Abstract
Somatic structural variants undoubtedly play important roles in driving tumourigenesis. This is evident despite the substantial technical challenges that remain in accurately detecting structural variants and their breakpoints in tumours and in spite of our incomplete understanding of the impact of structural variants on cellular function. Developments in these areas of research contribute to the ongoing discovery of structural variation with a clear impact on the evolution of the tumour and on the clinical importance to the patient. Recent large whole genome sequencing studies have reinforced our impression of each tumour as a unique combination of mutations but paradoxically have also discovered similar genome-wide patterns of single-nucleotide and structural variation between tumours. Statistical methods have been developed to deconvolute mutation patterns, or signatures, that recur across samples, providing information about the mutagens and repair processes that may be active in a given tumour. These signatures can guide treatment by, for example, highlighting vulnerabilities in a particular tumour to a particular chemotherapy. Thus, although the complete reconstruction of the full evolutionary trajectory of a tumour genome remains currently out of reach, valuable data are already emerging to improve the treatment of cancer.
Keywords: Structural variation, cancer, tumourigenesis, DNA double-strand breaks, mutational signatures, whole genome sequencing
Introduction
Tumours arise when normal cells accumulate enough genetic alterations to affect normal regulatory control systems 1. These alterations usually include somatic structural variants (SVs) which are large-scale changes to the genome, ranging from 50 nucleotides to many megabases in length 2. SVs can occur as a result of improper DNA double-strand break (DSB) repair 3 and are traditionally classified into five different types: insertions, deletions, duplications, inversions, and translocations 2. Although all tumours accumulate both somatic single-nucleotide variant (SNV) mutations and SVs, certain tumour types such as high-grade serous ovarian cancer (HGSOC) and invasive breast cancer 4 are dominated by SVs. Furthermore, in HGSOC 5 and pancreatic neuroendocrine tumours 6, there is good evidence for SVs playing prominent roles in driving tumourigenesis. Various sequencing strategies, particularly whole genome sequencing (WGS), can be used to identify structural variation in tumours by comparing the pattern of structural variation in tumour tissue with that observed in normal cells 2, 7. A range of computational tools are then used to detect these variants from sequencing data, but detection algorithms perform very differently in identifying particular SV classes, and none of the existing tools are effective in detecting all SVs on their own 8. Copy number alteration (CNA) detection in cancer samples has been reviewed extensively 9– 11 because of its important role in tumourigenesis 12, 13. For example, it has been suggested that HLA allele loss in 40% of non-small cell lung cancers enables immune escape 14. The detection of CNA is a more tractable problem than the detection of many other SVs, as CNAs can be inferred purely on the basis of changes in read depth over a region. Regions with lower read depth suggest loss of genetic material, whereas regions of higher read depth suggest gain. Aside from detecting copy number changes, analyses of SVs in the past have focused on investigating simpler rearrangements of known cancer genes or identifying fusion genes 15– 19; however, the focus is now shifting to considering more complex events and SVs as part of broader, genome-wide patterns of mutation 20. Our present knowledge of structural aberrations is clearly far from complete, such that although aneuploidy is well studied 21– 23, large-scale phenomena such as whole genome doubling have only recently been discovered to be a common event in many cancers and to have important implications for clinical prognoses 24.
Even when an SV is associated with patient survival, it is challenging to uncover the underlying mechanism (that is, how the SV impacts tumour function). It is known that SVs can alter the expression of oncogenes or tumour suppressor genes and that these changes in expression may directly contribute to tumourigenesis 3, 8, 25– 27. It also seems that SVs with breakpoints in the main body of a gene may often be associated with increased expression of the gene but that SVs with breakpoints within 50 kb upstream or downstream of a gene are correlated with decreased expression 25. Similarly, it has been observed that amplified regions of the genome are enriched for oncogenes but that deleted regions are enriched for tumour suppressors 1. However, SVs may alter the expression of genes through various mechanisms, and multiple SVs may occur at the same locus, so the impact of a particular SV on nearby genes is often difficult to predict 25. SVs have long been known to result in the formation of functional fusion genes by translocations 1. For example, the BCR1-ABL fusion gene is formed by a t(9;22)(q34;q11) translocation which leads to increased tyrosine kinase activity, uncontrolled proliferation, and ultimately chronic myeloid leukaemia (CML). This type of gene fusion can be targeted with kinase inhibitors, resulting in an effective treatment for CML. Rearrangements can also reposition regulatory elements such as enhancers next to oncogenes, which has been observed in Burkitt’s lymphoma and results in increased expression of the MYC oncogene 1, 25. Recent studies have suggested that enhancer adoption by oncogenes following the disruption of regulatory domains is fairly common in tumourigenesis, occurring at rates comparable to those of recurrent in-frame gene fusions 28, although the domain architectures themselves may simply be prone to higher rates of mutation 29. Finally, there appear to be SVs with breakpoints that do not have direct effects on either genes or regulatory elements and yet their presence is associated with patient survival rates 1. Beyond gene expression, the effects of SVs are not well studied, but notably modest correlations have been reported between the extent of tumour duplications and deletions involving a gene and the levels of the protein it encodes 30. In summary, although simpler SVs may tend to have intuitive effects on gene expression (for example, deletions leading to lower expression and amplifications leading to higher expression), it is generally not straightforward to interpret the functional effects of a given SV within a given tumour.
Identifying structural variation
Historically, chromosomal aberrations were detected by using microscopy and traditional karyotyping methods, which then were improved with the development of fluorescent in situ hybridisation techniques 31. Modern high-throughput sequencing technologies are capable of detecting a wider variety of structural variation at much higher resolution. The most widely used sequencing technologies generate billions of short reads about 100 nucleotides in length from sequencing libraries 8. The short lengths of the reads, sequencing platform error rates, and the sequencing libraries used can all present challenges for SV detection. Some reads map ambiguously to many different regions in the reference genome, which makes it difficult to determine the true location of the corresponding SV. This is particularly true of reads in repetitive regions 8. Smaller library insert sizes enable the precise identification of small SVs such as insertions and deletions; however, smaller insert sizes will cover only short stretches of sequence, making it hard to cover large SVs 2. Contrastingly, larger insert sizes allow the identification of large SVs but may have insufficient coverage to determine the location of breakpoints at base-pair resolution. Given the high cost of conducting sequencing experiments using multiple library sizes, it is important to use the appropriate strategy for the biological hypothesis under study. Chimeric reads are artificial sequences formed during the amplification step of polymerase chain reaction (PCR) and can also result in artefacts during analysis. The effect of chimeric reads on SV prediction can be mitigated by using multiple libraries per sample and filtering out novel sequences that are in only one library; however, this comes with the risk of filtering out low-frequency SVs 2. Alternatively, there are tools such as ChimeraChecker 32 and ChimeraSlayer 33 which are designed to remove chimeric reads. Finally, the genomic context of the sequence also impacts SV calling. Regions that are poorly characterised in the reference genome such as pericentromeric and telomeric regions present a challenge to the identification of SVs. In addition, repetitive or GC-rich regions are often problematic for confident read mapping, which can be overcome by sequencing technologies generating longer reads 2.
Several long-range technologies have been developed to try to address these challenges 34. PacBio 35 and Oxford Nanopore 36 sequencing platforms both produce long reads that can be used for SV detection. The resultant reads from these technologies average about 10 kb in length and some read lengths recorded up to 100 kb for PacBio and 1 Mb for Oxford Nanopore. However, these technologies are more expensive than standard Illumina short-read sequencing and have higher error rates. An alternative approach is to use a technology based on linked short reads, such as the platforms developed by 10X Genomics 37. This enables the identification of large SVs at a lower cost than for the long-read technologies; however, sequencing coverage can be relatively sparse, limiting the resolution of locally repetitive sequences 34. Although they are established, these technologies are currently not widely used in cancer genomics and the majority of SV detection in tumours for the moment is still carried out on the basis of short-read sequencing data.
Most SV detection algorithms or “callers” identify SVs from paired-end sequencing reads that are anomalously mapped. Anomalous reads can be used to predict candidate SV loci because reads that do not contain an SV breakpoint are expected to map concordantly 2. Anomalously mapped reads fall into one of three categories: discordant reads, soft-clipped reads, and one-end-anchored reads. Discordant reads can be used in the direct discovery of SVs and are mapped to either different chromosomes (providing evidence for translocations) or incorrect strands (suggesting inversions) or are in incorrect orientation (suggesting duplications) or are an incorrect distance apart (indicating the presence of an insertion or deletion). Detecting SVs from discordant reads is more effective for the detection of larger SV events, as they are unable to estimate breakpoint locations precisely and small SVs may have insufficient discordant read coverage to be detectable 8. Soft-clipped and one-end-anchored reads can be used for indirect SV discovery. Soft-clipped reads are only partially mapped, and one-end-anchored reads have only one read of the pair mapped. In both cases, split-read mapping is needed to successfully map both ends of the SV. These categories of anomaly are better for detecting smaller events such as small insertions and deletions, as they are able to estimate the breakpoints at a higher resolution.
In practice, SV callers use one or more types of anomalous reads to predict candidate SV loci in the following ways 2: first, by clustering discordant reads that support the same type of SV together (for example, SVDetect 38); second, by using split-read mapping to map the other end of indirectly discovered SVs (for example, BreakSeq 39) or to refine the estimate of the breakpoint location in directly discovered SVs (for example, LUMPY 40); and, third, by using de novo contig assembly to stitch together short reads to make a longer segment that can be mapped back to the reference and improve the ability to locate breakpoints (for example, CREST 41). Some callers use combinations of all of these techniques, in addition to assessing changes in read depth, in order to improve their detection sensitivity (for example, DELLY 42).
Assessing the accuracy of SV callers has proven to be challenging. The International Cancer Genome Consortium–The Cancer Genome Atlas (ICGC-TCGA) DREAM SMC-DNA Challenge ( https://www.synapse.org/#!Synapse:syn312572/wiki/) was launched in 2013, aiming to benchmark somatic SNV and SV predictions from a wide variety of algorithms using simulated and real tumour-normal WGS pairs. The project revealed widespread biases in caller predictions and recommended the use of consensus calls made by multiple callers for both SNV and SV prediction 43. More detailed accounts of the SV callers’ performance showed that several algorithms (including Breakdancer, DELLY, Pindel, Manta, and novoBreak) could achieve high precision after stringent filtering of SV calls and substantial overlaps in the breakpoints called between them 44. Unfortunately, SV calling is a rapidly evolving field, and there is no ongoing attempt to comprehensively benchmark algorithms as they appear or are updated. In spite of this, multiple comparative reviews of the most popular SV detection tools and algorithms have emerged 8, 45, 46. The ensemble approach to both SNV and SV calling explored in the DREAM Challenge has since become the de facto standard operating procedure in the field, as reflected in the recent re-analyses of large TCGA 47 and ICGC 48 tumour sequencing studies. The choice of algorithms employed in such an ensemble is often influenced by practical considerations, such as computational demands and run times, but should also consider the shared assumptions and biases between tools 49. Because any ensemble is expected to generate false-positive and false-negative results, there is also a requirement for validation by orthogonal data for SVs of particular interest, and long-read sequencing technologies may often be useful in this respect 50.
Genesis and patterns of structural variation
SVs arise when DNA DSBs are improperly repaired 51. The three main DSB repair mechanisms are homologous recombination (HR), non-homologous end joining (NHEJ), and replication-based mechanisms 3, 52. HR is the most common DNA repair mechanism 3 and is accurate as long as there is a homologous sequence to use as a repair template. However, in tumour genomes, there are few overlapping sequences at the breakpoints 53, 54, so generally HR is not possible and the error-prone NHEJ is implemented instead. NHEJ requires no homology and sometimes can generate short segments of microhomology or small insertions at the breakpoints 3. Replication-based mechanisms of DNA DSB repair such as microhomology-mediated break-induced repair (MMBIR) have also been described 55. It is thought that in these cases the replication fork can stall and the polymerase can switch template sequence using microhomology to any nearby single-stranded DNA 3. This can result in a range of complex structural rearrangements 56, 57. In tumour genomes, which are often HR deficient, these last two mechanisms of repair dominate 3. A further mechanism behind structural variation in cancer is chromosome instability. This is defined as an increased rate of chromosomal change in comparison with normal cells. Nearly all cancer cells have some degree of chromosomal instability, but it is unclear whether this drives tumourigenesis or is a by-product of cancer evolution 58. There is evidence to suggest that a background of chromosomal instability accelerates the rate of tumourigenesis aided by the acquisition of mutations that promote tolerance of instability (for example, inactivation of TP53) 58. Furthermore, the role of recombination activating gene (RAG) and activation-induced cytidine deaminase (AID) in the diversification of immunoglobulin-encoding genes has been proposed as a driver of SVs in leukaemia. The RAG1 and RAG2 endonucleases introduce DSBs followed by the recombination of the immunoglobulin variable, diversity, and joining regions. AID deaminates cytosine residues in the immunoglobulin switch and variable regions, allowing somatic hypermutation. This genetic diversification of antibodies is activated in response to infectious or inflammatory stimuli and has been shown to contribute to the accumulation of somatic mutations necessary for clonal evolution in leukaemia 59, 60. The mechanisms underlying the generation of structural variation outlined above have been reviewed extensively elsewhere 1, 3, 51, 61, 62.
The traditional model of cancer development involves the gradual sequential accumulation of mutations, which leads to gradual progression of normal tissue through increasingly disordered clinical and pathological stages until it becomes a malignant tumour 54. An alternative model was suggested by Stephens et al. to explain complex patterns of genomic rearrangement 54. They suggest a catastrophe-like model where multiple SVs occur as a result of a single event 54.
It is likely that such a model of mutation is behind complex patterns of genomic rearrangements found in cancer, such as chromothripsis and chromoplexy 54, 63– 66. Chromothripsis is characterised by tens to hundreds of genomic rearrangements across an extended chromosomal region. A region that has undergone chromothripsis shows the following hallmarks: oscillating copy number between usually two copy number states; the retention of heterozygosity in the regions with the higher copy number state; and clustering of SV breakpoints to a greater extent than would be expected given a background model of the regional propensity to rearrangement 54. Chromothripsis appears to be present in 2% to 3% of all cancers and has a higher prevalence in bone cancers (25%) 54. It has been reported to be associated with the presence of TP53 mutations in tumours of diverse origins, including medulloblastomas, acute myeloid leukaemia, and pancreatic tumours 67– 73, suggesting a connection to general genome instability. Moreover, chromothripsis has been associated with poorer prognosis in neuroblastoma 74, acute myeloid leukaemia 70, 73, multiple myeloma 75, and malignant melanoma 76. In contrast, it has been linked with longer progression-free survival in colorectal cancer 77. This suggests that chromothripsis has the potential to be an informative biomarker in tumours but with implications that may differ by cancer type. A further complex pattern of structural rearrangements is chromoplexy 78– 80. This is characterised by chains of translocations and deletions that are interdependent. It differs from chromothripsis in that it involves fewer rearrangements per chain (3–40+) and the rearrangements can be spread across up to 10 chromosomes. It is thought that multiple chains of chromoplexy can be formed in successive cell cycles but that all of the SVs in each chain are likely to be formed by one event 63. A further sign of chromoplexy is the presence of small deletions between fusion junctions of some translocation rearrangements known as “deletion bridges” 63, 79. Chromoplexy is largely copy number neutral and has a high prevalence in prostate cancers (88%) 63, 78, 80.
Rates of structural mutation vary across the genome, and there are some regions of the genome that are more susceptible to recurrent structural mutation than others. Glodzik et al. 81 used a large WGS breast cancer cohort 20 to identify such regions that appear to harbour greater recurrent structural variation than expected given the genomic characteristics of the region. These characteristics included replication timing domains, gene-rich regions, background copy number, chromatin states, and repetitive sequences. Many of the rearrangement hotspots found were dominated by long (greater than 100 kb) tandem duplications. These hotspots often duplicated key driver genes and regulatory elements and were enriched for breast cancer susceptibility loci and tissue-specific super enhancers. Thus, based upon the functional regions included, some fraction of these hotspots may represent driver events under selection in tumours, but many hotspots may also be generated by mutational bias at these regions (such that they are more prone to mutation or less accessible to repair) mediated by their chromatin structure and replication timing 81. Regardless of the relative roles of selection or other processes in their origins, as we discuss below, these patterns of recurrent structural mutation may be clinically informative.
From sequence to data to treatment
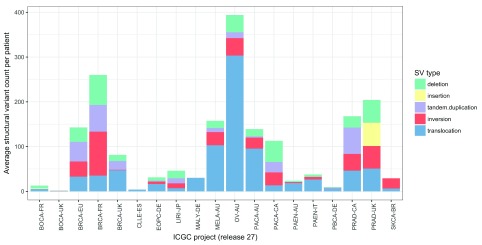
The role of genomics in precision therapy often focuses on using targeted sequencing technologies to test for the presence of clinically relevant SNVs. However, certain tumour types are characterised by modest SNV loads and instead show extensive structural rearrangements 4, 82, 83. Indeed, Ciriello et al. 82 suggest that there is an inverse relationship between the burden of SNVs and CNAs across tumour types (Figure 2a of 82). Figure 1 illustrates the average number of SVs per patient observed across 19 projects from ICGC 84 (release 27). Although it can be challenging to adjust for the inter-study differences highlighted below, it appears that certain (e.g. ovarian, breast, and prostate) tumours possess high frequencies of SVs but that other tumour types such as bone cancers and chronic lymphocytic leukaemia do not. However, there are also noticeable differences in the frequencies of mutations reported between cohorts of the same tumour type, which may reflect differences in cellularity, sequencing depth, and the SV callers used across studies. These differences emphasise the challenges that exist in measuring the degree of structural variation across studies with multiple confounding factors. Nevertheless, the clinical utility of certain SVs is already established. Currently, clinically actionable SVs include gene fusions 85 (for example, BCR-ABL), oncogene amplifications (for example, ERBB2), and tumour suppressor deletions (for example, BRCA1). Mertens et al. 85 discuss the impact of gene fusions in cancer and describe their utility as effective drug targets (Table 2 of 85). Further advances in technology will be necessary for the reliable identification of more complex SV patterns in a clinical setting where low-cost and rapid-turnaround assays are desirable 83.
Figure 1. Average number of structural variants per patient by cancer type.
The average number of structural variants per patient varies across 19 whole genome sequencing studies of cancer from the International Cancer Genomics Consortium (ICGC) (release 27). Structural variants were classified into five types: deletions, insertions, tandem duplications, inversions, and translocations. Some tumours such as ovarian, breast, and prostate have relatively large numbers of structural variants, whereas bone cancers and chronic lymphocytic leukaemias have relatively few. However, biological differences among tumour types can be confounded by technical differences among studies. SV, structural variant.
In the past, the analysis of somatic mutations in cancer has often focused on recurrent driver mutations located within the small fraction of the genome encoding proteins and thought to confer selective advantages to tumour cells 86, 87. However, it has been convincingly demonstrated that the total burden of mutations genome-wide in a tumour can represent the cumulative impact of mutagens, repair processes, and other influences along the path of tumourigenesis 88, 89. This overall burden of mutation can be disentangled to identify distinct and independent patterns of mutation which represent distinct mutational processes 90, 91. The associations seen between these patterns and particular DNA repair deficiencies were recently validated in vitro by using isogenic cell models 92. The computational analyses required to disentangle these patterns have been developed by using large pan-cancer cohorts of WGS/whole exome sequencing as well as for a large cohort of breast cancer WGS and have successfully identified over 30 patterns or signatures of SNVs classified by their substitution type and trinucleotide context 93. Analogous approaches have been proposed for the analyses of indels 94 and larger structural rearrangements 20; in the latter, SVs are classified on the basis of whether or not their breakpoints fall in a cluster of breakpoints in the genome. The rationale behind this separation is that clusters of breakpoints may be formed by mutational processes that are different from those that form dispersed breakpoints 20, 89. SVs are also classified by their type and size 20, and compound mutational signatures, including SNVs and SV breakpoints, can be constructed. This approach was successfully tested by using a large cohort of breast cancer WGS 20, resulting in the identification of characteristic signatures of rearrangement, reflecting HR deficiency, associated with the inactivating mutations of the BRCA1/2 genes. This signature of HR deficiency could then be sought in other tumour samples lacking mutated BRCA1/2 genes by using a new algorithm: HRDetect 95. This significantly expands the number of patients who have tumours showing HR deficiency (for example, due to epigenomic inactivation of BRCA1/2 or mutations in other HR genes) and who are expected to respond to therapies exploiting defective repair, such as poly (ADP-ribose) polymerase (PARP) inhibitors 95. The authors also demonstrated the success of HRDetect in predicting HR deficiency in ovarian and pancreatic tumours, suggesting broader potential for therapeutic stratification of patients. Another study has since validated these results, showing that HRDetect provides clinically relevant information independently of BRCA1/2 mutation status and that high HRDetect scores identified patients with good responses to platinum-based chemotherapy 96. However, the input to HRDetect relies on SV calls from a single SV caller. The incorporation of variant calls from additional callers to form consensus calls may be beneficial in ensuring the tool’s robustness. Beyond patient stratification, it has also been shown that SV mutational signatures can be used to predict overall survival in a cohort of HGSOC 97. Finally, it has been shown that the burden of SVs and SNVs in a tumour is associated with the degree of infiltration by host immune cells 98, linking the mutational state of a tumour to its tendency to elicit an immune response. At the cutting edge of cancer immunotherapy, efforts are under way to exploit such immunogenic tumour mutations and design individual anti-cancer vaccines, placing tumour genomics at the centre of oncology 99.
The last few decades have transformed our view of cancer to reveal each tumour as a largely unique constellation of SNV and SV mutations, and a tiny fraction of these mutations drive tumourigenesis 100. The complete reconstruction of the evolutionary trajectory travelled by a tumour could provide new biomarkers for rapid diagnosis and effective treatment and ultimately allow us to predict cancer progression. This comprehensive understanding is currently out of reach and, given the challenges involved, may remain so in the near future 101. However, in spite of our ignorance about the functional impacts of SVs and our modest success in detecting somatic mutations accurately, there are good reasons for optimism. Although we still lack the sample sizes required to detect all driver genes for most cancer types 102, there have already been unexpected benefits in amassing WGS tumour cohorts, with the genome-wide patterns of damage seen in a tumour providing novel biomarkers and guiding treatment. The next decade will see an explosion in the available data and improvements in analysis, making genomics a routine component of medical practice in oncology.
Abbreviations
AID, activation-induced cytidine deaminase; CML, chronic myeloid leukaemia; CNA, copy number alteration; DSB, double-strand break; HGSOC, high-grade serous ovarian cancer; HR, homologous recombination; ICGC, International Cancer Genome Consortium; NHEJ, non-homologous end joining; RAG, recombination activating gene; SNV, single-nucleotide variant; SV, structural variant; TCGA, The Cancer Genome Atlas; WGS, whole genome sequencing
Data and software availability
The data used to generate Figure 1 are available from the ICGC data portal at the following link: https://dcc.icgc.org/releases/current/Projects.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Javier Herrero, Bill Lyons Informatics Centre, UCL Cancer Institute, University College London, London, WC1E 6BT, UK
Jianmin Wang, Department of Biostatistics and Bioinformatics, Roswell Park Comprehensive Cancer Center, Buffalo, NY, USA
Funding Statement
The authors are supported by core funding of the UK Medical Research Council (MRC) to the MRC Human Genetics Unit and a UKRI Innovation Fellowship to Ailith Ewing.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; referees: 2 approved]
References
- 1. Inaki K, Liu ET: Structural mutations in cancer: mechanistic and functional insights. Trends Genet. 2012;28(11):550–9. 10.1016/j.tig.2012.07.002 [DOI] [PubMed] [Google Scholar]
- 2. Guan P, Sung WK: Structural variation detection using next-generation sequencing data: A comparative technical review. Methods. 2016;102:36–49. 10.1016/j.ymeth.2016.01.020 [DOI] [PubMed] [Google Scholar]
- 3. Yang L, Luquette LJ, Gehlenborg N, et al. : Diverse mechanisms of somatic structural variations in human cancer genomes. Cell. 2013;153(4):919–29. 10.1016/j.cell.2013.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bowtell DD, Böhm S, Ahmed AA, et al. : Rethinking ovarian cancer II: reducing mortality from high-grade serous ovarian cancer. Nat Rev Cancer. 2015;15(11):668–79. 10.1038/nrc4019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Patch AM, Christie EL, Etemadmoghadam D, et al. : Whole-genome characterization of chemoresistant ovarian cancer. Nature. 2015;521(7553):489–94. 10.1038/nature14410 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 6. Scarpa A, Chang DK, Nones K, et al. : Whole-genome landscape of pancreatic neuroendocrine tumours. Nature. 2017;543(7643):65–71. 10.1038/nature21063 [DOI] [PubMed] [Google Scholar]
- 7. Tubio JM: Somatic structural variation and cancer. Brief Funct Genomics. 2015;14(5):339–51. 10.1093/bfgp/elv016 [DOI] [PubMed] [Google Scholar]
- 8. Liu B, Conroy JM, Morrison CD, et al. : Structural variation discovery in the cancer genome using next generation sequencing: computational solutions and perspectives. Oncotarget. 2015;6(8):5477–89. 10.18632/oncotarget.3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu B, Morrison CD, Johnson CS, et al. : Computational methods for detecting copy number variations in cancer genome using next generation sequencing: principles and challenges. Oncotarget. 2013;4(11):1868–81. 10.18632/oncotarget.1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Beroukhim R, Mermel CH, Porter D, et al. : The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463(7283):899–905. 10.1038/nature08822 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 11. Shlien A, Malkin D: Copy number variations and cancer. Genome Med. 2009;1(6):62. 10.1186/gm62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Albertson DG, Collins C, McCormick F, et al. : Chromosome aberrations in solid tumors. Nat Genet. 2003;34(4):369–76. 10.1038/ng1215 [DOI] [PubMed] [Google Scholar]
- 13. Diskin SJ, Hou C, Glessner JT, et al. : Copy number variation at 1q21.1 associated with neuroblastoma. Nature. 2009;459(7249):987–91. 10.1038/nature08035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McGranahan N, Rosenthal R, Hiley CT, et al. : Allele-Specific HLA Loss and Immune Escape in Lung Cancer Evolution. Cell. 2017;171(6):1259–1271.e11. 10.1016/j.cell.2017.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Natrajan R, Wilkerson PM, Marchiò C, et al. : Characterization of the genomic features and expressed fusion genes in micropapillary carcinomas of the breast. J Pathol. 2014;232(5):553–65. 10.1002/path.4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu YM, Su F, Kalyana-Sundaram S, et al. : Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 2013;3(6):636–47. 10.1158/2159-8290.CD-13-0050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Karlsson J, Lilljebjörn H, Holmquist Mengelbier L, et al. : Activation of human telomerase reverse transcriptase through gene fusion in clear cell sarcoma of the kidney. Cancer Lett. 2015;357(2):498–501. 10.1016/j.canlet.2014.11.057 [DOI] [PubMed] [Google Scholar]
- 18. Giacomini CP, Sun S, Varma S, et al. : Breakpoint analysis of transcriptional and genomic profiles uncovers novel gene fusions spanning multiple human cancer types. PLoS Genet. 2013;9(4):e1003464. 10.1371/journal.pgen.1003464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Robinson DR, Kalyana-Sundaram S, Wu YM, et al. : Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nat Med. 2011;17(12):1646–51. 10.1038/nm.2580 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 20. Nik-Zainal S, Davies H, Staaf J, et al. : Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534(7605):47–54. 10.1038/nature17676 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 21. Gordon DJ, Resio B, Pellman D: Causes and consequences of aneuploidy in cancer. Nat Rev Genet. 2012;13(3):189–203. 10.1038/nrg3123 [DOI] [PubMed] [Google Scholar]
- 22. Naylor RM, van Deursen JM: Aneuploidy in Cancer and Aging. Annu Rev Genet. 2016;50:45–66. 10.1146/annurev-genet-120215-035303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grade M, Difilippantonio MJ, Camps J: Patterns of Chromosomal Aberrations in Solid Tumors. Recent Results Cancer Res. 2015;200:115–42. 10.1007/978-3-319-20291-4_6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bielski CM, Zehir A, Penson AV, et al. : Genome doubling shapes the evolution and prognosis of advanced cancers. Nat Genet. 2018;50(8):1189–95. 10.1038/s41588-018-0165-1 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 25. Zhang Y, Yang L, Kucherlapati M, et al. : A Pan-Cancer Compendium of Genes Deregulated by Somatic Genomic Rearrangement across More Than 1,400 Cases. Cell Rep. 2018;24(2):515–27. 10.1016/j.celrep.2018.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Alaei-Mahabadi B, Bhadury J, Karlsson JW, et al. : Global analysis of somatic structural genomic alterations and their impact on gene expression in diverse human cancers. Proc Natl Acad Sci U S A. 2016;113(48):13768–73. 10.1073/pnas.1606220113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Machiela MJ, Ho BM, Fisher VA, et al. : Limited evidence that cancer susceptibility regions are preferential targets for somatic mutation. Genome Biol. 2015;16(1):193. 10.1186/s13059-015-0755-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kaiser VB, Semple CA: When TADs go bad: chromatin structure and nuclear organisation in human disease [version 1; referees: 2 approved]. F1000Res. 2017;6: pii: F1000 Faculty Rev-314. 10.12688/f1000research.10792.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kaiser VB, Semple CA: Chromatin loop anchors are associated with genome instability in cancer and recombination hotspots in the germline. Genome Biol. 2018;19(1):101. 10.1186/s13059-018-1483-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu Y, Beyer A, Aebersold R: On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell. 2016;165(3):535–50. 10.1016/j.cell.2016.03.014 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 31. Speicher MR, Carter NP: The new cytogenetics: blurring the boundaries with molecular biology. Nat Rev Genet. 2005;6(10):782–92. 10.1038/nrg1692 [DOI] [PubMed] [Google Scholar]
- 32. Nilsson RH, Abarenkov K, Veldre V, et al. : An open source chimera checker for the fungal ITS region. Mol Ecol Resour. 2010;10(6):1076–81. 10.1111/j.1755-0998.2010.02850.x [DOI] [PubMed] [Google Scholar]
- 33. Haas BJ, Gevers D, Earl AM, et al. : Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21(3):494–504. 10.1101/gr.112730.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sedlazeck FJ, Lee H, Darby CA, et al. : Piercing the dark matter: bioinformatics of long-range sequencing and mapping. Nat Rev Genet. 2018;19(6):329–46. 10.1038/s41576-018-0003-4 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 35. Roberts RJ, Carneiro MO, Schatz MC: The advantages of SMRT sequencing. Genome Biol. 2013;14(7):405. 10.1186/gb-2013-14-6-405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jain M, Olsen HE, Paten B, et al. : The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol. 2016;17(1):239. 10.1186/s13059-016-1103-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zheng GX, Lau BT, Schnall-Levin M, et al. : Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nat Biotechnol. 2016;34(3):303–11. 10.1038/nbt.3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zeitouni B, Boeva V, Janoueix-Lerosey I, et al. : SVDetect: a tool to identify genomic structural variations from paired-end and mate-pair sequencing data. Bioinformatics. 2010;26(15):1895–6. 10.1093/bioinformatics/btq293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lam HY, Mu XJ, Stütz AM, et al. : Nucleotide-resolution analysis of structural variants using BreakSeq and a breakpoint library. Nat Biotechnol. 2010;28(1):47–55. 10.1038/nbt.1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Layer RM, Chiang C, Quinlan AR, et al. : LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 2014;15(6):R84. 10.1186/gb-2014-15-6-r84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang J, Mullighan CG, Easton J, et al. : CREST maps somatic structural variation in cancer genomes with base-pair resolution. Nat Methods. 2011;8(8):652–4. 10.1038/nmeth.1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rausch T, Zichner T, Schlattl A, et al. : DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28(18):i333–i339. 10.1093/bioinformatics/bts378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ewing AD, Houlahan KE, Hu Y, et al. : Combining tumor genome simulation with crowdsourcing to benchmark somatic single-nucleotide-variant detection. Nat Methods. 2015;12(7):623–30. 10.1038/nmeth.3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chong Z, Ruan J, Gao M, et al. : novoBreak: local assembly for breakpoint detection in cancer genomes. Nat Methods. 2017;14(1):65–7. 10.1038/nmeth.4084 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 45. Guan P, Sung WK: Structural variation detection using next-generation sequencing data: A comparative technical review. Methods. 2016;102:36–49. 10.1016/j.ymeth.2016.01.020 [DOI] [PubMed] [Google Scholar]
- 46. Escaramís G, Docampo E, Rabionet R: A decade of structural variants: description, history and methods to detect structural variation. Brief Funct Genomics. 2015;14(5):305–14. 10.1093/bfgp/elv014 [DOI] [PubMed] [Google Scholar]
- 47. Ellrott K, Bailey MH, Saksena G, et al. : Scalable Open Science Approach for Mutation Calling of Tumor Exomes Using Multiple Genomic Pipelines. Cell Syst. 2018;6(3):271–281.e7. 10.1016/j.cels.2018.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li Y, Roberts N, Weischenfeldt J, et al. : Patterns of structural variation in human cancer. bioRxiv. 2017. 10.1101/181339 [DOI] [Google Scholar]
- 49. Hofmann AL, Behr J, Singer J, et al. : Detailed simulation of cancer exome sequencing data reveals differences and common limitations of variant callers. BMC Bioinformatics. 2017;18(1):8. 10.1186/s12859-016-1417-7 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 50. Magi A, Semeraro R, Mingrino A, et al. : Nanopore sequencing data analysis: state of the art, applications and challenges. Brief Bioinform. 2017; bbx062. 10.1093/bib/bbx062 [DOI] [PubMed] [Google Scholar]
- 51. Currall BB, Chiang C, Talkowski ME, et al. : Mechanisms for Structural Variation in the Human Genome. Curr Genet Med Rep. 2013;1(2):81–90. 10.1007/s40142-013-0012-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Carvalho CM, Lupski JR: Mechanisms underlying structural variant formation in genomic disorders. Nat Rev Genet. 2016;17(4):224–38. 10.1038/nrg.2015.25 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 53. Stephens PJ, McBride DJ, Lin ML, et al. : Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462(7276):1005–10. 10.1038/nature08645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Stephens PJ, Greenman CD, Fu B, et al. : Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144(1):27–40. 10.1016/j.cell.2010.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 55. Lee JA, Carvalho CM, Lupski JR: A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131(7):1235–47. 10.1016/j.cell.2007.11.037 [DOI] [PubMed] [Google Scholar]
- 56. Hastings PJ, Ira G, Lupski JR: A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5(1):e1000327. 10.1371/journal.pgen.1000327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang F, Khajavi M, Connolly AM, et al. : The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat Genet. 2009;41(7):849–53. 10.1038/ng.399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lee JK, Choi YL, Kwon M, et al. : Mechanisms and Consequences of Cancer Genome Instability: Lessons from Genome Sequencing Studies. Annu Rev Pathol. 2016;11:283–312. 10.1146/annurev-pathol-012615-044446 [DOI] [PubMed] [Google Scholar]
- 59. Swaminathan S, Klemm L, Park E, et al. : Mechanisms of clonal evolution in childhood acute lymphoblastic leukemia. Nat Immunol. 2015;16(7):766–74. 10.1038/ni.3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Papaemmanuil E, Rapado I, Li Y, et al. : RAG-mediated recombination is the predominant driver of oncogenic rearrangement in ETV6-RUNX1 acute lymphoblastic leukemia. Nat Genet. 2014;46(2):116–25. 10.1038/ng.2874 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 61. Yi K, Ju YS: Patterns and mechanisms of structural variations in human cancer. Exp Mol Med. 2018;50(8):98. 10.1038/s12276-018-0112-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tubio JM: Somatic structural variation and cancer. Brief Funct Genomics. 2015;14(5):339–351. 10.1093/bfgp/elv016 [DOI] [PubMed] [Google Scholar]
- 63. de Pagter MS, Kloosterman WP: The Diverse Effects of Complex Chromosome Rearrangements and Chromothripsis in Cancer Development. Recent Results Cancer Res. 2015;200:165–93. 10.1007/978-3-319-20291-4_8 [DOI] [PubMed] [Google Scholar]
- 64. Holland AJ, Cleveland DW: Chromoanagenesis and cancer: mechanisms and consequences of localized, complex chromosomal rearrangements. Nat Med. 2012;18(11):1630–8. 10.1038/nm.2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rode A, Maass KK, Willmund KV, et al. : Chromothripsis in cancer cells: An update. Int J Cancer. 2016;138(10):2322–33. 10.1002/ijc.29888 [DOI] [PubMed] [Google Scholar]
- 66. Zhang CZ, Leibowitz ML, Pellman D: Chromothripsis and beyond: Rapid genome evolution from complex chromosomal rearrangements. Genes Dev. 2013;27(23):2513–30. 10.1101/gad.229559.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ding L, Wendl MC, McMichael JF, et al. : Expanding the computational toolbox for mining cancer genomes. Nat Rev Genet. 2014;15(8):556–70. 10.1038/nrg3767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rausch T, Jones DT, Zapatka M, et al. : Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148(1–2):59–71. 10.1016/j.cell.2011.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Poot M: Genes, Proteins, and Biological Pathways Preventing Chromothripsis. Methods Mol Biol. 2018;1769:231–51. 10.1007/978-1-4939-7780-2_15 [DOI] [PubMed] [Google Scholar]
- 70. Bochtler T, Granzow M, Stölzel F, et al. : Marker chromosomes can arise from chromothripsis and predict adverse prognosis in acute myeloid leukemia. Blood. 2017;129(10):1333–42. 10.1182/blood-2016-09-738161 [DOI] [PubMed] [Google Scholar]
- 71. Rücker FG, Dolnik A, Blätte TJ, et al. : Chromothripsis is linked to TP53 alteration, cell cycle impairment, and dismal outcome in acute myeloid leukemia with complex karyotype. Haematologica. 2018;103(1):e17–e20. 10.3324/haematol.2017.180497 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 72. Notta F, Chan-Seng-Yue M, Lemire M, et al. : A renewed model of pancreatic cancer evolution based on genomic rearrangement patterns. Nature. 2016;538(7625):378–82. 10.1038/nature19823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Fontana MC, Marconi G, Feenstra JDM, et al. : Chromothripsis in acute myeloid leukemia: Biological features and impact on survival. Leukemia. 2017. 10.1038/leu.2017.351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kloosterman WP, Koster J, Molenaar JJ: Prevalence and clinical implications of chromothripsis in cancer genomes. Curr Opin Oncol. 2014;26(1):64–72. 10.1097/CCO.0000000000000038 [DOI] [PubMed] [Google Scholar]
- 75. Magrangeas F, Avet-Loiseau H, Munshi NC, et al. : Chromothripsis identifies a rare and aggressive entity among newly diagnosed multiple myeloma patients. Blood. 2011;118(3):675–8. 10.1182/blood-2011-03-344069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hirsch D, Kemmerling R, Davis S, et al. : Chromothripsis and focal copy number alterations determine poor outcome in malignant melanoma. Cancer Res. 2013;73(5):1454–60. 10.1158/0008-5472.CAN-12-0928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Skuja E, Kalniete D, Nakazawa-Miklasevica M, et al. : Chromothripsis and progression-free survival in metastatic colorectal cancer. Mol Clin Oncol. 2017;6(2):182–6. 10.3892/mco.2017.1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Baca SC, Prandi D, Lawrence MS, et al. : Punctuated evolution of prostate cancer genomes. Cell. 2013;153(3):666–77. 10.1016/j.cell.2013.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 79. Shen MM: Chromoplexy: a new category of complex rearrangements in the cancer genome. Cancer Cell. 2013;23(5):567–9. 10.1016/j.ccr.2013.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Berger MF, Lawrence MS, Demichelis F, et al. : The genomic complexity of primary human prostate cancer. Nature. 2011;470(7333):214–20. 10.1038/nature09744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Glodzik D, Morganella S, Davies H, et al. : A somatic-mutational process recurrently duplicates germline susceptibility loci and tissue-specific super-enhancers in breast cancers. Nat Genet. 2017;49(3):341–8. 10.1038/ng.3771 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 82. Ciriello G, Miller ML, Aksoy BA, et al. : Emerging landscape of oncogenic signatures across human cancers. Nat Genet. 2013;45(10):1127–33. 10.1038/ng.2762 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 83. Macintyre G, Ylstra B, Brenton JD: Sequencing Structural Variants in Cancer for Precision Therapeutics. Trends Genet. 2016;32(9):530–42. 10.1016/j.tig.2016.07.002 [DOI] [PubMed] [Google Scholar]
- 84. International Cancer Genome Consortium, Hudson TJ, Anderson W, et al. : International network of cancer genome projects. Nature. 2010;464(7291):993–8. 10.1038/nature08987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Mertens F, Johansson B, Fioretos T, et al. : The emerging complexity of gene fusions in cancer. Nat Rev Cancer. 2015;15(6):371–81. 10.1038/nrc3947 [DOI] [PubMed] [Google Scholar]
- 86. Stratton MR, Campbell PJ, Futreal PA, et al. : The cancer genome. Nature. 2009;458(7329):719–24. 10.1038/nature07943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Stratton MR: Exploring the genomes of cancer cells: progress and promise. Science. 2011;331(6024):1553–8. 10.1126/science.1204040 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 88. Nik-Zainal S, Alexandrov LB, Wedge DC, et al. : Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149(5):979–93. 10.1016/j.cell.2012.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Nik-Zainal S, Morganella S: Mutational Signatures in Breast Cancer: The Problem at the DNA Level. Clin Cancer Res. 2017;23(11):2617–29. 10.1158/1078-0432.CCR-16-2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Alexandrov LB, Nik-Zainal S, Wedge DC, et al. : Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013;3(1):246–59. 10.1016/j.celrep.2012.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Alexandrov LB, Nik-Zainal S, Wedge DC, et al. : Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–21. 10.1038/nature12477 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 92. Zou X, Owusu M, Harris R, et al. : Validating the concept of mutational signatures with isogenic cell models. Nat Commun. 2018;9(1):1744. 10.1038/s41467-018-04052-8 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 93. Forbes SA, Beare D, Boutselakis H, et al. : COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45(D1):D777–D783. 10.1093/nar/gkw1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Helleday T, Eshtad S, Nik-Zainal S: Mechanisms underlying mutational signatures in human cancers. Nat Rev Genet. 2014;15(9):585–98. 10.1038/nrg3729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Davies H, Glodzik D, Morganella S, et al. : HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med. 2017;23(4):517–25. 10.1038/nm.4292 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 96. Zhao EY, Shen Y, Pleasance E, et al. : Homologous Recombination Deficiency and Platinum-Based Therapy Outcomes in Advanced Breast Cancer. Clin Cancer Res. 2017;23(24):7521–30. 10.1158/1078-0432.CCR-17-1941 [DOI] [PubMed] [Google Scholar]
- 97. Hillman RT, Chisholm GB, Lu KH, et al. : Genomic Rearrangement Signatures and Clinical Outcomes in High-Grade Serous Ovarian Cancer. J Natl Cancer Inst. 2018;110(3):265–272. 10.1093/jnci/djx176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Thorsson V, Gibbs DL, Brown SD, et al. : The Immune Landscape of Cancer. Immunity. 2018;48(4):812–830.e14. 10.1016/j.immuni.2018.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 99. Berger MF, Mardis ER: The emerging clinical relevance of genomics in cancer medicine. Nat Rev Clin Oncol. 2018;15(6):353–65. 10.1038/s41571-018-0002-6 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 100. Martincorena I, Raine KM, Gerstung M, et al. : Universal Patterns of Selection in Cancer and Somatic Tissues. Cell. 2017;171(5):1029–1041.e21. 10.1016/j.cell.2017.09.042 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 101. Jolly C, van Loo P: Timing somatic events in the evolution of cancer. Genome Biol. 2018;19(1):95. 10.1186/s13059-018-1476-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Lawrence MS, Stojanov P, Mermel CH, et al. : Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505(7484):495–501. 10.1038/nature12912 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation