Abstract
Evaluation of antigen-specific T-cell responses to viral antigens is frequently performed on IFN-γ secreting cells. However, T-cells are capable of producing many more functions than just IFN-γ, some of which, like Perforin, are associated with immune protection in HIV-1 disease elite controllers. We evaluated the extent of missed T-cell functions when IFN-γ secretion is used as a surrogate marker for further evaluation of T-cell functions. Intracellular cytokine staining assay and flow cytometry were used to assess peripheral blood mononuclear cells (PBMCs) from 31 HIV-infected ART-naive individuals for the extent to which gated CD4+ and CD8+ IFN-γ producing and non-producing T-cells also secreted IL-2, Perforin, and TNF-α functions. Similarly, the extent of missed virus-specific responses in IFN-γ ELISpot assay negative T-cells from 5 HIV-1 uninfected individuals was evaluated. Cells from HIV-infected individuals were stimulated with pooled consensus group M (Con M) peptides; and those from healthy individuals were stimulated with pooled adenovirus (Ad) peptides. Overall, frequencies of virus-specific IFN-γ secreting CD4+ and CD8+ cells were low. Proportions of IFN-γ negative CD4+ expressing IL-2, Perforin, or TNF-α to Con M were significantly higher (5 of 7 functional profiles) than the corresponding IFN-γ positive CD4+ (0 of 7) T-cell phenotype, p = 0.02; Fisher’s Exact test. Likewise, proportions of CD8+ T-cells expressing other functions were significantly higher in 4 of the 7 IFN-γ negative CD8+ T-cells. Notably, newly stimulated Perforin, identified as Perforin co-expression with IL-2 or TNF-α, was significantly higher in IFN-γ negative CD8+ T-cell than in the positive CD8+ T-cells. Using SEB, lower responses in IFN-γ positive cells were most associated with CD4+ than CD8+ T-cells. These findings suggest that studies evaluating immunogenicity in response to HIV and Adenovirus viral antigens should not only evaluate T-cell responsiveness among IFN-γ producing cells but also among those T-cells that do not express IFN-γ.
Keywords: HIV-1, IFN-γ negative T-cells, Vaccines, ELISpot assay, Flow cytometry, T-cell responses
1. Introduction
T-cells exert strong selective pressure on HIV replication [1]. In HIV-1 infected persons, their emergence coincides with reduced acute-phase plasma viremia, and their depletion is linked to loss of control of viral replication [1], [2]. Designing an effective T-cell based vaccine to prevent HIV acquisition requires understanding and detecting those T-cell functions that contribute to protection. The IFN-γ ELISpot assay is a cost-effective method for detecting HIV-specific T-cell responses [3], [4]. However, this assay was optimized to detect only IFN-γ production. Attempts to use ELISpot to distinguish dual cytokines detected significantly lower IFN-γ than when this function was evaluated alone [5]. While identifying T-cell responses by initially screening with the IFN-γ ELISpot assay is a robust and cost effective approach; it assumes that other virus-specific T-cell functions predominantly simultaneously express with IFN-γ. There are several limitations to using IFN-γ expression as a surrogate marker for further assessment of other T-cell responses to viral challenge. First, the detected IFN-γ responses are usually narrowly directed [6], [7]; in some cases, IFN-γ production positively correlates with enhanced viral replication [8], and its secretion does not always correlate with CD8+ T-cell cytolytic activity [9], [10]. Besides, most virus-specific IFN-γ producing cells are mono-functional, terminally differentiated T-cells that may be linked to poor clinical prognosis in HIV-infected patients [11], [12], [13], [14]. Finally, virus-specific IFN-γ expression failed to predict vaccine protection in a Phase III Step Study trial that evaluated efficacy of the MRKAd5 HIV-1 gag/pol/nef vaccine [4]. In that vaccine trial, T-cells isolated from 75% of the vaccinated individuals expressed IFN-γ [4], but the vaccine failed to protect them from acquiring HIV-infection. It remains unclear what the extent of missed detection is when you rely on IFN-γ expression as a representative surrogate for evaluating other co-expressed functional correlates of protection from HIV-1 disease.
On the other hand, expression of other T-cell functions, such as Perforin and MIP-1β, has been correlated with reduced viral load and slower disease progression in HIV-1 elite-controllers [15], [16]. Likewise, Interleukin 2 (IL-2) expression has been shown to activate natural killer (NK) cells leading to apoptosis of HIV-1 infected T-cells; and to enhance proliferation of HIV-1 specific CD8+ T-cells [17], [18]. Additionally, tumor necrosis factor-α (TNF-α) has been linked to protection by inducing apoptosis of virally infected target cells [19]. Therefore, many other cytokines are necessary for an effective host response to virus infection.
Evaluation of other cellular immune functions is commonly performed only among those T-cells initially identified to be IFN-γ secreting using the ‘back-gating’ procedure of flow cytometry analysis [20], or using ELISpot assay screening for individuals with positive IFN-γ secretion [21], [22], [23], [24], [25]. Proportions of CD4+ and CD8+ T-cells that lack expression of IFN-γ (IFN-γ negative) but secrete other T-cell functions in response to viral antigens have not been well studied. Thus, we have evaluated the potential extent of missing detection of virus-specific expression of IL-2, Perforin, and TNF-α by IFN-γ non-secreting T-cells, when IFN-γ is used as a surrogate for further T-cell functional evaluation. We evaluated the contribution of IFN-γ negative T-cells using a diseased state model represented by HIV-1 infected populations, and the extent of missed detection in a healthy immune system model represented by individuals that would ideally qualify for participation in vaccine clinical trials.
2. Materials and methods
2.1. Study participants
This study obtained residual PBMC specimens from two earlier MRC/UVRI cross-sectional cohorts, whose participants had previously consented to donate blood for use in future HIV-1 vaccine discovery research studies. One cohort comprised subjects who were chronically infected with HIV-1; these were used to model assessment of immune responses to disease. The second cohort comprised healthy HIV-uninfected volunteers selected according to the standard inclusion criteria for selecting healthy HIV-1 vaccine trials volunteers, to model assessment immune responses in healthy participants of clinical trials [26].
The HIV-1 infected cohort was recruited as part of a multi-center study to evaluate whether consensus group M peptides could be used to detect HIV-1 specific responses at similar frequencies across geographical regions of Uganda, South Africa, Cameroon, and Ethiopia diverse infecting HIV strains [27]. This was done because the different locations possessed completely different clades of circulating HIV-1 strains making it even more complicated to globally assess HIV-specific immune responses across sites with differing strains. This parent cohort enrolled 50 HIV-1 chronically infected, antiretroviral drug (ART) naïve adult men and women that were seeking voluntary HIV counseling and testing services at the Uganda Virus Research Institute (UVRI), and The AIDS Support Organization (TASO) clinics in Entebbe, Uganda, between August and September 2007.
The parent healthy cohort comprised 59 healthy HIV-uninfected adult men and women that were recruited from the UVRI clinic. The healthy volunteers were selected using similar criteria to that used for selecting HIV vaccine trial participants, as part of a multicenter trial (3 sites in Africa and one USA site) to evaluate extents of preexisting immune responses to chimpanzee Adenoviruses. They were previously shown to exhibit pre-existing antibodies to Adenoviruses, with median antibody titres of 190, IQR 20-330 [26], as evidence for prior exposure.
After obtaining signed informed consent, venous blood was obtained from each participant from both the cohorts. These study objectives were reviewed and approved by the institutional UVRI Science and Ethics Committee and the Uganda National Council of Science and Technology.
2.2. Study samples
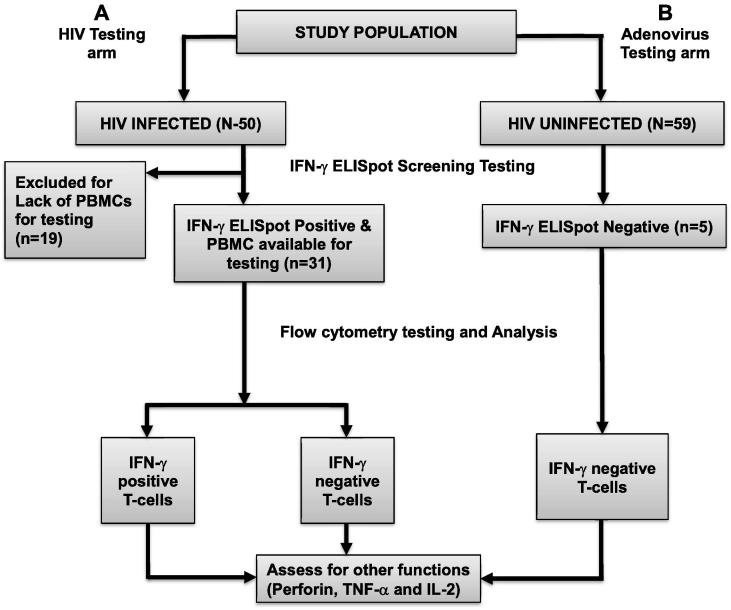
We used peripheral blood mononuclear cells (PBMCs) that were previously isolated from whole blood using Ficoll density gradient centrifugation, and cryopreserved as described before [27]. Here, cryopreserved PBMCs were thawed, rested overnight, and stimulated with either pooled Con M peptides (HIV-1 infected cohort), or Adenovirus 5, 6 or 7 peptides (healthy cohort) to quantify secretion of virus-specific IFN-γ using the IFN-γ ELISpot assay. PBMC specimens that secreted IFN-γ (IFN-γ positive) in response to Con M (n = 31), and those that lacked IFN-γ responses (IFN-γ negative) to Ad peptides (n = 5) were selected for further evaluation of other functions using flow cytometry, as summarized in Fig. 1.
Fig. 1.
Model for selection and evaluation of specimens for antigen-specific T-cell immune responses. This figure illustrates the screening for T-cell responses using the Interferon gamma (IFN-γ) ELISpot assay, and subsequent use of flow cytometry to evaluate co-expressing responses. Arm A denotes stimulation of PBMCs from HIV-1 infected individuals with consensus group M peptides. After flow cytometry, T-cells in Arm A were stratified into IFN-γ secreting (IFN-γ positive) and non-secreting (IFN-γ negative) populations. Secretion of other T-cell functions (Perforin, TNF-α and IL-2) was then compared across the two groups. Arm B comprised healthy HIV-1 uninfected individuals, serologically reactive to Adenovirus by antibody testing, but IFN-γ ELISpot negative to Adenovirus peptide stimulation. The IFN-γ negative T-cells were assessed for expression of Adenovirus-specific Perforin, TNF-α,and IL-2 using flow cytometry.
One of the inclusion criteria for HIV-infected volunteers at that time was having CD4 cell counts of ≥350 cell/μl. The 31 HIV-infected persons included in this analysis had median CD4 cell counts of 538 cell/μl (IQR 484–647), and median plasma viral loads of 15,550 RNA copies/ml (IQR 3450–52,400).
2.3. Use of IFN-γ ELISpot assay to screen for virus-specific IFN-γ responses to synthetic peptides
To quantify secretion of virus-specific IFN-γ response to Con M, we used 129 HIV-1 group M consensus peptides that were 14–16 amino acids long, overlapping by 11 amino acids, and spanning the entire length of the HIV Gag protein. These peptides were obtained from the USA NIH AIDS Reference Reagent Repository (Cat No. 11057; https://www.aidsreagentorg/Index.cfm). Con M peptides were selected because they were previously shown to detect comparable frequencies of HIV-specific CD4+ and CD8+ T-cell responses as consensus clade A and D peptides, in this population with mixed HIV clade A and D infection [27], supplementary figure 1A and 1B. Synthetic peptides were pooled and used in duplicate wells containing 100,000 PBMCs/well, with each individual peptide occurring at a final concentration of 2 μg/ml. A pool and matrix layout was used to ensure that each peptide occurred in two different wells, as previously described [27]. ELISpot plates were developed using Nova-Red substrate (Vector, Burlingame, CA) according to the manufacturer’s protocol. IFN-γ responses to peptide pools were quantified as spot-forming units per million PBMCs (SFU/106 PBMCs). The test acceptance criteria were ≥300 SFU/106 PBMCs in each phyto-hemagglutinin (PHA) positive control well; ≤100 SFU/106 PBMCs in all six-background wells; and ≤5 cumulative SFU/106 PBMCs in the two wells containing media only. Test wells with net responses ≥100 SFU/106 PBMC after subtracting three times the background SFU values were considered positive for IFN-γ production. Net responses below 100 SFU/106 PBMCs were considered negative for IFN-γ secretion, and were set to zero SFU/106 PBMC for purposes of easing data analysis.
To quantify Adenovirus-specific IFN-γ responses; synthetic adenovirus peptides that were 14–15 amino acids long overlapping by 10 amino acids were used. These peptides were obtained from Professor Ertl Hildegund C. J, Wistar Institute, Philadelphia, Pennsylvania, United States. Adenovirus (Ad) peptides traversed the conserved and variable hexon regions of Ad-serotype 5 (Ad5), Ad-serotype 6 (Ad6) and Ad-serotype 7 (Ad7). Up to 49 peptides were grouped by serotype, to generate four pools traversing the conserved hexon region, and one pool traversing the variable hexon region. A positive IFN-γ T-cell response was determined as described for HIV above.
2.4. PBMC stimulation and intracellular cytokine staining for flow cytometry
Cryopreserved PBMCs from previously defined IFN-γ ELISpot positive specimens to Con M (n = 31) and previously determined IFN-γ ELISpot negative specimens to Adenovirus (n = 5) were thawed, rested overnight and stimulated with Con M and Ad peptides, respectively, before they were further evaluated for other functions using flow cytometry. All PBMCs were stimulated with their respective antigens for 6 h at 37 °C in a 5% CO2 incubator; and stained with specified fluorochrome antibodies, as previously described [25]. Briefly, trypan blue exclusion counting was used to select groups of 500,000 live PBMCs. These were then incubated with: (1) media only as negative controls; (2) media containing pools of 2 μg/ml per peptide as test wells; or (3) media containing 2 μg/ml Staphylococcal enterotoxin B (SEB) (Sigma-Aldrich Logistic GmbH, Germany) as positive controls, in the presence of 10 μg/ml Golgi Plug™ and 1 μg/ml co-stimulatory antibodies CD28 and CD49 (BD Biosciences). After stimulation, the PBMCs were washed and stained with Aqua (L34957, Invitrogen), CD19 APC Alexafluor 750 (1072337A, Invitrogen), and CD14 APC Alexafluor750 antibodies (773927B, Invitrogen) to eliminate dead cells, B cells, and monocytes, respectively. The T-cells were defined using surface markers for CD3 (brilliant violet 570, B152103, Biolegend), CD8 (pacific blue, 22416, BD Bioscience) and CD4 (PE-Cy5.5, 1049514A, eBiosciences). Intracellular secretion of T-cells functions was quantified using Alexafluor 700, 21128, BD Bio- sciences (IFN-γ), APC, 341116BD Biosciences (IL-2), FITC B-D48 clone, F111124, Diaclone (Perforin) and PE-Cy7 antibodies, E07677-1632, ebiosciences (TNF-α). Up to 500,000 events were acquired on a custom-built, 18-color LSR II flow cytometer (BD Biosciences) using a threshold of 5000 on Forward Scatter Area (FSC-A).
2.5. Flow cytometry data analysis
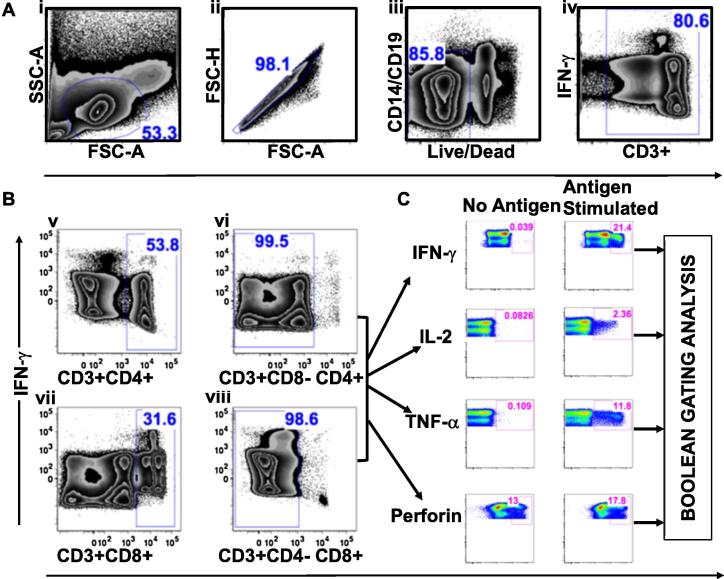
Flow cytometry datasets were analyzed with FlowJo software (FlowJo Treestar, version 9.7.4), using the gating strategy illustrated in Fig. 2. At least 300,000 lymphocytes (median 510,000; interquartile range [IQR] 430,000–520,000) were hierarchically gated to delineate CD4+ (CD3+CD8−CD4+) and CD8+ (CD3+CD4−CD8+) T-cells. Briefly, we first gated the lymphocytes, removed duplexes to select only single cells, and finally eliminated dead cells, B cells, and monocytes, Fig. 2A. IFN-γ secretions from non-CD3+ T-cells were eliminated, while all IFN-γ positive and negative CD3+ T-cells were selected for further stratifications. We then eliminated double positive (CD4+CD8+) T-cells to select pure singly stained CD4+ (CD3+CD8−CD4+) and CD8+ (CD3+CD4−CD8+) T-cells, Fig. 2B. CD4+ and CD8+ cells were then delineated with FlowJo Software to quantify proportions of functional cells. T-cell functions were defined using SEB stimulation guided gates (representing maximum activation), and unstimulated specimen-guided gates (representing background activation). Perforin lacked well-defined separation; therefore, we also used Flouresence minus One (FMO) controls and titration data to guide the gates for Perforin, supplementary Fig. 2. In a more stringent analysis, we perceived singly expressed Perforin responses as preformed and considered only Perforin co-expressed with another function as newly secreted. Boolean gating was used to distinguish the responding T-cells into 16 subsets of functions comprising polyfunctional, singly expressing, and non-functional subsets. Boolean gating delineated eight distinct subsets of IFN-γ positive co-expressing functions: IFNγ+IL2+Perf+TNFα+, IFNγ+IL2+Perf+TNFα−, IFNγ+IL2+Perf-TNFα+, IFNγ+IL2+Perf−TNFα−, IFNγ+IL2−Perf+TNFα+, IFNγ+IL2−Perf+TNFα−, IFNγ+IL2−Perf−TNFα+, IFNγ+IL2−Perf−TNFα−. Similarly, eight functional subsets of IFN-γ negative cells were delineated as follows: IFNγ−IL2+Perf+TNFα+, IFNγ−IL2+Perf+TNFα−, IFNγ−IL2+Perf−TNFα+, IFNγ−IL2+Perf−TNFα−, IFNγ−IL2−Perf+TNFα+, IFNγ−IL2−Perf+TNFα−, IFNγ−IL2−Perf−TNFα+, and IFNγ−IL2−Perf−TNFα−. Background fluorescence was deduced from unstimulated, media-only, and negative control wells, Fig. 2C. Pestle and SPICE software [28], version 5.1; http://exon.niaid.nih.gov/spice, was used to subtract background fluorescence and graphically summarize the T cells into IFN-γ secreting and non secreting subsets.
Fig. 2.
Gating strategy for defining antigen-specific T-cell functions using flow cytometry. This figure summarizes procedures for demarcating antigen-specific T-cells using FlowJo software, FlowJo, LLC. (i) First, lymphocytes were identified; (ii) plotted on forward scatter height (FSC-H) against forward scatter area (FSC-A) to select single cells, (iii) subjected to CD14, CD19, and Aqua antibody exclusion to remove B-cells, monocytes, and dead cells, respectively, (iv) before gating out CD3+ lymphocytes, A. Double positive T cells (CD3+CD4+CD8+) were excluded, while CD4+ (CD3+CD8−CD4+) and CD8+ (CD3+CD4−CD8+) T cells were selected for subsequent evaluations, B. Secretion of IFN-γ, IL-2, TNF-α, and Perforin was then quantified using cytokine-specific plots derived from FlowJo software, C. Finally, the co-expressed T cell functions were further distinguished into singly expressed, concurrently expressed, and non-functional subsets using FlowJo generated Boolean gating.
Tests were considered positive if at least 0.01% of gated T-cells were responsive after subtracting the background fluorescence. Poly-functionality was defined as the simultaneous secretion of at least three T-cell functions. Global T-cell responses were initially compared across gated IFN-γ positive and negative populations. To assess responses from only antigen-specific stimulation, and to clarify graphical illustrations of the data, subsets that lacked all evaluated T-cell functions (IFNγ−IL2−Perf−TNFα−) were sometimes not displayed on some graphs.
2.6. Statistical analysis
Frequency analyses and graphical illustrations of the expressed functions were performed using SPICE software version 5.1 and Prism 5 (Graph Pad Software, Inc., San Diego, CA, USA). SPICE automatically log-transformed the datasets, and used an in-built student’s T-test to compute statistical differences between expressed CD4 and CD8 functions across IFN-γ positive and IFN-γ negative T-cells. Proportions of responding subsets were compared across IFN-γ positive and IFN-γ negative T-cells using Fisher’s Exact test (Epi Info 7.2; https://www.cdc.gov/epiinfo/pc.html). Comparisons were considered significantly different if p-values were ≤0.05. When differences across the seven subgroups of co-expressed functions were compared, Bonferroni’s adjustment for multiple testing was applied, and only p-values ≤0.0071 were considered significant.
3. Results
3.1. IFN-γ negative CD4+ T-cells from HIV infected individuals can secrete IL-2, TNF-α, and Perforin
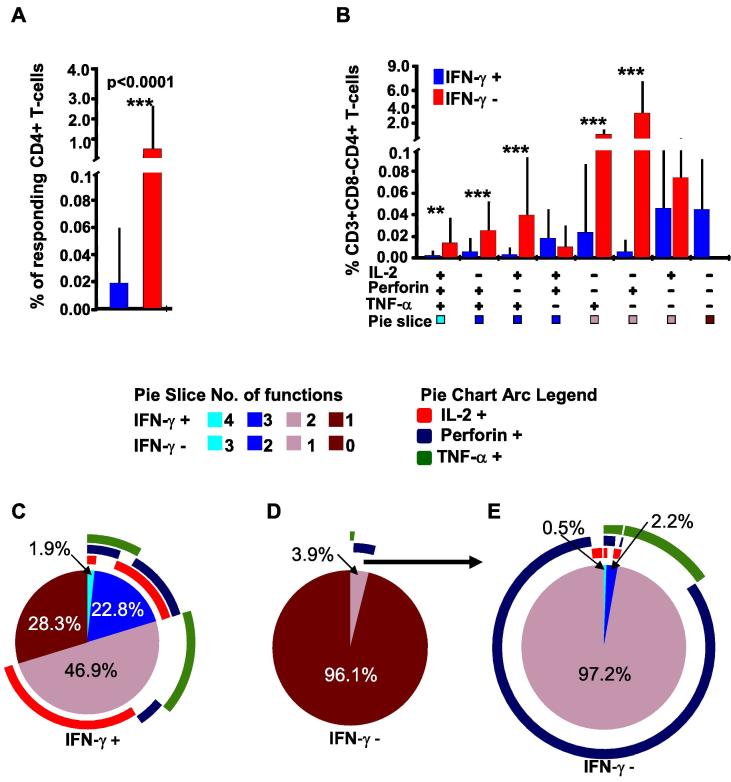
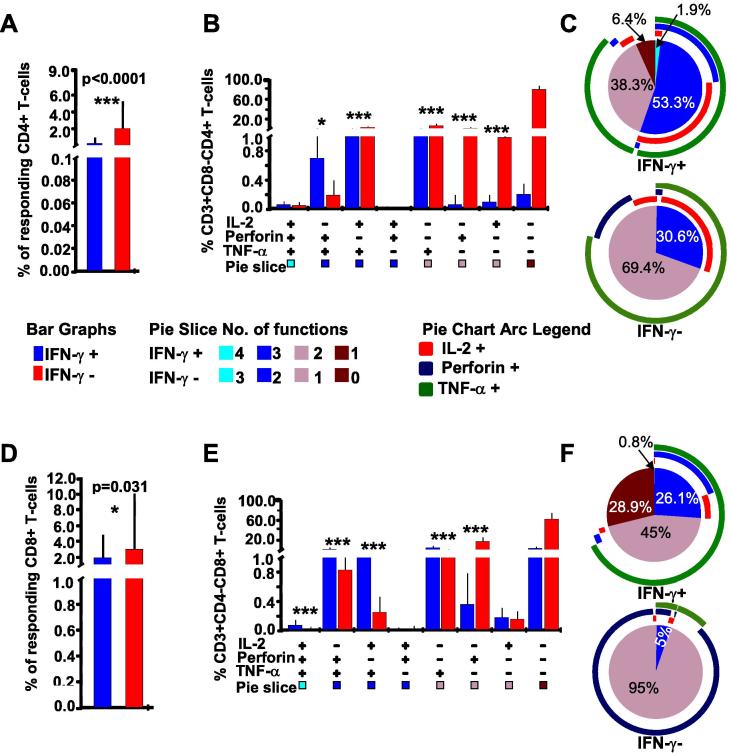
We first compared IFN-γ positive and negative CD4+ (CD3+CD8−CD4+) T-cells demarcated by hierarchical flow cytometry gating for the expression of HIV-specific IFN-γ, IL2, Perforin, and TNF-α. Overall, expression of other functions (IL2, Perforin or TNF-α) was significantly higher in IFN-γ negative CD4 T cells (0.58%) than in the IFN-γ positives (0.02%), p < 0.0001, in-built SPICE student’s T-test, Fig. 3A. Considering only the seven co-expressing T-cell functional profiles, responses were of significantly higher frequencies in five of seven IFN-γ negative CD4+ T-cells compared to none of the seven IFN-γ negative functional subsets; p = 0.02, Fisher’s Exact test the 3B. Considering the activated chronic status of the assessed population, and the indiscrete population of singly secreted Perforin, we adopted a more stringent approach to estimating newly secreted Perforin. Accordingly, singly expressed Perforin was ignored; and only Perforin co-expressed with another T-cell function was considered newly secreted. Correspondingly, Perforin co-expression with TNF-α (Perforin + TNF-α+) and IL-2 (Perforin + TNF-α+IL2+) was significantly higher in IFN-γ negative CD4+ T-cells subsets, p = 0.005 and 0.001, respectively, SPICE in-built T-test, Bonferroni adjusted significance of p = 0.0071 Fig. 3B.
Fig. 3.
IFN-γ negative T-cells from HIV infected individuals’ can produce detectable IL-2, TNF-α and/or Perforin. This figure compares IFN-γ positive and negative T-cells for frequencies of co-expressed T-cell functions after stimulation with HIV-1 consensus M (Con M) Gag antigens. FlowJo™ generated booleans were uploaded onto Pestle™ software to subtract backgrounds; and SPICE™ software, version 5.1 (NIAID) to stratify the CD4+ T-cells into IFN-γ positive and negative cells (IFN-γ overlaying). The T-cell responses were graphically summarized into eight functional combinations of IFN-γ secreting (red) and non-secreting (blue) cells using SPICE™, A, and B. Plus (+) and minus (−) signs below the x-axis indicate presence or absence of the specified function(s), respectively. Bars represent the frequencies (%) of expressed function(s) by the parent cells; and error bars represent their standard deviations. P values of ≤0.05 were considered statistically significant, and were denoted with * sign above the bars to indicate p-values ≤ 0.05, ** to indicate p-values ≤ 0.01, and *** to indicate p-values of ≤0.001. Bonferroni adjustment for multiple testing was applied when responses were compared across the functional combinations, in that case, P values of ≤0.0071 were considered significant. Cells that did not secrete any of the evaluated responses have been excluded to ease clarity. The y-axis has been cut for better visual presentation of the data, B. The pie charts summarize frequencies of indicated CD4+ T-cell functions in IFN-γ positive cells (C); in IFN-γ negative including non-secreting cells (D), and the IFN-γ negative without non-secreting cells (E). Pie arcs represent individual contributions of specified functions to the total T-cell response.
Of the responding IFN-γ positive CD4+ cells, 24.7% were polyfunctional; 28.3% secreted only IFN-γ, while the rest co-expressed IFN-γ responses with mainly IL2 or TNF-α, Fig. 3C. Among the IFN-γ negative CD4+ T-cells, 96% lacked all evaluated responses (IFNγ−IL2−Perf−TNFα− subset), Fig. 3D; this non-responsive subset was ignored in subsequent comparisons to ease statistical analysis and to clarify the graphical illustrations. Considering only the responsive IFN-γ negative CD4+ T-cells, only 0.5% were polyfunctional, 2.2% co-expressed two functions, and 97% predominantly secreted single functions of Perforin, Fig. 3E.
3.2. High proportions of IFN-γ negative CD8+ T-cells expressed Perforin, TNF-α and IL2 functions in response to HIV peptides
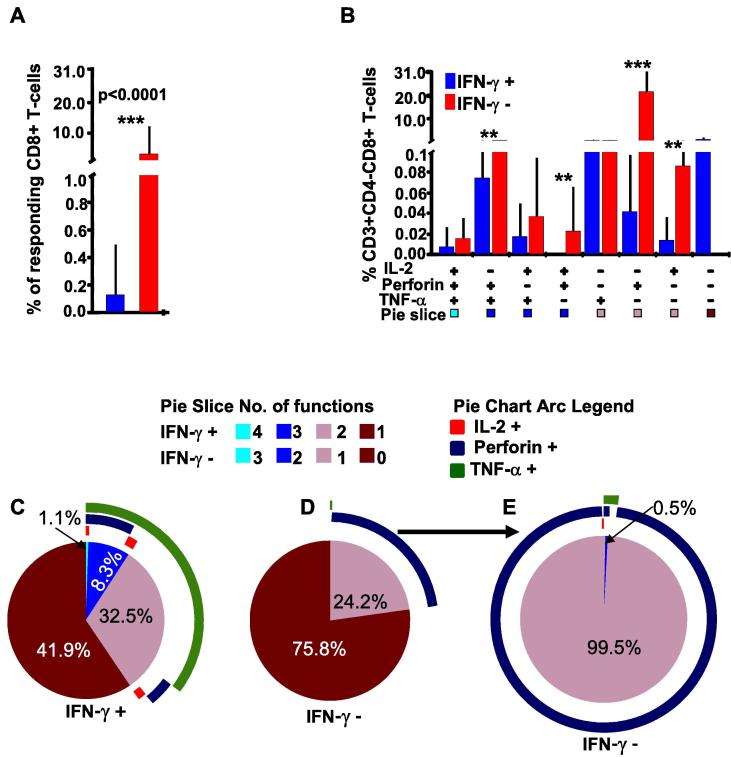
We next compared flow cytometry gated CD8+ (CD3 + CD4−CD8+) T-cells from HIV-infected individuals for virus-specific IL-2, Perforin and TNF-α expression across subsets of IFN-γ positive and IFN-γ negative T-cells. Similar to the CD4+ T-cells, IFN-γ negative CD8+ T-cells expressed other functions of IL-2, Perforin and TNF-α at higher frequencies (3.24%) than the IFN-γ positive T-cells (0.14%); p < 0.0001, SPICETM in-built T-test, Fig. 4A. Of the seven IFN-γ negative co-expressing CD8+ T-cell functional subsets, three had significantly higher CD8+ T-cell responses, p ≤ 0.0071, Bonferroni corrected, SPICE in-built T-test. Like with CD4+ cells, co-expression of Perforin and IL-2 or TNF-α, more representative of newly secreted HIV-specific Perforin, was significantly higher in three IFN-γ negative CD8+ T-cell subsets, Fig. 4B. Of the responding IFN-γ positive CD8+ cells, 42% secreted monofunctional IFN-γ: the rest co-expressed IFN-γ with mostly TNF-α, while 9.4% were polyfunctional, Fig. 4C. Of the IFN-γ negative CD8+ T-cells, 76% lacked all evaluated T-cell responses, Fig. 4D. When all non-responsive cells were removed, 99.5% of the functional IFN-γ negative CD8+ T-cells secreted singly secreted Perforin, Fig. 4E. Taken together, these data underscored the importance of comprehensively evaluating both IFN-γ positive and IFN-γ negative T-cells when assessing immune correlates of protection from infection.
Fig. 4.
Some IFN-γ negative CD8+ T-cells express high proportions of Perforin, TNF-α and/or IL2 functions. This figure compares frequencies of co-expressed CD8+ T-cell functions in IFN-γ positive and negative cells in response to HIV-1 Con M stimulation, A. FlowJo derived CD8+ T-cell booleans were stratified as 8 distinct combinations of IFN-γ secreting (red) and non-secreting (blue) functional profiles using SPICETM software, version 5.1 (NIAID). Positive signs (+) below the x-axis indicate presence of specified function(s), and negative signs indicate absence of that function. The bars represent frequencies (%) of parent T-cells expressing a specified combination of functions; error bars represent standard deviations. P values of ≤0.05 were considered statistically significant, and were denoted with a* sign above the bars to indicate p-values ≤0.05, ** to indicate p values ≤0.01, and *** to indicate p values ≤0.001. Bonferroni adjustment was applied to compare responses across subgroups, in such cases P values of ≤0.0071 were considered significant. Cells that did not secrete any functions have been excluded to improve clarity. The y-axis has been cut to improve visual presentation of the data, B. Cell populations were also summarized as pie charts with IFN-γ response overlaid, to illustrate frequencies of CD8+ T-cells that are secreting IFN-γ (C); CD8+ T –cells that are IFN-γ negative including non-secreting cells, (D), and IFN-γ negative CD8+ T-cells without the non-secreting cells E. Pie arcs represent the individual contribution of each function to the total T-cell response.
3.3. T-cells from HIV-1 infected individuals had a high intrinsic ability to secrete T-cell functions in response to polyclonal stimulation with SEB
We then evaluated if specimens from HIV infected individuals has lost their intrinsic capability to secrete T-cell responses. PBMCs from HIV-1 infected individuals were stimulated with SEB super antigen to determine the maximum secretion of functions in IFN-γ positive and negative T-cells. SEB-stimulated cells secreted all the evaluated functions. We found that both CD4+ and CD8+ T-cell responses were significantly increase when the specimens were stimulated with SEB, p < 0.0001 and p = 0.022, respectively, supplementary Fig. 3 TNF-α predominated in both IFN-γ positive and negative subsets. Like for HIV stimulation, SEB-stimulated, IFN-γ negative CD4+ T-cells secreted significantly higher responses (2.14%) than the IFN-γ positive cells (0.4%), p < 0.0001, SPICE in-built student’s T-test, Fig. 5A. Frequencies of responses were significantly higher in four IFN-γ negative CD4+ T-cell subsets (p ≤ 0.00071, Bonferroni adjusted), while three subsets lacked significant difference across the IFN-γ positive and negative phenotypes, Fig. 5B. Of the detectable responses from the IFN-γ positive CD4+ T-cells, 53% were polyfunctional (upper Fig. 5C). On the other hand, 69.4% of the IFN-γ negative responses were monofunctional, lower Fig. 5C.
Fig. 5.
T-cells from HIV-Infected individuals were capable of secreting high frequencies of SEB-stimulated T-cell responses. This figure summarizes the overall SEB-stimulated IFN-γ positive (blue) and negative (red) CD4+ T-cell responses, (A) The cells stratified into eight functional combinations of IFN-γ positive and negative cells, with bars representing frequencies (%) of expressed IL-2, TNF-α, and Perforin functions, and error bars the standard deviations. P values ≤ 0.05 are considered statistically significant, and are denoted with a * sign above the bars to indicate p-values ≤ 0.05; ** indicates p values ≤ 0.01 and *** indicates p values ≤ 0.001, (B) Pie charts summarize the proportions of functions in IFN-γ positive (upper C) and negative CD4+ T-cells (lower C). Pie arcs represent contributions of each function to the total response. Similarly, frequencies of IFN-γ positive and negative CD8+ T-cells are shown (D), distributed into eight profiles of functions (E), and summarized as pie charts and pie arcs, F.
Similar to CD4 cells, the overall SEB-stimulated responses were significantly higher in IFN-γ negative (3.0%) than in the positive (1.9%) CD8+ T-cells, p = 0.031, SPICE in-built student’s T-test, Fig. 5D. Contrary to the CD4+ cells, frequencies of SEB-stimulated CD8+ T-cell functions were significantly higher in four of seven IFN-γ positive functional subsets compared to one in the corresponding IFN-γ negative subsets, (p ≤ 0.0071, Bonferroni adjusted), Fig. 5E. Like in CD4+, the IFN-γ positive CD8+ T-cells were more polyfunctional (upper Fig. 5F), while 95% of the corresponding IFN-γ negative subsets expressed singly secreted Perforin, lower Fig. 5F. Taken together, these data exhibited a reduction in non-IFN-γ responses from the cryopreserved T-cells largely affecting the CD4+ T-cell subsets.
3.4. Virus-specific T-cell responses were detectable in ELISpot assay screen-out PBMCs from healthy volunteers
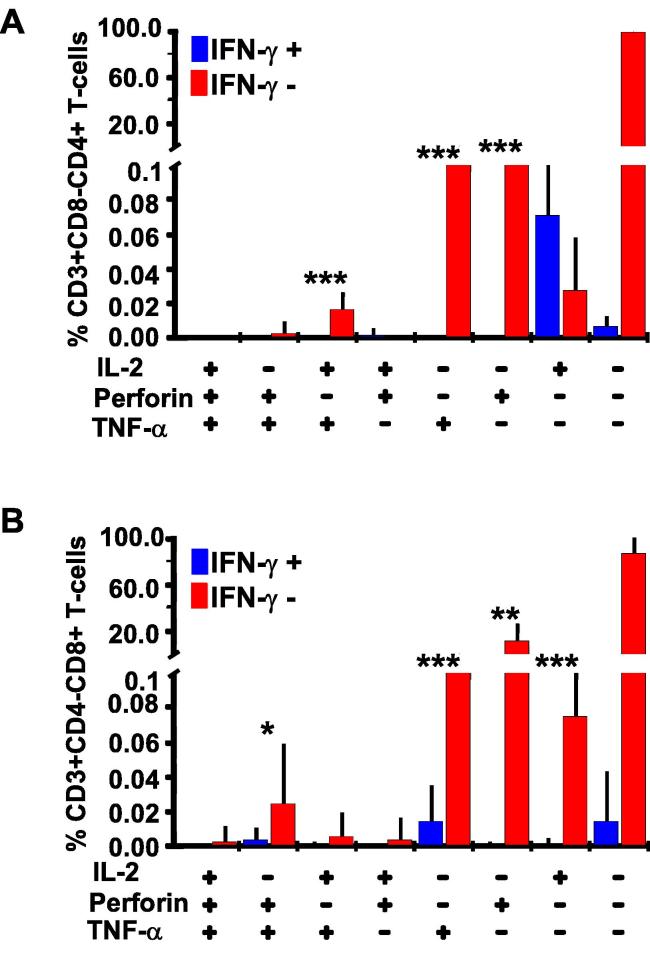
Finally, we assessed the extents of missed Adenovirus-specific IL-2, TNF-α and Perforin in PBMC specimens from healthy volunteers that had been screened as IFN-γ negative using ELISpot assay, and were pre-qualified for participating in vaccine trials. The ELISpot assay cut off for a positive response was 100 SFU/106 PBMCs after subtracting thrice the background. Thus, low levels of IFN-γ detection would be expected when such screen out specimens are subjected to flow cytometry. Generally, this population was less activated; T-cell responses were generally low compared to the HIV cohort. Overall, we found virus specific IL-2, TNF-a and Perforin responses in both CD4+ and CD8+ T-cells. Like with HIV, these responses were higher in the IFN-γ negative CD4+ (Fig. 6A) and CD8+ (Fig. 6B) T-cell subsets.
Fig. 6.
T-cell responses were detectable in some ELISpot assay screen-out IFN-γ negative cells from healthy volunteers. This figure illustrates T-cell responses detected in PBMCs from healthy individuals that were initially screened as IFN-γ ELISpot assay negative to Adenovirus peptides. These PBMCs were then stimulated with Adenovirus antigens for the co-expression of IL-2, TNF-α, and Perforin T-cell functions using flow cytometry. Bars indicate proportions of expressed IL-2, Perforin, and TNF-α in CD4+ T-cells, error bars indicate standard deviations (A). Positive signs below the x-axis indicate presence of specified function(s); and negative signs indicate absence of the specified function(s). Responses in IFN-γ positive and IFN-γ negative CD8+ T-cells are similarly shown in B.
4. Discussion
In this study, we evaluated the extent of missed T-cell responses when antigen-induced IFN-γ detection was used as the basis for determining subsequent estimation of co-expressed TNF-α, IL-2, Perforin T-cell responses in cryopreserved cells. Using cells from chronic HIV-1 infected individuals; we showed that although IFN-γ positive T-cells were more polyfunctional, HIV-specific IL-2, Perforin, and TNF-α responses also occurred at significantly higher proportions in the responsive IFN-γ negative T-cells. This was observed for both CD4+ and CD8+ T-cell phenotypes. The higher T-cell polyfunctionality among IFN-γ positive cells underscored their importance in assessing T-cell functional responses. However, the substantial levels of missed responses when IFN-γ negative cells are omitted highlights the importance of comprehensively evaluating all T-cell functions regardless of the IFN-γ ELISpot responsive status of the test PBMCs. Expression of TNF-α, IL-2, Perforin and/ or MIP-1β responses in IFN-γ negative T-cells has been reported before [29]. Here, we specifically compared the proportions of other T-cell functions concurrently expressed in IFN-γ secreting and non-secreting T-cells. A study conducted in HIV-1 chronically infected disease progressors, viremic controllers and elite controllers found that Perforin and MIP1α responses occurred at significantly higher proportions in IFN-γ negative CD8+ T-cells from elite controllers than in chronic progressors, supporting their importance [15]. The authors concluded that CD8+ T-cell expression of Perforin was a correlate of HIV control. In the same study, significantly more CD8+ T-cells from chronic progressors secreted IFN-γ compared to elite controllers; this implied that IFN-γ expression is not necessarily associated with controlling disease progression.
Within the scope of this study, we were not able to explain why measurable T-cell functions were much lower in IFN-γ secreting T-cells than in the corresponding IFN-γ non-secreting phenotype. We do not believe that these differences were due to sampling of an aging T-cell population. However, we used cryopreserved PBMCs in all our evaluations; and this could partly explain why responses in IFN-γ positive cells were quite low. A recent study on stored PBMCs showed that cryopreservation can result in up to five-fold reduction in antigen-specific IFN-γ producing CD4+ effector T-cell functionality while the IFN-γ negative phenotype remains relatively unaffected [30]. Also, greater expression of IFN-γ has been linked to the late stages of effector T-cell lifespan [12], and T-cell exhaustion [31], [32], [33]; the low levels of IFN-γ production may also suggest that our samples were not in a late stage. Unfortunately, due to specimen volume limitations, we were not able to specifically test for the presence of T-cell exhaustion. Although we lacked clinical information about the duration of HIV infection among the ART-naïve individuals from whom we isolated PBMCs, their median CD4 counts of >500 cells/μl, and viral load of about 15,000 copies/ml, did not imply T-cells exhaustion. Also, the high proportion of T-cells responding to SEB antigen indicated that our T-cells were immunologically competent and therefore able to optimally secret cytokines and functions when maximally stimulated with antigens.
Often, IFN-γ ELISpot assay is used to first identify individuals that produce IFN-γ in response to specified tested antigens; these are then assessed for production of other immune functions. We used flow cytometry to evaluate the extent of missed detection of specified T-cell responses by such screening approaches. Adenovirus-specific TNF-α, IL-2, and Perforin expression in IFN-γ negative CD4+ and CD8+ T-cells from healthy IFN-γ ELISpot negative individuals were quantified. We found that even cells from IFN-γ ELISpot negative individuals produce substantial T-cell functions that are detectable by intracellular cytokine staining flow cytometry. We showed that subsequent evaluations of other functions using only IFN-γ ELISpot assay positive specimens would significantly under estimate the detection of Perforin, a function established to be critical for viral control [15]. Two studies that compared use of ELISpot and flow cytometry assays for detection of responding T-cells showed that virus-specific T-cells were underestimated when immune responses were measured with IFN-γ ELISpot assay alone [31], [34]. Also, T-cells from HIV seropositive persons have been reported to respond differently to different HIV-1 epitopes, some of which can drive expression of immune functions other than IFN-γ [29], [35]. If the ELISpot assay is used to determine or screen for responses to these epitopes, then T-cells might be incorrectly identified as being non-responsive to antigen.
In HIV-infected individuals and healthy controls, higher proportions of IFN-γ negative CD4+ and CD8+ cells secreted mostly monofunctional Perforin. This predominance of singly expressed Perforin should be interpreted with caution. The ART-naïve cohort we evaluated was in a state of chronic activation due to continued presence of viral antigen. The evaluated HIV-specific T-cells were not in a true resting state, and possibly had accumulated levels of pre-existing Perforin in vivo. Perforin gates were particularly difficult to distinguish, suggesting Perforin was likely pre-existing. We used the anti-Perforin clone 48 antibody, which was previously shown to stain resting CD8+ T-cells equivalently to the δG9 Perforin clone [36]. Those authors discouraged use of δG9 Perforin because it could not recognize new Perforin that is trapped in the ER/Golgi compartment. They also found the B-D48 Perforin to be more associated with elite control of HIV-1. Clone B-D48 recognizes both pre-formed Perforin in cytotoxic granules and new Perforin secreted in response to antigenic stimulation, while clone δG9 recognizes Perforin within cytotoxic granules. Although the B-D48 Perforin clone does not specifically distinguish pre-formed from newly secreted Perforin, Hersperger et al, [15] showed that newly secreted Perforin co-expresses with other cytotoxic functions, and largely bypasses the cytotoxic granules. Therefore, focusing on only cells that have degranulated would most certainly underestimate the levels of secreted Perforin. Thus, we adopted a more stringent approach of assessing D48 Perforin co-expressing with another T-cell function as the one representing newly secreted, antigen-stimulated Perforin in activated T-cells. We still found that co-expressed Perforin occurred at significantly higher frequencies in both CD4+ and CD8+ IFN-γ negative T-cell subsets underscoring the potential for loss of critical cytolytic functions when IFN-γ negative cells are ignored in T-cell functional analyses.
We found qualitative differences in T-cell functions expressed by IFN-γ positive and negative T-cells. Poly-functionality was predominantly found in IFN-γ positive T-cells, and IFN-γ negative T-cells were largely mono-functional. Poly-functionality of virus-specific T-cell responses is associated with better HIV-1 disease control [37], [38], and CD8+ T-cells from HIV-1 disease elite-controllers are more likely to express single functions of either Perforin or MIP-1β, than CD8+ T-cells from HIV rapid disease progressors [15], [16]. Likewise, in vivo monofunctional Perforin secretion in CD4+ T-cells has been linked with control of viral infections in C57BL/6 mice [39]. Significantly higher proportions of IFN-γ negative CD4+ and CD8+ T-cells expressed single functions of TNF-α and IL-2 respectively compared to the IFN-γ secreting phenotype. Single function expression of IL-2 than IFN-γ was similarly found among T-cells isolated from adults who received a Hepatitis B booster vaccine [38] A study that examined whether CD8+ T-cells are helpful in preventing HIV viral replication showed that CD8+ T-cells that exhibited the strongest viral inhibition were sometimes monofunctional, and expressed functions other than IFN-γ [40].
Finally, IFN-γ secretion does not always correlate with cytotoxicity in virally infected T-cells in vitro [41]. These findings as well as our own indicate that evaluation of immunogenicity based on selection of IFN-γ secreting cells, in flow cytometry gating analysis and/ or ELISpot selection, may underestimate the potentially useful cytotoxic responses in both CD4+ and CD8+ T-cells. In our study, Perforin, a cytotoxic protein found in the granules of cytotoxic T-cells, was the primary function that was expressed alone and with other functions among IFN-γ negative T-cells. It has previously been shown that IL-2, which promotes proliferation of CD8+ and CD4+ T-cells and up-regulates the expression of Perforin are rarely co-expressed in the same cell [42], [43]. In one of these papers [42], the authors found co-expression of IL-2 and Perforin in IFN-γ+ CD8+ T-cells to be rare but to be associated with protection. In our study, CD8+ IFN-γ positive T-cells did not co-express IL-2 and Perforin in response to HIV-1 peptide stimulation. In contrast, we observed a significant proportion of IFN-γ negative T-cells co-expressing both these functions after HIV-1 Con M peptide stimulation further underscoring the importance of comprehensive evaluation of T-cells regardless of IFN-γ ELISpot status, especially when evaluating co-expression of Perforin.
This study had some limitations; apart from IFN-γ, we characterized polyfunctionality based on secretion of at least three functions. If we had measured the expression of more functions, some T-cells currently classified as mono-functional, may have been bi- or poly-functional. Also, we were unable to evaluate the presence of MIP1α or CD107α functions that commonly co-express with Perforin to effect cytolytic functions important for HIV-disease control. In addition, the overall sample size of persons from whom we isolated PBMCs was fairly small, and we did not have information on how long they had been HIV infected. Finally, we were unable to evaluate T-cell exhaustion, which would have helped estimate the stage of differentiation of our T-cells.
All the same, our findings show that some IFN-γ negative cells can produce cytokines. Therefore conventional approaches of analyzing flow cytometry data by setting cytokine gates based on IFN-γ,or use of IFN-γ ELISpot as a surrogate marker for further evaluation of virally induced immune responses may underestimate the extent of antigenic-specific T-cell immune response. Data from the healthy volunteers has implications for evaluating T-cell responses in vaccine trials. Evaluations of immunogenicity in response to viral antigens should not only evaluate T-cell responsiveness among IFN-γ producing cells but also among those T-cells that do not express IFN-γ. HIV vaccine development studies may benefit from expanding the evaluation of T-cell populations used to predict immunogenicity.
Author disclosure statement
The authors have declared that no competing interests exist.
Acknowledgements
We gratefully acknowledge all study participants and staff of The AIDS Support Organization and the Uganda Virus Research Institute in Entebbe. We thank the Medical Research Council (MRC UK) for co-funding this study and for enabling access to the study cohorts. Recruitment of study participants was funded by the International Atomic Energy Agency through grant RAF6/029 to the WHO/African AIDS Vaccine program. Flowcytometry reagents were secured using funds from The Wellcome Trust grant (087540) to the Training Health Researchers into Vocational Excellence (THRiVE) in East Africa program. HIV peptides were obtained through the NIH AIDS Research and Reference reagent program, Division of AIDS, NIAID, NIH, Adenovirus peptide pools were kindly provided by Professor Ertl Hildegund C. J from the Wistar Institute, Philadelphia, Pennsylvania, United States. All authors made substantial contributions to this work as follows: (i) Conceiving and designing the study: RN, JS, PK. Performing the flow cytometry experiments: RN, SM, BOA. Analyzing the data: RN, JS. Drafting and writing the manuscript: RN, JS. Revising and critical review of the manuscript: RN, JS, SM, BOA, CL, MBN, PK. Clinical evaluation, Informed consent and recruitment of study participants: MBN. Final approval of the version to be submitted RN, JS, and PK.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.vaccine.2018.11.024.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Goonetilleke N., Liu M.K., Salazar-Gonzalez J.F., Ferrari G., Giorgi E., Ganusov V.V. The first T-cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J Exp Med. 2009;206:1253–1272. doi: 10.1084/jem.20090365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jin X., Bauer D.E., Tuttleton S.E., Lewin S., Gettie A., Blanchard J. Dramatic rise in plasma viremia after CD8(+) T-cell depletion in simian immunodeficiency virus-infected macaques. J Exp Med. 1999;189:991–998. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altfeld M., Addo M.M., Shankarappa R., Lee P.K., Allen T.M., Yu X.G. Enhanced detection of human immunodeficiency virus type 1-specific T-cell responses to highly variable regions by using peptides based on autologous virus sequences. J Virol. 2003;77:7330–7340. doi: 10.1128/JVI.77.13.7330-7340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchbinder S.P., Mehrotra D.V., Duerr A., Fitzgerald D.W., Mogg R., Li D. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372:1881–1893. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boulet S., Ndongala M.L., Peretz Y., Boisvert M.P., Boulassel M.R., Tremblay C. A dual color ELISPOT method for the simultaneous detection of IL-2 and IFN-gamma HIV-specific immune responses. J Immunol Methods. 2007;320:18–29. doi: 10.1016/j.jim.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Streeck H., Jolin J.S., Qi Y., Yassine-Diab B., Johnson R.C., Kwon D.S. Human immunodeficiency virus type 1-specific CD8+ T-cell responses during primary infection are major determinants of the viral set point and loss of CD4+ T-cells. J Virol. 2009;83:7641–7648. doi: 10.1128/JVI.00182-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altfeld M., Kalife E.T., Qi Y., Streeck H., Lichterfeld M., Johnston M.N. HLA alleles associated with delayed progression to AIDS contribute strongly to the initial CD8(+) T-cell response against HIV-1. PLoS Med. 2006;3 doi: 10.1371/journal.pmed.0030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu T.M., Dubey S.A., Mehrotra D.V., Freed D.C., Trigona W.L., Adams-Muhler L. Evaluation of cellular immune responses in subjects chronically infected with HIV type 1. AIDS Res Hum Retroviruses. 2007;23:67–76. doi: 10.1089/aid.2006.0114. [DOI] [PubMed] [Google Scholar]
- 9.Varadarajan N., Julg B., Yamanaka Y.J., Chen H., Ogunniyi A.O., McAndrew E. A high-throughput single-cell analysis of human CD8(+) T-cell functions reveals discordance for cytokine secretion and cytolysis. J Clin Invest. 2011;121:4322–4331. doi: 10.1172/JCI58653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biswas P., Poli G., Kinter A.L., Justement J.S., Stanley S.K., Maury W.J. Interferon gamma induces the expression of human immunodeficiency virus in persistently infected promonocytic cells (U1) and redirects the production of virions to intracytoplasmic vacuoles in phorbol myristate acetate-differentiated U1 cells. J Exp Med. 1992;176:739–750. doi: 10.1084/jem.176.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Champagne P., Ogg G.S., King A.S., Knabenhans C., Ellefsen K., Nobile M. Skewed maturation of memory HIV-specific CD8 T lymphocytes. Nature. 2001;410:106–111. doi: 10.1038/35065118. [DOI] [PubMed] [Google Scholar]
- 12.Seder R.A., Darrah P.A., Roederer M. T-cell quality in memory and protection: implications for vaccine design. Nat RevImmunol. 2008;8:247–258. doi: 10.1038/nri2274. [DOI] [PubMed] [Google Scholar]
- 13.Burgers W.A., Riou C., Mlotshwa M., Maenetje P., de Assis Rosa D, Brenchley J. Association of HIV-specific and total CD8+ T memory phenotypes in subtype C HIV-1 infection with viral set point. J Immunol. 2009;182:4751–4761. doi: 10.4049/jimmunol.0803801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghiglione Y., Falivene J., Ruiz M.J., Laufer N., Socias M.E., Cahn P. Early skewed distribution of total and HIV-specific CD8+ T-cell memory phenotypes during primary HIV infection is related to reduced antiviral activity and faster disease progression. PLoS ONE. 2014;9:e104235. doi: 10.1371/journal.pone.0104235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hersperger A.R., Pereyra F., Nason M., Demers K., Sheth P., Shin L.Y. Perforin expression directly ex vivo by HIV-specific CD8 T-cells is a correlate of HIV elite control. PLoS Pathog. 2010;6:e1000917. doi: 10.1371/journal.ppat.1000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cocchi F., DeVico A.L., Yarchoan R., Redfield R., Cleghorn F., Blattner W.A. Higher macrophage inflammatory protein (MIP)-1alpha and MIP-1beta levels from CD8+ T-cells are associated with asymptomatic HIV-1 infection. Proc Natl Acad Sci U S A. 2000;97:13812–13817. doi: 10.1073/pnas.240469997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bell S.J., Cooper D.A., Kemp B.E., Doherty R.R., Penny R. Heterogeneous effects of exogenous IL-2 on HIV-specific cell-mediated immunity (CMI) Clin Exp Immunol. 1992;90:6–12. doi: 10.1111/j.1365-2249.1992.tb05823.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reier A., Mitsuyasu R.T. Use of interleukin-2 in immunotherapy of human immunodeficiency virus infection. BioDrugs. 1998;10:215–225. doi: 10.2165/00063030-199810030-00005. [DOI] [PubMed] [Google Scholar]
- 19.Talley A.K., Dewhurst S., Perry S.W., Dollard S.C., Gummuluru S., Fine S.M. Tumor necrosis factor alpha-induced apoptosis in human neuronal cells: protection by the antioxidant N-acetylcysteine and the genes bcl-2 and crmA. Mol Cell Biol. 1995;15:2359–2366. doi: 10.1128/mcb.15.5.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frederiksen J., Buggert M., Karlsson A.C., Lund O. NetFCM: a semi-automated web-based method for flow cytometry data analysis. Cytometry A. 2014;85:969–977. doi: 10.1002/cyto.a.22510. [DOI] [PubMed] [Google Scholar]
- 21.Hemelaar J., Gouws E., Ghys P.D., Osmanov S. Global trends in molecular epidemiology of HIV-1 during 2000–2007. Aids. 2011;25:679–689. doi: 10.1097/QAD.0b013e328342ff93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al-Sherbiny M., Osman A., Mohamed N., Shata M.T., Abdel-Aziz F., Abdel-Hamid M. Exposure to hepatitis C virus induces cellular immune responses without detectable viremia or seroconversion. Am J Trop Med Hyg. 2005;73:44–49. [PubMed] [Google Scholar]
- 23.Techakriengkrai N., Tansiri Y., Hansasuta P. Poor HIV control in HLA-B*27 and B*57/58 noncontrollers is associated with limited number of polyfunctional Gag p24-specific CD8+ T-cells. Aids. 2013;27:17–27. doi: 10.1097/QAD.0b013e32835ac0e1. [DOI] [PubMed] [Google Scholar]
- 24.Rutebemberwa A., Currier J.R., Jagodzinski L., McCutchan F., Birx D., Marovich M. HIV-1 MN Env 15-mer peptides better detect HIV-1 specific CD8 T-cell responses compared with consensus subtypes B and M group 15-mer peptides. Aids. 2005;19:1165–1172. doi: 10.1097/01.aids.0000176216.02743.98. [DOI] [PubMed] [Google Scholar]
- 25.Mugaba S., Nakiboneka R., Nanyonjo M., Bugembe-Lule D., Kaddu I., Nanteza B. Group M consensus Gag and Nef peptides are as efficient at detecting clade A1 and D cross-subtype T-cell functions as subtype-specific consensus peptides. Vaccine. 2014;32:3787–3795. doi: 10.1016/j.vaccine.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen H., Xiang Z.Q., Li Y., Kurupati R.K., Jia B., Bian A. Adenovirus-based vaccines: comparison of vectors from three species of adenoviridae. J Virol. 2010;84:10522–10532. doi: 10.1128/JVI.00450-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Serwanga J., Mugaba S., Pimego E., Nanteza B., Lyagoba F., Nakubulwa S. Profile of T-cell recognition of HIV Type 1 consensus group M Gag and Nef peptides in a clade A1- and D-infected ugandan population. AIDS Res Hum Retroviruses. 2012;28:384–392. doi: 10.1089/aid.2011.0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roederer M., Nozzi J.L., Nason M.C. SPICE: exploration and analysis of post-cytometric complex multivariate datasets. Cytometry A. 2011;79:167–174. doi: 10.1002/cyto.a.21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richmond M., McKinnon L.R., Kiazyk S.A., Wachihi C., Kimani M., Kimani J. Epitope mapping of HIV-specific CD8+ T-cell responses by multiple immunological readouts reveals distinct specificities defined by function. J Virol. 2011;85:1275–1286. doi: 10.1128/JVI.01707-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ford T., Wenden C., Mbekeani A., Dally L., Cox J.H., Morin M. Cryopreservation-related loss of antigen-specific IFNgamma producing CD4(+) T-cells can skew immunogenicity data in vaccine trials: Lessons from a malaria vaccine trial substudy. Vaccine. 2017;35:1898–1906. doi: 10.1016/j.vaccine.2017.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horton H., Russell N., Moore E., Frank I., Baydo R., Havenar-Daughton C. Correlation between interferon- gamma secretion and cytotoxicity, in virus-specific memory T-cells. J Infect Dis. 2004;190:1692–1696. doi: 10.1086/424490. [DOI] [PubMed] [Google Scholar]
- 32.Shankar P., Russo M., Harnisch B., Patterson M., Skolnik P., Lieberman J. Impaired function of circulating HIV-specific CD8(+) T-cells in chronic human immunodeficiency virus infection. Blood. 2000;96:3094–3101. [PubMed] [Google Scholar]
- 33.Kostense S., Vandenberghe K., Joling J., Van Baarle D., Nanlohy N., Manting E. Persistent numbers of tetramer+ CD8(+) T-cells, but loss of interferon-gamma+ HIV-specific T-cells during progression to AIDS. Blood. 2002;99:2505–2511. doi: 10.1182/blood.v99.7.2505. [DOI] [PubMed] [Google Scholar]
- 34.Riou C., Burgers W.A., Mlisana K., Koup R.A., Roederer M., Abdool Karim S.S. Differential impact of magnitude, polyfunctional capacity, and specificity of HIV-specific CD8+ T-cell responses on HIV set point. J Virol. 2014;88:1819–1824. doi: 10.1128/JVI.02968-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Obuku A.E., Bugembe D.L., Musinguzi K., Watera C., Serwanga J., Ndembi N. Macrophage inflammatory protein-1 beta and interferon gamma responses in ugandans with HIV-1 acute/early infections. AIDS Res Hum Retroviruses. 2016;32:237–246. doi: 10.1089/AID.2015.0157. [DOI] [PubMed] [Google Scholar]
- 36.Makedonas G., Banerjee P.P., Pandey R., Hersperger A.R., Sanborn K.B., Hardy G.A. Rapid up-regulation and granule-independent transport of perforin to the immunological synapse define a novel mechanism of antigen-specific CD8+ T-cell cytotoxic activity. J Immunol. 2009;182:5560–5569. doi: 10.4049/jimmunol.0803945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Betts M.R., Nason M.C., West S.M., De Rosa S.C., Migueles S.A., Abraham J. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T-cells. Blood. 2006;107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duvall M.G., Precopio M.L., Ambrozak D.A., Jaye A., McMichael A.J., Whittle H.C. Polyfunctional T-cell responses are a hallmark of HIV-2 infection. Eur J Immunol. 2008;38:350–363. doi: 10.1002/eji.200737768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fang M., Siciliano N.A., Hersperger A.R., Roscoe F., Hu A., Ma X. Perforin-dependent CD4+ T-cell cytotoxicity contributes to control a murine poxvirus infection. Proc Natl Acad Sci U S A. 2012;109:9983–9988. doi: 10.1073/pnas.1202143109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Freel S.A., Lamoreaux L., Chattopadhyay P.K., Saunders K., Zarkowsky D., Overman R.G. Phenotypic and functional profile of HIV-inhibitory CD8 T-cells elicited by natural infection and heterologous prime/boost vaccination. J Virol. 2010;84:4998–5006. doi: 10.1128/JVI.00138-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malyguine A.M., Strobl S., Dunham K., Shurin M.R., Sayers T.J. ELISPOT assay for monitoring cytotoxic T lymphocytes (CTL) activity in cancer vaccine clinical trials. Cells. 2012;1:111–126. doi: 10.3390/cells1020111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Makedonas G., Hutnick N., Haney D., Amick A.C., Gardner J., Cosma G. Perforin and IL-2 upregulation define qualitative differences among highly functional virus-specific human CD8 T-cells. PLoS Pathog. 2010;6:e1000798. doi: 10.1371/journal.ppat.1000798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cellerai C., Perreau M., Rozot V., Bellutti Enders F., Pantaleo G., Harari A. Proliferation capacity and cytotoxic activity are mediated by functionally and phenotypically distinct virus-specific CD8 T-cells defined by interleukin-7R{alpha} (CD127) and perforin expression. J Virol. 2010;84:3868–3878. doi: 10.1128/JVI.02565-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.