Abstract
Calpains are intracellular, calcium-activated cysteine proteases. Calpain-3 is abundant in skeletal muscle, where its mutation-induced loss of function causes limb-girdle muscular dystrophy type 2A. Unlike the small subunit–containing calpain-1 and -2, the calpain-3 isoform homodimerizes through pairing of its C-terminal penta-EF-hand domain. It also has two unique insertion sequences (ISs) not found in the other calpains: IS1 within calpain-3's protease core and IS2 just prior to the penta-EF-hand domain. Production of either native or recombinant full-length calpain-3 to characterize the function of these ISs is challenging. Therefore, here we used recombinant rat calpain-2 as a stable surrogate and inserted IS1 into its equivalent position in the protease core. As it does in calpain-3, IS1 occupied the catalytic cleft and restricted the enzyme's access to substrate and inhibitors. Following activation by Ca2+, IS1 was rapidly cleaved by intramolecular autolysis, permitting the enzyme to freely accept substrate and inhibitors. The surrogate remained functional until extensive intermolecular autoproteolysis inactivated the enzyme, as is typical of calpain-2. Although the small-molecule inhibitors E-64 and leupeptin limited intermolecular autolysis of the surrogate, they did not block the initial intramolecular cleavage of IS1, establishing its role as a propeptide. Surprisingly, the large-molecule calpain inhibitor, calpastatin, completely blocked enzyme activity, even with IS1 intact. We suggest that calpastatin is large enough to oust IS1 from the catalytic cleft and take its place. We propose an explanation for why calpastatin can inhibit calpain-2 bearing the IS1 insertion but cannot inhibit WT calpain-3.
Keywords: calpain, cysteine protease, muscular dystrophy, protease inhibitor, structure-function, protein engineering, calpain-2, calpain-3/p94, calpastatin, insertion sequence 1 (IS1), propeptide, p94, calpain inhibitor, limb-girdle muscular dystrophy, protease
Introduction
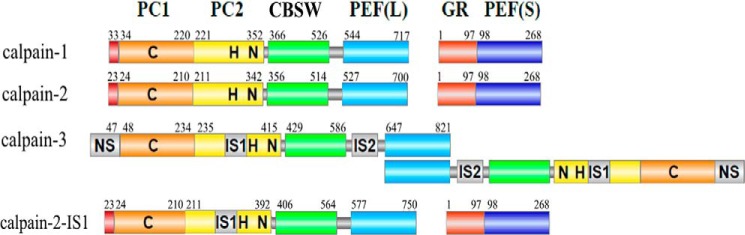
Calpains are intracellular cysteine proteases transiently activated as a result of calcium influx into the cell (1–3). Calpains play diverse roles in intracellular calcium signaling by catalyzing the limited cleavage of a wide range of protein substrates (4). In mammals, calpains exist as a family of over a dozen isoforms, all but one of which share a two-domain protease core (PC) 2 (5). Many of these isoforms have a C-terminal penta-EF hand (PEF) domain, which serves as a dimerization module via EF-hand pairing (6–9). Calpain-3 is a “tissue-specific” calpain abundant in skeletal muscle and is one of the PEF domain-containing calpain family members (10, 11). This calpain shares a similar domain organization and ∼50% sequence identity with the catalytic subunits of the ubiquitous calpain-1 and -2 (Fig. 1).
Figure 1.
Domain architecture of calpain-1, -2, and -3 and calpain-2–IS1. The domain nomenclature is based on the structures of calpain-1 and -2, where PC1 (orange) and PC2 (yellow) represent the protease core, CBSW (green) represents the calpain-type β-sandwich, and PEF (blue) represents the large (L) and small (S) subunit penta-EF hand domains. The catalytic triad residues of the cysteine protease core are indicated by C, H, and N. The unique insertion sequences of calpain-3 are shown in gray. The 48-residue IS1 was inserted in calpain-2 after Tyr250.
Calpain-3 has three unique sequences (NS, IS1, and IS2) that are not found in any of the other calpain family members and have no known homologs. These three extra sequences increase the molecular mass of calpain-3 from 80 to 94 kDa, which is why the enzyme is also referred to as p94 (10, 12). NS is a long N-terminal leader sequence, which replaces the N-terminal anchor helix of the large subunits of calpain-1 and -2. In the protease core, IS1 is a 48-residue insertion sequence. It occupies a region adjacent to the catalytic cleft, beginning after Asp267, within the PC2 domain (13). A second insertion sequence (IS2) lies in the region linking the calpain-type β-sandwich domain (CBSW) and the large-subunit PEF(L) domain (14). These distinctive regions are likely to be responsible for the unique characteristics of calpain-3.
Unlike the ubiquitously expressed calpain-1 and -2, which form heterodimers of a large and small subunit (8, 9), calpain-3 forms a homodimer via the pairing of its fifth EF-hand (15–19). Using the calpain-2 structure (8, 9, 20) as a guide, a model for the calpain-3 homodimer places a protease core at either end of the multidomain structure (17). Calpain-3 is encoded by a single gene (CAPN3), in which loss-of-function mutations have been linked to the progressive muscle degenerative disease limb girdle muscular dystrophy type 2A (LGMD2A) (21, 22). Although the functions of the enzyme remain poorly understood, calpain-3 is crucial for adult muscle physiology and homeostasis. Sarcomeric proteins such as nebulin (23), myosin (24), filamin C (25, 26), and titin (26, 27) have been shown to be in vivo substrates of calpain-3 and serve either a structural or contractile role in muscle fibers. In particular, calpain-3 has been reported to be tightly associated with the large sarcomeric protein, titin, via the enzyme's IS2 (28). It is speculated that one of calpain-3's primary functions in the sarcomere may be to rid the cell of damaged proteins and in this way contribute to muscular maintenance and integrity following eccentric, as opposed to exhaustive, exercise (29–37). Calpain-3 may also play a role in mechanical signal transduction and the promotion of muscle fiber elasticity (11, 38).
Another striking feature of calpain-3 is its rapid autolysis both in vivo and in vitro (39–41). With a half-life of less than 10 min, recovery of intact calpain-3 from muscle extracts has been difficult, even in the presence of the cysteine protease inhibitors E-64 and leupeptin (39, 41, 42). Attempts to produce recombinant whole calpain-3 have also been hampered by similar autoproteolytic degradation (16). The IS1 region in calpain-3 has been shown to be the principle site of autoproteolysis and reason for the instability of the enzyme. This has been interpreted as a degradative process. However, mounting evidence suggests that intramolecular autolysis of IS1 is required to activate calpain-3 (11, 16, 26, 43–46).
In the conventional calpain-1 and -2 the Ca2+-dependent alignment of the active site is an intrinsic part of the activation mechanism (47, 48). The same is true for calpain-3 (13). Characterization of the recombinant proteolytic core of calpain-3, containing both the NS and IS1 regions, demonstrated that, upon Ca2+ activation, rapid autolysis at a specific N-terminal site of IS1, located at or near Tyr274, is required before the enzyme can become available for substrate hydrolysis and/or accessible to inhibitors (16, 45, 46). Recently, Ye et al. (13) determined several X-ray crystal structures of the calpain-3 protease core. One of these structures showed that, in the Ca2+-bound form, IS1 is cleaved near its N terminus, leaving behind eight IS1 residues in the active-site cleft. A second intermolecular cleavage occurs sometime later, at Arg315 (45). Alanine substitution of Tyr274 has been shown to prevent both autolytic and proteolytic activity (26). Thus, the N-terminal region of IS1 appears to act as an internal propeptide, producing an activation mechanism unique to calpain-3. It is also a region that contains pathogenic mutations responsible for LGMD2A (49, 50).
In muscle tissue, the predominant form of calpain-3 is its intact form bound, presumably, to titin (29, 53). Rapid autolysis of calpain-3 can be prevented by the deletion of these insertion regions, as seen with the autoproteolytically resistant alternatively spliced variant, Lp82, lacking IS1 and IS2, which has been characterized in rat lens (54).
Calpastatin is a naturally occurring intrinsically disordered protein that has a complex inhibitory interaction with Ca2+-activated calpain. The calpain–calpastatin system protects the cell from pathological conditions, where unregulated calcium signals promote calpain overactivation (4, 55). Calpastatin consists of four homologous, tandemly repeating inhibitory domains (domains 1–4) and an N-terminal domain that lacks inhibitory function (domain L) (56–58). Therefore, whole calpastatin can simultaneously bind up to four calpains (59). Each inhibitory domain is subdivided into three subdomains, A, B, and C. Subdomains A and C form amphipathic α-helices upon binding to the PEF domains of Ca2+-activated calpain-2 (20, 60) and, by inference, to calpain-1. The B subdomain, equidistant from A and C, is responsible for the inhibitory activity of calpastatin. This region can be truncated to a 27-residue peptide (B-27), which interacts much more weakly with calpain than it does when the anchoring A and C regions are present but still inhibits the enzyme (61, 62). The mechanism of calpastatin inhibition, as demonstrated by the crystal structure of recombinant rat calpain-2 in complex with different calpastatin inhibitory domains (PDB entries 3BOW and 3DFO), revealed that calpastatin occupies both the prime and unprimed sides of the active-site cleft, and, although the calpastatin backbone passes through the active site, it escapes cleavage by looping out and around the active cysteine (20, 63). By recognizing multiple lower-affinity sites present on the calcium-bound calpain surface, in addition to the binding of the A and C helices to hydrophobic pockets in the PEF(L) and PEF(S) domains, a tight and specific calcium-dependent interaction is formed (20, 59). To date, calpain-3 has not been shown to be regulated by calpastatin inhibition, but rather the inhibitor has been speculated to be a calpain-3 substrate (64).
Here, we describe the expression, purification, and characterization of a chimeric rat calpain-2–IS1 construct as a surrogate for calpain-3 to characterize the unique functions of IS1. We demonstrated that, as with calpain-3, IS1 occupies the catalytic cleft of the enzyme, restricting the access of substrates and small-molecule inhibitors. At the higher calcium concentrations needed to activate calpain-2, IS1 undergoes rapid intramolecular cleavage, after which the enzyme can accept substrates and inhibitors. Treatment with small-molecule inhibitors prevents the intermolecular autoproteolysis typically seen with calpain-2, but does not prevent the initial intramolecular cleavage of IS1. Furthermore, we reveal that calpastatin completely inhibits calpain-2–IS1 activity even when IS1 is intact, suggesting that the B-peptide of calpastatin is able to displace IS1 from the catalytic cleft.
Results
Expression and purification of a recombinant calpain-2 containing IS1
To help deduce the role(s) of IS1, we inserted human calpain-3 IS1 into the corresponding region of recombinant rat calpain-2 (Fig. 1). This surrogate was chosen because it can be stably produced in Escherichia coli and requires higher calcium concentrations for activation than calpain-3. This made it less likely that the enzyme would be prematurely activated and autolyzed during production and purification. We found that inclusion of IS1 into calpain-2 did not significantly alter the enzyme's solubility, yield, or stability. Following production and purification, an intact large-subunit band was detected by SDS-PAGE, which was slightly larger than 80 kDa, by the size of IS1 (Fig. S1D). Thus, as predicted, the higher calcium concentrations required for activation of calpain-2 protected the IS1 insert from spurious autoproteolysis.
IS1 lies in the active-site cleft but does not impede the Ca2+-mediated conformational change needed for calpain activation
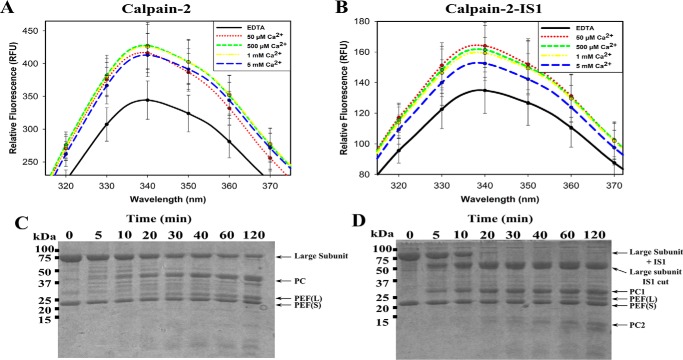
Having produced an intact calpain-2 containing IS1, we next examined the impact IS1 had on calpain-2 activation. During calpain-2 activation, Trp288 is displaced from its solvent-exposed position between the PC1 and PC2 domains into a hydrophobic pocket (47). This is a key step in permitting the two protease core domains to assume the catalytically competent arrangement of the active site. To make sure that IS1 does not impede this conformational change, tryptophan fluorescence of calpain-2 and calpain-2–IS1 was measured as a function of calcium concentration. The resting fluorescence emission spectrum of calpain-2 in EDTA produced a peak fluorescence of 344 RFU at 340 nm (Fig. 2A). In the presence of IS1, the peak tryptophan fluorescence at 340 nm was only 135 RFU, about 2.5 times less than that of calpain-2 (Fig. 2B). This suggests that IS1 lies within or in close proximity to the active-site cleft, thereby suppressing some of the tryptophan fluorescence from Trp288. Upon activation of calpain-2 with Ca2+, a 1.2-fold increase in fluorescence was produced. Although the resting tryptophan fluorescence of calpain-2–IS1 is considerably less than that of calpain-2, it also underwent a 1.2-fold increase in fluorescence when activated by Ca2+. This suggests that, although total tryptophan fluorescence is reduced in the calpain-2–IS1 surrogate, IS1 does not inhibit the conformational change associated with Ca2+ activation.
Figure 2.
Intrinsic tryptophan fluorescence and tryptic digestion of Ca2+-activated calpain-2 and calpain-2–IS1. A and B, the intrinsic tryptophan fluorescence emission spectra of 50 μm inactive C105S calpain-2 (A) and calpain-2–IS1 (B) titrated with 50 μm to 5 mm Ca2+. Measurements were made at an excitation wavelength of 280 nm and were carried out in 50 mm Tris-HCl (pH 7.4), 150 mm NaCl, and 0.1% β-mercaptoethanol. Error bars, S.D (n = 3). C and D, SDS-PAGE analysis of the tryptic digestion of 5 μm Ca2+-activated inactive C105S calpain-2 (C) and 5 μm inactive C105S calpain-2–IS1 (D). Reactions were carried out in 50 mm HEPES-NaOH (pH 7.4), 5 mm CaCl2, and 0.1% β-mercaptoethanol. Reactions were initiated by adding 0.1 μm trypsin and were stopped at each time point by the addition of SDS-PAGE loading buffer and heating at 95 °C.
To see if IS1 sits protected within the active-site cleft, we next compared the time-course tryptic digestion of calpain-2–IS1 with that of calpain-2. SDS-PAGE showed that approximately half of the 80-kDa catalytic subunit of calpain-2 was digested within 40 min, producing a 45-kDa fragment, which is the protease core (PC1 plus PC2). The ∼21-kDa PEF(S) small-subunit band was present at the start of digestion and remained constant. Just above it at ∼23 kDa, the PEF(L) domain was produced during digestion and increased in intensity due to cleavage of the linker region between the PEF(L) and CBSW domain (Fig. 2C). Calpain-2–IS1 generated a different profile when digested with trypsin. The catalytic subunit of calpain-2–IS1 was more susceptible to digestion and was cleaved to completion within 20 min, generating 55-kDa (large subunit distal to the IS1 cut), 30-kDa (PC1 domain), and 23-kDa (PEF(L) domain) fragments.
Although the tryptophan fluorescence results put part of IS1 in or within the vicinity of the active site, its high susceptibility to rapid proteolytic digestion by trypsin suggests that much of the remainder of IS1 is likely flexible and solvent-exposed.
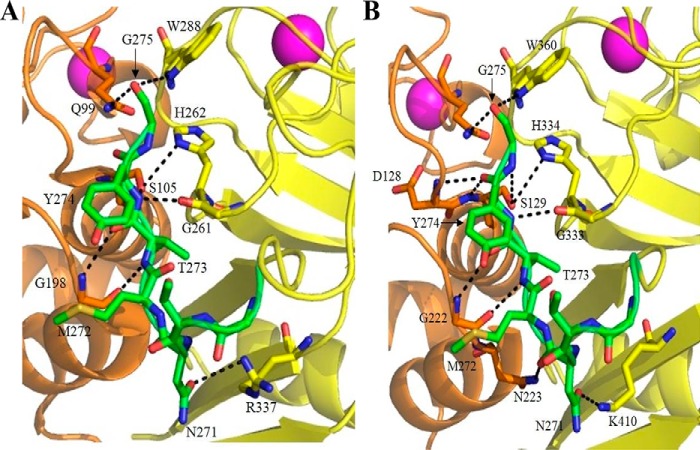
To evaluate how IS1 might bind the catalytic cleft in calpain-2, we used the recently solved crystal structure of the calpain-3 protease core (13) (PDB code 6BDT) as a guide to model the N-terminal region of IS1 into the active-site cleft of calcium-activated calpain-2 C105S (20) (PDB code 3BOW). These two crystal structures were overlapped by the PyMOL program to find the best possible three-dimensional match. In Fig. 3A, the calpain-3 residues, with the exception of the IS1 peptide, were removed from the match. No further analysis was necessary to see the interactions of IS1 with the catalytic cleft of calpain-2. Residues that belong to IS1 are shown in calpain-3 numbering and denoted with an asterisk. This model indicated that the calpain-2 active site can make several contacts with the IS1 backbone analogous to those made by calpain-3 (Fig. 3). However, four additional contacts are made at Asn271, Tyr274, and Gly275 by calpain-3, which are not present in the model (Table 1). Given that Asp104 and Ser105 occupy the same positions and similar orientations in calpain-2 as their corresponding residues, Asp128 and Ser129, in calpain-3, it is likely that similar contacts are generated with the IS1 peptide backbone at Tyr274* and Gly275*. Asp104 and Ser105 are within the range generally accepted for hydrogen bonding (Table 1). These and some other bonds are not shown in the calpain-2 model for clarity. The only significant difference between the contacts made by IS1 with the active sites of calpain-2 and -3 is a hydrogen bond from the Asn223 side chain of calpain-3, which is obviously not present in the corresponding residue (Ala199) of calpain-2.
Figure 3.
The N-terminal portion of IS1 modeled into the calpain-2 catalytic cleft produces interactions similar to those in the active site of the calpain-3 protease core. A, model of the N-terminal portion of IS1 (green) from Asn271* to Gly275* (calpain-3 numbering) in the active site of calpain-2 C105S protease core (PDB code 3BOW). B, X-ray crystal structure of the calpain-3 protease core C129S (PDB code 6BDT) (13). Residues belonging to the PC1 domain are shown in orange, and those belonging to the PC2 domain are shown in yellow. Dotted lines represent hydrogen bonding between IS1 and the calpain-2 and/or -3 catalytic cleft. Key binding residues are represented as sticks. Oxygen atoms are colored red, and nitrogen atoms are colored blue.
Table 1.
Hydrogen-bonding interactions of IS1 (in calpain-3 numbering and denoted by an asterisk) modeled into the calpain-2–IS1 model (PDB code 3BOW) compared with those in the crystal structure of the calpain-3 core (PDB code 6BDT)
For the calpain-2–IS1 model, the residues are given in calpain-2 numbering with their calpain-3 equivalents shown in parenthesis. NA, not applicable.
| IS1 (Asn271–Gly275) | Calpain-2–IS1 model | Distance | Calpain-3 structure | Distance |
|---|---|---|---|---|
| Å | Å | |||
| Side chain of Asn271* | Side chain of Arg337 (Lys410) | 3.7 | Side chain of Lys410 | 3.1 |
| C=O of Asn271* | NA | Side chain of Asn223 | 4.0 | |
| Met272* | NA | NA | ||
| NH of Thr273* | C = O of Gly198 (Gly222) | 3.0 | C = O of Gly222 | 3.3 |
| C=O of Thr273* | NH of Gly198 (Gly222) | 3.4 | NH of Gly222 | 3.6 |
| NH of Tyr274* | C = O of Gly261 (Gly333) | 2.8 | C = O of Gly333 | 3.2 |
| Side chain of Ser105 (Ser129) | 3.2 | Side chain of Ser129 | 3.5 | |
| C=O of Tyr274* | NH of Ser105 (Ser129) and | 3.0 | NH of Ser129 | 2.8 |
| NH of Asp104 (Asp128) | 3.5 | NH of Asp128 | 3.3 | |
| Gly275* Cα | Side chain of His262 (His334) | 3.3 (Van der Waals) | Side chain of His334 | 2.9 (Van der Waals) |
| NH of Gly275* | Side chain of Ser105 (Ser129) | 3.6 | Side chain of Ser129 | 3.6 |
| C=O of Gly275* | Side chain of Trp288 (Trp360) | 3.5 | Side chain of Trp360 | 3.4 |
| Side chain of Gln99 (Gln123) | 3.1 | Side chain of Gln123 | 3.2 |
Together, these results indicate that IS1 could occupy the calpain-2 catalytic cleft in a similar manner and location as it does in calpain-3, and likely with comparable affinity. This suggests that calpain-2 is a useful surrogate for the study of IS1 function.
Rapid autoproteolytic cleavage of IS1 activates calpain-2–IS1
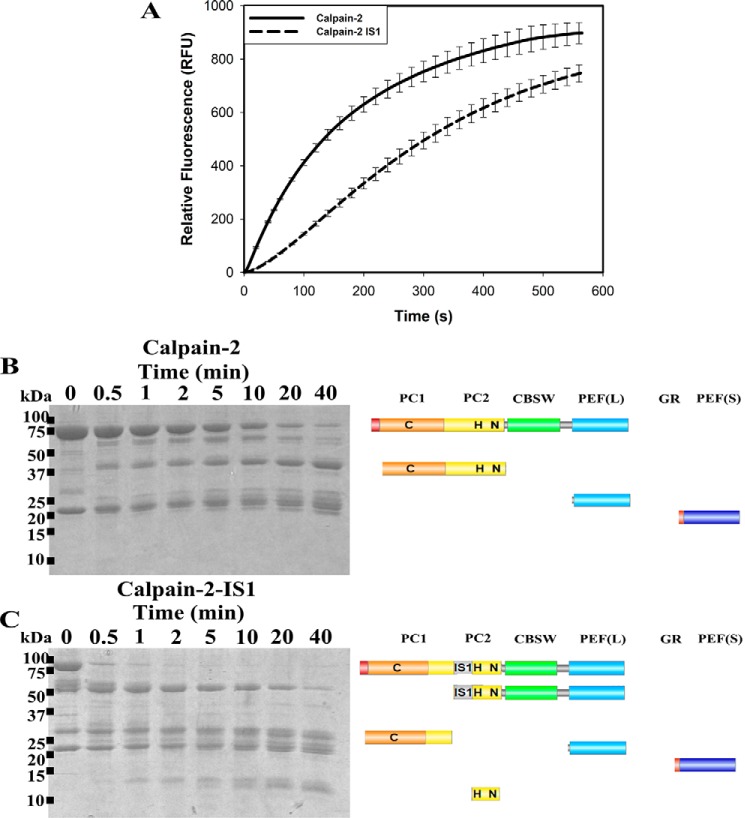
Because IS1 does not impede the calcium-mediated conformational change needed for calpain-2 activation, we next examined the impact of IS1 on substrate recognition and turnover. Substrate hydrolysis by calpain-2–IS1 was measured using an internally quenched FRET substrate (EDANS)-EPLFAERK-(DABCYL) (65). The rate of proteolysis was measured as the increase in fluorescence emission of EDANS at 500 nm (Fig. 4A). Upon the addition of 5 mm CaCl2, hydrolysis of the FRET substrate by calpain-2 produced a fluorescence progression curve with an initial rate of substrate turnover that began to diminish within a minute to form a characteristic rectangular hyperbola. The shape of the enzyme activity curve early in the reaction phase is attributed to ongoing enzyme depletion by intermolecular autoproteolysis rather than substrate depletion or end product inhibition. In contrast, calpain-2–IS1 showed a much slower initial rate of substrate hydrolysis, which then increased to produce a slight sigmoidal curvature within the first 300 s of the reaction. By the 300-s mark, the surrogate enzyme had produced 70% of the activity of calpain-2 but was continuing at a faster rate as it entered the hyperbolic phase. The gap in activity had narrowed by 600 s. Extrapolation predicts that the two reactions would reach a similar maximal fluorescence of ∼900 RFU. This is consistent with the activation profile of an enzyme containing a propeptide. IS1 appears to restrict the access of substrate to the catalytic site, indicating that autoproteolytic cleavage of IS1 is required before substrate binding. Similarly, IS1 must also interfere with calpain-2 autoproteolysis, resulting in a prolonged half-life of the surrogate as compared with calpain-2.
Figure 4.
The proteolytic and autoproteolytic behavior of calpain-2 and calpain-2–IS1. A, hydrolysis of 10 μm small molecule FRET substrate (EDANS)-EPLFAERK-(DABCYL) by 250 nm calpain-2 (solid black line) and calpain-2–IS1 (dotted black line). Substrate fluorescence was excited at 335 nm and emission was measured at 500 nm. Reactions were initiated by adding 5 mm CaCl2 and were carried out in 50 mm HEPES-NaOH (pH 7.4) and 0.1% β-mercaptoethanol at room temperature. Error bars, S.D. (n = 3). B and C, SDS-PAGE analysis of the autoproteolytic cleavage of 5 μm Ca2+-activated calpain-2 (B) and calpain-2–IS1 (C). Reactions were carried out under the same conditions as above and were stopped at each time point by the addition of SDS-PAGE loading buffer and heating at 95 °C. The resulting fragments were identified by in-gel tryptic digestion followed by LC-MS. The domain compositions of the autolytic fragments are shown in diagrammatic form on the right.
Because inclusion of IS1 seemed to extend the lifetime of the enzyme, we next investigated the effect of IS1 cleavage on the autolysis of calpain-2–IS1. Autolytic fragments were separated by SDS-PAGE and identified by LC-MS. We found that, upon the addition of Ca2+, the 80-kDa catalytic subunit of calpain-2 was rapidly autolyzed with a half-life of about 5 min (Fig. 4B). There was a shortening of the large subunit to ∼75 kDa, which overlapped with the release of the protease core at ∼45 kDa. Several smaller-molecular mass species between 20 and 25 kDa were also produced as the CBSW domain was digested to leave the PEF(L) domain, which was largely intact (Fig. 4B).
Similar to its tryptic digestion, calpain-2–IS1 produced a different autolytic fragmentation pattern as compared with calpain-2 (Fig. 4C). The intact large subunit of calpain-2–IS1 was rapidly cleaved within the first minutes of the reaction, similar in time scale with the sigmoidal region of the activity profile (Fig. 4A), generating a ∼30-kDa fragment that corresponded to the PC1 domain plus a small portion of the N-terminal region of the PC2 domain, to just inside IS1. This is consistent with what occurs in the protease core of calpain-3, where IS1 is cut at its N-terminal cleavage site in an intramolecular fashion (45, 46). A ∼55-kDa species, similar to the one produced by trypsin digestion (Fig. 2D), was also generated. This fragment matched the remainder of the catalytic subunit, including the ISI C-terminal region, to the PEF(L) domain. This fragment was cleaved again (Fig. 4B), with a half-life of about 5–10 min. Fragments between 20 and 25 kDa were produced from this digest and represent the PEF(L) and PEF(S) domains. Several small-molecular mass species between 10 and 15 kDa were also generated, which were from the remaining C-terminal portion of the PC2 domain up to the CBSW domain. The fact that autolytic activity of the surrogate enzyme persisted well after the completed cleavage of intact large subunit indicates that removal of IS1 does not disable the catalytic core; rather, it serves as an essential first step in enzyme activation.
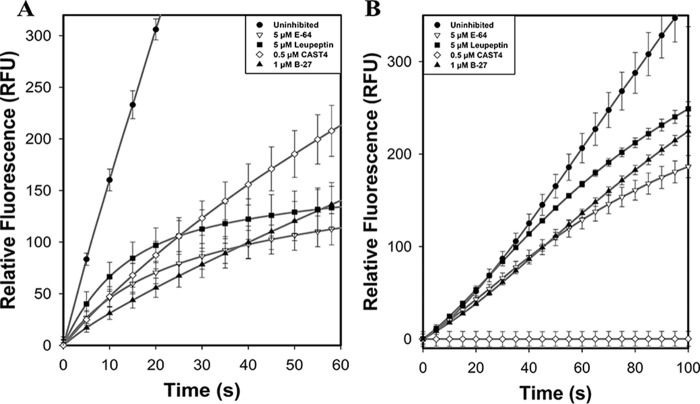
Inhibition of calpain by E-64 or leupeptin is limited by IS1
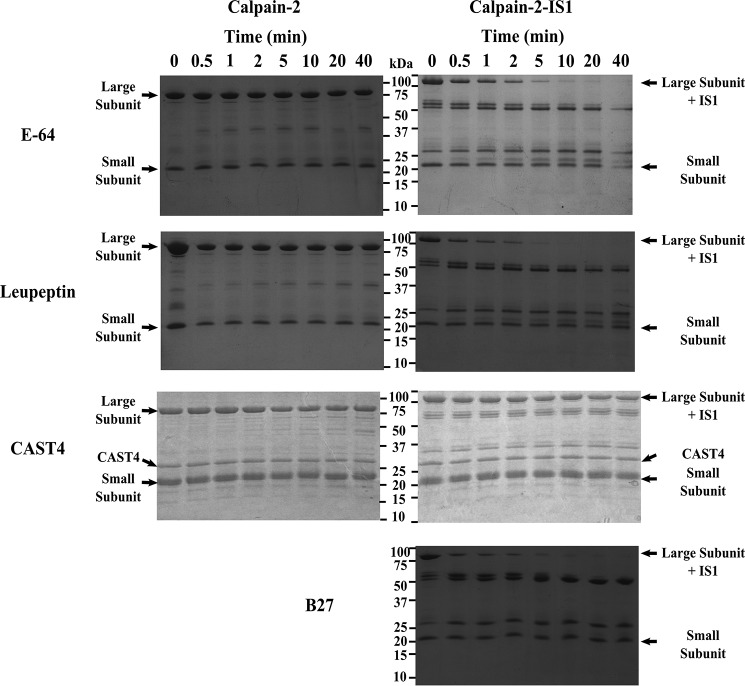
To determine whether intramolecular IS1 autolytic cleavage can be prevented, proteolytic activity of the calpain-2–IS1 surrogate was compared with that of calpain-2 in the presence of the small-molecule active site–directed calpain inhibitors E-64 or leupeptin. Inhibition by E-64 and leupeptin (Fig. 5A) reduced the initial rapid rate of reaction (within 5–10 s) of recombinant rat calpain-2 activity by 68 and 62%, respectively. An approach to plateau in the calpain-2 activity profile generated by these inhibitors was obvious by 60 s. In contrast, the initial rate of reaction of calpain-2–IS1 was not inhibited by leupeptin and was only marginally affected (5% less) by E-64 (Fig. 5B). However, an increase in inhibition was observed after the initial sigmoidal portion of the progress curve, where IS1 is cleaved. The characteristic approach to the plateau produced by these inhibitors was delayed. This retardation in the inhibition of calpain-2–IS1 was similar to the delayed onset of autoproteolysis found for uninhibited calpain-2–IS1 (Fig. 4A), indicating that IS1 also interferes with the binding of small molecule inhibitors to the active site. Our results show that E-64 and leupeptin have little to no effect on the intramolecular cleavage of IS1. This was confirmed by SDS-PAGE analysis of calpain-2–IS1 autolysis in the presence of each inhibitor (Fig. 6). Inhibition of calpain-2 autoproteolysis with E-64 and leupeptin yielded little to no noticeable change in the amount of intact catalytic subunit, showing that the covalent inhibitors can fully inactivate the enzyme (Fig. 6, left). However, E-64 and leupeptin treatment of calpain-2–IS1 were unable to prevent intramolecular IS1 cleavage. With E-64, the amount of intact calpain-2–IS1 large subunit was reduced by half after 1–2 min and was barely detectable by 10 min (Fig. 6, right). There was a concomitant rise in bands at 55 and 25 kDa, similar to those generated by the autolysis of untreated calpain-2–IS1 (Fig. 4). With leupeptin, half of the catalytic subunit was cleaved within 1 min, whereas full loss of the intact large subunit of calpain-2–IS1 had occurred by 5 min. A similar increase in bands at 55 and 25 kDa was generated.
Figure 5.
Calpain-2 and calpain-2–IS1 FRET substrate hydrolysis assay in the presences of inhibitors. A, the hydrolysis of 10 μm FRET substrate (EDANS)-EPLFAERK-(DABCYL) by 250 nm calpain-2 (black circles) in the presence of 5 μm E-64 (white triangles), 5 μm leupeptin (black squares), 500 nm CAST4 (white diamonds), and 1 μm B-27 (black triangles). Substrate fluorescence was excited 335 nm, and emission was measured at 500 nm. Reactions were initiated by adding 5 mm CaCl2 and were done in 50 mm HEPES-NaOH (pH 7.4) and 0.1% β-mercaptoethanol at room temperature. B, hydrolysis of (EDANS)-EPLFAERK-(DABCYL) by 250 nm calpain-2–IS1 in the presence of E-64 (white triangles), leupeptin (black squares), CAST4 (white diamonds), and B-27 (black triangles). Reactions were carried out under the same conditions as above. Error bars, S.D. (n = 3).
Figure 6.
Calpain-2 and calpain-2–IS1 autoproteolysis in the presence of inhibitors E-64, leupeptin, CAST4, and B-27. Left and right, SDS-PAGE analysis of the autoproteolytic cleavage of 5 μm Ca2+-activated calpain-2 (left) and calpain-2–IS1 (right) in the presence of 50 μm E-64, 50 μm leupeptin, 10 μm calpastatin CAST4, and 10 μm calpastatin B-27. Reactions were initiated by adding 5 mm CaCl2 and were carried out in 50 mm HEPES-NaOH (pH 7.4) and 0.1% β-mercaptoethanol at room temperature. Reactions were stopped at each time point by the addition of SDS-PAGE loading buffer and heating at 95 °C. Major bands in the digest are identified to the right of the gels. SDS-PAGE analysis of the uninhibited autolytic digestion of calpain-2 and calpain-2–IS1 is shown in Fig. 4.
Complete inhibition of calpain-2–IS1 by calpastatin
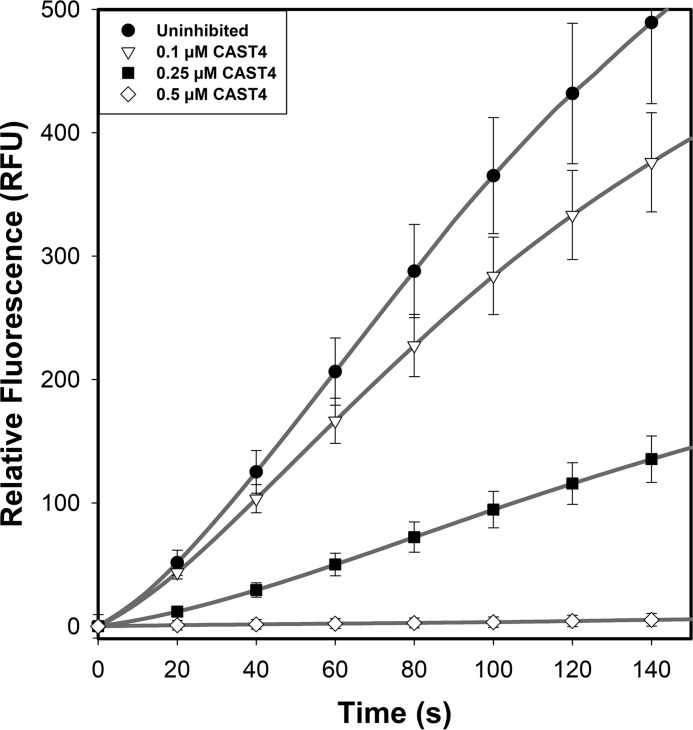
Given that the small-molecule inhibitors E-64 and leupeptin were insufficient to stop IS1 autolysis, a calpain-specific inhibitor with improved potency, like calpastatin, may be required. However, calpastatin has been shown not to inhibit calpain-3 (64); therefore, we set out to determine whether this lack of inhibitory effect on calpain-3 is due to the presence of IS1 interference with calpastatin binding to the active-site cleft. Substrate hydrolysis and autoproteolysis of calpain-2–IS1 and calpain-2 treated with the calpastatin inhibitory repeat CAST4 were compared. Treatment of calpain-2 at twice the molar ratio of CAST4 (Fig. 5A) reduced the initial rate of FRET substrate cleavage to one-third. Surprisingly, CAST4 addition to calpain-2–IS1 at the same molar ratio (Fig. 5B) resulted in complete knockout of calpain-2–IS1 activity. This inhibition was corroborated by SDS-PAGE analysis, where no autoproteolytic fragments were produced by either enzyme in the presence of CAST4 (Fig. 6). Reducing the CAST4 concentration to a 1:1 equivalency resulted in partial expression of calpain-2–IS1 activity and reproduced the typical sigmoidal curvature in the activity progress curve that is associated with IS1 cleavage (Fig. 7).
Figure 7.
Titration of calpain-2–IS1 with calpastatin CAST4. The hydrolysis of 10 μm FRET substrate (EDANS)-EPLFAERK-(DABCYL) by 250 nm calpain-2–IS1 (black line) in the presence of 0.1 μm (dotted line), 0.25 μm (dashed line), and 0.5 μm (dashed and dotted line) calpastatin CAST4 is shown. Substrate fluorescence was excited at 335 nm, and emission was measured at 500 nm. Reactions were initiated by adding 5 mm CaCl2 and were carried out in 50 mm HEPES-NaOH (pH 7.4) and 0.1% β-mercaptoethanol at room temperature. Error bars, S.D. (n = 3).
To determine whether the isolated calpastatin inhibitory B region could have the same effects as the full-length inhibitor, we treated calpain-2 and calpain-2–IS1 with the 27-residue B region peptide (B-27) at a molar ratio of 1:4. With calpain-2, this resulted in 80% inhibition when compared with the reaction rate of the enzyme alone (Fig. 5A). However, the shortened B-27 peptide proved considerably less effective than intact CAST4 at limiting IS1 autolysis, reducing calpain-2–IS1 activity by only 15% (Fig. 5B). As was observed with E-64 and leupeptin, increased inhibition of calpain-2–IS1 was only found following the sigmoidal portion of the progress curve, suggesting that B-27 does not inhibit IS1 cleavage. This was corroborated by SDS-PAGE analysis, which showed that half of the intact catalytic subunit of calpain-2–IS1 was cut after 1–2 min and the remainder was fully cleaved by 5 min (Fig. 6).
Calpastatin pulldown by calpain-2–IS1
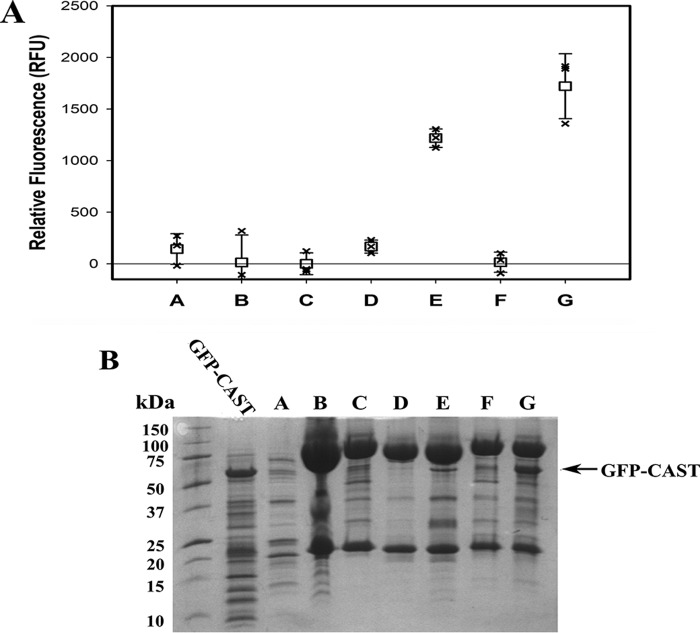
Pulldown experiments with N-terminally GFP-labeled CAST4 added to inactive C105S calpain-2 or inactive C105S calpain-2–IS1 were performed in the presence of Mg2+ or Ca2+ (Fig. 8) to assess calpastatin binding. Because it lacks a His6 tag, only trace amounts of GFP-CAST bound to the Ni-NTA affinity column, as indicated by low GFP fluorescence at 488 nm in sample A. Likewise, minimal fluorescence, ∼150 RFU, was observed when GFP-CAST was mixed with column-bound calpain-2 in the presence of Mg2+ (sample D). When mixed with column-bound Ca2+-activated calpain-2, a 7-fold increase in fluorescence was found (sample E). As expected, the pulldown experiment showed that calpastatin tightly associates with only the Ca2+-activated form of the enzyme. Pulldown of GFP-CAST with calpain-2–IS1 also showed little to no fluorescence in the presence of Mg2+ (sample F). When the enzyme was Ca2+-activated, a >30-fold increase in GFP fluorescence was detected (sample G). Column-bound protein was eluted and analyzed by SDS-PAGE, where a GFP-CAST band at 52 kDa was only found in the Ca2+-activated calpain-2 and calpain-2–IS1 lanes (Fig. 8B). GFP-CAST pulldowns showed that calpastatin can tightly bind calpain-2–IS1 C105S, when Ca2+-activated, even with IS1 intact.
Figure 8.
GFP-CAST is captured by His-tagged calpain. Pulldown experiments were performed by binding N-terminally GFP-tagged calpastatin CAST4 (GFP-CAST) (A), inactive C105S His6-tagged calpain-2 (B), or inactive C105S His6-tagged calpain-2–IS1 (C) to Ni-NTA–agarose resin. A 1 mg/ml aliquot of GFP-CAST was mixed with bound calpain-2 in the presence of 50 mm MgCl2 (D) or 50 mm CaCl2 (E). GFP-CAST was also mixed with bound calpain-2–IS1 in the presence of 50 mm MgCl2 (F) or 50 mm CaCl2 (G). Protein binding was carried out in, and washed with, buffer containing 50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 2% (v/v) glycerol, and 5 mm imidazole. A, immobilized calpain–CAST-GFP complex (50 μl) was diluted to 100 μl with wash buffer, and its fluorescence was measured with excitation at 488 nm and emission at 510 nm in triplicate in a 96-well plate. Error bars, S.D. (n = 3). B, SDS-PAGE analysis of the calpain–CAST-GFP complex. The black arrow indicates the GFP-CAST band pulled down by calpain binding.
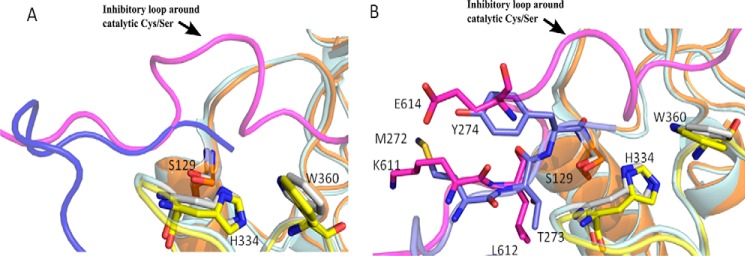
To characterize how calpastatin may outcompete IS1 for binding to the calpain-2 catalytic cleft, we overlapped the crystal structures of recombinant rat calpain-2 in complex with calpastatin (PDB code 3BOW) and the recently solved crystal structure of the calpain-3 protease core (PDB code 6BDT) (13, 20). We found that the backbone of the N-terminal portion of IS1 and the inhibitory B region of calpastatin occupy the same subsites within the active-site cleft and that their side chains project in similar orientations (Fig. 9). The overlapping structures show that the side chain of calpastatin Glu614, which is part of the inhibitory loop, projects away from the active-site cysteine (Ser129) and overlaps well with Tyr274*, the first residue upstream of the IS1 autoproteolytic cleavage site. The side chain of calpastatin Lys611 overlaps well with IS1 Met272*, also projecting away from the active site. Calpastatin Leu612 occupies the P2 subsite and overlaps with IS1 Thr273*.
Figure 9.
The N-terminal portion of IS1 shows partial overlap with part of the B-peptide of calpastatin in the active site of the calpain-3 protease core. Superposition of structures of calpain-3 protease core C129S (13) (PDB code 6BDT) and calpain-2 C129S in complex with calpastatin (PDB code 3BOW) shows that the backbone of the N-terminal portion of IS1 (blue) and the backbone of the B-peptide of calpastatin (magenta) partially overlap in the active site. The partial overlap of the backbones of IS1 and calpastatin is represented as a cartoon (A) and as sticks (B).
The results of the calpastatin pulldown experiments coupled with the high degree of similarity in active-site interactions of calpastatin and IS1, as indicated by the overlapped crystal structures, suggests that, in the presence of Ca2+, calpastatin forms tight interactions with calpain-2 that are sufficient to displace IS1 from its catalytic cleft. Taken together, these results demonstrate that calpastatin has the potential to be a potent calpain-3 inhibitor and that IS1 is not responsible for the failure of calpastatin to inhibit calpain-3.
Discussion
Recombinant rat calpain-2 serves as surrogate for the study of a calpain-3 insertion sequence
Efforts to recover whole calpain-3 have been hampered by rapid autodegradation of the enzyme, even in the presence of calpain inhibitors (16, 39–41). The IS1 region, located in the protease core of calpain-3, appears to be an obligate site of autoproteolysis for making the enzyme more accessible to substrates. Although this autolytic cleavage has been considered to be degradative, there would be no enzyme activity without it. Therefore, the difficulties attributed to autolytic degradation during the recovery of whole calpain-3 from tissue, or by recombinant methods, is likely due to intermolecular autolysis at other sites, like those in the CBSW domain or within IS2.
The recently solved crystal structure of the calpain-3 protease core (containing IS1) in the Ca2+-bound state has provided useful information for the design of isoform-specific inhibitors. However, one drawback of this structure is that it yields no information about the catalytic cleft subsites beyond the S4 area, where in the whole enzyme, contact with the CBSW domain would be made. Just looking at the core alone can potentially bias the development of substrates and/or inhibitors (13, 47, 66–70). To circumvent issues associated with the purification of labile intact whole calpain-3 and reliance on just the calpain-3 protease core, we used recombinant rat calpain-2 to mimic whole-calpain-3 activity and to act as a functional surrogate for the study of IS1.
The large catalytic subunit of calpain-2 shares a similar domain organization and ∼50% sequence identity with calpain-3 (10, 71), suggesting that calpain-2 can accommodate IS1 without disruption of its three-dimensional structure. Whole rat calpain-2 crystal structures of both the Ca2+-free (8, 9) and Ca2+-bound (20, 63) states have been solved, and the latter overlaps well with the recently solved calpain-3 protease core structure (13). The higher calcium requirement for half-maximal calpain-2 activation (200–1000 μm), as compared with calpain-1 (20–75 μm) and -3 (90–150 μm) (44, 72, 73), should help protect IS1 from spurious autolysis without requiring a change of the catalytic Cys to Ser or Ala (4, 44). Also, large quantities of recombinant rat calpain-2 enzyme have been easily produced in E. coli (74), which is not the case with calpain-1 and -3.
As with calpain-2, the calpain-2–IS1 surrogate was stably expressed and purified from E. coli, yielding similar quantities of pure protein. The absence of autoproteolytic degradation of IS1 in the apo state demonstrated that calpain-2 acts as a whole-calpain-3 mimic and surrogate host for the study of the structure and function of this unique calpain-3 insertion sequence. With successful production of this enzyme, there is also the potential to repeat this study with IS2, both alone and in combination with IS1.
IS1 occupies the catalytic cleft of the enzyme but does not inhibit Ca2+-mediated calpain activation
Ca2+-binding at two sites within the protease core act cooperatively to cause a large conformational change that realigns the catalytic triad residues. Realignment of the core causes Trp288 to shift from a solvent-exposed position to a hydrophobic patch, causing a change in calpain-2 fluorescence (8, 9, 47, 48). In C105S calpain-2–IS1, tryptophan fluorescence is muted, which we take as an indication that Trp288 is partially quenched by the nearby side chains or backbone of the IS1 segment, placing the insertion sequence within the catalytic cleft, even in the apo-form. Furthermore, upon Ca2+ addition, IS1 does not appear to interfere with the fluorescence change associated with calpain-2 activation.
In the crystal structure of the Ca2+-activated calpain-3 core, a remnant of the N-terminal region of IS1 was present in the catalytic cleft (13). Modeling this portion of IS1 into its equivalent position in the calpain-2 catalytic cleft established that calpain-2 mimics the subsite interactions made by calpain-3 with the backbone and side chains of IS1. Further still, the model predicted that the backbone of IS1 forms a hydrogen bond with the side chain of the key Trp288 residue, explaining how IS1 might suppress tryptophan fluorescence in the Ca2+-activated form.
Although the model and crystal structure reveal key insights about IS1 binding to the catalytic cleft, they do not provide information about the residues downstream of Gly275* or any potential contacts made by IS1 in the primed side of the active site. It is conceivable that IS1 may interact with the CBSW domain right next to PC2. However, we suspect that the central region of IS1 may interact with a binding partner in the sarcomere. The results of the trypsin proteolysis experiment described above can be used to explore this possibility. Trypsin digestion revealed that the central region of IS1 is flexible and solvent-exposed, making it highly susceptible to proteolytic digestion. This is consistent with what was described by the SAXS solution structures reported for the Ca2+-activated and Ca2+-free calpain-3 protease cores (13). Thus, most of IS1 is unstructured and unprotected even in the presence of neighboring calpain domains that are absent from the calpain-3 protease core structure, meaning that this part of IS1 likely does not bind to calpain itself. Therefore, the external binding partner scenario seems more likely. This would explain why the entire IS1 sequence is so well-conserved in mammals and why there are some LGMD2A mutations in the middle of the insertion sequence (Fig. S2). If IS1 was unstructured in vivo, it is unlikely that there would be much constraint on the sequence.
The N-terminal portion of IS1 plays an autoinhibitory propeptide role in the activation of calpain-3
As shown above, IS1 occupies the catalytic cleft of the calpain-2–IS1 protease core, which resulted in a slight sigmoidal shape in the early phase of the substrate hydrolysis reaction. We attributed this kinetic behavior to the autoinhibitory role of IS1 (46), where its position in the catalytic cleft interferes with substrate binding during the early phase of the enzyme reaction. Displacement of IS1 following intramolecular autoproteolysis was required before the enzyme could accept substrate and/or inhibitors, consistent with observations made using calpain-3 (46). Experiments with skinned muscle fibers have shown that calpain-3 is not activated following transient increases in Ca2+ concentration during normal muscle contraction; instead, calpain-3 activation is associated with prolonged increases in cytoplasmic Ca2+ levels, like those that follow eccentric exercise (29, 33, 34, 37). The autoinhibitory propeptide function of IS1 could possibly serve as a key protective feature against spurious calpain-3 activation during the brief Ca2+-signaling events associated with normal muscle contraction.
Being a multidomain heterodimer with exposed loops and linkers, calpain-2 is a prime target for digestion by proteases (75, 76), including by calpain itself (77, 78). We maintain that rapid autolysis of calpain-2 is primarily an artifact of its purification (2, 79). Similar intermolecular autolytic activity following enzyme activation of calpain-3 has been reported, primarily in the IS1, NS, and IS2 regions (45, 46, 51). Consistent with what has been previously reported, we found the C-terminal region of IS1 is a site of intermolecular autolysis. However, cuts here occur at a much slower rate than the intramolecular propeptide cleavage near the IS1 N-terminal end. Interestingly, autoinhibition of substrate hydrolysis by IS1 in the early stages of the reaction is accompanied by a reduced rate of intermolecular autoproteolysis, producing a less active but longer-lasting enzyme.
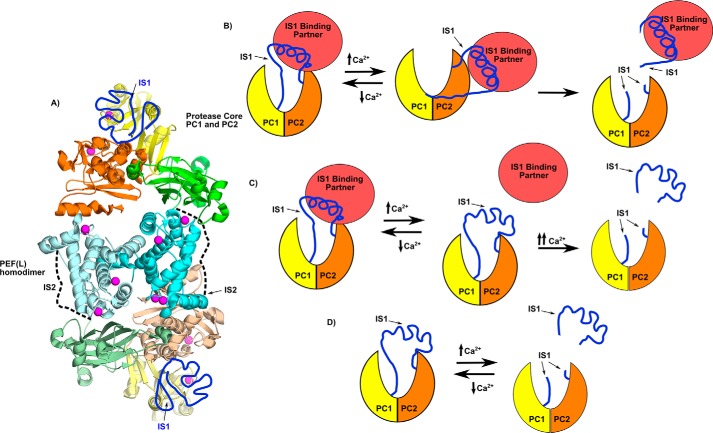
Sequence alignment of the central portion of IS1 between the intra- and intermolecular cut sites shows a high degree of conservation among mammals (Fig. S2). Also, the sequence of this region suggests that it has a high propensity to form a α-helix (46). However, as shown above and by Ye et al. (13), this region by itself is unstructured and solvent-exposed. We propose that the central region of IS1 adopts a α-helical structure only when bound to an as yet unknown partner. We hypothesize that an IS1-binding protein exists in the sarcomere and helps to protect calpain-3 from spurious activation (Fig. 10). This partner might be released from the Ca2+-activated enzyme via a conformational change or by IS1 cleavage at both the N and C termini, thereby opening up the catalytic cleft to substrates (Fig. 10B). But a more attractive possibility is that it could hold the N-terminal portion of the IS1 loop away from the active site, making it fully accessible to substrates without requiring its cleavage. The advantage of a nonproteolytic model of calpain-3 activation is that it can return to the inactive state following calcium signaling without damage or loss of the enzyme.
Figure 10.
Calpain-3 structural model and calpain-3 activation schemes. A, the calpain-3 whole-enzyme model based on the calpain-3 homodimer PEF(L) structure (PDB code 4OKH) (19) (cyan), calpain-3 protease core (PDB code 6BDT) (13), and calpain-2 (20). This model places a protease core at both ends of the protein, with the protease core domains PC1 and PC2 colored yellow and orange, respectively. The CBSW domain is colored green. Hypothetical positions of IS1 (dark blue) and IS2 (dotted line) are indicated. B, activation mechanism scheme where a binding partner (red) pulls IS1 away from the active site to allow for access of substrates and/or inhibitors, where IS1 remains intact. C, scheme where the binding partner (red) pulls on IS1 to draw it away from the active-site Cys, protecting it from autolytic cleavage. Following the conformation change required for activation, the partner releases IS1, so that it may be cleaved. D, the current in vitro model, were IS1 cleavage is required before substrate can access the active-site cleft.
Calpastatin as a novel guide to calpain-3 inhibition
LGMD2A is a progressive wasting disease that affects the proximal limb muscles, eventually leaving patients unable to walk. So far, around 500 different mutations in calpain-3 are known to cause LGMD2A (21, 32, 52, 80, 81); a subset of these predisposes the enzyme to increased autoproteolytic activity and subsequent inactivation (71, 82, 83). Above, we showed that autoinhibition of substrate hydrolysis by IS1 reduces the rate of calpain-2 autoproteolysis, generating a less active but longer-lasting enzyme. Therefore, we propose that treatment of calpain-3 with reversible inhibitors could prolong the half-life of the enzyme in patients exhibiting this category of mutation, thereby improving their outcome.
To date, nonspecific small-molecule calpain inhibitors like E-64 and leupeptin have not proven effective against the rapid autolysis of IS1 in calpain-3, requiring its removal before they can take effect (15, 43, 45, 46, 84, 85). Consistent with this, we found that treatment of calpain-2–IS1 with E-64 and leupeptin also proved unsuccessful at significantly reducing the rate of IS1 autolysis. Many of these established calpain inhibitors and their derivatives occupy the unprimed side of the active-site cleft, in the S1–S3 subsites (2), where, according to the calpain-3 protease core crystal structure (13) and calpain-2 model, the IS1 N-terminal region interacts with the catalytic cleft. This implies that inhibitors that bind to this region of the cleft, like E-64 and leupeptin, must outcompete the tight interactions made by IS1.
The primed side of the active-site cleft has not traditionally been targeted by inhibitors. In calpain-1 and -2, the flexibility of the gating loops is an encouragement to design compounds that extend into this side of the cleft (67, 69). Our results, consistent with what was shown by the crystal and SAXS solution structures of the calpain-3 protease core (13), show that the C-terminal portion of IS1 is highly flexible and may be much more easily pushed aside to make the primed side of the calpain-3 catalytic cleft more accessible to inhibitors. Tight-binding compounds that occupy both sides of the cleft, as with the naturally occurring calpain-1 and -2 inhibitor calpastatin, would be ideal.
The advantage of using whole calpain-2 as an IS1 surrogate is that it can accommodate inhibitors that extend beyond the catalytic core, into regions where contact with the CBSW domain and PEF domains are made. Inhibitors that make these additional contacts may be more capable of competing with IS1 for the active-site cleft. By superimposing the crystal structure of calpastatin-bound rat calpain-2 (20) with the calpain-3 protease core structure (13), we found that the backbone of the N-terminal portion of IS1 and the backbone of the corresponding region of the calpastatin B-peptide occupy nearly identical positions within the active-site cleft, with their side chains pointed in the same orientations and with similar subsite interactions, up until the point where calpastatin forms a loop just out of range of the catalytic Cys. Although calpastatin is not an endogenous inhibitor of calpain-3, we wanted to determine whether IS1 is responsible for preventing calpastatin inhibition of calpain-3 and whether inhibitors that span both sides of the active-site cleft could limit IS1 propeptide cleavage. We found that calpastatin completely stopped calpain-2–IS1 substrate turnover, leaving IS1 intact. This demonstrates that IS1 is not responsible for the failure of calpastatin to inhibit calpain-3 in vivo. However, using just the B region (B-27) peptide (61, 62), we were unable to inhibit IS1 propeptide cleavage, despite the fact that it also spans both the primed and unprimed sides of the catalytic cleft. We think the reason CAST is so much more effective than B-27 is that it is anchored at either end by binding to the PEF domains, which ensures the correct positioning of its inhibitory central section for binding the cleft. Therefore, we propose that calpastatin is indeed capable of inhibiting calpain-3, but does not do so in vivo because calpain-3 forms a homodimer with only one set of PEFs for the two enzymes (13, 19). Only one CAST can bind to the dimer and thus inhibit only one of the two protease cores. There would be no binding site for a second CAST.
Because calpastatin can effectively stop IS1 cleavage in calpain-2–IS1, we propose that the inhibitory B region could be used as a novel lead peptide for the design and development of calpain-3–specific inhibitors for the treatment of LGMD2A.
Conclusions
The rapid autoproteolytic activity within the unique NS, IS1, and IS2 regions of calpain-3 has confounded both the structural and functional study of this enzyme. Here, we have generated a stable calpain-2–IS1 surrogate for the mimicry of calpain-3 and for the study of its unique insertion sequences. We established that, in both calpain-2 and -3, IS1 plays a key autoinhibitory propeptide role. Its position in the catalytic cleft is responsible for the occlusion of substrates and inhibitors, rendering weak inhibitors ineffective against calpain-3 IS1 autolysis. However, the design of tight-binding calpain-specific inhibitors, like calpastatin and the N-terminal region of IS1, may exhibit sufficient calpain-3 inhibition to extend its half-life in vivo and ameliorate the effects of LGMD2A in some patients.
Experimental procedures
Materials
The FRET-based peptide substrates (EDANS)-EPLFAERK-(DABCYL) and (EDANS)-EEVYGMMK-(DABCYL) were synthesized by Biomer Technology (Pleasanton, CA). Recombinant active rat calpain-2 and inactive rat calpain-2 C105S were prepared as described previously (86). Calpastatin CAST4 was also prepared in-house as described previously (20). The calpastatin B-27 peptide calpain inhibitor was obtained from Calbiochem. Calpain inhibitors E-64 and leupeptin were purchased from BioShop (Ontario, CA).
Construction of calpain-2–IS1 surrogate expression vector
The DNA construct encoding the rat calpain-2 80-kDa large subunit, with the 48-residue human calpain-3 IS1 (YGTSPSGLNMGELIARMVRNMDNSLLQDSDLDPRGSDERPTRTIIPVQ) (Fig. S2) DNA sequence inserted after base 750, was synthesized by GeneArt (Life Technologies, Inc.) with a C-terminal His tag encoded. GeneOptimizer (Thermo Fisher Scientific) was used to codon-optimize the gene for improved expression in Escherichia coli. The DNA sequence was also optimized to reduce the formation of RNA secondary structure that might inhibit transcription and/or translation. The calpain-2–IS1 construct was inserted into pET-24a(+) between 5′ NdeI and 3′ XhoI restriction sites. Site-directed mutagenesis was performed to also make a C105S calpain-2–IS1 catalytically incompetent version of the surrogate enzyme. Positive clones were identified by restriction enzyme digestion and DNA sequencing (Robarts Research Institute, London, Canada).
Expression and purification of recombinant calpain-2–IS1 in E. coli
Plasmids from positive clones were electroporated into the E. coli BL-21 (DE3) (star) expression cell line transformed with pACpET-21k, which contains the cDNA encoding the rat calpain small subunit (CAPNS1) with a C-terminal truncation of the glycine-rich domain (86). Cultures were grown to A600 = 0.6 at 37 °C while shaking at 220 rpm in the presence of 100 μg/ml kanamycin and 100 μg/ml ampicillin. The culture was chilled to 23 °C, and protein production was induced by adding 0.1 mm isopropyl β-d-thiogalactoside, and expression was continued overnight at 23 °C with shaking at 220 rpm. The recombinant protein was purified following the protocols outlined by Elce (87) (Fig. S1). Concentrated samples were buffer-exchanged into 50 mm HEPES, pH 7.6, and 10 mm DTT, snap-frozen, and stored at −80 °C until needed. Total protein concentrations were measured by absorbance at 280 nm (ϵ = 1.353 m−1 cm−1), and purity was assessed by SDS-PAGE (10% gel).
Fluorescence-based measurements of calpain activity
Ca2+-induced activation, which causes a large conformational change in calpain-2 and calpain-2–IS1, was measured by intrinsic tryptophan fluorescence using excitation at 280 nm. The emission spectrum was recorded from 300 to 400 nm for 50 μm calpain-2 C015S and calpain-2–IS1 C105S inactive mutants titrated with 50 μm to 5 mm CaCl2 in buffer containing 50 mm Tris-HCl (pH 7.4), 150 mm NaCl, and 0.1% β-mercaptoethanol.
Hydrolysis of the small molecule FRET-based peptide (EDANS)-EPLFAERK-(DABCYL) (65) was measured in a 500-μl cuvette using an LS-55 fluorescence spectrometer (PerkinElmer Life Sciences) with excitation at 335 ± 10 nm and emission at 500 ± 10 nm. Measurements were performed in reaction buffer consisting of 50 mm HEPES-NaOH (pH 7.4) and 0.1% β-mercaptoethanol with 10 μm substrate. The solution was premixed with a stir bar for 30 s, and the reaction was initiated by the addition of 5 mm CaCl2. Assessment of calpain activity was done using 250 nm recombinant rat calpain-2 or calpain-2–IS1. The reactions were performed for 10 min at room temperature with constant stirring. Inhibition assays were conducted using 5 μm E-64, 5 μm leupeptin, 1 μm B-27 calpastatin peptide, or 0.5 μm whole-calpastatin CAST4.
Calpain autolysis SDS-polyacrylamide gels
Calpain autolysis was performed with 5 μm calpain-2 or calpain-2–IS1 in 200 μl of reaction buffer containing 50 mm HEPES-NaOH (pH 7.4) and 0.2 mm DTT. Reactions were activated with the addition of 5 mm CaCl2, and samples were drawn and the reactions stopped at each time point by adding SDS-PAGE loading buffer followed by heating at 95 °C. The calpain autoproteolytic fragments, separated by SDS-PAGE, were identified by in-gel trypsin digestion and LC-MS (SPARC BioCentre, The Hospital for Sick Children, Toronto, Canada). Inhibition of autoproteolysis was examined by adding 50 μm E-64, 50 μm leupeptin, 10 μm calpastatin B-27 peptide, or whole-calpastatin CAST4. Trypsin digestion of Ca2+-bound inactive C105S calpain-2 and calpain-2–IS1 was performed and analyzed following the same conditions outlined above.
GFP-CAST calpain-2 pulldown assay
Calpastatin CAST4 was N-terminally tagged with GFP by gene insertion at the 5′ NdeI restriction site of its DNA construct. GFP-CAST was produced in E. coli BL-21 (DE3) (star) expression cell line under kanamycin selection and was purified by heat denaturation at 68 °C and size-exclusion chromatography using a Superdex 75 10/300 GL column (GE Healthcare).
Calpastatin GFP-CAST pulldowns were performed by binding His6-tagged inactive calpain-2 or calpain-2–IS1 C105S mutants to 200 μl of packed Ni-NTA–agarose resin suspended in Ni-buffer containing 50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 2% (v/v) glycerol, 5 mm imidazole supplemented with either 10 mm CaCl2 or MgCl2. The Ni-NTA–agarose resin with bound calpain was washed three times with Ni-buffer. A 1-ml aliquot of 1 mg/ml GFP-CAST was applied to the immobilized calpain and followed by three washes with Ni-buffer. Measurement of GFP fluorescence, with excitation at 488 nm and emission at 510 nm, was carried out on 50-μl aliquots of the Ni-NTA–agarose resin suspension diluted to 100 μl with Ni-buffer on a Molecular Devices Spectra Max M2 fluorescent microplate reader using a 120-μl half-area 96-well Corning plate (black). Calpain–CAST-GFP complex bound to the Ni-NTA-agarose resin was mixed with SDS-PAGE loading buffer, heated to 95 °C, and assessed by SDS-PAGE
Author contributions
C.-S. E. M. designed and performed the experiments, analyzed data, and drafted the manuscript. Q. Y. provided molecular modeling support and structural analysis and revised the manuscript. P. L. D. and R. L. C. helped conceive the experiments, helped analyze and interpret the results, and edited the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
This work was supported by Canadian Institutes of Health Research Grant MOP 74681. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1 and S2.
- PC
- protease core
- PEF
- penta-EF hand
- NS
- N-terminal leader sequence
- IS
- insertion sequence
- CBSW
- calpain-type β-sandwich domain
- LGMD2A
- limb girdle muscular dystrophy type 2A
- RFU
- relative fluorescence units
- PDB
- Protein Data Bank
- EDANS
- 5-((2-aminoethyl)amino)naphthalene-1-sulfonic acid
- DABCYL
- 4-(4-dimethylaminophenylazo)benzoyl
- Ni-NTA
- nickel-nitrilotriacetic acid
- SAXS
- small-angle X-ray scattering.
References
- 1. Ono Y., Saido T. C., and Sorimachi H. (2016) Calpain research for drug discovery: challenges and potential. Nat. Rev. Drug. Discov. 15, 854–876 10.1038/nrd.2016.212 [DOI] [PubMed] [Google Scholar]
- 2. Campbell R. L., and Davies P. L. (2012) Structure-function relationships in calpains. Biochem. J. 447, 335–351 10.1042/BJ20120921 [DOI] [PubMed] [Google Scholar]
- 3. Croall D. E., and Ersfeld K. (2007) The calpains: modular designs and functional diversity. Genome Biol. 8, 218 10.1186/gb-2007-8-6-218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goll D. E., Thompson V. F., Li H., Wei W., and Cong J. (2003) The calpain system. Physiol. Rev. 83, 731–801 10.1152/physrev.00029.2002 [DOI] [PubMed] [Google Scholar]
- 5. Ono Y., and Sorimachi H. (2012) Calpains: an elaborate proteolytic system. Biochim. Biophys. Acta 1824, 224–236 10.1016/j.bbapap.2011.08.005 [DOI] [PubMed] [Google Scholar]
- 6. Maki M., Kitaura Y., Satoh H., Ohkouchi S., and Shibata H. (2002) Structures, functions and molecular evolution of the penta-EF-hand Ca2+-binding proteins. Biochim. Biophys. Acta 1600, 51–60 10.1016/S1570-9639(02)00444-2 [DOI] [PubMed] [Google Scholar]
- 7. Maki M., Maemoto Y., Osako Y., and Shibata H. (2012) Evolutionary and physical linkage between calpains and penta-EF-hand Ca2+-binding proteins. FEBS J. 279, 1414–1421 10.1111/j.1742-4658.2012.08560.x [DOI] [PubMed] [Google Scholar]
- 8. Hosfield C. M., Elce J. S., Davies P. L., and Jia Z. (1999) Crystal structure of calpain reveals the structural basis for Ca2+-dependent protease activity and a novel mode of enzyme activation. EMBO J. 18, 6880–6889 10.1093/emboj/18.24.6880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Strobl S., Fernandez-Catalan C., Braun M., Huber R., Masumoto H., Nakagawa K., Irie A., Sorimachi H., Bourenkow G., Bartunik H., Suzuki K., and Bode W. (2000) The crystal structure of calcium-free human m-calpain suggests an electrostatic switch mechanism for activation by calcium. Proc. Natl. Acad. Sci. U.S.A. 97, 588–592 10.1073/pnas.97.2.588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sorimachi H., Imajoh-Ohmi S., Emori Y., Kawasaki H., Ohno S., Minami Y., and Suzuki K. (1989) Molecular cloning of a novel mammalian calcium-dependent protease distinct from both m- and mu-types: specific expression of the mRNA in skeletal muscle. J. Biol. Chem. 264, 20106–20111 [PubMed] [Google Scholar]
- 11. Ono Y., Ojima K., Shinkai-Ouchi F., Hata S., and Sorimachi H. (2016) An eccentric calpain, CAPN3/p94/calpain-3. Biochimie 122, 169–187 10.1016/j.biochi.2015.09.010 [DOI] [PubMed] [Google Scholar]
- 12. Suzuki K. (1991) Nomenclature of calcium dependent proteinase. Biomed. Biochim. Acta 50, 483–484 [PubMed] [Google Scholar]
- 13. Ye Q., Campbell R. L., and Davies P. L. (2018) Structures of human calpain-3 protease core with and without bound inhibitor reveal mechanisms of calpain activation. J. Biol. Chem. 293, 4056–4070 10.1074/jbc.RA117.001097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ono Y., and Sorimachi H. (2015) Amino acid sequence alignment of vertebrate CAPN3/calpain-3/p94. Data Brief 5, 366–367 10.1016/j.dib.2015.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kinbara K., Sorimachi H., Ishiura S., and Suzuki K. (1998) Skeletal muscle-specific calpain, p49: structure and physiological function. Biochem. Pharmacol. 56, 415–420 10.1016/S0006-2952(98)00095-1 [DOI] [PubMed] [Google Scholar]
- 16. Kinbara K., Ishiura S., Tomioka S., Sorimachi H., Jeong S. Y., Amano S., Kawasaki H., Kolmerer B., Kimura S., Labeit S., and Suzuki K. (1998) Purification of native p94, a muscle-specific calpain, and characterization of its autolysis. Biochem. J. 335, 589–596 10.1042/bj3350589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ravulapalli R., Diaz B. G., Campbell R. L., and Davies P. L. (2005) Homodimerization of calpain 3 penta-EF-hand domain. Biochem. J. 388, 585–591 10.1042/BJ20041821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ravulapalli R., Campbell R. L., Gauthier S. Y., Dhe-Paganon S., and Davies P. L. (2009) Distinguishing between calpain heterodimerization and homodimerization. FEBS J. 276, 973–982 10.1111/j.1742-4658.2008.06833.x [DOI] [PubMed] [Google Scholar]
- 19. Partha S. K., Ravulapalli R., Allingham J. S., Campbell R. L., and Davies P. L. (2014) Crystal structure of calpain-3 penta-EF-hand (PEF) domain: a homodimerized PEF family member with calcium bound at the fifth EF-hand. FEBS J. 281, 3138–3149 10.1111/febs.12849 [DOI] [PubMed] [Google Scholar]
- 20. Hanna R. A., Campbell R. L., and Davies P. L. (2008) Calcium-bound structure of calpain and its mechanism of inhibition by calpastatin. Nature 456, 409–412 10.1038/nature07451 [DOI] [PubMed] [Google Scholar]
- 21. Richard I., Broux O., Allamand V., Fougerousse F., Chiannilkulchai N., Bourg N., Brenguier L., Devaud C., Pasturaud P., and Roudaut C. (1995) Mutations in the proteolytic enzyme calpain 3 cause limb-girdle muscular dystrophy type 2A. Cell 81, 27–40 10.1016/0092-8674(95)90368-2 [DOI] [PubMed] [Google Scholar]
- 22. Kramerova I., Beckmann J. S., and Spencer M. J. (2007) Molecular and cellular basis of calpainopathy (limb girdle muscular dystrophy type 2A). Biochim. Biophys. Acta 1772, 128–144 10.1016/j.bbadis.2006.07.002 [DOI] [PubMed] [Google Scholar]
- 23. Fanin M., Nascimbeni A. C., Fulizio L., Trevisan C. P., Meznaric-Petrusa M., and Angelini C. (2003) Loss of calpain-3 autocatalytic activity in LGMD2A patients with normal protein expression. Am. J. Pathol. 163, 1929–1936 10.1016/S0002-9440(10)63551-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cohen N., Kudryashova E., Kramerova I., Anderson L. V., Beckmann J. S., Bushby K., and Spencer M. J. (2006) Identification of putative in vivo substrates of calpain 3 by comparative proteomics of overexpressing transgenic and nontransgenic mice. Proteomics 6, 6075–6084 10.1002/pmic.200600199 [DOI] [PubMed] [Google Scholar]
- 25. Guyon J. R., Kudryashova E., Potts A., Dalkilic I., Brosius M. A., Thompson T. G., Beckmann J. S., Kunkel L. M., and Spencer M. J. (2003) Calpain 3 cleaves filamin C and regulates its ability to interact with γ- and δ-sarcoglycans. Muscle Nerve 28, 472–483 10.1002/mus.10465 [DOI] [PubMed] [Google Scholar]
- 26. Taveau M., Bourg N., Sillon G., Roudaut C., Bartoli M., and Richard I. (2003) Calpain 3 is activated through autolysis within the active site and lyses sarcomeric and sarcolemmal components. Mol. Cell. Biol. 23, 9127–9135 10.1128/MCB.23.24.9127-9135.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kramerova I., Kudryashova E., Tidball J. G., and Spencer M. J. (2004) Null mutation of calpain 3 (p94) in mice causes abnormal sarcomere formation in vivo and in vitro. Hum. Mol. Genet. 13, 1373–1388 10.1093/hmg/ddh153 [DOI] [PubMed] [Google Scholar]
- 28. Sorimachi H., Kinbara K., Kimura S., Takahashi M., Ishiura S., Sasagawa N., Sorimachi N., Shimada H., Tagawa K., and Maruyama K. (1995) Muscle-specific calpain, p94, responsible for limb girdle muscular dystrophy type 2A, associates with connectin through IS2, a p94-specific sequence. J. Biol. Chem. 270, 31158–31162 10.1074/jbc.270.52.31158 [DOI] [PubMed] [Google Scholar]
- 29. Murphy R. M., Snow R. J., and Lamb G. D. (2006) mu-Calpain and calpain-3 are not autolyzed with exhaustive exercise in humans. Am. J. Physiol. Cell Physiol. 290, C116–C122 10.1152/ajpcell.00291.2005 [DOI] [PubMed] [Google Scholar]
- 30. Kanzaki K., Kuratani M., Matsunaga S., Yanaka N., and Wada M. (2014) Three calpain isoforms are autolyzed in rat fast-twitch muscle after eccentric contractions. J. Muscle Res. Cell Motil. 35, 179–189 10.1007/s10974-014-9378-9 [DOI] [PubMed] [Google Scholar]
- 31. Kanzaki K., Watanabe D., Kuratani M., Yamada T., Matsunaga S., and Wada M. (2017) Role of calpain in eccentric contraction-induced proteolysis of Ca2+-regulatory proteins and force depression in rat fast-twitch skeletal muscle. J. Appl. Physiol. 122, 396–405 10.1152/japplphysiol.00270.2016 [DOI] [PubMed] [Google Scholar]
- 32. Hauerslev S., Sveen M. L., Duno M., Angelini C., Vissing J., and Krag T. O. (2012) Calpain 3 is important for muscle regeneration: evidence from patients with limb girdle muscular dystrophies. BMC Musculoskelet. Disord. 13, 43 10.1186/1471-2474-13-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Murphy R. M., Goodman C. A., McKenna M. J., Bennie J., Leikis M., and Lamb G. D. (2007) Calpain-3 is autolyzed and hence activated in human skeletal muscle 24 h following a single bout of eccentric exercise. J. Appl. Physiol. 103, 926–931 10.1152/japplphysiol.01422.2006 [DOI] [PubMed] [Google Scholar]
- 34. Murphy R. M., and Lamb G. D. (2009) Endogenous calpain-3 activation is primarily governed by small increases in resting cytoplasmic [Ca2+] and is not dependent on stretch. J. Biol. Chem. 284, 7811–7819 10.1074/jbc.M808655200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Murphy R. M., Vissing K., Latchman H., Lamboley C., McKenna M. J., Overgaard K., and Lamb G. D. (2011) Activation of skeletal muscle calpain-3 by eccentric exercise in humans does not result in its translocation to the nucleus or cytosol. J. Appl. Physiol. 111, 1448–1458 10.1152/japplphysiol.00441.2011 [DOI] [PubMed] [Google Scholar]
- 36. Murphy R. M., and Lamb G. D. (2009) Calpain-3 is activated following eccentric exercise. J. Appl. Physiol. 106, 2068; author reply 2069 10.1152/japplphysiol.00239.2009 [DOI] [PubMed] [Google Scholar]
- 37. Murphy R. M. (2010) Calpains, skeletal muscle function and exercise. Clin. Exp. Pharmacol. Physiol. 37, 385–391 10.1111/j.1440-1681.2009.05310.x [DOI] [PubMed] [Google Scholar]
- 38. Labeit S., Kolmerer B., and Linke W. A. (1997) The giant protein titin. Emerging roles in physiology and pathophysiology. Circ. Res. 80, 290–294 10.1161/01.RES.80.2.290 [DOI] [PubMed] [Google Scholar]
- 39. Sorimachi H., Toyama-Sorimachi N., Saido T. C., Kawasaki H., Sugita H., Miyasaka M., Arahata K., Ishiura S., and Suzuki K. (1993) Muscle-specific calpain, p94, is degraded by autolysis immediately after translation, resulting in disappearance from muscle. J. Biol. Chem. 268, 10593–10605 [PubMed] [Google Scholar]
- 40. Ono Y., Hayashi C., Doi N., Kitamura F., Shindo M., Kudo K., Tsubata T., Yanagida M., and Sorimachi H. (2007) Comprehensive survey of p94/calpain 3 substrates by comparative proteomics: possible regulation of protein synthesis by p94. Biotechnol. J. 2, 565–576 10.1002/biot.200700018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Federici C., Eshdat Y., Richard I., Bertin B., Guillaume J. L., Hattab M., Beckmann J. S., Strosberg A. D., and Camoin L. (1999) Purification and identification of two putative autolytic sites in human calpain 3 (p94) expressed in heterologous systems. Arch. Biochem. Biophys. 363, 237–245 10.1006/abbi.1998.1091 [DOI] [PubMed] [Google Scholar]
- 42. Sorimachi H., Ohmi S., Emori Y., Kawasaki H., Saido T. C., Ohno S., Minami Y., and Suzuki K. (1990) A novel member of the calcium-dependent cysteine protease family. Biol. Chem. Hoppe Seyler 371, 171–176 [PubMed] [Google Scholar]
- 43. Fukiage C., Nakajima E., Ma H., Azuma M., and Shearer T. R. (2002) Characterization and regulation of lens-specific calpain Lp82. J. Biol. Chem. 277, 20678–20685 10.1074/jbc.M200697200 [DOI] [PubMed] [Google Scholar]
- 44. García Díaz B. E., Gauthier S., and Davies P. L. (2006) Ca2+ dependency of calpain 3 (p94) activation. Biochemistry 45, 3714–3722 10.1021/bi051917j [DOI] [PubMed] [Google Scholar]
- 45. Rey M. A., and Davies P. L. (2002) The protease core of the muscle-specific calpain, p94, undergoes Ca2+-dependent intramolecular autolysis. FEBS Lett. 532, 401–406 10.1016/S0014-5793(02)03722-5 [DOI] [PubMed] [Google Scholar]
- 46. Diaz B. G., Moldoveanu T., Kuiper M. J., Campbell R. L., and Davies P. L. (2004) Insertion sequence 1 of muscle-specific calpain, p94, acts as an internal propeptide. J. Biol. Chem. 279, 27656–27666 10.1074/jbc.M313290200 [DOI] [PubMed] [Google Scholar]
- 47. Moldoveanu T., Hosfield C. M., Lim D., Elce J. S., Jia Z., and Davies P. L. (2002) A Ca2+ switch aligns the active site of calpain. Cell 108, 649–660 10.1016/S0092-8674(02)00659-1 [DOI] [PubMed] [Google Scholar]
- 48. Moldoveanu T., Jia Z., and Davies P. L. (2004) Calpain activation by cooperative Ca2+ binding at two non-EF-hand sites. J. Biol. Chem. 279, 6106–6114 10.1074/jbc.M310460200 [DOI] [PubMed] [Google Scholar]
- 49. Piluso G., Politano L., Aurino S., Fanin M., Ricci E., Ventriglia V. M., Belsito A., Totaro A., Saccone V., Topaloglu H., Nascimbeni A. C., Fulizio L., Broccolini A., Canki-Klain N., Comi L. I., Nigro G., Angelini C., and Nigro V. (2005) Extensive scanning of the calpain-3 gene broadens the spectrum of LGMD2A phenotypes. J. Med. Genet. 42, 686–693 10.1136/jmg.2004.028738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fanin M., Fulizio L., Nascimbeni A. C., Spinazzi M., Piluso G., Ventriglia V. M., Ruzza G., Siciliano G., Trevisan C. P., Politano L., Nigro V., and Angelini C. (2004) Molecular diagnosis in LGMD2A: mutation analysis or protein testing? Hum. Mutat. 24, 52–62 10.1002/humu.20058 [DOI] [PubMed] [Google Scholar]
- 51. Ono Y., Torii F., Ojima K., Doi N., Yoshioka K., Kawabata Y., Labeit D., Labeit S., Suzuki K., Abe K., Maeda T., and Sorimachi H. (2006) Suppressed disassembly of autolyzing p94/CAPN3 by N2A connectin/titin in a genetic reporter system. J. Biol. Chem. 281, 18519–18531 10.1074/jbc.M601029200 [DOI] [PubMed] [Google Scholar]
- 52. Beckmann J. S., and Spencer M. (2008) Calpain 3, the “gatekeeper” of proper sarcomere assembly, turnover and maintenance. Neuromuscul. Disord. 18, 913–921 10.1016/j.nmd.2008.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Branca D., Gugliucci A., Bano D., Brini M., and Carafoli E. (1999) Expression, partial purification and functional properties of themuscle-specific calpain isoform p94. Eur. J. Biochem. 265, 839–846 10.1046/j.1432-1327.1999.00817.x [DOI] [PubMed] [Google Scholar]
- 54. Ma H., Fukiage C., Azuma M., and Shearer T. R. (1998) Cloning and expression of mRNA for calpain Lp82 from rat lens: splice variant of p94. Invest. Ophthalmol. Vis. Sci. 39, 454–461 [PubMed] [Google Scholar]
- 55. Wendt A., Thompson V. F., and Goll D. E. (2004) Interaction of calpastatin with calpain: a review. Biol. Chem. 385, 465–472 [DOI] [PubMed] [Google Scholar]
- 56. Emori Y., Kawasaki H., Imajoh S., Minami Y., and Suzuki K. (1988) All four repeating domains of the endogenous inhibitor for calcium-dependent protease independently retain inhibitory activity: expression of the cDNA fragments in Escherichia coli. J. Biol. Chem. 263, 2364–2370 [PubMed] [Google Scholar]
- 57. Kawasaki H., Emori Y., Imajoh-Ohmi S., Minami Y., and Suzuki K. (1989) Identification and characterization of inhibitory sequences in four repeating domains of the endogenous inhibitor for calcium-dependent protease. J. Biochem. 106, 274–281 10.1093/oxfordjournals.jbchem.a122844 [DOI] [PubMed] [Google Scholar]
- 58. Takano E., Maki M., Mori H., Hatanaka M., Marti T., Titani K., Kannagi R., Ooi T., and Murachi T. (1988) Pig heart calpastatin: identification of repetitive domain structures and anomalous behavior in polyacrylamide gel electrophoresis. Biochemistry 27, 1964–1972 10.1021/bi00406a024 [DOI] [PubMed] [Google Scholar]
- 59. Hanna R. A., Garcia-Diaz B. E., and Davies P. L. (2007) Calpastatin simultaneously binds four calpains with different kinetic constants. FEBS Lett. 581, 2894–2898 10.1016/j.febslet.2007.05.035 [DOI] [PubMed] [Google Scholar]
- 60. Kiss R., Kovács D., Tompa P., and Perczel A. (2008) Local structural preferences of calpastatin, the intrinsically unstructured protein inhibitor of calpain. Biochemistry 47, 6936–6945 10.1021/bi800201a [DOI] [PubMed] [Google Scholar]
- 61. Betts R., and Anagli J. (2004) The β- and γ-CH2 of B27-WT's Leu11 and Ile18 side chains play a direct role in calpain inhibition. Biochemistry 43, 2596–2604 10.1021/bi0359832 [DOI] [PubMed] [Google Scholar]
- 62. Betts R., Weinsheimer S., Blouse G. E., and Anagli J. (2003) Structural determinants of the calpain inhibitory activity of calpastatin peptide B27-WT. J. Biol. Chem. 278, 7800–7809 10.1074/jbc.M208350200 [DOI] [PubMed] [Google Scholar]
- 63. Moldoveanu T., Gehring K., and Green D. R. (2008) Concerted multi-pronged attack by calpastatin to occlude the catalytic cleft of heterodimeric calpains. Nature 456, 404–408 10.1038/nature07353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ono Y., Kakinuma K., Torii F., Irie A., Nakagawa K., Labeit S., Abe K., Suzuki K., and Sorimachi H. (2004) Possible regulation of the conventional calpain system by skeletal muscle-specific calpain, p94/calpain 3. J. Biol. Chem. 279, 2761–2771 10.1074/jbc.M308789200 [DOI] [PubMed] [Google Scholar]
- 65. Cuerrier D., Moldoveanu T., and Davies P. L. (2005) Determination of peptide substrate specificity for mu-calpain by a peptide library-based approach: the importance of primed side interactions. J. Biol. Chem. 280, 40632–40641 10.1074/jbc.M506870200 [DOI] [PubMed] [Google Scholar]
- 66. Moldoveanu T., Campbell R. L., Cuerrier D., and Davies P. L. (2004) Crystal structures of calpain-E64 and -leupeptin inhibitor complexes reveal mobile loops gating the active site. J. Mol. Biol. 343, 1313–1326 10.1016/j.jmb.2004.09.016 [DOI] [PubMed] [Google Scholar]
- 67. Cuerrier D., Moldoveanu T., Inoue J., Davies P. L., and Campbell R. L. (2006) Calpain inhibition by α-ketoamide and cyclic hemiacetal inhibitors revealed by X-ray crystallography. Biochemistry 45, 7446–7452 10.1021/bi060425j [DOI] [PubMed] [Google Scholar]
- 68. Cuerrier D., Moldoveanu T., Campbell R. L., Kelly J., Yoruk B., Verhelst S. H., Greenbaum D., Bogyo M., and Davies P. L. (2007) Development of calpain-specific inactivators by screening of positional scanning epoxide libraries. J. Biol. Chem. 282, 9600–9611 10.1074/jbc.M610372200 [DOI] [PubMed] [Google Scholar]
- 69. Qian J., Cuerrier D., Davies P. L., Li Z., Powers J. C., and Campbell R. L. (2008) Cocrystal structures of primed side-extending α-ketoamide inhibitors reveal novel calpain-inhibitor aromatic interactions. J. Med. Chem. 51, 5264–5270 10.1021/jm800045t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Li Q., Hanzlik R. P., Weaver R. F., and Schönbrunn E. (2006) Molecular mode of action of a covalently inhibiting peptidomimetic on the human calpain protease core. Biochemistry 45, 701–708 10.1021/bi052077b [DOI] [PubMed] [Google Scholar]
- 71. Jia Z., Petrounevitch V., Wong A., Moldoveanu T., Davies P. L., Elce J. S., and Beckmann J. S. (2001) Mutations in calpain 3 associated with limb girdle muscular dystrophy: analysis by molecular modeling and by mutation in m-calpain. Biophys. J. 80, 2590–2596 10.1016/S0006-3495(01)76229-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cong J., Goll D. E., Peterson A. M., and Kapprell H. P. (1989) The role of autolysis in activity of the Ca2+-dependent proteinases (mu-calpain and m-calpain). J. Biol. Chem. 264, 10096–10103 [PubMed] [Google Scholar]
- 73. Yoshimura N., Kikuchi T., Sasaki T., Kitahara A., Hatanaka M., and Murachi T. (1983) Two distinct Ca2+ proteases (calpain I and calpain II) purified concurrently by the same method from rat kidney. J. Biol. Chem. 258, 8883–8889 [PubMed] [Google Scholar]
- 74. Graham-Siegenthaler K., Gauthier S., Davies P. L., and Elce J. S. (1994) Active recombinant rat calpain II. Bacterially produced large and small subunits associate both in vivo and in vitro. J. Biol. Chem. 269, 30457–30460 [PubMed] [Google Scholar]
- 75. Moldoveanu T., Hosfield C. M., Jia Z., Elce J. S., and Davies P. L. (2001) Ca2+-induced structural changes in rat m-calpain revealed by partial proteolysis. Biochim. Biophys. Acta 1545, 245–254 10.1016/S0167-4838(00)00286-7 [DOI] [PubMed] [Google Scholar]
- 76. Thompson V. F., Lawson K. R., Barlow J., and Goll D. E. (2003) Digestion of μ- and m-calpain by trypsin and chymotrypsin. Biochim. Biophys. Acta 1648, 140–153 10.1016/S1570-9639(03)00115-8 [DOI] [PubMed] [Google Scholar]
- 77. Crawford C., Willis A. C., and Gagnon J. (1987) The effects of autolysis on the structure of chicken calpain II. Biochem. J. 248, 579–588 10.1042/bj2480579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. DeMartino G. N., Huff C. A., and Croall D. E. (1986) Autoproteolysis of the small subunit of calcium-dependent protease II activates and regulates protease activity. J. Biol. Chem. 261, 12047–12052 [PubMed] [Google Scholar]
- 79. Chou J. S., Impens F., Gevaert K., and Davies P. L. (2011) m-calpain activation in vitro does not require autolysis or subunit dissociation. Biochim. Biophys. Acta 1814, 864–872 10.1016/j.bbapap.2011.04.007 [DOI] [PubMed] [Google Scholar]
- 80. Fanin M., and Angelini C. (2015) Protein and genetic diagnosis of limb girdle muscular dystrophy type 2A: the yield and the pitfalls. Muscle Nerve 52, 163–173 10.1002/mus.24682 [DOI] [PubMed] [Google Scholar]
- 81. Fanin M., Nascimbeni A. C., and Angelini C. (2007) Screening of calpain-3 autolytic activity in LGMD muscle: a functional map of CAPN3 gene mutations. J. Med. Genet. 44, 38–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Garnham C. P., Hanna R. A., Chou J. S., Low K. E., Gourlay K., Campbell R. L., Beckmann J. S., and Davies P. L. (2009) Limb-girdle muscular dystrophy type 2A can result from accelerated autoproteolytic inactivation of calpain 3. Biochemistry 48, 3457–3467 10.1021/bi900130u [DOI] [PubMed] [Google Scholar]
- 83. Pantoja-Melendez C. A., Miranda-Duarte A., Roque-Ramirez B., and Zenteno J. C. (2017) Epidemiological and molecular characterization of a Mexican population isolate with high prevalence of limb-girdle muscular dystrophy type 2A due to a novel calpain-3 mutation. PLoS One 12, e0170280 10.1371/journal.pone.0170280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Nakamura Y., Fukiage C., Ma H., Shih M., Azuma M., and Shearer T. R. (1999) Decreased sensitivity of lens-specific calpain Lp82 to calpastatin inhibitor. Exp. Eye Res. 69, 155–162 10.1006/exer.1998.0686 [DOI] [PubMed] [Google Scholar]
- 85. De Tullio R., Stifanese R., Salamino F., Pontremoli S., and Melloni E. (2003) Characterization of a new p94-like calpain form in human lymphocytes. Biochem. J. 375, 689–696 10.1042/bj20030706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Elce J. S., Hegadorn C., Gauthier S., Vince J. W., and Davies P. L. (1995) Recombinant calpain II: improved expression systems and production of a C105A active-site mutant for crystallography. Protein Eng. 8, 843–848 10.1093/protein/8.8.843 [DOI] [PubMed] [Google Scholar]
- 87. Elce J. S. (2000) Expression of m-calpain in Escherichia coli. Methods Mol. Biol. 144, 47–54 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.