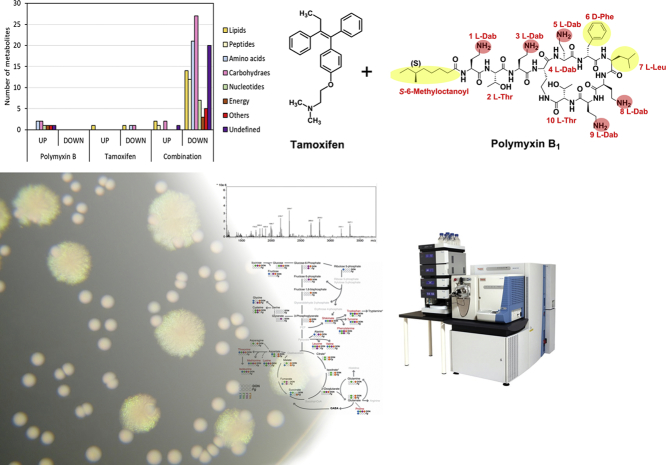
Abstract
Polymyxins are amongst the most important antibiotics in modern medicine, in recent times their clinical utility has been overshadowed by nosocomial outbreaks of polymyxin resistant MDR Gram-negative ‘superbugs’. An effective strategy to surmount polymyxin resistance is combination therapy with FDA-approved non-antibiotic drugs. Herein we used untargeted metabolomics to investigate the mechanism(s) of synergy between polymyxin B and the selective estrogen receptor modulator (SERM) tamoxifen against a polymyxin-resistant MDR cystic fibrosis (CF) Pseudomonas aeruginosa FADDI-PA006 isolate (polymyxin B MIC=8 mg/L , it is an MDR polymyxin resistant P. aeruginosa isolated from the lungs of a CF patient). The metabolome of FADDI-PA006 was profiled at 15 min, 1 and 4 h following treatment with polymyxin B (2 mg/L), tamoxifen (8 mg/L) either as monotherapy or in combination. At 15 min, the combination treatment induced a marked decrease in lipids, primarily fatty acid and glycerophospholipid metabolites that are involved in the biosynthesis of bacterial membranes. In line with the polymyxin-resistant status of this strain, at 1 h, both polymyxin B and tamoxifen monotherapies produced little effect on bacterial metabolism. In contrast to the combination which induced extensive reduction (≥ 1.0-log2-fold, p ≤ 0.05; FDR ≤ 0.05) in the levels of essential intermediates involved in cell envelope biosynthesis. Overall, these novel findings demonstrate that the primary mechanisms underlying the synergistic bactericidal effect of the combination against the polymyxin-resistant P. aeruginosa CF isolate FADDI-PA006 involves a disruption of the cell envelope biogenesis and an inhibition of aminoarabinose LPS modifications that confer polymyxin resistance.
Graphical Abstract
1. Introduction
Multi-drug resistant (MDR) P. aeruginosa is associated with life-threatening infections, commonly observed in cystic fibrosis (CF) lung, pneumonia, and surgical-site infections [1,2]. P. aeruginosa infections are notorious for developing high-level resistance towards a wide array of antibiotics, often leaving clinicians with few treatment options [[3], [4], [5]]. In 2015, the European Antimicrobial Resistance Surveillance Network (EARS-Net) reported that >13.7% of the P. aeruginosa isolates were MDR, showing resistance to most of the major antibiotic classes. The World Health Organization (WHO) has recently classified MDR P. aeruginosa as a top-priority critical pathogen, urgently requiring the development of new treatment options [6]. In patients suffering from CF, colonization of lungs with P. aeruginosa is associated with poorer quality of life and higher mortality rates [7,8].
Polymyxins (colistin and polymyxin B) are currently used as last-line treatment against MDR P. aeruginosa infections [9,10]. Although polymyxins exert their antibacterial activity by disorganizing the bacterial outer membrane (OM) via electrostatic interactions with the lipid A component of lipopolysaccharide (LPS), the exact antibacterial killing mechanism is still enigmatic [11]. The most common mechanism of polymyxin resistance seen in P. aeruginosa involves LPS modifications, including the addition of phosphoethanolamine (PEtN) and 4-amino-4-deoxy-L-arabinose (L-Ara4N) to its lipid A structure, or by deacylation, hydroxylation and palmitoylation of the lipid A fatty acyls [[12], [13], [14]]. These structural modifications reduce the overall net negative charge and hydrophobicity of the OM, thereby ablating the binding of the amphipathic polymyxin [2]. Treatment failure due to sub-optimal plasma drug concentrations or resistance to polymyxins in MDR P. aeruginosa can emerge after polymyxin monotherapy, which highlights the need for novel combination therapies e.g. with FDA-approved non-antibiotics [[15], [16], [17], [18]]. The use of synergistic combinations of antibiotics with FDA approved non-antibiotic drugs is a highly valuable and cost-effective strategy to enhance the clinical efficacy of currently used antibiotics against MDR bacterial pathogens [[19], [20], [21], [22]]. It has been proposed that polymyxins can increase the OM permeability and enable the non-antibiotic compounds to access their intracellular target(s) [19,21]. We recently showed that combining polymyxin B with selective estrogen receptor modulators (SERMs), namely tamoxifen, raloxifene and toremifene, resulted in synergistic bactericidal activity against a wide range MDR Gram-negative pathogens, particularly MDR P. aeruginosa CF isolates [21]. Polymyxin B in combination with toremifene synergistically caused a significant release in cytosolic green fluorescence protein, depolarizing the cytoplasmic membrane and permeabilizing activity in a nitrocefin assay. The combination also induced an increase of cellular reactive oxygen species and a distinctive damage to the outer membrane of P. aeruginosa cells. Given the importance of repurposing “off-the-shelf” non-antibiotics drugs to treat MDR P. aeruginosa, it is crucial to elucidate the mechanism(s) by which the polymyxin B-SERMs combinations achieve this powerful bacterial killing activity as well as their ability to suppress the emergence of polymyxin resistance.
Antibiotic combination therapies focus on phenotypic killing without systematic or mechanistic investigations [23]. Metabolomics represent a cutting-edge platform that is suitable for investigating the mode of action of and bacterial resistance to antibiotics through analysis of the metabolic response to drug treatment [24,25]. The profiling of significantly perturbed metabolites and reconstruction of their metabolic pathways provides a rapid and effective means for understanding how bacteria respond to the antimicrobials at the molecular level, the mechanism by which antibiotics kill bacterial cells and ultimately the identification of novel antibiotic targets [[26], [27], [28], [29], [30], [31]].
The present study employs a global metabolomics approach to decipher the complex interplay of multiple cellular pathways in a clinical MDR P. aeruginosa CF isolate in response to polymyxin B combination treatment with the SERM tamoxifen. The novel findings reveal for the first time that the synergistic bactericidal effect and suppression of polymyxin resistance of the combination involves the disruption of pathways that mediate bacterial cell envelope biogenesis and via inhibition of the OM remodeling processes that confer polymyxin resistance, such as modification of the lipid A phosphates with cationic aminoarabinose moieties.
2. Results
2.1. Multivariate and Univariate Analyses of the Metabolites Affected by Polymyxin B and Tamoxifen in P. aeruginosa.
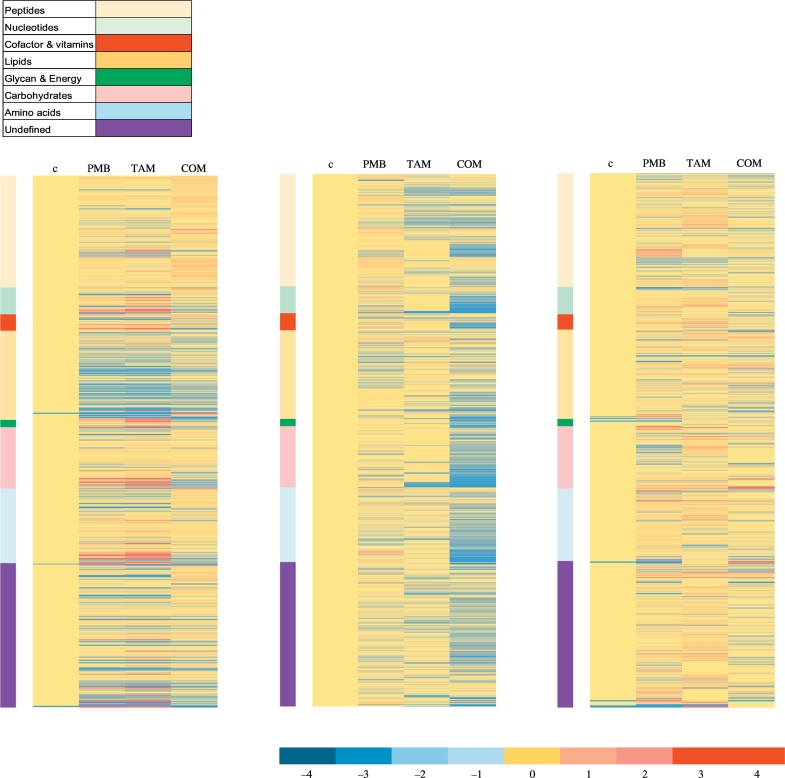
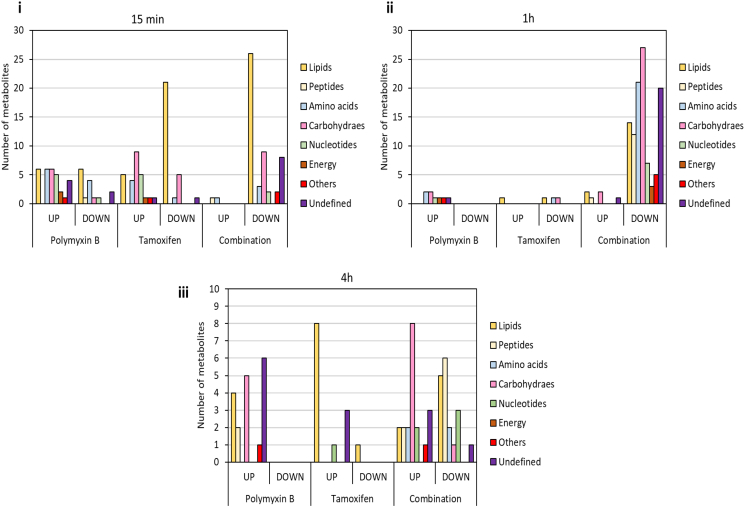
Multivariate data analysis using partial least squares discriminant analysis (PLS-DA), and univariate data analysis using one-way analysis of variance (ANOVA) followed by Fishers least square difference (LDS), were performed to determine the metabolites in P. aeruginosa FADDI-PA006 that were significantly affected by the polymyxin B and tamoxifen treatments (≥ 0.59 -log2-fold; p ≤ .05; FDR ≤ 0.05). The analytical reproducibility of the LC-MS analysis was determined by periodic analysis of a pooled biological quality control (PBQC) sample (Table S2), which demonstrated a median relative standard deviation (RSD) of 13%, which is well below the acceptable limits (< 20%) in standard metabolomics practice [32]. The reproducibility of all sample groups was acceptable across all time points (15 min, 1 and 4 h), where the median RSD across all time points was (20–29%) for untreated (control) groups and (20–28%) for the treated groups; consistent with some baseline variability in the dynamics of ordinary bacterial metabolism commonly seen with or without antibiotic treatments (Table S2) [33,34]. The multivariate analysis with PLS-DA revealed that the polymyxin B and tamoxifen monotherapy treatment groups did slightly overlap with untreated (control) groups at 15 min in the first two components, whereas a greater overall difference was observed between the combination treatment group and the untreated (control) group (Fig. S1A(i)). At 1 h, the PLS-DA scores plots showed a marked separation between the combination treatment group and all other treatment groups (i.e. the polymyxin B and tamoxifen monotherapy, and untreated control) in component 1, indicating that polymyxin-tamoxifen therapy at 1 h was more impactful on the bacterial metabolome (Fig. S1A(ii)). At 4 h, nearly all treatment samples clustered with minimal separation from the untreated group control. The effectiveness of all treatments (polymyxin B, tamoxifen monotherapy and their combination) to induce significant changes in bacterial metabolome was similar at 15 min; however, more bacterial metabolome perturbations were observed after polymyxin B-tamoxifen treatment in the subsequent time points (1 and 4 h), which induced perturbations of 115 and 38 metabolites in total, respectively (Fig. S2). Fig. S1B reveals that although most of the significantly affected metabolites (≥ 0.59 -log2-fold; p ≤ .05; FDR ≤ 0.05) were unique to the polymyxin B-tamoxifen combination treatment particularly at 1 and 4 h; polymyxin B monotherapy had more metabolites in common with the combination than tamoxifen monotherapy. A large proportion of affected metabolites following polymyxin B and tamoxifen monotherapy displayed a significant increase in levels at 15 min, whilst almost all significantly affected metabolites induced by combination therapy exhibited decreased levels (Fig. 1, Fig. 2(i)). Lipids, carbohydrates, and amino acids were the most commonly affected classes. The combination treatment induced more metabolome perturbations compared to polymyxin B and tamoxifen monotherapy at 1 h. Notably, nearly all of the significantly affected metabolites showed a remarkable decline in their levels after combination treatment (Fig. 1, Fig. 2(ii)), and a different distribution of metabolite classes was observed, where carbohydrate intermediates were primarily affected, followed by amino acids, unclassified metabolites and lipid metabolites (Fig. 2(ii)). Only a few metabolites showed were significantly affected following polymyxin B and tamoxifen monotherapy at 1 h (Fig. 2(ii)). At 4 h, different patterns were observed for the combination treatment in which fewer metabolites were impacted and the level ~50% of the metabolites increased dramatically. Similarly to the early time points (15 min and 1 h), most of the significantly affected metabolites following polymyxin B or tamoxifen monotherapy showed an increase (Fig. 2(iii)).
Fig. 1.
Monotherapy and combination of polymyxin B and tamoxifen induce global metabolic changes in P. aeruginosa FADDI-PA006. Heatmap profiles of all identified metabolites clustered by metabolite class after treatment with single and combination (COM) of polymyxin B (PMB) and tamoxifen (TAM) at 15 min, 1 h, and 4 h.
Fig. 2.
Total number of significant metabolite changes classified according to metabolite classes after antibiotic treatment of P. aeruginosa FADDI-PA006 at (i) 15 min, (ii) 1 h, and (iii) 4 h. (≥ 0.59 -log2-fold; p ≤ .05; FDR ≤ 0.05).
2.2. Impact of the Combination on Lipid Metabolism in P. aeruginosa
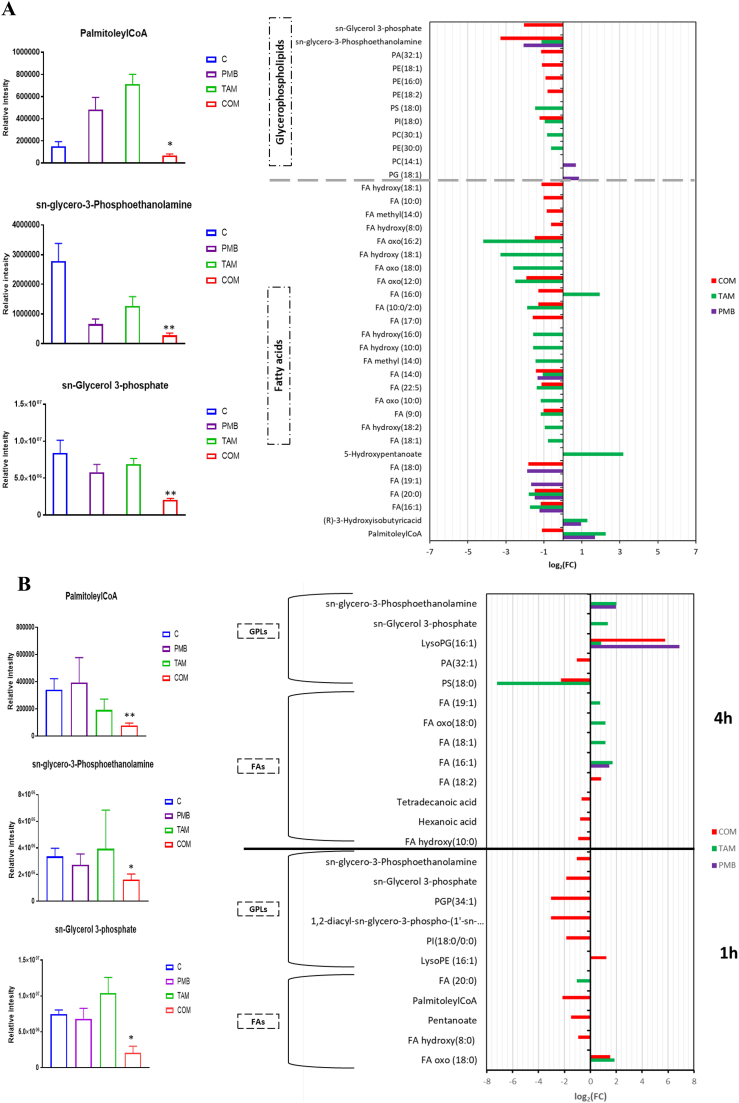
Lipids and lipid intermediates underwent markedly more perturbations at 15 min compared to later time points (1 and 4 h) across all treatment groups; notably, all treatments caused greater perturbations in fatty acids compared to glycerophospholipids (GPLs). Both tamoxifen alone and the combination therapy, caused a similar extent of lipid perturbations: all metabolites, primarily fatty acids and glycerophospholipids, were decreased following the combination therapy. Intriguingly, the combination caused a significant reduction in the levels of essential fatty acids involved in synthesis of outer membrane lipids FA (16:0) (hexadecanoic acid) and FA (14:0) (tetradecanoic acid) (≥ 1.0-log2-fold, p ≤ .05; FDR ≤ 0.05) (Fig. 3A). Furthermore, two fundamental phospholipid metabolites (sn-glycero-3-phosphoethanolamine and sn-glycerol 3-phosphate) were significantly reduced after combination treatment (≥ 2.0-log2-fold, p ≤ .05; FDR ≤ 0.05) (Fig. 3A). Importantly, the combination decreased palmitoleyl-CoA, a key intermediate involved in fatty acid elongation, at 15 min (≥ 1.0-log2-fold, p ≤ .05; FDR ≤ 0.05) (Fig. 3A).
Fig. 3.
Significantly impacted lipids in P. aeruginosa FADDI-PA006 following treatment with polymyxin B (PMB, purple), tamoxifen (TAM, green) and the combination (COM, red) at (A) 15 min, and (B) 1 h and 4 h. Lipid names are putatively assigned based on accurate mass. (** ≥ 2.0-log2-fold, p ≤ .05; FDR ≤ 0.05); (* ≥ 1.0-log2-fold, p ≤ .05; FDR ≤ 0.05).
Tamoxifen monotherapy had a considerable impact on lipid metabolites, in particular fatty acids, at 15 min (Fig. 3A). Although the levels of two important lipid metabolites, namely tetradecanoic acid and sn-glycero-3-phosphoethanolamine decreased after tamoxifen monotherapy (≥ 1.0-log2-fold, p ≤ .05; FDR ≤ 0.05), the levels of key intermediates in fatty acid synthesis, palmitoleyl-CoA and fatty acid (16:0) increased remarkably (≥ 2.0-log2-fold, p ≤ .05; FDR ≤ 0.05) (Fig. 3A). At 15 min, polymyxin B monotherapy only induced significant changes to a smaller number of fatty acids and glycerophospholipids (Fig. 3A).
On the other hand, the influence of all treatments (polymyxin B or tamoxifen monotherapy and their combination) was less pronounced on lipid pathways at the later time points (1 and 4 h) (Figs. 3B). However, combination therapy remained highly effective at reducing the levels of the aforementioned crucial lipid precursors at 1 h, including palmitoleyl-CoA, sn-glycero-3-phosphoethanolamine and sn-glycerol 3-phosphate (≥ 1.0-log2-fold, p ≤ .05; FDR ≤ 0.05) (Fig. 3B).
2.3. Impact of the Combination on Amino Sugar and Nucleotide Sugar Metabolism and Downstream Peptidoglycan, and Lipopolysaccharide Formation and Modification Pathways in P. aeruginosa.
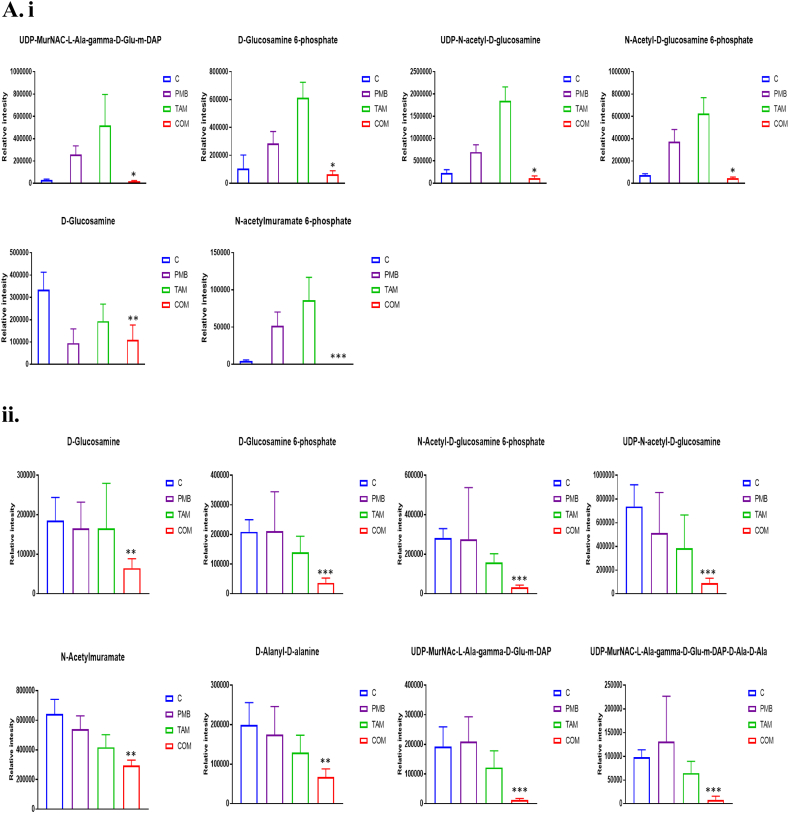
Several metabolites related to amino sugar and nucleotide sugar metabolism were significantly perturbed at 15 min across the treatment groups (Fig. 4A(i)). Tamoxifen monotherapy lead to a substantial increase in the levels of four essential amino- and nucleotide sugar metabolites that are directly related to interrelated peptidoglycan and LPS biosynthesis, namely UDP-N-acetyl-D-glucosamine (UDP-GlcNAc), N-Acetyl-D-glucosamine 6-phosphate, D-Glucosamine 6-phosphate, and N-acetylmuramate 6-phosphate (≥ 2.5-log2-fold, p ≤ .05; FDR ≤ 0.05)(Fig. 4B). Only one intermediate, namely D-Glucosamine, was slightly decreased following tamoxifen monotherapy (> 0.59-log2-fold, p ≤ .05; FDR ≤ 0.05) (Fig. 4A(i)). Likewise, four of these metabolites and one intermediate of peptidoglycan biosynthesis displayed a marked increase in their levels (except for D-Glucosamine) after polymyxin B monotherapy, namely UDP-GlcNAc, D-Glucosamine 6-phosphate, N-Acetyl-D-glucosamine 6-phosphate, N-acetylmuramate 6-phosphate, and UDP-MurNAC-L-Ala-gamma-D-Glu-m-DAP (≥ 1.5-log2-fold, p ≤ .05; FDR ≤ 0.05) (Fig. 4A(i)). In contrast, the combination treatment reduced the levels of six of these metabolites at 15 min, including D-Glucosamine, UDP-GlcNAc, D-Glucosamine 6-phosphate, N-Acetyl-D-glucosamine 6-phosphate, N-acetylmuramate 6-phosphate and UDP-MurNAC-L-Ala-gamma-D-Glu-m-DAP (> 0.59-log2-fold, p ≤ .05; FDR ≤ 0.05) (Fig. 4A(i)).
Fig. 4.
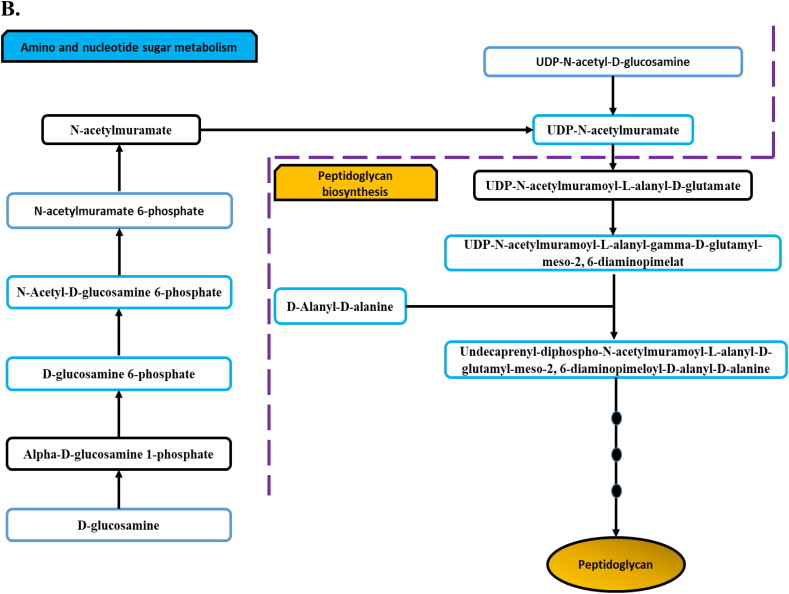
(A) Bar charts for the significantly impacted metabolites involved in amino sugar and nucleotide sugar and peptidoglycan biosynthesis of P. aeruginosa FADDI-PA006 following treatment with polymyxin B (PMB, purple), tamoxifen (TAM, green) monotherapy and in combination (COM, red) at (i) 15 min and (ii) 1 h (* > 0.59 -log2-fold; p ≤ .05; FDR ≤ 0.05), (** ≥ 1.0-log2-fold, p ≤ .05; FDR ≤ 0.05) & (*** ≥ 2.0-log2-fold, p ≤ .05; FDR ≤ 0.05). (B) Schematic diagram for amino sugar and nucleotide sugar metabolism and direct interrelated peptidoglycan biosynthetic pathway. Blue boxes indicated significantly impacted metabolites.
At the 1 h time point, both tamoxifen and polymyxin B monotherapy lacked effect on amino sugar and nucleotide sugar metabolites related to peptidoglycan biosynthesis, whilst the combination therapy induced significant changes in key peptidoglycan pathway intermediates (Fig. 4A(ii)). Specifically, three metabolites declined considerably following polymyxin B – tamoxifen combination treatment, namely D-alanyl-D-alanine, UDP-MurNAC-L-Ala-gamma-D-Glu-m-DAP, and UDP-MurNAC-L-Ala-gamma-D-Glu-m-DAP-D-Ala-D-Ala (≥ 1.5-log2-fold, p ≤ .05; FDR ≤ 0.05) (Fig. 4A(ii)). Moreover, five other essential precursors of amino sugar and nucleotide sugar metabolites were depleted, namely D-Glucosamine, D-Glucosamine 6-phosphate, N-Acetyl-D-glucosamine 6-phosphate, N-acetylmuramate, and UDP-GlcNAc (≥ 1.0-log2-fold, p ≤ .05; FDR ≤ 0.05) (Fig. 4). At 4 h, the impact of all treatments was minimal, with perturbations of only two intermediates for each of polymyxin B mono- and combination therapy, including N-Acetylmuramate and UDP-N-acetyl-D-glucosamine for polymyxin B monotherapy, and D-Glucosamine and N-acetylmuramate for the combination treatment. Tamoxifen monotherapy had no signifcant impact on amino sugar or nucleotide sugar metabolism at 4 h.
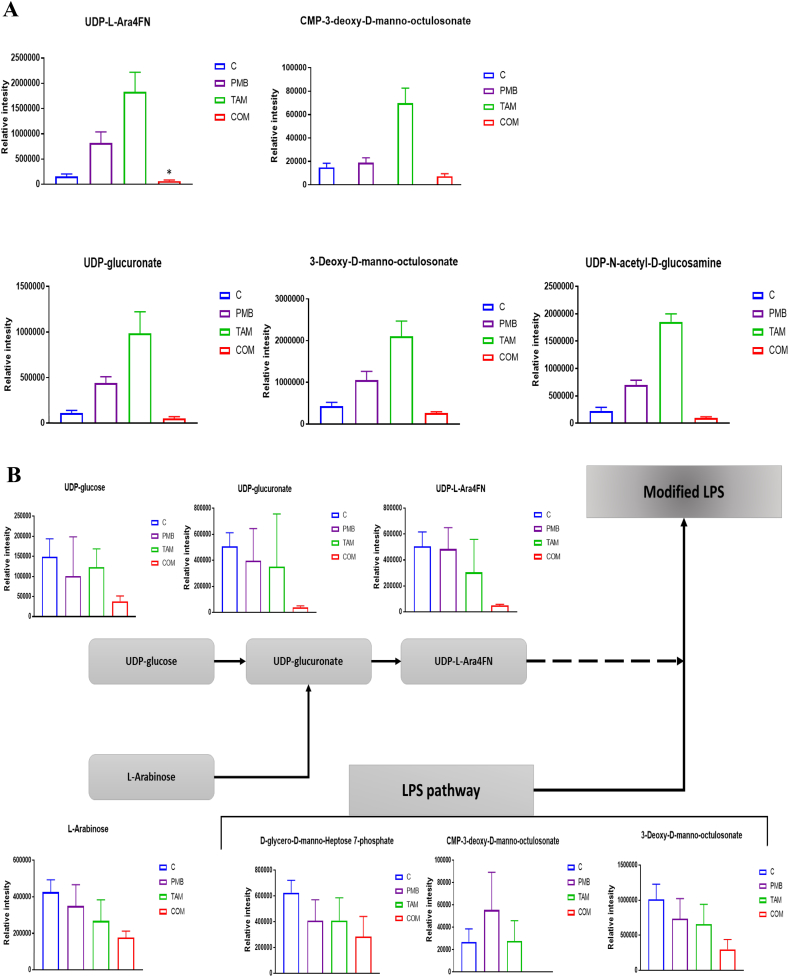
In addition to the aforementioned pathways, the combination treatment had a significant impact on LPS biosynthetic metabolites and the lipid A aminoarabinose modification pathway at both 15 min and 1 h. The concentrations of two crucial components of LPS biosynthesis 3-Deoxy-D-manno-octulosonate (KDO), and UDP-GlcNAc were significantly decreased after combination therapy at 15 min (> 0.59-log2-fold, p ≤ .05; FDR ≤ 0.05) (Figs. 5A). Furthermore, the combination treatment decreased the levels of two fundamental metabolites from the aminoarabinose lipid A modification pathway including UDP-L-Ara4FN (> 1.0-log2-fold, p ≤ .05; FDR ≤ 0.05) and UDP-glucuronate (> 0.59-log2-fold, p ≤ .05; FDR ≤ 0.05) at 15 min (Figs. 5A). In contrast, both tamoxifen and polymyxin B monotherapy increased the levels of these metabolites. Tamoxifen monotherapy induced a prominent increase in the concentrations of KDO and UDP-GlcNAc (> 2.0-log2-fold, p ≤ .05; FDR ≤ 0.05); and the levels of two lipid A aminoarabinose modification intermediates (UDP-glucuronate and UDP-L-Ara4FN) were also elevated (> 3.0-log2-fold, p ≤ .05; FDR ≤ 0.05) (Figs. 5A). Similarly, the UDP-glucuronate and UDP-L-Ara4FN levels were significantly elevated following polymyxin B monotherapy at 15 min (> 1.5-log2-fold, p ≤ .05; FDR ≤ 0.05).
Fig. 5.
(A) Bar charts for the significantly affected metabolites involved in LPS biosynthesis and its aminoarabinose modification following polymyxin B (PMB), tamoxifen (TAM), and combination (COM) treatment at 15 min *(≥ 1.0-log2-fold, p ≤ .05; FDR ≤ 0.05); (≥ 0.59-log2-fold, p ≤ .05; FDR ≤ 0.05). Control samples (blue), PMB (purple), TAM (green), and COM (red). (B) Bar charts and a diagram for the significantly impacted intermediates of LPS formation and its aminoarabinose modification after polymyxin B (PMB), tamoxifen (TAM), and the combination treatment at 1 h (≥ 1.0-log2-fold, p ≤ .05; FDR ≤ 0.05).
At 1 h, the combination treatment reduced the levels of three main intermediates of LPS biosynthesis including KDO, CMP-KDO, and D-glycero-D-manno-Heptose 7-phosphate (≥ 1.0-log2-fold, p ≤ .05; FDR ≤ 0.05) (Fig. 5B). The main precursors that are involved in the aminoarabinose modification of lipid A, were significantly decreased following the combination treatment at 1 h, particularly UDP-glucose, UDP-glucuronate, and UDP-L-Ara4FN (≥ 2.0-log2-fold, p ≤ .05; FDR ≤ 0.05) (Fig. 5B). By contrast, both polymyxin B and tamoxifen monotherapies did not influence LPS biosynthesis, nor lipid A aminoarabinose modification intermediates at 1 h (Fig. 5B). Notably, the levels of UDP-L-Ara4FN were greatly elevated following polymyxin B monotherapy at 4 h (≥ 3.0-log2-fold, p ≤ .05; FDR ≤ 0.05) while a much smaller increase was observed after the combination treatment at 4 h (> 0.59˂1.0-log2-fold, p ≤ .05; FDR ≤ 0.05) (Table S2). None of the treatments affected the LPS biosynthesis at 4 h (Table S2).
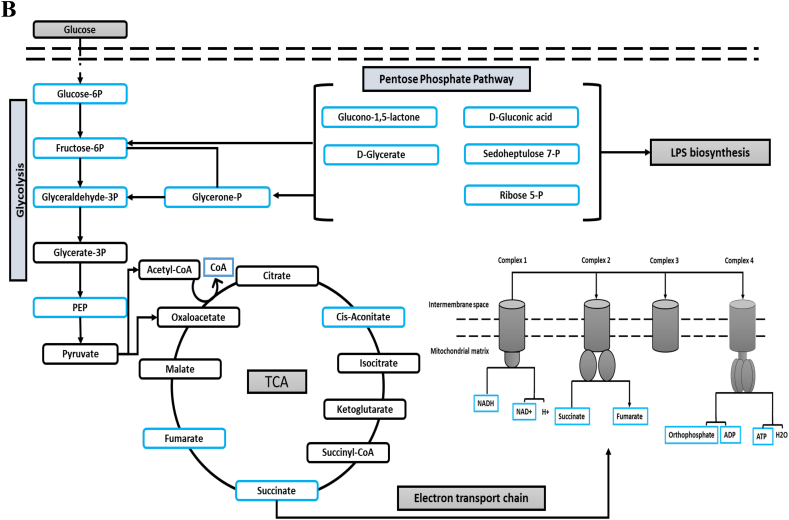
2.4. Impact of the Combination on Central Carbohydrate Metabolism and Energy Production Pathways in P. aeruginosa.
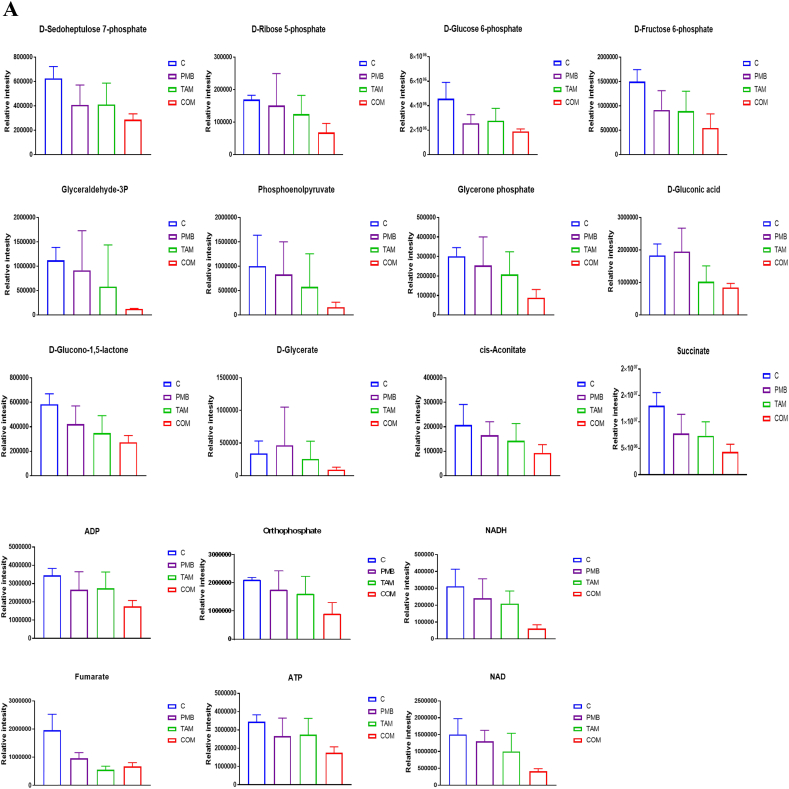
Central carbohydrate metabolism and energy production pathways were only affected at 1 h across all treatments. Both polymyxin B and tamoxifen monotherapies marginally affected only one metabolite, namely fumarate (Fig. 6A). The combination treatment induced marked changes of several intermediates involved in the main pathways of central carbohydrate metabolism, namely glycolysis, pentose phosphate pathway (PPP), tricarboxylic acid cycle (TCA) and the electron transport chain (ETC) (Fig. 6B). Interestingly, five intermediates of PPP decreased after combination treatment at 1 h, namely D-Glucono-1,5-lactone, D-Glycerate, D-Gluconic acid, D-Sedoheptulose 7-phosphate, and D-Ribose 5-phosphate (≥ 1.0-log2-fold, p ≤ .05; FDR ≤ 0.05) (Fig. 6A). Besides the PPP, another related pathway that was impacted by the combination therapy was glycolysis, of which five intermediates showed a significant reduction, namely D-Glucose 6-phosphate, D-Fructose 6-phosphate, glyceraldehyde-3-phosphate, phosphoenolpyruvate (PEP), and glycerone phosphate (≥ 1.0-log2-fold, p ≤ .05; FDR ≤ 0.05) (Figs. 6A & B). The TCA cycle also exhibited substantial changes for four intermediates following combination treatment: cis-aconitate, fumarate, CoA, and succinate (≥ 1.0-log2-fold, p ≤ .05; FDR ≤ 0.05) (Fig. 6B). Concomitantly, the concentration of four essential metabolites of the ETC was significantly decreased following the combination treatment, NADH, NAD+, ATP, and ADP (≥ 1.0-log2-fold, p ≤ .05; FDR ≤ 0.05) (Figs. 6A & B).
Fig. 6.
(A) Bar charts for the significantly affected intermediates of the interrelated pathways of glycolysis, pentose phosphate pathway (PPP), tricarboxylic acid cycle (TCA), and electron transport chain of P. aeruginosa FADDI-PA006 after polymyxin B (PMB, purple), tamoxifen (TAM, green), and the combination (COM, red) treatment at 1 h (≥ 1.0-log2-fold, p ≤ .05; FDR ≤ 0.05). (B) Schematic diagram for the impacted pathways (glycolysis, pentose phosphate pathway (PPP), tricarboxylic acid cycle (TCA), and electron transport) following treatment with polymyxin B, tamoxifen, and combination treatment at 1 h. Blue boxes indicated significantly impacted metabolites.
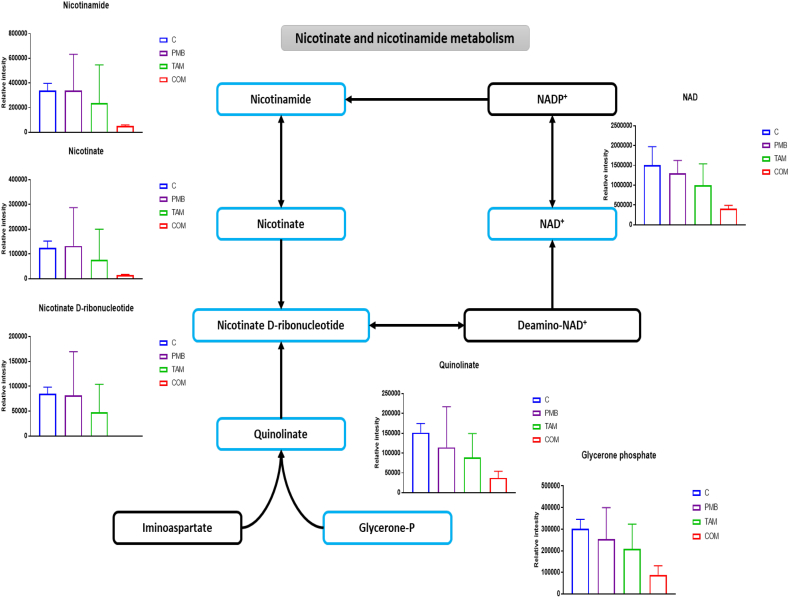
2.5. Effect of the Combination on Nicotinate and Nicotinamide Metabolism in P. aeruginosa.
Nicotinate and nicotinamide metabolism is vital for bacterial cell viability, as it generates the precursors of the coenzymes NAD+ and NADP+, which are crucial for cellular electron transfer reactions [35]. Strikingly, the combination treatment caused significant perturbations in the levels of six fundamental intermediates of nicotinate and nicotinamide metabolism, but only at 1 h (Fig. 7). The concentrations of glycerone phosphate, NAD+, nicotinate, nicotinamide, nicotinate D-ribonucleotide (NMN), and quinolinate (pyridine-2,3-dicarboxylate) were reduced following combination therapy (≥ 1.5-log2-fold, p ≤ .05; FDR ≤ 0.05) (Fig. 7). These pathways were not affected by either polymyxin B or tamoxifen monotherapy.
Fig. 7.
Schematic diagram and bar graphs for significantly affected metabolites of nicotinate and nicotinamide metabolism of P. aeruginosa FADDI-PA006 after polymyxin B (PMB, purple), tamoxifen (TAM, green), and the combination (COM, red) treatment at 1 h (≥ 1.0-log2-fold, p ≤ .05; FDR ≤ 0.05). Blue boxes indicated significantly affected metabolites.
3. Discussion
In recent years, combination therapies with FDA approved non-antibiotic drugs has emerged as a novel strategy for maintaining the clinical efficacy of polymyxins and suppressing outbreaks of polymyxin resistance [17,36,37]. We have previously shown that the combination of polymyxin B with a SERM produces a highly synergistic bactericidal effect against MDR CF P. aeruginosa isolates and prevents the emergence of polymyxin resistance [21]. SERMs are non-steroidal estrogen receptor modulators approved for chemotherapy and chemoprevention of breast cancers and osteoporosis [38]. Along with their antineoplastic activity, SERMs have been found to exhibit direct antimicrobial activities (i.e. antifungal, antiviral, antibacterial) [[39], [40], [41], [42]]. It has been proposed that SERMs operate via multiple mechanisms of antifungal activity, including membrane damage, inhibition of membrane peroxidation, cell cycle arrest, and interference with calmodulin [41]. However, their mechanism of antibacterial activity remains largely enigmatic. Understanding the biochemical mechanism(s) by which polymyxin B in combination with SERMs act synergistically against P. aeruginosa is vital for their potential future repurposing as antimicrobial agents. This report is the first to employ untargeted metabolomics to decipher the mechanisms of action of this novel combination against a MDR polymyxin resistant P. aeruginosa FADDI-PA006 isolated from the lungs of a CF patient. In order to ensure the study is clinically relevant, all experiments were performed using clinically achievable concentrations of polymyxin B (2 mg/L) and tamoxifen (8 mg/L) [2,38,43,44]. Three different drug exposure times, 15 min, 1 h and 4 h were examined.
It has been proposed that polymyxins exert their bactericidal activity via disorganization and perforation of the bacterial OM [45,46]. However, as the tested strain FADDI-PA006 is resistant to polymyxins, it is not surprising that polymyxin B monotherapy caused minimal perturbations of OM lipids (Fig. 3A & B). Although polymyxin B decreased the levels of essential membrane lipids such as the fatty acid (18:0) octadecanoic acid, a significant increase in the level of palmitoleyl-CoA was also reported. The saturated fatty acid (18:0) octadecanoic acid is one of the principal fatty acids making up the composition of the P. aeruginosa OM [47]; and a reduction in saturated fatty acids is usually associated with increased permeability and fluidity of the bacterial OM [48]. In addition to its estrogen receptor binding activity, tamoxifen is known to have multiple secondary cellular effects related to its direct molecular interactions with the membrane phospholipids and its ability to decrease membrane fluidity [49,50]. As illustrated in Fig. 3A, tamoxifen induced a remarkable increase in the levels of palmitic acid (FA (16:0)), an important membrane fatty acid which is known to decrease membrane fluidity and hence, reduce membrane permeability [51]. However, tamoxifen caused a significant reduction in the levels of key intermediates involved in LPS and phospholipid biosynthesis including tetradecanoic acid and sn-glycero-3-phosphoethanolamine [52]. In contrast, the combination treatment produced markedly different changes in lipid pathways, wherein most of the significantly perturbed lipids were decreased across all time points such as a significant decrease in the levels of sn-glycerol 3-phosphate and sn-glycero-3-phosphoethanolamine at 15 min and 1 h (Fig. 3A & B). Sn-glycerol 3-phosphate is a crucial intermediate required for the formation of membrane phospholipids, it undergoes acylation at the 1-position to form lysophosphatidic acid (LPA), and then a second acylation step produces phosphatidic acid (PA), the key intermediate in the synthesis of bacterial membrane glycerolipids [51,53]. Interestingly, in response to combination treatment, the levels of lysophosphatidylethanolamines (PE (18:1), PE (18:2) and LysoPE (16,0)) were significantly reduced, which indicates that the combination therapy potentially inhibits LPS modifications with ethanolamine compounds (Fig. 3A & B) [54]. The combination treatment also reduced the levels of fatty acids that are essential for LPS biosynthesis, tetradecanoic acid and palmitoleyl-CoA at early exposure times [52,55].
In addition to the effect on the lipid metabolism, pathway analysis revealed that polymyxin B monotherapy negatively impacted intermediates from the LPS and peptidoglycan biosynthetic pathways. Notably, our results are in line with a previous metabolomics study which revealed polymyxin B perturbs the main intermediates involved in LPS and peptidoglycan biosynthesis of the Gram-negative bacterium Acinetobacter baumannii [33]. Both Polymyxin B and tamoxifen monotherapy induced a significant increase in the concentrations of fundamental amino sugar and nucleotide sugar intermediates of the peptidoglycan biosynthetic pathway including N-Acetyl-D-glucosamine 6-phosphate, UDP-N-acetyl-D-glucosamine, and UDP-MurNAC-L-Ala-gamma-D-Glu-m-DAP (Fig. 4A). Furthermore, polymyxin B and tamoxifen monotherapy increased the levels of LPS biosynthetic pathway intermediates and the associated L-Ara4N lipid A modification intermediates, but only at 15 min. P. aeruginosa can develop polymyxin resistance via intrinsic (PmrB mutations) or extrinsic (mcr-1) mechanisms, which manifest the modification of LPS with the cationic moieties aminoarabinose (L-Ara4N) or phosphoethanolamine (PEtn) that act to repel the like-charged polymyxin [56,57].
The combination treatment induced a marked reduction of fundamental precursors (amino sugar and nucleotide sugar metabolism) of the interrelated peptidoglycan and LPS biosynthesis pathways; it is an important to note that this effect was more pronounced at 1 h than 15 min, opposite to the combination effect on lipid metabolism (Fig. 4, Fig. 5). For instance, the levels of UDP-N-acetyl-D-glucosamine (UDP-GlcNAc), N-Acetyl-D-glucosamine 6-phosphate (GlcNAc-6P), and D-Glucosamine 6-phosphate decreased dramatically (Fig. 4A & B). Amino sugars and nucleotide sugars form the essential components of cell surface structures of bacteria (peptidoglycan, LPS and the polysaccharide capsule). UDP-N-acetyl-D-glucosamine (UDP-GlcNAc) in particular plays an important role as amino sugar donor in many transferase reactions in the biogenesis of peptidoglycan, the core lipid A moieties of the LPS, and certain O-antigens of Gram-negative bacteria [[58], [59], [60], [61]]. It has been previously proposed that drugs interfering with bacterial nucleotide sugar biosynthetic pathways might be potential targets for the discovery of new therapeutics [62]. Our findings are in agreement with the previous metabolomics study in which the impact of a set of common antibiotics was tested against the Staphylococcus aureus metabolome [63]. The study revealed that vancomycin, a glycopeptide which acts via inhibition of bacterial cell wall synthesis, was greatly effective in reducing the amino sugar and nucleotide sugar intermediates after a 30 min treatment period [63]. Maifiah et al., found that colistin monotherapy and in combination with doripenem caused a significant depletion in the intracellular levels of several important metabolites associated with amino sugar and nucleotide sugar metabolism of A. baumannii at 1 and 4 h [33].
The bacterial cell envelope is composed of an inner cell membrane and a cell wall in Gram-positive bacteria, in addition to the outer membrane in Gram-negative bacteria. This structure provides structural integrity to the cell and protect these organisms from their unpredictable and often hostile environment [64]. Owing to its importance as a crucial cell wall structural component of bacteria, peptidoglycan biosynthesis has been extensively investigated as a target for developing of new antibacterial agents [65,66]. It is well known that several antibiotics exert their action by attacking peptidoglycan assembly, including vancomycin (which interferes with D-Alanyl-D-alanine) and fosfomycin [67,68]. A recent metabolomics study showed that fosfomycin and erythromycin significantly reduced levels of major precursors of cell wall biosynthesis in S. aureus63. Furthermore, it has been found that the combination of colistin and doripenem affected greatly the main intermediates of peptidoglycan biosynthesis of A. baumannii [33]. Our results are in agreement with these previous studies, where marked reductions were observed for key intermediates involved in the biosynthesis of peptidoglycan (Figs. 4A & B).
Importantly, our study is the first to demonstrate that combining tamoxifen with polymyxin B causes a remarkable decrease in the essential precursor metabolites of aminoarabinose (i.e. UDP-L-Ara4FN, UDP-glucuronate, and L-Arabinose), a major mechanism of polymyxin resistance (Figs. 5A & B) [56,57]. Although, the level of LPS components increased with polymyxin B and tamoxifen treatment, they display a considerable reduction after the combination treatment at 15 min and 1 h, in particular KDO, CMP-KDO, and D-glycero-D-manno-heptose 7-phosphate (Figs. 5A & B). The reduction in the concentrations of LPS and its aminoarabinose modification might result from either inhibition of amino sugar and nucleotide sugar metabolism, or from depletion of the main intermediates of PPP e.g., D-Ribose 5-phosphate and D-Sedoheptulose 7-phosphate (Figs. 6A & B). ADP-heptose, a key downstream metabolite of the heptose biosynthesis pathway, is an important component of the LPS inner core [69]. Mutations in the gene (GmhA) associated with ADP-glyceromannoheptose synthesis in Haemophilus influenza, which cause deficiencies in heptose biosynthesis, result in an a virulent phenotype, increased membrane permeability and increased susceptibility to antibiotics [70,71]. Additionally, KDO synthesis is initiated by activation of D-arabinose-5-phosphate isomerase (API), an enzyme that promotes the reversible isomerization of D-ribulose-5-phosphate (Ru5P) to D-arabinose-5-phosphate, a KDO precursor; inactivation of this enzyme causes death of E. coli [72]. Thus, the depletion in the level of main components of amino sugar and nucleotide sugar metabolism and its direct downstream pathways, peptidoglycan and LPS biosynthesis, suggest synergistic bactericidal activity of our combination against P. aeruginosa.
In addition to its potential impact on membrane structure, our combination treatment was effective in depleting several components of glycolysis and tricarboxylic acid cycle (TCA) and a directly connected pathway, the electron transport chain at 1 h (Figs. 6A & B). The levels of glyceraldehyde-3P, PEP, Fructose-6P, succinate, and fumarate significantly declined after combination therapy (Figs. 6A & B). Central carbohydrate metabolism is a metabolic network that involves glycolysis and the TCA cycle; it has been recently explored for new targets for antibacterial agents [73,74]. The significant decrease of fundamental intermediates of the TCA cycle, in particular succinate, likely results in a reduction of reducing equivalents for the electron transport chain. Succinate dehydrogenase is a vital enzyme that links the TCA cycle and the electron transport chain and is able to oxidize of succinate to fumarate [74]. The deletion of succinate dehydrogenase in S. enterica impaired its ability to colonize mice [75]. Succinate dehydrogenase has also been investigated in other bacteria such as P. aeruginosa as a promising next-generation drug target [76]. In addition to the TCA cycle defects, the polymyxin B – tamoxifen combination treatment resulted in depletion of several intermediates of the electron transport chain, including ATP, ADP, NAD+ and orthophosphate (Figs. 6A & B).
Nicotinate and nicotinamide metabolism was another strongly interrelated pathway that was extensively perturbed by the polymyxins B – tamoxifen combination at 1 h (Fig. 7). Six crucial precursors were significantly reduced following combination treatment e.g., quinolinate, NMN, and nicotinamide (Fig. 7). This biochemical pathway is a vital source of NAD+ (H) and NADP+ (H) cofactors, which are necessary for multiple complex reactions in several metabolic pathways including glycolysis, PPP, TCA cycle and fatty acid biosynthesis [35,77]. An investigation into the importance of bacterial NAD biosynthesis demonstrated that knock-down of the protein levels for the two genes, NadD and NadE, which are necessary for the last two steps of NAD biogenesis resulted in cell death in Mycobacteria [78]. Intriguingly, our combination produced a marked inhibition of nicotinate and nicotinamide biosynthesis (Fig. 7).
To sum up, the current study is the first to decipher the mechanism of synergistic bactericidal activity of a novel combination of a polymyxin with a SERM, using a metabolomics platform. Polymyxin B and tamoxifen monotherapy caused minor perturbations to the FADDI-PA006 metabolome at 15 min; which were mainly comprised of increased levels of selected metabolites. On the other hand, the combination treatment showed greater perturbation to the metabolome at 15 min, 1 and 4 h. The combination inhibited most affected pathways across all time exposures, with the greatest differences observed at 1 h. It is possible that the synergistic killing associated with this novel combination is largely due to inhibition of multiple complex interconnected pathways, in particular those related to bacterial cell envelope biosynthesis, and inhibition of bacterial resistance development via outer membrane modification. Additional synergistic effects were also noticed involving inhibition of key central metabolic pathways including glycolysis, PPP, TCA cycle and electron transport chain. Importantly, this study highlights the potential for repurposing of tamoxifen for an antimicrobial indication in combination with polymyxins. Multi-omics study along with further molecular work should be performed to validate the key pathways recognized from the metabolomics analysis. To do so, mutant libraries, in silico genome scale metabolic modelling and metabolite feeding experiments would be required. Further studies are warranted to examine other non-antibiotic drugs as antimicrobial agents in combination with the OM permeabilizing antimicrobial agents such as the polymyxins and aminoglycosides.
4. Methods
Detailed methods of all experimental procedures are provided in the supporting information
Acknowledgements
J.L. and T.V. are supported by research grants from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (R01 AI132681). J.L. and T.V. are also supported by the Australian National Health and Medical Research Council (NHMRC) as Senior Research and Career Development Level 2 fellows, respectively. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2018.11.001.
Contributor Information
Jian Li, Email: Jian.Li@monash.edu.
Tony Velkov, Email: Tony.Velkov@unimelb.edu.au.
Appendix A. Supplementary data
Supplementary material
References
- 1.Ventola C.L. The antibiotic resistance crisis: part 1: causes and threats. Pharm Ther. 2015;40:277. [PMC free article] [PubMed] [Google Scholar]
- 2.Velkov T., Roberts K.D., Nation R.L. Pharmacology of polymyxins: new insights into an ‘old'class of antibiotics. Future Microbiol. 2013;8:711–724. doi: 10.2217/fmb.13.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mesaros N., Nordmann P., Plésiat P. Pseudomonas aeruginosa: resistance and therapeutic options at the turn of the new millennium. Clin. Microbiol. Infect. 2007;13:560–578. doi: 10.1111/j.1469-0691.2007.01681.x. [DOI] [PubMed] [Google Scholar]
- 4.Hancock R.E., Speert D.P. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and impact on treatment. Drug Resist. Updat. 2000;3:247–255. doi: 10.1054/drup.2000.0152. [DOI] [PubMed] [Google Scholar]
- 5.Livermore D.M. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin. Infect. Dis. 2002;34:634–640. doi: 10.1086/338782. [DOI] [PubMed] [Google Scholar]
- 6.Johnson C. The 12 superbugs threatening human health.
- 7.Lyczak J.B., Cannon C.L., Pier G.B. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 2002;15:194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verkman A., Song Y., Thiagarajah J.R. Role of airway surface liquid and submucosal glands in cystic fibrosis lung disease. Am J Physiol-Cell Physiol. 2003;284:C2–C15. doi: 10.1152/ajpcell.00417.2002. [DOI] [PubMed] [Google Scholar]
- 9.Li J., Nation R.L., Turnidge J.D. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect. Dis. 2006;6:589–601. doi: 10.1016/S1473-3099(06)70580-1. [DOI] [PubMed] [Google Scholar]
- 10.Nation R.L., Li J., Cars O. Framework for optimisation of the clinical use of colistin and polymyxin B: the Prato polymyxin consensus. Lancet Infect. Dis. 2015;15:225–234. doi: 10.1016/S1473-3099(14)70850-3. [DOI] [PubMed] [Google Scholar]
- 11.Trimble M.J., Mlynárčik P., Kolář M. Polymyxin: alternative mechanisms of action and resistance. Cold Spring Harb. Perspect. Med. 2016;6:a025288. doi: 10.1101/cshperspect.a025288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ernst R.K., Guina T., Miller S.I. Salmonella typhimurium outer membrane remodeling: role in resistance to host innate immunity. Microbes Infect. 2001;3:1327–1334. doi: 10.1016/s1286-4579(01)01494-0. [DOI] [PubMed] [Google Scholar]
- 13.Raetz C.R., Reynolds C.M., Trent M.S. Lipid A modification systems in gram-negative bacteria. Annu. Rev. Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Llobet E., Campos M.A., Giménez P. Analysis of the networks controlling the antimicrobial-peptide-dependent induction of Klebsiella pneumoniae virulence factors. Infect. Immun. 2011;79:3718–3732. doi: 10.1128/IAI.05226-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J., Rayner C.R., Nation R.L. Heteroresistance to colistin in multidrug-resistant acinetobacter baumannii. Antimicrob. Agents Chemother. 2006;50:2946–2950. doi: 10.1128/AAC.00103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai Y., Chai D., Wang R. Colistin resistance of acinetobacter baumannii: clinical reports, mechanisms and antimicrobial strategies. J. Antimicrob. Chemother. 2012;67:1607–1615. doi: 10.1093/jac/dks084. [DOI] [PubMed] [Google Scholar]
- 17.Bergen P.J., Bulman Z.P., Saju S. Polymyxin combinations: pharmacokinetics and pharmacodynamics for rationale use. Pharmacotherapy. 2015;35:34–42. doi: 10.1002/phar.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henry R., Crane B., Powell D. The transcriptomic response of Acinetobacter baumannii to colistin and doripenem alone and in combination in an in vitro pharmacokinetics/pharmacodynamics model. J. Antimicrob. Chemother. 2015;70:1303–1313. doi: 10.1093/jac/dku536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ejim L., Farha M.A., Falconer S.B. Combinations of antibiotics and nonantibiotic drugs enhance antimicrobial efficacy. Nat. Chem. Biol. 2011;7:348. doi: 10.1038/nchembio.559. [DOI] [PubMed] [Google Scholar]
- 20.Schneider E.K., Reyes-Ortega F., Velkov T. Antibiotic–non-antibiotic combinations for combating extremely drug-resistant Gram-negative ‘superbugs’. Essays Biochem. 2017;61:115–125. doi: 10.1042/EBC20160058. [DOI] [PubMed] [Google Scholar]
- 21.Hussein M.H., Schneider E.K., Elliott A.G. From breast cancer to antimicrobial: combating extremely resistant Gram-negative “superbugs” using novel combinations of polymyxin B with selective estrogen receptor modulators. Microb. Drug Resist. 2017;23:640–650. doi: 10.1089/mdr.2016.0196. [DOI] [PubMed] [Google Scholar]
- 22.Schneider E.K., Azad M.A., Han M.-L. An “unlikely” pair: the antimicrobial synergy of polymyxin B in combination with the cystic fibrosis transmembrane conductance regulator drugs KALYDECO and ORKAMBI. ACS Infect Dis. 2016;2:478–488. doi: 10.1021/acsinfecdis.6b00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim T.-P., Lee W., Tan T.-Y. Effective antibiotics in combination against extreme drug-resistant Pseudomonas aeruginosa with decreased susceptibility to polymyxin B. PLoS One. 2011;6:e28177. doi: 10.1371/journal.pone.0028177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vincent I.M., Ehmann D.E., Mills S.D. Untargeted metabolomics to ascertain antibiotic modes of action. Antimicrob. Agents Chemother. 2016;60:2281–2291. doi: 10.1128/AAC.02109-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belenky P., Jonathan D.Y., Porter C.B. Bactericidal antibiotics induce toxic metabolic perturbations that lead to cellular damage. Cell Rep. 2015;13:968–980. doi: 10.1016/j.celrep.2015.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reiß S., Pané-Farré J., Fuchs S. Global analysis of the Staphylococcus aureus response to mupirocin. Antimicrob. Agents Chemother. 2012 Feb 1;56(2):787–804. doi: 10.1128/AAC.05363-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z.-Y., Zhu Q., Zhang H.-Y. Metabolite concentration as a criterion for antibacterial discovery. Curr. Comput. Aided Drug Des. 2013;9:412–416. doi: 10.2174/15734099113099990030. [DOI] [PubMed] [Google Scholar]
- 28.Okada B.K., Wu Y., Mao D. Mapping the trimethoprim-induced secondary metabolome of Burkholderia thailandensis. ACS Chem. Biol. 2016;11:2124–2130. doi: 10.1021/acschembio.6b00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lobritz M.A., Belenky P., Porter C.B. Antibiotic efficacy is linked to bacterial cellular respiration. Proc. Natl. Acad. Sci. 2015;112:8173–8180. doi: 10.1073/pnas.1509743112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zampieri M., Zimmermann M., Claassen M. Nontargeted metabolomics reveals the multilevel response to antibiotic perturbations. Cell Rep. 2017;19:1214–1228. doi: 10.1016/j.celrep.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 31.Maifiah M.H.M., Creek D.J., Nation R.L. Untargeted metabolomics analysis reveals key pathways responsible for the synergistic killing of colistin and doripenem combination against acinetobacter baumannii. Sci. Rep. 2017;7 doi: 10.1038/srep45527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirwan J.A., Weber R.J., Broadhurst D.I. Direct infusion mass spectrometry metabolomics dataset: a benchmark for data processing and quality control. Scientific Data. 2014;1:140012. doi: 10.1038/sdata.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maifiah M.H.M., Creek D.J., Nation R.L. Untargeted metabolomics analysis reveals key pathways responsible for the synergistic killing of colistin and doripenem combination against acinetobacter baumannii. Sci. Rep. 2017;7:45527. doi: 10.1038/srep45527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han M.-L., Zhu Y., Creek D.J. Alterations of metabolic and lipid profiles in polymyxin-resistant Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2018 May 25;62(6) doi: 10.1128/AAC.02656-17. (AAC. 02656–17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Begley T.P., Kinsland C., Mehl R.A. The biosynthesis of nicotinamide adenine dinucleotides in bacteria. Vitam. Horm. 2001;61:103–119. doi: 10.1016/s0083-6729(01)61003-3. [DOI] [PubMed] [Google Scholar]
- 36.Ly N.S., Bulitta J.B., Rao G.G. Colistin and doripenem combinations against Pseudomonas aeruginosa: profiling the time course of synergistic killing and prevention of resistance. J. Antimicrob. Chemother. 2015;70:1434–1442. doi: 10.1093/jac/dku567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bergen P.J., Bulman Z.P., Landersdorfer C.B. Optimizing polymyxin combinations against resistant gram-negative bacteria. Infect Dis Ther. 2015;4:391–415. doi: 10.1007/s40121-015-0093-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morello K.C., Wurz G.T., DeGregorio M.W. Pharmacokinetics of selective estrogen receptor modulators. Clin. Pharmacokinet. 2003;42:361–372. doi: 10.2165/00003088-200342040-00004. [DOI] [PubMed] [Google Scholar]
- 39.Butts A., Koselny K., Chabrier-Roselló Y. Estrogen receptor antagonists are anti-cryptococcal agents that directly bind EF hand proteins and synergize with fluconazole in vivo. MBio. 2014;5 doi: 10.1128/mBio.00765-13. (e00765–13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miguel D.C., Zauli-Nascimento R.C., Yokoyama-Yasunaka J.K. Tamoxifen as a potential antileishmanial agent: efficacy in the treatment of Leishmania braziliensis and leishmania chagasi infections. J. Antimicrob. Chemother. 2008;63:365–368. doi: 10.1093/jac/dkn509. [DOI] [PubMed] [Google Scholar]
- 41.Dolan K., Montgomery S., Buchheit B. Antifungal activity of tamoxifen: in vitro and in vivo activities and mechanistic characterization. Antimicrob. Agents Chemother. 2009;53:3337–3346. doi: 10.1128/AAC.01564-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johansen L., Brannan J., Delos S., Olinger GG FDA-approved selective estrogen receptor modulators inhibit Ebola virus infection. Sci. Transl. Med. 2013;5 doi: 10.1126/scitranslmed.3005471. (190ra79) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hochner-Celnikier D. Pharmacokinetics of raloxifene and its clinical application. Eur. J. Obstet. Gynecol. Reprod. Biol. 1999;85:23–29. doi: 10.1016/s0301-2115(98)00278-4. [DOI] [PubMed] [Google Scholar]
- 44.Taras T.L., Wurz G.T., Linares G.R. Clinical pharmacokinetics of toremifene. Clin. Pharmacokinet. 2000;39:327–334. doi: 10.2165/00003088-200039050-00002. [DOI] [PubMed] [Google Scholar]
- 45.Hancock R.E., Chapple D.S. Peptide antibiotics. Antimicrob. Agents Chemother. 1999;43:1317–1323. doi: 10.1128/aac.43.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hancock R.E. Cationic peptides: effectors in innate immunity and novel antimicrobials. Lancet Infect. Dis. 2001;1:156–164. doi: 10.1016/S1473-3099(01)00092-5. [DOI] [PubMed] [Google Scholar]
- 47.Cho K., Salton M. Fatty acid composition of bacterial membrane and wall lipids. Biochimica et Biophysica Acta (BBA)-Lipids and Lipid. Metabolism. 1966;116:73–79. doi: 10.1016/0005-2760(66)90093-2. [DOI] [PubMed] [Google Scholar]
- 48.Mansilla M.C., Cybulski L.E., Albanesi D. Control of membrane lipid fluidity by molecular thermosensors. J. Bacteriol. 2004;186:6681–6688. doi: 10.1128/JB.186.20.6681-6688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wiseman H., Quinn P., Halliwell B. Tamoxifen and related compounds decrease membrane fluidity in liposomes: mechanism for the antioxidant action of tamoxifen and relevance to its anticancer and cardioprotective actions? FEBS Lett. 1993;330:53–56. doi: 10.1016/0014-5793(93)80918-k. [DOI] [PubMed] [Google Scholar]
- 50.Custódio J.A., Almeida L.M., Madeira V.M. The anticancer drug tamoxifen induces changes in the physical properties of model and native membranes. Biochimica et Biophysica Acta (BBA)-Biomem. 1993;1150:123–129. doi: 10.1016/0005-2736(93)90080-j. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y.-M., Rock C.O. Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol. 2008;6:222. doi: 10.1038/nrmicro1839. [DOI] [PubMed] [Google Scholar]
- 52.Kusumoto S., Fukase K., Shiba T. Key structures of bacterial peptidoglycan and lipopolysaccharide triggering the innate immune system of higher animals: chemical synthesis and functional studies. Proc Jpn Acad Ser B. 2010;86:322–337. doi: 10.2183/pjab.86.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cronan J.E., Jr., Vagelos P.R. Metabolism and function of the membrane phospholipids of Escherichia coli. Biochimica et Biophysica Acta (BBA)-Rev Biomemb. 1972;265:25–60. doi: 10.1016/0304-4157(72)90018-4. [DOI] [PubMed] [Google Scholar]
- 54.Arroyo L.A., Herrera C.M., Fernandez L. The pmrCAB operon mediates polymyxin resistance in Acinetobacter baumannii ATCC 17978 and clinical isolates through phosphoethanolamine modification of Lipid A. Antimicrob. Agents Chemother. 2011 Aug;55(8):3743–3751. doi: 10.1128/AAC.00256-11. (AAC. 00256-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emiola A., Andrews S.S., Heller C. Crosstalk between the lipopolysaccharide and phospholipid pathways during outer membrane biogenesis in Escherichia coli. Proc. Natl. Acad. Sci. 2016;113:3108–3113. doi: 10.1073/pnas.1521168113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Olaitan A.O., Morand S., Rolain J.-M. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014;5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Raetz C.R. Bacterial lipopolysaccharides: a remarkable family of bioactive macroamphiphiles. Escherichia Coli Salmonella. 1996;1:1035–1063. [Google Scholar]
- 59.Van Heijenoort J. Murein synthesis. Escherichia Coli Salmonella Typhimurium. 1996:1025–1034. [Google Scholar]
- 60.Plumbridge J., Vimr E. Convergent pathways for utilization of the amino sugars N-acetylglucosamine, N-acetylmannosamine, and N-acetylneuraminic acid by Escherichia coli. J. Bacteriol. 1999;181:47–54. doi: 10.1128/jb.181.1.47-54.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mengin-Lecreulx D., Van Heijenoort J. Copurification of glucosamine-1-phosphate acetyltransferase and N-acetylglucosamine-1-phosphate uridyltransferase activities of Escherichia coli: characterization of the glmU gene product as a bifunctional enzyme catalyzing two subsequent steps in the pathway for UDP-N-acetylglucosamine synthesis. J. Bacteriol. 1994;176:5788–5795. doi: 10.1128/jb.176.18.5788-5795.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mäki M., Renkonen R. Biosynthesis of 6-deoxyhexose glycans in bacteria. Glycobiology. 2004;14:1R–15R. doi: 10.1093/glycob/cwh040. [DOI] [PubMed] [Google Scholar]
- 63.Dörries K., Schlueter R., Lalk M. Impact of antibiotics with various target sites on the metabolome of Staphylococcus aureus. Antimicrob. Agents Chemother. 2014;58:7151–7163. doi: 10.1128/AAC.03104-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bugg T.D., Braddick D., Dowson C.G. Bacterial cell wall assembly: still an attractive antibacterial target. Trends Biotechnol. 2011;29:167–173. doi: 10.1016/j.tibtech.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 66.Green D.W. The bacterial cell wall as a source of antibacterial targets. Expert Opin. Ther. Targets. 2002;6:1–20. doi: 10.1517/14728222.6.1.1. [DOI] [PubMed] [Google Scholar]
- 67.Barna J., Williams D. The structure and mode of action of glycopeptide antibiotics of the vancomycin group. Annu. Rev. Microbiol. 1984;38:339–357. doi: 10.1146/annurev.mi.38.100184.002011. [DOI] [PubMed] [Google Scholar]
- 68.Dijkmans A.C., Zacarías N.V.O., Burggraaf J. Fosfomycin: Pharmacological, clinical and future perspectives. Antibiotics. 2017;6:24. doi: 10.3390/antibiotics6040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matthaiou D.K., Michalopoulos A., Rafailidis P.I. Risk factors associated with the isolation of colistin-resistant gram-negative bacteria: a matched case-control study. Crit. Care Med. 2008;36:807–811. doi: 10.1097/CCM.0B013E3181652FAE. [DOI] [PubMed] [Google Scholar]
- 70.Taylor P.L., Blakely K.M., De Leon G.P. Structure and function of sedoheptulose-7-phosphate isomerase, a critical enzyme for lipopolysaccharide biosynthesis and a target for antibiotic adjuvants. J. Biol. Chem. 2008;283:2835–2845. doi: 10.1074/jbc.M706163200. [DOI] [PubMed] [Google Scholar]
- 71.Brooke J.S., Valvano M. Molecular cloning of the Haemophilus influenzae gmhA (lpcA) gene encoding a phosphoheptose isomerase required for lipooligosaccharide biosynthesis. J. Bacteriol. 1996;178:3339–3341. doi: 10.1128/jb.178.11.3339-3341.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gourlay L.J., Sommaruga S., Nardini M. Probing the active site of the sugar isomerase domain from E. coli arabinose-5-phosphate isomerase via X-ray crystallography. Protein Sci. 2010;19:2430–2439. doi: 10.1002/pro.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Murima P., McKinney J.D., Pethe K. Targeting bacterial central metabolism for drug development. Chem. Biol. 2014;21:1423–1432. doi: 10.1016/j.chembiol.2014.08.020. [DOI] [PubMed] [Google Scholar]
- 74.Oyedotun K.S., Lemire B.D. The quaternary structure of the Saccharomyces cerevisiae succinate dehydrogenase homology modeling, cofactor docking, and molecular dynamics simulation studies. J. Biol. Chem. 2004;279:9424–9431. doi: 10.1074/jbc.M311876200. [DOI] [PubMed] [Google Scholar]
- 75.Mercado-Lubo R., Gauger E.J., Leatham M.P. A Salmonella enterica serovar typhimurium succinate dehydrogenase/fumarate reductase double mutant is avirulent and immunogenic in BALB/c mice. Infect. Immun. 2008;76:1128–1134. doi: 10.1128/IAI.01226-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cook G.M., Greening C., Hards K. Energetics of pathogenic bacteria and opportunities for drug development. Adv. Microb. Physiol. 2014:1–62. doi: 10.1016/bs.ampbs.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 77.Boshoff H.I., Xu X., Tahlan K. Biosynthesis and recycling of nicotinamide cofactors in mycobacterium tuberculosis an essential role for nad in nonreplicating bacilli. J. Biol. Chem. 2008;283:19329–19341. doi: 10.1074/jbc.M800694200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rodionova I.A., Schuster B.M., Guinn K.M. Metabolic and bactericidal effects of targeted suppression of NadD and NadE enzymes in mycobacteria. MBio. 2014;5 doi: 10.1128/mBio.00747-13. (e00747–13) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material