Bifidobacterium animalis subsp. lactis is widely used in human food and dietary supplements. Although well documented to be safe, B. animalis subsp. lactis strains must not contain transferable antibiotic resistance elements. Many B. animalis subsp. lactis strains have different resistance measurements despite being genetically similar, and the reasons for this are not well understood. In the current study, we sought to examine how genomic differences between two closely related industrial B. animalis subsp. lactis strains contribute to different resistance levels. This will lead to a better understanding of resistance, identify future targets for analysis of transferability, and expand our understanding of tetracycline resistance in bacteria.
KEYWORDS: Bifidobacterium animalis subsp. lactis, antibiotic resistance, tetracycline, RNA sequencing, genomics, droplet digital PCR, ATP luminescence
ABSTRACT
Commercial probiotic bacteria must be tested for acquired antibiotic resistance elements to avoid potential transfer to pathogens. The European Food Safety Authority recommends testing resistance using microdilution culture techniques previously used to establish inhibitory thresholds for the Bifidobacterium genus. Many Bifidobacterium animalis subsp. lactis strains exhibit increased resistance to tetracycline, historically attributed to the ribosomal protection gene tet(W). However, some strains that harbor genetically identical tet(W) genes show various inhibition levels, suggesting that other genetic elements also contribute to observed differences. Here, we adapted several molecular assays to confirm the inhibition of B. animalis subsp. lactis strains Bl-04 and HN019 and employed RNA sequencing to assess the transcriptional differences related to genomic polymorphisms. We detected specific stress responses to the antibiotic by correlating ATP concentration to number of viable genome copies from droplet digital PCR and found that the bacteria were still metabolically active in high drug concentrations. Transcriptional analyses revealed that several polymorphic regions, particularly a novel multidrug efflux transporter, were differentially expressed between the strains in each experimental condition, likely having phenotypic effects. We also found that the tet(W) gene was upregulated only during subinhibitory tetracycline concentrations, while two novel tetracycline resistance genes were upregulated at high concentrations. Furthermore, many genes involved in amino acid metabolism and transporter function were upregulated, while genes for complex carbohydrate utilization, protein metabolism, and clustered regularly interspaced short palindromic repeat(s) (CRISPR)-Cas systems were downregulated. These results provide high-throughput means for assessing antibiotic resistances of two highly related probiotic strains and determine the genetic network that contributes to the global tetracycline response.
IMPORTANCE Bifidobacterium animalis subsp. lactis is widely used in human food and dietary supplements. Although well documented to be safe, B. animalis subsp. lactis strains must not contain transferable antibiotic resistance elements. Many B. animalis subsp. lactis strains have different resistance measurements despite being genetically similar, and the reasons for this are not well understood. In the current study, we sought to examine how genomic differences between two closely related industrial B. animalis subsp. lactis strains contribute to different resistance levels. This will lead to a better understanding of resistance, identify future targets for analysis of transferability, and expand our understanding of tetracycline resistance in bacteria.
INTRODUCTION
Bifidobacterium animalis subsp. lactis is a Gram-positive commensal species with a long history of safe use in food (1). B. animalis subsp. lactis is often used as an ingredient in yogurt and as a main component of probiotic dietary supplements (2, 3). Probiotics are defined as live microorganisms that, when administered in adequate amounts, confer a health benefit on the host (4, 5). Indeed, clinical studies assessing supplementation with B. animalis subsp. lactis have revealed probiotic effects, such as improved gastrointestinal transit and comfort (6–8), immune modulation (9, 10), and control of abdominal fat mass (11). More recently, microbiome analysis by DNA sequencing has demonstrated that B. animalis subsp. lactis also modulates gut and colorectal cancer-associated microbiota (12, 13). Genome sequencing of B. animalis subsp. lactis strains has determined genetic modes of action for various cellular functions and showed this subspecies to be somewhat monomorphic (14–17). Although all B. animalis subsp. lactis strains characterized so far are generally considered safe for consumption and show no toxic effects during in vivo animal studies (1), many exhibit tetracycline resistance, which is typically attributed to a tet(W) gene (18, 19). Furthermore, many strains have mobile genetic elements adjacent to the tet(W) gene, which may contribute to horizontal gene transfer, as demonstrated previously (20). Attempts to force translocation of tet(W) by conjugation have failed, which suggests that that this genetic combination does not pose a major transfer threat and may therefore be safe for consumption (18, 21). Interestingly, strains of B. animalis subsp. lactis with genetically identical tet(W) and transposase genes have different inhibition levels (18, 22, 23).
Tetracycline antibiotics were first developed in the 1940s and are used for many human ailments, such as pneumonia, cholera, and malaria (24), as well as in agriculture for growth promotion (25). Tetracyclines are a family of broad-spectrum, bacteriostatic drugs that prevent the aminoacyl-tRNA from binding to the A site on the 30S ribosome, thus inhibiting the elongation step in ribosomal protein synthesis (24, 26). Expanded- and broad-spectrum tetracycline families were developed to broaden the range of targets and circumvent resistance (27). There are four known tetracycline resistance mechanisms, as follows: factor-assisted ribosomal protection, point mutation of the ribosome, removal of the drug by efflux, and enzymatic inactivation of the drug (26–28). The tet(W) gene in B. animalis subsp. lactis is a GTPase that protects the ribosome by modifying the tetracycline target binding site (29). To test inhibition levels, the European Food Safety Authority (EFSA) recommends growing an individual strain in a concentration gradient of antibiotic until visible growth is inhibited (30). Here, we use microbiological and molecular methods to examine strain-specific responses to tetracycline in B. animalis subsp. lactis Bl-04 and HN019, commercial probiotic strains that have been confirmed to have few genomic differences, while likely having phenotypic differences (15).
RESULTS
In vitro antibiotic testing.
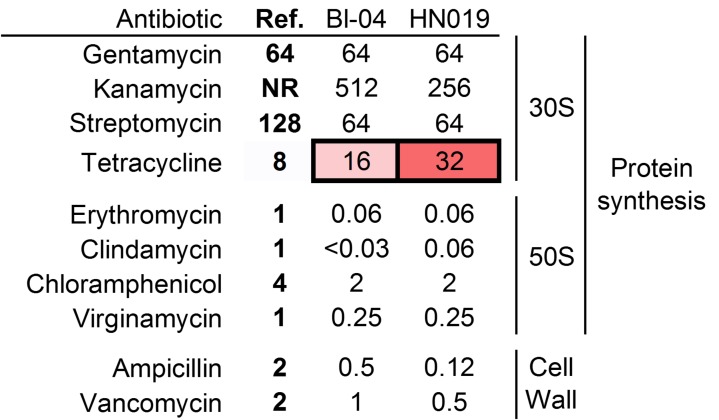
B. animalis subsp. lactis Bl-04 and HN019 were tested against the full complement of antibiotics outlined by the EFSA (Fig. 1). Interestingly, both strains have similar MIC levels for gentamicin and streptomycin, which target the 30S ribosome, at or below the recommended threshold. However, the MICs of the strains were above the recommended tetracycline threshold set for bifidobacteria (8 μg/ml), being 16 μg/ml and 32 μg/ml for Bl-04 and HN019, respectively. Furthermore, resistance to kanamycin was higher in Bl-04, although there is no recommended threshold. The strains were not above the threshold for any of the other antibiotics tested. These results demonstrate the specificity of resistance to individual drug molecules within strains.
FIG 1.
MIC profiles compared to the breakpoints for various antibiotics. Concentrations are in μg/ml. Boldface values are the thresholds for each antibiotic. Values in red are above the thresholds for bifidobacteria. Ref., threshold level for antibiotic. NR, testing was not required.
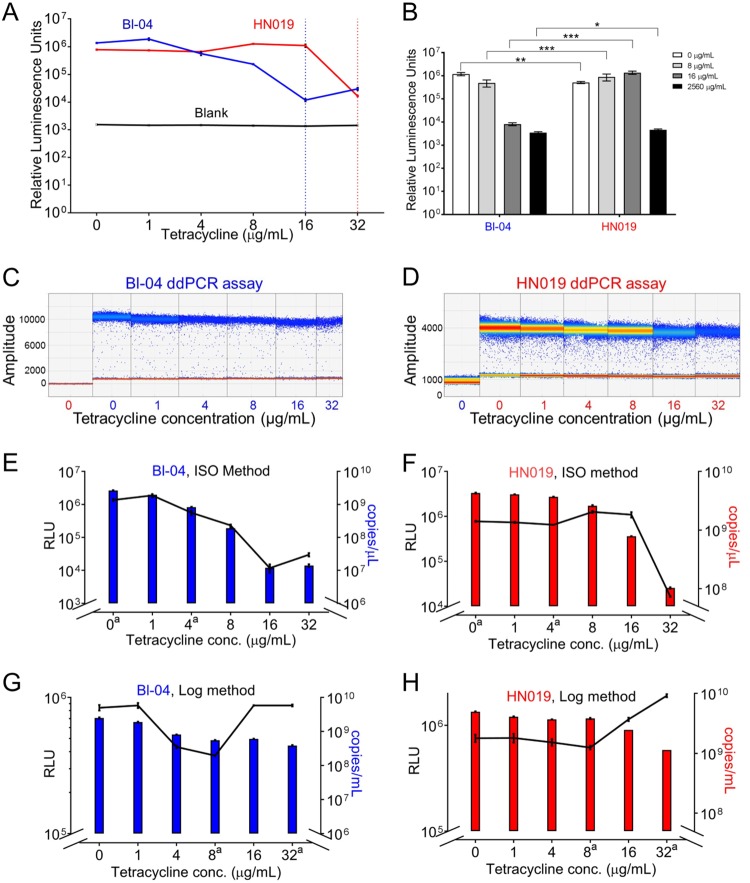
ATP concentrations were assayed to quantify any potential nonvisible growth over a range of tetracycline concentrations (Fig. 2A). ATP concentration was measured, as it is indicative of cellular metabolism and enzymatic processes (31). The tetracycline dilution series was aligned with the Bioo tetracycline detection assay standards to ensure correct amounts (Fig. S1). Both strains showed overall decreases in ATP levels at the respective MICs; however, the ATP levels did not reach the levels of the negative control. The strains were further tested at 8 and 16 μg/ml tetracycline and were significantly different at each concentration (2-sample t test; P < 0.001) (Fig. 2B). Surprisingly, the ATP concentration in HN019 increased with added tetracycline. Because ATP-activated efflux is a key resistance factor, we hypothesized that the increase in HN019 represents a stress response rather than growth. Digital PCR has recently been demonstrated to rapidly and accurately quantify probiotic cells (32), and it was used to quantify each strain separately (Fig. 2C and D). Comparing droplet digital PCR (ddPCR) to ATP concentrations (Fig. 2E and F) showed the concentration of both strains declining with increased tetracycline, which correlated with the ATP concentration for Bl-04 (P = 0.01) but not with that for HN019 (P = 0.569). We further hypothesized that measuring an acute exposure of the strains to tetracycline would demonstrate resistance more relevant for in vivo conditions, as the ISO method has a very small starting concentration of cells. The alternative method to the ISO procedure (denoted as LOG experiments) showed that ATP concentrations decreased in both strains until 8 μg/ml, followed by drastic increases in ATP compared to the slope of the ddPCR (Fig. 2G and H). Interestingly, after the initial decrease, the ATP concentrations peaked at 16 μg/ml for Bl-04 and 32 μg/ml for HN019, which are their respective MICs using the ISO method. This suggested that there are different cellular responses to tetracycline, and it might reflect normal growth behavior rather than persistence with a stress response at higher antibiotic concentrations.
FIG 2.
Assay of tetracycline response using ATP luminescence and droplet digital PCR. (A) ATP quantification using the microdilution procedure, with the dotted vertical lines showing the MIC levels for both strains. (B) The difference in ATP response between the two strains at the MIC thresholds (*, P < 0.01; **, P < 0.001; ***, P < 0.0001; paired t test). Droplet digital PCR results for the (C) Bl-04 and (D) HN019 assays show number of droplets per amplitude with a quantitative heat map applied. Sample colors in the x axes denote Bl-04 (blue) and HN019 (red) by tetracycline concentration. Droplet digital PCR assay results are shown as colored bars for (E) Bl-04 and (F) HN019 compared to ATP concentration (black lines). The experiment was repeated using acute exposure to tetracycline for (G) Bl-04 and (H) HN019. The y axes have been adjusted to better show overall correlation. aConditions used for the RNA-seq experiments.
Genome comparison.
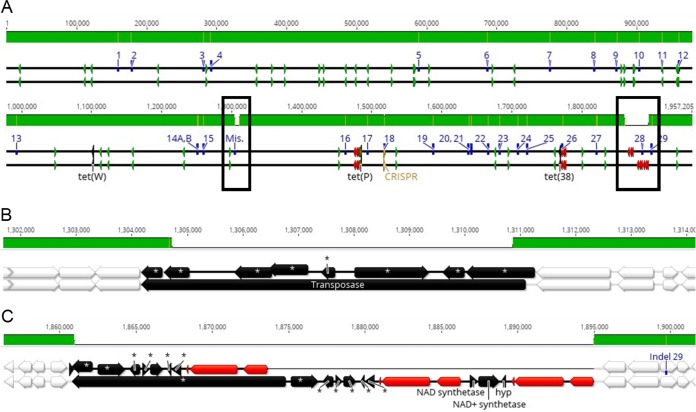
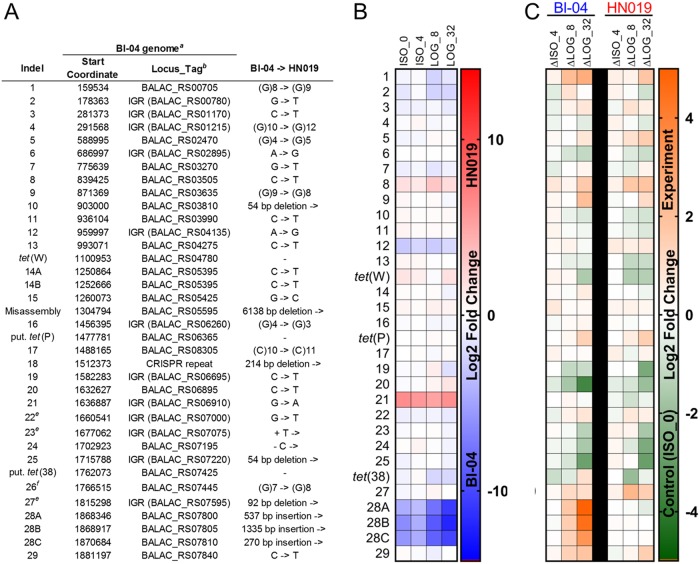
A hybrid sequencing approach for B. animalis subsp. lactis HN019 yielded a complete genome of 1,935,423 bp with an overall G+C content of 60.5%, and rapid annotation using subsystem technology (RAST) annotation predicted 1,620 coding sequences and 61 RNAs. While previous studies have compared the genomes of various B. animalis subsp. lactis strains, HN019 has been assessed either by targeted sequencing of specific genomic regions, or using a draft sequence (14, 15). Our genome sequence of HN019 showed 97.9% nucleotide pairwise identity to that of Bl-04 and was highly similar in structure and content (Fig. 3A). There were, however, genetic differences between the strains at 29 distinct genomic locations (Table S1), which were assessed using targeted PCR (Table S1). Previously reported deletions in a long-chain-fatty-acid–coenzyme A (CoA) ligase gene and clustered regularly interspaced short palindromic repeat (CRISPR) spacer region were confirmed (15). Insertion/deletion (indel) locations 22 and 23 were confirmed to be errors in the Bl-04 genome, and indels 26 and 28 could not be confirmed by PCR due to repeated nucleotide regions. There was a large difference at Bl-04 position 1304794, annotated as a transposase (BALAC_RS05595) (Fig. 3B). The HN019 genome instead encoded an ABC transporter and alpha-glucosidase that are located near indel 28 in the Bl-04 genome. Furthermore, the Bl-04 genome showed an additional rRNA operon near the same indel. Because repeated elements like transposons and rRNAs are notoriously difficult to assemble, especially without long-read sequencing technology (33), we assumed the Bl-04 genome had an assembly error and did not include genes from the misassemblies in the downstream indel analysis. Interestingly, the Bl-04 genome encoded an NAD synthetase, an NAD+ synthetase, and a hypothetical protein (predicted COG3077 DNA damage-inducible protein J) starting at position 1868346 in the Bl-04 genome that are not in the HN019 genome, although there was a different NAD synthetase in the HN019 genome at position 1244980 (Fig. 3C). The new HN019 assembly also showed two small intergenic insertions at positions 1721502 and 1820496 in the HN019 genome. Indels 14A and 14B occur in the same gene, and 14B alters a predicted restriction site found in B. animalis subsp. lactis (34). Comparing functional scenarios using RAST showed the same results in most categories, and all differences were due to the above polymorphisms. The additional stress response element was an error in the RAST prediction.
FIG 3.
Comparative genomics of B. animalis subsp. lactis Bl-04 and HN019. The HN019 genome (top black line) is aligned to Bl-04 (bottom black line) using progressiveMauve. The top green bar shows identity, with yellow and white areas showing polymorphisms and deletion events, respectively. The sequence lines show various annotations, as follows: blue, indels from Fig. 6; green, tRNAs; red, rRNAs; gold, CRISPR-Cas. The whole-genome alignment is divided into two continuous indel sequence lines (A) and zoomed in to the (B) possibly misassembled and (C) indel 28 regions. Annotations with asterisks are present elsewhere in the opposite genome. White annotations are identical between the strains. Unique gene names are annotated.
Reannotating both strains with RAST revealed seven genes related to antibiotic resistance, as follows: three for tetracycline, including tet(W), and four for fluoroquinolones. The genomes were compared to the Antibiotic Resistance Genes Database (ARDB), Comprehensive Antibiotic Resistance Database (CARD), and Antibiotic Resistance Gene-ANNOTation (Arg-Annot) using a blastx search, but no additional resistance genes met the 80% nucleotide identity threshold. The tet(W) gene and two putative tetracycline resistance genes had 100% DNA sequence identity between the two strains, and neither putative gene was in proximity (within 5 kb) to known mobile genetic elements. Both tetracycline resistance genes were further analyzed for protein function relative to tet(W), as antibiotic resistance genes share similar functionality with nonresistance genes that may be difficult to distinguish (35). The gene at Bl-04 position 1479751 was originally annotated as a translation elongation factor for GTPases (EF-G; locus tag BALAC_RS06365), and had a Tet(M) domain similar to that of the tet(W) gene. This is likely due to elongation factors binding to the same region of the ribosome as resistance proteins. The closest match to this gene in the ARDB was tet(P) from Clostridium perfringens, with a 30.1% amino acid match, which also has a 37.8% identity to the tet(W) amino acid sequence. The gene with locus tag BALAC_RS07425 at Bl-04 position 1763551 is a transport permease with amino acid regions that match antibiotic resistance transporters. The amino acid sequence is 99.8% identical to that of tet(36) in B. animalis subsp. lactis AD011. A tetR tetracycline repressor gene (BALAC_RS07655) was also homologous between the two strains. Other nonantibiotic resistance permeases are present in both strains, and efflux transport is a widespread feature, as there are 31 other membrane transport genes encoded in that genome (Table 1).
TABLE 1.
Metabolic responses to the tetracycline experiments
| RAST category |
B. lactis straina: |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bl-04 |
HN019 |
|||||||||||||
| Annotated genesb | ΔISO_4 |
ΔLOG_8 |
ΔLOG_32 |
Annotated genesc | ΔISO_4 |
ΔLOG_8 |
ΔLOG_32 |
|||||||
| ↓ | ↑ | ↓ | ↑ | ↓ | ↑ | ↓ | ↑ | ↓ | ↑ | ↓ | ↑ | |||
| Amino acids and derivatives | 200 | 1 | 17 | 13 | 24 | 200 | 1 | 1 | 17 | 11 | 18 | |||
| Carbohydrates | 158 | 15 | 12 | 41 | 23 | 163 | 20 | 12 | 59 | 9 | ||||
| Cell division and cell cycle | 24 | 1 | 2 | 24 | 1 | |||||||||
| Cell wall and capsule | 56 | 4 | 16 | 2 | 56 | 2 | 17 | 2 | ||||||
| Cofactors, etc. | 80 | 11 | 2 | 18 | 76 | 11 | 2 | 13 | ||||||
| DNA metabolism | 56 | 2 | 7 | 4 | 55 | 3 | 7 | 3 | ||||||
| Dormancy and sporulation | 1 | 1 | 1 | |||||||||||
| Fatty acids, lipids, and isoprenoids | 34 | 6 | 1 | 6 | 34 | 5 | 6 | 1 | 6 | |||||
| Iron acquisition and metabolism | 0 | 0 | ||||||||||||
| Membrane transport | 31 | 10 | 4 | 16 | 31 | 10 | 4 | 14 | ||||||
| Metabolism of aromatic compounds | 3 | 3 | ||||||||||||
| Miscellaneous | 13 | 1 | 1 | 1 | 13 | 1 | 7 | 1 | ||||||
| Motility and chemotaxis | 5 | 1 | 1 | 2 | 1 | 5 | 1 | 1 | 2 | |||||
| Nitrogen metabolism | 8 | 8 | 3 | |||||||||||
| Nucleosides and nucleotides | 67 | 5 | 6 | 67 | 2 | 8 | 1 | |||||||
| Transposable elements, etc. | 0 | 0 | ||||||||||||
| Phosphorus metabolism | 24 | 7 | 24 | 1 | 6 | |||||||||
| Photosynthesis | 0 | 0 | ||||||||||||
| Potassium metabolism | 11 | 1 | 2 | 6 | 11 | 2 | 3 | |||||||
| Protein metabolism | 176 | 1 | 30 | 4 | 176 | 4 | 12 | 6 | ||||||
| Regulation and cell signaling | 12 | 1 | 1 | 6 | 1 | 11 | 1 | 1 | 3 | |||||
| Respiration | 11 | 7 | 11 | 7 | 1 | |||||||||
| RNA metabolism | 63 | 1 | 1 | 6 | 63 | 1 | 1 | 4 | ||||||
| Secondary metabolism | 2 | 2 | 1 | 2 | ||||||||||
| Stress response | 30 | 1 | 3 | 1 | 5 | 4 | 31 | 1 | 1 | 5 | 6 | |||
| Sulfur metabolism | 18 | 2 | 1 | 2 | 7 | 18 | 3 | 1 | 2 | 8 | ||||
| Virulence, disease, and defense | 28 | 1 | 3 | 3 | 4 | 28 | 2 | 3 | 4 | 2 | ||||
| Not in subsystem | 867 | 31 | 27 | 103 | 122 | 219 | 286 | 872 | 12 | 1 | 134 | 108 | 234 | 289 |
The numbers of genes in each metabolic category that are significantly upregulated (↑) or downregulated (↓) are shown by strain and by results of each experiment compared to those of the control experiment (ISO_0).
Total number of genes annotated by RAST in the B. lactis Bl-04 genome.
Total number of genes annotated by RAST in the B. lactis HN019 genome.
Transcriptional response to tetracycline.
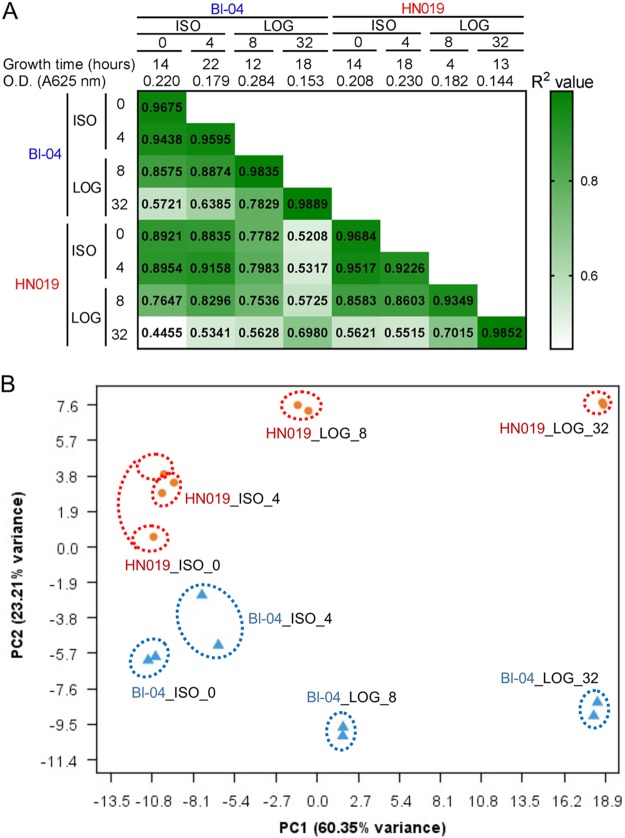
RNA transcript sequencing yielded an average of 6,594,563 paired-end reads per experiment. Although single reads were generated for each replicate, only paired-end reads were included in subsequent analysis. All read sets from both strains were mapped to the B. animalis subsp. lactis Bl-04 genome to directly analyze differences between the strains, with the Bl-04_ISO_0 experiment as the overall control. The read assemblies confirmed the PCR test findings, with indels 22, 23, and 27 incorrect in Bl-04. Reads could not be distinguished between the different RNA gene operons, so indel 26 could not be analyzed. None of the HN019 experiments resulted in reads for the NAD synthetase operon (indel 28), confirming the absence of the associated genes. The average correlation of the biological replicates was R2 = 0.9638, with the lowest correlation being R2 = 0.9226 for HN019_ISO_4 and the highest being R2 = 0.9889 for Bl-04_LOG_32 (Fig. 4A). Analysis of all replicates by principal-coordinate analysis (PCA) (Fig. 4B) showed replicates clustering better with the higher concentrations of tetracycline. Importantly, the experiments separated by concentration along the x axis and by strain along the y axis, indicating that antibiotic effect accounts for much of the observed differences, followed by strain. The overall gene expression differed from that of the control with increased tetracycline exposure (Fig. 5).
FIG 4.
Overview of the RNA sequencing results. (A) The test conditions and R2 correlations between different sample sets are shown with the heat map. Replicates are shown in the diagonal cells that match experiment names. (B) Principal-coordinate analysis shows the variation between strains determined using the DESeq2 method for Bl-04 (blue triangles) and HN019 (orange circles).
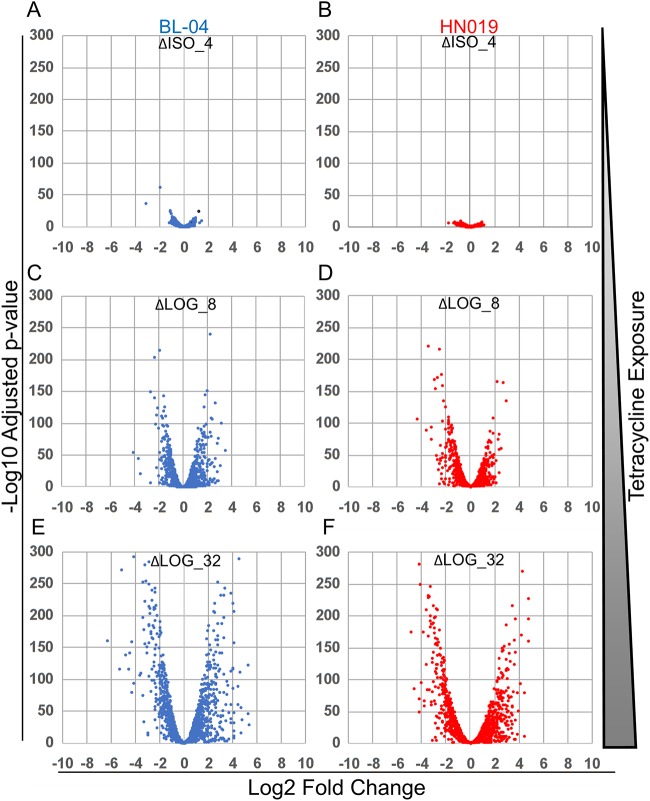
FIG 5.
Volcano plots of expression differences between Bl-04 and HN019 during different tetracycline exposures. Experimental names compared to the control ISO_0 experiment with no tetracycline are shown at the top of each plot and are as follows: (A) Bl-04 ΔISO_4, (B) HN019 ΔISO_4, (C) Bl-04 ΔLOG_8, (D) HN019 ΔLOG_8, (E) Bl-04 ΔLOG_32, and (F) HN019 ΔLOG_32. Negative log change represents higher expression in the control (ISO_0), and positive log change indicates higher expression in the respective treatments. Results are shown for Bl-04 (blue) and HN019 (red).
The assembly of reads to a single genome allowed for direct comparison of expression levels between experiments for each gene (Fig. 6). We first assessed the polymorphic differences between the two strains grown in ISO media without tetracycline (labeled ISO_0) to establish expression changes not due to tetracycline exposure. Genetic differences in intergenic regions were analyzed for potential promoters, and then the flanking genes were analyzed for transcription differences. Indels 2, 12, and 28 were significantly upregulated in Bl-04 cultures without tetracycline compared to in HN019 cultures. Indel 2 is a single-nucleotide transversion upstream of yihS, which isomerizes a range of sugars. Indel 12 is in the intergenic region upstream of a hypothetical gene related to a PAC2 family proteasome assembly chaperone and downstream of a putative promoter.
FIG 6.
Expression differences of the different polymorphic regions. (A) Overview of polymorphic regions. aIndel references are made to the Bl-04 genome. bLocus tags in parenthesis are genes associated with intergenic indels that were assessed in the expression data. eIndels are errors in the Bl-04 genome. fIndel could not be confirmed by PCR or transcript read analysis. (B) Expression differences at each locus between the strains showing upregulation in Bl-04 (blue) or HN019 (red). (C) Expression differences at each locus within the strains show upregulation in the control ISO_0 (green) or treatments (orange).
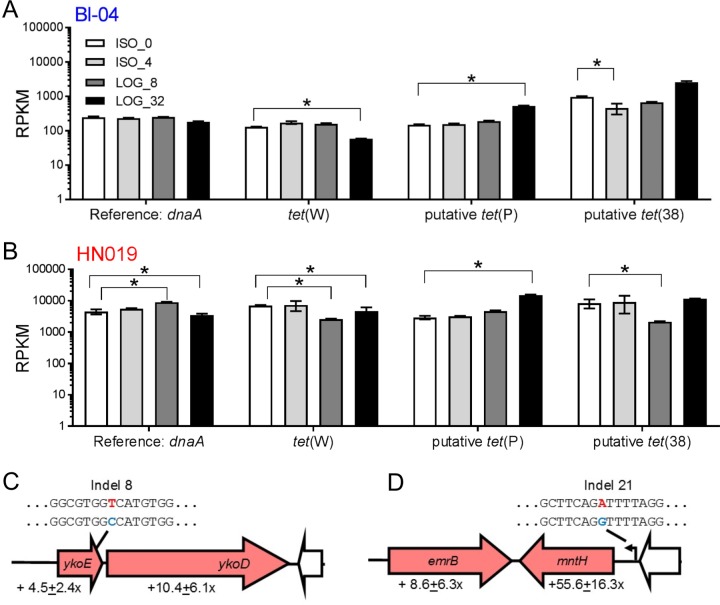
Indels 8 and 21 and tet(W) were significantly upregulated in HN019 compared to those in Bl-04 in experiment ISO_0 (Fig. 7). Indel 8 is a single-nucleotide polymorphism (SNP) that causes an alanine to valine mutation in ykoE, which transports hydroxymethylpyrimidine (HMP) and thiamine (vitamin B1). The adjacent ABC-binding protein is also upregulated in HN019. Indel 21 is in a putative promoter that may upregulate the upstream transport gene mntH (manganese, iron, and other metal transport), as well as an emrB multidrug transport gene previously annotated as a hypothetical gene. Interestingly, the mntH and emrB genes are oriented in opposite directions, yet similarly upregulated. Without the addition of tetracycline, tet(W) expression is higher in HN019, although there are no proximate genetic differences. This shows that tet(W) is constitutively expressed and may have further roles in general cell function. Furthermore, HN019 cultures generally reached the target optical density quicker than did Bl-04 cultures during each experiment (Fig. 4A), which indicates that these genetic polymorphisms provide phenotypic benefits.
FIG 7.
Differential expression of the three key genetic regions. Comparison of the raw reads per kilobase million (RPKM) for the three tetracycline resistance genes in (A) Bl-04 and (B) HN019 (*, adjusted P < 0.05 and fold change of >2×). Genomic representations of the key polymorphic regions (C) indel 8 and (D) indel 21, shared between the two strains, are represented by block arrows. Arrows are colored as follows: red, upregulated in HN019; white, identical genes; and black, putative promoter. Nucleotide identities for each strain are denoted by color, with Bl-04 and HN019 being blue and red, respectively.
The ISO_0 and ISO_4 experiments for both Bl-04 and HN019 were correlated within the range of the biological replicates (Bl-04 R2 = 0.9675; HN019 R2 = 0.9517). The expression profiles between the two strains correlated more during ISO_4 (R2 = 0.9158) than under normal growth conditions in ISO_0 (R2 = 0.9675). Twenty-seven genes were significantly upregulated in Bl-04 during exposure to ISO_4, seven of which are involved with transmembrane transport. Only one gene was assigned to a RAST category, the cold shock gene cspA, which was upregulated in the LOG experiments as well (Table 1).
Other genes, such as two cas genes in Bl-04, were significantly downregulated when there were subinhibitory levels of tetracycline, and all of the cas genes in the system were downregulated throughout the LOG tetracycline exposures (Fig. S4). The tet(W) gene expression only increased by 0.64-fold (adjusted P = 0.023). The putative tet(P) expression did not change and the putative tet(36) expression significantly decreased. Only two genes were significantly upregulated in HN019 during ISO_4, an isomerase involved in leucine biosynthesis and a transport component gene. Although tet(W) expression in HN019 was significantly higher in the ISO_0 experiment than Bl-04, it was not increased during the ISO_4 experiment. Indels 8 and 21 were also upregulated in HN019, which suggests contribution to tetracycline resistance as well as typical growth.
The in vitro testing showed Bl-04 and HN019 to have different tetracycline response thresholds, so results of the LOG_8 trial in HN019 were expected to be more related to those of the ISO trials, signifying resistance, while results of the LOG_8 trial in Bl-04 was expected to be more like those of LOG_32, since it had hit the threshold for the stress response. The PCA plot showed evidence of this, but the LOG_8 trials for the strains did not vary drastically (Fig. 4B). Nonetheless, the expression profiles for the higher concentrations were drastically different than those for the lower concentrations (Fig. 5).
The tet(W) expression decreased, while tet(P) and tet(36) expression increased with LOG exposure (Fig. 7A and B), which indicates that the latter two genes may provide better protection from the antibiotic to preserve cells in extreme exposure. Between the two strains, indels 19 and 20 showed expression differences only in the LOG experiments, which indicates tetracycline-dependent expression changes. Indel 19 is an intergenic SNP downstream of a galactokinase galK (Kyoto Encyclopedia of Genes and Genomes enzyme entry EC 2.7.1.6) that is upregulated in HN019 during LOG_8. Indel 20 is a SNP in the galactosyl transferase cpsD (EC 2.7.8.6) that was expressed higher in HN019 in LOG_8 and LOG_32. Indels 8 and 21 are again upregulated in HN019 for the LOG experiments, showing that both polymorphisms affect static expression of the associated genes (Fig. 7C and D).
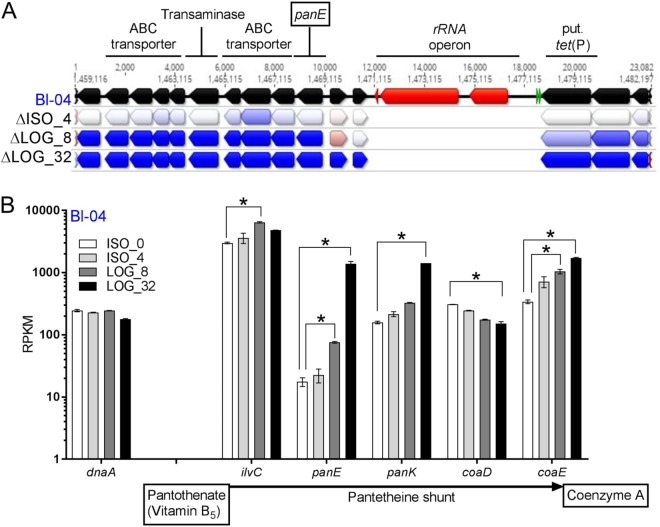
FIG 8.
Overview of upregulated gene cluster associated with coenzyme A biosynthesis in B. animalis subsp. lactis Bl-04. (A) The genetic region was annotated in the Bl-04 genome and colored as follows: black, coding sequences; red, rRNAs; and green, tRNAs. The arrows below each gene represent a log2 ratio fold change between ISO_4, LOG_8, and LOG_32 compared to ISO_0 in descending order, with heat coloring showing downregulation (red) and upregulation (blue) with tetracycline. (B) The raw RPKM expression in Bl-04 for various genes involved in coenzyme A biosynthesis from pantothenate. *, adjusted P < 0.05 and fold change of >2×. Expression of dnaA is shown for reference.
Genes from many different metabolic categories were significantly downregulated and upregulated in both strains during the LOG experiments (Table 1); however, most differences were seen in amino acid and carbohydrate transport and metabolism. Bl-04 and HN019 had upregulated genes for methionine biosynthesis. Alternatively, genes involved in glutamine, glutamate, aspartate, asparagine, threonine, homoserine, glycine, and alanine biosynthesis were downregulated in the LOG experiments for the two strains.
Carbohydrate utilization was also affected in the LOG experiments. Genes involved with lactose and galactose uptake and utilization were downregulated in LOG_8 and LOG_32, which is interesting, considering indels 19 and 20. Genes for fructoooligosaccharides, raffinose, maltose, and maltodextrin uptake were also downregulated. Interestingly, seven genes involved in the pentose phosphate pathway are downregulated in the LOG_32 experiments of both strains. A key indicator of metabolic status is the fructose-6-phosphate phosphoketolase gene xfp (EC 4.1.2.22), which is essential for saccharolytic fermentation as part of the “bifidus shunt” (37). The xfp gene is significantly downregulated in the two strains in the LOG_32 experiment. Conversely, genes involved with xylose utilization were upregulated in both strains across the LOG experiments, although xylose is not in either the MRS or ISO-Sensitest media. The cell wall and capsule metabolic category included genes downregulated in both strains, specifically in dTDP-rhamnose synthesis and rhamnose-containing glycans.
Of the cofactors, genes involved in riboflavin (vitamin B2), NAD, and coenzyme A (CoA) production were all upregulated in the LOG experiments in both strains. Interestingly, there is a cluster of 12 genes that are significantly upregulated in the LOG experiments upstream of an rRNA operon, two tRNAs (valine and glycine), and the putative tet(P) gene (Fig. 8A). One of the genes in the cluster is 2-dehydropantoate 2-reductase panE (EC 1.1.1.169), which is key in the biosynthesis of coenzyme A. Other genes that make up the pantetheine shunt are present in both strains, and most are similarly upregulated during the LOG experiments (Fig. 8B). Of the 12 genes clustered together, at least seven are involved with transport, one set for glutamate/aspartate and one for various metal ions. BLAST analysis failed to find the pantothenate transport genes panT and panF in the two strains, and neither have been reported in bifidobacteria, although there were transporter genes adjacent to the ilvC, panE, and panK genes.
DISCUSSION
Bacteria rapidly evolve countermeasures to resist increasingly overused and misused antibiotics, creating widespread resistance years after initial drug introduction (36, 38). The molecular benefits of developing resistance mechanisms far outweigh the costs (39, 40), so bacteria are constantly under pressure to acquire resistance elements (41). Monitoring antibiotic resistance and transferability is a concern in the food and dietary supplement industries to prevent the risk of industrial bacteria becoming reservoirs of resistance genes that can be transferred to pathogens (30, 42). Such products are most often tested using the ISO method for dairy products, which requires MIC measurements be compared to those for a representative species or an overall genus, as is the case of Bifidobacterium. However, due to the variation in test results, the method notes that it “should be used with caution when testing bifidobacteria” (43). The overall tetracycline limit of 8 μg/ml for bifidobacteria was derived from several studies that examined a variety of bifidobacterial species, resulting in a wide range of tetracycline resistances (30). However, Gueimonde et al. found unimodal resistance across 26 B. animalis subsp. lactis strains, with an average MIC of 16.9 μg/ml (18). Commercial B. animalis subsp. lactis strains Bl-04 and HN019 encompass the low level of genetic diversity within the subspecies (14–17), despite having distinct tetracycline resistances, which made them ideal subjects to locate key resistance elements. In this study, we used novel molecular strategies to confirm and further analyze the antibiotic resistance of the two strains.
The transcriptional response during subinhibitory tetracycline exposure revealed few significant changes in either strain, showing that overall functionality does not deviate from the normal metabolic processes until highly stressed. One interesting difference during the low-level exposure was the reduction of CRISPR-Cas-associated gene expression. CRISPR systems provide resistance against invasive genetic elements in bacteria (44) and have been harnessed to provide broad programmable genome editing capabilities (45). The downregulation of cas genes highlights the focus of the bacteria on hibernation, reducing the energy expended in nonessential pathways to survive like protection against phage predation. This supports recent findings that resistance to phage predation and antibiotics are inversely related (46). Recent efforts use endogenous CRISPR-Cas systems to modify a bacterium's own genome (47, 48); however, specificity is paramount, as self-targeting spacers can be fatal to a cell (49). With the variety of CRISPR-Cas systems in bifidobacteria and other organisms (50–52), these results suggest that concentrations of tetracycline or other antibiotics may be optimized to control nonspecific targeting and increase genome editing specificity using endogenous CRISPR in bacteria.
Acute exposure at higher concentrations of tetracycline caused substantial differences in gene expression. Protein, complex carbohydrate, and cell wall metabolisms were downregulated, notably in dTDP-rhamnose synthesis and rhamnose-containing glycans. Rhamnose polysaccharides have been previously described in B. animalis subsp. lactis (53) and are involved in cross-talk interaction with the host immune system (54). Moreover, they can be associated with human cell adherence and bacteriophage receptors (55). Alternatively, there was upregulation of transporter and amino acid metabolism, specifically that of methionine. This may be due to tetracycline inhibiting methionine, the typical starting amino acid in proteins, from incorporating with the ribosome. These findings are similar to other transcriptome and proteome experiments with tetracyclines (56, 57), even within a eukaryotic host (58). This is likely due to the cells entering the persistence stage, which is when cells become nondividing and dormant due to antibiotic exposure (59). Under these conditions, bacteria only perform essential functions, while simultaneously absorbing maximum amounts of environmental nutrients and exporting tetracycline. Persistence responses and thresholds are not well-studied in the dietary supplement industry, yet are important, since cells exposed to tetracycline levels above the MIC are still metabolically active.
A key metabolic pathway with consistent genetic upregulation was the pantothenate shunt, which synthesizes coenzyme A from vitamin B5 and is involved in metabolic reactions like lipid metabolism and respiration (60). Microbial cells uptake pantothenate, an ingredient in the ISO-Sensitest media, typically through the panT or panF transport genes (61). Although neither gene was located, pantothenate transporters are often colocalized with other pantothenate metabolic genes (62), and indeed, the ilvC, panE, and panK genes all were proximal to transporter genes upregulated in the LOG experiments. This indicates that environmental pantothenate and its derivative CoA are important for tetracycline tolerance. Along with being a key cofactor for many cellular processes, CoA is required to make nonribosomal peptides functional (63). Nonribosomal peptide synthesis is widespread in bacteria (64), and it could circumvent ribosome use to maintain key cellular functions during persistence. One of the genes in the pantothenate shunt was found in a cluster of upregulated genes adjacent to an rRNA operon and the putative tet(P) gene. This suggests that the rRNA may be directly protected due to the proximity of the tet(P) gene, which could then translate the adjacent CoA and transporter genes despite exposure to tetracycline.
Although the Bl-04 and HN019 genomes were considerably similar, there were several polymorphic regions, specifically indels 8 and 21, with expression differences that likely altered their phenotypes in each experiment. Small genetic differences contributing to additional tetracycline resistance are well established, as point mutations to the 16S rRNA gene can prevent tetracycline molecules from binding properly (26, 27, 65). Furthermore, an SNP in a tRNA dimethylallyltransferase (miaA) gene in B. animalis subsp. lactis was theorized to increase resistance levels (17). Indels 19 and 20, which are involved with lactose and galactose uptake, were upregulated in HN019 only during the LOG experiments. This may be beneficial, as both sugars are involved with exopolysaccharide biosynthesis and are key during stress conditions (54). Indel 15 is an SNP in the glcU gene that has been shown to increase glucose uptake several times over (15). If monosaccharide metabolism is preferred to disaccharide and polysaccharide metabolism during tolerance, then any increase in glucose intake would be greatly beneficial. We also noted that indel 14B altered the predicted restriction site in the gene, although the gene was not differentially expressed in our experiments. Nonetheless, epigenetic control of gene expression is common in bacteria, and epigenetic factors contributing to inherited tetracycline resistance have been demonstrated previously (66). Altogether, our results suggest that there are many genetic regions in B. animalis subsp. lactis besides tet(W) that combine to contribute to antibiotic resistance.
The tet(W) gene, assumed to provide resistance to tetracycline, is upregulated during the subinhibitory concentrations. Interestingly, the putative tet(P) in B. animalis subsp. lactis is a ribosomal protection GTPase similar to the tet(W), yet the tet(P) gene was upregulated during the LOG experiments, while tet(W) was not. The putative tet(36) is a tetracycline efflux pump related to genes previously reported in Bifidobacterium breve and Bifidobacterium longum (67, 68), and many other transporter genes were upregulated during tetracycline exposure. Indeed, the most apparent cause of additional resistance in HN019 is the emrB multidrug efflux gene, which is likely affected by indel 21 in the flanking promoter. Enzymatic protection genes, such as NADPH-requiring oxidoreductase tet(X), have not been reported in bifidobacteria (28). However, there were over 800 genes in the B. animalis subsp. lactis genomes, nearly 300 of which were upregulated in the LOG experiments, that were not assigned to a metabolic category, and novel antibiotic resistance genes in bifidobacteria are constantly being identified (69). Thus, there are many factors yet to be discovered that may also contribute broadly to antibiotic resistance in food and probiotic microorganisms.
Overall, we have shown that B. animalis subsp. lactis resistance to tetracycline does not just hinge on the tet(W) gene, but rather encompasses an orchestrated transcriptional network distributed across several genomic loci, in which even small polymorphisms may dramatically increase survival. Traditional microbiological methods have served to assess resistance in the past, but novel molecular assays and high-throughput nucleic acid sequencing provide a much greater resolution and should be used routinely to monitor and ensure safety for food and dietary supplement organisms.
MATERIALS AND METHODS
Bacterial cultures.
Vials of B. animalis subsp. lactis strain Bl-04 (ATCC SD-5219) and strain HN019 (ATCC SD-5674) culture were obtained from the DuPont global culture collection. Growth was generally obtained using De Man-Rogosa-Sharpe broth (part number [p/n] 288110; BD, Franklin Lakes, NJ) supplemented with cysteine-HCl (p/n C7880; Sigma-Aldrich, St. Louis, MO) (MRSC) at 0.05% (vol/vol) diluted in water (p/n 51200; Rockland, ME). Agar (p/n 214010; BD) was added to the MRSC broth as directed. The MIC testing also required ISO-Sensitest media (p/n CM0473; Oxoid, Basingstoke, Hampshire, UK). All samples in both media were grown at 37°C ± 1°C under anaerobic conditions using GasPacks (p/n 260001; BD).
MIC testing.
Initial tetracycline trials followed the ISO method (43). Briefly, strains were streaked for isolation using a three-phase pattern on MRSC and agar plates and incubated 16 to 24 h as described above. A solution of sterile lactic acid bacteria susceptibility test medium (LSM) broth (90% ISO-Sensitest [IST] broth and 10% MRSC) was aliquoted into 3-ml glass tubes suitable for spectrophotometry. Colonies from the streak plates were picked with sterile loops and transferred into the broth to an optical density (OD) of 0.16 to 0.20 at 625 nm (Genesys 20; Thermo). The inoculum was diluted 1:500 in LSM media prior to inoculation into the antibiotics.
Tetracycline-HCl (p/n T4062-5G; Sigma-Aldrich) was diluted in molecular-grade water to 5,120 μg/ml, based on the concentration per mg on the label. The stock solution was diluted 1:10 (vol/vol) and the dilution scheme in the ISO method was then used to make 50% serial dilutions (512 μg/ml, 256 μg/ml, 128 μg/ml, etc.) to 1 μg/ml. The stock solution and dilutions were made fresh with each experiment. The dilutions were added to the inoculum at equal amounts, thus halving the effective dilutions to a range of 256 μg/ml to 0.5 μg/ml. The combination antibiotic and inoculum were transferred to a clear PCR plate (p/n 14230232; Thermo) and sealed (p/n AB0558; Thermo). Plates were incubated for 48 h at 37°C ± 1°C in anaerobic conditions before being inspected for cellular material under a magnifying glass.
To test acute exposure, the inoculum described above was not finally diluted 1:500 but was added directly to the tetracycline gradient and only incubated for 16 h to remain in mid-log phase.
GlowMax Discover.
Concentrations of ATP were tested with the BacTiter-Glo microbial cell viability assay (p/n G8230; Promega, Madison, WI) on the GlowMax Discover microplate reader (p/n GM3000, Promega) following the manufacturer's instruction. Briefly, BacTiter-Glo buffer was added to the substrate and equilibrated to room temperature (22°C to 25°C) before use. The reagent was then added in equal parts to the samples in a black, opaque 96-well plate (p/n 3915; Costar, Corning, NY). The plate was placed on the GlowMax Discover, which was preprogrammed to mix with an orbital shaker, incubate at room temperature for 5 min, and then read the luminescence. Data were generated in relative luminescence units (RLU), which were not back calculated to ATP concentrations, although a standard of 10 mM rATP (p/n P113B; Promega) was added to each run to ensure run-to-run consistency. All samples were tested in duplicate, except for the retest that had six replicates per sample, and statistical comparisons were made using Prism v. 7.04 (GraphPad, La Jolla, CA).
The tetracycline standard dilutions were compared to the standards in the MaxSignal Tetracycline enzyme-limited immunosorbent assay (ELISA) kit (p/n 1016-04E; Bioo Scientific Corp., Austin, TX). The manufacturer's instructions for detection in milk/soured milk were used. The GlowMax instrument was programmed to make absorbance measurements of 450 nm (primary) and 600 nm (differential).
Droplet digital PCR.
Bl-04 and HN019 samples were diluted 1:10 by adding 120 μl of sample to 1,080 μl of Butterfield's phosphate buffer (p/n R23701; Fisher, Hampton, NH) in clear 1.5-ml centrifuge tubes (Fisher). Samples were then treated with the viability dye Pemax (p/n 4900013150; GenIUL, Barcelona, Spain) to nullify DNA associated with dead cells. A 500 μM stock solution of Pemax was created by adding 100 μl of standard buffer (p/n: 4900018000; GenIUL) to a monodose vial of Pemax. Four μl (final conc. 1.67 μM) and 12 μl (final concentration, 5 μM) of Pemax solution were added to Bl-04 and HN019 tubes, respectively. Tubes were incubated for 30 min at 37°C, protected from light, and gently shaken to facilitate Pemax reaction. After incubation, viability dye was permanently bound and further reaction halted via UV light activation for 15 min on a PMA-Lite device (Biotium, Fremont, CA). Cells were lysed by transferring 1 ml of treated samples to prefilled 2.0-ml tubes containing Triple-Pure high-impact 0.1-mm zirconium beads (D1032-01; Benchmark Scientific, Edison, NJ). Tubes were placed into a solid aluminum microvial holder pretempered to −20°C and shaken for 15 min on a Mini-Beadbeater-96 (120 V; BioSpec Products, Bartlesville, OK).
A PCR mixture was created by combining reagents in the following concentrations and volumes to create 25 μl reaction volume per replicate: molecular biology-grade water, 0.42 μl (p/n SH30538.03; Thermo Fisher); 5 μM forward primer, 4.5 μl; 5 μM reverse primer, 4.5 μl; 3 μM probe 2.08 μl (IDT, Coralville, IA); ddPCR Supermix for probes (no dUTP), 12.5 μl (p/n 1863024; Bio-Rad, Pleasanton, CA); and 1 μl of treated sample. The Bl-04 probe utilized 6-carboxyfluorescein (FAM) dye, and the HN019 probe utilized 6-carboxy-2,4,4,5,7,7-hexachlorofluorescein (HEX) dye. All oligonucleotide sequences are listed in Table 2.
TABLE 2.
Primers and probes used for the study
| Namea | Sequenceb |
|---|---|
| ddPCR | |
| Bl-04_F | 5′-CTT CCC AGA AGG CCG GGT-3′ |
| Bl-04_P | 5′-6-FAM/CGA AGA TGA/ZEN/TGT CGG AAC ACA AAC ACC CGG/3IABkFQ-3′ |
| Bl-04_R | 5′-CGA GGC CAC GGT GCT CAT ATA GA-3′ |
| HN019_F | 5′-TTC GAT GGT TCG CAC AGT GA-3′ |
| HN019_P | 5-6-FAM-AAA CAG GTC/ZEN/AAT CAG CGG CGC AGG GAG/3IABkFQ-3′ |
| HN019_R | 5′-GGT CTG ATG CCG CCT GAA AT-3′ |
| SNPs | |
| Indel_2_F | GCC GCA GAT CGA ATA CTG GG |
| Indel_2_R | AAG CGA CGA CCG AAT GCA AT |
| Indel_4_F | TCT GTT GCG GGA TGT CAT GC |
| Indel_4_R | GGC GAT TCA GGC GAA GTT CA |
| Indel_7_F | ACA GAA GTA GGC GAG GGG AC |
| Indel_7_R | AAC ATC ACC GCC GAT GAA CC |
| Indel_9_F | GGA ACT TGG CAG ACG TCT CG |
| Indel_9_R | CTG TTG ACT CCG GCT GCA TT |
| Indel_11_F | GCA GAT CGC CCC ATT GAA CA |
| Indel_11_R | TCG GGT CTG CTC GAC ATT CT |
| Indel_14A_F | CAC ACG ATC GGA ACC AGT GG |
| Indel_14A_R | ACG AAC GAA GTA GCC GAG GA |
| Indel_14B_F | CGC CGA ATC GCC ATA ATC CA |
| Indel_14B_R | CAG CCC GAA TCC ACT TGA CC |
| Indel_17_F | CTC AGT GTG CAC GCA CTC C |
| Indel_17_R | GAT GCA GGT TGA GCA AGG CG |
| Indel_19_F | CCT CGC TGT TTC GCT CTG AG |
| Indel_19_R | GGA AGG TGA CAT GCA GAC CG |
| Indel_20_F | AAC GAT CAT TTC CGC CAC CC |
| Indel_20_R | ATG CTG TTC GAT GCG TTG GT |
| Indel_22_F | CAT CCA CAG CAG CCA ACT CA |
| Indel_22_R | CCT GAA CCA GAT TGC CAC CG |
| Indel_23_F | TTC GTC ACT GGA TCG CAA GC |
| Indel_23_R | TCC AAC AAA CTC ACC GTG GC |
| Indel_27_F | GCT CTT CGT CTT CGC GGT AC |
| Indel_27_R | CCG ACA ATC TGC GGC AAT GA |
ddPCR, droplet digital PCR; SNP, single-nucleotide polymorphism.
ZEN, ZEN quencher (IDT); IABkFQ, Iowa Black fluorescence quencher.
Reactions were transferred to ddPCR 96-well plates. Plates were sealed using PX1 PCR plate sealer and pierceable foil heat seals (Bio-Rad). Samples were then transferred to an automated droplet generator (Bio-Rad), and droplets were formed per the manufacturer's instruction. The plate containing newly formed droplets were transferred to C1000 Touch thermal cycler with a 96-deep well reaction module (Bio-Rad). Thermocycling was completed under the following conditions: 95°C for 10 min, 95°C for 30 s, and 60°C for 1 min repeated for a total of 40 cycles, followed by 98°C for 10 min, then held at 10°C until transfer to a QX200 droplet reader (Bio-Rad). Droplets were analyzed utilizing QuantaSoft Software v. 1.7. The detection thresholds were adjusted as needed to meet the following criteria: at least 10,000 total droplets read, sample concentration between 100 and 2,000 copies per μl, and no major defects noted in 1-dimensional (1D) or 2-dimensional (2D) amplitude.
Genome sequencing and assembly.
HN019 was resequenced to generate a complete genome. Genomic DNA was prepared as described previously, except with MRSC (1). The shotgun libraries, sequencing, and assembly were carried out at the Roy J. Carver Biotechnology Center, University of Illinois at Urbana-Champaign (UIUC).
The shotgun genomic DNA libraries were constructed from 500 ng of DNA after sonication with an ME220 ultrasonicator (Covaris, Woburn, MA) to an average fragment size of 500 base pairs (bp) with the Hyper library preparation kit from (Kapa Biosystems, Roche, Basel, Switzerland). The individually barcoded libraries were amplified with 3 cycles of PCR and run on a fragment analyzer (AATI, Ankeny, IA) to confirm the absence of free primers and primer dimers and to confirm the presence of DNA of the expected size range. Libraries were pooled in equimolar concentration and size selected on a 2% agarose gel for fragments 500 bp to 800 bp in length. The pool was further quantitated by quantitative PCR on a CFX Connect real-time system (Bio-Rad, Hercules, CA).
The pooled shotgun libraries were sequenced on two MiSeq flow cells with paired-end reads 250 nucleotides in length using the v. 3 kits. The fastq read files were generated and demultiplexed with the bcl2fastq v. 2.17.1.14 conversion software (Illumina, San Diego, CA). For Nanopore sequencing, 1 μg of DNA was sheared in a g-Tube. Each fragment DNA sample was converted into a barcoded Nanopore library with the 1D Native barcoding genomic DNA kit (EXP-NBD103 and SQK-LSK108). Ten libraries were pooled and sequenced on a FLO-MIN106 R94.1 flow cell on a GridION X5 sequencer for 48 h. Base calling and barcode demultiplexing were performed with Albacore 2.17.
Initial Nanopore reads were base-called using Albacore v. 2.1.10 (Oxford Nanopore Technologies, Oxford, United Kingdom), retaining the original raw signal information in FAST5 format (for later assembly polishing) and producing sequence calls in FASTQ format for initial assembly steps. FASTQ data were initially assessed using FASTQC v. 0.11.5 (70). Reads were scanned and trimmed using the tools Porechop v. 0.2.3 (71) and seqtk, as described in Table S2A. Reads were then reassessed using FASTQC to ensure that trimming occurred and to establish sequence retention. These were used in downstream assembly steps.
Initial Illumina base calls were produced using Illumina's bcl2fastq, removing any residual sequence adapters. The resulting FASTQ data were initially assessed using FASTQC and then quality trimmed using Trimmomatic v. 0.36 (72), as described in Table S2B. Reads were then reassessed using FASTQC to ensure that any quality issues were addressed prior to use in downstream assembly steps.
A hybrid assembly was originally performed using the Unicycler assembler v 0.4.3 (73), as noted in Table S1C. QUAST (74) and MUMmer (75) were used to both generate some basic overall metrics of the assembly as well as to compare to the B. animalis subsp. lactis Bl-04 reference genome (Table S2D). Annotation was performed using Prokka (76; see also Table S2E) and rapid annotation by subsystem technology (RAST), using the default conditions (77, 78). Finally, Bandage (79) was utilized to assess completeness and potential assembly issues.
The Bl-04 and HN019 genomes were aligned and genetic differences were identified and characterized using progressiveMauve in Geneious (80). The indels were assessed by targeted PCR using primers in Table 2 designed by Primer3 in Geneious (81). Reaction mixtures were made as follows: molecular biology-grade water, 11 μl; 100 μM forward primer, 0.25 μl; 100 μM reverse primer, 0.25 μl; AmpliTaq Gold 360 (p/n N808024; ThermoFisher), 12.5 μl; and template, 1 μl. Template samples were prepared from processing 400 μl of the frozen culture vials with the Maxwell 16 cell DNA purification kit (p/n AS1020; Promega) on a Maxwell 16 (p/n AS2000, Promega), then diluted 1:10 (vol/vol) in 1× Tris-EDTA buffer at pH 8.0 (p/n BP2473-1; ThermoFisher). The thermocycler (MyCycler; Bio-Rad) was set as follows: step 1, 95°C for 10 min; step 2, 95°C for 30 s; step 3, 57°C for 30 s; step 4, 72°C for 1 min (with steps 2 to 4 repeated 34 cycles); step 5, 72°C for 5 min; and step 6, hold at 4°C. Amplicons were visualized using 2% agarose E-gels (p/n G601802; Thermo Fisher), purified using the Purelink PCR purification kit (p/n K310001; Thermo Fisher), and sequenced with Sanger technologies (Eurofins Genomics, Luxembourg). Resulting reads were aligned in Geneious.
The genomes for both strains were screened in Geneious v 11.0.4 (Biomatters, Auckland, New Zealand) for putative antibiotic resistance genes by comparing to known gene and protein functions in the Antibiotic Resistance Genes Database (82) and the Comprehensive Antibiotic Resistance Database (83), using previously described search parameters (1). Potential matches were considered relevant if the percent pairwise nucleotide match was >80%, the threshold for antibiotic resistance gene identity (24). Putative promoters were identified using the Neural Network Promoter Prediction online software (84).
RNA sequencing.
Four separate growth experiments were made with Bl-04 and HN019, in duplicate 3-ml amounts. First, experiment ISO_0 followed the ISO method with 0 μg/ml tetracycline, grown until the culture reached an OD of 0.14 to 0.30 (early log phase). Second, experiment ISO_4 was conducted as experiment ISO_0, except with a final concentration of 4 μg/ml tetracycline. Third, experiment LOG_8 followed the ISO method, except for the 1:500 dilution prior to inoculation. After combining to make the OD 0.16 to 0.20 inoculum, the culture was instead added directly to 16 μg/ml tetracycline, making the final tetracycline concentration 8 μg/ml. The culture was then grown until reaching OD 0.14 to 0.30. Lastly, experiment LOG_32 was conducted as ISO_8, except with a final concentration of 32 μg/ml tetracycline.
After reaching early log phase, cells were centrifuged for 10 min at 4,063 × g. The supernatant was removed, and 1 ml of TRIzol (p/n 15596026; Thermo) was added. The pellet was resuspended by vortexing, and the tubes were immediately frozen at −80°C. The cell pellets were later thawed, transferred to a Lysing Matrix B 2-ml tube (p/n 116911050; MPBio, Santa Ana, CA), and disrupted using a Mini-Beadbeater. The lysate was subjected to a chloroform organic extraction, followed by purification using an RNeasy minikit (p/n 74104; Qiagen, Hilden, Germany). Quality checks and quantifications of the isolated RNA were carried out on the Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA). rRNA was removed prior to library construction using a Ribo-Zero rRNA removal kit (Gram-positive bacteria) (p/n MRZGP126; Illumina). Stranded cDNA libraries were prepared using a TruSeq stranded mRNA kit (p/n 20020594; Illumina), quantitated by Agilent TapeStation, pooled equimolarly, and sequenced on one flow cell lane for 75 cycles using paired-end 75-base pair sequencing on an Illumina 2500 HiSeq rapid cluster kit v. 2 (p/n PE-402-4002; Illumina) and HiSeq rapid sequencing by synthesis (SBS) kit v. 2 (p/n FC-402-4021; Illumina).
Paired-end reads were imported and mapped to the B. animalis subsp. lactis Bl-04 genome using the Geneious for RNA Mapper with default settings in Geneious. The sequencing reads for all experiments and replicates of both strains are publicly available. Transcript levels in the resulting assemblies were calculated using the “calculate expression levels” function, and normalized for comparison of replicates and experiments using the “compare expression levels” function with the DESeq2 method and parametric fit type (85). The assemblies were exported as BAM files and imported into ArrayStar v. 1.2 (DNAStar, Madison, WI), processed using QSeq, and normalized by reads per kilobase of transcript per million mapped reads (RPKM). Regression analyses of the RNA data were made in ArrayStar software using the Student t test with false discovery rate (FDR) correction. Statistical analyses between sets of samples were analyzed using the DESeq2 method, as above. Differences in expression were considered significant if the absolute confidence (−log10 adjusted P value) was ± 1.00, and the log2 ratio was at ±1.00, representing a P value of ≤0.05 and a ≥2× fold change, respectively, after normalization. Nucleotide sequences of all significantly expressed genes were concatenated and uploaded in RAST to assign metabolic categories. Expression levels were visualized using Prism, and the volcano plots were generated using Excel 2016 (Microsoft, Redmond, WA).
Accession number(s).
The complete genome sequence for Bl-04 is publicly available (NCBI GenBank accession number CP001515). A draft genome sequence for HN019, consisting of 28 contigs, was published previously (NCBI accession number ABOT00000000.1). The new HN019 genome sequence is available in the NCBI databaseunder accession number CP031154. The paired-end reads of the RNA sequencing experiments are also available in the NCBI database under the Gene Expression Omnibus accession number GSE117878.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Chris Fields, Alvaro Hernandez, and Chris Wright at the Roy J. Carver Biotechnology Center at UIUC for sequencing, assembly, and annotation of the HN019 genome. Within DuPont Nutrition & Health, we thank Buffy Stahl and Martin Kullen for technical guidance and Véronique Laffitte for performing the initial ISO MIC testing. We also thank Moriah Nickerson and Laura Pérez-Brenner at Promega for technical and logistical support with the GlowMax Discover and associated assays.
Funding was provided by DuPont Nutrition & Health, which manufactures B. animalis subsp. lactis Bl-04 and HN019.
DuPont has a sponsored research agreement with North Carolina State University supporting C.H.C. and R.B. W.M., P.R., B.Z., and A.K. are employees of DuPont.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01999-18.
REFERENCES
- 1.Morovic W, Roper JM, Smith AB, Mukerji P, Stahl B, Rae JC, Ouwehand AC. 2017. Safety evaluation of HOWARU® Restore (Lactobacillus acidophilus NCFM, Lactobacillus paracasei Lpc-37, Bifidobacterium animalis subsp. lactis Bl-04 and B. lactis Bi-07) for antibiotic resistance, genomic risk factors, and acute toxicity. Food Chem Toxicol 110:316–324. doi: 10.1016/j.fct.2017.10.037. [DOI] [PubMed] [Google Scholar]
- 2.Morovic W, Hibberd AA, Zabel B, Barrangou R, Stahl B. 2016. Genotyping by PCR and high-throughput sequencing of commercial probiotic products reveals composition biases. Front Microbiol 7:1747. doi: 10.3389/fmicb.2016.01747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patro JN, Ramachandran P, Barnaba T, Mammel MK, Lewis JL, Elkins CA. 2016. Culture-independent metagenomic surveillance of commercially available probiotics with high-throughput next-generation sequencing. mSphere 1:e00057-16. doi: 10.1128/mSphere.00057-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S. 2014. Expert consensus document: the International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol 11:506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 5.WHO, FAO. 2002. Guidelines for the evaluation of probiotics in food. Joint FAO/WHO working group report. London, Ontario, Canada: http://www.who.int/foodsafety/fs_management/en/probiotic_guidelines.pdf. [Google Scholar]
- 6.Ibarra A, Latreille-Barbier M, Donazzolo Y, Pelletier X, Ouwehand AC. 2018. Effects of 28-day Bifidobacterium animalis subsp. lactis HN019 supplementation on colonic transit time and gastrointestinal symptoms in adults with functional constipation: a double-blind, randomized, placebo-controlled, and dose-ranging trial. Gut Microbes 9:236–251. doi: 10.1080/19490976.2017.1412908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ringel-Kulka T, Palsson OS, Maier D, Carroll I, Galanko JA, Leyer G, Ringel Y. 2011. Clinical trial: probiotic bacteria Lactobacillus acidophilus NCFM and Bifidobacterium lactis Bi-07 versus placebo for the symptoms of bloating in patients with functional bowel disorders-a double-blind study. J Clin Gastroenterol 45:518–525. doi: 10.1097/MCG.0b013e31820ca4d6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waller PA, Gopal PK, Leyer GJ, Ouwehand AC, Reifer C, Stewart ME, Miller LE. 2011. Dose-response effect of Bifidobacterium lactis HN019 on whole gut transit time and functional gastrointestinal symptoms in adults. Scand J Gastroenterol 46:1057–1064. doi: 10.3109/00365521.2011.584895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gill HS, Rutherfurd KJ, Cross ML, Gopal PK. 2001. Enhancement of immunity in the elderly by dietary supplementation with the probiotic Bifidobacterium lactis HN019. Am J Clin Nutr 74:833–839. doi: 10.1093/ajcn/74.6.833. [DOI] [PubMed] [Google Scholar]
- 10.Hidalgo-Cantabrana C, Nikolic M, López P, Suárez A, Miljkovic M, Kojic M, Margolles A, Golic N, Ruas-Madiedo P. 2014. Exopolysaccharide-producing Bifidobacterium animalis subsp. lactis strains and their polymers elicit different responses on immune cells from blood and gut associated lymphoid tissue. Anaerobe 26:24–30. doi: 10.1016/j.anaerobe.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Stenman LK, Lehtinen MJ, Meland N, Christensen JE, Yeung N, Saarinen MT, Courtney M, Burcelin R, Lähdeaho M-L, Linros J. 2016. Probiotic with or without fiber controls body fat mass, associated with serum zonulin, in overweight and obese adults-randomized controlled trial. EBioMedicine 13:190–200. doi: 10.1016/j.ebiom.2016.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hibberd AA, Lyra A, Ouwehand AC, Rolny P, Lindegren H, Cedgård L, Wettergren Y. 2017. Intestinal microbiota is altered in patients with colon cancer and modified by probiotic intervention. BMJ Open Gastroenterol 4:e000145. doi: 10.1136/bmjgast-2017-000145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hibberd AA, Yde CC, Ziegler ML, Honoré AH, Saarinen MT, Lahtinen S, Stahl B, Jensen HM, Stenman LK. Probiotic or synbiotic alters the gut microbiota and metabolism in a randomised controlled trial of weight management in overweight adults. Benef Microbes, in press. [DOI] [PubMed] [Google Scholar]
- 14.Barrangou R, Briczinski EP, Traeger LL, Loquasto JR, Richards M, Horvath P, Coûté-Monvoisin A-C, Leyer G, Rendulic S, Steele JL. 2009. Comparison of the complete genome sequences of Bifidobacterium animalis subsp. lactis DSM 10140 and Bl-04. J Bacteriol 191:4144–4151. doi: 10.1128/JB.00155-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Briczinski EP, Loquasto JR, Barrangou R, Dudley EG, Roberts AM, Roberts RF. 2009. Strain-specific genotyping of Bifidobacterium animalis subsp. lactis by using single-nucleotide polymorphisms, insertions, and deletions. Appl Environ Microbiol 75:7501–7508. doi: 10.1128/AEM.01430-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loquasto JR, Barrangou R, Dudley EG, Stahl B, Chen C, Roberts RF. 2013. Bifidobacterium animalis subsp. lactis ATCC 27673 is a genomically unique strain within its conserved subspecies. Appl Environ Microbiol 79:6903–6910. doi: 10.1128/AEM.01777-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Milani C, Duranti S, Lugli GA, Bottacini F, Strati F, Arioli S, Foroni E, Turroni F, van Sinderen D, Ventura M. 2013. Comparative genomics of Bifidobacterium animalis subsp. lactis reveals a strict monophyletic bifidobacterial taxon. Appl Environ Microbiol 79:4304–4315. doi: 10.1128/AEM.00984-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gueimonde M, Flórez AB, van Hoek AH, Stuer-Lauridsen B, Strøman P, Clara G, Margolles A. 2010. Genetic basis of tetracycline resistance in Bifidobacterium animalis subsp. lactis. Appl Environ Microbiol 76:3364–3369. doi: 10.1128/AEM.03096-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott KP, Melville CM, Barbosa TM, Flint HJ. 2000. Occurrence of the new tetracycline resistance gene tet(W) in bacteria from the human gut. Antimicrob Agents Chemother 44:775–777. doi: 10.1128/AAC.44.3.775-777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kazimierczak KA, Flint HJ, Scott KP. 2006. Comparative analysis of sequences flanking tet(W) resistance genes in multiple species of gut bacteria. Antimicrob Agents Chemother 50:2632–2639. doi: 10.1128/AAC.01587-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raeisi SN, Ghoddusi HB, Boll EJ, Farahmand N, Stuer-Lauridsen B, Johansen E, Sutherland JP, Ouoba LII. 2018. Antimicrobial susceptibility of bifidobacteria from probiotic milk products and determination of the genetic basis of tetracycline resistance in Enterococcus species after in vitro conjugation with Bifidobacterium animalis subsp. lactis. Food Control 94:205–211. doi: 10.1016/j.foodcont.2018.07.016. [DOI] [Google Scholar]
- 22.Aires J, Doucet-Populaire F, Butel M. 2007. Tetracycline resistance mediated by tet(W), tet(M), and tet(O) genes of Bifidobacterium isolates from humans. Appl Environ Microbiol 73:2751–2754. doi: 10.1128/AEM.02459-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flórez AB, Ammor MS, Álvarez-Martín P, Margolles A, Mayo B. 2006. Molecular analysis of tet(W) gene-mediated tetracycline resistance in dominant intestinal Bifidobacterium species from healthy humans. Appl Environ Microbiol 72:7377–7379. doi: 10.1128/AEM.00486-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chopra I, Roberts M. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev 65:232–260. doi: 10.1128/MMBR.65.2.232-260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dodds DR. 2017. Antibiotic resistance: A current epilogue. Biochem Pharmacol 134:139–146. doi: 10.1016/j.bcp.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 26.Wilson DN. 2014. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol 12:35–48. doi: 10.1038/nrmicro3155. [DOI] [PubMed] [Google Scholar]
- 27.Nguyen F, Starosta AL, Arenz S, Sohmen D, Dönhöfer A, Wilson DN. 2014. Tetracycline antibiotics and resistance mechanisms. Biol Chem 395:559–575. doi: 10.1515/hsz-2013-0292. [DOI] [PubMed] [Google Scholar]
- 28.Roberts MC. 2005. Update on acquired tetracycline resistance genes. FEMS Microbiol Lett 245:195–203. doi: 10.1016/j.femsle.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 29.The UniProt Consortium. 2017. UniProt: the universal protein knowledgebase. Nucleic Acids Res 45(D1):D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.EFSA. 2012. Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J 10:2740. [Google Scholar]
- 31.Fan F, Wood KV. 2007. Bioluminescent assays for high-throughput screening. Assay Drug Dev Technol 5:127–136. doi: 10.1089/adt.2006.053. [DOI] [PubMed] [Google Scholar]
- 32.Hansen SJ, Morovic W, DeMeules M, Stahl B, Sindelar CW. 2018. Absolute enumeration of probiotic strains Lactobacillus acidophilus NCFM® and Bifidobacterium animalis subsp. lactis Bl-04® via chip-based digital PCR. Front Microbiol 9:704. doi: 10.3389/fmicb.2018.00704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loman NJ, Quick J, Simpson JT. 2015. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat Methods 12:733. doi: 10.1038/nmeth.3444. [DOI] [PubMed] [Google Scholar]
- 34.O'Connell Motherway M, Watson D, Bottacini F, Clark TA, Roberts RJ, Korlach J, Garault P, Chervaux C, van Hylckama Vlieg JE, Smokvina T, van Sinderen D. 2014. Identification of restriction-modification systems of Bifidobacterium animalis subsp. lactis CNCM I-2494 by SMRT sequencing and associated methylome analysis. PLoS One 9:e94875. doi: 10.1371/journal.pone.0094875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arango-Argoty G, Garner E, Pruden A, Heath LS, Vikesland P, Zhang L. 2018. DeepARG: A deep learning approach for predicting antibiotic resistance genes from metagenomic data. Microbiome 6:23. doi: 10.1186/s40168-018-0401-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ventola CL. 2015. The antibiotic resistance crisis: part 1: causes and threats. Pharm Ther 40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 37.Scardovi V. 1965. The fructose-6-phosphate shunt as a peculiar pattern of hexose degradation in the genus Bifidobacterium. Ann Microbiol Enzymol 15:19–29. [Google Scholar]
- 38.Klein EY, Van Boeckel TP, Martinez EM, Pant S, Gandra S, Levin SA, Goossens H, Laxminarayan R. 2018. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc Natl Acad Sci U S A 115:E3463–E3470. doi: 10.1073/pnas.1717295115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lenski RE. 1998. Bacterial evolution and the cost of antibiotic resistance. Int Microbiol 1:265–270. [PubMed] [Google Scholar]
- 40.Andersson DI, Levin BR. 1999. The biological cost of antibiotic resistance. Curr Opin Microbiol 2:489–493. doi: 10.1016/S1369-5274(99)00005-3. [DOI] [PubMed] [Google Scholar]
- 41.Hwang TJ, Kim N, Kim HB, Lee BH, Nam RH, Park JH, Lee MK, Park YS, Lee DH, Jung HC. 2010. Change in antibiotic resistance of Helicobacter pylori strains and the effect of A2143G point mutation of 23S rRNA on the eradication of H. pylori in a single center of Korea. J Clin Gastroenterol 44:536–543. doi: 10.1097/MCG.0b013e3181d04592. [DOI] [PubMed] [Google Scholar]
- 42.Sanders ME, Akkermans LM, Haller D, Hammerman C, Heimbach JT, Hörmannsperger G, Huys G. 2010. Safety assessment of probiotics for human use. Gut Microbes 1:164–185. doi: 10.4161/gmic.1.3.12127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.ISO. 2010. ISO 10932:2010 (IDF 223:2010) Milk and milk products—determination of the minimal inhibitory concentration (MIC) of antibiotics applicable to bifidobacteria and non-enterococcal lactic acid bacteria (LAB). International Organization for Standards, Geneva, Switzerland. [Google Scholar]
- 44.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 45.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chan BK, Turner PE, Kim S, Mojibian HR, Elefteriades JA, Narayan D. 2018. Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol Med Public Health 2018:60–66. doi: 10.1093/emph/eoy005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hidalgo-Cantabrana C, O'Flaherty S, Barrangou R. 2017. CRISPR-based engineering of next-generation lactic acid bacteria. Curr Opin Microbiol 37:79–87. doi: 10.1016/j.mib.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 48.Luo ML, Mullis AS, Leenay RT, Beisel CL. 2015. Repurposing endogenous type I CRISPR-Cas systems for programmable gene repression. Nucleic Acids Res 43:674–681. doi: 10.1093/nar/gku971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paez-Espino D, Morovic W, Sun CL, Thomas BC, Ueda K-i, Stahl B, Barrangou R, Banfield JF. 2013. Strong bias in the bacterial CRISPR elements that confer immunity to phage. Nat Commun 4:1430. doi: 10.1038/ncomms2440. [DOI] [PubMed] [Google Scholar]
- 50.Briner AE, Lugli GA, Milani C, Duranti S, Turroni F, Gueimonde M, Margolles A, van Sinderen D, Ventura M, Barrangou R. 2015. Occurrence and diversity of CRISPR-Cas systems in the genus Bifidobacterium. PLoS One 10:e0133661. doi: 10.1371/journal.pone.0133661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hidalgo-Cantabrana C, Crawley AB, Sanchez B, Barrangou R. 2017. Characterization and exploitation of CRISPR loci in Bifidobacterium longum. Front Microbiol 8:1851. doi: 10.3389/fmicb.2017.01851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH. 2015. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol 13:722. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hidalgo-Cantabrana C, Sánchez B, Moine D, Berger B, Clara G, Gueimonde M, Margolles A, Ruas-Madiedo P. 2013. Insights into the ropy phenotype of the exopolysaccharide-producing strain Bifidobacterium animalis subsp. lactis A1dOxR. Appl Environ Microbiol 79:3870–3874. doi: 10.1128/AEM.00633-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hidalgo-Cantabrana C, Sánchez B, Milani C, Ventura M, Margolles A, Ruas-Madiedo P. 2014. Genomic overview and biological functions of exopolysaccharide biosynthesis in Bifidobacterium spp. Appl Environ Microbiol 80:9–18. doi: 10.1128/AEM.02977-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mistou M-Y, Sutcliffe IC, van Sorge NM. 2016. Bacterial glycobiology: rhamnose-containing cell wall polysaccharides in Gram-positive bacteria. FEMS Microbiol Rev 40:464–479. doi: 10.1093/femsre/fuw006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heo A, Jang H-J, Sung J-S, Park W. 2014. Global transcriptome and physiological responses of Acinetobacter oleivorans DR1 exposed to distinct classes of antibiotics. PLoS One 9:e110215. doi: 10.1371/journal.pone.0110215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang M, Qin H, Wang W, Zhang H, Long Y, Ye J. 2018. Global proteomic responses of Escherichia coli and evolution of biomarkers under tetracycline stress at acid and alkaline conditions. Sci Total Environ 627:1315–1326. doi: 10.1016/j.scitotenv.2018.01.342. [DOI] [PubMed] [Google Scholar]
- 58.Darby AC, Gill AC, Armstrong SD, Hartley CS, Xia D, Wastling JM, Makepeace BL. 2014. Integrated transcriptomic and proteomic analysis of the global response of Wolbachia to doxycycline-induced stress. ISME J 8:925. doi: 10.1038/ismej.2013.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Caskey JR, Embers ME. 2015. Persister development by Borrelia burgdorferi populations in vitro. Antimicrob Agents Chemother 59:6288–6295. doi: 10.1128/AAC.00883-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sibon OC, Strauss E. 2016. Coenzyme A: to make it or uptake it? Nat Rev Mol Cell Biol 17:605. doi: 10.1038/nrm.2016.110. [DOI] [PubMed] [Google Scholar]
- 61.Jaehme M, Slotboom DJ. 2015. Diversity of membrane transport proteins for vitamins in bacteria and archaea. Biochim Biophys Acta 1850:565–576. doi: 10.1016/j.bbagen.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 62.Rodionov DA, Hebbeln P, Eudes A, Ter Beek J, Rodionova IA, Erkens GB, Slotboom DJ, Gelfand MS, Osterman AL, Hanson AD. 2009. A novel class of modular transporters for vitamins in prokaryotes. J Bacteriol 191:42–51. doi: 10.1128/JB.01208-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goering AW, Li J, McClure RA, Thomson RJ, Jewett MC, Kelleher NL. 2017. In vitro reconstruction of nonribosomal peptide biosynthesis directly from DNA using cell-free protein synthesis. ACS Synth Biol 6:39–44. doi: 10.1021/acssynbio.6b00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang H, Fewer DP, Holm L, Rouhiainen L, Sivonen K. 2014. Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc Natl Acad Sci U S A 111:9259–9264. doi: 10.1073/pnas.1401734111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ross JI, Eady EA, Cove JH, Cunliffe WJ. 1998. 16S rRNA mutation associated with tetracycline resistance in a gram-positive bacterium. Antimicrob Agents Chemother 42:1702–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Adam M, Murali B, Glenn NO, Potter SS. 2008. Epigenetic inheritance based evolution of antibiotic resistance in bacteria. BMC Evol Biol 8:52. doi: 10.1186/1471-2148-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Margolles A, Florez AB, Moreno JA, Van Sinderen D, Clara G. 2006. Two membrane proteins from Bifidobacterium breve UCC2003 constitute an ABC-type multidrug transporter. Microbiology 152:3497–3505. doi: 10.1099/mic.0.29097-0. [DOI] [PubMed] [Google Scholar]
- 68.Price CE, Reid SJ, Driessen AJ, Abratt VR. 2006. The Bifidobacterium longum NCIMB 702259T ctr gene codes for a novel cholate transporter. Appl Environ Microbiol 72:923–926. doi: 10.1128/AEM.72.1.923-926.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang S, Gao X, Gao Y, Li Y, Cao M, Xi Z, Zhao L, Feng Z. 2017. Tetracycline resistance genes identified from distinct soil environments in China by functional metagenomics. Front Microbiol 8:1406. doi: 10.3389/fmicb.2017.01406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Retrieved February 2018. (Software.)
- 71.Wick RR. 2018. PoreChop. https://github.com/rrwick/Porechop. Retrieved February 2018. (Software.)
- 72.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Delcher AL, Salzberg SL, Phillippy AM. 2003. Using MUMmer to identify similar regions in large sequence sets. Curr Protoc Bioinformatics 1:10.3.1–10.3.18. doi: 10.1002/0471250953.bi1003s00. [DOI] [PubMed] [Google Scholar]
- 76.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 77.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M. 2014. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31:3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. 2012. Primer3-new capabilities and interfaces. Nucleic Acids Res 40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu B, Pop M. 2009. ARDB-antibiotic resistance genes database. Nucleic Acids Res 37:D443–D447. doi: 10.1093/nar/gkn656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, Lago BA, Dave BM, Pereira S, Sharma AN. 2017. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 45(D1):D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Reese MG. 2001. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput Chem 26:51–56. doi: 10.1016/S0097-8485(01)00099-7. [DOI] [PubMed] [Google Scholar]
- 85.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.