SUMMARY
Metastasizing breast carcinoma cells have been hypothesized to mobilize tissue-invasive activity by co-opting the proteolytic systems employed by normal mammary epithelial cells undergoing branching morphogenesis. However, the critical effectors underlying morphogenesis remain unidentified and their relationship to breast cancer invasion programs have yet to be established. Here we identify the membrane-anchored matrix metalloproteinase, Mmp14/MT1-MMP, but not the closely related proteinase, Mmp15/MT2-MMP, as the dominant proteolytic effector of both branching morphogenesis and carcinoma cell invasion in vivo. Unexpectedly, however, epithelial cell-specific targeting of Mmp14/MT1-MMP in the normal mammary gland fails to impair branching, whereas deleting the proteinase in carcinoma cells abrogates invasion, preserves matrix architecture and completely blocks metastasis. By contrast, in the normal mammary gland, extracellular matrix remodeling and morphogenesis are ablated only when Mmp14/MT1-MMP expression is specifically deleted from the periductal stroma. Together, these findings uncover the overlapping, but divergent strategies that underlie developmental versus neoplastic matrix remodeling programs.
Graphical Abstract
In brief:
Breast carcinoma cells are thought to co-opt proteolytic systems employed during developmental branching morphogenesis. Feinberg et al. show that these processes indeed both depend on the same protease, MT1-MMP, but that normal mammary gland development requires protease activity in the stroma, whereas in cancer, the carcinoma cells themselves drive the invasion program.
INTRODUCTION
In cancer, the deliberate, but inappropriate, re-activation of developmental programs normally silenced in adult tissues is thought to lend neoplastic cells their deadliest characteristics (Lambert et al., 2017). With regard to the expression of tissue-invasive activity, a property that endows carcinoma cells with metastatic potential, the co-opted developmental “homologue” has long been thought to be linked to branching morphogenesis (Cheung et al., 2013; Harper et al., 2016; Hosseini et al., 2016; Lambert et al., 2017). In this tightly regulated developmental process, epithelial rudiments activate a controlled form of tissue invasion wherein a network of tubular structures infiltrate surrounding stromal tissues (Cheung et al., 2013). In vitro studies suggest that both normal and neoplastic cells traverse extracellular matrix (ECM) barriers by either proteinase-dependent or independent mechanisms (Liu et al., 2015; Paul et al., 2017). However, the relative role of these pathways in vivo remains unclear and no single effector has been identified that co-regulates the invasion programs associated with branching morphogenesis and cancer progression.
In an effort to characterize the potentially unifying features by which normal and neoplastic cells traverse the ECM barriers encountered in vivo, we focused our attention on the mouse mammary gland wherein morphogenic and neoplastic programs can be compared directly. Unlike most developmental programs, the mammary gland confines the bulk of its tissue-invasive/tubulogenic activity to the postnatal period (Wang et al., 2017). Given this unique characteristic, the mammary gland provides a model wherein normal epithelial cells and carcinoma cells infiltrate tissues whose ECM composition and architecture are interchangeable. As such, we have compared the ECM remodeling programs mobilized during normal postnatal branching morphogenesis to those activated during breast carcinoma progression in a transgenic system that recapitulates key features of human breast carcinoma progression (Cheung et al., 2013; Ni et al., 2016; Wculek and Malanchi, 2015; Ye et al., 2015). In turn, we have identified a single proteolytic effector, the membrane-anchored metalloproteinase, Mmp14/MT1-MMP, as the upstream regulator of normal as well as neoplastic tissue-invasive activity. Unexpectedly, however, whereas targeting Mmp14 in mammary epithelial cells has no effect on the ECM remodeling programs associated with normal branching morphogenesis, extinguishing MT1-MMP expression in mammary carcinoma cells completely blocks invasion and metastasis. In marked contrast, branching morphogenesis is alternatively dependent on MT1-MMP-dependent proteolysis of the periductal ECM by the mammary gland stroma itself. Taken together, these studies identify a singular role for Mmp14/MT1-MMP in regulating developmental and neoplastic invasion programs by employing divergent cellular strategies to support these related, but distinctive ECM remodeling activities.
RESULTS
Differential Regulation of Normal versus Neoplastic Mammary Epithelial Cell Invasion Programs
Following the onset of puberty in female mice at approximately 3 wks of age, mammary epithelial cell rudiments engage a branching morphogenesis program that fills the stroma with a tubular network over the next 5–8 wks of life (Fig. 1A) (Wang et al., 2017). While several members of the matrix metalloproteinase (MMP) family, including the secreted MMPs, MMP-2, MMP-3 and MMP-13, have been implicated in mammary gland branching morphogenesis (Khokha and Werb, 2011), global knockout of each of these proteinases fails to inhibit tubulogenesis to a significant degree (Fig. S1A-C). Recent studies have, however, demonstrated that mammary epithelial cells also express at least 2 members of the membrane-anchored MMP family, i.e., Mmp14/MT1-MMP and Mmp15/MT2-MMP (Feinberg et al., 2016). Indeed, using Mmp14+/LacZ and Mmp15+/LacZ knock-in lines, both MT-MMPs are readily detected during morphogenesis (Fig. 1B,C). As such, we generated Mmp14f/f, Mmp15f/f and Mmp14f/f/Mmp15f/f mice that were then crossed with MMTV-Cre (line A) transgenics (Wagner et al., 2001) to target the mammary epithelium so as to monitor potential roles for these proteinases during branching morphogenesis.
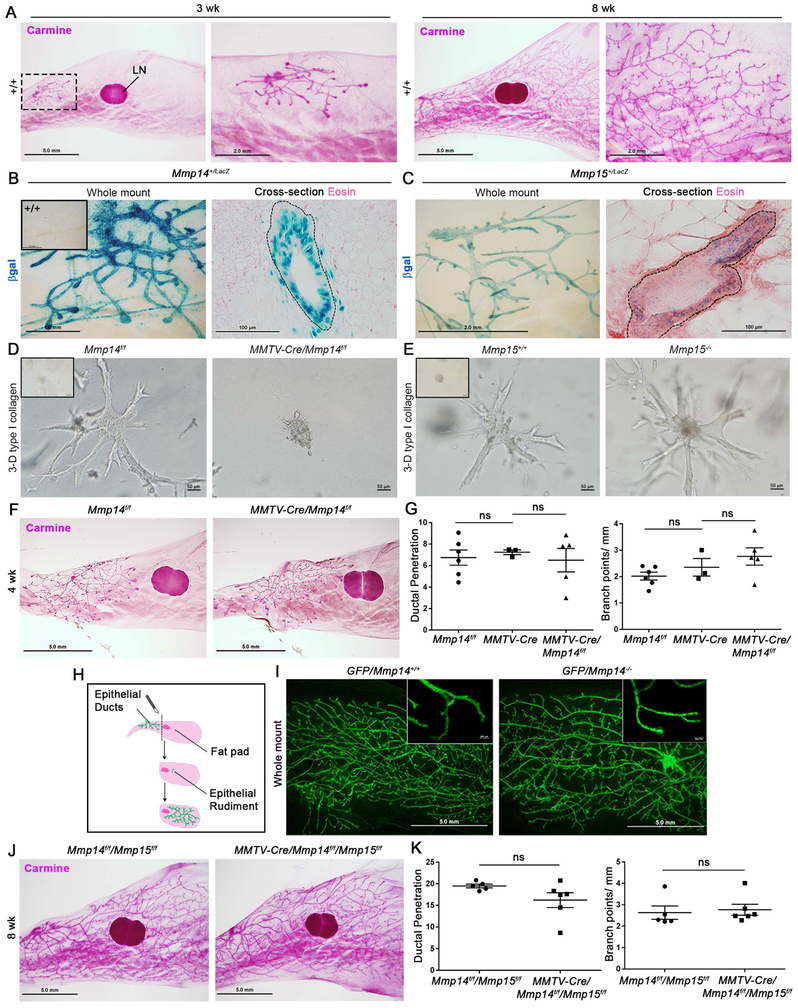
Figure 1. Postnatal mammary gland branching is independent of epithelial cell-derived MT1-MMP and MT2-MMP in vivo.
(A) Whole mounts from 3 wk-old and 8 wk-old wild-type mice. A dotted line box circumscribes the region for higher magnification (scale bars, 5.0 mm and 2.0 mm, respectively). “LN” marks the inguinal lymph node.
(B) LacZ staining of whole mount and cross-section from 4 wk-old Mmp14+/LacZ mouse (scale bars, 1.0 mm and 100 μm, respectively). Inset shows Mmp14+/+ whole mount.
(C) LacZ staining of whole mount and cross-section from 4 wk-old Mmp15+/LacZ mouse (scale bars, 2.0 mm and 100 μm, respectively).
(D,E) Bright-field micrographs of mammary epithelial organoids harvested from Mmp14f/f and MMTV-Cre +/−/Mmp14f/f mice (D) or Mmp15+/+ and Mmp15−/− mice (E), and embedded within 3-dimensional (3-D) type I collagen for 3–4 days with FGF-2. Insets show bright-field micrographs of organoids at time 0. (D) Organoid branch length is reduced from 117.2±5.4 μm with 9.1±0.6 branches/organoid in Mmp14f/f organoids (n=30) to 40.6±6.2 μm and 1.5±0.2 branches/organoid in MMTV-Cre +/−/Mmp14f/f organoids (n=33) (p<0.0001). (E) Organoid branch length is comparable at 133.5±11.1 μm with 7.0±0.4 branches/organoid in Mmp15 +/− organoids (n=10), with 142.7±12.1 μm and 6.9±0.4 branches/organoid in Mmp15−/− organoids (n=28) (p=0.805). Data are presented as mean ± SEM.
(F) Whole mounts from 4 wk-old Mmp14f/f and MMTV-Cre+/−/Mmp14f/f mice (scale bar, 5.0 mm).
(G) Quantifications of ductal penetration (mm) and branch points per mm duct for 4 wk-old Mmp14f/f (n=6), MMTV-Cre+/− (n=3), and MMTV-Cre+/−/Mmp14f/f (n=5) mice. Data are presented as mean ± SEM.
(H) Cartoon of mammary duct transplantation.
(I) Whole mounts of mammary tissue at 8 wks post-transplantation of Gfp +/−/Mmp14 +/− and Gfp +/−/Mmp14−/− ducts (scale bar, 5.0 mm).
(J) Whole mounts from 8 wk-old Mmp14f/f/Mmp15f/f and MMTV-Cre+/−/Mmp14f/f/Mmp15f/f mice (scale bar, 5.0 mm).
(K) Quantifications of ductal penetration (mm) and branch points per mm duct in Mmp14f/f/Mmp15f/f (n=5) and MMTV-Cre+/−/Mmp14f/f/Mmp15f/f (n=6) mice. Data are presented as mean ± SEM.
Given efficient targeting of Mmp14 and Mmp15 using the transgenic line (i.e., ≥90% excision; Fig. S1D), we first sought to assess the role of these proteinases in supporting branching morphogenesis in 3-dimensional (3-D) type I collagen hydrogels in vitro. Whereas Mmp14 targeting blocks branching with a 2.9-fold reduction in branch length and 5.9-fold reduction in branch number per organoid (p<0.0001), Mmp15-deleted epithelial cells undergo normal morphogenesis (Fig. 1D,E). By contrast, following Mmp14 targeting of the mammary epithelium in vivo, branching is unaffected as characterized by network complexity, duct elongation and branch point number through postnatal wks 4–8 (Fig. 1F,G; Fig. S1E). Despite the obvious contrast of this result with our ex vivo model, transcriptional profiling of Mmp14-targeted mammary epithelial cells versus Mmp14f/f littermate controls reveals only 86 gene changes in vivo with 62 genes down-regulated and 24 genes up-regulated using a 1.5-fold enrichment cutoff (n=3 per genotype). GO analyses of Functional Annotation clusters reveal differential expression of a suite of genes associated with epidermal and epithelial differentiation as well as development, but with no clear link to branching morphogenesis (Fig. S1F; Table S1). Nevertheless, we considered the possibility that small numbers of mammary epithelial cells that escape Cre-targeting might serve as pro-invasive “leader cells” in vivo (Lu et al., 2008). However, using the <5% subpopulation of Cre-escaped mammary epithelial cells found in MMTV-Cre+/−/Mmp14f/f/ROSA26R+/loxP transgenic mice to track Cre-targeted versus non-targeted epithelial cells, no competitive ‘leader cell’ advantage is observed for the MT1-MMP-expressing cells (Fig. S1G).
To stringently assess the ability of branching morphogenesis to proceed in an MT1-MMP-independent manner in vivo, ductal fragments were isolated from GFP+-expressing Mmp14+/− or Mmp14−/− donor mice and transplanted into the cleared mammary fat pads of wild-type recipients (Fig. 1H). Under these conditions, Mmp14-null mammary epithelial cells launch a branching program indistinguishable from control epithelial cells (Fig. 1I; Fig. S1H) and display normal epithelial cell organization as marked by the distribution of myoepithelial and luminal epithelial cell markers (i.e., CK5 and CK18, respectively) (Fig. S1I) as well as comparable distributions of E-cadherin (Fig. S1J), and zonula occludens (ZO)-1 (Fig. S1K). In addition, neither Mmp15 global knockout mice, MMTV-Cre+/−/Mmp15f/f conditional knockout mice nor mice engineered to undergo the combined targeting of epithelial cell Mmp14 and Mmp15 display mammary gland branching defects (Fig. 1J,K;Fig. S1L,M). Hence, despite an obvious dependence on MT1-MMP activity in supporting morphogenic activity ex vivo, neither epithelial cell-derived MT1-MMP nor MT2-MMP play required roles in this process in vivo.
Given the operating paradigm that cancer cell invasion programs inappropriately co-opt developmentally-related morphogenic activities (Cheung et al., 2013; Lambert et al., 2017), we next considered the possibility that Mmp14 and Mmp15 might likewise prove dispensable in MMTV-PyMT transgenic mice whose mammary gland carcinomas closely resemble human luminal B-type cancers (Cheung et al., 2013; Ni et al., 2016). As shown, both MMTV-PyMT+/−/Mmp14+/LacZ and MMTV-PyMT+/−/Mmp15+/LacZ mice register expression of each proteinase in primary tumors (Fig. 2A). However, following mammary epithelial cell targeting of Mmp14, Mmp15 or both Mmp14 and Mmp15 in tandem, neither tumor onset nor initial growth are affected (Fig. 2B-D). Independent of effects on tumor initiation, MMTV-PyMT carcinomas have recently been reported to infiltrate surrounding tissues by generating invasive strands of dual CK8/14-positive cells (Cheung et al., 2013). As such, we introduced the RosaRFP allele into the MMTV-PyMT/MMTV-Cre line to generate red fluorescent carcinoma cells whose invasive activity could be tracked directly. Under these conditions, RFP-positive carcinoma cells that co-express CK8 and CK14 are found invading the surrounding stromal tissues in both strand-like and single cell fashion (Fig. 2E,F and Fig. S2A; upper panels). As protease-dependent invasion programs would be predicted to correlate closely with ECM remodeling, the proteolysis of native collagen fibers was assessed in situ using carboxyfluorescein-labeled collagen hybridizing peptide (CF-CHP) that binds specifically to degraded collagen by folding into a triple helix (Hwang et al., 2017; Li et al., 2012). Importantly, labeling of primary MMTV-PyMT tumor tissue reveals dense arrays of remodeled collagen at the carcinoma-stromal tissue interface, but not at sites distant to the carcinoma cells (Fig. 2G; upper panels).
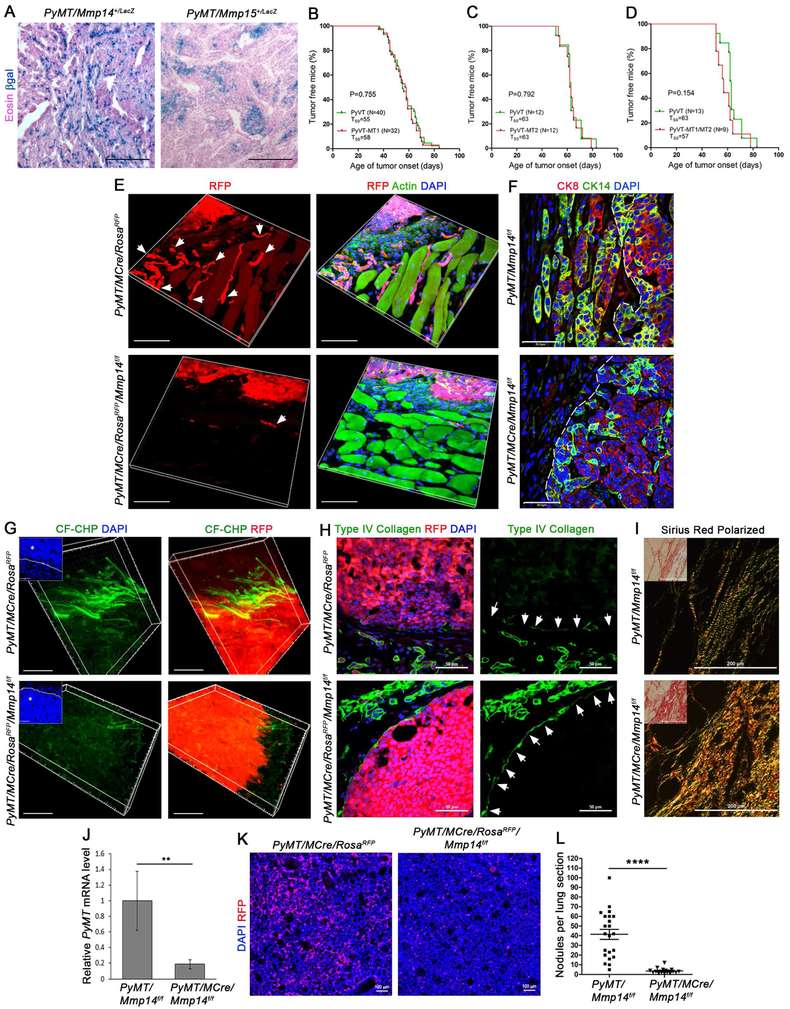
Figure 2. Mammary carcinoma-derived MT1-MMP dictates local invasion and metastasis.
(A) LacZ staining of cross-sections from MMTV-PyMT +/−/Mmp14+/LacZ (left panel) and MMTV-PyMT +/−/Mmp15+/LacZ (right panel) mammary tumors with Eosin counterstaining (scale bar, 200 μm).
(B-D) Kaplan-Meier plots depicting age of tumor onset (days) for (B) MMTV-PyMT +/−/Mmp14f/f (n=40) versus MMTV-PyMT +/−/MMTV-Cre +/−/Mmp14f/f (n=32) mice, (C) MMTV-PyMT +/−/Mmp15f/f (n=12) versus MMTV-PyMT +/−/MMTV-Cre +/−/Mmp15f/f (n=12) mice, and (D) MMTV-PyMT +/−/Mmp14f/f/Mmp15f/f (n=13) versus MMTV-PyMT +/−/MMTV-Cre +/−/Mmp14f/f/Mmp15f/f (n=9) mice.
(E) 3-D Reconstructions of RFP fluorescence with phalloidin (F-Actin) and DAPI staining in mammary tumor cross-sections from 3–4 month-old MMTV-PyMT +/−/MMTV-Cre +/−/RosaRFP and MMTV-PyMT +/−/MMTV-Cre +/−/RosaRFP/Mmp14f/f mice. Arrows mark strands of invasive tumor cells (scale bar, 50 μm).
(F) Cytokeratin (CK)-8 and CK14 immunofluorescence with DAPI staining in mammary tumor cross-sections from 3–4 month old MMTV-PyMT +/−/Mmp14f/f and MMTV-PyMT +/−/MMTV-Cre +/−/Mmp14f/f mice (scale bar, 50 μm). A dotted line marks the tumor-stromal interface.
(G) 3-D reconstructions of degraded collagen, labeled in situ with CF-CHP, in mammary tumors harvested from 3–4 month old MMTV-PyMT +/−/MMTV-Cre +/−/RosaRFP and MMTV-PyMT +/−/MMTV-Cre +/−/RosaRFP/Mmp14f/f mice. Inset shows DAPI staining with dotted line marking the tumor-stromal interface and an asterisk within the tumor. Right panels show CF-CHP immunofluorescence together with RFP fluorescence (scale bar, 30 μm). CF-CHP immunofluorescence is reduced from 40.0±3.1 total pixels/μm2 around MMTV-PyMT +/−/MMTV-Cre +/−/RosaRFP tumors (n=8) to 10.0±1.9 total pixels/μm2 around MMTV-PyMT +/−/MMTV-Cre +/−/RosaRFP/Mmp14f/f tumors (n=10), as normalized to scrambled CHP immunofluorescence (p<0.0001, mean ± SEM).
(H) Type IV collagen immunofluorescence and RFP fluorescence with DAPI staining in mammary tumor cross-sections from MMTV-PyMT +/−/MMTV-Cre +/−/RosaRFP and MMTV-PyMT +/−/MMTV-Cre +/−/RosaRFP/Mmp14f/f mice (left panels). Right panels show type IV collagen signal alone with arrows marking the tumor boundary (scale bar, 50 μm).
(I) Polarized light images of Sirius Red staining in 3–4 month-old MMTV-PyMT +/−/Mmp14f/f and MMTV-PyMT +/−/MMTV-Cre +/−/Mmp14f/f mammary tumors. Insets show bright-field images (scale bar, 200 μm).
(J) QPCR for PyMT transcripts in whole blood harvested from MMTV-PyMT +/−/Mmp14f/f and MMTV-PyMT +/−/MMTV-Cre +/−/Mmp14f/f mice (n=4 per genotype) (**p<0.001, mean ± SEM).
(K) RFP fluorescence and DAPI staining in lungs harvested from 2 month-old MMTV-PyMT +/−/MMTV-Cre +/−/RosaRFP and MMTV-PyMT +/−/MMTV-Cre +/−/RosaRFP/Mmp14f/f mice (scale bar, 100 μm).
(L) Quantification of lung nodules per cross-section in lungs harvested from MMTV-PyMT +/−/Mmp14f/f (n=22) and MMTV-PyMT +/−/MMTV-Cre +/−/Mmp14f/f (n=16) mice (****p<0.0001, mean ± SEM).
See also Figure S2.
To determine if either MT1-MMP or MT2-MMP plays a required role in driving carcinoma invasion, MMTV-PyMT+/−/MMTV-Cre+/−/Mmp14f/f or MMTV-PyMT +/−/MMTV-Cre +/−/Mmp15f/f were crossed into the RosaRFP line and local invasion assessed. Remarkably, targeting carcinoma-associated Mmp14, but not Mmp15, virtually abolishes tissue-invasive activity without affecting PyMT expression or downstream signaling cascades (Fig. 2E,F; lower panels, and Fig. S2A-C). As previously described (Cheung et al., 2013), E-cadherin expression is retained at the invasive edge of MT1-MMP-expressing carcinomas (Fig. S2D). We do, however, observe partial down-regulation of E-cadherin in regions of both Mmp14 wild-type and Mmp14-null carcinomas (Fig. S2D). Whereas targeting carcinoma cell Mmp14 does not affect the expression of the proteinase in surrounding stromal cells, or their expression of the mechanotransduction-sensitive transcriptional co-activator, YAP (Calvo et al., 2013) (Fig. S2E,F), local collagen degradation is reduced by 4-fold and largely abrogated in areas surrounding Mmp14-deleted carcinomas (p<0.0001) (Fig. 2G; lower panels). Furthermore, when RosaRFP+ tumor fragments isolated from control and Mmp14-targeted carcinomas are transplanted into wild-type recipients (Fig. S2G), only the wild-type fragments display tissue-invasive activity marked by invasive strands of RFP+/CK14+ carcinoma cells (Fig. S2H,I). Consistent with the unabated tissue-invasive activity of Mmp15-targeted carcinoma cells, local collagen remodeling is retained in these tumors (Fig. S2J). Confirming a dominant role for carcinoma, rather than stromal cell – derived MT1-MMP, targeting Mmp14 in the MMTV-PyMT stromal cell compartment with Fsp1-Cre (Fischer et al., 2015; Trimboli et al., 2009) does not affect tumor onset, growth or local invasion (Fig. S2K,L). Interestingly, the decrease in collagen degradation observed in MT1-MMP-deleted carcinomas is attended by the increased retention of type IV collagen staining in basement membrane zones as well as a marked increase in the levels of interstitial collagens in the stromal microenvironment [quantified as a 2.1-fold increase in total pixels of Sirius Red fluorescence per μm2 surrounding MMTV-PyMT +/−/MMTV-Cre +/−/Mmp14f/f tumors versus MMTV-PyMT +/−/Mmp14f/f tumors (n=14 fields and n=3 mice per genotype; p<0.0001)] (Fig. 2H,I). Increases in collagen content are observed in MT1-MMP-targeted carcinomas, despite preserved, if not reduced, expression of type I, III, V and VI collagen transcripts (Fig. S2M). Thus, whereas studies have emphasized the ability of increased collagen deposition to promote invasive activity (Kai et al., 2016), the loss of MT1-MMP activity undermines all pro-tumorigenic effects associated with these desmoplastic responses.
Given the loss of tissue-invasive activity in Mmp14-targeted carcinomas in vivo, we assessed the importance of MT1-MMP in controlling tumor intravasation and metastatic activity. Indeed, circulating carcinoma cell number is decreased by ~80% following Mmp14 excision, in tandem with a marked reduction in the number of carcinoma cells seeding the lung (Fig. 2J,K). More importantly, Mmp14 targeting in the carcinoma cells largely eliminates lung metastatic activity (Fig. 2L). Interestingly, the inhibition of metastatic activity appears to arise as a direct consequence of the loss of local tissue-invasive activity as intravenous injection of RFP-positive, MT1-MMP-null carcinoma cells generated lung metastases comparably to control carcinoma cells (data not shown). Of note, targeting of carcinoma cell-derived Mmp15 or stromal cell Mmp14 with Fsp1-Cre fails to block the development of lung metastases (Fig. S2N,O). Hence, in direct contrast to branching morphogenesis, MMTV-PyMT carcinoma cells display an absolute requirement for MT1-MMP activity in regulating invasion and metastasis.
MT1-MMP Directs Both Mouse and Human Breast Carcinoma Invasion Programs Ex Vivo
Unlike normal mammary epithelial cells whose intact basement membrane separates them from underlying interstitial tissues, invasive carcinoma cells actively embed themselves in type I collagen-rich stromal tissues (Nguyen-Ngoc et al., 2012; Sabeh et al., 2009). As such, tumor organoids isolated from MMTV-PyMT mice were examined in an ex vivo model where carcinoma organoids are embedded in native, cross-linked hydrogels of type I collagen (Nguyen-Ngoc et al., 2012; Sabeh et al., 2009). Under these conditions, primary MMTV-PyMT organoids isolated from control mice express MT1-MMP while CK8/14-dual positive cells invade the collagen hydrogel in a manner similar to that observed in vivo (Fig. 3A,B; upper panels). By contrast, organoids harvested from mice with Mmp14-targeted carcinoma cells, fail to express MT1-MMP or collagen-invasive activity ex vivo and display a 9.2-fold reduction in branch length as well as an 11.4-fold reduction in branch number (p<0.0001) (Fig. 3A,B; lower panels). Mmp14 wild-type and null organoids display no significant differences in proliferation or apoptosis (Fig. S3A). Nevertheless, Mmp14-deleted carcinoma organoids retain migratory activities comparable to wild-type carcinomas when seeded atop, as opposed to embedded within, type I collagen hydrogels (Fig. S3B-D). Ruling out unanticipated effects of Mmp14 expression on tumor initiation, when organoids from Mmp14f/f carcinomas are isolated and transduced with either adenoviral-GFP or adenoviral-Cre expression vectors, control transfectants fully retain 3-D invasive activity while acute Mmp14 deletion significantly inhibits collagen-invasive activity with a 4.5-fold reduction in branch length and a 5.5-fold decrease in branch number without affecting CK8/14 expression or localization (p<0.0001) (Fig. 3C,D). Conversely, organoids isolated from MT1-MMP-null carcinomas and transduced with either lentiviral-GFP or lentiviral-Mmp14 expression vectors, re-express MT1-MMP to wild-type levels and fully re-activate collagen-invasive activity (Fig. 3E,F). While cancer-associated fibroblasts (CAFs) can promote MMTV-PyMT carcinoma invasion in vitro (Corsa et al., 2016), wild-type CAFs fail to rescue the tissue-invasive activity of Mmp14-null carcinoma organoids (Fig. S3E,F). Conversely, tumor organoids harvested from MT2-MMP-targeted carcinomas retain full, if not enhanced, collagen-invasive activity (Fig. 3G).
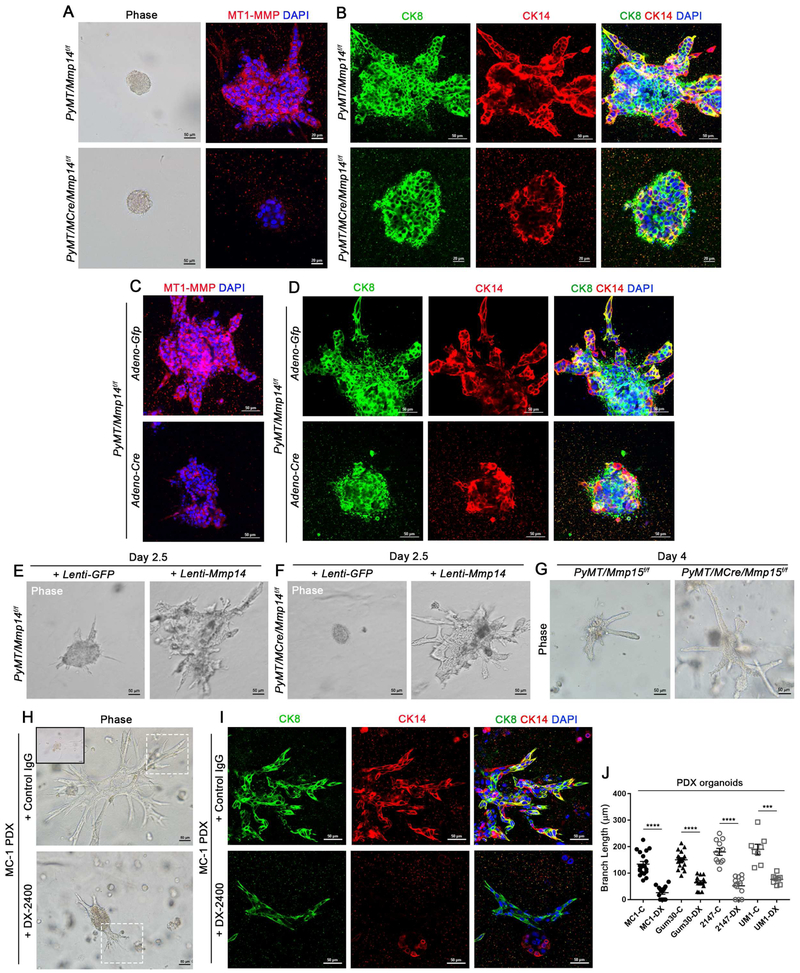
Figure 3. MT1-MMP Directs Mouse and Human Breast Carcinoma Invasion Programs Ex Vivo.
(A) Bright-field micrographs of mammary carcinoma-derived organoids harvested from 3 month-old MMTV-PyMT +/−/Mmp14f/f and MMTV-PyMT +/−/MMTV-Cre +/−/Mmp14f/f mice, embedded in type I collagen hydrogels for 1 hour (left panels; scale bar, 50 μm); and MT1-MMP immunofluorescence with DAPI staining after a 4 day-culture period in collagen with FGF-2 (right panels) (scale bar, 20μm). Tumor organoid branch length is reduced from 93.5±4.3 μm with 5.7±0.3 branches/organoid in MMTV-PyMT +/−/Mmp14f/f organoids (n=35) to 10.2±2.6 μm and 0.5±0.1 branches/organoid in MMTV-PyMT +/−/MMTV-Cre +/−/Mmp14f/f organoids (n=28) (p<0.0001, mean ± SEM).
(B) Immunofluorescence for CK8 and CK14 with DAPI staining in mammary carcinoma-derived organoids harvested from 3 month-old MMTV-PyMT +/−/Mmp14f/f and MMTV-PyMT +/−/MMTV-Cre +/−/Mmp14f/f mice and cultured within 3-D type I collagen for 4 days with FGF-2 (scale bars, 20 μm and 50 μm).
(C,D) MT1-MMP immunofluorescence with DAPI staining (C) as well as CK8 and CK14 with DAPI staining (D) in mammary carcinoma-derived organoids harvested from 3 month-old MMTV-PyMT +/−/Mmp14f/f mice and transduced with either Adenoviral-Gfp or Adenoviral-Cre prior to embedding in 3-D type I collagen for 4 days with FGF-2 (scale bars, 50 μm). Tumor organoid branch length is reduced from 95.0±5.8 μm and 6.1±0.4 branches/organoid with Adeno-Gfp (n=14) to 21.0±4.5 μm and 1.1±0.2 branches/organoid with Adeno-Cre (n=18) (p<0.0001, mean ± SEM).
(E,F) Bright-field micrographs of mammary carcinoma-derived organoids harvested from 3 month-old MMTV-PyMT +/−/Mmp14f/f (E) and MMTV-PyMT +/−/MMTV-Cre +/−/Mmp14f/f (F) mice and transduced with either Lentiviral-Gfp (left panel) or Lentiviral-Mmp14 (right panel) before embedding in 3-D type I collagen for 2.5 days with FGF-2 (scale bars, 50 μm). Lenti-Gfp-transduced MMTV-PyMT +/−/MMTV-Cre +/−/Mmp14f/f tumor organoids fail to branch (0±0μm branches, n=8) whereas MMTV-PyMT +/−/Mmp14f/f tumor organoid branches measure 59.3±3.0 μm with 6.4±0.4 branches/organoid, respectively (n=12) (p<0.0001). Lenti-Mmp14-transduced MMTV-PyMT +/−/Mmp14f/f tumor organoid branches measure 128.4±6.9 μm with 9.9±0.8 branches/organoid (n=16), while Lenti-Mmp14-transduced MMTV-PyMT +/−/MMTV-Cre +/−/Mmp14f/f tumor organoid branches measure 119.2±9.8 μm with 8.0±1.3 branches/organoid (n=10) (p>0.200). Data are presented as mean ± SEM.
(G) Bright-field micrographs of mammary carcinoma-derived organoids harvested from 3 month-old MMTV-PyMT +/−/Mmp15f/f and MMTV-PyMT +/−/MMTV-Cre +/−/Mmp15f/f mice and embedded in 3-D type I collagen for 4 days with FGF-2 (scale bar, 50 μm). MMTV-PyMT +/−/MMTV-Cre +/−/Mmp15f/f tumor organoids display increased branching, with 6.7±0.2 branches/organoid (n=11) versus the 4.7±0.7 branches/organoid present in MMTV-PyMT +/−/Mmp15f/f organoids (n=10) (p=0.023, mean ± SEM).
(H) Bright-field micrographs of organoids harvested from human patient-derived xenograft (PDX) MC-1 tumors and cultured in 3-D type I collagen for 4 days with FGF-2 in the presence of the anti-MT1-MMP antibody (DX-2400) or control IgG antibody (scale bar, 50 μm). Dotted line boxes indicate the branching structures examined in (I). Inset shows MC-1 organoid at time 0. Organoid branch length is reduced from 134.2±10.0 μm with 7.5±0.6 branches/organoid in the presence of control IgG (n=19) to 26.2±5.9 μm with 1.9±0.7 branches/organoid in the presence of DX-2400 (n=16) (p<0.0001, mean ± SEM).
(I) CK8 and CK14 immunofluorescence with DAPI staining in PDX MC-1 organoids cultured in 3-D type I collagen for 4 days with FGF-2 in the presence of DX-2400 or control IgG antibody (scale bar, 50 μm).
(J) Quantification of tumor organoid invasion (branch length, μm) from 4 triple-negative breast cancer PDX lines in the presence of control IgG (C) or DX-2400 (DX) (****p<0.0001, ***p=0.0004; mean ± SEM).
See also Figure S3.
Given links between MT1-MMP expression levels and poor clinical outcome in breast cancer patients (Lodillinsky et al., 2016; Rosse et al., 2014), we sought to determine whether a reliance on MT1-MMP-dependent invasion could be extended to human carcinomas. Indeed, tissues recovered from triple-negative breast cancer xenografts are strikingly MT1-MMP immunoreactive (Fig. S3G). As such, organoids isolated from 4 different patient-derived xenografts (PDXs) were embedded in collagen hydrogels in the absence or presence of a monoclonal antibody directed against the catalytic domain of human MT1-MMP (Devy et al., 2009). As observed with MMTV-PyMT-derived carcinomas, human breast cancer organoids display robust invasive activity while expressing CK8 and CK14 (Fig. 3 H-J; Fig. S3H,I; upper panels). In the presence of the MT1-MMP inhibitory monoclonal antibody, however, collagen-invasive activity is decreased by 2-to-5-fold in tandem with a general reduction in CK14 expression (Fig. 3H-J; Fig. S3H,I; lower panels). Hence, both mouse and human breast carcinomas are able to mobilize MT1-MMP to drive collagen-invasive programs.
Stromal cell-derived MT1-MMP regulates mammary gland ECM remodeling in vivo
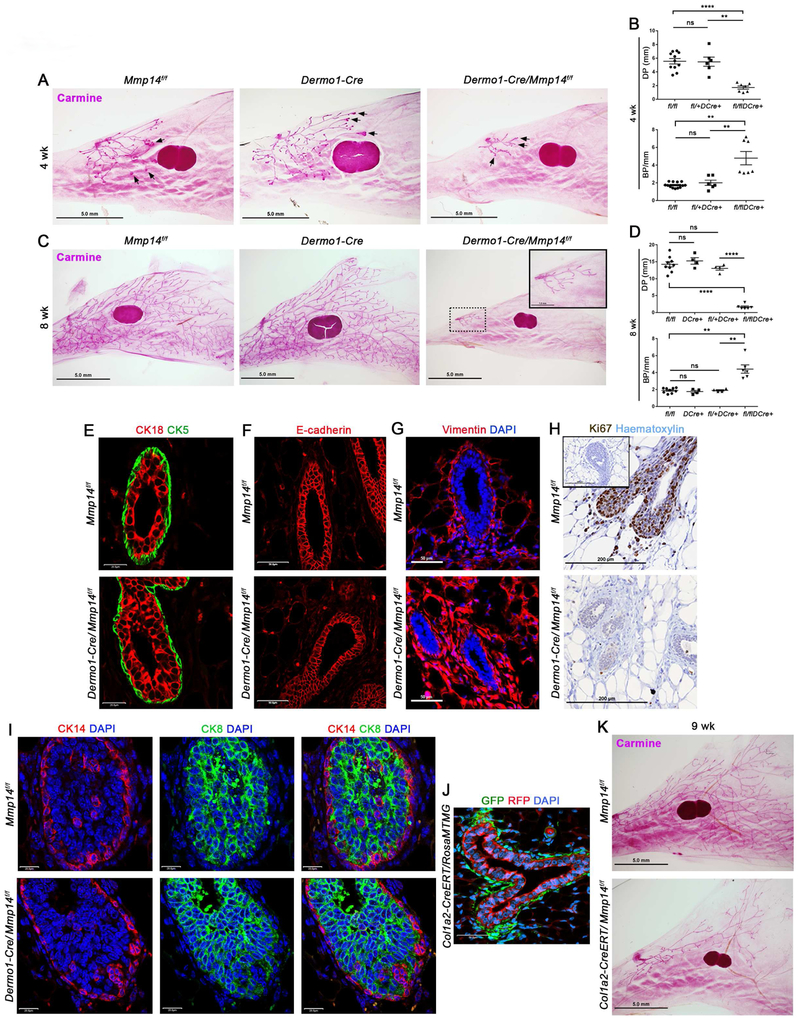
The inability of mammary epithelial cell MT1-MMP or MT2-MMP to support postnatal branching morphogenesis raises the possibility that normal ducts might invade stromal tissues by proteinase-independent mechanisms (Paul et al., 2017). However, using the collagen hybridizing peptide to probe for matrix remodeling events reveals discrete fields of degraded collagen along developing mammary epithelial ducts, with an 11.8-fold increase in fluorescence relative to a scrambled control peptide (p<0.0001) (Fig. 4A). Coincident with periductal collagen turnover, MT1-MMP protein expression likewise outlines the periductal space (Fig. 4B). Hence, to determine whether stromal cell-derived MT1-MMP alternatively acts as a collagenolytic effector during branching morphogenesis, Mmp14 was targeted in the mammary gland stroma by crossing Mmp14f/f mice with a Dermo1/Twist2-Cre knock-in line where Cre expression is largely confined to the mesenchyme (Tang et al., 2013). As expected, Dermo1-Cre-expressing mice crossed onto a RosamTmG background display widespread Cre expression in the mammary stromal cell compartment with green fluorescence highlighting the periductal fibroblasts as well as the surrounding mammary adipocytes, and red fluorescence restricted chiefly to the mammary epithelium (Fig. 4C). Confirming the efficiency of stromal cell targeting in Dermo1-Cre +/−/Mmp14f/f mice, epithelium-free mammary gland tissue displays a >90% decrease in Mmp14 expression (Fig. 4D) with a near complete loss of MT1-MMP at the protein level (Fig. 4E,F). Under these conditions, the formation of periductal collagen degradation products is nearly completely inhibited in Dermo1-Cre +/−/Mmp14f/f glands with a 10.1-fold reduction in CF-CHP immunofluorescence (p<0.0001) (Fig. 4G). In tandem with the observed block in collagen remodeling, periductal type I collagen protein levels are increased markedly as assessed by Sirius Red staining or transmission electron microscopy (TEM) (Fig. 4H-J). Likewise, the basement membrane displays a ~2-fold increase in type IV collagen immunostaining in tandem with a thickened basement membrane (Fig. 4J,K). Interestingly, despite the marked increases in type I and IV collagen levels, analyses of differentially expressed gene products in Dermo1-Cre +/−/Mmp14+/+ versus Dermo1-Cre +/−/Mmp14f/f mammary tissues demonstrate that stromal cell MT1-MMP targeting down-regulates, rather than upregulates, interstitial and basement membrane-associated transcripts (Fig. 4L).
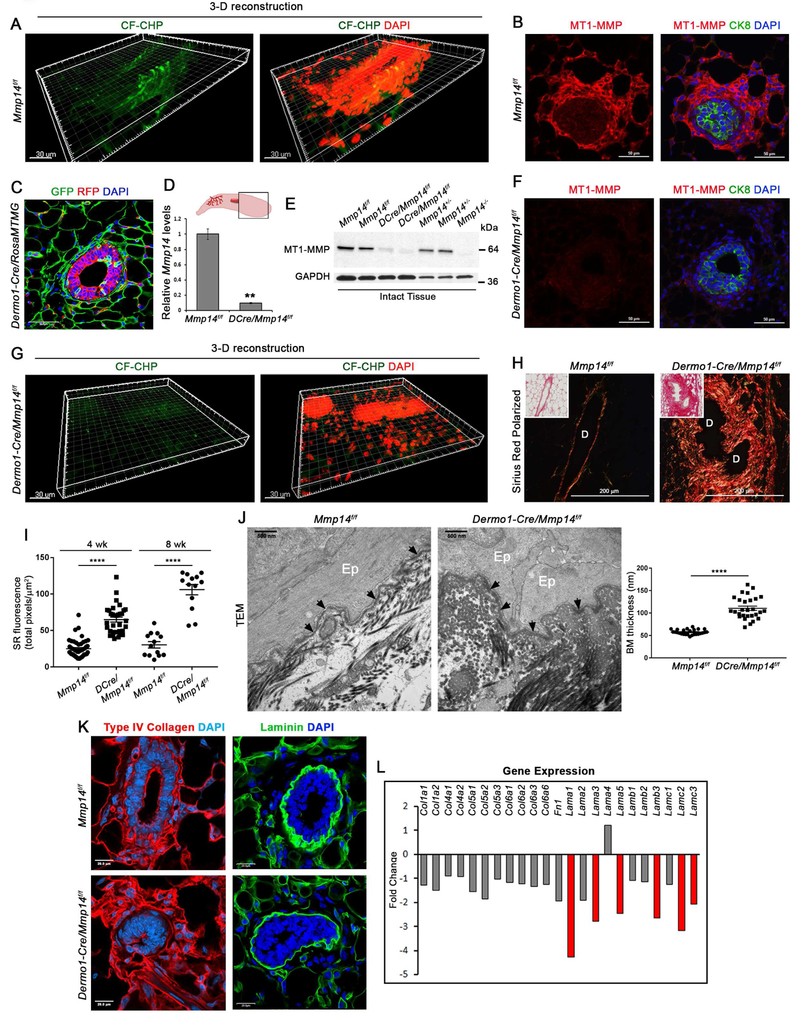
Figure 4. Stromal cell-derived MT1-MMP regulates extracellular matrix remodeling during branching morphogenesis in vivo.
(A) 3-D reconstruction of CF-CHP immunofluorescence in mammary gland tissue harvested from 4 wk-old Mmp14f/f mouse with DAPI staining (scale bar, 30 μm). Immunofluorescence increases from 2.7±0.5 total pixels/μm2 with scrambled CHP (n=6) to 31.8±2.0 total pixels/μm2 with CF-CHP (n=9) (p<0.0001, mean ± SEM).
(B) MT1-MMP and CK8 immunofluorescence with DAPI staining in mammary gland tissue harvested from 4 wk-old Mmp14f/f mouse (scale bar, 50 μm).
(C) Cross-section of mammary gland isolated from 4 wk-old Dermo1Cre +/−/RosaMTMGloxP/+ mouse with DAPI staining (scale bar, 50 μm).
(D,E) Mmp14 excision at the mRNA and protein levels as normalized to Gapdh (n=3 per genotype, **p<0.001). QPCR data are presented as mean ± SEM.
(F) MT1-MMP and CK8 immunofluorescence with DAPI staining in mammary gland harvested from 4 wk-old Dermo1-Cre +/−/Mmp14f/f mouse (scale bar, 50 μm).
(G) 3-D reconstruction of CF-CHP immunofluorescence in mammary gland harvested from 4 wk-old Dermo1-Cre +/−/Mmp14f/f mouse with DAPI staining (scale bar, 30 μm). CF-CHP immunofluorescence is reduced from 31.8±2.0 total pixels/μm2 in Mmp14f/f mammary glands (n=9) to 3.1±0.3 total pixels/μm2 in Dermo1-Cre +/−/Mmp14f/f mammary glands (n=8) (p<0.0001, mean ± SEM).
(H) Polarized light images of Sirius red staining of 8 wk-old Mmp14f/f and Dermo1-Cre +/−/Mmp14f/f mammary glands (scale bar, 200 μm). Insets show bright-field images. “D” marks the lumen of each mammary duct.
(I) Quantification of red fluorescence in polarized light images of Sirius Red staining in total pixels per surface area (μm2) in Mmp14f/f and Dermo1-Cre +/−/Mmp14f/f mammary glands at 4 wks (n>30 fields per genotype) and 8 wks (n=13 fields per genotype) (****p<0.0001; mean ± SEM).
(J) TEM of mammary glands isolated from 4 wk-old Mmp14f/f and Dermo1-Cre +/−/Mmp14f/f mice with quantification of basement membrane (BM) thickness (nm) in n≥26 fields per genotype (****p<0.0001; mean ± SEM) (scale bar, 500nm). “Ep” marks the mammary epithelial cells and arrows indicate the mammary epithelial basement membrane.
(K) Type IV collagen and laminin immunofluorescence in Mmp14f/f and Dermo1-Cre +/−/Mmp14f/f mammary gland sections with DAPI staining (scale bar, 20 μm).
(L) Changes in ECM transcriptomes in 4 wk-old Dermo1Cre +/−/Mmp14f/f and wild-type glands (n=3 per genotype).
Stromal cell-derived MT1-MMP regulates branching morphogenesis in vivo
Given MT1-MMP-dependent control of ECM remodeling during postnatal mammary gland development, mammary glands were recovered from 4–8 wk-old Dermo1-Cre +/−/Mmp14f/f conditional knockout mice as well as littermate controls, and whole mounts evaluated for effects on branching. Strikingly, targeting of stromal cell Mmp14 completely blocks postnatal mammary gland branching (Fig. 5A-D). Nevertheless, Dermo1-Cre +/−/Mmp14f/f ducts display unaltered luminal epithelial cell and myoepithelial cell organization, and retain comparable patterning of E-cadherin expression relative to littermate controls (Fig. 5E-F). By contrast, cross-sections of Dermo1-Cre +/−/Mmp14f/f mammary glands reveal an expansion of periductal fibroblast-like cells surrounding the truncated mammary epithelial ducts despite a substantive block in proliferation in both the epithelial cell and stromal cell compartments (Fig. 5G,H). To rule out possible confounding effects arising from Dermo1-Cre-mediated deletion of Mmp14 in stromal cell populations lying outside the mammary gland, intact glands recovered from Dermo1-Cre +/−/Mmp14f/f, Dermo1-Cre +/−/Mmp14+/+, and Mmp14f/f donors were transplanted into syngeneic, adult wild-type recipients, and mammary gland branching monitored (Fig. S4A). By 5 wks post-transplantation, while control Dermo1-Cre +/−/Mmp14+/+ or Mmp14f/f mammary glands branch effectively the Dermo1-Cre +/−/Mmp14f/f transplants remain unable to undergo branching (Fig. S4B), confirming an organ-autonomous role for stromal MT1-MMP.
Figure 5. Stromal cell-derived MT1-MMP controls mammary gland branching.
(A) Carmine-stained whole mounts of mammary glands isolated from Mmp14f/f, Dermo1-Cre +/− and Dermo1-Cre +/−/Mmp14f/f mice at 4 wks (scale bars, 5.0 mm). Arrows indicate representative terminal end bud structures.
(B) Quantification of ductal penetration (DP) (mm) and branch points per mm duct (BP/mm) in mammary glands harvested from 4 wk-old Mmp14f/f (n=11), Dermo1-Cre +/− (n=6), and Dermo1-Cre +/−/Mmp14f/f (n=7) mice (**p<0.01, ****p<0.0001; mean ± SEM).
(C) Carmine-stained whole mounts of mammary glands isolated from Mmp14f/f, Dermo1-Cre +/− and Dermo1-Cre +/−/Mmp14f/f mice at 8 wks (scale bars, 5.0 mm). Inset in right panel shows Dermo1-Cre +/−/Mmp14f/f gland at higher magnification (scale bar, 1.0 mm).
(D) Quantification of DP (mm) and BP/mm duct in mammary glands harvested from 8 wk-old Mmp14f/f (n=9), Dermo1-Cre +/− (n=4), Dermo1-Cre +/−/Mmp14f/+ (n=4) and Dermo1-Cre +/−/Mmp14f/f (n=6) mice (**p<0.004, ****p<0.0001; mean ± SEM).
(E-G) CK18 and CK5 immunofluorescence (E), E-cadherin immunofluorescence (F), and vimentin immunofluorescence with DAPI staining (G) in 4 wk-old Mmp14f/f and Dermo1-Cre +/−/Mmp14f/f mammary glands (scale bar, 50.0 μm).
(H) Ki67 staining with haematoxylin staining in 4 wk-old Mmp14f/f and Dermo1-Cre +/−/Mmp14f/f mammary glands (scale bar, 200 μm). Inset shows staining with Control IgG antibody.
(I) CK14 and CK8 immunofluorescence in the terminal end buds (TEBs) of Mmp14f/f and Dermo1-Cre +/−/Mmp14f/f mammary glands with DAPI staining (scale bar, 50.0 μm).
(J) GFP and RFP fluorescence with DAPI staining in 9 wk-old Col1a2-CreERT +/−/RosaMTMGloxP/+ mammary gland (scale bar, 50.0 μm).
(K) Carmine-stained mammary gland whole mounts from 9 wk-old Mmp14f/f and Col1a2-CreERT +/−/Mmp14f/f mice (scale bar, 5.0 mm).
See also Figure S4.
Mammary gland branching is thought to largely depend on the growth and branching of highly proliferative terminal end buds (TEBs) (Khokha and Werb, 2011). As such, Dermo1-Cre +/−/Mmp14-targeted mice were assayed for changes in TEB architecture. Though Dermo1-Cre +/−/Mmp14f/f mammary gland TEBs are 2-fold smaller in size and diameter relative to those in Dermo1-Cre +/−/Mmp14+/+ and Mmp14f/f mammary glands, Dermo1-Cre +/−/Mmp14f/f TEBs display a conserved distribution of basal-oriented CK14+ cells and a multilayered inner compartment of CK8+ luminal progenitor cells whose organization is comparable to that present in littermate control TEBs (Fig. 5I). Furthermore, mammary organoids recovered from stromal cell Mmp14-targeted mice branched comparably to controls when embedded in 3-D Matrigel (Fig. S4C). Nevertheless, gene expression profiling of tissues isolated from mammary glands of Dermo1-Cre +/−/Mmp14fl/f and Mmp14+/+ mice reveal ~1000 transcript changes (using a 2-fold enrichment cutoff; p<0.05; n=3 per genotype) with more than 750 genes down-regulated and 250 genes up-regulated in the Mmp14-targeted mice. Functional annotation clustering and enrichment analysis of these data (DAVID Bioinformatics Resource) indicate wide-ranging effects on Gene Ontology (GO) categories, including those associated with cell adhesion and chemotaxis, gland morphogenesis and epithelial cell development (Fig. S4D). In terms of stromal cell gene expression, however, growth factors and signaling molecules required for mammary gland branching, including cytokines, Wnt family members, and growth factor/signaling receptors (Robinson, 2007) are not altered significantly (data not shown). Likewise, when wild-type mammary epithelial organoids were co-cultured with fibroblasts harvested from Mmp14f/f or Dermo1-Cre +/−/Mmp14f/f mammary glands in vitro (Koledova et al., 2016), epithelial organoids undergo enhanced branching following co-culture with Dermo1-Cre +/−/Mmp14f/f fibroblasts relative to Mmp14f/f fibroblasts (Fig. S4E). Further, and in contrast with recent studies suggesting that MT1-MMP impacts tissue development by promoting senescence (Gutierrez-Fernandez et al., 2015), senescence-associated β-gal (SA-βgal) expression levels are globally reduced, rather than increased, in Dermo1-Cre +/−/Mmp14f/f mammary tissue relative to wild-type littermates (Fig. S4F). While other stromal cell populations have been shown to play critical roles in postnatal mammary gland branching, including macrophages, vascular endothelial cells and lymphatic endothelial cells (Inman et al., 2015), macrophage distribution is unaffected in Dermo1-Cre+/−/Mmp14f/f glands in tandem with normal, if not increased, numbers of blood vessels and lymphatics (Fig. S4G,H).
Though Dermo1-Cre is expressed throughout the mammary mesenchyme, including fibroblasts and adipocytes, branching is unaffected when adipocyte Mmp14 expression is targeted using an Adiponectin-Cre transgenic driver (data not shown). Consequently, efforts were directed towards targeting periductal fibroblasts in a more specific manner with a transgenic Col1a2-CreERT line that expresses a tamoxifen-inducible Cre recombinase driven by the mouse Col1a2 promoter (Zheng et al., 2002). Following tamoxifen administration at postnatal day (P)10 and P13 in Mmp14-wildtype Col1a2-CreERT +/− mice carrying a RosamTmG reporter, Cre recombination [marked by green fluorescence] is selectively induced in mammary periductal fibroblasts (Fig. 5J). Importantly, following tamoxifen administration, Mmp14-targeted mammary glands display a marked reduction in duct elongation at 9 wks in association with corresponding increases in interstitial collagen content (Fig. 5K; Fig. S5A,B).
A stromal MT1-MMP-type I collagen axis controls normal mammary gland branching morphogenesis.
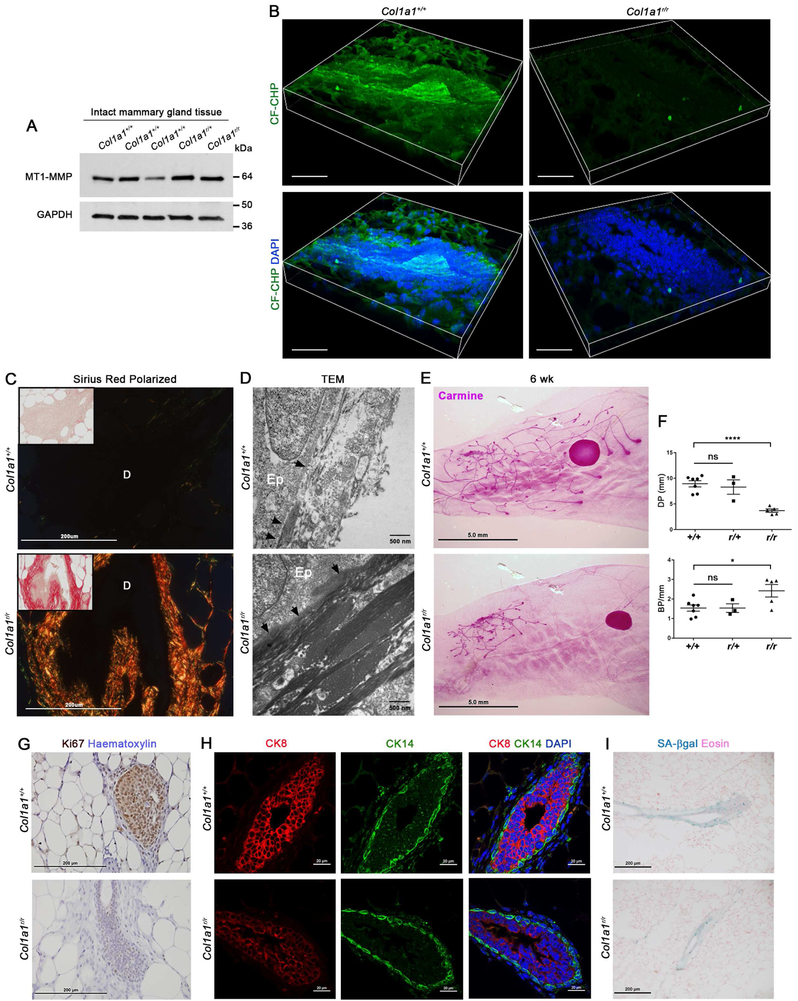
The increased collagen content of stromal Mmp14-targeted glands establishes a correlation, but not causal relationship, between MT1-MMP-dependent collagenolysis and branching. To establish a direct requirement for type I collagen remodeling in branching morphogenesis, we used a mouse type I collagen knock-in model where targeted substitutions around the collagenase cleavage site render the molecule resistant to collagenase attack (Liu et al., 1995). As expected, in situ labeling of degraded collagen in these knock-in mice reveals a near complete absence of collagenolytic activity with a 4.7-fold reduction in CF-CHP immunofluorescence despite normal Mmp14 expression levels (p<0.0001) (Fig. 6A,B). Consistent with this block in collagen remodeling, Sirius Red staining of interstitial collagens surrounding Col1a1r/r mammary ducts is increased relative to Col1a1+/+ or Col1a1r/+ mammary tissues [quantified as a 2.7-fold increase in total pixels of red fluorescence per μm2 in Col1a1r/r versus Col1a1+/+ gland cross-sections (n>16 fields per genotype; p<0.0001)] (Fig. 6C). Likewise, when assessed by TEM, whereas Col1a1+/+ mammary ducts are surrounded by bundles of loosely-packed interstitial fibrils (Fig. 6D), Col1a1r/r ducts are ensheathed by a striking increase in collagen bundles (Fig. 6D) similar to that observed in Dermo1-Cre +/−/Mmp14f/f mice (Fig. 4J). Interestingly, Col1a1r/r mammary ducts also display a ~5-fold increase in basement membrane width (Fig. 6D). Most importantly, coincident with changes in the periductal ECM, Col1a1r/r mice exhibit marked defects in ductal branching (Fig. 6E,F). As observed in stromal cell Mmp14-targeted mice, the block in branching morphogenesis occurs in tandem with attendant decreases in mammary epithelial cell and stromal cell proliferation without affecting mammary epithelial cell organization or cellular senescence (Fig. 6G-I). Hence, whereas mammary carcinoma cells themselves mobilize MT1-MMP to express collagen-invasive activity, the normal mammary gland alternatively relies on periductal stromal cell-derived MT1-MMP to control the developmentally-regulated invasion program associated with branching morphogenesis.
Figure 6. Postnatal mammary gland branching requires type I collagen remodeling.
(A) Western blot for MT1-MMP in mammary glands harvested from 5 wk-old Col1a1+/+, Col1a1r/+ and Col1a1r/r mice, as compared to GAPDH.
(B) 3-D reconstructions of CF-CHP immunofluorescence in mammary glands harvested from 6 wk-old Col1a1+/+ and Col1a1r/r mice with DAPI staining (scale bar, 30 μm). CF-CHP immunofluorescence is reduced from 116.5±6.3 total pixels/μm2 around Col1a1+/+ ducts (n=10) to 24.9±1.1 total pixels/μm2 around Col1a1r/r ducts (n=9) (p<0.0001, mean ± SEM).
(C) Polarized images of Sirius red staining in 5–6 wk-old Col1a1+/+ and Col1a1r/r glands. Insets show bright-field images (scale bar, 200 μm). “D” marks the lumen of each mammary duct.
(D) TEM of 5 wk-old Col1a1+/+ and Col1a1r/r glands (scale bar, 500 nm). “Ep” marks the mammary epithelial cells and arrows indicate the mammary epithelial basement membrane.
(E) Carmine staining of 6 wk-old Col1a1+/+ and Col1a1r/r glands (scale bar, 5.0 mm).
(F) Quantification of ductal penetration (DP) (mm) and branch points per mm duct (BP/mm) in 5–6 wk-old Col1a1+/+ (n=6), Col1a1r/+ (n=3), and Col1a1r/r (n=5) littermates (****p<0.0001, *p<0.05; mean ± SEM).
(G) Ki67 staining in 6 wk-old Col1a1+/+ and Col1a1r/r glands (scale bar, 200 μm).
(H) CK8 and CK14 immunofluorescence with DAPI staining in 6 wk-old Col1a1+/+ and Col1a1r/r glands (scale bar, 20 μm).
(I) Senescence-associated β-galactosidase (SA-βgal) staining (pH 6.0) with Eosin counter-staining in 6 wk-old Col1a1+/+ and Col1a1r/r glands (scale bar, 200 μm).
See also Figure S5.
DISCUSSION
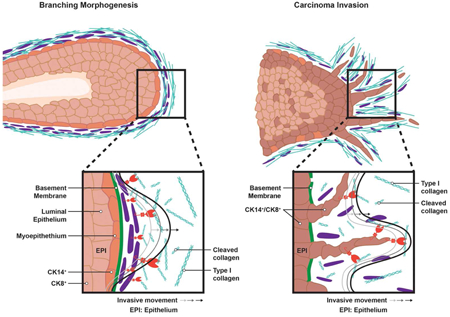
Normal and neoplastic mammary epithelial cells can each display single cell as well as collective cell patterns of migration as they advance through stromal tissues (Cheung et al., 2013; Lambert et al., 2017). However, during morphogenesis, migration proceeds in a highly regulated manner that contrasts with the apparently stochastic process that hallmarks cancer invasion (Cheung et al., 2013; Lambert et al., 2017). Further distinguishing these invasion programs, whereas normal epithelial cells are separated from the underlying interstitial matrix by the subtending basement membrane, carcinoma cells are able to penetrate this barrier, providing neoplastic cells with direct access to the type I collagen-rich stromal matrix (Nguyen-Ngoc et al., 2012; Rowe and Weiss, 2009). Consequently, the strategies employed by normal epithelial cells during branching morphogenesis, though potentially similar to those co-opted by invading cancer cells, necessarily involve distinct classes of epithelial cell-ECM interactions. Indeed, we now report that while both branching morphogenesis and cancer invasion rely on MT1-MMP to negotiate mammary gland stromal tissues, the effector cells and regulatory mechanisms responsible for these ECM remodeling programs are operationally distinct.
Divergent roles for epithelial cell- versus carcinoma cell- derived MT1-MMP
In vivo, mammary epithelial cells express MT1-MMP during branching morphogenesis (Feinberg et al., 2016; Mori et al., 2013). In a manner seemingly consistent with this observation, we and others have demonstrated a requirement for MT1-MMP in driving mammary epithelial cell tubulogenic programs in vitro when performed in type I collagen hydrogels (Feinberg et al., 2016; Mori et al., 2013). Unexpectedly, however Mmp14-null mammary epithelial cells readily mount an unaffected branching morphogenesis program in vivo. Similarly, while ex vivo studies of the salivary gland morphogenetic program have implicated MT2-MMP as a required effector of tubulogenic activity (Rebustini et al., 2009), neither the deletion of Mmp15 alone nor in combination with Mmp14 impairs mammary gland branching. Hence, previous studies emphasizing the role of mammary epithelial cell-derived proteinases in regulating branching in vitro fail to recapitulate the in vivo morphogenic program (Correia et al., 2013; Mori et al., 2013; Nelson et al., 2006; Simian et al., 2001). By contrast, mammary carcinoma cells mobilize MT1-MMP, perhaps as a consequence of neoplasia-associated losses in apical-based polarity (Halaoui et al., 2017), to drive invasion. While we observe partial defects in basement membrane integrity in MT1-MMP-deleted carcinomas in vivo, it should be stressed that proteolysis of the 300-nm thick basement membrane alone is not sufficient to support local invasion into the densely-structured surrounding stromal tissues. As such, the failure of MT1-MMP-targeted carcinoma cells to intravasate and metastasize is more closely linked to a requirement for the proteinase in allowing migrating cells to traverse the type I collagen-rich interstitial matrix. Underlining our in vivo findings, the loss of tissue-invasive activity observed at primary tumor sites is fully recapitulated when conditional knockout carcinoma cells or human breast cancer-derived PDXs are embedded in type I collagen hydrogels. Further, a requirement for carcinoma cell-mediated collagenolysis is supported by the inability of wild-type organoids to infiltrate 3-D hydrogels of collagenase-resistant, mutant type I collagen (Fig. S5C,D).
The ability of MT1-MMP targeting alone to block local tumor invasion and metastasis in vivo is, in several respects, surprising. First, in early clinical trials, pan-specific MMP inhibitors failed to exert therapeutic effects (Fingleton, 2008), but as we have previously reported, the peak plasma concentrations reached in patients fell far below those needed to inhibit the invasive activity of MT1-MMP-expressing carcinoma cells (Sabeh et al., 2009). Alternatively, the anti-MT1-MMP catalytic domain antibody used here has been shown to be effective in blocking cancer cell invasion in xenograft models (Ager et al., 2015; Devy et al., 2009). However, as the antibody targets MT1-MMP expression in all cell populations, MT1-MMP was proposed to exert its effect by regulating endothelial cell, macrophage and cancer cell functions (Ager et al., 2015; Devy et al., 2009). Instead, using a syngeneic model and specific cell-targeting strategies, we demonstrate that carcinoma cell-derived MT1-MMP serves as the dominant effector in vivo. Second, while recent studies have emphasized the ability of carcinoma cells to adopt an amoeboid phenotype that allows cells to negotiate ECM barriers independently of proteolytic activity, clear descriptions of this phenotype have largely been confined to in vitro models (Liu et al., 2015; Paul et al., 2017). By contrast, in the in vivo as well as ex vivo models employed here, proteinase-independent mechanisms are incapable of supporting tissue-invasive activity. As such, it seems likely that the desmoplastic response normally elicited in syngeneic tumor models creates collagen barriers that can only be negotiated by mobilizing the necessary proteolytic machinery. Third, the generally accepted tenet that dense collagen deposits engender cancer cells to assume a more aggressive behavior (Kai et al., 2016) may also require revisiting as the increase in local collagen deposits found in tissues surrounding MT1-MMP-deleted carcinoma cells do not support local invasion or metastasis. Apparently, carcinoma cells cannot assume a more aggressive phenotype in collagen-rich environments unless MT1-MMP is also expressed. Finally, while recent reports have highlighted the ability of cancer-associated fibroblasts to promote MMTV-PyMT-induced breast cancer invasion in vitro by remodeling the ECM (Calvo et al., 2013), we find that the MT1-MMP-positive fibroblasts found in association with MMTV-PyMT carcinomas are unable to rescue the invasion-null status of Mmp14-deleted cancer cells in vivo. Hence, at least in this model, cancer cell-, rather than fibroblast-, derived MT1-MMP serves as the dominant driver of tissue-invasive activity.
Fibroblast-mediated ECM remodeling: Development versus neoplasia
Our inability to implicate epithelial cell-derived Mmp14 or Mmp15 in branching morphogenesis, coupled with the fact that the leading edge of advancing epithelial ducts does not extend filipodia into the surrounding ECM in vivo, and is ensheathed by an unbroken basement membrane, raised the possibility that developmental morphogenesis proceeds independently of proteolytic remodeling (Nguyen-Ngoc et al., 2012; Williams and Daniel, 1983). Likewise, we have found that the early phases of mammary gland branching morphogenesis occurring between embryonic day 16 and postnatal day 10 proceed independently of MT1-MMP (Feinberg et al., 2016). Largely unappreciated, however, neither the basement membrane nor the interstitial matrix of most organ systems are fully mature at birth or in the early postnatal period when the ECM is dominated by hyaluronic acid and associated proteoglycans (Loganathan et al., 2016). With specific regard to type I collagen, major changes in collagen deposition and organization occur almost exclusively in the postnatal state, apparently in association with the animal’s adaptation to gravity-dependent mechanical stresses in the extra-uterine environment (Rowe and Weiss, 2009). However, by the onset of puberty, the mammary gland ECM is fully mature and the detection of collagenolytic activity generated during morphogenesis clearly establishes the fact that proteolytic remodeling is linked to postnatal branching activity.
Herein, we unexpectedly uncovered a required role for stromal cell MT1-MMP in driving morphogenesis. Not only does targeting of stromal cell Mmp14 completely block branching morphogenesis, but it also triggers a dramatic increase in ECM deposition, thereby demonstrating that periductal fibroblasts use MT1-MMP to both sculpt the basement membrane and regulate stromal matrix architecture. Consistent with studies highlighting the importance of type I collagen as a structural scaffold during morphogenesis (Piotrowski-Daspit et al., 2017), we find that mammary gland branching fails to proceed in mutant mice that express collagenase-resistant type I collagen, fully recapitulating the phenotype observed in the stromal Mmp14-targeted animals. Taken together, these findings serve to characterize a periductal fibroblast MT1-MMP/type I collagen axis as a definitive regulator of the postnatal branching morphogenesis program. Counterintuitively, our findings demonstrate that fast-moving mammary ducts (Williams and Daniel, 1983) actually elongate and branch in a proteolytically passive manner by “invading” a stromal microenvironment remodeled by periductal fibroblast-derived MT1-MMP.
In sum, despite the long held assumption that carcinoma cells purposefully reactivate morphogenesis-associated developmental programs to initiate tissue-invasive programs, the underlying processes that define these activities have remained the subject of conjecture. Here, we not only define the preeminence of ECM proteolysis over non-proteolytic mechanisms in regulating epithelial cell as well as carcinoma cell trafficking in vivo, but also identify MT1-MMP as a key effector of these matrix-remodeling processes. However, rather than mobilize MT1-MMP in an identical fashion, we highlight the existance of the distinct strategies that distinguish physiologic from pathologic tissue-invasive programs in vivo.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to, and will be fulfilled by, the Lead Author, Dr. Stephen J. Weiss (sjweiss@umich.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Mmp14lacZ/+ mice (Tang et al., 2013) were bred onto a C57BL/6J background. Mmp15lacZ/+ mice were generated as described (Feinberg et al., 2016) from C57BL/6J Mmp15lacZ/+ embryonic stem cell (ESC) clones from the UC Davis KOMP Repository. Mmp15lacZ/+ mice were bred and maintained on a C57BL/6J background or mated onto a FVB/NJ background. Mmp14 +/− mice (Feinberg et al., 2016) were bred with a transgenic mouse line expressing enhanced GFP under the control of a chicken β-actin promoter and mated onto a FVB/NJ background. Mmp3−/− mice (C57BL/6J background), Mmp13−/− (C57BL/6J background), and Mmp2−/− mice (129/SVE background) were genotyped as described (Feinberg et al., 2016). Mmp15 global knockout (Mmp15−/−) mice were generated as described (Feinberg et al., 2016) wherein exons 4 and 5 of the catalytic domain were floxed by loxP sites. Offspring carrying the Mmp15flox allele were either backcrossed to C57BL/6J or bred to C57BL/6J EIIA-Cre mice for germline deletion of Mmp15. Both Mmp15f/f mice and Mmp15+/− mice were backcrossed onto a FVB/NJ background (n>10 generations). Mmp14f/f mice were generated as described previously (Tang et al., 2013) wherein exons 2 through 4 of the catalytic domain were floxed by loxP sites, and were maintained on either a C57BL/6J or FVB/NJ background (n>10 generations). MMTV-Cre transgenic mice (line A) (Wagner et al., 2001) on a congenic FVB/NJ background (n>10 generations) were mated with Mmp14f/f mice, Mmp15f/f mice or Mmp14f/fMmp15f/f mice (FVB/NJ background, n>5 generations). Transgenic MMTV-PyMT mice (Cheung et al., 2013; Ni et al., 2016) were maintained on a FVB/NJ background. Hemizygous PyMT +/− male mice were bred with MMTV-Cre +/− or Fsp1-Cre +/− female mice (Trimboli et al., 2009) and double hemizygous males were bred with Mmp14f/f, Mmp15f/f or Mmp14f/fMmp15f/f females (FVB/NJ) and maintained on a congenic FVB/NJ background. Dermo1/Twist2-Cre mice (Tang et al., 2013) were bred with Mmp14f/f mice (C57BL/6J background) and maintained on a congenic C57BL/6J background (n>10 generations) or crossed onto a FVB/NJ background (n>5 generations). Tamoxifen-inducible Col1a2-CreERT transgenic mice were maintained on a C57BL/6J background and bred with Mmp14f/f mice on a C57BL/6J background. Tamoxifen was dissolved in ethanol at 80mg/ml and further diluted in sunflower seed oil (S5007, Sigma-Aldrich) to 1mg/ml prior to IP injections of 50ul at postnatal day (P)10 and P13. Col1a1r/r mice (Liu et al., 1995) were maintained on a 129/Sv background. All mouse work was performed with prior approval of the University of Michigan Institutional Animal Care and Use Committee (IACUC) (Animal Protocols: PRO00006828, PRO00006783, PRO00008101), in accordance with the guidelines established by the National Research Council Guide for the Care and Use of Laboratory Animals (NIH). Weaned (3 week-old to 5 month-old) female mice were housed in the University of Michigan Animal Facility, in an environment controlled for light (12 hours on/off) and temperature (21–23°C), with open access to water and food (Lab Diet, Cat#5LOD). For detailed mouse strain information, see Key Resources Table.
Patient-Derived Xenograft (PDX) Tumor Model
Patient-derived xenograft (PDX) tumor models were established as described (Peng et al., 2016). In brief, serially passaged PDX tumors were harvested at 700 to 1000 mm3 from severe combined immunodeficient mice (CB-17 SCID), minced into ~4mm pieces and washed in high-glucose DMEM (Invitrogen) containing 1% L-glutamine (Invitrogen), 10% Fetal Bovine Serum (FBS, Corning), 20 ng/ml rhEGF (Peprotech), 400 ng/ml hydrocortisone (Sigma-Aldrich), 5 g/ml insulin (Sigma-Aldrich), 50 mg/ml Nystatin (Sigma-Aldrich), antibiotic-antimycotic (GIBCO) and amphotericin-B (Sigma-Aldrich). After washing, tumor fragments were transplanted into the subcutaneous space of the dorsal region of CB-17 SCID recipient mice. All experiments included here were performed in PDX tumors at in vivo passage 2 or lower. The collection and handling of these specimens were performed under signed, informed consent and approval from appropriate institutional review boards.
Cells
Mammary epithelial organoids were isolated from inguinal mammary glands as described (Feinberg et al., 2016; Mori et al., 2013). In brief, mammary tissues harvested from 4–8 week old mice were minced and digested in DMEM/F12 containing 2.5 mg/ml BSA Fraction V (Sigma-Aldrich) and 0.2% collagenase III (Worthington Biochemical Corp) for 1–2 hours at 37°C with rotation. Mammary carcinoma cell organoids were harvested as described (Cheung et al., 2013), wherein MMTV-PyMT mammary tumors from 3–4 month-old mice were minced and digested in DMEM/F12 containing 0.2% trypsin-EDTA (GIBCO), 0.2% collagenase III (Worthington Biochemical Corp) for 30 minutes at 37°C with rotation. Human mammary PDX tumors were minced and digested at 37°C in DMEM/F12 containing 0.2% collagenase III (Worthington Biochemical Corp) for 1 hour. Both mouse and human mammary tumor organoid suspensions were applied to 100 μm cell strainers and washed in serum-free DMEM/F12. Organoids were separated from single cells with differential centrifugations prior to infection or resuspension in ECM proteins for in vitro culture. Normal mammary fibroblasts were isolated as described previously (Koledova et al., 2016), for which the supernatant fractions from the differential centrifugations were collected and the corresponding cell pellets were re-suspended for culture in high-glucose DMEM containing 10% FBS, 10 μg/ml insulin (GIBCO), Insulin-Transferrin-Selenium (ITS, Corning) and antibiotic-antimycotic (GIBCO). MMTV-PyMT cancer-associated fibroblasts (CAFs) were isolated from primary tumors as described (Sharon et al., 2013). In brief, MMTV-PyMT mammary tumors were minced and digested in 0.25% collagenase II (Worthington Biochemical Corp), 0.25% collagenase IV (Worthington Biochemical Corp) and deoxyribonuclease (Sigma-Aldrich) in FACS buffer (calcium- and magnesium-free PBS containing 0.5% BSA Fraction V) for 15 minutes at 37°C with rotation. Following the addition of pre-chilled high-glucose DMEM containing 10% FBS, the tissue/media mixture was filtered across a 70 μm cell strainer, centrifuged, and the red blood cells were lysed with 0.64% NH4Cl. The cell suspension was then washed with FACS buffer, filtered across a 40 μm cell strainer, centrifuged, and re-suspended in FACS buffer containing anti-mouse (CD16/CD32) Fc Block before incubation with PE-conjugated anti-PDGFRα and FITC-conjugated anti-F4/80 antibodies. The flow cytometry-sorted PDGFRα+ (F4/80-negative) cell population was expanded in vitro before co-culture with primary MMTV-PyMT mammary tumor organoids.
METHOD DETAILS
Genotyping
Genotyping was performed on purified chromosomal DNA prepared from tail tips. Mmp13 genotyping was carried out as described (Feinberg et al., 2016). For Col1a1r genotyping, the PCR product was digested with SphI for 3 hours at 37°C before gel electrophoresis. For genotyping primers, see Table S2.
Mammary gland whole mount preparation and imaging
Inguinal mammary glands were harvested, mounted on glass slides, stained with Carmine Alum, and processed as described previously (Feinberg et al., 2016) with a graded ethanol/xylene series. Whole mounts were imaged with a Leica MZFLIII dissecting microscope and branching was quantified with ImageJ software.
Mammary organoid transduction and 3-D culture
As modified from (Welm et al., 2008), freshly isolated mammary tumor organoids were infected with 1×107 TU lentivirus expressing Gfp or Mmp14 (Shimizu-Hirota et al., 2012) per ~2×106 cells for 17 hours at 37°C in Ultra-low attachment 24-well plates (Corning) . Lentivirus was prepared in DMEM/F12 containing ITS (Corning), 8 μg/ml polybrene (Sigma-Aldrich) and antibiotic-antimycotic (GIBCO). Alternatively, mammary tumor organoids were infected with 200-to-500 MOI Ad5 CMV-Cre or Ad5 CMV-eGFP adenovirus in DMEM/F12 containing ITS (Corning), 8 μg/ml polybrene (Sigma-Aldrich) and antibiotic-antimycotic (GIBCO) for ~24 hours at 37°C in Ultra-low attachment 24-well plates. Transduced mammary organoids were washed with DMEM/F12 and re-plated in Ultra-low attachment plates for overnight recovery before further differential centrifugations and resuspension in ECM. Both normal epithelial organoids and tumor organoids were embedded in acid-extracted rat-tail type I collagen at 2.2 mg/ml final concentration, as described (Feinberg et al., 2016; Sabeh et al., 2009). For mammary tumor organoid/CAF co-culture experiments, CAFs were added at a ratio of 3 CAFs per 1 organoid. Normal mammary epithelial organoids were alternatively embedded in growth-factor reduced, phenol red-free Matrigel (Corning). Mammary carcinoma-derived organoids were also embedded in acid-extracted type I collagen isolated from 4–6 month-old Col1a1+/+ and Col1a1r/r mouse tails. All hydrogels were assembled in 24-well plates. Normal mammary epithelial organoids and MMTV-PyMT mammary tumor organoids were cultured in DMEM/F12 containing ITS (Corning), 10 μg/ml insulin (GIBCO), 50 ng/μl FGF-2 (Sigma-Aldrich) and antibiotic-antimycotic (GIBCO). Human mammary PDX organoids were cultured in high-glucose DMEM (Invitrogen) containing 10% FBS, 1 μg/ml hydrocortisone (Sigma-Aldrich), 10 μg/ml insulin (GIBCO), ITS (Corning), 50 ng/ml FGF-2 (Sigma-Aldrich) and antibiotic-antimycotic (GIBCO) in the presence or absence of human isotype control IgG antibody (Invitrogen) or anti-MT1-MMP antibody (DX-2400) (Ager et al., 2015) at 100 μg/ml. DX-2400 was provided by the Kadmon Corporation. In vitro branching and invasion of mammary organoids was quantified from bright-field micrographs with ImageJ software.
Isolation of circulating tumor cells
Circulating tumor cells were isolated as described previously (Hosseini et al., 2016). In brief, 500 μl of whole blood was harvested from the right ventricle of each mouse using a 25 G × 5/8 needle (BD) and a 1 ml syringe (BD) containing heparin (Sigma-Aldrich). Blood was mixed with 0.64% NH4Cl, centrifuged at 1500 rpm for 5 minutes, and the pellet was washed twice with PBS before lysis in TRIzol (Ambion, Life Technologies) and RNA purification.
RNA extraction and gene expression analysis
For gene expression analysis of normal mammary gland tissue, inguinal mammary glands were flash frozen in liquid nitrogen and homogenized in TRIzol (Ambion, Life Technologies). Total RNA was extracted and purified using QIAGEN RNeasy Mini-kit columns (QIAGEN, 74104). RNA quality was confirmed using an Agilent 2100 Bioanalyzer and samples profiled on Affymetrix Mouse Gene 2.1 ST expression arrays. Expression values for each probe set were calculated using a robust multi-array average (RMA) (Irizarry et al., 2003) and filtered for genes with a greater than 1.5-fold change. Isolated normal mammary epithelial organoids, isolated mammary carcinoma cell (tumor) organoids and intact mammary tumor fragments were similarly lysed in TRIzol and processed for total RNA extraction and gene expression analysis. The Affymetrix microarray data have been deposited in the NCBI Gene Expression Omnibus (GEO) database (Edgar et al., 2002) and are available through accession numbers: GSE79088 (array of stromal cell-targeted mammary glands, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE79088), GSE118506 (array of Mmp14-targeted mammary epithelial cells; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE118506) and GSE118466 (array of Mmp14-targeted MMTV-PyMT mammary tumors, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE118466). Complementary cDNA was prepared from total RNA with the Invitrogen SuperScript II First-Strand Synthesis System (11904–018). Quantitative reverse-transcription PCR was performed with SYBR Green (Applied Biosystems) using Gapdh or Arbp/Rplp0 for reference gene expression. For quantitative real-time PCR primers, see Table S3.
Mammary tissue and whole gland transplants
Mammary tissue fragment transplants were carried out as described previously (Robinson et al., 2000). In brief, inguinal mammary glands of recipient mice (postnatal day 21–22, 10–11 grams body weight) were “cleared” of their endogenous epithelium by surgical removal of the mammary gland tissue between the cauterized femoral artery and the inguinal lymph node. The isolated epithelium-containing mammary tissue was mounted on glass slides for whole mount Carmine staining to confirm clearing. Single 1–2 mm fragments of donor mammary epithelium-containing tissue from 2-wk-old mice were transplanted into the remaining epithelium-free recipient mammary gland for in vivo growth. Transplants were allowed to branch in vivo for 6–8 wks prior to whole mount and histological assessments. For transplant of mammary tumor fragments, single 1–2 mm RFP+ fragments of MMTV-PyMT tumor tissue were isolated from 2–3 month old MMTV-PyMT +/−/MMTV-Cre+/−/RosaRFP/Mmp14f/+ and MMTV-PyMT +/−/MMTV-Cre+/−/RosaRFP/ Mmp14f/f mice and transplanted into the “cleared” mammary glands of syngeneic FVB/NJ recipient mice (Jackson Laboratory). For transplant of whole mammary glands, the inguinal mammary glands from 6–8 wk-old recipient mice were excised in entirety following cauterization of the femoral artery, and the isolated tissue was mounted on glass slides for whole mount Carmine staining to confirm clearing. Intact whole mammary glands from 5–10 day-old donor mice were sutured onto the gland-free subcutaneous surface. The transplants were allowed to branch in vivo for up to 5 wks prior to whole mount assessments. Athymic nude mice (Harlan Laboratories) were used as recipients for the transplant of Gfp +/−/Mmp14 +/− and Gfp +/−/Mmp14−/− ductal fragments. Syngeneic C57Bl/6J mice (Jackson Laboratory) were used as recipients for the transplant of Mmp14f/f and Dermo1-Cre +/−/Mmp14f/f mammary glands.
Western Blotting
Inguinal mammary gland tissue and MMTV-PyMT mammary tumor tissue was flash frozen in liquid nitrogen and homogenized in RIPA lysis buffer containing protease inhibitors (Roche, 11836153001) and phosphatase inhibitors (Thermo Scientific, 1862495). Protein concentrations were determined with the Pierce BCA Protein Assay Kit (Thermo Scientific), and extracts were analyzed by standard techniques with horseradish peroxidase–conjugated secondary antibodies (Cell Signaling).
LacZ and cellular senescence staining
For LacZ staining, inguinal mammary glands and mammary tumors were fixed at room temperature for 30–45 minutes in 4% paraformaldehyde (Electron Microscopy Sciences). Tissues were either transferred to 30% sucrose/PBS (w/v) for frozen sections, or incubated for 18–24 hours in PBS containing 4 mM potassium hexacyanoferrate (III) (Sigma-Aldrich), 4 mM potassium hexacyanoferrate (II) trihydrate (Sigma-Aldrich), 2 mM MgCl2 (Fisher Scientific), 0.2% NP-40 (Fisher Scientific), 0.1% sodium deoxycholic acid (Fisher Scientific) and 1 mg/ml Xgal (Invitrogen). Stained tissues and whole mounts were post-fixed with 10% formalin-phosphate (Fisher Scientific). Senescence-associated β-gal staining was carried out on mammary gland cryo-sections as described for the Senescence β-galactosidase Staining Kit (Cell Signaling). Sections were stained overnight (12–15 hours) in a non-humidified 37°C incubator, counter-stained with Eosin, and dehydrated with a graded ethanol/xylene series before mounting with Permount Toluene Solution (Fisher Scientific).
Immunofluorescence and immunohistochemistry
Mammary gland and mammary tumor tissue was fixed in 10% formalin-phosphate at 4° C overnight and either dehydrated in an ethanol/paraffin series for paraffin embedding or transferred to 30% (w/v) sucrose/PBS for OCT embedding. Following rehydration with a xylene/ethanol series and microwave-mediated antigen retrieval, paraffin sections were permeabilized with 0.3% Triton X-100/PBS, and blocked for 2 hours prior to primary antibody addition. Samples were incubated overnight at 4°C with primary antibodies. Frozen sections (10–30 μm) were used for immunostaining of MT1-MMP, basement membrane, macrophages (F4/80), blood vessels (CD-31) and lymphatics (Lyve-1). For MT1-MMP staining, frozen sections were washed in 1X PBS, permeabilized in 0.3% Triton X-100/PBS, blocked in 3% BSA, and incubated with primary antibody diluted in 3% BSA/PBS overnight at 4° C. For basement membrane staining, frozen sections were blocked in 5% FBS/1% BSA/PBS for 2 hours before overnight incubations with primary antibodies directed against type IV collagen or laminin. Following all primary antibody incubations, Alexa-488 or Alexa-594-conjugated secondary antibodies (Invitrogen Molecular Probes) were added in blocking buffer for 2–4 hours. Neoplastic mammary epithelial organoids were alternatively stained with HCS CellMask (Invitrogen Molecular Probes) following in vitro culture in 3-D type I collagen, fixation in 4% PFA and permeabilization with 0.3% Triton X-100/PBS. Nuclear counter-staining was carried out with either Toto3 (Invitrogen Molecular Probes) or 4, 6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich). All fluorescence images were acquired with an Olympus FluoView FV500 laser scanning confocal microscope or a Nikon A1/A1R confocal laser microscope. For immunohistochemistry, endogenous peroxidase activity was blocked with H2O2 and tissues sections developed with the Vectastain ABC (Vector Laboratories, PK-6100) and DAB kits (Vector Laboratories, SK-4100). Immunofluorescence was quantified from split-color channel images of confocal Z-slices using ImageJ software.
CHP peptide assay
Mammary gland cryosections (20–30 μM thickness) were washed in 1X PBS and blocked in 3% BSA/ 5% goat serum/ PBS at room temperature for 1 hour. During blocking, heat-activated fluorescently labeled CHP stock solution (CF-CHP [CF-G-(GPO)9] from 3Helix) and sequence-scrambled control peptide stock solution (Li et al., 2012) were diluted to 10–30 μM in PBS and incubated at 70°C for 10 minutes, followed by a brief (10–30 second) incubation on ice before immediate application to blocked tissue for overnight incubation at 4°C while protected from light. Following washes with PBS, nuclei were counterstained with DAPI and sections were mounted with Prolong Gold anti-fade reagent (Invitrogen). CHP immunofluorescence was quantified from split-color channel images of confocal Z-slices and confocal Z-stacks using ImageJ software.
Tissue preparation for TEM
Samples were fixed (1% glutaraldehyde/1% tannic acid/0.1 M Sorensen’s buffer, pH 7.2) and post-fixed with 1% osmium tetroxide prior to embedding in EMbed 812 epoxy resin (Electron Microscopy Sciences). Thin sections (70 nm) were post-stained with uranyl acetate and Reynolds Lead Citrate, and imaged with a JE0L JEM-1400 Plus electron microscope. Images were recorded digitally using a Hamamatsu ORCA-HR digital camera system, operated using AMT software (Advanced Microscopy Techniques Corp., Danvers, MA). Basement membrane thickness was quantified from n>7 regions per high magnification field and from n≥26 fields per genotype with ImageJ software.
Data and software availability
Microarray data are available at Gene Expression Omnibus (GEO) under accession numbers: GSE79088, GSE118506 and GSE118466.
QUANTIFICATION AND STATISTICAL ANALYSIS
All data from the in vivo and in vitro studies are shown as mean values with error bars representing the standard error (SEM). Replicates are indicated in the figure legends. Adobe Photoshop, ImageJ (NIH) and GraphPad Prism 7 software were used to process and analyze images and data. For each comparison between two groups, statistical analysis was performed and p values were calculated with an unpaired two-tailed Student’s t test with Welch’s correction using GraphPad Prism 7 software. For all tests, p<0.05 was considered statistically significant.
Supplementary Material
Highlights:
-
(1)
MT1-MMP governs both normal and neoplastic mammary epithelial cell invasion in vivo.
-
(2)
Mammary gland branching morphogenesis requires stromal – but not epithelial – MT1-MMP.
-
(3)
Mammary carcinoma cell-mobilized MT1-MMP dictates local invasion and metastasis.
-
(4)
MT1-MMP-dependent collagenolytic activity underpins mammary epithelial cell invasion.
ACKNOWLEDGEMENTS
We thank Gertraud Robinson (NIH, NIDDK) for transplant guidance and training. We thank Yang Li (University of Utah), Hemant Bid (Resonant Therapeutics) and Yuexian Hu (University of Michigan) for technical support. We thank David Ginsburg (University of Michigan), Xiao-Yan Li (University of Michigan), Yi Tang (University of Michigan) and Yang Li (University of Utah) for helpful discussions. We thank the Kadmon Corporation for providing the anti-MT1-MMP antibody, licensed to the Kadmon Corporation by the Dyax Corporation. This work was supported by grants from the NIH/NCI to SJW (CA071699), and grants from NIH RO1AR060484 and DOD W81XWH-12-1-0555 to SMY and the Breast Cancer Research Foundation to SJW.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental information includes fives figures and three tables.
DECLARATION OF INTERESTS
S.M.Yu. is the co-founder of 3Helix which commercializes the collagen hybridizing peptide.
REFERENCES
- Ager EI, Kozin SV, Kirkpatrick ND, Seano G, Kodack DP, Askoxylakis V, Huang Y, Goel S, Snuderl M, Muzikansky A, et al. (2015). Blockade of MMP14 activity in murine breast carcinomas: implications for macrophages, vessels, and radiotherapy. J Natl Cancer Inst 107, djv017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo F, Ege N, Grande-Garcia A, Hooper S, Jenkins RP, Chaudhry SI, Harrington K, Williamson P, Moeendarbary E, Charras G, et al. (2013). Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat Cell Biol 15, 637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung KJ, Gabrielson E, Werb Z, and Ewald AJ (2013). Collective invasion in breast cancer requires a conserved basal epithelial program. Cell 155, 1639–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia AL, Mori H, Chen EI, Schmitt FC, and Bissell MJ (2013). The hemopexin domain of MMP3 is responsible for mammary epithelial invasion and morphogenesis through extracellular interaction with HSP90beta. Genes Dev 27, 805–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsa CA, Brenot A, Grither WR, Van Hove S, Loza AJ, Zhang K, Ponik SM, Liu Y, DeNardo DG, Eliceiri KW, et al. (2016). The Action of Discoidin Domain Receptor 2 in Basal Tumor Cells and Stromal Cancer-Associated Fibroblasts Is Critical for Breast Cancer Metastasis. Cell Rep 15, 2510–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devy L, Huang L, Naa L, Yanamandra N, Pieters H, Frans N, Chang E, Tao Q, Vanhove M, Lejeune A, et al. (2009). Selective inhibition of matrix metalloproteinase-14 blocks tumor growth, invasion, and angiogenesis. Cancer Res 69, 1517–1526. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, and Lash AE (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg TY, Rowe RG, Saunders TL, and Weiss SJ (2016). Functional roles of MMP14 and MMP15 in early postnatal mammary gland development. Development 143, 3956–3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingleton B (2008). MMPs as therapeutic targets--still a viable option? Semin Cell Dev Biol 19, 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, Choi H, El Rayes T, Ryu S, Troeger J, et al. (2015). Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 527, 472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Fernandez A, Soria-Valles C, Osorio FG, Gutierrez-Abril J, Garabaya C, Aguirre A, Fueyo A, Fernandez-Garcia MS, Puente XS, and Lopez-Otin C (2015). Loss of MT1-MMP causes cell senescence and nuclear defects which can be reversed by retinoic acid. EMBO J 34, 1875–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halaoui R, Rejon C, Chatterjee SJ, Szymborski J, Meterissian S, Muller WJ, Omeroglu A, and McCaffrey L (2017). Progressive polarity loss and luminal collapse disrupt tissue organization in carcinoma. Genes Dev 31, 1573–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper KL, Sosa MS, Entenberg D, Hosseini H, Cheung JF, Nobre R, Avivar-Valderas A, Nagi C, Girnius N, Davis RJ, et al. (2016). Mechanism of early dissemination and metastasis in Her2+ mammary cancer. Nature 540, 552–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini H, Obradović MMS, Hoffmann M, Harper KL, Sosa MS, Werner-Klein M, Nanduri LK, Werno C, Ehrl C, Maneck M, et al. (2016). Early dissemination seeds metastasis in breast cancer. Nature 540, 552–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J, Huang Y, Burwell TJ, Peterson NC, Connor J, Weiss SJ, Yu SM, and Li Y (2017). In situ imaging of tissue remodeling with collagen hybridizing peptides. ACS Nano 11, 9825–9835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inman JL, Robertson C, Mott JD, and Bissell MJ (2015). Mammary gland development: cell fate specification, stem cells and the microenvironment. Development 142, 1028–1042. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, and Speed TP (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264. [DOI] [PubMed] [Google Scholar]
- Kai F, Laklai H, and Weaver VM (2016). Force matters: biomechanical regulation of cell invasion and migration in disease. Trends Cell Biol 26, 486–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokha R, and Werb Z (2011). Mammary gland reprogramming: metalloproteinases couple form with function. Cold Spring Harb Perspect Biol 3, a004333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koledova Z, Zhang X, Streuli C, Clarke RB, Klein OD, Werb Z, and Lu P (2016). SPRY1 regulates mammary epithelial morphogenesis by modulating EGFR-dependent stromal paracrine signaling and ECM remodeling. Proc Natl Acad Sci U S A 113, E5731–5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert AW, Pattabiraman DR, and Weinberg RA (2017). Emerging biological principles of metastasis. Cell 168, 670–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Foss CA, Summerfield DD, Doyle JJ, Torok CM, Dietz HC, Pomper MG, and Yu SM (2012). Targeting collagen strands by photo-triggered triple-helix hybridization. Proc Natl Acad Sci U S A 109, 14767–14772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wu H, Byrne M, Jeffrey J, Krane S, and Jaenisch R (1995). A targeted mutation at the known collagenase cleavage site in mouse type I collagen impairs tissue remodeling. J Cell Biol 130, 227–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YJ, Le Berre M, Lautenschlaeger F, Maiuri P, Callan-Jones A, Heuze M, Takaki T, Voituriez R, and Piel M (2015). Confinement and low adhesion induce fast amoeboid migration of slow mesenchymal cells. Cell 160, 659–672. [DOI] [PubMed] [Google Scholar]
- Lodillinsky C, Infante E, Guichard A, Chaligne R, Fuhrmann L, Cyrta J, Irondelle M, Lagoutte E, Vacher S, Bonsang-Kitzis H, et al. (2016). p63/MT1-MMP axis is required for in situ to invasive transition in basal-like breast cancer. Oncogene 35, 344–357. [DOI] [PubMed] [Google Scholar]
- Loganathan R, Rongish BJ, Smith CM, Filla MB, Czirok A, Benazeraf B, and Little CD (2016). Extracellular matrix motion and early morphogenesis. Development 143, 2056–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Ewald AJ, Martin GR, and Werb Z (2008). Genetic mosaic analysis reveals FGF receptor 2 function in terminal end buds during mammary gland branching morphogenesis. Dev Biol 321, 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H, Lo AT, Inman JL, Alcaraz J, Ghajar CM, Mott JD, Nelson CM, Chen CS, Zhang H, Bascom JL, et al. (2013). Transmembrane/cytoplasmic, rather than catalytic, domains of Mmp14 signal to MAPK activation and mammary branching morphogenesis via binding to integrin beta1. Development 140, 343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CM, Vanduijn MM, Inman JL, Fletcher DA, and Bissell MJ (2006). Tissue geometry determines sites of mammary branching morphogenesis in organotypic cultures. Science 314, 298–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Ngoc KV, Cheung KJ, Brenot A, Shamir ER, Gray RS, Hines WC, Yaswen P, Werb Z, and Ewald AJ (2012). ECM microenvironment regulates collective migration and local dissemination in normal and malignant mammary epithelium. Proc Natl Acad Sci U S A 109, E2595–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni T, Li XY, Lu N, An T, Liu ZP, Fu R, Lv WC, Zhang YW, Xu XJ, Grant Rowe R, et al. (2016). Snail1-dependent p53 repression regulates expansion and activity of tumour-initiating cells in breast cancer. Nat Cell Biol 18, 1221–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul CD, Mistriotis P, and Konstantopoulos K (2017). Cancer cell motility: lessons from migration in confined spaces. Nat Rev Cancer 17, 131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng D, Tanikawa T, Li W, Zhao L, Vatan L, Szeliga W, Wan S, Wei S, Wang Y, Liu Y, et al. (2016). Myeloid-derived suppressor cells endow stem-like qualities to breast cancer cells through IL6/STAT3 and NO/NOTCH cross-talk signaling. Cancer Res 76, 3156–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski-Daspit AS, Nerger BA, Wolf AE, Sundaresan S, and Nelson CM (2017). Dynamics of tissue-induced alignment of fibrous extracellular matrix. Biophys J 113, 702–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebustini IT, Myers C, Lassiter KS, Surmak A, Szabova L, Holmbeck K, Pedchenko V, Hudson BG, and Hoffman MP (2009). MT2-MMP-dependent release of collagen IV NC1 domains regulates submandibular gland branching morphogenesis. Dev Cell 17, 482–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson GW (2007). Cooperation of signalling pathways in embryonic mammary gland development. Nat Rev Genet 8, 963–972. [DOI] [PubMed] [Google Scholar]
- Robinson GW, Accili D, and Hennighausen L (2000). Rescue of mammary epithelium of early lethal phenotypes by embryonic mammary gland transplantation as exemplified with insulin receptor null mice In Methods in Mammary Gland Biology and Breast Cancer Research, Ip MM, and Asch BB, eds. (Boston, MA: Springer US; ), pp. 307–316. [Google Scholar]
- Rosse C, Lodillinsky C, Fuhrmann L, Nourieh M, Monteiro P, Irondelle M, Lagoutte E, Vacher S, Waharte F, Paul-Gilloteaux P, et al. (2014). Control of MT1-MMP transport by atypical PKC during breast-cancer progression. Proc Natl Acad Sci U S A 111, E1872–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe RG, and Weiss SJ (2009). Navigating ECM barriers at the invasive front: the cancer cell-stroma interface. Annu Rev Cell Dev Biol 25, 567–595. [DOI] [PubMed] [Google Scholar]
- Sabeh F, Shimizu-Hirota R, and Weiss SJ (2009). Protease-dependent versus -independent cancer cell invasion programs: three-dimensional amoeboid movement revisited. J Cell Biol 185, 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon Y, Alon L, Glanz S, Servais C, and Erez N (2013). Isolation of normal and cancer-associated fibroblasts from fresh tissues by Fluorescence Activated Cell Sorting (FACS). J Vis Exp, e4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu-Hirota R, Xiong W, Baxter BT, Kunkel SL, Maillard I, Chen XW, Sabeh F, Liu R, Li XY, and Weiss SJ (2012). MT1-MMP regulates the PI3Kdelta.Mi-2/NuRD-dependent control of macrophage immune function. Genes Dev 26, 395–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simian M, Hirai Y, Navre M, Werb Z, Lochter A, and Bissell MJ (2001). The interplay of matrix metalloproteinases, morphogens and growth factors is necessary for branching of mammary epithelial cells. Development 128, 3117–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Rowe RG, Botvinick EL, Kurup A, Putnam AJ, Seiki M, Weaver VM, Keller ET, Goldstein S, Dai J, et al. (2013). MT1-MMP-dependent control of skeletal stem cell commitment via a beta1-integrin/YAP/TAZ signaling axis. Dev Cell 25, 402–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimboli AJ, Cantemir-Stone CZ, Li F, Wallace JA, Merchant A, Creasap N, Thompson JC, Caserta E, Wang H, Chong JL, et al. (2009). Pten in stromal fibroblasts suppresses mammary epithelial tumours. Nature 461, 1084–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner KU, McAllister K, Ward T, Davis B, Wiseman R, and Hennighausen L (2001). Spatial and temporal expression of the Cre gene under the control of the MMTV-LTR in different lines of transgenic mice. Transgenic Res 10, 545–553. [DOI] [PubMed] [Google Scholar]
- Wang S, Sekiguchi R, Daley WP, and Yamada KM (2017). Patterned cell and matrix dynamics in branching morphogenesis. J Cell Biol 216, 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wculek SK, and Malanchi I (2015). Neutrophils support lung colonization of metastasis-initiating breast cancer cells. Nature 528, 413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welm BE, Dijkgraaf GJ, Bledau AS, Welm AL, and Werb Z (2008). Lentiviral transduction of mammary stem cells for analysis of gene function during development and cancer. Cell Stem Cell 2, 90–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JM, and Daniel CW (1983). Mammary ductal elongation: differentiation of myoepithelium and basal lamina during branching morphogenesis. Dev Biol 97, 274–290. [DOI] [PubMed] [Google Scholar]
- Ye X, Tam WL, Shibue T, Kaygusuz Y, Reinhardt F, Ng Eaton E, and Weinberg RA (2015). Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature 525, 256–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Zhang Z, Black CM, de Crombrugghe B, and Denton CP (2002). Ligand-dependent genetic recombination in fibroblasts : a potentially powerful technique for investigating gene function in fibrosis. Am J Pathol 160, 1609–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.