Abstract
Despite their potent antimicrobial activity, the usefulness of antimicrobial peptides (AMPs) as antibiotics has been limited by their toxicity to eukaryotic cells and a lack of stability in vivo. In the present study we examined the effects of introducing D-lysine residues into a 15-residue hybrid AMP containing residues 1–7 of cecropin A and residues 2–9 of melittin (designated CM15). Diastereomeric analogs of CM15 containing between two and five D-lysine substitutions were evaluated for their antimicrobial activity, lysis of human erythrocytes, toxicity to murine macrophages, ability to disrupt cell membranes, and protease stability. All of the analogs caused rapid permeabilization of the Staphylococcus aureus cell envelope, as indicated by uptake of SYTOX green. CM15 also permeabilized the plasma membrane of RAW264.7 macrophages, but this was substantially diminished for the D-lysine containing analogs. The introduction of D-lysine caused moderate decreases in antimicrobial activity for all analogs studied. However, D-Lys substitution produced a much more pronounced reduction in toxicity to eukaryotic cells, leading to marked improvements in antimicrobial efficacy for some analogs. Circular dichroism studies indicated a progressive loss of helical secondary structure upon introduction of D-lysine residues, and there was a good correspondence between helical content and eukaryotic cell cytotoxicity. Overall, these studies show that the biological activity of CM15 analogs containing D-lysine depends on both the number and position of D-Lys substitutions, and that such substitutions can dramatically lower toxicity to eukaryotic cells with only minimal decreases in antimicrobial activity.
The extensive use of classical antibiotics over the past sixty years has led to an increased prevalence of antibiotic resistance in both hospital- and community-acquired infections (1–6), giving rise to a critical need for the development of new antibiotics. In recent years there has been considerable interest in the development of antimicrobial peptides (AMPs) as novel antibiotics (7–11). These peptides occur naturally within all kingdoms of life, from humans to prokaryotes, and constitute an important part of the innate immune response. AMPs are typically 12–50 amino acids in length, contain from 2–9 positively charged residues, and adopt amphipathic secondary structures upon binding to membranes (9, 12–14). Their positive charge allows AMPs to interact more strongly with the highly negatively-charged membranes of bacteria as opposed to the nearly neutral plasma membranes of eukaryotic cells (9, 10, 12–17), and their amphipathic secondary structure facilitates partitioning into the membrane bilayer (18). For most AMPs, their mechanism of cell killing is believed to involve disruption of the membrane permeability barrier, either through the formation of discrete pores, detergent-like action, or perturbation of lipid packing (13, 19–21). AMPs exhibit broad specificity against both Gram-positive and Gram-negative bacteria, including strains that are multi-drug resistant. Importantly, because modification of the cell membrane is energetically costly (22), AMPs do not readily elicit the development of resistance.
Despite their potent antimicrobial activity, the usefulness of AMPs as antibiotics has been limited by their toxicity to eukaryotic cells and a lack of stability in vivo. To overcome issues with toxicity and stability there have been many approaches to peptide modification, including disruption of secondary structure (16, 23, 24), introduction of β- or D-amino acids (12, 23, 25–31), and increasing or decreasing peptide charge and/or hydrophobicity (12, 32–34). In particular, the introduction of D-amino acids is an approach that allows disruption of secondary structure while conserving overall charge and hydrophobicity. Several peptides containing a mixture of D- and L-amino acids have been shown to maintain antibacterial activity while having decreased toxicity to eukaryotic cells (12, 23, 27).
Of the many known natural and designed AMPs (34, 35), synthetic hybrids of the insect peptide cecropin and the bee venom peptide mellitin are among the most potent and well-studied (14, 28, 36–40). In particular, a hybrid containing residues 1–7 of cecropin and residues 2–9 of melittin (designated CM15) was the smallest construct found to maintain the antimicrobial activity of cecropin while decreasing the hemolytic effects of melittin (40). CM15, in which five of the fifteen residues are lysine, forms α-helical secondary structure upon binding to membranes (19, 28, 40–43) and exhibits potent, broad-spectrum antimicrobial activity (28, 32, 40). CM15 intercalates into lipid bilayers that mimic the bacterial plasma membrane and disrupts membrane structure (19, 32, 41, 42). Because of its small size, CM15 is readily amenable to manipulation of its structure and composition. In these studies we examined the effects of modifying CM15 by substitution of D-lysine for L-lysine. It was anticipated that these modifications would disrupt the secondary structure of the peptide, in this case lowering α-helical content, without changing the overall charge or ratio of hydrophilic to hydrophobic amino acids. Analogs of CM15 containing between two and five D-lysine substitutions were evaluated for their antimicrobial activity, toxicity towards human erythrocytes and murine macrophages, ability to permeabilize cell membranes, and protease stability. We find that changes in the biological activity of CM15 depend on both the number and position of D-lysine substitutions, and that such D-amino acid substitutions can dramatically lower toxicity to eukaryotic cells with only minimal decreases in antimicrobial activity.
Materials and Methods
Materials
Human serum albumin (HSA), Coomassie Blue G250, Triton X100, Sodium pyruvate, Penicillin/Streptomycin, 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), trypsin, and chymotrypsin were obtained from Sigma-Aldrich (St. Louis, MO). LB agar was obtained from Fischer Scientific (Pittsburgh, PA). Tris/Tricine gels were obtained from BioRad. Sytox SYTOX green, fetal bovine serum (FBS), and DMEM (12100-046) were obtained from Invitrogen and Mueller-Hinton broth (MHB) was obtained from Difco.
Peptide synthesis and purification
Peptides with amidated C-termini and acetylated N-termini were obtained commercially (Biosynthesis Inc., Lewisville, TX) or were synthesized on Rink amide MBHA resin using Fmoc (n-(9-fluorenyl)methoxycarbonyl) chemistry as previously described (41) on a CEM Discover microwave-assisted peptide synthesizer (CEM Biosciences, Matthews, NC). Following precipitation of crude peptides with ice cold diethyl ether, the peptides were resuspended in water and purified by reverse phase HPLC on a 10-μm, 1.0 × 25 cm C8 column (Vydac, Hesperia, CA) using a linear gradient of 20 – 80% acetonitrile in water/0.1% TFA over 40 min (41). Purified peptides were lyophilized and stored under vacuum until use. Peptide purity was assessed by analytical reverse-phase HPLC, and molecular mass was verified by MALDI-TOF mass spectrometry at the Protein and Nucleic Acid Shared Facility of the Medical College of Wisconsin. All peptides used in this study were > 95% pure.
Liposome preparation
Large unilameller vesicles (LUV) were prepared by extrusion using a mini-extrusion syringe device (Avanti Polar Lipids). Briefly, phosphatidylethanolamine (PE), phosphatidylglycerol (PG) and cardiolipin (CL) (molar ratio 70:25:5) were dried down under nitrogen and stored under vacuum overnight. Lipids were rehydrated in H2O, freeze-thawed using liquid nitrogen (5x), and extruded through a 0.1 nm filter (15x). Lipid concentration was determined by Stewart’s assay as previously described (41).
Circular Dichroism
Circular dichroism (CD) spectra were obtained in buffer, in the presence of 50% trifluoroethanol (TFE), and in the presence of liposomes. For liposome samples, peptide and LUVs composed of PEPGCL were diluted in H2O (350 μL) to give a final concentration of 60 μM peptide at a lipid-to-peptide molar ratio of 100:1. Samples were equilibrated overnight at 4°C and CD was performed at room temperature on a Jasco J-710 spectrapolarimeter (Tokyo, Japan) at a scan rate of 20 nm/min, 2 sec response time, and 1 nm bandwidth. For liposome samples, spectra were truncated at 200 nm due to excessive light scattering at lower wavelengths.
Antimicrobial Activity Assay
To assess antimicrobial activity, minimum inhibitory concentrations (MIC) were determined by broth-dilution assay as previously described (32). Briefly, bacteria were grown at 37°C overnight from frozen stock on LB or Pseudomonas-selective Vogel-Bonner minimal (VBM) agar plates with no antibiotic. Single colonies were inoculated into Mueller-Hinton broth (MHB), grown (37°C, shaking at 150rpm) until the OD at 600nm was approximately 0.5–0.8, and then diluted in MHB to a working concentration of 2×105 cells/mL. In a clear bottom 96-well plate, we first added 100uL MHB/0.1%HSA to each well. Peptide (100 μL, 256 μM in PBS) was then added to the first well in each row, followed by eight 2-fold serial dilutions. Finally, 100 μL of the bacterial working solution described above was added to each well, giving final concentrations of 64 – 0.5 μM peptide and 1 × 105 bacterial cells/ml. The plate was incubated overnight (37°C, 150 rpm) and the OD600 values were measured using a SpectraMax M2 plate reader (Molecular Devices, Mountain View, CA). MIC values are the lowest concentration of peptide at which there was no bacterial growth.
Hemolysis Assay
Outdated whole blood was obtained from the Blood Bank of Southeast Wisconsin. Red blood cells (RBC) were separated by centrifugation (5000g, 10 min, 4°C), washed 3 times with cold PBS, and diluted to give a working stock of 10% (v/v) in PBS containing 5 mM glucose. Peptide samples (100 μl) in PBS were prepared in Eppendorf tubes by making 2-fold serial dilutions starting at 256 μM, followed by the addition of an equal volume of the RBC working stock solution to give a final concentration of 5% RBC by volume. Following a 1 hr incubation (37°C, with shaking at 150 rpm) samples were centrifuged (3 min, 1000 rpm), the supernatants were transferred to a 96-well plate, and the OD at 543 nm was measured. Percent hemolysis was calculated relative to a negative control containing only PBS and a positive control containing 1% Triton X100.
Macrophage Toxicity by MTT Assay
The effects of peptides on macrophage viability were examined as previously described (32). RAW264.7 murine macrophages were cultured from frozen stock on tissue culture dishes (Sarstedt, 100×20 mm) in DMEM supplemented with sodium bicarbonate (44.05 mM), sodium pyruvate (1 mM), FBS (10%) and Penicillin/Streptomycin (1%) and maintained at 37°C, 5% CO2 until confluent. Media was aspirated and cells were rinsed with PBS to remove dead cells and waste. Fresh media was added and cells were scraped, counted using a hemacytometer, and seeded into a 96-well plate at a concentration of 10,000 cells/well. Cells were allowed to attach overnight (37°C, 5% CO2) and peptides (2 fold dilutions from 120 μM to 0.94 μM, final concentration) were added to each well. Plates were incubated for 1hr (37°C, 5% CO2) and MTT (20 μL of a 5mg/mL stock solution in PBS) was added to each well. After a 4 hour incubation (37°C, 5% CO2) the media was removed and the plate was allowed to dry. The resulting formazan dye, formed by the active metabolism of MTT in viable cells, was resuspended in 100uL of isopropanol and the OD at 570 nm was measured. Fraction of macrophage survival was calculated using the following equation:
Macrophage Toxicity by Sytox Green Assay
Cultured RAW264.7 macrophages were grown and plated in a 96-well format as described above and incubated overnight (37°C, 5% CO2). An appropriate concentration of peptide in 50 μL PBS was added to achieve a final concentration of four times the MIC value against S. aureus. Immediately following the addition of peptide, 100 μl of a working solution of Sytox green (3 μl/5 mL DMEM, prepared according to the manufacturer’s instructions) was added. Measurement of time course data was initiated immediately using a plate reader (Beckman Coulter DTX880) with excitation filter F485/20 and emission filter F535/25. Positive and negative controls without peptide contained 1% Triton X100 or PBS, respectively. To visualize Sytox green uptake by fluorescence microscopy, cells were treated in the same manner as above and images were acquired every 15 min using a Nikon Eclipse TE200 microscope. Bright field and fluorescence images were obtained at 10× magnification.
S. aureus Membrane Permeability by Sytox Uptake
Bacterial cells were diluted to 2×107 cells per well in a 96 well plate (in 50 μL PBS). Peptide in PBS was added to give a final peptide concentration equal to the MIC value. 100μL Sytox green (2 μL/mL in PBS) was added to each well and a 2 hr time course was obtained using a SpectraMax M2 plate reader (Molecular Devices, Mountain View, CA) with excitation at 488 nm, emission at 530 nm and cutoff of 495 nm. The fractional fluorescence was quantitated relative to 100% live and dead cell suspensions prepared by a modified Live/Dead Baclight protocol (Invitrogen). Briefly, 30 mL cultures of S. aureus 6538p were grown in MHB to late log phase. Cells were pelleted via centrifugation (10,000g, 15min), the supernatant discarded, and the pellet resuspended in sterile water (2 mL). Half of this preparation was added to 20 mL of sterile water while the other half was added to 20 mL of 70% isopropyl alcohol (IPA), incubated for 1hr with mixing every 15 min, pelleted via centrifugation (10,000g, 15 min), and washed with 20 mL sterile water.
Protease Susceptibility
Peptide (2.7 μg) and protease (10 ng, trypsin or chymotrypsin) were mixed in 10 μL of 10 mM Tris (pH7.5 or pH8, respectively). A proteolysis time course (1, 5, 10, 20, 30, and 60 min) was performed by removing aliquots at the designated times and quenching the protease with the addition of an equal volume of SDS PAGE sample buffer (200 mM Tris-HCl pH6.8/2% SDS/40% glycerol/0.04% Coomassie Blue). Samples were loaded into wells of a 16.5% Tris/Tricine gel (BioRad) and electrophoresed at room temperature (100V, 75 min). Peptide bands were fixed in a solution of 40% methanol/10% acetic acid for 1 hr, after which they were stained for 1 hr (10% acetic acid/0.025% Coomassie Blue G-250). Gels were destained in 10% acetic acid to visualize the bands. Wells containing standard concentrations of intact peptide were included in each gel as controls. For quantitation, gels were imaged using an Alpha Imager (Alpha Innotech, Santa Clara, CA).
Results
Peptide design and secondary structure
In an attempt to decrease the toxicity of CM15 to eukaryotic cells, as well as increase stability in the presence of common proteases, we designed a set of peptides containing D-amino acid substitutions (Table 1). These peptides retain the same charge and relative numbers of hydrophilic and hydrophobic amino acids as their parent peptide, but are expected to have disrupted secondary structure. By substituting D-lysine for L-lysine we sought to protect against proteases such as trypsin that cleave at cationic amino acid residues (i.e. lysine, arginine), and placement of D-amino acids near the ends of the peptide should provide protection against carboxy- and aminopeptidases.
Table 1.
Peptide designations and sequences.a
| Peptide Designation | Peptide Sequence |
|---|---|
| CM15 | KWKLFKKIGAVLKVL |
| D1,13 | kWKLFKKIGAVLkVL |
| D3,13 | KWkLFKKIGAVLkVL |
| D3,14 | KWkLFKKIGAVLKkL |
| D3,7,13 | KWkLFKkIGAVLkVL |
| D3,7,14 | KWkLFKkIGAVLKkL |
| D3,6,7,13,14 | KWkLFkkIGAVLkkL |
D-amino acids are represented in lower case and underlined.
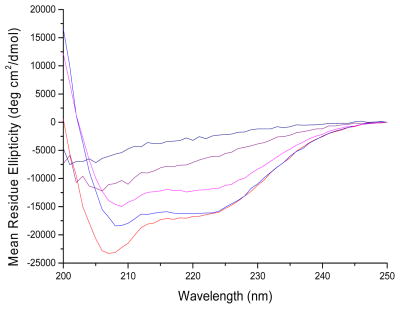
To evaluate anticipated changes in secondary structure of the D-lysine containing peptides, we examined their circular dichroism (CD) spectra. Previous studies of CM15 indicated α-helical contents of approximately 85% and 60% in TFE and membranes, respectively (42), with a well-defined alpha helix extending from residues 4 – 14 in the membrane-bound form (41, 43). All of the peptides were unstructured in buffer (data not shown). In the presence of liposomes mimicking the composition of the bacterial inner membrane, peptides CM15, D1,13, D3,13, and D3,14 all show prominent features at 208 and 222 nm, indicative of right-handed helical structure (Figure 1). Of the D-lysine containing peptides, D1,13 retained the most helical content (~ 84% of that observed for CM15, based on ellipticity at 208 nm), followed by D3,13 and D3,14 which gave virtually identical CD spectra with ~ 69% of the helical content observed for CM15. These results indicate that the α-helical structure in the central region of CM15 is largely preserved when D-amino acid substitutions are made near the ends of the peptide. In contrast, the CD spectrum of D3,7,13 bound to PE:PG:CL liposomes no longer exhibits minima at 208 and 222 nm, and peptide D3,6,7,13,14 exhibits a CD spectrum consistent with an extended or random coil structure (Figure 1). Thus, as anticipated, there is an increasing loss of helical secondary structure as additional D-amino acid residues are introduced.
Figure 1.
Circular dichroism of peptides in the presence of PE:PG:CL LUVs at a lipid/peptide ratio of 100:1. Peptides were at a concentration of 60 μM. Red line represents CM15, blue line is D1,13, magenta line is D3,13 purple line is D3,7,13 and black line is D3,6,7,13,14.
Antimicrobial activity
Broth dilution assays were used to compare antimicrobial activities of D-amino acid containing peptides to the parent peptide, CM15. Assays were performed against two Gram-positive bacterial species, Staphylococcus aureus and Staphylococcus epidermidis; and two Gram-negative species, Escherichia coli (strain BL21) and Pseudomonas aeruginosa (strains PA103 and PAO1). Results are summarized in Table 2. We began by testing the three di-substituted CM15 analogs, each containing two D-lysine residues near the ends of the peptide. Replacement of L-lysine with D-lysine at the first and thirteenth positions (D1,13) resulted in a small decrease in antibacterial activity, with two to four-fold increases in MIC values against all strains tested. Similarly, an analog containing L- to D-lysine substitutions at the third and thirteenth positions (D3,13) also showed a slight increases in MIC values as compared to the parent peptide. However, both of these peptides retain good antimicrobial activity in general, particularly against E. coli and the two strains of Staphylococcus. We also tested an analog containing D-lysine substitutions at positions three and fourteen (D3,14), based on our previous studies indicating that an analog of CM15 containing a V14K substitution retained good antimicrobial activity and had significantly reduced hemolytic activity and toxicity to murine macrophages(32). Surprisingly, this analog had substantially reduced activity against the E. coli and S. aureus test strains, although it retained reasonably good activity against S. epidermidis and the two test strains of P. aeruginosa (Table 2).
Table 2.
Minimal inhibitory concentrations (μM) of CM15 and its D-lysine diasteromers.
| Peptide | E. coli BL21 DE3 | Staph aureus 6835p | Staph epidermidis | P. aeruginosa PA103 | P. aeruginosa PAO1 |
|---|---|---|---|---|---|
| CM15 | 0.5 | 0.5 | 0.5 | 8 | 4 |
| D1,13 | 1 | 2 | 1 | 16 | 16 |
| D3,13 | 4 | 4 | 2 | 16 | 32 |
| D3,14 | 16 | 32 | 2 | 8 | 16 |
| D3,7,13 | 8 | 8 | 4 | 32 | 32 |
| D3,7,14 | >128 | 64 | 4 | 16 | 32 |
| D3,6,7,13,14 | >128 | >128 | >128 | >128 | >128 |
We also examined two peptides having a third D-lysine substitution near the center of the sequence (D3,7,13 and D3,7,14). Antimicrobial activity for D3,7,13 was decreased by 4 to 16-fold as compared to CM15 (2-fold relative to D3,13). Nonetheless, MIC values for D3,7,13 remained less than 8 μM for E. coli, S. aureus, and S. epidermidis. In contrast, D3,7,14 had an unexpectedly large decrease in activity against S. aureus and E. coli. Finally, we examined a peptide containing five D-lysine residues, with D-lysine replacing all L-lysine residues except at the first position (which is N-acetylated). D3,6,7,13,14 had little or no antibacterial activity, even at high peptide concentrations (up to 128 μM), against any of the bacteria tested.
Erythrocyte lysis and macrophage toxicity
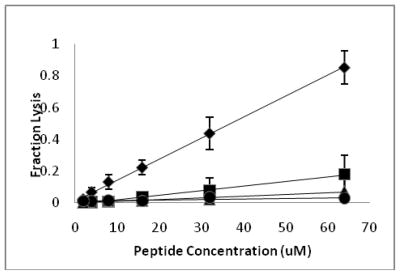
Having established that the majority of the D-amino acid containing peptides, with the exception of D3,6,7,13,14, retain antimicrobial activity, we next examined the effects of these peptides on eukaryotic cells. Lysis of red blood cells is commonly used to assess the cytotoxicity of antimicrobial peptides. Although originally reported to be non-hemolytic when tested against sheep erythrocytes (44), we previously observed significant hemolysis of human RBC by CM15 when tested at relatively high peptide concentrations (32). The fractional hemolysis observed for a 5% suspension of human RBC following a 1 hour incubation at 37°C is shown as a function of peptide concentration in Figure 2. RBC lysis increased approximately linearly with peptide concentration. As observed previously (32), significant lysis was observed with CM15. D1,13 was the next most hemolytic of the tested peptides, lysing approximately 25% of the red cell suspension at a peptide concentration of 64 μM (although it should be noted that this concentration is well above the MIC for D1,13 against any of the tested bacterial strains). The remaining peptides were essentially non-hemolytic. D3,13 and D3,14 caused less than 6% hemolysis at 64 μM, while less than 1% hemolysis was observed for peptides D3,7,13, D3,7,14, and D3,6,7,13,14. Thus, the introduction of D-lysine substitutions substantially reduces the hemolysis of human RBC by CM15 analogs.
Figure 2.
Hemolytic activity of all L-CM15 and CM15 analogs containing D-lysine. Shown is the percent red blood cell hemolysis after 1 hour incubation with each respective peptide (mean and standard deviation of at least three independent experiments). Diamonds represent CM15, squares represent D1,13, triangles represent D3,13, and circles represent D3,7,13. Not represented in the graph are D3,14, D3,7,14 and D3,6,7,13,14, all of which produced less than 5% hemolysis at concentrations up to 64 μM.
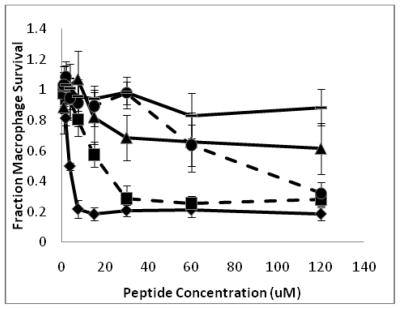
Although erythrocyte hemolysis is an important consideration and a convenient method for testing peptide toxicity to eukaryotic cells, human RBCs contain a relatively high cholesterol content, which may increase their resistance to AMP induced lysis (15, 17). Thus, to further examine the toxicity of these peptides to eukaryotic cells we used murine RAW264.7 macrophages. Figure 3 shows the effects of CM15 and its D-lysine containing analogs on macrophage survival, as indicated by their ability to metabolize MTT. As with RBC hemolysis, CM15 significantly decreased macrophage viability even at relatively low concentrations, inhibiting mitochondrial respiration with a LD50 of 3.8 μM (Table 3). D1,13 was again the next most cytotoxic of the peptides tested, decreasing macrophage viability by approximately 80% at concentrations ≥ 32 μM and having a LD50 of 5.4 μM. The remainder of the peptides had substantially lowered toxicity to macrophages, with LD50 values ranging from 78 to 130 μM (Table 3). Notably, peptides D3,13 and D3,7,13, which retain strong antimicrobial activity, exhibited LD50 values of 78 μM and 98 μM, respectively. Thus, as with RBC lysis, D-lysine substitution dramatically lowered toxicity towards cultured murine macrophages.
Figure 3.
Peptide toxicity to macrophages represented as the fraction of macrophage survival, as compared to a PBS control sample, versus peptide concentration. Survival is based on the mitochondrial conversion of MTT to formazan. Diamonds represent CM15, squares represent D1,13, triangles represent D3,13, circles represent D3,7,13, and dashes represent D3,6,7,13,14.
Table 3.
Effects of CM15 and its D-lysine diasteromers on the viability of RAW264.7 murine macrophages.
| Peptide | LD50 (μM) |
|---|---|
| CM15 | 3.8 ± 0.3 |
| D1,13 | 5.4 ± 0.8 |
| D3,13 | 78 ± 13 |
| D3,14 | 84 ± 14 |
| D3,7,13 | 98 ± 8.4 |
| D3,7,14 | 126 ± 11 |
| D3,6,7,13,14 | 131 ± 15 |
Peptide-induced membrane permeabilization
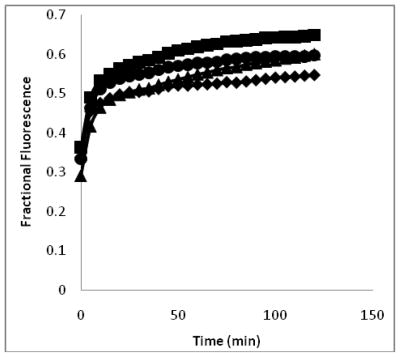
Membrane permeabilization is considered to be a key mechanism of bacterial cell killing by amphipathic α-helical AMPs (12, 13, 17, 19, 20). To investigate the effects of these peptides on cell membrane integrity in both bacterial and eukaryotic cells, we examined their ability to facilitate uptake of the membrane-impermeable dye SYTOX green. Upon membrane disruption, SYTOX green diffuses into cells and binds DNA, producing an approximately 500-fold enhancement of its fluorescence (45). As shown in Figure 4, CM15, D1,13, D3,13 and D3,7,13 all produced an immediate (< 2 minutes from time of peptide addition) permeabilization of the S. aureus cell envelope to SYTOX green, consistent with membrane disruption as a prominent mechanism of bactericidal activity.
Figure 4.
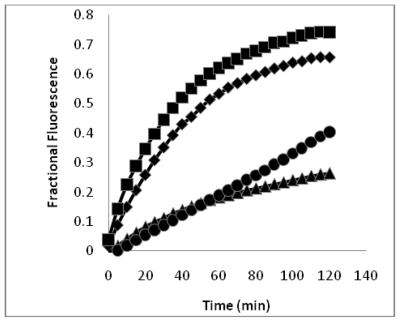
Peptide-induced uptake of SYTOX green by S. aureus. Fractional fluorescence is relative to isopropanol-killed cells (see Methods). Peptides were at concentrations equal to their respective MIC for S. aureus. Diamonds represent CM15 (0.5 μM), squares D1,13 (2 μM), triangles D3,13 (4 μM), and circles D3,7,13 (8 μM).
These peptides also caused permeabilization of the cell membrane in RAW264.7 macrophages, but at a slower rate and to a lesser degree than observed with S. aureus. Fluorescence microscopy images of macrophages exposed to CM15 or the D-lysine containing analogs (e.g. D3,7,13, Figure 5) demonstrated a time-dependent increase in SYTOX-positive cells. As indicated by the representative data shown in Figure 5, little or no SYTOX fluorescence was observed immediately after peptide addition. Consistent with their relative effects on macrophage survival as assessed by MTT metabolism, the increase in SYTOX uptake was much greater for CM15 than for D3,7,13 (Figure 5) or for the other D-lysine containing peptides. Kinetic curves for the increase in SYTOX fluorescence produced by exposure of macrophages to CM15, D1,13, D3,13 and D3,7,13, each tested at a concentration 4 times its respective MIC value for S. aureus, are shown in Figure 6. High levels of SYTOX uptake (60 – 80% of the values obtained by complete membrane permeabilization with Triton X100) were observed for CM15 (2 μM) and D1,13 (4 μM), while substantially lower levels of SYTOX uptake were observed for D3,13 (16 μM) and D3,7,13 (32 μM) despite the higher concentrations of the latter two peptides. Thus, these results parallel those based on MTT metabolism indicating the substantially reduced cytotoxicity of D3,13 and D3,7,13.
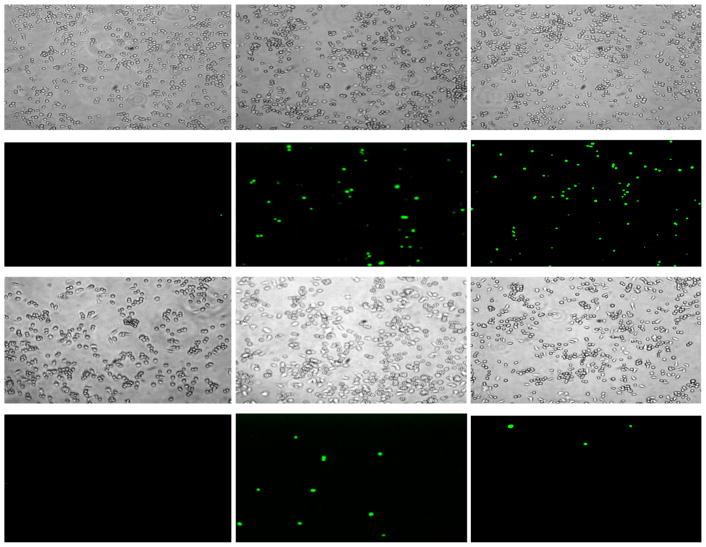
Figure 5.
Fluorescence microscopy (10× magnification) showing bright field (panels 1–3 and 7–9) and fluorescent images (panels 4–6 and 10–12) of macrophages in the presence of either 2 μM CM15 (panels 1–6) or 32 μM D3,7,13 (panels 7–12) immediately after peptide addition (column 1), and after incubation at 37°C for 15min (column 2), or 30min (column 3).
Figure 6.
Macrophage sytox uptake. The uptake of SYTOX green is measured by the enhancement of fluorescence upon SYTOX binding to DNA. Peptides were at concentrations equal to four times their MIC values for S. aureus. Diamonds represent CM15 (2 μM), squares D1,13 (8μM), triangles D3,13 (16 μM), and circles D3,7,13 (32 μM).
Selectivity indices
Antimicrobial efficacy is a combination of the ability to kill or inhibit growth of the pathogen and a lack of toxicity to host eukaryotic cells. As a comparative measure of in vitro efficacy, we calculated selectivity indices based on macrophage toxicity (Table 4) by taking the ratio of the macrophage LD50 to the MIC value for each peptide against each bacterial strain. Since lower toxicity to eukaryotic cells (reflected by an increased macrophage LD50) and greater antimicrobial activity (reflected by decreased MIC values) both increase this parameter, higher values of the selectivity index indicate a greater selectivity towards bacterial versus eukaryotic cells. Due to their substantially decreased macrophage toxicity, selectivity indices for D-lysine containing peptides were generally improved relative to CM15. Notably, peptides D3,13 and D3,7,13 had greater selectivity indices than CM15 against all of the tested bacteria (Table 4). Comparison of selectivity indices indicates that the impact of D-amino acid substitution within CM15 is sequence dependent. Among those peptides containing two D-lysine substitutions, D3,13 had selectivity indices greater than D1,13 against all bacterial strains tested, and substantially greater than D3,14 against E. coli and S. aureus. Addition of a third D-lysine generally decreased the selectivity of D3,7,13 as compared to D3,13 (with the exception of P. aeruginosa PAO1, for which they were essentially equal). Addition of D-lysine at position 7 also decreased the selectivity of D3,7,14 as compared to D3,14. Selectivity indices were not calculated for D3,6,7,13,14, since it had little or no antimicrobial activity. These results indicate that both the position and number of D-amino acid substitutions influence the relative selectivity of these diasteromeric CM15 analogs.
Table 4.
Selectivity indicesa
| Peptide | E. coli BL21 DE3 | Staph aureus 6835p | Staph epidermidis | P. aeruginosa 103 | P. aeruginosa 01 |
|---|---|---|---|---|---|
| CM15 | 7.6 | 7.6 | 7.6 | 0.48 | 0.95 |
| D1,13 | 5.4 | 2.7 | 5.4 | 0.34 | 0.34 |
| D3,13 | 19 | 19 | 39 | 4.8 | 2.4 |
| D3,14 | 5.2 | 2.6 | 42 | 10 | 5.2 |
| D3,7,13 | 12 | 12 | 24 | 3 | 3 |
| D3,7,14 | * | 2 | 31 | 7.9 | 3.9 |
Selectivity indices were calculated by dividing the LD50 for RAW264.7 macrophages by the MIC for each respective bacterial strain. Higher values indicate greater antimicrobial selectivity.
Not calculated due to low antimicrobial activity (MIC > 128 μM).
Protease Susceptibility
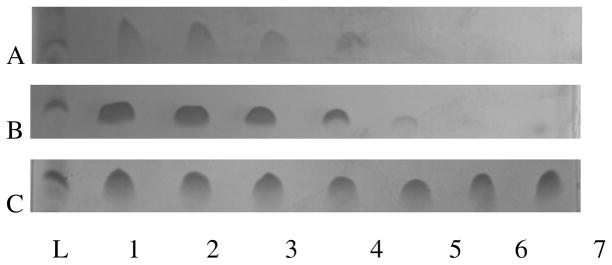
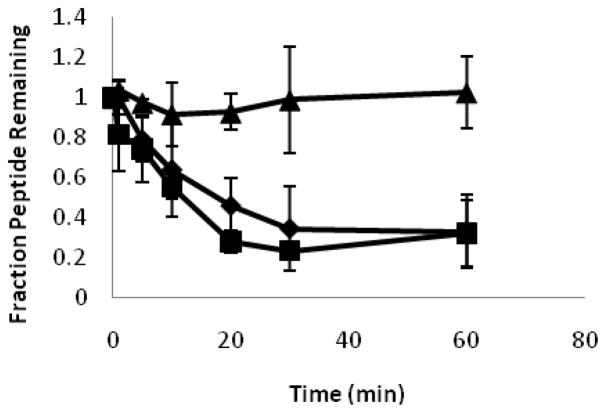
Introducing β- or D-amino acids into a peptide can increase its biological half-life by making it less susceptible to protease digestion (23, 25, 29, 30, 46–48). We used SDS PAGE to determine the relative susceptibility of CM15, D3,13 and D3,7,13 to trypsin and chymotrypsin. Representative gels from trypsin peptide digestion are shown in Figure 7. CM15 was readily degraded, with almost all of the peptide being digested within the first 20 minutes of incubation. Trypsin digestion of D3,13 was slightly reduced but overall remained comparable to CM15, while D3,7,13 exhibited minimal digestion under our experimental conditions. These observations were confirmed by densitometric analysis (Figure 8). As shown in Figure 8, there was no significant difference in the fractions of CM15 and D3,13 remaining at any given time point during trypsin digestion. In contrast, D3,7,13 exhibited minimal degradation throughout the 60 min incubation time. Although neither of the aromatic residues (Trp2 or Phe5) were replaced with D-amino acids, a similar trend was observed with chymotrypsin (data not shown), suggesting that the presence of nearby D-lysine residues (i.e. at positions 3 and 7) impaired degradation by this protease.
Figure 7.
Trypsin susceptibility. Representative SDS page gels showing degradation of (A) CM15, (B) D3,13, and (C) D3,7,13 upon incubation with trypsin. (L) 2 kDa band from peptide ladder, (1) peptide with no protease, (2 – 7) incubation times of 1 min, 5 min, 10 min, 20 min, 30 min, and 60 min.
Figure 8.
Peptide susceptibility to trypsin. Fraction of peptide remaining based on densitometric scans are shown versus the trypsin incubation time. Data (± s.d.) are from at least three independent experiments. Diamonds represent CM15, triangles D3,13, and circles D3,7,13.
Discussion
Clinically-useful AMPs must have not only good antimicrobial efficacy, but must also be minimally toxic to the host. Although numerous AMPs with outstanding antimicrobial activity have been identified, in order to further develop these peptides as useful antibiotics it will be essential to address their toxicity to eukaryotic cells and in vivo stability. Several studies have indicated that interruption of the hydrophobic surface and/or disruption of amphipathic secondary structure can lower the toxicity of AMPs to eukaryotic cells, while maintaining all or most of their antimicrobial activity (16, 23, 31, 46–48). The disruption of amphipathic secondary structure can be accomplished by the inclusion of polar or charged residues in the non-polar surface of helical AMPs (16, 23), or by the introduction of β-amino (25, 31, 46) or D-amino acids (47–49). The inclusion of β- or D-amino acids has the additional benefit of providing stability against protease digestion (23, 25, 25, 29, 46, 46, 47, 47, 48, 50). The goal of the present study was to determine the impact of introducing D-amino acids into CM15, a well-established representative of the linear, amphipathic α-helical class of AMPs. Our results show that selective D-lysine substitution can indeed substantially decrease peptide toxicity to eukaryotic cells and prevent degradation by common proteases, with the retention of good antimicrobial activity.
Remarkably, the introduction of only two D-lysine residues into CM15 was sufficient to produce substantial decreases in RBC hemolysis and macrophage cytotoxicity. The incorporation of D-lysine at the extreme ends of CM15 (in peptide D1,13), which decreased helical content only slightly, had the least impact on hemolysis and cytotoxicity. However, D-lysine substitution at positions 3 and 13 or at positions 3 and 14, either of which caused an approximately 30% reduction in helical content of the membrane-bound peptide, resulted in a 20-fold reduction in macrophage toxicity (i.e. a 20-fold increase in LD50). Similar reductions in hemolysis were observed, with peptides D3,13 and D3,14 being much less hemolytic than either CM15 or D1,13. These results are consistent with previous studies showing that introduction of a single charged residue on the hydrophobic face of amphipathic helical peptides (16, 23) or a single D-amino acid into a series of small β-sheet peptides (48) was sufficient to significantly attenuate hemolytic activity. Although low hemolytic activity can be misleading with regard to the general toxicity of AMPs to eukaryotic cells (10, 46) we observed a good correlation between hemolysis and toxicity to cultured macrophages for all of the peptides in this study.
The introduction of additional D-lysine residues further decreased helical content and eukaryotic cell toxicity, along with a concomitant decrease in antimicrobial activity. Indeed, all of the D-lysine containing analogs had some degree of reduction in antimicrobial activity when compared to the parent peptide, CM15, suggesting that the amphipathic α-helical structure of CM15 contributes to its potent antimicrobial effects. MIC values for D3,7,13 increased 2-fold relative to D3,13 against all bacterial species tested, and a 2-fold increase in MIC values for D3,7,14 relative to D3,14 was also observed against each of the bacterial test strains except E. coli (against which D3,7,14 was essentially inactive).
Although D3,13 exhibited the greatest antimicrobial selectivity, the addition of two D-amino acid residues near the ends of CM15 did not substantially increase resistance to proteolytic digestion. In contrast, D3,7,13 was not degraded by either trypsin or chymotrypsin under the defined experimental conditions. Notably, the presence of D-lysine at the 3 and 7 positions was evidently sufficient to protect against trypsin digestion at Lys6, or against chymotrypsin digestion at Trp2 or Phe5. Thus, it was not necessary to replace all of the L-lysine residues in CM15 with D-amino acids.
Numerous studies have shown that, for most AMPs, there is a strong correlation between antimicrobial activity and the ability of a given peptide to disrupt the bacterial membrane permeability barrier. All of the peptides in this study permeabilized membranes to the cationic cyanine dye, SYTOX green. Permeabilization of S. aureus cell membranes occurred rapidly, with significant levels of SYTOX uptake observed immediately upon addition of peptide. The rapid disruption of the membrane permeability barrier is consistent with the very rapid bacterial cell killing (e.g. an approximately 5-log reduction in CFU within 15 minutes) observed for CM15 (51). In contrast, permeabilization of the macrophage cell membrane to SYTOX occurred on a much slower time scale, and the extent of SYTOX uptake into macrophages was markedly decreased for the relatively non-cytotoxic peptides D3,13 and D3,7,13. Thus, although there are many other potential mechanisms may that contribute, these studies suggest that disruption of the plasma membrane may also play an important role in the eukaryotic cell cytotoxicity of CM15 and related AMPs.
In summary, the strong correlation between helical content and macrophage toxicity observed in these studies supports the general view that, within a given set of comparable AMPs, highly structured peptides with a well-defined amphipathic distribution of amino acid side chains have the greatest antimicrobial activity, but also the greatest eukaryotic cell cytotoxicity. Disruption of amphipathic secondary structure in CM15 produced modest reductions in antibacterial activity, but far more substantial decreases in toxicity to eukaryotic cells. Peptides D3,13 and D3,7,13 were non-hemolytic, and approximately 20 – 25 fold less cytotoxic to cultured murine macrophages than all-L CM15. Additionally, D3,7,13 was resistant to digestion by both trypsin and chymotrypsin. Thus, the selective introduction of D-amino acid residues into CM15 is a viable approach for enhancing antimicrobial selectivity and increasing resistance to proteolytic degradation.
Footnotes
This work was supported by the National Institutes of Health, National Institute of General Medical Sciences grant GM068829.
Reference List
- 1.Alekshun MN, Levy SB. Molecular mechanisms of antibacterial multidrug resistance. Cell. 2007;128:1037–1050. doi: 10.1016/j.cell.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007;298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 3.Levy SB, Marshall B. Antibacterial resistance worldwide: causes, challenges and responses. Nat Med. 2004;10:S122–S129. doi: 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- 4.Martinez JL. The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proc Biol Sci. 2009;276:2521–2530. doi: 10.1098/rspb.2009.0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Veesenmeyer JL, Hauser AR, Lisboa T, Rello J. Pseudomonas aeruginosa virulence and therapy: evolving translational strategies. Crit Care Med. 2009;37:1777–1786. doi: 10.1097/CCM.0b013e31819ff137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wright GD. The antibiotic resistome: the nexus of chemical and genetic diversity. Nat Rev Microbiol. 2007;5:175–186. doi: 10.1038/nrmicro1614. [DOI] [PubMed] [Google Scholar]
- 7.Hancock RE. Peptide antibiotics. Lancet. 1997;349:418–422. doi: 10.1016/S0140-6736(97)80051-7. [DOI] [PubMed] [Google Scholar]
- 8.Hancock RE, Sahl HG. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 9.Jenssen H, Hamill P, Hancock RE. Peptide antimicrobial agents. Clin Microbiol Rev. 2006;19:491–511. doi: 10.1128/CMR.00056-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuzaki K. Control of cell selectivity of antimicrobial peptides. Biochim Biophys Acta. 2009;1788:1687–1692. doi: 10.1016/j.bbamem.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 11.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 12.Oren Z, Shai Y. Selective lysis of bacteria but not mammalian cells by diastereomers of melittin: structure-function study. Biochemistry. 1997;36:1826–1835. doi: 10.1021/bi962507l. [DOI] [PubMed] [Google Scholar]
- 13.Huang HW. Action of antimicrobial peptides: two-state model. Biochemistry. 2000;39:8347–8352. doi: 10.1021/bi000946l. [DOI] [PubMed] [Google Scholar]
- 14.Cao Y, Yu RQ, Liu Y, Zhou HX, Song LL, Cao Y, Qiao DR. Design, recombinant expression, and antibacterial activity of the cecropins-melittin hybrid antimicrobial peptides. Curr Microbiol. 2010;61:169–175. doi: 10.1007/s00284-010-9592-7. [DOI] [PubMed] [Google Scholar]
- 15.Glukhov E, Stark M, Burrows LL, Deber CM. Basis for selectivity of cationic antimicrobial peptides for bacterial versus mammalian membranes. J Biol Chem. 2005;280:33960–33967. doi: 10.1074/jbc.M507042200. [DOI] [PubMed] [Google Scholar]
- 16.Hawrani A, Howe RA, Walsh TR, Dempsey CE. Origin of low mammalian cell toxicity in a class of highly active antimicrobial amphipathic helical peptides. J Biol Chem. 2008;283:18636–18645. doi: 10.1074/jbc.M709154200. [DOI] [PubMed] [Google Scholar]
- 17.Yeaman MR, Yount NY. Mechanisms of antimicrobial peptide action and resistance. Pharmacol Rev. 2003;55:27–55. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- 18.Fernandez-Vidal M, Jayasinghe S, Ladokhin AS, White SH. Folding amphipathic helices into membranes: amphiphilicity trumps hydrophobicity. J Mol Biol. 2007;370:459–470. doi: 10.1016/j.jmb.2007.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sato H, Feix JB. Peptide-membrane interactions and mechanisms of membrane destruction by amphipathic alpha-helical antimicrobial peptides. Biochim Biophys Acta. 2006;1758:1245–1256. doi: 10.1016/j.bbamem.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 20.Lee MT, Hung WC, Chen FY, Huang HW. Mechanism and kinetics of pore formation in membranes by water-soluble amphipathic peptides. Proc Natl Acad Sci U S A. 2008;105:5087–5092. doi: 10.1073/pnas.0710625105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 22.Nizet V. Antimicrobial peptide resistance mechanisms of human bacterial pathogens. Curr Issues Mol Biol. 2006;8:11–26. [PubMed] [Google Scholar]
- 23.Chen Y, Mant CT, Farmer SW, Hancock RE, Vasil ML, Hodges RS. Rational design of alpha-helical antimicrobial peptides with enhanced activities and specificity/therapeutic index. J Biol Chem. 2005;280:12316–12329. doi: 10.1074/jbc.M413406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dempsey CE, Hawrani A, Howe RA, Walsh TR. Amphipathic antimicrobial peptides--from biophysics to therapeutics? Protein Pept. Lett. 2010;17:1334–1344. doi: 10.2174/0929866511009011334. [DOI] [PubMed] [Google Scholar]
- 25.Schmitt MA, Weisblum B, Gellman SH. Interplay among folding, sequence, and lipophilicity in the antibacterial and hemolytic activities of alpha/beta-peptides. J Am Chem Soc. 2007;129:417–428. doi: 10.1021/ja0666553. [DOI] [PubMed] [Google Scholar]
- 26.Pag U, Oedenkoven M, Papo N, Oren Z, Shai Y, Sahl HG. In vitro activity and mode of action of diastereomeric antimicrobial peptides against bacterial clinical isolates. J Antimicrob Chemother. 2004;53:230–239. doi: 10.1093/jac/dkh083. [DOI] [PubMed] [Google Scholar]
- 27.Oren Z, Hong J, Shai Y. A repertoire of novel antibacterial diastereomeric peptides with selective cytolytic activity. J Biol Chem. 1997;272:14643–14649. doi: 10.1074/jbc.272.23.14643. [DOI] [PubMed] [Google Scholar]
- 28.Merrifield RB, Juvvadi P, Andreu D, Ubach J, Boman A, Boman HG. Retro and retroenantio analogs of cecropin-melittin hybrids. Proc Natl Acad Sci U S A. 1995;92:3449–3453. doi: 10.1073/pnas.92.8.3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamamoto K, Kida Y, Zhang Y, Shimizu T, Kuwano K. Antimicrobial activity and stability to proteolysis of small linear cationic peptides with D-amino acid substitutions. Microbiol Immunol. 2002;46:741–749. doi: 10.1111/j.1348-0421.2002.tb02759.x. [DOI] [PubMed] [Google Scholar]
- 30.Frackenpohl J, Arvidsson PI, Schreiber JV, Seebach D. The outstanding biological stability of beta- and gamma-peptides toward proteolytic enzymes: an in vitro investigation with fifteen peptidases. Chembiochem. 2001;2:445–455. doi: 10.1002/1439-7633(20010601)2:6<445::aid-cbic445>3.3.co;2-i. [DOI] [PubMed] [Google Scholar]
- 31.Epand RF, Schmitt MA, Gellman SH, Epand RM. Role of membrane lipids in the mechanism of bacterial species selective toxicity by two alpha/beta-antimicrobial peptides. Biochim Biophys Acta. 2006;1758:1343–1350. doi: 10.1016/j.bbamem.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 32.Sato H, Feix JB. Lysine-enriched cecropin-mellitin antimicrobial peptides with enhanced selectivity. Antimicrob Agents Chemother. 2008;52:4463–4465. doi: 10.1128/AAC.00810-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giangaspero A, Sandri L, Tossi A. Amphipathic alpha helical antimicrobial peptides. Eur J Biochem. 2001;268:5589–5600. doi: 10.1046/j.1432-1033.2001.02494.x. [DOI] [PubMed] [Google Scholar]
- 34.Zelezetsky I, Tossi A. Alpha-helical antimicrobial peptides--using a sequence template to guide structure-activity relationship studies. Biochim Biophys Acta. 2006;1758:1436–1449. doi: 10.1016/j.bbamem.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 35.Wang Z, Wang G. APD: the Antimicrobial Peptide Database. Nucleic Acids Res. 2004;32:D590–D592. doi: 10.1093/nar/gkh025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piers KL, Hancock RE. The interaction of a recombinant cecropin/melittin hybrid peptide with the outer membrane of Pseudomonas aeruginosa. Mol Microbiol. 1994;12:951–958. doi: 10.1111/j.1365-2958.1994.tb01083.x. [DOI] [PubMed] [Google Scholar]
- 37.Friedrich C, Scott MG, Karunaratne N, Yan H, Hancock RE. Salt-resistant alpha-helical cationic antimicrobial peptides. Antimicrob Agents Chemother. 1999;43:1542–1548. doi: 10.1128/aac.43.7.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fantner GE, Barbero RJ, Gray DS, Belcher AM. Kinetics of antimicrobial peptide activity measured on individual bacterial cells using high-speed atomic force microscopy. Nat Nanotechnol. 2010;5:280–285. doi: 10.1038/nnano.2010.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boman HG, Wade D, Boman IA, Wahlin B, Merrifield RB. Antibacterial and antimalarial properties of peptides that are cecropin-melittin hybrids. FEBS Lett. 1989;259:103–106. doi: 10.1016/0014-5793(89)81505-4. [DOI] [PubMed] [Google Scholar]
- 40.Andreu D, Ubach J, Boman A, Wahlin B, Wade D, Merrifield RB, Boman HG. Shortened cecropin A-melittin hybrids. Significant size reduction retains potent antibiotic activity. FEBS Lett. 1992;296:190–194. doi: 10.1016/0014-5793(92)80377-s. [DOI] [PubMed] [Google Scholar]
- 41.Bhargava K, Feix JB. Membrane binding, structure, and localization of cecropin-mellitin hybrid peptides: a site-directed spin-labeling study. Biophys J. 2004;86:329–336. doi: 10.1016/S0006-3495(04)74108-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pistolesi S, Pogni R, Feix JB. Membrane insertion and bilayer perturbation by antimicrobial peptide CM15. Biophys J. 2007;93:1651–1660. doi: 10.1529/biophysj.107.104034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Respondek M, Madl T, Gobl C, Golser R, Zangger K. Mapping the orientation of helices in micelle-bound peptides by paramagnetic relaxation waves. J Am Chem Soc. 2007;129:5228–5234. doi: 10.1021/ja069004f. [DOI] [PubMed] [Google Scholar]
- 44.Wade D, Andreu D, Mitchell SA, Silveira AM, Boman A, Boman HG, Merrifield RB. Antibacterial peptides designed as analogs or hybrids of cecropins and melittin. Int J Pept Protein Res. 1992;40:429–436. doi: 10.1111/j.1399-3011.1992.tb00321.x. [DOI] [PubMed] [Google Scholar]
- 45.Lebaron P, Catala P, Parthuisot N. Effectiveness of SYTOX Green stain for bacterial viability assessment. Appl Environ Microbiol. 1998;64:2697–2700. doi: 10.1128/aem.64.7.2697-2700.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olsen CA, Ziegler HL, Nielsen HM, Frimodt-Moller N, Jaroszewski JW, Franzyk H. Antimicrobial, hemolytic, and cytotoxic activities of beta-peptoid-peptide hybrid oligomers: improved properties compared to natural AMPs. Chembiochem. 2010;11:1356–1360. doi: 10.1002/cbic.201000232. [DOI] [PubMed] [Google Scholar]
- 47.Papo N, Oren Z, Pag U, Sahl HG, Shai Y. The consequence of sequence alteration of an amphipathic alpha-helical antimicrobial peptide and its diastereomers. J Biol Chem. 2002;277:33913–33921. doi: 10.1074/jbc.M204928200. [DOI] [PubMed] [Google Scholar]
- 48.Rathinakumar R, Walkenhorst WF, Wimley WC. Broad-spectrum antimicrobial peptides by rational combinatorial design and high-throughput screening: the importance of interfacial activity. J Am Chem Soc. 2009;131:7609–7617. doi: 10.1021/ja8093247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li X, Li Y, Han H, Miller DW, Wang G. Solution structures of human LL-37 fragments and NMR-based identification of a minimal membrane-targeting antimicrobial and anticancer region. J Am Chem Soc. 2006;128:5776–5785. doi: 10.1021/ja0584875. [DOI] [PubMed] [Google Scholar]
- 50.Chen Y, Vasil AI, Rehaume L, Mant CT, Burns JL, Vasil ML, Hancock RE, Hodges RS. Comparison of biophysical and biologic properties of alpha-helical enantiomeric antimicrobial peptides. Chem Biol Drug Des. 2006;67:162–173. doi: 10.1111/j.1747-0285.2006.00349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sato H, Feix JB. Osmoprotection of bacterial cells from toxicity caused by antimicrobial hybrid peptide CM15. Biochemistry. 2006;45:9997–10007. doi: 10.1021/bi060979m. [DOI] [PubMed] [Google Scholar]