roles in gene regulation during cardiac development and pathophysiology, particularly via epigenomic modulations, as showcased by the prototypic lncRNA H19 in the regulation of Igf2 gene imprinting 39, 40.
In the past 5 years, there has been an explosion of new discoveries of epigenomic regulatory lncRNAs, also referred to as epi-lncRNAs, in cardiac development and diseases 41, 42. For example, Mhrt and Chaer are reported to regulate chromatin modifications by direct interactions with histone modifiers, such as Brg1 and PRC2 complexes, respectively. The epigenomic impact of such interactions can be either local or more global. For example, Upperhand is a lncRNA that regulates neighboring hand2 gene expression in the developing heart in an allele-specific and cis-regulatory manner 43. In contrast, Chaer modulates global histone modifications as an epigenetic check-point for a large number of hypertrophic genes 44. Ultimately, the functional outcome of these epi-lncRNAs affects chromatin accessibility for transcription factors, Pol II, and other RNA-processing machinery. In addition to these relatively well-characterized epigenetic modulatory lncRNAs, lncRNAs are also reported to modulate RNA splicing, transportation, and translation 38. For example, CHRF and Miat can regulate cardiac gene expression and hypertrophy by binding to and interfering with miRNA functions, serving as so-called miRNA sponges 45, 46.
Given the role for lncRNAs in cardiac epigenetic regulation, interfering with lncRNA expression and function can have a significant impact on cardiac pathophysiology. For example, Mhrt expression can block, while Chaer and CHRF inactivation attenuates, pathological cardiac hypertrophy. Manipulation of the expression of the Wisper, Meg3, and MIAT lncRNAs also affects cardiac fibrosis and pathological remodeling 47– 49. Tissue and plasma lncRNAs have been identified as potential biomarkers to predict the disease outcome for heart failure 32, 50– 52. These studies highlight the therapeutic potential for lncRNA-targeted treatment for heart failure.
LncRNAs are clearly central to the epigenomic network in the heart. However, many challenging issues remain with regard to understanding lncRNA-mediated cardiac epigenetic regulation and translating lncRNA-targeted therapies to the clinic. First, given the fact that the vast majority of lncRNA species have not been functionally annotated, new high-throughout phenotypic screening approaches will be needed to systematically identify functionally important lncRNA species in heart diseases. Second, the mechanisms of lncRNA-mediated epigenetic regulation are extremely diverse and much remains to be discovered. Other than a few exceptions, the structural basis of lncRNA function is still poorly understood 53, 54. The detailed molecular processes involved in lncRNA-mediated regulation of histone modifications are largely unknown. In particular, the molecular link between lncRNA function and pathophysiological signaling is still elusive. Finally, many lncRNAs are species specific, leaving concerns about the translational relevance of studies conducted in rodents or other animal models 55, 56. Therefore, there are pressing needs to develop novel analytic tools using integrated approaches combined with human datasets to identify and characterize functionally important lncRNAs and to develop novel experimental approaches to characterize lncRNA function in epigenomic regulation by revealing the hidden code of lncRNA structure and identifying the intersections between lncRNAs and epigenomic modulators.
Chromatin architecture
To get the same genome to behave differently in the presence of identical transcriptional machinery, the hundreds of cell types in mammalian systems must change the interface between these tiers of molecules (i.e. between DNA and everything that regulates it). One manner in which this interface is altered is via structure and accessibility (two separate concepts) of chromatin, in large part through the actions of histone-modifying enzymes as discussed in the first part of this essay. Recent technologies that enable direct measurements of accessibility and structure of the genome reveal how these features are modified in a global, coordinated manner and implicate changes in chromatin architecture as a fundamental driver of disease.
Global changes in histone marks have been observed in multiple studies of animal models of heart failure, including a general trend of increased euchromatic marks and/or decreased heterochromatic marks (early observations from human hearts supported a similar trend 57). In mouse models of pressure overload, ChIP-seq has been used to map global deposition of various histone marks associated with transcriptional activation, repression, and enhancer formation in the basal and hypertrophied heart 58. Human 57, 59 and mouse 60 studies of DNA methylation, which is associated with gene expression and may participate in chromatin structure, have also revealed widespread changes during the development of heart failure, with specific localization around genes involved in the disease’s pathogenesis and related to chromatin structural features, such as topologically associated domains 61. Chromatin accessibility—that is, the local density of nucleosomes (i.e. how many occupy a fixed region of DNA) and the decoration of nucleosomes with post-translational modifications that facilitate (such as lysine acetylation in particular) or inhibit (such as trimethylation of histone H3 K9 or K27, for instance) transcriptional activity—can be globally assayed by various techniques, which report on the cumulative effects of multiple histone modifications, DNA modifications, and other protein binding rather than being dependent on assumptions based on ChIP-seq data for any single modification. Accessibility is a prerequisite for protein binding and hence transcription. Although it is beyond the scope of this review to comprehensively address this concept, there are scores of histone-modifying enzymes that modulate chromatin in any given cell type while, at the same time, DNA itself is modified, most notably through methylation and hydroxymethylation. These processes do not exist in isolation, and indeed some aspects of DNA methylation can be influenced by chromatin structure, as has been recently reported in cardiac myocytes 62. Indeed, the shift at specific loci across the genome between hydroxymethylation and methylation at cytosines has been shown to play a role in pathologic transcriptional changes in cardiac hypertrophy and failure 63. Furthermore, heart failure is associated with changes in DNA methylation, which appear to coordinate with chromatin features defined by histone modifications in both mice and humans 57, 60.
Chromatin structure, on the other hand, is a three-dimensional problem that incorporates localization within the nucleus (i.e. how close a given region of the genome is to nuclear features such as membranes, nucleoli, or transcription factories), nearness to other regions of the same or different chromosomes (in particular for the actions of transcriptional enhancers, which can act at a distance to regulate transcription during heart failure), and local architectural features that insulate against aberrant transcription while facilitating the appropriate variety. Recent investigations have shown cardiomyocyte chromatin to undergo selective, locus-specific, and highly tuned structural reorganization in the setting of pressure overload hypertrophy in animal models 64. Different cardiac cell genes exhibit distinct temporal schemes of chromatin remodeling after pathologic stress 58, 65, which have been attributed to underlying chromatin features, such as gene looping, in addition to the actions of enhancer elements. Histone modifications (and the suite of proteins that add, remove, and read these modifications, as well as ATP-dependent chromatin remodelers, which consume ATP to reposition nucleosomes along the genome) and chromatin-binding proteins such as CTCF and high mobility group proteins all contribute to chromatin structure by influencing nucleosome positioning, although a clear rubric for how nucleosome positioning and histone modification directly influence the folding of the genome remains to be determined (the highest resolution studies of chromatin structure across cell types reveal minor differences but reflect that topologically associating domains are largely conserved between lineages 66). Indeed, how local chromatin modifications determine local and global epigenomic structure is a fascinating frontier of basic and translational chromatin research. The most compelling evidence for lncRNA-mediated chromatin organization comes from the field of X chromosome inactivation, where the lncRNA Xist has been shown to govern inactivation-associated folding of the X chromosome based on its interaction with key sites on the chromosome and the recruitment of inactivated histone modifiers and DNA methylation machinery 67, 68.
Concluding remarks
New classes of therapeutic targets for heart failure require an appreciation of both the multicellular nature of the disease and the distinct pathophysiological mechanisms underpinning the diversity of symptoms in afflicted human populations. As put forth in this essay, we identify the interface among lncRNAs, histone modifications, and chromatin architecture as a key nodal point at which heart failure processes intersect, thus representing a ripe target for novel pharmacologic targeting. Challenges that remain include dissecting the roles of these different epigenetic regulators in different cell types in the heart and determining the optimum chromatin targets for a range of clinical heart failure phenotypes.
Acknowledgements
We thank the members of our laboratories for discussions related to the concepts presented in this essay.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Gianluigi Condorelli, Department of Cardiovascular Medicine, Humanitas Research Hospital, Humanitas University, Rozzano, Milan, Italy
Assam El-Osta, Epigenetics in Human Health and Disease Laboratory, Baker IDI Heart and Diabetes Institute, Melbourne, Victoria, Australia
Funding Statement
T.A.M. was supported by the National Institutes of Health (HL116848 and HL127240) and the American Heart Association (16SFRN31400013). Research in the Vondriska and Wang laboratories is supported by grants from the National Heart, Lung, and Blood Institute, the American Heart Association, and the David Geffen School of Medicine at UCLA.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; referees: 2 approved]
References
- 1. Benjamin EJ, Blaha MJ, Chiuve SE, et al. : Heart Disease and Stroke Statistics-2017 Update: A Report From the American Heart Association. Circulation. 2017;135(10):e146–e603. 10.1161/CIR.0000000000000485 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 2. Tan M, Luo H, Lee S, et al. : Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146(6):1016–28. 10.1016/j.cell.2011.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 3. Miyamoto S, Kawamura T, Morimoto T, et al. : Histone acetyltransferase activity of p300 is required for the promotion of left ventricular remodeling after myocardial infarction in adult mice in vivo. Circulation. 2006;113(5):679–90. 10.1161/CIRCULATIONAHA.105.585182 [DOI] [PubMed] [Google Scholar]
- 4. Wei JQ, Shehadeh LA, Mitrani JM, et al. : Quantitative control of adaptive cardiac hypertrophy by acetyltransferase p300. Circulation. 2008;118(9):934–46. 10.1161/CIRCULATIONAHA.107.760488 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 5. Wapenaar H, Dekker FJ: Histone acetyltransferases: challenges in targeting bi-substrate enzymes. Clin Epigenetics. 2016;8:59. 10.1186/s13148-016-0225-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lasko LM, Jakob CG, Edalji RP, et al. : Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature. 2017;550(7674):128–32. 10.1038/nature24028 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 7. Michaelides MR, Kluge A, Patane M, et al. : Discovery of Spiro Oxazolidinediones as Selective, Orally Bioavailable Inhibitors of p300/CBP Histone Acetyltransferases. ACS Med Chem Lett. 2018;9(1):28–33. 10.1021/acsmedchemlett.7b00395 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 8. Manning ET, Ikehara T, Ito T, et al. : p300 forms a stable, template-committed complex with chromatin: role for the bromodomain. Mol Cell Biol. 2001;21(12):3876–87. 10.1128/MCB.21.12.3876-3887.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ragvin A, Valvatne H, Erdal S, et al. : Nucleosome binding by the bromodomain and PHD finger of the transcriptional cofactor p300. J Mol Biol. 2004;337(4):773–88. 10.1016/j.jmb.2004.01.051 [DOI] [PubMed] [Google Scholar]
- 10. Hammitzsch A, Tallant C, Fedorov O, et al. : CBP30, a selective CBP/p300 bromodomain inhibitor, suppresses human Th17 responses. Proc Natl Acad Sci U S A. 2015;112(34):10768–73. 10.1073/pnas.1501956112 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 11. Picaud S, Fedorov O, Thanasopoulou A, et al. : Generation of a Selective Small Molecule Inhibitor of the CBP/p300 Bromodomain for Leukemia Therapy. Cancer Res. 2015;75(23):5106–19. 10.1158/0008-5472.CAN-15-0236 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 12. Weinert BT, Narita T, Satpathy S, et al. : Time-Resolved Analysis Reveals Rapid Dynamics and Broad Scope of the CBP/p300 Acetylome. Cell. 2018;174(1):231–244.e12. 10.1016/j.cell.2018.04.033 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 13. Roche J, Bertrand P: Inside HDACs with more selective HDAC inhibitors. Eur J Med Chem. 2016;121:451–83. 10.1016/j.ejmech.2016.05.047 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 14. Gregoretti IV, Lee YM, Goodson HV: Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004;338(1):17–31. 10.1016/j.jmb.2004.02.006 [DOI] [PubMed] [Google Scholar]
- 15. Antos CL, McKinsey TA, Dreitz M, et al. : Dose-dependent blockade to cardiomyocyte hypertrophy by histone deacetylase inhibitors. J Biol Chem. 2003;278(31):28930–7. 10.1074/jbc.M303113200 [DOI] [PubMed] [Google Scholar]
- 16. Bradner JE, West N, Grachan ML, et al. : Chemical phylogenetics of histone deacetylases. Nat Chem Biol. 2010;6(3):238–43. 10.1038/nchembio.313 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 17. Kook H, Lepore JJ, Gitler AD, et al. : Cardiac hypertrophy and histone deacetylase-dependent transcriptional repression mediated by the atypical homeodomain protein Hop. J Clin Invest. 2003;112(6):863–71. 10.1172/JCI19137 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 18. Yoshida M, Kijima M, Akita M, et al. : Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem. 1990;265(28):17174–9. [PubMed] [Google Scholar]
- 19. McKinsey TA: Therapeutic potential for HDAC inhibitors in the heart. Annu Rev Pharmacol Toxicol. 2012;52:303–19. 10.1146/annurev-pharmtox-010611-134712 [DOI] [PubMed] [Google Scholar]
- 20. Ooi JY, Tuano NK, Rafehi H, et al. : HDAC inhibition attenuates cardiac hypertrophy by acetylation and deacetylation of target genes. Epigenetics. 2015;10(5):418–30. 10.1080/15592294.2015.1024406 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 21. Brown JD, Lin CY, Duan Q, et al. : NF-κB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol Cell. 2014;56(2):219–31. 10.1016/j.molcel.2014.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 22. Li G, Ruan X, Auerbach RK, et al. : Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148(1–2):84–98. 10.1016/j.cell.2011.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 23. Zhang Y, Wong CH, Birnbaum RY, et al. : Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature. 2013;504(7479):306–10. 10.1038/nature12716 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 24. Anand P, Brown JD, Lin CY, et al. : BET bromodomains mediate transcriptional pause release in heart failure. Cell. 2013;154(3):569–82. 10.1016/j.cell.2013.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 25. Duan Q, McMahon S, Anand P, et al. : BET bromodomain inhibition suppresses innate inflammatory and profibrotic transcriptional networks in heart failure. Sci Transl Med. 2017;9(390): pii: eaah5084. 10.1126/scitranslmed.aah5084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haldar SM, McKinsey TA: BET-ting on chromatin-based therapeutics for heart failure. J Mol Cell Cardiol. 2014;74:98–102. 10.1016/j.yjmcc.2014.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Spiltoir JI, Stratton MS, Cavasin MA, et al. : BET acetyl-lysine binding proteins control pathological cardiac hypertrophy. J Mol Cell Cardiol. 2013;63:175–9. 10.1016/j.yjmcc.2013.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stratton MS, Lin CY, Anand P, et al. : Signal-Dependent Recruitment of BRD4 to Cardiomyocyte Super-Enhancers Is Suppressed by a MicroRNA. Cell Rep. 2016;16(5):1366–78. 10.1016/j.celrep.2016.06.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shortt J, Ott CJ, Johnstone RW, et al. : A chemical probe toolbox for dissecting the cancer epigenome. Nat Rev Cancer. 2017;17(3):160–83. 10.1038/nrc.2016.148 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 30. Müller S, Ackloo S, Arrowsmith CH, et al. : Donated chemical probes for open science. eLife. 2018;7: pii: e34311. 10.7554/eLife.34311 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 31. Reid BG, Stratton MS, Bowers S, et al. : Discovery of novel small molecule inhibitors of cardiac hypertrophy using high throughput, high content imaging. J Mol Cell Cardiol. 2016;97:106–13. 10.1016/j.yjmcc.2016.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bär C, Chatterjee S, Thum T: Long Noncoding RNAs in Cardiovascular Pathology, Diagnosis, and Therapy. Circulation. 2016;134(19):1484–99. 10.1161/CIRCULATIONAHA.116.023686 [DOI] [PubMed] [Google Scholar]
- 33. Lee JH, Gao C, Peng G, et al. : Analysis of transcriptome complexity through RNA sequencing in normal and failing murine hearts. Circ Res. 2011;109(12):1332–41. 10.1161/CIRCRESAHA.111.249433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li D, Chen G, Yang J, et al. : Transcriptome analysis reveals distinct patterns of long noncoding RNAs in heart and plasma of mice with heart failure. PLoS One. 2013;8(10):e77938. 10.1371/journal.pone.0077938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ounzain S, Micheletti R, Beckmann T, et al. : Genome-wide profiling of the cardiac transcriptome after myocardial infarction identifies novel heart-specific long non-coding RNAs. Eur Heart J. 2015;36(6):353–68a. 10.1093/eurheartj/ehu180 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 36. He C, Hu H, Wilson KD, et al. : Systematic Characterization of Long Noncoding RNAs Reveals the Contrasting Coordination of Cis- and Trans-Molecular Regulation in Human Fetal and Adult Hearts. Circ Cardiovasc Genet. 2016;9(2):110–8. 10.1161/CIRCGENETICS.115.001264 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 37. Yang KC, Yamada KA, Patel AY, et al. : Deep RNA sequencing reveals dynamic regulation of myocardial noncoding RNAs in failing human heart and remodeling with mechanical circulatory support. Circulation. 2014;129(9):1009–21. 10.1161/CIRCULATIONAHA.113.003863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Beermann J, Piccoli MT, Viereck J, et al. : Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol Rev. 2016;96(4):1297–325. 10.1152/physrev.00041.2015 [DOI] [PubMed] [Google Scholar]
- 39. Pant V, Kurukuti S, Pugacheva E, et al. : Mutation of a single CTCF target site within the H19 imprinting control region leads to loss of Igf2 imprinting and complex patterns of de novo methylation upon maternal inheritance. Mol Cell Biol. 2004;24(8):3497–504. 10.1128/MCB.24.8.3497-3504.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu L, An X, Li Z, et al. : The H19 long noncoding RNA is a novel negative regulator of cardiomyocyte hypertrophy. Cardiovasc Res. 2016;111(1):56–65. 10.1093/cvr/cvw078 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 41. Liu J, Wang DZ: An epigenetic "LINK(RNA)" to pathological cardiac hypertrophy. Cell Metab. 2014;20(4):555–7. 10.1016/j.cmet.2014.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Greco CM, Condorelli G: Epigenetic modifications and noncoding RNAs in cardiac hypertrophy and failure. Nat Rev Cardiol. 2015;12(8):488–97. 10.1038/nrcardio.2015.71 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 43. Anderson KM, Anderson DM, McAnally JR, et al. : Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature. 2016;539(7629):433–6. 10.1038/nature20128 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 44. Wang Z, Zhang XJ, Ji YX, et al. : The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat Med. 2016;22(10):1131–9. 10.1038/nm.4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang K, Liu F, Zhou LY, et al. : The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ Res. 2014;114(9):1377–88. 10.1161/CIRCRESAHA.114.302476 [DOI] [PubMed] [Google Scholar]
- 46. Zhu XH, Yuan YX, Rao SL, et al. : LncRNA MIAT enhances cardiac hypertrophy partly through sponging miR-150. Eur Rev Med Pharmacol Sci. 2016;20(17):3653–60. [PubMed] [Google Scholar]; F1000 Recommendation
- 47. Micheletti R, Plaisance I, Abraham BJ, et al. : The long noncoding RNA Wisper controls cardiac fibrosis and remodeling. Sci Transl Med. 2017;9(395): pii: eaai9118. 10.1126/scitranslmed.aai9118 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 48. Piccoli MT, Gupta SK, Viereck J, et al. : Inhibition of the Cardiac Fibroblast-Enriched lncRNA Meg3 Prevents Cardiac Fibrosis and Diastolic Dysfunction. Circ Res. 2017;121(5):575–83. 10.1161/CIRCRESAHA.117.310624 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 49. Qu X, Du Y, Shu Y, et al. : MIAT Is a Pro-fibrotic Long Non-coding RNA Governing Cardiac Fibrosis in Post-infarct Myocardium. Sci Rep. 2017;7: 42657. 10.1038/srep42657 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 50. Kumarswamy R, Bauters C, Volkmann I, et al. : Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ Res. 2014;114(10):1569–75. 10.1161/CIRCRESAHA.114.303915 [DOI] [PubMed] [Google Scholar]
- 51. de Gonzalo-Calvo D, Kenneweg F, Bang C, et al. : Circulating long-non coding RNAs as biomarkers of left ventricular diastolic function and remodelling in patients with well-controlled type 2 diabetes. Sci Rep. 2016;6: 37354. 10.1038/srep37354 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 52. Fan Z, Gao S, Chen Y, et al. : Integrative analysis of competing endogenous RNA networks reveals the functional lncRNAs in heart failure. J Cell Mol Med. 2018;22(10):4818–4829. 10.1111/jcmm.13739 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 53. Yan K, Arfat Y, Li D, et al. : Structure Prediction: New Insights into Decrypting Long Noncoding RNAs. Int J Mol Sci. 2016;17(1): pii: E132. 10.3390/ijms17010132 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 54. Sallam T, Sandhu J, Tontonoz P: Long Noncoding RNA Discovery in Cardiovascular Disease: Decoding Form to Function. Circ Res. 2018;122(1):155–66. 10.1161/CIRCRESAHA.117.311802 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 55. Freedman JE, Miano JM, National Heart, Lung, and Blood Institute Workshop Participants*: Challenges and Opportunities in Linking Long Noncoding RNAs to Cardiovascular, Lung, and Blood Diseases. Arterioscler Thromb Vasc Biol. 2017;37(1):21–5. 10.1161/ATVBAHA.116.308513 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 56. Lucas T, Bonauer A, Dimmeler S: RNA Therapeutics in Cardiovascular Disease. Circ Res. 2018;123(2):205–20. 10.1161/CIRCRESAHA.117.311311 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 57. Movassagh M, Choy MK, Knowles DA, et al. : Distinct epigenomic features in end-stage failing human hearts. Circulation. 2011;124(22):2411–22. 10.1161/CIRCULATIONAHA.111.040071 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 58. Papait R, Cattaneo P, Kunderfranco P, et al. : Genome-wide analysis of histone marks identifying an epigenetic signature of promoters and enhancers underlying cardiac hypertrophy. Proc Natl Acad Sci U S A. 2013;110(50):20164–9. 10.1073/pnas.1315155110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Meder B, Haas J, Sedaghat-Hamedani F, et al. : Epigenome-Wide Association Study Identifies Cardiac Gene Patterning and a Novel Class of Biomarkers for Heart Failure. Circulation. 2017;136(16):1528–44. 10.1161/CIRCULATIONAHA.117.027355 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 60. Chen H, Orozco LD, Wang J, et al. : DNA Methylation Indicates Susceptibility to Isoproterenol-Induced Cardiac Pathology and Is Associated With Chromatin States. Circ Res. 2016;118(5):786–97. 10.1161/CIRCRESAHA.115.305298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gilsbach R, Schwaderer M, Preissl S, et al. : Distinct epigenetic programs regulate cardiac myocyte development and disease in the human heart in vivo. Nat Commun. 2018;9(1): 391. 10.1038/s41467-017-02762-z [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 62. Nothjunge S, Nührenberg TG, Grüning BA, et al. : DNA methylation signatures follow preformed chromatin compartments in cardiac myocytes. Nat Commun. 2017;8(1): 1667. 10.1038/s41467-017-01724-9 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 63. Greco CM, Kunderfranco P, Rubino M, et al. : DNA hydroxymethylation controls cardiomyocyte gene expression in development and hypertrophy. Nat Commun. 2016;7: 12418. 10.1038/ncomms12418 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 64. Rosa-Garrido M, Chapski DJ, Schmitt AD, et al. : High-Resolution Mapping of Chromatin Conformation in Cardiac Myocytes Reveals Structural Remodeling of the Epigenome in Heart Failure. Circulation. 2017;136(17):1613–25. 10.1161/CIRCULATIONAHA.117.029430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sayed D, He M, Yang Z, et al. : Transcriptional regulation patterns revealed by high resolution chromatin immunoprecipitation during cardiac hypertrophy. J Biol Chem. 2013;288(4):2546–58. 10.1074/jbc.M112.429449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Schmitt AD, Hu M, Jung I, et al. : A Compendium of Chromatin Contact Maps Reveals Spatially Active Regions in the Human Genome. Cell Rep. 2016;17(8):2042–59. 10.1016/j.celrep.2016.10.061 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 67. Lee JT: Epigenetic regulation by long noncoding RNAs. Science. 2012;338(6113):1435–9. 10.1126/science.1231776 [DOI] [PubMed] [Google Scholar]
- 68. Engreitz JM, Pandya-Jones A, McDonel P, et al. : The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341(6147):1237973. 10.1126/science.1237973 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation