Abstract
More than 50 cytokines signal via the JAK/STAT pathway to orchestrate hematopoiesis, induce inflammation and control the immune response. Cytokines are secreted glycoproteins that act as intercellular messengers, inducing proliferation, differentiation, growth, or apoptosis of their target cells. They act by binding to specific receptors on the surface of target cells and switching on a phosphotyrosine‐based intracellular signaling cascade initiated by kinases then propagated and effected by SH2 domain‐containing transcription factors. As cytokine signaling is proliferative and often inflammatory, it is tightly regulated in terms of both amplitude and duration. Here we review molecular details of the cytokine‐induced signaling cascade and describe the architectures of the proteins involved, including the receptors, kinases, and transcription factors that initiate and propagate signaling and the regulatory proteins that control it.
Keywords: cytokine, Cytokine Signaling, JAK/STAT, SOCS, cytokine receptor, hematopoiesis
Abbreviations
- Note that a full list of the abbreviations for all cytokines is given in Table 1. Receptors for each cytokine are denoted by the cytokine abbreviation followed by “R”. For example, TpoR
Tpo Receptor
- AKT
protein kinase B
- AML
acute myeloid leukemia
- APS
SH2B adaptor protein 2
- ATP
adenosine triphosphatse
- B‐ALL
B cell lymphocytic leukemia
- CBP
CREB‐binding protein
- CD45
cluster of differentiation 45
- CHR
cytokine receptor homology region
- Elk
ETS domain containing protein
- ER
endoplasmic reticulum
- FERM
band 4.1, ezrin, radixin, moesin
- FnIII
fibronectin type III
- FRET
fluorescence resonance energy transfer
- Gp130
glycoprotein 130
- Grb2
growth factor receptor‐bound protein 2
- HP1
heterochromatin protein 1
- Ig
immunoglobulin
- IRF9
interferon response factor 9
- ISGF3
IFN‐stimulated gene factor 3
- JAK
Janus Kinase
- JH
JAK homology domain
- LNK
lymphocyte adaptor protein or SH2B adaptor protein 3
- MAM
meprin, A‐5 protein, and receptor protein phosphatase mu
- MAPK
mitogen‐activated protein kinases
- mTOR
mammalian target of rapamycin
- NK
natural killer
- P300
E1A binding protein p300
- PH
pleckstrin homology
- PI(3)K
phosphatidylinositol‐4,5‐bisphosphate 3 kinase
- PTP
protein tyrosine phosphatase
- PTP‐RT
protein tyrosine phosphatase, receptor type
- PTP1b
protein tyrosine phosphatase 1B
- pTyr
phosphotyrosine
- SH2
Src homology 2
- SH2B
SH2B adaptor protein 1
- SHP1
Src homology region 2 domain‐containing phosphatase 1
- SHP2
Src homology region 2 domain‐containing phosphatase 1
- SOCS
suppressor of cytokine signaling
- STAT
signal transducer and activator of transcription
- TC‐PTP
T cell protein tyrosine phosphatase
- TM
transmembrane
- TYK
tyrosine kinase
- βc
beta common
- γc
gamma common
Introduction
Cytokines are secreted glycoproteins that act as intercellular messengers to control the hematopoietic and immune systems and the inflammatory response. The term “cytokine” arises from the Greek κύτος and κίνησις (cell and movement) consistent with their ability to mobilize cells to sites of infection and inflammation. Historically, the plethora of different cytokines have been divided into five groups: (A) TNF‐alpha and related molecules, (B) IL‐1 family members, (C) TGF‐Betas, (D) factors that signal through receptor tyrosine kinases such as M‐CSF, (D) chemokines, and (E) cytokines that signal through the JAK/STAT pathway (Fig. 1). The latter group is perhaps the largest and comprises both hematopoietic growth factors such as EPO as well as immunomodulatory cytokines such as IL‐2 and inflammatory cytokines such as interferon gamma (Table 1). This review will focus entirely on this class and henceforth we use the term cytokine to describe only those that signal via the JAK/STAT cascade.
Figure 1.
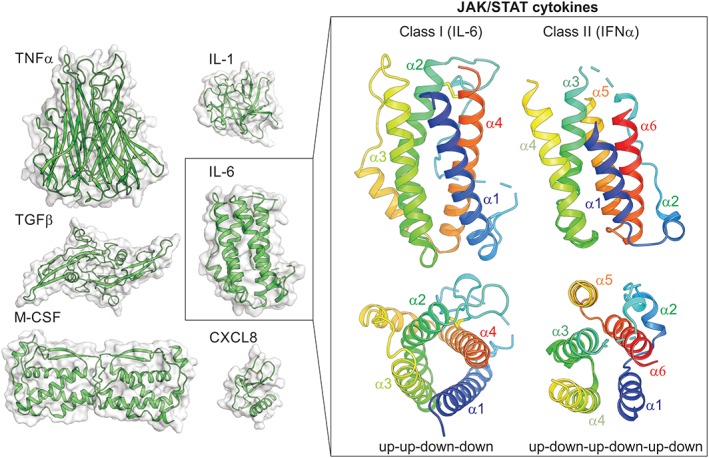
Cytokines . Structures of members of the TNFα‐family, TGFβ‐family, IL‐1‐like cytokines, chemokines (CXCL8), cytokines that signal through receptor tyrosine‐kinases (M‐CSF) or the JAK/STAT pathway (IL‐6) are shown on the left. JAK/STAT cytokines are helical bundle cytokines and can be divided into two classes. Examples of these two classes are shown on the right.
Table 1.
List of Cytokines that Signal through the JAK/STAT Pathway
| Abbreviation | Name | Major Functions |
|---|---|---|
| Class I cytokines | ||
| IL‐2 family | ||
| IL‐2 | Interleukin‐2 | Immune response, T‐cell differentiation |
| IL‐4 | Interleukin‐4 | TH2 differentiation |
| IL‐7 | Interleukin‐7 | T‐, B‐cell growth factor |
| IL‐9 | Interleukin‐9 | Pleiotropic, Stimulates, T‐, B‐ and NK cells |
| IL‐15 | Interleukin‐15 | Stimulates T‐ and NK‐cells |
| IL‐21 | Interleukin‐21 | Stimulates, T‐, B‐ and NK cells |
| IL‐3 family | ||
| IL‐3 | Interleukin‐3 | Multi‐lineage haematopoietic growth factor |
| IL‐5 | Interleukin‐5 | B‐cell development, eosinophils |
| GM‐CSF | Granulocyte/Macrophage Colony Stimulating Factor | Multi‐lineage haematopoietic growth factor, especially monocytes, neutrophils, eosinophils and basophils |
| IL‐6 family | ||
| IL‐6 | Interleukin‐6 | Pleiotropic, haematopoiesis, acute phase response, lymphoid differentiation |
| LIF | Leukemia Inhibitory Factor | Pleiotropic, blastocyst implantation, bone remodeling, CNS |
| CNTF | Ciliary NeuroTrophic growth Factor | Neuronal growth factor |
| CT1 | Cardiotrophin 1 | Cardiac myocytes growth factor |
| CLC | Cardiotrophin‐like cytokine | Neurological growth factor |
| OSM | Oncostatin M | Pleiotropic, bone formation |
| IL‐31 | Interleukin‐31 | Inflammatory, cell‐mediated immunty |
| NP | Neuropoietin | Neural growth factor |
| Homodimeric | ||
| G‐CSF | Granulocyte Colony Stimulating Factor | Stimulates granulocyte production, mobilises stem cells |
| EPO | Erythropoietin | Stimulates formation of erthrocytes |
| TPO | Thrombopoietin | Stimulates formation of megakaryocytes/platelets |
| GH | Growth Hormone | Growth |
| PRL | Prolactin | Milk production |
| LEP | Leptin | Regulates appetite |
| Others | ||
| IL‐12 | Interleukin‐12 | Stimulates T‐ and NK‐cells |
| IL‐13 | Interleukin‐13 | Pleiotropic, airway epithelia, allergic response |
| IL‐23 | Interleukin‐23 | Inflammation |
| TSLP | Thymic Stromal LymphoPoietin | Inflammatory, stimulates T‐ and B‐cells |
| Class II cytokines | ||
| Type I interferon | ||
| IFNα | Interferon alpha (23 subtypes) | Anti‐viral, secreted by lymphocytes, fibroblasts and monocytes |
| IFNβ | Interferon beta | Anti‐viral, ubiquitously expressed |
| IFNε | Interferon epsilon | Anti‐viral, expressed in female reproductive tract |
| IFNκ | Interferon kappa | Anti‐viral, expressed by keratinocytes |
| IFNω | Interferon omega | Anti‐viral, secreted by leukocytes |
| Type II interferon | ||
| IFNγ | Interferon gamma | Pro‐inflammatory, secreted by T‐ and NK‐cells, activates macrophages/monocytes |
| Type III interferon | ||
| IFNλ1 | Interferon lambda1 | Anti‐viral, similar to type I but acts on fewer cell‐types |
| IFNλ2 | Interferon lambda2 | Anti‐viral, similar to type I but acts on fewer cell‐types |
| IFNλ3 | Interferon lambda3 | Anti‐viral, similar to type I but acts on fewer cell‐types |
| IL‐10 family | ||
| IL‐10 | Interleukin‐10 | Anti‐inflammatory, inhibits macrophage activation |
| IL‐19 | Interleukin‐19 | Inflammatory, acts on dermal cells |
| IL‐20 | Interleukin‐20 | Inflammatory, acts on dermal cells |
| IL‐22 | Interleukin‐22 | Inflammatory, secreted by Th1 cells, acts on dermal cells |
| IL‐24 | Interleukin‐24 | Inflammatory, acts on dermal cells |
| IL‐26 | Interleukin‐26 | Antimicrobial, TH17 cytokine |
JAK/STAT signaling: from the cell‐surface to the nucleus
The molecular details of the JAK/STAT pathway were largely uncovered in a series of ground breaking studies from the laboratories of James Darnell, George Stark and Ian Kerr more than two decades ago.1 It is an elegantly simple signaling cascade in which the cytokine requires only three components (receptor, kinase, and transcription factor) to elicit a response (Fig. 2). Each cytokine binds to a specific receptor on the surface of its target cell. These receptors contain intracellular domains which are constitutively associated with members of the JAK (Janus Kinase) family of tyrosine kinases.2, 3, 4, 5, 6 JAKs are inactive prior to cytokine exposure however binding of cytokine to its receptor induces their auto‐activation by transphosphorylation. 7 Once activated, JAKs phosphorylate the intracellular tails of the receptors on specific tyrosines which in turn act as docking sites for members of the Signal Transducers and Activators of Transcription (STAT) family of transcription factors (Fig. 2).8 Receptor‐localized STATs are then phosphorylated by JAK9, 10 which leads to their disassociation from the receptor and translocation to the nucleus, where they drive the expression of cytokine‐responsive genes,11 often leading to proliferation and/or differentiation. To ensure that signaling is switched off appropriately, a number of proteins act to attenuate cytokine signaling at multiple levels of the pathway. Notably, the suppressors of cytokine signaling (SOCS) family are negative feedback inhibitors of the signaling cascade.12, 13 Although there are exceptions, a general rule of cytokine signaling is that each cytokine binds to a specific receptor, this induces activation of specific JAK(s) and STAT(s) and signaling is switched off by a particular SOCS protein (Fig. 3).
Figure 2.
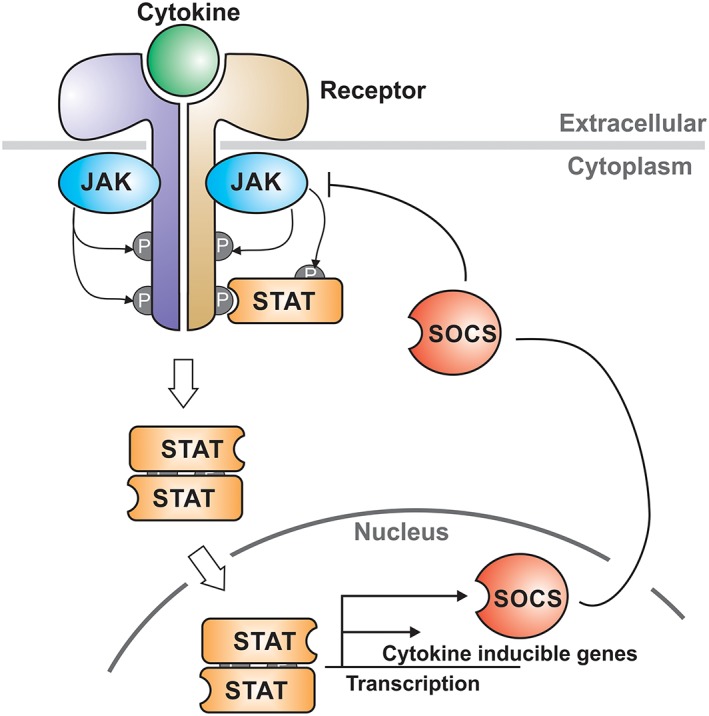
The JAK/STAT pathway. Schematic of the signaling cascade induced by cytokines that signal via the JAK/STAT pathway. Cytokine binds to a specific receptor and allows transactivation of the associated Janus Kinases (JAKs). Activated JAKs then phosphorylate tyrosines on the intracellular domains of the receptor which recruit the Signal Transducers and Activators of Transcription (STAT) transcription factors. STATs are translocated into the nucleus and upregulate the transcription of cytokine‐responsive genes. SOCS proteins are direct targets of STAT and act as negative‐feedback inhibitors to switch off the signaling cascade.
Figure 3.

Class I and Class II cytokines. Families of cytokines and the receptors they bind to are shown above the JAK‐, STAT‐, and SOCS‐family members they signal through.
Evolutionarily, the JAK/STAT pathway first arose in Bilateria; Drosophila for example contains the complete set of pathway components (cytokine, receptor, JAK, STAT). Although the simplicity of the system's architecture has been maintained, there has been a large expansion in the numbers of each of the components throughout the course of evolution. For example, Drosophila has three cytokine‐like molecules (unpaired, unpaired‐2, unpaired‐3), a single receptor (domeless), one JAK (hopscotch), one STAT (marelle/STAT92E), and three SOCS. In contrast, the human genome encodes >50 cytokines, a roughly equivalent number of receptor chains, four JAKs, seven STATs and eight SOCS (Table 2) and these numbers are not dramatically different in any vertebrates so far examined.45
Table 2.
JAK/STAT/SOCS Family Members
| Gene | Knockout mouse phenotype | Cytokines dysregulated | Reference |
|---|---|---|---|
| Jak1 | Early post‐natal lethality, neurological defects; SCID | IL‐2, IL‐6 family cytokines, IFNs | 14 |
| Jak2 | Embryonic lethal, failure of definitive erythropoiesis. | EPO, TPO, GH, PRL, IL‐3, IL‐5, GM‐CSF and IFNγ | 15, 16, 17, 18 |
| Jak3 | SCID | IL‐2 family cytokines | 19, 20, 21 |
| Tyk2 | Resistance to LPS, reduced IL‐12 response | IL‐12 | 22, 23, 24 |
| Stat1 | Impaired antiviral response | Type I/II/III interferons | 25 |
| Stat2 | Impaired antiviral response | Type I/III interferon | 26 |
| Stat3 | Embryonic lethal | IL‐6, IL‐10 family cytokines, G‐CSF | 27 |
| Stat4 | Impaired Th1 development | IL‐12 | 28 |
| Stat5a/b | Impaired mammary gland development, infertility, no NK cells | GH, PRL, IL‐3 family cytokines | 29, 30 |
| Stat6 | Impaired Th2 development | IL‐4/13 | 31 |
| Cish | Increased susceptibility to allergic asthma. Increased tumor immunity | IL‐2, IL‐4, IL‐15 | 32, 33 |
| Socs1 | neonatal lethality | IFNγ/α/β IL‐12/23, IL4/13 IL‐2 family cytokines | 34, 35, 36, 37, 38, 39, 40, 41 |
| Socs2 | gigantism, | GH | 42 |
| Socs3 | embryonic lethality; placental defects. | IL‐6 family cytokines, G‐CSF, Leptin | 43, 44 |
Cytokines and their receptors
Most cytokines are small helical‐bundle proteins usually ca. 150–200 amino acids in length. They are divided into two classes based on motifs found in their receptors (see below). Class I cytokines consist of four α‐helices in a characteristic up‐up‐down‐down configuration. Some of these, such as IL‐5 exist as dimers but the topology is conserved. The unusual up‐up‐down‐down configuration necessitates two long loops to connect the up‐up and down‐down pairs. In class II cytokines, one or both of these loops is replaced by an extra α‐helix resulting in 5–6 helices in total arranged in an anti‐parallel fashion. Again, some (such as IFNγ and IL‐10) function as dimers, where the 2 C‐terminal helices of one molecule are domain‐swapped into a second.
Cytokine receptors consist of multiple (usually two) protein chains. These receptor chains are type I single‐pass transmembrane proteins with conserved intracellular and extracellular features. The extracellular domains contain a region termed the hemopoietin domain or cytokine receptor homology region (CHR),46 formed by a pair of Fibronectin type III (FnIII) domains oriented at nearly right angles to one‐another (Fig. 4). FnIII domains are small ca. 100 residue domains that form a β sandwich comprised of a three‐ and a four‐stranded β‐sheet. The principal binding site for cytokines is at the junction between the two FnIII domains within the CHR, and flexible, variable loops from each domain determine specificity at this binding site.47, 48
Figure 4.
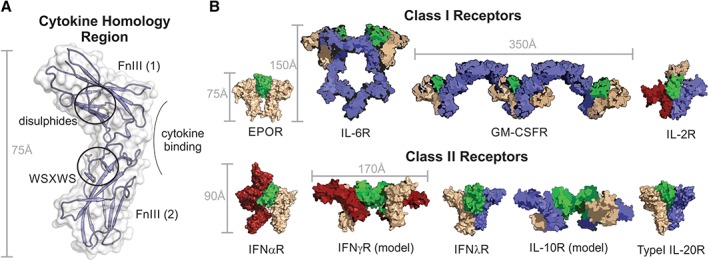
The cytokine homology region (CHR) forms the basis of all cytokine receptors. (A) The CHR from a Class I receptor (Growth Hormone Receptor) is shown with the two FnIII domains, disulfide bonds and WSXWS motif highlighted. All receptors contain a CHR however many receptors, especially those that recognize Class I cytokines, have additional FnIII and Ig domains and this results in a large variety of receptor architectures and stoichiometries. (B) Structures and models of a diverse range of cytokine:receptor complexes.
The CHRs of the receptors for Class I cytokines contain four conserved cysteines arranged in a CX‐(9–10)‐CXWX‐(26–32)‐CX‐(10–15)‐C sequence within the first FnIII domain (forming two intra‐domain disulfide bonds) and a “WSXWS motif” in the second FnIII domain.49 The CHRs of Class II receptors do not have the WSXWS motif and although they too share conserved cysteines, they are arranged differently. Intracellularly, both Class I and Class II receptors share sequences that allow for recruitment of JAKs and STATs.
Class I cytokine receptors
Class I receptors represent the largest group, with 34 Class I receptor chains encoded in the human genome.50 All Class I receptors contain a CHR but many also contain additional extracellular domains such as Ig domains, extra FnIII domains or even a second CHR. Class I receptors bind to a broad array of interleukins, hematopoietins, and growth factors whilst Class II receptors are more restricted, recognizing only interferons and IL‐10 family cytokines (Fig. 3).
As shown in Figure 3, there are three major shared chains utilized by Class I cytokines. These are gp130, beta common and gamma common, utilized by IL‐6, IL‐3, and IL‐2 family cytokines, respectively. In addition to these, there are two other shared chains used by cytokines in the IL4/13 and IL‐12/23 subgroups. Finally, there are the homodimeric receptors, consisting of two identical chains such as those used by EPO, TPO, GH, PRL, Leptin, and G‐CSF. Within each of these classes, the cytokine IL‐12Rβ2 receptor stoichiometry and organization can differ. A common theme within the non‐homodimeric receptors is that there will be a cytokine‐specific chain (nominally the “alpha” chain) that recognizes cytokine with high affinity, and the resulting dimer will then recruit a “shared” chain in order to initiate signaling. The alpha‐chain may or may not contain the intracellular motifs required to recruit a JAK kinase. Although there are differences in the number of individual chains that comprise a Class I receptor, receptors usually contain precisely two signaling chains (those whose cytoplasmic domain binds to a JAK family member to initiate signaling).
Homodimeric receptors
The homodimeric cytokine receptors are a family of structurally diverse receptors that are categorized by their use of two identical receptor chains. Some, like EPOR,51 GHR47 and PRLR52 are the most simple of all receptors in terms of architecture, the ectodomain of each receptor chain consisting only of a single CHR unit. Studies on EPOR and GHR in particular have formed the basis of the general paradigm for cytokine signaling. These three receptors, alongside the larger TPOR (which contains two CHRs per ectodomain chain), bind a single molecule of cytokine to form a signaling‐competent ternary complex.
Although these complexes are by nature asymmetric, it is the hinge regions between the two FnIII domains of the CHRs on both chains that participate in cytokine binding. One chain binds to the cytokine with high‐affinity, and the other chain binds to a lower‐affinity site on the cytokine. This phenomenon is observed for all classes of cytokine receptor.
The Leptin and G‐CSF receptors are also homodimeric but structurally have more in common with the IL‐6 receptor family (see below and Fig. 3). They are referred to as “tall receptors” as they contain “legs,” composed of two or three FnIII domains that link their cytokine‐binding region to the membrane. This places the cytokine binding region of these receptors as far as 120 Å from the cell‐surface. The leptin receptor is the largest of all cytokine receptors, consisting of (from N‐ to C‐terminus): Ig‐CHR‐Ig‐CHR‐FnIII‐FnIII. Both the leptin and G‐CSF receptors bind two molecules of cytokine, forming signaling competent tetramers with a similar architecture to that seen in many IL‐6 family receptors53 (Figs. 3 and 4).
IL‐6 family cytokine receptors
The IL‐6 family of cytokines includes IL‐6, IL‐11, IL‐27, LIF, OSM, CNTF, CT‐1, and CLC. The majority of these cytokines act via receptors that include the common gp130 chain. Like the G‐CSF and Leptin receptors, gp130 is also a tall receptor chain. Its extracellular domain consists of an Ig domain, a CHR, followed by three FnIII domains that form the legs. Within the IL‐6 family are two other tall receptor chains which are shared between cytokines on a more limited basis, these are LIFRβ and OSMR.
The signaling competent complex between IL‐6 or IL‐11 and their receptors are hexamers. These consist of two molecules each of cytokine, gp130, and a cytokine‐specific alpha chain (IL‐6Rα or IL‐11Rα).54, 55, 56 Two copies of gp130 are required because the intracellular domain of the cytokine‐specific alpha chains of these receptors do not bind JAK. Signal transduction is, therefore, achieved by transactivation of two JAK molecules bound to the two gp130 receptor chains.
Whilst the IL‐6 and IL‐11 signaling complexes are similar in structure and composition, other gp130 cytokines signal via different topological assemblies. For example, the complexes between CNTF, CLC, or CT‐1 and their receptors are tetramers. These cytokine/receptor complexes are not built around two gp130 chains but instead are built around a heterodimer between gp130 and another JAK‐binding tall receptor called LIFRβ.57, 58 As is the case for IL‐6, these cytokines first bind to cytokine‐specific non‐signaling alpha receptor chains and the resulting complexes then recruit gp130 or LIFRβ. Finally, LIF, OSM, and IL‐31 signal via trimeric complexes with the receptor being a heterodimer of two tall chains without the requirement for an alpha chain.
IL‐12/23 receptors
IL‐12 and IL‐23 also signal via tall receptors. Although there is no structure of the full receptors for these cytokines they are likely to be of similar architecture to the LIF receptor59, 60 (Fig. 4). IL‐12 and IL‐23 are unusual cytokines as they are both composed of two disulfide‐linked subunits that arise from different genes.61 The first of these subunits (IL‐12p35 and IL‐23p19) are classic four‐helix bundle cytokines. The second subunit (IL‐12p40, which is shared by both IL‐12 and IL‐23) is more like a receptor in structure, consisting of a CHR, replete with WSXWS motif, and an N‐terminal Ig domain, but lacking a transmembrane domain. Indeed, the p40 receptor‐like subunit associates with the 4‐helix bundle subunit in a classic receptor‐like manner, using the hinge region between the two FnIII domains of the CHR.62, 63 The receptors for IL‐12 and IL‐23 are also heterodimers which share a common chain, the interleukin‐12 receptor subunit β1 (IL‐12Rβ1). The other monomer in each receptor is a cytokine‐specific alpha chain (IL‐12Rβ2 or IL‐23R). As one would expect, the common receptor subunit recognizes the p40 subunit whilst the specific alpha chains recognize the classic 4‐helix bundle moieties within each cytokine heterodimer.64
IL‐3 family cytokines
The IL‐3 family (IL‐3,‐5, GMCSF) all signal via receptors that contain the common beta chain (beta‐common, βc).65 The extracellular region of βc is comprised of two CHRs (i.e. four FnIII domains). βc has no measurable affinity for any of the three cytokines alone, instead each of the three cytokines within this family has a specific alpha‐chain that facilitates a high affinity interaction with βc.66 The primary site of interaction with the alpha‐chain is the usual hinge region between the two FnIII domains of the CHR however these alpha‐chains also contain a N‐terminal Ig domain which wedges over the top of the cytokine, providing extra affinity. It is this binary complex which associates with βc with high affinity.
βc forms a closely associated dimer with the individual FnIII domains of each monomer drawn out like beads on a string, allowing the dimer to associate in an almost domain‐swapped fashion.67 The two cytokine binding sites on the βc dimer are thus formed at the spatial junction between an FnIII domain from both monomers. The structure of the signaling competent GM‐CSF/receptor complex shows the most surprising stoichiometry of all, forming dodecamers consisting of four cytokines, four βcs, and four alpha receptors.67 This dodecameric structure is the minimum structure that can allow juxtaposition of the intracellular JAKs, leading to transactivation and downstream signaling.
IL‐2 family cytokines
Finally, the IL‐2 family cytokines (IL‐2, ‐4, ‐7, ‐9, ‐15, ‐21) all signal via receptors that contain the common gamma chain (gamma common, γc), a JAK3‐associated receptor subunit. The extracellular region of γc is much smaller than that of gp130 or βc, consisting of just a single CHR. Most members of this family of cytokines signal via heterodimeric receptors (γc plus a specific alpha receptor), thus forming ternary signaling complexes upon the addition of cytokine. IL‐4, IL‐7, IL‐9, and IL‐21 all signal in this manner. The general paradigm for signaling by these cytokines is that γc (like gp130 or βc) has little affinity for the cytokines alone but binds strongly to a complex between the cytokine and the alpha receptor chain.
Despite being the defining family member of this group of cytokines, IL‐2 itself, along with IL‐15, are unusual in that they also require a third receptor subunit called IL2Rβ. IL‐2Rβ is a JAK1‐associated receptor subunit and is essential for signaling. The alpha receptors for IL‐2 and IL‐15 are non‐signaling chains distinct from other cytokine receptors as they consist of sushi domains (another beta‐sandwich fold) rather than a CHR. The full IL‐2 receptor is found on activated T‐ and NK‐cells, and binds IL‐2 with extremely high affinity (10pM). IL‐15 signals similarly to IL‐2 but IL‐15Rα usually presents IL‐15 from a different cell (i.e. in trans) thus IL‐15 requires cell–cell contact in order for signaling to occur.68
Just as is seen for the gp130 family, the common gamma receptor family also contains chains that are shared on a more limited basis. For example, IL‐7Rα can either heterodimerize with γc to form the IL‐7 receptor or with TSLPR to form the receptor for TSLP. Likewise, IL‐4Rα can heterodimerize with γc to form the Type I IL4R (found on hematopoietic cells); however, it can also heterodimerize with IL‐13Rα1 to form the Type II IL4R (found on non‐hematopoietic cells). Both IL‐4 and IL‐13 can signal via this complex and this is one of only a few examples of different Class I cytokines signaling via the same receptor, a phenomenon that is common for Class II cytokines as we shall now discuss.
Class II cytokine receptors
The Type II family cytokines encompass the interferons (IFN α,β,γ,λ,ε,κ,ω) and IL‐10 family cytokines.69 Signaling via Class II cytokine receptors (unlike Class I) adheres to a more common set of rules regarding stoichiometry and receptor assembly. Each Class II receptor is a heterodimer and each of these receptors associate with one molecule of cytokine to initiate signaling. The only exceptions to this rule are IL‐10 (and possibly IL‐26) and IFNγ which are dimeric cytokines and the stoichiometry of the entire signaling complex is, therefore, doubled (Figs. 3 and 4). All Type II cytokine receptor chains bind to JAK, unlike many Type I receptor alpha chains. Finally, the ectodomain architecture of all Class II receptors consist of just a single CHR (with the sole exception of IFNαR1 which has two) and are not decorated by extra Ig or FnIII domains.
Similar to Class I, the Class II receptor family consists of both shared chains and cytokine‐specific chains. However, a characteristic of class II cytokine signaling is the plasticity seen within the system, in many cases a single receptor can bind multiple cytokines and a single cytokine can in some cases bind multiple receptors.
The Type I interferon receptor (IFNα/β receptor)
The Type I interferon receptor is a heterodimer consisting of IFNαR1 and IFNαR2. IFNαR1 has a large extracellular domain that consists of two CHRs while IFNαR2, similar to all other Class II cytokine receptors has only a single CHR. IFNαR2 is the high affinity chain, interacting with ligand with sub‐nanomolar affinity whilst IFNαR1 binds with an affinity approximately two orders of magnitude lower.70
There is a vast array of type I IFNs, including IFN‐α (which itself has at least 13 different subtypes), IFN‐β, IFN‐ε, IFN‐κ, and IFN‐ω. Other interferons are found in different mammalian species. Remarkably, these all bind to the same receptor, even though the downstream consequences elicited by each Type I interferon differs. Structural studies have shown that each ligand binds the receptor at the same position; however, the stability of the resultant complexes differ.71 Thus, the half‐life of the cytokine/receptor complex appears to determine the downstream signaling outcomes.
The Type II interferon receptor (IFNγ receptor)
The interferon gamma receptor does not consist of any shared receptor chains and interacts only with a single ligand, interferon gamma. Likewise, IFNγ does not bind any other receptors. IFNγ acts as a homodimer and binds two copies of IFNγR1 with sub‐nanomolar affinity. The active signaling complex is then formed by recruiting two copies of IFNγR2,72 making the signaling complex hexameric.
The Type III interferon receptor (IFNλ receptor)
The Type III cytokines IFN‐λ1, ‐λ2, and ‐λ3 (also known as IL‐29, IL‐28A, and IL‐28B, respectively) signal via a single receptor. This Type III receptor consists of two chains, IL‐28R and IL‐10Rβ, the latter of which is shared with receptors for the IL‐10 family. The structure of IFNλ bound to its receptor is reminiscent of the GH:GHR complex structure with the ligand occupying a specially similar position albeit with a very different angle of occupation.73 The IL‐28R chain binds cytokine with high affinity and this binary complex then recruits IL10Rβ.
IL‐10 family receptors
IL‐10 family cytokines (IL‐10, IL‐19, IL‐20, IL‐22, IL‐24, and IL‐26) can be subdivided into two classes. Those that use the shared IL10Rβ chain (IL‐10, 22, 26) and those that use the shared IL20Rβ chain (IL‐19, 20, 24).
Like IFNγ, IL‐10 (and probably IL‐26) are homodimeric cytokines and their receptors likewise contain two copies of each of the two individual chains. The shared chain for these two receptors is IL10Rβ and the cytokine‐specific chains are IL‐10Rα and IL‐20Rα, respectively. In each case, the cytokine‐specific chains bind with high affinity to ligand (sub‐nanomolar), while IL‐10Rβ is the lower affinity (high micromolar – mM) receptor.74, 75 IL10Rβ also forms half of the receptor for IL‐22, a monomeric cytokine.75 The cytokine specific chain can be IL22Rα1, or alternatively it can recruit a soluble receptor (IL22BP) that can mediate its biological effects.
The remaining IL‐10 family cytokines (IL‐19, 20, 24) bind to two distinct receptors. The so‐called Type II IL‐20 receptor consists of a shared IL20Rβ and a cytokine‐specific subunit IL22R. The type I receptor contains the same shared IL20Rβ chain along with the IL20Rα subunit. IL‐20 and IL‐24 can signal via both receptors whereas IL‐19 binds to the type I receptor only. In the Type I receptor, IL20Rβ is the high affinity subunit.76 Overall the structure of the cytokine:receptor complex is similar to that of IFNλ. Modeling of the type II receptor suggests a similar overall architecture.77
Cytokine receptor cytoplasmic domains
Both Class I and II cytokine receptors are complexes of single pass transmembrane‐domain containing proteins. Many cytokine‐specific alpha receptor chains contain a short cytoplasmic region with no known function, but each functional receptor complex always consists of at least two (most often precisely two) individual receptor chains with long intracellular regions (several hundred amino acids in length) that are the scaffolds upon which signaling is initiated. These unstructured59, 60 cytoplasmic domains exist to provide sequence‐specific docking sites for JAKs and STATs. The JAK‐binding regions are known historically as the Box 1 and Box 2 motifs and are membrane proximal whilst the STAT binding motifs are located towards the C‐terminus, distal to the membrane. In some cases, in between these two motifs are additional binding sites for negative‐regulators such as the SOCS proteins.
The JAK binding motif: Box 1/2
Mutagenesis studies first identified two regions on the cytoplasmic tail of receptors, termed Box 1 and Box 2, critical for the association of JAKs with receptor.78 Box 1 is proline rich and is located approximately 10 residues from the C‐terminus of the transmembrane region of the receptor, whilst Box 2 is about 10–50 residues further downstream and is rich in hydrophobic residues. Apart from these features, these regions share low sequence homology between different receptors. Moving the Box 1/2 motif further from the membrane abolishes the ability of JAK to associate79 suggesting that membrane proximity is important for cytokine inducible activation. Specific receptors bind to specific JAKs, although some receptors (most notably the G‐CSF and IL‐6 family of receptors) can bind multiple JAKs.80 It is sequence differences within the Box 1 and Box 2 motifs of different receptors that determine which JAK (JAK1, JAK2, TYK2, or JAK3) is bound. For example, structural studies identified a PxxLxF sequence in JAK1‐binding Class II receptors as the key motif required for JAK1 interaction.81 No obvious motifs have yet been defined for other receptors. Very recently, it has been shown for two homodimeric receptors (EPOR and LeptinR) that the small region between the membrane and the Box 1 motif coordinates JAK homodimerization and is necessary for efficient signaling.82 It may be assumed (but has not yet been shown) that the same region of other receptors may mediate JAK dimerization as well.
STAT‐binding motifs
Once JAKs are activated (see below) they phosphorylate distal tyrosines on the receptor intracellular domains and these sites act as docking sites for the signal transducer and activator of transcription (STAT) transcription factors. Thus, all receptors contain conserved tyrosines that fulfill this function. Just as different receptors bind different JAKs, so they also bind different STATs.83 The ability of a certain cytokine to induce activation of a particular STAT is driven purely by the STAT‐binding sites contained within its receptor. STAT‐binding sites from one receptor can be replaced with binding sites for different STATs from other receptors and thereby activate non‐physiological STATs.84, 85 In many cases, a single pTyr can recruit multiple members of the STAT family, albeit with differing affinity.86 It is likely that the affinity for one STAT over another is purely a function of the sequence immediately surrounding the phosphotyrosine, for example pYxxP,87 pYxxQ,85 pYxxL,88 and pYxxF89 sequences are associated with recruitment of STAT1, STAT3, STAT5, and STAT6, respectively.
Some receptors contain multiple STAT binding sites, for example the IL‐6 receptor signaling chain (gp130) contains four STAT3 binding motifs90 and EPOR contains four STAT5 binding motifs on each chain.88 In other cases, such as in IFNγR, only a single STAT binding site is found.87
Other stimulatory sites
In addition to stimulating signaling by STATs, many cytokines also induce additional signaling pathways via the same receptors,91, 92 such as the MAPK and PI(3)K pathways. For example, IL‐6 family cytokines stimulate both these pathways. Although the mechanism of PI(3)K stimulation is unclear, the MAPK pathway is activated via the phosphatase SHP2:93 SHP2 binds to phosphotyrosine 759 on the gp130 subunit of the IL‐6 receptor; cytokine exposure activates SHP2; and this leads to Ras/Raf signaling which stimulates the MAPK cascade and ultimately transcriptional activators such as Elk.
Negative‐regulatory sites
In addition to stimulatory sites on the intracellular domains of cytokine receptors, there are often sites for regulatory proteins that inhibit signaling. In general, these inhibitory proteins interact with phosphotyrosine motifs on the receptors via SH2 (Src‐homology 2) domains (as do the STATs) and hence they are only recruited once the receptors are phosphorylated. The SOCS proteins are a family of negative‐regulatory proteins that all contain SH2 domains and many bind to specific receptor sites to inhibit signaling.94 Particularly well‐characterized sites are found on the IL‐6 and G‐CSF receptors (for SOCS3) and the GHR (for SOCS2). In all cases these SOCS binding sites are located C‐terminal to the JAK‐binding region of the receptor.
Janus Kinases (JAKs)
There are four members of the JAK family found in all vertebrates: JAK1, JAK2, JAK3, and TYK23, 4, 5, 95 (see Table 2). Each JAK is ca. 1000 residues in length and consists of four distinct domains: An N‐terminal FERM (band 4.1, Ezrin, Radixin, Moesin) domain followed by an SH2 domain and two kinase domains (Fig. 5). The first of these kinase domains is catalytically‐inactive and is therefore a pseudokinase domain (also termed the JAK Homology 2 or JH2 domain). The C‐terminal kinase domain is the catalytic domain in each JAK, historically termed the JH1 domain.
Figure 5.
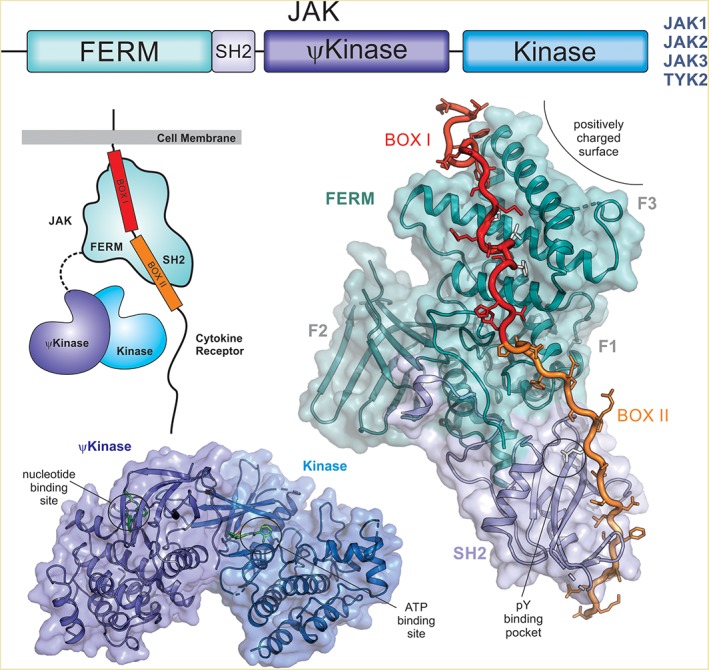
Janus kinases (JAKs). There are four members of the JAK family (JAK1, JAK2, JAK3, and TYK2) and all share similar domain architecture (top). The FERM and SH2 domains tether JAK to the receptor, binding Box I and Box II respectively (structure shown on the right, PDB ID: http://firstglance.jmol.org/fg.htm?mol=5L04)). The pseudokinase (ψkinase) regulates the activity of the catalytically active kinase domain (bottom, PDB ID: http://firstglance.jmol.org/fg.htm?mol=4OLI) via a mechanism that is unclear. There is no structure of a full‐length JAK protein and hence the relative orientation of the N‐ and C‐terminal halves of the protein is unknown (indicated schematically on the left).
FERM/SH2 domains
The FERM and SH2 domains together are responsible for binding to the receptor. This provides the specificity required to target a particular JAK to a particular receptor chain. With a few exceptions, a receptor chain only binds to a single, specific member of the JAK family. The FERM and SH2 domains are closely associated and form a single structural unit.81, 96, 97 Together they provide the binding site for the Box 1 and Box 2 motifs found in all cytokine receptors. The FERM domain is a three‐lobed structure consisting of an F1 lobe (ubiquitin‐like), an F2 lobe (acyl‐CoA‐binding protein‐like), and an F3 lobe (pleckstrin homology domain). The F2 lobe is primarily responsible for binding the membrane‐proximal Box 1 motif on the receptor and in addition contains a large surface with significant positive charge that probably interacts with the cell membrane.81, 96, 97 The SH2 domain interacts with the Box 2 motif. Interestingly, it coordinates a glutamate in the same way that other SH2 domains coordinate phosphotyrosine. Together, the Box 1 and Box 2 motifs form a long (85 Å), largely extended epitope that buries over 3000 Å2 of JAK.97 The relevant contributions of Boxes I and II toward overall affinity differ between receptors. For example, in the IFNλ receptor, Box 1 provides most of the affinity whilst the addition of Box 2 adds a further 10‐fold increase.81 In contrast, TYK2 binding to the IFNα receptor appears to be dominated by Box 2.96
Pseudokinase domain
The pseudokinase domain, critical for modulating the activity of the C‐terminal tyrosine kinase domain,98, 99, 100 is the most enigmatic of the JAK domains. Although it adopts a typical kinase fold,100, 101, 102, 103, 104, 105, 106 it is catalytically defective. The pseudokinase domain of JAK2 shows some residual activity that may be responsible for autophosphorylation in cis on two auto‐inhibitory phosphorylation sites, Ser523 and Tyr570100; however, it is likely to be completely inactive in other members of the JAK family,103 despite maintaining the ability to bind ATP.107 An emerging idea is that ATP binding by pseudokinase domains functions as a “molecular switch”107; however, this remains to be established for the JAK family.
The ability of the pseudokinase domain to regulate the activity of the kinase domain has been recognized for some time, since the observation that a mutation in the pseudokinase domain of the Drosophila JAK homolog, hop, resulted in hyperactive kinase activity and a leukemia‐like disease.108 Importantly, deletion of the pseudokinase domain increases the basal level of kinase activity but prevents a further increase in activity in response to cytokine.98, 99 The importance of the pseudokinase domain was further highlighted in 2006 when it was discovered that a V617F point mutation in human JAK2 was causative of a range of myeloproliferative neoplasms.109, 110, 111 This group of diseases, which includes Polycythemia Vera, Essential Thrombocythemia and the more severe Primary Myelofibrosis display aberrantly high levels of myeloid cells such as erythrocytes and platelets. The V617F mutation leads to an increased basal activity of JAK2 and cytokine‐independent signaling through the EPO and TPO receptors. The analogous mutation in other JAK family members has also been shown to cause aberrant signaling.103 Further mutations in the linker between the SH2 and pseudokinase domains, the so‐called exon 12 mutations, have since been discovered to lead to the same diseases.112, 113 Given that the pseudokinase domain plays a role in both switching on the kinase domain and controlling its maximal activity and that mutations within this domain cause disease there have been many efforts to understand this at a molecular level. To date, there is only a single experimentally‐determined structure of the tandem pseudokinase‐kinase domains (from Tyk2).102 This structure showed that the two domains adopt a back‐to‐back orientation, and activating mutations cluster together at the interface between these domains indicating that a physical interaction between the pseudokinase and kinase domains is required for correct regulation of JAK activation. Although equivalent structures of other JAKs have not been forthcoming, sophisticated modeling of the analogous construct from JAK2 gave a near identical prediction of the overall structure114 even prior to the TYK2 structure being solved.
Kinase domain
The kinase domain of JAK is the domain required for phosphorylation of receptor and subsequently the STAT transcription factors. All JAKs are tyrosine kinases and their kinase domains display a typical fold.115, 116, 117, 118, 119 Tyrosine kinases catalyze the transfer of a phosphate from ATP to a tyrosine‐containing protein. The general organization of a kinase includes two lobes: an N‐terminal lobe consisting mostly of β‐strands and a larger, mostly α‐helical C‐terminal lobe. The ATP‐binding site (and thus the active site) exists between these lobes. The conserved residues for ATP‐binding lie mainly on the N‐terminal lobe whilst the motifs for tyrosine binding, phosphotransfer and Mg2+‐binding are found mostly on the C‐terminal lobe. Magnesium is an essential factor for kinases, aiding the co‐ordination of ATP. A number of important motifs are characteristic of all kinases and these include (i) the VAIK motif of the β3 strand in the N‐lobe, in which the lysine anchors and orients the α‐ and β‐phosphates of ATP; (ii) an aspartic acid in a HRD motif of the catalytic loop (β6–β7) which provides the catalytic base; (iii) an aspartic acid in the DFG motif within the activation loop (β8–β9) that binds the Mg2+ coordinating the β‐ and γ‐phosphates of ATP120; and (iv) a glycine‐rich loop (G‐loop; GXGXXG) between the β1 and β2 strands of the N‐lobe. The latter is a key factor in binding ATP through hydrogen‐bond formation between the backbone of the loop and the ATP γ‐phosphate.121 In addition to these, two hydrophobic regulatory “spines” determine whether a kinase is capable of enzymatic activity, the Regulatory‐spine and the Catalytic‐spine.122, 123
Another important motif is the activation loop. This loop contains two consecutive tyrosines and the phosphorylation‐state of the first of these determines whether the enzyme is active or inactive. JAKs phosphorylate one‐another at this position in trans upon cytokine stimulation.7 Although many structures have been solved of JAK kinases domains with their activation loop in the phosphorylated (active) conformation,115, 116, 117, 118, 119 the inactive conformation has not been observed structurally but can be inferred from studies on other tyrosine kinases, in particular the insulin and IGF receptor kinases, in work led by Stevan Hubbard.124, 125, 126, 127 In the inactive conformation, the activation loop forces the N‐ and C‐lobes apart and blocks the ATP binding site. In addition, the substrate‐binding site (which is partly formed by residues within the loop) is not present and the orientation of the DFG motif does not allow magnesium binding.124 Phosphorylation results in reorientation of the activation loop such that it swings out of the ATP‐binding site and lies flat against the solvent exposed surface of the C‐lobe. This allows ATP and substrate to bind and catalysis to occur.127 Structurally trapping a tyrosine kinase in the process of auto‐activation (in trans) has only been successfully performed for the IGF1 receptor and the activation loop in this conformation is highly extended, allowing the first tyrosine to access the active site of a second kinase molecule and become phosphorylated.126 The second tyrosine within the activation loop has been found fully or partially phosphorylated in a number of JAK structures116, 117, 118, 119; however, its importance in terms of catalytic activity is unclear. In our studies on JAK2, we observe no difference in the activity of the kinase domain when this residue is mutated to phenylalanine (unpublished data).
The final motif of interest in the JAK kinase domains is the JAK insertion loop that is peculiar to this family.117 This loop links the αH and αI helices in the C‐lobe of the kinases and in JAK1, JAK2, and TYK2 is capped by a “GQM” motif that allows them to bind to SOCS1 and SOCS3, two regulatory proteins that can inhibit the catalytic activity of these kinases. JAK3 does not contain a GQM motif in its JAK insertion loop and is, therefore, immune to SOCS‐mediated inhibition.128
Signal Transducers and Activators of Transcription (STAT) proteins
The STATs are a family of proteins named for their dual roles of (1) transducing signals from cytokines and (2) promoting transcription of specific genes. The STATs predominantly reside in the cytoplasm as inactive dimers but are rapidly activated upon initiation of cytokine signaling and translocate into the nucleus.129, 130, 131 There are seven mammalian STATs (STAT1‐4, STAT5a, STAT5b, and STAT6)132, 133, 134 and each contains several conserved features; an N‐terminal region followed by a coiled‐coil domain, a DNA binding domain, a linker region, an SH2 domain, and a C‐terminal transactivation domain (Fig. 6). Located between the SH2 domain and the transactivation domain is a single conserved tyrosine residue which is the site at which the STAT proteins are phosphorylated by the JAKs and is essential for their activation.137 STATs exist as dimers both in their active and inactive forms, but the structural arrangement of the two dimeric species is very different. Most STATs function primarily as homodimers; however, heterodimeric complexes do occur and are particularly important for STAT2, which only acts as a heterodimer. STAT2 acts downstream of Type I and III interferons but it does so as part of a complex called ISGF3 (IFN‐stimulated gene Factor 3). ISGF3 is a three‐protein complex that contains STAT2, STAT1 and a third transcription factor called IRF9 which binds the STAT2 coiled‐coil domain.138 Figure 3 highlights the dominant STAT family members activated in response to individual cytokines; however, it should be noted that there is often low‐level activation of additional STATs.
Figure 6.
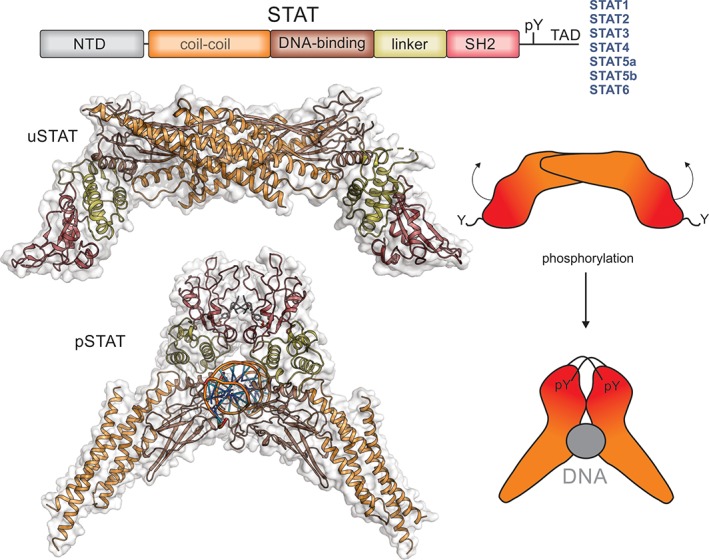
STATs. The Signal Tranducers and Activators of Transcription (STATs) are a family of latent transcription factors that are activated by phosphorylation following cytokine exposure. The same domain architecture is shared by all STAT proteins and is shown schematically above. Unphosphorylated STAT (uSTAT) exists as an antiparallel dimer in the cytoplasm (upper). The SH2 domain (red) of uSTAT binds to phosphotyrosines in cytokine receptors which allows JAK to phosphorylate a specific tyrosine located between the SH2 and transactivation domain (TAD). This phosphotyrosine is then targeted by the SH2 domain of the other monomer inducing a large rotation between the two subunits of the dimer and allowing phosphorylated STAT (pSTAT) to occupy its DNA‐binding competent dimeric structure (lower). The structures shown here are of STAT1 (PDB ID: http://firstglance.jmol.org/fg.htm?mol=1YVL,135 http://firstglance.jmol.org/fg.htm?mol=1BF5 136) with the colors matching the schematic representation above. The N‐terminal domain of STAT does not appear to form a stable interaction with the rest of the molecule and is not shown here. The transactivation domain (TAD) is unstructured but allows binding of accessory factors.
N‐terminal region
The N‐terminal domain of the STATs is largely conserved between all seven proteins and forms a bundle of alpha‐helices oriented roughly at right angles to one‐another.139, 140 Generation of chimeric STAT molecules where the N‐terminal domain of STAT1 was replaced with that of STAT2 or STAT5 revealed a role for the N‐terminal region in nuclear translocation and deactivation.141 The N‐terminal domain of the STAT proteins also plays a role in cooperative DNA binding142 between STAT dimers in regions of DNA where there are clusters of STAT binding sites, perhaps accounting for some of the specificity of cellular response to different cytokines.139
Coiled‐coil domain
The coiled‐coil domain of the STAT proteins is a region of approximately 180 amino acids immediately following the N‐terminal domain. It comprises four antiparallel alpha‐helices which form a bundle in a down‐up‐down‐up topology that is a major site of dimerization in the inactive form but then projects outwards from the core of the protein after activation. Allowing DNA binding and providing a surface for other proteins such as transcription factors to bind.136
DNA binding domain
The DNA binding domain allows STATs to function as transcription factors and targets specific DNA sequences.143 All STATs recognize palindromic DNA sequences with a TTCN2‐4GAA motif.144 While all STATs bind this motif, their sequence preferences differ. STAT1 and STAT5 show preference for sites with a three‐base pair spacer between the C and G (N3), with STAT1 also displaying preference for binding sites with a C at the ‐7 position, relative to the palindrome centre.145 STAT6, unlike the other STAT proteins, binds to sites with a four‐base pair spacer (N4)145, 146; however, STAT5147, 148 has also been shown to bind weakly to N4 sequences.145 STAT4 prefers the palindromic sequence (T/A)TTCC(C/G)GGAA(T/A) where the first and last T/A sites outside of the usual motif are also necessary for binding.149
Linker and SH2 domain
Immediately downstream of the DNA‐binding domain are the linker and SH2 domains. SH2 domains are modules that bind phosphotyrosine (pTyr) when it is embedded in a particular amino acid sequence motif. Each SH2 domain will have its own preferred sequence surrounding the pTyr, usually dictated by residues in the +1 and + 3 positions (relative to the pTyr). Once JAK is activated it immediately phosphorylates tyrosine residues in the receptor to which it is bound and the presence of an SH2 domain allows STATs to bind to those phosphorylated cytokine receptors. Thus which STATs will be activated by a particular cytokine depends on which receptor(s) their SH2 domain will bind.150, 151 Different STATs can bind to the same sequence with different affinities which accounts for some of the pleiotropy of the STAT family and redundancy of biological outcomes.152
STAT activation and transcription
Prior to activation, the STATs are found as inactive dimers in the cytoplasm. The DNA‐binding and coiled‐coil domains of two STAT monomers interact to form a reciprocal dimer (Fig. 6).135, 153, 154 STATs are activated by JAK‐catalyzed phosphorylation of a specific tyrosine between the SH2 and transactivation domain. Instead of binding the phosphorylated receptor, the SH2 domain of each STAT monomer now binds the newly created pTyr in the other monomer resulting in dissociation from the receptor and re‐orientation to form the active conformation with an exposed DNA‐binding domain. These dimers then translocate into the nucleus and induce transcription of genes whose promoters contain the appropriate STAT binding sites. The STATs form a scissor‐like structure around the DNA136 (stabilized by the reciprocal interactions between the SH2 domain of one monomer and the pTyr of the other) and transcription is facilitated by the recruitment of transcriptional co‐activators such as CBP/P300. Determining the complete set of genes upregulated by each of the STAT proteins has been difficult due to activation of other transcription factors by pathway crosstalk; however, several hundred to several thousand genes appear to be activated by each activated STAT155, 156, 157, 158 as well as a number of genes being downregulated.159
The dephosphorylation of STAT by phosphatases in the nucleus allows them to shuttle back into the cytoplasm for further rounds of activation.160, 161, 162 While STATs are known to be active transcription factors in their phosphorylated state, there are some cases where unphosphorylated STATs also appear to play a role in gene transcription.163, 164 It has also been suggested that unphosphorylated STATs are important for maintaining heterochromatin stability by associating with HP1, a heterochromatin protein necessary for heterochromatin formation.165, 166 Unphosphorylated STAT5 has an important role as a transcriptional repressor in megakaryocytes where it blocks differentiation.167
Negative regulation of JAK–STAT signaling
SOCS proteins
Signaling via the JAK–STAT signaling pathway is a dynamic process that involves the rapid transmission of signal from the cell membrane to the nucleus followed by a highly organized response and subsequent controlled downregulation and attenuation of the initial signal.137, 168 SOCS proteins are the primary drivers of signal attenuation, they are induced by cytokine exposure (via STAT) and then act as negative‐feedback inhibitors to switch off the signaling cacade.13, 94, 169, 170 There are eight SOCS proteins encoded in the human genome, SOCS1‐7 and CIS.13, 169, 170, 171 All eight contain N‐terminal domains of various length and often unknown function but are defined by the presence of a central SH2 domain and a short, C‐terminal domain called the SOCS box.171 CIS and SOCS1/2/3 are the members of the family associated with inhibiting signaling by JAK/STAT‐inducing cytokines while other SOCS proteins appear to regulate signaling by factors such as EGF and insulin.
Although induced by many cytokines,170 and potent inhibitors of many cytokine signaling pathways when artificially overexpressed, knockout studies have shown that each SOCS protein is specific for only a subset of cytokines.172, 173 For example SOCS3 is induced by IL‐2, IL‐3, IL‐4, IL‐6, IL‐7, IL‐9, IL‐10, IL‐11, IL‐12, IL‐13, IL‐21, IL‐22, G‐CSF, GM‐CSF, LIF, PRL, IFNα, IFNγ, GH, EPO, TPO, OSM, CT1, CNTF, and leptin170, 174, 175, 176, 177, 178, 179, 180, 181, 182, 183, 184, 185; however knockout studies have shown that only IL‐6 family cytokines,172 G‐CSF,173 and Leptin186 are aberrantly regulated in its absence. This specificity is provided by the SH2 domains of SOCS proteins which in general bind to phosphotyrosine‐containing motifs in specific cytokine receptors and therefore only inhibit signaling by cytokines that act via those receptors. For example the SH2 domains of CIS, SOCS2, and SOCS3 bind to motifs in the IL‐2,32 GH,187 and IL‐692, 188, 189 receptors, respectively.
Once bound to cytokine receptors, SOCS proteins induce their ubiquitination and degradation through the activity of their SOCS box domain. The SOCS box is structurally similar to a domain first described in VHL190 and has two distinct binding properties. The first is a short motif that allows it to bind to elonginBC,191, 192 an adapter complex that is also recruited by VHL. Once the ternary SOCS/elonginBC complex has formed the second motif can then recruit Cullin5, a 100 kDa E3 ligase scaffold.193 This differs from VHL which uses its SOCS box‐like domain to recruit Cullin2.194 Cullin5 is constitutively associated with a RING domain protein, Rbx2,194, 195 and this provides the E3 ubiquitin ligase activity by recruiting E2 ubiquitin conjugating enzymes to catalyze the ubiquitination of protein substrates. Therefore, SOCS proteins act as the substrate recruitment modules for Cullin5‐based E3 ligases and proteins bound by the SOCS SH2 domains (usually receptors) are ubiquitinated and subsequently degraded by the proteasome.
SOCS1 and SOCS3
SOCS1 and SOCS3 are unique amongst the SOCS family in containing a short motif called the Kinase Inhibitory Region (KIR).196, 197 This allows these two SOCS proteins to bind directly to the JAK kinase domain and inhibit its catalytic activity. The KIR is unstructured in the absence of JAK198, 199 but upon binding adopts an extended conformation that sits in the substrate binding groove of the kinase.200 This occludes the substrate‐binding site and prevents JAK from phosphorylating downstream substrates. SOCS1 and SOCS3 inhibit JAK1, JAK2, and TYK2 but not JAK3 due to the absence of a “GQM” sequence in the latter kinase.128 SOCS1 is approximately 10‐fold more potent than SOCS3 at inhibiting JAK, mainly due to sequence differences in the KIR.128, 201
In vivo, SOCS3 is a specific inhibitor of IL‐6 family cytokines,43, 202, 203, 204 G‐CSF,172, 173 and leptin186 as these cytokines all signal via receptors that contain binding sites for the SOCS3 SH2 domain.205 SOCS3 engages the receptor with its SH2 and simultaneously inhibits the JAK1, JAK2 or TYK2 associated with that receptor.200 SOCS1, on the other hand, is a potent inhibitor of interferon, IL‐12/23, IL‐4/13 and IL‐2 family cytokine signaling34, 35, 36, 37, 38, 39, 40, 41, 206, 207, 208 however, does not target the receptors for those cytokines with its SH2 domain. The target of the SOCS1 SH2 domain is unclear, it binds tightly to the activation loop sequence of all four JAKs when they are synthesized as synthetic peptides in vitro, however, appears to be sterically hindered from doing so when the activation loop is part of an intact kinase domain.201 Further work is needed to clarify to this issue.
Phosphatases
Association of cytokine with the appropriate receptor triggers a cascade of intracellular tyrosine phosphorylation: JAKs are auto‐phosphorylated in trans, receptors are then phosphorylated by these activated JAKs and finally the STATs are phosphorylated allowing them to adopt their active conformation. Therefore it is no surprise that phosphotyrosine phosphatases (PTPs) play a crucial role in regulating these signaling pathways.209, 210 Six phosphotyrosine phosphatases in particular have been shown to regulate JAK/STAT signaling: the receptor tyrosine phosphatases CD45 and PTP‐RT, two related cytoplasmic phosphatases PTP1b and TC‐PTP, and the SH2 domain containing phosphatases SHP1 and SHP2.211 These phosphatases are constitutively expressed and are, therefore, not feedback‐inhibitors. As such, they tend to restrain the amplitude of the signaling cascade rather than controlling its duration. It is the balance between the action of these phosphatases and the activity of the JAKs that determines the flux through the pathway. Determining the true targets of these phosphatases (JAKs, STATs, or receptors) has been difficult and at times contentious.
The receptor phosphatases CD45 and PTPRT
CD45 and PTPRT are both receptor phosphatases comprising an extracellular receptor‐like region, a transmembrane domain and two intracellular tandem phosphatase domains. For both CD45 and PTPRT the first phosphatase domain is the catalytically active domain, whereas the second is a catalytically dead pseudophosphatase domain that is thought to have regulatory roles in both proteins. Full length CD45 is ~140 kDa; however, alternative splicing gives rise to several different sized isoforms. The extracellular region of CD45 is comprised of three FnIII domains (Fig. 7). CD45 is highly expressed in hematopoietic cells and thought to dephosphorylate all four JAK proteins.13, 212 Cells deficient in CD45 display extended signaling in response to IL‐7, EPO, and interferon stimulation. PTPRT is a large protein containing an N‐terminal MAM domain followed by an Ig domain, four FnIII domains, a transmembrane domain and two tandem PTP domains. PTPRT directly interacts with and dephosphorylates the critical tyrosine residue in STAT3, pY705.213
Figure 7.
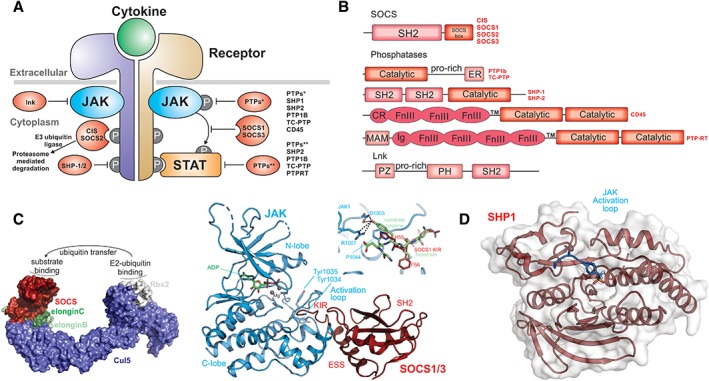
Regulation of cytokine signaling. (A) Schematic diagram showing regulators of cytokine signaling and where they act. (B) Domain architecture of the proteins indicated in A. (C) The primary negative feedback regulators of cytokine signaling are a subset of the SOCS (Suppressors of Cytokine Signaling) family, CIS, SOCS1, SOCS2, and SOCS3. These proteins function as the substrate recruitment modules of an E3 ubiquitin ligase (model structure shown in surface representation) and promote the ubiquitination and degradation of cytokine receptors and potentially other substrates. Substrates bind to the SH2 domain of SOCS proteins (red) and ubiquitin is transferred via an E2 ubiquitin‐conjugating enzyme that docks onto the RING‐domain protein Rbx2 (white). SOCS1 and SOCS3 (right) can also directly inhibit the JAK kinase domain by using their kinase inhibitory region (KIR) to block the substrate binding site of the kinase (PDB ID: http://firstglance.jmol.org/fg.htm?mol=6C7Y) (model of a substrate overlay shown inset). (D) Six tyrosine phosphatases have been shown to be important regulators of cytokine‐pathway activity, acting by dephosphorylating JAKs, STATs, or receptors. The structure of one of these, SHP1, has been solved in complex with the JAK activation loop of JAK2 (PDB ID: http://firstglance.jmol.org/fg.htm?mol=4GSO).
The SH2 domain containing phosphatases SHP1 and SHP2
SHP1 and SHP2 are cytoplasmic SH2 domain containing phosphatases. They are approximately 70 kDa, and composed of two SH2 domains and a single PTP domain that is negatively regulated by interactions with the SH2 domains.
The expression of SHP1 is limited to the hematopoietic lineage, where it regulates IL‐3, EPO, IFNα, and potentially other cytokine‐induced signaling by dephosphorylating JAK1, JAK2, and TYK2.214, 215, 216, 217, 218 SHP2 is ubiquitously expressed, but also plays an essential role in the regulation of hematopoiesis. Knockout of shp2 leads to increased JAK1 autophosphorylation and upregulation of interferon signaling217, 219 implying a role as a negative regulator. However, SHP2 is better characterized as a positive regulator of cytokine signaling. For example, it binds to pY759 on gp130 and activates the MAPK signaling cascade in response to IL‐6 and LIF. In fact, SHP2 was the first tyrosine phosphatase to be identified as a proto‐oncogene and somatic activating mutations of SHP2 have been identified in acute myeloid leukemia (AML) and B cell acute lymphoblastic leukemia (BALL).
The cytoplasmic phosphatases PTP1B and TC‐PTP
PTP1B and TC‐PTP are two highly related, ~40 kDa phosphatases that are tethered to the cytoplasmic face of the endoplasmic reticulum (ER).220, 221, 222 Substrate‐trapping mutants of PTP1B have been shown to interact directly with the activation loop of JAK2 and TYK2 suggesting these as the targets for its catalytic activity however it may also directly dephosphorylate STAT3.223, 224, 225, 226 PTP1B is a powerful regulator of leptin signaling and knockout mice show increased JAK2 phosphorylation in response to that cytokine.225 TC‐PTP is also tethered to the ER, however, a different isoform which lacks the ER‐targeting motif is found in the nucleus and can dephosphorylate STAT3. Both JAK1 and JAK3 are dephosphorylated by TC‐PTP and its knockout leads to increased IL‐2, IFNα and IFNγ signaling.227, 228
The adaptor protein, LNK
The lymphocyte adaptor protein, LNK, also known as SH2B3, is a member of the SH2 domain containing adaptor protein family which also comprises APS (SH2B1) and SH2B (SH2B2). This family of proteins contains three distinct domains: a dimerization domain (phenylalanine zipper) which allows homo‐dimerization, a Pleckstrin Homology (PH) domain and an SH2 domain.229 While APS and SH2B appear to activate cytokine signaling, LNK is a negative regulator of cytokines that signal via JAK2, particularly EPO and TPO.230, 231 LNK knockout mice have enhanced numbers of hematopoietic stem cells and are hyperresponsive to EPO and TPO and over‐expression of LNK inhibits megakaryocyte development. Consistent with its suppressive role, inactivation mutations in LNK are found in ca. 5% of MPNs and also in rare cases of idiopathic erythrocytosis. The SH2 domain of LNK binds directly to JAK2 (at pTyr813, located between the kinase and pseudokinase domains)232; however, it is unclear how this regulates signaling.
Case study: IL‐6 signaling
IL‐6 represents perhaps the archetypal cytokine, it being the closest homologue to cytokines present in extant insect species. IL‐6 (and related family members, Fig. 3) are all highly pleiotropic with roles in hematopoiesis, the acute phase response, development and both pro and anti‐inflammatory processes.233, 234, 235 Here we provide a summary of the molecular events involved in IL‐6 signaling.
IL‐6 production can be induced by a variety of stimuli and by many different cell types. The receptor for IL‐6, as discussed above, consists of the gp130 shared signaling chain and the non‐signaling cytokine‐specific IL‐6Rα chain. Although gp130 is found ubiquitously expressed, IL‐6Rα is found only on hepatocytes and some leukocytes.236 However, soluble IL‐6Rα is released from the liver and this allows IL‐6 to signal to many different cell‐types in a process called trans‐signaling.237 Although IL‐6 on its own has little affinity for gp130, it binds tightly to IL‐6Rα (either membrane bound or soluble). This dimeric complex can then bind with high affinity to gp130, using regions on both the cytokine and the alpha chain. The resulting ternary complex then dimerizes to form the signaling hexamer.54, 238 Thus, the formation of the signaling competent IL‐6 receptor complex is an ordered process. The molecular details of IL‐6 binding to its receptor were first described by Boulanger et al., who solved the structure of IL‐6 bound to the CHR of IL6Rα and the first three domains of gp130.54 Combined with the full structure of gp130239 these studies allow a full model of the IL‐6/IL‐6 receptor complex to be constructed (Fig. 8).239
Figure 8.
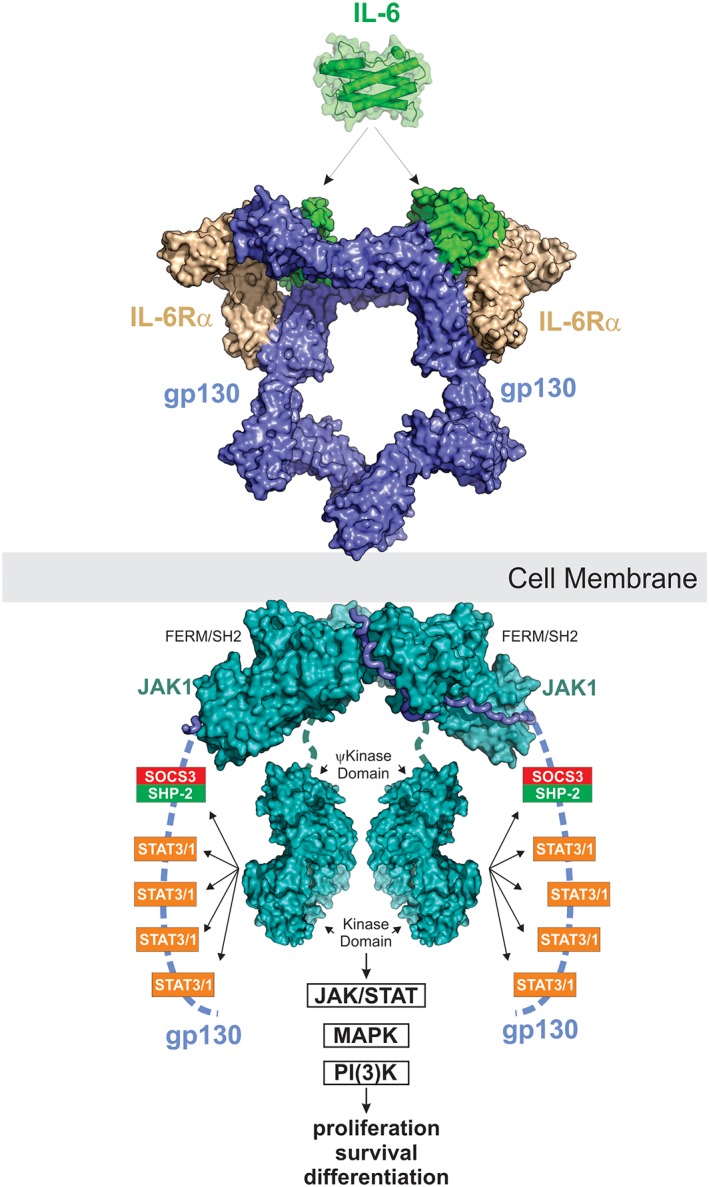
IL‐6 signaling. IL‐6 signals via a 2:2:2 complex between itself, gp130 and either membrane‐bound IL‐6Rα (classic signaling) or soluble IL‐6Rα (trans‐signaling). JAK1, JAK2 and TYK2 can all bind the intracellular domain of gp130; however, JAK1 appears to be the dominant kinase. The structure of JAK1 bound to the gp130 cytoplasmic domain is a model based on the structures of JAK1/IFNλR (PDB ID: http://firstglance.jmol.org/fg.htm?mol=5L04) and the JAK2/EPOR dimeric structure (coordinates kindly provided by R. Ferrao and P. Lupardus). JAK is activated by trans‐phosphorylation and then phosphorylates five tyrosine residues on the receptor intracellular domain. The four distal tyrosines are docking sites for STAT3 and to a lesser degree STAT1. Activated STAT3 is then phosphorylated by JAK and translocates into the nucleus to drive the biological response. The MAPK pathway is also stimulated by IL‐6 via SHP2 which binds to pY759 using its SH2 domain. The PI(3)K pathway is also activated in response to IL‐6. SOCS3 is a direct STAT3 target gene and binds to the SHP2‐binding site on the receptor via its SH2 domain. This inhibits MAPK signaling via displacement of SHP2 and also inhibits further STAT3 activation by direct inhibition of JAK catalytic activity.
Once IL‐6 has bound to its receptor, bringing together the two JAK‐associated gp130 chains, this allows transactivation of the associated JAKs. JAK1, JAK2, and TYK2 can all be found associated with gp130 although knockout studies suggest JAK1 may be dominant.14 The ability of gp130 to associate with different members of the JAK family is unusual (Fig. 3). Upon activation, these JAKs then phosphorylate five tyrosine residues in the gp130 cytoplasmic domain.90 The four distal sites (Y767, Y814, Y905, and Y915) are motifs targeted by the STAT3 (and to a lesser extent, STAT1) SH2 domain whilst the proximal phosphotyrosine (Y759) binds the negative regulatory protein SOCS3 and the phosphatase SHP2. STAT3 exists as a pre‐formed dimer; however, once it is docked onto the receptor it is phosphorylated by JAK on Y705 and pY705 is then bound by the SH2 domain of the opposing monomers. This results in a re‐orientation of STAT3 from an anti‐parallel to a parallel dimer (Fig. 6) and subsequent translocation to the nucleus where it induces the transcription of target genes.11 Activated STAT3 is observed within 15 min of IL‐6 exposure and STAT3 target genes are observed almost immediately after240 highlighting a rapid transcriptional response. Importantly, STAT1 is also activated alongside STAT3.
A well characterized activity of IL‐6 is its ability to induce the differentiation of monocytes into macrophages. An important component of this activity is the ability of IL‐6 to induce the master transcription factor PU.1 alongside hundreds of other genes (RM, unpublished data). One of the most important of the early genes upregulated by IL‐6 is SOCS3.171 SOCS3 mRNA and protein are observed within 30 min of IL‐6 exposure and it is one of the most highly expressed early response genes (RM, unpublished data). SOCS3 protein binds to pY759 on gp130 via its SH2 domain and then uses its kinase inhibitory region to inhibit the associated JAK1 (or JAK2, TYK2). This switches JAK off and prevents any further STAT3 phosphorylation. STAT3 is dephosphorylated in the nucleus by TC45, the nuclear isoform of the T‐cell protein tyrosine phosphatase (TC‐PTP)162 and is then shuttled into the cytoplasm by exportin‐1 to allow for subsequent activation cycles.241 SOCS3 inhibits the signaling cascade as described above but may also play a role in shaping the cellular response. Genetic deletion of SOCS3 leads to a wider transcriptional response after IL‐6 exposure, in particular there is increased expression of a number of genes associated with IFNγ (STAT1) signaling. Under normal conditions, therefore, SOCS3 appears responsible for dampening STAT1 transcriptional programs and allowing STAT3 to dominate,242 while eventually inhibiting both pathways.
Alongside activation of STAT3 (and STAT1), IL‐6 stimulates two other signaling cascades: the MAPK and PI(3)K pathways. The phosphatase, SHP2 binds to pY759 on gp130 and promotes activation of the MAPK cascade via a mechanism that is not completely understood but may involve Grb2.243 SOCS3 also binds to this site and can thereby inhibit both STAT3 and MAPK induced transcriptional responses. How IL‐6 induces the PI(3)‐kinase pathway is less clear but the end result is activation of the serine/threonine kinase AKT (protein kinase B) at the cell membrane and stimulation of downstream signaling including mTOR.
Unanswered questions
The most important unanswered question in the field is how the activation of JAK (by trans‐phosphorylation) is induced by cytokine binding and how this process goes awry in the presence of the activating mutations seen in the pseudokinase domain in human myeloproliferative diseases. The classical explanation given for the process of JAK activation was that simple dimerization of the receptor chains (by cytokine) brought the JAKs into close‐enough proximity for their kinase domains to phosphorylate one‐another. However it is now clear that many receptors exist as pre‐formed dimers even in the absence of cytokine244 and that it is rather a re‐orientation of these chains that allows JAK auto‐phosphorylation. In fact, in 2014, Brooks et al. performed a series of FRET‐based analyses to show that Growth Hormone induced a separation of the intracellular receptor domains and this led to a geometry where the kinase domains of the two JAK molecules were juxtaposed.245 Such a model supported their earlier analyses which showed that the GHR could be activated by tuning the relative orientation of the TM and juxtamembrane regions even in the absence of cytokine.246
This model suggests that prior to cytokine stimulation the pseudokinase domain from one JAK interacts with (and inhibits) the kinase domain from the other. After cytokine stimulation this inhibition is released. The importance of the pseudokinase domain in regulating the kinase domain is of course well‐established as described above and by the existence of activating mutations within this domain. The important structure of the TYK2 pseudokinase‐kinase domain pair highlighted that activating mutations tend to cluster near the interacting surface between the two domains however did not provide a molecular mechanism for what the pseudokinase domain was actually doing. The only structural information available for transphosphorylation of a tyrosine kinase was provided by crystallographic studies from the Hubbard laboratory of the IGF1 receptor kinase126; however, as far as JAK kinases are concerned, we have no picture of what allows their kinase domains to adopt this position and what prevents them from doing so in the first place. It seems clear that only structures of complete JAK proteins (with and without bound receptor) will provide a full molecular description of how JAK is activated and why this process goes awry upon mutation in human disease.
Disclosure statement
The authors declare no conflicts of interest.
Acknowledgements
This work was supported by the Cancer Council Victoria (Grant‐in‐aid 1065180) and the National Health and Medical Research Council (NHMRC) Australia(Project grant no. 1122999, Program grant no. 1113577), an NHMRC IRIISS Grant 9000220, and a Victorian State Government Operational Infrastructure Scheme grant. J. J. B. is supported by an NHMRC fellowship.
References
- 1. Stark GR, Darnell JE Jr (2012) The JAK‐STAT pathway at twenty. Immunity 36:503–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Velazquez L, Fellous M, Stark GR, Pellegrini S (1992) A protein tyrosine kinase in the interferon‐alpha/beta signaling pathway. Cell 70:313–322. [DOI] [PubMed] [Google Scholar]
- 3. Wilks AF (1989) Two putative protein‐tyrosine kinases identified by application of the polymerase chain reaction. Proc Natl Acad Sci U S A 86:1603–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wilks AF, Harpur AG, Kurban RR, Ralph SJ, Zurcher G, Ziemiecki A (1991) Two novel protein‐tyrosine kinases, each with a second phosphotransferase‐related catalytic domain, define a new class of protein kinase. Mol Cell Biol 11:2057–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Firmbach‐Kraft I, Byers M, Shows T, Dalla‐Favera R, Krolewski JJ (1990) tyk2, prototype of a novel class of non‐receptor tyrosine kinase genes. Oncogene 5:1329–1336. [PubMed] [Google Scholar]
- 6. Kawamura M, McVicar DW, Johnston JA, Blake TB, Chen YQ, Lal BK, Lloyd AR, Kelvin DJ, Staples JE, Ortaldo JR (1994) Molecular cloning of L‐JAK, a Janus family protein‐tyrosine kinase expressed in natural killer cells and activated leukocytes. Proc Natl Acad Sci U S A 91:6374–6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Feng J, Witthuhn BA, Matsuda T, Kohlhuber F, Kerr IM, Ihle JN (1997) Activation of Jak2 catalytic activity requires phosphorylation of Y1007 in the kinase activation loop. Mol Cell Biol 17:2497–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Argetsinger LS, Campbell GS, Yang X, Witthuhn BA, Silvennoinen O, Ihle JN, Carter‐Su C (1993) Identification of JAK2 as a growth hormone receptor‐associated tyrosine kinase. Cell 74:237–244. [DOI] [PubMed] [Google Scholar]
- 9. Schindler C, Shuai K, Prezioso VR, Darnell JE (1992) Interferon‐dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science 257:809–813. [DOI] [PubMed] [Google Scholar]
- 10. Shuai K, Schindler C, Prezioso VR, Darnell JE (1992) Activation of transcription by Ifn‐gamma – tyrosine phosphorylation of a 91‐Kd DNA‐binding protein. Science 258:1808–1812. [DOI] [PubMed] [Google Scholar]
- 11. Schindler C, Darnell J Jr (1995) Transcriptional responses to polypeptide ligands: the JAK‐STAT pathway. Annu Rev Biochem 64:621–652. [DOI] [PubMed] [Google Scholar]
- 12. Yoshimura A, Ohkubo T, Kiguchi T, Jenkins NA, Gilbert DJ, Copeland NG, Hara T, Miyajima A (1995) A novel cytokine‐inducible gene CIS encodes an SH2‐containing protein that binds to tyrosine‐phosphorylated interleukin 3 and erythropoietin receptors. EMBO J 14:2816–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Endo TA, Masuhara M, Yokouchi M, Suzuki R, Sakamoto H, Mitsui K, Matsumoto A, Tanimura S, Ohtsubo M, Misawa H, Miyazaki T, Leonor N, Taniguchi T, Fujita T, Kanakura Y, Komiya S, Yoshimura A (1997) A new protein containing an SH2 domain that inhibits JAK kinases. Nature 387:921–924. [DOI] [PubMed] [Google Scholar]
- 14. Rodig SJ, Meraz MA, White JM, Lampe PA, Riley JK, Arthur CD, King KL, Sheehan KC, Yin L, Pennica D, Johnson EM Jr, Schreiber RD (1998) Disruption of the Jak1 gene demonstrates obligatory and nonredundant roles of the Jaks in cytokine‐induced biologic responses. Cell 93:373–383. [DOI] [PubMed] [Google Scholar]
- 15. Neubauer H, Cumano A, Muller M, Wu H, Huffstadt U, Pfeffer K (1998) Jak2 deficiency defines an essential developmental checkpoint in definitive hematopoiesis. Cell 93:397–409. [DOI] [PubMed] [Google Scholar]
- 16. Parganas E, Wang D, Stravopodis D, Topham DJ, Marine JC, Teglund S, Vanin EF, Bodner S, Colamonici OR, van Deursen JM, Grosveld G, Ihle JN (1998) Jak2 is essential for signaling through a variety of cytokine receptors. Cell 93:385–395. [DOI] [PubMed] [Google Scholar]
- 17. Krempler A, Qi Y, Triplett AA, Zhu J, Rui H, Wagner KU (2004) Generation of a conditional knockout allele for the Janus kinase 2 (Jak2) gene in mice. Genesis 40:52–57. [DOI] [PubMed] [Google Scholar]
- 18. Wagner KU, Krempler A, Triplett AA, Qi Y, George NM, Zhu J, Rui H (2004) Impaired alveologenesis and maintenance of secretory mammary epithelial cells in Jak2 conditional knockout mice. Mol Cell Biol 24:5510–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nosaka T, van Deursen JM, Tripp RA, Thierfelder WE, Witthuhn BA, McMickle AP, Doherty PC, Grosveld GC, Ihle JN (1995) Defective lymphoid development in mice lacking Jak3. Science 270:800–802. [DOI] [PubMed] [Google Scholar]
- 20. Thomis DC, Gurniak CB, Tivol E, Sharpe AH, Berg LJ (1995) Defects in B lymphocyte maturation and T lymphocyte activation in mice lacking Jak3. Science 270:794–797. [DOI] [PubMed] [Google Scholar]
- 21. Park SY, Saijo K, Takahashi T, Osawa M, Arase H, Hirayama N, Miyake K, Nakauchi H, Shirasawa T, Saito T (1995) Developmental defects of lymphoid cells in Jak3 kinase‐deficient mice. Immunity 3:771–782. [DOI] [PubMed] [Google Scholar]
- 22. Shimoda K, Kato K, Aoki K, Matsuda T, Miyamoto A, Shibamori M, Yamashita M, Numata A, Takase K, Kobayashi S, Shibata S, Asano Y, Gondo H, Sekiguchi K, Nakayama K, Nakayama T, Okamura T, Okamura S, Niho Y, Nakayama K (2000) Tyk2 plays a restricted role in IFN alpha signaling, although it is required for IL‐12‐mediated T cell function. Immunity 13:561–571. [DOI] [PubMed] [Google Scholar]
- 23. Karaghiosoff M, Neubauer H, Lassnig C, Kovarik P, Schindler H, Pircher H, McCoy B, Bogdan C, Decker T, Brem G, Pfeffer K, Muller M (2000) Partial impairment of cytokine responses in Tyk2‐deficient mice. Immunity 13:549–560. [DOI] [PubMed] [Google Scholar]
- 24. Karaghiosoff M, Steinborn R, Kovarik P, Kriegshauser G, Baccarini M, Donabauer B, Reichart U, Kolbe T, Bogdan C, Leanderson T, Levy D, Decker T, Muller M (2003) Central role for type I interferons and Tyk2 in lipopolysaccharide‐induced endotoxin shock. Nat Immunol 4:471–477. [DOI] [PubMed] [Google Scholar]
- 25. Durbin JE, Hackenmiller R, Simon MC, Levy DE (1996) Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell 84:443–450. [DOI] [PubMed] [Google Scholar]
- 26. Park C, Li S, Cha E, Schindler C (2000) Immune response in Stat2 knockout mice. Immunity 13:795–804. [DOI] [PubMed] [Google Scholar]
- 27. Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, Kishimoto T, Akira S (1997) Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci U S A 94:3801–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thierfelder WE, van Deursen JM, Yamamoto K, Tripp RA, Sarawar SR, Carson RT, Sangster MY, Vignali DA, Doherty PC, Grosveld GC, Ihle JN (1996) Requirement for Stat4 in interleukin‐12‐mediated responses of natural killer and T cells. Nature 382:171–174. [DOI] [PubMed] [Google Scholar]
- 29. Liu XW, Robinson GW, Wagner KU, Garrett L, WynshawBoris A, Hennighausen L (1997) Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev 11:179–186. [DOI] [PubMed] [Google Scholar]
- 30. Teglund S, McKay C, Schuetz E, van Deursen JM, Stravopodis D, Wang DM, Brown M, Bodner S, Grosveld G, Ihle JN (1998) Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell 93:841–850. [DOI] [PubMed] [Google Scholar]
- 31. Akimoto T, Numata F, Tamura M, Takata Y, Higashida N, Takashi T, Takeda K, Akira S (1998) Abrogation of bronchial eosinophilic inflammation and airway hyperreactivity in signal transducers and activators of transcription (STAT)6‐deficient mice. J Exp Med 187:1537–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Delconte RB, Kolesnik TB, Dagley LF, Rautela J, Shi W, Putz EM, Stannard K, Zhang JG, Teh C, Firth M, Ushiki T, Andoniou CE, Degli‐Esposti MA, Sharp PP, Sanvitale CE, Infusini G, Liau NP, Linossi EM, Burns CJ, Carotta S, Gray DH, Seillet C, Hutchinson DS, Belz GT, Webb AI, Alexander WS, Li SS, Bullock AN, Babon JJ, Smyth MJ, Nicholson SE, Huntington ND (2016) CIS is a potent checkpoint in NK cell‐mediated tumor immunity. Nat Immunol 17:816–824. [DOI] [PubMed] [Google Scholar]
- 33. Yang XO, Zhang H, Kim BS, Niu X, Peng J, Chen Y, Kerketta R, Lee YH, Chang SH, Corry DB, Wang D, Watowich SS, Dong C (2013) The signaling suppressor CIS controls proallergic T cell development and allergic airway inflammation. Nat Immunol 14:732–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Starr R, Metcalf D, Elefanty AG, Brysha M, Willson TA, Nicola NA, Hilton DJ, Alexander WS (1998) Liver degeneration and lymphoid deficiencies in mice lacking suppressor of cytokine signaling‐1. Proc Natl Acad Sci U S A 95:14395–14399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Naka T, Matsumoto T, Narazaki M, Fujimoto M, Morita Y, Ohsawa Y, Saito H, Nagasawa T, Uchiyama Y, Kishimoto T (1998) Accelerated apoptosis of lymphocytes by augmented induction of Bax in SSI‐1 (STAT‐induced STAT inhibitor‐1) deficient mice. Proc Natl Acad Sci U S A 95:15577–15582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marine JC, Topham DJ, McKay C, Wang D, Parganas E, Stravopodis D, Yoshimura A, Ihle JN (1999) SOCS1 deficiency causes a lymphocyte‐dependent perinatal lethality. Cell 98:609–616. [DOI] [PubMed] [Google Scholar]
- 37. Alexander WS, Starr R, Metcalf D, Nicholson SE, Farley A, Elefanty AG, Brysha M, Kile BT, Richardson R, Baca M, Zhang JG, Willson TA, Viney EM, Sprigg NS, Rakar S, Corbin J, Mifsud S, DiRago L, Cary D, Nicola NA, Hilton DJ (1999) Suppressors of cytokine signaling (SOCS): negative regulators of signal transduction. J Leukoc Biol 66:588–592. [DOI] [PubMed] [Google Scholar]
- 38. Gingras S, Parganas E, de Pauw A, Ihle JN, Murray PJ (2004) Re‐examination of the role of suppressor of cytokine signaling 1 (SOCS1) in the regulation of toll‐like receptor signaling. J Biol Chem 279:54702–54707. [DOI] [PubMed] [Google Scholar]
- 39. Eyles JL, Metcalf D, Grusby MJ, Hilton DJ, Starr R (2002) Negative regulation of interleukin‐12 signaling by suppressor of cytokine signaling‐1. J Biol Chem 277:43735–43740. [DOI] [PubMed] [Google Scholar]
- 40. Naka T, Tsutsui H, Fujimoto M, Kawazoe Y, Kohzaki H, Morita Y, Nakagawa R, Narazaki M, Adachi K, Yoshimoto T, Nakanishi K, Kishimoto T (2001) SOCS‐1/SSI‐1‐deficient NKT cells participate in severe hepatitis through dysregulated cross‐talk inhibition of IFN‐gamma and IL‐4 signaling in vivo. Immunity 14:535–545. [DOI] [PubMed] [Google Scholar]
- 41. Davey GM, Starr R, Cornish AL, Burghardt JT, Alexander WS, Carbone FR, Surh CD, Heath WR (2005) SOCS‐1 regulates IL‐15‐driven homeostatic proliferation of antigen‐naive CD8 T cells, limiting their autoimmune potential. J Exp Med 202:1099–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Metcalf D, Greenhalgh CJ, Viney E, Willson TA, Starr R, Nicola NA, Hilton DJ, Alexander WS (2000) Gigantism in mice lacking suppressor of cytokine signalling‐2. Nature 405:1069–1073. [DOI] [PubMed] [Google Scholar]
- 43. Roberts AW, Robb L, Rakar S, Hartley L, Cluse L, Nicola NA, Metcalf D, Hilton DJ, Alexander WS (2001) Placental defects and embryonic lethality in mice lacking suppressor of cytokine signaling 3. Proc Natl Acad Sci U S A 98:9324–9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Takahashi Y, Carpino N, Cross JC, Torres M, Parganas E, Ihle JN (2003) SOCS3: an essential regulator of LIF receptor signaling in trophoblast giant cell differentiation. EMBO J 22:372–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liongue C, Sertori R, Ward AC (2016) Evolution of cytokine receptor signaling. J Immunol 197:11–18. [DOI] [PubMed] [Google Scholar]
- 46. Bazan JF (1990) Structural design and molecular evolution of a cytokine receptor superfamily. Proc Natl Acad Sci U S A 87:6934–6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. de Vos AM, Ultsch M, Kossiakoff AA (1992) Human growth hormone and extracellular domain of its receptor: crystal structure of the complex. Science 255:306–312. [DOI] [PubMed] [Google Scholar]
- 48. Yawata H, Yasukawa K, Natsuka S, Murakami M, Yamasaki K, Hibi M, Taga T, Kishimoto T (1993) Structure–function analysis of human IL‐6 receptor: dissociation of amino acid residues required for IL‐6‐binding and for IL‐6 signal transduction through gp130. EMBO J 12:1705–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bazan JF (1990) Shared architecture of hormone binding domains in type I and II interferon receptors. Cell 61:753–754. [DOI] [PubMed] [Google Scholar]
- 50. Boulay JL, O'Shea JJ, Paul WE (2003) Molecular phylogeny within type I cytokines and their cognate receptors. Immunity 19:159–163. [DOI] [PubMed] [Google Scholar]
- 51. Syed RS, Reid SW, Li CW, Cheetham JC, Aoki KH, Liu BS, Zhan HJ, Osslund TD, Chirino AJ, Zhang JD, Finer‐Moore J, Elliott S, Sitney K, Katz BA, Matthews DJ, Wendoloski JJ, Egrie J, Stroud RM (1998) Efficiency of signalling through cytokine receptors depends critically on receptor orientation. Nature 395:511–516. [DOI] [PubMed] [Google Scholar]
- 52. Elkins PA, Christinger HW, Sandowski Y, Sakal E, Gertler A, de Vos AM, Kossiakoff AA (2000) Ternary complex between placental lactogen and the extracellular domain of the prolactin receptor. Nat Struct Biol 7:808–815. [DOI] [PubMed] [Google Scholar]
- 53. Tamada T, Honjo E, Maeda Y, Okamoto T, Ishibashi M, Tokunaga M, Kuroki R (2006) Homodimeric cross‐over structure of the human granulocyte colony‐stimulating factor (GCSF) receptor signaling complex. Proc Natl Acad Sci U S A 103:3135–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Boulanger MJ, Chow DC, Brevnova EE, Garcia KC (2003) Hexameric structure and assembly of the interleukin‐6/IL‐6 alpha‐receptor/gp130 complex. Science 300:2101–2104. [DOI] [PubMed] [Google Scholar]
- 55. Chow D, Ho J, Nguyen Pham TL, Rose‐John S, Garcia KC (2001) In vitro reconstitution of recognition and activation complexes between interleukin‐6 and gp130. Biochemistry 40:7593–7603. [DOI] [PubMed] [Google Scholar]
- 56. Matadeen R, Hon WC, Heath JK, Jones EY, Fuller S (2007) The dynamics of signal triggering in a gp130‐receptor complex. Structure 15:441–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Huyton T, Zhang JG, Luo CS, Lou MZ, Hilton DJ, Nicola NA, Garrett TP (2007) An unusual cytokine: Ig‐domain interaction revealed in the crystal structure of leukemia inhibitory factor (LIF) in complex with the LIF receptor. Proc Natl Acad Sci U S A 104:12737–12742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gearing DP, Thut CJ, VandeBos T, Gimpel SD, Delaney PB, King J, Price V, Cosman D, Beckmann MP (1991) Leukemia inhibitory factor receptor is structurally related to the IL‐6 signal transducer, gp130. EMBO J 10:2839–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Skiniotis G, Lupardus PJ, Martick M, Walz T, Garcia KC (2008) Structural organization of a full‐length gp130/LIF‐R cytokine receptor transmembrane complex. Mol Cell 31:737–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lupardus PJ, Skiniotis G, Rice AJ, Thomas C, Fischer S, Walz T, Garcia KC (2011) Structural snapshots of full‐length Jak1, a transmembrane gp130/IL‐6/IL‐6Rα cytokine receptor complex, and the receptor‐Jak1 holocomplex. Structure 19:45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Oppmann B, Lesley R, Blom B, Timans JC, Xu YM, Hunte B, Vega F, Yu N, Wang J, Singh K, Zonin F, Vaisberg E, Churakova T, Liu MR, Gorman D, Wagner J, Zurawski S, Liu YJ, Abrams JS, Moore KW, Rennick D, de Waal‐Malefyt R, Hannum C, Bazan JF, Kastelein RA (2000) Novel p19 protein engages IL‐12p40 to form a cytokine, IL‐23, with biological activities similar as well as distinct from IL‐12. Immunity 13:715–725. [DOI] [PubMed] [Google Scholar]
- 62. Lupardus PJ, Garcia KC (2008) The structure of interleukin‐23 reveals the molecular basis of p40 subunit sharing with interleukin‐12. J Mol Biol 382:931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yoon C, Johnston SC, Tang J, Stahl M, Tobin JF, Somers WS (2000) Charged residues dominate a unique interlocking topography in the heterodimeric cytokine interleukin‐12. EMBO J 19:3530–3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bloch Y, Bouchareychas L, Merceron R, Skladanowska K, Van den Bossche L, Detry S, Govindarajan S, Elewaut D, Haerynck F, Dullaers M, Adamopoulos IE, Savvides SN (2018) Structural activation of pro‐inflammatory human cytokine IL‐23 by cognate IL‐23 receptor enables recruitment of the shared receptor IL‐12Rbeta1. Immunity 48:45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kitamura T, Sato N, Arai K, Miyajima A (1991) Expression cloning of the human IL‐3 receptor cDNA reveals a shared beta subunit for the human IL‐3 and GM‐CSF receptors. Cell 66:1165–1174. [DOI] [PubMed] [Google Scholar]
- 66. Hayashida K, Kitamura T, Gorman DM, Arai K, Yokota T, Miyajima A (1990) Molecular cloning of a second subunit of the receptor for human granulocyte‐macrophage colony‐stimulating factor (GM‐CSF): reconstitution of a high‐affinity GM‐CSF receptor. Proc Natl Acad Sci U S A 87:9655–9659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hansen G, Hercus TR, McClure BJ, Stomski FC, Dottore M, Powell J, Ramshaw H, Woodcock JM, Xu Y, Guthridge M, McKinstry WJ, Lopez AF, Parker MW (2008) The structure of the GM‐CSF receptor complex reveals a distinct mode of cytokine receptor activation. Cell 134:496–507. [DOI] [PubMed] [Google Scholar]
- 68. Dubois S, Mariner J, Waldmann TA, Tagaya Y (2002) IL‐15Ralpha recycles and presents IL‐15 In trans to neighboring cells. Immunity 17:537–547. [DOI] [PubMed] [Google Scholar]
- 69. Spangler JB, Moraga I, Mendoza JL, Garcia KC (2015) Insights into cytokine‐receptor interactions from cytokine engineering. Annu Rev Immunol 33:139–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kalie E, Jaitin DA, Podoplelova Y, Piehler J, Schreiber G (2008) The stability of the ternary interferon–receptor complex rather than the affinity to the individual subunits dictates differential biological activities. J Biol Chem 283:32925–32936. [DOI] [PubMed] [Google Scholar]
- 71. Thomas C, Moraga I, Levin D, Krutzik PO, Podoplelova Y, Trejo A, Lee C, Yarden G, Vleck SE, Glenn JS, Nolan GP, Piehler J, Schreiber G, Garcia KC (2011) Structural linkage between ligand discrimination and receptor activation by Type I interferons. Cell 146:621–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bach EA, Tanner JW, Marsters S, Ashkenazi A, Aguet M, Shaw AS, Schreiber RD (1996) Ligand‐induced assembly and activation of the gamma interferon receptor in intact cells. Mol Cell Biol 16:3214–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mendoza JL, Schneider WM, Hoffmann HH, Vercauteren K, Jude KM, Xiong A, Moraga I, Horton TM, Glenn JS, de Jong YP, Rice CM, Garcia KC (2017) The IFN‐lambda‐IFN‐lambdaR1‐IL‐10Rbeta complex reveals structural features underlying Type III IFN functional plasticity. Immunity 46:379–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Logsdon NJ, Jones BC, Josephson K, Cook J, Walter MR (2002) Comparison of interleukin‐22 and interleukin‐10 soluble receptor complexes. J Interferon Cytokine Res 22:1099–1112. [DOI] [PubMed] [Google Scholar]
- 75. Jones BC, Logsdon NJ, Walter MR (2008) Structure of IL‐22 bound to its high‐affinity IL‐22R1 chain. Structure 16:1333–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Logsdon NJ, Deshpande A, Harris BD, Rajashankar KR, Walter MR (2012) Structural basis for receptor sharing and activation by interleukin‐20 receptor‐2 (IL‐20R2) binding cytokines. Proc Natl Acad Sci U S A 109:12704–12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bleicher L, de Moura PR, Watanabe L, Colau D, Dumoutier L, Renauld JC, Polikarpov I (2008) Crystal structure of the IL‐22/IL‐22R1 complex and its implications for the IL‐22 signaling mechanism. FEBS Lett 582:2985–2992. [DOI] [PubMed] [Google Scholar]
- 78. Murakami M, Narazaki M, Hibi M, Yawata H, Yasukawa K, Hamaguchi M, Taga T, Kishimoto T (1991) Critical cytoplasmic region of the interleukin 6 signal transducer gp130 is conserved in the cytokine receptor family. Proc Natl Acad Sci U S A 88:11349–11353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Greiser JS, Stross C, Heinrich PC, Behrmann I, Hermanns HM (2002) Orientational constraints of the gp130 intracellular juxtamembrane domain for signaling. J Biol Chem 277:26959–26965. [DOI] [PubMed] [Google Scholar]
- 80. Stahl N, Boulton TG, Farruggella T, Ip NY, Davis S, Witthuhn BA, Quelle FW, Silvennoinen O, Barbieri G, Pellegrini S (1994) Association and activation of Jak‐Tyk kinases by CNTF‐LIF‐OSM‐IL‐6 beta receptor components. Science 263:92–95. [DOI] [PubMed] [Google Scholar]
- 81. Ferrao R, Wallweber HJ, Ho H, Tam C, Franke Y, Quinn J, Lupardus PJ (2016) The structural basis for class II cytokine receptor recognition by JAK1. Structure 24:897–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ferrao RD, Wallweber HJ, Lupardus PJ (2018) Receptor‐mediated dimerization of JAK2 FERM domains is required for JAK2 activation. Elife 7:e38089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Murray PJ (2007) The JAK‐STAT signaling pathway: input and output integration. J Immunol 178:2623–2629. [DOI] [PubMed] [Google Scholar]
- 84. Woldman I, Varinou L, Ramsauer K, Rapp B, Decker T (2001) The Stat1 binding motif of the interferon‐gamma receptor is sufficient to mediate Stat5 activation and its repression by SOCS3. J Biol Chem 276:45722–45728. [DOI] [PubMed] [Google Scholar]
- 85. Stahl N, Farruggella TJ, Boulton TG, Zhong Z, Darnell JE Jr, Yancopoulos GD (1995) Choice of STATs and other substrates specified by modular tyrosine‐based motifs in cytokine receptors. Science 267:1349–1353. [DOI] [PubMed] [Google Scholar]
- 86. Demoulin JB, Uyttenhove C, VanRoost E, deLestre B, Donckers D, VanSnick J, Renauld JC (1996) A single tyrosine of the interleukin‐9 (IL‐9) receptor is required for STAT activation, antiapoptotic activity, and growth regulation by IL‐9. Mol Cell Biol 16:4710–4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Greenlund AC, Farrar MA, Viviano BL, Schreiber RD (1994) Ligand‐induced IFN gamma receptor tyrosine phosphorylation couples the receptor to its signal transduction system (p91). EMBO J 13:1591–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Klingmuller U, Bergelson S, Hsiao JG, Lodish HF (1996) Multiple tyrosine residues in the cytosolic domain of the erythropoietin receptor promote activation of STAT5. Proc Natl Acad Sci U S A 93:8324–8328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hou J, Schindler U, Henzel WJ, Ho TC, Brasseur M, McKnight SL (1994) An interleukin‐4‐induced transcription factor: IL‐4 Stat. Science 265:1701–1706. [DOI] [PubMed] [Google Scholar]
- 90. Haan S, Hemmann U, Hassiepen U, Schaper F, Schneider‐Mergener J, Wollmer A, Heinrich PC, Grotzinger J (1999) Characterization and binding specificity of the monomeric STAT3‐SH2 domain. J Biol Chem 274:1342–1348. [DOI] [PubMed] [Google Scholar]
- 91. Bone H, Dechert U, Jirik F, Schrader JW, Welham MJ (1997) SHP1 and SHP2 protein‐tyrosine phosphatases associate with betac after interleukin‐3‐induced receptor tyrosine phosphorylation. Identification of potential binding sites and substrates. J Biol Chem 272:14470–14476. [DOI] [PubMed] [Google Scholar]
- 92. Lehmann U, Schmitz J, Weissenbach M, Sobota RM, Hortner M, Friederichs K, Behrmann I, Tsiaris W, Sasaki A, Schneider‐Mergener J, Yoshimura A, Neel BG, Heinrich PC, Schaper F (2003) SHP2 and SOCS3 contribute to Tyr‐759‐dependent attenuation of interleukin‐6 signaling through gp130. J Biol Chem 278:661–671. [DOI] [PubMed] [Google Scholar]
- 93. Anhuf D, Weissenbach M, Schmitz J, Sobota R, Hermanns HM, Radtke S, Linnemann S, Behrmann I, Heinrich PC, Schaper F (2000) Signal transduction of IL‐6, leukemia‐inhibitory factor, and oncostatin M: structural receptor requirements for signal attenuation. J Immunol 165:2535–2543. [DOI] [PubMed] [Google Scholar]
- 94. Kershaw NJ, Murphy JM, Lucet IS, Nicola NA, Babon JJ (2013) Regulation of Janus kinases by SOCS proteins. Biochem Soc Trans 41:1042–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Harpur AG, Andres AC, Ziemiecki A, Aston RR, Wilks AF (1992) JAK2, a third member of the JAK family of protein tyrosine kinases. Oncogene 7:1347–1353. [PubMed] [Google Scholar]
- 96. Wallweber HJ, Tam C, Franke Y, Starovasnik MA, Lupardus PJ (2014) Structural basis of recognition of interferon‐alpha receptor by tyrosine kinase 2. Nat Struct Mol Biol 21:443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Zhang D, Wlodawer A, Lubkowski J (2016) Crystal structure of a complex of the intracellular domain of interferon lambda receptor 1 (IFNLR1) and the FERM/SH2 domains of human JAK1. J Mol Biol 428:4651–4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Saharinen P, Takaluoma K, Silvennoinen O (2000) Regulation of the Jak2 tyrosine kinase by its pseudokinase domain. Mol Cell Biol 20:3387–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Saharinen P, Silvennoinen O (2002) The pseudokinase domain is required for suppression of basal activity of Jak2 and Jak3 tyrosine kinases and for cytokine‐inducible activation of signal transduction. J Biol Chem 277:47954–47963. [DOI] [PubMed] [Google Scholar]
- 100. Ungureanu D, Wu J, Pekkala T, Niranjan Y, Young C, Jensen ON, Xu CF, Neubert TA, Skoda RC, Hubbard SR, Silvennoinen O (2011) The pseudokinase domain of JAK2 is a dual‐specificity protein kinase that negatively regulates cytokine signaling. Nat Struct Mol Biol 18:971–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Silvennoinen O, Ungureanu D, Niranjan Y, Hammaren H, Bandaranayake R, Hubbard SR (2013) New insights into the structure and function of the pseudokinase domain in JAK2. Biochem Soc Trans 41:1002–1007. [DOI] [PubMed] [Google Scholar]
- 102. Lupardus PJ, Ultsch M, Wallweber H, Bir Kohli P, Johnson AR, Eigenbrot C (2014) Structure of the pseudokinase‐kinase domains from protein kinase TYK2 reveals a mechanism for Janus kinase (JAK) autoinhibition. Proc Natl Acad Sci U S A 111:8025–8030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Toms AV, Deshpande A, McNally R, Jeong Y, Rogers JM, Kim CU, Gruner SM, Ficarro SB, Marto JA, Sattler M, Griffin JD, Eck MJ (2013) Structure of a pseudokinase‐domain switch that controls oncogenic activation of Jak kinases. Nat Struct Mol Biol 20:1221–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Tokarski JS, Zupa‐Fernandez A, Tredup JA, Pike K, Chang C, Xie D, Cheng L, Pedicord D, Muckelbauer J, Johnson SR, Wu S, Edavettal SC, Hong Y, Witmer MR, Elkin LL, Blat Y, Pitts WJ, Weinstein DS, Burke JR (2015) Tyrosine kinase 2‐mediated signal transduction in T lymphocytes is blocked by pharmacological stabilization of its pseudokinase domain. J Biol Chem 290:11061–11074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Bandaranayake RM, Ungureanu D (2012) Crystal structures of the JAK2 pseudokinase domain and the pathogenic mutant V617F. Nat Struct Mol Biol 19:754–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Min X, Ungureanu D, Maxwell S, Hammaren H, Thibault S, Hillert EK, Ayres M, Greenfield B, Eksterowicz J, Gabel C, Walker N, Silvennoinen O, Wang Z (2015) Structural and functional characterization of the JH2 pseudokinase domain of JAK family tyrosine kinase 2 (TYK2). J Biol Chem 290:27261–27270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Murphy JM, Zhang Q, Young SN, Reese ML, Bailey FP, Eyers PA, Ungureanu D, Hammaren H, Silvennoinen O, Varghese LN, Chen K, Tripaydonis A, Jura N, Fukuda K, Qin J, Nimchuk Z, Mudgett MB, Elowe S, Gee CL, Liu L, Daly RJ, Manning G, Babon JJ, Lucet IS (2014) A robust methodology to subclassify pseudokinases based on their nucleotide‐binding properties. Biochem J 457:323–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Luo H, Rose P, Barber D, Hanratty WP, Lee S, Roberts TM, D'Andrea AD, Dearolf CR (1997) Mutation in the Jak kinase JH2 domain hyperactivates Drosophila and mammalian Jak‐Stat pathways. Mol Cell Biol 17:1562–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, Passweg JR, Tichelli A, Cazzola M, Skoda RC (2005) A gain‐of‐function mutation of JAK2 in myeloproliferative disorders. N Engl J Med 352:1779–1790. [DOI] [PubMed] [Google Scholar]
- 110. Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJ, Boggon TJ, Wlodarska I, Clark JJ, Moore S, Adelsperger J, Koo S, Lee JC, Gabriel S, Mercher T, D'Andrea A, Frohling S, Dohner K, Marynen P, Vandenberghe P, Mesa RA, Tefferi A, Griffin JD, Eck MJ, Sellers WR, Meyerson M, Golub TR, Lee SJ, Gilliland DG (2005) Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell 7:387–397. [DOI] [PubMed] [Google Scholar]
- 111. James C, Ugo V, Le Couedic JP, Staerk J, Delhommeau F, Lacout C, Garcon L, Raslova H, Berger R, Bennaceur‐Griscelli A, Villeval JL, Constantinescu SN, Casadevall N, Vainchenker W (2005) A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature 434:1144–1148. [DOI] [PubMed] [Google Scholar]
- 112. Scott LM, Tong W, Levine RL, Scott MA, Beer PA, Stratton MR, Futreal PA, Erber WN, McMullin MF, Harrison CN, Warren AJ, Gilliland DG, Lodish HF, Green AR (2007) JAK2 exon 12 mutations in polycythemia vera and idiopathic erythrocytosis. N Engl J Med 356:459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Pietra D, Li S, Brisci A, Passamonti F, Rumi E, Theocharides A, Ferrari M, Gisslinger H, Kralovics R, Cremonesi L, Skoda R, Cazzola M (2008) Somatic mutations of JAK2 exon 12 in patients with JAK2 (V617F)‐negative myeloproliferative disorders. Blood 111:1686–1689. [DOI] [PubMed] [Google Scholar]
- 114. Shan Y, Gnanasambandan K, Ungureanu D, Kim ET, Hammaren H, Yamashita K, Silvennoinen O, Shaw DE, Hubbard SR (2014) Molecular basis for pseudokinase‐dependent autoinhibition of JAK2 tyrosine kinase. Nat Struct Mol Biol 21:579–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Knighton DR, Zheng JH, Ten Eyck LF, Ashford VA, Xuong NH, Taylor SS, Sowadski JM (1991) Crystal structure of the catalytic subunit of cyclic adenosine monophosphate‐dependent protein kinase. Science 253:407–414. [DOI] [PubMed] [Google Scholar]
- 116. Williams NK, Bamert RS, Patel O, Wang C, Walden PM, Wilks AF, Fantino E, Rossjohn J, Lucet IS (2009) Dissecting specificity in the Janus kinases: the structures of JAK‐specific inhibitors complexed to the JAK1 and JAK2 protein tyrosine kinase domains. J Mol Biol 387:219–232. [DOI] [PubMed] [Google Scholar]
- 117. Lucet IS, Fantino E, Styles M, Bamert R, Patel O, Broughton SE, Walter M, Burns CJ, Treutlein H, Wilks AF, Rossjohn J (2006) The structural basis of Janus kinase 2 inhibition by a potent and specific pan‐Janus kinase inhibitor. Blood 107:176–183. [DOI] [PubMed] [Google Scholar]
- 118. Boggon TJ, Li Y, Manley PW, Eck MJ (2005) Crystal structure of the Jak3 kinase domain in complex with a staurosporine analog. Blood 106:996–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Chrencik JE, Patny A, Leung IK, Korniski B, Emmons TL, Hall T, Weinberg RA, Gormley JA, Williams JM, Day JE, Hirsch JL, Kiefer JR, Leone JW, Fischer HD, Sommers CD, Huang HC, Jacobsen EJ, Tenbrink RE, Tomasselli AG, Benson TE (2010) Structural and thermodynamic characterization of the TYK2 and JAK3 kinase domains in complex with CP‐690550 and CMP‐6. J Mol Biol 400:413–433. [DOI] [PubMed] [Google Scholar]
- 120. Hanks SK, Quinn AM, Hunter T (1988) The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 241:42–52. [DOI] [PubMed] [Google Scholar]
- 121. Scheeff ED, Eswaran J, Bunkoczi G, Knapp S, Manning G (2009) Structure of the pseudokinase VRK3 reveals a degraded catalytic site, a highly conserved kinase fold, and a putative regulatory binding site. Struct Fold Des 17:128–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Kornev AP, Haste NM, Taylor SS, Eyck LF (2006) Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc Natl Acad Sci U S A 103:17783–17788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Kornev AP, Taylor SS, Ten Eyck LF (2008) A helix scaffold for the assembly of active protein kinases. Proc Natl Acad Sci U S A 105:14377–14382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Hubbard SR, Wei L, Ellis L, Hendrickson WA (1994) Crystal structure of the tyrosine kinase domain of the human insulin receptor. Nature 372:746–754. [DOI] [PubMed] [Google Scholar]
- 125. Wei L, Hubbard SR, Hendrickson WA, Ellis L (1995) Expression, characterization, and crystallization of the catalytic core of the human insulin receptor protein‐tyrosine kinase domain. J Biol Chem 270:8122–8130. [DOI] [PubMed] [Google Scholar]
- 126. Wu J, Li W, Craddock BP, Foreman KW, Mulvihill MJ, Ji QS, Miller WT, Hubbard SR (2008) Small‐molecule inhibition and activation‐loop trans‐phosphorylation of the IGF1 receptor. EMBO J 27:1985–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Hubbard SR (1997) Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. EMBO J 16:5572–5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Babon JJ, Kershaw NJ, Murphy JM, Varghese LN, Laktyushin A, Young SN, Lucet IS, Norton RS, Nicola NA (2012) Suppression of cytokine signaling by SOCS3: characterization of the mode of inhibition and the basis of its specificity. Immunity 36:239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Darnell JE (1997) STATs and gene regulation. Science 277:1630–1635. [DOI] [PubMed] [Google Scholar]
- 130. Shuai K, Schindler C, Prezioso VR, Darnell JE Jr (1992) Activation of transcription by IFN‐: tyrosine phosphorylation of a 91‐kD DNA binding protein. Science 258:1812. [DOI] [PubMed] [Google Scholar]
- 131. Darnell JE Jr, Kerr IM, Stark GR (1994) Jak‐STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415–1420. [DOI] [PubMed] [Google Scholar]
- 132. Zhong Z, Wen Z, Darnell JE (1994) Stat3 and Stat4: members of the family of signal transducers and activators of transcription. Proc Natl Acad Sci U S A 91:4806–4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Shuai K, Stark GR, Kerr IM, Darnell JE (1993) A single phosphotyrosine residue of Stat91 required for gene activation by interferon‐gamma. Science 261:1744–1746. [DOI] [PubMed] [Google Scholar]
- 134. Copeland NG, Gilbert DJ, Schindler C, Zhong Z, Wen Z, Darnell JE, Mui AL‐F, Miyajima A, Quelle FW, Ihle JN (1995) Distribution of the mammalian Stat gene family in mouse chromosomes. Genomics 29:225–228. [DOI] [PubMed] [Google Scholar]
- 135. Mao X, Ren Z, Parker GN, Sondermann H, Pastorello MA, Wang W, McMurray JS, Demeler B, Darnell JE Jr, Chen X (2005) Structural bases of unphosphorylated STAT1 association and receptor binding. Mol Cell 17:761–771. [DOI] [PubMed] [Google Scholar]
- 136. Chen X, Vinkemeier U, Zhao Y, Jeruzalmi D, Darnell JE Jr, Kuriyan J (1998) Crystal structure of a tyrosine phosphorylated STAT‐1 dimer bound to DNA. Cell 93:827–839. [DOI] [PubMed] [Google Scholar]
- 137. Shuai K, Stark GR, Kerr IM, Darnell JE Jr (1993) A single phosphotyrosine residue of Stat91 required for gene activation by interferon. Science 261:1744–1746. [DOI] [PubMed] [Google Scholar]
- 138. Rengachari S, Groiss S, Devos JM, Caron E, Grandvaux N, Panne D (2018) Structural basis of STAT2 recognition by IRF9 reveals molecular insights into ISGF3 function. Proc Natl Acad Sci U S A 115:E601–E609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Vinkemeier U, Moarefi I, Darnell JE, Kuriyan J (1998) Structure of the amino‐terminal protein interaction domain of STAT‐4. Science 279:1048–1052. [DOI] [PubMed] [Google Scholar]
- 140. Hu T, Yeh JE, Pinello L, Jacob J, Chakravarthy S, Yuan G‐C, Chopra R, Frank DA (2015) Impact of the N‐terminal domain of STAT3 in STAT3‐dependent transcriptional activity. Mol Cell Biol 35:3284–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Strehlow I, Schindler C (1998) Amino‐terminal signal transducer and activator of transcription (STAT) domains regulate nuclear translocation and STAT deactivation. J Biol Chem 273:28049–28056. [DOI] [PubMed] [Google Scholar]
- 142. Xu X, Sun Y‐L, Hoey T (1996) Cooperative DNA binding and sequence‐selective recognition conferred by the STAT amino‐terminal domain. Science 273:794–797. [DOI] [PubMed] [Google Scholar]
- 143. Horvath CM, Wen Z, Darnell JE (1995) A STAT protein domain that determines DNA sequence recognition suggests a novel DNA‐binding domain. Genes Dev 9:984–994. [DOI] [PubMed] [Google Scholar]
- 144. Decker T, Kovarik P, Meinke A (1997) GAS elements: a few nucleotides with a major impact on cytokine‐induced gene expression. J Interferon Cytokine Res 17:121–134. [DOI] [PubMed] [Google Scholar]
- 145. Ehret GB, Reichenbach P, Schindler U, Horvath CM, Fritz S, Nabholz M, Bucher P (2001) DNA binding specificity of different STAT proteins: comparison of in vitro specificity with natural target sites. J Biol Chem 276:6675–6688. [DOI] [PubMed] [Google Scholar]
- 146. Li J, Rodriguez JP, Niu F, Pu M, Wang J, Hung L‐W, Shao Q, Zhu Y, Ding W, Liu Y (2016) Structural basis for DNA recognition by STAT6. Proc Natl Acad Sci U S A 113:13015–13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Gupta S, Yan H, Wong LH, Ralph S, Krolewski J, Schindler C (1996) The SH2 domains of Stat1 and Stat2 mediate multiple interactions in the transduction of IFN‐alpha signals. EMBO J 15:1075–1084. [PMC free article] [PubMed] [Google Scholar]
- 148. Yan H, Krishnan K, Greenlund A, Gupta S, Lim J, Schreiber R, Schindler C, Krolewski J (1996) Phosphorylated interferon‐alpha receptor 1 subunit (IFNaR1) acts as a docking site for the latent form of the 113 kDa STAT2 protein. EMBO J 15:1064–1074. [PMC free article] [PubMed] [Google Scholar]
- 149. Yamamoto K, Miura O, Hirosawa S, Miyasaka N (1997) Binding sequence of STAT4: STAT4 complex recognizes the IFN‐γ activation site (GAS)‐like sequence (T/A) TTCC (C/G) GGAA (T/A). Biochem Biophys Res Commun 233:126–132. [DOI] [PubMed] [Google Scholar]
- 150. Heim MH, Kerr IM, Stark GR, Darnell JE (1995) Contribution of STAT SH2 groups to specific interferon signaling by the Jak‐STAT pathway. Science 267:1347–1349. [DOI] [PubMed] [Google Scholar]
- 151. Shuai K, Horvath CM, Huang LHT, Qureshi SA, Cowburn D, Darnell JE Jr (1994) Interferon activation of the transcription factor Stat91 involves dimerization through SH2‐phosphotyrosyl peptide interactions. Cell 76:821–828. [DOI] [PubMed] [Google Scholar]
- 152. Lin J‐X, Migone T‐S, Tseng M, Friedmann M, Weatherbee JA, Zhou L, Yamauchi A, Bloom ET, Mietz J, John S (1995) The role of shared receptor motifs and common Stat proteins in the generation of cytokine pleiotropy and redundancy by IL‐2, IL‐4, IL‐7, IL‐13, and IL‐15. Immunity 2:331–339. [DOI] [PubMed] [Google Scholar]
- 153. Ren Z, Mao X, Mertens C, Krishnaraj R, Qin J, Mandal PK, Romanowski MJ, McMurray JS, Chen X (2008) Crystal structure of unphosphorylated STAT3 core fragment. Biochem Biophys Res Commun 374:1–5. [DOI] [PubMed] [Google Scholar]
- 154. Neculai D, Neculai AM, Verrier S, Straub K, Klumpp K, Pfitzner E, Becker S (2005) Structure of the unphosphorylated STAT5a dimer. J Biol Chem 280:40782–40787. [DOI] [PubMed] [Google Scholar]
- 155. Good SR, Thieu VT, Mathur AN, Yu Q, Stritesky GL, Yeh N, O'Malley JT, Perumal NB, Kaplan MH (2009) Temporal induction pattern of STAT4 target genes defines potential for Th1 lineage‐specific programming. J Immunol 183:3839–3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Satoh J‐i, Tabunoki H (2013) A comprehensive profile of ChIP‐Seq‐based STAT1 target genes suggests the complexity of STAT1‐mediated gene regulatory mechanisms. Gene Regul Syst Biol 7:S11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Alvarez JV, Frank DA (2004) Genome‐wide analysis of STAT target genes: elucidating the mechanism of STAT‐mediated oncogenesis. Cancer Biol Ther 3:1045–1050. [DOI] [PubMed] [Google Scholar]
- 158. Tripathi SK, Chen Z, Larjo A, Kanduri K, Nousiainen K, Äijo T, Ricaño‐Ponce I, Hrdlickova B, Tuomela S, Laajala E (2017) Genome‐wide analysis of STAT3‐mediated transcription during early human Th17 cell differentiation. Cell Rep 19:1888–1901. [DOI] [PubMed] [Google Scholar]
- 159. Villarino AV, Sciumè G, Davis FP, Iwata S, Zitti B, Robinson GW, Hennighausen L, Kanno Y, O'Shea JJ (2017) Subset‐and tissue‐defined STAT5 thresholds control homeostasis and function of innate lymphoid cells. J Exp Med 214:2999–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Yamamoto T, Sekine Y, Kashima K, Kubota A, Sato N, Aoki N, Matsuda T (2002) The nuclear isoform of protein‐tyrosine phosphatase TC‐PTP regulates interleukin‐6‐mediated signaling pathway through STAT3 dephosphorylation. Biochem Biophys Res Commun 297:811–817. [DOI] [PubMed] [Google Scholar]
- 161. Haspel RL, Darnell JE (1999) A nuclear protein tyrosine phosphatase is required for the inactivation of Stat1. Proc Natl Acad Sci U S A 96:10188–10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. ten Hoeve J, de Jesus Ibarra‐Sanchez M, Fu Y, Zhu W, Tremblay M, David M, Shuai K (2002) Identification of a nuclear Stat1 protein tyrosine phosphatase. Mol Cell Biol 22:5662–5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Chatterjee‐Kishore M, Wright KL, Ting JPY, Stark GR (2000) How Stat1 mediates constitutive gene expression: a complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. EMBO J 19:4111–4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Yang J, Liao X, Agarwal MK, Barnes L, Auron PE, Stark GR (2007) Unphosphorylated STAT3 accumulates in response to IL‐6 and activates transcription by binding to NFκB. Genes Dev 21:1396–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Yan S‐J, Lim SJ, Shi S, Dutta P, Li WX (2011) Unphosphorylated STAT and heterochromatin protect genome stability. FASEB J 25:232–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Shi S, Larson K, Guo D, Lim SJ, Dutta P, Yan S‐J, Li WX (2008) Drosophila STAT is required for directly maintaining HP1 localization and heterochromatin stability. Nat Cell Biol 10:489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Park HJ, Li J, Hannah R, Biddie S, Leal‐Cervantes AI, Kirschner K, Flores Santa Cruz D, Sexl V, Gottgens B, Green AR (2016) Cytokine‐induced megakaryocytic differentiation is regulated by genome‐wide loss of a uSTAT transcriptional program. EMBO J 35:580–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Shuai K, Ziemiecki A, Wilks AF, Harpur AG, Sadowski HB, Gilman MZ, Darnell JE (1993) Polypeptide signalling to the nucleus through tyrosine phosphorylation of Jak and Stat proteins. Nature 366:580–583. [DOI] [PubMed] [Google Scholar]
- 169. Naka T, Narazaki M, Hirata M, Matsumoto T, Minamoto S, Aono A, Nishimoto N, Kajita T, Taga T, Yoshizaki K, Akira S, Kishimoto T (1997) Structure and function of a new STAT‐induced STAT inhibitor. Nature 387:924–929. [DOI] [PubMed] [Google Scholar]
- 170. Starr R, Willson TA, Viney EM, Murray LJ, Rayner JR, Jenkins BJ, Gonda TJ, Alexander WS, Metcalf D, Nicola NA, Hilton DJ (1997) A family of cytokine‐inducible inhibitors of signalling. Nature 387:917–921. [DOI] [PubMed] [Google Scholar]
- 171. Hilton DJ, Richardson RT, Alexander WS, Viney EM, Willson TA, Sprigg NS, Starr R, Nicholson SE, Metcalf D, Nicola NA (1998) Twenty proteins containing a C‐terminal SOCS box form five structural classes. Proc Natl Acad Sci U S A 95:114–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Croker BA, Krebs DL, Zhang JG, Wormald S, Willson TA, Stanley EG, Robb L, Greenhalgh CJ, Forster I, Clausen BE, Nicola NA, Metcalf D, Hilton DJ, Roberts AW, Alexander WS (2003) SOCS3 negatively regulates IL‐6 signaling in vivo. Nat Immunol 4:540–545. [DOI] [PubMed] [Google Scholar]
- 173. Croker BA, Metcalf D, Robb L, Wei W, Mifsud S, DiRago L, Cluse LA, Sutherland KD, Hartley L, Williams E, Zhang JG, Hilton DJ, Nicola NA, Alexander WS, Roberts AW (2004) SOCS3 is a critical physiological negative regulator of G‐CSF signaling and emergency granulopoiesis. Immunity 20:153–165. [DOI] [PubMed] [Google Scholar]
- 174. Hamanaka I, Saito Y, Yasukawa H, Kishimoto I, Kuwahara K, Miyamoto Y, Harada M, Ogawa E, Kajiyama N, Takahashi N, Izumi T, Kawakami R, Masuda I, Yoshimura A, Nakao K (2001) Induction of JAB/SOCS‐1/SSI‐1 and CIS3/SOCS‐3/SSI‐3 is involved in gp130 resistance in cardiovascular system in rat treated with cardiotrophin‐1 in vivo. Circ Res 88:727–732. [DOI] [PubMed] [Google Scholar]
- 175. Bjorbaek C, Elmquist JK, El‐Haschimi K, Kelly J, Ahima RS, Hileman S, Flier JS (1999) Activation of SOCS‐3 messenger ribonucleic acid in the hypothalamus by ciliary neurotrophic factor. Endocrinology 140:2035–2043. [DOI] [PubMed] [Google Scholar]
- 176. Magrangeas F, Boisteau O, Denis S, Jacques Y, Minvielle S (2001) Negative regulation of onconstatin M signaling by suppressor of cytokine signaling (SOCS‐3). Eur Cytokine Netw 12:309–315. [PubMed] [Google Scholar]
- 177. Adams TE, Hansen JA, Starr R, Nicola NA, Hilton DJ, Billestrup N (1998) Growth hormone preferentially induces the rapid, transient expression of SOCS‐3, a novel inhibitor of cytokine receptor signaling. J Biol Chem 273:1285–1287. [DOI] [PubMed] [Google Scholar]
- 178. Kotenko SV, Izotova LS, Mirochnitchenko OV, Esterova E, Dickensheet H, Donnelly RP, Pestka S (2001) Identification, cloning, and characterization of a novel soluble receptor that binds IL‐22 and neutralizes its activity. J Immunol 166:7096–7103. [DOI] [PubMed] [Google Scholar]
- 179. Strengell M, Lehtonen A, Matikainen S, Julkunen I (2006) IL‐21 enhances SOCS gene expression and inhibits LPS‐induced cytokine production in human monocyte‐derived dendritic cells. J Leukoc Biol 79:1279–1285. [DOI] [PubMed] [Google Scholar]
- 180. Auernhammer CJ, Melmed S (1999) Interleukin‐11 stimulates proopiomelanocortin gene expression and adrenocorticotropin secretion in corticotroph cells: evidence for a redundant cytokine network in the hypothalamo‐pituitary‐adrenal axis. Endocrinology 140:1559–1566. [DOI] [PubMed] [Google Scholar]
- 181. Shen X, Hong F, Nguyen VA, Gao B (2000) IL‐10 attenuates IFN‐alpha‐activated STAT1 in the liver: involvement of SOCS2 and SOCS3. FEBS Lett 480:132–136. [DOI] [PubMed] [Google Scholar]
- 182. Lejeune D, Demoulin JB, Renauld JC (2001) Interleukin 9 induces expression of three cytokine signal inhibitors: cytokine‐inducible SH2‐containing protein, suppressor of cytokine signalling (SOCS)‐2 and SOCS‐3, but only SOCS‐3 overexpression suppresses interleukin 9 signalling. Biochem J 353:109–116. [PMC free article] [PubMed] [Google Scholar]
- 183. Bjorbaek C, Elmquist JK, Frantz JD, Shoelson SE, Flier JS (1998) Identification of SOCS‐3 as a potential mediator of central leptin resistance. Mol Cell 1:619–625. [DOI] [PubMed] [Google Scholar]
- 184. Pezet A, Favre H, Kelly PA, Edery M (1999) Inhibition and restoration of prolactin signal transduction by suppressors of cytokine signaling. J Biol Chem 274:24497–24502. [DOI] [PubMed] [Google Scholar]
- 185. Zimmerer JM, Lesinski GB, Kondadasula SV, Karpa VI, Lehman A, Raychaudhury A, Becknell B, Carson WE 3rd (2007) IFN‐alpha‐induced signal transduction, gene expression, and antitumor activity of immune effector cells are negatively regulated by suppressor of cytokine signaling proteins. J Immunol 178:4832–4845. [DOI] [PubMed] [Google Scholar]
- 186. Mori H, Hanada R, Hanada T, Aki D, Mashima R, Nishinakamura H, Torisu T, Chien KR, Yasukawa H, Yoshimura A (2004) Socs3 deficiency in the brain elevates leptin sensitivity and confers resistance to diet‐induced obesity. Nat Med 10:739–743. [DOI] [PubMed] [Google Scholar]
- 187. Greenhalgh CJ, Rico‐Bautista E, Lorentzon M, Thaus AL, Morgan PO, Willson TA, Zervoudakis P, Metcalf D, Street I, Nicola NA, Nash AD, Fabri LJ, Norstedt G, Ohlsson C, Flores‐Morales A, Alexander WS, Hilton DJ (2005) SOCS2 negatively regulates growth hormone action in vitro and in vivo. J Clin Invest 115:397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Fairlie WD, De Souza D, Nicola NA, Baca M (2003) Negative regulation of gp130 signalling mediated through tyrosine‐757 is not dependent on the recruitment of SHP2. Biochem J 372:495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Schmitz J, Weissenbach M, Haan S, Heinrich PC, Schaper F (2000) SOCS3 exerts its inhibitory function on interleukin‐6 signal transduction through the SHP2 recruitment site of gp130. J Biol Chem 275:12848–12856. [DOI] [PubMed] [Google Scholar]
- 190. Stebbins CE, Kaelin WG Jr, Pavletich NP (1999) Structure of the VHL‐ElonginC‐ElonginB complex: implications for VHL tumor suppressor function. Science 284:455–461. [DOI] [PubMed] [Google Scholar]
- 191. Zhang JG, Farley A, Nicholson SE, Willson TA, Zugaro LM, Simpson RJ, Moritz RL, Cary D, Richardson R, Hausmann G, Kile BJ, Kent SB, Alexander WS, Metcalf D, Hilton DJ, Nicola NA, Baca M (1999) The conserved SOCS box motif in suppressors of cytokine signaling binds to elongins B and C and may couple bound proteins to proteasomal degradation. Proc Natl Acad Sci U S A 96:2071–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Babon JJ, Sabo JK, Soetopo A, Yao S, Bailey MF, Zhang JG, Nicola NA, Norton RS (2008) The SOCS box domain of SOCS3: structure and interaction with the elonginBC‐cullin5 ubiquitin ligase. J Mol Biol 381:928–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Babon JJ, Sabo JK, Zhang JG, Nicola NA, Norton RS (2009) The SOCS box encodes a hierarchy of affinities for Cullin5: Implications for ubiquitin ligase formation and cytokine signalling suppression. J Mol Biol 387:162–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Kamura T, Maenaka K, Kotoshiba S, Matsumoto M, Kohda D, Conaway RC, Conaway JW, Nakayama KI (2004) VHL‐box and SOCS‐box domains determine binding specificity for Cul2‐Rbx1 and Cul5‐Rbx2 modules of ubiquitin ligases. Genes Dev 18:3055–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Mahrour N, Redwine WB, Florens L, Swanson SK, Martin‐Brown S, Bradford WD, Staehling‐Hampton K, Washburn MP, Conaway RC, Conaway JW (2008) Characterization of Cullin‐box sequences that direct recruitment of Cul2‐Rbx1 and Cul5‐Rbx2 modules to Elongin BC‐based ubiquitin ligases. J Biol Chem 283:8005–8013. [DOI] [PubMed] [Google Scholar]
- 196. Sasaki A, Yasukawa H, Suzuki A, Kamizono S, Syoda T, Kinjyo I, Sasaki M, Johnston JA, Yoshimura A (1999) Cytokine‐inducible SH2 protein‐3 (CIS3/SOCS3) inhibits Janus tyrosine kinase by binding through the N‐terminal kinase inhibitory region as well as SH2 domain. Genes Cells 4:339–351. [DOI] [PubMed] [Google Scholar]
- 197. Yasukawa H, Misawa H, Sakamoto H, Masuhara M, Sasaki A, Wakioka T, Ohtsuka S, Imaizumi T, Matsuda T, Ihle JN, Yoshimura A (1999) The JAK‐binding protein JAB inhibits Janus tyrosine kinase activity through binding in the activation loop. EMBO J 18:1309–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Babon JJ, Yao S, DP DS, Harrison CF, Fabri LJ, Liepinsh E, Scrofani SD, Baca M, Norton RS (2005) Secondary structure assignment of mouse SOCS3 by NMR defines the domain boundaries and identifies an unstructured insertion in the SH2 domain. FEBS J 272:6120–6130. [DOI] [PubMed] [Google Scholar]
- 199. Babon JJ, McManus EJ, Yao S, DeSouza DP, Mielke LA, Sprigg NS, Willson TA, Hilton DJ, Nicola NA, Baca M, Nicholson SE, Norton RS (2006) The structure of SOCS3 reveals the basis of the extended SH2 domain function and identifies an unstructured insertion that regulates stability. Mol Cell 22:205–216. [DOI] [PubMed] [Google Scholar]
- 200. Kershaw NJ, Murphy JM, Liau NP, Varghese LN, Laktyushin A, Whitlock EL, Lucet IS, Nicola NA, Babon JJ (2013) SOCS3 binds specific receptor‐JAK complexes to control cytokine signaling by direct kinase inhibition. Nat Struct Mol Biol 20:469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Liau NPD, Laktyushin A, Lucet IS, Murphy JM, Yao S, Whitlock E, Callaghan K, Nicola NA, Kershaw NJ, Babon JJ (2018) The molecular basis of JAK/STAT inhibition by SOCS1. Nat Commun 9:1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Yasukawa H, Ohishi M, Mori H, Murakami M, Chinen T, Aki D, Hanada T, Takeda K, Akira S, Hoshijima M, Hirano T, Chien KR, Yoshimura A (2003) IL‐6 induces an anti‐inflammatory response in the absence of SOCS3 in macrophages. Nat Immunol 4:551–556. [DOI] [PubMed] [Google Scholar]
- 203. Kinjyo I, Inoue H, Hamano S, Fukuyama S, Yoshimura T, Koga K, Takaki H, Himeno K, Takaesu G, Kobayashi T, Yoshimura A (2006) Loss of SOCS3 in T helper cells resulted in reduced immune responses and hyperproduction of interleukin 10 and transforming growth factor‐beta 1. J Exp Med 203:1021–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Ogata H, Chinen T, Yoshida T, Kinjyo I, Takaesu G, Shiraishi H, Iida M, Kobayashi T, Yoshimura A (2006) Loss of SOCS3 in the liver promotes fibrosis by enhancing STAT3‐mediated TGF‐beta 1 production. Oncogene 25:2520–2530. [DOI] [PubMed] [Google Scholar]
- 205. Babon JJ, Nicola NA (2013) The biology and mechanism of action of suppressor of cytokine signaling 3. Growth Factors 30:207–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Alexander WS, Starr R, Fenner JE, Scott CL, Handman E, Sprigg NS, Corbin JE, Cornish AL, Darwiche R, Owczarek CM, Kay TW, Nicola NA, Hertzog PJ, Metcalf D, Hilton DJ (1999) SOCS1 is a critical inhibitor of interferon gamma signaling and prevents the potentially fatal neonatal actions of this cytokine. Cell 98:597–608. [DOI] [PubMed] [Google Scholar]
- 207. Bullen DV, Darwiche R, Metcalf D, Handman E, Alexander WS (2001) Neutralization of interferon‐gamma in neonatal SOCS1−/− mice prevents fatty degeneration of the liver but not subsequent fatal inflammatory disease. Immunology 104:92–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Sporri B, Kovanen PE, Sasaki A, Yoshimura A, Leonard WJ (2001) JAB/SOCS1/SSI‐1 is an interleukin‐2‐induced inhibitor of IL‐2 signaling. Blood 97:221–226. [DOI] [PubMed] [Google Scholar]
- 209. Tonks NK, Neel BG (2001) Combinatorial control of the specificity of protein tyrosine phosphatases. Curr Opin Cell Biol 13:182–195. [DOI] [PubMed] [Google Scholar]
- 210. Hunter T (1995) Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell 80:225–236. [DOI] [PubMed] [Google Scholar]
- 211. Neel BG (1993) Structure and function of SH2‐domain containing tyrosine phosphatases. Semin Cell Biol 4:419–432. [DOI] [PubMed] [Google Scholar]
- 212. Irie‐Sasaki J, Sasaki T, Matsumoto W, Opavsky A, Cheng M, Welstead G, Griffiths E, Krawczyk C, Richardson CD, Aitken K (2001) CD45 is a JAK phosphatase and negatively regulates cytokine receptor signalling. Nature 409:349–354. [DOI] [PubMed] [Google Scholar]
- 213. Zhang X, Guo A, Yu J, Possemato A, Chen Y, Zheng W, Polakiewicz RD, Kinzler KW, Vogelstein B, Velculescu VE (2007) Identification of STAT3 as a substrate of receptor protein tyrosine phosphatase T. Proc Natl Acad Sci U S A 104:4060–4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Frank DA, Mahajan S, Ritz J (1997) B lymphocytes from patients with chronic lymphocytic leukemia contain signal transducer and activator of transcription (STAT) 1 and STAT3 constitutively phosphorylated on serine residues. J Clin Invest 100:3140–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. David M, Chen HE, Goelz S, Larner AC, Neel BG (1995) Differential regulation of the alpha/beta interferon‐stimulated Jak/Stat pathway by the SH2 domain‐containing tyrosine phosphatase SHPTP1. Mol Cell Biol 15:7050–7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Yetter A, Uddin S, Krolewski JJ, Jiao H, Yi T, Platanias LC (1995) Association of the interferon‐dependent tyrosine kinase Tyk‐2 with the hematopoietic cell phosphatase. J Biol Chem 270:18179–18182. [DOI] [PubMed] [Google Scholar]
- 217. Bone H, Dechert U, Jirik F, Schrader JW, Welham MJ (1997) SHP1 and SHP2 protein‐tyrosine phosphatases associate with βc after interleukin‐3‐induced receptor tyrosine phosphorylation. Identification of potential binding sites and substrates. J Biol Chem 272:14470–14476. [DOI] [PubMed] [Google Scholar]
- 218. Chim C‐S, Fung T‐K, Cheung W‐C, Liang R, Kwong Y‐L (2004) SOCS1 and SHP1 hypermethylation in multiple myeloma: implications for epigenetic activation of the Jak/STAT pathway. Blood 103:4630–4635. [DOI] [PubMed] [Google Scholar]
- 219. Hayakawa F, Towatari M, Iida H, Wakao H, Kiyoi H, Naoe T, Saito H (1998) Differential constitutive activation between STAT‐related proteins and MAP kinase in primary acute myelogenous leukaemia. Br J Haematol 101:521–528. [DOI] [PubMed] [Google Scholar]
- 220. Guan K, Haun RS, Watson SJ, Geahlen RL, Dixon JE (1990) Cloning and expression of a protein‐tyrosine‐phosphatase. Proc Natl Acad Sci U S A 87:1501–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Brown‐Shimer S, Johnson KA, Lawrence JB, Johnson C, Bruskin A, Green NR, Hill DE (1990) Molecular cloning and chromosome mapping of the human gene encoding protein phosphotyrosyl phosphatase 1B. Proc Natl Acad Sci U S A 87:5148–5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Frangioni JV, Beahm PH, Shifrin V, Jost CA, Neel BG (1992) The nontransmembrane tyrosine phosphatase PTP‐1B localizes to the endoplasmic reticulum via its 35 amino acid C‐terminal sequence. Cell 68:545–560. [DOI] [PubMed] [Google Scholar]
- 223. Takemoto S, Mulloy JC, Cereseto A, Migone T‐S, Patel BK, Matsuoka M, Yamaguchi K, Takatsuki K, Kamihira S, White JD (1997) Proliferation of adult T cell leukemia/lymphoma cells is associated with the constitutive activation of JAK/STAT proteins. Proc Natl Acad Sci U S A 94:13897–13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Salmeen A, Andersen JN, Myers MP, Tonks NK, Barford D (2000) Molecular basis for the dephosphorylation of the activation segment of the insulin receptor by protein tyrosine phosphatase 1B. Mol Cell 6:1401–1412. [DOI] [PubMed] [Google Scholar]
- 225. Zabolotny JM, Bence‐Hanulec KK, Stricker‐Krongrad A, Haj F, Wang Y, Minokoshi Y, Kim Y‐B, Elmquist JK, Tartaglia LA, Kahn BB (2002) PTP1B regulates leptin signal transduction in vivo. Dev Cell 2:489–495. [DOI] [PubMed] [Google Scholar]
- 226. Myers MP, Andersen JN, Cheng A, Tremblay ML, Horvath CM, Parisien J‐P, Salmeen A, Barford D, Tonks NK (2001) TYK2 and JAK2 are substrates of protein‐tyrosine phosphatase 1B. J Biol Chem 276:47771–47774. [DOI] [PubMed] [Google Scholar]
- 227. Kleppe M, Soulier J, Asnafi V, Mentens N, Hornakova T, Knoops L, Constantinescu S, Sigaux F, Meijerink JP, Vandenberghe P (2011) PTPN2 negatively regulates oncogenic JAK1 in T‐cell acute lymphoblastic leukemia. Blood 117:7090–7098. [DOI] [PubMed] [Google Scholar]
- 228. Simoncic PD, Lee‐Loy A, Barber DL, Tremblay ML, McGlade CJ (2002) The T cell protein tyrosine phosphatase is a negative regulator of janus family kinases 1 and 3. Curr Biol 12:446–453. [DOI] [PubMed] [Google Scholar]
- 229. Dhe‐Paganon S, Werner ED, Nishi M, Hansen L, Chi Y‐I, Shoelson SE (2004) A phenylalanine zipper mediates APS dimerization. Nat Struct Mol Biol 11:968–974. [DOI] [PubMed] [Google Scholar]
- 230. Tong W, Lodish HF (2004) Lnk inhibits Tpo–mpl signaling and Tpo‐mediated megakaryocytopoiesis. J Exp Med 200:569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Tong W, Zhang J, Lodish HF (2005) Lnk inhibits erythropoiesis and Epo‐dependent JAK2 activation and downstream signaling pathways. Blood 105:4604–4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Bersenev A, Wu C, Balcerek J, Tong W (2008) Lnk controls mouse hematopoietic stem cell self‐renewal and quiescence through direct interactions with JAK2. J Clin Invest 118:2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Chomarat P, Banchereau J, Davoust J, Palucka AK (2000) IL‐6 switches the differentiation of monocytes from dendritic cells to macrophages. Nat Immunol 1:510–514. [DOI] [PubMed] [Google Scholar]
- 234. McLoughlin RM, Jenkins BJ, Grail D, Williams AS, Fielding CA, Parker CR, Ernst M, Topley N, Jones SA (2005) IL‐6 trans‐signaling via STAT3 directs T cell infiltration in acute inflammation. Proc Natl Acad Sci U S A 102:9589–9594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Kopf M, Baumann H, Freer G, Freudenberg M, Lamers M, Kishimoto T, Zinkernagel R, Bluethmann H, Köhler G (1994) Impaired immune and acute‐phase responses in interleukin‐6‐deficient mice. Nature 368:339–342. [DOI] [PubMed] [Google Scholar]
- 236. Scheller J, Chalaris A, Schmidt‐Arras D, Rose‐John S (2011) The pro‐ and anti‐inflammatory properties of the cytokine interleukin‐6. Biochim Biophys Acta 1813:878–888. [DOI] [PubMed] [Google Scholar]
- 237. Waetzig GH, Rose‐John S (2012) Hitting a complex target: an update on interleukin‐6 trans‐signalling. Expert Opin Ther Targets 16:225–236. [DOI] [PubMed] [Google Scholar]
- 238. Boulanger MJ, Bankovich AJ, Kortemme T, Baker D, Garcia KC (2003) Convergent mechanisms for recognition of divergent cytokines by the shared signaling receptor gp130. Mol Cell 12:577–589. [DOI] [PubMed] [Google Scholar]
- 239. Xu Y, Kershaw NJ, Luo CS, Soo P, Pocock MJ, Czabotar PE, Hilton DJ, Nicola NA, Garrett TP, Zhang JG (2010) Crystal structure of the entire ectodomain of gp130: insights into the molecular assembly of the tall cytokine receptor complexes. J Biol Chem 285:21214–21218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Wormald S, Zhang JG, Krebes DL, Mielke LA, Silver J, Alexander WS, Speed TP, Nicola NA, Hilton DJ (2006) The comparative roles of suppressor of cytokine signaling‐1 and‐3 in the inhibition and desensitization of cytokine signaling. J Biol Chem 281:11135–11143. [DOI] [PubMed] [Google Scholar]
- 241. Herrmann A, Vogt M, Monnigmann M, Clahsen T, Sommer U, Haan S, Poli V, Heinrich PC, Muller‐Newen G (2007) Nucleocytoplasmic shuttling of persistently activated STAT3. J Cell Sci 120:3249–3261. [DOI] [PubMed] [Google Scholar]
- 242. Lang R, Pauleau AL, Parganas E, Takahashi Y, Mages J, Ihle JN, Rutschman R, Murray PJ (2003) SOCS3 regulates the plasticity of gp130 signaling. Nat Immunol 4:546–550. [DOI] [PubMed] [Google Scholar]
- 243. Neel BG, Gu H, Pao L (2003) The 'Shp'ing news: SH2 domain‐containing tyrosine phosphatases in cell signaling. Trends Biochem Sci 28:284–293. [DOI] [PubMed] [Google Scholar]
- 244. Livnah O, Stura EA, Middleton SA, Johnson DL, Jolliffe LK, Wilson LA (1999) Crystallographic evidence for preformed dimers of erythropoietin receptor before ligand activation. Science 283:987–990. [DOI] [PubMed] [Google Scholar]
- 245. Brooks AJ, Dai W, O'Mara ML, Abankwa D, Chhabra Y, Pelekanos RA, Gardon O, Tunny KA, Blucher KM, Morton CJ, Parker MW, Sierecki E, Gambin Y, Gomez GA, Alexandrov K, Wilson IA, Doxastakis M, Mark AE, Waters MJ (2014) Mechanism of activation of protein kinase JAK2 by the growth hormone receptor. Science 344:1249783. [DOI] [PubMed] [Google Scholar]
- 246. Brown RJ, Adams JJ, Pelekanos RA, Wan Y, McKinstry WJ, Palethorpe K, Seeber RM, Monks TA, Eidne KA, Parker MW, Waters MJ (2005) Model for growth hormone receptor activation based on subunit rotation within a receptor dimer. Nat Struct Mol Biol 12:814–821. [DOI] [PubMed] [Google Scholar]


