Abstract
Post-translational modification of the adeno-associated virus capsids is a poorly understood factor in the development of these viral vectors into pharmaceutical products. Here we report the extensive capsid deamidation of adeno-associated virus serotype 8 and seven other diverse adeno-associated virus serotypes, with supporting evidence from structural, biochemical, and mass spectrometry approaches. The extent of deamidation at each site depended on the vector’s age and multiple primary-sequence and three-dimensional structural factors. However, the extent of deamidation was largely independent of the vector recovery and purification conditions. We demonstrate the potential for deamidation to impact transduction activity and, moreover, correlate an early time point loss in vector activity to rapidly progressing spontaneous deamidation at several adeno-associated virus 8 asparagines. We explore mutational strategies that stabilize side-chain amides, improving vector transduction and reducing the lot-to-lot molecular variability that presents a key concern in biologics manufacturing. This study illuminates a previously unknown aspect of adeno-associated virus capsid heterogeneity and highlights its importance in the development of these vectors for gene therapy.
Keywords: post-translational modification, gene therapy, virus structure, structural biology, bioengineering, adeno-associated virus
Understanding and managing protein heterogeneity, often due to post-translational modifications, is critical for biologics development. Here we demonstrate widespread asparagine deamidation of adeno-associated virus gene therapy vectors and correlate its progress to a loss of transduction activity. We present mutagenic strategies that prevent deamidation, increase transduction, and improve manufacturing consistency.
Introduction
The adeno-associated virus (AAV) capsid is icosahedral in structure and is comprised of 60 viral protein (VP) monomers (VP1, VP2, and VP3) at a 1:1:10 ratio.1 All 519 amino acids in the VP3 protein sequence are contained within the C terminus of both VP1 and VP2. The shared VP3 sequences are primarily responsible for the overall capsid structure. Due to the structural flexibility of the VP1/2 unique regions and the low representation of VP1 and VP2 monomers relative to VP3 monomers in the assembled AAV capsid, VP3 is the only capsid protein that has been resolved via X-ray crystallography.2 VP3 contains nine hypervariable regions (HVRs) that are the primary source of sequence variation between AAV serotypes.3 Given their flexibility and location on the capsid surface, HVRs are largely responsible for interacting with target cells and the immune system.4, 5 Although the structures of several AAV serotypes are published (see PDB: 1LP3, 4RSO, 4V86, 3UX1, 3KIC, 2QA0, and 2G8G for the structure entries for AAV2, AAVrh.8, AAV6, AAV9, AAV3B, AAV8, and AAV4, respectively), we currently know very little about surface modifications on these capsids.1, 2, 6, 7, 8, 9, 10 Research suggests that intracellular phosphorylation of the capsid occurs at specific tyrosine residues.11 Despite putative glycosylation sites in the primary VP3 sequence, no glycosylation events have been identified in AAV2.12, 13 Furthermore, other AAV serotypes have not yet been evaluated for capsid glycosylation.
Post-translational modification (PTM) events on the AAV capsid, especially those that affect function, could impact the development of AAV as a pharmaceutical product for gene therapy applications. One such modification that has the potential to introduce vector heterogeneity is the deamidation of select amino acids. In some instances, this process is catalyzed by viral or bacterial deamidases to modulate host cell signaling pathways or innate immune responses.14, 15 More commonly, endogenous deamidation is an enzyme-independent spontaneous process. In some scenarios, this event serves as a molecular clock to indicate the relative age of a protein and regulate its turnover.16 Variations in PTMs of non-gene therapy protein therapeutics have complicated their development as drugs.17, 18 For example, deamidation of selected amino acids modulates the stability of and the immune response to the recombinant protective antigen-based anthrax vaccine.19, 20
Deamidation occurs when the amide group of an asparagine or, less frequently, a glutamine side chain is lost after nucleophilic attack from an adjacent main-chain amide. This process leads to a succinimidyl intermediate21 that, via hydrolysis, resolves into a mixture of aspartic acid and isoaspartic acid (or glutamic acid and isoglutamic acid22; see Figure 1A). Studies of short synthetic peptides estimate that this hydrolysis results in a 3:1 mixture of isoaspartic acid and aspartic acid.23 Several biophysical factors contribute to the kinetics of spontaneous deamidation including the identity of the N+1 amino acid, local flexibility of the peptide chain, overall protein structure, solvent accessibility, buffer identity, pH, and temperature.24, 25, 26 The N+1 residue is particularly important, because the size and charge of this residue influences the local flexibility of the peptide backbone; glycine in the N+1 position has the greatest effect on reducing the half-time of deamidation, followed by histidine and serine.16 The conformational flexibility and thermal vibration of the backbone are also important, because the deamidation rate of asparagine is greatly reduced when the residue is located in regions with ordered and rigid secondary and tertiary structures due to steric hindrance of the side chains of adjacent residues.
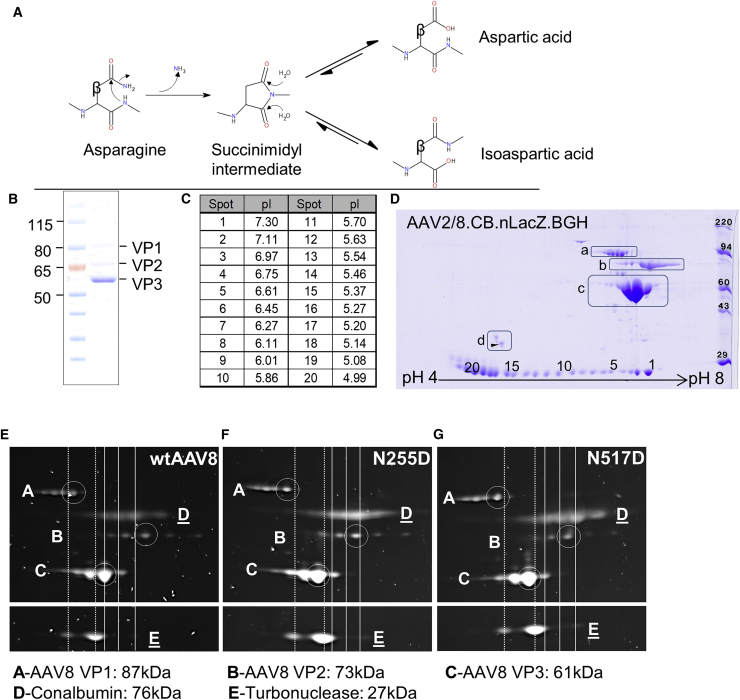
Figure 1.
Electrophoretic Analysis of AAV8 VP Isoforms
(A) Diagram illustrating the mechanism by which asparagine residues undergo nucleophilic attack by adjacent nitrogen atoms, forming a succinimidyl intermediate. This intermediate then undergoes hydrolysis, resolving into a mixture of aspartic acid and isoaspartic acid. The beta carbon is labeled as such. The diagram was generated in BIOVIA Draw 2018. (B) 1 μg of AAV8 vector was run on a denaturing one-dimensional SDS-PAGE. (C) Isoelectric points of carbonic anhydrase pI marker spots are shown. (D) 5 μg of AAV8 vector was analyzed by two-dimensional gel electrophoresis and stained with Coomassie blue. Spots 1–20 are carbamylated carbonic anhydrase pI markers. Boxed regions are as follows: a, VP1; b, VP2; c, VP3; and d, internal tropomyosin marker (arrow: tropomyosin spot of molecular weight [MW] = 33kDa, pI = 5.2). Isoelectric focusing was performed with a pI range of 4–8. 1 × 1011 GC of WT AAV8 (E) or mutant (F, N255D; and G, N517D) vector were analyzed by 2D gel electrophoresis and stained with SYPRO Ruby. Protein labeling: A, VP1; B, VP2; C, VP3; D, chicken egg white conalbumin marker; E, turbonuclease marker. Isoelectric focusing was performed with a pI range of 6–10. Primary VP1/2/3 isoform spots are circled, and migration distances of major spots of markers are indicated by vertical lines (turbonuclease, dashed; conalbumin, solid).
In this study, we characterized PTMs of the AAV8 vector capsid using gel electrophoresis, mass spectrometry, and de novo structural modeling. We extended this analysis to encompass representatives from seven other clinically relevant AAV clades and found broad agreement in the pattern and extent of capsid deamidation. We explore the functional consequences of vector deamidation and demonstrate strategies to mitigate its effects in order to improve vector performance and consistency in manufacturing.
Results
AAV8 Shows Substantial Charge Heterogeneity in Its Capsid Proteins
To qualitatively assess the presence of PTMs on the AAV8 vector capsid that affect vector performance, we analyzed AAV8 total capsid protein purified by iodixanol gradient both by one-dimensional (1D) and two-dimensional (2D) gel electrophoresis. In a 1D reducing SDS gel, VP1, VP2, and VP3 resolved as single bands at the appropriate molecular weights (Figure 1B).27 When further evaluated by 2D gel electrophoresis, which separates proteins based on charge (Figure 1C), each of the capsid proteins additionally resolved as a series of distinct spots with different isoelectric points (pIs) ranging from pH 6.3 to >7.0 depending on the VP isoform (Figure 1D). Individual spots on each VP were separated by discrete intervals of 0.1 pI units as measured as migration relative to the carbonic anhydrase isoform internal pI standards. The presence of these isoforms suggests that each VP has the potential to undergo multiple charge-altering modifications.
Deamidation, in which a fraction of side-chain amide groups, typically asparagine, is converted to carboxylic acid (Figure 1A), is a common source of charge heterogeneity in protein preparations. To determine whether deamidation could be responsible for the distinct population of VP charge isoforms, we mutated two AAV8 asparagine residues individually to aspartate. These capsid mutations should shift the charge by an amount equivalent to the complete deamidation of a single additional asparagine residue. Using 2D gel analysis of the mutants, we found that the major spots for VP1, VP2, and VP3 shifted one location more acidic (0.1 pH units) than the equivalent spots in wild-type (WT) AAV8 (Figures 1E–1G). The magnitude of this shift is equivalent to the spacing between the WT VP charge isoforms. Thus, the 2D gel patterning of AAV capsid proteins is consistent with multi-site deamidation.
Spontaneous Deamidation Occurs on the AAV8 Vector Capsid
To identify modifications responsible for the discrete spotting pattern for each capsid protein, we analyzed a panel of AAV8 vectors by mass spectrometry. Coverage of the AAV8 capsid protein averaged >95% of the total VP1 sequence (data not shown). We detected extensive deamidation of a subset of asparagine and glutamine residues by mass spectroscopy, which showed an increase of ∼1 Da in the observed mass of the individual peptides as compared with predicted values based on the sequence encoded by the DNA; we observed this pattern of deamidation in all preparations of AAV8 vectors (Figures 2A–2D).
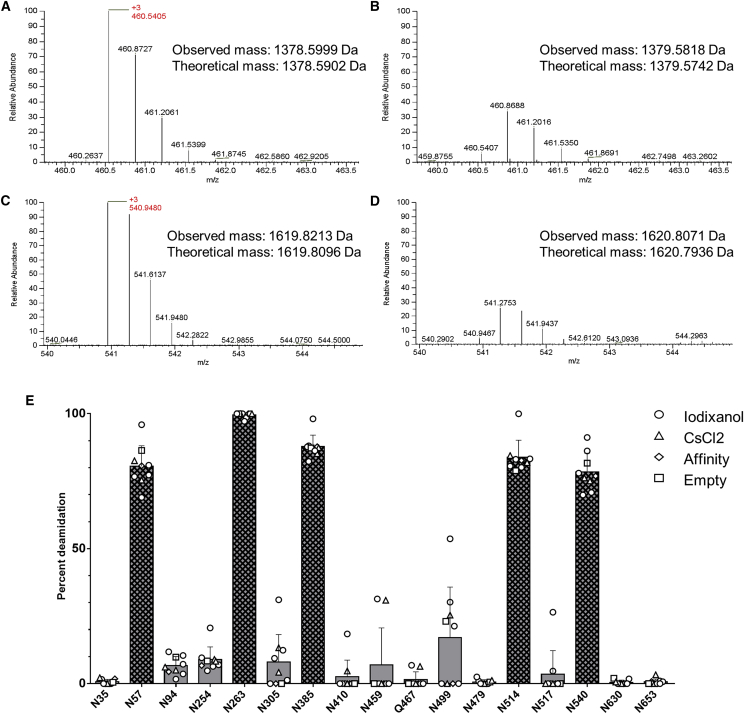
Figure 2.
Analysis of Asparagine and Glutamine Deamidation in AAV8 Capsid Proteins
(A and B) Electrospray ionization (ESI) mass spectrometry and theoretical and observed masses of the 3+ peptide (93–103) containing Asn-94 (A) and Asp-94 (B) are shown. (C and D) ESI mass spectrometry and theoretical and observed masses of the 3+ peptide (247–259) containing Asn-254 (C) and Asp-254 (D) are shown. The observed mass shifts for Asn-94 and Asn-254 were 0.982 and 0.986 Da, respectively, versus a theoretical mass shift of 0.984 Da. (E) Percent deamidation at specific asparagine and glutamine residues of interest are shown for AAV8 tryptic peptides purified by different methods. Bars indicating deamidation at asparagine residues with N+1 glycines are crosshatched. Residues determined to be at least 2% deamidated in at least one prep analyzed were included. Data are represented as mean ± SD.
To evaluate the global heterogeneity of deamidation between commonly used purification methods and to examine deamidation in the VP1 and VP2 unique regions, we selected nine lots of AAV8 produced by triple transfection in 293 cells and purified them by either cesium chloride gradient, iodixanol gradient, or affinity chromatography. Vectors also varied with respect to promoters and transgene cassettes. To determine whether the presence of the vector genome had an impact on deamidation, we evaluated an AAV8 preparation produced by triple transfection in 293 cells in the absence of cis plasmid (producing empty capsids only) and purified by iodixanol gradient.
A wide range of deamidation events was present across asparagine and glutamine residues of the AAV8 capsid, ranging from undetectable to over 99% of individual amino acids being deamidated (Figure 2E). The highest levels of deamidation (>75%) occurred at asparagine residues where the N+1 residue was glycine (i.e., NG pairs; Table 1). We detected lower levels of deamidation (i.e., up to 17%) at additional asparagine residues where the N+1 was not glycine. The average deamidation for asparagines was largely consistent between preparations. We also detected deamidation at glutamine residues but at a lower frequency than at asparagines; the highest percent we observed was <2% at Q467 (Figure 2E). This observation was inconsistent across preparations (data not shown). We observed the greatest preparation-to-preparation differences at residue N499 (N+1 residue is asparagine), with values ranging from <1% to over 50% deamidation. Regardless, the variations we observed in deamidation between preparations of vector did not appear to be related to purification method, transgene identity, or the presence of vector genome, suggesting that these factors do not impact deamidation rates.
Table 1.
Characteristics of Deamidated Residues of Interest
| N+1 Residue | Structural Topology | Structural Motif | Average % Deamidation | Temperature Factor (Å2) | |
|---|---|---|---|---|---|
| N35 | Q | N/A | N/A | 1 | N/A |
| N57 | G | N/A | N/A | 80 | N/A |
| N94 | H | N/A | N/A | 7 | N/A |
| N254* | N | surface exposed | not assigned | 9 | 35 |
| N255* | H | surface exposed | not assigned | N/A | 42 |
| N263 | G | surface exposed | HVR I | 99 | 51 |
| N305 | N | buried | alpha helix | 8 | 33 |
| N385 | G | surface exposed | HVR III | 88 | 41 |
| N410 | N | buried | not assigned | 3 | 33 |
| N459 | T | surface exposed | HVR IV | 7 | 65 |
| N499 | N | surface exposed | HVR V | 17 | 45 |
| N514* | G | surface exposed | HVR V | 84 | 36 |
| N517* | S | surface exposed | HVR V | 4 | 40 |
| N540* | G | buried | HVR VII | 79 | 40 |
| N630* | F | buried | not assigned | 1 | 32 |
| N653 | T | surface exposed | HI loop | 1 | 35 |
Asterisks represent residues selected for further analysis. NA, not applicable.
Next, we ran a series of experiments to determine whether sample handling contributed to the observed levels of deamidation in AAV8. Extreme temperature (70°C for 7 days) or pH (pH 2 or 10 for 7 days) did not significantly induce additional deamidation in the AAV8 capsid (Figures S1A and S1B) even though the conditions were harsh enough to partially denature some of the vector (Figure S1D). Given this resistance, we reasoned that the observed deamidation was unlikely to occur only in the purification phase, which was shorter and relatively mild in comparison. We attempted to perform mass spectrometry analysis on unpurified vector to determine the extent of deamidation before and after purification, but we were unsuccessful. Likewise, heavy water controls indicate that processing specific to our mass spectrometry workflow does not contribute to additional deamidation events28 (Figure S1C).
To validate our mass spectrometry workflow, we examined two recombinant proteins that have been evaluated previously for deamidation; our findings (Figures S2A and S2B) agree with the published results.29, 30 Additionally, we engaged a secondary institution to evaluate our raw data from AAV8. This independent analysis identified the same sites as deamidated, with minimal variation in the extent of modification at each site attributable to software-to-software variations in peak detection and area calculation (Figure S3).
Structural Topology, Temperature Factor, and the Identity of the N+1 Amino Acid Contribute to Deamidation Frequency
Because the structure of AAV8 has been solved and published (PDB: 2QA0),31 we next examined the AAV8 capsid structure for evidence of favorable conditions for non-enzymatic deamidation and to correlate percent deamidation with established structural features.2 We focused on asparagine residues exclusively, because the factors influencing asparagine deamidation are better characterized in the literature and asparagine deamidation events are far more common than glutamine deamidation events.16 We also determined the temperature (or B) factor for each of these residues from the AAV8 crystal structure; temperature factor is a measure of the displacement of an atom from its mean position, with higher values indicating a larger displacement, higher thermal vibration, and therefore increased flexibility.32 The majority of asparagines of interest were located in or near the surface-exposed HVRs (Table 1), which are structurally favorable for deamidation and provide a solvent-exposed environment.3 We found that residues located in these flexible loop regions were, on average, more frequently deamidated than residues in less flexible regions such as beta strands and alpha helices. For example, the NG residue at position N263 is part of HVR I, has a high temperature factor, and was >98% deamidated on average (Table 1). N514, which was deamidated ∼85% of the time (Table 1), is also in an HVR (HVR V) with an N+1 glycine; however, the local temperature factor is relatively low in comparison with that of N263 due to its interaction with residues on other VP monomers at the 3-fold axis. Less favorable +1 residues and lower temperature factors correlated with lower deamidation, even for HVR residues. For example, N517 was on average only 4% deamidated (Table 1); this residue has an equivalent temperature factor to the highly deamidated N514, but its N+1 residue is a serine, decreasing the likelihood of deamidation events due to steric hindrance. This demonstrates that a number of factors cumulatively determine the extent of deamidation at a given capsid position, although the identity of the +1 residue is apparently the most influential factor.
To test the role of the +1 residue in asparagine deamidation, we generated mutant vectors in which we individually mutated AAV8 NG sites at the +1 position to either alanine or serine. Model peptide studies indicate that NG peptides deamidate with a half-life as short as 1 day, whereas NA or NS peptides typically deamidate 25- or 16-fold more slowly, respectively.25 Mass spectrometry analysis of the vector mutants confirmed the central role of the +1 site in determining the extent of vector deamidation. NG sites in this set (>80% deamidation in WT) showed selective stabilization of the adjacent asparagine when the +1 site was changed to alanine (<5% deamidation) or serine (<14% deamidation) (Table 2).
Table 2.
Extent of Deamidation (%) at Five AAV8 NG Sites in WT and Six +1 Site Mutants
| Amino Acid Identity | WT (Average) | G58S | G58A | G264A | G386S | G386A | G515A |
|---|---|---|---|---|---|---|---|
| N57 | 81.8 | 8.4 | 1.9 | 89.7 | 89.7 | 91.6 | 93.6 |
| N263 | 99.3 | 98.2 | 98.9 | 4.8 | 100.0 | 94.5 | 97.2 |
| N385 | 89.1 | 96.3 | 94.8 | 97.1 | 13.5 | 2.5 | 97.0 |
| N514 | 85.2 | 100.0 | 98.0 | 98.8 | 100.0 | 100.0 | 2.2 |
| N540 | 84.5 | 95.0 | 92.6 | 97.9 | 96.9 | 86.1 | 89.5 |
Structural Modeling of AAV8 VP3 Confirms Deamidation Events
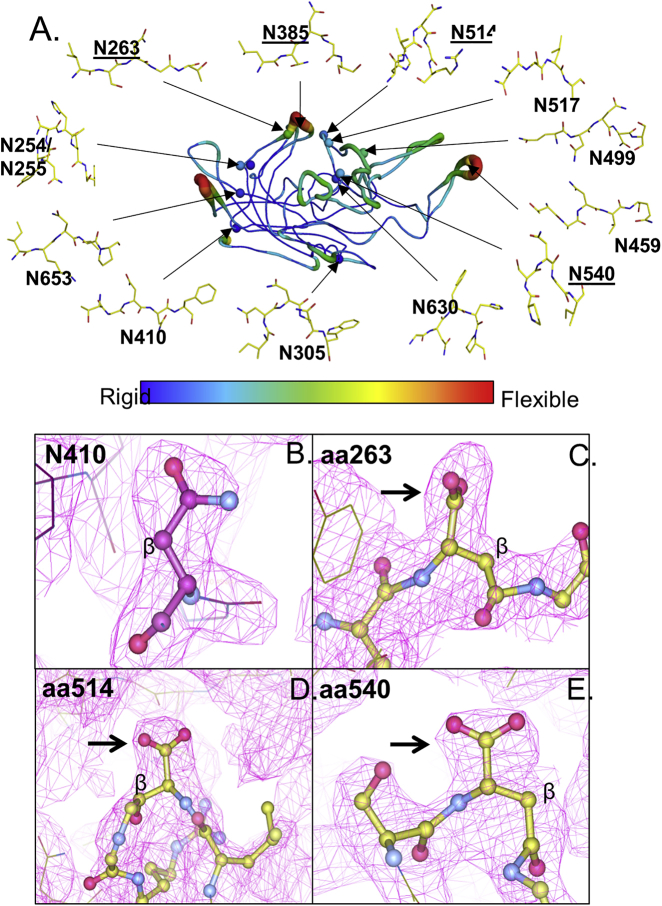
To provide direct evidence of deamidation in the context of an assembled capsid, we evaluated the crystal structure of AAV8.31 The resolution of the available crystal structure (i.e., 2.7 Å) of this serotype is not high enough to identify the terminal atoms in the R groups and, therefore, is insufficient to directly distinguish between asparagine, aspartic, and isoaspartic acid residues. Other aspects of the structure of the isomer of aspartic acid that forms under these conditions provided us an opportunity to determine deamidation from the 2.7 Å structure. This analysis was based on two assumptions: (1) the predominant product of spontaneous deamidation of an asparagine is isoaspartic rather than aspartic acid, which is generated at a 3:1 ratio23; and (2) we can differentiate between an asparagine or aspartic acid and an isoaspartic acid because the electron density map (magenta grid) corresponding to the R group of isoaspartic acid is shorter in length. This shorter R group is created when the beta carbon from the R group of isoaspartic acid is lost when it is incorporated into the main chain of the AAV8 VP3 capsid protein backbone following resolution of the succinimidyl intermediate during the deamidation reaction.
We first refined the AAV8 structure itself, generating an AAV8 capsid electron density that was not biased by the known AAV8 VP3 sequence. We then examined the refined AAV8 crystal structure for evidence of deamidation based on the presence of a shorter R group associated with isoaspartic acid (Figures 3A–3E). The electron density map confirmed a shorter R group for the highly deamidated N+1 glycine residues at positions 263 (Figure 3C), 385 (data not shown), 514 (Figure 3D), and 540 (Figure 3E) when compared with the asparagine at position 410, which had no deamidation detected by mass spectrometry (Figure 3B). The deamidation indicated by the electron density map is therefore consistent with the data generated by mass spectrometry at these sites with >75% deamidation. The resulting isoaspartic acid models were comparable with isoaspartic acid residues observed in the crystal structures of other known deamidated proteins, supporting the validity of our analysis of AAV8.33, 34, 35 This structural analysis serves as an independent confirmation of the deamidation phenomena observed when analyzing the AAV8 capsid via mass spectrometry.
Figure 3.
Structural Modeling of the AAV8 VP3 Monomer and Analysis of Deamidated Sites
(A) The AAV8 VP3 monomer (PDB: 3RA8) is shown in a coil representation. The color of the ribbon indicates the relative degree of flexibility (blue = most rigid/normal temperature factor; red = most flexible/high temperature factor). Spheres indicate residues of interest. Expanded diagrams are ball-and-stick representations of residues of interest and their surrounding residues to demonstrate local protein structure (blue, nitrogen; red, oxygen). Underlined residues are those in NG motifs. (B–E) Isoaspartic models of deamidated asparagines with N+1 glycines are shown. The 2FoFc electron density map (1 sigma level) generated from refinement of the AAV8 crystal structure (PDB: 3RA8) with (B) an asparagine model of N410 in comparison with isoaspartic acid models of (C) N263, (D) N514, and (E) N540. Electron density map is shown in the magenta grid. All atoms are colored by atom type: carbon, purple (B)/gree6n (C–E); nitrogen, blue; oxygen, pink. The beta carbon is labeled as such. The arrows indicate electron density corresponding to the R group of the residue of interest.
Deamidation of the AAV Capsid Is Not Serotype Specific
We investigated serotypes beyond AAV8 for evidence of capsid deamidation. We examined AAV9 vector preparations using 2D gel electrophoresis (Figure S4A) and mass spectrometry (Figure S4B), including controls for potential vector-processing effects (Figure S5). The pattern and extent of AAV9 deamidation were similar to that of AAV8. All four AAV9 NG sites were >85% deamidated; 13 non-NG sites were deamidated to a lesser extent, with a few sites showing high lot-to-lot variability in percent deamidation. Next, we applied our structural analysis workflow and refit existing AAV9 crystallographic data (Figure S4C; Table S1). As with AAV8, isoaspartic acid fit better into the electron density of several NG sites in the AAV9 crystal structure. We extended our 2D gel analysis (data not shown) and mass spectrometry (summarized in Table S2) to five additional evolutionarily diverse serotypes (rh32.33, AAV7, AAV5, AAV4, AAV3B, and AAV1). All of the capsids we examined contained a similar pattern and extent of deamidation, indicating that this modification is widespread in clinically relevant AAV vectors and is determined by similar underlying primary-sequence and structural factors.
Deamidation Events Can Affect Capsid Assembly and Transduction Efficiency
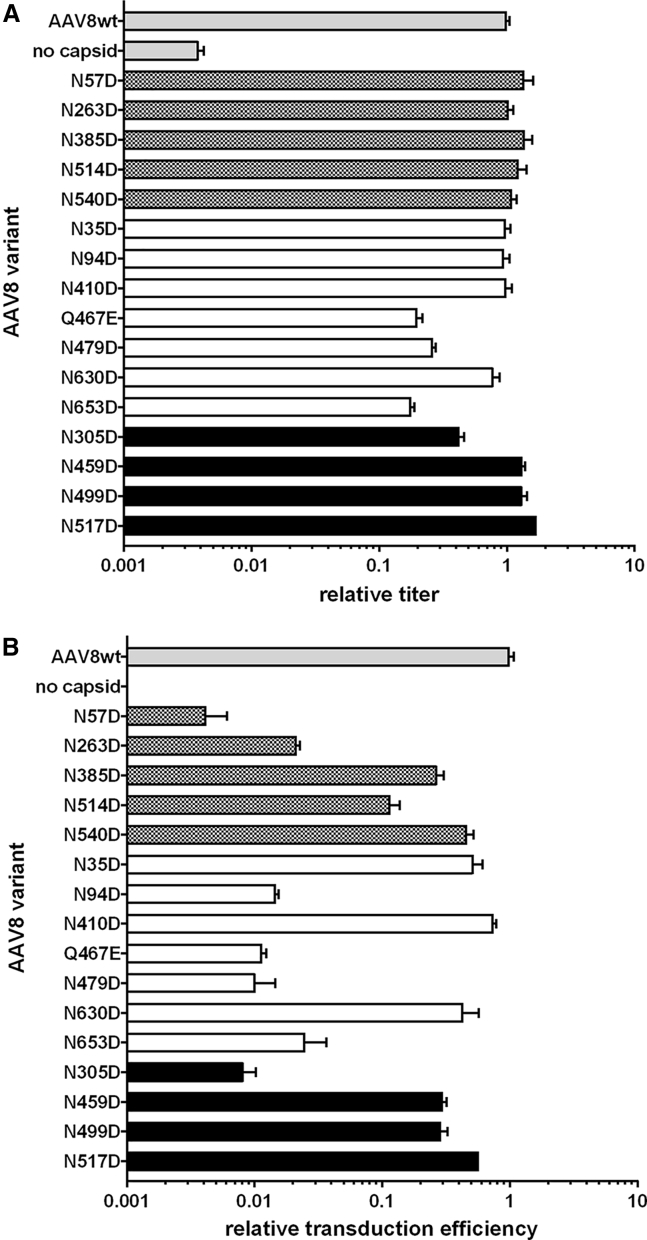
One approach to testing the functional impact of deamidation is by substituting asparagine with aspartate by genetic mutation. We generated an aspartate mutant vector encoding a luciferase reporter for each deamidated AAV8 asparagine by small-scale triple transfection of 293 cells, and titered the vectors by qPCR of DNase I-resistant genome copies (GCs) (Figure 4A). The mutations rarely affected capsid assembly relative to WT AAV8, and effects were limited to mostly buried, non-NG sites with low overall deamidation in the WT vector. Next, we assessed the mutant panel for in vitro transduction efficiency of human liver-derived Huh7 cells (Figure 4B). Several mutants showed impaired transduction efficiency, with positions N57, N94, N263, N305, Q467, N479, and N653 exhibiting >10-fold transduction loss. We observed a similar number of sensitive sites for AAV9 (Figure S6). Because typically only a fraction of residues at a given position are deamidated endogenously, this approach has the potential to overestimate functional loss for proteins such as capsids where the functional unit is a homomeric assembly; endogenous modification at one capsid site may be compensated for by a neighboring subunit with an intact residue. Nonetheless, we reasoned that this method could help prioritize deamidated residues for future monitoring during manufacturing or mutational stabilization. Functional data from populations of endogenously deamidating vectors will be required to place this loss-of-function mutagenesis data in the proper context.
Figure 4.
In Vitro Analysis of the Impact of Genetic Deamidation on Vector Performance
(A) Titers of WT AAV8 and genetic deamidation mutant vectors were produced by small-scale triple transfection in 293 cells, as measured by qPCR. Titers are reported relative to the WT AAV8 control. NG sites with high deamidation (patterned bars), sites with low deamidation (white bars), and highly variable sites (black bars) are presented with WT AAV8 and a negative control. (B) The transduction efficiency of mutant AAV8 vectors producing firefly luciferase is reported relative to the WT AAV8 control. Transduction efficiency was measured in luminescence units generated per GC added to HUH7 cells and was determined by performing transductions with crude vector at multiple dilutions. Transduction efficiency data were normalized to the WT reference. All data are represented as mean ± SD.
Vector Activity Loss over Time Is Correlated with Progressive Deamidation
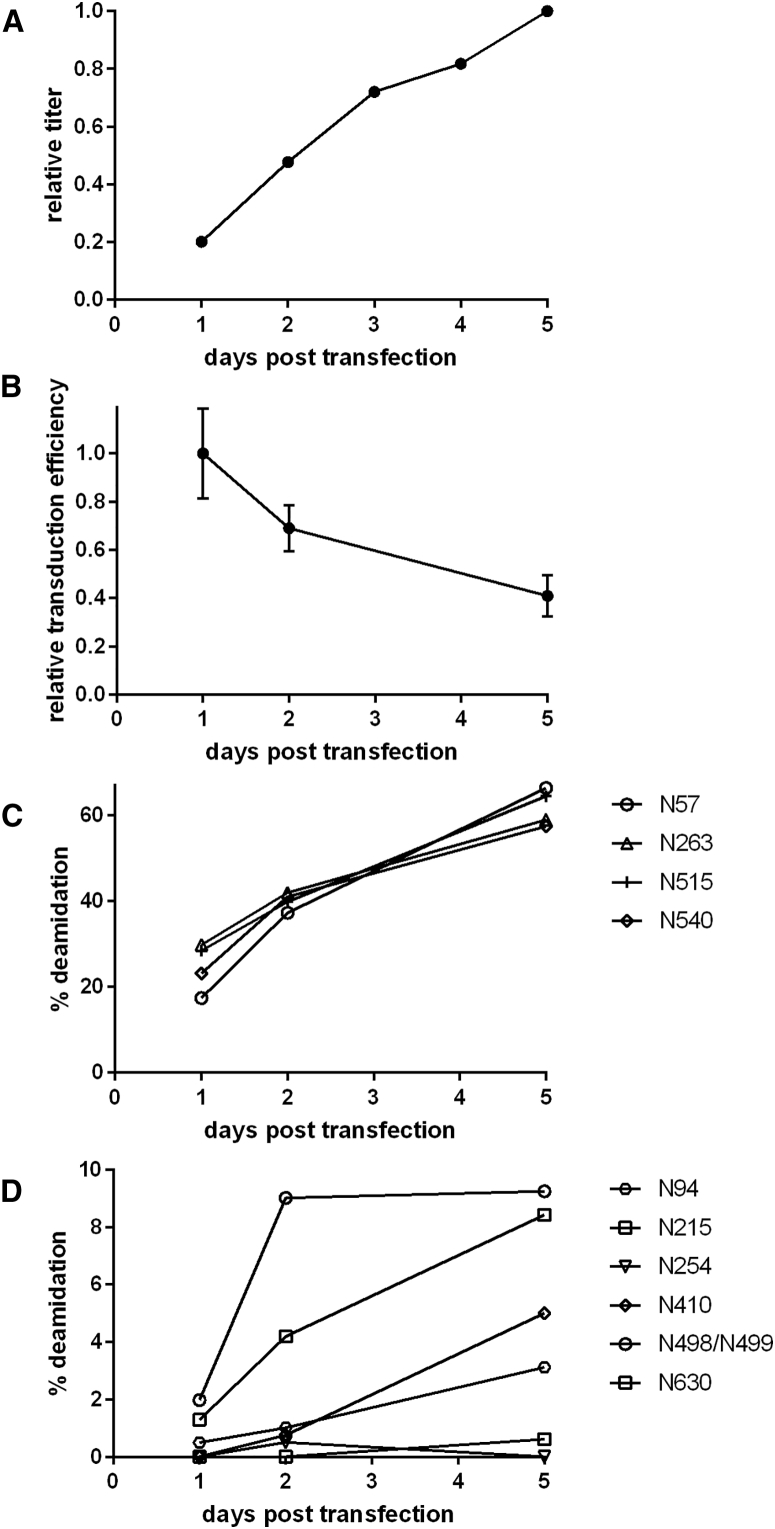
Given the apparently short half-life of NG deamidation, we reasoned that vector samples differing in age by as little as 1 day could show distinct deamidation profiles, thus providing an opportunity to correlate endogenous deamidation to function. Our large-scale vector preparation protocol calls for triple transfection of 293 cells followed by 5 days of incubation for vector production and 1–2 days for vector purification. To approximate this process, we prepared medium-scale triple transfections (10 × 15-cm cell culture dishes each) of 293 cells with WT AAV8. We collected vector (2 × 15-cm cell culture dishes/day) at 1-day intervals for 5 days, preserving the time points until the end of the fifth day by freezing vector at −80°C. Next, we assessed crude vector titer as described above. As expected, the number of assembled, DNaseI-resistant GCs increased over time (Figure 5A). We then quickly processed crude vector for early (days 1 and 2) and late (day 5) time points by affinity purification and measured in vitro transduction efficiency of huh7 cells. Relative transduction efficiency of the vector dropped progressively over time (Figure 5B). In terms of transgene expression per GC added to target cells, day 5 vector was only 40% as efficient as day 1 material. We observed this activity drop for crude material as well, indicating a change in molecular composition before purification (Figure S7). A similar trend in activity loss occurred for AAV9 over 5 days, with approximately 40% reduction in vector potency (Figure S7).
Figure 5.
Vector Activity Loss through Time Is Correlated to Progressive Deamidation
(A) Vector production (DNase I-resistant GCs) for a time course of triple-transfected HEK293 cells producing AAV8 vector packaging a luciferase reporter gene. GC levels are normalized to the maximum observed value. (B) Purified time-course vector was used to transduce Huh7 cells. Transduction efficiency (luminescence units per GC added to target cells) was measured as in Figure 4 using multiple dilutions of purified time-course vector samples. Error bars represent the SD of at least 10 technical replicates for each sample time. Deamidation of AAV8 NG sites (C) and non-NG sites (D) for vector collected 1, 2, and 5 days post-transfection.
Next, we measured deamidation of the time-course samples by mass spectrometry. NG site deamidation progressed substantially over every interval, with an average of 25% deamidation at day 1 and >60% of sites converted by day 5 (Figure 5C). Non-NG site deamidation generally progressed over 5 days, although at much lower levels and with less consistency between days 2 and 5 (Figure 5D). The data correlate endogenous vector deamidation to an early time point decay in specific activity and highlight a potential opportunity to capture more active vector by shortening the production cycle or finding capsid mutations that stabilize asparagines.
We note that the material used for mass spectrometry analysis in Figure 2 was at least 7 days post-transfection, due to an additional 2 days for purification. The higher NG site deamidation in these samples (>80%) indicates that deamidation likely continues after the period of expression and during the recovery and purification processes at approximately the same rates until NG sites are completely deamidated or the vector sample is frozen. Thus, deamidation is largely determined by the age of the vector and is not a process that is exclusive to or caused by the recovery and purification process. The much lower deamidation values in the day 1 compared with the day 5 material, which are both affinity purified, underscore this point.
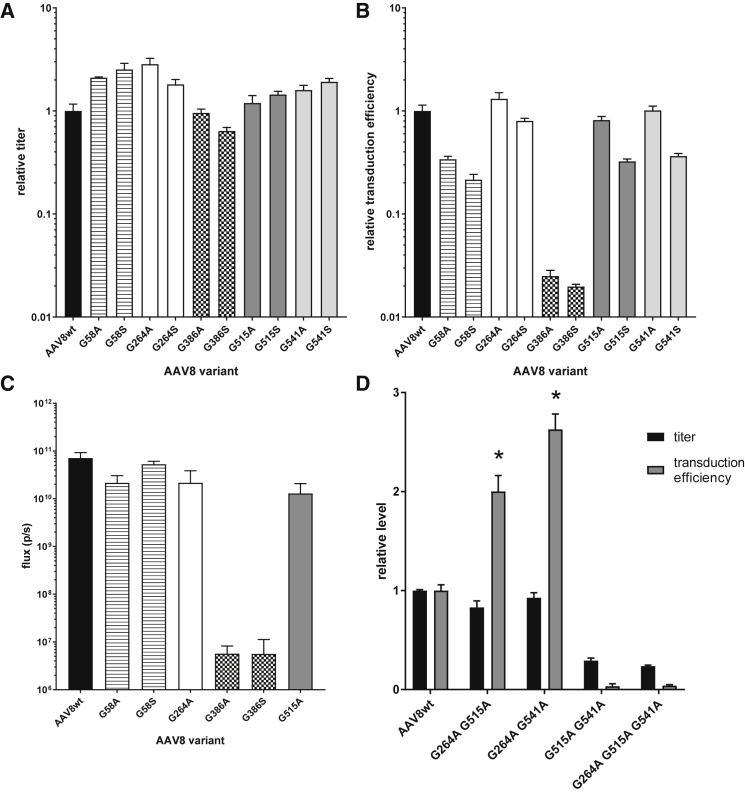
Stabilizing NG Asparagines Can Improve Vector Performance
Given the correlation between vector NG deamidation and transduction efficiency loss, we reasoned that stabilizing NG amides by +1 site mutagenesis may improve vector function. We produced vector in small scale for AAV8 NG site mutants in which we individually converted each +1 residue to alanine or serine. Single +1 mutants were well tolerated in terms of vector assembly (Figure 6A) and transduction efficiency (Figure 6B). G386 substitutions, located near a previously defined “dead zone” on the capsid surface,36 were defective for in vitro transduction. The loss of function in G386 mutants could indicate a preference for a deamidated asparagine at N385. Alternatively, the additional side-chain bulk at the +1 position may have a negative impact on function that is independent of amide-group stabilization. No single-site mutants significantly improved in vitro transduction, in spite of dramatic stabilization of their neighboring asparagines (Table 2). Because in vitro and in vivo transduction activities can be discordant, we tested a subset of the single-site +1 mutants for liver transduction in C57BL/6 mice. We performed intravenous tail vein injection (n = 3–5) and examined luciferase expression by imaging weekly for 2 weeks (Figure 6C). In vivo and in vitro transduction data were in agreement with each assay (i.e., within the error range). G386 substitutions were defective for transduction, whereas +1 site mutations at other positions were largely tolerated and allowed liver transduction at levels equivalent to but not exceeding WT AAV8.
Figure 6.
The Impact of Stabilizing Asparagines on Vector Performance
(A) Titers of WT AAV8 and +1 position mutant vectors were produced by small-scale triple transfection in 293 cells, as measured by qPCR. Titers are reported relative to the WT AAV8 control. (B) The transduction efficiency of mutant AAV8 vectors producing firefly luciferase is reported relative to the WT AAV8 control. Transduction efficiency was measured as in Figure 4 using crude vector material. (C) Luciferase expression on day 14 of the study period in the liver region from C57BL/6 mice injected intravenously with WT AAV8 or mutant vectors (n = 3–5) was measured by luciferase imaging and reported in total flux units. (D) The titers and transduction efficiency of multi-site AAV8 mutant vectors producing firefly luciferase are reported relative to the WT AAV8 control. All data are represented as mean ± SD. A two-sample t test (∗p < 0.005) was run to determine significance between WT AAV8 and mutant transduction efficiency for G264A/G515A and G264A/G541A.
Because stabilizing the amide at any one NG site may be necessary but not sufficient for functional restoration, we next evaluated vector variants with combinations of +1 site alanine substitutions. We recombined all three AAV8 NG sites for which the +1 alanine was highly functional (N263, N514, and N540). Some combinations, including the triple-mutant G264A/G515A/G541A, were assembled poorly and were dysfunctional for transduction. However, both pairwise combinations involving N263 (G264A/G515A and G264A/G541A) improved in vitro transduction efficiency (2.0- and 2.6-fold over WT AAV8, respectively) with no loss of titer (Figure 6D). Because these mutations introduce at least two changes (N-amide stabilization and a +1 residue side-chain substitution) these data do not conclusively link NG deamidation to functional loss. However, the data are consistent with the model established in the time-course study in which NG site deamidation can impact in vitro transduction efficiency.
Functional Asparagine Substitutions Improve Lot-to-Lot Reproducibility in Vector Manufacturing
Another potentially problematic aspect of the vector deamidation profiles we report is the high lot-to-lot variability in deamidation at some positions. For WT AAV8, this variability is most pronounced for N459 (observed deamidation ranging from 0% to 31%) and N499 (observed deamidation ranging from 0% to 53%). Variability in PTMs is typically de facto avoided during biologics development, either by avoiding clones altogether that exhibit this variability, carefully monitoring and controlling production strains and conditions, or by protein engineering the affected candidate.
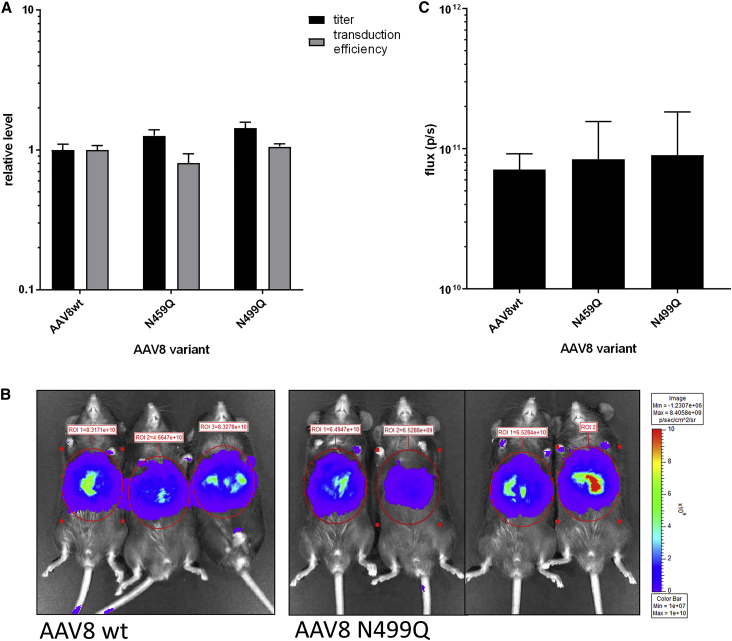
Because we were unable to determine the production or processing factors contributing to N459 and N499 deamidation variability (Figure 2E), we sought functional amino acid substitutions at these positions. We first evaluated small-scale vector preparations for conservative substitutions to glutamine at each position individually. Both N459Q and N499Q were assembled efficiently into vector and were equivalent to the WT AAV8 reference for in vitro transduction efficiency (Figure 7A). Next, we produced the mutants in large scale and performed mass spectrometry. Consistent with our observations of extremely rare glutamine deamidation, we observed selective and complete stabilization of the glutamine amides at positions 459 or 499 in these mutants (data not shown). We evaluated these mutant lots in vivo as above for liver transduction after tail vein injection in C57BL/6 mice (Figures 7B and 7C). The WT AAV8 vector lot used as a control in this experiment was deamidated 16.8% at N499, but was not deamidated at N459 (data not shown). Liver transduction at day 14 for both mutants was equivalent to WT AAV8. These data demonstrate the potential for a protein engineering approach to address the molecular variability associated with deamidation in manufactured AAV vectors.
Figure 7.
Functional Asparagine Substitutions at Non-NG Sites with High Variability between Lots
(A) Titers of WT AAV8 and mutant vectors were produced by small-scale triple transfection in 293 cells, as measured by qPCR. Transduction efficiencies for crude vector material were measured as described in Figure 4. Titers and transduction efficiencies were normalized to the value of the WT AAV8 control. (B) Representative luciferase images at day 14 post-injection are shown for mice receiving WT AAV8.CB7.ffluc and N499Q capsid mutant vector. (C) Luciferase expression on day 14 of the study period from C57BL/6 mice injected intravenously with WT AAV8 or mutant vectors (n = 3 or 4) was measured by luciferase imaging and reported in total flux units. WT control shared with Figure 6C. All data are represented as mean + SD.
Discussion
AAV gene therapy vectors have undergone less of the molecular-level scrutiny that typically accompanies the development and manufacturing of recombinant protein therapeutics. For example, monitoring and control of PTMs is an important aspect of monoclonal antibody development because of the impact that PTM profiles, including deamidation,37 can have on activity in vivo. Therapeutic antibody lots must conform to a reference lot in terms of PTM distributions as determined by mass spectrometry and other bioanalytical methods. AAV capsid PTMs have largely been unexplored; therefore, little is known about their potential to impact function or about strategies to control PTM levels in manufactured AAV therapies.
Here we investigated the PTM landscape of recombinant AAVs, beginning in depth with AAV8. We found widespread asparagine and occasional glutamine deamidation affecting 17 residues. Factors controlling AAV8 deamidation, principally primary-sequence and three-dimensional (3D) structural constraints, are likely conserved across the entire AAV phylogeny, as all serotypes analyzed by us to date exhibit a strikingly similar pattern of modification. Thus, deamidation is potentially a critical factor in the development of all future AAV therapeutics.
With this discovery, we were motivated to explore the functional impacts of AAV deamidation. The multimeric nature of AAV vector capsids, the extent and number of modified capsid residues, and the resulting mosaic diversity in vector particle compositions presented unique challenges to this analysis. The experimental repertoire that sufficiently parameterizes PTM impact in a simpler protein context is not directly applicable to AAV capsid analysis. For example, it would be impossible to purify or even enrich a preparation for a particular deamidated vector species to test its function directly and in isolation.
One approach that we tried to force an approximation of the modification at a given site is genetic substitution to aspartate. Beyond the previously noted differences between the distributions of position-specific modification on capsid assemblies with endogenous (mosaic) versus genetic (complete) deamidation, our results highlight additional considerations for interpreting this type of data. For example, we observed >50-fold transduction loss for the N263D mutant relative to WT AAV8 (Figure 4B). This was surprising given that the change in aspartate content at this position upon genetic conversion would be marginal; N263 is 99% deamidated in WT AAV8. One explanation for this discrepancy is that genetically encoded aspartate and the product of asparagine deamidation are molecularly distinct (L-aspartate versus a presumed 3:1 mixture of L/D-isoaspartate: L/D-aspartate). Thus, the genetic approximation may be insufficient at some positions. Another residue, the highly conserved N57, was also not tolerant of substitution to aspartate, although it was on average 80% and 97% deamidated in AAV8 and AAV9, respectively (Figures 4B and S6). Here, the residual intact amides may buffer the activity of the WT preparations through mosaic effects. However, we also detected the potential for cross-talk with other asparagines, confounding analysis of N57; the neighboring N66 became significantly deamidated when the position 57 amide was preserved mutagenically (N57Q, G58A, and G58S for AAV8; N57Q and G58A for AAV9; data not shown). Although this was the only case of cross-talk that we detected from mass spectrometry analysis of our mutants, it highlights another complication of interpreting our loss-of-function mutagenic data.
Given these caveats, we evaluated the impact of deamidation through time-course and gain-of-function mutagenesis experiments. Our data are consistent with a role for a subset of NG sites in the functional loss associated with very early time point deamidation. To our knowledge, this phenomenon has not been previously reported. Indeed, the experimental procedure we used to identify this decay was informed by the novel observation of very short half-life vector NG deamidation; storing early samples for even a single day in the refrigerator would likely diminish their distinction from late time point samples given the speed of spontaneous deamidation that we observed. Storage stability experiments comparing the activity of vector preparations over days or weeks after processing are routine in our lab and other manufacturing groups, but these comparisons are almost always made with vector material that is at least 7 days old, when most or all of the activity decay (and NG site deamidation) is complete. The data highlight an opportunity for process interventions or N-stabilizing mutagenic approaches to yield improved capsids. From a broader perspective, it is of interest to consider the role of a “deamidation clock” in the natural ecology of AAVs, where this phenomenon would presumably provide an advantage to the most recently translated virus particles from an infected cell for the next round of infection.
We did not explore the mechanistic underpinnings of NG deamidation-induced functional loss, although some prominent possibilities exist. All of the NG motifs in AAV8 and AAV9 VP3 are located in surface HVR loops. In AAV8, NG 514 and 540 are located near the 3-fold axis in an area known to play a significant role in transduction due to interactions with cellular receptors. Although no AAV8 receptor binding site has been fully interrogated, the LamR receptor has been implicated in AAV8 transduction.38 These studies identify amino acids (aa) 491–557 as important for these interactions. The functional implications of deamidation at sites in this region should be further evaluated.
In summary, we showed that AAV vector deamidation can impact transduction efficiency, and demonstrated strategies to stabilize amides and improve vector performance. A key future goal will be to extend these findings to appropriate animal model systems and consider the impact of deamidation and the performance of our stabilized variants in more complex functional contexts. Deamidation could influence tissue tropism and interactions of the capsid with the immune system, which should be evaluated carefully. Because these complex effects will likely be very difficult to determine conclusively for every deamidated residue in the capsid, it may be prudent to target the limited number of residues with high lot-to-lot variability in deamidation for stabilization through mutagenesis, as we have demonstrated successfully for the variable AAV8 asparagines 459 and 499. Additionally, deamidation analysis of vector preparations using our mass spectrometry workflow may prove beneficial in achieving functional consistency in manufactured lots of AAV gene therapy pharmaceuticals.
Materials and Methods
1D and 2D Gel Electrophoresis
For 1D SDS-PAGE analysis, we first denatured AAV vectors at 80°C for 20 min in the presence of lithium dodecyl sulfate and reducing agent. Then we ran them on a 4%–12% Bis-Tris gel for 90 min at 200 V and stained with Coomassie blue. For the data in Figure 1, Kendrick Laboratories (Madison, WI, USA) performed the 2D gel electrophoresis. For subsequent experiments, we performed 2D SDS-PAGE in-house. For this, we combined 3e11 GCs of AAV vector with 500 U turbonuclease marker (Accelagen, San Diego, CA, USA) in 150 μL of PBS with 35 mM NaCl and 1 mM MgCl2, and incubated at 37°C for 10 min. We next added 9 vol of absolute ethanol, vortexed the samples, and incubated them at −80°C for at least 2 hr followed by incubation on ice for 5 min and centrifugation at maximum speed for 30 min at 15°C. We decanted the supernatant and air-dried the pellet, which we then resuspended in resuspension buffer 1 (0.15% SDS, 50 mM DTT, 10 mM Tris [pH 7.5], and 1 μL [pH 6–9] ampholytes [Thermo Fisher] ZM0023, added day-of, in double distilled water [ddH2O]) and incubated undisturbed at room temperature. After 30 min, we flicked the sample tubes to mix them, added 1 μg of chicken conalbumin marker (Sigma-Aldrich, St. Louis, MO, USA), and incubated the samples at 37°C for 30 min, flicking to mix at 15 min. Samples were then transferred to 50°C for 15–20 min, vortexed, incubated at 95°C for 2.5 min, and allowed to cool before being centrifuged at maximum speed for 1 min and briefly vortexed. We then mixed 10 μL of each sample with 140 μL of resuspension buffer 2 (9.7 M urea, 2% (3-((3-cholamidopropyl) dimethylammonio)-1-propanesulfonate) [CHAPS], 0.002% bromophenol blue, and 0.05% ampholytes, described above, added day-of, in ddH2O) and incubated at room temperature for 10 min. We then applied the mixtures to pH 6–10 immobilized pH gradient (IPG) strips (Thermo Fisher, Waltham, MA, USA) and ran them on the ZOOM IPGRunner system according to the manufacturer’s instructions. We used the following isoelectric focusing parameters: 100–1,000 V for 120 min, 1,000–2,000 V for 120 min, 2,000 V for 120 min, limits of 0.1 W and 0.05 mA per strip run. IPG strips were then reduced and loaded in a single-well 4%–12% Bis-Tris gel and run in 1D as described above. We determined the relative migration of AAV VPs by comparison with internal control proteins turbonuclease (27 kDa; Accelagen) and chicken egg white conalbumin (76 kDa, pI 6.0–6.6; Sigma-Aldrich).
Vector Production
The University of Pennsylvania Vector Core produced recombinant AAV vectors for 1D and 2D gel electrophoresis and mass spectrometry experiments, and purified them by cesium chloride or iodixanol gradients as previously described.39, 40 We produced the affinity-purified vectors as follows: we grew HEK293 cells in ten 36-layer hyperstack vessels (Corning), co-transfected them with a mixture of vector genome plasmid (pAAV-LSP-IVS2.hFIXco-WPRE-bGH), trans plasmid containing AAV2 rep and AAV8 cap genes, and adenovirus helper plasmid. We used PEIpro (PolyPlus) as the transfection reagent. Five days post-transfection, the supernatant was harvested, clarified through Sartoguard PES Midicap filters (Sartorious Stedim), and treated with benzonase (Millipore), after which we added salt to bring it to 0.6 M. The clarified bulk harvest material was concentrated 10-fold by tangential flow filtration (TFF) and then diafiltered against 4 vol of affinity column loading buffer. We captured vectors on a POROS CaptureSelect (Thermo Fisher) affinity column and eluted the vector peak at low pH directly into neutralization buffer. We diluted the neutralized eluate into a high-pH binding buffer and loaded it onto an anion exchange polishing column (Cimultus QA-8; Bia Separations), where the preparation was enriched for genome-containing (full) particles. The full vector particles were eluted with a shallow salt elution gradient and neutralized immediately. Finally, we subjected the vector to a second round of TFF for final concentration and buffer exchange into formulation buffer (PBS + 0.001% Pluronic F-68).
We produced mutant vectors for in vitro assays by small-scale triple transfection of HEK293 cells in six-well plates. We mixed 5.6 μL of a 1 mg/mL polyethylenimine solution in 90 μL of serum-free media with plasmid DNA (0.091 μg of cis plasmid, 0.91 μg of trans plasmid, 1.82 μg of deltaF6 Ad-helper plasmid, in 90 μL of serum-free media), incubated it at room temperature for 15 min, and added it to cells in an additional 0.8 mL of fresh serum-free media. The next day, we replaced 0.5 mL of the top media with full serum media. We harvested vector 3 days post-transfection by freezing and thawing three times followed by centrifugation to remove cell debris and supernatant harvest. Cis plasmid contained a transgene cassette encoding the firefly luciferase transgene under the control of the chicken-beta actin (CB7) promoter with the Promega chimeric intron and rabbit beta-globin (RBG) polyadenylation signal. Trans plasmid encoded the WT AAV8 cap gene; to generate mutant AAV8 cap variants, we used the QuikChange Lightning Mutagenesis kit (Agilent Technologies, Wilmington, DE, USA). Vector was titered as previously described.
For time-course vector production experiments, we generated vector by medium-scale triple transfection of HEK293 cells in 15-cm tissue culture dishes. Per plate, we mixed 36 μL of a 1 mg/mL polyethylenimine solution in 2 mL of serum-free media with plasmid DNA (0.6 μg of cis plasmid, 5.8 μg of trans plasmid, 11.6 μg of deltaF6 Ad-helper plasmid), incubated it at room temperature for 15 min, and added it to cells at approximately 60% confluency on plates refreshed with 14 mL of serum-free media. The following day, we replaced 8 mL of the top media with fresh, full serum media. We harvested vector by collecting all top media, scraping cells from the dish, and freezing it at −80°C. We recovered crude vector from the mixture of supernatant and cells by applying three freeze/thaw cycles and clarifying the lysate by centrifugation. We purified and concentrated the vector for mass spectrometry analysis by adding benzonase, 1 M Tris (pH 7.5), and 5 M NaCl to the clarified lysate to final concentrations of 20 mM Tris and 360 mM NaCl. We captured vectors on a 1 mL POROS CaptureSelect affinity column and eluted the vector peak at low pH directly into neutralization buffer. Fractions were analyzed by absorbance at 280 nm, and the most concentrated fraction was subjected to mass spectrometry analysis.
For in vivo experiments, we produced vectors as previously described with a WT AAV8 capsid or with one of the deamidation mutants; the transgene cassette included a CB7 promoter, promega intron, firefly luciferase transgene, and RBG polyadenylation signal.
Mass Spectrometry Running, Digestion, and Analysis
Materials
We purchased ammonium bicarbonate, DTT, iodoacetamide (IAM), and 18O-enriched water (97.1% purity) from Sigma (St. Louis, MO, USA), and acetonitrile, formic acid, trifluoroacetic acid (TFA), 8 M guanidine hydrochloride (GndHCl), and trypsin from Thermo Fischer Scientific (Rockford, IL, USA).
Trypsin Digestion
We prepared stock solutions of 1 M DTT and 1.0 M IAM. Capsid proteins were denatured and reduced at 90°C for 10 min in the presence of 10 mM DTT and 2 M GndHCl. We allowed the samples to cool to room temperature and then alkylated them with 30 mM IAM at room temperature for 30 min in the dark. We quenched the alkylation reaction with the addition of 1 mL of DTT. We added 20 mM ammonium bicarbonate (pH 7.5–8) to the denatured protein solution at a volume that diluted the final GndHCl concentration to 200 mM. We added trypsin solution for a 1:20 trypsin-to-protein ratio and incubated at 37°C overnight. After digestion, we added TFA to a final concentration of 0.5% to quench the digestion reaction.
Liquid Chromatography Tandem-Mass Spectrometry
We performed online chromatography with an Acclaim PepMap column (15 cm long, 300 μm inner diameter) and a Thermo UltiMate 3000 RSLC system (Thermo Fisher Scientific) coupled to a Q Exactive HF with a NanoFlex source (Thermo Fisher Scientific). During online analysis, the column temperature was maintained at a temperature of 35°C. We separated peptides with a gradient of mobile phase A (MilliQ water with 0.1% formic acid) and mobile phase B (acetonitrile with 0.1% formic acid). We ran the gradient from 4% B to 6% B over 15 min, to 10% B for 25 min (40 min total), and then to 30% B for 46 min (86 min total). We loaded the samples directly to the column. The column size was 75 cm × 15 μm identifier (I.D.) and was packed with 2 μm of C18 media (Acclaim PepMap). Due to the loading, lead-in, and washing steps, the total time for each liquid chromatography tandem-mass spectrometry run was about 2 hr.
We acquired mass spectrometry data using a data-dependent top-20 method on a Q Exactive HF mass spectrometer, dynamically choosing the most abundant not-yet-sequenced precursor ions from the survey scans (200–2,000 m/z). We performed sequencing via higher-energy collisional dissociation fragmentation with a target value of 1e5 ions determined with predictive automatic gain control; we performed isolation of precursors with a window of 4 m/z. We acquired survey scans at a resolution of 120,000 at 200 m/z. We set the resolution for higher-energy collisional dissociation (HCD) spectra to 30,000 at 200 m/z with a maximum ion injection time of 50 ms and a normalized collision energy of 30. We set the S-lens radio frequency (RF) level to 50, which gave optimal transmission of the m/z region occupied by the peptides from our digest. We excluded precursor ions with single, unassigned, or six and higher charge states from fragmentation selection.
Data Processing
We used BioPharma Finder 1.0 software (Thermo Fischer Scientific) to analyze all data acquired. For peptide mapping, we performed searches using a single-entry protein Federal Acquisition Streamlining Act (FASTA) database with carbamidomethylation set as a fixed modification, and oxidation, deamidation, and phosphorylation set as variable modifications. We used a 10 ppm mass accuracy, a high protease specificity, and a confidence level of 0.8 for tandem-mass spectrometry spectra. Mass spectrometric identification of deamidated peptides is relatively straightforward; deamidation adds to the mass of intact molecule +0.984 Da (the mass difference between -OH and -NH2 groups). We determined the percent deamidation of a particular peptide by dividing the mass area of the deamidated peptide by the sum of the area of the deamidated and native peptides. Considering the number of possible deamidation sites, isobaric species that are deamidated at different sites may co-migrate at a single peak. Consequently, fragment ions originating from peptides with multiple potential deamidation sites can be used to locate or differentiate multiple sites of deamidation. In these cases, the relative intensities within the observed isotope patterns can be used to specifically determine the relative abundance of the different deamidated peptide isomers. This method assumes that the fragmentation efficiency for all isomeric species is the same and independent of the site of deamidation. This approach allows us to define the specific sites involved in deamidation and the potential combinations involved in deamidation.
Structural Analysis of the AAV Capsid
We obtained the AAV8 atomic coordinates, structural factors, and associated capsid model from the RCSB PDB (PDB: 3RA8). We performed structure refinement and generated an electron density independent of the primary aa sequence of AAV8 VP3 for use in 3D structural analysis of the capsid. We performed this analysis in order to observe the isoaspartic acid electron density in the AAV8 capsid that was not biased by the expected primary sequence of AAV8 VP3. Using the resulting structure, we modeled the four asparagines in the AAV8 VP3 primary sequence with N+1 glycines as isoaspartic acids and then refined the AAV8 capsid structure using Crystallography and NMR System software by strictly imposing the icosahedral non-crystallographic matrices using the standard refinement protocol.41 We obtained a structural model of isoaspartic acid from the HIC-Up database, followed by generation of a molecular dictionary in PRODRG for structure refinement.42 We then calculated the average electron density map of the AAV8 capsid (also in Crystallography and NMR System) and visualized it using Coot software, followed by minor adjustments of the resulting model to fit the modeled isoaspartic acid residues into the electron density map. We repeated this protocol to additionally model N512 in the AAV9 VP3 primary sequence with N+1 glycines (PDB: 3UX1). We generated all figures using Coot, PyMol, and UCSF Chimera.43, 44, 45 We obtained a number of structures of previously identified deamidated proteins (PDB: 1DY5, 4E7G, 1RTU, 1W9V, 4E7D, and 1C9D) for comparison of their electron density map for deamidated isoaspartic acid residues with our modeled isoaspartic acid residues from AAV8 and AAV9.33, 34, 35
We determined temperature factors for deamidated residues by averaging the temperature factors for each atom of each asparagine residue reported in the AAV8 or AAV9 crystal structure atomic coordinates (PDB: 3RA8, 3UX1).
Animal Studies
The Institutional Animal Care and Use Committee of the University of Pennsylvania approved all animal procedures. To evaluate vector performance, we injected 8-week-old male C57BL/6 mice intravenously via tail vein injection with 3e10 GCs of WT AAV8 or capsid mutant vector in a volume of 100 μL. All mice were sacrificed at day 14. For in vivo evaluation of luciferase expression, mice (∼20 g) were anesthetized and injected intraperitoneally with 200 μL of 15 mg/mL luciferin substrate (Perkin Elmer, Waltham, MA, USA). Mice were imaged 5 min after luciferin administration and imaged via an IVIS Xenogen In Vivo Imaging System. We used Living Image 3.0 software to quantify signal in the described regions of interest. We took measurements at days 7 and 14.
Evaluation of Mutant Vector Titer and Transduction Efficiency In Vitro
We determined vector titers by qPCR of the DNase I-resistant genomes. The qPCR primers anneal to the polyadenylation sequence of the packaged transgene. For in vitro evaluation of vector transduction efficiency by luciferase expression, we seeded 0.9e5 Huh7 cells/well in a black-walled 96-well plate in complete DMEM (10% fetal bovine serum, 1% penicillin and streptomycin). The next day, we removed the media and replaced it with 50 μL of crude or purified vector diluted in complete media. We tested four dilutions in a 3-fold dilution series for each vector sample. After 48 hr, we prepared luciferin (Promega, Madison, WI, USA) in complete media at 0.3 μg/μL and added it to transduced cells in a volume of 50 μL. Results were read on a Biotek Clarity luminometer. We found that luciferase activity/GC added to target cells was constant over a wide range of GCs, but could become saturated at high MOIs. Thus, we inspected the dilution series data (luminescent units versus GCs) for linearity, excluded the highest point if saturation was evident, and calculated an average luciferase/GC for values in the linear range of each assay for each variant. This yielded a transduction efficiency value. The data were normalized to simplify comparison by setting the WT control to a value of 1.
Author Contributions
Conceptualization, A.R.G., J.J.S., and J.M.W.; Methodology, A.R.G., J.J.S., M.R.A., M.L., and J.M.W.; Resources, M.R.A. and M.L.; Investigation, A.R.G., J.J.S., K.B.T., and L.G.; Formal Analysis, K.B.T. and L.G.; Writing – Original Draft, A.R.G. and J.J.S.; Writing – Review and Editing, A.R.G., J.J.S., K.B.T., L.G., M.L., and J.M.W.
Conflicts of Interest
J.M.W. is an advisor to, holds equity in, and has a sponsored research agreement with REGENXBIO and Scout Bio; he also has a sponsored research agreement with Ultragenyx, Biogen, and Janssen, which are licensees of Penn technology. In addition, he has sponsored research agreements with Precision Biosciences and Moderna Therapeutics. J.M.W. is an inventor on patents that have been licensed to various biopharmaceutical companies.
Acknowledgments
We would like to acknowledge Jenny Greig, Melanie Smith, Joanna Chorazeczewski, Deirdre McMenamin, Christine Draper, and the Gene Therapy Program in Comparative Medicine at the University of Pennsylvania for procedural assistance; Mingyao Li and Yan Che for assistance with statistical analyses; and Joshua Wilhide at the University of Maryland Baltimore County for secondary analysis of AAV8 mass spectrometry data. Funding for this project was provided by a National Heart, Lung, and Blood Institute grant (P01HL059407) as well as the University of Pennsylvania Perelman School of Medicine.
Footnotes
Supplemental Information includes seven figures, two tables, and Supplemental Materials and Methods and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.09.013.
Supplemental Information
References
- 1.Xie Q., Bu W., Bhatia S., Hare J., Somasundaram T., Azzi A., Chapman M.S. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. USA. 2002;99:10405–10410. doi: 10.1073/pnas.162250899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nam H.J., Lane M.D., Padron E., Gurda B., McKenna R., Kohlbrenner E., Aslanidi G., Byrne B., Muzyczka N., Zolotukhin S., Agbandje-McKenna M. Structure of adeno-associated virus serotype 8, a gene therapy vector. J. Virol. 2007;81:12260–12271. doi: 10.1128/JVI.01304-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Govindasamy L., DiMattia M.A., Gurda B.L., Halder S., McKenna R., Chiorini J.A., Muzyczka N., Zolotukhin S., Agbandje-McKenna M. Structural insights into adeno-associated virus serotype 5. J. Virol. 2013;87:11187–11199. doi: 10.1128/JVI.00867-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang L.Y., Patel A., Ng R., Miller E.B., Halder S., McKenna R., Asokan A., Agbandje-McKenna M. Characterization of the adeno-associated virus 1 and 6 sialic acid binding site. J. Virol. 2016;90:5219–5230. doi: 10.1128/JVI.00161-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raupp C., Naumer M., Müller O.J., Gurda B.L., Agbandje-McKenna M., Kleinschmidt J.A. The threefold protrusions of adeno-associated virus type 8 are involved in cell surface targeting as well as postattachment processing. J. Virol. 2012;86:9396–9408. doi: 10.1128/JVI.00209-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halder S., Van Vliet K., Smith J.K., Duong T.T., McKenna R., Wilson J.M., Agbandje-McKenna M. Structure of neurotropic adeno-associated virus AAVrh.8. J. Struct. Biol. 2015;192:21–36. doi: 10.1016/j.jsb.2015.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie Q., Lerch T.F., Meyer N.L., Chapman M.S. Structure-function analysis of receptor-binding in adeno-associated virus serotype 6 (AAV-6) Virology. 2011;420:10–19. doi: 10.1016/j.virol.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DiMattia M.A., Nam H.J., Van Vliet K., Mitchell M., Bennett A., Gurda B.L., McKenna R., Olson N.H., Sinkovits R.S., Potter M. Structural insight into the unique properties of adeno-associated virus serotype 9. J. Virol. 2012;86:6947–6958. doi: 10.1128/JVI.07232-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lerch T.F., Xie Q., Chapman M.S. The structure of adeno-associated virus serotype 3B (AAV-3B): insights into receptor binding and immune evasion. Virology. 2010;403:26–36. doi: 10.1016/j.virol.2010.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Govindasamy L., Padron E., McKenna R., Muzyczka N., Kaludov N., Chiorini J.A., Agbandje-McKenna M. Structurally mapping the diverse phenotype of adeno-associated virus serotype 4. J. Virol. 2006;80:11556–11570. doi: 10.1128/JVI.01536-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhong L., Li B., Jayandharan G., Mah C.S., Govindasamy L., Agbandje-McKenna M., Herzog R.W., Weigel-Van Aken K.A., Hobbs J.A., Zolotukhin S. Tyrosine-phosphorylation of AAV2 vectors and its consequences on viral intracellular trafficking and transgene expression. Virology. 2008;381:194–202. doi: 10.1016/j.virol.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murray S., Nilsson C.L., Hare J.T., Emmett M.R., Korostelev A., Ongley H., Marshall A.G., Chapman M.S. Characterization of the capsid protein glycosylation of adeno-associated virus type 2 by high-resolution mass spectrometry. J. Virol. 2006;80:6171–6176. doi: 10.1128/JVI.02417-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin X., Liu L., Nass S., O’Riordan C., Pastor E., Zhang X.K. Direct liquid chromatography/mass spectrometry analysis for complete characterization of recombinant adeno-associated virus capsid proteins. Hum. Gene Ther. Methods. 2017;28:255–267. doi: 10.1089/hgtb.2016.178. [DOI] [PubMed] [Google Scholar]
- 14.Zhao J., Li J., Xu S., Feng P. Emerging roles of protein deamidation in innate immune signaling. J. Virol. 2016;90:4262–4268. doi: 10.1128/JVI.01980-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao J., Zeng Y., Xu S., Chen J., Shen G., Yu C., Knipe D., Yuan W., Peng J., Xu W. A viral deamidase targets the helicase domain of RIG-I to block RNA-induced activation. Cell Host Microbe. 2016;20:770–784. doi: 10.1016/j.chom.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson N.E., Robinson A.B. Molecular clocks. Proc. Natl. Acad. Sci. USA. 2001;98:944–949. doi: 10.1073/pnas.98.3.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jenkins N., Murphy L., Tyther R. Post-translational modifications of recombinant proteins: significance for biopharmaceuticals. Mol. Biotechnol. 2008;39:113–118. doi: 10.1007/s12033-008-9049-4. [DOI] [PubMed] [Google Scholar]
- 18.Houde D., Peng Y., Berkowitz S.A., Engen J.R. Post-translational modifications differentially affect IgG1 conformation and receptor binding. Mol. Cell. Proteomics. 2010;9:1716–1728. doi: 10.1074/mcp.M900540-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Powell B.S., Enama J.T., Ribot W.J., Webster W., Little S., Hoover T., Adamovicz J.J., Andrews G.P. Multiple asparagine deamidation of Bacillus anthracis protective antigen causes charge isoforms whose complexity correlates with reduced biological activity. Proteins. 2007;68:458–479. doi: 10.1002/prot.21432. [DOI] [PubMed] [Google Scholar]
- 20.Verma A., Ngundi M.M., Burns D.L. Mechanistic analysis of the effect of deamidation on the immunogenicity of anthrax protective antigen. Clin. Vaccine Immunol. 2016;23:396–402. doi: 10.1128/CVI.00701-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang H., Zubarev R.A. Mass spectrometric analysis of asparagine deamidation and aspartate isomerization in polypeptides. Electrophoresis. 2010;31:1764–1772. doi: 10.1002/elps.201000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Catak S., Monard G., Aviyente V., Ruiz-López M.F. Deamidation of asparagine residues: direct hydrolysis versus succinimide-mediated deamidation mechanisms. J. Phys. Chem. A. 2009;113:1111–1120. doi: 10.1021/jp808597v. [DOI] [PubMed] [Google Scholar]
- 23.Geiger T., Clarke S. Deamidation, isomerization, and racemization at asparaginyl and aspartyl residues in peptides. Succinimide-linked reactions that contribute to protein degradation. J. Biol. Chem. 1987;262:785–794. [PubMed] [Google Scholar]
- 24.Robinson N.E., Robinson A.B. Deamidation of human proteins. Proc. Natl. Acad. Sci. USA. 2001;98:12409–12413. doi: 10.1073/pnas.221463198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson N.E., Robinson A.B. Prediction of protein deamidation rates from primary and three-dimensional structure. Proc. Natl. Acad. Sci. USA. 2001;98:4367–4372. doi: 10.1073/pnas.071066498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson A.B., Rudd C.J. Deamidation of glutaminyl and asparaginyl residues in peptides and proteins. Curr. Top. Cell. Regul. 1974;8:247–295. doi: 10.1016/b978-0-12-152808-9.50013-4. [DOI] [PubMed] [Google Scholar]
- 27.Rose J.A., Maizel J.V., Jr., Inman J.K., Shatkin A.J. Structural proteins of adenovirus-associated viruses. J. Virol. 1971;8:766–770. doi: 10.1128/jvi.8.5.766-770.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du Y., Wang F., May K., Xu W., Liu H. Determination of deamidation artifacts introduced by sample preparation using 18O-labeling and tandem mass spectrometry analysis. Anal. Chem. 2012;84:6355–6360. doi: 10.1021/ac3013362. [DOI] [PubMed] [Google Scholar]
- 29.Henderson L.E., Henriksson D., Nyman P.O. Primary structure of human carbonic anhydrase C. J. Biol. Chem. 1976;251:5457–5463. [PubMed] [Google Scholar]
- 30.Carvalho R.N., Solstad T., Bjørgo E., Barroso J.F., Flatmark T. Deamidations in recombinant human phenylalanine hydroxylase. Identification of labile asparagine residues and functional characterization of Asn → Asp mutant forms. J. Biol. Chem. 2003;278:15142–15152. doi: 10.1074/jbc.M212180200. [DOI] [PubMed] [Google Scholar]
- 31.Nam H.-J., Gurda B.L., McKenna R., Potter M., Byrne B., Salganik M., Muzyczka N., Agbandje-McKenna M. Structural studies of adeno-associated virus serotype 8 capsid transitions associated with endosomal trafficking. J. Virol. 2011;85:11791–11799. doi: 10.1128/JVI.05305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parthasarathy S., Murthy M.R. Analysis of temperature factor distribution in high-resolution protein structures. Protein Sci. 1997;6:2561–2567. doi: 10.1002/pro.5560061208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Esposito L., Vitagliano L., Sica F., Sorrentino G., Zagari A., Mazzarella L. The ultrahigh resolution crystal structure of ribonuclease A containing an isoaspartyl residue: hydration and sterochemical analysis. J. Mol. Biol. 2000;297:713–732. doi: 10.1006/jmbi.2000.3597. [DOI] [PubMed] [Google Scholar]
- 34.Noguchi S., Satow Y., Uchida T., Sasaki C., Matsuzaki T. Crystal structure of Ustilago sphaerogena ribonuclease U2 at 1.8 A resolution. Biochemistry. 1995;34:15583–15591. doi: 10.1021/bi00047a025. [DOI] [PubMed] [Google Scholar]
- 35.Rao F.V., Houston D.R., Boot R.G., Aerts J.M., Hodkinson M., Adams D.J., Shiomi K., Omura S., van Aalten D.M. Specificity and affinity of natural product cyclopentapeptide inhibitors against A. fumigatus, human, and bacterial chitinases. Chem. Biol. 2005;12:65–76. doi: 10.1016/j.chembiol.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 36.Aydemir F., Salganik M., Resztak J., Singh J., Bennett A., Agbandje-McKenna M., Muzyczka N. Mutants at the 2-fold interface of adeno-associated virus type 2 (AAV2) structural proteins suggest a role in viral transcription for AAV capsids. J. Virol. 2016;16:7196–7204. doi: 10.1128/JVI.00493-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nebija D., Noe C.R., Urban E., Lachmann B. Quality control and stability studies with the monoclonal antibody, trastuzumab: application of 1D- vs. 2D-gel electrophoresis. Int. J. Mol. Sci. 2014;15:6399–6411. doi: 10.3390/ijms15046399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akache B., Grimm D., Pandey K., Yant S.R., Xu H., Kay M.A. The 37/67-kilodalton laminin receptor is a receptor for adeno-associated virus serotypes 8, 2, 3, and 9. J. Virol. 2006;80:9831–9836. doi: 10.1128/JVI.00878-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lock M., Alvira M., Vandenberghe L.H., Samanta A., Toelen J., Debyser Z., Wilson J.M. Rapid, simple, and versatile manufacturing of recombinant adeno-associated viral vectors at scale. Hum. Gene Ther. 2010;21:1259–1271. doi: 10.1089/hum.2010.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao G.P., Alvira M.R., Wang L., Calcedo R., Johnston J., Wilson J.M. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl. Acad. Sci. USA. 2002;99:11854–11859. doi: 10.1073/pnas.182412299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brünger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 42.Kleywegt G.J. Crystallographic refinement of ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 2007;63:94–100. doi: 10.1107/S0907444906022657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 44.DeLano W.L. PyMOL: an open-source molecular graphics tool. CCP4 Newsl Protein Crystallogr. 2002;40:82–92. [Google Scholar]
- 45.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.