Inhibition of fatty acid synthase in Chlamydomonas activates major catabolic pathways, including autophagy, through a mechanism that involves the activation of a chloroplast quality control system.
Abstract
Fatty acids are synthesized in the stroma of plant and algal chloroplasts by the fatty acid synthase complex. Newly synthesized fatty acids are then used to generate plastidial lipids that are essential for chloroplast structure and function. Here, we show that inhibition of fatty acid synthesis in the model alga Chlamydomonas reinhardtii activates autophagy, a highly conserved catabolic process by which cells degrade intracellular material under adverse conditions to maintain cell homeostasis. Treatment of Chlamydomonas cells with cerulenin, a specific fatty acid synthase inhibitor, stimulated lipidation of the autophagosome protein ATG8 and enhanced autophagic flux. We found that inhibition of fatty acid synthesis decreased monogalactosyldiacylglycerol abundance, increased lutein content, down-regulated photosynthesis, and increased the production of reactive oxygen species. Electron microscopy revealed a high degree of thylakoid membrane stacking in cerulenin-treated cells. Moreover, global transcriptomic analysis of these cells showed an up-regulation of genes encoding chloroplast proteins involved in protein folding and oxidative stress and the induction of major catabolic processes, including autophagy and proteasome pathways. Thus, our results uncovered a link between lipid metabolism, chloroplast integrity, and autophagy through a mechanism that involves the activation of a chloroplast quality control system.
Photosynthetic organisms including algae and higher plants undergo profound metabolic arrangements under stress conditions such as nutrient starvation or high-light irradiance. As a primary response to stress, cells synthesize and accumulate high amounts of fatty acids and triacylglycerols (TAGs) as energy-rich reserves. The metabolism of TAGs has been investigated mainly in yeast and land plants, although substantial progress has been made recently in algae due to the biotechnological potential of these organisms as biofuel producers (Hu et al., 2008; Merchant et al., 2012; Liu and Benning, 2013; Li-Beisson et al., 2015). Upon stress, eukaryotic cells also activate autophagy, a major catabolic pathway by which cells degrade and recycle intracellular material. During autophagy, a portion of the cytoplasm that may include proteins, membranes, ribosomes, or even entire organelles is engulfed by a double membrane structure that grows around the cargo and forms an autophagosome. This double membrane vesicle is delivered to the vacuole (or lysosomes), where the cargo is degraded and recycled (He and Klionsky, 2009; Mizushima et al., 2011; Liu and Bassham, 2012; Marshall and Vierstra, 2018). Autophagy can be nonselective or highly selective depending on the nature of the cargo, and several types of selective autophagy have been reported, including mitophagy, proteaphagy, pexophagy, or chlorophagy, for the removal of mitochondria, proteasomes, peroxisomes, or chloroplasts, respectively (Floyd et al., 2012; Schreiber and Peter, 2014; Marshall et al., 2015; Young and Bartel, 2016; Izumi et al., 2017). Under normal growth conditions, there is a constitutive or basal level of autophagy in the cell that clears away damaged or unnecessary cytosolic material. However, upon stress, cells increase their autophagic degradation activity to eliminate damaged or toxic components and recycle cell contents in order to provide essential building blocks (e.g. amino acids and fatty acids) and energy sources that promote cell homeostasis and survival.
Autophagy is mediated by autophagy-related (ATG) genes, which have been identified in most eukaryotes, including algae and plants (Thompson and Vierstra, 2005; Bassham et al., 2006; Díaz-Troya et al., 2008; Shemi et al., 2015). A subset of ATG proteins constitutes the autophagy core machinery and is essential for the formation of the autophagosome and its fusion to the vacuole. Among them, the ATG8 and ATG12 ubiquitin-like conjugation systems participate in the binding of ATG8 to phosphatidylethanolamine, a key step in autophagosome formation (Mizushima et al., 2011; Feng et al., 2014). Core ATG proteins are highly conserved in the model unicellular green alga Chlamydomonas reinhardtii (Díaz-Troya et al., 2008; Pérez-Pérez and Crespo, 2014; Shemi et al., 2015). Unlike in plants, ATG genes are single copy in the Chlamydomonas genome, which facilitates the study of autophagy in this alga. Our current knowledge about autophagy in algae is still limited compared with other organisms, although the recent development of specific autophagy markers in Chlamydomonas has been fundamental to investigate this catabolic process (Pérez-Pérez et al., 2017). By monitoring the abundance, lipidation state, and cellular localization of ATG8, it has been reported that autophagy is activated in response to nitrogen, carbon, or phosphate limitation, stationary growth phase, oxidative stress, metal toxicity, or endoplasmic reticulum stress (Pérez-Pérez et al., 2010, 2012a; Davey et al., 2014; Goodenough et al., 2014; Pérez-Martín et al., 2014, 2015; Couso et al., 2018). Transcriptional activation of ATG genes also has been shown in Chlamydomonas cells subjected to different stress signals (Goodenough et al., 2014; Pérez-Martín et al., 2014, 2015; Ramundo et al., 2014; Schmollinger et al., 2014). Mounting evidence indicated that autophagy is regulated by the formation of reactive oxygen species (ROS) in algae (Pérez-Pérez et al., 2012b). Photooxidative damage of the chloroplast caused by the absence of protective carotenoids or exposure to high light resulted in the activation of autophagy in Chlamydomonas, suggesting that chloroplast activity might be linked to the control of autophagy (Pérez-Pérez et al., 2012a). Accordingly, the loss of chloroplast integrity in Chlamydomonas mutant cells lacking the essential stromal ClpP protease leads to enhanced autophagy (Ramundo et al., 2014).
Autophagy plays an important role in the control of lipid metabolism in animals and yeast. In mammals, autophagy is needed for the differentiation of adipocytes and the accumulation of lipid droplets (LDs) in hepatocytes, but this catabolic process also contributes to the selective degradation of LDs via lipophagy, pointing to a complex link between the metabolism of LDs and autophagy in these systems (for review, see Elander et al., 2018). Recent studies provided experimental evidence connecting lipid metabolism to autophagy in plants and algae (Elander et al., 2018). Autophagy-deficient rice (Oryza sativa) plants lacking ATG7 displayed an absence of LDs in tapetal cells, decreased TAG content in pollen grains, and complete sporophytic male sterility (Kurusu et al., 2014). In Arabidopsis (Arabidopsis thaliana), overexpression of ATG5 or ATG7 stimulates autophagic flux and increases the seed fatty acid content, whereas atg5 or atg7 knockout plants display decreased levels and a different composition of fatty acids in seeds (Minina et al., 2018). In Chlamydomonas, inhibition of autophagic flux blocks the synthesis of TAGs and the formation of LDs in nitrogen- or phosphate-starved cells, suggesting that the autophagic turnover of cellular material under nutrient stress is required for TAG synthesis (Couso et al., 2018). It is currently unknown whether the degradation of LDs by lipophagy is conserved in plants, although localization of these lipidic particles in the vacuole has been shown in the unicellular green algae Auxenochlorella protothecoides and Micrasterias denticulata. A. protothecoides cells have the ability to grow autotrophically in the light or heterotrophically by assimilating Glc from the medium. The transition from heterotrophic to autotrophic growth activates autophagy and seems to stimulate the direct engulfment of LDs by the vacuole through a microlipophagy-like process (Zhao et al., 2014). In M. denticulata cells subjected to carbon starvation, plastid lipid bodies are released to the cytoplasm, where they are engulfed by autophagosome-like structures and delivered to the vacuole for degradation (Schwarz et al., 2017). However, it is still unknown if the autophagy machinery mediates the degradation of these lipid bodies.
In this study, we investigated the relationship between lipid metabolism and autophagy in Chlamydomonas by inhibiting the synthesis of fatty acids in the chloroplast with cerulenin, a specific inhibitor of the keto-acyl-ACP synthase domain of type II fatty acid synthase (FAS) that forms a covalent bond with the catalytic Cys of this enzyme (Moche et al., 1999; Johansson et al., 2008). We found that FAS inhibition activates autophagy in Chlamydomonas by a mechanism that involves the up-regulation of a chloroplast quality control system. An in-depth analysis of cerulenin-treated cells indicated that FAS inhibition decreased the content of monogalactosyldiacylglycerol (MGDG), the most abundant plastid lipid, and altered the ultrastructure and function of the chloroplast, leading to photooxidative damage and the activation of major catabolic pathways in the cell.
RESULTS
Inhibition of FAS Triggers Autophagy in Chlamydomonas
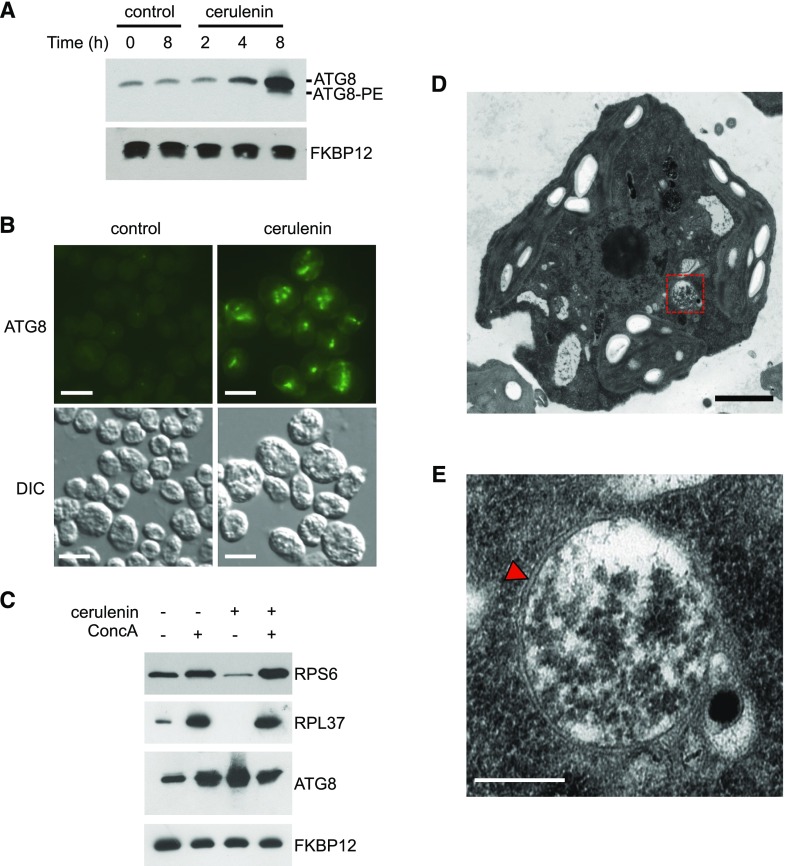
Most of the lipids found in Chlamydomonas cells are derived from the synthesis of fatty acids in the chloroplast (Li-Beisson et al., 2015). To investigate a possible link between lipid metabolism and autophagy in Chlamydomonas, we analyzed the role of fatty acid synthesis in the regulation of this catabolic process. To this aim, we blocked the de novo synthesis of fatty acids in the chloroplast using cerulenin as a specific inhibitor of FAS activity. Cerulenin has been widely used to inhibit FAS activity from different organisms, including bacteria, yeasts, plants, and algae, among others (D’Agnolo et al., 1973; Packter and Stumpf, 1975; Koo et al., 2005; Liu et al., 2012; Shpilka et al., 2015; Yang et al., 2015). In Chlamydomonas, it was shown previously that 10 µm cerulenin is the optimal concentration to prevent TAG synthesis in nitrogen-limited cells (Fan et al., 2011). We found that 10 µm cerulenin inhibits Chlamydomonas cell growth (Supplemental Fig. S1), so we decided to use this concentration of cerulenin to block fatty acid synthesis in our experiments unless indicated. Chlamydomonas cells growing exponentially were treated with cerulenin, and the lipidation state and abundance of ATG8 were examined by western blot at different times. Interestingly, the addition of cerulenin resulted in a substantial increase in ATG8 levels as well as the detection of lipidated ATG8 after 8 h compared with untreated cells (Fig. 1A). A moderate but reproducible effect on ATG8 abundance was visible after 4 h of cerulenin treatment (Fig. 1A). We also examined the cellular localization of ATG8 by immunofluorescence microscopy in cerulenin-treated cells, since activation of autophagy in Chlamydomonas results in the detection of this protein at punctate structures with an intense signal that likely corresponds to autophagosomes (Pérez-Pérez et al., 2010, 2017). The ATG8 signal was more intense, and several spots per cell were detected in cerulenin-treated cells compared with untreated control cells (Fig. 1B). The expression of some ATG genes from the core autophagy machinery is up-regulated when Chlamydomonas cells are exposed to different stress conditions (Goodenough et al., 2014; Pérez-Martín et al., 2014, 2015; Ramundo et al., 2014; Schmollinger et al., 2014). In close agreement, reverse transcription quantitative PCR (RT-qPCR) assays revealed that mRNA levels of ATG3, ATG8, and ATG12 genes increased in cerulenin-treated cells (Supplemental Fig. S2). To further demonstrate the activation of autophagy in these experiments, we determined the autophagic flux in Chlamydomonas cells upon cerulenin treatment. We recently showed that concanamycin A (ConcA), an inhibitor of vacuolar ATPase, blocks autophagic flux and prevents the degradation of the ribosomal proteins RPS6 and RPL37 under nitrogen or phosphate limitation in Chlamydomonas (Couso et al., 2018). We analyzed the levels of these proteins in cerulenin-treated cells in the absence or presence of ConcA. As shown previously (Couso et al., 2018), the abundance of RPS6 and RPL37 increased in Chlamydomonas cells treated with ConcA (Fig. 1C). We found that the levels of RPS6 and RPL37 decreased in cells treated with cerulenin, and the presence of ConcA largely prevented the degradation of these proteins following cerulenin treatment (Fig. 1C). Further evidence of autophagy activation in cerulenin-treated cells came with the detection by electron microscopy of double membrane vesicles, a distinctive feature of autophagosomes (Fig. 1, D and E). Altogether, these data demonstrated that the inhibition of fatty acid synthesis activates autophagy in Chlamydomonas.
Figure 1.
Activation of autophagic flux in Chlamydomonas cells treated with cerulenin. A, Chlamydomonas cells growing in the exponential phase were treated with 10 μm cerulenin for 0, 2, 4, and 8 h. Untreated cells at the initial (0 h) and the final (8 h) time points were used as a control. Fifteen micrograms of total extracts was resolved by 15% SDS-PAGE followed by immunoblotting with ATG8 antibodies (1:3,000 dilution). Anti-FKBP12 antibodies (1:5,000 dilution) were used as a loading control. B, Chlamydomonas cells treated with 10 μm cerulenin for 8 h were collected and processed for immunofluorescence microscopy to analyze ATG8 localization. Untreated cells were used as a control. DIC, Differential interference contrast. Bars = 10 μm. C, Chlamydomonas cells were treated for 8 h with 10 μm cerulenin and/or 0.1 μm ConcA. Fifteen micrograms of total extracts was resolved by 12% (RPS6) or 15% (RPL37, ATG8, and FKBP12) SDS-PAGE followed by western blotting with anti-OLLAS (1:1,000 dilution), anti-RPL37 (1:10,000 dilution), anti-ATG8 (1:3,000 dilution), and anti-FKBP12 (1:5,000 dilution) antibodies. D, Ultrastructure of a Chlamydomonas cell treated with 10 μm cerulenin for 8 h. The dashed-line red square indicates the autophagosome-like vesicle shown in E. Bar = 1 µm. E, Detail of an autophagosome from a cerulenin-treated cell shown in D. The characteristic double membrane of the autophagosome is indicated with a red arrowhead. Bar = 500 nm.
Inhibition of FAS Results in Decreased MGDG Levels
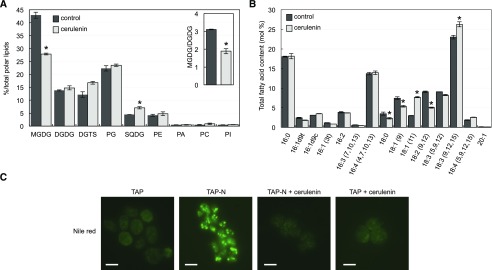
To find out the mechanism by which the synthesis of fatty acids regulates autophagy in Chlamydomonas, we first investigated whether inhibition of this process by cerulenin may affect the content of polar membrane lipids. Our results indicated that treatment of Chlamydomonas cells with cerulenin resulted in a pronounced decrease in the relative abundance of MGDG, the major lipid in thylakoid membranes (Gounaris and Barber, 1983; Moellering and Benning, 2011), and a relative increase in sulfoquinovosyl diacylglycerol level (Fig. 2A). No significant effect was observed in the digalactosyl diacylglycerol (DGDG) content following cerulenin treatment, which decreased the MGDG/DGDG ratio (Fig. 2A), a critical parameter for thylakoid membrane stability and the normal operation of photosystems (Dörmann and Benning, 2002; Moellering and Benning, 2011). We also determined the fatty acid profile of Chlamydomonas cells treated with cerulenin. The relative abundance of C16 fatty acids was not altered in cerulenin-treated cells, although some fluctuations were detected in C18 fatty acids. Specifically, the relative content of C18:1Δ9 and C18:2∆9,12 decreased, whereas C18:1∆11 and C18:3∆9,12,15 were enriched (Fig. 2B). To confirm that cerulenin is blocking the synthesis of fatty acids in our experiments, we analyzed the formation of LDs in Chlamydomonas cells subjected to nitrogen limitation in the presence or absence of the inhibitor, since TAG accumulation in nitrogen-starved cells requires the de novo synthesis of fatty acids (Fan et al., 2011; Liu et al., 2012). In close agreement with these studies, our results indicated that cerulenin fully prevented the detection of LDs in nitrogen-limited cells (Fig. 2C).
Figure 2.
Effects of FAS inhibition on lipids and fatty acid contents in Chlamydomonas. Cells growing exponentially in Tris-acetate phosphate (TAP) medium were treated with 10 μm cerulenin for 8 h. A and B, Quantification of polar lipids (A) and fatty acid levels (B) from Chlamydomonas cells in the absence (black bars) or presence (gray bars) of cerulenin. The inset in A shows the MGDG/DGDG ratio in control and cerulenin-treated cells. Three biological replicates were analyzed for each condition. *, Differences were significant at P < 0.05 according to Student’s t test. Error bars indicate sd. SQDG, Sulfoquinovosyl diacylglycerol. C, Cerulenin prevents the formation of LDs under nitrogen starvation. Chlamydomonas cells in the exponential growth phase were washed twice with nitrogen-free (TAP-N) or nitrogen-containing (TAP; control) medium and grown under these conditions for 24 h in the absence or presence of 10 μm cerulenin. Lipid bodies were stained with Nile Red and visualized by fluorescence microscopy. DGTS, diacylglycerol-N,N,N-trimethylhomoserine; PG, phosphatidylglycerol; PE, phosphatidylethanolamine; PA, phosphatidic acid; PC, phosphatidylcholine; PI, phosphatidylinositol. Bars = 10 μm.
Treatment of Chlamydomonas Cells with Cerulenin Enhances the Level of Lutein
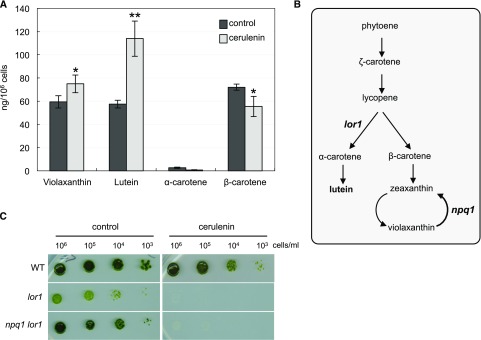
Carotenoids have essential functions in photosynthesis and photoprotection, as they can quench toxic ROS generated as by-products of photosynthesis (Li et al., 2009). Given that carotenoid deficiency has been linked to autophagy activation in Chlamydomonas (Pérez-Pérez et al., 2012a), we analyzed the effect of FAS inhibition on the carotenoid profile in Chlamydomonas. To this aim, cells growing at the exponential phase were treated with cerulenin and the level of prominent carotenoids was determined by HPLC analysis. Cerulenin drastically increased the content of lutein and produced a modest accumulation of violaxanthin (Fig. 3A). In contrast, the levels of α- and β-carotenes decreased following cerulenin treatment (Fig. 3A). To investigate if carotenoids, and in particular lutein, may play an important role in the cellular response to FAS inhibition, we analyzed the sensitivity of lutein-deficient mutants to cerulenin. We compared the growth of wild-type cells and the mutant strains lor1 and npq1 lor1 in the presence of a sublethal concentration of cerulenin (2.5 µm), since these mutants are unable to synthesize lutein (Niyogi et al., 1997; Fig. 3B). The growth of both lor1 and npq1 lor1 cells was extremely sensitive to cerulenin (Fig. 3C), suggesting that lutein may have a protective function in the cellular response to FAS inhibition. Lutein is a structural component of the light-harvesting complexes and contributes to the protection from photooxidative damage (Müller et al., 2001). Thus, the hypersensitivity of lutein-deficient cells to cerulenin and the enhanced production of lutein detected in cerulenin-treated cells strongly suggested that this carotenoid has a protective function against the cell damage caused by the inhibition of fatty acid synthesis in the chloroplast.
Figure 3.
Effects of FAS inhibition on carotenoid levels in Chlamydomonas. A, Changes in carotenoid contents in response to cerulenin treatment. Prominent carotenoids (violaxanthin, lutein, α-carotene, and β-carotene) were determined in Chlamydomonas cells untreated (black bars) or treated (gray bars) with 10 μm cerulenin for 8 h. Results are means of three independent experiments. Differences were significant according to Student’s t test: *, P < 0.05 and **, P < 0.01. B, Schematic representation of the carotenoid biosynthesis pathway in Chlamydomonas. Mutations in lor1 and npq1 lor1 strains are indicated. C, Chlamydomonas wild type (WT) and mutant strains lor1 and npq1 lor1 were subjected to 10-fold serial dilutions and spotted onto TAP plates containing a sublethal concentration (2.5 μm) of cerulenin. TAP plates were used as a control. Plates were incubated at 25°C under continuous illumination at 10 µE m−2 s−1.
Treatment of Chlamydomonas Cells with Cerulenin Causes Chloroplast Shrinkage and Hyperstacking of Thylakoid Membranes
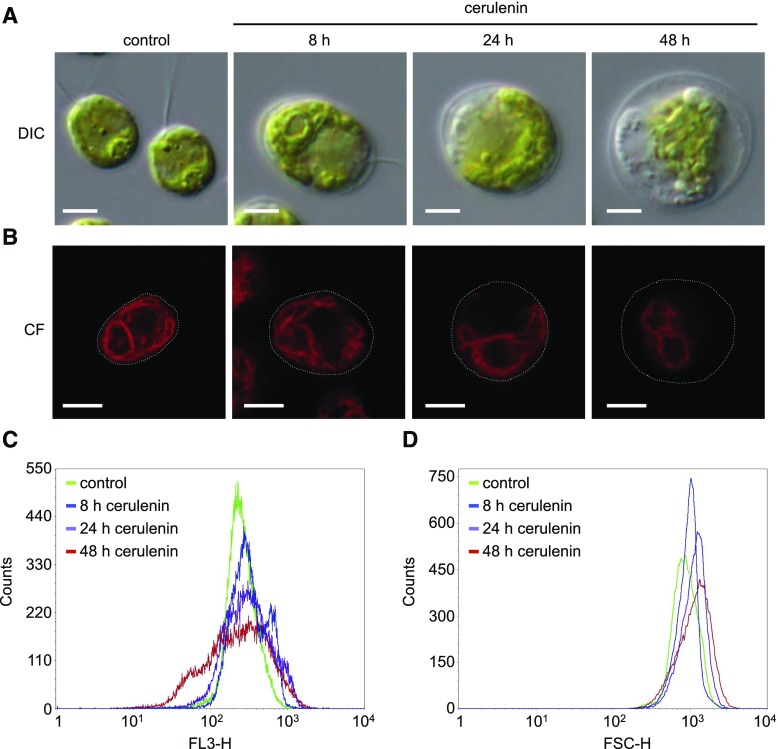
To gain further insight on the effect of FAS inhibition at the cellular level, we carried out a detailed microscopic study of Chlamydomonas cells treated with cerulenin at 8, 24, and 48 h. DIC analysis with an optical microscope revealed a gradual increase in cell size that was apparent after 8 h of treatment (Fig. 4A). A quantitative analysis of cell size by flow cytometry indicated that cerulenin indeed leads to a progressive enlargement of cells (Fig. 4C). We also observed with the light microscope a marked shrinkage of the chloroplast and bleaching of cells treated with cerulenin at 24 h that was more evident at 48 h (Fig. 4A). Fluorescence confocal microscopy was used to visualize the chloroplast in cells treated with cerulenin, and our results indicated that the fluorescence and size of the chloroplast decreased following cerulenin treatment (Fig. 4B). Quantification of chloroplast fluorescence by flow cytometry confirmed that cells treated with cerulenin display a significant loss of autofluorescence (Fig. 4D). Taken together, these results showed that the stress caused by the inhibition of FAS activity in Chlamydomonas had a strong impact on the morphology of the cell after 24 and 48 h of cerulenin treatment, although these effects were detectable within 8 h.
Figure 4.
Effects of cerulenin on Chlamydomonas cell morphology. A, DIC microscopy images of log-phase Chlamydomonas cells treated with 10 μm cerulenin for 8, 24, and 48 h. Untreated cells were used as a control. Bars = 5 μm. B, Chlorophyll fluorescence (CF) from individual cells treated with 10 µm cerulenin for 8, 24, and 48 h was visualized by confocal microscopy. Bars = 5 μm. C and D, Chlorophyll fluorescence (C) and size (D) of 5 × 104 cells treated with 10 µm cerulenin for 8, 24, and 48 h were analyzed and quantified by flow cytometry. FL3-H, Fluorescence (red channel); FSC-H, Forward-scattered light (size).
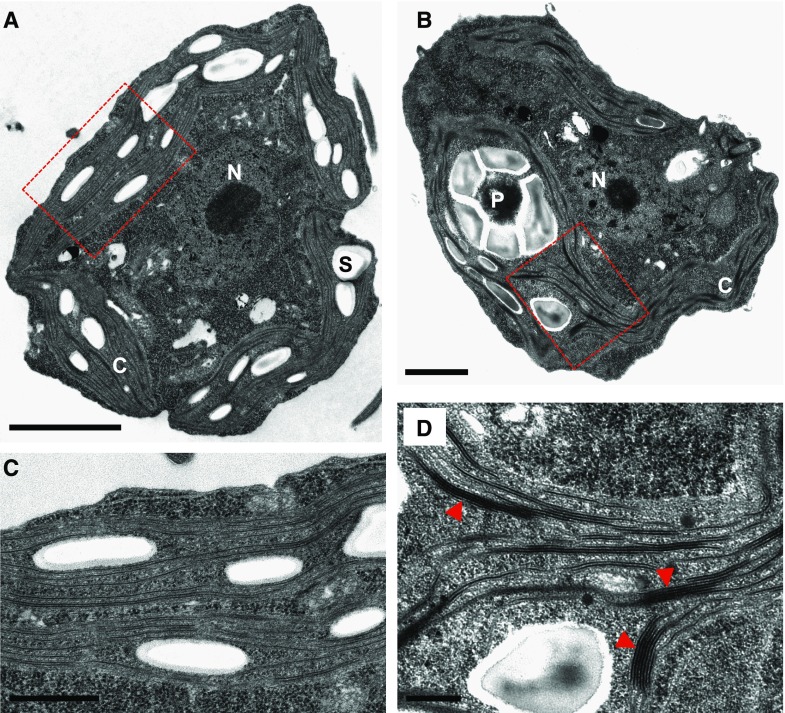
In order to identify early effects of FAS inhibition in the cell, we next performed an ultrastructural analysis of Chlamydomonas cells treated with cerulenin for 8 h using transmission electron microscopy. Control cells displayed a typical architecture with a single chloroplast that occupies a large portion of the cell (Fig. 5A). The thylakoid membranes from the chloroplast of control cells appear as flat vesicles or discs that can be either single or arranged in stacks of discs (Fig. 5, A and C; Goodenough and Levine, 1969). The arrangement of thylakoid membranes in Chlamydomonas is very different from the chloroplast membrane system of land plants, in which short segments of many stacked discs (grana) are interconnected by a network of long, single discs. Strikingly, the ultrastructural analysis of Chlamydomonas cells treated with cerulenin uncovered the presence of multiple regions in the chloroplast with a high degree of thylakoid membrane stacking (Fig. 5, B and D; Supplemental Fig. S3). The number of thylakoid membranes that pile up in cerulenin-treated cells was significantly higher compared with untreated cells, and up to 18 membranes were detected in some stacks (Supplemental Fig. S4). Such structures were barely found in control cells, which suggested that hyperstacking of thylakoid membranes might be an early effect of FAS inhibition.
Figure 5.
Ultrastructural analysis of Chlamydomonas cells treated with cerulenin. A and B, Electron microcopy images from Chlamydomonas cells growing in the absence (A) or presence (B) of 10 μm cerulenin for 8 h. C, Chloroplast; N, nucleus; P, pyrenoid; S, starch granule. C and D, Enlargement of A and B showing thylakoid membranes of untreated cells (C) and cerulenin-treated cells (D), respectively. Red arrowheads indicate groups of hyperstacked thylakoids in cerulenin-stressed cells (D). Dashed-line boxes in A and B show the enlarged areas in C and D, respectively. Bars = 2 µm (A), 1 µm (B), and 500 nm (C and D).
Photosynthetic Efficiency Is Negatively Regulated in Cells Treated with Cerulenin
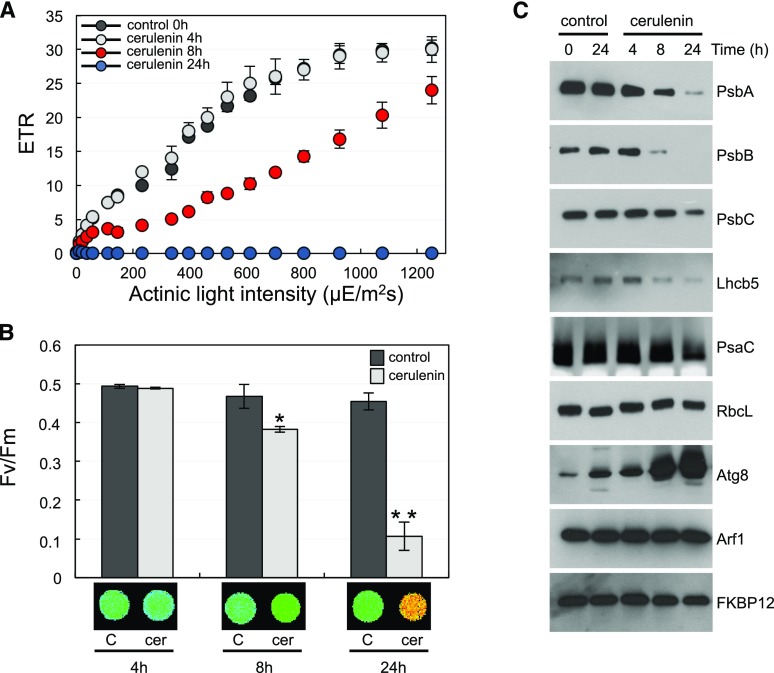
Our results indicated that treatment of Chlamydomonas cells with cerulenin had a deep impact on the chloroplast, including a decreased level of MGDG (Fig. 2A), increased lutein content (Fig. 3A), chloroplast shrinkage (Fig. 4), and hyperstacking of thylakoid membranes (Fig. 5). Based on these observations, we next analyzed whether the inhibition of FAS activity by cerulenin may have any effect on the photosynthetic activity. To this aim, we determined the photosynthetic efficiency of Chlamydomonas cells treated with cerulenin at different times using pulse amplitude-modulated (PAM) fluorometry. The analysis of the apparent electron transport rate (ETR) in cerulenin-treated cells revealed that the photosynthetic activity decreased after 8 h and was almost undetectable after 24 h of treatment (Fig. 6A). No significant effect was detected in cells treated for 4 h (Fig. 6A). We also determined the maximum photochemical efficiency of PSII, measured as the variable-to-maximal chlorophyll fluorescence ratio (Fv/Fm), in cells treated with cerulenin. These experiments showed a decrease of PSII efficiency at 8 h of treatment that was much stronger at 24 h (Fig. 6B). The low photosynthetic efficiency detected in cerulenin-treated cells might be due to an alteration in the abundance of photosynthetic proteins. To test this possibility, we analyzed the level of PSII and PSI components as well as some cytosolic proteins in cerulenin-treated cells. Western-blot analysis revealed a gradual decline in the abundance of PSII (PsbA, PsbB, PsbC, and Lhcb5) and PSI (PsaC) proteins following cerulenin treatment (Fig. 6C). No significant change was observed in RbcL abundance in these cells. The level of cytosolic proteins such as the ARF1 GTPase or the FKBP12 immunophilin remained stable following FAS inhibition, whereas, as expected, the autophagy protein ATG8 was strongly up-regulated (Fig. 6C). Together, these results indicated that the inhibition of FAS activity by cerulenin down-regulated the photosynthetic activity in Chlamydomonas.
Figure 6.
Photosynthetic properties of Chlamydomonas cells in response to cerulenin. A, Light response curve of untreated cells (black circles) or cerulenin-treated cells (gray circles, 4 h; blue circles, 8 h; red circles, 24 h). Cells were normalized to the same OD750nm values. The apparent ETR relative to the actinic light intensity (µE m−2 s−1) was determined as described in “Materials and Methods.” Error bars indicate sd. B, Fv/Fm in cells treated with cerulenin (cer) for 4, 8, or 24 h (gray columns) compared with untreated cells (black columns) at the same time points. Values are means of three independent experiments including at least three technical replicates. Differences were significant according to Student’s t test: *, P < 0.05 and **, P < 0.01. Error bars indicate sd. False color images of Fv/Fm from a typical experiment in cerulenin-treated cells are shown at bottom. C, Western-blot analysis of different plastid and cytosolic proteins following cerulenin treatment. Chlamydomonas cells in the exponential phase were treated with 10 µm cerulenin, and samples were collected at the indicated times. Twenty micrograms of total protein extracts was resolved by SDS-PAGE followed by immunoblotting with the indicated antibodies, except for PsbA and RbcL, where 5 µg of protein was analyzed.
Treatment of Chlamydomonas Cells with Cerulenin Activates a Chloroplast Stress Response and Increases ROS Production
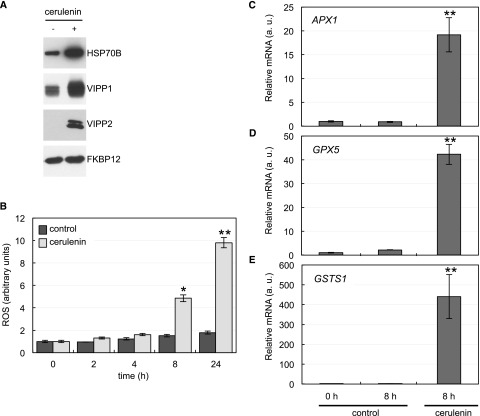
The finding that cerulenin treatment had negative effects in the chloroplast, including shrinkage, hyperstacking of thylakoid membranes, and impaired photosynthesis (Figs. 4–6), prompted us to analyze the level of some chloroplast-targeted proteins that respond to stress in cerulenin-treated cells. We first tested the level of the plastidic chaperone HSP70B and VIPP1 and VIPP2 proteins, which are induced under stress conditions including high light in Chlamydomonas (Nordhues et al., 2012). Compared with control cells, the abundance of HSP70B, VIPP1, and VIPP2 increased in cerulenin-treated cells, in particular VIPP2, which, as described previously, is barely detected under normal growth (Nordhues et al., 2012) but is strongly induced by cerulenin treatment (Fig. 7A). In close agreement, we also found that the expression of VIPP1 and VIPP2 genes was notably up-regulated in cerulenin-treated cells (Supplemental Fig. S5). These results suggested that FAS inhibition causes chloroplast damage that likely activates these genes in order to maintain the integrity of plastid membranes, as shown in Chlamydomonas and Arabidopsis for VIPP1 (Nordhues et al., 2012; Zhang et al., 2012).
Figure 7.
Impact of FAS inhibition on chloroplast stress proteins and ROS production. A, Western-blot analysis of chloroplast stress proteins. Log-phase Chlamydomonas cells were treated with 10 μm cerulenin for 8 h. Samples of nontreated cells were taken at the same time and used as a control. Five micrograms of total extracts was resolved by SDS-PAGE followed by immunoblotting with primary antibodies against HSP70B (1:10,000 dilution) and VIPP1 and VIPP2 (1:1,000 dilution). Anti-FKBP12 antibodies (1:5,000 dilution) were used as a loading control. B, Induction of ROS in response to cerulenin treatment. The content of ROS in cells treated with cerulenin for 2, 4, 8, and 24 h was determined. Untreated cells at the same time were used as a control. Values are means of three biological replicates. Differences were significant according to Student’s t test: *, P < 0.05 and **, P < 0.01. C to E, Analysis of APX1 (C), GPX5 (D), and GSTS1 (E) mRNA abundance by RT-qPCR in Chlamydomonas cells treated as indicated in A (a.u., arbitrary units). Three biological replicates with three technical replicates were analyzed for each condition. Differences were significant according to Student’s t test: **, P < 0.01.
Given that the plastid is one of the main sources of ROS in plants and algae, we next investigated whether cerulenin treatment may produce oxidative stress at this organelle. To this aim, we measured the production of ROS in cerulenin-treated cells in a time-course experiment. Our results indicated that the level of ROS increased after 8 h of treatment and was even higher after 24 h (Fig. 7B). To further show a link between chloroplast damage, oxidative stress, and cerulenin-induced stress, we analyzed the expression of the oxidative stress-related genes Ascorbate Peroxidase1 (APX1), Glutathione Peroxidase Homologous (GPX5), and Glutathione-S-Transferase1 (GSTS1) in cerulenin-stressed cells. These genes play an important role in the detoxification of ROS, and their expression responds to the presence of different forms of ROS (Fischer et al., 2007, 2012; Ledford et al., 2007). RT-qPCR assays revealed a strong increase in APX1, GPX5, and GSTS1 gene expression in cerulenin-treated cells compared with control cells (Fig. 7, C–E). Taken together, these results indicated that FAS inhibition by cerulenin increases the production of ROS and triggers a stress response in the chloroplast of Chlamydomonas cells.
Transcriptome Analysis of Cerulenin-Treated Cells
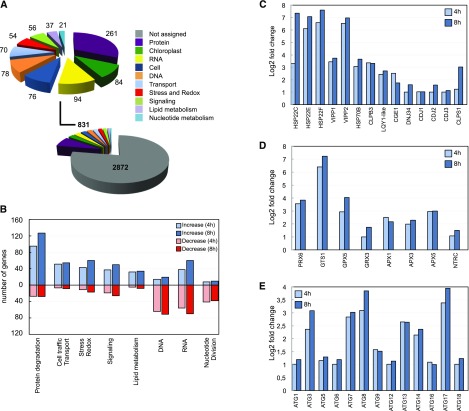
To understand the global impact of FAS inhibition, we undertook a genome-wide transcriptome analysis of Chlamydomonas cells treated with cerulenin. In these experiments, samples were taken at 0, 4, and 8 h following the addition of cerulenin in order to capture the primary responses to FAS inhibition. Based on the low effect of cerulenin on photosynthetic efficiency and ROS production at 4 h of cerulenin treatment (Figs. 6 and 7B), we reasoned that the primary transcriptional response might occur around this time. Three biological replicates were used for each time point, and according to principal component analysis, all samples clustered correctly with their respective groups (Supplemental Fig. S6). The sequence reads were aligned to the version 5.5 assembly of the Chlamydomonas genome (Merchant et al., 2007), and expression estimates were established for 17,741 genes. Differential expression analysis was performed, and only changes of 2-fold or greater and statistically significant were considered. The abundance of 4,030 transcripts changed between 0 and 4 h, and that of 5,503 transcripts changed between 0 and 8 h. The lists of up- and down-regulated transcripts identified in this way for the comparative analysis from 0 to 4 h and 0 to 8 h are shown in Supplemental Tables S1 to S4. Genes were categorized with the Algal Functional Annotation Tool (http://pathways.mcdb.ucla.edu/algal/index.html) and manual annotation. MapMan ontology analysis based on Chlamydomonas annotation revealed that the highest enriched categories between 0 and 4 h included transcripts encoding proteins putatively involved in protein metabolism, RNA and DNA metabolism, and transport (Fig. 8A). Chloroplast, cell traffic, stress, and redox also were among the most represented functional groups (Fig. 8A). The same categories were found between 0 and 8 h, although the number of genes in each group was higher, indicating that the effect of cerulenin at 8 h increased compared with 4 h (Supplemental Fig. S7). A closer examination showed an enrichment of up-regulated genes in categories related to protein degradation, membrane trafficking, chloroplast, redox/stress, and lipid metabolism, whereas down-regulated genes were preferentially associated with DNA, nucleotide metabolism, and cell division (Fig. 8B). Overall, these results reflect that treatment of Chlamydomonas cells with cerulenin promoted a stress response and blocked cell division.
Figure 8.
Functional classification of transcripts involved in the cellular response to cerulenin treatment. A, Functional characterization of genes differentially expressed between 0 and 4 h of cerulenin treatment (genes differentially expressed between 0 and 8 h are shown in Supplemental Fig. S7). The different categories are indicated with a color code. The number of transcripts in each category is shown. B, Number of genes in each category after 4 or 8 h of cerulenin treatment. Increased and decreased mRNA abundances are shown in blue (light for 4 h and dark for 8 h) and red (light for 4 h and dark for 8 h), respectively. C to E, Relative transcript abundance of genes encoding components of the chloroplast chaperones and stress proteins (C), chloroplast redox markers (D), and autophagy core machinery proteins (E), expressed as log2 fold change normalized to untreated cells (0 h). The principal component analysis corresponding to the gene expression of the three groups of samples is shown in Supplemental Figure S6. The genes up-regulated and down-regulated after 4 h of cerulenin treatment are shown in Supplemental Tables S1 and S2, respectively. The genes up-regulated and down-regulated after 8 h of cerulenin treatment are shown in Supplemental Tables S3 and S4, respectively.
Consistent with the activation of autophagy (Fig. 1), our genome-wide study revealed an up-regulation of transcripts encoding proteins that participate in the two major degradative pathways in eukaryotic cells, autophagy and the proteasome. Twelve ATG genes from the core autophagy machinery were up-regulated in cerulenin-treated cells (Fig. 8C), confirming that FAS inhibition activates autophagy in Chlamydomonas. The most abundant transcripts among the ATG genes encoded proteins involved in the formation of autophagosomal precursor structures (ATG13, ATG14, and ATG17) and the expansion and completion of the autophagosome (ATG3, ATG7, and ATG8; Fig. 8C). In addition to autophagy genes, we found 29 transcripts encoding putative subunits of the 26S proteasome and 22 genes related to ubiquitination (Supplemental Fig. S8), pointing to the activation of the ubiquitin-proteasome pathway upon FAS inhibition in Chlamydomonas.
Remarkably, genes encoding chloroplast chaperones and stress-related proteins were among the highest induced transcripts following cerulenin treatment. These genes included the small heat shock proteins HSP22C, HSP22E, and HSP22F, the stromal chaperone HSP70B, the Zn-finger domain LYQ1-like protein, and the chloroplast-targeted CLPB3 and CLPS2 chaperones (Fig. 8D). The HSP70B cochaperones CGE1, CDJ1, CDJ2, and CDJ3 and substrates VIPP1 and VIPP2 also were strongly up-regulated in cerulenin-treated cells (Fig. 8D).
A number of transcripts encoding redox-related proteins involved in ROS detoxification and predicted to localize to the plastid were induced following cerulenin treatment. This group of genes included GST (GSTS1), glutathione peroxidase (GPX5), glutaredoxin (GRX3), ascorbate peroxidases (APX1, APX2, and APX3), and peroxiredoxin (PRX6; Fig. 8E). Some of these genes, such as GSTS1 and GPX5, were shown previously to be induced by different stress conditions, including high-light stress (Fischer et al., 2007, 2012; Ledford et al., 2007). Interestingly, the transcript level of NTRC, which plays an important role in the protection of the chloroplast against oxidative damage (Pérez-Ruiz et al., 2006), also increased in cerulenin-treated cells (Fig. 8E). The identification of stress- and redox-related genes in this transcriptomic analysis was in close agreement with the harmful effects of cerulenin on chloroplast fitness (Figs. 4–6) and confirmed that FAS inhibition causes massive damage in the chloroplast of Chlamydomonas.
DISCUSSION
The de novo synthesis of fatty acids in algae and plants takes place in the chloroplast, and partial or total blockage of this vital process may have deleterious effects in the cell. In this study, we performed a detailed analysis of the primary response to FAS inhibition in Chlamydomonas using cerulenin to specifically block fatty acid synthesis. Our results indicated that treatment of Chlamydomonas cells with cerulenin activates autophagy, the major catabolic pathway in eukaryotic cells. Monitoring of specific autophagy markers confirmed the activation of autophagy in cerulenin-treated cells. First, FAS inhibition promoted the lipidation of ATG8 and the degradation of cytosolic ribosomal proteins RPS6 and RPL37, both landmarks of autophagic flux activation in Chlamydomonas (Pérez-Pérez et al., 2010, 2017; Couso et al., 2018). Second, FAS inhibition promoted the localization of ATG8 to punctate structures, similar to what has been described under autophagy-activating conditions (Pérez-Pérez et al., 2010, 2012a, 2017; Pérez-Martín et al., 2015). Third, double membrane vesicles that likely correspond to autophagosomes were identified in cerulenin-treated cells. Finally, RNA sequencing (RNA-Seq) data showed an up-regulation of ATG genes from the core autophagy machinery as well as genes from the ubiquitin-proteasome pathway. The activation of autophagy and ubiquitin-proteasome pathways in these cells strongly suggests that FAS inhibition caused substantial damage in the chloroplast that must be contained to prevent cell death. In Chlamydomonas, stress conditions that cause photooxidative damage of the chloroplast, such as carotenoid depletion or high-light stress, trigger autophagy (Pérez-Pérez et al., 2012a). Moreover, repression of the essential chloroplast gene clpP1, encoding the catalytic subunit of the stromal ClpP protease, activates autophagy in Chlamydomonas (Ramundo et al., 2014), further supporting a link between chloroplast damage and autophagy. The activation of autophagy by photooxidative stress in Chlamydomonas has been connected to the generation of ROS (Pérez-Pérez et al., 2012a) in a process that may imply the oxidation and inhibition of the ATG4 protease (Pérez-Pérez et al., 2016). In close agreement with these studies, our results indicate that the activation of autophagy in cerulenin-treated cells is likely associated with the chloroplast damage and the high level of ROS found in these cells (Fig. 9). Vacuolar transport and the degradation of entire chloroplasts by chlorophagy, a selective form of autophagy, has been shown in Arabidopsis plants exposed to UV-B damage or high-light stress (Izumi et al., 2017), but a similar mechanism cannot operate in Chlamydomonas due to the presence of a single chloroplast per cell. In yeast, cerulenin blocks the activation of autophagy by nitrogen starvation (Shpilka et al., 2015), but the underlying mechanism by which fatty acid synthesis regulates this degradative process might be different in nonphotosynthetic organisms.
Figure 9.
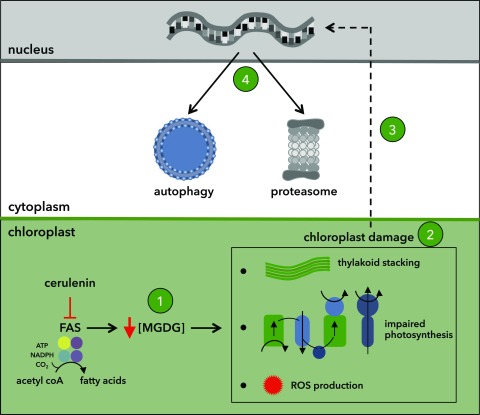
Hypothetical model depicting the cellular effect of cerulenin in Chlamydomonas. Inhibition of FAS by cerulenin results in a drop of MGDG levels (step 1). The change in the MGDG/DGDG ratio alters thylakoid structure, inhibits photosynthesis, and increases the production of ROS. Collectively, these alterations cause chloroplast damage (step 2), which then activates chloroplast-to-nucleus retrograde signaling (dashed line; step 3). Finally, the retrograde signal up-regulates the expression of components from degradative pathways including the autophagy and proteasome machineries in order to maintain cell homeostasis (step 4).
How does the inhibition of fatty acid synthesis generate photooxidative damage in the chloroplast? Our results revealed that proper synthesis of fatty acids and lipids is needed to maintain chloroplast integrity and function under normal growth conditions. Treatment of Chlamydomonas cells with cerulenin resulted in a significant decrease of MGDG (Fig. 2A), the most abundant lipid of thylakoids in the chloroplasts of algae and plant cells (Gounaris and Barber, 1983; Moellering and Benning, 2011). The negative impact of cerulenin on MGDG content is consistent with a recent study showing that incubation of Chlamydomonas cells with cerulenin under nitrogen-limiting conditions diminished the content of polar membrane lipids, including MGDG (Liu et al., 2016a). A similar negative effect of cerulenin on chloroplast lipids was reported earlier in land plants, since this drug markedly decreased the rate of accumulation of MGDG and DGDG during the greening period of barley (Hordeum vulgare) seedlings (Laskay et al., 1985). We also found that treatment of Chlamydomonas cells with cerulenin led to a pronounced increase in the level of lutein. Moreover, a hypersensitive phenotype to cerulenin was observed in lor1 and npq1 lor1 strains, as these two mutants were unable to grow in the presence of a sublethal concentration of this drug (Fig. 3C). lor1 and npq1 lor1 mutants are defective in the carotenoid biosynthetic pathway and cannot synthesize lutein (Niyogi et al., 1997). These mutant strains display a growth rate similar to wild-type cells under low light, but the npq1 lor1 double mutant is sensitive to high light due to the absence of the photoprotective carotenoids zeaxanthin, antheraxanthin, and lutein (Niyogi et al., 1997). The increased production of lutein in cerulenin-treated cells, together with the hypersensitivity of lutein-deficient mutants to this drug, strongly suggest that this carotenoid might play an important role in the protection against the cell damage triggered by cerulenin. A link between FAS inhibition and carotenoid metabolism has been shown in algae but with contradictory results. In Chlorella zofingiensis, cerulenin promotes astaxanthin accumulation under stress conditions (Liu et al., 2016b), whereas the opposite effect was reported in Haematococcus pluvialis, since cerulenin appears to abolish astaxanthin accumulation in light-stressed cells (Chen et al., 2015).
The inhibition of fatty acid synthesis caused pronounced changes in the morphology of Chlamydomonas cells, including enlarged cell size, chloroplast shrinkage, and loss of fluorescence (Fig. 4). Transmission electron microscopy analysis of Chlamydomonas cells treated with cerulenin also revealed defects in chloroplast structure, suggesting that FAS inhibition may lead to specific damage in this organelle. Thylakoid membranes from cerulenin-treated cells tend to form clusters, and hyperstacking of these membranes was easily detected (Fig. 5; Supplemental Figs. S3 and S4). Given that MGDG is the most abundant component of thylakoids and a structural lipid of thylakoid membrane protein complexes, the hyperstacking of thylakoids might be due to the drop of this lipid in cerulenin-treated cells. Supporting this model, previous studies in plants and algae have shown that MGDG deficiency affects the ultrastructure of chloroplasts. Treatment of barley seedlings with cerulenin during the greening period decreased the level of MGDG and prevented the formation of grana (Laskay et al., 1985). A mutation in Arabidopsis of the MGD1 gene encoding an MGDG synthase led to a severe underdevelopment of chloroplasts and the absence of internal membrane structures (Jarvis et al., 2000; Kobayashi et al., 2007). Long-term treatment (3–4 weeks) of Arabidopsis seedlings with galvestine-1, an inhibitor of MGDG synthase activity, lowered the MGDG content and impaired chloroplast development (Botté et al., 2011). Nannochloropsis mutants defective in a fatty acid elongase exhibited a specific decrease in MGDG and an altered ultrastructure of thylakoid membranes (Dolch et al., 2017). Interestingly, a loss-of-function Chlamydomonas mutant deficient in the MGDG-specific lipase PGD1 showed a relative increase in MGDG abundance and hyperstacking of the thylakoid grana (Du et al., 2018). The ratio of MGDG to DGDG in the chloroplast is critical for the proper structure and function of thylakoid membranes, and it must be finely regulated (Dörmann and Benning, 2002). Due to its biophysical properties, MGDG does not form bilayers (Gounaris and Barber, 1983). This is in contrast to DGDG, which is a bilayer-forming lipid (Webb and Green, 1991). Therefore, we hypothesize that the high degree of thylakoid stacking detected in Chlamydomonas cells treated with cerulenin might be related to a decrease in the MGDG/DGDG ratio, which affects the stability and fluidity of photosynthetic membranes (Gounaris and Barber, 1983; Webb and Green, 1991; Dörmann and Benning, 2002).
Global transcriptomic analysis of Chlamydomonas cells treated with cerulenin revealed a sharp activation of genes encoding chloroplast proteins involved in protein folding, oxidative stress, and ROS detoxification. Some chaperones and stress-related proteins were among the highest induced transcripts following cerulenin treatment. These genes included the small heat shock proteins HSP22C, HSP22E, and HSP22F and the stromal chaperone HSP70B. HSP22E/F proteins localize to the chloroplast (Rütgers et al., 2017) and participate in the response to severe stress, such as heat shock, oxidative stress, and prolonged sulfur or phosphorus starvation (Zhang et al., 2004; Fischer et al., 2005; Moseley et al., 2006; Schroda and Vallon, 2009). Transcriptomic and proteomic analyses of Chlamydomonas cells lacking the chloroplast ClpP protease showed a massive increase in HSP22C/E/F and other chloroplast chaperones such as HSP70B (Ramundo et al., 2014). Stromal HSP70B plays a fundamental role in the refolding of stress-denatured chloroplast proteins and the protection and repair of PSII when cells are exposed to excess light (Schroda and Vallon, 2009). In Chlamydomonas, HSP70B was found to interact with VIPP1 and the cochaperones CGE1, CDJ1, CDJ2, and CDJ3 (Schroda and Vallon, 2009), which participate in the biogenesis of thylakoid membranes (Nordhues et al., 2012). Chlamydomonas and other volvocean algae contain two VIPP paralogs, VIPP1 and VIPP2, and both proteins increased strongly during exposure to high light (Nordhues et al., 2012). Remarkably, all these genes were highly induced by cerulenin (Fig. 8), suggesting that FAS inhibition activates a chloroplast quality control system that is involved in the repair and turnover of damaged plastid proteins (Rochaix and Ramundo, 2018). Transcripts encoding redox-related proteins involved in ROS detoxification also were up-regulated following cerulenin treatment. This group of genes included GSTS1, GPX5, GRX3, APX1, APX2, APX3, and PRX6. In Chlamydomonas, peroxiredoxins and glutathione peroxidases play major roles in the protection of the cell from ROS-induced damage and oxidative stress (Dayer et al., 2008). Accordingly, GPX5 and GSTS1 expression was reported to be activated by different stress conditions, including high light (Fischer et al., 2007, 2012; Ledford et al., 2007). The plastid-localized NTRC also was up-regulated by cerulenin. In plants, NTRC plays a protective role in the chloroplast against oxidative damage by reducing 2-Cys peroxiredoxins (Pérez-Ruiz et al., 2006, 2017). Whether NTRC may have a similar function in algae is currently unknown, but the identification of this NADPH-dependent thioredoxin reductase among the genes induced in response to cerulenin in Chlamydomonas suggests a role in the redox protection of the chloroplast.
The thylakoid membrane defects found in Chlamydomonas cells treated with cerulenin correlated with a down-regulation of photosynthesis and an increase in the production of ROS. Under stress conditions, cells normally adapt their photosynthetic activity by remodeling photosynthetic membranes. As a result, excess light cannot be properly absorbed into the photosynthetic apparatus, favoring the production of ROS that cause oxidative damage and arrest cell growth. We hypothesize that the generation of ROS in cerulenin-treated cells is linked to the remodeling of thylakoid membranes and the defective assimilation of light by PSII and PSI. This is consistent with our results showing that FAS inhibition decreased the amount of PSII and PSI components and increased the level of ROS. A connection between impaired photosynthesis and the production of ROS has been reported in the npq1 lor1 double mutant. Upon high-light stress, PSII proteins are degraded and photosynthesis is down-regulated in this mutant, leading to high ROS production (Baroli et al., 2004). In close agreement with our results, it has been shown that long-term exposure to cerulenin increases the level of ROS in C. zofingiensis cells (Liu et al., 2016b). The structural defects in thylakoid membranes recently reported in a Chlamydomonas mutant lacking the MGDG lipase PGD1 also have been linked to an enhanced production of ROS in the chloroplast during nitrogen deprivation or high-light exposure (Du et al., 2018). Collectively, these observations indicate that ROS are generated in response to FAS inhibition or thylakoid membrane remodeling in algae. Given that ROS are established components of chloroplast-nucleus retrograde signaling (Dietz et al., 2016; Leister, 2017), the question arises whether ROS may cause the retrograde signal leading to autophagy activation upon FAS inhibition. We have shown previously that ROS are involved in the control of autophagy in Chlamydomonas (Pérez-Pérez et al., 2012a) and regulate the activity of the ATG4 protease in algae and yeast (Pérez-Pérez et al., 2014, 2016). Our results indicated that the up-regulation of autophagy and other stress-related genes in cerulenin-treated cells takes place within 4 h, when photosynthesis and ROS were not yet affected significantly (Figs. 6 and 7). Since FAS inhibition has a direct impact on plastid membrane composition (Fig. 2), it might be possible that the loss of integrity of thylakoid membranes is signaled to the nucleus by a ROS-independent mechanism in order to activate the expression of autophagy, proteasome, chloroplast chaperones, and other stress-related genes needed to maintain cell homeostasis (Fig. 9). However, it cannot be ruled out that a low abundance of ROS that was below the detection level in our study might act as signaling molecules activating retrograde signaling in cerulenin-treated cells. The cellular response of cerulenin-treated and ClpP-depleted cells is remarkably similar, since both conditions activate a chloroplast stress response and autophagy (Ramundo et al., 2014). Whether FAS inhibition and ClpP depletion operate through the same chloroplast-nucleus-cytosol signaling network remains to be investigated. The FAS inhibitor cerulenin might be a valuable tool with which to dissect retrograde signaling from the plastid to the nucleus and to identify selective substrates of autophagy in chloroplast-stressed cells.
MATERIALS AND METHODS
Strains, Media, and Growth Conditions
Chlamydomonas reinhardtii lor1 (CC-4100) and npq1 lor1 (CC-4108) mutants and the isogenic wild-type 4A+ strain (CC-4051) were obtained from the Chlamydomonas Resource Center (http://www.chlamycollection.org). Chlamydomonas strain OL-Rps6 expressing OLLAS (Escherichia coli OmpF Linker and mouse Langerin fusion sequence)-tagged RPS6 was described previously (Couso et al., 2018). The cell wall-deficient strain cw15 4B+ was obtained from the laboratory of Jean-David Rochaix (University of Geneva). Chlamydomonas cells were grown under standard illumination (40–50 µE m−2 s−1) at 25°C in TAP medium as described previously (Harris, 1989). The lor1 and npq1 lor1 mutants were maintained and grown in dim light (5–10 µE m−2 s−1) to prevent any light damage. When required, cells in the exponential growth phase (∼106 cells mL–1) were treated with cerulenin (Sigma-Aldrich; C2389) or ConcA (Santa Cruz Biotechnology; sc-202111A) at the indicated concentrations or subjected to nitrogen limitation. Stock solutions of cerulenin (20 mm) and ConcA (100 µm) were prepared in ethanol and dimethyl sulfoxide, respectively. For growth on plates, 1.2% (w/v) bacto agar (Difco; 214010) was added to the medium.
Lipid Analysis
Total lipids were extracted as described (Couso et al., 2018). For lipid fractionation, total lipid extracts were evaporated under nitrogen and the residue was dissolved in 5 mL of chloroform. The resulting solution was fractionated in a Lichrolut 0.5-g silica gel cartridge (Merck) using a vacuum manifold and then equilibrated with 2 mL of chloroform as described previously (Nash and Frankel, 1986). The solution of total lipids was loaded on the column, which was then washed with another 15 mL of chloroform to elute neutral lipids from the column. Subsequently, the column was washed with 10 mL of methanol to recover the polar lipids quantitatively. The polar lipid fraction was first evaporated to dryness under nitrogen and then dissolved in 1.5 mL of hexane:2-propanol (3:2). Polar lipids were analyzed and quantified by HPLC (Salas et al., 2006). Separation by HPLC was carried out in a Waters 2695 Module equipped with a Waters 2420 ELSD. Polar and neutral lipids were separated at 30°C using a Lichrospher 100 Diol 254-4 (5 μm) column (Merck) or a normal-phase Lichrocart 250-4 (5 μm) column (Merck). The data were processed using Empower software, and the ELSD was regularly calibrated using commercial high-purity standards for each lipid.
The total fatty acid composition was determined by adding a volume of 3.3 mL of methanol:toluene:dimethoxypropane:sulfuric acid (39:20:5:2) and 1.7 mL of heptane to the sample, and the mixture was heated at 80°C for 1 h. After cooling, the upper phase containing the fatty acid methyl esters was transferred to a fresh tube, washed with 6.7% sodium sulfate, and evaporated to dryness with nitrogen. The methyl esters were dissolved in an appropriate volume of heptane and analyzed by gas-liquid chromatography (Serrano-Vega et al., 2005). The different methyl esters were identified by comparing their retention times with those of known standards.
Nile Red Staining
Lipid body staining was performed with Nile Red as reported previously (Wang et al., 2009). Cells from Chlamydomonas cultures growing under rich or nitrogen starvation conditions in the absence or presence of cerulenin were fixed on ice for 20 min with 2% paraformaldehyde (Sigma-Aldrich; 158127) and then washed twice with phosphate-buffered saline (PBS). Microscopy was performed with a Leica DM6000B using a 100× oil-immersion objective with DIC optics or wide-field fluorescence equipped with a Leica L5 filter cube (excitation bandpass, 480/40 nm; dichroic, 505 nm; emission bandpass, 527/30 nm) and an ORCA-ER camera (Hamamatsu).
Carotenoid Extraction Analysis
Samples from Chlamydomonas cultures treated with cerulenin were 30-fold concentrated and used for carotenoid extraction with 80% (v/v) acetone as described (Pérez-Pérez et al., 2012a). Samples were analyzed by HPLC using a Waters Spherisorb ODS2 column (4.6 × 250 mm, 5-µm particle size; Waters; PSS831915). The chromatographic method used was reported previously (Baroli et al., 2003). Pigments were detected at 450 nm using a Waters 2996 photodiode array detector. Finally, carotenoids were identified by comparison of the individual characteristic absorption spectrum and the retention times of known standards. The quantification was performed using a calibration curve generated with commercially available carotenoid standards from Sigma-Aldrich. The quantity of carotenoids was relativized to the Chlamydomonas cell number.
Flow Cytometry
Wild-type Chlamydomonas cells treated with cerulenin for 8, 24, and 48 h were fixed on ice for 20 min with 2% (v/v) paraformaldehyde (Sigma-Aldrich; 158127) as described above for Nile Red staining. Samples were then subjected to analysis using a flow cytometer (BD FACSCalibur Cytometry System). The relative fluorescence intensity was detected by a photomultiplier tube using excitation at 488 nm and emission at 670 nm. Forward scatter analysis was performed to measure cell surface area and size. Data were processed with CellQuest ProV5.2.1 software (BD).
PAM/Fluorescence-Based Measurements
Chlamydomonas cells treated with cerulenin for 4, 8, and 24 h or the drug vehicle (control) were collected by centrifugation (4,000g, 5 min), washed once in fresh TAP medium, and ∼50-fold concentrated. Cells were normalized at OD750nm. Fluorescence-based photosynthetic parameters were measured with an Imaging-PAM Chlorophyll Fluorometer (model IMAG-MAX/L) with a camera (IMAG-K7; Heinz Walz) using a 96-microwell plate (Thermo Scientific; 137101). First, the Fv/Fm parameter, which defines the maximal PSII quantum yield and serves for definition of the sample limits, was measured. Then, the relative apparent photosynthetic ETR was calculated as follows: ETR = Yield × PAR × 0.5 × Absorptivity. ETR was measured at different actinic lights (photosynthetically active radiation [PAR]) from 0 to 1,251 µE m−2 s−1. Before measurements, cells were acclimated to dark for 10 min.
ROS Determination
Samples from Chlamydomonas cultures treated with cerulenin for 2, 4, 8, and 24 h were taken to measure ROS with a 2′,7′-dichlorodihydrofluorescein diacetate (Invitrogen; D-399) probe. The 2′,7′-dichlorodihydrofluorescein diacetate probe is converted into fluorescent dichlorofluorescein by ROS. The analysis of ROS was performed as described previously (Pérez-Pérez et al., 2012a), following the instructions of Joo et al. (2005). Cells treated with the drug vehicle at the same time points were used as a control. Each experiment was performed at least three independent times with three technical replicates each time.
Protein Preparation
Chlamydomonas cells from liquid cultures were collected by centrifugation (4,000g, 5 min), washed once in lysis buffer (50 mm Tris-HCl, pH 7.5), and resuspended in a minimal volume of the same buffer. Cells were lysed by two cycles of slow freezing to −80°C followed by thawing to room temperature. The soluble cell extract was separated from the insoluble fraction by centrifugation (15,000g, 20 min) at 4°C. For chloroplast protein analysis, total protein extracts were obtained by collecting cells by centrifugation (4,000g, 5 min). Cells were then washed once in the lysis buffer described above, and pellets were frozen in liquid nitrogen and immediately stored at −80°C until use. Cells were lysed by resuspending the pellet in the desired volume of lysis buffer 2 (50 mm Tris-HCl [pH 7.5], 2% [w/v] SDS [Sigma-Aldrich; 05030], 10 mm EDTA, and 1× protease inhibitor cocktail [Sigma-Aldrich; 11836153001]) and incubated at 37°C for 30 min. Then, the total protein extract was separated from unbroken membranes by centrifugation (15,000g, 20 min) at 4°C. Proteins were quantified with the Coomassie Blue dye-binding method (Bio-Rad; 500-0006) or with the bicinchoninic acid solution (Sigma-Aldrich; B9643) as described by the manufacturers.
Western-Blot Analysis
For immunoblot analyses, total protein extracts (5–15 μg) were subjected to 12% or 15% (v/v) SDS-PAGE and then transferred to nitrocellulose membranes (Bio-Rad; 162-0115). Primary antibodies and dilutions used were as follows: HSP70B (Agrisera; AS06175; 1:10,000), VIPP1 (1:1,000; Liu et al., 2005), VIPP2 (1:1,000; Nordhues et al., 2012), FKBP12 (1:5,000; Crespo et al., 2005), ATG8 (1:3,000; Pérez-Pérez et al., 2010), OLLAS (Thermo Scientific; MA5-16125; 1:1,000), Rpl37 (Agrisera; AS122115; 1:10,000), PsbA (Agrisera; AS01016; 1:20,000), PsbB (Agrisera; AS04038; 1:2,000), PsbC (Agrisera; AS111787; 1:8,000), Lhcb5 (Agrisera; AS09407; 1:10,000), RbcL (Agrisera; AS03037; 1:10,000), PsaC (Agrisera; AS10939; 1:1,000), and ARF1 (Agrisera; AS08325; 1:2,000). Secondary anti-rabbit (Sigma-Aldrich; A6154) and anti-rat (Thermo Scientific; A18866) antibodies were diluted 1:10,000 and 1:5,000, respectively. The incubation with antibodies was in PBS containing 0.1% (v/v) Tween 20 (Applichem; A4974) and 5% (w/v) milk powder. The Luminata Crescendo Millipore immunoblotting detection system (Millipore; WBLUR0500) was used to detect the proteins.
Optical and Confocal Microscopy
Chlamydomonas cells treated with cerulenin at the indicated times were collected by centrifugation and resuspended in 1 mL of PBS buffer. Cells were fixed with 2.5% (v/v) glutaraldehyde (Sigma-Aldrich; G5882) in 50 mm Tris-HCl (pH 7.5) for 1 h at 25°C. After fixing, cells were washed four times with 50 mm Tris-HCl (pH 7.5) and resuspended in the same buffer. Cells were examined with an optical microscope (AXIO Scope A1; Zeiss) equipped with DIC optics. Images were registered with an Axiocam 105 camera (Zeiss) and processed with ZEN 2.3 software (Zeiss). Chloroplast autofluorescence was detected with a confocal laser scanning microscope (LSM 7 DUO; Zeiss) using excitation at 633 nm and emission at 670 nm. Confocal micrographs were processed with ZEN2011 software (Zeiss).
Immunofluorescence Microscopy
Chlamydomonas-untreated (control) or cerulenin-treated cells were fixed and stained for immunofluorescence microscopy as described previously (Crespo et al., 2005). Affinity-purified anti-ATG8 antibody (1:500) was used as reported previously (Pérez-Pérez et al., 2010). For signal detection, fluorescein isothiocyanate-labeled goat anti-rabbit antibody (1:500 dilution; Sigma-Aldrich; F4890) was used. Cells were photographed on a DM6000B microscope (Leica) with an ORCA-ER camera (Hamamatsu) and processed with the Leica Application Suite Advanced Fluorescence software package. For comparative analysis, the same acquisition time was fixed for the fluorescein isothiocyanate signal.
Electron Microscopy
Chlamydomonas cells (∼2 × 106 cells mL–1) treated with cerulenin for 8 h were fixed with 2.5% glutaraldehyde (Sigma-Aldrich; G5882) in 0.1 m sodium-cacodylate buffer (pH 7.4) for 2 h at 25°C. After fixing, cells were washed five times with the same buffer at 25°C. Then, samples were postfixed in 1% osmium tetraoxide in 0.1 m cacodylate buffer (pH 7.4) for 1 h at 4°C. After washing, samples were immersed in 2% uranyl acetate, dehydrated through a gradient acetone series (50%, 70%, 90%, and 100%, v/v), and embedded in Spurr resin (Spurr, 1969). Semithin sections (300 nm thickness) were obtained with a glass knife and stained with 1% (w/v) Toluidine Blue for cell localization and reorientation using a conventional optical microscope. Once a suitable block face of the selected area was trimmed, several ultrathin sections (70 nm) were obtained using an ultramicrotome (Leica; UC7) equipped with a diamond knife (Diatome) and collected on 200-mesh copper grids. Sections were examined in a Zeiss Libra 120 transmission electron microscope and digitized (2,048 Å ∼ 2,048 Å ∼ 16 bits) using an on-axis mounted TRS camera.
RNA Isolation and RT-qPCR Analysis
Chlamydomonas cells treated with cerulenin or the drug vehicle were collected by centrifugation (4,000g, 5 min), washed once in 50 mm Tris-HCl (pH 7.5) buffer, and then pellets were frozen in liquid nitrogen and immediately stored at −80°C until use. Total RNA was isolated from frozen cell pellets as described previously (Crespo et al., 2005). First-strand cDNA was produced in a 50-µL reaction mixture containing 2 µg of total RNA, an oligo(dT) primer, and 100 units of SuperScript II RNase H-reverse transcriptase (Invitrogen; 18064-014). RT-qPCR was performed on an iCycler apparatus (Bio-Rad). The PCR mixtures were performed in a final volume of 20 µL, containing 10 µL of FastStart Universal SYBR Green master mix (Roche; 04913850001), 1 µL of cDNA dilution, 250 nm each primer, and redistilled water. The data were normalized to CBLP expression, a constitutively expressed gene encoding a protein homologous to the β-subunit of a G protein that is used as an internal control (Pootakham et al., 2010). The primer pairs used in this study are listed in Supplemental Table S5. All reactions were performed in triplicate with three biological replicates.
RNA-Seq Analysis
C. reinhardtii 4A+ strain (CC-4051) was grown in TAP medium as described above. Total RNA from Chlamydomonas cells treated with 10 µm cerulenin for 0, 4, and 8 h was isolated from frozen cell pellets using a phenol-chloroform procedure as described previously (Crespo et al., 2005). RNA-Seq data were generated by STABvida (http://www.stabvida.com) according to the following procedure. Quality control of RNA was confirmed by analyzing the RNA integrity number and the concentration with a Bioanalyzer (Bio-Rad). cDNA libraries were prepared using the Kapa Stranded mRNA Library Preparation Kit (Roche) and sequenced in the Illumina HiSeq 4000 platform using 150-bp paired-end sequencing reads. The analysis of the generated sequence raw data was carried out using CLC Genomics Workbench 10.1.1 (Qiagen). High-quality sequencing reads were generated after trimming and then mapped to the version 5.5 assembly of the C. reinhardtii genome (Merchant et al., 2007). Gene expression estimates were obtained for each individual sample in units of reads per kilobase of exon model per million mapped reads, as described previously (Mortazavi et al., 2008). Principal component analysis was applied to identify outlier samples for quality control. Differential expression analysis was performed using a generalized linear model approach influenced by the multifactorial edgeR method (Robinson et al., 2010). Functional analysis of the data was done using the Algal Functional Annotation Tool (http://pathways.mcdb.ucla.edu/algal/index.html) and manual annotation.
Statistical Analyses
All statistical analyses include arithmetic means and sd, and parametric two-tailed Student’s t tests were applied in all cases.
Accession Numbers
Sequence data from this article are deposited in the National Center for Biotechnology Information Sequence Read Archive database (https://www.ncbi.nlm.nih.gov/sra/) under accession number SRP155181.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Effect of cerulenin on Chlamydomonas cell growth.
Supplemental Figure S2. Effect of cerulenin on ATG mRNA abundance.
Supplemental Figure S3. Ultrastructural analysis of Chlamydomonas cells treated with cerulenin.
Supplemental Figure S4. Thylakoid hyperstacking in cerulenin-treated cells.
Supplemental Figure S5. Effect of cerulenin on VIPP1 and VIPP2 mRNA abundance.
Supplemental Figure S6. Principal component analysis corresponding to gene expression data of Chlamydomonas cells treated with cerulenin.
Supplemental Figure S7. Functional characterization of genes differentially expressed between 0 and 8 h following cerulenin treatment.
Supplemental Figure S8. Effect of cerulenin on the expression of proteasome and ubiquitin genes.
Supplemental Table S1. Genes induced greater than 2 times after 4 h of cerulenin treatment.
Supplemental Table S2. Genes repressed greater than 2 times after 4 h of cerulenin treatment.
Supplemental Table S3. Genes induced greater than 2 times after 8 h of cerulenin treatment.
Supplemental Table S4. Genes repressed greater than 2 times after 8 h of cerulenin treatment.
Supplemental Table S5. Primer pairs used for RT-qPCR.
Acknowledgments
We thank Dr. Michael Schroda (University of Kaiserslautern) for the generous gift of the anti-VIPP1 and anti-VIPP2 antibodies. We also thank Juan-Luis Ribas (Microscopy Service of the Centro de Investigación, Tecnología e Innovación, University of Seville) and M. Soledad Parra Camacho (Instituto de la Grasa, Consejo Superior de Investigaciones Científicas) for technical assistance with transmission electron microscopy assays and lipid analysis, respectively.
Footnotes
This work was supported in part by the Ministerio de Economía y Competitividad grant BFU2015-68216-P and Junta de Andalucía CVI-7336 (to J.L.C.), and BIO2015-74432-JIN (to M.E.P.-P.). L.G.H.-M. is a recipient of a fellowship from Ministerio de Economía y Competitividad (BES-2016-077314).
Articles can be viewed without a subscription.
References
- Baroli I, Do AD, Yamane T, Niyogi KK (2003) Zeaxanthin accumulation in the absence of a functional xanthophyll cycle protects Chlamydomonas reinhardtii from photooxidative stress. Plant Cell 15: 992–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroli I, Gutman BL, Ledford HK, Shin JW, Chin BL, Havaux M, Niyogi KK (2004) Photo-oxidative stress in a xanthophyll-deficient mutant of Chlamydomonas. J Biol Chem 279: 6337–6344 [DOI] [PubMed] [Google Scholar]
- Bassham DC, Laporte M, Marty F, Moriyasu Y, Ohsumi Y, Olsen LJ, Yoshimoto K (2006) Autophagy in development and stress responses of plants. Autophagy 2: 2–11 [DOI] [PubMed] [Google Scholar]
- Botté CY, Deligny M, Roccia A, Bonneau AL, Saïdani N, Hardré H, Aci S, Yamaryo-Botté Y, Jouhet J, Dubots E, et al. (2011) Chemical inhibitors of monogalactosyldiacylglycerol synthases in Arabidopsis thaliana. Nat Chem Biol 7: 834–842 [DOI] [PubMed] [Google Scholar]
- Chen G, Wang B, Han D, Sommerfeld M, Lu Y, Chen F, Hu Q (2015) Molecular mechanisms of the coordination between astaxanthin and fatty acid biosynthesis in Haematococcus pluvialis (Chlorophyceae). Plant J 81: 95–107 [DOI] [PubMed] [Google Scholar]
- Couso I, Pérez-Pérez ME, Martínez-Force E, Kim HS, He Y, Umen JG, Crespo JL (2018) Autophagic flux is required for the synthesis of triacylglycerols and ribosomal protein turnover in Chlamydomonas. J Exp Bot 69: 1355–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo JL, Díaz-Troya S, Florencio FJ (2005) Inhibition of target of rapamycin signaling by rapamycin in the unicellular green alga Chlamydomonas reinhardtii. Plant Physiol 139: 1736–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Agnolo G, Rosenfeld IS, Awaya J, Omura S, Vagelos PR (1973) Inhibition of fatty acid synthesis by the antibiotic cerulenin: specific inactivation of beta-ketoacyl-acyl carrier protein synthetase. Biochim Biophys Acta 326: 155–156 [DOI] [PubMed] [Google Scholar]
- Davey MP, Horst I, Duong GH, Tomsett EV, Litvinenko AC, Howe CJ, Smith AG (2014) Triacylglyceride production and autophagous responses in Chlamydomonas reinhardtii depend on resource allocation and carbon source. Eukaryot Cell 13: 392–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayer R, Fischer BB, Eggen RI, Lemaire SD (2008) The peroxiredoxin and glutathione peroxidase families in Chlamydomonas reinhardtii. Genetics 179: 41–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Troya S, Pérez-Pérez ME, Florencio FJ, Crespo JL (2008) The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy 4: 851–865 [DOI] [PubMed] [Google Scholar]
- Dietz KJ, Turkan I, Krieger-Liszkay A (2016) Redox- and reactive oxygen species-dependent signaling into and out of the photosynthesizing chloroplast. Plant Physiol 171: 1541–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolch LJ, Rak C, Perin G, Tourcier G, Broughton R, Leterrier M, Morosinotto T, Tellier F, Faure JD, Falconet D, et al. (2017) A palmitic acid elongase affects eicosapentaenoic acid and plastidial monogalactosyldiacylglycerol levels in Nannochloropsis. Plant Physiol 173: 742–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörmann P, Benning C (2002) Galactolipids rule in seed plants. Trends Plant Sci 7: 112–118 [DOI] [PubMed] [Google Scholar]
- Du ZY, Lucker BF, Zienkiewicz K, Miller TE, Zienkiewicz A, Sears BB, Kramer DM, Benning C (2018) Galactoglycerolipid lipase PGD1 is involved in thylakoid membrane remodeling in response to adverse environmental conditions in Chlamydomonas. Plant Cell 30: 447–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elander PH, Minina EA, Bozhkov PV (2018) Autophagy in turnover of lipid stores: trans-kingdom comparison. J Exp Bot 69: 1301–1311 [DOI] [PubMed] [Google Scholar]
- Fan J, Andre C, Xu C (2011) A chloroplast pathway for the de novo biosynthesis of triacylglycerol in Chlamydomonas reinhardtii. FEBS Lett 585: 1985–1991 [DOI] [PubMed] [Google Scholar]
- Feng Y, He D, Yao Z, Klionsky DJ (2014) The machinery of macroautophagy. Cell Res 24: 24–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer BB, Krieger-Liszkay A, Eggen RI (2005) Oxidative stress induced by the photosensitizers neutral red (type I) or rose bengal (type II) in the light causes different molecular responses in Chlamydomonas. Plant Sci 168: 747–759 [Google Scholar]
- Fischer BB, Krieger-Liszkay A, Hideg E, Snyrychová I, Wiesendanger M, Eggen RI (2007) Role of singlet oxygen in chloroplast to nucleus retrograde signaling in Chlamydomonas reinhardtii. FEBS Lett 581: 5555–5560 [DOI] [PubMed] [Google Scholar]
- Fischer BB, Ledford HK, Wakao S, Huang SG, Casero D, Pellegrini M, Merchant SS, Koller A, Eggen RI, Niyogi KK (2012) SINGLET OXYGEN RESISTANT 1 links reactive electrophile signaling to singlet oxygen acclimation in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 109: E1302–E1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd BE, Morriss SC, Macintosh GC, Bassham DC (2012) What to eat: evidence for selective autophagy in plants. J Integr Plant Biol 54: 907–920 [DOI] [PubMed] [Google Scholar]
- Goodenough UW, Levine RP (1969) Chloroplast ultrastructure in mutant strains of Chlamydomonas reinhardi lacking components of the photosynthetic apparatus. Plant Physiol 44: 990–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U, Blaby I, Casero D, Gallaher SD, Goodson C, Johnson S, Lee JH, Merchant SS, Pellegrini M, Roth R, et al. (2014) The path to triacylglyceride obesity in the sta6 strain of Chlamydomonas reinhardtii. Eukaryot Cell 13: 591–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gounaris K, Barber J (1983) Monogalactosyldiacylglycerol: the most abundant polar lipid in Nature. Trends Biochem Sci 8: 378–381 [Google Scholar]
- Harris EH. (1989) The Chlamydomonas Sourcebook. Academic Press, San Diego [Google Scholar]
- He C, Klionsky DJ (2009) Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 43: 67–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Sommerfeld M, Jarvis E, Ghirardi M, Posewitz M, Seibert M, Darzins A (2008) Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J 54: 621–639 [DOI] [PubMed] [Google Scholar]
- Izumi M, Ishida H, Nakamura S, Hidema J (2017) Entire photodamaged chloroplasts are transported to the central vacuole by autophagy. Plant Cell 29: 377–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P, Dörmann P, Peto CA, Lutes J, Benning C, Chory J (2000) Galactolipid deficiency and abnormal chloroplast development in the Arabidopsis MGD synthase 1 mutant. Proc Natl Acad Sci USA 97: 8175–8179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson P, Wiltschi B, Kumari P, Kessler B, Vonrhein C, Vonck J, Oesterhelt D, Grininger M (2008) Inhibition of the fungal fatty acid synthase type I multienzyme complex. Proc Natl Acad Sci USA 105: 12803–12808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo JH, Wang S, Chen JG, Jones AM, Fedoroff NV (2005) Different signaling and cell death roles of heterotrimeric G protein alpha and beta subunits in the Arabidopsis oxidative stress response to ozone. Plant Cell 17: 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Kondo M, Fukuda H, Nishimura M, Ohta H (2007) Galactolipid synthesis in chloroplast inner envelope is essential for proper thylakoid biogenesis, photosynthesis, and embryogenesis. Proc Natl Acad Sci USA 104: 17216–17221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo AJ, Fulda M, Browse J, Ohlrogge JB (2005) Identification of a plastid acyl-acyl carrier protein synthetase in Arabidopsis and its role in the activation and elongation of exogenous fatty acids. Plant J 44: 620–632 [DOI] [PubMed] [Google Scholar]
- Kurusu T, Koyano T, Hanamata S, Kubo T, Noguchi Y, Yagi C, Nagata N, Yamamoto T, Ohnishi T, Okazaki Y, et al. (2014) OsATG7 is required for autophagy-dependent lipid metabolism in rice postmeiotic anther development. Autophagy 10: 878–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskay G, Farkas T, Lehoczki E (1985) Cerulenin-induced changes in lipid and fatty acid content of chloroplasts in detached greening barley leaves. J Plant Physiol 118: 267–275 [DOI] [PubMed] [Google Scholar]
- Ledford HK, Chin BL, Niyogi KK (2007) Acclimation to singlet oxygen stress in Chlamydomonas reinhardtii. Eukaryot Cell 6: 919–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister D. (2017) Piecing the puzzle together: the central role of reactive oxygen species and redox hubs in chloroplast retrograde signaling. Antioxid Redox Signal doi.org/10.1089/ars.2017.7392 [DOI] [PubMed] [Google Scholar]
- Li Z, Wakao S, Fischer BB, Niyogi KK (2009) Sensing and responding to excess light. Annu Rev Plant Biol 60: 239–260 [DOI] [PubMed] [Google Scholar]
- Li-Beisson Y, Beisson F, Riekhof W (2015) Metabolism of acyl-lipids in Chlamydomonas reinhardtii. Plant J 82: 504–522 [DOI] [PubMed] [Google Scholar]
- Liu B, Benning C (2013) Lipid metabolism in microalgae distinguishes itself. Curr Opin Biotechnol 24: 300–309 [DOI] [PubMed] [Google Scholar]
- Liu Y, Bassham DC (2012) Autophagy: pathways for self-eating in plant cells. Annu Rev Plant Biol 63: 215–237 [DOI] [PubMed] [Google Scholar]
- Liu C, Willmund F, Whitelegge JP, Hawat S, Knapp B, Lodha M, Schroda M (2005) J-domain protein CDJ2 and HSP70B are a plastidic chaperone pair that interacts with vesicle-inducing protein in plastids 1. Mol Biol Cell 16: 1165–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Sun Z, Zhong Y, Huang J, Hu Q, Chen F (2012) Stearoyl-acyl carrier protein desaturase gene from the oleaginous microalga Chlorella zofingiensis: cloning, characterization and transcriptional analysis. Planta 236: 1665–1676 [DOI] [PubMed] [Google Scholar]
- Liu J, Han D, Yoon K, Hu Q, Li Y (2016a) Characterization of type 2 diacylglycerol acyltransferases in Chlamydomonas reinhardtii reveals their distinct substrate specificities and functions in triacylglycerol biosynthesis. Plant J 86: 3–19 [DOI] [PubMed] [Google Scholar]
- Liu J, Mao X, Zhou W, Guarnieri MT (2016b) Simultaneous production of triacylglycerol and high-value carotenoids by the astaxanthin-producing oleaginous green microalga Chlorella zofingiensis. Bioresour Technol 214: 319–327 [DOI] [PubMed] [Google Scholar]
- Marshall RS, Vierstra RD (2018) Autophagy: the master of bulk and selective recycling. Annu Rev Plant Biol 69: 173–208 [DOI] [PubMed] [Google Scholar]
- Marshall RS, Li F, Gemperline DC, Book AJ, Vierstra RD (2015) Autophagic degradation of the 26S proteasome is mediated by the dual ATG8/ubiquitin receptor RPN10 in Arabidopsis. Mol Cell 58: 1053–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Kropat J, Liu B, Shaw J, Warakanont J (2012) TAG, you’re it! Chlamydomonas as a reference organism for understanding algal triacylglycerol accumulation. Curr Opin Biotechnol 23: 352–363 [DOI] [PubMed] [Google Scholar]
- Minina EA, Moschou PN, Vetukuri RR, Sanchez-Vera V, Cardoso C, Liu Q, Elander PH, Dalman K, Beganovic M, Lindberg Yilmaz J, et al. (2018) Transcriptional stimulation of rate-limiting components of the autophagic pathway improves plant fitness. J Exp Bot 69: 1415–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y (2011) The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 27: 107–132 [DOI] [PubMed] [Google Scholar]
- Moche M, Schneider G, Edwards P, Dehesh K, Lindqvist Y (1999) Structure of the complex between the antibiotic cerulenin and its target, beta-ketoacyl-acyl carrier protein synthase. J Biol Chem 274: 6031–6034 [DOI] [PubMed] [Google Scholar]
- Moellering ER, Benning C (2011) Galactoglycerolipid metabolism under stress: a time for remodeling. Trends Plant Sci 16: 98–107 [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Moseley JL, Chang CW, Grossman AR (2006) Genome-based approaches to understanding phosphorus deprivation responses and PSR1 control in Chlamydomonas reinhardtii. Eukaryot Cell 5: 26–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller P, Li XP, Niyogi KK (2001) Non-photochemical quenching: a response to excess light energy. Plant Physiol 125: 1558–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash AM, Frankel EN (1986) Limited extraction of soybean with hexane. J Am Oil Chem Soc 63: 244–246 [Google Scholar]
- Niyogi KK, Björkman O, Grossman AR (1997) The roles of specific xanthophylls in photoprotection. Proc Natl Acad Sci USA 94: 14162–14167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordhues A, Schöttler MA, Unger AK, Geimer S, Schönfelder S, Schmollinger S, Rütgers M, Finazzi G, Soppa B, Sommer F, et al. (2012) Evidence for a role of VIPP1 in the structural organization of the photosynthetic apparatus in Chlamydomonas. Plant Cell 24: 637–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packter NM, Stumpf PK (1975) Fat metabolism in higher plants: the effect of cerulenin on the synthesis of medium- and long-chain acids in leaf tissue. Arch Biochem Biophys 167: 655–667 [DOI] [PubMed] [Google Scholar]
- Pérez-Martín M, Pérez-Pérez ME, Lemaire SD, Crespo JL (2014) Oxidative stress contributes to autophagy induction in response to endoplasmic reticulum stress in Chlamydomonas reinhardtii. Plant Physiol 166: 997–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Martín M, Blaby-Haas CE, Pérez-Pérez ME, Andrés-Garrido A, Blaby IK, Merchant SS, Crespo JL (2015) Activation of autophagy by metals in Chlamydomonas reinhardtii. Eukaryot Cell 14: 964–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez ME, Crespo JL (2014) Autophagy in algae. Perspectives in Phycology 1: 93–102 [Google Scholar]
- Pérez-Pérez ME, Florencio FJ, Crespo JL (2010) Inhibition of target of rapamycin signaling and stress activate autophagy in Chlamydomonas reinhardtii. Plant Physiol 152: 1874–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez ME, Couso I, Crespo JL (2012b) Carotenoid deficiency triggers autophagy in the model green alga Chlamydomonas reinhardtii. Autophagy 8: 376–388 [DOI] [PubMed] [Google Scholar]
- Pérez-Pérez ME, Lemaire SD, Crespo JL (2012a) Reactive oxygen species and autophagy in plants and algae. Plant Physiol 160: 156–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez ME, Zaffagnini M, Marchand CH, Crespo JL, Lemaire SD (2014) The yeast autophagy protease Atg4 is regulated by thioredoxin. Autophagy 10: 1953–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez ME, Lemaire SD, Crespo JL (2016) Control of autophagy in Chlamydomonas is mediated through redox-dependent inactivation of the ATG4 protease. Plant Physiol 172: 2219–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez ME, Couso I, Heredia-Martínez LG, Crespo JL (2017) Monitoring autophagy in the model green microalga Chlamydomonas reinhardtii. Cells 6: E36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Ruiz JM, Spínola MC, Kirchsteiger K, Moreno J, Sahrawy M, Cejudo FJ (2006) Rice NTRC is a high-efficiency redox system for chloroplast protection against oxidative damage. Plant Cell 18: 2356–2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Ruiz JM, Naranjo B, Ojeda V, Guinea M, Cejudo FJ (2017) NTRC-dependent redox balance of 2-Cys peroxiredoxins is needed for optimal function of the photosynthetic apparatus. Proc Natl Acad Sci USA 114: 12069–12074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pootakham W, Gonzalez-Ballester D, Grossman AR (2010) Identification and regulation of plasma membrane sulfate transporters in Chlamydomonas. Plant Physiol 153: 1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramundo S, Casero D, Mühlhaus T, Hemme D, Sommer F, Crèvecoeur M, Rahire M, Schroda M, Rusch J, Goodenough U, et al. (2014) Conditional depletion of the Chlamydomonas chloroplast ClpP protease activates nuclear genes involved in autophagy and plastid protein quality control. Plant Cell 26: 2201–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix JD, Ramundo S (2018) Chloroplast signaling and quality control. Essays Biochem 62: 13–20 [DOI] [PubMed] [Google Scholar]
- Rütgers M, Muranaka LS, Mühlhaus T, Sommer F, Thoms S, Schurig J, Willmund F, Schulz-Raffelt M, Schroda M (2017) Substrates of the chloroplast small heat shock proteins 22E/F point to thermolability as a regulative switch for heat acclimation in Chlamydomonas reinhardtii. Plant Mol Biol 95: 579–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas JJ, Martinez-Force E, Garces R (2006) Phospholipid and glycolipid accumulation in seed kernels of different sunflower mutants (Helianthus annuus). J Am Oil Chem Soc 83: 539–545 [Google Scholar]
- Schmollinger S, Mühlhaus T, Boyle NR, Blaby IK, Casero D, Mettler T, Moseley JL, Kropat J, Sommer F, Strenkert D, et al. (2014) Nitrogen-sparing mechanisms in Chlamydomonas affect the transcriptome, the proteome, and photosynthetic metabolism. Plant Cell 26: 1410–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber A, Peter M (2014) Substrate recognition in selective autophagy and the ubiquitin-proteasome system. Biochim Biophys Acta 1843: 163–181 [DOI] [PubMed] [Google Scholar]
- Schroda M, Vallon O (2009) Chaperones and proteases. In Stern DB, ed, The Chlamydomonas Sourcebook, Ed 2 Academic Press, Canada, pp 671–729 [Google Scholar]
- Schwarz V, Andosch A, Geretschläger A, Affenzeller M, Lütz-Meindl U (2017) Carbon starvation induces lipid degradation via autophagy in the model alga Micrasterias. J Plant Physiol 208: 115–127 [DOI] [PubMed] [Google Scholar]
- Serrano-Vega MJ, Martínez-Force E, Garcés R (2005) Lipid characterization of seed oils from high-palmitic, low-palmitoleic, and very high-stearic acid sunflower lines. Lipids 40: 369–374 [DOI] [PubMed] [Google Scholar]
- Shemi A, Ben-Dor S, Vardi A (2015) Elucidating the composition and conservation of the autophagy pathway in photosynthetic eukaryotes. Autophagy 11: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpilka T, Welter E, Borovsky N, Amar N, Mari M, Reggiori F, Elazar Z (2015) Lipid droplets and their component triglycerides and steryl esters regulate autophagosome biogenesis. EMBO J 34: 2117–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurr AR. (1969) A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res 26: 31–43 [DOI] [PubMed] [Google Scholar]
- Thompson AR, Vierstra RD (2005) Autophagic recycling: lessons from yeast help define the process in plants. Curr Opin Plant Biol 8: 165–173 [DOI] [PubMed] [Google Scholar]
- Wang ZT, Ullrich N, Joo S, Waffenschmidt S, Goodenough U (2009) Algal lipid bodies: stress induction, purification, and biochemical characterization in wild-type and starchless Chlamydomonas reinhardtii. Eukaryot Cell 8: 1856–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb MS, Green BR (1991) Biochemical and biophysical properties of thylakoid acyl lipids. Biochim Biophys Acta 1060: 133–158 [Google Scholar]
- Yang Y, Munz J, Cass C, Zienkiewicz A, Kong Q, Ma W, Sanjaya, Sedbrook J, Benning C (2015) Ectopic expression of WRINKLED1 affects fatty acid homeostasis in Brachypodium distachyon vegetative tissues. Plant Physiol 169: 1836–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young PG, Bartel B (2016) Pexophagy and peroxisomal protein turnover in plants. Biochim Biophys Acta 1863: 999–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Kato Y, Otters S, Vothknecht UC, Sakamoto W (2012) Essential role of VIPP1 in chloroplast envelope maintenance in Arabidopsis. Plant Cell 24: 3695–3707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Shrager J, Jain M, Chang CW, Vallon O, Grossman AR (2004) Insights into the survival of Chlamydomonas reinhardtii during sulfur starvation based on microarray analysis of gene expression. Eukaryot Cell 3: 1331–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Dai J, Wu Q (2014) Autophagy-like processes are involved in lipid droplet degradation in Auxenochlorella protothecoides during the heterotrophy-autotrophy transition. Front Plant Sci 5: 400. [DOI] [PMC free article] [PubMed] [Google Scholar]