Abstract
In the Cairns–Foster adaptive mutation system, a +1 lac frameshift mutant of Escherichia coli is plated on lactose medium, where the nondividing population gives rise to Lac+ revertant colonies during a week under selection. Reversion requires the mutant lac allele to be located on a conjugative F′lac plasmid that also encodes the error-prone DNA polymerase, DinB. Rare plated cells with multiple copies of the mutant F′lac plasmid initiate the clones that develop into revertants under selection. These initiator cells arise before plating, and their extra lac copies allow them to divide on lactose and produce identical F′lac-bearing daughter cells that can mate with each other. DNA breaks can form during plasmid transfer and their recombinational repair can initiate rolling-circle replication of the recipient plasmid. This replication is mutagenic because the amplified plasmid encodes the error-prone DinB polymerase. A new model proposes that Lac+ revertants arise during mutagenic over-replication of the F′lac plasmid under selection. This mutagenesis is focused on the plasmid because the cell chromosome replicates very little. The outer membrane protein OmpA is essential for reversion under selection. OmpA helps cells conserve energy and may stabilize the long-term mating pairs that produce revertants.
Keywords: mutagenesis, copy number variation, selective gene amplification, rolling-circle replication, recombination, DNA repair, conjugation, plasmid transfer, adaptive mutation
THERE is a controversy surrounding the question, “Do cells possess mechanisms to generate mutations in response to growth limitation?” This question was answered 60 years ago when classic experiments showed that stringent bacterial selections do not cause the mutations they detect (Luria and Delbrück 1943; Lederberg and Lederberg 1952). While this conclusion has been generalized broadly, it was based on use of stringent selections that can only detect preexisting mutants and therefore could not have revealed mutations induced by selection.
Cairns and Foster reopened the basic question by subjecting an Escherichia coli lac mutant to a less stringent selection (Cairns and Foster 1991). The strain they used carries a leaky lac frameshift mutation on an F′lac plasmid. Growth of this strain is prevented (just barely) by the lac mutation, but can be restored by even a small increase in lac function. The plated population (108 cells) does not grow on lactose, but gives rise to Lac+ revertant colonies that accumulate linearly at a rate of ∼10–20 colonies per day. After 5–6 days under selection, the revertant yield is roughly 100-fold higher than that predicted by the reversion rate of the lac mutation during unrestricted growth (10−9/cell per division) (Foster and Trimarchi 1994). Since the plated population does not grow under selection, revertants appear to be produced by mutagenesis without replication. The starved nongrowing cell population does not experience genome-wide mutagenesis whereas the Lac+ revertants show associated genomic mutations, suggesting an unevenly distributed level of genome-wide mutagenesis that is insufficient to have caused reversion (Torkelson et al. 1997; Rosche and Foster 1999; Godoy et al. 2000; Slechta et al. 2002). The behavior of this system has been explained in two general ways.
Stress-induced mutagenesis models suggest that cells possess evolved mechanisms to generate mutations when growth is blocked, and these mechanisms may direct genetic change preferentially to sites that improve growth in nondividing cells (Bjedov et al. 2003; Foster 2007; Galhardo et al. 2007). Supporters of these models have tried to define the mutagenic mechanism, which involves the error-prone repair polymerase DinB and homologous recombination (Cairns and Foster 1991; Harris et al. 1994; McKenzie et al. 2001). These models have been reviewed extensively (Foster 2007; Galhardo et al. 2007).
Selection models propose that there is no programmed mutagenic mechanism. Instead, the plated population of mutant cells (testers) includes rare cells with multiple copies of the mutant F′lac plasmid (initiator cells). Evidence was presented previously that each revertant is derived from one of these initiator cells, which arise before plating and cannot be stress-induced (Sano et al. 2014). Because of their extra copies of the leaky lac allele, the preexisting initiator cells can divide on selective medium and develop into lac+ revertants. Selection acts on the plasmid population within initiator cells by a multistep process that involves very few divisions of the plated cell population (Roth et al. 2006; Wrande et al. 2008; Yamayoshi et al. 2018). The problem is to understand the process by which selection acts on the plasmid population within an initiator cell (Maisnier-Patin and Roth 2015, 2016).
Attempts to decide between mutagenesis and selection have generated a body of data that is generally agreed upon but has been interpreted in conflicting ways. Both sides agree on the following points. The mutant lac allele carried by the plated tester cells does not support cell division on lactose, but retains some residual function (∼1% of normal) that supplies the energy necessary for reversion under selection. Residual growth of tester cells is prevented by a 10-fold excess of lac-deletion mutant cells (scavengers) that consume any carbon sources other than lactose that might contaminate the medium. Reversion requires the leaky lac allele to be located on a conjugative F′lac plasmid that also carries the dinB+ gene, encoding an error-prone DNA repair polymerase. Very few revertants appear when the mutant lac allele is located at its standard chromosomal position (Foster and Trimarchi 1995a; Radicella et al. 1995). The tester strain bearing the mutant F′lac plasmid must be capable of homologous recombination (RecA-RecBCD) (Cairns and Foster 1991; Harris et al. 1994). This strain must also possess two global control systems that affect dinB transcription: the SOS DNA repair system, which is derepressed in response to DNA damage (McKenzie et al. 2000), and the RpoS response, which induces dinB during the stationary phase (Lombardo et al. 2004).
Two types of Lac+ revertants appear during 5 days under selection. About 90% are stably Lac+ and have acquired a compensating frameshift mutation that creates a functional lacZ coding sequence. The remaining 10% carry a tandem amplification of the leaky mutant plasmid lac region. These unstable revertants grow by increasing their dosage of the leaky lac allele. In strains lacking the error-prone DNA polymerase DinB, the yield of stable revertants drops ∼10-fold but is not eliminated. Unstable Lac+ revertants form at the same rate with or without DinB (McKenzie et al. 2001; Yamayoshi et al. 2018). The above results have been interpreted either as support for stress-induced mutagenesis (Rosenberg et al. 2012) or as evidence that selection favors improved lactose degradation by acting through standard functions involved in plasmid transfer and DNA repair (Maisnier-Patin and Roth 2015, 2016). The results have not allowed for a decision between the two explanations of the adaptive mutation phenomenology.
A controversy regarding plasmid transfer
Early in the history of the adaptive mutation controversy, it was found that reversion is eliminated by conditions or mutations that prevent conjugational transfer of the F′lac plasmid between cells (Galitski and Roth 1995; Radicella et al. 1995; Peters et al. 1996; Godoy and Fox 2000). It was suggested that transfer functions might provide the DNA replication and mutagenesis needed for reversion in a nondividing cell population.
Supporters of stress-induced mutagenesis argued against mating. They pointed out that the number of revertants increases linearly with the number of plated parent cells (called “testers” here). If revertants were initiated by two mating cells, their number should increase exponentially (Foster and Trimarchi 1995a). They also showed that addition of F− cells had little effect on revertant yield. More directly, they found that revertants produced from mixtures of genetically marked tester strains rarely had plasmids that had been transferred from one tester type to another (Foster and Trimarchi 1995b). Since recombination functions (RecA-RecBCD) are essential for reversion but not transfer, they concluded that plasmid conjugation functions contributed to reversion only by making DNA nicks at the plasmid transfer origin (oriT). It was suggested that mutagenic repair of these nicks by homologous recombination stimulated reversion under selective conditions. This interpretation has been maintained by supporters of stress-induced mutagenesis (Rosenberg et al. 2012). The controversy over whether actual plasmid transfer is required for reversion is central to deciding whether selection or stress-induced mutation models best explain adaptive mutation.
The need to reexamine plasmid transfer was suggested by more recent results that show an intimate involvement of the F′lac plasmid in the reversion process.
First, revertants are derived from rare preexisting initiator cells (105) (Sano et al. 2014). Initiators can divide on lactose after plating because of their unusually high copy number (>10) of the F′lac plasmid. Initiators cannot be stress-induced since they arise in the tester population prior to plating on selective medium. Each revertant is derived from a single initiator cell, consistent with the linear dependence of revertant number on the number of plated cells. The bulk of the 108 plated cells cannot divide on lactose and do not contribute to reversion.
Second, the revertant yield does not fluctuate from one culture to the next. Initially this was taken as evidence that revertants arose in the nongrowing population after plating and might be stress-induced (Cairns and Foster 1991). However if preexisting initiator cells have extra plasmid copies, the fluctuation in their frequency is minimized by the mechanism that maintains copy number at an average of 1–2 per cell (Frame and Bishop 1971). The frequency of rare plasmid copy number variants (initiators) is also maintained at a steady state by balanced rates of copy number increase and loss (Reams et al. 2010).
Third, formation of stable Lac+ revertants under selection requires that the F′lac plasmid carrying the lac allele must also carry a functional copy of the dinB+ gene, which encodes an error-prone DNA repair polymerase (PolIV). A chromosomal copy of the dinB+ gene is not sufficient to support a high level of reversion (Slechta et al. 2003; Yamayoshi et al. 2018). Selection favors increases in the copy number of a plasmid that includes the lac and dinB+ genes (Yamayoshi et al. 2018). Induced transcription of the dinB+ gene by global regulatory systems is not sufficient for mutagenesis, but provides a level of DinB that can become mutagenic if amplified by plasmid copy number increases. General mutagenesis by DinB was shown previously to require an increase in the dinB copy number (Kim et al. 1997; Wagner et al. 1999).
A new model for reversion under selection in the Cairns system
The above observations showed a direct role of the F′lac plasmid in reversion and suggested a new model involving conjugation in the reversion process. Most of the 108 plated tester cells can neither divide nor conjugate under the stringent selection conditions. Despite their large population, these cells do not give rise to revertants. The new model focuses on the 105 initiator cells that arise prior to plating on selective medium (Sano et al. 2014). These cells can divide under selection because of their unusually high number of F′lac plasmids (>10). Each revertant clone is initiated by a single initiator cell, which divides to produce identical daughter cells. These daughter cells can mate with each other due to their proximity on the plate and because both can obtain energy from their extra copies of the partially functional lac allele (see Figure 1A). Noninitiator tester cells do not have sufficient energy to mate either with each other or with scavenger cells (the lac-deletion mutants that are plated at a 10-fold excess over testers to consume contaminating nutrients). Two independently plated initiator cells are unlikely to mate because only ∼105 such cells are plated, making it unlikely for two to contact each other on the plate. However, daughters produced by division of a single initiator cell lie immediately adjacent to each other and have extra copies of the F′lac plasmid.
Figure 1.
Mating between daughters of a single plated initiator cell. (A) The majority tester cell type carries one to two copies of an F′lac plasmid. Copy number is subject to stochastic variation, with an estimated one cell in a 1000 having a copy number >10. Rare cells with a high plasmid copy number can divide on selective lactose plates. Mating between daughter cells initiates a Lac+ revertant. (B and C) A single DNA strand transferred gains a complement in the recipient. Double-strand breaks can be repaired by recombination, using a single plasmid DNA template and very little replication (B). Alternatively the ends can be repaired using different template plasmids (C) and initiating rolling-circle replication.
When daughters of a single initiator cell mate, a single strand of plasmid DNA moves from the donor into the recipient cell, where a complementary strand is synthesized. Double-strand breaks formed during or after transfer can be repaired by recombination if the ends make convergent forks on a single template. Alternatively the two ends can recombine with different template plasmids and initiate rolling-circle replication as diagrammed in Figure 1C.
The increased plasmid copy number in the recipient cell facilitates rolling-circle replication because two ends are more likely to initiate independent forks on different circular templates instead of converging forks on a single template (see Figure 1, B and C). Repeated rolling-circle replication of the recipient plasmid provides multiple opportunities for reversion of the lac allele and also amplifies the dinB+ gene. Reversion to lac+ is made more likely by repeated replication of the lac allele and by overproduction of the mutagenic DinB polymerase. Mutagenic over-replication of the F′lac plasmid enhances the likelihood of a stable lac reversion mutation occurring in a cell that is not replicating its chromosome or dividing. Later steps in the reversion process will be described in the Discussion.
Support for plasmid transfer during reversion
Evidence described here suggests that actual plasmid transfer is central to the process by which selection drives formation of adaptive Lac+ revertants. This evidence supports the role of transfer presented in the above model and explains why early experiments missed the importance of conjugation. Also described are roles of the outer membrane protein OmpA, which is needed for reversion under selection. The OmpA protein is essential for mating in liquid but not on solid medium. We provide evidence that OmpA contributes to conservation of the energy needed for conjugation in nondividing cells. OmpA may also be needed for long-term stability of mating pairs that transfer DNA continuously for several days. Later steps in the reversion process suggested by the new model will be outlined in the Discussion section.
Materials and Methods
Bacterial strains
Strains and PCR primers used here are listed in Tables S1 and S2, which can be found in the Supplementary Material. All the E. coli strains are derivatives of strain P9OC (Coulondre and Miller 1977). Strain FC36 (TT24787) is a rifampicin-resistant (RifR) derivative of P9OC (Cairns and Foster 1991) and TT27036 is a streptomycin-resistant (SmR) derivative of P90C. Standard genetic techniques were used in strain constructions (Miller 1992). Insertions with antibiotic resistance markers were added to plasmid F′lac128 (from strain FC40/TR7178) by P22-mediated crosses done in Salmonella enterica. The plasmid was then transferred to the E. coli F− strains FC36 (TT24787) or TT27036, selecting for proline prototrophy. Chromosomal mutations in E. coli were moved between strains using P1-mediated transductions (Miller 1992). Mutations from the E. coli Keio collection were added to the chromosome by replacing the recipient locus with a KanR determinant flanked by FRT sites. To make double mutant strains, the first introduced KanR cassette was removed by deletion between FRT sites using the FLP recombinase expressed from pCP20 (Datsenko and Wanner 2000).
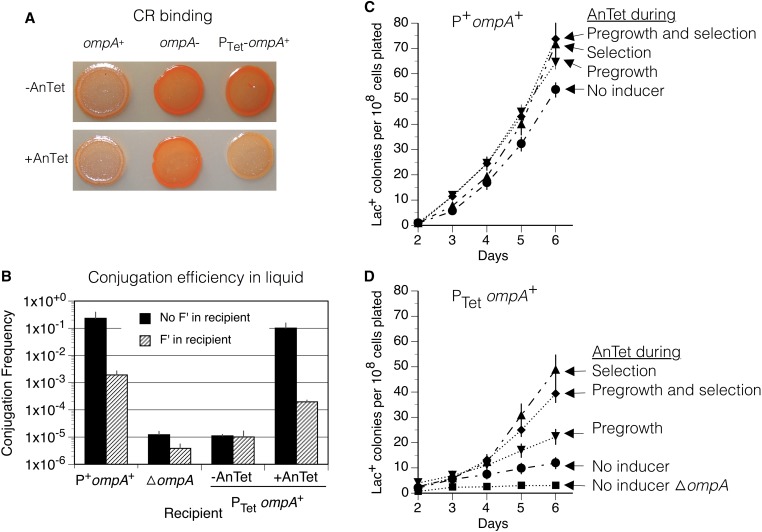
The tetA ompA operon fusion was created by λRed recombineering (Datsenko and Wanner 2000). A tetA-tetR cassette was PCR amplified from a Tn10 transposon using PCR primers TP3351 and TP3352 (Table S2). These primers produce a fragment with the tetR-tetA cassette flanked by sequence from just upstream of a functional ompA gene. Insertion of this fragment immediately upstream of the chromosomal ompA gene created the fusion operon. Junctions were confirmed by PCR amplification using the diagnostic primers listed in Table S2. The phenotype of recA mutant strains was confirmed by testing UV sensitivity; tra mutants were tested for conjugation efficiency and gal mutants for ability to grow on galactose. The ompA-deletion mutants were tested for conjugation efficiency in liquid medium and production of cellulose (See legend for Figure 6). To detect cellulose production, 5 µl of an overnight culture were spotted onto lysogeny broth (LB) plates containing 50 µg/ml of Congo Red. Anhydrotetracycline (AnTet) was used at 0.1 µg/ml to induce expression of the tetA-ompA fusion.
Figure 6.
Evidence that OmpA contributes to reversion during selection. (A) Appearance of colonies on Congo Red (CR) plates with or without the tetA gene inducer anhydrotetracycline, AnTet (216 nM). The strains are the wild-type ompA strain TR7178 (P+ ompA+), a strain with ompA deleted (TT27065, ΔompA), and a strain with ompA under tetA promoter control (TT27100, PTet ompA+). (B) Conjugation efficiency in liquid medium. The donor strain TT27083 (RifR/F′lac CamR) was mixed with one of the recipient strains TT27036 (P+ ompA+ StrR/F−), TT27037 (P+ ompA+ StrR/F′lac), TT27063 (ΔompA StrR/F−), TT27067 (ΔompA StrR/F′lac), TT27271 (PTet ompA+ StrR/F−), or TT27272 (PTet ompA+ StrR/F′lac). The inducer AnTet was added during pregrowth and mating in liquid LB medium. Conjugation efficiency is the number of transconjugants StrR CamR per donor cell. (C and D) Total cumulative numbers of revertant Lac+ colonies per 108 cells of the tester strain plated. Cells were pregrown to stationary phase in minimal glycerol with or without AnTet (216 nM) and then washed and plated on selective lactose plates with or without AnTc. Strains are TR7178 (P+ ompA+), TT27100 (PTet ompA+), and TT27065 (ΔompA). Each data point represents mean ± SEM (error bars) of five independent cultures of each strain.
Media and growth conditions
Bacterial strains were routinely grown in (LB), containing 10g tryptone, 5g yeast extract and 5g NaCl. Antibiotics were added at the following concentrations: tetracycline, 20 µg/ml; chloramphenicol, 20 µg/ml; kanamycin, 50 µg/ml; streptomycin, 200 µg/ml; and rifampicin, 100 µg/ml. Selection plates were no-citrate E medium (NCE), with appropriate additions.
Adaptive reversion experiments
Prior to plating of the tester strains, selection plates were prescavenged to remove contaminating carbon sources. A strain such as TR7177 (FC29; Table S1), whose plasmid carried a nonrevertible Lac− mutant (109 cells), was plated and incubated overnight. Tester strains with F′lac were grown to saturation at 37° in minimum NCE supplemented with 10 µg/ml thiamine (vitamin B1) and 0.1% glycerol, and these cultures were diluted 100-fold into the same medium and allowed to reach saturation again. About 108 washed tester cells were then spread on scavenged selection plate containing 0.1% lactose, thiamine, and the chromogenic β-galactosidase substrate 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal) at a concentration of 25 µg/ml. These reversion plates were incubated for 5–6 days, and new Lac+ colonies were counted every day.
Growth of the tester cell population in the lawn was determined by removing agar plugs from areas between Lac+ colonies on the selection plates. Cells from these plugs were resuspended in NCE medium, diluted, and plated for single colonies on LB plates containing rifampicin. To assess F′lac transfer in the lawn between testers cells or between tester and scavenger cells, lawn plugs were diluted and plated on rich media with antibiotics that detect cells with one chromosomal genotype that have acquired a plasmid from a different donor cell.
To assess F′lac transfer in the course of reversion, Lac+ colonies from day 3 to 5 were purified by streaking on lactose plates. One to ten single colonies from each Lac+ isolate were then tested for drug resistances by replica plating on rich medium containing rifampicin or streptomycin, to detect revertants in one tester or the other or in scavenger cell background. These revertants were also tested for resistances that might have been obtained with a plasmid from a different donor cell.
Quantitative conjugation
To determine mating efficiency on solid medium, Lac− donor and recipient cells were plated on lactose plates under reversion conditions at a ratio of 1:10. After 1 day of incubation, lawn samples were removed from the lactose plates as agar plugs (using a sterile glass tube). Appropriate dilutions were plated on LB medium containing streptomycin (to select recipient cells) and either tetracycline, chloramphenicol, or kanamycin to identify acquisition of the TetR, CamR, or KanR conferred by the transferred F′ factor.
To measure mating efficiency in liquid, donor and recipient cells were grown to midlog phase in LB, then mixed at a 1:10 ratio and allowed to mate for 30 min at 37° without shaking. Serial dilutions of the mating mix in NCE were then plated on selective media as described above for assay of mating on solid medium.
Determination of growth rates
Cells were grown in NCE medium supplemented with 10 µg/ml thiamine and a carbon source at 37°. Optical density (550 nm) was measured at constant time intervals for 16 hr using a Synergy HT Multi-Detection Microplate Reader (BioTek Instruments). Growth rates were determined using the slope of the kinetic curve in exponential phase.
Plasmid copy number
Genomic DNA from E. coli cells grown overnight in glycerol minimal medium was isolated using the Qiagen DNeasy Blood and Tissue Kit according to the manufacturer’s instructions. The relative copy number of the pck and lacZ gene was determined by quantitative PCR using an ABI 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA). Amplification reactions, containing SYBR Green PCR master mix (Applied Biosystems) and primers (listed in Table S2), were performed in a 384-well microtiter plate. All DNA samples were amplified in four independent reactions. The cycling conditions were: 50° for 2 min and 95° for 2 min, followed by 40 cycles of 95° for 15 sec and 60° for 1 min, and then 95° for 15 sec, 60° for 15 sec, and 95° for 15 sec. Relative gene copy numbers were determined using the SDS RQ manager v 1.2 software (Applied Biosystems).
Data availability
Strains generated in this study are available upon request. Table S1 in File S1 contains genotypes of the strains. Primer sequences are available in Table S2 in File S1. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7051757.
Results
Plasmid transfer under selection
The importance of plasmid transfer was first suggested by the severe reduction in revertant yield caused by mutations or experimental conditions that reduced conjugation (Galitski and Roth 1995; Radicella et al. 1995; Peters et al. 1996; Godoy and Fox 2000). Later, supporters of stress-induced mutation rejected the importance of plasmid transfer (Foster and Trimarchi 1995a,b) because revertant cells isolated from mixtures of genetically marked tester strains rarely show plasmid transfer. If mating were essential, it was reasoned that half of the revertants would be initiated by genetically distinct tester pairs and show transfer. The fact that only ∼4% showed transfer was interpreted as evidence that transfer is not required (Foster and Trimarchi 1995b). Unfortunately, no controls were done to test the frequency of plasmid transfer between tester cells not involved in reversion. In addition, these tests were done without scavenger cells, so selection was less stringent than reversion.
To test mating under reversion conditions, genetically marked tester cells were plated on lactose medium containing a 10-fold excess of scavenger cells to consume trace nutrients. Two tester strains (a and b) were plated at a ratio of 1:1 (108 cells of each) with a 10-fold excess (109) of scavenger cells. One strain (a = TT27274) carried a chromosomal RifR marker and a F′lac plasmid with TetR-, CamR-, and KanR-resistance determinants. The other tester strain (b = TT27037) carried a chromosomal StrR marker and a plasmid with no resistance determinants. Scavenger cells (TR7177) were used to prevent both growth and reversion on lactose. These cells carried an F′lac plasmid with a lac deletion and no resistance markers. The 1:1:10 cell mixture (a:b:scavenger) was plated on selective lactose plates and incubated for 5 days. Cells from Lac+ revertant colonies were tested for evidence of plasmid transfer between strains (a) and (b). Cells from the nonrevertant lawn were tested for evidence of plasmid transfer from strain (a) to strain (b), generating StrR cells that have acquired TetR, CamR, or KanR.
Transfer between F’-bearing strains under starvation conditions proved to be very rare (10−4–10−5). This is expected since mating is reduced by the plasmid exclusion systems (Novick 1969). In contrast, the Lac+ revertant colonies showed 100-fold more frequent transfer (3.6% of 280 late Lac+ revertants analyzed). The Lac+ revertants that revealed transfer were RifR and carried a revertant lac+ plasmid with no resistance determinants (60%) or with only one or two resistances (40%). This suggests that reversion was associated with transfer of an F′lac plasmid from a StrR tester (no plasmid resistances) to a RifR tester with occasional recombination between donor and recipient F′lac plasmids. The 100-fold higher frequency of revertants in cells showing plasmid transfer suggests an association between reversion and plasmid transfer that was not noted in previous tests.
The frequency of transfer among revertants in the above experiment (3.6%) is comparable to the 3.7% found previously (Foster and Trimarchi 1995b). From this seemingly low frequency of transfer, Foster and Trimarchi concluded that mating was not required for reversion. However, they did not test mating between F′lac tester cells under stringent selection conditions and thus failed to see that the frequency of transfer in Lac+ revertants was 100-fold higher than in parent lac cells. We interpret the association between reversion and plasmid transfer as support for the new model in which transfer is essential to initiate reversion.
The new model suggests that all revertants in a Cairns–Foster experiment arise during plasmid transfer, but this transfer is usually not apparent because it occurs between the identical daughters of a single initiator cell. This model predicts that the few revertants showing transfer between different genetically marked cells probably result from rare matings between different plated parent cells that happen to contact each other on the selection plate. Such contact is rare because only ∼105 initiator cells of each genotype type were plated. Thus, the model predicts that mating between plated cells is rare and the majority of revertants arise during mating between the genetically identical daughters of a single plated initiator cell. This transfer cannot be detected genetically.
Another way to explain the rarely detected transfer seen in Lac+ revertants is to propose that reversion occurred during mating between daughters of a single tester type but was later followed by transfer of the completed lac+ plasmid into a nonrevertant tester with a different genotype. If this occurred, one would expect the Lac+ revertant colony to be a mixture of cell types, some carrying the revertant plasmid in a cell with the donor genotype and some in a cell with a recipient genotype. This is not seen. The revertant colonies with a transferred plasmid are essentially homogeneous. Most revertant cells carry a revertant F′lac+ plasmid from the donor in a cell with the recipient genotype. This finding was previously reported by Foster and Trimarchi (1995a). It suggests that reversion occurs during transfer between cells as suggested by the new model. Evidence of plasmid transfer was also pursued by examining the scavenger cells.
In a standard Cairns–Foster reversion experiment, a few of the Lac+ revertants (4–8%) are scavenger cells that carry a plasmid with a revertant Lac+ allele (Foster and Trimarchi 1995b; Rosenberg et al. 1995). Since plated scavenger cells carry a plasmid with a nonrevertable lac deletion, any lac+ revertants found in this genetic background must have experienced plasmid transfer. The new model predicts that revertant scavengers form during transfer. That is, a mutant F′lac plasmid is transferred from a plated initiator into a scavenger cell, where it recombines with the recipient plasmid and initiates reversion. This mating is rare because scavenger cells, like standard tester cells, lack the energy to support mating. However, scavengers are present at a 10-fold excess, making mating more likely.
It is also possible that reversion occurs first by mating between daughters of one tester initiator cell, after which a fully revertant lac+ plasmid is transferred to a scavenger. Again, this would predict scavenger revertant colonies that include a mixture of chromosomal genotypes, some donor type and some recipient type. In practice, however, scavenger revertant colonies include almost exclusively cells with the chromosomal genotype of a scavenger, as expected for reversion during transfer.
The origin of scavenger revertants was examined using a recA mutation in tester or scavenger cells. If a fully revertant plasmid forms by mating between daughters of one tester cell or between a tester and a scavenger cell, the cell in which reversion occurs must be capable of homologous recombination (RecA+), which is essential for reversion under selection. The revertant plasmid could later transfer into either a recA+ or recA− scavenger since simple plasmid transfer does not require recombination.
Figure 2 shows standard reversion experiments with a recA mutation in either testers or scavengers. Only recA+ testers showed a full yield of Lac+ revertants and this yield is unaffected by a recA mutation in the scavengers. With a recA+ tester, very few of the total revertants (6.5 or 3.7%) carry the lac+ plasmid in a scavenger cell and a recA mutation in the scavenger does not reduce this percentage. Formation of these few scavenger revertants must involve F′lac+ plasmid transfer since the original scavenger cell carried a plasmid with a lac deletion, which cannot revert.
Figure 2.
Effect of recA mutations on acquisition of a revertant lac+ allele by scavenger cells. The graph describes a standard reversion experiment using tester and scavenger cells with or without a recA mutation. Tester cells are either TT27274 (recA+) or TT23247 (recA−), both of which carry a chromosomal rifampicin-resistance (RifR) mutation. Scavenger cells are either TR7177 (recA+) or TT23248 (recA−), both of which are RifS and carry an F’ plasmid with a lac deletion. The table shows the percentage of revertants with their F′lac+ alleles in a scavenger cell. Revertants (Lac+) in tester cells are RifR and those in scavenger cells are RifS. Following the percentage of Lac+ revertants in scavenger cells is the number of day 5 Lac+ revertants tested for antibiotic resistance, as described in Materials and Methods.
Tester cells with a recA mutation cannot form a Lac+ plasmid under selection and thus cannot complete a revertant plasmid that might be transferred to a scavenger. As seen in Figure 2, a recA mutant tester produces very few revertants, regardless of whether the scavenger is rec+ or recA−, but the fraction of revertant alleles found in recA+ scavenger cells increases to 90%. Thus revertants appear to form during the process of transferring a mutant plasmid from a tester initiator cell (where revertants cannot form) into a scavenger, as suggested in the new model. This dependence on RecA is seen (Figure 2) in that very few revertants appear when the tester is recA, and the vast majority of those residual revertants form in recA+ scavenger cells. That is, when the tester cell is recA, formation of scavenger revertants requires RecA+ function in the scavenger suggesting that reversion occurs after transfer of a mutant plasmid from a recA− tester into a recA+ scavenger.
When RecA function is absent from both tester and scavenger cells, revertants arise at a very low rate (2–3 per 108 plated cells after 5 days). This low revertant yield is about the same as that seen (with or without RecA) when the mutant lac allele is in the chromosome and reversion cannot be enhanced by selection (Foster and Trimarchi 1995a). This suggests that enhancement of reversion under selection requires both transfer and recombination.
Evidence for homologous recombination between plasmids during F’ × F’ mating
In the new model, recombination between donor and recipient plasmids initiates rolling-circle replication (see Figure 1C). Occasional recombination between plasmids was seen during the transfer tests described above and was confirmed in the scavenger revertant tests. To test recombination between plasmids more directly, crosses were performed under starvation conditions. As described above, donors carried a chromosomal RifR mutation and a plasmid that conferred resistance to other antibiotics (see Figure 3). Recipients carried a chromosomal StrR mutation and a plasmid with no resistance markers. The strains were mixed at a 1:10 ratio (donor:recipient) and plated together on selective lactose medium, where neither could grow. After 1 day at 37°C, the lawn was sampled and cells were plated on rich medium containing streptomycin to select for the recipient chromosomal resistance, and a second antibiotic to select one of the donor plasmid resistance determinants (see Figure 3). Recipient cells that received the selected donor plasmid marker (∼10−4/donor cell, as described above) were then tested by replica-plating for the presence of other plasmid marker(s).
Figure 3.
Recombination between F′lac plasmids following transfer under starvation. (A) The cross used a RifR donor strain TT27043 with two plasmid markers (TetRCamR) and a StrR recipient with no plasmid drug markers. The RecA+ recipient (left column) was TT27037 and the RecA− recipient (right column) was TT27048. (B) The donor strain TT27274 carried three plasmid markers (RifR/F’ TetRCamRKanR) and the RecA+ recipient TT27037 carried a chromosomal StrR and no plasmid markers (StrR/F’ no resistance). Recipient cells (StrR) that acquired one F′lac marker were selected and then tested for the other donor marker(s). The number n indicates the number of transconjugants analyzed.
As seen in panel A of Figure 3, the recA+ recipient cell (left column) frequently acquired both donor plasmid markers, but in 12 or 23% of the selected crosses, recombination occurred between the TetR and CamR markers and only one was inherited. The exchanges in panel A reflect homologous recombination since virtually no such exchanges occurred using the isogenic recA recipient (right column). In these experiments, the recipient was plated at a 10-fold excess over donor and transfer of the selected marker is only about twofold lower in RecA− than in RecA+ recipients (data not shown). Panel B describes a cross using a recA+ donor with three plasmid markers and a recA+ recipient with no plasmid markers. As in panel A, roughly 10–20% of transconjugants showed internal recombination within the plasmid. That is, the starvation conditions of a standard Cairns–Foster reversion experiment (RecA+) support frequent homologous recombination between donor and recipient plasmids. The RecA− conditions that eliminate plasmid–plasmid recombination in this experiment (panel A) also eliminate reversion in the standard reversion experiment. The plasmid recombination events observed during mating between two F’ cells (F’ × F’) resemble those formed following chromosome transfer from an Hfr strain to an F− recipient (Lloyd and Buckman 1995).
Effects of tra mutations on transfer and reversion under selection
Classic experiments demonstrated that the ability of F’ plasmids to transfer requires the products of the multiple plasmid tra genes [reviewed by Frost et al. (1994)]. Transfer (Tra) functions are also needed for reversion in the Cairns–Foster system (Foster and Trimarchi 1995a; Galitski and Roth 1995; Radicella et al. 1995; Peters et al. 1996). While the importance of Tra functions for reversion is widely accepted, supporters of stress-induced mutagenesis later rejected the importance of actual transfer, suggesting that conjugative functions contributed only by making nicks in DNA that lead to double-strand breaks when vegetative replication is occasionally initiated (Foster and Trimarchi 1995a). The tests that led to this conclusion were not done under actual starvation conditions and included no controls to show the frequency of transfer between nonrevertant cells (see first paragraph of Results.).
The effects of tra mutations on plasmid transfer and reversion were retested under the selection conditions of the Cairns–Foster experiment. In plasmid transfer tests, donors were tra mutant Lac− tester strains with a RifR mutation in their chromosome and either a TetR or CamR marker in their F′lac plasmid. Recipients were Lac− testers with a chromosomal StrR mutation and an F′lac plasmid with no added markers. Donors (RifR) and recipients (StrR) were mixed at a 1:10 ratio and plated together on standard lactose reversion medium where neither can grow but both can use their leaky lac alleles to extract minimal energy from lactose. After 1 day at 37°, lawn cells were removed from the lactose plate and replated on rich medium with streptomycin (to select recipients) and either tetracycline or chloramphenicol (to select cells that acquired a donor plasmid). Conjugation efficiency was defined as the number of StrR recipient cells that had acquired either TetR or CamR normalized to the number of RifR donor cells.
Figure 4 describes mating on solid medium of strains with a tra mutation in either the donor or recipient. Previous tests of tra mutants were done in liquid medium with the tra mutation in the donor and no F or F’ plasmid in the recipient. All the strains in the lac reversion experiment carry an F′lac plasmid, and matings involve crosses on solid medium between two F’-carrying strains. Unlike the highly efficient crosses between an F’ donor and F− recipient cells, plasmid transfer between two F’-bearing strains under reversion conditions is very inefficient (6 × 10−4 per donor cell). Most tra mutations reduce mating efficiency and have their effect only when carried by the donor. See traI, traD, traG, and traN in Figure 4.
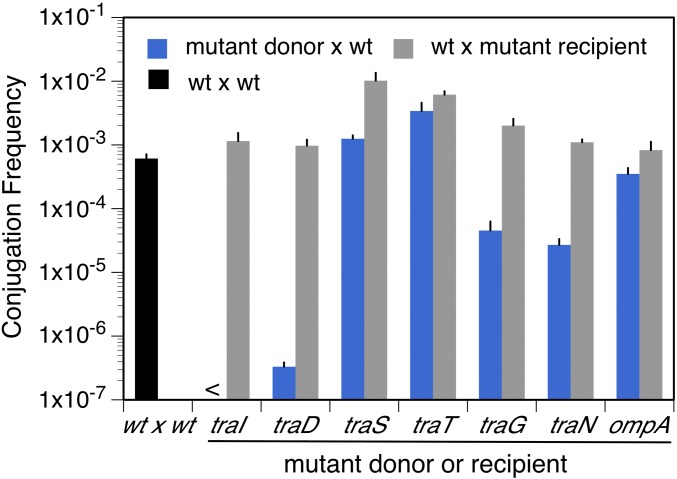
Figure 4.
Effect of tra and ompA mutations on plasmid transfer between F’-bearing strains on solid lactose medium. The conjugation frequency was measured between Lac− cells on lactose medium where neither cell can grow (see Materials and Methods). Donor and recipient cells were plated at a 1:10 ratio (108:109) on lactose and incubated for 1 day at 37°C. Conjugation frequency is defined as the number of recipients that have received a plasmid (StrR TetR or StrR CamR) divided by the total number of donor cells (RifR TetR or RifR CamR). Each number presented in the graph corresponds to at least three measurements from independent crosses ± SEM (error bars). The tra+ donor strains [wild type (wt)] are TT26307 (RifR/F′lac TetR) or TT27083 (RifR/F′lac CamR) and the tra+ recipient (wt) is TT27037 (StrR/F′lac), respectively. Each mutation tested is indicated near the x-axis, and a complete description of the genotypes is given in Table S1. The mutant donor strains used are TT27061 (traI), TT27089 (traD), TT27093 (traS), TT27092 (traT), TT27091 (traG, Tn10d-Tet insertion in the distal part of the gene), TT27090 (traN), and TT27066 (ompA). The recipients are TT27062 (traI), TT27095 (traD), TT27099 (traS), TT27098 (traT), TT27097 (traG), TT27096 (traN), and TT27067 (ompA). < indicates a conjugation frequency <10−7 transconjugants/donor cell. The black bar indicates mating between Tra+ donor and recipient cells.
Exceptional behavior is seen for genes controlling the plasmid exclusion system. The traS mutation stimulates mating 17-fold when carried in a recipient F’ plasmid and the traT mutation, stimulates mating 6- to 10-fold when carried by either the donor or recipient. Figure 5 shows that these mutations have parallel effects on reversion and transfer. The roles of these Tra proteins on transfer and reversion are described below. OmpA will be discussed later.
Figure 5.
Effect of tra and ompA mutations on lac reversion under selection. (A) Total numbers of accumulated Lac+ colonies per 108 cells plated at day 0 are shown for strains TR7178 (wild-type tra+ ompA+, solid circles), TT27084 (traD, open squares), TR7319 (traI::Tn5, solid triangles), and TT26893 (traI, solid inverted triangles). Lawn growth is shown for all strains except TR7319 (traI::Tn5), which was not determined. (B) Revertants and lawn growth for strains TT27088 (traS, open inverted triangles), TT27087 (traT, solid squares) and TR7178 (wild type tra+ ompA+, solid circles). (C) Strains TT27085 (traN, open diamonds), TT27086 (traG, open triangles), TT27065 (ompA, solid diamonds) and TR7178 (wild-type tra+ ompA+, solid circles). Each data point represents mean ± SEM (error bars) of at least eight independent cultures of each strain from two or more experiments. Lawn growth is the relative number of viable Lac− cells determined by sampling agar plugs removed from selection plate every day from day 0 to day 5. The cell number was normalized to the first day’s count for each strain. Values are means ± SEM for data from at least six samplings from independent selection plates.
Roles of Tra proteins in conjugation
Activities of the bifunctional TraI protein:
The N-terminal portion of the TraI protein has a single-strand endonuclease (nickase) activity that cleaves a site within oriT, the origin of plasmid transfer replication. The C-terminal region of TraI has an ATP-dependent 5′ to 3′ helicase activity that separates the 5′ end from the uncut strand at the nick (Traxler and Minkley 1988). TraI nicks DNA at oriT as part of a protein complex (relaxosome) that includes TraY and TraM proteins and the host factor IHF (Di Laurenzio et al. 1992). The nick made by TraI is a reversible trans-esterification in which the phosphate at the 5′ end forms a covalent bond with the hydroxyl group of Tyr22 of the TraI protein (Matson et al. 1993; Lanka and Wilkins 1995). The plasmid DNA remains at equilibrium between a cleaved and uncleaved state until the coupling protein TraD interacts with the relaxosome and initiates transfer of the 5′ end (Lanka and Wilkins 1995). When a donor and recipient cell form a mating pair, TraM protein stimulates movement of the single-strand DNA end to the membrane and induces a change in TraI conformation so that its helicase domain loads at the nick site [reviewed in de la Cruz et al. (2010)].
The DNA unwinding activity of the TraI helicase and the TraD ATPase act together to transport a single-strand DNA loop across the cell envelope (de la Cruz et al. 2010). TraI mutants are essentially devoid of transfer ability (Figure 4) and are extremely defective in reversion under selection (Figure 5A).
A particular traI::Tn5 insertion eliminates only the helicase, but leaves nickase intact (Carter et al. 1992). This insertion mutant lacks both transfer and reversion ability, suggesting that reversion can be eliminated without loss of nickase. As for most traI mutants, strains lacking traD function are defective in both transfer and reversion (Figure 4 and Figure 5A). The TraD motor protein brings plasmid DNA to the conjugation pore, but does not affect nicking (de la Cruz et al. 2010). In the absence of TraD, cells still nick DNA, make F-pilus structures, and form mating aggregates (Kingsman and Willetts 1978; Everett and Willetts 1980). The primary TraD defect is in moving DNA into the recipient. Thus, mutants lacking either the TraI helicase or the TraD motor protein lack reversion ability but both are still able to nick DNA. This is contrary to the idea that conjugation functions contribute to reversion only by making DNA breaks (Foster and Trimarchi 1995a; Rosenberg et al. 1995). Reversion appears to require transfer of a plasmid copy between cells as posited by the new model.
Effects of traT and traS mutations:
The TraT and TraS proteins are part of a mechanism that minimizes mating between two male cells (F’ × F’), so these proteins inhibit an activity that is required for reversion according to the new model. The TraT protein prevents interaction between the donor membrane protein TraN and the recipient outer membrane protein OmpA, and thus act to decrease stability of mating aggregates (Minkley and Willetts 1984). The TraS protein is located in the recipient inner membrane, where it interacts with TraG to block the transfer of DNA into an F’-bearing recipient (Audette et al. 2007). The TraS protein of the recipient has the biggest effect on reduction in mating and reversion. That is, normal TraT and TraS proteins act together to reduce transfer between F’ strains (F’ × F’) ∼100-fold (Achtman et al. 1977).
Loss of traS or traT functions is thus expected to stimulate plasmid transfer between two F’ cells. This is seen in Figure 4, which shows that each single mutation increases transfer frequency at least 2- to 17-fold. This increase in transfer frequency parallels an increase in the yield of Lac+ revertants (Figure 5). The revertant yield increases ∼1.5-fold in a traT mutant and 2.5-fold in a traS mutant. Thus, two mutations that enhance interactions between donor and recipient cell surfaces also stimulate both plasmid transfer and reversion under selection. Neither traS nor traT mutations are expected to affect nicking of plasmid DNA.
Effects of traN, traG, and ompA mutations:
Before plasmid transfer is initiated, cells form mating pairs or aggregates that consist of two or more tightly associated cells (Achtman 1975). These close contacts facilitate formation of mating pores or DNA conduits. In liquid medium, a donor cell pilus filament first adheres to the surface of a recipient cell. A donor cell moves closer to a recipient by retracting a pilus bound to the recipient cell. The N-terminal domain of the donor TraG protein is involved in pilus biosynthesis (Manning et al. 1981; Firth and Skurray 1992), while the C-terminal end interacts with the recipient LPS and OmpA outer membrane protein to stabilize a mating pair (Manning et al. 1981; Firth and Skurray 1992; Klimke and Frost 1998). Under normal mating conditions in liquid medium, the stabilizing effect of TraN, TraG, and OmpA proteins appears to overcome shear forces that act to separate pairs. These proteins facilitate formation of the tight associations that generate pores through which DNA moves.
A traN and traG mutation in the donor plasmid or an ompA mutation in the recipient chromosome severely reduces transfer from F+ to F− strains in liquid medium (>5000-fold) (Manning et al. 1981; Manoil and Rosenbusch 1982; Firth and Skurray 1992; Maneewannakul et al. 1992). However, the liquid mating defect of traN and traG (C-terminal domain) mutants is corrected ∼100-fold on solid medium such as that used in a reversion experiment (Manning et al. 1981; Manoil and Rosenbusch 1982). The increase in transfer ability on solid medium can be seen in Figure 4 by comparing the transfer ability of traN, traG, and ompA mutants to those of traI and traD mutants, which are completely transfer defective in both liquid and solid medium. Note that on solid medium the transfer ability of traN and traG mutants rises to about a 10th that of wild type. The transfer ability of an ompA mutant increases on solid medium to equal that of wild type (Figure 4).
Given the ability of ompA mutants to form mating pairs on solid medium (Figure 4), it is unclear why these mutants fail to produce lac+ revertants on selective plates (Figure 5C). On solid medium, traN and traG mutants show a 10-fold defect in transfer and a revertant yield that is about one fourth that of wild type. The ompA mutants are completely normal for mating on solid medium, but still extremely defective in reversion. These reversion phenotypes may be explained by the new model, which suggests that mating pairs or aggregates may allow continuous DNA transfer for several days on the selective plate. Thus, traN and traG mutants may form pairs on solid medium and allow nicking and DNA synthesis in the donor cells (Kingsman and Willetts 1978), but their mating pairs may be too unstable to allow the long-term transfer required for a full revertant yield under selection. Below, we will suggest two roles for the OmpA protein.
OmpA contributes to reversion during the selection period
In liquid medium, the OmpA protein (like TraG and TraN) is thought to help nascent mating pairs resist shear (Manoil and Rosenbusch 1982). On solid medium, the traG and traN mutants mate somewhat better but still show a defect in both transfer and reversion. The ompA mutants mate as well as wild type on solid medium, but they still show no reversion on a selection plate. This raises the question of how an ompA mutation blocks reversion on solid medium if it does not impair plasmid transfer. We suggest two roles for OmpA. First, it may be required for the long-term persistence of mating pairs under selection. Second, it may help cells conserve energy as described in the last section of Results.
The reversion process may require mating pairs to persist on the plate and continue transfer replication for several days. This long-term pair stability may require the OmpA protein. That is, cells may require OmpA to form mating pairs in liquid, but not on solid medium. However mating pairs that form on solid medium without OmpA may fail to continue transfer replication for the 5–6 day period of a reversion experiment. This idea predicts that OmpA protein function might be required throughout the several days of selection.
To test the time at which OmpA contributes to reversion, a standard chromosomal ompA gene was placed under control of a repressor that normally regulates transcription of the divergent tetA and tetR genes within transposon Tn10. The ompA gene was fused to the tetA operon such that transcription is induced by tetracycline or by its nontoxic analog AnTet. This foreign control of OmpA production was demonstrated by staining cells with Congo Red dye which detects the increase in extracellular cellulose production that is caused in the absence of the outer membrane protein OmpA (Ma and Wood 2009). Induced expression of an ompA+ allele by AnTet reduces Congo Red staining (Figure 6A).
Expression of OmpA from the tetA promoter allowed both F− and F’ cells to serve as recipients in liquid crosses (Figure 6B). A strain with a tetA-ompA fusion operon inserted at its chromosomal ompA locus could serve as a conjugational recipient only if ompA expression was induced by the tetracycline analog AnTet. Figure 6B describes the mating of a donor with an F′lac plasmid carrying a CamR-resistance marker. The recipient strain has a chromosomal SmR marker and either no F plasmid or an F′lac plasmid with no resistance marker. Cells were allowed to mate in liquid LB medium and then plated on rich plates with streptomycin and chloramphenicol to select recipient cells that had received the donor plasmid. Notice that in Figure 6B the presence of an F′lac plasmid in the recipient reduces transfer efficiency ∼100-fold (due to TraS and TraT functions). Deletion of the recipient ompA gene reduces transfer by roughly four orders of magnitude, to a level approximating that of a strain with an uninduced tetA-ompA transcription fusion. Induction of OmpA production by AnTet fully corrects this transfer defect.
The effect of OmpA on reversion was assessed using two tester strains, one ompA+ and the other with the chromosomal ompA+ replaced by a tetA-ompA fusion. Figure 6C shows that the normal revertant yield of ompA+ strain is unaffected by addition of inducer added at any time in the experiment. Figure 6D shows that a tetA-ompA fusion strain is nearly as defective as an ompA-deletion mutant. Reversion is restored to this strain when AnTet is included to the selection medium, suggesting that OmpA protein is necessary during the course of revertant development. Providing AnTet only during nonselective pregrowth stimulates reversion slightly, probably because OmpA is an abundant outer membrane protein that is expected to persist for several divisions after removal of inducer. We conclude that OmpA is not needed during pregrowth or the initial selection period, but is essential for later development of revertants under selection. This could reflect a role in long-term stabilization of mating pairs or in energy conservation, as described in the last section of Results.
Reversion and lawn growth in a recD mutant is reduced by both ompA and traI mutations
While both ompA and traI mutations reduce reversion in the Cairns–Foster system (see Figure 5), their effect is even more impressive in recD mutants, which have a 30-fold higher revertant yield and more extensive lawn growth than wild-type testers (Figure 7). The ompA mutation eliminates reversion and lawn growth on lactose both with and without scavengers. A traI mutation reduces reversion in recD but has a minimal effect on lawn growth. Interpreting these results requires understanding the role of RecD.
Figure 7.
Effect of ompA and traI mutations on reversion and lawn growth in a recD mutant. Total cumulative numbers of Lac+ colonies per 108 cells plated (A) in presence of scavenger cells or (B) without scavengers. The strains used are TR7178 (wild-type tra+ ompA+ recD+, solid circles), TT27058 (recD, solid squares), TT27113 (recD traI, open inverted triangles), TT27112 (recD ompA, solid triangles), TT27111 (ompA, solid diamonds), and TT26893 (traI, solid inverted triangles). There are no data for the recD strain plated without scavengers on lactose medium because the tester cells form a heavy blue lawn and no individual Lac+ colonies are visible. Each data point represents mean ± SEM (error bars) of at least six independent cultures of each strain from two or more experiments. Lawn growth is the relative number of viable lac– cells determined every day from day 0 to day 5. The cell number was normalized to the first day’s count for each strain. Values are means ± SEM for data from at least six samplings from independent selection plates.
The RecD protein is part of the RecBCD helicase-nuclease complex, which acts at a double-strand DNA end to unwind and then degrade both strands. After this complex contacts a chi site in DNA, it continues separating strands and degrading the 5′-ended single strand, leaving a 3′ overhang that is handed off to the strand exchange protein RecA, which initiates double-strand-break repair (Kowalczykowski 2015). In the absence of RecD, the RecBC protein separates strands from each other, but does not degrade the 5′ end or produce the 3′ overhang needed for recombination. The 3′ overhang is produced when the nuclease RecJ degrades the 5′ single-strand end. Repair is then initiated when RecA protein uses the 3′ end to invade a template duplex. Thus, in the absence of RecD, the time needed for RecJ to create the 3′ overhang delays double-strand-break repair and allows plasmid replication to proceed, increasing the F′lac plasmid copy number (Foster and Rosche 1999). The increased yield of revertants seen in a recD mutant has been attributed to the higher plasmid copy number and consequent increase in residual growth of the tester cell population (Foster and Rosche 1999). In the light of the new model, presented here, the increase in revertant yield could also result from reduced degradation of double-strand breaks and an increase in recombination between transferred material and the recipient plasmid.
Since traI and ompA mutations strongly reduce reversion in a recD strain, which has a high plasmid copy number, it seemed possible that these mutations might act by reducing plasmid copy number. This was tested by quantitative PCR, as seen in Figure 8. The standard F′lac copy number is one to two times that of the chromosome, while a recD mutation increases plasmid copy number about fivefold. Removal of TraI nickase or OmpA in an otherwise normal strain has no effect on copy number.
Figure 8.
Effects of ompA, traI, and recD mutations on F′lac copy number. Cells were grown to stationary phase in glycerol minimal medium and allowed to complete all rounds of replication. The F’ plasmid copy number was estimated by quantitative PCR of a segment internal to the lacZ gene and to the chromosomal pck gene. F’ plasmid copy number is expressed as relative copy number of lacZ to the reference pck gene. A wild-type (wt) E. coli strain (TR6968) with lacZ at its normal chromosomal position was used as control and set to one copy per cell (black bar). The strains carrying F′lac are TR7178 (wild-type rec+ tra+ ompA+), TT27058 (recD), TT26893 (traI), TT27113 (recD traI), TT27065 (ompA), and TT27112 (recD ompA). The error bars indicate the variations in relative copy number, with confidence level set at 95%.
The DNA nicks made by TraI at oriT are essential for plasmid transfer, but do not seem to increase plasmid copy number (Figure 8). TraI nicks may not affect copy number because they are rarely made or are rarely converted to double-strand breaks. When DNA is nicked at oriT, the 5′ single-strand end is covalently attached to the TraI protein. This attachment is reversible so, in the absence of transfer, the nick may be rehealed before a replication fork converts it to a break (Matson et al. 1993; Lanka and Wilkins 1995). In a recD mutant, the small drop in plasmid copy number caused by a traI or ompA mutation may reflect the small contribution of conjugation to DNA breaks. Therefore, it seems likely that the fivefold increase in plasmid copy number seen in a recD mutant results from spontaneous nicks and breaks that form at many positions, independent of conjugation functions. These results argue against the idea that TraI activity at oriT is a major source of nicks and breaks for stimulating reversion, as proposed by stress-induced mutation models (Foster 2007; Galhardo et al. 2007). Consistent with the new model, the drop in revertant number caused by a traI mutation in a recD mutant is probably due primarily to reduced transfer, not to a reduced plasmid copy number.
An ompA mutation lowers plasmid copy number only 25% in a recD mutant (Figure 8). Thus, the extreme reduction in Lac+ revertant yield caused by an ompA mutation seems unlikely to reflect reduced plasmid copy number (Figure 7). Since ompA mutations neither impair mating on solid medium nor reduce plasmid copy number, it seems that there must be a different reason for the striking reduction they cause in revertant number and lawn growth.
As seen in Figure 7A, a recD mutant produces 30-fold more revertants and shows roughly 15-fold more extensive lawn growth than wild-type cells under standard selection conditions. The increased number of revertants has been attributed to more growth of the tester population allowed by higher plasmid copy number (Foster and Rosche 1999). While the higher lac plasmid number may increase lawn growth slightly, we suggest that its huge effect on revertant number is due primarily to increasing the concentration of initiator cells in the pregrowth culture. Evidence for this conclusion is seen in the 100-fold drop in yield caused by adding a traI mutation to the recD background. The large TraI effect suggests that mating is required for the increased revertant number seen in a recD mutant. This large effect of a traI mutation on reversion in a recD mutant was reported previously, but was not interpreted in terms of plasmid transfer (Foster and Rosche 1999). Thus, a recD mutation seems to stimulate reversion as part of a process that requires plasmid transfer as proposed by the new reversion model. We suggest lack of RecD may increase the level of initiator cells and may increase the rate of recombination between plasmids.
Viewed in another way, a simple traI mutation essentially eliminates reversion (see Figure 5A and Figure 7A), but this reduced revertant yield is raised fivefold by an added recD mutation (Figure 7A). We suggest that when conjugation is blocked in a traI mutant, the small improvement provided by a recD mutation is due to increased growth as suggested previously (Foster and Rosche 1999). The effect of a recD mutation on lawn growth is most striking in the absence of scavengers as seen in the lower part of Figure 7B (compare reduced growth of the traI strain to that of the recD traI double mutant). We suggest that the extremely high revertant yield seen in a simple recD mutant (Figure 7A) is due primarily to an increase in initiator cell number caused by increased plasmid copy number. The small contribution from improved growth is more apparent in the absence of plasmid transfer.
Thus the recD mutation seems to increase revertant yield in two ways. First, it creates more initiator cells prior to selection, and second, it allows improved growth of the plated population to grow under selection. Surprisingly, both effects of a recD mutation are reversed by an ompA mutation, which eliminates both reversion and lawn growth under selection. Again the effect on lawn growth is most striking without scavengers as seen in the bottom panels of Figure 7, A and B (compare growth of recD to that of recD ompA double mutant). The OmpA protein thus seems to contribute to reversion in part by affecting the mating process, perhaps by extending the life of mating pairs. However the large effect of an ompA mutation on growth of a recD mutant under selection suggest that OmpA also helps cells extract energy from selection medium as described below.
The OmpA protein helps tester cells conserve energy
In considering how OmpA might contribute to energy for reversion, consider first how nongrowing cells extract energy from lactose under selection. Reversion in the Cairns–Foster system relies on the residual function of the mutant lac allele, which provides ∼1–2% of the β-galactosidase level seen in a fully induced lac+ strain (Cairns and Foster 1991). This residual function cannot support cell division on lactose (in the presence of scavengers) but is essential for reversion under selection (Andersson et al. 1998).
In stress-induced mutagenesis models, the energy provided by the residual LacZ function is used for the occasional firing of the plasmid vegetative origin, which converts nicks at oriT into double-strand DNA breaks and mutagenesis occurs during repair of these breaks (Foster and Trimarchi 1995a; Rosenberg et al. 1995). We argue above that this nicking makes little contribution to plasmid copy number and should have little effect on plasmid copy number without plasmid transfer. In the new model described here, multiple copies of the mutant lac allele provide energy to allow initiator cell division, conjugation between daughter cells, and plasmid over-replication after transfer. Many of the mutations that reduce revertant yield impair mechanisms required to amplify lac (e.g., the rec and tra mutations described above). One might expect therefore that revertant yield would also be reduced by mutations that limit the ability of cells to harvest energy from lactose metabolism or cause a waste of energy that could otherwise support reversion. We propose that OmpA helps conserve energy in nongrowing tester cells. Since OmpA is a major outer membrane protein, OmpA-deficient cells might be subject to envelop damage whose repair is energetically costly (Wang 2002).
To test effects of OmpA on energy metabolism, we first measured effects of ompA mutations on growth rate in liquid medium and found no statistically significant effect on growth of Lac− or Lac+ strains on glucose, galactose, lactose, or glycerol (data not shown). Similarly, Lac+ revertants arising in the Cairns–Foster system grow equally well on lactose with and without OmpA. However, ompA mutations did reduce residual growth of Lac− tester cells under selection conditions, suggesting that these tests are more sensitive. The recD mutation stimulated lawn growth ∼2.5- to 15-fold, with or without scavenger cells (Figure 7, bottom panels) and this growth was significantly reduced by an ompA mutation. As seen above, growth inhibition does not seem to result from reduced plasmid copy number. Thus, ompA mutations appear to reduce the ability of tester cells to derive or retain energy from lactose during starvation.
To understand how an outer membrane protein might affect sources of energy under selection, it is necessary to understand how the Cairns–Foster system manipulates metabolism and poises cells on the brink of growth. The LacZ activity splits lactose to glucose plus galactose. When wild-type cells grow rapidly on excess lactose, they use glucose preferentially and release galactose into the medium. However, in the Cairns–Foster selection system, the mutant lac allele is so severely compromised that starved mutant tester cells split traces of lactose and use both glucose and galactose to support reversion. Preventing use of galactose eliminates reversion in the Cairns–Foster system. This was seen first in Salmonella version of the system, where the residual LacZ activity allows testers to divide once per day; this growth and all reversion is prevented by blocking galactose metabolism with a galactose kinase (galK) mutation (Andersson et al. 1998).
In the E. coli system, residual growth is blocked by scavenger cells that are Lac− but can consume any excreted galactose. These scavenger cells essentially compete with testers for access to residual galactose. Growth of plated testers is prevented when the scavengers consume a critical portion of the galactose. We retested these effects in the E. coli system using a galK mutation to prevent use of galactose and reduce energy obtainable from lactose by about twofold (one of two sugars). When testers are GalK−, both reversion and residual lawn growth are eliminated (Figure 9).
Figure 9.
Effect of gal mutations on reversion in the Cairns–Foster experiment. The top graph presents the accumulation of Lac+ colonies per 108 plated cells seen in the tester strains TR7178 [wild-type (wt) gal+, circles], TT27330 (galK, triangles), TT27333 (galP, inverted triangles), TT27337 (mglB, squares), and TT27238 (galP mglB, diamonds). Scavengers used in 10-fold excess were galK+ (TR7177, solid symbols) or galK− (TT27331, open symbols). Each data point represents the mean ± SEM (error bars) of 10 independent cultures of each strain. The bottom graph shows the lawn growth of tester strains TR7178 (wt gal+, solid circles), TT27330 (galK, solid triangles), TT27238 (galP mglB, solid diamonds) plated with GalK+ scavenger cells and TR7178 (wt* gal+, open circles) plated with galK scavengers. The number of viable testers cells was determined every day from day 0 to day 5 and the cell number was normalized to the first day’s count for each strain. Lawn growth for TT27333 (galP) and TT27337 (mglB) was not determined. Values are means ± SEM for data from at least six samplings from independent selection plates.
In a standard reversion experiment, both tester and scavengers are Gal+ and compete for galactose (solid circles in Figure 9). When scavengers have a galK mutation, all galactose is left for use or reassimilation by the Gal+ testers. This stimulates reversion twofold by day 6 (open circles in Figure 9). Reassimilation of galactose by testers requires one of the multiple galactose transporters (Ganesan and Rotman 1966; Rotman et al. 1968; Wilson 1974). Most important is the high-affinity MglABCD transporter, whereas the low affinity GalP permease does not seem to contribute to reversion (Figure 9).
In summary, the Cairns–Foster selection poises cells on the brink of growth by setting up a competition between tester and scavenger cells for use of the traces of galactose produced from lactose by the residual activity of a mutant lacZ enzyme. This galactose can be used in the tester cell or it can escape to the outside medium. Escaped galactose can either be consumed by scavengers or reassimilated and used by the testers. We suggest that the OmpA protein may influence rates of galactose loss and reassimilation. That is, cells lacking OmpA may lose more galactose to outward diffusion or may be impaired for its reassimilation.
Discussion
The system of Cairns and Foster has been used to support the idea that cells might upregulate their genome-wide mutation rate in response to growth limitation (Foster 2007; Fitzgerald et al. 2017). We have argued against such programmed mutagenesis and favor a model in which natural selection acts on a population of replicating F′lac plasmids in cells that divide and replicate their chromosome very little (Maisnier-Patin and Roth 2015, 2016). The central feature of the new model is transfer of the mutant F′lac plasmid to an identical plasmid-bearing recipient, where recombination between plasmids initiates rolling-circle replication. This recombination is not mutagenic per se, but it initiates repeated plasmid replication, which provides energy, adds opportunities for mutation, and increases dosage of the dinB gene, encoding a mutagenic DNA polymerase. We suggest that in the Cairns–Foster system, selected increases in gene dosage (lac and dinB) give the appearance of purposefully directing mutagenesis to useful targets in a nongrowing cell population.
Resurrecting plasmid transfer as a central feature of reversion under selection
Reversion under selection in the Cairns–Foster system requires that the F′lac plasmid is able to transfer from one cell to another (Galitski and Roth 1995; Radicella et al. 1995; Peters et al. 1996; Godoy and Fox 2000). This suggested that the act of conjugation might be central to the reversion process. Supporters of stress-induced mutagenesis argued that transfer was not required and conjugation functions might be needed only to nick DNA at the plasmid transfer origin (oriT) (Foster and Trimarchi 1995a,b). These nicks could lead to breaks whose repair is mutagenic without involvement of transfer. In support of this idea, they showed that Lac+ revertants could form under selection without conjugation, when foreign nucleases were used to nick sites on the F′lac plasmid (Rodriguez et al. 2002; Ponder et al. 2005; Shee et al. 2011). The same enzymes had a very small effect on mutation when the nick is made near a chromosomal target gene, where rolling-circle replication is unlikely to accompany break repair (Shee et al. 2011). These experiments showed that enzymatically induced DNA damage could replace plasmid transfer and increase the frequency of selected Lac+ revertants on the F′lac plasmid (Ponder et al. 2005). Here, we provide evidence that plasmid transfer is still essential in the original Cairns–Foster system, but can be replaced by foreign nucleases that cut DNA more frequently.
Selection underlies reversion both in the original Cairns–Foster system (using conjugation) and when nucleases replace plasmid transfer. In both situations, recombination repairs DNA damage to the F′lac plasmid and initiates repeated replication of the included lac and dinB genes. Selection for local lac amplification directs mutagenesis to the over-replicated region. Reversion in the original Cairns–Foster system exploits an elegant, evolved DNA transfer system that makes occasional DNA nicks and directs a single DNA strand to a recipient, where it can initiate rolling-circle replication. The same result can be achieved without transfer by foreign endonucleases that make such frequent and repeated damage to the plasmid that transfer is not required.
The behavior of the Cairns–Foster system with induced endonucleases has been interpreted as evidence for mutagenic recombination and has been termed mutagenic break repair (Ponder et al. 2005). We submit the recombination per se is not mutagenic with or without conjugation. Instead recombination supports selected plasmid over-replication (rolling circle) during damage repair. Mutagenesis occurs because over-replication of the lac is accompanied by overproduction of the error-prone DinB polymerase. In both situations, it is the selection for more copies of lac that is responsible for the increase in mutation rate on the F′lac plasmid. These situations may be specific examples of the more general phenomenon of chromosome replication complexity, which elevates copy number of local genomic regions (Kuzminov 2016). Recombination can act on these regions to expand the copy number differences and increase the likelihood of mutations without increasing the error-rate of replication.
Conjugation in the Cairns–Foster system
Evidence presented here shows that the rare nicks made by TraI at oriT seldom lead to breaks in the absence of transfer. This may be because TraI actually catalyzes a reversible trans-esterification that attaches a 5′ DNA end covalently to the TraI protein, which conducts a plasmid copy into a recipient cell (Matson et al. 1993; Lanka and Wilkins 1995). In the absence of transfer, this trans-esterification (and nick) is reversible and the single-strand ends may not be subject to forming double-strand breaks. After transfer, the single strand is copied and subject to breaks, whose repair can initiate rolling-circle replication that amplifies both the lac and dinB+ genes.
In contrast to the frequent breaks made by the foreign nucleases, the rare nicking by TraI is not by itself sufficient for reversion, but can initiate the plasmid transfer that leads to plasmid over-replication under selection. Mating between F′lac initiator cells is extremely rare under selective conditions (10−4–10−5) as described above, but is 100-fold more frequent among revertants that arise from this population (Foster and Trimarchi 1995b; Godoy and Fox 2000). Several tra mutation types reduce revertant yield and block transfer without impairing ability to nick DNA at oriT. These include traD, traN, and traG mutations and the traI mutations that eliminate only the helicase (Kingsman and Willetts 1978). Conversely, both traS and traT mutations stimulate the frequency of reversion and mating, but neither is expected to affect nicking. Therefore, TraI nicking at oriT is not sufficient to stimulate reversion and does not correlate well with revertant yield.
Nicks made by TraI at oriT are essential to initiate transfer of F′lac into a recipient cell, where reversion then occurs during rolling-circle replication initiated by RecA-dependent homologous recombination between donor and recipient plasmids. Repeated transfer between members of one mating aggregate has been suggested before (Achtman et al. 1978; Peters et al. 1996; Godoy and Fox 2000). The amount of plasmid over-replication sufficient to produce a revertant may require a mating aggregate to continue transferring for several days, necessitating the pair stabilization provided by TraN, TraG, and OmpA (Arutyunov and Frost 2013).
Involvement of SOS and RpoS in the Cairns–Foster system
Formation of stable lac+ revertants in the Cairns system is seen only if both the lac and the dinB+ genes are located on the same F′lac plasmid (Slechta et al. 2003; Yamayoshi et al. 2018), where both genes can be over-replicated under selection. Induced transcription of the dinB+ gene by global regulatory systems SOS and RpoS is necessary but not sufficient for mutagenesis (Lombardo et al. 2004; Galhardo et al. 2009). In the new model described here, mutagenesis results from selected increases in the copy number of the F′lac plasmid with its dinB+ gene. We suggest that the global regulatory systems are needed only to assure a level of dinB transcription sufficient to cause mutagenesis, when the dinB gene is amplified with lac under selection.
How selection generates Lac+ revertants during plasmid transfer: later steps in the reversion process
The new model proposes that selection acts in several steps to favor localized over-replication of the growth-limiting mutant lac genes. Plasmid over-replication provides energy by increasing the copy number of the leaky mutant lac gene and increases the mutation rate by coamplification of dinB+. The combination of lac over-replication and plasmid mutagenesis increases the likelihood of a lac+ reversion event. The selective steps are as follows.
First, selection acts on rare initiator cells that have an unusually high (>10 copies) number of plasmid copies (Sano et al. 2014). The normal plasmid copy number averages between one and two (Frame and Bishop 1971), but may increase in individual cells due to occasional failures of the copy number control mechanism. Alternatively, the mechanism may dictate a plasmid copy number of 1–2 that is subject to stochastic variation from one cell to another. In either case, roughly 1 in 1000 plated cells has 10 or more plasmid copies. These rare initiator cells arise during nonselective pregrowth and initiate clones on lactose that give rise to lac+ revertant cells (Sano et al. 2014).
Second, selection prevents division of most of the 108 plated cells but allows the rare initiator cells (105) to divide on lactose. Cell division is allowed by multiple copies of the partially functional mutant lac allele (Slechta et al. 2003; Sano et al. 2014). The ability of initiator cells to divide on lactose is key to allowing the conjugation that is central to reversion.
The third selection step favors an increase in lac allele number that occurs following conjugational plasmid transfer between the daughters produced by division of a single initiator cell. The bulk of the plated population cannot mate due to a lack of energy. The plated rare initiator cells have enough energy and could, in principle, mate with each other, but rarely do so because these cells are unlikely to contact each other since only 105 are plated. However division of a single plated initiator cells can produce daughters that are able to mate with each other. Their energy problem is solved by a somewhat elevated number of plasmid copies. Their proximity problem is solved because daughters of a single initiator cell are located close together on the plate. The early tests of plasmid transfer could only detect transfer if independently plated initiator cells happen to contact each other (Foster and Trimarchi 1995a). Since few such cells are plated, they rarely contacted each other and mating was not detected. However daughters of a single initiator could contact each other and mate, leading to reversion.
The fourth selection step is the improved energy production caused by plasmid over-replication following transfer of an F′lac plasmid from one initiator daughter to another. Recombination between donor and recipient plasmids can initiate rolling-circle plasmid replication. Plasmid amplification provides energy by adding copies of the mutant lac allele. Plasmid over-replication also adds more dinB+ copies and increases the mutation rate. The model proposes that broken double-strand ends of the transferred plasmid are repaired by recombination with the recipient plasmid (see Figure 1, B and C). Normally, such a break would be repaired by convergent forks on a single plasmid template. Such repair replicates a very short region that is unlikely to include lac especially if initiated at oriT. However, when the recipient daughter cell has multiple F′lac plasmid copies, the broken ends of the donated plasmid are likely to recombine with different template plasmids and start unopposed replication forks that achieve rolling-circle replication of the entire recipient plasmid. This may be a specific example of recombinational misrepair that leads to creation of structures that can amplify under selection and provide local over-replication (Khan et al. 2016; Kuzminov 2016).
The fifth and final selection step is formation of a lac+ allele by a frameshift mutation occurring during repeated mutagenic rolling-circle plasmid replication. As soon as a stable lac+ allele forms, energy is restored, cells divide, and plasmid copy number drops to the normal 1–2 copies per cell. The new reversion model must still solve some numerological problems.
Linear accumulation of revertants
In the course of the Cairns–Foster reversion experiment, revertant colonies accumulate linearly with time above a nongrowing cell lawn. The bulk of the plated population neither divides nor mates under selection and is irrelevant to the reversion process. The critical replication occurs within the clones derived from initiator cells. If developing clones expanded by exponential growth, one would expect the number of revertant colonies to accumulate exponentially over time. That is, an exponentially increasing number of cells in each clone would be at risk to experience a reversion event. This is not the case. The number of stable revertant colonies increases linearly with time, suggesting that each clone includes a constant number of replicating lac alleles that are subject to reversion. Rolling-circle replication produces a linearly expanding number of lac alleles from a single lac template and explains the linear increase in lac copies on the plate. However, there is still the problem of cell division.
The new model proposes that the linear concatemer of plasmid copies produced by rolling-circle replication is cut to individual circular plasmids by the F-plasmid resolution system [reviewed by Summers (1996)]. Thus the number of individual plasmid copies increases linearly until it is sufficient to allow cell division (∼10 copies per cell). The first cell division after mating generates two daughter cells that may have very different prospects. One daughter inherits a small number of plasmids whose replication is blocked by the plasmid number control mechanism (Nordström and Austin 1989). This cell partitions its plasmid copies at each division. Ultimately copy number drops to the maintenance level (1–2) and further cell division is impossible: these cells become indistinguishable from the bulk of the plated population of noninitiator cells. The other cell produced by division following a mating can continue plasmid replication as suggested below.
Two ways to maintain a single replicating “mother cell”
We suggest that the linear production of revertants from a particular clone can be achieved in either of two ways—one is statistical and one is essentially hardwired.
By the statistical route, division interrupts the mating and neither of the produced cells is able to continue replicating its plasmid. The copy number would drop in both cell lineages and the clone would become extinct as a source of revertant cells. Plasmid replication could restart only if another mating occurred within the clone. In this case, DNA transferred from the new donor would initiate a new rolling-circle replication fork. The clone would see a repeated series of events—mating, plasmid amplification, and division—over 5 days. If the probability of a new mating is low, some clones would go extinct and the others would show a constant rate of lac replication per unit time. The collective population of developing clones would produce a linearly increasing number of lac revertant alleles. This would lead to a linear increase in revertant colony number over time.
By the hardwired route, the first mating pair might remain stably associated and act as a structurally stable “mother cell” that continuously transfers plasmid DNA until the recipient has sufficient copies to divide. At division, the recipient member of the mating pair divides and produces a daughter cell that is eliminated as a source of revertants. The other daughter is part of the original mating pair, which remains stably engaged in transferring plasmid material and doing rolling-circle replication of the recipient plasmid. No secondary mating events need to be initiated. We suggested above that ompA, traN, or traG mutants might fail to produce revertants because their initial mating pair is too unstable. That is, mating pairs lacking stabilization systems might not continue transferring for the 5-day period of the reversion experiment.
In either of the two suggested processes, a revertant is initiated by a single cell that divides and produces daughter cells that are able to conjugate. This initial mating pair continuously throws off daughters that may divide a few times, but ultimately become equivalent to the bulk of the plated population. Only the cell engaged in plasmid transfer replication has a chance of producing a revertant allele. Because of this cell, the clone as a whole has a constant chance of reversion per unit time. This explains the linear increase in revertant colony number over the course of a reversion experiment.
Numerology of the reversion process
During nonselective growth in liquid medium, the reversion rate of the lac mutation is 10−9/cell per generation (Cairns and Foster 1991). Under selection, 105 initiator cells give rise to 50 revertant colonies over the course of 5 days with very little growth of the lawn population. Each revertant cell arises within a clone initiated by one of the 105 plated initiator cells. With an unenhanced mutation rate, each of the 105 initiator clones would have to replicate a lac region 100,000 times per day to produce the 10 new revertants per day that appear somewhere on the selection plate (50 revertants over 5 days). If DinB overproduction increases the mutation rate 100-fold, each clone would have to replicate lac 1000 times per day. Even less replication would be needed if the DNA synthesis associated with the RecA-mediated double-strand-break repair is the equivalent of “break-induced replication,” which is less accurate than normal vegetative replication (Kuzminov 1995; Malkova and Ira 2013).
It should be noted that the process described here directs mutagenesis to the F′lac plasmid (Foster 1997; Bull et al. 2001; Stumpf et al. 2007) because only this element is replicating. The cellular chromosome replicates very little before a reversion event restores a lac+ allele on the plasmid. Thus one expects mutagenesis to be focused on the F′lac plasmid. However, after a lac+ allele forms, a host cell may initially replicate its chromosome in the presence of a high level of DinB polymerase. This model makes predictions regarding the distribution of general mutagenesis in final revertants. These predictions have been tested previously by scoring associated mutations in Lac+ cells (Rosche and Foster 1999) and are currently being tested using full genome sequencing (A. Nguyen, I. Yamayoshi, E. Kofoid, S. Maisnier-Patin, and J. R. Roth, personal communication).
Role of plasmid transfer in the origin of unstable Lac+ revertants under selection
The new model and data that support it have been described here in terms of stable revertants, which are 90% of the total at day 5. However the model also explains the unstable revertants (10%), which do not have stably corrected lac alleles and owe their growth ability to a tandem amplified array of the partially functional mutant lac allele within the parental F′lac plasmid. Formation of unstable revertants also requires the lac allele to be located somewhere on the F′lac plasmid and to be transfer between cells, but does not require the DinB polymerase.
The new model suggests that unstable revertants are started by initiator cells that carry multiple copies of an F′lac plasmid, each of which has a short internal duplication of the lac region. Under selection, these initiator cells divide and their daughter cells mate. However with an internal lac duplication, the initial mating sets up recombination between donor and recipient daughter cells, both carrying a plasmid with a lac duplication. This mating provides an opportunity for unequal plasmid–plasmid recombination and that can expand or contract the amplified lac region. In these developing revertant clones, selection favors both increases in plasmid copy number and increases in the size each of the tandem lac amplifications. The intermediates in this reversion process are quite different from those for stable revertants.
Increases in F’ plasmid copy number are not heritable. That is, when a cell with an elevated plasmid copy divides, its copy number drops twofold. Replication of the plasmid is prevented until the copy number approaches the maintained minimal level. However, expansions of a tandem amplification of lac are heritable in that cell division generates two cells, each of which can inherit plasmid copies with the expanded amplification. Thus, while an excessive plasmid copy number drops two fold at each cell division, an excessive number of lac alleles within a tandem array does not (the size of a tandem array is subject to expansion and contraction caused by much rarer events, typically occurring at a rate of ∼0.1 per generation). Therefore once the size of a tandem lac array reaches 10 or more copies per plasmid, cell division produces daughters that are able to divide exponentially on lactose. The number of cells in the colony then expands exponentially and selection to maintain a rolling circle is relaxed. Cells in these colonies are unstably Lac+ because during nonselective growth, they can lose tandem lac copies and their ability to grow on lactose. That is, while the tandem array is heritable over a few generations, the number of copies in a tandem array can vary due to unequal recombination between sister chromosomes over several generations. The formation of unstable Lac+ revertant cells therefore requires initiator cells with a preexisting lac duplication and recombination ability, but it does not require the plasmid dinB gene or an increased mutation rate.
Channeling metabolic energy without cell division
The Cairns–Foster system poses a metabolic problem in that reversion requires residual function of the mutant lac operon, even though very few cell divisions occur during the reversion process. The model described here proposes that the residual function of the mutant lac operon extracts energy that is insufficient for growth on lactose, but can support replication of the F′lac plasmid during plasmid transfer from one cell to another. Selection favors cells in which the residual lac activity is amplified. Only these cells continue to replicate lac and have a chance to revert. Thus selection acts on a population of replicating lac alleles within cells that are not dividing extensively. The variants come from stochastic fluctuations in plasmid copy number caused by conjugational transfer replication, vegetative plasmid replication, and plasmid segregation. Production of these variants may also be mediated by host repair functions such as recombination and DNA damage repair.
The evidence for stress-induced mutagenesis is flawed
The idea of a molecular mutagenesis mechanism that is induced in response to growth limitation (stress) has been suggested for many organisms. While we doubt that such models are correct for any system, we are strongly convinced that the behavior of the Cairns–Foster system does not support this general idea.
Stress-induced mutagenesis models attempt to explain the behavior of the Cairns–Foster system by proposing a general mechanism that mutagenizes the entire genome of a subpopulation in response to growth-limiting conditions (Foster 2007; Galhardo et al. 2007). This mechanism is proposed to have evolved to help populations adapt to unfavorable conditions. We suggest here that, in fact, the physiological systems required for reversion are a chance combination of functions produced by an infective agent (the conjugative F-plasmid), functions that mediate DNA break repair (recombination) and an error-prone DNA repair polymerase that supports replication of damaged DNA (DinB). The behavior of the Cairns–Foster system relies on the ability of natural selection to weave a path that integrates these various contributions.
The theoretical problem with an inducible mechanism that purposefully generates mutations is that bacterial growth is often limited by external problems that cannot be solved by mutation, e.g., lack of nutrients. Under such conditions, mutagenesis imposes a cost with no chance of a solution (Roth et al. 2003; Andersson et al. 2011). Upregulation of mutagenesis has also been invoked to explain initiation of cancers in response to cell growth limitation by metazoan developmental mechanisms (Fitzgerald et al. 2017). Improved growth circumvents these controls. It seems unlikely that survival of metazoans has been enhanced by malignancy, but this conclusion is implied by the suggestion that stress-induced mutagenic mechanisms evolved to circumvent normal cell control mechanisms.
Acknowledgments
We would like to thank Susan K. Amundsen, Gerald Smith, Patricia Foster, Jun Lu, Laura S. Frost, Thomas J. Silhavy, Mike Syvanen, and Beth A. Traxler for sending us strains. This work benefitted from helpful suggestions from colleagues Wolf Heyer, Steve Kowalczykowski, Andrei Kuzminov, and Frank Stahl; and from members of this laboratory, Eric Kofoid, Carla Ladd, Natalie Mari, Amanda Nguyen, Oyang Teng, and Itsugo Yamayoshi. The project was supported by National Institutes of Health grant GM-27068 (to JRR).
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7051757.
Communicating editor: D. Bishop
Literature Cited
- Achtman M., 1975. Mating aggregates in Escherichia coli conjugation. J. Bacteriol. 123: 505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achtman M., Kennedy N., Skurray R., 1977. Cell–cell interactions in conjugating Escherichia coli: role of TraT protein in surface exclusion. Proc. Natl. Acad. Sci. USA 74: 5104–5108. 10.1073/pnas.74.11.5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achtman M., Morelli G., Schwuchow S., 1978. Cell-cell interactions in conjugating Escherichia coli: role of F pili and fate of mating aggregates. J. Bacteriol. 135: 1053–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson D. I., Slechta E. S., Roth J. R., 1998. Evidence that gene amplification underlies adaptive mutability of the bacterial lac operon. Science 282: 1133–1135. 10.1126/science.282.5391.1133 [DOI] [PubMed] [Google Scholar]
- Andersson, D. I., D. Hughes, and J. R. Roth, 2011 The origin of mutants under selection: interactions of mutation, growth, and selection., in EcoSal-Escherichia Coli and Salmonella: Cellular and Molecular Biology, Ed. 3, edited by A. Böck, R. Curtiss, Ed. 3, J. B. Kaper, P. D. Karp, F. C. Neidhardt et al. ASM Press, Washington, D. C: 10.1128/ecosalplus.5.6.6 [DOI] [PubMed] [Google Scholar]
- Arutyunov D., Frost L. S., 2013. F conjugation: back to the beginning. Plasmid 70: 18–32. 10.1016/j.plasmid.2013.03.010 [DOI] [PubMed] [Google Scholar]
- Audette G. F., Manchak J., Beatty P., Klimke W. A., Frost L. S., 2007. Entry exclusion in F-like plasmids requires intact TraG in the donor that recognizes its cognate TraS in the recipient. Microbiology 153: 442–451. 10.1099/mic.0.2006/001917-0 [DOI] [PubMed] [Google Scholar]
- Bjedov I., Tenaillon O., Gerard B., Souza V., Denamur E., et al. , 2003. Stress-induced mutagenesis in bacteria. Science 300: 1404–1409. 10.1126/science.1082240 [DOI] [PubMed] [Google Scholar]
- Bull H. J., Lombardo M.-J., Rosenberg S. M., 2001. Stationary-phase mutation in the bacterial chromosome: recombination protein and DNA polymerase IV dependence. Proc. Natl. Acad. Sci. USA 98: 8334–8341. 10.1073/pnas.151009798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J., Foster P. L., 1991. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics 128: 695–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter J. R., Patel D. R., Porter R. D., 1992. The role of oriT in tra-dependent enhanced recombination between mini-F-lac-oriT and lambda plac5. Genet. Res. 59: 157–165. 10.1017/S0016672300030433 [DOI] [PubMed] [Google Scholar]
- Coulondre C., Miller J. H., 1977. Genetic studies of the lac repressor. IV. Mutagenic specificity in the lacI gene of Escherichia coli. J. Mol. Biol. 117: 577–606. 10.1016/0022-2836(77)90059-6 [DOI] [PubMed] [Google Scholar]
- Datsenko K. A., Wanner B. L., 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97: 6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz F., Frost L. S., Meyer R. J., Zechner E. L., 2010. Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol. Rev. 34: 18–40. 10.1111/j.1574-6976.2009.00195.x [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L., Frost L. S., Paranchych W., 1992. The TraM protein of the conjugative plasmid F binds to the origin of transfer of the F and ColE1 plasmids. Mol. Microbiol. 6: 2951–2959. 10.1111/j.1365-2958.1992.tb01754.x [DOI] [PubMed] [Google Scholar]
- Everett R., Willetts N., 1980. Characterisation of an in vivo system for nicking at the origin of conjugal DNA transfer of the sex factor F. J. Mol. Biol. 136: 129–150. 10.1016/0022-2836(80)90309-5 [DOI] [PubMed] [Google Scholar]
- Firth N., Skurray R., 1992. Characterization of the F plasmid bifunctional conjugation gene, traG. Mol. Gen. Genet. 232: 145–153. 10.1007/BF00299147 [DOI] [PubMed] [Google Scholar]
- Fitzgerald D. M., Hastings P. J., Rosenberg S. M., 2017. Stress-induced mutagenesis: implications in cancer and drug resistance. Annu. Rev. Cancer Biol. 1: 119–140. 10.1146/annurev-cancerbio-050216-121919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P. L., 1997. Nonadaptive mutations occur on the F′ episome during adaptive mutation conditions in Escherichia coli. J. Bacteriol. 179: 1550–1554. 10.1128/jb.179.5.1550-1554.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P. L., 2007. Stress-induced mutagenesis in bacteria. Crit. Rev. Biochem. Mol. Biol. 42: 373–397. 10.1080/10409230701648494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P. L., Rosche W. A., 1999. Increased episomal replication accounts for the high rate of adaptive mutation in recD mutants of Escherichia coli. Genetics 152: 15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P. L., Trimarchi J. M., 1994. Adaptive reversion of a frameshift mutation in Escherichia coli by simple base deletions in homopolymeric runs. Science 265: 407–409. 10.1126/science.8023164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P. L., Trimarchi J. M., 1995a Adaptive reversion of an episomal frameshift mutation in Escherichia coli requires conjugal functions but not actual conjugation. Proc. Natl. Acad. Sci. USA 92: 5487–5490. 10.1073/pnas.92.12.5487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P. L., Trimarchi J. M., 1995b Conjugation is not required for adaptive reversion of an episomal frameshift mutation in Escherichia coli. J. Bacteriol. 177: 6670–6671. 10.1128/jb.177.22.6670-6671.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame R., Bishop J. O., 1971. The number of sex-factors per chromosome in Escherichia coli. Biochem. J. 121: 93–103. 10.1042/bj1210093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost L. S., Ippen-Ihler K., Skurray R. A., 1994. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 58: 162–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galhardo R. S., Hastings P. J., Rosenberg S. M., 2007. Mutation as a stress response and the regulation of evolvability. Crit. Rev. Biochem. Mol. Biol. 42: 399–435. 10.1080/10409230701648502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galhardo R. S., Do R., Yamada M., Friedberg E. C., Hastings P. J., et al. , 2009. DinB upregulation is the sole role of the SOS response in stress-induced mutagenesis in Escherichia coli. Genetics 182: 55–68. 10.1534/genetics.109.100735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galitski T., Roth J. R., 1995. Evidence that F plasmid transfer replication underlies apparent adaptive mutation. Science 268: 421–423. 10.1126/science.7716546 [DOI] [PubMed] [Google Scholar]
- Ganesan A. K., Rotman B., 1966. Transport systems for galactose and galactosides in Escherichia coli. I. Genetic determination and regulation of the methyl-galactoside permease. J. Mol. Biol. 16: 42–50. 10.1016/S0022-2836(66)80261-9 [DOI] [PubMed] [Google Scholar]
- Godoy V. G., Fox M. S., 2000. Transposon stability and a role for conjugational transfer in adaptive mutability. Proc. Natl. Acad. Sci. USA 97: 7393–7398. 10.1073/pnas.130186597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy V. G., Gizatullin F. S., Fox M. S., 2000. Some features of the mutability of bacteria during nonlethal selection. Genetics 154: 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R. S., Longerich S., Rosenberg S. M., 1994. Recombination in adaptive mutation. Science 264: 258–260. 10.1126/science.8146657 [DOI] [PubMed] [Google Scholar]
- Khan S. R., Mahaseth T., Kouzminova E. A., Cronan G. E., Kuzminov A., 2016. Static and dynamic factors limit chromosomal replication complexity in Escherichia coli, avoiding dangers of runaway overreplication. Genetics 202: 945–960. 10.1534/genetics.115.184697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. R., Maenhaut-Michel G., Yamada M., Yamamoto Y., Matsui K., et al. , 1997. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: an overexpression of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc. Natl. Acad. Sci. USA 94: 13792–13797. 10.1073/pnas.94.25.13792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsman A., Willetts N., 1978. The requirements for conjugal DNA synthesis in the donor strain during Flac transfer. J. Mol. Biol. 122: 287–300. 10.1016/0022-2836(78)90191-2 [DOI] [PubMed] [Google Scholar]
- Klimke W. A., Frost L. S., 1998. Genetic analysis of the role of the transfer gene, traN, of the F and R100–1 plasmids in mating pair stabilization during conjugation. J. Bacteriol. 180: 4036–4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski S. C., 2015. An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 7: a016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov A., 1995. Collapse and repair of replication forks in Escherichia coli. Mol. Microbiol. 16: 373–384. 10.1111/j.1365-2958.1995.tb02403.x [DOI] [PubMed] [Google Scholar]
- Kuzminov A., 2016. Chromosomal replication complexity: a novel DNA metrics and genome instability factor. PLoS Genet. 12: e1006229 10.1371/journal.pgen.1006229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanka E., Wilkins B. M., 1995. DNA processing reactions in bacterial conjugation. Annu. Rev. Biochem. 64: 141–169. 10.1146/annurev.bi.64.070195.001041 [DOI] [PubMed] [Google Scholar]
- Lederberg J., Lederberg E. M., 1952. Replica plating and indirect selection of bacterial mutants. J. Bacteriol. 63: 399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd R. G., Buckman C., 1995. Conjugational recombination in Escherichia coli: genetic analysis of recombinant formation in Hfr x F- crosses. Genetics 139: 1123–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo M. J., Aponyi I., Rosenberg S. M., 2004. General stress response regulator RpoS in adaptive mutation and amplification in Escherichia coli. Genetics 166: 669–680. 10.1534/genetics.166.2.669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luria S. E., Delbrück M., 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28: 491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q., Wood T. K., 2009. OmpA influences Escherichia coli biofilm formation by repressing cellulose production through the CpxRA two-component system. Environ. Microbiol. 11: 2735–2746. 10.1111/j.1462-2920.2009.02000.x [DOI] [PubMed] [Google Scholar]
- Maisnier-Patin S., Roth J. R., 2015. The origin of mutants under selection: how natural selection mimics mutagenesis (adaptive mutation), pp. 97–115 in Microbial Evolution, edited by Ochman H. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 10.1101/cshperspect.a018176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisnier-Patin S., Roth J. R., 2016. The adaptive mutation controversy, pp. 26–36 in Encyclopedia of Evolutionary Biology, edited by Kliman R. M. Academic Press, Oxford: 10.1016/B978-0-12-800049-6.00229-8 [DOI] [Google Scholar]
- Malkova A., Ira G., 2013. Break-induced replication: functions and molecular mechanism. Curr. Opin. Genet. Dev. 23: 271–279. 10.1016/j.gde.2013.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maneewannakul S., Kathir P., Ippen-Ihler K., 1992. Characterization of the F plasmid mating aggregation gene traN and of a new F transfer region locus trbE. J. Mol. Biol. 225: 299–311. 10.1016/0022-2836(92)90923-8 [DOI] [PubMed] [Google Scholar]
- Manning P. A., Morelli G., Achtman M., 1981. TraG protein of the F sex factor of Escherichia coli K-12 and its role in conjugation. Proc. Natl. Acad. Sci. USA 78: 7487–7491. 10.1073/pnas.78.12.7487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoil C., Rosenbusch J. P., 1982. Conjugation-deficient mutants of Escherichia coli distinguish classes of functions of the outer membrane OmpA protein. Mol. Gen. Genet. 187: 148–156. 10.1007/BF00384398 [DOI] [PubMed] [Google Scholar]
- Matson S. W., Nelson W. C., Morton B. S., 1993. Characterization of the reaction product of the oriT nicking reaction catalyzed by Escherichia coli DNA helicase I. J. Bacteriol. 175: 2599–2606. 10.1128/jb.175.9.2599-2606.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie G., Lee P., Lombardo M.-J., Hastings P., Rosenberg S., 2001. SOS mutator DNA polymerase IV functions in adaptive mutation and not adaptive amplification. Mol. Cell 7: 571–579. 10.1016/S1097-2765(01)00204-0 [DOI] [PubMed] [Google Scholar]
- McKenzie G. J., Harris R. S., Lee P. L., Rosenberg S. M., 2000. The SOS response regulates adaptive mutation. Proc. Natl. Acad. Sci. USA 97: 6646–6651. 10.1073/pnas.120161797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. H., 1992. A Short Course in Bacterial Genetics. Cold Spring Harbor Laboratory Press, New York. [Google Scholar]
- Minkley E. G., Jr., Willetts N. S., 1984. Overproduction, purification and characterization of the F TraT protein. Mol. Gen. Genet. 196: 225–235. 10.1007/BF00328054 [DOI] [PubMed] [Google Scholar]
- Nordstrom K., Austin S. J., 1989. Mechanisms that contribute to the stable segregation of plasmids. Annu. Rev. Genet. 23: 37–69. 10.1146/annurev.ge.23.120189.000345 [DOI] [PubMed] [Google Scholar]
- Novick R. P., 1969. Extrachromosomal inheritance in bacteria. Bacteriol. Rev. 33: 210–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J. E., Bartoszyk I. M., Dheer S., Benson S. A., 1996. Redundant homosexual F transfer facilitates selection-induced reversion of plasmid mutations. J. Bacteriol. 178: 3037–3043. 10.1128/jb.178.11.3037-3043.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponder R. G., Fonville N. C., Rosenberg S. M., 2005. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol. Cell 19: 791–804. 10.1016/j.molcel.2005.07.025 [DOI] [PubMed] [Google Scholar]
- Radicella J. P., Park P. U., Fox M. S., 1995. Adaptive mutation in Escherichia coli: a role for conjugation. Science 268: 418–420. 10.1126/science.7716545 [DOI] [PubMed] [Google Scholar]
- Reams A. B., Kofoid E., Savageau M., Roth J. R., 2010. Duplication frequency in a population of Salmonella enterica rapidly approaches steady state with or without recombination. Genetics 184: 1077–1094. 10.1534/genetics.109.111963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez C., Tompkin J., Hazel J., Foster P. L., 2002. Induction of a DNA nickase in the presence of its target site stimulates adaptive mutation in Escherichia coli. J. Bacteriol. 184: 5599–5608. 10.1128/JB.184.20.5599-5608.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosche W. A., Foster P. L., 1999. The role of transient hypermutators in adaptive mutation in Escherichia coli. Proc. Natl. Acad. Sci. USA 96: 6862–6867. 10.1073/pnas.96.12.6862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg S. M., Harris R. S., Torkelson J., 1995. Molecular handles on adaptive mutation. Mol. Microbiol. 18: 185–189. 10.1111/j.1365-2958.1995.mmi_18020185.x [DOI] [PubMed] [Google Scholar]
- Rosenberg S. M., Shee C., Frisch R. L., Hastings P. J., 2012. Stress-induced mutation via DNA breaks in Escherichia coli: a molecular mechanism with implications for evolution and medicine. BioEssays 34: 885–892. 10.1002/bies.201200050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth J. R., Kofoid E., Roth F. P., Berg O. G., Seger J., et al. , 2003. Regulating general mutation rates: examination of the hypermutable state model for cairnisian adaptive mutation. Genetics 163: 1483–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth J. R., Kugelberg E., Reams A. B., Kofoid E., Andersson D. I., 2006. Origin of mutations under selection: the adaptive mutation controversy. Annu. Rev. Microbiol. 60: 477–501. 10.1146/annurev.micro.60.080805.142045 [DOI] [PubMed] [Google Scholar]
- Rotman B., Ganesan A. K., Guzman R., 1968. Transport systems for galactose and galactosides in Escherichia coli. II. Substrate and inducer specificities. J. Mol. Biol. 36: 247–260. 10.1016/0022-2836(68)90379-3 [DOI] [PubMed] [Google Scholar]
- Sano E., Maisnier-Patin S., Aboubechara J. P., Quinones-Soto S., Roth J. R., 2014. Plasmid copy number underlies adaptive mutability in bacteria. Genetics 198: 919–933. 10.1534/genetics.114.170068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shee C., Ponder R., Gibson J. L., Rosenberg S. M., 2011. What limits the efficiency of double-strand break-dependent stress-induced mutation in Escherichia coli? J. Mol. Microbiol. Biotechnol. 21: 8–19. 10.1159/000335354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slechta E. S., Liu J., Andersson D. I., Roth J. R., 2002. Evidence that selected amplification of a bacterial lac frameshift allele stimulates Lac(+) reversion (adaptive mutation) with or without general hypermutability. Genetics 161: 945–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slechta E. S., Bunny K. L., Kugelberg E., Kofoid E., Andersson D. I., et al. , 2003. Adaptive mutation: general mutagenesis is not a programmed response to stress but results from rare coamplification of dinB with lac. Proc. Natl. Acad. Sci. USA 100: 12847–12852. 10.1073/pnas.1735464100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf J. D., Poteete A. R., Foster P. L., 2007. Amplification of lac cannot account for adaptive mutation to Lac+ in Escherichia coli. J. Bacteriol. 189: 2291–2299. 10.1128/JB.01706-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers D. K., 1996. The Biology of Plasmids. Blackwell Science, Oxford: 10.1002/9781444313741 [DOI] [Google Scholar]
- Torkelson J., Harris R. S., Lombardo M. J., Nagendran J., Thulin C., et al. , 1997. Genome-wide hypermutation in a subpopulation of stationary-phase cells underlies recombination-dependent adaptive mutation. EMBO J. 16: 3303–3311. 10.1093/emboj/16.11.3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxler B. A., Minkley E. G., Jr., 1988. Evidence that DNA helicase I and oriT site-specific nicking are both functions of the F TraI protein. J. Mol. Biol. 204: 205–209. 10.1016/0022-2836(88)90609-2 [DOI] [PubMed] [Google Scholar]
- Wagner J., Gruz P., Kim S. R., Yamada M., Matsui K., et al. , 1999. The dinB gene encodes a novel E. coli DNA polymerase, DNA pol IV, involved in mutagenesis. Mol. Cell 4: 281–286. 10.1016/S1097-2765(00)80376-7 [DOI] [PubMed] [Google Scholar]
- Wang Y., 2002. The function of OmpA in Escherichia coli. Biochem. Biophys. Res. Commun. 292: 396–401. 10.1006/bbrc.2002.6657 [DOI] [PubMed] [Google Scholar]
- Wilson D. B., 1974. The regulation and properties of the galactose transport system in Escherichia coli K12. J. Biol. Chem. 249: 553–558. [PubMed] [Google Scholar]
- Wrande M., Roth J. R., Hughes D., 2008. Accumulation of mutants in “aging” bacterial colonies is due to growth under selection, not stress-induced mutagenesis. Proc. Natl. Acad. Sci. USA 105: 11863–11868. 10.1073/pnas.0804739105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamayoshi I., Maisnier-Patin S., Roth J. R., 2018. Selection-enhanced mutagenesis of lac genes is due to their coamplification with dinB encoding an error-prone DNA polymerase. Genetics 208: 1009–1021. 10.1534/genetics.117.300409 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains generated in this study are available upon request. Table S1 in File S1 contains genotypes of the strains. Primer sequences are available in Table S2 in File S1. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7051757.