Abstract
Meiotic recombination ensures faithful segregation of homologous chromosomes during meiosis and generates genetic diversity in gametes. MEIOB (meiosis specific with OB domains), a meiosis-specific single-stranded DNA-binding homolog of replication protein A1 (RPA1), is essential for meiotic recombination. Here, we investigated the molecular mechanisms of MEIOB by characterizing its binding partners spermatogenesis associated 22 (SPATA22) and RPA. We find that MEIOB and SPATA22 form an obligate complex and contain defined interaction domains. The interaction between these two proteins is unusual in that nearly any deletion in the binding domains abolishes the interaction. Strikingly, a single residue D383 in MEIOB is indispensable for the interaction. The MEIOB/SPATA22 complex interacts with the RPA heterotrimeric complex in a collaborative manner. Furthermore, MEIOB and SPATA22 are recruited to induced DNA double-strand breaks (DSBs) together but not alone. These results demonstrate the cooperative property of the MEIOB-SPATA22 complex in its interaction with RPA and recruitment to DSBs.
Keywords: meiosis, RPA, MEIOB, SPATA22, DNA double-strand breaks
Summary Sentence
MEIOB and SPATA22 function as a novel meiosis-specific heterocomplex.
Introduction
Meiosis consists of two cell divisions that produce haploid gametes from diploid progenitors. The first meiotic division (Meiosis I) features a prolonged prophase I that coordinates homologous chromosome pairing and reciprocal exchange of chromosomal DNA in a process termed meiotic recombination [1–3]. This leads to the generation of physical links between homologous chromosomes and ensures their faithful segregation at the end of Meiosis I. The second meiotic division (Meiosis II), like mitosis, involves segregation of sister chromatids. Meiotic recombination and random segregation of homologous chromosomes generate genetic diversity within a population, providing a framework for genome evolution and natural selection [4]. Importantly, errors in meiosis are leading causes of aneuploidy, birth defects, and infertility [5].
Meiotic recombination is initiated by formation of SPO11-catalyzed DNA double-strand breaks (DSBs) [6–8]. DSB ends are resected in a 5΄ to 3΄ manner to generate single-stranded DNAs (ssDNA). The ∼1000 base pair long 3΄ ssDNA tails are bound by the RPA (replication protein A) complex (a heterotrimer of RPA1, RPA2, and RPA3). The binding of RPA to ssDNA prevents degradation and secondary structure formation [9]. Resected ssDNA ends are docking sites for ssDNA-binding recombinases RAD51 and DMC1 [10,11]. RAD51 and DMC1 initiate the 3΄ single-strand invasion into the DNA duplex of the homologous chromosome [12]. Displacement of one strand of the homolog results in the formation of the single-stranded D loop. RAD51/DMC1-mediated strand invasion provides a basis for repair of DSBs as well as synapsis of homologous chromosomes, the latter of which requires the synaptonemal complex [13,14]. Following strand invasion, recombination intermediates are repaired towards either crossover or noncrossover pathways [1]. A number of proteins participate in the processing of recombination intermediates, including MSH4, MSH5, and TEX11 [15–18]. MLH1/MLH3 are endonucleases responsible for the formation of the majority of crossovers [19–22]; whereas, MUS81 produces a subset of MLH1/MLH3-independent crossovers [23,24]. In addition, ubiquitin E3 ligase HEI10, SUMO E3 ligase RNF212, and CNTD1 regulate crossover formation [25–27]. Meiotic recombination is tightly regulated by these meiosis-specific factors as well as proteins involved in general DSB repair. Completion of at least one crossover per homologous chromosome pair ensures the accurate connection and faithful segregation of maternal and paternal chromosomes during meiosis I. Failure or reduction in crossover causes meiotic arrest and thus infertility.
Although a large number of meiotic recombination proteins have been identified, the mechanisms of recombination intermediate processing remains elusive. We and others have discovered a meiosis-specific protein called MEIOB (meiosis specific with OB domains) that is required for meiotic recombination [28,29]. MEIOB is a meiosis-specific paralog of RPA1, which is ubiquitously expressed. MEIOB binds specifically to ssDNA in vitro and localizes as foci on meiotic chromosomes. We previously found that MEIOB associates with SPATA22 (spermatogenesis associated 22) and RPA2 in vivo [28]. Consistently, MEIOB colocalizes with SPATA22 and RPA on meiotic chromosomes [28–31]. Inactivation of Meiob leads to infertility in both sexes. In Meiob-deficient germ cells, DSBs are formed but not repaired, resulting in a lack of crossover formation. Interestingly, Meiob mouse mutant phenocopies Spata22 mutant [28,30–32]. The localization of MEIOB and SPATA22 on meiotic chromosomes are interdependent; however, localization of RPA is independent of MEIOB and SPATA22 [28,31,33]. Despite these findings, the molecular interaction between MEIOB, SPATA22, and RPA and the impact of these interactions on their function remain unknown. Here, we report that MEIOB and SPATA22 form an obligate complex and harbor discrete interaction domains. Our results show that the MEIOB-SPATA22 interaction is essential for their association with the RPA complex and recruitment to DSBs.
Materials and methods
Expression constructs
Mouse Meiob cDNA was subcloned into the pcDNA6 vector (Invitrogen) harboring an N-terminal Flag tag and a C-terminal Myc tag. Mouse Spata22 cDNA was subcloned into the pcDNA6 vector harboring an N-terminal Flag tag and a C-terminal V5 tag. All subsequent Meiob or Spata22 truncation fragments and point mutations were subcloned into the above Flag-Myc or Flag-V5 vectors, respectively. The following 26 “alanine-scanning” Meiob point mutations were generated: D298A, Y300A, K306A, P318A, F319A, Y320A, Y324A, I327A, D333A, D334A, E335A, F372A, L373A, F375A, D383A, H384A, T385A, F409A, L410A, M412A, K421A, W422A, L424A, H439A, R440A, R442A. Two additional mutations (D383E and D383N) were made at D383. Mouse Rpa1 (isoform 2), Rpa2, and Rpa3 cDNAs were amplified from bulk testis cDNAs prepared from postnatal day 22 mice on a mixed C57BL/6J × 129 background and subcloned into the pcDNA6 vector. Green fluorescent protein (GFP)-MEIOB, GFP-SPATA22, and GFP-RPA1 constructs were generated by subcloning into the pEGFP-C1 vector (Clontech). All the constructs were verified by Sanger sequencing on an ABI 3730 DNA analyzer.
Cell culture and transfections
All cells were maintained in DMEM/high glucose (Mediatech) supplemented with 10% FBS (Sigma) and 1× penicillin-streptomycin (Invitrogen). U2OS-DSB reporter cell line was described previously [34]. Plasmid DNA transfections in HEK 293T cells were carried out using a standard calcium phosphate method. Transfections in U2OS cells were performed with Lipofectamine 3000 (Invitrogen) according to the manufacturer's instructions.
Antibodies
The following primary antibodies were used in this study: anti-Myc monoclonal (1:4000, catalog number 631206, Clontech), anti-V5 monoclonal (1:4000, catalog number R960-25, Invitrogen), anti-β-actin (1:10 000, catalog number A5441, Sigma), anti-GFP (1:10 000, catalog number 11814460001, Roche) and anti-Flag monoclonal (1:10 000, catalog number F1804, Sigma) (Supplementary Table S1).
Co-immunoprecipitation and immunoblotting assays
A total of 1 × 107 cells were harvested at 24–36 h post transfection and lysed in 1 ml NETN buffer (100 mM NaCl, 1 mM EDTA, 20 mM Tris-HCl, pH 8.0, and 0.5% Nonidet P-40) supplemented with 1× protease inhibitor cocktail (Sigma). Benzonase (Sigma) was added to the buffer at a concentration of 90 U/μl. For immunoprecipitation (IP), 1.5% of the lysates were used as inputs. About 2–3 mg of the lysates were incubated with 1–2 μl of primary antibodies for 1 h at 4°C, followed by incubation with protein G Dynabeads (Thermo Fisher Scientific) overnight. The immunoprecipitated complexes were washed with the NETN buffer four times and boiled in 30 μl 2× SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis) loading buffer for 5 min. About 10–20 μl of supernatants were resolved by SDS-PAGE, transferred onto nitrocellulose membranes using iBlot (Invitrogen), and immunoblotted with indicated antibodies. Images and band quantifications were analyzed using the ImageJ v1.44 software. All co-IP and quantification experiments were performed at least three times.
DNA double-strand break recruitment assay
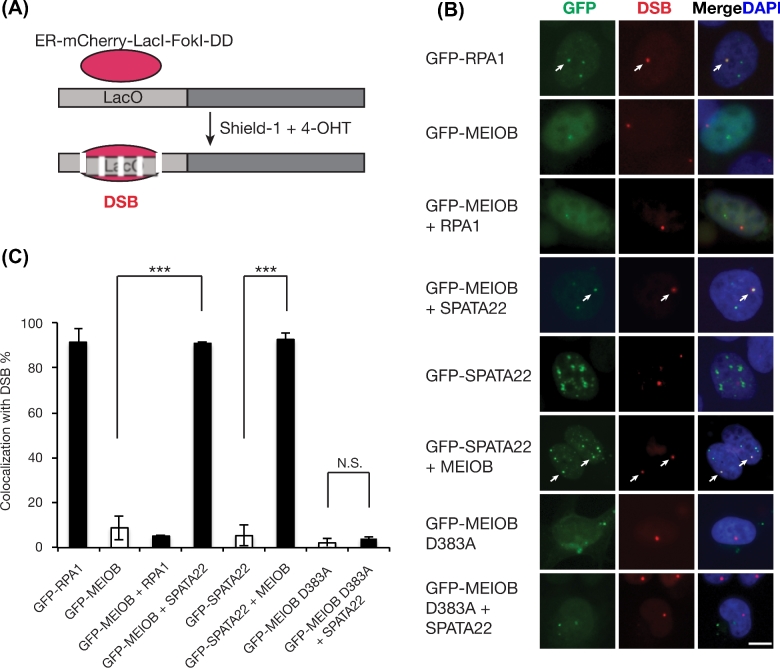
U2OS-DSB reporter cells were grown on glass coverslips and transfected with plasmids as indicated. Thirty-six hours posttransfection, cells were treated with 1 μM Shield-1 (Cheminpharma) and 1 μM 4-hydroxytamoxifen (4-OHT) (Sigma) for 4 h. Following treatment, cells were fixed in 2% paraformaldehyde for 10 min. Coverslips were then mounted onto glass slides with Vectashield mounting medium containing DAPI (Vector Laboratories). Images were captured with a Hamamatsu C11440 digital camera connected to a Leica DM5500B microscope equipped with a 40×/0.85 dry immersion lens. Images were analyzed using the LAS X software. The percentage of colocalization with DSB was calculated as the number of colocalization positive cells divided by the number of DSB and GFP-protein expression double-positive cells. A minimum of 30 (MEIOB only and SPATA22 only) or 100 GFP and DSB double-positive cells were counted per experiment. The experiments were performed three times. Statistical analysis was performed with Student t-test. The values were presented as mean ± s.d. (Figure 5C).
Figure 5.
DSB recruitment of MEIOB and SPATA22 depends on their coexpression and interaction. (A) Schematic of the inducible DSB reporter in U2OS cells. ER, estrogen receptor; DD, destabilization domain; 4-OHT, 4-hydroxytamoxifen; Shield-1, stabilizing ligand of DD-tagged protein. (B) Representative images of the colocalization between GFP-tagged proteins and induced DSBs (red). Reporter U2OS cells were transfected with indicated plasmids before administration of Shield-1 and 4-OHT. Scale bar, 10 μm. (C) Percentage of the indicated GFP-tagged proteins that colocalize with DSBs. More than 30 GFP and DSB double-positive cells were counted per experiment. The experiments were performed three times. ***, P < 0.005; N.S., not significant.
Structure modeling
The MEIOB structure was predicted using the Phyre2 protein structure prediction server [35]. The modeled structure was visualized using Pymol.
Results
Complex formation of MEIOB and SPATA22 enhances protein abundance
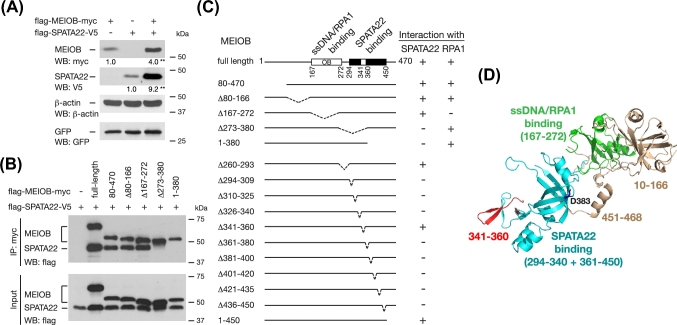
Our previous study suggests that the stability of MEIOB and SPATA22 in germ cells is dependent on each other [28]. We tested this possibility in a somatic cell line—HEK 293T cells, which do not express MEIOB or SPATA22. HEK 293T cells were transfected with mouse MEIOB alone, or mouse SPATA22 alone, or both. Strikingly, coexpression of MEIOB and SPATA22 increased MEIOB and SPATA22 protein levels by 4-fold and 9-fold, respectively (Figure 1A). Furthermore, co-IP assay showed that MEIOB and SPATA22 formed a complex when expressed in HEK 293T cells, supporting that SPATA22 is a bona fide binding partner of MEIOB (Figure 1B). These results are consistent with our previous finding of complex formation in testis and indicate that the interaction between these two meiosis-specific proteins dramatically stabilizes both of them.
Figure 1.
MEIOB forms an obligate complex with SPATA22. (A) Western blot analysis of MEIOB and SPATA22 protein levels in cells transfected with indicated plasmids. β-actin serves as a loading control and GFP serves as a transfection control. The numbers below the bands indicate normalized fold increases of MEIOB and SPATA22 protein levels. **, P < 0.01. (B) Co-IP analysis of MEIOB deletions and SPATA22. (C) Schematic diagram of MEIOB deletions and results of interactions with SPATA22 or RPA1, respectively. +, positive co-IP; –, negative co-IP. Western blots (WB) of the first five MEIOB deletions are shown in panel B and the remaining 11 deletions are shown in Supplementary Figure S1A and S1B. The OB fold (aa 167–272) is also shown. Western blots concerning RPA1 interaction are shown in Figure 4A. (D) Modeling of mouse MEIOB structure based on the fungus RPA heterotrimer structure by Phyre2 algorithm [19,20]. Residues 10–468 of MEIOB were modeled. Defined interaction domains and various regions are color-coded. The critical Asp383 residue is shown in blue.
The SPATA22-interacting domain in MEIOB is distinct from its OB single-stranded DNA-binding domain
Given the strong association of MEIOB and SPATA22, we made a series of deletion mutants of MEIOB to map its SPATA22-binding domain by transfection and co-IP. We first found that the oligonucleotide/oligosaccharide-binding (OB)-fold domain of MEIOB (aa 167–272), which binds to ssDNA, was dispensable for interaction with SPATA22 (Figure 1B and C). Further deletion studies showed that the SPATA22-binding domain was located in the C-terminal region (aa 273–470), because two MEIOB fragments (1-380 and Δ273-380) failed to interact with SPATA22 (Figure 1B and C).
To pinpoint the SPATA22-binding domain, we generated a series of 15–20 aa deletions within the 273–470 region (Figure 1C; Supplementary Figure S1A and S1B). Co-IP assays revealed that the region of aa 294–450 was required for its interaction with SPATA22 and thus constituted the SPATA22-binding domain, which does not overlap with the MEIOB OB domain. Remarkably, any of these deletions within the binding domain (aa 294–450) except aa 341–360 abolished the interaction, suggesting a unique interaction property between MEIOB and SPATA22.
Aspartic acid D383 in MEIOB is required for interaction with SPATA22
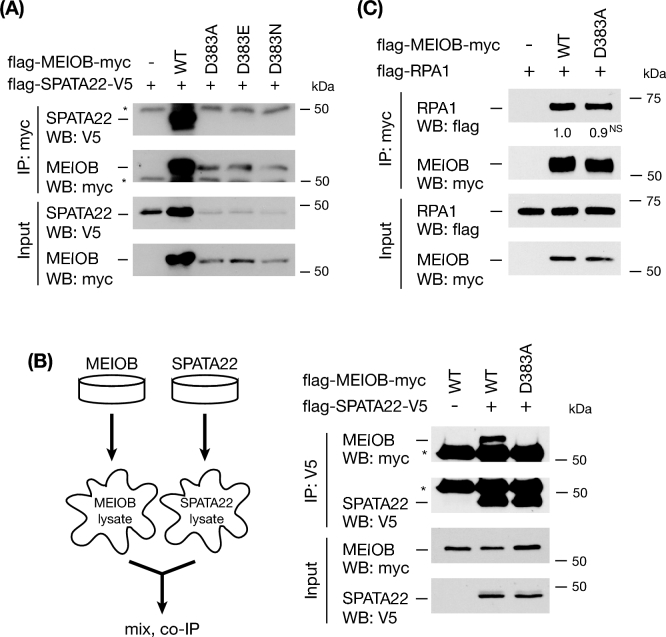
Given the unusual interaction property between MEIOB and SPATA22, we hypothesized the existence of residues critical for their interaction. To screen for such residues, we performed “alanine scanning” mutagenesis on 26 evolutionarily conserved residues within the SPATA22-binding domain. We identified conserved residues by alignment of MEIOB protein sequences from six species (mouse, human, chicken, dog, zebrafish, and Xenopus). Out of 26 residues tested (see Materials and Methods), only one residue—D383 was found to be essential for MEIOB-SPATA22 interaction, while mutation in each of the remaining 25 residues did not disrupt the interaction. D383A mutation completely blocked the interaction (Figure 2A). Substitution of the aspartic acid (D383) residue for glutamic acid (E, the same charge but different size) or asparagine (N, same size but no charge) still disrupted the interaction (Figure 2A), suggesting that both charge and size of D383 are required for the interaction. We observed a simultaneous decrease in the protein abundance of MEIOB and SPATA22 when D383 was mutated, likely due to the lack of interaction (Figure 2A).
Figure 2.
D383A mutation in MEIOB abolishes its interaction with SPATA22 but not with RPA1. (A) Mutations of D383 in MEIOB disrupt interaction with SPATA22. MEIOB WT serves as a positive co-IP control. *, antibody heavy chain. (B) Left, schematic of the co-IP analysis using separate MEIOB- or SPATA22-containing cell lysates. Right, co-IP analysis of the interaction between SPATA22 and WT MEIOB or mutant MEIOB (D383A) by mixing of separate protein lysates. *, antibody heavy chain. (C) Co-IP analysis of the interaction between RPA1 and WT MEIOB or mutant MEIOB (D383A). The numbers indicate a normalized RPA1/MEIOB ratio from the IP blots. NS, not significant.
To rule out the possibility that the abolished binding was caused by reduced protein levels of MEIOB and SPATA22 and thus reduced sensitivity of co-IP assay, we expressed wild-type MEIOB, mutant MEIOB (D383A), and SPATA22 in HEK 293T cells separately (Figure 2B). We then mixed equal amounts of MEIOB and SPATA22 cell lysates and performed the co-IP assay. These results showed that wild-type MEIOB was robustly associated with SPATA22 but MEIOB (D383A) was not (Figure 2B). Therefore, our data demonstrate that D383 in MEIOB is required for MEIOB-SPATA22 interaction.
Modeled MEIOB structure reveals spatially discrete domains
The Phyre2 web portal was employed to generate a three-dimensional homology model of MEIOB using the known fungus RPA heterotrimer structure as a template (Figure 1D) [35,36]. However, modeling of the SPATA22 structure was not reliable since more than 50% of the sequence was predicted to be disordered. The predicted MEIOB structure model suggests that the SPATA22-binding domain is distinct from its ssDNA-binding OB fold (Figure 1D). Consistent with its lack of effect on SPATA22-binding, the 20-aa region (aa 341–360) comprising two β-strands is located outside the SPATA22-binding domain. Likewise, the C-terminal α-helix (aa 451–468) has no effect on SPATA22 binding and is also located outside the binding domain. The critical Asp 383 is situated at a β-turn within the SPATA22-binding domain. In conclusion, the predicted MEIOB structure is supported by our biochemical data.
C-terminal mapping of the MEIOB-binding domain in SPATA22
Using the same deletion strategy as described above, we defined the MEIOB-binding domain in SPATA22 (Figure 3A). We initially mapped the binding domain to the C-terminal region (aa 204–358) of SPATA22 (Figure 3A and B). The binding domain was finally narrowed down to aa 224–340 by a series of deletions of 20 aa or fewer (Figure 3A; Supplementary Figure S2A and S2B). This region was highly conserved and corresponded to the second conserved region (CR2: aa 238 - 332) in SPATA22 previously defined by multiple sequence alignment [32]. Like the SPATA22-binding domain of MEIOB, any of these 20-aa deletions in the MEIOB-binding domain of SPATA22 disrupted the interaction, further supporting the unique interaction pattern between these two proteins. The original Spata22 mutant contained a nonsense mutation at amino acid 275 (Y275X) in the MEIOB-binding domain, presumably abolishing MEIOB-SPATA22 interaction [32].
Figure 3.
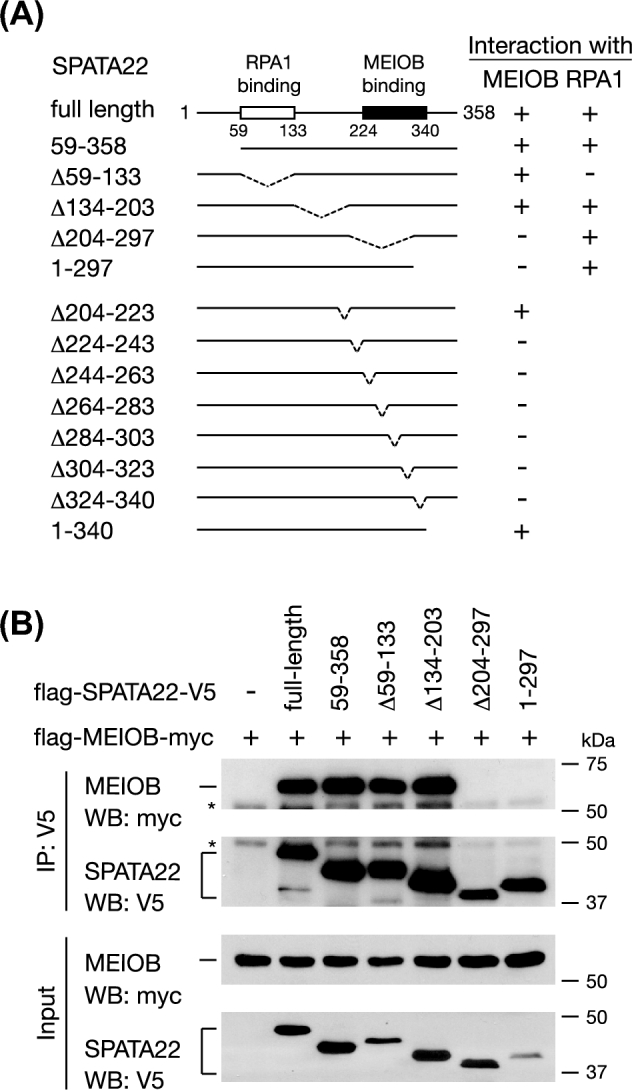
Identification of the MEIOB-binding domain in SPATA22. (A) Schematic diagram of SPATA22 deletions and co-IP results of interactions with MEIOB. +, positive co-IP; –, negative co-IP. (B) Western blots of co-IP analysis of the first five SPATA22 deletions. The remaining eight deletions are shown in Supplementary Figure S2A and S2B. *, antibody heavy chain. Co-IP results of SPATA22 interaction with RPA1 are shown in panel A but relevant western blots are shown in Supplementary Figure S3A.
Cooperative interaction of MEIOB and SPATA22 with the replication protein A complex
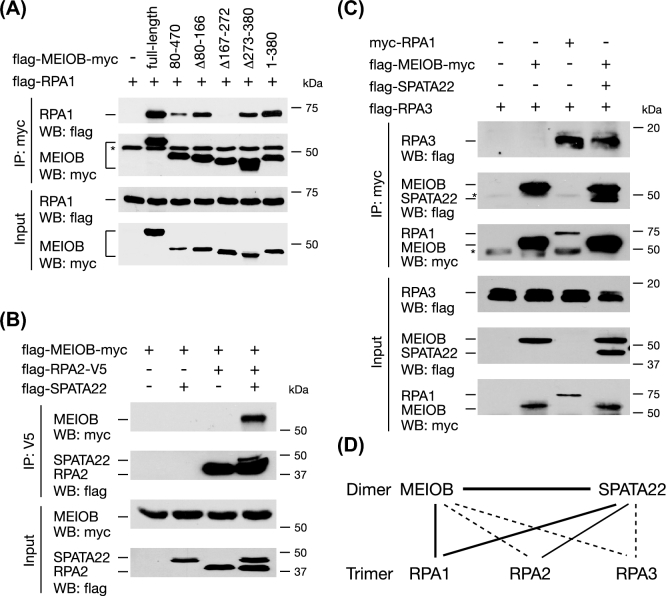
RPA is a ubiquitous ssDNA-binding heterotrimeric complex [9]. Three lines of evidence support the notion that MEIOB functions in close relationship with the RPA complex [28]. First, MEIOB colocalizes with RPA1 and RPA2 as foci in spermatocytes. Second, RPA2 is associated with MEIOB by co-IP in testes. Third, the OB-fold domain of MEIOB is homologous to that of RPA1 [33]. This raises an important question: how is MEIOB associated with the RPA complex? First, by cotransfection in HEK 293T cells followed by co-IP, we found that MEIOB was strongly associated with RPA1 (Figure 4A). Interaction analysis using MEIOB deletion mutants showed that this association was dependent on the OB-fold domain of MEIOB but not the SPATA22-binding domain (Figures 1C and 4A). As expected, the D383A mutation in MEIOB did not block its interaction with RPA1 (Figure 2C). Second, MEIOB was associated with RPA2 in the presence of SPATA22 but not without SPATA22 (Figure 4B). This in vitro result recapitulated the previously reported in vivo MEIOB-RPA2 association in testes, where both MEIOB and SPATA22 were expressed [28]. Finally, MEIOB alone was not associated with RPA3; however, SPATA22 promoted a strong MEIOB-RPA3 interaction (Figure 4C). Taken together, these results demonstrate that MEIOB directly associates with RPA1 through its OB-fold domain but requires SPATA22 to interact with RPA2 and RPA3 (Figure 4D).
Figure 4.
MEIOB and SPATA22 cooperatively interact with RPA. (A) Mapping of the RPA1-binding domain in MEIOB. Co-IP analysis of RPA1 and MEIOB deletions. (B) Co-IP analysis of MEIOB and RPA2 interaction in the presence or absence of SPATA22. (C) Co-IP analysis of MEIOB and RPA3 interaction in the presence or absence of SPATA22. *, antibody heavy chain. (D) Diagram of interactions among MEIOB, SPATA22, and the three RPA subunits. Solid lines indicate strong interactions as assayed by co-IP. Dashed lines indicate interactions only in the presence of both MEIOB and SPATA22. The interactions among RPA1, RPA2, and RPA3 are not shown. RPA1/RPA2/RPA3 or RPA2/RPA3 form soluble complexes when coexpressed in bacteria; however, RPA1/RPA2 or RPA1/RPA3 do not form soluble complexes in bacteria [43].
We next examined the association of SPATA22 with the RPA complex. Like MEIOB, SPATA22 was directly associated with RPA1 (Supplementary Figure S3A). The RPA1-binding domain (aa 59–133) of SPATA22 was different from its MEIOB-binding domain (aa 224–340) (Figure 3A). SPATA22 clearly formed a complex with RPA2 (Supplementary Figure S3B) but did not associate with RPA3 (Supplementary Figure S3C). As neither MEIOB nor SPATA22 alone interacts with RPA3 but the MEIOB-SPATA22 complex does, it is possible that the RPA3-binding interface is only formed in the complex. Even though untagged endogenous RPA1/RPA2/RPA3 are constitutively expressed in HEK 293T cells, the endogenous RPA proteins do not appear to interfere with our co-IP results possibly due to their much lower abundance than the tagged overexpressed RPA proteins. The fact that MEIOB or SPATA22 forms a complex with the tagged RPA1 but not with the tagged RPA3 in transfected cells (Figure 4D) suggests that the endogenous RPA1 could not bridge the complex formation with the overexpressed tagged RPA3.
Recruitment of MEIOB and SPATA22 to double-strand break sites requires their complex formation
Meiotic recombination is initiated by SPO11-catalyzed DSBs [6,7]. Like other recombination proteins, MEIOB forms foci on meiotic chromosomes in early prophase I meiocytes, suggesting that MEIOB localizes to DSBs [28,29]. However, direct evidence of MEIOB recruitment to the DSB sites is lacking. To address this question, we utilized an inducible DSB reporter system in U2OS cells, which consists of a mCherry-LacI-fused FokI endonuclease and a LacO-repeat containing transgene element [34]. Upon administration of 4-OHT and Shield-1, FokI is expressed, activated, recruited to the LacO-repeat locus, and generates DSB, which is manifested as a single red DSB focus per nucleus (Figure 5A). As a positive control, GFP-RPA1 was mostly (90% of the time) recruited to the single DSB in U2OS cells (Figure 5B and C). MEIOB alone rarely formed foci in U2OS cells, and the rare MEIOB foci did not colocalize with the induced DSBs (Figure 5B and C). Although RPA1 was clearly recruited to DSBs, coexpression of RPA1 and MEIOB did not restore the MEIOB foci formation or recruitment of MEIOB to DSBs, suggesting that MEIOB-RPA1 interaction is not sufficient for DSB recruitment (Figure 5B and C). However, when SPATA22 was coexpressed, GFP-MEIOB formed discrete foci in cells, one of which nearly always (90%) overlapped with the DSB focus (Figure 5B and C). Likewise, GFP-SPATA22 alone formed aggregates that did not colocalize with the DSB focus. With the coexpression of MEIOB, GFP-SPATA22 formed discrete foci, one of which overlapped with the DSB focus (Figure 5C). These results demonstrate that MEIOB and SPATA22 are robustly recruited to induced DSBs when coexpressed. MEIOB or SPATA22 interacts with RPA1 but neither alone is recruited to DSBs in U2OS cells, suggesting that the endogenous RPA1 in U2OS cells is not sufficient to recruit either protein to DSBs.
We examined the effect of the interaction-blocking mutation in MEIOB on DSB recruitment. GFP-MEIOB D383A failed to form DSB-colocalizing foci by itself or with the coexpression of SPATA22 (Figure 5B and C). This result shows that the complex formation of MEIOB and SPATA22 is required for recruitment to DSB sites.
Discussion
In this study, we have comprehensively characterized the interactions between MEIOB and SPATA22 as well as with the RPA complex, providing mechanistic insights into their role in meiotic recombination. Our results demonstrate that MEIOB and SPATA22, two meiosis-specific proteins, strongly interact with each other, suggesting that they form a heterodimer or a hetero-oligomer. Multiple lines of evidence support the heterodimer/oligomer formation of MEIOB and SPATA22: co-IP in testes [28], interdependent protein stability in testes [28,31], colocalization on meiotic chromosomes [28,30,31], co-IP and interdependent stability in a heterologous system (HEK 293T somatic cells, this study). Meiotic recombination proteins often function as heterodimers such as RAD51/DMC1 [11], HOP2/MND1 [37], MSH4/MSH5 [16], and MLH1/MLH3 [22]. RAD51/DMC1 recombinases form filaments on resected 3΄ ssDNA ends and drive strand invasion into the homologous chromosome [11,38]. The HOP2/MND1 heterodimer interacts with RAD51/DMC1 and augments their recombinase activities [37]. MSH4/MSH5 stabilizes invasion intermediates and Holliday junctions [16]. MLH1/MLH3 is a MSH2/MSH3-stimulated endonuclease and is required for formation of most meiotic crossovers [17,22]. Our previous studies suggest that MEIOB/SPATA22 may function in the second end capture after strand invasion [28]. Study of a Spata22 rat mutant suggests that SPATA22 is important for the maintenance of RAD51 foci on meiotic chromosomes [30]. Our data support that MEIOB and SPATA22 form a new meiosis-specific heterodimer. Future biochemical experiments will be needed to determine the role of the MEIOB/SPATA22 heterodimer during meiotic recombination and its relationship with the above mentioned other meiosis-specific heterodimers.
The MEIOB-SPATA22 interaction exhibits unique characteristics. We have identified the discrete interaction domains in both proteins. Unusually, a series of small deletions within the interaction domains of either protein abolished their interaction, suggesting that their binding is synergistic and that disruption anywhere along the binding interface may lead to the collapse of the complex. In support of this notion, strikingly, a single amino acid D383 in the SPATA22-binding domain of MEIOB is essential for the interaction. Mutation studies showed that both charge and structure at residue D383 are important. The majority of binding energy required for protein-protein complex formation usually involves only a few key residues within an interaction hot spot [39]. Therefore, residue D383 is a critical residue presumably located in such an interaction hot spot. As an alternative explanation, the small deletions in the binding domains and the D383A mutation could result in misfolding of the respective binding domains. The precise role of D383 and the boundaries of the binding domains may be revealed once the crystal structure of the MEIOB-SPATA22 complex is solved in the future.
Both MEIOB and SPATA22 interact with RPA1. Notably, the RPA1-binding domains in MEIOB and SPATA22 are separate from their dimerization domain, suggesting that they are capable of interacting with RPA1 as a heterocomplex (Figures 1C and 3A). Furthermore, neither MEIOB nor SPATA22 alone interacts with RPA3, but they do when both are present. These results support the notion that MEIOB and SPATA22 interact with the RPA complex in a cooperative manner. Based on these extensive interaction studies, we hypothesize that the MEIOB-SPATA22 heterodimer interacts with the RPA heterotrimeric complex in vivo (Figure 4D). Interestingly, RPA still forms foci on meiotic chromosomes in the absence of MEIOB or SPATA22, showing that the formation and localization of the RPA complex are independent of MEIOB and SPATA22 [28–31]. To address whether the focal localization of MEOB/SPATA22 requires RPA, conditional inactivation of RPA1 in meiotic germ cells is required in future studies, since global inactivation of Rpa1 is embryonic lethal [40]. It is also possible that there are in vivo subcomplexes of different combinations such as MEIOB/RPA2/RPA3, MEIOB/SPATA22/RPA2, MEIOB/SPATA22/RPA3, etc. Future biochemical experiments on testes and an Rpa1 conditional knockout mutant will be needed to test this possibility. Both MEIOB and RPA1 bind to ssDNA. We find that the RPA1-binding domain and ssDNA-binding domain in MEIOB overlap with each other. This raises the possibility that binding of RPA with the MEIOB/SPATA22 complex may dislodge the complex from ssDNA or vice versa.
Biochemically, MEIOB possesses ssDNA-binding activity [28,29]. Many ssDNA-binding proteins are recruited to the DSB sites in germ cells, including RPA, RAD51, and DMC1 [41]. MEIOB colocalizes with RPA as foci on meiotic chromosomes, showing that MEIOB is recruited to DSBs in germ cells. Here we show that, in human bone osteosarcoma (U2OS) cell line (somatic cells), MEIOB and SPATA22 are recruited to DSBs as a complex but not alone. The MEIOB-SPATA22 complex localizes to DSBs possibly through direct binding to ssDNA or interaction with other proteins at DSBs. Furthermore, our result from U2OS cells suggests that the recruitment of the MEIOB-SPATA22 complex to DSBs may not require additional meiosis-specific proteins in vivo.
Study of MEIOB/SPATA22 may have translational implications. MEIOB is a cancer-testis gene and plays a role in tumorigenesis in human lung adenocarcinoma [42]. MEIOB could be a cancer immunotherapy target, since it is not present in normal somatic cells. Given the specific requirement of MEIOB and SPATA22 for fertility in rodents (mouse and rat) [28–32], mutations in either gene in humans are expected to cause sterility. The unusual interaction properties between MEIOB and SPATA22 may provide opportunities for the development of protein–protein interaction inhibitors with potential as male contraceptive agents. Such contraceptives are expected to be reversible with minimal side effects, since both MEIOB and SPATA22 are only present in meiotic germ cells.
Supplementary data
Supplementary data are available at BIOLRE online.
Supplementary Figure S1. Additional western blot images for identification of the SPATA22-interaction domain in MEIOB. (A and B) Co-IP analysis of the remaining 11 MEIOB deletions (Figure 1C) and SPATA22. *, antibody heavy chain.
Supplementary Figure S2. Additional western blot images for identification of the MEIOB-interaction domain in SPATA22. (A and B) Co-IP analysis of the remaining eight SPATA22 deletions (Figure 3A) and MEIOB.
Supplementary Figure S3. Associations between SPATA22 and the RPA1-RPA2-RPA3 trimer. (A–C) Co-IP analysis of the interactions between SPATA22 and RPA1 (A), RPA2 (B), and RPA3 (C). Co-IP results of interactions between SPATA22 variants and RPA1 are summarized in Figure 3A. *, antibody heavy chain. **, this band (∼37 kDa) is likely due to translation initiation from an internal ATG codon in SPATA22. This band also appears in Figure 3B.
Table S1. List of antibodies.
Supplementary data are available at BIOLRE online.
Supplementary Figure S1. Additional western blot images for identification of the SPATA22-interaction domain in MEIOB. (A and B) Co-IP analysis of the remaining 11 MEIOB deletions (Figure 1C) and SPATA22. *, antibody heavy chain.
Supplementary Figure S2. Additional western blot images for identification of the MEIOB-interaction domain in SPATA22. (A and B) Co-IP analysis of the remaining eight SPATA22 deletions (Figure 3A) and MEIOB.
Supplementary Figure S3. Associations between SPATA22 and the RPA1-RPA2-RPA3 trimer. (A–C) Co-IP analysis of the interactions between SPATA22 and RPA1 (A), RPA2 (B), and RPA3 (C). Co-IP results of interactions between SPATA22 variants and RPA1 are summarized in Figure 3A. *, antibody heavy chain. **, this band (∼37 kDa) is likely due to translation initiation from an internal ATG codon in SPATA22. This band also appears in Figure 3B.
Table S1. List of antibodies.
Acknowledgments
We thank members of the Wang laboratory for comments on the manuscript.
Authors’ contributions: YX carried out the experiments. ES performed the structure modeling. YX, RAG, and PJW analyzed data. PJW and YX wrote the manuscript. All the authors were involved in the discussion and commented on the manuscript.
References
- 1. Baudat F, Imai Y, de Massy B. Meiotic recombination in mammals: localization and regulation. Nat Rev Genet 2013; 14:794–806. [DOI] [PubMed] [Google Scholar]
- 2. Handel MA, Schimenti JC. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat Rev Genet 2010; 11:124–136. [DOI] [PubMed] [Google Scholar]
- 3. Zickler D, Kleckner N. Recombination, Pairing, and synapsis of homologs during meiosis. Cold Spring Harb Perspect Biol 2015; 7:10.1101/cshperspect.a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Buard J, de Massy B. Playing hide and seek with mammalian meiotic crossover hotspots. Trends Genet 2007; 23:301–309. [DOI] [PubMed] [Google Scholar]
- 5. Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet 2001; 2:280–291. [DOI] [PubMed] [Google Scholar]
- 6. Baudat F, Manova K, Yuen JP, Jasin M, Keeney S. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol Cell 2000; 6:989–998. [DOI] [PubMed] [Google Scholar]
- 7. Romanienko PJ, Camerini-Otero RD. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell 2000; 6:975–987. [DOI] [PubMed] [Google Scholar]
- 8. Lange J, Yamada S, Tischfield SE, Pan J, Kim S, Zhu X, Socci ND, Jasin M, Keeney S. The landscape of mouse meiotic double-strand break formation, processing, and repair. Cell 2016; 167:695–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem 1997; 66:61–92. [DOI] [PubMed] [Google Scholar]
- 10. Neale MJ, Keeney S. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature 2006; 442:153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cloud V, Chan YL, Grubb J, Budke B, Bishop DK. Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science 2012; 337:1222–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brown MS, Bishop DK. DNA strand exchange and RecA homologs in meiosis. Cold Spring Harb Perspect Biol 2014; 7:a016659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang F, Wang PJ. The Mammalian synaptonemal complex: a scaffold and beyond. Genome Dyn 2009; 5:69–80. [DOI] [PubMed] [Google Scholar]
- 14. Fraune J, Schramm S, Alsheimer M, Benavente R. The mammalian synaptonemal complex: protein components, assembly and role in meiotic recombination. Exp Cell Res 2012; 318:1340–1346. [DOI] [PubMed] [Google Scholar]
- 15. Kneitz B, Cohen PE, Avdievich E, Zhu L, Kane MF, Hou H Jr, Kolodner RD, Kucherlapati R, Pollard JW, Edelmann W. MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev 2000; 14:1085–1097. [PMC free article] [PubMed] [Google Scholar]
- 16. Snowden T, Acharya S, Butz C, Berardini M, Fishel R. hMSH4-hMSH5 recognizes Holliday Junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol Cell 2004; 15:437–451. [DOI] [PubMed] [Google Scholar]
- 17. Kolas NK, Svetlanov A, Lenzi ML, Macaluso FP, Lipkin SM, Liskay RM, Greally J, Edelmann W, Cohen PE. Localization of MMR proteins on meiotic chromosomes in mice indicates distinct functions during prophase I. J Cell Biol 2005; 171:447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang F, Gell K, van der Heijden GW, Eckardt S, Leu NA, Page DC, Benavente R, Her C, Hoog C, McLaughlin KJ, Wang PJ. Meiotic failure in male mice lacking an X-linked factor. Genes Dev 2008; 22:682–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Baker SM, Plug AW, Prolla TA, Bronner CE, Harris AC, Yao X, Christie DM, Monell C, Arnheim N, Bradley A, Ashley T, Liskay RM. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat Genet 1996; 13:336–342. [DOI] [PubMed] [Google Scholar]
- 20. Edelmann W, Cohen PE, Kane M, Lau K, Morrow B, Bennett S, Umar A, Kunkel T, Cattoretti G, Chaganti R, Pollard JW, Kolodner RD et al. Meiotic pachytene arrest in MLH1-deficient mice. Cell 1996; 85:1125–1134. [DOI] [PubMed] [Google Scholar]
- 21. Lipkin SM, Moens PB, Wang V, Lenzi M, Shanmugarajah D, Gilgeous A, Thomas J, Cheng J, Touchman JW, Green ED, Schwartzberg P, Collins FS et al. Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat Genet 2002; 31:385–390. [DOI] [PubMed] [Google Scholar]
- 22. Rogacheva MV, Manhart CM, Chen C, Guarne A, Surtees J, Alani E. Mlh1-Mlh3, a meiotic crossover and DNA mismatch repair factor, is a Msh2-Msh3-stimulated endonuclease. J Biol Chem 2014; 289:5664–5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. de los Santos T, Hunter N, Lee C, Larkin B, Loidl J, Hollingsworth NM. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics 2003; 164:81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Holloway JK, Booth J, Edelmann W, McGowan CH, Cohen PE. MUS81 generates a subset of MLH1-MLH3-independent crossovers in mammalian meiosis. PLoS Genet 2008; 4:e1000186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ward JO, Reinholdt LG, Motley WW, Niswander LM, Deacon DC, Griffin LB, Langlais KK, Backus VL, Schimenti KJ, O’Brien MJ, Eppig JJ, Schimenti JC. Mutation in mouse hei10, an e3 ubiquitin ligase, disrupts meiotic crossing over. PLoS Genet 2007; 3:e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Qiao H, Prasada Rao HB, Yang Y, Fong JH, Cloutier JM, Deacon DC, Nagel KE, Swartz RK, Strong E, Holloway JK, Cohen PE, Schimenti J et al. Antagonistic roles of ubiquitin ligase HEI10 and SUMO ligase RNF212 regulate meiotic recombination. Nat Genet 2014; 46:194–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Holloway JK, Sun X, Yokoo R, Villeneuve AM, Cohen PE. Mammalian CNTD1 is critical for meiotic crossover maturation and deselection of excess precrossover sites. J Cell Biol 2014; 205:633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Luo M, Yang F, Leu NA, Landaiche J, Handel MA, Benavente R, La Salle S, Wang PJ. MEIOB exhibits single-stranded DNA-binding and exonuclease activities and is essential for meiotic recombination. Nat Commun 2013; 4:2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Souquet B, Abby E, Herve R, Finsterbusch F, Tourpin S, Le Bouffant R, Duquenne C, Messiaen S, Martini E, Bernardino-Sgherri J, Toth A, Habert R et al. MEIOB targets single-strand DNA and is necessary for meiotic recombination. PLoS Genet 2013; 9:e1003784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ishishita S, Matsuda Y, Kitada K. Genetic evidence suggests that Spata22 is required for the maintenance of Rad51 foci in mammalian meiosis. Sci Rep 2014; 4:6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hays E, Majchrzak N, Daniel V, Ferguson Z, Brown S, Hathorne K, La Salle S. Spermatogenesis associated 22 is required for DNA repair and synapsis of homologous chromosomes in mouse germ cells. Andrology 2017; 5:299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. La Salle S, Palmer K, O’Brien M, Schimenti JC, Eppig J, Handel MA. Spata22, a novel vertebrate-specific gene, is required for meiotic progress in mouse germ cells. Biol Reprod 2012; 86:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ribeiro J, Abby E, Livera G, Martini E. RPA homologs and ssDNA processing during meiotic recombination. Chromosoma 2016; 125:265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, Mer G, Greenberg RA. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat Struct Mol Biol 2013; 20:317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 2015; 10:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fan J, Pavletich NP. Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev 2012; 26:2337–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Petukhova GV, Pezza RJ, Vanevski F, Ploquin M, Masson JY, Camerini-Otero RD. The Hop2 and Mnd1 proteins act in concert with Rad51 and Dmc1 in meiotic recombination. Nat Struct Mol Biol 2005; 12:449–453. [DOI] [PubMed] [Google Scholar]
- 38. Bishop DK. RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell 1994; 79:1081–1092. [DOI] [PubMed] [Google Scholar]
- 39. Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science 1995; 267:383–386. [DOI] [PubMed] [Google Scholar]
- 40. Wang Y, Putnam CD, Kane MF, Zhang W, Edelmann L, Russell R, Carrion DV, Chin L, Kucherlapati R, Kolodner RD, Edelmann W. Mutation in Rpa1 results in defective DNA double-strand break repair, chromosomal instability and cancer in mice. Nat Genet 2005; 37:750–755. [DOI] [PubMed] [Google Scholar]
- 41. Moens PB, Kolas NK, Tarsounas M, Marcon E, Cohen PE, Spyropoulos B. The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA-DNA interactions without reciprocal recombination. J Cell Sci 2002; 115:1611–1622. [DOI] [PubMed] [Google Scholar]
- 42. Wang C, Gu Y, Zhang K, Xie K, Zhu M, Dai N, Jiang Y, Guo X, Liu M, Dai J, Wu L, Jin G et al. Systematic identification of genes with a cancer-testis expression pattern in 19 cancer types. Nat Commun 2016; 7:10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Henricksen LA, Umbricht CB, Wold MS. Recombinant replication protein A: expression, complex formation, and functional characterization. J Biol Chem 1994; 269:11121–11132. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data are available at BIOLRE online.
Supplementary Figure S1. Additional western blot images for identification of the SPATA22-interaction domain in MEIOB. (A and B) Co-IP analysis of the remaining 11 MEIOB deletions (Figure 1C) and SPATA22. *, antibody heavy chain.
Supplementary Figure S2. Additional western blot images for identification of the MEIOB-interaction domain in SPATA22. (A and B) Co-IP analysis of the remaining eight SPATA22 deletions (Figure 3A) and MEIOB.
Supplementary Figure S3. Associations between SPATA22 and the RPA1-RPA2-RPA3 trimer. (A–C) Co-IP analysis of the interactions between SPATA22 and RPA1 (A), RPA2 (B), and RPA3 (C). Co-IP results of interactions between SPATA22 variants and RPA1 are summarized in Figure 3A. *, antibody heavy chain. **, this band (∼37 kDa) is likely due to translation initiation from an internal ATG codon in SPATA22. This band also appears in Figure 3B.
Table S1. List of antibodies.