Abstract
Despite major advances in our understanding of TGF-β signaling in multiple cell types, little is known about the direct target genes of this pathway in human eosinophils. These cells constitute the major inflammatory component present in the sputum and lung of active asthmatics and their numbers correlate well with disease severity. During the transition from acute to chronic asthma, TGF-β levels rise several fold in the lung which drives fibroblasts to produce extracellular matrix (ECM) and participate in airway and parenchymal remodeling. In this report, we use purified blood eosinophils from healthy donors and analyze baseline and TGF-β responsive genes by RNA Seq, and demonstrate that eosinophils (PBE) express 7,981 protein-coding genes of which 178 genes are up-regulated and 199 genes are down-regulated by TGF-β. While 18 target genes have been previously associated with asthma and eosinophilic disorders, the vast majority have been implicated in cell death and survival, differentiation, and cellular function. Ingenuity pathway analysis revealed that 126 canonical pathways are activated by TGF-β including iNOS, TREM1, p53, IL-8 and IL-10 signaling. As TGF-β is an important cytokine for eosinophil function and survival, and pulmonary inflammation and fibrosis, our results represent a significant step toward the identification of novel TGF-β responsive genes and provide a potential therapeutic opportunity by selectively targeting relevant genes and pathways.
Keywords: RNA-Seq, TGF-β, eosinophils, human, asthma, gene expression, blood
Introduction
Pro-inflammatory cell infiltration of the bronchial airway, mucosa and wall in response to allergen is pathogneumonic of all forms of asthma (1). While all lineages are present, eosinophils typically comprise >50% of the airway inflammatory infiltrate despite being a minor (1–3%), and rapidly turning over constituent of the circulating white blood cell population. Tissue entry is unidirectional with eosinophilic inflammation lasting for weeks to months as the cells resist apoptosis through exposure to prosurvival signals induced by IL-3, IL-5, GM-CSF and other agonists. While in the lung, eosinophils can present antigen, enhance inflammation and fibrosis by the release of cytokines and chemokines, and drive airway remodeling by the release of metalloproteinases (MMP) and TGF-β as well as other critical functions.
TGF-β is generally considered anti-inflammatory however its levels are increased (2) in the bronchoalveolar lavage (BAL) of patients with allergic asthma and correlates well with airway and parenchymal remodeling (3). While pulmonary eosinophils release and are exposed to TGF-β, it remains unclear what genes are induced by TGF-β nor how they are regulated. In fibroblasts, TGF-β modulates the expression of a variety of ECM genes including collagens, MMPs, and tissue inhibitors of MMP (TIMP) (4). Most of these genes are related to tissue morphology and fibrosis, cancer development, cell growth and differentiation, and response to injury. Thus TGF-β responsive ECM genes are major contributors to both normal and pathological tissue fibrosis in the lung, liver, kidney as well as in cancer. The role of TGF-β in the immune system is broad, and plays fundamental roles in the regulation of cytokine gene expression during immune cell development and response to microbe infection as well as in many immune disorders (5).
Intracellular pathways mediating TGF-β signaling include Smad2/3, MAPKs, and PI3Ks, all of which have been implicated in the profibrotic response of fibroblasts (6). Despite the presence of TGF-β receptors (type I and II) and Smad2/3, little is known of the signaling or gene expression of eosinophils after exposure to TGF-β. In this study, we conducted RNA Seq analysis with control and TGF-β treated eosinophils from peripheral human blood and identified several hundred differentially regulated genes. Pathway analysis revealed that TGF-β activates ~126 canonical pathways including those implicated in carcinogenesis, nitric oxide biology, TREM1, and p53, IL-8, and IL-10 signaling. Since TGF-β is an important factor for cell survival, and pulmonary inflammation and fibrosis, our data provide new information to understand TGF-β signaling at the molecular level in eosinophils and possibly help identify novel targets for therapeutic intervention.
Methods
Subjects and eosinophil preparation
Peripheral blood was obtained by venipuncture from healthy or mildly atopic donors. Eosinophils from the blood were purified as described (7). Only populations >96% eosinophils were used for the studies. Eosinophils were cultured at a density of 1×106 cells/ml in RPMI-1640 medium, 10% FBS and gentamycin (50 mg/ml). All participants have a clinical record at the University of Wisconsin Hospital and written informed consent was obtained according to an approved protocol of the University of Wisconsin Hospital Institutional Review Board. The review board also specifically approved this study before initiation. The viability of cells was determined after each cell isolation and cell lysates/RNA were generated if the viability was > 96% at day 0 and >80% at day 4 after incubation with 200 pM IL-5.
Sample preparation and RNA-Seq
Freshly purified eosinophils (5×106 cells) were cultured in the presence and absence of TGF-β (1 ng/ml) for 5 h. Total RNA were isolated using RNAeasy Mini Kit (Qiagen) and the amount and purity were determined by NANODROP 2000c Spectrophotometer (Thermo Scientific). The quality of extracted total RNA was measured on an Agilent RNA 6000 Nano Chip and Agilent 2100 Bioanalyzer (Agilent Technologies). Samples were used for sequencing if the RNA Integrity Number (RIN) was > 7.5. One microgram of RNA was subjected to polyA-tailed cDNA library construction using TruSeq RNA Sample Preparation Kit v2 (Illumina). Sequencing was performed in the Genomics Core at UT-Southwestern Medical Center on an Illumina HiSeq™ 2500 (Illumina Inc.) according to the manufacturer’s protocol. Mapping reads and algorithm for RPKM (reads per kilobase per million mapped reads) were refined with the CLC Bioinformatics Database. The cutoff value for gene transcriptional activity was determined based on a 95% confidence interval for all RPKM values. RPKM >2.00 was used as the cutoff for positive expression (8).
Pathway Analysis
Lists of differentially expressed genes (DEG) from RNA-Seq analysis were generated using a cut-off of >1.5 fold between control and treated cells and used as the input for Ingenuity Pathway Analysis (IPA) software (www.ingenuity.com). Canonical pathways that were enriched in the DEG datasets were determined. Overrepresented pathways measure the likelihood of association between an experimental gene set and molecules in reference gene sets for a specific process/pathway. In general, p-values less than 0.05 by right-tailed Fisher’s Exact Test indicate a statistically significant, non-random association. Ingenuity uses public databases and performs in-house curation to formulate and update signaling pathways.
Reverse Transcription and Real-time PCR
After total RNA was reverse-transcribed, PCR was performed with a SYBR PCR master mix with the primers shown (Table 1S). An ABI 7500 thermocycler (Applied Biosystems) was used for 40 cycles of PCR. ΔCT calculates the differences between target CT values and the normalizer (housekeeping gene) for each sample: ΔCT = CT (target) − CT (normalizer). The comparative ΔΔCT calculates the differences between each sample ΔCT value and the baseline ΔCT. The comparative expression level (fold changes) was obtained transforming the logarithmic values to absolute values using 2−ΔΔCT. All data from untreated control cells was normalized to 100%.
Results
Identification and transcriptional profile of TGF-β target genes
In order to identify TGF-β signaling pathways and downstream genes, we performed RNA-Seq with subsequent pathway analysis. RNA was isolated from freshly purified, untreated eosinophils or cells treated with 1 ng/ml TGF-β for 5 h. This time point was chosen to minimize eosinophil apoptosis. Cells from 3 donors were subjected to quality control and single donor that met all the quality control metrics of donor health, percent cell survival and RNA integrity were employed. These included eosinophils with > 80% viability, 72 h after 200 uM of IL-5 and an RNA Integrity Number (RIN) >7.5 (Figure 1A). PolyA+ mRNA was purified from total RNA and used for cDNA library construction. Our sequencing detected 17,052 protein-coding genes (expression level: 0.001–7135) in untreated eosinophils. Using expression levels of > 2 as an arbitrary cutoff (8) for authentic gene expression (signal above background), 7,981 genes are expressed (Supplementary Table 2S) in resting eosinophils. Among these, 178 genes were increased (1.5 – 6.7 fold) (Figure 1B, Table 1) while 199 genes decreased (1.5 – 7.6 fold) (Table 2) (Figure 1B-C), indicating ~5% of the expressed genes were regulated by TGF-β.
Figure 1. Expression of TGF-β target genes by human eosinophils.
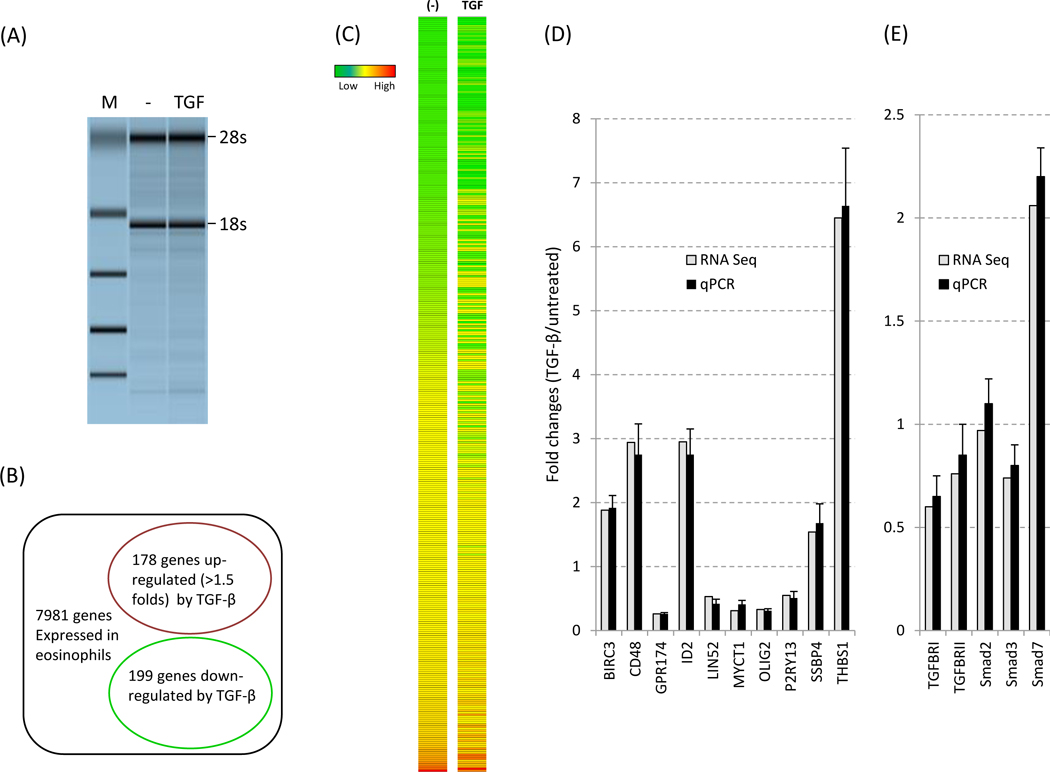
(A) Cells were left untreated or treated with TGF-β (1ng/ml) for 5 h before harvest. RNA quality was determined as described in Methods before RNA-Seq anaysis. M: size marker. (B) Genes altered (cut off: 1.5 fold) by TGF-β are shown in the circles. The expression level of 7981 genes are greater than 2.00 (PKAM). (C) Heat map of the TGF-β responsive genes (377 genes total) is shown. (D) Validation of RNA-Seq by qPCR with the 10 select transcripts. qPCR data were obtained from 13 eosinophil donors and normalized to GAPDH expression. (E) Validation of RNA-Seq by qPCR with the 6 transcripts associated with canonical TGF-β signaling. Smad6 was undetectable (not shown). The fold change for each gene was calculated and compared between the two detection methods.
Table 1. Genes upregulated by TGF-β in eosinophils.
Genes with fold changes > 1.5 are shown in the table.
| Genes | Folds | Genes | Folds | Genes | Folds | Genes | Folds |
|---|---|---|---|---|---|---|---|
| ABCA2 | 1.52 | GRASP | 2.72 | NLRC3 | 2.22 | TMBIM1 | 1.62 |
| ABHD15 | 1.62 | GRN | 1.67 | NR2F6 | 1.75 | TMEM120B | 1.53 |
| ACOT7 | 2.18 | GZF1 | 1.78 | NTMT1 | 1.51 | TMEM8 | 1.53 |
| ACP5 | 1.54 | HES1 | 2.08 | NXT1 | 1.54 | TNFSF14 | 2.04 |
| AGPAT2 | 1.56 | HES6 | 1.69 | ODC1 | 1.55 | TNK2 | 1.56 |
| ANXA2R | 1.76 | HMGA1 | 1.65 | OPRL1 | 1.58 | TRIM36 | 2.31 |
| ARHGEF19 | 1.67 | HMGN1 | 1.51 | ORC1L | 1.54 | TRIM8 | 1.71 |
| ARID5A | 1.61 | HMOX1 | 2.09 | OSM | 2.20 | TRIOBP | 1.16 |
| ARRDC2 | 1.87 | HOMER3 | 2.40 | P2RY8 | 1.83 | TSPAN14 | 1.52 |
| ASCL2 | 2.04 | ICAM1 | 1.67 | PIEZO1 | 1.71 | TTF2 | 1.52 |
| BBC3 | 1.73 | ID2 | 2.95 | PIM1 | 1.74 | TUBB8 | 2.03 |
| BCL9L | 1.96 | IER2 | 1.88 | PLEC1 | 1.65 | UAP1L1 | 1.63 |
| BCOR | 1.93 | IER3 | 2.43 | PLK3 | 3.74 | UBA5 | 1.55 |
| BLMH | 1.84 | IRAK1 | 1.69 | POLR2J3 | 1.54 | UBR7 | 1.68 |
| C20orf112 | 2.42 | IRAK2 | 1.66 | PPIF | 1.70 | UBTF | 1.84 |
| C9orf142 | 1.84 | IRF5 | 1.73 | PRKD3 | 1.62 | VDAC1 | 1.59 |
| CABIN1 | 1.53 | ITGA5 | 1.99 | PTPRCAP | 1.56 | VEGFA | 1.57 |
| CARD9 | 1.75 | JDP2 | 1.55 | QPCT | 1.87 | VIM | 1.80 |
| CCDC85B | 2.32 | JHDM1D | 1.56 | RASSF7 | 1.68 | VKORC1L1 | 1.55 |
| CD151 | 1.78 | JUN | 2.06 | RD3 | 2.14 | WIZ | 1.70 |
| CD69 | 1.72 | KCTD11 | 2.15 | RDX | 1.63 | XXYLT1 | 2.15 |
| CD83 | 1.66 | KCTD15 | 1.92 | RFX1 | 1.53 | YWHAH | 1.87 |
| CDCA4 | 1.61 | KCTD5 | 1.55 | RFX2 | 1.98 | ZBTB4 | 1.51 |
| CDKN1A | 1.83 | KDM2B | 1.57 | RGCC | 1.74 | ZC3H12A | 1.68 |
| CERK | 1.56 | KLF10 | 2.28 | RGS3 | 1.81 | ZFP36 | 1.58 |
| CIB1 | 1.55 | LAT2 | 1.66 | RHOF | 1.61 | ZNF124 | 1.85 |
| CISH | 2.11 | LDHA | 1.50 | RP11–395P17.3 | 1.78 | ZNF219 | 1.67 |
| DKFZp761E198 | 1.66 | LIMK1 | 1.55 | S1PR2 | 3.64 | ZNF469 | 1.60 |
| DNAJA2 | 1.50 | LMO7 | 1.78 | SAMD1 | 2.61 | ||
| DNMT1 | 1.66 | LOC100128071 | 1.52 | SCAMP4 | 1.57 | ||
| EFEMP2 | 1.78 | LOC100287078 | 1.54 | SDCBP2 | 1.67 | ||
| EGFL7 | 2.11 | LOC100287351 | 1.57 | SERINC2 | 1.83 | ||
| EIF2C1 | 1.57 | LOC100287578 | 2.35 | SLC18A2 | 1.92 | ||
| ELMSAN1 | 1.70 | LPPR2 | 1.66 | SLC1A4 | 1.96 | ||
| ENC1 | 1.55 | LRIG1 | 1.62 | SLC1A5 | 2.47 | ||
| ENO3 | 2.20 | LTC4S | 1.51 | SLC7A5 | 2.53 | ||
| EPOR | 2.04 | LYL1 | 1.56 | SMAD7 | 2.06 | ||
| ETS2 | 1.65 | MAPK13 | 1.52 | SNAI3 | 1.65 | ||
| FAM43A | 2.39 | MARCH3 | 1.55 | SNRNP25 | 1.54 | ||
| FAM65A | 1.72 | MICAL1 | 1.61 | SPAG1 | 1.93 | ||
| FAM78A | 1.72 | MICALL1 | 1.58 | SRXN1 | 1.52 | ||
| FASN | 1.85 | MKI67IP | 1.54 | SSBP4 | 1.54 | ||
| FBXL14 | 1.91 | MOB2 | 1.80 | ST6GALNAC6 | 1.98 | ||
| FCGR3A | 1.58 | MUM1 | 1.67 | STAMBPL1 | 1.54 | ||
| FHOD1 | 1.53 | MXRA7 | 1.63 | STARD10 | 1.68 | ||
| FLJ14213 | 1.58 | NAPRT1 | 1.56 | STK39 | 2.03 | ||
| FMNL3 | 1.52 | NFAM1 | 1.55 | TBC1D2 | 1.51 | ||
| GADD45G | 1.61 | NFKB1 | 1.51 | TBC1D9B | 1.66 | ||
| GATSL3 | 1.53 | NFKBIE | 1.88 | TCF7 | 1.74 | ||
| GPR153 | 1.78 | NKD1 | 1.87 | TIAM2 | 1.66 |
Table 2. Genes downregulated by TGF-β in eosinophils.
Genes with fold changes > 1.5 are shown in the table.
| Genes | Folds | Genes | Folds | Genes | Folds | Genes | Folds |
|---|---|---|---|---|---|---|---|
| ACAT2 | 1.51 | DDX21 | 1.78 | LGALS12 | 1.86 | PSTPIP2 | 1.53 |
| ACP6 | 1.63 | DUSP6 | 3.28 | LILRB2 | 1.81 | PTGER4 | 2.25 |
| ADAM19 | 1.90 | E2F1 | 1.72 | LIN52 | 1.88 | PTPN22 | 1.54 |
| ADORA2B | 1.93 | ECHDC3 | 1.68 | LIPA | 1.69 | PTPRE | 1.96 |
| ADRB2 | 5.03 | EEPD1 | 1.55 | LOC100286999 | 2.05 | PTPRJ | 1.79 |
| AHR | 2.42 | EHD4 | 1.55 | LOC100287172 | 2.16 | PXMP4 | 1.53 |
| AJAP1 | 1.60 | EMR3 | 1.52 | LOC100287759 | 1.72 | PYGL | 1.80 |
| ALAS1 | 2.04 | EVI2B | 1.67 | LOC100287887 | 1.55 | RABEPK | 1.56 |
| ALDH6A1 | 2.84 | FAM101B | 2.18 | LOC100288921 | 1.84 | RDH11 | 1.67 |
| AMICA1 | 2.01 | FAM110A | 1.62 | LRRC33 | 1.52 | RGS2 | 1.54 |
| ANKRD37 | 1.53 | FAM117A | 1.54 | LYSMD4 | 1.67 | RHOQ | 1.64 |
| ANKRD55 | 1.86 | FAM127B | 1.57 | LYZ | 1.57 | RIPK2 | 1.82 |
| ANXA1 | 1.73 | FAM65B | 1.61 | MARCH1 | 3.50 | RPAP3 | 1.59 |
| AP4B1 | 1.50 | FAM98A | 1.55 | MCART1 | 1.54 | SFMBT2 | 1.72 |
| AREG | 2.10 | FAS | 1.52 | MCCC1 | 1.61 | SGPP1 | 1.60 |
| ARL4C | 2.03 | FBN1 | 1.62 | MEGF9 | 1.69 | SIGMAR1 | 2.01 |
| ARL6IP6 | 1.80 | FCGR2B | 1.78 | METTL7A | 1.94 | SIRPD | 1.89 |
| ARRDC4 | 1.65 | FH | 1.56 | MICU1 | 1.51 | SLAMF6 | 2.22 |
| ASMTL | 2.02 | FLVCR1 | 1.66 | MPHOSPH6 | 1.75 | SLC20A1 | 2.38 |
| ATP10D | 1.80 | FOPNL | 1.52 | MPZL3 | 2.25 | SLC39A3 | 1.67 |
| BIRC3 | 1.88 | FRMD4B | 1.70 | MRPS6 | 1.72 | SMAD5 | 1.52 |
| BOLA2B | 1.51 | FRRS1 | 1.57 | MSTO1 | 1.52 | SMN1 | 1.82 |
| C12orf65 | 1.51 | FYB | 1.56 | MT1X | 1.59 | SNRPF | 1.61 |
| C3AR1 | 1.78 | GALC | 1.62 | MTHFD2 | 1.56 | SORD | 2.24 |
| C4orf33 | 1.63 | GAS7 | 1.53 | MYB | 3.71 | SP4 | 1.95 |
| C7orf25 | 1.53 | GNG2 | 1.56 | MYCT1 | 3.26 | SPTB | 1.80 |
| C9orf40 | 1.61 | GOLGA8N | 1.64 | MYD88 | 1.55 | STK17B | 1.84 |
| CA5BP1 | 2.23 | GPR174 | 3.75 | NAMPT | 2.24 | SUCNR1 | 2.21 |
| CACNG6 | 1.52 | GPR65 | 2.05 | NIACR2 | 2.62 | SULT1A3 | 1.56 |
| CACNG8 | 1.58 | HAS1 | 1.87 | NIN | 1.59 | TBC1D8 | 1.90 |
| CACUL1 | 1.54 | HAUS8 | 1.54 | NOV | 3.20 | TCP11L1 | 1.86 |
| CASP3 | 2.11 | HELB | 2.18 | NUCB2 | 1.50 | TFDP2 | 1.57 |
| CBLB | 2.22 | HILPDA | 4.74 | NUDT4 | 1.64 | TGFBR1 | 1.82 |
| CCDC146 | 1.56 | HIPK2 | 1.56 | NUDT5 | 1.52 | THBS1 | 6.45 |
| CCL4L1 | 1.66 | HLX | 7.58 | OLIG1 | 2.06 | TKTL1 | 1.60 |
| CD200R1 | 2.54 | HLX-AS1 | 3.90 | OLIG2 | 3.02 | TMEM2 | 1.83 |
| CD244 | 1.96 | HRASLS5 | 1.86 | OXER1 | 2.32 | TMEM55A | 1.87 |
| CD48 | 2.94 | HRH4 | 1.92 | P2RY13 | 1.83 | TMEM60 | 1.53 |
| CD84 | 2.27 | IDO1 | 2.46 | P2RY14 | 3.13 | TNFSF13B | 1.53 |
| CENPBD1 | 1.61 | IKZF2 | 1.65 | PAPSS1 | 1.51 | TRIM5 | 1.52 |
| CLC | 1.97 | IL18RAP | 2.25 | PDZD11 | 1.69 | TRPC6 | 2.74 |
| CLEC12A | 1.58 | IL1RL1 | 1.63 | PECI | 1.56 | TSEN34 | 1.71 |
| CNPY4 | 1.53 | IL34 | 1.80 | PGM2 | 1.51 | TUBA1A | 1.70 |
| CREG1 | 1.50 | IL5RA | 2.15 | PHB | 1.58 | UBASH3B | 1.67 |
| CSAR2 | 1.70 | IMPACT | 1.56 | PHTF2 | 1.68 | UGT2B28 | 1.58 |
| CXorf21 | 1.60 | INPP1 | 1.55 | PIK3R3 | 2.10 | XK | 2.08 |
| CXorf65 | 2.22 | INTS7 | 1.51 | PM20D2 | 1.51 | YEATS4 | 1.69 |
| CYP51A1 | 1.65 | IPCEF1 | 2.08 | PMAIP1 | 1.97 | ZNF524 | 1.92 |
| DAPK1 | 1.70 | KIAA1024 | 1.61 | PRDM1 | 1.60 | ZSCAN16 | 1.53 |
| DAPP1 | 2.01 | KLF6 | 2.08 | PRIMPOL | 2.07 |
Validation of the RNA-Seq data by qPCR
To validate the DEGs observed, 10 affected genes were randomly picked for qPCR analysis using eosinophils from 13 additional donors (see primers in Table 1S). We also included canonical TGF-β signaling components TGFBR1, TGFBR2, Smad2, Smad3, • •Smad6 and Smad7 in validation panel. The results (Figure 1D, 1E) revealed that the expression profiles of all these genes were entirely consistent with the RNA-Seq findings. Smad6 was undetectable neither by RNA Seq nor by qPCR. In addition, the expression profiles of entire genes by the RNA Seq were strongly correlated with proteomic data from independent donors (Shen et al. Manuscript in preparation). Thus, RNA-Seq can be performed with eosinophil RNA and the quality and coverage of sequencing yields valid and reproducible results.
Classification of the TGF-β target genes based on biological function
We classified the 377 genes (affected by TGF-β) into 33 categories in terms of biological and cellular function (Table 3) using IMT software. Despite a substantial overlap, many DEGs were unique to specific categories. Somewhat surprisingly for non-dividing, terminally differentiated eosinophils, many of the 377 genes have been implicated in cancer, cell death and survival, cell-to-cell signaling and interactions, along with the expected genes controlling cellular function, movement and maintenance. These data suggest that TGF-β may play important roles in the regulation of eosinophil function in tumor immunity, cell death, cell-to-cell interaction, and pulmonary migration.
Table 3. Expression of TGF-β target genes categorized by biological function.
Numbers in parenthesis next to the name of each category indicates the number of identified, related genes and shown in the right column.
| Cancer (44) | ADAM19, AGPAT2, ARRDC2, ARRDC4, ASCL2, BCL9L, CD151, CIB1, CREG1, DNAJA2, DNMT1, ECHDC3, EGFL7, FAM110A, FAM65B, FCGR2B, FRRS1, GALC, HMGN1, IKZF2, IL34, IMPACT, ITGA5, LGALS12, LRIG1, NKD1, NOV, OLIG1, PHB, PIK3R3, POLR2J3, RPAP3, SERINC2, SMN1, SNRPF, SULT1A3, THBS1, TMEM2, TNK2, TUBA1A, TUBB8, UBASH3B, VDAC1, WIZ |
| Carbohydrate metabolism (34) | ADORA2B, ADRB2, ALDH6A1, AREG, BLMH, CASP3, CCL4L1, CYP51A1, EMR3, EVI2B, FAS, GNG2, GPR65, GPR153, GPR174, HOMER3, HRH4, ICAM1, INTS7, OPRL1, OSM, P2RY13, P2RY14, PTGER4, PYGL, S1PR2, SORD, SUCNR1, TMBIM1, TSEN34, VEGFA, VIM, YWHAH, ZNF124 |
| Cardiovascular disease (20) | ACP5, ARID5A, ARL4C, CABIN1, CNPY4, MARCH3, MRPS6, MT1X, NUDT4, NUDT5, PSTPIP2, PTPN22, PTPRJ, RGS2, RGS3, SLC18A2, SRXN1, STARD10, TRPC6, XK |
| Cell cycle (29) | AHR, ARHGEF19, ATP10D, BOLA2B, CDCA4, E2F1, FAM101B, FAM98A, FHOD1, HAUS8, HELB, HMGA1, IER2, IER3, KCTD5, MICALL1 NIN, NLRC3, NXT1,ODC1, PAPSS1, PDZD11, SIGMAR1, SP4, SPAG1, STAMBPL1, STK39, TBC1D8, TMEM55A |
| Cell death and survival (77) | ADORA2B, ADRB2, ALDH6A1, AREG, ASMTL, BLMH, BIR3, C9orf40, CASP3, CCL4L1, CD84, CD200R1, CYP51A1, DUSP6, EEPD1, EFEMP2, EHD4, EMR3, ENC1, ENO3, ETS2, FAM127B, FAM65A, FAM78A, FAS, FBN1, FCGR3A, FH, GAS7, GNG2, HAS1, HIPK2, HMOX1, ICAM1, IDO1, IL18RAP, IL1RL1, IL5RA, INPP1, IRAK1, IRAK2, IRF5, KLF10, LAT2, LDHA, LILRB2, LIPA, LMO7, LYZ, METTL7A, MSTO1, MXRA7, MYB, MYD88, MTHFD2, PM20D2, PMAIP1, PRDM1, PRKD3, PTPRE, PXMP4, QPCT, RFX1, RFX2, SLC39A3, TGFBR1, TIAM2, TNFSF13B, TNFSF14, TTF2, UBA5, VEGFA, VIM, YWHAH, ZBTB4, ZC3H12A, ZNF124 |
|
Cell-To-Cell signaling and interaction (15) |
ACP6, UBR7, CD151, CIB1, CREG1, FAM117A, FCGR2B, ITGA5, LRIG1, MCCC1, NOV, PIK3R3, RDH11, THBS1, TNK2 |
|
Cellular assembly and Organization (44) |
ACP6, ANXA1, ARHGEF19, ARL6IP6, ATP10D, BOLA2B, C12orf65, UBR7, C7orf25, C9orf50, C9orf142, CCDC85B, DAPK1, FAM101B, FAM117A, FAM98A, FMNL3, GADD45G, GZF1, HAUS8, HRASLS5, KCTD5, LIMK1, MCCC1, MICALL1, NFAM1, NIN, NLRC3, NXT1, PAPSS1, PTPRCAP, RABEPK, RASSF7, RD3, RDH11, RDX, RHOF, RHOQ, SFMBT2, SPAG1, STAMBPL1, TCP11L1, TMEM55A, ZSCAN16 |
| Cellular compromise (12) | ANXA1, DAPK1, DAPP1, GADD45G, LIMK1, NFAM1, PTPRCAP, RABEPK, RASSF7, RDX, RHOF, RHOQ |
| Cellular development (93) | ACOT7, AMICA1, ANKRD37, ANKRD55, BIR3, BLMH, C3AR1, CD84, CD200R1, CD83, CISH, CLC, CYP51A1, DUSP6, EFEMP2, EHD4, EMR3, ENC1, ENO3, EPOR, ETS2, FAS, FBN1, FCGR3A, FH, FLVCR1, GRASP, HAS1, HIPK2, HMOX1, IDO1, IER2, IL18RAP, IL1RL1, IL5RA, INPP1, IPCEF1, IRAK1, IRAK2, IRF5, KCTD11, KCTD15, KLF10, LAT2, LDHA, LILRB2, LIPA, LMO7, LYL1, LYZ, MAPK13, MARCH1, MEGF9, METTL7A, MTHFD2, MYB, MYD88, NAMPT, NFKB1, NKD1, NUCB2, OLIG1, OLIG2, PGM2, PIM1, PMAIP1, PRDM1, PRKD3, PTPRE, QPCT, RFX1, RFX2, SGPP1, SIRPD, SNRNP25, ST6GALNAC6, TGFBR1, TIAM2, TKTL1, TNFSF13B, TNFSF14, TRIM36, TRIM5, TRIOBP, TSPAN14, TTF2, UBA5, UGT2B28, ZBTB4, ZC3H12A, ZFP36, ZNF219, ZNF524 |
|
Cellular function and Maintenance (94) |
ACOT7, ACP6, AJAP1, AMICA1, ANKRD37, ANKRD55, ANXA1, AP4B1, ARHGEF19, ATP10D, BBC3, BLMH, BOLA2B, C14orf142, C3AR1, CACNG6, CACNG8, CARD9, CBLB, CD244, CD48, CD69, CD84, CD200R1, CERK, CLC, CYP51A1, DAPK1, DAPP1, DUSP6, EFEMP2, EHD4, EMR3, ENO3, FAM101B, FAM117A, FAM98A, FAS, FBN1, FYB, GADD45G, GRASP, GRN, HAUS8, HIPK2, IDO1, INPP1, KCTD15, KCTD5, KLF10, LAT2, LIMK1, LTC4S, MCCC1, MICALL1, MPHOSPH6, MYB, NFAM1, NFKBIE, NLRC3, NXT1, PAPSS1, PLK3, PTPRCAP, QPCT, RABEPK, RASSF7, RDH11, RDX, RHOF, RHOQ, RIPK2, SDCBP2, SIRPD, SLAMF6, SLC1A4, SLC7A5, SNAI3, SPAG1, SSBP4, STAMBPL1, STK17B, TBC1D2, TBC1D9B, TGFBR1, TIAM2, TMEM55A, TRIM5, TRIM8, TSPAN14, TTF2, UAP1L1, ZBTB4, ZNF524 |
|
Cellular growth and Proliferation (62) |
AHR, BIR3, CDCA4, E2F1, ECHDC3, ENC1, ETS2, FCGR3A, FH, FHOD1, FLVCR1, GAS7, HAS1, HELB, HMGA1, HMOX1, IER2, IER3, IL18RAP, IL1RL1, IL5RA, IPCEF1, IRAK1, IRAK2, KCTD11, IRF5, LDHA, LILRB2, LIPA, LMO7, LYZ, MEGF9, METTL7A, MTHFD2, MYD88, NAMPT, NKD1, NUCB2,ODC1, OLIG1, OLIG2, PDZD11, PGM2, PMAIP1, PRKD3, PTPRE, RFX1, RFX2, SGPP1, SIGMAR1, SNRNP25, SP4, ST6GALNAC6, STK39, TBC1D8, TKTL1, TNFSF13B, TNFSF14, UBA5, UGT2B28, ZC3H12A, ZNF219 |
| Cellular movement (15) | BCL9L, EVI2B, ECHDC3, FAM110A, FAM65B, FRRS1, GALC, IKZF2, IL34, IMPACT, NKD1, OLIG1, SERINC2, TMEM2, UBASH3B |
| Developmental disorder (10) | CCDC146, FBXL14, FRMD4B, MUM1, MYCT1, PPIF, SAMD1, SCAMP4, SLC7A5, TMEM60 |
|
Digestive system development and function (21) |
ACAT2, ALAS1, BCOR, DDX21, FASN, HES1, HES6, HLX, ID2, JDP2, JUN, KDM2B, KLF6, LIN52, NR2F6, ORC1L, SLC20A1, TFDP2, UBTF, VKORC1L1, YEATS4 |
| Embryonic development (21) | ACAT2, ALAS1, BCOR, DDX21, FASN, HES1, HES6, HLX, ID2, JDP2, JUN, KDM2B, KLF6, LIN52, NR2F6, ORC1L, SLC20A1, TFDP2, UBTF, VKORC1L1, YEATS4 |
| Endocrine system disorders (14) | ASMTL, C9orf40, EEPD1, FAM127B, FAM65A, FAM78A, MSTO1, MXRA7, NAPRT1, P2RY8, PHTF2, PM20D2, PXMP4, SLC39A3 |
| Gene expression (29) | AJAP1, ASMTL, C3AR1, C9orf40, CACUL1, CACNG6, CACNG8, CARD9, EEPD1, FAM127B, FAM65A, FAM78A, MPHOSPH6, MSTO1, MXRA7, NAPRT1, P2RY8, PHTF2, PM20D2, PXMP4, SDCBP2, SLAMF6, SLC1A4, SLC39A3, SNAI3, SSBP4, TBC1D2, TBC1D9B, UAP1L1 |
|
Hair and skin development and function (1) |
MPZL3 |
| Hematological disease (20) | ACP5, ARID5A, ARL4C, CABIN1, CNPY4, MARCH3, MRPS6, MT1X, NUDT4, NUDT5, PSTPIP2, PTPN22, PTPRJ, RGS2, RGS3, SLC18A2, SRXN1, STARD10, TRPC6, XK |
|
Hematological system development and function (21) |
ACOT7, AMICA1, ANKRD37, ANKRD55, AP4B1, BBC3, C3AR1, CARD9, CBLB, CD48, CD69, CD244, CD83, CERK, CISH, CLC, EMR3, EPOR, FYB, GRASP, GRN, KCTD15, LTC4S, LYL1, MAPK13, MARCH1, NFKB1, NFKBIE, PIM1, PLK3, RIPK2, SIRPD, SLC7A5, STK17B, TRIM36, TRIOBP, TRIM5, TRIM8, TSPAN14, ZFP36, ZNF524 |
| Hematopoiesis (11) | CD83, CISH, EPOR, LYL1, MAPK13, MARCH1, NFKB1, PIM1, TRIM36, TRIOBP, ZFP36 |
|
Hepatic system development and function (21) |
ACAT2, ALAS1, BCOR, DDX21, FASN, HES1, HES6, HLX, ID2, JDP2, JUN, KDM2B, KLF6, LIN52, NR2F6, ORC1, SLC20A1, TFDP2, UBTF, VKORC1L1, YEATS4 |
| Hereditary disorder (20) | ACP5, ARID5A, ARL4C, CABIN1, CNPY4, MARCH3, MRPS6, MT1X, NUDT4, NUDT5, PSTPIP2, PTPN22, PTPRJ, RGS2, RGS3, SLC18A2, SRXN1, STARD10, TRPC6, XK |
| Molecular transport (21) | BLMH, CYP51A1, EMR3, FAS, GPR65, GPR153, GPR174, HOMER3, HRH4, INTS7, OPRL1, OSM, P2RY13, P2RY14, PTGER4, PYGL, S1PR2, SORD, SUCNR1, TMBIM1, TSEN34 |
|
Nervous system development and function (14) |
ARL6IP6, C12orf65, C7orf25, NTMT1, UBR7, CCDC85B, FMNL3, GZF1, HRASLS5, RASSF7, RD3, SFMBT2, TCP11L1, ZSCAN16 |
|
Organismal injury and abonormalities (29) |
ADAM19, AGPAT2, ARRDC2, ARRDC4, ASCL2, CCDC146, DNAJA2, DNMT1, EGFL7, FBXL14, FRMD4B, HMGN1, LGALS12, MUM1, MYCT1, PHB, POLR2J3, PPIF, RPAP3, SAMD1, SCAMP4, SLC7A5, SMN1, SNRPF, SULT1A3, TMEM60, TUBB8, VDAC1, WIZ |
|
Reproductive system development and function (13) |
ENC1, FLVCR1, IER2, IPCEF1, KCTD11, MEGF9, NUCB2, PGM2, SNRNP25, ST6GALNAC6, TKTL1, UGT2B28, ZNF219 |
| Reproductive system disease (10) | CCDC146, FBXL14, FRMD4B, MUM1, MYCT1, PPIF, SAMD1, SCAMP4, SLC7A5, TMEM60 |
|
Skeletal and muscular Disorders (15) |
AJAP1, C3AR1, C5AR2, CACNG6, CACNG8, CARD9, MPHOSPH6, SDCBP2, SLAMF6, SLC1A4, SNAI3, SSBP4, TBC1D2, TBC1D9B, UAP1L1 |
| Small molecule biochemistry (33) | ADORA2B, ADRB2, ALDH6A1, AREG, BLMH, CASP3, CCL4L1, CYP51A1, EMR3, FAS, GNG2, GPR65, GPR153, GPR174, HOMER3, HRH4, ICAM1, INTS7, OPRL1, OSM, P2RY13, P2RY14, PTGER4, PYGL, S1PR2, SORD, SUCNR1, TMBIM1, TSEN34, VEGFA, VIM, YWHAH, ZNF124 |
| Tissue development (35) | AHR, BCL9L, CD151, CDCA4, CIB1, E2F1, FAM110A, FAM65B, FCGR2B, FHOD1, FRRS1, GALC, HELB, HMGA1, IER2, IER3, IKZF2, IL34, IMPACT, ITGA5, LRIG1, NKD1, NOV, ODC1, PDZD11, PIK3R3, SERINC2, SIGMAR1, SP4, STK39, TBC1D8, THBS1, TMEM2, TNK2, UBASH3B |
| Tissue morphology (52) | ADAM19, AGPAT2, AP4B1, ARL6IP6, ARRDC2, ARRDC4, ASCL2, BBC3, C12orf65, C7orf25, NTMT1, C9orf142, CARD9, CBLB, CCDC85B, CD48, CD69, CD244, CEDKN1A, CERK, DNAJA2, DNMT1, EGFL7, FMNL3, FYB, GZF1, GRN, HMGN1, HRASLS5, LGALS12, LTC4S, NFKBIE, PHB, PLK3, POLR2J3, RASSF7, RD3, RIPK2, RPAP3, SFMBT2, SLC7A5, SMN1, SNRPF, STK17B, SULT1A3, TCP11L1, TRIM8, TUBA1A, TUBB8, VDAC1, WIZ, ZSCAN16 |
Eosinophils have significant anti-tumor activity (9) and based on the data above, several genes including ADAM19, CD151 and VDAC1 may be involved. The function of these genes in asthma and their modulation by TGF-β signaling has not been investigated. ADAM19 (adamalysin19), is a cell surface glycoprotein that was up-regulated in the most invasive human brain tumors (astrocytoma and glioblastoma) (10) and inflamed and fibrotic lung and kidney (11). The membrane associated CD151 (tetraspanin) is frequently overexpressed on cancer cells and functionally linked to early steps of tumor growth, migration and metastasis although its mechanism of action remains obscure (12). VDAC1, a mitochondrial voltage-dependent anion channel is a Bcl-2 dependent pro-apoptotic protein (13).
TGF-β antagonizes IL-5 pro-survival signaling by inhibiting Akt phosphorylation, leading to calpain cleavage and activation in eosinophils (14). However, neither Akt nor calpain expression were affected by TGF-β (Table 1 and 2), underscoring the multiple levels (transcriptional and post-translational) through which this cytokine modulates eosinophil biology. We found 77 TGF-β target genes are related to cell death and survival (Table 3) and include BIRC3, CASP3, FAS, HIPK2, IL5RA, and MYB, all of which were down-regulated by TGF-β (Table 2), while the threonine-serine kinase Pim1 was upregulated (Table 1). Pim1 is overexpressed in a range of hematologic malignancies and solid tumors (15) and regulates apoptosis, metabolism, cell cycle, self-renewal and migration as well as lymphocyte responses and hematopoietic lineage development. BIRC3 (cIAP2) inhibits apoptosis by binding to TRAF1/2 (16) and blocks the spontaneous formation of the ripoptosome, a large multi-protein pro-apoptotic complex. While the effector CASP3 and death receptor FAS mediate proapoptotic signaling, IL5RA transduces pro-survival signals to eosinophils (17). HIPK2, a serine/threonine kinase, interacts with many transcription factors such as p53, CREB1 and PDX1 (18). HIPK2 inhibits cell growth and promotes apoptosis through activation of p53/TP53 both at the transcriptional and protein level. The proto-oncogene MYB encodes a transcription factor, plays an important role in the control of proliferation and differentiation of hematopoietic progenitor cells (19). To date, there are no any reported data regarding the role of BIR3, HIPK2 or MYB in eosinophils.
Only a small number of genes (Table 3) were functionally clustered within the cell-to-cell signaling and interaction space despite the important role of these functions in allergic inflammation. Eosinophils interactions with other cells can be modulated by prostaglandin EP4 receptors (endothelial-eosinophils), chymase and prostaglandin (mast cells-eosinophils), house dust mite (epithelial-eosinophils), phytohemagglutinin/PMA and cytokine/chemokines (lymphocytes-eosinophils). In concordance, there have been few functional studies to determine if these TGF-β responsive genes have a role in cell-to-cell interactions. Based on these data, we conclude that TGF-β does not significantly contribute to cell-cell communication in asthma.
Surprisingly, there are many TGF-β target genes (Table 3) categorized to cellular development (93 genes), cellular growth and proliferation (62 genes) and cellular function and maintenance (94 genes), and include CD69, FAS, and MYB. CD69 is expressed on the surface of activated Tregs and eosinophils, induced by TGF-β (Table and elevated on BAL eosinophils from patients with asthma or blood eosinophils activated in vitro by IL3, IL5 or GM-CSF (20). Forty-one genes (Table 3) are involved in diseases, development and function of the hematologic system. To date, the role of TGF-β in the regulation of hematopoietic cell (e.g. eosinophils, red blood cells, and lymphocytes) development and differentiation has been largely unknown. However, disruption of the TGF-β pathway has been associated with hematopoietic tumors, eosinophilia and abnormal cytokine production (21, 22).
Pathway analysis
In order to identify potential signaling pathways that regulate the TGF-β target genes, we employed Ingenuity Pathway Analysis (IPA). We found that TGF-β significantly (p<0.05) (Table 3S) activates 124 of the 362 canonical pathways. Representative pathways (top 5) include molecular mechanisms of cancer, iNOS, TREM1, p53, and IL-8 signaling. Twenty-one genes were connected to the molecular mechanisms of cancer (top 1, Table 3S). Unexpectedly, canonical TGF-β/Smad signaling (Table 3S), while significant (p=0.0123027) was well down the list (rank 68) but included several interacting and regulated transcripts (JUN, TGFBR1, SMAD7, MAPK13 and SMAD5). CCR3 receptor-mediated migration signaling was ranked similarly (p=0.0446684) despite a similar number of interconnected and modulated genes (PIK3R3, GNG2, MAPK13, PRKD3 and LIMK1). These data suggest that TGF-β plays a minor role in the regulation of ECM production and cell migration by eosinophils. Indeed, there are no supportive data published regarding TGF-β involvement in these pathways in eosinophils. Collectively, this analysis provides valuable information identifying how the growth factor modulates its target genes through multiple signaling cascades.
Figure 2 shows the network of cellular processes potentially influenced by TGF-β. The network includes 27 hub genes (green and orange colors) for the major signaling pathways identified. Several interacting genes are common among the pathways identified. For example, JUN (increased by 2.06 fold by TGF-β) is involved in 63 pathways (out of 126) (Table 3S) and NFKB1 (increased by 1.51 fold) in 87 pathways. E2F1 was down-regulated (1.72 fold) (Table 2) by TGF-β and seen only in cancer-related pathways (Table 3S). E2F1 along with JUN plays a critical role in controlling both cell cycle progression and apoptotic cell death in response to DNA damage and oncogene activation. These genes could directly or indirectly regulate some of downstream TGF-β targets among the 377 genes. Due to space limitation, we did not further query the RNA-Seq dataset (containing all TGF-β responsive genes) to map the potential downstream transcripts in detail using IPA’s Grow Pathway algorithm.
Figure 2. Analysis of pathways responsible for the expression of TGF-β target genes.
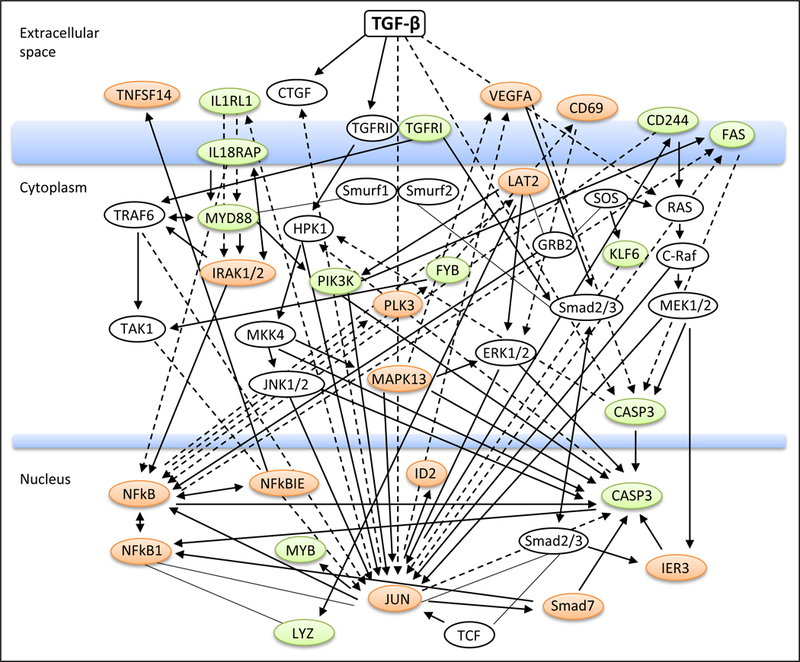
The annotated interactions and regulatory relationships were generated using IPA’s (Ingenuity Pathway Analysis) connectivity analysis. Hub genes (in white circles) were defined as those regulating or interacting with ≥5 TGF-β targeted transcripts. Representative genes immediately downstream of hub genes are shown (orange, upregulated; green, downregulated).
TGF-β target genes associated with asthma and eosinophilic diseases
In order to further explore the significance of the TGF-β target genes in asthma and eosinophil-associated disorders, we performed database search (MalaCards and NCBI) for the 377 genes. The search yielded 18 asthma-related genes. Important genes include AHR, CD69, FAS, and JUN (Table 4). AHR (arylhydrocarbon receptor) plays an important role in airway inflammation via its effect on bronchial epithelia and immune cells (23). After ligation, the AHR-dioxin complex induces the transcription of detoxification enzymes to detoxify dioxin-like compounds into physiologic metabolites and influence immune responsiveness and mucosal barrier function of epithelium in the lung. Genetic knock-down of AHR increased the level of TGF-β while TGF-β treatment decreased AHR (2.42 fold, Table 2) suggesting AHR may repress TGF-β signaling in early stage asthma, resulting in a pro-inflammatory effect. CD69 (increased by TGF-β, Table 1), an early activation marker antigen of lymphocytes, eosinophils and neutrophils, is a target gene of TGF-β in monocytes (24). TGF-β rapidly upregulated CD69 transcription via TAK1-mediated, p38 MAPK activation (25). CD69 expression on eosinophils was also significantly increased in asthmatic patients after exposure to allergen (26) whereas knock-down or administration of anti-CD69 dramatically increased airway eosinophils recruitment and enhanced Th2/Th17 response in the BAL (27). Thus, CD69 activation may inhibit the exacerbation of allergic asthma by limiting the recruitment of activated immune cells or/and modulating the immune cell function in the lung. JUN (increased by TGF-β, Table 1) is an important regulator of pro-inflammatory genes, tissue remodeling and apoptosis. Multiple studies (28–30) demonstrated that JUN activation is essential for eosinophils and lymphocyte accumulation, airway smooth muscle cell proliferation, bronchial hyper-responsiveness and airway remodeling following acute or chronic allergen challenge. During airway remodeling JNK1 played a major role in augmenting the profibrotic effects of TGF-β, which included epithelial-to-mesenchymal transition (EMT) of airway epithelial cells (28).
Table 4.
TGF-β target genes that have been associated with asthma and eosinophilic diseases.
| Symbol | Description | Major function and implication in Asthma |
|---|---|---|
| ADRB2 | Adrenoceptor beta 2, surface | A member of the adrenergic receptor group of G protein-coupled receptor; mediate catecholamine-induced activation of adenylate cyclase; associated with nocturnal asthma |
| AHR | Aryl hydrocarbon receptor | A transcription factor; mediates biochemical and toxic effects of halogenated aromatic hydrocarbons; regulate cell cycle; associated with eosinophilic fascilitis. |
| CACNG6 | Calcium channel, gamma subunit 6 | A voltage-dependent calcium channel; associated with aspirin-intolerant asthma |
| CD244 | Natural killer cell receptor 2B4 | A cell surface receptor; modulate NK-cell cytolytic activity and leukocyte activation; involved in the allergic asthma |
| CD69 | CD69 molecule | A member of calcium dependent lectin superfamily of type II transmembrane receptors; involved in lymphocyte proliferation and activation; implicated in the allergic airway inflammation and hyperresponsiveness. |
| CLC | Charcot-Leyden crystal galectin | An enzyme which acts on membranes to regulate multifunctional lysophospholipids; regulates immune response; essential for the anergy and suppressive function of Treg; associate with some myeloid leukemias; Involved in the recruitment of immune cells in chronic asthma. |
| FAS | Fas cell surface death receptor | A membe of the TNF-receptor superfamily; regulates programmed cell death; implicated in the pathogenesis of various malignancies and diseases of immune system; transduce proliferating signals in fibroblasts and T cells; important in eosinophil and T lymphocyte apoptosis in allergic asthma. |
| HRH4 | Histamine receptor H4 | Play a role in inflammation and allergy response; mediates chemotaxis of mast cells and eosinophils; controls cytokine release from dendritic and T cells; play a role in the differentiation of myeloblasts and promyelocytes; likely influence the pathogenesis of infection-induced asthma. |
| ICAM1 | Intercellular Adhesion Molecule 1 | A cell surface glycoprotein; ligand for leukocyte adhesion protein LFA-1; play a role during leukocyte trans-endothelial migration; receptor for rhinovirus; binds to integrins of type CD11a/CD18 or CD11b/CD18; associated with childhood asthma. |
| IDO1 | Indoleamine 2,3-dioxygenase 1 | Play a role in antimicrobial and antitumor defense, neuropathology, immunoregulation and antioxidant activity; modulates T-cell behavior; immunomodulator in allergy and asthma. |
| IL18RAP | IL18 receptor accessory protein | Enhances IL18-bining activity of the IL18 receptor; mediates IL18-dependent activation of NF-kappa-B and JNK; polymorphisms of the gene were associated with asthma and atopy. |
| IL1RL1 | Interleukin 1 receptor-like 1 | A receptor for IL33; likely involved in the function of helper T cells; genetic variation of the gene was associated with asthma pathophysiology. |
| IL5RA | Interleukin 5 receptor, alpha | An IL5-specific subunit of a heterodimeric cytokine receptor; binds IL5 and mediate its signaling; contribute to the pathogenesis of bronchial asthma; the receptor mediated signaling is important in maintaining the survival and funcions of B cells and eosinophils. |
| IRF5 | Interferon regulatory factor 5 | A transcription factor involved in the induction of interfereons IFNA and INFB and inflammatory cytokines upon virus infection; modulate cell growth, differentiation, apoptosis, and immune system activity. Haplotype of the gene was associated with asthma and severity of asthmatic symptoms. |
| JUN | Jun proto-oncogene | A transcription factor which is highly similar to the viral protein; promotes activity of NR5A1 when phosphorylated by HIPK3 leading to increased steroidogenic gene expression upon cAMP signaling stimulation; associated with airway remodeling and corticosteroids resistance in the asthma. |
| LTC4S | Leukotriene C4 synthase | Involved in the production of leukotrienes which are mediators of anaphylaxis and inflammatory conditions such as asthma; localizes to the nuclear envelope and adjacent endoplasmic reticulum. |
| OSM | Oncostatin M | A cytokine which inhibits proliferation of a number of tumor cell lines; regulates cytokine production (IL-6, G-CSF and GM-CSF) from endothelial cells; involved in the maturation of fetal hepatocytes, thereby promoting liver development and regeneration; relevant to airway remodeling and pathogenesis of asthma. |
| SMAD7 | SMAD family member 7 | An antagonist of TGF-beta receptor superfamily signaling; inhibits the signaling by associating with their receptors thus preventing SMAD2 access; expression is induced by TGFBR1; associated with airway inflammation and reactivity in asthma |
Discussion
Resting and activated eosinophils synthesize and release a variety of (over 30) cytokines, growth factors, and chemokines. Eosinophils are the predominant source of TGF-β in the airway and bronchial tissues of asthmatics. The cells are concomitantly exposed to many chemokines and cytokines in asthmatic lungs which may synergistically increase or suppress TGF-β’s effects. Freshly purified blood eosinophils can secrete > 120 pg/ml of TGF-β which was increased slightly in the presence of survival cytokines after 24 h culture (31). In addition, TGF-β can overcome IL-5 signaling and induces eosinophils apoptosis after 3 days of in vitro culture (14). These, and other observations demonstrated (14, 32) that eosinophils possess an intact TGF-β/Smad signaling apparatus, however, the functionality of the system and the affected target genes are largely unknown. Therefore, we performed RNA-Seq with purified blood eosinophils to identify direct TGF-β target genes and characterize their potential relevance to asthma pathology.
Previously, using an oligonucleotide-based microarray on the total RNA from resting and IL-5/GM-CSF-treated eosinophils (33), we detected ~3,000 transcripts in resting cells of which 264 (~9%) were altered by prosurvival cytokines. RNA-Seq technology identified ~17,000 poly A-tailed transcripts from resting cells and the differential expression of 377 (~2%) genes between control and TGF-β treated eosinophils. Our data demonstrate the feasibility and sensitivity of this approach for eosinophils. RNA-Seq also offers the potential for precise demarcation of intron-exon boundaries, 5′/3′-UTR regions, splice variants, single nucleotide polymorphisms (SNPs), and potentially, new transcripts with low copy number. In this study, we harvested cells at 5 h (34) to identify primarily intermediate and late response transcripts. The cutoff for differential expression was set at 1.5 fold because many TGF-β targets, particularly ECM genes, are only moderately induced (~3 fold). qPCR validation of 16 selected transcripts covering a wide range of expression showed good agreement with sequence data. Overall, RNA-Seq produced a comprehensive list of total and differential eosinophil gene expression responsive to TGF-β.
Analysis of DEG profiles (Table 1 and Table 2) showed a similar numbers of transcripts were increased (178 genes) or decreased (199 genes) by TGF-β treatment compared to control. Cluster analysis further identified that most of the genes are implicated in cellular function and maintenance (94 genes), cellular development (93 gene), cell death and survival (77 genes), cellular growth and proliferation (62 genes), hematological disease/system development (41 genes), and tissue development (35 genes) although similar gene sets were overrepresented between some clusters. Moreover, pathway analysis showed that TGF-β activates various canonical signaling which culminated in 124 major pathways such as those involved in molecular mechanisms of cancer, iNOS signaling, TREM1 signaling, p53 signaling and IL-8 signaling, providing greater biological insights in unraveling the potential molecular mechanisms exerted by TGF-β effects. While the network of the pathways reported here is contextualized to TGF-β exposure, it could form the basis for further hypothesis testing of other TGF-β family members for similar transcriptional changes, signaling pathways and activation of kinases within the cells.
The sensitivity of cDNA microarray has often yielded conflicting results. For example, in human keratinocytes and mouse breast cells (35, 36), TGF-β modulated genes predominantly involved in EMT, whereas in fibroblasts and A549 cancer cells TGF-β dramatically induced genes (up to 265 ECM genes) responsible for tissue fibrosis (34, 37, 38). Using RNA-Seq we found that the pattern of cancer-related gene alterations are similar to the TGF-β target genes reported in some cancer models (39–42). The representative genes include ADAM19, DNMT1, GALC, ITGA5, LRIG1, NOV, PHB, PIK3R3, and THBS1. Moreover, a subset of cell survival and death-related genes (CASP3, FAS, BIR3, HIPK2, IL5RA, and MYB) found in our study were also described in neurons, dendritic cells, epithelial cells, and smooth muscle treated with TGF-β (43–45). Among the canonical TGF-β signaling components, only TGFBR1 (Table 2) and inhibitory Smad7 (Table 1) were downregulated and upregulated, respectively, whereas the TGF-β itself was not changed, indicating a negative feedback mechanism via modulation of intracellular signaling molecules. Collectively, these observations suggest that TGF-β presumably utilizes evolutionally conserved phosphorylation signaling (e.g. Smad2/3) and feedback mechanisms that mayfunction independent of cell types while certain downstream target genes are cell type specific. We detected rapid (30 min-1 h) Smad3 phosphorylation after TGF-β treatment in these terminally differentiated immune cells (unpublished data). Further work is required to confirm the biological response and significance of each gene in the asthma pathogenesis and eosinophil-related disorders.
It is becoming clear that TGF-β signaling is increased in the lung of asthmatics. The major function of the cytokine is to initiate ECM production by parenchymal fibroblasts. We found that eosinophils express a few ECM genes such as COL18A1 (RPKM=13), COL9A2 (RPKM=33), COL9A3 (RPKM=6.5), MMP25 (RPKM=292), and TIMP2 (RPKM=164) but none of them were altered by TGF-β. Instead, a number of apoptosis-related genes were highly expressed and significantly modulated by TGF-β, strongly supporting a role for TGF-β in the induction of eosinophil apoptosis (14, 46).
Database search revealed that 18 genes (out of the 377) (Table 4) have been previously associated with asthma although we can’t rule out the possibility that additional genes are also involved in the disease. 11 (out of the 18) genes code for cell surface receptors, implying eosinophils may actively engage in fibrosis development through cell-cell contact or intracellular signaling. The other genes code for transcription factors (3 genes), intracellular enzymes (3 genes) and one cytokine. The latter, Oncostatin M (OSM) (increased 2.2 fold by TGF-β), is known to inhibit the proliferation of tumor cell lines and regulate cytokine production (IL-6 and GM-CSF) from endothelial cells. Resting eosinophils expressed high levels of OSM (RPKM=51) (Table 2S) but not its receptor (OSMR) (RPKM=0). The protein level of Oncostatin M was significantly increased in the lung of asthmatics and predominantly localized to airway neutrophils and macrophages (47), indicating eosinophils may augment airway inflammation through paracrine OSM release. Of particular interest among the 11 receptors are AHR, CD69 and FAS (Table 4) and the intracellular enzymes CLC, IDO1 and LTC4S. CLC (charcot-leyden crystal protein) is a lysophospholipase, comprising approximately 7–10% of the total cellular protein and expressed exclusively in human eosinophils and basophils (48). It is also found in body fluids/secretions and eosinophilic tissues in the form of hexagonal bipyramidal crystals (49) whose levels were significantly elevated in patients with asymptomatic or acute asthma (50). IDO1 is another candidate molecule broadly expressed during infection, transplantation, pregnancy, autoimmunity and neoplasia (51). Expression of the IDO1 was regulated by AHR via an autocrine AHR-IL6-STAT3 signaling loop (52). AHR is a ligand-activated transcription factor and plays an important role in controlling cellular response to small environmental molecules (diet, flora and metabolism). Interestingly, TGF-β treatment significantly increased both IDO1 and AHR transcripts to the similar levels (2.4 folds) (Table 1), an event consistent with their mutual interaction but respective roles in cellular response. IDO1 inhibits the proliferation and activation of antigen-specific T lymphocytes and induces immune tolerance, which is considered to be the result from a cellular depletion of tryptophan by IDO1 activity (53).
In summary, we have shown that eosinophil RNA can be isolated and used for RNA-Seq analysis. We have applied this technology to assess differential gene expression induced by TGF-β, a critical pro-fibrotic cytokine, highly expressed by pulmonary eosinophils during asthma exacerbations. Our data demonstrate that hundreds of genes are modulated by this cytokine with many implicated in asthma, eosinophil biology or function. The elucidation of these genes will hopefully provide new targets that can be leveraged for therapeutic development against asthma.
Supplementary Material
Acknowledgments
Grant support: This study was supported by P01 HL088594 to J.S.M.
Abbreviations used:
- TGF
transforming growth factor
- MMP
metalloproteinases
- ECM
extracellular matrix
- TIMP
tissue inhibitors of MMP
- RPKM
(reads per kilobase per million mapped reads)
- DEG
differentially expressed genes
- BAL
bronchoalveolar lavage
- IPA
Ingenuity Pathway Analysis
- RIN
RNA Integrity Number
- RT-PCR
Reverse transcription polymerase chain reaction
- GM-CSF
Granulocyte-macrophage colony-stimulating factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Footnotes:
Conflict of interest: The authors declare that they have no competing interest.
References
- 1.Kim HY, DeKruyff RH, Umetsu DT (2010) The many paths to asthma: phenotype shaped by innate and adaptive immunity. Nat Immunol 11:577–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batra V, Musani AI, Hastie AT, Khurana S, Carpenter KA, Zangrilli JG, Peters SP (2004) Bronchoalveolar lavage fluid concentrations of transforming growth factor (TGF)-beta1, TGF-beta2, interleukin (IL)-4 and IL-13 after segmental allergen challenge and their effects on alpha-smooth muscle actin and collagen III synthesis by primary human lung fibroblasts. Clin Exp Allergy 34:437–444. [DOI] [PubMed] [Google Scholar]
- 3.Halwani R, Al-Muhsen S, Al-Jahdali H, Hamid Q (2011) Role of transforming growth factor-β in airway remodeling in asthma. Am J Respir Cell Mol Biol 44:127–133. [DOI] [PubMed] [Google Scholar]
- 4.Makinde T, Murphy RF, Agrawal DK (2007) The regulatory role of TGF-beta in airway remodeling in asthma. Immunol Cell Biol 85:348–356. [DOI] [PubMed] [Google Scholar]
- 5.Travis MA, Sheppard D (2014) TGF-β activation and function in immunity. Annu Rev Immunol 32:51–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leask A, Abraham DJ (2004) TGF-beta signaling and the fibrotic response. FASEB J 18:816–827 [DOI] [PubMed] [Google Scholar]
- 7.Hansel TT, De Vries IJ, Iff T, Rihs S, Wandzilak M, Betz S, Blaser K, Walker C (1991) An improved immunomagnetic procedure for the isolation of highly purified human blood eosinophils. J Immunol Methods 145:105–110. [DOI] [PubMed] [Google Scholar]
- 8.Dozmorov I and Lefkovits I (2009) Internal standard-based analysis of microarray data. Part 1: analysis of differential gene expressions. Nucleic Acids Res 37: 6323–6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tepper RI, Coffman RL, Leder P (1992) An eosinophil-dependent mechanism for the antitumor effect of interleukin-4. Science 257:548–551. [DOI] [PubMed] [Google Scholar]
- 10.Wildeboer D, Naus S, Amy Sang QX, Bartsch JW, Pagenstecher A (2006) Metalloproteinase disintegrins ADAM8 and ADAM19 are highly regulated in human primary brain tumors and their expression levels and activities are associated with invasiveness. J Neuropathol Exp Neurol 65:516–527. [DOI] [PubMed] [Google Scholar]
- 11.Qi B, Newcomer RG, Sang QX. ADAM19/adamalysin 19 structure, function, and role as a putative target in tumors and inflammatory diseases (2009) Curr Pharm Des 15:2336–2348. [DOI] [PubMed] [Google Scholar]
- 12.Sadej R, Grudowska A, Turczyk L, Kordek R, Romanska HM (2014) CD151 in cancer progression and metastasis: a complex scenario. Lab Invest 94:41–51. [DOI] [PubMed] [Google Scholar]
- 13.Brahimi-Horn MC, Mazure NM (2014) Hypoxic VDAC1: a potential mitochondrial marker for cancer therapy. Adv Exp Med Biol 772:101–110. [DOI] [PubMed] [Google Scholar]
- 14.Xie Q, Shen ZJ, Oh J, Chu H, Malter JS (2011) Transforming Growth Factor-β1 Antagonizes Interleukin-5 Pro-Survival Signaling by Activating Calpain-1 in Primary Human Eosinophils. J Clin Cell Immunol Suppl 1 pii: 003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merkel AL, Meggers E, Ocker M (2012) PIM1 kinase as a target for cancer therapy. Expert Opin Investig Drugs 21:425–436. [DOI] [PubMed] [Google Scholar]
- 16.Zheng C, Kabaleeswaran V, Wang Y, Cheng G, Wu H (2010) Crystal structures of the TRAF2: cIAP2 and the TRAF1: TRAF2: cIAP2 complexes: affinity, specificity, and regulation. Mol Cell 38:101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JH, Chang HS, Kim JH, Park SM, Lee YM, Uh ST, Rhim T, Chung IY, Kim YH, Park BL, Park CS, Shin HD (2007) Genetic effect of CCR3 and IL5RA gene polymorphisms on eosinophilia in asthmatic patients. J Allergy Clin Immunol 120:1110–1117. [DOI] [PubMed] [Google Scholar]
- 18.Sombroek D, Hofmann TG (2009) How cells switch HIPK2 on and off. Cell Death Differ 16:187–194. [DOI] [PubMed] [Google Scholar]
- 19.Ramsay RG, Gonda TJ. MYB function in normal and cancer cells (2008) Nat Rev Cancer 8:523–534. [DOI] [PubMed] [Google Scholar]
- 20.Morii T, Nishikawa K, Ako H, Narita N (1993) [Expression of activation antigen CD69 on eosinophils]. Nihon Rinsho 51:588–592. [PubMed] [Google Scholar]
- 21.Ogawa T, Uemura K (1993) [CT and MRI diagnosis of hemorrhagic infarction]. Nihon Rinsho 51 Suppl:800–805. [PubMed] [Google Scholar]
- 22.Gatault S, Legrand F, Delbeke M, Loiseau S, Capron M (2012) Involvement of eosinophils in the anti-tumor response. Cancer Immunol Immunother 61:1527–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beamer CA, Shepherd DM (2013) Role of the aryl hydrocarbon receptor (AhR) in lung inflammation. Semin Immunopathol 35:693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marzio R, Mauël J, Betz-Corradin S (1999) CD69 and regulation of the immune function. Immunopharmacol Immunotoxicol 21:565–582. [DOI] [PubMed] [Google Scholar]
- 25.Wöbke TK, von Knethen A, Steinhilber D, Sorg BL (2013) CD69 is a TGF-β/1α,25-dihydroxyvitamin D3 target gene in monocytes. PLoS One 8:e64635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Julius P, Luttmann W, Knoechel B, Kroegel C, Matthys H, Virchow JC Jr (1999) CD69 surface expression on human lung eosinophils after segmental allergen provocation. Eur Respir J 13:1253–1259. [DOI] [PubMed] [Google Scholar]
- 27.Martín P, Gómez M, Lamana A, Matesanz Marín A, Cortés JR, Ramírez-Huesca M, Barreiro O, López-Romero P, Gutiérrez-Vázquez C, de la Fuente H, Cruz-Adalia A, Sánchez-Madrid F (2010) The leukocyte activation antigen CD69 limits allergic asthma and skin contact hypersensitivity. J Allergy Clin Immunol 126:355–365. [DOI] [PubMed] [Google Scholar]
- 28.van der Velden JL, Hoffman SM, Alcorn JF, Tully JE, Chapman DG, Lahue KG, Guala AS, Lundblad LK, Aliyeva M, Daphtary N, Irvin CG, Janssen-Heininger YM (2014) Absence of c-Jun NH2-terminal kinase 1 protects against house dust mite-induced pulmonary remodeling but not airway hyperresponsiveness and INFLAMMATION. Am J Physiol Lung Cell Mol Physiol 306:L866–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nath P, Eynott P, Leung SY, Adcock IM, Bennett BL, Chung KF (2005) Potential role of c-Jun NH2-terminal kinase in allergic airway INFLAMMATION and remodelling: effects of SP600125. Eur J Pharmacol 506:273–283. [DOI] [PubMed] [Google Scholar]
- 30.Eynott PR, Nath P, Leung SY, Adcock IM, Bennett BL, Chung KF (2003) Allergen-induced INFLAMMATION and airway epithelial and smooth muscle cell proliferation: role of Jun N-terminal kinase. Br J Pharmacol 140:1373–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohkawara Y, Tamura G, Iwasaki T, Tanaka A, Kikuchi T, Shirato K (2000) Activation and transforming growth factor-beta production in eosinophils by hyaluronan. Am J Respir Cell Mol Biol 23:444–451. [DOI] [PubMed] [Google Scholar]
- 32.Kanzaki M, Shibagaki N, Hatsushika K, Mitsui H, Inozume T, Okamoto A, Dobashi Y, Ogawa H, Shimada S, Nakao A (2007) Human eosinophils have an intact Smad signaling pathway leading to a major transforming growth factor-beta target gene expression. Int Arch Allergy Immunol 142:309–317. [DOI] [PubMed] [Google Scholar]
- 33.Bates ME, Liu LY, Esnault S, Stout BA, Fonkem E, Kung V, Sedgwick JB, Kelly EA, Bates DM, Malter JS, Busse WW, Bertics PJ (2004) Expression of interleukin-5- and granulocyte macrophage-colony-stimulating factor-responsive genes in blood and airway eosinophils. Am J Respir Cell Mol Biol 30:736–743. [DOI] [PubMed] [Google Scholar]
- 34.Verrecchia F, Chu ML, Mauviel A (2001) Identification of novel TGF-beta /Smad gene targets in dermal fibroblasts using a combined cDNA microarray/promoter transactivation approach. J Biol Chem 276:17058–17062. [DOI] [PubMed] [Google Scholar]
- 35.Xie L, Law BK, Aakre ME, Edgerton M, Shyr Y, Bhowmick NA, Moses HL (2003) Transforming growth factor beta-regulated gene expression in a mouse mammary gland epithelial cell line. Breast Cancer Res 5:R187–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zavadil J, Bitzer M, Liang D, Yang YC, Massimi A, Kneitz S, Piek E, Bottinger EP (2001) Genetic programs of epithelial cell plasticity directed by transforming growth factor-beta. Proc Natl Acad Sci U S A 98:6686–6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Renzoni EA, Abraham DJ, Howat S, Shi-Wen X, Sestini P, Bou-Gharios G, Wells AU, Veeraraghavan S, Nicholson AG, Denton CP, Leask A, Pearson JD, Black CM, Welsh KI, du Bois RM (2004) Gene expression profiling reveals novel TGFbeta targets in adult lung fibroblasts. Respir Res 5:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keating DT, Sadlier DM, Patricelli A, Smith SM, Walls D, Egan JJ, Doran PP (2006) Microarray identifies ADAM family members as key responders to TGF-beta1 in alveolar epithelial cells. Respir Res 7:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lei J, Hasegawa H, Matsumoto T, Yasukawa M (2010) Peroxisome proliferator-activated receptor α and γ agonists together with TGF-β convert human CD4+CD25- T cells into functional Foxp3+ regulatory T cells. J Immunol 185: 7186–7198. [DOI] [PubMed] [Google Scholar]
- 40.Lin HY, Yang LT (2013) Differential response of epithelial stem cell populations in hair follicles to TGF-β signaling. Dev Biol 373:394–406. [DOI] [PubMed] [Google Scholar]
- 41.Lafont J, Jacques C, Le Dreau G, Calhabeu F, Thibout H, Dubois C, Berenbaum F, Laurent M, Martinerie C (2005) New target genes for NOV/CCN3 in chondrocytes: TGF-beta2 and type X collagen. J Bone Miner Res 20:2213–2223. [DOI] [PubMed] [Google Scholar]
- 42.Ranganathan P, Agrawal A, Bhushan R, Chavalmane AK, Kalathur RK, Takahashi T, Kondaiah P (2007) Expression profiling of genes regulated by TGF-beta: differential regulation in normal and tumour cells. BMC Genomics 8:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tzai TS, Shiau AL, Liu LL, Wu CL (2000) Immunization with TGF-beta antisense oligonucleotide-modified autologous tumor vaccine enhances the antitumor immunity of MBT-2 tumor-bearing mice through upregulation of MHC class I and Fas expressions. Anticancer Res 20:1557–1562. [PubMed] [Google Scholar]
- 44.Shang Y, Doan CN, Arnold TD, Lee S, Tang AA, Reichardt LF, Huang EJ (2013) Transcriptional corepressors HIPK1 and HIPK2 control angiogenesis via TGF-β-TAK1-dependent mechanism. PLoS Biol 11:e1001527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gadson PF Jr, Dalton ML, Patterson E, Svoboda DD, Hutchinson L, Schram D, Rosenquist TH (1997) Differential response of mesoderm- and neural crest-derived smooth muscle to TGF-beta1: regulation of c-myb and alpha1 (I) procollagen genes. Exp Cell Res 230:169–180. [DOI] [PubMed] [Google Scholar]
- 46.Alam R, Forsythe P, Stafford S, Fukuda Y (1994) Transforming growth factor beta abrogates the effects of hematopoietins on eosinophils and induces their apoptosis. J Exp Med 179:1041–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simpson JL, Baines KJ, Boyle MJ, Scott RJ, Gibson PG (2009) Oncostatin M (OSM) is increased in asthma with incompletely reversible airflow obstruction. Exp Lung Res 35:781–794. [DOI] [PubMed] [Google Scholar]
- 48.Dvorak AM, Letourneau L, LOGIN GR, Weller PF, Ackerman SJ (1988) Ultrastructural localization of the Charcot-Leyden crystal protein (lysophospholipase) to a distinct crystalloid-free granule population in mature human eosinophils. Blood 72:150–158. [PubMed] [Google Scholar]
- 49.Dvorak AM, Letourneau L, Login GR, Weller PF, Ackerman SJ (1988) Ultrastructural localization of the Charcot-Leyden crystal protein (lysophospholipase) to a distinct crystalloid-free granule population in mature human eosinophils. Blood 72:150–158. [PubMed] [Google Scholar]
- 50.Dor PJ, Ackerman SJ, Gleich GJ (1984) Charcot-Leyden crystal protein and eosinophil granule major basic protein in sputum of patients with respiratory diseases. Am Rev Respir Dis 130:1072–1077. [DOI] [PubMed] [Google Scholar]
- 51.Jaronen M, Quintana FJ (2014) Immunological Relevance of the Coevolution of IDO1 and AHR. Front Immunol 5: 521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Litzenburger UM, Opitz CA, Sahm F, Rauschenbach KJ, Trump S, Winter M, Ott M, Ochs K, Lutz C, Liu X, Anastasov N, Lehmann I, Höfer T, von Deimling A, Wick W, Platten M (2014) Constitutive IDO expression in human CANCER is sustained by an autocrine signaling loop involving IL-6, STAT3 and the AHR. Oncotarget 5:1038–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cesario A, Rocca B, Rutella S (2011) The interplay between indoleamine 2, 3-dioxygenase 1 (IDO1) and cyclooxygenase (COX)-2 in chronic inflammation and cancer. Curr Med Chem 18:2263–2271. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


