Abstract
Motivation: Supervised machine learning is widely applied to transcriptomic data to predict disease diagnosis, prognosis or survival. Robust and interpretable classifiers with high accuracy are usually favored for their clinical and translational potential. The top scoring pair (TSP) algorithm is an example that applies a simple rank-based algorithm to identify rank-altered gene pairs for classifier construction. Although many classification methods perform well in cross-validation of single expression profile, the performance usually greatly reduces in cross-study validation (i.e. the prediction model is established in the training study and applied to an independent test study) for all machine learning methods, including TSP. The failure of cross-study validation has largely diminished the potential translational and clinical values of the models. The purpose of this article is to develop a meta-analytic top scoring pair (MetaKTSP) framework that combines multiple transcriptomic studies and generates a robust prediction model applicable to independent test studies.
Results: We proposed two frameworks, by averaging TSP scores or by combining P-values from individual studies, to select the top gene pairs for model construction. We applied the proposed methods in simulated data sets and three large-scale real applications in breast cancer, idiopathic pulmonary fibrosis and pan-cancer methylation. The result showed superior performance of cross-study validation accuracy and biomarker selection for the new meta-analytic framework. In conclusion, combining multiple omics data sets in the public domain increases robustness and accuracy of the classification model that will ultimately improve disease understanding and clinical treatment decisions to benefit patients.
Availability and Implementation: An R package MetaKTSP is available online. (http://tsenglab.biostat.pitt.edu/software.htm).
Contact: ctseng@pitt.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
High-throughput experimental techniques, including microarray and massively parallel sequencing, have been widely applied to discover underlying biological processes and to predict the multi-causes of complex diseases (e.g. cancer diagnosis, Ramaswamy et al., 2001), prognosis (van de Vijver et al., 2002) and therapeutic outcomes, Ma et al., 2004). The associated data analysis has brought new statistical and bioinformatic challenges and many new methods have been developed in the past 15 years. In particular, methods for classification and prediction analysis (a.k.a. supervised machine learning) are probably the most relevant tools towards translational and clinical applications. Take breast cancer as an example, many expression-based biomarker panels have been developed [e.g. MammaPrint (van ’t Veer et al., 2002), Oncotype DX (Paik et al., 2004), Breast Cancer Index BCI (Zhang et al., 2013) and PAM50 (Parker et al., 2009)] for classification/prediction of survival, recurrence, drug response and disease subtype. Reproducibility analysis of these markers and classification models has been a major concern and has drawn significant attention to ensure clinical applicability of these panels (Garrett-Mayer et al., 2008; Kuo et al., 2006; MAQC Consortium et al., 2006; Mitchell et al., 2004; Sato et al., 2009; Marchionni et al., 2013; Ma et al., 2014). Many articles have focused on normalization, reproducibility of marker detection, inter-lab or inter-platform correlation concordance. For direct clinical utilities, more attention have shifted towards cross-study validation or inter-study prediction (i.e. a prediction model is established in one study and validated independently in a test study (Bernau et al., 2014; Cheng et al., 2009; Mi et al., 2010; Xu et al., 2008). Such an issue is critical for translating models from transcriptomic studies into a practical clinical tool. For example, the training cohort may have utilized an old Affymetrix U133 platform. A biomarker panel and a model are constructed and a test study from a different medical center using an RNA-seq platform is available. A successful machine learning model should retain high prediction accuracy in such inter-lab and inter-platform validation. We note that many normalization methods have been developed to adjust for systematic biases across studies, including distance weighted discrimination (Benito et al., 2004), cross-platform normalization (Shabalin et al., 2008) and Knorm correlation (Teng et al., 2007). But the normalization performance largely depends on whether the observed data structure fits the model assumptions. In most applications, researchers have applied meta-analysis methods and have avoided relying on effectiveness of normalization (Tseng et al., 2012). To compare the meta-analysis methods with mega-analysis (i.e. normalize across studies and directly merge data for inference) in this article, we only perform simple quantile normalization within each study and then standardize each sample to mean zero and unit SD before we adopt mega-analysis.
In addition to the issue of cross-study validation, it’s critical to select a robust and accurate machine learning method. In the literature, many supervised machine learning methods have been proposed and applied to high-throughput experimental data. For example, the CMA package allows easy implementation of 21 popular classification methods such as linear or quadratic discriminant analysis, lasso, elastic net, support vector machines (SVMs), random forest, PAM etc (Slawski et al., 2008). In addition to these popular methods, the top scoring pair (TSP) method (Afsari et al., 2014; Geman et al., 2004; Tan et al., 2005) is a straightforward prediction rule utilizing building blocks of rank-altered gene pairs in case and control comparison (see Section 2.1 for more details). The method is mostly rank-based without any model parameter. It is invariant to monotone data transformation and the feature selection and the model are more transparent for biological interpretation. Although TSP and its variant are robust methods that do not require normalization in cross-study validation, we have found that some of the selected TSPs from the training study may not reproduce in the test study possibly due to platform differences.
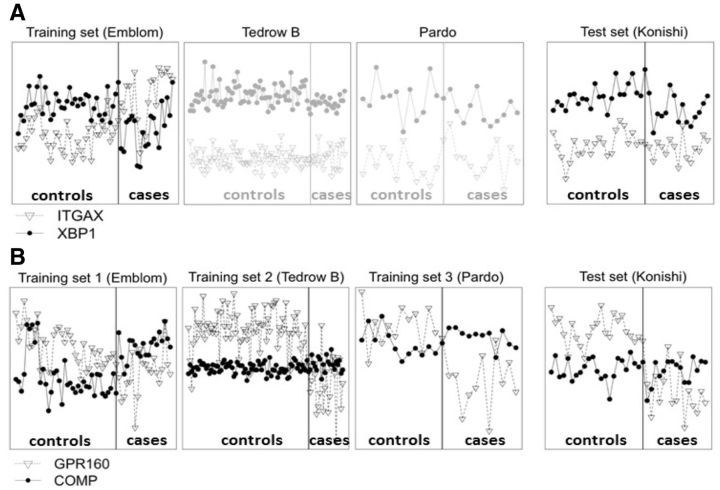
Figure 1A illustrates the expression levels of a good TSP gene pair, ITGAX and XBP1, identified from the first IPF (idiopathic pulmonary fibrosis) training study Emblom (see data descriptions in Supplementary Table S1). XBP1 is over-expressed than ITGAX in control samples but under-expressed in cases. If we use this TSP to validate in the test study Konishi, we find that XBP1 is over-expressed than ITGAX in both cases and controls and we obtain 0% sensitivity and 100% specificity (i.e. Youden index = sensitivity + specificity − 1 = 0). We found similar poor performance in two other studies Tedrow B and Pardo, showing that the TSP is likely a false positive. In Figure 1B, GPR160 is over-expressed than COMP in controls and under-expressed in cases for all three studies Emblom, Tedrow B and Pardo. It is a more reliable TSP across three studies and conceptually is less likely a false positive. Indeed, the cross-study validation in Konishi shows good performance with 80% Youden index. The two real examples in Figure 1 argue the potential of a meta-analytic approach by combining multiple training transcritomic studies to identify reliable TSPs so the resulting model has enhanced cross-study validation performance.
Fig. 1.
Two TSP examples from real data to show advantage of MetaTSP. X-axis and Y-axis refer to sample indices and gene expression levels, respectively. (A) Gene pair ITGAX/XBP1 has high TSP score (XBP1 > ITGAX in controls but ITGAX > XBP1 in cases) in the training ‘Emblom’ study but fail to replicate in the testing ‘Konishi’ study as well as the other two Tedrow B and Pardo studies. (B) Gene pair GPR160/COMP has high TSP scores (GPR160 > COMP in controls and COMP > GPR160 in cases) in all three training studies ‘Emblom’, ‘Tedrow B’ and ‘Pardo’. The gene pair is successfully validated in the testing ‘Konishi’ study
In this article, we propose three meta-analytic approaches for TSP method (MetaTSP) by combining information across multiple training studies using (i) averaged TSP scores (ii) combining P-values via Fisher’s method (Fisher 1925; 1948) (iii) combining P-values via Stouffers method (Stouffer 1949). To decide the number of TSPs used for model construction, a classical cross validation (CV) method and a variance optimization (VO) (Afsari et al., 2014) method are applied and compared. Simulations and three real omics data sets (two gene expression data on breast cancer and IPF, and one pan-cancer methylation data) are used to benchmark the cross-study validation performance.
2 Methods
2.1 TSP algorithm and kTSP
The original TSP algorithm was first proposed by Geman et al. (2004). Denote by data matrix the gene expression intensity of gene g (1 ≤ g ≤ G) in sample n (1 ≤ n ≤ N) and yn the class label of sample n. Particularly, we consider , representing controls and cases for binary classification in this article. For any gene pair i and j (1 ≤ i, j ≤ G), define the conditional ordering probability score for , where Xi and Xj are gene expression intensities of gene i and j. Intuitively, Tij(0) is the probability in controls that gene j has larger expression intensity than that of gene i and similarly Tij(1) is for cases. Given observed expression profile data matrix X, the probability scores can be estimated as , where I(⋅) is an indicator function that generates value one if the statement inside the parenthesis is true and zero otherwise. The discriminant score of the gene pair is defined as . Note that −1 ≤ Sij ≤ 1 always holds. When Sij = 1, expression of gene j is always greater than that in gene i in cases and expression of gene j is always smaller than that in gene i among controls. As a result, the ordering of gene i and gene j expression is predictive to the class label. On the contrary, if Sij = −1, gene j always has a smaller expression than gene i in cases and the relation is reversed in controls. In summary, the absolute value of Sij reflects the predictive value of the gene pair. The TSP algorithm seeks the best gene pair as the classifier. When multiple gene pairs give the same highest absolute score, the best pair that gives the largest differential magnitude Dij is chosen, where Dij = |dij(1) − dij (0)| and for , where Rin is the rank of the ith gene in the nth sample. When a new test sample is encountered in the future, the class prediction is determined by
By construction, the TSP classifier above is based on only one TSP (two genes) and the method can be very sensitive to slight noise perturbations (Geman et al., 2004). To circumvent this issue, Tan et al. (2005) introduced kTSP to combine multiple TSPs for a more stable algorithm. The method identified the sorted TSPs similar to above. Instead of choosing only the best TSP, it selected the top K (K is a parameter to be tuned) TSPs to construct the model. The TSPs were selected from the sorted list such that the genes in the TSPs had no overlap otherwise the latter TSPs containing overlapping genes would be skipped and the next TSP in the sorted list would be considered. In other words, the selected top K TSPs always contain 2K distinct genes. Suppose represents the K selected TSPs. The kTSP algorithm makes a prediction for a new test sample by . In a sense, the k-TSP is an ensemble classifier that aggregates multiple weak classifiers by majority vote (Opitz and Maclin, 1999). To avoid ties, we usually select odd numbers for K in binary classification.
The TSP algorithms have the following advantages for omics prediction analysis: (i) The method is non-parametric since the method is constructed based on the relative ranking of gene pairs. Since different transcriptomic studies are usually conducted in different labs and in different platforms, the applicability of non- parametric nature facilitates cross-study validation that we aim in this article. (ii) The method is based on one or a few gene pairs. The biological interpretation of the model and the translational application are more straightforward. It is more likely to succeed by designing a reproducible commercial assay for wider clinical applications, such as the 21-gene RT-PCR-based Oncotype DX test for breast cancer (Paik et al., 2004). (iii) Researchers have repeatedly found that the family of TSP algorithms provides good prediction performance in many transcriptomic data (Price et al., 2007; Raponi et al., 2008; Xu et al., 2005).
2.2 Estimate K for kTSP
To estimate the best K in the kTSP algorithm, we will apply and compare the following two methods.
2.2.1 Cross-validation (CV) method
In Tan et al. (2005), leave-one-out CV was used to determine K in kTSP. In each iteration, one sample was left out as the test sample. The remaining samples were used to construct a prediction model and apply to the test sample. The procedure was repeated until each sample was left out as the test sample once. The cross-validated error rates were then calculated for different selections of K and the best K that produced the smallest CV error rate was chosen.
2.2.2 VO method
Afsari et al. (2014) recently developed a VO method to estimate K in kTSP. Recall that , where . The kTSP algorithm searches for the optimized TSPs without overlapping genes:
Define the t-statistics of the target function:
K is chosen by the value that maximizes tkTSP (i.e. K* = arg maxKtkTSP). The VO procedure greatly reduced high computational demand in CV.
2.3 MetaKTSP algorithms
As mentioned in the introduction section, cross-study validation via MegaKTSP (i.e. naively combine multiple normalized data sets and apply kTSP) may not be suitable to identify a robust prediction gene pair. Alternatively, we propose a MetaKTSP framework below. Denote by the expression profile of study m, where represents the gene expression intensity of gene g (1 ≤ g ≤ G), sample n (1 ≤ n ≤ N(m)) in study m (1 ≤ m ≤ M). Let the discriminant score for gene i and j in study m take the difference of two averages of Bernoulli random variables. We started by developing three meta-analytic approaches (by Fisher score, Stouffer score and mean score) to choose the K non-overlapping TSPs for prediction model construction (denoted as ). When a new test sample, is encountered in the future, the class prediction by the kth TSP and study m is:
The final meta-analyzed class prediction is determined by
Below we introduce the three meta-analytic approaches to select the top K TSPs. In meta-analysis, test statistics (e.g. it t-statistics) across studies are not comparable and combining P-values has become a popular practice. Under the null hypothesis that gene i and j are not discriminant, can be well-approximated by Gaussian distribution since is the difference of two averages of independent Bernoulli trials. The two-sided P-value of is calculated as . Alternatively, one-sided p-values can be calculated as for left-sided P-value and for right-sided P-value.
2.3.1 Select K TSPs by Fisher’s method
The Fisher’s method combines P-values across studies by , where is the two-sided P-value of the discriminant score of gene i and j in study m. Under null hypothesis that gene i and j have no discriminant power in all studies, . This classical P-value combination procedure has a well-known problem that the discriminant scores across studies may have discordant signs but all with small two-sided P-values that generate a significant meta-analyzed P-value. To circumvent this discordant problem, we apply a one-sided test modification technique discussed in Owen (2009). Define and , where and are the left and right one-sided P-values of discriminant score of gene i and j in study m. The modified one-sided corrected Fisher’s statistic is . The top K gene pairs with the largest meta-analyzed Fisher score (i.e. ) and with no overlapping genes are selected.
2.3.2 Select K TSPs by Stouffer’s method
Instead of using log-transformation in Fisher’s method, Stouffer’s method applies an inverse normal transformation by . Under null hypothesis that gene i and j have no discriminant power in all studies, . The top K gene pairs with the smallest meta-analyzed two-sided P-values and with no overlapping genes are selected for prediction. Note that Stouffer’s method has an advantage over Fisher’s method that one-sided concordance correction is not necessary if one-sided P-values are input in the inverse normal transformation.
2.3.3 Select K TSPs by mean score
Because the discriminant score is difference of two conditional probabilities, the scores are directly comparable across studies and can be directly combined. We define the mean score to combine M studies. The top K gene pairs with the largest absolute value of the meta-analyzed scores (i.e. ) and with no overlapping genes are selected for prediction model construction. In addition, we propose the weighted mean score , where is sample size of study m. It is commonplace that each study has a range of sample size, and the variance of S(m) is increasingly influenced as sample size rises. Therefore, it is worth to adjust sample size to the total discriminant score.
2.4 Estimate K for MetaKTSP
Similar to Section 2.2, cross-validation and VO methods can be extended to estimate K for MetaKTSP.
2.4.1 Cross-validation
Each of the M studies are firstly split into V equal-sized subgroups. In each cross-validation, one subgroup of samples in each study is left out as the testing samples. The remaining (V − 1) subgroups are used as training samples to construct the classifier and then apply to the test sample. We choose the optimal K such that the highest average Youden index over M studies is obtained. In this article, we adopted 5-fold cross-validation.
2.4.2 Variance optimization
Motivated by Afsari et al. (2014), we define the following target function:
K is chosen by the value that maximizes (i.e. . We will show its equal or slightly improved performance compared with CV in our proposed meta-analytic scheme and this estimation method will be recommended in practice.
3 Results
3.1 Simulations
We hypothesize that if gene pairs are consistently identified with strong TSP scores over multiple training studies such gene pairs outperform original TSPs from a single study. We tested this hypothetical argument using simulated data sets. Below we describe simulated expression profiles under correlated gene structures to mimic real data sets. We performed a smaller scale of simulation with G = 200 genes and M = 4 transcriptomic studies, where the number of samples is randomly generated; (for study m (1 ≤ m ≤ M = 4) of sample subgroup j (i.e. j = 1 for controls and j = 2 for cases). Denote expression data matrix by for gene and 1 ≤ m ≤ M = 4.
3.1.1 Step 1. Simulate consensus predictive genes
(i) Consider consensus predictive genes that are expressed with a crossover pattern across two subgroups for all studies. For each of the two clusters c (1 ≤ c ≤ 2) in study m(1 ≤ m ≤ M) that contains consensus predictive genes, sample gene correlation structure for every gene cluster c and sample subgroup j of study m, where denotes inverse Wishart distribution, I is the identity matrix, and J is the matrix with all the entries being 1. Set vector as the square roots of the diagonal elements in . Calculate such that .
(ii) We simulate two clusters of consensus predictive genes, each containing 20 genes. The first down-regulated gene cluster is generated from , where sample u belongs to class j in study m and for j = 1 (controls) and for j = 2 (cases). This is a smaller effect size simulation. We also simulate a strong effect size simulation by for controls and cases. Similarly, the second up-regulated gene cluster is simulated from , where for controls and cases in weak signal scenario and and 1 in strong signal scenario. These 40 consensus predictive genes are the basis to aggregate predictive power across studies (red dotted rectangle in Supplementary Figure S1).
3.1.2 Step 2. Simulate study-specific predictive genes
We next simulate four clusters (m′ = 1,2,3,4) of study specific genes, each containing 10 genes. Each gene cluster has specific predictive power to the corresponding study m. The down-regulated genes are simulated from , where (1 ≤ m ≤ 4) are simulated similar to (1) of Step 1 and or –4 for controls and cases. For up-regulated predictive genes, we randomly sample from and for controls and cases. When , the gene cluster m′ has no predictive power in study m and is randomly sampled from N(0,1) (blue dotted rectangle in Supplementary Figure S1). These study-specific genes are a main source of errors in cross-study validation.
3.1.3 Step 3. Simulate non-informative genes
Finally, the remaining 80 non-informative genes are simulated by for 121 ≤ g ≤ 200.
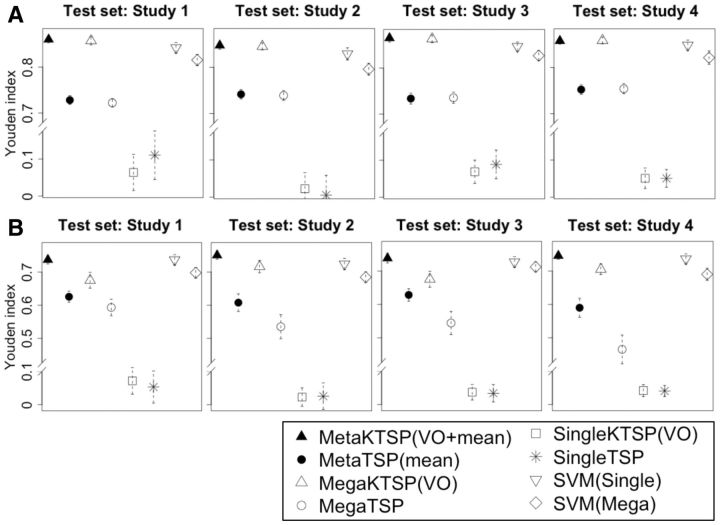
We repeated simulations for 50 times, and the results are benchmarked by averaged Youden index. Figure 2 shows the simulation evaluation for different methods using Youden index, and we tested MetaKTSP (VO + mean) and MetaTSP (mean). In each meta-analysis evaluation, we take one study out as the test study, combine the remaining three studies to select the TSPs and construct the model, and finally use the model to predict samples in the test study. The result of Figure 2A in the weaker signal setting () shows that the MetaKTSP (VO + mean) method performed well (Youden Index = 0.857–0.865). The MetaTSP (mean) performed slightly worse (Youden Index = 0.734–0.752). In mega-analysis approaches, the three training studies are normalized and combined into one study to construct the prediction model and evaluate in the test study. In single study analysis, the accuracy was evaluated by averaging inter-study accuracy from each of the three training studies to the test study. The result of Figure 2B (; weak signal scenario) clearly shows inferior performance of MegaKTSP and MegaTSP approaches, and poor performance of single study KTSP and TSP approaches. The single study SVM and mega-analysis of SVM also performed slightly worse than MetaKTSP in Figure 2A and B. Taken together, this confirms our hypothesis that prediction model from a single study may not be robust and accurate. Proper meta-analysis by combining multiple training studies improves the stability and accuracy of the model to predict an independent test study. Supplementary Figure S2A and B contain simulation results of all meta- and mega-analytic methods in strong and weak signal cases. In the weaker signal case in Supplementary Figure S2B (), we found that MetaKTSP using Fisher’s selecting approach often has inferior performance than Stouffer and mean methods. This is probably because of the nature of heavy tail log-transformation in the Fisher’s method. A P-value close to 0 (e.g. 1E-20) can contribute a very large score in Fisher’s method and can easily dominate the analysis. The inverse transformation in Stouffer’s method and the mean score approach somewhat alleviated the problem. From Supplementary Figure S2A and B, it is evidently shown that MetaKTSP (mean) is superior (or equal at least) to weighted MetaKTSP (weighted.mean). Interestingly, even if the parameter for mean () decreases in value (1–0.8), the order of Youden Index largely remains the same. We conclude that MetaKTSP (VO + mean) generally outperformed the other methods, and so chose to apply this method in the following real applications.
Fig. 2.
(A,B) show results of inter-study prediction using four simulated data sets (A: , B: ; and ) Y-axis represents the average Youden index. The bar plots indicate the standard error of estimated Youden index
3.2 Application to genomic data sets
Below we demonstrate application of MetaKTSP methods to three real omics examples of breast cancer expression profiles (1658 samples in seven studies), IPF expression profiles (IPF; 291 samples in six studies) and The Cancer Genome Atlas multi-cancer methylation profiles (TCGA, http://cancergenome.nih.gov/; 1785 samples in six studies). Supplementary Table S1 provides detailed data description of all 19 studies and their data sources. Genes and methylation probes were matched across studies. Non-expressed and/or non-informative genes were filtered according to the rank sum of mean intensities and variances across studies. Note that this filtering procedure has been used in a previous meta-analysis work (Wang et al., 2012) and the filtering is unbiased in the prediction accuracy estimate since class labels are not used in the procedure. This generated 3035 genes in breast cancer, 3010 genes in IPF and 3061 methylation probes in TCGA for down-stream prediction analysis.
From simulation, VO feature selection method performed slightly better than CV method so it was applied to all TSP methods to determine K in real data. We tested Meta-KTSP (mean), Meta-KTSP (Stouffer), and single- and mega-variations of KTSP and five popular machine learning methods, including linear discriminant analysis, CART, K-nearest-neighbor, random forest and SVMs. The complete result is shown in Supplementary Table S4. Figure 3 shows the inter-study prediction performance of selected methods of the three real examples (A, breast cancer ER+ versus ER− prediction by expression profiles; B, IPF versus controls prediction by expression profiles; C, cancer versus adjacent normal prediction by methylation profiles). Mega-SVM was the best performer among the five existing machine learning methods tested so we chose to present Mega-SVM and Single-SVM in Figure 3. For single study analysis, we performed all pairs of cross-study validation and averaged the performance. For mega-analysis, each sample was standardized to mean zero and unit variance and multiple studies were merged for analysis. Finally, we aggregated Youden indexes of all studies using weighted average by sample size (last plot in each row). In Figure 3, MetaKTSP (VO + mean) obviously best performed inter-study prediction of all three examples, whereas mega-analysis methods had worse performance and single study analysis without combining information across studies performed the worst. In the example of breast- and pan-cancer analysis, the performance of single study analysis was below random guess (Youden index < 0). This suggests that prediction models from single study analysis mostly reflected study-specific (cancer-specific) signature that could not be generalized to other cancers. In addition, to assess robustness of MetaKTSP (VO + mean), we performed 50 simulations of bootstrapped samples and applied Meta-KTSP and single study kTSP and calculated the degree of robustness by calculating the number of overlapping TSPs between bootstrapped data analysis and whole data analysis divided by the number TSPs detected by whole data analysis. Supplementary Figure S4 and Supplementary Table S2A and B clearly showed greater robustness of MetaKTSP than individual study kTSP analysis in selecting top gene pairs. Supplementary Figure S6 provides further insight on this concept. In Supplementary Figure S6A, nine TSPs were selected in individual training studies (Breast invasive carcinoma, Colon adenocarcinoma, Kidney renal clear cell carcinoma, Lung adenocarcinoma and Stomach adenocarcinoma), respectively. When these TSPs were evaluated in the ovarian cancer (OV) study, the absolute discriminant scores dropped significantly, many of which dropped from close to 1 to below 0.5. On the contrary, the nine TSPs selected by meta-analysis shared universally large discriminant scores for all five training studies (Supplementary Figure S6B) and the discriminant scores were mostly maintained in the test OV study. Supplementary Figure S3A–C provides the full results of all 15 methods comparison in the 3 examples.
Fig. 3.
Three examples of Inter-study prediction with applications to real data sets (A, breast cancer: ER+ versus ER−; B, Idiopathic pulmonary fibrosis; C, Six differents cancers in TCGA. Y-axis represents the average Youden index
It is interesting to note that Emblom and Larsson studies in the IPF examples had almost none predictive value (Youden index near 0), while the other four studies performed well. This argues that the two studies might have heterogeneous cohorts from the other four studies or they may have worse experimental quality [see similar quality control result in (Kang et al., 2012) for the same data sets]. In practice, one may perform such CV to exclude potential ‘outlier’ studies before implementing MetaKTSP.
Below we explore biological validation of detected gene pairs from MetaKTSP using existing literature. We first applied MetaKTSP (VO + mean) to all seven breast cancer studies and identified nine TSPs. For the 18 genes in the 9 detected TSPs, 12 of them were found to associate with ER expression in previous publications and all of them had consistent differential expression direction compared with the microarray data (Table 1). For the pan-cancer methylation result, we also identified 9 TSPs and 15 of the 18 genes have been previously indicated as cancer related (Supplementary Table S3). For example, the PCDH8 gene from the fourth gene pair was previously confirmed as a candidate tumor suppressor regulated by methylation in multiple cancers: (i) Kidney cancer: frequent promoter region methylation (58%) in primary renal cell carcinoma tumor samples (Morris et al., 2011). (ii) Breast cancer: either mutation or epigenetic silencing in a high fraction of breast carcinomas inactivates PCDH8 that leads to oncogenesis in cancers (Yu et al., 2008) (iii) Stomach cancer: tumor suppressor function in gastric cancer (Zhang et al., 2012).
Table 1.
The list of nine identified gene pairs of Average MetaKTSP and the existing breast cancer gene signatures
| Label | Gene1 | Gene2 | Averaged scores | References |
|---|---|---|---|---|
| Pair 1 | E2F3 (ER−) | GATA3 (ER+) | −0.710 | Tordai et al. (2008, E2F3, ER−), Usary et al. (2004, GATA3, ER+) |
| Pair 2 | ODC1 (ER−) | DNALI1 (ER+) | −0.669 | Parris et al. (2010, DNALI1, ER+) |
| Pair 3 | LAD1 (ER−) | SCCPDH (ER+) | −0.656 | Dvorkin-Gheva and Hassell (2011, SCCPDH, ER+), Smith et al. (2008, LAD1, ER−) |
| Pair 4 | SRPK1 (ER−) | MYB (ER+) | −0.649 | van Roosmalen et al. (2015, SRPK1, ER−) |
| Pair 5 | DACH1 (ER+) | FOXC1 (ER−) | 0.644 | Powe et al. (2014, DACH1, ER+), Ray et al. (2010, FOXC1, ER−) |
| Pair 6 | WARS (ER−) | FBP1 (ER+) | −0.637 | van’t Veer et al. (2002, FBP1, ER+) |
| Pair 7 | RNASEH1 (ER−) | MAGED2 (ER+) | −0.632 | Thakkar et al. (2010, MAGED2, ER+) |
| Pair 8 | CDCA8 (ER−) | AFF3 (ER+) | −0.629 | Thakkar et al. (2010, AFF3, ER+) |
| Pair 9 | MRFAP1L1 (ER+) | KCMF1 (ER−) | 0.625 | Symmans et al. (2010, KCMF1, ER−) |
4 Conclusion and discussion
As high-throughput experimental data become more and more prevalent and publicly available, integrative methods to fully utilize information from the abundant multi-lab data sets have become critical. Generating predictive biomarkers and classification model from a single study often suffer from limited sample size and possibly study-specific biases. The resulting models are often found with poor performance in cross-study validation (Correa and Reis-Filho, 2009; McShane et al., 2013; Kern, 2012; Reid et al., 2005). To improve translational and clinical utility of the biomarker discovery and classification model construction, combining information from multiple studies provide a promising opportunity. In this article, we seek to improve a TSP method that is a non-parametric, accurate and easily interpretable model that likely will succeed in cross-study validation for clinical applications. We developed three MetaKTSP approaches that combine multiple omics data sets to improve the credibility of TSP biomarker selection. Using simulations and real transcriptome and methylome data sets, we demonstrate its improved performance on cross-study validation. We compared two methods, CV and VO, to decide the number of TSPs used in the model construction. The result showed similar performance of the two model selection methods. Since VO does not involve repeated subsampling and is computationally faster, we recommend to use VO for future applications.
There are a few limitations and future directions to consider. First, our method and evaluation focus on binary case-control classification. The method could be extended to multi-class classification scenario. Second, biological knowledge such as pathways or known disease relevant genes can be incorporated to enhance the TSP discovery accuracy. For example, Oncotype DX started with 250 breast cancer related genes to identify the 21 predictive genes in their panel. Although this runs the risk to miss understudied but significant biomarkers, this approach can potentially improve cross-study validation in well-studied diseases. Third, we may take into account the original differences across platforms to pursue more accurate meta-analysis. In particular, gene expression platforms may measure different genes on different scales. Therefore, it is worth to match up genes across platforms by mapping onto identical exon sites and probes. Finally, the current TSP approaches may be extended towards module-based prediction scheme where TSPs of gene modules are sought to provide extra redundancy and robustness (Mi et al., 2010). The ‘MetaKTSP’ R package is available on the authors website and is part of MetaOmics, a software suite for omics data meta-analysis of differentially expressed gene detection, pathway, prediction, clustering, classification and network analyses.
Supplementary Material
Funding
The authors are supported by R01: RO1CA190766 and the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (NRF-2013R1A1A2008619).
Conflict of Interest: none declared.
References
- Afsari B. et al. (2014) Rank discriminants for predicting phenotypes from RNA expression. Ann. Appl. Stat., 8, 1469–1491. [Google Scholar]
- Benito M. et al. (2004) Adjustment of systematic microarray data biases. Bioinformatics, 20, i105–i114. [DOI] [PubMed] [Google Scholar]
- Bernau C. et al. (2014) Cross-study validation for the assessment of prediction algorithms. Bioinformatics, 30, i105–i112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C. et al. (2009) Ratio adjustment and calibration scheme for gene-wise normalization to enhance microarray inter-study prediction. Bioinformatics, 25, 1655–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa G., Reis-Filho J. (2009) Microarray-based gene expression profiling as a clinical tool for breast cancer management: are we there yet? Int. J. Surg. Pathol., 17, 285–302. [DOI] [PubMed] [Google Scholar]
- Dvorkin-Gheva A., Hassell J. (2011) Hormone receptor and ERBB2 status in gene expression profiles of human breast tumor samples. Plos One, 6, e26023.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R.A. (1925) Statistical Methods for Research Workers. Oliver and Boyd (Edinburgh). [Google Scholar]
- Fisher R. (1948) Questions and answers #14. Am. Stat., 2, 30–31. [Google Scholar]
- Garrett-Mayer E. et al. (2008) Cross-study validation and combined analysis of gene expression microarray data. Biostatistics, 9, 333–354. [DOI] [PubMed] [Google Scholar]
- Geman D. et al. (2004) Expression Profiles from Pairwise mRNA Comparisons. Stat. Appl. Genet. Mol. Biol., 3, article 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern S. (2012) Why your new cancer biomarker may never work: recurrent patterns and remarkable diversity in biomarker failures. Cancer Res., 72, 6097–6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D. et al. (2012) MetaQC: objective quality control and inclusion/exclusion criteria for genomic meta-analysis. Nucleic Acids Res., 40, e15.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo W. et al. (2006) A sequence-oriented comparison of gene expression measurements across different hybridization-based technologies. Nat. Biotechnol., 24, 832–840. [DOI] [PubMed] [Google Scholar]
- Ma S. et al. (2014) Measuring the effect of inter-study variability on estimating prediction error. PLoS One, 9, e110840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X. et al. (2004) A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell, 5, 607–616. [DOI] [PubMed] [Google Scholar]
- Marchionni L. et al. (2013) A simple and reproducible breast cancer prognostic test. BMC Genomics, 17, 336.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAQC Consortium. et al. (2006) The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat. Biotechnol., 24, 1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McShane L., Polley M. Development of omics-based clinical tests for prognosis and therapy selection: the challenge of achieving statistical robustness and clinical utility. Clin. Trials, 10, 653–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi Z. et al. (2010) Module-based prediction approach for robust inter-study predictions in microarray data. Bioinformatics, 26, 2586–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell S. et al. (2004) Inter-platform comparability of microarrays in acute lymphoblastic leukemia. BMC Genomics, 5, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris M. et al. (2011) Genome-wide methylation analysis identifies epigenetically inactivated candidate tumour suppressor genes in renal cell carcinoma. Oncogene, 30, 1390–1401. [DOI] [PubMed] [Google Scholar]
- Opitz D., Maclin R. (1999) Popular ensemble methods: an empirical study. J. Artif. Intell. Res., 11, 169–198. [Google Scholar]
- Owen A.B. (2009) Karl Pearson’s meta-analysis revisited. Ann. Stat., 37, 3867–3892. [Google Scholar]
- Paik S. et al. (2004) A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med., 351, 2817–2826. [DOI] [PubMed] [Google Scholar]
- Parker J. et al. (2009) Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol., 27, 1160–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parris T. et al. (2010) Clinical implications of gene dosage and gene expression patterns in diploid breast carcinoma. Clin. Cancer Res., 16, 3860–3874. [DOI] [PubMed] [Google Scholar]
- Powe D. et al. (2014) DACH1: its role as a classifier of long term good prognosis in luminal breast cancer. PLoS One, 9, e84428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price N. et al. (2007) Highly accurate two-gene classifier for differentiating gastrointestinal stromal tumors and leiomyosarcomas. Proc. Natl. Acad. Sci. USA, 104, 3414–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S. et al. (2001) Multiclass cancer diagnosis using tumor gene expression signatures. Proc. Natl. Acad. Sci. USA, 15149–15154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raponi M. et al. (2008) A 2-gene classifier for predicting response to the farnesyltransferase inhibitor tipifarnib in acute myeloid leukemia. Blood, 5, 2589–2596. [DOI] [PubMed] [Google Scholar]
- Ray P. et al. (2010) FOXC1 is a potential prognostic biomarker with functional significance in basal-like breast cancer. Cancer Res., 70, 3870–3876. [DOI] [PubMed] [Google Scholar]
- Reid J. et al. (2005) Limits of predictive models using microarray data for breast cancer clinical treatment outcome. J. Natl. Cancer Inst., 97, 927–930. [DOI] [PubMed] [Google Scholar]
- Sato F. et al. (2009) Intra-platform repeatability and inter-platform comparability of microRNA microarray technology. PLoS One, 4, e5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalin A. et al. (2008) Merging two gene-expression studies via cross-platform normalization. Bioinformatics, 24, 1154–1160. [DOI] [PubMed] [Google Scholar]
- Stouffer S. (1949) The American Soldier: Adjustment during Army Life, Vol. 1. Princeton: Princeton University Press. [Google Scholar]
- Slawski M. et al. (2008) CMA: a comprehensive Bioconductor package for supervised classification with high dimensional data. BMC Bioinformatics, 9, 439.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. et al. (2008) Meta-analysis of breast cancer microarray studies in conjunction with conserved cis-elements suggest patterns for coordinate regulation. BMC Bioinformatics, 28, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symmans W. et al. (2010) Genomic index of sensitivity to endocrine therapy for breast cancer. J. Clin. Oncol., 28, 4111–4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan A. et al. (2005) Simple decision rules for classifying human cancers from gene expression profiles. Bioinformatics, 21, 3896–3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S. et al. (2007) A statistical framework to infer functional gene associations from multiple biologically interrelated microarray experiments. J. Am. Stat. Assoc., 104, 465–473. [Google Scholar]
- Thakkar A. et al. (2010) Identification of gene expression signature in estrogen receptor positive breast carcinoma. Biomark. Cancer, 2, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tordai A. et al. (2008) Evaluation of biological pathways involved in chemotherapy response in breast cancer. Breast Cancer Res., 10, R37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng G. et al. (2012) Comprehensive literature review and statistical considerations for microarray meta-analysis. Nucleic Acids Res., 40, 3785–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usary J. et al. (2004) Mutation of GATA3 in human breast tumors. Oncogene, 23, 7669–7678. [DOI] [PubMed] [Google Scholar]
- van de Vijver M. et al. (2002) A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med., 347, 1999–2009. [DOI] [PubMed] [Google Scholar]
- van Roosmalen W. et al. (2015) Tumor cell migration screen identifies SRPK1 as breast cancer metastasis determinant. J. Clin. Invest., 125, 1648–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van’t Veer L. et al. (2002) Gene expression profiling predicts clinical outcome of breast cancer. Nature, 415, 530–536. [DOI] [PubMed] [Google Scholar]
- Wang X. et al. (2012) Detecting disease-associated genes with confounding variable adjustment and the impact on genomic meta-analysis: With application to major depressive disorder. BMC Bioinformatics, 13, 13–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L. et al. (2005) Robust prostate cancer marker genes emerge from direct integration of inter-study microarray data. Bioinformatics, 20, 3905–3911. [DOI] [PubMed] [Google Scholar]
- Xu L. et al. (2008) Merging microarray data from separate breast cancer studies provides a robust prognostic test. BMC Bioinformatics, 9, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. et al. (2008) PCDH8, the human homolog of PAPC, is a candidate tumor suppressor of breast cancer. Oncogene., 27, 4657–4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D. et al. (2012) Frequent silencing of protocadherin 8 by promoter methylation, a candidate tumor suppressor for human gastric cancer. Oncol. Rep., 28, 1785–1791. [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. (2013) Breast cancer index identifies early-stage estrogen receptor-positive breast cancer patients at risk for early- and late-distant recurrence. Clin. Cancer Res., 19, 4196–4205. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.