Abstract
Background:
The major mode of metabolism of nicotine is by hydroxylation via cytochrome P450 (CYP) 2A6, but it can also undergo glucuronidation by UDP-glucuronosyltransferases and oxidation by flavin monooxygenases (FMOs). The goal of this study was to examine the potential importance of FMOs in nicotine metabolism and assess the potential impact of missense polymorphisms in active FMOs on nicotine-N’-oxide formation.
Methods:
Urine samples from 106 current Chinese smokers were analyzed for nicotine metabolites by mass spectrometry (MS). Wild-type FMOs 1 through 5 and their most prevalent non-synonymous variants were cloned and over-expressed in HEK293 cells, and were tested in oxidation reactions against nicotine.
Results:
A strong inverse correlation was observed between the ratio of urinary 3’-hydroxycotinine/cotinine, a measure of CYP2A6 activity, and the urinary levels of nicotine-N’-oxide alone (r=−0.383, p <0.001) or nicotine-N’-oxide measured as a ratio of total nicotine metabolites (r=−0.414, p <0.001) in smokers. In addition to FMO1 and FMO3, the functional FMO2427Q isoform was active against nicotine, while FMO4 and FMO5 exhibited low activity against nicotine (KM>5.0 mM). Significant (p <0.05) decreases in N’-oxidation activity (Vmax/KM) were observed for the FMO1I303V, FMO3N61S, FMO3D132H, FMO3V257M, and FMO3E308G variants in vitro when compared to their respective wild-type isoforms; the truncated FMO2Q472stop isoform exhibited no enzyme activity.
Conclusions:
These data indicate that increases in nicotine-N’-oxidation occurs in subjects with deficient CYP2A6 activity, that several FMO enzymes are active in nicotine-N’-oxidation.
Impact:
Several common missense FMO variants are associated with altered enzyme activity against nicotine and may play an important role in nicotine metabolism in low-CYP2A6 activity subjects.
Keywords: Flavin monooxygenase, FMO, nicotine-N’-oxide, CYP2A6, nicotine metabolism, nicotine dependence, non-synonymous polymorphisms, pharmacogenetics
Introduction
Currently, nearly 480,000 deaths in the United States (US) are attributable to cigarette smoking and second hand smoke exposure annually, and tobacco use is the leading cause of preventable deaths worldwide. (1,2) Approximately 90% of lung cancer in humans are attributed to cigarette smoking, and lung cancer results in more deaths each year than any other cancer type. (3,4)
Nicotine is abundant in tobacco and tobacco smoke, activating the nicotinic acetylcholine receptors (nAChRs) in the brain, resulting in tobacco addiction. (5) Nicotine is rapidly metabolized in vivo, with a half-life of approximately 2 h. (6) The major metabolic pathway for nicotine is hydroxylation by cytochrome P450 (CYP) 2A6. Nicotine also exhibits metabolism through either glucuronidation, catalyzed by the UDP-glucuronosyltransferases (UGTs), or oxygenation by flavin monooxygenases (FMOs; see Figure 1). Besides nicotine, the FMO family catalyzes the oxidation of heteroatoms (such as nitrogen, sulfur, phosphorus, and selenium), xenobiotics (such as drugs and pesticides), (7) and endogenous compounds [such as cysteamine, (8) trimethylamine, (9) phenethylamine and tyramine (10)]. The human family of FMOs includes 5 known functional genes - FMO1, FMO2, FMO3, FMO4 and FMO5. The FMO1, FMO2, FMO3, and FMO4 genes are clustered in chromosome 1, region q24.3, while the FMO5 gene is located at q21 of the same chromosome. (11) These enzymes exhibit a tissue-specific expression pattern in adults. FMO1 is mainly expressed in the kidney and FMO3-FMO5 in the liver, with FMO4 and FMO5 expressed at lower levels than FMO3. (12) FMO2 is mainly expressed in the lung, (13) but its expression in other organ tissues has not been thoroughly investigated. The most common FMO2*1 allele (rs6661174) acts as a pseudogene, coding for an early stop codon (Q472Stop) resulting in a truncated protein without the last 64 C-terminal amino acids. (13) This common variant accounts for >95% of all FMO2 alleles in all populations combined. The remaining 5% of FMO2 alleles encode a full-length, active FMO2 protein, with African Americans exhibiting a minor allele frequency (MAF) of 14% for these functional alleles. (14) The high overall allelic prevalence corresponding to the truncated form of this protein has likely resulted in the null expression of this enzyme observed in most human studies.
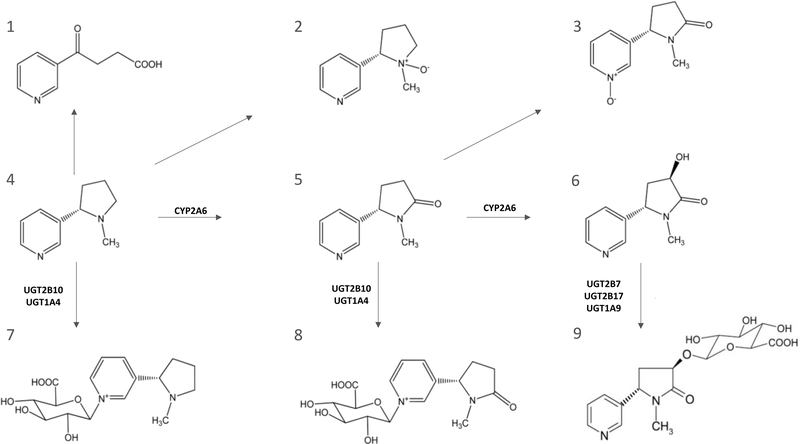
Figure 1. Metabolic scheme for nicotine.
(1) 4-hydroxy-4-(3-pyridyl)-butanoid acid, (2) nicotine-N’-oxide, (3) cotinine-N-oxide, (4) nicotine, (5) cotinine, (6) trans-3’-hydroxycotinine, (7) nicotine-N-glucuronide, (8) cotinine-N-glucuronide, and (9) cotinine-3’-O-glucuronide. Adapted from reference (17).
It has been reported that FMOs are the enzymes responsible for the stereoselective N’-oxidation of nicotine. (15) Nicotine-N’-oxide (NOX) formation accounts for 4–7% of total urinary nicotine metabolites in humans under normal circumstances, (16,17) In a study of five subjects, when the CYP2A6-mediated hydroxylation pathway is diminished or impaired, urinary levels of NOX are increased to up to 31% of total nicotine metabolites. (18) While some studies have shown that FMO1 and FMO3 may also play a role in NOX formation, (15,19) a comprehensive analysis of all five FMO enzymes on the impact of nicotine oxidation has not yet been performed.
Studies have shown a marked difference in tobacco addiction profiles among different populations. (20) This could be due to several factors including genetic differences in the enzymes that metabolize nicotine. (21–23) Functional polymorphisms in CYP2A6 have been correlated with differences in cotinine and 3-hydroxycotinine formation. (22) Similarly, genetic variants in several UGTs are correlated with differences in the glucuronidation of urinary nicotine, cotinine and 3-hydroxycotinine. (24,25) Genome-wide association studies (GWAS) have identified associations between FMO1 and FMO3 single nucleotide polymorphisms (SNPs) and nicotine dependence.(7,19,26–28) Previous studies have shown that genetic variations in FMO3 in different smoker cohorts were associated with nicotine dependence (28) while a recent in vivo study demonstrated an association between the FMO3 E308G/E158K haplotype with nicotine N’-oxidation and smoking behavior. (29) However, the mechanism behind the functional role of those polymorphisms in nicotine metabolism and addiction was not addressed in these studies.
The goal of the present study was to examine the variability of NOX formation in smoker’s urine, fully characterize FMO involvement in NOX formation, and assess the impact of the most prevalent non-synonymous polymorphisms in active FMO enzymes on nicotine metabolite production in vitro.
Materials and Methods
Chemicals and Materials.
Pooled human liver, lung, esophagus or kidney RNA was purchased from Biochain (Newark, CA). Platinum Pfx DNA polymerase, the pcDNA3.1/V5-His-TOPO mammalian expression vector, Lipofectamine 3000, One Shot TOP10 competent E. coli, Superscript II RT kits, SuperScript VILO synthesis kits, anti-V5-HRP antibody, and Luria broth base were obtained from Invitrogen (Carlsbad, California, USA). The anti-calnexin-HRP antibody was purchased from Abcam (Cambridge, UK) while the QuikChange II Site-Directed Mutagenesis Kit used to make FMO variant vectors was acquired from Agilent (Santa Clara, California, USA). Oligonucleotides used for site-directed mutagenesis were manufactured by Integrated DNA Technologies (Coralville, Iowa, USA). RNeasy kits and QIAquick gel extraction kits were purchased from Qiagen (Valencia, California, USA) while the GeneJet plasmid mini and midipreps were purchased from Thermo Fisher Sci (Whaltman, MA, USA). The Ambion PureLink RNA Mini kit was purchased from Life Technologies (Carlsbad, CA, USA) while the BCA protein assays used in protein assessment were purchased from Pierce (Rockford, Illinois, USA). Dulbecco’s Modified Eagles Medium, Dulbecco’s phosphate-buffered saline, fetal bovine serum, and geneticin (G418) were purchased from Gibco (Grand Island, New York, USA). The NADPH regeneration system was purchased from Corning (Corning, New York, USA). Nicotine tartrate and benzydamine hydrochloride used as substrates for N’-oxidation activity assays were purchased from Sigma-Aldrich (St. Louis, Missouri, USA). Pooled human liver microsomes were purchased from Sekisui XenoTech (Kansas City, Kansas, USA). TaqMan qPCR probes were purchased from AB Applied Biosystems (Foster City, CA, USA). High performance liquid chromatography-grade ammonium acetate and acetonitrile were purchased from Fisher Scientific (Pittsburgh, Pennsylvania, USA) while the ACQUITY UPLC BEH-HILIC (1.7μm 2.1 × 100 mm) column was purchased from Waters (Milford, Massachusetts, USA). The internal standard d3-NOX was purchased from Toronto Research Chemicals (Ontario, Canada).
Nicotine metabolites in the urine of smokers.
Urine samples from 106 Chinese smokers who participated in two cancer epidemiology cohorts were analyzed. All 106 subjects were free of cancer at the latest follow-up in 2015. Among them, 63 were from the Shanghai Cohort Study, a prospective cohort of 18,244 Chinese men in Shanghai, China, when they were 45–64 years of age at enrollment during 1986–1989. (30,31) The mean (SD) age at the time of urine sample collection was 56.6 (5.5) y. The remaining 43 subjects were from the Singapore Chinese Health Study, a prospective cohort of 63,257 Chinese men and women in Singapore at 45–74 years of age when they were recruited into the study in 1993–1998. (32,33) Of the 43 subjects, 7 (16%) were women and the average age (SD) at urine collection was 67.0 (6.4) y. The Shanghai cohort subjects smoked an average of 15.3 cigarettes/day for 46.5 y as compared to 17.2 cigarettes/day and 31.7 y for the Singapore cohort subjects. In addition to nicotine, eight of its major metabolites (nicotine-N’-oxide, cotinine-N-oxide, trans-3-hydroxycotinine, nicotine-N-glucuronide, cotinine-N-glucuronide, trans-3′-hydroxycotinine-O-glucuronide and 4-hydroxy-4-(3-pyridyl)-butanoid acid) were quantified in urine samples of the 106 smokers using a previously reported liquid chromatography (LC)-mass spectrometry (MS) method. (17)
Generation of FMO over-expressing cell lines.
Pooled RNA (2 μg) from lung, liver, esophagus or kidney was used to generate corresponding cDNAs using the SuperScript II RT-PCR kit, and cDNA corresponding to 10 ng RNA was used as template for PCR amplification of wild-type (wt) FMOs 1–5 using 1 U of Pfx polymerase and FMO gene-specific primers (see Supplemental Table 1). PCR was performed with an initial denaturation temperature of 94ºC for 2 min, followed by 40 cycles of 94ºC for 30 s, 45 s at the specific Tm for each FMO (see Supplemental Table 1), and 68ºC for 105 s, and a final cycle of 10 min at 68ºC. Sequences of each PCR-amplified wt FMO product were verified by Sanger sequencing, and each were cloned into the pcDNA 3.1/V5-His-TOPO vector in-frame with the plasmid V5 epitope tag at the C-terminus 3’-end prior to transformation into One Shot TOP10 competent E. coli using standard protocols. Insert orientation and sequences of plasmid DNA prepared from individual clones were confirmed by a second round of Sanger sequencing. Lipofectamine 3000 reagent was used to transfect HEK293 cells with 2.5 μg of each FMO-containing pcDNA 3.1/V5-His-TOPO vector, with cells grown in Dulbecco’s Modified Eagles Medium supplemented with 10% fetal bovine serum and 1.5% geneticin (G418). The parent cell line used in this study, HEK293, was purchased from ATCC in 2015 and was authenticated by ATCC in 2017 using short-tandem repeat polymorphisms analysis. No test was performed to detect Mycoplasma in these cells.
FMO variants were created by site-directed mutagenesis of wt FMO-containing pcDNA3.1/ V5-His-TOPO plasmids using QuikChange II XL kits and specific mutagenic FMO primers (see Supplemental Table 2). All FMO variants were confirmed by Sanger sequencing and transfected into HEK293 cells as described above. For cloning of the FMO2 full-length isoform, we used the high-prevalence FMO2*Q472Stop variant as template for site directed mutagenesis to change the codon from the premature stop to a glutamine residue.
Enzyme kinetic assays and UPLC-MS/MS analysis.
Microsomal membrane fractions of FMO over-expressing cell lines were prepared by differential centrifugation as previously described (34,35) and used as the enzyme source for all activity assays. For relative FMO quantification, equal amounts of microsomal protein (20 μg) were loaded on 10% SDS-polyacrylamide gels and FMO protein quantity determined by Western blot analysis using an anti-V5-HRP antibody in a 1:2,500 dilution. As a loading control for microsomal fractions, an anti-calnexin-HRP antibody was used in a 1:5,000 dilution for all Western blots. Image J software (http://rsb.info.nih.gov/ij; National Institutes of Health, Bethesda, MD, USA) was used to perform densitometry analysis, and the relative expression of each FMO-containing microsomal preparation was used for normalization in N’-oxidation activity assays.
N’-oxidation reactions (final volume = 25 μL) contained 25 μg of total microsomal protein, 50 mM potassium phosphate, a NADPH regenerating system (1.55 mmol/L NADP+, 3.3 mmol/L glucose-6-phosphate, 3.3 mmol/L MgCl2, 0.5 of 40 U/mL glucose-6-phosphate dehydrogenase) and varying concentrations of nicotine tartrate (0.05 – 5,000 μM). For reactions with benzydamine, assays were performed as described above for nicotine using benzydamine hydrochloride at concentrations equivalent to the reported KM’s for each FMO for this substrate. (36) All incubations were performed for 60 min at 37°C with shaking at 300 rpm, with reactions terminated by the addition of 25 μL ice-cold acetonitrile containing internal standard (d3-NOX). Supernatants were collected after centrifugation at 16,100 g for 10 min at 4°C prior to subsequent UPLC-MS/MS analysis. Pooled human liver microsomes and untransfected parent HEK293 cell microsomes were used as the protein source for positive and negative assay controls, respectively.
UPLC-MS/MS was performed with mobile phases that consisted of 5 mmol/L of NH4AC (pH 5.7) in either 50% acetonitrile (v/v) (buffer A) or 90% acetonitrile (buffer B). An UPLC-BEH-HILIC column (2.1 × 100 mm, 1.7 μm) was used for separation of NOX from nicotine as follows: 20% buffer A for 1.5 min, a linear gradient to 100% buffer A from 1.5–2.5 min, maintenance of 100 % buffer A for 3 min, and a re-equilibrium step to the initial 20% buffer A conditions from 5.5 to 7 min (flow rate 0.4 mL.min−1). The injection volume was 2 μL using a column temperature of 30ºC. MS/MS detection was performed in a Waters ACQUITY XEVO TQD instrument in MRM ESI+ mode. The MS/MS scan was performed using the following mass transitions: m/z 179.1→117.1, 182.1→117.1 and 163.1→105 and to monitor NOX, d3-NOX and nicotine, respectively. The desolvation temperature was 500°C, with 800 L/h of nitrogen gas. The collision energy was optimized at 28 V, 27 V and 28 V for NOX, nicotine, and d3-NOX, respectively. A cone voltage of 25 V and 0.5 s dwell time resulted in high-sensitivity detection of NOX and d3-NOX. Metabolite retention times observed in the enzymatic incubations were compared with the retention times of d3 internal standard metabolites. UPLC-MS/MS detection for benzydamine and benzydamine-N-oxide was performed using a previously reported separation method. (37)
mRNA expression analysis.
Total RNA was isolated from wt FMO3 (FMO3158E/308E), FMO3158K/308E, FMO3158E/308G and FMO3158K/308G expressing HEK293 cells using the Ambion PureLink RNA Mini kit according to the manufacturer’s protocol. Total RNA (1 μg) was used to synthesize cDNA using the SuperScript VILO synthesis kit following the manufacturer’s protocol. qPCR analysis was performed using a TaqMan probe specific for the FMO3 human gene (Hs00199368_m1) while a TaqMan probe specific for the RPLP0 gene (Hs00420895_gH) was used as a housekeeping gene. Expression of the FMO3 gene was normalized to the internal control RPLP0 and the fold-change was calculated using the delta-delta Ct method. (38) All experiments were performed in triplicate.
Statistical analysis.
To compare the levels of NOX formation in subjects stratified by the nicotine metabolic ratio (NMR), statistical stratifications by median and quartiles were performed. Once the samples were stratified, relative FMO activity was determined by the levels of NOX/total nicotine equivalents (TNE, equivalent to nicotine plus its eight measured metabolites) in the stratified groups. The distributions of these urinary biomarkers measured or their ratios were markedly skewed toward high values, which were normalized to a large extent by transformation to logarithmic values. Therefore, formal statistical testing was performed on logarithmically transformed values, and geometric (as opposed to arithmetic) means are presented. Statistical analyses were conducted using the statistics program R v.3.4 (Vienna, Austria). All analyses were two-sided and considered significant if p <0.05 for all tests. Continuous outcome variables were analyzed using Student’s t-test or two-way analysis of variance (ANOVA) using a Tukey post-hoc test for multiple corrections for all possible pairwise comparisons. (39)
Kinetic parameters were determined from the Michaelis–Menten equation using GraphPad Prism version 6.01 (GraphPad Software, San Diego, CA). Relative maximum reaction rates (Vmax) were calculated as pmol NOX.L−1.min−1, with values normalized to wt FMO-V5 microsomal protein as determined by Western blot analysis using Image J software as described above. All reported values represent the results [e.g., mean + standard deviation (SD)] of three independent experiments, with triplicates run for each experiment. The activity of each variant enzyme was compared to its corresponding wt isoform using the Student’s t-test. A two-tailed P-value of less than 0.05 was considered the threshold for statistical significance.
Results
Nicotine metabolites in smokers.
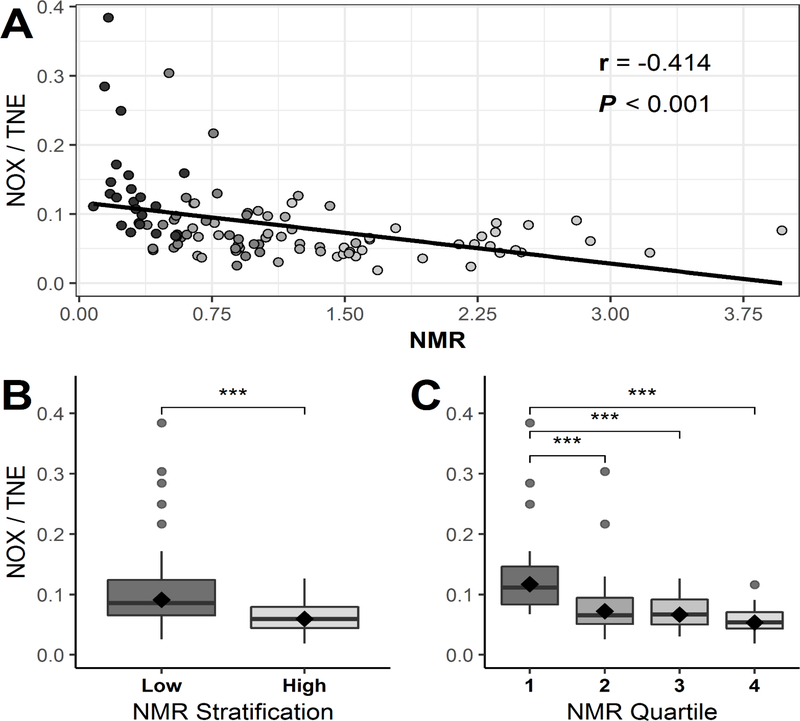
Table 1 shows the levels of nicotine and its eight major metabolites in the urine of Chinese smokers. NOX accounted for a mean of 7.4% (95% CI, 6.7% - 8.2%) of the urinary TNE in the 106 smokers examined. To examine whether NOX formation differed based on CYP2A6 activity, subjects were stratified based on their NMR, which is the ratio of urinary 3’-hydroxycotinine/cotinine in smokers and which has been shown to be an effective marker of CYP2A6 activity. (40) A strong negative correlation was observed between the urinary NMR ratio and both urinary NOX levels alone (r = −0.383, p<0.001) and urinary NOX measured as a ratio of TNE (r = −0.414, p<0.001; Figure 2, panel A). When stratifying urine samples at the median value of NMR, the mean percentage of the urinary NOX/TNE ratio for each stratified group was 9.2% for the low CYP2A6 activity group and 6.0% for the high activity group, a significant (p<0.001) 1.53-fold increase in NOX levels between groups (Figure 2, panel B). When stratifying samples by quartiles for NMR, the mean % urinary NOX/TNE for each group (low to high NMR) was 11.8%, 7.3%, 6.7% and 5.4%, respectively, corresponding to a significant trend (p < 0.001) with a 2.2-fold difference between the lowest vs highest NMR quartiles (Figure 2, panel C). Furthermore, when subjects (n=5) with the lowest 5% urinary NMR were compared to the highest 5% urinary NMR samples (n=5), the percent urinary NOX/TNE were 18.8 in the former compared with 5.5 in the latter, respectively (Supplementary Figure 2).
Table 1.
Urinary nicotine metabolite profile in Chinese smokers.
| Metabolite | Geometric mean (95% CI) (nmol/mg creatinine) |
|---|---|
| trans-3’-Hydroxycotinine | 6.19 (4.94, 7.76) |
| 4-HPBAa | 9.81 (8.21, 11.7) |
| 3’-Hydroxycotinine-O-Glucuronide | 3.49 (2.85, 4.28) |
| Cotinine | 7.77 (6.19, 9.76) |
| Nicotine | 3.96 (2.93, 5.36) |
| Cotinine-N-Glucuronide | 3.30 (2.64, 4.12) |
| Nicotine-N’-Oxide | 3.70 (3.03, 4.52) |
| Nicotine-N-Glucuronide | 2.89 (2.32, 3.60) |
| Cotinine-N-Oxide | 1.81 (1.54, 2.13) |
| TNEb | 50.1 (41.9, 59.9) |
| NOX/TNEc | 0.074 (0.067, 0.082) |
| NMRd | 0.80 (0.69, 0.92) |
HPBA, 4-hydroxy-4-(3-pyridyl)-butanoic acid
TNE, total nicotine equivalents (nicotine + trans-3’-Hydroxycotinine + 4-HPBA + 3HC-O-Glucuronide + cotinine + cotinine-N-Glucuronide + nicotine-N’-Oxide + nicotine-N-Glucuronide + cotinine-N-Oxide)
Ratio of nicotine-N’-oxide (NOX) / TNE
Ratio of trans-3’-hydroxycotinine / cotinine
Figure 2. Analysis of urinary NOX levels in Chinese smokers.
Panel A, Scatterplot of urinary NMR (3’-hydroxycotinine/cotinine) and urinary NOX/TNE in Asian smokers. Black line indicates the linear regression line of the data. The Pearson correlation coefficient (r-value) is adjusted for study site. Panel B, urinary NOX/TNE shown for samples stratified by the median for urinary NMR. Panel C, urinary NOX/TNE shown for samples stratified by quartiles of urinary NMR. The black diamonds within the boxplots (panels B and C) indicate the geometric mean of each group, the horizontal line of the boxplots are median values, while individual dots are considered outliers. *** Indicates statistical significance p ≤ 0.001.
FMO activities against nicotine.
HEK293 cell lines expressing V5-tagged FMO enzymes were created and FMO expression in microsomal fractions verified by Western blot using an anti-V5 antibody (Supplemental Figure 1). Wt FMOs 1–5 were successfully expressed in HEK293 cells, with the relative level of FMO expression calculated based on the relative band intensities of the V5 signal and were used to calculate relative Vmax’s obtained in N’-oxidation activity assays. Catalytic activities for each of the FMO enzymes was verified using benzydamine as a probe substrate, with all wt and variant FMO enzymes demonstrating benzydamine N’-oxidation activity. The KM values found for each FMO against benzydamine (60 ± 8 μM, 440 ± 65 μM, 80 ± 6 μM, >3 mM, and >2mM for FMOs 1, 2, 3, 4, and 5, respectively) were similar to those reported previously. (36)
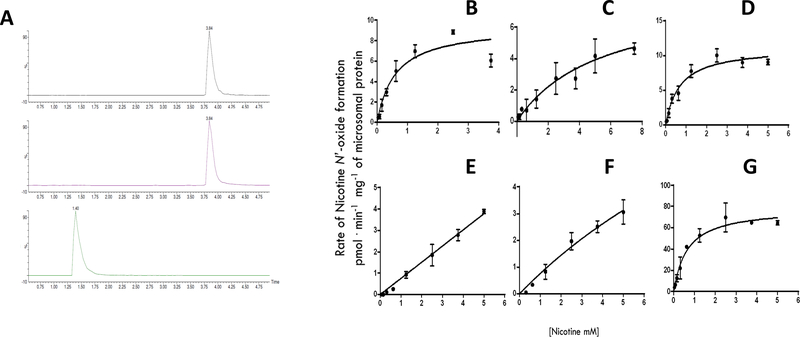
Using microsomes isolated from FMO over-expressing HEK293 cell lines, in vitro assays demonstrated that all of the wt FMOs (FMO1, FMO2, FMO3, FMO4 and FMO5) exhibited activity against nicotine. As shown by UPLC-MS/MS, a NOX peak was observed at a retention time of 3.84 min that was easily discernable from the nicotine substrate peak at 1.40 min (Figure 3, Panel A). Representative kinetics plots of NOX formation for individual FMO enzymes are shown in Figure 3, panels B-F. Enzyme kinetic analysis suggested that both FMOs 1 and 3 exhibited the highest overall activity for nicotine (Vmax/KM = 16 nL•min−1•mg−1 for both FMO1 and FMO3; Table 2). The KM values of 0.75 and 0.70 mM for FMO1 and FMO3, respectively, were similar to that observed for human liver microsomes (KM = 0.55; Table 2 and Figure 3, panel G). While full-length FMO2 exhibited less overall activity (Vmax/KM = 1.8 nL•min−1•mg−1) than either FMO1 or FMO3, the activities of FMO4 and FMO5 were very low, with KM values >5 mM (Table 2 and Figure 3, panels E and F, respectively).
Figure 3. Representative UPLC-MS/MS chromatograms and Michaelis-Menten kinetic curves of nicotine-N’-oxide (NOX) formation.
Panel A shows representative UPLC-MS/MS chromatograms of NOX formation. Top panel, NOX-d3 internal standard [retention time (RT) = 3.84 min, m/z: 182.1 > 117.1]; middle panel, NOX standard (RT = 3.84 min, m/z: 179.1 > 117.1); bottom panel, nicotine standard (RT = 1.4 min, m/z: 163.1 > 106.1). For each compound, the ratio of analyte peak area to that of the deuterated standard was used for quantification. Panels B-F show Michaelis-Menten curves for wild-type FMOs. Panel B, wt FMO1; Panel C, wt FMO2; Panel D, wt FMO3; Panel E, wt FMO4; Panel F, wt FMO5; and Panel G, HLM. Each curve represents a set of three different repetitions. The rate of NOX formation was adjusted per μg of corresponding FMO protein expressed in each cell line, as determined by Western blot analysis using a V5 antibody.
Table 2.
Kinetic analysis of nicotine N’-oxidation by wt and variant FMOs.a
| Enzyme | KM | Vmax | Vmax/KM | Relative activity d |
|---|---|---|---|---|
| (mM) | (pmol • min−1 • mg−1) b | (nL • min−1 • mg−1) b | ||
| FMO1 | 0.8 ± 0.2 | 11.0 ± 1.5 | 15.5 ± 3.1 | 1.0 |
| FMO1I303V | 1.8 ± 0.9 | 6.9 ± 1.4 | 4.5 ± 1.4 | 0.3* |
| FMO2 | 4.5 ± 2.7 | 6.5 ± 1.6 | 1.8 ± 0.7 | |
| FMO2Q472stop | Inactive | |||
| FMO3 | 0.7 ± 0.1 | 11.2 ± 0.5 | 16.1 ± 1.4 | 1.0 |
| FMO3N61S | 4.7 ± 0.7 | 18.2 ± 2.7 | 3.9 ± 0.2 | 0.2* |
| FMO3D132H | 2.1 ± 0.5 | 7.9 ± 1.1 | 3.9 ± 0.5 | 0.2* |
| FMO3E158K | 1.6 ± 0.4 | 9.8 ± 3.5 | 6.2 ± 1.1 | 0.4* |
| FMO3V257M | 1.8 ± 0.2 | 11.8 ± 0.3 | 6.8 ± 0.8 | 0.4* |
| FMO3V277A | 0.7 ± 0.3 | 6.7 ± 1.5 | 9.8± 2.1 | 0.6 |
| FMO3E308G | 1.7 ± 0.7 | 13.0 ± 2.1 | 8.9 ± 2.7 | 0.5* |
| FMO3158K/308G | 2.2 ± 0.5 | 31.6 ± 3.3 | 14.9 ± 2.2 | 0.9 |
| FMO4 | > 5 | ND | ND | |
| FMO5 | > 5 | ND | ND | |
| HLM c | 0.5 ± 0.6 | 79 ± 2.6 | 158 ± 24.3 |
Data are expressed as the mean ± S.D. of three independent experiments. Nicotine concentrations of 0.005-5 mM were used for kinetic analysis.
Vmax was calculated per total microsomal protein levels after normalization based on microsomal FMO expression levels as determined by Western blot analysis as described in the Materials and Methods.
Pooled human liver microsomes (HLM) from 20 individuals.
Relative activities show the Vmax/KM ratio of each FMO variant versus its corresponding wild-type FMO.
p < 0.05 vs corresponding wild-type FMO.
Effects of FMO1 and FMO3 nonsynonymous polymorphisms on N’-oxidation activity.
Prevalent missense polymorphisms for the three most active FMO enzymes (FMO1, FMO2, and FMO3) were identified using the National Center for Biotechnology Information (NCBI) SNP database (dbSNP). For this study, only non-synonymous polymorphisms with a MAF >1% in the 1000 Genomes Project were used. (41) Non-synonymous SNPs were identified for FMO1 (I303V, rs16864314) and FMO3 (N61S, rs72549322; D132H, rs12072582; E158K, rs2266782; V257M, rs1736557; and V277A, rs2066530; Table Supplemental TS2). An E158K/E308G haplotype allele with a MAF ≥ 1% was also identified for FMO3. Other than the truncated FMO2472Stop variant, no independent missense polymorphisms with a MAF ≥ 1% were identified for this gene. Variants were cloned and over-expressed in the HEK293 cell line as described for wt FMOs.
The FMO1303V variant exhibited a significantly (p =0.023) higher KM (1.8 mM) and a significant (p =0.023) 3.5-fold lower overall catalytic activity (Vmax/KM = 4.5 nL•min−1•mg−1) as compared to wt FMO1 (KM = 0.8 mM; Vmax/KM = 16 nL•min−1•mg−1; Table 2). The FMO2 protein encoded by the common FMO2Q472stop variant exhibited no activity against nicotine or benzydamine in vitro using up to 5 mM benzydamine. The FMO361S, FMO3132H, FMO3158K and FMO3257M variants all exhibited significantly (p = 0.006, 0.003, 0.002, and 0.003, respectively) lower overall catalytic efficiencies as determined by Vmax/KM (ranging from 3.9 to 8.9 nL•min−1•mg−1) as compared to its wt FMO3 counterpart (Vmax/KM = 16 nL•min−1•mg−1). While the KM was identical for the wt FMO3 and FMO3277A isoforms (0.7 mM for both), the Vmax for the FMO3277A variant (6.7 pmol•min−1•mg−1) was 40% (p =0.029) lower than that observed for wt FMO3 (11 pmol•min−1•mg−1). The KM values for all the other FMO3 variants were significantly (p < 0.05) higher than that observed for wt FMO3, with the FMO361S variant exhibiting a KM that was 6.7-fold higher (4.7 mM). While microsomes containing the FMO3158K/308G haplotype variant exhibited a KM that was 3.1-fold higher than the wt isoform (2.2 mM vs 0.7 mM, respectively), there was no significant (p =0.565) difference in overall catalytic activity as determined by Vmax/KM (15 vs. 16 nL•min−1•mg−1, respectively).
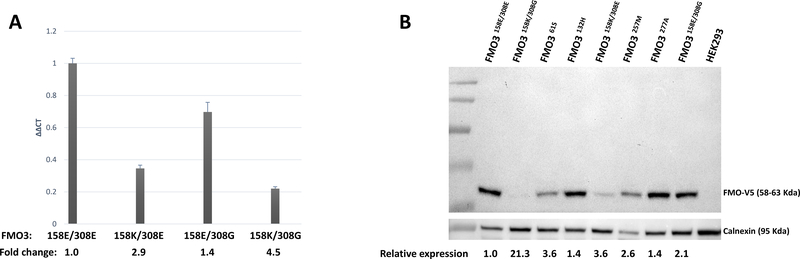
Previous studies suggested that the FMO3158K/308G allele was associated with decreased FMO3 protein expression. (42) In the present analysis, FMO3 mRNA expression was determined in the HEK293 cell lines overexpressing wt FMO3 (FMO3158E/308E), or FMO3158K/308G variants. Similarly, significant (p < 0.05) decreases in both mRNA (2.9- and 1.4-fold) and protein (3.6- and 2.1-fold) expression were observed for the FMO3158K/308E and FMO3158E/308G over-expressing HEK293 cell lines, respectively, when compared to cells over-expressing wt FMO3 (Figure 4). In contrast, while a 4.5-fold decrease in mRNA expression was observed for the FMO3158K/308G variant vs. wt FMO3, a >21-fold lower level of FMO3 protein was observed for cells over-expressing this variant as compared to cells over-expressing wt FMO3.
Figure 4. Expression analysis for FMO3 isoforms over-expressed in HEK293 cells.
Panel A, qPCR results for wt FMO3158E/308E, FMO3158K/308E, FMO3158E/308G, and FMO3158K/308G; Panel B, The relative expression of each FMO enzyme is shown at the bottom, with the expression of wt FMO3158E/308E designated as 1.0. Calnexin was used as the loading control and non-transfected HEK293 cells were used as the negative control.
Discussion
The levels of urinary NOX as a percentage of total nicotine metabolites for 106 Chinese smokers showed high variability between subjects. When samples were stratified by CYP2A6 activity (determined by the urinary NMR), the levels of urinary NOX were significantly different between high vs. low CYP2A6 activity groups. NOX formation comprised ≥19% of total nicotine metabolites in subjects exhibiting the lowest CYP2A6 activities. These data suggest that nicotine N’-oxidation is a key metabolism pathway for smokers, particularly those individuals whose CYP2A6 activity is diminished or deleted. For example, the NOX/TNE ratio was 18.8% in smokers with the lowest 5% NMR group whereas the corresponding NOX/TNE ratio was 5.5% in smokers with the highest 5% NMR. These results are consistent with a previous report where NOX levels were as high as 31% in two subjects whose CYP2A6 gene was deleted. (18) This is particularly important for Asian populations where the MAF of the CYP2A6 deletion allele is high (15–20%). (43) While the CYP2A6 deletion allele (CYP2A6 *4) is much less prevalent in other populations including Caucasians and African Americans,(44) the prevalence of other alleles that code for CYP2A6 variants with decreased enzyme function is still high in these populations. (45,46) Therefore, the impact of FMO variants on nicotine metabolism needs to be examined in other racial groups.
In the present study, the five human FMO enzymes were tested for N’-oxidation activity against nicotine in vitro. While all of the recombinant FMO isoforms mediated NOX formation, FMO1 and FMO3 exhibited the highest activity, with the KM for FMO1 (0.8 mM) similar to that reported previously for this enzyme (1.2 mM). (19) While FMO3 exhibited a KM (0.8 mM) that was similar to that observed for FMO1 in the present study, it was significantly lower than that observed in previous studies where the rate of NOX formation was still linear at 2 mM nicotine. (19) These differences in kinetics for FMO3 is likely due to the fact that the insect baculosomal system was used as the FMO over-expression system in previous studies, while the FMOs (including FMO3) were over-expressed in the human HEK293 cell line in the present study.
FMOs exhibit tissue-specific expression. FMO1 is an extra-hepatic drug metabolizing enzyme expressed in kidney, intestine and possibly the brain, (47) whereas FMO3 is the FMO isoform most highly expressed in liver. (12) Given that the hepatic enzymes FMO4 and FMO5 exhibited minimal levels of activity against nicotine in the present study, the data presented in this study suggest that FMO3 may be the primary hepatic enzyme involved in NOX formation, with FMO1 potentially more important for extra-hepatic nicotine metabolism.
The FMO2 variant that expresses a functional full-length protein is observed primarily in people of African descendent ancestry (MAF = 0.133), with most of the remaining human population expressing the FMO2472stop allele which codes for a truncated non-functional enzyme.(14,48) The present study is the first to demonstrate that the functional full-length FMO2 variant has activity against nicotine. While its KM (4.5 mM) was about 5–6-fold higher than that observed for either FMO1 or FMO3, this enzyme has been reported to be primarily expressed in human lung, with FMO2 mRNA detected in human lung specimens from people of African descent. (14) Given the low prevalence of the full-length functional FMO2 allele in the Asian population (0%), (41) FMO2 is, however, unlikely to play a role in variability in NOX formation in the Asian populations examined in the present study.
Given that FMOs 1, 2 and 3 were the main catalysts for nicotine-N’-oxidation in the present study, the potential role of prevalent genetic variants (MAF ≥ 1%) in these enzymes on nicotine metabolism was investigated. The FMO1303V variant exhibited a significant (p = 0.02) decrease in activity when compared with its wt counterpart, showing a ~3-fold decrease in overall NOX formation as determined by Vmax/KM. In previous studies, Furnes et al. (49) tested this variant against several substrates and did not observe differences in sulfoxidation activity against methyl-p-tolyl sulfide, fenthion and methamizole when compared to the wt FMO1 isoform. However, in the same study, N-oxygenation activity was decreased against imipramine, decreasing both its affinity and maximum velocity, supporting the theory that differences observed in catalytic activity could be substrate dependent. The FMO1303V variant is present in African Americans and in the Chinese population (MAF = 10 and 4%, respectively) but not in Caucasians (MAF < 0.0.01%). Since smoking related GWAS studies have been performed in primarily Caucasian populations, the effect of this variant on nicotine dependence and/or cigarette consumption had not been well studied. (19,26,28) Interestingly, several non-coding region variants in FMO1 have also been shown to be correlated with nicotine dependence in Caucasian populations. This includes several FMO1 promoter variants which were associated with differences in FMO1 expression and protein levels, subsequently resulting in differences in overall activity between individuals. (19)
The FMO2472stop variant was tested using nicotine as a substrate, and as expected, exhibited no activity against nicotine. This finding agrees with previously reported data where the truncated protein is catalytically inactive against a variety of substrates. (13,14,50)
The FMO361S, FMO3132H, FMO3158K, and FMO3257M variants showed statistically significant (p <0.05) decreases in overall catalytic activity as compared to the wt FMO3 isoform, suggesting that individuals with any of these genetic variants could impair nicotine transformation to NOX in vivo. This decrease was as high as 4-fold for both the FMO361S and FMO3132H variants. The FMO361S variant results in an amidic to hydroxylic amino acid change, and, consistent with the present study, previous reports on this variant showed abolished activity towards trymethylamine N-oxidation. (51) The FMO3132H variant has one of the most drastic changes in terms of amino acid structure, changing from an acidic to a basic amino acid. Similar to the decreases in nicotine N’-oxidation activity observed in the present study, the FMO3132H variant was associated with 30 and 60% decreases in activity against methimazole and trimethylamine, respectively, in previous studies. (52)
In the present study, the FMO3158K and FMO3257M variants exhibited 50–60% decreases in overall activity when compared to wt FMO3. The FMO3158K variant was shown in different studies to exhibit similar decreases in catalytic activity towards a variety of other substrates(14,52–54) This variant is widely distributed among different ethnicities (MAF: CEU, 41%; ASW, 33%; MXL, 43%; CDX, 13%; (41)), and consistent with the lower activity observed for this variant against nicotine in the present study, this polymorphism has been associated with nicotine dependence and/or cigarette consumption in previous studies. (26,28,29)
For the FMO3257M variant, an aliphatic amino acid is changed to a sulfur-containing amino acid. This polymorphism is widely distributed among different populations (MAF: CEU 7%, ASW, 9%; MXL, 5%; CDX, 21%) (41). Similar to that observed for the FMO3158K variant, a significant 2.6-fold increase in KM (but no significant alteration in Vmax) was observed as compared to wt FMO3, resulting in significant decreases in overall catalytic activity against nicotine. Interestingly, previous studies reported no significant difference in activity between the FMO3257M variant and wt FMO3 on trimethylamine-N-oxidation, suggesting substrate specificity for this SNP. (51,53)
The FMO3277A variant, which retains the aliphatic nature of the amino acid at position 277, exhibited a minimal change in overall activity against nicotine (Vmax = 6.7 vs 11.2 pmol.min−1.mg−1, for FMO3277A and wt FMO3, respectively). This is similar to that observed in previous studies against other compounds including trimethylamine-N-oxidation. (55)
The FMO3158K/308G haplotype variant occurs with high frequency among different populations (MAF: CEU, 22%; ASW, 9%; MXL, 5%; CDX, 11%). The linkage disequilibrium for the two SNPs at codons 158 and 308 is 98% and the frequency of this haplotype is defined by the MAF of the E308G variant. (41) Results from the present study indicated no significant difference in overall activity of this haplotype variant against nicotine. However, in vivo studies have shown that individuals with this FMO3 variant exhibit impaired activity towards a number of FMO3 substrates including nicotine (28,29,56) A recent study showed that this haplotype was correlated with a 50% reduction in FMO3 protein levels in a panel of human adult liver samples, a pattern not observed for mRNA levels measured in the same samples. (42) It was suggested that overall protein instability, resulting in an early ubiquitin-proteasome pathway, was potentially responsible for the in vivo reduced enzymatic activity of this haplotype. Consistent with these results, the HEK293 cell line over-expressing the FMO3158K/308G variant exhibited decreases in FMO3 protein levels that were >4.5-fold less than the decreases observed for FMO3 mRNA as compared to HEK293 cells that over- expressed wt FMO3. This differential pattern of expression was not observed when examining the effects on FMO3 protein and mRNA expression of cells over-expressing each of the corresponding missense SNPs at codons 158 and 308 individually and suggest a direct effect on protein stability by the FMO3158K/308G haplotype variant.
While not observed as an independent SNP in the population, the FMO3308G variant has been previously tested against a variety of substrates in vitro, showing a trend of decreased activity. (53,57) This is consistent with the results obtained in the present study where this SNP is associated with decreased in vitro NOX formation. In studies implicating the FMO3158K/308G haplotype, due to the high linkage disequilibrium between the FMO3 codons 158 and 308 SNPs, the FMO3308G variant has been significantly correlated with nicotine dependence (time to first cigarette in the morning). (29) In the same study, this variant was associated with aberrant mRNA splicing in the liver and brain, suggesting that this SNP could also potentially be affecting overall FMO protein levels by multiple mechanisms.
The present study is, to our knowledge, the most comprehensive study examining nicotine-N’-oxidation by the FMO family of enzymes. Nicotine-N’-oxidation is one of the direct detoxification pathways for nicotine, and while it accounts for a mean of 4 – 7% of total urinary nicotine metabolites across multiple studies, (16,17) data from the present study demonstrate that NOX formation may be a critical pathway for nicotine elimination in people with diminished or deficient CYP2A6 activity. Further, results from the present study suggest that functional SNPs in FMOs 1, 2 and 3 may play an important role in nicotine metabolism variability in people with diminished or deficient CYP2A6 activity. Studies in larger populations will be necessary to better establish the role of FMO genetic variants in overall nicotine metabolism.
Supplementary Material
Relative quantification by Western blot analysis of FMO-V5 wt over-expressed enzymes in HEK293 cells. The relative expression of each FMO enzyme is shown at the bottom, with the expression of FMO2 (full-length) designated as 1.0. Calnexin was used as the loading control and non-transfected HEK293 cells were used as the negative control.
Urinary NOX/TNE stratified by NMR level. The geometric mean of NOX/TNE was calculated for twenty groups stratified by 5 percentiles of NMR.
Acknowledgements
The authors thank the Mass Spectrometry Core facility at Washington State University Spokane for their help with LC/MS.
Sources of funding:
-
(1)
National Institutes of Health, National Institutes of Environmental Health Sciences [Grant R01-ES025460] to P. Lazarus
-
(2)
The Fulbright-Garcia Robles Program to Y.X. Perez-Paramo.
-
(3)
Health Sciences and Services Authority of Spokane, WA [Grant WSU002292] to WSU College of Pharmacy and Pharmaceutical Sciences
-
(4)
National Institutes of Health, National Cancer Institute [Grants R01-CA1440324, UM1-CA182876] to J.-M. Yuan
Footnotes
Conflicts of interest: None declared
References
- 1.Smoking Facts | American Lung Association [Internet]. 2017. [cited 2017 Sep 12]. Available from: http://www.lung.org/stop-smoking/smoking-facts/
- 2.U.S. Department of Health and Human Services. The Health Consequences of Smoking—50 Years of Progress. A Report of the Surgeon General. [Internet]. 2017. [cited 2017 Nov 30]. Available from: https://www.cdc.gov/tobacco/data_statistics/sgr/50th-anniversary/index.htm
- 3.Hecht SS, Kassie F, Hatsukami DK. Chemoprevention of lung carcinogenesis in addicted smokers and ex-smokers. Nat Rev Cancer. 2009;9:476–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.American Cancer Society. Key Statistics for Lung Cancer [Internet]. 2017. [cited 2017 Aug 23]. Available from: https://www.cancer.org/cancer/non-small-cell-lung-cancer/about/key-statistics.html
- 5.Mansvelder HD, McGehee DS. Cellular and synaptic mechanisms of nicotine addiction. J Neurobiol. Wiley Subscription Services, Inc., A Wiley Company; 2002;53:606–17. [DOI] [PubMed] [Google Scholar]
- 6.Benowitz NL, Jacob P. Nicotine and cotinine elimination pharmacokinetics in smokers and nonsmokers. Clin Pharmacol Ther. 1993;53:316–23. [DOI] [PubMed] [Google Scholar]
- 7.Krueger SK, Williams DE. Mammalian flavin-containing monooxygenases: Structure/function, genetic polymorphisms and role in drug metabolism. Pharmacol Ther. 2005;106:357–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poulsen L Organic sulfur substrates for the microsomal flavin- containing monooxygenase. Rev Biochem Toxicol. 1981;3:33–49. [Google Scholar]
- 9.Gut I, Conney AH. Trimethylamine N-oxygenation and N-demethylation in rat liver microsomes. Biochem Pharmacol. 1993;46:239–44. [DOI] [PubMed] [Google Scholar]
- 10.Lin J, Berkman CE, Cashman JR. N-Oxygenation of Primary Amines and Hydroxylamines and Retroreduction of Hydroxylamines by Adult Human Liver Microsomes and Adult Human Flavin-Containing Monooxygenase 3. Chem Res Toxicol. 1996;9:1183–93. [DOI] [PubMed] [Google Scholar]
- 11.Phillips IR, Dolphin CT, Clair P, Hadley MR, Hutt AJ, McCombie RR, et al. The molecular biology of the flavin-containing monooxygenases of man. Chem Biol Interact. 1995;96:17–32. [DOI] [PubMed] [Google Scholar]
- 12.FMO - The Human Protein Atlas [Internet]. [cited 2017 Apr 4]. Available from: http://www.proteinatlas.org/search/FMO
- 13.Dolphin CT, Beckett DJ, Janmohamed A, Cullingford TE, Smith RL, Shephard EA, et al. The flavin-containing monooxygenase 2 gene (FMO2) of humans, but not of other primates, encodes a truncated, nonfunctional protein. J Biol Chem. 1998;273:30599–607. [DOI] [PubMed] [Google Scholar]
- 14.Whetstine JR, Yueh MF, McCarver DG, Williams DE, Park CS, Kang JH, et al. Ethnic differences in human flavin-containing monooxygenase 2 (FMO2) polymorphisms: detection of expressed protein in African-Americans. Toxicol Appl Pharmacol. 2000;168:216–24. [DOI] [PubMed] [Google Scholar]
- 15.Park SB, Jacob P, Benowitz NL, Cashman JR. Stereoselective metabolism of (S)-(−)-nicotine in humans: Formation of trans-(S)-(−)-nicotine N-1’-oxide. Chem Res Toxicol. 1993;6:880–8. [DOI] [PubMed] [Google Scholar]
- 16.Hukkanen J, Jacob P, Benowitz NL. Metabolism and disposition kinetics of nicotine. Pharmacol Rev. 2005;57:79–115. [DOI] [PubMed] [Google Scholar]
- 17.Chen G, Giambrone NE Jr, Dluzen DF, Muscat JE, Berg A, Gallagher CJ, et al. Glucuronidation genotypes and nicotine metabolic phenotypes: Importance of UGT2B10 and UGT2B17 knock-out polymorphisms. Cancer Res. 2010;70:7543–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamanaka H, Nakajima M, Nishimura K, Yoshida R, Fukami T, Katoh M, et al. Metabolic profile of nicotine in subjects whose CYP2A6 gene is deleted. Eur J Pharm Sci. 2004;22:419–25. [DOI] [PubMed] [Google Scholar]
- 19.Hinrichs AL, Murphy SE, Wang JC, Saccone S, Saccone N, Steinbach JH, et al. Common polymorphisms in FMO1 are associated with nicotine dependence. Pharmacogenet Genomics. 2011;21:397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.American Lung Association. Research and Program Services Epidemiology and Statistics Unit Trends in Tobacco Use July 2011. [Google Scholar]
- 21.Caraballo RS, Giovino GA, Pechacek TF, Mowery PD, Richter PA, Strauss WJ, et al. Racial and Ethnic Differences in Serum Cotinine Levels of Cigarette Smokers. JAMA. 1998;280:135–9. [DOI] [PubMed] [Google Scholar]
- 22.Pianezza ML, Sellers EM, Tyndale RF. Nicotine metabolism defect reduces smoking. Nature. 1998;393:750. [DOI] [PubMed] [Google Scholar]
- 23.Moolchan ET, Franken FH, Jaszyna-Gasior M. Adolescent nicotine metabolism: ethnoracial differences among dependent smokers. Ethn Dis. 2006;16:239–43. [PubMed] [Google Scholar]
- 24.Chen G, Dellinger RW, Sun D, Spratt TE, Lazarus P. Glucuronidation of Tobacco-Specific Nitrosamines by UGT2B10. Drug Metab Dispos. 2008;36:824–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen G, Blevins-Primeau AS, Dellinger RW, Chen G, Blevins-primeau AS, Dellinger RW, et al. Glucuronidation of Nicotine and Cotinine by UGT2B10 : Loss of Function by the UGT2B10 Codon 67 ( Asp > Tyr ) Polymorphism Function by the UGT2B10 Codon 67 ( Asp > Tyr ) Polymorphism. Cancer Res. 2007;67:9024–9. [DOI] [PubMed] [Google Scholar]
- 26.Chenoweth MJ, Zhu AZX, Sanderson L, Ahluwalia JS, Benowitz NL, Tyndale RF. Variation in P450 oxidoreductase ( POR ) A503V and flavin-containing monooxygenase ( FMO ) −3 E158K is associated with minor alterations in nicotine metabolism , but does not alter cigarette consumption. Pharmacogenet Genomics. 2013;24:172–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chenoweth MJ, Sylvestre MP, Contreras G, Novalen M, O’Loughlin J, Tyndale RF. Variation in CYP2A6 and tobacco dependence throughout adolescence and in young adult smokers Drug Alcohol Depend. Elsevier Ireland Ltd; 2016;12/09:139–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bloom AJ, Murphy SE, Martinez M, von Weymarn LB, Bierut LJ, Goate A. Effects upon in-vivo nicotine metabolism reveal functional variation in FMO3 associated with cigarette consumption. Pharmacogenet Genomics. 2013;23:62–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teitelbaum AM, Murphy SE, Akk G, Baker TB, Germann A, von Weymarn LB, et al. Nicotine dependence is associated with functional variation in FMO3, an enzyme that metabolizes nicotine in the brain. Pharmacogenomics J. 2017;1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan J-M, Ross RK, Wang X-L, Gao Y-T, Henderson BE, Yu MC. Morbidity and Mortality in Relation to Cigarette Smoking in Shanghai, China JAMA. American Medical Association; 1996;275:1646. [PubMed] [Google Scholar]
- 31.Yuan JM, Ross RK, Chu XD, Gao YT, Yu MC. Prediagnostic levels of serum beta-cryptoxanthin and retinol predict smoking-related lung cancer risk in Shanghai, China Cancer Epidemiol Biomarkers Prev. American Association for Cancer Research; 2001;10:767–73. [PubMed] [Google Scholar]
- 32.Hankin JH, Stram DO, Arakawa K, Park S, Low SH, Lee HP, et al. Singapore Chinese Health Study: development, validation, and calibration of the quantitative food frequency questionnaire. Nutr Cancer. 2001;39:187–95. [DOI] [PubMed] [Google Scholar]
- 33.Yuan J, Stram DO, Arakawa K, Lee H, Yu MC. Dietary Cryptoxanthin and Reduced Risk of Lung Cancer : The Singapore Chinese Health Study 1. Cancer Epidemiol Biomarkers Prev. 2003;12:890–8. [PubMed] [Google Scholar]
- 34.Dellinger RW, Fang JL, Chen G, Weinberg R, Lazarus P. Importance of UDP-glucuronosyltransferase 1A10 (UGT1A10) in the detoxification of polycyclic aromatic hydrocarbons: Decreased glucuronidative activity of the UGT1A10139LYS isoform. Drug Metab Dispos. 2006;34:943–9. [DOI] [PubMed] [Google Scholar]
- 35.Peterson A, Xia Z, Chen G, Lazarus P. Exemestane potency is unchanged by common nonsynonymous polymorphisms in CYP19A1: results of a novel anti-aromatase activity assay examining exemestane and its derivatives. Pharmacol Res Perspect. 2017;5:e00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Störmer E, Roots I, Brockmöller J. Benzydamine N-oxidation as an index reaction reflecting FMO activity in human liver microsomes and impact of FMO3 polymorphisms on enzyme activity. Br J Clin Pharmacol. 2000;50:553–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taniguchi-Takizawa T, Shimizu M, Kume T, Yamazaki H. Benzydamine N-oxygenation as an index for flavin-containing monooxygenase activity and benzydamine N-demethylation by cytochrome P450 enzymes in liver microsomes from rats, dogs, monkeys, and humans Drug Metab Pharmacokinet. Elsevier; Ltd; 2015;30:64–9. [DOI] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- 39.R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2017. [Google Scholar]
- 40.Dempsey D, Tutka P, Iii PJ, Allen F, Tyndale RF, Benowitz NL, et al. Nicotine metabolite ratio as an index of cytochrome P450 2A6 metabolic activity. Clin Pharmacol Ther. 2004;450:64–72. [DOI] [PubMed] [Google Scholar]
- 41.1000 genomes. 1000 Genomes browser: Homo sapiens FMO [Internet]. 2018. [cited 2018 Apr 5]. Available from: http://phase3browser.1000genomes.org/Homo_sapiens/Search/Results?site=ensembl&q=FMO
- 42.Xu M, Bhatt DK, Yeung CK, Claw KG, Chaudhry AS, Gaedigk A, et al. Genetic and Non-genetic Factors Associated with Protein Abundance of Flavin-containing Monooxygenase 3 in Human Liver. J Pharmacol Exp Ther. 2017;363:265–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oscarson M Genetic polymorphisms in the cytochrome P450 2A6 (CYP2A6) gene: Implications for interindividual differences in nicotine metabolism. Drug Metab Dispos. 2001;29:91–5. [PubMed] [Google Scholar]
- 44.Nakajima M, Kuroiwa Y, Yokoi T. Interindividual differences in nicotine metabolism and genetic polymorphisms of human CYP2A6. Drug Metab Rev. 2002;34:865–77. [DOI] [PubMed] [Google Scholar]
- 45.Haberl M, Anwald B, Klein K, Weil R, Fuß C, Gepdiremen A, et al. Three haplotypes associated with CYP2A6 phenotypes in Caucasians. Pharmacogenet Genomics. 2005;15:609–24. [DOI] [PubMed] [Google Scholar]
- 46.Fukami T, Nakajima M, Higashi E, Yamanaka H, McLeod HL, Yokoi T. A novel CYP2A6*20 allele found in African-American population produces a truncated protein lacking enzymatic activity. Biochem Pharmacol. 2005;70:801–8. [DOI] [PubMed] [Google Scholar]
- 47.Yeung CK, Lang DH, Thummel KE, Rettie AE. Immunoquantitation of FMO1 in human liver, kidney, and intestine. Drug Metab Dispos. 2000;28:1107–11. [PubMed] [Google Scholar]
- 48.Bekele E, Mendell NR, Shephard EA. The potentially deleterious functional variant flavin-containing monooxygenase 2*1 is at high frequency throughout sub-Saharan Africa. Pharmacogenet Genomics. 2009;18:877–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Furnes B, Schlenk D. Evaluation of xenobiotic N- and S-oxidation by variant flavin-containing monooxygenase 1 (FMO1) enzymes. Toxicol Sci. 2004;78:196–203. [DOI] [PubMed] [Google Scholar]
- 50.Furnes B, Feng J, Sommer SS, Schlenk D. Identification of novel variants of the flavin-containing monooxygenase gene family in African Americans. Drug Metab Dispos. 2003;31:187–93. [DOI] [PubMed] [Google Scholar]
- 51.Dolphin CT, Janmohamed A, Smith RL, Shephard EA, Phillips IR. Compound heterozygosity for missense mutations in the flavin-containing monooxygenase 3 (FM03) gene in patients with fish-odour syndrome. Pharmacogenetics. 2000;10:799–807. [DOI] [PubMed] [Google Scholar]
- 52.Lattard V, Zhang JUN, Tran Q, Furnes B, Schlenk D, Cashman JR, et al. Two New Polymorphisms of the Fmo3 Gene in Caucasian and African- American Populations : Comparative Genetic and Functional Studies Abstract : Drug Metab Dispos. 2003;31:854–60. [DOI] [PubMed] [Google Scholar]
- 53.Cashman JR, Bi Y, Lin J, Youil R, Knight M, Forrest S, et al. Human flavin-containing monooxygenase form 3: cDNA expression of the enzymes containing amino acid substitutions observed in individuals with trimethylaminuria. Chem Res Toxicol. 1997;10:837–41. [DOI] [PubMed] [Google Scholar]
- 54.Cashman JR, Zhang J. Interindividual differences of human flavin-containing monooxygenase 3: Genetic polymorphisms and functional variation. Drug Metab Dispos. 2002;30:1043–52. [DOI] [PubMed] [Google Scholar]
- 55.Cashman JR. Human flavin-containing monooxygenase (form 3): polymorphisms and variations in chemical metabolism. Pharmacogenomics. 2002;3:325–39. [DOI] [PubMed] [Google Scholar]
- 56.Park C-S, Kang J, Chung W, Yi H, Pie J, Park D, et al. Ethnic differences in allelic frequency of two flavin-containing monooxygenase 3 (FMO3) polymorphisms: linkage and effects on in vivo and in vitro FMO activities. Pharmacogenetics. 2002;12:77–80. [DOI] [PubMed] [Google Scholar]
- 57.Park CS, Chung WG, Kang JH, Roh HK, Lee KH, Cha YN. Phenotyping of flavin-containing monooxygenase using caffeine metabolism and genotyping of FMO3 gene in a Korean population. Pharmacogenetics. 1999;9:155–64. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relative quantification by Western blot analysis of FMO-V5 wt over-expressed enzymes in HEK293 cells. The relative expression of each FMO enzyme is shown at the bottom, with the expression of FMO2 (full-length) designated as 1.0. Calnexin was used as the loading control and non-transfected HEK293 cells were used as the negative control.
Urinary NOX/TNE stratified by NMR level. The geometric mean of NOX/TNE was calculated for twenty groups stratified by 5 percentiles of NMR.