Abstract
Raf1/c-Raf is a well-characterized serine/threonine–protein kinase that links Ras family members with the MAPK/ERK signaling cascade. We have identified a novel splice isoform of human Raf1 that causes protein truncation and loss of the C-terminal kinase domain (Raf1-tr). We found that Raf1-tr has increased nuclear localization compared with full-length Raf1, and this finding was secondary to reduced binding of Raf1-tr to the cytoplasmic chaperone FK506 binding protein 5. We show that Raf1-tr has increased binding to DNA-dependent protein kinase (DNA-PK), which inhibits DNA-PK function and causes amplification of irradiation- and bleomycin-induced DNA damage. We found that the human colorectal cancer cell line, HCT-116, displayed reduced expression of Raf1-tr, and reintroduction of Raf1-tr sensitized the cells to bleomycin-induced apoptosis. Furthermore, we identified differential Raf1-tr expression in breast cancer cell lines and showed that breast cancer cells with increased Raf1-tr expression become sensitized to bleomycin-induced apoptosis. Collectively, these results demonstrate a novel Raf1 isoform in humans that has a unique noncanonical role in regulating the double-stranded DNA damage response pathway through modulation of DNA-PK function.—Nixon, B. R., Sebag, S. C., Glennon, M. S., Hall, E. J., Kounlavong, E. S., Freeman, M. L., Becker, J. R. Nuclear localized Raf1 isoform alters DNA-dependent protein kinase activity and the DNA damage response.
Keywords: nuclear translocation, cancer biology, signal transduction
The Ras-Raf-ERK pathway is a highly conserved cellular signaling cascade that is important in regulating differentiation, growth, and survival in a variety of different cell types. Under physiologic conditions, growth factor–induced activation of Ras recruits and activates Raf kinase family members (A-Raf, B-Raf, or Raf1/C-Raf), which in turn induce activation of MEK and subsequently ERK (1). In addition to its role in normal cellular physiology, dysregulated activation of the Ras-Raf-ERK pathway leads to the development and/or progression of different malignant cell types (2). Due to its integral role in Ras-Raf-ERK signaling, stringent regulation of Raf1 kinase activity is essential to maintaining normal cellular homeostasis.
In addition to the well-defined canonical role for kinases in modulating phosphorylation of protein targets, there are also multiple examples of noncanonical roles for many different kinases. For example, Janus kinases, while normally phosphorylating transcription factors to induce activation, have been shown to alter gene transcription through histone phosphorylation (3). Furthermore, some kinases have also been found to participate in DNA binding and protein scaffolding, which highlights the diversity of noncanonical kinase functions (4). Similar to other kinases, Raf1 reportedly exhibits noncanonical functions. For example, in a MAPK-independent manner, Raf1 has been shown to play a role in tumor progression through association with mitotic spindles in proliferating oncogenic cells (5).
In the present study, we identified a novel splice isoform of human Raf1 that causes truncation of the protein and loss of the kinase domain, truncated Raf1 (Raf1-tr). We discovered that Raf1-tr preferentially localizes to the nucleus secondary to alterations in cytoplasmic chaperone binding and preferentially interacts with unique binding partners compared with full-length Raf1 (Raf1-fl). We then show that Raf1-tr displays increased binding to DNA-dependent protein kinase (DNA-PK) and alters the function of this protein in regulating the DNA damage response in normal and malignant cells. Collectively, our results define a unique noncanonical role for a novel human Raf1 isoform in regulating the DNA damage response through modulation of DNA-PK function.
MATERIALS AND METHODS
Cell lines
Human embryonic kidney (HEK) 293T and mammary adenocarcinoma cells (lines MCF7 and MDA-MB-231) were cultured in DMEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37°C in a humidified 5% CO2 atm. Human colorectal carcinoma cells (HCT-116) were cultured in McCoy’s 5A (modified) Medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37°C in a humidified 5% CO2 atm. HAP1 parental cells that express Raf1 (C631; Horizon Discovery, Waterbeach, United Kingdom) or HAP1 cells containing a 22 bp deletion in exon 2 (HZGHC002809c001; Horizon Discovery) were cultured in Iscove’s Modified Dulbecco’s Medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37°C in a humidified 5% CO2 atm.
Plasmids
Reverse transcription of human peripheral blood mononuclear cell RNA was performed by using oligo DT primers to select poly-adenylated RNA species. Using this cDNA library, PCR was then performed using the following primer pair: hRAF1_cDNA_F_FLAG: 5′-GCCACCATGGATTACAAGGATGACGATGACAAGGAGCACATACAGGGAGCTTG-3′; hRAF1_cDNA_R: 5′-GGTGCAAAGTCAACTAGAAGACAGG-3′. These PCR products were TA cloned into pCR8/GW/TOPO and then sequenced. The Raf1 cDNA clone predicted to produce a full-length 648 aa Raf1 protein was named Raf1-fl, and the Raf1 cDNA predicted to produce a truncated 382 aa Raf1 protein was named Raf1-tr. LR clonase–mediated recombination was used to insert the N-terminal FLAG tagged Raf1-fl or Raf1-tr cDNA isoforms into a gateway-modified version of pEF1α internal ribosome entry site (IRES)-AcGFP1 vector to create pEF1α-FLAGhRaf-1(FL)-IRES-AcGFP1 (Raf1-fl) or pEF1α-FLAGhRafIn9-IRES-AcGFP1 (Raf1-tr) constructs. In addition, the pEF1α-IRES-AcGFP1 (empty) construct alone was used to verify no adverse effects as a result of plasmid transfection.
Transient transfection
HEK or HCT-116 cells were transfected by using the LipoD293 (SL100668; SignaGen, Rockville, MD, USA) transfection reagent according to the manufacturer’s instructions. Briefly, culture medium was replaced 1 h before transfection with fresh complete medium. Transfection complexes were formed by using a LipoD293:DNA ratio of 3:1. First, 1 µg of Raf1-fl, Raf1-tr, or Empty plasmid was added to high-glucose serum-free medium with subsequent mixing followed by the addition of LipoD293. The DNA-lipid complex was allowed to form for 10 min at room temperature and was then added dropwise to wells. After overnight incubation, medium containing liposomal complexes was removed and replaced with complete medium to allow the cells to recover from transfection. Transfection efficiency was verified by using immunofluorescence microscopy to identify green fluorescent protein (GFP)-positive cells indicating successful transfection.
For FK506 binding protein 5 (FKBP5) siRNA exposure, HEK cells were exposed to 50 nM FKBP5 siRNA (siRNA ID s5214; Thermo Fisher Scientific) for 72 h at 37°C using the Lipofectamine 3000 Reagent (Thermo Fisher Scientific). For MYC siRNA exposure, MDA-MB-231 cells were exposed to 25 nM MYC siRNA (Thermo Fisher Scientific; siRNA ID s9130) for 96 h at 37°C using the Lipofectamine 3000 Reagent (Thermo Fisher Scientific). Nontargeting siRNA (D-001810-10-05; Dharmacon, Lafayette, CO, USA) was used as a control for all siRNA experiments.
Cell treatments
To measure phosphorylation of ERK and for coimmunoprecipitation, transfected HEK cells were maintained in serum-free medium for 24 h followed by exposure to 5 ng/ml human epidermal growth factor (hEGF) (R&D Systems, Minneapolis, MN, USA) for 5 min at 37°C. In addition, transfected HEK cells were maintained in complete serum with subsequent ERK phosphorylation analysis. To measure DNA-PK phosphorylation and γ-H2AX, HEK cells were incubated for 2 h at 37°C with 0.05 mg/ml bleomycin (13877; Cayman Chemicals, Ann Arbor, MI, USA) in complete medium. To measure RPA32 phosphorylation, HEK cells were incubated for 1 h at 37°C with 1 µM camptothecin (11694; Cayman Chemicals) in complete medium. To measure DNA fragmentation, HEK cells were incubated for 30 min at 37°C with 0.05 mg/ml bleomycin in complete medium. To measure apoptosis, HCT-116 or MCF7 and MB-231 cells were incubated for 72 h at 37°C with 0.150 or 0.05 mg/ml bleomycin in complete medium, respectively. MB-231 were also exposed to 0.05 mg/ml bleomycin with concurrent exposure to 0.5 µM DNA-PK inhibitor NU-7441 (S2638; Selleck Chemicals, Houston, TX, USA) or 25 nM MYC siRNA as previously described. In addition, HEK cells were exposed to 1 Gy of γ-rays ([137Cs] at 2 Gy/min) and allowed to recover for 2 h. To measure nuclear translocation, HEK cells were incubated at 37°C with 1 µM geldanamycin for 8 h.
qRT-PCR
Human RNA from various tissues and cell lines underwent reverse transcription (QuantiTect Reverse Transcription Kit; Qiagen, Hilden, Germany) to generate cDNA. For quantitative RT-PCR, Sybr Green reagent (Thermo Fisher Scientific) was used to perform the PCR portion of the experiment. The 2-ΔΔCt method was used to normalize the gene of interest to the endogenous housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and fold change was relative to indicated group in the figure legends. Genes with Ct values >35 were considered undetectable. Supplemental Table 1 presents the oligonucleotide sequences.
Coimmunoprecipitation
HEK cells were seeded into 60-mm dishes at 106 cells per well and transfected as described earlier. After transfection, cells were lysed with Cell Lytic MT buffer (MilliporeSigma, Burlington, MA, USA), and cell lysate was then incubated with FLAG M2 (MilliporeSigma) antibody while rocking for 1 h at 4°C. Immune complexes were captured by incubation with protein A/G plus agarose beads (Santa Cruz Biotechnology, Dallas, TX, USA) by rocking overnight at 4°C. The following day, beads were washed 4 times with lysis buffer, and proteins were eluted with 150 ng/µl 3X FLAG peptide (MilliporeSigma) per manufacturer’s instructions or eluted directly with 2X Laemmli sample buffer (Bio-Rad, Hercules, CA, USA) followed by heating at 80°C for 5 min.
Mass spectrometry
For mass spectrophotometric analysis, HEK lysates were incubated with anti-FLAG M2 magnetic beads (MilliporeSigma) for 90 min at 4°C. Beads were then washed 4 times, and proteins were eluted by using 3X FLAG peptide (MilliporeSigma). Eluate then underwent tryptic digestion with subsequent multidimensional protein identification technology (MudPIT) mass spectrometry conducted in the Vanderbilt Mass Spectrometry Research Center Proteomics Core, essentially as previously described (6, 7). All MS/MS samples were analyzed using Sequest (v.27, rev. 12; Thermo Fisher Scientific). Sequest searched the uniprot-human-reference-canonical_20121108_rev database with a fragment ion mass tolerance of 0.00 Da and a parent ion tolerance of 2.5 Da. Oxidation of methionine and carbamidomethyl of cysteine were specified in Sequest as variable modifications. Scaffold (v.Scaffold_4.4.6; Proteome Software, Portland, OR, USA) was used to validate MS/MS–based peptide and protein identifications. Peptide identifications were accepted if they could be established at >98.0% probability to achieve a false discovery rate <0.1% according to the Peptide Prophet algorithm (8). Protein identifications were accepted if they could be established at >99.0% probability to achieve a false discovery rate <1.0% and contained at least 3 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm (9). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Proteins were annotated with GO terms from the National Center for Biotechnology Information (10). To determine the types of differential Raf1-fl and Raf1-tr binding partners, gene symbols for the identified proteins were input to the Protein Analysis Through Evolutionary Relationships (PANTHER) classification system (11) to generate protein classifications for proteins with increased binding to Raf1-tr and decreased binding to Raf1-tr.
Immunoblot analysis
Cells were lysed by using RIPA buffer (MilliporeSigma) with protease and phosphatase inhibitors (Halt inhibitor cocktail; Thermo Fisher Scientific) followed by solubilization in 2X Laemmli sample buffer. Normal and malignant colon tissue samples were provided by the National Cancer Institute’s Cooperative Human Tissue Network and were homogenized in Cell Lytic MT lysis buffer with protease and phosphatase inhibitors followed by solubilization in 2× Laemmli sample buffer. All samples were heated at 80°C for 5 min with subsequent centrifugation for 5 min for clarity. Protein concentration was determined by using the DC Protein Assay Kit (Bio-Rad) and separated by SDS-PAGE using a 10 or 4–20% Tris-glycine gel. Separate proteins were then transferred to a 0.45 µm low-fluorescence PVDF membrane (MilliporeSigma) or 0.2 µm nitrocellulose membrane (Bio-Rad) at 100 V for 90 min at 4°C. Membranes were subsequently blocked with 5% bovine serum albumin (BSA) or 5% nonfat dry milk. Identification of phospho ERK1/2 (4370; Cell Signaling Technology, Danvers, MA, USA), total ERK1/2 (9107; Cell Signaling Technology), B-Raf (sc-9002; Cell Signaling Technology), Ras (MABS195; MilliporeSigma), N-terminal Raf-1 #1 (MAB6771; Abnova, Taipei City, Taiwan), N-terminal Raf-1 #2 (610151; BD Biosciences, San Jose, CA, USA), C-terminal Raf-1 (04-739; MilliporeSigma), FLAG M2 (F1804; Sigma), FKBP5 (AF4094; R&D Systems), phospho DNA-PK Ser2056 (ab18192; Abcam, Cambridge, United Kingdom), phospho DNA-PK Thr2609 (GTX24194; GeneTex, Irvine, CA, USA), DNAPK (A300-518A; Bethyl Laboratories, Montgomery, TX, USA), γH2AX (A300-081A-M; Bethyl Laboratories), H2AX (A303-837A; Bethyl Laboratories), RPA32/RPA2 (2208; Cell Signaling Technology), heat shock protein 90 (Hsp90) α (ab133491; Abcam), MYC (sc-40; SCBT), or β-actin (8457; Cell Signaling Technology) was performed with overnight membrane incubation at 4°C. Membranes were washed with 0.1% Tween-20 (Thermo Fisher Scientific) and exposed for 1 h to goat anti-rabbit IRDye 800CW (925-32211; Li-Cor, Lincoln, NE, USA)/goat-anti-mouse IRDye 680LT (925-68020; Li-Cor) for fluorescent imaging, or goat anti-mouse horseradish peroxidase (HRP) (sc-516102; SCBT), goat anti-rabbit HRP (7074; Cell Signaling Technology), or donkey anti-goat HRP (sc-2020; SCBT) for chemiluminescent detection. Membranes were washed again and fluorescently detected by using an Odyssey infrared imaging system (LI-COR) or exposed to film after membrane incubation with Western Lightning Plus-ECL (PerkinElmer, Waltham, MA, USA). Electronic images obtained from Odyssey imaging or 600 dpi scanned film images were analyzed by using Image Studio Lite (v.5.2; Li-Cor) to quantitate band intensity.
Immunofluorescence
Cells were fixed with 4% paraformaldehyde and washed with PBS. After washes, cells were then permeabilized with 0.2% Triton X-100 with subsequent PBS washes. Fixed cells were then incubated with 1% BSA in PBS for 1 h at room temperature. Incubation with N-terminal Raf-1 #2 (610151; BD Biosciences) or FLAG M2 (F1804; MilliporeSigma) was performed at 4°C overnight. Sections were washed with PBS and then incubated with fluorescent secondary goat anti-rabbit Alexa Fluor 594 (A-11005; Thermo Fisher Scientific) or goat anti-mouse Alexa Fluor 488 (A-10667; Thermo Fisher Scientific) antibodies for 1 h in the dark at room temperature. After PBS washes, sections were counterstained and mounted with ProLong Gold Antifade mountant with DAPI (Thermo Fisher Scientific) and allowed to cure overnight in the dark at room temperature. Slides were imaged on an LSM 710 META inverted confocal microscope (Carl Zeiss, Oberkochen, Germany) performed in part through the use of the Vanderbilt Cell Imaging Shared Resource. Nuclear fluorescence was determined by using the following equation: corrected nuclear fluorescence = integrated density – (area of nuclei × mean fluorescence of background readings), in which integrated density is fluorescence intensity of the defined region of interest, area of nuclei is the size of the defined region of interest, and mean fluorescence of background readings is the average intensity of 10 background regions of interest.
Proximity ligation assay
HEK cells were transfected as described earlier and exposed to 5 ng/ml hEGF for 5 min at 37°C. Cells were subsequently washed with ice-cold PBS and fixed with 4% paraformaldehyde. Fixed cells were then blocked with 1% BSA and incubated with FLAG M2 (MilliporeSigma), N-terminal Raf-1 #2 (BD Biosciences), or DNA-PK (Bethyl Laboratories) antibodies overnight at 4°C. Duolink Proximity Ligation Assay (MilliporeSigma) was performed per manufacturer’s instructions. Briefly, secondary antibodies labeled with proximity ligation assay (PLA) probes were diluted 1:5 in 1% BSA buffer, and cells were incubated with the probes in the dark for 1 h at 37°C. A PLA was performed by addition of ligase solution to cells in the dark for 30 min at 37°C. Amplification was conducted after addition of polymerase to cells in the dark for 100 min at 37°C. After PBS washes, sections were counterstained and mounted with ProLong Gold Antifade mountant with DAPI and allowed to cure overnight in the dark at room temperature. Slides were imaged at ×20 magnification on an inverted wide-field fluorescent microscope (Olympus, Tokyo, Japan), and interactions per total cells was manually counted.
Comet assay
HEK cells were transfected and exposed to bleomycin as described earlier. A neutral comet assay (Trevigen, Gaithersburg, MD, USA) was then performed according to the manufacturer’s instructions. Briefly, cells were scraped in ice-cold PBS, centrifuged at 1000 rpm for 5 min, and resuspended in ice-cold PBS. Cell number was obtained and subsequently diluted to 1 × 105 cells/ml in ice-cold PBS. Cells were then diluted 1:10 in warm, low-melting agarose and pipetted onto slides, which were then incubated flat at 4°C for 30 min. Slides were then placed in ice-cold lysis buffer and incubated at 4°C overnight. The following day, slides were placed in an electrophoresis unit with neutral electrophoresis buffer (50 mM Tris, 150 mM sodium acetate, pH 9) and ran at 20 V for 45 min at 4°C. After electrophoresis, slides were incubated with DNA precipitation buffer (1 mM ammonium acetate in 95% ethanol) for 30 min at room temperature and then 70% ethanol for 30 min. Slides were then dried at 45°C for 10–15 min. Nuclei embedded in agarose were incubated with 2 µg/ml ethidium bromide in the dark for 30 min at room temperature. Slides were then rinsed in water and allowed to dry in the dark at room temperature. Slides were imaged at ×10 magnification on an inverted wide-field fluorescent microscope (Olympus), and comets were analyzed by using Comet Score software (http://www.autocomet.com). The Olive tail moment, defined according to the percentage of DNA in the tail multiplied by the length between the center of the comet head and tail, was used as the metric of fragmentation (12). A minimum of 50 comets were analyzed per group.
TUNEL assay
Cells were exposed to small molecules as previously described. Identification of apoptotic cells was then conducted by using TUNEL Andy Fluor 594 Apoptosis Detection Kit (GeneCopoeia, Rockville, MD, USA) according to the manufacturer’s instructions. Briefly, cells were washed with ice-cold PBS and fixed with 4% paraformaldehyde. Cells were then permeabilized with 0.2% Triton X-100 (Thermo Fisher Scientific) and exposed to terminal deoxynucleotidyl transferase reagent for 60 min at 37°C. Cells were then washed with a 3% BSA solution and incubated with streptavidin-Andy Fluor 594 for 30 min at room temperature. After additional washing with 3% BSA, cells were incubated with goat anti-rabbit GFP Tag Alexa Fluor 488 (Thermo Fisher Scientific) in the dark for 1 h at room temperature to identify transfected cells if necessary and subsequently washed with 3% BSA solution. Slides were then sealed with DAPI-containing mountant and allowed to cure overnight at room temperature in the dark. The following day, cells were imaged with a wide-field inverted fluorescent microscope (Olympus) and quantified in ImageJ software (National Institutes of Health, Bethesda, MD, USA) to identify GFP-positive, TUNEL-positive cells.
Quantification and statistical analysis
All data are displayed as means ± sem unless otherwise noted. Data were analyzed by using Student’s t test for comparisons between 2 groups or 1-way ANOVA for comparisons across multiple groups by using GraphPad Prism (GraphPad Software, La Jolla, CA, USA). Significance was defined as P < 0.05. No statistical methods or criteria were used to estimate sample size or to include/exclude samples. All experiments were performed at least 3 times, with representative experiments depicted.
RESULTS
Alternative splicing results in a novel truncated human Raf1 protein isoform
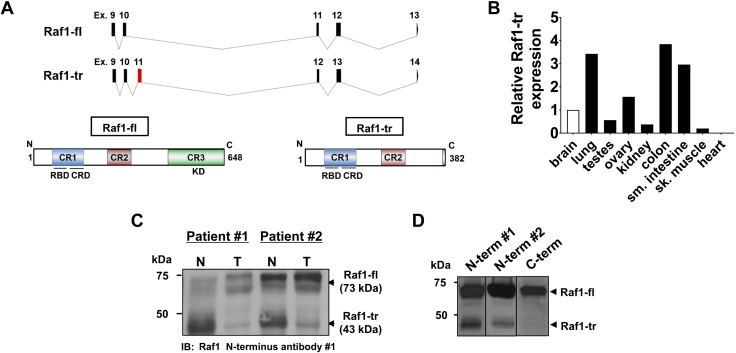
We identified a Raf1 splice isoform from cDNA generated from human peripheral blood mononuclear cell RNA. The incorporation of the new exon 11 in the Raf1 splice isoform mRNA causes a frame shift and the introduction of 3 premature stop codons to appear in frame (Fig. 1A, top and Supplemental Fig. S1). Compared with Raf1-fl, which is a 648-aa protein (Fig. 1A, bottom left), the premature stop codons in the novel splice isoform are predicted to cause truncation of Raf1 and produce a 382-aa protein that lacks the C-terminal kinase domain (Raf1-tr) (Fig. 1A, bottom right). Expression of the Raf1-tr mRNA was evaluated across multiple different human tissues, and expression was highest in the colon, small intestine, and lung; the lowest expression was observed in striated muscle tissue (Fig. 1B). Because the highest expression of Raf1-tr transcript was observed in normal colon tissue, we then compared Raf1-tr protein expression in matched human normal colon and colorectal adenocarcinoma tumors. We discovered that Raf1-tr protein expression was reduced in colon tumors compared with normal colon tissue from the same subject (Fig. 1C).
Figure 1.
Alternative splicing results in a Raf1-tr protein isoform: A) Exon/intron map of Raf1-fl and the identified splice isoform (Raf1-tr) (top). New exon in Raf1-tr is highlighted in red. Protein product of Raf1-fl (bottom left) showing conserved regions containing a Ras-binding domain and cysteine-rich domain (blue), serine-threonine–rich domain (red), and kinase domain (green). Raf1-tr protein product (bottom right) is truncated and has all conserved regions except for the kinase domain. B) Measurement of Raf1-tr gene expression in a panel of human tissues. The gene of interest was normalized to GAPDH expression and represented as fold change relative to brain tissue. C) Western blot of Raf1 in matched normal (N) and adenocarcinoma colon (T) lysate showing that Raf1-tr protein is differentially expressed in normal and colon tumor tissue. D) Probing for Raf1 with 2 unique N-terminal targeted antibodies demonstrates a 73 kDa Raf1-fl species and a 43 kDa Raf1-tr species in HEK cell lysate. Probing for Raf1 with a C-terminal targeted antibody detects only the 73 kDa Raf1-fl species.
Similar to colon tissue, we also detected expression of Raf1-fl and Raf1-tr in HEK293T cells. To confirm that Raf1-tr was not a degradation product, antibodies targeted to the N-terminal and C-terminal regions of Raf1 were used. Two different N-terminal antibodies detected both Raf1-fl protein and Raf1-tr protein. In contrast, an antibody that targets the C-terminal kinase domain of Raf1 could only detect Raf1-fl protein and not Raf1-tr (Fig. 1D). Importantly, we validated the specificity of all 3 Raf1 antibodies by comparing a cell line that expresses Raf1 vs. a Raf1-null mutant of the same cell line (Supplemental Fig. S2A). Taken together, these results confirm that the Raf1-tr transcript produces a truncated 43 kDa Raf1 protein isoform and demonstrate a correlation between Raf1-tr transcript and protein levels.
Effect of Raf1-tr on canonical Ras-ERK signaling
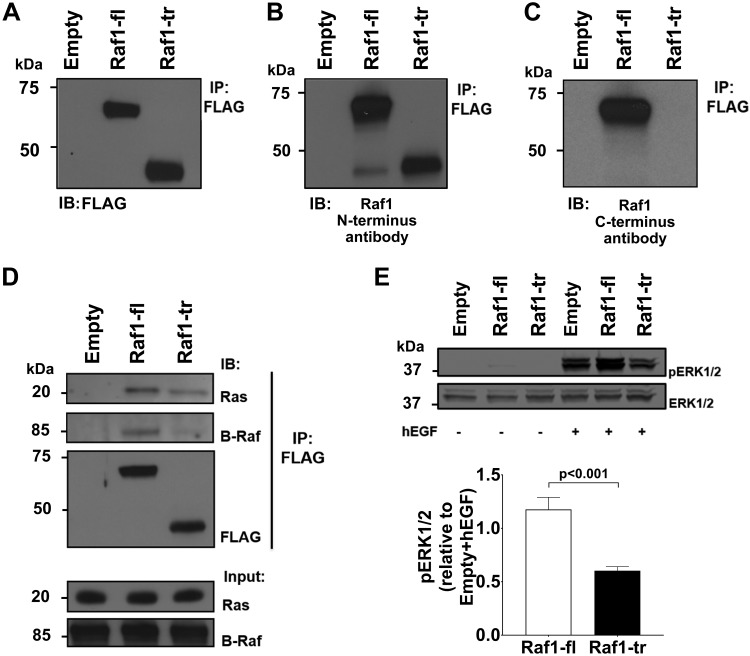
To determine the effect of Raf1-tr on canonical Ras-ERK signaling, N-terminal FLAG-tagged Raf1-fl or Raf1-tr expression constructs were created by using the cDNA sequences for each Raf1 isoform. After transfection of these constructs into HEK cells and verification of similar transfection efficiency (Supplemental Fig. S2B), immunoprecipitation was performed followed by Western blotting. First, we used an antibody targeted to FLAG to show that our Raf1-fl and Raf1-tr constructs result in the appropriate sized protein products (Fig. 2A). Next, we verified the expression of Raf1-fl and Raf1-tr by using the N-terminal antibody (Fig. 2B). Interestingly, endogenous Raf1-tr bound to Raf1-fl. We also confirmed that the antibody targeting the C-terminal region of Raf1-fl did not detect the overexpressed Raf1-tr protein (Fig. 2C).
Figure 2.
The effect of Raf1-tr on Ras-ERK signaling: FLAG-tagged Raf1-fl or Raf1-tr was expressed in HEK cells with subsequent FLAG immunoprecipitation and Western blot. Empty vector was used as a negative control. A) Probing for FLAG identified 2 protein species with the predicted MW for Raf1-fl (top band) and Raf1-tr (lower band). B) Probing for N-terminal Raf1 shows both the full-length and truncated Raf1 proteins. C) Probing for C-terminal Raf1 only results in the identification of the Raf1-fl protein. D) Probing for Ras and B-Raf shows that both Raf1-fl and Raf1-tr bind with Ras while Raf1-tr exhibited minimal binding to B-Raf compared with Raf1-fl. E) Western blot of phosphorylated ERK1/2 in HEK cells expressing empty vector, Raf1-fl, or Raf1-tr exposed to vehicle or 5 ng/ml hEGF for 5 min. Total ERK1/2 was used as a loading control. Blot is representative of 6 experiments. The results are shown as means ± sem.
We then exposed Raf1-fl– or Raf1-tr–expressing cells to hEGF to activate Ras-Raf-MEK signaling and performed coimmunoprecipitation for FLAG. Similar to Raf1-fl, Raf1-tr bound Ras (pan Ras antibody). In contrast, Raf1-tr displayed minimal binding with B-Raf (Fig. 2D). These results are supported by previous research showing the requirement of the Raf1 C-terminal kinase domain in dimerization with B-Raf for mitogen-activated signal transduction (13, 14). At baseline, we found no difference in ERK1/2 phosphorylation in serum-starved cells expressing Raf1-fl or Raf1-tr. Conversely, Raf1-tr–expressing starved cells displayed a 40% reduction in ERK1/2 phosphorylation after hEGF stimulation relative to Raf1-fl (Fig. 2E). The observed decrease in ERK1/2 phosphorylation by Raf1-tr was also confirmed in the presence of complete medium without hEGF stimulation (Supplemental Fig. S2C, D). These results confirm that Raf1-tr interacts with Ras, but because it lacks the C-terminal kinase domain, it does not participate in the downstream phosphorylation of canonical MEK-ERK signaling.
Raf1-tr has novel protein binding partners
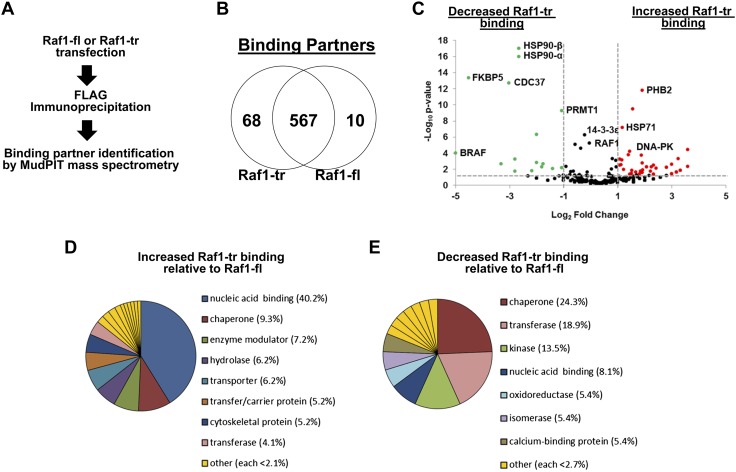
We performed MudPIT mass spectrometry on FLAG immunoprecipitated Raf1-fl or Raf1-tr eluate to determine if the protein-binding partners differ between Raf1-fl and Raf1-tr (Fig. 3A). Importantly, there were a large number of protein-binding partners that were shared between Raf1-fl and Raf1-tr, and many of these protein interactions have been previously reported for Raf1. Of the most abundant 12 Raf1-fl–binding partners, 11 had been previously identified to interact with Raf1 (Supplemental Table 2). However, both protein isoforms also exhibited a number of unique binding partners (Fig. 3B). We grouped proteins based on if they had significantly increased or decreased binding to Raf1-tr (Fig. 3C). Importantly, our proteomic assay confirmed our earlier Western blot results that B-Raf had minimal binding to Raf1-tr. We then used the PANTHER classification system (15) to group these Raf1-tr binding partners according to protein class. Interestingly, we observed increased association of Raf1-tr to proteins localized to the nucleus and decreased association of Raf1-tr to cytoplasmic chaperone proteins (Fig. 3D, E). Overall, these results show that Raf1-fl and Raf1-tr share many binding partners, but there is a clear shift in specific classes of binding partners between the 2 Raf1 isoforms.
Figure 3.
Raf1-tr exhibits unique protein binding partners: A) Outline of experimental procedure for mass spectrometry identification of Raf1-binding partners. B) Venn diagram of proteins identified to be unique to Raf1-tr (left) and Raf1-fl (right). Proteins that bound both Raf1 species fall in the middle. C) Volcano plot of proteins that exhibit decreased (left) or increased (right) binding to Raf1-tr relative to Raf1-fl binding. Thresholds of P < 0.05 (y axis) and 2-fold change (x axis) are shown as dashed lines. D) Pie chart showing breakdown of protein classification of identified binding partners that exhibit increased binding to Raf1-tr. E) Pie chart showing breakdown of protein classification that exhibit decreased binding to Raf1-tr relative to Raf1-fl binding.
Raf1-tr has reduced binding to chaperone FKBP5, which increases its nuclear localization
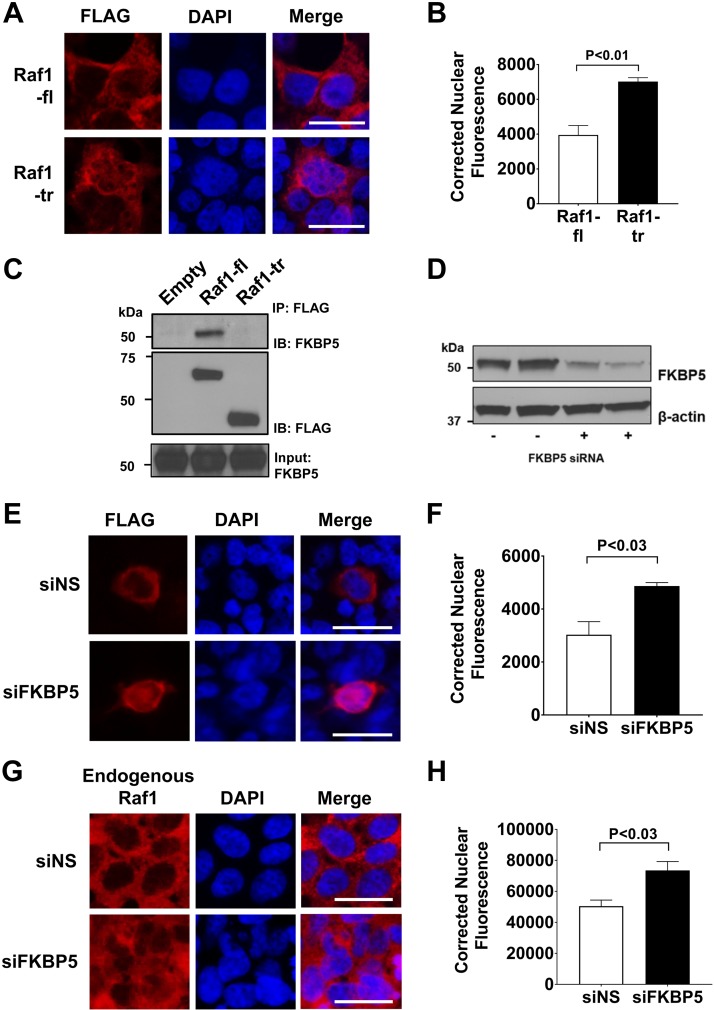
Our proteomic analysis suggested an increase in nuclear protein interactions for Raf1-tr compared with Raf1-fl. We therefore compared the cellular localization of each protein isoform and found that Raf1-tr had increased nuclear localization compared with Raf1-fl (Fig. 4A, B). Our proteomic analysis identified reduced binding of the cytoplasmic chaperone protein FKBP5 to Raf1-tr. Immunoprecipitation and Western blotting were therefore performed to confirm the reduced binding of FKBP5 to Raf1-tr (Fig. 4C). When FKBP5 protein expression was reduced (Fig. 4D), we found that Raf1-fl protein had increased cytoplasmic to nuclear translocation, which was similar to Raf1-tr (Fig. 4E, F). Similar to Raf1-fl, increased nuclear translocation of endogenous Raf1 was also observed after FKBP5 knockdown (Fig. 4G, H).
Figure 4.
Truncated Raf1 has increased nuclear localization: A) Measurement of cellular localization of FLAG tagged Raf1-fl (top) or Raf1-tr (bottom) that was expressed in HEK cells. FLAG immunofluorescence is shown in red with nuclei (blue) counterstained with DAPI. Scale bars, 25 µm. Images are representative of experiments run in triplicate. B) Quantitation of nuclear Raf1 localization by corrected nuclear fluorescence calculation. A minimum of 280 cells per group were analyzed. C) FLAG-tagged Raf1 (Raf1-fl) or Raf1-tr was expressed in HEK cells with subsequent FLAG immunoprecipitation and Western blot for FKBP5. D) Western blot of FKBP5 after siRNA knockdown of FKBP5 in HEK cells. β-actin was used as a loading control. E) Measurement of FLAG-tagged Raf1-fl localization (red) after control nonspecific siRNA (top, siNS) or FKBP5 siRNA (bottom, siFKBP5) by immunofluorescence. Nuclei (blue) were counterstained with DAPI. Scale bar, 25 µm. Images are representative of experiments run in triplicate. F) Quantitation of nuclear Raf1-fl localization by corrected nuclear fluorescence calculation. A minimum of 200 cells per group were analyzed. G) Measurement of endogenous Raf1 localization (red) after control nonspecific siRNA (top, siNS) or FKBP5 siRNA (bottom, siFKBP5) by immunofluorescence. Nuclei (blue) were counterstained with DAPI. Scale bar, 25 µm. Images are representative of experiments run in triplicate. H) Quantitation of nuclear Raf1 localization by corrected nuclear fluorescence calculation. A minimum of 300 cells per group were analyzed. The results are shown as means ± sem.
Another chaperone protein, Hsp90, also had decreased binding to Raf1-tr in our proteomic assay. Therefore, this result was first confirmed by using immunoprecipitation and Western blot Supplemental Fig. S3A). We then exposed Raf1-fl–expressing cells to geldanamycin, which has been previously shown to inhibit the association of Raf1 with Hsp90 isoforms (16). In contrast to the results after knockdown of FKBP5, no increase was observed in Raf1-fl nuclear translocation with inhibition of Hsp90 (Supplemental Fig. S3B, C). These results show that the association of Raf1-fl with FKBP5, and not Hsp90, helps to prevent the nuclear translocation of this protein. Likewise, the reduced binding of Raf1-tr to FKBP5 contributes to the increased nuclear localization of this protein.
Raf1-tr has increased binding to the nuclear localized protein DNA-PK and modulates it function
Our proteomic assay identified DNA-PK as the most abundant nuclear binding partner with Raf1-tr. To confirm this interaction, we performed a PLA (17). This method allowed us to confirm this protein–protein interaction in the endogenous state and to determine the cellular localization of this interaction. First, we validated a Raf1 antibody for cell immunofluorescence (Supplemental Fig. S4A, B). We then conducted several PLA control reactions to test for off-target PLA signal production and observed minimal nonspecific interactions (Supplemental Fig. S5A–C).
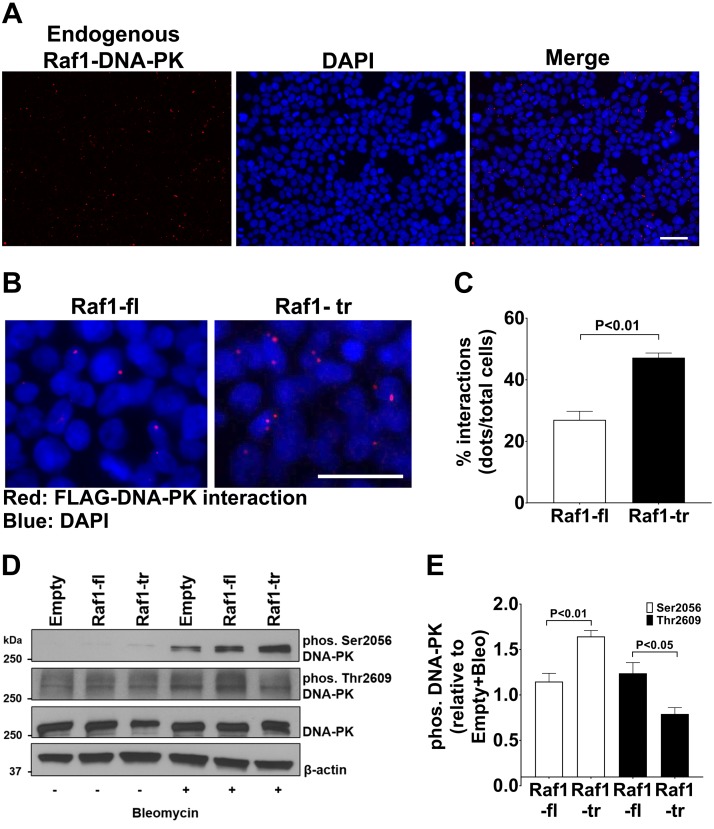
We then performed PLA between Raf1 and DNA-PK and discovered a robust interaction between the proteins that was primarily localized to the nucleus of the cell (Fig. 5A). We then expressed FLAG-tagged Raf1-fl or Raf1-tr to determine if there was a difference between the two Raf1 isoforms in their interactions with DNA-PK. Similar to the proteomic results, a significant increase was observed in Raf1-tr and DNA-PK interactions compared with Raf1-fl (Fig. 5B, C). Importantly, the PLA results also confirmed that the DNA-PK and Raf1 interactions were primarily localized to the nucleus of the cell.
Figure 5.
Truncated Raf1 preferentially binds DNA-PK and alters DNA-PK phosphorylation: A) PLA for endogenous Raf1 and DNA-PK in HEK cells. Interactions between Raf1 and DNA-PK are shown in red with nuclei (blue) counterstained with DAPI. B) PLA for FLAG and DNA-PK in HEK cells expressing FLAG-tagged Raf1-fl or Raf1-tr. Interactions between FLAG-tagged Raf1-fl or Raf1-tr and DNA-PK are shown in red with nuclei (blue) counterstained with DAPI. Images are representative of experiments run in triplicate. C) Quantitation of Raf1-fl and Raf1-tr interactions with DNA-PK. A minimum of 150 cells per group were analyzed. D) Western blot of DNA-PK Ser 2056 and Thr 2609 phosphorylation, and total DNA-PK in HEK cells expressing empty vector, Raf1-fl, or Raf1-tr exposed to vehicle (−) or 0.05 mg/ml bleomycin (+) for 2 h. β-actin was used as a loading control. Blot is representative of experiments run in quadruplicate. E) Quantitation of phosphorylated DNA-PK normalized to empty vector exposed to bleomycin (Bleo). The results are shown as mean ± sem. Scale bars, 50 µm.
Next, we wanted to determine if Raf1-fl or Raf1-tr altered the function of DNA-PK. The phosphorylation of DNA-PK is an important regulator of its function in repairing DNA damage (18). Therefore, we investigated if Raf1-fl or Raf1-tr could alter the phosphorylation of DNA-PK serine-2056 (Ser2056) and threonine-2609 (Thr2609). The Ser2056 autophosphorylation site has been shown to inhibit efficient DNA repair by altering DNA-PK function. In the absence of DNA damage, there were no observable effects of Raf1-fl or Raf1-tr on Ser2056 phosphorylation (Fig. 5D). In contrast, after exposure to the DNA damage–inducing agent bleomycin, DNA-PK Ser2056 phosphorylation increased in the Raf1-tr expressing cells compared with Raf1-fl expressing cells (Fig. 5D, E). In contrast to DNA-PK Ser2056, DNA-PK Thr2609 phosphorylation has been shown to promote DNA repair (19, 20). We observe a decrease in DNA-PK threonine 2609 phosphorylation in Raf1-tr–expressing cells. Taken together, these results show that Raf1-tr has increased binding to the nuclear localized protein DNA-PK and causes alterations in DNA-PK phosphorylation, which should lead to impaired DNA repair by DNA-PK.
Raf1-tr amplifies DNA damage
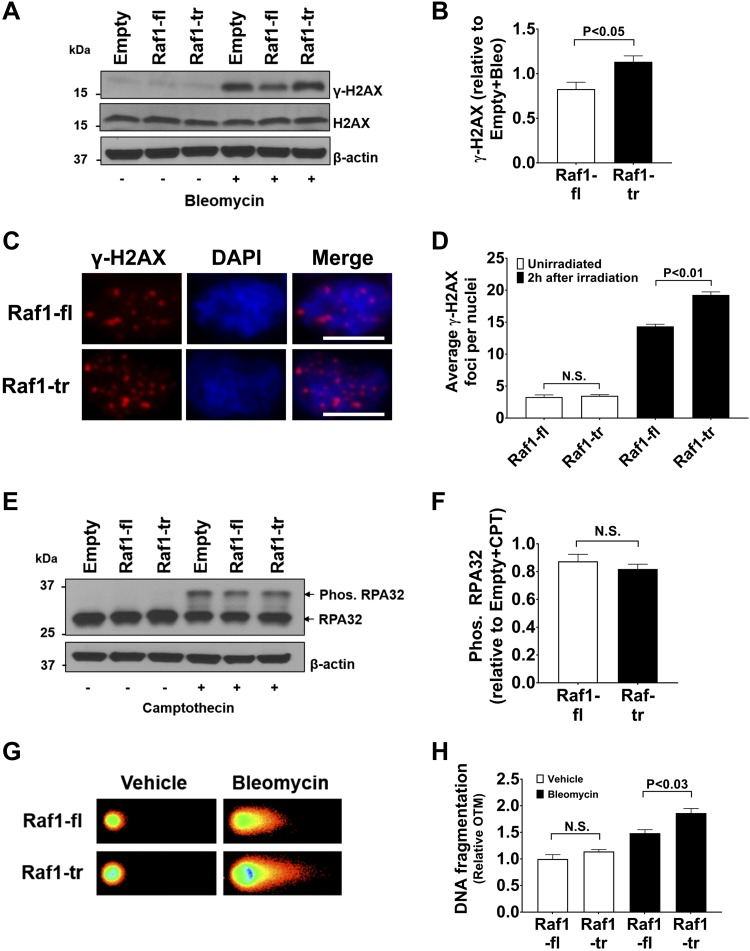
Given the changes in DNA-PK phosphorylation, we wanted to determine if cells expressing Raf1-tr had alterations in their response to DNA damage. Measurement of γ-H2AX provides a useful metric to assess DNA damage because this protein functions to recruit repair proteins to the site of double-stranded DNA damage (21, 22). Raf1-fl– or Raf1-tr–expressing cells exhibit little γ-H2AX at baseline, as shown by Western blot. However, Raf1-tr–expressing cells displayed an increase in γ-H2AX after exposure to bleomycin, which induces double-stranded DNA lesions (Fig. 6A, B). DNA damage induced by irradiation exhibited a similar response to bleomycin, with elevated levels of γ-H2AX in HEK cells expressing Raf1-tr compared with HEK cells expressing Raf1-fl (Fig. 6C, D). Next, we used camptothecin to induce single-stranded DNA lesions and measured RPA32 phosphorylation. Phosphorylation of RPA32 occurs upon RPA32 binding with single-stranded DNA (23). No significant difference was observed in RPA32 phosphorylation between Raf1-fl– and Raf1-tr–expressing cells after exposure to camptothecin (Fig. 6E, F). These results suggest that Raf1-tr alters the double-stranded DNA damage response pathway.
Figure 6.
Raf1-tr increases DNA damage: A) Western blot of γ-H2AX and H2AX in HEK cells expressing empty vectorRaf1-fl, or Raf1-tr exposed to vehicle (−) or 0.05 mg/ml bleomycin (+) for 2 h. β-actin was used as a loading control. Blot is representative of experiments run in triplicate. B) Quantitation of γ-H2AX normalized to empty vector exposed to bleomycin (Bleo). C) HEK cells expressing Raf-fl or Raf1-tr were irradiated with 1 Gy of radiation and allowed to recover in complete medium for 2 h. γ-H2AX immunofluorescence is shown in red with nuclei (blue) counterstained with DAPI. Scale bars, 10 µm. Images are representative of experiments run in triplicate. D) Quantitation of γ-H2AX foci. A minimum of 180 nuclei per group were analyzed. E) Western blot of RPA32 phosphorylation in HEK cells expressing empty vector, Raf1-fl, or Raf1-tr exposed to vehicle (−) or 1 µM camptothecin (+) for 1 h. Exposure to camptothecin results in a non-phosphorylated RPA32 band (lower) and a slower migrating phosphorylated band (upper). β-actin was used as a loading control. Blot is representative of experiments run in triplicate. F) Quantitation of RPA32 phosphorylation normalized to empty vector exposed to camptothecin (CPT). G) Neutral comet assay of nuclei from HEK cells expressing Raf1-fl or Raf1-tr exposed to vehicle or 0.05 mg/ml bleomycin for 30 min. Images are representative of experiments run in triplicate. H) Quantitation of DNA fragmentation by Olive tail moment (OTM). A minimum of 200 nuclei per group were analyzed. The results are shown as means ± sem. N.S., not significant.
Because γ-H2AX is an indirect measure of DNA damage, we then directly assessed DNA damage by using a neutral Comet assay that primarily measures double-stranded DNA fragmentation (24). Concurrent with increased γ-H2AX, a significant increase was observed in DNA fragmentation in cells expressing Raf1-tr compared with Raf1-fl after exposure to bleomycin (Fig. 6G, H). These results confirm that Raf1-tr amplifies the formation of double-stranded DNA lesions induced by bleomycin through inhibition of the DNA damage response protein DNA-PK.
Raf1-tr increases the apoptotic response of malignant cells with DNA damage
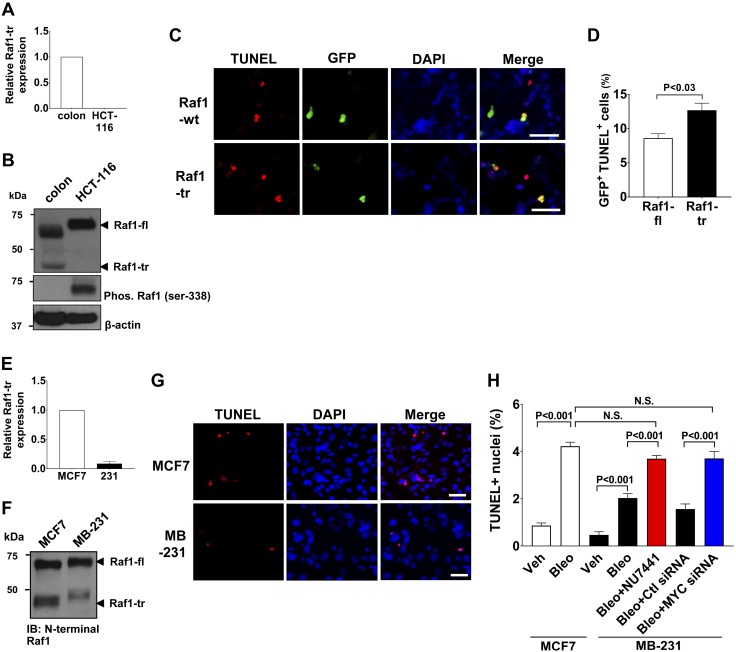
We had previously identified differential expression of Raf1-tr in colon tissue compared with malignant colon tissue (Fig. 1C). We then compared normal colon tissue with the colorectal carcinoma cell line HCT-116 and found an absence of Raf1-tr in the cancer cell line (Fig. 7A, B). Raf1-fl is slower migrating in HCT-116 cells compared with normal colon tissue secondary to increased phosphorylation (Fig. 7B). We ectopically expressed Raf1-tr in HCT-116 cells and evaluated their apoptotic response to bleomycin. We found that HCT-116 cells expressing Raf1-tr had increased levels of apoptosis after exposure to bleomycin compared with HCT-116 cells transfected with Raf1-fl expression (Fig. 7C, D). These results show that re-expression of the Raf1-tr isoform in a cell normally deficient of this protein sensitizes the cell to bleomycin-induced apoptosis.
Figure 7.
Raf1-tr increases the apoptotic response of cancer cells exposed to DNA damage. A) Measurement of Raf1-tr gene expression in colon tissue and the HCT-116 colorectal carcinoma cell line. The gene of interest was normalized to GAPDH expression and represented as fold change relative to colon tissue. B) Western blot of Raf1 in colon and HCT-116 lysate showing that Raf1-tr protein expression is absent in HCT-116 cells. Phosphorylation of Raf1-fl in HCT-116 cells results in a slower migrating species compared with normal colon. C) TUNEL stain (red) of HCT116 cells expressing Raf1-fl or Raf1-tr exposed to 0.150 mg/ml bleomycin for 72 h. Transfected cells were identified by coexpression of Raf1-fl or Raf1-tr and GFP (green) using IRES. Nuclei were counterstained with DAPI (blue). D) Quantitation of GFP-positive apoptotic cells. A minimum of 250 cells per group were analyzed. E) Measurement of Raf1-tr gene expression in MCF7 and MB-231 mammary adenocarcinoma cell lines. The gene of interest was normalized to GAPDH expression and represented as fold change relative to MCF7 cells. F) Western blot of Raf1 in MCF7 and MB-231 cell lysate demonstrating decreased Raf1-tr expression in MB-231 cells relative to MCF7 cells. G) TUNEL stain (red) of MCF7 and MB-231 cells exposed to 0.05 mg/ml bleomycin for 72 h. Nuclei were counterstained with DAPI (blue). H) Quantitation of TUNEL-positive nuclei in MCF7 cells exposed to vehicle (Veh) or bleomycin (Bleo) compared with MB-231 cells exposed to Veh alone, Bleo alone, Bleo + DNA-PK inhibitor NU7441 (0.5 μM) and Bleo + control (Ctl) or MYC siRNA (25 nM). A minimum of 250 cells per group were analyzed. The results are shown as means ± sem. N.S., not significant. Scale bars, 50 µm. Images are representative of experiments run in triplicate.
To determine if other types of cancers have differential expression of Raf1-tr, we examined Raf1-tr expression in multiple different human breast cancer cell lines and identified differential transcript expression (Fig. 7E). A Western blot was then performed to confirm that the high-expressing MCF7 cell line had increased Raf1-tr protein expression compared with the low-expressing MDA-MB-231 (MB-231) cell line (Fig. 7F). We then exposed the MCF7 and MB-231 cells to the DNA damage agent bleomycin and measured the apoptotic response of the cells. A significant increase in apoptosis was found in MCF7 cells relative to MB-231 cells after exposure to bleomycin (Fig. 7G, H). We then performed selective inhibition of DNA-PK in MB-231 cells using NU7441 to mimic the effect that increased Raf1-tr expression would have in the MB-231 cells (25). DNA-PK inhibition resulted in increased bleomycin-induced apoptosis that approached levels originally seen in the MCF7 cells (Fig. 7H and Supplemental Fig. S6A).
It had been previously shown that MYC can suppress the expression of a truncated A-Raf splice isoform, and knock down of MYC could increase expression of that truncated A-Raf splice isoform (26). We therefore evaluated if MYC was also negatively regulating the expression of Raf1-tr. First, we found that MB-231 cells had elevated expression of MYC compared with MCF-7 cells (Supplemental Fig. S7A). We then performed siRNA knockdown of MYC in the MB-231 cells and found that we could successfully induce expression of Raf1-tr by reducing MYC expression (Supplemental Fig. S7B–F). The MYC-deficient MB-231 cells that had increased Raf1-tr levels were then exposed to bleomycin. We found that MB-231 cells with increased expression of Raf1-tr had increased bleomycin-induced apoptosis compared with MB-231 cells with lower expression of Raf1-tr (Fig. 7H and Supplemental Fig. S6B). Taken together, these results suggest that MYC is a negative regulator of Raf1-tr expression and further confirm that increased expression of Raf1-tr sensitizes cells to DNA damage–induced apoptosis.
DISCUSSION
The present study identified a novel human Raf1 splice variant that is differentially expressed in multiple human tissues and produces a truncated protein product. We discovered that this Raf1-tr has increased nuclear localization secondary to decreased binding to the cytoplasmic chaperone protein FKBP5. In addition, Raf1-tr has increased association with a DNA damage–regulating protein, DNA-PK, and inhibits the function of this protein through altering its phosphorylation. This results in increased double-stranded DNA damage after the exposure to bleomycin and radiation. Likewise, inducing the expression of Raf1-tr in a colorectal cancer cell line deficient of this protein increases the apoptotic response of these malignant cells to double-stranded DNA damage. Collectively, our study presents a novel noncanonical signaling mechanism through which alternative Raf1 splicing modifies the DNA damage response of human cells.
We showed that Raf1-tr modulates the function of DNA-PK by increasing Ser2056 phosphorylation and decreasing Thr2609 phosphorylation, resulting in increased DNA damage after genotoxic insult. DNA-PK is known to play an important role in sensing DNA damage and the subsequent repair process (27). As such, DNA-PK activity is tightly regulated by clusters of phosphorylation sites that are required for efficient repair after DNA damage (18, 28). Autophosphorylation of DNA-PK at Ser2056 has been shown to reduce the ability of DNA-PK to repair DNA damage (18). In contrast, phosphorylation of DNA-PK Thr2609 has been shown to enhance DNA repair (19, 20). Our data show that Raf1-tr increases phosphorylation at DNA-PK Ser2056 and decreases phosphorylation at Thr2609 after exposure to DNA double-stranded breaks, and this process is the mechanism through which Raf1-tr increases DNA damage and apoptosis in cells exposed to genotoxic stress. Previously, it has been shown that alterations in wild-type Raf1 phosphorylation can modulate radiation-induced DNA damage through altering Chk2 activity (29). Our proteomic assay did not uncover an interaction of Raf1-tr or Raf1-fl with Chk2, which may be explained by the fact that our assay was not performed using irradiated cells.
The present study found that Raf1-tr has an increased propensity for nuclear localization relative to Raf1-fl. Previous studies have shown that Raf1 is capable of translocating to the nucleus (5, 30, 31). Our research has extended these findings by uncovering a novel role for the chaperone protein FKBP5 in regulating the cellular localization of Raf1. Although chaperone proteins play an important role in aiding efficient protein folding and stability, they can also regulate protein localization (32, 33). FKBP5 has been shown to retain proteins targeted to the nucleus in the cytoplasm (34, 35). Assembly of FKBP5 protein complexes includes interactions with chaperone protein Hsp90 (36, 37), suggesting that Hsp90 could also participate in Raf1 cytoplasmic sequestration. However, we did not detect a role for Hsp90 is regulating Raf1 cellular localization. In addition to our identified interaction with Raf1, FKBP5 has also been shown to associate with B-Raf (38); however, the functional consequence of this interaction remains unknown. Continued investigation will be required to fully understand the regulatory capacity of the FKBP5 chaperone on the Raf family member proteins.
We identified an alternatively spliced Raf1 transcript that was differentially expressed in multiple different human tissues and cell lines. Interestingly, alternative splicing has been previously reported for murine A-Raf, and the resultant truncated protein product was shown to attenuate Ras-ERK signaling and promote myogenic differentiation (39). MYC regulation of splicing factors has been found to modulate splicing of human A-Raf with increased MYC levels resulting in reduced truncated A-Raf expression (26). Our data also showed that MYC serves as a negative regulator of Raf1-tr expression. This finding highlights a role for MYC in regulating the alternative splicing of multiple Raf family members and that alternative splicing of Raf family members is an evolutionarily conserved mechanism to modify both canonical and noncanonical Raf signaling.
Alternative splicing has also been implicated in regulating multiple different malignant pathways (40). Interestingly, we found that some cancer cell lines and human tumors had reduced expression of Raf1-tr. It has been shown that that many different types of malignant cells can alter transcript splicing to sustain proliferation, decrease apoptosis, and promote drug resistance (41). We suspect that reduction of Raf1-tr expression in malignant cells serves as a protective mechanism to promote cell survival in the setting of DNA damage. The clinical efficacy of multiple different chemotherapeutic agents and radiation therapy depends on DNA damage–induced malignant cell apoptosis. In addition, heterodimer formation between B-Raf and Raf1 can occur, and these heterodimers have increased kinase activity compared with homodimers (13, 14). We did not detect Raf1-tr binding to B-Raf, and this lack of heterodimer formation may suggest another potential mechanism through which increased Raf1-tr expression could negatively modulate the growth of malignant cells. It will be important to further understand how alterations in Raf1 splicing modify malignant cell responses and if these pathways can be targeted therapeutically.
In summary, we identified a novel human Raf1 splice isoform that preferentially translocates to the nucleus because of reduced association with the cytoplasmic chaperone protein FKBP5. This Raf1 isoform associates with nuclear DNA-PK and inhibits the function of this DNA damage response protein. This action causes cells expressing this Raf1 splice isoform to have increased levels of DNA damage. Importantly, this truncated Raf1 protein is differentially expressed in normal and malignant cells. This study uncovered a novel human Raf1 isoform that has a noncanonical role in regulating the cellular DNA damage response and modifies the apoptotic response of multiple different cell types to genotoxic stress.
Supplementary Material
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
ACKNOWLEDGMENTS
Mass spectrometry was performed in the Vanderbilt Mass Spectrometry Research Center Proteomics Core Laboratory, and the authors thank Hayes McDonald (Vanderbilt University School of Medicine) for his advice and technical support. The authors also thank David Cortez (Vanderbilt University School of Medicine) for thoughtful discussion and review of the manuscript. This work was supported by the U.S. National Institutes of Health, National Heart, Lung, and Blood Institute (Grants K08HL116803 and R01HL136824 to J.R.B., and T32HL105334 to B.R.N.); the PhRMA Foundation (Postdoctoral Fellowship to S.C.S.); and the American Heart Association (Grant PTF29620009 to B.R.N.). The authors declare no conflicts of interest.
Glossary
- BSA
bovine serum albumin
- DNA-PK
DNA-dependent protein kinase
- FKBP5
FK506 binding protein 5
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GFP
green fluorescent protein
- hEGF
human epidermal growth factor
- HRP
horseradish peroxidase
- Hsp90
heat shock protein 90
- IRES
internal ribosome entry site
- PANTHER
Protein Analysis Through Evolutionary Relationships
- PLA
proximity ligation assay
- Raf1-fl
full-length Raf1
- Raf1-tr
truncated Raf1
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
B. R. Nixon, S. C. Sebag, M. L. Freeman, and J. R. Becker developed the research study design; B. R. Nixon, S. C. Sebag, M. S. Glennon, E. J. Hall, E. S. Kounlavong, and J. R. Becker conducted experiments and data analysis; B. R. Nixon and J. R. Becker prepared the manuscript; and all authors edited the manuscript.
REFERENCES
- 1.Chang F., Steelman L. S., Lee J. T., Shelton J. G., Navolanic P. M., Blalock W. L., Franklin R. A., McCubrey J. A. (2003) Signal transduction mediated by the Ras/Raf/MEK/ERK pathway from cytokine receptors to transcription factors: potential targeting for therapeutic intervention. Leukemia 17, 1263–1293 10.1038/sj.leu.2402945 [DOI] [PubMed] [Google Scholar]
- 2.Roberts P. J., Der C. J. (2007) Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene 26, 3291–3310 10.1038/sj.onc.1210422 [DOI] [PubMed] [Google Scholar]
- 3.Rui L., Drennan A. C., Ceribelli M., Zhu F., Wright G. W., Huang D. W., Xiao W., Li Y., Grindle K. M., Lu L., Hodson D. J., Shaffer A. L., Zhao H., Xu W., Yang Y., Staudt L. M. (2016) Epigenetic gene regulation by Janus kinase 1 in diffuse large B-cell lymphoma. Proc. Natl. Acad. Sci. USA 113, E7260–E7267 10.1073/pnas.1610970113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kung J. E., Jura N. (2016) Structural basis for the non-catalytic functions of protein kinases. Structure 24, 7–24 10.1016/j.str.2015.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mielgo A., Seguin L., Huang M., Camargo M. F., Anand S., Franovic A., Weis S. M., Advani S. J., Murphy E. A., Cheresh D. A. (2011) A MEK-independent role for CRAF in mitosis and tumor progression. Nat. Med. 17, 1641–1645 10.1038/nm.2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacCoss M. J., McDonald W. H., Saraf A., Sadygov R., Clark J. M., Tasto J. J., Gould K. L., Wolters D., Washburn M., Weiss A., Clark J. I., Yates J. R., III (2002) Shotgun identification of protein modifications from protein complexes and lens tissue. Proc. Natl. Acad. Sci. USA 99, 7900–7905 10.1073/pnas.122231399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Panaccione A., Zhang Y., Mi Y., Mitani Y., Yan G., Prasad M. L., McDonald W. H., El-Naggar A. K., Yarbrough W. G., Ivanov S. V. (2017) Chromosomal abnormalities and molecular landscape of metastasizing mucinous salivary adenocarcinoma. Oral Oncol. 66, 38–45 10.1016/j.oraloncology.2016.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 10.1021/ac025747h [DOI] [PubMed] [Google Scholar]
- 9.Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 10.1021/ac0341261 [DOI] [PubMed] [Google Scholar]
- 10.Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G.; The Gene Ontology Consortium (2000) Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mi H., Muruganujan A., Casagrande J. T., Thomas P. D. (2013) Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 8, 1551–1566 10.1038/nprot.2013.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olive P. L., Banáth J. P., Durand R. E. (1990) Heterogeneity in radiation-induced DNA damage and repair in tumor and normal cells measured using the “comet” assay. Radiat. Res. 122, 86–94 10.2307/3577587 [DOI] [PubMed] [Google Scholar]
- 13.Rushworth L. K., Hindley A. D., O’Neill E., Kolch W. (2006) Regulation and role of Raf-1/B-Raf heterodimerization. Mol. Cell. Biol. 26, 2262–2272 10.1128/MCB.26.6.2262-2272.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weber C. K., Slupsky J. R., Kalmes H. A., Rapp U. R. (2001) Active Ras induces heterodimerization of cRaf and BRaf. Cancer Res. 61, 3595–3598 [PubMed] [Google Scholar]
- 15.Mi H., Lazareva-Ulitsky B., Loo R., Kejariwal A., Vandergriff J., Rabkin S., Guo N., Muruganujan A., Doremieux O., Campbell M. J., Kitano H., Thomas P. D. (2005) The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 33, D284–D288 10.1093/nar/gki078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schulte T. W., Blagosklonny M. V., Ingui C., Neckers L. (1995) Disruption of the Raf-1-Hsp90 molecular complex results in destabilization of Raf-1 and loss of Raf-1-Ras association. J. Biol. Chem. 270, 24585–24588 10.1074/jbc.270.41.24585 [DOI] [PubMed] [Google Scholar]
- 17.Thymiakou E., Episkopou V. (2011) Detection of signaling effector-complexes downstream of bmp4 using PLA, a proximity ligation assay. J. Vis. Exp. 3, pii: 2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui X., Yu Y., Gupta S., Cho Y. M., Lees-Miller S. P., Meek K. (2005) Autophosphorylation of DNA-dependent protein kinase regulates DNA end processing and may also alter double-strand break repair pathway choice. Mol. Cell. Biol. 25, 10842–10852 10.1128/MCB.25.24.10842-10852.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan D. W., Chen B. P., Prithivirajsingh S., Kurimasa A., Story M. D., Qin J., Chen D. J. (2002) Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 16, 2333–2338 10.1101/gad.1015202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding Q., Reddy Y. V., Wang W., Woods T., Douglas P., Ramsden D. A., Lees-Miller S. P., Meek K. (2003) Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase is required for efficient end processing during DNA double-strand break repair. Mol. Cell. Biol. 23, 5836–5848 10.1128/MCB.23.16.5836-5848.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma A., Singh K., Almasan A. (2012) Histone H2AX phosphorylation: a marker for DNA damage. Methods Mol. Biol. 920, 613–626 10.1007/978-1-61779-998-3_40 [DOI] [PubMed] [Google Scholar]
- 22.Paull T. T., Rogakou E. P., Yamazaki V., Kirchgessner C. U., Gellert M., Bonner W. M. (2000) A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10, 886–895 10.1016/S0960-9822(00)00610-2 [DOI] [PubMed] [Google Scholar]
- 23.Wold M. S. (1997) Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66, 61–92 10.1146/annurev.biochem.66.1.61 [DOI] [PubMed] [Google Scholar]
- 24.Collins A. R. (2004) The comet assay for DNA damage and repair: principles, applications, and limitations. Mol. Biotechnol. 26, 249–261 10.1385/MB:26:3:249 [DOI] [PubMed] [Google Scholar]
- 25.Zhao Y., Thomas H. D., Batey M. A., Cowell I. G., Richardson C. J., Griffin R. J., Calvert A. H., Newell D. R., Smith G. C., Curtin N. J. (2006) Preclinical evaluation of a potent novel DNA-dependent protein kinase inhibitor NU7441. Cancer Res. 66, 5354–5362 10.1158/0008-5472.CAN-05-4275 [DOI] [PubMed] [Google Scholar]
- 26.Rauch J., Moran-Jones K., Albrecht V., Schwarzl T., Hunter K., Gires O., Kolch W. (2011) c-Myc regulates RNA splicing of the A-Raf kinase and its activation of the ERK pathway. Cancer Res. 71, 4664–4674 10.1158/0008-5472.CAN-10-4447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson S. P., Jeggo P. A. (1995) DNA double-strand break repair and V(D)J recombination: involvement of DNA-PK. Trends Biochem. Sci. 20, 412–415 10.1016/S0968-0004(00)89090-8 [DOI] [PubMed] [Google Scholar]
- 28.Neal J. A., Sugiman-Marangos S., VanderVere-Carozza P., Wagner M., Turchi J., Lees-Miller S. P., Junop M. S., Meek K. (2014) Unraveling the complexities of DNA-dependent protein kinase autophosphorylation. Mol. Cell. Biol. 34, 2162–2175 10.1128/MCB.01554-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Advani S. J., Camargo M. F., Seguin L., Mielgo A., Anand S., Hicks A. M., Aguilera J., Franovic A., Weis S. M., Cheresh D. A. (2015) Kinase-independent role for CRAF-driving tumour radioresistance via CHK2. Nat. Commun. 6, 8154 10.1038/ncomms9154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang S., Ghosh R. N., Chellappan S. P. (1998) Raf-1 physically interacts with Rb and regulates its function: a link between mitogenic signaling and cell cycle regulation. Mol. Cell. Biol. 18, 7487–7498 10.1128/MCB.18.12.7487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geil W. M., Yen A. (2014) Nuclear Raf-1 kinase regulates the CXCR5 promoter by associating with NFATc3 to drive retinoic acid-induced leukemic cell differentiation. FEBS J. 281, 1170–1180 10.1111/febs.12693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hartl F. U., Bracher A., Hayer-Hartl M. (2011) Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332 10.1038/nature10317 [DOI] [PubMed] [Google Scholar]
- 33.Young J. C., Barral J. M., Ulrich Hartl F. (2003) More than folding: localized functions of cytosolic chaperones. Trends Biochem. Sci. 28, 541–547 10.1016/j.tibs.2003.08.009 [DOI] [PubMed] [Google Scholar]
- 34.Lukic I., Mitic M., Soldatovic I., Jovicic M., Maric N., Radulovic J., Adzic M. (2015) Accumulation of cytoplasmic glucocorticoid receptor is related to elevation of FKBP5 in lymphocytes of depressed patients. J. Mol. Neurosci. 55, 951–958 10.1007/s12031-014-0451-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wochnik G. M., Rüegg J., Abel G. A., Schmidt U., Holsboer F., Rein T. (2005) FK506-binding proteins 51 and 52 differentially regulate dynein interaction and nuclear translocation of the glucocorticoid receptor in mammalian cells. J. Biol. Chem. 280, 4609–4616 10.1074/jbc.M407498200 [DOI] [PubMed] [Google Scholar]
- 36.Chen M. S., Silverstein A. M., Pratt W. B., Chinkers M. (1996) The tetratricopeptide repeat domain of protein phosphatase 5 mediates binding to glucocorticoid receptor heterocomplexes and acts as a dominant negative mutant. J. Biol. Chem. 271, 32315–32320 10.1074/jbc.271.50.32315 [DOI] [PubMed] [Google Scholar]
- 37.Davies T. H., Ning Y. M., Sánchez E. R. (2005) Differential control of glucocorticoid receptor hormone-binding function by tetratricopeptide repeat (TPR) proteins and the immunosuppressive ligand FK506. Biochemistry 44, 2030–2038 10.1021/bi048503v [DOI] [PubMed] [Google Scholar]
- 38.Eisenhardt A. E., Sprenger A., Röring M., Herr R., Weinberg F., Köhler M., Braun S., Orth J., Diedrich B., Lanner U., Tscherwinski N., Schuster S., Dumaz N., Schmidt E., Baumeister R., Schlosser A., Dengjel J., Brummer T. (2016) Phospho-proteomic analyses of B-Raf protein complexes reveal new regulatory principles. Oncotarget 7, 26628–26652 10.18632/oncotarget.8427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yokoyama T., Takano K., Yoshida A., Katada F., Sun P., Takenawa T., Andoh T., Endo T. (2007) DA-Raf1, a competent intrinsic dominant-negative antagonist of the Ras-ERK pathway, is required for myogenic differentiation. J. Cell Biol. 177, 781–793 10.1083/jcb.200703195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oltean S., Bates D. O. (2014) Hallmarks of alternative splicing in cancer. Oncogene 33, 5311–5318 10.1038/onc.2013.533 [DOI] [PubMed] [Google Scholar]
- 41.Sveen A., Kilpinen S., Ruusulehto A., Lothe R. A., Skotheim R. I. (2016) Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene 35, 2413–2427 10.1038/onc.2015.318 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.