Abstract
Calcineurin (CaN) is a serine/threonine phosphatase that regulates a variety of physiological and pathophysiological processes in mammalian tissue. The calcineurin (CaN) regulatory domain (RD) is responsible for regulating the enzyme’s phosphatase activity, and is believed to be highly-disordered when inhibiting CaN, but undergoes a disorder-to-order transition upon diffusion-limited binding with the regulatory protein calmodulin (CaM). The prevalence of polar and charged amino acids in the regulatory domain (RD) suggests electrostatic interactions are involved in mediating calmodulin (CaM) binding, yet the lack of atomistic-resolution data for the bound complex has stymied efforts to probe how the RD sequence controls its conformational ensemble and long-range attractions contribute to target protein binding. In the present study, we investigated via computational modeling the extent to which electrostatics and structural disorder co-facilitate or hinder CaM/CaN association kinetics. Specifically, we examined several RD constructs that contain the CaM binding region (CAMBR) to characterize the roles of electrostatics versus conformational diversity in controlling diffusion-limited association rates, via microsecond-scale molecular dynamics (MD) and Brownian dynamic (BD) simulations. Our results indicate that the RD amino acid composition and sequence length influence both the dynamic availability of conformations amenable to CaM binding, as well as long-range electrostatic interactions to steer association. These findings provide intriguing insight into the interplay between conformational diversity and electrostatically-driven protein-protein association involving CaN, which are likely to extend to wide-ranging diffusion-limited processes regulated by intrinsically-disordered proteins.
1. Introduction
Calcineurin (CaN) is a ubiquitously expressed protein that regulates myriad developmental and signaling processes (1; 2). It is chiefly regulated by calmodulin (CaM), one of the most prolific proteins in terms of its role in shaping intracellular signal transduction cascades. Despite the fundamental importance of CaM-regulated CaN phosphatase activity in organism physiology, the molecular mechanisms governing this process are incompletely understood. Given that CaM/CaN is a prototypical example of a protein/protein complex involving a globular protein (CaM) and an intrinsically disordered binding domain (CaN) (3; 4), structural details of the protein/protein complex are restricted to intact CaM bound to a small fragment of the CaN regulatory peptide. In this regard, the CaM/CaN complex is similar to the tens of CaM/protein target complexes (5) that have resisted structure determination methods beyond the binding of short peptides. Remarkably, despite the CaN regulatory domain presenting little stabilized secondary structure, the CaM/CaN complex binds with picomolar affinity (6), afforded in part by rapid, diffusion-limited binding.
CaN is heterodimeric protein consisting of two domains: chain A (57-61 kDa) and chain B (19 kDa) (2; 7), while CaM (17 kDa) is comprised of two alpha-helix rich domains capable of binding Ca2+. At Ca2+ concentrations typical of resting cells (50 to 100 nM) (8), CaN phosphatase activity is negligible, while CaM is believed to be in Ca2+-free state (9). Under these conditions, the CaN catalytic domain is autoinhibited by the protein’s auto-inhibitory domain (AID). At rising Ca2+ concentrations, the CaN AID is removed from the catalytic domain. CaM binding to the AID-containing CaN regulatory domain (RD) (Ser373 to Thr468) is a critical determinant of this process (4; 10). Interestingly, like many intrinsically disordered peptide (IDP)-containing complexes, well-defined secondary structure is observed only upon binding a protein target (11; 12; 13; 14).
In absence of hydrophobic residues (15) that would otherwise promote collapse of protein into a molten apolar core, many IDPs such as the CaN RD are polyampholytic (16). Metrics like the net charge per residue (NCPR) have been proposed to relate charge density in IDPs to ensemble properties including compactness and shape (17; 18). Formally, NCPR = ∣f+ – f−∣ where f+ and f− are fractions of positively and negatively charged residues, respectively, and fraction of charged residues (FCR) is calculated as where Nc is the number of charged residues and Nt is the total number of residues. Generally, IDPs with large NCPR values (> 0.25) tend to adopt more extended conformations due to repulsive intra-molecular electrostatic interactions (17; 19). However, the distribution of charged residues in the CaN RD is heterogeneous, thus such sequence-dependent metrics have limited utility in determining localized properties, such as the availability of binding motifs to target proteins.
We hypothesized therefore that RD sequence charge composition (as measured by NCPR) and ionic strength influence the dynamic availability of conformations amenable to CaM binding, while long-range electrostatic interactions drive diffusion-limited association (see Fig. 1). To investigate this hypothesis, we utilize long-timescale MD simulations to probe the highly dynamic conformational ensembles comprising the RD constructs, toward delineating the extent to which conformational gating kinetics and long-range electrostatic interactions govern IDP/protein association. A chief outcome of this work is that charge-sensitive ’local’ and long range factors, namely IDP conformational dynamics and IDP/target electrostatic interactions, can jointly facilitate diffusion-limited target association.
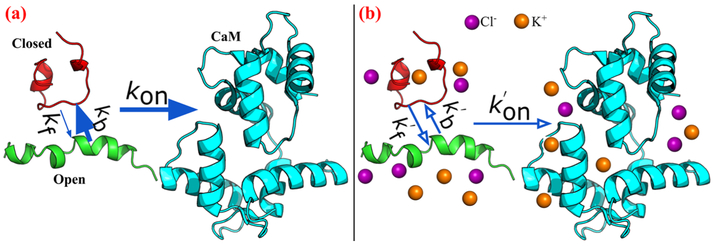
Figure 1.
Schematic of CaN peptide binding to CaM. The green and red peptides represent CaM conformations which are capable and incapable of binding to CaM (colored in cyan). The association rates between intrinsically-disordered CaN peptides and CaM are controlled by open/closed state gating kinetics (depicted by kb and kf) and the CaM/CaN diffusional encounter rate (depicted by kon). Our study demonstrates that low (a) and high (b) ionic strengths influence both contributions to afford diffusion limited encounters
2. Methods
The initial structures of three CaN RD constructs predicted by Rosetta (20) (see Sect. 2.1) were followed by extensive microsecond scaled MD via Amber14 (21) (see Sect. 2.2). The 2D REUS potential of mean force (PMF) calculations of CaN RD constructs were performed by NAMD2.11 (22) (see Sect. 2.3). The MD trajectories were used to characterize the conformational dynamic of CaN RD constructs through Markov state model (MSM) via Aqualab (23) (see Sect. 2.4). The diffusional encounter rates of CaN RD constructs with CaM were simulated by BrownDye package (24) (see Sect. 2.5) and the effective association rates with conformational dynamics taken into account were given in Sect. 2.6.
2.1. Structure preparation
The N-domain (residue numbers 3 to 75) and C-domain (residue numbers 76 to 147) of CaN were extracted from the crystal structure (PDB ID: 4Q5U (25)). For CaN peptides, three different peptides with varying lengths and charge distributions were considered: 1) pCaN: native binding region for CaM. 2) lpCaN: elongated pCaN with five additional residues added to two ends of pCaN, respectively. 3) lpcCaN: charge mutated lpCaN having EESE to KKSK mutations at the C-terminal end. Since diffusion limited binding between CaM and CaN are suggested for both intact CaN and its regulatory domain (arXiv:1611.04080v1), we postulated that these comparatively shorter constructs could capture key factors governing diffusion limited association in a computationally-tractable manner. Rosetta (20) was used to model initial conformations for the CaN peptides.
The number of output conformations was set to ten. No pre-defined secondary structure file was specified. The rest parameters were not explicitly specified (namely using their default values, as that listed in (26)). According to the energy score, for each CaN peptide, the conformation with lowest energy was picked out for further extensive MD sampling. Although just one conformations was selected for each CaN peptide, it was expected that the following microsecond MD ensure adequate sampling.
2.2. Molecular dynamic simulation
We next performed MD simulations to extensively explore the conformational space of the CaN peptides. The Amber ff99SB-ILDN (27) force field was chosen, given its improved recapitulation of experimentally-observed IDP ensembles, in contrast to common forcefields that tend to predict overly collapsed states for IDPs (28). The MD was performed by using Amber14 (21). The implicit solvent model (igb = 2 with salt concentration = 0.15 M) was used. The reason for choosing implicit solvent model was as following: 1) According to a recent study (29), a combination of ff99SB-ILDN with implicit solvent model could achieve reasonably accurate sampling for IDPs; 2) Implicit solvent model would enable more sufficient sampling with affordable time cost. The cutoff value for non-bond interactions was set as 999 A. The starting structure was first subjected to 50000 steps of energy minimization. The minimized structure was slowly heated from 1 to 298.15 K by using the Berendsen Thermostat within 800 ps. During the MD process, the time interval was set to 2 fs and the SHAKE (30) constraints were applied on bonds involving hydrogen atoms. By setting the initial temperature in the heating stage equal to 1 instead of 0 and ig = −1 would generate different initial velocity distributions for the system, thus independent simulations can be achieved. For each peptide, three independent MDs were performed to ensure the reliability of the sampling (total 15 μs production run for each peptide). To study the effect of ionic strength on sampling, we ran analogous simulation with salt concentration = 1.5 M, resulting in a total 30 μs production run for each peptide (15 μs at 0.15 M and 15 μs at 1.5 M ionic strength). Although 1.5 M ionic strength is non-physiological, however, this high ionic strength would significantly increase the electrostatic-screening effect and facilitates more pronounced changes in CaN peptide conformational properties under a different electrostatic environment as compared to the physiological 0.15 M ionic strength. In RMSF calculations, for each peptide, we first performed rms-fitting of trajectory frames to the first frame of trajectory. The rms-fitting would eliminate the translational and rotational effect and ensure the RMSF reflect the fluctuation of atoms. Contact map data of heavy atoms was collected via CPPTRAJ in Amber with distance cutoff as 7 Å and only residue pairs which are at least 5 residues apart (i and i + 5) in sequence are considered.
2.3. Two-dimensional replica-exchange umbrella sampling (REUS) PMF calculation
Two-dimensional PMF calculations were performed to characterize the free energy profile associated with conformational space of each peptide. Two reaction coordinate (RC)s were defined: 1) α which describes the α-helical content of the peptide (ranging from 0.1 to 0.9) and 2) radius of gyration (Rg) of the peptide (ranging from 5 to 32 Å). Each RC range was divided into nine bins resulting in total 81 windows (with interval being 0.1 and 3 Å for α and Rg, respectively). The two force constants of the harmonic potentials imposed on these two RCs are 1.000 × 103 kcal mol−1 U−2 for α and 2.5 kcal mol−1
Å−2 for Rg. For each peptide, the representative structure from the most populated cluster was chosen as the starting structure. NAMD2.11 (22) was chosen to perform the 2D REUS calculations due to it’s colvar module which supports various user-defined collective variables. The CHARMM36 (31; 32) force field was used in the 2D REUS calculations. For each window, the simulation length was set to 20 ns and only the last 15 ns data was used to calculate free energy by WHAM (33).
2.4. Markov state model (MSM) analysis via Aqualab
For each peptide, a 1D kinetic trajectory was created from the 15 μs MD trajectory describing the state change along simulation time. Open states were defined based on examining Browndye-predicted association rates as a function of root mean squared deviations (RMSD); the RMSD value below which the association rate rapidly increased above negligible values was utilized as the open/closed state cutoff criterion. We note the ’closed states’ are not necessarily precluded from binding, but we assume that the timescale for assuming a conformation compatible with the complex via induced fit is slow. These criteria differed among the three peptides, based on our Fig. S10 and Fig. S9. For pCaN, the open state was defined as RMSD < 7 Å while for lpCaN and lpcCaN the open state are defined as RMSD < 5 Å. Using these cutoffs, Markovian networks were created based on the 1D trajectory (rates of open to closed and vice versa) via Aqualab (23), for which conditions such as detailed balance (34) were imposed to define P, the equilibrium probability matrix and T, the transition probability matrix.
2.5. Brownian dynamic (BD) simulations
The binding of CaN peptide and N/C terminal domains of CaM are treated as two independent events and simulated separately by using the BrownDye package (24). For each peptide, ten conformations for each RMSD cluster were randomly selected to perform BD simulations with N/C-domain of CaM. The PDB2PQR (35) was first used to generate the pqr files for CaM N/C domains and the selected conformations of CaN peptides from MD trajectory with radii and point charge parameters adapted from the AMBER99 (36) force field. The generated pqr files were then passed into APBS (37) to evaluate the electrostatic potential of these structures. APBS was used to numerically solve the linearized Poisson-Boltzmann equation assuming an ionic strength of 0.15 M and 1.5 M NaCl:
| [1] |
where ψ is the electrostatic potential, ρiqi is the charge distribution of fixed charge i, and κ is the inverse of Debye length. The Debye length reflects the scale over which mobile charges could screen out electric potential fields.
In present BD simulation, the reaction criterion was chosen to be six pairs of contacts with distance of contact being less than 10 Å. The contact list was created via the make_rxn_pairs routine in Browndye package based on the pCaN-CaM complex crystal structure (PDB ID: 4Q5U (25)) with distance cutoff being 5 Å. 10000 single trajectory simulations for each system were conducted on 10 parallel processors using nam-simulation. Thus for each peptide, the total number of BD trajectories was about 1 million. The reaction rate constants were calculated using compute-rate-constant from the BrownDye package.
To estimate the association rate and its sensitivity to ionic strength, we computed association rates for the terminal domains separately, assuming that both components bind independently,
| [2] |
where the rates in the right hand side correspond to the association rates for the C and N terminal domains, respectively. We anticipate that this expression under-approximates the rate of complex formation, given that tethered binding partners generally exhibit higher efficiencies for forming intact complexes (38; 39).
2.6. Effective association rate combined with gating kinetics
The effective association rate constant after taking conformational dynamics into account was given by Szabo et al (40):
| [3] |
where
| [4] |
where KD is the association rate when the peptide is always in open state and in present study, KD is the BD simulated association rate constant of the open state CaN peptides with CaM (e.g., KD = kon, open). Keq is characteristic constant indicating the extent to which the association is diffusion-controlled (see (40) for more details). In present study, we set Keq = 1 × 1020M−1s−1 and justified by showing in Fig. S8 the sensitivity of keff to Keq, as we can see that keff values for all peptides become fixed after Keq is larger than 1 × 106 M−1 s−1 (see also Sect. S.1). kf and kb are the conversion rate between the open and closed state determined from MSM. R is the contact distance at which the transient complex formed and in present study we set R equal to the average b-radius values from BD simulations. D is the relative translational diffusional constant and was calculated via (24; 41).
| [5] |
where f∞ is the reaction probability which was at the order of 1 × 10−4 given by BD simulations.
3. Results and Discussion
3.1. Molecular simulations confirm the intrinsically-disordered structure of the CaN regulatory domain
Several studies using circular dichroism spectroscopy, hydrogen-deuterium exchange mass spectrometry, Fourier transform infrared spectroscopy and X-ray crystallography indicate that the nearly one-hundred amino acids of the CaN RD domain (Ser373 to Thr468 (3; 4)) form an intrinsically disordered ensemble (42; 43; 44; 4). Of these, approximately twenty amino acids (Ala391-Arg414) comprising the CAMBR adopt an alpha-helix in the presence of CaM (25). Here we examine three RD constructs (pCaN, lpCaN and lpcCaN, see Fig. 2) that present diffusion-limited association with CaM (arXiv:1611.04080v1). pCaN spans residues A391 to R414 and has been co-crystallized with CaM (PDB ID: 4Q5U (25)). lpCaN includes three additional polar residues (Ser3, Ser32 and Ser34) and four additional charged residues (Asp1, Glu30, Glu31 and Glu33) while lpcCaN is a peptide of the same length with substitutions of three glutamic acids with lysines at the C-termini of lpCaN. The set of constructs considered here span a range of charge densities that we later demonstrate tune CaM/CaN association kinetics. pCaN and lpcCaN have similar NCPR values of 0.291 and 0.264, which are considerably larger than the value for lpCaN (0.088); previous works (17; 19), suggest NCPR scores above 0.25 reflect extended IDP conformations given the propensity for repulsive intramolecular interactions, whereas those below this threshold are comparatively compact. We expected therefore that 1) the CaN peptides lack well-resolved secondary structure characteristic of a folded protein and 2) the ensemble of lpCaN should be modestly more compact than that of lpcCaN, given that latter has higher charge density.
Figure 2:
Amino acid sequences of three CaN peptide constructs examined here, including their respective FCR and NCPR scores. pCaN: native CAMBR of CaN (Ala391- Arg414). lpCaN: five predominantly negatively charged residues affixed to the pCaN termini. lpcCaN: lpCaN construct with three positively-charged substitutions at the C-terminus. In present study, the residue numbers of lpCaN and lpcCaN are counted from 1 to 34 while pCaN has the residue numbers from 6 to 29.
To investigate the hypothesis, we performed 5 μs MD simulations in triplicate (total 15 μs) at 0.15 M and 1.5 M ionic strength, respectively. The choice of physiological (0.15 M) and high ionic strength was intended to probe the contribution of intra-peptide electrostatic interactions to ensemble properties, as such interactions would be screened at high ionic strength. While simulations of IDPs of up to 100 residues have been reported elsewhere (19), the breadth of simulations used in this study restricted our construct sizes to 24 to 34 a.a. Our MD simulations indicate that the heavy atom root mean squared fluctuations (RMSF) for each residue in Fig. S1(e-f) are shown to be larger than 5 Å for all three peptides at both ionic strengths, which is consistent with the high mobility loop scores reported in Fig. S1(b). These data suggest that the peptides do not form stable folded structures in solution regardless of ionic strength.
The MD-generated structures present a multitude of conformations, ranging from loosely-formed, hairpin-like configurations to extended structures. lpCaN presents perhaps the most hairpin character, as corroborated by intramolecular contacts reported through contact map analyses in Fig. S2. Among these contacts are prominent interactions between Arg12-Glu30, Arg23-Glu30 and Arg13-Glu32, which we attribute to transient salt-bridge formation. For lpcCaN, the mutation of negative residues (Glu30, Glu31 and Glu33) to the positively charged residues (Lys30, Lys31 and Lys33) appears to disrupt these intramolecular contacts, thereby yielding a more extended conformational ensemble relative to lpCaN. Given the similar NCPR values of pCaN and lpcCaN, we expected pCaN would similarly present fewer intramolecular contacts than lpCaN. Surprisingly, pCaN has similar contact map features as lpCaN, that is, both peptides have comparable intra-contacts. Later we will demonstrate that it is in fact the interconversion kinetics, not the average structural properties, of lpCaN and pCaN that dictate binding kinetics. Additionally, we found that increasing ionic strength to 1.5 M screens the electrostatic interactions between residues comprising the reported salt bridges. As a result, we observe for pCaN and lpcCaN that the structures become modestly more extended on average.
It is important to acknowledge that while implicit solvent simulations permit greater degrees of conformation sampling compared to their all-atom counterparts (45), the utilization of an implicit solvent comes with certain limitations. Among these include the propensity to overestimate alpha-helical character and to alter the interaction strengths of salt-bridging amino acid pairs (46). Similarly, in the event that charged amino acids might chelate counterions in solution, as exemplified by acidic EF-hands in diverse Ca2+ binding proteins (47), implicit solvent may underestimate the strength of such interactions. It would therefore be of interest in future studies to assess the significance of these limitations in IDPs using explicit all-atom simulations of comparable length to implicit solvent trajectories.
To support the formation of the CaN/CaM protein-protein interaction (PPI), the CaN CAMBR must be revealed to the solvent-exposed CaM surface. The exposure of the CAMBR could occur spontaneously, which would promote binding by presenting mutually compatible conformations independent of the complementary species, or via an induced-fit mechanism when the binding partner, CaM, is present. In the previous section, we indicated that the peptides have considerable structural variability, therefore here we determine whether this variability confers greater access to the CAMBR independent of CaM.
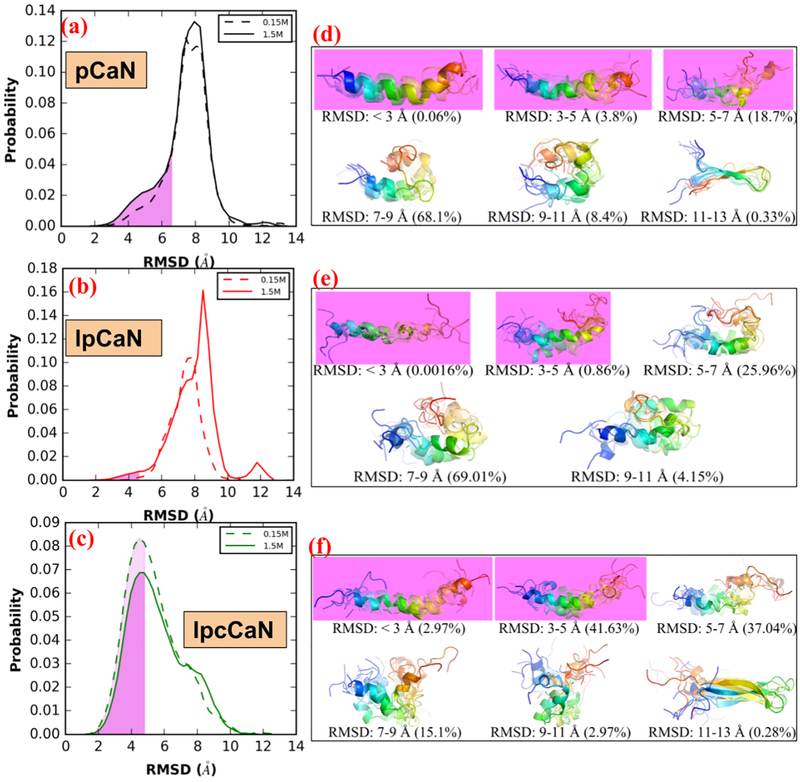
In Fig. 3(a-c), we report the RMSD of the CAMBR binding region for each configuration from the MD simulations, relative to the extended, alpha-helical pCaN conformation that is compatible with the CaM binding surface. From these simulations, we identify conformations that are amenable for CaM binding (”open” state) and those unsuitable for CaM binding (”closed” state), using a cutoff of pCaN: 7 Å, lpCaN and lpcCaN: 5 Å. We utilized a more restrictive criterion for the longer constructs, as the 7 Å cutoff assumed for pCaN yielded structures that were incompatible with CaM. RMSD values below the cutoff more closely resemble the fully-extended reference structure, whereas values above this cutoff are more compact. As shown in Fig. 3(a-c), all three peptides adopt a small percentage of CaM-compatible configurations as measured by RMSD and the percentages appear to be insensitive to ionic strength. These data additionally indicate that lpCaN (NCPR = 0.088) has the smallest percentage of CaM-campatible structures as assessed by RMSD compared with the bound CaN complex, relative to lpcCaN (NCPR = 0.264) and pCaN (NCPR = 0.291).
Figure 3:
Distribution of RMSD (with respect to bound-pCaN crystal structure in PDB 4Q5U) in the MD of each CaN peptide at 0.15 M ionic strength and 1.5M ionic strength, respectively(a-c). The shaded area colored in violet denotes the open state-like conformation. The representative structures (colored in rainbow with N-termini as blue and C-termini as red) for each RMSD range and percentage of conformations within this RMSD range were also shown(d-f).
In Fig. 3(d-f), we present the structures of the most probable conformations based on RMSD clustering analysis. Significantly, each peptide was observed to partially fold into an α-helix, indicating that bound-like ’residual’ structures can spontaneously form in the absence of the binding partner, as has been reported for other IDPs (48; 49; 50; 51). The open state probabilities we determined represented a small, but significant fraction (pCaN: ~20%; lpCaN: ~1%; lpcCaN: ~45%) of the conformations sampled. Importantly, these data indicate extended CaM-compatible conformations form spontaneously and in a charge density (NCPR)-dependent manner (see Fig. 3(a-c)). We speculate that the tendency for a percentage of the conformational ensemble to assume an extended pose relative to a hairpin fold suggests that intra-molecular repulsion may partially destabilize the formation of loose hairpins. This effect would be exacerbated with charge densities of increasing magnitude, such as those reflected in the NCPR values for pCaN and lpcCaN, and relatively diminished for low NCPR peptides like lpCaN.
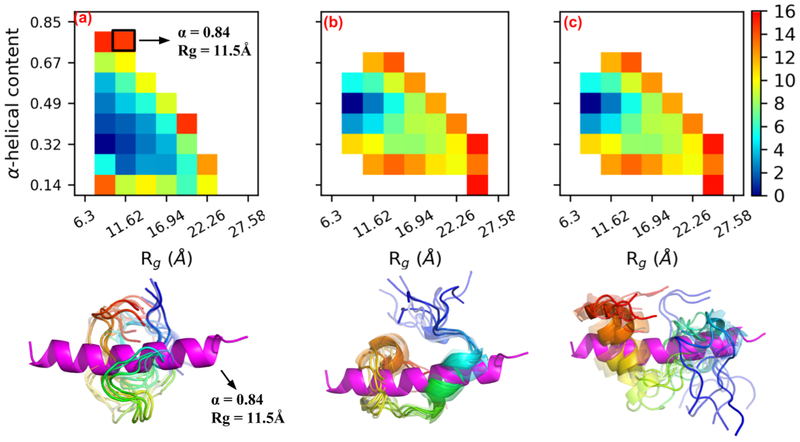
To establish a thermodynamic basis for the trends of greater conformational diversity for the high NCPR cases (pCaN and lpcCaN) relative to the low NCPR case (lpCaN), we report potential of mean force (PMF) calculations for these peptides as a function of α-helical character, a measure of secondary structure formation, and Rg, a measure of compactness (see Fig. 4). Such PMFs have been used to characterize the propensity for IDPs to assume specific ensemble characteristics, including IDP compactness (52; 53). Each construct preferentially adopted smaller α-helical character than the 84% reflected in pCaN when bound to CaM. Lacking CaM, unfolded CaN RD states dominate the conformational distribution, thus CaM is apparently necessary to shift the IDP ensemble toward the bound state via an induced-fit mechanism.
Figure 4:
Two dimensional PMFs for pCaN (a), lpCaN (b) and lpcCaN (c) at 0.15 M ionic strength. The x and y axis depict α-helical and Rg reaction coordinates, respectively. The x axis of (b) and (c) are hidden for clarity. For each peptide, ten randomly selected structures (colored in rainbow with N-termini as blue and C-termini as red) from lowest energy area are compared against bound state pCaN conformation (colored in magenta) from PDB 4Q5U (α = 0.844, Rg = 11.54 Å). The unit of color bar is kbT where kb is Boltzmann constant and T = 298 K is temperature.
Interestingly, we observe that the range of Rg and α-helical values within a few kbT of the energy minima (0 kbT) are larger for the high NCPR cases compared to lpCaN. These data mirror our findings for the histogram of RMSD distributions in Fig. 3, with the low NCPR case presenting a narrower distribution relative to the high NCPR cases. Further, the PMF data support the observation for the lpCaN and lpcCaN peptides that the former structure assumes a more compact, hairpin-like configuration relative to the latter, as we observed in Fig. 3(d-f). This indicates that the high NCPR cases access a larger range of conformations in their IDP ensembles that overlap with the CaM-bound structures, albeit in contrast to the more narrowly peaked distributions presented for the low NCPR (lpCaN) configuration. Our results are consistent with the work done by Mao et al (17) for protamine IDPs demonstrating that globule-to-coil transitions were more favored with increasing of NCPR values.
3.2. CaN regulatory domain ensemble conformational dynamics are rapid and have modest ionic-strength dependence
Our unconstrained MD and PMF calculations both indicate that the CaN RD peptides do not readily assume an open-state compatible with CaM, although there exist some infrequent, CaM-compatible configurations. In this regard, one can view the accessibility of the prefolded CAMBR domain to CaM as a ’gating’ event, which in principle could control the apparent binding rate for this process (54; 55). Given that our previous work (arXiv:1611.04080v1) in which CaN peptides are assumed to have fully CaM-compatible CAMBR conformations demonstrated that all three peptides are capable of binding CaM, albeit at substantially different rates. We hypothesized that the appearance of bound-like structures before binding serves to nucleate loosely-associated CaM-compatible transient encounter states with low alpha-helical character, which permits ’induced folding ‘ in the presence of CaM to access alpha-helix rich bound-states.
As a first step towards probing this hypothesis, we first estimated the transition kinetics between open and closed states identified in Sect. 3.1.2 using Markov state model (MSM) analysis. Intuitively, we would expect that higher rates of accessing CaM-compatible open states would maximize the CaM/CaN association rate. We note here that we defined the open state as consisting of conformations below the 7 Å (for pCaN) and 5 Å (for lpCaN and lpcCaN) RMSD cutoff used in Fig. S10 (the red dash lines depict the RMSD of open and closed state for each peptide), while all conformations with dissimilar RMSDs were lumped into a single closed state. We verify that the states are essentially Markovian as the correlation times become negligible beyond roughly tens of nanoseconds (see Fig. S4) which is faster than the diffusion encounter time. Overall, based on this partition of MD data, the transition rates between closed and open states are rapid (the slowest rate is at 1 × 107 s−1 order, see Table S1) and lead to the short-lived open states shown in Fig. S5 (average life times of open state for all three peptides under both ionic strengths are around 0.2 ns).
Ionic strength was shown to have negligible impact on the RMSD of our predicted peptide structures relative to CaM-bound conformation. However, given the pronounced role of electrostatics in facilitating protein/protein association rates and protein folding (56; 57), we sought to determine whether transition kinetics between conformations were influenced by ionic strength. Hence, we compared MSM rate predictions for MD generated structures at low (0.15 M) and high (1.5 M) ionic strength. Here we found that for pCaN and lpcCaN, increasing ionic strength does not affect the gating rates between open and closed states. However, for lpCaN, increasing ionic strength increased kf from 1 × 107 s−1 to 1 × 108 s−1 order. As a result the lifetime of its closed states decreased from 12.83 to 4.42 ns, as shown in Fig. S5. Hence, the open and closing kinetics of peptides with high NCPR are appear to be less sensitive to ionic strength, compared to structures with low NCPR. These results concur with findings from Liu et al (58), for which they demonstrated that the fast-phase structural fluctuations as measured by Fluorescence correlation spectroscopy (FCR) for the IDP Sic1 disappeared with decreasing ionic strength. At first glance, it is surprising that the ionic strength did not appreciably alter the average properties of the conformation ensemble. However, we anticipate that the change in ionic strength, while significant, was insufficient to strongly disfavor the desolvation of the charged groups in order to drive hydrophobic collapse. In contrast, a less hydrophilic solvent, e.g. one with a low dielectric constant, would strongly reduce the desolvation energy and thereby is expected to lead to a hydrophobic collapse.
3.3. Long-range interactions promote rapid CaM/CaN association
Our results thus far indicate that the CaN RD peptides adopt CaM-compatible conformations in the absence of CaM frequently, albeit transiently. Here we determine the compatibility of these transient states with the CaM/CaN binding interface using Brownian dynamic (BD) simulations. Specifically, we sought to evaluate two hypotheses: 1) that frequent presentation of CaN open states promote near diffusion-limited association rates and 2) that long-range electrostatic interactions are exploited in PPIs involving IDPs. Motivating our first hypothesis are recent indications that target-compatible residual structures of the isolated p53 up-regulated modulator of apoptosis (PUMA) IDP form spontaneously as a function of ionic strength and electrolyte composition (59). For the latter hypothesis, we adopt the paradigm of electrostatically-driven association of globular proteins (60; 61; 62; 41), which depends critically on the notion of a transient encounter complex (63; 64). The encounter complex serves as the rate determining step in PPI formation, whereby a protein loosely binds to its protein target, before adopting the fully-formed bound configuration. However, unlike PPIs involving globular partners that typically feature regions of complementarily-charged hydrophilic patches (65; 66), such patches may only be transiently presented in IDPs. Though these results offer specific insights into the ubiquitous Ca2+ signaling partners CaM and CaN, these trends may vary in importance depending on the IDP sequence, particularly those with vastly different amino acid charge densities and distributions.
We tested these hypotheses by assuming each peptide must achieve a minimal number of ’native contacts’ with the CaM N-terminal and C-terminal domains to constitute a transient encounter complex. The native contacts are obtained by analyzing the crystal structure of CaM-pCaN complex, in which key interactions between CaM and pCaN were extracted to guide the BD simulations. From this standpoint, the lenient conditions for association are tantamount to the notion of a transient encounter complex (67; 68), which is formed upon association of two binding partners prior to forming the fully-bound complex. Because we test the first hypothesis using conformations generated from the MD simulations without CaM, this test bears similarity to the conformational selection paradigm (69), though we emphasize CaM is likely required to completely form the bound complex from the transient encounter state. However, the abundance IDP-target protein complexes that exhibit diffusion-limited association suggest that the folding rate from the encounter state to the bound complex is rapid, as elaborated in Sect. S.1. In Fig. S10 and Fig. S9 we show that the MD-generated open states presented in each of the peptide configurations are compatible with both the N- and C-terminal CaM domains to varying degrees, as the open state of each peptide gives BD-simulated kons in the diffusion-limited regime (> 1 × 107 M−1 s−1). Notably, all peptides considered here are capable of forming the transient encounter complex with CaM at rates that decrease with increasing ionic strength (see also Sect. S. 4.1). While not explored in this study, binding affinities via methods including Molecular Mechanics-Generalized Born Surface area (MM-GBSA) techniques (70; 71) could help rank configurations most compatible with the bound complex.
3.4. Rapid IDP conformational ensemble dynamics promote rapid association
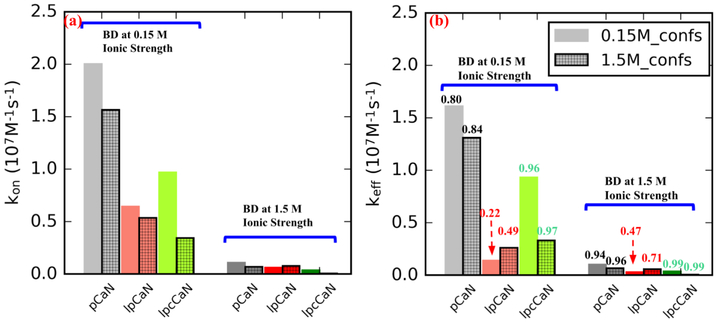
Lastly, we investigated the role of conformational gating rates on the effective association rates based on the stochastic gating model postulated by Szabo et al (40). In this gating model, kon is the expected ’ideal’ association rate between CaN peptide and CaM. By ’ideal’ it means that the CaN peptide is aways in CaM-compatible open state and the concentration of CaN peptide and CaM are sufficiently large, thus kon here reflect the diffusion timescale of CaN peptide to CaM. By considering this, kon can be compared with kb/kf which reflect the conversion time scale between open/closed states of CaN peptide. In the gating model, there are two limits that bound the effective rates: 1) given gating rates that are significantly faster than diffusional encounter rate (kf + kb ⪢ kon), the effective association rate keff is equivalent to the rate associated with the open state, that is, keff = kon. 2) given gating rates significantly smaller than the diffusional encounter rate (kf + kb ⪡ kon), keff is given by the weighted average of the association rates for all accessed states, that is keff = ⟨kon⟩.
Rates associated with intermediate regimes are obtained by evaluating Eq. 3 using the MSM-estimated gating rates. Based on the data in Table S2 we show in Fig. 5 for pCaN and lpcCaN that keff and kon are comparable (e.g. keff / kon → 1), indicating a marginal effect of conformational gating on the association rate. This arises because the conformational transition rates are of the order 1 × 109 s−1, roughly 100 times faster than diffusional encounter rate, based on our BD simulated kons of 1 × 107 M−1 s−1 order (see Table S2). In contrast, the slower transition kinetics for lpCaN yield a keff that is about 50% of the maximal kon, albeit it is still in a diffusion-limited regime. Moreover, the rates are strongly attenuated at 1.5 M relative to low ionic strength conditions of 0.15 M, which suggest the strong role of long-range electrostatic interactions in promoting association. These data indicate that diffusion-limited association kinetics are realized in the CaN IDP constructs, though the effective rate depends both on ensemble gating kinetics and long-range electrostatic interactions.
Figure 5:
Association rate constants between CaN peptide and CaM before (a) and after (b) taking CaN peptide’s conformational dynamic (from MSM modeling) into con-sideration using Eq. 3. The bars without grids and with grids depict results in which CaN peptide conformations were sampled at 0.15 M and 1.5 M ionic strength, respectively. In (a), kon was calculated via Eq. 2 where kCterm and KNterm are the average values of ten randomly selected conformations from each peptide’s open state. In (b) the numbers above each bar represent the ratios of keff to kon.
An important factor to consider in this model is that the inability of the closed state to bind as rapid as the open state does not render the former ’unsuitable’ for binding. Rather, we anticipate its binding rate will be much slower relative to the open states, as the latter of which requires lesser structural reorganization to assume the correct binding pose. We support this speculation based on Go model predictions summarized in Sect. S4, for which we demonstrated a greater degree of frustration of transition from closed-like conformations to open-like. Namely, for closed state pCaN conformations, the BD-simulated complex presents an average fraction of native contact as Qn = 0.24 and Qc = 0.22, for the percent native contacts of the N and C terminal CaM domains, respectively while the open state pCaN conformations yield, Qn = 0.56 and Qc = 0.59 (see Fig. S12 and Fig. S11). Since lower native contact values will likely encounter greater frustration in folding landscapes compared to near native states, and reduced frustration results in increased rates of folding kinetics (72), we anticipate that induced-fit binding occurs more slowly than the binding of open-state conformations.
We acknowledge that experimental validation would have nicely complemented the model predictions. However, given the availability of experimental kinetics data from stopped-flow measurements of pCaN/CaN binding to CaM (arXiv: 1611.04080v1), a computational model consistent with these data offers experimentally-verifiable insights into molecular mechanisms governing their binding. Of these, the couplings between peptide charge distribution and ionic strength on the dynamics of an IDP conformational ensemble is one that could be examined through spectroscopic techniques including time-resolved Forster resonance energy transfer (25) or spin-relaxation times from nuclear magnetic resondance spectroscopy (73). As with many techniques used to resolve structural characteristics of IDP conformational ensembles, the measurements present ensemble-averaged information, therefore atomistic-scale, computational models will continue to be a strong complement to such probes for characterization of IDP behavior.
4. Conclusions
Our studies of CaN conformational dynamics and CaM/CaN association reveal several interesting features. While the role of charge distribution in IDPs has been shown to be a strong predictor of ensemble structure including compactness (17; 19), our simulations reveal that measures such as NCPR may offer predictive estimates for the ionic strength sensitivity of conformation transition kinetics. Namely, higher NCPR structures are more likely to adopt conformations that complement their binding target, and are less sensitive to changes in ionic strength that may influence gating kinetics. However, it is important to note that this trend may not generalize to necessarily all IDPs, given the wide range of protein/protein association rates (< 1 × 103 to > 1 × 109 M−1 s−1 (41)) reported in the literature, which hints at the possibility of different assembly mechanisms. Second, we demonstrate that long-range electrostatic interactions can play a paramount role in determining the kinetics of forming PPIs involving intrinsically-disordered partners, while protein-solvent and protein-protein electrostatic interactions govern the kinetics of presenting target-compatible binding motifs. Together, these factors suggest that IDPs can achieve diffusion-limited association by controlling conformational gating, so long as a conformation amenable to association is rapidly sampled. Overall our findings build upon the growing understanding of the roles of conformation selection and induced fit in dictating PPIs, both identifying how conformational selection can accelerate association, despite potential requirements for induced fitting in order to adopt the final binding pose.
Our study focused on CaN ‘s binding interaction with CaM, of which the latter regulates a staggering array of eukaryotic signaling cascades through forming PPIs with target protein (5). What sets CaM apart from other such hubs is the surprisingly diverse variety of targets it regulates, despite presenting a single isoform across all mammalian species (74). In part, its ability to regulate this diversity is attributed to the conformational heterogeneity of the CaM binding interface (75) it is capable of forming. These findings provide intriguing insight into the interplay between conformational diversity and electrostatically-driven protein-protein association involving CaN, which are likely to extend to wide-ranging processes regulated by intrinsically-disordered proteins. As such, exploiting IDP composition to tune PPI kinetics could offer new tools to probe and modulate important biochemical signal transduction pathways.
Supplementary Material
Table S1 Average life times and gating rates between CaN peptides’ open and closed conformations sampled at 0.15 M and 1.5 M ionic strength (sate definition is based on RMSD, see red dash lines in Fig. S10 and Fig. S9).
Table S2 BD-simulated encounter rates (kon) under 0.15 M and 1.5 M ionic strength for the open state CaN peptides sampled the same two ionic strengths. The effective association rates (keff) calculated via Eq. 3 are also shown.
Figure S1: (a) Sequences of the three CaN peptides studied in the present work. The positively charged residues and negatively charged residues are colored in blue and red, respectively, along with FCR and NCPR scores. (b) Disorder probabilities predicted by DisEMBL [83]. The shown curves are scores based on "hot-loops" which is reported to be a good criterion to define disorder [83, 84]. The blue dash line denotes random expectation values. (c) Mean hydropathy score and mean net charges of the three peptides and their locations in the Uversky diagram [85] (d) Locations of the three CaN peptides and CaN RD in the Das-Pappu diagram [86]. Figures (c) and (d) are generated by localCIDER [87]. (e-f) RMSF of each residue during the 15 μs MD at 0.15M and 1.5M ionic strength, respectively.
Figure S2: Contact map analysis of 15 μs MD trajectory of three CaN peptide under 0.15 M and 1.5 M ionic strength, respectively. Contact data was collected via CPPTRAJ in Amber with distance cutoff as 7 Å and only residue pairs which are at least 5 residues apart (i and i+5) in sequence are considered. The unit of numbers on color bar is number of average contacts for each pair over the simulation time.
Figure S3: Contact map analysis of each single trajectory of pCaN triplicate simulations at 0.15 M ionic strength. The three trajectories show consistent contact features, implying that our μs scale simulation of CaN peptide are converged.
Figure S4: Life time distribution of open and closed states for each CaN peptide calculated from the 15 μs MD trajectory (0.15 M ionic strength) by Aqualab using the MSM model.
Figure S5: Average life time of open and closed states of CaN peptides determined by MSM at 0.15 M (faded color) and 1.5 M (dark color), respectively. The specific life time and corresponding gating rates are listed in Table S1.
Figure S6: Average Rg of CaN peptides at 0.15 M (faded color) and 1.5 M (dark color), respectively.
Figure S7: BD calculated association kinetics between rigid CaN peptides and CaM. The electrostatic interaction was turned off by setting CaN peptides charges to be zero. Specifically, after turning of electrostatic interaction, lpCaN retains 50% above association rates while lpCaN and lpcCaN reduce to much smaller kons, implying that electrostatic interaction has larger impacts on lpCaN and lpcCaN than pCaN.
Figure S8: Sensitivity of keff to Keq for three CaN peptides sampled at 0.15 M and 1.5 M ionic strength, respectively..
Figure S9: BD-simulated separate association rate constants with CaM C domain vs. RMSD under 0.15 M and 1.5 M ionic strength with CaN peptides conformations sampled at the same two ionic strengths, respectively. The red dash line designates the border of open and closed states based on RMSD.
Figure S10: BD-simulated separate association rate constants with CaM N domain vs. RMSD under 0.15 M and 1.5 M ionic strength with CaN peptides conformations sampled at the same two ionic strengths, respectively. The red dash line designates the border of open and closed states based on RMSD.
Figure S11: Fraction of native contact (Q) and free energy profile in CG simulation with G−o model. A: Fractions of native contact between N/C-domain of CaM and pCaN (denoted as Qn and Qc, respectively) along CG simulation time (only the first 300 ns of 1 μs is shown for clarity). B: 2D free energy profile projected along Qn and Qc in CG simulation. The shaded areas colored in light red and blue depict the ranges of Qn and Qc values in BD simulations, respectively. The last frame of each BD trajectory of the 10 pCaN open conformations (RMSD < 3 Å) with CaM N/C domain were used to calculate average and standard deviation of Qn and Qc.
Figure S12: Fractions of native contact between N/C-domain of CaM and closed state pCaN in the BD-simulated pCaN-CaM complex.
Acknowledgments
This article is dedicated to the memory of late Professor Jeffry A. Madura. Research reported in this publication was supported by the Maximizing Investigators’ Research Award (MIRA) (R35) from the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health (NIH) under grant number R35GM124977. This work used the Extreme Science and Engineering Discovery Environment (XSEDE) (76), which is supported by National Science Foundation grant number ACI-1548562
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Jain J, McCafffrey PG, Miner Z, Kerppola TK, Lambert JN, Verdine GL, Curran T, Rao A, The t-cell transcription factor nfatp is a substrate for calcineurin and interacts with fos and jun, Nature 365 (1993) 353–355. [DOI] [PubMed] [Google Scholar]
- [2].Rusnak F, Mertz P, Calcineurin: Form and function, Physiological Reviews 80 (4) (2000)1483–1521. [DOI] [PubMed] [Google Scholar]
- [3].Yang S-A, Klee CB, Low affinity ca2+-binding sites of calcineurin b mediate conformational changes in calcineurin a, Biochemistry 39 (51) (2000) 16147–16154, pMID: 11123943. arXiv: 10.1021/bi001321q, doi: 10.1021/bi001321q. URL 10.1021/bi001321q [DOI] [PubMed] [Google Scholar]
- [4].Rumi-Masante J, Rusinga FI, Lester TE, Dunlap TB, Williams TD, Dunker AK, Weis DD, Creamer TP, Structural basis for activation of calcineurin by calmodulin, Journal of Molecular Biology 415 (2) (2012) 307–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kursula P, The many structural faces of calmodulin: a multitasking molecular jackknife, Amino Acids 46 (10) (2014) 2295–2304. [DOI] [PubMed] [Google Scholar]
- [6].Quintana AR, Wang D, Forbes JE, Waxham MN, Kinetics of calmodulin binding to calcineurin, Biochemical and Biophysical Research Communications 334 (2) (2005) 674–680. [DOI] [PubMed] [Google Scholar]
- [7].Klee CB, Ren H, Wang X, Regulation of the calmodulin-stimulated protein phosphatase, calcineurin, Journal of Biological Chemistry 273 (22) (1998) 13367–13370. arXiv: http://www.jbc.org/content/273/22/13367.full.pdf+html, doi: 10.1074/jbc.273.22.13367. URL http://www.jbc.org/content/273/22/13367.short [DOI] [PubMed] [Google Scholar]
- [8].Lewis RS, Calcium signaling mechanisms in t lymphocytes, Annual Review of Immunology 19 (1) (2001) 497–521, pMID: 11244045. doi: 10.1146/annurev.immunol.19.1.497. [DOI] [PubMed] [Google Scholar]
- [9].Persechini A, Cronk B, The relationship between the free concentrations of ca2+ and ca2+-calmodulin in intact cells, Journal of Biological Chemistry 274 (11) (1999) 6827–6830. [DOI] [PubMed] [Google Scholar]
- [10].Dunlap TB, Cook EC, Rumi-Masante J, Arvin HG, Lester TE, Creamer TP, The distal helix in the regulatory domain of calcineurin is important for domain stability and enzyme function, Biochemistry 52 (48) (2013) 8643–8651. [DOI] [PubMed] [Google Scholar]
- [11].Wright PE, Dyson H, Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm, Journal of Molecular Biology 293 (2) (1999) 321–331. [DOI] [PubMed] [Google Scholar]
- [12].Dyson H, Wright PE, Coupling of folding and binding for unstructured proteins, Current Opinion in Structural Biology 12 (1) (2002) 54–60. [DOI] [PubMed] [Google Scholar]
- [13].Dyson HJ, Wright PE, Intrinsically unstructured proteins and their functions (2005). doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- [14].Sugase K, Dyson HJ, Wright PE, Mechanism of coupled folding and binding of an intrinsically disordered protein, Nature 447 (7147) (2007) 1021–1025. [DOI] [PubMed] [Google Scholar]
- [15].Marsh JA, Forman-Kay JD, Sequence determinants of compaction in intrinsically disordered proteins, Biophysical Journal 98 (10) (2010) 2383–2390. doi: 10.1016/j.bpj.2010.02.006. URL http://www.sciencedirect.com/science/article/pii/S0006349510002341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Dobrynin AV, Colby RH, Rubinstein M, Polyampholytes, Journal of Polymer Science Part B: Polymer Physics 42 (19) (2004) 3513–3538. [Google Scholar]
- [17].Mao AH, Crick SL, Vitalis A, Chicoine CL, Pappu RV, Net charge per residue modulates conformational ensembles of intrinsically disordered proteins, Proceedings of the National Academy of Sciences 107 (18) (2010) 8183–8188. arXiv:http://www.pnas.org/content/107/18/8183.full.pdf, doi: 10.1073/pnas.0911107107. URL http://www.pnas.org/content/107/18/8183.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Müller-Späth S, Soranno A, Hirschfeld V, Hofmann H, Rüegger S, Reymond L, Nettels D, Schuler B, Charge interactions can dominate the dimensions of intrinsically disordered proteins, Proceedings of the National Academy of Sciences 107 (33) (2010) 14609–14614. arXiv:http://www.pnas.org/content/107/33/14609.full.pdf, doi: 10.1073/pnas.1001743107. URL http://www.pnas.org/content/107/33/14609.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sherry KP, Das RK, Pappu RV, Barrick D, Control of transcriptional activity by design of charge patterning in the intrinsically disordered ram region of the notch receptor, Proceedings of the National Academy of Sciences 114 (44) (2017) E9243–E9252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Simons KT, Bonneau R, Ruczinski I, Baker D, Ab initio protein structure prediction of casp iii targets using rosetta, Proteins: Structure, Function, and Bioinformatics 37 (S3) (1999)171–176. [DOI] [PubMed] [Google Scholar]
- [21].Case DA, Babin V, Berryman JT, Betz RM, Cai Q, Cerutti DS, Cheatham TE, Darden TA, Duke RE, Gohlke H, Goetz AW, Gusarov S, Homeyer N, Janowski P, Kaus J, Kolossváry I, Kovalenko A, Lee TS, LeGrand S, Luchko T, Luo R, Madej B, Merz KM, Paesani F, Roe DR, Roitberg A, Sagui C, Salomon-Ferrer R, Seabra G, Simmerling CL, Smith W, Swails J, Walker, Wang J, Wolf RM, Wu X, Kollman PA, Amber 14, University of California, San Francisco: (2014). [Google Scholar]
- [22].Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K, Scalable molecular dynamics with namd, Journal of Computational Chemistry 26 (16) (2005) 1781–1802. doi: 10.1002/jcc.20289. URL 10.1002/jcc.20289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Berezovska G, Prada-Gracia D, Mostarda S, Rao F, Accounting for the kinetics in order parameter analysis: Lessons from theoretical models and a disordered peptide, The Journal of Chemical Physics 137 (19) (2012) 194101 arXiv: 10.1063/1.4764868, doi: 10.1063/1.4764868. URL 10.1063/1.4764868 [DOI] [PubMed] [Google Scholar]
- [24].Huber GA, McCammon JA, Browndye: A software package for brownian dynamics, Computer Physics Communications 181 (11) (2010) 1896–1905. doi: 10.1016/j.cpc.2010.07.022. URL http://www.sciencedirect.com/science/article/pii/S0010465510002559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Dunlap TB, Guo H-F, Cook EC, Holbrook E, Rumi-Masante J, Lester TE, Colbert CL, Vander Kooi CW, Creamer TP, Stoichiometry of the calcineurin regulatory domain–calmodulin complex, Biochemistry 53 (36) (2014) 5779–5790, pMID: 25144868. arXiv: 10.1021/bi5004734, doi: 10.1021/bi5004734. URL 10.1021/bi5004734 [DOI] [PubMed] [Google Scholar]
- [26].Xiong P, Wang M, Zhou X, Zhang T, Zhang J, Chen Q, Liu H, Protein design with a comprehensive statistical energy function and boosted by experimental selection for foldability, Nature Communications 5 (2014) 5330. [DOI] [PubMed] [Google Scholar]
- [27].Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE, Improved side-chain torsion potentials for the amber ff99sb protein force field, Proteins: Structure, Function, and Bioinformatics 78 (8) (2010) 1950–1958. doi: 10.1002/prot.22711. URL 10.1002/prot.22711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Henriques J, Cragnell C, Skepö M, Molecular dynamics simulations of intrinsically disordered proteins: Force field evaluation and comparison with experiment, Journal of Chemical Theory and Computation 11 (7) (2015) 3420–3431, pMID: 26575776. arXiv: 10.1021/ct501178z, doi: 10.1021/ct501178z. URL 10.1021/ct501178z [DOI] [PubMed] [Google Scholar]
- [29].Maffucci I, Contini A, An updated test of amber force fields and implicit solvent models in predicting the secondary structure of helical, β-hairpin, and intrinsically disordered peptides, Journal of Chemical Theory and Computation 12 (2) (2016) 714–727. [DOI] [PubMed] [Google Scholar]
- [30].Ryckaert J-P, Ciccotti G, Berendsen HJ, Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes, Journal of Computational Physics 23 (3) (1977) 327–341. doi: 10.1016/0021-9991(77)90098-5. URL http://www.sciencedirect.com/science/article/pii/0021999177900985 [DOI] [Google Scholar]
- [31].MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiórkiewicz-Kuczera J, Yin D, Karplus M, All-atom empirical potential for molecular modeling and dynamics studies of proteins, The Journal of Physical Chemistry B 102 (18) (1998) 3586–3616. [DOI] [PubMed] [Google Scholar]
- [32].MacKerell AD, Feig M, Brooks CL, Improved treatment of the protein backbone in empirical force fields, Journal of the American Chemical Society 126 (3) (2004) 698–699. [DOI] [PubMed] [Google Scholar]
- [33].Grossfield A, Wham: the weighted histogram analysis method. URL http://membrane.urmc.rochester.edu/content/wham [Google Scholar]
- [34].Bowman GR, Beauchamp KA, Boxer G, Pande VS, Progress and challenges in the automated construction of Markov state models for full protein systems, The Journal of chemical physics 131 (12) (2009) 124101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA, Pdb2pqr: expanding and upgrading automated preparation of biomolecular structures for molecular simulations, Nucleic Acids Research 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wang J, Cieplak P, Kollman PA, How well does a restrained electrostatic potential (resp) model perform in calculating conformational energies of organic and biological molecules?, Journal of Computational Chemistry 21 (12) (2000) 1049–1074. [Google Scholar]
- [37].Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA, Electrostatics of nanosystems: Application to microtubules and the ribosome, Proceedings of the National Academy of Sciences 98 (18) (2001) 10037–10041. arXiv:http://www.pnas.org/content/98/18/10037.full.pdf, doi: 10.1073/pnas.181342398. URL http://www.pnas.org/content/98/18/10037.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Van Valen D, Haataja M, Phillips R, Biochemistry on a Leash: The Roles of Tether Length and Geometry in Signal Integration Proteins, Biophysical Journal 96 (4) (2009) 1275–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Shoemaker BA, Portman JJ, Wolynes PG, Speeding molecular recognition by using the folding funnel: the fly-casting mechanism, in: Proceedings of the …, Vol. 97, National Acad Sciences, 2000, p. 8868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Szabo A, Shoup D, Northrup SH, McCammon JA, Stochastically gated diffusion influenced reactions, The Journal of Chemical Physics 77 (9) (1982) 4484–4493. arXiv: 10.1063/1.444397, doi: 10.1063/1.444397. URL 10.1063/1.444397 [DOI] [Google Scholar]
- [41].Schreiber G, Haran G, Zhou H-X, Fundamental aspects of protein-protein association kinetics, Chemical Reviews 109 (3) (2009) 839–860, pMID: 19196002. doi: 10.1021/cr800373w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kissinger CR, Parge HE, Knighton DR, Lewis CT, Pelletier LA, Tempczyk A, Kalish VJ, Tucker KD, Showalter RE, Moomaw EW, Gastinel LN, Habuka N, Chen X, Maldonado F, Barker JE, Bacquet R, Villafranca JE, Crystal structures of human calcineurin and the human FKBP12-FK506-calcineurin complex, Nature 378 (6557) (1995) 641–644. [DOI] [PubMed] [Google Scholar]
- [43].Manalan AS, Klee CB, Activation of calcineurin by limited proteolysis, Proceedings of the National Academy of Sciences 80 (14) (1983) 4291–4295. arXiv:http://www.pnas.org/content/80/14/4291.full.pdf URL http://www.pnas.org/content/80/14/4291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Shen X, Li H, Ou Y, Tao W, Dong A, Kong J, Ji C, Yu S, The secondary structure of calcineurin regulatory region and conformational change induced by calcium/calmodulin binding, Journal of Biological Chemistry 283 (17) (2008) 11407–11413. arXiv:http://www.jbc.org/content/283/17/11407.full.pdf+html, doi: 10.1074/jbc.M708513200. URL http://www.jbc.org/content/283/17/11407.abstract [DOI] [PubMed] [Google Scholar]
- [45].Onufriev A, Chapter 7 - implicit solvent models in molecular dynamics simulations: A brief overview, Vol. 4 of Annual Reports in Computational Chemistry, Elsevier, 2008, pp. 125–137. doi: 10.1016/S1574-1400(08)00007-8. URL http://www.sciencedirect.com/science/article/pii/S1574140008000078 [DOI] [Google Scholar]
- [46].Ruhong Z, Free energy landscape of protein folding in water: Explicit vs. implicit solvent, Proteins: Structure, Function, and Bioinformatics 53 (2) 148–161. doi: 10.1002/prot.10483. [DOI] [PubMed] [Google Scholar]
- [47].Grabarek Z, Structural Basis for Diversity of the EF-hand Calcium-binding Proteins, Journal Of Molecular Biology 359 (3) (2006) 509–525. [DOI] [PubMed] [Google Scholar]
- [48].Fuxreiter M, Simon I, Friedrich P, Tompa P, Preformed structural elements feature in partner recognition by intrinsically unstructured proteins, Journal of Molecular Biology 338 (5) (2004) 1015–1026. doi: 10.1016/j.jmb.2004.03.017. URL http://www.sciencedirect.com/science/article/pii/S0022283604003079 [DOI] [PubMed] [Google Scholar]
- [49].Sivakolundu SG, Bashford D, Kriwacki RW, Disordered p27kip1 exhibits intrinsic structure resembling the cdk2/cyclin a-bound conformation, Journal of Molecular Biology 353 (5) (2005) 1118–1128. doi: 10.1016/j.jmb.2005.08.074. URL http://www.sciencedirect.com/science/article/pii/S0022283605010545 [DOI] [PubMed] [Google Scholar]
- [50].Yoon M-K, Venkatachalam V, Huang A, Choi B-S, Stultz CM, Chou JJ, Residual structure within the disordered c-terminal segment of p21waf1/cip1/sdi1 and its implications for molecular recognition, Protein Science 18 (2) (2009) 337–347. doi: 10.1002/pro.34. URL 10.1002/pro.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kjaergaard M, Teilum K, Poulsen FM, Conformational selection in the molten globule state of the nuclear coactivator binding domain of cbp, Proceedings of the National Academy of Sciences 107 (28) (2010) 12535–12540. doi: 10.1073/pnas.1001693107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Song D, Wang W, Ye W, Ji D, Luo R, Chen H-F, ff14idps force field improving the conformation sampling of intrinsically disordered proteins, Chemical Biology and Drug Design 89 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Guo X, Han J, Luo R, Chen H-F, Conformation dynamics of the intrinsically disordered protein c-myb with the ff99idps force field, RSC Adv. 7 (2017) 29713–29721. doi: 10.1039/C7RA04133K. URL 10.1039/C7RA04133K [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Zhou H-X, Wlodek ST, McCammon JA, Conformation gating as a mechanism for enzyme specificity, Proceedings of the National Academy of Sciences 95 (16) (1998) 9280–9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Zhou H-X, From induced fit to conformational selection: A continuum of binding mechanism controlled by the timescale of conformational transitions, Biophysical Journal 98 (6) (2010) L15–L17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Roca M, Messer B, Warshel A, Electrostatic contributions to protein stability and folding energy, FEBS Letters 581 (10) (2007) 2065–2071. doi: 10.1016/j.febslet.2007.04.025. URL http://www.sciencedirect.com/science/article/pii/S0014579307004103 [DOI] [PubMed] [Google Scholar]
- [57].Zhou H-X, Pang X, Electrostatic interactions in protein structure, folding, binding, and condensation, Chemical Reviews 0 (0) (2018) null. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Liu B, Chia D, Csizmok V, Farber P, Forman-Kay JD, Gradinaru CC, The effect of intrachain electrostatic repulsion on conformational disorder and dynamics of the sic1 protein, The Journal of Physical Chemistry B 118 (15) (2014) 4088–4097. [DOI] [PubMed] [Google Scholar]
- [59].Wicky BIM, Shammas SL, Clarke J, Affinity of idps to their targets is modulated by ion-specific changes in kinetics and residual structure, Proceedings of the National Academy of Sciences 114 (37) (2017) 9882–9887. doi: 10.1073/pnas.1705105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Schreiber G, Fersht AR, Rapid, electrostatically assisted association of proteins, Nature Structural Biology 3 (427). [DOI] [PubMed] [Google Scholar]
- [61].Sheinerman FB, Norel R, Honig B, Electrostatic aspects of protein-protein interactions, Current Opinion in Structural Biology 10 (2) (2000) 153–159. doi: 10.1016/S0959-440X(00)00065-8. URL http://www.sciencedirect.com/science/article/pii/S0959440X00000658 [DOI] [PubMed] [Google Scholar]
- [62].Vijayakumar M, Wong K-Y, Schreiber G, Fersht AR, Szabo A, Zhou H-X, Electrostatic enhancement of diffusion-controlled protein-protein association: comparison of theory and experiment on barnase and barstar11edited by b. honig, Journal of Molecular Biology 278 (5) (1998) 1015–1024. doi: 10.1006/jmbi.1998.1747. URL http://www.sciencedirect.com/science/article/pii/S0022283698917478 [DOI] [PubMed] [Google Scholar]
- [63].Gabdoulline R, Wade R, Simulation of the diffusional association of barnase and barstar, Biophysical Journal 72 (5) (1997) 1917–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Alsallaq R, Zhou H-X, Electrostatic rate enhancement and transient complex of protein–protein association, Proteins: Structure, Function, and Bioinformatics 71 (1) (2008) 320–335. doi: 10.1002/prot.21679. URL 10.1002/prot.21679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Lee L-P, Tidor B, Optimization of binding electrostatics: Charge complementarity in the barnase-barstar protein complex, Protein Science 10 (2) (2001) 362–377. doi: 10.1110/ps.40001. URL 10.1110/ps.40001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Green DF, Tidor B, Design of improved protein inhibitors of hiv-1 cell entry: Optimization of electrostatic interactions at the binding interface, Proteins: Structure, Function, and Bioinformatics 60 (4) (2005) 644–657. doi: 10.1002/prot.20540. URL 10.1002/prot.20540 [DOI] [PubMed] [Google Scholar]
- [67].Gabdoulline RR, Wade RC, On the protein–protein diffusional encounter complex, Journal of Molecular Recognition 12 (4) (1999) 226–234. doi: URL [DOI] [PubMed] [Google Scholar]
- [68].Tang C, Iwahara J, Clore MG, Visualization of transient encounter complexes in protein-protein association, Nature 444 (2006) 383–386. [DOI] [PubMed] [Google Scholar]
- [69].Boehr DD, Nussinov R, Wright PE, The role of dynamic conformational ensembles in biomolecular recognition, Nature Chemical Biology 5 (11) (2009) 789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Onufriev A, Case DA, Bashford D, Effective Born Radii in the Generalized Born Approximation: The Importance of Being Perfect, J Comput Chem 23 (2002) 1297–1304. [DOI] [PubMed] [Google Scholar]
- [71].Onufriev A, Bashford D, Case DA, Exploring protein native states and large-scale conformational changes with a modified generalized born model, Proteins-Structure Function And Bioinformatics 55 (2) (2004) 383–394. [DOI] [PubMed] [Google Scholar]
- [72].Tzul FO, Schweiker KL, Makhatadze GI, Modulation of folding energy landscape by charge-charge interactions: Linking experiments with computational modeling, Proceedings of the National Academy of Sciences 112 (3) (2015) 201410424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Markwick PRL, Malliavin T, Nilges M, Structural Biology by NMR: Structure, Dynamics, and Interactions, PLoS Computational Biology 4 (9) (2008) e1000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Chin D, Means AR, Calmodulin: a prototypical calcium sensor, Trends in Cell Biology 10 (8) (2000) 322–328. [DOI] [PubMed] [Google Scholar]
- [75].Shukla D, Peck A, Pande VS, Conformational heterogeneity of the calmodulin binding interface, Nature Communications 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Towns J, Cockerill T, Dahan M, Foster I, Gaither K, Grimshaw A, Hazlewood V, Lathrop S, Lifka D, Peterson GD, Roskies R, Scott JR, Wilkins-Diehr N, Xsede: Accelerating scientific discovery, Computing in Science & Engineering 16 (5) (2014) 62–74. [Google Scholar]
- [77].Price ES, DeVore MS, Johnson CK, Detecting intramolecular dynamics and multiple forster resonance energy transfer states by fluorescence correlation spectroscopy, The Journal of Physical Chemistry B 114 (17) (2010) 5895–5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Negi S, Aykut AO, Atilgan AR, Atilgan C, Calmodulin readily switches conformation upon protonating high pka acidic residues, The Journal of Physical Chemistry B 116 (24) (2012) 7145–7153. [DOI] [PubMed] [Google Scholar]
- [79].Brooks CL, Helix-coil kinetics: Folding time scales for helical peptides from a sequential kinetic model, The Journal of Physical Chemistry 100 (7) (1996) 2546–2549. [Google Scholar]
- [80].De Sancho D, Best RB, What is the time scale for alpha-helix nucleation?, Journal of the American Chemical Society 133 (17) (2011) 6809–6816. [DOI] [PubMed] [Google Scholar]
- [81].Clarke DT, Doig AJ, Stapley BJ, Jones GR, The a-helix folds on the millisecond time scale, Proceedings of the National Academy of Sciences 96 (13) (1999) 7232–7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Tzul FO, Vasilchuk D, Makhatadze GI, Evidence for the principle of minimal frustration in the evolution of protein folding landscapes, Proceedings of the National Academy of Sciences 114 (9) (2017) E1627–E1632. arXiv:http://www.pnas.org/content/114/9/E1627.full.pdf, doi: 10.1073/pnas.1613892114. URL http://www.pnas.org/content/114/9/E1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB, Protein disorder prediction: Implications for structural proteomics, Structure 11 (11) (2003) 1453–1459. doi: 10.1016/j.str.2003.10.002. URL http://www.sciencedirect.com/science/article/pii/S0969212603002351 [DOI] [PubMed] [Google Scholar]
- [84].Peti W, Page R, Strategies to maximize heterologous protein expression in escherichia coli with minimal cost, Protein Expression and Purification 51 (1) (2007) 1–10. doi: 10.1016/j.pep.2006.06.024. URL http://www.sciencedirect.com/science/article/pii/S1046592806001951 [DOI] [PubMed] [Google Scholar]
- [85].Uversky VN, Gillespie JR, Fink AL, Why are “natively unfolded” proteins unstructured under physiologic conditions?, Proteins: Structure, Function, and Bioinformatics 41 (3) (2000) 415–427. doi: URL [DOI] [PubMed] [Google Scholar]
- [86].Das RK, Pappu RV, Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues, Proceedings of the National Academy of Sciences 110 (33) (2013) 13392–13397. arXiv:http://www.pnas.org/content/110/33/13392.full.pdf, doi: 10.1073/pnas.1304749110. URL http://www.pnas.org/content/110/33/13392.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Holehouse AS, Das RK, Ahad JN, Richardson MO, Pappu RV, Cider: Resources to analyze sequence-ensemble relationships of intrinsically disordered proteins, Biophysical Journal 112 (1) (2017) 16–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Karanicolas J, Brooks CL, Improved gō-like models demonstrate the robustness of protein folding mechanisms towards non-native interactions, Journal of Molecular Biology 334 (2) (2003) 309–325. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Average life times and gating rates between CaN peptides’ open and closed conformations sampled at 0.15 M and 1.5 M ionic strength (sate definition is based on RMSD, see red dash lines in Fig. S10 and Fig. S9).
Table S2 BD-simulated encounter rates (kon) under 0.15 M and 1.5 M ionic strength for the open state CaN peptides sampled the same two ionic strengths. The effective association rates (keff) calculated via Eq. 3 are also shown.
Figure S1: (a) Sequences of the three CaN peptides studied in the present work. The positively charged residues and negatively charged residues are colored in blue and red, respectively, along with FCR and NCPR scores. (b) Disorder probabilities predicted by DisEMBL [83]. The shown curves are scores based on "hot-loops" which is reported to be a good criterion to define disorder [83, 84]. The blue dash line denotes random expectation values. (c) Mean hydropathy score and mean net charges of the three peptides and their locations in the Uversky diagram [85] (d) Locations of the three CaN peptides and CaN RD in the Das-Pappu diagram [86]. Figures (c) and (d) are generated by localCIDER [87]. (e-f) RMSF of each residue during the 15 μs MD at 0.15M and 1.5M ionic strength, respectively.
Figure S2: Contact map analysis of 15 μs MD trajectory of three CaN peptide under 0.15 M and 1.5 M ionic strength, respectively. Contact data was collected via CPPTRAJ in Amber with distance cutoff as 7 Å and only residue pairs which are at least 5 residues apart (i and i+5) in sequence are considered. The unit of numbers on color bar is number of average contacts for each pair over the simulation time.
Figure S3: Contact map analysis of each single trajectory of pCaN triplicate simulations at 0.15 M ionic strength. The three trajectories show consistent contact features, implying that our μs scale simulation of CaN peptide are converged.
Figure S4: Life time distribution of open and closed states for each CaN peptide calculated from the 15 μs MD trajectory (0.15 M ionic strength) by Aqualab using the MSM model.
Figure S5: Average life time of open and closed states of CaN peptides determined by MSM at 0.15 M (faded color) and 1.5 M (dark color), respectively. The specific life time and corresponding gating rates are listed in Table S1.
Figure S6: Average Rg of CaN peptides at 0.15 M (faded color) and 1.5 M (dark color), respectively.
Figure S7: BD calculated association kinetics between rigid CaN peptides and CaM. The electrostatic interaction was turned off by setting CaN peptides charges to be zero. Specifically, after turning of electrostatic interaction, lpCaN retains 50% above association rates while lpCaN and lpcCaN reduce to much smaller kons, implying that electrostatic interaction has larger impacts on lpCaN and lpcCaN than pCaN.
Figure S8: Sensitivity of keff to Keq for three CaN peptides sampled at 0.15 M and 1.5 M ionic strength, respectively..
Figure S9: BD-simulated separate association rate constants with CaM C domain vs. RMSD under 0.15 M and 1.5 M ionic strength with CaN peptides conformations sampled at the same two ionic strengths, respectively. The red dash line designates the border of open and closed states based on RMSD.
Figure S10: BD-simulated separate association rate constants with CaM N domain vs. RMSD under 0.15 M and 1.5 M ionic strength with CaN peptides conformations sampled at the same two ionic strengths, respectively. The red dash line designates the border of open and closed states based on RMSD.
Figure S11: Fraction of native contact (Q) and free energy profile in CG simulation with G−o model. A: Fractions of native contact between N/C-domain of CaM and pCaN (denoted as Qn and Qc, respectively) along CG simulation time (only the first 300 ns of 1 μs is shown for clarity). B: 2D free energy profile projected along Qn and Qc in CG simulation. The shaded areas colored in light red and blue depict the ranges of Qn and Qc values in BD simulations, respectively. The last frame of each BD trajectory of the 10 pCaN open conformations (RMSD < 3 Å) with CaM N/C domain were used to calculate average and standard deviation of Qn and Qc.
Figure S12: Fractions of native contact between N/C-domain of CaM and closed state pCaN in the BD-simulated pCaN-CaM complex.