Abstract
Retrotransposons have played an important role in the evolution of host genomes1,2. Their impact is mainly deduced from the composition of DNA sequences that have been fixed over evolutionary time 2. Such studies provide important “snapshots” reflecting the historical activities of transposons but do not predict current transposition potential. We previously reported Sequence-Independent Retrotransposon Trapping (SIRT) as a method that, by identification of extrachromosomal linear DNA (eclDNA), revealed the presence of active LTR retrotransposons in Arabidopsis3. However, SIRT cannot be applied to large and transposon-rich genomes, as found in crop plants. We have developed an alternative approach named ALE-seq (amplification of LTR of eclDNAs followed by sequencing) for such situations. ALE-seq reveals sequences of 5’ LTRs of eclDNAs after two-step amplification: in vitro transcription and subsequent reverse transcription. Using ALE-seq in rice, we detected eclDNAs for a novel Copia family LTR retrotransposon, Go-on, which is activated by heat stress. Sequencing of rice accessions revealed that Go-on has preferentially accumulated in indica rice grown at higher temperatures. Furthermore, ALE-seq applied to tomato fruits identified a developmentally regulated Gypsy family of retrotransposons. A bioinformatic pipeline adapted for ALE-seq data analyses is used for the direct and reference-free annotation of new, active retroelements. This pipeline allows assessment of LTR retrotransposon activities in organisms for which genomic sequences and/or reference genomes are either unavailable or of low quality.
Chromosomal copies of activated retrotransposons containing long terminal repeats (LTRs) are transcribed by RNA polymerase II, followed by reverse transcription of transcripts to extrachromosomal linear DNAs (eclDNA); these integrate back into host chromosomes3. Because of the two obligatory template switches during reverse transcription, the newly synthetized eclDNA is flanked by LTRs of identical sequence. Their subsequent divergence due to the accumulation of mutations correlates well with length of time since the last transposition, and thus transposon age4. However, the age of LTR retrotransposons cannot be used to predict their current transpositional potential. Moreover, predictions are further complicated by recombination events that occur with high frequency between young and old members of a retrotransposon family5; thus old family members also contribute to the formation of novel recombinant elements that insert into new chromosomal positions5. Although, retrotransposon activities can be relatively easily measured at the transcriptional level6, the presence of transcripts is a poor predictor of transpositional potential due to posttranscriptional control of this process7,8. In addition, direct detection of transposition by genome-wide sequencing to identify new insertions is too expensive and time-consuming to be applied as a screening method. Clearly, the development of an expeditious approach to identify active retrotransposons that predict their transposition potential would be welcomed. We previously described the SIRT strategy for Arabidopsis that led to the identification of eclDNA of a novel retroelement and subsequent detection of new insertions3. Thus, the presence of eclDNAs, the last pre-integration intermediate, was shown to be a good predictor of retrotransposition potential.
Results
Development of ALE-seq
Retrotransposons include a conserved sequence known as the primer binding site (PBS), where binding of the 3’ end of cognate tRNA initiates the reverse transcription reaction3. Met-iCAT (Methionine tRNA-CAT anticodon) PBS was chosen for SIRT as it is the site present in the majority of annotated Arabidopsis retrotransposons3. To examine whether Met-iCAT PBS sequences are also predominant in LTR retrotransposons of other plants, we used the custom-made software LTRpred for de novo annotation of LTR retrotransposons in rice and tomato genomes (see Materials and methods). Young retroelements were selected by filtering for at least 95% identity between the two LTRs and subsequently examined for their cognate tRNAs (Supplementary Figure 1). As in Arabidopsis, around 80% of LTR retrotransposons in the tomato genome contained Met-iCAT PBS (Supplementary Figure 1). In contrast, only 30% harboured Met-iCAT PBS in rice, and Arg-CCT (Arginine tRNA-CCT anticodon) PBS was found in 60% of young LTR retrotransposons (Supplementary Figure 1). Nonetheless, we used Met-iCAT PBS in our initial experiments because most retrotransposons known to be active in rice callus (e.g. Tos17 and Tos19) contain Met-iCAT PBS. Initially, SIRT was performed on DNA extracted from rice leaves and calli; however, we did not detect eclDNAs for Tos17 and Tos19 in rice tissues by this method (Supplementary Figure 2). We reasoned that the short stretch of PBS used for primer design in SIRT may have impaired PCR efficiency due to the many PBS-related sequences present in larger genomes containing a high number of retroelements, as is the case in rice.
To counter this problem, we developed an alternative method, named ALE-seq, with significantly improved selectivity and sensitivity of eclDNA detection. A crucial difference to SIRT is that ALE-seq amplification of eclDNA is separated into two reactions: in vitro transcription and reverse transcription (Figure 1a). This decoupling of the use of the two priming sequences followed by the digestion of non-templated DNA and RNA is significantly more selective and efficient than the single PCR amplification in SIRT.
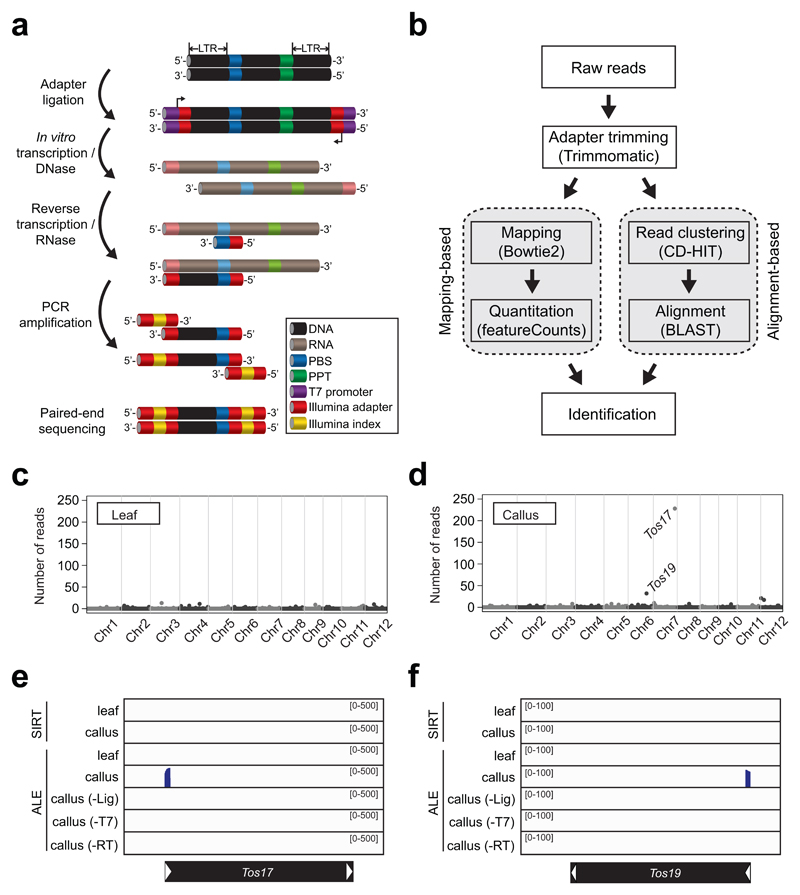
Figure 1. Detection of eclDNA by ALE-seq.
a, The workflow of ALE-seq. The colour code is indicated in a box. b, Analysis pipeline of ALE-seq results. The sequenced reads can be mapped to the reference genome or aligned to each other to obtain a cluster consensus. c and d, Genome-wide plots of rice ALE-seq results from leaf (c) and callus (d). The levels are shown as number of reads mapped to each retrotransposon. Dots represent annotated retrotransposons; those corresponding to Tos17 and Tos19 are indicated. e and f, Read coverage plots mapped to Tos17 (e) and Tos19 (f). The black bars represent retrotransposons and white arrowheads indicate LTRs.
ALE-seq starts with ligation to the ends of eclDNA of an adapter containing a T7 promoter sequence at its 5’ end and subsequent in vitro transcription with T7 RNA polymerase. The synthesized RNA is then reverse transcribed using the primer that binds the transcripts at the PBS site. The adapter and the oligonucleotides priming reverse transcription are anchored with partial Illumina adapter sequences (Supplementary Table 1), which allows the amplified products to be directly deep-sequenced in a strand-specific manner. The ALE-seq-sequences derived from retrotransposon eclDNAs are predicted to contain the intact 5’ LTR up to the PBS site, flanked by Illumina paired-end sequencing adapters. We used the Illumina MiSeq platform for sequencing because its long reads of 300 bp from both ends cover the entire LTR lengths of most potentially active elements. It is worth noting that the Illumina adapters were tagged to the intact LTR DNA without fragmentation of the amplicons. This together with the long reads of MiSeq allowed us to reconstitute the complete LTR sequences, even in the absence of the reference genome sequence. The reconstituted LTRs were analysed using the alignment-based approach that complements the mapping-based approach when the reference genome is incomplete (Figure 1b).
First, we tested ALE-seq on Arabidopsis by examining heat-stressed Col-0 Arabidopsis plants9, met1-1 mutant3 and epi128, a met1-derived epigenetic recombinant inbred line. ALE-seq cleanly and precisely recovered sequences of complete LTRs for Onsen, Copia21 and Evade in samples containing their respective eclDNA (Supplementary Figure 3)3,8,9. Due to priming of the reverse transcription reaction at PBS, the reads were explicitly mapped to the 5’ but not to the 3’ LTR, although the two LTRs have identical sequences. The ALE-seq reads have well-defined extremities, starting at the position marking the start of LTRs and finishing at the PBS, which is consistent with their eclDNA origin. The ends of LTRs can also be inspected for conserved sequences that would further confirm their eclDNA origin (Supplementary Figure 4). This reduced ambiguity of read mapping in ALE-seq analysis, combined with the clear-cut detection of LTR ends, allows for explicit and precise assignment of ALE-seq results to active LTR retrotransposons.
Since SIRT failed to detect eclDNAs of rice retrotransposons known to be activated in rice callus, we examined whether ALE-seq would identify their eclDNAs. As shown in Figure 1c to f, ALE-seq unambiguously detected eclDNAs of Tos17 and Tos19 in rice callus, but not in leaf samples. To test whether detection of 5’ LTR sequences requires the entire ALE-seq procedure, we performed control experiments with depleted ALE-seq reactions, for example, in the absence of enzymes for either ligation, in vitro transcription, or reverse transcription. All incomplete procedures failed to produce sequences containing 5’ LTRs derived from eclDNAs (Figure 1e and f). Taken together, the data show that ALE-seq can detect eclDNAs of LTR retrotransposons in Arabidopsis as well as in rice with considerably greater efficiency than the SIRT method.
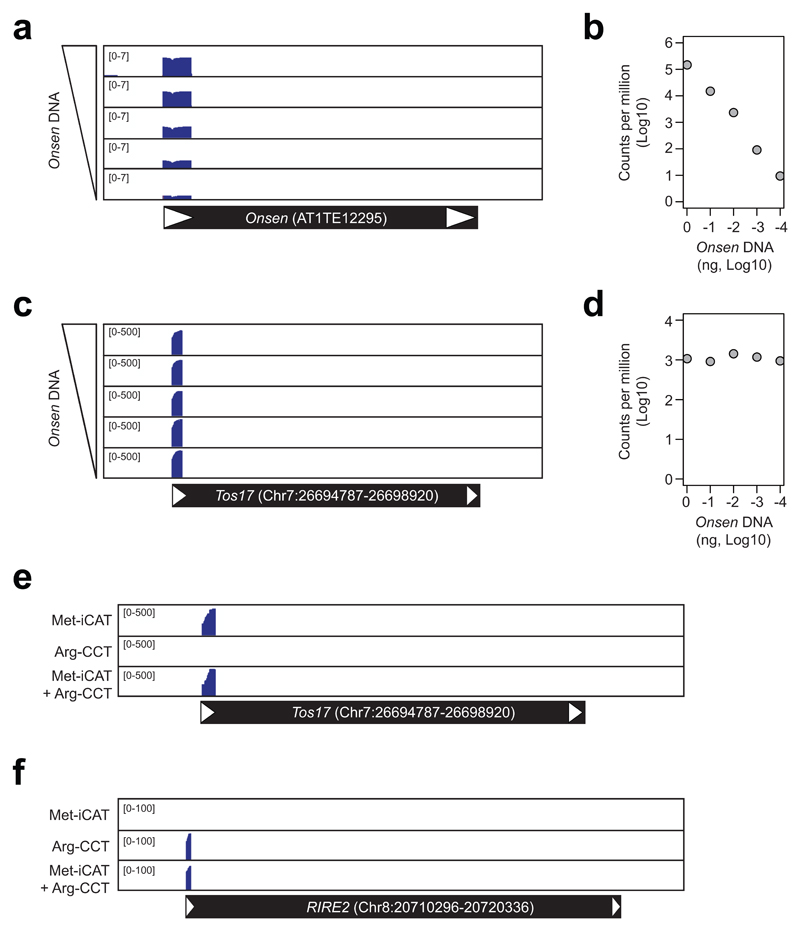
To examine the suitability of ALE-seq for quantitative determination of eclDNA levels, we carried out a reconstruction experiment spiking 100 ng of genomic DNA from rice callus with differing amounts of PCR-amplified full-length Onsen DNA from 1 ng to 100 fg (Figure 2a to d). The results in Figure 2a and b show that the readouts of ALE-seq for Onsen correlate well with the input amounts (R2=0.99). The initial ALE-seq steps of ligation and in vitro transcription impinged proportionally on the input DNA, resulting in unbiased quantification of the eclDNA and minimal quantitative distortion of the final ALE-seq data. Noticeably, the levels of Tos17 were similar in all the spiked samples, indicating that addition of Onsen DNA did not influence the detection sensitivity of Tos17, at least for the amounts tested (Figure 2c and d). Thus, ALE-seq can be used to accurately determine eclDNA levels.
Figure 2. Sensitivity and specificity of eclDNA detection by ALE-seq.
a-d, ALE-seq reconstruction experiment with varying amounts of PCR-amplified Onsen DNA added to rice callus DNA. Genome browser image with the read coverage (a and c) and quantitated read counts (b and d) for Onsen (a and b) and Tos17 (c and d) loci. The amounts of Onsen DNA added were 1 ng, 100 pg, 10 pg, 1 pg or 100 fg; 100 ng of rice callus DNA was used. Note that read coverage values are log10-converted in a. For b and d, values are shown as log10-converted counts per million sequenced reads. e and f, Read coverage plots for the ALE-seq of rice callus using different RT primers. Tos17 and RIRE2 transposons are depicted below the plots as in Figure 1.
Most rice retrotransposons harbour Arg-CCT PBS (Supplementary Figure 1). We tested whether the reverse transcription reaction can be multiplexed to capture both types of retrotransposons (containing Arg-CCT or Met-iCAT PBS) and whether multiplexing of the reverse transcription primers compromises the sensitivity of the procedure. ALE-seq was performed on DNA from rice callus, testing each of the reverse transcription primers separately or as a mixture of both primers in a single reaction. As shown in Figure 2e and Supplementary Figure 5, the levels of Tos17 recorded in the samples with both primers were similar to the Met-iCAT primer alone. Importantly, we also detected the eclDNAs of the RIRE2 element containing Arg-CCT PBS (Figure 2f), which was known to be transpositionally active in rice callus7.
Identification of Go-on retrotransposon using ALE-seq
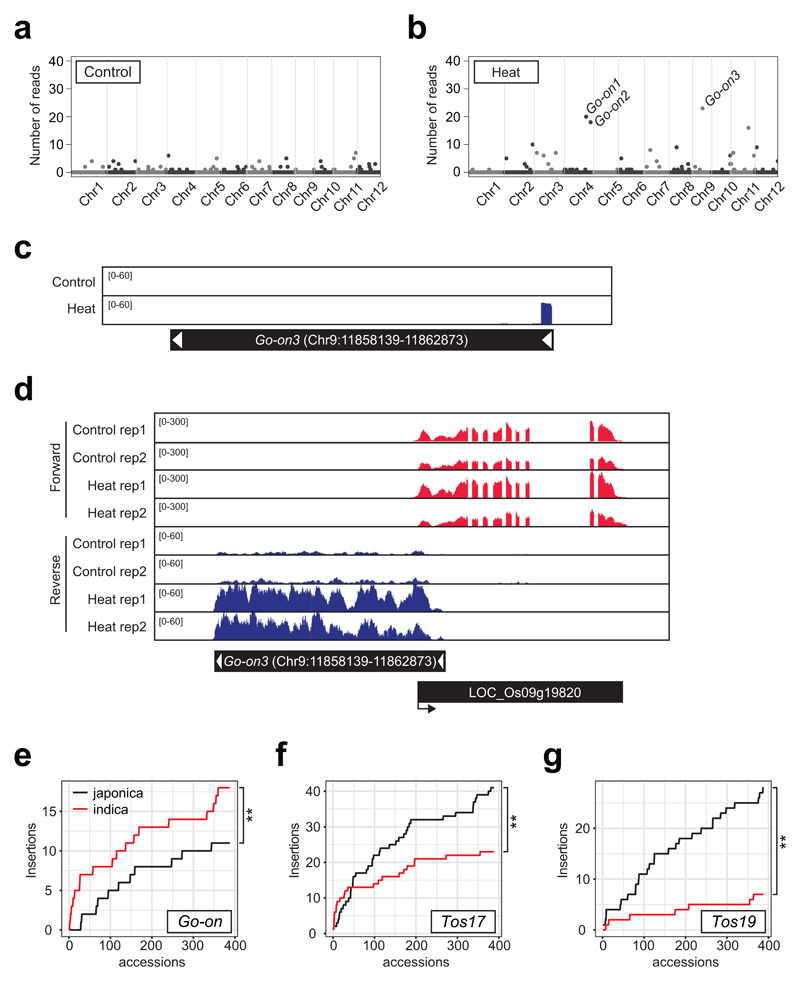
We next used ALE-seq to search for novel active rice retrotransposons. Since many plant retrotransposons are transcriptionally activated by abiotic stresses9,10, we subjected rice plants to heat stress before subjecting them to ALE-seq. In this way we identified a Copia-type retrotransposon able to synthetize eclDNA in the heat-stressed plants (Figure 3a to c) and named this element Go-on (the Korean for ‘high temperature’). The three retrotransposons with the highest eclDNA levels in heat-stress conditions all belong to the Go-on family (Figure 3b and Supplementary Figure 6). Although, eclDNAs were detected for all three copies, Go-on3 seems to be the youngest and, thus, possibly the most active family member, containing identical LTRs and a complete ORF (Supplementary Figure 6). As depicted in Supplementary Figure 6, the 5’ LTR sequences of the three Go-on copies are identical; thus the ALE-seq reads derived from Go-on3 LTR were also cross-mapped to other copies that are possibly inactive or have reduced activities. To further determine whether sequences of Go-on LTRs recovered by ALE-seq are indeed derived from Go-on3 or also from other family members, we performed an ALE-seq experiment using RT primers located further downstream of the PBS, including sequences specific for each Go-on family member (Supplementary Figure 6). The amplified ALE-seq products revealed that the eclDNAs produced in heat-stressed rice originated only from Go-on3 (Supplementary Figure 6). We validated the production of eclDNAs of Go-on3 by sequencing the junction of the adapter and the 5’ end of LTR (Supplementary Figure 6) and by qPCR (Supplementary Figure 7).
Figure 3. Identification of a novel heat-activated retrotransposon in rice.
a and b, Genome-wide plots of rice ALE-seq results as in Figure 1. Control (a) and heat-stressed (b) rice plants were used. One-week-old seedlings were subjected to heat stress (44°C) for 3 days. Met-iCAT PBS primer was used in RT. The levels are shown as the number of reads mapped to retroelements. Three Go-on copies are indicated in b. c, Read coverage plot for Go-on3. d, RNA-seq data showing Go-on3 and a neighbouring gene. RNA-seq data were generated using the same plant materials as in a and b. The experiment was repeated independently two times with similar results. e-g, Cumulative plots for the number of non-reference insertions of Go-on (e), Tos17 (f), and Tos19 (g) in the genomes of 388 japonica and indica rice accessions. The statistical difference was determined by iterating random selection of 200 accessions out of 388 and performing the two-tailed Wilcoxon test. ** P =2.2e-16.
Next, we examined whether Go-on3 is transcriptionally activated in rice subjected to heat stress. RNA-seq and the RT-qPCR data clearly showed that Go-on is strongly activated in heat-stress conditions (Figure 3d and Supplementary Figure 7). Similar to many other retrotransposons including ONSEN of Arabidopsis9,11,12, the LTR sequence of Go-on3 contains cis-acting regulatory element such as the heat shock transcription factor HSFC1-binding sequence motif (Supplementary Figure 7), which is suggestive to its heat stress-mediated transcriptional activation (Figure 3d). To determine whether Go-on is also activated in indica rice, we heat-stressed plants of IR64 for three days and examined Go-on RNA and DNA levels. Similar to japonica rice, Go-on RNA and DNA accumulated markedly under heat stress (Supplementary Figure 8), suggesting that the trigger for Go-on activation is conserved in both of these evolutionarily distant rice genotypes. Analysis of the RNA-seq data from the heat-stressed rice plants revealed a poor correlation between the mRNA and eclDNA levels of retrotransposons (Supplementary Figure 9). Given that eclDNAs captured by ALE-seq in Arabidopsis and rice (Figure 1c to f and Supplementary Figure 3) are all known well for their transposition competence, this possibly agrees with the notion that the eclDNA level is a better predictor of retrotransposition than the RNA level.
To possibly relate accumulation of Go-on copies in plant populations grown in different temperatures, we analysed the historical retrotransposition of Go-on using the genome resequencing data of rice accessions from the 3,000 Rice Genome Project13. First, we retrieved the raw sequencing data for all 388 japonica rice accessions and the same number of randomly selected sequences of indica rice accessions. Using the Transposon Insertion Finder (TIF) tool14, japonica and indica sequences were analysed for the number of Go-on copies and their genome-wide distribution. Only non-reference insertions that were absent in the reference genome were scored and the cumulative number of new insertions was plotted (Figure 3e to g). Figure 3e shows that the indica rice population grown in a warmer climate15 accumulated significantly more Go-on copies than the japonica population. As controls, we also examined the accumulation of Tos17 and Tos19, which were not activated by heat stress in our ALE-seq profile (Figure 3a and b). Both retrotransposons showed more transposition events in japonica than in indica rice (Figure 3f, g and Supplementary Figure 10). Therefore, the copy number of Go-on in rice accessions correlated with their growth temperatures, which could possibly be related to occasional Go-on activation in elevated ambient temperatures.
Identification of FIRE retrotransposon using ALE-seq
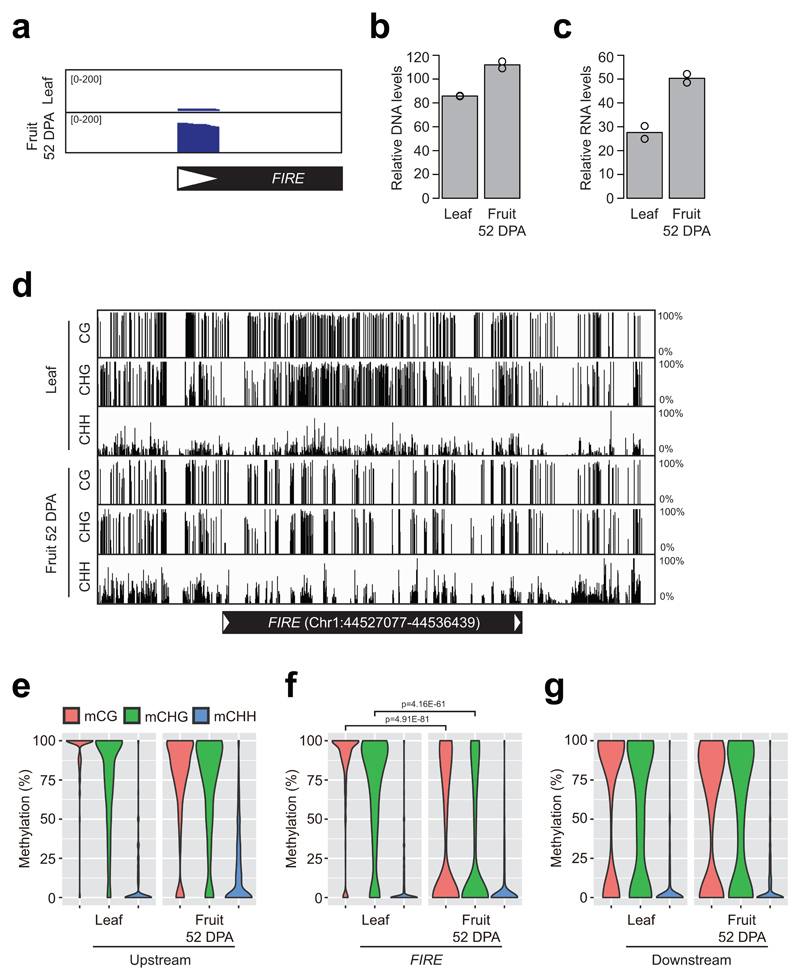
It was reported previously that the tomato genome (Solanum lycopersicum) experiences a significant loss of DNA methylation in fruits during their maturation, which leads to transcriptional activation of retrotransposons16. However, it was not known whether these transcriptionally activated tomato transposons synthesise eclDNA. It was questionable whether the ALE-seq strategy is sensitive enough to detect eclDNA in the ~950 Mb tomato genome, which is almost three times as large as ~400 Mb of rice17. To address these questions, ALE-seq was carried out on DNA samples from fruits at 52 days post anthesis (DPA), when the loss of DNA methylation is most pronounced16, and from leaves as a control. It is important to note that we used tomato cultivar (cv.) M82 for these experiments, as it is commonly used for genetic studies18,19, and that the sequence of the current tomato reference genome is based on cv. Heinz 170617. Since retrotransposon sequences and their chromosomal distributions differ largely between genomes of different varieties within the same plant species20–22, we could not use the standard mapping-based annotation of the ALE-seq results. As a consequence, we developed a reference-free and alignment-based approach that adopts the clustering of reads based on their sequence similarities (Figure 1b). Briefly, the reads from both samples were pooled and then clustered by sequence homology (See Materials and methods). The consensus of each cluster was determined and used as the reference in paired-end mapping. Subsequently, the consensus sequences were used for a BLAST search against the reference genome for the closest homologues. In this way, the BLAST search was able to map the clustered ALE-seq output to reference genome annotated retrotransposons, which are most similar to the ALE-seq recovered sequences. Applying this strategy, we identified a retroelement belonging to a Gypsy family (FIRE, Fruit-Induced RetroElement) that produces significant amounts of eclDNA at 52 DPA during fruit ripening (Figure 4a and b). We also determined the transcript levels of the FIRE element in leaves and 52 DPA fruit samples. As shown in Figure 4c, fruit RNA levels were enhanced twofold compared to leaves, where FIRE eclDNA was barely detectable (Figure 4a). Finally, we found that the DNA methylation status of the FIRE element was lower in fruits than leaves in all three sequence contexts (Figure 4d and f). In contrast, the DNA methylation levels of sequences directly flanking FIRE were similar in leaves and fruits (Figure 4e to g).
Figure 4. Identification of a tomato retrotransposon activated in fruit pericarp.
a, Read coverage plot for the FIRE retrotransposon identified in tomato fruit pericarp by ALE-seq. Met-iCAT PBS primer was used in RT. b and c, The DNA (b) and RNA (c) levels of FIRE in leaves and fruits determined by qPCR. The levels are means of two biological replicates. Normalization was done against SlGAPDH (Solyc03g111010) and SlCAC (Solyc08g006960) for DNA and RNA analyses, respectively. d, Genome browser image for the DNA methylation levels at FIRE element in leaves and fruits of tomato. The levels are shown as percent methylation of each cytosine. e-g, Violin plots for DNA methylation levels at the upstream (e), FIRE (f) and downstream (g) regions. Only cytosines supported by at least three reads in both samples were considered. In FIRE locus, for example, 4,032 out of 4,078 cytosines in both strands were analysed. The upstream and downstream regions are immediate flanking sequences taken for the same length as FIRE of 9.362 kb. P-values were determined by a two-sided Fisher’s t-test using 558 CG and 717 CHG sites at FIRE locus. Other samples with insignificant statistical difference are not shown for the p-values.
Discussion
Recently, a novel active retrotransposon was identified in rice by sequencing extrachromosomal circular DNA (eccDNA) produced as a by-product of retrotransposition or by nuclear recombination reactions of eclDNAs23,24. Although the method of eccDNA sequencing has certain advantages over SIRT, such as increased sensitivity and the recovery of sequences of the entire element, it also has certain limitations. For example, the method requires relatively large amounts of starting material but still shows serious limits in efficiency and indicative power for retrotransposition. The method did not detect the eccDNA of Tos19 in rice callus, where this transposon is known to move23, however, direct comparison of both methods on the same biological samples was not performed. More importantly, eccDNAs may also be the result of genomic DNA recombination25 and these background products may be misleading when extrapolating to the transpositional potential of a previously unknown element. In this respect, ALE-seq is a significantly improved tool that largely overcomes the above-mentioned limitations of previous methods and requires only 100 ng of plant DNA.
The heat-responsiveness of Go-on, the novel heat-activated Copia family retrotransposon of rice detected using ALE-seq, seems to be conferred by cis-acting DNA elements embedded in the LTR, which are similar to the heat-activated Onsen retrotransposon in Arabidopsis 11,12. Although heat stress can induce production of mRNA and eclDNA of Onsen, its retrotransposition is tightly controlled by the small interfering RNA pathway9. Given that real-time transposition of rice retrotransposons has only been detected in epigenetic mutants26,27 and triggered by tissue culture conditions causing vast alterations in the epigenome7,or as a result of interspecific hybridization28, an altered epigenomic status seems to be an important prerequisite for retrotransposition. In fact, we failed to detect transposed copies of Go-on in the progeny of heat-stressed rice plants. Thus, although Go-on produces eclDNAs after heat stress, it may be mobilized only at low frequency in wild type rice due to epigenetic restriction of retrotransposition. Nevertheless, on an evolutionary scale, the higher number of new insertions of Go-on in indica rice populations grown at elevated temperatures might suggest its potential mobility.
Many retrotransposons are transcriptionally reactivated during specific developmental stages or in particular cell types29,30. In tomato, fruit pericarp exhibits a reduction in DNA methylation during ripening16. This is largely attributed to higher transcription of the DEMETER-LIKE2 DNA glycosylase gene31. Despite massive transcriptional reactivation of retrotransposons in tomato fruits, it has been difficult to determine whether further steps toward transposition also take place. Using ALE-seq, we identified eclDNA that we annotated using a reference-free and alignment-based approach to a novel FIRE element. FIRE has 164 copies in the reference tomato genome and in a conventional mapping-based approach the ALE-seq reads of FIRE cross-mapped to multiple copies, making it difficult to assign eclDNA levels to particular family members (Supplementary Figure 11). Therefore, our strategy can be used in situations where sequence of the reference genome is unavailable or the mapping of reads is hindered by the high complexity and multiplicity of the retrotransposon population.
ALE-seq could also be applied to non-plant systems. For example, numerous studies in various eukaryotes, including mammals, found that retrotransposons are transcriptionally activated by certain diseases or at particular stages during embryo development32,33. It was also suggested that retrotransposition might be an important component of disease progression34. Given that the direct detection of retrotransposition is challenging, it would be interesting to use ALE-seq to determine whether such temporal relaxations of epigenetic transposon silencing also result in the production of the eclDNAs, as the direct precursor of the chromosomal integration of a retrotransposon.
Materials and methods
Plant materials
Seeds of Oryza sativa ssp. japonica cv. Nipponbare and Oryza sativa ssp. indica cv. IR64 were surface-sterilized in 20% bleach for 15 min, rinsed three times with sterile water and germinated on ½-MS media. Rice plants were grown in 10 h light / 14 h dark at 28°C and 26°C, respectively. For heat-stress experiments, 1-week-old rice plants were transferred to a growth chamber at 44°C and 28°C in light and dark, respectively. Rice callus was induced by the method used for rice transformation as previously described35.
Tomato plants (Solanum lycopersicum cv. M82) were grown under standard greenhouse conditions (16 h supplemental lighting of 88 w/m2 at 25°C and 8 h at 15°C). Tomato leaf tissue samples were taken from 2-month-old plants. Tomato fruit pericarp tissues were harvested at 52 days post anthesis (DPA).
Annotation of LTR retrotransposons
Functional de novo annotation of LTR retrotransposons for the genomes of TAIR10 (Arabidopsis), MSU7 (rice) and SL2.50 (tomato) was achieved by the LTRpred pipeline (https://github.com/HajkD/LTRpred) using the parameter configuration: minlenltr = 100, maxlenltr = 5000, mindistltr = 4000, maxdisltr = 30000, mintsd = 3, maxtsd = 20, vic = 80, overlaps = “no”, xdrop = 7, motifmis = 1, pbsradius = 60, pbsalilen = c(8,40), pbsoffset = c(0,10), quality.filter = TRUE, n.orf = 0. The plant-specific tRNAs used to screen for primer binding sites (PBS) were retrieved from GtRNAdb36 and plantRNA37 and combined in a custom fasta file. The hidden Markov model files for gag and pol protein conservation screening were retrieved from Pfam38 using the protein domains RdRP_1 (PF00680), RdRP_2 (PF00978), RdRP_3 (PF00998), RdRP_4 (PF02123), RVT_1 (PF00078), RVT_2 (PF07727), Integrase DNA binding domain (PF00552), Integrase zinc binding domain (PF02022), Retrotrans_gag (PF03732), RNase H (PF00075) and Integrase core domain (PF00665). Computationally reproducible scripts for generating annotations can be found at http://github.com/HajkD/ALE.
ALE-seq library preparation
Genomic DNA was extracted using a DNeasy Plant Mini Kit (Qiagen) following the manufacturer’s instruction. Genomic DNA (100 ng) was used for adapter ligation with 4 µl of 50 µM adapter DNA. After an overnight ligation reaction at 4°C, the adapter-ligated DNA was purified by AMPure XP beads (Beckman Coulter) at a 1:0.5 ratio. In vitro transcription reactions were performed using a MEGAscript RNAi kit (Thermo Fisher) with minor modifications. Briefly, the reaction was carried out for 4 h at 37°C and the template DNA was digested prior to RNA purification. Purified RNA (3 µg) was subjected to reverse transcription (RT) using a Transcriptor First Strand cDNA Synthesis Kit (Roche). Transcriptor First Strand cDNA Synthesis Kit was chosen because the RTase of the kit is thermostable. This allowed the RT reaction at higher temperature (55°C) that reduces the RT-inhibiting RNA secondary structure formation. The custom RT primers were added as indicated for each experiment. After the RT reaction, 1 µl of RNase A/T1 (Thermo Fisher) was added to digest non-templated RNA and the reaction mixture was incubated at 37°C for at least 30 min. Single-stranded first strand cDNA was PCR-amplified by 25 cycles using Illumina TruSeq HT dual adapter primers and the PCR product was purified by AMPure XP beads (Beckman Coulter) at a 1:1 ratio. After purification, the eluted DNA was quantified using a KAPA Library Quantification Kit (KAPA Biosystems) and run on the MiSeq v3 2 X 300 bp platform in the Department of Pathology of the University of Cambridge. Due to the nature of ALE-seq that specifically amplifies ecDNAs, some ecDNA-free samples did not produce enough library DNAs which, although suboptimal loading, were nevertheless sequenced. It is advisable to spike in PCR-amplified retrotransposon DNA as described below. The oligonucleotide sequences are provided in Supplementary Table 1.
Preparation of full-length Onsen DNA
The full-length Onsen copy (AT1TE12295) was amplified using Phusion High-Fidelity DNA polymerase (New England Biolabs). PCR products were run on 1% agarose gels. The full-length fragment was then purified by QIAquick Gel Extraction (Qiagen) and its concentration was measured using the Qubit Fluorometric Quantitation system (Thermo Fisher). Primers used for amplification are listed in Supplementary Table 1.
RT-qPCR analyses
Samples were ground in liquid nitrogen using mortar and pestle. An RNeasy Plant Mini Kit (Qiagen) was used to extract total RNA following the manufacturer’s instructions. The amount of extracted RNA was estimated using the Qubit Fluorometric Quantitation system (Thermo Fisher). cDNAs were synthesized using a SuperScript VILO cDNA Synthesis Kit (Invitrogen). Real-time quantitative PCR was performed in the LightCycler 480 system (Roche) using primers listed in Supplementary Table 1. LightCycler 480 SYBR green I master premix (Roche) was used to prepare the reaction mixture in a volume of 10 µl. The results were analysed by the ΔΔCt method.
RNA-seq library construction
Total RNA was prepared as described above. An Illumina TruSeq Stranded mRNA Library Prep kit (Illumina) was used according to the manufacturer’s instructions. The resulting library was run on an Illumina NextSeq 500 machine (Illumina) in the Sainsbury Laboratory at the University of Cambridge.
Analysis of next-generation sequencing data
For RNA-seq data analysis, the adapter and the low-quality sequences were removed by Trimmomatic software39. The cleaned reads were mapped to the MSU7 version of the rice reference genome (http://rice.plantbiology.msu.edu) using TopHat240. The resulting mapping files were processed to the Cufflinks/Cuffquant/Cuffnorm pipeline41 guided by the annotation file which includes the MSU7 reference gene annotation (http://rice.plantbiology.msu.edu/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/pseudomolecules/version_7.0/all.dir/) and our custom retrotransposon annotation. Visualization of sequencing data was performed using an Integrative Genomics Viewer (IGV)42.
For ALE-seq data analysis, the adapter sequence was removed from the raw reads using Trimmomatic software. For the mapping-based approach, paired-end reads were mapped to the reference genomes (Arabidopsis, TAIR10; rice, MSU7; tomato, SL2.50) using Bowtie243 with minor optimization. In most short-read sequencing platforms, it is often difficult to assign the multi-mapped reads of TEs to precise genomic location. However, as MiSeq outputs relatively longer reads, we presumed that ALE-seq reads have less ambiguity than other sequencing platforms and set the parameters dealing with multi-mappers to default. It is only the maximum fragment length option which is set to 500 by default that was manipulated to 3000 (-X 3000). The numbers of reads mapped throughout each retrotransposon were counted by the featureCounts tool of the SubRead package44 using the custom annotation file created by LTRpred. Since featureCounts recognizes multi-mappers by SAM file’s NH tag that bowtie2 does not generate, multi-mapped reads are counted as one read aligned to a single genomic location, which reduces quantitation bias that often happens to multi-mappers. IGV was used to visualize the sequencing data. For the alignment-based approach, the forward and reverse reads were merged to yield the full-length fragment sequences and converted to fasta files using the BBTools (https://jgi.doe.gov/data-and-tools/bbtools/). The fasta files created for all the samples were concatenated to get a master fasta file that is later inputted to CD-HIT software45 to cluster the reads by sequence similarity with the following options: -c 0.95, -ap 1, -g 1. CD-HIT outputs a fasta file of representative reads for each cluster. The resulting fasta file was used as reference for paired-end mapping of initial fastq files. The mapped reads were counted with the featureCounts tool. Those clusters that significantly differed in the number of mapped reads in different samples were further analysed for their identities using BLAST search.
For Bisulfite sequencing analysis, raw sequenced reads derived from tomato fruits (52 DPA) and leaves were downloaded from the public repository (SRP008329)16 and re-analysed as previously described46, with minor modifications. Briefly, high-quality sequenced reads were mapped with Bismark47 on the cv. Heinz 1706 reference genome (https://solgenomics.net), including a chloroplast sequence obtained from GenBank database (NC_007898.3) to estimate the conversion rate. After methylation call and correction for unconverted cytosines, the methylation proportions at each cytosine position with a coverage of at least 3 reads were used to generate a bedGraph file for each cytosine context, using the R Bioconductor packages DMRCaller48 and Rtracklayer49. The IGV browser was used to visualize the methylation profiles.
Detection of retrotransposon insertions
The insertions of selected retrotransposons were detected from the genome resequencing data of japonica and indica rice accessions downloaded from the 3,000 rice genome project (PRJEB6180). The Transposon Insertion Finder (TIF) program14 was used to identify the split reads in the fastq files and detect newly integrated copies. We used MSU7 (http://rice.plantbiology.msu.edu) and ShuHui498 (http://www.mbkbase.org) for the reference of japonica and indica rice, respectively. Only non-reference insertions were considered and common insertions found in multiple accessions were counted as a single retrotransposition event.
Supplementary Material
Acknowledgements
This work was supported by European Research Council (EVOBREED) [322621]; Gatsby Fellowship [AT3273/GLE].
Footnotes
Data availability
The next generation sequencing data that support the findings of this study are available in the Sequence Read Archive (SRA) repository with the identifier SRP155920.
Code availability
The custom scripts used in this study are available in http://github.com/HajkD/ALE.
Author Contributions
J.C. and J.P. conceived the research. J.C., M.B., M.C., H.-G.D., A.B. and M.O. performed experiment. J.C., M.B., M.C. and H.-G.D. analysed data. J.C. and J.P. wrote and revised the manuscript.
Competing Interests
The authors declare that no competing interests exist.
References
- 1.Lisch D. How important are transposons for plant evolution? Nat Rev Genet. 2012;14:49–61. doi: 10.1038/nrg3374. [DOI] [PubMed] [Google Scholar]
- 2.Chuong EB, Elde NC, Feschotte C. Regulatory activities of transposable elements: from conflicts to benefits. Nat Rev Genet. 2017;18:71–86. doi: 10.1038/nrg.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Griffiths J, Catoni M, Iwasaki M, Paszkowski J. Sequence-Independent Identification of Active LTR Retrotransposons in Arabidopsis. Mol Plant. 2017;11:508–511. doi: 10.1016/j.molp.2017.10.012. [DOI] [PubMed] [Google Scholar]
- 4.Ma J, Bennetzen JL. Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci U S A. 2004;101:12404–12410. doi: 10.1073/pnas.0403715101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanchez DH, Gaubert H, Drost H, Zabet NR, Paszkowski J. High-frequency recombination between members of an LTR retrotransposon family during transposition bursts. Nat Commun. 2017;8:1–6. doi: 10.1038/s41467-017-01374-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Picault N, et al. Identification of an active LTR retrotransposon in rice. Plant J. 2009;58:754–765. doi: 10.1111/j.1365-313X.2009.03813.x. [DOI] [PubMed] [Google Scholar]
- 7.Sabot F, et al. Transpositional landscape of the rice genome revealed by paired-end mapping of high-throughput re-sequencing data. Plant J. 2011;66:241–246. doi: 10.1111/j.1365-313X.2011.04492.x. [DOI] [PubMed] [Google Scholar]
- 8.Mirouze M, et al. Selective epigenetic control of retrotransposition in Arabidopsis. Nature. 2009;461:427–430. doi: 10.1038/nature08328. [DOI] [PubMed] [Google Scholar]
- 9.Ito H, et al. An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature. 2011;472:115–119. doi: 10.1038/nature09861. [DOI] [PubMed] [Google Scholar]
- 10.Paszkowski J. Controlled activation of retrotransposition for plant breeding. Curr Opin Biotechnol. 2015;32:200–206. doi: 10.1016/j.copbio.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Cavrak VV, et al. How a Retrotransposon Exploits the Plant’s Heat Stress Response for Its Activation. PLoS Genet. 2014;10:1–12. doi: 10.1371/journal.pgen.1004115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pietzenuk B, et al. Recurrent evolution of heat-responsiveness in Brassicaceae COPIA elements. Genome Biol. 2016:1–15. doi: 10.1186/s13059-016-1072-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The 3, 000 rice genomes project. The 3, 000 rice genomes project. Gigascience. 2014;3:1–6. [Google Scholar]
- 14.Nakagome M, et al. Transposon Insertion Finder (TIF): a novel program for detection of de novo transpositions of transposable elements. BMC Bioinformatics. 2014;15:1–9. doi: 10.1186/1471-2105-15-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiong ZY, et al. Latitudinal Distribution and Differentiation of Rice Germplasm: Its Implications in Breeding. Crop Sci. 2011;51:1050–1058. [Google Scholar]
- 16.Zhong S, et al. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat Biotechnol. 2013;31:154–159. doi: 10.1038/nbt.2462. [DOI] [PubMed] [Google Scholar]
- 17.Consortium T. tomato genome. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635–641. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eshed Y, Zamir D. An Introgression Line Population of Lycopersicon pennellii in the Cultivated Tomato Enables the Identification and Fine Mapping of Yield-Associated QTL. Genetics. 1995;141:1147–1162. doi: 10.1093/genetics/141.3.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eshed Y, Zamir D. Less-Than-Additive Epistatic Interactions of Quantitative Trait Loci in Tomato. Genetics. 1996;143:1807–1817. doi: 10.1093/genetics/143.4.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quadrana L, et al. The Arabidopsis thaliana mobilome and its impact at the species level. Elife. 2016;5:1–25. doi: 10.7554/eLife.15716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stuart T, et al. Population scale mapping of transposable element diversity reveals links to gene regulation and epigenomic variation. Elife. 2016;5:1–27. doi: 10.7554/eLife.20777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei B, et al. Genome-wide characterization of non-reference transposons in crops suggests non-random insertion. BMC Genomics. 2016;17:1–13. doi: 10.1186/s12864-016-2847-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lanciano S, et al. Sequencing the extrachromosomal circular mobilome reveals retrotransposon activity in plants. PLOS Genetics. 2017;13 doi: 10.1371/journal.pgen.1006630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Møller HD, et al. Formation of Extrachromosomal Circular DNA from Long Terminal Repeats of Retrotransposons in Saccharomyces cerevisiae. G3 (Bethesda) 2015;6:453–462. doi: 10.1534/g3.115.025858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Møller HD, Parsons L, Jørgensen TS, Botstein D, Regenberg B. Extrachromosomal circular DNA is common in yeast. Proc Natl Acad Sci U S A. 2015;112:3114–3122. doi: 10.1073/pnas.1508825112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng C, et al. Loss of function mutations in the rice chromomethylase OsCMT3a cause a burst of transposition. Plant J. 2015;83:1069–1081. doi: 10.1111/tpj.12952. [DOI] [PubMed] [Google Scholar]
- 27.Cui X, et al. Control of transposon activity by a histone H3K4 demethylase in rice. Proc Natl Acad Sci U S A. 2013;110:1953–1958. doi: 10.1073/pnas.1217020110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang ZH, et al. Genomewide Variation in an Introgression Line of Rice-Zizania Revealed by Whole-Genome re-Sequencing. PLoS One. 2013;8:1–12. doi: 10.1371/journal.pone.0074479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Freeling M, Lisch D. Epigenetic reprogramming during vegetative phase change in maize. Proc Natl Acad Sci U S A. 2010;107:22184–22189. doi: 10.1073/pnas.1016884108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slotkin RK, et al. Epigenetic Reprogramming and Small RNA Silencing of Transposable Elements in Pollen. Cell. 2009;136:461–472. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu R, et al. A DEMETER-like DNA demethylase governs tomato fruit ripening. Proc Natl Acad Sci. 2015;112:10804–10809. doi: 10.1073/pnas.1503362112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goodier JL. Retrotransposition in tumors and brains. Mobile DNA. 2014;5:1–6. doi: 10.1186/1759-8753-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baillie JK, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mullins CS, Linnebacher M. Human endogenous retroviruses and cancer : Causality and therapeutic possibilities. World J Gastroenterol. 2012;18:6027–6035. doi: 10.3748/wjg.v18.i42.6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho J, Paszkowski J. Regulation of rice root development by a retrotransposon acting as a microRNA sponge. Elife. 2017;6:1–21. doi: 10.7554/eLife.30038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan PP, Lowe TM. GtRNAdb 2 . 0 : an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2016;44:184–189. doi: 10.1093/nar/gkv1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daujat M, et al. PlantRNA, a database for tRNAs of photosynthetic eukaryotes. Nucleic Acids Res. 2012;41:273–279. doi: 10.1093/nar/gks935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finn RD, et al. Pfam : the protein families database. Nucleic Acids Res. 2014;42:222–230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolger AM, Lohse M, Usadel B. Trimmomatic : a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim D, et al. TopHat2 : accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:1–13. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:516–520. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robinson JT, et al. Integrative Genomics Viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2013;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liao Y, Smyth GK, Shi W. The Subread aligner : fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:1–17. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li W, Godzik A. Cd-hit : a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 46.Catoni M, et al. DNA sequence properties that predict susceptibility to epiallelic switching. EMBO. 2017;36:617–628. doi: 10.15252/embj.201695602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krueger F, Andrews SR. Bismark : a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Catoni M, Tsang JMF, Greco AP, Zabet NR. DMRcaller : a versatile R / Bioconductor package for detection and visualization of differentially methylated regions in CpG and non-CpG contexts. Nucleic Acids Res. 2018:1–11. doi: 10.1093/nar/gky602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lawrence M, Gentleman R, Carey V. rtracklayer : an R package for interfacing with genome browsers. Bioinformatics. 2009;25:1841–1842. doi: 10.1093/bioinformatics/btp328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.