Abstract
Chronic exposure to cortisol is associated with cardiovascular, metabolic, and psychiatric disorders. Although cortisol can be readily measured from peripheral sources such as blood, urine, or saliva, multiple samplings spanning several days to weeks are necessary to reliably assess chronic cortisol exposure levels (referred to as cortisol load). Although cortisol levels in hair have been proposed as a measure of cortisol load, measurement is cumbersome and many people are not candidates due to short hair length and use of hair dyes. To date, there are no blood biomarkers that capture cortisol load.
To identify a blood biomarker capable of integrating one-month cortisol exposure levels, 75 healthy participants provided 30+ days of awakening and bedtime saliva cortisol and to completed psychosocial measures of anxiety, depression, and stress. Mean daily awakening and bedtime cortisol levels were then compared to CpG methylation levels, gene expression, and genotypes of the stress response gene FKBP5 obtained from blood drawn on the last day of the study.
We found a correlation between FKBP5 methylation levels and mean 30+day awakening and bedtime cortisol levels (|r|≥0.32, p≤0.006). We also observed a sex-specific correlation between bedtime cortisol levels and FKBP5 mRNA expression in female participants (r=0.42, p=0.005). Dividing the 30-day sampling period into four weekly bins showed that the correlations for both methylation and expression were not being driven by cortisol levels in the week preceding the blood draw. We also identified a female-specific association between FKBP5 mRNA expression and scores on the Beck Anxiety Inventory (r=0.37, p=0.013) and Beck Depression Inventory-II (r=0.32, p=0.033). Finally, DNA was genotyped at four SNPs, and variation in rs4713902 was shown to have an effect on FKBP5 expression under a codominant model (f=3.41, p=0.048) for females only.
Our results suggest that blood FKBP5 DNA methylation and mRNA expression levels may be a useful marker for determining general or sex-specific 30-day cortisol load and justifies genome-wide approaches that can potentially identify additional cortisol markers with broader clinical utility.
Keywords: Stress, Cortisol, Allostatic Load, FKBP5, gene expression, DNA methylation
Introduction
Allostatic load is a theoretical construct representing the sum of detrimental consequences of stress accumulated over time.(Ogden, 2004) High allostatic load is a major contributor to poor health outcomes. Chronic exposure to the stress hormone cortisol is a significant component of allostatic load, and is associated with cardiovascular and metabolic disorders.(Bose et al., 2009; Brydon et al., 2006; Hackett and Steptoe, 2016; Kelly and Ismail, 2015) The brain is especially susceptible to cortisol, as prolonged exposure is associated with an increase in neuropsychiatric disorders,(Fardet et al., 2012) decrease in hippocampal volume,(Rahman et al., 2016) and cognitive decline.(Arnsten, 2015; McKlveen et al., 2016) Given the detrimental impact of chronic exposure to cortisol, an easily accessible biological measurement capable of capturing long term cortisol exposure would be immensely useful for testing the association of cortisol load and disease susceptibility.
While cortisol can be measured from various peripheral sources such as urine, blood, saliva, and hair, measurements from these sources either do not adequately reflect long-term cortisol exposure or are burdensome to perform. Lack of accuracy of these measurements is attributed to the pulsatile nature of cortisol secretion. Cortisol measured in urine, blood, and saliva tends to only reflect the most recent circadian and stressful events. While the assessment of 24-hour urine free cortisol levels provides a metric for a single day’s cortisol exposure, other cortisol measures in urine, saliva, or blood provide even less time-based information. Unless multiple samplings are measured, an accurate determination of cortisol load is not possible. Although measurement of cortisol in hair samples is thought to indicate long-term exposure to cortisol, sampling is cumbersome and not always possible especially in individuals with short hair, recent hair shampooing and/or who use hair dyes.(Heinze et al., 2016; Rippe et al., 2016)
As a motivating example, the field of diabetes management was greatly advanced when it was determined that a single glycated hemoglobin level was a surrogate that integrated the previous 90 days of glucose exposure.(Saudek et al., 2008) A single blood test that integrates long-term exposure to cortisol would permit assessment of inter-individual differences in cortisol load thus providing a tool to predict future health consequences and treatment options.
Although blood cortisol measurements suffer from significant moment-to-moment variability, other molecules found in blood may be able to successfully capture more long-term cortisol exposure. For proof-of-concept, we designed a study to determine if the mean cortisol values collected over a month correlated with FKBP5 mRNA expression and methylation in blood obtained following 30+ days of saliva collection. We selected these measurements as our initial test biomarkers because dose-dependent changes in gene expression and methylation were observed following exposure to glucocorticoids.(Lee et al., 2011; Yang et al., 2012) In these earlier preclinical studies, we observed that the level of rodent Fkbp5 gene methylation and expression in blood samples measured following a month of corticosterone exposure reflected the mean glucocorticoid concentration measured during the prior 4-week treatment period. In a human study of Cushing’s Disease (tumor-induced hypercortisolism), we showed glucocorticoid-dependent changes in FKBP5 DNA methylation.(Resmini et al., 2016) Recently, Bali et al. showed that the glucocorticoid prednisone administered to healthy subjects increased FKBP5 mRNA expression in blood.(Bali et al., 2016) FKBP5 gene is also notable for its association with neuropsychiatric disorders.(Binder et al., 2004; Klengel et al., 2013; Willour et al., 2009)
This study was conducted as a proof-of-principle to determine if we could identify a blood marker for cortisol load. We hypothesized that FKBP5 DNA methylation and gene expression derived at the end of a 30-day cortisol collection period would negatively and positively correlate, respectively, with the mean of the cortisol levels collected in the month prior to the molecular assessments. We further hypothesized that self-reported anxiety/depression symptoms and perceived stress levels would be associated with FKBP5 methylation or gene expression.
Methods
Subjects
We recruited 18 – 60 year old participants of self-reported European ancestry using local flyers and newspaper advertisements. Three hundred and six individuals completed a brief telephone screen; 113 persons who appeared to be eligible were invited to provide informed consent and participate in a comprehensive assessment at Johns Hopkins School of Medicine. In-person assessments were conducted by a Master’s-level interviewer and included contact and demographic information (i.e., date of birth, race/ethnicity, education level, employment status), recent alcohol and drug use assessed using the 90-day Timeline Follow-back (Sobell and Sobell, 1992) and breathalyzer and urine toxicology screens, and past year mental health symptoms obtained using the MINI International Neuropsychiatric Interview for DSM-IV. (Sheehan et al., 1998) Each participant also completed a health checklist that obtained self-reported information on medical history, recent hospitalizations, current prescribed and OTC medications. Persons were excluded from study participation if they reported: using chewing tobacco or a similar product; had a serious medical condition; had taken a medication in the past 3 months that could affect cortisol assay measurements (including antidepressants and anti-anxiety medications); met criteria for a current DSM-5 diagnosis; tested positive for or self-reported recent drug use; exceeded NIAAA guidelines for social drinking; or were currently or had been pregnant or breast feeding within the past 3 months. We also excluded persons who were not going to be continuously available to complete daily study procedures over the 30 days following the assessment. Of the 113 participants who provided informed consent, 24 met exclusion criteria, and 2 were discontinued due to difficulties with the required blood draws. One subject did not complete the daily data collection procedures and dropped out during the study. This resulted in 86 participants who were eligible and provided daily and study visit data. All study procedures were reviewed and approved by the Johns Hopkins School of Medicine Institutional Review Board.
Procedures
The study protocol consisted of an in-person study visit 1, followed by approximately 30 days of at-home data collection, and then an in-person study visit 2. Eligibility was determined immediately after the assessment interviews and self-report documents were completed at study visit 1. Eligible persons then completed the remainder of the visit 1 procedures, received written instructions and all materials for daily data collection, and had an appointment scheduled for visit 2 which occurred as close to but no less than 30 days following visit 1.
Psychosocial Measurements
At visits 1 and 2, psychosocial measures included the Perceived Stress Scale (PSS),(Cohen et al., 1983) Beck Anxiety Inventory (BAI),(Beck et al., 1988) and the Beck Depression Inventory II (BDI-II).(Beck and Beamesderfer, 1974; Beck et al., 1996) Each day between study visits 1 and 2, participants received telephone, text and/or email prompts for saliva and psychosocial data collection at awakening and at bedtime; subjects were able to select their preferred method and time of reminder contact. Participants were contacted every 15 minutes for a total of 3 contacts or until the Interactive Voice Response (IVR) system was activated. Subjects also had the opportunity to call into the IVR system and avoid the reminders. During the morning call, participants were instructed to enter their actual awakening and bed times and saliva collection times using the telephone key pad. During the evening call, subjects completed the telephone Stress Assessment Procedure (SAP), which included several items to measure daily stress exposure. Specifically, participants rated their emotional (mental stress, upset, overwhelmed feelings), physical (muscular stress, physical tiredness, fatigue), and overall (combined impact of both emotional and physical stress) stress as well as their ability to manage these stressors. SAP items were rated on an analog scale between 0 (no stress/successfully managed) – 9 (worst ever/unable to manage) using the telephone key pad. Five paper copies of the SAP were supplied to the participant in the event that the telephone system malfunctioned or became unusable for any reason. To promote accuracy and timeliness, data were collected via real-time methods using Survey Monkey and CallFire Interactive Voice Response programming. Participants received a bonus payment if they submitted at least 90% of specimens. At visit 2, participants also completed the health checklist to determine if they had experienced any recent health problems that would confound data collection.
Measurement of cortisol
Between visits 1 and 2, salivettes were used to collect saliva twice daily for cortisol measurement, and specimens were mailed to the lab weekly in preprinted, prepaid envelopes. Participants were instructed to collect the morning sample within the first hour after awakening and the evening sample within one hour of bedtime, and to label the tube using a preprinted sticker with the date and morning/evening. They were also given clear instructions not to “make up” for missed samples by collecting a specimen outside the collection windows. Typed procedures for saliva collection were pasted to the front of the bag containing the salivettes to standardize procedures. Participants received an extra 7-day supply of salivettes in case they had to reschedule visit 2; this ensured that data collection continued until the morning of the visit. Salivary cortisol assays were performed by radioimmunoassay (MP Biomedicals LLC, Solon, OH) using a model 1470 counter (PerkinElmer, Shelton, CT). The inter- and intra-assay coefficients of variation for all assays are less than 10%.
We selected awakening and bedtime salivary sampling as our cortisol metric because we previously showed in 935 participants drawn from the Multi-Ethnic Study of Atherosclerosis (MESA) database a correlation between awakening and bedtime salivary cortisol vs. area under the cortisol curve determined during the wake cycle.(Golden et al., 2013) Thus awakening and bedtime cortisol is a reasonable surrogate for cortisol exposure that begins at awakening and terminates at bedtime.
Extraction of genomic DNA and messenger RNA from blood
Blood samples for RNA on visit 2 were collected between 11am-2pm to minimize the effect of the circadian rhythm on plasma cortisol levels and FKBP5 expression. Immediately after blood collection, red blood cells were lysed using 3.5 times the volume of ACK Lysing Buffer (Quality Biological, Gaithersburg, MD). Remaining white blood cells were pelleted by centrifugation at 300 x g for 5 minutes, resuspended in ice cold PBS, and separated into two equal aliquots. Genomic DNA was extracted from the other aliquot using the MasterPure Complete DNA Extraction kit (Epicentre, Madison, WI) according to the manufacturer’s protocol. DNA concentrations were determined by Qubit 2.0 Fluorometer (ThermoFisher, Waltham, MA). Messenger RNA was extracted from one aliquot of white blood cells using the RNeasy Mini Kit (Qiagen, Germantown, MD) according to the manufacturer’s protocol. Concentrations and RNA integrity numbers (RIN) were determined from each mRNA sample by TapeStation 2200 (Agilent Technologies, Santa Clara, CA). Samples with RIN<8.0 were excluded.
Bisulfite pyrosequencing
Twenty nanograms (ng) of the extracted DNA were used for bisulfite conversion according to the manufacturer’s protocol (EZ DNA Methylation Gold Kit; Zymo Research, Irvine, CA). Percent DNA methylation at 7 targeted intronic CpGs, chosen based on their association with childhood trauma exposure and Cushing’s Disease,(Klengel et al., 2013; Resmini et al., 2016) was determined by bisulfite pyrosequencing, which measures methylation variation at >90% precision. Briefly, two sets of bisulfite PCR primers were designed to target two intronic CpGs within the human FKBP5 gene. Two rounds of PCR amplification were performed using 3.5 μL of the bisulfite-converted DNA for the outer PCR and 2 μL of the outer PCR reaction for the nested PCR. One of the nested bisulfite primers was biotinylated and HPLC-purified, allowing it to bind to sepharose beads and become single-stranded, in preparation for bisulfite pyrosequencing. The single-stranded amplicons were annealed to pyrosequencing primers and subjected to primer extension and nucleotide incorporation using the PyroMark Q96 MD pyrosequencer (Qiagen). The pyrosequencer QCpG program determines percent DNA methylation at all of the CpG dinucleotides downstream of the annealed primer. Genomic coordinates of each of the CpG are provided in Supplementary Table 5.
Reverse transcription and real-time quantitative PCR (qPCR)
Total RNA (~100 ng) was reverse transcribed in a 20 µL reaction volume using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA) with random hexamers. The reaction mixture was incubated for 10 minutes at 25°C, 120 minutes at 37°C, and finally for 5 minutes at 85°C, according to the manufacturer’s instructions. Synthesized cDNA was subsequently diluted 10-fold prior to use in real-time qPCR. Real-time qPCR experiments were performed in a 96-well plate using the Applied Biosystems StepOnePlus real-time PCR instrument. TaqMan gene expression assays consisted of a mix of unlabeled PCR primers for human FKBP5 (Applied Biosystems; Hs01561006_m1) and Taq-Man MGB probe (FAM dye labeled). The reaction mixture for real-time qPCR contained 9.0 µL cDNA solutions (~10 ng), each of the forward and reverse primers and the MGB probe used at 0.9 µM and 0.25 µM, respectively, and 1x Taqman Universal Master Mix II with UNG (Applied Biosystems; 4440038). The mixture was activated (2 min, 50°C), denatured (10 min, 95°C) and subjected to 40 amplification cycles (15 sec, 95°C; 1 min, 60°C) with a single measurement of fluorescence for the FKBP5 primer set.
PCR Efficiency/Analytical Sensitivity
A plasmid DNA containing the full coding sequence for human FKBP5 was obtained from OriGene (Rockville, MD). The concentration of the plasmid DNA was measured using a fluorometer, and the corresponding gene copy number was determined using the following equation (Whelan et al., 2003):
This formula yields 1.5 × 1011 FKBP5 mRNA copies per µg of plasmid DNA. A stock solution of linearized plasmid DNA, containing 1.0 × 1010 copies of FKBP5 mRNA per µl was prepared and serially diluted over 9 orders of magnitude. Standard curves were constructed via qPCR to calculate the efficiency of PCR, linear range and the limits of detection, and quantification for the assay. PCR efficiency was calculated from the slope of the standard curve that was run in nine replicates at each concentration incorporating a total of 81 data points as follows (Rasmussen, 2001):
An interplate calibrator (TATAA IPC250PFAM) positive control was also included with each run to monitor inter-assay variations. A reproducible Cq for the interplate calibrator amplification with standard deviation ≤ 0.2 was required as a pass criterion for the test plates.
qPCR Data Analysis
TaqMan qPCR data analysis was carried out using the StepOne Plus software V 2.3. Amplification plots were visualized across the entire 96 well plate for FKBP5. Fractional cycle (Cq) values were returned by manually setting the threshold to intersect at the exponential phase of amplification plots (defined at 0.1). Cq values generated for the test samples, tested in triplicate, were preprocessed using GenEx (MultiD Analysis) software. FKBP5 copy numbers per ng of total RNA from test samples and the associated 95% confidence intervals were then estimated by entering the standard curve at the preprocessed Cq and reading out the log of the concentration on the x-axis.
Genotyping
Genotyping of four SNPs within the FKBP5 gene (UCSC Genome Browser: rs1360780, rs9470080, rs9296158, rs4713902) was performed using the genotyping function on the PyroMark Q96 MD. One round of PCR was performed using 20 ng of gDNA and two PCR primers flanking the SNP, one of which was biotinylated and HPLC-purified. In a similar procedure as bisulfite pyrosequencing, this time with non-bisulfite converted DNA, genotypes were determined by primer extension and nucleotide incorporation.
Assessment of Cellular Composition of Blood
Cellular composition of blood was deconvoluted using an epigenetic approach to determine whether there were differences in the whole blood cell populations in high vs. low cortisol groups. Five pyrosequencing assays representing CD8+ T-cells, natural killer (NK) cells, B-cells, monocytes, and granulocytes were designed against 5 CpGs (CG25939861, CG27582527, CG19276014, CG23244761, and CG05398700, respectively) assessed on the Illumina BeadChip 450K platform.(Houseman et al., 2012; Jaffe and Irizarry, 2014) Each CpG was chosen for low levels of methylation in its corresponding cell type compared to methylation levels of others. For example, Jaffe and Irizarry reported a beta value (~percent methylation) of 0.13 for CG19276014 in B-cells but 0.99, 0.97, 0.98, and 0.99 for T-cells, NK cells, monocytes, and granulocytes, respectively.(Jaffe and Irizarry, 2014) Regression analysis was performed between the CpG percent methylation determined from bisulfite pyrosequencing and saliva cortisol levels. We observed no evidence of significant differences in cellular composition with respect to the two saliva cortisol measurements (r2<0.1, p>0.4). As a result, cell type methylation values were omitted from subsequent downstream analyses.
Statistics
All data were examined for possible errors or outliers. Five individuals were excluded whose daily cortisol values were more than three standard deviations from the mean. In addition, four participants with awakening cortisol values equal to their bedtime cortisol values were removed (no circadian rhythm). Two participants had FKBP5 mRNA expression values greater than 2,000 copies per ng of total RNA and were removed from the analysis. Thus, 75 participants’ data were analyzed.
Mean awakening and bedtime salivary cortisol were calculated as the mean of the 30+ daily measures of awakening and bedtime cortisol, respectively. Similarly, weekly cortisol metrics were calculated as the mean value over each week. Given that participants were in the study for anywhere from 30–37 days, the weekly metrics were calculated only for the last four full weeks a participant was in the study. T-tests were used to test for differences in demographics, psychometric measures and cortisol metrics in males and females. A variance ratio test was used to examine differences in variability of the awakening and bedtime cortisol values. Intraclass correlation coefficients (ICC) were calculated to test for stability of cortisol values over 30+ days. Daily stability was calculated as the ICC across all 30+ daily values of awakening or bedtime cortisol. Weekly stability was calculated as the ICC across the four weekly means of awakening or bedtime cortisol.
For our analysis of FKBP5 mRNA expression, Pearson’s correlation coefficients were used to examine correlation between expression levels and mean 30+ day awakening and bedtime cortisol. Partial correlations were computed adjusting for sex and age. In an exploratory analysis, we tested for interactions between the cortisol metrics and sex. This was assessed using a likelihood ratio test (LRT) comparing a linear regression model of FKBP5 mRNA expression on cortisol including an interaction term (cortisol x sex) to a model without the term for the interaction. Similar analyses were performed for FKBP5 methylation. Other exploratory analyses included testing for correlations among the expression levels, methylation levels, and psychometric scales, again using Pearson’s correlations and partial correlations adjusting for sex and age. To account for the primary expression analyses (2 mean cortisol measures – bedtime and awakening) and primary methylation analyses (7 independent CpGs x 2 mean cortisol measures), we computed the number of effectively independent tests, taking into consideration correlation between variables. (Li and Ji, 2005) For methylation, we considered a p≤0.004 (p=0.05 / 14 effectively independent tests) to be evidence of a significant correlation. For exploratory analyses, p<0.05 was considered a significant preliminary correlation. All analyses were performed using STATA version 14 software.
Results
Psychosocial, FKBP5, and endocrine measurements
The final sample consisted of 75 healthy, White male (N=31) and female (N=44) participants with a mean age of 32.6 (s.d.=11.6). There were no significant sex differences observed in the scores of the Beck Depression Inventory-II, Beck Anxiety Inventory, Perceived Stress Scale, emotional stress scores, or FKBP5 mRNA levels, although all five measures were slightly higher in the female participants (Table 1). Also, no biologically significant methylation differences were observed by sex at the 7 CpGs interrogated in this study (differences <4.7%, data shown for CpG-1 only).
Table 1.
Demographics, cortisol metrics, FKBP5 expression and psychosocial measures by sex
| Females N=44) |
Males (N=31) |
Total (N=75) |
T-statistic, p-value |
|
|---|---|---|---|---|
| Age (yrs) | 34.5 (12.9) | 30.0 (9.1) | 32.6 (11.6) | t73=1.64, p=0.1056 |
| Awakening cortisol, day 1–30+, (ng/mL) | 9.2 (2.5) | 9.5 (2.2) | 9.3 (2.3) | t73=−0.53, p=0.5960 |
| Bedtime cortisol, day 1–30+ (ng/mL) | 2.1 (0.9) | 2.3 (1.3) | 2.2 (1.1) | t73=−0.78, p=0.4387 |
| FKBP5 expression (copies/ng total RNA) | 895.5 (479.0) | 737.4 (321.9) | 830.2 (426.0) | t73=1.60, p=0.1140 |
| FKBP5 methylation at CpG-1 | 94.1% (2.8) | 93.9% (2.4) | 94.0% (2.6) | t73=0.36 p=0.7168 |
| BAI | 5.0 (4.0) | 3.7 (3.9) | 4.5 (4.0) | t73=1.40, p=0.1645 |
| BDI-II | 5.6 (6.4) | 5.2 (4.6) | 5.4 (5.7) | t73=0.30, p=0.7629 |
| PSS | 14.3 (7.2) | 13.0 (6.7) | 13.7 (7.0) | t73=0.76, p=0.4508 |
| Emotional stress | 2.8 (1.5) | 2.6 (1.2) | 2.7 (1.3) | t73=0.54, p=0.5908 |
Shown are mean (s.d.).
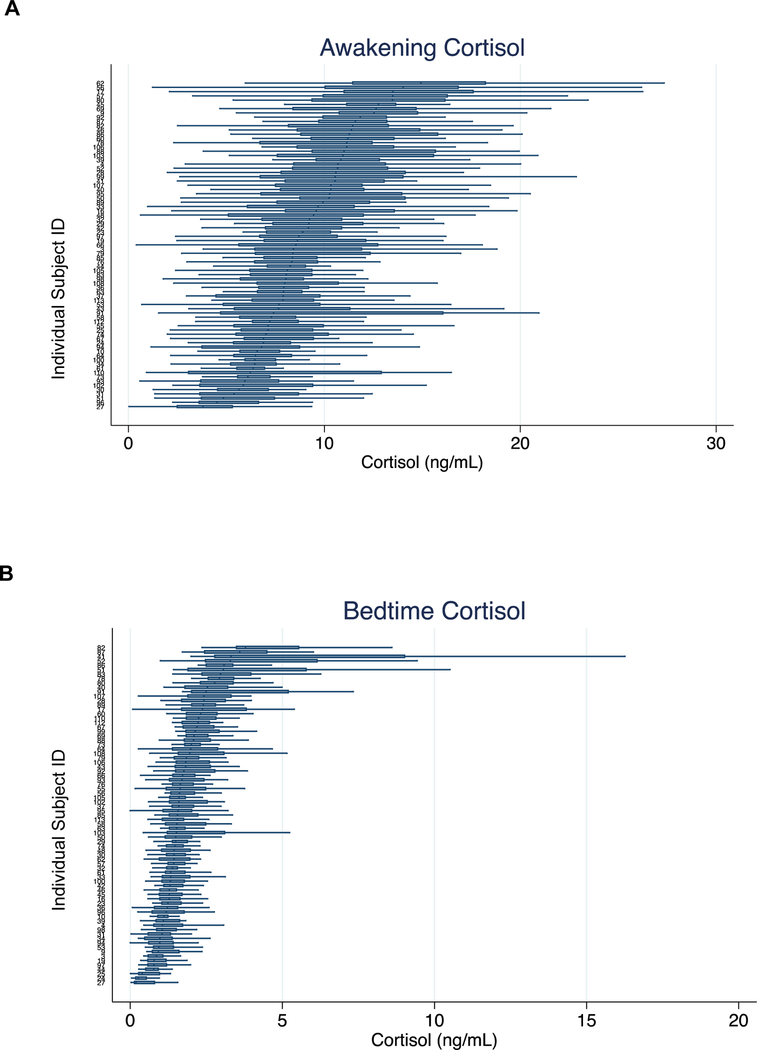
Awakening and bedtime saliva cortisol levels are presented in Table 1 and Figure 1, with horizontal boxplots denoting each individual’s distribution of 30+ day cortisol measurements. The boxplots are sorted such that participants showing the highest median cortisol levels are at the top and those with the lowest median levels at the bottom. In general, we observed greater variation in the awakening cortisol levels (Figure 1A), with most of the median levels ranging between 5 and 15 ng/mL, than in the bedtime cortisol levels (Figure 1B), with most of the values falling between 0 and 5 ng/mL (f2458,2578=0.30, p<0.0001). The cortisol metrics did not differ by sex (p>0.44) (Table 1) or age (p>0.14).
Figure 1A and 1B. Distribution of 30+ day cortisol values, by subject.
Shown are A) awakening cortisol and B) bedtime cortisol. In each figure the median (vertical bar), 25th and 75th percentiles (box), and maximum and minimum values (horizontal bars) are shown for each subject.
Stability of cortisol values over the study period is reported using Intraclass Correlation Coefficients (ICC, Supplementary Table 1). The daily stability of cortisol over 30+ days was moderate and was greater for awakening cortisol (ICC=0.25) than for bedtime cortisol (ICC=0.15). In order to mitigate the potential effects of one or two days of extreme cortisol values on our measure of stability, we next computed the mean cortisol value over each week. The stability of weekly mean cortisol was higher, and again showed greater stability for awakening cortisol (ICC=0.59) than for bedtime cortisol (ICC=0.53).
Comparison of saliva cortisol levels and FKBP5 methylation in the blood
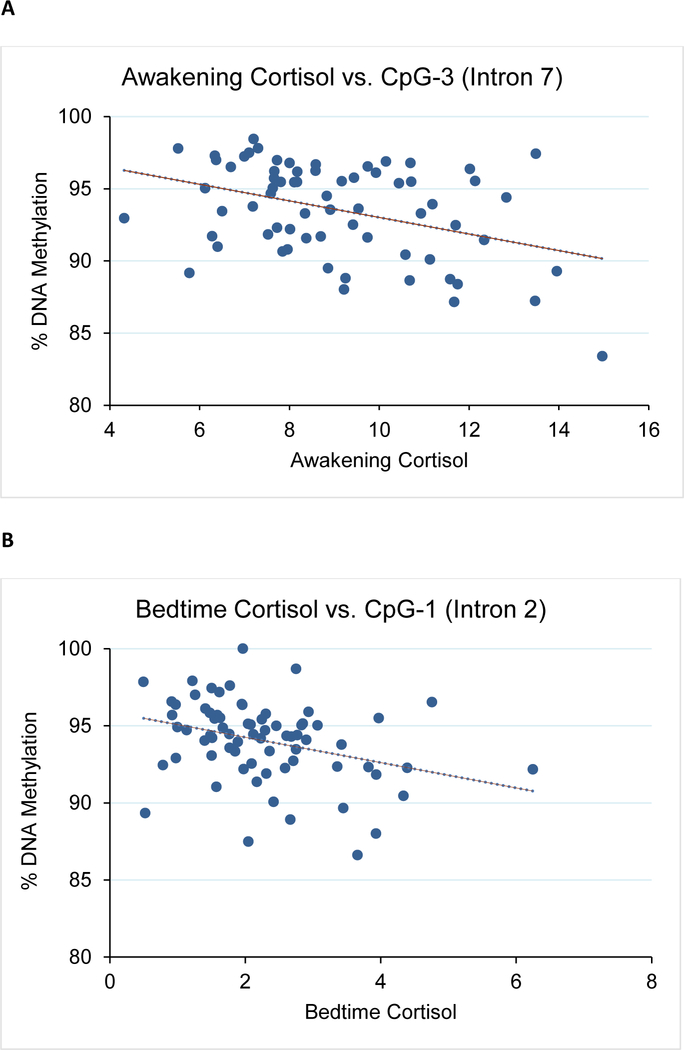
We first asked whether CpG methylation levels correlated with the observed mean 30+ day cortisol levels. We performed bisulfite pyrosequencing on a total of 7 candidate CpGs from the two intronic regions previously shown to differ in patients with Cushing’s Disease (a high cortisol state) compared to healthy controls with normal cortisol exposure.(Klengel et al., 2013; Resmini et al., 2016) A significant correlation was observed between awakening cortisol levels and CpG-3 methylation in both males and females (intron 7, r=−0.39, p=0.001, Figure 2A and Supplementary Table 2). We also observed a similar correlation between bedtime cortisol and CpG-1 (intron 2, r=−0.34, p=0.004, Figure 2B and Supplementary Table 2). For CpG-3, its correlation to awakening cortisol levels was more significant for females (r=−0.45, p=0.004) than males (r=−0.35, p=0.062). For CpG-1, its correlation to bedtime cortisol levels was similar for both males (r=−0.40, p=0.031) and females (r=−0.33, p=0.040). No significant relationships were observed between DNA methylation and emotional stress or between DNA methylation and FKBP5 expression for all samples (p>0.05, data not shown).
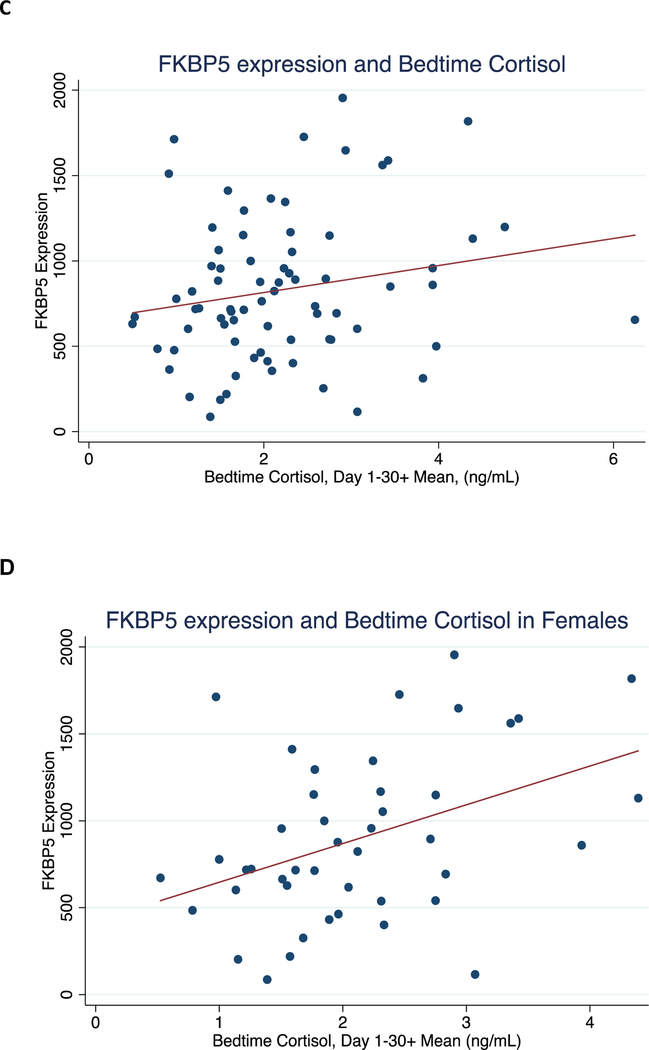
Figure 2. Correlation between mean 30+ day bedtime cortisol with FKBP5 methylation levels and expression.
Shown are: A) CpG-3 methylation vs. awakening cortisol for all subjects, B) CpG-1 methylation vs. bedtime cortisol for all subjects, C) FKBP5 expression vs. bedtime cortisol for all subjects, and D) FKBP5 expression vs. bedtime cortisol for females.
To confirm that methylation levels did not reflect only the most recent cortisol levels during the last week of collection, methylation levels of CpG-1 and CpG-3 were compared against each of the four weekly means for bedtime and awakening cortisol levels. We found significant correlations during the first three weeks of the 30-day study for both bedtime (CpG-1: |r|≥0.28, p≤0.012) and awakening cortisol levels (CpG-3: (|r|≥0.26, p≤0.025, Table 2). Interestingly, there were no significant correlations with the last week before the blood draw (CpG-1 vs. bedtime: r=−0.18, p=0.119 and CpG-3 vs. awakening: (r=−0.09, p=0.440). Further, both CpG-1 and CpG-3 were compared against the visit 2 BAI, BDI-II, and PSS measures, and no significant relationships were observed (|r|<0.1 and p>0.4).
Table 2.
Correlation between FKBP5 methylation (males and females) and expression (females only) vs. mean weekly awakening and bedtime cortisol.
| Awakening Cortisol vs. CpG-3 | Bedtime Cortisol vs. CpG-1 | Awakening Cortisol vs. Expression | Bedtime Cortisol vs. Expression | |
|---|---|---|---|---|
| Week 1 | r=−0.09 p=0.4400 |
r=−0.18 p=0.1194 |
r=−0.02 p=0.8754 |
r=0.12 p=0.4548 |
| Week 2 |
r=−0.26 p=0.0247 |
r=−0.33 p=0.0040 |
r=0.03 p=0.8303 |
r=0.37 p=0.0135 |
| Week 3 |
r=−0.35 p=0.0023 |
r=−0.29 p=0.0115 |
r=0.11 p=0.4823 |
r=0.50 p=0.0006 |
| Week 4 |
r=−0.34 p=0.0034 |
r=−0.28 p=0.0158 |
r=−0.08 p=0.6171 |
r=0.28 p=0.0692 |
Shown are Pearson’s coefficients and p-values. Weeks represent the last four weeks of the study period, with Week 1 representing the final week before the study completion.
Comparison of saliva cortisol levels and FKBP5 expression in the blood
We also examined FKBP5 expression levels, which ranged from 86.4 to 1954.7 and did not differ by sex (t73=1.60, p=0.11) or age (t73=−1.73, p=0.089, see Table 1). Blood samples for FKBP5 mRNA expression were collected between 11am-2pm; mean FKBP5 levels measured from samples collected in the morning did not differ from levels measured in samples collected in the afternoon. We did not observe a significant correlation between FKBP5 mRNA levels and either mean 30+ day bedtime cortisol (r=−0.20, p=0.093; Figure 2C and Table 3) or mean 30+ day awakening cortisol (r=−0.03, p=0.78), in contrast to the observations made with DNA methylation. We then tested whether sex modified the relationship between FKBP5 mRNA levels and cortisol. There was no evidence of an interaction between mean awakening cortisol and sex (LRT χ2=0.14, p=0.71). However, we did detect an interaction between mean bedtime cortisol and sex (LRT χ2=7.01, p=0.008). This finding prompted us to examine sex differences. There was a significant correlation between FKBP5 mRNA and mean bedtime cortisol levels in females only (r=0.42, p=0.005, Figure 2D) and not in males (r=−0.05, p=0.79, Table 3). A linear regression model indicated that 17.3% of the variance in FKBP5 mRNA expression in females was explained by mean bedtime cortisol levels.
Table 3.
Correlation between FKBP5 expression and mean cortisol metrics, by sex
| Females and Males (N=75) | Females (N=44) | Males (N=31) | |
|---|---|---|---|
| Awakening cortisol, day 1–30+ | r=−0.03 p=0.7836 |
r=0.01 p=0.9523 |
r=−0.10 p=0.6079 |
| Bedtime Cortisol, day 1–30+ | r=−0.20 p=0.0928 |
r=0.42 p=0.0050 |
r=−0.05 p=0.7892 |
Shown are Pearson’s coefficients and p-values.
To assess whether the relationship between FKBP5 mRNA expression and bedtime cortisol in females might be driven primarily by more recent cortisol exposure, we examined the correlation of each of the four weekly means with FKBP5 mRNA levels. Moderate correlations were again observed across only the first three weeks of the study (r=0.28–0.50, Table 2).
FKBP5 mRNA expression levels were also compared against the three psychosocial instruments from visit 2: BAI, BDI-II, and PSS. As with bedtime cortisol levels, significant female-specific associations were observed with BAI (r=0.37, p=0.013) and BDI-II (r=0.32, p=0.033; Supplementary Table 3). Mean daily emotional stress measures did not correlate with FKBP5 mRNA expression or the cortisol metrics. Further, we examined whether the use of birth control affected FKBP5 expression levels in females and found that its use did not significantly affect expression levels (t=−1.49, p=0.143).
Comparison of saliva cortisol levels and FKBP5 SNPs
We also obtained genotype information for four SNPs (rs1360780, rs9470080, rs9296158, and rs4713902) previously associated with childhood trauma exposure, Cushing’s Disease, and the Trier Social Stress Test.(Klengel et al., 2013; Mahon et al., 2013; Resmini et al., 2016) We observed no significant deviations from Hardy Weinberg equilibrium (HWE) in the full sample of 75 individuals (p-value for HWE > 0.1). We assessed the effect of genetic variation on bedtime and awakening cortisol levels, emotional stress, and FKBP5 expression using a codominant model. Interestingly, only variation in rs4713902 in females was shown to have significant effects (f=3.41, p=0.048) on FKBP5 expression (Supplementary Table 4). No significant genotype effect was observed on DNA methylation levels of CpG-1 or CpG-3 (data not shown).
Cellular composition of blood
Finally, we asked whether the association between the mean cortisol levels and FKBP5 gene expression was driven by cortisol-induced changes in the cell types from blood. Bisulfite pyrosequencing of CpGs that were used (Jaffe and Irizarry, 2014) to characterize five different cell types in blood samples (T-cells, B-cells, NK cells, monocytes, and granulocytes) showed no significant association between DNA methylation levels of these CpGs and mean saliva cortisol levels (r<0.3, p>0.4).
Discussion
In this study, the relationship between awakening and bedtime cortisol levels sampled over a month or more were compared to blood FKBP5 DNA methylation and mRNA expression levels drawn at the end of the cortisol collection period. Awakening and bedtime salivary sampling was chosen as the cortisol metric stemming from our previous study, where over 900 participants drawn from the Multi-Ethnic Study of Atherosclerosis (MESA) database showed a moderate correlation between awakening and bedtime salivary cortisol vs. the area under the cortisol curve determined during the wake cycle (Golden et al., 2013).
The FKBP5 gene was selected as a potential biomarker because of findings in our rodent model showing it to be a glucocorticoid-sensitive gene whose expression and methylation status responds in a dose-dependent manner to glucocorticoids. In the present study, we identified significant correlations between the means of bedtime and awakening cortisol levels and intronic methylation status of FKBP5. Also, there was a sex-specific correlation between the mean of bedtime cortisol levels and FKBP5 mRNA levels in female participants but not males. We did not identify a correlation of awakening cortisol with FKBP5 mRNA levels in either sex. Female FKBP5 mRNA levels also correlated with scores on the BDI-II and BAI. Interestingly, genetic variations, expression, and/or methylation status in the FKBP5 gene is associated with depression and antidepressant response,(Binder et al., 2004) bipolar disorder,(Willour et al., 2009) and exposure to childhood trauma.(Klengel et al., 2013) This finding is particularly interesting in light of the role of FKBP5 in affective disorders and given we enrolled healthy, non-clinical participants. The correlations might be stronger in a clinical cohort. There were no significant sex differences in mean awakening and bedtime salivary cortisol measures, FKBP5 methylation and expression levels, and psychosocial instrument scores.
We then analyzed the data to determine if the FKBP5 DNA methylation and mRNA levels measured at the end of the one-month collection period reflected the most recent cortisol levels or cortisol levels measured over the entire month. We found that the correlative relationship held when FKBP5 methylation levels were compared to cortisol levels averaged at successively earlier periods of time (e.g., 7-day blocks) from the termination of the month-long study. In fact, the correlation was strongest with cortisol measurements obtained during the first three weeks of the study, suggesting that FKBP5 methylation levels were capturing chronic cortisol levels. Similarly, we observed stronger correlations between FKBP5 expression and bedtime cortisol levels for the second and third weeks in females. This finding was noteworthy given the transient nature of mRNA expression and therefore the likelihood of expression levels to only reflect the last days of cortisol exposure. We reason that the final week of cortisol measurements was the weakest contributor to the correlations because insufficient time had passed during this period for the cortisol levels to be adequately reflected in the methylation and expression levels. If this is true, then we may observe a stronger correlation by extending the study for an additional week or two, so that the cortisol levels during this week became “chronic.”
Certain studies have suggested that cortisol metrics taken daily for 1–3 days and then assessed for AUC, slope, or average, such as the cortisol awakening response (CAR), may be a stable phenotype with power to predict the development of cardiovascular disorders,(Kumari et al., 2011; Rosmond and Bjorntorp, 2000) diabetes mellitus (Bruehl et al., 2009), and neuropsychiatric illness.(Huber et al., 2006; Rohleder et al., 2004; Vreeburg et al., 2013; Vreeburg et al., 2009) However, other studies do not support this contention.(Hajat et al., 2013; Kuehl et al., 2015; Spanakis et al., 2016) The discrepancies between studies may stem from the limitations of the cortisol metric employed. Bedtime cortisol is a valid metric for a single day’s cortisol exposure.(Golden et al., 2013) However, as shown by the ICC calculations in our study, there is considerable within-subject, day-to-day variability over the course of a month. Most cortisol metrics have only been studied over a few days and are thus less representative of cortisol load. The usefulness of any cortisol metric, including the CAR as an indicator of glucocorticoid load or to predict future cortisol-related illness is hampered when the collection occurs only over a few days. Moreover, collections that could be more accurate require several cortisol samplings per day over many days and are not practical for epidemiologic studies of cortisol load or, for that matter, to be employed in clinical use.(DeSantis et al., 2015) Given that bedtime cortisol is associated with cortisol exposure during a single wake cycle, and measurement of FKBP5 methylation and expression (for women only) reflects a month’s summation of bedtime cortisol levels, it is possible that such measurements from a blood sample can function as a moderate measure of one month of cortisol load. Further interrogation of the expression levels of other RNAs by RNA-Seq, additional genetic variations by genotyping, or epigenetic differences by epigenomic platforms in our sample may uncover a much stronger measure of cortisol load for both sexes. Additional studies in a larger cohort are needed to reproduce the current findings and to delineate the contribution of acute and chronic stressors and other clinical factors to FKBP5 methylation, expression, and cortisol levels.
It is curious why the cortisol vs. FKBP5 expression correlation was present in women but not in men. The FKBP5 gene not only contains glucocorticoid response elements but also consensus hormone response elements capable of binding sex hormones.(Hubler and Scammell, 2004; Magee et al., 2006) It is known that estrogen as well as cortisol activate FKBP5 mRNA expression.(Malviya et al., 2013) Many studies have shown glucocorticoids increase FKBP5 gene expression in a dose dependent manner in various tissues.(Guess et al., 2010; Lee et al., 2011; Yang et al., 2012) A recent study has shown that treatment of hippocampal cells with increasing doses of corticosterone increased the expression of FKBP5, and this effect was potentiated by estrogen exposure.(Malviya et al., 2013) Therefore, differences in sex hormone levels between males and females may play a role in the association. In the female subjects, it is unlikely that fluctuations in sex hormone levels, such as that of estrogen during the menstrual cycle, would significantly contribute to the observed correlation between FKBP5 expression and bedtime cortisol levels, since each female participant would cycle through both phases of the menstrual cycle (follicular and luteal) in random order over the 30+-day period.
We observed a broader utility for DNA methylation as an indicator of cortisol exposure than for gene expression. Methylation levels of intronic CpGs were correlated with both awakening and bedtime cortisol levels for both sexes, with similar statistical significance as that derived from comparing FKBP5 expression and bedtime cortisol levels in females. It is interesting to note that CpG-1 in the second intron of FKBP5 correlated with bedtime cortisol levels was previously associated with loss of methylation and hippocampal volume in Cushing’s Disease.(Resmini et al., 2016). Despite the promising finding with CpG-1 methylation and bedtime cortisol levels, we did not observe a stronger correlation in females that could account for the sex-specific correlation between bedtime cortisol and gene expression, nor did we observe a significant correlation between FKBP5 expression and CpG-1 methylation for both sexes. A clear biological or technical explanation remains elusive. As with FKBP5 expression and cortisol levels, we speculate that FKBP5 methylation occurs as the result of physical interactions among multiple GREs, EREs (estrogen response elements), AREs (androgen response elements), and additional response elements.(Hubler and Scammell, 2004; Klengel et al., 2013; Magee et al., 2006; Malviya et al., 2013) Therefore, methylation levels of one CpG may not reflect the expression profile of each individual. However, CpG-1 can indicate exposure to glucocorticoids because this CpG is near a GRE. In our previous study, we showed that methylation changes in CpGs near a GRE can capture the history of glucocorticoid exposure for a one-month period and is superior to gene expression levels in this regard.(Ewald et al., 2014; Lee et al., 2010; Lee et al., 2011)
We also observed a subtle relationship between rs4713902 and FKBP5 expression in females only. However, the correlation was relatively weak, and it is not clear at this time whether this particular SNP can explain the female-specific expression correlation with bedtime cortisol levels. We were surprised that we were not able to observe a significant correlation between rs1360780 and FKBP5 expression, given the location of the SNP near a GRE in the second intron and the association of its minor TT genotype with higher FKBP5 expression levels.(Binder et al., 2004; Klengel et al., 2013) Nevertheless, we note that rs4713902 is located in intron 1 and nearby a poorly conserved region in the mouse blood demonstrated to undergo substantial GC-induced loss of DNA methylation.(Ewald et al., 2014) In vitro studies are necessary to clearly establish the role of this SNP on gene expression.
Finally, we found no evidence of substantial changes in the cellular composition of blood, at least based on several representative CpGs that have been used to characterize the different cell types in the blood.(Jaffe and Irizarry, 2014) Despite the wide inter-individual range for bedtime and awakening cortisol levels, the absence of such a change is not surprising given that all of our subjects are healthy and thus may be subject to only minor fluctuations in the composition of blood cell types.
Our current study has several notable features. To our knowledge, this is the first time that 30+ consecutive days of cortisol were collected in the same participants examining the stability and usefulness of this metric. Second, participants received real-time reminders for collection of saliva, which helps to minimize noncompliance and faulty data collection. Third, quality control measures showed that 90% of participants collected all their cortisol samples, and the awakening/bedtime cortisol ratios strongly suggested that collection occurred at the proper time of day reflecting the circadian rhythm. Fourth, healthy, unmedicated, racially homogenous participants were recruited to minimize confounders in this proof-of-principle study. Fifth, our hypothesis was based on strong preclinical literature showing the utility of measuring FKBP5 expression and methylation as surrogates for 30-day glucocorticoid exposure.(Ewald et al., 2014; Lee et al., 2010; Lee et al., 2011) Sixth, the FKBP5 mRNA and methylation assays utilized in this study are well characterized and highly quantitative.(Rasmussen, 2001; Resmini et al., 2016; Whelan et al., 2003) Lastly, we adapted a candidate epigenetic approach to demonstrate that individual differences in the awakening and bedtime cortisol levels did not significantly affect cell composition of blood that could have contributed to the observed variations in FKBP5 expression levels. All of these strengths lend support to the relevance and reliability of our findings. Nonetheless, our study also has several limitations. First, our study only recruited healthy individuals. Although the range in the cortisol metrics was robust, it most likely would have been larger and provided more opportunities to find stronger correlations if the sample included participants with DSM-5 anxiety or depression diagnoses or who suffered from chronic somatic diseases. Second, we enrolled only White participants and do not know if the findings generalize to an ethnically diverse population. For example, racial and ethnic differences that exist in daily cortisol profiles (DeSantis et al., 2015; Hajat et al., 2010) may confound the relationship between FKBP5 expression and methylation vs. mean cortisol levels. Third, we relied on each participant to routinely collect awakening and bedtime saliva for cortisol measurements. Despite digital reminders for different measurements, noncompliance with the timing of sampling could be a possible source of error in this study. However, solid evidence for a robust circadian rhythm was evident in all subject data when comparing awakening and bedtime cortisol levels suggesting good time compliance. Also, mean awakening and bedtime cortisol values were similar to the values in the much larger MESA sample.(Golden et al., 2013) Fourth, the FKBP5 methylation-cortisol correlation was of only moderate strength and FKBP5 expression-cortisol correlation was sex-specific. However, as a proof-of-concept, it is encouraging that a biomarker exists that may have some usefulness in measuring cortisol load in the healthy population. Fifth, there was an absence of a significant correlation between DNA methylation and gene expression. Finally, blood measurements of DNA methylation and gene expression raise the question whether they directly regulate present cortisol levels rather than reflect past cortisol exposure. To answer this adequately, assessment of Visit 1 blood is necessary to establish causality, where, for instance, a stronger correlation between methylation expression levels at Visit 1 and ensuing 4-week cortisol levels would suggest that methylation levels are directly responsible for regulating cortisol levels. However, we measured FKBP5 methylation and expression in white blood cells and not in a CNS site known for regulating glucocorticoid negative feedback and cortisol secretion. We are unaware of any mechanism whereby lymphocytes regulate the HPA axis on a daily basis, except during acute illness. This fact in itself strongly suggests that methylation and expression levels only reflect past exposure history.
In summary, we collected daily saliva samples for one month or more and blood samples at the study completion and tested the utility of one type of biomolecule as an indicator of cortisol exposure. We found that the expression levels of the stress-response gene FKBP5 was a sex-specific indicator of cortisol exposure over a one-month period, whereas DNA methylation levels were applicable to both sexes. Given these initial findings, there is great possibility for identifying more robust and non sex-specific surrogates for cortisol exposure in genome-wide studies of DNA methylation or RNA expression. When identified, these biomarkers would have the potential to assess stress-induced risk for cardiovascular, metabolic, and psychiatric disorders.
Supplementary Material
Acknowledgements
This study was funded by the Rales Family Foundation (GSW). Corcept Therapeutics arranged and funded the measurement of FKBP5 mRNA expression.
Footnotes
Conflict of interest: None
Financial disclosure: None
References
- Arnsten AF, 2015. Stress weakens prefrontal networks: molecular insults to higher cognition. Nat Neurosci 18, 1376–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bali U, Phillips T, Hunt H, Unitt J, 2016. FKBP5 mRNA Expression Is a Biomarker for GR Antagonism. J Clin Endocrinol Metab 101, 4305–4312. [DOI] [PubMed] [Google Scholar]
- Beck AT, Beamesderfer A, 1974. Assessment of depression: the depression inventory. Mod Probl Pharmacopsychiatry 7, 151–169. [DOI] [PubMed] [Google Scholar]
- Beck AT, Epstein N, Brown G, Steer RA, 1988. An inventory for measuring clinical anxiety: psychometric properties. J Consult Clin Psychol 56, 893–897. [DOI] [PubMed] [Google Scholar]
- Beck AT, Steer RA, Ball R, Ranieri W, 1996. Comparison of Beck Depression Inventories -IA and -II in psychiatric outpatients. J Pers Assess 67, 588–597. [DOI] [PubMed] [Google Scholar]
- Binder EB, Salyakina D, Lichtner P, Wochnik GM, Ising M, Putz B, Papiol S, Seaman S, Lucae S, Kohli MA, Nickel T, Kunzel HE, Fuchs B, Majer M, Pfennig A, Kern N, Brunner J, Modell S, Baghai T, Deiml T, Zill P, Bondy B, Rupprecht R, Messer T, Kohnlein O, Dabitz H, Bruckl T, Muller N, Pfister H, Lieb R, Mueller JC, Lohmussaar E, Strom TM, Bettecken T, Meitinger T, Uhr M, Rein T, Holsboer F, Muller-Myhsok B, 2004. Polymorphisms in FKBP5 are associated with increased recurrence of depressive episodes and rapid response to antidepressant treatment. Nat Genet 36, 1319–1325. [DOI] [PubMed] [Google Scholar]
- Bose M, Olivan B, Laferrere B, 2009. Stress and obesity: the role of the hypothalamic-pituitary-adrenal axis in metabolic disease. Curr Opin Endocrinol Diabetes Obes 16, 340–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruehl H, Wolf OT, Convit A, 2009. A blunted cortisol awakening response and hippocampal atrophy in type 2 diabetes mellitus. Psychoneuroendocrinology 34, 815–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brydon L, Magid K, Steptoe A, 2006. Platelets, coronary heart disease, and stress. Brain Behav Immun 20, 113–119. [DOI] [PubMed] [Google Scholar]
- Cohen S, Kamarck T, Mermelstein R, 1983. A global measure of perceived stress. J Health Soc Behav 24, 385–396. [PubMed] [Google Scholar]
- DeSantis AS, Adam EK, Hawkley LC, Kudielka BM, Cacioppo JT, 2015. Racial and ethnic differences in diurnal cortisol rhythms: are they consistent over time? Psychosom Med 77, 6–15. [DOI] [PubMed] [Google Scholar]
- Ewald ER, Wand GS, Seifuddin F, Yang X, Tamashiro KL, Potash JB, Zandi P, Lee RS, 2014. Alterations in DNA methylation of Fkbp5 as a determinant of blood-brain correlation of glucocorticoid exposure. Psychoneuroendocrinology 44, 112–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fardet L, Petersen I, Nazareth I, 2012. Suicidal behavior and severe neuropsychiatric disorders following glucocorticoid therapy in primary care. Am J Psychiatry 169, 491–497. [DOI] [PubMed] [Google Scholar]
- Golden SH, Sanchez BN, Wu M, Champaneri S, Diez Roux AV, Seeman T, Wand GS, 2013. Relationship between the cortisol awakening response and other features of the diurnal cortisol rhythm: the Multi-Ethnic Study of Atherosclerosis. Psychoneuroendocrinology 38, 2720–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guess A, Agrawal S, Wei CC, Ransom RF, Benndorf R, Smoyer WE, 2010. Dose- and time-dependent glucocorticoid receptor signaling in podocytes. Am J Physiol Renal Physiol 299, F845–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett RA, Steptoe A, 2016. Psychosocial Factors in Diabetes and Cardiovascular Risk. Curr Cardiol Rep 18, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajat A, Diez-Roux A, Franklin TG, Seeman T, Shrager S, Ranjit N, Castro C, Watson K, Sanchez B, Kirschbaum C, 2010. Socioeconomic and race/ethnic differences in daily salivary cortisol profiles: the multi-ethnic study of atherosclerosis. Psychoneuroendocrinology 35, 932–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajat A, Diez-Roux AV, Sanchez BN, Holvoet P, Lima JA, Merkin SS, Polak JF, Seeman TE, Wu M, 2013. Examining the association between salivary cortisol levels and subclinical measures of atherosclerosis: the Multi-Ethnic Study of Atherosclerosis. Psychoneuroendocrinology 38, 1036–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinze K, Lin A, Reniers RL, Wood SJ, 2016. Longer-term increased cortisol levels in young people with mental health problems. Psychiatry Res 236, 98–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT, 2012. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber TJ, Issa K, Schik G, Wolf OT, 2006. The cortisol awakening response is blunted in psychotherapy inpatients suffering from depression. Psychoneuroendocrinology 31, 900–904. [DOI] [PubMed] [Google Scholar]
- Hubler TR, Scammell JG, 2004. Intronic hormone response elements mediate regulation of FKBP5 by progestins and glucocorticoids. Cell Stress Chaperones 9, 243–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AE, Irizarry RA, 2014. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol 15, R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly SJ, Ismail M, 2015. Stress and type 2 diabetes: a review of how stress contributes to the development of type 2 diabetes. Annu Rev Public Health 36, 441–462. [DOI] [PubMed] [Google Scholar]
- Klengel T, Mehta D, Anacker C, Rex-Haffner M, Pruessner JC, Pariante CM, Pace TW, Mercer KB, Mayberg HS, Bradley B, Nemeroff CB, Holsboer F, Heim CM, Ressler KJ, Rein T, Binder EB, 2013. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat Neurosci 16, 33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehl LK, Hinkelmann K, Muhtz C, Dettenborn L, Wingenfeld K, Spitzer C, Kirschbaum C, Wiedemann K, Otte C, 2015. Hair cortisol and cortisol awakening response are associated with criteria of the metabolic syndrome in opposite directions. Psychoneuroendocrinology 51, 365–370. [DOI] [PubMed] [Google Scholar]
- Kumari M, Shipley M, Stafford M, Kivimaki M, 2011. Association of diurnal patterns in salivary cortisol with all-cause and cardiovascular mortality: findings from the Whitehall II study. J Clin Endocrinol Metab 96, 1478–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RS, Tamashiro KL, Yang X, Purcell RH, Harvey A, Willour VL, Huo Y, Rongione M, Wand GS, Potash JB, 2010. Chronic corticosterone exposure increases expression and decreases deoxyribonucleic acid methylation of Fkbp5 in mice. Endocrinology 151, 4332–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RS, Tamashiro KL, Yang X, Purcell RH, Huo Y, Rongione M, Potash JB, Wand GS, 2011. A measure of glucocorticoid load provided by DNA methylation of Fkbp5 in mice. Psychopharmacology (Berl) 218, 303–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ji L, 2005. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity (Edinb) 95, 221–227. [DOI] [PubMed] [Google Scholar]
- Magee JA, Chang LW, Stormo GD, Milbrandt J, 2006. Direct, androgen receptor-mediated regulation of the FKBP5 gene via a distal enhancer element. Endocrinology 147, 590–598. [DOI] [PubMed] [Google Scholar]
- Mahon PB, Zandi PP, Potash JB, Nestadt G, Wand GS, 2013. Genetic association of FKBP5 and CRHR1 with cortisol response to acute psychosocial stress in healthy adults. Psychopharmacology (Berl) 227, 231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malviya SA, Kelly SD, Greenlee MM, Eaton DC, Duke BJ, Bourke CH, Neigh GN, 2013. Estradiol stimulates an anti-translocation expression pattern of glucocorticoid co-regulators in a hippocampal cell model. Physiol Behav 122, 187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKlveen JM, Morano RL, Fitzgerald M, Zoubovsky S, Cassella SN, Scheimann JR, Ghosal S, Mahbod P, Packard BA, Myers B, Baccei ML, Herman JP, 2016. Chronic Stress Increases Prefrontal Inhibition: A Mechanism for Stress-Induced Prefrontal Dysfunction. Biol Psychiatry [DOI] [PMC free article] [PubMed]
- Ogden J, 2004. Health Psychology: A Textbook, 3rd Edition ed. Open University Press - McGraw-Hill Education, New York, NY. [Google Scholar]
- Rahman MM, Callaghan CK, Kerskens CM, Chattarji S, O’Mara SM, 2016. Early hippocampal volume loss as a marker of eventual memory deficits caused by repeated stress. Sci Rep 6, 29127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen R, 2001. Quantification on the LightCycler Springer Berlin; Heidelberg. [Google Scholar]
- Resmini E, Santos A, Aulinas A, Webb SM, Vives-Gilabert Y, Cox O, Wand G, Lee RS, 2016. Reduced DNA methylation of FKBP5 in Cushing’s syndrome. Endocrine 54, 768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippe RC, Noppe G, Windhorst DA, Tiemeier H, van Rossum EF, Jaddoe VW, Verhulst FC, Bakermans-Kranenburg MJ, van IMH, van den Akker EL, 2016. Splitting hair for cortisol? Associations of socio-economic status, ethnicity, hair color, gender and other child characteristics with hair cortisol and cortisone. Psychoneuroendocrinology 66, 56–64. [DOI] [PubMed] [Google Scholar]
- Rohleder N, Joksimovic L, Wolf JM, Kirschbaum C, 2004. Hypocortisolism and increased glucocorticoid sensitivity of pro-Inflammatory cytokine production in Bosnian war refugees with posttraumatic stress disorder. Biol Psychiatry 55, 745–751. [DOI] [PubMed] [Google Scholar]
- Rosmond R, Bjorntorp P, 2000. The hypothalamic-pituitary-adrenal axis activity as a predictor of cardiovascular disease, type 2 diabetes and stroke. J Intern Med 247, 188–197. [DOI] [PubMed] [Google Scholar]
- Saudek CD, Herman WH, Sacks DB, Bergenstal RM, Edelman D, Davidson MB, 2008. A new look at screening and diagnosing diabetes mellitus. J Clin Endocrinol Metab 93, 2447–2453. [DOI] [PubMed] [Google Scholar]
- Sheehan DV, Lecrubier Y, Sheehan KH, Amorim P, Janavs J, Weiller E, Hergueta T, Baker R, Dunbar GC, 1998. The Mini-International Neuropsychiatric Interview (M.I.N.I.): the development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. J Clin Psychiatry 59 Suppl 20, 22–33;quiz 34–57. [PubMed] [Google Scholar]
- Sobell L, Sobell M, 1992. Timeline follow-back: a technique for assessing self-reported alcohol consumption., in: Litten R, Allen J (Eds.), Measuring Alcohol Consumption: Psychosocial and Biochemical Methods Humana Press, Totowas, NJ. [Google Scholar]
- Spanakis EK, Wang X, Sanchez BN, Diez Roux AV, Needham BL, Wand GS, Seeman T, Golden SH, 2016. Lack of significant association between type 2 diabetes mellitus with longitudinal change in diurnal salivary cortisol: the multiethnic study of atherosclerosis. Endocrine 53, 227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreeburg SA, Hoogendijk WJ, DeRijk RH, van Dyck R, Smit JH, Zitman FG, Penninx BW, 2013. Salivary cortisol levels and the 2-year course of depressive and anxiety disorders. Psychoneuroendocrinology 38, 1494–1502. [DOI] [PubMed] [Google Scholar]
- Vreeburg SA, Hoogendijk WJ, van Pelt J, Derijk RH, Verhagen JC, van Dyck R, Smit JH, Zitman FG, Penninx BW, 2009. Major depressive disorder and hypothalamic-pituitary-adrenal axis activity: results from a large cohort study. Arch Gen Psychiatry 66, 617–626. [DOI] [PubMed] [Google Scholar]
- Whelan JA, Russell NB, Whelan MA, 2003. A method for the absolute quantification of cDNA using real-time PCR. J Immunol Methods 278, 261–269. [DOI] [PubMed] [Google Scholar]
- Willour VL, Chen H, Toolan J, Belmonte P, Cutler DJ, Goes FS, Zandi PP, Lee RS, MacKinnon DF, Mondimore FM, Schweizer B, Bipolar Disorder Phenome G, Consortium NGIBD, DePaulo JR Jr., Gershon ES, McMahon FJ, Potash JB, 2009. Family-based association of FKBP5 in bipolar disorder. Mol Psychiatry 14, 261–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Ewald ER, Huo Y, Tamashiro KL, Salvatori R, Sawa A, Wand GS, Lee RS, 2012. Glucocorticoid-induced loss of DNA methylation in non-neuronal cells and potential involvement of DNMT1 in epigenetic regulation of Fkbp5. Biochem Biophys Res Commun 420, 570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.