The long Read Annotation (LoReAn) pipeline provides automated genome annotation with superior performance by incorporating single-molecule cDNA sequencing data.
Abstract
Single-molecule full-length complementary DNA (cDNA) sequencing can aid genome annotation by revealing transcript structure and alternative splice forms, yet current annotation pipelines do not incorporate such information. Here we present long-read annotation (LoReAn) software, an automated annotation pipeline utilizing short- and long-read cDNA sequencing, protein evidence, and ab initio prediction to generate accurate genome annotations. Based on annotations of two fungal genomes (Verticillium dahliae and Plicaturopsis crispa) and two plant genomes (Arabidopsis [Arabidopsis thaliana] and Oryza sativa), we show that LoReAn outperforms popular annotation pipelines by integrating single-molecule cDNA-sequencing data generated from either the Pacific Biosciences or MinION sequencing platforms, correctly predicting gene structure, and capturing genes missed by other annotation pipelines.
Genome sequencing has advanced nearly every discipline within the biological sciences, as the ongoing decreasing sequencing costs and increasing computational capacity allows many laboratories to pursue genomics-based answers to biological questions. New sequencing technologies designed to sequence longer contiguous DNA molecules, such as Pacific Biosciences’ (PacBio) single-molecule real-time sequencing (SMRT) and Oxford Nanopore Technologies’ (ONT) MinION, have ushered in the most recent genomics revolution (Koren and Phillippy, 2015). These advances are further enhancing the ability to generate high-quality genome assemblies of large, complex eukaryotic genomes (Faino et al., 2015; Chin et al., 2016; Davey et al., 2016; Jiao and Schneeberger, 2017).
A high-quality genome assembly, represented by (near-)chromosome completion, helps address many biological questions but often requires functional features to be further defined (Thomma et al., 2016). The process of genome annotation, i.e. the identification of protein-coding genes and their structural features, such as intron-exons boundaries, is important to capture biological values of a genome assembly (Yandell and Ence, 2012). Genomes can be annotated using computer algorithms in so-called ab initio gene predictions and using wet-lab-generated data, such as complementary DNA (cDNA) or protein datasets for evidence-based predictions, and current annotation pipelines typically incorporate both types of data (Cantarel et al., 2008; Yandell and Ence, 2012). Ab initio gene prediction tools are based on statistical models, most often hidden Markov models, which are trained using known proteins, and typically perform well at predicting conserved or core genes (Goodswen et al., 2012; Yandell and Ence, 2012). However, the ab initio prediction accuracy decreases for organism-specific genes, for genes encoding small proteins and across organisms with differing intron-exon features (Ter-Hovhannisyan et al., 2008; Hoff et al., 2016). Furthermore, ab initio annotation of nonmodel genomes remains challenging as appropriate training data are not always available, and genome characteristics across organisms can vary (Reid et al., 2014). To improve genome annotations, cDNA sequencing (RNA-seq) data can be incorporated to train ab initio software (Hoff et al., 2016) and to provide additional evidence for defining accurate gene models (Wang et al., 2009). Most genome annotations to date rely on a combination of short-read mapping data and ab initio gene prediction. However, errors occur during this process because short-read RNA-seq data cannot always be unequivocally mapped, because a single read does not span a gene’s full length, and because of differences in evidence weighting leading to gene prediction. LoReAn was developed to also use information from long-read sequencing data to help address issues of mapping and gene structure, with greater emphasis given to empirical mapping data during the gene prediction process.
Current annotation pipelines use a combination of ab initio and evidence-based predictions to generate accurate consensus annotations. MAKER2 is a user friendly, fully automated annotation pipeline that incorporates multiple sources of gene prediction information and has been extensively used to annotate eukaryotic genomes (Cantarel et al., 2008; Holt and Yandell, 2011; Smith et al., 2011, 2013; Amemiya et al., 2013; Ming et al., 2015; Lamichhaney et al., 2016). The Broad Institute Eukaryotic Genome Annotation Pipeline (here referred to as BAP) has mainly been used to annotate fungal genomes (Linde et al., 2015; Muñoz et al., 2015; Ma et al., 2016) and integrates multiple programs and evidences for genome annotation (Haas et al., 2008, 2011). BRAKER1 and CodingQuarry are two gene-prediction software packages that utilize RNA-seq data and genome sequence to predict gene models. BRAKER1 is a pipeline for unsupervised RNA-seq-based genome annotation that combines the advantages of GeneMark-ET and Augustus. BRAKER1 is a two-step software that combines GeneMark-ET and intron-hints, derived from mapped RNA-seq, to generate a species-specific database that Augustus can use for gene prediction (Hoff et al., 2016). CodingQuarry is a pipeline for RNA-Seq assembly-supported training and gene prediction, which is only recommended for application to fungi. In this tool, Cufflink’s assembled RNA-seq is used to build a hidden Markov model that is used for gene prediction (Testa et al., 2015). A limitation of these annotation pipelines is that experimental evidence from short-read RNA-seq mapping can be lost due to evidence weighting, and the pipelines cannot natively exploit gene structure information from single-molecule cDNA sequencing.
In addition to improving genome assembly (Phillippy, 2017), long-read sequencing data can be used to improve genome annotation. The use of single-molecule cDNA sequencing can increase the accuracy of automated genome annotation by improving genome mapping of sequencing data, correctly identifying intron-exon boundaries, directly identifying alternatively spliced transcripts, identifying transcription start and end sites, and providing precise strand orientation to single-exon genes (Minoche et al., 2015; Abdel-Ghany et al., 2016; Wang et al., 2016). However, several hurdles limit the implementation of long-read sequencing data into automated genome annotation, such as the higher per-base costs when compared to short-read data, the relatively high error rates for long-read sequencing technologies, and the lack of bioinformatics tools to integrate long-read data into current annotation pipelines (Faino and Thomma, 2014; Laver et al., 2015). The first two limitations are addressed by the continual reduction in sequencing cost and improved base calling by long-read sequencing providers and the development of bioinformatics methods to correct for sequencing errors (Loman et al., 2015; Laehnemann et al., 2016). To address the disconnect between genome annotation pipelines and the latest sequencing technologies, we developed the long-read annotation (LoReAn) pipeline. LoReAn is an automated annotation pipeline that takes full advantage of MinION or PacBio SMRT long-read sequencing data in combination with protein evidence and ab initio gene predictions for full-genome annotation. Short-read RNA-seq can be used in LoReAn to train ab initio software. Based on the reannotation of two fungal and two plant species, we demonstrate that LoReAn can provide annotations with increased accuracy by incorporating single-molecule cDNA sequencing data from different sequencing platforms.
RESULTS
LoReAn Design and Implementation
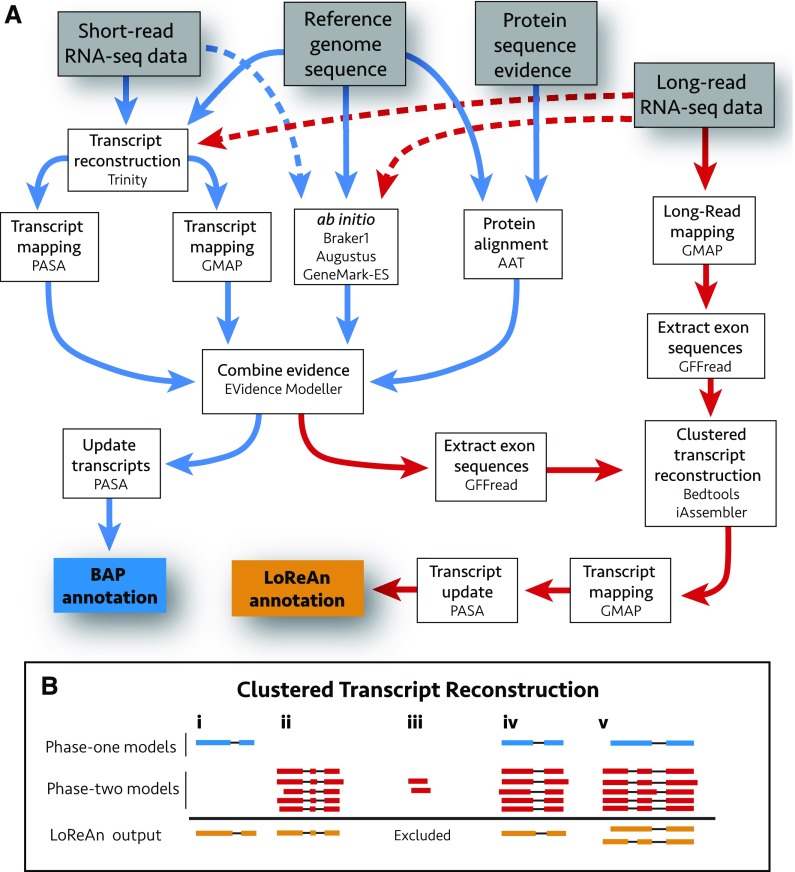
The LoReAn pipeline can be conceptualized in two phases. The first phase involves genome annotation based on ab initio and evidence-based predictions (Fig. 1A; blue arrows) and largely follows the workflow previously described in the BAP (Haas et al., 2008, 2011). This first phase produces a full-genome annotation and requires the minimum input of a reference genome, protein sequence of known and, possibly, related species, and a species name from the Augustus prediction software database (Stanke et al., 2008). We implemented two changes into the first phase of LoReAn, which we refer to as BAP+. First, LoReAn used RNA-seq reads as input in combination with the BRAKER1 software (Hoff et al., 2016) to produce a species-specific database for the Augustus prediction software. Additionally, RNA-seq data were assembled into full-length cDNA using Trinity software (Grabherr et al., 2011), and the assembled transcripts were aligned to the genome using both the program to assemble spliced alignments (PASA; Haas et al., 2008) and the genomic mapping and alignment program (GMAP; Wu and Watanabe, 2005). The output of PASA software was passed to the Evidence modeler (EVM; Haas et al., 2008) as cDNA evidence while the output of GMAP was given to EVM as ab initio evidence.
Figure 1.
Schematic overview of the LoReAn pipeline and clustered transcript reconstruction. A, Illustration of the computational workflow for the LoReAn pipeline. Gray boxes represent input data, and each white box represents a step in the annotation process with mention of the specific software. The boxes connected by blue arrows integrate the steps from the previously described BAP, described in the text as phase one of LoReAn (Haas et al., 2008). The LoReAn pipeline (boxes connected by red arrows) integrates the BAP workflow but additionally incorporates long-read sequencing data, described in the text as phase two of LoReAn. The blue box, “BAP annotation,” represents the annotation results from the BAP pipeline used for comparison in this study, while the orange box “LoReAn annotation” represents the annotation results from the LoReAn pipeline using long-read sequencing data. Dashed arrows represent optional steps for the pipeline. B, Illustration of the clustered transcript reconstruction. Gene models are depicted as exons (boxes) and connecting introns (lines). Blue models represent BAP annotations, while red models represent hypothetical long reads mapped to the genome. Orange models represent consensus annotations reported in the final LoReAn output. Various scenarios can occur: (i) High-confidence predictions from the BAP are kept regardless of whether they are supported by long reads. (ii and iii) Clusters of mapped long reads are used to generate a consensus prediction model, unless the model is supported by less than a user-defined minimum depth. (iv) Overlapping BAP and mapped long reads are combined to a consensus model. (v) Two annotations are reported if no consensus can be reached for the BAP and clustered long-read data.
The second phase of LoReAn incorporates single-molecule cDNA sequencing with the annotation results of the first phase by utilizing an alternative approach to reconstruct full-length transcripts (Fig. 1A, red arrows). Single-molecule long-read sequencing reads are mapped to the genome using GMAP, which allows the determination of transcript structure (i.e. start, stop, and exon boundaries) from a single cDNA molecule (Križanovic et al., 2018). The underlying reference sequence is extracted to overcome sequence errors associated with long-read sequencing, and these sequences are combined with the gene models from the first phase in a process we refer to as “clustered transcript reconstruction” (Fig. 1, A and B). Through this process, consensus gene models are built by combining the first- and second-phase gene models that cluster at the same locus. Optionally, model clustering can be conducted in a strand-specific manner (LoReAn stranded, see Supplemental Data for details), where only gene models mapping on the same DNA coding strand are used to build a consensus model. These high-confidence models are mapped back to the reference using GMAP to correct open reading frames, and subsequently, PASA is used to update the gene models by identifying untranslated regions (UTRs) and alternatively spliced transcripts to generate a final annotation. Sequence-based support for the final gene models (Fig. 1B, orange models) can come from the first phase annotation alone (Fig. 1B, i), the second phase, given a sufficient level of support (Fig. 1B, ii and iii), or through a combination of the two phases (Fig. 1B, iv and v). If a single consensus annotation cannot be reached between the two phases, both annotations are kept in the final output (Fig. 1B, v).
LoReAn Produces High-Quality Gene Predictions Based on Gene, Exon, and Intron Metrics Compared to the Reference Annotation and Empirical Data
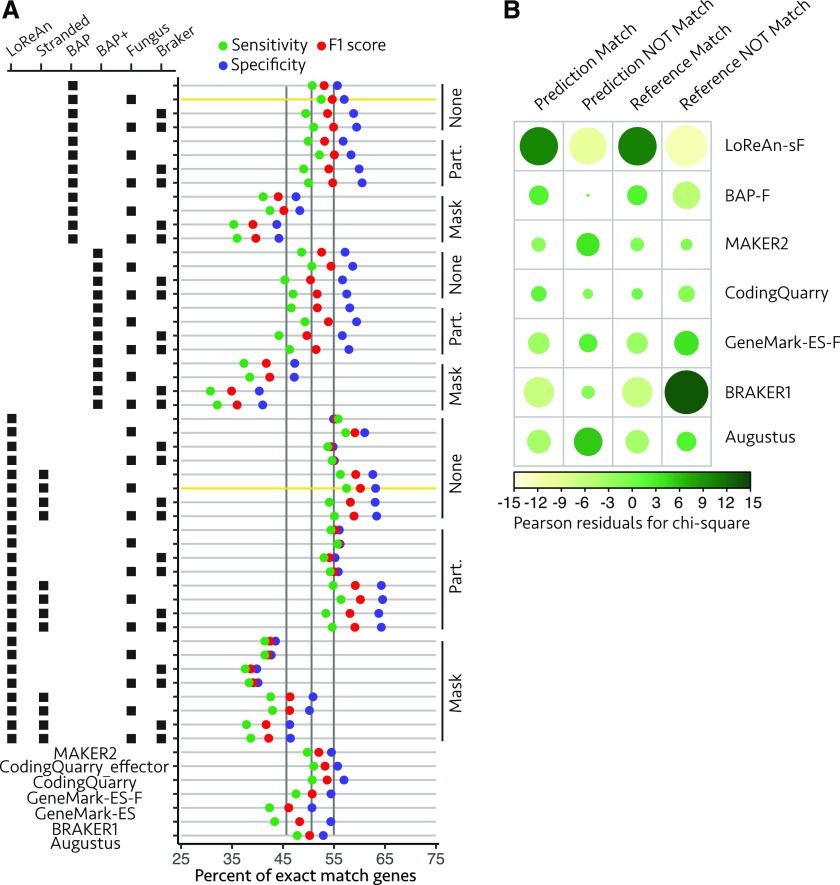
To test the performance of LoReAn, we reannotated the genome sequence of the haploid fungus Verticillium dahliae, an important pathogen of hundreds of plant species, including many crops (Fradin and Thomma, 2006; Klosterman et al., 2009). The genome of V. dahliae strain JR2 was used for testing LoReAn because it is assembled into complete chromosomes and has a manually curated annotation, providing a high-confidence resource for reference (Faino et al., 2015). A total of 55 annotations were generated and compared, of which 24 were produced using LoReAn, 12 using BAP and 12 using BAP+ with different genome masking and ab initio options (description in Supplemental Data), along with output from the annotation software MAKER2, BRAKER1, Augustus, and two each from CodingQuarry and GeneMark-ES. We determined the quality of annotation outputs by comparing each to the reference annotation for exact matches to either genes or exon locations (Fig. 2A; Supplemental Tables S1 and S2; reference annotation contains 11,385 gene models and 28,142 exons). These comparisons were used to calculate sensitivity (how much of the reference is correctly predicted), specificity (how much of the prediction is in the reference), and F1 score (the harmonic mean of sensitivity and specificity). These metrics were calculated based on commonly described methods used within the gene prediction community (see "Materials and Methods" and Keibler and Brent [2003], Yandell and Ence [2012], and Chan et al. [2017]). Hard masking, where short DNA repeat (>10 bp) sequences are removed from the genome prior to annotation, significantly affected the quality of predicted gene and exon models, with partially masked or nonmasked genome inputs producing significantly improved annotations (Supplemental Figs. S1A and S2A; Supplemental Tables S3 and S4). On average, the "fungus" option (-f) of the ab initio software GeneMark-ES produced the best gene and exon predictions (Supplemental Figs. S1B and S2B; Supplemental Tables S3 and S4). Gene predictions from LoReAn using coding strand information (LoReAn-s) produced statistically similar results to LoReAn for exact match genes and similar results to LoReAn and BAP for exact match exons (Supplemental Tables S2 and S3). However, the F1 score for exact match gene and exon predictions were significantly higher for LoReAn stranded compared to the other three outputs (Supplemental Figs. S1C and S2C), indicating that using strand information improves the overall quality of the annotation.
Figure 2.
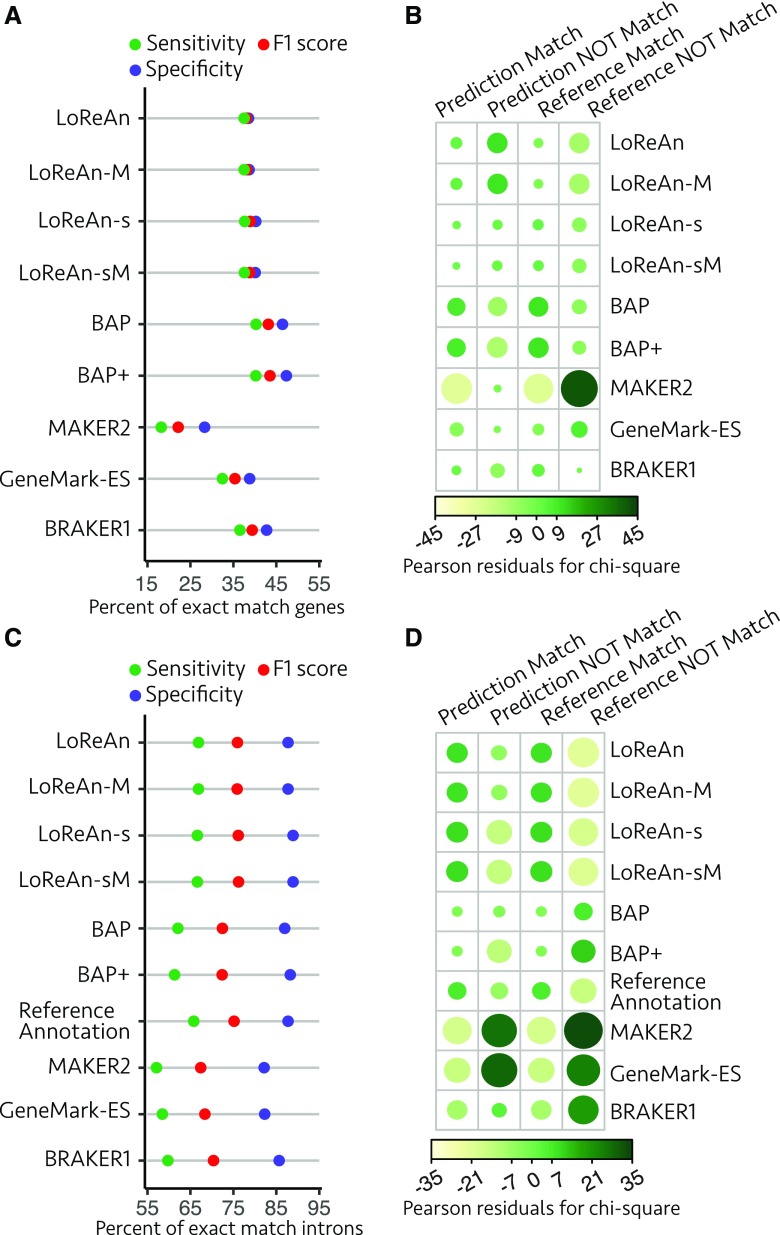
Annotation quality summary for exact match genes to the reference. A, Gene annotation quality summary, where each horizontal bar represents an annotation output and each colored dot represents the sensitivity (green), specificity (purple), and F1 score (red). The annotations are labeled using the left grid, where the group of horizontal black dots defines the parameters used in the annotation. Possible parameters include using LoReAn, BAP, or BAP+ pipeline, stranded mode for LoReAn (Stranded), the fungus option for GeneMark-ES (Fungus), or the BRAKER1 program for Augustus (BRAKER1). Annotation options are grouped by the level of reference masking—partially masked (Part.), nonmasked (None), or fully masked (Mask). The results from additionally tested annotation pipelines are shown at the bottom. Three vertical gray lines represent the first quantile, median, and third quantile for the F1 score. The two annotations highlighted with a yellow horizontal bar were used for subsequent analysis. B, The proportion of exact match to nonmatching gene predictions (specificity) and exact match to missing gene predictions (sensitivity) were compared using a χ2 test of independence. The residuals from the analysis are shown with the size and color representing the magnitude and direction of the association between rows and columns. GeneMark-ES-F, GeneMark gene prediction software using the "fungus" option. LoReAn-sF, LoReAn using strand information and the "fungus" option of GeneMark-ES. BAP-F, The Broad Institute eukaryotic genome annotation pipeline described in the text and using the "fungus" option of GeneMark-ES.
A single output from LoReAn and BAP were selected for further comparison to the outputs from the other annotation software (i.e. MAKER2, CodingQuarry, GeneMark, BRAKER1, and Augustus). The LoReAn-stranded run using the "fungus" option of GeneMark-ES (referred to as LoReAn-sF throughout) and the BAP run using the "fungus" option of GeneMark-ES (referred to as BAP-F throughout) using a nonmasked genome were selected because they had the highest F1 scores and used similar settings to the other pipelines, thereby enabling comparisons (Fig. 2A, highlighted by horizontal yellow lines). The seven annotation pipelines were compared by computing the number of predicted genes that matched the reference and the number of reference genes that were matched (i.e. numbers for specificity and sensitivity). A χ2 test of independence indicated a significant, nonequal association between the seven annotation pipelines and the tested annotation metrics (Pearson’s χ2 test of independence, χ2 = 913.61, P value < 2.2e-16). Plotting the residuals of the χ2 test indicated that LoReAn-sF had the largest positive association for correctly predicting gene models (columns 1 and 3, Fig. 2B) and the largest negative association for predicting wrong gene models or missing gene models (columns 2 and 4, Fig. 2B). These results show that there are statistically significant differences between the tested pipelines for annotation quality, and the residual analysis indicates that LoReAn-sF has the best association with desirable annotation metrics.
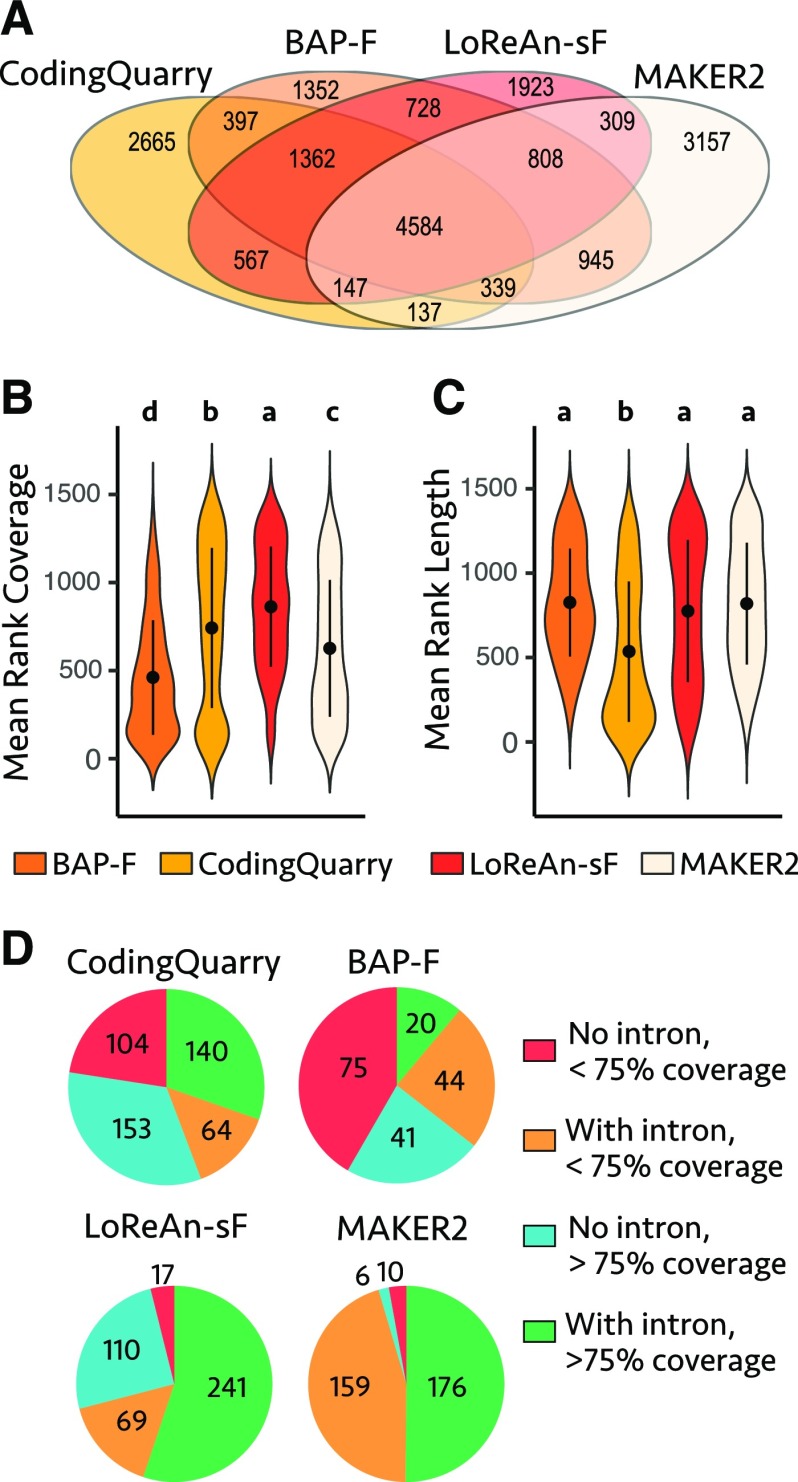
To further characterize gene prediction differences between annotation pipelines, four outputs were selected for comparison. MAKER2 and CodingQuarry were selected as they represent popular annotation choices and performed well, along with the output of LoReAn-sF and BAP-F, as they are the focus of the study. The gene predictions were compared head-to-head in the absence of a reference annotation by determining the overlap between exact match genes. There were 4,584 genes with the same predicted structure (i.e. start, stop, intron position) from the four pipelines, equivalent to approximately 40% of the genes in the reference annotation (Fig. 3A). BAP-F predicted the fewest unique genes (1,352), while MAKER2 predicted the most (3,157; Fig. 3A). However, the use of exact match gene structure to identify unique coding sequences is potentially misleading, as two gene predictions can code for the same or a similar protein without the exact same structure. To generate a more biologically relevant comparison of unique protein coding differences, we grouped translated protein sequences of each annotation into homologous groups using orthoMCL (Li et al., 2003; Chen et al., 2006). Using these groups, we identified protein coding sequences that were unique to a single annotation pipeline, referred to as singletons. We identified 1,429 singletons across the four annotations, with CodingQuarry predicting the most (461) and BAP-F predicting the fewest (180; Fig. 3D). To validate the predicted singletons, the percent coverage of the predicted transcripts was calculated using short-read RNA-seq data. Singletons from LoReAn-sF were covered on average across 80% of their length by mapped RNA-seq reads, while the next highest was CodingQuarry with 63% average coverage. Nonparametric statistical test for the rank of the mean coverage shows that singletons predicted by LoReAn-sF had significantly higher RNA-seq coverage compared to the other pipelines (Fig. 3B; Kruskal-Wallis test). Analyzing the length of the predicted singletons indicated that CodingQuarry predicted the shortest singletons, while the other pipelines were not statistically different than one another (Fig. 3C; Kruskal-Wallis test). The singletons were further grouped by the presence/absence of introns and RNA-seq coverage, as gene models with introns and RNA-seq support are more likely to be true genes. Singletons that contained at least one intron and had RNA-seq reads covering at least 75% of their length were considered the highest-confidence models. Pairwise, two-sample test for equal proportions indicated the outputs of LoReAn-sF and MAKER2 were statistically similar for the proportion of singletons in this high-confidence group, but both were significantly higher than the other two pipelines (Supplemental Table S5). The collective, comprehensive comparison of LoReAn versus other commonly used annotation software shows that LoReAn outperforms these for many gene prediction quality metrics.
Figure 3.
Analysis of predicted singletons across four pipelines. A, Venn diagram showing the overlap and uniqueness of predicted genes based on genomic location. The Venn diagram shows that 4,584 genes were annotated with the exact same features across all four pipelines. The numbers captured by only a single annotation pipeline are considered singletons—genes whose structure is uniquely annotated by a given pipeline. Note, these singletons do not necessarily represent unique loci. B, Short-read RNA-seq data were mapped to the genome, and the percent length coverage of each gene annotation was calculated. The data were not completely normally distributed, so a nonparametric Kruskal-Wallis test was used to rank the mean of the coverage. Data are shown as violin plots, with the tails representing the data range and the mean and sd are shown as a black point and black vertical lines, respectively. Letters shown above each violin plot represent post-hoc statistical groupings where plots with the same letter are statistically indistinguishable. Multiple comparisons were made using the nonparametric Kruskal-Wallis rank of means, and post-hoc differences were determined using Fisher’s least significant difference, P < 0.05 with Bonferroni correction. C, Same as in B except the mean rank of the singleton length is analyzed. D, The orthoMCL singletons from each pipeline were grouped into one of four categories shown in the key representing if the singleton contained an intron or not and if the singleton’s length was covered by over 75% with RNA-seq data. The number of singletons within each of the four categories is shown.
LoReAn Predicts the Most Accurate Intron Locations Compared to Empirical RNA-seq Predictions
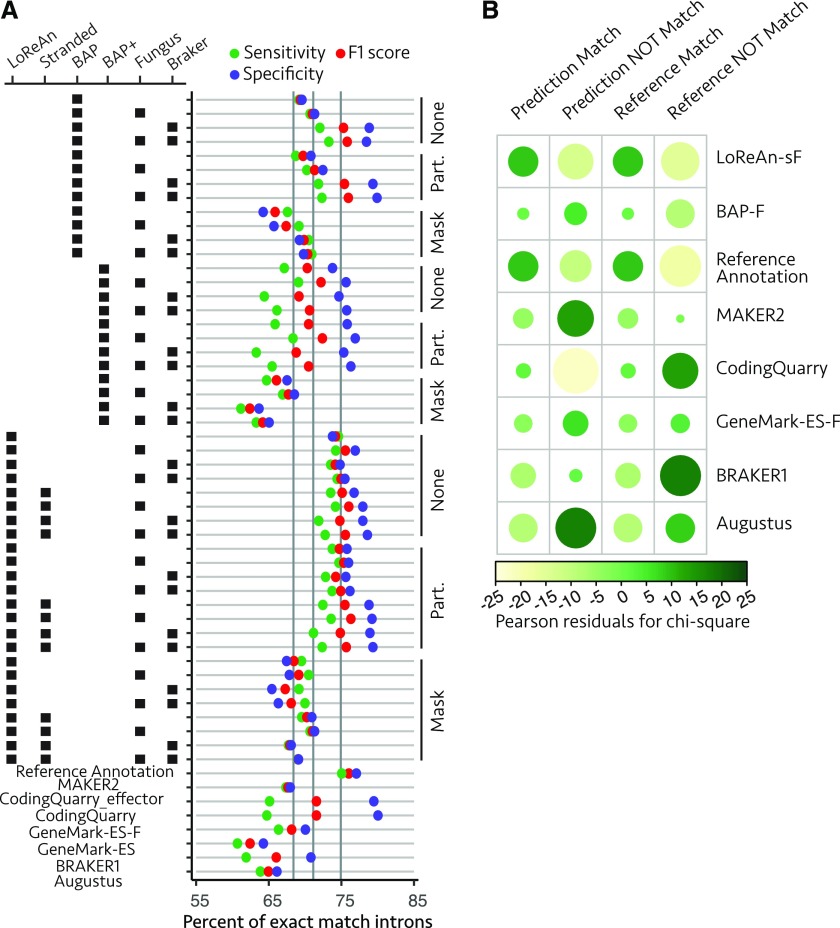
Annotation comparisons are commonly made against a reference annotation. This carries inherent problems, however, as many organisms do not have a high-confidence reference annotation. Additionally, the overlap between the software used to generate a given reference annotation and that being tested introduces bias in the analysis. To evaluate the annotation outputs produced here in the absence of a reference annotation, we devised an approach to quantify annotation quality based on empirical RNA-seq output rather than a reference annotation. The locations of predicted introns from the annotation outputs were compared to the locations of the inferred introns from long- and short-read mapped data, using the same annotation outputs compared earlier for gene and exon locations (Fig. 4A). This analysis also allowed the reference annotation to be compared to the intron locations inferred from the RNA-seq data. To formally test for statistical differences, the same annotation pipelines compared earlier were compared using a χ2 test of independence, which indicated a significant, nonequal association between the annotation pipelines and intron predictions (Pearson’s χ2 test of independence, χ2 = 3,220.7, P value < 2.2e-16). The residual plot of the χ2 test shows the magnitude and direction of the association between the pipelines and the predictions (Fig. 4B). Multiple two-sample tests of proportions between the annotation predictions and the reference annotation, testing if the annotation prediction showed improved metrics when compared with the reference, showed that only CodingQuarry outperformed the reference annotation for the specificity metric (Supplemental Table S6). This indicated the reference annotation was the most similar to the RNA-seq data, which is not surprising given the high-quality reference annotation. The analysis was rerun to test if the LoReAn-sF outperformed the other pipelines, and LoReAn-sF outperformed the other software in nearly every instance (Table1). This further indicates that the LoReAn software offers improved annotation performance when evaluated against RNA-seq data in the absence of a reference annotation.
Figure 4.
Annotation quality summary for predicted introns exactly matching RNA-seq-inferred introns. A, Predicted intron quality summary, where each horizontal bar represents an annotation output, and each colored dot represents the sensitivity (green), specificity (purple), and F1 score (red) as described in Figure 2A. B, The proportion of exact match to nonmatching intron predictions (specificity) and exact match to missing intron predictions (sensitivity) were compared using a χ2 test of independence as described in Figure 2B. GeneMark-ES-F, GeneMark gene prediction software using the "fungus" option. LoReAn-sF, LoReAn using strand information and the "fungus" option of GeneMark-ES.
Table 1. χ2 test of proportions for predicted exact match introns inferred from RNA-seq mapping data in V. dahliae.
| Pipeline | Predictions Matching RNA-seq | Predictions Not Matching | Predictions Missing | Specificity Less Than LoReAn-sFa | Sensitivity Less Than LoReAn-sFb |
|---|---|---|---|---|---|
| BAP-F | 13,128 | 5,284 | 5,446 | <0.0001 | <0.0001 |
| MAKER2 | 12,509 | 5,903 | 6,065 | <0.0001 | <0.0001 |
| CodingQuarry | 12,021 | 3,001 | 6,553 | N.S.c | <0.0001 |
| GeneMark-ES-F | 12,323 | 5,274 | 6,251 | <0.0001 | <0.0001 |
| BRAKER1 | 11,491 | 4,739 | 7,083 | <0.0001 | <0.0001 |
| Augustus | 11,857 | 6,079 | 6,717 | <0.0001 | <0.0001 |
| Reference annotation | 13,935 | 4,145 | 4,639 | N.S.c | N.S.c |
| LoReAn-sF | 13,780 | 3,899 | 4,794 | Not applicable | Not applicable |
Column reports P values for the χ2 test of proportions for the specificity metric
Column reports P values for the χ2 test of proportions for the sensitivity metric
N.S., not significant with a P value greater than 0.05
Only the LoReAn Pipeline Correctly Annotates the Ave1 Effector Locus
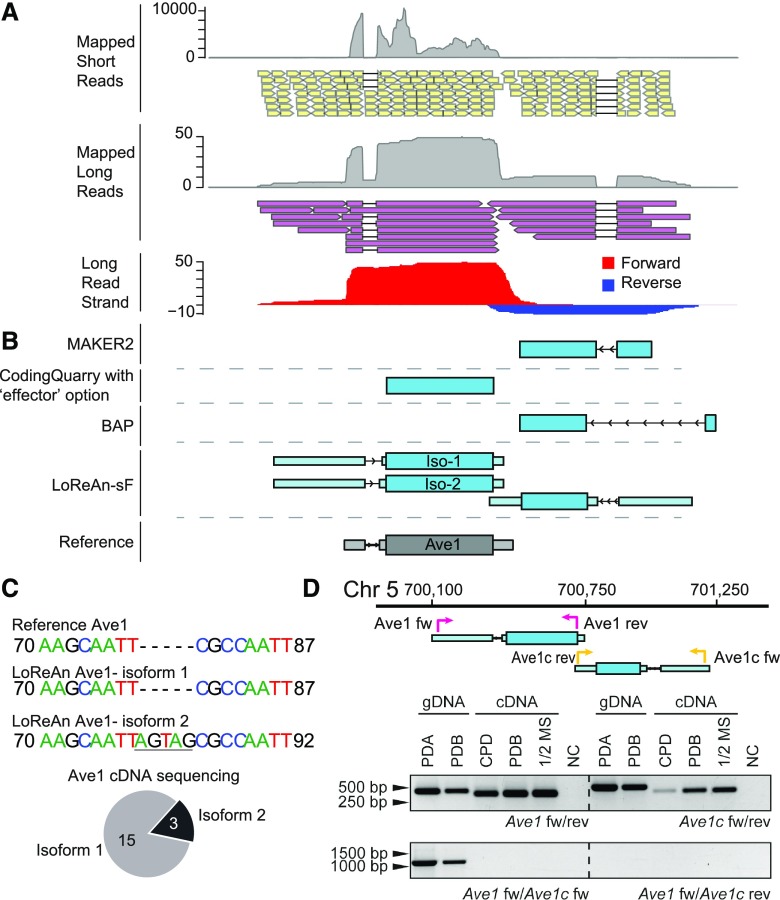
Plant-pathogenic fungi encode in planta-secreted proteins, termed effectors, which serve to facilitate infection (Cook et al., 2015; Lo Presti et al., 2015). Effectors are generally characterized as lineage-specific small, secreted, Cys-rich proteins with generally no characterized protein domains or homology, characteristics which can make effectors difficult to predict with automated annotation (Sperschneider et al., 2015). To test how LoReAn and the other annotation pipelines performed at a specific effector locus, we detailed the annotation results for the V. dahliae Ave1 locus, which encodes a small-secreted protein that functions to increase virulence during plant infection (de Jonge et al., 2012). As previously reported, a considerable number of short RNA-seq reads uniquely map to the Ave1 locus (de Jonge et al., 2012), along with single-molecule cDNA reads identified here (Fig. 5A). Interestingly, MAKER2, BAP, Augustus, GeneMark-ES, and CodingQuarry (default) each failed to predict the previously characterized Ave1 gene despite the abundance of uniquely mapped reads (Fig. 5B; Supplemental Fig. S3). The MAKER2, BAP, and Augustus pipelines predicted a separate gene on the opposite strand located to the 3′ end of the Ave1 gene that is absent in the reference annotation. The CodingQuarry software run with the "effector" option predicted the correct coding sequence model for Ave1. The LoReAn-sF, LoReAn-s, PASA, GMAP, and BAP+ software predicted two genes at the locus, one corresponding to the known Ave1 gene, and an additional gene to the 3′ end of Ave1 (called Ave1c), similar to the gene model identified by MAKER2 (Fig. 5B; Supplemental Fig. S3).
Figure 5.
The LoReAn pipeline most accurately annotates a specific fungal locus encoding a strain-specific gene. A, Short-read RNA-seq data mapped to the locus are shown as a coverage plot (gray peaks) and as representative individual reads (yellow boxes). Long reads from single-molecule cDNA data mapped to the locus are shown as a coverage plot (gray peaks) and representative reads (purple boxes). Thick black lines linking mapped reads represent gaps in the mapped reads and are indicative of introns. The long-read data were split by mapping strand and coverage plots for forward (red) and reverse (blue) coverage plots. B, Gene model predictions from four annotation pipelines are illustrated. Light blue boxes represent untranslated regions (5′ and 3′ UTR), dark blue boxes represent coding sequence boundaries, and thin black lines depict introns. Arrows in the introns indicate the direction of transcription. The MAKER2 and BAP pipelines predict a single transcript coded on the reverse strand at the 3′ end of the known Ave1 transcript. LoReAn-sF predicts two transcripts corresponding to the Ave1 gene along with the similar transcript predicted by MAKER2. The reference Ave1 transcript is shown in gray. C, To confirm the presence of an alternative splice site in the 5′ UTR of the Ave1 transcript, 18 cDNA clones were randomly chosen and sequenced. Isoform 1 sequence is identical to the reference Ave1 sequence and was identified in 15 of the 18 clones. Isoform 2 has a 5-bp insertion in the 5′ UTR resulting from an alternative exon splice site and was identified in 3 of the 18 sequenced clones. The Ave1 reference sequence is shown from bases 71 through 86. D, The presence of Ave1 and the additional gene transcribed to the 3′ end of Ave1, termed Ave1close (Ave1c), was confirmed using PCR on genomic DNA (gDNA) and cDNA. PCR using gene specific primers, termed Ave1 fw + rev (pink arrows) or Ave1c fw + rev (yellow arrows), shows that both genes are expressed in either potato dextrose broth (PDB) Czapek-dox (CPD) or half-strength Murashige-Skoog (1/2 MS) media. The inverse orientation of the two genes was confirmed using forward primers only, which amplified the entire locus, resulting in a band of approximately 1,118 bp, but does not amplify product using cDNA as the template.
LoReAn-sF additionally predicted two mRNAs corresponding to the previously characterized Ave1 gene, termed isoform-1 and -2 (Fig. 5B). To confirm the presence of two Ave1 isoforms, cDNAs were amplified and cloned into vectors, and 18 clones were randomly selected for sequencing. A majority of the sequenced transcripts, 15 of 18, had a sequence corresponding to isoform-1, the known Ave1 transcript, while the other three were the isoform-2 sequence (Fig. 5C). The isoform-2 transcript is the result of an alternative splice junction 5 bp upstream of the previously identified splice site in the Ave1 5′ UTR intron and is not predicted to alter the protein coding sequence. The accuracy of the new gene prediction at the Ave1 locus (two Ave1 isoforms and one additional gene model) was additionally tested by showing the expression of the Ave1c gene. Two sets of primers (Ave1 and Ave1c fw and rev) amplified bands of the expected sizes, confirming the expression of both genes across various V. dahliae growth conditions (Fig. 5D, top amplification panel). We also attempted to amplify a specific product from both genomic DNA and cDNA to confirm the orientation and rule out a transcriptional fusion. Consistent with the annotation, the amplification using a genomic DNA template was successful, while the cDNA template failed to amplify a product (Fig. 5D, bottom amplification panel, primers Ave1 fw + Ave1c fw). Collectively, these results confirm that LoReAn predicts the most accurate gene models at the Ave1 locus, including a splice-variant of Ave1.
LoReAn Produces the Most Accurate Annotation of a Second Fungal Genome Using PacBio Iso-seq Reads
The basidiomycete Plicaturopsis crispa, mostly known for its wood-degrading abilities, has a relatively complex transcriptome with high levels of exons per gene—5.6 exons per gene compared to V. dahliae’s 2.5 exons per gene (Gordon et al., 2015). A total of nine annotations of the P. crispa genome from five pipelines were generated using publicly available short-read Illumina RNA-seq and single-molecule PacBio Iso-seq data (Kohler et al., 2015). The LoReAn annotations predicted the greatest number of genes, transcripts, and exons, while BAP and BAP+ had the greatest number of genes, transcripts, and exons exactly matching the reference (Table 2; Supplemental Table S7). The F1 scores for exact match genes was highest for the BAP outputs, but overall similar between the annotations (Fig. 6A). Testing the exact match gene proportions used for sensitivity and specificity indicated significant association between annotation pipelines and the metrics (Fig. 6B; Pearson’s χ2 test of independence, χ2 = 3,220.7, P value <2.2e-16). LoReAn-sF scored significantly higher than MAKER2 for sensitivity and specificity of exact match gene proportions and higher than GeneMark-ES for exact match sensitivity (Supplemental Table S8). However, these comparisons depended on the starting reference annotation.
Table 2. χ2 test of proportions for predicted exact match introns inferred from RNA-seq mapping data in P. crispa.
| Pipeline | Predictions Matching RNA-seq | Predictions Not Matching | Predictions Missing | Specificity Less Than Reference Annotationa | Sensitivity Less Than Reference Annotationb |
|---|---|---|---|---|---|
| LoReAn | 58,831 | 8,235 | 29,119 | N.S.c | <0.001 |
| LoReAn-M | 58,798 | 8,219 | 29,152 | N.S.c | <0.001 |
| LoReAn-s | 58,556 | 7,318 | 29,394 | <0.001 | <0.001 |
| LoReAn-sM | 58,622 | 7,326 | 29,328 | <0.001 | <0.001 |
| BAP | 54,565 | 8,204 | 33,385 | N.S.c | N.S.c |
| BAPplus | 53,943 | 7,159 | 34,007 | <0.001 | N.S.c |
| Augustus | 52,525 | 8,773 | 35,425 | N.S.c | N.S.c |
| GeneMark-ES-F | 51,450 | 11,085 | 36,500 | N.S.c | N.S.c |
| MAKER2 | 50,228 | 10,881 | 37,722 | N.S.c | N.S.c |
| Reference annotation | 57,837 | 80,54 | 30,113 | Not applicable | Not applicable |
Column reports P values for the χ2 test of proportions for the specificity metric
Column reports P values for the χ2 test of proportions for the sensitivity metric
N.S., not significant with a P value greater than 0.05
Figure 6.
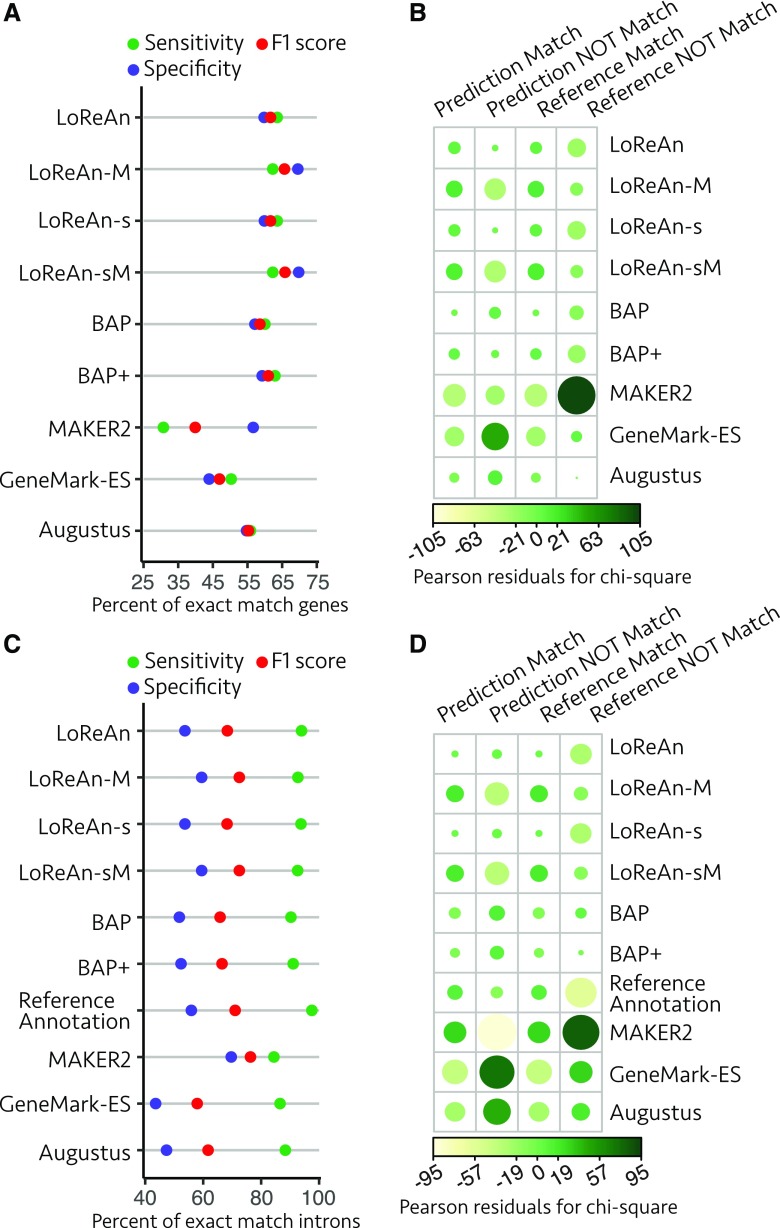
Assessment of gene and intron predictions from P. crispa. A, Annotation quality metrics are shown for exact match genes as detailed for Figure 2A. LoReAn, LoReAn in nonstranded mode using a nonmasked genome; LoReAn-M, LoReAn in nonstranded mode using a masked genome; LoReAn-s, LoReAn in stranded mode using a nonmasked genome; LoReAn-sM, LoReAn in stranded mode using a masked genome; BAP, broad annotation pipeline; BAP+, broad annotation pipeline plus additional modifications described in text. B, The proportion of exact match to nonmatching gene predictions (specificity) and exact match to missing gene predictions (sensitivity) compared using a χ2 test of independence as described in Figure 2B. C and D are for exact match intron analysis, represented as in A and B, respectively.
To better understand the differences between the outputs, we applied the empirical intron analysis and calculated the annotation quality metrics (Fig. 6C). The sensitivity and specificity proportions for exact match introns indicated significant differences across the pipelines (Fig. 6D, Pearson’s χ2 test of independence, χ2 = 8,583, P value <2.2e-16). Using χ2 tests of proportions between the individual annotation outputs to that of the reference annotation showed that LoReAn in stranded mode using a masked or nonmasked genome produces significantly improved intron location estimates than the current reference (Table 2). LoReAn stranded outperformed all other pipelines for exact intron specificity and sensitivity (Supplemental Table S9). These results indicate the LoReAn pipeline produces an improved annotation compared to the current reference and produces results as good and, under some metrics, better than other annotation options.
LoReAn Produces High-Quality Annotations for Larger Plant Genomes Using PacBio Iso-seq Data
To further test LoReAn, we reannotated the 135-megabase (Mb) Arabidopsis and 375-Mb rice (Oryza sativa) genomes using PacBio Iso-seq data. These genomes are larger and contain a higher percentage of repetitive elements than the two fungal genomes tested. The Arabidopsis annotations generated here were compared to the reference annotation, TAIR10, which is highly curated and represents one of the most complete plant genome annotations (Lamesch et al., 2012; Berardini et al., 2015). The LoReAn outputs using a nonmasked genome had the highest number of genes and transcripts exactly matching the reference, while BAP+ had the highest number of exact match exons (Supplemental Table S10). We conducted similar analyses as used for the fungal genomes for exact match genes compared to the reference annotation (Fig. 7, A and B). There was a significant difference across the pipelines for sensitivity and specificity of exact match genes (Fig. 7B; Pearson’s χ2 test of independence, χ2 = 22,393, P value <2.2e-16). Two-sample proportion testing showed LoReAn-sF outperformed MAKER2, GenMark-ES, and Augustus for predicting genes based on exact match analysis (Supplemental Table S11). Similar results were seen for the inferred intron analysis (Fig. 7, C and D). The pipelines were not equal for exact intron sensitivity and specificity (Fig. 7D; Pearson’s χ2 test of independence, χ2 = 38,926, P value <2.2e-16). The results of the pipelines were compared to those from the reference annotation, and LoReAn using a masked genome in standard or stranded mode and MAKER2 outperformed the current reference annotation for matching intron location specificity (Table 3). None of the pipelines outperformed the reference annotation for sensitivity, reflecting the high quality of the TAIR10 annotation.
Figure 7.
Assessment of gene and intron predictions from Arabidopsis. A, Annotation quality metrics are shown for exact-match genes as detailed for Figure 2A. LoReAn, LoReAn in nonstranded mode using a nonmasked genome; LoReAn-M, LoReAn in nonstranded mode using a masked genome; LoReAn-s, LoReAn in stranded mode using a nonmasked genome; LoReAn-sM, LoReAn in stranded mode using a masked genome; BAP, broad annotation pipeline; BAP+, broad annotation pipeline plus additional modifications described in text. B, The proportion of exact match to nonmatching gene predictions (specificity) and exact match to missing gene predictions (sensitivity) compared using a χ2 test of independence as described in Figure 2B. C and D are for exact match intron analysis, represented as in A and B, respectively.
Table 3. χ2 test of proportions for predicted exact match introns inferred from RNA-seq mapping data in Arabidopsis.
| Pipeline | Predictions Matching RNA-seq | Predictions Not Matching | Predictions Missing | Specificity Less Than Reference Annotationa | Sensitivity Less Than Reference Annotationb |
|---|---|---|---|---|---|
| LoReAn | 67,064 | 57,969 | 4,376 | N.S.c | N.S.c |
| LoReAn-M | 66,199 | 45,073 | 5,241 | <0.001 | N.S.c |
| LoReAn-s | 67,003 | 57,897 | 4,437 | N.S.c | N.S.c |
| LoReAn-sM | 66,133 | 44,987 | 5,307 | <0.001 | N.S.c |
| BAP | 64,444 | 59,868 | 6,996 | N.S.c | N.S.c |
| BAPplus | 65,037 | 59,153 | 6,403 | N.S.c | N.S.c |
| Augustus | 63,137 | 70,107 | 8,303 | N.S.c | N.S.c |
| GeneMark-ES | 61,815 | 80,017 | 9,625 | N.S.c | N.S.c |
| MAKER2 | 60,289 | 26,238 | 11,151 | <0.001 | N.S.c |
| Reference annotation | 69,657 | 54,910 | 1,783 | Not applicable | Not applicable |
Column reports P values for the χ2 test of proportions for the specificity metric
Column reports P values for the χ2 test of proportions for the sensitivity metric
N.S., not significant with a p-value greater than 0.05
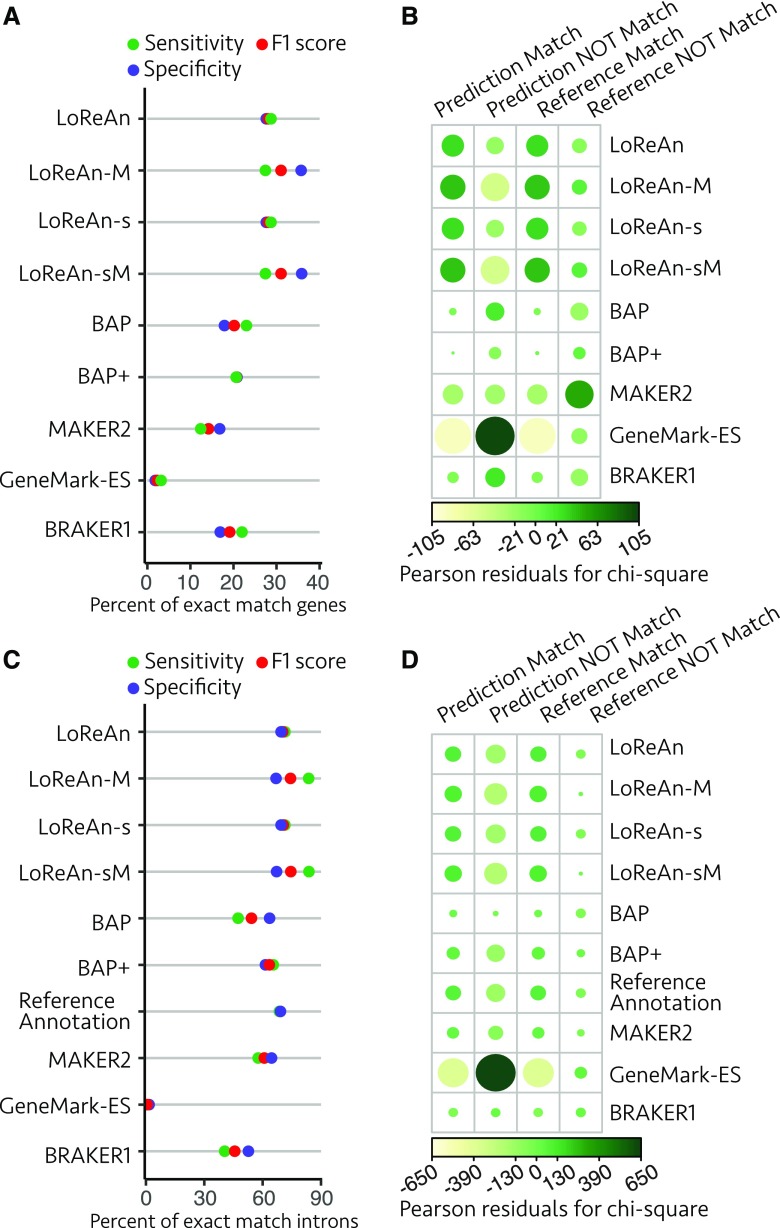
The same analysis procedures for the rice genome showed LoReAn performed as well as or better than the other tested pipelines for gene and intron predictions (Fig. 8; Supplemental Table S12). There was a significant difference between the pipelines for exact match gene sensitivity and specificity (Pearson’s χ2 test of independence, χ2 = 50,019, P value <2.2e-16) with the LoReAn outputs having strong associations to positive annotation metrics (Fig. 8B). Indeed, pairwise two-sample proportion testing of the other pipelines against LoReAn-s showed that LoReAn-s was significantly better for both gene prediction sensitivity and specificity (Supplemental Table S13). LoReAn also performed well for predicting exact intron locations (Fig. 8, C and D), and again, there was a significant difference across the pipelines for gene sensitivity and specificity (Pearson’s χ2 test of independence, χ2 = 984,030, P value <2.2e-16). All LoReAn pipelines outperformed the reference annotation for intron match specificity, and LoReAn using the full-genome and strand information outperformed the reference annotation for intron match sensitivity (Table 4). These data show that LoReAn provides robust results across genomes of varying features and sizes and, in many instances, outperformed other currently used annotation software.
Figure 8.
Assessment of gene and intron predictions from O. sativa. A, Annotation quality metrics are shown for exact match genes as detailed for Figure 2A. LoReAn, LoReAn in nonstranded mode using a nonmasked genome; LoReAn-M, LoReAn in nonstranded mode using a masked genome; LoReAn-s, LoReAn in stranded mode using a nonmasked genome; LoReAn-sM, LoReAn in stranded mode using a masked genome; BAP, broad annotation pipeline; BAP+, broad annotation pipeline plus additional modifications described in text. B, The proportion of exact match to nonmatching gene predictions (specificity) and exact match to missing gene predictions (sensitivity) compared using a χ2 test of independence as described in Figure 2B. C and D are for exact match intron analysis, represented as in A and B, respectively.
Table 4. χ2 test of proportions for predicted exact match introns inferred from RNA-seq mapping data in O. sativa.
| Pipeline | Predictions Matching RNA-seq | Predictions Not Matching | Predictions Missing | Specificity Less Than Reference Annotationa | Sensitivity Less Than Reference Annotationb |
|---|---|---|---|---|---|
| LoReAn | 84,679 | 33,790 | 37,383 | <0.001 | N.S.c |
| LoReAn-M | 81,707 | 15,929 | 40,355 | <0.001 | N.S.c |
| LoReAn-s | 84,880 | 33,749 | 37,182 | <0.001 | 0.007 |
| LoReAn-sM | 81,870 | 15,892 | 40,192 | <0.001 | N.S.c |
| BAP | 77,609 | 85,999 | 44,453 | N.S.c | N.S.c |
| BAPplus | 75,048 | 39,740 | 47,014 | N.S.c | N.S.c |
| Augustus | 64,279 | 94,836 | 57,783 | N.S.c | N.S.c |
| GeneMark-ES | 1,958 | 447,395 | 120,104 | N.S.c | N.S.c |
| MAKER2 | 78,819 | 57,887 | 43,243 | N.S.c | N.S.c |
| Reference annotation | 84,325 | 38,746 | 37,737 | Not applicable | Not applicable |
Column reports P values for the χ2 test of proportions for the specificity metric
Column reports P values for the χ2 test of proportions for the sensitivity metric
N.S., not significant with a P value greater than 0.05
DISCUSSION
High-throughput sequencing continues to have profound impacts on biological systems and the questions researchers are addressing. The technical improvements and associated reduction in cost have resulted in a deluge of high-quality model and nonmodel genomes from across the kingdoms of life. To capture the value of these assembled genomes, equal advances are needed in defining the functional elements of the genome. One such technical advance is the ability to sequence full-length single-molecule cDNAs that directly contain information on transcript structure and alternative forms. This information has previously helped identify alternatively spliced transcripts (Au et al., 2013; Abdel-Ghany et al., 2016), but single-molecule long-reads have not been systematically incorporated into annotation pipelines. The newly developed LoReAn pipeline integrates both short-read RNA-seq and long-read single-molecule cDNA sequencing with ab initio gene prediction to generate high-accuracy gene predictions. In total, three separate analyses using a reference annotation, head-to-head comparison, and comparison to empirical data indicate that LoReAn produces high-quality annotations of the four genomes tested. These results show that LoReAn has improved performance for predicting gene structures and intron locations.
Whereas several genome annotation tools use experimental data (i.e. RNA-seq) for gene prediction, none of them fully utilize this information. This is apparent for genes such as Ave1, where there is ample RNA-seq evidence supporting the gene model, but prediction software, including MAKER2, GeneMark-ES, and BAP, do not predict the gene. The annotation pipeline CodingQuarry also does not predict the Ave1 transcript when run in the default mode but does predict the transcript when run using the "effector" option. LoReAn correctly predicts the Ave1 transcript, plus an additional new transcript at the locus. The ability to correctly annotate genes with unique features or restricted taxonomic distribution, such as effectors, is relevant to many biological questions and will aid comparative genomic studies. LoReAn was designed to incorporate information from both short- and long-read RNA-seq data, as we believe with increasing sequencing depth, length, and accuracy, this significant source of empirical evidence will greatly improve gene prediction.
The technical and biological characteristics of a genome will impact the annotation options needed to achieve high-quality gene predictions. Genome masking significantly affected the gene prediction output of the V. dahliae annotation. From a technical aspect, genome masking prior to annotation can have a large impact when annotating highly contiguously assembled genomes. Fragmented genome assemblies often lack repetitive regions and are de facto masked. Masking the telomere-to-telomere complete V. dahliae strain JR2 genome resulted in gene predictions that were fragmented because of coding regions overlapping masked regions. Our results indicate that genome masking of short repetitive DNA decreases the quality of the genome annotation and that using a partially (only masking repeats >400 bp) or nonmasked genome may improve annotation results. From a biological perspective, our results show that strand information had a significant impact on annotation quality for the V. dahliae genome. Compact fungal genomes have genes with overlapping UTRs, which make gene prediction difficult. Using strand information, LoReAn can assign transcripts to the correct coding strand and avoid the prediction of fused genes. Additionally, strand information is used to assign single-exon genes to the correct strand. These results need to be confirmed on a greater number of genomes with diverse characteristics before being fully generalizable. Collectively, our results suggest that both technical and biological information, such as assembly completeness and coding sequence overlap, can impact genome annotation quality and should be considered early during project design.
Our results show that LoReAn can successfully use single-molecule cDNA sequencing data from different platforms to produce high-quality genome annotations, similar to or better than the current community references for four diverse genomes. This suggests robust performance of LoReAn across sequencing platforms and for annotating small fungal genomes of 35 Mb to the rice genome of ∼375 Mb. We speculate that the use of annotation software such as LoReAn, which incorporates single-molecule cDNA sequencing into the annotation process, will significantly improve genome annotation and aid in answering biological questions across all domains of life.
MATERIALS AND METHODS
Growth Conditions and RNA Extraction
Verticillium dahliae strain JR2 (Faino et al., 2015) was maintained on potato dextrose agar plates grown at approximately 22°C and stored in the dark. Conidiospores were collected from 2-week-old potato dextrose agar plates using half-strength potato dextrose broth (PDB), and subsequently 1 × 106 spores were inoculated in glass flasks containing 50 mL of either PDB, half-strength Murashige and Skoog (MS) medium supplemented with 3% Suc, or xylem sap collected from greenhouse-grown tomato (Solanum lycopersicum) plants of the cultivar Moneymaker. For analysis of Ave1 transcription, V. dahliae strain JR2 was additionally grown in 50 mL of Czapek-dox media following the manufacturer’s guidelines (Oxoid Microbiology Products, Thermo Scientific). The cultures were grown for 4 d in the dark at 22°C and 160 rpm. The cultures were strained through miracloth (22 μm; EMD Millipore), pressed to remove liquid, and flash frozen in liquid nitrogen. Next, the cultures were to ground to powder with a mortar and pestle using liquid nitrogen to ensure samples remained frozen.
RNA extraction was carried out using TRIzol (Thermo Fisher Scientific) following manufacturer guidelines. Following RNA resuspension, contaminating DNA was removed using the TURBO DNA-free kit (Ambion, Thermo Fisher Scientific), and the RNA was checked for integrity by separating 2 μL of each sample on a 2% agarose gel. RNA samples were quantified using a Nanodrop (Thermo Fisher Scientific) and stored at −80°C.
Library Preparation and Sequencing—Illumina
Each RNA sample from V. dahliae strain JR2 grown in PDB, half-strength MS, and xylem sap was used to construct an Illumina sequencing library for RNA-sequencing by the Beijing Genomics Institute following manufacturer guidelines (Illumina). In brief, mRNA was enriched using oligo(dT) magnetic beads. The RNA was then fragmented and double stranded cDNA synthesized following manufacturer guidelines (Illumina). The fragments were then end-repaired and poly-adenylated to allow for the addition of sequencing adapters, followed by fragment enrichment using PCR amplification. Library quality was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies). Qualified libraries were sequenced on an Illumina HiSeq-2000 (Illumina) at the Beijing Genomics Institute.
cDNA Synthesis and Normalization, Library Preparation, and Sequencing—ONT
For the synthesis of single-stranded cDNA, 1 μg of each RNA sample was reverse-transcribed using the Mint-2 cDNA synthesis kit as described by the manufacturer (Evrogen), using the primers PlugOligo-1 (5′ end) and CDS-1 (3′ end). For each sample, 1 μL of cDNA was amplified with PCR for 15 cycles (95°C for 15 s, 66°C for 20 s, and 72°C for 3 min) to generate double-stranded cDNA and purified with 1.8× volume Agencourt AMPure XP magnetic beads (Beckman Coulter).
Three cDNA samples were normalized with the Trimmer-2 cDNA normalization kit following the manufacturer’s guidelines (Evrogen). The cDNA was precipitated, denatured, and hybridized for 5 h. Next, the double-stranded cDNA fraction was cleaved and the remaining single stranded cDNA amplified with PCR for 18 cycles (95°C for 15 s, 66°C for 20 s, and 72°C for 3 min).
Library preparation for the three samples was performed using the Nanopore Sequencing Kit (v. SQK-MAP006) following the manufacturer’s guidelines (ONT). The cDNA was end-repaired and dA-tailed using the NEBNext End Repair and NEBNext dA-Tailing Modules following the manufacturer’s instructions (New England BioLabs [NEB]). The reactions were cleaned using an equal volume of Agencourt AMPure XP magnetic beads (Beckman Coulter), followed by ONT adapter ligation using Blunt/TA Ligation Master Mix (NEB). The adapter-ligated fragments were purified using Dynabeads MyOne Streptavidin C1 (Thermo Fisher Scientific).
Sequencing was performed on three different MinION flow cells (v. FLO-MAP103, ONT). After priming the flow cells with sequencing buffer, 6 μL of the library preparation was added. Additional library preparation (6 μL) was added to the flow cells at 3, 17, and 24 h after the run was started. Base-calling was performed using the Metrichor app (v. 2.39.1, ONT), and Poretools (v. 0.5.1) was used to generate FASTQ files from the Metrichor produced FAST5 files (Loman and Quinlan, 2014).
Software in LoReAn Pipeline
LoReAn is implemented in Python3. Usage and parameters to run LoReAn, including default settings, are detailed at https://github.com/lfaino/LoReAn/blob/master/OPTIONS.md. Mandatory parameters are protein sequences of related organisms, a reference genome sequence, and an identification name for the species form the Augustus database. Other inputs are as follows: short-reads (i.e. Illumina RNA-seq), which may be single or paired-end, and long-reads from either MinION or SMRT sequencing platforms. LoReAn outputs a GFF3 file with genome annotations.
The most convenient way to install and run LoReAn is by using the Docker (https://www.docker.com/) image. Information about the software and how to use it can be found at the https://github.com/lfaino/LoReAn repository. Additional information regarding the settings used for programs in this work can be found in the Supplemental Data. The following programs and versions were used for LoReAn: for read mapping, STAR (version 2.5.3a; Dobin et al., 2013) and GMAP (v. 2017-06-20; Wu and Watanabe, 2005); to assemble and reconstruct transcripts from short reads, Trinity (v. 2.2.0; Grabherr et al., 2011) run on “genome-guided mode,” followed by PASA (v. 2.1.0; Haas et al., 2008); to map protein sequences, AAT (v. 03-05-2011; Huang et al., 1997); for gene prediction, GeneMark-ES (v4.34; Lomsadze et al., 2014) and Augustus (v3.3; Stanke et al., 2008) as ab initio software but BRAKER1 (v. 2; Hoff et al., 2016) in substitution of Augustus to generate ab initio gene prediction for organisms not present in the Augustus catalog when RNA-seq is supplied; GMAP (v. 2017-06-20; Wu and Watanabe, 2005) for long-read mapping and for assembled ESTs after Trinity assembly; EVM (v. 1.1.1; Haas et al., 2008) to combine the output from the previous tools to generate a combined annotation model. For EVM, evidence weights were set to 1, and default options were used. Bedtools suite (v. 2.21.0; Quinlan and Hall, 2010) was used to extract the genomic sequence, merge, and cluster the long-reads. iAssembler (v. 1.32; Zheng et al., 2011) was used to call a consensus on the clusters (i.e. the process of transcript reconstruction). GenomeTools (v. 1.5.9) software was used at several stages in the LoReAn pipeline (Gremme et al., 2013). Additional information about the tools used can be found at https://github.com/lfaino/LoReAn/blob/master/README.md.
Genome Masking
To study the effect of genome masking on automated genome annotation with LoReAn, the pipeline was run on stranded mode using three reference genomes with different levels of repetition masking: a fully masked genome with all repetitive sequences masked, a partially masked genome where only repetitions larger than 400 bps were masked, and a full genome with no repetition masking. Repeats were masked using RepeatMasker software as previously described (Faino et al., 2016).
LoReAn Stranded Mode
To use the software in strand mode efficiently, sequences from the same transcript need to have the same strand. However, sequencing is random and, depending from which fragment sequencing starts, fragments from the same transcript could be sequenced in forward or reverse orientation compared to the transcription direction. Unlike in DNA sequencing, the direction of the cDNA long-read sequencing can be inferred by localizing only one or both directions between the 3′ adapter or the 5′ adapter used during the cDNA production. Using the Smith-Waterman alignment, the location of the adapter/s in the sequenced fragments can be identified and the sequencing orientation adjusted based on the adapter alignment onto the fragments. For the MinION data generated, the 5′ PlugOligo-1 AAGCAGTGGTATCAACGCAGAGTACGCGGG and 3′-CDS AAGCAGTGGTATCAACGCAGAGTACTGGAG primer sequences associated with the cDNA synthesis and normalization process were used to identify the coding strand for each long read. For the PacBio Arabidopsis (Arabidopsis thaliana) experiment, primers AAGCAGTGGTATCAACGCAGAGTACGCGGG and AAGCAGTGGTATCAACGCAGAGTACTTTTT were used for the correction of the transcript orientation. Rice (Oryza sativa) and Plicaturopsis crispa PacBio transcripts were oriented by using the sequence AAAAAAAAAAAAAAAAAAAAAAAAAAAAGTACTCTGCGTTGATACCACTGCTT.
Annotation Quality Definitions
The common metrics sensitivity, specificity, and accuracy were used to compare the annotation features. These metrics have been previously discussed in the context of annotations (Yandell and Ence, 2012). In brief, sensitivity is a measure of how well an annotation identifies the known features of a reference, also called a true positive rate. Here, sensitivity was calculated as ([Annotation matching reference/total Reference] * 100) for a specific feature of interest and represented the percentage of known reference features captured. Specificity is a measure of how many of the annotated features were in the reference, also called positive predictive value. Here, specificity was calculated as ([Annotation matching reference/total Annotation] * 100) for a specific feature of interest and represented the percentage of all the annotation features that matched the reference. These comparisons can be for any annotation feature, such as genes, transcripts, or individual exons for exact matches or for a specified overlap to a reference. The F1 score accounted for both sensitivity and specificity to measure annotation quality in a single number, calculated as the harmonic mean of sensitivity and specificity ([Sensitivity * Specificity]/[Sensitivity + Specificity]) * 2.
Head-to-Head Comparisons between Annotations
To determine the unique protein-coding genes annotated between LoReAn-sF, BAP-F, MAKER2, and CodingQuarry, the annotations were compared using orthoMCL (Li et al., 2003). OrthoMCL was downloaded from https://github.com/apetkau/orthomcl-pipeline and run using default settings.
Intron Analysis
Introns were extracted from mapped reads using the same methodology from BRAKER1 (Hoff et al., 2016). Introns supported from at least two reads were extracted and used in the intron set. Genome tool software (Gremme et al., 2013) was used to annotate introns in the gff3 file. Custom scripts were used to identify exact match intron coordinates from the annotation files that overlapped with the intron coordinates from the RNA-seq data. Sensitivity, specificity, and F1 score were calculated as described before.
Statistical Comparisons between Annotation Outputs
Statistical comparisons were made using the R software package (R Core Team, 2016). χ2 tests of independence (chisq.test in R) were computed to test for association between the annotation pipelines and the sensitivity and specificity metrics. The residuals of the test were used to assess the direction and magnitude of the associations across the data, but no formal post-hoc testing was performed on this data. Two-proportion z-tests (prop.test in R) were used to compare individual annotation results against the reference annotation or against a LoReAn output. These tests were conducted for both gene and intron features using the number of matched features and nonmatched features or the number of matched and missing features (i.e. specificity and sensitivity). The two-sample tests were conducted as one-tailed to determine the difference compared to the reference, and Bonferroni multiple testing correction was applied to adjust the P value needed to reject the null hypothesis. For singleton analysis, the read coverage and length data were compared using the nonparametric Kruskal-Wallis test (kruskal in R from the agricolae [de Mendiburu, 2016] package) to avoid the assumption of equal distribution and variance of the data. The proportion of highest-quality singletons from each pipeline were compared against the results of LoReAn-sF using the two-proportions z-test. The effect of genome masking, ab initio options and pipeline options were tested using ANOVA, and Tukey’s honestly significant post-hoc test (alpha = 0.05) was used to determine statistical grouping.
Ave1 Isoform Analysis
Ave1 isoforms were confirmed using cDNA-PCR of infected plant material with V. dahliae strain JR2. Specific primers for the Ave1 gene (F-TTTAACACTTCACTCTGCTCTCG; R-CCTTGTGTGCTGCTTTGGTA) and for Ave1c gene (F-CGCCGGCAATACTATCTCAA; R-ATCCTGTGGGCAACAATAGC) were used to identify the two Ave1 isoforms.
Availability
The LoReAn source code is available at https://github.com/lfaino/LoReAn/ and provided under an MIT license, available at https://github.com/lfaino/LoReAn/blob/master/LICENSE. Documentation and software are available at https://github.com/lfaino/LoReAn. The software can run on all platforms when deployed via Docker (https://www.docker.com/).
All genome annotations, scripts, and additional files generated and/or analyzed in the paper can be found at https://github.com/lfaino/files-paper-LoReAn.git.
A dataset to test the correct installation of the tool can be found at https://github.com/lfaino/LoReAn-Example.git. This dataset contains all the data to annotate a single chromosome of V. dahliae strain JR2.
Accession Numbers
The V. dahliae strain JR2 reference annotation version 5 was used in the analysis. Version 5 was generated by comparing the concordance of all gene models of version 4 with the long-read information. Subsequently, the improved version 5 was deposited at ENSEMBL fungi database and can be downloaded at http://fungi.ensembl.org/Verticillium_dahliaejr2/Info/Index.
The P. crispa reference genome and annotation were downloaded from JGI (http://genome.jgi.doe.gov/pages/dynamicOrganismDownload.jsf?organism=Plicr1). The Arabidopsis genome sequence and reference annotation were downloaded from the TAIR database (ftp://ftp.arabidopsis.org/home/tair/Sequences/whole_chromosomes/; https://www.arabidopsis.org/download_files/Genes/TAIR10_genome_release/TAIR10_gff3/TAIR10_GFF3_genes.gff). The rice genome sequence and annotation were retrieved from the ENSEMBL plant database (http://plants.ensembl.org/Oryza_sativa/Info/Index). The sequencing data are accessible at the NCBI SRA database. The short-read Arabidopsis dataset is deposited under SRA accession number SRR5446746 and the PacBio dataset under SRA accession number SRR5445910. The V. dahliae Illumina transcriptome is deposited under accession number SRR5440696, while the Nanopore transcriptome data are deposited as SRR5445874. The P. crispa PacBio reads were downloaded from the publicly accessible NCBI SRA site, runs SRR5077068 to SRR5077144 and Illumina data from run SRR1577770. The O. sativa data were downloaded from the European Nucleotide Archive under runs ERR91110 and ERR911111 and the Illumina data from run ERR748773.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Annotation quality metrics and summary statistics for predicted genes
Supplemental Figure S2. Annotation quality metrics and summary statistics for predicted exons
Supplemental Figure S3. Ave1 gene model predictions from different software used during annotation
Supplemental Table S1. Number of predicted genes and exact match predictions to the JR2 reference
Supplemental Table S2. Number of predicted exons and exact match predictions to the JR2 reference
Supplemental Table S3. Gene prediction summaries for annotation options
Supplemental Table S4. Exon prediction summaries for annotation options
Supplemental Table S5. Two-sample test of proportions for high-confidence singleton predictions
Supplemental Table S6. Two-samples test of proportions for exact match intron based on empirical RNA-seq intron identification in V. dahliae
Supplemental Table S7. Predicted features for P. crispa annotation analysis
Supplemental Table S8. P. crispa χ2 test of exact match gene proportions against LoReAn_sF
Supplemental Table S9. P. crispa χ2 test of exact match intron proportions against LoReAn_sF
Supplemental Table S10. Predicted features for Arabidopsis annotation analysis
Supplemental Table S11. Arabidopsis χ2 test of exact match gene proportions against LoReAn_s
Supplemental Table S12. Predicted features for O. sativa annotation analysis
Supplemental Table S13. O. sativa χ2 test of exact match gene proportions against LoReAn_s
Acknowledgments
We thank Jordi Coolen for his assistance in writing the LoReAn software and Michael Seidl for reading the manuscript.
Footnotes
Funding was provided to D.E.C. by the Human Frontiers in Science Program (HFSP) long-term fellowship (LT000627/2014-L). Work in the laboratory of B.P.H.J.T. is supported by a Vici grant of the Research Council for Earth and Life Sciences (ALW) of the Netherlands Organization for Scientific Research (NWO). The Arabidopsis long-read sequencing was performed within the ZonMw-project number 435003020 entitled “Arabidopsis transcript isoform identification using PacBio sequencing technology.”
Articles can be viewed without a subscription.
Senior author.
References
- Abdel-Ghany SE, Hamilton M, Jacobi JL, Ngam P, Devitt N, Schilkey F, Ben-Hur A, Reddy ASN (2016) A survey of the sorghum transcriptome using single-molecule long reads. Nat Commun 7: 11706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amemiya CT, Alföldi J, Lee AP, Fan S, Philippe H, Maccallum I, Braasch I, Manousaki T, Schneider I, Rohner N, et al. (2013) The African coelacanth genome provides insights into tetrapod evolution. Nature 496: 311–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au KF, Sebastiano V, Afshar PT, Durruthy JD, Lee L, Williams BA, van Bakel H, Schadt EE, Reijo-Pera RA, Underwood JG, et al. (2013) Characterization of the human ESC transcriptome by hybrid sequencing. Proc Natl Acad Sci USA 110: E4821–E4830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini TZ, Reiser L, Li D, Mezheritsky Y, Muller R, Strait E, Huala E (2015) The Arabidopsis information resource: making and mining the “gold standard” annotated reference plant genome. Genesis 53: 474–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Korf I, Robb SMC, Parra G, Ross E, Moore B, Holt C, Sánchez Alvarado A, Yandell M (2008) MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res 18: 188–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K-L, Rosli R, Tatarinova TV, Hogan M, Firdaus-Raih M, Low EL (2017) Seqping: gene prediction pipeline for plant genomes using self-training gene models and transcriptomic data. BMC Bioinformatics 18(Suppl 1): 1426–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Mackey AJ, Stoeckert CJ Jr., Roos DS (2006) OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res 34: D363–D368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C-S, Peluso P, Sedlazeck FJ, Nattestad M, Concepcion GT, Clum A, Dunn C, O’Malley R, Figueroa-Balderas R, Morales-Cruz A, et al. (2016) Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods 13: 1050–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DE, Mesarich CH, Thomma BPHJ (2015) Understanding plant immunity as a surveillance system to detect invasion. Annu Rev Phytopathol 53: 541–563 [DOI] [PubMed] [Google Scholar]
- Davey JW, Chouteau M, Barker SL, Maroja L, Baxter SW, Simpson F, Merrill RM, Joron M, Mallet J, Dasmahapatra KK, et al. (2016) Major improvements to the Heliconius melpomene genome assembly used to confirm 10 chromosome fusion events in 6 million years of butterfly evolution. G3 (Bethesda) 6: 695–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge R, van Esse HP, Maruthachalam K, Bolton MD, Santhanam P, Saber MK, Zhang Z, Usami T, Lievens B, Subbarao KV, Thomma BP (2012) Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc Natl Acad Sci USA 109: 5110–5115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mendiburu F. (2016) agricolae: Statistical procedures for agricultural research.https://CRAN.R-project.org/package=agricolae
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faino L, Thomma BPHJ (2014) Get your high-quality low-cost genome sequence. Trends Plant Sci 19: 288–291 [DOI] [PubMed] [Google Scholar]
- Faino L, Seidl MF, Datema E, van den Berg GCM, Janssen A, Wittenberg AHJ, Thomma BPHJ (2015) Single-molecule real-time sequencing combined with optical mapping yields completely finished fungal genome. MBio 6: e00936-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faino L, Seidl MF, Shi-Kunne X, Pauper M, van den Berg GCM, Wittenberg AHJ, Thomma BPHJ (2016) Transposons passively and actively contribute to evolution of the two-speed genome of a fungal pathogen. Genome Res 26: 1091–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin EF, Thomma BPHJ (2006) Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol Plant Pathol 7: 71–86 [DOI] [PubMed] [Google Scholar]
- Goodswen SJ, Kennedy PJ, Ellis JT (2012) Evaluating high-throughput ab initio gene finders to discover proteins encoded in eukaryotic pathogen genomes missed by laboratory techniques. PLoS One 7: e50609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon SP, Tseng E, Salamov A, Zhang J, Meng X, Zhao Z, Kang D, Underwood J, Grigoriev IV, Figueroa M, et al. (2015) Widespread polycistronic transcripts in fungi revealed by single-molecule mRNA sequencing. PLoS One 10: e0132628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremme G, Steinbiss S, Kurtz S (2013) GenomeTools: a comprehensive software library for efficient processing of structured genome annotations. IEEE/ACM Trans Comput Biol Bioinform 10: 645–656 [DOI] [PubMed] [Google Scholar]
- Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, Orvis J, White O, Buell CR, Wortman JR (2008) Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol 9: R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Zeng Q, Pearson MD, Cuomo CA, Wortman JR (2011) Approaches to fungal genome annotation. Mycology 2: 118–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff KJ, Lange S, Lomsadze A, Borodovsky M, Stanke M (2016) BRAKER1: unsupervised RNA-Seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics 32: 767–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C, Yandell M (2011) MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12: 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Adams MD, Zhou H, Kerlavage AR (1997) A tool for analyzing and annotating genomic sequences. Genomics 46: 37–45 [DOI] [PubMed] [Google Scholar]
- Jiao W-B, Schneeberger K (2017) The impact of third generation genomic technologies on plant genome assembly. Curr Opin Plant Biol 36: 64–70 [DOI] [PubMed] [Google Scholar]
- Keibler E, Brent MR (2003) Eval: a software package for analysis of genome annotations. BMC Bioinformatics 4: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klosterman SJ, Atallah ZK, Vallad GE, Subbarao KV (2009) Diversity, pathogenicity, and management of verticillium species. Annu Rev Phytopathol 47: 39–62 [DOI] [PubMed] [Google Scholar]
- Kohler A, Kuo A, Nagy LG, Morin E, Barry KW, Buscot F, Canbäck B, Choi C, Cichocki N, Clum A, et al. ; Mycorrhizal Genomics Initiative Consortium (2015) Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat Genet 47: 410–415 [DOI] [PubMed] [Google Scholar]
- Koren S, Phillippy AM (2015) One chromosome, one contig: complete microbial genomes from long-read sequencing and assembly. Curr Opin Microbiol 23: 110–120 [DOI] [PubMed] [Google Scholar]
- Križanovic K, Echchiki A, Roux J, Šikic M (2018) Evaluation of tools for long read RNA-seq splice-aware alignment. Bioinformatics 34: 748–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laehnemann D, Borkhardt A, McHardy AC (2016) Denoising DNA deep sequencing data-high-throughput sequencing errors and their correction. Brief Bioinform 17: 154–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, et al. (2012) The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res 40: D1202–D1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhaney S, Fan G, Widemo F, Gunnarsson U, Thalmann DS, Hoeppner MP, Kerje S, Gustafson U, Shi C, Zhang H, et al. (2016) Structural genomic changes underlie alternative reproductive strategies in the ruff (Philomachus pugnax). Nat Genet 48: 84–88 [DOI] [PubMed] [Google Scholar]
- Laver T, Harrison J, O’Neill PA, Moore K, Farbos A, Paszkiewicz K, Studholme DJ (2015) Assessing the performance of the Oxford Nanopore Technologies MinION. Biomol Detect Quantif 3: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ Jr., Roos DS (2003) OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13: 2178–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linde J, Duggan S, Weber M, Horn F, Sieber P, Hellwig D, Riege K, Marz M, Martin R, Guthke R, et al. (2015) Defining the transcriptomic landscape of Candida glabrata by RNA-Seq. Nucleic Acids Res 43: 1392–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loman NJ, Quinlan AR (2014) Poretools: a toolkit for analyzing nanopore sequence data. Bioinformatics 30: 3399–3401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loman NJ, Quick J, Simpson JT (2015) A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat Methods 12: 733–735 [DOI] [PubMed] [Google Scholar]
- Lomsadze A, Burns PD, Borodovsky M (2014) Integration of mapped RNA-Seq reads into automatic training of eukaryotic gene finding algorithm. Nucleic Acids Res 42: e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Presti L, Lanver D, Schweizer G, Tanaka S, Liang L, Tollot M, Zuccaro A, Reissmann S, Kahmann R (2015) Fungal effectors and plant susceptibility. Annu Rev Plant Biol 66: 513–545 [DOI] [PubMed] [Google Scholar]
- Ma L, Chen Z, Huang W, Kutty G, Ishihara M, Wang H, Abouelleil A, Bishop L, Davey E, Deng R, et al. (2016) Genome analysis of three Pneumocystis species reveals adaptation mechanisms to life exclusively in mammalian hosts. Nat Commun 7: 10740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R, VanBuren R, Wai CM, Tang H, Schatz MC, Bowers JE, Lyons E, Wang M-L, Chen J, Biggers E, et al. (2015) The pineapple genome and the evolution of CAM photosynthesis. Nat Genet 47: 1435–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoche AE, Dohm JC, Schneider J, Holtgräwe D, Viehöver P, Montfort M, Sörensen TR, Weisshaar B, Himmelbauer H (2015) Exploiting single-molecule transcript sequencing for eukaryotic gene prediction. Genome Biol 16: 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz JF, Gauthier GM, Desjardins CA, Gallo JE, Holder J, Sullivan TD, Marty AJ, Carmen JC, Chen Z, Ding L, et al. (2015) The dynamic genome and transcriptome of the human fungal pathogen blastomyces and close relative emmonsia. PLoS Genet 11: e1005493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillippy AM. (2017) New advances in sequence assembly. Genome Res 27: xi–xiii [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2016) R: A Language and Environment for Statistical Computing.https://www.R-project.org/
- Reid I, O’Toole N, Zabaneh O, Nourzadeh R, Dahdouli M, Abdellateef M, Gordon PMK, Soh J, Butler G, Sensen CW, et al. (2014) SnowyOwl: accurate prediction of fungal genes by using RNA-Seq and homology information to select among ab initio models. BMC Bioinformatics 15: 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CD, Zimin A, Holt C, Abouheif E, Benton R, Cash E, Croset V, Currie CR, Elhaik E, Elsik CG, et al. (2011) Draft genome of the globally widespread and invasive Argentine ant (Linepithema humile). Proc Natl Acad Sci USA 108: 5673–5678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Kuraku S, Holt C, Sauka-Spengler T, Jiang N, Campbell MS, Yandell MD, Manousaki T, Meyer A, Bloom OE, et al. (2013) Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat Genet 45: 415–421, 421e1–421e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperschneider J, Dodds PN, Gardiner DM, Manners JM, Singh KB, Taylor JM (2015) Advances and challenges in computational prediction of effectors from plant pathogenic fungi. PLoS Pathog 11: e1004806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Diekhans M, Baertsch R, Haussler D (2008) Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24: 637–644 [DOI] [PubMed] [Google Scholar]
- Ter-Hovhannisyan V, Lomsadze A, Chernoff YO, Borodovsky M (2008) Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res 18: 1979–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa AC, Hane JK, Ellwood SR, Oliver RP (2015) CodingQuarry: highly accurate hidden Markov model gene prediction in fungal genomes using RNA-seq transcripts. BMC Genomics 16: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BPHJ, Seidl MF, Shi-Kunne X, Cook DE, Bolton MD, van Kan JAL, Faino L (2016) Mind the gap; seven reasons to close fragmented genome assemblies. Fungal Genet Biol 90: 24–30 [DOI] [PubMed] [Google Scholar]
- Wang B, Tseng E, Regulski M, Clark TA, Hon T, Jiao Y, Lu Z, Olson A, Stein JC, Ware D (2016) Unveiling the complexity of the maize transcriptome by single-molecule long-read sequencing. Nat Commun 7: 11708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Watanabe CK (2005) GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21: 1859–1875 [DOI] [PubMed] [Google Scholar]
- Yandell M, Ence D (2012) A beginner’s guide to eukaryotic genome annotation. Nat Rev Genet 13: 329–342 [DOI] [PubMed] [Google Scholar]
- Zheng Y, Zhao L, Gao J, Fei Z (2011) iAssembler: a package for de novo assembly of Roche-454/Sanger transcriptome sequences. BMC Bioinformatics 12: 453. [DOI] [PMC free article] [PubMed] [Google Scholar]