Abstract
Over the past few years, symptoms akin to late blight disease have been reported on a variety of crop plants in South America. Despite the economic importance of these crops, the causal agents of the diseases belonging to the genus Phytophthora have not been completely characterized. In this study, a new Phytophthora species was described in Colombia from tree tomato (Solanum betaceum), a semi-domesticated fruit grown in northern South America. Comprehensive phylogenetic, morphological, population genetic analyses, and infection assays to characterize this new species, were conducted. All data support the description of the new species, Phytophthora betacei sp. nov. Phylogenetic analyses suggest that this new species belongs to clade 1c of the genus Phytophthora and is a close relative of the potato late blight pathogen, P. infestans. Furthermore, it appeared as the sister group of the P. andina strains collected from wild Solanaceae (clonal lineage EC-2). Analyses of morphological and physiological characters as well as host specificity showed high support for the differentiation of these species. Based on these results, a complete description of the new species is provided and the species boundaries within Phytophthora clade 1c in northern South America are discussed.
Keywords: host specificity, microsatellites, oomycetes, species delimitation, tree tomato
INTRODUCTION
Oomycetes represent a diverse group of fungus-like eukaryotic microorganisms widely distributed in nature. Their ecological characteristics have been extensively studied to reveal a wide diversity of ecological niches (e.g., Soanes et al. 2007). Particular emphasis has been placed on the study of plant pathogens for which ecological speciation seems a common process due to specialization to particular host species (Harrington et al. 2002, Tellier et al. 2010). Furthermore, oomycetes show an unprecedented plasticity in terms of genome size and ploidy (Haas et al. 2009, Yoshida et al. 2013), which could influence rates of speciation and extinction (Santini et al. 2009, Wood et al. 2009, Muir & Hahn 2015, Puttick et al. 2015). Recently, new hybrid species were described showing that interspecific hybridisation also plays a major role in adaptation and speciation of Phytophthora species (Brasier et al. 2004, Jung et al. 2017).
Molecular taxonomy can use DNA sequences to identify and delimitate species that are not amenable to genetic crosses (Roe et al. 2010). In combination with classical taxonomy, molecular studies are useful in describing allopolyploid interspecific Phytophthora hybrids as distinct species (Brasier et al. 2004, Man in ’t Veld et al. 2012, Bertier et al. 2013, Husson et al. 2015, Jung et al. 2017). The premise of these approaches is to identify discrete genetic groups that have ceased genetic exchanges with other groups. The number of studies defining species in this way has increased recently, mainly due to the ease of obtaining information on population-level DNA variation (Roe et al. 2010, Singh et al. 2015). However, this approach has inherent limitations. Gene genealogies tend to overestimate the number of species as the population structure within a species may be mistaken for species boundaries (Dettman et al. 2003). Furthermore, multi-locus species delimitation, relying on reciprocal monophyly and strict genealogical congruence, may fail to differentiate among recently diverged lineages (Hickerson et al. 2006, Knowles et al. 2007, Shaffer & Thomson 2007).
The most notable genus within oomycetes, Phytophthora, includes pathogens that infect a broad range of hosts in both agricultural and natural environments, causing adverse economic consequences (Erwin & Ribeiro 1996, Duncan 1999, Fry 2008, Forbes et al. 2013). To date, the genus Phytophthora comprises more than 150 recognised species, classified into 10 phylogenetic clades (Blair et al. 2008, Kroon et al. 2012, Martin et al. 2014). Over the past few decades, the numbers of recognised species within most divisions of the genus Phytophthora have nearly tripled within 20 years since Erwin and Ribeiro in 1996 listed 52 species, and greater species diversity has been considerably reported in all Phytophthora clades (Cooke et al. 2000, Kroon et al. 2012, Martin et al. 2012, Forbes et al. 2013). However, defining clear and objective species boundaries, as is the case for most oomycetes, remain a challenge in all Phytophthora clades.
Within the genus, P. infestans has become a ‘model system’ because of its undoubted economic impact. This pathogen affects important crops, such as potatoes (Solanum tuberosum) and tomatoes (Solanum lycopersicum) (Haverkort et al. 2008, 2009), making it one of the most threatening plant disease agents in the world. Although P. infestans was once considered a single species (henceforth referred to as P. infestans s.lat.), it has been shown to be a species complex (Forbes et al. 2013). Four other species related to P. infestans s.str. have been identified over the last 35 years. Phytophthora mirabilis has been found in Central America (Galindo & Hohl 1985) infecting only Mirabilis jalapa, an ornamental and medicinal plant in the region. Phytophthora ipomoeae (Flier et al. 2002) infects two morning glory species endemic to the highlands of central Mexico, Ipomoea longipedunculata and I. purpurea (Flier et al. 2002, Badillo-Ponce et al. 2004). Phytophthora phaseoli, initially classified as P. infestans (Thaxter 1889), is distributed globally but infects only lima beans (Phaseolus lunatus). Due to host-preference studies, P. phaseoli was described as a different species and for over 60 years thought to be the closest relative of P. infestans. Genetic comparisons have also revealed the existence of a separate group composed of strains from Ecuador and Peru that are collectively called P. andina (Oliva et al. 2010). To date, host range and distribution of P. andina is restricted to the Andean region of Ecuador and Peru where it has been reported in tree tomato crops (these isolates belong to the EC-3 clonal lineage) and wild solanaceous species (isolates belonging to the EC-2 clonal lineage) (Oliva et al. 2010). These observations, along with previous reports based on phylogenetic analyses using both nuclear and mitochondrial markers, reveal this species as polyphyletic, suggesting it might be a species complex comprising at least two genetically distinct lineages (Adler et al. 2002, Kroon et al. 2004, Chacón et al. 2006, Oliva et al. 2010, Cárdenas et al. 2012, Forbes et al. 2013, Goss et al. 2014, Lassiter et al. 2015, Forbes et al. 2016).
As in Ecuador and Peru, the southern region of Colombia has suffered a recent increase of late blight, leading to devastating declines in and disease outbreaks on tree tomato (Solanum betaceum) crops caused by Phytophthora. However, these have not been formally described. In this study, an integrative approach that leveraged phylogenetic study, morphological assays, population genetics and host specificity was used to characterise them. Here, a new species of Phytophthora infecting the tree tomato was found and defined. Morphological, physiological and molecular characteristics were used to provide a complete description of this species. Finally, this new species was formally described as Phytophthora betacei sp. nov. Furthermore, as this new species was described, the boundaries of other species within clade 1c were re-defined. For convenience, this new species will be referred by the proposed name throughout this article.
MATERIALS AND METHODS
Sampling and Phytophthora isolation
All samples were collected in southern Colombia between 2008 and 2009. Three to four leaves from 10 randomly selected tree tomato (Solanum betaceum) plants per plantation that showed symptoms resembling late blight were collected from 34 locations from two Colombian states, Nariño and Putumayo. The initial collections comprised over 970 putatively infected leaves. One to three lesions per leaf were excised (~0.5 to 1cm2) from the margin between necrotic and healthy tissues. Excised leaf pieces were surface-sterilised by submerging them in 70 % ethanol for 20 to 30 s and then washing them with sterile distilled water to remove the excess ethanol (~10s). The leaf pieces were dried on a sterile paper towel and subsequently transferred to a tree tomato agar (TTA) medium. The medium was prepared with tree tomato fruit (0.25 g of CaCO3, 0.5g of yeast extract, 25g of sucrose, 15g of agar and 100mL of tree tomato extract, composed of 550g of tree tomato fruit per litre of water). Subsequently, single zoospores were isolated from sporangia washed from the tree tomato medium with sterile distilled water. In total, 128 P. betacei strains were successfully isolated. All single zoospore isolates were cultured in a tree tomato medium for seven to 15d at 18 °C and then stored in the Phytophthora collection of the Museum of Natural History at Universidad de los Andes. Isolate P8084 was selected as the type culture for further taxonomic description. All P. betacei isolates collected in this study are deposited in the Museum of Natural History under accession numbers Andes-F 1081 to Andes-F 1207.
Phylogenetic analyses and population genetics
DNA extraction and sequencing
The mycelia of each Phytophthora strain were grown in a liquid Plich medium (Van der Lee et al. 1997) for 15d at 20°C. Subsequently, mycelia were washed with sterile distilled water and macerated thoroughly in liquid nitrogen using a cooled mortar and pestle. The macerated mycelia (0.1g) were immediately transferred into a micro-centrifuge tube (1.5mL) and DNA was extracted using the DNA kit OmniPrep (G-Biosciences) while following the manufacturer's instructions. The DNA was suspended in Tris-EDTA buffer (pH8.0) and was treated with RNAse. The quality and quantity of the DNA were measured, using NanoDrop ND-1000.
Whole genome sequencing and mitochondrial genome assembly
Two P. betacei isolates (P8084 and N9022) were sequenced using Illumina sequencing. A standard shotgun library (1× 200bp) was constructed and sequenced by Beijing Genomic Institute (BGI, Hong Kong, China) using an Illumina Hiseq2000 platform with paired-ends chemistry and 100 cycles. In total 40Gb of 96–100bp paired-end reads from two libraries with insert lengths of 200bp was generated. Furthermore, 22Gb of Illumina mate-pair libraries (6kb insert size) for each of the P. betacei isolates was also generated. Read mapping was done with BWA-MEM 0.7.12 (Li 2013) with the parameter k = 10 using P. infestans as a reference (Haas et al. 2009). Variants were called with GATK 3.2-2 (McKenna et al. 2010) using default parameters.
Phylogenetic analysis using mitochondrial genomes
Two newly assembled mitochondrial genomes of P. betacei (data taken from the whole genome sequences) were compared to data from Martin et al. (2016) in order to infer their phylogenetic position within clade 1c. MITObim (Hahn et al. 2013) was used to assemble the mitochondrial genome of each isolate. The process included 30 iterations using the quick approach with T30-4 haplotype Ia as the reference genome. Individual gene regions were aligned using MAFFT v.7.187 (Katoh & Toh 2010). Then, maximum likelihood (ML) analyses were performed using RAxML v.7.6.3 (Stamatakis 2006), as implemented on the CIPRES portal (Miller et al. 2010). The sequence alignment was partitioned into five subsets (rRNA genes, tRNA genes, first and second codon positions, third codon positions and intergenic regions) according to Martin et al. (2016). The GTRGAMMA (GTR + G) model for nucleotide substitution was used but allowed for the estimation of different shapes, GTR rates, and base frequencies for each partition. The majority rule criterion implemented in RAxML (-autoMRE) was used to assess clade support.
Phylogenetic analyses of SNP data using genotyping-by-sequencing
Genomic DNA was isolated with the DNeasy® Plant Mini Kit (QIAGEN, Germany). Genotyping-by-sequencing (GBS) was performed (Elshire et al. 2011) at the Institute of Genomic Diversity (Cornell University) for a total of 70 Phytophthora strains in total. These included 12 isolates (10 from Colombia and two reference strains from the United States, US-8 and US-12). For P. betacei, 35 isolates were included (clonal lineage EC-3), one P. andina (clonal lineage EC-3) and five P. andina (clonal lineage EC-2). Finally, three P. andina isolates of unknown clonal lineage, as well as five P. mirabilis, eight P. ipomoeae, and one P. phaseoli isolates were also included (Table 1). Briefly, genome complexity was reduced by digesting the total genomic DNA from individual samples with the type II restriction endonuclease ApeKI, which recognises a degenerate 5-bp sequence (GCWGC, where W is A or T) and creates a 5’ overhang (3bp). The digested products were then ligated to adapter pairs with enzyme-compatible overhangs; one adapter contained the barcode sequence and the other the binding site for the Illumina sequencing primer. The GBS library fragment-size distributions were checked on a BioAnalyzer (Agilent Technologies, Inc., USA). The PCR products were quantified and diluted for sequencing on an Illumina HiSeq 2500 sequencer (Illumina Inc., USA). A 96-well plate, comprising 70 samples and one blank, was multiplexed on a single Illumina flow cell lane.
Table 1.
Description of isolates of P. infestans, P. andina and P. betacei used for the morphological, physiological, phylogenetic and host preference assays.
| Species | Sample ID | Locality (Country/State/Locality) | Original host | Year | Locus lineage | Mitochondrial haplotype | Mating type | Assay1 | Source |
|---|---|---|---|---|---|---|---|---|---|
| P. infestans T-30-4 strain 2 | 1826 | Scotland (SCRI) | NA | NA | NA | NA | NA | C | Grünwald lab culture collection |
| P. infestans T-30-4 strain 3 | 4392 | Scotland | NA | 2007 | NA | NA | NA | C | Grünwald lab culture collection |
| P. infestans | C0008S | Colombia/Putumayo/Sibundoy | S. tuberosum | 2013 | NA | NA | NA | C | LAMFU4 |
| C003210 | Colombia /Nariño | S. tuberosum | 2013 | NA | NA | NA | C | LAMFU4 | |
| C003B21 | Colombia/Nariño | S. tuberosum | 2013 | NA | NA | NA | C | LAMFU4 | |
| PUA10096 | Colombia/Cundinamarca/Guasca | S. phureja | 2013 | NA | NA | A1 | C | LAMFU4 | |
| RB003 | Colombia /Nariño | S. tuberosum | 2013 | NA | NA | NA | C | LAMFU4 | |
| RB005 | Colombia /Nariño | S. tuberosum | 2013 | NA | NA | NA | C, D | LAMFU4 | |
| RC1-6 | Colombia/Cundinamarca/Rosal | S. tuberosum | 2015 | EC-1 | IIa | A1 | A, B | LAMFU4 | |
| SP03562 | Colombia /Putumayo/Sibundoy | S. tuberosum | 2013 | NA | NA | NA | C | LAMFU4 | |
| STG100 | Colombia/Nariño/Guachucal | S. tuberosum | 2013 | EC-1 | IIa | A1 | A, B | LAMFU4 | |
| STT161 | Colombia/Nariño/Túquerres | S. tuberosum | 2013 | EC-1 | IIa | A1 | A, B | LAMFU4 | |
| US040009 | USA/New York | S. tuberosum | NA | US-8 | NA | A2 | A, B | Fry lab culture collection | |
| US940480 | NA | NA | NA | US-8 | Ia | A2 | C | Fry lab culture collection | |
| US940494 | USA | S. lycopersicum | NA | US-12 | NA | A1 | C | Fry lab culture collection | |
| US970001 | USA/Florida | S. lycopersicum | 1997 | US-17 | NA | A1 | A, B | Fry lab culture collection | |
| VPC7-10 | Colombia/Cundinamarca/Villa Pinzon | S. tuberosum | 2015 | EC-1 | IIa | A1 | A, B | LAMFU4 | |
| Z3-2 | Colombia /Cundinamarca/Zipacón | S. phureja | 2007 | NA | NA | A1 | A, B, C, D | Vargas et al. (2009) | |
| P. andina5 | EC 3163 | Ecuador | Anarrhichomenum | NA | EC-2 | Ic | A1 | C | Oliva et al. (2010), Goss et al. (2011) |
| EC 3189 | Ecuador | Anarrhichomenum | NA | EC-2 | Ic | A2 | C | Oliva et al. (2010), Goss et al. (2011) | |
| EC 3399 | Ecuador | Anarrhichomenum | NA | EC-2 | Ia | A2 | A, B, D | Oliva et al. (2010), Goss et al. (2011) | |
| EC 3399 | Ecuador | Anarrhichomenum | NA | EC-2 | Ia | A1 | C | Oliva et al. (2010), Goss et al. (2011) | |
| EC 3510 | Ecuador | S. betaceum | NA | EC-3 | Ia | A1 | A, B, D | Goss et al. (2011) | |
| EC 3510 | Ecuador | S. betaceum | NA | EC-3 | Ia | A1 | C | Goss et al. (2011) | |
| EC 3563 | Ecuador | S. quitoense | NA | NA | Ia | A1 | C | Goss et al. (2011) | |
| EC 3678 | Ecuador | Anarrhichomenum | NA | EC-2 | Ic | A1 | C | Goss et al. (2011) | |
| EC 3780 | Ecuador | S. hispidum | NA | NA | Ic | NA | C | Goss et al. (2011) | |
| EC 3818 | Ecuador | Anarrhichomenum | NA | EC-2 | Ia | A2 | A, B, D | Oliva et al. (2010), Goss et al. (2011) | |
| EC 3818 | Ecuador | Anarrhichomenum | NA | EC-2 | Ia | A2 | C | Oliva et al. (2010), Goss et al. (2011) | |
| EC 3821 | Ecuador | Anarrhichomenum | NA | NA | Ia | NA | C | Goss et al. (2011) | |
| EC 3836 | Ecuador | S. betaceum | 2008 | EC-3 | Ia | A1 | D | Goss et al. (2011) | |
| P. betacei | A01492 | Colombia/Antioquia | S. betaceum | 2012 | EC-3 | Ia | ND | C | This study |
| CO1298 | Colombia/Putumayo/Sibundoy | S. betaceum | 2012 | EC-3 | Ia | ND | C | This study | |
| MFM-N9012 | Colombia/Nariño/Buesaco | S. betaceum | 2009 | EC-3 | Ia | ND | C | This study | |
| MFM-N9022 | Colombia/Nariño/Buesaco | S. betaceum | 2009 | EC-3 | Ia | ND | A, B, C, D | This study | |
| MFM-N9025 | Colombia/Nariño/Buesaco | S. betaceum | 2009 | EC-3 | Ia | ND | C | This study; Forbes et al. (2016) | |
| MFM-N9039 | Colombia/Nariño/Buesaco | S. betaceum | 2009 | EC-3 | Ia | ND | A, B, C | This study | |
| MFM-N9041 | Colombia/Nariño/Consaca | S. betaceum | 2009 | EC-3 | Ia | ND | C | This study | |
| MFM-N9046 | Colombia/Nariño/Pasto | S. betaceum | 2009 | EC-3 | Ia | ND | C | This study | |
| MFM-N9056 | Colombia/Nariño/Consaca | S. betaceum | 2009 | EC-3 | Ia | ND | C | This study | |
| MFM-N9057 | Colombia/Nariño/Consaca | S. betaceum | 2009 | EC-3 | Ia | ND | C | This study | |
| MFM-N9065 | Colombia/Nariño/Iles | S. betaceum | 2009 | EC-3 | Ia | ND | C | This study | |
| MFM-N9071 | Colombia/Nariño/Iles | S. betaceum | 2009 | EC-3 | Ia | ND | A, B, C | This study | |
| MFM-P8012 | Colombia/Putumayo/Colon | S. betaceum | 2008 | EC-3 | Ia | ND | C | This study | |
| MFM-P8029 | Colombia/Nariño/Buesaco | S. betaceum | 2008 | EC-3 | Ia | ND | C | This study | |
| MFM-P8050 | Colombia/Putumayo/Colon | S. betaceum | 2008 | EC-3 | Ia | ND | C | This study | |
| MFM-P8064 | Colombia/Putumayo/Santiago | S. betaceum | 2008 | EC-3 | Ia | ND | C | This study | |
| MFM-P8071 | Colombia/Putumayo/Colon | S. betaceum | 2008 | EC-3 | Ia | ND | C | This study | |
| MFM-P8075 | Colombia/Putumayo/Colon | S. betaceum | 2008 | EC-3 | Ia | ND | C | This study | |
| MFM-P8077 | Colombia/Putumayo/Colon | S. betaceum | 2008 | EC-3 | Ia | ND | A, B, C | This study | |
| MFM-P8084 | Colombia/Putumayo/Colon | S. betaceum | 2008 | EC-3 | Ia | ND | A, B, C, D | This study | |
| MFM-P8093 | Colombia/Putumayo | S. betaceum | 2008 | EC-3 | Ia | ND | C | This study | |
| MFM-P8096 | Colombia/Putumayo/San Francisco | S. betaceum | 2008 | EC-3 | Ia | ND | C | This study | |
| MFM-P8099 | Colombia/Putumayo/San Francisco | S. betaceum | 2008 | EC-3 | Ia | ND | C | This study | |
| MFM-P9105 | Colombia/Putumayo/San Francisco | S. betaceum | 2009 | EC-3 | Ia | ND | C | This study | |
| MFM-P9127 | Colombia/Putumayo/San Francisco | S. betaceum | 2009 | EC-3 | Ia | ND | C | This study | |
| MFM-P9128 | Colombia/Putumayo/Sibundoy | S. betaceum | 2009 | EC-3 | Ia | ND | C | This study | |
| MFM-P9129 | Colombia/Putumayo/Sibundoy | S. betaceum | 2009 | EC-3 | Ia | ND | C | This study | |
| MFM-P9146 | Colombia/Putumayo/Sibundoy | S. betaceum | 2009 | EC-3 | Ia | ND | C | This study | |
| MFM-P9147 | Colombia/Putumayo/Sibundoy | S. betaceum | 2009 | EC-3 | Ia | ND | C | This study | |
| MFM-P9151 | Colombia/Putumayo/Sibundoy | S. betaceum | 2009 | EC-3 | Ia | ND | C | This study | |
| MFM-P9153 | Colombia/Putumayo/Sibundoy | S. betaceum | 2009 | EC-3 | Ia | ND | C | This study | |
| S00321 | Colombia/Putumayo/Sibundoy | S. betaceum | 2012 | EC-3 | Ia | ND | C | This study | |
| S06298 | Colombia/Putumayo/Santiago | S. betaceum | 2012 | EC-3 | Ia | ND | C | This study | |
| S07198 | Colombia/Putumayo/Sibundoy | S. betaceum | 2012 | EC-3 | Ia | ND | C | This study | |
| S07398 | Colombia/Putumayo/Sibundoy | S. betaceum | 2012 | EC-3 | Ia | ND | C | This study | |
| P. mirabilis | 1260 | Mexico/Texcoco6 | NA | NA | NA | NA | NA | C | Grünwald lab culture collection |
| 1276 | Mexico | M. jalapa | 1998 | NA | NA | NA | C | Grünwald lab culture collection | |
| 1991 | Mexico/Coyoacán | M. jalapa | 2000 | NA | NA | NA | C | Grünwald lab culture collection | |
| 1992 | Mexico/Coyoacán | M. jalapa | 2000 | NA | NA | NA | C | Grünwald lab culture collection | |
| 1994 | Mexico/Coyoacán | M. jalapa | 2007 | NA | NA | NA | C | Grünwald lab culture collection | |
| P. ipomoeae | 1270 | Mexico | I. longipedunculata | 1999 | NA | NA | NA | C | Grünwald lab culture collection |
| 1271 | Mexico | I. longipedunculata | 1999 | NA | NA | NA | C | Grünwald lab culture collection | |
| 1989 | Mexico | I. longipedunculata | 2000 | NA | NA | NA | C | Grünwald lab culture collection | |
| 1990 | Mexico/Michoacan | Ipomoea spp. | 1999 | NA | NA | NA | C | Grünwald lab culture collection | |
| 4666 | Mexico | I. longipedunculata | 1999 | NA | NA | NA | C | Grünwald lab culture collection | |
| 4667 | Mexico | I. longipedunculata | 1999 | NA | NA | NA | C | Grünwald lab culture collection | |
| 4669 | Mexico | I. longipedunculata | 1999 | NA | NA | NA | C | Grünwald lab culture collection | |
| 4670 | Mexico | I. longipedunculata | 1999 | NA | NA | NA | C | Grünwald lab culture collection | |
| P. phaseoli | 5134 | USA | Phaseolus lunatus | 2003 | NA | NA | NA | C | Grünwald lab culture collection |
NA data not available.
ND Not determined; Oospores formed showed abnormal appearance in mating crosses between of P. betacei (isolate Andes-F 1172) and P. infestans strain of the A2 mating type (US040009).
1 Isolates used for A = morphological, B = Physiological, C = Phylogenetic analysis with GBS data, D = Host preference assay.
2 F1 of a cross between two aggressive strains of P. infestans originally isolated from potato in The Netherlands. Isolate used for genome sequence.
3 Duplicate of #1826.
4 LAMFU: The Mycology and Plant Pathology Laboratory at Universidad de los Andes, Bogotá, Colombia.
5 Isolates classified as P. andina according to Oliva et al. (2010).
6 From CBS in The Netherlands, ATCC 64130.
To sort each of the GBS barcode samples into separate fastq files, Phytophthora samples were demultiplexed using sabre (https://github.com/najoshi/sabre), allowing no mismatches within the barcode. In total, 1 992 701 tags were analysed and mapped against the P. infestans T30-4 reference genome (Haas et al. 2009), using Bowtie v.2.2.3 (Langmead 2010). Of this total number of reads, 917 890 (46.1%) were aligned to unique positions, 573 880 (28.8%) were aligned to multiple positions and 500 931 (25.1%) could not be aligned to the reference genome. For SNP calling, each SAM sample alignment file was converted into a BAM file, followed by sorting and indexing using SAMtools. SNPs and indels were called simultaneously using the variant caller GATK v.4.3.10 (McKenna et al. 2010). The final dataset consisted of 70 samples with 23 480 SNPs obtained from GATK. To discard the presence of sequencing errors in the data, all samples that did not fulfil the following criteria were filtered out: mapping QUAL >30, an overall coverage between 8 and 32X (cutoff values between 5% and 95% coverage) and a minor allele frequency (MAF) >0.05. To infer the phylogenetic relationships of species from Phytophthora clade 1c (Table 1), a matrix was created with all high-quality SNP loci obtained from the GBS analyses concatenated into a single alignment. An ML phylogenetic tree was generated using RAxML (Stamatakis 2006) under the general reversible nucleotide substitution model (GTR) with 1 000 bootstrap replicates to quantify branch support. The software jModelTest v.2.1.7 was used to select the best-fit substitution model, while P. phaseoli was used as an outgroup. The phylogenetic tree was visualised using Figtree v.1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/) (Rambaut 2009).
Restriction fragment length polymorphism analysis using mitochondrial haplotyping and probe RG57
Phytophthora lineages have been characterised by using mitochondrial haplotyping (Carter et al. 1990, Griffith & Shaw 1998) and a restriction fragment length polymorphism (RFLP) analysis with the highly polymorphic probe RG57 (Goodwin et al. 1992). The same approach to compare P. betacei with previously reported lineages within the genus Phytophthora was used in this study. The mitochondrial haplotype was determined using the PCR-RFLP method with reference strains US-1 and US-8 included as controls (Griffith & Shaw 1998, Ordoñez et al. 2000, Adler et al. 2002, Gavino & Fry 2002).
Population genetic analyses using microsatellite data
The Phytophthora isolates used in this study were genotyped and compared with the closely related species P. infestans and P. andina using simple sequence repeats (SSRs). All SSRs were sequenced and analysed using the protocols developed previously by Lees et al. (2006) and described in the Eucablight Network's Protocol section dated March 2008 (www.eucablight.org). In total 116 P. betacei isolates obtained in this study, 117 P. infestans isolates and 17 P. andina isolates reported in Goss et al. (2014) were included in the analysis. Among the 117 P. infestans isolates, there were 17 distinct clonal lineages, as well as genotypically diverse isolates from Mexico and Northern Europe. A principal component analysis (PCA) was conducted for the combined P. betacei, P. infestans and P. andina microsatellite data implemented in the adegenet package (Jombart et al. 2008). The allele frequencies at bi-allelic sites for the triploid P. infestans isolates (1/3 or 2/3) were unknown. To account for this uncertainty, the alleles at each locus for each isolate were subsampled. Because adegenet treats ploidy as a global parameter, resampled datasets for the strains from all species were generated, assuming all the individuals across species had the same ploidy. To account for both the uncertainty in allele frequencies at bi-allelic sites in triploid P. infestans isolates, as well as for the fact that adegenet would require all samples to have the same ploidy, each 100 independent diploid and 100 independent triploid resampled datasets for the PCA were generated (i.e., within each subsampled dataset, all individuals were diploids or triploids), and adegenet was run independently on each of them.
To estimate the number of populations that would best explain the genetic variance in the group of isolates studied, the Bayesian model-based clustering program STRUCTURE v.2.3 (Pritchard et al. 2000) was used. To account for allele frequency uncertainty at the bi-allelic triploid P. infestans loci and because ploidy is a global parameter in STRUCTURE, the same 200 resampled datasets for the PCA were used. STRUCTURE was run in total 32 000 times: (2 ploidies) × (100 resampled datasets) × (8 populations, K = 1 to 8) × (20 repetitions for K selection). Each run involved 1 000 000 MCMC steps with a burn-in of 100 000 and the following parameters were used: NOADMIX = 0, LINKAGE = 0, INFERALPHA = 1, ALPHA = 1.0, UNIFPRIORALPHA = 1, ALPHAMAX = 10.0 and FREQSCORR = 0. The ΔK method (Evanno et al. 2005) was used to infer the most likely number of clusters by evaluating the rate of change in the log probability of data between successive K values for each resampled dataset.
Population genetics analyses using genotyping-by-sequencing data
To corroborate the results of the population structure analyses obtained by using 11 microsatellite loci, a PCA was conducted based on the 23 480 high-quality SNP markers obtained from the GBS analyses. High quality was defined as SNPs with MAF >0.05 and with less than 20% missing data, that is, SNPs that were present in at least 80 % of the strains assessed. Genetic structure was also estimated using the Bayesian assignment test implemented in the program STRUCTURE v.2.3 (Pritchard et al. 2000) for high-quality SNP markers. These are defined as SNPs with MAF >0.05 and with less than 10% missing data. In total, 48 samples were used: 12 for P. infestans, 29 for P. betacei and 7 for P. andina (EC-2). The run parameters were as follows: 24 runs with four repetitions with 100 000 MCMC steps and a burn-in period of 10 000 for six populations (K = 1 to 6), under the NOADMIX ancestry model and allele frequencies correlated. The ΔK of Evanno (Evanno et al. 2005) was calculated using the application Structure Harvester v.0.6.94 (Earl & vonHoldt 2012) to infer the most likely number of clusters.
Nuclear genome size determination
The nuclear genome size of P. betacei was assessed using flow cytometry. Nuclear DNA was isolated from one strain of each species (P. infestans: T30-4, P. betacei: P8084 and P. andina: EC3510). Sample preparation and laser flow cytometry conditions were set according to Catal et al. (2010). Approximately 200 mg of fresh mycelium was grown in a liquid Plich medium for 10–15d at 20°C. The grown mycelia were washed by centrifugation, and sliced for 3–4 min in 1mL of PBS containing 0.1% Triton X-100 at pH7.4. The processed samples were subsequently maintained on ice for 4h. The solution was then filtered through a nitrocellulose membrane (0.45 μm; diam 25 mm; Bio-Rad). The filtrate was then pelleted by centrifugation at 5 000 × g for 10min at 4°C and it was washed with sterile PBS. Afterwards, nuclei isolated from samples were stained with propidium iodide (PI) using the BD cycle test plus DNA Reagent kit (BD Biosciences, Heidelberg, Germany) according to the manufacturer's instructions. Briefly, 1mL of buffer solution was added to the pellet obtained from previous centrifugation, and cells were subsequently resuspended at a low speed. The experiment was run twice. A volume of 250μL of Solution A (Trypsin) was added and mixed by tapping and then incubated for 10 min at room temperature. Next, 200μL of solution B (RNase A and trypsin inhibitor) was added and mixed by tapping and was again incubated for 10 min at room temperature. Finally, 200μL of solution C (PI) at 4°C was added to each tube, mixed by tapping, and then incubated for 10min in the dark on ice. The sample was filtered and run on a FACS Canto II cytometer. Results were visualised using the FACSDiva software (BD Bioscience) and data were recorded using the FlowJo v.7.5.5 software (Tree Star, Inc. Ashland, OR, USA). Diploid cells from avian erythrocytes were used as standards for instrument calibration. The measurements of relative fluorescence intensity were performed on 5 000 to 20 000 nuclei for each sample (Galbraith et al. 1983). Three independent samples per isolate were prepared and run separately. The estimation of DNA content was calculated as done by Dolezel et al. (2007). Heterogeneity across runs was minimised by calculating the genome size of each of the Phytophthora isolates using the internal reference standard (chicken; 2C = 2.4 pg; Nakamura et al. 1990). Genome size differences were compared across species by fitting a one-way ANOVA where species was the only factor and genome size was the response. Pairwise comparisons were done using Tukey's honest significant difference (HSD) test with the R library ‘multcomp’ (function ‘glht’).
Morphological characterization of P. betacei isolates
Morphological traits, including sporangial morphology, the presence of hyphal swelling or chlamydospores, oogonia, oospores and antheridia were examined to determine whether isolates of P. betacei presented morphological differences with isolates of the close species, P. andina and P. infestans (Table1). Sporangia morphology was evaluated on four culture media V8 juice agar (V8, 1.0g CaCO3/100 ml V8 Juice, 15g bacteriological agar), Potato Dextrose Agar (PDA, Oxoid Ltd, UK), Corn Meal Agar (CMA, Oxoid Ltd, UK) and TTA (described above), and quantitative measurements of sporangial sizes were scored on each isolate-medium combination. Sporangia were collected from a Petri dish incubated at 20°C in the dark for 2wk (optimum growth temperature for all isolates; see results), and the length (μm), breadth (μm) and length : breadth ratio (μm) of sporangia were measured. Furthermore, the three-dimensional volume of each sporangia was calculated according to Seidl Johnson et al. (2014). Briefly, the volume of each sporangia was calculated based on a standard equation: Volume = (4/3)×π ×A×B2, where A is half of the major axis and B is half of the minor axis (assuming that each sporangium has a spherical shape).
To harvest sporangia, each Petri plate was flooded with sterile distilled water, and colonies were gently scraped with a sterile glass rod to dislodge sporangia. The resulting sporangia suspension was placed in sterile plastic 2-mL microcentrifuge tubes and it was examined using the 40X oil objective in a fully motorized Olympus IX81 microscope (40X/1.3 N.A objective). Measurements of 30 randomly selected sporangia were taken for each isolate-medium combination described above. The resulting transmitted light images were processed and were merged using the Helicon Focus software (http://www.heliconsoft.com/). Pictures of sporangia were further analysed and processed using the ImageJ software. Two biological replicates were conducted for each isolate-medium combination.
The shape, type of apex, papillation, caducity and special qualitative features of sporangia were also recorded. The presence of hyphal swelling or aggregations were evaluated using mycelia from 15-d-old actively growing colony margins collected using a scalpel and immediately suspended in a drop (~50μL) of sterile distilled water. Furthermore, differences in microscopic hyphal characteristics were detected among P. betacei, P. infestans and P. andina. The width of the primary hyphae was also included as a character for identification (Fichtnera et al. 2011). Briefly, the width of the primary hyphae was determined by growing each isolate in the four-culture medium described above. The primary hyphae for each isolate-medium combination were evaluated using mycelia from 15-d-old actively growing colony margins collected using a scalpel and immediately suspended in a drop (~50μL) of sterile distilled water. Pictures of the primary hyphae were processed as described above and were further analysed using the ImageJ software. In total, 20 measurements of hyphal width were taken of each colony, and the average width of the primary hyphae per colony was recorded.
The production of gametangia structures (oogonia and antheridia) was evaluated for all isolates of P. betacei (Table 1). Oogonia were produced by growing each P. betacei isolate with a known isolate of P. infestans, either the A1 mating type (US970001 US-17 genotype) or A2 mating type (US040009, US-8 genotype), on Petri plates containing V8-Rye Agar (50% clarified V8 (10%), 50% clarified rye (10%), 1.5% agar, and 1mL of β-sitosterol per litre, adjusted to pH 8.5) (Mayton et al. 2000). Polycarbonate membrane test was used to separate physically P. betacei isolates from the P. infestans A2 mating type (Ko 1978, Gallegly & Hong 2008). Petri plates were kept at 18°C for 15–30d in the dark. To evaluate the number of oospores produced, 1cm2 of agar in the hyphal interface of the two colonies was excised using a scalpel. Samples were fixed for microscopic examination of the diameter and length, gametangial morphology, and wall morphology of oogonia/oospore.
Colony morphology, radial growth and cardinal temperature
To assess the effect of temperature and culture media on the colony morphology and on radial growth rate of P. betacei isolates (Table 1), four distinct media and eight different incubation temperatures were used. The four culture media tested were: V8, PDA (Oxoid Ltd, UK), CMA and TTA. Nine representative isolates were selected: three isolates of P. betacei (P8084, N9022, P8077 (EC-3)), three isolates of P. andina (EC3510 (EC-3, Ia), EC3399 (EC-2, Ia), EC3818 (EC-2, Ia)) and three isolates of P. infestans (Z3-2 (EC-1), US040009 (US-8), US970001 (US-17)) (Table 1). One circular inoculum plug (~ 5 mm diam) of an actively growing culture of a 15-d-old culture on V8 medium was placed in the centre of each Petri dish (90 mm diam). The colony morphology and radial growth rate for each isolate-media combination were evaluated after 15d of incubation at different temperatures in a dark chamber with constant humidity. Colony morphology was described according to Erwin & Ribeiro (1996) and Gallegly & Hong (2008). The mean characteristics, including texture, density and growth were identified by visual observations while the radial growth rate was measured by taking pictures using a Canon Digital EOS Rebel T3i / 600D camera (Tokyo, Japan) and importing the images into the ImageJ software (rsb.info.nih.gov/ij/).
To evaluate the optimum temperature for mycelial growth, all isolate-medium combinations previously described were incubated at 4, 10, 15, 20, 25, 30, and 35°C. Before evaluating each temperature, all isolates (Table 1) were sub-cultured onto V8 agar and incubated at 18°C in a dark chamber with constant humidity for growth stimulation (Jung et al. 2002, 2017). All combinations of isolates and media were tested in two independent blocks (biological replicates) with two technical replicates per combination. For the temperatures 10, 15, 20, 25, 30 and 35°C, identical incubators were used. The refrigeration equipment was used to keep the isolates at 4°C. The radial growth rate was measured at 3, 7, 11 and 15 d post inoculation (dpi) using the same procedure described above.
Statistical analyses to determine the radial growth rate were only conducted for isolates incubated at the optimum temperature (see results). The normality of the residuals of the linear models for each trait measured was assessed. In all cases, they were not normally distributed (Shapiro-Wilk test; P<0.05) and thus, differences in the radial growth rate of the three species at different temperatures and different media were assessed.
The observations of PDA were excluded because P. betacei did not grow on this medium. The observations obtained were pooled from all other media and a fitted linear model was created where the radial mycelial growth rate was the response, and the interaction between temperature and species was the only effect of the model. Pairwise comparisons were done using Tukey's HSD (honest significant difference) test with the R library ‘multcomp’ (function ‘glht’).
Quantitative morphological variation among Phytophthora species
Quantitative variation in terms of morphological traits among species was evaluated using a linear model approach, where the measurements were the response variables and the species was the only fixed effect. The normality of the residuals of each linear model was assessed using the Shapiro-Wilk test (function ‘shapiro.test’, package ‘stats’; R Core team 2013). Based on the Shapiro-Wilk test, residuals were not normally distributed in any of the linear models (P<0.05). Thus, the non-parametric Kruskal-Wallis test was used. To identify which groups of isolates differed from the others, multiple comparisons using non-parametric Nemenyi post hoc tests were performed. All Kruskal-Wallis tests were conducted using the R package ‘stats’ (R Core team 2013) and the Nemenyi and Tukey post hoc tests were conducted using the Pairwise Multiple Comparison of Mean Ranks Package (‘pmcmr’) implemented in R (Pohlert 2014).
Next, whether the morphology of P. betacei and other Phytophthora species differed was established by visualising all the morphological traits in a bi-dimensional plane using a discriminant function analysis (DA) based on the linear combination of morphological variables. To this end, a matrix with six traits (radial mycelial growth rate, primary hyphal width, sporangia length, sporangia breadth, sporangia volume and the sporangia length-to-breadth ratio) and a total of 15 isolates (five isolates for P. betacei, seven isolates for P. infestans, one isolate for P. andina clonal lineage EC-3 and two isolates for P. andina clonal lineage EC-2) was generated. Analyses were conducted using the ‘lda’ function from the package ‘mass’ in R (Venables & Ripley 2002).
Host pathogenicity assays and evaluation of host preference
A group of six isolates was used to estimate the effect of host specialization among P. infestans, P. andina and P. betacei. Two isolates of each species were used for all infection assays: strains P8084 and N9022 for P. betacei, strains Z3-2 and RB005 for P. infestans and strains EC3510 (EC-3, Ia) and EC3836 (EC-3, Ia) for P. andina, all isolated from tree tomatoes (Table 1). The P. andina strains EC3399 (EC-2, Ia) and EC3818 (EC-2, Ia), isolated from hosts in the Anarrichomenum complex (Table 1) were also included. Each Phytophthora isolate was inoculated onto three different Solanum host species: S. tuberosum group phureja (yellow potato), S. lycopersicum (tomato) and the S. betaceum variety Común (tree tomato). For isolates EC3399 and EC3818, no symptoms of infection were detected in inoculations done on S. tuberosum, S. betaceum and S. lycopersicum. Thus, these isolates were excluded from the final analysis.
Plants were grown in a greenhouse (17–19°C) and leaves or leaflets were harvested after 8–10 wk. Detached leaves were placed abaxial side up, on the base of 90-mm Petri plates containing moist paper towels. Three leaves were used per isolate as technical replicates. Each leaf was inoculated at four points with two 20μL droplets of a sporangial suspension (3.5 × 104 sporangia/mL) on each side of the main vein. The Petri dishes were sealed with Parafilm and incubated at 15°C with a 16h light period. Each experiment consisted of four hosts, three genotypes, two isolates per genotype, three leaflets per isolate and four inoculation points per leaflet. The whole experiment was repeated three times.
The latent period, total lesion area and number of sporangia produced were documented by taking daily pictures of the inoculated leaves from day 1 to day 9. The latent period was scored as the number of days it took from inoculations until sporangia were observed. The lesion area was scored as the necrotic area around the inoculation site at 9d post-inoculation (dpi), and it was measured using ImageJ (rsb.info.nih.gov/ij/). The number of sporangia produced on each leaf nine dpi was assessed by pooling individual lesions into 15mL disposable polypropylene culture tubes with 3mL of sterile distilled water. After vortexing for 10s sporangial numbers were counted at least twice using a haemocytometer. The total number of sporangia was calculated by averaging the sporangia counts per aliquot, and then multiplying the result by the dilution factor.
The total number of sporangia per day was calculated by dividing the total number of sporangia produced after 9d by the number of days when sporangia were visible (9d–latent period). The number of sporangia produced was calculated by subtracting the total number of sporangia produced (9dpi) from the sporangial concentration in the original inoculum suspension (2 800 sporangia per leaflet).
A fitness parameter for each replicate was calculated as the reproductive rate of each genotype on each host as follows:
| (1) |
To quantify the variation in fitness, a full factorial linear mixed model was fitted with the R package ‘nlme’ (function ‘lme’; Pinheiro et al. 2013). In the linear model, fitness was the response variable, both genotype (P. infestans, P. betacei or P. andina) and host (capiro potato, yellow potato, tomato or tree tomato) were fixed effects and strain (two independent isolates per genotype) was a random effect nested within a genotype. The significance of all interactions was assessed with Crawley's (1993, 2002) ML approach, in which the full model containing all factors and interactions was fitted and then simplified by a series of stepwise deletions, starting with the fixed-effect interaction and progressing to the interaction terms. The critical probabilities for retaining factors and determining whether effects or interactions were significant were 5% for main effects and 1% for two-way interactions. The linear model followed the formula:
| (2) |
Because the residuals of this linear model were not normally distributed (Shapiro-Wilk normality test, W = 0.8342; P<1 × 10−15) they were analysed in a nonparametric framework. Fitness calculations of each host were compared among genotypes, using a Kruskal-Wallis test with the R package ‘stats’ (R Core team 2013), followed by pairwise comparisons with a Nemenyi test with a Tukey-Dist approximation for independent samples, using the R package ‘pmcmr’ (Pohlert 2014).
Molecular diagnosis of P. betacei based on SNP data
To distinguish among P. betacei, P. andina (EC-2) and P. infestans, SNPs obtained from GBS data were analysed for 55 Phytophthora isolates in total (12 P. infestans, 35 P. betacei and eight P. andina (EC-2); Table 1). Potentially diagnostic SNPs were selected by calculating the allele frequencies and allele counts of each SNP for the entire dataset (55 samples and 22 788 SNPs). Samples belonging to each species were separated into three different files (P. betacei, P. andina (EC-2) and P. infestans) and allele counts were calculated for each dataset. SNPs with changes in the major allele in P. betacei were selected as candidates of differentiations relative to P. infestans and P. andina (EC-2) isolates.
RESULTS
Phylogenetic relationships of the Phytophthora clade 1c species using nuclear and mitochondrial genomes
A phylogenetic reconstruction using 23 480 nuclear SNPs showed P. betacei, P. andina and P. infestans as more closely related to each another than to P. ipomoeae or P. mirabilis (Fig.1a). The former three species formed a monophyletic group. All P. infestans clonal lineages (EC-1, US-8 and US-12) formed a monophyletic group. Phytophthora betacei appeared as the sister group of the P. andina strains collected from other wild plants of the Solanaceae family. This P. andina group comprised the EC-2 clonal lineage with mitochondrial haplotypes Ia and Ic and some isolates of unknown clonal lineage. The two clades, P. betacei and P. andina (EC-2 clonal lineage), were reciprocally monophyletic, providing evidence of the divergence of the two species. The only isolate formerly identified as P. andina clonal lineage EC-3 that was included in the analysis grouped together with P. betacei. This P. andina strain was also isolated from S. betaceum (Fig. 1a). A phylogenetic analysis of the mitochondrial genome sequences did not differentiate among the three species of the P. infestans s.lat. complex, with one notable exception: the P. andina clonal lineage EC-2 with the Ic mtDNA type appeared as the sister species of P. mirabilis (Fig. 1b).
Fig. 1.
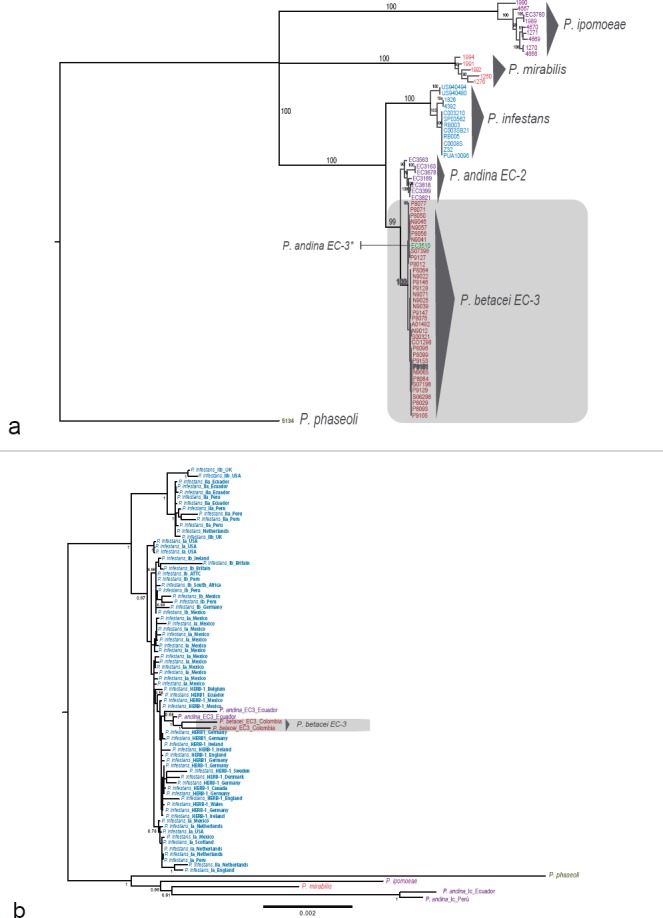
Phylogenetic relationships of the Phytophthora betacei isolates and the closely related species, based on genotyping by sequencing (GBS) data and phylogeny of complete mitochondrial genomes. a. The tree was inferred using maximum likelihood (ML) with P. phaseoli as an outgroup. Support values associated with branches correspond to ML bootstrap support values (BS); b. ML phylogeny of complete mitochondrial genomes showing the phylogenetic position of P. betacei sp. nov. Only bootstrap support values above 50% are shown.
Nuclear genome size determination
Genome sizes differed significantly among the three species (F2,103 = 6.09, P<0.01). Post-hoc comparisons revealed that P. betacei has the largest genome among the three species (mean size genome P. betacei = 1.13pg). The P. infestans’ genome was almost half the size of that of P. betacei (mean size genome P. infestans = 0.67pg). Differences in genome size between P. betacei and P. infestans were significant (Tukey HSD test comparing P. betacei and P. infestans: t-value = 3.45, P<0.01). The mean genome size of P. andina seems to intermediate between P. betacei and P. infestans (mean size genome P. andina = 0.97). Differences in genome size between P. andina and P. betacei, or between P. andina and P. infestans were not significant (Tukey HSD test comparing P. andina and P. infestans: t-value = 0.08213; Tukey HSD test comparing P. andina and P. betacei:0.48; P>0.05 in both comparisons) (data not shown).
Population structure analyses
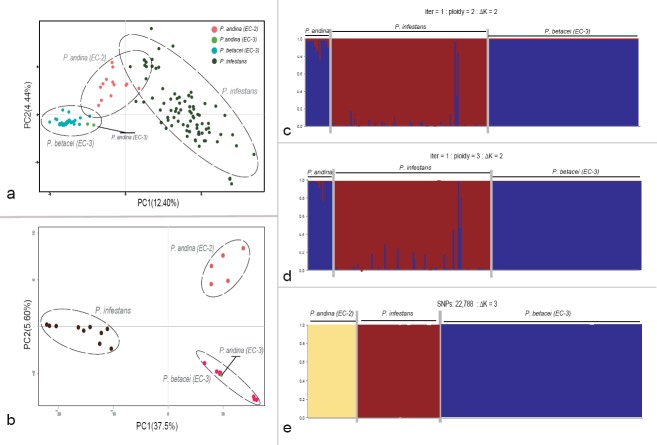
Microsatellite and whole-genome SNP data differed among the populations of P. infestans, P. betacei and P. andina (Fig. 2a–e). The results obtained from the PCA, using microsatellite data, suggested three genetic groups (Fig. 2a). The PC1 separated P. infestans and P. betacei (mean variance explained = 12.40%), indicating that a large proportion of the genetic variation is explained by the genetic difference between isolates belonging to these two species. The PC2 (mean variance explained = 4.44%) showed an intraspecific variation within P. infestans that was larger than the variations within P. betacei or P. andina. The two P. andina strains of the EC-3 clonal lineage were grouped closely with P. betacei. For this analysis, PCs 3, 4 and 5 primarily represent intraspecific variation within P. infestans (data not shown). Similar results were obtained for the GBS data supporting the clustering of the SSR analysis (Fig. 2b). The PCA on the GBS data including 23 480 SNPs showed strong genetic difference among strains of P. infestans, P. betacei and P. andina (Fig. 2b). The PC1 accounted for 37.5% of the total variation and separated P. infestans from the P. andina/P. betacei clades. The PC2 identified 5.8% of the variation between the strains and separated P. betacei and P. andina (Fig. 2b).
Fig. 2.
Principal component analysis (PCA) and STRUCTURE results for microsatellite and Genotyping-by-sequencing data showing the genetic structure for P. betacei, P. andina and P. infestans. a–b. Results for PCA analysis for SSR and GBS data, respectively. Principal components (PC) 1 and 2 are shown and the percentage of variance explained by each eigenvalue is shown within parentheses on each axis. Individuals of P. infestans are shown in blue, P. andina in orange and P. betacei in green; c–d. STRUCTURE results for SSR data for resampled diploid and triploid datasets, respectively (ΔK = 2); e. STRUCTURE results for Genotyping-by-sequencing data. data (ΔK = 3). Classification of Phytophthora samples was established according to the optimal population number (Evanno's method). The distribution of the individuals in different populations is indicated by the different colours on each plot.
The Bayesian assignment test conducted in STRUCTURE for the SSR and GBS data showed similar results (Fig. 2c–e). The STRUCTURE analysis showed two genetic clusters or populations that could explain the genetic variance for the SSR data (ΔK = 2; Fig. 2c–d). The two genetic clusters matched P. infestans s.str. and P. betacei. The third species, P. andina (both lineages EC-2 and EC-3) could not be assigned and seemed to cluster with P. betacei but not with P. infestans (Fig. 2c–d). Population assignment was robust to changes in ploidy; the most likely clustering was with two populations across diploid and triploid subsample datasets (ΔK = 2; Fig. 2c–d). The STRUCTURE analysis supports genetic differentiation between P. infestans and P. betacei, with uncertain clustering for P. andina. For the GBS data, the STRUCTURE analysis showed that the most likely clustering was three populations (ΔK = 3; Fig. 2e). The first genetic clusters matched with P. infestans s.str. and the second matched with P. betacei. The third cluster was assigned to P. andina samples (lineage EC-2) supporting the genetic differentiation among P. infestans, P. andina and P. betacei (Fig. 2e).
TAXONOMY
Phytophthora betacei Mideros, L.E. Lagos & S. Restrepo, sp. nov.—MycoBank MB815748; Fig. 3
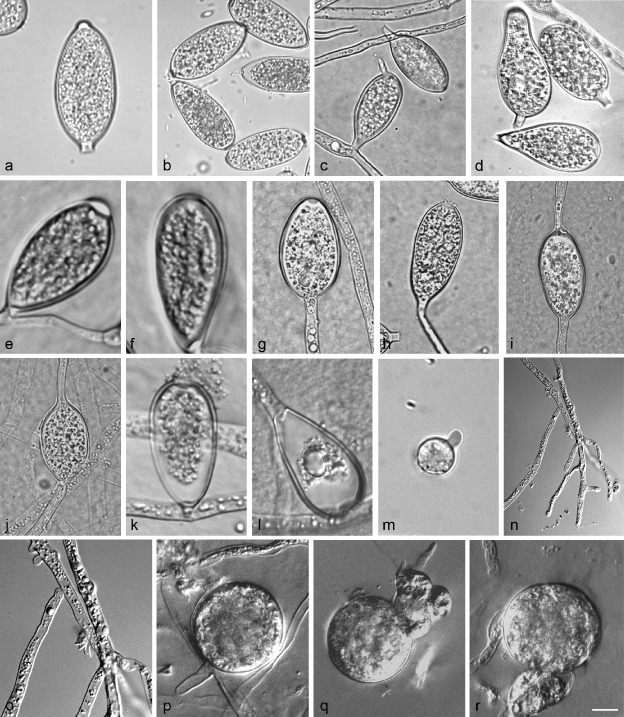
Fig. 3.
Phytophthora betacei morphology.a–c. Typical P. betacei sporangia. a. Limoniform; b–c. elongated ellipsoid; d. distorted or irregular shape; e. sporangia with papillate; f. semi-papillate or non-papillate sporangia; g–j. sporangia proliferation; k–m. zoospores cysts diameter; n–o. absence of hyphal swelling or chlamydospores and characteristics of primary hyphae; p–r. Phytophthora betacei gametangia. Abnormal oospores detected for P. betacei isolates (MFM-N9022, MFM-P8084, MFM-P8077).—Scale bar = 10 μm.
Etymology. Name refers to Solanum betaceum, the host plant from which the isolates were obtained.
Typus. Colombia, Putumayo, Colon, San Pedro locality, isolated from small pieces of infected leaves from S. betaceum plants, 2008, M.F. Mideros (Andes-F 1172 holotype, culture on TTA, MFM-P8084 ex-type culture).
Sporangia (Fig. 3a–d)—Phytophthora betacei produce sporangia typically borne terminally with limoniform (Fig. 3a), elongated ellipsoid (Fig. 3b–c) and distorted or irregular shape (Fig. 3d). No relationship between sporangia shape and culture medium was found. Sporangia were caducous with papillate (Fig. 3e; 75% on average) and semi-papillate or non-papillate sporangia (Fig. 3f; 25% on average) with short pedicels (2.6 ± 0.5 μm length). Typically, sporangia proliferated terminally in a simple sympodium with external proliferation close to the sporangial base and occasionally intercalary (Fig.3g–j). Zoospores of P. betacei were discharged directly through a small exit pore 4.2–4.6 μm wide (overall average 4.4 ± 0.2 μm) (Fig. 3k–l). They were spherical and motile. The average diameter of the cysts was 7.74 ± 0.9 μm (Fig. 3m). Sporangia from five isolates of P. betacei on V8 agar averaged 36.3 ± 6.0 μm in length × 17.3 ± 2.9 μm in breadth with a range of isolate means of 24.27–40.45 × 11.8–23.1 μm. The average length-to-breadth ratio on V8 was 2.1 ± 0.4. Sporangia from the same isolates on CMA averaged 37.6 ± 5.4 μm of length × 16.9 ± 2.3 μm of breadth with a range of isolate means of 27.6–47.8 × 12.25–22.25 μm. The average length-to-breadth ratio on CMA was 2.2 ± 0.2. Finally, on TTA sporangia of all isolates averaged 39.3 ± 4.8 μm in length × 15.8 ± 5.7 μm in breadth with a range of isolate means of 31.6–49.2 × 12.13–19.23 μm. The average length-to-breadth ratio on TTA was 2.6 ± 0.2. Chlamydospores absent. Catenulate, ellipsoid or globose hyphal swellings were not observed (Fig. 3n–o). The primary hyphae of P. betacei varied from 2.6–3.5 μm in width with a range of isolate means of 2.8 ± 0.7 μm.
Oogonia, oospores and antheridia (Fig. 3p–r)—Phytophthora betacei is heterotallic with a low oospore production (15.4 ± 10.9 oospores per mm2) and it has an abnormal appearance when crossed with a P. infestans strain of the A2 mating type (US040009). Some isolates never formed oospores and the abortion rate was very high (60–80% on average; Fig. 3q–r). In the polycarbonate membrane test, oogonia from four isolates were borne terminally or laterally and not ornamented. The average oogonial diameter among the isolates was 22.2–26.1 μm (overall average 25.0 ± 0.9 μm). Oogonia were usually globose to subglobose with a smooth wall (Fig. 3p). Oospores were produced within 25 to 30 days after pairing with the P. infestans isolate A2 mating type (US040009) on the rye agar medium. Oogonia were often turned golden-brown on maturity. Antheridia were amphigynous with mean dimensions of 12.0 ± 0.3 × 9.0 ± 0.5 μm (Fig. 3q–r).
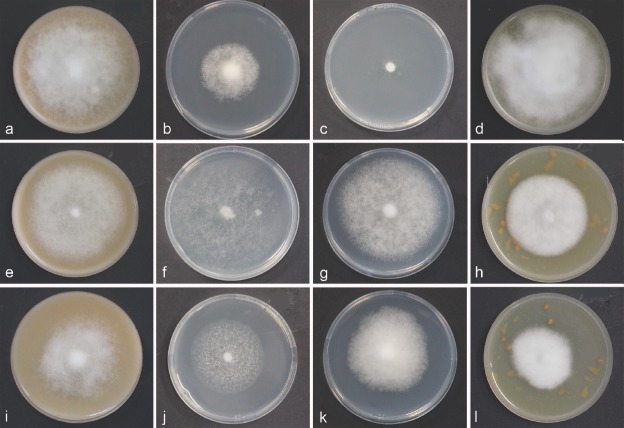
Colony morphology, growth rate and cardinal temperatures (Fig. 4a–l)—All P. betacei isolates tested formed colonies with different growth pattern and irregular margins on V8, CMA, TTA and PDA (Fig. 4a–l). Phytophthora betacei isolates grew on V8 juice agar (V8) producing fluffy colonies with non-defined margin. Aerial mycelium is sparse and velvety with no discernible pattern (Fig. 4a). The optimum temperature for the growth of P. betacei on V8 was between 10 and 20°C with an average radial growth rate of 1.19 ± 0.6 mm/d. On CMA, P. betacei isolates produced colonies that were fluffy and smooth, similar to V8 medium with irregular margins (Fig.4b). The optimum temperature for the growth of P. betacei on CMA was 20°C with an average radial growth rate of 0.65 ± 0.3mm/d (Fig. 5). The growth of P. betacei isolates on PDA was limited (Fig. 4d). The radial growth rate was on average 0.02 ± 0.01 mm/d (Fig. 5). No sporulation was detected on PDA. Finally, on TTA, P. betacei produced white colonies with sparse and with dense, fluffy aerial mycelium. The radial growth rate and sporulation of P. betacei isolates were more abundant on the TTA medium than on any of the other three media tested (Fig. 4d). The average radial growth rate at the optimum temperature of 20°C was 2.38 ± 0.19mm/d. No growth was recorded below 10°C or above 25°C, as these temperatures were lethal to most isolates, thus, the maximum temperature for growth was 20°C (Fig. 5).
Fig. 4.
Colony morphology, of Phytophthora betacei, P. infestans and P. andina. a–l. Colony morphology and radial growth for P. betacei (ex-type culture Andes-F 1172), P. infestans (isolate VPC7-10) and P. andina (isolate EC 3818) (from top to bottom) after 15d incubation at 18°C on V8 juice agar (V8), corn meal agar (CMA), potato dextrose agar (PDA), and tree tomato agar (TTA) (from left to right).
Fig. 5.
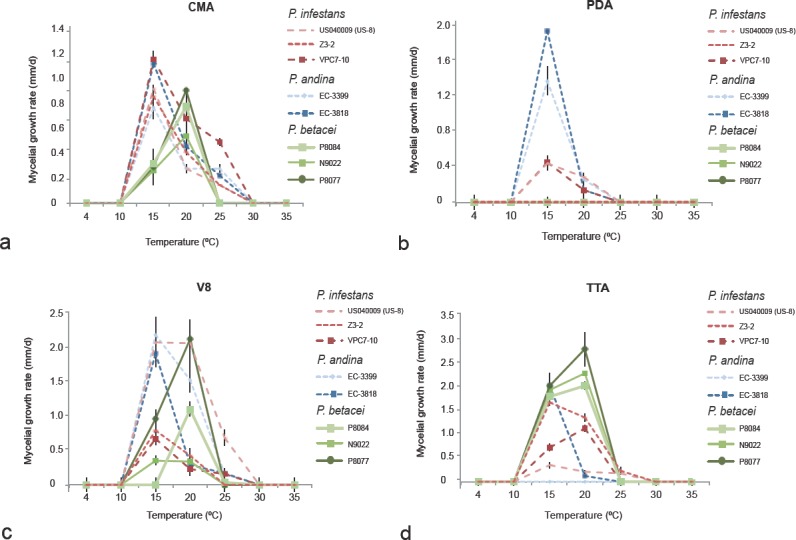
Radial growth rate of Phytophthora betacei on four culture media at different temperatures. Means and standard errors were calculated from three isolates: green lines = P. betacei isolates (ex-type Andes-F 1172, isolate P8077 and N9039), blue lines = P. andina (isolates EC 3818 and EC 3399), and red lines = P. infestans (isolates VPC7-10, Z-32 and US040009), on: a. Corn meal agar (CMA); b. potato dextrose agar (PDA); c. V8 juice agar (V8); d. tree tomato agar (TTA).
Additional specimens examined. Table 1.
Notes—Disease caused by P. betacei can lead to the complete loss of the S. betaceum crops (tree tomato) 5–10 d after the first symptoms are detected. In the field, the pathogen is able to defoliate the tree completely in approximately 1wk. The symptoms of P. betacei on tree tomatoes differed from those generated by P. infestans on potatoes in forming concentric blighted areas that produce sporangia, and covered large areas of the leaves and petioles. In the field, no symptoms were observed on fruits, and the disease was rarely found on stems. From the GBS data, we identified 22 788 SNPs in total, 150 of which were able to discriminate P. betacei from P. andina (EC-2) and P. infestans. Although all 150SNPs were classified as potentially diagnostic SNPs, this set of markers should then be validated using a larger P. betacei collection to confirm their robustness to diagnose this species. Phytophthora betacei can be morphologically distinguished from the P. andina or P. betacei species by sporangia size, with P. betacei isolates showing a greater length-to-breadth ratio of sporangia (ranges from 2.13–2.56) than P. infestans and P. andina isolates. Phytophthora betacei also showed an elongated ellipsoid or limoniform sporangia shape in contrast to P. infestans or P. andina, which showed ovoid sporangia on all isolates. In addition, the new species can be distinguished from P. andina (EC-2) and P. infestans isolates using a combination of morphological and physiological traits. A discriminant analysis showed that length-to-breadth ratio of sporangia and the width of the primary hyphae are the morphological traits that explain the 99% of the variance of the data (Fig. 6). The first function (LD1) characterized the groups based mostly on the length-to-breadth ratio of sporangia. For the second function (LD2), the primary hyphal width and the length-to-breadth ratio of sporangia helped discriminate among these groups of isolates. Sporangial measurements (length (μm), breadth (μm), length-to-breadth ratio) differed significantly among all species (Kruskal-Wallis rank sum test, P<0.01), confirming that this morphological character can be used to separate P. betacei from the other two related species. However, some differences were more striking between species depending on the medium used (comparisons of P. betacei with either P. infestans or P. andina). The optimal and lethal temperatures for P. betacei, P. infestans and P. andina (EC-2) were similar on all media tested. However, at the optimal temperature (20°C), P. betacei grew more rapidly than the other two species (linear coefficients: P. betacei vs P. infestans: 12.36; P. betacei vs P. andina: 9.37, both P<0.001). Isolates of P. infestans and P. andina were also able to grow at 25°C in at least one of the four media tested. In contrast, none of the P. betacei isolates was able to grow at 25°C (Fig 5). The type of culture media (V8, PDA, TTA and CMA) affected the radial growth rate of the different species at 15°C. No significant differences were found when comparing isolates of P. betacei with those of P. infestans and P. andina (EC-2) growing on the CMA culture medium. However, a notable exception for P. betacei isolates was detected on the PDA. Phytophthora betacei showed low rates of growth on the PDA and a complete absence of sporulation. Significant differences were also detected among all isolates of P. betacei, P. andina and P. infestans growing on the V8 and TTA culture media (Kruskal-Wallis χ2 = 80.703, df = 3, P<0.001). The original descriptions of the P. infestans and P. andina isolates were congruent with the data obtained in the present study. The host range of P. betacei is only restricted to S. betaceum, showing a strong host-plant specialisation in reciprocal inoculation experiments. Phytophthora betacei could not infect either tomatoes or potatoes but showed the highest fitness on S. betaceum (Fig. 7).
Fig. 6.
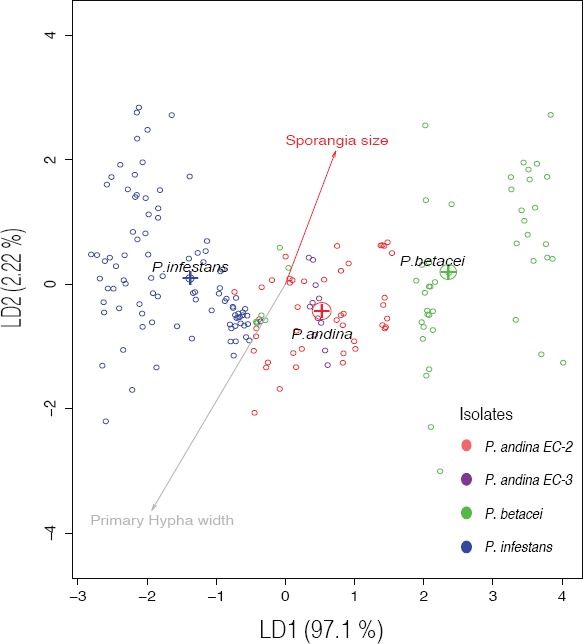
Plot of the morphological traits of Phytophthora species using a discriminant analysis (DA) showing the first and second discriminant components. Phytophthora betacei strains are shown in blue, P. infestans in green and P. andina in red.
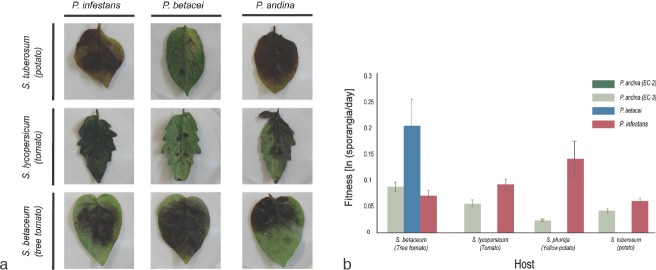
Fig. 7.
Host specialisation results in strong premating reproductive isolation of P. infestans, P. betacei and P. andina isolates. a. Fitness obtained from reciprocal infection assays. Results of infection for P. infestans, P. betacei and P. andina (EC-3 clonal lineage) on the main hosts evaluated: potatoes (S. tuberosum), tomatoes (S. lycopersicum) and tree tomatoes (S. betaceum) after 9d post inoculation (dpi). The isolates of P. andina of the EC-2 clonal lineage did not produce any symptoms on any of the hosts; b. values of fitness (sporangia per day) for P. infestans, P. betacei and P. andina (EC-2 and EC-3 clonal lineages). On tree tomato, isolates of P. betacei showed significantly higher fitness than P. infestans and P. andina (EC-3 clonal lineage). Conversely, isolates of P. betacei are unable to infect other hosts where P. infestans thrives. Additionally, P. andina (EC-3 clonal lineage) shows significantly lower fitness on all hosts tested.
Host pathogenicity assays and evaluation of host preference
Because the P. infestans, P. betacei and P. andina (EC-2) strains were isolated from different hosts, the hypothesis that the three species were host specialised or had a reduced fitness on their alternate host was tested. Isolates of P. andina of the EC-2 and EC-3 clonal lineages were included to make all possible pairwise comparisons. Isolates of P. andina of the EC-2 clonal lineage did not produce any symptoms on any of the hosts used. Phytophthora infestans had a higher fitness on tomatoes and yellow potatoes compared to P. betacei and P. andina (Table 2), while P. betacei could not infect neither tomatoes nor potatoes but showed the highest fitness on tree tomatoes (Table 2). The formerly P. andina (EC-3) strains assayed here were able to infect all hosts but showed a lower fitness than P. infestans on the three hosts (tomatoes, yellow potatoes and tree tomatoes). They also displayed a lower fitness on tree tomatoes compared to P. betacei. All pairwise comparisons indicated that strains of P. infestans and P. betacei displayed different fitness properties on every host assessed (Fig. 7a–b).
Table 2.
Pairwise comparisons of overall fitness on four different hosts. Pairwise comparisons were made using a Kruskal Wallis rank sum test followed by a Nemenyi test with multiple comparisons. Statistically significant P-values (P<0.05) are shown in bold.
| Host | Mean fitness (Standard Deviation) |
Kruskal Wallis rank sum test |
Nemenyi pairwise comparisons |
|||||
|---|---|---|---|---|---|---|---|---|
| P. infestans | P. andina (EC-3) | P. betacei | χ2, df = 2 | P-value | P. infestans vs P. andina (EC-3) | P. infestans vs P. betacei | P. betacei vs P. andina (EC-3) | |
| Tomato (S. lycopersicum) | 0.0481 (± 0.0589) | 0.0421 (± 0.1055) | 0 (± 0) | 18.0462 | 1.206 × 10−4 | 0.5063 | 0.0097 | 0.2034 |
| Potato (S. tuberosum var. capiro) | 0.0146 (± 0.0341) | 0.0267 (± 0.0486) | 0 (± 0) | 4.7732 | 0.09194 | 0.89 | 0.76 | 0.45 |
| Yellow potato (S. phureja) | 0.0604 (± 0.0450) | 0.0545 (± 0.0574) | 0 (± 0) | 14.6133 | 6.711 × 10−4 | 0.999 | 0.019 | 0.021 |
| Tree tomato (S. betaceum) | 0.0659 (± 0.0622) | 0.0298 (± 0.0462) | 0.1025 (± 0.0583) | 28.9611 | 5.143 × 10−7 | 0.036 | 0.033 | 5.7 × 10−7 |
DISCUSSION
Here, a new species of P. betacei that is closely related to P. infestans and P. andina is described. This finding provides novel insights into the evolutionary history of the Irish famine pathogen P. infestans and its close relatives. Furthermore, the species boundaries within the complex of P. andina, originally described as a polyphyletic taxon, are refined. These two sets of results are discussed in the following two subsections.
Phytophthora betacei is a new species
In this study, the new taxon, P. betacei was described based on physiological, morphological, population genetic and phylogenetic analyses, as well as differences in host specificity. All these analyses strongly support the designation of the new species P. betacei within Phytophthora clade 1c.
The first line of evidence for the distinction between P. betacei and the other species of Phytophthora clade 1c is the high genetic differentiation among the genetic groups. Nuclear phylogenies indicate that the triad P. infestans, P. betacei and P. andina form a monophyletic clade whose closest known relatives are other members of the Phytophthora clade 1c (i.e., P. ipomoeae, P. phaseoli and P. mirabilis). The three species, P. betacei, P. infestans and P. andina (from the clonal lineage EC-2; see below) are reciprocally monophyletic, suggesting they lack recent gene flow and can be considered different species. Interestingly, the mitochondrial markers do not separate the three species. This result is consistent with a scenario of speciation with secondary contact and mitochondrial introgression, a phenomenon common across the tree of life (Funk & Omland 2003).
A second line of evidence for the existence of P. betacei as a separate species from P. infestans s.str. involves differences in allele frequencies in each of these genetic groups. All analyses using both SSR loci and SNP markers suggest the existence of two discrete genetic clusters that correspond to P. infestans s.str. and P. betacei. Phytophthora andina has a less clear origin and this is discussed below. The data further suggests that P. infestans and P. betacei are isolated genetic groups with little detectable nuclear gene flow between them.
In addition to genetic variation, morphological differences among P. betacei, P. andina and P. infestans were determined. Four morphological characteristics (primary hyphal width and sporangial length, breadth, and length-to-breadth ratio) and mycelial radial growth on four different media traits were measured. The discriminant analysis clearly separated P. betacei from P. infestans in all media tested. The most striking morphological differences between P. betacei and P. infestans are the length-to-breadth ratio of sporangia, the primary hyphal width (μm) and the radial mycelial growth rate. Differences between P. andina and P. infestans or P. betacei are not as clear as the statistical differences between P. andina and P. infestans and between P. andina and P. betacei, and they are dependent on the medium tested. Combining all the morphological variables, it is evident that the P. betacei strains collected in Colombia comprise a well-differentiated group of strains (Fig. 6).
The final line of evidence comes from infection assays on the native host range of the three species and from observations in nature. The host pathogenicity assays indicate that P. betacei is a tree tomato specialist unable to colonise potatoes and tomatoes (Fig. 7). Conversely, P. infestans has a low fitness on tree tomatoes, the only known host of P. betacei. These reciprocal differences in host pathogenicity represent a strong reproductive isolating mechanism between the two species (Restrepo et al. 2014). Host specificity is considered one of the most important isolating mechanisms between species of plant pathogens (reviewed in Harrington & Rizzo 1999, Coyne & Orr 2004). In asexual populations, host specialisation could be associated with a strong niche partition, which is common in species with asexual reproduction and strong local adaptation to the host (Poulin 2005, Halkett et al. 2006). Plant pathogens are commonly restricted to their hosts; thus, host specialisation can result in a strong premating barrier (Stukenbrock 2013, Vialle et al. 2013, Restrepo et al. 2014).
Notably, all isolates of P. betacei belonging to the EC-3 clonal lineage are closely related to the isolates previously described as P. andina EC-3 (Oliva et al. 2010, Goss et al. 2011, 2014, Lassiter et al. 2015, Martin et al. 2016), which in turn should be considered P. betacei. All EC-3 isolates form a monophyletic group differentiated from P. infestans and P. andina EC-2. The results support a finer definition of the species boundary of P. andina.
Phytophthora andina previously characterized as a polyphyletic group, includes only EC-2
In the literature, P. andina has been reported to be polyphyletic and include the following three clonal lineages: the P. andina EC-2 mitochondrial haplotype Ia, the P. andina EC-2 mitochondrial haplotype Ic, and P. andina EC-3 (Adler et al. 2004, Gómez-Alpizar et al. 2008). This species has been controversial since its erection, as species are expected to be monophyletic with the expectation of descent from one common ancestor. Phytophthora andina was shown to be a hybrid based on cloning nuclear haplotypes from several loci showing that one ancestor is P. infestans while the other ancestor remains to be described (Goss et al. 2011). Later, it has been hypothesized to have arisen from hybridization based on the conflicting phylogenetic information of mitochondrial and nuclear genealogies (Martin et al. 2016). Based on the observed hybrid nature and the polyphyletic mitochondrial phylogenies in P. andina, P. andina was not appropriately described as a new species (Cárdenas et al. 2012). The identification of P. betacei as a new species, distinct from P. infestans, sheds some light on the origin of P. andina.
The results support a monophyletic grouping of the EC-2 P. andina clonal lineages of mitochondrial haplotypes Ia and Ic that are closely related and that form a monophyletic group distinct from P. betacei and P. infestans. Supported by phylogenetic and population genetic analyses, P. andina EC-3 should now be considered P. betacei. These results indicate that the initial definition of P. andina included isolates that were either P. betacei or were closely related to P. betacei, namely, the EC-3 clonal lineage. However, further work is needed to determine whether all isolates of P. andina of the EC-3 clonal lineage collected from tree tomato in Ecuador (mitochondrial haplotype Ia, reported in Oliva et al. 2010, Goss et al. 2011, 2014, Lassiter et al. 2015, Martin et al. 2016) indeed cluster with isolates of P. betacei. Generally, the results confirmed previous observations that P. andina, as currently described, is a polyphyletic group that requires redefinition (Gómez-Alpizar et al. 2008, Cárdenas et al. 2012, Forbes et al. 2012). Redefining P. andina including lineages of clonal lineage EC-2 makes this group monophyletic and provides a biologically rigorous species definition. Thus, P. andina s.lat. is proposed to be the proper description of P. andina EC-2.
Whether there is reciprocal host specificity between P. betacei and P. andina (EC-2) remains an open question, yet some predictions can be formulated. Because all known EC-2 P. andina isolates have been collected in Anarrichomenum and other wild species, it is possible the species will be specialised on these plants. This study showed that strains of P. andina of the EC-2 clonal lineage could not infect S. betaceum, the only known host of P. betacei. Several authors have documented host specificity between the P. andina EC-2 and EC-3 clonal lineages in nature (Adler et al. 2004, Gómez-Alpizar et al. 2007, Oliva et al. 2010). Isolates of the EC-3 clonal lineage have always been collected from S. betaceum plants, again suggesting a strong isolating mechanism between P. betacei and P. andina (EC-2). However, it is important to mention that a group of strains referred to as P. andina has recently been reported as infecting S. betaceum in Peru (lineage PE-8) (Forbes et al. 2016). Thus, further analyses and a greater number of isolates are needed to determine the host range and fitness of isolates belonging to the PE-8 clonal lineage.
Conclusions
This study provided several lines of evidence supporting the claim that P. betacei is a distinct, previously undescribed species within Phytophthora clade 1c. The findings also resolve the polyphyletic nature of P. andina. Together, the evidence shows that the dyad P. andina/P. betacei is a sister clade of the potato blight pathogen, P. infestans. The strong host specialisation of P. infestans and P. betacei may act as a premating barrier that restricts gene flow between these two species in nature. It remains unclear whether host specialisation facilitated or initiated the speciation process in the P. infestans s.lat. complex. However, in this report, it was shown that ecological differences are important in the persistence of P. infestans and P. betacei as genetically isolated units across an overlapping area in the northern Andes. More studies are needed to further characterise the evolution of the closely related species and to understand the process of divergence in this group. In general, the results obtained have implications for the understanding of how new plant pathogen species originate and persist. The findings also highlight the importance of sampling plant pathogens of semi-domesticated or undomesticated hosts.
Acknowledgments
This work was supported by the Department of Biological Sciences at Universidad de los Andes. Additional funding for this research was provided by the Research Fund of the School of Sciences and the Office of the Vice President for Research from Universidad de los Andes.
REFERENCES
- Adler NE, Chacon G, Flier WG, et al. 2002. The andean fruit crop, pear melon (Solanum muricatum) is a common host for A1 and A2 strains of Phytophthora infestans in Ecuador. Plant Pathology 51: 802 [Google Scholar]
- Adler NE, Erselius LJ, Chacón MG, et al. 2004. Genetic diversity of Phytophthora infestans sensu lato in Ecuador provides new insight into the origin of this important plant pathogen. Phytopathology 94: 154–162. [DOI] [PubMed] [Google Scholar]
- Badillo-Ponce G, Fernández-Pavía SP, Grünwald NJ, et al. 2004. First report of blight on Ipomoea purpurea caused by Phytophthora ipomoeae. Plant Disease 88: 1283. [DOI] [PubMed] [Google Scholar]
- Bertier L, Leus L, D'hondt L, et al. 2013. Host adaptation and speciation through hybridization and polyploidy in Phytophthora. PLoS ONE 8: e85385. doi: 10.1371/journal.pone.0085385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair JE, Coffey MD, Park SY, et al. 2008. A multi-locus phylogeny for Phytophthora utilizing markers derived from complete genome sequences. Fungal Genetics and Biology 45: 266–277. [DOI] [PubMed] [Google Scholar]
- Brasier CM, Kirk SA, Delcan J, et al. 2004. Phytophthora alni sp. nov. and its variants: Designation of emerging heteroploid hybrid pathogens spreading on Alnus trees. Mycological Research 108: 1172–1184. [DOI] [PubMed] [Google Scholar]
- Cárdenas M, Tabima J, Fry WE, et al. 2012. Defining species boundaries in the genus Phytophthora: the case of Phytophthora andina A response to ‘Phytophthora andina sp. nov., a newly identified heterothallic pathogen of solanaceous hosts in the Andean highlands’ (Oliva et al. 2010). Plant Pathology 61: 215–220. [Google Scholar]
- Carter DA, Archer SA, Buck KW, et al. 1990. Restriction fragment length polymorphisms of mitochondrial DNA of Phytophthora infestans. Mycological Research 94: 1123–1128. [Google Scholar]
- Catal M, King L, Tumbalam P, et al. 2010. Heterokaryotic nuclear conditions and a heterogeneous nuclear population are observed by flow cytometry in Phytophthora infestans. Cytometry Part A 77: 769–775. [DOI] [PubMed] [Google Scholar]
- Chacón MG, Adler NE, Jarrin F, et al. 2006. Genetic structure of the population of Phytophthora infestans attacking Solanum ochranthum in the highlands of Ecuador. European Journal of Plant Pathology 115: 235–245. [Google Scholar]
- Cooke DEL, Drenth A, Duncan JM, et al. 2000. A molecular phylogeny of Phytophthora and related oomycetes. Fungal Genetics and Biology 30: 17–32. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. 2004. Speciation. 1st edn Sinauer Associates, USA. [Google Scholar]
- Crawley MJ. 1993. GLIM for Ecologists. (Ecological Methods & Concepts). Methods in Ecology Series. Blackwell Scientific, UK. [Google Scholar]
- Crawley MJ. 2002. Statistical computing: an introduction to data analysis using S-Plus. Wiley, USA. [Google Scholar]
- Dettman JR, Jacobson DJ, Taylor JW. 2003. A multilocus genealogical approach to phylogenetic species recognition in the model eukaryote Neurospora. Evolution 57: 2703–2720. [DOI] [PubMed] [Google Scholar]
- Dolezel J, Greilhuber J, Suda J. 2007. Estimation of nuclear DNA content in plants using flow cytometry. Nature protocols 2: 2233–2244. [DOI] [PubMed] [Google Scholar]
- Duncan J. 1999. Phytophthora – an abiding threat to our crops. Microbiology Today 26: 114–116. [Google Scholar]
- Earl DA, vonHoldt BM. 2012. “STRUCTURE HARVESTER: a website and program for visualizing structure output and implementing the Evanno method. Conservation Genetics Resources 4: 359–361. [Google Scholar]
- Elshire RJ, Glaubitz JC, Sun Q, et al. 2011. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PloS One 6: e19379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin DC, Ribeiro OK. 1996. Phytophthora Diseases Worldwide. St Paul, American Phytopathological Society (APS) Press, USA. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Molecular Ecology 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- Fichtnera EJ, Rizzoa DM, Kirkb SA, et al. 2011. Infectivity and sporulation potential of Phytophthora kernoviae to select North American native plants. Plant Pathology 61: 224–233. [Google Scholar]
- Flier WG, Grunwald NJ, Kroon LPNM, et al. 2002. Phytophthora ipomoeae sp. nov., a new homothallic species causing leaf blight on Ipomoea longipedunculata in the Toluca Valley of Central Mexico. Mycological Research 106: 848–856. [Google Scholar]
- Forbes GA, Gamboa S, Lindqvist-Kreuze H, et al. 2016. Identification of an A2 population of Phythophthora andina attacking tree tomato in Peru indicates a risk of sexual reproduction in this pathosystem. Plant Pathology 65: 1109–1117. [Google Scholar]
- Forbes GA, Morales JG, Restrepo S, et al. 2013. Phytophthora infestans and Phytophthora andina on Solanaceous hosts in South America. In: Lamour K. (ed), Phytophthora: a global perspective: 48–58. CABI Plant Protection Series, No. 2, UK. [Google Scholar]
- Forbes GA, Ristaino JB, Oliva RF, et al. 2012. A rebuttal to the letter to the editor concerning 'Defining species boundaries in the genus Phytophthora: the case of Phytophthora andina'. Plant Pathology 61: 221–223. [Google Scholar]
- Fry W. 2008. Phytophthora infestans: The Plant (and R Gene) Destroyer. Molecular Plant Pathology 9: 385–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk DJ, Omland KE. 2003. Species-level paraphyly and polyphyly: frequency, and consequences, with insights from animal mitochondrial DNA. Annual Review of Ecology and Systematics 34: 397–423. [Google Scholar]
- Galbraith DW, Harkins KR, Maddox JM, et al. 1983. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220: 1049–1051. [DOI] [PubMed] [Google Scholar]
- Galindo J, Hohl RH. 1985. Phytophthora mirabilis, a new species of Phytophthora. Sydowia 38: 87–96. [Google Scholar]
- Gallegly ME, Hong C. 2008. Phytophthora: Identifying species by morphology and DNA fingerprints. American Phytopathological Society (APS) Press, USA. [Google Scholar]
- Gavino PD, Fry WE. 2002. Diversity in and evidence for selection on the mitochondrial genome of Phytophthora infestans. Mycologia 94: 781–793. [PubMed] [Google Scholar]
- Gómez-Alpizar L, Carbone I, Ristaino JB. 2007. An Andean origin of Phytophthora infestans inferred from mitochondrial and nuclear gene genealogies. Proceedings of the National Academy of Sciences of the United States of America 104: 3306–3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Alpizar L, Hu CH, Oliva R, et al. 2008. Phylogenetic relationships of Phytophthora andina, a new species from the highlands of Ecuador that is closely related to the Irish potato famine pathogen Phytophthora infestans. Mycologia 100: 590–602. [DOI] [PubMed] [Google Scholar]
- Goodwin SB, Drenth A, Fry WE. 1992. Cloning and genetic analyses of two highly polymorphic, moderately repetitive nuclear DNAs from Phytophthora infestans. Current Genetics 22: 107–115. [DOI] [PubMed] [Google Scholar]
- Goss EM, Cardenas ME, Myers K, et al. 2011. The plant pathogen Phytophthora andina emerged via hybridization of an unknown Phytophthora species and the Irish potato famine pathogen, P. infestans. PLoS One 6: e24543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss EM, Tabima JF, Cooke DE, et al. 2014. The Irish potato famine pathogen Phytophthora infestans originated in Central Mexico rather than the Andes. Proceedings of the National Academy of Sciences of the United States of America 111: 8791–8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith GW, Shaw DS. 1998. Polymorphisms in Phytophthora infestans: Four mitochondrial haplotypes are detected after PCR amplification of DNA from pure cultures or from host lesions. Applied and Environmental Microbiology 64: 4007–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Kamoun S, Zody MC, et al. 2009. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461: 393–398. [DOI] [PubMed] [Google Scholar]
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – A baiting and iterative mapping approach. Nucleic Acids Research 41: e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkett F, Kindlmann P, Plantegenest M, et al. 2006. Temporal differentiation and spatial coexistence of sexual and facultative asexual lineages of an Aphid species at mating sites. Journal of Evolutionary Biology 19: 809–815. [DOI] [PubMed] [Google Scholar]
- Harrington TC, Pashenova NV, McNew DL, et al. 2002. Species delimitation and host specialization of Ceratocystis laricicola and C. polonica to larch and spruce. Plant Disease 86: 418–422. [DOI] [PubMed] [Google Scholar]
- Harrington TC, Rizzo DM. 1999. Defining species in the fungi. In: Worrall JJ. (ed), Structure and dynamics of fungal populations: 43–70. Kluwer Academic, The Netherlands. [Google Scholar]
- Haverkort AJ, Boonekamp PM, Hutten R, et al. 2008. Societal costs of late blight in potato and prospects of durable resistance through cisgenic modification. Potato Research 51: 47–57. [Google Scholar]
- Haverkort AJ, Struik PC, Visser RGF, et al. 2009. Applied biotechnology to combat late blight in potato caused by Phytophthora infestans. Potato Research 52: 249–264. [Google Scholar]
- Hickerson MJ, Meyer CP, Moritz C. 2006. DNA barcoding will often fail to discover new animal species over broad parameter space. Systematic Biology 55: 729–739. [DOI] [PubMed] [Google Scholar]
- Husson C, Aguayo J, Revellin C, et al. 2015. Evidence for homoploid speciation in Phytophthora alni supports taxonomic reclassification in this species complex. Fungal Genetics Biology 77: 12–21. [DOI] [PubMed] [Google Scholar]
- Jombart T, Devillard S, Dufour AB, et al. 2008. Revealing cryptic spatial patterns in genetic variability by a new multivariate method. Heredity 101: 92–103. [DOI] [PubMed] [Google Scholar]
- Jung T, Hansen EM, Winton L, et al. 2002. Three new species of Phytophthora from European oak forests. Mycological Research 106: 397–411. [Google Scholar]
- Jung T, Jung MH, Scanu B, et al. 2017. Six new Phytophthora species from ITS clade 7a including two sexually functional heterothallic hybrid species detected in natural ecosystems in Taiwan. Persoonia 38: 100–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Toh H. 2010. Parallelization of the MAFFT Multiple Sequence Alignment Program. Bioinformatics 26: 1899–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles LL, Carstens BC, Weins J. 2007. Delimiting species without monophyletic gene trees. Systematic Biology 56: 887–895. [DOI] [PubMed] [Google Scholar]
- Kroon LP, Bakker FT, Van den Bosch GB, et al. 2004. Phylogenetic analysis of Phytophthora species based on mitochondrial and nuclear DNA sequences. Fungal Genetics and Biology 41: 766–782. [DOI] [PubMed] [Google Scholar]
- Kroon LP, Brouwer H, De Cock AW, et al. 2012. The genus Phytophthora anno 2012. Phytopathology 102: 348–364. [DOI] [PubMed] [Google Scholar]
- Ko WH. 1978. Heterothallic Phytophthora-evidence for hormonal regulation of sexual reproduction. Journal General Microbiology 107: 15–18. [Google Scholar]
- Langmead B. 2010. Aligning short sequencing reads with Bowtie. Current Protocols in Bioinformatics 11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassiter ES, Russ C, Nusbaum C, et al. 2015. Mitochondrial genome sequences reveal evolutionary relationships of the Phytophthora 1c Clade species. Current Genetics 61: 567–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees AK, Wattier R, Shaw DS, et al. 2006. Novel microsatellite markers for the analysis of Phytophthora infestans populations. Plant Pathology 55: 311–319. [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997 [q-bio.GN]. [Google Scholar]
- Man in 't Veld WA, Rosendahl KCHM, Hong C. 2012. Phytophthora × serendipita sp. nov. and P. × pelgrandis, two destructive pathogens generated by natural hybridization. Mycologia 104: 1390–1396. [DOI] [PubMed] [Google Scholar]
- Martin FN, Abad ZG, Balci Y, et al. 2012. Identification and detection of Phytophthora: reviewing our progress, identifying our needs. Plant Disease 96: 1080–1103. [DOI] [PubMed] [Google Scholar]
- Martin FN, Blair JE, Coffey MD. 2014. A combined mitochondrial and nuclear multilocus phylogeny of the genus Phytophthora. Fungal Genetics and Biology 66: 19–32. [DOI] [PubMed] [Google Scholar]
- Martin MD, Vieira FG, Ho SY, et al. 2016. Genomic characterization of a South American Phytophthora hybrid mandates reassessment of the geographic origins of Phytophthora infestans. Molecular Biology and Evolution 33: 478–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayton H, Smart C, Moravec B, et al. 2000. Oospore survival and pathogenicity of single oospore recombinant progeny from a cross involving US-17 and US-8 genotypes of Phytophthora infestans. Plant Disease 84: 1190–1196. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, et al. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. In: Gateway Computing Environments Workshop (GCE): 1–8. USA. [Google Scholar]
- Muir CD, Hahn MW. 2015. The limited contribution of reciprocal gene loss to increased speciation rates following whole-genome duplication. The American Naturalist 185: 70–86. [DOI] [PubMed] [Google Scholar]
- Nakamura D, Tiersch TR, Douglass M, et al. 1990. Rapid identification of sex in birds by flow cytometry. Cytogenetics and Cell Genetics 53(4): 201–205. [DOI] [PubMed] [Google Scholar]
- Oliva RF, Kroon LPNM, Chacón G, et al. 2010. Phytophthora andina sp, nov., a newly identified heterothallic pathogen of solanaceous hosts in the Andean highlands. Plant Pathology 59: 613–625. [Google Scholar]
- Ordoñez ME, Hohl HR, Velasco JA, et al. 2000. A novel population of Phytophthora, similar to P. infestans, attacks wild solanum species in Ecuador. Phytopathology 90: 197–202. [DOI] [PubMed] [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, et al. 2013. Nlme: Linear and nonlinear mixed effects models. R package version 3.1: 131 [Google Scholar]
- Pohlert T. 2014. The pairwise multiple comparison of mean ranks package (PMCMR)s. R package version 3.1: 1–9. [Google Scholar]
- Poulin R. 2005. Relative infection levels and taxonomic distances among the host species used by a parasite: insights into parasite specialization. Parasitology 130: 109–115. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttick MN, Clark J, Donoghue PC. 2015. Size is not everything: rates of genome size evolution, not C-value, correlate with speciation in angiosperms. Proceedings of the Royal Society B 282: 20152289. doi: 10.1098/rspb.2015.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org/ [accessed October 2013]. [Google Scholar]
- Rambaut A. 2009. FigTree, a graphical viewer of phylogenetic trees. Tree figure drawing tool version 1.3.1. Institute of Evolutionary Biology, University of Edinburgh, UK. [Google Scholar]
- Restrepo S, Tabima JF, Mideros MF, et al. 2014. Speciation in fungal and oomycete plant pathogens. Annual Review of Phytopathology 52: 289–316. [DOI] [PubMed] [Google Scholar]
- Roe AD, Rice AV, Bromilow SE, et al. 2010. Multilocus species identification and fungal DNA barcoding: Insights from blue stain fungal symbionts of the Mountain Pine Beetle. Molecular Ecology Resources 10: 946–959. [DOI] [PubMed] [Google Scholar]
- Santini F, Harmon LJ, Carnevale G, et al. 2009. Did genome duplication drive the origin of teleosts? A comparative study of diversification in ray-finned fishes. BMC Evolutionary Biology 9: 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidl Johnson A, Frost KE, Rouse DI, et al. 2014. Effect of temperature and sporulation of US-22, US-23, and US-24 Clonal lineages of Phytophthora infestans and implications for late blight epidemiology. Ecology and Epidemiology: Phytopathology 105: 449–459. [DOI] [PubMed] [Google Scholar]
- Shaffer HB, Thomson RC. 2007. Delimiting species in recent radiations. Systematic Biology 56: 896–906. [DOI] [PubMed] [Google Scholar]
- Singh G, Dal Grande F, Divakar PK, et al. 2015. Coalescent-based species delimitation approach uncovers high cryptic diversity in the cosmopolitan lichen-forming fungal genus Protoparmelia (Lecanorales, Ascomycota). PLoS One 10: e0124625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soanes DM, Thomas AR, Talbot NJ. 2007. Insights from sequencing fungal and oomycete genomes: What can we learn about plant disease and the evolution of pathogenicity? The Plant Cell 19: 3318–3326. doi: 10.1105/tpc.107.056663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- Stukenbrock EH. 2013. Evolution, selection and isolation: a genomic view of speciation in fungal plant pathogens. New Phytologist 199: 895–907. [DOI] [PubMed] [Google Scholar]
- Tellier A, De Vienne DM, Giraud T, et al. 2010. Theory and examples of reciprocal influence between hosts and pathogens, from short-term to long-term interactions: Coevolution, cospeciation and pathogen speciation following host shifts. In: Barton AW. (ed), Host-pathogen interactions: genetics, immunology and physiology: 37–77. Nova Science Publishers, USA. [Google Scholar]
- Thaxter R. 1889. A new American Phytophthora. Botanical Gazette Crawfordsville 14: 273–274. [Google Scholar]
- Van der Lee T, De Witte I, Drenth A, et al. 1997. AFLP linkage map of the oomycete Phytophthora infestans. Fungal Genetics and Biology 21: 278–291. [DOI] [PubMed] [Google Scholar]
- Vargas AM, Quesada-Ocampo LM, Céspedes MC, et al. 2009. Characterization of Phytophthora infestans populations in Colombia: first report of the A2 mating type. Phytopathology 99: 82–88. [DOI] [PubMed] [Google Scholar]
- Venables WN, Ripley BD. 2002. Modern applied statistics with S. 4nd edn Springer, USA. [Google Scholar]
- Vialle A, Feau N, Frey P, et al. 2013. Phylogenetic species recognition reveals host-specific lineages among poplar rust fungi. Molecular Phylogenetics and Evolution 66: 628–644. [DOI] [PubMed] [Google Scholar]
- Wood TE, Takebayashi N, Barker MS, et al. 2009. The frequency of polyploid speciation in vascular plants. Proceedings of the National Academy of Sciences of the United States of America 106: 13875–13879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Schuenemann VJ, Cano LM, et al. 2013. The rise and fall of the Phytophthora infestans lineage that triggered the Irish potato famine. eLife 2: e00731. [DOI] [PMC free article] [PubMed] [Google Scholar]