Abstract
Several neurodegenerative diseases including Alzheim er’s Disease (AD) are characterized by ubiquitin-positive pathological protein aggregates. Here, an immunoaffinity approach is utilized to enrich ubiquitylated isopeptides after trypsin digestion from five AD and five age-matched control postmortem brain tissues. Label-free MS-based proteomic analysis identified 4,291 unique ubiquitylation sites mapping to 1,682 unique proteins. Differential enrichment analysis showed that over 800 ubiquitylation sites were significantly altered between AD and control cases. Of these, approximately 80% were increased in AD, including seven polyubiquitin linkages, which is consistent with proteolytic stress and high burden of ubiquitylated pathological aggregates in AD. The microtubule associated protein Tau, the core component of neurofibrillary tangles, had the highest number of increased sites of ubiquitylation per any protein in AD. Tau polyubiquitylation from AD brain homogenates was confirmed by reciprocal co-immunoprecipitation and by affinity capture using tandem ubiquitin binding entities (TUBEs). Co-modified peptides, with both ubiquitylation and phosphorylation sites, were also enriched in AD. Notably, many of the co-modified peptides mapped to Tau within KXGS motifs in the microtubule binding repeat suggesting that cross-talk between phosphorylation and ubiquitylation occurs on Tau in AD. Overall, these findings highlight the utility of MS to map ubiquitylated substrates in human brain and provides insight into mechanisms underlying pathological protein posttranslational modification in AD.
Keywords: mass spectrometry, neurodegeneration, post-translational modifications, protein aggregation, proteomics, ubiquitylation
1. Introduction
Ubiquitylation is a posttranslational modification (PTM) modulating multiple cellular pathways including maintenance of cellular protein homeostasis, cell signaling, transcription, DNA repair and cell division[1, 2]. The covalent conjugation of ubiquitin to primarily the ε amino group of lysine residues is achieved by sequential activities of ubiquitin activating (E1), ubiquitin conjugating (E2) and ubiquitin ligase (E3) enzymes and scaffolds[3, 4]. Ubiquitin signaling is highly conserved among eukaryotes[5] and, to date, 3 E1s, over 40 E2s and more than 600 E3 ligases have been described[4]. Ubiquitylation regulates cellular processes by modulating protein turnover, activity, protein-protein interactions and sub-cellular localization[6]. Both the proteasome and autophagy degradation pathways rely on ubiquitin signaling for selectively targeting substrates for degradation[7–9]. The importance of ubiquitin signaling is illustrated by impairment in protein homeostasis which is linked with protein aggregation in proteinopathies[1, 10]. Namely, several neurodegenerative diseases including Alzheimer’s Disease (AD), frontotemporal lobar degeneration (FTD), Parkinson’s disease (PD), Huntington’s disease (HD) and corticobasal degeneration (CBD) have characteristic ubiquitin positive pathological protein aggregates [8, 10, 11]. One of the core features of T auopathies including AD is the accumulation of neurofibrillary tangles (NFTs) composed of the microtubule-associated protein Tau (MAPT)[12–14]. A key physiological functions of Tau is to stabilize microtubules, however, in pathological conditions, Tau is heavily phosphorylated and forms insoluble NFTs that no longer stabilize microtubules[14, 15]. Although Tau phosphorylation is a robust marker of NFT pathology, recent studies emphasize the role of other PTMs such as ubiquitylation[12, 16], acetylation[17, 18], and O-GlcNAcetylation[19]. However, global ubiquitylation substrates and ubiquitylation sites in AD brain is lacking. Thus, mapping the brain ubiquitylome associated in AD brain will contribute towards our understanding of the underlying proteostasis mechanisms as well as reveal potential diagnostic biomarkers that translate into cerebrospinal fluid (CSF).
MS based quantitative proteomics is a powerful tool for the identification and quantification of proteins and their PTMs from complex protein mixtures[20, 21–23]. Mapping of ubiquitin sites by MS relies on identification of the last two remnant glycine residues (di-Gly) of ubiquitin on lysine residues following trypsin digestion1[21, 22]. A di-Gly remnant on lysine blocks trypsin cleavage producing miscleaved isopeptides with a mass shift of 114.043 Da, which is resolved by high resolution MS[21, 22]. However, the identification of ubiquitin substrates using MS is challenging due to the low stoichiometry of ubiquitin modified isopeptides in complex protein extracts such as brain homogenate[23, 24]. To overcome this limitation, many enrichment techniques have been developed including expression of epitope tagged ubiquitin[25], ubiquitin binding antibodies and ubiquitin binding domains[26], which enrich both conjugated and free ubiquitin[25, 27]. However, a major breakthrough was reached with the development of antibodies specific to peptides carrying the di-Gly remnant (K-ε-GG) that enabled robust and reliable identification and quantification of ubiquitylated isopeptides from cells and tissue extracts[23, 28].
Here, we analyzed human control and AD postmortem brain tissue samples to assess both total changes to the proteome and ubiquitylome associated with AD neuropathology. Coupling immunoaffinity enrichment of ubiquitylated isopeptides using a di-Gly remnant specific antibody[23] with label-free MS-based proteomics revealed 4291 unique ubiquitin modified sites mapping to 1682 unique proteins. Differential enrichment analysis revealed that over 800 di-Gly sites were significantly altered between AD and control cases. Of these, approximately 80% were increased in AD, which included the seven poly-ubiquitin linkages. We also mapped 28 sites of ubiquitylation on Tau, the core component of neurofibrillary tangles in AD, which also had the highest number of increased di-Gly isopeptides. Finally, additional database search results identified enrichment of co-modified isopeptides, having both ubiquitylation and phosphorylation sites, in AD. Notably, many of the co-modified Tau isopeptides carry KXGS motifs that influence Tau microtubule binding propensity and aggregation[29]. Overall, the study highlights the utility of MS to perform comprehensive mapping of the human brain ubiquitylome changes in AD brain, which provides insight into underlying proteostasis pathways and targets that may have potential as novel diagnostic biomarkers.
2. Experimental Section
2.1. Human postmortem brain tissues
All human postmortem brain tissues were obtained from Emory’s Alzheim er’s Disease Research Center (ADRC) Brain Bank. Frontal cortex brain tissue samples from five AD and five control cases, matched for age, gender and post-mortem interval (PMI), were selected for the study. Cases were selected based on Braak and CERAD scores which are neuropathologic measures of NFTs and amyloid plaque burden, respectively. Clinical and pathologic traits for all subjects are provided in Table S1.
2.2. Tissue homogenization and enzymatic digestion
Tissue samples were prepared essentially as described before with slight modifications[30]. Postmortem human frontal cortex brain samples from five AD cases and five healthy controls were homogenized in 8M urea lysis buffer (8M urea, 100 mM NaHPO4 buffer system, pH 8.5, supplemented with HALT protease and phosphatase cocktail inhibitors (Thermo Fisher Scientific, # 78440) using a Bullet Blender (Next Advance) per manufacturer’s protocol. Each tissue sample was placed in 1.5 ml Rino tube containing 750 mg stainless steel beads (0.9–2 mm in diameter). 500 μl 8M urea lysis buffer was added to each tissue and blended twice for 5 min at 4°C. Homogenates were transferred to clean Eppendorf tubes and centrifuged at 10,000×g for 5 min and sonicated (Sonic Dismembrator, Fisher Scientific) 3 times for 5 s with 15 s intervals of rest at 30% amplitude to disrupt nucleic acids. Protein concentration was determined by the bicinchoninic acid (BCA) method. Protein homogenates were diluted with 50 mM NH4HCO3 to approximately 2 M urea concentration and reduced using 1 mM 1,4-dithiothreitol (DTT) for 30 min and alkylated with 5 mM iodoacetamide (IAA) for 30 min in the dark. Proteins were digested with Lys-C (Wako; 1:100 enzyme: substrate ratio) at room temperature for 3 h followed by further overnight digestion with trypsin (Promega; 1:50 enzyme: substrate ratio) at room temperature. Tryptic peptides were subsequently acidified using 1% formic acid (FA) and 0.1% trifluoroacetic acid (TFA) before desalting and purification using Sep-Pak C18 columns (Waters) followed by peptide elution in 50% acetonitrile.
2.3. Immunoaffinity enrichment of di-Gly isopeptides
Ubiquitin modified isopeptides were enriched with a di-Gly remnant specific antibody (PTMScan® Ubiquitin Remnant Motif (K-ε-GG) Kit; Cell Signaling, # 5562) according to manufacturer’s protocol. The specificity of this antibody for K-ε-GG isopeptide in the absence or presence of proteasome inhibition has been described[31]. Briefly, 10 mg of urea homogenates from control (n=5) and AD (n=5) cases were enzymatically digested and desalted as described above. Peptides eluted in 50% acetonitrile were frozen at −80°C overnight followed by drying using lyophilization. Peptides were resuspended in immunoaffinity purification (IAP) buffer (50 mM MOPS, pH 7.2, 10 mM sodium phosphate, 50 mM NaCl) followed by incubation with K-ε-GG antibody conjugated beads for 2 hr at 4°C. Beads were washed with IAP buffer four times and affinity enriched peptides were eluted using 50 μl of 0.15% TFA (v/v) twice followed by desalting using C18 StageTips before LC-MS/MS analysis.
2.4. Liquid Chromatography Tandem Mass Spectrometry
Peptides were reconstituted in peptide loading buffer (0.1% TFA) containing 0.2 pmol of isotopically labeled peptide calibrants (Thermo Fisher Scientific, #88321). Approximately 1 μg of peptide was separated on homemade C18 fused silica column (75 μm internal diameter). Elution was performed over a 140-min gradient at a rate of 400 nL/min with a solvent ranging from 3% to 80% (buffer A: 0.1% formic acid in water, buffer B: 0.1% formic acid in acetonitrile). Peptides were analyzed on an Orbitrap Fusion Tribrid Mass Spectrometer (ThermoFisher Scientific). The mass spectrometer duty cycle was programmed to collect at top speed with 3 second cycles. The MS scans (400–1500 m/z range, 50 ms maximum injection time) were collected at a resolution of 120,000 and ACG target of 200,000 ion counts in profile mode. Higher-energy collision dissociation (HCD) MS/MS spectra (1.6 m/z isolation window, 0.5 m/z offset, 30% collision energy, 10,000 ACG target, 35 ms maximum ion time) were detected in the ion trap. Dynamic exclusion was set to exclude previously sequenced precursor ions for 30 s within a 10 ppm window. Precursor ions with charge states 2–7 were included. Each di-Gly isopeptide enriched sample was analyzed in technical replicate.
2.5. Label-Free Protein Quantification
Raw files for total proteome and ubiquitylome (di-Gly enriched) were searched using MaxQuant integrated Andromeda search engine (versions 1.6.0.1 or 1.5.3.3[32]). A separate search was performed for doubly modified peptides with both ubiquitylation and phosphorylation sites using MaxQuant (version 1.5.5.1). UniProt protein sequences containing both Swiss-Prot and TrEMBL human protein sequences (90,411 target sequences downloaded April 21, 2015), were duplicated into a reverted (decoy) peptide database, searched, and used to control peptide and razor protein false discovery rate (FDR) at 1% within MaxQuant. Methionine oxidation (+15.9949 Da), asparagine and glutamine deamidation (+0.9840 Da), N-terminal acetylation (+42.0106 Da) and cysteine carbamidomethylation (+57.0215 Da) were assigned as fixed modifications. Ubiquitylation (+114.043Da) was assigned as a variable modification for di-Gly peptide search. For doubly modified peptide searches, both ubiquitylation and phosphorylation (+79.9663Da) were included as variable modifications on lysine and serine, threonine, or tyrosine, respectively. Tryptic peptides with 2 or 5 miscleavages were included for total proteome or ubiquitin-modified proteome database search, respectively. A precursor mass tolerance of ±20 ppm was applied prior to mass accuracy calibration and ±4.5 ppm after internal MaxQuant calibration. Other search settings included a maximum peptide mass of 6,000 Da, a minimum peptide length of 6 residues, 0.6 Da tolerance for low resolution MS/MS scans obtained in the linear ion trap. The false discovery rate (FDR) for peptide spectral matches, proteins, and site decoy fraction were all set to 1 %. The label-free (LFQ) algorithm in MaxQuant was used for protein quantitation as previously described[33]. Missing values were imputed assuming informative missingness such that missing values were replaced with a left Gaussian tail random distribution per parameters previously determined ideal for LFQ based studies[34]. All raw files and MaxQuant search output data files are available on Synapse https://www.synapse.org/#!Synapse:syn11931357 (doi: 10.7303/syn11931344).
2.6. Gene Ontology (GO) Enrichment and Hierarchical Clustering Analysis
Functional enrichment of differentially expressed proteins was determined using the GO-Elite (v1.2.5) python package[35]. GO-Elite Hs (human) databases were downloaded on or after June 2016. The set of all proteins identified in the proteomic analyses was used as the background. Z score and one tailed Fisher’s exact test (Benjamini-Hochberg FDR corrected) was used to assess the significance of the Z score. Peptides were collapsed to one gene symbol and each gene symbol was equally weighted regardless of the number of matching peptides. Z score cut-off of 1.96, P value cut off 0.01 and a minimum of 5 genes per ontology were used as filters prior to pruning the ontologies. Clustering analysis on differentially abundant ubiquitin modified peptides was performed with the R NMF package in Microsoft R open V3.3. Age, sex and PMI-regressed log2 abundances were converted to Z scores (mean centered abundance, fold of SD) and clustered with Euclidian distance metric, complete method of the hclust function called from NMF package aheatmap function.
2.7. Validation of Tau Ubiquitylation
Healthy control (n=4) and AD (n=4) postmortem frontal cortex brain tissues were homogenized in NP-40 lysis buffer (25 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 5% glycerol, HALT cocktail protease and phosphatase inhibitors, 5 mM iodoacetamide, pH 7.5) using a bullet blender according to manufacturer’s instructions followed by centrifugation at 10,000 × g for 10 min at 4°C. For immunoprecipitation assay, 1 mg of protein from each brain sample was first pre-cleared using Protein A-Sepharose beads (Invitrogen #101041). Samples were incubated with a primary antibody (Tau5, Invitrogen (MA5–12808) or FK2, Enzo (BML-PW8810)) overnight at 4°C. Purified Mouse IgG2a K isotype (BD pharmingen # 550339) was used as an isotype-matched negative control. Lysates were subsequently incubated with DynaBeads-Protein G (Invitrogen #1003D) beads for 1 hr. Beads were then washed 3 times using wash buffer (50 mM Tris HCl, pH 8, 150 mM NaCl and 1% NP-40). Bound protein complexes were eluted by boiling in Laemmli sample buffer (65.8 mM Tris-HCL, 26.3% Glycerol, 2% SDS, 0.02% bromophenol blue) at 98°C for 5 min followed by immunoblot analysis. For polyubiquitylated proteins pull-down assay, 1 mg of proteins from each control and AD (n=2 each) brain homogenate were pre-cleared as described above. Pre-cleared brain homogenates were incubated with 2 μg of purified tandem ubiquitin binding entities (Biotin-TUBE1, LifeSensors, cat #UM301) overnight at 4°C followed by incubation with streptavidin-magnetic beads (Invitrogen, 11205D) for 1 h at room temperature. Beads were then washed with wash buffer followed by 5 min boiling in Laemmli sample buffer for immunoblot analysis.
2.8. Immunoblot Analysis
Immunoblotting was prepared essentially as described with slight modifications[36]. Protein extracts boiled in Laemmli sample buffer were resolved on Bolt 4–12% Bis-Tris gels (ThermoFisher Scientific) followed by transfer to nitrocellulose membrane using iBlot 2 dry blotting system (ThermoFisher Scientific). Membranes were incubated with blocking buffer for 30 min followed by overnight primary antibody incubation, washed with TBST (Tris-buffered saline, 0.1% Tween 20) and incubated with fluorophore-conjugated AlexaFluor-680 or AlexaFluor-800 secondary antibodies for 1 hr at room temperature and scanned using an Odyssey Infrared Imaging System (LI-COR Biosciences).
3. Results and Discussion
3.1. Quantitative Proteomic Analysis of Human Brain Homogenates
In this study we assessed both the total proteome as well as ubiquitylome by LC-MS/MS analysis (Figure 1) from the same individual control and AD human postmortem frontal cortex brain tissues (n=5 each). Cases were matched for age, gender and post-mortem interval (PMI) and selected based on CERAD and Braak scores, which measure β-amyloid (Aβ) plaque burden and pathological Tau distribution, respectively[37]. AD cases had an average Braak score of 6 (highest), indicating that Tau pathology had spread to the frontal cortex, compared to the controls (average=1.4) (Table S1).
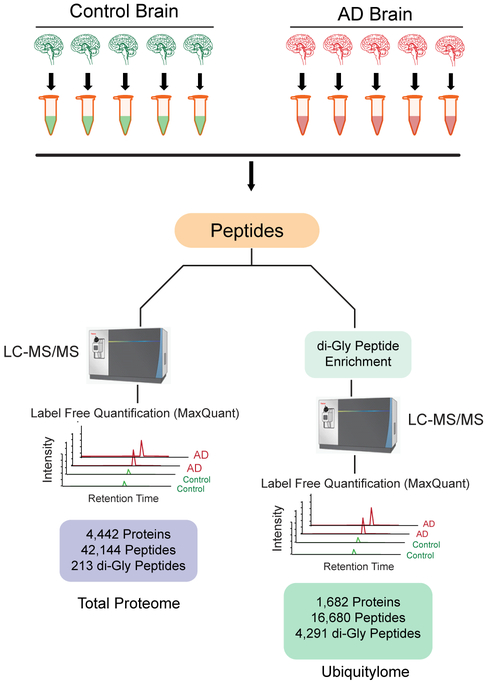
Figure 1.
Experiment workflow. A) Control and AD human postmortem frontal cortex brain tissues (n=5 each group) were homogenized in 8M urea lysis buffer. For both global and ubiquitylome analysis, brain protein extracts from each sample was first denatured, alkylated and proteolytically digested followed by peptide desalting as described in methods. For global proteome analysis, peptides were analyzed by LC-MS/MS on Orbitrap Fusion Tribrid mass spectrometer. For brain ubiquitylome analysis, peptides were subjected to di-Gly affinity enrichment as described in methods followed by LC-MS/MS analysis in replicate on an Orbitrap Fusion Tribrid mass spectrometer. Protein abundance was calculated by peptide ion-intensity measurements across LC-MS runs using the label free quantification (LFQ) algorithm in MaxQuant.
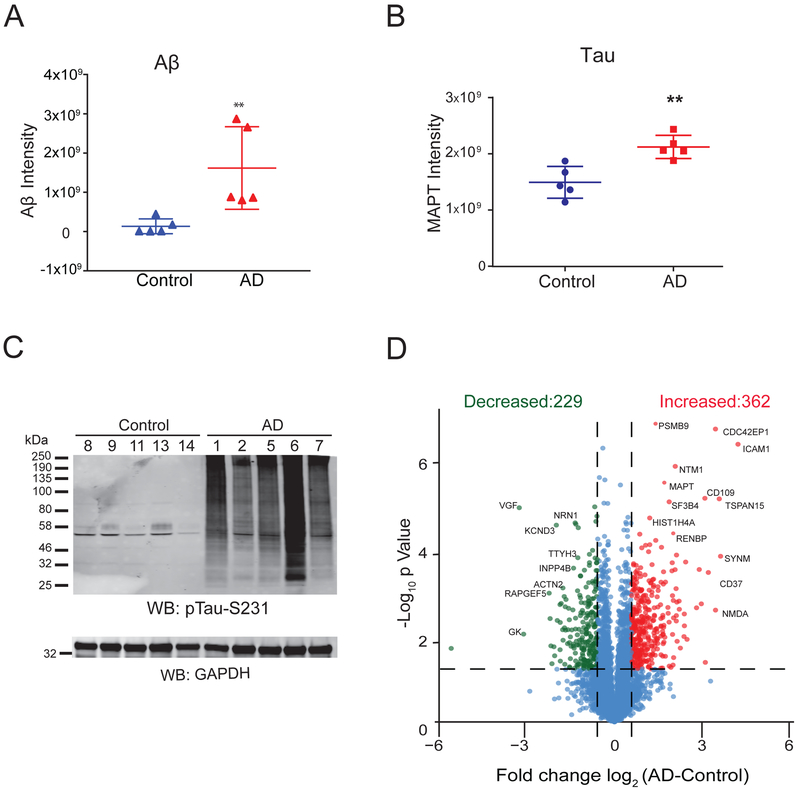
Label-free proteomic analysis was first performed on total brain homogenates to validate signature changes associated with AD pathology and, as described below, to assess the level of ubiquitin enrichment following di-Gly peptide affinity capture. In the total proteome analysis, we identified 42,144 peptides mapping to 4,443 protein groups across all samples analyzed. Aβ levels, quantified by summing ion intensities of two amyloid precursor protein (APP) peptides corresponding to the Aβ peptide residues 6–16 (HDSGYEVHHQK) and 17–28 (LVFFAEDVGSNK)[30], were significantly increased in AD cases compared to controls consistent with high CERAD neuropathology scores (Figure 2A and Table S1). Furthermore, LFQ measurements for Tau were also increased in AD cases (Figure 2B), which is confirmed by Tau phosphorylation levels by immunoblot (Figure 2C). Differential expression analysis revealed 591 proteins that were significantly changed in AD compared to control cases (1.5-fold-change, p<0.05) (Table S2). Of these, 362 and 229 proteins were increased and decreased, respectively (Figure 2D). Proteins decreased in AD were enriched for gene ontology (GO) terms linked to nerve growth factor receptor signaling, actin cytoskeleton organization, and synapse processes consistent with the down-regulation of proteins associated with neurons and synapses[30] (Figure S1). Conversely, GO analysis of the proteins increased in AD showed enrichment for cell adhesion, nucleosome assembly, and complement activation pathways. Increased complement activation markers in AD is consistent with the enhanced inflammation described in AD[38]. Therefore, label-free proteomic analysis, prior to ubiquitin enrichment, of the selected AD cases revealed significant increase of Aβ and Tau as well as changes in expected disease signatures associated with AD neuropathological burden.
Figure 2.
Quantitative analysis of total brain proteome. A) Comparison of Aβ peptide levels measured by LFQ ion intensities in AD and control brain samples showed a significant increase in Aβ peptide levels in AD. Error bars represent ± SD (**p<0.05). B) Comparison of Tau protein levels in AD vs control cases measured by LFQ ion intensities showed a significant increase in Tau measures in AD. Error bars represent ± SD (**p<0.05). C) AD and control brain samples homogenized in 8M Urea lysis buffer were resolved on SDS-PAGE followed by western blot analysis using anti-pTau-S231 antibody. D) Volcano plot displays individual proteins (log2 fold-change) vs. t test significance [-log10(p value)]. Statistically significant proteins [-log10(p value) ≥ 1.30 and ±1.5-fold-change) are colored as red (increased) or green (decreased) whereas proteins that did not meet criteria are colored in blue. Among these, 362 proteins (red) showed a 1.5-fold-increase in AD samples and 229 proteins (green) showed a 1.5-fold-decrease in AD cases compared to controls.
3.2. Identification and Quantification of Ubiquitylated Isopeptides
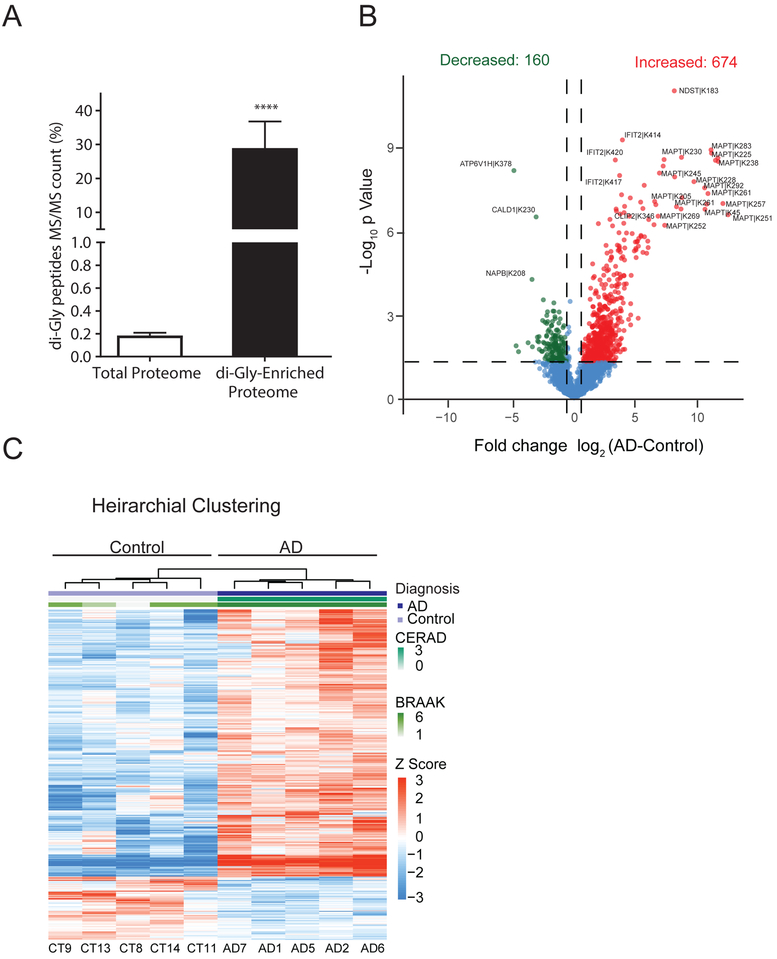
The major goal of this study was to identify and quantify global ubiquitylome changes in brain associated with AD. However, identification of di-Gly isopeptides in the total proteome is challenging due to the low stoichiometry of ubiquitin modified peptides in the total proteome[21, 23, 27]. Indeed, following database searches we observed a very low percentage of di-Gly isopeptides (0.18% of total peptide spectral matches identified) in the total proteome in control and AD cases further justifying the need for ubiquitin enrichment approaches. To this end, di-Gly isopeptides were enriched using a di-Gly remnant specific antibody from each control and AD case (Figure 1). All samples were analyzed as technical replicates, which were significantly correlated (R2>0.95, p<0.01), indicating high reproducibility in peptide quantification (Figure S2). These LFQ replicate measurements of di-Gly isopeptide intensities for each sample were then averaged for subsequent analyses. In total, we identified and quantified 16,680 peptides mapping to 1,682 proteins, of which 4,291 peptides were ubiquitylated. Comparison of di-Gly isopeptides in the total proteome versus ubiquitin-enriched proteome indicated a significant increase in the percentage of di-Gly peptides identified (Figure 3A). The total percent of peptide spectral matches (MS/MS count) increased from 0.18% (55/29322) in the total proteome to 28.9% (3411/11768) following ubiquitin enrichment. This fold enrichment (156.4 ±42.71) was significant (p<0.0001) across all cases and highlights the effectiveness of di-Gly specific antibody in enriching previously ubiquitylated isopeptides from human postmortem brain tissue.
Figure 3.
Quantitative analysis of AD brain ubiquitylome following di-Gly peptide enrichment. A) Comparison of the percentage of di-Gly peptide spectral matches (MS/MS count) in total proteome vs GG-enriched proteome. In the total brain proteome only 0.18% (55/29322) of matched peptides were di-Gly modified versus 28.9% (3411/11768) of matched peptides in immunoaffinity (di-Gly specific antibody) enriched proteome. Error bars represent ± SD (***p<0.0001). B) Volcano plots displays individual di-Gly peptides log2-fold-change vs. t test significance [-log10(p value)] for AD vs Control. Red (n=674) and green (n=160) points represent significantly (-log10(p value) ≥ 1.30, at least ±1.5-fold change) differentially increased or decreased di-Gly peptides in the AD compared to controls, respectively. Blue points are di-Gly peptides identified that are not significant. C) Hierarchical clustering analysis using di-Gly peptide intensities segregates AD cases based on ubiquitylation pattern. Log2 di-Gly peptide intensities were converted to Z scores (mean centered abundance, fold of standard deviation) for clustering.
3.3. Differential Ubiquitylation in AD brain
Differential enrichment analysis resulted in 834 di-Gly peptides that were at least 1.5-fold higher or lower in AD cases compared to controls (p < 0.05) (Table S3). The majority of the di-Gly peptides (~80%) were significantly enriched in AD, whereas only 20% of di-Gly peptides were decreased in AD compared to controls (Figure 3B). This bias towards increased ubiquitylation in AD supports the high burden of ubiquitin positive pathological protein aggregates in AD and is also reflective of proteolytic stress induced in cells[39, 40] . Consistently, the microtubule associated proteins Tau (MAPT) had the most increased sites of ubiquitylation per any protein (~5%) (Figure 3B, Table S3, Supporting Information). Of note, we did not observe an increase in di-Gly isopeptides mapping to the Aβ sequence in amyloid precursor protein (APP) or apolipoprotein (APOE), the latter representing the gene product for the major genetic risk factor for AD that associates with Aβ senile plaques in AD brain[41]. This indicates that the majority of the ubiquitylated substrates in AD brain are associated with NFT pathology or intracellular processes potentially related to protein homeostasis. Towards this end, GO analysis of increased ubiquitylated substrates were enriched in pathways and processes related to axonal function, microtubule associated complex, membrane ion transport and ATPase activity (Figure S3). For example, increased ubiquitylation of cytoskeleton associated proteins included, FLOT1, GAPDH, GNAZ, GSN, MAP4, MAP6, MAPT, PPIA, TUBB4B, VIM, YWHAB, YWHAE, YWHAG, YWHAH, YWHAQ, YW HAZ suggesting the potential impact of ubiquitylation on cytoskeletal structure and integrity in AD (Table S4, Supporting Information). Transporting ATPases are also highly ubiquitylated in AD and include the subunits of the Na+/K+ pump (Table S4, Supporting Information). Na+/K +- ATPases function is altered in activated microglia in AD[42] suggesting a potential role for ubiquitylation in regulating microglia Na+/K+- ATPase activity in AD. Proteins that were significantly less ubiquitylated in AD compared to controls were enriched for GO terms related to nucleoside phosphate and small molecule processes (Figure S3, Table S4). This included the vacuolar ATPases ATP6V1H and ATP6V1B2, which are subunits of a multi-subunit enzyme involved in acidification of intracellular organelles, critical in protein sorting, receptor mediated endocytosis and in generating synaptic vesicle proton gradient[43]. Thus, the loss of ubiquitylation of subunits within this pathway in AD implies potential dysregulation in protein sorting and organelle acidification. Finally, to determine if AD cases could be classified based solely on brain protein ubiquitylation pattern, a supervised hierarchical clustering analysis was performed using the 834 di-Gly isopeptides that were significantly altered in AD. As the resulting dendrogram indicates (Figure 3C), ubiquitylation signals are sufficient to stratify AD cases from controls. Together these findings highlight the increased burden of protein ubiquitylation in AD brain, which is likely related to intracellular AD pathology (i.e., Tau NFTs) and/or proteolytic stress linked to the dysfunctional ubiquitin-proteasome system (UPS) and autophagy pathways[7, 44].
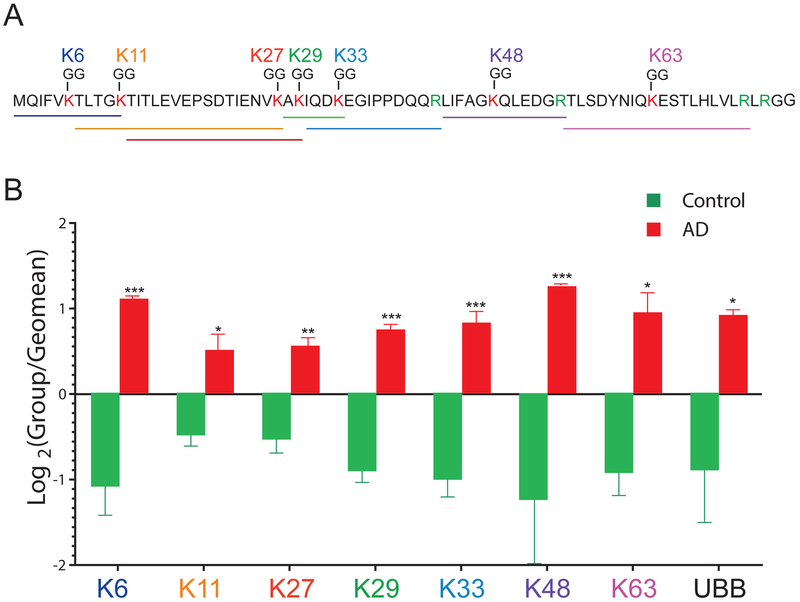
3.4. Polyubiquitin Linkage Analysis in AD
A polyubiquitin chain is assembled by ubiquitin conjugation to one of the seven lysine side chains (K6, K11, K27, K33, K48 and K63) within ubiquitin itself[45] (Figure 4A). Each polyubiquitin chain has unique topology adding complexity to the ubiquitin code, which is read by the ubiquitin binding domains (UBDs) of ubiquitin receptors[46]. K48-linked-polyubiqutin chains primarily serve as signals for degradation by the proteasome, whereas mono-ubiquitylation and K63-linked-polyubiquitin linkages serve as non-degradative signals for highly regulated cellular processes such as endocytosis, lysosomal degradation and DNA repair[7, 39]. Although studies predominantly focus on K48-linked polyubiquitin chains in proteasomal degradation, the role of the remaining polyubiquitin linkages is increasingly being recognized. For instance, the K29-linkage is reported to have roles in the proteasomal and autophagy pathways and K6-, K27-, K33-linkages are shown to play roles in DNA repair pathways[45]. MS analysis of polyubiquitin linkages in total human brain homogenates by Dammer et.al [39] identified K11-, K29-, K48- and K63-linked polyubiquitin chains without ubiquitin affinity enrichment. Here we observed all seven polyubiquitin linkage types including atypical polyubiquitin linkages such as K6-, K27-and K33-linked chains that have not been identified in human brain tissues (Figure 4B and Table S5). Total ubiquitin protein (UBB) levels were also measured from LFQ intensities obtained from total proteome analysis. Notably, each of the linkage type as well as UBB itself were significantly enriched in AD cases (Figure 4B). This is consistent the significant increase in ubiquitylated protein substrates in AD brain quantified by MS (Figure 4B), which is likely caused by dysfunctional protein degradation pathways in AD [10]. Contrary to Dammer et al, we observed increased K29-linked polyubiquitin chains in AD cases compared to controls[39]. This discrepancy is likely due to enriching minor ubiquitin peptides using the di-gly isopeptide specific antibodies compared to Dammer et al[39], which quantified the polyubiquitin linkages without pre-enrichment. Ultimately, to resolve the precise temporal sequence of polyubiquitin chain enrichment in the progression of AD, CSF biomarker studies that target the specific polyubiquitin chains are necessary.
Figure 4.
Polyubiquitin-linkage analysis in AD brain. A) Amino acid sequence of a 76 amino acid ubiquitin, depicting the seven internal lysine residues (red) that are sites of polyubiquitin attachment. Arginine is highlighted (green) as a site of trypsin cleavage. Peptide lengths of each linkage are underlined and color-coded B) A bar graph depicting increased polyubiquitin linkage peptide intensities (K6, K11, K27, K29, K33, K48, K63) and total ubiquitin protein (UBB) levels measured by LFQ ion intensities. Each log2 GG-peptide intensity was normalized to the geometric mean across all AD and control samples (n=10). Significance was measured using one-way non-parametric ANOVA (*p<0.05, **p<0.001, ***p<0.0001).
3.5. Identification of Tau Ubiquitylation Sites In AD
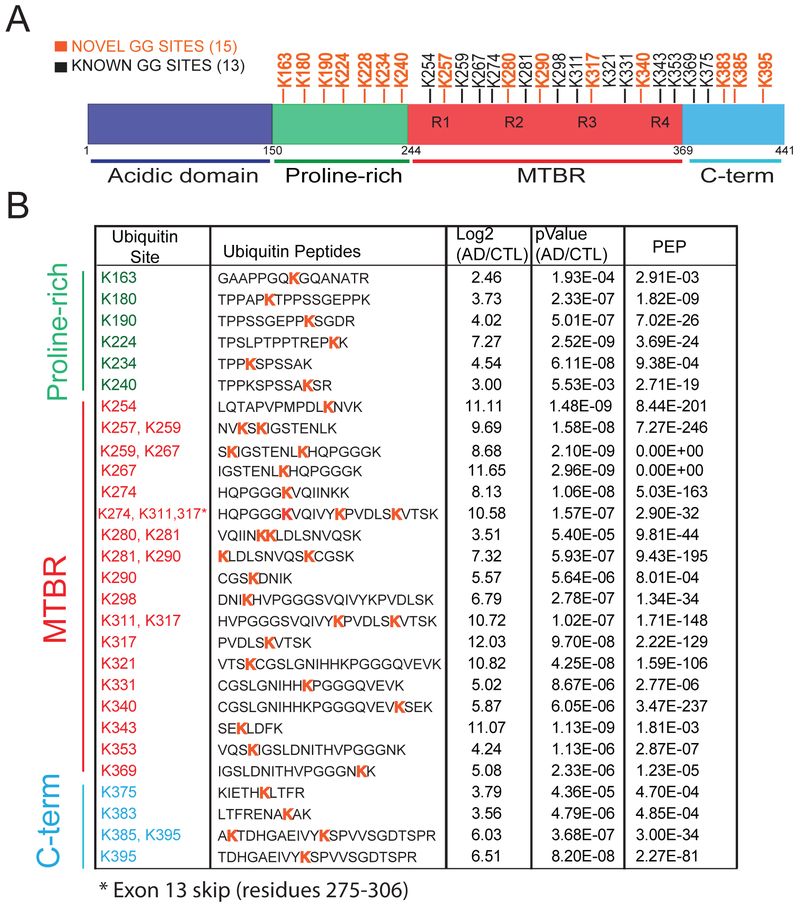
Although the role of Tau phosphorylation in promoting dissociation from microtubules and aggregation has been well described [47], sites of Tau ubiquitylation associated with AD have not been entirely mapped. Pioneering proteomic analysis of Tau from AD brain, enriched using paired helical filament (PHF) specific antibodies, identified ubiquitylation on three lysine residues, K254, K311 and K352[48]. However, Tau (2N4R isoform; 441 total residues) has 44 lysine residues that are potential ubiquitin conjugation sites. Here we identified a total of 28 Tau ubiquitylation sites in human AD brain samples (Figure 5). Of these, 15 sites (Figure 5A; labelled in orange) have not been identified previously[48, 49]. We also mapped 6 ubiquitylation sites (K259, K267, K281, K290, K298 and K311) previously identified in mouse Tau from transgenic mice over-expressing human APP (hAPP)9. Notably, the majority of these ubiquitylation sites on Tau are located in the proline-rich domain and microtubule binding region (MTBR); the latter forms the amyloid core of NFTs[50] (Figure 5A). The proline-rich domain in Tau has Pro-XX-Pro (PXXP) motifs predicted to serve as recognition sites for SH3-domain containing proteins such as kinases[13, 51] that act on Tau. Modifications within the MTBR are also predicted to impact Tau microtubule binding causing microtubule destabilization[15, 52]. For example, phosphorylation of Tau at S262 within the MTBR is reported to disrupt Tau microtubule binding and cause microtubule depolymerization[15, 52].
Figure 5.
Quantification of Tau ubiquitin sites in AD Brain. A) schematic representation of Tau protein domains and ubiquitylation sites identified in this study. Residues are numbered according to Tau 441 isoform (P10636–8). A total of 15 novel ubiquitylation sites are indicated in orange, whereas previously known sites are in black. B) Statistical analysis (Student’s t test) indicates a significant fold change increase in Tau ubiquitin site intensities in AD compared to controls (p<0.05). PEP (posterior error probability) measures a probability of a peptide to be a false hit. All ubiquitin sites had localization probabilities above 90 percent, as assessed on the fragmentation spectra by MaxQuant. *Exon 13 skip event (amino acid 275–306 in Tau 441 isoform).
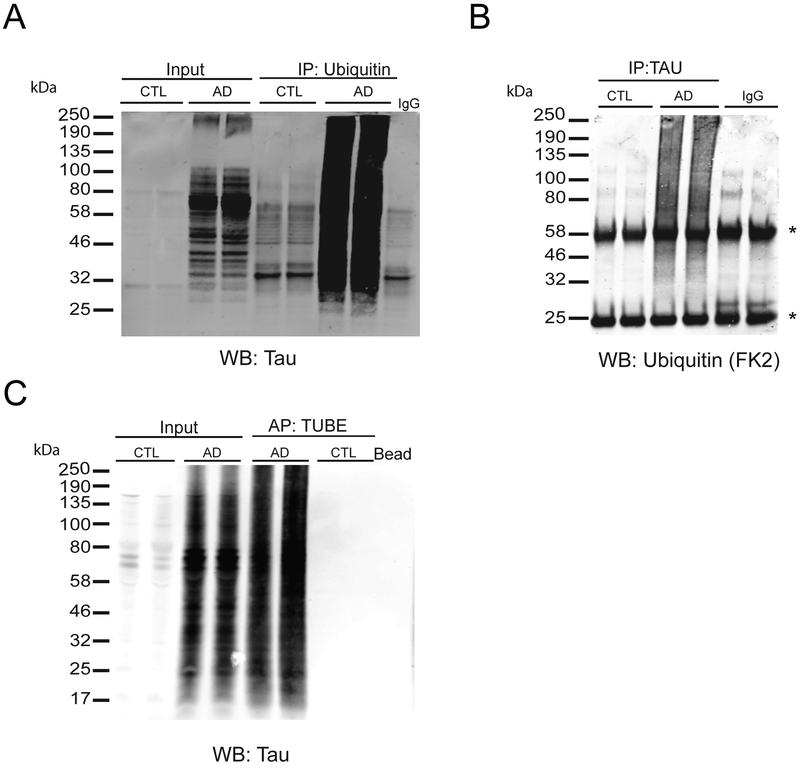
To validate Tau ubiquitylation identified by MS, we performed reciprocal co-immunoprecipitation (co-IP) experiments from control and AD brain tissues (Figure 6A). Polyubiquitin immunoprecipitations (FK2 antibody) were enriched with high molecular weight (HMW) Tau immunoreactivity from AD brain extracts, whereas IPs from control extracts showed little detectable HMW Tau signal. Conversely, Tau IP from AD brain extracts were selectively enriched with HMW polyubiquitin compared to Tau IPs from controls (Figure 6B). Negative control IPs using non-specific IgG failed to enrich appreciable levels of Tau or poly-ubiquitin. Finally, we affinity captured poly-ubiquitylated proteins from AD and control brain extracts using Tandem Ubiquitin Binding Entities (TUBEs), which are tandem UBDs with dissociation constants for poly-ubiquitin in the nanomolar range[53]. Consistent with the antibody co-IP results, TUBE captured ubiquitylated substrates from AD brain extracts were enriched with HMW Tau immunoreactivity compared to substrates captured from controls extracts (Figure 6C). Collectively, these findings demonstrate that Tau is polyubiquitylated in AD brain, likely via K6-, K11-, K48- linkages[48], and further reinforce the quantitative proteomics, which show that Tau is a significantly ubiquitylated substrate in AD brain.
Figure 6.
Validation of Tau ubiquitylation in AD brain. A) Validation of Tau polyubiquitylation in AD. Immunoprecipitation (IP) analysis performed on control and AD brain extracts (n=2, each) using anti-ubiquitin antibody (FK2) followed by immunoblotting using tau antibody. B) Reciprocal IP was performed on AD and control brain extracts using anti-Tau antibody followed by immunoblotting using anti-ubiquitin antibody (FK2) shows high molecular weight immunoreactivity in AD cases. *Indicates light and heavy immunoglobulin chains. C) Affinity precipitation (AP) of polyubiquitylated proteins using TUBE-biotin followed by western blot analysis using anti-tau antibody to detect polyubiquitylated Tau. Lysates incubated with beads alone were used as negative controls. Inputs refer to protein lysates prior immunoprecipitation.
3.6. Co-Modification of Tau by Phosphorylation and Ubiquitylation In AD Brain
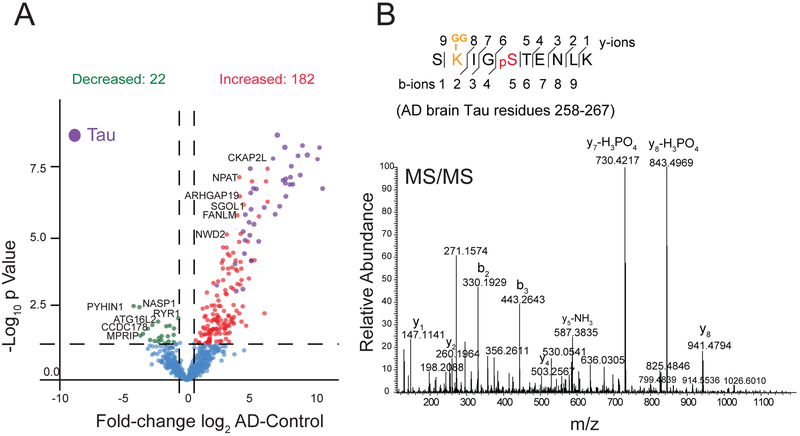
Cross-talk between PTMs is hypothesized to add complexity to cellular homeostasis regulatory processes[54]. The role of complex PTMs on Tau protein aggregation is evidenced by cross-talk between phosphorylation and acetylation[15, 55]. The MTBR of Tau has four KXGS motifs and competition between phosphorylation (on serine) and acetylation (on lysine) within this motif has been reported[17]. Hypoacetylation within KXGS motifs is associated with hyperphosphorylation and increased Tau aggregation. However, the PTM cross-talk between ubiquitylation and phosphorylation on Tau or other proteins in human brain is not well established. Here, we identified a total of 2,780 human brain peptides that were co-modified by both phosphorylation and ubiquitylation (Table S6). Differential enrichment analysis revealed significant changes in 204 co-modified peptides in AD compared to control samples (Table S7). Of these, 162 peptides were increased and 62 were decreased in AD cases (Figure 7A). Of note, 25% of these co-modified peptides mapped to Tau, with the majority harboring the KXGS motif. This indicates that crosstalk between phosphorylation and ubiquitylation also occurs within this motif on Tau (Figure 7B). Besides phosphorylation and ubiquitylation, lysine residues in Tau have been reported to undergo multiple post-translational modifications including acetylation[18, 56], glycation[57], methylation[58], and SUMOlyation[55]. Future studies are needed to characterize potential relationship between phosphorylation and these lysine modifications on Tau protein function. GO enrichment analysis on co-modified gene products mapped to a wide variety of biological processes including: ubiquitin-specific protease activity, G1/S transition of mitotic cell cycle, and macromolecule catabolic processes (Table S8). Thus, cross-talk between ubiquitylation and phosphorylation, which can occur in cells following proteolytic stress[59], may influence a diverse number of proteins and pathways in AD brain.
Figure 7.
Co-modification of Tau by phosphorylation and ubiquitylation in AD brain A) Volcano plot displays log2 fold change of intensities in AD versus control of co-modified peptides with both ubiquitylation and phosphorylation sites (minimum ±1.5-fold change, and Student’s t test significance, p<0.05). Red dots represent 182 dually modified peptides significantly increased in AD cases compared to controls and green dots represent 22 dually modified peptides decreased in AD compared to controls. Co-modified peptides from Tau protein are highlighted in purple. B) A representative MS/MS spectrum of a Tau peptide SK-GGIGPSTENLK (Residue 258–267) with ubiquitylation site at K259 and phosphorylation site at S262.
4. Conclusions
Using an immunoaffinity capture approach we mapped changes in AD brain ubiquitylome revealing a global increase in Tau ubiquitylation and other novel targets in AD. Overall, these findings will contribute towards our understanding of the underlying proteostasis pathways altered in AD and the potential identification of novel diagnostic biomarkers in CSF or other biofluids.
Supplementary Material
Figure S1. Gene Ontology (GO) analysis of total proteome changes in AD. The list of GO terms and associated Z-scores for 362 AD increased and 229 decreased proteins are listed. Green, blue and red bars represent cellular components, molecular function, and biological processes, respectively. Z score cut-off of 1.96, P value cut off 0.01 and a minimum of 5 gene products per ontology were used as filters prior to pruning the ontologies.
Figure S2. Pearson's correlation plots of technical replicates. Di-Gly peptides from 10 samples were run on LC-MS/MS as technical replicates. LFQ intensities were used for Pearson's correlation coefficient analysis to calculate rho (cor) and P-values.
Figure S3. GO analysis of AD associated brain ubiquitylome changes. GO terms associated with 674 proteins with increased ubiquitylation and 160 proteins with decreased ubiquitylation measures in AD cases compared to controls and Z-scores are listed. Green, blue and red bars represent cellular components, molecular function, and biological processes, respectively. Z score cut-off of 1.96, P value cut off 0.01 and a minimum of 5 genes per ontology were used as filters prior to pruning the ontologies.
Table S1. Case IDs and Trait Information
Table S2. Total Proteome Differential Expression Analysis
Table S3. GlyGly IP Differential Expression Analysis
Table S4. Ubiquitylome GO Terms
Table S5. Ubiquitin linkages calculations
Table S6. GlyGly and Phospho Peptides
Table S7. Co-modified peptides differential Expression Analysis
Table S8. GO terms for co-modified peptides
Significance Statement.
Ubiquitin signaling plays critical roles in maintaining cellular protein homeostasis through clearing protein aggregates via the autophagy and proteasomal degradation pathways. One of the hallmarks of Alzheim er’s Disease (AD) and related neurodegenerative diseases is dysregulation of cellular protein homeostasis and accumulation of ubiquitin-positive protein aggregates in brain. Although previous studies have described the association of ubiquitin signaling defects with AD, site-specific changes in the ubiquitin-modified AD brain proteome have not been characterized. Here, we mapped the ubiquitin-modified proteome in AD brain using immunoaffinity enrichment coupled with high resolution mass spectrometry. A quantitative proteomic analysis identified changes in AD brain ubiquitylation patterns compared to control brain tissues. We also report novel ubiquitylation sites in the microtubule binding repeats and other domains of Tau, the core component of neurofibrillary tangles in AD. Finally, co-modified peptides, containing both ubiquitylation and phosphorylation, were also enriched in AD brain, suggesting that cross-talk between phosphorylation and ubiquitylation occurs in disease. Our findings highlight the utility of an immunoaffinity enrichment approach coupled to MS analysis for mapping AD associated ubiquitylation changes in human brain, which provides insights into underlying proteostasis pathways altered in AD.
Acknowledgements
This study was provided by grants from the Accelerating Medicine Partnership AD (U01AG046161–02, the National Institute on Aging (R21AG054206, 5R01AG053960, RF1AG057470, and RF1AG057471, the NINDS Emory Neuroscience Core (P30NS055077), and the Emory Alzheim er’s Disease Research Center (P50AG025688. N.T.S. was supported in part by a Biomarkers Across Neurodegenerative Grant (11060) funded by the Alzheim er’s Association (ALZ), Alzheim er’s Research UK (ARUK), The Michael J. Fox Foundation for Parkinson’s Research (MJFF), and the W eston Brain Institute. M.H.A. was supported by an NIH training grant T32 NS00748018. We would like to thank Dr. Tauseef Butt (Life Sensors) for providing the Biotin-TUBE1 reagent.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
References:
- [1].Bence NF, Sampat RM, Kopito RR, Science 2001, 292, 1552. [DOI] [PubMed] [Google Scholar]
- [2].Gilberto S, Peter M, The Journal of cell biology 2017, 216, 2259; [DOI] [PMC free article] [PubMed] [Google Scholar]; Grillari J, Grillari Voglauer R, Jansen-Durr P, Advances in experimental medicine and biology 2010, 694, 172; [DOI] [PubMed] [Google Scholar]; Muratani M, Tansey WP, Nature reviews Molecular cell biology 2003, 4, 192; [DOI] [PubMed] [Google Scholar]; Ciechanover A, The EMBO journal 1998, 17, 7151; [DOI] [PMC free article] [PubMed] [Google Scholar]; Hershko A, Ciechanover A, Annual Reviews 4139 El Camino Way, PO Box 10139, Palo Alto, CA 94303–0139, USA, 1998. [Google Scholar]
- [3].Scheffner M, Nuber U, Huibregtse JM, Nature 1995, 373, 81. [DOI] [PubMed] [Google Scholar]
- [4].Weissman AM, Nature reviews Molecular cell biology 2001, 2, 169; [DOI] [PubMed] [Google Scholar]; Scheffner M, Huibregtse JM, Vierstra RD, Howley PM, Cell 1993, 75, 495. [DOI] [PubMed] [Google Scholar]
- [5].Kerscher O, Felberbaum R, Hochstrasser M, Annu. Rev. Cell Dev. Biol 2006, 22, 159. [DOI] [PubMed] [Google Scholar]
- [6].Gendron JM, Webb K, Yang B, Rising L, Zuzow N, Bennett EJ, Mol Cell Proteomics 2016, 15, 2576; [DOI] [PMC free article] [PubMed] [Google Scholar]; Wickner S, Maurizi MR, Gottesman S, Science 1999, 286, 1888. [DOI] [PubMed] [Google Scholar]
- [7].Kirkin V, McEwan DG, Novak I, Dikic I, Molecular cell 2009, 34, 259. [DOI] [PubMed] [Google Scholar]
- [8].Tan JM, Wong ES, Kirkpatrick DS, Pletnikova O, Ko HS, Tay S-P, Ho MW, Troncoso J, Gygi SP, Lee MK, Human molecular genetics 2007, 17, 431. [DOI] [PubMed] [Google Scholar]
- [9].Ciechanover A, Nature reviews Molecular cell biology 2005, 6, 79. [DOI] [PubMed] [Google Scholar]
- [10].Rubinsztein DC, Nature 2006, 443, 780. [DOI] [PubMed] [Google Scholar]
- [11].Prusiner SB, New England Journal of Medicine 2001, 344, 1516; [DOI] [PubMed] [Google Scholar]; Koo EH, Lansbury PT, Kelly JW, Proceedings of the National Academy of Sciences 1999, 96, 9989; [DOI] [PMC free article] [PubMed] [Google Scholar]; Kopito RR, Trends in cell biology 2000, 10, 524; [DOI] [PubMed] [Google Scholar]; Ross CA, Poirier MA, 2004.
- [12].Cripps D, Thomas SN, Jeng Y, Yang F, Davies P, Yang AJ, J Biol Chem 2006, 281, 10825. [DOI] [PubMed] [Google Scholar]
- [13].Guo T, Noble W, Hanger DP, Acta Neuropathol 2017, 133, 665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lee VM, Goedert M, Trojanowski JQ, Annual review of neuroscience 2001, 24, 1121. [DOI] [PubMed] [Google Scholar]
- [15].Iqbal K, Alonso A. d. C., Chen, Chohan MO, El-Akkad E, Gong C-X, Khatoon, Li B, Liu F, Rahman A, Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease 2005, 1739, 198. [DOI] [PubMed] [Google Scholar]
- [16].Petrucelli L, Dickson D, Kehoe K, Taylor J, Snyder H, Grover A, De Lucia M, McGowan E, Lewis J, Prihar G, Human molecular genetics 2004, 13, 703. [DOI] [PubMed] [Google Scholar]
- [17].Cook C, Carlomagno Y, Gendron TF, Dunmore J, Scheffel K, Stetler C, Davis M, Dickson D, Jarpe M, DeTure M, Human molecular genetics 2013, 23, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Min S-W, Cho S-H, Zhou Y, Schroeder S, Haroutunian V, Seeley WW, Huang EJ, Shen Y, Masliah E, Mukherjee C, Neuron 2010, 67, 953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Liu F, Iqbal K, Grundke-Iqbal I, Hart GW, Gong CX, Proc Natl Acad Sci U S A 2004, 101, 10804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chen PC, Na CH, Peng J, Amino acids 2012, 43, 1049; [DOI] [PMC free article] [PubMed] [Google Scholar]; Na CH, Peng J, Methods in molecular biology (Clifton, N.J.) 2012, 893, 417; [DOI] [PMC free article] [PubMed] [Google Scholar]; Ren RJ, Dammer EB, Wang G, Seyfried NT, Levey AI, Transl Neurodegener 2014, 3, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP, Nature biotechnology 2003, 21, 921. [DOI] [PubMed] [Google Scholar]
- [22].Udeshi ND, Mertins P, Svinkina T, Carr SA, Nat Protoc 2013, 8, 1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Xu G, Paige JS, Jaffrey SR, Nature biotechnology 2010, 28, 868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, Harper JW, Gygi SP, Mol Cell 2011, 44, 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Danielsen JM, Sylvestersen KB, Bekker-Jensen S, Szklarczyk D, Poulsen JW, Horn H, Jensen LJ, Mailand N, Nielsen ML, Mol Cell Proteomics 2011, 10, M110003590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Aillet F, Lopitz-Otsoa F, Hjerpe R, Torres-Ramos M, Lang V, Rodríguez MS, Ubiquitin Family Modifiers and the Proteasome: Reviews and Protocols 2012, 173. [DOI] [PubMed] [Google Scholar]
- [27].Matsumoto M, Hatakeyama S, Oyamada K, Oda Y, Nishimura T, Nakayama KI, Proteomics 2005, 5, 4145. [DOI] [PubMed] [Google Scholar]
- [28].Udeshi ND, Svinkina T, Mertins P, Kuhn E, Mani DR, Qiao JW, Carr SA, Mol Cell Proteomics 2013, 12, 825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Cook C, Carlomagno Y, Gendron TF, Dunmore J, Scheffel K, Stetler C, Davis M, Dickson D, Jarpe M, DeTure M, Petrucelli L, Hum Mol Genet 2014, 23, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Seyfried NT, Dammer EB, Swarup V, Nandakumar D, Duong DM, Yin L, Deng Q, Nguyen T, Hales CM, Wingo T, Glass J, Gearing M, Thambisetty M, Troncoso JC, Geschwind DH, Lah JJ, Levey AI, Cell Syst 2017, 4, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, Molecular cell 2011, 44, 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Cox J, Mann M, Nature biotechnology 2008, 26, 1367. [DOI] [PubMed] [Google Scholar]
- [33].Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M, Molecular & cellular proteomics 2014, 13, 2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J, Nature methods 2016. [DOI] [PubMed] [Google Scholar]
- [35].Zambon AC, Gaj S, Ho I, Hanspers K, Vranizan K, Evelo CT, Conklin BR, Pico AR, Salomonis N, Bioinformatics 2012, 28, 2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Dammer EB, Fallini C, Gozal YM, Duong DM, Rossoll W, Xu P, Lah JJ, Levey AI, Peng J, Bassell GJ, Seyfried NT, PLoS One 2012, 7, e38658; [DOI] [PMC free article] [PubMed] [Google Scholar]; Diner I, Hales CM, Bishof I, Rabenold L, Duong DM, Yi H, Laur O, Gearing M, Troncoso J, Thambisetty M, Lah JJ, Levey AI, Seyfried NT, J Biol Chem 2014, 289, 35296; [DOI] [PMC free article] [PubMed] [Google Scholar]; Donovan LE, Higginbotham L, Dammer EB, Gearing M, Rees HD, Xia Q, Duong DM, Seyfried NT, Lah JJ, Levey AI, PROTEOM ICS-Clinical Applications 2012, 6, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Braak H, Alafuzoff I, Arzberger T, Kretzschmar H, Del Tredici K, Acta neuropathologica 2006, 112, 389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Rogers J, Cooper NR, Webster S, Schultz J, McGeer PL, Styren SD, Civin WH, Brachova L, Bradt B, Ward P, Proceedings of the National Academy of Sciences 1992, 89, 10016; [DOI] [PMC free article] [PubMed] [Google Scholar]; Yasojima K, Schwab C, McGeer EG, McGeer PL, The American journal of pathology 1999, 154, 927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Dammer EB, Na CH, Xu P, Seyfried NT, Duong DM, Cheng D, Gearing M, Rees H, Lah JJ, Levey AI, Rush J, Peng J, J Biol Chem 2011, 286, 10457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Udeshi ND, Mani D, Eisenhaure T, Mertins P, Jaffe JD, Clauser KR, Hacohen N, Carr SA, Molecular & Cellular Proteomics 2012, mcp. M111. 016857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Saunders AM, Strittmatter WJ, Schmechel D, George-Hyslop PS, Pericak-Vance M, Joo S, Rosi B, Gusella J, Crapper-MacLachlan D, Alberts M, Neurology 1993, 43, 1467; [DOI] [PubMed] [Google Scholar]; Yu J-T, Tan L, Hardy J, Annual review of neuroscience 2014, 37, 79. [DOI] [PubMed] [Google Scholar]
- [42].Vitvitsky VM, Garg SK, Keep RF, Albin RL, Banerjee R, Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease 2012, 1822, 1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Mellman I, Fuchs R, Helenius A, Annual review of biochemistry 1986, 55, 663. [DOI] [PubMed] [Google Scholar]
- [44].Kerscher O, Felberbaum R, Hochstrasser M, Annual review of cell and developmental biology 2006, 22, 159; [DOI] [PubMed] [Google Scholar]; Graham SH, Liu H, Ageing Research Reviews 2017, 34, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Rajalingam K, Dikic I, Cell 2016, 164, 1074. [DOI] [PubMed] [Google Scholar]
- [46].Pickart CM, Fushman D, Current opinion in chemical biology 2004, 8, 610. [DOI] [PubMed] [Google Scholar]
- [47].Lindwall G, Cole RD, Journal of Biological Chemistry 1984, 259, 5301. [PubMed] [Google Scholar]
- [48].Cripps D, Thomas SN, Jeng Y, Yang F, Davies P, Yang AJ, Journal of Biological Chemistry 2006, 281, 10825. [DOI] [PubMed] [Google Scholar]
- [49].Morris M, Knudsen GM, Maeda S, Trinidad JC, Ioanoviciu A, Burlingame AL, Mucke L, Nat Neurosci 2015, 18, 1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Fitzpatrick AW, Falcon B, He S, Murzin AG, Murshudov G, Garringer HJ, Crowther RA, Ghetti B, Goedert M, Scheres SH, Nature 2017, 547, 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Bhaskar K, Yen S-H, Lee G, Journal of Biological Chemistry 2005, 280, 35119. [DOI] [PubMed] [Google Scholar]
- [52].Mandelkow EM, Mandelkow E, Cold Spring Harb Perspect Med 2012, 2, a006247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Hjerpe R, Aillet F, Lopitz-Otsoa F, Lang V, England P, Rodriguez MS, EMBO reports 2009, 10, 1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Altelaar AM, Munoz J, Heck AJ, Nature Reviews Genetics 2013, 14, 35; [DOI] [PubMed] [Google Scholar]; Hunter T, Molecular cell 2007, 28, 730; [DOI] [PubMed] [Google Scholar]; Venne AS, Kollipara L, Zahedi RP, Proteomics 2014, 14, 513. [DOI] [PubMed] [Google Scholar]
- [55].Kontaxi C, Piccardo P, Gill AC, Frontiers in molecular biosciences 2017, 4, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Min S-W, Chen X, Tracy TE, Li Y, Zhou Y, Wang C, Shirakawa K, Minami SS, Defensor E, Mok SA, Nature medicine 2015, 21, 1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Du Yan S, Yan SF, Chen X, Fu J, Chen M, Kuppusamy P, Smith MA, Perry G, Godman GC, Nawroth P, Nature medicine 1995, 1, 693; [DOI] [PubMed] [Google Scholar]; Ledesma MD, Bonay P, Colaco C, Avila J, Journal of Biological Chemistry 1994, 269, 21614. [PubMed] [Google Scholar]
- [58].Thomas SN, Funk KE, Wan Y, Liao Z, Davies P, Kuret J, Yang AJ, Acta neuropathologica 2012, 123, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Swaney DL, Beltrao P, Starita L, Guo A, Rush J, Fields S, Krogan NJ, Villen J, Nat Methods 2013, 10, 676. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Gene Ontology (GO) analysis of total proteome changes in AD. The list of GO terms and associated Z-scores for 362 AD increased and 229 decreased proteins are listed. Green, blue and red bars represent cellular components, molecular function, and biological processes, respectively. Z score cut-off of 1.96, P value cut off 0.01 and a minimum of 5 gene products per ontology were used as filters prior to pruning the ontologies.
Figure S2. Pearson's correlation plots of technical replicates. Di-Gly peptides from 10 samples were run on LC-MS/MS as technical replicates. LFQ intensities were used for Pearson's correlation coefficient analysis to calculate rho (cor) and P-values.
Figure S3. GO analysis of AD associated brain ubiquitylome changes. GO terms associated with 674 proteins with increased ubiquitylation and 160 proteins with decreased ubiquitylation measures in AD cases compared to controls and Z-scores are listed. Green, blue and red bars represent cellular components, molecular function, and biological processes, respectively. Z score cut-off of 1.96, P value cut off 0.01 and a minimum of 5 genes per ontology were used as filters prior to pruning the ontologies.
Table S1. Case IDs and Trait Information
Table S2. Total Proteome Differential Expression Analysis
Table S3. GlyGly IP Differential Expression Analysis
Table S4. Ubiquitylome GO Terms
Table S5. Ubiquitin linkages calculations
Table S6. GlyGly and Phospho Peptides
Table S7. Co-modified peptides differential Expression Analysis
Table S8. GO terms for co-modified peptides