Abstract
Nuclear architecture is decisive for the assembly of transcriptional responses. However, how chromosome organization is dynamically modulated to permit rapid and transient transcriptional changes in response to environmental challenges remains unclear. Here we show that hyperosmotic stress disrupts different levels of chromosome organization, ranging from A/B compartment changes to reduction in the number and insulation of topologically associating domains (TADs). Concomitantly, transcription is greatly affected, TAD borders weaken, and RNA Polymerase II runs off from hundreds of transcription end sites. Stress alters the binding profiles of architectural proteins, which explains the disappearance of local chromatin organization. These processes are dynamic, and cells rapidly reconstitute their default chromatin conformation after stress removal, uncovering an intrinsic organization. Transcription is not required for local chromatin reorganization, while compartment recovery is partially transcription-dependent. Thus, nuclear organization in mammalian cells can be rapidly modulated by environmental changes in a reversible manner.
Chromosome organization plays a key role in controlling biological processes such as responses to hormones, cell differentiation, and inactivation of the X Chromosome in mammalian systems, which indicates its importance for gene transcription regulation (Nora et al. 2012; de Laat and Duboule 2013; Le Dily et al. 2014; Lupiáñez et al. 2015; Bonev et al. 2017). Hi-C and related techniques have emerged as powerful tools for addressing chromosome folding genome-wide and have revealed different levels of chromatin organization. Initially, low-resolution maps allowed the definition of A and B compartments as regions spanning several megabases that are related to active and inactive transcriptional regions, respectively (Lieberman-Aiden et al. 2009). Later on, enhanced-resolution maps enabled the identification of topologically associating domains (TADs) (Dixon et al. 2012; Nora et al. 2012) that are proposed to be one of the basic units of chromosomal organization and are important for transcription regulation and replication (Pope et al. 2014; Lupiáñez et al. 2015). TADs are discrete units that exhibit a high frequency of interactions within each domain and contain sharp boundaries that isolate them from adjacent domains. Subsequent studies with higher resolution defined sub-TAD structures such as loops (Phillips-Cremins et al. 2013; Rao et al. 2014). TAD boundaries as well as loop anchors are enriched with architectural proteins such as the transcription factor CCCTC-binding factor (CTCF) and the ring-shaped multiprotein complex cohesin. Moreover, both CTCF and cohesin play key roles in establishing TADs and loops, as degradation of either CTCF or a member of the cohesin complex (RAD21) resulted in the disappearance of these structures (Nora et al. 2017; Rao et al. 2017). In contrast, CTCF degradation had no effect on compartment organization, whereas cohesin has been shown to antagonize compartment segregation (Schwarzer et al. 2017).
In the context of stress and chromosome organization, a study in Drosophila described that, upon heat-stress, architectural proteins are redistributed from TAD borders to intra-TAD regions, which facilitates long inter-TAD promoter–enhancer interactions of heat-stress response genes. Moreover, polycomb-group proteins play an important role in this reorganization by establishing new long-range interactions that propagate transcription silencing upon heat-stress (Li et al. 2015). However, the dynamics of these changes in chromatin structure in mammals in response stimuli and its reversibility remain unexplored. In mammalian cells, hyperosmotic stress impinges on different aspects of cellular physiology such as the cell cycle and transcription modulation and is therefore a suitable model for studying corresponding cellular responses such as transcription regulation (Ferreiro et al. 2010b; de Nadal et al. 2011). Moreover, microscopy studies revealed that hyperosmotic shock promotes rapid and reversible chromatin condensation (Albiez et al. 2006; Irianto et al. 2013) and a reduction in nuclear volume (Finan et al. 2009; Finan et al. 2011) and inhibits both RNA and DNA synthesis (Albiez et al. 2006). Thus, both adaptive transcription regulation and chromatin condensation occur concomitantly upon hyperosmotic stress. Here, we aimed to unravel the effect of osmostress on chromosome folding dynamics and its impact on transcription.
Results
Hyperosmotic stress perturbs A/B compartment organization
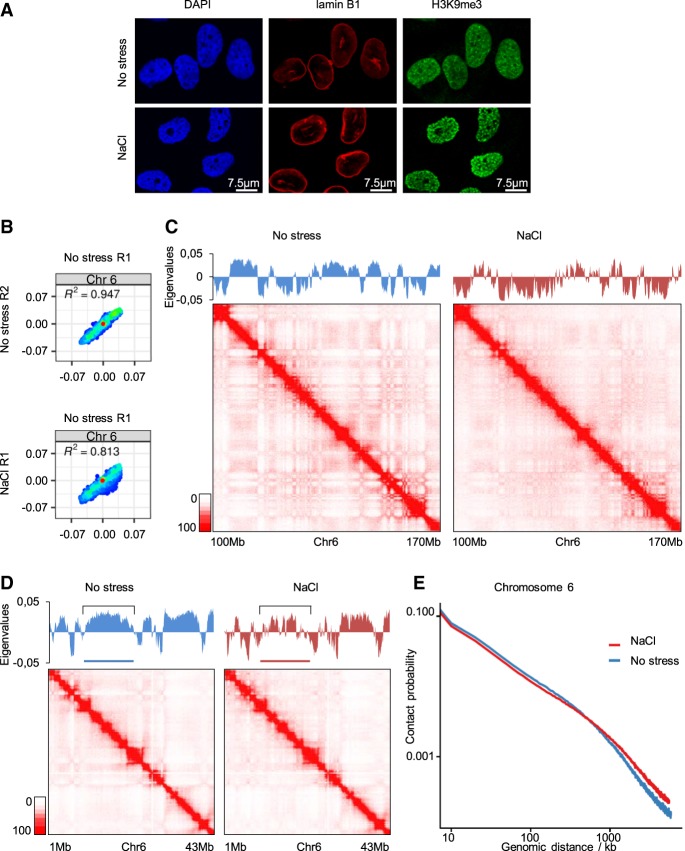
We initially tested the effects of hyperosmotic stress (60 min in 110 mM NaCl, 488 mOsm; conditions with maximal activation of stress-responsive kinases and still maximal cell survival; see Methods) (Supplemental Fig. S8A) on the nucleus by immunofluorescence on G0/1 arrested T47D cells. Staining of DNA with 4′,6-diamidino-2-phenylindole (DAPI) or H3K9me3 using antibodies indicated a condensation of chromatin and shrinking of the nuclei upon stress; however, osmostress did not compromise nuclear integrity as the nuclear lamina remained intact (Fig. 1A). Thus, an increase of osmolarity has a rapid strong effect on chromatin organization. To assess chromosome organization, we harvested T47D cells that were maintained in isotonic medium or were exposed to mild hyperosmotic stress (110 mM NaCl) for 1 h, and performed in situ Hi-C. We used arrested T47D cells to eliminate possible effects of stress on cell cycle progression that could lead to indirect differences in chromosome organization (Reyes et al. 2014; Clare et al. 2016; Nagano et al. 2017). Of note, exposure to mild hyperosmolarity did not affect cell viability even at higher NaCl doses (up to 200 mM) and a longer exposure (Supplemental Fig. S1A).
Figure 1.
Hyperosmotic stress has a strong impact on A/B compartment organization. (A) Representative immunofluorescence images of no stress or 1 h NaCl-treated cells for lamin B1 and H3K9me3, as well as stained with DAPI. (B) Correlation of eigenvalues obtained from Hi-C data for Chromosome 6 (Chr 6), comparing the two nonstressed replicates (R1 and R2) and comparing the nonstressed (R1) with the NaCl-treated (1 h 110 mM NaCl; R1) cells. (C,D) Eigenvalues and Hi-C maps from different regions of Chromosome 6 of nonstressed or NaCl-treated cells. (E) Contact probability of pairs of 100-kb-bin-dependent genomic distance on Chromosome 6, with or without NaCl treatment.
We processed two independent biological Hi-C replicates and obtained more than 100 million valid contacts in each (Supplemental Fig. S1B). Compartment segregation and putative changes due to stress were examined using a principal component analysis (PCA) of normalized Hi-C matrices, which allows identification of the A and B compartments, corresponding to active and inactive transcriptional regions, respectively (Lieberman-Aiden et al. 2009; Rao et al. 2014). Comparing PCA-derived eigenvalues revealed that hyperosmotic stress changed the compartment organization of most, if not of all, chromosomes (Supplemental Fig. S1C). Taking Chromosome 6 as an example, the correlation of eigenvalues between both control samples was high (0.947) but was clearly lower when comparing the control to a NaCl-treated sample (0.813) (Fig. 1B; Supplemental Fig. S1D). Further analysis of the eigenvalues indicated that hyperosmotic stress promoted an increase in the amount of genomic sequences defined as B-like compartments as well as lower eigenvalues overall (Fig. 1B; Supplemental Fig. S1C,D). These changes could be confirmed quantitatively by calculating the compartment strength (Supplemental Fig. S1E) and were also visible as a shift from A–A to B–B interactions in saddle plots (Supplemental Fig. S1F; Nora et al. 2017; Schwarzer et al. 2017; Wutz et al. 2017). Representation of the Hi-C data of the control sample in a contact matrix displayed the classical plaid pattern as described by others (Lieberman-Aiden et al. 2009); however, this pattern was strongly affected by, and showed less segregation upon, hyperosmotic stress (Fig. 1C; Supplemental Fig. S1G). Of note, in regions where an A compartment spanned several megabases in control conditions, NaCl treatment promoted B-like insertions, leading to an increase in segregation (Fig. 1D, highlighted region). These changes in compartment organization were also reflected in an analysis of contact probability that was dependent on genomic distance (Fig. 1E). In this analysis, medium- to long-range interactions were increased under stress compared with nonstress conditions, while short-range contacts, corresponding to local chromatin organization, were diminished upon stress. This dual effect was also evident in a representation of a matrix in which the signal obtained under nonstressed conditions was subtracted from that obtained under NaCl treatment (Supplemental Fig. S2A). Therefore, hyperosmotic stress induces changes in compartment segregation, promoting B-like compartments and increasing interactions between them. Importantly, these changes occur rapidly (within 1 h) and in a nonrandom fashion as both biological replicates exhibited very similar eigenvalues (Fig. 1B; Supplemental Fig. S1D).
Stress leads to a decrease in the insulation and the number of TADs, and to a disappearance of loops
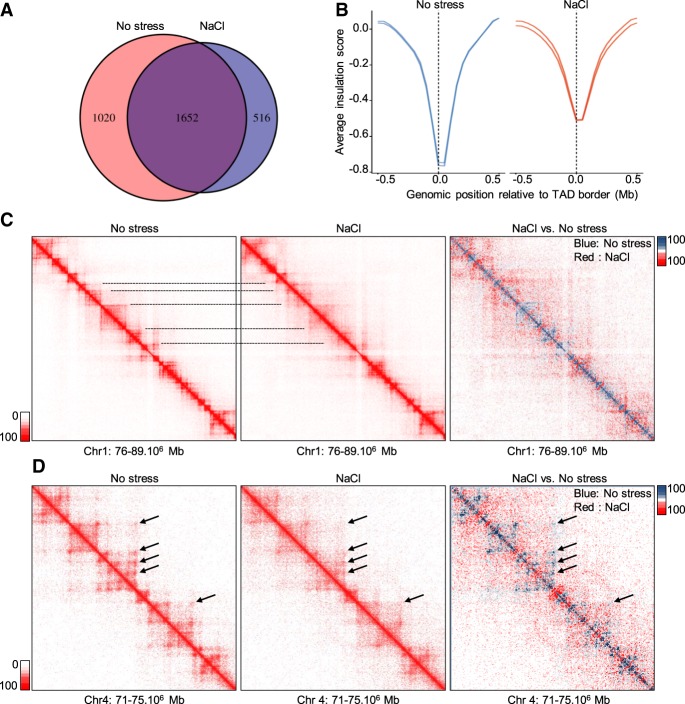
We next analyzed the effects of osmostress on a more local level of chromatin organization. By using TADbit (Serra et al. 2017), we identified around 2600 TADs in each of the nonstressed replicates, whereas only around 2100 TADs were detected upon hyperosmotic stress (Fig. 2A). The decrease in TAD number was accompanied by a significant reduction in their insulation level as was evident from an inspection of the insulation score around TAD borders (Fig. 2B; Crane et al. 2015). Likewise, an aggregated peak analysis revealed a marked decrease of interactions between the two borders of the same TAD (Supplemental Fig. S2B).
Figure 2.
Stress causes a decrease in the number of TADs and in their insulation. (A) Overlap of consensus TAD borders for nonstressed and NaCl-treated cells obtained using TADbit. Consensus borders were defined by allowing a shift of one bin (50 kb) in each direction between the two biological replicates. Similarly, TAD borders were overlapped, allowing a 50-kb shift in each direction. (B) The insulation score in a window of 1 Mb around consensus TAD borders in nonstressed and NaCl-treated cells (50-kb resolution). (C) Example of a Hi-C interaction matrix from a region of Chromosome 1 (Chr 1) (20-kb resolution) in unstressed and NaCl-treated cells and following subtraction of the signal under both conditions. Blue indicates higher signal in control; red, higher signal in stress conditions. Lines indicate different TADs. (D) Example of a Hi-C interaction matrix from a region of Chromosome 4 (Chr 4; 10-kb resolution) in unstressed and NaCl-treated cells and following subtraction of the signals (as above). Arrows indicate loops.
To visualize and better understand these effects, we pooled the two Hi-C replicates and generated a reference contact matrix for a region in which fewer TADs were called upon hyperosmotic stress (Fig. 2C). As predicted by TADbit, some TADs disappeared when cells were exposed to NaCl, although some borders were conserved, leading to bigger TADs (Fig. 2C). Indeed, both mean and median TAD size increased upon NaCl addition (Supplemental Fig. S2C). These effects were also evident following subtraction of the two contact matrices, which showed that TADs that were clearly defined in control conditions vanished upon stress (Fig. 2C, right panel). Of note, this Hi-C contact map also revealed that stress reduced intra-TAD interactions while promoting inter-TAD contacts (Fig. 2C), in accordance with the contact probability/genomic distance analysis (Fig. 1E; Supplemental Fig. S2D). We then investigated the effect of osmostress on local chromatin organization and found that osmostress completely abolished loop structures, indicating drastic osmostress-induced alterations on the short-range chromatin interactions (Fig. 2D). In summary, hyperosmotic stress strongly affected local chromatin organization by decreasing short-range interactions and weakening TAD borders, leading to a disappearance and/or fusion of TADs and loops.
Hyperosmotic stress significantly changes the transcriptome
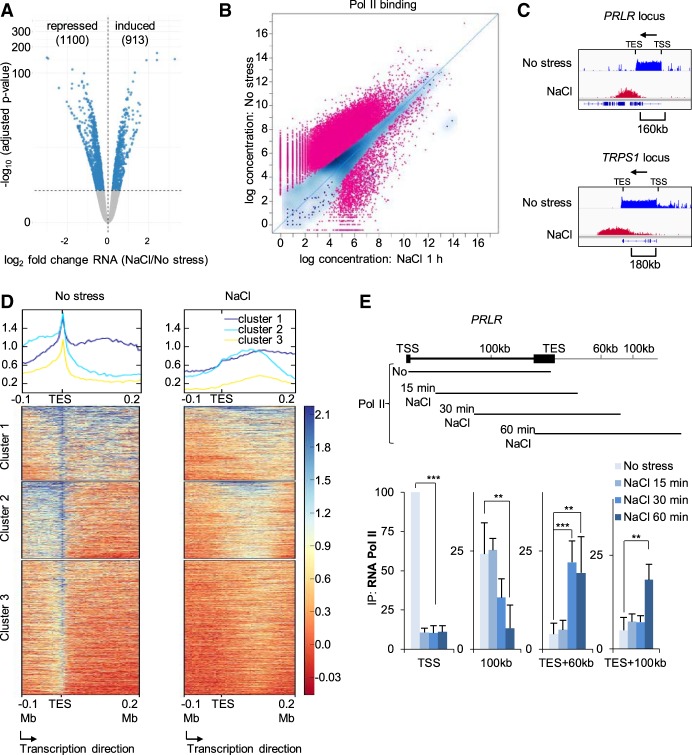
Since local chromatin structures were less defined and the segregation profile of compartments was changed upon hyperosmotic shock, we next investigated transcriptional modulation upon osmostress. First, we assessed global transcription by performing time-course experiments followed by RNA-seq. In these experiments, the mRNA levels of many genes were observed to change rapidly after osmostress, with 913 genes being up-regulated and 1100 down-regulated after 1 h of NaCl treatment (Fig. 3A; Supplemental Table S1). These numbers increased to 4224 and 4568 genes after 6 h of NaCl exposure (Supplemental Fig. S3A; Supplemental Table S1).
Figure 3.
Global transcriptional changes upon hyperosmotic shock. (A) Volcano plot for RNA-seq data after 1 h of osmostress. All genes that were significantly up- or down-regulated in stressed samples compared with the control appear as blue dots (repressed = 1100; induced = 913). (B) Pol II ChIP-seq signals of control cells and of cells stressed with NaCl for 1 h. Each dot corresponds to one peak, while red color indicates a significant change (35,340 peaks; FDR < 0.05). (C) IGV profiles of Pol II ChIP-seq data for the indicated genes in control and 1-h NaCl–treated cells. Arrows indicate gene directionality. (TSS) Transcription start site; (TES) transcription end site. (D) deepTools heatmaps obtained from k-means clustering of the Pol II ChIP-seq signal around the TES of genes with detected Pol II run-off. (E) (Top) Schematic of the PRLR locus. The lines below represent the expected Pol II position at the indicated time based on the elongation transcription rate in vivo. (Bottom) Pol II ChIP results of time-course experiments across the PRLR locus analyzed using qPCR (SEM of N = 3). (*) P < 0.05; (**) P < 0.005; (***) P < 0.0005; Student's t-test.
To gain a more direct view of transcription regulation beyond RNA-steady state levels, we assessed RNA polymerase II (Pol II) distribution genome-wide in the absence or after 1 h of osmostress using ChIP (using antibodies against the largest subunit, POLR2A) followed by deep sequencing (ChIP-seq). These experiments showed that genes that have been previously linked to stress and have been described as immediate early genes showed a significant increase in Pol II binding at their transcription start site (TSS) and gene body upon stress (e.g. NR4A1, FOS, FOSB, and ID2) (Supplemental Fig. S3B). Nevertheless, in clear contrast, osmostress strongly reduced Pol II binding genome-wide (Fig. 3B).
We further observed genomic regions with an increase in Pol II signals in response to stress that did not present the prototypical profile of Pol II distribution in mammalian cells, namely, high levels at the TSS, lower levels in the gene body, and a pause at the transcription end site (TES) (Jonkers et al. 2014). These Pol II–enriched genomic regions were found downstream from genes that showed a clear loss of a Pol II signal at their TSS and along most of the 5′ region of the gene body following osmostress (Fig. 3C; Supplemental Fig. S3C). Instead, an increase in the Pol II signal at ∼150–200 kb downstream from the TSS was observed after 60 min of stress (Fig. 3C; Supplemental Fig. S3C), which fits with the expected Pol II progression based on the in vivo transcription elongation rates (Ardehali and Lis 2009; Fuchs et al. 2014; Jonkers et al. 2014). We identified around 1500 genes that showed a similar profile (Fig. 3D; Supplemental Fig. S3D; Supplemental Table S2). Therefore, our data suggested a global Pol II run-off from transcribing genes upon stress. To confirm this possibility, we performed time-course experiments in which we osmostressed cells for different times and then assessed Pol II binding using ChIP-qPCR at different regions of two representative gene loci (PRLR and MED13L) (Fig. 3E; Supplemental Fig. S3E) and also a control locus (FOS) (Supplemental Fig. S3F), which is actually up-regulated due to osmostress and shows constant recruitment of Pol II at the TSS as long as stress is present. To estimate Pol II progression, we assumed an elongation rate between 2–3 kb/sec (Fuchs et al. 2014; Jonkers et al. 2014), which means that Pol II would travel around 37, 75, and 150 kb in samples stressed for 15, 30, and 60 min, respectively. As expected, the Pol II signal dramatically dropped at the TSS of both genes in all stressed compared with nonstressed samples, indicating that the loading of Pol II is impaired upon stress. In contrast, in a region 100 kb downstream from the PRLR TSS, we detected similar levels of Pol II in control, 15- and 30-min-stressed cells but lower levels than controls in 60-min-stressed cells, since Pol II would have already traveled further at this time point (>150 kb). When Pol II was monitored in a region 60 kb downstream from the TES, we only detected a signal in samples stressed for 30 or 60 min, whereas in a region 100 kb downstream, we only detected a signal at the 60-min time point (Fig. 3E). These findings were identical for MED13L (Supplemental Fig. S3E); are in line with the distances expected for the traveling of Pol II; and confirm that hyperosmotic stress indeed causes a lack of recruitment of Pol II to the TSS, except for osmostress-responsive genes such as FOS (Supplemental Fig. S3F), and induces Pol II to run-off beyond the TES. To determine whether Pol II located beyond the termination site was actively transcribing, we generated cDNA by a reverse transcriptase reaction using random hexamers and thereby detected stress-induced transcripts from downstream regions of PRLR and MED13L (Supplemental Fig. S3G) as described previously (Vilborg et al. 2015). These combined results indicate that hyperosmotic stress dramatically changes the transcriptome profile and induces widespread transcription beyond the TES due to Pol II read-through.
Pol II read-through coincides with changes in local chromatin organization
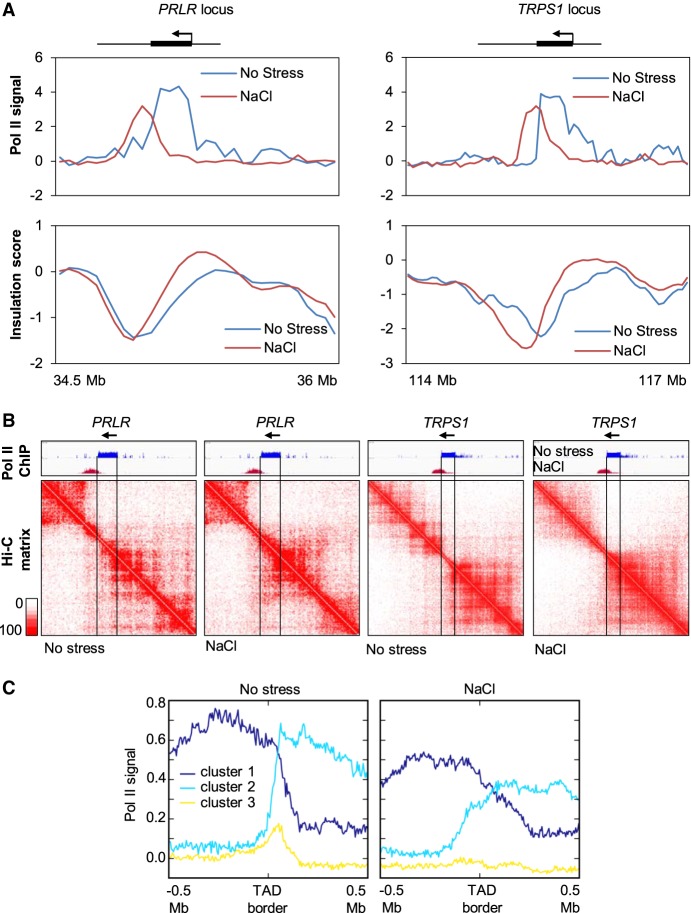
Next, we investigated whether the observed relocalization of Pol II upon osmostress might reflect local chromatin organization changes. We therefore selected two genes, PRLR and TRPS1, in which Pol II traveled far beyond the TES following osmostress, and assessed Pol II position and the insulation score metric (Crane et al. 2015). The most negative values of this insulation score corresponded to a TAD border domain that shifted after hyperosmotic shock, overlapping with osmostress-induced Pol II redistribution (Fig. 4A). Local chromatin organization changes were also clearly observed at 10-kb resolution in contact matrices from the same regions (Fig. 4B), suggesting that the presence of Pol II at the coding regions under normal conditions prevented interactions with neighboring regions that were consequently established upon Pol II run-off. To further investigate the interplay between TAD borders and Pol II, we visualized the effect of osmostress on Pol II distribution within 500 kb upstream of and downstream from each border (Fig. 4C). Two out of three groups of genes presented high levels of Pol II on one side of the TAD border and a dramatic drop in the signal following crossing of the border (dark and light blue) in the absence of stress. However, upon hyperosmotic stress, these profiles were markedly different as the reduction in the Pol II signal when crossing the border was much less pronounced and the signal progressively faded rather than dropping abruptly (Fig. 4C). These data indicated that, in the absence of stress, Pol II terminates close to the border in many TADs. In contrast, when cells are challenged with hyperosmotic stress, Pol II crosses those borders, which coincides with TAD border weakening. Thus, hyperosmotic Pol II redistribution and read-through correlates with changes in local chromatin organization.
Figure 4.
Pol II read-through coincides with local contact map changes. (A) The Pol II ChIP-seq signal and the insulation score around PRLR and TRPS1 loci in nonstressed and NaCl-treated cells. (B) Hi-C contact maps of the PRLR and TRPS1 loci under both conditions (10-kb resolution). (C) Profiles of Pol II occupation of consensus TAD borders in nonstressed and NaCl-treated cells. The TAD borders were defined under nonstress conditions (allowing a 50-kb shift between replicates). Binding profiles were clustered using k-means and signals of 500 kb upstream of or downstream from each TAD border. Genomic loci and clustering are independent of Figure 3C.
Chromatin binding profiles of architectural proteins are significantly altered upon hyperosmotic stress
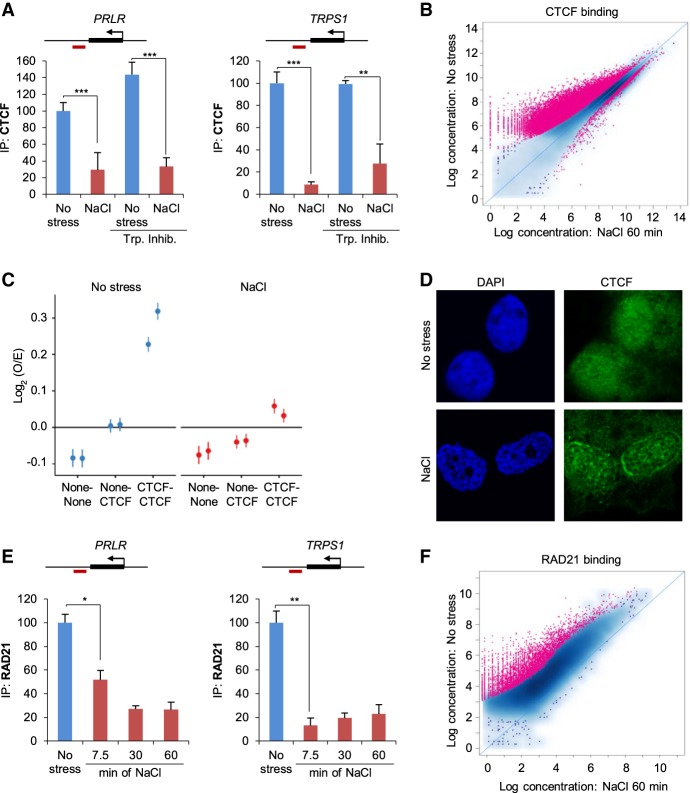
CTCF has been shown to be responsible for establishing loop domains and TADs in conjunction with cohesin, and degradation of CTCF or a component of the cohesin complex was shown to lead to the disappearance of such structures (Nora et al. 2017; Rao et al. 2017). We therefore sought to investigate whether CTCF binding to chromatin regions was affected by osmostress. Initially, we used qPCR to assess CTCF binding at sites close to genes such as PRLR and TRPS1 that had shown Pol II read-through and local organization rearrangements. A strong reduction in CTCF binding to these regions was observed upon osmostress (Fig. 5A). Of note, this decrease in CTCF binding was independent of ongoing transcription, as transcription inhibition did not prevent CTCF disengagement (Fig. 5A). Importantly, transcription inhibition per se did not influence CTCF binding (Fig. 5A).
Figure 5.
Architectural proteins are displaced from their normal chromatin binding sites after osmostress. (A) ChIP analysis of CTCF binding at previously described CTCF sites (Le Dily et al. 2014) in PRLR and TRPS1 loci in unstressed or 1-h NaCl–treated samples. Red boxes indicate amplified regions in the qPCR. When indicated (Trp. Inhib), a transcription inhibitor (200 nM flavopiridol) was added 2 h prior to stress (SEM of N = 3). (**) P < 0.005; (***) P < 0.0005; Student's t-test. (B) Global CTCF binding assessed by ChIP-seq, comparing control with 60-min NaCl treatment. Each dot represents one peak. A red color indicates a significant change (19,845 peaks, FDR < 0.05). (C) Aggregate short-range Hi-C signal (<2 Mb) between pairs of random genomic sites (none-none), between one CTCF and one random site (none-CTCF), or between pairs of CTCF sites (CTCF-CTCF) in unstressed and NaCl-treated samples. (D) Representative immunofluorescence images of no stress or 1-h NaCl–treated cells for CTCF and stained with DAPI. (E) ChIP-qPCR analysis of RAD21 binding at the indicated CTCF sites (red boxes indicate amplified region in the qPCR) in control cells or at the indicated times (minutes) of NaCl treatment (SEM of N = 3). (*) P < 0.05; (**) P < 0.005; Student's t-test. (F) Global RAD21 binding assessed by ChIP-seq, comparing control with 60-min NaCl treatment. Each dot represents one peak. A red color indicates a significant change (4536 peaks, FDR < 0.05).
We then extended the analysis of CTCF binding to a genome-wide scale by performing CTCF ChIP-seq in the absence or presence of stress for different time periods (7.5 and 60 min). Overall, CTCF binding was strongly reduced when cells were exposed to hyperosmotic shock; ∼50% of CTCF peaks showed a decrease in their signal when the cells were stressed (Fig. 5B). Moreover, this effect occurred rapidly, within 7.5 min (Supplemental Fig. S4A), and a longer treatment only slightly enhanced the CTCF dissociation (Supplemental Fig. S4B). Of note, we did not find newly established CTCF peaks at either of the stress time points. Additional analyses of sites in which the CTCF signal was reduced in stressed compared with control samples revealed an equal distribution of such sites throughout the genome (Supplemental Fig. S4C). However, these sites tended to be located more often within TADs rather than at TAD borders compared to unaffected peaks (Supplemental Fig. S4D). Since signal reduction coincided with loop disappearance, a direct link between a decrease in CTCF binding and local chromatin structure can be anticipated.
In addition, peaks that displayed a decrease in CTCF binding also exhibited a lower strength of their CTCF binding motif, which was evident on a genome-wide scale. Specifically, a HOMER analysis (Heinz et al. 2010) found a consensus CTCF motif in only 36.31% of affected sites compared with 53.52% of unaffected sites. Similarly, the de novo identified CTCF binding motif for unaffected peaks exhibited a clearer nucleotide preference than that of affected peaks as demonstrated by lower information content per base pair compared with sites without significant change in signal (Supplemental Fig. S4E,F). Moreover, unaffected peaks were more likely to overlap with basal RAD21 peaks and constitutive CTCF binding sites as defined by public data compared with peaks showing a significant decrease in the CTCF binding signal (Supplemental Fig. S4G). These combined results indicated that the CTCF that is recruited to weaker CTCF binding sites is more prone to dissociate upon hyperosmotic shock than is CTCF that is recruited to higher-affinity CTCF binding sites.
We then integrated the contact data with CTCF localization by aggregating the Hi-C signals for all short-range (<2 Mb) CTCF–CTCF combinations in control and stressed samples. This analysis revealed a strong decrease in Hi-C signals between pairs of CTCF sites upon NaCl treatment, whereas the contacts between CTCF sites and sites with random sequences did not differ from those of controls upon osmostress (Fig. 5C). Thus, osmostress-induced CTCF dissociation from its binding sites correlates with a prominent loss of contacts caused by hyperosmotic stress. To complement our sequencing-based results, we visualized CTCF localization by immunofluorescence. We observed that hyperosmotic stress promoted the formation of speckles as well as a strong perinuclear relocalization, indicating dissociation of CTCF from the chromatin, in line with our ChIP-seq results (Fig. 5D).
Next, we examined whether osmostress might also affect cohesin occupancy by performing ChIP analysis of the cohesin component RAD21 in several reference genes (i.e., PRLR, TRPS1). NaCl treatment reduced RAD21 levels at the selected sites (Fig. 5E), and the reduction was similar to those seen in similar experiments using CTCF. We then assessed genome-wide RAD21 localization by ChIP-seq in the absence of stress or in cells exposed to hyperosmotic stress for an hour. In accordance with the qPCR data, RAD21 association strongly diminished upon stress, with 25.6% of peaks exhibiting a significantly lower signal (Fig. 5F). Furthermore, we observed a significant association between sites with lower RAD21 signal and sites with lower CTCF occupancy, in line with current models of chromatin organization that propose CTCF as an anchor protein needed to fixate the cohesin ring to the chromatin (Supplemental Figs. S4H, S8B,C). Our combined data support the notion that hyperosmotic stress causes a rapid displacement of key architectural proteins from binding sites occupied in basal conditions, which might explain the weakening/disappearance of TADs and loops.
Re-establishment of nonstress conditions restores the “default” genome structure
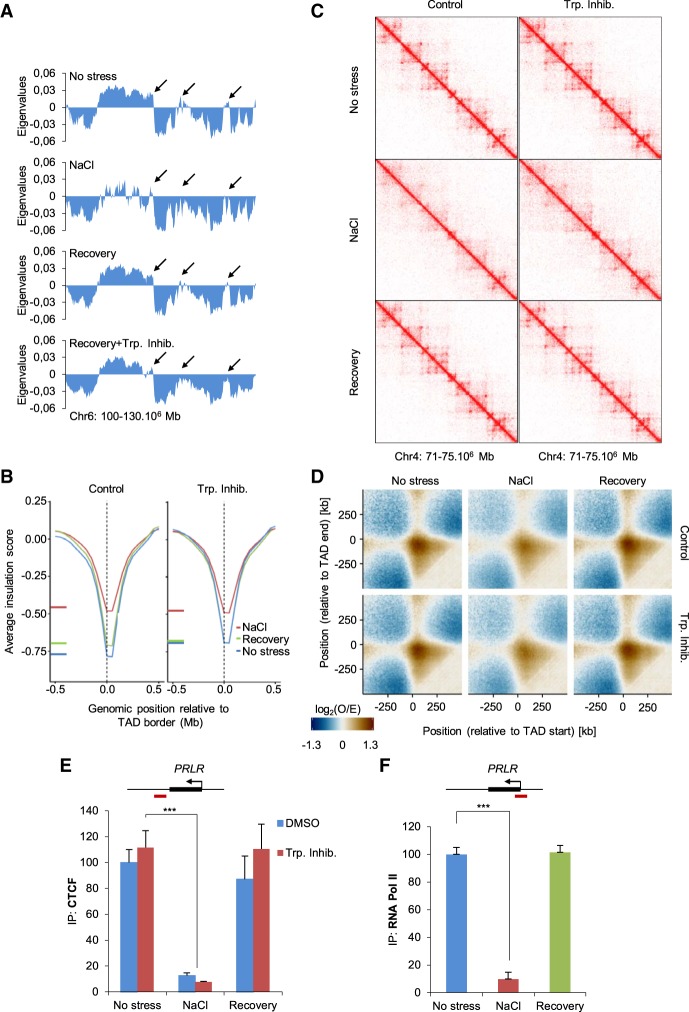
A key biological question is whether cells maintain the capacity to restore their “default” chromosome configuration when shifted back to nonstress conditions after stress treatment. To assess this possibility, we stressed cells with NaCl for 60 min and then let them recover for a further 60 min in an isotonic medium before performing Hi-C as well as ChIPs of CTCF and Pol II followed by qPCR. DAPI staining indicated that DNA condensation was fully recovered after this period of time (Supplemental Fig. S5A). To decipher the contribution of ongoing transcription to these processes, we included the transcription initiation inhibitor triptolide in our experimental set up (Supplemental Figs. S5B,C, S8D). The eigenvalues obtained in this new set of Hi-C experiments came from at least 100 million valid contacts per sample (Supplemental Fig. S1B). These experiments confirmed the effect of hyperosmotic stress on compartment organization (Fig. 6A; Supplemental Fig. S5D). Changing the cells back to isotonic conditions for 1 h was sufficient to restore their initial chromosome A/B compartment organization (Fig. 6A; Supplemental Fig. S5E,F, S6A). The inhibition of transcription initiation did not have a major impact on this level of chromosome organization by itself (Supplemental Figs. S5D, S6B,C), but recovery in the presence of triptolide was not complete (Fig. 6A; Supplemental Figs. S5F, S6C). Indeed, some chromatin regions did not recover their initial compartment organization (Fig. 6A; Supplemental Fig. S5F arrows). This phenomenon was also seen genome-wide; thus, although the correlation of eigenvalues between recovery plus transcription inhibition and control was high, it was virtually always lower than the correlation between recovery samples without inhibitor and control (Supplemental Fig. S6A). These differences were not as obvious when visualized as saddle plots; however, recovery of compartment strength with triptolide was lower compared with the absence of triptolide (Supplemental Fig. S6B,C).
Figure 6.
Stress response is a dynamic reversible process. (A) Eigenvalues from a region of Chromosome 6 under the indicated conditions: NaCl 60 min, recovery (NaCl 60 min followed by isotonic 60 min). When indicated, the transcription inhibitor (Trp. Inhib.) triptolide (500 nM) was added prior to stress. Arrows indicate regions that do not fully recover following inhibition of transcription. (B) Insulation score under the indicated condition in a 1-Mb window surrounding TAD borders defined in the absence of stress (50-kb resolution). (C) Example of Hi-C interaction matrices from a region of Chromosome 4 (15-kb resolution) under the indicated conditions. (D) Aggregated contact profiles centered on TAD borders representing the loss and recovery of interactions between both ends of TADs, both with and without triptolide. Data are presented as the log2 ratio of observed and expected contacts in 10-kb bins. (E) CTCF binding at the indicated CTCF binding site; red box indicates amplified region in the qPCR, as analyzed using qPCR in control, NaCl, and recovery samples (SEM of N = 3). (***) P < 0.005; Student's t-test. (F) Pol II binding at the PRLR TSS (red box indicates amplified region in the qPCR) was analyzed using ChIP-qPCR in control, NaCl, or recovery samples (SEM of N = 3). (***) P < 0.0001; Student's t-test.
Next, we analyzed TAD and loop organization when cells were shifted back to isotonic conditions after osmostress. Assessing TAD borders indicated that a 60-min recovery time after osmostress was sufficient to restore insulation scores to their nonstressed levels (Fig. 6B). Moreover, recovery of TADs and loops was observed when analyzing the contact matrix of a previously studied region in which hyperosmotic stress had promoted the disappearance of both. Notably, transcription inhibition per se did not have much influence on either the insulation score or the contact matrices. Thus, recovery of TADs and loops after stress occurred regardless of transcription inhibition and was almost indistinguishable to recovery without a transcription inhibitor (Fig. 6C,D; Supplemental Fig. S6D).
Consistent with the recovery of local chromatin organization following the removal of osmostress, qPCR analyses of ChIPs and immunofluorescence imaging confirmed that CTCF binding was also restored to preosmostress levels (Fig. 6E; Supplemental Figs. S7A,B,C, S8E). Moreover, NaCl-induced dissociation and de novo recruitment of CTCF during recovery were transcription independent and occurred rapidly, as increased ChIP-qPCR signal could be measured after 2.5 min (Supplemental Fig. S7C,D). As shown for CTCF, RAD21 localization and ChIP signal also recovered globally after 60 min (Supplemental Fig. S7E,G), while an additional ChIP-seq experiment confirmed that displacement and recovery of both CTCF and RAD21 were largely transcription independent (Supplemental Fig. S7C). Furthermore, we did not observe changes of overall RAD21 chromatin association as assessed by chromatin fractionation, indicating that cohesin mostly remains chromatin bound during osmostress (Supplemental Fig. S7F). Finally, we asked whether Pol II was recruited back to the TSS when cells were regrown in isotonic conditions after osmostress. Indeed, under these conditions, Pol II levels at the TSS of PRLR were indistinguishable from its levels under nonstressed (Fig. 6F). These data show that cells can restore their default chromosome organization and the initial distribution of CTCF and Pol II within 1 h after hyperosmotic stress removal. Transcription initiation does not seem to have major effects on the recovery of local organization of the genome but might be necessary for full compartment recovery.
Discussion
Nuclear architecture is decisive for transcription regulation in mammalian cells (Lupiáñez et al. 2015; Dekker and Mirny 2016). However, how chromosome organization is dynamically modulated to permit rapid and transient changes in transcription in response to environmental challenges remains unclear. Here, we show that hyperosmotic stress induces dynamic changes in chromosome organization at different levels that affect gene expression and Pol II distribution. Importantly, while a number of studies have described changes in compartment organization during differentiation or across cell types (Dixon et al. 2015; Bonev et al. 2017; Stadhouders et al. 2018), as well as during cell cycle progression (Nagano et al. 2017), our findings represent the first evidence of dynamic large-scale compartment reorganization that is triggered by an extracellular stimulus.
Osmostress strongly reduced TAD and loop-like structures, as would be expected if chromatin binding of architectural proteins was perturbed (Haarhuis et al. 2017; Nora et al. 2017; Rao et al. 2017; Schwarzer et al. 2017; Wutz et al. 2017). Indeed, osmostress promoted the dissociation of two key architectural proteins involved in local organization, CTCF and RAD21, from their respective chromatin binding sites. Neither of these proteins showed evidence of osmostress-induced post-translational modifications (Supplemental Figs. S7F, S8F). Although ablation of either CTCF or RAD21 did not appear to have much an effect on compartment organization (Nora et al. 2017; Rao et al. 2017), another study showed increased interactions between B compartments when the cohesin loading factor Nipbl was deleted (Schwarzer et al. 2017). Since this reported phenotype partially resembles the one we observed upon hyperosmotic stress, it might suggest that perturbations of the cohesin complex contribute to compartment organization. However, the compartment changes reported by Nipbl depletion were weaker than the ones induced upon osmostress, while Nipbl depletion simultaneously caused a total loss of TADs and loops, which we did not observe upon stress. Moreover, our chromatin fractionation experiment showed that the majority of cohesin seemed to remain chromatin-bound, indicating an unspecific distribution due to the lack of available CTCF anchors and the drastically altered Pol II landscape (Busslinger et al. 2017). Therefore, cohesin–chromatin binding and the resulting loop–extrusion model seem to be insufficient to explain all our observations, and it is reasonable to assume that other mechanisms participating in chromatin organization are perturbed due to osmostress. Such mechanisms could include the influence of histone marks on compartment segregation (Lieberman-Aiden et al. 2009; Rao et al. 2014) and/or changes in phase separation due to the efflux of water, which in turn increases intracellular protein and ion concentrations, altering phase separation (Keating 2012; Larson et al. 2017; Strom et al. 2017). Lastly, recent simulations suggested that compartment organization is determined by phase separation, while local chromatin structures depend on loop extrusion via CTCF and cohesin (Nuebler et al. 2018). In light of our findings, such a model appears to be plausible to explain the effects we observe on different levels of chromatin organization, although further research is required to clarify this hypothesis.
Our study also addressed the dynamics of the structural changes promoted by hyperosmotic stress and the relevance of transcription for these processes. Recovery of all levels of chromosome organization from compartments to loops occurred within a narrow window of time when cells were regrown in isotonic conditions after osmostress, in line with previous microscopy-based results (Albiez et al. 2006; Irianto et al. 2013). Thus, these changes represent a transient state until cells sense a favorable environment and restore their original 3D conformation, which could be driven by an underlying mechanism encoded within the chromatin. It is worth noting that transcription inhibition per se did not influence any of the chromatin levels tested. This result is in contrast with previous studies in Drosophila in which transcription inhibition decreased TAD insulation. Thus, transcription could play a different role in local chromatin organization in Drosophila and humans (Hug et al. 2017; Nora et al. 2017; Rao et al. 2017; Rowley et al. 2017). Here, we found that inhibition of transcription initiation did not have a major effect either on recovery of TAD and loop formation after reversal of osmostress or on de novo recruitment of architectural proteins. Thus, transcription might, for the most part, be dispensable for these levels of chromosome organization, and this notion is in accordance with previous studies (Hug et al. 2017; Ke et al. 2017). In contrast, compartments did not fully recover after reversal of osmostress when transcription initiation was inhibited, indicating that transcription participates in the re-establishment of these compartments. These effects of inhibition of transcription initiation on compartments were rather modest.
The reorganization of chromatin interactions coincides with massive changes in mRNA transcript levels and Pol II distribution as was indicated by previous microscopy results (Albiez et al. 2006). Following osmostress, many genes showed a clear reduction on Pol II loading, and moreover, Pol II traveled beyond the TES and actively transcribed those regions. The observed Pol II run-off is in accordance with a previous paper, which reported the production of long noncoding RNAs that were linked to upstream genes upon osmotic shock (Vilborg et al. 2015). Since, Pol II run-off and transcriptional read-through coincided with TAD border weakening and loss of intra-TAD domains, it is tempting to speculate that the observed aberrant transcription might have been a consequence of loss of chromatin structure. Consistent with this idea, a potential link between these two phenomena was outlined by the work of Schwarzer et al. (2017), which described an increase in exogenic transcription when Nipbl was deleted. Thus, TAD borders and other intra-TAD structures that are established by architectural proteins might play a role in containing widespread transcription.
In the context of stress, chromatin organization, and environmental insults, it has been shown that heat-shock in Drosophila cells causes TAD border weakening and an increase in inter-TAD interactions, similar to our observations using osmostress (Li et al. 2015). Despite these phenotypical similarities, we expect that the underlying mechanisms of these changes to differ. Specifically, heat-stress promoted a reorganization of several architectural proteins, whereas we observed a clear dissociation of architectural proteins from chromatin as our ChIP-seq data indicated no newly formed CTCF peaks after stress. Moreover, it was shown that depletion of Rad21 in Drosophila increased TAD border strength (Li et al. 2015), while our data indicated a decrease in TAD border strength following RAD21 delocalization due to hyperosmotic shock. However, although the mechanisms underlying chromosome reorganization following stress might differ between these two organisms, it is still possible that chromosome reorganization through changes in architectural proteins will be a common feature by which cells respond to dramatic changes in the environment.
Overall, our study highlights how different levels of chromosome conformation can undergo dramatic changes while retaining the ability to restore their initial status. These changes occur within minutes following exposure to abrupt alterations in the environment that are within a physiological range in the human body. This phenomenon would suggest that many cells of the human organism might undergo constant rearrangements of nuclear organization; in particular, kidney and gastrointestinal tract cells as well as chondrocytes (Irianto et al. 2013) are constantly challenged by changes in osmolarity. Lastly, given that an increasing number of studies have correlated hyperosmolarity with inflammation and other related diseases (Brocker et al. 2012), our results encourage further study of the possible contribution of chromatin rearrangements to such conditions.
Methods
Cell culture
T47D cells were grown for 48 h with RPMI containing charcoal-treated FBS as described previously (Reyes et al. 2014; Clare et al. 2016). Cells were then switched to a medium without serum (288 mOsm) for 18 h prior to treatment. Subsequently, NaCl (110 mM) was added directly to the medium (final osmolality, 488 mOsm), and the cells were harvested at the indicated times. Transcription inhibitors (500 nM of triptolide or 200 nM of flavopiridol) were added 2 h prior to stress. In the recovery experiments, cells were stressed for 1 h with NaCl and were then shifted to isotonic medium for a further hour.
Cell viability in response to stress was assessed by labeling living cells with propidium iodide (PI) followed by FACS analysis. The 1-h time point was chosen for the bulk of our experiments because the stress-induced kinase p38 (MAPK14) is at peak activity (Ferreiro et al. 2010a,b) and effects on transcription can be reliably followed.
ChIP and ChIP-seq
ChIPs were performed as previously described (Vicent et al. 2014). For specific details, see the Supplemental Methods. Antibodies used for ChIP experiments were as follows: anti-CTCF antibody (Millipore no. 07-729), anti-Rad21 (Abcam no. Ab992), anti-POLR2A (Millipore no. 05-623; Santa Cruz N-20 no. SC-899 discontinued; Cell Signaling no. 2629).
RNA and RNA-seq
RNA from control or NaCl-treated T47D cells was extracted at the indicated times using the RNeasy Plus Mini Kit (Invitrogen no. 74134). The qPCR analysis reverse transcriptase reaction was performed using 0.5–1 µg of RNA and the RevertAid first strand cDNA synthesis kit (Fermentas no. K1622). Expression of the indicated genes was assessed using qPCR and was referenced to GAPDH expression.
For RNA-seq experiments, stranded poly(A) selected libraries were prepared using the TruSeq stranded mRNA library prep kit (Illumina, RS-122-2101) according to the manufacturer's standard protocol. Libraries were subjected to 50-bp paired-end sequencing.
In situ Hi-C library preparation
In situ Hi-C experiments were performed as previously described (Rao et al. 2014) with some modifications. Briefly, adherent cells were crosslinked in 1% formaldehyde, quenched with glycine, and washed in 2 mL of PBS containing protease inhibitors. Cross-linked cells were incubated for 30 min on ice in 3C lysis buffer (10 mM Tris-HCl at pH 8, 10 mM NaCl, 0.2% NP-40), centrifuged for 5 min at 3500g, resuspended in 190 µL of NEBuffer2 1× (New England BioLabs [NEB]) and 10 µL of 10% SDS, and were then incubated for 10 min at 65°C. After the addition of Triton X-100 and 15-min incubation at 37°C, nuclei were centrifuged for 5 min at 3500g and were resuspended in 300 µL of NEBuffer2 1×. Nuclei were digested overnight using 400 U of the MboI restriction enzyme (NEB). Before fill-in with bio-dATP, nuclei were pelleted and resuspended in fresh 1× repair buffer (1.5 µL of 10 mM dCTP, 1.5 µL of 10 mM dGTP, 1.5 µL of 10 mM dTTP, 37.5 µL of 0.4 mM biotin-dATP, 50 U of DNA Polymerase I large [Klenow] fragment in 300 µL NEBuffer2 1×). Ligation was performed for 4 h at 16°C using 10,000 cohesive end units of T4 DNA ligase (NEB) in 1.2 mL of ligation buffer (120 µL of 10× T4 DNA ligase buffer, 100 µL of 10% Triton X-100, 12 µL of 10 mg/mL BSA, 963 µL of H2O). After reversion of the cross-link, DNA was purified using phenol/chloroform/isoamyl. Purified DNA was sonicated to an average size of 300–400 bp using a Bioruptor Pico (Diagenode; eight cycles of 20 sec on and 60 sec off), and 3 µg of sonicated DNA was used per in situ Hi-C library preparation. Briefly, biotinylated DNA was pulled down using 20 µL of Dynabeads MyOne T1 streptavidin beads in binding buffer (5 mM Tris-HCl at pH 7.5, 0.5 mM EDTA, 1 M NaCl). End-repair, A-tailing and Illumina adaptors ligation were performed on the beads using NEBNext library preparation end-repair and A-tailing modules (NEB). Libraries were amplified using eight cycles of PCR and were controlled for quality by low sequencing depth on a NextSeq 500 prior to higher sequencing depth on HiSeq 2000.
Normalization of Hi-C contact matrices
Hi-C data were processed using an in-house pipeline based on TADbit (Serra et al. 2017) and is available as Supplemental Code.
Processing of RNA-sequencing data
RNA-sequencing reads were mapped against hg38 (obtained via UCSC golden path) using TopHat2 v2.1.0 (settings: --max-multihits 1 --library-type fr-firststrand --b2 --very-sensitive --no-coverage-search) (Kim et al. 2013) and supplying a transcriptome index based on GENCODE v24 (Harrow et al. 2012; https://www.gencodegenes.org). Reads covering GENCODE v24 features were counted using featureCounts v1.5.1 in stranded paired-end fashion (-s 2 -p) (Liao et al. 2014). Differential expression analysis was performed using DESeq2 (v1.14.1) (Love et al. 2014) comparing treated and untreated samples per time point, while results were visualized using ggplot2 (v2.2.1) (Wickham 2016).
Processing of CTCF, RAD21, and Pol II ChIP-sequencing data
ChIP-sequencing reads were mapped against hg38 (obtained via UCSC golden path) using Bowtie 2 v2.2.6 (settings: --very-sensitive) (Langmead and Salzberg 2012). Narrow CTCF and RAD21 peaks were called using MACS2 (Zhang et al. 2008) callpeak (-g hs -B --SPMR), supplying the sequenced corresponding input DNA as background control. Differential binding of CTCF and RAD21 was assessed using DiffBind v2.4.6 (Zhang et al. 2008; Ross-Innes et al. 2012).
Since Pol II exhibits a broad binding pattern, broad Pol II peaks were called MACS2 callpeak and were later used to identify Pol II run-off regions (-g hs -B --SPMR --broad --broad-cutoff 0.1 --P-value 0.01). However, since multiple peaks can be called per gene, we assessed differential Pol II binding by calculating its occupancy of regions 5 kb downstream from the TSS and by subsequently supplying these raw counts to DESeq2. To focus on potentially interesting gene loci, we filtered the GENCODE v24 genes for the following categories prior to occupancy quantification: “protein coding, lincRNA, antisense, transcribed_unprocessed_pseudogene, transcribed_processed_pseudogene, processed_pseudogene, processed_transcript, macro_lncRNA, unitary_pseudogene, transcribed_unitary_pseudogene, translated_unprocessed_pseudogene, unprocessed pseudogene.” As the RAF2 sequencing sample had been sequenced to a significantly greater depth compared with the other Pol II ChIP-seq samples, we used “samtools view -s 0.5” to subsample reads from the BAM file and prevent sequencing depth-related biases (Li 2011).
Other relevant information such as protein extraction, chromatin fractionation, ChIP details, and detailed bioinformatics analysis such as Hi-C or identification of Pol II run-off locations can be found in the Supplemental Methods.
Data access
All Hi-C and RNA-sequencing data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE111904. The ChIP-seq data from this study have been submitted to GEO under accession number GSE111923. The pipeline for Hi-C data analysis is available as Supplemental Code.
Supplementary Material
Acknowledgments
We thank Marc Martí-Renom for helpful discussions. R.A. was a recipient first of a Beatriu de Pinós (Generalitat de Catalunya) and later of a Marie-Curie Cofund program fellowship (EU). R.B. is a recipient of a Juan de la Cierva fellowship. The study was supported by grants from the Spanish Ministry of Economy and Competitiveness (BFU2015-64437-P and FEDER, BFU2014-52125-REDT, and BFU2014-51672-REDC to F.P.; BFU2017-85152-P and FEDER to E.d.N.), the Catalan Government (2017 SGR 799), the Fundación Botín, and the Banco Santander through its Santander Universities Global Division to F.P. and the Unidad de Excelencia Maria de Maeztu, MDM-2014-0370. F.P. is recipient of an ICREA Acadèmia (Generalitat de Catalunya). M.B. received the support of the European Research Council (ERC Synergy Grant 4D Genome 609989) and CERCA.
Author contributions: R.A., F.L.D., and Y.C. designed and performed experiments. R.A., R.B., F.L.D., E.V., and J.Q. analyzed the data. R.A., R.B., F.L.D., M.B., E.d.N., and F.P. participated in experimental design, data analysis, and writing the article.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.238527.118.
Freely available online through the Genome Research Open Access option.
References
- Albiez H, Cremer M, Tiberi C, Vecchio L, Schermelleh L, Dittrich S, Küpper K, Joffe B, Thormeyer T, von HJ, et al. 2006. Chromatin domains and the interchromatin compartment form structurally defined and functionally interacting nuclear networks. Chromosome Res 14: 707–733. 10.1007/s10577-006-1086-x [DOI] [PubMed] [Google Scholar]
- Ardehali MB, Lis JT. 2009. Tracking rates of transcription and splicing in vivo. Nat Struct Mol Biol 16: 1123–1124. 10.1038/nsmb1109-1123 [DOI] [PubMed] [Google Scholar]
- Bonev B, Mendelson CN, Szabo Q, Fritsch L, Papadopoulos GL, Lubling Y, Xu X, Lv X, Hugnot JP, Tanay A, et al. 2017. Multiscale 3D genome rewiring during mouse neural development. Cell 171: 557–572. 10.1016/j.cell.2017.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocker C, Thompson DC, Vasiliou V. 2012. The role of hyperosmotic stress in inflammation and disease. Biomol Concepts 3: 345–364. 10.1515/bmc-2012-0001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busslinger GA, Stocsits RR, van der Lelij P, Axelsson E, Tedeschi A, Galjart N, Peters JM. 2017. Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nature 544: 503–507. 10.1038/nature22063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clare SE, Gupta A, Choi M, Ranjan M, Lee O, Wang J, Ivancic DZ, Kim JJ, Khan SA. 2016. Progesterone receptor blockade in human breast cancer cells decreases cell cycle progression through G2/M by repressing G2/M genes. BMC Cancer 16: 326 10.1186/s12885-016-2355-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane E, Bian Q, McCord RP, Lajoie BR, Wheeler BS, Ralston EJ, Uzawa S, Dekker J, Meyer BJ. 2015. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature 523: 240–244. 10.1038/nature14450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat W, Duboule D. 2013. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature 502: 499–506. 10.1038/nature12753 [DOI] [PubMed] [Google Scholar]
- de Nadal E, Ammerer G, Posas F. 2011. Controlling gene expression in response to stress. Nat Rev Genet 12: 833–845. 10.1038/nrg3055 [DOI] [PubMed] [Google Scholar]
- Dekker J, Mirny L. 2016. The 3D genome as moderator of chromosomal communication. Cell 164: 1110–1121. 10.1016/j.cell.2016.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W, et al. 2015. Chromatin architecture reorganization during stem cell differentiation. Nature 518: 331–336. 10.1038/nature14222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreiro I, Barragan M, Gubern A, Ballestar E, Joaquin M, Posas F. 2010a. The p38 SAPK is recruited to chromatin via its interaction with transcription factors. J Biol Chem 285: 31819–31828. 10.1074/jbc.M110.155846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreiro I, Joaquin M, Islam A, Gomez-Lopez G, Barragan M, Lombardia L, Domínguez O, Pisano DG, Lopez-Bigas N, Nebreda AR, et al. 2010b. Whole genome analysis of p38 SAPK-mediated gene expression upon stress. BMC Genomics 11: 144 10.1186/1471-2164-11-144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan JD, Chalut KJ, Wax A, Guilak F. 2009. Nonlinear osmotic properties of the cell nucleus. Ann Biomed Eng 37: 477–491. 10.1007/s10439-008-9618-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan JD, Leddy HA, Guilak F. 2011. Osmotic stress alters chromatin condensation and nucleocytoplasmic transport. Biochem Biophys Res Commun 408: 230–235. 10.1016/j.bbrc.2011.03.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs G, Hollander D, Voichek Y, Ast G, Oren M. 2014. Cotranscriptional histone H2B monoubiquitylation is tightly coupled with RNA polymerase II elongation rate. Genome Res 24: 1572–1583. 10.1101/gr.176487.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarhuis JHI, van der Weide RH, Blomen VA, Yáñez-Cuna JO, Amendola M, van Ruiten MS, Krijger PHL, Teunissen H, Medema RH, van SB, et al. 2017. The cohesin release factor WAPL restricts chromatin loop extension. Cell 169: 693–707. 10.1016/j.cell.2017.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. 2012. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 22: 1760–1774. 10.1101/gr.135350.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug CB, Grimaldi AG, Kruse K, Vaquerizas JM. 2017. Chromatin architecture emerges during zygotic genome activation independent of transcription. Cell 169: 216–228.e19. 10.1016/j.cell.2017.03.024 [DOI] [PubMed] [Google Scholar]
- Irianto J, Swift J, Martins RP, McPhail GD, Knight MM, Discher DE, Lee DA. 2013. Osmotic challenge drives rapid and reversible chromatin condensation in chondrocytes. Biophys J 104: 759–769. 10.1016/j.bpj.2013.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers I, Kwak H, Lis JT. 2014. Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. eLife 3: e02407 10.7554/eLife.02407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y, Xu Y, Chen X, Feng S, Liu Z, Sun Y, Yao X, Li F, Zhu W, Gao L, et al. 2017. 3D chromatin structures of mature gametes and structural reprogramming during mammalian embryogenesis. Cell 170: 367–381. 10.1016/j.cell.2017.06.029 [DOI] [PubMed] [Google Scholar]
- Keating CD. 2012. Aqueous phase separation as a possible route to compartmentalization of biological molecules. Acc Chem Res 45: 2114–2124. 10.1021/ar200294y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, Narlikar GJ. 2017. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547: 236–240. 10.1038/nature22822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Dily F, Baù D, Pohl A, Vicent GP, Serra F, Soronellas D, Castellano G, Wright RH, Ballare C, Filion G, et al. 2014. Distinct structural transitions of chromatin topological domains correlate with coordinated hormone-induced gene regulation. Genes Dev 28: 2151–2162. 10.1101/gad.241422.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993. 10.1093/bioinformatics/btr509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Lyu X, Hou C, Takenaka N, Nguyen HQ, Ong CT, Cubeñas-Potts C, Hu M, Lei EP, Bosco G, et al. 2015. Widespread rearrangement of 3D chromatin organization underlies Polycomb-mediated stress-induced silencing. Mol Cell 58: 216–231. 10.1016/j.molcel.2015.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, et al. 2015. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161: 1012–1025. 10.1016/j.cell.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Lubling Y, Várnai C, Dudley C, Leung W, Baran Y, Mendelson CN, Wingett S, Fraser P, Tanay A. 2017. Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature 547: 61–67. 10.1038/nature23001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485: 381–385. 10.1038/nature11049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Goloborodko A, Valton AL, Gibcus JH, Uebersohn A, Abdennur N, Dekker J, Mirny LA, Bruneau BG. 2017. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169: 930–944. 10.1016/j.cell.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuebler J, Fudenberg G, Imakaev M, Abdennur N, Mirny LA. 2018. Chromatin organization by an interplay of loop extrusion and compartmental segregation. Proc Natl Acad Sci 115: E6697–E6706. 10.1073/pnas.1717730115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, Ong CT, Hookway TA, Guo C, Sun Y, et al. 2013. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 153: 1281–1295. 10.1016/j.cell.2013.04.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope BD, Ryba T, Dileep V, Yue F, Wu W, Denas O, Vera DL, Wang Y, Hansen RS, Canfield TK, et al. 2014. Topologically associating domains are stable units of replication-timing regulation. Nature 515: 402–405. 10.1038/nature13986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huang SC, Glenn St HB, Engreitz JM, Perez EM, Kieffer-Kwon KR, Sanborn AL, Johnstone SE, Bascom GD, Bochkov ID, et al. 2017. Cohesin loss eliminates all loop domains. Cell 171: 305–320. 10.1016/j.cell.2017.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes D, Ballaré C, Castellano G, Soronellas D, Bagó JR, Blanco J, Beato M. 2014. Activation of mitogen- and stress-activated kinase 1 is required for proliferation of breast cancer cells in response to estrogens or progestins. Oncogene 33: 1570–1580. 10.1038/onc.2013.95 [DOI] [PubMed] [Google Scholar]
- Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, et al. 2012. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481: 389–393. 10.1038/nature10730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley MJ, Nichols MH, Lyu X, Ando-Kuri M, Rivera ISM, Hermetz K, Wang P, Ruan Y, Corces VG. 2017. Evolutionarily conserved principles predict 3D chromatin organization. Mol Cell 67: 837–852. 10.1016/j.molcel.2017.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer W, Abdennur N, Goloborodko A, Pekowska A, Fudenberg G, Loe-Mie Y, Fonseca NA, Huber W, Haering H, Mirny L, et al. 2017. Two independent modes of chromatin organization revealed by cohesin removal. Nature 551: 51–56. 10.1038/551S51a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra F, Baù D, Goodstadt M, Castillo D, Filion GJ, Marti-Renom MA. 2017. Automatic analysis and 3D-modelling of Hi-C data using TADbit reveals structural features of the fly chromatin colors. PLoS Comput Biol 13: e1005665 10.1371/journal.pcbi.1005665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadhouders R, Lubberts E, Hendriks RW. 2018. A cellular and molecular view of T helper 17 cell plasticity in autoimmunity. J Autoimmun 87: 1–15. 10.1016/j.jaut.2017.12.007 [DOI] [PubMed] [Google Scholar]
- Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH. 2017. Phase separation drives heterochromatin domain formation. Nature 547: 241–245. 10.1038/nature22989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicent GP, Nacht AS, Ballaré C, Zaurin R, Soronellas D, Beato M. 2014. Progesterone receptor interaction with chromatin. Methods Mol Biol 1204: 1–14. 10.1007/978-1-4939-1346-6_1 [DOI] [PubMed] [Google Scholar]
- Vilborg A, Passarelli MC, Yario TA, Tycowski KT, Steitz JA. 2015. Widespread inducible transcription downstream of human genes. Mol Cell 59: 449–461. 10.1016/j.molcel.2015.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York. [Google Scholar]
- Wutz G, Várnai C, Nagasaka K, Cisneros DA, Stocsits RR, Tang W, Schoenfelder S, Jessberger G, Muhar M, Hossain MJ, et al. 2017. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J 36: 3573–3599. 10.15252/embj.201798004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based Analysis of ChIP-Seq (MACS). Genome Biol 9: R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.