Abstract
ATM is a well-known master regulator of double strand break (DSB) DNA repair and the defective DNA repair has been therapeutically exploited to develop PARP inhibitors based on the synthetic lethality strategy. ATM mutation is found with increased prevalence in advanced metastatic castration resistant prostate cancer (mCRPC). However, the molecular mechanisms underlying ATM mutation-driving disease progression are still largely unknown. Here we report that ATM mutation contributes to the CRPC progression through a metabolic rather than DNA repair mechanism. We showed that ATM deficiency generated by CRISPR/Cas9 editing promoted CRPC cell proliferation and xenograft tumor growth. ATM deficiency altered cellular metabolism and enhanced Warburg effect in CRPC cells. We demonstrated that ATM deficiency shunted the glucose flux to aerobic glycolysis by upregulating LDHA expression, which generated more lactate and produced less mitochondrial ROS to promote CRPC cell growth. Inhibition of LDHA by siRNA or inhibitor FX11 generated less lactate and accumulated more ROS in ATM-deficient CRPC cells and therefore potentiated the cell death of ATM-deficient CRPC cells. These findings suggest a new therapeutic strategy for ATM-mutant CRPC patients by targeting LDHA-mediated glycolysis metabolism, which might be effective for the PARP inhibitor resistant mCRPC tumors.
Keywords: ATM, glycolysis, LDHA, castration-resistant
Introduction
ATM (Ataxia Telangiectasia Mutated) mutation that usually leads to loss of function is found in many types of cancer, ranging from hematological malignancies to solid tumors (Choi, et al. 2016). ATM protein kinase is best known as a chief guardian to protect genomic integrity through its DNA repair functions (Shiloh 2014). In regard to prostate cancer, ATM mutation is identified in ~2% of localized, non-indolent prostate cancer (Fraser, et al. 2017). In advanced metastatic castration resistant prostate cancer (mCRPC), the prevalence of ATM mutation increases to 5–11% (Abida, et al. 2017; Beltran, et al. 2013; Robinson, et al. 2015). Notably, ATM mutation is the second largest group, next to BRCA2 mutation, of DNA-repair gene (DRG) mutation that is found > 20% of mCRPC (Robinson et al. 2015). The identification of DRG mutation status in prostate cancer (most commonly BRCA2 and ATM) provides us not only the prognostic biomarkers to predict disease progression, but also new druggable targets for the treatment of mCRPC. Germline mutations of BRCA1/2 and ATM have been shown to distinguish the risk for lethal and indolent prostate cancer and are associated with shorter survival time (Na, et al. 2017). The clinical impact of DRG mutation on CRPC patients’ response to next-generation hormonal therapy (abiraterone, enzalutamide) remains uncertain as two studies show that CRPC patients with DRG mutations have worse response to abiraterone and enzalutamide (Annala, et al. 2018; Antonarakis, et al. 2018), whereas another two studies demonstrate that DRG mutation-bearing CRPC patients have superior response to enzalutamide and abiraterone (Antonarakis et al. 2018; Hussain, et al. 2018). Despite of the response discrepancy, the mutations of BRCA1/2 and ATM are known as highly promising prognostic biomarkers to predict the therapeutic response of CRPC patients to androgen receptor (AR)-targeted therapies. On the other hand, DRG mutations in mCRPC patients can be therapeutically exploited with PARP inhibitor based on the synthetic lethality strategy (Mateo, et al. 2015).
In addition to its hierarchical regulatory role in DNA repair, ATM has been shown to play a critical role in regulating cellular metabolism (Ambrose and Gatti 2013; Shiloh and Ziv 2013). Cancer cells can rewire their metabolic pathways to meet their needs for malignant transformation (Cairns, et al. 2011). A well-known metabolic phenotype is Warburg effect, which refers to the increased glucose consumption and lactate production in cancer cells even under normal oxygen concentration (Warburg 1956). The functions of Warburg effect remain unclear, but it is proposed to promote tumor progression through several mechanisms, such as increasing access to a limited energy source, enhancing immune cell evasion and allowing for signal transduction through ROS or chromatin modulation (Liberti and Locasale 2016). ATM-deficient human lymphoblastoid cells and neural stem cells have been shown to have intrinsic mitochondrial dysfunctions with defective respiration capacity (Ambrose, et al. 2007; Lee, et al. 2013). A recent study demonstrated that ATM inactivation decreases p53 and upregulates c-Myc to enhance glucose and glutamine consumption, which functions to bypass the nutrient deficiency-induced senescence (Aird, et al. 2015). These studies suggest that ATM function in metabolism is different from its classic role in DNA repair.
In this study, we report that ATM deficiency leads to enhanced Warburg effect in CRPC cells and as a consequence promotes the progression of CRPC. We demonstrated that ATM deficiency further diverted the glucose flux from mitochondria-mediated oxidative phosphorylation (OXPHOS) pathway to aerobic glycolysis by up-regulating lactate dehydrogenase A (LDHA) expression. Inhibition of LDHA by siRNA or inhibitor increased ROS level in cells and suppressed the cell growth of ATM-deficient CRPC cells, which suggests a potential alternative therapeutic strategy for ATM mutation-bearing CRPC patients.
Results
ATM deficiency promotes CRPC progression
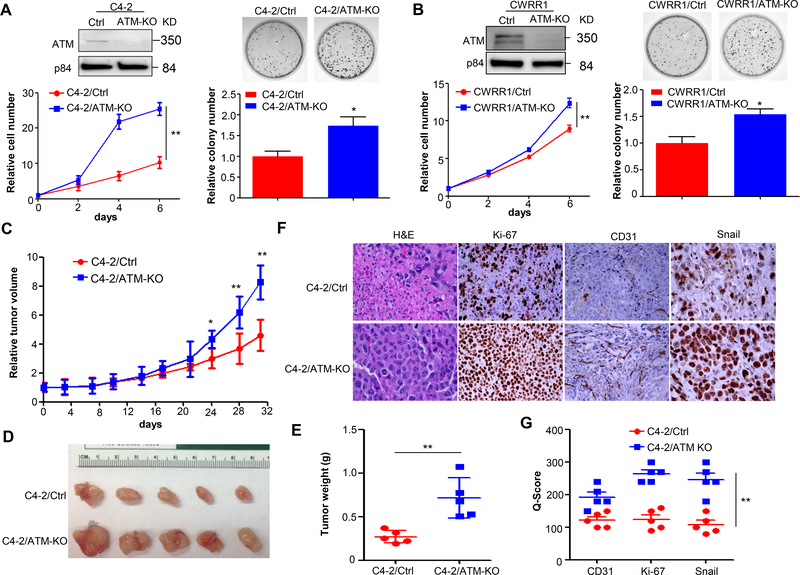
ATM mutations including missense mutations usually lead to loss of or reduced function of ATM kinase activity (Hu and Gatti 2008). To address whether ATM mutation affects CRPC progression, we established ATM knock out (ATM-KO) and corresponding control (Ctrl) stable cell lines by CRISPR/Cas9 editing for C4–2 and CWRR1, two CRPC cell lines. ATM knock-out unexpectedly increased cell proliferation and colony formation in C4–2 cells as shown by the cell growth curve and colony formation assay (Fig.1A). A similar result of cell proliferation and colony formation was observed for CWRR1 cells (Fig.1B), suggesting that ATM deficiency promotes CRPC cell growth, at least in the two examined CRPC cell lines. To confirm this observation in vivo, we subcutaneously injected C4–2/Ctrl and C4–2/ATM-KO cells into immunodeficient SCID/NSG mice and monitored the xenograft tumor formation. As shown in Fig.1C, ATM knock-out significantly promoted the xenograft tumor growth, which is consistent with the in vitro proliferation and colony formation results. The xenograft tumors were then dissected at the end of the in vivo experiment and subjected to immunohistochemistry (IHC) staining analysis. ATM-KO tumors were obviously larger and heavier than control tumors (Fig.1D &E). The H&E (hematoxylin and eosin) staining showed that ATM-KO tumors had less cell necrosis compared to that of control tumors (Fig.1F), suggestive of increased cell proliferation and decreased cell death in ATM-KO tumors. The increased proliferation was further validated by a robust Ki-67 (a proliferation marker) staining in ATM-KO tumors (Fig.1F &G). In addition, ATM-KO tumors showed a strong expression in both vasculature and epithelial-mesenchymal transition (EMT), determined by CD31 and Snail staining (Fig.1F&G), indicating that ATM-KO CRPC tumors are more aggressive than control tumors. Taken together, we showed that ATM deficiency promotes CRPC progression both in vitro and in vivo.
Figure 1.
ATM deficiency promotes CRPC progression. (A&B) Cell proliferation (left panel) and colony formation (right panel) of C4–2/Ctrl vs. C4–2/ATM-KO cells and CWRR1/Ctrl vs. CWRR1/ATM-KO cells generated by CRISPR/Cas9 technology. Top, nuclear lysates were extracted and ATM expression was determined by Western blot. p84 was used as a loading control. Bottom, cell proliferation was measured by MTS assay at each indicated time points and normalized to Day 0 (n = 3 replicates). Colony formation assay was performed for C4–2/Ctrl vs. C4–2/ATM-KO cells and CWRR1/Ctrl vs. CWRR1/ATM-KO cells. Top, representative images for formed colonies. Bottom, graphs showing the quantifications of colonies formed (n = 3 replicates). (C) Xenograft tumor growth of C4–2 stable cell lines generated in (A). Tumor volumes were measured twice a week and normalized to Day 0 (n = 5 mice per group). (D) Representative tumors in each group. (E) The weight of xenograft tumors obtained at the end of animal study (n = 5 mice per group). (F) Representative images of H&E and IHC analysis of CD31, Ki-67 and Snail in xenografts obtained from (E). Magnification: 200×. (G) A plot of Quick-score for IHC staining in (F). All data are depicted as mean±s.d. * p < 0.05 and ** p < 0.01 by two-tailed Student’s t-test.
ATM deficiency alters cellular metabolism and enhances Warburg effect in CRPC cells
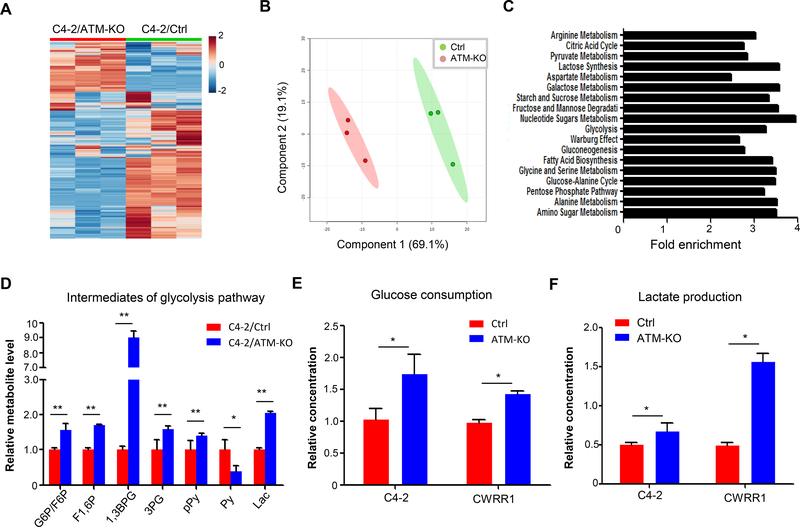
Cellular metabolic reprogramming has been shown to support their rapid proliferation and sustainable growth of cancer cells (Pavlova and Thompson 2016). To test whether ATM deficiency-induced CRPC progression is caused by alteration of cellular metabolism, we conducted a metabolite profiling for C4–2/Ctrl and C4–2/ATM-KO cells. Notably, ATM deficiency resulted in a significant change of global metabolites in C4–2 cells (Fig.2A, Supplementary table 1). This observation is supported by a principal component analysis in which C4–2/ATM-KO cells were segregated from C4–2/Ctrl cells on metabolite profiles (Fig.2B). We further analyzed the metabolic pathways associated with the changed metabolites and found that ATM deficiency significantly altered a large number of metabolism pathways, especially glucose-related energy pathways (Fig.2C). It is well known that cancer cells produce energy to support their growth through aerobic glycolysis instead of mitochondria-mediated OXPHOS (Warburg effect). To investigate whether ATM deficiency affects Warburg effect in CRPC cells, we compared the levels of intermediate metabolites during the aerobic glycolysis pathway from the profiling data. All glycolytic intermediates, except pyruvate, were shown significantly higher levels in C4–2/ATM-KO cells compared to that in C4–2/Ctrl cells (Fig.2D). In agreement with the profiling data, ATM deficiency led to an increase in both glucose consumption and lactate production in C4–2 cells (Fig.2E &F). A similar result of increased glucose consumption and lactate production was observed in another ATM-deficient CRPC CWRR1 cells (Fig.2 E&F).Therefore ATM-deficient CRPC cells have an altered metabolism profile with enhanced aerobic glycolysis, which suggests that it is highly likely that ATM deficiency promotes CRPC progression through enhanced aerobic glycolysis.
Figure 2.
ATM deficiency leads to an enhanced glycolytic utilization in CRPC cells. (A) Heat map to show levels of global metabolites in C4–2/Ctrl and C4–2/ATM-KO cells (n = 3 biological replicates). (B) Principal component analysis (PCA) of metabolic distributions in the indicated cell lines. (C) Enriched glucose-related metabolite pathways in C4–2/ATM-KO cells. Statistically significant differences were assessed by MetaboAnalyst software as indicated in the Materials and Methods. (D) LC-MS analysis of cell extractions to compare the levels of glycolytic intermediates between C4–2/Ctrl and C4–2/ATM-KO cells (n = 3). G6P, glucose 6-phosphate; F6P, fructose 6-phosphate; F1,6P, fructose 1,6-bisphosphate; 1,3BPG, 1,3-bisphosphoglycerate; 3PG, 3-phosphoglycerate; pPy, phosphoenolpyruvate; Py, pyruvate; Lac, lactate. (E & F) Glucose consumption and lactate production in paired C4–2/Ctrl and C4–2/ATM-KO, CWRR1/Ctrl and CWRR1/ATM-KO cells. Glucose consumption was calculated as indicated in the Materials and Methods. Results are normalized to the control group (n = 3). All data are depicted as mean ± s.d. * p < 0.05 and ** p < 0.01 by two-tailed Student’s t-test.
ATM deficiency detours glucose flux into aerobic glycolysis in CRPC cells
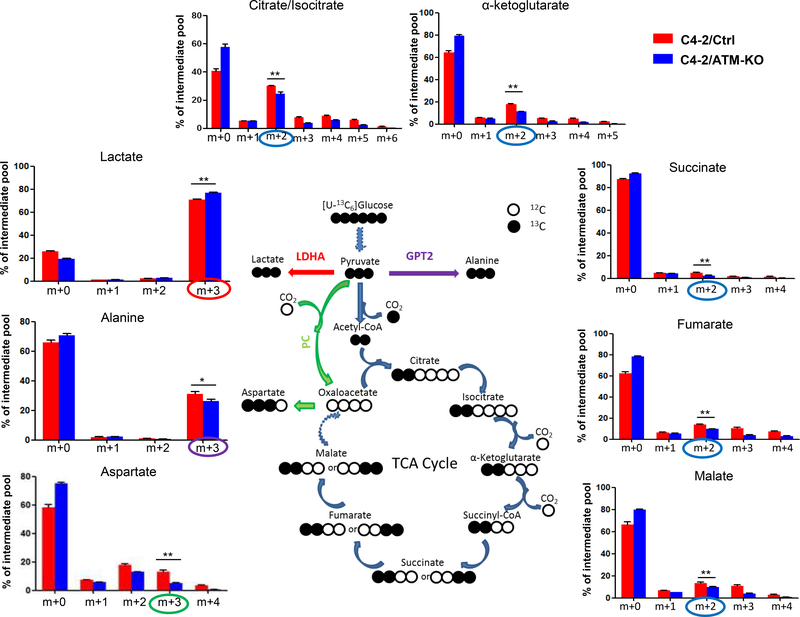
To further validate the enhanced Warburg effect in ATM-deficient CRPC cells, we used uniformly 13C-labeled glucose ([U-13C]-Glc) to trace the glucose influx in C4–2/Ctrl and C4–2/ATM-KO cells. Three different glucose-derived metabolite pathways were followed: 1) 13C3-lactate (m+3 form) that represents the endpoint of aerobic glycolysis; 2) intermediates of TCA (tricarboxylic acid) cycle (from 13C2-citrate to 13C2-malate) that represents the mitochondrial OXPHOS pathway; and 3) 13C3-Alanine and 13C3-Aspartate that indicate other glucose-derived pathways. As shown in Fig.3, the level of glucose-derived 13C3-lactate was significantly raised in ATM-KO C4–2 cells whereas the levels of glucose-derived intermediates of TCA cycle (13C2-citrate/isocitrate, 13C2-α-ketoglutarate, 13C2-succinate, 13C2-fumarate and 13C2-malate) were all reduced in ATM-KO C4–2 cells, suggesting that ATM deficiency shunts the glucose flux to glycolysis from OXPHOS. The augment of the entry of glucose to glycolysis is further supported by the observation that the production of glucose-derived 13C3-alanine and 13C3-aspartate was attenuated in C4–2/ATM-KO cells (Fig.3). A similar result was found in CWRR1 cells as well wherein the 13C3-lactate level was increased in CWRR1/ATM-KO cells while the concentrations of other representative glucose-derived metabolites were decreased in CWRR1/ATM-KO cells (Supplementary Fig.1). Taken together, the glucose tracing results suggest that ATM deficiency further diverts the glucose flux to aerobic glycolytic pathways, i.e. enhances Warburg effect, in CRPC cells.
Figure 3.
ATM deficiency detours the glucose flux in CRPC cells. C4–2/Ctrl and C4–2/ATM-KO cells were grown in uniformly 13C-labeled glucose medium. The incorporation of 13C atoms from 13C6-Glc in to citrate/isocitrate, α-ketoglutarate (α-KG), succinate, fumarate, malate, lactate, alanine and aspartate are denoted as m+n, where n is the number of 13C atoms. Blue, red, purple and green circles all indicate that 13C atoms are derived from 13C6-Glc but through different pathways. A scheme describing glucose flows is shown in the middle of the figure. LDHA, lactate dehydrogenase A; GPT2, glutamate-pyruvate transaminase; PC, pyruvate carboxylase; CO2 indicates where carbon dioxide is released. All data are depicted as mean ± s.d. * p < 0.05 and ** p < 0.01 by two-tailed Student’s t-test.
Upregulation of LDHA contributes to ATM deficiency-induced CRPC progression
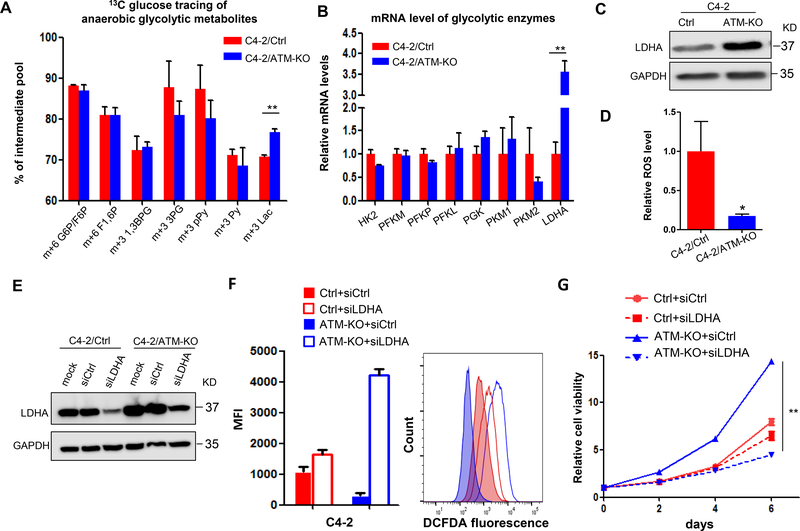
The enhanced Warburg effect as well as the increased production of lactate in ATM-deficient CRPC cells prompted us to determine which enzyme in the glycolysis pathway is regulated by ATM deficiency. We first compared the level of metabolites along the glycolysis pathway from the tracing data and found that only glucose-derived lactate, not other intermediate metabolites, was significantly increased in C4–2/ATM-KO cells (Fig.4A), suggesting that the enzyme LDHA, that is responsible for converting pyruvate to lactate, might be upregulated by ATM deficiency. Actually, we examined the mRNA level of LDHA as well as other enzymes along the glycolysis pathway by RT-qPCR in the two cell lines. Only LDHA was dramatically upregulated in C4–2/ATM-KO cells (Fig.4B) and this upregulation is further supported by the western blot showing ~3 fold increase of LDHA protein expression in ATM-KO cells (Fig.4C). To further confirm the upregulation of LDHA by ATM loss, we treated C4–2 cells with ATM inhibitor KU-60019 and found that the expression of LDHA increased after 72 hours treatment of cells with KU-60019 (Supplementary Fig.2).
Figure 4.
ATM deficiency upregulates LDHA to promote CRPC progression. (A) 13C-isotopomer tracing analysis of glucose-derived glycolytic metabolites in C4–2/Ctrl and C4–2/ATM-KO cells (n = 3). G6P, glucose 6-phosphate; F6P, fructose 6-phosphate; F1,6 P, fructose 1,6-bisphosphate; 1,3BPG, 1,3-bisphosphoglycerate; 3PG, 3-phosphoglycerate; pPy, phosphoenolpyruvate; Py, pyruvate; Lac, lactate. (B) RT-qPCR to determine the mRNA levels of glycolytic enzymes. Data are normalized to the control group (n = 3 independent experiments). HK, hexokinase; PFK, phosphofructokinase; PGK, phosphoglycerate kinase; PK, pyruvate kinase; LDHA, lactate dehydrogenase A. (C) Western blot to determine LDHA protein expression. GAPDH was used as a loading control. (D) Relative level of intracellular ROS. Results are normalized to control group (n = 3 independent experiments). (E) C4–2/Ctrl and C4–2/ATM-KO cells were transfected with siRNAs targeting either a scramble sequence (siCtrl) or LDHA (siLDHA). Western blot confirmed knockdown of LDHA expression by using siRNAs. GAPDH was used as a loading control. (F) Intracellular ROS production was detected with DCFDA fluorescence and monitored by flow cytometry at 24 hours post-transfection with siCtrl or siLDHA. Left, median fluorescent intensity. Right, the representative image of histograms of ROS (n = 3 independent experiments). (G) Cell viability of C4–2/Ctrl and C4–2/ATM-KO cells transfected with siCtrl and siLDHA was determined by MTS assay at the indicated time points (n = 3 replicates). All data are depicted as mean ± s.d. * p < 0.05 and ** p < 0.01 by two-tailed Student’s t-test.
The upregulation of LDHA by ATM deficiency facilitated the conversion of glucose-derived pyruvate to lactate (Fig.4A), which is also expected to decrease the entry of pyruvate to mitochondria for OXPHOS and thereby decrease the production of mitochondrial ROS. We therefore determined the ROS level in C4–2/Ctrl and C4–2/ATM-KO cells by DCFDA (5-(and-6)-carboxy-2’, 7’-dichlorodihydrofluorescein diacetate) fluorescence flow cytometry. As expected, we found that C4–2/ATM-KO cells produced much less ROS than C4–2/Ctrl cells (Fig.4D), indicating that the cellular oxidative stress is much less in C4–2/ATM-KO cells and consistent with its increased proliferation capacity. Reduction of LDHA is then expected to divert the pyruvate to mitochondria, increase the production of ROS and thereby induce the cell death (Le, et al. 2010). We used siRNA to knock down LDHA expression in both C4–2/Ctrl and C4–2/ATM-KO cells as well as CWRR1/Ctrl and CWRR1/ATM-KO cells (Fig.4E and Supplementary Fig.3A) and found that reduction of LDHA by siRNA significantly induced more ROS in C4–2/ATM-KO and CWRR1/ATM-KO cells than C4–2/Ctrl and CWRR1/Ctrl cells (Fig.4F and Supplementary Fig.3B) and as a consequence remarkably inhibited cell growth of C4–2/ATM-KO and CWRR1/ATM-KO cells but only caused a marginal inhibition on C4–2/Ctrl and CWRR1/Ctrl cell growth (Fig.4G and Supplementary Fig.3C).
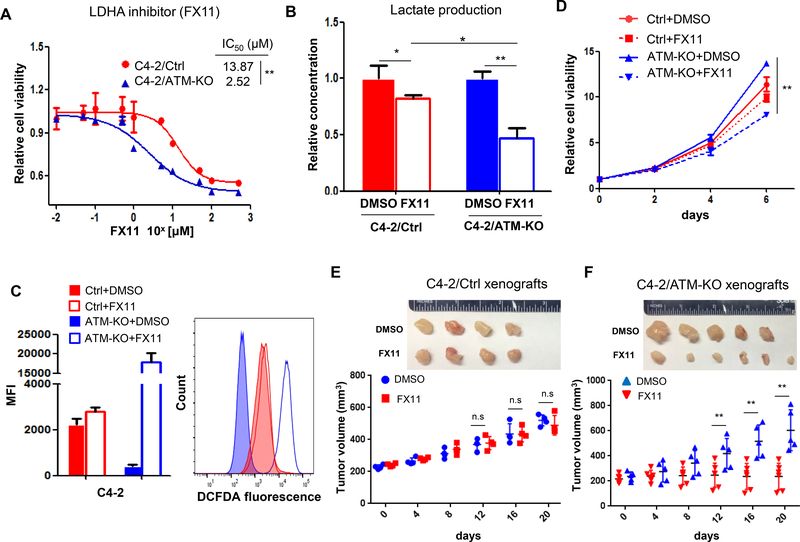
LDHA inhibitor FX11 suppresses ATM-deficient CRPC cells growth
Since inhibition of LDHA by siRNA suppressed the C4–2/ATM-KO cell growth by inducing oxidative stress, we next sought to determine whether LDHA inhibitor has the same inhibitory effect on ATM-deficient CRPC cells that may document a new therapeutic target for this subset of CRPC patients. We selected a small-molecule FX11, which has been shown to preferentially inhibit LDHA and suppress tumor progression (Le et al. 2010). First, we examined the IC50 of FX11 on the two C4–2 cell lines and found that C4–2/ATM-KO cells were more sensitive to the inhibitor than C4–2/Ctrl cells (IC50 = 2.52 μM for C4–2/ATM-KO versus IC50 = 13.87 μM for C4–2/Ctrl) (Fig.5A). Then, we examined the lactate production after cells being treated with FX11. An obvious reduction of lactate secretion was observed for both C4–2/Ctrl and C4–2/ATM-KO cells but FX11 treatment resulted in a further significant reduction of lactate production in C4–2/ATM-KO cells (Fig.5B), manifesting the efficacy of this LDHA inhibitor on ATM-deficient CRPC cells. To determine the effect of FX11 on cell biological functions, we measured the ROS level and cell growth after cells being treated with FX11. Similar to the effect of siLDHA, FX11 treatment led to more ROS production in C4–2/ATM-KO cells and consequently induced more cell death on C4–2/ATM-KO cells (Fig.5 C&D). Additionally, we employed FX11 in CWRR1 cells as well and found the similar results shown in C4–2 cells (Supplementary Fig.4). To confirm that FX11 also has the efficacy on the ATM-deficient xenograft tumors in vivo, we established the C4–2 xenograft tumors in SCID/NSG mice and intraperitoneally injected either FX11 or vehicle every other day after tumors reached the size of 200 mm3. As shown in Fig.5E & F, FX11 profoundly inhibited tumor growth of C4–2/ATM-KO xenografts but had a marginal insignificant inhibition on C4–2/Ctrl xenograft tumors, in a similar manner to the in vitro results (Fig.5D).
Figure 5.
LDHA inhibitor FX11 suppresses ATM-deficient CRPC cell growth. (A) IC50 values of FX11 were measured on C4–2/Ctrl and C4–2/ATM-KO cells treated with a series concentration of FX11 for 72 hours. (B) Lactate production of C4–2/Ctrl and C4–2/ATM-KO cells treated with either DMSO or FX11 for 72 hours. Results are normalized to DMSO group and set as “1”. (C) Intracellular ROS production was detected with DCFDA fluorescence and monitored by flow cytometry at 72 hours post-treatment with either DMSO or 3 μM FX11. Left, median fluorescent intensity. Right, the representative image of histograms of ROS (n = 3 independent experiments). (D) Cell viability of C4–2/Ctrl and C4–2/ATM-KO cells treated with either DMSO or FX11 was determined by MTS assay at the indicated time points (n = 3 replicates). (E & F) Tumor growth of C4–2/Ctrl and C4–2/ATM-KO xenografts treated intraperitoneally with either DMSO or FX11 when tumor reached ~200mm3. Tumor volumes were measured every four days. Top, Representative tumor images in each group. Bottom, The growth curve of xenograft tumors after treatment of vehicle (DMSO) or FX11. All data are depicted as mean ± s.d. * p < 0.05 and ** p < 0.01 by two-tailed Student’s t-test.
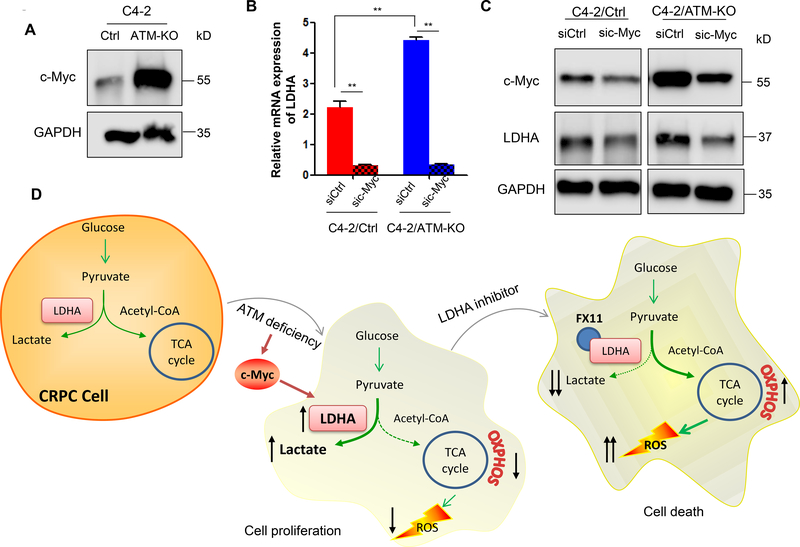
ATM deficiency upregulates LDHA expression via enhancing c-Myc level
Next, we sought to explore the potential mechanism how LDHA is regulated by ATM. The link between LDHA and c-Myc has been well established and LDHA is identified as a c-Myc-responsive gene (He, et al. 2015; Lewis, et al. 1997; Shim, et al. 1997). In addition, it has been shown that c-Myc is upregulated by ATM inactivation in response to replication stress (Aird et al. 2015). Hence, we wanted to know whether the upregulation of LDHA by ATM loss is mediated by c-Myc. First, we examined the c-Myc protein level in both C4–2/Ctrl and C4–2/ATM-KO cells and found that c-Myc protein level was dramatically increased in ATM-deficient C4–2 cells (Fig.6A). Then, we silenced c-Myc by siRNAs and found that knockdown of c-Myc resulted in reduced LDHA expression at mRNA and protein levels in both C4–2/Ctrl and C4–2/ATM-KO cells (Fig.6B &C), further confirming the ATM regulation of LDHA through c-Myc.
Figure 6.
ATM deficiency upregulates LDHA expression via enhancing c-Myc level. (A) Western blot to determine c-Myc protein expression in C4–2/Ctrl and C4–2/ATM-KO cells. GAPDH was used as a loading control. (B & C) C4–2/Ctrl and C4–2/ATM-KO cells were transfected with siRNAs targeting either a scramble sequence (siCtrl) or c-Myc (sic-Myc). RT-qPCR confirmed that LDHA mRNA was downregulated after c-Myc knockdown in both C4–2/Ctrl and C4–2/ATM-KO cells (B) and Western blot confirmed knockdown of c-Myc expression by siRNAs and the subsequent downregulation of LDHA protein expression (C). GAPDH was used as a loading control. (D) A working model of up-regulated LDHA contributing to the progression of ATM-deficient CRPC.
Discussion
Collectively, we demonstrated in this study that ATM deficiency promoted the cell proliferation of CRPC cells by enhancing Warburg effect. ATM deficiency induced LDHA upregulation that shunted the glucose-derived pyruvate into aerobic glycolysis to generate more lactate and simultaneously reduced the entry of pyruvate to mitochondria and therefore produced less ROS. Both increased lactate and reduced ROS cooperated to promote the progression of CRPC (Fig.6D). Further, we showed that inhibition of LDHA by siRNA or inhibitor FX11 decreased the lactate production and increased ROS level in ATM-deficient CRPC cells and therefore potentiated the cell death of ATM-deficient CRPC cells (Fig.6D). Additionally, we demonstrated that c-Myc mediates the upregulation of LDHA by ATM loss (Fig.6D). These findings for the first time reveal a novel non-DNA repair mechanism whereby ATM mutations contribute to the CRPC progression and suggest an alternative therapeutic target for the treatment of ATM mutant CRPC tumors in addition to PARP inhibitors.
Although the clinical impact of ATM mutation (and BRAC1/2 mutation) on CRPC patients’ response to AR-targeted therapies remains debated (Annala, et al. 2017; Annala et al. 2018; Antonarakis et al. 2018; Hussain et al. 2018), the increased prevalence of ATM mutation in therapeutic resistant mCRPC patients and the worse prognosis of ATM mutant prostate cancer patients implicate the important role of ATM deficiency in the disease progression of prostate cancer and therapeutic resistance development. ATM and BRCA2 are known to play a critical role in homologous recombination repair (HRR) of DNA double strand breaks. The clinical observation that ATM-mutated and BRCA2-mutated tumors are enriched in mCRPC suggests that ATM deficiency and/or BRCA2 deficiency should re-establish other pathways to drive the disease progression irrespective of defective HRR DNA repair. Actually, germline BRCA2 mutant prostate tumors have a mutational profile that more closely resembles metastatic and aggressive prostate tumors and deregulates MED12L/MED12 pathway (Taylor, et al. 2017). Unfortunately, the genomic and epi-genomic profiles of ATM-mutant prostate tumors are still not available. Here we used CRPC cell and xenograft models to demonstrate that ATM deficiency resulted in CRPC progression by enhancing Warburg effect, which may shed new light on the issue of how ATM mutation driving prostate cancer progression. Transgenic or prostate conditional Atm knockout mouse model may help to solve this issue and debate.
ATM can modulate carbon metabolism through many pathways such as glycolysis and pentose phosphate pathway (PPP) (Cosentino, et al. 2011; Kruger and Ralser 2011). In regard to glycolysis, ATM has been shown to transcriptionally modulate glucose transporter genes SLC2A1 and SLC2A4 which encode for GLUT1 and GLUT4, respectively (Schwartzenberg-Bar-Yoseph, et al. 2004). Here we demonstrated that ATM deficiency or inhibition led to upregulation of LDHA in both mRNA and protein level (Fig.4 and Supplementary Fig.2) and the potential underlying mechanism might be mediated by transcription factor c-Myc (Fig.6). Interestingly, these findings are quite consistent with a recent study that ATM inactivation enhances glucose consumption by upregulating c-Myc expression (Aird et al. 2015) and other previous studies that c-Myc is positively correlated with LDHA expression in lymphoid cells and pancreatic cancer cells (He et al. 2015; Lewis et al. 1997; Shim et al. 1997). LDHA is overexpressed in many cancers and high LDHA levels frequently correlate with poor prognosis and survival (Girgis, et al. 2014). More importantly, based on a most recent study, LDHA has been implicated as significant clinic biomarker since its genomic abnormalities would link to high pretreatment LDH level and poor response to docetaxel in mCRPC (Hiew, et al. 2018). Furthermore, 64.3% of the patients with high LDH level were determined to have DNA repair gene mutations such as BRCA1/2 and ATM by next-generation sequencing (Hiew et al. 2018). Inhibition of LDHA in multiple tumor models decreased tumor growth, emphasizing the importance of LDHA in tumor progression (Le et al. 2010; Xie, et al. 2014). Lactate, which has been merely considered as a metabolic by-product for decades, is now recognized as a primary source of carbon for energy production (Hui, et al. 2017) as well as a multi-functional metabolite that can promote cancer growth (San-Millan and Brooks 2017). Up-regulating LDHA expression also reduces the carbon flow from pyruvate into the TCA cycle, thereby minimizing ROS that is normally generated as byproducts of OXPHOS (Newington, et al. 2012). Our data revealed that C4–2/ATM-KO cells had less ROS and produced more lactate, both may cooperate together to promote CRPC cells growth (Fig.4). Interestingly, another study reported that inhibiting ATM by ATM inhibitor KU-55933 stimulates the glycolysis pathway while reducing mitochondrial membrane potential and coupled respiration (Zakikhani, et al. 2012), which is corroborating with our findings in ATM-depleted CRPC cells.
PARP inhibitors, based on the synthetic lethality strategy, have been shown effective to treat BRCA1/2 mutation –bearing ovarian cancer and breast cancer (George, et al. 2016). Mateo et al. also demonstrated that PARP inhibitor Olaparib as a monotherapy can lead to a high response rate in patients who had defects in DNA repair genes (mostly BRCA2 and ATM) (Mateo et al. 2015), which paves the way to use PARP inhibitor for the treatment of mCRPC patients with DRG mutations. However, PARP inhibitor resistance have been shown to develop in ovarian and breast cancer and the underlying resistant mechanisms are mostly related to the restoration of HRR repair capacity in these DRG mutant tumors (Lord, et al. 2015). In this study, we demonstrated that ATM deficiency promoted CRPC tumor growth through its non-DNA repair function and our data indicated that ATM-deficient CRPC cells were more sensitive to LDHA inhibitors, strongly suggesting an alternative therapeutic option for ATM mutant CRPC tumors by targeting LDHA, which may also be effective for the PAPR inhibitor resistant CRPC tumors.
Materials and Methods
Cell culture
C4–2 and CWRR1 human prostate cancer cell lines were obtained from American type Culture Collection (ATCC). All experiments were performed using cells that were <10 passages. C4–2 and CWRR1 cells were cultured in RPMI-1640 medium (ATCC) supplemented with 10% fetal bovine serum (FBS; Corning) and 100 units/ml penicillin/streptomycin (Gibco) in a 5% (vol/vol) CO2 and 95% (vol/vol) air incubator at 37 °C.
Colony formation assay
1×104 Cells were seeded into 6-cm dishes and grown for up to 14 days for colony formation. Cell culture media was refreshed every three days and colonies were fixed with cold methanol and then stained with 1% crystal violet. The number of colonies was counted and imaged with a VersaDoc Imaging System (BioRad).
CRISPR/Cas9-mediated ATM knockout
To establish ATM knockout stable cell line, we used the CRIPR/Cas9 technology. Single guided RNA (sgRNA) sequences were generated by CRISPR design tool (crispr.mit.edu). sgRNA sequences for ATM are available upon request. Annealed double stranded sgRNA oligos were ligated to the lentiCRISPR vector 2 (deposited by Dr. Feng Zhang to Addgene, Camridge, MA) at a ribonucleoprotein complex which expressed both Cas9 and sgRNA. To produce infectious transgenic lentivirus, the transfer plasmid was transfected into 293T cells together with packaging plasmid and envelope plasmid. After medium changed and a brief incubation period, supernatant containing the virus was collected and centrifuged to concentrate virus. Subsequently, cells were selected by puromycin for more than 14 days. Population that showed no target protein expression was confirmed by western blot.
RNA interference
siRNAs targeting human LDHA (ON-TARGETplus SMARTpool: L-008201–00-0005) and ON-TARGETplus Nontargeting pool were purchased from Dharmacon. siRNAs targeting human c-Myc (sc-29226) were purchased from Santa Cruz. 105/well C4–2/Ctrl (CWRR1/Ctrl) or C4–2/ATM-KO (CWRR1/ATM-KO) cells were seeded in 6-well plate. After 24 hours, 30 pmol siLDHA/sic-Myc or siCtrl was transfected into cells using Lipofectamine RNAiMAX (Invitrogen) in Opti-MEM reduced serum medium (Gibco) for 24h.
Cell viability assays
The MTS assay (Biovision) was employed to determine cell viability. 1000/well cells were seeded in 96-well plate. 10 μl /well MTS Reagent was added into each well and incubated for 1 hour at each time point. Absorbance was measured at 490nm using SpectraMax M3 reader with SoftMax Pro 6 software for data acquisition and analysis. For cells treated with siRNAs, 5 pmol siLDHA or siCtrl was transfected into each well and cell viability was measured at each time point. For cells treated with FX11 (Millopore), 3 μM FX11 was added in culture medium followed by cell viability measurement. For IC50 determination of FX11, cells were treated by a series of concentration for 72 hours. IC50 value was calculated by GraphPad Prism software.
Animal study
The animal experiment protocol (A092–16-04) was reviewed and approved by the Duke Institutional Animal Care and Use Committee. Immunocompromised NSG mice were purchased from Jackson Laboratories. 107 C4–2/Ctrl or C4–2/ATM-KO cells were resuspended in 100 μl of saline with 50% Matrigel (Corning) and injected subcutaneously into both sides of flank regions of 4–6 weeks old male NSG mice. When tumor volume reached a size of 100 mm3 which was set as Day 0, tumors were measured twice a week and tumor volumes were determined by caliper measurement of tumor length (L) and width (W) according to the formula (LxW2)/2. For FX11 in vivo experiment, when tumor volume reached 200 mm3, C4–2/Ctrl and C4–2/ATM-KO grafted mice were divided into two groups randomly. Each group of mice were injected with either control 2% (vol/vol) DMSO or 50 μg of FX11 intraperitoneally every other day. Tumor volumes were measured every four days.
Immunohistochemical staining and Quick-score system
Immunohistochemical staining (IHC) was performed as described in previous study (Park, et al. 2017). Xenograft tumor sections were deparaffinized and rehydrated. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide in methanol. Heat-induced antigen retrieval was carried out for all sections in 0.01 M citrate buffer, pH 6.0, using a steamer at 95oC. Primary antibodies were diluted with PBS containing 1% BSA to an optimal concentration and applied to the sections overnight. Slides were then incubated with rabbit anti-mouse or goat anti-rabbit IgG/HRP second antibody for 45 minutes. Diaminobenzidine (DAB) was then applied for 10 minutes. The sections were counterstained with hematoxylin, dehydrated, coverslipped and visualized. Primary antibodies used in this study are ant-CD31 (Abcam, Ab28364, 1:100), anti-Ki67 (ThermoFisher, RM-9106, 1:300) and anti-Snail (Abcam, Ab85936, 1:600). Quick-score was calculated as reported (Park et al. 2017), the intensity of staining is multiplied by the percentage of staining to derive a composite score (a range from 0 to 300).
Metabolite extraction and mass spectrometry
As described previously(Pan, et al. 2016), 105 /well cells were plated in 6-well plate. 24 hours later, medium was replaced freshly as equal volume (2 ml) per well and cells were incubated for another 24 hours. Next, medium was aspirated and 80% (vol/vol) HPLC grade methanol/water was added into each well. Plates were placed on dry ice immediately and then transferred in a −80°C freezer for 15 minutes for further enzyme inactivation. Cells were then scraped into extraction solvent. After centrifuge for 10 minutes, supernatant was transferred into a new Eppendorf tube. A speed vacuum was used to dry the samples at room temperate then the dry pellets were sent to LOCASALE LAB (Duke University) for further liquid chromatography mass spectrometry analysis (LC-MS). Data was normalized to cell numbers.
13C isotope tracing assay
105 /well cells were seeded in 6-well plate in 2 ml complete medium. After overnight incubation, medium was removed, and replaced by exact 2 ml fresh glucose-deprived media (Gibco 11966) containing 25 mmol/L uniformly 13C-labeled glucose isotope (Cambridge). After 24 hours incubation, metabolites were extracted and sent for further LC-MS analysis.
Glucose consumption assay
Glucose concentration in culture medium was determined by Glucose Colorimetric/Fluorometric Assay Kit (Biovision) following the manufacturer’s protocol. Glucose consumption was calculated as measured value subtracted by original concentration.
Lactate secretion assay
The lactate level in the medium was analyzed by Lactate Colorimetric/Fluorometric Assay Kit (Biovision). 10 μl of sample was added into 40 μl Lactate Assay Buffer, and then 50 μl Reaction Mix was added into each diluted sample. The reaction was incubated for 30 minutes at room temperature while being protected from light. Absorbance at OD570 nm was measured by SpectraMax M3 reader. All assays were performed in triplicate.
RNA extraction and quantitative real-time PCR
Total RNA was extracted using RNeasy mini kits (Qiagen, Valencia, CA). cDNA synthesis was performed by PrimeScript RT Master Mix (Takara). Quantitative real-time PCR (qRT-PCR) was applied on ABI 7500 (Applied Biosystems, Foster City, CA) using SYBR Green PCR Master Mix (Quanta Biosciences). Actin expression assay was used as an internal control. All qPCRs were determined in triplicate. Sequences of primers use in study are available upon request.
Immunoblotting
Total protein was collected by lysing adherent cells with RIPA buffer (Sigma) supplemented with phosphatase and protease inhibitor cocktail (Thermo Fisher). For nuclear protein extraction, nuclear lysates were isolated with an NE-PER Nuclear and Cytoplasmic Extraction Reagents Kit (Thermo) with phosphatase and protease inhibitor cocktail contained. Protein concentration was measured using the Bradford reagent (Bio-rad). Equal amount of protein was loaded for immunoblotting. After electrophoresis, the protein was transferred to polyvinylidene difuoride transfer membrane (PVDF) followed by 5% non-fat milk blocking buffer and incubated overnight with primary antibodies. After washing with TBST (TBS with 0.1% Tween), anti-mouse or anti-rabbit second antibodies conjugated with horseradish peroxidase-conjugated (HRP) were used to probe the membranes. Samples were developed by Chemiluminescent Substrate (Thermo Fisher) and exposed by Odyssey Imaging Systems (LI-COR). Densitometry was performed using ImageJ software (National Institutes of Health) and samples were normalized to internal controls. Primary antibodies used in this study are anti-ATM (Novus, NB100–104), anti-p84 (Thermo Fisher, PA4–27816), anti-LDHA (Cell Signaling, 2012S), anti-c-Myc (Abcam, 32072) and anti-GAPDH (Abcam, Ab9484).
ROS measurement
As previously described(Le et al. 2010), intracellular ROS production was determined by staining cells with 10 μM 5-(and-6)-carboxy-2’,7’-dichlorodihydrofluorescein diacetate (carboxy-H2DCFDA, Thermo Fisher) according to the manufacturer’s instructions. 105 cells/well (6-well plate) were treated with either 3 μM FX11 for 72 hours or 30 pmol siLDHA for 24 hours. Stained cells were analyzed by FACScan flow cytometers at excitation and emission wavelengths of 485 and 520 nm, respectively.
Statistical analysis
All experiments were performed at least three times and data was presented as the mean±standard deviation (SD). Statistically significant differences between two groups were assessed using a two-tail paired t-test. p<0.05 was considered significant. All statistical analyses were performed using GraphPad Prism software and Microsoft Excel. Principal component analysis (PCA) and enrichment of metabolite pathways were performed by MetaboAnalyst online software (http://www.metaboanalyst.ca/).
Supplementary Material
Acknowledgements
We thank Drs. Yanjing Li, Yu Yin and Yan Chang for their technical help; Dr. Yen-Nien Liu for his comments. We thank Drs. Juan Liu and Jason Locasale for their expertise in LC-MS analysis and tracing.
Funding
This study was supported by NIH grants R01 CA172603 (J. Huang), R01 CA205001 (J. Huang) and National Natural Science Foundation of China (#81630019).
Footnotes
Declaration of interest
No potential conflicts of interest are to be disclosure by authors.
References
- Abida W, Armenia J, Gopalan A, Brennan R, Walsh M, Barron D, Danila D, Rathkopf D, Morris M, Slovin S, et al. 2017. Prospective Genomic Profiling of Prostate Cancer Across Disease States Reveals Germline and Somatic Alterations That May Affect Clinical Decision Making. JCO Precis Oncol 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aird KM, Worth AJ, Snyder NW, Lee JV, Sivanand S, Liu Q, Blair IA, Wellen KE & Zhang R 2015. ATM couples replication stress and metabolic reprogramming during cellular senescence. Cell Rep 11 893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose M & Gatti RA 2013. Pathogenesis of ataxia-telangiectasia: the next generation of ATM functions. Blood 121 4036–4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose M, Goldstine JV & Gatti RA 2007. Intrinsic mitochondrial dysfunction in ATM-deficient lymphoblastoid cells. Hum Mol Genet 16 2154–2164. [DOI] [PubMed] [Google Scholar]
- Annala M, Struss WJ, Warner EW, Beja K, Vandekerkhove G, Wong A, Khalaf D, Seppälä I-L, So A & Lo G 2017. Treatment outcomes and tumor loss of heterozygosity in germline DNA repair–deficient prostate cancer. European urology 72 34–42. [DOI] [PubMed] [Google Scholar]
- Annala M, Vandekerkhove G, Khalaf D, Taavitsainen S, Beja K, Warner EW, Sunderland K, Kollmannsberger C, Eigl BJ & Finch D 2018. Circulating tumor DNA genomics correlate with resistance to abiraterone and enzalutamide in prostate cancer. Cancer discovery CD-17–0937. [DOI] [PubMed] [Google Scholar]
- Antonarakis ES, Lu C, Luber B, Liang C, Wang H, Chen Y, Silberstein JL, Piana D, Lai Z, Chen Y, et al. 2018. Germline DNA-repair Gene Mutations and Outcomes in Men with Metastatic Castration-resistant Prostate Cancer Receiving First-line Abiraterone and Enzalutamide. Eur Urol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran H, Yelensky R, Frampton GM, Park K, Downing SR, MacDonald TY, Jarosz M, Lipson D, Tagawa ST & Nanus DM 2013. Targeted next-generation sequencing of advanced prostate cancer identifies potential therapeutic targets and disease heterogeneity. European urology 63 920–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns RA, Harris IS & Mak TW 2011. Regulation of cancer cell metabolism. Nat Rev Cancer 11 85–95. [DOI] [PubMed] [Google Scholar]
- Choi M, Kipps T & Kurzrock R 2016. ATM Mutations in Cancer: Therapeutic Implications. Mol Cancer Ther 15 1781–1791. [DOI] [PubMed] [Google Scholar]
- Cosentino C, Grieco D & Costanzo V 2011. ATM activates the pentose phosphate pathway promoting anti-oxidant defence and DNA repair. Embo J 30 546–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser M, Sabelnykova VY, Yamaguchi TN, Heisler LE, Livingstone J, Huang V, Shiah YJ, Yousif F, Lin X, Masella AP, et al. 2017. Genomic hallmarks of localized, non-indolent prostate cancer. Nature 541 359–364. [DOI] [PubMed] [Google Scholar]
- George A, Kaye S & Banerjee S 2016. Delivering widespread BRCA testing and PARP inhibition to patients with ovarian cancer. Nat Rev Clin Oncol. [DOI] [PubMed] [Google Scholar]
- Girgis H, Masui O, White NM, Scorilas A, Rotondo F, Seivwright A, Gabril M, Filter ER, Girgis AH & Bjarnason GA 2014. Lactate dehydrogenase A is a potential prognostic marker in clear cell renal cell carcinoma. Molecular cancer 13 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He TL, Zhang YJ, Jiang H, Li XH, Zhu H & Zheng KL 2015. The c-Myc-LDHA axis positively regulates aerobic glycolysis and promotes tumor progression in pancreatic cancer. Med Oncol 32 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiew K, Hart CA, Ali A, Elliott T, Ramani V, Sangar V, Lau M, Maddineni S, Brown M & Clarke N 2018. Primary Mutational Landscape Linked with Pre-Docetaxel Lactate Dehydrogenase Levels Predicts Docetaxel Response in Metastatic Castrate-Resistant Prostate Cancer. Eur Urol Focus. [DOI] [PubMed] [Google Scholar]
- Hu H & Gatti RA 2008. New approaches to treatment of primary immunodeficiencies: fixing mutations with chemicals. Curr Opin Allergy Clin Immunol 8 540–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui S, Ghergurovich JM, Morscher RJ, Jang C, Teng X, Lu W, Esparza LA, Reya T, Zhan L & Guo JY 2017. Glucose feeds the TCA cycle via circulating lactate. Nature 551 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M, Daignault-Newton S, Twardowski PW, Albany C, Stein MN, Kunju LP, Siddiqui J, Wu YM, Robinson D, Lonigro RJ, et al. 2018. Targeting Androgen Receptor and DNA Repair in Metastatic Castration-Resistant Prostate Cancer: Results From NCI 9012. J Clin Oncol 36 991–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger A & Ralser M 2011. ATM is a redox sensor linking genome stability and carbon metabolism. Sci Signal 4 pe17. [DOI] [PubMed] [Google Scholar]
- Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM, Royer RE, Vander Jagt DL, Semenza GL & Dang CV 2010. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proceedings of the National Academy of Sciences 107 2037–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P, Martin NT, Nakamura K, Azghadi S, Amiri M, Ben-David U, Perlman S, Gatti RA, Hu H & Lowry WE 2013. SMRT compounds abrogate cellular phenotypes of ataxia telangiectasia in neural derivatives of patient-specific hiPSCs. Nat Commun 4 1824. [DOI] [PubMed] [Google Scholar]
- Lewis BC, Shim H, Li Q, Wu CS, Lee LA, Maity A & Dang CV 1997. Identification of putative c-Myc-responsive genes: characterization of rcl, a novel growth-related gene. Mol Cell Biol 17 4967–4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberti MV & Locasale JW 2016. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem Sci 41 211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord CJ, Tutt AN & Ashworth A 2015. Synthetic lethality and cancer therapy: lessons learned from the development of PARP inhibitors. Annu Rev Med 66 455–470. [DOI] [PubMed] [Google Scholar]
- Mateo J, Carreira S, Sandhu S, Miranda S, Mossop H, Perez-Lopez R, Nava Rodrigues D, Robinson D, Omlin A & Tunariu N 2015. DNA-repair defects and olaparib in metastatic prostate cancer. New England Journal of Medicine 373 1697–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na R, Zheng SL, Han M, Yu H, Jiang D, Shah S, Ewing CM, Zhang L, Novakovic K & Petkewicz J 2017. Germline mutations in ATM and BRCA1/2 distinguish risk for lethal and indolent prostate cancer and are associated with early age at death. European urology 71 740–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newington JT, Rappon T, Albers S, Wong DY, Rylett RJ & Cumming RC 2012. Overexpression of pyruvate dehydrogenase kinase 1 and lactate dehydrogenase A in nerve cells confers resistance to amyloid β and other toxins by decreasing mitochondrial respiration and reactive oxygen species production. Journal of Biological Chemistry 287 37245–37258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan M, Reid MA, Lowman XH, Kulkarni RP, Tran TQ, Liu X, Yang Y, Hernandez-Davies JE, Rosales KK & Li H 2016. Regional glutamine deficiency in tumours promotes dedifferentiation through inhibition of histone demethylation. Nature cell biology 18 1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JW, Lee JK, Witte ON & Huang J 2017. FOXA2 is a sensitive and specific marker for small cell neuroendocrine carcinoma of the prostate. Modern Pathology 30 1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlova NN & Thompson CB 2016. The Emerging Hallmarks of Cancer Metabolism. Cell Metab 23 27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D, Van Allen EM, Wu Y-M, Schultz N, Lonigro RJ, Mosquera J-M, Montgomery B, Taplin M-E, Pritchard CC & Attard G 2015. Integrative clinical genomics of advanced prostate cancer. Cell 161 1215–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San-Millan I & Brooks GA 2017. Reexamining cancer metabolism: lactate production for carcinogenesis could be the purpose and explanation of the Warburg Effect. Carcinogenesis 38 119–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzenberg-Bar-Yoseph F, Armoni M & Karnieli E 2004. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer research 64 2627–2633. [DOI] [PubMed] [Google Scholar]
- Shiloh Y 2014. ATM: expanding roles as a chief guardian of genome stability. Exp Cell Res 329 154–161. [DOI] [PubMed] [Google Scholar]
- Shiloh Y & Ziv Y 2013. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol 14 197–210. [PubMed] [Google Scholar]
- Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, Dalla-Favera R & Dang CV 1997. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci U S A 94 6658–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RA, Fraser M, Livingstone J, Espiritu SM, Thorne H, Huang V, Lo W, Shiah YJ, Yamaguchi TN, Sliwinski A, et al. 2017. Germline BRCA2 mutations drive prostate cancers with distinct evolutionary trajectories. Nat Commun 8 13671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O 1956. On the origin of cancer cells. Science 123 309–314. [DOI] [PubMed] [Google Scholar]
- Xie H, Hanai J-i, Ren J-G, Kats L, Burgess K, Bhargava P, Signoretti S, Billiard J, Duffy KJ & Grant A 2014. Targeting lactate dehydrogenase-a inhibits tumorigenesis and tumor progression in mouse models of lung cancer and impacts tumor-initiating cells. Cell metabolism 19 795–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakikhani M, Bazile M, Hashemi S, Javeshghani S, Avizonis D, St Pierre J & Pollak MN 2012. Alterations in cellular energy metabolism associated with the antiproliferative effects of the ATM inhibitor KU-55933 and with metformin. PloS one 7 e49513. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.