Abstract
Emerging evidence has shown that the hepatocyte growth factor (HGF) and its receptor, MET proto-oncogene, receptor tyrosine kinase (MET), promote cell proliferation, motility, morphogenesis, and angiogenesis. Whereas up-regulation of MET expression has been observed in aggressive and metastatic prostate cancer, a clear understanding of MET function in prostate tumorigenesis remains elusive. Here, we developed a conditional Met transgenic mouse strain, H11Met/+:PB-Cre4, to mimic human prostate cancer cells with increased MET expression in the prostatic luminal epithelium. We found that these mice develop prostatic intraepithelial neoplasia after HGF administration. To further assess the biological role of MET in prostate cancer progression, we bred H11Met/+/PtenLoxP/LoxP:PBCre4 compound mice, in which transgenic Met expression and deletion of the tumor suppressor gene Pten occurred simultaneously only in prostatic epithelial cells. These compound mice exhibited accelerated prostate tumor formation and invasion as well as increased metastasis compared with PtenLoxP/LoxP:PB-Cre4 mice. Moreover, prostatic sarcomatoid carcinomas and lesions resembling the epithelial-to-mesenchymal transition developed in tumor lesions of the compound mice. RNA-Seq and qRT-PCR analyses revealed a robust enrichment of known tumor progression and metastasis-promoting genes in samples isolated from H11Met/+/PtenLoxP/LoxP:PB-Cre4 compound mice compared with those from PtenLoxP/LoxP:PB-Cre4 littermate controls. HGF-induced cell proliferation and migration also increased in mouse embryonic fibroblasts (MEFs) from animals with both Met transgene expression and Pten deletion compared with Pten-null MEFs. The results from these newly developed mouse models indicate a role for MET in hastening tumorigenesis and metastasis when combined with the loss of tumor suppressors.
Keywords: prostate cancer, transgenic mice, gene knockout, tumor cell biology, androgen receptor, phosphatase and tensin homolog (PTEN), hepatocyte growth factor/scatter factor (HGF/SF), HGF/SF, Met, metastasis, tumorigenesis, PB-Cre4, prostatic intraepithelial neoplasia, epithelial-to-mesenchymal transition
Introduction
Prostate cancer is the most common malignancy and the second leading cause of cancer mortality in men in the United States (1). The androgen-signaling pathway plays an essential role in prostate tumorigenesis. Thus, androgen deprivation therapy results in a rapid tumor regression and has been routinely used in clinics for the treatment of advanced prostate cancer. However, androgen deprivation therapy eventually fails in nearly all cases of prostate cancer when patients develop castration-resistant prostate cancer (CRPC),3 which currently lacks curable treatment and claims more than 250,000 lives worldwide each year (2).
Hepatocyte growth factor (HGF) plays a critical role in the regulation of cell growth, cell motility, morphogenesis, and angiogenesis (3). It has been shown that HGF derived from prostate stroma significantly increases proliferation, motility, and invasion of malignant cells through its receptor, c-Met (4, 5). The c-Met receptor tyrosine kinase is encoded by MET, a proto-oncogene, and has been shown to play a promotional role in the proliferation and progression of a wide variety of human malignancies, including prostate cancer (4, 6). The aberrant expression of HGF and c-Met often correlates with poor prognosis in cancer patients (7). Specifically, up-regulation of c-Met expression was observed in a majority of the metastatic prostate cancer lesions (6, 8–11). Interestingly, an inverse correlation between the expression of AR and c-Met has been observed in prostate epithelium and prostate cancer cell lines (4, 6, 8, 12, 13). An increase of c-Met expression was also observed in castrated rat (4, 15) and mouse models (14). Moreover, it has been shown that the AR represses Met expression in prostate cancer cells (16). Accordingly, MET expression appears very limited in normal prostate luminal cells and primary prostate cancer cells (13). The above data imply a critical role of HGF/MET signaling pathways in prostate tumorigenesis.
Whereas overexpression of c-Met is frequently observed in metastatic prostate cancers and positively correlates with prostate cancer progression (17, 18), the biological consequence of aberrant Met up-regulation in disease progression and CRPC development is largely unknown. In addition, there is a lack of appropriate mouse models to directly assess an increased expression of c-Met in prostate cancer initiation and progression. In this study, we developed a conditional Met transgenic mouse strain in which the mouse Met transgene was specifically targeted into the H11b locus (19). By intercrossing the Met-floxed mice with PB-Cre4 mice (20), we generated a conditional Met transgenic mouse line, H11Met/+:PB-Cre4. Conditional expression of the Met transgene in prostatic luminal epithelial cells is achieved by Cre expression, mimicking the condition of human prostate cancer cells with increased MET expression. Intriguingly, H11Met/+:PB-Cre4 mice develop prostatic intraepithelial neoplasia (PIN) after HGF administration. To assess the effect of Met in prostate cancer progression, we generated H11Met/+/PtenLoxP/LoxP:PB-Cre4 compound mice, in which transgenic Met expression and deletion of the tumor suppressor Pten simultaneously co-occur in prostatic epithelial cells. We observed accelerated prostate cancer progression, aggressive tumor invasion, and increased metastasis in these compound mice. Moreover, development of prostatic sarcomatoid carcinomas and lesions resembling epithelial-to-mesenchymal transition is also observed in the prostate of the compound mice. RNA-Seq and qRT-PCR analyses showed a robust enrichment of known tumor progression and metastasis–promoting genes, including Met, Spp1, Fn1, and Cdh2, in samples isolated from H11Met/+/PtenLoxP/LoxP:PB-Cre4 compound mice compared with those from PtenLoxP/LoxP:PB-Cre4 littermate controls. Our study demonstrates a promotional role of Met in PTEN-mediated oncogenic transformation in prostate tumorigenesis and provides a series of novel and biologically relevant mouse models for investigating prostate cancer initiation and progression.
Results
Activation of HGF/Met signaling in prostatic epithelial cells induces oncogenic transformation in the mouse prostate
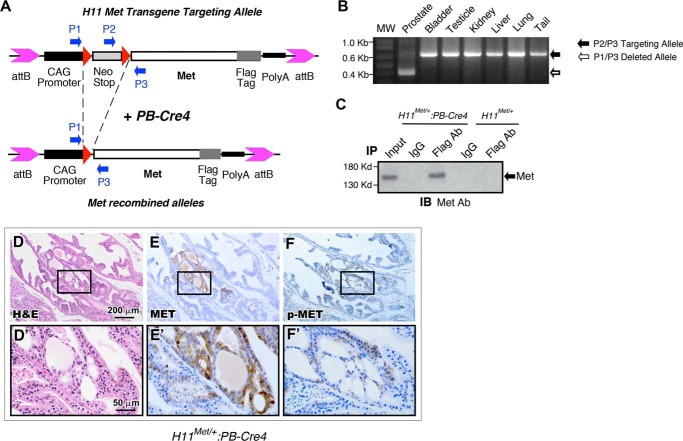
Increased c-Met expression has been observed frequently in advanced prostate cancers (4, 8). Unfortunately, there is no appropriate mouse model that can be used to directly assess the effect of increased expression of c-Met in prostate cancer initiation and progression. In this study, we generated a conditional Met transgenic line using integrase-mediated transgenic technology (19). A loxP-flanked transcriptional silencing element, LoxP-stop-loxP (LSL) cassette, was inserted between the CAG promoter (21, 22) and the FLAG-tagged mouse Met coding sequence (Fig. 1A). Because the CAG promoter is ubiquitously active in most mouse tissues (23), the Met transgene expression in this model can be achieved in a constitutive but tissue-specific manner through Cre recombinase-mediated removal of the LSL cassette (Fig. 1A). To generate prostate-specific expression of the Met transgene, H11Met/+ mice were bred with PB-Cre4 mice, in which Cre expression is regulated by a modified probasin promoter, ARR2PB, in prostatic luminal epithelium (20). To confirm PB-Cre4–mediated genomic recombination, genomic DNA samples were isolated from a variety of tissues of the H11Met/+:PB-Cre4 mice. Genetic recombination and removal of the LSL cassette from the Met transgene were detected specifically in the prostate, but not in the bladder, testes, kidney, liver, lungs, or tail (Fig. 1B). To detect the Met transgene expression, cell lysates from mouse prostate tissues of H11Met/+:PB-Cre4 or H11Met/+ only mice were immunoprecipitated with either a FLAG antibody or normal IgG as the control. Transgenic Met expression was detected in the sample immunoprecipitated with the FLAG antibody, but not with an IgG or in the tissue samples isolated from control mice (Fig. 1C). These data demonstrate the specific expression of the Met transgene in the prostate of H11Met/+:PB-Cre4 mice.
Figure 1.
Generation of mice with prostate-specific expression of Met. A, schematic of the conditional mouse Met transgene-targeting construct. Red triangles, LoxP sequences. B, genomic PCR analysis of prostate-specific recombination of Met transgene. C, anti-Met immunoblot (IB) of whole prostate extracts from 3-month-old H11Met/+ and H11Met/+:PB-Cre4 mice immunoprecipitated (IP) with FLAG antibody or normal IgG beads. D–F, histological and immunohistochemical analysis of 10-month-old H11Met/+:PB-Cre4 mouse 12-weeks post-HGF administration. D and D′, low and high magnification of H&E staining. Shown is immunohistochemical analysis of Met (E and E′) and phosphorylated Met (F and F′) expression on sequential sections.
H11Met/+:PB-Cre4 mice were born at the expected Mendelian ratios and appeared normal with no obvious differences from their WT littermates at birth. Per the recommendations of the Mouse Models of Human Cancers Consortium Prostate Pathology Committee (24), we assessed H11Met/+:PB-Cre4 mice from birth to 20 months of age and did not observe any obvious pathological changes in the mouse prostate tissues. To assess HGF-Met–mediated effects, we then administered recombinant HGF to 6-month-old H11Met/+:PB-Cre4 mice as well as age- and sex-matched WT controls and examined their prostates between 8 and 10 months of age (n = 4, each group). We observed pathological lesions of typical low-grade prostatic intraepithelial neoplasia in H11Met/+:PB-Cre4 mice but not controls (Fig. 1D). Immunohistochemical (IHC) analyses showed strong cytoplasmic immunoreactivity to a Met antibody in atypical cells within PIN lesions (Fig. 1E). Intriguingly, these cells are also reactive to phospho-Met antibody, providing a direct link between the activation of the Met transgene and PIN transformation in the mouse prostate (Fig. 1F). These data demonstrate that activating HGF-Met signaling induces oncogenic transformation in the mouse prostate. However, the transgenic mice did not reveal prostate carcinomas, suggesting that other additional factors may be required in initiating prostate tumor development.
Simultaneous expression of Met and deletion of Pten in the prostate induces earlier onset and more aggressive prostatic carcinomas
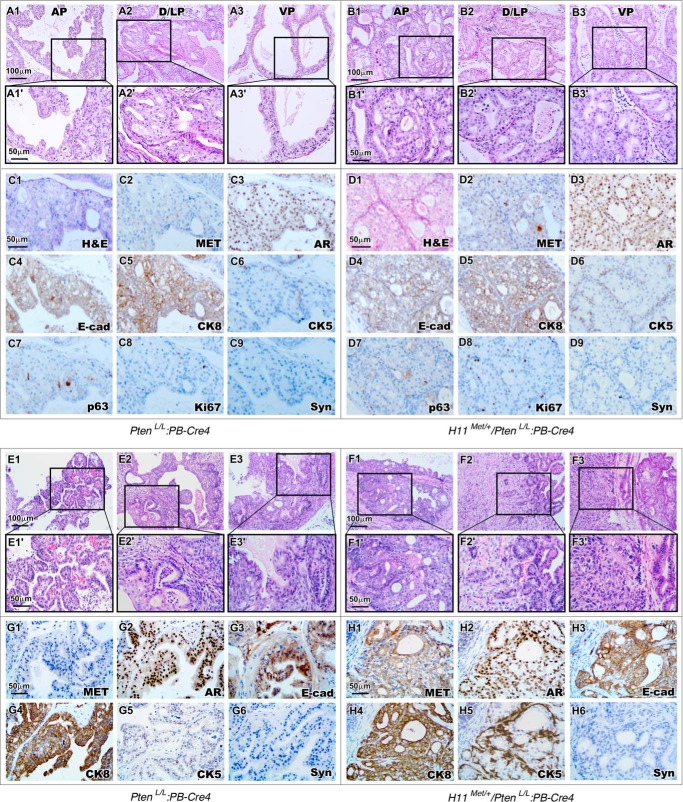
Both increased MET expression and aberrant phosphatidylinositol 3-kinase signaling alterations have been observed in advanced and metastatic prostate cancers (4, 8). Recent studies further suggest these two alterations may co-exist in prostate cancer (25). To directly assess the collaborative role of these alterations in prostate tumorigenesis, we generated H11Met/+/PtenLoxP/LoxP:PB-Cre4 (H11Met/+/PtenL/L:PB-Cre4) compound mice, in which conditional Met expression and Pten deletion co-occur in prostate epithelium. This new mouse model enables us to investigate the potential effect of increased MET expression and PTEN deletion in prostate tumorigenesis. We observed the pathologic changes that represent mouse PIN lesions in the four prostatic lobes in PtenL/L:PB-Cre4 mice up to 4 months of age. As shown in the representative images (Fig. 2, A1–A3), analysis of the prostates revealed cribriform structures along with stratification of cells, papilliferous structures, and tufts of cells. Lesions in both the dorsal and lateral lobes appear more severe than in other lobes (Fig. 2, A2 and A2′). Intriguingly, the age-matched H11Met/+/PtenL/L:PB-Cre4 mice developed much more drastic lesions (Fig. 2, B1–B3). Typical cribriform and papilliferous structures completely filled the lumen of prostatic glands (Fig. 2, B1–B3). Atypical epithelial cells lacking normal polarity appeared in all prostatic lobes (Fig. 2, B1′–B3′). IHC analyses of the PIN regions in both genotype mice showed similar immunoreactivities with AR, E-cadherin, cytokeratin-8 (CK8), or p63 antibody (Fig. 2, C3–C5, C7, D3–D5, and D7) but no staining with synaptophysin (Fig. 2, C9 and D9), a marker of prostatic neuroendocrine cells. Interestingly, more atypical cells showed positive staining of CK5, the prostatic basal cell marker, within PIN lesions of H11Met/+/PtenL/L:PB-Cre4 mice than those of PtenL/L:PB-Cre4 mice (Fig. 2 (C6 and D6) and Fig. S1C).
Figure 2.
Conditional expression of the mouse Met transgene and homozygous Pten deletion in mouse prostate leads to development of HGPIN and prostatic adenocarcinomas. A and B, histology of 3-month-old PtenL/L:PB-Cre4 (A) and H11Met/+/PtenL/L:PB-Cre4 (B) mouse prostates. ×200 magnification images of mPIN lesions in AP/DLP/VP are shown in A1–A3 and B1–B3. Boxes, ×400 magnification images (A1′–A3′ and B1′–B3′). C and D, immunohistochemical analysis of 4-month-old PtenL/L:PB-Cre4 (C1–C9) and H11Met/+/PtenL/L:PB-Cre4 (D1–D9) mouse prostates with different antibodies as labeled. E and F, histology of 12-month-old PtenL/L:PB-Cre4 (E) and H11Met/+/PtenL/L:PB-Cre4 (F) mouse prostates. ×200 magnification images of prostatic adenocarcinomas are shown in E1–E3 and F1–F3. Boxes, ×400 magnification images of invasive adenocarcinoma (E1′–E3′ and F1′–F3′), as well as an expansion of EMT pattern with nuclear atypia, polygonal cell pattern, and pleomorphism (F3′) in H11Met/+/PtenL/L:PB-Cre4 tumor. G and H, immunohistochemical analysis of PtenL/L:PB-Cre4 (G1–G6) and H11Met/+/PtenL/L:PB-Cre4 (H1–H6) adenocarcinoma with different antibodies as labeled.
We then continued our analysis of aged PtenL/L:PB-Cre4 and H11Met/+/PtenL/L:PB-Cre4 mice up to 24 months of age. We observed that PtenL/L:PB-Cre4 mice develop intracystic carcinomas between 4 and 10 months of age and adenocarcinoma and invasive carcinomas after 10 months (Table 1). The representative images show corresponding pathologic changes, with features typical of prostatic adenocarcinomas and invasive carcinomas as reported previously (26) (Fig. 2, E1–E3). In contrast, H11Met/+/PtenL/L:PB-Cre4 mice showed more aggressive tumor phenotypes than PtenL/L:PB-Cre4 littermates. Comparison of the pathologies of PtenL/L:PB-Cre4 and H11Met/+/PtenL/L:PB-Cre4 mice revealed an overall more invasive tumor phenotype in the latter (Fig. 2, E versus F). In addition to prostatic adenocarcinomas and invasive carcinomas (Fig. 2, F1–F3), we also observed pathological changes that are similar to prostatic sarcomatoid carcinomas as well as features of an epithelial–mesenchymal transition (EMT) indicating nuclear atypia, a polygonal cell pattern, and pleomorphism (Fig. 2, F3 and F3′). A significant increase in MET staining (Fig. 2H1) was observed in tumor lesions of the compound mice. Elevated staining of phospho-Met and phospho-Erk was also observed in adjacent sections of tumor tissues isolated from the compound mice compared with those from PtenL/L:PB-Cre4 only mice (see Fig. S1, A1–A3 and B1–B3), suggesting the activation of HGF/Met axis in the tumor lesions. Similar to the expression patterns observed in the HGPIN regions of younger H11Met/+/PtenL/L:PB-Cre4 mice, adenocarcinoma lesions from the compound mice also revealed an increase in the number of CK5-positive cells compared with those from PtenL/L:PB-Cre4 only mice (Fig. 2H5 and Fig. S1C). We did not observe any significant difference in the expression of AR, CK8, and E-cadherin between the above two groups of mice (Fig. 2, G2–G4 and H2–H4). Samples from both PtenL/L:PB-Cre4 and H11Met/+/PtenL/L:PB-Cre4 were negative for synaptophysin staining (Fig. 2, G6 and H6). The presence of increased CK5 expression in tumor cells of H11Met/+/PtenL/L:PB-Cre4 suggests a novel role of the Met transgene in inducing prostatic tumor cell transdifferentiation.
Table 1.
Pathological abnormalities of H11Met/+/PtenL/L:PB-Cre4 and PtenL/L:PB-Cre4 mice
| Genotype | <4 months | 4–10 months | >10 months |
|---|---|---|---|
| PtenL/L:PB-Cre4 | 4 of 4 LGPIN | 6 of 6 multifocal HGPIN | 6 of 8 intracystic carcinoma |
| 2 of 4 multifocal HGPIN | 2 of 6 intracystic carcinoma | 4 of 8 adenocarcinoma | |
| 3 of 8 invasive carcinoma | |||
| 1 of 8 invasive carcinoma with metastasisa | |||
| H11Met/+/PtenL/L:PB-Cre4 | 5 of 5 multifocal HGPIN | 5 of 5 multifocal HGPIN | 7 of 8 adenocarcinoma, 6 of 8 invasive carcinoma |
| 1 of 5 intracystic carcinoma | 6 of 8 invasive carcinoma | ||
| 2 of 5 adenocarcinomas | 5 of 8 invasive carcinoma with EMT | ||
| 1 of 5 invasive carcinoma | 5 of 8 invasive carcinoma with metastasisb |
a Metastatic sites include lymph node and lung tissues.
b Metastatic sites include lymph node, lung, and pancreas tissues.
Conditional expression of Met enhances tumor aggressiveness and induces an epithelial-to-mesenchymal transition in prostate tumors
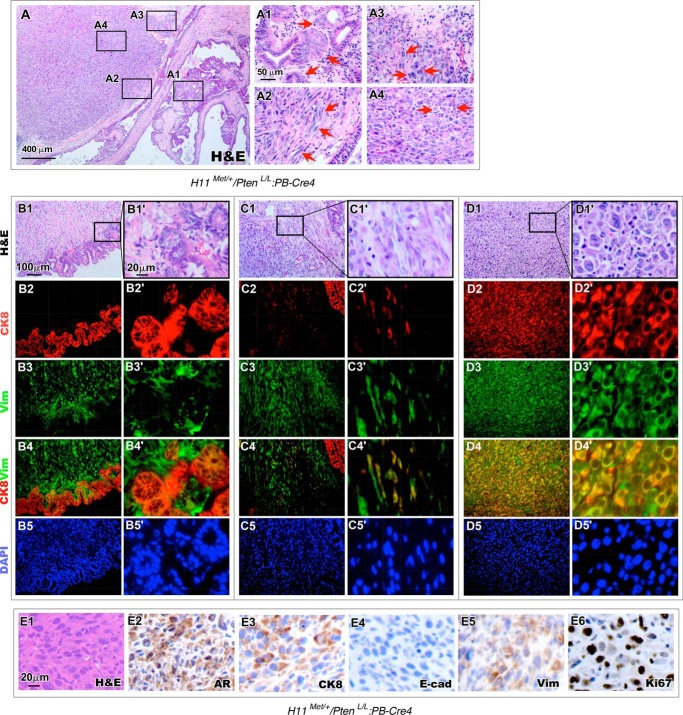
Scrutiny of H11Met/+/PtenL/L:PB-Cre4 compound mice revealed an aggressive tumor phenotype with sarcomatoid carcinoma development that did not occur in prostatic lesions of the PtenL/L:PB-Cre4 mice. Representative images from 12-month-old H11Met/+/PtenL/L:PB-Cre4 compound mice show poorly differentiated carcinomas with areas of invasion, where the epithelial tumor cells have permeated the basement membrane into the surrounding tissue (Fig. 3, A and A1). Cells located at sites of invasion maintain a columnar epithelial shape and some regions of pseudostratified epithelial layers (Fig. 3A1, arrows). A transition from adenocarcinoma and invasive carcinoma lesions to pathology resembling sarcomatoid carcinomas can be observed. Sarcomatoid carcinoma was classified as a biphasic malignant neoplasm, which is frequently observed during the process of EMT (27–29). We observed spindle-like tumor cells (Fig. 3A2, arrows), tumor cells with a multitude of mitotic figures (Fig. 3A3, arrows), and haphazard acini and lobules of pleomorphic cells (Fig. 3A4), indicating that the above lesions possess mesenchymal morphology and highly proliferative and invasive features.
Figure 3.
Epithelial-to-mesenchymal features are present in H11Met/+/PtenL/L:PB-Cre4 tumors. A, histology of a 12-month-old H11Met/+/PtenL/L:PB-Cre4 mouse prostate. High-magnification images showed invasive prostatic adenocarcinoma (A1) and significant expansion of sarcomatoid carcinoma with spindled, epithelioid features, nuclear atypia, atypical mitoses, and pleomorphism (A2–A4). B–D, low and high magnification of histology and immunofluorescence images of invasive adenocarcinoma (B1–B5′), EMT (C1–C5′), and sarcomatoid carcinoma (D1–D5′). The invasive adenocarcinoma showed an epithelial pattern characteristic of glandular structures (B1 and B1′) that was positive for CK8 but negative for vimentin (B2–B5′). The cells in the EMT region (C1 and C1′) were predominantly spindle-like cells, which were stained positive for CK8 and vimentin (C2–C5′). Sarcomatoid carcinoma regions showed a polygonal cell pattern, fusiform, and large pleomorphic nuclei (D1–D1′) and were positive for both CK8 and vimentin (D2–D5′). E, immunohistochemical analysis of AR (E2), CK8 (E3), E-cadherin (E4), vimentin (E5), and Ki67 (E6) in H11Met/+/PtenL/L:PB-Cre4 sarcomatoid carcinoma region.
Observation of the co-occurring prostatic adenocarcinoma, invasive carcinoma, and sarcomatoid carcinoma in the prostate of H11Met/+/PtenL/L:PB-Cre4 is new and has not been observed in PtenL/L:PB-Cre4 mice. It suggests an important role of Met in promoting tumor progression. To further characterize these lesions, we performed co-immunofluorescent assays to assess the cellular properties of the various regions within the tumor lesions, which include poorly differentiated and invasive carcinomas (Fig. 3, B1–B1′), sarcomatoid carcinomas (Fig. 3, D1–D1′), and the transition between these two phenotypes (Fig. 3, C1–C1′). Tumor cells in adenocarcinoma regions appeared positively stained for CK8, a prostatic luminal epithelial cell marker, and negatively stained for vimentin (Vim), a stromal cell marker (Fig. 3, B2–B4′). Within EMT “transition” areas (Fig. 3, C1–C1′), the majority of spindle-like tumor cells were immunoreactive for Vim, but not for the luminal cell marker CK8. Intriguingly, scattered tumor cells revealed double-positive staining for CK8 and Vim (Fig. 3, C4–C4′). Moreover, in sarcomatoid lesions, large, polygonal-shaped cells with little cytoplasm and broad pleomorphic nuclei (Fig. 3, D1–D1′) showed positive staining for both CK8 and Vim (Fig. 3, D2–D4). Using IHC approaches, we further assessed the cellular properties of sarcomatoid tumor cells (Fig. 3E1). The majority of tumor cells showed positive staining with the AR and CK8 (Fig. 3, E2 and E3) but not E-cadherin (Fig. 3E4). Positive staining of Vim appeared in the tumor cells. These expression patterns are consistent with the above co-immunofluorescent experiments and suggest that the process of EMT has occurred in those cells. Additionally, we also examined Ki67 expression and observed significant Ki67-positive cells in sarcomatoid tumor areas, demonstrating the highly proliferative feature of sarcomatoid carcinoma cells (Fig. 3E6). Development of EMT and sarcomatoid tumor lesions in the prostate of H11Met/+/PtenL/L:PB-Cre4 compound mice provides a direct line of evidence of the promotional role of Met in prostate cancer progression.
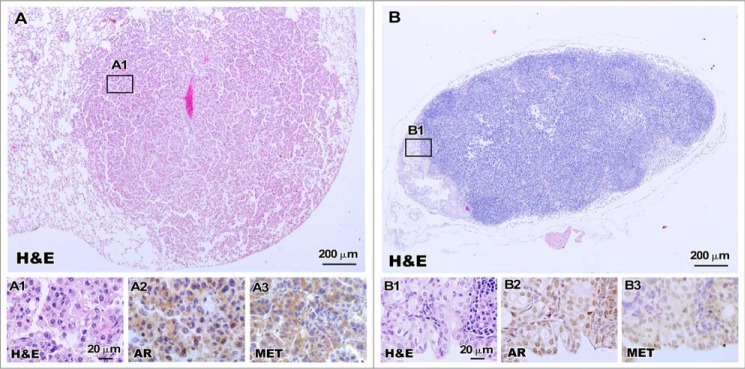
It has been shown that PtenL/L:PB-Cre4 mice develop metastatic diseases in the lung and lymphoid nodes (30). Given the more aggressive tumor phenotype seen in H11Met/+/PtenL/L:PB-Cre4 compound mice, we systematically examined the mice for the presence of metastases during this investigation. We observed prostate cancer metastasis in both PtenL/L:PB-Cre4 and H11Met/+/PtenL/L:PB-Cre4 mice after 10 months of age (Table 1). Five of eight H11Met/+/PtenL/L:PB-Cre4 mice (62.5%) showed metastatic lesions, but only one of eight PtenL/L:PB-Cre4 mice (12.5%) revealed lung and lymph node metastasis (Table 1). H11Met/+/PtenL/L:PB-Cre4 mice showed metastatic lesions in lungs and periprostatic lymph nodes (Fig. 4). Histopathological analysis of the lung tissues confirmed the presence of distal metastasis in H11Met/+/PtenL/L:PB-Cre4 mice, showing circumscribed nodules within the lung parenchyma (Fig. 4, A and A1). The metastatic foci in the lung were composed of solid nests (poorly differentiated; Fig. 4A1). Histological section of the lymph node demonstrated partial replacement of the cortex by metastatic adenocarcinoma. The lymph node metastasis showed prominent lumen formation (Fig. 4B1). The nuclei displayed a moderate degree of polymorphism, and cytoplasmic vacuoles were present in a subset of the tumor cells. Additionally, IHC analyses of the metastatic tumor cells showed positive staining for AR and Met (Fig. 4, A3–A4 and B3–B4).
Figure 4.
Lung and lymph node metastases develop in H11Met/+/PtenL/L:PB-Cre4 mice. Shown are representative images of H&E-stained lung (A) and lymph node (B) metastases. High-magnification images showing a solid growth pattern in the lung (A1) and a glandular pattern in the lymph node (B1). Immunohistochemical analysis of AR (A2 and B2) and Met (A3 and B3) showed positivity in both lung and lymph node metastases of H11Met/+/PtenL/L:PB-Cre4 mice.
Investigation of transcriptional profiles that contribute to prostate cancer progression and metastasis
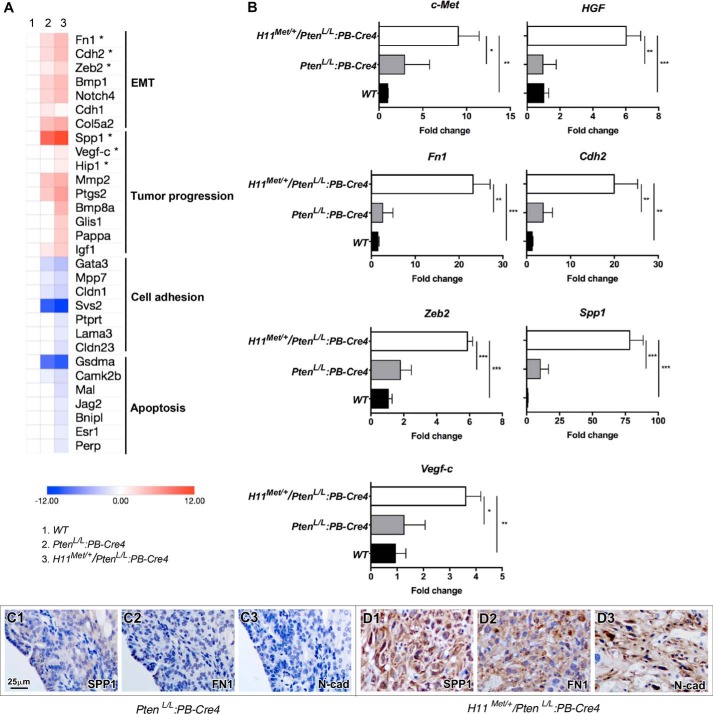
To determine the molecular mechanisms underlying early-onset Met-mediated tumor development and progression, we performed RNA-Seq approaches to search for the alterations between prostate tumor tissues isolated from PtenL/L:PB-Cre4 and H11Met/+/PtenL/L:PB-Cre4 compound mice. We microscopically confirmed that the tumor tissues used to prepare RNA samples for these analyses were composed of more than 80% tumor cells. Analyses of gene expression profiles yielded 1075 differentially expressed genes that were up-regulated >2-fold and 966 differentially expressed genes that were down-regulated >2-fold in H11Met/+/PtenL/L:PB-Cre4 when compared with the PtenL/L:PB-Cre4 mice. A heat map (Fig. 5A) depicts the potential targets that are associated with EMT, tumor progression, cell adhesion, and apoptosis within the context of prostate cancer (31–54). As shown in Fig. 5B, we observed a significant increase in the expression of both Met and Hgf in prostate tumor samples of H11Met/+/PtenL/L:PB-Cre4 compound mice compared with PtenL/L:PB-Cre4 mice. An increasing HGF expression in the above tumor tissues suggests an autocrine regulation of HGF/Met signaling in prostatic tumor growth and progression. Because the tumor samples from the H11Met/+/PtenL/L:PB-Cre4 compound mice showed the pathological changes of EMT and sarcomatoid tumors, we further validated the expression of EMT markers Fn1 (55), Cdh2 (56, 57), Zeb2 (56, 58), and Spp1 (59) (n = 4) (Fig. 5B). Quantitative PCR analyses showed a significant increase in the expression of the above EMT-related genes in the H11Met/+/PtenL/L:PB-Cre4 compound mice compared with the PtenL/L:PB-Cre4 mice (Fig. 5B). Using IHC approaches, we further confirmed elevated expression of Spp1, Fn1, and N-cadherin proteins in prostate tumor samples isolated from H11Met/+/PtenL/L:PB-Cre4 compound mice (Fig. 5, D1–D3 versus C1–C3). These data provide a molecular basis for HGF/Met signaling in promoting EMT lesions in H11Met/+/PtenL/L:PB-Cre4 compound mice.
Figure 5.
The conditional expression of mouse Met transgene and homozygous Pten deletion induces a gene signature pattern that is associated with tumor progression. A, heat map of representative gene sets that are altered in PtenL/L:PB-Cre4 and H11Met/+/PtenL/L:PB-Cre4 tumors. B, qRT-PCR validation of the gene expression in PtenL/L:PB-Cre4 and H11Met/+/PtenL/L:PB-Cre4 tumors. The data are presented as the mean ± S.D. (error bars) for three independent samples. *, p < 0.05; **, p < 0.01; ***, p < 0.001. C, immunohistochemical analysis of SPP1 (C1 and D1), FN1 (C2 and D2), and N-cadherin (C3 and D3) expression in prostate tumors of PtenL/L:PB-Cre4 and H11Met/+/PtenL/L:PB-Cre4 (D1–D3) mice.
Conditional expression of Met increases cellular proliferation, migration, and induction of EMT-associated gene expression
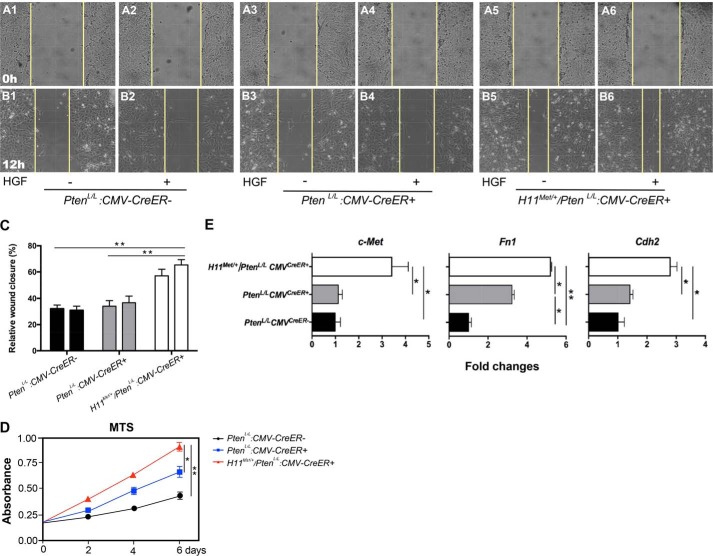
Development of EMT and sarcomatoid carcinomas in the prostate of H11Met/+/PtenL/L:PB-Cre4 mice suggests a promotional role of Met in cell proliferation, mobility, and migration (60). To test our hypothesis, we performed a set of proof-in-principle experiments using mouse embryonic fibroblasts (MEFs) that were prepared from PtenL/L, PtenL/L:CMVCreER+, and H11Met/+/PtenL/L:CMVCreER+ embryos (see “Materials and methods”). Using in vitro cell-wounding assays, we directly assessed the effect of Met expression in cell mobility and migration. MEFs of the various genotypes were cultured to 90% confluence and then wounded by a single scratch as described previously (61) and maintained in serum-free medium either in the presence or absence of HGF for 12 h. Images captured immediately after cell wounding demonstrated equal sized scratches across different genotypes (Fig. 6, A1–A6). Following 12 h of wound healing, significant differences in the wound area were visually evident between the various genotypes (Fig. 6, B1–B6). Whereas PtenL/L:CMVCreER+ and PtenL/L control MEFs exhibited slight migration toward the wound, MEFs from H11Met/+/PtenL/L:CMVCreER+ embryos (Fig. 6, B5–B6) demonstrated more closure of the wound, indicative of an increased ability to migrate. Whereas no significant difference was detected between the samples cultured in the absence or presence of HGF, we did observed a slight increase in wound healing from the H11Met/+/PtenL/L:CMVCreER+ MEFs in the presence of HGF, but not in other samples (Fig. 6C). In this study, we also assessed the proliferative ability of the above MEFs using MTS assays. Although, in agreement with previous studies (30), deletion of Pten increases cellular proliferation in MEFs (Fig. 6D, blue line), conditional expression of Met in combination with Pten deletion further enhances cellular proliferation (Fig. 6D, red line). Using qRT-PCR approaches, we examined the expression of Met, Fn1, and Cdh2 in MEFs within three different genotypes. We observed increased expression of Met, as well as Fn1 and Cdh2, in H11Met/+/PtenL/L:CMVCreER+ MEFs compared with both PtenL/L:CMVCreER+ and PtenL/L MEFs (Fig. 6D). Taken together, the above results demonstrate a promotional role of Met in cellular migration and proliferation, which is consistent with the aggressive tumor phenotype with EMT features as observed in our mouse models.
Figure 6.
The conditional expression of mouse Met transgene and homozygous Pten deletion increases cell proliferation and migration in MEFs. A and B, wound-healing assay of MEFs. Shown are before (A) and after (B) images of scratches in the monolayer of PtenL/L:CMVCreER-, PtenL/L:CMVCreER+, and H11Met/+/PtenL/L:CMVCreER+ MEFs cultured in the absence/presence of HGF for 12 h. C, graphical representation of relative wound closure. The data represent quantifications of multiple images from two independent experiments ± S.D. D, graphical representation of cellular proliferation of PtenL/L:CMVCreER−, PtenL/L:CMVCreER+, and H11Met/+/PtenL/L:CMVCreER+ MEFs measured by MTS reduction. The data represent the mean ± S.D. of three independent experiments. E, qRT-PCR analysis of Met, Fn1, and Cdh2 expression in PtenL/L:CMVCreER+ and H11Met/+/PtenL/L:CMVCreER+ MEFs. Bars, mean ± S.D. (error bars) of three independent experiments. *, p < 0.05; **, p < 0.01.
Discussion
Emerging evidence from basic, translational, and clinical studies has shown the significance of the HGF/Met signaling pathway in prostate cancer progression and CRPC development (62). Up-regulation of c-Met expression has been observed in a majority of advanced and metastatic prostate cancer lesions (6, 8–11). However, the biological role of Met in oncogenic transformation and prostate cancer initiation has not been investigated using biologically relevant models. In this study, we developed a novel mouse model, H11Met/+:PB-Cre4 mice, where expression of the mouse Met gene is specifically activated in prostatic luminal epithelial cells through Cre-LoxP–mediated recombination. We observed mouse prostatic intraepithelial neoplasia development in H11Met/+:PB-Cre4 mice following the administration of HGF but not in control mice that also received HGF treatments. These results represent the first line of evidence directly demonstrating that the activation of the HGF/Met signaling axis can initiate oncogenic transformation and PIN formation in the mouse prostate. In this study, we did not observe more severe pathologic changes in H11Met/+:PB-Cre4 mice, suggesting that the activation of the HGF/Met axis alone is not sufficient to induce PIN lesions to progress to prostate cancer. Although the precise mechanisms for the above observation are currently unclear, other oncogenic hits may be required in collaboratively enhancing Met-induced tumor formation in the mouse prostate. In addition, low and inconsistent levels of HGF have been reported in mice (63). Therefore, the H11Met/+:PB-Cre4 mice will be an experimental tool for further investigating the biological role of Met and other potential hits in prostate cancer initiation.
Multiple lines of evidence have shown the important roles of Met and phosphatidylinositol 3-kinase signaling pathways in prostate cancer progression and metastasis (3). However, the combinatorial effect of these two pathways in prostate tumorigenesis has not been fully evaluated using the biologically relevant mouse model. In this study, we directly addressed this question using newly generated H11Met/+/PtenL/L:PB-Cre4 mice. We observed the development of prostatic invasive carcinomas and metastatic diseases in lung and lymph node tissues in PtenL/L:PB-Cre4 mice as reported previously (30). Intriguingly, H11Met/+/PtenL/L:PB-Cre4 littermates exhibited more aggressive tumor phenotypes in this study (Table 1). In addition to an accelerated HGPIN and prostatic adenocarcinoma development, H11Met/+/PtenL/L:PB-Cre4 compound mice also showed dominant EMT and sarcomatoid carcinoma lesions with invasive carcinomas. Increased metastatic diseases in lungs and lymph nodes at earlier ages were also revealed in the compound mice compared with Pten null mice. All of these data support a promotional role of Met in prostate cancer progression and metastasis. It should be noted that PtenL/L:PB-Cre4 mice in this study developed prostate tumors relatively more slowly than those in the previous report (30). Actually, our above observation on PtenL/L:PB-Cre4 mice was in agreement with a few other studies (26, 64), although the precise reasons for the difference is not clear.
In this study, we assessed the cellular properties of atypical and tumor cells in PIN and cancer lesions in both PtenL/L:PB-Cre4 and H11Met/+/PtenL/L:PB-Cre4 mice, respectively. Interestingly, IHC analyses revealed an increase in cells that are immunoreactive to CK5, a marker of prostatic basal epithelial cells, within both PIN and prostatic adenocarcinoma lesions in H11Met/+/PtenL/L:PB-Cre4 compound mice. Because a modified probasin promoter was used to activate transgenic Met expression in prostatic luminal epithelial cells (20), increasing CK5-positive cells in PIN and tumor lesions suggest that atypical and tumor cells underwent transdifferentiation during the period of tumor development and progression. Development of EMT and sarcomatoid tumor lesions with invasive carcinomas in the compound mice supports the trend of tumor progression in the compound mice. Positive staining of AR in both EMT and sarcomatoid tumor cells further demonstrates their origin of prostatic luminal cells. These lines of evidence implicate a novel role of transgenic Met in enhancing tumor cell transdifferentiation and promoting EMT and tumor progression. In this study, we also examined the role of transgenic Met in cell proliferation and migration. In this set of proof-of-principle experiments, we generated MEFs from either H11Met/+/PtenL/L:CMVCreER+ or PtenL/L:CMVCreER+ embryos. Using MTS and monolayer wound-healing assays, we detected an increase in cellular proliferation and motility in MEFs possessing both transgenic Met expression and Pten deletion. These results are consistent with aggressive tumor phenotypes observed in the prostate of H11Met/+/PtenL/L:PB-Cre4. Taken together, using both in vivo and in vitro experimental approaches, we showed that increasing Met expression enhances cell proliferation and mobility, induces transdifferentiation, and promotes tumor progression and metastasis.
To further understand the molecular mechanism underlying Met-mediated tumor progression and metastasis, we examined the transcriptional profile in prostate tumor tissues isolated from both PtenL/L:PB-Cre4 and H11Met/+/PtenL/L:PB-Cre4 mice using RNA-Seq approaches. Increased expression of mesenchymal markers, such as vimentin, N-cadherin, fibronectin 1, and SPP1, was detected in tumor samples isolated from H11Met/+/PtenL/L:PB-Cre4 mice compared with those from PtenL/L:PB-Cre4 littermates. Identification of a subset of genes related to tumor transdifferentiation and EMT in the samples of H11Met/+/PtenL/L:PB-Cre4 mice is intriguing and provides a molecular basis for transgenic Met-induced tumor progression and metastasis in the compound mice. EMT has mostly been studied with regard to the acquisition of invasive characteristics and metastatic potential (60). Determining the molecular mechanisms by which transgenic Met increases the expression of genes that promote EMT and tumor progression is important and should be carried out in the future.
A significant challenge within the field of prostate cancer has been to develop biologically relevant models for studying metastatic bone lesions (65). The HGF-Met signaling axis is known to be important to bone remodeling; both the ligand and receptor are found to be expressed in the bone marrow and can induce cellular proliferation of both osteoclasts and osteoblasts (66, 67). Furthermore, an increase in expression of both the Met receptor and the HGF ligand has been observed at sites of prostate cancer bone metastasis, suggesting that this pathway may be active during bone metastasis (8). Despite the evidence indicating that c-Met expression is associated with bone metastases in human prostate cancer, we were only able to detect metastasis to the lung and lymphoid nodes in H11Met/+/PtenL/L:PB-Cre4 compound mice as reported in PtenL/L:PB-Cre4 mice (30). Although the reasons why prostate tumor cells with transgenic Met expression are unable to further advance to aggressive disease with bone metastasis in the above compound mice are unclear, data generated from this study have lead us to pursue a few new experiments. Increased activation of Met receptor by the HGF ligand may be necessary to achieve bone metastases using the new mouse model with both transgenic HGF and Met expression. Alternatively, modification of other signaling axes may be necessary for the dissemination of prostate cancer cells to the bones. In support of this hypothesis, a recent clinical trial using cabozantinib, a dual inhibitor of c-Met and VEGFR2, indicated a resolution of bone metastases in 12% of men treated (68). Therefore, development of new mouse models with additional genetic changes in addition to Met activation may be considered in the future.
In this study, we have developed a novel and biologically relevant model for examining the role of c-Met in prostate tumorigenesis. We have shown that c-Met can enhance PTEN-mediated prostate tumor progression by inducing an EMT tumor phenotype and increasing the incidence of metastasis. Given the importance of c-Met in human prostate cancer, we aim to use this model to understand c-Met signaling and its effects during prostate cancer development and progression.
Materials and methods
Generation of the Met transgene target vector
To generate the LSL-Met transgenic mouse line, we used integrase-mediated transgenesis technology (19). In brief, mouse Met cDNA was subcloned into the pB378 vector that contains an attB recombination site. A loxP-PGK-Neomycin-STOP-loxP cassette was inserted between the CMV early enhancer/chicken β-actin (CAG) promoter and a FLAG-tagged mouse Met coding sequence followed by a polyadenylation signal (Fig. 1A). The DNA was purified and microinjected along with φC31 integrase mRNA into zygotic pro-nuclei of an FVB mouse that contains an attP docking site at the H11 locus. The above targeted mice were screened by mouse tail tissue genomic PCR using P1 (5′-AGCGCATCGCCTTCTATCGCCTTC-3′) and P3 (5′-AAACAATCTGGGTGTTCC-3′) primers and further confirmed by DNA sequencing.
Mouse breeding and genotyping
The founder mice were bred with WT C57Bl6/J, and progenies were genotyped to confirm the presence of the transgene. To generate conditional Met transgenic mice, the LSL-Met transgenic mice (H11Met/+) were intercrossed with the PB-Cre4 strain, carrying the Cre transgene under the control of a modified probasin promoter (ARR2PB) (20). Mice homozygous for floxed Pten, PtenL/L, were obtained from the Jackson Laboratory (Bar Harbor, ME; strain 004597). All animals used in this study were on a C57BL/6 background, and all experiments were performed in accordance with animal care guidelines approved by the institutional animal care and use committee at Beckman Research Institute and City of Hope.
For genotyping, mouse tail tips were incubated in lysis buffer (catalog no. 102-T, VIAGEN Biotech, Los Angeles, CA) for genomic DNA. The conditional expression of H11Met/+ was detected with the forward 5′-AGCGCATCGCCTTCTATCGCCTTC-3′ and reverse 5′-AAACAATCTGGGTGTTCC-3′ primers. PCR was performed as follows: 5 min at 94 °C and then 34 cycles of 94 °C for 30 s, 62 °C for 30 s, and 72 °C for 70 s, followed by a final step at 72 °C for 5 min. For Pten allele, the forward 5′-TCCCAGAGTTCATACCAGGA-3′ and reverse 5′-AATCTGTGCATGAAGGGAAC-3′ primers were used to distinguish the WT and floxed alleles (69). For PB-Cre4, the forward 5′-GCAGGAAGCTACTCTGCACCTTG-3′ and reverse 5′-GATCCTGGCAATTTCGGCTAT-3′ primers were used (70).
To assess the genetic recombination of the transgene, genomic DNA samples were isolated from mice prostate, bladder, testicle, kidney, liver, lung, and tail, digested with lysis buffer (100 mm NaCl, 10 mm EDTA, 20 mm Tris, pH 7.5, 0.5% SDS, 0.2 mg/ml proteinase K). PCR was performed with different DNA samples using the primers P1 (5′- AGCGCATCGCCTTCTATCGCCTTC-3′), P2 (5′-TTCGGCTTCTGGCGTGTGAC-3′), and P3 (5′-AAACAATCTGGGTGTTCC-3′). Genomic DNA fragments from H11Met/+:PB-Cre4 mice were amplified at 94 °C for 5 min and then 36 cycles of 94 °C for 45 s, 58 °C for 30 s, and 72 °C for 60 s, followed by 72 °C for 5 min.
Mouse procedures
For HGF injection, recombinant human HGF (294-HG/CF, R&D Systems, Minneapolis, MN) was dissolved in sterile PBS at 100 μg/ml and stored at −80 °C. Three doses of HGF were given to 6-month-old H11Met/+:PB-Cre4 mice at 0.5 μg/g body weight via intraperitoneal injection for 3 consecutive weeks. Tissues were harvested 120 days after the initial HGF injection.
Immunoprecipitation and Western blotting
Prostates from H11Met/+ and H11Met/+:PB-Cre4 mice were cut into small pieces, homogenized, and used for protein collection with radioimmune precipitation buffer (0.5% Nonidet P-40, 0.3% Triton X-100, 15 mm MgCl2, 5 mm EDTA, 150 mm NaCl, 50 mm Tris-HCl, pH 7.8) as described previously (70, 71). Whole-cell lysates were precleared for 20 min with 10 μl of protein A/G–agarose beads (Pierce, Waltham, MA) and then incubated with pre-equilibrated anti-FLAG M2 magnetic beads (M8823, Sigma-Aldrich) at 4 °C with gentle rotation overnight. The beads were washed three times in lysis buffer, eluted by boiling in SDS-sample buffer, and then resolved on an 8% SDS-polyacrylamide gel. The proteins were transferred onto a nitrocellulose membrane and probed with anti-Met antibody (sc-162, Santa Cruz Biotechnology, Inc.; 1:50). Detection was performed with ECL reagents (Amersham Biosciences).
Histological analyses and immunostaining
Mouse tissues were fixed and processed as described in our previous study (72). For histological analysis, 5-μm serial sections were processed from Clearify (American MasterTech (Lodi, CA)) to water through a decreasing ethanol gradient, stained with 5% (w/v) Harris hematoxylin and eosin, and processed back to Clearify through an increasing ethanol gradient.
For immunohistochemical assays, 5-μm sections were boiled in 0.01 m citrate buffer (pH 6.0) for 20 min after redehydration from Clearify to water, placed in 0.3% H2O2/methanol for 15 min, and blocked by 5% goat serum or 5% donkey serum. Tissue slides were then exposed to first antibodies in PBS with 1% goat (or donkey serum) at 4 °C overnight. The following dilutions were used: 1:150 dilution of anti-MET (AF527, R&D Systems), 1:100 anti-phospho-MET (3077, Cell Signaling, Danvers, MA), 1:1000 anti-mouse/human AR (PA5-16750, Invitrogen), 1:200 anti-E-cadherin (c20820, BD Transduction Laboratories), 1:2000 anti-CK8 (MMS-162P, Covance, Brea, CA), 1:2600 anti-CK5 (PRB-160P, Covance), 1:75 anti-synaptophysin (08-1130, Invitrogen), 1:2000 anti-p63 (sc-8431, Santa Cruz Biotechnology), 1:400 anti-ki67 (D3B5, Cell Signaling), 1:2500 anti-vimentin (919101, Biolegend, San Diego, CA), 1:100 anti-fibronectin 1 (MA5-11981, Invitrogen), 1:200 anti-SPP1 (91655, Abcam, Cambridge, MA), 1:100 anti-N-cadherin (33-3900, Invitrogen), 1:200 anti-phospho-ERK1/2 (D13.14.3E, Cell Signaling). Tissues were then incubated with biotinylated goat anti-mouse, goat anti-rabbit (Vector Laboratories, BA-1000 or BA-9200) or donkey anti-goat (ab6987, Abcam) at 1:750 dilution for 1 h at room temperature followed by a 45-min incubation with horseradish peroxidase–conjugated streptavidin (Vector Laboratories, SA-5004). Immunostainings were visualized using a DAB kit (Vector Laboratories, SK-4100). Slides were counterstained with hematoxylin, and coverslips were mounted with Permount Mounting Medium (SP15-500, Fisher).
For immunofluorescent staining, mouse tissues were cut into 5-μm sections and then boiled in 0.01 m citrate buffer (pH 6.0) for 20 min after redehydration, blocked in 5% normal goat serum for 30 min, and incubated with primary antibodies diluted in 1% normal goat serum at 4 °C overnight. Slides were washed in PBS and then incubated with fluorescence-conjugated secondary antibodies for 1 h and mounted with VECTASHIELD Mounting Medium with 4′,6-diamidino-2-phenylindole (H-1200, Vector Laboratories).
Mouse embryonic fibroblasts, MTS cell viability assay, and wound-healing assay
H11Met/+/PtenL/L females were mated with PtenL/L:CMV CreER+ males. Pregnant females received two intraperitoneal injections of tamoxifen (Sigma) at E11.5 and E12.5 of 2 mg/25 g body weight and were sacrificed at E13.5. The embryos were isolated in cold PBS and then digested with 0.25% trypsin for 30 min at 37 °C, followed by the addition of Dulbecco's modified Eagle's medium containing 10% FBS and 1% penicillin/streptomycin. The cells were directly plated into 6-well plates and allowed to adhere overnight. To determine the genotypes, embryo yolk sacs isolated during the dissection were digested, and genomic DNA was extracted (73, 74) and used for genotyping with appropriate primers as mentioned above. For CMVCreER, the forward 5′-TTGCCTGCATTACCGGTCGATGCA-3′ and reverse 5′-GATCCTGGCAATTTCGGCTAT-3′ primers were used with the program listed as follows: 94 °C for 5 min and then 29 cycles of 94 °C for 20 s, 64 °C for 30 s, and 72 °C for 25 s, followed by 72 °C for 2 min.
For the MTS cell viability assay, ∼2000 cells/well were plated in 96-well plate and then harvested at different time points. Cell growth assays were carried out using an MTS cell proliferation assay kit (G5430, Promega, Madison, WI). Cell numbers were determined by absorbance at 490 nm, as suggested by the manufacturer.
For the wound-healing assay, 2 × 105 cells/well were seeded into a 12-well plate and cultured in the absence or presence of 10 nm recombinant human HGF to 90% confluence. The cells were then starved with 0.1% FBS overnight and scratched with a sterile 200-μl tip to form a straight wound. The cells were washed three times with PBS and cultured in normal medium for an additional 24 h. A Nikon ECLIPSE E800 microscope was used to measure the wound closure. Images were recorded at 0, 12, and 24 h after wounding with QImaging RETIGA EXi camera and QCapture software (QImaging, Surrey, Canada). The distances invaded by the cells at the front of the wound were measured from the control and the experimental samples.
RNA isolation, RNA-Seq, and qRT-PCR
RNA samples were isolated from age-matched mice of different genotypes. The prostate tissues were homogenized in RNA-Bee (TEL-TEST, Inc., Friendswood, TX), and total RNA was isolated as recommended by the manufacturer. The purified RNA libraries were then sequenced using the Illumina HiSeq 2000 at the City of Hope Integrative Genomics Core.
RT was carried out as described in our previous report (75). For quantitative PCR, cDNA samples were mixed with SYBR GreenER qPCR Super Mix Universal (11762, Invitrogen) and specific primers, and quantitative PCR was performed according to the manufacturer's protocol. Relative mRNA levels were calculated by the ΔΔC(T) method (76). Reactions were done in triplicate, and the values were normalized by Ppia (peptidylprolyl isomerase A) expression levels. Primers for Met (5′-CTGGTGCCCTTACACTAAAC-3′; 5′-GAGACCTTCCCTCACTTAGATA-3′), Hgf (5′-GATGAACTCCAGGGCATAATC-3′; 5′-CAAGCTGCATCCTCCTATTC-3′), Spp1 (5′-ATCTCACCATTCGGATGAGTCT-3′; 5′-TGTAGGGACGATTGGAGTGAAA-3′) (77), Fn1 (5′-GTGACACTTATGAGCGCCCTA-3′; 5′-CCACTTGTCGCCAATCTTGTA-3′) (78), Cdh2 (5′-CCAGCAGATTTCAAGGTGGAC-3′; 5′-TTACAGCTACCTGCCACTTTTC-3′) (79), Zeb2 (5′-CCACGCAGTGAGCATCGAA-3′; 5′-CAGGTGGCAGGTCATTTTCTT-3′) (80), Vegf-c (5′-AGACGGACACACATGGAGGT-3′; 5′-AAAGACTCAATGCATGCCAC-3′) (81), and Ppia (5′-TGTGCCAGGGTGGTGACTTT-3′; 5′-CGTTTGTGTTTGGTCCAGCAT-3′) (82) were synthesized and used in the qPCRs, respectively.
Statistical analyses
Data are shown as the mean ± S.D. Differences between groups were examined by two-tailed Student's t test or two-way analysis of variance for comparisons among multiple groups. For all analyses, p < 0.05 was considered statistically significant.
Author contributions
J. M., E. H., and Z. S. conceptualization; J. M., E. H., H. Z., H. W., D.-H. L., M. L. G., and Z. S. data curation; J. M., E. H., H. W., and Z. S. formal analysis; J. M., E. H., V. L., and Z. S. validation; J. M., E. H., S. B., D. T. J., Y. H., E.-J. Y., H. W., D.-H. L., and Z. S. investigation; J. M. and Z. S. visualization; J. M., S. B., H. Z., D. T. J., Y. H., E.-J. Y., V. L., D.-H. L., J. A., and Z. S. methodology; J. M., E. H., and Z. S. writing-original draft; J. M., E. H., J. A., and Z. S. writing-review and editing; S. B., H. Z., Y. H., E.-J. Y., V. L., J. A., M. L. G., and Z. S. resources; D. T. J. and Z. S. project administration; M. L. G. and Z. S. funding acquisition; Z. S. supervision.
Supplementary Material
This work was supported in part by National Institutes of Health Grants R01CA070297, R01CA151623, R01CA166894, R21CA190021, and R01DK104941. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Fig. S1.
- CRPC
- castration-resistant prostate cancer
- HGF
- hepatocyte growth factor
- AR
- androgen receptor
- PB
- probasin
- PIN
- prostate intraepithelial neoplasia
- HGPIN and LGPIN
- high-grade and low-grade PIN, respectively
- CK
- cytokeratin
- EMT
- epithelial-to-mesenchymal transition
- RT
- reverse transcription
- qRT-PCR
- quantitative RT-PCR
- qPCR
- quantitative PCR
- IHC
- immunohistochemical
- Vim
- vimentin
- MEF
- mouse embryo fibroblast
- CMV
- cytomegalovirus
- E
- embryonic day
- H&E
- hematoxylin and eosin.
References
- 1. American Cancer Society (2017) Cancer Facts & Figures 2017, American Cancer Society, Atlanta [Google Scholar]
- 2. Jemal A., Bray F., Center M. M., Ferlay J., Ward E., and Forman D. (2011) Global cancer statistics. CA Cancer J. Clin. 61, 69–90; Correction (2011) CA Cancer J. Clin.61, 134 10.3322/caac.20107 [DOI] [PubMed] [Google Scholar]
- 3. Matsumoto K., and Nakamura T. (1996) Emerging multipotent aspects of hepatocyte growth factor. J. Biochem. 119, 591–600 10.1093/oxfordjournals.jbchem.a021283 [DOI] [PubMed] [Google Scholar]
- 4. Humphrey P. A., Zhu X., Zarnegar R., Swanson P. E., Ratliff T. L., Vollmer R. T., and Day M. L. (1995) Hepatocyte growth factor and its receptor (c-MET) in prostatic carcinoma. Am. J. Pathol. 147, 386–396 [PMC free article] [PubMed] [Google Scholar]
- 5. Kasai S., Sugimura K., Matsumoto K., Nishi N., Kishimoto T., and Nakamura T. (1996) Hepatocyte growth factor is a paracrine regulator of rat prostate epithelial growth. Biochem. Biophys. Res. Commun. 228, 646–652 10.1006/bbrc.1996.1710 [DOI] [PubMed] [Google Scholar]
- 6. Pisters L. L., Troncoso P., Zhau H. E., Li W., von Eschenbach A. C., and Chung L. W. (1995) c-met proto-oncogene expression in benign and malignant human prostate tissues. J. Urol. 154, 293–298 10.1016/S0022-5347(01)67297-5 [DOI] [PubMed] [Google Scholar]
- 7. Birchmeier C., Birchmeier W., Gherardi E., and Vande Woude G. F. (2003) Met, metastasis, motility and more. Nat. Rev. Mol. Cell Biol. 4, 915–925 10.1038/nrm1261 [DOI] [PubMed] [Google Scholar]
- 8. Knudsen B. S., Gmyrek G. A., Inra J., Scherr D. S., Vaughan E. D., Nanus D. M., Kattan M. W., Gerald W. L., and Vande Woude G. F. (2002) High expression of the Met receptor in prostate cancer metastasis to bone. Urology 60, 1113–1117 10.1016/S0090-4295(02)01954-4 [DOI] [PubMed] [Google Scholar]
- 9. Watanabe M., Fukutome K., Kato H., Murata M., Kawamura J., Shiraishi T., and Yatani R. (1999) Progression-linked overexpression of c-Met in prostatic intraepithelial neoplasia and latent as well as clinical prostate cancers. Cancer Lett. 141, 173–178 10.1016/S0304-3835(99)00102-0 [DOI] [PubMed] [Google Scholar]
- 10. van Leenders G. J., Sookhlall R., Teubel W. J., de Ridder C. M., Reneman S., Sacchetti A., Vissers K. J., van Weerden W., and Jenster G. (2011) Activation of c-MET induces a stem-like phenotype in human prostate cancer. PLoS One 6, e26753 10.1371/journal.pone.0026753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Russo A. L., Jedlicka K., Wernick M., McNally D., Kirk M., Sproull M., Smith S., Shankavaram U., Kaushal A., Figg W. D., Dahut W., Citrin D., Bottaro D. P., Albert P. S., Tofilon P. J., and Camphausen K. (2009) Urine analysis and protein networking identify met as a marker of metastatic prostate cancer. Clin. Cancer Res. 15, 4292–4298 10.1158/1078-0432.CCR-09-0599 [DOI] [PubMed] [Google Scholar]
- 12. Schalken J. A., and van Leenders G. (2003) Cellular and molecular biology of the prostate: stem cell biology. Urology 62, 11–20 10.1016/S0090-4295(03)00758-1 [DOI] [PubMed] [Google Scholar]
- 13. van Leenders G., van Balken B., Aalders T., Hulsbergen-van de Kaa C., Ruiter D., and Schalken J. (2002) Intermediate cells in normal and malignant prostate epithelium express c-MET: implications for prostate cancer invasion. Prostate 51, 98–107 10.1002/pros.10073 [DOI] [PubMed] [Google Scholar]
- 14. Hu P., Chu G. C., Zhu G., Yang H., Luthringer D., Prins G., Habib F., Wang Y., Wang R., Chung L. W., and Zhau H. E. (2011) Multiplexed quantum dot labeling of activated c-Met signaling in castration-resistant human prostate cancer. PLoS One 6, e28670 10.1371/journal.pone.0028670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nishi N., Oya H., Matsumoto K., Nakamura T., Miyanaka H., and Wada F. (1996) Changes in gene expression of growth factors and their receptors during castration-induced involution and androgen-induced regrowth of rat prostates. Prostate 28, 139–152 10.1002/(SICI)1097-0045(199603)28:3%3C139::AID-PROS1%3E3.0.CO%3B2-A [DOI] [PubMed] [Google Scholar]
- 16. Verras M., Lee J., Xue H., Li T. H., Wang Y., and Sun Z. (2007) The androgen receptor negatively regulates the expression of c-Met: implications for a novel mechanism of prostate cancer progression. Cancer Res. 67, 967–975 10.1158/0008-5472.CAN-06-3552 [DOI] [PubMed] [Google Scholar]
- 17. Gupta S., Li J., Kemeny G., Bitting R. L., Beaver J., Somarelli J. A., Ware K. E., Gregory S., and Armstrong A. J. (2017) Whole genomic copy number alterations in circulating tumor cells from men with abiraterone or enzalutamide-resistant metastatic castration-resistant prostate cancer. Clin. Cancer Res. 23, 1346–1357 10.1158/1078-0432.CCR-16-1211 [DOI] [PubMed] [Google Scholar]
- 18. Cannistraci A., Federici G., Addario A., Di Pace A. L., Grassi L., Muto G., Collura D., Signore M., De Salvo L., Sentinelli S., Simone G., Costantini M., Nanni S., Farsetti A., Coppola V., et al. (2017) C-Met/miR-130b axis as novel mechanism and biomarker for castration resistance state acquisition. Oncogene 36, 3718–3728 10.1038/onc.2016.505 [DOI] [PubMed] [Google Scholar]
- 19. Tasic B., Hippenmeyer S., Wang C., Gamboa M., Zong H., Chen-Tsai Y., and Luo L. (2011) Site-specific integrase-mediated transgenesis in mice via pronuclear injection. Proc. Natl. Acad. Sci. U.S.A. 108, 7902–7907 10.1073/pnas.1019507108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu X., Wu J., Huang J., Powell W. C., Zhang J., Matusik R. J., Sangiorgi F. O., Maxson R. E., Sucov H. M., and Roy-Burman P. (2001) Generation of a prostate epithelial cell-specific Cre transgenic mouse model for tissue-specific gene ablation. Mech. Dev. 101, 61–69 10.1016/S0925-4773(00)00551-7 [DOI] [PubMed] [Google Scholar]
- 21. Alexopoulou A. N., Couchman J. R., and Whiteford J. R. (2008) The CMV early enhancer/chicken β actin (CAG) promoter can be used to drive transgene expression during the differentiation of murine embryonic stem cells into vascular progenitors. BMC Cell Biol. 9, 2 10.1186/1471-2121-9-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kosuga M., Enosawa S., Li X. K., Suzuki S., Matsuo N., Yamada M., Roy-Chowdhury J., Koiwai O., and Okuyama T. (2000) Strong, long-term transgene expression in rat liver using chicken β-actin promoter associated with cytomegalovirus immediate-early enhancer (CAG promoter). Cell Transplant. 9, 675–680 10.1177/096368970000900513 [DOI] [PubMed] [Google Scholar]
- 23. Okabe M., Ikawa M., Kominami K., Nakanishi T., and Nishimune Y. (1997) “Green mice” as a source of ubiquitous green cells. FEBS Lett. 407, 313–319 10.1016/S0014-5793(97)00313-X [DOI] [PubMed] [Google Scholar]
- 24. Ittmann M., Huang J., Radaelli E., Martin P., Signoretti S., Sullivan R., Simons B. W., Ward J. M., Robinson B. D., Chu G. C., Loda M., Thomas G., Borowsky A., and Cardiff R. D. (2013) Animal models of human prostate cancer: the consensus report of the New York meeting of the Mouse Models of Human Cancers Consortium Prostate Pathology Committee. Cancer Res. 73, 2718–2736 10.1158/0008-5472.CAN-12-4213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Armenia J., Wankowicz S. A. M., Liu D., Gao J., Kundra R., Reznik E., Chatila W. K., Chakravarty D., Han G. C., Coleman I., Montgomery B., Pritchard C., Morrissey C., Barbieri C. E., Beltran H., et al. (2018) The long tail of oncogenic drivers in prostate cancer. Nat. Genet. 50, 645–651 10.1038/s41588-018-0078-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Su Q., Zhang B., Zhang L., Dang T., Rowley D., Ittmann M., and Xin L. (2017) Jagged1 upregulation in prostate epithelial cells promotes formation of reactive stroma in the Pten null mouse model for prostate cancer. Oncogene 36, 618–627 10.1038/onc.2016.232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sung C. O., Choi H., Lee K. W., and Kim S. H. (2013) Sarcomatoid carcinoma represents a complete phenotype with various pathways of epithelial mesenchymal transition. J. Clin. Pathol. 66, 601–606 10.1136/jclinpath-2012-201271 [DOI] [PubMed] [Google Scholar]
- 28. de Reyniès A., Jaurand M. C., Renier A., Couchy G., Hysi I., Elarouci N., Galateau-Sallé F., Copin M. C., Hofman P., Cazes A., Andujar P., Imbeaud S., Petel F., Pairon J. C., Le Pimpec-Barthes F., et al. (2014) Molecular classification of malignant pleural mesothelioma: identification of a poor prognosis subgroup linked to the epithelial-to-mesenchymal transition. Clin. Cancer Res. 20, 1323–1334 10.1158/1078-0432.CCR-13-2429 [DOI] [PubMed] [Google Scholar]
- 29. Conant J. L., Peng Z., Evans M. F., Naud S., and Cooper K. (2011) Sarcomatoid renal cell carcinoma is an example of epithelial–mesenchymal transition. J. Clin. Pathol. 64, 1088–1092 10.1136/jclinpath-2011-200216 [DOI] [PubMed] [Google Scholar]
- 30. Wang S., Gao J., Lei Q., Rozengurt N., Pritchard C., Jiao J., Thomas G. V., Li G., Roy-Burman P., Nelson P. S., Liu X., and Wu H. (2003) Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell 4, 209–221 10.1016/S1535-6108(03)00215-0 [DOI] [PubMed] [Google Scholar]
- 31. Mulholland D. J., Kobayashi N., Ruscetti M., Zhi A., Tran L. M., Huang J., Gleave M., and Wu H. (2012) Pten loss and RAS/MAPK activation cooperate to promote EMT and metastasis initiated from prostate cancer stem/progenitor cells. Cancer Res. 72, 1878–1889 10.1158/0008-5472.CAN-11-3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ding Z., Wu C. J., Chu G. C., Xiao Y., Ho D., Zhang J., Perry S. R., Labrot E. S., Wu X., Lis R., Hoshida Y., Hiller D., Hu B., Jiang S., Zheng H., et al. (2011) SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature 470, 269–273 10.1038/nature09677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kozakowski N., Hartmann C., Klingler H. C., Susani M., Mazal P. R., Scharrer A., and Haitel A. (2014) Immunohistochemical expression of PDGFR, VEGF-C, and proteins of the mToR pathway before and after androgen deprivation therapy in prostate carcinoma: significant decrease after treatment. Target Oncol. 9, 359–366 10.1007/s11523-013-0298-1 [DOI] [PubMed] [Google Scholar]
- 34. Rao D. S., Hyun T. S., Kumar P. D., Mizukami I. F., Rubin M. A., Lucas P. C., Sanda M. G., and Ross T. S. (2002) Huntingtin-interacting protein 1 is overexpressed in prostate and colon cancer and is critical for cellular survival. J. Clin. Invest. 110, 351–360 10.1172/JCI0215529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Barbieri A., Bimonte S., Palma G., Luciano A., Rea D., Giudice A., Scognamiglio G., La Mantia E., Franco R., Perdonà S., De Cobelli O., Ferro M., Zappavigna S., Stiuso P., Caraglia M., and Arra C. (2015) The stress hormone norepinephrine increases migration of prostate cancer cells in vitro and in vivo. Int. J. Oncol. 47, 527–534 10.3892/ijo.2015.3038 [DOI] [PubMed] [Google Scholar]
- 36. De Marzo A. M., Platz E. A., Sutcliffe S., Xu J., Grönberg H., Drake C. G., Nakai Y., Isaacs W. B., and Nelson W. G. (2007) Inflammation in prostate carcinogenesis. Nat. Rev. Cancer 7, 256–269 10.1038/nrc2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Singh V., Singh L. C., Vasudevan M., Chattopadhyay I., Borthakar B. B., Rai A. K., Phukan R. K., Sharma J., Mahanta J., Kataki A. C., Kapur S., and Saxena S. (2015) Esophageal cancer epigenomics and integrome analysis of genome-wide methylation and expression in high risk Northeast Indian population. OMICS 19, 688–699 10.1089/omi.2015.0121 [DOI] [PubMed] [Google Scholar]
- 38. Vadnais C., Shooshtarizadeh P., Rajadurai C. V., Lesurf R., Hulea L., Davoudi S., Cadieux C., Hallett M., Park M., and Nepveu A. (2014) Autocrine activation of the Wnt/β-catenin pathway by CUX1 and GLIS1 in breast cancers. Biol. Open 3, 937–946 10.1242/bio.20148193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bischof P., and Mégevand M. (1986) Pregnancy-associated plasma protein-A concentrations in men with testicular and prostatic tumors. Arch. Androl. 16, 155–160 10.3109/01485018608986936 [DOI] [PubMed] [Google Scholar]
- 40. Chen C., Zhang Q., Liu S., Parajuli K. R., Qu Y., Mei J., Chen Z., Zhang H., Khismatullin D. B., and You Z. (2015) IL-17 and insulin/IGF1 enhance adhesion of prostate cancer cells to vascular endothelial cells through CD44-VCAM-1 interaction. Prostate 75, 883–895 10.1002/pros.22971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bhardwaj A., Rao M. K., Kaur R., Buttigieg M. R., and Wilkinson M. F. (2008) GATA factors and androgen receptor collaborate to transcriptionally activate the Rhox5 homeobox gene in Sertoli cells. Mol. Cell. Biol. 28, 2138–2153 10.1128/MCB.01170-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stucke V. M., Timmerman E., Vandekerckhove J., Gevaert K., and Hall A. (2007) The MAGUK protein MPP7 binds to the polarity protein hDlg1 and facilitates epithelial tight junction formation. Mol. Biol. Cell 18, 1744–1755 10.1091/mbc.e06-11-0980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hahn-Strömberg V., Askari S., Befekadu R., Matthiessen P., Karlsson S., and Nilsson T. K. (2014) Polymorphisms in the CLDN1 and CLDN7 genes are related to differentiation and tumor stage in colon carcinoma. APMIS 122, 636–642 10.1111/apm.12211 [DOI] [PubMed] [Google Scholar]
- 44. Chang W. C., Chou C. K., Tsou C. C., Li S. H., Chen C. H., Zhuo Y. X., Hsu W. L., and Chen C. H. (2010) Comparative proteomic analysis of proteins involved in the tumorigenic process of seminal vesicle carcinoma in transgenic mice. Int. J. Proteomics 2010, 726968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee J. W., Jeong E. G., Lee S. H., Nam S. W., Kim S. H., Lee J. Y., Yoo N. J., and Lee S. H. (2007) Mutational analysis of PTPRT phosphatase domains in common human cancers. APMIS 115, 47–51 10.1111/j.1600-0463.2007.apm_554.x [DOI] [PubMed] [Google Scholar]
- 46. Sathyanarayana U. G., Padar A., Suzuki M., Maruyama R., Shigematsu H., Hsieh J. T., Frenkel E. P., and Gazdar A. F. (2003) Aberrant promoter methylation of laminin-5-encoding genes in prostate cancers and its relationship to clinicopathological features. Clin. Cancer Res. 9, 6395–6400 [PubMed] [Google Scholar]
- 47. Katoh M., and Katoh M. (2003) CLDN23 gene, frequently down-regulated in intestinal-type gastric cancer, is a novel member of CLAUDIN gene family. Int. J. Mol. Med. 11, 683–689 [PubMed] [Google Scholar]
- 48. Saeki N., Usui T., Aoyagi K., Kim D. H., Sato M., Mabuchi T., Yanagihara K., Ogawa K., Sakamoto H., Yoshida T., and Sasaki H. (2009) Distinctive expression and function of four GSDM family genes (GSDMA-D) in normal and malignant upper gastrointestinal epithelium. Genes Chromosomes Cancer 48, 261–271 10.1002/gcc.20636 [DOI] [PubMed] [Google Scholar]
- 49. Kim J. H., Kim T. W., and Kim S. J. (2011) Downregulation of ARFGEF1 and CAMK2B by promoter hypermethylation in breast cancer cells. BMB Rep. 44, 523–528 10.5483/BMBRep.2011.44.8.523 [DOI] [PubMed] [Google Scholar]
- 50. Kurashige J., Sawada G., Takahashi Y., Eguchi H., Sudo T., Ikegami T., Yoshizumi T., Soejima Y., Ikeda T., Kawanaka H., Uchiyama H., Yamashita Y., Morita M., Oki E., Saeki H., et al. (2013) Suppression of MAL gene expression in gastric cancer correlates with metastasis and mortality. Fukuoka Igaku Zasshi 104, 344–349 [PubMed] [Google Scholar]
- 51. Sasnauskienė A., Jonušienė V., Krikštaponienė A., Butkytė S., Dabkevičienė D., Kanopienė D., Kazbarienė B., and Didžiapetrienė J. (2014) NOTCH1, NOTCH3, NOTCH4, and JAG2 protein levels in human endometrial cancer. Medicina (Kaunas) 50, 14–18 10.1016/j.medici.2014.05.002 [DOI] [PubMed] [Google Scholar]
- 52. Xie L., Qin W. X., He X. H., Shu H. Q., Yao G. F., Wan D. F., and Gu J. R. (2004) Differential gene expression in human hepatocellular carcinoma Hep3B cells induced by apoptosis-related gene BNIPL-2. World J. Gastroenterol. 10, 1286–1291 10.3748/wjg.v10.i9.1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chimento A., Sirianni R., Casaburi I., Ruggiero C., Maggiolini M., Andò S., and Pezzi V. (2012) 17β-Estradiol activates GPER- and ESR1-dependent pathways inducing apoptosis in GC-2 cells, a mouse spermatocyte-derived cell line. Mol. Cell. Endocrinol. 355, 49–59 10.1016/j.mce.2012.01.017 [DOI] [PubMed] [Google Scholar]
- 54. Chen K., Luo Z., Li Z., Liu Y., and Zhao Q. (2011) PERP gene therapy attenuates lung cancer xenograft via inducing apoptosis and suppressing VEGF. Cancer Biol. Ther. 12, 1114–1119 10.4161/cbt.12.12.18435 [DOI] [PubMed] [Google Scholar]
- 55. Thiery J. P. (2002) Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2, 442–454 10.1038/nrc822 [DOI] [PubMed] [Google Scholar]
- 56. Lamouille S., Xu J., and Derynck R. (2014) Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15, 178–196 10.1038/nrm3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Liu G. L., Yang H. J., Liu T., and Lin Y. Z. (2014) Expression and significance of E-cadherin, N-cadherin, transforming growth factor-β1 and Twist in prostate cancer. Asian Pac. J. Trop. Med. 7, 76–82 [DOI] [PubMed] [Google Scholar]
- 58. Korpal M., Lee E. S., Hu G., and Kang Y. (2008) The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 283, 14910–14914 10.1074/jbc.C800074200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Xu C., Sun L., Jiang C., Zhou H., Gu L., Liu Y., and Xu Q. (2017) SPP1, analyzed by bioinformatics methods, promotes the metastasis in colorectal cancer by activating EMT pathway. Biomed. Pharmacother. 91, 1167–1177 10.1016/j.biopha.2017.05.056 [DOI] [PubMed] [Google Scholar]
- 60. Nauseef J. T., and Henry M. D. (2011) Epithelial-to-mesenchymal transition in prostate cancer: paradigm or puzzle? Nat. Rev. Urol. 8, 428–439 10.1038/nrurol.2011.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yan G., Chen V., Lu X., and Lu S. (2017) A signal-based method for finding driver modules of breast cancer metastasis to the lung. Sci. Rep. 7, 10023 10.1038/s41598-017-09951-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Varkaris A., Corn P. G., Gaur S., Dayyani F., Logothetis C. J., and Gallick G. E. (2011) The role of HGF/c-Met signaling in prostate cancer progression and c-Met inhibitors in clinical trials. Expert Opin. Investig. Drugs 20, 1677–1684 10.1517/13543784.2011.631523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jeffers M., Rong S., and Vande Woude G. F. (1996) Hepatocyte growth factor/scatter factor-Met signaling in tumorigenicity and invasion/metastasis. J. Mol. Med. (Berl.) 74, 505–513 10.1007/BF00204976 [DOI] [PubMed] [Google Scholar]
- 64. Adissu H. A., McKerlie C., Di Grappa M., Waterhouse P., Xu Q., Fang H., Khokha R., and Wood G. A. (2015) Timp3 loss accelerates tumour invasion and increases prostate inflammation in a mouse model of prostate cancer. Prostate 75, 1831–1843 10.1002/pros.23056 [DOI] [PubMed] [Google Scholar]
- 65. Berish R. B., Ali A. N., Telmer P. G., Ronald J. A., and Leong H. S. (2018) Translational models of prostate cancer bone metastasis. Nat. Rev. Urol. 15, 403–421 10.1038/s41585-018-0020-2 [DOI] [PubMed] [Google Scholar]
- 66. Boccaccio C., and Comoglio P. M. (2006) Invasive growth: a MET-driven genetic programme for cancer and stem cells. Nat. Rev. Cancer 6, 637–645 10.1038/nrc1912 [DOI] [PubMed] [Google Scholar]
- 67. Grano M., Galimi F., Zambonin G., Colucci S., Cottone E., Zallone A. Z., and Comoglio P. M. (1996) Hepatocyte growth factor is a coupling factor for osteoclasts and osteoblasts in vitro. Proc. Natl. Acad. Sci. U.S.A. 93, 7644–7648 10.1073/pnas.93.15.7644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Daudigeos-Dubus E., Le Dret L., Bawa O., Opolon P., Vievard A., Villa I., Bosq J., Vassal G., and Geoerger B. (2017) Dual inhibition using cabozantinib overcomes HGF/MET signaling mediated resistance to pan-VEGFR inhibition in orthotopic and metastatic neuroblastoma tumors. Int. J. Oncol. 50, 203–211 10.3892/ijo.2016.3792 [DOI] [PubMed] [Google Scholar]
- 69. Kwak M. K., Johnson D. T., Zhu C., Lee S. H., Ye D. W., Luong R., and Sun Z. (2013) Conditional deletion of the Pten gene in the mouse prostate induces prostatic intraepithelial neoplasms at early ages but a slow progression to prostate tumors. PLoS One 8, e53476 10.1371/journal.pone.0053476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yu E. J., Hooker E., Johnson D. T., Kwak M. K., Zou K., Luong R., He Y., and Sun Z. (2017) LZTS2 and PTEN collaboratively regulate ss-catenin in prostatic tumorigenesis. PLoS One 12, e0174357 10.1371/journal.pone.0174357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sharma M., Chuang W. W., and Sun Z. (2002) Phosphatidylinositol 3-kinase/Akt stimulates androgen pathway through GSK3β inhibition and nuclear β-catenin accumulation. J. Biol. Chem. 277, 30935–30941 10.1074/jbc.M201919200 [DOI] [PubMed] [Google Scholar]
- 72. Zhu C., Luong R., Zhuo M., Johnson D. T., McKenney J. K., Cunha G. R., and Sun Z. (2011) Conditional expression of the androgen receptor induces oncogenic transformation of the mouse prostate. J. Biol. Chem. 286, 33478–33488 10.1074/jbc.M111.269894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Beliakoff J., Lee J., Ueno H., Aiyer A., Weissman I. L., Barsh G. S., Cardiff R. D., and Sun Z. (2008) The PIAS-like protein Zimp10 is essential for embryonic viability and proper vascular development. Mol. Cell. Biol. 28, 282–292 10.1128/MCB.00771-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Peng Y., Clark C., Luong R., Tu W. H., Lee J., Johnson D. T., Das A., Carroll T. J., and Sun Z. (2011) The leucine zipper putative tumor suppressor 2 protein LZTS2 regulates kidney development. J. Biol. Chem. 286, 40331–40342 10.1074/jbc.M111.302059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hohaus S., Petrovick M. S., Voso M. T., Sun Z., Zhang D. E., and Tenen D. G. (1995) PU.1 (Spi-1) and C/EBP α regulate expression of the granulocyte-macrophage colony-stimulating factor receptor α gene. Mol. Cell. Biol. 15, 5830–5845 10.1128/MCB.15.10.5830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Livak K. J., and Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔC(T) method. Methods 25, 402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 77. Lee S. H., Luong R., Johnson D. T., Cunha G. R., Rivina L., Gonzalgo M. L., and Sun Z. (2016) Androgen signaling is a confounding factor for β-catenin-mediated prostate tumorigenesis. Oncogene 35, 702–714 10.1038/onc.2015.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Das D. K., Naidoo M., Ilboudo A., Park J. Y., Ali T., Krampis K., Robinson B. D., Osborne J. R., and Ogunwobi O. O. (2016) miR-1207-3p regulates the androgen receptor in prostate cancer via FNDC1/fibronectin. Exp. Cell Res. 348, 190–200 10.1016/j.yexcr.2016.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Nalla A. K., Estes N., Patel J., and Rao J. S. (2011) N-cadherin mediates angiogenesis by regulating monocyte chemoattractant protein-1 expression via PI3K/Akt signaling in prostate cancer cells. Exp. Cell Res. 317, 2512–2521 10.1016/j.yexcr.2011.07.024 [DOI] [PubMed] [Google Scholar]
- 80. Hu Z., and Wang J. (2014) Histone deacetylase inhibitor induces the expression of select epithelial genes in mouse utricle sensory epithelia-derived progenitor cells. Cell Reprogram 16, 266–275 10.1089/cell.2013.0086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lee A. S., Lee J. E., Jung Y. J., Kim D. H., Kang K. P., Lee S., Park S. K., Lee S. Y., Kang M. J., Moon W. S., Kim H. J., Jeong Y. B., Sung M. J., and Kim W. (2013) Vascular endothelial growth factor-C and -D are involved in lymphangiogenesis in mouse unilateral ureteral obstruction. Kidney Int. 83, 50–62 10.1038/ki.2012.312 [DOI] [PubMed] [Google Scholar]
- 82. Gong Z. K., Wang S. J., Huang Y. Q., Zhao R. Q., Zhu Q. F., and Lin W. Z. (2014) Identification and validation of suitable reference genes for RT-qPCR analysis in mouse testis development. Mol. Genet. Genomics 289, 1157–1169 10.1007/s00438-014-0877-6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.