Summary
In the developing pancreas, transient Neurog3-expressing progenitors give rise to four major islet-cell types, α, β, δ, and γ; when and how the Neurog3+ cells choose cell-fate is unknown. Using single-cell RNAseq, trajectory analysis, and combinatorial lineage tracing, we showed here that the Neurog3+ cells co-expressing Myt1 (i.e., Myt1+Neurog3+) were biased towards β-cell fate; while those not simultaneously expressing Myt1 (Myt1-Neurog3+) favored α-fate. Myt1 manipulation only marginally affected α- vs. β-cell specification, suggesting Myt1 as a marker but not determinant for islet-cell type specification. The Myt1+Neurog3+ cells displayed higher Dnmt1 expression and enhancer methylation at Arx, an α-fate-promoting gene. Inhibiting Dnmts in pancreatic progenitors promoted α-cell specification, while Dnmt1 over-expression or Arx enhancer hyper-methylation favored β-cell production. Moreover, the pancreatic progenitors contained distinct Arx enhancer methylation states without transcriptionally definable sub-populations, a phenotype independent of Neurog3 activity. These data suggest that Neurog3-independent methylation on fate-determining gene-enhancers specifies distinct endocrine-cell programs.
eTOC:
During embryogenesis, seemingly equivalent progenitors produce distinct cell types. The mechanisms driving this divergence are unknown. Liu et al. show that the pancreatic progenitors before endocrine determination are transcriptionally uniform but epigenetically distinct pools with different DNA methylation in gene enhancers. This epigenetic variation biases progenitors toward specific islet-cell fate.
Introduction
Producing specialized cell types from common progenitors is a fundamental process in organ development and regeneration. Although deterministic cues and stochastic gene expression are known to govern cell-type specification in neurogenesis and retinogenesis (Doe, 2017; Wang and Cepko, 2016), the mechanisms regulating cell specification in most other organs remain poorly understood. These include the pancreatic endocrine islets, which comprise four major cell types – the α, β, δ, and γ cells that secrete glucagon (Gcg), insulin (Ins), somatostatin (Sst), pancreatic polypeptide (Ppy), respectively, and a minor ε cell type that secrete ghrelin (Ghrl) (Bastidas-Ponce et al., 2017; Jennings et al., 2015; Larsen and Grapin-Botton, 2017; Romer and Sussel, 2015).
During embryogenesis, a group of seemingly equivalent pancreatic progenitor cells (PPCs) transiently activate Neurog3 to differentiate into hormone+ islet cells. This need the function of a set of transcription factors (TFs), including those depend on Neurog3 (e.g. Arx, NeuroD1, Pax4, Pax6, and Runx1t1), those do not (e.g. Glis3 and Prdm16), and those depends on Neurog3 for high but not for low expression (e.g. Nkx2.2, Nkx6.1, Pdx1, and Rfx6) [(Romer and Sussel, 2015) and references therein]. Inactivating any of these above genes perturbs islet-cell allocation but does not eliminate any single cell type, highlighting the combinatorial nature of endocrine islet cell-type specification.
In mice, endocrine differentiation preferentially gives rise to α cells before E12, followed by α, β, and γ cells between E12.5 to E16.5, and then δ and γ cells afterwards (Johansson et al., 2007; Kopp et al., 2011). This temporal competence depends on the properties of the PPCs (Johansson et al., 2007). These findings, combined with the fact that each Neurog3+ cell produces one islet-cell type (Desgraz and Herrera, 2009), suggest that Neurog3+ progenitors are differentially configured to adopt a particular fate. Indeed, Neurog3+ progenitors were found to heterogeneously express a variety of TFs, including Myt1, Rfx6 and Nkx6.1 (Nelson et al., 2007; Smith et al., 2010; Soyer et al., 2010; Wang et al., 2007). The origin and functional significance of the Neurog3+ cell heterogeneity, however, remain unknown.
Cell heterogeneity can originate from stochastic gene expression or predetermined events (Simon et al., 2018). In the former case, identical cells can produce variable levels of proteins due to stochastic interactions between TFs and DNA elements or mRNA and ribosome (Dar et al., 2012; Harper et al., 2011; Raj and van Oudenaarden, 2008; Salas et al., 2016; Tang et al., 2011). In the latter model, differential environmental cues, cell cycle, and asymmetry of cell division can alter the genetic and/or epigenetic landscapes of otherwise identical cells. In both cases, challenge remains to determine if the differentially expressed factors have functional consequences of only represent biological noise.
Using islet cells as examples, the transient presence of the TFs in a subset of progenitors could initiate fate choice. By far, none of the tested genes fit into this initiator role. For example, while high Nkx6.1 is only detected in a subset of Neurog3+ cells, its transient over-expression in all Neurog3+ cells did not re-allocate cell type (Nelson et al., 2007) and only sustained Nkx6.1 overexpression favored the production of β over α cells (Schaffer et al., 2013). These results, corroborated by recent findings that Nkx6.1-negative PPCs activate Neurog3 and then Nkx6.1 to produce insulin+ cells (Petersen et al., 2017), suggest that Nkx6.1 promotes β-cell (trans)differentiation rather than specification. Sustained overexpression of Arx, Pax4, and Pdx1 in pancreatic or endocrine progenitors can also alter islet cell-type allocation (Collombat et al., 2007; Collombat et al., 2009; Yang et al., 2011). Yet, Arx and Pax4 were detected in the same set of endocrine progenitors, suggesting unknown mechanisms to resolve Arx+Pax4+ progenitors to different lineages (Collombat et al., 2005; Collombat et al., 2003). Pdx1 overexpression in endocrine progenitors promotes β-like cell production via trans differentiation (Yang et al., 2011). Thus, other instructive mechanisms that funnel portions of the Neurog3+ cells towards different islet-cell types remain to be discovered.
Epigenetic regulators could direct cell-type specification by enabling future TF binding and transcriptional modulation (Xie et al., 2015). A particularly relevant finding is that β-cell fate maintenance depends on DNA methylation, a process modulated by DNA methyltransferases (Dnmts, with Dnmt1, Dnmt3a, and Dnmt3b) and demethylating enzymes (Tets, short for Ten Eleven Translocation with three paralogs, Tet1, Tet2, and Tet3) (Wu and Zhang, 2017). DNA methylation of Arx enhancers, including UR2, represses its expression in islet cells to maintain β-cell fate (Dhawan et al., 2011). Moreover, Nkx2.2, a key determinant of β-cell differentiation and maintenance (Gutierrez et al., 2017; Mastracci et al., 2011; Sussel et al., 1998; Wang et al., 2004), was shown to recruit Dnmt3a to the Arx promoter to repress its expression, suggesting Nkx2.2-Dnmt3a-mediated DNA methylation in β-cell fate decision and/or maintenance (Papizan et al., 2011).
Our previous studies showed that although Myt1 expression is eventually activated in all differentiated hormone+ islet cells, the transient Neurog3+ progenitors in produced different Myt1 levels (Wang et al., 2007). Here we dissect the functional implication of the heterogeneous Myt1 production in Neurog3+ cells. We identify the earliest detectable mark that divides the PPCs into functionally distinct sub-populations and further uncover different epigenetic makeups developed in PPCs in the absence of Neurog3 as an initiator that primes progenitors toward specific islet-cell lineages.
Results
Increased proportion of Myt1-expressing Neurog3+ endocrine progenitors correlated with β-cell production
At E10.5, when most newly derived islet cells become glucagon-expressing α cells, ~8% Neurog3+ cells expressed Myt1; at E14.5 and E16.5, when β and γ cells are produced in prominent numbers, Myt1 was detected in >40% Neurog3+ cells (Figure S1A-C). Interestingly, high levels of Pax6 and Pdx1, other Neurog3 targets, were barely detectable in Neurog3+ cells at any stage examined (Figure S1C-E). These results are consistent with the idea that Myt1 expression in a subset of Neurog3+ cells is correlated with birth of specific islet cell types.
Trajectory analysis revealed β-cell biasing of Myt1-expressing endocrine progenitors
We used single-cell RNA-seq (scRNA-seq) and trajectory analysis to compare the transcriptome and potential fates of Myt1+Neurog3+ (referred to as M+N+ hereafter) and Myt1-Neurog3+ (referred to as M-N+ hereafter) cells. High-quality expression profiles were obtained for 1,635 flow-sorted eGFP+ cells from E14.5 Neurog3eGFP/+ pancreata in two biological replicates (Figure S1F-H). Because eGFP is long-lived, the Neurog3eGFP positive cells include both endocrine progenitors and newly differentiated islet cells. Leveraging the existence of these cell derivatives allowed us to construct trajectories from progenitors to differentiated cells.
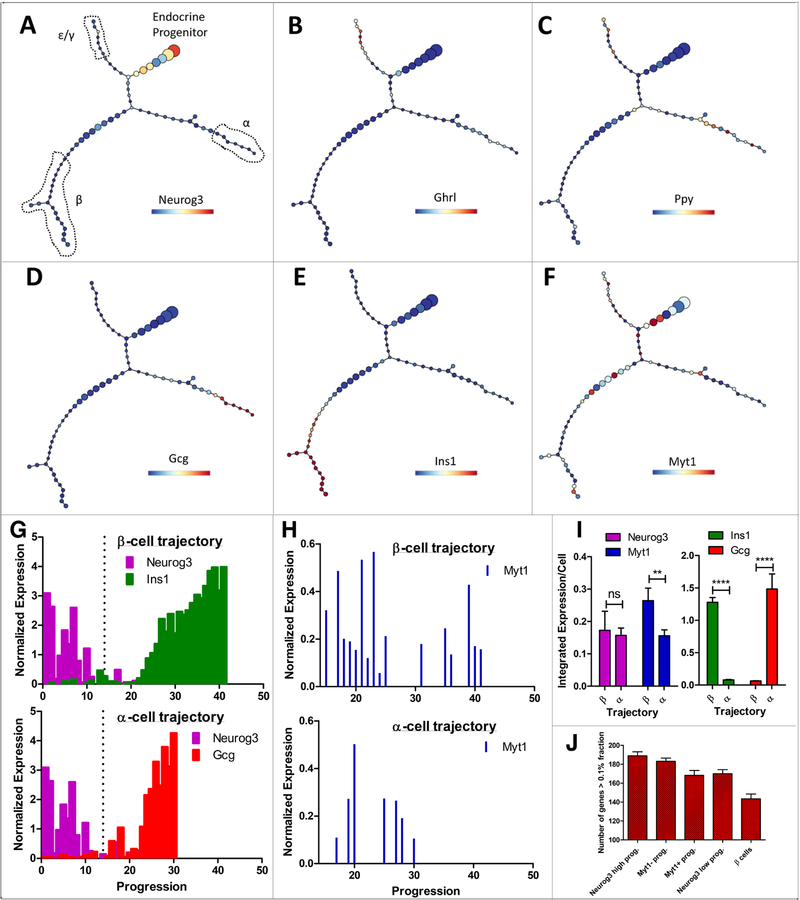
After initial quality control, processing, and gating on islet cells (Figure S1I), unsupervised p-Creode trajectory analysis revealed four cell populations connected by a multi-branching trajectory (Figure 1A and Figure S1J). Supporting published results (Johansson et al., 2007; Prado et al., 2004), an endocrine progenitor pool with high Neurog3 transcription was observed to first branch into a mixed pool of Ghrl+ ε and Ppy+ γ cells (Figure, 1A-C). Progenitors then further branched into Gcg+ α cells (Figure 1D) and Ins1+/Ins2+ β cells (Figure 1E). Because δ cells are rare at E14.5, the absence of enough intermediate cell states prevented making relevant trajectory connections and led to their exclusion (Figure S1J5).
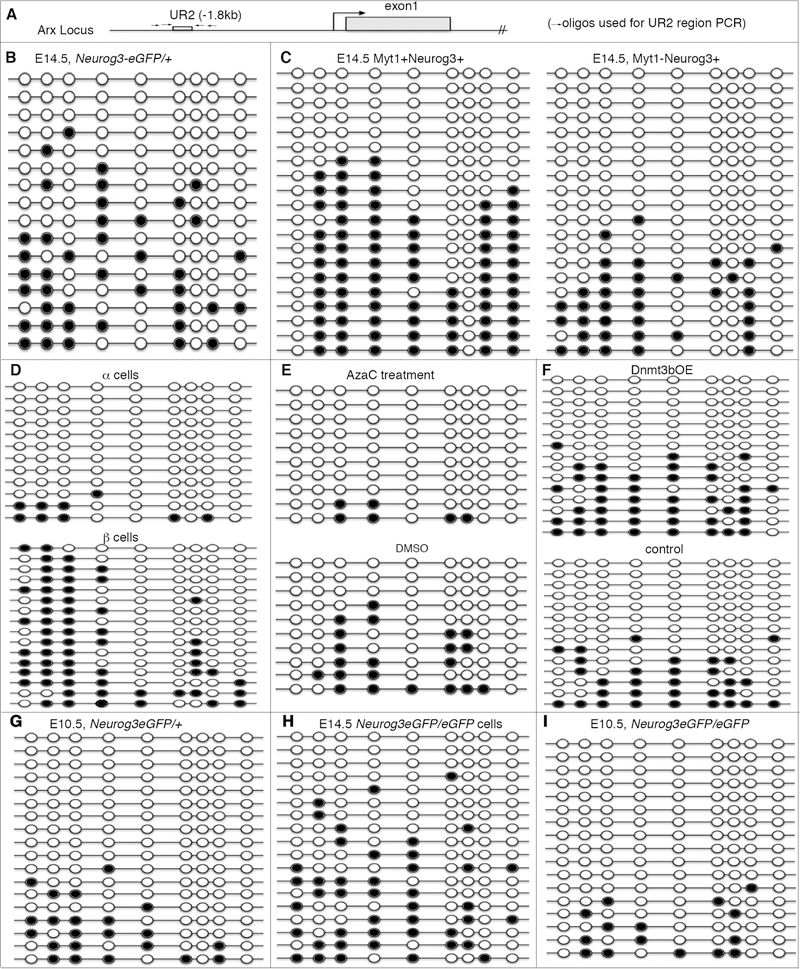
Figure 1. Single-cell gene expression projects Myt1-expressing endocrine progenitors preferentially towards the β-cell fate.
(A-F) p-Creode trajectory analysis of scRNA-seq data from E14.5 PPCs depicting α-, β-, ε-, and γ-cell differentiation from Neurog3+ progenitors. Overlays represent gene expression levels on a variance normalized (Asinh) scale. Shown are representative graphs from N=100 resampled runs on n=2 biological replicates. (G) Expression of Neurog3, Ins1, and Gcg plotted as a function of progression on p-Creode trajectories in A. X-axis represents cell states traversed from progenitor to differentiated cell states. Y-axis represents gene expression levels on a variance normalized (Asinh) scale. Dotted lines represent the bifurcation point from a common progenitor (left) and cells projecting into β- (top) or α-branches (bottom) (right). (H) Myt1 expression in β- (top, 306 cells) and α- (bottom, 114 cells) trajectory as they exit the common progenitor pool and progress toward β- and α-cell fates. (I) Integrated expression of selected markers in cells of α- or β-biased trajectories, presented as per cell levels only including cells after the bifurcation point. Error bars, SEM from n=5 p-Creode trajectories generated by resampled runs. ** p<0.01, **** p<0.0001 by t-test. (J) Number of genes expressed in different sub-cell populations at high levels. Only genes with levels >0.1% of the total transcripts were counted. Error bars, SEM. Also see Figure S1 and Table S1.
p-Creode allows for pseudo-temporal ordering that depicts progression relationships (Herring et al., 2018). This analysis revealed that progenitors progressively lose Neurog3 expression while gaining β- or α-cell features in two separate trajectories (Figure 1G). By comparing the transient Neurog3+ progenitors of the β- with the α-trajectory, we observed enriched Myt1 transcripts in the β-cell trajectory (Figure 1F, H, I), consistent with the idea that Neurog3+ endocrine progenitor cells with higher Myt1 expression are biased towards β-cell fate. To corroborate this conclusion, we examined the gene expression complexity of each subset of progenitor cells. As progenitors differentiate into specialized cell type, their number of expressed genes tends to decrease (Wang et al., 2011). The number of threshold genes (e.g., those accounting for more than 0.1% fraction of total transcript detected), progressively decreased as Neurog3+ progenitors transition into Neurog3- cells and finally to β cells (Figure 1J). Similarly, when we parsed progenitors into Myt1+ and Myt1- biased populations, the number of threshold genes decreased as well (Figure 1J), consistent with them being partitioned into specialized fate.
Biased emergence of β-cells from the Myt1+Neurog3+ pool
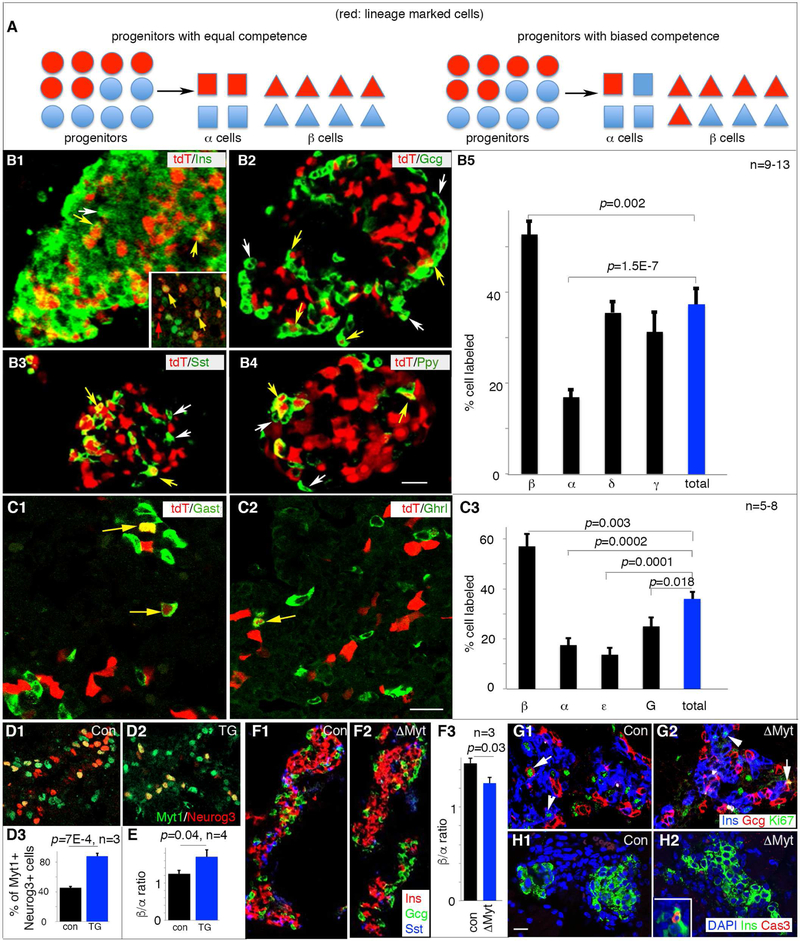
The fates of Myt1+Neurog3+ progenitors was examined using a bipartite-Cre-based lineage tracing (Xu et al., 2007). Knock-in or BAC-based transgenic mice were derived to drive expression of inactive nCre or cCre in Neurog3+ (Neurog3nCre) or Myt1+ cells (Myt1cCre), respectively. Cre reconstituted from the co-expressed nCre and cCre in M+N+ cells irreversibly activates tdTomato (tdT) expression in the Ai9 [Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J] Cre-reporter mice for lineage tracing. The Myt1cCre; Neurog3nCre; Ai9 (hereafter “MNA”) mice showed no detectable defects in pancreatic structure, β-cell mass, blood glucose level, and glucose-clearing capability (p=0.36) (Figure S2A-D).
We quantified the relative proportions of α/β/δ/γ cells derived from the M+N+ cells in P1 (postnatal day 1) MNA mice. The ε cells were not examined because they are rare in postnatal stages. If M+N+ and M-N+ cells were functionally identical, we expected the same proportion of each cell type to come from M+N+ progenitors, assayed by the proportion of each cell type expressing tdT (Figure 2A). If M+N+ progenitors were biased for a particular cell type, we expected that a higher portion of that particular cell-type to express tdT than the overall islet cell population would do (Figure 2A). In P1 MNA pancreata, we obtained a ~38% total islet cell marking with tdT activation (Figure 2B), in line with the 40–50% Neurog3+ cells that co-express Myt1 (Figure S1C). This finding suggests that nCre produced in the transient Neurog3+ cells were not retained over long-period of time. Otherwise, the nCre retained long-term in the Myt1-Neurog3+ cells would have recombined with cCre produced later to label most of the endocrine islet cells, because all hormone+ cells eventually activate Myt1.
Figure 2. The Myt1+Neurog3+ progenitors are biased toward β-cell fate with Myt1 serving marginal instructive roles.
For all panels with quantification, marked p-values were from t-Test. (A) Diagrammatic explanation of the quantification process. The Myt1+Neurog3+ islet progenitors cells were marked as red (tdT). If the Myt1+ (red dots) and Myt1- (blue dots) Neurog3+ progenitors have equal competence, we expect the portion of each islet cell type expressing tdT to be equal (left half of panel A). If the Myt1+Neurog3+ progenitors are biased toward one cell fate, e.g., β-cell fate, the portion of tdT+ β cells will be over-represented. (B1-5) Hormone staining of tdT+ islets and quantification of tdT+ islet cell types at P1. White arrows, hormone-tdT+ cells. Yellow arrows, hormone+tdT+ cells. Inset in B1, Pdx1 staining in several tdT+ cells. Scale bar, 20 μm. Panel B5 showed (mean + SEM). (C1-3) Hormone staining in tdT+ islets and quantification at E16.5. Yellow arrows, hormone+tdT+ cells. Scale bar, 20 μm. Panel C3 showed (mean + SEM). (D1-3) Myt1-Neurog3 co-expression in Neurog3Myt1OE transgenic (TG) mice at E14.5. Panel D3 showed (mean + SEM). (E) β/α cell ratio in E16.5 control and TG pancreata, presented as (mean + SEM). (F1-3) Ins/Gcg/Sst staining and quantification of β/α cell ratio in E16.5 islets of control and ΔMyt pancreata. (G, H) Ki67 (G) and cleaved-Cas3 (H) labeling of islet cells in E16.5 control or ΔMyt pancreata. Note the cycling Gcg+ or Ins+ cells in both control (G1) and mutant islets (G2) (arrows and arrowheads, respectively). Inset in H2, positive control for Cas3 staining. Images from D-H used identical scales, with bar in H1=20 μm. Also see Figure S2 and Table S3.
P1 MNA islets had 53%, 17%, 35%, and 31% of β, α, δ, and γ cells expressing tdT, respectively (Figure 2B). Thus, the β cells are over-represented while the α cells were under-represented amongst the tdT+ islet cells, suggesting that the M+N+ progenitors are biased towards β-cell fate (P=0.002) and away from the α-cell fate (p=1.5E-7). We did not detect strong bias of M+N+ for or against the δ- or γ-cell fate (p=0.57 and 0.19, respectively) (Figure 2B5).
We also examined if this biased M+N+ cell fate was observed at early embryonic stages, when α, β, ε, and G (gastrin-producing) cells were all present at substantial numbers (Figure 2C and data not shown)(Arnes et al., 2012; Suissa et al., 2013): E16.5 MNA islets had an overall ~36% islet cells expressing tdT. Yet ~57% insulin+, ~16% glucagon+, ~13% ghrelin+, and 27% gastrin+ cells expressed tdT (Figure 2C3). These findings suggest that the M+N+ cells, before E16.5, are also biased for β-cell fate, strongly away from α- and/or ε-cell fate, and weakly away from the G-cell fate. This biased lineage allocation was not altered by Neurog3 dosage, because a Neurog3BAC- based nCre transgene, when combined with Myt1cCre, led to a similar cell type allocation (Figure S2E-G).
Myt1 is only marginally involved in modulating islet-cell type allocation
We tested if high Myt1 production in Neurog3+ progenitors was sufficient for β- cell fate choice, driving transgenic Myt1 expression in all Neurog3+ cells with a 6.7 kb Neurog3 promoter (Gu et al., 2002), yielding (Neurog3Myt1OE) mice. Three G0 (founder generation) Neurog3Myt1OE mice were born. They all died perinatally, associated with impaired movement and growth retardation (data not shown). We therefore derived several more G0 Neurog3Myt1OE transgenic embryos and analyzed their pancreatic endocrine cells. In three independent G0 E14.5 Neurog3Myt1OE embryos, we found that ~90% of Neurog3+ cells co-expressed Myt1 (Figure 2D1-D3). This Myt1 over-expression was associated with a minor but statistically significant increased β- to α-cell representation in another three E16.5 Neurog3Myt1OE embryos (p=0.04. Figure 2E). Because the conditions were different for hormone and Myt1 immunodetection and we fixed the E16.5 Neurog3Myt1OE pancreata for hormone expression assays, we could not directly examine the degree of Myt1-Neurog3 overlap in the E16.5 Neurog3Myt1OE embryos.
To investigate if Myt1 is required for β- vs. α-cell specification, we inactivated the three Myt genes in the PPCs using Pdx1Cre to create the ΔMyt condition (Huang et al., 2018). In E16.5 ΔMyt islets, single hormone-expressing α, β, and δ cells were produced in large numbers as in controls, but there was a small decrease in β- to α-cell ratio in ΔMyt islets (Figure 2F). There was no change in islet-cell proliferation or death (Figure 2G, H). These findings suggest that the Myt-mediated processes have marginal roles in β- vs. α-cell allocation. Myt-independent mechanisms, differentially present between M+N+ and M-N+ cells, likely play major roles in regulating such allocation.
Single-cell profiling distinguishes biological processes in M+N+ cells
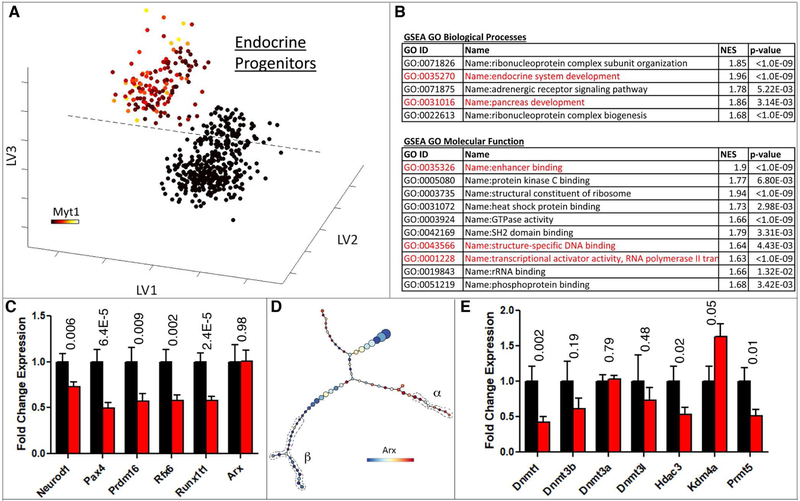
Supervised transcriptome analysis was used to compare the gene expression in M+N+ and M-N+ cells. We used Partial Least Squares Discriminant Analysis (PLSDA) (Lau et al., 2012; Lau et al., 2013) to separate these two progenitor populations from the entire Neurog3eGFP-expressing cell pool. Progenitor cells were identified on the basis of the lack of gene expression signatures indicative of more mature stages of the islet-cell differentiation pathway. This exclusion left 151 M+N+ and 399 M-N+ undifferentiated endocrine progenitors (Figure 3A). A model with three latent variables captured the maximum amount of variance while returning the least amount of calibration and cross-validation error (Figure S3A-C). By these criteria, differentially expressed genes were used for GO term and gene-set enrichment analyses (GSEA). Upregulated genes in the Myt1+ progenitor population were enriched for several Biological Processes and Molecular Function GO terms (Figure 3B). GSEA on both the upregulated genes in Myt1+ progenitors and the differentially expressed genes between the two populations identified enriched gene sets that were altered upon epigenetic perturbations (Figure S3D). These combined analyses suggest that one principal difference between the two subsets of Neurog3+ cells was centered on regulation at the epigenetic level and subsequent transcriptional effects.
Figure 3. Epigenetic and DNA methylation processes are enriched in Myt1+ endocrine progenitors.
(A) PLSDA of scRNA-Seq data to identify Myt1+ against Myt1- endocrine progenitors. X,Y,Z axes are the latent variables generated by PLSDA. Overlays represent Myt1 expression levels on a variance normalized (Asinh) scale. (B) GO term enrichment analysis of genes upregulated in Myt1+ endocrine progenitor cells compared with Myt1-cells. Highlighted terms in red are of interest to this study. (C) Relative expression of elected TF genes in Myt1+ (black) and Myt1-(red) endocrine progenitor cells. Error bars represent SEM from n³150 cells. (D) Arx expression overlaid on p-Creode trajectory in Figure 1. Cells with recognizable β- and α-cell features were circled. (E) Relative epigenetic genes expression in Myt1+ (black) and Myt1-(red) endocrine progenitor cells. Error bars represent SEM from n≥151 cells. In panels C and E, p-values (t-Test) were placed on top of each gene. Also see Figure S3.
In order to identify the factors that functionally bias M+N+ cells to the β-cell lineage, we focused on TFs and epigenetic factors, because they together dictate the gene expression networks specific for each islet cell type. The top TFs upregulated in M+N+ progenitors include NeuroD1, Pax4, Prdm16, Rfx6, and Runx1T1 (Figure 3C. Table S1), all having proved roles most connected to β-cell production (Benitez et al., 2014; Mastracci et al., 2013; Sosa-Pineda et al., 1997; Sugiyama et al., 2013).
Although Arx is an instructive factor in α-cell specification, low and similar Arx expression levels were detected in M+N+ and M-N+ cells (Figure 3C and Figure S3E). Differential Arx expression was observed only after cells gained recognizable β- or α-cell features on p-Creode maps, with β cells suppressing Arx expression (Figure 3D). The findings indicate that high Arx levels allow a quick development of α-cell features. Moreover, the p-Creode analysis detected Pax4 differences prior to Arx (Figure S3F), implying different mode of regulation for Pax4 or Arx.
Several other TFs were differentially expressed between the two sub-cell populations. These factors, including Tox3, Zfp85, Hoxb2, and Oligo2 (Table S1), have not been studied in islet development and function. They will not be pursued here due to the need of deriving loss-of-function alleles. We focused on epigenetic factors because the availability of chemical inhibitors that enable the ready loss-of-function studies.
Epigenetic factors regulates α- vs. β-cell fate choice
DNA methyltransferase 1 (Dnmt1), histone-deacetylase 3 (Hdac3), and protein arginine N-methyltransferase 5 (Prmt5) were up-regulated in M+N+ compared to M-N+ progenitor cells, while lysine-specific demethylase 4a (Kdm4a) was down-regulated in M+N+ cells (Figure 3E). The enrichment of Hdac3 in the β-biased M+N+ progenitors is consistent with a report that inhibiting the class I Hdacs (Hdac1, 2, 3, and 8) with valproic acid (VPA) nearly abolished β-cell differentiation in rat pancreatic culture (Haumaitre et al., 2008). We verified these findings in mouse pancreatic cells (data not shown). However, VPA also inhibited the differentiation of Neurog3+ progenitors to α cells: VPA-treatment induced ~14-fold increase in Neurog3 expression, but only ~3-fold increase of α-cell production (Haumaitre et al., 2008). Thus, Hdac3 is likely involved in the differentiation, instead of fate allocation of α vs. β cells.
Kdm4a demethylates lysine-9 and lysine-36 in histone H3 (Cardamone et al., 2014), while Prmt5 methylates arginines of histone H4 (Fabbrizio et al., 2002), to regulate gene expression. Because the expression of Kdm4a was lower while Prmt5 was higher in β-biased progenitors (Figure 3E), a straightforward model is that higher histone methylation favors the β-cell production. Contrary to this expectation, inhibiting overall methylation with adenosine dialdehyde (Adox) in pancreatic bud culture significantly increased the production of insulin expressing cells (Figure S4A-G). These findings underscore the importance of methylation events in α- vs. β-cell differentiation but suggest that Kdm4a and/or Prmt5-regulated methylation events are not direct determinants of α- vs. β-cell fate choice.
We focused on Dnmt1, because it is the top differentially expressed epigenetic modifier between the M+N+ and M-N+ progenitors (P=0.002, Table S1), and previous studies have indicated the involvement of DNA methylation in islet cell-type specification and maintenance (Dhawan et al., 2011; Papizan et al., 2011).
Inhibiting DNA methylation in pancreatic progenitors promotes α-cell production
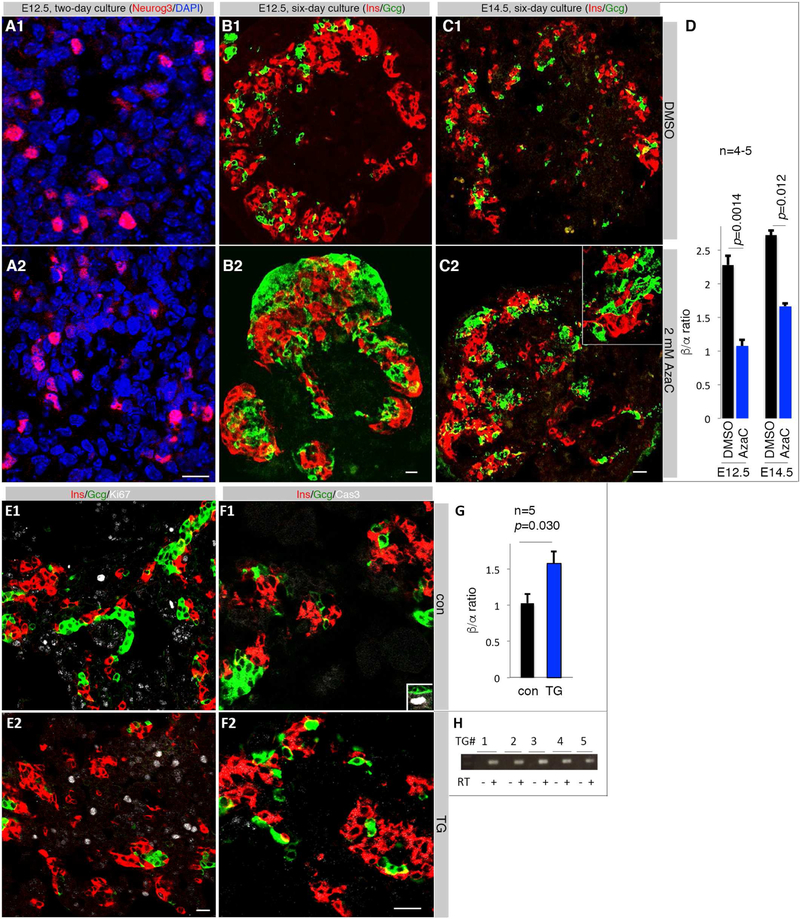
We tested the effects of Dnmt inhibition on islet-cell production in pancreatic explant cultures. To avoid potential redundancy amongst the Dnmts (Elliott et al., 2016), we used 5-azacytidine (AzaC) that inhibits all Dnmts. When E12.5 or E14.5 pancreatic buds were cultured with 2.0 μM AzaC, endocrine differentiation continued as shown by sustained Neurog3 expression (Figure 4A). The ratio of β/α cells under AzaC was substantially decreased after six days of culture (Figure 4D, Figure S4H-J). We observed no significant difference in Gcg+Ins+ double-positive cells 2, 4, or 6 days after culture with AzaC (Figure 4B, C and data not shown), nor islet-cell death or proliferation (Figure S4K-N). These findings suggest that the decreased β/α ratio induced by AzaC arose from altered progenitor allocation.
Figure 4. Manipulating DNA methylation promotes α-cell differentiation.
Scale bars = 20 μm for all images. (A) Neurog3 staining in E12.5 pancreatic buds two days after culture. (B, C) Ins and Gcg expression after 6 days in culture of E12.5 and E14.5 pancreata. Inset in C2 is an enlarged region showing the lack of overlapping between Ins and Gcg immunosignals. (D) Quantification of β/α cell ratios. Error bars are SEM. Marked p-values were from t-test. (E) Co-staining for Ins, Gcg, and Ki67 in E15.5 control and a Pdx1DNMT1 TG pancreata. (F) Co-staining for Ins, Gcg, and activated Caspase 3 (Cas3). Inset in F1 showed a Cas3+ cells in the duct, serving as control. (G) Quantification of β/α cell ratios in five independent Pdx1DNMT1 transgenic pancreata. Error bars are SEM. Presented p-value was from t-Test. (H) RT-PCR detection of DNMT1 mRNA in transgenic pancreatic cells. Also see Figure S4 and S4.
DNMT1 overexpression in pancreatic progenitors promotes β-cell production
We tested if increased Dnmt1 activity is sufficient to alter endocrine-cell-type specification. A 6.3 kb Pdx1 promoter was used to drive the expression of human DNMT1 cDNA in transgenic mice (Pdx1DNMT1). Five transient G0 Pdx1DNMT1 embryos were obtained at E15.5. There were no recognizable differences in gross pancreatic differentiation between these TG embryos over control littermates (Figure 4E, F), nor changed islet-cell proliferation (Figure 4E) or death (Figure 4F). Yet, the β/α cell ratios were significantly increased in the TG pancreata (p=0.03, Figure 4G) that expressed DNMT1 mRNA (Figure 4H). Interestingly, overexpression of Dnmt3b did not alter islet cell-type allocation. This was shown using TetODnmt3b and Rosa26rTTA-ires-eGFP; Pdx1Cre mice in the presence of Doxycycline from E10-E14.5 (Figure S4O-Q)(Linhart et al., 2007). Thus, Dnmt1 but not Dnmt3b is the limiting factor for DNA methylation-mediated α- vs. β-cell fate determination, consistent with the finding that Dnmt1 but not Dnmt3a or Dnmt3b was enriched in the β-biased Neurog3+ progenitors (Figure 3E).
Neurog3-independent methylation in Arx cis-regulatory region correlates with α-cell specification
The above findings suggest that differential DNA-methylation of key endocrinegene enhancers could specify α- vs. β-cell allocation. An ideal study would be purifying the M+N+ and M-N+ progenitor cells and determining their methylome differences to infer candidate genes. Yet the technical challenges in purifying these cells in high numbers have prevented us from this analysis. We therefore combined a candidate and genome-wide methylome assays in overall Neurog3-expressing cells to determine how DNA methylation regulates α- vs. β-cell allocation.
DNA methylation within the regulatory elements of known α- (Arx and Pax6) or β-promoting genes (Myt1, Nkx2.2, Nkx6.1, Pax4, and Pdx1) was assayed with PCR after bisulfide conversion. The relevant cis-regulatory elements of these genes have been reported (Brink et al., 2001; Dhawan et al., 2011; Gerrish et al., 2000; Watada et al., 2000; Watada et al., 2003; Xu et al., 1999). No CpG island was predicted in the reported cis-regulatory regions of Myt1 (Wang et al., 2008). None of the CpG islands present in the tested regulatory regions of Nkx2.2, Nkx6.1, Pax4, and Pax6 were significantly methylated in Neurog3+ endocrine progenitors (Figure S5A). A Pdx1 CpG island showed differential methylation in two of the nine CpG di-nucleotides in half of the DNA templates (Figure S5A). This low methylation level is poorly correlated with Pdx1 expression (Yang et al., 2012) and was not further pursued.
A well-established DNA-methylated regulatory region is UR2 of Arx (1.8 kb upstream of the transcription initiation site, Figure 5A). UR2 methylation status in individual DNA molecules from Neurog3+ cells varied greatly, with some analyte copies showing most of the CpG dinucleotides as methylated yet none in others (Figure 5B). This finding, together with the known correlation between UR2-methylation and Arx expression (Dhawan et al., 2011), led us to postulate that UR2 methylation state can distinguish α- and β-cell progenitors. Consistent with this hypothesis, the β-cell-biased M+N+ progenitors showed a significantly higher UR2 methylation frequency than the α- cell-biased M-N+ cells (Figure 5C. p=0.025). Moreover, embryonic β cells have higher levels of UR2 methylation than α-cells (Figure 5D). AzaC treatment, which increased α-cell specification (reported above), significantly reduced UR2 methylation in Neurog3+progenitor cells (Figure 5E); while Dnmt3bOE, which failed to alter islet cell type specification, did not affect UR2 methylation in Neurog3+ cells (p=0.44, Figure 5F). Moreover, Neurog3eGFP expressing heterozygous progenitors at E10.5, which predominantly give rise to α cells, had significantly lower DNA methylation than E14.5 cells (Figure 5G vs. 5A; p=0.03). These combined results support the idea that Arx cis-regulatory regions are target of cell-type-determining methylation in sub-populations of endocrine progenitor cells.
Figure 5. High methylation in the UR2 enhancer of Arx in PPCs correlates with the production of β cells.
In all panels, black and open circles indicated methylated or non-methylated CpG dinucleotides, respectively. (A) A diagram showing the location of UR2 in Arx. (B-I) UR2 methylation in different cell populations, the eGFP+ cells of E14.5 Neurog3eGFP/+ pancreata (B), E14.5 Myt1+Neurog3+ and Myt1-Neurog3+ cells (C), purified E16.5 α and β cells (D); AzaC treated and control Neurog3eGFP/+ cells (E); Neurog3eGFP+ cells with or without Dnmt3b overexpression (F); E10.5 Neurog3eGFP + heterozygous cells (G); Neurog3-transcribing cells of E14.5 Neurog3eGFP/eGFP null pancreas (H), and E10.5 Neurog3-transcribing Neurog3eGFP/eGFP null cells (I). Also see Figure S5, S6 and Table S2.
We next determined the cell types when differential UR2-methylation was generated. Using E14.5 Neurog3eGFP/eGFP (Neurog3-null) pancreata, we flow-sorted cells that were primed towards the endocrine pathway by activating Neurog3 transcription but failed to enter the endocrine lineage due to the lack of Neurog3 protein (referred as “endo-ready PPCs” hereafter). The distribution of the methylation heterogeneity in the UR2 region in Neurog3eGFP/eGFP cells (compare Figure 5A and 5H) was similar to Neurog3eGFP/+ cells. The inactivation of Neurog3 changed little on the UR2 methylation patterns of E10.5 pancreatic progenitors (Figure 5G and 5I). These data suggest that the UR2 methylation patterns established before Neurog3 expression is a key factor determining islet cell type.
Genome-wide methylome assays uncovered both Neurog3-dependent and independent hypomethylated DNA regions
In order to corroborate the above candidate gene-based data, we compared the methylome of E14.5 endo-ready PPCs and Neurog3eGFP/+ endocrine-producing cells. Global analysis revealed 4,712 differentially methylated regions (DMRs) between the two cell types, with a majority of them located in introns and distal or intergenic regions (Figure S5B, C). The majority (~90%) of DMRs were hypomethylated in Neurog3eGFP/+ cells compared with endo-ready PPCs, implying that activation of Neurog3 results in demethylation at these putative enhancer regions. Associating these DMRs with closest genes (Table S2) provided a list of candidates for future methylation-related studies.
Focusing on the methylation states of several functionally tested loci, we identified DMRs that could potentially regulate islet cell differentiation. For example, methylome assays detected several DMRs ~60 kilo base 3’ to Nkx6.1 (Figure S6A). We only observed low-level methylation in the 5’ regulatory region of Nkx6.1, consistent with our candidate PCR-based assays (Figure S6B). Examining the Arx locus yielded similar findings: there were several DMRs between the endo-ready PPCs and Neurog3eGFP/+ cells (Figure S6C); but we did not observe differential methylation in the UR2 regions (Figure S6D). Because proximal elements such as UR2 and that in the 5’ end of Nkx6.1 have been shown to regulate gene expression (Dhawan et al., 2011; Watada et al., 2000) and they were not included in DMRs, we conclude that the PPCs before Neurog3 activation have already been divided into sub-populations, with different methylation states in regulatory elements poising cells for different levels of gene expression. The methylation landscape was further modified after Neurog3 activation, which likely regulates the differentiation and function of islet cells.
Targeted UR2 methylation favors β-cell specification
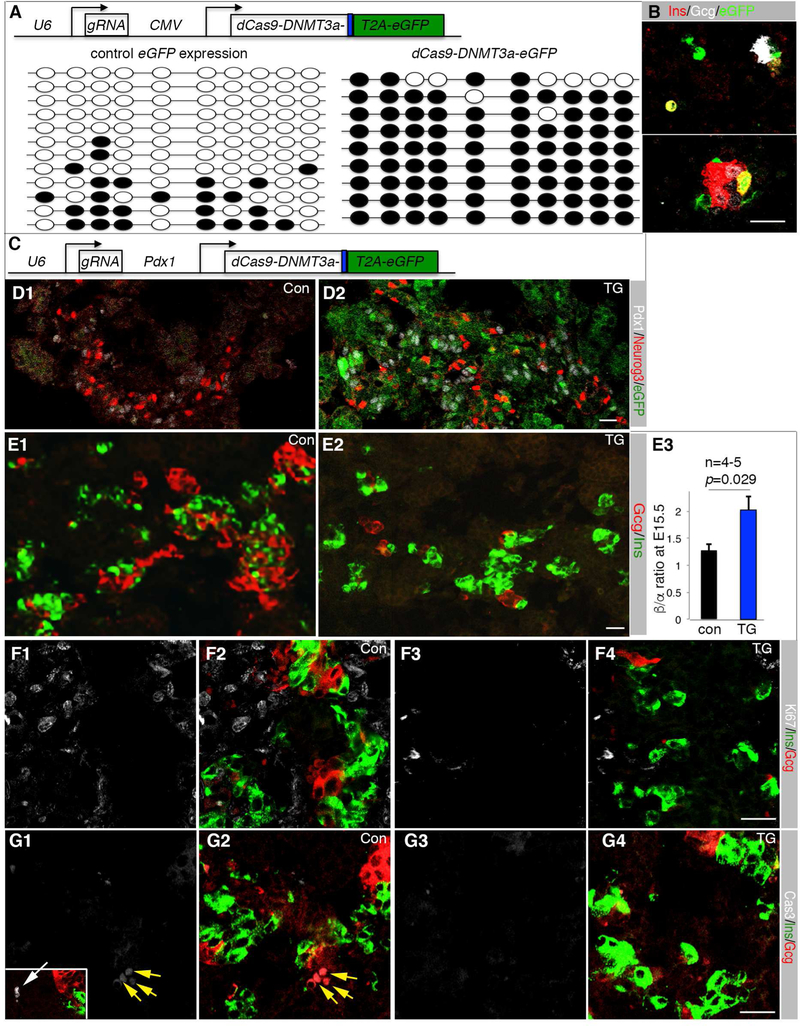
We examined if UR2 methylation is sufficient to alter α- vs. β-cell fate choice. A dCas9-DNMT3a-T2A-EGFP producing construct together with a guide RNA transcribing construct was used to ectopically methylate UR2 (Figure 6A). This construct produced eGFP and a dCas9-Dnmt3a fusion protein that could not cleave DNA but could methylate nearby CpG islands (Stepper et al., 2017). When electroporated into pancreatic cells and cultured in vitro in hang-drop culture, these constructs methylated nearly all the CpG dinucleotides in the UR2 region in eGFP+ cells (Figure 6A), which correlated with the eGFP+ cells expressing insulin but not glucagon (Figure 6B).
Figure 6. UR2 hyper-methylation promotes β-cell production.
(A) The top depicts the DNA construct electroporated into E12.5 pancreatic cells. eGFP+ cells were sorted after three-day hang-drop culture and used for UR2 methylation assays (bottom). Dark circles indicated methylated CpG dinucleotide. (B) Ins/Gcg expression status of electroporated pancreatic cells 4-days after hang-drop culture. (C) The DNA construct used for transgenic (TG) mouse derivation. dCas9 and the DNMT3a activation domain were produced as a fusion protein. eGFP was produced as a separate protein via a T2A peptide. (D) Neurog3 and eGFP expression in E15.5 control (con) and TG pancreatic sections. (E) Ins/Gcg staining and quantification. The p-value in C3 is from t-Test. Error bars in E3 represent SEM. (F) Ki67 detection in E15.5 Ins+ or Gcg+ cells. F1 and F2, control sections. F3 and F4, TG sections. Note that single Ki67 (F1, F3) and merged Ki67, Ins, Gcg channels (F2, F4) were shown. (G) Same as F, except activated Cas3 was assayed. Inset in G1 is a Cas3+ cell (white arrow). Scale bars in all panels, 20 μm. Also see Table S3.
Transient transgenic mice were derived using a 6.3 kb Pdx1 promoter to drive the expression of dCas9-Dnmt3a-T2A-eGFP and an U6 promoter to drive the expression of the guide RNA (Figure 6C). Five independent transgenic E15.5 embryos were derived (Figure 6D and data not shown). Endocrine commitment, assayed by Neurog3 expression, was largely normal in these TG pancreata (Figure 6D). There was a significant increase in β to α cell ratio in the transgenic over control pancreata (Figure 6E), without altering islet-cell proliferation or apoptosis (Figure 6F, G) or producing Gcg+Ins+ transient cells (Figure 6F, G). These data suggest that UR2 hyper-methylation in the Pdx1+ progenitor cells promotes β-cell allocation from progenitor cells.
Neurog3-null PPCs constitute a transcriptionally uniform population despite their differential UR2 methylation
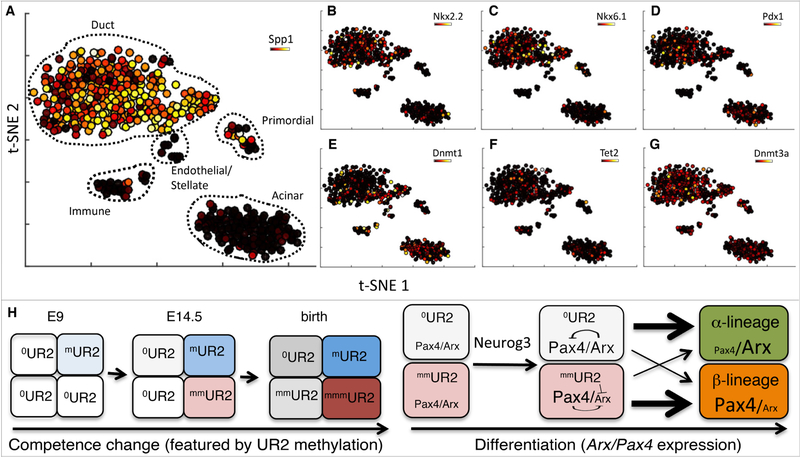
Lastly, we determined how the differential Dnmt1 expression and/or UR2 methylation were initiated in the pancreatic progenitors. t-SNE analysis of scRNA-seq profiles of flow-sorted E14.5 endo-ready PPCs showed that Neurog3-null progenitors largely adopt duct and acinar identities (Figure 7A, Figure S7A-C). Neurog3-dependent genes, including Arx, Pax4, and Myt1, were suppressed in these Neurog3-null cells (Figure S7D-F). Transcription of several Neurog3-independent genes known to contribute to β-cell development – including Glis3, Nkx2.2, Nkx6.1, Prdm16, and Pdx1 – was unaffected (Figure 7B-D and Figure S7G-I).
Figure 7: Epigenetic, but not transcriptional, heterogeneity was detected in endo-ready PPCs.
(A) t-SNE analysis of scRNA-seq data generated from E14.5 Neurog3eGFP/eGFP cells overlaid with the duct marker Spp1. Cell populations expressing characteristic markers are labeled. (B-G) Overlays of several genes over the t-SNE map in A. Overlays represent gene expression levels on a variance normalized (Asinh) scale. (H) A model to explain the temporal competence changes in PPCs (left) and the derivation of multiple islet cell types (right). Rectangles of different color represent cells of different competence state. The number of “m” next to “UR2” indicates the degree of UR2 methylation, which increases over embryogenesis under the influence of maybe Dnmts/Tets. This methylation increase changes the temporal competence of the PPCs. UR2 methylation is asynchronous, producing PPCs with different levels of methylation. If a PPC has no/low levels of UR methylation, it can activate both Arx and Pax4 upon high Neurog3 production. High levels of Arx immediately activate Gcg expression to bias for α-fate selection. If a PPC has high UR2 methylation, it can only activate Pax4 but not Arx to favor β-cell fate.
Surprisingly, the t-SNE clustering patterns of cells expressing DNA-methylation genes, including Dnmts and Tets were distributed uniformly amongst the Neurog3-null duct cell populations (Figure 7E-G, Figure S7J-L), with the t-SNE plots showing the main duct and acinar cell populations distributed cleanly into separable groups, without evident sub-clusters (Figure 7A). Thus, these combined findings raised the possibility that the differential Dnmt1 expression and/or DNA methylation in progenitor cells have likely occurred prior to these cells adopting divergent endocrine transcriptional identities.
Discussion
This study determines when and how pancreatic progenitor cells (PPCs) begin to enter specific islet cell-type differentiation programs. We showed that at transcription level, the PPCs prior to Neurog3-induced commitment (endo-ready PPCs) comprise a uniform cell pool. At the epigenetic level, the endo-ready PPCs are subpopulations with different levels of methylation in enhancers of key cell-type promoting genes such as Arx. This differential epigenetic marking is a major initiator in α- vs. the β-cell lineage determination by differentially poising gene transcription. Given the ubiquitous nature of Dnmt expression and DNA methylation in all cells, it is possible that this methylation-dependent cell specification mechanism can be extended to other organs. It could also direct α- vs. the β-cell fate choice in human in vivo pancreatogenesis or in vitro islet cell derivation from ES or iPS cell culture.
Our combined findings are consistent with a model that the transcriptionally uniform endo-ready PPCs pre-configure their epigenetic landscape by gradually and asynchronously increasing UR2 methylation. Lower UR2 methylation primes PPCs for higher Arx expression to adopt α-cell fate, while those with higher UR2 methylation suppressed Arx expression to lead to β-cell fate (Figure 7H). This model is consistent with reports that UR2 methylation can repress Arx transcription (Dhawan et al., 2011) and that Arx promotes α-cell development (Collombat et al., 2003). Our model also highlights two previously unknown features: one is the Neurog3-independent and asynchronous competence change that creates an epigenetically heterogeneous population primed for different fates (Figure 7H, left); the other is the Neurog3-dependent transcription that reads the epigenetic state of each endo-ready PPC to activate differential gene expression to partition cells into different lineages (Figure 7H, right).
DNA methylation-regulated Arx expression is unlikely the only major pathway that allocate islet-cell types. For one, the targeted over methylation assay was limited to UR2. Yet our genome-wide methylome assays revealed tens of thousands of hypomethylated regions with different level of methylation between different DNA molecules. Additionally, we identified several thousand DMRs between the Neurog3 null and Neurog3 heterozygous endocrine cells, highlighting both Neurog3 dependent and independent methylation events. Thus, future tests will be needed to verify the roles of each DMR in islet cell allocation, differentiation, and function. To this end, scoring genome-wide DMRs between M+N+ and M-N+ cells will likely help to narrow down the candidates that regulate islet-cell type determination, with potential future technical breakthroughs in methylome assays with limited number of DNA molecules.
DNA-methylation is unlikely the only biochemical changes that occur in endo-ready PPCs to promote specific cell fate choice either. Neither Dnmt inhibition nor UR2 hypermethylation can completely prevent the production of multiple islet cell types. Thus, non-epigenetic factors (including the few TFs differentially expressed in the subsets of Neurog3+ cells) could also account for the different properties of PPCs that prime progenitors towards different fates.
How the variable Dnmt1 expression and UR2 methylation patterns originate in the endo-ready PPCs remains unclear. One possibility is that the PPC pool consists of cell subsets that may have escaped our scRNA-seq assays. Improved scRNA-seq, expanded to include non-coding RNAs, could provide clarification. Alternatively, the stochastic nature of the gene expression and methylation event could lead to variable Dnmt1 expression and/or UR2 methylation (Vogt, 2015). To this end, an exciting concept emerged over the past decade is that noise-like stochastic gene expression can create cellular asymmetry to influence cell differentiation (Abranches et al., 2014; Chatfield et al., 2014; Stumpf et al., 2017; Subramanian et al., 2005), which has been proposed to explain the random nature of Neurog3 activation in, and proliferation/differentiation capabilities of, PPCs (Kim et al., 2015; Larsen et al., 2017). Thus, we regard stochasticity as a likely possibility until other relevant deterministic factors become identified, i.e. pre-existing methylation heterogeneity, probably generated stochastically, is key to later differentiation into multiple cell types, orchestrated by permissive executors.
A notable finding in our trajectory study is the lack of higher Arx transcription in the α-biased Myt1-Neurog3+ progenitors, until Gcg transcription was detected. It is possible that the Neurog3-enhanced Arx expression is a relatively slow process, while the activation of Gcg transcription by Arx is a fast one. This combination would have substantially reduced the detectable Myt1-Neurog3+Arx+Gcg- intermediate population. A slow Arx activation can occur if unknown co-factors or epigenetic modifications need to occur for Neurog3 to activate high Arx expression. Supporting the involvement of other factors in Arx expression, we detected low Arx expression in Neurog3−/− progenitor cells, in both single cells (Figure S7), concomitant with the detection of low levels of Gcg expression (Wang et al., 2008). Thus, identifying the factor(s) beside Neurog3 that potentiate Arx transcription is another key future study.
Star Method
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by Guoqiang Gu (Guoqiang.gu@vanderbilt.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice derivation and usage:
Mouse production and usage followed protocol approved by the Vanderbilt IACUC for Dr. Gu, in compliance with the policies of AALAC. For all studies, both male and females were included, at equal ratios when possible. The embryonic stages/ages used were discussed and detailed in each result description. All mice were group-housed (when possible) in standard ventilated cages with free access to water and food. Purchased mice from commercial vendors were acclimated for >one week in facility before use.
The Neurog3nCre knock-in mouse was derived by gene targeting. A vector was constructed to contain the following elements from 5’ to 3’: a 3.5 kb 5’ arm, Lox71, nCre-T2A-Neurog3, FRT flanked hygromycin cassette, Lox2272, Neurog3 polyA signal, and 1.8 kb 3’ arm. The presence of nCre-T2A-Neurog3 allows the co-production of nCre and Neurog3 to avoid the haploinsufficiency of Neurog3. Targeted ES cell clones were verified with both PCR and Southern blot before blastocyst injection (data not shown). Chimeras were crossed with Flpe mice to delete the FRT-flanked selection cassette for deriving the Neurog3nCre allele. The Myt1cCre allele was generated through BAC recombineering followed by pronuclear injection. A targeting vector containing ~250bp 5’ arm, cCre coding sequence directly following the Myt1 start codon, FRT-flanked Neo/Kan selection cassette, and ~350bp 3’ arm was constructed. The targeting vector was then recombined into a BAC (RP23–381D14) containing Myt1 (including ~80 kb upstream and ~20 kb downstream of Myt1). Two transgenic founders were derived. One line was used for all experiments, while the other was not pursued due to poor fertility. Derivation of Neurog3tgnCre mice was performed with a similar strategy as Myt1cCre, except BAC RP23–121F10 was used.
Neurog3Myt1OE, Pdx1DNMT1 and Pdx1dCas9-DNMT3a-eGFP-gRNA transgenic mice were derived via pronuclear injection as well. The 6.7 kb Neurog3 promoter and the 6.3 kb Pdx1 promoter were reported in (Gu et al., 2002). The Myt1 cDNA was reported in (Wang et al., 2008). cDNA clones for DNMT1 or dCas9-DNMT3a-eGFP were purchased from Addgene (clone #36939 and #71666). For Neurog3Myt1OE plasmid construction, the Myt1 coding region was isolated and inserted between the Neurog3 promoter and a SV40 PolyA sequence. For Pdx1DNMT1 or Pdx1dCas9-DNMT3a-eGFP plasmid construction, the DNMT1 or dCas9-DNMT3a-eGFP coding region was isolated and inserted between the Pdx1 promoter and a SV40 PolyA sequence, respectively. To obtain the final UR2 methylation construct Pdx1dCas9-DNMT3a-eGFP-gRNA, a DNA fragment with U6 promoter and gRNA full sequences were biochemically synthesized and inserted into the 5’ end of the Pdx1dCas9-DNMT3a-eGFP plasmid. The vector sequence were then removed from the end constructs via restriction digestion and used for pronuclear injection. The verification of transgene expression was done with immunostaining (Neurog3Myt1OE and Pdx1dCas9- DNMT3a-eGFP-gRNA) or RT-PCR (Pdx1DNMT1). In the latter case, ~1/10th of the pancreatic tissue were used for RNA isolation before tissue fixation and section.
The Rosa26tdTomato/+ (Ai9) and TetODnmt3b; Rosa26rTTA-ires-eGFP mice were purchased from the Jackson Laboratory (Belteki et al., 2005; Linhart et al., 2007; Madisen et al., 2010). Neurog3EGFP/+ mouse was a gift from Dr. Kaestner (Lee et al., 2002). All oligos used for genotyping were listed in S3. Routine crosses utilized outbred wild-type CD1 mice from Charles River. Myt1F, Myt1LF, and St18F allele was described in (Wang et al., 2007) and (Huang et al., 2018). The Pdx1Cre line was described in (Gu et al, 2002).
Primary pancreatic cell culture
For bud culture, freshly E12.5 or E14.5 embryonic pancreata were cultured in RPMI1066 supplemented with 10% FBS on 0.45 micrometer Millicell filters (Millipore) in an air-liquid interface as described in (Haumaitre et al., 2008), with DMSO, 2 μM AzaC, or 5 μM Adox added into media. Briefly, staged embryonic pancreata were dissected from ICR mice and placed into Millicell filters. Intact E12.5 pancreatic buds were directly used. E14.5 pancreatic buds were cut into 4–5 pieces and randomly distributed onto different filters for treatment with control or different inhibitors. The filters were placed into wells of 24-well plates. 0.5 ml media (with inhibitors or DMSO control) were added for culture at 37 °C with 5% carbon dioxide. Media were changed once every other day until tissue collection. For recovering the buds, they could be directly fixed on filter or removed from filter then frozen, section, and fix for subsequent staining. For examining cell-type specification, pancreata from CD1 embryos were used. For examining the DNA methylation alterations by AzaC, Neurog3eGFP/+ pancreata were used so that Neurog3 expressing cells can be flow sorted for downstream analyses.
For hang drop culture, E12.5 pancreata were dissociated with trypsin into single cells. The cells were then electroporated with CMVeGFP control or CMVdCas9-DNMT3a-eGFP- gRNA plasmids, respectively. Cells were then cultured in RPMI1066 supplemented with 10% FBS as 40 microlitter hang drops for four days before spun onto slides for immune assays following the procedure in (Foty, 2011) at 37 °C with 5% carbon dioxide. To test the effect of dCAS9-DNMT3a expression on DNA methylation, eGFP+ cells were FACS-enriched four days after electroporation and used for bi-sulfite sequencing, using the PCR-based approach.
METHOD DETAILS
Immunostaining, imaging, and cell counting
Immunostaining with tissue sections followed routine protocols, with all antibodies described in Key Resource Table. Both tissue sections and dissociated cells were used. For cell counting in Myt1 overexpression, Myt mutants, and Pdx1DNMT1, and Pdx11dCas9-DNMT3a-eGFP cases, tissue sections were used, counted in double blind settings. Briefly, pancreata of specific genotype and stage were dissected, fixed in 4% paraformaldehype (in PBS with Ca2+ and Mg2+) at 4 °C overnight. The tissues were then washed 3X in PBS and prepared as frozen section in OCT. The sections were then stained as the following: sections were left on 37 °C heat block for ~30 minutes, washed 3X 5minutes in 1X PBS, permeabilized in 0.2% triton 100 in 1XPBS for 30 minutes at room temperature. The slides were then covered with 0.1 ml (1XPBS+0.1%BSA+5% donkey serum+0.1% tween-20+0.1% Triton-100, called basal solution A from now on) at room temperature for 30 minutes. The basal solution was then switched to primary antibodies diluted in basal solution at 1:50 to 1:5000 ratio (depending on antibody used, see key reagent table) and incubated at 4 °C overnight, washed 3X in 1XPBS +0.1% Tween-20, covered with 0.1 ml fluorophore-conjugated secondary antibodies diluted 1:1000 in basal solution, incubated at room temperature for another 30 minutes, and then washed 3X with 1XPBS+0.1% Tween-20 for observation.
For cell counting in combinatorial lineage tracing, pancreata of desired age and genotype were partially dissociated with trypsin into small tissue clusters (5–100 cell clusters) and stained using whole mount methods. Briefly, the dissociated cells were fixed in 4% paraformaldehype (in PBS with Ca2+ and Mg2+) at 4 °C overnight, washed 3X with PBS after 1000 rpm spinning in microfuges, blocked with 1 ml basal solution for 30 minutes at room temperature, switched to 1 ml primary antibodies diluted in basal solution, incubated at 4 °C overnight, washed 3X in 1XPBS +0.1% Tween-20. The lysates were then switched to 1ml fluorophore-conjugated secondary antibodies diluted 1:1000 in basal solution, incubated at room temperature for another 30 minutes, and then washed 3X with 1XPBS+0.1% Tween-20. The cells were then spread onto slides for imaging and counting. The stained cells were imaged using a confocal microscope and counted. A Leica TCS-SP5 scanning and an Olympus FV-1000 confocal microscope were used. All cell counting data were presented in Table S3.
Cell isolation
For flow-sorting of Neurog3eGFP-expressing cells, pancreata of desired stage and genotype were dissected in Ca2+/Mg2+-free PBS. 0.25% trypsin was used to dissociate the pancreatic tissues into single cells at 37 °C, usually complete within 10 minutes (Gu et al., 2004). Cells were then stained with propydium iodide and sorted using BD FACSAriaTMIII with professional help. RNA and DNA were then purified and used for gene expression analysis or bisulfite sequencing using Zymo-RNA purification (Zymo Research) or EZ DNA Methylation-Direct Kit (Zymo Research, D5020) following recommended procedures. In order to avoid sequencing multiple clones from a single PCR template for DNA methylation assays, PCR amplification was done in 4–8 separate tubes and the fragments were cloned separately. The oligos used are listed in Key Resource table. Note that for hard-to-amplify fragments, such as UR2, two rounds of PCR reactions were performed. For isolating Myt1+Neurog3+ cells in bisulfite sequencing, E14.5 pancreatic buds were frozen, sectioned, fixed, and stained for Neurog3 and Myt1 expression. Tissue sections were then collected from slides and mechanically dissociated into single cells and used for sorting and DNA preparation.
Bisulfite sequencing
Whole genome bisulfite libraries were constructed using tagmentation-based methods as previously described (Schlesinger et al., 2013; Wang et al., 2013b). Briefly, eGFP+ cells (~100,000 cells) were isolated from E14.5 Neurog3eGFP/eGFP and Neurog3eGFP/+ pancreata via FACS. Genomic DNA was prepared using a Qiagen genomic DNA isolation kit. Transposomes were assembled with Tn5 enzyme purified in house using a plasmid construct kindly provided by the Sandberg lab and according to protocols previously described (Picelli et al., 2014). Tagmentation, oligonucleotide replacement and gap repair were carried out on 75ng of genomic DNA. Bisulfite conversion of DNA was performed using the EZ DNA Methylation Lightning kit (Zymo cat # D5030) according to the manufacturers recommendations. After desulphonation and purification of sodium bisulfite treated libraries, samples were amplified for 12 cycles with the Kapa HiFi HotStart Uracil+ Ready Mix according to the manufacturers instructions. WGBS libraries were sequenced on the Illumina Novaseq (paired end 150bp read lengths), obtaining approximately 400 million reads per library. Sequenced reads were trimmed using Trimgalore (https://github.com/FelixKrueger/TrimGalore) and mapped to the mouse genome mm10 assembly using the WALT aligner (Chen et al., 2016). We achieved 9–10x coverage of symmetric CpG sites and subsequently processed mapped reads using MethPipe methylation analysis pipeline in order to identify hypomethylated and differentially methylated regions (Song et al., 2013).
Single-cell RNA-sequencing
FACS-purified Neurog3eGFP+ cell suspensions from E14.5 pancreata were washed and then checked for viability with Trypan Blue. Samples with >95% viability were counted, mixed with 10% human K562 cells to evaluate doublet rate, and then encapsulated and barcoded using the inDrop platform (1CellBio) (Klein et al., 2015). After CEL-Seq-based library preparation, samples were sequenced using Nextseq 500 (Illumina) in a customized format. All four samples from this report were multiplexed in a single sequencing run. Overall, 3,368 cells were sequenced with, on average, 5,886 unique transcripts per cell (as measured by number of UMIs) identified from, on average, 56,343 reads per cell. Sequencing data were mapped, quantified, and normalized following the protocol of our previous report (Herring et al., 2018).
Single-cell data analysis
For p-Creode analysis (Herring et al., 2018), a Neurog3-dependent gene set (Gu et al., 2004) was used in place of the routine Neighborhood Variance Ratio select gene procedure. p-Creode parameters were selected according to documented sources (https://github.com/KenLauLab/pCreode), and the representative trajectory from N=100 runs were used. PLSDA (Lau et al., 2012; Lau et al., 2013) and t-SNE (van der Maaten and Hinton, 2013) analyses were performed in Matlab (Mathworks), and GO term enrichment was performed in WebGestallt (http://www.webgestalt.org/) (Wang et al., 2013a). GSEA was performed (http://www.broad.mit.edu/gsea/) by pre-ranking differentially expressed genes between the cell populations by 1) their magnitude of overexpression and 2) their p-value of change based on t-test (Subramanian et al., 2005).
QUANTIFICATION AND STATISTICAL ANALYSIS
Data in the figures were represented as mean ± SEM (standard error of the mean). Student’s t-test was used for statistical comparison between two groups that are normally distributed using the built-in function of Microsoft-Excel program. p-values of less than 0.05 were considered significant.
DATA ACCESSIBILITY
The raw Sc-RNA-seq and Methylome data for all samples have been deposited in Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) under ID code: GSE118122, GSE117616, AND GSE118120 for scRNAseq data and GSE119110 for methylome data. The processed single cell data are also available on www.flowrepository.org (flowrepository), with accession FR-FCM-ZYRT.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies (with fold of dilution indicted) | ||
| 488-donkey anti Guinea Pig (1:1000) |
Jackson Immunoresearch | 706-545-148 RRID:AB_2340472 |
| 488-donkey anti goat igG (1:1000) | Jackson Immunoresearch | 705-545-147 RRID:AB_2336933 |
| Alexa Fluor® 647 AffiniPure Donkey Anti-Guinea Pig IgG (H+L) (1:1000) |
Jackson Immunoresearch | 706-605-148 RRID:AB_2340476 |
| Alexa Fluor® 488 AffiniPure Donkey Anti-Rabbit IgG (H+L) (1:1000) |
Jackson Immunoresearch | 711-545-152 RRID:AB_2313584 |
| Alexa Fluor® 647 AffiniPure Donkey Anti-Rabbit (1:1000) |
Jackson Immunoresearch | 711-605-152 RRID:AB_2492288 |
| Alexa Fluor® 647 AffiniPure Donkey Anti-Mouse IgG (H+L) (1:1000) |
Jackson Immunoresearch | 715-605-150 RRID:AB_2340862 |
| Alexa Fluor® 594 AffiniPure Donkey Anti-Mouse (1:1000) |
Jackson Immunoresearch | 715-585-150 RRID:AB_2340854 |
| Alexa Fluor® 594 AffiniPure Donkey Anti-Goat IgG (H+L) (1:1000) |
Jackson Immunoresearch | 705-585-003 RRID:AB_2340432 |
| Alexa 647 AffiniPure Rabbit Anti- Syrian Hamster IgG (H+L) (1:1000) |
Jackson Immunoresearch | 307-605-003 RRID:AB_2339601 |
| Biotin-sp anti rabbit (1:1000) | Jackson Immunoresearch | 711065-152 RRID:AB_2340593 |
| biotin anti Rat (1:1000) | Jackson Immunoresearch | 712-065-151 (RRID: N/A) |
| Cy3-Donkey anti-mouse (1:1000) | Jackson Immunoreserach | 715-165-150 RRID:AB_2340813 |
| Cy3-streptavidin (1:1000) | Vector laboroatories | SA-1300 (RRID: N/A) |
| Cy5- streptavidin (1:1000) | Vector laboroatories | SA-1500 (RRID: N/A) |
| FITC anti Rabbit (1:1000) | Jackson Immunoresearch | 711-095-152 RRID:AB_2315776 |
| Goat anti-Pdx1 (1:2000) | ABcam | AB47383 RRID:AB_2162359 |
| Goat anti-somatostatin (1:500) | ABcam | AB30788 RRID:AB_778010 |
| Goat anti-insulin (1:500) | Santa Cruz | sc-7839 RRID:AB_2296108 |
| Guinea pig anti insulin (1:1000) | Dako | A0564 RRID: N/A |
| Rabbit anti-Myt1 (1:1000) | This paper | N/A |
| Mouse anti-glucagon (1:5000) | Millipore | MabN238 RRID:NA |
| Goat anti-Neurog3 (1:1000) Rabbit anti-Pdx1 (1:5000) |
This lab | NA |
| ABcam | AB47267 RRID:AB_777179 |
|
| Rabbit anti-Nkx6.1 (1:5000) | Gift of P. Serup | NA |
| Mouse anti-chicken Pax6(1:50) | Hybridoma bank | AB_528427 RRID: N/A |
| Rabbit anti-MafB (1:1000) | Gift of R. Stein | NA |
| Rabbit anti-Cleaved Caspase 3 (1:500) |
SYSY | #105173 RRID:AB_887838 |
| Rabbit anti-Ghrelin (1:1000) | Antibody-Online | ABIN223664 RRID:AB_10844168 |
| Rabbit anti-glucagon (1:100) | ABcam | AB92517 RRID:AB_10561971 |
| Rabbit anti-somatostatin (1:2000) | ABcam | AB6741 RRID:AB_955424 |
| Rabbit anti-Ki67(1:500) | Abcam | Ab 15580 RRID:AB_443209 |
| Guinea pig anti-Ppy (1:5000) | Millipore | 4041-01 RRID:AB_433709 |
| Rabbit anti-Gastrin (1:500) | Antibody-Online | ABIN734318 RRID:AB_11209678 |
| Bacterial and Virus Strains | ||
| Bl-21 | Vanderbilt MPB Core | NA |
| DH5-alpha | Vanderbilt MPB Core | NA |
| Xl-1 | Vanderbilt MPB Core | NA |
| Biological Samples | ||
| Bovine serum albumin | Sigma | A9418 |
| Donkey Serum | Jackson Immunoresearch | 017-000-121 RRID:AB_2337258 |
| Fetal Bovine Serum | Atlanta Biologicals | S10250 |
| Chemicals, Peptides, and other key reagents | ||
| Collagenase from Clostridium histolyticum |
Sigma | C5138 |
| DMSO | Sigma | D2650 |
| Doxycycline | Sigma | D9891 |
| Dapi | Sigma | D9542 |
| Adenosine, periodate oxidized | Sigma | 7154 |
| 5-Azacytidine | Sigma | A2385 |
| Paraformaldehyde | Sigma | P6148 |
| Millicell | Millipore | PIHA01250 |
| Trypsin | Sigma | C5138-5G |
| SuperScript III Reverse Transcriptase |
Thermo | 18080085 |
| RNaseOUT Recombinant Ribonuclease Inhibitor |
Thermo | 10777019 |
| Barcoded hydrogel microspheres (BHM) |
1cell | |
| Droplet Stabilization Oil | Droplet Genomics | DG-DSO-20 |
| Cell Barcoding Chip V2 | Droplet Genomics | DG-CBC2-80 |
| 1H,1H,2H,2H-perfluoro-1-octanol, 97% ; used at 20% in HFE |
Sigma | 370533-5g |
| Exonuclease I | NEB | M0293L |
| FastDigest HinFI restriction endonuclease |
Thermo | FD0804 |
| Agencourt AMPure XP magnetic beads |
Beckman Coulter | A63881 |
| Agencourt AMPure XP magnetic beads |
Beckman Coulter | A63881 |
| NEBNext® mRNA Second Strand Synthesis Module |
NEB | E6111L |
| HiScribe T7 High Yield RNA Synthesis Kit |
NEB | E2040S |
| RNA Fragmentation Reagents | Thermo | AM8740 |
| PrimeScript Reverse Transcriptase | Takara | 2680B |
| Deoxynucleotide (dNTP) Solution Mix (10 mM each) |
NEB | N0447L |
| RNaseOUT Recombinant Ribonuclease Inhibitor |
Thermo | 10777019 |
| Kapa 2x HiFi Hot Start PCR mix | VWR | KK2601 |
| Eva Green Dye | VWR | 31000-T |
| Corning Costar Spin-X | Thermo | CLS8162 |
| Qiagen genomic DNA isolation | Qiagen | 69504 |
| EZ DNA Methylation Lightning kit | Zymo | D5030 |
| Tn5 enzyme for methylome DNA tagmentation |
Home made | |
| 10X Ampligase Buffer for methylome library construction |
Epicentre | A1905b |
| dNTP for methylome library construction |
Thermo | R72501 |
| T4 DNA Polymerase | NEB | M0203S |
| Ampligase | Lucigen | A3210K |
| DNA oligos used (Synthesized by IDT) | ||
| Ai9-1 | GCGGCCACTACCTGGTGGAG T |
Ai9 genotyping |
| Ai9-2 | CCACGCCACGTTGCCTGACAA | |
| ngn3ncre5s | CCAAAGGGTGGATGAGGGG CG |
Ngn3nCre genotyping |
| ngn3ncre5a | ATGTGGCGTCCACGGGGAGT | |
| myt1cCre5s | GGCAAACTTCTGACCCAGAG GT |
Myt1cCre genotyping |
| myt1cCre5a | GTTGGTCCATCCGCCAGCCTG CA |
|
| DNMT1 | TACCCACCATGACAGGAAGA | Pdx1-DNMT1 |
| DNMT2 | GGGATGAGGGTGTTGAACTG | genotyping |
| 3bs | CCCTGACCTACGGCGTGCAG TGC |
TetO-Dnmt3b genotyping |
| 3ba | CTCGATGTTGTGGCGGATCTT G |
|
| P5 | TTGAAACAAGTGCAGGTGTT CG |
Pdx1-dCas9 genotyping |
| 1843 | CCATCTTATCGTCATCGTCTTT G |
|
| rTTA-s | CCAGAAGCTTGGTGTAGAGC AG |
RiprTTA genotyping |
| rTTA-a | GGCTGGCTCTGCACCTTGGT GATC |
|
| P5 | TTGAAACAAGTGCAGGTGTT CG |
Pdx1-Cre genotyping |
| ngn3ncre5a | ATGTGGCGTCCACGGGGAGT | |
| 801 | TCCACCTGTCCGTTCAGTG | Myt1F genotyping |
| 806 | AGATCCTTCCAGGGTGGAGA | |
| 858 | GTATGGGGAAACTGCTGAAT GAA |
Myt1L genityping |
| 661 | GCATCCAGACAGACTGCGGT GA |
|
| Myt3B | TGAGACTGAGACTACTTGTTA GC |
Myt3F/F genotyping |
| Myt3C | GCTTTCTGGGTTCATTTCTG | |
| ngn3ncre5s | CCAAAGGGTGGATGAGGGG CG |
Ngn3-Myt1 genotyping |
| Myt1a | GGTAACCCTCATCCAGAGCC AG |
|
| eGFPa | GCTGAACTTGTGGCCGTTTAC GT |
Ngn3-eGFP genotyping |
| Ngn3c | TGCAGTGACCTCTAAGTCAG AGGCT |
|
| 3bs | CCCTGACCTACGGCGTGCAG TGC |
Dnmt3t genotyping |
| 3ba | CTCGATGTTGTGGCGGATCTT G |
|
| Arxs | ACGACTTCCTCCAGGACTATA C |
Arx real time RT-PCR |
| Arxa | GGCTGTCACCAACTAC TTCAA |
|
| Gcgs | TGCAATGGTTAATGAGCACT AAAAG |
Gcg real time RT-PCR |
| Gcga | GATCCGGGAATTTGTCATTCT C |
|
| UR-1086 | GGATTTATTTTTATTTTATTTT TTGTGTGTG |
UR2 CpG island PCR |
| UR-1215 | TTTCCCTCAAACTTCCAAACA AAAACCCTC |
|
| UR-1818 | GTGTGGAGTTTATTTTGAAGT TA |
|
| UR-1819 | CAAACTTCCAAACAAAAACCC TC |
|
| Pax4-1083 | GAAATTGGTATTATTTTTTTTA AAAGGTGTTAAGAG |
Pax4 CpG island PCR |
| Pax4-1041 | TAATAATTAAAATTCCTCCCT ACTTCCTTC |
|
| Pax6-1078 | GTGGTTATGTTTATTAGGTGG TGATTTTA |
Pax6 CpG island PCR |
| Pax6-1089 | CTCATTAAAACCAAATAATAA TAAAAAC |
|
| Pdx1-1278 | TAGATTATTTGTGAGGGTTAA TATTTTGT |
Pdx1 CpG island PCR |
| Pdx1-1279 | CCTCAATAAATCCATTATTCA AAATAACC |
|
| Myt1-1079 | AAGGGTTTATGGGTAGTGTA TTTATAAAG |
Myt1 CpG island PCR |
| Myt1-1007 | TAAATTTCAATTAAATATCTTC TCCCCTCA |
|
| Nkx2.2-997 | TGGAGTTTTAGTTTGAAGTTG GGAGGAGGG |
Nkx2.2 CpG island PCR |
| Nkx2.2-1085 | AAAACTCTAAACTCCCAACTC CTTCTACAACCC |
|
| Nkx6.1-1080 | AAGGTGTGGTGTTTTTAGGT GGGTGTGTTTTAGGAG |
Nkx6.1 CpG island PCR |
| Nkx6.1-1081 | AAAAAAAATAAAAAACCAAA ATAAAAACTTTC |
|
| Tn5mC-Apt1 | T/iMe-dC/GT/iMe-dC/GG/iMe- dC/AG/iMe-dC/GT/iMe- dC/AGATGTGTATAAGAGA/iMe -dC/AG |
Methylation assay adaptor |
| Tn5mC1.1-A1block | 5Phos/CTGTCTCTTATA CA/3ddC |
|
| Critical Commercial Assays | ||
| High capacity cDNA synthesis | Applied BIosystems | 4368814 |
| SYBER Green qpcr mix | Biorad | 1705060 |
| Deposited Data | ||
| Methylome of Ngn3+/− cells (read 1) |
1747-GG1-AGGCAGAAAT- TATCCTCTTC_S1_R1_001. fastq.gz |
GSE119110- SM3358197 |
| Methylome of Ngn3+/− cells (read 2) |
1747-GG1-AGGCAGAAAT- TATCCTCTTC_S1_R2_001. fastq.gz |
GSE119110- GSM3358197 |
| Methylome of Ngn3−/− cells (read 1) |
1747-GG2-TCCTGAGCAT- AGAGTAGATC_S2_R1_00 1.fastq.gz |
GSE119110- GSM3358198 |
| Methylome of Ngn3−/− cells (read 2) |
1747-GG1-TCCTGAGCAT- AGAGTAGATC_S2_R2_00 1.fastq.gz |
GSE119110- GSM3358198 |
| ScRNAseq-Ngn3+/−___sample one | 431-GG-1 |
GSE118122 GSM3305228 |
| ScRNAseq-Ngn3+/−___sample two | 431-GG-2 |
GSE118122 GSM3305229 |
| ScRNAseq-Ngn3−/−___sample one | 431-GG-3 | GSE118120 |
| ScRNAseq-Ngn3−/−___sample two | 431-GG-4 | GSE118120 |
Supplementary Material
Highlights:
Neurog3-null cells have distinct DNA methylation states but uniform transcriptome
Neurog3+ cells have distinct transcriptome, DNA methylation state, and cell fate
β-biasing Neurog3+ progenitors co-express Myt1 and have higher Arx enhancer methylation
Increased methylation in Arx promoter drives progenitors to the β-cell lineage
Acknowledgement
We thank Dr. Wright of Vanderbilt for intellectual input and help with the manuscript. This study is supported by NIDDK grants (DK065949 for GG, DK103831 and CA095103 for KSL, AB, and AJS, and U54MD007586 for BL). C.A.H. is funded by a training grants HD007502 and GM120940. S. W. is supported by the American Diabetes Association Pathway to Stop Diabetes Grant 1-26-INI-16. Confocal imaging in VUMC Cell Imaging Shared Resource was supported by NIH grants CA68485, DK20593, DK58404, DK59637 and EY08126. Cell sorting was performed in VUMC [supported by the Vanderbilt Ingram Cancer Center (P30 CA68485) and the Vanderbilt Digestive Disease Research Center (DK058404)] and Meharry Medical College (NIH grant U54MD007586 ).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of Interests:
The authors declare no competing interests.
References:
- Abranches E, Guedes AM, Moravec M, Maamar H, Svoboda P, Raj A, and Henrique D (2014). Stochastic NANOG fluctuations allow mouse embryonic stem cells to explore pluripotency. Development 141, 2770–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnes L, Hill JT, Gross S, Magnuson MA, and Sussel L (2012). Ghrelin expression in the mouse pancreas defines a unique multipotent progenitor population. PLoS One 7, e52026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastidas-Ponce A, Scheibner K, Lickert H, and Bakhti M (2017). Cellular and molecular mechanisms coordinating pancreas development. Development 144, 2873–2888. [DOI] [PubMed] [Google Scholar]
- Belteki G, Haigh J, Kabacs N, Haigh K, Sison K, Costantini F, Whitsett J, Quaggin SE, and Nagy A (2005). Conditional and inducible transgene expression in mice through the combinatorial use of Cre-mediated recombination and tetracycline induction. Nucleic Acids Res 33, e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez CM, Qu K, Sugiyama T, Pauerstein PT, Liu Y, Tsai J, Gu X, Ghodasara A, Arda HE, Zhang J, et al. (2014). An integrated cell purification and genomics strategy reveals multiple regulators of pancreas development. PLoS Genet 10, e1004645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink C, Chowdhury K, and Gruss P (2001). Pax4 regulatory elements mediate beta cell specific expression in the pancreas. Mech Dev 100, 37–43. [DOI] [PubMed] [Google Scholar]
- Cardamone MD, Tanasa B, Chan M, Cederquist CT, Andricovich J, Rosenfeld MG, and Perissi V (2014). GPS2/KDM4A pioneering activity regulates promoter-specific recruitment of PPARgamma. Cell Rep 8, 163–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatfield J, O’Reilly MA, Bachvarova RF, Ferjentsik Z, Redwood C, Walmsley M, Patient R, Loose M, and Johnson AD (2014). Stochastic specification of primordial germ cells from mesoderm precursors in axolotl embryos. Development 141, 2429–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Smith AD, and Chen T (2016). WALT: fast and accurate read mapping for bisulfite sequencing. Bioinformatics 32, 3507–3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collombat P, Hecksher-Sorensen J, Broccoli V, Krull J, Ponte I, Mundiger T, Smith J, Gruss P, Serup P, and Mansouri A (2005). The simultaneous loss of Arx and Pax4 genes promotes a somatostatin-producing cell fate specification at the expense of the (alpha)- and (beta)-cell lineages in the mouse endocrine pancreas. Development 132, 2969–2980. [DOI] [PubMed] [Google Scholar]
- Collombat P, Hecksher-Sorensen J, Krull J, Berger J, Riedel D, Herrera PL, Serup P, and Mansouri A (2007). Embryonic endocrine pancreas and mature beta cells acquire alpha and PP cell phenotypes upon Arx misexpression. J Clin Invest 117, 961–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collombat P, Mansouri A, Hecksher-Sorensen J, Serup P, Krull J, Gradwohl G, and Gruss P (2003). Opposing actions of Arx and Pax4 in endocrine pancreas development. Genes Dev 17, 2591–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collombat P, Xu X, Ravassard P, Sosa-Pineda B, Dussaud S, Billestrup N, Madsen OD, Serup P, Heimberg H, and Mansouri A (2009). The ectopic expression of Pax4 in the mouse pancreas converts progenitor cells into alpha and subsequently beta cells. Cell 138, 449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar RD, Razooky BS, Singh A, Trimeloni TV, McCollum JM, Cox CD, Simpson ML, and Weinberger LS (2012). Transcriptional burst frequency and burst size are equally modulated across the human genome. Proc Natl Acad Sci U S A 109, 17454–17459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desgraz R, and Herrera PL (2009). Pancreatic neurogenin 3-expressing cells are unipotent islet precursors. Development 136, 3567–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawan S, Georgia S, Tschen SI, Fan G, and Bhushan A (2011). Pancreatic beta cell identity is maintained by DNA methylation-mediated repression of Arx. Dev Cell 20, 419–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CQ (2017). Temporal Patterning in the Drosophila CNS. Annu Rev Cell Dev Biol 33, 219–240. [DOI] [PubMed] [Google Scholar]
- Elliott EN, Sheaffer KL, and Kaestner KH (2016). The ‘de novo’ DNA methyltransferase Dnmt3b compensates the Dnmt1-deficient intestinal epithelium. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbrizio E, El Messaoudi S, Polanowska J, Paul C, Cook JR, Lee JH, Negre V, Rousset M, Pestka S, Le Cam A, et al. (2002). Negative regulation of transcription by the type II arginine methyltransferase PRMT5. EMBO Rep 3, 641–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foty R (2011). A simple hanging drop cell culture protocol for generation of 3D spheroids. J Vis Exp. [DOI] [PMC free article] [PubMed]
- Gerrish K, Gannon M, Shih D, Henderson E, Stoffel M, Wright CV, and Stein R (2000). Pancreatic beta cell-specific transcription of the pdx-1 gene. The role of conserved upstream control regions and their hepatic nuclear factor 3beta sites. J Biol Chem 275, 3485–3492. [DOI] [PubMed] [Google Scholar]
- Gu G, Dubauskaite J, and Melton DA (2002). Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development 129, 2447–2457. [DOI] [PubMed] [Google Scholar]
- Gu G, Wells JM, Dombkowski D, Preffer F, Aronow B, and Melton DA (2004). Global expression analysis of gene regulatory pathways during endocrine pancreatic development. Development 131, 165–179. [DOI] [PubMed] [Google Scholar]
- Gutierrez GD, Bender AS, Cirulli V, Mastracci TL, Kelly SM, Tsirigos A, Kaestner KH, and Sussel L (2017). Pancreatic beta cell identity requires continual repression of non-beta cell programs. J Clin Invest 127, 244–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper CV, Finkenstadt B, Woodcock DJ, Friedrichsen S, Semprini S, Ashall L, Spiller DG, Mullins JJ, Rand DA, Davis JR, et al. (2011). Dynamic analysis of stochastic transcription cycles. PLoS Biol 9, e1000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haumaitre C, Lenoir O, and Scharfmann R (2008). Histone deacetylase inhibitors modify pancreatic cell fate determination and amplify endocrine progenitors. Mol Cell Biol 28, 6373–6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring CA, Banerjee A, McKinley ET, Simmons AJ, Ping J, Roland JT, Franklin JL, Liu Q, Gerdes MJ, Coffey RJ, et al. (2018). Unsupervised Trajectory Analysis of Single-Cell RNA-Seq and Imaging Data Reveals Alternative Tuft Cell Origins in the Gut. Cell Syst 6, 37–51 e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Walker EM, Dadi PK, Hu R, Xu Y, Zhang W, Sanavia T, Mun J, Liu J, Nair GG, et al. (2018). Synaptotagmin 4 Regulates Pancreatic beta Cell Maturation by Modulating the Ca(2+) Sensitivity of Insulin Secretion Vesicles. Dev Cell 45, 347–361 e345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings RE, Berry AA, Strutt JP, Gerrard DT, and Hanley NA (2015). Human pancreas development. Development 142, 3126–3137. [DOI] [PubMed] [Google Scholar]
- Johansson KA, Dursun U, Jordan N, Gu G, Beermann F, Gradwohl G, and Grapin-Botton A (2007). Temporal control of neurogenin3 activity in pancreas progenitors reveals competence windows for the generation of different endocrine cell types. Dev Cell 12, 457–465. [DOI] [PubMed] [Google Scholar]
- Kim YH, Larsen HL, Rue P, Lemaire LA, Ferrer J, and Grapin-Botton A (2015). Cell cycle-dependent differentiation dynamics balances growth and endocrine differentiation in the pancreas. PLoS Biol 13, e1002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, and Kirschner MW (2015). Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp JL, Dubois CL, Schaffer AE, Hao E, Shih HP, Seymour PA, Ma J, and Sander M (2011). Sox9+ ductal cells are multipotent progenitors throughout development but do not produce new endocrine cells in the normal or injured adult pancreas. Development 138, 653–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen HL, and Grapin-Botton A (2017). The molecular and morphogenetic basis of pancreas organogenesis. Semin Cell Dev Biol 66, 51–68. [DOI] [PubMed] [Google Scholar]
- Larsen HL, Martin-Coll L, Nielsen AV, Wright CVE, Trusina A, Kim YH, and Grapin-Botton A (2017). Stochastic priming and spatial cues orchestrate heterogeneous clonal contribution to mouse pancreas organogenesis. Nat Commun 8, 605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau KS, Cortez-Retamozo V, Philips SR, Pittet MJ, Lauffenburger DA, and Haigis KM (2012). Multi-scale in vivo systems analysis reveals the influence of immune cells on TNF-alpha-induced apoptosis in the intestinal epithelium. PLoS Biol 10, e1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau KS, Schrier SB, Gierut J, Lyons J, Lauffenburger DA, and Haigis KM (2013). Network analysis of differential Ras isoform mutation effects on intestinal epithelial responses to TNF-alpha. Integr Biol (Camb) 5, 1355–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Perreault N, Brestelli JE, and Kaestner KH (2002). Neurogenin 3 is essential for the proper specification of gastric enteroendocrine cells and the maintenance of gastric epithelial cell identity. Genes & development 16, 1488–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhart HG, Lin H, Yamada Y, Moran E, Steine EJ, Gokhale S, Lo G, Cantu E, Ehrich M, He T, et al. (2007). Dnmt3b promotes tumorigenesis in vivo by gene-specific de novo methylation and transcriptional silencing. Genes Dev 21, 3110–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature Neuroscience 13, 133–U311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastracci TL, Anderson KR, Papizan JB, and Sussel L (2013). Regulation of Neurod1 contributes to the lineage potential of Neurogenin3+ endocrine precursor cells in the pancreas. PLoS Genet 9, e1003278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastracci TL, Wilcox CL, Arnes L, Panea C, Golden JA, May CL, and Sussel L (2011). Nkx2.2 and Arx genetically interact to regulate pancreatic endocrine cell development and endocrine hormone expression. Dev Biol 359, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SB, Schaffer AE, and Sander M (2007). The transcription factors Nkx6.1 and Nkx6.2 possess equivalent activities in promoting beta-cell fate specification in Pdx1+ pancreatic progenitor cells. Development 134, 2491–2500. [DOI] [PubMed] [Google Scholar]
- Papizan JB, Singer RA, Tschen SI, Dhawan S, Friel JM, Hipkens SB, Magnuson MA, Bhushan A, and Sussel L (2011). Nkx2.2 repressor complex regulates islet beta-cell specification and prevents beta-to-alpha-cell reprogramming. Genes Dev 25, 2291–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen MBK, Azad A, Ingvorsen C, Hess K, Hansson M, Grapin-Botton A, and Honore C (2017). Single-Cell Gene Expression Analysis of a Human ESC Model of Pancreatic Endocrine Development Reveals Different Paths to beta-Cell Differentiation. Stem Cell Reports 9, 1246–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Bjorklund AK, Reinius B, Sagasser S, Winberg G, and Sandberg R (2014). Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res 24, 2033–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado CL, Pugh-Bernard AE, Elghazi L, Sosa-Pineda B, and Sussel L (2004). Ghrelin cells replace insulin-producing beta cells in two mouse models of pancreas development. Proc Natl Acad Sci U S A 101, 2924–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, and van Oudenaarden A (2008). Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135, 216–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romer AI, and Sussel L (2015). Pancreatic islet cell development and regeneration. Curr Opin Endocrinol Diabetes Obes 22, 255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas EN, Shu J, Cserhati MF, Weeks DP, and Ladunga I (2016). Pluralistic and stochastic gene regulation: examples, models and consistent theory. Nucleic Acids Res 44, 4595–4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer AE, Taylor BL, Benthuysen JR, Liu J, Thorel F, Yuan W, Jiao Y, Kaestner KH, Herrera PL, Magnuson MA, et al. (2013). Nkx6.1 controls a gene regulatory network required for establishing and maintaining pancreatic Beta cell identity. PLoS Genet 9, e1003274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger F, Smith AD, Gingeras TR, Hannon GJ, and Hodges E (2013). De novo DNA demethylation and noncoding transcription define active intergenic regulatory elements. Genome Res 23, 1601–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon CS, Hadjantonakis AK, and Schroter C (2018). Making lineage decisions with biological noise: Lessons from the early mouse embryo. Wiley Interdiscip Rev Dev Biol, e319. [DOI] [PMC free article] [PubMed]
- Smith SB, Qu HQ, Taleb N, Kishimoto NY, Scheel DW, Lu Y, Patch AM, Grabs R, Wang J, Lynn FC, et al. (2010). Rfx6 directs islet formation and insulin production in mice and humans. Nature 463, 775–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q, Decato B, Hong EE, Zhou M, Fang F, Qu J, Garvin T, Kessler M, Zhou J, and Smith AD (2013). A reference methylome database and analysis pipeline to facilitate integrative and comparative epigenomics. PLoS One 8, e81148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa-Pineda B, Chowdhury K, Torres M, Oliver G, and Gruss P (1997). The Pax4 gene is essential for differentiation of insulin-producing beta cells in the mammalian pancreas. Nature 386, 399–402. [DOI] [PubMed] [Google Scholar]
- Soyer J, Flasse L, Raffelsberger W, Beucher A, Orvain C, Peers B, Ravassard P, Vermot J, Voz ML, Mellitzer G, et al. (2010). Rfx6 is an Ngn3-dependent winged helix transcription factor required for pancreatic islet cell development. Development 137, 203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepper P, Kungulovski G, Jurkowska RZ, Chandra T, Krueger F, Reinhardt R, Reik W, Jeltsch A, and Jurkowski TP (2017). Efficient targeted DNA methylation with chimeric dCas9-Dnmt3a-Dnmt3L methyltransferase. Nucleic Acids Res 45, 1703–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf PS, Smith RCG, Lenz M, Schuppert A, Muller FJ, Babtie A, Chan TE, Stumpf MPH, Please CP, Howison SD, et al. (2017). Stem Cell Differentiation as a Non-Markov Stochastic Process. Cell Syst 5, 268–282 e267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Benitez CM, Ghodasara A, Liu L, McLean GW, Lee J, Blauwkamp TA, Nusse R, Wright CV, Gu G, et al. (2013). Reconstituting pancreas development from purified progenitor cells reveals genes essential for islet differentiation. Proc Natl Acad Sci U S A 110, 12691–12696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suissa Y, Magenheim J, Stolovich-Rain M, Hija A, Collombat P, Mansouri A, Sussel L, Sosa-Pineda B, McCracken K, Wells JM, et al. (2013). Gastrin: a distinct fate of neurogenin3 positive progenitor cells in the embryonic pancreas. PLoS One 8, e70397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussel L, Kalamaras J, Hartigan-O’Connor DJ, Meneses JJ, Pedersen RA, Rubenstein JL, and German MS (1998). Mice lacking the homeodomain transcription factor Nkx2.2 have diabetes due to arrested differentiation of pancreatic beta cells. Development 125, 2213–2221. [DOI] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Nordman E, Bao S, Lee C, Wang X, Tuch BB, Heard E, Lao K, and Surani MA (2011). Deterministic and stochastic allele specific gene expression in single mouse blastomeres. PLoS One 6, e21208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten L, and Hinton G (2013). Visualizing Data using t-SNE. Journal of Machine Learning Research 9, 2579–2605. [Google Scholar]
- Vogt G (2015). Stochastic developmental variation, an epigenetic source of phenotypic diversity with far-reaching biological consequences. J Biosci 40, 159–204. [DOI] [PubMed] [Google Scholar]
- Wang J, Duncan D, Shi Z, and Zhang B (2013a). WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res 41, W77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Elghazi L, Parker SE, Kizilocak H, Asano M, Sussel L, and Sosa-Pineda B (2004). The concerted activities of Pax4 and Nkx2.2 are essential to initiate pancreatic beta-cell differentiation. Dev Biol 266, 178–189. [DOI] [PubMed] [Google Scholar]
- Wang J, Zhang K, Xu L, and Wang E (2011). Quantifying the Waddington landscape and biological paths for development and differentiation. Proc Natl Acad Sci U S A 108, 8257–8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Gu L, Adey A, Radlwimmer B, Wang W, Hovestadt V, Bahr M, Wolf S, Shendure J, Eils R, et al. (2013b). Tagmentation-based whole-genome bisulfite sequencing. Nat Protoc 8, 2022–2032. [DOI] [PubMed] [Google Scholar]
- Wang S, and Cepko CL (2016). Photoreceptor Fate Determination in the Vertebrate Retina. Invest Ophthalmol Vis Sci 57, ORSFe1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Hecksher-Sorensen J, Xu Y, Zhao A, Dor Y, Rosenberg L, Serup P, and Gu G (2008). Myt1 and Ngn3 form a feed-forward expression loop to promote endocrine islet cell differentiation. Dev Biol 317, 531–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zhang J, Zhao A, Hipkens S, Magnuson MA, and Gu G (2007). Loss of Myt1 function partially compromises endocrine islet cell differentiation and pancreatic physiological function in the mouse. Mech Dev 124, 898–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watada H, Mirmira RG, Leung J, and German MS (2000). Transcriptional and translational regulation of beta-cell differentiation factor Nkx6.1. J Biol Chem 275, 34224–34230. [DOI] [PubMed] [Google Scholar]
- Watada H, Scheel DW, Leung J, and German MS (2003). Distinct gene expression programs function in progenitor and mature islet cells. J Biol Chem 278, 17130–17140. [DOI] [PubMed] [Google Scholar]
- Wu X, and Zhang Y (2017). TET-mediated active DNA demethylation: mechanism, function and beyond. Nat Rev Genet 18, 517–534. [DOI] [PubMed] [Google Scholar]
- Xie R, Carrano AC, and Sander M (2015). A systems view of epigenetic networks regulating pancreas development and beta-cell function. Wiley Interdiscip Rev Syst Biol Med 7, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu PX, Zhang X, Heaney S, Yoon A, Michelson AM, and Maas RL (1999). Regulation of Pax6 expression is conserved between mice and flies. Development 126, 383–395. [DOI] [PubMed] [Google Scholar]
- Xu Y, Xu G, Liu B, and Gu G (2007). Cre reconstitution allows for DNA recombination selectively in dual-marker-expressing cells in transgenic mice. Nucleic Acids Res 35, e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang BT, Dayeh TA, Volkov PA, Kirkpatrick CL, Malmgren S, Jing X, Renstrom E, Wollheim CB, Nitert MD, and Ling C (2012). Increased DNA methylation and decreased expression of PDX-1 in pancreatic islets from patients with type 2 diabetes. Mol Endocrinol 26, 1203–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YP, Thorel F, Boyer DF, Herrera PL, and Wright CV (2011). Context-specific alpha-to-beta-cell reprogramming by forced Pdx1 expression. Genes Dev 25, 1680–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw Sc-RNA-seq and Methylome data for all samples have been deposited in Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) under ID code: GSE118122, GSE117616, AND GSE118120 for scRNAseq data and GSE119110 for methylome data. The processed single cell data are also available on www.flowrepository.org (flowrepository), with accession FR-FCM-ZYRT.