Summary
The exact identity of castrate-resistant (CR) cells and their relation to CR prostate cancer (CRPC) is unresolved. We use single-cell gene profiling to analyze the molecular heterogeneity in basal and luminal compartments. Within the luminal compartment, we identify a subset of cells intrinsically resistant to castration with a bi-lineage gene expression pattern. We discover LY6D as a marker of CR prostate progenitors with multipotent differentiation and enriched organoid-forming capacity. Lineage tracing further reveals that LY6D+ CR luminal cells can produce LY6D− luminal cells. In contrast, in luminal cells lacking PTEN, LY6D+ cells predominantly give rise to LY6D+ tumor cells, contributing to high-grade PIN lesions. Gene expression analyses in patients’ biopsies indicate that LY6D expression correlates with early disease progression, including progression to CRPC. Our studies thus identify a subpopulation of luminal progenitors characterized by LY6D expression and intrinsic castration resistance. LY6D may serve as a prognostic maker for advanced prostate cancer.
Keywords: castration-resistant prostate cancer, LY6D, luminal progenitor, tumor-initiating cells, intrinsic resistance
Graphical Abstract
Highlights
-
•
Single-cell analysis identifies a highly heterogeneous luminal compartment
-
•
LY6D marks luminal cells that are resistant to castration with bi-lineage capacity
-
•
LY6D correlates with prostate cancer development from luminal lineage
-
•
LY6D expression in human prostate cancer correlates with early disease progression
To address the identity of castration-resistant prostate cells in vivo, Barros-Silva et al. coupled single-cell analysis with organoid culture and in situ lineage tracing. They identify LY6D as a progenitor marker that liaises intrinsically castration-resistant luminal cells and castration-resistant prostate tumor growth.
Introduction
Prostate cancer (PCa) remains the second most lethal malignancy among Western men (Siegel et al., 2014) and is treated mainly with androgen deprivation therapy (ADT). Although therapy is initially effective, most patients inevitably progress to castration-resistant PCa (CRPC). Such patients may benefit from new-generation drugs targeting the androgen receptor (AR) axis (e.g., enzalutamide and abiraterone) (de Bono et al., 2011, Scher et al., 2012). However, therapeutic options for CRPC remain largely limited (Watson et al., 2015). Two models have been proposed to explain the mechanism of castration resistance (Isaacs, 2008, Shen and Abate-Shen, 2010). The prevailing model posits that androgen-dependent cancer cells adapt to ADT by acquiring castration-resistant (CR) properties through genetic/epigenetic changes. Alternatively, a subset of PCa cells may be intrinsically CR and are selected for growth following ADT.
The prostate gland is composed of basal and luminal epithelial cells (of note, these are positional terms and do not fully reflect the cellular subtypes), as well as rare neuroendocrine cells (Shen and Abate-Shen, 2010). Unlike basal cells, which are largely insensitive to androgen deprivation (Bauman et al., 2014, Isaacs, 2008, Lin-Tsai et al., 2014), the majority of luminal cells undergo apoptosis during castration, although a small proportion remains CR (Rane et al., 2014, Wang and Shen, 2011). Prostate basal cells possess multipotent stem cell activity and can differentiate into luminal cells and serve as a cell-of-origin of PCa, as revealed by transplantation (Goldstein et al., 2010, Lawson et al., 2010) and injury repair (Kwon et al., 2014, Toivanen et al., 2016) assays. However, lineage-tracing studies using inducible luminal-specific mouse models, such as Nkx3.1-CreER (Wang et al., 2009), PSA-CreER (Liu et al., 2011), and K8-CreER (Choi et al., 2012, Ousset et al., 2012), demonstrate that the prostate luminal lineage in adults is largely self-sustained by luminal cells. In particular, these studies support the existence of CR multipotent and unipotent luminal progenitor (LP) cells that repopulate the luminal lineage upon androgen-induced regeneration (Choi et al., 2012, Ousset et al., 2012, Wang et al., 2009, Wang et al., 2013, Wang et al., 2014). Lineage-tracing experiments reveal that PCa may have a basal origin; however, luminal cells have been shown as the preferred cell-of-origin (Choi et al., 2012, Ousset et al., 2012, Wang et al., 2009, Wang et al., 2013, Wang et al., 2014). Moreover, the recently developed organoid system has allowed detection of multipotent or unipotent LPs in vitro from both human and mouse origins (Agarwal et al., 2015, Chua et al., 2014, Karthaus et al., 2014, Kwon et al., 2016). Despite these efforts, the identity of CR prostate cells in vivo, particularly CR luminal cells, and their contribution to CRPC remain largely unresolved. To address these, we utilized a Fluidigm multiplex qPCR-based single-cell expression analysis platform (Guo et al., 2013) to interrogate expression profiles of individual prostate cells sorted from hormone-naive (HN) and castrated mice, and coupled the analysis with organoid culture and in situ lineage tracing. With this multidisciplinary approach, here we report the heterogeneity within the luminal lineage, and identification of LY6D as a progenitor marker that is linked to CR luminal cells and CRPC.
Results
Heterogeneity within the Prostate Luminal Lineage
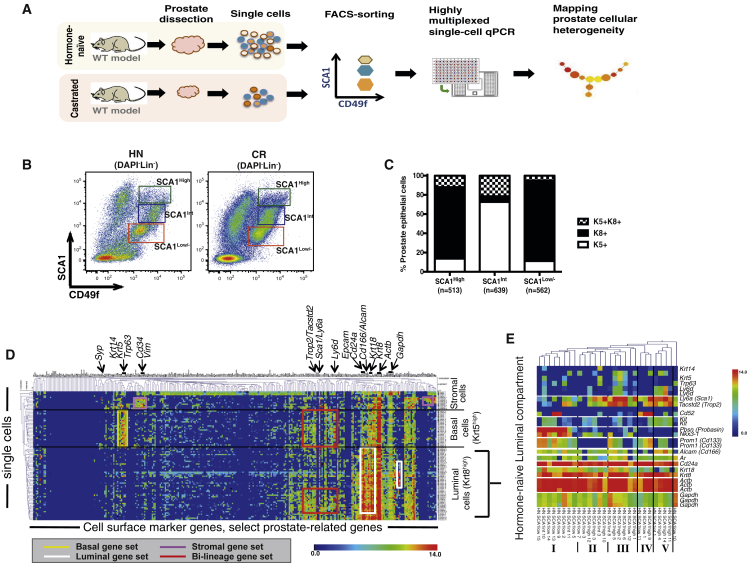
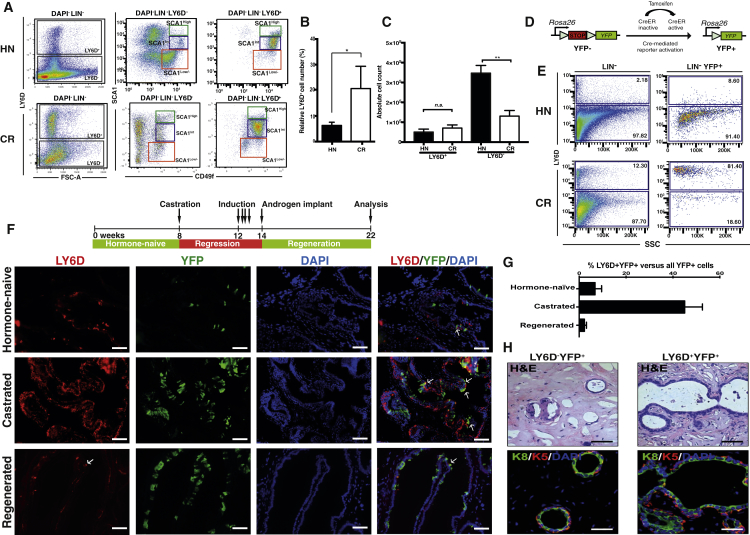
Using a Fluidigm multiplex qPCR-based approach (Guo et al., 2013), we interrogated expression levels of ∼300 genes, including most CD (cluster of differentiation) markers, as well as prostate-related genes (Table S1), in individual prostate cells sorted from HN or castrated mice (Figure 1A). Our goal was to identify prostate epithelial subpopulations intrinsically resistant to androgen deprivation based on profiling of cell surface markers. To isolate single prostate cells, we utilized fluorescence-activated cell sorting (FACS) based on cell surface profiles of lineage markers (Lin, including CD45, CD31, TER119), SCA1, and CD49f, which separates prostate cells into the three subpopulations (referred to as “LSC subpopulations”): basal cells (Lin−SCA1+CD49f+), luminal cells (Lin−SCA1low/−CD49flow; hereafter SCA1low/−), and stromal cells (Lin−SCA1+CD49f−) (Lawson et al., 2007, Lawson et al., 2010). Although the Lin−SCA1+CD49f+ gate has been reported to contain predominantly basal cells (Lawson et al., 2010), we found that this population could be further separated into two subpopulations based on high or intermediate levels of SCA1 expression (hereafter referred to as SCA1high or SCA1int, respectively; Figure 1B). Immunofluorescent (IF) and FACS intracellular staining of these LSC subpopulations for the luminal marker Keratin 8 (K8) and basal marker Keratin 5 (K5) revealed that the SCA1high subpopulation consisted predominantly of K8+ luminal cells, rather than K5+ basal cells (which is the predominant cell type within SCA1int) (Figures 1C, S1A, and S1B). Upon castration, both SCA1high and SCA1int subpopulations were enriched, possibly due to more extensive loss of SCA1low/− luminal cells (Figure 1B). Of note, several studies have observed high levels of SCA1 expression in proximal luminal cells (Korsten et al., 2009, Leong et al., 2008). Furthermore, a recent study described a similar subpopulation of FACS-sorted SCA1-high cells, which are localized in the proximal prostatic ducts and represent an androgen-independent subpopulation of LPs (Kwon et al., 2016). Upon castration, we also observed that in all three subpopulations (i.e., SCA1high, SCA1int, SCA1low/−), the percentages of K5+K8+ cells were notably increased compared to those of HN mice (Figure S1D). The percentages of K5+K8+ cells we detected in prostate subpopulations, sorted from both HN and CR mice, were higher (5%–20%) than those identified by IHC in situ (<5% of prostate cells) (Ousset et al., 2012, Wang et al., 2013), which may be due to single-cell isolation process or sensitiveness of different antibody-staining techniques. The presence of K5/K8 double-positive prostate cells was unlikely due to a sorting artifact from cell doublets, as our sorting strategy and assessment by microscopy assured that the majority of sorted cells were single cells (Figures S1E–S1H).
Figure 1.
Single-Cell Expression Profiling of Prostate Cells from HN and Castrated Mice
(A) Schematic diagram demonstrating experimental setup for single-cell expression profiling.
(B) FACS gating strategy used to sort single prostate cells, from Lineage-negative cells (Lin−: CD31−CD45−TER119−).
(C) Quantification of keratin expression profiles by IF based on three FACS-sorted populations as in (B). The number of cells counted in each group is shown from five mice. See also Figures S1A–S1C.
(D) Hierarchical clustering of single prostate cells from HN and castrated (CR) mice showing separation of luminal, basal, and stromal cells, as well as the luminal (white), basal (yellow), bi-lineage (red), and stromal (purple) gene sets. Color scale is indicated. See also Table S2.
(E) Enlarged view of select genes from the heatmap in Figure S1I clustering prostate cells in the luminal lineage from HN mice into five subsets. Color scale is indicated.
See also Figure S1.
Upon confirming individual cell sorting from the SCA1low/−, SCA1high, and SCA1int gates from both HN and age-matched castrated (i.e., CR) male mice (Figures 1B and S1E–S1H), we performed multiplex qPCR-based single-cell gene profiling. Unsupervised hierarchical clustering of the entire dataset (HN + CR) classified single cells into three major groups, including those expressing luminal (e.g., Krt8, Krt18, and Cd24a) and basal (e.g., Krt5, Krt14, and Trp63) gene sets at higher levels, which may represent prostate epithelial cells in the luminal and basal lineages, respectively (Figure 1D; Table S2). In support of this conclusion, we noticed that most single cells clustered into these two groups were originally sorted from the SCA1low/− and high and SCA1int gates (Figure 1D), which are enriched with K8+ luminal and K5+ basal cells (Figure 1C), respectively. In addition, we observed a small number of cells, largely from the SCA1high gate, that expressed high levels of stromal cell marker genes such as Vimentin (Vim) (Lawson et al., 2010) and Cd34 (Lawson et al., 2007), but very low levels of Keratin genes (Figure 1D). Since the SCA1high gate is in close proximity to the stromal cell gate (Figure 1B), these cells were probably “contaminating” stromal cells (which also express SCA1 highly) sorted as SCA1high cells, although we cannot fully exclude the possibility that these were epithelial cells undergoing mesenchymal differentiation.
By unsupervised clustering, we observed that prostate epithelial cells within the luminal lineage appeared more heterogeneous than those in the basal lineage (Figure 1D). To understand prostate epithelial cell heterogeneity pre-existing in HN mice, we first focused on the expression data of HN single cells (Figure S1I). We found that cells of the luminal lineage (i.e., Krt8high) from HN mice could be separated into at least five subsets (Figure 1E). Krt8high-subset I, characterized by mRNA expression of Pbsn and Nkx3-1 at the highest levels, likely represents terminally differentiated luminal cells. Krt8high-subsets III and V both exhibit expression of prostate stem/progenitor marker genes (i.e., Sca1/Ly6a [Xin et al., 2005], Trop2/Tacstd2 [Goldstein et al., 2008], Cd166/Alcam [Jiao et al., 2012]); they may denote distinct subpopulations of multipotent or unipotent LPs, as some cells within Krt8high-subset III express basal genes (e.g., Krt5, Trp63), whereas all cells within Krt8high-subset V express Kit (encoding C-KIT), an established marker of prostate stem/progenitor cells (Leong et al., 2008). We identified expression of the lymphocyte antigen 6 complex, locus D gene (Ly6d, encoding a cell surface marker that has not been implicated in prostate biology) specifically in Krt8high-subsets III and V, raising a possibility for Ly6d as a potential marker for prostate stem/progenitor cells. Of note, the marker Ly6d, as well as the majority of known prostate stem/progenitor marker genes, are within a bi-lineage gene set that is expressed at medium or high levels in many basal cells and a subset of luminal cells (e.g., Krt8high-subsets III and V) (Figures 1D and 1E).
CR Luminal Cells Exhibit Bi-lineage Expression Pattern
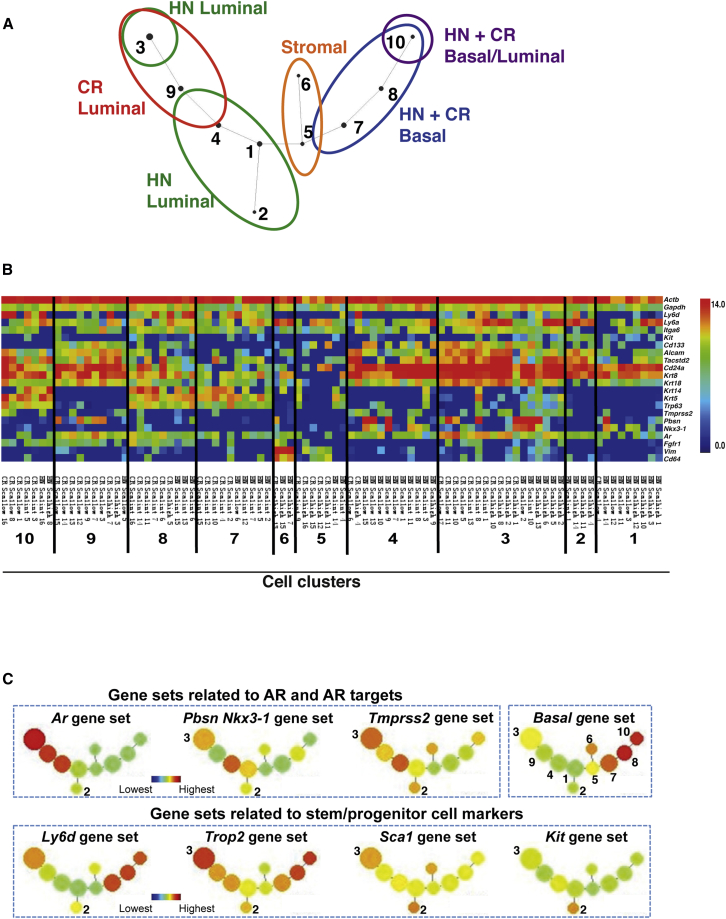
In light of heterogeneity within the luminal lineage, we hypothesized that any similarity of single-cell signatures between HN and CR cells might form a foundation for an in silico strategy to identify one or more pre-existing prostate subpopulations(s) intrinsically resistant to androgen deprivation. To test this hypothesis, we mapped the lineage relationship between HN and CR prostate cells (i.e., cells with similar expression signatures) using SPADE (spanning-tree progression analysis of density-normalized events) analysis, a method originally developed to extract cellular hierarchy from high-dimensional mass cytometry data (Bendall et al., 2011, Qiu et al., 2011). We adapted this approach recently to analyze high-dimensional qPCR data by using a dimension reduction strategy (Guo et al., 2013) and applied it here. We first reduced data dimensions from ∼300 genes to 23 gene sets with similar expression patterns (Table S3), followed by redundancy removal by extracting the average value of each gene set. We subjected this reduced gene expression data to the SPADE algorithm to distill the 23 dimensional single-cell data down to an undirected, acyclic graph (assignment of each single cell to a cell cluster in Table S4) that assesses the relationship between different prostate cell subpopulations. Different cell lineages are readily separated into distinct branches (Figure 2A). As anticipated, since basal cells are largely insensitive to androgen deprivation (Shen and Abate-Shen, 2010), most HN and CR cells in the basal lineage were intermingled in two cell clusters (#7–8; Figures 2A and 2B). Similarly, Vimentin and Cd34-expressing cells (i.e., “contaminating” stromal cells) from both types of mice were also mixed together in two cell clusters (#5–6; Figures 2A and 2B). HN and CR prostate cells in the luminal lineage were distributed mainly into five cell clusters with partial overlap (#1–4 and 9; Figures 2A and 2B). Among these, only cluster #3 contained comparable numbers of luminal-lineage cells from both HN and CR samples. These cells expressed the marker Ly6d and high mRNA levels for multiple prostate stem/progenitor markers (e.g., Sca1, Trop2, and Cd166) with the exception of Kit. Furthermore, they expressed a bi-lineage gene expression profile characterized by high levels of luminal genes (e.g., Krt8, Krt18, Cd24a) and the Ar gene, low or intermediate expression of one or more basal-lineage associated genes (e.g., Krt5, Krt14, Trp63), as well as intermediate or high expression of several AR target genes (e.g., Pbsn, Nkx3-1, and Tmprss2 [Wang et al., 2007]) (Figures 2B, 2C, and S2A). In addition, a new luminal cell cluster was detected in CR samples (cluster #9; Figures 2A and 2B). CR cells in this cluster were obtained mainly from the SCA1low/− gate and expressed stem/progenitor marker genes Trop2 and Cd166, as well as Ar at high levels. Interestingly, most CR cells in this cluster also highly expressed Fgf1r (Figures 2B and S2A), a gene principally expressed in some basal cells in HN mice (Figure 2B). Last, although cell cluster #10 positioned closest to basal cell clusters #7 and #8 (Figure 2A), CR and HN cells in this cluster often expressed both basal (e.g., Krt5, Trp63) and luminal genes (e.g., Krt8, Cd24a) at high levels (Figures 2B and S2A), raising a possibility that they represent basal cells committed to the luminal lineage (Rane et al., 2014, Uzgare et al., 2004, Wang et al., 2013, Xin et al., 2007). Overall, our data reveal that CR prostate cells in both the basal and luminal lineages remain heterogeneous; among them, most CR luminal-lineage cells exhibit a bi-lineage gene expression profile at varying degrees (Figures 1D and 2B). Notably, this CR expression pattern includes high-level expression of multiple prostate stem/progenitor genes and the Ar gene.
Figure 2.
Mapping Lineage Relation of CR Prostate Luminal and Basal Cells to Their Counterparts from HN Mice
(A) SPADE analysis based on 23 gene sets (Tables S3 and S4) showing relationship of luminal and basal cells subsets from hormone-naive (HN) and castrated (CR) mice. #1∼10 indicate cells with similar expression patterns of the 23 gene sets grouped as 10 cell clusters.
(B) Heatmaps showing expression levels of select genes in each single cells grouped based on their cell cluster assignment (as in A). Color scale is indicated.
(C) Average expression levels of select gene sets in each cluster of single cells (as in A). See also Figure S2A for expression levels of all 23 gene sets.
See also Figure S2.
LY6D Marks Subsets of Prostate Basal and Luminal Cells
Changes in the expression pattern of each gene in CR versus HN luminal cells at the single-cell level could reflect enrichment (or loss) of a pre-existing HN subset expressing that gene upon castration, and/or upregulation or downregulation of that gene in response to castration. We reasoned that if we could identify a cell surface marker that is widely expressed in CR luminal cells, but only expressed in a subset of HN luminal cells (with expression level comparable to that of CR luminal cells), we might be able to define intrinsically CR luminal cells and meanwhile provide support to the notion of pre-existing CR cells.
We analyzed the single-cell dataset, focusing on cells sorted from the SCA1low/− gate (i.e., luminal cells) and marker genes that are expressed in the candidate LP subsets (Figure 1E). Although many of the previously defined prostate stem/progenitor cell-related genes exhibited upregulation in CR luminal cells, they were widely expressed in HN luminal cells (e.g., Trop2, Cd166, Cd133, Cd49f; Figure S2B); others exhibited either no expression change (e.g., Sca1; Figure S2C) or downregulation (e.g., Kit, Nkx3.1; Figure S2D) in CR luminal cells compared to HN luminal cells. However, one of these genes, Ly6d, exhibited upregulation in CR luminal cells and was only expressed in a subset of HN luminal cells (Figure S2E). Of note, Ly6d has a human ortholog, whereas its close family member Sca1/Ly6a does not, thus providing a potential opportunity for clinical translation. Therefore, we focused subsequent analysis on this marker.
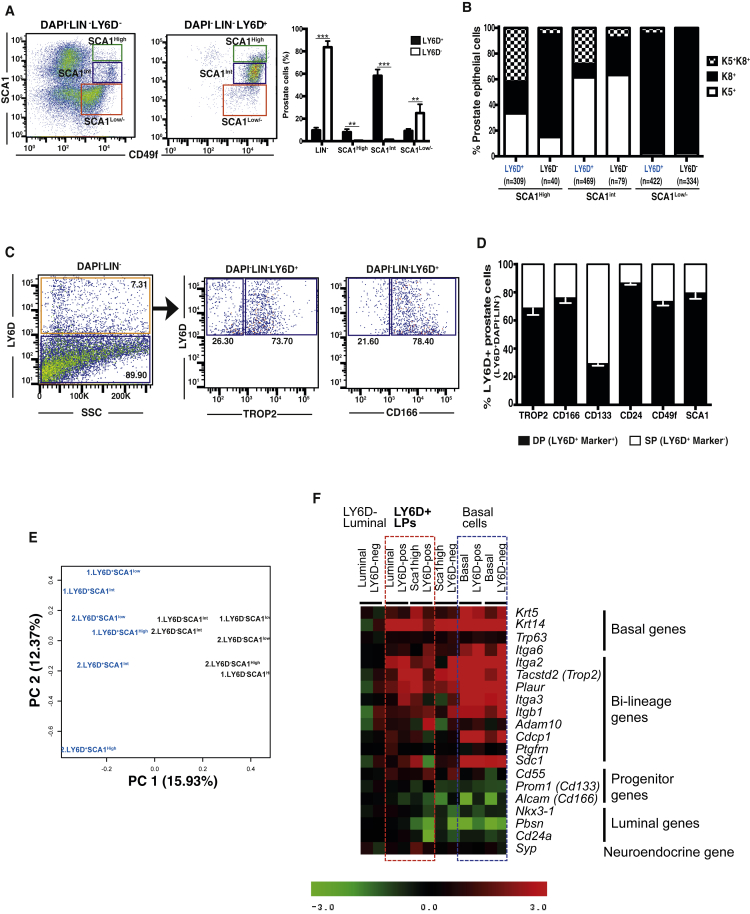
We first examined the expression of LY6D protein in the 3 subsets of HN prostate cells, defined by SCA1high, SCA1int, and SCA1low/−. By FACS analysis, we found the majority of LY6D+ prostate cells within the SCA1int basal gate. However, smaller subsets of LY6D+ cells were also present in both the SCA1high and SCA1low/− gates (which are enriched with K8+ luminal cells) (Figure 3A). Quantification of the LY6D+ and LY6D− subsets revealed that the majority of prostate cells in the SCA1int (mainly K5+ basal) and SCA1high (mainly K8+ luminal) gates were LY6D+, whereas only a small subset of luminal cells in the SCA1low/− gate was also LY6D+ (Figure 3A). IF staining showed that the majority of LY6D+ cells in the SCA1high, or SCA1int, or SCA1low/− gate were K5+K8+ and K8+, or K5+-only, or K8+-only prostate cells, respectively (Figures 3B and S3A). Next, we examined the relation of LY6D to other prostate cell surface markers. We found that LY6D exhibited the largest overlapping staining pattern with TROP2 (i.e., most LY6D+ cells were TROP2+ and vice versa), followed by SCA1 and CD166; LY6D staining had the least overlap with CD133 (Figures 3C, 3D, S3B, and S3C).
Figure 3.
Characterization of LY6D+ Prostate Epithelial Subpopulations from HN Mice
(A) FACS analysis of Lin− cells from prostates (from HN mice) confirmed LY6D expression in basal (SCA1int), intermediate (SCA1high), and a small portion of luminal (SCA1low/−) cells. n = 3 for each sorted subpopulation. p values: ∗∗p < 0.01 and ∗∗∗p < 0.001. Error bars represent mean ± SEM.
(B) Quantification of keratin expression by IF staining of the FACS-sorted subpopulations as in (A). The number of cells counted in each group is shown from four mice. See also Figure S3A.
(C) FACS analysis of LY6D+ cells from HN prostates for their staining patterns of select prostate stem/progenitor and luminal surface markers.
(D) Quantification of marker profiles based on FACS subpopulations as in (C). The average of frequency of the indicated populations is from three mice.
(E) Comparison of the gene expression profiles from LY6D+ and LY6D− subpopulations (based on RNA-seq data) by principal-component analysis (PCA).
(F) Heatmap of differentially expressed genes in sorted LY6D+ and LY6D− subsets from the SCA1high or int or low/− populations. Log2 expression values (based on microarray) were normalized to luminal (SCA1low/−) LY6D− subset for each gene (=0).
See also Figure S3.
To further assess whether LY6D+ and LY6D− subpopulations, in particular those within the SCA1low/− luminal gate, represent separate entities, we performed gene expression profiling, by both RNA sequencing (RNA-seq) and microarray, on sorted LY6D+ and LY6D− subsets from the SCA1high, SCA1int, and SCA1low/− gates, respectively. By principal-component analysis (PCA), we found that the LY6D+ cells from all 3 subsets clustered together and separated from the LY6D− subsets (Figure 3E). Moreover, the LY6D+ subsets from the SCA1high and SCA1low/− gates (i.e., representing cells in the luminal lineage) exhibited higher expression levels of many bi-lineage genes (based on Figure 1D) as well as basal genes (e.g., Krt5, Krt14) than LY6D− luminal cells from the SCA1low/− gate (Figure 3F). Together, these data suggest that LY6D selectively marks prostate cells in the luminal lineages with a bi-lineage, stem/progenitor-like gene expression signature.
LY6D+ Prostate Cells Are Organoid-Forming Progenitor Cells
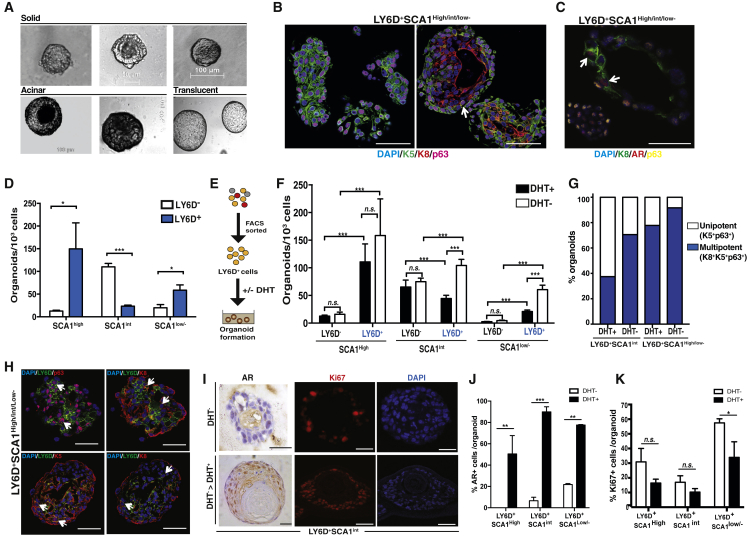
Our single-cell and LY6D+ subpopulations expression data, supported by staining pattern of LY6D in HN prostate cells, raised the possibility that LY6D marks prostate basal and luminal cells with stem/progenitor properties. To test this, we sorted LY6D+ and LY6D− prostate epithelial cells from HN mice for organoid culture, a recently developed approach for in vitro characterization of stem/progenitor cells, particularly LPs (Chua et al., 2014, Gao et al., 2014, Karthaus et al., 2014). We found that sorted LY6D+ cells formed organoids with distinct morphologies, unrelated to the expression level of SCA1. The resulting organoids exhibited a morphology of solid multilayer mass (Figure 4A, 50–100 μm), or acinar morphology (>100 μm) composed of a lumen surrounded by multiple layers, or translucent appearance (>100 μm) with a large and hollow lumen surrounded by a thin layer of cells resembling prostate ducts (Figures 4A and S4). Further characterization of LY6D+ cell-derived organoids by IF staining revealed primarily two types, including organoids composed of only K5+p63+cells (unipotent basal) as well as multipotent organoids, with cells types of diverse lineages, mostly composed of a peripheral layer of basal (K5+ and p63+) and K8+ (luminal) cells surrounding the lumen (Figures 4B and S4A). The multi-lineage organoids derived from all LY6D+ subpopulations contained AR+ cells (Figures 4C and S4B), similar to what has been reported for multipotent organoids derived from CD49f+ and CD26+ prostate cells (Karthaus et al., 2014). Notably, we did not observe unipotent luminal (K8+) organoids from untransformed mouse prostate cells, as reported previously (Agarwal et al., 2015, Karthaus et al., 2014, Liu et al., 2016). Importantly, in a small portion (<10%) of multipotent organoids derived from LY6D+ subpopulations, we observed K8+ cells co-expressing both K5 and p63 (i.e., K5+p63+K8+ cells), in addition to double- or single-positive cells for these three markers (Figures 4B and S4A). To further assess the stem/progenitor properties of LY6D+ cells, organoids from SCA1high, SCA1int, and SCA1low populations were quantified. The sorted LY6D+SCA1high subpopulation exhibited by far the highest organoid-forming efficiency (Figure 4D). Of note, within the SCA1high and SCA1low/− luminal-enriched subpopulations, almost all cells with organoid-forming ability were derived from the LY6D+ subset (Figure 4D). Organoid formation was maintained for at least three passages (Figure S4D). Moreover, the SCA1high and low/− subpopulations formed increasing numbers of organoids with large lumen or translucent appearance over serial passages, suggesting an enrichment of multipotent organoids under this culture condition (Figure S4E).
Figure 4.
LY6D+ Prostate Cells Are Enriched for Organoid-Forming Potential
(A) Representative phase images of solid, acinar, and translucent organoids from FACS-sorted Lin−SCA1highLY6D+, Lin−SCA1intLY6D+, and Lin−SCA1low/−LY6D+ prostate cells.
(B) Co-IF analysis of organoids derived from FACS-sorted Lin−SCA1highLY6D+, Lin−SCA1intLY6D+, and Lin−SCA1low/−LY6D+ prostate cells for Keratins and p63 expression. Arrow depicts K5+K8+p63+ triple-positive cells. See Figure S4A for individual channels. Scale bars: 50 μm.
(C) AR expression in K8+ cells in representative multipotent organoids derived from FACS-sorted Lin−SCA1high LY6D+, Lin−SCA1intLY6D+, or Lin−SCA1low/−LY6D+ prostate cells. Arrows depict AR+K8+ double-positive cells. See Figure S4B for individual channels. Scale bars: 50 μm.
(D) Quantification of organoids formed from FACS-sorted LY6D+ and LY6D− (Lin−SCA1high, Lin−SCA1int, and Lin−SCA1low/−) prostate cell subsets (1,000–3,000 cells/well) from HN mice. n = 5 for each sorted subpopulation. p values: ∗p < 0.05 and ∗∗∗p < 0.001. Error bars represent mean ± SEM.
(E) Schematic diagram showing organoid culture in the presence or absence of androgen (DHT, dihydrotestosterone).
(F) Quantification of organoids derived from FACS-sorted LY6D+ and LY6D− (Lin−SCA1high, Lin−SCA1int, and Lin−SCA1low/−) prostate cell subsets (1,000–3,000 cells/well) from HN mice, in the presence (+) or absence (−) of DHT. n = 5 for each sorted subpopulation. p value: ∗∗∗p < 0.001. n.s., nonsignificant. Error bars represent mean ± SEM.
(G) Quantification of unipotent (K5+p63+) and multipotent (K8+ K5+p63+) organoids formed from FACS-sorted LY6D+ (Lin−SCA1high, Lin−SCA1int, and Lin−SCA1low/−) prostate cell subsets (1,000–3,000 cells/well) from HN mice, in the presence (+) or absence (−) of DHT. n = 3 for each sorted subpopulation.
(H) Representative co-IF analysis of LY6D, p63, and keratin markers for organoids derived from FACS-sorted LY6D+ (Lin−SCA1high, Lin−SCA1int, and Lin−SCA1low/−) prostate cells. Arrows depict double-positive cells. See Figure S4C for individual channels. Scale bars: 50 μm.
(I) Androgen-response analysis of CR organoid outgrowth from FACS-sorted Lin−SCA1highLY6D+ subpopulation, in the absence of DHT, only or followed by DHT stimulation, measured by nuclear AR and Ki67 staining. See also Figure S4F. Scale bars: 50 μm.
(J) Quantification of nuclear AR expression on LY6D+ (Lin−SCA1high, Lin−SCA1int, and Lin−SCA1low/−) derived organoids from castrated mice. Organoids were cultured in the absence of DHT for 14 days, and then stimulated for 7 days with 100 nM DHT. n = 3 for each sorted subpopulation. p values: ∗∗∗p < 0.001 and ∗∗p < 0.01. Error bars represent mean ± SEM.
(K) Quantification of Ki67 expression on LY6D+ (Lin−SCA1high, Lin−SCA1int, and Lin−SCA1low/−) derived organoids from castrated mice. Organoids were cultured in the absence of DHT for 14 days, and then stimulated for 7 days with 100 nM DHT. n = 3 for each sorted subpopulation. p value: ∗ p < 0.05. n.s., non-significant. Error bars represent mean ± SEM.
See also Figures S3 and S4.
To assess androgen dependency of organoid formation from LY6D+ prostate epithelial cells, we cultured the sorted cells in the presence or absence of dihydrotestosterone (DHT) (Figure 4E). Strikingly, we found that LY6D+ cells from all three LSC gates formed organoids in an androgen-independent manner. Of note, LY6D+ prostate cells from the SCA1low/− and SCA1int gates formed significantly more organoids in the absence of DHT, compared to LY6D− subpopulations counterparts (Figure 4F). Moreover, the LY6D+SCA1high and low/− subpopulations formed increasing numbers of multipotent organoids that expanded under androgen deprivation (Figure 4G). In the organoids derived from sorted LY6D+ cells, IF analysis confirmed the presence of LY6D-expressing cells, which often overlapped with K5+p63+K8+ cells (Figures 4H and S4C). We also observed LY6D staining in organoids originated from sorted LY6D− cells, by both IF and FACS analyses (Figures 4H, S3D, and S3E). This observation suggests that the organoid culture condition, which favors growth of stem/progenitor cells, may also trigger expression of stem/progenitor marker genes (e.g., LY6D), as described previously for other stem/progenitor marker genes (e.g., SCA1 and TROP2) (Goldstein et al., 2008, Kwon et al., 2016).
To further characterize the CR properties of LY6D+ cells, we sorted LY6D+ LSC subpopulations from castrated mice. After 14 days in androgen-deprived conditions, we stimulated LY6D+ CR organoids with 100 nM DHT for 7 days. After androgen stimulation, these organoids exhibited a phenotype of enhanced luminal differentiation with increased luminal layers, strong nuclear expression of AR (in contrast to the cytoplasmic low expression of AR under the androgen-deprived conditions), and a slight reduction in Ki67+ proliferating cells (Figures 4I, 4K, and S4F). These data indicate that the organoid-forming properties of LY6D+ cells are maintained in castrated animals.
CR LY6D+ Cells in Luminal Lineage Have Regenerative Capacity
Our data suggest that LY6D+ cells are intrinsically resistant to androgen deprivation and have multi-lineage organoid-forming properties. To provide further evidence for this hypothesis, we analyzed the relative and absolute number of LY6D+ cells in HN and CR animals. As expected from our single-cell profiling (Figure 2B), the relative frequencies of LY6D+ cells were increased in prostates from castrated mice compared to those from HN mice (Figures 5A and 5B). Importantly, the absolute numbers of the total LY6D+ prostate cells from castrated mice remained largely unchanged compared to those from HN mice, indeed suggesting the presence of an intrinsically CR stem/progenitor cell population (Figure 5C). Co-IF staining of prostate sections from castrated mice revealed the presence of K8+ CR prostate cells that were also positive for both LY6D and K5. Similarly, such LY6D+K8+K5+ cells could also been found in HN mice (Figure S5A). To determine whether LY6D+ prostate cells in the luminal lineage possess stem/progenitor cell activity in vivo, we utilized a genetic marking approach to label the K8+ prostate luminal lineage and correlate luminal-lineage cells with LY6D expression (Figure 5D). For this, K8-CreER mice (Van Keymeulen et al., 2011) were crossed into a conditional Cre-reporter mouse strain Rosa26-loxPStoploxP-YFP (R26Y) (Srinivas et al., 2001) to generate K8-CreER;R26Y double-transgenic mice. Administration of tamoxifen to K8-CreER;R26Y mice transiently activates Cre-recombinase, leading to activation of the YFP reporter in K8-CreER-expressing cells and their progeny (Figure 5D). Upon tamoxifen, we found that YFP+ prostate epithelial cells from both HN and castrated mice were K8+ luminal, but not K5+ basal cells (Figure S5B). Consistent with previous luminal lineage-tracing results (Choi et al., 2012, Liu et al., 2011), we also observed a similar frequency of YFP-labeled luminal cells from HN or castrated mice (Figure S5C). Using this genetic model, we validated that LY6D+ YFP-marked luminal cells were indeed enriched in prostates from castrated mice compared to HN mice by FACS, as the majority (>75%) of YFP+ (K8+) CR luminal-lineage cells were LY6D+ (Figure 5E). IF staining of HN prostates confirmed that YFP+LY6D+ luminal cells were indeed rare in the distal regions of the prostate (Figures 5F and S5E), but abundant in proximal regions with high-level expression of nuclear AR (Figure S5D; compared to rare AR+ YFP+LY6D+ cells in the distal region, Figure S5E). YFP+LY6D+ luminal cells dramatically increased after castration both in proximal and distal regions (Figure 5F) and exhibited cytoplasmic AR expression (Figure S5E). Moreover, tamoxifen-induced K8-CreER;R26Y mice, resupplied with testosterone to induce prostate regeneration, exhibited a LY6D expression pattern similar to HN mice; that is, progeny of labeled LY6D+ luminal cells did not appear to retain LY6D expression after regeneration, while retaining the YFP label. Only rare LY6D+ cells were observed in the regenerated prostate luminal epithelium (Figures 5F and S5E). Quantification of percentages of YFP+LY6D+ cells in all YFP-marked cells in prostate sections from HN, castrated, and regenerated mice further confirmed the dynamics of the YFP+LY6D+ luminal population (Figure 5G). Together, these results suggest that CR LY6D+ luminal-lineage cells are derived from enriched, pre-existing LY6D+ luminal-lineage cells in HN mice, although we cannot fully exclude the possibility of a LY6D− luminal cell of origin (upon castration-induced upregulation of Ly6d expression). Sorting of YFP+ cells from HN mice further confirmed the organoid-forming, multi-lineage potential of luminal LY6D+ cells in the absence or presence of androgen (Figures S5F and S5G).
Figure 5.
LY6D Marks CR Luminal Cells that May Possess Regeneration Capacity
(A) FACS plots comparing LY6D staining of total Lin− and LY6D+ or − SCA1High or int or Low/− prostate cells from hormone-naive (HN) and castrated (CR) males.
(B) Quantification of LY6D expression profiles based on FACS subpopulations as in (A) (HN versus castrated). The average of frequency of the indicated populations is from five mice. p value: ∗p < 0.05. Error bars represent mean ± SEM.
(C) Quantification of the absolute numbers of LY6D+ or − subpopulations based on FACS subpopulations as in (A) (HN versus castrated). The average of frequency of the indicated populations is from five mice. p value: ∗∗p < 0.01. n.s., non-significant. Error bars represent mean ± SEM.
(D) Schematic diagram showing genetic marking by YFP via tamoxifen-induced activation of CreER.
(E) Representative FACS plots showing LY6D staining from tamoxifen-induced K8-CreER;R26Y prostates for Lin− or Lin−YFP+ populations. Tamoxifen induction performed on HN or fully regressed males (after castration) (CR).
(F) Representative IF staining results showing YFP and LY6D staining, as well as the co-IF patterns in HN, castrated, and regenerated prostates from K8-CreER;R26Y mice after tamoxifen. Nuclei, DAPI. Arrows depict YFP+LY6D+ double-positive cells. Scale bars: 50 μm.
(G) Quantification of the percentage of LY6D+YFP+ per total YFP+ cells from tamoxifen-induced K8-CreER;R26Y prostates. The average of frequency of the indicated populations is from three mice. Error bars represent mean ± SEM.
(H) Representative subcutaneous outgrowths from FACS-sorted YFP-marked prostate cells separated as LY6D+ and LY6D− subsets. Note engrafted YFP+LY6D+ cells could generate much larger outgrowth composed of both K5+ (red) and K8+ (green) prostate cells. Scale bars: 50 μm. See also Figures S5H and S5I.
See also Figure S5.
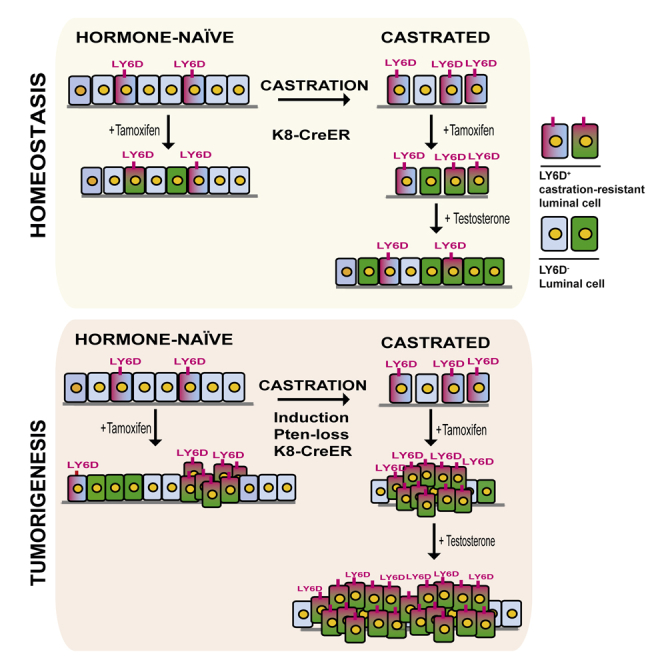
Our data suggest that, upon regeneration, LY6D+ CR luminal-lineage cells can give rise to LY6D− daughter cells. In order to evaluate this hypothesis, we performed a prostate epithelial reconstitution assay (Lukacs et al., 2010) for FACS-sorted subsets of prostate cells from tamoxifen-induced K8-CreER;R26Y males. Compared to YFP+LY6D− cells, which only formed very small outgrowths (1–2 ducts/explant), YFP+LY6D+ cells formed large ductal outgrowths (5–7 ducts/explant) composed of K8+ luminal cells surrounded by K5+ basal cells, resembling normal prostate ducts, as well as K8+ luminal-only ducts (Figures 5H and S5I). In situ Ly6d expression analysis on explants derived from YFP+LY6D+ cells confirmed the presence of luminal cells negative for Ly6d intermingled with Ly6d-expressing prostate cells (Figure S5H).
Collectively, our data show that LY6D is a marker of LP cells, which are intrinsically CR and possess prostate regenerative capacity.
LY6D Correlates with PCa Development from Luminal Lineage
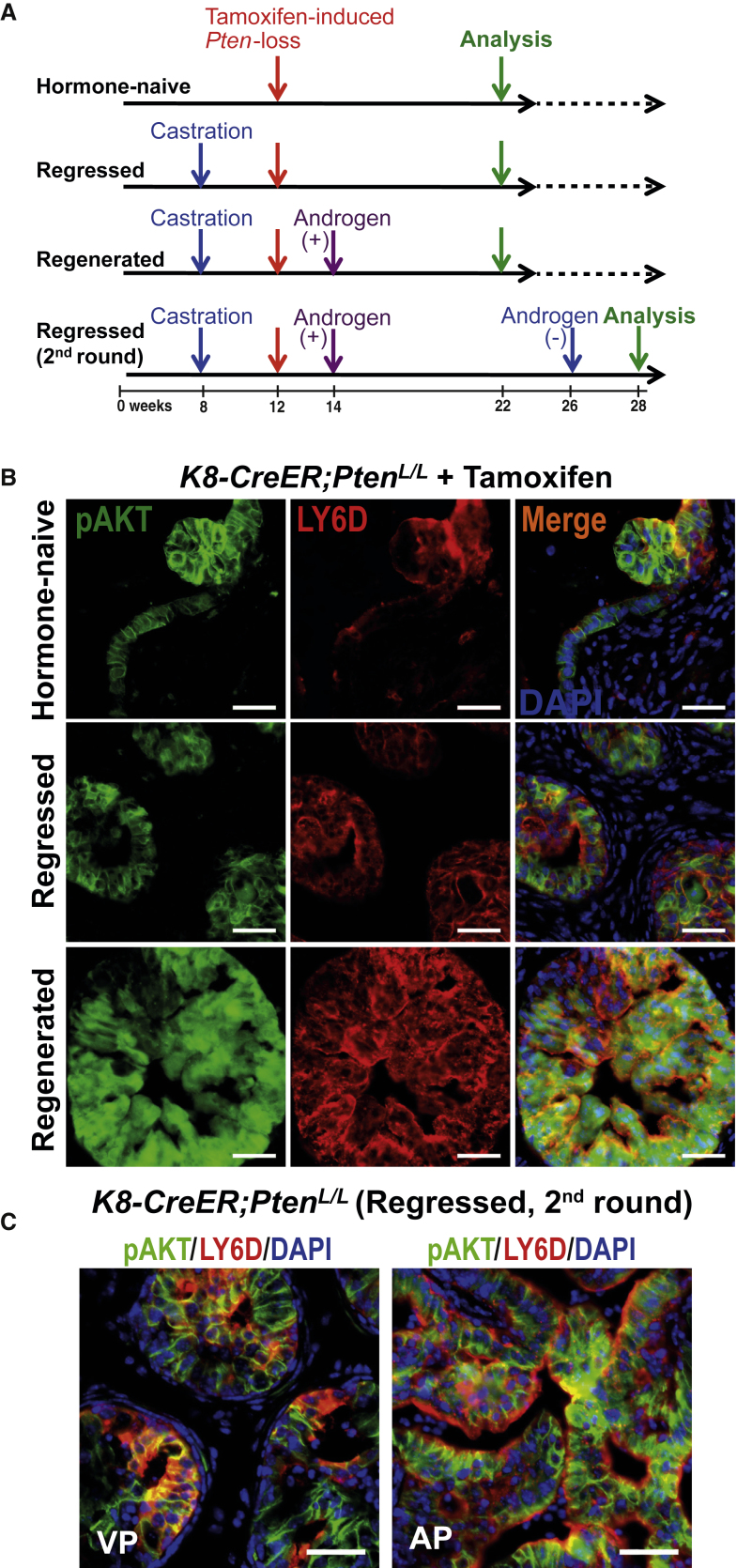
To determine whether LY6D+ prostate luminal cells contribute to PCa initiation, we utilized an inducible PCa murine model initiated from K8+ luminal-lineage cells upon Pten loss (K8-CreER;PtenL/L; Figure 6A). PCa initiation and progression in all induced K8-CreER;PtenL/L males was tracked by immunostaining for phosphorylated AKT (pAKT), as pAKT can be detected in Pten-null cells even before the onset of prostatic intraepithelial neoplasia (PIN) lesions (Figure S6A) (Shappell et al., 2004). This allows visualization of both “primed” preneoplastic and progressive lesions. In HN K8-CreER;PtenL/L males, loss of Pten in luminal-lineage cells led to development of pAKT+ preneoplastic lesions and low-grade PINs (LG-PINs) (Ittmann et al., 2013, Park et al., 2002), which were largely LY6D+ (Figure 6B). Co-IF staining showed that the majority of these LY6D+ cells were indeed K8+ (Figure S6B). In castrated K8-CreER;PtenL/L males, Pten inactivation in CR K8+ luminal-lineage cells led to the development of PIN lesions in all lobes with an abundancy of pAKT+LY6D+ cells (Figure 6B). Co-IF staining revealed that the LY6D+ cells included both K8+ and K8+K5+ cells (Figure S6B). Following testosterone replacement in castrated K8-CreER;PtenL/L males with Pten inactivation, the regenerated prostate epithelia predominantly exhibited large areas of pAKT+LY6D+ high-grade PIN (HG-PIN) (Ittmann et al., 2013, Park et al., 2002, Shappell et al., 2004) lesions (Figure 6B). Co-IF staining showed that the majority of these LY6D+ cells were K8+, with a smaller subset of K8/K5 double-positive cells (Figure S6B). Such HG-PIN lesions were maintained in prostates undergoing a second round of regression (i.e., CR HG-PINs; Figure 6C).
Figure 6.
LY6D+ Prostate Cells Are Involved in Prostate Cancer Initiation and Progression
(A) Experimental scheme for the lineage-tracing experiment in (B) and (C) showing the time points when K8-CreER;PtenL/L males were castrated, injected with tamoxifen (to induce Pten inactivation), regenerated with androgen, and analyzed.
(B) Co-IF staining of HN, regressed, and regenerated K8-CreER;PtenL/L prostates showing overlap of pAKT (green) and LY6D (red) in HG-PIN lesions. Nuclei, DAPI (blue). Scale bars: 50 μm.
(C) Co-IF staining showing abundant pAKT+ LY6D+ prostate cancer cells in CR HG-PIN lesions observed in K8-CreER;PtenL/L mice at the second round of regression, with Pten inactivation induced upon surgical castration (i.e., first round regression). VP, ventral prostate; AP, anterior prostate. Scale bars: 50 μm.
See also Figure S6.
Next, we examined AR expression in the K8-CreER;PtenL/L model. In prostates from castrated K8-CreER;PtenL/L males in which Pten inactivation occurred at the regressed stage, we observed abundant LY6D+ cells expressing cytoplasmic AR (Figure S6C), a staining pattern similar to that of castrated WT mice (Figure S5E). Upon regeneration, K8-CreER;PtenL/L prostate lesions were composed mainly of LY6D+ luminal cells with strong nuclear AR expression (Figure S6C), while in the regenerated prostates of WT mice, LY6D+AR+ cells were rare (Figure S5E). Taken together, these findings suggest that LY6D expression correlates with PCa initiation and progression to CR growth from the luminal lineage.
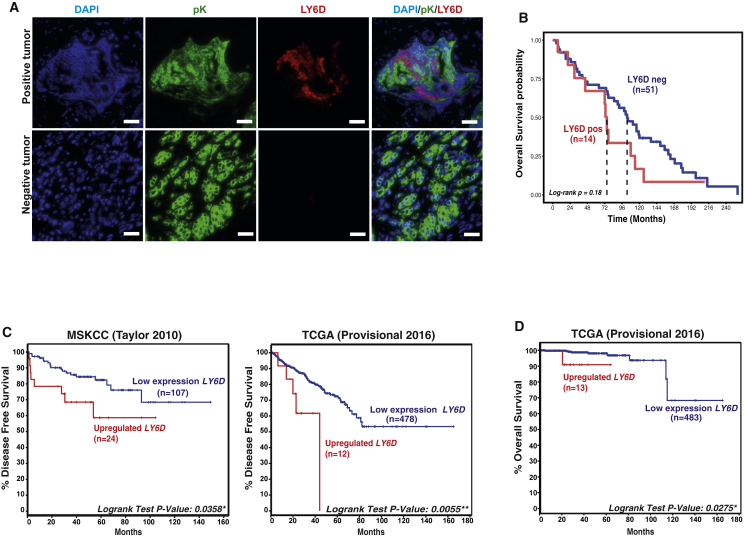
LY6D Correlates with Advanced PCa in Patients
Encouraged by these results, we investigated whether LY6D expression correlates with PCa patient outcomes. First, LY6D protein expression was assessed in tissue microarrays from the Christie Hospital collection (containing 2–3 tumor cores and 1–2 normal-adjacent cores per patient). Analyses of samples from 65 patients indicated that LY6D-positive cases exhibited a trend toward worse overall survival, although statistical significance was not reached, possibly due to a small sample size (Figures 7A, 7B, and S7A). To obtain additional evidence for correlation of LY6D expression with advanced PCa, we analyzed several publicly available genomic datasets. We first analyzed the well-annotated Memorial Sloan Kettering Cancer Center (MSKCC) (Taylor et al., 2010) and The Cancer Genome Atlas (TCGA) (Cancer Genome Atlas Research Network, 2015) cohorts and observed significant association of biochemical relapse of patients with upregulated LY6D expression (Figure 7C); in contrast, correlation of upregulated expression of several known stem/progenitor markers (e.g., TROP2/TACSTD2 [Goldstein et al., 2008] and CD166/ALCAM [Jiao et al., 2012]) with patient outcomes is not significant or exhibits conflicting levels of significance between the MSKCC (Taylor et al., 2010) and TCGA (Cancer Genome Atlas Research Network, 2015) cohorts (Figure S7B). Of note, PCa patients with high levels of LY6D showed a reduced overall survival (Figure 7D), despite the relatively short follow-up of PCa patients in the TCGA cohort. Moreover, an inferior clinical outcome of patients with LY6D expression was maintained when we restricted our analyses only to patients with Gleason score 7 (GS7) (Figure S7C).
Figure 7.
LY6D Is Associated with Advanced Human PCa
(A) Representative IF staining results showing pan-Keratin (pK) (epithelial marker) and LY6D staining in human prostate cancer samples. Scale bars: 20 μm.
(B) Kaplan-Meier curve of tissue microarray showing association of LY6D positivity (based on protein) with patient outcomes. The red line depicts patients with positive LY6D (LY6Dpos) expression (n = 14), whereas the blue line patients with negative LY6D (LY6Dneg) expression (n = 51).
(C) Kaplan-Meier curve of human prostate cancer (PCa) cohorts (Cancer Genome Atlas Research Network, 2015, Taylor et al., 2010) analyzing time to biochemical recurrence from diagnosis. The red line depicts patients with high LY6D expression (mRNA [z score > 1.3]), whereas the blue line patients with low LY6D expression. Patients with missing disease-free survival status in TCGA; n = 6 are excluded.
(D) Kaplan-Meier curve of human TCGA PCa cohort (Cancer Genome Atlas Research Network, 2015) analyzing patients’ overall survival.
See also Figure S7.
Comparative copy number analysis revealed that LY6D amplification is much more frequent in metastatic CRPC patient samples than in primary prostate tumors or non-castrated metastatic cancers, and is particularly high in CRPC metastatic patients with neuroendocrine phenotype (Figure S7D). Since LY6D maps ∼16 Mb apart from MYC in the chromosome 8q24 region, most of the cases patients showed a co-amplification of LY6D and MYC. To exclude the possibility that the association of LY6D expression with aggressive PCa is related to MYC amplifications, we examined the association of LY6D and MYC mRNA expression and disease outcomes, in the above-analyzed MSKCC and TCGA cohorts. We did not observe a correlation between MYC and LY6D mRNA expression (Figure S7E). In addition, comparison of patients with high to those with low MYC mRNA expression levels did not show differences in the frequency of biochemical relapses (Figure S7F). Moreover, we also found no correlation between MYC mRNA expression or amplifications and LY6D mRNA expressions in primary PCa tumors (Figures S7E–S7G), suggesting that the negative prognosis of patients with LY6D+ PCa is not attributed to MYC amplifications.
Taken together, data from both animal models and patient cohort analyses suggest that LY6D+ luminal cells are intrinsically androgen-resistant and possess stem/progenitor cell properties, features that may contribute significantly to prostate carcinogenesis. Furthermore, LY6D may be used as a prognostic marker of aggressive disease and disease recurrence, and may serve as a predictive marker for development of CRPC.
Discussion
Our single-cell expression analysis reveals molecular heterogeneity in the prostate luminal and, to a lesser degree, basal lineages. We found that a subset of prostate cells in the luminal lineage co-express multiple basal (e.g., Krt5, Krt14, Trp63) and luminal (e.g., Krt8, Krt18, and Ar) markers together with prostate stem/progenitor marker genes (e.g., Ly6d, Trop2, Sca1, Cd133, Cd166). Many of these stem/progenitor genes are found within a bi-lineage gene set expressed in both basal cells and a portion of luminal cells. Intermediate prostate cells have been associated with luminal differentiation of basal cells (Ousset et al., 2012, Wang et al., 2009, Wang et al., 2013). Whether these cells are uniquely derived from basal cells or from luminal committed progenitors remains an open question. An important observation from our study is that many CR prostate cells in the luminal lineage exhibit a similar bi-lineage expression signature to that of intermediate cells, raising a possibility that such intermediate cells may be intrinsically CR.
We identified LY6D as a marker that links intermediate cells to prostate stem/progenitor cells and CR prostate cells. From our organoid culture analysis, we observed LY6D+p63+K8+, as well as LY6D+p63−K8+, and LY6D+p63+K8− cells, suggesting that cells from these LY6D+ subpopulations contain multipotent progenitor cells. Further supporting this, we have observed LY6D+K5+p63+K8+ cells in a subset of multipotent organoids derived from LY6D+ subpopulations, although we cannot entirely rule out the possibility that differentiated progenitors derived from those multipotent cells temporally maintain the expression of LY6D.
Our in vitro and in vivo data also support that LY6D+ cells in the luminal lineage represent multipotent and/or unipotent LPs inherently resistant to androgen deprivation and with regenerative capacity. We found that LY6D+ prostate cells from the SCA1high and SCA1low/− gates were enriched with organoid-forming multipotent LPs and formed increasing numbers of multipotent organoids in the absence of androgen, which has been further validated using organoid outgrowth of luminal YFP+LY6D+ cells. In addition, CR LY6D+ cell-derived organoids underwent nuclear AR localization and luminal differentiation upon androgen stimulation. Consistent with our observations, it was reported that a label-retaining AR+ progenitor population that expresses multiple prostate stem/progenitor markers was present in the castrated prostate and exhibited sign of expansion following castration (Shi et al., 2014). However, we also cannot entirely exclude the possibility that luminal cells can also adapt to castration and subsequently upregulate expression of some of the stem/progenitor genes (e.g., Ly6d) (Zhang et al., 2016).
LY6D is a gene with no established role in prostate development or cancer. It is a member of the Ly6/uPAR family characterized by their roles in cell proliferation, cell-cell interaction, immune cell maturation, and cytokine production, which are all essential components of tumor initiation and progression (Loughner et al., 2016). Under physiological conditions, LY6D has been used as a marker of early B cell specification (Inlay et al., 2009), although its role in B cell development is unclear. In malignancies, however, LY6D expression is induced on the surface of various cancer cells after genotoxic stress, which was proposed as a marker to stratify patients with increased risk for developing distant metastases (Kurosawa et al., 2012, Rubinfeld et al., 2006). Consistent with this notion, a recent study revealed positive correlations of increased expression of multiple Ly6 family members, including LY6D, with poor patient outcomes in multiple cancer types (Luo et al., 2016). LY6D plays an important role in adhesion of head and neck cancer cells to endothelial cells, which is essential during the metastatic extravasion process (Eshel et al., 2002). Disruption of its closely related members (i.e., LY6E/K) in human breast cancer cell lines has been reported to modulate transforming growth factor β (TGF-β) signaling (AlHossiny et al., 2016). However, in PCa, the functional role of LY6D for tumorigenesis and tumor maintenance, if any, remains unknown and awaits further investigations.
A major challenge in the management of patients with PCa is the distinction of indolent from aggressive disease. Hence, markers to predict clinical outcomes are urgently needed. Human LY6D is located on chromosome 8q24, a region that is frequently amplified in PCa and contains numerous risk loci (Brakenhoff et al., 1995, Nakagawa et al., 2012). Analysis of human PCa cohorts revealed that higher LY6D expression levels, after removing cases with MYC amplification or potential MYC overexpression (due to close distance between these two genes and amplification of this genomic region), appeared to be associated with more aggressive disease and worse outcomes, suggesting that LY6D may serve as a prognostic biomarker for advanced PCa. Further studies are warranted to determine the precise role of LY6D in prostate epithelial heterogeneity, PCa initiation, and progression to adenocarcinoma, to validate its utility as a biomarker for patient stratification and to assess the impact of CR LY6D+ cells as therapeutic targets for patients with lethal PCa.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal, CK5 | Covance | Cat#PRB-160P |
| Mouse monoclonal, 1E8 CK8 | Covance | Cat#MMS-162P |
| Guinea pig, polyclonal, CK8 | Abcam | Cat#ab194130 |
| Chicken polyclonal, YFP | Abcam | Cat#ab13970 |
| Rabbit monoclonal, D5.1 GFP | Cell Signaling | Cat#2956 |
| Rabbit polyclonal, AR | Santa Cruz | Cat#sc-816 |
| Rabbit polyclonal, KI67 | Bethyl Laboratories | Cat#IHC-00375 |
| Mouse monoclonal, 4A4, p63 | Millipore | Cat#MAB4135 |
| Rabbit polyclonal, aSMA | Abcam | Cat#ab5694 |
| Rabbit monoclonal, 736E11 pAKT | Cell Signaling | Cat#3787 |
| Rabbit monoclonal, D9E pAKT | Cell Signaling | Cat#4060 |
| Rabbit monoclonal, EP1601Y, CK5-AF647 | Abcam | Cat#ab193895 |
| Rabbit monoclonal, EP1628Y, CK8-AF488 | Abcam | Cat#ab192467 |
| Rat monoclonal, 49-H4, LY6D | BD | Cat#BD-557360 |
| Rabbit polyclonal, LY6D | Sigma | Cat#HPA024755 |
| Mouse monoclonal, AR441, AR | DAKO | Cat#M3562 |
| Mouse monoclonal, C-11, pan-cytokeratin | Sigma | Cat#C2931 |
| Rabbit monoclonal, EP1601Y, CK5 | Abcam | Cat#ab52635 |
| Rabbit monoclonal, EP1628Y, CK8 | Abcam | Cat#ab53280 |
| Mouse monoclonal, 4A4, p63 | Abcam | Cat#ab735 |
| Rabbit monoclonal, D5.1, GFP | Cell Signaling | Cat#2956 |
| RNAscope® 2.5 LS Probe_Mm-Ly6d | ACD | Cat#532071 N/A |
| RNAscope® 2.5 LS Probe- Hs-LY6D | ACD | Cat#484688 |
| SCA1-PE | eBioscience | Cat#12-5981 |
| SCA1-PE-Cy7 | eBioscience | Cat#25-5981 |
| CD49f-APC | eBioscience | Cat#17-0495 |
| LY6D-PE | eBioscience | Cat#12-5974 |
| CD133-FITC | eBioscience | Cat#11-1331 |
| CD24-FITC | eBioscience | Cat#11-0242 |
| CD166-PE | eBioscience | Cat#12-1668 |
| SCA1-APC-Cy7 | Biolegend | Cat#108126 |
| CD31 | Thermo Fisher Scientific | Cat#13-0311/480311 |
| CD45 | Thermo Fisher Scientific | Cat#13-0451/480451 |
| TER119 | Thermo Fisher Scientific | Cat#13-5921/485921 |
| Mouse TROP-2 Biotinylated Antibody | R&D Systems | Cat#BAF1122 |
| Streptavidin-PE Cy7 | eBioscience | Cat#25-4317 |
| Streptavidin-APC | eBioscience | Cat#17-4317 |
| Streptavidin-eFluor 450 | eBioscience | Cat#48-4317 |
| Mouse Monoclonal Anti-Cytokeratin, PanCK | Sigma | Cat#C2931 |
| Biological Samples | ||
| Tissue-microarray, FFPE transurethral resection of the prostate (TURP) samples, patient specimens | MCRC Biobank | Bioethics 10_NOCL_02; #14ESBA01MTA |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DAPI | Sigma | Cat#D9542-10MG |
| DAPI | Life Technologies | Cat#D3571 |
| BD Perm/Wash buffer | BD Biosciences | Cat#554723 |
| Tamoxifen | Sigma | Cat#T5648 |
| Matrigel | Corning/BD | Cat#354230 |
| BME-2 | AMSBIO | Cat#3533-010-02 |
| B-27 | Life Technologies | Cat#17504-044 |
| EGF | PeproTech | Cat#AF-100-15 |
| R-spondin 1 | R&D Systems | Cat#4645-RS-025 |
| Noggin | PeproTech | Cat#120-10C |
| Y-27632 dyhydrochloride | Sigma-Aldrich | Cat#Y0503 |
| A83-01 | Tocris Bioscience | Cat#2939 |
| Dihydrotestosterone DHT | Sigma | Cat#730637 |
| TrypLE | Life technologies | Cat#12563011 |
| Richard-Allan Scientific HistoGel | Thermo Scientific | Cat#HG-4000-012 |
| Tissue-Tek® O.C.T. Compound | Sakura® Finetek | Cat#4583 |
| Opal 7-Color Automation IHC Kit | Perkin Elmer | Cat#NEL794001KT |
| Goat serum | DAKO | Cat#X0907 |
| Hoechst 33342 | Thermo Fisher | Cat#R37605 |
| Live/Dead cells exclusion dye | Thermo Fisher | Cat#L10119 |
| Collagenase, Type I, powder | GIBCO | Cat#17018-029 |
| Dispase | Life Technologies | Cat#17105041 |
| Critical Commercial Assays | ||
| GeneChip Mouse Gene 2.0 ST Array | Affymetrix/Thermo Fisher | Cat#902119 |
| NORGEN RNA clean-up and Concentration Micro kit | Norgen | Cat#236000 |
| Lexogen QuantSeq 3′ mRNA-Seq Library Prep Kit | FWD | Cat#015 |
| Deposited Data | ||
| Microarray data | This paper | GSE92473 |
| RNA-seq data | This paper | GSE92622 |
| Experimental Models: Organisms/Strains | ||
| C57BL/6 | The Jackson Laboratory | 000664 | Black 6 |
| Oligonucleotides | ||
| Primers for β-actin gene, ACTB, Forward: TTCACCACCACAGCTGAGAG; Reverse: ATAGTGATGACCTGGCCGTC | Sigma | N/A |
| Primers used for single cell expression profiling: see Table S5. | N/A | N/A |
| Software and Algorithms | ||
| ImageJ/Fiji software | NIH Image | https://fiji.sc |
| Columbus Image Analysis system | Perkin Elmer | http://www.perkinelmer.com/product/image-data-storage-and-analysis-system-columbus |
| FlowJo | FlowJo LLC | https://www.flowjo.com/solutions/flowjo |
| SPADE algorithm | https://www.nature.com/articles/nbt.1991 | https://www.nature.com/articles/nbt.1991 |
| Graphpad Prism version 6 | GraphPad Software | https://www.graphpad.com |
| Definiens Tissue Phenomics Software | Definiens | https://www.definiens.com |
| Multiple Experiment Viewer (MeV) | Center for Cancer Computational Biology (CCCB), Dana-Farber Cancer Institute | http://www.tm4.org |
| GenePattern | Broad Institute of MIT & Harvard | http://www.broadinstitute.org/cancer/software/genepattern/ |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Esther Baena (esther.baena@cruk.manchester.ac.uk).
Experimental Model and Subject Details
Animal models
For animal studies, K8-CreER transgenic mouse (Van Keymeulen et al., 2011), R26Y reporter mouse (C57BL/6) (Srinivas et al., 2001), and Pten conditional knockout mouse (PtenL/L) (C57BL/6) (Lesche et al., 2002) mice were obtained from The Jackson Laboratory (JAX). Surgical castrations and subcutaneous implantation of 60-day release 12.5mg testosterone pellets (Innovative Research of America) were performed on mature males (at least 8 weeks of age). Lineage tracing was performed by intraperitoneal injection of 100μl tamoxifen (Sigma, 20mg ml-1). All animal procedures were approved by the corresponding Institutional Animal Care and Use Committees (IACUCs) or Animal Welfare and Ethical Review Bodies (AWERB) of the CRUK Manchester Institute, in accordance with ARRIVE guidelines and National Home Office regulations under the Animals (Scientific Procedures) Act 1986. Tissues used for histological analysis were fixed in 10% formalin (Fisher Scientific) for 1 hr, washed in PBS, then saturated in 30% sucrose overnight at 4°C. Organoids were fixed in 2% PFA for 20mins, then washed in PBS, and saturated in sucrose. Tissues and organoids were then embedded in OCT compound (Sakura) and stored at −80°C prior to cryosectioning. Cryosections were subsequently used for immunofluorescence (IF) and immunohistochemistry (IHC) staining.
Patients Tissue microarrays (TMA)
The Manchester Cancer Research Centre (MCRC) human prostate tissue collection was constructed with archival transurethral resection of the prostate (TURP) samples from prostate cancer patients collected within the Salford NHS Foundation Trust with informed subject consent for construction of tissue microarrays [built by the Genito[HYPHEN]Urinary cancer research group at the Christie Hospital (Manchester, UK)], and under the approval from the MCRC Biobank Review Board (10_NOCL_02). The section of TMA slides for the evaluation of the LY6D protein expression by multiplexed IF staining was assessed by the MCRC Biobank Review Board (14_ESBA_01). All patients evaluated in the current study presented with localized prostate cancer at diagnosis (M0 patients).
Organoid-formation assay
In vitro organoids formation were performed as described below [and also reported in (Chua et al., 2014, Karthaus et al., 2014)]. Mouse prostate single cells suspensions were isolated by digestion of mouse prostate with collagenase/dispase (1mg ml-1) for 1h at 37°C. Single cells suspensions were embedded in growth factor reduced Matrigel (Corning/BD, 354230) or BME-2 (AMSBIO, 3533-010-02) and plated a 40 μL drop/well (1,000-3,000 cells/drop) in the center of 96-well optical bottom plates. Organoid culture medium was composed of ADMEM/F12 medium with B27 (Life Technologies, 17504-044), 50mg ml-1 EGF (PeproTech, AF-100-15), 500ng ml-1 recombinant R-spondin 1 (R&D Systems, 4645-RS-025), 100ng ml-1 Noggin (PeproTech, 120-10C), 10μM Y-27632 dyhydrochloride (Sigma-Aldrich, Y0503), 200nM A83-01 (Tocris Bioscience, 2939) and plus/minus 1nM Dihydrotestosterone (DHT, Sigma 730637); as described in (Chua et al., 2014, Karthaus et al., 2014)); the medium was changed once every 4 days and organoids outgrowths were counted with a Zeiss Axiovert 200M inverted microscope at 10X magnification approximately 7-9 days post-seeding. Organoids were measured using AxioVision 4.9 image acquisition software. Cells from organoids used for serial passages were recovered from Matrigel or BME2 with Cell Recovery Solution (Corning, 354253), dissociated into single cells with TrypLE (Life technologies, 12563011), and replated. Murine prostates of hormone-naive and castrated mice were dissected and dissociated into prostate single cells and FACS sorted for epithelial LY6D+/− SCA1high or int or low/- subpopulations. For regeneration assays, each sorted subpopulation from HN or castrated mice was seeded in the absence of DHT for 14 days, then the growing media was supplemented with DHT (100nM) for up to 7 days. Organoids were imaged and counted at day 7 and 14 organoids under androgen-deprived conditions, as described above. Experimental endpoint was 7 days after adding DHT to the media, when organoids were imaged and fixed.
Prostate regeneration assay
Prostate regeneration assay was performed as previously described (Lukacs et al., 2010). Briefly, after FACS, the sorted YFP+ LY6D+ or - luminal cells were seeded in PrEGM (Lonza, Cat# CC-3165) with Matrigel for 9-10 days. Cells were then collected, and approximately 2.5 × 105 cells were mixed with 2.5 × 105 Urogenital sinus mesenchyme (UGSM) cells and subcutaneously injected with Matrigel into flanks of Rag2−/− mice. Outgrowths were collected 8 weeks post implantation.
Method Details
IF and IHC staining and histology
Cryosections of prostate tissues were cut at 8μm, blocked in 2.5% goat serum, and incubated with primary antibodies overnight at 4°C. The Matrigel drops containing organoids were embedded in histogel (Thermo Scientific, Richard-Allan Scientific HistoGel) or suspended in PBS as previously described (Broutier et al., 2016). They were then fixed in 4% PFA overnight at 4°C, and embedded in paraffin following standard protocols. For frozen sections, organoids were fixed in 2% PFA for 20min at room temperature, washed in PBS, and saturated in 30% sucrose overnight at 4°C. Tissues were then embedded in OCT compound (Sakura) and stored at −80°C prior to cryosectioning. FFPE- or cryo- sections (5 μm) were subsequently used for IF and IHC staining. Antibodies were used to detect Keratin 5 (K5, Covance PRB-160P; Abcam # ab193895), Keratin 8 (K8, Covance MMS-162P; Abcam ab194130; Abcam # ab193895), YFP (Abcam 13970), LY6D (BD 557360), AR (Santa Cruz sc-816), KI67 (IHC-00375, Bethyl Laboratories), p63 (Millipore clone4A4 MAB4135) or pAKT (Cell Signaling 4060). Alexa Fluor-conjugated secondary antibodies (Life Technologies) were incubated for 1 hr at room temperature. Nuclei were counterstained with DAPI and slides were sealed with Vectashield mounting media (Vector). A similar IF staining procedure was also performed for FACS-sorted cells cytospin onto glass slides. For multiplexed staining, the Opal protocol (Perkin Elmer, NEL794001KT) was used following the manufacturers’ instructions. Briefly, the slides were deparaffinized in xylene and rehydrated in ethanol. Antigen retrieval was performed in citrate buffer (pH 6.0) in microwave oven. Primary rabbit antibodies for K5 (1:1000) were incubated for 1 h in a humidified chamber at room temperature, followed by detection using the rabbit Detection HRP kit. Visualization of K5 was accomplished using fluorescein TSA Plus (1:50). In serial fashion, the slides were then retrieved in citrate buffer (pH 6.0) and then incubated with primary antibodies for K8 (1:1000), and Ki67 (1:1000). The in situ hybridization to detect Ly6d (RNAscope probe Ms-Ly6d, ACD #532071) was done following the manufacturers’ instructions. Briefly, FFPE sections were deparaffinized in xylene and rehydrated in ethanol, then incubated with protease K for 15min. The sections were then washed, and hybridized with the Ly6d probe followed by immunostaining with K8 antibody (1/200, ab53280) for 1h at room temperature. Cryosections used for histological examination (H&E stained) and IHC staining were mounted in Permount mounting media (Fisher). Quantification of IF staining was performed by ImageJ software (NIH Image, Maryland, USA).
For whole-mount IF staining, organoids were dissociated from the 3D matrix (Matrigel or BME 2) and fixed with PFA 4% for 20 min on ice. After washing in PBS, organoids were permeabilized with PBS-T (0.4% Triton X-100 in PBS) for 15 min and blocked using 10% goat serum (Goat serum, DAKO #X0907) for 1h at room temperature. Primary antibodies staining for K5 (1/100, ab193895), K8 (1/100, ab192467) and LY6D (1/100) was performed by overnight incubation at 4°C. Next day organoids were washed and incubated with the fluorescently labeled secondary antibody for 2-4 hr at room temperature. Nuclei were counterstained with DAPI (2 μg/ml, Sigma #D9542-10MG). IF images were acquired using high content screening system Opera Phenix (Perkin Elmer) and evaluated with the Columbus Image Analysis system.
FACS cell sorting
FACS analyses and sorting were performed as described in (Lukacs et al., 2010). Mouse prostate single cells suspensions were isolated by digestion of mouse prostate with collagenase/dispase (1mg ml-1) for 1 hr at 37°C followed by 0.25% Tripsin/EDTA (2min at 37°C) and Dispase/DNaseI (5mg/ml Dispase, 1U/μL DNase, incubation for 5min at 37°C) treatments. Dissociated prostate epithelial cells were stained with specified fluorochrome-labeled antibodies (eBioscience) for 15 min on ice, washed, filtered (0.50μm cell strainer, Corning) and analyzed and/or sorted using BD FACS Aria II/III flow cytometers. FACS analysis was performed using FlowJo CE software. Sorting based on DAPI, Lineage (Lin: CD31, CD45, TER119), SCA1, and CD49f was used to separate viable prostate epithelial cells from stromal cells (Goldstein et al., 2008, Lawson et al., 2007). Intracellular K5 and K8 analysis were performed as previously described in (Goldstein et al., 2010). Briefly, following staining with cell surface markers, prostate cells were fixed with 1% PFA, permeabilised (BD Perm/Wash buffer, BD Biosciences #554723), incubated with K5-AF645 (Abcam, #ab52635) or K8-AF488 (Abcam, ab53280) for 30min on ice, washed and analyzed using BD Fortessa. DAPI (Life Technologies, D3571) was used for cell viability. Antibodies used for FACS were purchased from eBiosciences (San Diego, CA) and included SCA1-PE (12-5981), SCA1-PE-Cy7 (25-5981), CD49f-APC (17-0495), LY6D-PE (12-5974), CD133-FITC (11-1331), CD24-FITC (11-0242) or CD166-PE (12-1668). SCA1-APC-Cy7 (108126) was purchased from Biolegend (San Diego, CA). Biotinylated or e-Fluor-450 conjugated CD31, CD45, TER119 (13-0311/48-0311, 13-0451/48-0451, 13-5921/48-5921) or biotinylated TROP2 (R&D BAF1122) were also used in combination with streptavidin-PE Cy7 (25-4317), -APC (17-4317) or -eFluor 450 (48-4317).
Single cell sorting quality assessment
Mouse prostate single-cell suspension was filtered and stained with Hoechst 33342 (ThermoFisher, R37605) and dead cells exclusion dye (Thermo Fisher, L10119) according to manufacturer’s protocol. Doublets were excluded by successive gating of live cells by FSC-H versus FSC-A and SSC-A versus SSC-W plot. Live single cells (Hoechst+) were sorted into 96-well optical-bottom plates (Nunc, 152036). Plates were scanned at the focal plane on Opera Phenix High Content Screening System (HH14000000). Two 96-well plates from independent sortings were analyzed using the Columbus 2.3 Image Analysis System (Perkin Elmer).
Single cell expression profiling and data analysis
Multiplexed primer design, pre-amplification, and high-throughput microfluidic real-time PCR were carried out as described in (Guo et al., 2013) and detailed below. Mouse prostate single cells suspensions were filtered and doublets were gated out by FSC-H versus FSC-A and subsequently SSC-W versus SSC-A plot in FACS, a strategy that leads to a very low doublet rate. We then FACS-sorted prostate cells of interest into 96-well plates. After single-cell sorting, we ran RT-qPCR for the β-actin gene in each sorted single cells. We selected those cells with β-actin CT values within 10-17 cycles in the qRT-PCR reaction, as different single cells have different RNA contents. Cells with β-actin CT values outside of this range were considered to be cell clumps or degraded cells and were discarded. This strategy allowed us to assure one single cell per well for further single cell analysis. Hierarchical clustering and heatmaps were generated using MeV (Saeed et al., 2003) with default parameters, and as described (Guo et al., 2013). SPADE analysis was performed as described below (and also in (Guo et al., 2013)). ∼300 genes were divided into 23 gene sets of similar expression patterns by hierarchical clustering. The gene set-averaged expression data were analyzed by using the SPADE algorithm (Qiu et al., 2011). Raw data form MSKCC (GSE21032) and TCGA (dbGap: phs000915.v1.p1) cohorts were downloaded and patients were subdivided based on their LY6D levels. Statistical significance of Gleason score, T stage and Lymph node positivity was calculated using the Student t test.
Gene expression profiling and data analysis
For RNA-seq, RNA isolation from two biological replicates was performed using NORGEN RNA clean-up and Concentration Micro kit (Norgen, 236000) following manufacturer’s instructions. Lexogen QuantSeq 3′ mRNA-Seq Library Prep Kit (FWD, Cat. No. 015) for Illumina was used for the construction of sequencing libraries from 10-20 ng of total RNA, and then sequenced in Illumina NextSeq500 at the CRUK-Manchester Institute Molecular Biology Core facility. The fastq read files for the mouse QuantSeq sequencing were trimmed with BBDUK from the BBMAP tools software, to remove the first 12 bases of each read and to remove contaminates as suggested by the lexogen documentation, trailing polyG and polyA tails were removed with fqtrim and cutadapt. After trimming the reads were aligned to the mouse MM38 genome reference downloaded from ENSEMBL with STAR (Dobin et al., 2013). The aligned reads were then allocated to genetic features in the MM38 v86 annotation using featureCounts from the Subread software (Liao et al., 2013). The featureCounts count matrix was then read into the R bioconductor package edgeR (Robinson et al., 2010), and Counts Per Million reads were used to generate heatmaps. The differential expression analysis was performed in edgeR using TMM normalization and generalized linear model differential expression test.
For microarray, total RNAs from sorted LY6D+ and LY6D- prostate epithelial subpopulations were prepared by the RNeasy kit (QIAGEN, Valencia, CA). The NuGen (San Carlos, CA) Ovation Pico WTA System V2 kit was used with 500 pg of starting RNA. 5 μg of DNA from the Ovation protocol was labeled with the Encore® Biotin Module (NuGen). 34 μl (782 ng) of fragmented and labeled DNA (in 116 μl of Hybridization cocktail) was loaded on the Mouse Gene 2.0 ST gene chip (Affymetrix, Santa Clara, CA) and the chip was hybridized for 16-18 hr in a 45°C Affymetrix Gene Chip Hybridization Oven 645. The chip was stained and washed on an Affymetrix GeneChip Fluidics Station 450 using wash protocol FS450_0002. The chip was scanned on an Affymetrix Genechip Scanner 7G Plus at Molecular Biology Core Facilities at Dana-Farber Cancer Institute (DFCI). All arrays were normalized and processed in GenePattern (https://www.broadinstitute.org/cancer/software/genepattern/). Multiple Experiment Viewer (MeV) program (http://www.tm4.org/) was used to visualize the expression data.
IF for Tissue microarrays (TMA)
The Manchester Cancer Research Centre (MCRC) human prostate tissue microarray collection, constructed with FFPE transurethral resection of the prostate (TURP) samples (Bioethics 10_NOCL_02) was used to evaluate the LY6D protein expression by multiplexed IF staining. All patients evaluated in this cohort presented with localized PCa at diagnosis (M0 patients). The Opal protocol (Perkin Elmer, NEL794001KT) was used following the manufacturers’ instructions, and run on the Leica BOND RX automated system. Briefly, the slides were deparaffinized in xylene and rehydrated in ethanol. Antigen retrieval was performed in citrate buffer (pH 6.0) in microwave oven. Endogenous peroxidase blocking was performed of 10min incubation in 10% H2O2, followed by 10% casein treatment for 10min. Primary rabbit antibodies for LY6D (1:100) were incubated for 30min in a humidified chamber at room temperature, followed by detection using the rabbit Detection HRP kit. Visualization of LY6D was accomplished using fluorescein TSA Plus (1:50). In serial fashion, the slides were then retrieved in citrate buffer (pH 6.0) and then incubated with primary antibody for mouse anti-human panKeratin (1:10000), and counterstain with DAPI. Scanned tissue microarray slides were blindly analyzed and quantified with Definiens software. Overall survival after radical prostatectomy/diagnosis was assessed by Kaplan-Meier method and log-rank test in Graphpad Prism version 6 (GraphPad Software, La Jolla California USA).
We selected epithelial regions by Keratin staining in each core and scored the LY6D positive cells, using anti-human LY6D antibody (Sigma, HPA 024755). We used the Manchester Cancer Research Centre (MCRC) archival diagnostic prostate tissue collection from PCa patients collected within the Salford NHS Foundation Trust with informed subject consent for construction of tissue microarrays [built by the Genito-Urinary cancer research group at the Christie Hospital (Manchester, UK)], and under the approval from the MCRC Biobank Review Board (10_NOCL_02; #14_ESBA_01). Tissue microarray slides were scanned with the Mirax automated imaging system (Zeiss) using a 20x magnification objective and the image analysis and quantification was performed using Definiens Tissue Phenomics Software. This is a software program for the automated image analysis of tissue samples. We evaluated immunostaining blinded to clinicopathological information and scored the cores as negative (0), or positive (1). We used univariate Cox regression analysis (R packages: ‘survminer’ and ‘survival’) to assess the ability of LY6D expression to predict overall survival after radical prostatectomy/diagnosis.
Quantification and Statistical Analysis
Continuous data are reported as mean ± SEM. Categorical data are reported as frequencies and percentages. Categorical data were evaluated using chi-square test. Comparison of disease free survival and overall survival between groups was done using Kaplan-Meier method and log-rank test. Student’s t test was used to compare means between two groups. Statistical significance was set at p < 0.05. Data for evaluating pathological features was downloaded from cBioportal (http://www.cbioportal.org/). Disease free survival and overall survival for Taylor and TCGA cohorts were evaluated online on cBioportal. Statistical analysis was done using IBM SPSS version 23 (IBM corp., Armonk, New York, USA) unless stated otherwise. For LY6D expression Kaplan-Meier analysis, the optimal z-score cut-off was calculated using the R2: genomics analysis and visualization platform (https://hgserver1.amc.nl:443/) in the TCGA cohort, and independently validated the same z-score (> 1.3) cut-off in the MSKCC cohort. For animal model studies, no statistical method was used to pre-determine the sample size for mice. No randomization or blinding was used in the in vivo studies.
Data and Software Availability
The authors declare that all data supporting the findings of this study are available within the article and its Supplemental information files or from the lead contact upon reasonable request. The expression profiling data have been deposited in the GenBank GEO database (https://www.ncbi.nlm.nih.gov/geo/) under accession codes GSE92473 (microarray) and GSE92622 (RNA-seq).
Acknowledgments
We thank all members of Prostate Oncobiology Lab and Prostate Club for helpful discussions and, in particular, V. Ubertini and R. Mevel. We thank I.G. Mills, T.C. Somervaille, and I. Ringshausen for their critical reading of this manuscript. We thank the BWH and CRUK-MI Flow Cytometry Cores, and, in particular, Y. Qiu, J. Barry, and A. Banyard. We thank the CRUK-MI Molecular Biology and Computational Cores, and, in particular, W. Breitwieser and P. Schofield. We thank the MCRC Biobank, CRUK-MI Histology and Imaging Units, and P. March from the Manchester University Imaging Facility. This research was supported by start-up fund from CRUK Manchester Institute to E.B. (C5759/A20971); by a Project Development Award from A. David Mazzone Awards Program (2012_PD_156), a US Department of Defense Idea Development Award (W81XWH-15-1-0546), and start-up fund from BWH to Z.L.; and by funds from NIH and Harvard Stem Cell Institute to S.H.O., who is an Investigator of the Howard Hughes Medical Institute.
Author Contributions
J.D.B.-S., I.S., and E.B. performed single-cell FACS-sorting, FACS, surgical mouse procedures, and RNA-seq analyses. D.E.L. performed initial FACS, mouse lineage tracing, and cancer induction experiments. J.D.B.-S. and I.S. performed FACS-sorting, cytospin, mouse lineage tracing, and organoid analyses. G.G. performed single-cell Fluidigm qPCR. G.-C.Y., Z.L., and E.B. performed bioinformatics analyses. A.A. and E.B. performed human patient cohort analysis. I.S. and I.P. analyzed TMA. G.A. provided expertise on histology. H.P. performed RNA microarrays and histology. M.B. and N.W.C. built the Christie TMA. R.T.B. evaluated pathology of mice. E.B., Z.L., and S.H.O. conceived and supervised the study. E.B. and Z.L. wrote the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: December 18, 2018
Footnotes
Supplemental Information includes seven figures and five tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.11.069.
Contributor Information
Stuart H. Orkin, Email: stuart_orkin@dfci.harvard.edu.
Zhe Li, Email: zli4@rics.bwh.harvard.edu.
Esther Baena, Email: esther.baena@cruk.manchester.ac.uk.
Supplemental Information
References
- Agarwal S., Hynes P.G., Tillman H.S., Lake R., Abou-Kheir W.G., Fang L., Casey O.M., Ameri A.H., Martin P.L., Yin J.J. Identification of different classes of luminal progenitor cells within prostate tumors. Cell Rep. 2015;13:2147–2158. doi: 10.1016/j.celrep.2015.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AlHossiny M., Luo L., Frazier W.R., Steiner N., Gusev Y., Kallakury B., Glasgow E., Creswell K., Madhavan S., Kumar R., Upadhyay G. Ly6E/K signaling to TGFβ promotes breast cancer progression, immune escape, and drug resistance. Cancer Res. 2016;76:3376–3386. doi: 10.1158/0008-5472.CAN-15-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauman T.M., Vezina C.M., Huang W., Marker P.C., Peterson R.E., Ricke W.A. Beta-catenin is elevated in human benign prostatic hyperplasia specimens compared to histologically normal prostate tissue. Am. J. Clin. Exp. Urol. 2014;2:313–322. [PMC free article] [PubMed] [Google Scholar]
- Bendall S.C., Simonds E.F., Qiu P., Amir A.D., Krutzik P.O., Finck R., Bruggner R.V., Melamed R., Trejo A., Ornatsky O.I. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakenhoff R.H., Gerretsen M., Knippels E.M., van Dijk M., van Essen H., Weghuis D.O., Sinke R.J., Snow G.B., van Dongen G.A. The human E48 antigen, highly homologous to the murine Ly-6 antigen ThB, is a GPI-anchored molecule apparently involved in keratinocyte cell-cell adhesion. J. Cell Biol. 1995;129:1677–1689. doi: 10.1083/jcb.129.6.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broutier L., Andersson-Rolf A., Hindley C.J., Boj S.F., Clevers H., Koo B.-K., Huch M. Culture and establishment of self-renewing human and mouse adult liver and pancreas 3D organoids and their genetic manipulation. Nat. Protoc. 2016;11:1724–1743. doi: 10.1038/nprot.2016.097. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi N., Zhang B., Zhang L., Ittmann M., Xin L. Adult murine prostate basal and luminal cells are self-sustained lineages that can both serve as targets for prostate cancer initiation. Cancer Cell. 2012;21:253–265. doi: 10.1016/j.ccr.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua C.W., Shibata M., Lei M., Toivanen R., Barlow L.J., Bergren S.K., Badani K.K., McKiernan J.M., Benson M.C., Hibshoosh H., Shen M.M. Single luminal epithelial progenitors can generate prostate organoids in culture. Nat. Cell Biol. 2014;16:951–961. doi: 10.1038/ncb3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bono J.S., Logothetis C.J., Molina A., Fizazi K., North S., Chu L., Chi K.N., Jones R.J., Goodman O.B., Jr., Saad F., COU-AA-301 Investigators Abiraterone and increased survival in metastatic prostate cancer. N. Engl. J. Med. 2011;364:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshel R., Zanin A., Kapon D., Sagi-Assif O., Brakenhoff R., van Dongen G., Witz I.P. Human Ly-6 antigen E48 (Ly-6D) regulates important interaction parameters between endothelial cells and head-and-neck squamous carcinoma cells. Int. J. Cancer. 2002;98:803–810. doi: 10.1002/ijc.10301. [DOI] [PubMed] [Google Scholar]
- Gao D., Vela I., Sboner A., Iaquinta P.J., Karthaus W.R., Gopalan A., Dowling C., Wanjala J.N., Undvall E.A., Arora V.K. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159:176–187. doi: 10.1016/j.cell.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A.S., Lawson D.A., Cheng D., Sun W., Garraway I.P., Witte O.N. Trop2 identifies a subpopulation of murine and human prostate basal cells with stem cell characteristics. Proc. Natl. Acad. Sci. USA. 2008;105:20882–20887. doi: 10.1073/pnas.0811411106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A.S., Huang J., Guo C., Garraway I.P., Witte O.N. Identification of a cell of origin for human prostate cancer. Science. 2010;329:568–571. doi: 10.1126/science.1189992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G., Luc S., Marco E., Lin T.-W., Peng C., Kerenyi M.A., Beyaz S., Kim W., Xu J., Das P.P. Mapping cellular hierarchy by single-cell analysis of the cell surface repertoire. Cell Stem Cell. 2013;13:492–505. doi: 10.1016/j.stem.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inlay M.A., Bhattacharya D., Sahoo D., Serwold T., Seita J., Karsunky H., Plevritis S.K., Dill D.L., Weissman I.L. Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes Dev. 2009;23:2376–2381. doi: 10.1101/gad.1836009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs J.T. Prostate stem cells and benign prostatic hyperplasia. Prostate. 2008;68:1025–1034. doi: 10.1002/pros.20763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ittmann M., Huang J., Radaelli E., Martin P., Signoretti S., Sullivan R., Simons B.W., Ward J.M., Robinson B.D., Chu G.C. Animal models of human prostate cancer: the consensus report of the New York meeting of the Mouse Models of Human Cancers Consortium Prostate Pathology Committee. Cancer Res. 2013;73:2718–2736. doi: 10.1158/0008-5472.CAN-12-4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao J., Hindoyan A., Wang S., Tran L.M., Goldstein A.S., Lawson D., Chen D., Li Y., Guo C., Zhang B. Identification of CD166 as a surface marker for enriching prostate stem/progenitor and cancer initiating cells. PLoS One. 2012;7:e42564. doi: 10.1371/journal.pone.0042564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthaus W.R., Iaquinta P.J., Drost J., Gracanin A., van Boxtel R., Wongvipat J., Dowling C.M., Gao D., Begthel H., Sachs N. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell. 2014;159:163–175. doi: 10.1016/j.cell.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsten H., Ziel-van der Made A., Ma X., van der Kwast T., Trapman J. Accumulating progenitor cells in the luminal epithelial cell layer are candidate tumor initiating cells in a Pten knockout mouse prostate cancer model. PLoS One. 2009;4:e5662. doi: 10.1371/journal.pone.0005662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosawa M., Jeyasekharan A.D., Surmann E.-M., Hashimoto N., Venkatraman V., Kurosawa G., Furukawa K., Venkitaraman A.R., Kurosawa Y. Expression of LY6D is induced at the surface of MCF10A cells by X-ray irradiation. FEBS J. 2012;279:4479–4491. doi: 10.1111/febs.12034. [DOI] [PubMed] [Google Scholar]
- Kwon O.-J., Valdez J.M., Zhang L., Zhang B., Wei X., Su Q., Ittmann M.M., Creighton C.J., Xin L. Increased Notch signalling inhibits anoikis and stimulates proliferation of prostate luminal epithelial cells. Nat. Commun. 2014;5:4416. doi: 10.1038/ncomms5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon O.-J., Zhang L., Xin L. Stem cell antigen-1 identifies a distinct androgen-independent murine prostatic luminal cell lineage with bipotent potential. Stem Cells. 2016;34:191–202. doi: 10.1002/stem.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson D.A., Xin L., Lukacs R.U., Cheng D., Witte O.N. Isolation and functional characterization of murine prostate stem cells. Proc. Natl. Acad. Sci. USA. 2007;104:181–186. doi: 10.1073/pnas.0609684104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson D.A., Zong Y., Memarzadeh S., Xin L., Huang J., Witte O.N. Basal epithelial stem cells are efficient targets for prostate cancer initiation. Proc. Natl. Acad. Sci. USA. 2010;107:2610–2615. doi: 10.1073/pnas.0913873107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong K.G., Wang B.-E., Johnson L., Gao W.-Q. Generation of a prostate from a single adult stem cell. Nature. 2008;456:804–808. doi: 10.1038/nature07427. [DOI] [PubMed] [Google Scholar]
- Lesche R., Groszer M., Gao J., Wang Y., Messing A., Sun H., Liu X., Wu H. Cre/loxP-mediated inactivation of the murine Pten tumor suppressor gene. Genesis. 2002;32:148–149. doi: 10.1002/gene.10036. [DOI] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Tsai O., Clark P.E., Miller N.L., Fowke J.H., Hameed O., Hayward S.W., Strand D.W. Surgical intervention for symptomatic benign prostatic hyperplasia is correlated with expression of the AP-1 transcription factor network. Prostate. 2014;74:669–679. doi: 10.1002/pros.22785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Pascal L.E., Isharwal S., Metzger D., Ramos Garcia R., Pilch J., Kasper S., Williams K., Basse P.H., Nelson J.B. Regenerated luminal epithelial cells are derived from preexisting luminal epithelial cells in adult mouse prostate. Mol. Endocrinol. 2011;25:1849–1857. doi: 10.1210/me.2011-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Grogan T.R., Hieronymus H., Hashimoto T., Mottahedeh J., Cheng D., Zhang L., Huang K., Stoyanova T., Park J.W. Low CD38 identifies progenitor-like inflammation-associated luminal cells that can initiate human prostate cancer and predict poor outcome. Cell Rep. 2016;17:2596–2606. doi: 10.1016/j.celrep.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughner C.L., Bruford E.A., McAndrews M.S., Delp E.E., Swamynathan S., Swamynathan S.K. Organization, evolution and functions of the human and mouse Ly6/uPAR family genes. Hum. Genomics. 2016;10:10. doi: 10.1186/s40246-016-0074-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacs R.U., Goldstein A.S., Lawson D.A., Cheng D., Witte O.N. Isolation, cultivation and characterization of adult murine prostate stem cells. Nat. Protoc. 2010;5:702–713. doi: 10.1038/nprot.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L., McGarvey P., Madhavan S., Kumar R., Gusev Y., Upadhyay G. Distinct lymphocyte antigens 6 (Ly6) family members Ly6D, Ly6E, Ly6K and Ly6H drive tumorigenesis and clinical outcome. Oncotarget. 2016;7:11165–11193. doi: 10.18632/oncotarget.7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa H., Akamatsu S., Takata R., Takahashi A., Kubo M., Nakamura Y. Prostate cancer genomics, biology, and risk assessment through genome-wide association studies. Cancer Sci. 2012;103:607–613. doi: 10.1111/j.1349-7006.2011.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ousset M., Van Keymeulen A., Bouvencourt G., Sharma N., Achouri Y., Simons B.D., Blanpain C. Multipotent and unipotent progenitors contribute to prostate postnatal development. Nat. Cell Biol. 2012;14:1131–1138. doi: 10.1038/ncb2600. [DOI] [PubMed] [Google Scholar]
- Park J.-H., Walls J.E., Galvez J.J., Kim M., Abate-Shen C., Shen M.M., Cardiff R.D. Prostatic intraepithelial neoplasia in genetically engineered mice. Am. J. Pathol. 2002;161:727–735. doi: 10.1016/S0002-9440(10)64228-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu P., Simonds E.F., Bendall S.C., Gibbs K.D., Jr., Bruggner R.V., Linderman M.D., Sachs K., Nolan G.P., Plevritis S.K. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat. Biotechnol. 2011;29:886–891. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane J.K., Droop A.P., Pellacani D., Polson E.S., Simms M.S., Collins A.T., Caves L.S.D., Maitland N.J. Conserved two-step regulatory mechanism of human epithelial differentiation. Stem Cell Reports. 2014;2:180–188. doi: 10.1016/j.stemcr.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinfeld B., Upadhyay A., Clark S.L., Fong S.E., Smith V., Koeppen H., Ross S., Polakis P. Identification and immunotherapeutic targeting of antigens induced by chemotherapy. Nat. Biotechnol. 2006;24:205–209. doi: 10.1038/nbt1185. [DOI] [PubMed] [Google Scholar]
- Saeed A.I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Scher H.I., Fizazi K., Saad F., Taplin M.-E., Sternberg C.N., Miller K., de Wit R., Mulders P., Chi K.N., Shore N.D., AFFIRM Investigators Increased survival with enzalutamide in prostate cancer after chemotherapy. N. Engl. J. Med. 2012;367:1187–1197. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- Shappell S.B., Thomas G.V., Roberts R.L., Herbert R., Ittmann M.M., Rubin M.A., Humphrey P.A., Sundberg J.P., Rozengurt N., Barrios R. Prostate pathology of genetically engineered mice: definitions and classification. The consensus report from the Bar Harbor meeting of the Mouse Models of Human Cancer Consortium Prostate Pathology Committee. Cancer Res. 2004;64:2270–2305. doi: 10.1158/0008-5472.can-03-0946. [DOI] [PubMed] [Google Scholar]
- Shen M.M., Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev. 2010;24:1967–2000. doi: 10.1101/gad.1965810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Gipp J., Dries M., Bushman W. Prostate progenitor cells proliferate in response to castration. Stem Cell Res. (Amst.) 2014;13:154–163. doi: 10.1016/j.scr.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R., Ma J., Zou Z., Jemal A. Cancer statistics, 2014. CA Cancer J. Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- Srinivas S., Watanabe T., Lin C.S., William C.M., Tanabe Y., Jessell T.M., Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor B.S., Schultz N., Hieronymus H., Gopalan A., Xiao Y., Carver B.S., Arora V.K., Kaushik P., Cerami E., Reva B. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toivanen R., Mohan A., Shen M.M. Basal progenitors contribute to repair of the prostate epithelium following induced luminal anoikis. Stem Cell Reports. 2016;6:660–667. doi: 10.1016/j.stemcr.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzgare A.R., Xu Y., Isaacs J.T. In vitro culturing and characteristics of transit amplifying epithelial cells from human prostate tissue. J. Cell. Biochem. 2004;91:196–205. doi: 10.1002/jcb.10764. [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A., Rocha A.S., Ousset M., Beck B., Bouvencourt G., Rock J., Sharma N., Dekoninck S., Blanpain C. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- Wang Z.A., Shen M.M. Revisiting the concept of cancer stem cells in prostate cancer. Oncogene. 2011;30:1261–1271. doi: 10.1038/onc.2010.530. [DOI] [PubMed] [Google Scholar]
- Wang Q., Li W., Liu X.S., Carroll J.S., Jänne O.A., Keeton E.K., Chinnaiyan A.M., Pienta K.J., Brown M. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol. Cell. 2007;27:380–392. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Kruithof-de Julio M., Economides K.D., Walker D., Yu H., Halili M.V., Hu Y.-P.H., Price S.M., Abate-Shen C., Shen M.M. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009;461:495–500. doi: 10.1038/nature08361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.A., Mitrofanova A., Bergren S.K., Abate-Shen C., Cardiff R.D., Califano A., Shen M.M. Lineage analysis of basal epithelial cells reveals their unexpected plasticity and supports a cell-of-origin model for prostate cancer heterogeneity. Nat. Cell Biol. 2013;15:274–283. doi: 10.1038/ncb2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.A., Toivanen R., Bergren S.K., Chambon P., Shen M.M. Luminal cells are favored as the cell of origin for prostate cancer. Cell Rep. 2014;8:1339–1346. doi: 10.1016/j.celrep.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson P.A., Arora V.K., Sawyers C.L. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat. Rev. Cancer. 2015;15:701–711. doi: 10.1038/nrc4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin L., Lawson D.A., Witte O.N. The Sca-1 cell surface marker enriches for a prostate-regenerating cell subpopulation that can initiate prostate tumorigenesis. Proc. Natl. Acad. Sci. USA. 2005;102:6942–6947. doi: 10.1073/pnas.0502320102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin L., Lukacs R.U., Lawson D.A., Cheng D., Witte O.N. Self-renewal and multilineage differentiation in vitro from murine prostate stem cells. Stem Cells. 2007;25:2760–2769. doi: 10.1634/stemcells.2007-0355. [DOI] [PubMed] [Google Scholar]
- Zhang B., Kwon O.-J., Henry G., Malewska A., Wei X., Zhang L., Brinkley W., Zhang Y., Castro P.D., Titus M. Non-cell-autonomous regulation of prostate epithelial homeostasis by androgen receptor. Mol. Cell. 2016;63:976–989. doi: 10.1016/j.molcel.2016.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.