Abstract
IFNγ is a key regulator of inflammatory responses but its role in influenza A virus (IAV) pathogenesis is unclear. Our studies show that infection of mice lacking the IFNγ receptor (IFNγR-/-) at a dose which caused severe disease in wild type 129 Sv/Ev (WT) mice resulted in milder clinical symptoms and significantly lower lung virus titers by 6 days post-infection (dpi). Viral spread was reduced in IFNγR-/- lungs at 2 and 4 dpi. Levels of inflammatory cytokines and chemokines were lower in IFNγR-/- mice at 2 dpi and there was less infiltration of monocyte/macrophage lineage cells than in WT mice. There was no difference in CD4+ and CD8+ T cells and alveolar macrophages in the bronchoalveolar lavage fluid (BALF) at 2 and 4 dpi but by 4 dpi IFNγR-/- mice had significantly higher percentages of neutrophils. Our data strongly suggest that IAV can use the inflammatory response to promote viral spread.
Abbreviations: BALF, Bronchoalveolar lavage fluid; IAV, Influenza A virus; AEC, Alveolar epithelial cells; AM, Alveolar macrophages; WT, Wild type 129 Sv/Ev; IFNγR-/-, 129 Sv/Ev mice lacking the interferon gamma receptor; dpi, Days post-infection; EGFP, Enhanced green fluorescent protein; IFN, Interferon; BMDM, Bone marrow derived macrophages; SDHA, Succinate dehydrogenase complex A
Keywords: Influenza A virus, Lung inflammation, IFNγ, Host response
1. Introduction
Influenza A virus (IAV) infection leads to an inflammatory response characterized by the initiation of an antiviral interferon response and the release of inflammatory mediators which signal leukocyte recruitment to the infected site. Immune cell infiltration is crucial for control of virus replication and resolution of infection. However, this response often contributes to pathogenesis and morbidity and, in the case of highly pathogenic IAVs such as the 1918 H1N1 pandemic strain and the recent avian H5N1 and H7N9 strains, an excessive inflammatory response (Chen et al., 2013, Perrone et al., 2008, Tumpey et al., 2005) can cause irreparable damage to the lungs resulting in high mortality rates (To et al., 2001).
IFNγ is a major driver of inflammatory responses. It is produced by many different cell types including NK, NK T cells, macrophages and T cells and coordinates a broad range of biological activities (Akdis et al., 2011). A major role of IFNγ is activation of a classical pro-inflammatory macrophage response characterized by secretion of inflammatory cytokines such as TNFα and the production of chemokines CCL2 and CCL3 which lead to recruitment of monocytes. IFNγ also induces synthesis of the chemokines CXCL9 and CXCL10 which recruit T cells to sites of infection (Taub et al., 1993) and has a major role in linking the innate and adaptive immune responses. Given the role of IFNγ in driving inflammation, we hypothesized that it would contribute to the excessive inflammatory response associated with severe IAV infections. Supporting this hypothesis, our in vitro studies on bone marrow-derived macrophages (BMDMs) showed that WT BMDMs activated with IFNγ produced significantly higher levels of pro-inflammatory cytokines that IFNγR-/- BMDMs following infection with IAV suggesting a potential impact on IAV infection in vivo (Campbell et al., 2015).
A number of studies have used mice lacking the IFNγ receptor or in which IFNγ is depleted by antibody to look at the role of IFNγ in IAV infections. Whilst there are differences in T cell responses in primary (Turner et al., 2007) and secondary infections (Bot et al., 1998) in the absence of an IFNγ response, most studies have concluded that IFNγ does not have a significant impact on the outcome of IAV infection (Baumgarth and Kelso, 1996, Graham et al., 1993, Turner et al., 2007). However, decreased cellular infiltration was noted in the lungs of IFNγ depleted mice infected with an H3N1 reassortant virus (Baumgarth and Kelso, 1996, Turner et al., 2007). More recently, Califano et al. (2017) showed that IFNγ-/- mice infected with the H1N1 pandemic virus A/California/04/2009 had decreased immunopathology and enhanced survival, an outcome attributed to suppression of group II innate lymphoid cells by IFNγ. It is worth noting that differences in outcome may be due to use of different strains of virus or mouse (Pica et al., 2011, Srivastava et al., 2009, Trammell et al., 2012) and that the dose of virus delivered can affect the activation of the immune response (Pang et al., 2013). Together, these factors can alter the inflammatory response and subsequent immunopathology, making the overall effect of IFNγ in IAV infection difficult to interpret.
In this study, infection of WT or IFNγR-/- mice with the H1N1 virus A/WSN/33 demonstrated that lack of IFNγ signaling resulted in more rapid control of virus replication and reduced disease symptoms in a dose dependent manner. IFNγR-/- mice had attenuated viral spread within the lungs and a lowered inflammatory response, in particular reduced infiltration of monocyte/macrophage lineage cells correlating with the lack of IFNγ response. Together, our data demonstrate that IFNγ-dependent promotion of the classical inflammatory response enhances IAV spread thus promoting infection.
2. Materials and methods
2.1. Mouse model of influenza A virus infection
129 Sv/Ev (WT) mice and IFNγR-/- mice on the 129 Sv/Ev background (Huang et al., 1993) were purchased from B&K Universal and bred in-house. WT and IFNγR-/- mice expressing enhanced green fluorescent protein (EGFP) under control of the Csfr1 promoter, herein referred to as MacGreen, were derived by crossing MacGreen mice on the C57BL/6 background (Sasmono et al., 2003) with WT and IFNγR-/- mice and then back-crossing MacGreen offspring at least seven times with WT and IFNγR-/- mice. MacGreen animals were maintained as heterozygous for the EGFP reporter and homozygous for the IFNγ receptor deletion. All animal work was carried out under the authority of a UK Home Office Project Licence within the terms and conditions of the UK Home Office “Animals (Scientific Procedures) Act 1986” and subject to local ethical review. Appropriate numbers of animals (sample size) were determined using formal power analysis, based on pilot in vivo experiments. Predicted outcomes and variation in assay techniques were used to determine the power of the experiment, allowing for statistical judgements that are accurate and reliable and responsible hypothesis testing. Six- to 8-week-old female mice were used in all experiments. Mice were anesthetized using isoflurane (Merial, Boehringer Ingelheim Animal Health, UK) and infected intranasally with 104 pfu H1N1 A/WSN/33 IAV in 40 µl volume, unless otherwise stated. Mice were weighed daily and assessed for visual signs of clinical disease, including inactivity, ruffled fur, and laboured breathing. At various times after infection, mice were euthanized by CO2 asphyxiation, and tissues collected. By 8 dpi, WT animals had lost 25–30% of their original body weight and were euthanized for humane reasons. No IFNγR-/- mice lost more than 11% body weight.
2.2. Cells and virus
Madin Darby canine kidney epithelial cells (MDCK, ATCC), were maintained in Dulbecco's modified Eagle's medium (ThermoFisher Scientific UK) supplemented with 10% fetal bovine serum (ThermoFisher Scientific UK), 50 U/ml penicillin and 50 µg/ml streptomycin (Thermofisher Scientific, UK). Influenza A virus strain A/WSN/33 was obtained from Dr D Jackson, University of St Andrews. Virus was propagated in MDCK cells and titered by standard plaque assays as previously described (Nicol et al., 2012).
2.3. Histopathology
Lungs were inflation-fixed with neutral buffered formalin, embedded in paraffin, processed to 10 µm sections and stained with hematoxylin and eosin (H&E) using routine methods. Sections were scored blinded using a semi-quantitative system to examine the following criteria: epithelial and interstitial necrosis, peribronchial inflammation, perivascular inflammation, interstitial inflammation and, for day 8, perivascular and peribronchial oedema, using a score varying from 0 to 3. Scores were calculated from the sum of the scores for the five individual criteria determined for each mouse.
2.4. Quantitation of virus spread
Paraffin embedded sections were deparaffinized, rehydrated in graded alcohol solutions to water, followed by heat-induced antigen retrieval in Tris/EDTA pH 9.0 buffer. IAV was detected using a polyclonal goat anti-influenza A virus antibody (USSR (H1N1), AbD Serotec). Endogenous peroxidase was blocked with 3% hydrogen peroxide, followed by secondary antibody donkey anti-goat HRP (AbD Serotec) and DAB chromogen staining (SIGMAFastTM 3,3′-DAB tablets, Sigma-Aldrich, UK). Tissue was counterstained with haematoxylin, blued with Scott's tap water, rinsed in water, dehydrated through graded alcohols, cleared and mounted with Mowiol. DAB stained sections were used to map sites of active virus infection within lung sections using NanoZoomer Digital Pathology Viewing software (NDP.view, Hamamatsu). Area (µm2) of infection and total lung area were exported to and analysed by Excel.
2.5. Quantitation of monocyte/macrophage infiltration
MacGreen WT and IFNγR-/- lungs were inflation-fixed with 4% paraformaldehyde for 4 h, followed by 24 h in 18% (w/v) sucrose and mounted with Tissue-Tek OCT medium in isopentane cooled over dry ice. Frozen sections were cut at 8–10 µm using a cryostat, air dried and stored at −70 °C. Sections were air dried, blocked with CAS block (Invitrogen) and stained with goat anti-GFP (AHP975; AbD Serotec, UK) followed by anti-goat Alexa Fluor-488 (Invitrogen). Cells were counterstained with DAPI to visualize nuclei and mounted with ProLong Gold (Invitrogen). Sections were analysed using Leica DMLB, Zeiss LSMpascal confocal microscopy and ImageJ software.
2.6. Bronchoalveolar lavage fluid (BALF) preparation and flow cytometry analysis
BALF was collected from control and IAV infected WT and IFNγR-/- mice. Lungs were flushed multiple times with 1 ml ice-cold sterile HBSS with 3 mM EDTA, pH 7.2. BALF was pooled and maintained on ice until centrifugation at 350×g for 10 min, 4 °C, and treated with red blood cell lysis buffer prior to further analysis. Cells were resuspended in FACS buffer (PBS/FCS) with Fc block (CD16/32, 0.5 mg/ml; BD Biosciences, UK) and stained for 1 h at 4 °C using the following antibodies: anti-CD11b-APC (M1/70.11.5; Miltenyi Biotec Ltd., UK), anti-Ly6G-PE/Cy7 (BD Biosciences), anti-CD4-PE (RM4–5; BD Biosciences), anti-CD8-FITC (53–6.7; BD Biosciences), anti-CD11c-PE/Cy7 (N418; BioLegend, UK), anti-Siglec-F BV421 (E50–2440; BD Biosciences), and appropriate isotype/FMO controls. Cell viability was determined using Zombie fixable dyes during staining to exclude dead cells (Zombie Aqua or Violet; BioLegend). Cells were fixed with BD Cytofix (BD Biosciences) and analysed using the BD LSR Fortessa and FlowJo V10 software (FlowJo LLC, USA) using a gating strategy adapted from Misharin et al. (2013) (Supplementary Fig. S4).
2.7. Determination of cytokine protein production by array and ELISA
WT and IFNγR-/- mice were challenged with 1 × 104 PFU A/WSN/33 influenza virus and, at appropriate times, lungs were harvested and homogenised in serum free DMEM medium using the TissueLyser II system (Qiagen, UK). Lysate was clarified and supernatant was used in the Proteome Profiler, mouse cytokine array panel A kit (R&D Systems, USA), following manufacturer's guidelines, developed with IRDye 800CW Streptavidin and analysed using the LI-COR Odyssey imaging system (LI-COR, USA) and Image Studio Lite software (LI-COR). ELISAs were performed on clarified supernatant. IFNβ ELISAs used anti-mouse IFNβ capture antibody (clone RMMB-1), a rabbit anti-mouse IFNβ detection antibody (R&D Systems), and goat anti-rabbit-HRP (Abcam, UK) and were developed using DuoSet ancillary substrate reagent pack 2 (R&D Systems). IFNλ was detected using murine IL-28A/B (IFNλ2/3) DuoSet ELISA kit and ancillary reagent kit 2, following the manufacturer's guidelines (R&D Systems). ELISA optical densities determined using a microplate reader set at 450 nm, with wavelength correction. Standard curves were created and four parameter logistic curve-fit used, and absolute quantities of analytes obtained in pg/ml.
2.8. qRT-PCR
Lungs from WT mice and IFNγR-/- mice were harvested into RNALater and after 24 h RNA was extracted using RNeasy kits (Qiagen), with on column DNase treatment (Qiagen). cDNA was synthesized with SuperScript II RT kit (ThermoFisher Scientific) and Oligo(dT)15 primers. qRT-PCR was performed using SYBRGreen I (Biogene) and FastStart Taq (Roche). Primers for succinate dehydrogenase complex A (SDHA) and CD206 were as previously described (Campbell et al., 2015). The specificity of the amplicons was verified by melt curve analysis, and PCR efficiencies determined. All primers had an efficiency >95%. The threshold cycles (Ct) were determined in triplicate, normalized to levels of a constitutive housekeeping gene and used to obtain the relative levels of genes of interest using the 2-∆Ct method. Alternatively, absolute copy number was calculated from standard curves and normalized to levels of expression of the housekeeping gene SDHA.
2.9. Statistical analyses
All statistical analyses were carried out in R (Version 3.4.2 © 2017 R Foundation for Statistical Computing). Viral titer data, lung section areas, protein array, gene expression, IFNγ, IFNλ and gene copy/reaction data were log10 transformed prior to analyses to normalise the residuals. Back-transformed least square mean ± 95% confidence intervals for summary measures were obtained (not reported). As different experiments had been used to generate the data, where possible (days 1–4) experiment number was added as a fixed effect in all analyses. For post day 4, either only a single experiment was carried out, or only one mouse strain was used in that experiment, so no experiment effect was added. Statistical significance was taken when p was < 0.05. Differences in weight, virus load and lung section data on different days post-infection between mouse strains were evaluated with analysis of variance (ANOVA). Log10 total lung area was added as a covariate in the analyses of lung section areas. Overall differences between mouse strains in protein array and gene expression data were also evaluated with ANOVA, with two-sample t-tests used to look at individual cytokine and chemokines. For histopathology score data ordinal logistic regression (OLR) were carried out. Binary logistic regression models (BLR) were used to evaluate differences between mouse strains in the percentage of CD4+ and CD8+ T cells, neutrophils and alveolar macrophages in BALF. Where there was over dispersion in these BLR models, this was adjusted for by using a ‘quasibinomial’ error structure. Differences in gene copies between mouse strains was evaluated using two-sample t-tests.
3. Results
3.1. Absence of a type II IFN receptor confers resistance to severe influenza A virus infection
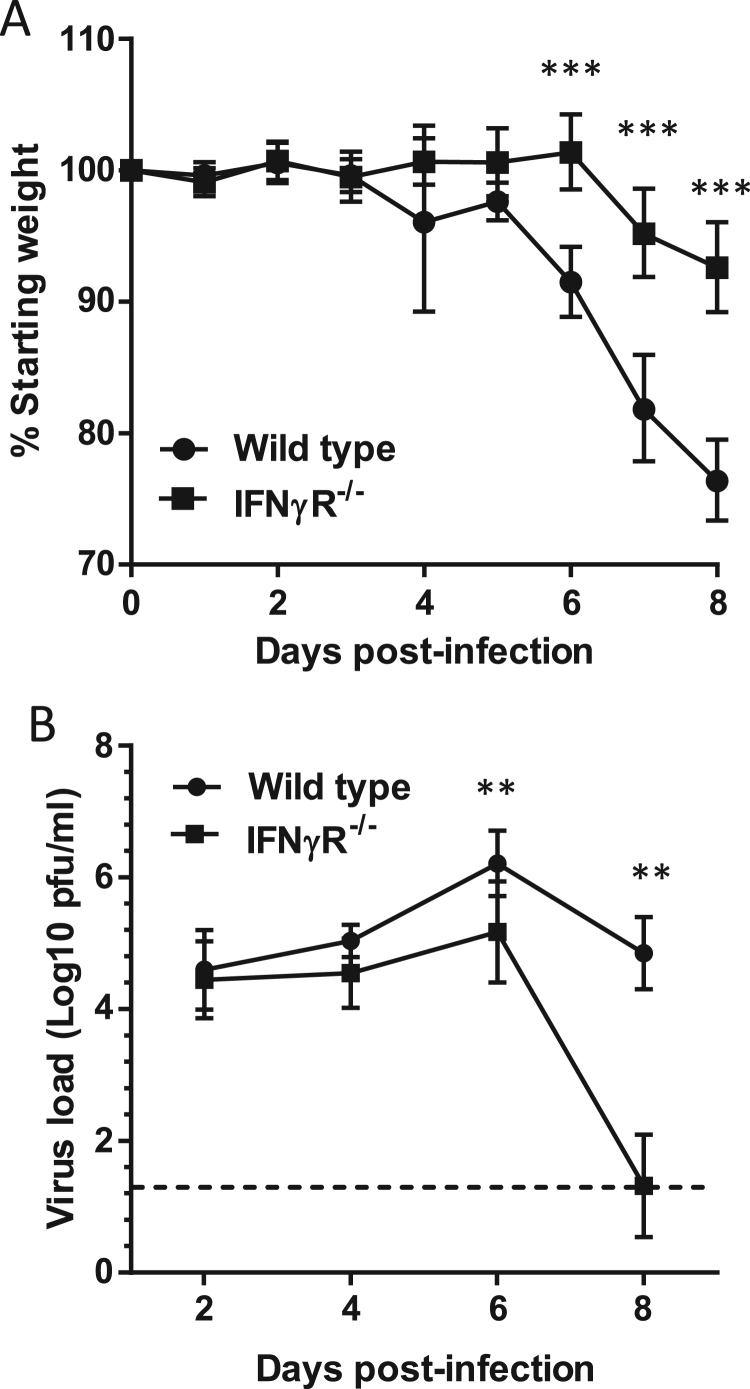
In order to investigate the role of IFNγ in IAV infection, we infected WT and IFNγR-/- mice with the H1N1 IAV strain A/WSN/33. Infection of WT mice resulted in progressive weight loss from 6 days post-infection (Fig. 1A) and an increase in clinical symptoms characterized by hunching, ruffled fur, decreased mobility (data not shown). In contrast, IFNγR-/- mice had no weight loss at day 6 and lost significantly less weight at 7 and 8 dpi (Fig. 1A, p < 0.001). When virus loads in these mice were quantified by plaque assay, there was no difference in total lung virus loads at 2 or 4 dpi (p > 0.174) but at 6 and 8 dpi IFNγR-/- mice had significantly less virus (p < 0.002) and by 8 dpi the majority of animals had cleared the virus (Fig. 1B). These results were highly reproducible with both clinical signs and virus loads indicating that IFNγR-/- mice developed disease of reduced severity compared to WT mice.
Fig. 1.
IFNγR-/-mice are more resistant to severe IAV infection than wild type mice. Wild type and IFNγR-/- mice were infected with A/WSN/33 and monitored daily for weight loss (A); at appropriate time points, lungs were harvested and assayed for infectious virus (B). Graphs show mean with 95% CI, data are from 6 experiments in total with between 29 (weight day 0 wild type) and 6 (days 7–8 IFNγR-/-) mice per time point. Data were analysed by ANOVA, with experiment added as a fixed effect for days 1–4. ***p < 0.001, **p < 0.01.
3.2. Comparative histopathology of infection in WT and IFNγR-/- mice
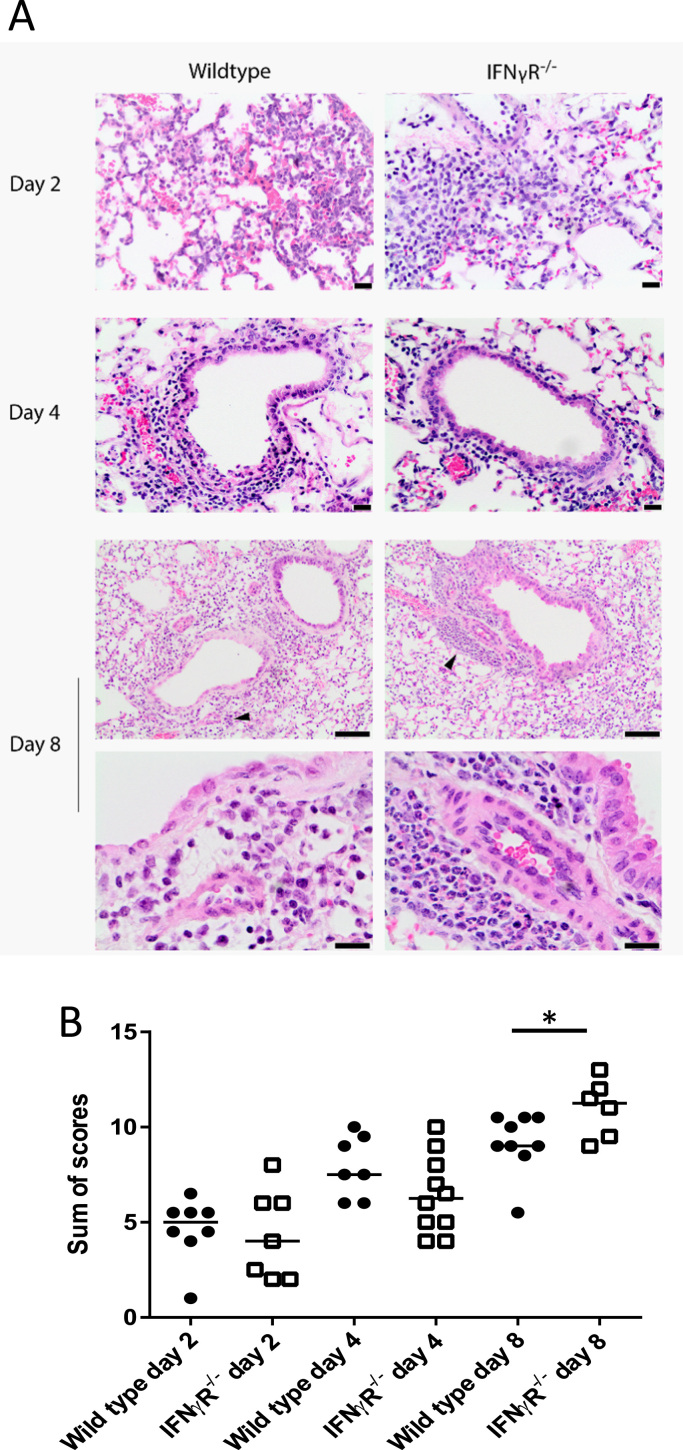
To determine whether the enhanced survival of IFNγR-/- mice following IAV infection was associated with decreased pathology, lung H&E sections were blind scored for evidence of tissue damage (Fig. 2A). The pathology identified in the lungs was characterized by mild to severe, focal to coalescing, acute to subacute bronchointerstitial pneumonia with epithelial cell necrosis, consistent with infection with influenza A virus. As expected, the severity of the histopathological changes increased over time (day 2 < day 4 < day 8). Sections were scored on a scale of 0–3 for epithelial and interstitial necrosis as well as peribronchial, perivascular and interstitial inflammation (Supplementary Fig. S1). At 2 and 4 dpi, there were no significant differences in overall scores between WT and IFNγR-/- mice (p = 0.084, Fig. 2B), however, the IFNγR-/- mice showed a trend towards reduced levels of epithelial necrosis (days 2 and 4) and interstitial inflammation (day 2) compared to the WT mice (Supplementary Fig. S1). At 8 dpi the IFNγR-/- mice had fewer clinical symptoms and lower virus loads, but higher overall pathology scores compared to WT mice (Fig. 2B) due to increased perivascular and interstitial infiltration by neutrophils. Interestingly, the extent of necrosis at 8 dpi was similar in both strains (Supplementary Fig. 1). The increased necrosis and inflammation in the WT mice at early times was identified as a key finding, potentially providing an indicator as to the role played by IFNγ in infection.
Fig. 2.
Histopathological changes within the lungs of wild type and IFNγR-/-mice infected with IAV. Wild type and IFNγR-/- mice were infected with IAV A/WSN/33 and, at the indicated times post-infection, mice were euthanized, lungs were inflation-fixed and H&E sections were prepared. Sections were scored blind for epithelial and interstitial necrosis, peribronchial, perivascular and interstitial inflammation and perivascular and peribronchial oedema using a score from 0 to 3. (A) Representative H&E sections. Arrow heads indicate perivascular cuffing and these areas are shown at higher magnification in the bottom panels. Scale bars are 50 µm, top 4 panels and bottom 2 panels and 20 µm for day 8 low magnification. (B). Sum of pathology scores from individual mice with median. Data represent four independent experiments, with 6–10 mice per time point. *p < 0.05 by ordinal logistic regression, with experiment added as a fixed effect for days 2 and 4.
3.3. Viral spread is controlled in the lungs of IFNγR-/- mice at early time points
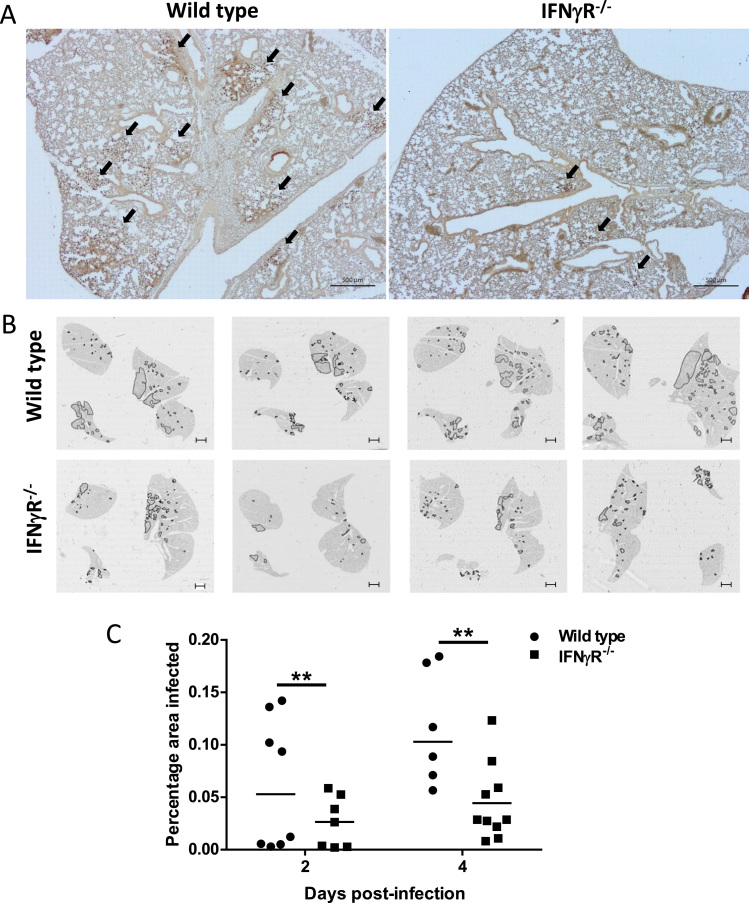
To determine if the absence of the IFNγR influenced the spread of IAV infection in the lungs, lung sections were stained with antibody to H1N1 virus and examined by microscopy. Low power images (Fig. 3A) showed that there was less virus antigen in the IFNγR-/- lungs than in the WT lungs. In order to quantitate this, whole lung sections stained with antibody to H1N1 and were scanned by NanoZoomer allowing us to measure the total infected area per lung section. Whole lung section maps (Fig. 3B) showed that WT mice had a greater overall spread of the virus than IFNγR-/- mice and more foci of active infection. As there was some mouse to mouse variation, we quantitated the infected areas and calculated the percentage of total lung area infection at 2 and 4 dpi (Fig. 3C). This showed that the IFNγR-/- mice had a significantly (p = 0.001) lower mean area of infection than WT mice (Fig. 3C), indicating that spread of infection was already controlled at these early time points.
Fig. 3.
Virus infection is more widespread in the lungs of wild type mice compared with those of IFNγR-/-mice. Whole lung sections were paraffin-embedded, stained with antibody to H1N1 virus and imaged by light microscopy or scanned for virus antigen using the NanoZoomer Digital Slide Scanner. (A) Representative lung Section 2 dpi. Arrows indicate virus antigen positive areas. (B) Map of lung sections from individual mice at 2 dpi showing sites of active virus infection. Areas positive for viral antigen are outlined in black. Images are from four individual mice. Slide bar is 1 mm. (C) Virus antigen positive areas at 2 and 4 dpi were quantitated using NDP.view software (Hamamatsu). Graph shows the percentage of total lung area positive for virus antigens per mouse with median. **p < 0.01, (taking into account total lung area imaged as a covariate, and experiment as a fixed effect). Data are from two independent experiments, n = 4 mice per group, per experiment.
3.4. Comparative expression of cytokines and chemokines in IFNγR-/- mice
As type I and type III interferons mediate control of IAV infection (Crotta et al., 2013), we hypothesized that differences in type I and/or type III interferon responses might account for the reduced viral spread in the lungs of IFNγR-/- mice. However, there was no statistically significant difference between mouse strains observed (p > 0.254) by ELISAs for IFNβ and IFNλ (Supplementary Fig. S2) or qRT-PCR for IFNβ and IFNα4 transcripts (Supplementary Fig. S3). This indicated that control of spread was not likely to be due to differential expression of these cytokines early after infection (Supplementary Fig. S3). WT mice did, however, have greater levels of IFNλ by 6 dpi, though this difference was not statistically significant (Supplementary Fig. S2).
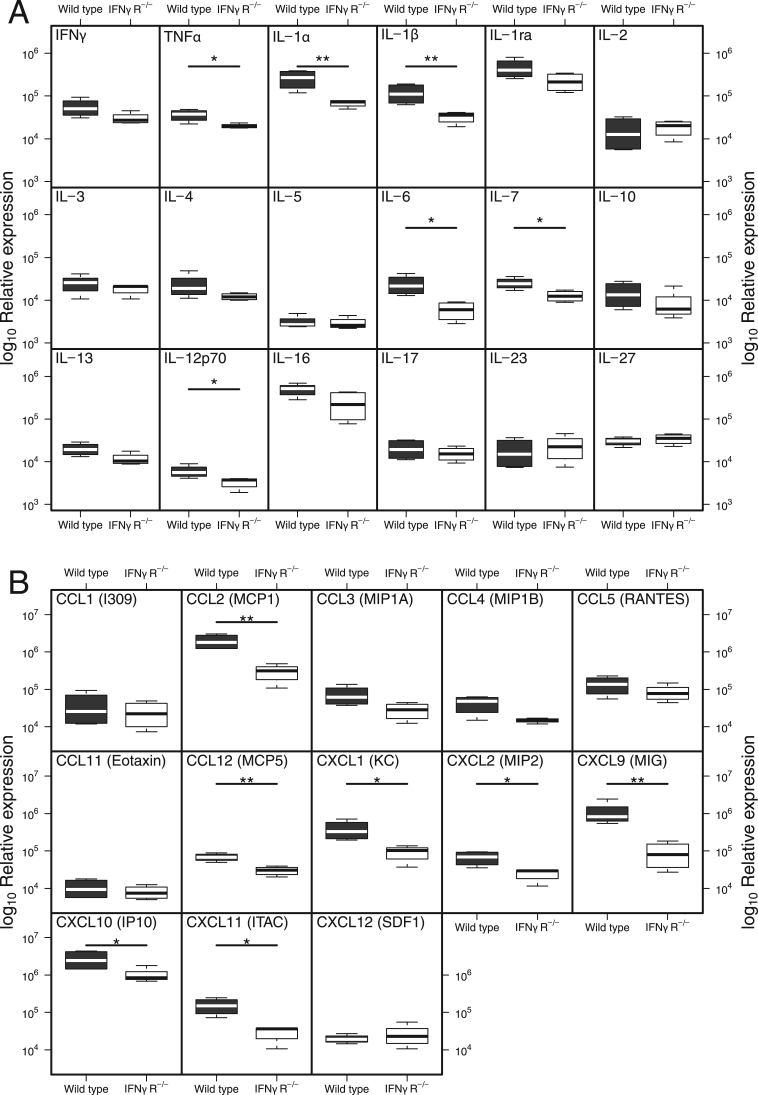
Given the role of IFNγ as a major pro-inflammatory mediator, we asked if there were changes in other cytokines or chemokines which would allow us to understand how the lack of IFNγ was protective. We therefore compared expression of pro-inflammatory mediators in IFNγR-/- and WT mice in lung homogenate using protein arrays. A number of inflammatory cytokines, including TNFα, IL-1α, IL-1β, IL-6, IL-7 and IL-12p70, were elevated in the lungs of WT mice at 2 dpi when compared with IFNγR-/- mice (p < 0.05, Fig. 4A). In contrast, twelve other inflammatory cytokines showed no significant differences between mouse strains at this early time (Fig. 4A, p > 0.06).
Fig. 4.
Differences in cytokine and chemokine expression in wild type and IFNγR-/-mice can be detected two days after infection. Wild type and IFNγR-/- mice were infected with IAV. Lungs were harvested 2dpi and homogenates were assayed for expression of cytokines (A) or chemokines (B) by protein array. Data show min/max with median, n = 4 mice per group. **p < 0.01, *p < 0.05 by two sample t-tests on log10 transformed data.
The array analysis revealed higher levels of the chemoattractants MCP-1 (CCL2), MCP-5 (CCL12), KC (CXCL1), MIP-2 (CXCL2), MIG (CXCL9), IP-10 (CXCL10) and I-TAC (CXCL11) (p < 0.044, Fig. 4B) in WT mice when compared to IFNγR-/- mice at 2 dpi. Increased expression of MCP-1 (CCL2), MCP-5 (CCL12), MIG (CXCL9) and I-TAC (CXCL11) was maintained in WT mice at 6 dpi (p < 0.024) and, in addition, levels of MIP1α (CCL3), MIP1β (CCL4), RANTES (CCL5) were elevated at this later time point (Supplementary Fig. S4). Overall, the expression of inflammatory mediators in the IFNγR-/- mice was lower at 2 dpi and by 6 dpi there was further evidence of a greater inflammatory response in the WT mice. The muted inflammatory response to IAV infection in the IFNγR-/- mice correlates with the lack of clinical symptoms and decreased weight loss.
3.5. Comparative lung cell populations in WT and IFNγR-/- mice
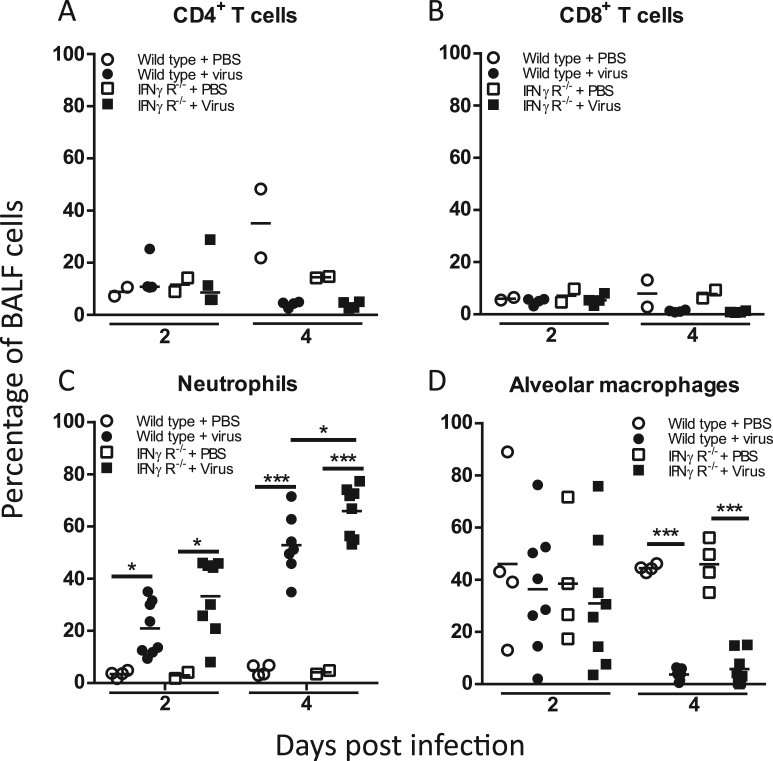
We next examined whether these altered cytokine and chemokine profiles in IFNγR-/- mice following IAV infection influenced the lung cellular environment and whether differences in immune cell recruitment might account for the differential control of infection. Flow cytometric analysis demonstrated that the BALF from IAV-infected WT and IFNγR-/- mice 2 or 4 dpi contained similar percentages of T cells to the mock-infected mice and, importantly, that percentages of CD4+ T or CD8+ T cells were similar in both strains (p > 0.422, Fig. 5A, B). By 2 dpi, however, the BALF cell population was comprised of over 20% CD11b+ Ly6G+ neutrophils. The percentage of neutrophils increased between 2 and 4 dpi and by 4 dpi there were significantly more neutrophils in the IFNγR-/- mice (p = 0.013, Fig. 5C, gating strategy Supplementary Fig. S5). Thus, although there was no difference in T cell recruitment at early time points, lack of IFNγR resulted in differences in the neutrophil percentages.
Fig. 5.
Immune cell populations in BALF. Wild type and IFNγR-/- mice were infected with IAV and BALF collected at indicated times post-infection. Frequency of CD4+ T cells (A), CD8+ T cells (B), neutrophils (C), and alveolar macrophages (AMs) (D) were quantified by FACS. CD4+ and CD8+ T cells were detected with directly conjugated antibodies to CD4 and CD8. Neutrophils were CD11b+Ly6G+ and AMs were CD11b-,CD11c+,SiglecF+ populations. Graphs show individual mice from two independent experiments, n = 2 mice per PBS group and n = 4 per virus group. Bars show mean values. ***p < 0.001, **p < 0.01, *p < 0.05 by binary logistic regression, with experiment added as a fixed effect for neutrophil and AM analyses.
We next considered whether early clearance of virus in the IFNγR-/- mice might be related to differences in lung macrophage populations. Firstly, we asked if there were differences in the percentages of alveolar macrophage (AM) as these are the first immune cells which encounter virus within the lung. We found that percentages of AMs (defined as CD11b- CD11c+ Siglec-F+, gating strategy Supplementary Fig. S5) (Misharin et al., 2013) were similar in the lungs of mock-infected and IAV-infected WT and IFNγR-/- mice at 2 dpi (p < 0.778). By 4 dpi, however, as has been previously noted for IAV infections (Ghoneim et al., 2013, LeMessurier et al., 2016), the AM population was severely depleted in both strains (p < 0.001, Fig. 5D). Thus, there was no apparent significant difference in the percentages of AMs or the kinetics of AM loss between the WT and IFNγR-/- mice (p = 0.062).
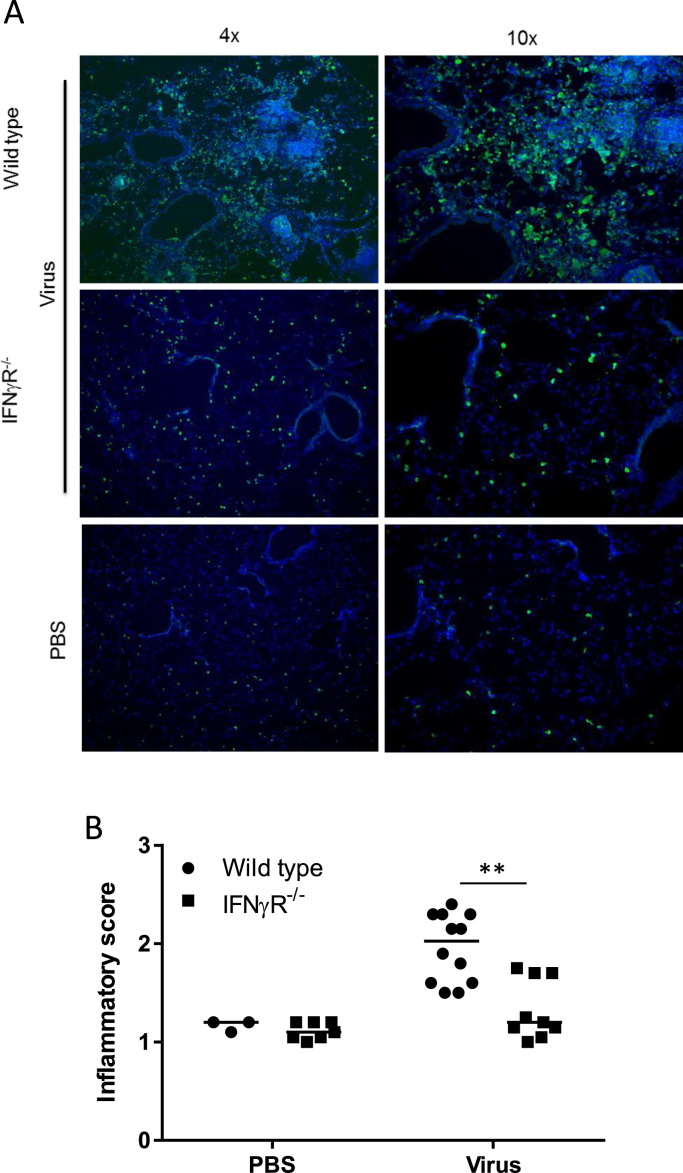
Alveolar macrophages, however, represent only one of the lung macrophage populations. Interstitial macrophages are found within the lung parenchyma and, in the infected lung, monocytes are recruited and differentiate into exudate derived macrophages. In order to look at the overall macrophage population in the lung, we utilized the MacGreen mouse model where all cells of macrophage lineage express EGFP (Sasmono et al., 2003). WT and IFNγR-/- mice were backcrossed with MacGreen mice and infected with IAV and EGFP expression in lung sections was visualized and quantitated. Comparison with mock-infected mice demonstrated that there was less recruitment of EGFP positive cells into the lungs of MacGreen IFNγR-/- mice at 2 dpi, when compared to the MacGreen WT controls (Fig. 6A, B), a difference that was statistically significant (p < 0.003). Thus there were significant differences in both the neutrophil and macrophage populations following IAV infection of IFNγR-/- mice.
Fig. 6.
Decreased infiltration of inflammatory cells in IFNγR-/-mice. Wild type MacGreen and IFNγR-/- MacGreen mice were infected with IAV and lung sections analysed for presence of EGFP expressing cells 2 dpi by staining with goat anti-GFP followed by Alexa Fluor-488. Cells were counterstained with DAPI (blue). Ten fields were imaged for each mouse and the distribution of green cells blind scored as follows: 0 = no green cells; 1 = green cells present; 2 = moderate focal or multifocal; 3 = marked EGFP expression apparent on majority of area. Graph represents three independent experiments. Solid line shows median inflammatory score. **p < 0.01 from ordinal logistic regression.
4. Discussion
Our in vitro studies on infection of BMDMs showed that IFNγR-/- macrophages had a muted inflammatory response following IAV infection (Campbell et al., 2015). Based on these studies, we set out to investigate in detail how a deficiency in the ability to mount a classical macrophage response would impact the outcome of infection in vivo by comparing the kinetics of infection in IFNγR-/- mice with those in WT mice. Importantly, we used the same strain of virus (A/WSN/33) and the same genetic background of mice (129 Sv/Ev) as in our in vitro studies, as both virus strain and mouse genetic background can have a significant effect on the outcome of infection (Pica et al., 2011, Srivastava et al., 2009, Trammell et al., 2012).
At virus doses that caused severe clinical disease in WT mice, IFNγR-/- mice were less susceptible to IAV infection. From 2 dpi virus spread was attenuated in IFNγR-/- mice and there was decreased recruitment of monocyte/macrophage lineage cells. Although there was no difference in virus titers or clinical signs at early time points (2 and 4 dpi), at later times differences were found in both parameters. By 8 dpi, WT mice had severe disease and were culled for humane reasons while IFNγR-/- animals had fewer clinical signs and weight loss at day 8 and no signs of infection by day 15 (data not shown).
Our data contrast with other studies which have concluded that IFNγ plays no role in mouse survival or morbidity and does not affect virus clearance (Baumgarth and Kelso, 1996, Graham et al., 1993, Price et al., 2000, Turner et al., 2007). However, as noted above, both virus and mouse strain can affect outcome. Interestingly, these studies showed that although IFNγ did not have a major role in the development of antibody and/or CD8 T cell responses to IAV infection, there was reduced cellular infiltration in the absence of IFNγ (Baumgarth and Kelso, 1996, Turner et al., 2007). Our studies also show reduced cellular infiltration and explore how events early in infection influence the longer term outcome. The data provide new insight into the role of IFNγ in inflammatory cell recruitment and its importance in IAV pathogenesis.
The reduced inflammatory response in the IFNγR-/- mice could result from the inability of macrophages and other cells to respond to IFNγ. Production of pro-inflammatory cytokines correlates with clinical symptoms and weight loss in IAV infection (Cheung et al., 2002, Peiris et al., 2009) and hence the lack of such symptoms in IFNγR-/- mice likely reflects the lack of pro-inflammatory response. Similarly, the reduced macrophage/monocyte infiltration observed 2 dpi correlates with the lower production of chemokines such as MCP1/CCL2 and CCL12 both of which are produced by inflammatory macrophages and are chemoattractants for cells of this lineage. Importantly, our findings show that an aggressive inflammatory response is not essential for control of infection.
The mechanism by which virus spread is controlled at early times and how this relates to the decreased lung virus loads at day 6 and more rapid clearance of the virus remains to be determined. Interplay between type I and type II interferons has been documented (Stifter et al., 2016) and we hypothesized that the lack of IFNγ might result in an enhanced type I or type III IFN response which would account for the reduced spread of virus at days 2 and 4 and the lower viral load at day 6. However, we could find no evidence for elevated levels of IFNβ or IFNλ in the lungs of IFNγR-/- mice at early time points indicating that control of infection is unlikely to be due to these cytokines.
T cell responses to IAV infections are detected by 6 dpi and hence it is also unlikely that enhanced T cell responses account for the decreased spread at 2 and 4 dpi. In agreement with this, we did not find increased T cell infiltration into the BALF of IFNγR-/- mice. Hence it is unlikely that early recruitment of T cells accounts for the control of virus spread.
In contrast, neutrophil populations were increased in the IFNγR-/- mice by 2 dpi and by day 4 this was statistically significant. At day 8, neutrophil infiltration represented the major component of histopathology in IFNγR-/- mice. Similar increases in neutrophil numbers have been reported at late time points (day 7) in C57/BL6 IFNγR-/- mice infected with H1N1 A/PR8/34 (Stifter et al., 2016). Production of chemokines that drive neutrophil recruitment is stimulated by IL-17 (Linden et al., 2005) which is, in turn, under control of IFNγ (Nandi and Behar, 2011). In addition, IFNγ directly impairs survival of polymorphonuclear cells (Nandi and Behar, 2011), suggesting that, in the absence of IFNγ, neutrophils are likely to be recruited in higher numbers and to survive for longer. Neutrophil infiltration has been documented in animal models of severe IAV infection (Graham et al., 1993, Hussell et al., 2001, Tate et al., 2010, Taub et al., 1993, To et al., 2001) and they contribute to pathogenesis in response to other infections. However, our data indicate that neutrophils are likely to have diverse roles in IAV pathogenesis, highlighting a potentially important role in early control of infection. Further studies are required to determine whether the early increase in neutrophil numbers in the absence of IFNγR signaling contributes to the control of spread and clearance of virus and how the high neutrophil numbers at later times relate to virus pathogenesis.
Macrophages can be infected by IAV and our in vitro studies on BMDMs showed that alternatively activated BMDMs are more readily infected and more quickly killed by IAV (Campbell et al., 2015). Interestingly, we found that the alternative macrophage marker CD206 is expressed at significantly higher levels in lungs of IFNγR-/- mice (Supplementary Fig. S6) suggesting that macrophages in the lungs of these mice have a less pro-inflammatory phenotype than WT mice. CD206 has been shown to be important in virus uptake into macrophages (Reading et al., 2000, Upham et al., 2010) and hence, it is possible that macrophages in IFNγR-/- mice are more readily infected with IAV and thus act as a “sink” (Nicol and Dutia, 2014, Schneider et al., 2014) to remove infectious virus. As IAV replicates poorly in macrophages, this would result in less virus to infect epithelial cells and thus reduced virus spread in the IFNγR-/- mice. AMs are the first macrophages to encounter virus, hence such a scenario might result in more rapid loss of AMs in IFNγR-/- mice. However, we did not observe differences in kinetics of AM loss in WT and IFNγR-/- mice and similar percentages of AMs were positive for viral antigen 24 h post-infection (data not shown). Hence, additional studies are needed to determine whether infection of AMs or other lung macrophage populations contributes to the control of virus spread in IFNγR-/- mice.
The phenotype of AM may affect IAV infection in other ways. AMs have recently been shown to protect type I alveolar epithelial cells (AECs) from IAV infection by a previously unrecognized mechanism which is independent of type I and type II IFN (Cardani et al., 2017). These studies showed that specific depletion of AM resulted in enhanced viral titers and viral spread and that control of titer and spread was restored by transfer of AM. Reduced susceptibility was suggested to be due to suppression of the cysteinyl leukotriene pathway enzymes in type I AECs. The phenotype of the AM may alter its ability to suppress this pathway and the alternative phenotype in IFNγR-/- mice might be more efficient at suppressing viral replication in AECs.
An interesting possibility to consider is that the virus has found ways to exploit the IFNγ response to enhance infection and hence replication and spread is attenuated in its absence. Such dependence has been described in Tlr7-/-Mavs-/- mice where infection with sub-lethal doses of virus leads to lower lung virus titers than in WT mice. In these knockout mice, there is reduced recruitment of monocytes leading to lower numbers of infected monocyte derived DCs (Pang et al., 2013) and the decrease in target cells has been proposed to account for the decrease in viral titers. Interestingly, this effect is overcome when Tlr7-/-Mavs-/- mice are infected with higher doses. Similarly, in our model, the absence of an innate inflammatory response protects mice from lethal infection at lower doses of virus but not at higher doses (data not shown). We also found decreased recruitment of monocyte lineage cells and it is possible that a similar mechanism, i.e. reduced numbers of target cells, is responsible for the control of spread and lower virus titers. Such dependence on host responses may be crucial in human infections. Although lack of an IFNγ response is rare in humans, respiratory diseases or other infections may alter the host's ability to mount an efficient inflammatory response. Our data suggest this may impact virus amplification. This may be particularly important where the infective dose is low, as may occur in a natural infection, and the virus must be rapidly amplified to establish an infection before secondary immune responses are activated.
In conclusion, our data show that lack of an IFNγ response leads to an attenuated IAV infection characterized by fewer clinical signs, decreased viral titers and a decreased inflammatory response including lower levels of inflammatory cytokines/chemokine and less infiltration of monocyte/macrophage lineage cells. Attenuation of infection is dependent on the dose of virus and did not occur when high doses were used. The data presented here provide evidence that IFNγ responsiveness plays a crucial indirect role in early control of virus spread and shed further light on how the inflammatory response is an integral part of IAV pathogenesis. Understanding the precise mechanisms by which IAV interacts with the innate immune system, particularly in the context of low, physiological doses of virus will aid in designing new prophylactic and therapeutic strategies for prevention and control of IAV infections.
Acknowledgements
This work was supported by the Biotechnology and Biological Sciences Research Council Institute Strategic Program Grants BB/J004324/1 and BB/P013740/1 to The Roslin Institute; the Scottish Higher Education Funding Council grant to the Interdisciplinary Centre for Human and Avian Influenza Research (ICHAIR); and GMC was funded by a joint ICHAIR-BBSRC Studentship. The authors would like to thank Professor Paul Digard for critical reading of the manuscript.
Acknowledgments
Author contributions
MQN, GMC, YL performed experiments. DJS carried out statistical analyses. PB carried out histological analysis. MQN, ID AAN and BMD planned and supervised the research and MQN, DJS and BMD prepared the figures and wrote the manuscript.
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2018.10.017.
Appendix A. Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
References
- Akdis M., Burgler S., Crameri R., Eiwegger T., Fujita H., Gomez E., Klunker S., Meyer N., O'Mahony L., Palomares O., Rhyner C., Quaked N., Schaffartzik A., Van De Veen W., Zeller S., Zimmermann M., Akdis C.A. Interleukins, from 1 to 37, and interferon-gamma: receptors, functions, and roles in diseases. J. Allergy Clin. Immunol. 2011;127:701–721. doi: 10.1016/j.jaci.2010.11.050. (e701-e770) [DOI] [PubMed] [Google Scholar]
- Baumgarth N., Kelso A. In vivo blockade of gamma interferon affects the influenza virus-induced humoral and the local cellular immune response in lung tissue. J. Virol. 1996;70:4411–4418. doi: 10.1128/jvi.70.7.4411-4418.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bot A., Bot S., Bona C.A. Protective role of gamma interferon during the recall response to influenza virus. J. Virol. 1998;72:6637–6645. doi: 10.1128/jvi.72.8.6637-6645.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Califano D., Furuya Y., Roberts S., Avram D., McKenzie A.N.J., Metzger D.W. IFN-gamma increases susceptibility to influenza A infection through suppression of group II innate lymphoid cells. Mucosal Immunol. 2017 doi: 10.1038/mi.2017.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell G.M., Nicol M.Q., Dransfield I., Shaw D.J., Nash A.A., Dutia B.M. Susceptibility of bone marrow-derived macrophages to influenza virus infection is dependent on macrophage phenotype. J. Gen. Virol. 2015;96:2951–2960. doi: 10.1099/jgv.0.000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardani A., Boulton A., Kim T.S., Braciale T.J. Alveolar macrophages prevent lethal influenza pneumonia by inhibiting infection of Type-1 alveolar epithelial cells. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006140. (e1006140) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liang W., Yang S., Wu N., Gao H., Sheng J., Yao H., Wo J., Fang Q., Cui D., Li Y., Yao X., Zhang Y., Wu H., Zheng S., Diao H., Xia S., Zhang Y., Chan K.H., Tsoi H.W., Teng J.L., Song W., Wang P., Lau S.Y., Zheng M., Chan J.F., To K.K., Chen H., Li L., Yuen K.Y. Human infections with the emerging avian influenza A H7N9 virus from wet market poultry: clinical analysis and characterisation of viral genome. Lancet. 2013;381:1916–1925. doi: 10.1016/S0140-6736(13)60903-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung C.Y., Poon L.L.M., Lau A.S., Luk W., Lau Y.L., Shortridge K.F., Gordon S., Guan Y., Peiris J.S.M. Induction of proinflammatory cytokines in human macrophages by influenza A (H5N1) viruses: a mechanism for the unusual severity of human disease? Lancet. 2002;360:1831–1837. doi: 10.1016/s0140-6736(02)11772-7. [DOI] [PubMed] [Google Scholar]
- Crotta S., Davidson S., Mahlakoiv T., Desmet C.J., Buckwalter M.R., Albert M.L., Staeheli P., Wack A. Type I and type III interferons drive redundant amplification loops to induce a transcriptional signature in influenza-infected airway epithelia. PLoS Pathog. 2013;9:e1003773. doi: 10.1371/journal.ppat.1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoneim H.E., Thomas P.G., McCullers J.A. Depletion of alveolar macrophages during influenza infection facilitates bacterial superinfections. J. Immunol. 2013;191:1250–1259. doi: 10.4049/jimmunol.1300014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham M.B., Dalton D.K., Giltinan D., Braciale V.L., Stewart T.A., Braciale T.J. Response to influenza infection in mice with a targeted disruption in the interferon gamma gene. J. Exp. Med. 1993;178:1725–1732. doi: 10.1084/jem.178.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Hendriks W., Althage A., Hemmi S., Bluethmann H., Kamijo R., Vilcek J., Zinkernagel R.M., Aguet M. Immune response in mice that lack the interferon-gamma receptor. Science. 1993;259:1742–1745. doi: 10.1126/science.8456301. [DOI] [PubMed] [Google Scholar]
- Hussell T., Pennycook A., Openshaw P.J. Inhibition of tumor necrosis factor reduces the severity of virus-specific lung immunopathology. Eur. J. Immunol. 2001;31:2566–2573. doi: 10.1002/1521-4141(200109)31:9<2566::aid-immu2566>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- LeMessurier K.S., Lin Y., McCullers J.A., Samarasinghe A.E. Antimicrobial peptides alter early immune response to influenza A virus infection in C57BL/6 mice. Antivir. Res. 2016;133:208–217. doi: 10.1016/j.antiviral.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden A., Laan M., Anderson G.P. Neutrophils, interleukin-17A and lung disease. Eur. Respir. J. 2005;25:159–172. doi: 10.1183/09031936.04.00032904. [DOI] [PubMed] [Google Scholar]
- Misharin A.V., Morales-Nebreda L., Mutlu G.M., Budinger G.R., Perlman H. Flow cytometric analysis of macrophages and dendritic cell subsets in the mouse lung. Am. J. Respir. Cell Mol. Biol. 2013;49:503–510. doi: 10.1165/rcmb.2013-0086MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi B., Behar S.M. Regulation of neutrophils by interferon-gamma limits lung inflammation during tuberculosis infection. J. Exp. Med. 2011;208:2251–2262. doi: 10.1084/jem.20110919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol M.Q., Dutia B.M. The role of macrophages in influenza A virus infection. Future Virol. 2014;9:847–862. [Google Scholar]
- Nicol M.Q., Ligertwood Y., Bacon M.N., Dutia B.M., Nash A.A. A novel family of peptides with potent activity against influenza A viruses. J. Gen. Virol. 2012;93:980–986. doi: 10.1099/vir.0.038679-0. [DOI] [PubMed] [Google Scholar]
- Pang I.K., Pillai P.S., Iwasaki A. Efficient influenza A virus replication in the respiratory tract requires signals from TLR7 and RIG-I. Proc. Natl. Acad. Sci. USA. 2013;110:13910–13915. doi: 10.1073/pnas.1303275110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Cheung C.Y., Leung C.Y., Nicholls J.M. Innate immune responses to influenza A H5N1: friend or foe? Trends Immunol. 2009;30:574–584. doi: 10.1016/j.it.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone L.A., Plowden J.K., Garcia-Sastre A., Katz J.M., Tumpey T.M. H5N1 and 1918 pandemic influenza virus infection results in early and excessive infiltration of macrophages and neutrophils in the lungs of mice. PLoS Pathog. 2008;4:e1000115. doi: 10.1371/journal.ppat.1000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pica N., Iyer A., Ramos I., Bouvier N.M., Fernandez-Sesma A., Garcia-Sastre A., Lowen A.C., Palese P., Steel J. The DBA.2 mouse is susceptible to disease following infection with a broad, but limited, range of influenza A and B viruses. J. Virol. 2011;85:12825–12829. doi: 10.1128/JVI.05930-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price G.E., Gaszewska-Mastarlarz A., Moskophidis D. The role of alpha/beta and gamma interferons in development of immunity to influenza A virus in mice. J. Virol. 2000;74:3996–4003. doi: 10.1128/jvi.74.9.3996-4003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reading P.C., Miller J.L., Anders E.M. Involvement of the mannose receptor in infection of macrophages by influenza virus. J. Virol. 2000;74:5190–5197. doi: 10.1128/jvi.74.11.5190-5197.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasmono R.T., Oceandy D., Pollard J.W., Tong W., Pavli P., Wainwright B.J., Ostrowski M.C., Himes S.R., Hume D.A. A macrophage colony-stimulating factor receptor-green fluorescent protein transgene is expressed throughout the mononuclear phagocyte system of the mouse. Blood. 2003;101:1155–1163. doi: 10.1182/blood-2002-02-0569. [DOI] [PubMed] [Google Scholar]
- Schneider C., Nobs S.P., Heer A.K., Kurrer M., Klinke G., van Rooijen N., Vogel J., Kopf M. Alveolar macrophages are essential for protection from respiratory failure and associated morbidity following influenza virus infection. PLoS Pathog. 2014;10:e1004053. doi: 10.1371/journal.ppat.1004053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava B., Blazejewska P., Hessmann M., Bruder D., Geffers R., Mauel S., Gruber A.D., Schughart K. Host genetic background strongly influences the response to influenza a virus infections. PLoS One. 2009;4:e4857. doi: 10.1371/journal.pone.0004857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stifter S.A., Bhattacharyya N., Pillay R., Florido M., Triccas J.A., Britton W.J., Feng C.G. Functional interplay between Type I and II interferons is essential to limit influenza A virus-induced tissue inflammation. PLoS Pathog. 2016;12:e1005378. doi: 10.1371/journal.ppat.1005378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate M.D., Pickett D.L., van Rooijen N., Brooks A.G., Reading P.C. Critical role of airway macrophages in modulating disease severity during influenza virus infection of mice. J. Virol. 2010;84:7569–7580. doi: 10.1128/JVI.00291-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taub D.D., Conlon K., Lloyd A.R., Oppenheim J.J., Kelvin D.J. Preferential migration of activated CD4+ and CD8+ T cells in response to MIP-1 alpha and MIP-1 beta. Science. 1993;260:355–358. doi: 10.1126/science.7682337. [DOI] [PubMed] [Google Scholar]
- To K.F., Chan P.K., Chan K.F., Lee W.K., Lam W.Y., Wong K.F., Tang N.L., Tsang D.N., Sung R.Y., Buckley T.A., Tam J.S., Cheng A.F. Pathology of fatal human infection associated with avian influenza A H5N1 virus. J. Med. Virol. 2001;63:242–246. doi: 10.1002/1096-9071(200103)63:3<242::aid-jmv1007>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Trammell R.A., Liberati T.A., Toth L.A. Host genetic background and the innate inflammatory response of lung to influenza virus. Microbes Infect./Inst. Pasteur. 2012;14:50–58. doi: 10.1016/j.micinf.2011.08.008. [DOI] [PubMed] [Google Scholar]
- Tumpey T.M., Garcia-Sastre A., Taubenberger J.K., Palese P., Swayne D.E., Pantin-Jackwood M.J., Schultz-Cherry S., Solorzano A., Van Rooijen N., Katz J.M., Basler C.F. Pathogenicity of influenza viruses with genes from the 1918 pandemic virus: functional roles of alveolar macrophages and neutrophils in limiting virus replication and mortality in mice. J. Virol. 2005;79:14933–14944. doi: 10.1128/JVI.79.23.14933-14944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner S.J., Olivas E., Gutierrez A., Diaz G., Doherty P.C. Disregulated influenza A virus-specific CD8+ T cell homeostasis in the absence of IFN-gamma signaling. J. Immunol. 2007;178:7616–7622. doi: 10.4049/jimmunol.178.12.7616. [DOI] [PubMed] [Google Scholar]
- Upham J.P., Pickett D., Irimura T., Anders E.M., Reading P.C. Macrophage receptors for influenza A virus: role of the macrophage galactose-type lectin and mannose receptor in viral entry. J. Virol. 2010;84:3730–3737. doi: 10.1128/JVI.02148-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material