Abstract
Numerous studies have investigated individual biomarkers in relation to risk of type 2 diabetes. However, few have considered the interconnectivity of these biomarkers in the etiology of diabetes as well as the potential changes in the biomarker correlation network during diabetes development. We conducted a secondary analysis of 27 plasma biomarkers representing glucose metabolism, inflammation, adipokines, endothelial dysfunction, IGF axis, and iron store plus age and BMI at blood collection from an existing case-control study nested in the Nurses’ Health Study (NHS), including 1,303 incident diabetes case subjects and 1,627 healthy women. A correlation network was constructed based on pairwise Spearman correlations of the above factors that were statistically different between case and noncase subjects using permutation tests (P < 0.0005). We further evaluated the network structure separately among diabetes case subjects diagnosed <5, 5–10, and >10 years after blood collection versus noncase subjects. Although pairwise biomarker correlations tended to have similar directions comparing diabetes case subjects to noncase subjects, most correlations were stronger in noncase than in case subjects, with the largest differences observed for the insulin/HbA1c and leptin/adiponectin correlations. Leptin and soluble leptin receptor were two hubs of the network, with large numbers of different correlations with other biomarkers in case versus noncase subjects. When examining the correlation network by timing of diabetes onset, there were more perturbations in the network for case subjects diagnosed >10 years versus <5 years after blood collection, with consistent differential correlations of insulin and HbA1c. C-peptide was the most highly connected node in the early-stage network, whereas leptin was the hub for mid- or late-stage networks. Our results suggest that perturbations of the diabetes-related biomarker network may occur decades prior to clinical recognition. In addition to the persistent dysregulation between insulin and HbA1c, our results highlight the central role of the leptin system in diabetes development.
Introduction
Biomarkers are widely used in molecular epidemiologic research to understand the etiology of chronic diseases and to assist in risk prediction for disease prevention and early detection (1). Traditional studies typically focus on a single or several related biomarkers involved in the same biologic pathway (e.g., inflammatory pathway) or reflecting one underlying exposure (e.g., endothelial dysfunction). However, as the causes of human diseases are commonly multifactorial, elucidating the interdependency and interconnectivity among different biomarkers and pathways may provide a more comprehensive view of and insight into the pathogenic process. Network-based approaches have only recently been used in epidemiologic research (2,3) but provide the opportunity to systematically interrogate individual biomarkers and pathways to uncover new links among them.
Type 2 diabetes is a chronic, multisystem, and complex metabolic disorder with rapidly rising burden during the past two decades (4). It is characterized by impaired glucose metabolism and insulin resistance, coupled with dysregulation of multiple biologic pathways. We and others have shown that inflammatory biomarkers (5), adipokines (6,7), IGF axis (8), biomarkers of endothelial dysfunction (9), and body iron stores (10), among other circulating biomarkers (11–13), are predictive of future risk of diabetes. These studies, although providing important evidence for the underlying etiology, examined different groups of biomarkers in isolation. It remains unclear at the system level how one group of biomarkers may interact or connect with biomarkers in other biologic pathways to contribute to diabetes development.
Therefore, we conducted a secondary data analysis that leveraged existing prediagnostic plasma biomarker data from a case-control study of type 2 diabetes nested in the Nurses’ Health Study (NHS) cohort, including 27 circulating biomarkers among 2,930 women (1,303 incident diabetes case subjects and 1,627 healthy women). We used these data to identify the perturbed biomarker correlation network in women who developed diabetes over follow-up versus those who did not. We further used the longitudinal study design of our cohort to characterize whether the correlation network patterns changed with increasing time between blood collection and diagnosis to examine patterns in progression to type 2 diabetes.
Research Design and Methods
Study Population
The NHS was established in 1976 among 121,700 U.S. female registered nurses, ages 30–55 years (14). All women completed a baseline questionnaire, and their health conditions and lifestyle factors have been updated biennially by follow-up questionnaires. Between 1989 and 1990, 32,826 women who were free of cancer provided a heparin blood sample. A prospective, nested case-control study has been conducted to examine individual plasma biomarkers in relation to diabetes risk using incident cases diagnosed after blood collection (5–12). For each case subject, one to two control subjects were randomly sampled from those who were free of type 2 diabetes, cardiovascular disease, and cancer at the time of the case diagnosis and were matched on age at blood draw, date of blood draw, race, and fasting status of the blood sample.
Ascertainment of Incident Diabetes Case Subjects
On each biennial questionnaire, participants reported diagnoses of type 2 diabetes, which were further ascertained by a supplementary questionnaire querying information on symptoms, diagnostic tests, and relevant treatment. For cases diagnosed through 1997, a confirmed type 2 diabetes case subject was required to meet the following criteria according to the National Diabetes Data Group: 1) elevated plasma glucose levels (fasting glucose ≥140 mg/dL, random glucose ≥200 mg/dL, or glucose ≥200 mg/dL after an oral glucose test) with presence of at least one symptom (polydipsia, polyuria, polyphagia, weight loss, or coma), 2) elevated plasma glucose on at least two occasions with no symptoms, and 3) hypoglycemic therapy with insulin or oral medications. For cases diagnosed after 1997, the confirmation was based on the American Diabetes Association recommendations, which used an updated cutoff of 126 mg/dL for fasting glucose. In a validation study of 62 NHS participants ascertained to have diabetes through the supplementary questionnaire, 61 (98%) were confirmed by review of medical records (15).
Measurement of Plasma Biomarkers
The assay details for measuring each biomarker have been described previously (5–12). In brief, leptin, soluble leptin receptor (sOB-R), and interleukin 6 (IL-6) were measured by an ultrasensitive ELISA assay from R&D Systems (Minneapolis, MN). Total and high-molecular-weight adiponectin were assayed with a quantitative monoclonal sandwich ELISA (ALPCO Diagnostics Inc., Salem, NH). Resistin was measured by ELISA (Linco Research, St. Charles, MO) with a minimum detectable limit of 0.16 ng/mL. Fasting insulin concentrations were determined by an electrochemiluminescence immunoassay using the Roche E modular system (Roche Diagnostics, Indianapolis, IN), and hs-CRP was measured via an immunoturbidimetric assay (Denka Seiken, Tokyo, Japan). All assays followed the NHS protocol, including 10% blinded quality control replicates. The mean interassay CVs were generally <10% for various biomarkers. The laboratory was blinded to the sample status (i.e., case subjects, control subjects, or quality control subjects), and case subjects and the matched control subjects were assayed together in the same batch.
Statistical Analyses
Due to the nested case-control design, women who were free of diabetes at the time they were sampled as control subjects may have developed diabetes at a later time. As the objective of this study was to identify biomarker networks associated with diabetes status (vs. estimate diabetes incidence rate ratio as in the original case-control study), the incidence density sampling/matching design was not considered and the control women who later developed diabetes (n = 129) were considered as case subjects in this analysis. Our comparison was women who developed diabetes at any time from blood draw to June 2013 (∼23 years) versus those who did not. The diabetes status was termed as case subjects versus noncase subjects to distinguish from the original case-control status. As all available biomarkers have been identified in previous studies as potential risk factors to predict diabetes risk (5–12), we considered all biomarkers in the analysis; we also included age and BMI, which are established diabetes risk factors. We calculated the pairwise Spearman correlations between age, BMI, and 27 plasma biomarkers in case and noncase subjects separately. Each correlation was calculated among women with nonmissing data for that pairwise comparison. To quantify the difference in the correlations between case and noncase subjects, we evaluated the difference in correlation between the two groups and assessed the statistical significance of the difference using permutation tests that randomly assigned the case and noncase status and calculated the correlations between the reassigned groups (2,3). The process was repeated 1,000 times to obtain the distribution of the correlation differences under the null hypothesis that the connectivity between biomarkers was not associated with case status. Based on this distribution, a standardized correlation difference (the z statistic) was calculated. Selection criteria for correlations to evaluate in a network analysis were as follows: 1) an absolute correlation difference of |Δr| > 0.15 and 2) a corresponding standardized correlation difference of |z| > 3.5 (approximately equivalent to a two-sided P value <0.0005). The selected connections were plotted as an undirected network graph, with the network hub being the biomarker with the largest number of connections (edges) with other biomarkers. The width of the edges was proportional to the absolute difference in the correlations between case and noncase subjects. The type of the edges (i.e., solid vs. dashed) was used to indicate whether the magnitude of the correlations was greater in case or noncase subjects, and the color of the edges was used to indicate the direction of the correlations. Sensitivity analyses were conducted to evaluate the potential impact of loosening (i.e., |Δr| > 0.1 and |z| > 3) or tightening (i.e., |Δr| > 0.2 and |z| > 4) the criteria for entry into the network structure analysis.
Next, to understand the dynamic biomarker network patterns in the pathogenesis of diabetes, we constructed three networks in parallel by comparing diabetes case subjects diagnosed in different periods of time relative to the blood collection with the noncase subjects, including 1) case subjects diagnosed <5 years after blood collection, 2) case subjects diagnosed 5–10 years after blood collection, and 3) case subjects diagnosed >10 years after blood collection. Similar methods as described above were used to derive the correlation network.
Several sensitivity analyses were conducted to address the potential impact of age and BMI on the correlation network. First, we calculated pairwise partial Spearman correlations adjusted for age and BMI and compared them with crude correlations. We also calculated differences in partial correlations between case and noncase subjects and compared them with differences in crude correlations. Second, given that BMI was one of the strongest risk factors for diabetes, we conducted “block” permutation tests within quintiles of BMI (i.e., permuted case status within the same BMI category) to evaluate its impact on the network structure. Third, to exclude the potential age effects that may be associated with the length of intervals between blood collection and diabetes diagnosis, we constructed the networks by comparing each case group (as described above) to the age-matched noncase subjects (as opposed to all noncase subjects). All analyses were performed using R statistical packages (version 3.2.5), and the network structure was visualized in Cytoscape (16).
Results
There were 1,627 women who developed diabetes by the end of follow-up (June 2013) and 1,303 noncase subjects included in the analysis (Table 1). The number of women with various biomarkers ranged from 433 for high-molecular-weight adiponectin to 2,361 for proinsulin. Among diabetes case subjects, 311 were diagnosed <5 years after blood collection, 491 5–10 years, and 501 >10 years. Most biomarkers had significantly different levels between case and noncase subjects (P < 0.05), with expected trends by time to diabetes diagnosis (P-trend < 0.05). Due to matched design, case and noncase subjects were similar regarding age distribution.
Table 1.
Distribution of biomarkers, age, and BMI by diabetes status and by time to diabetes diagnosis
| Time to diabetes diagnosis after blood collection |
|||||||
|---|---|---|---|---|---|---|---|
| Noncase subjects2 | Case subjects | P value | >10 years | 5–10 years | <5 years | P-trend | |
| Total n1 | 1,627 | 1,303 | 501 | 491 | 311 | ||
| Biomarkers of glucose metabolism | |||||||
| Proinsulin, pmol/L | 9.8 (5.6, 19.6) | 18.6 (7.7, 54.7) | <0.0001 | 12.0 (6.6, 23.0) | 17.0 (7.8, 38.4) | 21.0 (8.1, 61.4) | <0.0001 |
| C-peptide, nmol/L | 0.5 (0.3, 0.9) | 0.8 (0.5, 1.6) | <0.0001 | 0.6 (0.4, 1.2) | 0.8 (0.5, 1.3) | 0.9 (0.5, 1.7) | <0.0001 |
| Insulin, μU/mL | 6.1 (2.6, 14.5) | 8.6 (3.5, 20.6) | <0.0001 | 6.7 (3.2, 17.2) | 8.7 (3.5, 19.5) | 13.3 (6.1, 28.0) | <0.0001 |
| HbA1c, % | 5.4 (4.9, 5.8) | 5.7 (5.1, 6.6) | <0.0001 | 5.3 (5.0, 5.8) | 5.6 (5.1, 6.2) | 6.4 (5.5, 7.6) | <0.0001 |
| HbA1c, mmol/mol | 36 (30, 40) | 39 (32, 49) | <0.0001 | 34 (31, 40) | 38 (32, 44) | 46 (37, 60) | <0.0001 |
| Inflammatory biomarkers | |||||||
| CRP, mg/L | 1.4 (0.3, 6.1) | 3.1 (0.8, 10.8) | <0.0001 | 2.5 (0.7, 8.2) | 3.1 (0.8, 10.2) | 4.0 (1.3, 14.7) | <0.0001 |
| IL-6, pg/mL | 1.8 (0.9, 4.2) | 2.5 (1.2, 5.2) | <0.0001 | 2.4 (1.3, 4.5) | 2.2 (1.1, 4.4) | 3.0 (1.4, 7.0) | <0.0001 |
| IL-18, pg/mL | 267 (159, 447) | 319 (189, 535) | <0.0001 | 304 (183, 495) | 322 (192, 562) | 331 (194, 542) | 0.06 |
| sTNFR2, pg/mL | 2,292 (1,681, 3,430) | 2,606 (1,861, 3,858) | <0.0001 | 2,595 (1,796, 3,780) | 2,556 (1,829, 3,770) | 2,683 (1,921, 4,109) | 0.11 |
| Adipokines | |||||||
| Leptin, ng/mL | 17.4 (6.1, 42.9) | 25.4 (11.7, 48.3) | <0.0001 | 24.4 (10.6, 49.0) | 26.7 (12.3, 48.4) | 24.6 (11.7, 47.9) | 0.85 |
| sOB-R, ng/mL | 32.1 (22.0, 46.9) | 27.0 (19.2, 39.2) | <0.0001 | 27.5 (19.9, 38.9) | 26.5 (18.9, 38.2) | 27.3 (19.2, 40.4) | 0.43 |
| ADPN, µg/mL | 17.0 (9.4, 28.7) | 10.7 (4.9, 21.1) | <0.0001 | 12.5 (6.3, 23.0) | 10.4 (4.7, 20.8) | 9.5 (4.6, 19.7) | <0.0001 |
| HMW ADPN, µg/mL | 6.5 (3.1, 14.7) | 3.5 (1.4, 8.3) | <0.0001 | 4.2 (1.8, 9.3) | 3.3 (1.3, 7.9) | 3.1 (1.2, 7.8) | <0.0001 |
| Resistin, ng/mL | 15.7 (8.9, 28.9) | 17.8 (9.9, 34.8) | <0.0001 | 18.9 (10.1, 37.3) | 17.3 (9.7, 32.1) | 17.5 (9.6, 34.5) | 0.09 |
| Biomarkers of endothelial dysfunction | |||||||
| E-sel, ng/mL | 43.6 (24.5, 72.2) | 60.3 (34.0, 100.2) | <0.0001 | 52.4 (30.6, 85.9) | 57.7 (33.6, 91.2) | 66.3 (37.1, 130.5) | <0.0001 |
| ICAM, ng/mL | 247 (195, 318) | 274 (213, 378) | <0.0001 | 265 (197, 337) | 267 (213, 346) | 286 (213, 422) | <0.0001 |
| VCAM, ng/mL | 529 (392, 721) | 547 (394, 762) | 0.01 | 512 (371, 714) | 528 (386, 726) | 585 (404, 823) | <0.0001 |
| Biomarkers of the IGF axis | |||||||
| IGF-1, ng/mL | 143 (91, 220) | 143 (93, 221) | 0.97 | 149 (99, 221) | 143 (95, 227) | 133 (83, 211) | 0.001 |
| Free IGF, ng/mL | 0.4 (0.2, 0.9) | 0.4 (0.2, 0.9) | 0.45 | 0.4 (0.2, 0.9) | 0.5 (0.2, 1.0) | 0.4 (0.2, 0.8) | 0.11 |
| IGFBP-1, ng/mL | 33.4 (11.1, 80.4) | 16.5 (4.4, 52.4) | <0.0001 | 17.4 (4.3, 52.8) | 14.5 (3.8, 50.3) | 18.1 (6.7, 57.0) | 0.89 |
| IGFBP-2, ng/mL | 493 (210, 1,042) | 282 (134, 622) | <0.0001 | 304 (139, 700) | 273 (134, 578) | 257 (125, 551) | 0.004 |
| IGFBP-3, ng/mL | 4,537 (3,451, 5,941) | 4,917 (3,626, 6,678) | <0.0001 | 4,929 (3,781, 6,589) | 4,930 (3,655, 6,876) | 4,872 (3,283, 6,633) | 0.88 |
| Biomarkers for iron status | |||||||
| Ferritin, ng/mL | 45.7 (11.0, 144.4) | 67.6 (17.2, 239.1) | <0.0001 | 61.7 (20.6, 173.5) | 58.8 (12.4, 209.9) | 84.4 (20.6, 285.4) | <0.0001 |
| sTfR, mg/L | 3.1 (2.2, 4.3) | 3.3 (2.3, 4.6) | <0.0001 | 3.3 (2.3, 5.1) | 3.3 (2.3, 4.5) | 3.3 (2.3, 4.7) | 0.67 |
| Other biomarkers | |||||||
| Vitamin D, ng/mL | 22.5 (13.7, 33.8) | 20.2 (12.7, 30.3) | <0.0001 | 21.9 (14.4, 34.3) | 20.6 (12.9, 30.7) | 19.6 (12.6, 29.2) | 0.11 |
| HGB, g/dL | 14.7 (11.9, 18.1) | 14.8 (12.2, 18.2) | 0.39 | 14.5 (11.4, 18.3) | 15.4 (12.7, 18.7) | 14.6 (12.0, 17.7) | 0.61 |
| HCO3, mmol/L | 22.1 (18.8, 25.7) | 21.8 (18.6, 25.6) | 0.09 | 21.9 (18.9, 24.7) | 21.9 (18.7, 25.8) | 21.8 (18.4, 25.2) | 0.77 |
| Creatinine, mg/dL | 0.7 (0.6, 0.9) | 0.7 (0.5, 0.9) | 0.002 | 0.7 (0.6, 0.9) | 0.7 (0.5, 0.9) | 0.7 (0.5, 0.9) | 0.53 |
| Other factors at blood collection | |||||||
| Age, years | 56.1 (6.9) | 56.0 (6.9) | 0.87 | 55.1 (6.8) | 56.1 (6.8) | 57.4 (6.9) | <0.0001 |
| BMI, kg/m2 | 26.5 (5.6) | 29.9 (5.5) | <0.0001 | 29.2 (5.3) | 30.1 (5.4) | 30.8 (5.8) | <0.0001 |
Data are n or geometric means (10th, 90th percentiles), except for age and BMI, which are presented as mean (SD).
ADPN, adiponectin; E-sel, E-selectin; HCO3, bicarbonate; HGB, hemoglobin; HMW, high molecular weight; ICAM, intercellular adhesion molecule; sTfR, soluble transferrin receptor; sTNFR2, soluble tumor necrosis factor receptor type 2.
1Actual sample size varied by biomarkers.
2One hundred twenty-nine women sampled as control subjects in original nested case-control studies later developed diabetes and were considered as case subjects in the analysis.
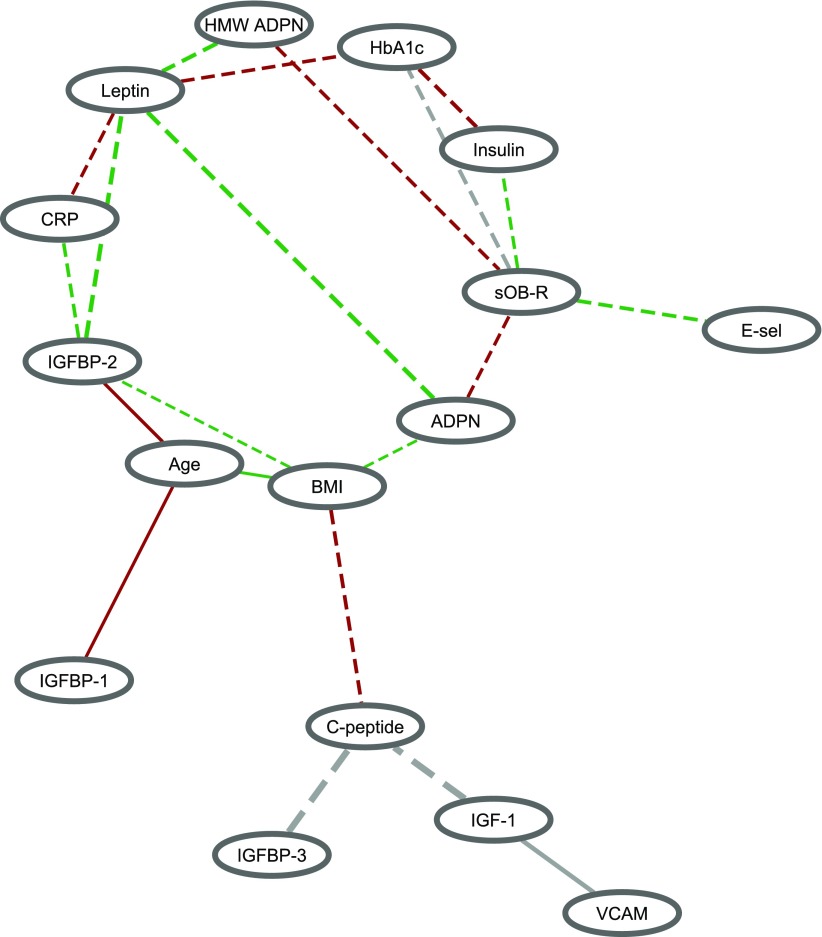
Most of the correlations between biomarkers were in the same direction between case and noncase subjects (Supplementary Fig. 1), but these correlations were, in general, stronger in noncase subjects than in case subjects (i.e., more dashed lines compared with solid lines) (Fig. 1). For example, insulin was more strongly positively correlated with HbA1c in noncase subjects (r = 0.62) than in case subjects (r = 0.41), and the inverse association between leptin and total adiponectin was also stronger in noncase subjects (r = −0.26) than in case subjects (r = −0.07). Leptin appeared to be the hub of the network, with connections with five other biomarkers that differed significantly between case and noncase subjects, including total adiponectin, high-molecular-weight adiponectin, CRP, HbA1c, and IGF binding protein 2 (IGFBP-2). sOB-R was another important node in the network, and also had connections with five other biomarkers, including insulin, HbA1c, total adiponectin, high-molecular-weight adiponectin, and E-selectin. In addition, BMI had differential connections with age, adiponectin, C-peptide, and IGFBP-2 between case and noncase subjects. Notably, the correlation between sOB-R and HbA1c and the relationships involving IGFBP-3, IGF-1, vascular cell adhesion molecule (VCAM), and C-peptide were in the opposite direction by diabetes status (gray edges). When the threshold for selecting edges was lowered (Supplementary Fig. 2A), leptin (seven edges) and sOB-R (nine edges) were still the biomarkers with the most connections (highest degree); BMI also had seven edges. By contrast, when a more strict threshold was applied (Supplementary Fig. 2B), only five edges remained, including the connections of leptin with total adiponectin, high-molecular-weight adiponectin, and IGFBP-2, as well as the connections of HbA1c with insulin and sOB-R.
Figure 1.
Biomarker correlation network comparing 1,303 diabetes case subjects vs. 1,627 noncase subjects. Significant edges were identified by the threshold of both |Δr| > 0.15 and z > 3.5 (equivalent to a two-sided P value <0.0005). The width of the edge is proportional to the corresponding difference in the biomarker correlation between case and noncase subjects. A solid edge indicates that the absolute value of the correlation is stronger in case than in noncase subjects, and a dashed edge indicates the opposite. The color of the edge indicates the direction of the correlation in case and noncase subjects: red, positive correlations in both case and noncase subjects; green, negative correlations in both case and noncase subjects; gray, opposite correlations between case and noncase subjects. ADPN, adiponectin; E-sel, E-selectin; HMW, high molecular weight.
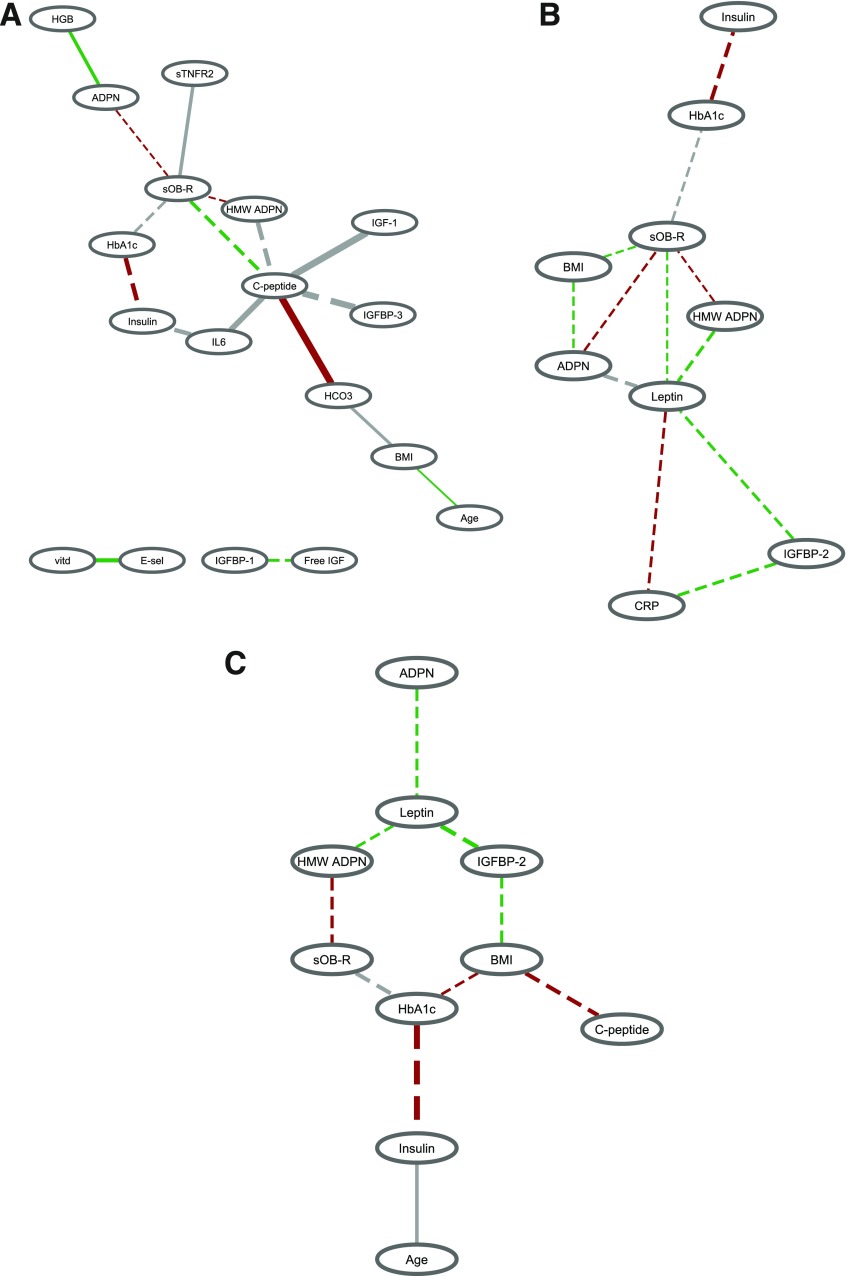
Overall, compared with noncase subjects, there were more perturbations in the biomarker correlation structure for diabetes case subjects diagnosed many years after blood collection than case subjects diagnosed shortly after blood collection (Fig. 2 and Supplementary Fig. 3). The number of significant edges was 17 for case subjects diagnosed >10 years after blood collection versus noncase subjects, 12 for case subjects diagnosed 5–10 years after blood collection, and 10 for case subjects diagnosed <5 years after blood collection. Comparison of case subjects diagnosed >10 years after blood collection versus healthy women showed a central role of C-peptide in the early pathogenesis of diabetes (Fig. 2A). The correlations of C-peptide with sOB-R, high-molecular-weight adiponectin, IL-6, IGF-1, IGFBP-3, and bicarbonate were significantly different between case and noncase subjects. Particularly, the associations with C-peptide were in the opposite direction between case and noncase subjects for high-molecular-weight adiponectin (r = 0.08 in case subjects and r = −0.36 in noncase subjects), IL-6 (r = −0.31 in case subjects and r = 0.24 in noncase subjects), IGF-1 (r = −0.45 in case subjects and r = 0.21 in noncase subjects), and IGFBP-3 (r = −0.28 in case subjects and r = 0.34 in noncase subjects). A large difference in the correlation between case versus noncase subjects was observed for HbA1c and insulin (r = 0.19 in case subjects and r = 0.63 in noncase subjects). For case subjects diagnosed 5–10 years after blood collection (Fig. 2B), leptin was the hub of the biomarker network connected to CRP, sOB-R, adiponectin, high-molecular-weight adiponectin, and IGFBP-2. Notably, the correlation between leptin and adiponectin was inverse among noncase subjects (r = −0.26) but slightly positive among case subjects (r = 0.06). Similar to case subjects diagnosed >10 years after blood draw, a stronger positive relationship between HbA1c and insulin for noncase subjects (r = 0.63) than case subjects (r = 0.31) was also observed. Finally, for the network considering diabetes case subjects shortly diagnosed after blood collection (Fig. 2C), HbA1c had three differential connections with insulin, sOB-R, and BMI between case and noncase subjects. Again, HbA1c and insulin was the pair that showed the largest correlation difference by diabetes status (r = 0.15 in case subjects and r = 0.63 in noncase subjects).
Figure 2.
Biomarker correlation network comparing diabetes case subjects diagnosed during different time periods after blood collection vs. noncase subjects. A: Case subjects (n = 501) diagnosed >10 years after blood collection vs. 1,627 noncase subjects. B: Case subjects (n = 491) diagnosed 5–10 years after blood collection vs. 1,627 noncase subjects. C: Case subjects (n = 311) diagnosed <5 years after blood collection vs. 1,627 noncase subjects. Significant edges were identified by the threshold of both |Δr| > 0.15 and z > 3.5 (equivalent to a two-sided P value <0.0005). The width of the edge is proportional to the corresponding difference in the biomarker correlation between case and noncase subjects. A solid edge indicates that the absolute value of the correlation is stronger in case than in noncase subjects, and a dashed edge indicates the opposite. The color of the edge indicates the direction of the correlation in case and noncase subjects: red, positive correlations in both case and noncase subjects; green, negative correlations in both case and noncase subjects; gray, opposite correlations between case and noncase subjects. ADPN, adiponectin; E-sel, E-selectin; HCO3, bicarbonate; HGB, hemoglobin; HMW, high molecular weight; sTNFR2, soluble tumor necrosis factor receptor type 2; vitd, vitamin D.
Despite moderate attenuations after adjustment for age and BMI using partial Spearman correlations in both case and noncase subjects, most correlations remained statistically significant after multiple comparison correction (false discovery rate corrected P value <0.05) (Supplementary Fig. 4). Differences in partial correlations between case and noncase subjects were similar to differences in crude correlations (Supplementary Fig. 5), suggesting that the differential correlation network observed in the primary analysis was not entirely explained by differences in age or BMI between case and noncase subjects. When the permutation tests were conducted within quintiles of BMI as a way to control for the effect of adiposity, we observed a similar correlation network structure (Supplementary Fig. 6). Additional sensitivity analyses comparing case subjects diagnosed in different time periods after blood draw versus their age-matched noncase subjects yielded similar results (data not shown).
Discussion
In this secondary analysis of biomarker correlation networks for diabetes, we observed significant differences between diabetes case and noncase subjects for correlations involving biomarkers of inflammation, adipokines, IGF axis, and endothelial dysfunction. Importantly, our results indicate that the biomarker correlation structure was disturbed many years before clinical diagnosis of diabetes, with more differences observed in early versus late development of diabetes. Highly connected biomarkers varied across stages of diabetes development, as measured by time between blood collection and diagnosis, including C-peptide for >10 years before diagnosis, leptin for 5–10 years before diagnosis, and HbA1c for <5 years before diagnosis. By contrast, the insulin/HbA1c correlation was consistently weaker in case versus noncase subjects across the entire course of diabetes development.
The pairwise biomarker correlations, either positive or negative, were in general stronger in noncase than in case subjects, suggesting that different pathways and their interdependence were more tightly regulated in healthy women. The overall network that we observed when considering all case subjects highlights leptin as a highly connected node with differential associations to multiple markers spanning different biologic axes, including adipose secretion (adiponectin), inflammation (CRP), IGF (IGFBP-2), and glucose regulation (HbA1c). Notably, both leptin and adiponectin are adipokines secreted by adipose tissues and exhibit opposite trends with adiposity, with a higher leptin-to-adiponectin ratio being strongly associated with insulin resistance and increased diabetes risk (17,18). In addition, experimental evidence demonstrates that circulating CRP may bind to leptin to reduce its affinity to leptin receptor and impair downstream signaling, leading to leptin resistance (19). Emphasizing the central nature of leptin in diabetes development, there are known interrelationships of leptin resistance with energy intake, glucose homeostasis, and adipogenesis (20). Further, administration of leptin to leptin-deficient morbidly obese adults has been shown to induce a significant elevation in IGFBP-2 (21). However, our study observed an inverse association between leptin and IGFBP-2 that was stronger in noncase than case subjects. The potential association between leptin and IGFBP-2 in normal individuals or in individuals with leptin resistance and its relevance to diabetes warrant further investigation. The biologic activity of leptin is also regulated by sOB-R, the primary leptin-binding protein in circulation, which is another important node in the diabetes biomarker network. Although both leptin and sOB-R are hubs in the overall biomarker network, sOB-R seems to be an earlier marker than leptin (i.e., only sOB-R, but not leptin, is “visible” on the network corresponding to case subjects diagnosed >10 years after blood collection) (7,22). Taken together, our findings support a central role of the leptin system on multiple biologic systems that may act synergistically in the pathogenesis of diabetes.
The difference in the HbA1c/insulin relationship by diabetes status was consistently observed between case and noncase subjects, even more than a decade before development of diabetes. This suggests that glucose dysregulation and insulin resistance, which are hallmarks of diabetes, may emerge many years before clinical diagnosis and persist throughout the pathogenic progression. Currently, HbA1c is widely used as a clinical marker for glycemic control among individuals with diabetes. Consideration of how to assess this dysregulation could present an opportunity for early intervention and prevention of full diabetes.
However, the network structure and hub appeared to change over time, suggesting that the progression to diabetes begins many years before overt presentation through different stages. Over a decade before diabetes diagnosis, the relationship between C-peptide and markers of adipose secretion, inflammation, and IGF activity were different from those who did not develop diabetes. C-peptide, which is cleaved from proinsulin to form insulin, is a better indicator for pancreatic β-cell function than insulin, as insulin may be rapidly metabolized in liver (23). The high degree of differential connectedness of C-peptide suggests that changes in insulin secretion due to impaired β-cell function or insulin resistance and subsequent alterations in various physiologic pathways may be an early event in diabetes development. Interestingly, in type 1 diabetes, there are two distinct phases of C-peptide decline, an early exponential decline shortly after diagnosis followed by a prolonged stable phase (24). Notably, one strong edge that was uniquely observed in the early-stage network was the difference in the correlation between C-peptide and bicarbonate, which may reflect the disrupted acid-base balance due to glucose dysregulation (25). Given that exocrine secretion of the pancreas is a major source for endogenous bicarbonate, this may also suggest an overall decline in pancreatic functions during the early stage of diabetes development (26,27). Finally, genetic loci identified for diabetes susceptibility have been shown to act primarily through β-cell dysfunction and insulin secretion (28).
With the progression of metabolic abnormalities, the network hub shifted to leptin. Given the central role of leptin in regulating appetite, food intake, and body weight (20,29,30) and its strong associations with diet quality (31), physical activity (32), and sleep (33,34), our results provide additional evidence that altered behavioral and lifestyle factors may be key players in accelerating the onset of diabetes. Finally, the network observed close to the clinical presentation centered around HbA1c, reflecting the consequences of long-term suboptimally controlled blood glucose on multiple biologic pathways. Intriguingly, the differences in the biomarker correlations in the early development of diabetes tended to be in opposite directions (e.g., case subjects had a negative correlation whereas control subjects had a positive correlation or vice versa, represented by gray edges), whereas the correlation differences at a later stage of diabetes development were more likely to be stronger in noncase than case subjects, but in the same direction. This suggests that there may be adaptive mechanisms of the human body to metabolic dysregulation. This is also supported by the reduced number of significant edges in the network with progression to diabetes.
Our analysis represents efficient use of existing data that shed light on the pathogenesis of diabetes from a network perspective untapped in prior studies. The network findings integrated multiple biologic pathways, revealed their potential interdependencies and mutual influences in diabetes development, and expanded our knowledge of the pathogenic process on a systemic level that cannot be generated from single-biomarker studies. Our results also suggest that similar network-based approaches may be implemented in epidemiologic studies to provide new insights relevant to the etiology of other diseases. Other strengths of the study included availability of a large number of biomarkers from archived prediagnostic blood samples and long follow-up for diabetes incidence after blood collection. Ascertainment of diabetes diagnoses through supplemental questionnaires and medical record review, including abstraction of the exact date of diagnosis, reduced potential misclassification and allowed assessment of biomarker networks by the interval between blood draw and diabetes diagnosis.
However, due to the nature of secondary analysis, several limitations should be acknowledged. First, as biomarkers were not measured on each participant, we constructed the correlation network using the complete-subject analysis approach. Thus, the precision of the estimate as well as the associated potential bias may vary by each pair of the correlation. Similarly, we were not able to calculate partial Spearman correlations simultaneously adjusted for other biomarkers to identify the independent associations. Second, our analysis by time to diabetes diagnosis were based on case groups independent from each other, and we cannot rule out the possibility that the differences we observed across time to diabetes diagnosis may be attributed to certain sample differences across case groups. Analyses using repeated blood collections on the same individuals over the course of diabetes development would be ideal to elucidate the research question and should be explored in future studies. In addition, we were not able to differentiate whether changes in the biomarker correlation network over time fully reflected the pathophysiologic progression to diabetes or were partly due to changes in BMI or other lifestyle factors during diabetes development. Third, interpretation of our results requires caution as evidence of correlation does not necessarily imply causation. Future work mechanistically linking these biomarkers will be necessary in order to fully interpret these results and inform potential intervention strategies. Finally, our study focused on predominantly Caucasian women, and whether the findings can be generalized to men or other racial/ethnic groups requires additional investigation.
In summary, this network analysis of diabetes biomarkers highlights the central role of the leptin system in connection with other biologic pathways in promoting the clinical onset of diabetes, as well as the decade-long, persistent dysregulation between insulin and HbA1c throughout the development of diabetes. Biomarker networks featuring C-peptide, leptin, and HbA1c may mark different stages of diabetes pathogenesis, and additional studies are needed to confirm these findings and understand their potential preventive and therapeutic implications. Future epidemiologic studies may also leverage network-based approaches to advance the current etiologic knowledge for other diseases.
Supplementary Material
Article Information
Acknowledgments. The authors thank the participants and staff of the NHS for their valuable contributions.
Funding. This work was supported by the National Institutes of Health (grants UM1-CA-186107, R01-CA-49449, P30-DK-46200, and R01-DK-112940). T.H. is a recipient of the American Heart Association Postdoctoral Fellowship (Founders Affiliate) Award (16POST27480007). K.G. is supported by National Institutes of Health grant K25-HL-133599. K.L.I. is supported by a National Health and Medical Research Foundation fellowship.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. T.H. conducted the analyses, researched data, contributed to the discussion and interpretation of the data, drafted the manuscript, and critically revised the manuscript for important intellectual content. K.G. conceived the study, conducted the analyses, contributed to the discussion and interpretation of the data, and critically revised the manuscript for important intellectual content. O.A.Z. conducted the analyses, contributed to the discussion and interpretation of the data, and critically revised the manuscript for important intellectual content. J.H.K. and K.L.I. researched data, contributed to the discussion and interpretation of the data, and critically revised the manuscript for important intellectual content. A.R.S., B.M.B., and F.B.H. contributed to the discussion and interpretation of the data and critically revised the manuscript for important intellectual content. C.P.H. and S.S.T. conceived the study, contributed to the discussion and interpretation of the data, and critically revised the manuscript for important intellectual content. T.H. and S.S.T. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db18-0892/-/DC1.
References
- 1.Schulte PA, Perera FP. Molecular Epidemiology: Principles and Practices. San Diego, CA, Academic Press, 2012 [Google Scholar]
- 2.Chu JH, Hersh CP, Castaldi PJ, et al. . Analyzing networks of phenotypes in complex diseases: methodology and applications in COPD. BMC Syst Biol 2014;8:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nishihara R, Glass K, Mima K, et al. . Biomarker correlation network in colorectal carcinoma by tumor anatomic location. BMC Bioinformatics 2017;18:304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng Y, Ley SH, Hu FB. Global aetiology and epidemiology of type 2 diabetes mellitus and its complications. Nat Rev Endocrinol 2018;14:88–98 [DOI] [PubMed] [Google Scholar]
- 5.Hu FB, Meigs JB, Li TY, Rifai N, Manson JE. Inflammatory markers and risk of developing type 2 diabetes in women. Diabetes 2004;53:693–700 [DOI] [PubMed] [Google Scholar]
- 6.Heidemann C, Sun Q, van Dam RM, et al. . Total and high-molecular-weight adiponectin and resistin in relation to the risk for type 2 diabetes in women. Ann Intern Med 2008;149:307–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun Q, van Dam RM, Meigs JB, Franco OH, Mantzoros CS, Hu FB. Leptin and soluble leptin receptor levels in plasma and risk of type 2 diabetes in U.S. women: a prospective study. Diabetes 2010;59:611–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajpathak SN, He M, Sun Q, et al. . Insulin-like growth factor axis and risk of type 2 diabetes in women. Diabetes 2012;61:2248–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meigs JB, Hu FB, Rifai N, Manson JE. Biomarkers of endothelial dysfunction and risk of type 2 diabetes mellitus. JAMA 2004;291:1978–1986 [DOI] [PubMed] [Google Scholar]
- 10.Jiang R, Manson JE, Meigs JB, Ma J, Rifai N, Hu FB. Body iron stores in relation to risk of type 2 diabetes in apparently healthy women. JAMA 2004;291:711–717 [DOI] [PubMed] [Google Scholar]
- 11.Mandel EI, Curhan GC, Hu FB, Taylor EN. Plasma bicarbonate and risk of type 2 diabetes mellitus. CMAJ 2012;184:E719–E725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pittas AG, Sun Q, Manson JE, Dawson-Hughes B, Hu FB. Plasma 25-hydroxyvitamin D concentration and risk of incident type 2 diabetes in women. Diabetes Care 2010;33:2021–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song Y, Wang L, Pittas AG, et al. . Blood 25-hydroxy vitamin D levels and incident type 2 diabetes: a meta-analysis of prospective studies. Diabetes Care 2013;36:1422–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colditz GA, Hankinson SE. The Nurses’ Health Study: lifestyle and health among women. Nat Rev Cancer 2005;5:388–396 [DOI] [PubMed] [Google Scholar]
- 15.Manson JE, Rimm EB, Stampfer MJ, et al. . Physical activity and incidence of non-insulin-dependent diabetes mellitus in women. Lancet 1991;338:774–778 [DOI] [PubMed] [Google Scholar]
- 16.Shannon P, Markiel A, Ozier O, et al. . Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finucane FM, Luan J, Wareham NJ, et al.; (on behalf of the European Group for the Study of Insulin Resistance: Relationship Between Insulin Sensitivity and Cardiovascular Disease Risk Study Group ). Correlation of the leptin:adiponectin ratio with measures of insulin resistance in non-diabetic individuals. Diabetologia 2009;52:2345–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thorand B, Zierer A, Baumert J, Meisinger C, Herder C, Koenig W. Associations between leptin and the leptin/adiponectin ratio and incident type 2 diabetes in middle-aged men and women: results from the MONICA/KORA Augsburg study 1984-2002. Diabet Med 2010;27:1004–1011 [DOI] [PubMed] [Google Scholar]
- 19.Chen K, Li F, Li J, et al. . Induction of leptin resistance through direct interaction of C-reactive protein with leptin. Nat Med 2006;12:425–432 [DOI] [PubMed] [Google Scholar]
- 20.Myers MG Jr., Leibel RL, Seeley RJ, Schwartz MW. Obesity and leptin resistance: distinguishing cause from effect. Trends Endocrinol Metab 2010;21:643–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Licinio J, Caglayan S, Ozata M, et al. . Phenotypic effects of leptin replacement on morbid obesity, diabetes mellitus, hypogonadism, and behavior in leptin-deficient adults. Proc Natl Acad Sci USA 2004;101:4531–4536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Owecki M, Nikisch E, Miczke A, Pupek-Musialik D, Sowiński J. Free leptin index as a marker for leptin action in diabetes and obesity: leptin and soluble leptin receptors relationship with HbA1c. Neuroendocrinol Lett 2010;31:679–683 [PubMed] [Google Scholar]
- 23.Horwitz DL, Starr JI, Mako ME, Blackard WG, Rubenstein AH. Proinsulin, insulin, and C-peptide concentrations in human portal and peripheral blood. J Clin Invest 1975;55:1278–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shields BM, McDonald TJ, Oram R, et al.; TIGI Consortium . C-peptide decline in type 1 diabetes has two phases: an initial exponential fall and a subsequent stable phase. Diabetes Care 2018;41:1486–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palmer BF, Clegg DJ. Electrolyte and acid-base disturbances in patients with diabetes mellitus. N Engl J Med 2015;373:548–559 [DOI] [PubMed] [Google Scholar]
- 26.Lima MJ, Muir KR, Docherty HM, et al. . Suppression of epithelial-to-mesenchymal transitioning enhances ex vivo reprogramming of human exocrine pancreatic tissue toward functional insulin-producing β-like cells. Diabetes 2013;62:2821–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Q, Melton DA. Pancreas regeneration. Nature 2018;557:351–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voight BF, Scott LJ, Steinthorsdottir V, et al.; MAGIC Investigators; GIANT Consortium . Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet 2010;42:579–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenbaum M, Leibel RL. The role of leptin in human physiology. N Engl J Med 1999;341:913–915 [DOI] [PubMed] [Google Scholar]
- 30.Schwartz MW, Woods SC, Porte D Jr., Seeley RJ, Baskin DG. Central nervous system control of food intake. Nature 2000;404:661–671 [DOI] [PubMed] [Google Scholar]
- 31.Huang T, Tobias DK, Hruby A, Rifai N, Tworoger SS, Hu FB. An increase in dietary quality is associated with favorable plasma biomarkers of the brain-adipose axis in apparently healthy US women. J Nutr 2016;146:1101–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bouassida A, Chamari K, Zaouali M, Feki Y, Zbidi A, Tabka Z. Review on leptin and adiponectin responses and adaptations to acute and chronic exercise. Br J Sports Med 2010;44:620–630 [DOI] [PubMed] [Google Scholar]
- 33.Taheri S, Lin L, Austin D, Young T, Mignot E. Short sleep duration is associated with reduced leptin, elevated ghrelin, and increased body mass index. PLoS Med 2004;1:e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spiegel K, Leproult R, L’hermite-Balériaux M, Copinschi G, Penev PD, Van Cauter E. Leptin levels are dependent on sleep duration: relationships with sympathovagal balance, carbohydrate regulation, cortisol, and thyrotropin. J Clin Endocrinol Metab 2004;89:5762–5771 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.