Meisner and Albrechtsen present two methods for inferring population structure and admixture proportions in low depth next-generation sequencing (NGS). NGS methods provide large amounts of genetic data but are associated with statistical uncertainty, especially for low-depth...
Keywords: Population structure, PCA, admixture, ancestry, next-generation sequencing, genotype likelihoods, low depth
Abstract
We here present two methods for inferring population structure and admixture proportions in low-depth next-generation sequencing (NGS) data. Inference of population structure is essential in both population genetics and association studies, and is often performed using principal component analysis (PCA) or clustering-based approaches. NGS methods provide large amounts of genetic data but are associated with statistical uncertainty, especially for low-depth sequencing data. Models can account for this uncertainty by working directly on genotype likelihoods of the unobserved genotypes. We propose a method for inferring population structure through PCA in an iterative heuristic approach of estimating individual allele frequencies, where we demonstrate improved accuracy in samples with low and variable sequencing depth for both simulated and real datasets. We also use the estimated individual allele frequencies in a fast non-negative matrix factorization method to estimate admixture proportions. Both methods have been implemented in the PCAngsd framework available at http://www.popgen.dk/software/.
POPULATION genetic studies often consist of individuals of diverse ancestries, and inference of population structure therefore plays an important role in population genetics and association studies. Population stratification can act as a confounding factor in association studies as it can lead to spurious associations (Marchini et al. 2004). Principal component analysis (PCA) has been used in genetics for a long time, such as in Menozzi et al. (1978) where synthetic maps were produced in an exploratory analysis of genetic variation. PCA is now a common tool in population genetic studies, where its dimension reduction properties can be used to visualize population structure by summarizing the genetic variation through principal components (Novembre and Stephens 2008), correct for population stratification in association studies, and investigate demographic history (Patterson et al. 2006; Price et al. 2006; Fumagalli et al. 2013) as well as perform genome selection scans (Hao et al. 2015; Galinsky et al. 2016; Luu et al. 2017). PCA is an appealing approach to infer population structure as the aim is not to classify the individuals into discrete populations, but instead to describe continuous axes of genetic variation such that heterogeneous populations and admixed individuals can be better represented (Patterson et al. 2006). Another successful approach in modeling complex population structure is to estimate admixture proportions based on clustering-based methods (Pritchard et al. 2000; Tang et al. 2005; Alexander et al. 2009; Skotte et al. 2013), such as the popular software ADMIXTURE, which have also been used for correction of population stratification in association studies (Price et al. 2010).
Next-generation sequencing (NGS) methods (Metzker 2010) produce a large amount of DNA sequencing data at low cost and are commonly used in population genetic studies (Nielsen et al. 2012). But NGS methods are associated with high error rates usually caused by several factors such as sampling, alignment, and sequencing errors. Many NGS studies are based on medium (<15×) and low (<5×) depth data due to the demand for large sample sizes as seen in large-scale sequencing studies, e.g., 1000 Genomes Project Consortium (2010, 2012). However, the use of medium-, and, especially, low-depth sequencing data introduces challenges rooted in the statistical uncertainty induced when calling genotypes and variants in these scenarios (Nielsen et al. 2012). The statistical uncertainty increases for low-depth samples due to the increased difficulty of distinguishing a variable site from a sequencing error with the information provided. Problems can arise due to chromosomes being sampled with replacement in the sequencing process, and both alleles may not have been sampled for a heterozygous individual in low-depth scenarios. Homozygous genotypes may also be wrongly inferred as heterozygous due to sequencing errors. Thus, genotype calling will associate individuals with a statistical uncertainty that should be taken into account (Nielsen et al. 2011, 2012).
To overcome these problems related to NGS data and genotype calling, probabilistic methods have been developed to take use of genotype likelihoods in combination with external information for various population genetic parameters (Kim et al. 2011; Nielsen et al. 2012; Fumagalli et al. 2013; Skotte et al. 2013; Vieira et al. 2013; Korneliussen et al. 2014; Kousathanas et al. 2017), such that posterior genotype probabilities can be used to model the related uncertainty. Genotype likelihoods can be estimated to incorporate errors of the sequencing process such as the base quality scores as well as the allele sampling (McKenna et al. 2010). These posterior genotype probabilities have also been used to call genotypes with a higher accuracy than previous methods for low-depth NGS data (Nielsen et al. 2011, 2012).
We present two new methods for low-depth NGS data using genotype likelihoods to model complex population structure that connect the results of PCA with the admixture proportions of clustering-based approaches. One method performs a variant of PCA using an iterative heuristic approach of estimating individual allele frequencies to compute a covariance matrix, while the other uses the estimated individual allele frequencies in an accelerated non-negative matrix factorization (NMF) algorithm to estimate admixture proportions. The performances of the two methods are assessed on both simulated and real datasets in regards to existing methods for both low-depth NGS and genotype data. The methods have been implemented in a framework called PCAngsd (PCA of NGS data).
Materials and Methods
We will analyze NGS data of n diploid individuals across m variable sites. These sites will either be known or called single-nucleotide polymorphisms (SNPs), which are assumed to be diallelic such that the major and minor allele of each SNP have been inferred. This can either be done from sequencing reads (Kim et al. 2011) or from genotype likelihoods (Korneliussen et al. 2014) and only three different genotypes will be possible. Thus, we assume that a genotype G can be seen as a binomial random variable with realizations 0, 1, and 2 that represent the number of copies of the minor allele in a site for a given individual in the absence of population structure. The expectation and variance of G can therefore be defined as and with p representing the allele frequency of a population, which we also refer to as population allele frequency.
However, genotypes are not observed in NGS data and we will instead work on genotype likelihoods that also include information of the sequencing process. The genotype likelihoods are the probability of the observed sequencing data X given the three different possible genotypes, for One method to compute genotype likelihoods from sequencing reads is described in the supplemental material based on the simple GATK model (McKenna et al. 2010).
External information can be incorporated to define posterior genotype probabilities using Bayes’ theorem in combination with genotype likelihoods (Nielsen et al. 2011). The population allele frequency is often used as information in the estimation of prior genotype probability for an individual i in site s (Kim et al. 2011; Nielsen et al. 2012; Fumagalli et al. 2013; Vieira et al. 2013). Assuming the population is in Hardy-Weinberg equilibrium (HWE) for a site s, the prior genotype probability is then given as and for the three different possible genotypes. As defined in Kim et al. (2011), using the estimated population allele frequency the posterior genotype probability is computed as follows for individual i in site s:
| (1) |
PCA
The standard way of performing PCA in population genetics and using it to infer population structure is based on the method defined in Patterson et al. (2006). For a genotype matrix of n individuals and m variable sites, the covariance matrix , also known as the genetic relationship matrix (GRM), is computed as follows for two individuals i and j:
| (2) |
Here, is the observed genotype for individual i in site s, to distinguish it from G defined above for unobserved genotypes, and is the estimated population allele frequency. The principal components are then inferred by performing an eigendecom-position of the covariance matrix, such that with being the matrix of eigenvectors and the diagonal matrix of the corresponding eigenvalues. Principal components and eigenvectors will be used interchangeably throughout this study. The top principal components capture most of the population structure as they represent the projection of the individuals on axes of genetic variation in the dataset (Patterson et al. 2006; Engelhardt and Stephens 2010).
This method has been extended to NGS data in Fumagalli et al. (2013), as well as in Skotte et al. (2012), using the probabilistic framework described in Equation 1, by summing over the genotypes of each individual weighted by the joint posterior genotype probabilities under the assumption of HWE in the whole sample. The method has been implemented in the ngsTools framework (Fumagalli et al. 2014). The covariance matrix is estimated as follows for NGS data using only known variable sites for two individuals i and j:
| (3) |
ngsTools splits up the joint posterior probability, into for by assuming conditional independence between individuals given the estimated population allele frequencies. The nondiagonal entries in the covariance matrix are now directly estimated from the posterior expectations of the genotype instead of the observed genotypes as described in Equation 2. The original method weighs each site by its probability of being a variable site such that SNP calling is not needed prior to the covariance matrix estimation. This is not taken into account in this study as we are using called variable sites to infer population structure. The population allele frequencies are estimated from the genotype likelihoods using an expectation maximization (EM) algorithm (Kim et al. 2011) as described in the supplemental material.
The problem with this approach is that the assumption of conditional independence between individuals given the population allele frequency is only valid when there is no population structure. Here, we propose a novel approach of estimating the covariance matrix using iteratively estimated individual allele frequencies to update the prior information of the posterior genotype probability. Thereby, we condition on the individual allele frequencies as in the clustering-based approaches such as Pritchard et al. (2000), Tang et al. (2005), Alexander et al. (2009), Skotte et al. (2013).
Individual allele frequencies
A model for estimating individual allele frequencies based on population structure was introduced in STRUCTURE (Pritchard et al. 2000), as later described in Equation 13. Hao et al. (2015) proposed a different model for estimating individual allele frequencies by using the information in the principal components instead of having an assumption of K ancestral populations. The model is defined as the matrix product,
| (4) |
where represents the population structure such that represents the mapping of the population structure to the allele frequencies. Hao et al. (2015) estimated the individual allele frequencies through a singular value decomposition (SVD) method, where genotypes are reconstructed using only the top D principal components such that they will be modeled by population structure. A similar approach has been proposed by Conomos et al. (2016), where the inferred principal components are used to estimate individual allele frequencies in a simple linear regression model. However, due to working on NGS data and not knowing the genotypes, we are extending the method of Hao et al. (2015) to NGS data by using posterior expectations of the genotypes, referred to as genotype dosages, instead of genotypes. Thus, we will be using,
| (5) |
for individual i in site s.
The individual allele frequencies are then estimated by performing a SVD on the centered genotype dosages, and reconstructing them using only the top D principal components. is then added to the reconstruction and scaled by based on a binomial distribution assumption of for and to produce the individual allele frequencies. Since SVD is a method that takes real-valued input, we will have to truncate the estimated individual allele frequencies in order to constrain them in the range However, Hao et al. (2015) showed that the resulting estimates were still very accurate for common variants considering this limitation.
For ease of notation, let be the matrix of genotype dosages, for and The following steps for estimating the individual allele frequencies are adopted from the SVD method (Hao et al. 2015) to work on NGS data:
| Algorithm 1: SVD method for estimating individual allele frequencies. |
| 1. The centered genotype dosages are constructed as for |
| 2. Perform SVD on the centered genotype dosages, where will represent population structure similarly to |
| 3. Define to be the prediction of the centered genotype dosages using only the top D principal components, |
| 4. Estimate by adding to row-wise and scaling by based on |
For matrix notations, define and all representing column vectors, such that Equation 4 can be approximated as Finally, is truncated to constrain allele frequency estimates in a range based on a small value γ , such that for and
We now incorporate the individual allele frequencies into the estimation of posterior genotype probabilities. The estimated individual allele frequencies are used as updated prior information instead of the population allele frequencies, and will be able to model missing data with the inferred population structure of the individuals. Thus, the posterior genotype probabilities are estimated as follows for individual i in site s:
| (6) |
Each individual is now seen as a single population with allele frequency where as the prior genotype probability are estimated assuming HWE, such that and An updated definition of the posterior expectations of the genotypes is then given as:
| (7) |
This procedure of updating the prior information can be iterated to estimate new individual allele frequencies on the basis of updated population structure. Therefore, we propose the following algorithm for an iterative procedure of estimating the individual allele frequencies.
| Algorithm 2: Iterative estimation of individual allele frequencies. |
| 1. Estimate population allele frequencies from genotype likelihoods (see supplemental material). |
| 2. Estimate posterior genotype probabilities and genotype dosages based on genotype likelihoods and . |
| 3. Estimate using the SVD based method on as described in Algorithm 1. |
| 4. Estimate posterior genotype probabilities and genotype dosages using updated prior information, |
| 5. Repeat steps 3 and 4 until individual allele frequencies have converged. |
Convergence of our iterative method is defined as when the root-mean-square deviation (RMSD) of the inferred population structure in the SVD is smaller than a value between two successive iterations. The RMSD of iteration for D principal components is given as,
| (8) |
Covariance matrix
We now use the final set of individual allele frequencies to estimate an updated covariance matrix in a similar model as in Equation 3, but incorporating the individual allele frequencies into the joint posterior probability. The entries of the covariance matrix are now defined as follows for individuals i and j:
| (9) |
For the joint posterior probability can be computed as since, in contrast to the assumption made in the model of Fumagalli et al. (2013) using population allele frequencies, the individuals are conditionally independent given the individual allele frequencies. The above equation can be expressed in terms of the genotype dosages for ease of notation and computation for
| (10) |
However, for (diagonal of the covariance matrix), the joint posterior probability is simplified to such that the estimation of the diagonal covariance entries is given as:
| (11) |
An eigendecomposition of the updated estimated covariance matrix is then performed to obtain the principal components as described earlier, Note that and from algorithm 1 are not the same even though they both represent population structure through axes of genetic variation in the dataset. This is due to a different scaling, and the joint posterior probability of Equation 11 is not taken into account in for
Number of principal components
It can be hard to determine the optimal number of principal components that represent population structure. In our method, we are using Velicier’s minimum average partial (MAP) test as proposed by Shriner (2011) to automatically detect the number of top principal components D used for estimating the individual allele frequencies. Shriner showed that the test based on a Tracy-Widom distribution (Patterson et al. 2006) systematically overestimates the number of significant principal components, and performs even worse for datasets including admixed individuals. However, in order to be able to perform the MAP test and detect the optimal D, an initial covariance matrix is estimated based on the model in Equation 3.
The MAP test is performed on the estimated initial covariance matrix for NGS data as an approximation of the Pearson correlation matrix used by Shriner. Using the notation of Shriner, is defined as the matrix of partial correlations after having partialed out the first d principal components. Velicer (1976) proposed the summary statistic where represents the entry in for individuals i and j. Thus, the test statistic represents the average squared correlation after partialing out the top d principal components. The number of top principal components that represent population structure is then chosen as for We have used the same implementation of the MAP test as Shriner.
The MAP test, and the preceding estimation of the initial covariance matrix, can be avoided by having prior knowledge of an optimal D for the dataset being analyzed and manually selecting D.
Genotype calling
As previously shown in Nielsen et al. (2012) and Fumagalli et al. (2013), genotypes can be called from posterior genotype probabilities to achieve higher accuracy in low-depth NGS scenarios. We can adapt this concept to our posterior genotype probabilities based on individual allele frequencies, such that genotypes can be called at a higher accuracy in structured populations from low-depth NGS data. The genotype for individual i in site s is called as follows:
| (12) |
Admixture proportions
Based on the likelihood model defined in STRUCTURE (Pritchard et al. 2000), individual allele frequencies can be estimated using admixture proportions and population-specific allele frequencies (Alexander et al. 2009), such that:
| (13) |
for an individual i in a variable site s. This is based on an assumption of K ancestral populations where and Here and must be inferred in order to estimate the individual allele frequencies, whereas K is assumed to be known. One probabilistic approach for inferring population structure through admixture proportions for low-depth NGS data has been implemented in the NGSadmix software (Skotte et al. 2013). Here both parameters, and are jointly estimated in an EM algorithm using genotype likelihoods.
In our case, we have already estimated the individual allele frequencies based on our iterative procedure using PCA described above. K can be chosen as the number of principal components since it would explain the number of distinct ancestral population from which the individual allele frequencies have been estimated. There is, however, not always a direct interpretation between principal components and admixture proportions (Alexander et al. 2009; Engelhardt and Stephens 2010). Therefore, we propose an approach based on NMF to infer and using only our estimated individual allele frequencies as information for low depth NGS data. NMF has previously been applied directly on genotype data to infer population structure and admixture proportions by Frichot et al. (2014), where their method showed comparable accuracy and faster runtime in comparison to ADMIXTURE.
NMF is a dimension reduction and factor analysis method for finding a low-rank approximation of a matrix, which is similar to PCA, but NMF is constrained to find non-negative low dimensional matrices. For an non-negative matrix the goal of NMF is to find an approximation of based on two non-negative factor matrices and such that:
| (14) |
will consist of columns of non-negative basis vectors such that linear combinations of these approximates through Thus, based on the non-negative nature of our parameters, we can apply the ideas of NMF to infer admixture proportions and population-specific allele frequencies from our individual allele frequencies. We use a combination of recent research in NMF to minimize the following least squares problem with a sparseness constraint on
| (15) |
for and Here is the Frobenius norm of a matrix and α is the regularization parameter controlling the sparseness enforced as also introduced in Frichot et al. (2014).
Lee and Seung (1999, 2001) proposed an multiplicative update (MU) algorithm to solve the standard NMF problem without the sparseness constraint included above. Their update rules can be seen as conservative steps in a gradient descent optimization problem for updating and which ensure that the non-negative constraint holds for each update. Hoyer (2002) extended the MU to incorporate the sparseness constraint described in Equation 15 for For the regularization parameter is used to reduce noise, especially induced by the uncertainty of low-depth NGS data, in the estimated admixture proportions by enforcing sparseness in the solution. An iteration of using the MU rules is then described as follows:
| (16) |
| (17) |
where represents element-wise multiplication, and the division operator is element-wise as well.
However, MU has been shown to have a slow convergence rate, especially for dense matrices, and our approach is therefore to accelerate MU by combining two different techniques. We propose an algorithm of combining the acceleration scheme described by Gillis and Glineur (2012) with the asymmetric stochastic gradient descent algorithm (ASG-MU) of Serizel et al. (2016) for updating and in a fast approach. The acceleration scheme of Gillis and Glineur (2012) updates each matrix and a fixed number of times at a lower computational cost without losing the convergence properties of MU. We simply incorporate this acceleration scheme inside ASG-MU that works by randomly assigning the columns of into a set of B mini-batches, which are then updated sequentially in a permuted order to improve the convergence rate and performance of MU (Serizel et al. 2016). After each update, we truncate the entries of both and to be in range and normalize the rows of to sum to one. The concept of combining an acceleration scheme with a stochastic gradient descent approach for MU has also been explored in Kasai (2017).
The algorithm is iterated until the admixture proportions has converged. Convergence is defined as when the RMSD of estimated admixture proportions of two successive iterations are smaller than a value φ (). The RMSD of iteration is given as,
| (18) |
The α parameter enforcing sparseness in the estimated solution of is arbitrarily specified. However the use of the likelihood measure in the NGSdamix (Skotte et al. 2013) model can be used to determine the α parameter fitting the dataset. The likelihood measure is defined as:
| (19) |
where Based on the fast estimation of admixture proportions using our NMF algorithm, an appropriate α can easily be found by scanning a specified interval in an automated fashion based on the likelihood measure. This can be performed without sacrificing significant runtime compared to NGSadmix due to already having estimated the individual allele frequencies for a particular K.
Implementation
Both presented methods have been implemented in a Python framework named PCAngsd. The framework is freely available at http://www.popgen.dk/software/.
The memory requirements of PCAngsd is as the entire matrix of genotype likelihoods needs to be stored in memory for both methods. The most computationally expensive step is the estimation of individual allele frequencies and covariance matrix . However, a fast SVD method for only computing the top D eigenvectors, implemented in the Scipy library (Jones et al. 2014) using ARPACK (Lehoucq et al. 1998) as an eigensolver, has been used to speed up the iterative estimations of the individual allele frequencies. PCAngsd is also multithreaded to take advantage of several cores, and the backbone of the framework is based on Numpy data structures (van der Walt et al. 2011) using the Numba library (Lam et al. 2015) to speed up bottlenecks with just-in-time (JIT) compilation.
Simple simulation of genotypes and sequencing data
To test the capabilities of our two presented methods, we simulated low-depth NGS data and generated genotype likelihoods. Allele frequencies of the reference panel of the Human Genome Diversity Project (HGDP) (Cann et al. 2002) were used to generate a total of 380 individuals from three distinct populations (French, Han Chinese, Yoruba) including admixed individuals in ∼0.4 million SNPs across all autosomes. As the allele frequencies are known for each population, the genotypes of each individual can be sampled from a binomial distribution for each diallelic SNP, using the population-specific allele frequency or an admixed allele frequency as parameter. No linkage disequilibrium (LD) was simulated. The genotypes are therefore known and are used in the evaluation of our methods in our low-depth scenarios. The number of reads in each SNP were sampled from a Poisson distribution with a mean parameter resembling the average sequencing depth of the individual, and the genotype was used to sample the number of derived alleles from a binomial distribution using the sampled depth as parameter. The average sequencing depth of each individual was sampled uniformly random from a range of Sequencing errors were incorporated by sampling each read with a probability of being an error. The genotype likelihoods were then finally generated from the probability mass function of a binomial distribution using the sampled parameters and ϵ. This approach of genotype likelihood simulation has previously been used in Kim et al. (2011), Skotte et al. (2013), and Vieira et al. (2013).
A complex admixture scenario was constructed to test the capabilities of our methods; 100 individuals were sampled directly from each of the population-specific allele frequencies (nonadmixed), while 50 individuals were sampled to have equal ancestry from each of the three distinct populations (three-way admixture). Finally, 30 individuals were sampled from a gradient of ancestry between all pairs of the ancestral populations (two-way admixture).
1000 Genomes low-depth sequencing data
We also analyzed human low-coverage NGS data of 193 individuals from the 1000 Genomes Project Consortium et al. (2010, 2012). The individuals were from four different populations consisting of 41 from CEU (Utah residents with Northern and Western European ancestry), 40 from CHB (Han Chinese in Beijing), 48 from YRI (Yoruba in Ibadan), and 64 individuals from MXL (Mexican ancestry in Los Angeles), representing an admixed scenario of European and Native American ancestry. The individuals from the low-coverage datasets have a varying sequencing depth from 1.5× to 12.5× after site filtering. An advantage of using the low-coverage data of the 1000 Genomes Project data are that reliable genotypes are available that can be used for validation purposes.
SNP calling and estimation of genotype likelihoods of the 1000 Genomes dataset was performed in ANGSD (Korneliussen et al. 2014) using simple read quality filters. A significance threshold of was used for SNP calling alongside a MAF threshold of 0.05 to remove rare variants. A total number of 8 million variable sites across all autosomes was used in the analyses. The full ANGSD command used to generate the genotype likelihoods is provided in the supplemental material.
Waterbuck low-depth sequencing data
Lastly, an animal dataset (nonmodel organism) as also included in our study. A reduced low-depth NGS dataset of the waterbuck (Kobus ellipsiprymnus) originating from C. Pedersen et al. (University of Copenhagen, unpublished data) was analyzed. The dataset consists of 73 samples that were sampled at five different sites in Africa with a varying sequencing depth from 2.2× to 4.7× aligned to 88,935 scaffolds. The dataset was reduced to only include sampling sites with >10 samples such that the inferred axes of genetic variation will reflect true population structure. As performed for the 1000 Genomes dataset, genotype likelihoods were estimated in ANGSD with the same SNP and MAF filters. A total number of 9.4 million SNPs across the autosomes of the waterbuck was analyzed in this study.
Data availability
The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. The waterbuck dataset analyzed in our study is publicly available in the European Nucleotide Archive (ENA) repository (PRJEB28089). Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6953243.
Results
For the simulated and 1000 Genomes datasets, results estimated in PCAngsd on low-depth NGS data were evaluated against the results estimated from genotype data, as well as naively called genotypes from genotype likelihoods. The model in Equation 2 was used to perform PCA, while ADMIXTURE was used to estimate admixture proportions on the “true” genotype datasets. The performance of PCAngsd was also compared to existing genotype likelihood methods, with the ngsTools model (Equation 3) for performing PCA, and NGSadmix (Equation 19) for estimating admixture proportions. In all the following cases of admixture plots estimated by PCAngsd, we used and α was chosen as the one maximizing the likelihood measure described above (Equation 19), also shown in Supplemental Material, Figure S5.
RMSD was used to evaluate the performances of both NGS methods for estimating admixture proportions in terms of accuracy:
| (20) |
where and represent the estimated admixture proportion for individual i in ancestral population k from known genotypes and NGS data, respectively. The accuracy of the inferred PCA plots of both NGS methods was also compared to the PCA plots of known genotypes for the simulated and 1000 Genomes datasets using RMSD. However, a Procrustes analysis (Wang et al. 2010; Fumagalli et al. 2013) had to be performed prior to the comparison as the direction of the principal components can differ based on the eigendecomposition of the covariance matrices.
All tests in this study were performed server-side using 32 threads (Intel Xeon CPU E5-2690) for both PCAngsd and NGSadmix.
Simulation
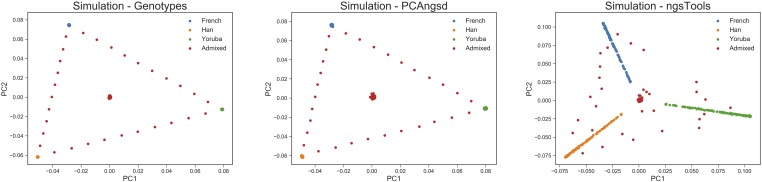
The results of performing PCA on the simulated dataset based on frequencies from three human populations are displayed in Figure 1, where we simulated unadmixed, two-way admixed and three-way admixed individuals. The MAP test reported two significant principal components, which was also expected for individuals simulated from three distinct populations. The inferred principal components clearly show the importance of taking individual allele frequencies into account in the probabilistic framework. Here, PCAngsd was able to infer the population structure of individuals from distinct populations and admixed individuals nicely, as also verified by a Procrustes analysis obtaining a RMSD of 0.00121, when compared to the PCA inferred from the true genotypes. There is clear bias in the results of the ngsTools model, where the patterns represent sequencing depth rather than population structure, as seen in Figure S1. The individuals are acting as a gradient toward the origin due to their varying sequencing depth. The biased performance of ngsTools was also reflected in the corresponding Procrustes analysis, with a RMSD of 0.0174.
Figure 1.
PCA plots of the top two principal components in the simulated dataset consisting of 380 individuals and 0.4 million variable sites. The left-hand plot shows the PCA performed on the known genotypes using Equation 2. The middle plot shows the PCA performed by PCAngsd, and the right-hand plot displays the PCA performed by the ngsTools model (Equation 3).
To ensure that the individual allele frequencies estimated using PCAngsd are representative estimates, we compared them to the allele frequencies of the HGDP reference panel from which the genotypes of each individual has been sampled. Sampling errors were therefore not taken into account in the comparison. The estimates obtained from NGSadmix were also compared. The estimates of PCAngsd obtain a RMSD value of 0.0330, and the estimates of NGSadmix a value of 0.0327 based on low-depth NGS data. The results of PCAngsd are displayed in Figure S9.
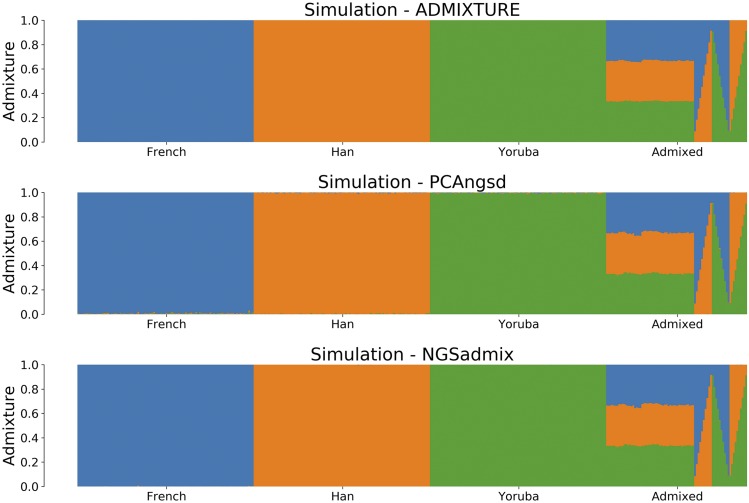
The estimated admixture proportions of the simulated dataset are displayed in Figure 2. PCAngsd estimated the admixture proportions well with a RMSD of 0.00476 compared to the ADMIXTURE estimates of the known genotypes, but was, however, outperformed by NGSadmix with a RMSD of 0.00184. For the 380 individuals and 0.4 million SNPs using PCAngsd had an average runtime of only 2.9 min while NGSadmix had an average runtime of 7.9 min (Table 1).
Figure 2.
Admixture plots for of the simulated dataset where each bar represents a single individual and the different colors reflect each of the K components. The first plot is the admixture proportions estimated in ADMIXTURE using the known genotypes, which we use as the ground-truth in our simulation studies. The second plot shows admixture proportions estimated using PCAngsd with parameter and the bottom plot using NGSadmix.
Table 1. Average runtimes of 10 initializations for both PCAngsd and NGSadmix.
| Dataset | PCAngsd | NGSadmix (min) | Depth (×) | |||
|---|---|---|---|---|---|---|
| Simulated | 380 | 0.4 million | 3 | 2.9 min (2.1 min) | 7.9 | |
| 1000 Genomes | 193 | 8 million | 4 | 27.3 min (19.5 min) | 424.9 | |
| Waterbuck | 73 | 9.4 million | 5 | 14.5 min (9.3 min) | 192 |
The runtimes reported for PCAngsd include reading of data and estimation of covariance matrix and admixture proportions, while runtimes listed in parentheses only include estimation of admixture proportions, when parsing previously estimated individual allele frequencies. All tests have been performed server-side using 32 threads.
1000 Genomes
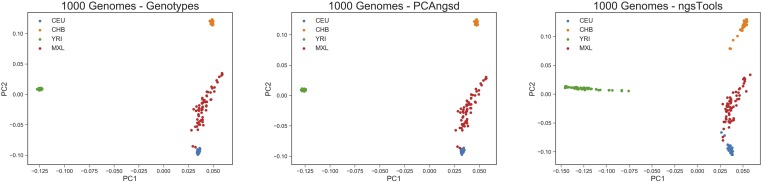
We also applied the methods of PCAngsd to the CEU (European ancestry), CHB (Chinese ancestry), YRI (Nigerian ancestry), and MXL (Mexican ancestry) populations of the low-coverage 1000 Genomes dataset. The MAP test indicated evidence of three significant principal components, meaning that the Native American ancestry explains enough genetic variance in the dataset to represent an axis of its own. The results of the PCA are displayed in Figure 3. As was also seen for the simulated dataset, PCAngsd is able to cluster all individuals almost perfectly, while the ngsTools model is only able to capture some of the same population structure patterns with some of the populations looking admixed. Its results are still biased by the variable sequencing depth, as also seen in Figure S2. The RMSD values of the Procrustes analyses verify the observations, where PCAngsd has a RMSD of 0.00182 compared to ngsTools with a RMSD of 0.0075.
Figure 3.
PCA plots of the top two principal components for the 1000 Genomes dataset with 193 individuals and 8 million variable sites. The left-hand plot is based on the reliable genotypes of the overlapping variable sites in the low depth NGS data, the middle plot is performed by PCAngsd and the right-hand plot is performed by the ngsTools model.
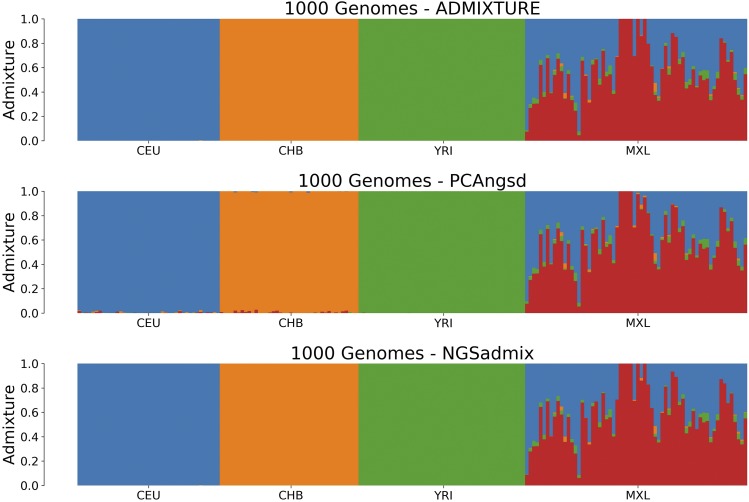
The admixture plots are displayed in Figure 4. was is not able to outperform NGSadmix in terms of accuracy; however, it as still able to estimate a very similar result. PCAngsd has some issues with noise in its estimation, but is, however, able to reduce it with the use of the sparseness parameter, The likelihood measure in Equation 19 was used to easily find an optimal α, as seen in Figure S10. PCAngsd estimates the admixture proportions with a RMSD of 0.0108 compared to NGSadmix with a RMSD of 0.007148. The average runtime for 193 individuals and 8 million SNPs using was 27.3 min, for PCAngsd, and 7.1 hr for NGSadmix, making PCAngsd >15× faster than NGSadmix while both performing PCA and estimating admixture proportions.
Figure 4.
Admixture plots for of the 1000 Genomes dataset, where each bar represents a single individual and the different colors reflect each of the K components. The first plot is the admixture proportions estimated in ADMIXTURE using the reliable genotypes, the second plot shows admixture proportions estimated in PCAngsd with parameter and the last plot is the admixture proportions estimated in NGSadmix.
Waterbuck
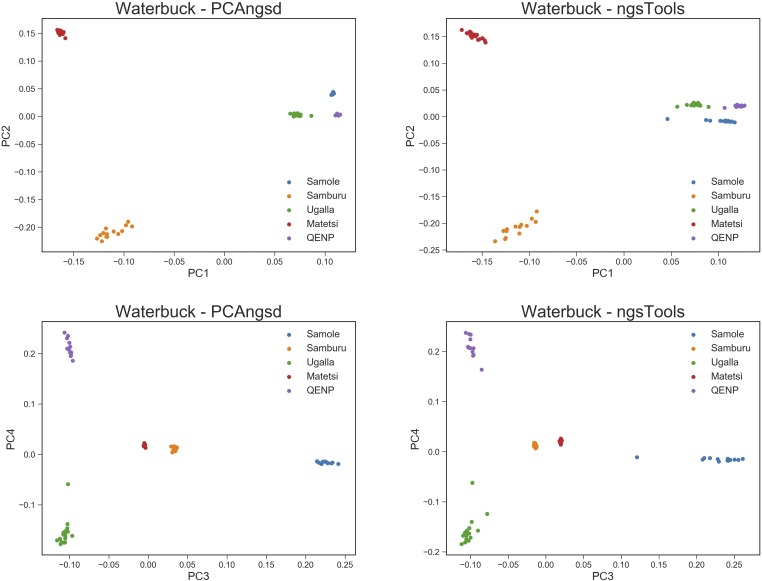
Lastly, we analyzed the low-depth whole genome sequencing waterbuck dataset consisting of 73 individuals from five localities. The MAP test reported four significant principal components explaining the genetic variation in the dataset, which also fits with having five distinct waterbuck sampling sites. The PCA plots are visualized in Figure 5, where the top four principal components for each method are plotted. Once again, PCAngsd is able to cluster the populations much better than the ngsTools model; however, the effect is not as apparent as for the other datasets. Interestingly, populations can switch positions between the two methods, as seen with Samole on the second principal component, and Samburu and Matetsi on the third principal component.
Figure 5.
PCA plots of the top four principal components for the waterbuck dataset with 73 individuals and 9.4 million variable sites. The first row displays the plots of the first and second principal components for PCAngsd and the ngsTools model, respectively, while the second row displays the plots of the third and fourth principal components.
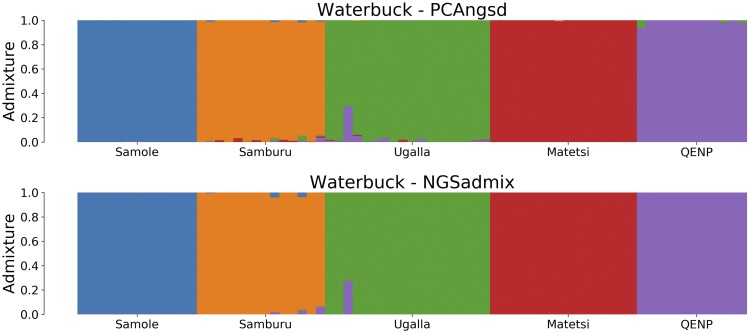
As a few clusters are not so well defined, they will affect the admixture plots seen in Figure 6, where the increased level of noise is hard to remove without also affecting the true ancestry signals. Still, PCAngsd is capturing the same ancestry signals as NGSadmix with the use of the sparseness parameter. It is worth noting that an admixed individual of Ugalla and QENP was captured in both PCA and admixture estimation of PCAngsd, as also verified by the NGSadmix method. The runtime for the waterbuck dataset consisting of 73 samples and 9.4 million SNPs using was an average of 14.5 min for PCAngsd, while NGSadmix had an average runtime of 3.2 hr, thus making PCAngsd >13× faster.
Figure 6.
Admixture plots for of the waterbuck dataset where each bar represents a single individual and the different colors reflect each of the K components. The first plot is the admixture proportions estimated in PCAngsd with parameter and the second plot shows the admixture proportions estimated in NGSadmix.
Naively called genotypes
We lso inferred population structure from naively called genotypes of the simulated and 1000 Genomes datasets, and the results are visualized in Figures S7 and S8. Genotypes were called by choosing the genotypes with the highest genotype likelihoods. No filters were applied in the genotype calling, since Skotte et al. (2013) showed that naively called genotypes had higher accuracy of inferred admixture proportion when no filters were used. The Procrustes analyses report RMSD values of 0.0123 and 0.00310 for performing PCA on the simulated and the 1000 Genomes dataset, respectively (cf. RMSD values of 0.00121 and 0.00182 using PCAngsd). Here, the naively called genotypes performed slightly better than ngsTools in both cases, but the results were still biased by sequencing depth. ADMIXTURE estimates admixture proportions from the called genotypes, with RMSD values of 0.00995 and 0.00865 for the two datasets, respectively, thus performing slightly better than PCAngsd for the 1000 Genomes dataset.
Discussion
We have presented two methods for inferring population structure and admixture proportions in low-depth NGS data, and both methods have been implemented in a framework named PCAngsd. We developed a method to iteratively estimate individual allele frequencies based on PCA using genotype likelihoods in a heuristic approach. We connected principal components to admixture proportions such that we are able to infer and estimate both in a very fast approach, making it feasible to analyze large datasets.
Based on the results when inferring population structure using PCA, it is clear that the increased uncertainty of low-depth sequencing data biases the clustering of populations using the ngsTools model, which also takes genotype uncertainty into account. Contrary to PCAngsd, population structure is not taken into account when using the posterior genotype probabilities to estimate the covariance matrix. The ngsTools model uses population allele frequencies as prior information for all individuals, such that individuals are assumed to be sampled from a homogeneous population. This assumption is, of course, violated when individuals are sampled from structured populations with diverse ancestries. Missing data are therefore modeled by population allele frequencies that resemble an average across the entire sample, which is similar to setting standardized genotypes to 0 in the estimation of the covariance matrix for genotype data. As an effect of this, the low-depth individuals are modeled by sequencing depth instead of population structure. These results may lead to misinterpretations of population structure or admixture only due to low and variable sequencing depth. But the bias is not seen for individuals with equal sequencing depth, as shown in Figure S4 for the ngsTools model. Here, all individuals have been simulated with an average sequencing depth of 2.5×, such that individuals will inherit approximately the same amount of missing data. However, PCAngsd is able to overcome the observed bias of low and variable sequencing depth by using individual allele frequencies as prior information, which leads to more accurate results in all datasets of the study, as missing data are modeled accounting for inferred population structure. The assumption of conditional independence between individuals in the estimation of the covariance matrix (Equation 10) also holds for structured populations by conditioning on individual allele frequencies.
The number of significant eigenvectors used in the estimation of individual allele frequencies is determined by the MAP test. The MAP test is performed on the covariance matrix estimated from the ngsTools model. Thus, in cases of complex population structure, and low and variable sequencing depth, it is possible that the MAP test will not find a suitable number of significant eigenvectors to represent the genetic variation of the dataset. It could, therefore, be more relevant to use prior information regarding the number of eigenvectors needed for the dataset instead. However, for each of the cases analyzed in this study, the MAP test inferred the expected number of significant eigenvectors to describe the population structure.
PCAngsd is able to approximate the results of NGSadmix to a high degree when estimating admixture proportions using solely the estimated individual allele frequencies. However, although PCAngsd is not able to outperform NGSadmix in terms of accuracy, it is able to capture the exact same ancestry patterns as the clustering-based methods in a much faster approach, as shown by the runtimes of each method. Another advantage of PCAngsd is that the estimated individual allele frequencies need to be computed only once for a specific K, thus multiple different random seeds can be tested in the same run for an even greater speed advantage over NGSadmix, as the iterative estimation of individual allele frequencies is the most computational expensive step in PCAngsd. A proper α value, controlling the sparseness enforced in the estimated admixture proportions, can also be found through an automated scan implemented in our framework based on the likelihood measure of NGSadmix. PCAngsd is therefore an appealing alternative for estimating admixture proportions for low-depth NGS data as convergence and runtime can be a problem for a large number of parameters in NGSadmix. PCAngsd was only seen to converge to a single solution for all our practical tests, where we used five batches for all analyses ().
Both methods of the PCAngsd framework rely on an representative set of individual allele frequencies, which we model using the inferred principal components of the SVD on the genotype dosages. The number of individuals representing each population or subpopulation is essential for inferring principal components that describe true population structure, as each individual will contribute to the construction of these axes of genetic variation. This particular effect can be seen in the PCA results of the waterbuck dataset where the populations are described only by a low number of individuals, such that some of the clusters are not as well defined as for the other datasets. The admixture proportions estimated from the waterbuck dataset are therefore affected as well, which can be seen by the additional noise in the admixture plots.
The PCAngsd framework may be able to push the lower boundaries of sequencing depth required to perform population genetic analyses on NGS data in large-scale genetic studies. This is also demonstrated by downsampling the 1000 Genomes dataset in Figures S5 and S6, which display the robustness of PCAngsd in fairly low sequencing depth. However when down-sampling to only 1% of the reads, the PCA and admixture results become very noisy. PCAngsd also demonstrates an effective approach for dealing with merged datasets of various sequencing depths, as missing data will be modeled by population structure. Further, the estimated individual allele frequencies open up the development and extension of population genetic models based on a similar probabilistic framework, such that population structure can be taken into account in heterogeneous populations.
Acknowledgments
This project was funded by the Lundbeck foundation (R215-2015-4174).
Footnotes
Communicating editor: J. Novembre
Literature Cited
- Alexander D. H., Novembre J., Lange K., 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19: 1655–1664. 10.1101/gr.094052.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann H. M., De Toma C., Cazes L., Legrand M.-F., Morel V., et al. , 2002. A human genome diversity cell line panel. Science 296: 261–262. 10.1126/science.296.5566.261b [DOI] [PubMed] [Google Scholar]
- Conomos M. P., Reiner A. P., Weir B. S., Thornton T. A., 2016. Model-free estimation of recent genetic relatedness. Am. J. Hum. Genet. 98: 127–148. 10.1016/j.ajhg.2015.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt B. E., Stephens M., 2010. Analysis of population structure: a unifying framework and novel methods based on sparse factor analysis. PLoS Genet. 6: e1001117 10.1371/journal.pgen.1001117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frichot E., Mathieu F., Trouillon T., Bouchard G., François O., 2014. Fast and efficient estimation of individual ancestry coefficients. Genetics 196: 973–983. 10.1534/genetics.113.160572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli M., Vieira F. G., Korneliussen T. S., Linderoth T., Huerta-Sánchez E., et al. , 2013. Quantifying population genetic differentiation from next-generation sequencing data. Genetics 195: 979–992. 10.1534/genetics.113.154740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli M., Vieira F. G., Linderoth T., Nielsen R., 2014ngstools: methods for population genetics analyses from next-generation sequencing data. Bioinformatics 30: 1486–1487. 10.1093/bioinformatics/btu041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinsky K. J., Bhatia G., Loh P.-R., Georgiev S., Mukherjee S., et al. , 2016. Fast principal-component analysis reveals convergent evolution of adh1b in Europe and East Asia. Am. J. Hum. Genet. 98: 456–472. 10.1016/j.ajhg.2015.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium, . G. R. Abecasis, Altshuler D., Auton A., Brooks L. D., et al. , 2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073. 10.1038/nature09534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium, . G. R. Abecasis, Auton A., Brooks L. D., DePristo M. A., et al. , 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65. 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis N., Glineur F., 2012. Accelerated multiplicative updates and hierarchical ALS algorithms for nonnegative matrix factorization. Neural Comput. 24: 1085–1105. 10.1162/NECO_a_00256 [DOI] [PubMed] [Google Scholar]
- Hao W., Song M., Storey J. D., 2015. Probabilistic models of genetic variation in structured populations applied to global human studies. Bioinformatics 32: 713–721. 10.1093/bioinformatics/btv641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer P. O., 2002. Non-negative sparse coding, pp. 557–565 in Proceedings of the 2002 12th IEEE Workshop on Neural Networks for Signal Processing IEEE, Martigny, Switzerland. [Google Scholar]
- Jones, E., T. Oliphant, P. Peterson et al., 2014 SciPy: Open Source Scientific Tools for Python, 2001–, http://www.scipy.org/
- Kasai, H., 2017 Stochastic variance reduced multiplicative update for nonnegative matrix factorization. arXiv:1710.10781.
- Kim S. Y., Lohmueller K. E., Albrechtsen A., Li Y., Korneliussen T., et al. , 2011. Estimation of allele frequency and association mapping using next-generation sequencing data. BMC Bioinformatics 12: 231 10.1186/1471-2105-12-231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneliussen T. S., Albrechtsen A., Nielsen R., 2014. Angsd: analysis of next generation sequencing data. BMC Bioinformatics 15: 356 10.1186/s12859-014-0356-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousathanas A., Leuenberger C., Link V., Sell C., Burger J., et al. , 2017. Inferring heterozygosity from ancient and low coverage genomes. Genetics 205: 317–332. 10.1534/genetics.116.189985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S. K., Pitrou A., Seibert S., 2015. Numba: a llvm-based python jit compiler, pp. 7 in Proceedings of the Second Workshop on the LLVM Compiler Infrastructure in HPC ACM, New York. [Google Scholar]
- Lee D. D., Seung H. S., 1999. Learning the parts of objects by non-negative matrix factorization. Nature 401: 788–791. 10.1038/44565 [DOI] [PubMed] [Google Scholar]
- Lee D. D., Seung H. S., 2001. Algorithms for non-negative matrix factorization, pp. 556–562 in Advances in Neural Information Processing Systems, edited by Leen T. K., Dietterich T. G., Tresp V. MIT Press, Cambridge, MA. [Google Scholar]
- Lehoucq R. B., Sorensen D. C., Yang C., 1998. ARPACK Users’ Guide: Solution of Large-Scale Eigenvalue Problems with Implicitly Restarted Arnoldi Methods, Vol. 6 Siam, Philadelphia: 10.1137/1.9780898719628 [DOI] [Google Scholar]
- Luu K., Bazin E., Blum M. G., 2017. pcadapt: an R package to perform genome scans for selection based on principal component analysis. Mol. Ecol. Resour. 17: 67–77. 10.1111/1755-0998.12592 [DOI] [PubMed] [Google Scholar]
- Marchini J., Cardon L. R., Phillips M. S., Donnelly P., 2004The effects of human population structure on large genetic association studies. Nat. Genet. 36: 512–517. 10.1038/ng1337 [DOI] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., et al. , 2010. The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menozzi P., Piazza A., Cavalli-Sforza L., 1978. Synthetic maps of human gene frequencies in Europeans. Science 201: 786–792. 10.1126/science.356262 [DOI] [PubMed] [Google Scholar]
- Metzker M. L., 2010. Sequencing technologies–the next generation. Nat. Rev. Genet. 11: 31–46. 10.1038/nrg2626 [DOI] [PubMed] [Google Scholar]
- Nielsen R., Paul J. S., Albrechtsen A., Song Y. S., 2011. Genotype and SNP calling from next-generation sequencing data. Nat. Rev. Genet. 12: 443–451. 10.1038/nrg2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R., Korneliussen T., Albrechtsen A., Li Y., Wang J., 2012. SNP calling, genotype calling, and sample allele frequency estimation from new-generation sequencing data. PLoS One 7: e37558 10.1371/journal.pone.0037558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre J., Stephens M., 2008. Interpreting principal component analyses of spatial population genetic variation. Nat. Genet. 40: 646–649. 10.1038/ng.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N., Price A. L., Reich D., 2006. Population structure and eigenanalysis. PLoS Genet. 2: e190 10.1371/journal.pgen.0020190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A. L., Patterson N. J., Plenge R. M., Weinblatt M. E., Shadick N. A., et al. , 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38: 904–909. 10.1038/ng1847 [DOI] [PubMed] [Google Scholar]
- Price A. L., Zaitlen N. A., Reich D., Patterson N., 2010. New approaches to population stratification in genome-wide association studies. Nat. Rev. Genet. 11: 459–463. 10.1038/nrg2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M., Donnelly P., 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizel R., Essid S., Richard G., 2016. Mini-batch stochastic approaches for accelerated multiplicative updates in nonnegative matrix factorisation with beta-divergence, pp. 1–6 in IEEE 26th International Workshop on Machine Learning for Signal Processing (MLSP) IEEE, Piscataway, NJ. [Google Scholar]
- Shriner D., 2011. Investigating population stratification and admixture using eigenanalysis of dense genotypes. Heredity 107: 413–420. 10.1038/hdy.2011.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skotte L., Korneliussen T. S., Albrechtsen A., 2012. Association testing for next-generation sequencing data using score statistics. Genet. Epidemiol. 36: 430–437. 10.1002/gepi.21636 [DOI] [PubMed] [Google Scholar]
- Skotte L., Korneliussen T. S., Albrechtsen A., 2013. Estimating individual admixture proportions from next generation sequencing data. Genetics 195: 693–702. 10.1534/genetics.113.154138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Peng J., Wang P., Risch N. J., 2005. Estimation of individual admixture: analytical and study design considerations. Genet. Epidemiol. 28: 289–301. 10.1002/gepi.20064 [DOI] [PubMed] [Google Scholar]
- van der Walt S., Colbert S. C., Varoquaux G., 2011. The NumPy array: a structure for efficient numerical computation. Comput. Sci. Eng. 13: 22–30. 10.1109/MCSE.2011.37 [DOI] [Google Scholar]
- Velicer W. F., 1976. Determining the number of components from the matrix of partial correlations. Psychometrika 41: 321–327. 10.1007/BF02293557 [DOI] [Google Scholar]
- Vieira F. G., Fumagalli M., Albrechtsen A., Nielsen R., 2013Estimating inbreeding coefficients from NGS data: impact on genotype calling and allele frequency estimation. Genome Res. 23: 1852–1861. 10.1101/gr.157388.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Szpiech Z. A., Degnan J. H., Jakobsson M., Pemberton T. J., et al. , 2010. Comparing spatial maps of human population-genetic variation using procrustes analysis. Stat. Appl. Genet. Mol. Biol. 9: 13 10.2202/1544-6115.1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. The waterbuck dataset analyzed in our study is publicly available in the European Nucleotide Archive (ENA) repository (PRJEB28089). Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6953243.