Abstract
Background
Despite extensive efforts on the genomic characterization of gliomas, very few studies have reported the genetic alterations of cerebellar glioblastoma (C-GBM), a rare and lethal disease. Here, we provide a systematic study of C-GBM to better understand its specific genomic features.
Methods
We collected a cohort of C-GBM patients and compared patient demographics and tumor pathologies with supratentorial glioblastoma (S-GBM). To uncover the molecular characteristics, we performed DNA and mRNA sequencing and DNA methylation arrays on 19, 6, and 4 C-GBM cases, respectively. Moreover, chemical drug screening was conducted to identify potential therapeutic options for C-GBMs.
Results
Despite differing anatomical origins of C-GBM and S-GBM, neither histological, cytological, nor patient demographics appeared significantly different between the 2 types. However, we observed striking differences in mutational patterns, including frequent alterations of ATRX, PDGFRA, NF1, and RAS and absence of EGFR alterations in C-GBM. These results show a distinct evolutionary path in C-GBM, suggesting specific therapeutic targeted options. Targeted-drug screening revealed that C-GBMs were more responsive to mitogen-activated protein kinase kinase (MEK) inhibitor and resistant to epidermal growth factor receptor inhibitors than S-GBMs. Also, differential expression analysis indicated that C-GBMs may have originated from oligodendrocyte progenitor cells, suggesting that different types of cells can undergo malignant transformation according to their location in brain. Master regulator analysis with differentially expressed genes between C-GBM and proneural S-GBM revealed NR4A1 as a potential therapeutic target.
Conclusions
Our results imply that unique gliomagenesis mechanisms occur in adult cerebellum and new treatment strategies are needed to provide greater therapeutic benefits for C-GBM patients.
Key Points
1. Distinct genomic profiles of 19 adult cerebellar GBMs were characterized.
2. MEK inhibitor was highly sensitive to cerebellar GBM compared with supratentorial GBM.
3. Master regulator analysis revealed NR4A1 as a potential therapeutic target in cerebellar GBM.
Keywords: brain location, cancer genomics, cerebellar glioblastoma, glioblastoma subclassification
Importance of the study
Only a few studies have reported the molecular characteristics of adult C-GBM, a subtype comprising 1% of glioblastoma cases located within the infratentorial brain region due to its rarity. By identifying genomic profiles from 19 adult C-GBM samples, we revealed the genetic intertumoral heterogeneity in C-GBM as well as distinct genomic characteristics from those of S-GBMs, emphasizing the need of individualized therapies for C-GBM patients. Furthermore, we performed chemical drug screening to examine drug sensitivity, which enabled us to provide potential targeted therapy options for C-GBM patients.
To understand the molecular mechanisms influencing oncogenesis, progression, and response to specific cancer therapies, researchers have published systematic analyses of genomic and transcriptomic datasets from patient tumors and deposited results in publicly accessible archives such as The Cancer Genome Atlas (TCGA). Glioblastoma (GBM) was in the vanguard of those genomic studies and represents one of the most extensively studied cancers to date.1,2 Yet, nearly all studies have focused on the most sought-after gliomas that originate in the supratentorial region of the brain and have omitted the small fraction (0.9%) of adult GBMs located within the brain’s infratentorial region, such as cerebellar glioblastomas (C-GBMs).3 Owing to the rarity of samples, only a few characteristics of C-GBMs have been described using immunohistochemistry, PCR amplification, and various sequencing methods.4–6 Although these sample sizes were small, they indicated that specific tumorigenic mechanisms, including immunonegativity for epidermal growth factor receptor (EGFR) prevail in C-GBMs. Recently, 17 diffuse cerebellar gliomas (DCGs) were characterized by next-generation sequencing (NGS), and they shared similar molecular characteristics with H3 K27M-mutant midline gliomas, including oligodendroglial differentiation as well as H3 K27M mutations.7 However, GBMs are notorious for their intertumoral heterogeneity, by showing various genetic alterations across tumor samples.1 This high level of intertumoral heterogeneity emphasizes the importance of personalized therapies in GBM. Therefore, we additionally characterized the molecular mechanisms of C-GBM by performing sequencing and analyses of targeted-, whole-exome, and mRNA data, as well as methylation arrays in a total of 19 C-GBM cases. In addition to revealing genetic features of C-GBMs, chemical drug screening was conducted on C-GBM patient-derived cells to examine therapeutic responses. We also compared C-GBM with the most common supratentorial glioblastomas (S-GBMs), by systematically reviewing clinical information in 853 GBM cases to identify whether C-GBMs are demographically and pathologically different from S-GBMs or not.
Materials and Methods
Patients and Specimens
Our study was approved by the institutional review boards of Samsung Medical Center (#2016-08-031) and Seoul National University Hospital (#H-1404-056-572). We retrospectively reviewed all patients who were diagnosed with either cerebellar or supratentorial GBMs and underwent craniotomies at Samsung Medical Center from 1996 to 2016. Clinical information such as sex, age, postoperative treatment modality, and survival time was collected. Tumor location was confirmed from preoperative magnetic resonance (MR) images. All cases were histologically confirmed as primary GBM by experienced pathologists. We received clinical information, MR images, and genetic profiles of the SNU001, SNU002, and SNU006 cases from Seoul National University. In total, 176 S-GBM and 19 C-GBM patients underwent sequencing. Among the 19 C-GBM cases, genomic DNA was extracted from 10 fresh-frozen tumor samples, and in 9 cases genomic DNA was isolated from formalin-fixed paraffin wax-embedded tumor samples. The isolated genomic DNA was subjected to exome sequencing and DNA methylation array analyses. Total RNA from 6 tumor samples was isolated and then processed for RNA sequencing.
Data Generation from DNA and RNA Sequencing
Using data from whole-exome and targeted-DNA sequencing, we detected single-nucleotide variants and indels and estimated copy number in C-GBM samples. RNA sequencing data were used to determine gene expression, GBM subtype, and gene fusion events. Detailed methods are described in the Supplementary material. The sequencing data have been deposited in the European Genome-phenome Archive (accession number: EGAS00001002517) for controlled access.
Master Regulator Analysis
Master regulator analysis was performed using the R package RTN, following the main vignette.8 The list of transcription factors was downloaded from Fantom5 (http://fantom.gsc.riken.jp/5; Accessed August 7, 2018) and used as an input to construct the transcriptional network.
Topological Data Analysis
We built topological representations of gene expression data using the Mapper algorithm, as implemented by Ayasdi.9 The output of Mapper is a low-dimensional network representation of the data, where nodes represent sets of samples with similar global transcriptional profiles as measured by correlation of the expression levels of the genes with the highest variance across samples (SD > 1 was used as cutoff).
Public Microarray Data Processing
For public microarray data, raw CEL files were downloaded from the Gene Expression Omnibus under accession numbers GSE36245 (Strum et al), GES5675 (Sharma et al), GSE34824 (Schwartzentruber et al), GSE44971 (Lambert et al), and GSE50161 (Griesinger et al), and normalized together using the RMA function (Affy package) to reduce batch effects.10–14 For data of Roth et al and Gravendeel et al, we downloaded the processed expression data from accession numbers GSE3526 and GSE16011, respectively.15,16 For the Repository of Molecular Brain Neoplasia Data (REMBRANDT) dataset, CEL files and a corresponding clinical information file were available from REMBRANDT data portal in 2011 and now available from ArrayExpress under accession number E-MTAB-207.17
DNA Methylation Subtyping Classification
The Illumina MethylationEPIC BeadChip kit was used to estimate the DNA methylation levels of C-GBM samples. The resulting IDAT files were used to determine their methylation classes, and MolecularNeuropathology.org (https://www.molecularneuropathology.org/mnp; Accessed August 7, 2018) provided the brain tumor methylation classification tool (Classifier: 11b2).
Clinical Statistics
Clinical characteristics of supratentorial and cerebellar GBMs were compared statistically using Fisher’s exact test. The log-rank test was used to compare overall survival, estimated using Kaplan–Meier curves, between the groups.
Results
Clinical Analysis for 853 Glioblastoma Patients
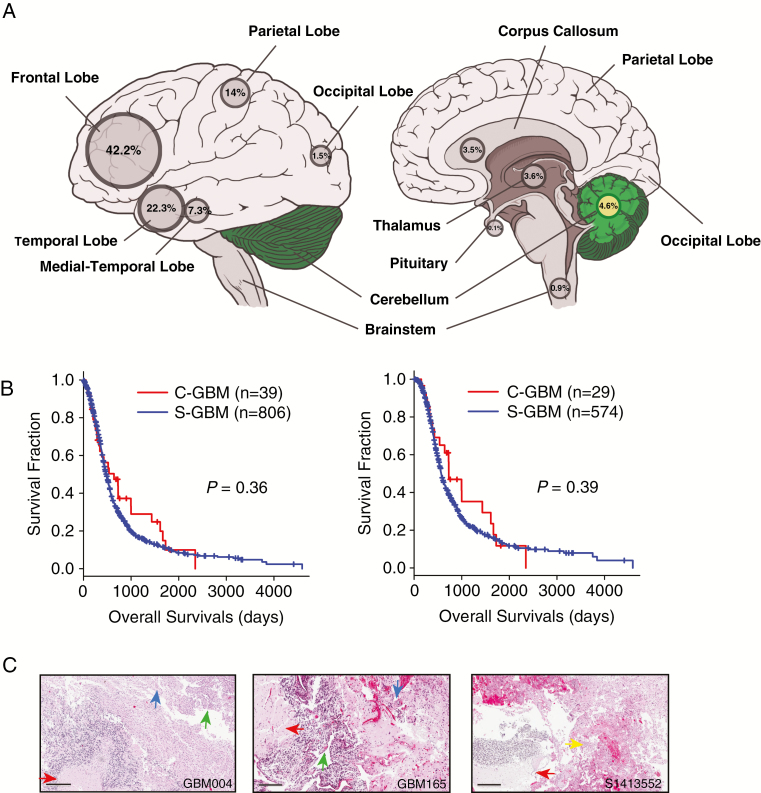
We determined tumor locations from MR images and analyzed the clinical information of 853 primary GBM patients older than 18 years of age diagnosed between 1996 and 2016. To remove any bias for the clinical analysis, secondary GBM cases were excluded. Among this cohort, 94.5% were located in the supratentorial region and 39 cases (4.6%) were found in the cerebellum—a greater incidence rate than in previous studies (Fig. 1A).3 There were no significant differences in demographic features or treatment regimens between C-GBM and S-GBM (Table 1). Although the median survival was longer in the C-GBM than in S-GBM patients, it was not statistically significant regardless of usage of temozolomide (Fig. 1B). Evaluation of histological features (eg, necrosis) in all C-GBMs were typical of S-GBMs (Fig. 1C).
Fig. 1.
C-GBMs among 853 glioblastoma patients. (A) Location information of 853 glioblastoma patients. Among them, 39 GBMs (4.6%) were located in the cerebellum. (B) Kaplan–Meier survival plots between C-GBMs and S-GBMs for all patients (left) and temozolomide-treated patients (right). P-values were estimated by log-rank tests. (C) Histological features of C-GBM. Glioblastoma features were evident, such as tumor necrosis (red arrow), endovascular proliferation (blue arrow), hypercellularity (green arrow), and glomeruloid endovascular proliferation (yellow arrow). Scale bars, 300 μm (GBM004), 400 μm (GBM165), and 200 μm (S1413552).
Table 1.
Clinical differences between supratentorial and cerebellar glioblastomas
| S-GBM | C-GBM | P-value | |
|---|---|---|---|
| Cases, n | 806 (94.49%) | 39 (4.57%) | |
| Age, y, at diagnosis, median (range) | 55.32 (19–89) | 56.90 (24–89) | 0.54 |
| Sex, n | 0.25 | ||
| Male | 439 (54.45%) | 25 (64.10%) | |
| Female | 367 (45.53%) | 14 (35.90%) | |
| Treatment, n | 0.86 | ||
| CCRT | 503 (62.41%) | 21 (53.85%) | |
| TMZ alone | 71 (8.81%) | 8 (20.21%) | |
| Other | 232 (28.78%) | 10 (25.63%) | |
| Overall survival (95% CI), days | |||
| All | 480 (445–521) | 641 (344–1433) | 0.36 |
| CCRT/TMZ-treated | 569 (535–621) | 728 (535–1666) | 0.39 |
Eight cases of brainstem glioblastomas were excluded; CCRT = concurrent chemoradiation therapy; TMZ = temozolomide.
Distinct Genomic Profiles of Cerebellar Glioblastoma
To reveal the genomic characteristics of C-GBMs, we performed whole-exome sequencing (WES), targeted-DNA sequencing (GliomaSCAN), and mRNA sequencing (RNA-seq) for 9, 10, and 6 samples, respectively—in total 19 patients (Supplementary Table 3). The analyzed MR images of C-GBMs show tumors located within diverse areas of the cerebellar hemisphere without any consistent characteristics (Supplementary Figure 2A–C).
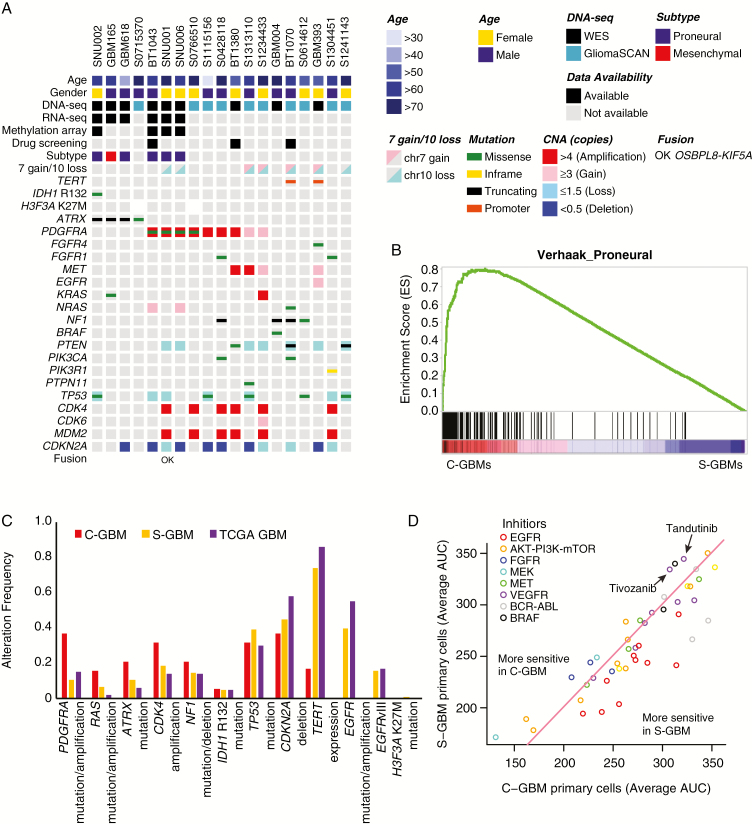
From WES data, we annotated the mutational status of 9 C-GBMs; additionally, glioma-specific mutational status of 10 C-GBMs was annotated from GliomaSCAN data (Fig. 2A; Supplementary Figures 3, 4A–C). Consistent with previous C-GBM studies, no EGFR-activating alterations, including EGFR variant III, existed in our C-GBM cohort, whereas EGFR alterations are among the most frequent alterations in S-GBM (Fig. 2C).1,4,5,7 On the other hand, our data showed that mutually exclusive alterations in alpha thalassemia/mental retardation syndrome X-linked (ATRX) or platelet derived growth factor receptor A (PDGFRA) occur more frequently in C-GBMs than in S-GBMs (58%, 11/19 vs 20.1%, 30/139, Fisher’s exact test, P = 0.001; Fig. 2A). Especially, 4 of 19 (21.1%) samples carried a mutation in the ATRX gene (2 stop codons, 1 missense, and another frameshift), while 3 were wild type for both isocitrate dehydrogenase (IDH1) and TP53, unlike adult supratentorial gliomas (Fig. 2A, Supplementary Table 4); in both our S-GBM and TCGA GBM cohorts, ATRX mutations occurred in only 10.8% and 5.8% of samples, and most (76.5%) were accompanied by IDH1 and TP53 mutations (Fig. 2C).1 Significance of copy number alterations C-GBMs was calculated by GISTIC and it revealed that the 4q12 region, covering PDGFRA, was most significantly amplified in C-GBMs (q-value = 10−6; Supplementary Figure 4A).18 GISTIC also identified 12q14.1 and 12q15 amplification in C-GBM tumors where CDK4 and MDM2 are located. Gene expression profiling also represents genomic characteristics, as a gene set enrichment analysis (GSEA) found that the proneural glioblastoma subtype markers were enriched in C-GBMs versus S-GBMs, and 5 of 6 C-GBMs were classified as tumor-intrinsically proneural subtype, which is characterized by IDH1, ATRX, TP53 mutation, CDK4 amplification, or PDGFRA amplification/mutation (Fig. 2A and B, Supplementary Table 3).1,19,20 Also, mitogen-activated protein kinase (MAPK) pathway–associated genes, RAS and NF1 alteration, were notable in C-GBM with 32% of incidence (Fig. 2A and C). Unlike pancreatic, colon, or lung cancer, RAS alterations are not commonly reported in GBMs.1,21 However, mutational analysis revealed that 3 of 19 (15.8%) C-GBMs possessed either RAS hotspot mutation (Q61H/K) or amplification (Fig. 2A and C).
Fig. 2.
Genomic characteristics of C-GBMs. (A) Summary of genomic alterations in 19 C-GBM samples with WES and GliomaSCAN data. RNA-seq results were used to detect gene fusion. The TERT promoter could not be covered by WES and thus TERT promoter mutation status was estimated by single nucleotide polymorphism analysis. The H3F3A region was not covered by GliomaSCAN, so H3F3A K27M mutation was confirmed by Sanger sequencing. (B) GSEA plot for Verhaak’s proneural gene set between 6 C-GBMs and 160 S-GBMs. (C) Alteration frequencies of GBM driver genes in C-GBM, S-GBM, and TCGA GBM data. (D) Drug response plot of 20 S-GBM and 3 C-GBM cells for 45 targeted drugs. x-Axis indicates average AUC values of C-GBM cells and y-axis indicates those of S-GBM cells. Circle dots above pink line are more sensitive in C-GBM and below pink line are more sensitive in S-GBM. Color of dots indicates primary target gene of inhibitors.
Since histone H3 mutation is a hallmark mutation for pediatric glioma and diffuse intrinsic pontine glioma (DIPG), and H3 K27M mutation was also recurrently found in a recent DCG study, we evaluated the mutational state of histone H3 genes in our C-GBM tumors and detected no mutations within H3 genes (Fig. 2A; Supplementary Table 4).7,12,22 Furthermore, Sanger sequencing and immunohistochemistry were also unable to detect H3F3A K27M (Fig. 2A; Supplementary Figure 5).
Telomerase reverse transcriptase (TERT) promoter mutations with chromosome 7 gain and chromosome 10 loss (chr7 gain/chr10 loss) are frequent events in IDH1 wild-type S-GBMs.23 However, only 2 of 19 C-GBMs (10.5%) harbored TERT promoter mutations with chr7 gain/chr10 loss, and another 2 samples contained chr7 gain/chr10 loss without the TERT promoter mutations. Consequently, TERT expression was significantly infrequent in C-GBM compared with S-GBM (P = 0.007; Fig. 2A and C; Supplementary Figure 4D; Supplementary Table 5).
Drug Response in Cerebellar Glioblastomas
To investigate chemical drug sensitivity associated with genetic alterations in C-GBMs, we compared average drug responses in 3 C-GBMs with those in 20 S-GBMs for 45 gene-targeted chemical drugs (Fig. 2D). As we expected based on the mutational profiles, C-GBMs were less sensitive than S-GBMs in all EGFR-targeted drugs and showed higher sensitivity for mitogen-activated protein kinase kinase (MEK) inhibitors, which are known to be effective for neurofibromatosis 1 (NF1) loss and Ras-activated tumors by regulating the MAPK pathway.24 We also evaluated the efficacy of various vascular endothelial growth factor receptor and Abl inhibitors, since these target PDGFR, and PDGFR-specific drugs are not yet available.25 Of these, tivozanib and tandutinib reduced cell viability more in C-GBMs than in S-GBMs. The drug response data emphasized the importance of personalized therapy according to genomic alterations and the efficacy of MEK inhibitors for C-GBM patients.
Gene Fusion in Cerebellar Glioblastoma
Gene fusions have become dominant cancer driver mutations, and many novel cancer-related gene fusions were recently discovered in part due to improved algorithms in NGS.26 Also in GBM, oncogene activation was achieved through gene fusion and resulted in tumorigenesis.27,28 We discovered one in-frame gene fusion, OSBPL8-KIF5A, in SNU001 in our C-GBM cases as well as confirmed the fusion by reverse transcription PCR (Fig. 2A, Supplementary Figure 6A and C). We then compared OSBPL8 and KIF5A expression levels among both S-GBMs and C-GBMs and found that both genes had upregulated expression levels in sample SNU001 (Supplementary Figure 6B). In particular, KIF5A was the most expressed in our fusion case among all GBM samples. As a member of the kinesin family, KIF5A has been reported as a putative driver gene in GBM and amplified in ~10% of GBM cases, which implies to us that KIF5A could be one of the potential therapeutic targets for this patient.29
Transcriptome and DNA Methylome Similarity of Cerebellar Glioblastomas
Pediatric brain tumors, such as medulloblastoma, ependymoma, and pilocytic astrocytoma (PA), usually arise in the infratentorial region of the brain; however, adult brain tumors are more commonly located within the supratentorial region. To understand whether transcriptomic patterns were associated with location rather than disease type, we first performed hierarchical clustering to compare the expression pattern similarity of C-GBMs with public microarray datasets, including adult and pediatric GBMs, as well as other brain tumors.10–14 Although all sets were profiled with the same microarray platform (Affymetrix U133 Plus 2.0 array), we normalized all public data together to reduce the possibility of batch effects due to different data sources, and our results showed that brain tumors appeared clustered by their disease types under principal component analysis (Supplementary Figure 7A and B).
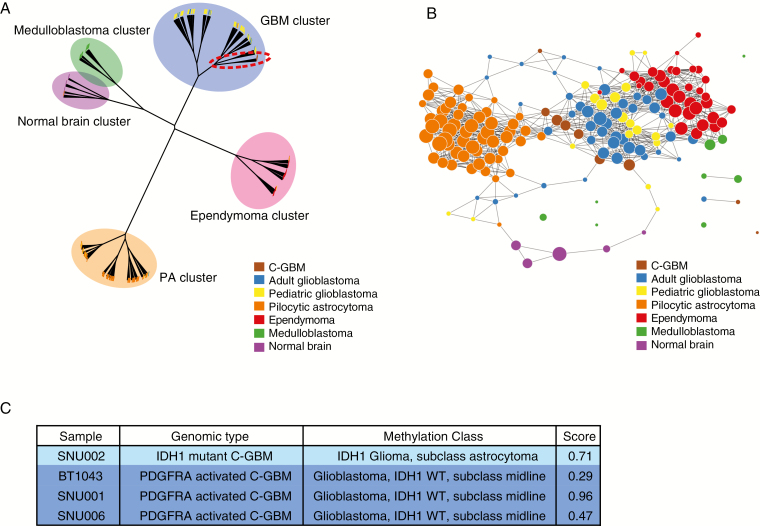
C-GBMs clustered near pediatric and adult GBMs, implying that transcriptomes of C-GBMs are more similar to S-GBMs than other brain tumors usually located in the infratentorial brain region (Fig. 3A). In addition to hierarchical clustering analysis, a topological data analysis revealed expression similarities, but more fine-grained relationships could be discerned between adult C-GBM and GBM clusters, including pediatric GBM and PA clusters (Fig. 3B). In order to confirm there was no bias due to a different platform measuring gene expression again, we performed clustering using RNA-seq data of PA, ependymoma, and medulloblastoma originated from cerebellum with microarray data of brain tumors. The hierarchical clustering result showed that PA, ependymoma, and medulloblastoma were clustered according to their disease type but not platform, indicating there was no platform bias (Supplementary Figure 7C).
Fig. 3.
Global transcriptomic and methylomic patterns of C-GBMs. (A) Hierarchical clustering tree of C-GBMs with normal brains and multiple brain tumors based on mRNA expression levels. Samples were clustered by diseases, and C-GBMs were gathered with adult and pediatric GBM in the GBM cluster. (B) A topological data analysis of C-GBMs with normal brain and multiple brain tumor samples. C-GBMs were close to adult GBMs. (C) Methylation classification of C-GBMs based on calibrated score. The classification results are provided by MolecularNeuropathology.org.
Besides RNA-seq, DNA methylation profiling for 4 C-GBMs was performed, and then their DNA methylation patterns were compared with over 2800 other brain tumors on MolecularNeuropathology.org. All 3 PDGFRA-activated C-GBMs were classified as IDH1 wild-type GBM, subclass midline lacking H3 K27M mutation, while the IDH1-mutant C-GBM was classified as IDH1 glioma, subclass astrocytoma (Fig. 3C). As observed in transcriptome analysis, we confirmed that C-GBMs share similar methylation patterns with those of other GBM tumors and found that there are infratentorial brain region–specific methylation patterns for C-GBM tumors.
Location-Specific and -Independent RNA Expression Patterns in Cerebellar Glioblastoma
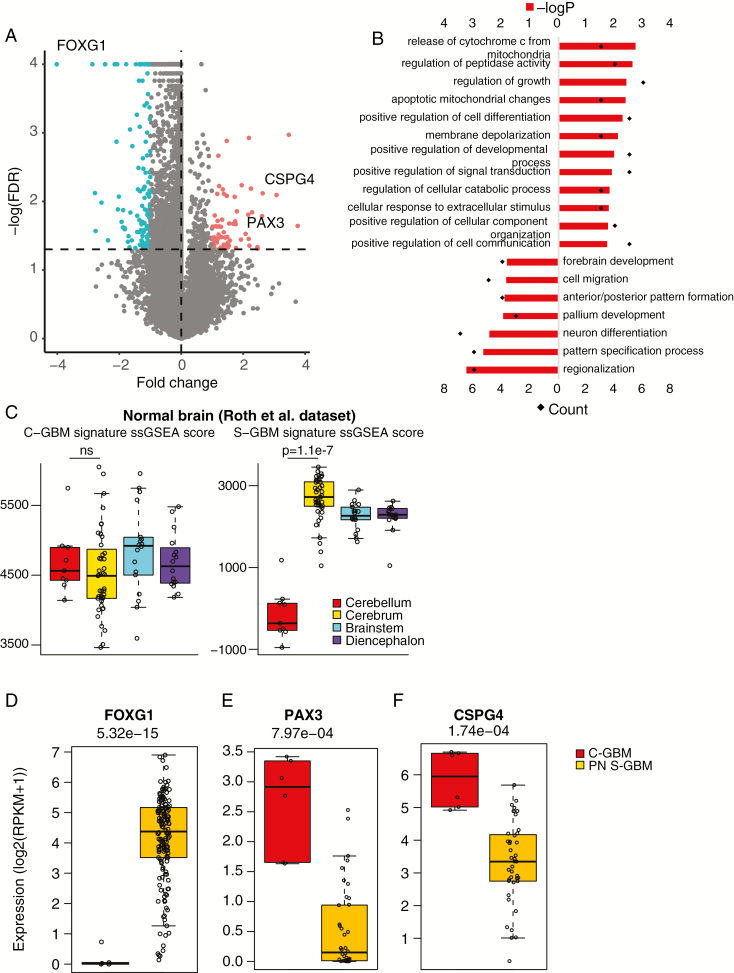
From genomic, transcriptomic, and methylomic profiles, we confirmed that C-GBMs shared common GBM genetic features, but also had unique mutational profiles and location-specific methylation patterns. Previous PA studies found differentially expressed and differentially methylated genes depending on their origin within the brain.11,13 We assessed whether any location-specific expression patterns arose in C-GBM from 6 C-GBM samples containing IDH1, ATRX, or PDGFRA alterations. Since C-GBM tumors were transcriptomically similar to proneural GBMs, we compared gene expression levels between these 6 samples and 40 proneural S-GBMs. FOXG1, known as a ventral telencephalon marker and a GBM-promoting factor, was identified as the most differentially expressed gene in S-GBMs (Fig. 4A and D; Supplementary Table 6). FOXG1 expression is abundant in GBM, except H3 K27M GBM tumors, which are located mostly in the brain midline.10 Using publicly available normal brain expression profiles, we confirmed that FOXG1 was expressed only in the cerebrum (Supplementary Figure 8B). Next, to define both a signature of C-GBM (C-GBM signature) and a signature of S-GBM (S-GBM signature), we selected for genes having expression level 2-fold greater in one than in the other (Student’s t-test < 0.05 and false discovery rate [FDR] <5%). Gene Ontology of these 2 signatures from DAVID (Database for Annotation, Visualization and Integrated Discovery) found that the S-GBM signature was related to forebrain development and single-sample GSEA score levels of S-GBM were significantly higher than in the normal brain dataset (Fig. 4B and C; Supplementary Figure 8A). The C-GBM signature was associated with cell communication, differentiation, apoptosis, and developmental processes. Although this signature does not appear to be restricted to the cerebellum, it was enriched in the normal cerebellum and can distinguish normal cerebellum expression patterns from other normal brain regions (Fig. 4B).16 A subset of the signature included PAX3, which appeared in an infratentorial-specific manner in a previous PA study as well (Fig. 4E).11 We also confirmed that the C-GBM and S-GBM signatures, which generated from C-GBM versus proneural S-GBM comparison, were downregulated and upregulated, respectively, even in other subtypes of GBMs (Supplementary Figure 8D).
Fig. 4.
Genes differentially expressed between C-GBMs and proneural S-GBMs. (A) A volcano plot showing genes differentially expressed between C-GBMs and proneural (PN) S-GBMs. Red and green dots indicate significantly upregulated genes in C-GBMs (C-GBM signature) and PN S-GBMs (S-GBM signature), respectively. (B) Gene Ontology results for C-GBM signature and S-GBM signature. (C) Single-sample GSEA scores of C-GBM (left) and S-GBM signatures (right) in the normal brain expression data of Roth et al. (D–F) Expression levels of FOXG1 (D), PAX3 (E), and CSPG4 (F) in C-GBM and PN S-GBM samples.
In addition to location-dependent genes, location-independent genes, such as CSPG4 (also known as NG2), appeared to be upregulated in C-GBM versus proneural S-GBM, while the expression level of CSPG4 in normal cerebellum was comparable to that of normal cerebrum (Fig. 4F; Supplementary Figure 8C). CSPG4 is known to be expressed in oligodendrocyte progenitor cells (OPCs) and important for GBM proliferation.30,31 Glioblastomas could arise from lineage-restricted progenitors, and their subtypes are determined by the cell of origin.32 Enrichment of the proneural gene set in C-GBM tumors and overexpression of the OPC marker may suggest that C-GBMs originated from OPCs, consistent with a previous report.7
Master Regulator Analysis of Cerebellar Glioblastoma
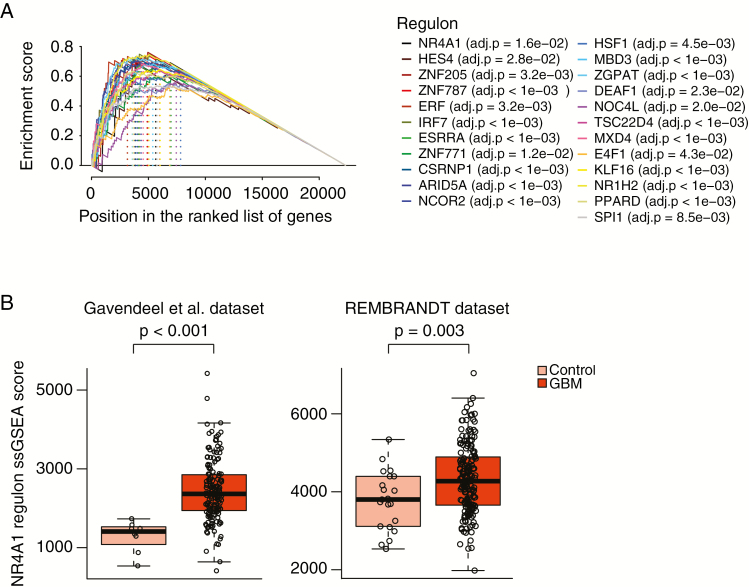
To obtain a better understanding of the pathways involved in the regulation of C-GBM signature genes, we constructed a transcriptional network (TN) based on C-GBM and proneural S-GBM samples using the ARACNe algorithm.33 In this network, each transcription factor (TF) is assigned a list of candidate target genes, referred to as its regulon. The final TN was composed of 81635 transcriptional interactions between 1634 TFs and 20428 potential target genes. Next, we applied the master regulator analysis (MRA) algorithm to this network. In MRA, the statistical significance of overlap between the regulon of each TF and the C-GBM signature genes is assessed using GSEA. At FDR < 5%, MRA inferred 23 C-GBM–specific TFs as master regulators (Fig. 5A).
Fig. 5.
Master regulator analysis of C-GBMs versus PN S-GBMs. (A) GSEA analysis showing genes in 23 master regulator regulons ranked by their differential expression. (B) Single-sample GSEA scores of NR4A1 regulons in GBM and nontumor samples in Gavendeel et al (left) and REMBRANDT (right) dataset.
Among these 23 master regulators, nuclear receptor subfamily 4 group A member 1 (NR4A1) could be both tumor suppressive and pro-oncogenic depending on tumor types and context.34 Inhibition of nuclear NR4A1 and nuclear export of NR4A1 suppressed tumor cell proliferation and promoted apoptosis.34 Expression level of NR4A1 regulons was increased in GBMs compared with normal brain control as well as increased in C-GBM compared with all S-GBM. These results indicated that nuclear NR4A1-mediated transactivation played an oncogenic role in GBM, and nuclear export might be needed to encourage tumor cell apoptosis (Fig. 5B).
Discussion
Unlike pediatric brain tumors, adult brain tumors are located mostly in supratentorial regions, and this appears to be associated with the lack of oncogenic molecules such as substance P in the adult infratentorial brain region.35 To date, about 1% of GBMs have been reported to be located in the cerebellum.3,35 In this study, 853 adult GBM locations were analyzed, and as expected, most of them originated from the supratentorial brain, with the exception of 39 cerebellar and 8 brainstem GBMs. Analysis of our cohort shows C-GBMs were not demographically and pathologically different from S-GBMs.
Owing to limited numbers of C-GBM cases, genomic characterization is lacking compared with the many studies published on S-GBMs.1,19 Recently, 17 DCGs were systematically characterized, and it was found that genes related with chromatin regulation and p53 functional disruption were frequently altered in DCGs.7 However, genetic diversity among GBM patients is one of the obstacles to improving survival benefits of GBM patients and therefore individualized treatment is required. Thus, we additionally analyzed genomic profiles for 19 C-GBM samples via whole-exome sequencing and GliomaSCAN.
As previous cerebellar glioma studies indicated, EGFR alterations such as EGFR mutation, focal amplification, and structural variants were not detected in our C-GBM cohort, while EGFR is often hyperactivated in S-GBMs.1,4,7EGFR activation is one of the characteristics of classical GBM, and consistently we could not find the classical subtype in our C-GBM tumors.19
Alterations in the PDGFRA gene were the most common events in our C-GBM cohort (36.8%) and in contrast to S-GBMs (10%) (P = 0.007). Also, CDK4 amplification tended to occur more in C-GBMs than in S-GBMs. Surprisingly, such mutations are more often found in proneural GBM, a disease whose cells resemble oligodendrocytes.19 In addition, CSPG4, one of the OPC markers, was differentially expressed in C-GBM compared with S-GBM. A recent study revealed that CSPG4-Cre could generate mouse brain tumors with expression of OPC markers and that these frequently occur in the ventral region.32 These results imply that our C-GBM samples possibly originate from the lineage-restricted progenitor, OPC. Previously, upregulated SOX10 in DCG epigenetically resulted in enrichment of PDGFRA-related genes rather than PDGFRA amplification.7 However, the high alteration frequency of PDGFRA amplification and mutation in our C-GBM samples indicates that PDGFRA activation already occurs at the genetic level and leads to oligodendroglial differentiation.
ATRX mutations were also frequent events (21.1%) in C-GBM tumors, while only 10% of S-GBMs possessed ATRX mutations. Interestingly, 3 of 4 (75%) ATRX mutant C-GBMs did not harbor IDH1 mutations, whereas only 3% of GBMs acquired ATRX mutations without IDH1 mutations.36ATRX and TERT promoter mutations are mutually exclusive and both contribute to maintaining telomeres by alternative lengthening of telomeres (ALT) or telomerase overexpression.2TERT promoter mutations occurred in only 10.5% of C-GBMs (2/19), although they were detectable in ~80% of overall GBMs.23 In addition to TERT promoter mutations, among 6 C-GBM samples with gene expression profiling, only 1 C-GBM sample (16.7%) showed a low level of TERT expression, while the rest of the samples did not express TERT. These high incidences of ATRX mutation and low frequencies of TERT promoter mutation/expression indicate that C-GBMs tend to maintain telomeres by ALT, as observed in IDH1-mutant astrocytoma or pediatric GBMs, rather than telomerase activation.12,23 However, in spite of the high incidence of ATRX mutation (22.2%) in C-GBM tumors, 13/19 (68.4%) tumors harbored neither ATRX mutation nor TERT promoter mutation. Through either telomerase activation or ALT, telomeres are longer in glioma than in normal tissues or other cancer.37 It would be interesting to identify whether C-GBMs have shorter telomeres than expected or if telomeres are maintained via other mechanisms, such as TP53 and RB1 alterations.37
IDH1-wild type ATRX and PDGFRA alterations are also common in pediatric or young adult glioma patients.12,38 H3 mutations are also usually detected in DIPG and pediatric gliomas.10,22,39 Surprisingly, we, however, did not detect any H3 mutations within our WES data cohort, and none of an additional 15 C-GBM tumors showed H3F3A K27M mutation when validated by Sanger sequencing or immunohistochemistry. These results imply that C-GBM is a distinct tumor type from pediatric and midline gliomas as well as S-GBM.
Moreover, we conducted chemical drug screenings on C-GBM primary cells and identified that they were more responsive to MEK inhibitors compared with S-GBM cells. Also, genomic alterations in MAPK pathway–associated genes, such as RAS and NF1, were remarkable in C-GBMs. MAPK activation in tumorigenesis was usually found in PA and pediatric gliomas whose major lesion is cerebellum.40,41 Therefore, we assumed that brain location–related MAPK activation could play a role in adult C-GBM, and targeting MAPK therapy is considerable for C-GBM patients.
In addition, location-specific gene expression and methylation patterns were revealed as observed in PA.11,13 We identified 23 master regulators whose regulons are upregulated in C-GBM compared with S-GBM. Among the 23 master regulators, NR4A1 was interesting, since it is a pro-apoptotic molecule in cytoplasm, but at the same time nuclear NR4A1-mediated transactivation is pro-oncogenic.34 NR4A1 regulons were revealed as upregulated in GBMs compared with normal brain and especially in C-GBMs. These results demonstrate that NR4A1 is a pro-oncogenic molecule as a transcription factor in GBM and can be a potential target for C-GBM tumor in particular.
Although we reconfirmed that C-GBM has an oligodendrocyte lineage, the same as a previous study, our systematic genomic analysis of C-GBM revealed different genomic characteristics from the previous report.7 In particular, absence of histone 3 mutations in this study cohort was remarkable compared with the previous study, which included 3 H3F3A K27M-mutant samples (11%). A recent study reported that the median age of H3 K27M-mutant adult glioma patients was 32, much younger than H3/IDH wild-type gliomas.42 Consistently, 2 of 3 DCG patients with H3F3A K27M mutation were younger than 30 years from the previous study.7 However, there were no subjects under 30 years among our IDH1 wild-type C-GBM patients whose H3F3A K27M status had been identified, and lack of young adult patients in our C-GBM cohort might result in absence of H3F3A K27M mutation. Besides H3F3A, SETD2, and PPM1D genes were not detected, but instead ATRX, PDGFRA, NF1, and RAS alteration were notable in our C-GBM dataset. Lack of young adult C-GBM patients could not fully account for the genomic inconsistency between our C-GBM cohort and the previous study. The genomic distinction indicates that there is intertumoral heterogeneity of C-GBM genome, and more C-GBM samples need to be characterized to provide more confident therapeutic strategies for C-GBM patients.
Funding
This research was supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI14C3418). Additional support was provided by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Ministry of Science & ICT (NRF-2018M3A9H3021707). E.L. is supported by NIH (F99 CA212478).
Supplementary Material
Acknowledgments
We thank Seung Won Choi for discussion. The biospecimens for this study were provided by Samsung Medical Center BioBank.
Conflict of interest statement. None declared.
Authorship statement. D-H.N., R.R., and C-K.P. supervised the entire project. H.J.C, J.Z., S.W.J., Y.L, D.G.K., H.J.K., M.B, S.H.A., and Y.J.S. performed the majority of experiments and analyses. D-S.K., J-I.L., J.W.C., H.J.S., C-K.P., and D-H.N. provided surgical specimens. S.W.J., D-S.K., D-H.N., S-Y.L., S.T.K., D.H.L., Y-S.D., J-I.L., H.J.S., and J.W.C. assisted with the interpretation of clinical results. D-H.N., C-K.P., and W-Y.P. organized and processed genome data of the specimens. H.J.C., E.L., J.Z., J.S.K., and S.W.J. wrote the manuscript with feedback from D-H.N., R.R., and C-K.P.
References
- 1. Brennan CW, Verhaak RG, McKenna A, et al. ; TCGA Research Network The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ceccarelli M, Barthel FP, Malta TM, et al. ; TCGA Research Network Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Adams H, Chaichana KL, Avendano J, Liu B, Raza SM, Quinones-Hinojosa A. Adult cerebellar glioblastoma: understanding survival and prognostic factors using a population-based database from 1973 to 2009. World Neurosurg. 2013; 80(6):e237–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Utsuki S, Oka H, Miyajima Y, Kijima C, Yasui Y, Fujii K. Adult cerebellar glioblastoma cases have different characteristics from supratentorial glioblastoma. Brain Tumor Pathol. 2012;29(2):87–95. [DOI] [PubMed] [Google Scholar]
- 5. Takahashi Y, Makino K, Nakamura H, et al. . Clinical characteristics and pathogenesis of cerebellar glioblastoma. Mol Med Rep. 2014;10(5):2383–2388. [DOI] [PubMed] [Google Scholar]
- 6. Milinkovic VP, Skender Gazibara MK, Manojlovic Gacic EM, Gazibara TM, Tanic NT. The impact of TP53 and RAS mutations on cerebellar glioblastomas. Exp Mol Pathol. 2014;97(2):202–207. [DOI] [PubMed] [Google Scholar]
- 7. Nomura M, Mukasa A, Nagae G, et al. . Distinct molecular profile of diffuse cerebellar gliomas. Acta Neuropathol. 2017;134(6):941–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fletcher MN, Castro MA, Wang X, et al. . Master regulators of FGFR2 signalling and breast cancer risk. Nat Commun. 2013;4:2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Singh G, Mémoli F, Carlsson GE. Topological methods for the analysis of high dimensional data sets and 3d object recognition. Paper presented at: SPBG2007. [Google Scholar]
- 10. Sturm D, Witt H, Hovestadt V, et al. . Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22(4):425–437. [DOI] [PubMed] [Google Scholar]
- 11. Sharma MK, Mansur DB, Reifenberger G, et al. . Distinct genetic signatures among pilocytic astrocytomas relate to their brain region origin. Cancer Res. 2007;67(3):890–900. [DOI] [PubMed] [Google Scholar]
- 12. Schwartzentruber J, Korshunov A, Liu XY, et al. . Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482(7384):226–231. [DOI] [PubMed] [Google Scholar]
- 13. Lambert SR, Witt H, Hovestadt V, et al. . Differential expression and methylation of brain developmental genes define location-specific subsets of pilocytic astrocytoma. Acta Neuropathol. 2013;126(2):291–301. [DOI] [PubMed] [Google Scholar]
- 14. Griesinger AM, Birks DK, Donson AM, et al. . Characterization of distinct immunophenotypes across pediatric brain tumor types. J Immunol. 2013;191(9):4880–4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gravendeel LA, Kouwenhoven MC, Gevaert O, et al. . Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009;69(23):9065–9072. [DOI] [PubMed] [Google Scholar]
- 16. Roth RB, Hevezi P, Lee J, et al. . Gene expression analyses reveal molecular relationships among 20 regions of the human CNS. Neurogenetics. 2006;7(2):67–80. [DOI] [PubMed] [Google Scholar]
- 17. Madhavan S, Zenklusen JC, Kotliarov Y, Sahni H, Fine HA, Buetow K. Rembrandt: helping personalized medicine become a reality through integrative translational research. Mol Cancer Res. 2009;7(2):157–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Verhaak RG, Hoadley KA, Purdom E, et al. ; Cancer Genome Atlas Research Network Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Q, Hu B, Hu X, et al. . Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell. 2018;33(1):152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72(10):2457–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nikbakht H, Panditharatna E, Mikael LG, et al. . Spatial and temporal homogeneity of driver mutations in diffuse intrinsic pontine glioma. Nat Commun. 2016;7:11185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Killela PJ, Reitman ZJ, Jiao Y, et al. . TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110(15):6021–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. See WL, Tan IL, Mukherjee J, Nicolaides T, Pieper RO. Sensitivity of glioblastomas to clinically available MEK inhibitors is defined by neurofibromin 1 deficiency. Cancer Res. 2012;72(13):3350–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Heldin CH. Targeting the PDGF signaling pathway in tumor treatment. Cell Commun Signal. 2013;11:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mertens F, Johansson B, Fioretos T, Mitelman F. The emerging complexity of gene fusions in cancer. Nat Rev Cancer. 2015;15(6):371–381. [DOI] [PubMed] [Google Scholar]
- 27. Singh D, Chan JM, Zoppoli P, et al. . Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337(6099): 1231–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim J, Lee Y, Cho HJ, et al. . NTRK1 fusion in glioblastoma multiforme. PLoS One. 2014;9(3):e91940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chandrasekaran G, Tátrai P, Gergely F. Hitting the brakes: targeting microtubule motors in cancer. Br J Cancer. 2015;113(5):693–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Polito A, Reynolds R. NG2-expressing cells as oligodendrocyte progenitors in the normal and demyelinated adult central nervous system. J Anat. 2005;207(6):707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Al-Mayhani MT, Grenfell R, Narita M, et al. . NG2 expression in glioblastoma identifies an actively proliferating population with an aggressive molecular signature. Neuro Oncol. 2011;13(8):830–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Alcantara Llaguno SR, Wang Z, Sun D, et al. . Adult lineage-restricted CNS progenitors specify distinct glioblastoma subtypes. Cancer Cell. 2015;28(4):429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Margolin AA, Nemenman I, Basso K, et al. . ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7(Suppl 1):S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee SO, Li X, Khan S, Safe S. Targeting NR4A1 (TR3) in cancer cells and tumors. Expert Opin Ther Targets. 2011;15(2):195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kast RE. Why cerebellar glioblastoma is rare and how that indicates adjunctive use of the FDA-approved anti-emetic aprepitant might retard cerebral glioblastoma growth: a new hypothesis to an old question. Clin Transl Oncol. 2009;11(7):408–410. [DOI] [PubMed] [Google Scholar]
- 36. Pekmezci M, Rice T, Molinaro AM, et al. . Adult infiltrating gliomas with WHO 2016 integrated diagnosis: additional prognostic roles of ATRX and TERT. Acta Neuropathol. 2017;133(6):1001–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Barthel FP, Wei W, Tang M, et al. . Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nat Genet. 2017;49(3):349–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Koschmann C, Zamler D, MacKay A, et al. . Characterizing and targeting PDGFRA alterations in pediatric high-grade glioma. Oncotarget. 2016;7(40):65696–65706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu G, Diaz AK, Paugh BS, et al. . The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet. 2014;46(5):444–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jones DT, Hutter B, Jäger N, et al. ; International Cancer Genome Consortium PedBrain Tumor Project Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet. 2013;45(8):927–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang J, Wu G, Miller CP, et al. ; St Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet. 2013;45(6):602–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Meyronet D, Esteban-Mader M, Bonnet C, et al. . Characteristics of H3 K27M-mutant gliomas in adults. Neuro Oncol. 2017;19(8):1127–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.