Abstract
Globalized infectious diseases are causing species declines worldwide, but their source often remains elusive. We used whole-genome sequencing to solve the spatiotemporal origins of the most devastating panzootic to date, caused by the fungus Batrachochytrium dendrobatidis, a proximate driver of global amphibian declines. We traced the source of B. dendrobatidis to the Korean peninsula, where one lineage, BdASIA-1, exhibits the genetic hallmarks of an ancestral population that seeded the panzootic. We date the emergence of this pathogen to the early 20th century, coinciding with the global expansion of commercial trade in amphibians, and we show that intercontinental transmission is ongoing. Our findings point to East Asia as a geographic hotspot for B. dendrobatidis biodiversity and the original source of these lineages that now parasitize amphibians worldwide.
The discovery of the amphibian-killing fungus B. dendrobatidis (1, 2) was a turning point in understanding why amphibian species worldwide are in steep decline. Although amphibian declines and extinctions had been recorded by herpetologists as early as the 1970s, they were only recognized in 1990 as a global phenomenon that could not be explained by environmental changes and anthropogenic factors alone (3). The emergence of B. dendrobatidis and the disease that it causes, amphibian chytridiomycosis, as a causative agent of declines has been documented across six different regions: Australia (~1970s and 1990s) (4), Central America (~1970s) (5), South America (~1970s and 1980s) (6, 7), the Caribbean islands (~2000s) (8), the North American Sierra Nevada (~1980s and 1990s) (9), and the Iberian Peninsula (~1990s) (10). The panzootic has been attributed to the emergence of a single B. dendrobatidis lineage, known as BdGPL (Global Panzootic Lineage) (11). However, 20 years after identification of the disease, the timing of its worldwide expansion remains unknown and previous estimates for time to most recent common ancestor (TMRCA) for BdGPL span two orders of magnitude, from 100 years before the present (11) to 26,000 years before the present (12). The geographic origin of the pathogen is similarly contested, with the source of the disease variously suggested to be Africa (13), North America (14), South America (15), Japan (16), and East Asia (17).
Global diversity of B. dendrobatidis
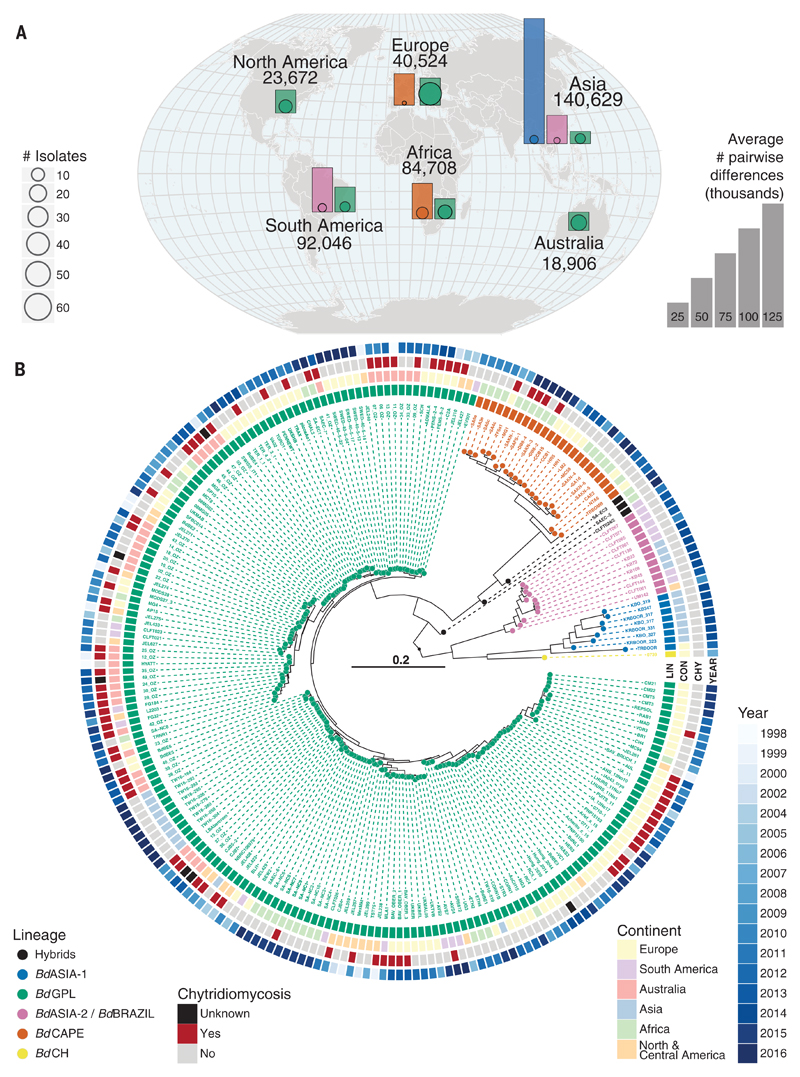
To resolve these inconsistencies, we isolated B. dendrobatidis from all the candidate source continents and sequenced the genomes of 177 isolates to high depth, then combined our data with published genomes from three prior studies (11, 12, 18) to generate a globally representative panel of 234 isolates (Fig. 1A and fig. S1). This data set covers all continents from which B. dendrobatidis has been detected to date, and spans infections of all three extant orders of Amphibia (fig. S1 and table S1). Mapped against the B. dendrobatidis reference genome JEL423, our sequencing recovered 586,005 segregating single-nucleotide polymorphisms (SNPs). Phylogenetic analysis recovered all previously detected divergent lineages (Fig. 1B and fig. S2). The previously accepted lineages BdGPL (global), BdCAPE (African), BdCH (European), and BdBRAZIL (Brazilian) were all detected (19), but our discovery of a new hyperdiverse lineage in amphibians native to the Korean peninsula (BdASIA-1) redefined these lineages and their relationships. The BdCH lineage, which was previously thought to be enzootic to Switzerland (11), now groups with the BdASIA-1 lineage. A second Asian-associated lineage (BdASIA-2) was recovered from invasive North American bullfrogs in Korea and is closely related to the lineage that is enzootic to the Brazilian Atlantic forest (BdBRAZIL) (20). It was not possible to infer the direction of intercontinental spread between isolates within this lineage, so it was named BdASIA-2/BdBRAZIL. Conditional on the midpoint rooting of the phylogeny in Fig. 1B, we now define the main diverged lineages as BdGPL, BdCAPE, BdASIA-1 (which includes the single BdCH isolate), and BdASIA-2/BdBRAZIL. Previous phylogenetic relationships developed using the widely used ribosomal intragenic spacer ITS-1 region do not accurately distinguish B. dendrobatidis lineages (fig. S3), and this likely explains much of the place-of-origin conflict in the literature (15–17).
Fig. 1. Genetic diversity and phylogenetic tree of a global panel of 234 B. dendrobatidis isolates.
(A) Map overlaid with bar charts showing the relative diversity of isolates found in each continent and by each major lineage (excluding isolates from traded animals). The bar heights represent the average numbers of segregating sites between all pairwise combinations of isolates of each lineage in each continent (therefore, only lineages with two or more isolates from a continent are shown). Outlined points at the base of each bar are scaled by the number of isolates for each lineage in that continent. The numbers around the outside of the globe are the average number of segregating sites between all pairwise combinations of isolates grouped by continent. Colors denote lineage as shown in (B). (B) Midpoint rooted radial phylogeny supports four deeply diverged lineages of B. dendrobatidis: BdASIA-1, BdASIA-2/BdBRAZIL, BdCAPE, and BdGPL. All major splits within the phylogeny are supported by 100% of 500 bootstrap replicates. See fig. S2 for tree with full bootstrap support values on all internal branches.
Pairwise comparisons among isolates within each lineage show that the average number of segregating sites is greater for BdASIA-1 than for any other lineage by a factor of 3 (Fig. 1A and Table 1) and that nucleotide diversity (π; fig. S4) is greater by a factor of 2 to 4. Seven of our eight BdASIA-1 isolates were recently cultured from wild South Korean frogs, and the other came from the pet trade in Belgium; all eight were aclinical infections. These isolates show that the Korean peninsula is a global center of B. dendrobatidis diversity and that East Asia may contain the ancestral population of B. dendrobatidis, as suggested by Bataille et al. (17).
Table 1. Comparison of common genetic diversity measures among B. dendrobatidis lineages.
Total segregating sites for each lineage include all segregating sites where genotype calls were made in at least half of the isolates. Average pairwise-segregating sites are the average numbers of sites with different genotypes between all pairs of isolates within a lineage. Total homozygous segregating sites include all sites within a lineage where there is at least one homozygous difference between isolates. Average pairwise-homozygous segregating sites are the average numbers of sites with different homozygous genotypes between all pairs of isolates within a lineage. Nucleotide diversity (π) is the mean of the per-site nucleotide diversity. Tajima’s D is reported as the mean over 1-kbp bins.
| Lineage | Number of isolates | Total segregating sites | Average pairwise-segregating sites | Total homozygous segregating sites | Average pairwise-homozygous segregating sites | π | Tajima’s D |
|---|---|---|---|---|---|---|---|
| BdASIA-1 | 8 | 327,996 | 142,437 | 108,353 | 21,716 | 0.0044 | 0.2540 |
| BdASIA-2/BdBRAZIL | 12 | 148,021 | 51,069 | 48,722 | 6,216 | 0.0018 | 0.9825 |
| BdCAPE | 24 | 146,466 | 38,881 | 53,884 | 4,977 | 0.0016 | 0.3143 |
| BdGPL | 187 | 127,770 | 26,546 | 68,493 | 3,101 | 0.0009 | 0.9792 |
We investigated this hypothesis further using Bayesian-based haplotype clustering (21) and found the greatest haplotype sharing among isolates within BdASIA-1 and between BdASIA-1 and all other lineages (fig. S5). This provides direct genetic evidence that BdASIA-1 shares more diversity with the global population of B. dendrobatidis than any other lineage. In an independent test of ancestry, we used OrthoMCL (22) to root a B. dendrobatidis phylogeny to its closest known relative, B. salamandrivorans, which currently threatens salamanders (23). This tree indicates that the Asian and Brazilian isolates of B. dendrobatidis lie outside a clade comprising all other isolates (fig. S6 and table S2). To identify the signature of demographic histories across lineages, we used Tajima’s D (24). Genome scans of most lineages showed highly variable positive and negative values of D with maximum amplitude exhibited by BdGPL (–2.6 to +6.2; Fig. 2F), indicating that these lineages (BdASIA-2/BdBRAZIL, BdCAPE, and BdGPL) have undergone episodes of population fluctuation and/or strong natural selection that are consistent with a history of spatial and host radiations. In striking contrast, BdASIA-1 shows a flat profile for Tajima’s D (Fig. 2F) indicating mutation-drift equilibrium likely reflective of pathogen endemism in this region.
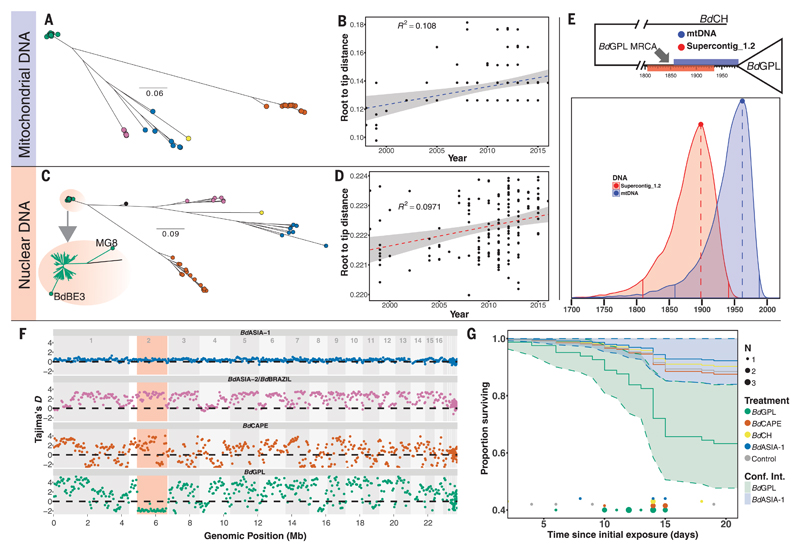
Fig. 2. Dating the emergence of BdGPL.
(A) Maximum likelihood (ML) tree constructed from 1150 high-quality SNPs found within the 178-kbp mitochondrial genome. (B) Linear regression of root-to-tip distance against year of isolation for BdGPL isolates in mitochondrial DNA phylogeny in (A), showing a significant temporal trend (F = 14.35, P = 0.00024). (C) ML tree constructed from a 1.66-Mbp region of low recombination in Supercontig_1.2. Two BdGPL isolates, BdBE3 and MG8, fall on long branches away from the rest of the BdGPL isolates (see inset zoom) as a result of introgression from another lineage (BdCAPE; see Fig. 3B) and were excluded from the dating analysis. (D) Linear regression of root-to-tip distance against year of isolation for BdGPL isolates from phylogeny in (C), with a significant temporal trend (F = 15.92, P = 0.0001). (E) Top: BdGPL and outgroup BdCH, with the 95% HPD estimates for MRCA for BdGPL from mtDNA dating (blue) and nuclear DNA dating (red). Bottom: Full posterior distributions from tip-dating models for mtDNA (blue) and partial nuclear DNA (red) genomes. Solid vertical lines are limits of the 95% HPD. Dashed vertical lines denote the maximal density of the posterior distributions. (F) Sliding 10-kb, nonoverlapping window estimates of Tajima’s D for each of the main B. dendrobatidis lineages. The region highlighted in red is the low-recombination segment of Supercontig_1.2. (G) Survival curves for Bufo bufo metamorphs for different B. dendrobatidis treatment groups: BdASIA-1 (blue), BdCAPE (orange), BdCH (yellow), BdGPL (green), and control (gray). Confidence intervals are shown for BdGPL and BdASIA-1, showing no overlap by the end of the experiment. Instances of mortalities in each treatment group are plotted along the x axis, with points scaled by number of mortalities at each interval (day).
Dating the emergence of BdGPL
The broad range of previous estimates for the TMRCA of BdGPL spanning 26,000 years (11, 12) can be explained by two sources of inaccuracy: (i) unaccounted recombination and (ii) the application of unrealistic evolutionary rates. To address these, we first interrogated the 178,280-kbp mitochondrial genome (mtDNA), which has high copy number and low rates of recombination relative to the nuclear genome. To resolve the structure of the mtDNA genome, we resorted to long-read sequencing using a MinION device (Oxford Nanopore Technologies, Cambridge, UK), which allowed us to describe this molecule’s unusual configuration; B. dendrobatidis carries three linear mitochondrial segments, each having inverted repeats at the termini with conserved mitochondrial genes spread over two of the segments (fig. S7). Additionally, we sought regions of the autosomal genome with low rates of recombination to obtain an independent estimate of the TMRCA of BdGPL.
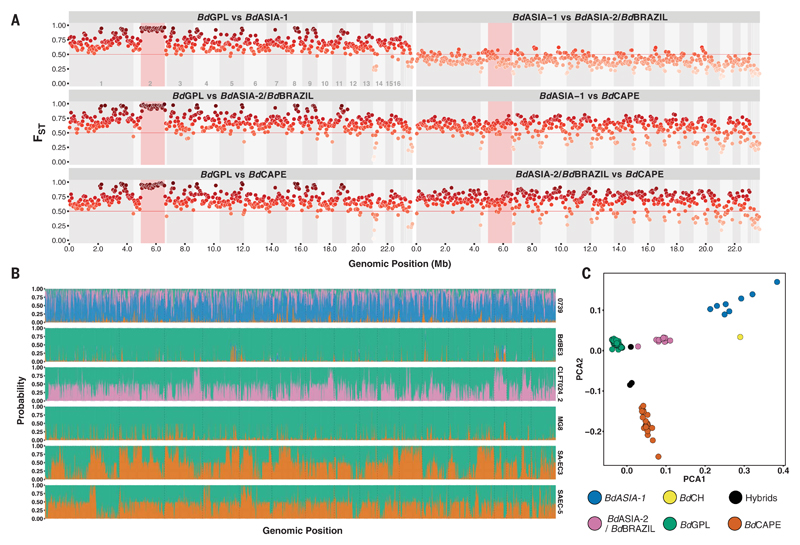
Detection of crossover events in the B. dendrobatidis autosomal genome (18) using a subset of the isolates in this study revealed a large (1.66 Mbp) region of Supercontig_1.2 in BdGPL that exhibits several features that identified it as a recombination “coldspot”: (i) a continuous region of reduced Tajima’s D (Fig. 2F), (ii) sustained high values of population differentiation as measured by the fixation index (FST) when compared with all other lineages (Fig. 3A), (iii) a continuous region of reduced nucleotide diversity (π; fig. S4), and (iv) shared loss of heterozygosity (fig. S8). We expanded sampling to infer the temporal range of pathogen introductions using a broad panel of isolates with known date of isolation (n = 184, ranging from 1998 to 2016) and whole-genome RNA baiting to obtain reads from preserved amphibians that had died of chytridiomycosis. We then investigated whether our data set contained sufficient signal to perform tip-dating inferences by building phylogenetic trees using PhyML (25) (Fig. 2, A and C). We fitted root-to-tip distances to collection dates both at the whole-tree and within-lineage scales. We observed a positive and significant correlation within BdGPL only, for both the mitochondrial and nuclear genomes, demonstrating sufficient temporal signal to perform thorough tip-dating inferences at this evolutionary scale (Fig. 2, B and D).
Fig. 3. FST and site-by-site STRUCTURE analysis.
(A) Non-overlapping 10-kb sliding window of FST between lineages. The region highlighted in red is the Supercontig_1.2:500,000–2,160,000 low-recombination region. (B) Site-by-site analysis of population ancestry for a random selection of 9905 SNPs. Results show those isolates to be either hybrid (SA-EC3, SA-EC5, and CLFT024/2) or with significant introgression from nonparental lineages (isolates BdBE3 and MG8) or a chimera of unsampled diversity, likely originating from East Asia (0739, the BdCH isolate). Each column represents a biallelic SNP position. The columns are colored according to the joint probability of either allele copy arising from one of four distinct populations. Colors represent assumed parental lineages as given in Fig. 3C. (C) Principal components analysis of 3900 SNPs in linkage equilibrium. Each point represents an isolate, colored by phylogenetic lineage. The isolates separate into clearly defined clusters. The axes plot the first and second principal components, PCA1 and PCA2.
Tip-dating in BEAST was used to co-estimate ancestral divergence times and the rate at which mutations accumulate within the BdGPL lineage. The mean mitochondrial substitution rate was 1.01 × 10−6 substitutions per site per year [95% highest posterior density (HPD), 4.29 × 10−7 to 1.62 × 10−6]. The mean nuclear substitution rate was 7.29 × 10−7 substitutions per site per year (95% HPD, 3.41 × 10−7 to 1.14 × 10−6), which is comparable to a recent report of an evolutionary rate of 2.4 × 10−6 to 2.6 × 10−6 substitutions per site per year for another unicellular yeast, Saccharomyces cerevisiae beer strains (26). These rate estimates are faster by a factor of >300 than the rate used in a previous study (12) to obtain a TMRCA of 26,400 years for BdGPL. Accordingly, we estimate that the ancestor of the amphibian panzootic BdGPL originated between 120 and 50 years ago (Fig. 2E), with HPD estimates of 1898 (95% HPD, 1809 to 1941) and 1962 (95% HPD, 1859 to 1988) for the nuclear and mitochondrial dating analyses, respectively (Fig. 2E).
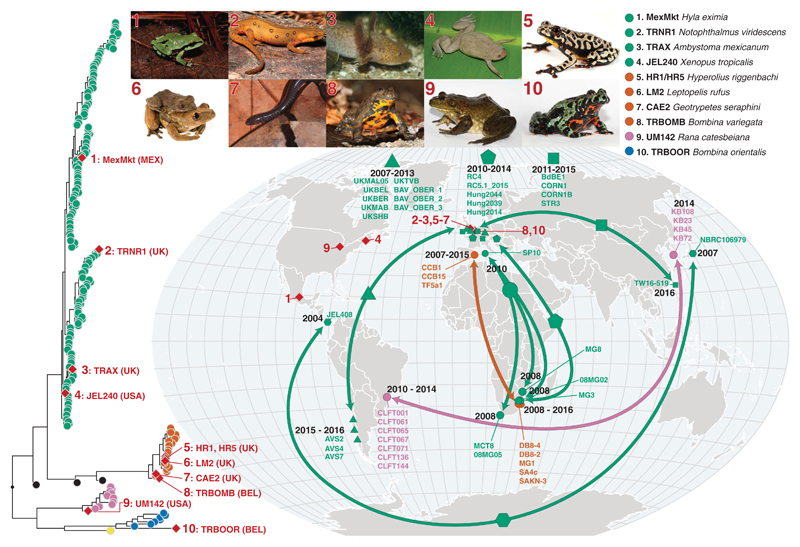
We considered an additional calibration approach for the TMRCA of the mitochondrial genome where we included informative priors on nodes around the dates for the first historical descriptions of BdGPL detection in Australia (1978), Central America (1972), Sierra de Guadarrama (Europe) (1997), and the Pyrenees (Europe) (2000). We did not include priors for nodes where observed declines have been reported but where the lineage responsible for those declines is unknown. This mixed dating method based on tip and node calibration yielded very similar estimates [TMRCA estimates of 1975 (95% HPD, 1939 to 1989) (fig. S9)], further strengthening our confidence in a recent date of emergence for BdGPL. An expansion of BdGPL in the 20th century coincides with the global expansion in amphibians traded for exotic pet, medical, and food purposes (27, 28). Within our phylogeny, we found representatives from all lineages among traded animals (figs. S10 to S14) and identified 10 events where traded amphibians were infected with non-enzootic isolates (Fig. 4). This finding demonstrates the ongoing failure of international biosecurity despite the listing of B. dendrobatidis by the World Organisation for Animal Health (OIE) in 2008.
Fig. 4. Genotypes of Bd isolated from infected amphibians in the international trade and phylogenetically linked genotypes from segregated geographic localities.
The red diamonds on the phylogeny indicate isolates recovered from traded animals. Their geographic location is displayed by the red diamonds on the map. The red numbers link each trade isolate to the relevant picture of the donor host species atop the figure and their placement in the phylogeny. The arrows on the map link geographically separated isolates that form closely related phylogenetic clades with high bootstrap support (≥90%). Each clade is denoted by a different-shaped point on the map; names of isolates within each clade are displayed on the map. The dates displayed indicate the sampling time frame for each clade. The phylogenetic position of each clade is displayed in figs. S10 to S14. The colors of points and arrows on the map indicate lineage according to Fig. 1. A browsable version of this phylogeny can be accessed at https://microreact.org/project/GlobalBd. [Photo credits: (1) Hyla eximia, Ricardo Chaparro; (2) Notophthalmus viridescens, Patrick Coin/CC-BY-SA 2.5; (3) Ambystoma mexicanum, Henk Wallays; (4) Xenopus tropicalis, Daniel Portik; (5) Hyperolius riggenbachi and (6) Leptopelis rufus, Brian Freiermuth; (7) Geotrypetes seraphini, Peter Janzen; (8) Bombina variegata, (9) Rana catesbeiana, and (10) Bombina orientalis, Frank Pasmans]
Hybridization between recontacting lineages of B. dendrobatidis
To determine the extent to which the four main lineages of B. dendrobatidis have undergone recent genetic exchange, we used the site-by-site–based approach implemented in STRUCTURE (29). Although most isolates could be assigned unambiguously to one of the four main lineages, we identified three hybrid genotypes (Fig. 3B), including one previously reported hybrid (isolate CLFT024/2) (20), and discovered two newly identified hybrids of BdGPL and BdCAPE in South Africa. Furthermore, BdCH (isolate 0739) appears to be a chimera of multiple lineages that may represent unsampled genomic diversity residing in East Asia, rather than true hybridization. These hybrid genomes demonstrate that B. dendrobatidis is continuing to exchange haplotypes among lineages when they interact after continental invasions, generating novel genomic diversity. We analyzed isolate clustering using principal components analysis on a filtered subset of 3900 SNPs in linkage equilibrium, revealing an overall population structure that is consistent with our phylogenetic analyses (Fig. 3C). In addition, the putatively identified hybrid isolates of B. dendrobatidis were shown to fall between main lineage clusters (Fig. 3C), further strengthening our hypothesis of haplotype exchange occurring during secondary contact between lineages.
Associations among lineage, virulence, and declines
Genotypic diversification of pathogens is commonly associated with diversification of traits associated with host exploitation (30) and is most commonly measured as the ability to infect a host and to cause disease post-infection. We tested for variation of these two phenotypic traits across four B. dendrobatidis lineages by exposing larval and postmetamorphic common toads (Bufo bufo). Larvae are highly susceptible to infection but do not die before metamorphosis, in contrast to postmetamorphic juveniles, which are susceptible to infection and fatal chytridiomycosis (31). In tadpoles, both BdGPL and BdASIA-1 were significantly more infectious than BdCAPE and BdCH (fig. S15 and tables S3 and S4). In metamorphs, BdGPL was significantly more infectious than the other treatments, relative to the control group, and was significantly more lethal in experimental challenge than the geographically more restricted BdCAPE, BdASIA-1, and BdCH (Fig. 2G). We further tested for differences in virulence among lineages by using our global data set to examine whether chytridiomycosis was nonrandomly associated with B. dendrobatidis lineage. We detected a significant difference (P < 0.001) in the proportion of isolates associated with chytridiomycosis among the three parental lineages (BdASIA-1 and BdASIA-2/BdBRAZIL were grouped because of low sample sizes), and post hoc tests indicated significant excess in virulence in both BdGPL and BdCAPE lineages relative to the combined BdASIA-1 and BdASIA-2/BdBRAZIL (all P < 0.05). However, we did not detect a significant difference between BdGPL and BdCAPE (fig. S16 and table S5). These data suggest that although BdGPL is highly virulent, population-level outcomes are also context-dependent (32); under some conditions, other lineages can also be responsible for lethal amphibian disease and population declines (33).
Historical and contemporary implications of panzootic chytridiomycosis
Our results point to endemism of B. dendrobatidis in Asia, out of which multiple panzootic lineages have emerged. These emergent diasporas include the virulent and highly transmissible BdGPL, which spread during the early 20th century via a yet unknown route to infect close to 700 amphibian species out of ~1300 thus far tested (34). With more than 7800 amphibian species currently described, the number of affected species is likely to rise. The international trade in amphibians has undoubtedly contributed directly to vectoring this pathogen worldwide (Fig. 4) (35, 36), and within our phylogeny we identified many highly supported (≥90% bootstrap support) clades on short branches that linked isolates collected from wild amphibian populations across different continents (Fig. 4 and figs. S10 to S14). However, the role of globalized trade in passively contributing to the spread of this disease cannot be ruled out. It is likely no coincidence that our estimated dates for the emergence of BdGPL span the globalization “big bang”—the rapid proliferation in intercontinental trade, capital, and technology that started in the 1820s (37). The recent invasion of Madagascar by Asian common toads hidden within mining equipment (38) demonstrates the capacity for amphibians to escape detection at borders and exemplifies how the unintended anthropogenic dispersal of amphibians has also likely contributed to the worldwide spread of pathogenic chytrids.
The hyperdiverse hotspot identified in Korea likely represents a fraction of the Batrachochytrium genetic diversity in Asia, and further sampling across this region is urgently needed because the substantial global trade in Asian amphibians (39) presents a risk of seeding future outbreak lineages. Unique ribosomal DNA haplotypes of B. dendrobatidis have been detected in native amphibian species in India (40, 41), Japan (16), and China (42). Although caution should be observed when drawing conclusions about lineages based on short sequence alignments (fig. S3), other endemic lineages probably remain undetected within Asia. It is noteworthy that the northern European countryside is witnessing the emergence of B. salamandrivorans, which also has its origin in Asia. The emergence of B. salamandrivorans is linked to the amphibian pet trade (43), and the broad expansion of virulence factors that are found in the genomes of these two pathogens is testament to the evolutionary innovation that has occurred in these Asian Batrachochytrium fungi (23).
Our findings show that the global trade in amphibians continues to be associated with the translocation of chytrid lineages with panzootic potential. Ultimately, our work confirms that panzootics of emerging fungal diseases in amphibians are caused by ancient patterns of pathogen phylogeography being redrawn as largely unrestricted global trade moves pathogens into new regions, infecting new hosts and igniting disease outbreaks. Within this context, the continued strengthening of transcontinental biosecurity is critical to the survival of amphibian species in the wild (44).
Supplementary Material
Acknowledgments
DNA sequencing was carried out in the NBAF GenePool genomics facility at the University of Edinburgh, and we thank the GenePool staff for their assistance. This work used the computing resources of the UK MEDical BIOinformatics partnership - aggregation, integration, visualization and analysis of large, complex data (UK MED-BIO) which is supported by the Medical Research Council (grant number MR/L01632X/1). We thank J. Rhodes for the provision of flow cells and reagents for MinION sequencing, the staff at Oxford Nanopore Technologies for admission to the MinION Early Access Programme, and three anonymous reviewers for constructive comments and suggestions.
Funding: S.J.O., T.W.J.G., L.Br., A.Lo., A.A.C., D.S.S., E.A.C., C.M., J.B., D.M.A., F.C., and M.C.F. were supported through NERC (standard grant NE/K014455/1). S.J.O. acknowledges a Microsoft Azure for Research Sponsorship (subscription ID: ab7cd695-49cf-4a83-910a-ef71603e708b). T.W.J.G., A.Lo., A.A.C., D.S.S., E.A.C., C.M., J.B., D.M.A., F.C., and M.C.F. were also supported by the EU BiodivERsA scheme (RACE, funded through NERC directed grant NE/G002193/1 and ANR-08-Biodiversa-002-03) and NERC (standard grant NE/K012509/1). M.C.F., E.A.C., and C.M. acknowledge the Nouragues Travel Grant Program 2014. R.A.F. was supported by an MIT/Wellcome Trust Fellowship. T.W.J.G. was supported by the People’s Trust for Endangered Species and the Morris Animal Foundation (D12ZO-002). J.M.G.S. and M.C.F. were supported by the Leverhulme Trust (RPG-2014-273) and the Morris Animal Foundation (D16ZO-022). F.B. was supported by the ERC (grant ERC 260801–Big_Idea). D.M.A. was funded by Wellcome Trust grant 099202. J.V. was supported by the Hungarian Scientific Research Fund (OTKA K77841) and Bolyai János Research Scholarship, Hungarian Academy of Sciences (BO/00579/14/8). D.J.G. was supported by the Conservation Leadership Programme (grant 0134010) with additional assistance from F. Gebresenbet, R. Kassahun, and S. P. Loader. C.S.-A. was supported by Fondecyt Nº11140902 and 1181758. T.M.D.-B. was supported by the Royal Geographical Society and the Royal Zoological Society of Scotland with assistance from M. Hirschfeld and the Budongo Conservation Field Station. B.W. was supported by the National Research Foundation of Korea (2015R1D1A1A01057282). L.F.T. was supported by FAPESP (#2016/25358-3) and CNPq (#300896/2016-6). L.Be., L.F.S., and R.J.W. were supported by the Australian Research Council (FT100100375, DP120100811). A.A.C. was supported by a Royal Society Wolfson Research Merit award. J.H., A.La., and S.M. were funded by the Swedish Research Council Formas (grant no. 2013-1389-26445-20). C.W. was funded by the National Research Foundation, South Africa. T.Y.J. and T.S.J. acknowledge NSF grant DEB-1601259. W.E.H. was funded by the NSERC Strategic and Discovery grant programs.
Footnotes
Author contributions: All authors contributed ideas, data, and editorial advice. S.J.O., A.R., R.A.F., K.A.M., B.B., and M.C.F. conducted analyses. G.M.R., T.W.J.G., and L.Br. conducted disease experiments. S.J.O., F.B., T.W.J.G., and M.C.F. wrote the paper with input from all authors.
Competing interests: K.A.M. sits on an expert panel at the European Food Safety Authority addressing the risks of importation and spread of the salamander chytrid Batrachochytrium salamandrivorans, a species of fungus that is the closest known relative to the pathogen addressed in this manuscript.
Data and materials availability: Sequences have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA). All sequences are available from NCBI BioProject accession PRJNA413876 (www.ncbi.nlm.nih.gov/bioproject/PRJNA413876). The supplementary materials contain additional data. Phylogenetic trees are available from TreeBASE, project accession URL http://purl.org/phylo/treebase/phylows/study/TB2:S22286. A browsable version of the phylogeny and metadata in Fig. 1B is accessible at https://microreact.org/project/GlobalBd.
References
- 1.Fisher MC, et al. Nature. 2012;484:186–194. doi: 10.1038/nature10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger L, et al. Proc Natl Acad Sci USA. 1998;95:9031–9036. doi: 10.1073/pnas.95.15.9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaustein AR, Wake DB. Trends Ecol Evol. 1990;5:203–204. [Google Scholar]
- 4.Skerratt LF, et al. EcoHealth. 2007;4:125–134. [Google Scholar]
- 5.Cheng TL, Rovito SM, Wake DB, Vredenburg VT. Proc Natl Acad Sci USA. 2011;108:9502–9507. doi: 10.1073/pnas.1105538108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lips KR, Diffendorfer J, Mendelson JR, Sears MW. PLOS Biol. 2008;6:e72. doi: 10.1371/journal.pbio.0060072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carvalho T, Becker CG, Toledo LF. Proc R Soc B. 2017;284 doi: 10.1098/rspb.2016.2254. 20162254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hudson MA, et al. Sci Rep. 2016;6 30772. [Google Scholar]
- 9.Rachowicz LJ, et al. Ecology. 2006;87:1671–1683. doi: 10.1890/0012-9658(2006)87[1671:eidaap]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 10.Bosch J, Martinez-Solano I, Garcia-Paris M. Biol Conserv. 2001;97:331–337. [Google Scholar]
- 11.Farrer RA, et al. Proc Natl Acad Sci USA. 2011;108:18732–18736. doi: 10.1073/pnas.1111915108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenblum EB, et al. Proc Natl Acad Sci USA. 2013;110:9385–9390. doi: 10.1073/pnas.1300130110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weldon C, du Preez LH, Hyatt AD, Muller R, Spears R. Emerg Infect Dis. 2004;10:2100–2105. doi: 10.3201/eid1012.030804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Talley BL, Muletz CR, Vredenburg VT, Fleischer RC, Lips KR. Biol Conserv. 2015;182:254–261. [Google Scholar]
- 15.Rodriguez D, Becker CG, Pupin NC, Haddad CFB, Zamudio KR. Mol Ecol. 2014;23:774–787. doi: 10.1111/mec.12615. [DOI] [PubMed] [Google Scholar]
- 16.Goka K, et al. Mol Ecol. 2009;18:4757–4774. doi: 10.1111/j.1365-294X.2009.04384.x. [DOI] [PubMed] [Google Scholar]
- 17.Bataille A, et al. Mol Ecol. 2013;22:4196–4209. doi: 10.1111/mec.12385. [DOI] [PubMed] [Google Scholar]
- 18.Farrer RA, et al. PLOS Genet. 2013;9:e1003703. doi: 10.1371/journal.pgen.1003703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Argimón S, et al. Microb Genomics. 2016;2:e000093. doi: 10.1099/mgen.0.000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schloegel LM, et al. Mol Ecol. 2012;21:5162–5177. doi: 10.1111/j.1365-294X.2012.05710.x. [DOI] [PubMed] [Google Scholar]
- 21.Lawson DJ, Hellenthal G, Myers S, Falush D. PLOS Genet. 2012;8:e1002453. doi: 10.1371/journal.pgen.1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L, Stoeckert CJ, Jr, Roos DS. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farrer RA, et al. Nat Commun. 2017;8 doi: 10.1038/ncomms14742. 14742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tajima F. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guindon S, et al. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 26.Gallone B, et al. Cell. 2016;166:1397–1410.e16. doi: 10.1016/j.cell.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fisher MC, Garner TWJ. Fungal Biol Rev. 2007;21:2–9. [Google Scholar]
- 28.Carpenter AI, Andreone F, Moore RD, Griffiths RA. Oryx. 2014;48:565–574. [Google Scholar]
- 29.Pritchard JK, Stephens M, Donnelly P. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price SJ, et al. Curr Biol. 2014;24:2586–2591. doi: 10.1016/j.cub.2014.09.028. [DOI] [PubMed] [Google Scholar]
- 31.Garner TWJ, et al. Oikos. 2009;118:783–791. [Google Scholar]
- 32.Bates KA, et al. Nat Commun. 2018;9:693. doi: 10.1038/s41467-018-02967-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doddington BJ, et al. Ecology. 2013;94:1795–1804. doi: 10.1890/12-1270.1. [DOI] [PubMed] [Google Scholar]
- 34.Olson DH, Ronnenberg KL. FrogLog. 2014;22:17–21. [Google Scholar]
- 35.Walker SF, et al. Curr Biol. 2008;18:R853–R854. doi: 10.1016/j.cub.2008.07.033. [DOI] [PubMed] [Google Scholar]
- 36.Wombwell EL, et al. EcoHealth. 2016;13:456–466. doi: 10.1007/s10393-016-1138-4. [DOI] [PubMed] [Google Scholar]
- 37.O’Rourke KH, Williamson JG. Eur Rev Econ Hist. 2002;6:23–50. [Google Scholar]
- 38.Kolby JE. Nature. 2014;509:563. doi: 10.1038/509563a. [DOI] [PubMed] [Google Scholar]
- 39.Herrel A, van der Meijden A. Herpetol J. 2014;24:103–110. [Google Scholar]
- 40.Dahanukar N, et al. PLOS ONE. 2013;8:e77528. doi: 10.1371/journal.pone.0077528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Molur S, Krutha K, Paingankar MS, Dahanukar N. Dis Aquat Organ. 2015;112:251–255. doi: 10.3354/dao02804. [DOI] [PubMed] [Google Scholar]
- 42.Bai C, Liu X, Fisher MC, Garner WJT, Li Y. Divers Distrib. 2012;18:307–318. [Google Scholar]
- 43.Martel A, et al. Science. 2014;346:630–631. doi: 10.1126/science.1258268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roy HE, et al. Conserv Lett. 2017;10:477–484. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.