Abstract
Aberrant DNA methylation is a distinguishing feature of cancer. Yet, how methylation affects immune surveillance and tumor metastasis remains ambiguous. We introduce a novel method, Guide Positioning Sequencing (GPS), for precisely detecting whole-genome DNA methylation with cytosine coverage as high as 96% and unbiased coverage of GC-rich and repetitive regions. Systematic comparisons of GPS with whole-genome bisulfite sequencing (WGBS) found that methylation difference between gene body and promoter is an effective predictor of gene expression with a correlation coefficient of 0.67 (GPS) versus 0.33 (WGBS). Moreover, Methylation Boundary Shift (MBS) in promoters or enhancers is capable of modulating expression of genes associated with immunity and tumor metabolism. Furthermore, aberrant DNA methylation results in tissue-specific enhancer switching, which is responsible for altering cell identity during liver cancer development. Altogether, we demonstrate that GPS is a powerful tool with improved accuracy and efficiency over WGBS in simultaneously detecting genome-wide DNA methylation and genomic variation. Using GPS, we show that aberrant DNA methylation is associated with altering cell identity and immune surveillance networks, which may contribute to tumorigenesis and metastasis.
DNA methylation of cytosines in the metazoan genome is a stable epigenetic mark and has been intimately linked to cancer (Feinberg et al. 2016). Aberrant DNA methylation patterns can be found in cancer genomes and related to cancer progression (Spencer et al. 2017). In addition, it has been shown that a high level of methylation in promoters is likely to induce gene silencing, whereas a high level of methylation in the gene body is linked to increased gene expression (Neri et al. 2017). However, the relationship between DNA methylation in the gene body and promoter as well as how aberrant DNA methylation influences gene expression during tumorigenesis and metastasis require further exploration.
The interaction between cancer and the host immune system is complicated, and the immune system plays a critical role in the surveillance against cancer (Grivennikov et al. 2010; Schreiber et al. 2011; Vinay et al. 2015). Recently, emerging evidence shows that T cell activation by interrupting the interaction between PDCD1 and CD274 (also known as PD-1 and PD-L1) can disrupt tumor cell growth; meanwhile DNA methylations are involved in the transition between effector and memory CD8 T cell (Akondy et al. 2017; Youngblood et al. 2017). The interruption can be mediated by the alteration of DNA methylation (Goltz et al. 2016; Ghoneim et al. 2017). The immune system was shown to be capable of suppressing tumor growth by destroying cancer cells, while simultaneously promoting tumor progression by establishing a favorable microenvironment for tumor outgrowth (Eggert et al. 2016; Mohme et al. 2017). We postulated that epigenetic aberrations such as aberrant DNA methylation could potentially affect immune genes through immune-epigenetic interactions and instigate cancer cell evasion from the host immune system.
Bisulfite conversion of genomic DNA coupled with next-generation sequencing is generally considered the “gold standard” in genome-wide base-resolution methylome detection (Rivera and Ren 2013). There is, however, a built-in limitation of the bioinformatics analysis of bisulfite sequencing, whose mapping strategy is based either on wild-card aligners or three-letter aligners (Krueger et al. 2012). Each has its set of restrictions. Wild-card aligners achieve a higher genomic coverage, but do so at the expense of a preference toward increased DNA methylation levels, because the extra cytosines (Cs) in a methylated read can raise the sequence complexity to a sufficient level such that unique alignment to the genome is maintained, while the corresponding, unmethylated, T-containing read is discarded due to nonunique alignment. On the other hand, three-letter aligners eliminate remaining Cs from bisulfite sequencing reads, which consequently diminishes sequence complexity in that a greater percentage of reads is discarded due to ambiguous alignment positions (Bock 2012). In light of these limitations, there is an urgent need for further developing and optimizing the current genome-wide DNA methylation profiling method with a focus on improving mapping accuracy and cost-efficiency.
To solve these problems, we developed a novel method, Guide Positioning Sequencing (GPS) for genome-wide DNA methylation detection. By applying GPS to detect DNA methylation in normal liver cells and hepatoma cell lines 97L and LM3 (Li et al. 2003; Zhang et al. 2012), we investigate aberrant DNA methylation patterns in regulating gene expression related to tumorigenesis and metastasis.
Results
Accurate detection of genome-wide DNA methylation by Guide Positioning Sequencing
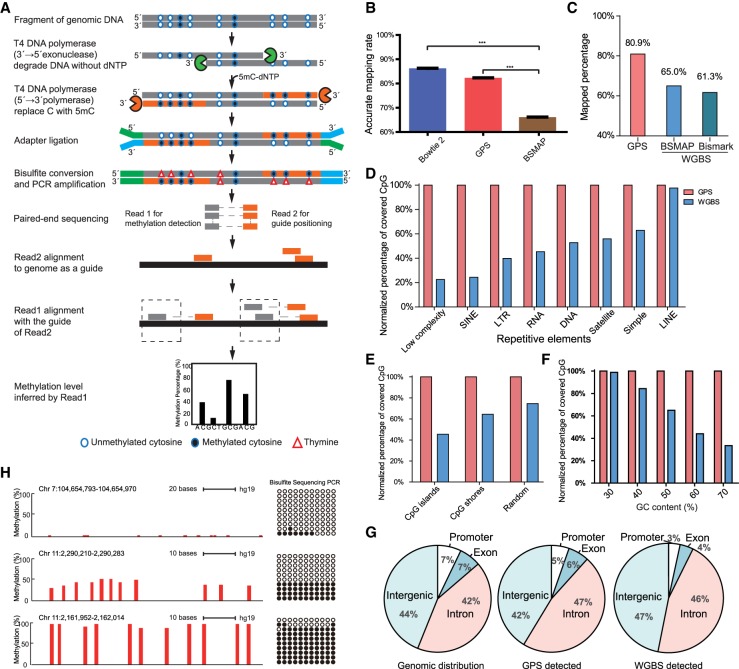
In this section, we present our novel method for genome-wide DNA methylation detection. Taking advantage of both 3′→5′ exonuclease and 5′→3′ polymerase activities of the T4 DNA polymerase, we were able to obtain a DNA fragment integrated with methylcytosine. The 3′ end of the DNA fragment with nonconvertible methylcytosine after bisulfite treatment can act as a guide for calculating DNA methylation of the 5′ end through pair-end sequencing (Fig. 1A). This Guide Positioning Sequencing (GPS) method enables us to detect genome-wide DNA methylation precisely with a high cytosine coverage rate. As shown in Supplemental Figure S1A, T4 DNA polymerase synthesizes chimera fragments whose 3′ end sequence with dmCTP integration matched perfectly with the reference genome, whereas the 5′ end sequence without dmCTP integration showed several C→T mismatches due to considerable C→T conversion. To evaluate the accuracy and performance of GPS alignment, we randomly generated one million pair-end reads from the reference human genome to simulate pair-end sequencing results. With the help of the already known genome position derived from simulated reads, we can evaluate the accuracy of DNA methylation detection strategies by comparing their accurate alignment rate. We observed that the accurate alignment rate of WGBS aligned by BSMAP (Xi and Li 2009) can be as low as 66.2%, suggesting that approximately one-third of the reads had been discarded during alignment. The rate of accurate alignment for GPS, on the other hand, can reach as high as 82.3%, closer to that of the widely used genome DNA alignment tool Bowtie 2 (Langmead and Salzberg 2012) at 86.3%. With Bowtie 2, both ends are kept to its original genome sequence (P-value <0.001, one-tailed paired t-test) (Fig. 1B). We further observed that GPS consistently performed better than WGBS aligned by BSMAP, even with the increased rate of mismatches/indels (Supplemental Fig. S1B).
Figure 1.
Guide Positioning Sequencing (GPS) detects genome-wide DNA methylation accurately with high coverage rate. (A) Schematic of GPS workflow for DNA methylation detection. The gray line represents original DNA sequence, and the orange line represents DNA treated by T4 DNA polymerase, which replaces cytosine with 5′-methylcytosine at 3′ end of DNA fragment. The solid circle (●) represents methylated cytosine, and the open circle (○) represents unmethylated cytosine, whereas the triangle (Δ) represents thymine. Blue and green short lines represent the NGS linker. Read1 represents the bisulfite-converted 5′ end of fragments, whereas Read2 represents the 3′ end of fragments, which is the same as the genome sequence due to 5′-methylcytosine replacement. (B) The accurate alignment rate of Bowtie 2 and GPS is obviously higher than that in BSMAP based on simulated data: (***) P < 0.001, one-tailed paired t-test. (C) Alignment efficiency of GPS compared with WGBS. DNA methylation of 97L cells was detected in parallel by GPS and WGBS, and mapped percentage of paired reads in GPS is 80.9%, 15%–20% higher than two popular WGBS alignment tools, BSMAP and Bismark. (D) Performance of GPS in detecting DNA methylation of repetitive elements. GPS had advantage over WGBS in detecting CpG sites of repetitive elements, especially in Low complexity, SINE, and LTR. The y-axis indicates the ratio (%) of WGBS/GPS-covered CpG sites. (E) Performance of GPS in CpG islands and CpG shores compared with WGBS. WGBS only detected ∼43% CpGs of those by GPS in CpG islands under similar sequencing depth for methylation detection. (F) Performance of GPS in different GC content. (G) Distributions of original genomic, GPS-detected, and WGBS-detected CpG sites. Genomic distribution shows total CpG sites in the genome. GPS detected that distribution of CpG sites is more similar to the original genomic CpG distribution compared with WGBS. (H) GPS-detected DNA methylation verified by Sanger sequencing from bisulfite-treated DNA. The red bar represents a single cytosine site, and the height of the red bar represents the methylation level. Each column of circles represents a single cytosine corresponding to the red bar. The solid circle (●) represents methylated cytosine, and the open circle (○) represents unmethylated cytosine.
Further side-by-side experiments of GPS and WGBS show that, in hepatoma cell line 97L cells, the alignment rate of GPS is 80.9%, almost 15%–20% higher than that of WGBS analyzed by either BSMAP or Bismark (Krueger and Andrews 2011) with about 0.4 billion reads (Fig. 1C; Supplemental Table S1). DNA methylation is highly correlated on the commonly detected CpG sites (r = 0.89, P-value <2.2 × 10−16, Pearson correlation coefficient) with both methods (Supplemental Fig. S1C,D). Further analyses demonstrate that GPS has higher efficiency when detecting DNA methylation in the repetitive elements, CpG islands, and GC-rich region such as promoter regions (Fig. 1D–F; Supplemental Fig. S2). Moreover, the methylation sites detected by GPS have no distribution bias in promoter and functional genome elements (Fig. 1G), and GPS is more accurate than WGBS as verified by bisulfite pyrosequencing (Supplemental Fig. S1E). GPS is more cost-effective compared with WGBS (Supplemental Table S2). Screenshots of UCSC Genome Browser showed that GPS detected more CpG sites than public WGBS data sets in CpG islands and repetitive regions (Supplemental Fig. S1F,G). Taken together, GPS is a powerful tool with significant advantages over WGBS in detecting genome-wide DNA methylation.
To investigate the effects of DNA methylation patterns on tumorigenesis, we applied GPS to detect global DNA methylation in normal liver and compared it with two hepatoma cell lines, 97L and LM3 (Supplemental Methods). In human normal liver cells (Liver, for short), GPS detected 54,853,393 of the total 56,434,896 (97%) CpG sites. As for all the cytosines in Liver, 1,123,233,333 of the total 1,170,378,405 cytosines (96%) are covered by at least one read (Supplemental Table S3). This indicates that GPS is an effective method for detecting both CpG and non-CpG methylation. In addition, GPS detected 99.66% (7348/7373) of the cytosines in the mitochondrial genome. The coverage rate of autosome is higher than 96%, whereas that of the X and Y Chromosome is ∼90% (Supplemental Fig. S3A). GPS detected more CpG sites than WGBS in each chromosome with the same number of bisulfite-converted reads in 97L (Supplemental Fig. S3B). To validate the GPS-detected methylation, we selected regions with distinct methylation levels that were then confirmed by bisulfite TA clone sequencing (Fig. 1H). Moreover, we validated 13 additional regions using bisulfite pyrosequencing, the results of which continue to demonstrate the efficiency of GPS in detecting global DNA methylation (Supplemental Fig. S3C,D).
We next compared GPS with WGBS in detecting genetic variants and found that more genetic variants were detected by GPS than by WGBS with the same raw reads. We found 91% of the 2,296,462 variations detected in Liver cells overlap with those in the dbSNP database. By comparing Liver and two hepatoma cell lines with metastasis potential to the lung, we also found that three potential mutants in hepatoma cell lines are located in the lung-related gene CAV2, which were validated by bisulfite pyrosequencing (Supplemental Fig. S4). This suggests that GPS can simultaneously obtain genome as well as epigenome information in one go, and can, therefore, assist in investigating the crosstalk between genome and epigenome such as allele specific methylation (ASM). For example, we identified 1820 ASMs by GPS as compared to 135 ASMs by WGBS with the same amount of data. Two ASMs detected by GPS were verified by TA clone bisulfite sequencing in hepatoma 97L cells, which are located within the CCDC97 and TOP1MT gene enriched with transcription factor and DNase I hypersensitive sites (Supplemental Fig. S5). As such, GPS is able to accurately detect not only DNA methylation but also genetic variation.
Methylation of gene body difference to promoter correspondingly predicts gene expression related to tumor metabolism and immune surveillance network
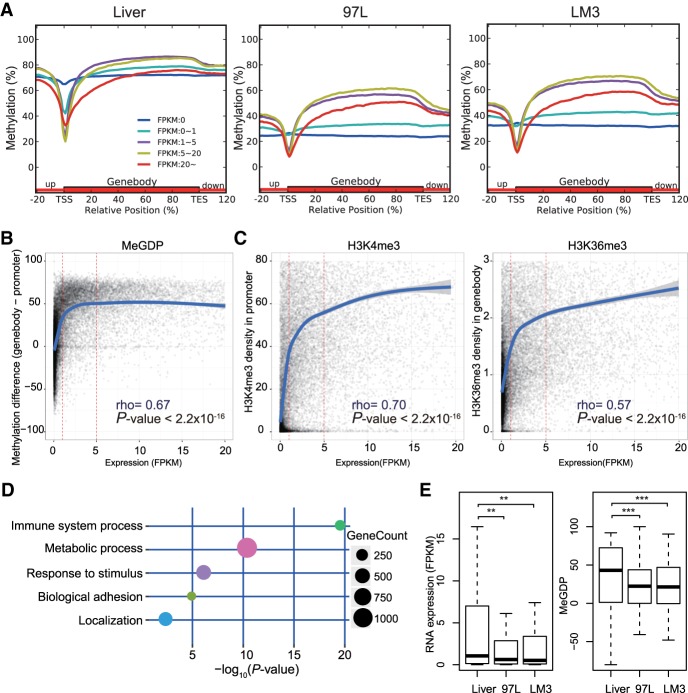
To investigate the effects of DNA methylation patterns on tumorigenesis, we grouped genes by their expression (FPKM) and found that in general, gene expression is associated with DNA methylation in the promoter (±1 kbp around TSS from RefSeq) as well as in the gene body (Fig. 2A). Genes with higher expression usually acquire lower promoter methylation and higher gene body methylation. However, we found that when gene expression FPKM is over 20, DNA methylation in the gene body is no longer positively related to gene expression. Genes with expression over 20 FPKM are more hypomethylated in the gene body, shorter in length, more conserved, and enriched in metabolic processes (Supplemental Fig. S6). Since DNA methylation level in either promoter or gene body independently is not significantly correlated to gene expression, we determined to investigate whether combining the DNA methylation of gene body and promoter can serve as a better predictor for gene expression.
Figure 2.
DNA methylation difference between gene body and promoter coordinates in regulating gene expression related to tumor-immune surveillance network. (A) Distribution of methylation level on promoter and gene body according to gene expression of FPKM (Fragments Per Kilobase of exon per Million fragments mapped) in Liver and hepatoma cell line 97L and LM3. Grouped genes are based on FPKM as FPKM = 0, 0–1, 1–5, 5–20, and more than 20. Highly expressed genes exhibit gene body hypermethylation and promoter hypomethylation and vice versa, except genes with FPKM more than 20 (red line). (B) Correlation between DNA methylation and gene expression FPKM in 97L with a Spearman's rank correlation coefficient of 0.67. The y-axis represents the numeral methylation difference between gene body and promoter, and the x-axis represents the gene FPKM. (C) Correlation between gene expression and H3K4me3 in promoter (left) or H3K36me3 in gene body (right) in 97L. Both H3K4me3 and H3K36me3 are positively correlated with gene expression when FPKM is more than 5. (D) Immune surveillance and tumor metabolism-related genes associated with aberrant DNA methylation. Decreased expressed genes with MeGDP reduction in 97L compared with Liver are enriched in GO terms correlated with tumorigenesis, like immune system process, response to stimulus as well as metabolic process and biological adhesion. (E) Gene expression of 102 interferon gamma pathway genes related to immune surveillance is down-regulated in hepatoma cells compared with Liver, where MeGDP is decreased accordingly. Wilcoxon matched pairs test: (**) P-value <0.01; (***) P-value <0.001.
We plotted a scatter diagram to show the numerical DNA methylation difference between gene body and promoter regions (MeGDP, i.e., methylation of gene body difference to promoter) in correlation with gene expression in 97L cells (Fig. 2B; Supplemental Table S4). The fitted curve shows the following trends: Within expression range FPKM 0–1, MeGDP increases at a steep rate; within expression range FPKM 1–5, the curve levels off with a much slower rate of increase; and within expression range FPKM 5–20, there is little if any rate of change. We found similar results when we grouped genes according to FPKM by bar plots (Supplemental Fig. S7A). Collectively, these data indicate that the MeGDP may be an on/off switch (FPKM 0–1) for gene expression. Furthermore, we found that MeGDP and gene expression are correlated with rho as high as 0.67 (P-value <2.2 × 10−16, Spearman's rank correlation), suggesting that MeGDP is a considerably useful predictor for gene expression. Meanwhile, the coefficient calculated by WGBS data set is 0.33 (Supplemental Fig. S7B), which may be due to the limitations of WGBS, such as inaccuracy and lower coverage in GC-rich regions and repetitive elements compared to GPS.
In addition, H3K4me3 and H3K36me3 enrichment also show high correlation with various levels of gene expression, even when FPKM is above 20 (Supplemental Fig. S8). When gene expression FPKM is greater than 5, H3K4me3 or H3K36me3 enrichment is still positively correlated with gene expression (Fig. 2C). We made similar observations in Liver and LM3 cells (Supplemental Fig. S7C,D). These results demonstrate the effect of histone methylations on gene regulation.
To further investigate the relationship between gene expression and MeGDP in tumorigenesis, we selected down-regulated genes in 97L cells that exhibit a decrease in MeGDP as compared to normal liver cells to perform Gene Ontology (GO) analysis (Fig. 2D). We found that this set of genes is enriched in the immune system process and metabolic process, which have been shown to be involved in tumorigenesis. These results may help us to understand the mechanism of cancer cell escape from the immune surveillance system in an alternative way, by which internal immune-related molecules might be epigenetically silenced by aberrant DNA methylation during tumor development. It is known that DNA methyltransferase inhibitors can up-regulate immune signaling in cancer through the viral defense pathway and reverse tumor-immune evasion, which may also modulate the epigenetic states of T cell phenotypes, as DNA methylation can act as a regulator for programming of T cell exhaustion and rejuvenation (Chiappinelli et al. 2015; Ghoneim et al. 2017; Topper et al. 2017). Recently, it has been reported that mutation or copy number loss of immune surveillance-related interferon gamma (IFNG) pathway genes is interpreted as a failure to respond to anti-CALA4 in melanoma patients (Gao et al. 2016). Similarly, we found that IFNG pathway genes are down-regulated in hepatoma cell lines, and MeGDP is reduced accordingly (P-value <0.001, one-tail Wilcoxon signed-rank test) (Fig. 2E), where expression and MeGDP are highly correlated (ρ = 0.62, P-value = 5.8 × 10−11, Spearman's rank correlation). Accordingly, we found that immune-related genes EDNRB, ACP5, and BST2 were all up-regulated by about two- to 75-fold after 5-AZA demethylation treatment (Supplemental Fig. S9). We also selected up-regulated genes in 97L cells that exhibit an increase in MeGDP as compared to normal liver cells to perform GO analysis (Supplemental Fig. S10). Therefore, we conclude that the reactivation of immunological surveillance genes in tumor cells by selective DNA methylation inhibition may be an alternative strategy in anti-tumor immunotherapy.
Methylation Boundary Shift in the promoter region modulates tumor-related ribosomal gene expression
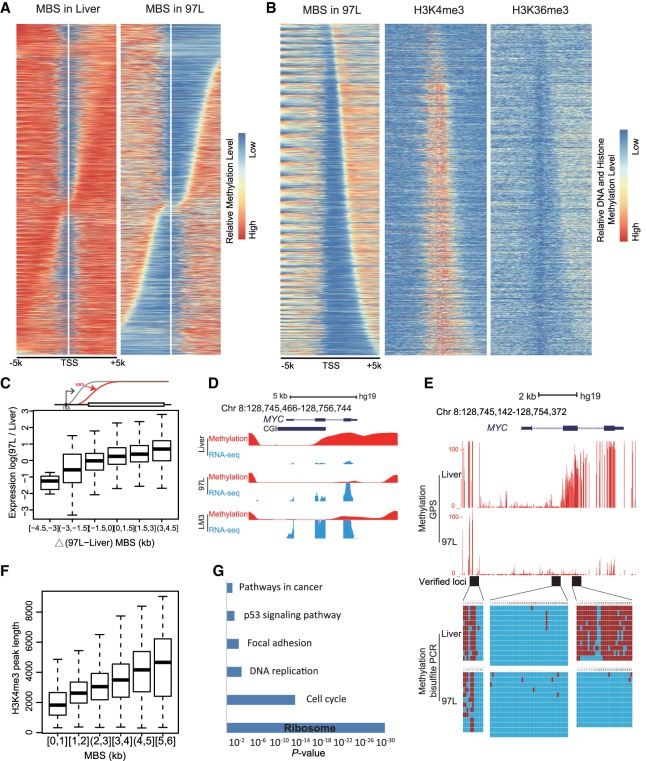
Although the pattern of DNA methylation in the promoter region is known to be in a “V” shape, the groove of the hematoma cell lines has shown a much wider opening than that of normal liver cells, suggesting that tumor cells may have much broader hypomethylation in the promoter region. We termed the DNA methylation boundary extension around transcription start site (TSS) as the Methylation Boundary Shift (MBS). To further investigate the extension of MBS in promoter, we selected genes with broadened MBS in 97L as compared to Liver cells and arranged the genes according to the length and direction of the DNA methylation extension from TSS (Fig. 3A). We categorized two groups of genes based on the direction of the MBS extension, direction to downstream, and direction to upstream of TSS. A similar MBS pattern can also be found in LM3 and primary liver cancer cells (Supplemental Fig. S11A,B). We observed that H3K4me3 and H3K36me3 are mutually exclusive within the MBS region, consistent with their role as the marker for active genes. Meanwhile, the boundary of the MBS extension also highly coincides with H3K4me3 and H3K36me3 patterns (Fig. 3B).
Figure 3.
Methylation Boundary Shift (MBS) in the promoter region modulates gene expression. (A) MBS in 97L cells displaying MBS extension to the upstream or downstream from TSS compared with Liver cells. Genes are arranged from top to bottom according to the length and direction of MBS. (B) H3K4me3 and H3K36me3 in 97L display a mutually exclusive enrichment within the MBS region. H3K4me3 enrichment and length are in agreement with MBS, where H3K36me3 is mutually exclusive. (C) MBS extension is positively related to gene expression in the box plot. The x-axis is the delta MBS (difference between 97L and Liver), and the y-axis is the 97L/Liver expression ratio. Gene expression is up-regulated with the MBS increase. Schematic MBS is displayed in the upper panel. (D) MBS is related to MYC up-regulation in hepatoma cells. Smoothed methylation (in red) shows the MBS to downstream in hepatoma cells, whereas MYC expression (in blue) is increased compared with liver cells. (E) MBS in the promoter of MYC is observed and verified by bisulfite sequencing PCR in three sequential loci (black block). Each red line represents a CpG site, and its height represents methylation level (upper). Each column represents a CpG site, and each line represents a picked TA clone by Sanger sequencing. Red blocks represent methylated CpGs, and blue blocks represent unmethylated CpGs (lower). (F) Length of the H3K4me3 peak is positively correlated with MBS downstream from TSS. The y-axis represents the length of the H3K4me3 peak, and the x-axis is the MBS extension downstream from TSS. (G) Ribosomal genes are significantly enriched in genes whose up-regulation is accompanied by MBS. Compared with liver cells, KEGG analysis displays that up-regulated genes with MBS extension downstream from TSS in 97L are enriched in terms such as ribosome and cell cycle, which are important for tumorigenesis.
To investigate the role of MBS in gene regulation, we analyzed the length of the MBS extension with corresponding gene expression level. By comparing 97L with Liver on highly expressed genes, we found that the length of the MBS extension downstream from TSS is positively correlated with the increase of gene expression (Fig. 3C), whereas the MBS extension to the upstream or promoter methylation alteration shows no obvious correlation with gene expression (Supplemental Fig. S11C). We observed similar results when comparing LM3 or primary liver cancer cells to Liver (Supplemental Fig. S11D,E). For example, oncogene MYC expression increased with the MBS extension downstream from TSS (Fig. 3D) and the MBS extension had been further verified by Sanger bisulfite sequencing (Fig. 3E). Moreover, MBS extension downstream from TSS coincides with the H3K4me3 peak length correspondingly (Fig. 3F), and the H3K4me3 peak length was reported to be involved in gene expression related to cell identity and tumorigenesis (Benayoun et al. 2014; Chen et al. 2015; Dahl et al. 2016). Here, we demonstrated that both MBS and H3K4me3 peak length is in fact positively correlated with gene expression (Supplemental Fig. S11F). Taken together, these results indicate that MBS in the promoter region may play an important role in modulating gene expression.
Next, we performed KEGG analysis for up-regulated genes in 97L cells with downstream MBS extension and found that these selected genes are enriched in pathways including ribosome and cell cycle (Fig. 3G; Supplemental Fig. S12A). We found that 48 of 60 up-regulated ribosomal genes with MBS were identified by GPS, but only seven by WGBS (Supplemental Fig. 12B,C). Therefore, MBS may contribute to the increased expression of ribosome biogenesis genes and consequently promote cell proliferation and transformation.
To further validate the correlation of gene expression and MeGDP or MBS, we performed GPS in two breast cell lines, MCF-10A and MCF-10A-1H (Zheng et al. 2018). We found the correlation between gene expression and MeGDP or MBS were consistent with the previous results (Supplemental Fig. S13A–C), which were also verified by other public WGBS data (Supplemental Fig. S13D–G).
Methylation Boundary Shift in enhancers promotes gene expression related to tumorigenesis
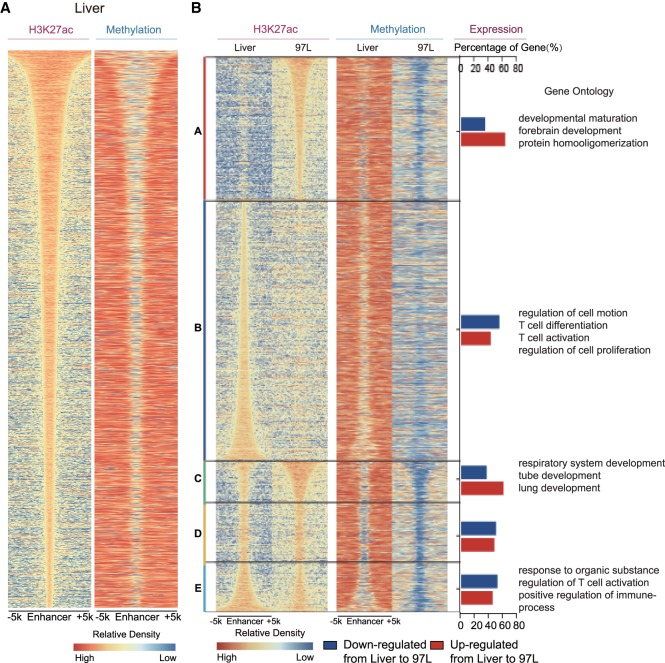
Like the promoter, the enhancer is another kind of regulatory element that correlates with DNA methylation (Aran and Hellman 2013; Hon et al. 2013). There might be several millions of enhancer elements embedded in the human genome, in which H3K27ac is a notable histone modification that marks the active enhancer (Creyghton et al. 2010; Shen et al. 2012). Given that MBS extension in the promoter region is clearly correlated with gene expression, we decided to explore whether MBS may also occur in the enhancer region during tumorigenesis. We sorted the predicted enhancers according to the length of H3K27ac peaks and compared them with the corresponding DNA methylation pattern. The width of H3K27ac peaks also coordinated with MBS in Liver as well as in 97L and LM3 cells (Fig. 4A; Supplemental Fig. S14A), indicating that MBS may also have regulatory effects on enhancers as well as the shaping of promoter activity to regulate gene expression.
Figure 4.
MBS is in accordance with enhancer alteration. (A) Methylation boundary shift in enhancer region. Enhancer is defined as the H3K27ac peak except those located in promoter region (TSS ±1 kb). Enhancers are arranged according to the H3K27ac peak length in Liver, and methylation level of the enhancer is shown in the same order as H3K27ac peaks. (B) General enhancer alteration including gain/loss and length variation coordinately with aberrant DNA methylation in 97L cells. A–E are classified as follows: A (97L-specific enhancer); B (Liver-specific enhancer); C (enhancer in 97L cells with width at least 500 bp more than that in Liver); D (enhancer in 97L or Liver cells extends <500 bp); and E (enhancer in Liver with width at least 500 bp more than that in 97L cells). A–E represent enhancer gain, loss, extension, no extension, and reverse extension, respectively, in 97L compared with Liver cells with corresponding aberrant DNA methylation in the right column. Percentage of up-regulated (red bar) and down-regulated genes (blue bar) is significantly correlated with enhancer altering pattern (P-value = 7.565 × 10−13, Pearson's χ2 test). Gene Ontology analyses show that genes coordinated with enhancer alteration are enriched in terms related to cell identity transformation, immune, and T cell activation.
Compared with the normal liver cells, we discovered that 97L cells gained a set of enhancers and lost another set, and the changes of enhancers are coupled with DNA methylation alteration (Fig. 4B). GO analysis showed that genes that gain enhancers in 97L are enriched in developmental maturation and forebrain development, whereas genes that lose enhancers are enriched in the regulation of cell motion as well as T cell differentiation and activation, both of which are closely associated with tumorigenesis. Apart from enhancer gain or loss above, we also grouped enhancers by enhancer length extension, no extension, and reverse extension in 97L compared to Liver (Fig. 4B). Corresponding methylation levels for each enhancer show the consistency between enhancer length and MBS. GO analysis shows that genes with enhancer extension are enriched in lung development, and genes with enhancer reverse extension are enriched in the regulation of T cell activation, which are important to cell identity and tumorigenesis, respectively. In addition, five enhancer patterns show significant differences in the up- and down-regulated genes comparing 97L to Liver (P-value = 7.6 × 10−13, χ2 test) (Fig. 4B), where gained/extended enhancers tend to have more up-regulated genes than lost/reversely extended enhancers. Moreover, analysis of enhancers in comparing Liver versus LM3 also shows similar results (Supplemental Fig. S14B). Seeing that LM3 is derived from 97L and can easily metastasize to lung, gain or loss of enhancers is significantly less when comparing 97L versus LM3 (Supplemental Fig. S14C), indicating that 97L and LM3 are more similar except for their metastasis properties. Furthermore, genes that gained or lost enhancers between 97L and LM3 are enriched in lymphocyte proliferation and response to drugs, respectively. Meanwhile, genes with broadening or shortening of the enhancer are enriched for positive regulation of cell differentiation as well as glucose metabolic process, which contributes to our understanding of the metastasis properties of LM3. Together, these results suggest that aberrant DNA methylation pattern in the enhancer region may modulate enhancer features and regulate gene expression, which further shapes tumor cell behavior during tumorigenesis and metastasis.
Aberrant DNA methylation patterns induce gain or loss of cell identity through enhancer switching, resulting in tumorigenesis and metastasis
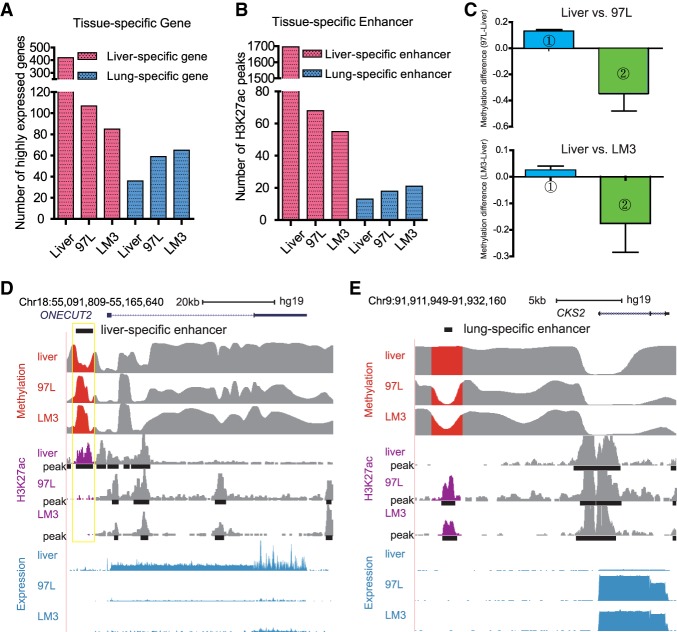
Human cells and tissues maintain cell identity by expressing cell-type–specific genes, which are controlled by tissue-specific enhancers or other related epigenetic components. In turn, cells can lose their identity once the expression of their own cell-type–specific gene expression is reduced, or when other cell-type–specific genes are expressed during tumor development (Heintzman et al. 2009; Roadmap Epigenomics Consortium et al. 2015). Knowing this, we focused on the analysis of the tissue-specific genes and enhancers. We found that the number of highly expressed liver-specific genes in 97L and LM3 is reduced by 74% and 80%, respectively, as compared to Liver. Meanwhile, the number of lung-specific genes (ICAM1 for instance) as well as testis (COIL), stomach (ZDHHC18), heart (MAPKAPK2), and colon (MAX) tissue-specific genes (Pan et al. 2013) increased in 97L and LM3 cells, indicating that hepatoma cells have lost their identity and adopted characteristics of other cell types during the tumor development process. We found that there are more lung-specific genes in LM3 than in 97L cells, consistent with the fact that LM3 is more prone to lung metastasis (Fig. 5A; Supplemental Fig. S14D). Thus, the lung-specific genes expressed in LM3 might aid the LM3 cells in adopting features of the lung and surviving in the lung environment.
Figure 5.
Aberrant DNA methylation pattern is correlated with enhancer alteration and cell identity. (A) 97L and LM3 lose the identity of liver by losing the liver-specific gene expression while they gain lung cell identity by expressing the lung-specific gene. The y-axis shows the number of highly expressed genes (FPKM ≥5). There are more lung-specific genes in LM3 cells compared with 97L cells. (B) Tissue-specific enhancers altered accordingly in 97L and LM3 with preferential metastasis to lung. For liver-specific enhancers, the number of overlapped H3K27ac peaks sequentially decrease in Liver, 97L, and LM3, which is related to loss of liver cell identity, whereas lung-specific enhancer is increased, especially in LM3 cells, which may promote expression of lung-specific genes, further helping LM3 cells metastasize to lung. (C) Loss or gain of tissue-specific enhancer is correlated with DNA methylation. Column 1 (blue bar) shows liver tissue-specific enhancers lost in hepatoma cells are accompanied by increased DNA methylation, and Column 2 (green bar) shows lung tissue-specific enhancers that are gained in hepatoma cells might be induced by largely decreased DNA methylation. The y-axis represents normalized and comparable methylation difference. Liver versus 97L is displayed in the upper panel, whereas the Liver versus LM3 is in the lower panel. (D) Tissue-specific enhancer gain or loss with aberrant DNA methylation is correlated with tissue-specific gene expression. Liver-specific gene ONECUT2 is down-regulated in 97L and LM3 with the loss of liver-specific enhancer where DNA methylation is increased. (E) Lung-related gene CKS2 is up-regulated in 97L and LM3 cells with gain of lung-specific enhancer where DNA methylation is decreased.
Since it is well known that enhancers are tissue-specific, we conducted analysis on the tissue-specific enhancers and aberrant DNA methylation in hepatoma cells. Accordingly, we found that the number of liver-specific enhancers was reduced in 97L and LM3 cells compared to Liver, whereas the number of lung-specific enhancers increased in 97L and LM3 cells. As expected, there are more lung-specific enhancers in LM3 than in 97L cells, consistent with the lung-specific genes (Fig. 5B). Moreover, the gain of lung-specific enhancers is associated with DNA methylation reduction, and the loss of liver-specific enhancers is associated with DNA methylation increase in 97L and LM3 cells (Fig. 5C), suggesting that aberrant DNA methylation is correlated with cell identity changes by altering the tissue-specific enhancer. Meanwhile, the loss or gain of cell identities accompanied by aberrant DNA methylation is an alternative process during tumorigenesis, and hepatoma metastasis is associated with the expression of lung-specific genes, which facilitates the adaptation of liver cancerous cells in the lung environment. For example, ONECUT2, a liver-specific gene, is silenced in 97L and LM3 cells and is coupled with a loss of H3K27ac peak and gain of DNA methylation in a nearby liver-specific enhancer region (Fig. 5D). On the other hand, we observed that increased expression of CKS2 in 97L and LM3, a lung-specific gene, is coupled with an increased level of H3K27ac and decreased DNA methylation in a lung-specific enhancer (Fig. 5E). Hence, these results imply that aberrant DNA methylation pattern in the enhancer region may modulate enhancer switching, which in turn affect its target gene expression and further shape tumor cell behavior during tumorigenesis and metastasis. This scenario is an elegant illustration for interpreting tumor metastasis.
Our findings here demonstrate that lung-specific genes expressed in LM3 cells may have aided them to better adapt and survive in the lung environment, indicating that the loss of cell type identity and gain of other cell identity is a critical turning point during tumor development, which also provide an alternative way to understand organ-specific tumor metastasis.
Discussion
The diploid human epigenome contains more than 109 cytosines, of which approximately one-twentieth are CpGs. Methylated cytosines, in particular, are stable epigenetic marks that can be passed onto the next generation. Furthermore, DNA methylation patterns have been regarded as a transcript of environmental exposures in one's lifetime and have been used as biomarkers for disease diagnosis and risk detection. Dynamic DNA methylation can occur at either distinct regions of the genome or the same region but with varying levels. It is known that transcription factors tend to bind at Low Methylated Regions (LMRs), which display tissue-specific or developmental stage–specific patterns (Feldmann et al. 2013). The interplay between LMRs and TF is usually associated with changes in enhancer function, which further influences gene expression and cell state.
Variation in DNA methylation can take place at single cytosine sites, known as a methylation variable position (MVP), or single methylation polymorphism (SMP), the epigenetic equivalent of SNP. Accurately capturing SMP or low-level DNA methylation patterns is critical for studying their impact on human diseases. Current strategies for detecting DNA methylation genome-wide include (1) using a restriction enzyme to differentiate and recognize methylated versus unmethylated cytosine bases; (2) immunoprecipitation of methylated DNA; and (3) bisulfite treatment that converts unmethylated cytosines to uracils while leaving methylated cytosines unchanged. Among them, WGBS is currently the most popular method; however, as we mentioned before, WGBS bioinformatics analysis is usually based on either wild-card or three-letter aligners and tends to waste a large proportion of the sequencing reads (Kunde-Ramamoorthy et al. 2014; Plongthongkum et al. 2014; Marx 2016). To reconcile this problem, we developed the GPS method, in which the first part of the pair-ended read is the original DNA sequence and the second part of the read pair is bisulfite-converted sequence. Since the first read can be accurately mapped to the reference genome, it can serve as guide for the second read. GPS has high DNA methylation coverage rate, at nearly 96% of the human liver genome and can also detect strand-specific whole-genome methylome. Compared with WGBS, the advantages of GPS include higher cytosine coverage, especially in CpG islands and repetitive elements, and higher accuracy in DNA methylation detection as verified by bisulfite pyrosequencing. Moreover, it can detect both genetic information and epigenetic information in a single experiment, which is important for samples with limited DNA, such as single cells. GPS is a powerful tool for studying global DNA methylation patterns and their effects on human diseases.
Extensive literature has shown that the accumulation of genetic and epigenetic abnormalities is related to cancer. Through reverse genetic methods, the role of genetic alterations in cancer development has been well illustrated. However, the role of epigenetic alterations in cancer has not been shown equally well. For example, little is known about the interactions between the promoter and gene body methylation. Our results indicate that gene expression is correlated with the methylation level of gene body difference to promoter (MeGDP), suggesting that methylation in promoter and gene body may work in a collaborative manner to activate gene expression. Further analysis shows that genes associated with MeGDP are involved in the immune system process, in which the equilibrium of cancer cells versus host immune surveillance may be disrupted, resulting in edited immunogenicity by immunoepigenetics to weaken immune responses and promote clinically apparent cancers, which may help us to understand the limited response of current immune therapy by blocking PD-1/PD-L1, and DNMT inhibitors may reverse the immune signals of cancer cell for the basis of epigenetic therapy in clinic trials (Jones et al. 2016).
Furthermore, Methylation Boundary Shift in the promoter and enhancer regions as a specific pattern of DNA methylation may alter gene expression, especially in highly expressed genes. We found that MBS expansion in cancer cells is closely associated with ribosomal gene expression, which is related to nucleolus function and involved in tumorigenesis. MBS is also supported by the finding that hypomethylated regions expand and contract with lineage specificity in the adult hematopoietic compartment (Hodges et al. 2011). Meanwhile, the liver cancer–specific shortening of MBS are accompanied with expression reduction of immune and immune escape–related genes, which can also play an essential role in immune surveillance.
Tumor metastasis is the process by which malignant tumor cells penetrate the wall of lymphatic or blood vessels and circulate to other sites through the bloodstream and eventually form another clinically detectable tumor. In this scenario, it is difficult for cancer cells to survive outside their region of origin, and therefore the metastasizing tumor cells must either find a location with similar characteristics or remodel and adapt themselves to the new environment for assimilated symbiosis. In support of this “Assimilated Symbiosis” theory, we showed that LM3 cells, which can easily metastasize to lung, have altered DNA methylation patterns with reduced liver-specific genes and increased lung-specific genes. We also analyzed DNA methylation in the liver- and lung-specific enhancer regions in 97L and LM3 cells, which altered accordingly as well, suggesting DNA methylation mediates cancer metastasis by shaping tumor cell behavior to environment. Our results suggest that liver cells may lose their identity and gain the lung cell identity to adapt to the lung environment through aberrant DNA methylation, which provide a new insight for understanding the mechanism of tumor metastasis.
In summary, GPS can unbiasedly and precisely detect the DNA methylome, especially in GC-rich and repetitive regions as compared to WGBS. MeGDP and MBS may act as effective parameters in gene regulation related to immunity and tumor metabolism. Meanwhile, aberrant DNA methylation in tissue-specific enhancers may contribute to altering cell identity and help us to further understand DNA methylation during tumor development and metastasis.
Methods
GPS library construction
Genomic DNA from tissues and cell lines was extracted using QIAamp DNA Mini Kit (Qiagen, 51306). Genomic DNA was fragmented into 300–500 bp by sonication using bioruptor (Diagenode, Bioruptor plus). Thirty units of T4 DNA polymerase (New England BioLabs, M0203L) was used to perform 3′→5′ digestion of the DNA fragments for 100 min at 12°C followed by adding 10 µL dNTP mix which contained dATP, dTTP, dGTP, and 5′-methyl-dCTP nucleotide (final concentration 0.5 mM) and incubating for 30 min at 37°C. Then, A-tailing was performed by klenow fragment (3′→5′ exo-) from NEB (M0212L), and methylated adapter was ligated to DNA fragments using T4 DNA ligase from NEB (M0203L) according to the manufacturer's instruction. The fragments were size-selected and processed to bisulfite conversion, CT-transformed DNA was amplified with the KAPA HiFi Uracil + DNA Polymerase (KAPA Biosystems, KM2801) using Illumina TruSeq primers, fragments between 400 and 500 bp were selected for high-throughput sequencing on Hi Seq 2500 (Illumina). Spike-in lambda DNA (Promega, D150A) was added at a mass ratio of 1/200 for bisulfite conversion testing.
Adjustment for GPS Read2 and Read1
We first performed quality control and adapter trimming using NGS QC Toolkit v2.3.3. The GPS library was sequenced using Illumina HiSeq 2500 with pair end (2 × 100 bp). According to the GPS library construction, the unmethylated cytosine (C) in Read1 was converted into thymine (T) by bisulfite treatment coupled with PCR amplification, while the C in Read2 was not converted into T, so that the sequence in Read2 was the same as the sequence of the reference genome. However, due to possible insufficiency in T4 DNA polymerase treatment, there may exist the conversions of C→T in 3′-end of Read2. These conversions in Read2 could influence the efficiency of Read2 mapping onto the reference genome.
To minimize such influence, we searched the sequencing boundary between treated and not treated by T4 DNA polymerase (Supplemental Fig. S15). If the T4 DNA polymerase sufficiently treated the 3′-end of the fragment, the cytosines (Cs) in the 3′-end of the fragment could be all methylated. Otherwise, the unmethylated Cs could exist in the fragment, especially proximate to the 5′-end, and these unmethylated Cs could be converted into Ts after bisulfite treatment. The complement nucleobase of the C-converted T in the opposite strand was G-converted A in Read2. According to the Illumina pair-end sequencing principle, the Read2 is located in the 3′-end opposite strand of the fragment. So the methylated and unmethylated C were presented as G and A in Read2, respectively, due to the complement of C = G and T = A. Due to the methylation level of C in CH (H is A, T, C), context is generally very low, if the methylated CH was found, it suggests the T4 polymerase treatment was sufficient, otherwise it was insufficient. The corresponding sequences of the methylated CH in Read2 were 3′-G[A/T/G]-5′. So, we scanned Read2 from 5′- to 3′-end to find the last 3′-G[A/T/G]-5′, which was treated as the boundary between treated and not treated by T4 polymerase. The Read2 sequence between the boundary and the 3′-end was discarded, and the remaining was used for aligning onto the genome. If the length of the remaining read was <35 bp, the read would also be discarded. We also noticed that, in the case of samples with high methylated CH, such as neuronal tissue, the CHs were substantially methylated and would remain cytosine after bisulfite treatment whether T4 polymerase-treated or not. However, analysis showed high efficiency of T4 polymerase in GPS experiment, so that high methylated CHs would have a minor effect on the mapping efficiency.
Conversely, there also exists overtreatment of T4 polymerase, which also possibly induced over methylation in the 3′-end of Read1. To overcome such potential bias, we scanned Read1 from the 3′- to the 5′-end to find the first conversion of C/T, which was defined as a boundary between nontreatment and treatment of T4 polymerase. The sequence between the boundary and 5′-end of Read1 was used for methylation calling (Supplemental Fig. S15).
GPS alignment
Adjusted Read2s were aligned onto the reference genome (hg19) using Bowtie 2 (v2.2.3) (Langmead and Salzberg 2012) with default parameters except “-k 20”, which allows to output up to 20 candidate mapping positions. Statistically, there are 56,434,896 CpG sites in the human reference assembly GRCh37 (hg19) and 58,690,664 CpG sites in GRCh38 (hg38), with 96% common CpG sites in both references. Thus, realigning the reads to GRCh38 (hg38) would not significantly affect the conclusions.
After the alignment of Read2, the fragments were anchored into the genome. According to the Illumina sequencing specification, the Read1 is located in the opposite strand downstream from the corresponding Read2. The fragment size in the GPS library was approximately 400–500 bp. We used the Smith-Waterman, an algorithm of local sequence alignment, to map the Read1 to the reference target sequence that was located within 1 kb of the opposite strand downstream from the Read2. The algorithm first built a score matrix as follows:
where S = sequence from Read1; T = sequence from reference genome; S[i],T[j] = ith nucleobase in S, and jth in T; m = length of S; and n = length of T.
To obtain the optimum local alignment, our script started with the highest value in the matrix F(i, j). Then, go backward to one of positions (i−1, j), (i, j−1), and (i−1, j−1), depending on the direction of movement used to construct the matrix. This methodology is maintained until a matrix cell with zero value is reached. Finally, the alignment is reconstructed as follows: Starting with the highest value, reach (i, j) using the previously calculated path. A diagonal jump implies there is an alignment (either a match or a mismatch). A bottom-up jump implies there is an insertion. A right-left jump implies there is a deletion. The Smith-Waterman allows exactly aligning Read1 onto the reference genome with tolerance of C/T mismatch. For each cytosine, we counted the number of “T” (NT) and the number of “C” (NC). The methylation percentage in the cytosine is defined as NC/(NC + NT) × 100%.
Data access
Raw and aligned sequencing data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www. ncbi.nlm.nih.gov/geo/) under accession number GSE92328. The GPS method is available on GitHub (https://github.com/lijinbella/GPS) and as a Supplemental Code.
Competing interest statement
The authors declare the following competing interests:
Patent 1
Patent applicant (institution): Fudan University
Name of inventor(s): Wenqiang Yu, Yan Li, Feizhen Wu
Application number: ZL 2013 1 0572289.X
Manuscript aspect: DNA methylation detection method GPS
Patent 2 (international patent)
Patent applicant (institution): Fudan University
Name of inventor(s): Wenqiang Yu, Yan Li, Feizhen Wu
Application number: PCT/CN2014/090979
Manuscript aspect: DNA methylation detection method GPS
Supplementary Material
Acknowledgments
We thank Guoming Shi for supplying hepatoma cell lines 97L and LM3. We thank Yue Yu for manuscript revision and careful reading of the manuscript. We thank Yao Xiao, Min Xiao, Xiaoguang Ren, Lan Zhang, Liping Zhao, RuKui Zhang, and Shuzheng Song for editorial help and comments on the manuscript. This work was supported by the Ministry of Science and Technology (Grant Nos. 2016YFC0900303 and 2018YFC1005004), Major Special Projects of Basic Research of Shanghai Science and Technology Commission (Grant No. 18JC1411101), the Science and Technology Innovation Action Plan of Shanghai (Grant No. 17411950900), the National Natural Science Foundation of China (Grant Nos. 31671308, 31872814, and 81272295), the Shanghai Science and Technology Committee (Grant No. 12ZR1402200), the National High-tech R&D program, 863 Program (Grant No. 2015AA020108), the National Key Research and Development Program of China (Grant No. 2016YFC1000500), the Ministry of Education of the People's Republic of China (Grant No. 2009CB825600), and the National Key R&D Program of China (Grant No. 2018YFC0910405).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.240606.118.
References
- Akondy RS, Fitch M, Edupuganti S, Yang S, Kissick HT, Li KW, Youngblood BA, Abdelsamed HA, McGuire DJ, Cohen KW, et al. 2017. Origin and differentiation of human memory CD8 T cells after vaccination. Nature 552: 362–367. 10.1038/nature24633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran D, Hellman A. 2013. DNA methylation of transcriptional enhancers and cancer predisposition. Cell 154: 11–13. 10.1016/j.cell.2013.06.018 [DOI] [PubMed] [Google Scholar]
- Benayoun BA, Pollina EA, Ucar D, Mahmoudi S, Karra K, Wong ED, Devarajan K, Daugherty AC, Kundaje AB, Mancini E, et al. 2014. H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell 158: 673–688. 10.1016/j.cell.2014.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C. 2012. Analysing and interpreting DNA methylation data. Nat Rev Genet 13: 705–719. 10.1038/nrg3273 [DOI] [PubMed] [Google Scholar]
- Chen K, Chen Z, Wu D, Zhang L, Lin X, Su J, Rodriguez B, Xi Y, Xia Z, Chen X, et al. 2015. Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor-suppressor genes. Nat Genet 47: 1149–1157. 10.1038/ng.3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, Hein A, Rote NS, Cope LM, Snyder A, et al. 2015. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 162: 974–986. 10.1016/j.cell.2015.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci 107: 21931–21936. 10.1073/pnas.1016071107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl JA, Jung I, Aanes H, Greggains GD, Manaf A, Lerdrup M, Li G, Kuan S, Li B, Lee AY, et al. 2016. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 537: 548–552. 10.1038/nature19360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggert T, Wolter K, Ji J, Ma C, Yevsa T, Klotz S, Medina-Echeverz J, Longerich T, Forgues M, Reisinger F, et al. 2016. Distinct functions of senescence-associated immune responses in liver tumor surveillance and tumor progression. Cancer Cell 30: 533–547. 10.1016/j.ccell.2016.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Koldobskiy MA, Göndör A. 2016. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet 17: 284–299. 10.1038/nrg.2016.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann A, Ivanek R, Murr R, Gaidatzis D, Burger L, Schubeler D. 2013. Transcription factor occupancy can mediate active turnover of DNA methylation at regulatory regions. PLoS Genet 9: e1003994 10.1371/journal.pgen.1003994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Shi LZ, Zhao H, Chen J, Xiong L, He Q, Chen T, Roszik J, Bernatchez C, Woodman SE, et al. 2016. Loss of IFN-γ pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell 167: 397–404.e9. 10.1016/j.cell.2016.08.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoneim HE, Fan Y, Moustaki A, Abdelsamed HA, Dash P, Dogra P, Carter R, Awad W, Neale G, Thomas PG, et al. 2017. De novo epigenetic programs inhibit PD-1 blockade-mediated T cell rejuvenation. Cell 170: 142–157.e19. 10.1016/j.cell.2017.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goltz D, Gevensleben H, Dietrich J, Ellinger J, Landsberg J, Kristiansen G, Dietrich D. 2016. Promoter methylation of the immune checkpoint receptor PD-1 (PDCD1) is an independent prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients following radical prostatectomy. Oncoimmunology 5: e1221555 10.1080/2162402X.2016.1221555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Greten FR, Karin M. 2010. Immunity, inflammation, and cancer. Cell 140: 883–899. 10.1016/j.cell.2010.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112. 10.1038/nature07829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges E, Molaro A, Dos Santos CO, Thekkat P, Song Q, Uren PJ, Park J, Butler J, Rafii S, McCombie WR, et al. 2011. Directional DNA methylation changes and complex intermediate states accompany lineage specificity in the adult hematopoietic compartment. Mol Cell 44: 17–28. 10.1016/j.molcel.2011.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon GC, Rajagopal N, Shen Y, McCleary DF, Yue F, Dang MD, Ren B. 2013. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nature Genetics 45: 1198–1206. 10.1038/ng.2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Issa JP, Baylin S. 2016. Targeting the cancer epigenome for therapy. Nat Rev Genet 17: 630–641. 10.1038/nrg.2016.93 [DOI] [PubMed] [Google Scholar]
- Krueger F, Andrews SR. 2011. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27: 1571–1572. 10.1093/bioinformatics/btr167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Kreck B, Franke A, Andrews SR. 2012. DNA methylome analysis using short bisulfite sequencing data. Nat Methods 9: 145–151. 10.1038/nmeth.1828 [DOI] [PubMed] [Google Scholar]
- Kunde-Ramamoorthy G, Coarfa C, Laritsky E, Kessler NJ, Harris RA, Xu M, Chen R, Shen L, Milosavljevic A, Waterland RA. 2014. Comparison and quantitative verification of mapping algorithms for whole-genome bisulfite sequencing. Nucleic Acids Res 42: e43 10.1093/nar/gkt1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Tang Y, Ye L, Liu B, Liu K, Chen J, Xue Q. 2003. Establishment of a hepatocellular carcinoma cell line with unique metastatic characteristics through in vivo selection and screening for metastasis-related genes through cDNA microarray. J Cancer Res Clin Oncol 129: 43–51. 10.1007/s00432-002-0396-4 [DOI] [PubMed] [Google Scholar]
- Marx V. 2016. Genetics: profiling DNA methylation and beyond. Nat Methods 13: 119–122. 10.1038/nmeth.3736 [DOI] [PubMed] [Google Scholar]
- Mohme M, Riethdorf S, Pantel K. 2017. Circulating and disseminated tumour cells – mechanisms of immune surveillance and escape. Nat Rev Clin Oncol 14: 155–167. 10.1038/nrclinonc.2016.144 [DOI] [PubMed] [Google Scholar]
- Neri F, Rapelli S, Krepelova A, Incarnato D, Parlato C, Basile G, Maldotti M, Anselmi F, Oliviero S. 2017. Intragenic DNA methylation prevents spurious transcription initiation. Nature 543: 72–77. 10.1038/nature21373 [DOI] [PubMed] [Google Scholar]
- Pan JB, Hu SC, Shi D, Cai MC, Li YB, Zou Q, Ji ZL. 2013. PaGenBase: a pattern gene database for the global and dynamic understanding of gene function. PLoS One 8: e80747 10.1371/journal.pone.0080747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plongthongkum N, Diep DH, Zhang K. 2014. Advances in the profiling of DNA modifications: cytosine methylation and beyond. Nat Rev Genet 15: 647–661. 10.1038/nrg3772 [DOI] [PubMed] [Google Scholar]
- Rivera CM, Ren B. 2013. Mapping human epigenomes. Cell 155: 39–55. 10.1016/j.cell.2013.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. 2015. Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330. 10.1038/nature14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber RD, Old LJ, Smyth MJ. 2011. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science 331: 1565–1570. 10.1126/science.1203486 [DOI] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. 2012. A map of the cis-regulatory sequences in the mouse genome. Nature 488: 116–120. 10.1038/nature11243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer DH, Russler-Germain DA, Ketkar S, Helton NM, Lamprecht TL, Fulton RS, Fronick CC, O'Laughlin M, Heath SE, Shinawi M, et al. 2017. CpG island hypermethylation mediated by DNMT3A is a consequence of AML progression. Cell 168: 801–816.e13. 10.1016/j.cell.2017.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topper MJ, Vaz M, Chiappinelli KB, DeStefano Shields CE, Niknafs N, Yen RC, Wenzel A, Hicks J, Ballew M, Stone M, et al. 2017. Epigenetic therapy ties MYC depletion to reversing immune evasion and treating lung cancer. Cell 171: 1284–1300.e21. 10.1016/j.cell.2017.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinay DS, Ryan EP, Pawelec G, Talib WH, Stagg J, Elkord E, Lichtor T, Decker WK, Whelan RL, Kumara HM, et al. 2015. Immune evasion in cancer: mechanistic basis and therapeutic strategies. Semin Cancer Biol 35(Suppl): S185–S198. 10.1016/j.semcancer.2015.03.004 [DOI] [PubMed] [Google Scholar]
- Xi Y, Li W. 2009. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics 10: 232 10.1186/1471-2105-10-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngblood B, Hale JS, Kissick HT, Ahn E, Xu X, Wieland A, Araki K, West EE, Ghoneim HE, Fan Y, et al. 2017. Effector CD8 T cells dedifferentiate into long-lived memory cells. Nature 552: 404–409. 10.1038/nature25144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Sun HC, Wang WQ, Zhang QB, Zhuang PY, Xiong YQ, Zhu XD, Xu HX, Kong LQ, Wu WZ, et al. 2012. Sorafenib down-regulates expression of HTATIP2 to promote invasiveness and metastasis of orthotopic hepatocellular carcinoma tumors in mice. Gastroenterology 143: 1641–1649.e5. 10.1053/j.gastro.2012.08.032 [DOI] [PubMed] [Google Scholar]
- Zheng YZ, Xue MZ, Shen HJ, Li XG, Ma D, Gong Y, Liu YR, Qiao F, Xie HY, Lian B, et al. 2018. PHF5A epigenetically inhibits apoptosis to promote breast cancer progression. Cancer Res 78: 3190–3206. 10.1158/0008-5472.CAN-17-3514 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.