Summary
Human-induced environmental change and habitat fragmentation pose major threats to biodiversity and require active conservation efforts to mitigate their consequences. Genetic rescue through translocation and the introduction of variation into imperiled populations has been argued as a powerful means to preserve, or even increase, the genetic diversity and evolutionary potential of endangered species [1, 2, 3, 4]. However, factors such as outbreeding depression [5, 6] and a reduction in available genetic diversity render the success of such approaches uncertain. An improved evaluation of the consequence of genetic restoration requires knowledge of temporal changes to genetic diversity before and after the advent of management programs. To provide such information, a growing number of studies have included small numbers of genomic loci extracted from historic and even ancient specimens [7, 8]. We extend this approach to its natural conclusion, by characterizing the complete genomic sequences of modern and historic population samples of the crested ibis (Nipponia nippon), an endangered bird that is perhaps the most successful example of how conservation effort has brought a species back from the brink of extinction. Though its once tiny population has today recovered to >2,000 individuals [9], this process was accompanied by almost half of ancestral loss of genetic variation and high deleterious mutation load. We furthermore show how genetic drift coupled to inbreeding following the population bottleneck has largely purged the ancient polymorphisms from the current population. In conclusion, we demonstrate the unique promise of exploiting genomic information held within museum samples for conservation and ecological research.
Keywords: conservation genomics, population genomics, endangered species, extinction, demography, inbreeding, mutation load, genetic recovery, ancient genomics, ornithology
Highlights
-
•
Recent population decline of crested ibis in 10 kya was likely shaped by human activity
-
•
Modern group has lost almost half of genetic variations in pre-bottleneck ancestors
-
•
Modern group suffers from a high inbreeding coefficient and deleterious mutation load
-
•
Ancestral balancing selection is outweighed by genetic drift in the modern group
Feng et al. use whole-genome sequencing of contemporary and historic crested ibis, an iconic endangered bird species, to explore how their genetic diversity has changed through time. Their analyses reveal the roles of genetic drift and intensive inbreeding on the loss of genetic diversity in today’s population.
Results
Historical and Contemporary Population Structure of the Crested Ibis
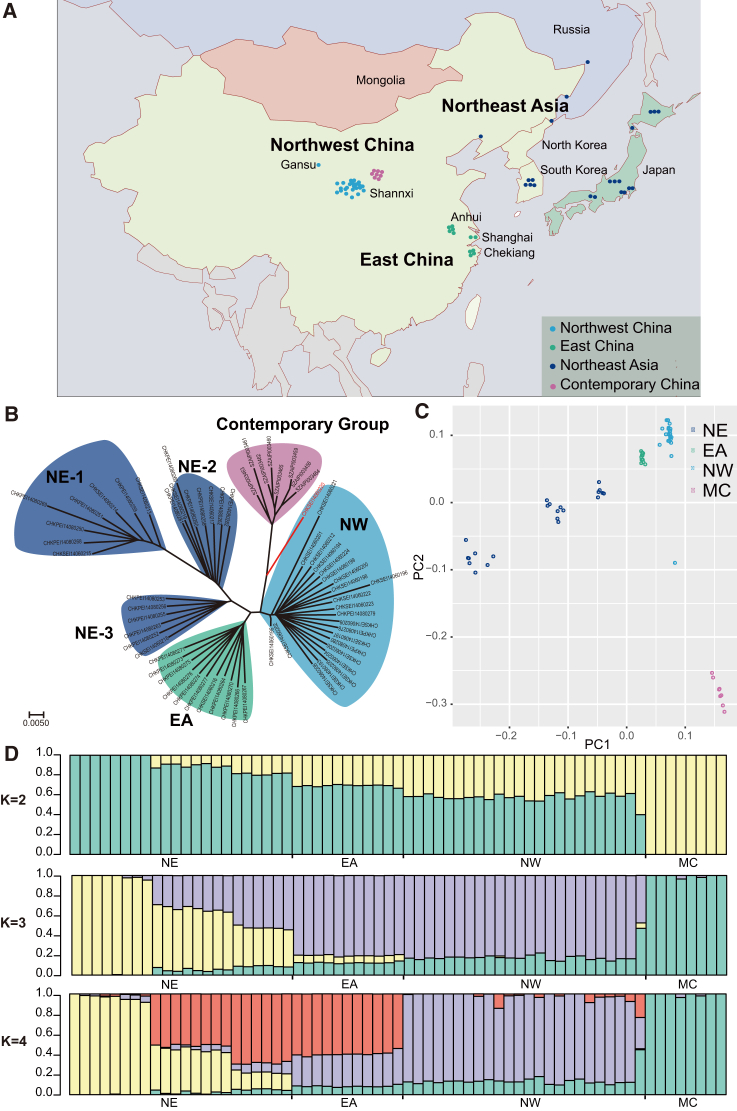
We collected 57 historic crested ibis samples dating to between 1841 and 1922. The geographic origin of these samples broadly covers the historical distribution of the species, including east China, northwest China, northeast China, Korea, Japan, and Russia (Figure 1A). We initially generated an average of ∼179 million (M) paired-end short sequencing reads for historic samples (49 base pairs [bp] for most samples and 100 bp for two; Data S1). After filtering to remove contaminants and other sequencing artifacts, we obtained final genomic coverage at ca. 7-fold for each sample. Analysis with mapDamage2.0 revealed signs of cytosine deamination at the ends of the sequences, which are normally expected from the historic DNA sequences (Data S1). We combined these data with previously published sequencing data for eight contemporary individuals [10] and used ANGSD v0.615 [11] to identify 5,268,206 SNPs in total. On average, each historical sample contained 1,088,820 (±112,638) heterozygous sites, while each contemporary sample contained 788,162 (±35,747) heterozygous sites.
Figure 1.
Sampling Map and Population Structure of the Crested Ibis
(A) Map of sampling locations.
(B) NJ phylogeny of all 65 individuals (57 historic and 8 contemporary) correlates with geographic distribution in (A), except for the one (marked in red) collected from the NW, which is closely related to the contemporary group. Three NE subgroups (NE-1, NE-2, and NE-3), EA, NW, and contemporary groups (MC) are labeled by different background colors.
(C) Principal component analysis for all 65 individuals. The observed result is consistent with that of the NJ phylogeny.
(D) Population structure of all 65 individuals (K = 2, 3, and 4). The population origin of each individual is indicated on x axis. Each individual is represented by a bar that is segmented into colors based on the ancestry proportions given the assumption of K populations.
After excluding linked SNP loci that could potentially bias clustering results (see STAR Methods), we built a neighbor-joining (NJ) tree [12] using all 65 samples (Figure 1B). The NJ tree assigned the historical samples to three major groups, consistent with their original geographic provenance. A northeast Asian group (NE) exhibits the most diversity and can be further divided into three subgroups (NE-1, NE-2, and NE-3) according to their genetic distances. The remaining samples fall into two groups: east China (EA) and northwest China (NW). All contemporary samples (MC) are from northwest China and form a monophyletic group. These clustering results were also supported by principal component analysis (PCA) [13] (Figure 1C).
We ran STRUCTURE v2.3.4 [14] to explore the genetic composition of each group and subgroup after initially removing potential bias caused by the missing loci (Figure 1D; Data S2). Sample clusters were evaluated using the ad hoc statistic (ΔK) [15]. The contemporary group separated from the historical populations when setting the clusters K as 2. The ΔK value reaches its maximum at K = 3, indicating the uppermost level of structure. At K = 4, the clusters reflect the geographic distribution of historical samples. Overall the population structuring indicates that while the contemporary group is genetically distinct to all historical groups, it shares a moderate level of genetic variation with the EA and NW groups.
Population Size Dynamics of the Crested Ibis
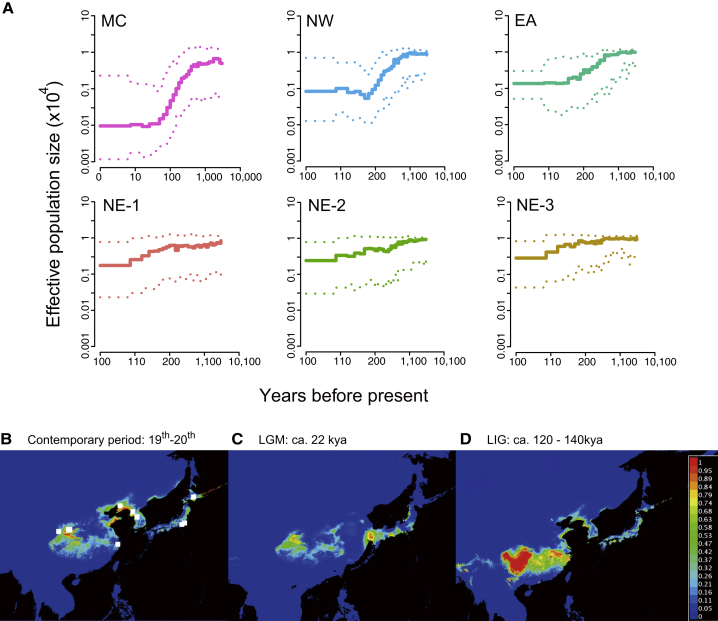
We used the historic genomic data to confirm the decline of the effective population size (Ne) reported in a previous study [10] (Figure S1). Because the power of PSMC (pairwise sequentially Markovian coalscent) to reconstruct demography is poor over recent history due to the limited number of recombination events [16], we used an alternate method based on approximate Bayesian computation, PopSizeABC v2.1 [17], to better characterize the Ne dynamics over the last 10,000 years, during which the climate has been relatively stable [18]. We found that all groups declined at different rates over the last 10,000 years, and such decline was followed by a short period of relative population stability before the near extinction of the crested ibis in the 20th century (Figure 2A). The recent demographic pattern implies that the impact of human activity on the population decline may have already commenced up to 600 years ago (the starting point of decline in the NW group).
Figure 2.
Inference of Recent Population Size Tendency and Prediction of Breeding Ranges for the Crested Ibis at the Contemporary, the Last Glacial Maximum, and the Last Interglacial Periods
(A) The recent effective population size (Ne) for each group is inferred using PopSizeABC. A 90% confidence interval is indicated by dotted lines.
(B–D) Reconstruction of ecological niche models for the contemporary (19th and 20th centuries, B), last glacial maximum (LGM, approximately 22 kya, C) and the last interglacial (LIG, approximately 120–140 kya, D) periods. Pictures show the point-wise mean of 10 replicates using 19 environmental layers. The colors indicate the predicted probability that conditions are suitable, with red indicating high and blue indicating low probabilities. The white square points in (B) indicate the geographic coordinates of the crested ibis samples.
Environmental suitability is an important factor that influences population fluctuation. Therefore, we used ecological niche models (ENMs) [19] to reconstruct the suitable niche for crested ibis during the contemporary period (19th and 20th centuries), the last glacial maximum (LGM, approximately 22 thousand years ago [kya]) and the last interglacial period (LIG, approximately 120–140 kya) based on 35 occurrences of crested ibis samples associated with a latitude and longitude record during the 19th and 20th centuries in the VertNet database (Figures 2B–2D) (see STAR Methods). During the LIG period, the potential breeding range of the crested ibis covered broad areas across south China, with extensive hotspots of habitat (Figure 2D). During the LGM, their potential breeding range showed extensive contraction, corresponding to the climate change during this period [20] (Figure 2C). Today, our simulations predict that the suitable breeding range would largely resemble that for the LGM, with multiple distribution hotspots with suitable probability higher than 50% (Figure 2B). Nevertheless, our data shows that Ne continued to decline in the contemporary period (Table S1), consistent with the fact that the historical groups went extinct across most of the breeding range during the 19th and 20th centuries [9]. The reconstructed suitable niche pattern implies that the population collapse during the late-19th and early-20th centuries was more likely a result of human activity rather than climatic factors. This is consistent with previous analyses on the availability of habitat suitable for crested ibis, which demonstrated how radical land-cover changes (e.g., through urban development, increased cultivation of land, and increased crop diversification) between the 1970s and 2000s reduced the swamp environments that used to be their feeding areas [21, 22].
Severe Genetic Diversity Loss in the Contemporary Population Caused by Inbreeding
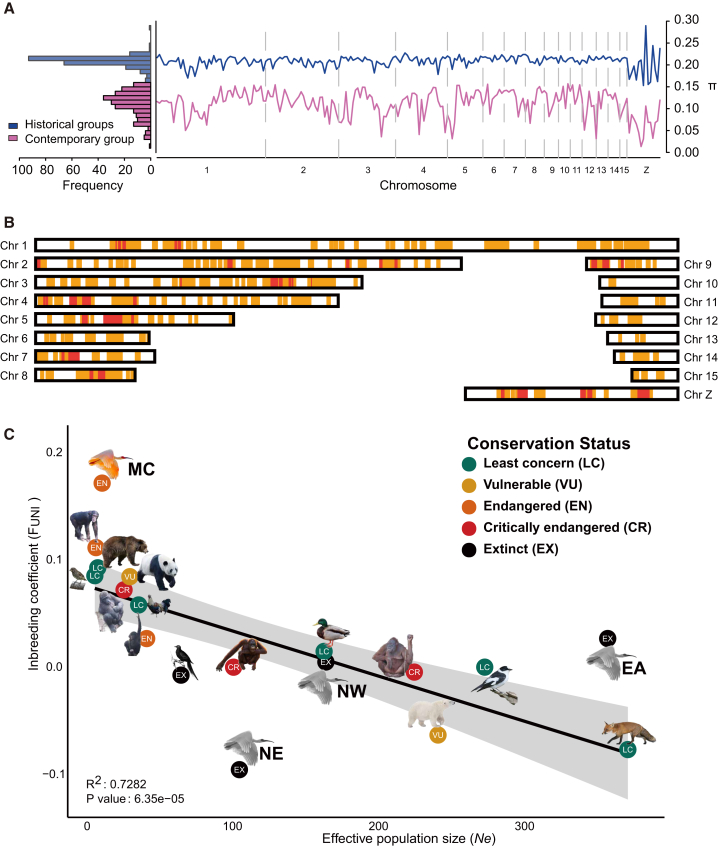
Although the size of the contemporary population has been steadily increasing owing to recent conservation efforts, its nucleotide diversity (π) is considerably lower than that of the historic population (mean value is 0.112, while mean value of the historic samples is 0.208; Figure 3A). We also found accelerated fixation of polymorphisms in the contemporary population, with ca. 70% of the SNPs that were polymorphic in the historical groups having been fixed in the contemporary group (see STAR Methods). We further identified the identical by descent (IBD) regions, which means the alleles derive from the ancestor and maintain in descendent populations [23]. We obtained a total of 444.5 megabases (Mb) merged excess IBD regions in the contemporary group (Figure 3B), 31.8 Mb for the NW, 31.0 Mb for the NE, and 0 Mb for the EA groups.
Figure 3.
Loss of Genetic Diversity and Elevated Inbreeding in the Crested Ibis
(A) Left: A histogram describing mean π for 5 Mb windows across the crested ibis genome in the historical and contemporary groups. Right: Genomic distribution of individual pairwise estimates of mean π in 5 Mb windows across the crested ibis genome in historical and contemporary groups. Chromosome boundaries are indicated as vertical dashed lines.
(B) The chromosomal distribution of IBD regions in the contemporary population. IBD regions are marked in orange, with additional red highlighting if supported by more than half samples.
(C) Linear model of inbreeding coefficient (FUNI) and effective population size (Ne) based on 14 animal species using the published population datasets. R-squared value and the p value of F test indicated a significant positive correlation between FUNI and Ne, while the contemporary population (labeled as MC) is a significant outlier. The historical crested ibis populations are indicated in gray.
Such dramatic loss of genetic diversity could be caused by the unavoidable effects of inbreeding. To assess the extent of inbreeding, we estimated FUNI based on the correlation between uniting gametes following Wright [24]. A lower (i.e., negative) value indicates an excess of heterozygosity and little inbreeding effect, while a positive value indicates an excess of homozygosity and strong inbreeding effect. We subsequently calculated FUNI and Ne for 14 animal species spanning different conservation statuses. A regression correlation including the crested ibis and other species clearly shows that the contemporary population is a significant outlier, highlighting the severity of its inbreeding (Figure 3C; Table S2; see STAR Methods). This finding is consistent with the fact that the gene pool of all contemporary individuals was inherited from only two breeding pairs.
Patterns of Deleterious Mutations across Populations Support Ne Collapse, Bottlenecks, and Inbreeding in the Contemporary Population
Recent conservation genetic studies have demonstrated that small populations are susceptible to allelic drift, leading to allele loss or fixation, which can be evaluated using the site-frequency-spectrum (SFS) [25]. In our analysis, all historic populations showed a general pattern in which most mutations occurred at low frequencies, while few mutations segregated at high frequencies (Figures S2A–S2C). However, in the contemporary population, we observed a reduced number of low-frequency mutations and a relatively higher fraction of mutations that segregate at medium or higher frequencies (Figure S2D). We hypothesize that these mutations occur at low rates and that relaxed purifying selection has probably led to random fixation, likely due to the high level of inbreeding in the contemporary population. An alternative possibility might be that the contemporary individuals sampled were from a sub-structured population; however, we find no such evidence in the results of the admixture analysis to support this hypothesis.
We further used the Grantham score [26] as an index to identify potential deleteriousness of missense mutations in the coding regions of historic and contemporary populations. The proportional excess of homozygous deleterious-derived mutations could be likely due to the varying efficacy of purifying selection, resulting from differences in the demographic histories of the populations [27, 28]. We found that the ratio of homozygous sites to homo- and heterozygous sites in the contemporary population is significantly higher than that in the historic populations (0.328 ± 0.041 in contemporary group, 0.207 ± 0.039 in historical group, Welch two-sample t test, 2.524e−05) (Figure S2E). A similar difference could also be seen when using loss of function (LOF) as an index (Figure S2F; see STAR Methods). Despite only ∼100 years difference between the collection dates of historic and contemporary samples, the number of homozygous deleterious mutations has doubled in the contemporary population, showcasing the severity of the bottleneck.
Role of Natural Selection in Shaping the Genetic Diversity of the Crested Ibis
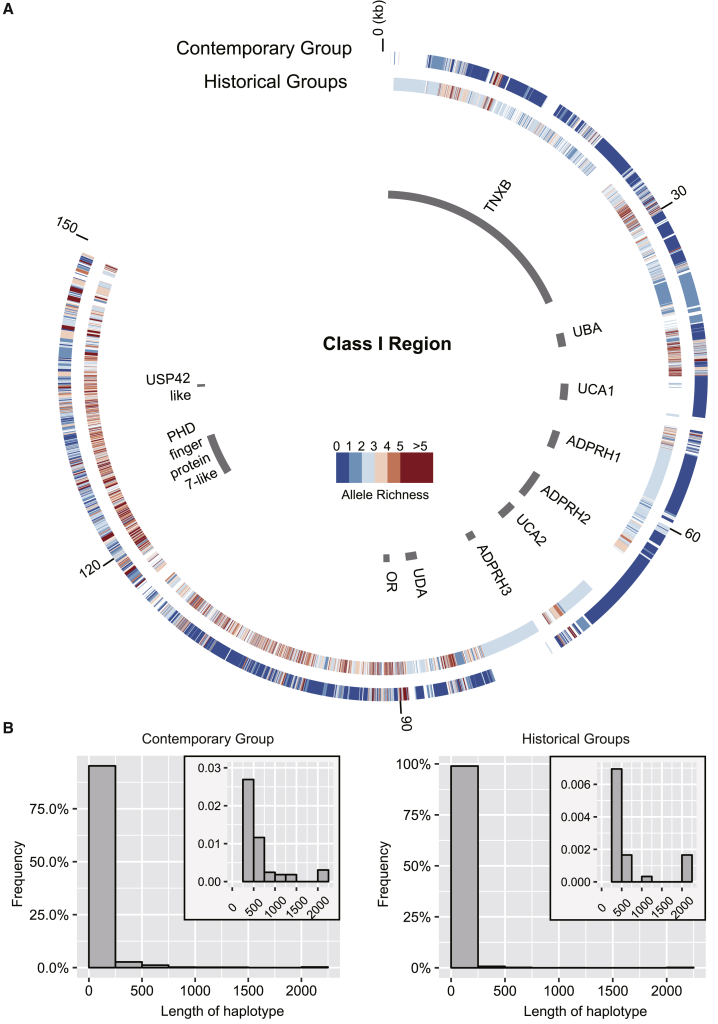
The effect of selection is considered to be limited in contrast to that of genetic drift in populations with a small Ne [29]. Immune genes are believed to be under pathogen-mediated balancing selection, thus maintaining high polymorphism levels in wild populations [30, 31]. However, we found that the genetic diversity of the major histocompatibility complex (MHC) has experienced drastic haplotype loss in the contemporary group. The overall allelic richness (AR) of the entire MHC region in the contemporary group is significantly lower than that shown in the historical groups (meanAR of the contemporary group is 2.44, meanAR of the historical group is 2.92, Welch two-sample t test, p < 2.2e−16; see STAR Methods). Furthermore, haplotype blocks in the contemporary group were much longer and exhibited lower diversity (Figures 4 and S3). This dramatically reduced diversity of the MHC in the contemporary population may confer a high risk of increasing the burden of transmissible pathogens when captive animals are released into the wild [32].
Figure 4.
Haplotype Structure and Length Distribution of the Class I Region in Historical and Contemporary Groups
(A) Haplotype blocks are detected based on the SNP datasets for historical and contemporary groups, respectively. Colored circles show the haplotype structure (each box indicates the start and end SNPs of one haplotype) and the diversity along the class I region (color indicates the allelic richness). The allelic richness of historical groups is estimated in the random way (see STAR Methods). Gaps in the colored circles are the intervals between two neighboring haplotype blocks. Genomic structures of the class I region are drawn as gray bars, with different sizes showing different gene loci of varied sizes.
(B) Length distribution of haplotype blocks. Haplotype blocks with lengths larger than 2,000 bp are grouped into the last bar. The plotting area for the bin of 250–2,000 bp is enlarged in the top right corner.
See also Figure S3.
We subsequently performed a broader scan across the coding region to identify genomic loci that were previously under balancing selection but have been fixed in the current population (see STAR Methods). In total we found 53 genes with high Tajima’s D values in at least two of the historic groups, implying that they were once under balancing selection. Of these, nine genes exhibit a negative Tajima’s D value in the contemporary group that might have been substantially affected by bottleneck effects during the breeding program. Several of these genes are involved in processes relating to immunity, the nervous system, and reproduction. For example, polymorphism has been completely lost in half of the SNP loci in gene KPNA1, a member of the importin or karyopherin alpha family that has been reported to be involved in the development and/or repair processes of the nervous system [33].
Discussion
Thanks to the development of paleogenomic sequencing techniques and the recovery of genetic data from museum samples collected up to 100 years before the introduction of the modern management program, our study has reconstructed a much-improved demographic history of the crested ibis over the last 10,000 years. The continued population decline of this species in this period therefore not only leads to the extreme bottleneck that started in the late-19th century but also provides new insights into the negative effect of human activities. Given that crested ibis did not appear to favor colder climates, the warmer climate of the past 10,000 years should have stopped the prior decline and incurred a recovery—something we see no evidence of. One explanation for this discrepancy might be that the crested ibis had entered into an evolutionary dead end from which they were unable recover, because of their small Ne and the low genetic diversity. However, given the success of the current management program in increasing their population size to ca. 2,600 individuals from only seven founding individuals, it is clear that the species still maintains a robust reproductive rate. An alternate explanation for their population decline might therefore relate to human interference. Human activities, like over-hunting and development of farm land, might have led to the fragmentation of niches for the crested ibis and ultimately increased the inbreeding coefficient within the isolated subpopulations. Consistently, recent research on crested ibis notes how high human-induced mortality risks play a significantly negative effect on the population growth and range expansion of the surviving population [34].
One factor that contributed to the crested ibis’s recent demography is the effect of inbreeding. There is little doubt that inbreeding reduces the fitness of a population [35, 36, 37], particularly for critically endangered species with small population sizes. However, previous studies that have argued that inbreeding depression can elevate extinction risk are based on simulations [38], and thus there is a need for actual evidence with which to verify whether the consequences of inbreeding, such as the accumulation of deleterious alleles, will lead to and even accelerate the loss of diversity. Our findings indicate that the contemporary crested ibis population has dramatically reduced the ancestral genetic diversity, undergone higher allelic fixation, and carried an elevated load of homozygous deleterious mutations that associated with a raised inbreeding coefficient. Importantly, the inclusion of samples spanning 1841–2011 CE enabled us to accurately quantify the extent to which the most recent demographic decline has led to a loss in their genetic variation as well as increased inbreeding levels and genetic load, all of which are regarded as useful genomic parameters for endangered species [39]. Our analyses showed how the overall nucleotide diversity of the contemporary group is only 53.85% of that which was present ca. 100 years ago. This has been accompanied by an almost 4-fold elevation in their inbreeding coefficient and a doubling of their homozygous deleterious mutation load.
The conservation program has had remarkable success in increasing the size of the crested ibis population, from the seven individuals during their near extinction to currently ca. 2,600 individuals. Nevertheless, the contemporary population suffers from dramatic diversity loss across the genome. This overall reduced genetic diversity represents a risk when they are introduced to the natural environment as part of the conservation program. We acknowledge, however that it may be, that the protected environment of the management program will help them to overcome the potential harmful effects from the reduced diversity, thus enabling them to survive into the future with low genetic diversity, as for example has been the case for the endangered island fox over the past millennia [40]. In that example, it is speculated that their isolated island habitat might provide a friendly environment, given its lack of competitors and predators, thus to some extent reducing the negative effect of genetic load.
Ultimately, recent successful re-introduction of the crested ibis into the wild has provided an important step forward in this regard [41]. Long-term monitoring of their genetic diversity in the wild will provide a crucial index for their long-term persistence and management. Recently, efforts have been commenced to establish a DNA identification profiling (DIP) platform using short tandem repeat markers to help avoid close crosses [42]. Given that our analyses identified several candidate genes that no longer exhibit high polymorphism, we suggest it may be useful to incorporate them into the existing DIP for monitoring and preserving genomic diversity. Lastly, our study highlights how integrating genomic information from the modern and historic samples can help to reveal the evolutionary history of endangered species and in doing so provide guiding information of benefit to future conservation programs.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological Samples | ||
| 27 Nipponia nippon tissue samples | American Museum of Natural History | Data S1 |
| 19 Nipponia nippon tissue samples | Natural History Museum (London) | Data S1 |
| 2 Nipponia nippon tissue samples | World Museum, Liverpool | Data S1 |
| 1 Nipponia nippon tissue sample | Naturalis Biodiversity Center Leiden, the Netherlands | Data S1 |
| 1 Nipponia nippon tissue sample | Senckenberg Natural History Museum | Data S1 |
| 2 Nipponia nippon tissue samples | Stuttgart State Museum of Natural History | Data S1 |
| 2 Nipponia nippon tissue samples | Smithsonian Institute National Museum of Natural History | Data S1 |
| 2 Nipponia nippon tissue samples | Natural History Museum Vienna | Data S1 |
| 1 Nipponia nippon tissue sample | Natural History Museum of Denmark | Data S1 |
| Deposited Data | ||
| Whole-genome sequencing data of 57 historical Nipponia nippon samples | This paper | NCBI Project ID: PRJNA481495; CNSA: CNP0000084 |
| Whole-genome sequencing data of 8 contemporary Nipponia nippon samples | [10] | NCBI Project ID: PRJNA232572 |
| Nipponia nippon reference genome | [10] | NCBI Project ID: PRJNA232572 |
| Whole-genome sequencing data of 13 Pan paniscus | [43] | SRA: SRP018689 |
| Whole-genome sequencing data of 5 Pongo abelii | [43] | SRA: SRP018689 |
| Whole-genome sequencing data of 5 Pongo pygmaeus | [43] | SRA: SRP018689 |
| Whole-genome sequencing data of 25 Pan troglodytes | [43] | SRA: SRP018689 |
| Whole-genome sequencing data of 31 Gorilla gorilla | [43] | SRA: SRP018689 |
| Whole-genome sequencing data of 10 Vulpes vulpes | [44] | SRA: SRP100625 |
| Whole-genome sequencing data of 18 Ursus maritimus | [45] | SRA: SRA092289 |
| Whole-genome sequencing data of 10 Ursus arctos | [45] | SRA: SRA092289 |
| Whole-genome sequencing data of 14 Anas platyrhynchos | [46] | SRA: SRP125660 |
| Whole-genome sequencing data of 6 Tibetan fowl from Qinghai | [47] | SRA: SRP040477 |
| Whole-genome sequencing data of 5 Platyspiza crassirostris | [48] | SRA: SRP048643 |
| Whole-genome sequencing data of 20 Ficedula albicollis | [49] | SRA: ERA362744, ERA362838, ERA391986, ERA391991 |
| Whole-genome sequencing data of 34 Ailuropoda melanoleuca | [50] | SRA: SRP013618 |
| Whole-genome sequencing data of 6 Ectopistes migratorius | [51] | SRA: SRP042357, SRP103042 |
| MHC class 1 region sequences of Nipponia nippon | NCBI | KP182409.1 |
| MHC class 2 region sequences of Nipponia nippon | NCBI | KP182408.1 |
| MHC extended region sequences of Nipponia nippon | NCBI | KP182407.1 |
| Locality data of crested ibis fromm American Museum of Natural History. AMNH Bird Collection (published on 2013-10-30) | VertNet database | http://ipt.vertnet.org:8080/ipt/resource.do?r=amnh_birds |
| Locality data of crested ibis from California Academy of Sciences. CAS Ornithology (ORN) (published on 2016-10-31) | VertNet database | http://ipt.calacademy.org:8080/ipt/resource.do?r=orn |
| Locality data of crested ibis from Museum of Comparative Zoology, Harvard University (published on 2017-07-11) | VertNet database | http://digir.mcz.harvard.edu/ipt/resource.do?r=mcz_subset_for_vertnet |
| Locality data of crested ibis from Museum of Vertebrate Zoology, UC Berkeley. MVZ Bird Collection (Arctos) (published on 2015-10-27) | VertNet database | http://ipt.vertnet.org:8080/ipt/resource.do?r=mvz_bird |
| Locality data of crested ibis from National Museum of Natural History, Smithsonian Institution. NMNH Birds (published on 2016-07-21) | VertNet database | https://collections.nmnh.si.edu/ipt/resource?r=nmnh_extant_dwc-a |
| Locality data of crested ibis (1842.1.19.90) from Natural History Museum. Natural History Museum (London) Collection Specimens (published on 2015-10-10) | VertNet database | http://data.nhm.ac.uk/object/7e36ff64-1cf0-44f6-b30d-ae0715a85a53 |
| Locality data of crested ibis (1852.2.5.12) from Natural History Museum. Natural History Museum (London) Collection Specimens (published on 2015-10-10) | VertNet database | http://data.nhm.ac.uk/object/5a4a8a93-ead3-42ce-9229-a8d34a8d0a0e |
| Locality data of crested ibis (1891.10.19.20) from Natural History Museum. Natural History Museum (London) Collection Specimens (published on 2015-10-10) | VertNet database | http://data.nhm.ac.uk/object/b1a0a925-5132-40c7-a9c3-2a999d336660 |
| Locality data of crested ibis (1892.4.2.492) from Natural History Museum. Natural History Museum (London) Collection Specimens (published on 2015-10-10) | VertNet database | http://data.nhm.ac.uk/object/1a589acb-26c9-48c0-9365-6a7620d12e5d |
| Locality data of crested ibis (1892.4.2.493) from Natural History Museum. Natural History Museum (London) Collection Specimens (published on 2015-10-10) | VertNet database | http://data.nhm.ac.uk/object/7be93e6c-f56d-4922-95ce-31635a6e3c30 |
| Locality data of crested ibis (1897.10.30.2) from Natural History Museum. Natural History Museum (London) Collection Specimens (published on 2015-10-10) | VertNet database | http://data.nhm.ac.uk/object/f7ca4de8-0772-4743-9c82-59656231504d |
| Locality data of crested ibis (1897.10.30.3) from Natural History Museum. Natural History Museum (London) Collection Specimens (published on 2015-10-10) | VertNet database | http://data.nhm.ac.uk/object/f9b28058-f974-40c1-9300-0248271cf128 |
| Locality data of crested ibis (1897.10.30.4) from Natural History Museum. Natural History Museum (London) Collection Specimens (published on 2015-10-10) | VertNet database | http://data.nhm.ac.uk/object/927fb79d-4100-49a0-894a-3a8cfa644e1f |
| Locality data of crested ibis (1897.10.30.5) from Natural History Museum. Natural History Museum (London) Collection Specimens (published on 2015-10-10) | VertNet database | http://data.nhm.ac.uk/object/218dc4cd-6aa4-4349-956b-9bd253b1e036 |
| Locality data of crested ibis (1897.10.30.6) from Natural History Museum. Natural History Museum (London) Collection Specimens (published on 2015-10-10) | VertNet database | http://data.nhm.ac.uk/object/69828014-8f2c-4e67-8fc2-f1ac1e623b63 |
| Locality data of crested ibis (1900.9.9.12) from Natural History Museum. Natural History Museum (London) Collection Specimens (published on 2015-10-10) | VertNet database | http://data.nhm.ac.uk/object/e04fe7fe-46c3-43e2-89d4-cb106558181f |
| Locality data of crested ibis (1900.9.9.13) from Natural History Museum. Natural History Museum (London) Collection Specimens (published on 2015-10-10) | VertNet database | http://data.nhm.ac.uk/object/1e5fed44-06b0-4cfe-959c-e1d3902947f3 |
| Locality data of crested ibis (1908.1.5.20) from Natural History Museum. Natural History Museum (London) Collection Specimens (published on 2015-10-10) | VertNet database | http://data.nhm.ac.uk/object/3ff29518-a451-416a-9e56-42992ea304fb |
| Locality data of crested ibis (1908.1.5.21) from Natural History Museum. Natural History Museum (London) Collection Specimens (published on 2015-10-10) | VertNet database | http://data.nhm.ac.uk/object/1a3f8ba6-4d7f-45fe-b165-07fd1ef26207 |
| Locality data of crested ibis (1908.1.5.22) from Natural History Museum. Natural History Museum (London) Collection Specimens (published on 2015-10-10) | VertNet database | http://data.nhm.ac.uk/object/786144ad-774e-49d8-9ed9-b6378db8cdfe |
| Locality data of crested ibis (1908.1.5.23) from Natural History Museum. Natural History Museum (London) Collection Specimens (published on 2015-10-10) | VertNet database | http://data.nhm.ac.uk/object/7a9df17a-c7d3-4d73-8ddf-e72c3ece16e6 |
| Locality data of crested ibis (1908.1.5.24) from Natural History Museum. Natural History Museum (London) Collection Specimens (published on 2015-10-10) | VertNet database | http://data.nhm.ac.uk/object/365367ee-fd32-4981-a62d-853df63b0a35 |
| Locality data of crested ibis (1908.1.5.25) from Natural History Museum. Natural History Museum (London) Collection Specimens (published on 2015-10-10) | VertNet database | http://data.nhm.ac.uk/object/35a1776c-7568-4876-b800-753f527569ec |
| Locality data of crested ibis (1908.1.5.26) from Natural History Museum. Natural History Museum (London) Collection Specimens (published on 2015-10-10) | VertNet database | http://data.nhm.ac.uk/object/d580fe99-1e9d-4540-9ca0-2e10ab18dcc3 |
| Locality data of crested ibis (1912.9.23.14) from Natural History Museum. Natural History Museum (London) Collection Specimens (published on 2015-10-10) | VertNet database | http://data.nhm.ac.uk/object/ec073508-ceae-4688-880c-3fd00f855aaf |
| Locality data of crested ibis (1988.17.1) from Natural History Museum. Natural History Museum (London) Collection Specimens (published on 2015-10-10) | VertNet database | http://data.nhm.ac.uk/object/141067d4-2105-438d-b128-2388ffd3670d |
| Locality data of crested ibis (1988.17.2) from Natural History Museum. Natural History Museum (London) Collection Specimens (published on 2015-10-10) | VertNet database | http://data.nhm.ac.uk/object/21cd5738-ae0f-496c-b798-8fa4b60715f7 |
| Locality data of crested ibis from University Museum of Zoology Cambridge (Zoology). UMZC Zoological Specimens (published on 2015-02-16) | VertNet database | http://ipt.vertnet.org:8080/ipt/resource.do?r=umzc_vertebrates |
| Locality data of crested ibis from University of Michigan Museum of Zoology. UMMZ Birds Collection (published on 2015-10-28) | VertNet database | http://ipt.vertnet.org:8080/ipt/resource.do?r=ummz_birds |
| WorldClim v1.4 | [52] | http://www.worldclim.org/current |
| Software and Algorithms | ||
| PALEOMIX | [53] |
https://geogenetics.ku.dk/publications/paleomix; RRID: SCR_015057 |
| Picard v1.92 | N/A |
http://broadinstitute.github.io/picard/; RRID:SCR_006525 |
| ANGSD v0.615 | [11] | https://github.com/ANGSD/angsd |
| RapidNJ v2.3.2 | [54] | http://birc.au.dk/software/rapidnj/ |
| R package ‘pcadapt’ | [13] | https://cran.r-project.org/web/packages/pcadapt/index.html |
| STRUCTURE v2.3.4 | [14] |
https://web.stanford.edu/group/pritchardlab/structure.html; RRID:SCR_002151 |
| PSMC | [16] | https://github.com/lh3/psmc |
| PopSizeABC v2.1 | [17] | https://forge-dga.jouy.inra.fr/projects/popsizeabc/ |
| Maxent v3.4.0 | [55] | https://github.com/mrmaxent/Maxent |
| GRASS GIS v7.2.1 | N/A | https://grass.osgeo.org/grass7/ |
| R v3.2.2 | [56] | https://www.r-project.org |
| NeEstimator v2.1 | [57] | http://www.molecularfisherieslaboratory.com.au/neestimator-software/ |
| Genome-wide Complex Trait Analysis (GCTA) v1.91.4 | [58] | http://cnsgenomics.com/software/gcta/ |
| PLINK v1.07 | [59] |
http://zzz.bwh.harvard.edu/plink/; RRID:SCR_001757 |
| GERMLINE v1.5.1 | [60] |
http://gusevlab.org/software/germline/; RRID:SCR_001720 |
| R package ‘pegas’ v0.10 | [61] | https://cran.r-project.org/web/packages/pegas/index.html |
| SnpEff v4.3 | [62] |
http://snpeff.sourceforge.net/SnpEff.html; RRID:SCR_005191 |
| GERBIL v1.1 | [63] | https://github.com/dice-group/gerbil |
| R package ‘PopGenome’ v2.6.0 | [64] | https://cran.r-project.org/web/packages/PopGenome/index.html |
Contact for Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Guojie Zhang (guojie.zhang@bio.ku.dk).
Experimental Model and Subject Details
The current study compares whole genome sequencing data generated from 57 historic crested ibis (22 samples from Northeast Asia, 11 samples from East China, and 24 from Northeast China) against 8 previously published contemporary genomes, to characterize the temporal changes of genetic diversity for this iconic endangered species. All known information on the context and sequencing information of the samples is provided in Data S1. All researches are under the oversight of the institutional review board.
Method Details
Whole-genome sequencing of museum samples
DNA was extracted at the Centre for GeoGenetics from preserved historic skin and toepad samples from 57 crested ibis. Tissue was macerated in extraction buffer and digested overnight according to a modified silica extraction protocol of Rohland [65]. Briefly, buffer (pH8.0, 0.5M EDTA, 1% SDS, 50mM DTT, 1mg/μl, ProteinaseK) was used with samples and negative controls (in a ratio of ∼16:1) for overnight digestion at 55°C. Samples and controls were then spun down and the supernatant was transferred to filter columns (Amicon®, 30KDa cutoff). After concentration to sub 1ml volumes, concentrates were passed through MinElute columns (QIAGEN®, Venlo, Netherlands) according to the manufacturer’s protocol, and finally eluted in 60μl EB buffer. This DNA was converted into Illumina sequencing libraries through blunt-end ligation using the NEB E6070 kit and the slightly modified version of the protocol of Vilstrup et al. [66], reported by Paijmans et al. [67]. Each DNA tissue extract was used to create one library. Each unique library included individual barcodes and were sequenced by Illumina HiSeq2000 using a TruSeq SBS sequencing kit version 3 (Illumina®, San Diego, CA) for 100 cycles from each end of the fragments. Reads were analyzed with Casava1.8.2 (Illumina®, San Diego, CA). In summary, we obtained 10∼28 Gb of sequencing for the 57 historical samples (Data S1).
Read alignment and SNP calling
We used the identical bioinformatical procedures for the raw reads of all 65 study individuals, including 57 museum individuals in this study and 8 contemporary samples obtained from the previous study (PRJNA308878) [10] (Data S1). All raw sequencing reads were put into PALEOMIX [53] for the adaptor removal, the crested ibis reference genome mapping (PRJNA308878) [10], duplicate filtering, initial BAM rescaled, etc. In order to improve the quality of the initial alignments, the PALEOMIX pipeline incorporates several useful steps: Picard tools for filtering, mapDamage2.0 for rescaling (only applied for the museum samples), and local re-alignment. After that, the mean sequencing depth estimated from the BAM files, ranged from 3.83-10.13x (museum samples) to 20.05-26.71x (contemporary samples), with an average depth of 7x over museum individuals and 21.64x over contemporary individuals (Data S1). Further to avoid any bias in SNP calling due to the different sequencing depths, we randomly extracted data to a coverage of only 7x from the original BAM files of the contemporary samples.
We identified SNPs using Analysis of Next Generation Sequencing Data (ANGSD) v0.615 [11] based on a BAM list containing 57 BAM files of the museum samples and 8 extracted BAM files of the contemporary samples with the following parameters:
doMajorMinor 1 -GL 2 -doMaf 2 -doGeno 7 -SNP_pval 1e-6 -doPost 1 -doCounts 1 -doglf 4 -dumpCounts 4 -HWE_pval 1 -minMaf 0.01
Considering the low depth of sequencing data caused by the features of museum samples, we further added two steps to reduce the false positives:
-
1.
ANGSD provides the genotypes for each individual. We removed the singleton SNPs.
-
2.
If the genotype of one individual is assigned as major/major, we marked its genotype as NN when the depth of its major allele is less than 2. If genotype of one individual is assigned as minor/minor, we will mark its genotype to NN when the depth of its minor allele is less than 2. If the genotype of one individual is assigned as major/minor, we will mark its genotype to NN when neither the depth of its major nor minor allele is less than 2.
Post-processing, a total of 5,268,206 SNPs were retained for further analysis.
Quantification and Statistical Analysis
Data pre-processing
Since background linkage disequilibrium (LD) generated by genetic drift could generate spurious clustering [14, 68], we first need to avoid such bias. Given that the high missing ratio of the loci will influence the accuracy of the LD value estimation, in the following analyses we only used the loci for which the missing ratio is less than 20%. We first ran a pilot test to select the appropriate distance cutoff for pairwise LD calculation using PLINK v1.07 [59]. In our dataset, when the distance of neighboring SNPs is beyond 500k, 97.2% LD values are less than 0.2 (very weak LD values), which suggests there are very few localized SNP clusters on this scale. Furthermore, following published guidance [68], we set the cutoff for the high LD value to 0.5. We used the following steps to filter the data. On each chromosome/scaffold, we started at the first SNP locus, measured the pairwise LD successively, until we reached the first SNP for which LD was less than 0.5. We then retained the first and last SNP loci, then started this process again from the last SNP until all SNP loci had been scanned. We finally obtained a dataset of 3,246,559 SNPs to be used in the population structure analyses.
Population structure
We used the processed SNP set to construct a phylogenetic tree for all 65 sequenced samples using RapidNJ v2.3.2 [54] based on the neighbor-joining method [12]. The core of this method is to calculate the distance matrix of Dij between each pair of individuals (i and j) as the following formula:
M is the number of segregating sites in i and j
L is the length of regions
dij is the distance between individuals i and j at site
dij = 0, when individuals i and j are both homozygous for the same allele (AA/AA)
dij = 0.5, when at least one of the genotypes of an individual i or j is heterozygous (Aa/AA, AA/Aa or Aa/Aa)
dij = 1, when individuals i and j are both homozygous but for different alleles (AA/aa or aa/AA)
We also identified the groups/subgroup structure of all 65 individuals by principal component analysis (PCA) of biallelic SNPs using the R package ‘pcadapt’ v3.0.4 [13] in R v3.2.2 [56]. We chose 4 as the optimal K from the PCA by scree plot, because the eigenvalue departs from the straight line. PCA result is consistent with that observed in the NJ tree.
The individual ancestries of all 65 samples were inferred by the Bayesian inference program STRUCTURE v2.3.4 [14] after removing the missing loci. Analyses were run five times for each level of K (from 2 through 10), with a 40,000 Burn-in period and 15,000 MCMC repetitions using the admixture model without prior information about populations. The ad hoc statistic ΔK, based on the rate of change in the log probability of data between successive K values, was used to find the optimal number of subpopulations [15] (Data S2).
Population demographic analysis
To gain insights into the demographic history of the historical and contemporary populations, we first performed the PSMC analysis [16]. We used the heterozygous SNP loci with MAF > = 0.2 and sequencing depth > 2X and < 30X on autosomal sequences. According to our previous experience [69], we set the parameters for the PSMC to be “N30 –t5 –r5 –p 4+30∗2+4+6+10,” and performed bootstraps (100 times) to represent the variance of results. To further reconstruct recent demographic history (from the sampling time points to 10 kya), we ran an approximate Bayesian computation approach named PopSizeABC v2.1 [17] using the same input of the PSMC analysis, with the following parameters: mac (minor allele count threshold for AFS and IBS statistics computation) is 0; mac_ld (minor allele count threshold for LD statistics computation) are 2,3,5,9 separately; L (size of each segment, in bp) is 4000000; nb_rep (number of simulated datasets) is 500; nb_seg (number of independent segments in each dataset) is 30; all others as default. Ne of the three groups (EA, NW and contemporary group) at the sampling time point was estimated by NeEstimator v2.1 [57], and according to the NJ tree, the Ne of three NE subgroups were calculated respectively using the same method (Table S1).
Ecological niche models analysis
There are 35 occurrences of crested ibis assigned to clear locality in the 19th and 20th centuries in the VertNet database, which can be used for the breeding ranges predication. We put the latitude and longitude to these 35 occurrences based on their identified localities into this analysis. Subsequently these geographic coordinates were used to construct ecological niche models (ENMs) for the contemporary period (19th-20th centuries), the Last Glacial Maximum period (LGM, approximately 22 kya) and the Last Interglacial period (LIG, approximately 120-140 kya). To build the ENMs, we also used the bioclimatic variables from WorldClim v1.4 [52] as the environmental layers in Maxent v3.4.0 [55]. The spatial resolution for all bioclimatic variables was unified to 2.5 arc-minutes (approximately 4 km × 4 km). For the LIG period, we aggregated the original 30 arc-second data to above unified resolution by function r.resamp.stats in GRASS GIS v7.2.1 with new cell size 0.041666666667 and average aggregation method. For all 19 bioclimatic variables, the predictions were performed in 10 replicates using Maxent, with extrapolation disabled and the other parameters set to default.
Temporal population polymorphism
In order to comprehensively characterize the population polymorphism, we used three methods. First, we estimated nucleotide diversity (π) for all 65 individuals in each 5Mb window across the chromosomes using the nuc.div function within the R package ‘pegas’ v0.10 [61]. Second, we measured the extent of the polymorphism fixation process in the contemporary group, when restricting the number of individuals with missing genotypes to be fewer than 5 in the historical groups, and no missing individuals in the contemporary group. Third, we identified the Identity by Descent (IBD) regions using GERMLINE v1.5.1 [60] on each chromosome in the historical and contemporary groups respectively.
Correlation between inbreeding coefficient and effective population size (Ne)
As we know that populations with smaller Ne tend to have proportionally more individuals with higher inbreeding coefficients than populations with larger Ne, we explored whether the historical groups with the relative larger Ne had also suffered from inbreeding, and whether the inbreeding coefficient of the contemporary group is significantly higher than that of other species who have a similar Ne. We therefore collected 14 published population SNP datasets [43, 44, 45, 46, 47, 48, 49, 50, 51], estimated their inbreeding coefficients and Ne, and used these data to build the correlation relationship. The inbreeding coefficient (FUNI) was estimated based on the correlation between uniting gametes following Wright [24] using Genome-wide Complex Trait Analysis (GCTA) v1.91.4 [58]. The Ne was estimated by NeEstimator v2.1 [57]. All results can be found in Table S2. Furthermore, the linear regression function (lm) in R v3.2.2 was applied to build and examine the correlation between FUNI and Ne, and was also used to determine whether the values of the contemporary group deviated significantly from the regression equation. The F-statistic of the coefficient indicated that FUNI has a significant correlation with Ne (p value = 0.0000697), and the adjusted R−squared (0.7891) also supported the high credibility of the regression equation. By using this regression equation, we found that when Ne was set to 10.7 (Ne of the contemporary group), the FUNI fit was 0.07 (95% CIs: 0.05-0.10). However, the FUNI of the contemporary group was 0.17, verifying its significant deviation from the regression equation.
Patterns of deleterious mutations across populations
We used two different measures for estimating the deleterious load: deleterious missense mutations, and loss of function (LOF) mutations. Since these two measures need the ancestral state for the crested ibis for comparison, we used the genotypes of major alleles in the historical groups to represent the ancestral state. In each measure, we compared the counts of 1) only homozygous sites (2 per each site) and 2) both homozygous and heterozygous sites (2 per homo- and 1 per heterozygous site).
The first measure we used to diagnose the deleteriousness of missense mutations is the Grantham Score [26], a measure of physical/chemical properties of amino acid changes. The score ranges from 5 to 215, with scores bigger than 150 designated as radical or deleterious [70]. We further categorized coding mutations into 1) deleterious, 2) benign, and 3) synonymous mutations as follows:
-
1)
deleterious: missense mutations with Grantham Scores equal to or more than 150.
-
2)
benign: missense mutations with Grantham Scores less than 150.
-
3)
synonymous: synonymous mutations, which do not cause amino acid sequence change.
The second measure is the LOF mutations inferred for the coding region SNPs of each sample using the SnpEff v4.3 algorithm [62]. We built a database from the crested ibis annotation and the reference crested ibis genome sequence, and used the input files in VCF format to annotate SNPs and assigned a SnpEff category to the input SNPs per individual. Briefly, two categories, stop codon (nonsense) and splice site disrupting single-nucleotide variants (SNVs), are considered as the LOF mutations. The ratio of homo- to homo- and heterozygous LOF sites in the contemporary population is significantly higher than that in the historic populations (0.336 ± 0.042 in contemporary group, 0.253 ± 0.038 in historical group, Welch Two Sample t test, p = 0.0005386, Figure S2F).
MHC diversity analysis
A previous study exploring the MHC genomic organization and structure showed that the crested ibis contains 54 genes within three regions spanning 500 kb [71]. We downloaded the following three region sequences from NCBI (Class I 150 kb KP182409.1, Class II 85 kb KP182408.1, and Extended Region 265 kb KP182407.1). We produced the MHC SNPs dataset for all 65 individuals by mapping the raw sequencing data against three regions using the same method as hereinbefore described. Since the genome is organized into blocks of haplotypes [72, 73], we compared the extent of the diversity changes in MHC region based on the haplotype block composition. The software GERBIL v1.1, a tool in the GEVALT software suite [63], was used to capitalize on the greater resolution of genomic markers, and to aggregate SNPs into discrete haplotype blocks. Haplotype blocks were detected based on the MHC SNPs dataset of 57 historical samples after filtering the loci with more than half missing individuals. The historical MHC SNPs dataset was first converted to the format described in the GERBIL software manual. Briefly, individual tab-delimited files were produced for each of the scaffolds, with columns representing SNPs and every two consecutive rows corresponding to one sample. SNPs were represented in 0, 1 format or a “”? if missing. Re-formatted files were analyzed in an iterative fashion using default software parameters (sample code: gerbil.exe < input filename > <output filename > ). We also identified the haplotype blocks based on the SNPs dataset of 8 contemporary samples after filtering the loci with more than half missing individuals using the same way. In this way, haplotype structures in two groups were independent, and we used the length and the allelic richness of haplotype blocks as the indexes to feature the haplotype structures and the genetic diversity, respectively. Since the different sample size should be taken into account when comparing the genetic diversity between the historical and contemporary groups, we picked up 8 samples from the historical groups 10 times randomly, counted the number of alleles for each time and used the average values to represent the genetic diversity of the historical groups (Figure 4, Figure S3).
Natural selection analysis
To measure the selection across the coding regions, we used the R package ‘PopGenome’ v2.6.0 [64] to calculate Tajima’s D value [74], since a positive value is indicative of balancing selection or population contraction. We divided all SNPs into the 3 historical groups (NW, NE and EA) and the contemporary group, and only chose the loci without missing data in each group as input data. To reduce the influence of demographic history when interpreting the Tajima’s D results, for each group we selected genes with the top 5% highest Tajima’s D values, and considered those genes that were present in at least two of the historic populations as candidate genes that were under balancing selection at least 100 years ago.
Data and Software Availability
Raw sequence read data have been deposited into the study accession number PRJNA481495 and are also available in the CNGB Nucleotide Sequence Archive under accession number CNSA: CNP0000084.
Acknowledgments
We are grateful to the National Museum of Natural History (USNM) (Gary Graves, Chris Milensky); Senckenberg Museum of Natural History (Gerald Mayr); Natural History Museum, Tring (Hein van Grouw); Department of Zoology, National Museums Liverpool (Tony Parker, Clemency Fisher); Center for Natural History/Zoological Museum Hamburg (Alexander Has); Naturalis Biodiversity Center Leiden, the Netherlands (Pepijn Kamminga); NRM Stockholm (Ulf Johansson); and Natural History Museum of Denmark (Jon Fjeldså) for providing access to museum specimens and Nigel Collar for providing background information critical to locating the samples. This work was supported by the Strategic Priority Research Program of Chinese Academy of Sciences (XDB31020000, XDB13000000), Carlsberg Foundation grant to G.Z. (CF16-0663), ERC Consolidator Grant 681396 ‘Extinction Genomics’ (M.T.P.G.), BFU2017-86471-P (MINECO/FEDER, UE) (T.M.-B.), U01 MH106874 grant (T.M.-B.), Howard Hughes International Early Career (T.M.-B.), Obra Social “La Caixa” and Secretaria d’Universitats i Recerca (T.M.-B.), CERCA Programme del Departament d’Economia i Coneixement de la Generalitat de Catalunya (T.M.-B.), and Deutsche Forschungsgemeinschaft (DFG) fellowship (KU 3467/1-1) (M.K.).
Author Contributions
C.L., M.T.P.G. and G.Z. conceived the study; M.T.P.G. collected museum samples, which were prepared into sequencing libraries by R.B.; C.L. managed sample sequencing; C.L., Y.D., L.Z., and H.P. developed, tested, and implemented the detailed SNPs calling pipeline; S.F., Q.F., C.L., and G.C. carried out genetic analyses described in the manuscript; S.H., M.K. and T.M.-B. assisted with the mutation load analysis; S.F., Q.F., C.L., T.M.-B., M.T.P.G. and G.Z. wrote and edited the manuscript; All authors read and approved the final manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: January 10, 2019
Footnotes
Supplemental Information includes three figures, two tables, and two data files and can be found with this article online at https://doi.org/10.1016/j.cub.2018.12.008.
Supplemental Information
References
- 1.Weeks A.R., Sgro C.M., Young A.G., Frankham R., Mitchell N.J., Miller K.A., Byrne M., Coates D.J., Eldridge M.D., Sunnucks P. Assessing the benefits and risks of translocations in changing environments: a genetic perspective. Evol. Appl. 2011;4:709–725. doi: 10.1111/j.1752-4571.2011.00192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong D.P., John L.C. Effects of familiarity on the outcome of translocations, II. A test using New Zealand Robins. Biol. Conserv. 1995;71:281–288. [Google Scholar]
- 3.Whiteley A.R., Fitzpatrick S.W., Funk W.C., Tallmon D.A. Genetic rescue to the rescue. Trends Ecol. Evol. 2015;30:42–49. doi: 10.1016/j.tree.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 4.Weeks A.R., Heinze D., Perrin L., Stoklosa J., Hoffmann A.A., van Rooyen A., Kelly T., Mansergh I. Genetic rescue increases fitness and aids rapid recovery of an endangered marsupial population. Nat. Commun. 2017;8:1071. doi: 10.1038/s41467-017-01182-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olden J.D., Leroy Poff N., Douglas M.R., Douglas M.E., Fausch K.D. Ecological and evolutionary consequences of biotic homogenization. Trends Ecol. Evol. 2004;19:18–24. doi: 10.1016/j.tree.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 6.Leimu R., Fischer M. Between-population outbreeding affects plant defence. PLoS ONE. 2010;5:e12614. doi: 10.1371/journal.pone.0012614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Draheim H.M., Baird P., Haig S.M. Temporal analysis of mtDNA variation reveals decreased genetic diversity in least terns. Condor. 2012;114:145–154. [Google Scholar]
- 8.Wandeler P., Hoeck P.E., Keller L.F. Back to the future: museum specimens in population genetics. Trends Ecol. Evol. 2007;22:634–642. doi: 10.1016/j.tree.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 9.Collar N.J., Andreev A., Chan S., Crosby M., Subramanya S., Tobias J. Fifth Edition. BirdLife International; Cambridge: 2001. Threatened birds of Asia: the BirdLife International red data book. [Google Scholar]
- 10.Li S., Li B., Cheng C., Xiong Z., Liu Q., Lai J., Carey H.V., Zhang Q., Zheng H., Wei S. Genomic signatures of near-extinction and rebirth of the crested ibis and other endangered bird species. Genome Biol. 2014;15:557. doi: 10.1186/s13059-014-0557-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korneliussen T.S., Albrechtsen A., Nielsen R. ANGSD: Analysis of Next Generation Sequencing Data. BMC Bioinformatics. 2014;15:356. doi: 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 13.Luu K., Bazin E., Blum M.G. pcadapt: an R package to perform genome scans for selection based on principal component analysis. Mol. Ecol. Resour. 2017;17:67–77. doi: 10.1111/1755-0998.12592. [DOI] [PubMed] [Google Scholar]
- 14.Falush D., Stephens M., Pritchard J.K. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evanno G., Regnaut S., Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 16.Li H., Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475:493–496. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boitard S., Rodríguez W., Jay F., Mona S., Austerlitz F. Inferring Population Size History from Large Samples of Genome-Wide Molecular Data - An Approximate Bayesian Computation Approach. PLoS Genet. 2016;12:e1005877. doi: 10.1371/journal.pgen.1005877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kindler P., Guillevic M., Baumgartner M., Schwander J., Landais A., Leuenberger M. Temperature reconstruction from 10 to 120 kyr b2k from the NGRIP ice core. Clim. Past. 2014;10:887–902. [Google Scholar]
- 19.Hung C.M., Shaner P.J., Zink R.M., Liu W.C., Chu T.C., Huang W.S., Li S.H. Drastic population fluctuations explain the rapid extinction of the passenger pigeon. Proc. Natl. Acad. Sci. USA. 2014;111:10636–10641. doi: 10.1073/pnas.1401526111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian Z., Jiang D. Revisiting last glacial maximum climate over China and East Asian monsoon using PMIP3 simulations. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2016;453:115–126. [Google Scholar]
- 21.Mochizuki, S., and Murakami, T. (2010). Temporal change of crested ibis habitat in Shaanxi Province, China. Proceedings of the 31st Asian Conference on Remote Sensing.
- 22.Li X.H., Tian H.D., Li D.M. Why the crested ibis declined in the middle twentieth century. Biol. Conserv. 2009;18:2165–2172. [Google Scholar]
- 23.Albrechtsen A., Moltke I., Nielsen R. Natural selection and the distribution of identity-by-descent in the human genome. Genetics. 2010;186:295–308. doi: 10.1534/genetics.110.113977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright S. Coefficients of inbreeding and relationship. Am. Nat. 1922;56:330–338. [Google Scholar]
- 25.Bouzat J.L. Conservation genetics of population bottlenecks: the role of chance, selection, and history. Conserv. Genet. 2010;11:463–478. [Google Scholar]
- 26.Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 27.Lohmueller K.E. The distribution of deleterious genetic variation in human populations. Curr. Opin. Genet. Dev. 2014;29:139–146. doi: 10.1016/j.gde.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 28.Lohmueller K.E., Indap A.R., Schmidt S., Boyko A.R., Hernandez R.D., Hubisz M.J., Sninsky J.J., White T.J., Sunyaev S.R., Nielsen R. Proportionally more deleterious genetic variation in European than in African populations. Nature. 2008;451:994–997. doi: 10.1038/nature06611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woolfit M. Effective population size and the rate and pattern of nucleotide substitutions. Biol. Lett. 2009;5:417–420. doi: 10.1098/rsbl.2009.0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reed D.H., Frankham R. Correlation between fitness and genetic diversity. Conserv. Biol. 2003;17:230–237. [Google Scholar]
- 31.Messaoudi I., Guevara Patiño J.A., Dyall R., LeMaoult J., Nikolich-Zugich J. Direct link between mhc polymorphism, T cell avidity, and diversity in immune defense. Science. 2002;298:1797–1800. doi: 10.1126/science.1076064. [DOI] [PubMed] [Google Scholar]
- 32.Grenfell B.T., Dobson A.P., Moffatt H.K.E. Cambridge University Press; 1995. Ecology of Infectious Diseases in Natural Populations. [Google Scholar]
- 33.Wang B., Li Z., Xu L., Goggi J., Yu Y., Zhou J. Molecular cloning and characterization of rat karyopherin alpha 1 gene: structure and expression. Gene. 2004;331:149–157. doi: 10.1016/j.gene.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 34.Sun Y., Wang T., Skidmore A.K., Palmer S.C.F., Ye X., Ding C., Wang Q. Predicting and understanding spatio-temporal dynamics of species recovery: implications for Asian crested ibis Nipponia nippon conservation in China. Divers. Distrib. 2016;22:893–904. [Google Scholar]
- 35.DeRose M.A., Roff D.A. A Comparison of Inbreeding Depression in Life-History and Morphological Traits in Animals. Evolution. 1999;53:1288–1292. doi: 10.1111/j.1558-5646.1999.tb04541.x. [DOI] [PubMed] [Google Scholar]
- 36.Keller L.F., Waller D.M. Inbreeding effects in wild populations. Trends Ecol. Evol. 2002;17:230–241. [Google Scholar]
- 37.Charlesworth D., Charlesworth B. Inbreeding depression and its evolutionary consequences. Annu. Rev. Ecol. Syst. 1987;18:237–268. [Google Scholar]
- 38.O’Grady J.J., Brook B.W., Reed D.H., Ballou J.D., Tonkyn D.W., Frankham R. Realistic levels of inbreeding depression strongly affect extinction risk in wild populations. Biol. Conserv. 2006;133:42–51. [Google Scholar]
- 39.Frankham R. Genetics and extinction. Biol. Conserv. 2005;126:131–140. [Google Scholar]
- 40.Robinson J.A., Ortega-Del Vecchyo D., Fan Z., Kim B.Y., vonHoldt B.M., Marsden C.D., Lohmueller K.E., Wayne R.K. Genomic flatlining in the endangered island fox. Curr. Biol. 2016;26:1183–1189. doi: 10.1016/j.cub.2016.02.062. [DOI] [PubMed] [Google Scholar]
- 41.Yu X.P., Chang X.Y., Li X., Chen W.G., Shi L. Return of the Crested Ibis Nipponia nippon: a reintroduction programme in Shaanxi province, China. BirdingASIA. 2009;11:80–82. [Google Scholar]
- 42.Oldoni F., Castella V., Hall D. A novel set of DIP-STR markers for improved analysis of challenging DNA mixtures. Forensic Sci. Int. Genet. 2015;19:156–164. doi: 10.1016/j.fsigen.2015.07.012. [DOI] [PubMed] [Google Scholar]
- 43.Prado-Martinez J., Sudmant P.H., Kidd J.M., Li H., Kelley J.L., Lorente-Galdos B., Veeramah K.R., Woerner A.E., O’Connor T.D., Santpere G. Great ape genetic diversity and population history. Nature. 2013;499:471–475. doi: 10.1038/nature12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kukekova A.V., Johnson J.L., Xiang X., Feng S., Liu S., Rando H.M., Kharlamova A.V., Herbeck Y., Serdyukova N.A., Xiong Z. Red fox genome assembly identifies genomic regions associated with tame and aggressive behaviours. Nat Ecol Evol. 2018;2:1479–1491. doi: 10.1038/s41559-018-0611-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu S., Lorenzen E.D., Fumagalli M., Li B., Harris K., Xiong Z., Zhou L., Korneliussen T.S., Somel M., Babbitt C. Population genomics reveal recent speciation and rapid evolutionary adaptation in polar bears. Cell. 2014;157:785–794. doi: 10.1016/j.cell.2014.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Z., Jia Y., Almeida P., Mank J.E., van Tuinen M., Wang Q., Jiang Z., Chen Y., Zhan K., Hou S. Whole-genome resequencing reveals signatures of selection and timing of duck domestication. Gigascience. 2018;7:giy027. doi: 10.1093/gigascience/giy027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li D., Che T., Chen B., Tian S., Zhou X., Zhang G., Li M., Gaur U., Li Y., Luo M. Genomic data for 78 chickens from 14 populations. Gigascience. 2017;6:1–5. doi: 10.1093/gigascience/gix026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lamichhaney S., Berglund J., Almén M.S., Maqbool K., Grabherr M., Martinez-Barrio A., Promerová M., Rubin C.J., Wang C., Zamani N. Evolution of Darwin’s finches and their beaks revealed by genome sequencing. Nature. 2015;518:371–375. doi: 10.1038/nature14181. [DOI] [PubMed] [Google Scholar]
- 49.Nadachowska-Brzyska K., Burri R., Smeds L., Ellegren H. PSMC analysis of effective population sizes in molecular ecology and its application to black-and-white Ficedula flycatchers. Mol. Ecol. 2016;25:1058–1072. doi: 10.1111/mec.13540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao S., Zheng P., Dong S., Zhan X., Wu Q., Guo X., Hu Y., He W., Zhang S., Fan W. Whole-genome sequencing of giant pandas provides insights into demographic history and local adaptation. Nat. Genet. 2013;45:67–71. doi: 10.1038/ng.2494. [DOI] [PubMed] [Google Scholar]
- 51.Murray G.G.R., Soares A.E.R., Novak B.J., Schaefer N.K., Cahill J.A., Baker A.J., Demboski J.R., Doll A., Da Fonseca R.R., Fulton T.L. Natural selection shaped the rise and fall of passenger pigeon genomic diversity. Science. 2017;358:951–954. doi: 10.1126/science.aao0960. [DOI] [PubMed] [Google Scholar]
- 52.Hijmans R.J., Cameron S.E., Parra J.L., Jones P.G., Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 2005;25:1965–1978. [Google Scholar]
- 53.Schubert M., Ermini L., Der Sarkissian C., Jónsson H., Ginolhac A., Schaefer R., Martin M.D., Fernández R., Kircher M., McCue M. Characterization of ancient and modern genomes by SNP detection and phylogenomic and metagenomic analysis using PALEOMIX. Nat. Protoc. 2014;9:1056–1082. doi: 10.1038/nprot.2014.063. [DOI] [PubMed] [Google Scholar]
- 54.Simonsen M., Mailund T., Pedersen C.N. Springer; 2008. Rapid neighbour-joining. [Google Scholar]
- 55.Phillips S.J., Anderson R.P., Schapire R.E. Maximum entropy modeling of species geographic distributions. Ecol. Modell. 2006;190:231–259. [Google Scholar]
- 56.R Core Development Team . R Foundation for Statistical Computing; 2016. R: a language and environment for statistical computing. [Google Scholar]
- 57.Do C., Waples R.S., Peel D., Macbeth G.M., Tillett B.J., Ovenden J.R. NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne ) from genetic data. Mol. Ecol. Resour. 2014;14:209–214. doi: 10.1111/1755-0998.12157. [DOI] [PubMed] [Google Scholar]
- 58.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gusev A., Lowe J.K., Stoffel M., Daly M.J., Altshuler D., Breslow J.L., Friedman J.M., Pe’er I. Whole population, genome-wide mapping of hidden relatedness. Genome Res. 2009;19:318–326. doi: 10.1101/gr.081398.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Paradis E. pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics. 2010;26:419–420. doi: 10.1093/bioinformatics/btp696. [DOI] [PubMed] [Google Scholar]
- 62.Cingolani P., Platts A., Wang L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kimmel G., Shamir R. GERBIL: Genotype resolution and block identification using likelihood. Proc. Natl. Acad. Sci. USA. 2005;102:158–162. doi: 10.1073/pnas.0404730102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pfeifer B., Wittelsbürger U., Ramos-Onsins S.E., Lercher M.J. PopGenome: an efficient Swiss army knife for population genomic analyses in R. Mol. Biol. Evol. 2014;31:1929–1936. doi: 10.1093/molbev/msu136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rohland N., Hofreiter M. Ancient DNA extraction from bones and teeth. Nat. Protoc. 2007;2:1756–1762. doi: 10.1038/nprot.2007.247. [DOI] [PubMed] [Google Scholar]
- 66.Vilstrup J.T., Seguin-Orlando A., Stiller M., Ginolhac A., Raghavan M., Nielsen S.C., Weinstock J., Froese D., Vasiliev S.K., Ovodov N.D. Mitochondrial phylogenomics of modern and ancient equids. PLoS ONE. 2013;8:e55950. doi: 10.1371/journal.pone.0055950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paijmans J.L.A., Barnett R., Gilbert M.T.P., Zepeda-Mendoza M.L., Reumer J.W.F., de Vos J., Zazula G., Nagel D., Baryshnikov G.F., Leonard J.A. Evolutionary History of Saber-Toothed Cats Based on Ancient Mitogenomics. Curr. Biol. 2017;27:3330–3336.e5. doi: 10.1016/j.cub.2017.09.033. [DOI] [PubMed] [Google Scholar]
- 68.Kaeuffer R., Réale D., Coltman D.W., Pontier D. Detecting population structure using STRUCTURE software: effect of background linkage disequilibrium. Heredity (Edinb) 2007;99:374–380. doi: 10.1038/sj.hdy.6801010. [DOI] [PubMed] [Google Scholar]
- 69.Nadachowska-Brzyska K., Li C., Smeds L., Zhang G., Ellegren H. Temporal Dynamics of Avian Populations during Pleistocene Revealed by Whole-Genome Sequences. Curr. Biol. 2015;25:1375–1380. doi: 10.1016/j.cub.2015.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li W.H., Wu C.I., Luo C.C. Nonrandomness of point mutation as reflected in nucleotide substitutions in pseudogenes and its evolutionary implications. J. Mol. Evol. 1984;21:58–71. doi: 10.1007/BF02100628. [DOI] [PubMed] [Google Scholar]
- 71.Chen L.C., Lan H., Sun L., Deng Y.L., Tang K.Y., Wan Q.H. Genomic organization of the crested ibis MHC provides new insight into ancestral avian MHC structure. Sci. Rep. 2015;5:7963. doi: 10.1038/srep07963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Taillon-Miller P., Bauer-Sardiña I., Saccone N.L., Putzel J., Laitinen T., Cao A., Kere J., Pilia G., Rice J.P., Kwok P.Y. Juxtaposed regions of extensive and minimal linkage disequilibrium in human Xq25 and Xq28. Nat. Genet. 2000;25:324–328. doi: 10.1038/77100. [DOI] [PubMed] [Google Scholar]
- 73.Daly M.J., Rioux J.D., Schaffner S.F., Hudson T.J., Lander E.S. High-resolution haplotype structure in the human genome. Nat. Genet. 2001;29:229–232. doi: 10.1038/ng1001-229. [DOI] [PubMed] [Google Scholar]
- 74.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence read data have been deposited into the study accession number PRJNA481495 and are also available in the CNGB Nucleotide Sequence Archive under accession number CNSA: CNP0000084.