Abstract
The accurate and quantitative detection of microRNAs (miRNAs) as next-generation, reliable biomarkers will provide vital information for cancer research and treatment. However, their unique, intrinsic features pose quite a challenge for miRNA profiling, especially for multiplexed detection. Thus, there is a strong and an ever-growing need to develop an accurate, simple, sensitive and specific miRNA sensing method.
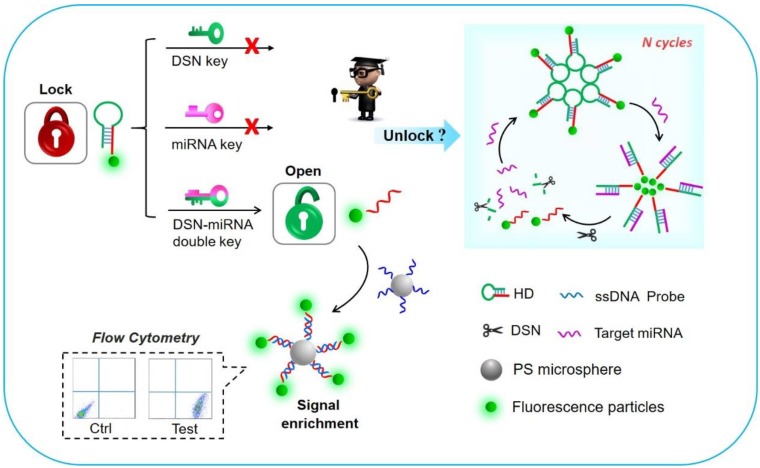
Methods: In this study, a simple and novel sensor is presented that uses a flow cytometry (FCM) method based on the double key “unlocked mechanism” and a fluorescence enrichment signal amplification strategy. The “unlocked mechanism” was cleverly designed via using hairpin DNA probes (HDs) labeled by fluorescent particles (FS) as the lock to block part of them, which can specifically hybridize with the probe on polystyrene microparticles (PS). The target miRNA and duplex-specific nuclease (DSN) forming the double key can specifically open the HDs and cleave a single-stranded DNA (ssDNA) into DNA/RNA dimers circularly in order to unlock the special part of the HDs to be specially enriched further on the PS.
Results: The designed sensor with a hairpin structure and DSN special performance was found to have a high specificity. The circularly unlocking fluorescent probes and fluorescent signal enrichment can be beneficial for achieving a high sensitivity with a detection limit of 3.39 fM for miRNA-21. Meanwhile, the performance of multiplexing was estimated by simultaneous detection of miR-21 and miR-141, and the method also allowed for miR-21 detection in breast cancer blood samples.
Conclusion: The designed sensor based on an “unlocked mechanism” and a signal enrichment strategy resulted in a one-pot, highly specific and sensitive detection of multiplex miRNAs. The whole detection without the need for a complex purification process is based on a FCM and is expected to have a great value in cancer diagnosis and biomedical research.
Keywords: miRNA, “unlocked mechanism”, DSN, signal enrichment
Introduction
MicroRNAs (miRNAs) are single-stranded, endogenous, noncoding RNAs (approximately 22 nucleotides in length) that play important roles in living organisms 1-3. It has been well recognized that an abnormal abundance of miRNAs is closely associated with carcinogenesis 4, 5. However, the unique, intrinsic features of miRNAs, such as their small size, low abundance and high homology, pose quite a challenge for miRNA profiling 6-8. At present, conventional miRNA detection strategies, including quantitative reverse transcriptase PCR (qRT-PCR) 9, 10, northern blotting technology 11, 12 and oligonucleotide microarrays 13, 14, have been widely applied for miRNA sensing as the gold-standard strategies. Nevertheless, these strategies suffer from shortcomings such as the requirement of tedious procedures, long assay times, expensive reagents and sophisticated primers, and poor detection sensitivity or low specificity 15-19. Thus, there is a strong and an ever-growing need to develop an accurate, simple, sensitive and specific miRNA sensing method.
Recently, fluorescence technology has been widely used for fabricating multiple miRNAs sensors due to its inherent advantages of low sample volume, simple operations and easy readout 20-22. However, most of the homogeneous fluorescent assays are not quite suitable for complex miRNAs detection in clinical samples owing to the coincidence/interference between different fluorescence signals and a strong matrix background. To overcome this problem, microbeads have been widely employed in multiplexed biosensing platforms 23-25, especially when combined with analytical flow cytometry (FCM) techniques, which have become a powerful tool for quantitative detection of analytes with multifarious analysis parameters. This also opens up a new avenue for multiplex target miRNAs detection. It has achieved a rapid measurement of thousands of microbeads per second in complex substances. Their flexibility, high throughput, and multiplexing compatibility make them quite suitable for a simultaneous multiplex detection 26-28.
Also attracting much attention for target miRNAs detection is the duplex-specific nuclease (DSN), which aids in target recycling and signal amplification. DSN is a promising tool due to its inherent and specific preference for only cleaving DNA in DNA/RNA and DNA/DNA dimers while having no effect on single-stranded DNA (ssDNA) and leaving RNA strands intact 29. Their selective-cutting ability enables the target miRNA to remain intact, which assists in target recycling and triggers circular reactions to actualize signal amplification. DSN-aided target recovery has been widely employed for miRNA detection coupled with different platforms, such as fluorescence-based assays 30, electrochemical strategies 31, chemiluminescence sensors 32 and magnetic relaxation switch sensors 33.
In this work, we present an innovative “unlocked mechanism” based on DSN-miRNA forming a double key. Working together, the double key circularly unlocks closed DNA probes, which can then be further enriched on the surface of microbead probes to enable fluorescence signal amplification, finally achieving a one-pot, highly specific and sensitive detection of multiplex miRNAs based on FCM. The “unlocked mechanism” was cleverly designed by using the hairpin structure as the lock and DSN-miRNA as the double key. They are both committed to high specificity and selectivity. Importantly, the DSN-assisted target recovery technique is devoted to recycling tiny amounts of miRNAs, which can effectively extend the detection limit and enable a large analytical range. Furthermore, the large surface area of the probe-modified polystyrene microparticles (PS) allows for a high immobilization density of DNA and, as the signal enrichment carriers, enables them to collect the unlocked, free fluorescent particles (FS), leading to further signal amplification. Thus, high sensitivity can be achieved for miRNA sensing. Additionally, this detection platform has the advantages of flexibility, high-reusability and high-throughput by suspension array assays 34, 35. The whole detection can be performed in one-pot, without any complex purification process. Therefore, the proposed FCM platform, which is based on the aforementioned “unlocked mechanism” and signal enrichment carriers, demonstrates a novel, rapid, sensitive and specific avenue for the simultaneous detection of multiplex miRNAs.
Methods
Carboxyl-modified PS (10 μm, 2.5% w/v) were obtained from BaseLine (Tianjin, China). DSN was purchased from Newborn (Shenzhen, China). Carboxyl fluorescent particles (YG, 2.5% w/v, 0.049 μm, cat. no. QDSCF20050) and fluorescent carboxyl particles (pink, 2.5% w/v, 0.05 μm, cat. no. QDSCF10050) were both provided by HyperCyte (Beijing, China). Total miRNA extraction kits, cDNA synthesis and qPCR detection kits (cat. no. B1803, D1801, AP01108) were provided by HaiGene (Harbin, China). All of the DNA/RNA oligonucleotides sequences (HPLC purified) used in our research were provided by Sangon Biotech (Shanghai, China). RNAase-free water was-purchased from Sangon Biotech (Shanghai, China) and the N3-dimethylpropane-1,3-diamine (EDC) activator was purchased from Aladdin (Shanghai, China). Phosphate-buffered saline (PBS, 0.01 M, pH 7.2-7.4) was acquired from Solarbio (Tianjin, China). FCM analyses were detected by a flow cytometer (BD FACSCalibur, USA). Fluorescence images were acquired using a fluorescence microscope (Olympus, Japan). The concentration of cDNA (reverse transcribed by miRNA extracted from biological samples) was detected by LightCycler®96 (Roche, Switzerland).
Preparation of FS-HD probes
The amine-modified hairpin DNA probes (HDS) were coupled to the surface of carboxyl FS by an EDC active method for the preparation of FS-HD probes. First, 5.6 μL of 2.5% solids of 0.05 μm FS was suspended in 2-Morpholinoethanesulfonic acid, monohydrate (MES) buffer (50 mM, pH 6). Then, the HDs were incubated at 95 °C and 2 μL at 100 μM was added after being reduced to room temperature by a gradual freezing method. After simply mixing the microspheres and probes, a covalent reaction was initiated by adding 10 µL of 16 mg/mL EDC and the volume of the solution was then adjusted to 280 µL. Next, in order to achieve an effective coupling effect, the mixture was incubated in the dark overnight. Finally, the HD-functionalized nanospheres were washed with PBS by ultracentrifuge (47500 × g at 30 min) and stored at 4 °C.
Preparation of PS-ssDNA probes
The EDC active method was also carried out in order to make PS-ssDNA probes by coupling amine-modified ssDNA probes to the carboxyl PS. First, PS (25 mg/mL, 10 μL) were resuspended in PBS buffer (200 µL, pH 7.2-7.4, 0.01 M) before conjugation. Then, varying amounts of ssDNA followed by 8 μL of 16 mg/mL EDC were added for the covalent reaction. Next, the mixture was incubated overnight in the dark at room temperature. Several washings with PBS were used for the removal of uncombined ssDNA probes. Finally, the PS-ssDNA probe conjunctions were dispersed in PBS. The probes were stored at 4 °C.
Target miRNA detection
The FS-HD probes (20 µL), DSN (1 U/µL, 0.5 μL), and DSN buffer (4.5 μL, pH 8.0, 10×, 50 mM Tris-HCl, 1 mM DTT, 5 mM MgCl2) were prepared for the miRNA detection mixture. After adding 10 µL of different concentrations of miRNA, the mixture reagents were incubated at 45 °C for 90 min in the dark for miRNA recycling. After the incubation, DSN stop solution (2×, 5 μL, 10 mM Ethylene Diamine Tetraacetic Acid (EDTA)) was added, mixed and incubated for 5 min to inactivate the enzyme. Then, 20 μL of PS-ssDNA probes was added into each tube and incubated for 60 min at 37 °C. Finally, after centrifugation and purification (12000 rpm, 5 min), the PS were directly resuspended into 450 µL of PBS and immediately analyzed by FCM.
FCM analysis and multiplex miRNA detection
The FCM analysis of miR-21 was carried out with a 488-nm laser excitation by using the fluorescence 1 (FL1) channel while the analysis of miR-141 was carried out with a 538-nm laser excitation by using the fluorescence 2 (FL2) channel. A total of 3000 PS were collected for each sample, and the amount of miRNA could be faithfully reflected by the mean fluorescence intensity (MFI) of the PS.
The simultaneous multiplex miRNA detection was employed in the mixture containing 20 µL FS-HD probes (10 µL was modified with HD-21, 10 µL was modified with HD-141), 4.5 μL DSN buffer, 0.5 μL DSN, and four associations of target miRNAs: (a) 0 µL miR-21 + 0 µL miR-141, (b) 10 µL miR-21 + 0 µL miR-141, (c) 0 µL miR-21 + 10 µL miR-141, and (d) 10 µL miR-21 + 10 µL miR-141. These mixtures were incubated for 90 min at 45 °C. Then, the fluorescent intensity of these microbeads was immediately detected by FCM.
Specificity of the miRNA detection assay
To evaluate the specificity and selectivity of the detection sensor, the assay's specificity was evaluated by analyzing samples including target miR-21 and different mismatched miRNAs (negative control miRNA (NC), miR-200b, miR-141, Let-7d, and single mismatched miRNA (SM)). The concentration of miRNAs used in the assay was 1 nM.
miRNA extraction
The General Hospital, affiliated with Tianjin Medical University, provided blood samples after obtaining informed consent. This work was also approved by the Hospital's Ethics Committee, according to the ethical standards of the Helsinki Declaration. The extraction kit, mentioned earlier in this section, was used for total miRNA extraction. The extracted miRNA was quantified by FCM and the qRT-PCR method.
miRNA detection in biological samples
The standard addition method was carried out to study the applicability of our double key unlocking method for miRNA detection in biological samples. According to the preceding miRNA detection procedure in the section of “Target miRNA detection”, the standard miR-21 (50 × 10-12 M) was separately analyzed in dilute human serum samples.
Comparing the double key unlocking method with qRT-RCR
We extracted the total miRNA from the whole blood samples by using the aforementioned miRNA extraction kit. For miR-21 detection by our double key unlocking method, we added 10 µL of extracted miRNA into a 25-µL mixture solution containing 20 µL of FS-HDs conjugates, 4.5 µL of 10× DSN buffer, and 0.5 µL DSN (1 U/µL). Then, we incubated it at 45 °C for 90 min. The PS-ssDNA probes (20 µL) were mixed and incubated together for signal enrichment at 37 °C for 60 min after inactivating the DSN by a stop solution. Finally, we recorded fluorescence by FCM.
For the investigation of miR-21 by the traditional qRT-PCR method, we first needed to reverse transcribe miR-21 from the total miRNA extraction into cDNA at 37 °C for 60 min and 95 °C for 5 min (containing a tag with a 3' poly (A) tail during the subsequent reverse transcription) according to the instructions for the cDNA synthesis kit. The acquired cDNA was amplified according to the manufacturer's instructions for the HG miRNA SYBR Green PCR kit. A titration experiment with serial dilutions of cDNA, converted from standard miRNAs with known concentrations, was carried out to acquire a linear relationship of miR-21 concentrations and the threshold cycle by qRT-PCR. Therefore, we could calculate the concentration of miR-21 according to the linear regression equation by the qRT-PCR method.
Results and discussion
The detection principle of the double key unlocking assay
The working principle of the detection platform based on the DSN-miRNA double key “unlocked mechanism” and fluorescence signal enrichment strategy is schematically shown in Figure 1. In this work, the FS were introduced as the signal reporter and were labeled on the HDs, forming the FS-HDs structure, which as the locked and inactivated structure can't anchor on the DNA modified PS without the existence of target miRNA. The “unlocked mechanism” could not work well without the DSN-miRNA double key due to the lack of effect of target miRNA pairing with the hairpin as well as the lack of the degradation effect of the DSN. So, the FS of the HDs could not trigger the target recycles or perform the following signal enrichment events. However, after the introduction of target miRNAs and DSN, the “unlocked mechanism” was activated by the DSN-miRNA double key. The hairpin structure was opened and the DSN specifically recognized and digested the DNA in DNA/RNA duplex, but left the target miRNA intact, which assisted with target miRNA recycling and initiating more cycles of unlocked events. Thus, the remaining FS-ssDNA probes were unlocked and activated, which could then hybridize with the PS-ssDNA probes to realize the fluorescence enrichment and signal amplification. Therefore, the fluorescence intensity of the FS assembled to the capture probe and the PS as the fluorescence enrichment carriers could be facilely and reliably collected and evaluated by simultaneous FCM, which ensured high sensitivity as well as multidetection.
Figure 1.
Schematic illustration of the “unlocked mechanism” detection avenue based on the DSN-miRNA double key and the fluorescence signal enrichment carriers for signal amplification.
Preparation and characterization of probes
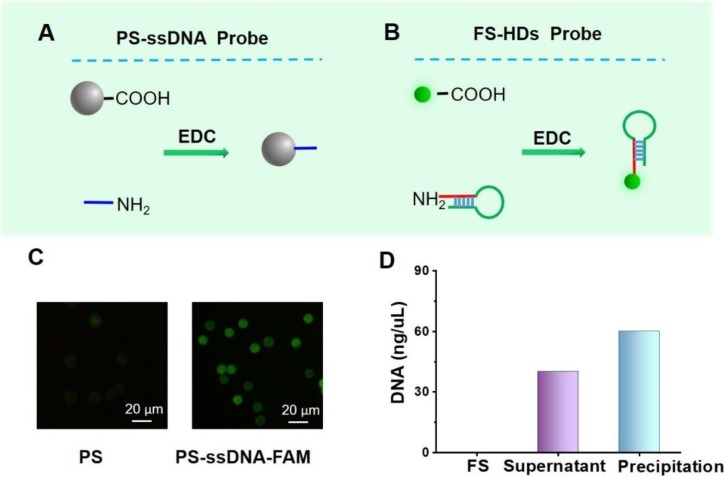
The PS-ssDNA and FS-HD probes were prepared according to the principles shown in Figure 2A-B. The oligonucleotide sequences are listed in Table S1. To characterize the successful combination of aminated ssDNA and carboxyl-coated PS, the FAM-labeled ssDNA with the same sequence (FAM-ssDNA, 3' modified by an amino group and 5' modified by FAM) was adopted to make the PS-ssDNA-FAM probes following the EDC method. Confocal laser scanning microscopy was performed to characterize these functional PS. As shown in Figure 2C, the PS modified with FAM-ssDNA presented a significant fluorescence signal enhancement compared with PS alone, suggesting the successful preparation of PS-ssDNA probes. Furthermore, the PS-ssDNA probes were also confirmed by UV-vis absorption spectra (Figure S1). For the FS-HD probes, which were characterized by the NanoDrop spectrophotometer, we suggested that the HDs were successfully labeled on FS (Figure 2D).
Figure 2.
Preparation and characterization of probes. (A, B) A schematic of the preparation the PS-ssDNA and FS-HD probes. (C) Images of PS coated with and without FAM-ssDNA probes detected by confocal laser scanning microscopy (excitation wavelength: 495 nm). (D) The general evaluation of DNA content by the NanoDrop spectrophotometer.
The “unlocked mechanism” and signal enrichment strategy
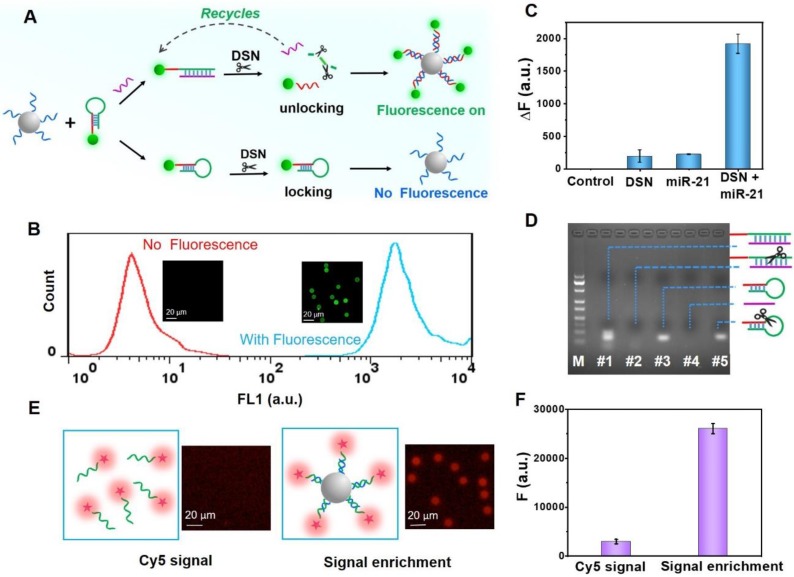
We then studied the necessity of the double key, as illustrated in Figure 3C. It is evident that the “unlocked mechanism” only works well by integrating DSN and miRNA, which work together as the DSN-miRNA double key, demonstrating that DSN-miRNA specifically initiated the unlocked events. In addition, even if miRNA could open the HDs without DSN, because of the steric stabilization of the FS and DNA/RNA duplex, there was a negligible fluorescence intensity on the PS. The electrophoresis experiment further confirmed the results as shown in Figure 3D, where the bands of the DNA/RNA duplex and HDs can be observed clearly (lanes #1 and #3) in the absence of DSN. However, after the introduction of DSN, the DNA/RNA duplex bands began to fade and nearly disappeared after a period of incubation (lane #2), indicating that DSN exhibited a strong preference for cleaving DNA only in the DNA/RNA duplexes group, but had a negligible effect on RNA and HDs. Therefore, the DSN-miRNA double key is a necessary element for the “unlocked mechanism” to work well in the assay.
Figure 3.
The evaluation of “unlocked mechanism” and signal enrichment amplification strategy. (A) The principle of the double key unlocking detection assay with and without target miRNAs. The “unlocked mechanism” based on the DSN-miRNA double key and the mechanism of DSN-assisted target recycling and signal amplification are illustrated in the presence of target miRNAs. (B) The effect of signal enrichment events triggered by the “unlocked mechanism”. FCM detection and fluorescent images of PS-ssDNA before (no fluorescence) and after (with fluorescence) reactions. (C) The “unlocked mechanism” only works well on the target miR-21 and DSN, forming the DSN-miRNA double key. The concentration of miR-21 was 1 nM. (D) Agarose gel electrophoresis (3%) used for the exploration of the DSN cleaving function on DNA/RNA dimers. The left lane is the DNA ladder marker (20 bp to 200 bp). Lane #1 is HDs+ miRNA, lane #2 is HDs + miRNA + DSN, lane #3 is HDs, lane #4 is miRNA and lane #5 is HDs + DSN. (E) Fluorescent confocal images of the Cy5 signal in the homogeneous phase and anchored on the PS. (F) The fluorescent intensity of the Cy5 signal in the homogeneous phase and on the microbeads by a signal enrichment strategy. Error bars show standard deviations (n = 3).
Moreover, in order to identify the effect of the signal enrichment strategy, an experiment was performed in which the control group contained 5 × 10-8 M of Cy5 labeled ssDNA (1 mL) and the experimental group had the same volume and concentration of Cy5 labeled ssDNA (Cy5-ssDNA) and 0.025 mg/mL PS-ssDNA probes. Both groups were characterized by fluorescence microscopy under the same conditions followed by analysis of the fluorescence intensity by Azure C600. The results indicated that the fluorescent intensity on the microbeads was nearly 8.77-fold higher than that in the homogeneous phase, indicating the feasibility of signal enhancement by the fluorescence enrichment carriers (Figure 3E-F).
Finally, FCM was introduced to characterize the detection based on the “unlocked mechanism” and signal enrichment strategy. As shown in Figure 3B, the PS-ssDNA fluorescence signal significantly increased after reacting with the unlocked and remaining FS-ssDNA probes. A comparison of the fluorescence microscopy images of PS-ssDNA with and without the addition of target miRNA shows a significant enhancement of the fluorescence signal, indicating the successful and efficient consequence of the “unlocked mechanism” and signal enrichment strategy.
Optimization of the double key unlocking detection assay
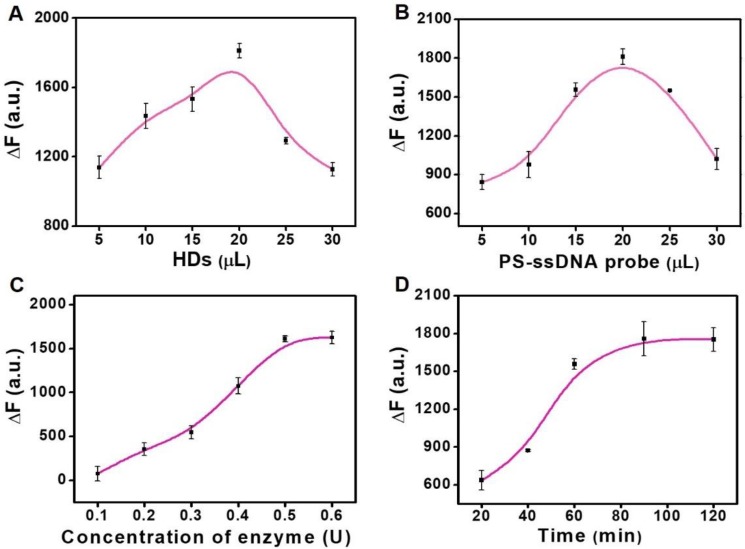
We further explored the optimum experimental factors to achieve a higher sensitivity based on the double key unlocking assay. First, we studied the concentration of HDs and PS-ssDNA probes for the miRNA assay. The value of ∆F (∆F = F1 - F0, where F1 represent the fluorescence intensities of PS enrichment carriers for the positive groups, and F0 represent the fluorescence intensities of PS enrichment carriers for the negative groups) was calculated for performance evaluation. The concentration of HDs loaded on the FS-HDs has a strong effect on the DSN-aided target recycling. With this in mind, we studied the FS-HD probes with different volumes of HDs (Figure 4A). It can be seen that it achieved its best cyclic amplification with 20 μL of HDs (10 μM). Therefore, this volume was adopted in the following assay. Then, we also explored the volume of PS-ssDNA probes (1.25 mg/mL) and found that 20 μL showed the best signal enrichment amplification (Figure 4B).
Figure 4.
Optimization assays. (A) Effect of the concentration of HDs loaded on the FS-HD probes for DSN-aided target recycling. (B) Effect of the concentration of PS-ssDNA probes for signal enrichment. (C) Optimization of the DSN concentration. (D) Optimization of the DSN cleaving time. Error bars show standard deviation (n = 3).
As for the miRNA detection assay, the concentration of DSN and reaction time were both critical parameters for DSN performance, as well as for the high performance of DNA/RNA hybrids, where what matters is the “unlocked mechanism”. As shown in Figure 4C, the ∆F value increased progressively with DSN concentration, but it was almost near plateau after 0.5 U of DSN. Thus, the enzyme concentration of 0.5 U was chosen for the following assays. The reaction time was also optimized; the value of ∆F increased progressively with a prolonged incubation time and nearly reached a plateau after a 90-min incubation with 1 nM of target miRNA (Figure 4D). Therefore, in the subsequent assays, we used 90 min as an optimized incubation time.
Sensitivity and specificity of miRNA detection
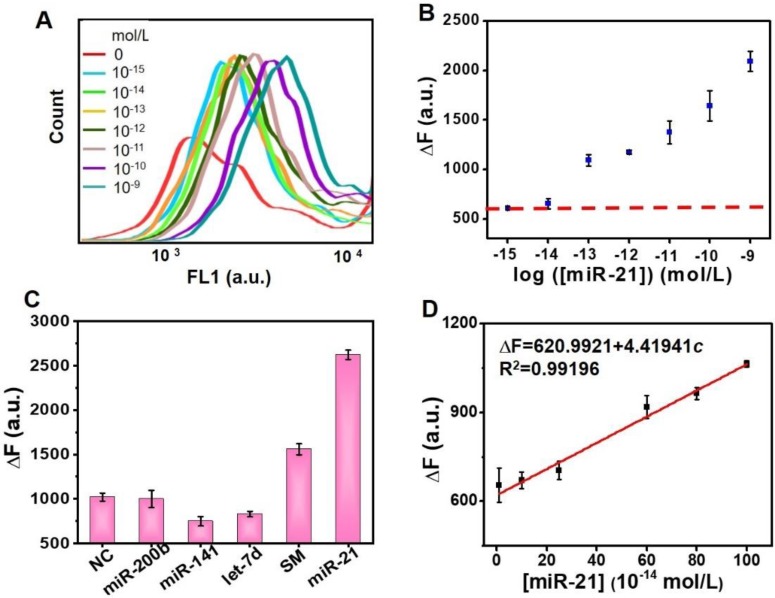
The sensitivity of the proposed detection sensor was investigated in the optimized reaction. Figure 5A shows the corresponding fluorescence spectra of the PS-ssDNA probes in which an increase in the target miR-21 concentration-responsive signal could also be clearly observed in FL1 (488 nm) channel of FCM. The results also support the feasibility of the DSN-miRNA assisted “unlocked mechanism” and the signal enrichment strategy, since the unlocked probes with FS could successfully anchor on the PS. The MFI of each PS was recorded for the quantitative analysis of the target miR-21. The dependence of the ∆F value on miR-21 concentrations is plotted in Figure 5B and 5D. It can be seen that the ∆F value of the PS increases progressively with the target concentrations, and the ∆F value is linearly proportional to the concentration of miR-21 in the range from 0.01 pM and 1 pM (∆F = 620.9921 + 4.41941c, R2 = 0.99196). The value of the limit of detection (LOD) is calculated as: LOD = 3S/M (S represents the standard deviation of blank samples, and M represents the slope of the standard curve). The LOD for this assay is 3.39 fM.
Figure 5.
Sensitivity and specificity of miRNA detection. (A) Spectra of the fluorescence response of the probe 2 with different miR-21 concentrations (0 mol/L, 10-15 mol/L, 10-14 mol/L, 10-13 mol/L, 10-12 mol/L, 10-11 mol/L, 10-10 mol/L, and 10-9 mol/L). (B) Quantitative determination of target miR-21 under optimized conditions. (C) Specificity evaluation: ΔF in response to evaluating NC, miR-200, miR-141, let-7d, SM and miR-21 samples. Error bars show the standard deviation of three independent assays. (D) Linear region for target miRNA detection. Each assay repeats at least three times.
In addition to the necessary high sensitivity, a high specificity is also a critical factor for practical miRNA analysis. To evaluate the specificity of this assay, different miRNA sequences (NC, miR-200b, miR-141, Let-7d, and SM) mismatched with the hairpin sequence were chosen to compare with the perfectly matched target miR-21 (see Table S1 for sequences details). Figure 5C shows that the ∆F value of the target miR-21 performs much higher than the mismatched miRNAs, indicating that our detection sensor has a high specificity for discriminating target miRNA.
Practicability of the miRNA detection platform
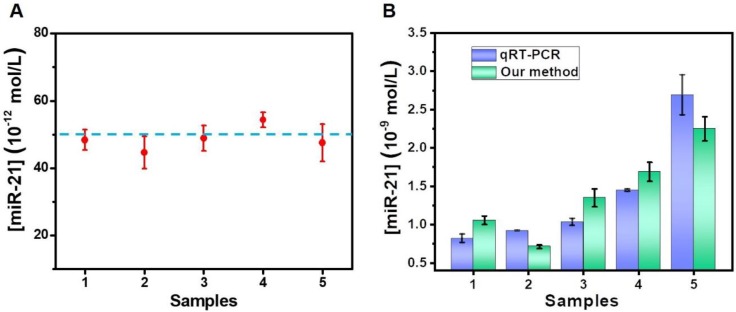
To evaluate the practicability of the proposed method in biological samples, the target miR-21 was detected using a standard addition to spiked human serum samples, as performed in previous reports 36, 37. A final concentration (50 × 10-12 M) of standard miRNA was added into five healthy serum samples for evaluation by our proposed method. A calibration curve of miR-21, shown in Figure S9, was also produced for the calculation of determined concentrations. As displayed in Figure 6A, the analytical results were basically consistent with the theoretical value. Moreover, we also determined the accuracy of our proposed method by measuring the recovery. As shown in Table S2, the method reveals good recovery rates in the range of 89.29-108.65%, and the relative standard deviation (RSD) for all of the samples was below 11.75%.
Figure 6.
Analysis of serum samples. (A) Results for the determination of miR-21 in human serum samples by a standard addition method. (B) The results of breast cancer blood samples comparing qRT-PCR and our sensor. Error bars show standard deviation (n = 3).
Finally, the detection platform was also used to determine miR-21 levels in breast cancer blood samples. After extracting the total miRNA from the blood samples, the abundance of miR-21 was determined. The concentration of miR-21 was acquired by testing the ∆F value. A further measurement of the samples was also carried out by qRT-PCR. Figure 6B shows that the results of the qRT-PCR and our method were in good concordance, indicating the potential of the proposed double key unlocking method for miRNA detection in biological samples.
Simultaneous detection of multiplex miRNAs
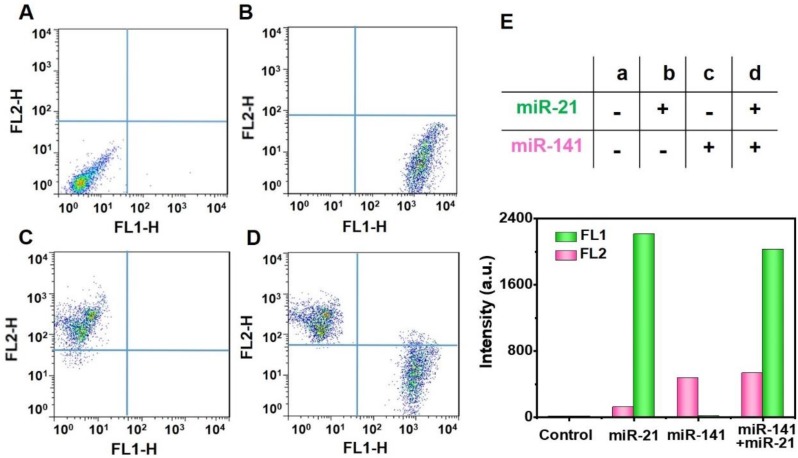
At present, the FCM strategy has become a strong technology for simultaneous multidetection 38. It is well known that cancers are often related to multiplex miRNA markers, which have proven valuable for the early-phase diagnosis of tumors in asymptomatic individuals 39, and the targets miR-21 and miR-141 have been reported as epithelial-associated miRNAs that are overexpressed in a wide range of common human cancers, including breast, lung, and prostate cancer 40, 41. We, next, explored the potential for simultaneous multiple miRNA detection by using two randomly-selected targets and two different kinds of FS (YG for miR-21 by FL1 and pink for miR-141 by FL2). The analyses of the targets miR-21 and miR-141 were carried out by using the FL1 and FL2 channels, respectively. In the presence of different experimental combinations, the two different FS can simultaneously detect the targets miR-21 and miR-141 with negligible cross-talk. We found that there was no obvious fluorescence of capture probes in the lack of target miRNAs (Figure 7A). Furthermore, the introduction of only one single target, miR-21 or miR-141, only resulted in a fluorescence signal increase in the FL1 channel (Figure 7B) or the FL2 channel (Figure 7C). With the addition of both miR-21 and miR-141, the fluorescence in both channels was observed to enhance greatly (Figure 7D). The histograms of the fluorescence response of the multiplex analysis of miR-21 and miR-141 are illustrated in Figure S8. In addition, we can clearly see these concrete results from Figure 7E, indicating the potential of the proposed double key unlocking method for simultaneous multidetection.
Figure 7.
Multiplex miRNAs detection. Multiplex analysis of miR-21 (FL1-H) and miR-141 (FL2-H) by using yellowish green (YG) and pink fluorescence microspheres. YG (Ex 488 nm, Em 509 nm), pink (Ex 538 nm, Em 584 nm). Experimental combinations: (A) 0 µL miR-21 + 0 µL miR-141, (B) 10 µL miR-21 + 0 µL miR-141, (C) 0 µL miR-21 + 10 µL miR-141, and (D) 10 µL miR-21 + 10 µL miR-141. (E) The evaluation of multiplex miRNAs detection by Fluorescence intensity. The fluorescence intensity values are the statistical and quantitative results of A to D.
Conclusions
In conclusion, a novel assay based on an “unlocked mechanism” with a double key and fluorescence enrichment with signal amplification has been proposed for multiplex miRNAs detection by FCM. The target miRNA and DSN act together as a double key to induce the effect of the “unlocked mechanism”, increasing the specificity of detection. Both the recycling cutting effect of DSN and fluorescence enrichment of PS contributes to the high sensitivity for detecting the target miR-21. Meanwhile, with the help of FCM, the proposed detection platform enabled a simultaneous detection of multiplex miRNAs with high specificity. Overall, the proposed strategy shows a great potential for early clinical diagnosis.
Supplementary Material
Supplementary figures and tables.
Acknowledgments
The authors gratefully acknowledge that this work was financially supported by the National Natural Science Foundation of China (31600800), the National Key Research and Development Plan “nanotech” key project (2017YFA0205104), and the Natural Science Foundation of Tianjin, China (17JCQNJC09100, 16ZXMJSY00010, 17ZXMFSY00210).
Abbreviations
- Ctrl
control
- DSN
duplex-specific nuclease
- EDC
N3-dimethylpropane-1,3-diamine
- EDTA
ethylene diamine tetraacetic acid
- FCM
flow cytometry
- FL
fluorescence
- FL-H
fluorescence height
- FS
fluorescent particles
- HD
hairpin DNA
- HDs
hairpin DNA probes
- LOD
limit of detection
- MES
2-Morpholinoethanesulfonic acid, monohydrate
- MFI
mean fluorescence intensity
- miRNA
microRNA
- NC
negative control miRNA
- PBS
phosphate-buffered saline
- PS
polystyrene microparticles
- qRT-PCR
quantitative reverse transcriptase PCR
- RSD
relative standard deviation
- SM
single mismatched miRNA
- ssDNA
single-stranded DNA
- YG
yellowish green.
References
- 1.Jin Z, Geissler D, Qiu X, Wegner KD, Hildebrandt N. A rapid, amplification-free, and sensitive diagnostic assay for single-step multiplexed fluorescence detection of microRNA. Angew Chem Int Ed Engl. 2015;54:10024–10029. doi: 10.1002/anie.201504887. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol. 2010;11:252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 4.Su X, Xing J, Wang Z, Chen L, Cui M, Jiang B. microRNAs and ceRNAs: RNA networks in pathogenesis of cancer. Chin J Cancer Res. 2013;25:235–239. doi: 10.3978/j.issn.1000-9604.2013.03.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiu HS, Martinez MR, Komissarova EV, Llobet-Navas D, Bansal M, Paull EO, The number of titrated microRNA species dictates ceRNA regulation. Nucleic Acids Res; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen P, Li W, Liu Y, Ding Z, Deng Y, Zhu X. et al. High-throughput low-background g-quadruplex aptamer chemiluminescence assay for ochratoxin A using a single photonic crystal microsphere. Anal Chem. 2017;89:11862–11868. doi: 10.1021/acs.analchem.7b03592. [DOI] [PubMed] [Google Scholar]
- 7.Robles G, Fresno JM, Martinez-Tarifa JM, Ardila-Rey JA, Parrado-Hernandez E. Partial discharge spectral characterization in HF, VHF and UHF bands using particle swarm optimization. Sensors. 2018;18:746. doi: 10.3390/s18030746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu W, Wang J, Wu Q, Tang W, Feng Q, Sun J. et al. High-throughput sample-to-answer detection of DNA/RNA in crude samples within functionalized micro-pipette tips. Biosens Bioelectron. 2016;75:28–33. doi: 10.1016/j.bios.2015.08.016. [DOI] [PubMed] [Google Scholar]
- 9.Wang P, Jing F, Li G, Wu Z, Cheng Z, Zhang J. et al. Absolute quantification of lung cancer related microRNA by droplet digital PCR. Biosens Bioelectron. 2015;74:836–842. doi: 10.1016/j.bios.2015.07.048. [DOI] [PubMed] [Google Scholar]
- 10.Benes V, Castoldi M. Expression profiling of microRNA using real-time quantitative PCR, how to use it and what is available. Methods. 2010;50:244–249. doi: 10.1016/j.ymeth.2010.01.026. [DOI] [PubMed] [Google Scholar]
- 11.Varallyay E, Burgyan J, Havelda Z. MicroRNA detection by northern blotting using locked nucleic acid probes. Nat Protoc. 2008;3:190–196. doi: 10.1038/nprot.2007.528. [DOI] [PubMed] [Google Scholar]
- 12.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 13.Li W, Ruan K. MicroRNA detection by microarray. Anal Bioanal Chem. 2009;394:1117–1124. doi: 10.1007/s00216-008-2570-2. [DOI] [PubMed] [Google Scholar]
- 14.Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nat Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- 15.Pomponi D, Bernardi ML, Liso M. et al. Allergen micro-bead array for IgE detection: a feasibility study using allergenic molecules tested on a flexible multiplex flow cytometric immunoassay. PLoS One. 2012;7:e35697. doi: 10.1371/journal.pone.0035697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Gao Z, Ye H, Wan S, Pierce M, Tang D. et al. A non-enzyme cascade amplification strategy for colorimetric assay of disease biomarkers. Chem Commun (Camb) 2017;53:9055–9058. doi: 10.1039/c7cc04521b. [DOI] [PubMed] [Google Scholar]
- 17.Valoczi A, Hornyik C, Varga N, Burgyan J, Kauppinen S, Havelda Z. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 2004;32(e):175. doi: 10.1093/nar/gnh171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wark AW, Lee HJ, Corn RM. Multiplexed detection methods for profiling microRNA expression in biological samples. Angew Chem Int Ed Engl. 2008;47:644–652. doi: 10.1002/anie.200702450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012;13:358–369. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou W, Li D, Xiong C, Yuan R, Xiang Y. Multicolor-Encoded Reconfigurable DNA Nanostructures Enable Multiplexed Sensing of Intracellular MicroRNAs in Living Cells. ACS Appl Mater Interfaces. 2016;8:13303–13308. doi: 10.1021/acsami.6b03165. [DOI] [PubMed] [Google Scholar]
- 21.Cui L, Lin X, Lin N, Song Y, Zhu Z, Chen X. et al. Graphene oxide-protected DNA probes for multiplex microRNA analysis in complex biological samples based on a cyclic enzymatic amplification method. Chem Commun (Camb) 2012;48:194–196. doi: 10.1039/c1cc15412e. [DOI] [PubMed] [Google Scholar]
- 22.Li X, Song J, Xue Q, Zhao H, Liu M, Chen B. et al. Sensitive and selective detection of the p53 gene based on a triple-helix magnetic probe coupled to a fluorescent liposome hybridization assembly via rolling circle amplification. Analyst. 2017;142:3598–3604. doi: 10.1039/c7an01255a. [DOI] [PubMed] [Google Scholar]
- 23.Bing T, Liu X, Cheng X, Cao Z, Shangguan D. Bifunctional combined aptamer for simultaneous separation and detection of thrombin. Biosens Bioelectron. 2010;25:1487–1492. doi: 10.1016/j.bios.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 24.Mei H, Bing T, Qi C, Zhang N, Liu X, Chang T. et al. Rational design of Hg2+ controlled streptavidin-binding aptamer. Chem Commun (Camb) 2013;49:164–166. doi: 10.1039/c2cc36416f. [DOI] [PubMed] [Google Scholar]
- 25.Butterworth P, Baltar HT, Kratzmeier M, Goldys EM. Simple bead assay for detection of live bacteria. Anal Chem. 2011;83:1443–1447. doi: 10.1021/ac103109v. [DOI] [PubMed] [Google Scholar]
- 26.Qi Y, Qiu L, Fan W, Liu C, Li Z. An enzyme-free flow cytometric bead assay for the sensitive detection of microRNAs based on click nucleic acid ligation-mediated signal amplification. Analyst. 2017;142:2967–2973. doi: 10.1039/c7an00989e. [DOI] [PubMed] [Google Scholar]
- 27.Ren W, Liu H, Yang W, Fan Y, Yang L, Wang Y. et al. A cytometric bead assay for sensitive DNA detection based on enzyme-free signal amplification of hybridization chain reaction. Biosens Bioelectron. 2013;49:380–386. doi: 10.1016/j.bios.2013.05.055. [DOI] [PubMed] [Google Scholar]
- 28.Xu J, Wang Y, Yang L, Gao Y, Li B, Jin Y. A cytometric assay for ultrasensitive and robust detection of human telomerase RNA based on toehold strand displacement. Biosens Bioelectron. 2017;87:1071–1076. doi: 10.1016/j.bios.2016.08.038. [DOI] [PubMed] [Google Scholar]
- 29.Qiu X, Zhang H, Yu H, Jiang T, Luo Y. Duplex-specific nuclease-mediated bioanalysis. Trends Biotechnol. 2015;33:180–188. doi: 10.1016/j.tibtech.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 30.Degliangeli F, Kshirsagar P, Brunetti V, Pompa PP, Fiammengo R. Absolute and direct microRNA quantification using DNA-gold nanoparticle probes. J Am Chem Soc. 2014;136:2264–2267. doi: 10.1021/ja412152x. [DOI] [PubMed] [Google Scholar]
- 31.Tian B, Ma J, Qiu Z, Zardan GDLT, Donolato M, Hansen M.F. et al. Optomagnetic Detection of MicroRNA Based on Duplex-Specific Nuclease-Assisted Target Recycling and Multilayer Core-Satellite Magnetic Superstructures. ACS Nano. 2017;11:1798–1806. doi: 10.1021/acsnano.6b07763. [DOI] [PubMed] [Google Scholar]
- 32.Wang Q, Yin BC, Ye BC. A novel polydopamine-based chemiluminescence resonance energy transfer method for microRNA detection coupling duplex-specific nuclease-aided target recycling strategy. Biosens Bioelectron. 2016;80:366–372. doi: 10.1016/j.bios.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 33.Lu W, Chen Y, Liu Z. et al. Quantitative detection of microRNA in one step via next generation magnetic relaxation witch sensing. ACS Nano. 2016;10:6685–6692. doi: 10.1021/acsnano.6b01903. [DOI] [PubMed] [Google Scholar]
- 34.Nolan JP, Sklar LA. Suspension array technology: evolution of the flat-array paradigm. Trends Biotechnol. 2002;20:9–12. doi: 10.1016/s0167-7799(01)01844-3. [DOI] [PubMed] [Google Scholar]
- 35.Nolan JP, Mandy FF. Suspension array technology: new tools for gene and protein analysis. Cell Mol Biol. 2001;47:1241–1256. [PubMed] [Google Scholar]
- 36.Zhao Q, Piao J, Peng W, Wang Y, Zhang B, Gong X. et al. Simple and sensitive quantification of microRNAs via PS@Au microspheres-based DNA probes and DSN-assisted signal amplification platform. ACS Appl Mater Interfaces. 2018;10:3324–3332. doi: 10.1021/acsami.7b16733. [DOI] [PubMed] [Google Scholar]
- 37.Zhang J, Zhao Q, Wu Y, Zhang B, Peng W, Piao J. et al. The construction of a novel nucleic acids detection microplatform based on the NSET for one-step detecting TK1-DNA and microRNA-21. Biosens Bioelectron. 2017;97:26–33. doi: 10.1016/j.bios.2017.05.039. [DOI] [PubMed] [Google Scholar]
- 38.Pomponi D, Bernardi ML, Liso M, Palazzo P, Tuppo L, Rafaiani C. et al. Allergen micro-bead array for IgE detection: a feasibility study using allergenic molecules tested on a flexible multiplex flow cytometric immunoassay. PLoS One. 2012;7:e35697. doi: 10.1371/journal.pone.0035697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yin BC, Liu YQ, Ye BC. One-step, multiplexed fluorescence detection of microRNAs based on duplex-specific nuclease signal amplification. J Am Chem Soc. 2012;134:5064–5067. doi: 10.1021/ja300721s. [DOI] [PubMed] [Google Scholar]
- 40.Yaman AF, Kovancilar M, Dizdar Y, Darendeliler E, Holdenrieder S, Dalay N. et al. Investigation of miR-21, miR-141, and miR-221 in blood circulation of patients with prostate cancer. Tumour Biol. 2011;32:583–588. doi: 10.1007/s13277-011-0154-9. [DOI] [PubMed] [Google Scholar]
- 41.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S. et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables.