Despite the extensive literature on methods for assessing interactions between genetic and environmental factors, approaches for the joint analysis of multiple G-E interactions are surprisingly lacking. Kim et al. compare the power and robustness....
Keywords: gene and environment interaction, joint test analysis, Score test statistic, genetic risk score, environment risk score
Abstract
With growing human genetic and epidemiologic data, there has been increased interest for the study of gene-by-environment (G-E) interaction effects. Still, major questions remain on how to test jointly a large number of interactions between multiple SNPs and multiple exposures. In this study, we first compared the relative performance of four fixed-effect joint analysis approaches using simulated data, considering up to 10 exposures and 300 SNPs: (1) omnibus test, (2) multi-exposure and genetic risk score (GRS) test, (3) multi-SNP and environmental risk score (ERS) test, and (4) GRS-ERS test. Our simulations explored both linear and logistic regression while considering three statistics: the Wald test, the Score test, and the likelihood ratio test (LRT). We further applied the approaches to three large sets of human cohort data (n = 37,664), focusing on type 2 diabetes (T2D), obesity, hypertension, and coronary heart disease with smoking, physical activity, diets, and total energy intake. Overall, GRS-based approaches were the most robust, and had the highest power, especially when the G-E interaction effects were correlated with the marginal genetic and environmental effects. We also observed severe miscalibration of joint statistics in logistic models when the number of events per variable was too low when using either the Wald test or LRT test. Finally, our real data application detected nominally significant interaction effects for three outcomes (T2D, obesity, and hypertension), mainly from the GRS-ERS approach. In conclusion, this study provides guidelines for testing multiple interaction parameters in modern human cohorts including extensive genetic and environmental data.
GENE and environment (G-E) interaction has been studied for a wide range of human traits using both genome-wide scale interaction screening (Hamza et al. 2011; Hancock et al. 2012; Wei et al. 2012; Wu et al. 2012; Siegert et al. 2013) and targeted analyses focusing on sets of genes or single nucleotide polymorphisms (SNPs) (Mahdi et al. 2009; Risch et al. 2009; Nickels et al. 2013; Dashti et al. 2015). In regards to the limited success, a number of statistical methods have been developed to improve the detection of G-E interaction effects (Thomas 2010a; Aschard et al. 2012; Gauderman et al. 2013). In particular, statistics based on aggregated genetic information have been shown to be a promising path forward (Manning et al. 2011; Hutter et al. 2012; Ma et al. 2013; Courtenay et al. 2014; Qi et al. 2014; Jiao et al. 2015; Aschard et al. 2017). In practice, the most common strategy consists in testing for genetic risk score (GRS)-by-exposure interaction using SNPs previously identified in marginal genetic effect screenings (Ripatti et al. 2010; Salvatore et al. 2014; Pisanu et al. 2017), although the approach is applicable to any sets of SNPs [e.g., gene-level sets, pathway- or network-level sets, or polygenic set (Thomas 2010b; Meyers et al. 2013)]. Basically, the GRS-based method aggregates genetic information by summing risk alleles (alleles associated with increased value of quantitative traits or greater risk of disease traits). Potential gain in power for such approaches comes from circumventing a penalty for multiple testing [a single 1 degree-of-freedom (df) test rather than one test per SNP]. However, the main limitation is that the power gain relies on an assumption that interaction effects, if present, are highly correlated with the marginal genetic effects (i.e., the risk alleles of SNPs in the GRS have G-E interaction effects in the same direction). Note that this is very similar to the burden test assumption for rare variants analysis (Lee et al. 2014). When this concordance assumption does not hold, the standard individual SNP-based interaction approach can outperform the GRS-based interaction approach.
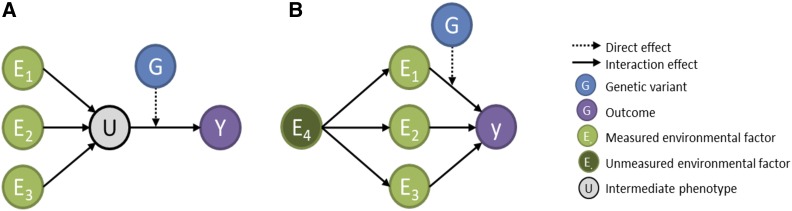
Given that a growing amount of extensive phenotypic and epidemiological data in human genetic cohorts becomes available, joint interaction tests involving multiple SNPs and multiple exposures have been seldom considered, although a linear mixed model approach using a random effect for multiple interactions has been recently described (Moore et al. 2019). There are several arguments in favor of applying joint interaction approach for multiple interactions. First, multiple environmental factors might influence a disease through the same intermediate mechanisms. For example, exposure to various carcinogens increases the risk of cancers by increasing the risk of deleterious genetic mutations (Kawaguchi et al. 2006; Ferreccio et al. 2013). Similarly, shared intermediate phenotypes (e.g., atherosclerosis) for heart attack and stroke are known to be associated with multiple lifestyle factors (e.g., smoking, diet, and alcohol consumption) (Massin et al. 2007; Rafieian-Kopaei et al. 2014). In such situations, one can hypothesize that nongenetic risk factors may also have shared interaction effects with genetic variants of the disease in question (Figure 1A). Second, because most exposures associated with human diseases display modest effect sizes, risk score approaches integrating all effects of risk factors (McClelland et al. 2015; Merchant 2017) can potentially lead to increased power as done for GRS. Moreover, some exposures have strong correlations with one another [e.g., cigarette smoking and alcohol consumption (Fisher and Gordon 1985), or diet and socio-economic status (Darmon and Drewnowski 2008)]. Correlations among exposures, if induced by an unmeasured variable, can be used to improve power to detect interactions though only a part of exposures interact with genetic variants (Aschard et al. 2014) (Figure 1B).
Figure 1.
Hypothetical causal model. In (A), multiple exposures (E1, E2, and E3) influence an intermediate phenotype (U), which effect on the outcome (Y) depends on a genetic variant (G). This scenario induces multiple interaction effects between G and the multiple exposures on the Y. In (B), multiple exposures (E1, E2, and E3) are influenced by another unmeasured variable, inducing a correlation between them. However, only one of these exposures interacts with G. In such case, the joint test of all interactions is more powerful than the test of E1xG only.
As environmental data are increasingly common in large-scale human genetic studies, interaction analyses including multiple SNPs and multiple exposures might be performed systematically on behalf of standard G-E interaction screenings. However, despite a few recent works published (Casale et al. 2017), our knowledge of the strengths and limitations of joint analysis approach for multiple G-E interactions is still limited. Here, we addressed a part of this question and explored the relative performance of four joint G-E interaction test approaches for both quantitative and binary trait models: (1) a joint test for multiple single SNP-by-single exposure interaction effects (omnibus test), (2) an interaction test between (weighted or unweighted) GRS and multiple exposures, (3) an interaction test for multiple SNPs and an environmental risk score (ERS), and (4) a GRS-by-ERS interaction test. Specifically, we assessed their robustness and relative power through simulations using three different statistical tests [Wald test, Score test and likelihood ratio test (LRT)] and varying a range of parameters including the total number of SNPs and exposures considered, the presence of correlation between exposures, dependence between SNPs and exposures, and the pattern of G-E interactions in regards of the marginal genetic and environmental effects. We further demonstrated the relevance of the proposed approaches in three large sets of population-based cohort data focusing on four common complex traits [coronary heart disease (CHD), type 2 diabetes (T2D), obesity, and hypertension] and four environmental risk factors (total energy intake, diet quality, physical activity, and smoking status).
Materials and Methods
Model overview
Consider the following generalized linear model, including main effects of genetic and exposure risk factors and G-E interaction effects:
| (1) |
where are SNPs, are exposures, and the link function is either the identity when the outcome is continuous or expit() when is a disease probability. In this model, is the main effect of (, where is the total number of SNPs), is the main effect of (, where is the total number of exposures), and is the interaction effect between and .
We aim at assessing the relative performances of joint interaction tests, where multiple interaction parameters are tested jointly in a fixed effect model, whether or not some of the predictors are aggregated into summary variables (see next section about GRS and ERS). For mathematical convenience, we present here three joint tests using the Wald, LRT, and Score statistics. The multivariate Wald statistics is defined as:
| (2) |
where is the vector of the estimated interaction effect parameters tested jointly and is the estimated variance-covariance matrix of these parameters, i.e.:
Under the null hypothesis of no interaction effect ( = … = = 0), follows a chi-squared distribution with df equal to , the total number of interaction terms tested jointly .
The LRT statistics is defined as:
| (3) |
where is the likelihood of the model when the estimated interaction coefficients using Maximum Likelihood Estimators (MLE) or Ordinary Least Square. The follows a chi-squared distribution with df under the null.
The Score test statistics is defined as:
| (4) |
where is the value of the derivative of the log-likelihood when , and is the Fisher Information. Under the null hypothesis, the follows a chi-square distribution with df. Note that an important difference between the Score test and the other tests is the total number of parameters estimated. Indeed, the Wald test requires the unrestricted estimates of the parameter (i.e., the model including interactions, so that the total number of parameters equals M + K + M×K), while the LRT requires quires both the restricted (without interaction) and unrestricted estimates of the parameter [i.e., (M+K)×2 + M×K], and the Score test requires restricted estimates of the parameter (i.e., M + K). More details on these test statistics are described in Supplemental Note (see Supplemental Material).
Interaction tests considered
In the standard omnibus test, all interaction effects, from Equation 1 are estimated and tested jointly, so the test statistic . For GRS-based interaction tests, we consider both weighted and unweighted forms, where a GRS is built as the (weighted or unweighted) sum of risk alleles of the SNPs. Explicitly, , and , where is commonly defined as marginal genetic risk estimates from . We first use GRS in a multi-exposure by GRS (multiE-GRS) model:
| (5) |
so that the corresponding combined test of the interaction terms is: . As for GRS, we also consider the use of ERS to capture a global effect of multiple exposures. The ERS is built similarly to the using weights from marginal environmental models (i.e., ). The multi-SNP by ERS (multiSNP-ERS) model is then defined as:
| (6) |
and the corresponding combined test of the interaction terms is defined as . Finally, we consider the GRS-by-ERS interaction (GRS-ERS) approach:
| (7) |
in which test statistics of can be defined as
Simulation study
Unless otherwise stated, we simulated series of 10,000 replicates each including N = 20,000 samples using Equation 1 with K = [2–10] correlated exposures and M = [10, 100, 300] independent SNPs, while varying the distribution of exposures data (normal/non-normal, correlated strongly/moderately), and allowing for dependence between SNPs and exposures. We also varied the parameters of the model , but always assumed nonzero main effects of the genetic variants and environmental factors that increased risk of diseases (i.e., and ). In each series, we explored the performance of six joint test approaches (i.e., omnibus, multiE-uGRS, multiE-wGRS, multiSNP-ERS, uGRS-ERS, and wGRS-ERS) in null models and alternative models for robustness and power, respectively.
For null simulation series, we calculated genomic inflation factor as the ratio of the median value of observed chi-square statistics over the median value of expected chi-square statistics. Power was estimated under the alternative models for two main scenarios, (1) different percentages of true G-E interaction (i.e., 20, 40, or 60%), and (2) in the presence or absence of correlation between marginal effects and interaction effects. When comparing power, we also performed a standard univariate model testing each G-E pairwise interaction independently (e.g., 100 univariate models for testing 10 SNP and 10 exposures). The significance threshold for the six interaction approaches was 0.05, while the significance threshold for the univariate model was adjusted for multiple testing by using Bonferroni correction (= 0.05/, where is the total number of interaction parameters tested). Binary traits were analyzed using logistic regression, while quantitative traits were analyzed using standard linear regression. For hypothesis testing, we considered three test statistics in the joint test: Wald test, Score test, and LRT.
Genetic variants were drawn independently of each other from a binomial distribution with n = 2 and using the coded allele as the risk allele. We considered two scenarios including only common variants [risk allele frequency (RAF) of 1–99%] or only rare variants (RAF of 0.1–1% or 99–99.9%). To mimic the genome-wide significant SNPs, the main effects of SNPs were drawn from a left truncated normal distribution, mean of 0, and variance equals to /, where , the trait heritability, equals 0.3 and , the number of causal SNPs, equals 10,000. Then, actual SNPs coefficients were derived by rescaling the main genetic effects based on the expected probability of allele frequencies , where is RAF. We generated exposure values from a multivariate normal distribution with mean 0 and a covariance matrix set for the presence of relatively strong (mean pairwise r-squared equals 0.10, Supplemental Material, Figure S6A) or moderate correlation (mean pairwise r-squared equals 0.02, Figure S6B). For non-normal exposures, we randomly selected 50% of the exposures and squared all values, resulting in a chi-squared distribution of the exposure. Exposure effects on the outcome were drawn from absolute values of a normal distribution and assuming the total outcome variance explained by all exposures ranged between 0.02 and 0.05.
When assuming G-E correlations, we randomly selected 50% of the exposures and a random set of associated SNPs and added genetic effects drawn from a normal distribution with mean 0 and variance 0.005 (so that each associated SNP explained on average 0.5% percent of the variance of the exposure) to the selected exposures. For power, we generated G×E interactions using standardized SNPs in order to ensure that the interaction terms do not modify the marginal effects of SNPs (i.e., adding interactions of unstandardized SNPs could change the direction of SNP marginal effects in the model) (Aschard 2016). For linear outcome, G-E interaction effects were generated from uniform distribution in the range [0.001, 0.002] when assuming correlation between interaction effects and marginal effects, and in the range [−0.007, 0.007] when assuming no correlation between those. For binary outcome, were also drawn from uniform using the ranges [0.002, 0.002] and [−0.01, 0.01] in the presence or absence of the correlation, respectively. Finally, for logistic models, we defined the intercept so that the expected baseline prevalence equaled to 30%.
Real data application
We applied our interaction test approaches to three large sets of human cohort data, the Nurses’ Health Study (NHS I), NHS II, and Health Professional Follow-up Study (HPFS). The total sample size available with genetic data in the three cohorts was 37,664. Though all analyses were conducted in the combined cohort data, each analysis included different numbers of cohort participants depending upon the availability of disease and exposure data considered. All disease and exposure data were drawn from self-reported biannual questionnaires of each cohort.
We focused on four binary traits, T2D, CHD, obesity, and hypertension. For each trait, we excluded all individuals who had the disease prior to each cohort inception (baseline). Then, we defined cases as individuals who had reported to have the disease since the baseline and controls were defined as individuals who had never reported it between the baseline and their last time of follow-up. Exposure variables we considered were established risk factors of the four diseases, such as smoking status (ever smoker vs. never smoker), physical activity [measured as Metabolic Equivalent of Task (MET) hours per week], diet quality [Healthy Eating Index (HEI) (range: 1–100) that indicates healthier dietary intake with higher score)], and total energy intake (kcal/week). To avoid potential reverse causation, we used exposure data measured at the earliest time point in the follow-up (i.e., baseline of each cohort). Since all the traits were binary, and based on our simulation results, we used a logistic regression model with Score test statistic.
To build G-E interactions, we included sets of SNPs previously identified to be associated with the four traits from large-scale GWAS (International Consortium for Blood Pressure Genome-Wide Association Studies et al. 2011; Morris et al. 2012; Locke et al. 2015; Nikpay et al. 2015). We also included the following covariates: age (when exposures were measured), study (NHS, NHSII, and HPFS), genotyping platforms (Affymetrix, IIllumina, Omniexpress, Oncoarray, and Humancore exomchip) (Lindström et al. 2017), and principal components computed from the full sets of genotypes (top three principal components of each platform).
Data availability
The authors affirm that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6849047.
Results
Validity of the statistical approaches for multiple interactions
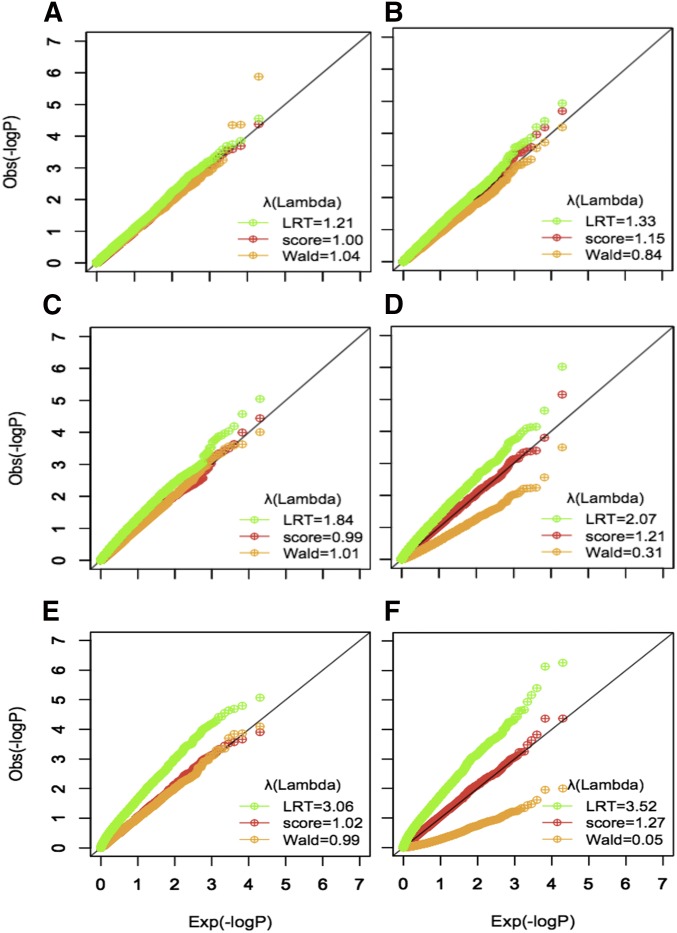
We first compared the robustness of three test statistics (Wald test, Score test, and LRT) when testing multiple parameters jointly in linear and logistic regression models (Figure 2). Using null models of no interaction effect (but assuming the presence of marginal genetic and environmental effects), we simulated series of 20,000 individuals with = 100 independent common SNPs and = [2, 6, 10] normally distributed and correlated exposures. For binary outcomes, we considered a prevalence of 30%. The Wald statistics showed strong robustness in the linear regression but severe deflation in logistic regression as the number of interactions tested jointly increases. The standard LRT statistics, derived from (MLE), showed strong inflation with increasing number of interactions in both linear and logistic regression models. Note that inflation for the linear model could be easily fixed by substituting MLE with ordinary least squares estimates (Supplemental Note and Figure S1), but such a fix is not possible for logistic model. The Score statistics showed the highest robustness in the both linear and logistic regression, although we noted some non-negligible inflation for logistic regression as the number of interactions increased. As discussed in the Supplemental Note, the better calibration of the Score test is likely explained by the smaller number of parameters that have to be estimated (i.e., conversely, the LRT and Wald test face instability because of the many interaction parameters that the tests have to estimate).
Figure 2.
Q-Q plots of Wald test, Score test, and LRT. To evaluate robustness of the three test statistics, we generated 10,000 series of 20,000 samples with 100 common SNPs and up to 10 correlated and normally distributed exposures under the null hypothesis of no interaction effect but in the presence of main genetic and environmental effects. For logistic model, we considered a disease prevalence of 30%. To compare the robustness, we simulated the data with different number of exposures 2, 6, and 10 corresponding to the number of event per variable (EVP) of 30, 10, and 6. (A) Q-Q plot of linear regression with EPV of 30. (B) Q-Q plot of logistic regression with EPV of 30. (C) Q-Q plot of linear regression with EPV of 10. (D) Q-Q plot of logistic regression with EPV of 10. (E) Q-Q plot of linear regression with EPV of 6. (F) Q-Q plot of logistic regression with EPV of 6.
Because the bias was more obvious in logistic regression, we further examined whether the bias was influenced by the modeling of G-E interaction effects or simply due to the number of outcome events per predictor variables (EPV) (Peduzzi et al. 1996) (Figure S2). We compared chi-squared statistic distributions of the three test statistics of omnibus test under a complete null model (i.e., no interaction and no main effects) for a fixed EPV (e.g., EPV = 5), while assessing marginal genetic effects only, or interaction effects only ,with the same number of parameters in testing jointly. More precisely, we compared two scenarios: (1) draw independent SNPs and tested SNPs, only a subset of parameters jointly (e.g., M = 120, L = 100); or (2) draw independent SNPs and correlated exposures, and tested interaction parameters jointly (e.g., M = 10, K = 10, L = 100). As shown in Figure S2 for = [100, 400], we observed trends similar to those from Figure 1 in marginal effect models (inflation for LRT and deflation for Wald test). Although we noticed the bias might be slightly larger for G-E interaction models, we did not observe any major qualitative difference between interaction models and SNP only models, suggesting the bias is driven mostly by the small number of EPV (e.g., EPV < 10).
Exploring further the impact of different numbers of EPV on multivariable interaction tests, we found that joint analysis of multiple parameters (in our case multiple interaction terms) tended to be dramatically more sensitive to EPV than standard univariate test (Figure S3). As expected based on existing literature, univariate tests were robust across different test statistics once the test achieved the rule of thumb of EPV = 10 (Vittinghoff and McCulloch 2007), regardless of the total sample size. Conversely, in omnibus tests for a fixed EPV, Wald test and LRT statistics showed increasing deflation and inflation, respectively, as the sample size increased. For example, whereas an EPV of 10 might be sufficient to have a calibrated LRT for a sample size of 1000, increasing the sample size to 5000 required to reach an EPV of 50 to have a valid test. Again, only the Score test showed good calibration across the different numbers of EPV and sample size, highlighting this should be the preferred statistics for testing multiple interactions jointly in modern genetic datasets including hundreds of thousands of individuals.
Robustness comparison in joint analysis approaches
Based on the results above, we examined the six G-E interaction test strategies (i.e., omnibus, multiE-uGRS, multiE-wGRS, multiSNP-ERS, uGRS-ERS, and wGRS-ERS) under the null using Score test for linear and logistic regression. Table 1 shows type I error rates calculated as a genomic inflation factor for normally distributed and highly correlated exposures while varying three parameters: the RAF, the number of SNP analyzed jointly, and the presence or absence of G-E dependence. In linear models, all six approaches showed consistent and strong robustness regardless of RAF, the number of SNPs tested, and dependence between SNPs and exposures. Similarly, logistic models for GRS-related approaches (multiE-uGRS, multiE-wGRS, uGRS-ERS, and wGRS-ERS) showed consistent robustness. Conversely, multi-SNP approaches (omnibus and multi SNP-ERS) showed moderate to strong inflated statistics with increasing number of variants and decreasing RAF (Figure S4 and Table 1), highlighting the limitation of the Score test in logistic regression when the number of parameters becomes too large in the baseline model (i.e., the model without interaction). Such inflation was also found in type I error rates for omnibus tests with 300 common SNPs (Table S1). When performing the same simulations but using moderately correlated (Table S2) or non-normally distributed exposures with moderate correlation (Table S3), we observed similar findings of consistent robustness in linear models and GRS-based approaches of logistic models, and inflation in multi-SNP approaches of logistic models.
Table 1. Genomic inflation factors under the null hypothesis of no interaction for six interaction tests with normally distributed and highly correlated exposures.
| Independent G-E | Dependent G-E | ||||||
|---|---|---|---|---|---|---|---|
| SNP (N) | SNP (N) | ||||||
| 10 | 100 | 300 | 10 | 100 | 300 | ||
| Linear regression | |||||||
| Rare SNPa | Omnibus | 1.01 | 1.02 | 0.92 | 0.99 | 0.97 | 1.03 |
| MultiE-uGRS | 0.99 | 0.96 | 0.98 | 1.05 | 1.02 | 1.01 | |
| MultiE-wGRS | 0.96 | 0.96 | 0.96 | 0.93 | 0.94 | 0.95 | |
| MultiSNP-ERS | 1.00 | 1.00 | 1.01 | 1.03 | 1.00 | 1.02 | |
| uGRS-ERS | 1.01 | 1.01 | 1.02 | 1.01 | 0.99 | 1.02 | |
| wGRS-ERS | 0.96 | 0.98 | 0.99 | 1.00 | 0.98 | 1.04 | |
| Common SNPb | Omnibus | 0.99 | 0.99 | 0.99 | 1.03 | 1.02 | 1.02 |
| MultiE-uGRS | 0.99 | 0.98 | 1.05 | 1.01 | 0.99 | 1.05 | |
| MultiE-wGRS | 1.01 | 0.98 | 1.02 | 0.98 | 1.00 | 1.01 | |
| MultiSNP-ERS | 1.01 | 1.05 | 1.00 | 0.99 | 1.00 | 0.99 | |
| uGRS-ERS | 0.97 | 1.01 | 1.00 | 1.01 | 1.01 | 1.01 | |
| wGRS-ERS | 0.99 | 1.01 | 0.99 | 1.02 | 0.98 | 0.98 | |
| Logistic regression | |||||||
| Rare SNPa | Omnibus | 1.09 | 1.60 | 3.49c | 1.13 | 1.65 | 3.63c |
| MultiE-uGRS | 1.02 | 1.01 | 1.04 | 1.00 | 1.02 | 1.05 | |
| MultiE-wGRS | 1.02 | 1.00 | 0.98 | 0.94 | 0.93 | 0.98 | |
| MultiSNP-ERS | 1.05 | 1.20 | 1.58 | 1.05 | 1.17 | 1.57 | |
| uGRS-ERS | 0.99 | 1.00 | 0.98 | 1.01 | 0.98 | 1.01 | |
| wGRS-ERS | 1.03 | 0.98 | 0.99 | 1.02 | 0.96 | 1.01 | |
| Common SNPb | Omnibus | 1.03 | 1.32 | 2.74c | 1.02 | 1.31 | 2.81c |
| MultiE-uGRS | 0.98 | 1.01 | 0.98 | 0.98 | 1.05 | 0.96 | |
| MultiE-wGRS | 0.99 | 0.99 | 0.99 | 0.96 | 1.00 | 1.02 | |
| MultiSNP-ERS | 0.86 | 1.04 | 1.40 | 0.98 | 1.07 | 1.41 | |
| uGRS-ERS | 0.98 | 1.04 | 0.99 | 1.01 | 0.97 | 0.98 | |
| wGRS-ERS | 0.99 | 1.01. | 1.01 | 0.96 | 0.98 | 1.06 | |
Rare SNP: RAF = [0.1–1% or 99–99.9%].
Common SNP: RAF = [1–99%].
5000 replicates.
Power comparison for joint analysis approaches
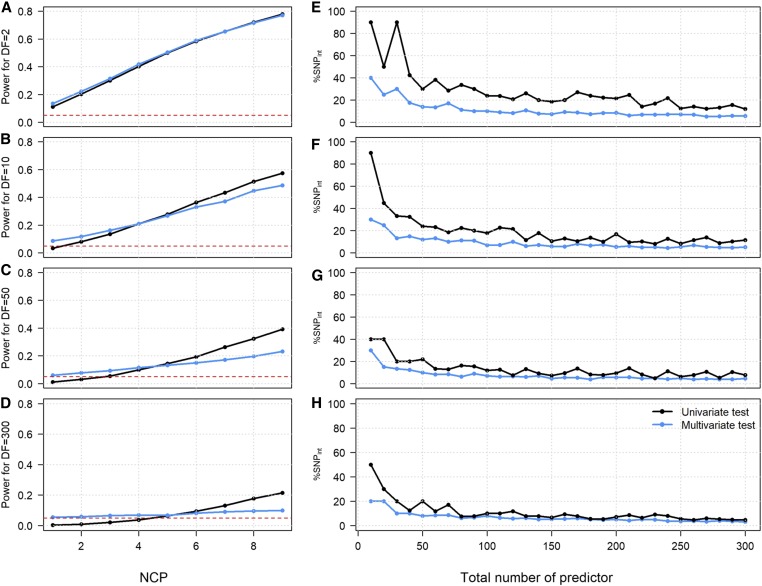
We aimed first at understanding the potential benefit of testing jointly multiple interaction parameters, as opposed to testing them separately and correcting for multiple testing using a Bonferroni adjustment. To address this question, we considered a set of predictor (here interaction effects) and derived the theoretical power of the two aforementioned approaches, i.e., the joint test of all predictors vs. the test of each single predictor followed by correction of the P-values for the tests performed, while assuming a subset of the predictor are associated with the outcome. Unsurprisingly, as showed in Figure 3, A–D we first found that, when , there are at best small gains from using multiple interaction tests. The single predictor approach tends to have higher power as the effect of the variants increased and K increased (e.g., noncentrality parameter (ncp) = 9, , Figure 3C), while the multivariate approach performs slightly better for small effect. Conversely when there are multiple associated predictors (i.e., as increases relative to ), the multivariate approach performs in general better than the univariate approach (Figure 3, E–H). For example, to achieve 80% power at a nominal level of 5% while analyzing =100 predictors, and assuming very small effect (ncp = 1), the univariate test requires up to 20% of the predictors to be associated with the outcome, while the multivariate test would achieve the same power if 10% of the predictors are associated (Figure 3E).
Figure 3.
Degrees of freedom vs. correction for multiple testing. In (A–D) we considered a single associated predictor with noncentrality parameter (ncp) in [1–9] and compared the cost of adding K equals 1 (A), 9 (B), 49 (C), 299 (D) null statistics, to form a K+1 degree of freedom test vs. correcting the univariate test of that predictors for a total of K+1 tests. We plotted the power of the resulting univariate test (black line) and multivariate test (blue line) as a function of the ncp. The red dashed line represents the 5% threshold. In (E–H), we have drawn series of K in [10–300] chi-squared statistics with 1 d f, representing single predictor (e.g., SNPs) tests, while varying the proportion of chi-squared under the null and chi-squared under the alternative. Under the alternative we chi-squared were drawn from a noncentral chi-squared distribution with ncp equals to 1 (E), 2 (F), 3 (G), and 4 (H), while under the null, chi-squared were drawn from a central chi-squared. For each series, we derived the minimum proportion of associated predictors (%SNPint) required to achieve 80% power at a significance threshold of 0.05 with a multivariate test of all K terms (blue lines), and with a univariate test (black lines). For the latter, we considered the null hypothesis that none of the predictor tested reaches the significance threshold after correcting for the K tests performed.
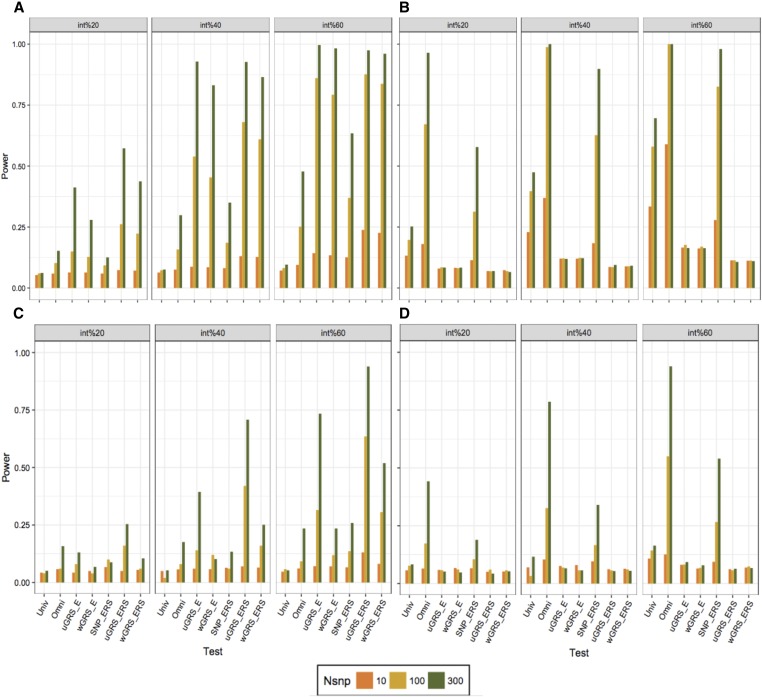
To compare the relative power of the six G-E interaction strategies, we next performed a series of simulations using linear and logistic regression models (Figure 4). We used, as in Table 1, normally distributed and highly correlated exposures. As expected, the power of all the approaches increased with increasing number SNPs tested and increasing proportion of true effects among the interactions. However, we found that the relative gain in power relied on the correlation between interactions and marginal effects and the correlation across exposures. In linear models, score-based approaches (i.e., using GRS and ERS) were the most powerful when simulating G-E interaction effects correlated with marginal genetic and environmental effects. Conversely, when simulating G-E interaction effects uncorrelated to marginal effects, the omnibus test was the most powerful approach. Logistic models showed qualitatively similar power results. Using normally distributed exposures with moderate correlation appeared to have limited impact on the power results (Figures S5 and S6). However, we noticed that power advantages of the GRS-based approaches over multi-SNP approaches in the presence of correlation between interactions and marginal effects tended to decrease with increasing correlation between exposures (i.e., coefficients: 0.02–0.20) (Figure S7). When there was no correlation between interactions and marginal effects, we observed that the power of multi-SNP approaches tended to increase with increasing exposure correlation, particularly univariate test and multiSNP-ERS test in linear models.
Figure 4.
Power comparison of G-E interaction approaches with normal distributed and highly correlated exposures. We derived series of 10,000 simulated replicates (except for univariate and omnibus tests in logistic models using 1000 replicates) and each included 20,000 samples, 10 exposures, and a varying number of SNP (n = 10, 100, and 300). (A) presents results for linear models assuming all G-E interactions effects correlated with marginal effects, (B) linear models assuming no correlations between G-E interaction effects and marginal effects, (C) logistic models assuming correlations between G-E interaction effects and marginal effects, and (D) logistic models assuming no correlations between G-E interaction effects and marginal effects.
Application to data from large population-based cohorts
Using the Score test statistic in logistic regression, we conducted six interaction analyses per each trait, testing jointly for multiple interactions between SNPs known to be associated with four traits (number of SNPs included = 65, 76, 27, and 48 for T2D, obesity, hypertension, and CHD, respectively) and four exposures (HEI as a measure of diet quality, total calorie intake, MET-hour per week as a measure of physical activity, and ever smoking) (Table S4). Details of the SNPs are described in Tables S7–S10. Prior to the interaction tests, we assessed marginal effects of GRS and exposures with the traits. T2D and obesity showed associations with all the four exposures, whereas hypertension was associated with HEI, MET-hour, and smoking ever, and CHD was associated with HEI and ever smoking (Table S5). For each trait, we constructed ERS including only exposures that were marginally associated with the trait. We observed only limited correlation between the four exposures in our data, such as the maximum correlation coefficient was 0.17 between HEI and MET-hour per week (Table S6). Results of joint interaction tests are presented in Table 2. We found that 7 (29%) out of the 24 interaction tests performed showed nominal significance. The most significant interaction was found in omnibus test for obesity with all four exposures (P = 0.003). Their interactions were also detected with GRS-ERS approaches (P = 0.037 and 0.028 for uGRS-ERS and wGRS-ERS, respectively). Nominally significant interactions were also observed between hypertension and three exposures (HEI, MET-hour, and smoking) through multiSNP-ERS (P = 0.006), uGRS-ERS (P = 0.046), and wGRS-ERS (P = 0.021). Lastly, omnibus test detected a nominally significant interaction effect of T2D and the four exposures (P = 0.015). Note that we kept the omnibus in this analysis despite type I error rate is not fully controlled. However, based on simulations presented in Table S1, we expect any bias to be minimal (e.g., type I error rate at 5% equals 0.059 for the same number predictors in simulated data).
Table 2. Joint analysis approaches for multiple G-E interactions in NHS I, NHS II, and HPFS cohorts.
| T2D | Obesity | Hypertension | CHD | |
|---|---|---|---|---|
| (P-value) | (P-value) | (P-value) | (P-value) | |
| Univariatea | 0.433 | 0.686 | 0.096 | 1.000 |
| Omnibus | 0.015b | 0.003b | 0.355 | 0.501 |
| MultiE-uGRS | 0.244 | 0.511 | 0.101 | 0.391 |
| MultiE-wGRS | 0.544 | 0.563 | 0.117 | 0.540 |
| MultiSNP-ERS | 0.051 | 0.208 | 0.006b | 0.567 |
| uGRS-ERS | 0.473 | 0.037b | 0.046b | 0.576 |
| wGRS-ERS | 0.680 | 0.028b | 0.021b | 0.698 |
T2D, type 2 diabetes; CHD, coronary heart disease; uGRS, unweighted genetic risk score; wGRS, weighted genetic risk score; ERS, environmental risk score.
Reported P-values were corrected for multiple testing by multiplying the total number of G-E interactions using the minimum P-value among the interactions: 0.002 (T2D), 0.002 (obesity), 0.0009 (hypertension), and 0.018 (CHD).
Nominally significant interactions.
Discussion
Recent advances in big data era enable us to jointly assess multiple G-E interaction effects; however, the statistical challenges and benefits of such approaches remain unknown. To address this gap, we defined a set of parsimonious approaches for multiple interactions, and explored their statistical properties in simulations and real data application. Our results indicate that joint test approaches, in particular aggregating multiple marginal effects such as GRS or ERS, offer both robustness and power gain, potentially allowing for the identification of G-E interactions missed by a standard univariate test. However, we also found several important issues that should be considered for the joint analysis approaches.
First, contrary to univariate test, the choice of the statistics was crucial in the joint test for multiple interactions (i.e., omnibus) especially for binary traits. In logistic models, Wald test and LRT statistics were deflated and inflated, respectively, with increasing number of interaction parameters, whereas Score statistic was consistently more robust in the same scenarios. Consistent with findings from previous studies, the type I error rate was larger as the EPV decreases (Peduzzi et al. 1996; Vittinghoff and McCulloch 2007). Furthermore, the two previous studies using Wald test reported that, for the low EPV (e.g., EPV < 10), sample variance estimates were not robust (Peduzzi et al. 1996) and bias of regression coefficients increased in both positive and negative directions (Vittinghoff and McCulloch 2007). Because the biased coefficients often lead to extreme values of MLE, we observed highly inflated LRT statistics when EPV is < 10 (Supplemental Note). Although the previous studies did not evaluate Score test, we found that the Score test statistics were substantially robust even with EPV <10, suggesting it should be preferred when testing multiple parameters jointly in logistic regression. Permutations might be an alternative to control type I error rates of the three test statistics. Permutation would be computationally demanding and might break down part of the structure in the data, making interpretation more difficult. However, it might be applicable in some case, and preliminary analyses we conducted showed encouraging results (Tables S11 and S12).
In contrast of the logistic regression, all approaches showed strong robustness in linear models. In general, linear models do not face issues similar to small EPV. For example, prior studies assessing the number of subjects per variable (SPV, similar in principle to the EPV, but applied to linear regression) showed that two SPV were enough to have adequate estimates on regression coefficients, SE, and confidence intervals in linear regression (Austin and Steyerberg 2015). In our simulations, this was true for Wald and Score test statistics with the lowest SPV of 6.7 (= 20,000/3000) but not for LRT. However, slight inflation in LRT statistics can be easily corrected by using estimates from ordinary least squares instead of using maximum likelihood estimates (Supplemental Note).
Another important limitation of multi-SNP approaches was minor/risk allele frequency of SNPs when testing for binary traits. Our simulations showed severe type I error rates increase with increasing the number of variants in logistic models and the bias was worse with rare variants (RAF < 1%) included in the analysis. Although we did not assess type I error rates with non-normally distributed continuous outcomes, previous works highlighted that special caution is required for rare variants analysis, especially when analyzing non-normally distributed traits (e.g., traits from gamma or log normal distributions) (Schwantes-An et al. 2016). We only considered independent variants in all our analyses. Using correlated variants for GRS-based approaches would require estimating weights from e.g., multiple regressions or penalized models to avoid redundancy of genetic information. For SNP-based approaches, correlation between SNPs might increase instability of all approaches and is therefore not recommended (Table S13). While the Score test performed again better, analyzing large number of correlated variants might induce substantial inflation of the type I error rate.
Lastly, the presence or absence of correlation between interaction and marginal effects played a substantial role to gain power in jointly testing multiple interactions. As seen in Figure 4, the relative power gain of the six approaches was highly sensitive to the correlation. When the interaction effects are correlated to marginal effects, GRS-based approaches outperformed the others in most of scenarios. However, when the correlation is absent, SNP-based approaches (i.e., omnibus or multiSNP-ERS) had better performance than the GRS-based approaches. Similar trends of power gain have been discussed in previous studies on rare-variant association tests. For example, burden tests, which are very similar to our aggregating methods (e.g., GRS or ERS), examine genetic associations by aggregating effects of a set of rare variants into a genetic score (Wu et al. 2011). Because the test requires a strong assumption of the same direction and magnitude of effects, the test is obviously less powerful when the assumption is violated (Lee et al. 2014). As an alternative, researchers have proposed variance-component tests that are powerful with different directions of marginal effects. Such an approach has been recently proposed for GxE (Moore et al. 2019) and might be compared against our fixed effect approach in the future.
We found that joint analysis approaches, especially joint tests with aggregated effects (e.g., GRS or ERS), could detect more multiple G-E interaction effects with strong robustness, and, further, much power with correlation between marginal effects and G-E interaction effects. Because of computational convenience, these score-based approaches would be easily applicable to other interaction tests, such as gene-by-gene (G-G) interactions or multivariate interaction tests for multiple traits in future research. Although our simulations used aggregating approaches for both marginal effects and interaction effects as Equation 7, the benefits of the score-based approaches might be achieved even if one tests aggregating effects of interactions (e.g., GRS-ERS) in models with multiple marginal effects (e.g., , ). Because our joint approaches test the interaction effect only (e.g., ), increase in the number of total parameters would not have influence on detecting interaction effects as long as using the Score test statistic.
Our joint analysis approaches also have some limitations. First, because joint test approaches examine whether any of multiple interactions have a signal or not, it does not provide evidence for any specific interaction effects of SNPs and the exposures that are the main drivers of the interaction signals. Instead, joint test approaches offer an opportunity to gain insights into global interaction patterns. For example, significant GRS-based interaction would indicates an overall decrease or increase of the genetic effect with exposure, while the Omnibus and SNP-ERS tests would indicate more diffuse G × E interactions with limited structure. Second, in practice, it might not be easy to generate ERS where environmental factors are coded as being categorical or have different units of measurements. Also, it is still an open question what, and how many, environmental factors should be tested jointly for G-E interactions. Our recommendation is to include risk factors that have strong biological evidence on shared mechanisms, because, otherwise, interpretation can be challenging. Third, when applying the standard univariate approach, we used a nominal significance threshold of 5% after correction for multiple testing without accounting for correlations between SNPs and exposures or between exposures. This is the most stringent approach, and more advanced strategies might be considered in future (Sun and Lin 2017).
In summary, our study shows that approaches allowing for the joint analysis of multiple G-E interaction effects outperform standard pairwise interaction test in many scenarios. Particularly, GRS-based approaches in conjunction with a Score test showed both strong robustness and some of the largest gain in power, although alternatives approaches might be considered depending on the investigator hypotheses about correlation between GxE effects and marginal effects. Overall, this study provides the community guidelines for testing multiple interaction parameters in modern human cohorts including extensive genetic and environmental data.
Acknowledgments
This work was supported by National Institutes of Health National Human Genome Research Institute grant R21HG007687 to H.A. and National Institutes of Health grants P30 DK46200 and DK112940.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6849047.
Communicating editor: E. Eskin
Literature Cited
- Aschard H., 2016. A perspective on interaction effects in genetic association studies. Genet. Epidemiol. 40: 678–688. 10.1002/gepi.21989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschard H., Lutz S., Maus B., Duell E. J., Fingerlin T. E., et al. , 2012. Challenges and opportunities in genome-wide environmental interaction (GWEI) studies. Hum. Genet. 131: 1591–1613. 10.1007/s00439-012-1192-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschard H., Vilhjalmsson B. J., Greliche N., Morange P. E., Tregouet D. A., et al. , 2014. Maximizing the power of principal-component analysis of correlated phenotypes in genome-wide association studies. Am. J. Hum. Genet. 94: 662–676. 10.1016/j.ajhg.2014.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschard H., Tobin M. D., Hancock D. B., Skurnik D., Sood A., et al. , 2017. Evidence for large-scale gene-by-smoking interaction effects on pulmonary function. Int. J. Epidemiol. 46: 894–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin P. C., Steyerberg E. W., 2015. The number of subjects per variable required in linear regression analyses. J. Clin. Epidemiol. 68: 627–636. 10.1016/j.jclinepi.2014.12.014 [DOI] [PubMed] [Google Scholar]
- Casale F. P., Horta D., Rakitsch B., Stegle O., 2017. Joint genetic analysis using variant sets reveals polygenic gene-context interactions. PLoS Genet. 13: e1006693 10.1371/journal.pgen.1006693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtenay M. D., Cade W., Schwartz S. G., Kovach J. L., Agarwal A., et al. , 2014. Set-based joint test of interaction between SNPs in the VEGF pathway and exogenous estrogen finds association with age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 55: 4873–4879. 10.1167/iovs.14-14494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmon N., Drewnowski A., 2008. Does social class predict diet quality? Am. J. Clin. Nutr. 87: 1107–1117. 10.1093/ajcn/87.5.1107 [DOI] [PubMed] [Google Scholar]
- Dashti H. S., Follis J. L., Smith C. E., Tanaka T., Garaulet M., et al. , 2015. Gene-environment interactions of Circadian-related genes for cardiometabolic traits. Diabetes Care 38: 1456–1466 [corrigenda: Diabetes Care 40: 1420 (2017)]. 10.2337/dc14-2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreccio C., Yuan Y., Calle J., Benitez H., Parra R. L., et al. , 2013. Arsenic, tobacco smoke, and occupation: associations of multiple agents with lung and bladder cancer. Epidemiology 24: 898–905. 10.1097/EDE.0b013e31829e3e03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher M., Gordon T., 1985. The relation of drinking and smoking habits to diet: the lipid research clinics prevalence study. Am. J. Clin. Nutr. 41: 623–630. 10.1093/ajcn/41.3.623 [DOI] [PubMed] [Google Scholar]
- Gauderman W. J., Zhang P., Morrison J. L., Lewinger J. P., 2013. Finding novel genes by testing G × E interactions in a genome-wide association study. Genet. Epidemiol. 37: 603–613. 10.1002/gepi.21748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamza T. H., Chen H., Hill-Burns E. M., Rhodes S. L., Montimurro J., et al. , 2011. Genome-wide gene-environment study identifies glutamate receptor gene GRIN2A as a Parkinson’s disease modifier gene via interaction with coffee. PLoS Genet. 7: e1002237 10.1371/journal.pgen.1002237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock D. B., Soler Artigas M., Gharib S. A., Henry A., Manichaikul A., et al. , 2012. Genome-wide joint meta-analysis of SNP and SNP-by-smoking interaction identifies novel loci for pulmonary function. PLoS Genet. 8: e1003098 10.1371/journal.pgen.1003098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter C. M., Chang-Claude J., Slattery M. L., Pflugeisen B. M., Lin Y., et al. , 2012. Characterization of gene-environment interactions for colorectal cancer susceptibility loci. Cancer Res. 72: 2036–2044. 10.1158/0008-5472.CAN-11-4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Consortium for Blood Pressure Genome-Wide Association Studies. Ehret G. B., Munroe P. B., Rice K. M., Bochud M., et al. , 2011. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 478: 103–109. 10.1038/nature10405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao S., Peters U., Berndt S., Bezieau S., Brenner H., et al. , 2015. Powerful set-based gene-environment interaction testing framework for complex diseases. Genet. Epidemiol. 39: 609–618. 10.1002/gepi.21908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi I., Doi M., Kakinuma S., Shimada Y., 2006. Combined effect of multiple carcinogens and synergy index. J. Theor. Biol. 243: 143–151. 10.1016/j.jtbi.2006.05.027 [DOI] [PubMed] [Google Scholar]
- Lee S., Abecasis G. R., Boehnke M., Lin X., 2014. Rare-variant association analysis: study designs and statistical tests. Am. J. Hum. Genet. 95: 5–23. 10.1016/j.ajhg.2014.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindström S., Loomis S., Turman C., Huang H., Huang J., et al. , 2017. A comprehensive survey of genetic variation in 20,691 subjects from four large cohorts. PLoS One 12: e0173997 10.1371/journal.pone.0173997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke A. E., Kahali B., Berndt S. I., Justice A. E., Pers T. H., et al. , 2015. Genetic studies of body mass index yield new insights for obesity biology. Nature 518: 197–206. 10.1038/nature14177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Clark A. G., Keinan A., 2013. Gene-based testing of interactions in association studies of quantitative traits. PLoS Genet. 9: e1003321 10.1371/journal.pgen.1003321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdi H., Fisher B. A., Kallberg H., Plant D., Malmstrom V., et al. , 2009. Specific interaction between genotype, smoking and autoimmunity to citrullinated alpha-enolase in the etiology of rheumatoid arthritis. Nat. Genet. 41: 1319–1324. 10.1038/ng.480 [DOI] [PubMed] [Google Scholar]
- Manning A. K., LaValley M., Liu C. T., Rice K., An P., et al. , 2011. Meta-analysis of gene-environment interaction: joint estimation of SNP and SNP × environment regression coefficients. Genet. Epidemiol. 35: 11–18. 10.1002/gepi.20546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massin M. M., Hovels-Gurich H., Seghaye M. C., 2007. Atherosclerosis lifestyle risk factors in children with congenital heart disease. Eur. J. Cardiovasc. Prev. Rehabil. 14: 349–351. 10.1097/01.hjr.0000224483.72726.1a [DOI] [PubMed] [Google Scholar]
- McClelland R. L., Jorgensen N. W., Budoff M., Blaha M. J., Post W. S., et al. , 2015. 10-year coronary heart disease risk prediction using coronary artery calcium and traditional risk factors: derivation in the MESA (multi-ethnic study of atherosclerosis) with validation in the HNR (Heinz Nixdorf Recall) study and the DHS (Dallas heart study). J. Am. Coll. Cardiol. 66: 1643–1653. 10.1016/j.jacc.2015.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant A. T., 2017. The INTERSTROKE study on risk factors for stroke. Lancet 389: 35–36. 10.1016/S0140-6736(16)32613-7 [DOI] [PubMed] [Google Scholar]
- Meyers J. L., Cerda M., Galea S., Keyes K. M., Aiello A. E., et al. , 2013. Interaction between polygenic risk for cigarette use and environmental exposures in the Detroit neighborhood health study. Transl. Psychiatry 3: e290 10.1038/tp.2013.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R., Casale F. P., Bonder M. J., Horta D., BIOS consortium et al. , 2019. A linear mixed model approach to study multivariate gene-environment interactions. Nat. Genet. 51: 180–186. 10.1038/s41588-018-0271-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris A. P., Voight B. F., Teslovich T. M., Ferreira T., Segrè A. V., et al. , 2012. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 44: 981–990. 10.1038/ng.2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickels S., Truong T., Hein R., Stevens K., Buck K., et al. , 2013. Evidence of gene-environment interactions between common breast cancer susceptibility loci and established environmental risk factors. PLoS Genet. 9: e1003284 10.1371/journal.pgen.1003284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikpay M., Goel A., Won H. H., Hall L. M., Willenborg C., et al. , 2015. A comprehensive 1,000 genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 47: 1121–1130. 10.1038/ng.3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peduzzi P., Concato J., Kemper E., Holford T. R., Feinstein A. R., 1996. A simulation study of the number of events per variable in logistic regression analysis. J. Clin. Epidemiol. 49: 1373–1379. 10.1016/S0895-4356(96)00236-3 [DOI] [PubMed] [Google Scholar]
- Pisanu C., Preisig M., Castelao E., Glaus J., Pistis G., et al. , 2017. A genetic risk score is differentially associated with migraine with and without aura. Hum. Genet. 136: 999–1008. 10.1007/s00439-017-1816-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Q., Kilpelainen T. O., Downer M. K., Tanaka T., Smith C. E., et al. , 2014. FTO genetic variants, dietary intake and body mass index: insights from 177,330 individuals. Hum. Mol. Genet. 23: 6961–6972. 10.1093/hmg/ddu411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafieian-Kopaei M., Setorki M., Doudi M., Baradaran A., Nasri H., 2014. Atherosclerosis: process, indicators, risk factors and new hopes. Int. J. Prev. Med. 5: 927–946. [PMC free article] [PubMed] [Google Scholar]
- Ripatti S., Tikkanen E., Orho-Melander M., Havulinna A. S., Silander K., et al. , 2010. A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet 376: 1393–1400. 10.1016/S0140-6736(10)61267-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N., Herrell R., Lehner T., Liang K. Y., Eaves L., et al. , 2009. Interaction between the serotonin transporter gene (5-HTTLPR), stressful life events, and risk of depression: a meta-analysis. JAMA 301: 2462–2471. 10.1001/jama.2009.878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvatore J. E., Aliev F., Edwards A. C., Evans D. M., Macleod J., et al. , 2014. Polygenic scores predict alcohol problems in an independent sample and show moderation by the environment. Genes (Basel) 5: 330–346. 10.3390/genes5020330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwantes-An T. H., Sung H., Sabourin J. A., Justice C. M., Sorant A. J. M., et al. , 2016. Type I error rates of rare single nucleotide variants are inflated in tests of association with non-normally distributed traits using simple linear regression methods. BMC Proc. 10: 385–388. 10.1186/s12919-016-0060-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegert S., Hampe J., Schafmayer C., von Schonfels W., Egberts J. H., et al. , 2013. Genome-wide investigation of gene-environment interactions in colorectal cancer. Hum. Genet. 132: 219–231. 10.1007/s00439-012-1239-2 [DOI] [PubMed] [Google Scholar]
- Sun, R., and X. Lin, 2017 Set-based tests for genetic association using the generalized Berk-Jones statistic. arXiv: 1710.02469. [DOI] [PMC free article] [PubMed]
- Thomas D., 2010a Gene–environment-wide association studies: emerging approaches. Nat. Rev. Genet. 11: 259–272. 10.1038/nrg2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D., 2010b Methods for investigating gene-environment interactions in candidate pathway and genome-wide association studies. Annu. Rev. Public Health 31: 21–36. 10.1146/annurev.publhealth.012809.103619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vittinghoff E., McCulloch C. E., 2007. Relaxing the rule of ten events per variable in logistic and Cox regression. Am. J. Epidemiol. 165: 710–718. 10.1093/aje/kwk052 [DOI] [PubMed] [Google Scholar]
- Wei S., Wang L. E., McHugh M. K., Han Y., Xiong M., et al. , 2012. Genome-wide gene-environment interaction analysis for asbestos exposure in lung cancer susceptibility. Carcinogenesis 33: 1531–1537. 10.1093/carcin/bgs188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Kraft P., Zhai K., Chang J., Wang Z., et al. , 2012. Genome-wide association analyses of esophageal squamous cell carcinoma in Chinese identify multiple susceptibility loci and gene-environment interactions. Nat. Genet. 44: 1090–1097 [corrigenda: Nat. Genet. 46: 1040–1041 (2014)]. 10.1038/ng.2411 [DOI] [PubMed] [Google Scholar]
- Wu M. C., Lee S., Cai T., Li Y., Boehnke M., et al. , 2011. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 89: 82–93. 10.1016/j.ajhg.2011.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors affirm that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6849047.