Abstract
Background
There are clear differences in embryo development between angiosperm and gymnosperm species. Most of the current knowledge on gene expression and regulation during plant embryo development has derived from studies on angiosperms species, in particular from the model plant Arabidopsis thaliana. The few published studies on transcript profiling of conifer embryogenesis show the existence of many putative embryo-specific transcripts without an assigned function. In order to extend the knowledge on the transcriptomic expression during conifer embryogenesis, we sequenced the transcriptome of zygotic embryos for several developmental stages that cover most of Pinus pinaster (maritime pine) embryogenesis.
Results
Total RNA samples collected from five zygotic embryo developmental stages were sequenced with Illumina technology. A de novo transcriptome was assembled as no genome sequence is yet published for Pinus pinaster. The transcriptome of reference for the period of zygotic embryogenesis in maritime pine contains 67,429 transcripts, which likely encode 58,527 proteins. The annotation shows a significant percentage, 31%, of predicted proteins exclusively present in pine embryogenesis. Functional categories and enrichment analysis of the differentially expressed transcripts evidenced carbohydrate transport and metabolism over-representation in early embryo stages, as highlighted by the identification of many putative glycoside hydrolases, possibly associated with cell wall modification, and carbohydrate transport transcripts. Moreover, the predominance of chromatin remodelling events was detected in early to middle embryogenesis, associated with an active synthesis of histones and their post-translational modifiers related to increased transcription, as well as silencing of transposons.
Conclusions
Our results extend the understanding of gene expression and regulation during zygotic embryogenesis in conifers and are a valuable resource to support further improvements in somatic embryogenesis for vegetative propagation of conifer species. Specific transcripts associated with carbohydrate metabolism, monosaccharide transport and epigenetic regulation seem to play an important role in pine early embryogenesis and may be a source of reliable molecular markers for early embryogenesis.
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1564-2) contains supplementary material, which is available to authorized users.
Keywords: Zygotic embryo, Pinus pinaster, Embryogenesis, RNA-seq, Developmental stages, Transcript profiling
Background
In higher plants, embryogenesis starts with the zygote formation and comprehends the whole developmental process that leads to a full mature and dormant embryo, enclosed by the seed tissues [1]. Most of current knowledge about plant embryogenesis derived from studies on angiosperm species, in particular from the model organism Arabidopsis thaliana (reviewed in [2]). However, gymnosperm and angiosperm lineages are estimated to have driven apart over 300 million years ago [3] and their differences, in particular at the embryogenic phase, are well known (reviewed by [4]). Molecular studies of embryogenesis in gymnosperms, and especially in conifers, have gained interest in the last few years (reviewed in [5, 6]). This has been driven by a better understanding of how the characteristic differences in embryo development between angiosperms and gymnosperms are established at the molecular level, and their evolutionary implications. Moreover, further improvement of somatic embryogenesis, an attractive technology for large scale vegetative propagation of economically important conifers, is largely dependent on additional knowledge about the basic processes controlling embryo development.
Next-generation sequencing (NGS) technologies applied to mRNA discover and profiling (RNA-seq) have proved useful to study plant gene regulation, in particular for the non-model species still missing a genome of reference (reviewed by [7, 8]). Large RNA sequencing projects such as the 1KP project alone achieved the transcriptomic sequencing in over 1000 different plant species which represents a huge effort with high impact in phylogenetic and land plant evolution studies [http://www.onekp.com; [9]]. RNA-seq data have considerably advanced our knowledge about the regulation of plant stress responses [10], plant development [11–13], synthesis of commercially/biotechnologically relevant plant products [14] or even the evolution of specific genes [15]. The embryo mRNA transcriptomes of several plant species, including rice (Oryza sativa) [16], maize (Zea mays) [17, 18], canola (Brassica napus) [19] and radish (Raphanus sativus L.) [20], have been generated by RNA-seq technology. In conifers, the transcriptome of early developmental stages of Scots pine (Pinus sylvestris) [21] dominant embryo has been profiled using high-throughput sequencing. Additionally, other studies addressing gymnosperm embryogenesis using NGS (reviewed by [22]) include the reports by Yakovlev et al. (2014) on embryo transcriptome changes in Picea abies under different temperature conditions [23], on the transcriptomes of embryogenic and non-embryogenic tissues of Picea balfouriana [24] on transcriptome comparative analysis of early somatic embryo formation and seed development in Araucaria angustifolia [25], on a comprehensive transcriptome survey of several Pinus lambertiana tissue types including embryos [26], on somatic embryo transcriptome profiling in Picea abies and [27] on the identification of carbohydrate-mediated responses associated with Araucaria angustifolia embryo formation.
Previously, a time-course transcriptomic study in Pinus pinaster pointed out the relevance of epigenetic regulators and specific transcription factors during the development of the embryo [28]. In such study, a cross-species microarray hybridization approach was followed, limiting the identification of candidate transcripts to the set of array probes derived from Pinus taeda root and needle tissues. Building upon that study, we extend here the scope of the transcriptomic analysis by using a high-throughput sequencing approach, with its known benefits over microarrays (reviewed in [29]), including the capacity to retrieve novel and/or lowly expressed transcripts, or alternative splice variants that might have been missed by microarray analysis [12]. In this work, we generated a species-specific transcriptome of the developing embryo aiming to have a significantly extended catalogue of maritime pine transcripts expressed during embryogenesis, targeting those transcripts with higher differences in expression during embryo development. By following this approach, we have identified over-represented processes, namely carbohydrate metabolism and epigenetic regulation, in specific phases of embryo development as well as specific transcripts involved. This unique resource in maritime pine further contributes for deepening our knowledge of the transcriptional activity during embryogenesis in conifers.
Results
Comprehensive transcriptome assembly
We performed RNA-seq on five embryogenesis stages (Day0, Day5, Day11, Day15 and Day25) according to a previously reported staging system [30], which cover the whole developmental period of the zygotic embryo of P. pinaster, up to the maturation stage. To capture the transcriptome landscape of the embryo and major differences in gene expression throughout development Illumina short-reads technology was used to sequence the RNA-seq libraries. In the absence of a published reference genome for P. pinaster, a comprehensive transcriptome assembly approach ([31]; reviewed in [32]) was adopted, which combines a de novo assembly of the reads using Trinity (version 2.0.6) [33] and a guided-assembly by mapping both the reads and de novo assembled transcripts against P. taeda genome (version 1.01) [34]. A total of ca. 319 M read-pairs were obtained, with an average of 63.8 M read-pairs per sample and each sample contributing from 44.9 M (14.1%) to 75.4 M (23.6%) read-pairs. A 5.3% of the reads was removed by the filtering steps. The resulting ca. 302 M read-pairs were subsequently used for de novo transcriptome assembly and mapping to the P. taeda genome. After obtaining the comprehensive transcriptome assembly, a total of 183.4 M read-pairs mapped in the correct distance and orientation (Properly paired) to this reference for expression analysis, which represents 57.5% of the raw reads (Table 1).
Table 1.
RNA-seq and mapping statistics of P. pinaster embryo developmental stages
| Embryo developmental stage | Day0 | Day5 | Day11 | Day15 | Day25 |
|---|---|---|---|---|---|
| Read lengtha | PE 50 bp | PE 50 bp | PE 50 bp | PE 100 bp | PE 100 bp |
| Raw pairs of reads | 72,632,308 | 55,550,140 | 44,898,746 | 75,433,086 | 70,463,384 |
| Clean pairs of reads | 72,472,236 | 55,413,990 | 44,803,454 | 66,982,962 | 62,388,454 |
| Pairs of reads mapping in correct distance and orientation on P. taeda genome (Properly paired) used for assembly | 61,626,536 | 45,975,573 | 37,763,000 | 60,334,604 | 55,692,971 |
| Pairs of reads properly mapping on P. pinaster comprehensive transcriptome used for expression | 44,178,974 | 33,660,866 | 28,187,082 | 41,345,428 | 35,994,032 |
aPE = paired-end
The final reference transcriptome of maritime pine zygotic embryogenesis contains 67,429 transcripts (deposited at DDBJ/ENA/GenBank under the accession GGEX01000000, https://www.ebi.ac.uk/ena/data/view/GGEX01000000), varying in length from 148 bp to 12,752 bp and with a mean length of 999 bp (Table 2).
Table 2.
Statistics of the assembled transcriptome from P. pinaster embryo development
| Reference transcriptome | |
|---|---|
| Total assembled transcripts | 67,429 |
| ExN50a | 1653 bp |
| Shorter assembled transcript length (transcript) | 148 bp (Pp11025) |
| Longer assembled transcript length (transcript) | 12,752 bp (Pp28188) |
| Mean length | 999 bp |
| Median length | 627 bp |
| Transcripts without N’s bases | 65,852 |
| Mean gap percentage per transcript length | 0.2% |
| Predicted coding transcripts | 58,527 |
| Predicted non-coding transcripts | 8896 |
aExN50 = transcript length metric that considers the top most highly expressed transcripts and means that at least 50% of the assembled transcript nucleotides were found in transcripts that were at least this length
Transcriptome annotation
All ORF possibilities were generated from the newly assembled transcriptome, and only one per transcript (the longest one in case of multiple possibilities) was translated to generate the final proteome containing 58,527 proteins (Additional file 1). The annotation of the assembled developing embryo transcriptome was done using two sources of information: best reciprocal hits (BRH) to the proteomes of P. taeda, P. lambertiana and A. thaliana, and homology to proteins in NCBI databases (Additional file 2). The function and gene ontology (GO) terms from annotated BRHs were assigned back to the P. pinaster protein, to annotate as result 14,211 P. pinaster proteins. Blast2GO (version 3.1) was used to annotate the transcripts, starting from the BLASTX alignments of the transcriptome to the NCBI non-redundant proteins database (E-value < 10− 3, Additional file 3). Nearly 30,000 sequences were annotated with at least one GO term, and around 7500 transcripts had a homologous in the NCBI database but could not clearly be associated to a GO term. There were 28,780, 26,585 and 24,241 transcripts with at least one F:GO, P:GO or C:GO term, respectively. There are 16,056 transcripts with at least one GO term from each of the three categories. Over 20,000 P. pinaster protein sequences did not align to any protein in the database (Additional file 4). The homologous proteins presented a mean similarity of 71%, while 54% of the BLAST hits had a sequence similarity over 70% (Additional file 5). The analysis of the highest scoring homologous sequence to each P. pinaster transcripts showed that Picea sitchensis, a close relative of P. pinaster, is the most represented species by far, with over 16,000 BLAST Top-Hits. The other homologous belonged to species from different plant taxonomic groups, including gymnosperms, angiosperms and mosses. Four other Pinus species were represented, namely P. taeda, P. radiata, P. sylvestris and P. monticola (Additional file 6). The comparison with EBI’s InterPro database (IPS) for protein sequences and functions revealed about 37,500 P. pinaster sequences with a IPS result; IPS analysis contributed GO terms to over 20,000 P. pinaster sequences (Additional file 7). Mapping results revealed that UniProtKB and TAIR were the two main source databases of GO terms associated to P. pinaster sequences (Additional file 8).
Functional regulation during embryo development
The proteins from P. pinaster, P. taeda and P. lambertiana were clustered together according to the eggNOG group of their respective best orthologous sequenced in EMBL’s eggNOG database of functionally annotated proteins (Additional file 9). When comparing the number of groups in the three species, P. pinaster had the highest number of exclusive groups (4355). Still, most of the groups, in a total of 5698, had proteins from the three species (Fig. 1). The groups with more protein members (Additional file 10) are common to the three conifer species, and were annotated as containing “pentatricopeptide -PPR- repeats”, either implicated in replication, recombination and repair or with a function unknown, “terminal inverse repeats -TIR-”, “leucine rich repeats” or “NB-ARC domains”, which are molecular switches implicated in signal transduction mechanisms. On the other hand, the groups exclusive of P. pinaster that include at least 10 proteins were annotated as “zinc finger proteins” (19 members), “sister chromatid cohesion protein PDS5” (13 members), “GDP-L-galactose phosphorylases” and “zinc ion binding proteins” (11 members each), and several clusters annotated as “retrotransposon proteins”. However, most of the groups exclusive of P. pinaster contained only one protein.
Fig. 1.
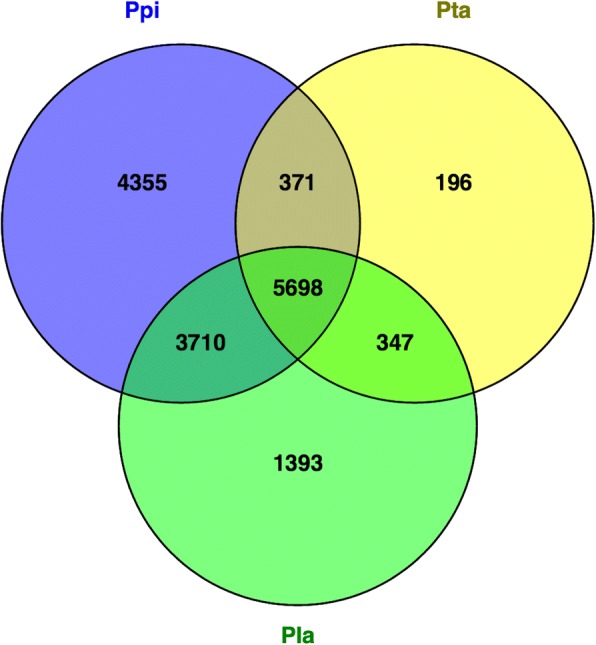
Venn diagram of the number of eggNOGs found with the predicted proteins of P. pinaster (Ppi) and two other relative conifer species, P. taeda (Pta) and P. lambertiana (Pla). Proteins were first annotated with the eggNOG numbers of their best homolog and then those sharing the same eggNOG number were concatenated. The numbers in the intersections represent the eggNOGs these conifers have in common
Since each protein group was classified into a functional eggNOG annotation [35], it was possible to condense the functional information to 24 categories. Most of the predicted proteins and associated eggNOGs had function unknown (5684 from the total 14,134 eggNOGS found in P. pinaster) (Additional file 10). For the remaining, a heatmap of functional categories expressed throughout P. pinaster embryo development (Fig. 2) shows three major clusters of functional categories predominantly expressed at the early (Day0), middle (Day5 and Day11) or late (Day15 and Day25) embryogenesis stages. At the early embryo stages, functions associated to the cytoskeleton, energy production and conversion, carbohydrate transport and metabolism, amino acid transport and metabolism and intracellular trafficking, secretion and vesicular transport are prevalent, with the first three being just up-regulated at these stages. In contrast, replication, recombination and repair, and cell cycle control, cell division, chromosome partitioning, and chromatin structure and dynamics follow the opposite trend and are up-regulated in late embryo stages.
Fig. 2.
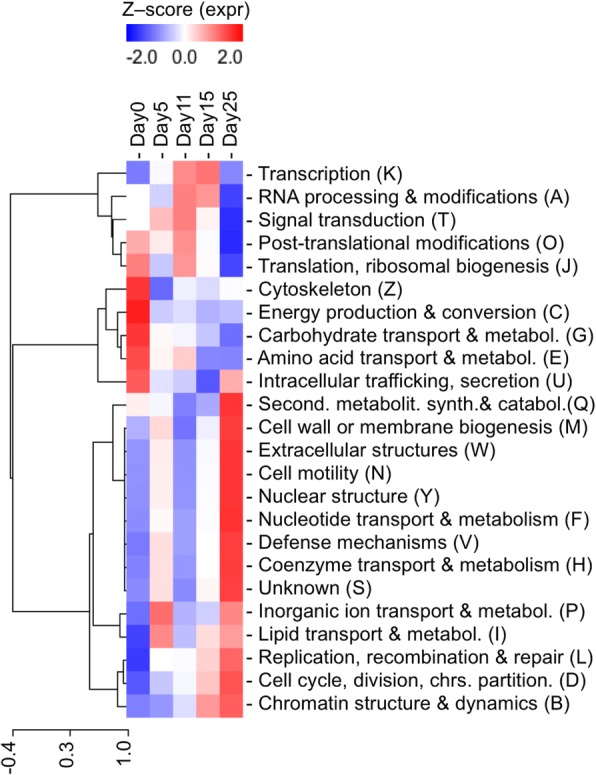
Heatmap of the eggNOG functional categories expressed throughout P. pinaster embryo developmental stages. The heatmap was built after the counts of each transcript belonging to a protein group in such category were added up, and later transformed in Z-scores. The capital letters in between brackets originate in the eggNOGs database and are specific to the functional categories. The hierarchical clustering on the left side of the image represents the correlation of the expression levels associated with distinct functional categories
Regarding the overall metabolic activity of the developing embryo, carbohydrate transport and metabolism is upregulated at early embryo stages decreasing towards the mature embryo, amino acid transport and metabolism is predominant in early embryo to early cotyledonary embryo stages, while secondary metabolites biosynthesis, transport, and catabolism peak at the mature embryo stage. Nucleotide transport and metabolism and coenzyme transport and metabolism show highest expression at mature embryo stage, whereas lipid transport and metabolism is relevant both in the pre-cotyledonary and mature embryo stages.
Differentially expressed transcripts along embryo development
From the 67,429 assembled transcripts, 64,766 are clearly expressed in at least one developmental stage and 39,838 are expressed in all five stages (Additional file 11). The number of transcripts expressed in each stage is equivalent, over 10,000 expressed transcripts per stage. A total of 4953 transcripts (7.3%) are expressed in only one stage of embryo development: 848, 338, 282, 1703, 1782 are exclusively expressed in Day0, Day5, Day11, Day15, and Day25, respectively.
A differential expression analysis (FDR < 0.05) between each pair of consecutive stages identified 1738 transcripts (2.6%) differentially expressed in at least one transition (Additional file 12). A total of 798, 383, 591 and 568 transcripts were differentially expressed in the first (from Day0 to Day5), second (from Day5 to Day11), third (from Day11 to Day15), and fourth (from Day15 to Day25) transition, respectively. Up-regulation of differentially expressed transcripts is predominant in the first transition, accounting for 594 of the 1185 up-regulated differentially expressed transcripts (Fig. 3) specific for first transition, while down-regulation of differentially expressed transcripts is more abundant in the last transition (Fig. 4), representing approximately 38% of the total down-regulated transcripts. Each developmental transition shares few up- or down-regulated transcripts with the consecutive transition, supporting that the developmental stages selected for this study are clearly differentiated in terms of ongoing transcriptional activity.
Fig. 3.
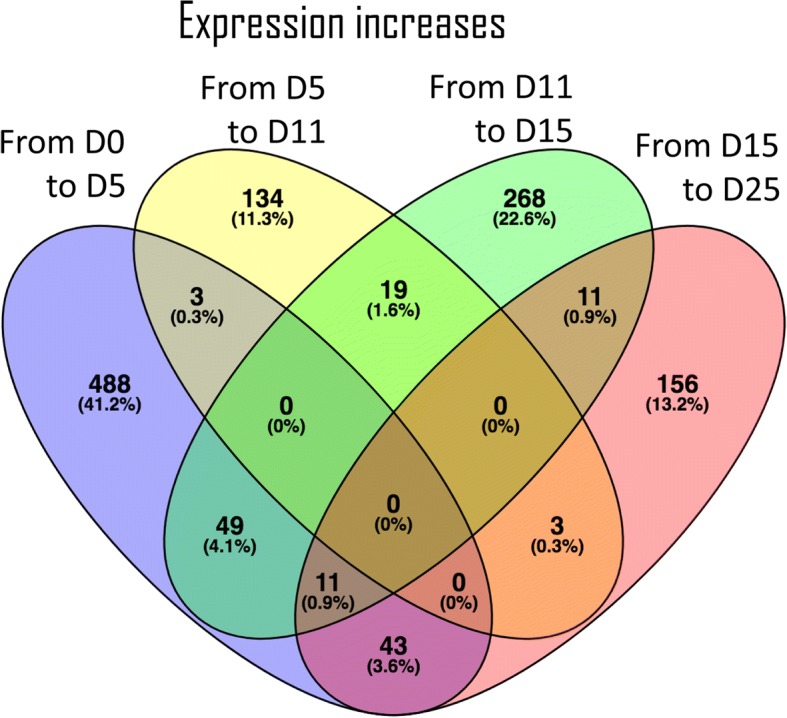
Venn diagram of the 1185 differentially expressed transcripts found up-regulated between two consecutive embryo developmental stages. The number of transcripts and respective percentage (relative to the total aforementioned 1185 transcripts) are represented for each transition. The numbers in the intersections represent transcripts found up-regulated in more than one developmental transition
Fig. 4.
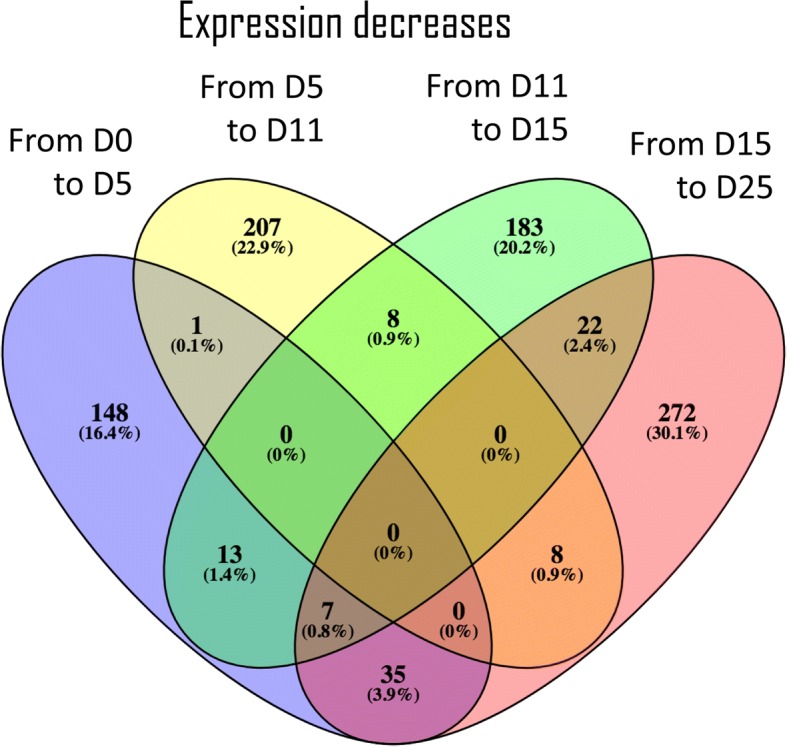
Venn diagram of the 904 differentially expressed transcripts found down-regulated between two consecutive embryo developmental stages. The number of transcripts and respective percentage (relative to the total aforementioned 904 transcripts) are represented for each transition. The numbers in the intersections represent transcripts found down-regulated in more than one developmental transition
A gene enrichment analysis of the differentially expressed transcripts retrieved over-represented GO terms associated with the first and last stage transitions (see Additional files 13, 14 and 15). Both transitions share an enrichment of down-regulated transcripts associated with regulation of cell cycle and cell division. Moreover, the first transition seems to be characterized by down-regulation of transcripts involved in monosaccharide transport and plant-type cell wall cellulose metabolism. The last transition is characterized by down-regulated transcripts related with epigenetics and annotated with the DNA-dependent DNA replication GO term. As for the up-regulated transcripts, those found in the first transition are associated with terpenoid catabolism and tertiary alcohol metabolism, while the only enriched GO term found in last transition is the molecular function nutrient reservoir activity (data not shown). Overall, the terms carbohydrate transport and metabolism (Table 3) and epigenetics related terms (Table 4) are highly represented in the list of differentially expressed transcripts.
Table 3.
Differentially expressed transcripts related to carbohydrate transport and metabolism
| Clustera | Transcript | At Locus | Pta Locus | Pla Locus | Annotationb |
|---|---|---|---|---|---|
| k1 | Pp10265 | #N/A | #N/A | #N/A | phosphoenolpyruvate carboxykinase |
| k1 | Pp1126 | #N/A | #N/A | #N/A | mannan endo- -beta-mannosidase |
| k1 | Pp19154 | #N/A | #N/A | #N/A | cytosolic triosephosphate isomerase |
| k1 | Pp28580 | AT4G37870.1 | 2A_all_VO_L_6958_T_66/101|m.15610 | V1_2kb/polished_high/000086|m.144 | phosphoenolpyruvate carboxykinase |
| k1 | Pp42692 | #N/A | #N/A | #N/A | udp-d-glucuronate 4-epimerase 2 |
| k1 | Pp45907 | #N/A | #N/A | #N/A | triose phosphate phosphate non-green precursor |
| k1 | Pp45908 | #N/A | #N/A | #N/A | phosphate phosphoenolpyruvate translocator precursor |
| k2 | Pp11437 | #N/A | #N/A | #N/A | xyloglucan endotransglucosylase hydrolase |
| k2 | Pp11438 | #N/A | #N/A | P/miseq/c27580_g1_i1|m.27950 | PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 26 |
| k2 | Pp12819 | AT1G76160.1 | 2A_all_VO_L_1_T_165541/181832|m.14942 | E1_2kb_2/polished_high/000754|m.928 | L-ascorbate oxidase homolog SKU5 SIMILAR 5 (SKS5) |
| k2 | Pp1567 | #N/A | #N/A | #N/A | multicopper oxidase |
| k2 | Pp18330 | #N/A | 6A_all_VO_L_6290_T_21/157|m.53348 | #N/A | PREDICTED: sugar transport protein 13 |
| k2 | Pp18567 | #N/A | #N/A | DCR/hiseq/c107286_g1_i1|m.126449 | PREDICTED: probable pectinesterase 68 |
| k2 | Pp1892 | #N/A | #N/A | #N/A | aldose 1-epimerase |
| k2 | Pp20813 | #N/A | #N/A | #N/A | hexose transporter |
| k2 | Pp2456 | #N/A | #N/A | #N/A | cellulose synthase |
| k2 | Pp27962 | #N/A | 5A_I15_VO_L_1793_T_10/15|m.46809 | #N/A | PREDICTED: CMP-sialic acid transporter 2 |
| k2 | Pp28117 | #N/A | #N/A | #N/A | alcohol dehydrogenase |
| k2 | Pp28964 | #N/A | 2A_all_VO_L_8976_T_26/41|m.15937 | #N/A | Xyloglucan endotransglucosylase/hydrolase protein A precursor, putative |
| k2 | Pp30302 | #N/A | #N/A | #N/A | glyoxalase i |
| k2 | Pp32670 | AT5G26340.1 | #N/A | JASS/hiseq/c63525_g1_i1|m.64429 | SUGAR TRANSPORT PROTEIN 13 (STP13), ATSTP13, MSS1 |
| k2 | Pp32960 | #N/A | #N/A | #N/A | probable inositol transporter 2-like |
| k2 | Pp34009 | #N/A | 3A_I18_VO_L_2_T_2368/136250|m.32448 | Basket/c19128_g1_i1|m.26924 | PREDICTED: L-ascorbate oxidase homolog |
| k2 | Pp36526 | #N/A | #N/A | #N/A | L-ascorbate oxidase-like protein |
| k2 | Pp37397 | #N/A | #N/A | #N/A | nadp-dependent malic enzyme |
| k2 | Pp37548 | AT4G39770.1 | #N/A | M_S1/c25586_g1_i4|m.129341 | trehalose-6-phosphate phosphatase |
| k2 | Pp38427 | #N/A | #N/A | #N/A | pyruvate kinase |
| k2 | Pp38450 | AT2G01850.1 | 2A_I15_VO_L_1_T_124932/133144|m.21738 | V1_1kb_1/015976|m.720027 | ENDOXYLOGLUCAN TRANSFERASE A3 (EXGT-A3), XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 27 (XTH27), ATXTH27 |
| k2 | Pp38781 | AT4G02290.1 | 2A_I15_VO_L_39_T_24/48|m.21781 | S_2kb/polished_high/000346|m.477 | glycosyl hydrolase 9B13, endoglucanase 17 |
| k2 | Pp38924 | #N/A | #N/A | #N/A | xylose isomerase |
| k2 | Pp39507 | AT5G13870.1 | 6A_I20_VO_L_1_T_64241/133533|m.55411 | E1_2kb_2/polished_high/002288|m.2304 | ENDOXYLOGLUCAN TRANSFERASE A4 (EXGT-A4), XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 5 (XTH5) |
| k2 | Pp42916 | #N/A | 5A_I12_VO_L_2_T_51143/53062|m.43159 | #N/A | phosphoenolpyruvate carboxykinase |
| k2 | Pp43330 | #N/A | #N/A | #N/A | pectin methylesterase (pectinesterase) |
| k2 | Pp43761 | #N/A | #N/A | #N/A | glycosyl hydrolase-like protein |
| k2 | Pp46170 | AT3G13790.1 | 6A_all_VO_L_6326_T_78/89|m.53354 | #N/A | Arabidopsis thaliana CELL WALL INVERTASE 1 (ATCWI1), ATBFRUCT1, ATCWINV1, CWI1 |
| k2 | Pp47826 | AT5G03630.1 | 5A_all_VO_L_2839_T_31/70|m.40851 | Wound/hiseq/c70186_g2_i1|m.47659 | Pyridine nucleotide-disulphide oxidoreductase family protein (MDAR2), monodehydroascorbate reductase |
| k2 | Pp6019 | AT1G77210.1 | #N/A | #N/A | SUGAR TRANSPORT PROTEIN 14 (STP14), ATSTP14 |
| k2 | Pp6337 | #N/A | #N/A | #N/A | multicopper oxidase |
| k2 | Pp8252 | #N/A | #N/A | #N/A | hexose transporter |
| k2 | Pp8434 | AT1G68560.1 | #N/A | S_2kb/polished_high/000741|m.810 | ALPHA-XYLOSIDASE 1 (XYL1), ALTERED XYLOGLUCAN 3 (AXY3), THERMOINHIBITION RESISTANT GERMINATION 1 (TRG1), ATXYL1, GH31 |
| k2 | Pp8435 | #N/A | #N/A | #N/A | alpha-xylosidase precursor |
| k2 | Pp8535 | #N/A | #N/A | #N/A | endoglucanase |
| k2 | Pp9898 | AT3G59480.1 | #N/A | S_2kb_2/020091|m.629185 | pfkB-like carbohydrate kinase family protein |
| k3 | Pp15644 | #N/A | #N/A | #N/A | beta glucosidase 43 |
| k3 | Pp21641 | #N/A | #N/A | #N/A | mannan endo- -beta-mannosidase 7 |
| k3 | Pp23792 | #N/A | 6A_all_VO_L_13474_T_9/19|m.54026 | V1_1kb_3/polished_high/001086|m.1085 | senescence-associated protein 29 |
| k4 | Pp16423 | #N/A | #N/A | #N/A | protein |
| k4 | Pp24290 | #N/A | #N/A | #N/A | beta-amylase 7-like |
| k4 | Pp27124 | #N/A | #N/A | #N/A | o-glycosyl hydrolases family 17 protein |
| k4 | Pp34845 | #N/A | #N/A | #N/A | phosphatidylinositol 4-kinase |
| k4 | Pp9901 | #N/A | #N/A | #N/A | alkaline alpha galactosidase i |
| k5 | Pp14379 | #N/A | #N/A | #N/A | O-Glycosyl hydrolases family 17 protein |
| k5 | Pp1469 | #N/A | #N/A | #N/A | unknown; unknown [Picea sitchensis] |
| k5 | Pp32140 | #N/A | #N/A | #N/A | brassinosteroid-regulated protein bru1 |
| k5 | Pp32141 | #N/A | #N/A | SDN/miseq/c29601_g2_i2|m.39898 | xyloglucan endotransglucosylase/hydrolase protein 24 |
| k5 | Pp32144 | #N/A | #N/A | P/miseq/c33524_g1_i3|m.50829 | PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23-like |
| k5 | Pp39583 | #N/A | #N/A | #N/A | thermostable beta-glucosidase |
| k5 | Pp41586 | #N/A | #N/A | #N/A | probable glycerophosphoryl diester phosphodiesterase 3-like |
| k5 | Pp4948 | #N/A | #N/A | #N/A | catalase |
| k5 | Pp7568 | #N/A | #N/A | #N/A | alcohol dehydrogenase |
| k5 | Pp9008 | AT1G22170.1 | #N/A | E1/hiseq/c25131_g1_i3|m.5975 | Phosphoglycerate mutase-like protein |
| k6 | Pp23909 | #N/A | #N/A | #N/A | mannan endo- -beta-mannosidase 7 |
| k6 | Pp876 | #N/A | #N/A | #N/A | neurofilament protein h form h2 |
| k6 | Pp9495 | #N/A | #N/A | #N/A | phospholipase c |
| k7 | Pp26503 | #N/A | #N/A | #N/A | galactinol synthase |
| k7 | Pp2858 | #N/A | #N/A | #N/A | succinate dehydrogenase |
| k7 | Pp34900 | #N/A | #N/A | #N/A | malate synthase |
| k7 | Pp34906 | AT5G03860.1 | 2A_I2_OT_comp27109_c0_seq3|m.23989 | Neg_S1/c31207_g1_i2|m.108321 | malate synthase |
| k7 | Pp40679 | #N/A | 6A_I23_VO_L_1_T_44295/165398|m.58874 | BRN/hiseq/c65594_g2_i2|m.37248 | galactinol synthase |
| k7 | Pp40719 | AT3G24090.1 | 5A_all_VO_L_2_T_192437/409051|m.39335 | E1/hiseq/c40068_g1_i1|m.27499 | glucosamine-fructose-6-phosphate aminotransferase |
| k8 | Pp14503 | #N/A | #N/A | #N/A | beta-amylase 7-like |
| k8 | Pp44718 | #N/A | #N/A | #N/A | beta-amylase 7-like |
| k9 | Pp32321 | #N/A | #N/A | P/miseq/c26472_g1_i4|m.24888 | endoxyloglucan transferase A4 |
| k9 | Pp39715 | #N/A | 3A_I18_VO_L_2_T_63095/136250|m.32723 | #N/A | aldose 1-epimerase family protein |
| k9 | Pp40720 | #N/A | #N/A | #N/A | glucosamine--fructose-6-phosphate aminotransferase |
| k10 | Pp240 | #N/A | #N/A | #N/A | beta-glucosidase 44-like |
| k10 | Pp30121 | AT4G38970.1 | 3A_I16_VO_L_2_T_48696/60890|m.30958 | V_1kb/017920|m.889708 | fructose-bisphosphate aldolase 2 |
| k11 | Pp14541 | #N/A | #N/A | #N/A | aldehyde dehydrogenase family 2 member mitochondrial-like |
| k11 | Pp15288 | #N/A | #N/A | #N/A | pyruvate decarboxylase isozyme |
| k11 | Pp18297 | #N/A | 5A_I14_VO_L_947_T_33/56|m.45171 | DCS1kb_1/003053|m.3985 | PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 8-like |
| k11 | Pp23089 | #N/A | #N/A | #N/A | polygalacturonase |
| k11 | Pp35326 | #N/A | #N/A | #N/A | protein |
| k11 | Pp9001 | #N/A | #N/A | #N/A | probable xyloglucan endotransglucosylase hydrolase protein 23 |
| k11 | Pp995 | #N/A | #N/A | #N/A | myo-inositol-1-phosphate synthase |
| k11 | Pp9981 | AT1G32860.1 | #N/A | #N/A | Glycosyl hydrolase superfamily protein |
| k12 | Pp46622 | AT4G25000.1 | #N/A | SDN/miseq/c15856_g1_i1|m.11496 | alpha-amylase-like |
aNumber of the cluster of expression that the transcript is associated with
bOrder of preference for annotating each P. pinaster transcript after its homologs is: A. thaliana, P. taeda, P. lambertiana, and Blast2Go annotation against NCBI
Table 4.
Differentially expressed transcripts involved in epigenetic regulation (with BRH found)
| Clustera | Transcript | At Locus | Pta Locus | Pla Locus | Annotationb |
|---|---|---|---|---|---|
| DNA modification | |||||
| 3 | Pp11214 | AT1G57820.1 | #N/A | V_2kb_2/polished_high/000175|m.181 | VARIANT IN METHYLATION 1 (VIM1), ORTHRUS 2 (ORTH2) |
| 5 | Pp3794 | #N/A | #N/A | P/miseq/c15561_g1_i1|m.8297 | PREDICTED: DNA (cytosine-5)-methyltransferase DRM2-like |
| Histone modification | |||||
| 1 | Pp30887 | #N/A | #N/A | SDN/miseq/c40928_g1_i1|m.61118 | ubiquitin-conjugating enzyme 28, E2 |
| 3 | Pp44003 | AT5G24330.1 | 6A_all_VO_L_4227_T_27/112|m.53170 | DCS/hiseq/c48519_g1_i1|m.18331 | ARABIDOPSIS TRITHORAX-RELATED PROTEIN 6 (ATXR6), SET DOMAIN PROTEIN 34 (SDG34) |
| 10 | Pp33894 | #N/A | 5A_I16_NT_comp45588_c0_seq2|m.47551 | S/hiseq/c42737_g1_i1|m.36406 | PREDICTED: histone-lysine N-methyltransferase SUVR5 |
| Chromatin formation or chromatin remodelling | |||||
| 3 | Pp26994 | AT5G22750.1 | #N/A | S/hiseq/c37932_g2_i1|m.21712 | RAD5, RAD5A |
| 3 | Pp34781 | AT1G65470.1 | 6A_I23_VO_L_4689_T_39/51|m.59955 | RF-S_3/polished_high/005053|m.5101 | FASCIATA 1 (FAS1), FUGU 2, FUGU2, NFB2, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP B |
| 7 | Pp30270 | AT5G37055.1 | #N/A | BRN/hiseq/c66142_g2_i1|m.38827 | SERRATED LEAVES AND EARLY FLOWERING (SEF), ATSWC6 |
| 8 | Pp14163 | AT1G05490.1 | #N/A | S/hiseq/c33164_g1_i2|m.13589 | CHROMATIN REMODELING 31 (CHR31) |
| RNA silencing | |||||
| 1 | Pp12441 | #N/A | 5A_I13_OT_comp19461_c0_seq4|m.44088 | #N/A | PREDICTED: protein argonaute 2-like |
| Histones | |||||
| 3 | Pp36206 | #N/A | #N/A | E1_2kb_2/polished_high/001369|m.1490 | histone H3 |
| 3 | Pp38724 | #N/A | #N/A | DCS1kb_3/polished_high/000803|m.897 | histone H2A 12 |
| 6 | Pp46360 | #N/A | 2A_all_VO_L_1_T_65055/181832|m.14308 | #N/A | PREDICTED: histone H1-like |
| 10 | Pp14332 | AT2G30620.1 | #N/A | M_S1/c4520_g1_i1|m.114500 | HISTONE 1.2 (H1.2) |
| 10 | Pp46359 | #N/A | #N/A | SDN/miseq/c31005_g5_i1|m.46415 | histone H1.2 |
| Others | |||||
| 3 | Pp41359 | #N/A | 5A_all_VO_L_69333_T_11/13|m.42398 | V_3_3-6 kb/016515|m.360307 | Telomere-associated protein RIF1 |
aNumber of the cluster of expression that the transcript is associated with
bOrder of preference for annotating each P. pinaster transcript after its homologs is: A. thaliana, P. taeda, and P. lambertiana
Clustering of the differentially expressed transcripts
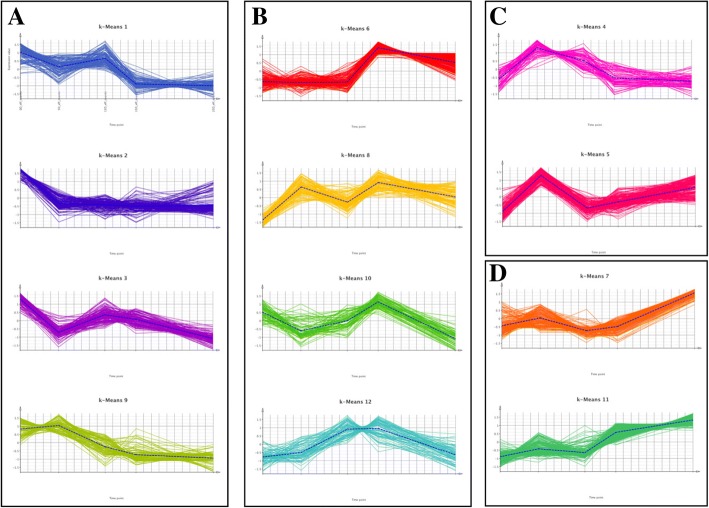
The 1738 differentially expressed transcripts throughout embryo development could be grouped into 12 distinct clusters (K) with similar patterns of expression by k-means clustering analysis (Fig. 5). Overall, the clusters could be further divided in four groups (A to D) according to the embryo stage in which the expression peaks: (A) early embryo stages (clusters 1, 2, 3, 9), (B) early-cotyledonary and/or cotyledonary stages (clusters 6, 8, 10, 12), (C) pre-cotyledonary embryo stages (clusters 4, 5), and (D) mature embryo stage (clusters 7, 11). Following an enrichment analysis performed in each cluster, several processes and functions were found over-represented in different phases of embryo development (Fig. 5). The transcripts in group A are enriched in several GO terms including carbohydrate metabolism, monosaccharide transport, sterol metabolism, cell wall organization or biogenesis, DNA-dependent DNA replication, cell cycle, regulation of gene expression, epigenetic, among others (Fig. 5). In cluster 2, processes related to carbohydrate metabolism and monosaccharide transport and to the cell wall are highly represented (Additional file 16). In cluster 3, which differs from cluster 2 mainly because the expression stays relatively high at mid-embryo stages before decreasing towards the mature embryo stage, DNA-dependent DNA replication is particularly relevant in terms of the number of contributing transcripts (89 transcripts) and includes chromatin organization, DNA modification, DNA methylation, histone lysine methylation, DNA packaging, histone methylation, regulation of gene expression, epigenetic (Additional file 17). The transcripts in group B are enriched in GO terms such as steroid dehydrogenase activity, sesquiterpenoid catabolism, release of seed from dormancy and tertiary alcohol metabolism. In group D, the most represented terms are nutrient reservoir activity, carbon-oxygen lyase activity, alpha-bisabolene synthase activity and terpene synthase activity.
Fig. 5.
K-means clustering of differentially expressed transcripts along P. pinaster embryo development. Transcripts were clustered together according to their expression profiles and a representative mean expression profile (dashed line) was represented for every cluster. The 12 k-means clusters generated were further divided in four groups (A to D) depending on the embryo developmental stage in which the expression peaks. The inset displays the processes and functions found over-represented among the transcripts from the different clusters (and groups). a Clusters K1, K2, K3, K9, showing decreasing expression along development. Cellular carbohydrate metabolism, monosaccharide transport, carbohydrate metabolism, sterol metabolism, cell wall organization or biogenesis, external encapsulating structure organization,cell wall macromolecule metabolism, DNA-dependent DNA replication, biological regulation, organic cyclic compound metabolism, cellular aromatic compound metabolism, cellular component organization or biogenesis, cell cycle,developmental process, shoot system development, multicellular organismal process, mitotic cell cycle process, methylation, cell proliferation, microtubule-based process. b Clusters K6, K8, K10, K12, with expression peaking at Day11 and/or Day15. Steroid dehydrogenase activity, acting on the CH-CH group of donors, 3-oxo-5-alpha-steroid 4-dehydrogenase activity, abscisic acid catabolism, release of seed from dormancy. c Clusters K4, K5, with expression peaking at Day5. No GO-terms. d Clusters K7, K11, with increasing expression along development. Nutrient reservoir activity, carbon-oxygen lyase activity, acting on phosphates, carbon-oxygen lyase activity, alpha-bisabolene synthase activity, terpene synthase activity
Validation by qPCR
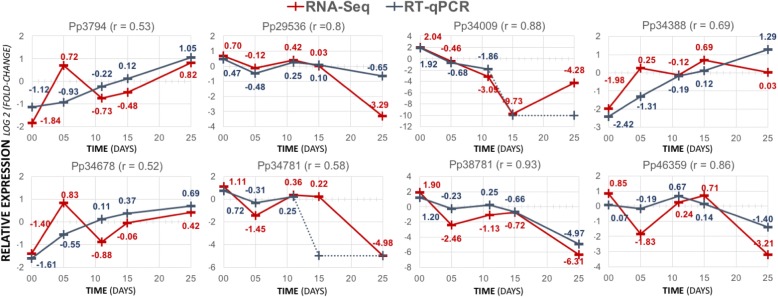
A subset of eight transcripts was selected based on their expression profile and putative involvement in carbohydrate metabolism (Table 3) or epigenetic regulation (Table 4), to independently validate the RNA-seq results using RT-qPCR (information about the primers can be found in Additional file 18). Five of these genes are differentially expressed transcripts throughout embryo development (FDR < 0.05) and are included in different transcription profiles (clusters 2, 3, 5, 10). The RT-qPCR and RNA-seq expression results are generally in good agreement as demonstrated by the values of Pearson correlation, which ranged between 0.52 (Pp34678) and 0.93 (Pp38781), confirming the peaks of expression associated with specific developmental stages (Fig. 6). There is also a good agreement between RT-qPCR and RNA-seq for transcripts which had not been predicted as differentially expressed (Pp29536, Pp34388 and Pp34678).
Fig. 6.
Validation of P. pinaster transcripts profile obtained by RNA-seq (red line) with RT-qPCR (blue line). Pearson correlation values (r) between the two technologies are shown. Fold-change values are also shown for each developmental stage. Dotted lines connect relative expression values calculated for Cq values detected in the last 5 cycles of the qPCR amplification program
Discussion
In this work, RNA-seq is used to provide a comprehensive overview of the transcriptome of the maritime pine developing embryo. Previously, the first transcriptomic analysis of the maritime pine embryo using the same developmental stages as those analysed here, has relied on the use of a loblolly pine (P. taeda) microarray to identify transcripts present during embryogenesis which are conserved between the two species [36]. Although the microarray had been successfully used for cross-species hybridization [37, 38], it contained approximately 25,000 unique cDNAs from P. taeda representative of cDNA libraries exclusively prepared from root and needle tissues, limiting the scope of the analysis considering our main focus on embryo development. In the present study, such limitation is overcome and the transcriptomic landscape during embryogenesis has been significantly expanded. Over 300 M read-pairs obtained from embryos at five stages of development were assembled using a comprehensive strategy to produce a reference transcriptome for maritime pine zygotic embryogenesis with 67,429 transcripts. This number is three times the number of expressed transcripts that had been previously identified with the P. taeda microarray [36], corresponding to a similar increase in the number of annotated proteins. Nevertheless, Picea sitchensis, Vitis vinifera and Ricinus communis remain the top three most represented species in the distribution of all Top-BLAST hits.
The annotation of P. pinaster, P. taeda and P. lambertiana predicted proteins with the eggNOG number of its best homolog revealed that most annotated proteins are shared by the three species (40.3% in P. pinaster, 51.1% in P. lambertiana, and 86.2% in P. taeda). P. pinaster and P. lambertiana predicted proteomes have approximately 10 times more proteins in common than those shared with P. taeda, while the latter shares roughly the same percentage of proteins with either species. Moreover, the percentage of exclusive predicted proteins was higher in P. pinaster (30.8%), against 3.0% in P. taeda. The analysis of orthologous groups highlighted some eggNOGs, shared by the three species, with a high number of protein members, which include “PPR repeat” and “Retrotransposon protein” involved in replication, recombination and repair, or with function unknown, and several eggNOGs described as “Leucine Rich Repeat (LRR)”, and “nucleotide binding domain with an ARC motif (NB-ARC domain)” involved in signaling transduction mechanisms, and “toll/interleukin-1 receptor (TIR)” with function unknown. These results are in close agreement with the reported annotation of the shoot transcriptome of Pinus patula in which the largest family that was identified, including 1794 members, contained LRR, TIR, NB-ARC, Golgi transport complex 5 (COG5) and poxvirus A32 protein motifs [31].
Carbohydrate metabolism and transport in early embryogenesis
This work gathered several evidences, both from the analysis of functional categories represented throughout embryo development and from the analysis of specific transcripts, pointing to a prominent role of carbohydrate transport and metabolism early in P. pinaster embryogenesis. The analysis of functional categories assigned to eggNOGs showed up-regulation of carbohydrate transport and metabolism in early embryo stages and a reduction in expression towards later stages of embryogenesis. Also, the enrichment analysis of all the differentially expressed transcripts revealed a group of highly expressed transcripts peaking at Day0 (cluster 2 profile) enriched in carbohydrate metabolism, including cellular carbohydrate metabolism. In the same profile, cellular carbohydrate metabolism appears associated to the metabolism of the primary cell wall components cellulose, pectin, and glucan. Additionally, the 204 differentially expressed transcripts down-regulated from Day0 to Day5 were enriched in monosaccharide transport and plant-type cell wall cellulose metabolism (see Additional file 13). A recent study in P. sylvestris seed development has also pointed out to an over-representation of carbohydrate metabolic process and cell wall modification terms among the differentially expressed transcripts over-represented at early developmental stages [21].
Many glycoside hydrolases encoding transcripts were found more expressed in the first embryo stage (cluster 2), including the Pp38781 homolog of AT4g02290 (glycosyl hydrolase 9 family) whose expression profile was successfully validated by RT-qPCR. In particular, the presence of several pine homologs of α-xylosidase and xyloglucan endotransglycosylase (previously named xyloglucan-endo-β-glucanase) within this expression profile points out the importance of xyloglucan mobilization and/or degradation in pine early embryogenesis [39]. Xyloglucan is the main hemicellulose constituent of the primary cell walls of spermatophytes except for grasses (reviewed by [40]), and the glycosidases capable of trimming the xyloglucan side chains are expected to act in the turnover or recycling of xyloglucan during cell wall expansion (reviewed in [41]). The Arabidopsis ALPHA-XYLOSIDASE 1/ ALTERED XYLOGLUCAN 3/ THERMOINHIBITION RESISTANT GERMINATION 1 (XYL1/ AXY3/ TRG1), whose putative maritime pine homolog (Pp8434) was also up-regulated in early embryogenesis, codes for an enzyme involved in xyloglucan degradation into free monosaccharides [42] and the only α-xylosidase active against xyloglucan [43]. Germinating seeds of Arabidopsis xyl1 loss-of-function mutants display cell wall loosening and reduced seed dormancy due to alterations in primary cell wall integrity [44]. Also a putative homolog (Pp38450) of the Arabidopsis ENDOXYLOGLUCAN TRANSFERASE A3/ XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 27 (EXGT-A3/ XTH27), involved in the differentiation of tracheary elements through the degradation of the xyloglucan deposited in the cell walls [45], was identified with the same expression profile (cluster 2), together with a putative homolog of XTH5, a gibberellin (GA)-inducible gene expressed in the embryonic axis and in the radicle of seeds, involved in germination [46]. Overall, it seems that the glycoside hydrolases involved in cell-wall modification during germination, are also relevant for the control of early stages of pine embryogenesis. In fact, enzymes responsible for xyloglucan degradation have already been found associated to somatic embryogenesis induction in divergent species (reviewed by [47, 48]), including conifers such as P. radiata where the up-regulation of α-D-galactosidase (SEPR1) was detected [49]. In Picea abies somatic embryogenesis a Xyloglucan:xyloglucosyl transferase and a Glycosyl transferase family 1 protein were found differentially expressed and down-regulated in the transitions from proliferation to differentiation of early embryos and from early embryos to development of late embryos [38].
Other members of the glycosyl hydrolase family were found differentially expressed in our results, including a putative homolog (Pp46170) of CELL WALL INVERTASE 1 (CWINV1/CWI1), described as playing a role in plant sink tissues where it performs the hydrolysis of sucrose, unloaded from the phloem via apoplast, into fructose and glucose (reviewed in [50]). Studies in cotton and Arabidopsis seeds revealed that CWINV is expressed all over the globular embryo but seems to be restricted to the central zone of the torpedo embryo [51]. In a comparison of the response to somatic embryogenesis induction in shoot primordia derived from adult trees of Picea glauca, up-regulation of CWINV1 was detected only in non-responsive genotype, and a possible link with biotic stress response was discussed [52]. There are also many evidences that associate CWINV-mediated sucrose hydrolysis with the sugar signaling that promotes cell division in early embryogenesis (reviewed by [50, 53]).
Many transcripts coding for putative carbohydrate transporters also peaked at the first embryo stage (Day0), most of them hexose (or monosaccharide) transporters involved in transport and intake into the cytoplasm, possibly after CWIN has cleaved the sucrose present in the apoplast [51]. Although still very little is known about the role played by these hexose transporters specifically in early plant embryogenesis (reviewed in [54]), previous work on other sink tissues that, like the developing embryo, are also symplastically isolated, had shown their dependence on transport proteins to uptake the sucrose-derivatives through the plasma membrane (reviewed by [55]). A putative pine homolog (Pp6019) of the Arabidopsis SUGAR TRANSPORT PROTEIN 14 (AtSTP14) belonging to the AtSTPs family, but specific for galactose transport and expressed both in source (green leaves) and in sink tissues (seed endosperm and cotyledons) [56], was also detected. There are several evidences pointing to a role of AtSTP14 in cell wall recycling, namely in the transport of the cell wall-derived galactose released upon cell wall degradation performed by β-galactosidases (glycosyl hydrolases) and other enzymes [56]. Yet another putative pine homolog (Pp32670) of the Arabidopsis AtSTP13 was found in cluster 2, likely involved in the transport of monosaccharides derived from the cell wall (reviewed in [54]).
The GO enrichment results from clusters 2 and 3, which gather several glycosyl hydrolases, carbohydrate transporters and kinases, as well as cell cycle related transcripts, seem to support an active communication between regulators of carbohydrate metabolism and cell cycle in P. pinaster early embryogenesis. So far, the impact on plant cell cycle control and cell division derived from the interaction between the nutritional state and genetic control has been elucidated only in post-embryonic development studies [57]. Observations in Arabidopsis show that carbohydrates availability plays a role on cell decision over G2/M transition by sugar signaling interaction with specific key cell cycle regulators, such as CYCB1;1 and CDKB1;1, which directly impacts proliferation of meristematic tissues [58]. Among the five cyclin dependent kinase b transcripts found among clusters 2 and 3 there is Pp42651, a putative pine homolog of the Arabidopsis CYCLIN-DEPENDENT KINASE B2;2 (CDKB2;2) which has been shown to be a regulator of cell cycle progression and SAM organization, and involved in hormone signaling [59].
Epigenetics associated transcripts in early to middle embryogenesis
Many transcripts associated with different components of epigenetic regulation are found differentially expressed during maritime pine embryogenesis. Most of them are included in cluster 3 profile, characterized by a peak of expression at early embryogenesis.
Five putative histone subunits homologs are differentially expressed across pine embryo development and follow different expression profiles. Pp36206 and Pp38724, putatively encoding core H3 and H2A histone subunits, respectively, show a higher abundance in early embryogenesis and generally decreasing towards late embryogenesis (with a second minor peak at Day11). Also in Picea abies somatic embryogenesis a Histone 3 was found differentially expressed and down-regulated in the late embryogeny phase [60]. Histone H3 is a known phosphorylation target, in a cell cycle-dependent manner, of all three Arabidopsis Aurora kinases [61, 62]. Interestingly, a pine homolog of Aurora-2 (Pp32543) is present in the same cluster. Additionally, the H1 (linker) histones, putatively encoded by Pp46360, Pp14332 and Pp46359 pine transcripts following an overall profile presenting a peak of expression at D11/D15 stages (cluster 6 or 10), have been reported as involved in DNA methylation and demethylation, cell-cycle progression, and plant development (reviewed by [63]). A microarray analysis of somatic embryogenesis material from Picea abies revealed up-regulation of Histone H1 in proembryogenic masses (PEMs) one day after withdrawal of plant growth regulators, when PEM-to-embryo transition is induced, and in the transition from proliferation to differentiation of early embryos [38].
Genes associated with chromatin formation or remodelling also appeared differentially expressed across pine embryo development. A Serrated and early flowering/ SWR1 complex subunit 6 (SEF/SWC6) pine homolog (Pp30270) increased its expression throughout embryo development (cluster 7). SEF encodes a subunit of SWR1 chromatin-remodelling complex, which is responsible for the ATP-dependent replacement of histone H2A by H2A.Z variant, and is associated with flowering repression in Arabidopsis by means of positive regulation of the flowering repressors FLOWERING LOCUS C (FLC) and MADS-AFFECTING FLOWERING 4 (MAF4) [64–66]. Two other SNF2-related chromatin remodelling putative transcripts follow different expression profiles. Pp14163, the putative pine homolog of CHROMATIN REMODELING 31 (CHR31) peaked at Day5 and Day15, while the homolog of RAD5 (Pp26994), was over-represented at early embryogenesis, pointing to a developmental stage dependent expression of these genes. Also peaking in early embryos was Pp34781, a putative FASCIATA 1 (FAS1) pine homolog encoding one of the three subunits of the histone chaperone Chromatin Assembly Factor-1 (CAF-1). Its Arabidopsis counterpart is required during post-embryonic development, for proper organization and function of both apical meristems, however it appears not to be needed during embryo development, at least during Arabidopsis late embryogenesis [67].
The transcriptome of P. pinaster embryogenesis is abundant in transcripts possibly encoding enzymes for post-translational modification of histone subunits, in particular E2 ubiquitin-conjugating enzymes. Pp30887, a putative pine homolog of E2 ubiquitin-conjugating enzyme 28 gene, is differentially expressed and up-regulated in early and middle embryogenesis (cluster 1). These proteins are known to perform H2B monoubiquitination that has been associated with transcriptional activation (reviewed in [68]). Histone lysine methylation also plays a relevant role in pine embryogenesis considering the identification of the differentially expressed transcripts Pp44003 and Pp33894, putatively coding for an ARABIDOPSIS TRITHORAX-RELATED PROTEIN 6 (ATXR6) (in cluster 3) and a SU(VAR)3–9-RELATED protein 5 (SUVR5) (in cluster 10), respectively. ATXR5 and ATXR6, involved in the repressive chromatin modification H3K27me1, contribute to keep the constitutive heterochromatin status, in most cases of transposons and other repetitive and silent elements, and to prevent re-replication to occur in the same cell cycle [69, 70]. On the other hand, SUVR5 is responsible for the repressive chromatin modification H3K9me2, independently of the presence of DNA methylation [71], whose presence is usually associated to transposon silencing and DNA methylation control, being predominant in pericentromeric/centromeric regions [72].
A putative homolog of VARIANT IN METHYLATION 1/ORTHRUS 2 (VIM1/ORTH2) gene, a methylcytosine-binding protein that collaborates with DNA METHYLTRANSFERASE 1 (MET1) to promote CpG methylation and centromeres organization [73, 74], was also found in our data (Pp11214) showing a maximum expression in the earlier embryo stages (cluster 3) and decreasing towards the mature embryo. In Picea abies, VIM1 was found differentially expressed between somatic embryos in the early morphogenesis stage grown under two distinct temperature conditions (18 °C and 30 °C), being up-regulated at higher temperature [23]. However, Pp3794, which is an homolog of the plant DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2), a major de novo DNA methyltransferase gene responsible for DNA methylation in all sequence contexts (CG, CHG and CHH) (reviewed by [75]), followed an expression profile peaking at Day5, but increasing from middle embryogenesis towards maturation (cluster 5). DRM2 maintains CHH methylation through de novo methylation, typically within the RNA-directed DNA methylation (RdDM) pathway (reviewed in [76]). Although the expression of the pine putative DRM2 peaks before reaching maturation, a tendency for a steady increase from the middle embryo stages up to the mature embryo was detected. This observation is in agreement with studies in Arabidopsis showing that mature embryos exhibit saturation of the CHH methylation sites, and a higher activity of RdDM and expression of DRM2 when comparing with early embryos [77, 78].
Finally, many transcripts associated with RNA silencing have been found in the pine embryo transcriptome. Within this group, it is worth highlighting Pp12441, a putative pine homolog of the RNA silencing player ARGONAUTE 2 which has been associated with biotic stress response [79], up-regulated during early and middle embryogenesis (cluster 1).
Conclusions
This work provides an additional resource to help understand the gene regulation and major events associated with embryogenesis progression in conifers. By using RNA-seq technology to access the genes being expressed at specific embryo developmental stages, we have extended the previously published transcriptome profiling of maritime pine zygotic embryogenesis which had been obtained with DNA microarray hybridization technology. One such important outcome is that carbohydrate transport and metabolism was found clearly over-represented in early embryo stages. Either the analysis of functional categories assigned to eggNOGs, or the enrichment analysis of the differentially expressed transcripts and identification of many putative glycoside hydrolases and carbohydrate transport genes, point towards their relevant role in pine embryo development. Another relevant outcome providing strong support to previous studies is the predominance, during early and middle embryogenesis, of several events of chromatin remodelling evidenced by an active synthesis of histones and their post-translational modifiers associated to increased transcription, as well as silencing of transposons.
While there is no genome published for P. pinaster, this transcriptome of reference for pine zygotic embryogenesis is useful to the plant research community focused on the improvement of the vegetative propagation of conifers through somatic embryogenesis.
Methods
Plant material
Immature female cones were collected from open-pollinated P. pinaster Ait. trees growing in a clonal seed orchard at Mata Nacional do Escaroupim, Portugal (Longitude 8°44’W, latitude 39°4’N). This seed orchard was established by top grafting of clones genetically selected in a half-sib progeny test. The plus trees were originally selected in Mata Nacional de Leiria (Portugal) in 1963/64 [80]. The trees are part of an experimental plantation established for research on land of the Portuguese government. The cones were obtained from INIAV (Oeiras, Portugal), Ministério da Agricultura, Florestas e Desenvolvimento Rural, and were provided upon permission by the forest engineers Alexandre Aguiar and Isabel Carrasquinho, complying with institutional and national guidelines.
The collection period occurred between mid June and end of July. Seeds were removed and used to isolate embryos as previously described in [36]. Each embryo was quickly evaluated for developmental stage following the staging system described by [30], the suspensor was removed, and the embryo immediately frozen in liquid nitrogen into different pools according to the stage. Five different embryo developmental stages were considered as follows: Day0 included the early embryo stages T0, T1 and T2; Day5 included the pre-cotyledonary embryo stages T3 and T4; Day11 included the early cotyledonary embryo stage T4B; Day15 included the cotyledonary embryo stage T5; and Day25 included the mature embryo stage T7. Depending on the embryo stages, each pool contained 20–65 zygotic embryos. Several separate pools were prepared for each stage and samples were stored at − 80 °C until further analysis.
RNA extraction and sequencing
RNA extraction from each embryo pool was performed with RNeasy Plant Mini kit (Qiagen, Valencia CA, USA), using buffer RLC, according to the manufacturer’s instructions. RNA yield and purity were determined using ND-1000 spectrophotometer (NanoDrop, Wilmington DE, USA), and integrity was checked by electrophoresis in 0.8% agarose gel and staining with RedSafe™ Nucleic Acid Staining Solution (iNtRON Biotechnology). RNA samples were cleaned from DNA contamination using RNase-Free DNase I (Qiagen), according to manufacturer’s instructions. Total RNA samples from the five embryo stage pools (one biological replicate), were sent to the sequencing service provider where Illumina RNA-seq libraries were prepared and sequenced using the HiSeq 2000 platform.
RNA-seq data pre-processing and comprehensive assembly
A comprehensive transcriptome assembly approach [31] reviewed in [32] was adopted to generate the reference transcriptome (Software was run with default parameters unless otherwise indicated): The raw reads were filtered with Trimmomatic (v 0.32; [81]) using the default options for paired-end (PE) reads to remove Illumina adaptor sequences, reads with low quality or complexity (SLIDINGWINDOW:4:5), 5 bp from both ends (LEADING:5 TRAILING:5) and reads shorter than 25 bp (MINLEN:25). All the clean reads were used for de novo assembly with Trinity, but only pairs where both reads remained were used for the guided assembly or expression analysis. Trinity (v 2.0.6; [33]) was used to generate a de novo assembly with default parameters, plus “--min_glue 4 --CuffFly --group_pairs_distance 600 --genome_guided_max_intron 10000”. The P. taeda genome and annotations (v 1.01) were downloaded from the genome project at the University of California [34]. Clean pairs of reads from each embryo stage were independently aligned to this P. taeda genome using GSNAP without gene annotation (v 2014-08-04; [82]). We only used “concordant paired” alignments where both reads in a pair align with a minimum length of 25 bp (50 bp per alignment), in the right forward-reverse orientation and insert length distance. The read alignments from all the stages, together with the P. taeda gene annotation, were used as input in Cufflinks (v. 2.2.1; [83]) to reconstruct another set of transcripts. The final step involves combining both set of transcripts, from Trinity and Cufflinks: These transcripts were aligned to the P. taeda genome with GMAP (v 2014-12-22, [84]), and the aligning transcripts were clustered by locus with PASA (release 20,140,417; [85]). However, the transcripts that did not align on the P. taeda genome were clustered by sequence using CD-HIT with “-pid 1” (v 4.6.3; [86]), later validated by checking the presence of a clear ORF within each of them with TransDecoder (v 2.0.1; [87]) using all the Viridiplantae proteins in UniProt as reference, and finally concatenated to the previous transcripts (those clustered by locus). To produce the final comprehensive transcriptome, we filtered out the 602 short transcripts without an ORF and shorter than 200 bp. These 602 transcripts are listed in Additional file 2.
Functional annotation
Transcripts were compared with the NCBI non-redundant (nr) and Arabidopsis TAIR protein databases using NCBI BLASTX with an E-value of 1e-10. Results were imported in Blast2GO [88] to annotate the GO terms, enzymatic protein codes and KEGG pathways. The conserved motifs and structures in the transcripts were identified by comparison against the motifs databases in EBI InterPro (http://www.ebi.ac.uk/interpro/interproscan.html). We used the Plant Transcription Factor database (PLNTFDB, http://plntfdb.bio.uni-potsdam.de/) as reference to identify the TFs and other transcriptional regulators in our transcriptome. The database contains close to 30,000 protein sequences of experimentally-identified elements from diverse plant species, and their classification in families according to their protein domains by HMM methods. The sequences of the differentially expressed transcripts were aligned to the PLNTFDB using BLASTX and a minimum E-value of 1e-10. We considered any transcript with a result under that threshold as a TF/transcriptional regulator, and annotated it within the family of the homologous with a lower E-value.
All ORF possibilities were generated from the newly assembled transcriptome using TransDecoder as previously described, but only the longest ORF per transcript was retained in the final P. pinaster proteome. We classified any assembled transcript where TransDecoder could not identify an ORF as non-coding. The transcriptomes of other conifers were downloaded from the PineRefSeq project at University of Davis. The proteome for each of them was built in a similar way as for P. pinaster with TransDecoder. Best reciprocal hits (BRH) were identified by aligning all the proteins against each other with BLASTP with an E-value of 1e-5. Any annotation from the close relatives was assigned back to the original P. pinaster transcript. The proteins from P. pinaster, P. taeda and P. lambertiana were clustered together according the eggNOG group of their respective best orthologous sequenced in EBI’s eggNOG database of functionally annotated proteins. Each protein was firstly aligned to a database of proteins that have already been categorized, and then annotated with the eggNOG number of its best homolog. Clusters of proteins were made among the three conifer species by concatenating the proteins annotated with the same eggNOG number. The annotated database of protein sequences and descriptions is available to download at EMBL (http://eggnogdb.embl.de/).
Analysis of expression, gene enrichment and clustering
The clean reads from each of the developing stages were aligned to the comprehensive transcriptome with Bowtie (v 2.2.5; [89]) and retaining only pairs of reads mapping in the right distance and orientation (−-very-sensitive -X 1000 --no-mixed --no-discordant). From these alignments, the abundance of each transcript was quantified in each stage using eXpress (v. 1.5.1; [90]) to produce a table of counts that was used downstream in edgeR [91]. Transcripts with less than 10 counts were discarded at this point. Due to the technical difficulties in isolating enough amounts of zygotic embryos at the very early stages of development, we do not have replicated libraries. To minimize the impact of the lack of replicates, we followed the protocol recommended by the developers in such cases [92]: a common dispersion was calculated for the whole dataset with “x = DGEList(round(read.delim(‘eXpress.eff_counts’)),group=c(1,1,1,1,1); x = calcNormFactors(x); x = estimateDisp(x);”, and later differentially expressed transcripts were identified between each pair of consecutive stages at FDR < 0.05 using edgeR’s exact test “exactTest()”, which allows both common dispersion and single factor experiments (time in ours).
Differentially expressed transcripts were divided in clusters according to the normalized number of aligned reads in each stage by K-means clustering implemented in Mayday [93] based on Euclidian correlation between expression values. The list of transcripts in each cluster was used in Blast2GO to identify the enriched GO terms. The enrichment analysis was based on a F-fisher test (FDR < 0.05). The relation among GO terms was assigned using REVIGO with the Resvik algorithm option [94] and plotted in R with the Treemap library (github.com/mtennekes/treemap.git). To build the expression heatmap by functional categories, the counts of each transcript belonging to a protein group in such category were added up, and later transformed in Z-scores, clustered, and plotted in a heatmap using Mayday [93].
Expression validation by RT-qPCR
A subset of genes from the de novo assembled P. pinaster transcriptome was selected, based on differential expression and/or epigenetic-related annotation, to validate the RNA-seq results by RT-qPCR. Primer3Plus (http://primer3plus.com/cgi-bin/dev/primer3plus.cgi) was used to design the RT-qPCR primers, with the following conditions: 50–60% of GC content and Tm between 50 and 65 °C (according to Nearest Neighbor Tm); should bind the 5′ or 3′ less conserved regions of the transcript; the amplicon size of 75–200 bp (Additional file 18). The quality of the primers was verified with PCR Primer Stats (http://www.bioinformatics.org/sms2/pcr_primer_stats.html) and OligoAnalyzer 3.1 (https://eu.idtdna.com/calc/analyzer).
RNA samples were first quantified in Qubit 3.0 Fluorimeter using the RNA BR Assay kit (Thermo Fisher Scientific). The cDNA synthesis was performed using the Transcriptor High Fidelity cDNA Synthesis Kit (Roche Diagnostics), according to the manufacturer’s instructions in the Standard Procedure for Quantitative RT-PCR, adding 1000 ng of total RNA per 20 uL reaction mix. Three biological replicates were prepared for each embryo stage pool.
All qPCR experiments were performed in a LightCycler 480 (Roche Diagnostics) with 96-well white plates (Roche Diagnostics), where each 20 uL qPCR reaction mixture included 1X SYBR Green I Master (Roche Diagnostics), 500 nM of each primer and 2 uL of 1:20 diluted cDNA. Three technical replicates were prepared for each biological replicate. The amplification program was the same for all genes, with slight differences in the annealing temperatures: 95 °C for 10 min, 40 cycles of 10 s at 95 °C, 20 s at annealing temperature and 8 s at 72 °C (annealing temperatures in Additional file 18). To check for the presence of a single peak upon amplification, melting curves were obtained. In order to discard any possibility of remnant gDNA contamination, control samples were prepared from the same RNAs but in the absence of the Reverse Transcriptase enzyme, and used as templates in qPCR amplification with the ELF1A primers, where the absence of signal proved the effectiveness of the previous RNase-free DNase I (Qiagen) treatment. As additional controls, non-template controls (NTC) and positive controls were included in all runs. Additionally, three biological replicates of a pool sample, prepared from equal amounts of total RNA from each of the five embryo stage pools, were also included in all runs. The efficiency of each pair of primers was manually calculated according to [95], using the Cp values retrieved by the ROCHE LC480 software (Additional file 18). The relative expression of each gene of interest (GOI) was efficiency corrected as described in [95], using ELF1A, ATUB and Histo3 as reference genes, which showed up as reliable reference genes in a previous study on P. pinaster somatic embryogenesis [96].
The data obtained from the RNA-seq experiment and the RT-qPCR were compared. From the RNA-seq a logarithmic ratio of base 2 between the counts (from eXpress) of a gene in each developmental stage and the mean counts of the same gene in all developmental stages were made. A similar approach was followed for the data obtained by RT-qPCR by doing a logarithmic ratio of base 2 between the normalized quantities (delta-delta-Ct) of the gene of interest in each developmental stage and the mean normalized quantities of the same gene in all developmental stages in analysis. If expression could not be detected by RT-qPCR after 35 cycles, we assigned an expression value lower than the previous detected.
Additional files
List of all sequences of proteins in the final proteome encoded by the reference transcriptome of P. pinaster zygotic embryogenesis. Only the longest ORF possibility per transcript was translated into protein sequence to generate the final proteome. (FASTA 14164 kb)
Reference transcriptome of P. pinaster zygotic embryogenesis with annotations from BRH to the proteomes of P. taeda, P. lambertiana and A. thaliana, and from homology to proteins in NCBI databases. Table S1: List of assembled transcripts, longest ORFs, and corresponding predicted proteins. Table S2: Annotation information for the predicted proteins derived from the longest ORFs. Table S3: List of short non-coding transcripts without an ORF that were excluded. (XLSX 11164 kb)
E-value distribution of the BLAST hits resulting from the BLASTX alignment of the P. pinaster transcriptome to the NCBI non-redundant proteins database. (PNG 74 kb)
The functional annotation of P. pinaster transcriptome done with Blast2GO generated different charts. The “data distribution” chart shows the distribution of un-blasted (with BLAST (without hits)), blasted (with BLAST hits), mapped (with mapping) and annotated (with GO annotation) transcripts over the whole transcriptome. The “GO mapping distribution” chart is a representation of the amount of GO terms assigned to each sequence during the GO Mapping step. The third chart represents the number of annotations achieved at distinct GO levels (0-to-15), listing the GO terms by biological process (P), molecular function (F) and cellular component (C). The “annotation distribution” chart shows the number of sequences annotated with different amounts of GO terms. (PDF 2905 kb)
Distribution of the sequence similarities (percentage) that were calculated for the BLAST hits. (PNG 79 kb)
Distribution of species to which most transcripts were aligned when only considering the Top-BLAST hits. (PNG 103 kb)
InterProScan (IPS) results showing the number of transcripts with and without IPS as well as with GO terms retrieved by this annotation step. (PNG 45 kb)
Distribution of the number of GO terms, retrieved by the Blast2GO mapping step, per database resource. (PNG 24 kb)
List of proteins from P. pinaster, P. taeda and P. lambertiana clustered together according to the eggNOG group of their respective best orthologous sequenced in EMBL’s eggNOG database of functionally annotated proteins. (TXT 3127 kb)
Number of proteins from P. pinaster, P. taeda and P. lambertiana clustered together according to the eggNOG group of their respective best orthologous sequenced in EMBL’s eggNOG database of functionally annotated proteins. Information on each eggNOG group is complemented by a general category and its description. (XLSX 844 kb)
Transcripts abundance per developmental time point (0D_eff_counts to 25D_eff_counts). (XLSX 4407 kb)
Results from the edgeR analysis for the identification of differentially expressed transcripts between each pair of consecutive stages at FDR < 0.05. Fold-change (FC), counts per million (CPM) and p-value data are shown per developmental transition and transcript. Table S4: Complete list of transcripts analysed with edgeR. Table S5: Exclusive list of 1738 differentially expressed transcripts along embryo development, including the number of the cluster of expression profile. (XLSX 2943 kb)
REVIGO TreeMap representation of GO terms enrichment analysis associated with biological process GO terms found in the list of 204 differentially expressed transcripts down-regulated in transition from Day0 to Day5. (PNG 62 kb)
REVIGO TreeMap representation of GO terms enrichment analysis associated with biological process GO terms found in the list of 594 differentially expressed transcripts up-regulated in transition from Day0 to Day5. (PNG 50 kb)
REVIGO TreeMap representation of GO terms enrichment analysis associated with biological process GO terms found in the list of 344 differentially expressed transcripts down-regulated in transition from Day15 to Day25. (PNG 167 kb)
REVIGO TreeMap representation of GO terms enrichment analysis associated with biological process GO terms found in the list of differentially expressed transcripts included in cluster 2. (PNG 53 kb)
REVIGO TreeMap representation of GO terms enrichment analysis associated with biological process GO terms found in the list of differentially expressed transcripts included in cluster 3. (PNG 75 kb)
Transcripts with BRHs validated by relative RT-qPCR. The order of preference for annotating each P. pinaster transcript after its homologs is: A. thaliana, P. taeda, and P. lambertiana. (DOCX 15 kb)
Acknowledgments
Isabel Carrasquinho and Alexandre Aguiar from INIAV are acknowledged for provision of plant material. Marta Simões is acknowledged for participation in the preparation of plant material.
Funding
This work was supported through projects funded by (1) the European Commission Seventh Framework Programme (FP7, Grant Agreement N°289841-PROCOGEN), and (2) Fundação para a Ciência e a Tecnologia (FCT), through grants GREEN-it (UID/Multi/04551/2013), IF/01168/2013 and the doctoral fellowship SFRH/BD/79779/2011 (to ASR). JDV was supported by an Institute Programme Grant to Earlham Institute (BBS/E/T/000PR9818) from the Biotechnology and Biological Science Research Council (BBSRC), UK. The funding bodies had no role in the design of the study, collection, analysis, and interpretation of data, or in writing the manuscript.
Availability of data and materials
The data were deposited in the European Nucleotide Archive (ENA) under the study PRJEB21602, with the run accessions ERR2238559 (stages T0/T1/T2), ERR2238560 (stages T3/T4), ERR2238561 (stage T4B), ERR2021873 (stage T5) and ERR2021874 (stage T7) [https://www.ebi.ac.uk/ena/data/view/PRJEB21602]. The Transcriptome Shotgun Assembly project has been deposited at DDBJ/ENA/GenBank under the accession GGEX00000000. The version described here is the first version, GGEX01000000 [https://www.ebi.ac.uk/ena/data/view/GGEX01000000].
Abbreviations
- BRH
Best reciprocal hits
- GO
Gene ontology
- IPS
InterPro database
- NGS
Next-generation sequencing
- ORF
Open reading frame
- RNA-seq
RNA sequencing
Authors’ contributions
This study was conceived and directed by CMM. ASR did the experimental work, including preparation of total RNA for sequencing and gene expression validation by RT-qPCR. JDV performed the bioinformatics analysis. ASR; JDV and CMM participated in the analysis of results and their biological interpretation. ASR, JDV and CMM wrote the paper. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Andreia S. Rodrigues, Email: sofiasantos@itqb.unl.pt
José J. De Vega, Email: Jose.DeVega@earlham.ac.uk
Célia M. Miguel, Phone: +351 214469627, Email: cmiguel@itqb.unl.pt
References
- 1.Goldberg RB, de Paiva G, Yadegari R. Plant embryogenesis: zygote to seed. Science (80- ). 1994;266:605–614. doi:10.1126/science.266.5185.605. [DOI] [PubMed]
- 2.De Smet I, Lau S, Mayer U, Jürgens G. Embryogenesis - the humble beginnings of plant life. Plant J. 2010;61:959–970. doi: 10.1111/j.1365-313X.2010.04143.x. [DOI] [PubMed] [Google Scholar]
- 3.Smith SA, Beaulieu JM, Donoghue MJ. An uncorrelated relaxed-clock analysis suggests an earlier origin for flowering plants. Proc Natl Acad Sci U S A. 2010;107:5897–5902. doi: 10.1073/pnas.1001225107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raghavan V, Sharma KK. Zygotic Embryogenesis in Gymnosperms and Angiosperms. In: Thorpe TA, editor. In Vitro Embryogenesis in Plants. 1995. pp. 73–115. [Google Scholar]
- 5.Miguel CM, Rupps A, Raschke J, Rodrigues AS, Trontin JF. Impact of molecular studies on somatic embryogenesis development for implementation in conifer multivarietal forestry. In: Park YS, Bonga JM, Moon HK, editors. Vegetative propagation of Forest trees. Seoul: National Institute of Forest Science; 2016. pp. 373–421. [Google Scholar]
- 6.Lelu-Walter MA, Klimaszewska K, Miguel C, Aronen T, Hargreaves C, Teyssier C, et al. Somatic embryogenesis for more effective breeding and deployment of improved varieties in Pinus spp.: bottlenecks and recent advances. In: Somatic Embryogenesis - Fundamental Aspects and Applications. Loyola-Vargas VM, Ochoa-Alejo N, editors. Springer International Publishing; 2016. p. 319–365.
- 7.Bräutigam A, Gowik U. What can next generation sequencing do for you? Next generation sequencing as a valuable tool in plant research. Plant Biol (Stuttg) 2010;12:831–841. doi: 10.1111/j.1438-8677.2010.00373.x. [DOI] [PubMed] [Google Scholar]
- 8.Strickler SR, Bombarely A, Mueller LA. Designing a transcriptome next-generation sequencing project for a nonmodel plant species. Am J Bot. 2012;99:257–266. doi: 10.3732/ajb.1100292. [DOI] [PubMed] [Google Scholar]
- 9.Wickett NJ, Mirarab S, Nguyen N, Warnow T, Carpenter E, Matasci N, et al. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc Natl Acad Sci U S A. 2014;111:E4859–E4868. doi: 10.1073/pnas.1323926111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun H, Paulin L, Alatalo E, Asiegbu FO. Response of living tissues of Pinus sylvestris to the saprotrophic biocontrol fungus Phlebiopsis gigantea. Tree Physiol. 2011;31:438–451. doi: 10.1093/treephys/tpr027. [DOI] [PubMed] [Google Scholar]
- 11.Miguel A, de Vega-Bartol J, Marum L, Chaves I, Santo T, Leitão J, et al. Characterization of the cork oak transcriptome dynamics during acorn development. BMC Plant Biol. 2015;15:158. doi: 10.1186/s12870-015-0534-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narsai R, Gouil Q, Secco D, Srivastava A, Karpievitch YV, Liew LC, et al. Extensive transcriptomic and epigenomic remodelling occurs during Arabidopsis thaliana germination. Genome Biol. 2017;18:172. doi: 10.1186/s13059-017-1302-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zenoni S, Ferrarini A, Giacomelli E, Xumerle L, Fasoli M, Malerba G, et al. Characterization of transcriptional complexity during berry development in Vitis vinifera using RNA-Seq. Plant Physiol. 2010;152:1787–1795. doi: 10.1104/pp.109.149716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao M, Zhang Y, Chen X, Lee E-J, Barber CJS, Chakrabarty R, et al. Transcriptome analysis based on next-generation sequencing of non-model plants producing specialized metabolites of biotechnological interest. J Biotechnol. 2013;166:122–134. doi: 10.1016/j.jbiotec.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 15.Jayasena AS, Secco D, Bernath-Levin K, Berkowitz O, Whelan J, Mylne JS. Next generation sequencing and de novo transcriptomics to study gene evolution. Plant Methods. 2014;10:34. doi: 10.1186/1746-4811-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu H, Gao Y, Wang J. Transcriptomic analysis of rice (Oryza sativa) developing embryos using the RNA-Seq technique. PLoS One. 2012;7:e30646. doi: 10.1371/journal.pone.0030646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J, Zeng B, Zhang M, Xie S, Wang G, Hauck A, et al. Dynamic transcriptome landscape of maize embryo and endosperm development. Plant Physiol. 2014;166:252–264. doi: 10.1104/pp.114.240689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teoh KT, Requesens DV, Devaiah SP, Johnson D, Huang X, Howard JA, et al. Transcriptome analysis of embryo maturation in maize. BMC Plant Biol. 2013;13:19. doi: 10.1186/1471-2229-13-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng W, Yan F, Zhang X, Tang Y, Yuan Y. Transcriptional profiling of canola developing embryo and identification of the important roles of BnDof5.6 in embryo development and fatty acids synthesis. Plant Cell Physiol. 2015;56:1624–1640. doi: 10.1093/pcp/pcv074. [DOI] [PubMed] [Google Scholar]
- 20.Zhai L, Xu L, Wang Y, Zhu X, Feng H, Li C, et al. Transcriptional identification and characterization of differentially expressed genes associated with embryogenesis in radish (Raphanus sativus L.) Sci Rep. 2016;6:21652. doi: 10.1038/srep21652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merino I, Abrahamsson M, Sterck L, Craven-Bartle B, Canovas F, von Arnold S. Transcript profiling for early stages during embryo development in scots pine. BMC Plant Biol. 2016;16:255. doi: 10.1186/s12870-016-0939-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trontin J-F, Klimaszewska K, Morel A, Hargreaves C, Lelu-Walter M-A. Molecular aspects of conifer zygotic and somatic embryo development: a review of genome-wide approaches and recent insights. Methods Mol Biol. 2016;1359:167–207. doi: 10.1007/978-1-4939-3061-6_8. [DOI] [PubMed] [Google Scholar]
- 23.Yakovlev IA, Lee Y, Rotter B, Olsen JE, Skrøppa T, Johnsen Ø, et al. Temperature-dependent differential transcriptomes during formation of an epigenetic memory in Norway spruce embryogenesis. Tree Genet Genomes. 2014;10:355–366. doi: 10.1007/s11295-013-0691-z. [DOI] [Google Scholar]
- 24.Li Q, Zhang S, Wang J. Transcriptome analysis of callus from Picea balfouriana. BMC Genomics. 2014;15:553. doi: 10.1186/1471-2164-15-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elbl P, Lira BS, Andrade SCS, Jo L, dos Santos ALW, Coutinho LL, et al. Comparative transcriptome analysis of early somatic embryo formation and seed development in Brazilian pine, Araucaria angustifolia (Bertol.) Kuntze. Plant Cell Tissue Organ Cult. 2015;120:903–915. doi: 10.1007/s11240-014-0523-3. [DOI] [Google Scholar]
- 26.Gonzalez-Ibeas D, Martinez-Garcia PJ, Famula RA, Delfino-Mix A, Stevens KA, Loopstra CA, et al. Assessing the Gene Content of the Megagenome: Sugar Pine (Pinus lambertiana) G3 (Bethesda) 2016;6:3787–3802. doi: 10.1534/g3.116.032805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dobrowolska I, Businge E, Abreu IN, Moritz T, Egertsdotter U. Metabolome and transcriptome profiling reveal new insights into somatic embryo germination in Norway spruce (Picea abies) Tree Physiol. 2017;37:1752–1766. doi: 10.1093/treephys/tpx078. [DOI] [PubMed] [Google Scholar]
- 28.Navarro BV, Elbl P, De Souza AP, Jardim V, de Oliveira LF, Macedo AF, et al. Carbohydrate-mediated responses during zygotic and early somatic embryogenesis in the endangered conifer. Araucaria angustifolia PLoS One. 2017;12:e0180051. 10.1371/journal.pone.0180051. [DOI] [PMC free article] [PubMed]
- 29.De Vega-Bartol JJ, Simões M, Lorenz WW, Rodrigues AS, Alba R, Dean JFD, et al. Transcriptomic analysis highlights epigenetic and transcriptional regulation during zygotic embryo development of Pinus pinaster. BMC Plant Biol. 2013;13:123. doi: 10.1186/1471-2229-13-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonçalves S, Cairney J, Maroco J, Oliveira MM, Miguel C. Evaluation of control transcripts in real-time RT-PCR expression analysis during maritime pine embryogenesis. Planta. 2005;222:556–563. doi: 10.1007/s00425-005-1562-0. [DOI] [PubMed] [Google Scholar]
- 32.Visser EA, Wegrzyn JL, Steenkmap ET, Myburg AA, Naidoo S. Combined de novo and genome guided assembly and annotation of the Pinus patula juvenile shoot transcriptome. BMC Genomics. 2015;16:1057. doi: 10.1186/s12864-015-2277-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin JA, Wang Z. Next-generation transcriptome assembly. Nat Rev Genet. 2011;12:671–682. doi: 10.1038/nrg3068. [DOI] [PubMed] [Google Scholar]
- 34.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wegrzyn JL, Liechty JD, Stevens KA, Wu L-S, Loopstra CA, Vasquez-Gross HA, et al. Unique features of the loblolly pine (Pinus taeda L.) megagenome revealed through sequence annotation. Genetics. 2014;196:891–909. doi: 10.1534/genetics.113.159996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Powell S, Szklarczyk D, Trachana K, Roth A, Kuhn M, Muller J, et al. eggNOG v3.0: orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Res. 2012;40(Database issue):D284–D289. doi: 10.1093/nar/gkr1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Zyl L, von Arnold S, Bozhkov P, Chen Y, Egertsdotter U, Mackay J, et al. Heterologous array analysis in Pinaceae: hybridization of Pinus taeda cDNA arrays with cDNA from needles and embryogenic cultures of P. Taeda, P. Sylvestris or Picea abies. Comp Funct Genomics. 2002;3:306–318. doi: 10.1002/cfg.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vestman D, Larsson E, Uddenberg D, Cairney J, Clapham D, Sundberg E, et al. Important processes during differentiation and early development of somatic embryos of Norway spruce as revealed by changes in global gene expression. Tree Genet Genomes. 2011;7:347–362. doi: 10.1007/s11295-010-0336-4. [DOI] [Google Scholar]
- 39.Edwards M, Dea IC, Bulpin PV, Reid JS. Xyloglucan (amyloid) mobilisation in the cotyledons of Tropaeolum majus L. seeds following germination. Planta. 1985;163:133–140. doi: 10.1007/BF00395907. [DOI] [PubMed] [Google Scholar]
- 40.Scheller HV, Ulvskov P. Hemicelluloses. Annu Rev Plant Biol. 2010;61:263–289. doi: 10.1146/annurev-arplant-042809-112315. [DOI] [PubMed] [Google Scholar]
- 41.Park YB, Cosgrove DJ. Xyloglucan and its interactions with other components of the growing cell wall. Plant Cell Physiol. 2015;56:180–194. doi: 10.1093/pcp/pcu204. [DOI] [PubMed] [Google Scholar]
- 42.Sampedro J, Sieiro C, Revilla G, González-Villa T, Zarra I. Cloning and expression pattern of a gene encoding an alpha-xylosidase active against xyloglucan oligosaccharides from Arabidopsis. Plant Physiol. 2001;126:910–920. doi: 10.1104/pp.126.2.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sampedro J, Pardo B, Gianzo C, Guitián E, Revilla G, Zarra I. Lack of α-xylosidase activity in Arabidopsis alters xyloglucan composition and results in growth defects. Plant Physiol. 2010;154:1105–1115. doi: 10.1104/pp.110.163212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shigeyama T, Watanabe A, Tokuchi K, Toh S, Sakurai N, Shibuya N, et al. α-Xylosidase plays essential roles in xyloglucan remodelling, maintenance of cell wall integrity, and seed germination in Arabidopsis thaliana. J Exp Bot. 2016;67:5615–5629. doi: 10.1093/jxb/erw321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsui A, Yokoyama R, Seki M, Ito T, Shinozaki K, Takahashi T, et al. AtXTH27 plays an essential role in cell wall modification during the development of tracheary elements. Plant J. 2005;42:525–534. doi: 10.1111/j.1365-313X.2005.02395.x. [DOI] [PubMed] [Google Scholar]
- 46.Ogawa M. Gibberellin biosynthesis and response during Arabidopsis seed germination. THE PLANT CELL ONLINE. 2003;15:1591–1604. doi: 10.1105/tpc.011650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malinowski R, Filipecki M. The role of cell wall in plant embryogenesis. Cell Mol Biol Lett. 2002;7:1137–1151. [PubMed] [Google Scholar]
- 48.Smertenko A, Bozhkov PV. Somatic embryogenesis: life and death processes during apical-basal patterning. J Exp Bot. 2014;65:1343–1360. doi: 10.1093/jxb/eru005. [DOI] [PubMed] [Google Scholar]
- 49.Aquea F, Arce-Johnson P. Identification of genes expressed during early somatic embryogenesis in Pinus radiata. Plant Physiol Biochem. 2008;46:559–568. doi: 10.1016/j.plaphy.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 50.Ruan Y-L. Sucrose metabolism: gateway to diverse carbon use and sugar signaling. Annu Rev Plant Biol. 2014;65:33–67. doi: 10.1146/annurev-arplant-050213-040251. [DOI] [PubMed] [Google Scholar]
- 51.Wang L, Ruan Y-L. New insights into roles of cell wall invertase in early seed development revealed by comprehensive spatial and temporal expression patterns of GhCWIN1 in cotton. Plant Physiol. 2012;160:777–787. doi: 10.1104/pp.112.203893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rutledge RG, Stewart D, Caron S, Overton C, Boyle B, MacKay J, et al. Potential link between biotic defense activation and recalcitrance to induction of somatic embryogenesis in shoot primordia from adult trees of white spruce (Picea glauca) BMC Plant Biol. 2013;13:116. doi: 10.1186/1471-2229-13-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weber H, Borisjuk L, Wobus U. Molecular physiology of legume seed development. Annu Rev Plant Biol. 2005;56:253–279. doi: 10.1146/annurev.arplant.56.032604.144201. [DOI] [PubMed] [Google Scholar]
- 54.Slewinski TL. Diverse functional roles of monosaccharide transporters and their homologs in vascular plants: a physiological perspective. Mol Plant. 2011;4:641–662. doi: 10.1093/mp/ssr051. [DOI] [PubMed] [Google Scholar]
- 55.Büttner M, Sauer N. Monosaccharide transporters in plants: structure, function and physiology. Biochim Biophys Acta. 2000;1465:263–274. doi: 10.1016/S0005-2736(00)00143-7. [DOI] [PubMed] [Google Scholar]
- 56.Poschet G, Hannich B, Büttner M. Identification and characterization of AtSTP14, a novel galactose transporter from Arabidopsis. Plant Cell Physiol. 2010;51:1571–1580. doi: 10.1093/pcp/pcq100. [DOI] [PubMed] [Google Scholar]
- 57.Peng L, Skylar A, Chang PL, Bisova K, Wu X. CYCP2;1 integrates genetic and nutritional information to promote meristem cell division in Arabidopsis. Dev Biol. 2014;393:160–170. doi: 10.1016/j.ydbio.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 58.Skylar A, Sung F, Hong F, Chory J, Wu X. Metabolic sugar signal promotes Arabidopsis meristematic proliferation via G2. Dev Biol. 2011;351:82–89. doi: 10.1016/j.ydbio.2010.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Andersen SU, Buechel S, Zhao Z, Ljung K, Novák O, Busch W, et al. Requirement of B2-type cyclin-dependent kinases for meristem integrity in Arabidopsis thaliana. Plant Cell. 2008;20:88–100. doi: 10.1105/tpc.107.054676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stasolla C, Bozhkov PV, Chu TM, van Zyl L, Egertsdotter U, Suarez MF, et al. Variation in transcript abundance during somatic embryogenesis in gymnosperms. Tree Physiol. 2004;24:1073–1085. doi: 10.1093/treephys/24.10.1073. [DOI] [PubMed] [Google Scholar]
- 61.Demidov D, Van Damme D, Geelen D, Blattner FR, Houben A. Identification and dynamics of two classes of aurora-like kinases in Arabidopsis and other plants. Plant Cell. 2005;17:836–848. doi: 10.1105/tpc.104.029710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kawabe A, Matsunaga S, Nakagawa K, Kurihara D, Yoneda A, Hasezawa S, et al. Characterization of plant Aurora kinases during mitosis. Plant Mol Biol. 2005;58:1–13. doi: 10.1007/s11103-005-3454-x. [DOI] [PubMed] [Google Scholar]
- 63.Over RS, Michaels SD. Open and closed: the roles of linker histones in plants and animals. Mol Plant. 2014;7:481–491. doi: 10.1093/mp/sst164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.March-Díaz R, García-Domínguez M, Florencio FJ, Reyes JC. SEF, a new protein required for flowering repression in Arabidopsis, interacts with PIE1 and ARP6. Plant Physiol. 2007;143:893–901. doi: 10.1104/pp.106.092270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Choi K, Park C, Lee J, Oh M, Noh B, Lee I. Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development. 2007;134:1931–1941. doi: 10.1242/dev.001891. [DOI] [PubMed] [Google Scholar]
- 66.Lázaro A, Gómez-Zambrano A, López-González L, Piñeiro M, Jarillo JA. Mutations in the Arabidopsis SWC6 gene, encoding a component of the SWR1 chromatin remodelling complex, accelerate flowering time and alter leaf and flower development. J Exp Bot. 2008;59:653–666. doi: 10.1093/jxb/erm332. [DOI] [PubMed] [Google Scholar]
- 67.Kaya H, Shibahara KI, Taoka KI, Iwabuchi M, Stillman B, Araki T. FASCIATA genes for chromatin assembly factor-1 in arabidopsis maintain the cellular organization of apical meristems. Cell. 2001;104:131–142. doi: 10.1016/S0092-8674(01)00197-0. [DOI] [PubMed] [Google Scholar]
- 68.Berr A, Shafiq S, Shen W-H. Histone modifications in transcriptional activation during plant development. Biochim Biophys Acta. 1809;2011:567–576. doi: 10.1016/j.bbagrm.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 69.Jacob Y, Feng S, LeBlanc CA, Bernatavichute YV, Stroud H, Cokus S, et al. ATXR5 and ATXR6 are H3K27 monomethyltransferases required for chromatin structure and gene silencing. Nat Struct Mol Biol. 2009;16:763–768. doi: 10.1038/nsmb.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jacob Y, Stroud H, Leblanc C, Feng S, Zhuo L, Caro E, et al. Regulation of heterochromatic DNA replication by histone H3 lysine 27 methyltransferases. Nature. 2010;466:987–991. doi: 10.1038/nature09290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Caro E, Stroud H, Greenberg MVC, Bernatavichute YV, Feng S, Groth M, et al. The SET-domain protein SUVR5 mediates H3K9me2 deposition and silencing at stimulus response genes in a DNA methylation-independent manner. PLoS Genet. 2012;8:e1002995. doi: 10.1371/journal.pgen.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bernatavichute YV, Zhang X, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS One. 2008;3:e3156. doi: 10.1371/journal.pone.0003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Woo HR, Pontes O, Pikaard CS, Richards EJ. VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization. Genes Dev. 2007;21:267–277. doi: 10.1101/gad.1512007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Woo HR, Dittmer TA, Richards EJ. Three SRA-domain methylcytosine-binding proteins cooperate to maintain global CpG methylation and epigenetic silencing in Arabidopsis. PLoS Genet. 2008;4:e1000156. doi: 10.1371/journal.pgen.1000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pikaard CS, Mittelsten SO. Epigenetic regulation in plants. Cold Spring Harb Perspect Biol. 2014;6:a019315. doi: 10.1101/cshperspect.a019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bouyer D, Kramdi A, Kassam M, Heese M, Schnittger A, Roudier F, et al. DNA methylation dynamics during early plant life. Genome Biol. 2017;18:179. doi: 10.1186/s13059-017-1313-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kawakatsu T, Nery JR, Castanon R, Ecker JR. Dynamic DNA methylation reconfiguration during seed development and germination. Genome Biol. 2017;18:171. doi: 10.1186/s13059-017-1251-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fátyol K, Ludman M, Burgyán J. Functional dissection of a plant Argonaute. Nucleic Acids Res. 2016;44:1384–1397. doi: 10.1093/nar/gkv1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Aguiar A, Almeida MH. Borralho N. Silva Lusitana: Genetic Control of Growth, Wood Density and Stem Characteristics of Pinus pinaster in Portugal; 2003. [Google Scholar]
- 81.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu TD, Watanabe CK. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21:1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- 85.Haas BJ, Delcher AL, Mount SM, Wortman JR, Smith RK, Hannick LI, et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003;31:5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 87.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, et al. De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 89.Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Roberts A, Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat Methods. 2013;10:71–73. doi: 10.1038/nmeth.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen Y, McCarthy D, Ritchie M, Robinson M, Smyth GK. edgeR: differential expression analysis of digital gene expression data User’s Guide. 2008. [DOI] [PMC free article] [PubMed]
- 93.Battke F, Symons S, Nieselt K. Mayday--integrative analytics for expression data. BMC Bioinformatics. 2010;11:121. doi: 10.1186/1471-2105-11-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pfaffl MW. Relative quantification. In: Real-time PCR. Dorak MT, editor. Garland Science; 2007. p. 63–82.
- 96.De Vega-Bartol JJ, Santos RR, Simões M, Miguel CM. Normalizing gene expression by quantitative PCR during somatic embryogenesis in two representative conifer species: Pinus pinaster and Picea abies. Plant Cell Rep. 2013;32:715–729. doi: 10.1007/s00299-013-1407-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of all sequences of proteins in the final proteome encoded by the reference transcriptome of P. pinaster zygotic embryogenesis. Only the longest ORF possibility per transcript was translated into protein sequence to generate the final proteome. (FASTA 14164 kb)
Reference transcriptome of P. pinaster zygotic embryogenesis with annotations from BRH to the proteomes of P. taeda, P. lambertiana and A. thaliana, and from homology to proteins in NCBI databases. Table S1: List of assembled transcripts, longest ORFs, and corresponding predicted proteins. Table S2: Annotation information for the predicted proteins derived from the longest ORFs. Table S3: List of short non-coding transcripts without an ORF that were excluded. (XLSX 11164 kb)
E-value distribution of the BLAST hits resulting from the BLASTX alignment of the P. pinaster transcriptome to the NCBI non-redundant proteins database. (PNG 74 kb)
The functional annotation of P. pinaster transcriptome done with Blast2GO generated different charts. The “data distribution” chart shows the distribution of un-blasted (with BLAST (without hits)), blasted (with BLAST hits), mapped (with mapping) and annotated (with GO annotation) transcripts over the whole transcriptome. The “GO mapping distribution” chart is a representation of the amount of GO terms assigned to each sequence during the GO Mapping step. The third chart represents the number of annotations achieved at distinct GO levels (0-to-15), listing the GO terms by biological process (P), molecular function (F) and cellular component (C). The “annotation distribution” chart shows the number of sequences annotated with different amounts of GO terms. (PDF 2905 kb)
Distribution of the sequence similarities (percentage) that were calculated for the BLAST hits. (PNG 79 kb)
Distribution of species to which most transcripts were aligned when only considering the Top-BLAST hits. (PNG 103 kb)
InterProScan (IPS) results showing the number of transcripts with and without IPS as well as with GO terms retrieved by this annotation step. (PNG 45 kb)
Distribution of the number of GO terms, retrieved by the Blast2GO mapping step, per database resource. (PNG 24 kb)
List of proteins from P. pinaster, P. taeda and P. lambertiana clustered together according to the eggNOG group of their respective best orthologous sequenced in EMBL’s eggNOG database of functionally annotated proteins. (TXT 3127 kb)
Number of proteins from P. pinaster, P. taeda and P. lambertiana clustered together according to the eggNOG group of their respective best orthologous sequenced in EMBL’s eggNOG database of functionally annotated proteins. Information on each eggNOG group is complemented by a general category and its description. (XLSX 844 kb)
Transcripts abundance per developmental time point (0D_eff_counts to 25D_eff_counts). (XLSX 4407 kb)
Results from the edgeR analysis for the identification of differentially expressed transcripts between each pair of consecutive stages at FDR < 0.05. Fold-change (FC), counts per million (CPM) and p-value data are shown per developmental transition and transcript. Table S4: Complete list of transcripts analysed with edgeR. Table S5: Exclusive list of 1738 differentially expressed transcripts along embryo development, including the number of the cluster of expression profile. (XLSX 2943 kb)
REVIGO TreeMap representation of GO terms enrichment analysis associated with biological process GO terms found in the list of 204 differentially expressed transcripts down-regulated in transition from Day0 to Day5. (PNG 62 kb)
REVIGO TreeMap representation of GO terms enrichment analysis associated with biological process GO terms found in the list of 594 differentially expressed transcripts up-regulated in transition from Day0 to Day5. (PNG 50 kb)
REVIGO TreeMap representation of GO terms enrichment analysis associated with biological process GO terms found in the list of 344 differentially expressed transcripts down-regulated in transition from Day15 to Day25. (PNG 167 kb)
REVIGO TreeMap representation of GO terms enrichment analysis associated with biological process GO terms found in the list of differentially expressed transcripts included in cluster 2. (PNG 53 kb)
REVIGO TreeMap representation of GO terms enrichment analysis associated with biological process GO terms found in the list of differentially expressed transcripts included in cluster 3. (PNG 75 kb)
Transcripts with BRHs validated by relative RT-qPCR. The order of preference for annotating each P. pinaster transcript after its homologs is: A. thaliana, P. taeda, and P. lambertiana. (DOCX 15 kb)
Data Availability Statement
The data were deposited in the European Nucleotide Archive (ENA) under the study PRJEB21602, with the run accessions ERR2238559 (stages T0/T1/T2), ERR2238560 (stages T3/T4), ERR2238561 (stage T4B), ERR2021873 (stage T5) and ERR2021874 (stage T7) [https://www.ebi.ac.uk/ena/data/view/PRJEB21602]. The Transcriptome Shotgun Assembly project has been deposited at DDBJ/ENA/GenBank under the accession GGEX00000000. The version described here is the first version, GGEX01000000 [https://www.ebi.ac.uk/ena/data/view/GGEX01000000].