We present data that concurs with the reported geographical expansion of scrub typhus outside the “Tsutsugamushi Triangle” and addition of Orientia chuto as a second species in the Orientia genus. Wild rodents were caught in Marigat, Baringo County, Kenya, and ectoparasites, including chiggers, were recovered.
KEYWORDS: Orientia chuto, Orientia tsutsugamushi, scrub typhus, Neotrombicula and Microtrombicula chiggers, rodents, chiggers
ABSTRACT
We present data that concurs with the reported geographical expansion of scrub typhus outside the “Tsutsugamushi Triangle” and addition of Orientia chuto as a second species in the Orientia genus. Wild rodents were caught in Marigat, Baringo County, Kenya, and ectoparasites, including chiggers, were recovered. Rodent and chigger species were identified by taxonomic features. DNA was extracted from the chiggers and used to amplify and/or sequence the 47-kDa high temperature transmembrane protein (TSA47), the 56-kDa type-specific antigen (TSA56), and the 16S rRNA (rrs) Orientia genes. The main rodent hosts identified were Acomys wilsoni, Crocidura sp., and Mastomys natalensis, which accounted for 59.2% of the total collection. Of these, A. wilsoni and M. natalensis harbored most of the chiggers that belonged to the Neotrombicula and Microtrombicula genera. A pool of chiggers from one of M. natalensis was positive for Orientia by TSA47 PCR, but Orientia did not amplify with the TSA56 primers. On sequencing the 850 bp of the TSA47 gene, the closest phylogenetic relative was O. chuto, with 97.65% sequence homology compared to 84.63 to 84.76% for O. tsutsugamushi. 16S rRNA deep sequencing also revealed O. chuto as the closest phylogenetic relative, with 99.75% sequence homology. These results and the existing immunological and molecular reports are strongly suggestive of the existence of Orientia species in Kenya.
INTRODUCTION
Scrub typhus is caused by infection with a Gram-negative obligate intracellular bacterium of the genus Orientia. It is transmitted to humans through the bite of infected larval trombiculid mites, commonly referred to as chiggers, which serve as vectors and main reservoirs of this pathogen (1). Chiggers primarily feed on rodents, but occasionally on humans, to whom they may transmit the Orientia species. The disease is predominantly endemic in the Asia-Pacific region, an area commonly referred to as the “Tsutsugamushi Triangle” that extends from northern Japan in the east to Pakistan and Afghanistan in the west and northern Australia in the south (2). Approximately one million cases occur in the area where scrub typhus is endemic each year, and more than a billion people are at risk worldwide. For a long time, Orientia tsutsugamushi was the only member of the genus Orientia, until 2010, when a new species, Orientia chuto, was identified in Australia in a patient who contracted scrub typhus in Dubai (3).
Antigenically, the O. tsutsugamushi strains are very diverse, but three of them have been well characterized and are used as reference strains, namely Karp (New Guinea), Kato (Japan), and Gilliam (Burma) (4). By molecular methods, the observed genetic variability depends on the target molecular marker. For example, using the tremendously variable 56-kDa type-specific antigen (TSA56), Kelly et al. observed 9 groupings among 135 strains tested (4). Even greater sequence diversity (53.1%) was found with O. chuto (3). Using the more conserved 47-kDa high temperature transmembrane protein (TSA47) gene, Jiang et al. evaluated the genetic variability of recent clinical isolates of Orientia, including O. chuto. By both nucleotide and translated amino acid sequences, the O. chuto strain showed significant sequence diversity compared to that of other O. tsutsugamushi strains (5). Among the O. tsutsugamushi strains, the shared identity of the nucleotide and translated amino acid sequences was greater than 96%, compared to 82% for the O. chuto strain. On the basis of the 16S rRNA (rrs) gene, which evolves relatively slowly, the closest O. tsutsugamushi strains (Ikeda, Kato, and Karp) showed 98.5% homology (3) to O. chuto.
The clinical presentations of scrub typhus include high-grade fever, headache, and myalgia. These symptoms are not different from those exhibited in other infectious diseases, such as leptospirosis, murine typhus, malaria, and dengue fever. Although scrub typhus is easily treatable with doxycycline if diagnosed early, in the absence of specific symptoms, such as the pathognomonic eschar, the disease may be confused with a variety of other febrile illness, and treatment may not be appropriately administered. In such cases, the median case fatality rate is approximately 6%, but it can be up to 40% to 50% (6).
Very little information exists on scrub typhus or the probable vectors in Africa, although recent studies have found enhanced reactivity to scrub typhus antigens in patients in Kenya (7, 8) and DNA of Orientia from rodent samples collected in West Africa (9). These findings suggest that active transmission of this disease may be occurring in these regions. It remains to be determined whether these findings represent spread of Orientia species outside the Tsutsugamushi Triangle or identifies hitherto unknown disease endemic foci.
Following description of specific immunoglobulins against scrub typhus antigen in patients with acute febrile illness in Marigat, Kenya (7), we went back to the same areas and conducted surveillance for chiggers in wild-caught rodents and thereafter determined if the chiggers harbored Orientia. We now report on the identification and molecular characterization of O. chuto in trombiculid chigger mites collected from wild rodents in Kenya.
MATERIALS AND METHODS
Ethical statement.
The animal protocol used in this study was reviewed and approved by the Walter Reed Army Institute of Research under protocol number AP-12-001, KEMRI SSC #2208, KEMRI ACUC #2208, and National Museums of Kenya NMK/SCom2013/08.
Rodent collection sites.
Rodent surveys were conducted in June 2017 in the semi-arid Baringo County in Marigat area, located approximately 260 km north-west of Nairobi. Chigger development is temperature dependent and cooler months are ideal for their detection. In Kenya, cold months fall between May and August. A region in Marigat known as Perkerra was selected for surveillance, following earlier descriptions of specific immunoglobulins against scrub typhus group antigens in patients from this region (7). Five sites in Perkerra were selected for the installation of rodent traps (Fig. S1).
A total of 64 to 67 Sherman collapsible rodent traps per site was set up each night in a variety of habitats, including cropped fields, grass fields, gardens, orchards, and around buildings. Traps were placed in a grid format every 10 m or a transect, according to the land topography. Rodent traps were baited with appropriate foods, such as green nuts mixed with peanut butter, fried potato chips, sausages, or other local foods. Geographic coordinates of the sampling sites were recorded using a handheld GPS tracker (Garmin, Olathe, KS) and used to locate trapping sites on the map (Fig. S1). Rodent traps were set before sunset, and trapped rodents were collected early the following morning before 1000 h or checked two to three times throughout the daytime.
Rodent processing and ectoparasite collection.
Animal euthanasia was performed at the collection sites. A mixture of the anesthetic agents ketamine and xylazine at 40 mg/kg and 3 mg/kg, respectively, was injected intraperitoneally until safe anesthesia was achieved (10). Blood samples were collected immediately by cardiac exsanguination and aliquoted into Vacutainer tubes (Becton, Dickinson and Company, NJ, USA). Rodents were identified to species using taxonomic keys obtained from the Kingdon Field Guide to African Mammals (11).
Following euthanasia, individual rodent bodies were immediately placed in plastic bags to minimize the potential loss of ectoparasites. Cotton balls moistened with isoflurane (Forane; Abbott Laboratories, IL, USA) were then placed into the bags containing the dead rodents and left in the bags for 5 min. The rodent bodies were then transferred to plastic trays coated with petroleum jelly on all corners and edges. The trays were then placed over pans of water to prevent escape of ectoparasites. The rodents were thoroughly examined for presence of ectoparasites by brushing their bodies with fine combs. All ectoparasites, including chiggers, ticks, fleas, and lice, were collected using fine paintbrushes and preserved in 70% ethanol. Niches of chiggers aggregating in the ears or body were removed, together with a thin layer of the skin surface, using fine forceps and lancets in order to minimize the damage to chiggers. All ectoparasites and pieces of ear skin were preserved in 70% ethanol and labeled individually according to a number assigned to each of the rodent hosts. The ectoparasite specimens were stored at room temperature for a maximum of 5 days and later transferred to 4°C to be used for further investigations. In the current study, only the chigger data are reported.
For collection of tissue samples, the surface of the rodent abdomen was cleaned with 70% ethanol before necropsy. Approximately 50- to 100-mg size biopsy specimens were collected from each of the internal organs, including lung, liver, spleen, and kidney, and were stored in screw-cap plastic vials and then placed in liquid nitrogen charged containers. The tissue biopsy specimens were later transferred to a −80°C freezer to be used in the future for pathogen identification.
Identification of chigger mites.
Chiggers were individually separated from the skin tissues and/or hairs using sterile needles under an inverted microscope (EMZ; Meiji Techno Co. Ltd., Japan). The total number of chiggers recovered from rodent hosts was counted. General characteristics of chiggers, including shape of the body and variation of color, were recorded. A total number of 767 chiggers was recovered. Due to logistical difficulties of examining all 767 chiggers, a representative number (20%) was sampled and mounted with Hoyer’s mounting medium (Berlese’s fluid-gum chloral; TCS Biosciences Ltd., UK) on glass slides. Slides were dried at 50°C before sealing with nail polish. Slide-mounted chiggers were examined under a dissecting microscope (Carl Zeiss AG, Oberkochen, Germany) at ×400 magnification. Standard taxonomic keys described by Nadchatram and Alexander (12) were used to identify the genera of the chiggers. Chiggers were then pooled in pools of 5 according to their genera and rodent hosts.
Extraction of DNA from chiggers and rodent tissue samples.
A total of 470 chiggers in 5 pools of 94 specimens each was used for DNA extraction. DNA was extracted from both the chigger mites and rodent tissue samples using the QIAamp DNA minikit (Qiagen, CA, USA) as recommended by the manufacturer with some modifications. The chiggers were washed several times in distilled water to eliminate ethanol. Chiggers were then macerated in 90 µl of tissue lysis buffer with sterile micropestles under a microscope. The lysates were digested with proteinase K (2 mg/ml) at 56°C for 3 h, followed by nucleic acid purification per the manufacturer’s instructions. The DNA from the chiggers and tissue samples was eluted in 20 µl and 100 µl elution buffer, respectively. All samples were then aliquoted into 2 tubes, one for immediate pathogen detection and another for future use. The DNA extracts were then stored at −80°C until PCR was performed.
Orientia sp. qPCR screening.
To screen the chiggers and rodent tissues for Orientia, extracted DNA was used to amplify by qPCR the Orientia TSA47 gene using the following primers: 3′-AACTGATTTTATTCAAACTAATGCTGCT-5′ as forward, 3′-TATGCCTGAGTAAGATACRTGAATRGAATT-5′ as reverse, and FAM-TGGGTAGCTTTGGTGGACCGATGTTTAATCT-BHQ1 as probe (13). The PCR reaction was conducted in a total volume of 12.5 µl and contained 1× SensiFAST Probe Lo-ROX (Bioline, Australia), 1.5 mM MgCl2, 0.1 µM forward and reverse primers, 0.2 µM probe, and 1.5 µl of DNA and PCR-grade water. The PCR amplification consisted of an initial holding step at 50°C for 2 min and 95°C for 2 min, followed by 45 cycles at 95°C for 15 sec and 60°C for 30 sec. To monitor for cross-contamination between samples, template-free negative controls were included in all assays. The positive controls for the assays were DNA extracted from O. tsutsugamushi-infected Leptotrombidium chigger mites provided by the U.S. Army Medical Directorate of the Armed Forces Research Institute of Medical Sciences (USAMD-AFRIMS).
TSA47 and TSA56 amplification by conventional PCR.
DNA samples from chiggers and rodent tissues testing positive by qPCR for the TSA47 gene were used to amplify both the TSA47 and TSA56 genes by conventional PCR, using primers described in Table 1. To amplify the TSA47 gene, a nested PCR was performed as previously described (3). In brief, both the primary and secondary PCR was performed in a 25-µl reaction mixture containing 1× MyTaq buffer (Bioline, NSW, Australia), 0.04 U/µL of MyTaq polymerase, 0.3 µM forward and reverse primers, 2 µl of the DNA, and PCR-grade water. The PCR amplification conditions were also similar for both the first and second PCR and comprised an initial denaturation at 95°C for 2 min, followed by 94°C for 30 sec, annealing at 54°C for 30 sec, extension at 68°C for 2 min for 40 cycles, and a final extension at 72°C for 7 min. To amplify the TSA56 gene, a nested PCR was also performed, as previously described (3). In brief, the PCRs were performed in a 25-µl reaction volume containing 2 µl of DNA template, 5 µl of 5× MyTaq buffer (Bioline, NSW, Australia), 0.04 U/µL of MyTaq polymerase, 0.3 µM outer primer pair (RTS-8 and RTS-9), and PCR-grade water. The amplification conditions included heat denaturation at 94°C for 1 min, annealing at 55°C for 1.5 min, and polymerization at 72°C for 2 min. Amplification proceeded for 30 cycles. In the second PCR step, amplification conditions were similar but used the inner primer pair, RTS-6 and RTS-7 (Table 1). All amplicons were visualized on a 2% agarose gel stained with 2.5 µl of EZ-Vision in-gel solution (Amresco, TX, USA).
TABLE 1.
Primers used in PCR amplification and sequencing of TSA47, TSA56, and 16s rRNA genes of Orientia species
| Species | Target | Primer | Oligonucleotide sequence (5′–3′) | Amplicon size |
|---|---|---|---|---|
| O. tsutsugamushi | TSA47 | Ot-145Fa,c | ACAGGCCAAGATATTGGAAG | |
| Ot-1780Ra,c | AATCGCCTTTAAACTAGATTTACTTATTA | |||
| Ot-263Fb,c | GTGCTAAGAAARGATGATACTTC | 871 bp | ||
| Ot-1133Rb,c | ACATTTAACATACCACGACGAAT | |||
| O. chuto | Chur-627Fa,c | GCGGGATATAGGTAGTTCAA | 821 bp | |
| Chur-669Fb,c | TATTCAAACTAATGCTGTTGTGC | |||
| Ot-1404Ra ,b,c | GATTTACTTATTAATRTTAGGTAAAGCAATGT | |||
| Orientia spp. | TSA56 | RTS-8a,c | AGGATTAGAGTGTGGTCCTT | |
| RTS-9a,c | GTTGGAGGAATGATTACTGG | |||
| RTS-6b,c | GTTGGAGGAATGATTACTGG | 620 bp | ||
| RTS-7b,c | AGCGCTA GGTTTATTAGCAT | |||
| Bacteria | 16S rRNA V3-V4 | rrs-Forwardc | TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG | 441 bp |
| rrs-Reversec | GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC |
Primary conventional PCR primers.
Secondary conventional PCR primers.
Sequencing primers.
Gene sequencing.
Following gel electrophoresis, amplicons with visible bands were purified using Agencourt AMPure XP beads (Beckman Coulter, CA, USA) as recommended by the manufacturer and thereafter sequenced in both directions using the BigDye Terminator cycle sequencing kit v 3.1 (Applied Biosystems, CA, USA) with specific primers (Table 1). The cycle sequencing products were cleaned with Agencourt CleanSEQ beads (Beckman Coulter, CA, USA) and then sequenced on the ABI 3130 genetic analyzer (Applied Biosystems, CA, USA).
16S rRNA deep sequencing.
Rodent tissue samples and chigger mites that tested positive for Orientia by the TSA47 qPCR were also amplified using primers specific for the 441-bp (V3 to V4) variable fragment of the 16S rRNA (rrs) gene (14). This region is nested between base pairs 320 and 760 of the rrs gene of O. chuto that Izzard et al. described (3). The amplification was carried out in a 25-µl reaction volume consisting of 1× NEBNext mix (New England Biolabs, MA, USA), 1 µM each primer, and 2.5 µl of DNA template. The PCR amplification conditions were as follows: initial denaturation at 95°C for 3 min, followed by 25 cycles of 95°C for 30 sec, 55°C for 30 sec, and 72°C for 30 sec, with a final extension of 72°C for 5 min. Amplicons were purified using Agencourt AMPure XP beads (Beckman Coulter) according to the manufacturer’s instructions. Illumina barcode sequences were introduced through a limited cycle amplification step to generate indexed libraries, followed by library quantification on a Qubit fluorometer v 2.0 (Invitrogen, CA, USA). Samples were normalized to 8 pmol, pooled, and sequenced on the Illumina MiSeq platform (2 × 300-bp paired-end reads) (Illumina, USA).
Data analysis.
The epidemiological parameters calculated included rodent infestation rate (total number of captured rodents from all traps), mean abundance of chiggers (total number of chiggers collected divided by total number of captured rodents), chigger infestation rate (percentage of rodents infested with chiggers), and chigger index (total number of chiggers divided by total number of rodents infested with chiggers). Digital maps were created using qGIS v 2.18.16 (https://qgis.org/en/site/).
Nucleic acid sequence data of the TSA47 gene was checked for quality and then assembled into consensus sequences using CLC Main Workbench v 7 (CLC Inc, Aarhus, Denmark). The taxonomic identity of the sequences was confirmed by querying them against a nucleotide database (GenBank) using the nucleotide Basic Local Alignment Search Tool (BLASTn). Pairwise comparisons against validated Orientia species were performed within CLC Main Workbench v 7.
For the 16S rRNA deep sequencing, nucleotide sequences were processed with mothur v 3.9.2 pipeline (15). In brief, the sequences were assembled into contigs, then trimmed for errant contigs by discarding sequences with ambiguous bases, sequences shorter than 430 bp or longer than 500 bp, and sequences with homopolymer stretches greater than 8 bp. Sequences were checked for chimeras and singletons, then clustered into operational taxonomic units (OTUs) with a 97% similarity cutoff. Curated sequences were then taxonomically classified using the SILVA rRNA gene database at a confidence threshold of 80% (16). A 16S rRNA sequence of the Orientia genus was identified in the contig file and used for pairwise comparison with the validated Orientia species within CLC Main Workbench v 7.
For phylogenetic inference, MEGA v 7.0 was used to determine the best nucleotide substitution model, HKY+G, for the TSA47 and rrs genes. This model was used to infer maximum likelihood phylogenetic trees in MEGA. The bootstrap values were calculated from 1,000 replications, and only values higher than 50% were shown on the tree.
Accession numbers.
The sequence data of the TSA47 (807 nucleotide positions) and 16S rRNA (400 nucleotide positions) genes of O. chuto strain Marigat obtained in this study have been submitted to GenBank and given the accession numbers MH719012 and MH716016, respectively.
RESULTS
Acomys wilsoni, Crocidura spp., and Mastomys natalensis are the main rodent hosts in Marigat.
A total of 54 small rodents were captured during five trap-nights. Table 2 summarizes the rodents captured and species composition at the trapping sites. The highest number of rodents was recorded at site 5 (Fig. S1). No rodents were trapped at site 3. Rodent species composition was found to be relatively similar in all trapping surveillance sites (between 4 and 5). All trapped rodents were classified into eight species that belonged to three families and two orders (Table 2). Rodents from the order Rodentia, family Muridae, were the most predominant and accounted for 59.2% of the total collection. Among these, Acomys wilsoni was the most abundant species (38.9%) and was captured at all trap sites, followed by Crocidura sp. (25.9%), and Mastomys natalensis (14.8%).
TABLE 2.
Diversity of small rodents collected in Marigat
| Order | Family | Rodent species (no. of specimens) |
No of trapped rodents at each collection site (% of trap) |
No. of rodents with chiggers (% chigger infestation) |
No. of chiggers (chigger index) |
|||
|---|---|---|---|---|---|---|---|---|
| Site 1 | Site 2 | Site 4 | Site 5 | |||||
| Rodentia | Muridae | Acomys wilsoni (13) | 5 (23.8) | 3 (14.3) | 1 (4.8) | 12 (57.1) | 15 (71.4) | 440 (29.3) |
| Lophuromys sikapusi (1) | 0 | 1 (100) | 0 | 0 | 0 | 0 | ||
| Mastomys natalensis (8) | 2 (25) | 0 | 2 (25) | 4 (50) | 7 (87.5) | 322 (46) | ||
| Rattus rattus (2) | 0 | 1 (50) | 0 | 1 (50) | 1 (50) | 5 (5) | ||
| Murinae | Arvicanthis niloticus (6) | 0 | 4 (66.7) | 2 (33.3) | 0 | 0 | 0 | |
| Eulipotyphla | Soricidae | Crocidura sp. (24) | 1 (7.1) | 3 (21.4) | 3 (21.4) | 7 (41.2) | 0 | 0 |
| Crocidura olivieri (2) | 1 (50) | 0 | 0 | 1 (50) | 0 | 0 | ||
| Both orders | All families | Total (54) | 9 (16.7) | 12 (22.2) | 8 (14.8) | 25 (46.3) | 23 (42.6) | 767 (33.3) |
Marigat chiggers belong to Neotrombicula and Microtrombicula genera.
From the 54 rodents examined, 50% (27/54) were found to be infested with ectoparasites, and nearly all had chigger mites (85.2%; 23/27) (Fig. S2), with a mean abundance of 14.2. Trombiculid chigger mites were recovered from 3 of the 7 rodent species: 71.4% (15/21) for Acomys wilsoni, 87.5% (7/8) for Mastomys natalensis, and 50% (1/2) for Rattus rattus. The chigger indices by rodent species are shown in Table 2. A total number of 767 chigger mites was recovered from 23 rodent hosts, giving an overall chigger index of 33.3.
Rodents with chigger infestation were found at all trapping sites, suggesting an abundance of chiggers in the areas surveyed. The highest number of chigger mites was obtained from site 5, which accounted for 46.8% of the total recovery. As shown in Fig. S1, the highest chigger index (20.1; 161/8) was recorded at site 4, which is a National Irrigation Board Farm, followed by plantation site 1 in an apiculture center (19.6; 176/9), then site 5, which is a beehive experimental field (14.4; 359/25), and then plantation site 2 in the apiculture center (5.9; 71/12). Of the 767 chiggers, 57.3% (440) were from Acomys wilsoni, while 42% (322) were from Mastomys natalensis (Table 2). Less than 1% of chiggers were recovered from Rattus rattus, suggesting that Acomys wilsoni and Mastomys natalensis are the main rodent hosts in the surveyed areas. By morphological taxonomy, the chiggers belonged to two genera, the Microtrombicula and Neotrombicula (Fig. S3A and B, respectively). Of the 23 rodents with parasitic chiggers, 26.1% (6/23) had polyspecific parasitism, with Neotrombicula and Microtrombicula chiggers cofeeding on the same hosts. The highest number of rodents with polyspecific parasitism belonged to A. wilsoni (5/6). The remaining rodent hosts (17/23) were found to harbor chiggers from the Microtrombicula genus only.
Identification of O. chuto in rodent samples and trombiculid chiggers.
Of the chigger mites screened by the TSA47 gene qPCR, a pool of chigger mites recovered from one M. natalensis specimen was positive for Orientia. As shown in Fig. 1, an 850-bp fragment was obtained with O. chuto-specific primers (lane 4). The O. tsutsugamushi primers that target the TSA56 gene failed to amplify the chigger mite sample (lane 6) but amplified the O. tsutsugamushi positive control (lane 5).
FIG 1.
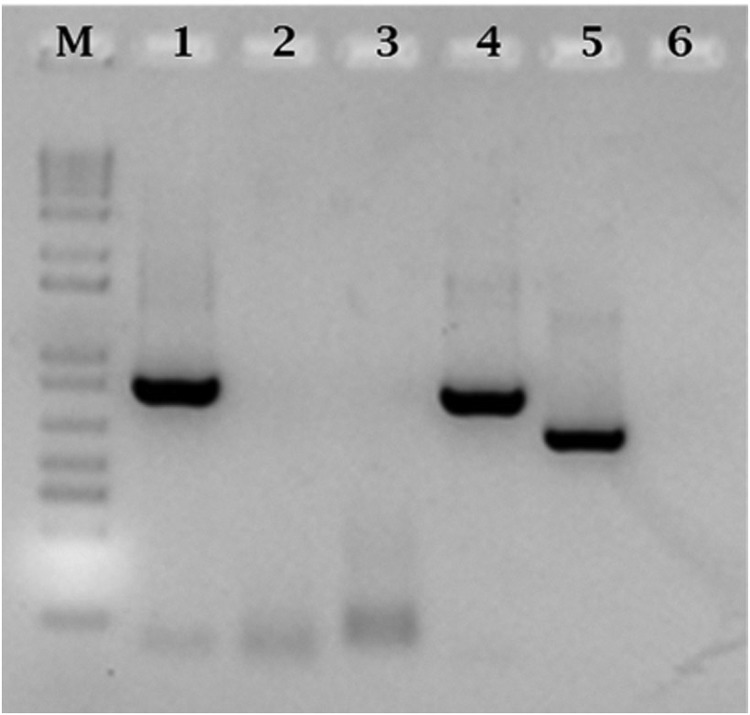
Agarose gel electrophoresis of TSA47 gene (lanes 1 to 4) amplified by nested PCR with species-specific primers. M: 1 Kb Plus DNA ladder (Invitrogen, CA, USA). Lane 1, O. tsutsugamushi positive control amplified with O. tsutsugamushi primers. Lane 2, lack of amplification of chigger mite sample by O. tsutsugamushi primers. Lane 3, lack of amplification of O. tsutsugamushi positive control by O. chuto primers. Lane 4, amplification of chigger mite sample by O. chuto primers. Lane 5, amplification of O. tsutsugamushi positive control with genus-specific TSA56 primers. The Orientia genus TSA56 primers did not amplify the chigger mite sample (lane 6).
Phylogenetic analysis of Orientia strain from Kenya.
Phylogenetic analysis of the 850-bp TSA47 gene against Orientia sequences available in GenBank suggest that the Orientia sequence amplified from our study was closely related to the O. chuto reference strain (accession number HM156063) with pairwise nucleotide identity of 97.65%. The next closest relatives were sequences from the O. tsutsugamushi group, with a percent homology range of 84.63% to 84.76% (Fig. S4A).
By pairwise nucleotide alignment of the 16S rRNA gene of the study OTU to available Orientia sequences in the GenBank, the closest phylogenetic relative was O. chuto (accession number NR_117903), with 99.75% sequence homology. The next closest relatives were sequences from the O. tsutsugamushi group, with a percent homology range of 98.50% to 97.75% (Fig. S4B).
From the 16S rRNA deep sequencing of the chigger samples, seven bacteria genera were identified, including Orientia, Yokenella, Borrelia, Caulobacter, Actinomycetospora, Bosea, and Coxiella (data not shown).
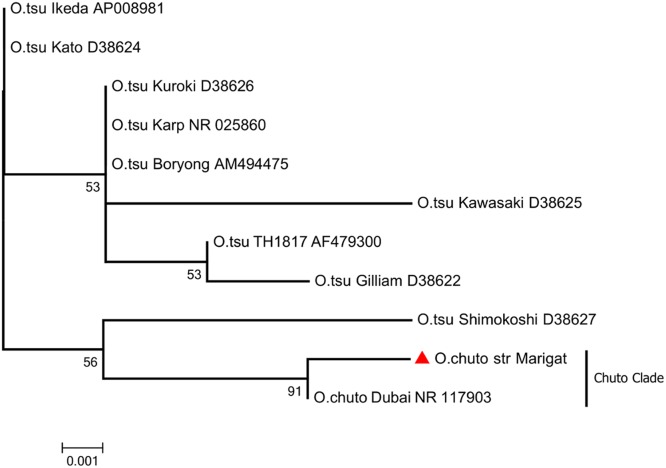
The topology of the TSA47 gene tree inferred with the maximum likelihood (ML) method (Fig. 2) shows two groupings, namely, the Tsutsugamushi clade and the Chuto clade, that are well supported with 100% bootstrap confidence values. The sequence obtained from the study OTU shares a common ancestor with the O. chuto strain Dubai (accession number HM156063) to form sister OTUs. Also, the sequence from the study OTU had a longer branch, indicating that it had more substitutions per nucleotide position.
FIG 2.
Phylogenetic tree of the study strain and prototype Orientia strains. The tree was constructed using the maximum likelihood method, based on 850 bp sequence of the TSA47 gene. The red triangle indicates the sequence from this study.
As with the TSA47 gene tree, the topology of the 16S rRNA gene tree inferred with the ML method (Fig. 3) shows the Tsutsugamushi and the Chuto clades, with the latter being well supported at 91% bootstrap confidence values. The sequence obtained from the study OTU delineated at the base of the unrooted phylogenetic tree and had long branches, indicating differences from other Orientia sequences.
FIG 3.
Evolutionary tree of Orientia species based on the 441-bp sequence of the 16S rRNA of the study OTU. The tree was generated using the maximum likelihood method in MEGA v 7.0 software. The red triangle corresponds to the sequence of the study OTU.
DISCUSSION
The data reported in this study and other recent reports provide evidence to indicate that scrub typhus is no longer restricted to the area once referred to as the Tsutsugamushi Triangle and that, apart from O. tsutsugamushi, other species of Orientia may exist (see reference 17 for a review). In the review by Jiang and Richards (17), it is clear that existence of scrub typhus outside the “triangle” is not a new finding. In eastern Africa, for example, there are records of cases resembling scrub typhus in native Africans that date to 1951 (18). Since then, serological and molecular evidence of scrub typhus in Africa has been accumulating (7–9, 19, 20). More recent studies have revealed existence of endemic scrub typhus in South America (21–23). Our study attests to these claims and provides molecular evidence for the existence of Orientia in Kenya that is more similar to O. chuto than to O. tsutsugamushi.
We have previously described the occurrence of specific immunoglobulins against Orientia group antigens in patients with acute febrile illness from several locations in Kenya, including Marigat (7). Molecular analysis was not carried out in these patient samples. To investigate the potential risk of scrub typhus in those areas, we went back to the Marigat area and conducted surveillance for Orientia in the chigger mites and their wild rodent hosts. In this study, an abundance of small rodents in the surveyed areas within Marigat is reported. We successfully trapped 54 rodents from only a five-night surveillance period. As shown in Table 2, a variety of rodent species were identified, but only those in the Muridae family (85%) were hosts to two genera of trombiculid chigger mites, the Neotrombicula and Microtrombicula (Fig. S2 and S3). Chiggers of these genera are commonly found in rodent hosts from many countries within the Tsutsugamushi Triangle (17). Furthermore, the overall chigger index determined by our study (33.3) was within the ranges of seasonal chigger indices reported from countries where scrub typhus is endemic (24, 25). It is known that chigger development is temperature dependent and cooler months are the most active for chiggers. The survey was thus carried out in the cold season of June. In the Muridae family, chiggers were found in A. wilsoni, M. natalensis, and R. rattus. Of these, M. natalensis had the highest prevalence of chigger infestation.
In the present study, we report on the identification and molecular characterization of O. chuto in trombiculid chigger mites collected from wild rodents in Kenya. To our best knowledge, O. chuto has never been identified in Africa. It is also the first time to confirm the presence of scrub typhus vector in the area outside the geography space where scrub typhus has been unequivocally identified. Molecular characterization results showed that lung tissue and a pool of chigger mites from one of M. natalensis were positive for Orientia by PCR targeting the TSA47 and 16S rRNA (rrs) genes, indicating that M. natalensis may be an important host for scrub typhus agent in Kenya. M. natalensis is a natal multimammate African rat that commonly occurs in agricultural fields, grasslands, and dry forest and has been confirmed as a paratenic host of Lassa fever virus in West Africa (26). The abundance of M. natalensis populations in Marigat indicates the potential risk of rodent-borne hemorrhagic fever virus and scrub typhus in this area.
To identify and characterize the Kenyan Orientia species, we used species-specific primers to amplify an 850-bp fragment of the TSA47 gene and a 650-bp fragment of the TSA56 gene that have been found to discriminate between the two species in the Orientia genus (3–5). Unexpectedly, the Orientia species in our sample turned out to be O. chuto (with TSA47 gene) and not O. tsutsugamushi. Primers that target the TSA56 gene of O. tsutsugamushi and O. chuto strain Dubai consistently failed to amplify the chigger mite sample, whereas it amplified the O. tsutsugamushi positive control sample (Fig. 1). The TSA56 gene is very variable, and a sequence diversity of 53% was found between O. tsutsugamushi and O. chuto (4). We are of the opinion that the TSA56 gene of our study OTU may be very different from that of O. chuto strain Dubai, described by Izzard et al. (3), and that this variability may have caused the amplification failure. On sequencing the TSA47 gene of the Kenyan Orientia sample, its closest homology was to O. chuto strain Dubai (accession number HM156063), with a nucleotide similarity of 97.65%. The nucleotide identity of O. chuto strain Marigat identified from our study showed 84.63% to 84.76% identity to published O. tsutsugamushi (Fig. S4A) but formed a sister OTU with O. chuto strain Dubai. The study OTU was, however, distinguishable from the Dubai strain by having a longer branch (Fig. 2).
To further confirm the finding of O. chuto in the Kenyan chigger sample, we used 16S rRNA deep sequencing as an alternative approach. Seven bacteria genera, including Orientia, were identified. The Orientia sequence had a 99.75% similarity to that of O. chuto strain Dubai (GenBank accession number NR117903), compared to 98.75% with the closest O. tsutsugamushi strain (Fig. S4B). Similar sequence homologies were reported between O. chuto strain Dubai and other O. tsutsugamushi strains (3).
Taken together, the data from the current study and those from previous serological surveys for scrub typhus in patients with acute febrile illness (7, 8) provide unequivocal evidence for occurrence of trombiculid chigger mites, the rodent hosts, and Orientia in Kenya. Our study identifies A. wilsoni, M. natalensis, and R. rattus as important host for chiggers. From the molecular data, the Orientia species identified in Kenyan chiggers is more closely related to O. chuto that was described in 2010 by Izzard’s study (3). Although the occurrence of scrub typhus disease remains to be proven, our data and the existing immunological and molecular evidence (7–9) are strongly suggestive of the disease, and the threat for scrub typhus must be considered real.
Supplementary Material
ACKNOWLEDGMENTS
Funding for this study was obtained from the Armed Forces Health Surveillance Branch (AFHSB) and its GEIS (Global Emerging Infections Surveillance and Response) Section.
Material has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation and/or publication. The opinions or assertions contained herein are the private views of the authors, and are not to be construed as official or as reflecting true views of the Department of the Army or the Department of Defense.
J.W. conceived and designed the experiments. C.M., P.L., S.L., S.Y., D.A., N.A., T.G., and E.W. collected the samples. C.M., B.M., and G.K. performed the experiments. G.K. analyzed the bioinformatics data. C.M., P.L., G.K., B.M., D.A., E.W., and J.W. wrote the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01124-18.
REFERENCES
- 1.Pham XD, Suzuki H, Takaoka H. 2001. Distribution of unengorged larvae of Leptotrombidium pallidum and other species in and around the rodent nest holes. Southeast Asian J Trop Med Public Health 32:553–557. [PubMed] [Google Scholar]
- 2.Fournier PE, Siritantikorn S, Rolain JM, Suputtamongkol Y, Hoontrakul S, Charoenwat S, Losuwanaluk K, Parola P, Raoult D. 2008. Detection of new genotypes of Orientia tsutsugamushi infecting humans in Thailand. Clin Microbiol Infect 14:168–173. doi: 10.1111/j.1469-0691.2007.01889.x. [DOI] [PubMed] [Google Scholar]
- 3.Izzard L, Fuller A, Blacksell SD, Paris DH, Richards AL, Aukkanit N, Nguyen C, Jiang J, Fenwick S, Day NPJ, Graves S, Stenos J. 2010. Isolation of a novel Orientia species (O. chuto sp. nov.) from a patient infected in Dubai. J Clin Microbiol 48:4404–4409. doi: 10.1128/JCM.01526-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelly DJ, Fuerst PA, Ching W, Richards AL. 2009. Scrub typhus: the geographic distribution of phenotypic and genotypic variants of Orientia tsutsugamushi. Clin Infect Dis 48:S203–S230. doi: 10.1086/596576. [DOI] [PubMed] [Google Scholar]
- 5.Jiang J, Paris DH, Blacksell SD, Aukkanit N, Newton PN, Phetsouvanh R, Izzard L, Stenos J, Graves SR, Day NPJ, Richards AL. 2013. Diversity of the 47-kD HtrA nucleic acid and translated amino acid sequences from 17 recent human isolates of Orientia. Vector Borne Zoonotic Dis 13:367–375. doi: 10.1089/vbz.2012.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor AJ, Paris DH, Newton PN. 2015. A systematic review of mortality from untreated scrub typhus (Orientia tsutsugamushi). PLoS Negl Trop Dis 9:e0003971. doi: 10.1371/journal.pntd.0003971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thiga JW, Mutai BK, Eyako WK, Ng’Ang’ AZ, Jiang J, Richards AL, Waitumbi JN. 2015. High seroprevalence of antibodies against spotted fever and scrub typhus bacteria in patients with febrile illness, Kenya. Emerg Infect Dis 21:688–691. doi: 10.3201/eid2104.141387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maina AN, Farris CM, Odhiambo A, Jiang J, Laktabai J, Armstrong J, Holland T, Richards AL, O’Meara WP. 2016. Q fever, scrub typhus, and rickettsial diseases in children, Kenya, 2011–2012. Emerg Infect Dis 22:883–886. doi: 10.3201/eid2205.150953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cosson JF, Galan M, Bard E, Razzauti M, Bernard M, Morand S, Brouat C, Dalecky A, Bâ K, Charbonnel N, Vayssier-Taussat M. 2015. Detection of Orientia sp. DNA in rodents from Asia, West Africa and Europe. Parasit Vectors 8:172. doi: 10.1186/s13071-015-0784-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wixson SK, Smiler KL. 1997. Anesthesia and analgesia in rodents, p. 165–203. In Kohn DF, Wixson SK, White WJ, Benson GJ, Anesthesia and analgesia in laboratory animals. Academic Press, Cambridge, MA. [Google Scholar]
- 11.Kingdon J. 2013. The Kingdon field guide to African mammals, 2nd ed A&C Black, London, UK. [Google Scholar]
- 12.Nadchatram M, Dohany AL. 1974. A pictorial key to the subfamilies, genera and subgenera of Southeast Asian chiggers (Acari, Prostigmata, Trombiculidae). Institute for Medical Research, Kuala Lumpur, Malaysia. [Google Scholar]
- 13.Jiang J, Chan T-C, Temenak JJ, Dasch GA, Ching W-M, Richards AL. 2004. Development of a quantitative real-time polymerase chain reaction assay specific for Orientia tsutsugamushi. Am Soc Trop Med Hyg 70:351–356. doi: 10.4269/ajtmh.2004.70.351. [DOI] [PubMed] [Google Scholar]
- 14.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO. 2013. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang J, Richards AL. 2018. Scrub typhus: no longer restricted to the Tsutsugamushi Triangle. Trop Med Infect Dis 3:E11. doi: 10.3390/tropicalmed3010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giroud P, Jadin J. 1951. Antibodies to Rickettsia orientalis in Africans and Asians living in Ruanda-Urundi (Belgian Congo). Bull Soc Pathol Exot Filiales 44:50–51. (In French.) [PubMed] [Google Scholar]
- 19.Horton KC, Jiang J, Maina A, Dueger E, Zayed A, Ahmed AA, Pimentel G, Richards AL. 2016. Evidence of Rickettsia and Orientia infections among abattoir workers in Djibouti. Am J Trop Med Hyg 95:462–465. doi: 10.4269/ajtmh.15-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolo AO, Sibeko-Matjila KP, Maina AN, Richards AL, Knobel DL, Matjila PT. 2016. Molecular detection of zoonotic Rickettsiae and Anaplasma spp. in domestic dogs and their ectoparasites in Bushbuckridge, South Africa. Vector Borne Zoonotic Dis 16:245–252. doi: 10.1089/vbz.2015.1849. [DOI] [PubMed] [Google Scholar]
- 21.Balcells ME, Rabagliati R, García P, Poggi H, Oddó D, Concha M, Abarca K, Jiang J, Kelly DJ, Richards AL, Fuerst PA. 2011. Endemic scrub typhus-like illness, Chile. Emerg Infect Dis 17:1659–1663. doi: 10.3201/eid1709.100960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weitzel T, Dittrich S, López J, Phuklia W, Martinez-Valdebenito C, Velásquez K, Blacksell SD, Paris DH, Abarca K. 2016. Endemic scrub typhus in South America. N Engl J Med 375:954–961. doi: 10.1056/NEJMoa1603657. [DOI] [PubMed] [Google Scholar]
- 23.Kocher C, Jiang J, Morrison AC, Castillo R, Leguia M, Loyola S, Ampuero JS, Cespedes M, Halsey ES, Bausch DG, Richards AL. 2017. Serologic evidence of scrub typhus in the peruvian Amazon. Emerg Infect Dis 23:1389–1391. doi: 10.3201/eid2308.170050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee SH, Lee YS, Lee IY, Lim JW, Shin HK, Yu JR, Sim S. 2012. Monthly occurrence of vectors and reservoir rodents of scrub typhus in an endemic area of Jeollanam-do, Korea. Korean J Parasitol 50:327–331. doi: 10.3347/kjp.2012.50.4.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Candasamy S, Ayyanar E, Paily K, Karthikeyan PA, Sundararajan A, Purushothaman J. 2016. Abundance & distribution of trombiculid mites & Orientia tsutsugamushi, the vectors & pathogen of scrub typhus in rodents & shrews collected from Puducherry & Tamil Nadu, India. Indian J Med Res 144:893–900. doi: 10.4103/ijmr.IJMR_1390_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lecompte E, Fichet-Calvet E, Daffis S, Koulémou K, Sylla O, Kourouma F, Doré A, Soropogui B, Aniskin V, Allali B, Kouassi Kan S, Lalis A, Koivogui L, Günther S, Denys C, ter Meulen J. 2006. Mastomys natalensis and Lassa fever, West Africa. Emerg Infect Dis 12:1971–1974. doi: 10.3201/eid1212.060812. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.