Abstract
Forward genetics remains a powerful method for revealing the genes underpinning organismal form and function, and for revealing how these genes are tied together in gene networks. In maize, forward genetics has been tremendously successful, but the size and complexity of the maize genome made identifying mutant genes an often arduous process with traditional methods. The next generation sequencing revolution has allowed for the gene cloning process to be significantly accelerated in many organisms, even when genomes are large and complex. Here, we describe a bulked-segregant analysis sequencing (BSA-Seq) protocol for cloning mutant genes in maize. Our simple strategy can be used to quickly identify a mapping interval and candidate single nucleotide polymorphisms (SNPs) from whole genome sequencing of pooled F2 individuals. We employed this strategy to identify narrow odd dwarf as an enhancer of teosinte branched1, and to identify a new allele of defective kernel1. Our method provides a quick, simple way to clone genes in maize.
Keywords: forward genetics, maize genetics, mutant genes, bulked-segregant analysis, next generation sequencing
Forward genetics remains a powerful way to ‘ask the plant’ which genes matter for a particular trait or phenotype (Mueller 2006). Because forward genetics relies on random mutagenesis, it presents an unbiased method for identifying novel genes that act in particular pathways. While forward genetic screens can reveal new genes, they can also reveal novel functions for known genes; providing a richer, more deeply nuanced view of gene function essential for a true understanding of how genes and gene networks contribute to building an organism (Nawy et al. 2010; Gallavotti et al. 2010; Vlad et al. 2014; Gillmor et al. 2016). After random mutagenesis, mutant genes are most often identified through linkage mapping.
Linkage mapping, using a combination of bulked-segregant analysis (BSA) and fine mapping, has been very successful for cloning maize genes (Gallavotti and Whipple 2015). The process starts when a mutant of interest is crossed to a wild-type individual in a contrasting genetic background. The resulting F1 individuals are selfed or backcrossed, and mutants are identified in the F2 or backcross population. These mutants can be used to identify a region of increased homozygosity in a chromosomal region physically linked to the lesion causing the mutant phenotype. This region of increased homozygosity can be rapidly detected using bulked-segregant analysis (Michelmore et al. 1991).
In bulked-segregant analysis, pools of wild-type and mutant individuals are genotyped at markers spread across the genome. In chromosomal regions not linked to the mutant lesion, markers will be segregating according to typical 1:2:1 (or 1:1 in backcross) segregation ratios. In a pooled BSA sample, these unlinked markers will all be genotypically heterozygous. Linked markers will be homozygous for the mutant parent genotype, unless F1 recombination has happened between the mutant lesion and a particular marker. A chromosomal region enriched for these linked markers represents a likely location for the mutant gene under study, and the coordinates of these linked markers define an initial mapping interval. Once this initial mapping interval is identified, F1 recombination is again used to place recombination breakpoints in individual mutants, and thus narrow the mapping interval using fine mapping (Gallavotti and Whipple 2015). The fine mapping process is often time consuming, and identifying causative lesions and mutant genes can take years.
The advent of next-generation sequencing (NGS) means that BSA can be used to very quickly identify a very small mapping interval, and even the causative lesion, without fine mapping. In BSA-Seq, whole genome shotgun sequencing using NGS can be used to quickly genotype mutant vs. non mutant BSA pools at many thousands of markers spread across the genome. These BSA-Seq data can reveal the genomic interval that contains the mutant gene of interest, and with enough coverage, the lesion itself (Zou et al. 2016). Since random mutagenesis produces lesions throughout the genome, and since NGS does not depend on established genotyping assays, BSA-Seq can be used to identify a chromosomal region using just the polymorphisms induced by mutagenesis. In this case, the contrasting genetic background used for the BSA can simply be that of an unmutagenized parent. This modification to BSA-Seq has been called MutMap (Abe et al. 2012). BSA-Seq (and MutMap) has been used to identify mutant loci in Arabidopsis thaliana, soybean, barley, Mimulus, rice, sorghum, and Brachypodium distachyon (Schneeberger and Weigel 2011; Abe et al. 2012; Mascher et al. 2014; Woods et al. 2014; Addo-Quaye et al. 2017; Ding et al. 2017; Song et al. 2017; Jiao et al. 2018). In maize, BSA coupled to RNA-Seq (BSR-Seq) has been used successfully to clone mutant genes (Liu et al. 2012; Li et al. 2013; Nestler et al. 2014; Tang et al. 2014). Conventional BSA-Seq has been used to identify genomic regions underlying variation in flowering time and plant height QTL in a maize population (Haase et al. 2015), but cloning mutant genes using BSA-Seq is not yet routine in maize.
Here, we report a user-friendly protocol for cloning mutant maize genes using BSA-Seq. We used this protocol to clone two genes recovered from EMS mutagenesis screens. We used conventional BSA-Seq in the case of one gene, and MutMap in the other. We identified narrow odd dwarf (nod) as an enhancer of teosinte branched1 (tb1) using conventional BSA-Seq, and identified a seedling lethal allele of defective kernel1 (dek1) using MutMap (Doebley et al. 1997; Lid et al. 2002; Becraft et al. 2002; Rosa et al. 2017). In addition, we used our tb1 enhancer data to design insertion-deletion (indel) markers for downstream fine mapping. This fine mapping to reduce the recombination interval is critical in cases where only a single allele for a particular mutant exists, and in cases where no clear candidate lesion is identified. Our method provides a quick, easy method for cloning mutant genes in maize.
Methods
Plant material and isolation of mutants
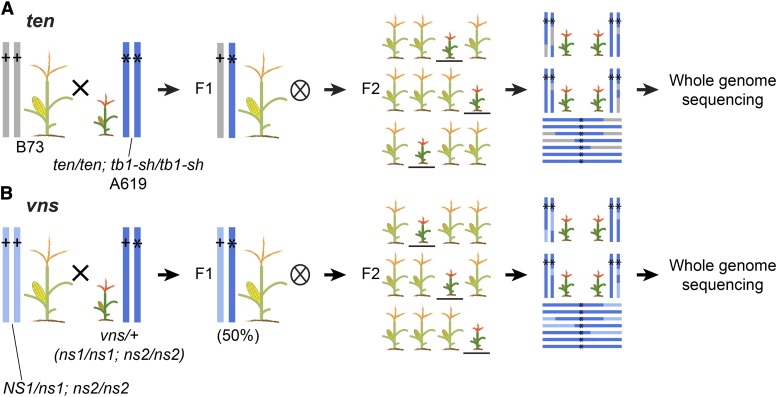
To identify enhancers and suppressors of the tb1 (Doebley et al. 1997) and narrow sheath (ns) (Scanlon et al. 1996) phenotypes, we performed EMS mutagenesis screens. tb1 encodes a TCP transcription factor with a well characterized role in suppressing axillary branching or tillering (Doebley et al. 1997). The ns loci regulate the initiation of lateral cells in the maize shoot apical meristem. ns1 and ns2 encode duplicate homeodomain transcription factors, homologous to PRESSED FLOWER in Arabidopsis thaliana (Nardmann et al. 2004). We mutagenized pollen homozygous for a weak allele of tb1 in the A619 inbred (tb1-sh), and ns in an unknown background obtained from Pioneer Hi-Bred Intl. (ns1;ns2), using established protocols (Neuffer et al. 1997). After mutagenesis, M1 progeny were selfed to generate M2 populations, where we identified the mutants tb1 enhancer (ten) and very narrow sheath (vns). ten, which arose in the A619 genetic background, was crossed to the B73 genetic background and then selfed to generate an F2 mapping population (Figure 1a). vns is seedling-lethal. Therefore a wild-type sibling, heterozygous at vns, was backcrossed to a ns1 heterozygote fixed for ns2 (ns1/NS1;ns2/ns2), and the progeny selfed to generate a mapping population (Figure 1b).
Figure 1.
Crossing scheme to generate ten and vns mapping populations. (A) For ten, we crossed a homozygous ten tb1-sh double mutant (in A619) to a wild-type individual of a widely divergent genotype (B73), then selfed many plants in the resulting F1. We selected ten mutants in the F2 generation, regardless of the tb1 phenotype. We pooled tissue from 101 of these mutants into a single DNA extraction and a single library for whole genome shotgun sequencing. (B) For vns, we backcrossed a plant heterozygous at vns to a wild-type individual of the unmutagenized parental genotype. We selfed many individuals in the resulting F1 (50% heterozygous at vns). We selected 9 vns mutants in the F2 generation, regardless of the ns phenotype, and pooled tissue into a single DNA extraction and NGS library.
For BSA-seq, tissue samples were collected from 101 ten mutant individuals, and 9 vns mutant individuals in the mapping populations. We used this small number of vns individuals because that was what we had available from a small mapping population. Mutants in the F2 mapping populations were sampled independent of their genotypes at tb1 or ns1 (i.e., neither ten nor vns was dependent on tb1 or ns, respectively). Because the vns genetic background is unknown, we also collected tissue samples for 9 wild-type siblings in the vns mapping population and for the parents of the EMS mutagenesis screen. To characterize the genetic interaction between tb1 and ten, we counted tillers in families segregating tb1-sh and ten.
DNA extractions and NGS sequencing
Tissue samples were pooled by hole punching each leaf twice to ensure equal representation of individuals. We extracted DNA from the pools of tissue using a CTAB method (Gallavotti and Whipple 2015). Libraries were prepared using the TruSeq DNA sample prep kit according to the manufacturer’s instructions (Illumina). We sequenced the ten mutant pool on an Illumina Hi-Seq 2500 at Brigham Young University. Sequenced reads were 125bp long with paired-ends. We sequenced vns mutants, vns wild-type siblings, and unmutagenized parents on an Illumina Hi-Seq 2500 at Cornell University. Sequenced reads from vns were 150bp long with paired ends. Different sequencing platforms were used because of differing availability at our respective institutions.
NGS quality control and read alignment
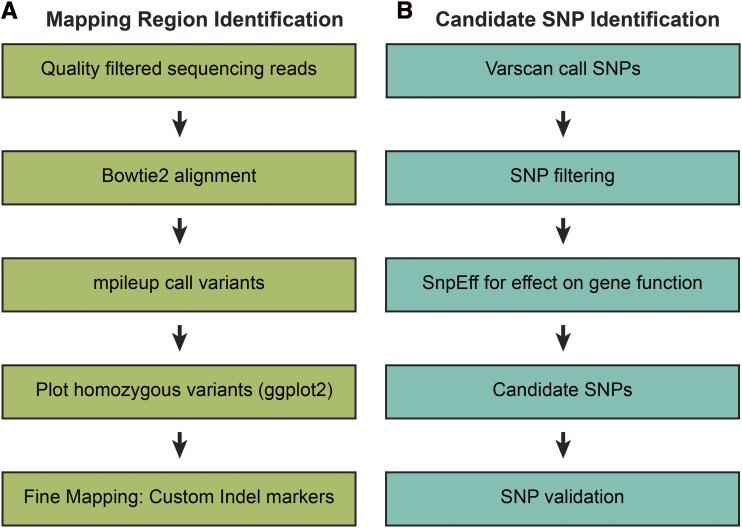
We used the genomic tools hosted on Galaxy web portal to process our Next-Gen sequencing data (Afgan et al. 2016). An overview of our process is shown in Figure 2. We used FastQC (v. 0.69) to determine the quality of our sequencing data, and established a PHRED quality cutoff of 20 based on this analysis (Ewing et al. 1998; Andrews 2014). We used FASTQ Groomer (v. 1.1.1) to convert our FASTQ files to Sanger format for input into downstream Galaxy tools (Blankenberg et al. 2010). Trimmomatic (v. 0.36.3) was used for sliding window trimming averaged across 4 base pairs with average quality >20 and for adapter sequence removal (Bolger et al. 2014). The Galaxy tool cat, which concatenates datasets tail-to-head (cat) (v. 0.1.0) was used to join sequencing files together if data were split from multiple Illumina sequencing runs (Afgan et al. 2018). We used Bowtie2 (v. 2.3.2.2) to align sequencing reads to the B73 reference genome version 4, release 56 and to generate an index (Langmead and Salzberg 2012; Jiao et al. 2017). Read alignment was assessed with Mtools Flagstat (v. 2.0) (Li et al. 2009).
Figure 2.
A simplified overview of our BSA-Seq pipeline. (A) We analyzed our NGS data in Galaxy and in R to identify chromosomal regions that contained the mutant genes. (B) SNP filtering to identify candidate (EMS) SNPs.
Variant calling
SAMtools mpileup (v. 2.1.3) was used to output variants from the indexed bam file into pileup or variant call format (vcf) files (Li et al. 2009). Pileup files were generated for identification of the mapping region and vcf files were generated for identification of candidate SNPs. To filter variant files for mapping region identification, we used SAMtools filter pileup (v. 1.0.2), a custom Galaxy tool, to filter based on read coverage and quality (Li et al. 2009). We filtered out non-variant positions, and focused only on variant positions with coverage of 8 or more reads, each with a quality score of 20 or more. To generate vcf files for candidate SNP identification, we used Varscan (v. 0.1). We used a minimum homozygous calling frequency of 0.99, and filtered out SNPs with low read coverage (coverage <8) (Koboldt et al. 2012). We used this coverage cutoff to reduce artifacts from sequencing and/or mapping error, and 8 was well below the coverage means for both of our datasets.
We also used our mapping data to generate an A619 variant file. We used Varscan (v. 0.1) to call SNPs in ten and 4 other A619 mutants. We did not assign a coverage cutoff for these datasets, and retained only SNPs that had a paired read. We used the function findCommonVariants in the Rsubread package (Shi et al. 2013) to identify SNPs common to all 5 datasets (File S1).
Data processing for plotting and SNP filtering
Each filtered pileup file was split into 10 chromosomes. At each nucleotide position in the pileup file, we calculated variant allele frequency by dividing the number of reads that differed from the B73 reference sequence by the quality adjusted total number of reads at that position. This variant allele frequency was added as a new column in the pileup file. Nucleotide position, reference allele, alternate allele, coverage, and variant allele frequency were extracted from the pileup file to create a variant file for downstream analysis in R (Wickham and Francois 2015; Wickham 2016).
Mapping region identification
We plotted our data against the B73 reference genome, to identify chromosomal regions enriched for homozygous SNPs, presumably in linkage with the causative lesions. Before plotting, we filtered positions with coverage >100 to remove highly enriched positions in our data. High coverage SNPs are likely in repeat rich regions and will not be informative for mapping region identification (Addo-Quaye et al. 2017). We used ggplot2 to plot the number of homozygous variant positions that differed from the B73 reference genome, per 1Mbp chromosomal bin (Wickham and Francois 2015; Wickham 2016). For these analyses, nucleotide positions that differed from the reference genome at a frequency greater than or equal to 0.99 were defined as homozygous variants. For vns, we went on to remove SNPs that were in the wild-type sibling and unmutagenized parent datasets, and plot only those homozygous SNPs that were unique to the mutant pool. For both ten and vns, we also plotted the number of homozygous canonical EMS variants (G to A and C to T transitions) per 1Mbp bin. We defined the limits of the ten and vns mapping intervals as those chromosomal coordinates where variant frequency returned to background levels.
SNP filtering for candidate SNP identification
Once we had identified likely mapping intervals for ten and vns, we searched for potentially causative lesions in each of these intervals. Here, we returned to the vcf file generated using Varscan. Although we could have investigated the filtered EMS SNPs from our pileup file used to determine the mapping region directly, we chose to use Varscan because it outputs a vcf file, needed for downstream candidate SNP analyses using SnpEff (Cingolani et al. 2012). We filtered out all background A619 SNPs (File S1) from the ten dataset, and all parental and homozygous wild-type SNPs from the vns dataset. We reasoned that it was highly unlikely that the causative SNPs could be any known SNP present in any maize inbred. Therefore, we applied an additional SNP filtering step, and removed all the maize Hapmap 3.2.1 SNPs from both datasets, and our A619 SNPs from vns (Merchant et al. 2016; Bukowski et al. 2018). Once we had a set of positions enriched for SNPs unique to each of our particular mutant pools, we filtered out SNPs that were not homozygous and were not canonical EMS changes (G to A or C to T) (Till et al. 2004). The final set of homozygous EMS SNPs unique to each dataset was the input for an analysis of likely SNP effects on gene function.
To identify SNPs in our filtered datasets that might negatively affect gene function, we used SnpEff (version 4.3a). SnpEff identifies nonsense SNPs and splice site mutations as likely to have highly deleterious effects on gene function, and all missense SNPs as likely to have moderate effects on gene function (Cingolani et al. 2012). Functional annotations for the genes disrupted by likely moderate- and high-effect candidate SNPs were obtained from Gramene (Tello-Ruiz et al. 2018). Candidate SNPs were validated through Sanger sequencing and/or complementation crosses.
Indel marker design and fine mapping
To refine our ten mapping interval, we designed custom markers for fine mapping. We used Varscan (v. 0.1) to identify insertion and deletion polymorphisms (indels) in our mapping interval that differentiated our mutant pool from the B73 reference genome. We chose indels that were 15 base pairs in length or longer, and designed primers to flank these indels by 100-150bp (Table S1). DNA was extracted from each of the 101 F2 mutants originally pooled into one NGS library and used for Indel PCR (Gallavotti and Whipple 2015). Size differences between the resulting PCR products were detected on 3.5% agarose gels.
Data availability
Raw sequencing data available at NCBI SRA (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA476333). A protocol for NGS data analysis and plotting in R is available at protocols.io (https://www.protocols.io/view/bsa-seq-in-maize-qyedxte). Supplemental material available at Figshare: https://doi.org/10.25387/g3.7014851.
Results
Mutant phenotypes
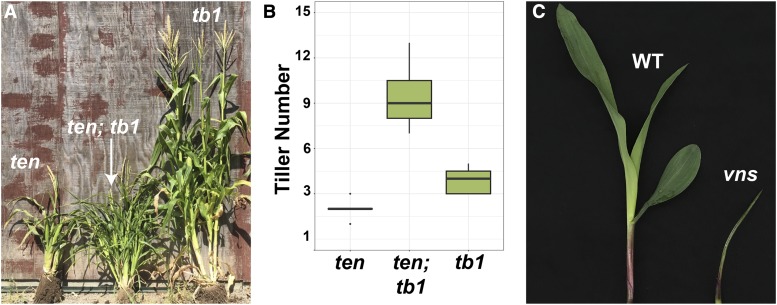
From an EMS mutagenesis screen of tb1-sh mutants in the A619 genetic background, we recovered a dwarf mutant with additional tillers that we called tb1 enhancer (ten) (Figure 3a). very narrow sheath (vns) was uncovered in an independent EMS mutagenesis screen. vns single mutants are seedling lethal and often fail to develop more than 3 leaves (Figure 3c). Although vns was isolated in an ns enhancer screen, vns did not enhance ns.
Figure 3.
ten and vns were identified in EMS mutagenesis screens. (A) ten (left), tb1-sh ten (middle), tb1-sh (right). (B) Quantification of tillers in a 9:3:3:1 population of ten and tb1-sh shows a synergistic interaction between ten and tb1-sh. (C) Wild-type (left) and vns mutant (right) exhibiting reduced growth at the seedling stage.
To determine the nature of the genetic interaction between ten and tb1, we counted tillers in a population segregating both tb1-sh and ten. Tiller number (per plant) was counted in 15 tb1-sh single mutants, 15 ten single mutants and 15 tb1-sh; ten double mutants. tb1-sh and ten single mutants had an average of 4 and 2 tillers respectively. ten tb1-sh double mutants had an average of 9 tillers, showing a synergistic interaction between ten and tb1-sh in tiller development (Figure 3b).
Both ten and vns are on chromosome one
To identify the causative lesions underlying the ten and vns mutant phenotypes, we sequenced pooled DNA from 101 ten individuals and 9 vns individuals from F2 mapping populations (Figure 1). We sequenced ten pools and vns pools on an Illumina platform with outputs of 125 bp and 150bp paired end reads, respectively. For ten, we recovered 450,665,452 reads, and for vns we recovered 311,711,046 reads. For ten, 97% of our reads (438,746,898) mapped to version 4 of the maize genome, and 91% were properly paired (409,548,662). For vns, 90% of our reads (279,239,524) mapped, and 77.5% were properly paired (241,580,812). The average coverage of all ten and vns mapped reads was 26-fold and 17-fold respectively (Table 1). The combined dataset that we used to generate the A619 SNP dataset included 1.1 billion mapped reads, for a mean coverage of 59-fold (File S1).
Table 1. Sequencing data summary for ten and vns.
| Measure | ten | vns |
|---|---|---|
| Reads | 450,665,452 | 311,711,046 |
| Mapped reads | 438,746,898 | 279,239,524 |
| % Mapped Reads | 97.36 | 89.58 |
| Paired reads | 409,548,662 | 241,580,812 |
| Coverage | 26.06 | 16.59 |
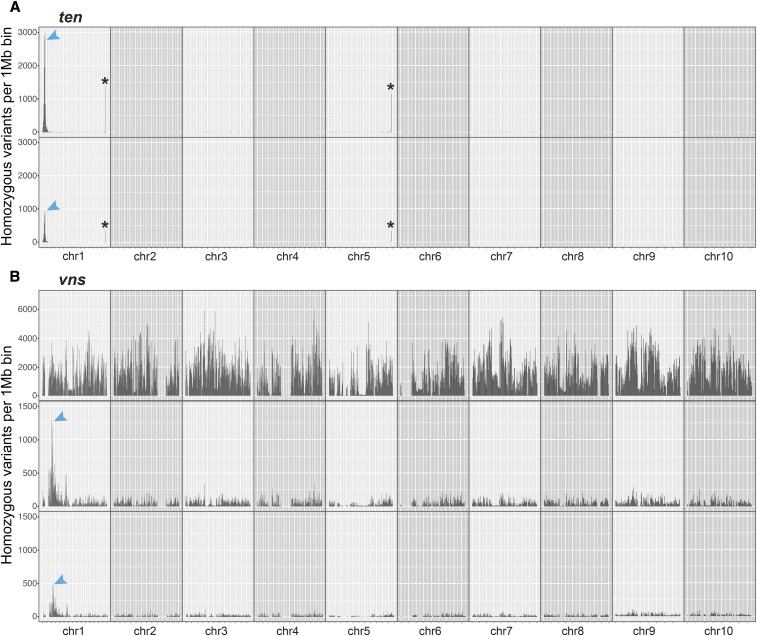
To identify the chromosomal regions corresponding to ten and vns, we searched for regions of the genome enriched for homozygous variants in the mutant pools. For ten, we plotted the number of homozygous SNPs in the NGS data that differed from B73 (variant frequency >=0.99), per 1 Mbp bin. This plotting quickly identified a tall peak on the short arm of chromosome 1, between 0 and 20 Mbp. Plotting only homozygous canonical EMS SNPs (G to A and C to T transitions) identified a peak at the same position, although this peak was shorter (Figure 4a). We also identified two much smaller peaks at the telomeres of chromosome 1 and 5 that contained regions with protein coding genes. These telomeric peaks, at regions of the genome where recombination is high (Gore et al. 2009), were very narrow, and had sharp boundaries. In contrast, the peak at the top of chromosome 1 was much wider and taller than any of the telomeric peaks, and had broad shoulders. These broad shoulders are what we expected from a segregating locus mapped by BSA, where each genome in the pool will have different recombination breakpoints surrounding the mutant lesion. We hypothesized that the telomeric peaks were likely caused by differences between our B73 stocks and those used to generate the B73 reference genome. These differences may be because of the histories of our B73 stocks in our own labs, or because of the stocks’ provenance (Liang and Schnable 2016). We focused on the 0-20 Mbp peak at the top of chromosome 1 as the initial ten mapping interval, which included 623 genes.
Figure 4.
Both ten and vns are on chromosome 1. (A) ten likely lies between 0 and 20 Mbp on chromosome 1. Plotting the number of homozygous positions that differ from the B73 reference genome (per 1 Mbp chromosomal bin) reveals a large peak on chromosome 1 (top). This peak is still visible, but smaller, if only homozygous EMS SNPs are plotted (bottom). (B) vns likely lies between 40 and 70 Mbp on chromosome 1. Plotting the number of homozygous positions that differ from the B73 reference genome (per 1 Mbp chromosomal bin) reveals no distinct peaks (top). After filtering out parental, wild-type sibling, Hapmap 3.2.1 and background SNPs, a large peak on the short arm of chromosome 1 is revealed (middle). This peak is still visible, but smaller, if only homozygous EMS SNPs are plotted (bottom). *indicates a region where our B73 stocks likely differed from those that were used to generate the B73 reference genome.
For vns, identifying a chromosomal location was not quite as simple because vns arose in an unknown genetic background, and a vns heterozygote was crossed to an unmutagenized parent, and not outcrossed to any reference genotype (e.g., B73) (Figure 1b). Thus, plotting only homozygous vns variants revealed, as expected, extensive homozygosity distinct from the B73 reference. There was still no clear peak when we plotted only canonical EMS SNPs. However, after removing all unmutagenized parental and wild-type sibling SNPs, as well as all the HapMap 3.2.1 and A619 SNPs, we found a region of high homozygosity on the short arm of chromosome 1 between 40 and 70 Mbp (Figure 4b). This vns interval was larger compared to the ten region, perhaps due to the small number of individuals (9) included in each vns pool, or the increased recombination at the end of chr.1 where ten is located. As with ten, plotting only EMS SNPs still revealed a (shorter) peak. The presence of many non-EMS SNPs in the vns mapping interval is likely a result of heterozygosity in the parental ns1 ns2 stock. The vns mapping interval included 651 genes.
Fine mapping of ten using custom indel markers quickly reduced the mapping interval
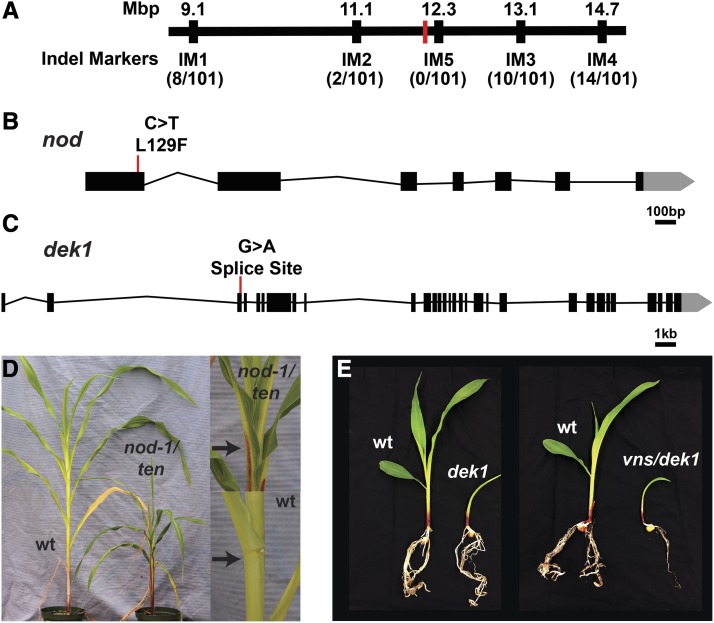
Both the ten and vns pools were sequenced fairly deeply (Table 1), and our sequencing data were thus likely to contain both causative lesions. However, in cases where mutant pools are sequenced to a shallower depth, and the causative lesions might be missed, we reasoned that the sequencing data could be used to design markers for fine mapping to reduce the mapping interval. This fine mapping will also be critical for reducing the mapping interval if there are multiple gene candidates in the mapping interval. Therefore, as a proof of concept, we used our NGS data to design five insertion/deletion (indel) markers to refine the ten interval (Gallavotti and Whipple 2015) (Table S1). We used these markers to genotype 101 ten individuals from the F2 mapping population and narrowed the ten interval to a region that includes 74 genes between 11.1 and 13.1 Mbp (Figure 4a). Thus, we were able to quickly reduce the ten mapping interval more than eightfold, and simplify the evaluation of candidate SNPs.
SNP filtering and candidate SNP identification
Next, we wanted to identify candidate SNPs in our BSA-seq mapping regions. We used a variant calling program, Varscan, to call genotypes at variant positions based on allele frequency (Koboldt et al. 2012). Varscan identified 36,186,873 and 36,790,695 SNPs genome-wide in the ten and vns datasets, respectively (minimum read depth of 8) (Table 2). We removed putative background and non-causative SNPs from these datasets, as described in the Methods. After SNP filtering, we were left with 50 homozygous EMS SNPs in the ten mapping interval, and 427 homozygous EMS SNPs in the vns interval (allele frequency > 0.99).
Table 2. Varscan SNP data summary for ten and vns (coverage > 8).
| SNPs | ten (ROI = 0–20Mbp) | vns (ROI = 40–70Mbp) |
|---|---|---|
| Genome-Wide | ||
| Total SNPs (genome wide) | 36,186,873 | 36,790,695 |
| Homozygous EMS SNPs after filtering | 148 | 7,899 |
| Region of Interest (ROI) | ||
| Total SNPs | 266,837 | 470,763 |
| Homozygous EMS SNPs after filtering | 50 | 427 |
| High effect homozygous EMS SNPs | 0 | 1 |
| Moderate effect homozygous EMS SNPs | 4 | 2 |
To identify which SNPs in these final sets might have deleterious effects on gene function, we ran these SNPs through SnpEff (Cingolani et al. 2012). SnpEff identified 4 missense mutations in our ten mapping interval. The vns mapping interval included 1 splice site mutation, and 2 missense mutations (Table 3).
Table 3. Candidate SNPs for ten and vns.
| Chr | Position | Ref | Alt | Gene ID | SnpEff Call | Effect | Functional Annotation |
|---|---|---|---|---|---|---|---|
| ten | |||||||
| 1 | 12172055 | C | T | Zm00001d027722 | Moderate | L129F | narrow odd dwarf (nod) |
| 1 | 12286537 | C | T | Zm00001d027725 | Moderate | P208L | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase 1 (DHS1) |
| 1 | 12631437 | C | T | Zm00001d027752 | Moderate | G139E | SH2 Domain Containing Protein |
| 1 | 13337447 | G | A | Zm00001d027775 | Moderate | D67N | Histone H2A 8 |
| vns | |||||||
| 1 | 47570165 | G | A | Zm00001d028818 | High | Splice Site | defective kernel1 (dek1) |
| 1 | 53173992 | G | A | Zm00001d028965 | Moderate | A38V | Protein of unknown function |
| 1 | 53174101 | G | A | Zm00001d028965 | Moderate | R2C | Protein of unknown function |
Candidate SNP validation
In our ten dataset, the four missense SNPs were in genes encoding a 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase 1 (DHS1) (Zm00001d027725); an SH2 domain protein (Zm00001d027752); Histone H2A 8 (Zm00001d027775); and a known maize gene - narrow odd dwarf (nod) (Zm00001d027722). nod encodes a maize MID-COMPLEMENTING ACTIVITY protein (Rosa et al. 2017). nod-1 is a loss-of-function allele of nod in the B73 background (Rosa et al. 2017). Compared to the B73 inbred, nod-1 exhibits several pleiotropic developmental defects, such as short stature, small organs, a lack of ligules, compromised apical dominance, and increased tiller growth (Rosa et al. 2017). This pleiotropic phenotype strongly resembled ten single mutants. Combined with our NGS data, this led us to speculate that ten could be an allele of nod.
To test whether ten was an allele of nod, we Sanger sequenced nod in one of our ten mutants, and performed complementation crosses. Sanger sequencing confirmed that ten harbored the same missense mutation in nod that was identified through NGS (Figure 5b). In our field, nod-1 homozygous individuals failed to produce mature tassels or ears by the time that ten plants were ready for crossing. Therefore, we crossed ten to heterozygous nod plants and examined the F1 progeny. Both ten and nod-1 are recessive mutations. Therefore, if ten were an allele of nod, we expected about half of the F1 progeny to show the nod phenotype, and half of the F1 progeny to show a wild-type phenotype. Otherwise, if ten were not an allele of nod, we expected all F1 progeny to show a wild-type phenotype. Indeed, we found that 12 of 26 F1 plants were short, and lacked ligules (P = 0.69, chi-squared test, one degree of freedom, Figure 5d), as is the case with nod mutants (Rosa et al. 2017). Therefore, we concluded that ten is an allele of nod, and henceforth will refer to it as nod-ten.
Figure 5.
ten and vns encode alleles of nod and dek1, respectively. (A) Fine mapping of ten using custom indel markers reduced the mapping interval to a region containing 74 genes. The approximate position of nod-ten is marked in red. (B) The ten lesion in nod is a missense mutation. (C) The vns lesion in dek1 is a splice site mutation. (D) ten fails to complement nod. As in nod-1 mutants (Rosa et al. 2017), the ligule and auricle are both absent in the leaves of ten/nod-1 F1 plants (top). The black arrow indicates the ligule in a wild-type plant (bottom), and the lack of a ligule in a ten/nod-1 F1 plant. (E) vns fails to complement dek1.
The single likely high effect SNP in the vns mapping interval was in a donor splice site of a known maize gene, defective kernel1 (dek1) (Zm00001d028818) (Figure 5c). Dek1 encodes a membrane-spanning protein with a calpain protease domain (Lid et al. 2002). dek1 mutants contain a root primordium but lack a shoot structure (Neuffer et al. 1997), similar to what was observed in vns mutants. This led us to suspect that vns might encode an allele of dek1.
To determine if the SNP in dek1 was responsible for the vns phenotype, we crossed heterozygous vns plants with heterozygous dek1 plants. If vns were an allele of dek1, we expected about one quarter of the F1 progeny to show the dek1 phenotype, and three quarters of the F1 progeny to show a wild-type phenotype. Otherwise, if vns were not an allele of dek1, we expected all F1 progeny to show a wild-type phenotype. In the F1 progeny, 9 of 36 plants were very small, and died shortly after producing only a small number of leaves (P = 1, chi-squared test, one degree of freedom, Figure 5e). Similarly, 7 of 36 F1 progeny of the heterozygous dek1 self exhibited the same phenotype (P = 0.44, chi-squared test, one degree of freedom). Thus, vns failed to complement the dek1 mutant phenotype, indicating that vns is an allele of dek1. Henceforth we will refer to vns as dek1-vns.
Discussion
Here, we report a BSA-Seq method for cloning EMS mutants in maize. Using this method, we identified and validated causative SNPs for two separate EMS-induced mutants. We have provided a detailed protocol for conducting these analyses using Galaxy, as well as a SNP variant file for the A619 genetic background (File S1). This has the potential to be useful for anyone working in A619 - including for making a synthetic A619 reference genome (McKenna et al. 2010). In addition, we showed an interaction between the known maize genes nod and tb1 (Doebley et al. 1997; Rosa et al. 2017).
We discovered a synergistic interaction between tb1 and nod in regulating tiller production (Figure 3). Although nod is predicted to encode a membrane-localized maize MID-COMPLEMENTING ACTIVITY homolog, and may function to coordinate plant development in response to intrinsic and extrinsic cues (Rosa et al. 2017), how exactly nod regulates plant development remains largely unknown. The homolog of NOD in Arabidopsis thaliana, MCA1, may be involved in Ca2+ uptake and the cell wall stress response pathway (Yamanaka et al. 2010). In maize, hormone metabolism and cell division are overrepresented GO categories in nod-1 mutant transcriptomes, relative to wild-type (Rosa et al. 2017). Interestingly, tb1 is downregulated (2.4-fold) in nod-1 shoot apices, relative to wild-type (Rosa et al. 2017), which suggests that nod might act upstream of tb1 to regulate its expression. nod could represent a sensor of positional and environmental cues that regulate tb1-mediated tiller outgrowth. The exact mechanism through which nod and tb1 synergistically regulate tiller outgrowth requires further investigation.
In any BSA-Seq experiment, the key goals are to define the smallest mapping interval possible, and to identify a small number of potentially causative lesions. To achieve these goals, both the size of the mutant pool and sequencing depth must be considered. In our experiments, the nod-ten BSA-Seq mapping interval encompassed ∼20Mbp on the short arm of chromosome 1, and included 623 genes. The dek1-vns mapping interval encompassed ∼30Mbp on the short arm of chromosome 1, and included 651 genes (Figure 4). Thus, despite a much smaller mutant pool (9 dek1-vns individuals vs. 101 nod-ten individuals), and a lower recombination rate where dek1-vns is located (Gore et al. 2009), the dek1-vns mapping interval included only 28 (4%) more genes than the nod-ten mapping interval. Mean coverage in both datasets was fairly high (26-fold for ten and 17-fold for vns), allowing for both causative lesions to be captured. To identify just the genomic interval that contains a particular gene, sequencing depth could be reduced, but the chances of catching the causative lesion will be similarly reduced. In a case where no clear candidate lesion is recovered, and/or there are many genes in a BSA-Seq mapping interval, fine mapping would become essential. Thus, our results indicate that sequencing to the deepest level affordable is more important than a big mutant pool for a successful BSA-Seq experiment.
The most important advantage of BSA-Seq over other methods is its simplicity, both in terms of sample collection and data analysis. While BSA-Seq is used extensively in other taxa (Schneeberger and Weigel 2011; Abe et al. 2012; Mascher et al. 2014; Woods et al. 2014; Ding et al. 2017; Song et al. 2017; Jiao et al. 2018), gene mapping via Bulked-Segregant RNA-Seq (BSR-Seq) is more often used for cloning maize genes (Liu et al. 2012; Li et al. 2013; Nestler et al. 2014; Tang et al. 2014). In BSR-Seq, RNA from a pool of mutants and RNA from a pool of non-mutants is used to make RNA-Seq libraries (Liu et al. 2012). These libraries are sequenced, and the resulting reads mapped to the maize reference genome. BSR-Seq offers a very good method for genome reduction, thus increasing sequencing depth without increasing cost. If the RNA-Seq is performed using the right tissue at the right developmental stage, BSR-Seq offers the advantage of differential gene expression data as well as mapping information. In contrast, since BSA-seq relies on DNA extraction, sample collection can be done at any developmental stage, and from any tissue. Although bulk RNA extractions are not extraordinarily challenging, DNA extractions are far simpler, and can be performed by relatively inexperienced trainees in the lab. This technical simplicity offers the advantage of being able to involve high school students and junior undergraduates in authentic research experiences (Lopatto et al. 2014). In addition, the data analysis for BSR-Seq is not as straightforward as it is for BSA-Seq. In BSR-Seq, allele-specific expression must be accounted for, as well as differential expression of genes not linked to the mutant gene in mutant vs. wild-type pools (Liu et al. 2012). Here, too, simplicity offers speed for the experienced researcher, and excellent training opportunities for beginning scientists.
For dek1-vns, we used a modification of BSA-Seq that has been called MutMap (Abe et al. 2012). In MutMap, instead of generating an F1 between contrasting genotypes, the F1 comes from a mutant backcrossed to a wild-type, unmutagenized individual in the same genetic background as the mutant. Thus, MutMap introduces no additional genetic variation in the initial F1 cross. Instead, MutMap relies on co-segregation of induced SNPs with the mutant phenotype, allowing for the identification of a region of increased homozygosity in the mutant pool. MutMap would likely also have been successful in identifying nod-ten. In both of our datasets, just mapping the EMS mutations reveals clear peaks corresponding to the mutant genes (Figure 4). One advantage that MutMap offers is that no additional genetic variation is introduced in the F1 cross. This genetic variation can suppress or enhance mutant phenotypes, which can make scoring F2 populations challenging. However, in these cases, conventional BSA-Seq could be used to identify both causative lesions and modifiers in one step (Song et al. 2017).
Another advantage of BSA-Seq over MutMap is that even if the lesion is not captured, or if there are many candidate lesions in a mapping interval, a researcher is immediately poised to design indel markers and commence fine-mapping in an identified genomic interval. Numerous candidate lesions are more likely when the mutant under study is not from an EMS mutagenesis experiment, which is fairly common in maize (Vollbrecht et al. 1991; Thompson et al. 2009; Whipple et al. 2011). In these cases, lesions are not necessarily of a defined type (e.g., G to A or C to T transitions), which makes SNP filtering challenging. However, high homozygosity that is polymorphic with a reference genome will still reveal the chromosomal location of the causative lesion, and streamline subsequent fine mapping.
Technical and conceptual advances will eliminate some, but not all, sources of uncertainty when it comes to capturing the candidate lesions underlying mutant phenotypes. With shallow read depths, causative lesions may not be captured; but deep coverage is likely to become ever more attainable as NGS costs drop, thus eliminating this source of uncertainty. There may be a number of potentially causative lesions in a candidate region; but as the maize pan-genome is resolved (Hirsch et al. 2014; Bukowski et al. 2018), the pool of background SNPs that can be eliminated gets ever-deeper when cloning clear null mutations unlikely to be present in natural variation. Other potential sources of uncertainty will likely never go away. For example, filtering out all of the HapMap and pan-genome SNPs will be less useful when trying to identify natural modifiers. BSA-Seq and fine mapping will be particularly helpful in narrowing down a candidate region for a natural modifier; when a causative lesion is not in the coding sequence of an annotated gene; or when mutant pools are contaminated with wild-type or heterozygous samples. Thus, BSA-Seq offers many advantages that are likely to prove useful for mapping and cloning many maize mutants.
Acknowledgments
The authors thank Courtney Babbitt and Beth Thompson for helpful discussion; Sarah Hake for the kind gift of the tb1-sh allele; and Ed Wilcox at the BYU DNA sequencing facility for technical assistance. This work was supported by a USDA-Hatch grant to M.E.B. (MAS00501); the Biology Department at the University of Massachusetts Amherst; an NSF PGRP grant to C.J.W. (IOS-1339332); and an NSF Postdoctoral Research Fellowship in Biology to P.A.C. (IOS-1612235).
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.7014851.
Communicating editor: J. Yang
Literature Cited
- Abe A., Kosugi S., Yoshida K., Natsume S., Takagi H., et al. , 2012. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 30: 174–178. 10.1038/nbt.2095 [DOI] [PubMed] [Google Scholar]
- Addo-Quaye C., Buescher E., Best N., Chaikam V., Baxter I., et al. , 2017. Forward Genetics by Sequencing EMS Variation-Induced Inbred Lines. G3: Genes, Genomes. Genetics 7: 413–425. 10.1534/g3.116.029660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afgan E., Baker D., Batut B., Van Den Beek M., Bouvier D., Čech M., et al. , 2018. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nuc. acids research, 46: W537–W544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afgan E., Baker D., van den Beek M., Blankenberg D., Bouvier D., et al. , 2016. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 44: W3–W10. 10.1093/nar/gkw343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S., 2014. FastQC: a quality control tool for high throughput sequence data. Version 0.11. 2. Babraham Institute, Cambridge, UK http://www. bioinformatics. babraham. ac.uk/projects/fastqc.
- Becraft P. W., Li K., Dey N., Asuncion-Crabb Y., 2002. The maize dek1 gene functions in embryonic pattern formation and cell fate specification. Development 129: 5217–5225. [DOI] [PubMed] [Google Scholar]
- Blankenberg D., Gordon A., Von Kuster G., Coraor N., Taylor J., et al. , 2010. Manipulation of FASTQ data with Galaxy. Bioinformatics 26: 1783–1785. 10.1093/bioinformatics/btq281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukowski R., Guo X., Lu Y., Zou C., He B., et al. , 2018. Construction of the third-generation Zea mays haplotype map. Gigascience 7: 1–12. 10.1093/gigascience/gix134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang L. L., Coon M., Nguyen T., et al. , 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B., Mou F., Sun W., Chen S., Peng F., et al. , 2017. A dominant-negative actin mutation alters corolla tube width and pollinator visitation in Mimulus lewisii. New Phytol. 213: 1936–1944. 10.1111/nph.14281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J., Stec A., Hubbard L., 1997. The evolution of apical dominance in maize. Nature 386: 485–488. 10.1038/386485a0 [DOI] [PubMed] [Google Scholar]
- Ewing B., Hillier L., Wendl M. C., Green P., 1998. Base-Calling of Automated Sequencer Traces UsingPhred. I. Accuracy Assessment. Genome Res. 8: 175–185. 10.1101/gr.8.3.175 [DOI] [PubMed] [Google Scholar]
- Gallavotti A., Long J. A., Stanfield S., Yang X., Jackson D., et al. , 2010. The control of axillary meristem fate in the maize ramosa pathway. Development 137: 2849–2856. 10.1242/dev.051748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A., Whipple C. J., 2015. Positional cloning in maize (Zea mays subsp. mays, Poaceae). Appl. Plant Sci. 3: apps.1400092 10.3732/apps.1400092 [DOI] [Google Scholar]
- Gillmor C. S., Roeder A. H. K., Sieber P., Somerville C., Lukowitz W., 2016. A Genetic Screen for Mutations Affecting Cell Division in the Arabidopsis thaliana Embryo Identifies Seven Loci Required for Cytokinesis. PLoS One 11: e0146492 10.1371/journal.pone.0146492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore M. A., Chia J.-M., Elshire R. J., Sun Q., Ersoz E. S., et al. , 2009. A first-generation haplotype map of maize. Science 326: 1115–1117. 10.1126/science.1177837 [DOI] [PubMed] [Google Scholar]
- Haase N. J., Beissinger T., Hirsch C. N., Vaillancourt B., Deshpande S., et al. , 2015. Shared Genomic Regions Between Derivatives of a Large Segregating Population of Maize Identified Using Bulked Segregant Analysis Sequencing and Traditional Linkage Analysis. G3 (Bethesda) 5: 1593–1602. 10.1534/g3.115.017665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch C. N., Foerster J. M., Johnson J. M., Sekhon R. S., Muttoni G., et al. , 2014. Insights into the maize pan-genome and pan-transcriptome. Plant Cell 26: 121–135. 10.1105/tpc.113.119982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Burow G., Gladman N., Acosta-Martinez V., Chen J., et al. , 2018. Efficient Identification of Causal Mutations through Sequencing of Bulked F2 from Two Allelic Bloomless Mutants of Sorghum bicolor. Front. Plant Sci. 8: 2267 10.3389/fpls.2017.02267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Peluso P., Shi J., Liang T., Stitzer M. C., et al. , 2017. Improved maize reference genome with single-molecule technologies. Nature 546: 524–527. 10.1038/nature22971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt D. C., Zhang Q., Larson D. E., Shen D., McLellan M. D., et al. , 2012. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22: 568–576. 10.1101/gr.129684.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z., Schnable J. C., 2016. RNA-Seq Based Analysis of Population Structure within the Maize Inbred B73. PLoS One 11: e0157942 10.1371/journal.pone.0157942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lid S. E., Gruis D., Jung R., Lorentzen J. A., Ananiev E., et al. , 2002. The defective kernel 1 (dek1) gene required for aleurone cell development in the endosperm of maize grains encodes a membrane protein of the calpain gene superfamily. Proc. Natl. Acad. Sci. USA 99: 5460–5465. 10.1073/pnas.042098799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Li D., Liu S., Ma X., Dietrich C. R., et al. , 2013. The maize glossy13 gene, cloned via BSR-Seq and Seq-walking encodes a putative ABC transporter required for the normal accumulation of epicuticular PLoS One 8: e82333 10.1371/journal.pone.0082333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G. K., Shi W., 2013. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nuc. Acids Res., 41: e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Yeh C.-T., Tang H. M., Nettleton D., Schnable P. S., 2012. Gene Mapping via Bulked Segregant RNA-Seq (BSR-Seq). PLoS One 7: e36406 10.1371/journal.pone.0036406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopatto D., Hauser C., Jones C. J., Paetkau D., Chandrasekaran V., et al. , 2014. A central support system can facilitate implementation and sustainability of a Classroom-based Undergraduate Research Experience (CURE) in Genomics. CBE Life Sci. Educ. 13: 711–723. 10.1187/cbe.13-10-0200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher M., Jost M., Kuon J.-E., Himmelbach A., Aßfalg A., et al. , 2014. Mapping-by-sequencing accelerates forward genetics in barley. Genome Biol. 15: R78 10.1186/gb-2014-15-6-r78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., et al. , 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant N., Lyons E., Goff S., Vaughn M., Ware D., et al. , 2016. The iPlant Collaborative: Cyberinfrastructure for Enabling Data to Discovery for the Life Sciences. PLoS Biol. 14: e1002342 10.1371/journal.pbio.1002342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore R. W., Paran I., Kesseli R. V., 1991. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 88: 9828–9832. 10.1073/pnas.88.21.9828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller R. J., 2006. Ask the plant: investigating and teaching plant structure. Bot. J. Linn. Soc. 150: 73–78. 10.1111/j.1095-8339.2006.00489.x [DOI] [Google Scholar]
- Nardmann J., Ji J., Werr W., Scanlon M. J., 2004. The maize duplicate genes narrow sheath1 and narrow sheath2 encode a conserved homeobox gene function in a lateral domain of shoot apical meristems. Development 131: 2827–2839. 10.1242/dev.01164 [DOI] [PubMed] [Google Scholar]
- Nawy T., Bayer M., Mravec J., Friml J., Birnbaum K. D., et al. , 2010. The GATA factor HANABA TARANU is required to position the proembryo boundary in the early Arabidopsis embryo. Dev. Cell 19: 103–113. 10.1016/j.devcel.2010.06.004 [DOI] [PubMed] [Google Scholar]
- Nestler J., Liu S., Wen T.-J., Paschold A., Marcon C., et al. , 2014. Roothairless5, which functions in maize (Zea mays L.) root hair initiation and elongation encodes a monocot-specific NADPH oxidase. Plant J. 79: 729–740. 10.1111/tpj.12578 [DOI] [PubMed] [Google Scholar]
- Neuffer M. G., Coe E. H., Wessler S. R., et al. , 1997. Mutants of maize. Cold Spring Harbor Laboratory Press, New York. [Google Scholar]
- Rosa M., Abraham-Juárez M. J., Lewis M. W., Fonseca J. P., Tian W., et al. , 2017. The Maize MID-COMPLEMENTING ACTIVITY Homolog CELL NUMBER REGULATOR13/NARROW ODD DWARF Coordinates Organ Growth and Tissue Patterning. Plant Cell 29: 474–490. 10.1105/tpc.16.00878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlon M. J., Schneeberger R. G., Freeling M., 1996. The maize mutant narrow sheath fails to establish leaf margin identity in a meristematic domain. Development 122: 1683–1691. [DOI] [PubMed] [Google Scholar]
- Schneeberger K., Weigel D., 2011. Fast-forward genetics enabled by new sequencing technologies. Trends Plant Sci. 16: 282–288. 10.1016/j.tplants.2011.02.006 [DOI] [PubMed] [Google Scholar]
- Song J., Li Z., Liu Z., Guo Y., Qiu L.-J., 2017. Next-Generation Sequencing from Bulked-Segregant Analysis Accelerates the Simultaneous Identification of Two Qualitative Genes in Soybean. Front. Plant Sci. 8: 919 10.3389/fpls.2017.00919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H. M., Liu S., Hill-Skinner S., Wu W., Reed D., et al. , 2014. The maize brown midrib2 (bm2) gene encodes a methylenetetrahydrofolate reductase that contributes to lignin accumulation. Plant J. 77: 380–392. 10.1111/tpj.12394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tello-Ruiz M. K., Naithani S., Stein J. C., Gupta P., Campbell M., et al. , 2018. Gramene 2018: unifying comparative genomics and pathway resources for plant research. Nucleic Acids Res. 46: D1181–D1189. 10.1093/nar/gkx1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B. E., Bartling L., Whipple C., Hall D. H., Sakai H., et al. , 2009. bearded-ear Encodes a MADS Box Transcription Factor Critical for Maize Floral Development. The Plant Cell Online 21: 2578–2590. 10.1105/tpc.109.067751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till B. J., Reynolds S. H., Weil C., Springer N., Burtner C., et al. , 2004. Discovery of induced point mutations in maize genes by TILLING. BMC Plant Biol. 4: 12 10.1186/1471-2229-4-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad D., Kierzkowski D., Rast M. I., Vuolo F., Dello Ioio R., et al. , 2014. Leaf Shape Evolution Through Duplication, Regulatory Diversification, and Loss of a Homeobox Gene. Science 343: 780–783. 10.1126/science.1248384 [DOI] [PubMed] [Google Scholar]
- Vollbrecht E., Veit B., Sinha N., Hake S., 1991. The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature 350: 241–243. 10.1038/350241a0 [DOI] [PubMed] [Google Scholar]
- Whipple C. J., Kebrom T. H., Weber A. L., Yang F., Hall D., et al. , 2011. grassy tillers1 promotes apical dominance in maize and responds to shade signals in the grasses. Proc. Natl. Acad. Sci. USA 108: E506–E512. 10.1073/pnas.1102819108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H., 2016. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag, New York. [Google Scholar]
- Wickham H., Francois R., 2015. dplyr: A grammar of data manipulation. R package version 0. 4 3.
- Woods D. P., Ream T. S., Minevich G., Hobert O., Amasino R. M., 2014. PHYTOCHROME C is an essential light receptor for photoperiodic flowering in the temperate grass, Brachypodium distachyon. Genetics 198: 397–408. 10.1534/genetics.114.166785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka T., Nakagawa Y., Mori K., Nakano M., Imamura T., et al. , 2010. MCA1 and MCA2 that mediate Ca2+ uptake have distinct and overlapping roles in Arabidopsis. Plant Physio., 152: 1284–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou C., Wang P., Xu Y., 2016. Bulked sample analysis in genetics, genomics and crop improvement. Plant Biotechnol. J. 14: 1941–1955. 10.1111/pbi.12559 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw sequencing data available at NCBI SRA (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA476333). A protocol for NGS data analysis and plotting in R is available at protocols.io (https://www.protocols.io/view/bsa-seq-in-maize-qyedxte). Supplemental material available at Figshare: https://doi.org/10.25387/g3.7014851.