Mammalian fertility is tightly regulated by a SHOC1-SPO16 complex, an XPF-ERCC1–like complex, to promote homologous recombination.
Abstract
Segregation of homologous chromosomes in meiosis I is tightly regulated by their physical links, or crossovers (COs), generated from DNA double-strand breaks (DSBs) through meiotic homologous recombination. In budding yeast, three ZMM (Zip1/2/3/4, Mer3, Msh4/5) proteins, Zip2, Zip4, and Spo16, form a “ZZS” complex, functioning to promote meiotic recombination via a DSB repair pathway. Here, we identified the mammalian ortholog of Spo16, termed SPO16, which interacts with the mammalian ortholog of Zip2 (SHOC1/MZIP2), and whose functions are evolutionarily conserved to promote the formation of COs. SPO16 localizes to the recombination nodules, as SHOC1 and TEX11 do. SPO16 is required for stabilization of SHOC1 and proper localization of other ZMM proteins. The DSBs formed in SPO16-deleted meiocytes were repaired without COs formation, although synapsis is less affected. Therefore, formation of SPO16-SHOC1 complex–associated recombination intermediates is a key step facilitating meiotic recombination that produces COs from yeast to mammals.
INTRODUCTION
Meiosis is a specialized type of cell division that happens in both males and females to give rise to haploid gametes through one round of DNA replication and two rounds of cell divisions. Precise segregation of homologous chromosomes (homologs) in meiosis I is ensured by the interhomolog crossovers (COs), which are selectively generated from DNA double-strand breaks (DSBs) by meiotic homologous recombination (1, 2). In mammalian meiosis, DSBs are generated by SPO11 early in meiotic prophase I and rapidly resected to form 3′ single-stranded DNA (3′-ssDNA) overhangs, which are coated with RAD51/DMC1 recombinases, as well as the ssDNA binding proteins, replication protein A (RPA) proteins (3, 4). RPA proteins interact with two additional meiosis-specific proteins, MEIOB and SPATA22, to promote meiotic recombination (5–7). Through complicated and strictly regulated processes of strand invasion (which generates displacement loops, or D loops), single-end invasion (SEI), second-end capture, and double Holliday junction (dHJ) formation and resolution, only a small proportion (approximately 10%) of DSBs finally become COs. However, each homologous chromosome pair acquires at least one CO (8). Errors in CO formation and distribution result in germline loss, aneuploidy, and embryonic development abnormalities, which contribute to diseases related to miscarriage and human infertility (9, 10).
ZMM proteins are a group of functionally related proteins known for their roles in promoting formation of COs and synaptonemal complex (SC) in budding yeast, mammals, and plants (1, 11). At least eight members of ZMM proteins have been identified in budding yeast (Saccharomyces cerevisiae): Zip1/2/3/4, Mer3, Msh4/5, and Spo16 (11, 12). Mutations involving these genes result in defects in the formation of both COs and SC, which result in zipper-like structures between homologous pairs. Except for the transverse filament Zip1, which assembles the central element of SC, ZMM proteins are localized to the designated COs and functions collaboratively to promote CO assurance (each homolog receives at least one CO) and interference (COs are properly distributed) in yeast (12, 13). Similarly, insufficient CO formation was also observed in mouse models lacking the evolutionally conserved Zmm orthologs: Shoc1 (ortholog of Zip2), Rnf212 (ortholog of Zip3), Hei10 (Rnf212 paralog with domain similarity), Tex11 (ortholog of Zip4), Hfm1 (ortholog of Mer3), Msh4, and Msh5.
In budding yeast, the CO/NCO (non-CO) decision occurs as early as at leptotene-zygotene transition, along with the formation of CO-correlated SEIs, which later become dHJs and exclusively give rise to COs, while the other DSBs are repaired through a synthesis-dependent strand annealing (SDSA) pathway (1, 14). Zip2, Zip4, and Spo16 form a trimeric ZZS complex in budding yeast (15). In this complex, Zip2 and Spo16 form an XPF-ERCC1–like dimeric complex, although without a functional endonuclease activity, and Zip2 interacts with a third protein, Zip4, to recruit additional factors to promote CO formation. During yeast meiosis, the ZZS complex binds and stabilizes early recombination intermediates to facilitate the formation of CO-specific SEIs (16). Similarly in thale cress, SHOC1-PTD complex, an XPF-ERCC1–like complex, regulates meiotic recombination (17). The mammalian ortholog of Zip2, or SHOC1/MZIP2, has been characterized recently (18, 19). However, the mammalian ortholog of Spo16 remains elusive.
In this study, we identified the mammalian ortholog of Spo16 and characterized its functions in complex with SHOC1 in meiotic recombination in mammals. Deletion of SPO16 leads to nonhomologous chromosome pairing and insufficient CO formation in both spermatocytes and female primordial germ cells (PGCs). SPO16 has an XPF-like domain and interacts with SHOC1. Similar to SHOC1, SPO16 forms discrete foci on chromosomal axes from leptotene stage to early-pachytene stage. However, compared to meiocytes null for SHOC1, SPO16-null meiocytes exhibited milder defects in DNA repair and synapsis, suggesting that SHOC1 alone is partially functional and DSBs are partially repaired. Together with previous studies, our results suggest that ZMM proteins and their functions in meiotic recombination are extremely conserved from yeast to humans.
RESULTS
Identification of mammalian SPO16
In an effort to gain understanding of meiotic prophase I, we analyzed the previously reported single-cell transcriptome profiling results obtained from human PGCs (20). We sorted a list of 342 genes by strict criteria for their specific and high expression in 17-week-old female PGCs, which correspond to meiotic prophase I (table S1). Among these, at least 68 genes are well characterized as their protein products being involved in meiosis, including components of the SC (SYCP1/2/3, SYCE1/2/3, and SIX6OS1), proteins in homologous recombination (SPO11, HORMAD1/2, RAD51, MEIOB, and SPATA22), ZMM proteins (HFM1, RNF212, and MSH5), and telomeric proteins such as SPDYA, MAJIN, and TERB2.
C1orf146 is one of the most differentially expressed genes and is conserved in almost all metazoans, except for nematodes (fig. S1A). 1700028k03rik is the mouse ortholog of human C1orf146 and encodes a protein of 186 amino acids. To gain insights into the possible functions of proteins encoded by C1orf146 orthologs, we did a homology search using HHpred. As shown in table S2, protein encoded by 1700028k03rik exhibited high similarities to a group of XPF-like proteins, such as HsFAAP24, ApXPF, HsFAMCM, HsERCC1, HsEME1, and ScZip2. These proteins have an active or inactive XPF-like domain to form heterodimers (MUS81-EME1/2 complexes, FANCM-FAAP24 complex, XPF-ERCC1 complex, and Zip2-Spo16 complex) or homodimer (ApXPF). Zip2-Spo16 complex is an XPF-ERCC1–like heterodimer that is specific to meiotic recombination in budding yeast. While the mouse ortholog of Zip2 (SHOC1) has been characterized recently, the ortholog of Spo16 remains elusive (18, 19). We postulated that protein encoded by 1700028k03rik might be the ortholog of Spo16, termed SPO16. Prediction of its secondary structure by online tools, PSIPRED and I-TASSER, showed that MmSPO16 had a secondary structure similar to ApXPF and ScSPO16 (fig. S1, B to D). Similar to the other XPF-like proteins, MmSPO16 contains a central domain and a HhH domain (16). The central domain of MmSPO16 had the same arrangement of α helices and β-strands as ApXPF and ScZip2, while the HhH domain was made up of three α helices, which is similar to ScSPO16 (fig. S1, B to D). When overexpressed in cell lines, SPO16 interacted with the XPF-like domain (amino acids 975 to 1156) of mouse SHOC1 (Fig. 1, A and B), strongly suggesting that SPO16 is the mammalian ortholog of ScSpo16 and AtPTD.
Fig. 1. Identification of mammalian SPO16.
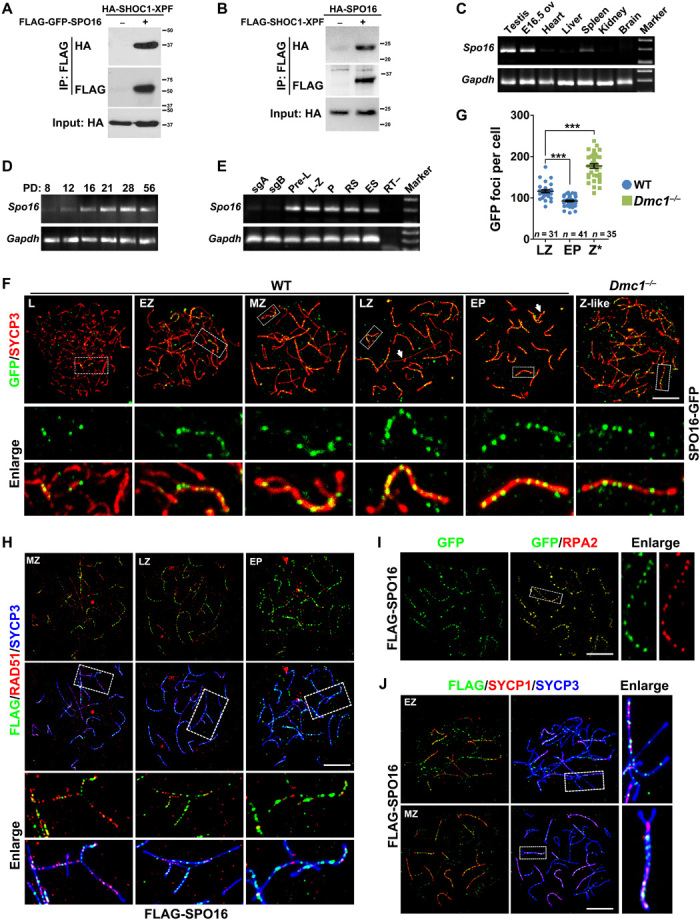
(A and B) Immunoprecipitation (IP) experiments showing the interaction of SPO16 with the XPF-like domain of SHOC1 (amino acids 975 to 1156). HA, hemagglutinin. (C to E) Levels of Spo16 mRNA in multiple mouse tissues (C), in developing testes (D), and in male germ cells at different stages (E). Gapdh served as the loading control. sgA, spermatogonia type A; sgB, spermatogonia type B; pre-L, preleptonema; L-Z, leptonema to zygonema; P, pachynema; RS, round spermatids; ES, elongated spermatids; ov, ovary. (F) Immunofluorescent staining of exogenously expressed, GFP-tagged SPO16 protein (green) and SYCP3 (red) on the nuclear surface spreads of WT or Dmc1−/− testes electroporated with plasmids encoding SPO16-GFP. SYCP3 marks the meiotic chromosome axes. Arrows indicate the sex bodies. Enlarged images show the partially or fully synapsed homologous pairs and the regions of which are bordered with a dashed line. L, leptotene; EZ, early zygonema; MZ, midzygonema; LZ, late zygonema; EP, early pachynema; Z-like, zygonema-like. Scale bar, 10 μm. (G) Quantification of GFP foci detected in WT and Dmc1−/− spermatocytes at indicated stages. Z*, zygonema-like. Numbers of spermatocytes analyzed (n) are indicated. Median focus numbers are marked. Error bar indicates SEM. ***P < 0.001 by two-tailed Student’s t tests. (H) Costaining of FLAG-SPO16 (green) with RAD51 (red) on the nuclear surface spreads of WT testes electroporated with plasmids encoding FLAG-SPO16. RAD51 marks early recombination nodules. Scale bar, 10 μm. (I) Costaining of SPO16-GFP (green) with RPA2 (red) on nuclear spreads prepared from WT testes electroporated with plasmids encoding SPO16-GFP. Scale bar, 10 μm. (J) Costaining of FLAG-SPO16 (green) with SYCP1 (red) on the nuclear surface spreads of WT testes electroporated with plasmids encoding FLAG-SPO16. Scale bar, 10 μm.
The mouse gene Spo16, or 1700028k03rik, is localized to chromosome 5 and predicted to be of six exons. Polymerase chain reaction (PCR) amplification and sequencing of the full-length mouse Spo16 complementary DNA (cDNA) from mouse testis samples and a testis cDNA library confirmed the predicted sequence. Real-time PCR results of multiple mouse tissues showed that Spo16 is specifically expressed in adult testes and in embryonic ovaries at embryonic day 16.5 (E16.5), which contain abundant meiotic cells (Fig. 1C). More specifically, the Spo16 mRNA was only expressed in spermatocytes that entered meiosis (Fig. 1, D and E).
SPO16 is located to chromosome axes
We cloned the cDNA encoding SPO16 and tagged it with the green fluorescent protein (GFP) or FLAG peptides. When overexpressed in wild-type (WT) testes, SPO16 localized to chromosome axes (shown by SYCP3 staining) as discrete foci and in a stage-dependent manner (Fig. 1, F and G, and fig. S2A). We detected very few foci in leptonema spermatocytes, but the number of foci gradually increased with the progression of synapsis from leptonema to late zygonema (Fig. 1G). An average number of 116 ± 3.39 (n = 31) foci was found in spermatocytes at late-zygotene stage and was slightly decreased to 92 ± 1.93 (n = 41) at early pachynema when full synapsis is achieved (Fig. 1, F and G). The immunostaining pattern of SPO16 appears later than that of early recombination markers (RAD51 and DMC1) but similarly to the known ZMM proteins such as TEX11 (5, 6, 21). Furthermore, SPO16 was partially colocalized with TEX11 in zygonema (55.3 ± 1.97% colocalization; n = 6) and in early-pachynema (63.9 ± 5.75% colocalization; n = 6) spermatocytes (fig. S2B).
Notably, we detected most of the SPO16 foci at synapsed regions (Fig. 1F and fig. S2A). Dmc1−/− spermatocytes were arrested at a zygotene-like stage without synapsis (4). In Dmc1−/− spermatocytes, we found SPO16 foci on the unsynapsed chromosome axes, the number of which was close to the number of initial DSBs and is greater than the number of ZMM-associated dHJs in WT spermatocytes (Fig. 1, F and G). During DNA repair in mitosis, which lacks DMC1, or in yeast with increased RAD51 activity, D loops are preferentially formed between sister chromatids (22, 23). We postulated that these SPO16 foci in Dmc1−/− spermatocytes might be the sites of inter–sister chromatid joint molecules. These SPO16 foci could not be resected DSBs because SPO16 was less colocalized with RAD51, which marks resected 3′-ssDNAs and the transiently formed D loops at unsynapsed regions (Fig. 1H). Consistent with these observations, SPO16 colocalized with RPA2, a component of RPA complex that marks the sites of middle recombination intermediates at the early-pachytene stage, in WT spermatocytes (Fig. 1I).
In WT testes, spermatocytes developed through leptotene, zygotene, pachytene, and diplotene stages, with the assembly and disassembly of SC from the paired homologs. SYCP1 is the central element of SC. When stained for SYCP1, the transverse filaments of SC central element that represents synapsed regions, we observed a portion of SPO16 foci on the regions without SYCP1 staining (Fig. 1J), indicating that SPO16 forms foci before completion of synapsis. While SPO16 foci were frequently observed at early pachynema on nuclear spreads of testes electroporated with GFP-SPO16, SPO16 foci were never seen in late-pachynema spermatocytes that were positive for MLH1 (n > 200), suggesting the removal of SPO16 from late recombination intermediates (fig. S2C).
Knockout of SPO16 causes sterility in both males and females
To investigate the physiological functions of SPO16 in mammalian meiosis, we generated Spo16 knockout using CRISPR-Cas9 techniques. As shown in Fig. 2A, the translation start site (ATG) of Spo16 is located in exon 2, while the stop codon is located in exon 6. We targeted the exon 3 by a single-guide RNA (sgRNA; Fig. 2A). Following microinjection and embryo transfer, we identified one null allele of Spo16. This allele contains a 19–base pair (bp) insertion in exon 3, which causes a frameshift and creates a premature stop codon (Fig. 2B and fig. S2D). To rule out the possibility of off-target effects, we backcrossed the founder male to WT females continuously for three generations. After that, heterozygous (Spo16+/−) males were crossed to heterozygous females to obtain homozygous (Spo16 −/−) mice. The mutation was further confirmed by sequencing of genomic DNA extracted from heterozygous (+/−) and homozygous knockout (−/−) mice (fig. S2E). We detected Spo16 mRNA at a relatively lower level in Spo16−/− testes (fig. S2F). Spo16+/− and Spo16−/− pups were obtained according to Mendelian ratios, and no overt defects were observed, suggesting that SPO16 is not required for viability or development in mice (fig. S2, G and H). However, both Spo16−/− males and females were sterile, whereas Spo16+/− mice exhibited normal fertility (fig. S2I).
Fig. 2. Deletion of Spo16 led to massive germline loss and infertility in both males and females.
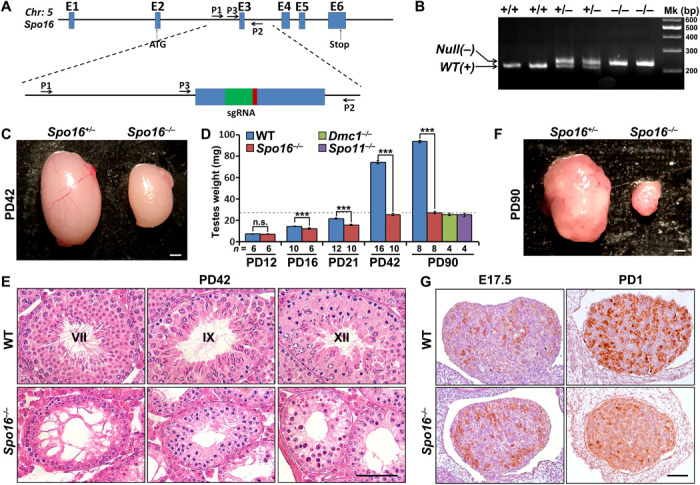
(A) Schematic diagram of the CRISPR-Cas9 strategy to generate null allele for Spo16. Exons, sgRNA, and primers are indicated. The detailed sequence is shown in fig. S2D. Chr, chromosomes. (B) Genotyping results to distinguish the WT (+) allele and the null allele (−), which contains 19-bp insertion. Mk, DNA marker. (C) Representative image of testes derived from Spo16+/− and Spo16−/− males at the age of PD42. (D) Weights of testes derived from WT and Spo16−/− males at indicated ages. Numbers of testes analyzed (n) are indicated. Error bars indicate SEM. n.s., not significant. Dashed line shows the weight of the knockout testes at PD90. ***P < 0.001 by two-tailed Student’s t tests. (E) H&E staining results of paraffin-embedded testes from WT and Spo16−/− males. Stages of seminiferous tubules in control testes are indicated. Scale bar, 50 μm. (F) Morphology of Spo16+/− and Spo16−/− ovaries at PD42. (G) Immunohistochemistry (IHC) staining of MVH showing the oocytes in WT and Spo16−/− ovaries at E17.5 and PD1. Scale bar, 50 μm.
Mature Spo16−/− males had smaller testes and did not produce sperm (Fig. 2, C and D, and fig. S3A). We observed the difference in testis size as early as postnatal day 16 (PD16), when the first wave of spermatocytes reached the early-pachytene stage (Fig. 2D). A reduction of Spo16−/− testis size was seen at PD42 and PD90, approximately 70% smaller than WT testes by PD90 (Fig. 2D). This size difference is similar to the meiosis initiation–defective Spo11−/− and the recombination-defective Dmc1−/− testes (Fig. 2D). Hematoxylin and eosin (H&E) staining of testes derived from WT and Spo16−/− males at PD42 showed arrest of spermatogenesis at meiotic prophase I, as well as massive apoptosis, which was confirmed by immunostaining of cleaved caspase 3 on sections prepared with PD21 testes (Fig. 2E and fig. S3, A to C). Immunostaining of PLZF (promyelocytic leukaemia zinc finger; a marker of undifferentiated spermatogonia) and MVH (mammalian homolog of vasa; a marker of germ cells) ruled out the possibility of spermatogonia loss or defective meiosis entry in Spo16 −/− testes (fig. S3, D and E). Smaller ovaries and premature ovarian failure were found in adult Spo16 −/− females, which indirectly caused obesity (six of eight) in 3-month-old females (Fig. 2F and fig. S2H). Massive oocyte loss was found in Spo16 −/− ovaries between E17.5 and PD1, due to ectopic DNA damage signaling as shown by γH2AX immunofluorescence staining (Fig. 2G and fig. S3, F to H).
SPO16 deficiency leads to asynapsis and nonhomologous pairing
Synapsis is one of the hallmarks of meiotic prophase I progression. We prepared nuclear surface spreads with adult testes (PD42) or embryonic ovaries (E17.5) derived from WT and Spo16−/− mice and stained these with the synapsed chromosome marker SYCP1 and the unsynapsed chromosome marker HORMAD1. At pachynema, SYCP1 assembled to the full length between homologs except for the unpaired regions in the X-Y chromosome pair (or sex body), whereas HORMAD1 was removed from chromosome axes and only retained on the unpaired regions of sex body (Fig. 3, A to C). Complete synapsis was never seen in Spo16−/− spermatocytes. Instead, incomplete synapsis in Spo16−/− spermatocytes resulted in two stages, equivalent to late-zygotene and early-pachytene stages, namely, zygotene-like and pachytene-like stages, respectively (Fig. 3, A to C). The zygotene-like stage was characterized by partial pairing and widely distributed unsynapsed zipper-like chromosome forks, while the pachytene-like stage represented spermatocytes with most of the chromosomes well paired except for one “multichromosome structure” (Fig. 3, B and C). Similarly, SPO16-deleted female PGCs were arrested at a zygotene-like stage due to failure in SC assembly to the full length along chromosome pairs (Fig. 3, D to F). As shown in fig. S4, A and B, the defects in synapsis were confirmed by staining of another central element of SC, SIX6OS1, on nuclear surface spreads, as well as SYCP1 staining on sections (24).
Fig. 3. Meiocytes null for SPO16 failed to achieve complete synapsis.
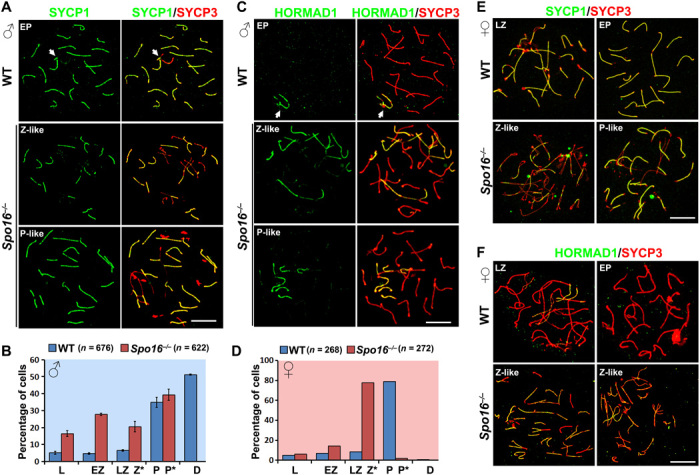
(A) Staining of the synapsed chromosome marker SYCP1 (green) with SYCP3 (red) on the nuclear surface spreads of spermatocytes derived from WT and Spo16−/− males at PD42. P-like, pachynema-like. Scale bar, 10 μm. (B) Meiotic prophase progression in WT and Spo16−/− testes. Error bars indicate SEM. P*, pachynema-like. The numbers of spermatocytes analyzed (n) are indicated. (C) Staining of the unsynapsed chromosome marker HORMAD1 (green) with SYCP3 (red) on the nuclear surface spreads of spermatocytes derived from WT and Spo16−/− males at PD42. Scale bar, 10 μm. (D) Meiotic prophase I progression of female PGCs in WT and Spo16−/− ovaries. The numbers of spermatocytes analyzed (n) are indicated. (E and F) SYCP1 (E) and HORMAD1 (F) were stained together with SYCP3 on the nuclear surface spreads of female PGCs derived from WT and Spo16−/− ovaries at E17.5. Scale bars, 10 μm.
The multichromosome structure was found in 98.9% of pachytene-like Spo16 −/− spermatocytes (n = 620). Chromosomes in this structure paired to more than one chromosome, which suggested nonhomologous pairing in spermatocytes null for SPO16. Nonhomologous pairing was also reported in Msh4 and Msh5 knockout mice, indicating functional relatedness of SPO16 and ZMM proteins (25, 26). We also stained nuclear surface spreads and testis sections for γH2AX, which is a marker of DNA damage signaling. γH2AX level detected in Spo16 −/− leptonema spermatocytes was comparable to that in WT leptonema sprmatocytes and was removed with the repair of DSBs and pairing of chromosomes in late-zygonema WT spermatocytes and zygonema-like Spo16 −/− spermatocytes (fig. S4, C and D). Similar to the localization of γH2AX only to sex body at pachynema, γH2AX staining was restricted to the multichromosome structure found in pachytene-like Spo16 −/− spermatocytes (Fig. 4A and fig. S4, C and D). This structure is similar to the “pseudosex body” (PSB) formed in Spo11−/− spermatocytes (27). We quantified an average number of 6.29 chromosomes within these PSBs (Fig. 4, A and B). Nonhomologous pairing and asynapsis in pachytene-like spermatocytes were further illustrated by staining of telomeric marker TRF1 and centromeric marker CREST, the numbers of which were notably increased in spermatocytes null for SPO16 (Fig. 4, C to E). However, SPO16-null spermatocytes were arrested before midpachytene stage, because H1T signal was not detected (fig. S4E) and Zfy1 and Zfy2 were not sufficiently silenced in Spo16 −/− testes, when compared to WT controls (fig. S2F).
Fig. 4. SPO16 deletion caused defects in homolog pairing and meiotic recombination.
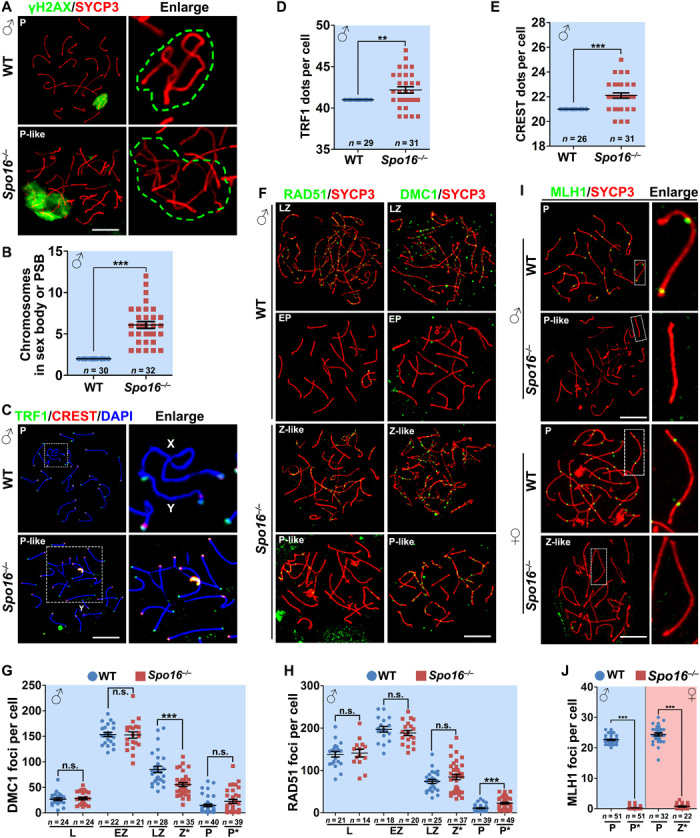
(A) γH2AX (green) staining marked the sex body of WT spermatocytes at pachytene stage and PSB of pachytene-like Spo16−/− spermatocytes. Green dashed lines in the magnified images indicate the area of γH2AX staining. Scale bar, 10 μm. (B) Quantification of chromosomes within the sex body or PSB. Numbers of spermatocytes analyzed (n) are indicated. ***P < 0.001 by two-tailed Student’s t tests. (C) Staining of TRF1 (marker of telomeres; green) and CREST (marker of centromeres; red) showing nonhomologous pairing and asynapsis in Spo16−/− spermatocytes. Scale bar, 10 μm. (D and E) Quantification of TRF1 (D) and CREST (E) dots in WT and Spo16−/− spermatocytes at pachytene and pachytene-like stages, respectively. Numbers of spermatocytes analyzed (n) are indicated. Median focus numbers are marked. Error bars indicate SEM. **P < 0.01 and ***P < 0.001 by two-tailed Student’s t tests. (F to H) Early recombination markers, RAD51 and DMC1, were detected on the nuclear surface spreads of WT and Spo16−/− spermatocytes (F), and the quantifications of RAD51 and DMC1 foci are shown in (G) and (H), respectively. Scale bar, 10 μm. Numbers of spermatocytes analyzed (n) are indicated. ***P < 0.001 by two-tailed Student’s t tests. (I and J) MLH1 (late recombination marker; green) was detected on the nuclear surface spreads of WT and Spo16−/− spermatocytes and PGCs (I) at indicated stages, and the quantification of MLH1 foci is shown in (J). Scale bar, 10 μm. ***P < 0.001 by two-tailed Student’s t tests.
SPO16 is required for meiotic recombination and crossing-over
Immunofluorescent staining of γH2AX on nuclear surface spreads and testis sections suggests that most of the DSBs generated at leponema are repaired in spermatocytes null for SPO16. We then dissected the progression of homologous recombination in Spo16 −/− meiocytes. Recombinases RAD51 and DMC1 bind to resected 3′-ssDNA, mediate strand invasion in meiotic prophase I, and therefore represent early homologous recombination events. Immunostaining and quantification showed comparable levels of RAD51 and DMC1 foci in Spo16 −/− spermatocytes at leptotene and early-zygotene stages, indicating that DSB generation and resection were not affected by SPO16 deletion (Fig. 4, F to H, and fig. S5A). The numbers of these foci were diminished with the progression of DSB repair (DSBR) through zygonema to early pachynema in WT spermatocytes or through zygotene-like to pachytene-like stages in Spo16 −/− spermatocytes (Fig. 4, F to H, and fig. S5A). Compared to the WT controls at the early-zygotene stage, we also noticed a slight increase of RAD51 and DMC1 foci in pachytene-like Spo16 −/− spermatocytes, which is consistent with the observation of asynapsis and formation of the PSB structure in these cells. These results revealed that most of the DSBs generated in leptonema spermatocytes were resected and repaired, except for the unrepaired ones forming in PSB structures. Incomplete DSBR and meiotic recombination were also found in female oocytes (Fig. 2G and fig. S5, B to D).
Because SPO16-deleted spermatocytes failed to progress to metaphase, we are not able to analyze the chiasmata directly. Moreover, Spo16 −/− spermatocytes failed to progress to the midpachytene stage (fig. S4E). Immunostaining of MLH1 showed that no late recombination nodules were detected in Spo16 −/− meiocytes (Fig. 4, I and J).
Assembly of ZMM foci during meiotic recombination
In budding yeast, the ZZS complex binds and stabilizes the first CO-specific recombination intermediates, SEIs. Western blot showed that SHOC1 level was decreased in Spo16 −/− testes at the age of PD21 (Fig. 5A). When stained on nuclear surface spreads prepared with WT and Spo16 −/− testes, we found that the number of SHOC1 foci is decreased in zygonema-like and pachynema-like Spo16 −/− spermatocytes (Fig. 5, B and C), suggesting that SPO16 is required for SHOC1 localization, probably through protein-protein interaction.
Fig. 5. Assembly of ZMM foci during meiotic recombination.
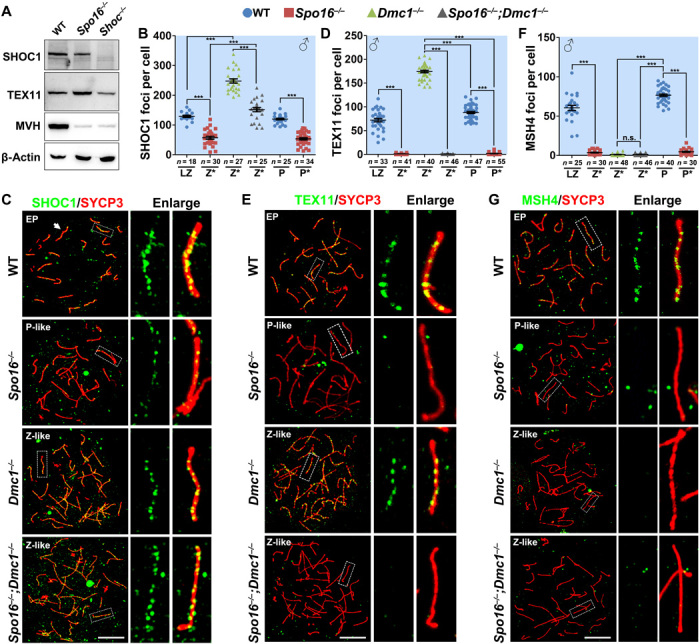
(A) Western blot showing the levels of SHOC1 and TEX11 in WT, Spo16−/−, and Shoc1−/− testes at the age of PD21. (B and C) SHOC1 was detected on the nuclear surface spreads of WT, Spo16−/−, Dmc1−/−, and Spo16−/−;Dmc1−/− spermatocytes at indicated stages (C), and the quantification of SHOC1 foci is shown in (B). Scale bar, 10 μm. ***P < 0.001 by two-tailed Student’s t tests. (D and E) Immunostaining of TEX11 on the nuclear surface spreads of WT, Spo16−/−, Dmc1−/−, and Spo16−/−;Dmc1−/− spermatocytes at indicated stages. The quantification of TEX11 foci is shown in (D). Scale bar, 10 μm. ***P < 0.001 by two-tailed Student’s t tests. (F and G) Immunostaining results and quantification of MSH4 foci were detected on the nuclear surface spreads of WT, Spo16−/−, Dmc1−/−, and Spo16−/−;Dmc1−/− spermatocytes at indicated stages. Scale bar, 10 μm. ***P < 0.001 by two-tailed Student’s t tests.
Zip4 is the third component of the ZZS complex and is believed to be recruited to early recombination intermediates by Zip2 (15). TEX11 is the mammalian ortholog of Zip4 and assembles discrete foci on chromosomal axes in WT spermatocytes at late-zygotene and early-pachytene stages (Fig. 5, D and E). However, in spermatocytes null for SPO16, TEX11 foci were abolished, although most of the chromosome pairs are synapsed (Fig. 5, D and E). The defects in TEX11 foci assembly was not because of decreased level of TEX11. Actually, the TEX11 level was increased in spermatocytes null for SPO16, but decreased in spermatocytes null for SHOC1 (Fig. 5A). Deletion of SPO16 also affected the localization of another ZMM protein, MSH4 (Fig. 5, F and G). Localization of MSH4 and TEX11 to the recombination sites was also totally abolished in female PGCs deficient for SPO16 (fig. S5, E to H). These results suggest that SPO16 is required for the stabilization of SHOC1 and localization of TEX11 and MSH4 to recombination intermediates.
In Dmc1−/− spermatocytes that were arrested at a zygotene-like stage, SHOC1 and TEX11 localized to the sites of presumed inter–sister chromatid joint molecules (Fig. 5, C and E). This is consistent with the SPO16 foci examined in DMC1-null spermatocytes (Fig. 1, D and E) and suggests that the mammalian ZZS complex is recruited to the sites of early recombination intermediates, regardless of whether the D loops are formed between homologs or between sister chromatids. However, MSH4 foci were not observed in Dmc1−/− spermatocytes, suggesting that the MSH4-MSH5 complex only assembles foci in the sites of interhomolog recombination intermediates that are stabilized by the ZZS complex (Fig. 5, F and G).
To investigate how the inter–sister chromatid recombination intermediates are processed in the absence of SPO16, we investigated the formation of ZMM foci in Spo16 −/−;Dmc1−/− double-knockout spermatocytes. In these spermatocytes, the number of SHOC1 foci was significantly decreased, when compared to the number of SHOC1 foci observed in Dmc1−/− spermatocytes (Fig. 5, B and C), further demonstrating that SPO16 is required for the stabilization of SHOC1. Similar to the phenotypes in Spo16 −/− spermatocytes, TEX11 foci were also abolished in Spo16 −/−;Dmc1−/− spermatocytes (Fig. 5, D and E).
DSBs are repaired without formation of COs in SPO16-null meiocytes
Noting that most of the DSBs formed in pachynema-like Spo16 −/− spermatocytes are repaired, as indicated by RAD51 foci and γH2AX immunostaining (Fig. 4, A to E), we are interested in how the DSBs are repaired in spermatocytes null for SPO16. As indicated by the immunostaining of ZMM proteins, middle recombination intermediates are not formed in these spermatocytes, suggesting that the DSBs might not have been repaired through the dHJ-forming DSBR pathway.
The RPA complex, which is composed of RPA1/2/3, MEIOB, and SPATA22, exhibits affinity to single-stranded and branched DNA and is localized to early and middle recombination nodules during meiosis (5, 6, 28, 29). Consistently, in WT spermatocytes, the number of SPATA22 foci peaked at the early-zygotene stage and was markedly decreased at the late-zygotene stage and the pachytene stage (Fig. 6, A and B, and fig. S6A). SPATA22 foci are specific to recombination intermediates, because in Spo11−/− or Spo16 −/−;Spo11−/− spermatocytes with reduced DSB generation, SPATA22 foci were not detected (fig. S6B), whereas in Dmc1−/− or Spo16 −/−;Dmc1−/− spermatocytes that failed to repair meiotic DSBs, the number of SPATA22 foci remained at a high level (Fig. 6, A and B). Therefore, we infer that the number of SPATA22 foci represents the number of early or middle recombination intermediates. As with the progression of meiotic recombination in meiotic prophase I, the RPA complex is removed from the sites of DSBs that are repaired without the formation of joint molecules.
Fig. 6. SPO16 was required for the stabilization for RPA-SPATA22-MEIOB complex.
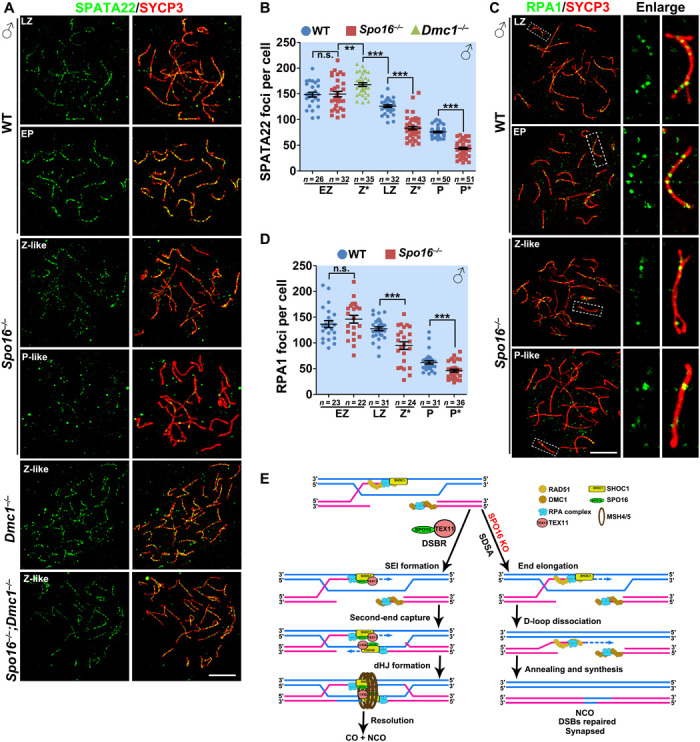
(A and B) SPATA22 was detected on the nuclear surface spreads of WT, Spo16−/−, Dmc1−/−, and Spo16−/−;Dmc1−/− spermatocytes (A), and the quantification is shown in (B). Scale bar, 10 μm. **P < 0. 01 and ***P < 0.001 by two-tailed Student’s t tests. (C and D) RPA1 was detected on the nuclear surface spreads of WT and Spo16−/− spermatocytes (C), and the quantification of RPA1 foci is shown in (D). Scale bar, 10 μm. ***P < 0.001 by two-tailed Student’s t tests. (E) Schematic diagram showing a proposed model of the roles of mammalian SHOC1-SPO16-TEX11 complex in meiotic recombination in meiotic prophase I. After strand invasion, SHOC1 is recruited to the joint molecules or D loops. This is a step crucial for the stabilization of these structures and promoting DNA synthesis and elongation of the DNA ends. After that, SHOC1 further recruits SPO16 and TEX11 to facilitate the repair of DSBs through a DSBR pathway with the formation of dHJs. In the absence of SPO16 or TEX11, the DSBs are repaired via a SDSA pathway. KO, knockout.
In Spo16 −/− spermatocytes, foci of SPATA22 were comparable at early zygonema (Fig. 6 and fig. S6), suggesting that the early steps of meiotic recombination were not affected by SPO16 deletion. The decrease in the number of SPATA22 foci was also observed in zygotene-like and pachytene-like Spo16 −/− spermatocytes (Fig. 6, A and B). However, the number of SPATA22 foci was significantly reduced in Spo16 −/− spermatocytes at zygotene-like and pachytene-like stages, when compared to the corresponding WT late-zygonema and early-pachynema spermatocytes (Fig. 6, A and B). This is different from the zygonema-like SHOC1-null spermatocytes in which the number of SPATA22 foci is higher than the WT late-zygonema and early-pachynema spermatocytes (19). The decrease in SPATA22 foci suggests fewer number of recombination intermediates in Spo16 −/− spermatocytes at zygotene-like and pachytene-like stages when compared to WT controls at similar stages, implying that Spo16 −/− spermatocytes have their DSBs repaired without the formation of joint molecules. Similarly, the numbers of RPA1 and RPA2 foci were also decreased in zygotene-like and pachytene-like Spo16 −/− spermatocytes, when compared to the WT spermatocytes at the corresponding stages (Fig. 6, C and D, and fig. S6).
DISCUSSION
Spo16 was identified here, together with a subset of well-established prophase I regulators and a list of new genes, in an attempt to gain insights into mammalian meiotic prophase I using the single-cell RNA sequencing data of female PGCs (20). During the preparation of this manuscript, several proteins on this list, such as SHOC1, MCMDC2, and SIX6OS1, were reported to regulate homologous recombination and synapsis in meiotic prophase I (18, 24, 30, 31). However, the function of SPO16 has never been investigated before the present study, although one article reported that the rat ortholog of Spo16, LOC689986, is specifically expressed in the neocortex (32). Spo16 mRNA was not detected in brain sample by reverse transcription PCR (RT-PCR; Fig. 1A), and Spo16 −/− mice are viable and have no appreciable phenotypes in terms of growth and aging. The only defect observed in Spo16 −/− mice is sterility due to problems in meiotic homologous recombination. On the basis of the ZMM-like localization pattern, the XPF-like domain, and its interaction with SHOC1, we conclude that the previously uncharacterized SPO16 is the mammalian ortholog of ScSpo16 and AtPTD and forms a dimeric complex with SHOC1 to participate in the processing of meiotic recombination intermediates (Fig. 6E). Identification of SPO16 strongly demonstrates that the machineries used in eukaryotic meiosis are conserved from budding yeast to mammals.
From yeast to mammals, repair of DSBs during homologous recombination involves two major pathways: the DSBR pathway and the SDSA pathway (33). While the SDSA pathway occurs in both mitotic and meiotic homologous recombinations and results only in NCOs, the DSBR pathway is strongly activated in meiotic recombination with the formation of dHJs, which are resolved as COs or NCOs in mammals. ZMM proteins are believed to facilitate the SEI-dHJ transition to promote crossing-over in these processes. Deletion of ZMM proteins causes severe defects in CO formation in yeast, mammals, and plants (1, 11, 17, 21). The dynamics of SPO16 foci on chromosomes are similar to that of the known ZMM proteins. SPO16 localizes to the recombination intermediates shortly after strand invasion (later than RAD51) but before the formation of dHJs (foci in DMC1-null spermatocytes). Consistent with the localization pattern, deletion of SPO16 abolished the assembly of ZMM protein–associated middle recombination intermediates, although full synapsis is achieved on most of the homologous pairs. In pachytene-like Spo16 −/− spermatocytes, RAD51 foci and γH2AX immunostaining are restricted only to the PSB region. These results indicate that the DSBs generated in Spo16 −/− spermatocytes are majorly repaired, however, without the formation of joint molecules.
Upon meiotic progression from early-zygotene stage to early-pachytene stage in WT spermatocytes, the numbers of RPA1 or SPATA22 foci are decreased (Fig. 6, B and D), indicating the first step of CO/NCO differentiation. Therefore, we infer that RPA proteins are retained on the sites of CO-prone joint molecules (SEIs and dHJs), whereas they dissociate from the recombination intermediates that are processed through the NCO-forming SDSA pathway. The numbers of RPA1, RPA2, and SPATA22 foci are decreased in zygonema-like and pachynema-like spermatocytes upon SPO16 deletion, implying that the D loops formed in Spo16 −/− meiocytes are dissociated and repaired through the SDSA pathway.
Together, our results suggest a possible model for the processing of recombination intermediates during meiotic recombination. After strand invasion, D loops are stabilized by binding of SHOC1, which promotes end elongation. SPO16 and TEX11 are then recruited to these structures to facilitate the formation of SEIs, which are CO-prone recombination intermediates. After that, MSH4-MSH5 foci are assembled to promote the formation of dHJs, along with the repair of DSBs.
MATERIALS AND METHODS
Mice
Dmc1−/− and Spo11−/− mice were previously reported and purchased from the Jackson Laboratory (3, 4). Null allele for Spo16 was generated using CRISPR-Cas9 technology according to standard protocols (34). A guide sequence of 5′-CCTAGAAAATCGAAGCCACA-3′ with a protospacer adjacent motif sequence was selected on the basis of the website prediction results (https://zlab.bio/guide-design-resources?). The guide sequence was cloned to the pUC57-sgRNA vector. Following linearization, sgRNA and Cas9 mRNA were in vitro transcribed and purified according to the manufacturer’s instructions (AM1345, AM1354, and AM1908, Ambion; and 74104, QIAGEN). Mouse zygotes were obtained by superovulation of 7- to 8-week-old females mating with males of the same strain. A mixture of Cas9 mRNA (40 ng/μl) and sgRNA (40 ng/μl) was injected into zygotes using the Eppendorf TransferMan NK2. Injected zygotes were transferred into pseudopregnant ICR (Institute of Cancer Research) female mice (15 to 25 zygotes per mouse) after 2-hour recovery culture in KSOM (potassium-supplemented simplex optimized medium).
All mice were maintained under specific pathogen–free conditions in a controlled environment of 20° to 22°C, with a 12-hour light/12-hour dark cycle, 50 to 70% humidity, and food and water provided ad libitum. Animal care and experimental procedures were conducted in accordance with the University of Gothenburg, Sweden. All mutant mouse strains had a C57BL/6 background. Genotyping primers used are listed in table S2. For each mouse experiment, at least three mice were analyzed, otherwise stated.
Homology searches and modeling
Homology search for MmSPO16 orthologs and related proteins was performed using PSI-BLAST and HHpred (http://toolkit.tuebingen.mpg.de/). Alignment of SPO16 orthologs were performed with Clustal-Omega (www.ebi.ac.uk/Tools/msa/clustalo/) and colored with ESPript (http://espript.ibcp.fr/) using the Risler homology matrix (with a global score of 0.5). Secondary structure prediction was performed using the PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/), while homology modeling was performed with I-TASSER (https://zhanglab.ccmb.med.umich.edu/I-TASSER/) (35–37). The C-score for the secondary structure of MmSPO16 is −1.67. Protein structure comparison was performed with FATCAT (http://fatcat.burnham.org/). The structure alignment for MmSPO16 and ApXPF has 177 equivalent positions with a root mean square deviation (RMSD) of 2.35, without twists (P = 2.22 × 10–16). The structure alignment for MmSPO16 and ScSPO16 has 126 equivalent positions with an RMSD of 3.07, with four twists (P = 0.0264).
Semiquantitative RT-PCR
Except for the embryonic ovary sample that was collected from embryonic females at E16.5, other tissues were from adult male mice. Male germ cells were prepared by a bovine serum albumin (BSA) gradient as previously described (38). Total RNA was extracted using the RNeasy Mini Kit (no. 74106, QIAGEN) according to the manufacturer’s instructions and reverse-transcribed to obtain cDNA (no. 1708890, Bio-Rad). PCR was performed with Taq DNA polymerase under standard conditions (PCR for 28 cycles). Gapdh served as the loading control. Primer sequences were listed in table S2.
Cloning and electroporation
cDNA encoding Spo16 was PCR-amplified from the mouse testis cDNA library (no. 9537, Takara Bio) and cloned to a pCAG-GFP vector (no. 11150, Addgene) for C-terminal fusion or a modified pCAG-FLAG for N-terminal fusion. Plasmids were maxi-prepared (no. 12163, QIAGEN) and injected into live mouse testes at PD18. The testes were subjected to electroporation as previously described (39). Preparation and immunofluorescent staining of nuclear surface spreads is detailed below.
Nuclear surface spreading
Seminiferous tubules were prepared from juvenile and adult testes, while female PGCs were obtained from embryonic ovaries at E17.5. Nuclear surface spreads were prepared as previously described (40). Briefly, seminiferous tubules or embryonic ovaries were treated with a hypotonic buffer [30 mM tris, 50 mM sucrose, 17 mM trisodium citrate dehydrate, 5 mM EDTA, and 0.5 mM dithiothreitol (pH 8.2)] for 30 min and subsequently smashed in 100 mM sucrose buffer (pH 8.2). The suspension was then added to a slides containing fixative buffer [1% paraformaldehyde and 0.15% Triton X-100 (pH 9.2)]. After at least 2 hours of incubation in a humidify box, the slides were air-dried and washed.
Immunofluorescent staining and imaging
After extensive washing in phosphate-buffered saline (PBS), slides were blocked with 1% BSA in PBST (PBS with 0.1% Tween 20) for 30 min and subsequently subjected to incubation with primary and secondary antibodies. The primary antibodies used were listed in table S3, while the secondary antibodies with minimal cross-reactivity were purchased from Jackson ImmunoResearch Laboratories. The signals were examined under a DeltaVision microscope, and images for quantification were taken using this microscope. Representative images were taken using a confocal laser scanning microscope (Zeiss LSM 700, Carl Zeiss AG, Germany).
Western blot analysis
Testes were lysed with SDS sample buffer and heated for 5 min at 95°C. Samples were separated by SDS–polyacrylamide gel electrophoresis and electrophoretically transferred to polyvinylidene difluoride membranes (Millipore Corp., Bedford, MA, USA), followed by blocking in tris-buffered saline with Tween 20 (TBST) containing 5% defatted milk (BD Biosciences, Franklin Lakes, NJ, USA) for 30 min. After probing with primary antibodies, the membranes were washed in TBST, incubated with a horseradish peroxidase–linked secondary antibody (Jackson ImmunoResearch Laboratories) for 1 hour, followed by three washes with TBST. Bound antibodies were detected using the SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific Inc., Waltham, MA, USA). The primary antibodies and dilution factors used are listed in table S3.
Cell lines and immunoprecipitation
HeLa and B16F1 cells were grown in Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin-streptomycin solution (Gibco) at 37°C in a humidified 5% CO2 incubator. Cell lines were in healthy conditions but were not tested for mycoplasma contamination. Transient plasmid transfection was performed using Lipofectamine 2000 (Invitrogen). After a 24-hour transfection, cells were lysed in lysis buffer [50 mM tris-HCl (pH 7.5), 150 mM NaCl, 10% glycerol, and 0.5% NP-40; protease and phosphatase inhibitors were added before use]. Cell lyses were incubated with FLAG agarose beads at 4°C for 4 hours, followed by washing. SDS sample buffer was added to the beads, and the eluates were used for Western blot analysis.
Histological analyses
PBS-buffered, formalin-fixed, paraffin-embedded samples were sectioned (5 μm thick) for H&E staining and immunohistochemistry (IHC). To gain better histology of testis seminiferous tubules, testes were fixed in Bouin’s solution and subjected to H&E staining. For IHC, sections were deparaffinized and rehydrated. Primary antibodies were applied at suitable dilutions (table S3) at room temperature for 1 hour and then incubated with biotinylated secondary antibodies for 30 min. Sections were then stained using Vectastain ABC (anti-biotin complex) and DAB (3, 3-diaminobenzidine) peroxidase substrate kits (Vector Laboratories, Burlingame, CA, USA). Antibodies used in this study are listed in table S3.
Statistics
Results are given as means ± SEM. Each experiment included at least three independent samples and was repeated at least three times. Results for two experimental groups were compared by two-tailed unpaired Student’s t tests. Statistically significant values are presented as *P < 0.05, **P < 0.01, and ***P < 0.001.
Supplementary Material
Acknowledgments
We thank M. Pilon and P. Carlsson for helpful comments on the manuscript. We thank M. A. Handel and E. Bolcun-Filas for the H1T antibody. Funding: We acknowledge K. Liu for substantial support in funds (VR: 621-2014-5830 and 521-2012-2841) and equipment at the early stage of this study and discussions. This study is partially supported by Sven and Lily Lawskis Foundation (to C.Y.). We also acknowledge the Centre for Cellular Imaging at the University of Gothenburg and the National Microscopy Infrastructure (NMI; VR-RFI 2016-00968) for providing assistance in microscopy. Author contribution: C.Y. identified the candidate gene. C.Y. and Q.Z. designed and performed experiments and analyzed data. S.-Y.J. did the microinjection to generate knockout alleles and performed the immunoprecipitation. K.B. generated and validated the SIX6OS1 antibody. C.Y. wrote the manuscript. All authors contributed to the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/1/eaau9780/DC1
Fig. S1. MmSPO16 has a conserved XPF-like domain.
Fig. S2. Localization of SPO16 and generation of knockout mice.
Fig. S3. SPO16 deletion leads to massive germline loss.
Fig. S4. Insufficient meiotic prophase progression in Spo16−/− testes.
Fig. S5. Insufficient meiotic recombination in SPO16-deleted spermatocytes and oocytes.
Fig. S6. Detection of RPA complex in WT and SPO16-deleted spermatocytes.
Table S1. Genes specifically expressed in meiotic prophase I.
Table S2. Homology of MmSPO16 to known proteins.
Table S3. Primer sequences.
Table S4. Antibody information.
REFERENCES AND NOTES
- 1.Hunter N., Meiotic recombination: The essence of heredity. Cold Spring Harb. Perspect. Biol. 7, a016618 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zickler D., Kleckner N., Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb. Perspect. Biol. 7, a016626 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baudat F., Manova K., Yuen J. P., Jasin M., Keeney S., Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell 6, 989–998 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Pittman D. L., Cobb J., Schimenti K. J., Wilson L. A., Cooper D. M., Brignull E., Handel M. A., Schimenti J. C., Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol. Cell 1, 697–705 (1998). [DOI] [PubMed] [Google Scholar]
- 5.La Salle S., Palmer K., O’Brien M., Schimenti J. C., Eppig J., Handel M. A., Spata22, a novel vertebrate-specific gene, is required for meiotic progress in mouse germ cells. Biol. Reprod. 86, 45 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo M., Yang F., Leu N. A., Landaiche J., Handel M. A., Benavente R., La Salle S., Wang P. J., MEIOB exhibits single-stranded DNA-binding and exonuclease activities and is essential for meiotic recombination. Nat. Commun. 4, 2788 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Souquet B., Abby E., Hervé R., Finsterbusch F., Tourpin S., Le Bouffant R., Duquenne C., Messiaen S., Martini E., Bernardino-Sgherri J., Toth A., Habert R., Livera G., MEIOB targets single-strand DNA and is necessary for meiotic recombination. PLOS Genet. 9, e1003784 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones G. H., The control of chiasma distribution. Symp. Soc. Exp. Biol. 38, 293–320 (1984). [PubMed] [Google Scholar]
- 9.Hassold T., Hall H., Hunt P., The origin of human aneuploidy: Where we have been, where we are going. Hum. Mol. Genet. 16, R203–R208 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Hassold T., Hunt P., To err (meiotically) is human: The genesis of human aneuploidy. Nat. Rev. Genet. 2, 280–291 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Lynn A., Soucek R., Börner G. V., ZMM proteins during meiosis: Crossover artists at work. Chromosome Res. 15, 591–605 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Shinohara M., Oh S. D., Hunter N., Shinohara A., Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nat. Genet. 40, 299–309 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Fung J. C., Rockmill B. M., Odell M. J., Roeder G. S., Imposition of crossover interference through the nonrandom distribution of synapsis initiation complexes. Cell 116, 795–802 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Hunter N., Kleckner N., The single-end invasion: An asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell 106, 59–70 (2001). [DOI] [PubMed] [Google Scholar]
- 15.De Muyt A., Pyatnitskaya A., Andréani J., Ranjha L., Ramus C., Laureau R., Fernandez-Vega A., Holoch D., Girard E., Govin J., Margueron R., Couté Y., Cejka P., Guérois R., Borde V., A meiotic XPF–ERCC1-like complex recognizes joint molecule recombination intermediates to promote crossover formation. Genes Dev. 32, 283–296 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.K. Arora, K. D. Corbett, The conserved Xpf:Ercc1-like Zip2:Spo16 complex controls meiotic crossover formation through structure-specific DNA binding. Nucleic Acids Res.; 10.1093/nar/gky1273 (2018). [DOI] [PMC free article] [PubMed]
- 17.Macaisne N., Vignard J., Mercier R., SHOC1 and PTD form an XPF–ERCC1-like complex that is required for formation of class I crossovers. J. Cell Sci. 124, 2687–2691 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Guiraldelli M. F., Felberg A., Almeida L. P., Parikh A., Castro R. O., Pezza R. J., SHOC1 is a ERCC4-(HhH)2-like protein, integral to the formation of crossover recombination intermediates during mammalian meiosis. PLOS Genet. 14, e1007381 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Q., Shao J., Fan H.-Y., Yu C., Evolutionarily-conserved MZIP2 is essential for crossover formation in mammalian meiosis. Commun. Biol. 1, 147 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo F., Yan L., Guo H., Li L., Hu B., Zhao Y., Yong J., Hu Y., Wang X., Wei Y., Wang W., Li R., Yan J., Zhi X., Zhang Y., Jin H., Zhang W., Hou Y., Zhu P., Li J., Zhang L., Liu S., Ren Y., Zhu X., Wen L., Gao Y. Q., Tang F., Qiao J., The transcriptome and DNA methylome landscapes of human primordial germ cells. Cell 161, 1437–1452 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Reynolds A., Qiao H., Yang Y., Chen J. K., Jackson N., Biswas K., Holloway J. K., Baudat F., Massy B., Wang J., Höög C., Cohen P. E., Hunter N., RNF212 is a dosage-sensitive regulator of crossing-over during mammalian meiosis. Nat. Genet. 45, 269–278 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y., Gaines W. A., Callender T., Busygina V., Oke A., Sung P., Fung J. C., Hollingsworth N. M., Down-regulation of Rad51 activity during meiosis in yeast prevents competition with Dmc1 for repair of double-strand breaks. PLOS Genet. 10, e1004005 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kadyk L. C., Hartwell L. H., Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132, 387–402 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gómez-H L., Felipe-Medina N., Sánchez-Martín M., Davies O. R., Ramos I., García-Tuñón I., Rooij D. G., Dereli I., Tóth A., Barbero J. L., Benavente R., Llano E., Pendas A. M., C14orf39/Six6os1 is a constituent of the synaptonemal complex and is essential for mouse fertility. Nat. Commun. 7, 13298 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edelmann W., Cohen P. E., Kneitz B., Winand N., Lia M., Heyer J., Kolodner R., Pollard J. W., Kucherlapati R., Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nat. Genet. 21, 123–127 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Kneitz B., Cohen P. E., Avdievich E., Zhu L., Kane M. F., Hou H. Jr., Kolodner R. D., Kucherlapati R., Pollard J. W., Edelmann W., MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev. 14, 1085–1097 (2000). [PMC free article] [PubMed] [Google Scholar]
- 27.Bellani M. A., Romanienko P. J., Cairatti D. A., Camerini-Otero R. D., SPO11 is required for sex-body formation, and Spo11 heterozygosity rescues the prophase arrest of Atm−/− spermatocytes. J. Cell Sci. 118, 3233–3245 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Ribeiro J., Abby E., Livera G., Martini E., RPA homologs and ssDNA processing during meiotic recombination. Chromosoma 125, 265–276 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu Y., Greenberg R. A., Schonbrunn E., Wang P. J., Meiosis-specific proteins MEIOB and SPATA22 cooperatively associate with the single-stranded DNA-binding replication protein A complex and DNA double-strand breaks. Biol. Reprod. 96, 1096–1104 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finsterbusch F., Ravindranathan R., Dereli I., Stanzione M., Tränkner D., Tóth A., Alignment of homologous chromosomes and effective repair of programmed DNA double-strand breaks during mouse meiosis require the minichromosome maintenance domain containing 2 (MCMDC2) protein. PLOS Genet. 12, e1006393 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McNairn A. J., Rinaldi V. D., Schimenti J. C., Repair of meiotic DNA breaks and homolog pairing in mouse meiosis requires a minichromosome maintenance (MCM) paralog. Genetics 205, 529–537 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ersland K. M., Håvik B., Rinholm J. E., Gundersen V., Stansberg C., Steen V. M., LOC689986, a unique gene showing specific expression in restricted areas of the rodent neocortex. BMC Neurosci. 14, 68 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wold M. S., Replication protein A: A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66, 61–92 (1997). [DOI] [PubMed] [Google Scholar]
- 34.Wang H., Yang H., Shivalila C. S., Dawlaty M. M., Cheng A. W., Zhang F., Jaenisch R., One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y., I-Tasser: Fully automated protein structure prediction in Casp8. Proteins 77 ( suppl. 9), 100–113 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roy A., Yang J., Zhang Y., COFACTOR: An accurate comparative algorithm for structure-based protein function annotation. Nucleic Acids Res. 40, W471–W477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang J., Zhang Y., I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 43, W174–W181 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gan H., Wen L., Liao S., Lin X., Ma T., Liu J., Song C.-X., Wang M., He C., Han C., Tang F., Dynamics of 5-hydroxymethylcytosine during mouse spermatogenesis. Nat. Commun. 4, 1995 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Shibuya H., Morimoto A., Watanabe Y., The dissection of meiotic chromosome movement in mice using an in vivo electroporation technique. PLOS Genet. 10, e1004821 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peters A. H., Plug A. W., Vugt M. J., Boer P., A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 5, 66–68 (1997). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/1/eaau9780/DC1
Fig. S1. MmSPO16 has a conserved XPF-like domain.
Fig. S2. Localization of SPO16 and generation of knockout mice.
Fig. S3. SPO16 deletion leads to massive germline loss.
Fig. S4. Insufficient meiotic prophase progression in Spo16−/− testes.
Fig. S5. Insufficient meiotic recombination in SPO16-deleted spermatocytes and oocytes.
Fig. S6. Detection of RPA complex in WT and SPO16-deleted spermatocytes.
Table S1. Genes specifically expressed in meiotic prophase I.
Table S2. Homology of MmSPO16 to known proteins.
Table S3. Primer sequences.
Table S4. Antibody information.


