Abstract
Although time-lapse analysis of early embryo cleavage parameters (morphokinetics) predicts blastocyst development, it has not been definitively linked to establishing pregnancy and live birth. For example, a direct comparison of the developmental potential of embryos with optimal kinetic parameters compared to suboptimal kinetics has not been performed with human embryos. To ascertain whether such a linkage exists, we developed a mouse model of morphokinetic analysis of early embryo cleavage using time-lapse microscopy to predict blastocyst formation and tested whether cleavage parameters predict pregnancy outcome by transferring morphokinetically optimal and suboptimal embryos into a single host. Using classification and regression trees, we established that the timing of the second and third mitotic divisions (division from two to three and three to four cells, respectively) predicts blastocyst development in the mouse. Using this prediction model, we found that the incidence of sustained implantation at mid-gestation was significantly higher for the optimal compared to suboptimal embryos. In addition, the incidence of resorption among implanted embryos was significantly higher in the suboptimal compared to the optimal group. Transcript profiling of optimal and suboptimal embryos revealed minimal differences between the two groups, suggesting that time-lapse imaging of early embryo cleavage events provides additional information regarding developmental competence apart from gene expression.
Keywords: blastocyst, embryo development, morphokinetics, pregnancy, time lapse
Introduction
Morphologic evaluation of embryos derived by in vitro fertilization (IVF) has been used for embryo selection since the establishment of IVF as a viable treatment for infertility. However, this method has limitations for predicting implantation and live birth [1-4]. Moreover, although the recent use of embryo biopsy with preimplantation genetic screening (PGS) has improved the incidence of pregnancy in single-embryo transfer cycles [5], PGS is an invasive technique, and its long-term effects on the embryo are unknown [1, 6]. An alternative to PGS, noninvasive embryo selection using time-lapse imaging and morphokinetic measurements of early embryonic cleavage events, has been proposed for selecting embryos with the greatest implantation potential [7, 8]. Through acquisition of images of the developing embryo at preset intervals, time-lapse imaging provides precise information about the timing of early cleavage events [9, 10]. Specifically, the duration and synchronization of the first three mitotic divisions of the embryo predict blastocyst development in human embryos [11]. The field of time-lapse imaging has rapidly expanded, with several studies demonstrating the ability of such methodology to predict embryos that will progress to the blastocyst stage [12-16].
There is increasing clinical evidence that early cleavage timing parameters predictive of blastocyst development also correlate to embryo implantation and establishment of pregnancy [17-19]. Retrospective studies suggest that morphokinetic parameters can be used to select embryos with higher implantation potential [14, 20-22]. However, several systematic reviews independently conclude that there is currently insufficient evidence to support the clinical use of time-lapse imaging data for predicting live birth [23-26]. The greatest limitations of many of these studies are differences in culture conditions between the imaged and nonimaged embryos and a nonrandomized experimental design [18, 19]. Randomized studies transferring embryos with optimal versus suboptimal cleavage parameters, preferably into the same individual, are necessary to conclusively prove that morphokinetics can improve embryo selection. Such studies, however, cannot ethically be performed in humans.
Mouse has proven to be an excellent model for studying early embryo development and implantation. In this study, we utilize a mouse model that identifies morphokinetic cleavage parameters derived from time-lapse imaging that not only predict development of early cleavage stage embryos to the blastocyst stage but also predict their potential to implant and establish a pregnancy.
Materials and Methods
Embryo Collection
All experiments and procedures were approved by the University of Pennsylvania Institutional Animal Care and Use Committee. Adult mice were obtained and housed in a temperature-controlled environment with a 12:12 light:dark cycle and fed food and water ad libitum. Six-week-old female CF1 mice (Harlan Laboratories, Indianapolis, IN) were superovulated with i.p. injections of 5 IU of equine chorionic gonadotropin (EMD Millipore, Billerica, MA) followed by 5 IU of human chorionic gonadotropin (hCG; Sigma-Aldrich, St. Louis, MO) 48 h later. Females were mated with B6D2F1/J males (Jackson Laboratory, Bar Harbor, ME). Zygotes with two pronuclei (PN) were collected approximately 22 h after hCG. Embryos were collected in HEPES-buffered Whitten Medium [27] and treated with hyaluronidase (1 mg/ml; Sigma-Aldrich) to disperse the cumulus cells. The embryos were then washed through a series of drops of culture medium prior to use.
Embryo Culture
Following cumulus cell removal, embryos were transferred to time-lapse culture dishes (Eeva Dish) for imaging using the Eeva System (Progyny, Menlo Park, CA). These dishes contain 20 individual wells that are connected within the same 40-μl drop of K+ simplex optimized medium with amino acids (KSOM+AA) (Specialty Media; EMD Millipore) under mineral oil. Twenty embryos were placed in each dish, and the dish was placed on the Eeva time-lapse camera in a humidified atmosphere at 37°C with 5% CO2 and either atmospheric (20%) oxygen (suboptimal culture condition) or low (5%) oxygen (optimal culture condition) for 4 days. Multiple mating experiments were performed and multiple mice used per experiment to limit intradam bias.
Time-Lapse Imaging and Model Building
Images were captured by the Eeva system every 5 min using a camera system that utilized bright-field microscopy. After 4 days of culture, the digital images were converted to video for analysis. The resulting videos were assessed manually for early cleavage kinetics at four time points: duration of first cytokinesis (C1; 1c–2c) and the time intervals between cytokinesis 1 and 2 (P2; 2c–3c), 2 and 3 (P3; 3c–4c), and 3 and 4 (P4; 4c–5c). Final morphologic stage was assessed on Day 4.5. Embryos were designated as cleavage, morula, early blastocyst (beginning of blastocele cavity forming), blastocyst (complete formation of blastocele cavity but without expansion of total embryo size), or expanded blastocyst (expansion of total embryo size). A model predicting expanded blastocyst formation from the morphokinetic time points was constructed using classification and regression trees (CART). The model begins by identifying the single best cutoff value (time point) that can be used to split the data into the two subgroups that differ most significantly in respect to the outcome (expanded blastocyst vs. other morphologic stage). The model then continues to identify cutoff points for each node (or cluster) until a set stopping point (a maximum number of steps or minimum subgroup size). To avoid overgrowth of the resulting tree, the model is pruned using cross validation. The CART approach is an alternative to the traditional methods for prediction [28, 29]. The 5% and 20% O2 cohorts were used as two independent populations for model building. The model was built using the 20% O2 cohort; the ability of the model to predict blastocyst formation in the 5% O2 cohort was then assessed.
Statistical Analysis
The predictive model was built using CART, as described above. Differences in cleavage time points between 5% and 20% O2 and between embryos that progressed to the expanded blastocyst stage and those that arrested were assessed using analysis of variance (ANOVA) and the Student t-test. Statistical analysis was performed using R Software (Vienna, Austria) and GraphPad Prism version 6 (San Diego, CA).
Embryo Transfer
Six-week-old CF1 females were superovulated as above and mated to transgenic males heterozygous for green fluorescent protein (GFP) (C57BL/6-Tg [CAG-EGFP] 1Osb/J; Jackson Laboratory). 2PN embryos were collected as above and cultured in 5% O2 in the Eeva system. The time-lapse videos were analyzed for the morphokinetic time points, and each embryo was assigned a status of “optimal” or “suboptimal” cleavage timing based on the CART algorithm described above. On Day 4.5, embryos were assessed for morphologic stage and, using fluorescent microscopy, for GFP expression status.
Ten blastocyst-stage embryos were transferred into a single horn of a pseudopregnant CF1 female on Day 4.5 (Postcoital Day 3.5 for the recipients) using the Non-Surgical Embryo Transfer Device (Paratechs, Lexington, KY) per the manufacturer's protocol. For each experiment, embryos were transferred into two recipients: one female received suboptimal GFP positive embryos and optimal GFP negative embryos, whereas the other female received optimal GFP positive embryos and suboptimal GFP negative embryos (Fig. 1). Embryos that had reached the expanded blastocyst stage were considered eligible for transfer and were transferred based solely on the morphokinetic classification established by CART analysis and GFP status without regard to morphology beyond the designation of expanded blastocyst.
Fig. 1.
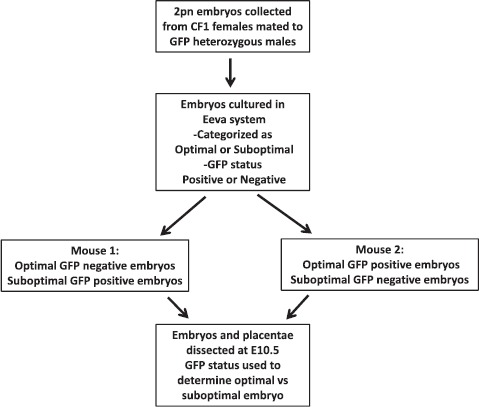
Design of transfer experiments.
Cell Counts
Optimal and suboptimal blastocysts were fixed on Day 4.5 in 3.7% paraformaldehyde for 1 h at room temperature (RT) and blocked overnight at 4°C in PBS + 0.3% bovine serum albumin (BSA). The blastocysts were permeabilized in 0.1% Triton X-100 (Sigma-Aldrich) for 15 min at RT, then incubated in PBS + 0.3% BSA containing a rabbit monoclonal anti-CDX2 antibody (1:100 dilution; Abcam, Cambridge, MA) for 1 h at RT. Following incubation in the presence of the primary antibody, the embryos were washed three times in PBS + 0.3% BSA for 15 min each and incubated in PBS + 0.3% BSA containing anti-rabbit alexaFluor 488 (1:500 dilution; Abcam), for 1 h at RT. The blastocysts were then washed three times in PBS + 0.3% BSA and mounted in Vectashield (Vector Laboratories, Burlingame, CA) plus TO-PRO-3-Iodide (Life Technologies, Grand Island, NY), 1:250. Z-stack images were obtained using confocal microscopy, and cells in both the inner cell mass (ICM) and the trophectoderm (TE) were manually counted. ICM and TE cells were distinguished based on differential staining for CDX2 (TE only) compared to TO-PRO-3-Iodide (both ICM and TE). Differences in cell numbers were assessed using the Student t-test.
Fetal Evaluation
Pregnant females were euthanized 7 days following embryo transfer (Day E10.5). Implantation sites, including fetus and placenta, were dissected from the uterine horn, and each was analyzed for GFP status using fluorescent microscopy to determine whether the implantation site arose from an optimal or a suboptimal embryo. The number of implantation sites (containing both embryo and placenta) and the number of implantation site resorptions (containing only a small amount of placental tissue and no embryo) were recorded. Differences in the incidence of implantation and ongoing pregnancy and the incidence of early loss (number of resorptions out of total implanted) were assessed using the Fisher exact test.
Microarray Analysis of Gene Expression
2PN embryos were collected following mating of superovulated CF1 female mice to transgenic GFP males as described above and cultured in 5% O2 in the Eeva time-lapse system. Cleavage parameters were analyzed, and each embryo was assigned an optimal or suboptimal status based on the CART algorithm. On Day 4.5, embryos that had reached the expanded blastocyst stage were snap frozen individually prior to storage at −80°C. Optimal (n = 5) and suboptimal (n = 5) blastocysts were selected for single-embryo microarray analysis of gene expression. RNA was isolated from each blastocyst using the PicoPure RNA isolation kit (Arcturus, Mountain View, CA) according to manufacturer's protocol. RNA was frozen at −80°C and submitted to the Molecular Profiling Facility of the University of Pennsylvania for GeneChip labeling and hybridization. RNA was converted to cDNA, amplified, and hybridized to Affymetrix Mouse Gene 1.0 ST arrays (Affymetrix, Santa Clara, CA) per the manufacturer's protocols as described in the Ovation Pico WTA system version 2 user guide (NuGEN, San Carlos, CA) and the Affymetrix GeneChip Expression Analysis Technical Manual. Arrays were scanned and processed using Affymetrix Command Console software yielding probe intensity files for each sample. Probe intensity files were normalized using robust multiarray averages (Partek Genomics Suite version 6.6; Partek, St. Louis, MO) yielding log2-normalized intensities for each transcript ID in each sample. Principal component analysis was used to visualize global variation among the samples. Statistical analysis was performed using Statistical Analysis of Microarrays (Stanford University, Palo Alto, CA) [30] to determine significant differences in gene expression in the two groups based on fold change and the false-discovery rate for multiple testing.
Results
Model Building
Prior to testing whether morphokinetics can predict the implantation potential of a mouse blastocyst, we first established the parameters associated with blastocyst development. To build a model of morphokinetic analysis, a total of 313 2PN embryos were collected and cultured using the time-lapse system; 180 embryos were cultured in 5% O2 and 133 in 20% O2. Because the incidence of development in mouse to the blastocyst stage is high relative to that in human, we employed suboptimal culture conditions, that is, 20% oxygen, for model building to achieve a comparable developmental profile in the mouse model. Progression to the expanded blastocyst was 89% (161/180) in 5% O2 and 70% (93/133) in 20% O2 (P < 0.0001). The outcomes of all cultured embryos are summarized in Table 1. As expected, embryos cultured in 20% O2 were more likely to arrest at the cleavage and early blastocyst stages compared to embryos cultured in 5% O2.
Table 1.
Developmental outcomes of 2PN embryos cultured in 5% and 20% O2.
| Embryo morphology on Day 4.5 | 5% O2 (n = 180) | 20% O2 (n = 133) | P-value |
|---|---|---|---|
| Cleavage | 7 (3.9%) | 14 (12.8%) | 0.02 |
| Morula | 6 (3.3%) | 3 (1.6%) | 0.74 |
| Early blastocyst | 6 (3.3%) | 23 (17.3%) | <0.0001 |
| Expanded blastocyst | 161 (89.4%) | 93 (69.9%) | <0.0001 |
Embryos (300) that progressed to at least the three-cell blastomere stage were included for cleavage parameter assessment and model building. All four cleavage parameters that were analyzed differed significantly between embryos that progressed to expanded blastocysts compared to those that did not; however, only the interval between the third and fourth cytokinesis (P4; 4c–5c) differed in the 5% compared to the 20% O2 cohorts (Table 2). A visual model of parameters C1, P2, and P3 is shown in Figure 2. The final CART model utilized two parameters: P2 (2c–3c) and P3 (3c–4c) (Fig. 3). Once the model was built with the 20% O2 cohort, it was applied to the 5% O2 cohort. In 20% O2, the model showed a sensitivity of 92% and a specificity of 60% for predicting expanded blastocyst formation. In 5% O2, the model showed even greater sensitivity and specificity, 98% and 63%, respectively, for predicting expanded blastocyst formation, corresponding to a positive predictive value of 96% and a negative predictive value of 71%.
Table 2.
Cleavage parameters of embryos by morphologic stage on Day 4 of culture in 5% and 20% O2.
| Cleavage parameter | 5% O2 | 20% O2 | P-value (two-way ANOVA) | |||
|---|---|---|---|---|---|---|
| Expanded blast (n = 161) | Other morphologic stage (n = 16) | Expanded blast (n = 93) | Other morphologic stage (n = 30) | 5% vs. 20% O2 | Expanded blast vs. other stage | |
| C1 (mean hours) | 0.09 | 0.18 | 0.17 | 0.21 | 0.097 | 0.040 |
| P2 (mean hours) | 20.65 | 22.81 | 21.1 | 23.7 | 0.058 | <0.0001 |
| P3 (mean hours) | 0.79 | 1.99 | 1.08 | 1.60 | 0.809 | <0.0001 |
| P4 (mean hours) | 10.12 | 11.38 | 11.26 | 12.32 | 0.033 | 0.017 |
Fig. 2.
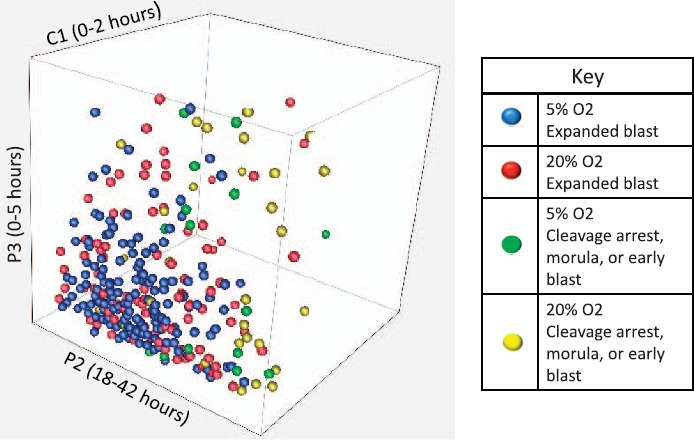
Three-dimensional plot of C1, P2, and P3 in 20% and 5% O2 by embryo morphology on Day 4 of culture. Each dot represents one embryo. Axes (not shown) are in hours.
Fig. 3.
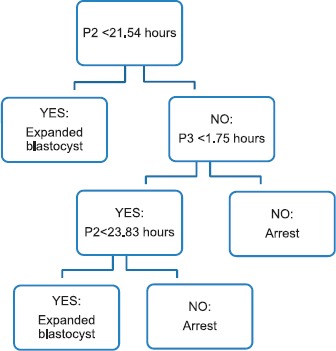
CART model to predict expanded blastocyst formation in 2PN embryos
Implantation and Developmental Potential of Optimal and Suboptimal Embryos
To determine whether our model's prediction of optimal and suboptimal embryos translated into a difference in a blastocyst's implantation potential, we performed a series of transfer experiments. A total of 100 blastocyst-stage embryos (75 optimal and 25 suboptimal) were transferred nonsurgically into 10 mice, with 10 embryos transferred per mouse. Embryos were classified as optimal or suboptimal based on cleavage parameters, and GFP status was used to label the embryos for identification posttransfer. Of the 10 mice, eight became pregnant, and two mice (both in the same transfer experiment) did not achieve pregnancy and were excluded from the analysis. Therefore, a total of 80 embryos (58 optimal and 22 suboptimal) in eight mice were included for analysis.
The outcomes of the transfer experiments are summarized in Figure 4. The overall incidence of implantation per embryo transferred was not statistically different between groups and was 86% (50/58) in the optimal embryo group and 77% (17/22) in the suboptimal embryo group (P = 0.3). The incidence of sustained implantation, defined as the number of complete implantation sites at Day E10.5 (embryo plus placenta) divided by the number of embryos transferred, was significantly higher for the optimal compared to suboptimal embryos: 60% (35/58) in the optimal group compared to 32% (7/22) in the suboptimal group (P = 0.03). The incidence of early pregnancy loss, defined as the number of resorption sites divided by the total number of implanted embryos, was significantly higher in the suboptimal compared to optimal group: 59% (10/17) in the suboptimal group compared to 30% (15/50) in the optimal group (P = 0.04).
Fig. 4.
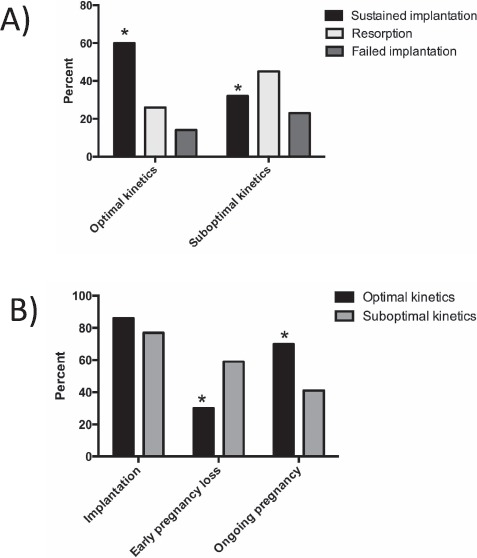
Outcomes of transfer experiments. A) Summary of outcomes for transfers of optimal and suboptimal blastocysts. B) There was no difference in implantation rate (percent of embryos implanted/total embryos transferred) in the two groups. Early pregnancy loss rate was higher and ongoing pregnancy rate lower (both expressed as percent of embryos/total number of implanted embryos) in mice with suboptimal embryos. *P < 0.05.
Transcript Profiling in Optimal and Suboptimal Blastocysts
To further our understanding of genes involved in embryo implantation and competence, we utilized our morphokinetic parameters to compare gene expression differences in expanded blastocysts with optimal and suboptimal timing using microarrays. Using a false discovery rate of less than 10%, only 13 genes showed differential expression between the optimal and suboptimal embryos. When the false discovery rate was expanded to 25%, differential gene expression was still seen in only 74 genes (Supplemental Table S1; Supplemental Data are available online at www.biolreprod.org) and included 35 genes up-regulated and 39 genes down-regulated in the suboptimal group. A heat map of these 74 genes is shown in Supplemental Figure S1A. Principal component analysis showed tight clustering of the embryos with optimal timing, whereas suboptimal embryos exhibited no discernible clustering pattern (Supplemental Fig. S1B). To minimize the possibility that the differences in gene expression between the two groups of embryos reflected differences in developmental stage, trophoblast and ICM cell number were counted. There was no difference in cell number in optimal compared to suboptimal blastocysts for either ICM (21.8 ± 4.4 vs. 20.4 ± 4.3, P = 0.63) or TE (56 ± 4.4 vs. 54.6 ± 7.6, P = 0.73) cells (Supplemental Fig. S2).
Discussion
In this study, we generated a mouse model of time-lapse microscopy with morphokinetic parameters that predicts blastocyst formation and pregnancy outcome. We utilized a transgenic mouse line to compare implantation potential of embryos with optimal and suboptimal cleavage parameters in a single host to gain insight into the potential of time-lapse imaging in improving embryo selection. To our knowledge, this is the first study that validates a morphokinetic model in mouse based on multiple parameters in different culture conditions and further validates the model with pregnancy outcome data.
The experimental design, utilizing GFP-labeled embryos, allows transfer of both optimal and suboptimal embryos into a single host via nonsurgical embryo transfer, controlling for host factors, even to the level of the uterine horn. In addition, in both groups, embryos remain undisturbed for the entire length of in vitro culture, eliminating fluctuations in temperature and pH that could be responsible for the altered developmental potential in the human studies of time-lapse imaging [18]. Utilizing our model, we show an increase in the ongoing incidence of pregnancy and a decrease in the incidence of early pregnancy loss among embryos with optimal early cleavage parameters while minimizing confounding factors.
Our findings are consistent with both human and animal data that show that early cleavage parameters predict blastocyst development [11-16, 31-34]. Prior mouse studies have utilized the first and second cleavage divisions [34] or timing of the first cleavage division [31, 33] and associated these timings with blastocyst development. Our study is the first, however, to incorporate multiple cleavage parameters into a statistical model to predict blastocyst development in mouse. Notably, our algorithm identified the same two parameters, P2 and P3, as correlating best with blastocyst development as are used in human embryos [12]. This finding reinforces decades of data demonstrating that mouse remains an appropriate model system for studying embryo development and reproduction and suggests that these finding are likely relevant to clinical care. Our model has a higher sensitivity and lower specificity than the equivalent human model used clinically (98% and 63% for mouse vs. 59% and 84% for human); these differences are likely due to the higher blastocyst formation rate seen in mouse compared to human embryo development.
Because the incidence of development in mouse to the blastocyst stage is higher than that observed in human, we employed suboptimal culture conditions, that is, 20% oxygen, to achieve a comparable incidence in the mouse model. Note that 20% oxygen is still used clinically in some human IVF. The differences in the incidence of blastocyst formation and the developmental potential of mouse compared to human embryos could, however, potentially limit conclusions drawn from the mouse model. In addition, because of differences in the efficiency of development to the blastocyst stage and a limitation in the number of embryos that could be imaged per experiment, fewer suboptimal embryos were obtained for each experiment than optimal embryos. Thus, we were not able to obtain sufficient suboptimal blastocysts to transfer with an equal number of optimal blastocysts. Consequently, the 1:3 ratio was determined by the number of suboptimal blastocysts and the minimum number required for embryo transfer. From our previous work, we determined that the optimal number of embryos to transfer per mouse when performing nonsurgical embryo transfer is 10; therefore, we transferred a total of 10 embryos per mouse combining optimal and suboptimal embryos. Finally, it should be noted that embryo coculture improves developmental potential [35-37]; therefore, enriching the transfer with more optimal embryos would be expected to positively affect the suboptimal embryos.
Mouse and human embryos also differ in that the incidence of aneuploidy is much higher in human embryos and can affect implantation, cause miscarriage, and affect cleavage parameters. Nevertheless, the ability of morphokinetics to predict implantation in mouse suggests that time-lapse imaging technology provides additional information on embryo developmental competence beyond that of traditional genetic screening. Additionally, given the minimal differences in transcript profiling observed between optimal and suboptimal embryos, morphokinetics may provide information regarding developmental competence beyond that of gene expression.
An additional difference between our model and human models is that embryos generated for our study were fertilized in vivo, which may change embryo kinetics. It should be noted, however, that following fertilization, all embryos are cultured in vitro. As our first measured parameter occurs after the beginning of the first cytokinesis, the kinetics are likely to mimic what is found following IVF.
Similar to our findings, other studies demonstrate an increased incidence of blastocyst development and ongoing pregnancy among mouse embryos with specific embryo cleavage timings, although these studies use imprecise timings, such as estimated time of fertilization [33, 34]. Our experimental design identified morphokinetic cleavage parameters that not only predict development of early cleavage stage embryos to the blastocyst stage but also predict their potential to implant and establish a pregnancy. As such, the mouse model provides an excellent tool to better understand events or factors associated with embryo developmental competence. Finally, our study provides further support for human data suggesting that embryo cleavage kinetics are beneficial for embryo selection, which will ultimately increase the chance of a successful pregnancy.
Supplementary Material
Acknowledgment
The authors thank Progyny for their support including their donation of the equipment used for the time-lapse experiments.
References
- 1. Montag M, Toth B, Strowitzki T.. New approaches to embryo selection. Reprod Biomed Online 2013; 27:539–546. [DOI] [PubMed] [Google Scholar]
- 2. Capalbo A, Rienzi L, Cimadomo D, Maggiulli R, Elliott T, Wright G, Nagy ZP, Ubaldi FM.. Correlation between standard blastocyst morphology, euploidy and implantation: an observational study in two centers involving 956 screened blastocysts. Hum Reprod 2014; 29:1173–1181. [DOI] [PubMed] [Google Scholar]
- 3. Guerif F, Lemseffer M, Leger J, Bidault R, Cadoret V, Chavez C, Gasnier O, Saussereau MH, Royere D.. Does early morphology provide additional selection power to blastocyst selection for transfer? Reprod Biomed Online 2010; 21:510–519. [DOI] [PubMed] [Google Scholar]
- 4. Racowsky C, Ohno-Machado L, Kim J, Biggers JD.. Is there an advantage in scoring early embryos on more than one day? Hum Reprod 2009; 24:2104–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Forman EJ, Hong KH, Ferry KM, Tao X, Taylor D, Levy B, Treff NR, Scott RT Jr. In vitro fertilization with single euploid blastocyst transfer: a randomized controlled trial. Fertil Steril 2013; 100:100–107. [DOI] [PubMed] [Google Scholar]
- 6. Wu Y, Lv Z, Yang Y, Dong G, Yu Y, Cui Y, Tong M, Wang L, Zhou Z, Zhu H, Zhou Q, Sha J.. Blastomere biopsy influences epigenetic reprogramming during early embryo development, which impacts neural development and function in resulting mice. Cell Mol Life Sci 2014; 71:1761–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Herrero J, Meseguer M.. Selection of high potential embryos using time-lapse imaging: the era of morphokinetics. Fertil Steril 2013; 99:1030–1034. [DOI] [PubMed] [Google Scholar]
- 8. Swain JE. Could time-lapse embryo imaging reduce the need for biopsy and PGS? J Assist Reprod Genet 2013; 30:1081–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wong C, Chen AA, Behr B, Shen S.. Time-lapse microscopy and image analysis in basic and clinical embryo development research. Reprod Biomed Online 2013; 26:120–129. [DOI] [PubMed] [Google Scholar]
- 10. Conaghan J. Time-lapse imaging of preimplantation embryos. Semin Reprod Med 2014; 32:134–140. [DOI] [PubMed] [Google Scholar]
- 11. Wong CC, Loewke KE, Bossert NL, Behr B, De Jonge CJ, Baer TM. Reijo Pera RA. Non-invasive imaging of human embryos before embryonic genome activation predicts development to the blastocyst stage. Nat Biotechnol 2010; 28:1115–1121. [DOI] [PubMed] [Google Scholar]
- 12. Conaghan J, Chen AA, Willman SP, Ivani K, Chenette PE, Boostanfar R, Baker VL, Adamson GD, Abusief ME, Gvakharia M, Loewke KE, Shen S.. Improving embryo selection using a computer-automated time-lapse image analysis test plus day 3 morphology: results from a prospective multicenter trial. Fertil Steril 2013; 100:412–419. [DOI] [PubMed] [Google Scholar]
- 13. Cruz M, Garrido N, Herrero J, Perez-Cano I, Munoz M, Meseguer M.. Timing of cell division in human cleavage-stage embryos is linked with blastocyst formation and quality. Reprod Biomed Online 2012; 25:371–381. [DOI] [PubMed] [Google Scholar]
- 14. Dal Canto M, Coticchio G. Mignini Renzini M, De Ponti E, Novara PV, Brambillasca F, Comi R, Fadini R. Cleavage kinetics analysis of human embryos predicts development to blastocyst and implantation. Reprod Biomed Online 2012; 25:474–480. [DOI] [PubMed] [Google Scholar]
- 15. Chamayou S, Patrizio P, Storaci G, Tomaselli V, Alecci C, Ragolia C, Crescenzo C, Guglielmino A.. The use of morphokinetic parameters to select all embryos with full capacity to implant. J Assist Reprod Genet 2013; 30:703–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kirkegaard K, Kesmodel US, Hindkjaer JJ, Ingerslev HJ.. Time-lapse parameters as predictors of blastocyst development and pregnancy outcome in embryos from good prognosis patients: a prospective cohort study. Hum Reprod 2013; 28:2643–2651. [DOI] [PubMed] [Google Scholar]
- 17. Yang Z, Zhang J, Salem SA, Liu X, Kuang Y, Salem RD, Liu J.. Selection of competent blastocysts for transfer by combining time-lapse monitoring and array CGH testing for patients undergoing preimplantation genetic screening: a prospective study with sibling oocytes. BMC Med Genomics 2014; 7:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rubio I, Galan A, Larreategui Z, Ayerdi F, Bellver J, Herrero J, Meseguer M.. Clinical validation of embryo culture and selection by morphokinetic analysis: a randomized, controlled trial of the EmbryoScope. Fertil Steril 2014; 102:1287–1294. [DOI] [PubMed] [Google Scholar]
- 19. VerMilyea MD, Tan L, Anthony JT, Conaghan J, Ivani K, Gvakharia M, Boostanfar R, Baker VL, Suraj V, Chen AA, Mainigi M, Coutifaris C et al. Computer-automated time-lapse analysis results correlate with embryo implantation and clinical pregnancy: a blinded, multi-centre study. Reprod Biomed Online 2014; 29:729–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Basile N, Vime P, Florensa M. Aparicio Ruiz B, Garcia Velasco JA, Remohi J, Meseguer M. The use of morphokinetics as a predictor of implantation: a multicentric study to define and validate an algorithm for embryo selection. Hum Reprod 2015; 30:276–283. [DOI] [PubMed] [Google Scholar]
- 21. Campbell A, Fishel S, Bowman N, Duffy S, Sedler M, Thornton S.. Retrospective analysis of outcomes after IVF using an aneuploidy risk model derived from time-lapse imaging without PGS. Reprod Biomed Online 2013; 27:140–146. [DOI] [PubMed] [Google Scholar]
- 22. Meseguer M, Herrero J, Tejera A, Hilligsoe KM, Ramsing NB, Remohi J.. The use of morphokinetics as a predictor of embryo implantation. Hum Reprod 2011; 26:2658–2671. [DOI] [PubMed] [Google Scholar]
- 23. Armstrong S, Arroll N, Cree LM, Jordan V, Farquhar C.. Time-lapse systems for embryo incubation and assessment in assisted reproduction. Cochrane Database Syst Rev 2015; 2:CD011320. [DOI] [PubMed] [Google Scholar]
- 24. Kaser DJ, Racowsky C.. Clinical outcomes following selection of human preimplantation embryos with time-lapse monitoring: a systematic review. Hum Reprod Update 2014; 20:617–631. [DOI] [PubMed] [Google Scholar]
- 25. Kirkegaard K, Ahlstrom A, Ingerslev HJ, Hardarson T.. Choosing the best embryo by time lapse versus standard morphology. Fertil Steril 2015; 103:323–332. [DOI] [PubMed] [Google Scholar]
- 26. Racowsky C, Kovacs P, Martins WP.. A critical appraisal of time-lapse imaging for embryo selection: where are we and where do we need to go? J Assist Reprod Genet 2015; 32:1025–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Whitten WK, Biggers JD.. Complete development in vitro of the pre-implantation stages of the mouse in a simple chemically defined medium. J Reprod Fertil 1968; 17:399–401. [DOI] [PubMed] [Google Scholar]
- 28. Breiman L. Classification and Regression Trees.Belmont, CA:Wadsworth International Group;1984. [Google Scholar]
- 29. Steinberg D, Colla P.. CART—Classification and Regression Trees.San Diego, CA:Salford Systems;1997. [Google Scholar]
- 30. Tusher VG, Tibshirani R, Chu G.. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 2001; 98:5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Arav A, Aroyo A, Yavin S, Roth Z.. Prediction of embryonic developmental competence by time-lapse observation and “shortest-half” analysis. Reprod Biomed Online 2008; 17:669–675. [DOI] [PubMed] [Google Scholar]
- 32. Hashimoto S, Kato N, Saeki K, Morimoto Y.. Selection of high-potential embryos by culture in poly(dimethylsiloxane) microwells and time-lapse imaging. Fertil Steril 2012; 97:332–337. [DOI] [PubMed] [Google Scholar]
- 33. Lee YS, Thouas GA, Gardner DK.. Developmental kinetics of cleavage stage mouse embryos are related to their subsequent carbohydrate and amino acid utilization at the blastocyst stage. Hum Reprod 2015; 30:543–552. [DOI] [PubMed] [Google Scholar]
- 34. Pribenszky C, Losonczi E, Molnar M, Lang Z, Matyas S, Rajczy K, Molnar K, Kovacs P, Nagy P, Conceicao J, Vajta G.. Prediction of in-vitro developmental competence of early cleavage-stage mouse embryos with compact time-lapse equipment. Reprod Biomed Online 2010; 20:371–379. [DOI] [PubMed] [Google Scholar]
- 35. Gopichandran N, Leese HJ.. The effect of paracrine/autocrine interactions on the in vitro culture of bovine preimplantation embryos. Reproduction 2006; 131:269–277. [DOI] [PubMed] [Google Scholar]
- 36. Ebner T, Shebl O, Moser M, Mayer RB, Arzt W, Tews G.. Group culture of human zygotes is superior to individual culture in terms of blastulation, implantation and life birth. Reprod Biomed Online 2010; 21:762–768. [DOI] [PubMed] [Google Scholar]
- 37. O'Doherty EM, Wade MG, Hill JL, Boland MP.. Effects of culturing bovine oocytes either singly or in groups on development to blastocysts. Theriogenology 1997; 48:161–169. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


